
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 27 April 2022
Sec. Invertebrate Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.863380
This article is part of the Research TopicEndocrine Regulation of Insect DiapauseView all 8 articles
 Satoshi Homma1†
Satoshi Homma1† Akihisa Murata1†
Akihisa Murata1† Masato Ikegami1
Masato Ikegami1 Masakazu Kobayashi1
Masakazu Kobayashi1 Maki Yamazaki1
Maki Yamazaki1 Kento Ikeda2
Kento Ikeda2 Takaaki Daimon3
Takaaki Daimon3 Hideharu Numata2
Hideharu Numata2 Akira Mizoguchi4
Akira Mizoguchi4 Kunihiro Shiomi1*
Kunihiro Shiomi1*The bivoltine strain of the domestic silkworm, Bombyx mori, exhibits a facultative diapause phenotype that is determined by maternal environmental conditions during embryonic and larval development. Although a recent study implicated a circadian clock gene period (per) in circadian rhythms and photoperiod-induced diapause, the roles of other core feedback loop genes, including timeless (tim), Clock (Clk), cycle (cyc), and cryptochrome2 (cry2), have to be clarified yet. Therefore, the aim of this study was to elucidate the roles of circadian clock genes in temperature-dependent diapause induction. To achieve this, per, tim, Clk, cyc, and cry2 knockout (KO) mutants were generated, and the percentages of diapause and non-diapause eggs were determined. The results show that per, tim, Clk, cyc, and cry2 regulated temperature-induced diapause by acting upstream of cerebral γ-aminobutyric acid (GABA)ergic and diapause hormone signaling pathways. Moreover, the temporal expression of the clock genes in wild-type (wt) silkworms was significantly different from that of thermosensitive transient receptor potential ankyrin 1 (TRPA1) KO mutants during embryonic development. Overall, the findings of this study provide target genes for regulating temperature-dependent diapause induction in silkworms.
The circadian clock is involved not only in the daily regulation of several physiological, metabolic, and behavioral activities, including egg hatching and adult eclosion, but also in time-compensated Sun compass navigation, photoperiodism, and dormancy in insects (Patke et al., 2020; Saunders, 2020; Ahmad et al., 2021; Brady et al., 2021). Certain clock gene members drive the core functioning of the circadian clock, forming a transcriptional feedback loop module referred to as the “core feedback loop” (Hardin, 2011). An insect circadian clock model has been developed in Drosophila melanogaster; the first clock gene period (per) identified in D. melanogaster mutants reared in constant darkness (DD) allows free-running circadian rhythms (Konopka and Benzer, 1971). Previous studies have demonstrated that the core feedback loop is composed of products of per, timeless (tim), Clock (Clk), and cycle (cyc) in D. melanogaster (Hardin, 2011; Patke et al., 2020). CLK and CYC heterodimers drive per and tim transcription through E-box enhancer elements. The resultant PER and TIM proteins form heterodimers that translocate back into the nucleus to repress their own transcription via inhibitory effects on CLK and CYC, resulting in circadian oscillation in the expression of several clocks and related genes. Moreover, Drosophila cryptochrome (Drosophila-type CRY is designated as CRY1) is co-localized in clock cells with PER and TIM and functions as a blue-light photoreceptor involved in photic entrainment. CRY1 disrupts PER and TIM heterodimers by directly interacting with TIM in a light-dependent manner and also participates in its own light-dependent degradation (Hardin, 2011; Patke et al., 2020).
Apart from CRY1, a vertebrate-like light-insensitive CRY designated as CRY2 has been identified in every non-drosophilid insect (Reppert, 2007). The monarch butterfly, Danaus plexippus, possesses two CRY proteins, one ortholog of the light-sensitive CRY1 and a second light-insensitive CRY designated as CRY2 (Yuan et al., 2007). CRY2, a clock factor that forms the core feedback loop, is the major transcriptional repressor of CLK/CYC-mediated transcription, whereas PER is associated with CRY2 stabilization rather than with active transcriptional repression (Reppert, 2007; Zhu et al., 2008; Zhan et al., 2011). These findings suggest that extensive functional diversity exists among species with regard to cry genes and that clock genes participating in the core feedback loop have been evolutionally selected in insects (Reppert, 2007; Tomioka and Matsumoto, 2015; Beer and Helfrich-Forster, 2020; Brady et al., 2021).
Over the years, clock genes, including per, tim, Clk, cyc, cry1, and cry2, have been identified in the domestic silkworm, Bombyx mori (Kawamoto et al., 2019). The expression of per, tim, Clk, cry1, and cry2 exhibits temporal changes during embryonic development under both light/dark and DD conditions (Tao et al., 2017; Ikeda et al., 2019; Nartey et al., 2021). Additionally, core clock gene mutations disrupt not only the daily rhythm in both egg-hatching and adult eclosion rhythms in each per and tim knockout (KO; Ikeda et al., 2019; Nartey et al., 2021) but also the photoperiodic diapause, as a specific subtype of dormancy disrupted in per KO (Ikeda et al., 2021). Overall, research findings suggest that these clock genes operate as components of the core feedback loop module in the circadian clock of B. mori.
The bivoltine Kosetsu B. mori strain exhibits embryonic diapause, which is induced transgenerationally as a maternal effect (Yamashita and Hasegawa, 1985; Denlinger et al., 2012; Yokoyama et al., 2021). Maternal environmental conditions during embryonic and larval development influence progeny diapause. Generally, the temperature of the maternal environment is the predominant factor determining the occurrence of embryonic diapause, regardless of the photoperiod during embryonic and larval development (Shiomi et al., 2015; Tsuchiya et al., 2021). When eggs are incubated at 25°C under continuous darkness (25DD), the resultant female moths lay nearly 100% diapause eggs. In contrast, incubation of eggs at 15°C under dark conditions (15DD) resulted in moths that laid nearly 100% non-diapause eggs, although photoperiodic conditions during larval stages affected diapause determination when eggs were incubated at 20°C under continuous light (Ikeda et al., 2021). Thus, Kosetsu is considered a typical strain that is susceptible to temperature-dependent diapause induction.
The molecular mechanisms underlying the induction of embryonic diapause in B. mori have been extensively investigated (Denlinger et al., 2012; Tsuchiya et al., 2021). Our previous study revealed that embryonic diapause is induced by the diapause hormone (DH) signaling pathway, which consists of a highly sensitive and specific interaction between the DH and DH receptor (DHR) during pupal-adult development (Shiomi et al., 2015). DH is released from the corpus cardiacum (Sato et al., 1998) into the hemolymph and acts on DHR in the ovary (Homma et al., 2006). Moreover, cerebral γ-aminobutyric acid (GABA)ergic and corazonin pathways modulate DH release during temperature-dependent expression of the plasma membrane GABA transporter [GAT] (Tsuchiya et al., 2021). Recent studies revealed that the transient receptor potential ankyrin 1 (TRPA1; BmoTRPA1) acts as a thermosensitive channel activated at temperatures above 21°C and affects diapause induction through DH release in Kosetsu. Thus, BmoTRPA1 acts as a molecular switch for temperature-dependent diapause induction in Kosetsu (Sato et al., 2014; Yokoyama et al., 2021). However, it is unknown whether clock genes are involved in temperature-dependent diapause induction.
Therefore, the aim of this study was to elucidate the involvement of clock genes in temperature-dependent diapause induction. To achieve this, we generated KO mutants of the clock genes and examined their expression and effects on diapause induction.
The bivoltine Kosetsu B. mori strain was used in this study. Kosetsu exhibits a typical temperature-dependent diapause induction under rearing conditions (Tsuchiya et al., 2021; Yokoyama et al., 2021). The eggs were incubated under two different conditions: 1) at 25°C under continuous darkness (25DD) to obtain diapause eggs from wild-type (wt) silkworms and 2) at 15°C under continuous darkness (15DD) to obtain non-diapause eggs from wt. Larvae were then reared under a 13-h light/11-h dark cycle (13L11D) at 25°C as previously described (Shiomi et al., 2015). The pupae used in this experiment were collected within 1 h after pupation (referred to as day 0) to synchronize their subsequent development. The pupae were kept at 25°C under 12-h light/12-h dark (12L12D) conditions (Shiomi et al., 2015). The percentages of diapause eggs laid by the adults were estimated by counting the number of eggs in diapause after non-diapause eggs hatched. The results are expressed as the average percent diapause in each egg batch (Shiomi et al., 2015).
KO mutants of per (per+13) and BmoTRPA1 (∆TRPA1_1,429) were constructed following previously described procedures (Ikeda et al., 2021; Yokoyama et al., 2021). The transcription activator-like effector nuclease (TALEN)-based mutant lines of five genes (tim, Clk, cyc, cry1, and cry2) were constructed according to previous methods (Takasu et al., 2014; Yokoyama et al., 2021). DNA constructs containing TAL segments were prepared using a Golden Gate TALEN kit (Addgene, Cambridge, MA, United States). To screen for germline mutagenesis, the G0 adults were mated with wt. The oviposited G1 eggs were collected, and approximately 10 eggs from each brood were pooled for genomic DNA extraction using DNAzol reagent (Thermo Fisher Scientific, Waltham, MA, United States). DNA fragments containing the targeted region of interest were amplified by PCR using Takara Ex Taq (Takara, Tokyo, Japan) and specific primers (Supplementary Table S1). To test for mutagenesis, each PCR product of tim, Clk, cyc, cry1, and cry2 was digested using the restriction enzymes StyI, BsaI, NcoI, BsrBI, and SalI, respectively. Mutated PCR products were verified by sequencing. The broods containing mutated sequences were reared, and mutated G1 adults were crossed with siblings carrying the same mutation. Homozygous and hemizygous mutants were obtained after confirmation by sequencing the target region in the G2 or G3 egg genome.
The plant alkaloid picrotoxin (PTX; 5 μg/μl; Sigma-Aldrich, St. Louis, United States), which is a widely used ionotropic GABA and glycine receptor antagonist (Masiulis et al., 2019), was dissolved in distilled water, and 10 µl was injected into pupae through the intersegmental membrane between the second and third abdominal segment at 1–6 h after pupation. DH of 95% purity (HPLC area percentage) was obtained from Operon Biotechnologies (Tokyo, Japan) and dissolved in peanut oil (Sigma-Aldrich); 10 µl solution at 10 pmol/μl was injected into the pupa, 3.5 days after pupation.
The hemolymph was collected at ZT6 (ZT = zeitgeber time, ZT = 0 corresponds to lights on). The DH extract was prepared from the hemolymph as described previously (Shiomi et al., 2015). DH levels were measured using a time-resolved fluoroimmunoassay as described previously (Shiomi et al., 2015).
The eggs were incubated at 25°C under 12L12D, entrained to 12L12D, and collected at 6-h intervals for a 1-day cycle (ZT0–24) and the following day in DD (circadian time; CT0∼24). The eggs were collected at the appropriate time point and stored at −80°C until RNA extraction. RNA extraction and first-strand DNA synthesis were performed using TRI Reagent® (Molecular Research Center, Inc, Cincinnati, OH, United States) and ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan) containing the random primers and oligo dT primer, respectively. qPCR analysis was performed for the four sets of eggs using TB Green Premix DimerEraser using the Thermal Cycler Dice® Real-Time System (TP800; Takara Bio, Tokyo, Japan) and specific primers shown in Supplementary Table S1. The qPCR conditions were as follows: 95°C for 30s and then 40 cycles of 95°C for 5 s, 55°C for 30 s, and 72°C for 30 s with 0.3 µM concentration of each primer. The efficiency of amplification and detection by all primer sets was validated by determining the slope of Ct versus dilution plot on serial dilution. Individual reactions were used to quantify each RNA level in a given cDNA sample, and the average Ct from four samples within the same run was used for quantification. For statistical analysis, the data for each gene were normalized to rp49 as an external standard and normalized to the average of all time points within a set.
Statistical parameters, including definitions and values of n, are provided in the relevant figures or corresponding figure legends. Statistical analyses were performed in Excel 2011 (Microsoft) using software add-in Toukei-Kaiseki Ver. 3.0 (Esumi). p-values were determined using the Steel–Dwass test in diapause-egg-inducing activity and by one-way analysis of variance (ANOVA), Tukey–Kramer, and Student’s t-tests in temporal expression change in qPCR analysis and others. Data are expressed as mean ± standard deviation (SD). The mean values were considered to be significantly different at *p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates no significant difference.
To determine the role of clock genes in temperature-induced diapause, TALEN-mediated genome editing technology was used for KO silkworm tim, Clk, cyc, cry1, and cry2 genes. Briefly, we determined the cDNA sequences containing the TALEN-target sequence of each gene in the Kosetsu strain and then designed a TALEN target for each gene (Figure 1). Thereafter, we isolated hemizygous or homozygous mutants in each gene. In tim mutants, a 7-base sequence was deleted, a 15-base sequence was inserted into the spacer region, and a 13-base sequence was deleted between two TALEN binding sites, designated as ∆tim2532 and ∆tim9536 (Figures 1A, B). ∆tim2532 and ∆tim9536 were considered null mutants, which could not translate full-length and lacked functional domains of TIM due to a frameshift of tim cDNA (Supplementary Figure S1). KO mutants of Clk (∆Clk 1922 and ∆Clk7851; Figures 1C,D), cyc (∆cyc3717 and∆cyc3718; Figures 1E,F), cry1 (∆cry1_5,317; Figures 1G,H), and cry2 (∆cry2_5,731; Figures 1I,J) were generated using the same procedure. These clock gene mutants and the previously constructed per+13 were used in subsequent experiments.
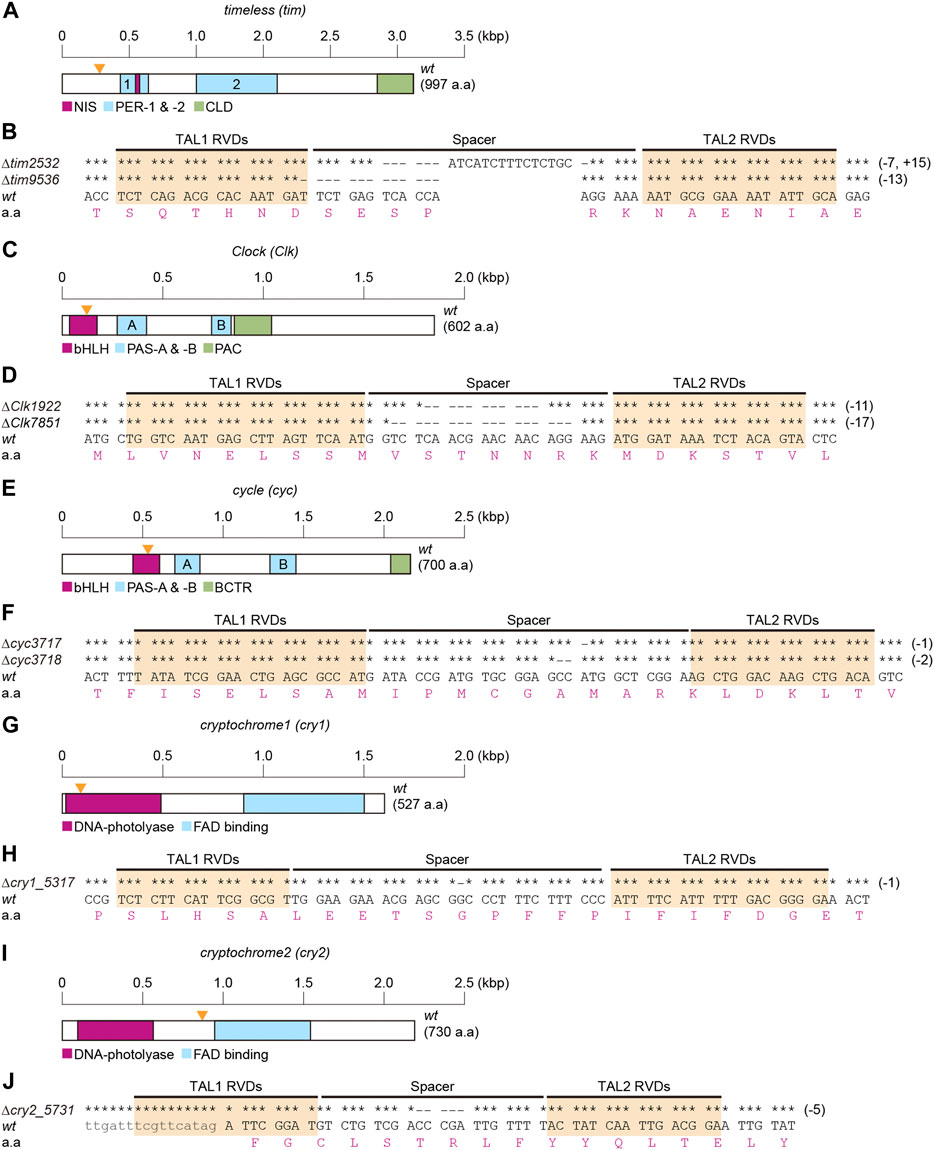
FIGURE 1. Construction of clock gene knockout mutants. Clock genes include timeless [tim] (A,B), Clock [Clk] (C,D), cycle [cyc] (E,F), cryptochrome1 [cry1] (G,H), and crtyptochrome2 [cry2] (I,J). Schematic representations of the coding regions of each cDNA structure are shown in A,C,E,G,I. (A) Functional domains of TIM, NIS (nuclear localization signal), PER (PER dimerization domain)-1 and -2, and CLD (cytoplasmic localization domain) are represented as referred in Iwai et al. (2006), and Tomioka and Matsumoto (2015). (C) Functional domains of CLK, bHLH (basic helix-loop-helix), PAS (Per-Arnt-Singleminded)-A and -B, and PAC are represented as referred in Ponting and Aravind (1997), Allada et al. (1998), and Moriyama et al. (2012). (E) Functional domains of CYC, bHLH, PAS-A and -B, and BCTR (BMAL1 C-terminal region) are represented according to Markova et al. (2003), Kamae et al., 2010, and Uryu et al. (2013). (G,I) Functional domains of CRY1 and CRY2, DNA-photolyase, and FAD (flavin adenine dinucleotide) binding are represented by searching the InterPro (http://www.ebi.ac.uk/interpro/) database and as referred in Tokuoka et al. (2017). The sizes of cDNA are indicated by scales in each map (A,C,E,G,I). Hyphens indicate the deleted bases in spacer and TAL RDV regions of each mutant, and identical bases are indicated by asterisks (B,D,F,H,J).
We investigated the phenotypes of the mutants with respect to diapause occurrence (Figure 2A). The results showed that wt moths laid 100% diapause eggs from 25DD silkworms (silkworm resulting from eggs incubated under 25DD condition), whereas per, tim, Clk, cyc, and cry2 KO silkworms laid varying percentages of non-diapause eggs. Particularly, knocking out per and tim remarkably reduced diapause egg-inducing activity. However, cry1 KO did not significantly affect embryonic diapause occurrence (Figure 2A). Additionally, we examined whether each clock gene acts hierarchically upstream of the GABAergic and DH signaling pathways (Figure 2A). The pupae of the per, tim, Clk, cyc, cry1, and cry2 KO mutants were injected with ionotropic GABA receptor antagonists PTX and DH since they are chemically potent diapause egg inducers (Shiomi et al., 2015; Tsuchiya et al., 2021). Both PTX and DH injection restored embryonic diapause in the pupae of per, tim, Clk, cyc, and cry2 mutants, which was comparable to those of 25DD wt silkworms (Figure 2A). However, compared with 15DD wt silkworms (silkworms resulting from eggs incubated under 15DD condition), there was no significant difference in the percentage of diapause eggs laid by the KO mutants (Figure 2B). Furthermore, PTX or DH injection into the pupae of KO mutants promoted diapause egg-inducing activity, suggesting that the GABAergic and DH signaling pathways were retained in the KO mutants as observed in 15DD silkworms (Figure 2B).
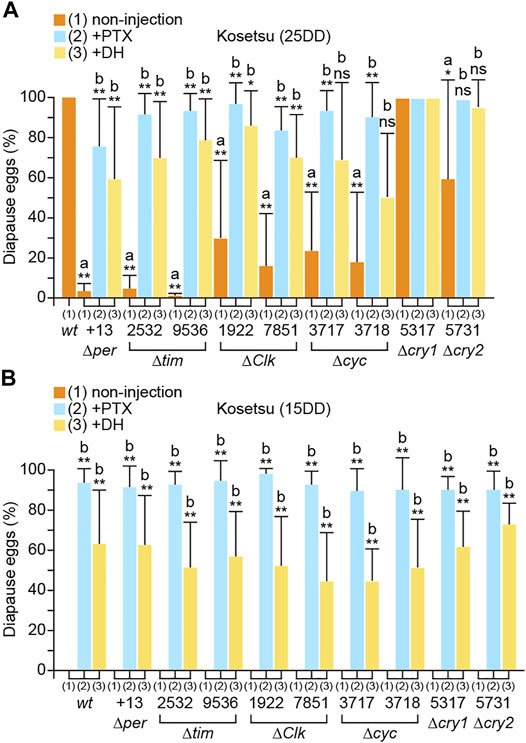
FIGURE 2. Disruption of temperature-dependent diapause induction in clock gene knockout mutants. wt eggs were incubated under constant darkness (DD) at 25°C [25DD] (A) or DD at 15°C [15DD] (B). Diapause egg-inducing activities were measured in progeny eggs (non-injection). For rescue experiments, both wt and knockout mutants were injected with picrotoxin (PTX; 50 μg; +PTX) 1 day after pupation or with diapause hormone (DH; 100 pmol; +DH) 3.5 days after pupation, and the percentages of diapause and non-diapause eggs were determined. Each bar represents mean ± standard deviation (SD) of 20–30 animals. Statistical differences were examined by the Steel–Dwass test between each non-injected knockout mutant and wt (A) and between injected and non-injected treatments in each knockout mutant (B). ns, non-significant; *, p < 0.05; **, p < 0.01.
Moreover, the hemolymph DH levels of KO mutants were measured at 2 and 4 days after pupation (Figure 3). At 2 days after pupation (P2), the DH levels of the per, Clk, cyc, and cry2 KO mutants were similar to those of 25DD and 15DD wt silkworms. However, there was a significant decrease in the DH level of tim KO silkworm. At 4 days after pupation (P4), there was a decrease in the DH levels of per, tim, Clk, and cyc KO silkworms compared with 25DD wt silkworms. However, the DH contents of cry1 and cry2 silkworms were not significantly affected (Figure 3).
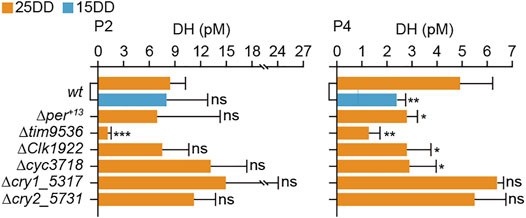
FIGURE 3. Diapause hormone (DH) concentration in hemolymph during pupal-adult development in clock gene knockout mutants. Both wt and knockout mutants’ eggs were incubated under constant darkness (DD) at 25°C (25DD), and only wt eggs were incubated under DD at 15°C (15DD). Hemolymph was collected at 2 (P2) and 4 (P4) days after pupation. Each bar represents mean ± standard deviation (SD) of five samples. Statistical differences were examined by Student’s t-test between wt at 15DD or each knockout mutant and wt at 25DD. ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To investigate whether only Bombyx clock genes are the core components of the circadian clock in B. mori or temperature signals via BmoTRPA1 expression also affect the expression of clock genes, we examined the expression profiles of the clock genes in wt and BmoTRPA1 KO mutants during 4–6 days after oviposition (E4–E6) at 6-h intervals under the 12L12D photoperiod using qPCR (Figure 4). Generally, there were significant changes in the expression of per in wt silkworms (p = 0.0095, one-way ANOVA), and they exhibited a weak daily rhythm with LD and zeitgeber time (ZT). Peak expressions were observed at ZT0 and ZT24, and the lowest expression was observed at ZT6, with the rhythm persisting in DD (Figure 4A; orange line). Similarly, there were significant changes in the mRNA levels of tim and cry2 (p = 0.013 and 0.046, respectively) in wt silkworms (Figures 4B, F). Although temporal changes were present in the expression profiles of cyc (p = 0.00092), Clk (p = 0.014), and cry1 (p = 0.0024), the expressions were arrhythmic during the tested period (Figures 4C–E). Overall, we observed daily expressional changes of per, tim, and cry2 in wt silkworms, with peak levels between ZT 16 and 24.
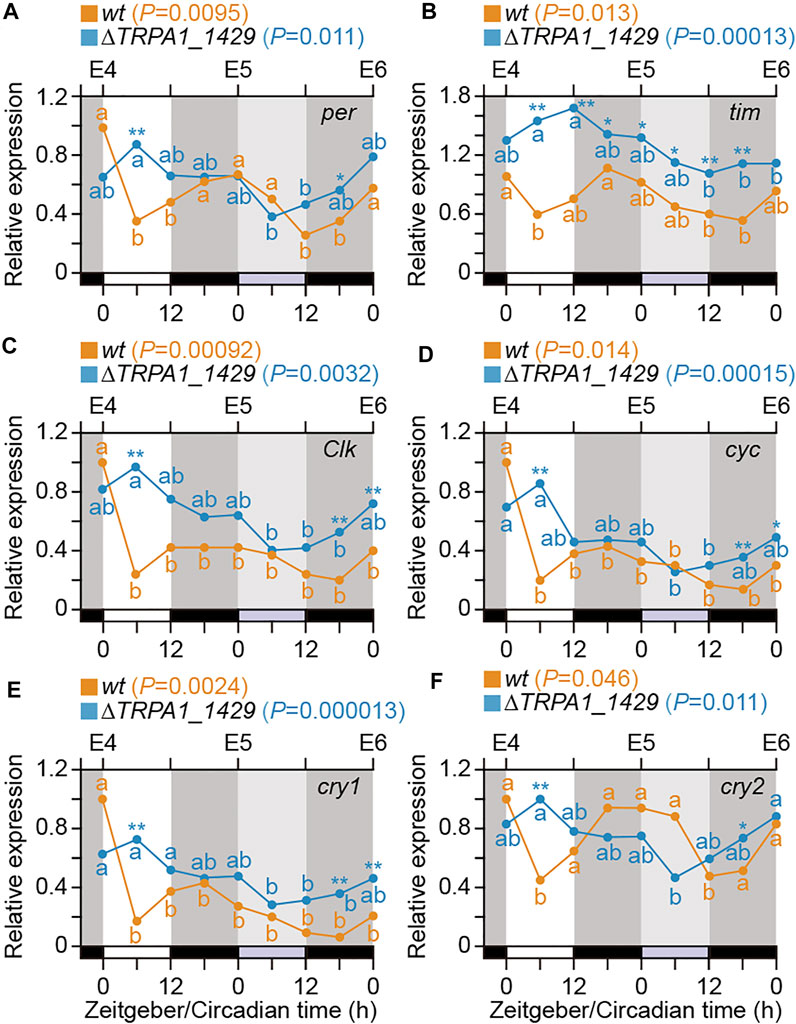
FIGURE 4. Temporal expression patterns of clock genes in eggs. Temporal profiles of clock gene expression in eggs of wt (orange line) and BmoTRPA1 (∆TRPA1_1,429) knockout mutants (blue line) during 4–6 days (E4∼E6) after oviposition. Eggs were incubated under 12-h light/12-h dark (12L12D) conditions at 25°C and collected at 6-h intervals for a 1-day cycle (ZT0–24) and the following day in constant darkness (DD; circadian time; CT0∼24). mRNA levels of period [per] (A), timeless [tim] (B), Clock [Clk] (C), cycle [cyc] (D), cryptochrome1 [cry1] (E), and crtyptochrome2 [cry2] (F) were determined by qPCR. Each value is the mean from four sets of eggs. Open bars, light; black bars, dark; gray bars, subjective day. Using one-way analysis of variance (ANOVA), p values are represented in each usage guide of wt and ∆TRPA1_1,429 in each panel. Different lower-case letters indicate values that are significantly different from each other in each wt (Orange) and ∆TRPA1_1,429 (Blue) [Tukey–Kramer test]. Significant differences (wt vs. ∆TRPA1_1,429 in each stage) are represented by an asterisk (Student’s t-test). *p < 0.05; **p < 0.01.
Moreover, although there were significant temporal changes in the expression profiles of the six genes in BmoTRPA1 (∆TRPA1_1,429) KO mutants during the timeframe examined, no daily rhythm was observed (Figures 4A, B; blue lines). Additionally, a comparison of the expression levels of the genes in both wt and BmoTRPA1 KO mutant silkworms showed significant differences in the expression profiles of the genes at the examined stages, especially tim (Figure 4; asterisk).
Over the years, studies have indicated the oscillating expression of core clock genes in lepidopterans, including various strains of B. mori, and in the monarch butterfly cell line, DpN1 (Zhu et al., 2008; Brady et al., 2021). The findings of the present study show that knocking out clock genes of per, tim, Clk, cyc, and cry2 disrupts temperature-dependent diapause induction in silkworms. Additionally, some core clock genes, including per, tim, and cry2, exhibited oscillating expression during embryonic development. Together, the results suggest that the per and tim KO strains do not show circadian rhythms both in behavior and in clock gene expression (Ikeda et al., 2019; Nartey et al., 2021). We suggested that per, tim, Clk, cyc, and cry2 are the core components of the circadian clock in B. mori. However, studies have revealed that the biological clocks of insects are not uniform among species. For instance, cry1 is absent in Tribolium castaneum (only cry2 exists), whereas cry2 is present (tim is absent) in Apis mellifera (Rubin et al., 2006; Yuan et al., 2007). Therefore, further investigations are required to verify the roles of clock genes in lepidopterans.
The present study’s findings indicate that the clock genes per, tim, Clk, cyc, and cry2 are involved in temperature-dependent diapause induction. Involvement of clock genes in photoperiodic induction of diapause has been shown in various insects, including B. mori (e.g., Pavelka et al., 2003; Sakamoto et al., 2009; Ikeno et al., 2010; Merlin et al., 2013; Meuti et al., 2015; Cui et al., 2021; Ikeda et al., 2021). However, the role of clock genes in temperature-dependent diapause induction has not been shown. Moreover, injection of DH or PTX induced diapause in clock gene KO mutants, and hemolymph concentrations of DH in 25DD were lower in clock gene KO mutants except for the cry2-KO mutant than those in wt. Therefore, these clock genes are involved in the upstream mechanism of DH secretion. We speculate that these genes, which probably constitute the core loop of the circadian clock, regulate temperature-dependent diapause induction as a circadian clock module (Emerson et al., 2009).
In the bivoltine strain of B. mori, the diapause/non-diapause phenotype is determined only during a temperature-sensitive period. This period lasts for 3 days under 25DD or 9 days under 15DD conditions, including the embryonic reversal stage, which corresponds to stages 20–23 of embryogenesis (Watanabe, 1924; Xu et al., 1995). A previous study indicated that BmoTRPA1 is expressed in a temperature-sensitive period and acts as a thermosensitive TRP channel, which is activated at a threshold of ∼21°C, and determines progeny diapause (Sato et al., 2014). In the present study, knocking out BmoTRPA1 disrupted the rhythmic expression of clock genes in silkworms at different stages, including the temperature-sensitive period. Moreover, studies have shown that orthologs of TRPA1 in D. melanogaster function in temperature control of circadian rhythm for daily behaviors (Lee and Montell, 2013; Green et al., 2015; Lamaze et al., 2017). Therefore, it is suggested that the Bombyx TRPA1-activated signaling pathway could be linked to circadian clock oscillation in temperature-dependent diapause induction.
Although the findings of this study show that core clock genes are involved in temperature-dependent diapause induction, the molecular mechanism of clock functions under different temperature conditions, especially 25DD and 15DD incubation conditions, is unknown. However, recent genetic and molecular studies of wild D. melanogaster populations revealed that polymorphisms at the tim locus facilitate seasonal adaptation (Tauber et al., 2007; Cai and Chiu, 2021), that is, tim produces four temperature-dependent splice variants in response to temperature changes. Since some splicing events generate truncated TIM proteins, this could differentially affect TIM functions, such as PER-binding and light sensitivity in the circadian clock (Cai and Chiu, 2021). Moreover, the core feedback loop of the circadian clock drives the interlocked negative feedback sub-loop. CLK/CYC activates the transcription of vrille (vri) and Par domain protein 1ε (Pdp1ε) and activates the transcription of clockwork orange (cwo), which regulates the amplitude of per and tim mRNA oscillations (Tomioka and Matsumoto, 2015). Overall, we speculated that there are differences in the expression levels, selective splicing, and functions in clock genes and/or clock-related genes under 25DD and 15DD incubation conditions, which could affect clock gene oscillations, such as stability and amplitude for diapause determination.
A previous study demonstrated that knocking out BmoTRPA1 can disrupt the TRPA1-activated pathway, which could influence temperature-dependent diapause induction and initiate photoperiodic diapause induction (Yokoyama et al., 2021). Therefore, it is suggested that the wt Kosetsu strain potentially possesses a signaling pathway involved in photoperiodic diapause induction that might be canceled by the strong linkage of BmoTRPA1-activated and DH signaling pathways. Consequently, the progeny diapause may be dependent on the photo- or scotophase length in BmoTRPA1 KO mutants. Therefore, we speculate that clock genes are engaged in both photoperiodic and temperature-dependent diapause induction and that the circadian clock is involved in the selection and integration of several environmental signals.
A previous study found that the expression of the plasma membrane GAT gene was 10–100-fold higher in the pupal brain–subesophageal ganglion complex under 25DD incubation condition than that under 15DD incubation condition (Tsuchiya et al., 2021). Under 25DD conditions, the female adults of the GAT KO mutant laid mostly non-diapause eggs. Therefore, we conclude that the GABAergic signaling system may be influenced by the temperature-dependent expression of the plasma membrane GAT since the plasma membrane GAT actively transports GABA from the synaptic cleft back into presynaptic neurons and glial cells to terminate GABA-stimulated responses (Tsuchiya et al., 2021). We demonstrated that clock genes are involved in temperature-dependent diapause induction upstream of GABAergic and DH signaling pathways. Therefore, clock genes may influence GABAergic signals directly and indirectly by regulating GAT expression, thereby inducing embryonic diapause in progenies. Overall, the findings of this study provide an excellent model for investigating relationships among the circadian clock, temperature, and photoperiodic response.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
KS designed the research. SH, AMu, MI, MK, MY, KI, TD, AMi, and KS performed the research. KS and AMi analyzed the data. HN and KS interpreted the data and supervised the study. KS wrote the manuscript with support from all authors.
This research was funded by the Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are indebted to the Division of Gene Research, Research Center for Human and Environmental Sciences, and Nouzyou for providing the facilities used in this.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.863380/full#supplementary-material
Ahmad M., Li W., Top D. (2021). Integration of Circadian Clock Information in the Drosophila Circadian Neuronal Network. J. Biol. Rhythms 36, 203–220. doi:10.1177/0748730421993953
Allada R., White N. E., So W. V., Hall J. C., Rosbash M. (1998). A Mutant Drosophila Homolog of Mammalian Clock Disrupts Circadian Rhythms and Transcription of Period and Timeless. Cell 93, 791–804. doi:10.1016/s0092-8674(00)81440-3
Beer K., Helfrich-Förster C. (2020). Model and Non-model Insects in Chronobiology. Front. Behav. Neurosci. 14, 601676. doi:10.3389/fnbeh.2020.601676
Brady D., Saviane A., Cappellozza S., Sandrelli F. (2021). The Circadian Clock in Lepidoptera. Front. Physiol. 12, 776826. doi:10.3389/fphys.2021.776826
Cai Y. D., Chiu J. C. (2021). Timeless in Animal Circadian Clocks and beyond. FEBS J. 1–17. doi:10.1111/febs.16253
Cui W.-Z., Qiu J.-F., Dai T.-M., Chen Z., Li J.-L., Liu K., et al. (2021). Circadian Clock Gene Period Contributes to Diapause via GABAeric-Diapause Hormone Pathway in Bombyx mori. Biology 10, 842. doi:10.3390/biology10090842
Denlinger D. L., Yocum G. D., Rinehart J. P. (2012). “Hormonal Control of Diapause,” in Hormonal Control of diapauseInsect Endocrinology. Editor L I Gilbert (San Diego: Academic Press), 430–463. doi:10.1016/b978-0-12-384749-2.10010-x
Emerson K. J., Bradshaw W. E., Holzapfel C. M. (2009). Complications of Complexity: Integrating Environmental, Genetic and Hormonal Control of Insect Diapause. Trends Genet. 25, 217–225. doi:10.1016/j.tig.2009.03.009
Green E. W., O’Callaghan E. K., Hansen C. N., Bastianello S., Bhutani S., Vanin S., et al. (2015). Drosophila Circadian Rhythms in Seminatural Environments: Summer Afternoon Component Is Not an Artifact and Requires TrpA1 Channels. Proc. Natl. Acad. Sci. U.S.A. 112, 8702–8707. doi:10.1073/pnas.1506093112
Hardin P. E. (2011). Molecular Genetic Analysis of Circadian Timekeeping in Drosophila. Adv. Genet. 74, 141–173. doi:10.1016/b978-0-12-387690-4.00005-2
Homma T., Watanabe K., Tsurumaru S., Kataoka H., Imai K., Kamba M., et al. (2006). G Protein-Coupled Receptor for Diapause Hormone, an Inducer of Bombyx Embryonic Diapause. Biochem. Biophysical Res. Commun. 344, 386–393. doi:10.1016/j.bbrc.2006.03.085
Ikeda K., Daimon T., Sezutsu H., Udaka H., Numata H. (2019). Involvement of the Clock Gene Period in the Circadian Rhythm of the Silkmoth Bombyx mori. J. Biol. Rhythms 34, 283–292. doi:10.1177/0748730419841185
Ikeda K., Daimon T., Shiomi K., Udaka H., Numata H. (2021). Involvement of the Clock Gene Period in the Photoperiodism of the Silkmoth Bombyx mori. Zoolog. Sci. 38, 523–530. doi:10.2108/zs210081
Ikeno T., Tanaka S. I., Numata H., Goto S. G. (2010). Photoperiodic Diapause under the Control of Circadian Clock Genes in an Insect. BMC Biol. 8, 116. doi:10.1186/1741-7007-8-116
Iwai S., Fukui Y., Fujiwara Y., Takeda M. (2006). Structure and Expressions of Two Circadian Clock Genes, Period and Timeless in the Commercial Silkmoth, Bombyx mori. J. Insect Physiol. 52, 625–637. doi:10.1016/j.jinsphys.2006.03.001
Kamae Y., Tanaka F., Tomioka K. (2010). Molecular Cloning and Functional Analysis of the Clock Genes, Clock and Cycle, in the Firebrat Thermobia domestica. J. Insect Physiol. 56, 1291–1299. doi:10.1016/j.jinsphys.2010.04.012
Kawamoto M., Jouraku A., Toyoda A., Yokoi K., Minakuchi Y., Katsuma S., et al. (2019). High-quality Genome Assembly of the Silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 107, 53–62. doi:10.1016/j.ibmb.2019.02.002
Konopka R. J., Benzer S. (1971). Clock Mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. doi:10.1073/pnas.68.9.2112
Lamaze A., Öztürk-Çolak A., Fischer R., Peschel N., Koh K., Jepson J. E. C. (2017). Regulation of Sleep Plasticity by a Thermo-Sensitive Circuit in Drosophila. Sci. Rep. 7, 40304. doi:10.1038/srep40304
Lee Y., Montell C. (2013). Drosophila TRPA1 Functions in Temperature Control of Circadian Rhythm in Pacemaker Neurons. J. Neurosci. 33, 6716–6725. doi:10.1523/jneurosci.4237-12.2013
Markova E. P., Ueda H., Sakamoto K., Oishi K., Shimada T., Takeda M. (2003). Cloning of Cyc (Bmal1) Homolog in Bombyx mori: Structural Analysis and Tissue Specific Distributions. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 134, 535–542. doi:10.1016/s1096-4959(03)00004-6
Masiulis S., Desai R., Uchański T., Serna Martin I., Laverty D., Karia D., et al. (2019). GABAA Receptor Signalling Mechanisms Revealed by Structural Pharmacology. Nature 565, 454–459. doi:10.1038/s41586-018-0832-5
Merlin C., Beaver L. E., Taylor O. R., Wolfe S. A., Reppert S. M. (2013). Efficient Targeted Mutagenesis in the Monarch Butterfly Using Zinc-finger Nucleases. Genome Res. 23, 159–168. doi:10.1101/gr.145599.112
Meuti M. E., Stone M., Ikeno T., Denlinger D. L. (2015). Functional Circadian Clock Genes Are Essential for the Overwintering Diapause of the Northern House Mosquito, Culex pipiens. Culex Pipiensj. Exp. Biol. 218, 412–422. doi:10.1242/jeb.113233
Moriyama Y., Kamae Y., Uryu O., Tomioka K. (2012). gb'clock Is Expressed in the Optic Lobe and Is Required for the Circadian Clock in the Cricket Gryllus Bimaculatus. J. Biol. Rhythms 27, 467–477. doi:10.1177/0748730412462207
Nartey M. A., Sun X., Qin S., Hou C. X., Li M. W. (2021). CRISPR/Cas9‐based Knockout Reveals that the Clock Gene Timeless Is Indispensable for Regulating Circadian Behavioral Rhythms in Bombyx mori. Insect Sci. 28, 1414–1425. doi:10.1111/1744-7917.12864
Patke A., Young M. W., Axelrod S. (2020). Molecular Mechanisms and Physiological Importance of Circadian Rhythms. Nat. Rev. Mol. Cel Biol 21, 67–84. doi:10.1038/s41580-019-0179-2
Pavelka J., Shimada K., Kostal V. (2003). TIMELESS: A Link between Fly's Circadian and Photoperiodic Clocks? Eur. J. Entomol. 100, 255–265. doi:10.14411/eje.2003.041
Ponting C. P., Aravind L. (1997). PAS: a Multifunctional Domain Family Comes to Light. Curr. Biol. 7, R674–R677. doi:10.1016/s0960-9822(06)00352-6
Reppert S. M. (2007). The Ancestral Circadian Clock of Monarch Butterflies: Role in Time-Compensated Sun Compass Orientation. Cold Spring Harbor Symposia Quantitative Biol. 72, 113–118. doi:10.1101/sqb.2007.72.056
Rubin E. B., Shemesh Y., Cohen M., Elgavish S., Robertson H. M., Bloch G. (2006). Molecular and Phylogenetic Analyses Reveal Mammalian-like Clockwork in the Honey Bee (Apis mellifera) and Shed New Light on the Molecular Evolution of the Circadian Clock. Genome Res. 16, 1352–1365. doi:10.1101/gr.5094806
Sakamoto T., Uryu O., Tomioka K. (2009). The Clock Gene Period Plays an Essential Role in Photoperiodic Control of Nymphal Development in the Cricket Modicogryllus Siamensis. J. Biol. Rhythms 24, 379–390. doi:10.1177/0748730409341523
Sato A., Sokabe T., Kashio M., Yasukochi Y., Tominaga M., Shiomi K. (2014). Embryonic Thermosensitive TRPA1 Determines Transgenerational Diapause Phenotype of the Silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U S A. 111, E1249–E1255. doi:10.1073/pnas.1322134111
Sato Y., Shiomi K., Saito H., Imai K., Yamashita O. (1998). Phe-X-Pro-Arg-Leu-NH2 Peptide Producing Cells in the central Nervous System of the Silkworm, Bombyx mori. J. Insect Physiol. 44, 333–342. doi:10.1016/s0022-1910(97)00140-6
Saunders D. S. (2020). Dormancy, Diapause, and the Role of the Circadian System in Insect Photoperiodism. Annu. Rev. Entomol. 65, 373–389. doi:10.1146/annurev-ento-011019-025116
Shiomi K., Takasu Y., Kunii M., Tsuchiya R., Mukaida M., Kobayashi M., et al. (2015). Disruption of Diapause Induction by TALEN-Based Gene Mutagenesis in Relation to a Unique Neuropeptide Signaling Pathway in Bombyx. Sci. Rep. 5, 15566. doi:10.1038/srep15566
Takasu Y., Tamura T., Sajwan S., Kobayashi I., Zurovec M. (2014). The Use of TALENs for Nonhomologous End Joining Mutagenesis in Silkworm and Fruitfly. Methods 69, 46–57. doi:10.1016/j.ymeth.2014.02.014
Tao H., Li X., Qiu J. F., Liu H. J., Zhang D. Y., Chu F., et al. (2017). The Light Cycle Controls the Hatching Rhythm in Bombyx mori via Negative Feedback Loop of the Circadian Oscillator. Arch. Insect Biochem. Physiol. 96, 1–14. doi:10.1002/arch.21408
Tauber E., Zordan M., Sandrelli F., Pegoraro M., Osterwalder N., Breda C., et al. (2007). Natural Selection Favors a Newly Derived Timeless Allele in Drosophila melanogaster. Science 316, 1895–1898. doi:10.1126/science.1138412
Tokuoka A., Itoh T. Q., Hori S., Uryu O., Danbara Y., Nose M., et al. (2017). Cryptochrome Genes Form an Oscillatory Loop Independent of the Per/tim Loop in the Circadian Clockwork of the Cricket Gryllus Bimaculatus. Zoolog. Lett 3, 5. doi:10.1186/s40851-017-0066-7
Tomioka K., Matsumoto A. (2015). Circadian Molecular Clockworks in Non-model Insects. Curr. Opin. Insect Sci. 7, 58–64. doi:10.1016/j.cois.2014.12.006
Tsuchiya R., Kaneshima A., Kobayashi M., Yamazaki M., Takasu Y., Sezutsu H., et al. (2021). Maternal GABAergic and GnRH/corazonin Pathway Modulates Egg Diapause Phenotype of the Silkworm Bombyx mori. Proc. Natl. Acad. Sci. U S A. 118, 1–9. doi:10.1073/pnas.2020028118
Uryu O., Karpova S. G., Tomioka K. (2013). The Clock Gene Cycle Plays an Important Role in the Circadian Clock of the Cricket Gryllus Bimaculatus. J. Insect Physiol. 59, 697–704. doi:10.1016/j.jinsphys.2013.04.011
Watanabe K. (1924). Studies on the Voltinism of the Silkworm, Bombyx mori. Bull. Sericult Exp. Sta (Tokyo) 6, 411–455.
Xu W.-h., Sato Y., Ikeda M., Yamashita O. (1995). Stage-dependent and Temperature-Controlled Expression of the Gene Encoding the Precursor Protein of Diapause Hormone and Pheromone Biosynthesis Activating Neuropeptide in the Silkworm, Bombyx mori. J. Biol. Chem. 270, 3804–3808. doi:10.1074/jbc.270.8.3804
Yokoyama T., Saito S., Shimoda M., Kobayashi M., Takasu Y., Sezutsu H., et al. (2021). Comparisons in Temperature and Photoperiodic-dependent Diapause Induction between Domestic and Wild mulberry Silkworms. Sci. Rep. 11, 8052. doi:10.1038/s41598-021-87590-4
Yuan Q., Metterville D., Briscoe A. D., Reppert S. M. (2007). Insect Cryptochromes: Gene Duplication and Loss Define Diverse Ways to Construct Insect Circadian Clocks. Mol. Biol. Evol. 24, 948–955. doi:10.1093/molbev/msm011
Zhan S., Merlin C., Boore J. L., Reppert S. M. (2011). The Monarch Butterfly Genome Yields Insights into Long-Distance Migration. Cell 147, 1171–1185. doi:10.1016/j.cell.2011.09.052
Keywords: clock gene, diapause, Bombyx mori, transcription activator-like effector nuclease (TALEN), circadian clock
Citation: Homma S, Murata A, Ikegami M, Kobayashi M, Yamazaki M, Ikeda K, Daimon T, Numata H, Mizoguchi A and Shiomi K (2022) Circadian Clock Genes Regulate Temperature-Dependent Diapause Induction in Silkworm Bombyx mori. Front. Physiol. 13:863380. doi: 10.3389/fphys.2022.863380
Received: 27 January 2022; Accepted: 23 March 2022;
Published: 27 April 2022.
Edited by:
Jin-Jun Wang, Southwest University, ChinaReviewed by:
Eran Tauber, University of Haifa, IsraelCopyright © 2022 Homma, Murata, Ikegami, Kobayashi, Yamazaki, Ikeda, Daimon, Numata, Mizoguchi and Shiomi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunihiro Shiomi, c2hpb21pQHNoaW5zaHUtdS5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.