
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Physiol. , 28 February 2022
Sec. Invertebrate Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.852357
Ants are highly diverse social insects living in colonies consisted of up to millions of individuals with reproductive division of labors. Due to the interests in disclosing the genetic and epigenetic regulation mechanisms underlying the distinct developmental trajectories between castes and division of labor in colonies, many ant species have recently been established as laboratory models for evolutionary development and social behavior studies. These functional studies often request a precise quantification of the relative gene expression level, which relies on a stably expressed reference genes for normalization. A core set of reliable reference genes for this purpose however has not been established yet in ants. In the present study, we tested the expression patterns and amplification efficiencies of 12 abundantly expressed candidate genes in Monomorium pharaonis, one of the few ant species that are suitable for laboratory rearing and experimentation. We quantified the expression levels of these genes by RT-qPCR in seven different conditions: embryo development, sexual development, worker development, adult phenotypes, tissues, and two abiotic manipulative treatments in pharaoh ant. Finally, five genes, elongation factor-1 alpha (EF1A), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), TATA-box-binding protein (TATA), tubulin gamma-2 chain-like (TBLg2), heat shock protein 67B2-like (HSP67) were found to be the most stable reference genes across seven conditions. We also identified the most stable reference genes applicable for each distinct condition and the optimal number of reference genes entailed were evaluated. Our study validates reliable reference genes for RT-qPCR analysis which lays the foundation for future studies in pharaoh ant.
Social insects live in complex societies with sophisticated social behavior and division of labor. How natural selection shape their caste system has long fascinated biologists (Hölldobler and Wilson, 1990; Trible and Kronauer, 2017). The differentiated castes vary in morphology, physiology, and behavior even though they have the same genome (Abouheif and Wray, 2002; Li et al., 2014). With the morphologically and behaviorally distinct individuals, ant colony organizes into a complicated society with intricate division of labor among different castes (Hölldobler and Wilson, 1990). Many studies suggested that the caste differentiation is regulated by differentially expressed genes among castes (Evans and Wheeler, 2002; Pereboom et al., 2005; Khila and Abouheif, 2010; Qiu et al., 2018). Thus, interrogating the molecular mechanism underlying caste differentiation through the gene expression analysis has been the major focus in recent studies in social insects (Gospocic et al., 2017; Chandra et al., 2018).
Pharaoh ant (Monomorium pharaonis) is of high fecundity, short lifespan and polygynous (multiple queens in the same colony) colony organization, making it easier for lab rearing and suitable as a model organism to study the molecular mechanism of caste differentiation (Peacock and Baxter, 1949; Schmidt et al., 2011). The reproductive and non-reproductive castes can be distinguished via morphological traits since the 2nd instar larval stage: the worker larvae are furnished with numerous, bifurcated hairs and sexual (female reproductive and male) larvae are almost hairless; the sexual larvae are almost twice the size of worker larvae (Edwards, 1991). In addition, many differentially expressed genes among castes or individuals with different social roles have been identified by high-throughput sequencing and bioinformatics analysis in pharaoh ant (Mikheyev and Linksvayer, 2015; Qiu et al., 2018; Warner et al., 2019). However, studies on validation of the functions of differentially expressed genes in regulating caste phenotypes are lagging behind. The causal relationship linking gene expression with particular caste traits remains poorly understood.
Real-time quantitative polymerase chain reaction (RT-qPCR) is a popular method for analyzing gene expression level (Nolan et al., 2006; Derveaux et al., 2010). This technique can evaluate the expression level of target genes effectively by comparing to stable reference genes or by depicting standard curve of target genes (Bustin, 2000), with a lower cost and less time-consumption than RNA-seq. Therefore, it is a method still widely used to quantify gene expression even in the post-genomic era when transcriptome sequencing is readily available due to its high sensitivity and accuracy (Kang et al., 2017). However, the pipetting errors and differences in amplification efficiency among different samples can cause error for the threshold cycle (Ct, also called Cq), which is an important value to calculate the expression level of target genes. To minimize the error, stably expressed reference genes are necessary for gene expression analysis.
Reference genes are often used to normalize the expression level of target genes in different samples in RT-qPCR (Radonić et al., 2004). These genes are usually chosen from housekeeping genes, which are typically constitutive genes required for maintaining basic cellular functions, and are expressed in all cells at relatively constant levels (Butte et al., 2001; Zhu et al., 2008). Therefore, reference genes could be used to normalize the expression level of target genes among different samples to reduce or even eliminate systematic errors. Nevertheless, no universal gene can be applied to all situations as reference even in the same species (Cheng et al., 2013; Gao et al., 2017; Kang et al., 2017; Moreira et al., 2017; do Livramento et al., 2018). Given reference gene can directly affect gene expression profile analysis (do Livramento et al., 2018), making a thorough reference gene selection and validation prior to RT-qPCR analysis of target genes is necessary.
To date, the reference genes were only described in fire ant Solenopsis invicta, leaf-cutting ant Atta sexdens rubropilosa in detail among ant species (Cheng et al., 2013). In this study, we selected 12 genes as potential reference genes: 11 from our RNA-seq data and one commonly used reference gene outside of our data—18S ribosomal RNA (18S). The gene fragments were cloned into vector, and the amplification efficiency was calculated by standard curve method. Then we examined the expression stability of these genes in different developmental stages and tissues: (1) embryo development, (2) worker development, (3) sexual development, (4) adult phenotypes, (5) gyne/queen tissues (brain, midgut, fat body, and ovary); and two abiotic manipulative treatments for gyne/queen: (6) injection of eGFP dsRNA and (7) starvation treatment. In addition, the RT-qPCR results were evaluated with four methods [geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and the comparative ΔCt method (Silver et al., 2006)]. Finally, the performance of these potential reference genes were integrated by a web-based comprehensive tool RefFinder (Xie et al., 2012). We validated the best reference genes and optimal gene numbers applicable to different developmental stages, adult phenotypes, tissues, and abiotic treatments for gene expression analysis in M. pharaonis, which provides a valuable guidance for future studies on functional validation of differentially expressed genes linking to caste phenotypes.
The samples (eggs, larvae, pupae, and adults) were collected from colonies reared at Kunming Institute of Zoology and kept under a constant temperature of 27°C and 65% humidity. The samples used for abiotic treatments consisted of heads, thorax, and abdomens. All samples were collected in triplicate, flash froze in liquid nitrogen and stored at −80°C until use.
2nd, 5th, and 8th day Eggs were collected. Since a single egg is too small for the RNA extraction, a pool of about 50 eggs was set as one sample to ensure high quality of RNA.
To obtain a stable reference gene during worker developmental stages, we collected 3rd instar worker larvae, young worker pupae, old worker pupae, and worker adult according to morphological features depicted by Berbdt and Kremer (1986). Given the tiny size of worker, 10 individuals were used for each worker sample.
We collected 3rd instar sexual larvae. As the sex can be distinguished since the pupae stage, we prepared male/gyne young pupae, male/gyne old pupae, and male/gyne/queen adults, respectively. Gyne specifically refers to the virgin queen whose gene expression pattern is different from the reproductive active queen. Two individuals were enough for sexual samples because of their big size.
Here, we classified adult pharaoh ants based on their caste, reproductive condition and sex (Smith and Suarez, 2010). Four adult phenotypes were analyzed: gyne (unmated reproductive female), queen (mated, reproductively active female), worker (un-reproductive female) and male.
Insemination induces drastic changes in physiology and behavior when a gyne becomes to a queen. Thus, tissue comparisons for reference genes between gyne and queen were also conducted. Individuals were dissected under a stereo microscope to obtain brain, midgut, fat body, and ovary. Each sample is consisted of a pool of twenty heads, thoraxes, and abdomens from gynes or queens, respectively.
RNA interference (RNAi) is a popular method to interrogate gene functions. RNAi components are frequently administered to insects through micro injection technique. Micro injection would cause tissue damage to insect thus can be considered as an abiotic stress condition. We tested reference genes performance under micro injection treatment in which 1,000 ng/μL control eGFP dsRNA was injected into adult bodies. After 48 h, heads, thoraxes, and abdomens of injected and non-injected groups were, respectively, collected and analyzed.
Starvation treatment is an abiotic stress condition which would elicit different behavioral responses in gyne and queen. After 5 days starvation treatment, heads, thoraxes, and abdomens of starved and control groups were, respectively, collected and analyzed.
Total RNA was extracted using TRIzol Reagent (Invitrogen, United States) following the manufacturer’s instruction. RNA was eluted in 10–20 μL RNase-free water depending on the size of precipitate. The quality and concentration of RNA were measured using NanoDrop (Thermo Fisher Scientific, United States). Then 500 ng RNA was used to synthesize the first strand cDNA with Primescript RT reagent kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). Finally, all the cDNA samples were diluted 10-folds to be used as templates of RT-qPCR.
The top 10 frequently used reference genes in insects were Actin, RPL, Tubulin, GAPDH, RPS, 18S, EF1A, TATA, HSP, and SDHA (Lü et al., 2018). We picked out the genes, annotated as the homologs of these 10 widely used reference genes, from our RNA-seq data of embryonic stages (unpublished data). The low expression genes (TPM < 5 in at least 2 stages) were ruled out. Then, variance stabilizing transformation (VST) in R software (Anders and Huber, 2010) was used to eliminate the bias caused by expression level. Finally, the standard deviation of each gene was calculated. Except genes from our data, 18S (18S ribosomal protein sequence, accession: JQ695802.1) and NADH (NADH-ubiquinone oxidoreductase subunit 8, accession: XM_012688246) were also added as potential candidate reference gene. Primers for each candidate gene were manually designed using Primer Premier 5.0 (Lalitha, 2002), and the quality of primers were evaluated by Oligo 7.0 (Rychlik, 2007; Table 1).
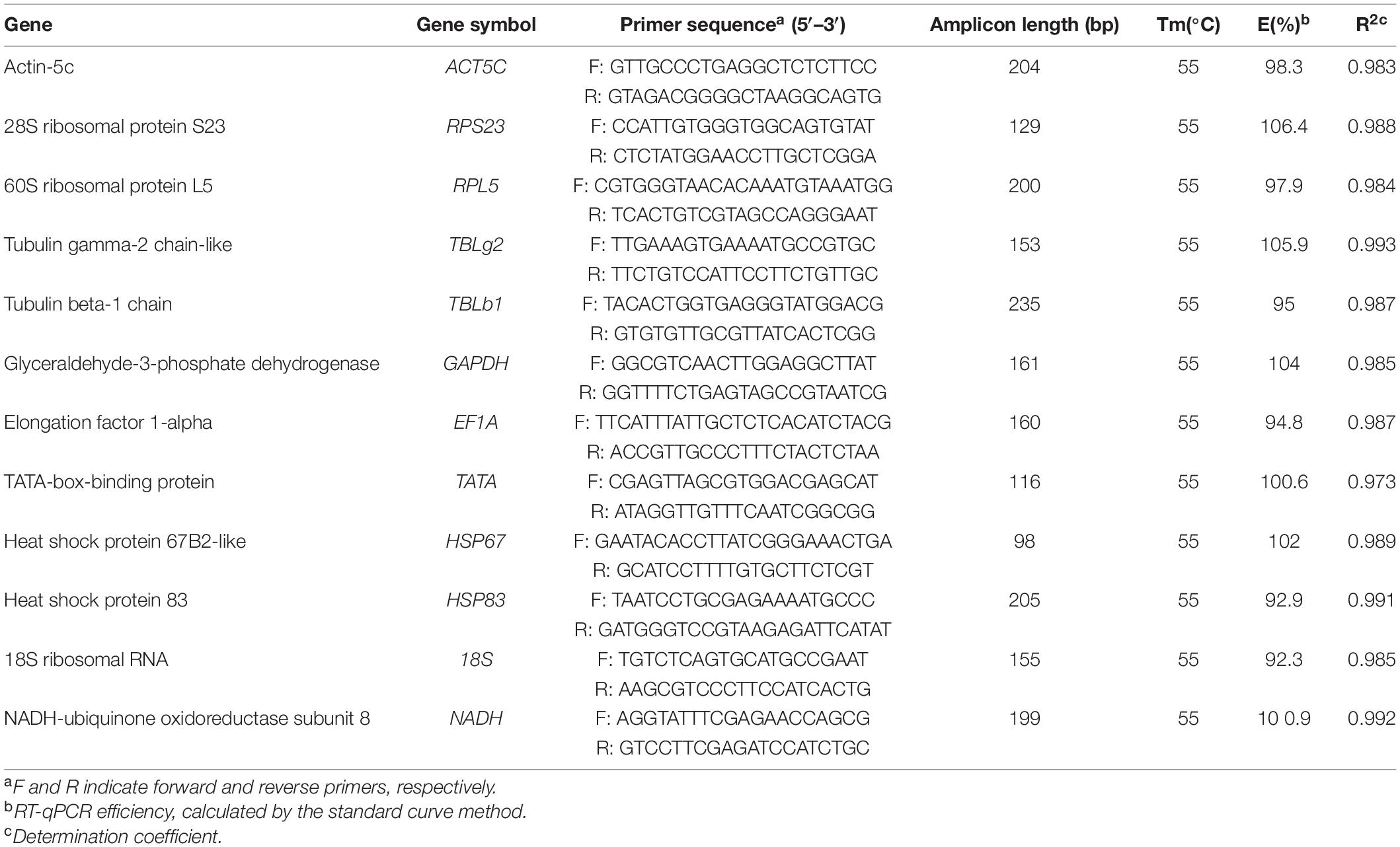
Table 1. The primer sets and amplification efficiency of the potential reference genes used for RT-qPCR.
RT-qPCR was conducted using CFX96 Touch Real-time PCR Detection System (Bio-Rad, United States) based on TB Green Premix Ex Taq (Tli RNaseH Plus) (TaKaRa, Japan). Amplifications were carried out under the following conditions: initial denaturation at 95°C for 5 min followed by 40 cycles of 20 s at 95°C, for 20 s at 55°C, and for 20 s at 72°C. This was followed by 1 min at 95°C and 1 min at 50°C. Finally, a melting curve analysis with the temperature raised from 65 to 95°C in sequential steps of 0.5°C for 10 s. The reaction system is 20 μL, including 10 μL TB Green Premix EX Taq polymerase, 0.5 μL of each primer, 2 μL cDNA and 7 μL ddH2O. For each biological sample, three technical replicates were performed and the average cycle quantification (Cq) value were calculated. To create the standard curve, the amplicons were cloned into pMD-19T vector (TaKaRa, Japan). Then six 10-fold serial dilutions (from 1e + 4 to 1e + 9 copy number per microliter) were made according to copy number of the plasmids. With the dilutions as templates to conduct RT-qPCR, the standard curves were made. The amplification efficiency (E) values were calculated based on the equation: E = [10(–1/slope)-1] × 100 (Radonić et al., 2004), where slope is the slope of the standard curve.
The average Cq values were used for data analysis. Four algorithms including geNorm, NormFinder, BestKeeper and comparative ΔCt method, were used to evaluate the expression stability of the 12 potential reference genes. Finally, RefFinder was used to conduct a comprehensive evaluation and screen the optimal reference genes. The optimal number of reference genes for normalization was determined by geNorm.
In our RNA-seq data, none of the genes was annotated as 18S and SDHA, two of the top 10 candidate genes in insects; 205 genes were annotated as the homologs of the other eight reference genes (Supplementary Table 1). After ruling out low expression genes according to the quantification with RNA-seq, 175 genes were left (Supplementary Table 2). Then we ranked the homologs of each gene according to their standard deviation across 16 embryonic stages (Supplementary Table 3). Finally, we chose potential reference genes with two criteria if one gene has more than one homolog: (1) have the low standard deviation among the homologs; (2) the TPM value should larger than 1,000 in at least eight stages. Addition to 18S, annotated in NCBI, and NADH, validated in ants (do Livramento et al., 2018; Rajakumar et al., 2018), we obtained a list of 12 candidate reference genes for the subsequent experiments (Table 1).
Firstly, the specificity of these reference genes was investigated by RT-PCR. All genes had a single band of the expected size on 1% agarose gel. Sequencing results showed that these amplicons were identical to corresponding gene sequences. Also, they had single peaks in melting-curve analysis in RT-qPCR (Supplementary Figure 1). The amplification efficiency for each gene ranged from 92.3 to 106.4%, showing a good amplification performance (Table 1). And all standard curves are shown in Supplementary Figure 2.
Among all samples for seven conditions, gene expression analysis of the 12 candidate reference genes presented a broad range of variance of Cq values from 9.57 (18S) to 33.03 (TATA) (Figure 1). Of the 12 reference genes, 18S was the most abundant transcripts, and TATA was expressed at the lowest level.
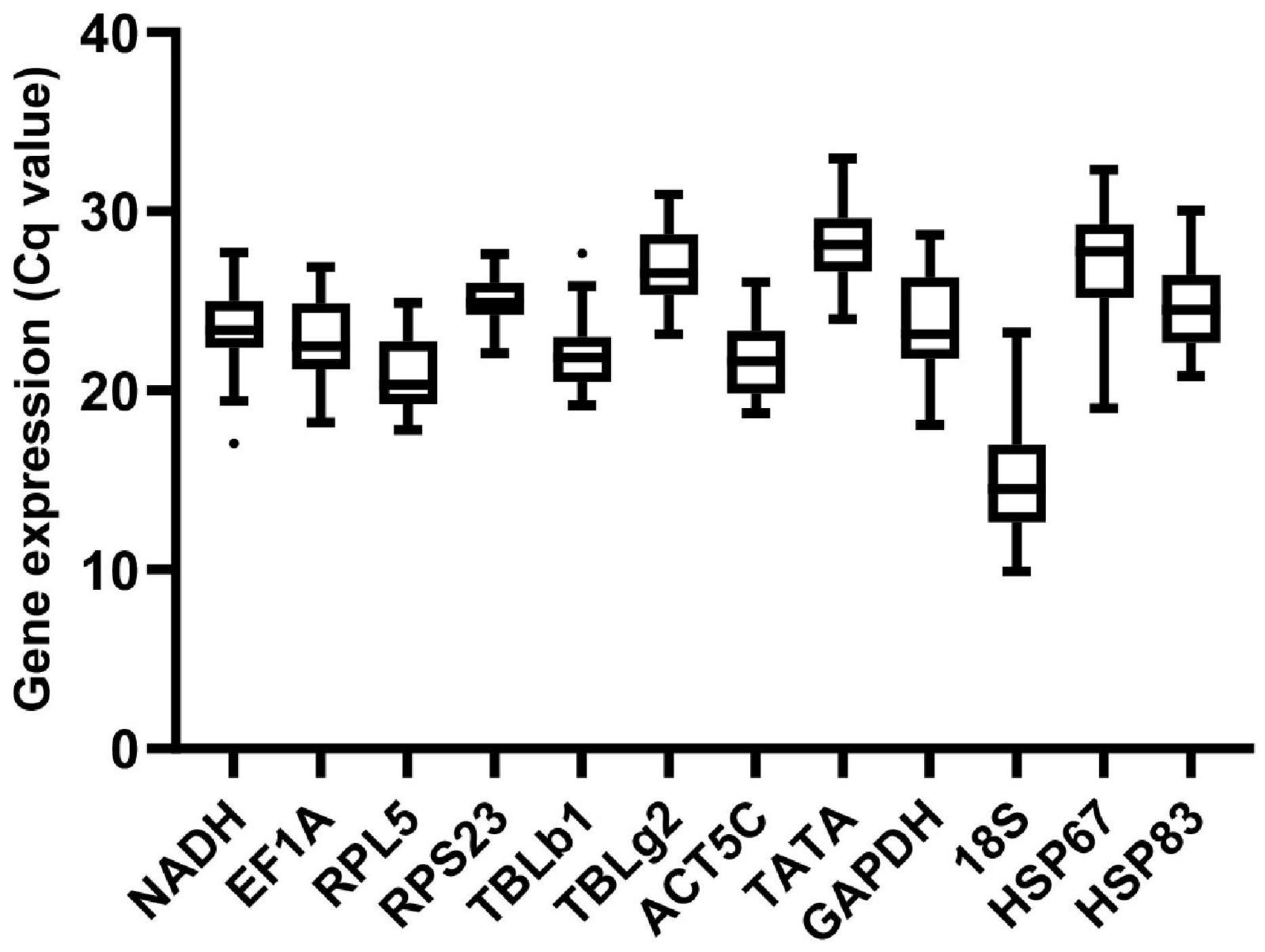
Figure 1. Expression profiles of reference genes in all 36 samples in M. pharaonis. The expression level of reference genes is presented with Cq value. The dots indicate outliers.
All analysis programs ranked TATA and 18S as the least stable genes. However, the programs indicated different candidates for the most stable gene. GAPDH was the most stable gene in comparative ΔCt and NormFinder. However, geNorm indicated both NADH and EF1A are the most stable gene. The most stable gene RPL5, calculated by BestKeeper, only ranked in the 10th position in comparative ΔCt and NormFinder (Table 2). Integration of the results by RefFinder gives an overall stability ranking: GAPDH > EF1A > ACT5C > NADH > RPS23 > HSP67 > RPL5 > TBLg2 > TBLb1 > HSP83 > TATA > 18S (Figure 2A). This order is different from that according to RNA-seq data: RPS23 > HSP67 > NADH > RPL5 > EF1A > TATA > TBLg2 > HSP83 > TBLb1 > GAPDH > ACT5C (Supplementary Table 3).
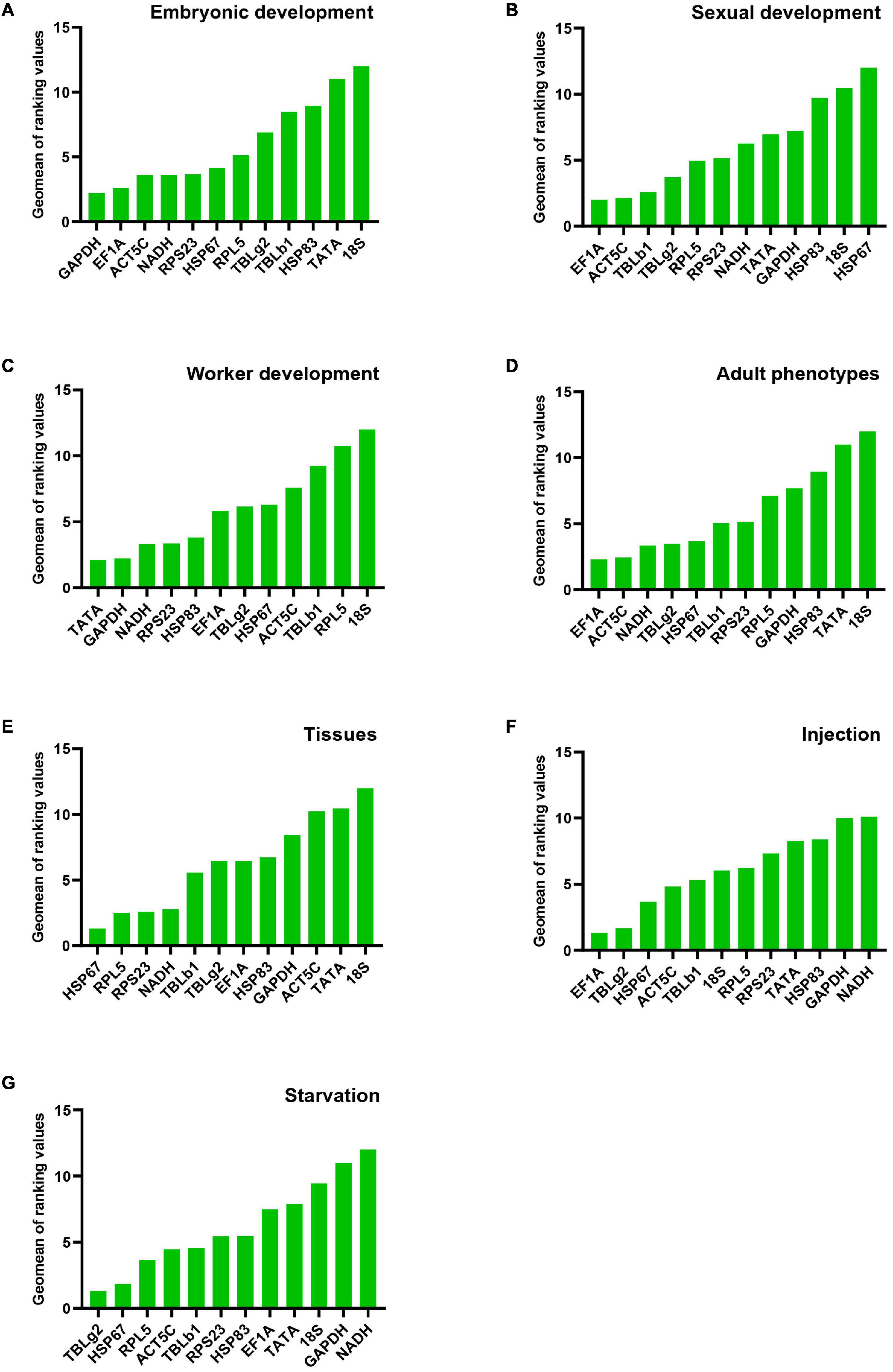
Figure 2. Stability of candidate reference gene expression in M. pharaonis in conditions of embryonic development (A), sexual development (B), worker development (C), adult phenotypes (D), tissues (E), injection (F), and starvation (G). The stability of reference gene expression was calculated by RefFinder, which summarizing four different algorithms (geNorm, Norm finder, BestKeeper, and comparitive ΔCt). A lower value means more stable expression.
For the stability ranking, four software showed distinct orders due to different algorithms. ACT5C was regarded the most stable reference gene in comparative ΔCt and NormFinder. However, it ranked 7th in BestKeeper. HSP67 was the least stable reference gene during sexual development in four programs (Table 2). RefFinder was used to integrate the results of the four programs. The overall stability ranking of the reference genes is: EF1A > ACT5C > TBLb1 > TBLg2 > RPL5 > RPS23 > NADH > TATA > GAPDH > HSP83 > 18S > HSP67 (Figure 2B).
Data from four developmental stages of worker, 3rd instar larvae, young pupae, old pupae, and adults, were used to screen the stable reference genes. NADH and HSP83, GAPDH, RPS23, TATA were considered as the most stable reference genes by geNorm, NormFinder, BestKeeper, and comparative ΔCt, respectively (Table 2). The overall stability ranking of the reference genes by RefFinder is: TATA > GAPDH > NADH > RPS23 > HSP83 > EF1A > TBLg2 > HSP67 > ACT5C > TBLb1 > RPL5 > 18S (Figure 2C).
All analysis programs indicated that 18S was the least stable gene. However, they showed different choices for the most stable gene. EF1A was regarded as the most stable reference gene in comparative ΔCt and NormFinder. However, it was ranked at 7th position in BestKeeper (Table 2). RefFinder was used to integrate the results of the four programs. The overall stability ranking of the reference genes by RefFinder is: EF1A > ACT5C > NADH > TBLg2 > HSP67 > TBLb1 > RPS23 > RPL5 > GAPDH > HSP83 > TATA > 18S (Figure 2D).
All analysis programs indicated 18S is the least stable genes among the brain, midgut, fat body, and ovary of gyne/queen, but they showed different results for the most stable gene. Comparative ΔCt, NormFinder, and BestKeeper suggested HSP67 is the most stable gene, while both RPS23 and RPL5 were presented as the most stable gene in geNorm (Table 2). According to RefFinder, the overall stability of the reference genes was ranked as follows: HSP67 > RPL5 > RPS23 > NADH > TBLb1 > TBLg2 > EF1A > HSP83 > GAPDH > ACT5C > TATA > 18S (Figure 2E).
Injection of eGFP dsRNA results in the change of stability ranking among these genes. All programs, except BestKeeper, suggested EF1A is the most stable gene. While BestKeeper indicated 18S is the most stable gene (Table 3). According to RefFinder, the overall stability ranking of the reference genes is EF1A > TBLg2 > HSP67 > ACT5C > TBLb1 > 18S > RPL5 > RPS23 > TATA > HSP83 > GAPDH > NADH (Figure 2F).
For starvation treatment, the stability ranking was also changed compared with non-starvation control group. All analysis programs, except BestKeeper, indicated TBLg2 was the most stable gene (Table 3). According to RefFinder, the overall stability ranking of the reference genes is TBLg2 > HSP67 > RPL5 > ACT5C > TBLb1 > RPS23 > HSP83 > EF1A > TATA > 18S > GAPDH > NADH (Figure 2).
Significant errors can be caused by using inappropriate number of reference genes (do Livramento et al., 2018). The minimal number of reference genes necessary for normalization is determined by V-value (pairwise variation) in geNorm program. V-value is calculated by two neighboring normalization factors (NF). Usually, a value below 0.15 is considered ideal but it depends on situation. However, 0.15 must not to be taken as a too strict cut-off. In some cases, the trend is also equally informative (Vandesompele et al., 2002).
In our study, the optimal numbers of reference genes are various under different experimental conditions. Using 0.15 as a threshold value, the optimal number of reference genes for worker development, tissues, injection, and starvation are 4, 4, 3, and 5, respectively. For gene expression analysis in embryonic development and adult phenotypes, the V values were all below 0.15, thus the observed trend (of changing V-values when using additional genes) can be used as guidance. Three and two reference genes are needed for embryonic development and adult phenotypes, respectively. As to sexual development, four reference genes are required for normalization. Therefore, we must make a balance between efficiency and accuracy (Figure 3).
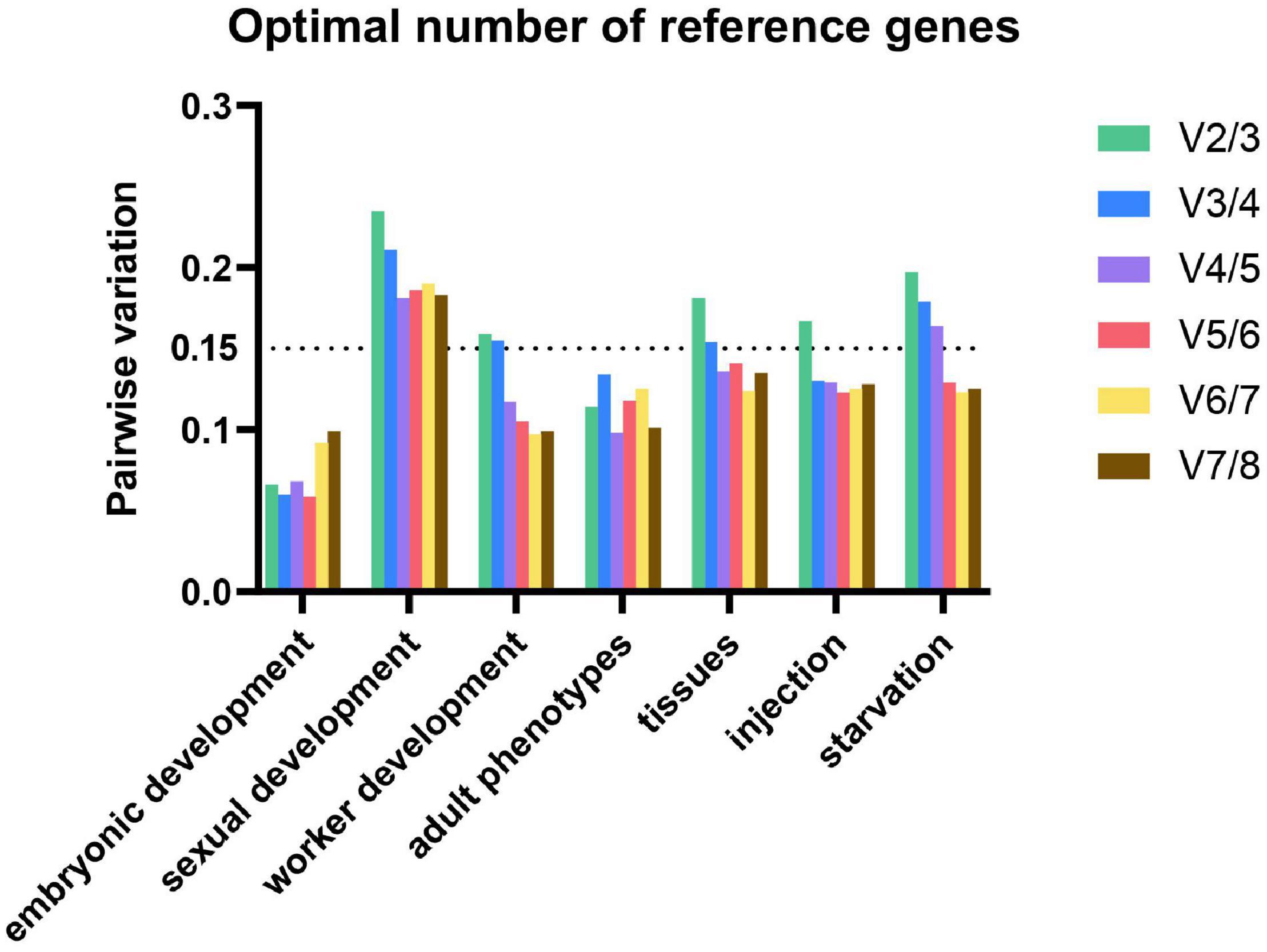
Figure 3. Optimal number of reference genes for normalization in M. pharaonis. The pairwise variation (Vn/Vn+1) was analyzed between the normalization factors NFn and NFn+1 by geNorm. Average value of pairwise variations (v) indicates the optimal number of reference genes. The values that above the threshold value of 0.15 means an extra reference gene should add to the stability of the normalization factor.
RT-qPCR is a widely used method to evaluate the gene expression levels because of its low cost and high sensitivity. Especially in the genomic era, differentially expressed genes identified by RNA-seq analysis need to be verified by RT-qPCR (Bonasio et al., 2010; Gospocic et al., 2017). In our study, the stability ranking of reference genes was calculated according to RNA-seq data and RT-qPCR data. The different stability ranking indicate that RT-qPCR verification is indispensable even that RNA-seq data can provide a wealth of quantitative gene expression information. On the other hand, RT-qPCR data can improve our efficiency to choose potential candidate genes. It is not easy to make choice manually among 205 potential candidate genes. To minimize the variation and errors in RT-qPCR, data normalization is essential to obtain accurate gene expression data.
Some genes have already been frequently used as reference genes in RT-qPCR experiment (Lü et al., 2018). However, it is impossible to find a universal reference gene applicable to all species since the gene can participate in different cellular functions in different species (Moreira et al., 2017; do Livramento et al., 2018). NADH has been validated as a stable reference gene for RT-qPCR in several hymenopteran species (do Livramento et al., 2018; Jia et al., 2018; Rajakumar et al., 2018), but our data shows that it is not suitable for some situation of M. pharaonis. Especially, for gene expression analysis of neural plasticity between gyne and queen tissues, NADH is the least stable gene among the 12 reference genes. Since reference gene directly affects the results of gene expression analysis, it is necessary to do a comprehensive selection and validation of reference genes in M. pharaonis in different conditions.
Previous studies showed that only one reference gene was used in some gene expression analysis in ant, such as ribosomal protein L32 in Camponotus floridanus (Kupper et al., 2016) and β-actin in Polyrhachis vicina Roger (Shumin et al., 2010; Liu et al., 2013). However, the amplification efficiency of single reference gene was not mentioned in these studies. Our study gives the first insights into the knowledge of reference genes in M. pharaonis, which provides a valuable guideline for future studies interrogating molecular basis underlying division of labor in ants.
Notably, this is the first study to evaluate reference genes under abiotic treatments in ants. The treatment of injection and starvation are often used to study the genes or nutritional factors influencing queen reproductive ability. Thus, Whether the expression of reference genes is influenced by the abiotic treatment need to be validated. Although the stability rankings under different treatments are not the same as the control, the orders do not change dramatically. The most stable genes (HSP67, RPL5, and ACT5C) in control group, still show good performance in abiotic treatment groups.
In conclusion, we evaluated performance of 12 candidate reference genes in M. pharaonis under different situations by using four different methods. Consensus lists for gene stability were obtained and the optimal reference gene numbers were recommended under different situations. The result presented here is essential for gene expression analysis and will guide future gene function studies in pharaoh ant.
The data presented in the study are deposited in the NCBI GEO repository (https://www.ncbi.nlm.nih.gov/geo/), accession number BioProject PRJNA767561.
GD, GZ, and WL designed the study. JZ collected the samples. GD, QG, and JC performed the experiments and collected the data. GD analyzed the data. GD, QG, and WL wrote the manuscript. All authors read and approved the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (No. 31900399) to WL.
GD and GZ were employed by BGI-Shenzhen.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Ruyan Li for offering NADH probe information.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.852357/full#supplementary-material
Abouheif, E., and Wray, G. A. (2002). Evolution of the gene network underlying wing polyphenism in ants. Science 297, 249–252. doi: 10.1126/science.1071468
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genom. Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Berbdt, K., and Kremer, G. (1986). Die Larvenmorphologie der Pharaoameise Monomorium pharaonis (L.) (Hymenoptera, Formicidae). Zool. Anz. 216, 305–320.
Bonasio, R., Zhang, G., Ye, C., Mutti, N. S., Fang, X., Qin, N., et al. (2010). Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071. doi: 10.1126/science.1192428
Bustin, S. A. (2000). Absolute quantification of mrna using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193. doi: 10.1677/jme.0.0250169
Butte, A. J., Dzau, V. J., and Glueck, S. B. (2001). Further defining housekeeping, or “maintenance,” genes Focus on “A compendium of gene expression in normal human tissues.”. Physiol. Genom. 7, 95–96. doi: 10.1152/physiolgenomics.2001.7.2.95
Chandra, V., Fetter-Pruneda, I., Oxley, P. R., Ritger, A. L., McKenzie, S. K., Libbrecht, R., et al. (2018). Social regulation of insulin signaling and the evolution of eusociality in ants. Science 361, 398–402. doi: 10.1126/science.aar5723
Cheng, D., Zhang, Z., He, X., and Liang, G. (2013). Validation of Reference Genes in Solenopsis invicta in Different Developmental Stages, Castes and Tissues. PLoS One 8:e57718. doi: 10.1371/journal.pone.0057718
Derveaux, S., Vandesompele, J., and Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. doi: 10.1016/j.ymeth.2009.11.001
do Livramento, K. G., Freitas, N. C., Máximo, W. P. F., Zanetti, R., and Paiva, L. V. (2018). Gene expression profile analysis is directly affected by the selected reference gene: the case of leaf-cutting atta sexdens. Insects 9:18. doi: 10.3390/insects9010018
Edwards, J. P. (1991). Caste regulation in the pharaoh’s ant Monomorium pharaonis: recognition and cannibalism of sexual brood by workers. Physiol. Entomol. 16, 263–271. doi: 10.1111/j.1365-3032.1991.tb00565.x
Evans, J. D., and Wheeler, D. E. (2002). Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl. Acad. Sci. 96, 5575–5580. doi: 10.1073/pnas.96.10.5575
Gao, X. K., Zhang, S., Luo, J. Y., Wang, C. Y., Lü, L. M., Zhang, L. J., et al. (2017). Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene 637, 211–218. doi: 10.1016/j.gene.2017.09.057
Gospocic, J., Shields, E. J., Glastad, K. M., Lin, Y., Penick, C. A., Yan, H., et al. (2017). The Neuropeptide Corazonin Controls Social Behavior and Caste Identity in Ants. Cell 170, 748.e–759.e. doi: 10.1016/j.cell.2017.07.014
Jia, L. Y., Chen, L., Keller, L., Wang, J., Xiao, J. H., and Huang, D. W. (2018). Doublesex evolution is correlated with social complexity in ants. Genom. Biol. Evol. 10, 3230–3242. doi: 10.1093/gbe/evy250
Kang, Z. W., Liu, F. H., Tian, H. G., Zhang, M., Guo, S. S., and Liu, T. X. (2017). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Khila, A., and Abouheif, E. (2010). Evaluating the role of reproductive constraints in ant social evolution. Philos. Trans. R. Soc. B Biol. Sci. 365, 617–630. doi: 10.1098/rstb.2009.0257
Kupper, M., Stigloher, C., Feldhaar, H., and Gross, R. (2016). Distribution of the obligate endosymbiont Blochmannia floridanus and expression analysis of putative immune genes in ovaries of the carpenter ant Camponotus floridanus. Arthropod Struct. Dev. 45, 475–487. doi: 10.1016/j.asd.2016.09.004
Lalitha, S. (2002). Primer Premier 5. Biotech. Softw. Internet Rep. 1, 270–272. doi: 10.1089/152791600459894
Li, Q., Wang, Z., Lian, J., Schiøtt, M., Jin, L., Zhang, P., et al. (2014). Caste-specific RNA editomes in the leaf-cutting ant Acromyrmex echinatior. Nat. Commun. 5:4943. doi: 10.1038/ncomms5943
Liu, L. P., Dang, L., Xi, G. S., and Wang, F. (2013). Molecular cloning, characterization and expression analysis of calreticulin gene in the ant polyrhachis vicina roger (hymenoptera: SsFormicidae). Sociobiology 60, 355–361. doi: 10.13102/sociobiology.v60i4.355-361
Lü, J., Yang, C., Zhang, Y., and Pan, H. (2018). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Mikheyev, A. S., and Linksvayer, T. A. (2015). Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. Elife 2015:e04775. doi: 10.7554/eLife.04775
Moreira, A. C., dos Santos, A. M., Carneiro, R. L., Bueno, O. C., and Souza, D. H. F. (2017). Validation of reference genes in leaf-cutting ant Atta sexdens rubropilosa in different developmental stages and tissues. Int. J. Environ. Agric. Biotechnol. 2, 743–755. doi: 10.22161/ijeab/2.2.23
Nolan, T., Hands, R. E., and Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Peacock, A., and Baxter, A. (1949). Studies in Pharaoh’s ant, Monomorium pharaonis (L.).I. The rearing of artificial colonies. Entomol. Mon. Mag. 85, 256–260.
Pereboom, J. J. M., Jordan, W. C., Sumner, S., Hammond, R. L., and Bourke, A. F. G. (2005). Differential gene expression in queen-worker caste determination in bumble-bees. Proc. R. Soc. B Biol. Sci. 272, 1145–1152. doi: 10.1098/rspb.2005.3060
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Qiu, B., Larsen, R. S., Chang, N. C., Wang, J., Boomsma, J. J., and Zhang, G. (2018). Towards reconstructing the ancestral brain gene-network regulating caste differentiation in ants. Nat. Ecol. Evol. 2, 1782–1791. doi: 10.1038/s41559-018-0689-x
Radonić, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862. doi: 10.1016/j.bbrc.2003.11.177
Rajakumar, R., Koch, S., Couture, M., Favé, M. J., Lillico-Ouachour, A., Chen, T., et al. (2018). Social regulation of a rudimentary organ generates complex worker-caste systems in ants. Nature 562, 574–577. doi: 10.1038/s41586-018-0613-1
Rychlik, W. (2007). OLIGO 7 primer analysis software. Methods Mol. Biol. 402, 35–59. doi: 10.1385/1-59745-528-8:35
Schmidt, A. M., Linksvayer, T. A., Boomsma, J. J., and Pedersen, J. S. (2011). No benefit in diversity? The effect of genetic variation on survival and disease resistance in a polygynous social insect. Ecol. Entomol. 36, 751–759. doi: 10.1111/j.1365-2311.2011.01325.x
Shumin, L., Ming, J., Cuiping, B., and Gengxi, S. (2010). Real-time quantitative RT-PCR detection for developmental expression of a muscarinic cholinergic receptor gene in the ant Polyrhachis vicina. J. Northwest A. F. Univ. 38, 161–166. doi: 10.1111/j.1744-7917.2010.01338.x
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Smith, C. R., and Suarez, A. V. (2010). The trophic ecology of castes in harvester ant colonies. Funct. Ecol. 24, 122–130. doi: 10.1111/j.1365-2435.2009.01604.x
Trible, W., and Kronauer, D. J. C. (2017). Caste development and evolution in ants: it’s all about size. J. Exp. Biol. 220, 53–62. doi: 10.1242/jeb.145292
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genom. Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Warner, M. R., Qiu, L., Holmes, M. J., Mikheyev, A. S., and Linksvayer, T. A. (2019). Convergent eusocial evolution is based on a shared reproductive groundplan plus lineage-specific plastic genes. Nat. Commun. 10:261. doi: 10.1038/s41467-019-10546-w
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2
Keywords: Monomorium pharaonis, real-time quantitative PCR, RNA-seq, reference gene, expression stability, optimal reference gene number
Citation: Ding G, Gao Q, Chen J, Zhao J, Zhang G and Liu W (2022) Validation of Potential Reference Genes for Real-Time qPCR Analysis in Pharaoh Ant, Monomorium pharaonis (Hymenoptera: Formicidae). Front. Physiol. 13:852357. doi: 10.3389/fphys.2022.852357
Received: 11 January 2022; Accepted: 27 January 2022;
Published: 28 February 2022.
Edited by:
Shun-Fan Wu, Nanjing Agricultural University, ChinaReviewed by:
Yixiang Qi, South China Agricultural University, ChinaCopyright © 2022 Ding, Gao, Chen, Zhao, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojie Zhang, Z3VvamllLnpoYW5nQGJpby5rdS5kaw==; Weiwei Liu, bGl1d2Vpd2VpQG1haWwua2l6LmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.