
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 06 April 2022
Sec. Aquatic Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.831226
This article is part of the Research TopicNew Insights into Mucosal-Associated Lymphoid Tissues in FishView all 16 articles
 Ming Li1,2†
Ming Li1,2† Xuyang Zhao1,2†
Xuyang Zhao1,2† Jiayuan Xie1,3
Jiayuan Xie1,3 Xinyu Tong4
Xinyu Tong4 Junwei Shan1,2
Junwei Shan1,2 Mijuan Shi1
Mijuan Shi1 Guangxin Wang1
Guangxin Wang1 Weidong Ye1,3
Weidong Ye1,3 Yuhang Liu1,2
Yuhang Liu1,2 Bruno Hamish Unger1
Bruno Hamish Unger1 Yingyin Cheng1
Yingyin Cheng1 Wanting Zhang1
Wanting Zhang1 Nan Wu1,3*
Nan Wu1,3* Xiao-Qin Xia1,3*
Xiao-Qin Xia1,3*To help prevent foodborne enteritis in aquaculture, several feed additives, such as herbal medicine, have been added to fish diets. Predictions of effective herb medicines for treating fish foodborne enteritis from key regulated DEGs (differentially expressed genes) in transcriptomic data can aid in the development of feed additives using the Traditional Chinese Medicine Integrated Database. Seabuckthorn has been assessed as a promising candidate for treating grass carp soybean-induced enteritis (SBMIE). In the present study, the SBMIE zebrafish model was used to assess seabuckthorn’s therapeutic or preventative effects. The results showed that intestinal and hepatic inflammation was reduced when seabuckthorn was added, either pathologically (improved intestinal villi morphology, less oil-drops) or growth-related (body fat deposition). Moreover, seabuckthorn may block the intestinal p53 signaling pathway, while activating the PPAR signaling pathway and fatty acid metabolism in the liver. 16S rRNA gene sequencing results also indicated a significant increase in OTU numbers and skewed overlapping with the fish meal group following the addition of seabuckthorn. Additionally, there were signs of altered gut microbiota taxa composition, particularly for reduced TM7, Sphingomonas, and Shigella, following the addition of seabuckthorn. Hindgut imaging of fluorescent immune cells in SBMIE larvae revealed the immune regulatory mechanisms at the cellular level. Seabuckthorn may significantly inhibit the inflammatory gathering of neutrophils, macrophages, and mature T cells, as well as cellular protrusions’ formation. On the other hand, in larvae, seabuckthorn inhibited the inflammatory aggregation of lck+ T cells but not immature lymphocytes, indicating that it affected intestinal adaptive immunity. Although seabuckthorn did not affect the distribution of intestinal CD4+ cells, the number of hepatic CD4+ cells were reduced in fish from the seabuckthorn supplementation group. Thus, the current data indicate that seabuckthorn may alleviate foodborne gut-liver symptoms by enhancing intestinal mucosal immunity and microbiota while simultaneously inhibiting hepatic adipose disposition, making it a potential additive for preventing fish foodborne gut-liver symptoms.
Due to the high cost and limited supply of fishmeal, the increased inclusion of plant proteins in aquafeeds has become a common practice (Li et al., 2019). Given the importance of plant proteins as fish meal substitutes in aquaculture, consequential side effects such as foodborne enteritis and hepatic symptoms should be avoided or treated. Though carnivorous fish species are much more sensitive to plant-sourced proteins like soybean meal (SBM) (Booman et al., 2018), herbivorous and omnivorous species like grass carp (Ctenopharyngodon idella) and zebrafish (Danio rerio) can also develop enteritis and liver pathologies (Wu et al., 2018; Wu et al., 2020). Recently, additives or probiotics have been used to prevent decreased digestion, enteritis, and poor growth performance caused by over-substitution with plant proteins.
With advancing transgenic techniques, fluorescent proteins can be used with zebrafish lines to label various immune cell populations, including T cells (Kasheta et al., 2017; Coronado et al., 2019; Li et al., 2020a), macrophages (Ellett et al., 2011) and neutrophils (Brugman et al., 2009; Oehlers et al., 2011; Okuda et al., 2015; Enya et al., 2018). At approximately 1 week of age (6 to 9 dpf), the early phases of intestinal innate immune cells, including neutrophils and macrophages, can be detected (Bravo-Tello et al., 2017; Ikeda-Ohtsubo et al., 2020). At an adaptive level, intestinal lymphocytes, including mainly T and B lymphocytes can also be observed at a very early age (5 dpf), and later the response related to T cell subpopulations could be observed in 3- to 4- week-old larva (before intestinal pigments cover) (Coronado et al., 2019). Thus, zebrafish may be an excellent model organism for screening and demonstrating the processes of innovative and effective additions.
Additionally, computational approaches to medicine selection have shed light on veterinary research and its applications. Herb medicines have been used to treat animal diseases and can occasionally be used in place of antibiotics to improve animal health. The bioinformatics database TCMID (Traditional Chinese Medicines Integrated Database) contains comprehensive information on the interactions of compounds, proteins and herbs (Qu et al., 2019a; Zhang et al., 2020a). The database can provide formula, herbal medicine, herbal ingredients, and herbal ingredients-target information. In aquaculture, herbs have also been used to enhance fish immunity and growth performance (Zhang et al., 2009; Abarike et al., 2019).
Dietary supplements containing Astragalus membranaceus, Angelica sinensis, and Crataegus hupehensis can increase the expression of immune genes such as beta-defensin, lysozyme, and heat shock protein 70 in both the intestine and the head-kidney, enhancing growth performance and disease resistance in tilapia (Oreochromis niloticus) (Abarike et al., 2019). Propolis and Herba Epimedii extracts may promote respiratory burst activity, phagocytosis of phagocytic cells in the blood, and lysozyme activity in plasma in Chinese suckerfish (Myxocyprinus asiaticus) (Zhang et al., 2009). As a result of its widespread usage in systematic pharmacology and the recent findings on effective herbs (Zhang et al., 2020b; Wang et al., 2021; Ye et al., 2021; Yu et al., 2021) for treating enteritis (Yu et al., 2021), TCMID may also serve as a platform for predicting successful medications or feed additives in aquaculture.
The protective effect of dietary ingredients affects both the gut and liver of fish via the biofluid network (Roques et al., 2020). This includes bile acid, intestinal mucus, and microbiota, as well as metabolites and immune cells found in the blood. Hepatic disease can result in dysfunction of the intestinal barrier and host-microbiome imbalances (Tranah et al., 2021). Thus, the fish gut-liver immune axis has been proposed (Wu et al., 2016) and has been demonstrated to be involved in the development of foodborne gut-liver symptoms (Wu et al., 2018) in a variety of commercial species. While zebrafish are a commonly used animal model in aquaculture research, it was discovered that hepatic health also affects the intestine mucosa. For instance, the hepatotoxicity of palmitic acid alters the intestinal microbiota (Ding et al., 2018).
Gut microbiota are essential for the gut-liver axis’ immune homeostasis (Yang et al., 2020). Diet may have a significant impact on the complex interplay between the gut microbiota, the intestinal barrier, the immune system, and the liver (Martín-Mateos and Albillos, 2021). In fish, the gut microbiota is involved in intestinal barrier function, such as intestinal immune reactions and intestinal integrity (Li et al., 2019). The fact that not only effective additives but also fermented soybean meal (FSBM) are utilized to complement the diet may help alleviate foodborne enteritis in the fish (Catalán et al., 2018; Li et al., 2019; Liu et al., 2019; Li et al., 2020b), indicating the bacteria’s functional role in intestinal homeostasis. FSBM was discovered to improve the health and growth physiology of salmon by increasing the growth of intestinal lactic acid bacteria, resulting in a prebiotic-like effect (Catalán et al., 2018). SBM fermented with Enterococcus faecium (Li et al., 2020b) or the addition of the commensal bacterial Shewanella sp. MR7 (Li et al., 2019) is effective against the typical SBMIE (soybean meal induced enteritis) in turbot. Supplementation of Bacillus’ metabolite sodium butyrate in a high-soybean meal fish diet (40% fish meal protein replaced by soy protein) may alter the composition of the intestinal microbiota whilst regulating intestinal gene expression, such as by downregulating TNF-alpha and NF-kappaB and upregulating genes involved in the intestinal epithelium’s tight junction (Liu et al., 2019).
Seabuckthorn (Hippophae rhamnoides), a traditional Mongolian and Tibetan medicine, has been shown to protect against liver disease by regulating the liver metabolome and abundance of gut microbiota (Ran et al., 2021). Its protective effect on the liver has been well established, not only in diet-induced but also in toxicant-induced hepatic symptoms (Kwon et al., 2017; Wang et al., 2018; Zhao et al., 2020). However, research on its intestinal effect is still in its infancy. As a result of the gut-liver axis, the gut microbiota may be influenced, when seabuckthorn supplementation in the diet increases the ratio of Firmicutes/Bacteroidetes (Guo et al., 2020). For treating intestinal damage or inflammation, seabuckthorn oil downregulated intestinal mRNA levels of inflammatory factors, including TNF-α, IL-1β, IL-6 and IL-8 in mice (Shi et al., 2017). Additionally, seabuckthorn berry juice may increase the antioxidant capacity of rats in a simulated gastrointestinal tract (Zhao et al., 2020). As a result, seabuckthorn was hypothesized to affect fish gut-liver symptoms. TCMID was used to screen drugs for SBMIE in this study, and it was predicted that seabuckthorn, as well as the target genes, may aid in the treatment of fish SBMIE.
The current study used the TCMID (http://www.megabionet.org/tcmid/) to predict the target drug to investigate in the treatment of fish enteritis. The STRING database was used to obtain information on protein-protein interactions. To identify key fish SBMIE genes, we used the Page Rank algorithm to access previously published grass carp SBMIE transcriptomic DEG data (Wu et al., 2018), which included gene ID, fold change, and expression level (Morrison et al., 2005). The importance of a gene in this algorithm was determined by the network’s structure and its initial importance. The structure of the network can be determined by the gene dependence network, and the initial significance of the gene can be determined by its differential expression.
Fold-change was used to calculate its differential expression: fd = log2 a − log2 b. Where “a” was the expressional value of a gene in the diseased intestine and “b” was the expressional value of the gene in the control. Therefore, when fd > 0, it showed that the gene was upregulated by fd times in disease tissues, and if fd < 0, it showed that the gene was downregulated by fd times. On this basis, the following formula was used to calculate the importance of each gene:
The formula indicated that the relevance of vertex “j” was dependent on two values: the initial importance of the gene (the differential expression value of gene I and the importance of all vertices pointing to vertex “j” (the second term on the right of the formula). If “d” was greater, it indicated that the genes’ importance was more dependent on gene dependence. When “d” was less than one, it indicated that the genes were more reliant on their initial importance. In this case, “d” prioritized 0.5. The DEGs (differentially expressed genes) of SBMIE can be sorted according to their relevance from highest to lowest using the above calculation.
The top 300 Generank-ranked genes in grass carp SBMIE DEGs (Wu et al., 2018) were used as the drug’s target genes, and the herbal components that can target these genes were determined using the TCMID database’s information on the relationships between effective compounds and genes (Huang et al., 2018). Herbal medicines containing the herbal ingredients discovered using the aforementioned methods were mined using the TCMID database’s information on herbal ingredients. The top herbal medicine was chosen for follow-up experimental verification based on the number of ingredients.
In the current study, wild-type zebrafish (AB strain), as well as several ZFIN (the Zebrafish Information Network, http://zfin.org/) included transgenic zebrafish lines, including Tg(lyz:DsRED2) (with neutrophils labeled, https://zfin.org/ZDB-TGCONSTRCT-071109-3), (Campbell et al., 2021), Tg(mpeg1:EGFP) (with macrophages labeled, http://zfin.org/ZDB-TGCONSTRCT-120117-1), (Li et al., 2020a), Tg(rag2:DsRed) (with lymphocytes labeled, http://zfin.org/ZDB-TGCONSTRCT-131022-4), (Cheng et al., 2020) and Tg(lck:lck-eGFP) (with T cells labeled, https://zfin.org/ZDB-TGCONSTRCT-070117-48), (Jerison and Quake, 2020), were purchased from CZRC (China Zebrafish Resource Center, http://en.zfish.cn/) and were maintained according to standard protocols (Renshaw et al., 2006). The transgenic zebrafish used in this study included innate immune cell labeled Tg(lyz:DsRED2); Tg(mpeg1:EGFP), as well as adaptive immune cell labeled Tg(rag2:DsRed) or Tg(lck:lck-eGFP). Specifically, since the zebrafish rag2+ cells could respond to inflammatory signals (Li et al., 2014) and the rag2E450fs mutant even lacked mature T cells (Moore et al., 2016), Tg(rag2:DsRed) was used to analyze the lymphocytes’ response with SBMIE modeling. The Institute of Hydrobiology of the Chinese Academy of Sciences approved the use of animals in this study. All experiments followed the committee’s guidelines.
Shaanxi Tengmai Biotechnology Co., Ltd. in China supplied the seabuckthorn fruit powder, which was made using a low-temperature continuous vacuum belt dryer. The frozen fruit was squeezed to extract juice after thawing and was then left to settle. Then the juice, excluding the oil, was concentrated and dried using the low-temperature continuous vacuum belt dryer. After this process, the seabuckthorn fruit powder was obtained. The negative (FM diet) and positive (SBM diet) zebrafish diets were created using a previously published zebrafish SBMIE modeling formula (Bravo-Tello et al., 2017). To test the modifying effect of seabuckthorn as an effective additive, 5,000 ppm of seabuckthorn was added to the positive control (SBM diet) in the diet formulation. A previously published method was used to conduct a feeding trial in adult fish to model soybean-induced enteritis (SBMIE) with modifications (Wu et al., 2020). In brief, 270 3-month old wide type zebrafish (with the initial weight 0.175 ± 0.05 g) were fed the FM diet for 2 weeks. Then, 6-week feeding trials of experimental diets, including FM (negative control), SBM (positive control), as well as the drug (seabuckthorn) groups (SBM +35ppm seabuckthorn fruit powder) (Table 1), were carried through feeding twice a day. The feeding trials for the SBMIE larvae model followed our recently published protocol (Xie et al., 2021). In brief, a 4-day feeding trial of experimental diets from 5 to 8 dpf using larvae of Tg(lyz:DsRED2); Tg(mpeg1:EGFP) was completed for examining the response of innate immune cells. A 10-day feeding trial of experimental diets from 17 to 27 dpf using Tg(rag2:DsRed) or Tg(lck:lck-eGFP) larvae was completed for adaptive immune cells.
Adult zebrafish hindgut, a third of the whole intestine, from bend 2 to the anus (Lickwar et al., 2017), were used for sampling. Tissue samples were collected for quantitative PCR, HE staining, oil-red staining, immunohistochemistry, and transcriptome analysis (Wu et al., 2018). We used the hindgut tissue from both female and male fish for the omics investigation to prevent sex-biased differences. The complete hindgut contents were collected in sterile tubes and preserved at −80°C for 16S rRNA gene sequencing (Deng et al., 2020). For the sampling of larvae, only the caudal part of 27 dpf larvae has been sampled for HE (Hematoxylin–eosin) staining, during the imaging of the adaptive immune cells.
To evaluate growth performance, the 3 months old zebrafish of wide type used for SBMIE modeling were randomly distributed and fed in 9 tanks. For every group, there were three repetitions and 30 fish in each tank. In the tank, the water temperature was 28 ± 0.5°C, and the pH was 7.2–7.6. Meanwhile, DO (dissolved oxygen) was kept above 5.0 mg/L. To obtain more accurate growth parameters, zebrafish were fed the experimental diet at a rate of 3% of their body weight per meal, with two meals per day and a half-hour satiety interval. Each meal was fed 6 times, with the dosage gradually decreasing to ensure that the feed for each meal was consumed by the experimental fish. During this process, we discovered that zebrafish could consume the feed without any waste.
As most of the ordinary parameters of growth performance were found to not be significantly changed (data not shown), a 3D micro-CT (Bruker SkyScan 1276 Micro-CT Scanner) was used to measure the volume of total adipocyte tissue in the zebrafish just after the feeding trial using the previously described method (Hasumura et al., 2012; Zang et al., 2018). To prevent motion interference, the zebrafish were anesthetized with tricaine methanesulfonate (MS222, Sigma-Aldrich, USA), before being put into the scanner (Xiong et al., 2017). Insta-Recon software was used to reconstruct and visualize the fish images, and CT analyzer software was used to analyze areas of fish adipose tissues and their entire trunk (Ondruš et al., 2021).
For all the histological and immunostaining analyses, the tested hindgut tissues were from three fish and there were at least 3 slice repetitions for the tissue from each fish. HE staining was used to show the structure of the mucosal fold in the hindgut (in both adult zebrafish and 27 dpf larvae) as well as the reticuloendothelial structure in the liver to demonstrate basic pathology (in adult zebrafish). According to previously published methods, oil-red staining was used to visualize the oil droplets in the liver (Wu et al., 2018). For both HE and oil-red staining analysis, the last third of the intestinal tissues from three fish in each group were used to make sections (10 μm), and at least ten sections per fish were observed in a bright field under a microscope. To observe the adult zebrafish intestine and liver tissue, a microscope (Olympus, BX53) with a magnification of 20-fold was used. Meanwhile, for the larva tissue, the Aperio VERSA Slide Scanner (Aperio VERSA 8, Leica) with a magnification of 40-fold was applied. The zebrafish anti-CD4-1 antibody (GeneTex, USA, GTX16589) was used in immunohistochemistry analysis to label mainly T helper cells in the gut and liver to further illustrate the inflammation at the lymphocyte level, as previously described (Wu et al., 2018). DAPI was also used to stain the nuclei in parallel.
The RNA extraction and qPCR analysis were carried out following the procedures outlined in our previously published paper (Wu et al., 2018; Wu et al., 2020). All RNA extractions were performed immediately following the sampling of either the hindgut or liver tissues or at the same time as the sampling for the transcriptomic study. Supplementary Table S1 lists the primers used to quantify intestinal or hepatic DEGs. The tested hindgut or liver tissues for qPCR analysis were from 6 fish which contained 3 repetitions. In each repetition, hindgut or liver tissues from a female fish and a male fish were mixed.
Transcriptomic analysis was used to determine the gene expression profile of distal intestine and hepatic RNA (n = 3) in adult fish from all groups, including the negative control (FM group), the positive control (SBM group), and the tested drug inclusion group (SB group). The process for preparing the gene library and sequencing the transcriptome was consistent with previously described methods (Johnson and Hofmann, 2016). To summarize, sequencing libraries were prepared using the NEBNext RUltraTM RNA Library Prep Kit for Illumina R (NEB, USA) according to the manufacturer’s guidelines, and the quality of the libraries was determined using the Agilent Bioanalyzer 2100 system. On an Illumina platform (NovaSeq 6000), the library preparations were sequenced and 150 bp paired-end reads were produced.
Hisat2 (version 2.0.4) (Kim et al., 2019) was used to map the clean reads to the zebrafish genome (GRCz11, https://www.ncbi.nlm.nih.gov/genome/?term=txid7955[orgn]). The number of reads that were mapped were counted verse by verse (version 0.1.5). The reads count was imputed into DESeq2 (version 1.24.0) to analyze DEGs. To select DEGs for analysis, DESeq2 (Love et al., 2014) was used with | log2FoldChange | > 1 and padj 0.05. For annotation, clusterProfiler (version 3.12.0) (Yu et al., 2012) was used to perform enrichment analysis of both GO (Gene Ontology) terms and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, with p < 0.05 considered significant enrichment. When the parameters used were not listed, default parameters were used.
The composition of bacterioplankton was analyzed using NovaSeq sequencing of 16S rRNA gene amplicons (n = 6) as previously published protocol (Xie et al., 2021). In brief, the V3V4 regions of the 16S rRNA gene were amplified. After construction of the libraries, the paired-end 250-nucleotide reads were obtained using the Illumina NovaSeq platform. Using the DADA2 plugin, raw sequences were trimmed, quality filtered, denoised, merged, chimera, and dereplicated (Callahan et al., 2016). Reads with 100 percent nucleotide sequence identity across all samples were assigned to operational taxonomic units (OTUs), and taxonomy was assigned to Non-singleton amplicon sequence variants (ASVs) using the classify-sklearn nave Bayes taxonomy classifier in the feature-classifier plugin (Bokulich et al., 2018) against the Greengenes reference database (DeSantis et al., 2006).
Following previously published methods for zebrafish larvae imaging (Bravo-Tello et al., 2017; Coronado et al., 2019; Ikeda-Ohtsubo et al., 2020), in vivo imaging of intestinal immune cells in zebrafish was conducted according to our recently published protocol (Li et al., 2021; Xie et al., 2021). As the innate cell’s response could be examined at a very early stage, the feeding trial was done from 5 dpf to 9 dpf, using the neutrophil and macrophage labeled Tg lines. At an adaptive aspect, soybean meal was fed after the grass carp’s body length was at least 10 cm making it a juvenile in aquaculture. The feeding protocol for live imaging of the lymphocytes was to mimic the commercial fish’s feeding strategy. Zebrafish intestinal adaptive immune function has been well developed after 18 dpf (the starting time point for feeding trial to test lymphocytes’ response) and up to 27 dpf when it is nearly an adult fish (the ending time point). Thus, the SBMIE modeling used to analyze the lymphocytes in the lymphocyte labeled larva was carried out in well-developed hindguts, allowing us to test the possibility of seabuckthorn mitigating the side effect of soybean meal diet on adaptive immunity.
Methodologically, before the feeding trial in larvae, to prepare the experimental feeds, the diets for the FM, SBM, and SB groups were crushed and passed through an 80-mesh sieve. 200 larvae were used for each group to analyze either innate or adaptive immune cells. The water temperature was kept at 28 ± 0.5°C. For each group, from 0 to 3 dpf, the larvae were kept in Danieau’s solution (http://cshprotocols.cshlp.org/content/2013/5/pdb.rec074260.full) with the anti-fungal agent methylene blue (MB, 0.0001%–0.0005%), later the larvae were kept in Danieau’s solution. During the feeding trial to analyze either innate or adaptive immune cells, larvae were fed three times per day. On the day for imaging, larvae were anesthetized with MS222 before being embedded in 1% LMP Agarose (Invitrogen, dissolved in Danieau’s solution). A Leica SP8 microscope was used to examine the experimental larvae. At least 10 larvae were observed under a microscope for each group. To generate a picture for the entire posterior part of the intestine, a composition of several stacks (n = 13) was merged for imaging the hindgut signals.
The feeding protocol is shown in Supplementary Figure S1. To analyze innate immune cells, between 5 and 8 dpf, the larvae of Tg(lyz:DsRED2); Tg(mpeg1:EGFP) were kept in Danieau’s solution and fed experimental diets. Upon imaging of innate immune cells, including neutrophils (lyz:DsRED2 labeled) and macrophages (mpeg1:EGFP labeled), was performed in the hindgut (posterior part) of the larvae of Tg(lyz:DsRED2); Tg(mpeg1:EGFP) at 9 dpf (Xie et al., 2021). Meanwhile, to test the cellular response of adaptive immune cells, the immature (rag2:DsRed labeled) (Zhang et al., 2018) and mature lymphocytes (lck-eGFP labeled) (Langenau and Zon, 2005), in the intestine (posterior part) of Tg(rag2:DsRed) and Tg(lck:lck-eGFP) larvae (Xie et al., 2021), were imaged. After 12 days (5–16 dpf) feeding of Larval AP100 Diet (Zeigler) as well as 10 days of experimental diets feeding from 17 dpf, the intestinal lymphocyte in larvae were imaged at 27 dpf.
The FIJI/ImageJ software (Póvoa et al., 2021) was used to calculate the area and intensity of HE stained mucosal folds, oil drops, fluorescent cells, and immunohistochemistry signals. In the oil-red staining result for hepatic images, the area ratio of oil drops vs. total area was then calculated. During the immunohistochemistry analysis, the ratio of CD4+ cells vs. total DAPI-stained nuclei was calculated to reveal the hepatic pathology. The fluorescent-labeled signals or immunostained signals for intestinal signals were calculated using previously published methods (Xie et al., 2021). Data were analyzed in EXCEL for the correlation analysis comparing fold changes between qPCR and RNA-Seq results. The R-value was calculated using Pearson’s correlation. The information was processed by the GraphPad Prism 5 software (GraphPad Software Inc. CA, United States). The bar diagram represented mean ± SEM. For the comparison between two groups, data were analyzed using a T-test. While for the comparison among multiple groups, data were analyzed using one-way ANOVA followed by a Duncan’s test. Differences were considered significant at p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***).
GeneRank selected the key 300 SBMIE DEGs based on SBMIE-related DEGs in grass carp and the protein-protein interaction network (Supplementary Table S2). Then, using the components targeting those key genes, a medicine containing effective components was chosen from TCMID. The results revealed that seabuckthorn contained five effective components, as well as the component’s key regulated genes (Table 2), based on TCMID correlations between drug components and responsive genes (Huang et al., 2018). Meanwhile, 33 DEGs from the transcriptome of grass carp with SBMIE were matched to the effective components (Table 3), potentially narrowing the drug range to seabuckthorn. As a result, seabuckthorn was chosen as a candidate to treat foodborne enteritis in fish.
When the SBM group (positive control) was compared to the FM group (negative control), HE staining revealed an intestinal mucosa lesion (Figure 1) and loosening of the hepatic reticuloendothelial structure (Supplemntary Figure S2) (negative control). The inclusion of seabuckthorn at a concentration of 35 ppm significantly reduced the appearance of such pathological change in the SBMIE adult model, as evidenced by a very significantly (p < 0.0001) decreased percentage of oil red-stained droplets (Figures 1A,a) and a more compact reticuloendothelial structure (Supplementary Figure S2) in the liver, as well as very significantly (p < 0.0001) shortened villi (Figures 1A,b). Both the amount and particle size of inflammation-related lipid droplets in the SBM group were reduced a lot in the SB group (Figures 1A,b). Additionally, in the SBMIE larvae model, the intestinal villi in 27 dpf larvae from the SB group were still within the normal length comparable to that in the FM group. However, in the SBM group the intestinal fold had significantly (p < 0.0001) shortened villi length in the SBM group (Figure 1B). Longer intestinal villi (p < 0.0001) were seen in the SB group compared to the SBM group at 27 dpf in the posterior part of the intestine (Figure 1B). Yet, the villi structure in the last portion of the hindgut (near the anus) did not differ much in length (Supplementary Figure S3).
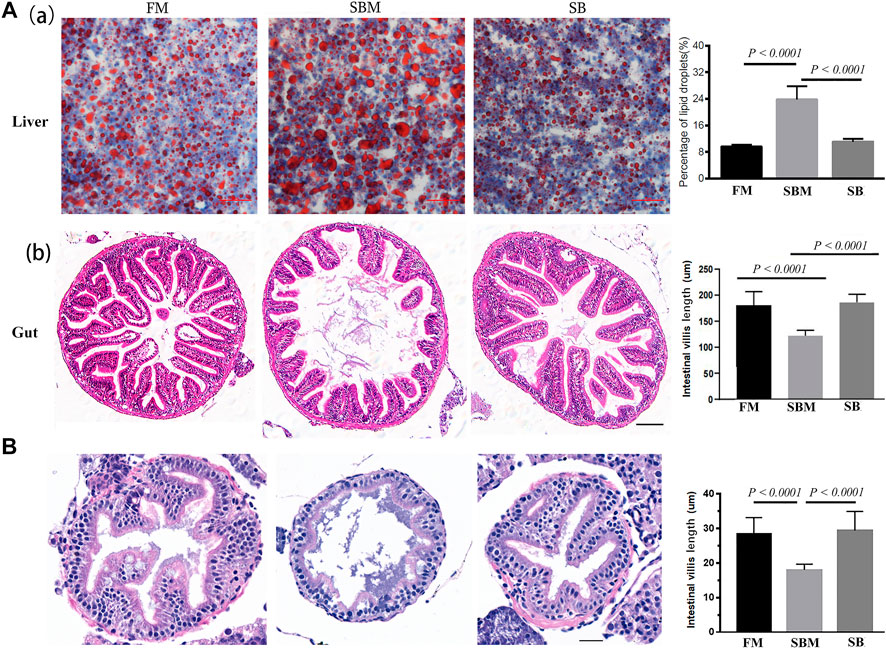
FIGURE 1. Histological analysis of seabuckthorn’s effect pathologically. (A) In the SBMIE adult model, both decreased hepatic oil-red stained droplets (a) and increased length of intestinal villi (b) were found relieved in wide type zebrafish from the SB group compared to the SBM group. The scale bar in hepatic and intestinal images of adult zebrafish indicated 50 μm. (B) HE staining of intestinal mucosa as well as quantitative analysis of intestinal villi length in 27 dpf wide type larva feed FM, SBM and SB diets. Scale bar: 20 μm. FM: fish meal diet; SBM: soybean meal diet; SB: seabuckthorn supplementary SBM diet. All bar diagrams of quantitative analysis for hepatic oil red-stained signals or the intestinal villi length were at the right side of the typical images.
The immunohistochemistry result for the CD4 protein indicated the presence of the intestinal T helper lymphocyte. This was reflected by the CD4 signals that were detected in the intestinal villi only in the LP layer, with very few at the base of the mucosal fold (Figure 2A). Quantitative analysis of the number of CD4+ cells divided by the length of intestinal villi revealed no significant difference in the ratio between the SB and SBM groups (data not shown). The SB group had a lower ratio of hepatic CD4+ cells (Figure 2B) compared to the SBM group. Quantitative analysis of signals revealed a significant decrease in the ratio of hepatic CD4+ cells to total cells, as indicated by the DAPI stained nucleus (Figure 2B). Additionally, it is interesting to note that some hepatic CD4+ cells had prolongations.
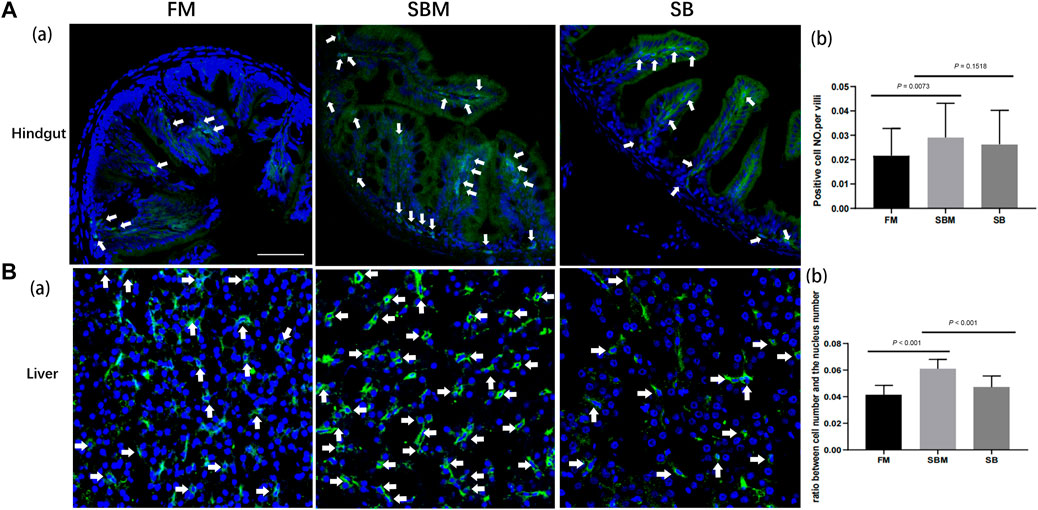
FIGURE 2. The effect of seabuckthorn on intestinal and hepatic T helper cells reflected by immunohistochemistry signals. (A) The representative images (a) and the bar diagram for quantitative analysis (b) of intestinal CD4 labeled Th cell signals as well as the quantitative analysis of signals in FM, SBM and SB groups; (B) The representative images (a) and the bar diagram for quantitative analysis (b) of hepatic CD4 labeled Th cell signals as well as the quantitative analysis of signals in FM, SBM and SB groups. The typical signals were indicated by arrows. Scale bar: 100 μm.
A 3D micro-CT scanner was used to determine body fat. After scanning anesthetic adult zebrafish, the image data was processed to create a 3D structural reconstruction and quantitatively analyzed. The results indicate that the percentages of body fat in the FM, SBM, and SB groups were 8.99 percent, 2.14%, and 6.83%, respectively. The reconstructed three-dimensional images may correspond to areas of body fat in zebrafish. The area of body fat in SBM fish was significantly less than that in FM and SB fish, whereas the area of body fat in SB fish appeared to be comparable to that in FM fish (Figure 3).
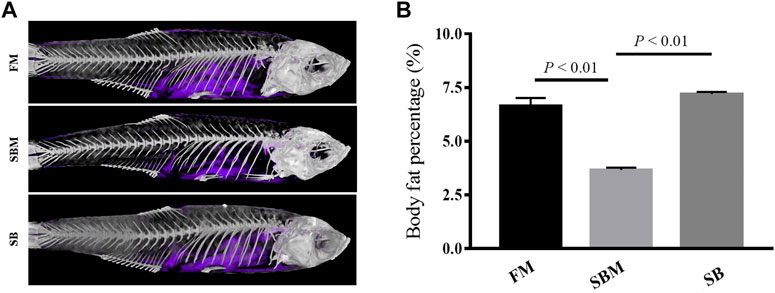
FIGURE 3. The reconstructed image of body fat revealed by 3D micro-CT in adult zebrafish from FM, SBM and SB groups. The body fat in the fish trunk was colored purple. (A) The images of body fat (in purple) within the fish from all groups; (B) The bar diagram to show percentages of body fat in whole fish trunk.
The qPCR validation result (Supplementary Table S1) demonstrated that the fold-changes between DGE and qPCR were highly correlated (R > 0.8) across 22 reactions encoding 9 genes, indicating that the current transcriptomic study successfully decoded gene expression. The results of the comparison between the SBM and FM groups (Supplementary Figure S4) revealed intestinal GO terms associated with immune responses, such as “humoral immune response” and “complement activation.” Between the SBM and FM groups, the KEGG pathways “lysosome,” “cytokine-cytokine receptor interaction”, and “ferroptosis” were identified for the hepatic inflammation, while the lipid metabolism-related pathways “PPAR signaling pathways” and “fatty acid elongation” were also enriched. This resulted from the successful transcriptomic modeling of SBMIE. Then, using intestinal DEGs, the drug-incorporated group could be analyzed for its moderating effect on SBMIE (Supplementary Table S3). Numerous terms related to cell proliferation were included in the enriched GO terms for biological processes, including “cell cycle checkpoint,” “DNA-dependent DNA replication,” “protein-DNA complex assembly”, “DNA replication,” “protein-DNA complex subunit organization,” “cell cycle regulation,” “negative regulation of cell cycle,” “DNA metabolic process,” “mitotic cell cycle,” “DNA replication checkpoint,” “mitotic metaphase plate congression,” and “attachment of spindle” (Figure 4A, left; Supplementary Table S4). In addition to the enriched GO terms, the enriched KEGG pathways (Figure 4, right; Supplementary Table S5) included cell division-related pathways such as “regulation of actin cytoskeleton,” “mismatch repair,” “DNA replication,” “nucleotide excision repair,” and “splice some.” Additionally, the pathways “primary bile acid biosynthesis,” “PPAR signaling pathway,” and “phagosome” were also enriched for intestinal DEGs.
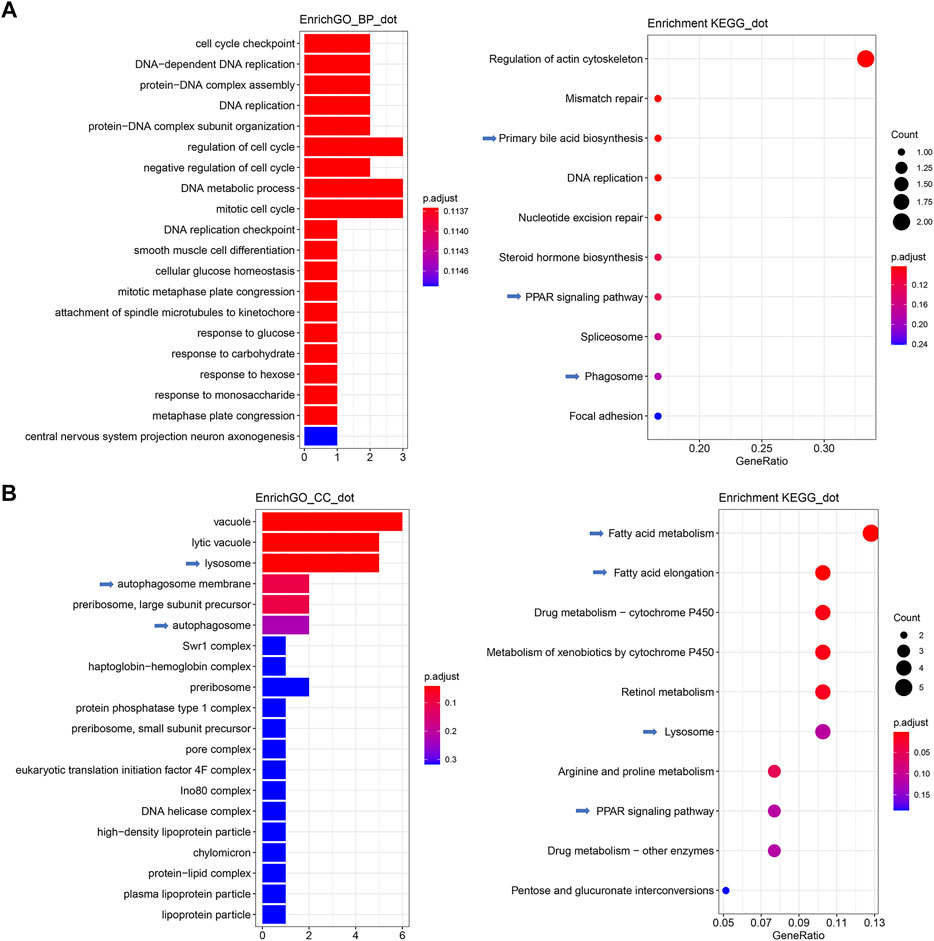
FIGURE 4. GO terms and KEGG pathways enriched for both intestinal and hepatic DEGs in comparison between SB and SBM groups in zebrafish SBMIE adult model. (A) Key intestinal terms biological processes (left) and KEGG pathways (right); (B) Key hepatic terms for cellular components (left) and KEGG pathways (right). The immune-related terms and pathways are labeled with arrows.
Meanwhile, for hepatic DEGs derived from the comparison of the SB vs. SBM groups (Supplementary Table S6), enriched GO terms for cellular components included terms, such as “lysosome,” “autophagosome membrane,” “autophagosome,” “high-density lipoprotein particle,” “protein-lipid complex,” “plasma lipoprotein particle,” and “lipoprotein” (Figure 4B, left; Supplementary Table S7). Additionally, the hepatic enrichment analysis (Figure 4B, right; Supplementary Table S8) also revealed the GO terms “Lysosome” and “PPAR signaling pathway” as well as KEGG pathways “fatty acid metabolism” and “fatty acid elongation.”
The raw data from the current transcriptome sequencing analysis has been submitted to the Genome Sequence Archive (GSA) database (http://gsa.big.ac.cn/index.jsp) under the BioProject identifier < PRJCA005917> and data ID < CRA004582>. Within this BioProject identifier, there was one data ID for the transcriptomic data and another for the 16s rRNA gene sequencing result, as described below. This study shared the same positive (SBM group) and negative (FM group) controls with a published study on sinomenine’s anti-enteritis effect (Xie et al., 2021), since both drugs were analyzed for the anti-SBMIE effect at the same time using the same batch of fish.
There were 8434, 5976, and 9027 OTUs in the hindgut of adult fish belonging to the FM, SBM, and SB groups, respectively. As illustrated in the Venn diagram, 799 OTUs overlapped between the SB and FM groups, but only 471 OTUs overlapped between the SB and SBM groups (Figure 5A). As illustrated in Figure 5A, the overlap between the seabuckthorn and FM groups covered 799 OTU, while the overlap between the seabuckthorn and SBM groups contained 471 OTU. The SB group-specific region contained 6827 OTU, which was slightly more than the FM group-specific region’s 6346 OTU and significantly more than the SBM group-specific region’s 4052 OTU. The OTUs identified a large number of phyla in the fish intestinal microbiota to estimate their abundance.
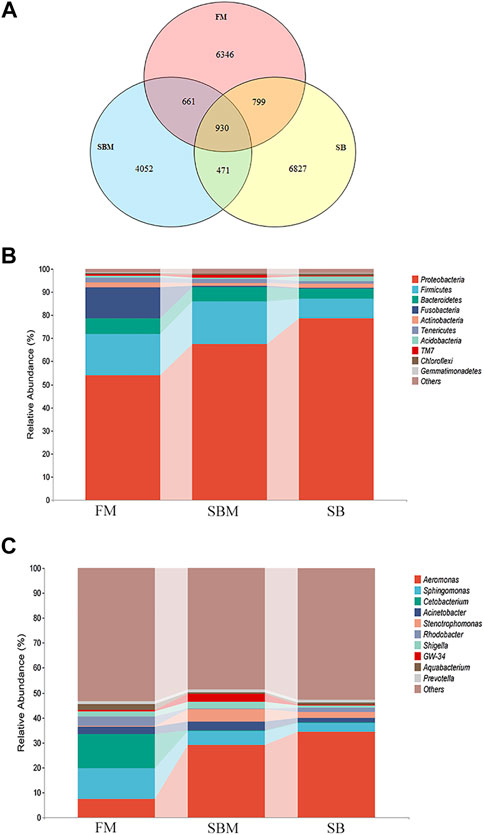
FIGURE 5. Analysis of intestinal microbiota OTU and composition influenced by seabuckthorn dietary inclusion in zebrafish SBMIE adult model. (A) Venn diagram of OTU in FM, SBM and SB groups; (B) hindgut bacteria composition at phylum level; (C) hindgut bacteria composition at the genus level. The alteration trends between adjacent bars for the current revealed taxa were indicated by the interval belts.
Among the 10 phyla with the most abundance, compared to the SBM groups, in the SB group, the increased phyla included Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, and Gemmatimonadetes, while decreased phyla included Bacteroidetes, Firmicutes, Fusobacteria, Tenericutes and TM7 (Figure 5B). At the genus level (Figure 5C), among the most abundant 10 genera, compared to the SBM groups, in the SB group, the increased genera included Aeromonas, Rhodobacter, Pelomonas and Aquabacterium, while decreased genera included Sphingomonas, Cetobacterium, Acinetobacter and Shigella. More specifically, the relative abundance of the 16S RNA gene was listed for each sample at both phylum and genus levels in Supplementary Table S9.
The raw data of the current 16S rRNA gene sequencing analysis was submitted to the Genome Sequence Archive (GSA) database (http://gsa.big.ac.cn/index.jsp) with the BioProject identifier < PRJCA005917> and data ID < CRA004611>. This study shared the same positive (SBM group) and negative (FM group) controls with the parallel analysis on the sinomenine test since seabuckthorn and sinomenine were analyzed for the anti-SBMIE effect at the same time using the same batch of fish.
At a cellular level, compared to the SBM group (positive control), seabuckthorn could significantly (p < 0.01) relieve inflammatory aggregation of innate immune cells, including lyz:DsRED2 labeled neutrophils and mpeg1:EGFP labeled macrophages (Figure 6; Supplementary Figure S5), whilst significantly (p = 0.03) reduced inflammatory aggregation of lck+ T cells (Figure 7; Supplementary Figure S5) in the intestine (posterior part) of zebrafish larvae in SB group. Morphologically, seabuckthorn could inhibit the formation of protrusions in macrophages in the hindgut (Figure 6). While, compared to the FM group (negative control), there were no significant changes in both number and morphology in the SB group for most observed immune cells, the differences of both number and morphology for rag2:DsRed labeled lymphocytes were observed in Figure 7. The factor that rag2:DsRed labeled lymphocytes in the SB group aggregated in the hindgut and still had cellular protrusions, was similar (p = 0.75) to that in the SBM group.
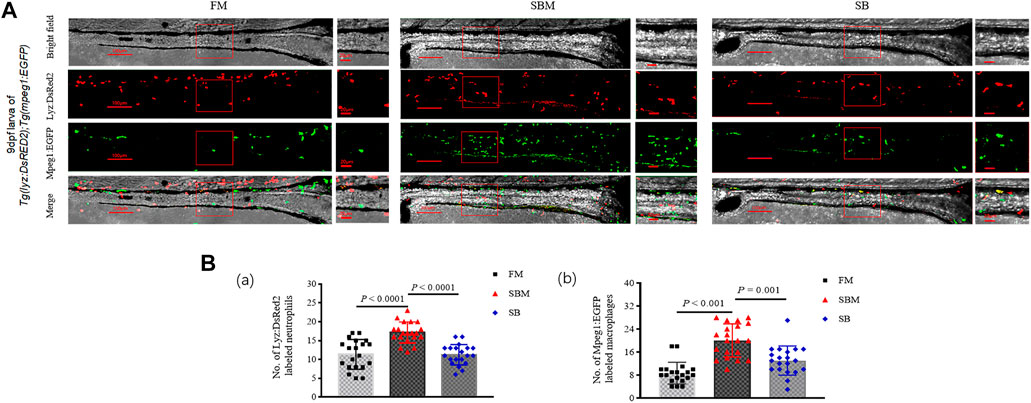
FIGURE 6. Imaging of innate immune cells and expressional analysis of immune-related genes in zebrafish SBMIE larva model. (A) lyz:DsRED2 labeled neutrophils and mpeg1:EGFP labeled macrophages imaging in the intestine (posterior part, n = 10) using 9 dpf larvae of Tg(lyz:DsRED2); Tg(mpeg1:EGFP). The square indicated the zoomed in region. The scale bar in whole pictures was 100 μm, while the scale bar in the enlarged view was 20 μm. (B) quantitative analysis of the signals using bar-diagrams. (a) Number of Lyz:DsRed2 labeled neutrophils in larvae from either FM, SBM, or SB groups; (b) No. of Mpeg1:EGFP labeled macrophages in larvae from either FM, SBM, or SB groups. The very significant differences (p < 0.01) of both Lyz:DsRed2 and Mpeg1:EGFP labeled cells between SB and SBM groups were indicated by p-value.
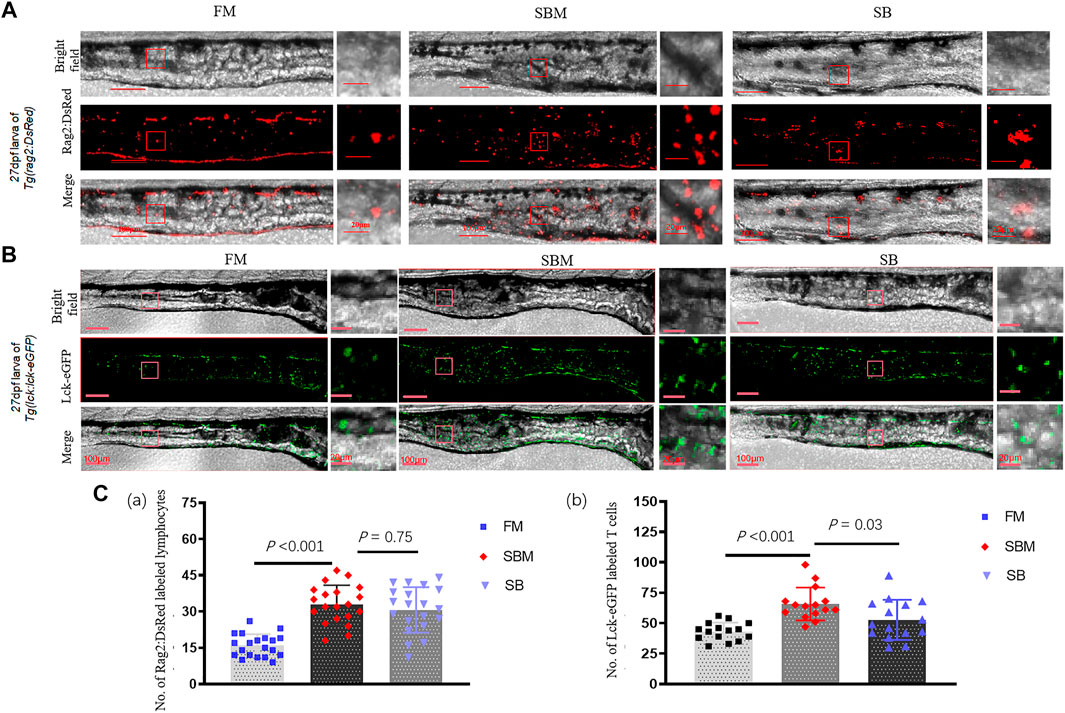
FIGURE 7. Imaging of lymphocytes and expressional analysis of immune-related genes in SBMIE larvae model. (A) rag2:DsRed labeled immature lymphocytes imaging in 27 dpf Tg(rag2:DsRed) larvae’s intestine (posterior part, n = 10); (B) lck-eGFP labeled mature T cells imaging in 27 dpf Tg(lck:lck-eGFP) larvae’s intestine (posterior part). The square indicated the zoomed in region. The scale bar in whole pictures was 100 μm, while the scale bar in the enlarged view was 20 μm. (C) quantitative analysis of the signals using bar-diagrams. (a) Number of Rag2:DsRed labeled lymphocytes in larvae from either FM, SBM, or SB groups; (b) Number of Lck-eGFP labeled T cells in larvae from either FM, SBM, or SB groups. The significant differences (p < 0.05) of Lck+ T cells between SB and SBM groups were indicated by the p-value.
Through molecular, cellular, and bio-informational approaches, this comprehensive characterization revealed the effects of dietary inclusion of seabuckthorn on fish gut-liver immunity as well as intestinal bacterial communities in the zebrafish SBMIE model. This has provided clues for developing effective herb-based feed additives to prevent foodborne disease in aquaculture.
Based on the successful modeling of SBMIE in either adult zebrafish or zebrafish larvae (Xie et al., 2021), we modeled in our positive control both the typical intestinal pathology and the typical inflammatory hepatic lipid deposition, similar to the gut-liver symptoms revealed in carp (Wu et al., 2018) (SBM group). The fact that the outcome of recovered body fat trunk in adult zebrafish from the SB group suggested a positive effect of seabuckthorn’s inclusion in alleviating SBMIE-related symptoms. The pathological data hinted that the SB group had less intestinal and hepatic inflammation. Seabuckthorn’s anti-inflammatory role may be reflected in both the liver and the gut, with fewer hepatic oil droplets and a more compact reticuloendothelial structure, as well as longer intestinal villi. Among the seabuckthorn’s target genes, mapk1 and mapk3 may reflect cell proliferation (Masselli et al., 2013), rspo4 might indicate the promotion of WNT signals, which was important for intestinal crypt formation and renewal (Kriz and Korinek, 2018).
In terms of immunity, the fact that the SB group could inhibit both the gathering of innate cells and the formation of protrusions on macrophages was echoed by seabuckthorn’s target gene plg. During SBMIE, zebrafish plasminogen may play a role in inflammation-stimulated macrophage recruitment as previously reviewed (Miles et al., 2012). Fewer protrusions may indicate less activation via forming phagocytic synapses (Goodridge et al., 2011; Li et al., 2020c). At an adaptive immunity level, seabuckthorn had less influence on lymphocyte differentiation, as evidenced by unaltered SBM-induced aggregation and protrusions formation of rag2:DsRed labeled lymphocytes as well as an insignificantly altered CD4+ cell intestinal location pattern. Nonetheless, the fact that fewer intestinal lck+ signals SB target gene zap70, a tyrosine kinase important for TCR signaling and late T cell activation (Carpino et al., 2004) suggests a possible inhibition of T cell maturation. However, as demonstrated by the effects on the gut-liver axis, our discovery that the inclusion of seabuckthorn reduced the number of CD4+ cells in the liver was consistent with seabuckthorn’s intestinal effect on T cells. In addition, the few hepatic CD4+ cells with prolongations might suggest the tissue-resident CD4+ monocytes as previously reported in zebrafish (Dee et al., 2016; Lin et al., 2019).
As a result, a systemic study at the omics level made sense to reveal the underlying molecular mechanisms. The DEGs obtained from the comparison of the SB and SBM groups already provided some hints for intestinal or hepatic immune reactions caused by seabuckthorn. For example, increased intestinal expression of c1qc may indicate improved complement system function, which may be beneficial to the zebrafish commensal microbiota (López Nadal et al., 2020). The downregulation of cdt1, chaf1b, nabp1b, and other genes in the SB group compared to the SBM group may indicate a reduction in intestinal cell proliferation. A higher level of apoa4b in the liver may indicate improved lipid absorption and metabolism (Qu et al., 2019b; Hu, 2020). The effect of SB on lipid deposition was consistent with the target gene acacb of seabuckthorn, which was recently proposed as a therapeutic target for metabolic syndrome (Chen et al., 2019).
By incorporating DEGs into enrichment analysis, the currently revealed GO terms and KEGG pathways, as well as clues for improved intestinal cell proliferation, such as DNA replication and cell cycle regulation, were discovered. As well, the possibility that seabuckthorn’s target gene p53 may suggest that seabuckthorn supplementation could reduce apoptosis. On the other hand, in terms of immunity, the KEGG pathway “phagosome” was discovered in the comparison of the SB and SBM groups, as phagocyte is an innate immune bioprocess primarily found in fish macrophages. Similar to the SBMIE larvae model, innate immune cells were found to be significantly regulated by SB inclusion in terms of number and cellular protrusions. As a result, seabuckthorn may significantly protect intestinal innate immunity by inhibiting inflammatory aggregation and activation of innate immune cells. Autophagy and lysosome-related GO terms and KEGG pathways in the liver were compared between the SB and SBM groups and demonstrated that the inclusion of seabuckthorn to the SBM diet restored dysregulated hepatic autophagy and lysosome function. The increased lysosome-based autophagy of lipid droplets in the hepatocytes (Schulze et al., 2020) could also explain the SB group’s reduced droplets.
Moreover, several KEGG pathways reflected seabuckthorn’s effect. The intestinal “primary bile acid biosynthesis” may indicate that intestinal epithelial function was regulated and might play a role in anti-inflammation (Pavlidis et al., 2015; Hegyi et al., 2018). Among the biological processes enhanced by seabuckthorn in mouse livers, peroxisome proliferator-activated receptor (PPAR) α and PPAR-γ were significantly increased (Pichiah et al., 2012). In zebrafish, the PPAR pathway was shown to be critical for intestinal homeostasis (Qin et al., 2018). Comparing the regulated PPAR signaling pathways between two control groups (SBM vs. FM), the PPAR signaling pathways revealed by both intestinal and hepatic DEGs may suggest a critical role for PPAR signaling in seabuckthorn’s effect on SBMIE alleviation. Additionally, because L-Arginine has been shown to modulate T cell metabolism (Geiger et al., 2016) and arginine has been shown to protect intestinal health (Zheng et al., 2017), the current study identified the hepatic KEGG pathway “Arginine and proline metabolism,” which may promote gut-liver immunity.
Additionally, as a result of the interaction between the gut microbiota and the liver (Deng et al., 2020), our findings indicated that seabuckthorn supplementation resulted in an increase in OTU numbers and an FM-biased bacterial community in the SB group compared to the SBM group. Taxonomic changes were observed at the phylum level, as Actinobacteria were found to produce active metabolites against pathogenic microorganisms (Jami et al., 2015), and the current study revealed an increase in Actinobacteria, suggesting a protective role for the SB group. When comparing the composition of the intestinal taxa of carnivorous and omnivorous fish, the fact that the SB group had higher levels of Acidobacteria and Chloroflexi may be related to improved protein utilization. This is because Acidobacteria and Chloroflexiact as major components of carnivorous fish’s intestinal microbiota (Wei et al., 2019). Reduced TM7 levels may indicate decreased intestinal inflammation, as intestinal TM7 bacterial phylogenies were previously identified as a pro-inflammatory factor in IBD (inflammatory bowel disease) (Kuehbacher et al., 2008). The levels of Bacteroidetes decreased upon inclusion of seabuckthorn, which may be a sign of recovery from SBMIE. In turbot, a carnivorous fish species, the SBM diet’s intestinal mucosal microbiota was dominated by Bacteroidetes (Li et al., 2019). Reduced Sphingomonas was identified as a disease biomarker in the zebrafish (Ma et al., 2020), as was Shigella, which was associated with enteritis (human Crohn’s disease) (Zhang et al., 2020c). Both of these observations suggested that the fish probably had recovered from enteritis. Increased levels of the probiotic Rhodobacter (Liu et al., 2012; Ye et al., 2019) and the butyrate-producing Aquabacterium (Gupta et al., 2019) may act as protective factors.
The effect of seabuckthorn on lipid metabolism and microbiota in zebrafish supported the notion that seabuckthorn could correct the lipid metabolism disease produced by a high-fat diet in mice by altering the gut microbiota (Guo et al., 2020). In the current results, decreased adipose tissue deposition suggested less inflammation in the inner organ. The total body fat in the fish trunk is a measure of growth performance. Thus, with fewer inflammatory droplets in the liver and improved gut shape and microbiota, the fish in the SB group were able to maintain body fat levels comparable to those in the FM group. As the possible effective components, the anticipated acids in Table 2 were consistent with previous findings that unsaturated fatty acids in seabuckthorn could protect against infections, prevent allergies, and reduce inflammation (Zielińska and Nowak, 2017). However, only flavonoid glycosides extracted from seabuckthorn leaves were found to be anti-inflammatory in mice with hepatic steatosis (Kwon et al., 2017). Thus, the delicate mechanisms underlying effective components and the routes implicated remain unknown.
In conclusion, by including seabuckthorn fruit powder in the SBM diet, we were able to alleviate the typical gut and liver pathology associated with SBMIE, via enhancing intestinal immunity and hepatic metabolic adjustments. The involved mechanisms may include inhibition of intestinal cell apoptosis, excessive immune cell aggregation, and microbiota dysbiosis in the gut, together with reduced Th cell aggregation and lipid metabolic burden in the liver. Our findings indicated that dietary seabuckthorn supplementation may protect fish from foodborne gut and liver symptoms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Institute of Hydrobiology of the Chinese Academy of Sciences approved the use of animals in this study.
NW conceived the project. XZ, NW and ML wrote the manuscript. ML, XZ, JX, JS, GW, YL and YC performed the experiments. ML, MS, WY, XZ, NW and WZ did data analysis. BU did language proofreading. The manuscript was read and approved by X-QX.
This research is supported by the National Natural Science Foundation of China (31872592).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Xing Wang from the Institute of Hydrobiology, Chinese Academy of Sciences for their assistance with micro-CT and confocal microscopy imaging, respectively, and Prof. Rui Wang from Wuhan Polytechnic University for providing trainee to assist with this study. Our thank also goes to Prof. Ming Jiang for his assistance in creating zebrafish feed. We would like to thank Dr. Luyuan Pan from CZRC for suggestions on the zebrafish husbandry. We would also like to thank the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China, for their assistance with the computation in this work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.831226/full#supplementary-material
Abarike E. D., Jian J., Tang J., Cai J., Yu H., Chen L. (2019). Traditional Chinese Medicine Enhances Growth, Immune Response, and Resistance to Streptococcus Agalactiae in Nile tilapia. J. Aquat. Anim. Health 31, 46–55. doi:10.1002/aah.10049
Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2's Q2-Feature-Classifier Plugin. Microbiome 6, 90. doi:10.1186/s40168-018-0470-z
Booman M., Forster I., Vederas J. C., Groman D. B., Jones S. R. M. (2018). Soybean Meal-Induced Enteritis in Atlantic salmon (Salmo salar) and Chinook salmon (Oncorhynchus tshawytscha) but Not in Pink salmon (O. Gorbuscha). Aquaculture 483, 238–243. doi:10.1016/j.aquaculture.2017.10.025
Bravo-Tello K., Ehrenfeld N., Solís C. J., Ulloa P. E., Hedrera M., Pizarro-Guajardo M., et al. (2017). Effect of Microalgae on Intestinal Inflammation Triggered by Soybean Meal and Bacterial Infection in Zebrafish. PLoS One 12, e0187696. doi:10.1371/journal.pone.0187696
Brugman S., Liu K. Y., Lindenbergh–Kortleve D., Samsom J. N., Furuta G. T., Renshaw S. A., et al. (2009). Oxazolone-induced Enterocolitis in Zebrafish Depends on the Composition of the Intestinal Microbiota. Gastroenterology 137, 1757–1767. doi:10.1053/j.gastro.2009.07.069
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 13, 581–583. doi:10.1038/nmeth.3869
Campbell C. A., Fursova O., Cheng X., Snella E., McCune A., Li L., et al. (2021). A Zebrafish Model of Granulin Deficiency Reveals Essential Roles in Myeloid Cell Differentiation. Blood Adv. 5, 796–811. doi:10.1182/bloodadvances.2020003096
Carpino N., Turner S., Mekala D., Takahashi Y., Zang H., Geiger T. L., et al. (2004). Regulation of ZAP-70 Activation and TCR Signaling by Two Related Proteins, Sts-1 and Sts-2. Immunity 20, 37–46. doi:10.1016/s1074-7613(03)00351-0
Catalán N., Villasante A., Wacyk J., Ramírez C., Romero J. (2018). Fermented Soybean Meal Increases Lactic Acid Bacteria in Gut Microbiota of Atlantic Salmon (Salmo salar). Probiotics Antimicro. Prot. 10, 566–576. doi:10.1007/s12602-017-9366-7
Chen L., Duan Y., Wei H., Ning H., Bi C., Zhao Y., et al. (2019). Acetyl-CoA Carboxylase (ACC) as a Therapeutic Target for Metabolic Syndrome and Recent Developments in ACC1/2 Inhibitors. Expert Opin. Investig. Drugs 28, 917–930. doi:10.1080/13543784.2019.1657825
Cheng B., Zhang H., Hu J., Peng Y., Yang J., Liao X., et al. (2020). The Immunotoxicity and Neurobehavioral Toxicity of Zebrafish Induced by Famoxadone-Cymoxanil. Chemosphere 247, 125870. doi:10.1016/j.chemosphere.2020.125870
Coronado M., Solis C. J., Hernandez P. P., Feijóo C. G. (2019). Soybean Meal-Induced Intestinal Inflammation in Zebrafish Is T Cell-dependent and Has a Th17 Cytokine Profile. Front. Immunol. 10, 610. doi:10.3389/fimmu.2019.00610
Dee C. T., Nagaraju R. T., Athanasiadis E. I., Gray C., Fernandez Del Ama L., Johnston S. A., et al. (2016). CD4-transgenic Zebrafish Reveal Tissue-Resident Th2- and Regulatory T Cell-like Populations and Diverse Mononuclear Phagocytes. J.I. 197, 3520–3530. doi:10.4049/jimmunol.1600959
Deng Y., Zhang Y., Chen H., Xu L., Wang Q., Feng J. (2020). Gut-Liver Immune Response and Gut Microbiota Profiling Reveal the Pathogenic Mechanisms of Vibrio Harveyi in Pearl Gentian Grouper (Epinephelus Lanceolatus♂ × E. Fuscoguttatus♀). Front. Immunol. 11, 607754. doi:10.3389/fimmu.2020.607754
DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi:10.1128/AEM.03006-05
Ding Q., Zhang Z., Ran C., He S., Yang Y., Du Z., et al. (2018). The Hepatotoxicity of Palmitic Acid in Zebrafish Involves the Intestinal Microbiota. J. Nutr. 148, 1217–1228. doi:10.1093/jn/nxy084
Ellett F., Pase L., Hayman J. W., Andrianopoulos A., Lieschke G. J. (2011). mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood 117, e49–e56. doi:10.1182/blood-2010-10-314120
Enya S., Kawakami K., Suzuki Y., Kawaoka S. (2018). A Novel Zebrafish Intestinal Tumor Model Reveals a Role for Cyp7a1-dependent Tumor-Liver Crosstalk in Tumor's Adverse Effects on Host. Dis. Model. Mech. 11, 32383. doi:10.1242/dmm.032383
Geiger R., Rieckmann J. C., Wolf T., Basso C., Feng Y., Fuhrer T., et al. (2016). L-arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842. doi:10.1016/j.cell.2016.09.031
Goodridge H. S., Reyes C. N., Becker C. A., Katsumoto T. R., Ma J., Wolf A. J., et al. (2011). Activation of the Innate Immune Receptor Dectin-1 upon Formation of a 'phagocytic Synapse'. Nature 472, 471–475. doi:10.1038/nature10071
Guo C., Han L., Li M., Yu L. (2020). Seabuckthorn (Hippophaë Rhamnoides) Freeze-Dried Powder Protects against High-Fat Diet-Induced Obesity, Lipid Metabolism Disorders by Modulating the Gut Microbiota of Mice. Nutrients 12, 265. doi:10.3390/nu12010265
Gupta S., Lokesh J., Abdelhafiz Y., Siriyappagouder P., Pierre R., Sørensen M., et al. (2019). Macroalga-derived Alginate Oligosaccharide Alters Intestinal Bacteria of Atlantic Salmon. Front. Microbiol. 10, 2037. doi:10.3389/fmicb.2019.02037
Hasumura T., Shimada Y., Kuroyanagi J., Nishimura Y., Meguro S., Takema Y., et al. (2012). Green tea Extract Suppresses Adiposity and Affects the Expression of Lipid Metabolism Genes in Diet-Induced Obese Zebrafish. Nutr. Metab. (Lond) 9, 73. doi:10.1186/1743-7075-9-73
Hegyi P., Maléth J., Walters J. R., Hofmann A. F., Keely S. J. (2018). Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev. 98, 1983–2023. doi:10.1152/physrev.00054.2017
Hu Y. (2020). Establishment of a Lipid-Lowering Drug Screaning Platform in Zebrafish by ApoEb Knockout. Shanghai: Naval Medical University.
Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., et al. (2018). TCMID 2.0: a Comprehensive Resource for TCM. Nucleic Acids Res. 46, D1117–D1120. doi:10.1093/nar/gkx1028
Ikeda-Ohtsubo W., López Nadal A., Zaccaria E., Iha M., Kitazawa H., Kleerebezem M., et al. (2020). Intestinal Microbiota and Immune Modulation in Zebrafish by Fucoidan from Okinawa Mozuku (Cladosiphon Okamuranus). Front. Nutr. 7, 67. doi:10.3389/fnut.2020.00067
Jami M., Ghanbari M., Kneifel W., Domig K. J. (2015). Phylogenetic Diversity and Biological Activity of Culturable Actinobacteria Isolated from Freshwater Fish Gut Microbiota. Microbiol. Res. 175, 6–15. doi:10.1016/j.micres.2015.01.009
Jerison E. R., Quake S. R. (2020). Heterogeneous T Cell Motility Behaviors Emerge from a Coupling between Speed and Turning In Vivo. Elife 9, e5393310. doi:10.7554/eLife.53933
Johnson K. M., Hofmann G. E. (2016). A Transcriptome Resource for the Antarctic Pteropod Limacina Helicina antarctica. Mar. Genomics 28, 25–28. doi:10.1016/j.margen.2016.04.002
Kasheta M., Painter C. A., Moore F. E., Lobbardi R., Bryll A., Freiman E., et al. (2017). Identification and Characterization of T Reg-like Cells in Zebrafish. J. Exp. Med. 214, 3519–3530. doi:10.1084/jem.20162084
Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Kriz V., Korinek V. (2018). Wnt, RSPO and Hippo Signalling in the Intestine and Intestinal Stem Cells. Genes 9, 20. doi:10.3390/genes9010020
Kuehbacher T., Rehman A., Lepage P., Hellmig S., Fölsch U. R., Schreiber S., et al. (2008). Intestinal TM7 Bacterial Phylogenies in Active Inflammatory Bowel Disease. J. Med. Microbiol. 57, 1569–1576. doi:10.1099/jmm.0.47719-0
Kwon E.-Y., Lee J., Kim Y., Do A., Choi J.-Y., Cho S.-J., et al. (2017). Seabuckthorn Leaves Extract and Flavonoid Glycosides Extract from Seabuckthorn Leaves Ameliorates Adiposity, Hepatic Steatosis, Insulin Resistance, and Inflammation in Diet-Induced Obesity. Nutrients 9, 569. doi:10.3390/nu9060569
Langenau D. M., Zon L. I. (2005). The Zebrafish: a New Model of T-Cell and Thymic Development. Nat. Rev. Immunol. 5, 307–317. doi:10.1038/nri1590
Li C., Zhang B., Liu C., Zhou H., Wang X., Mai K., et al. (2020). Effects of Dietary Raw or Enterococcus Faecium Fermented Soybean Meal on Growth, Antioxidant Status, Intestinal Microbiota, Morphology, and Inflammatory Responses in Turbot (Scophthalmus maximus L.). Fish Shellfish Immunol. 100, 261–271. doi:10.1016/j.fsi.2020.02.070
Li C., Zhang B., Wang X., Pi X., Wang X., Zhou H., et al. (2019). Improved Utilization of Soybean Meal through Fermentation with Commensal Shewanella Sp. MR-7 in Turbot (Scophthalmus maximus L.). Microb. Cel Fact 18, 214. doi:10.1186/s12934-019-1265-z
Li M., Xie J., Zhao X., Li X., Wang R., Shan J., et al. (2021). Establishing the Foodborne-Enteritis Zebrafish Model and Imaging the Involved Immune Cells’ Response. Acta Hydrobiologica Sinica 46, 104. doi:10.7541/2022.2021.104
Li M., Wang H., Li W., Xu X. G., Yu Y. (2020). Macrophage Activation on "phagocytic Synapse" Arrays: Spacing of Nanoclustered Ligands Directs TLR1/2 Signaling with an Intrinsic Limit. Sci. Adv. 6, 6. doi:10.1126/sciadv.abc8482
Li X., Zhang F., Wu N., Ye D., Wang Y., Zhang X., et al. (2020). A Critical Role of Foxp3a-Positive Regulatory T Cells in Maintaining Immune Homeostasis in Zebrafish Testis Development. J. Genet. Genomics 47, 547–561. doi:10.1016/j.jgg.2020.07.006
Li Y., Esain V., Teng L., Xu J., Kwan W., Frost I. M., et al. (2014). Inflammatory Signaling Regulates Embryonic Hematopoietic Stem and Progenitor Cell Production. Genes Dev. 28, 2597–2612. doi:10.1101/gad.253302.114
Lickwar C. R., Camp J. G., Weiser M., Cocchiaro J. L., Kingsley D. M., Furey T. S., et al. (2017). Genomic Dissection of Conserved Transcriptional Regulation in Intestinal Epithelial Cells. Plos Biol. 15, e2002054. doi:10.1371/journal.pbio.2002054
Lin X., Wen Z., Xu J. (2019). Tissue-resident Macrophages: from Zebrafish to Mouse. Blood Sci. 1, 57–60. doi:10.1097/BS9.0000000000000013
Liu W.-S., Chen M.-C., Chiu K.-H., Wen Z.-H., Lee C.-H. (2012). Amelioration of Dextran Sodium Sulfate-Induced Colitis in Mice by Rhodobacter Sphaeroides Extract. Molecules 17, 13622–13630. doi:10.3390/molecules171113622
Liu Y., Chen Z., Dai J., Yang P., Xu W., Ai Q., et al. (2019). Sodium Butyrate Supplementation in High-Soybean Meal Diets for Turbot (Scophthalmus maximus L.): Effects on Inflammatory Status, Mucosal Barriers and Microbiota in the Intestine. Fish Shellfish Immunol. 88, 65–75. doi:10.1016/j.fsi.2019.02.064
López Nadal A., Ikeda-Ohtsubo W., Sipkema D., Peggs D., McGurk C., Forlenza M., et al. (2020). Feed, Microbiota, and Gut Immunity: Using the Zebrafish Model to Understand Fish Health. Front. Immunol. 11, 114. doi:10.3389/fimmu.2020.00114
Love M. I., Huber W., Anders S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Ma C., Guo H., Chang H., Huang S., Jiang S., Huo D., et al. (2020). The Effects of Exopolysaccharides and Exopolysaccharide-Producing Lactobacillus on the Intestinal Microbiome of Zebrafish (Danio rerio). BMC Microbiol. 20, 300. doi:10.1186/s12866-020-01990-6
Martín-Mateos R., Albillos A. (2021). The Role of the Gut-Liver axis in Metabolic Dysfunction-Associated Fatty Liver Disease. Front. Immunol. 12, 660179. doi:10.3389/fimmu.2021.660179
Masselli E., Mecucci C., Gobbi G., Carubbi C., Pierini V., Sammarelli G., et al. (2013). Implication of MAPK1/MAPK3 Signalling Pathway in t(8;9)(p22;24)/PCM1-JAK2myelodysplastic/myeloproliferative Neoplasms. Br. J. Haematol. 162, 563–566. doi:10.1111/bjh.12392
Miles L. A., Lighvani S., Baik N., Andronicos N. M., Chen E. I., Parmer C. M., et al. (2012). The Plasminogen Receptor, , and Macrophage Function. J. Biomed. Biotechnol. 2012, 1–14. doi:10.1155/2012/250464
Moore F. E., Garcia E. G., Lobbardi R., Jain E., Tang Q., Moore J. C., et al. (2016). Single-cell Transcriptional Analysis of normal, Aberrant, and Malignant Hematopoiesis in Zebrafish. J. Exp. Med. 213, 979–992. doi:10.1084/jem.20152013
Morrison J. L., Breitling R., Higham D. J., Gilbert D. R. (2005). GeneRank: Using Search Engine Technology for the Analysis of Microarray Experiments. Bmc Bioinformatics 6, 233. doi:10.1186/1471-2105-6-233
Oehlers S. H., Flores M. V., Okuda K. S., Hall C. J., Crosier K. E., Crosier P. S. (2011). A Chemical Enterocolitis Model in Zebrafish Larvae that Is Dependent on Microbiota and Responsive to Pharmacological Agents. Dev. Dyn. 240, 288–298. doi:10.1002/dvdy.22519
Okuda K. S., Misa J. P., Oehlers S. H., Hall C. J., Ellett F., Alasmari S., et al. (2015). A Zebrafish Model of Inflammatory Lymphangiogenesis. Biol. Open 4, 1270–1280. doi:10.1242/bio.013540
Ondruš J., Hubatka F., Kulich P., Odehnalová N., Harabiš V., Hesko B., et al. (2021). A Novel Approach to Imaging Engorged Ticks: Micro-CT Scanning of Ixodes Ricinus Fed on Blood Enriched with Gold Nanoparticles. Ticks Tick-borne Dis. 12, 101559. doi:10.1016/j.ttbdis.2020.101559
Pavlidis P., Powell N., Vincent R. P., Ehrlich D., Bjarnason I., Hayee B. (2015). Systematic Review: Bile Acids and Intestinal Inflammation-Luminal Aggressors or Regulators of Mucosal Defence? Aliment. Pharmacol. Ther. 42, 802–817. doi:10.1111/apt.13333
Pichiah P. B. T., Moon H.-J., Park J.-E., Moon Y.-J., Cha Y.-S. (2012). Ethanolic Extract of Seabuckthorn (Hippophae Rhamnoides L) Prevents High-Fat Diet-Induced Obesity in Mice through Down-Regulation of Adipogenic and Lipogenic Gene Expression. Nutr. Res. 32, 856–864. doi:10.1016/j.nutres.2012.09.015
Póvoa V., Rebelo de Almeida C., Maia-Gil M., Sobral D., Domingues M., Martinez-Lopez M., et al. (2021). Innate Immune Evasion Revealed in a Colorectal Zebrafish Xenograft Model. Nat. Commun. 12, 1156. doi:10.1038/s41467-021-21421-y
Qin C., Xie Y., Wang Y., Li S., Ran C., He S., et al. (2018). Impact of Lactobacillus Casei BL23 on the Host Transcriptome, Growth and Disease Resistance in Larval Zebrafish. Front. Physiol. 9, 1245. doi:10.3389/fphys.2018.01245
Qu J., Ko C.-W., Tso P., Bhargava A., Apolipoprotein A-I. V. (2019). Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells 8, 319. doi:10.3390/cells8040319
Qu Y., Zhang Z., Lu Y., Zheng D., Wei Y. (2019). Network Pharmacology Reveals the Molecular Mechanism of Cuyuxunxi Prescription in Promoting Wound Healing in Patients with Anal Fistula. Evidence-Based Complement. Altern. Med. 2019, 1–9. doi:10.1155/2019/3865121
Ran B., Guo C. E., Li W., Li W., Wang Q., Qian J., et al. (2021). Sea Buckthorn ( Hippophae Rhamnoides L.) Fermentation Liquid Protects against Alcoholic Liver Disease Linked to Regulation of Liver Metabolome and the Abundance of Gut Microbiota. J. Sci. Food Agric. 101, 2846–2854. doi:10.1002/jsfa.10915
Renshaw S. A., Loynes C. A., Trushell D. M. I., Elworthy S., Ingham P. W., Whyte M. K. B. (2006). A Transgenic Zebrafish Model of Neutrophilic Inflammation. Blood 108, 3976–3978. doi:10.1182/blood-2006-05-024075
Roques S., Deborde C., Richard N., Skiba‐Cassy S., Moing A., Fauconneau B. (2020). Metabolomics and Fish Nutrition: a Review in the Context of Sustainable Feed Development. Rev. Aquacult 12, 261–282. doi:10.1111/raq.12316
Schulze R. J., Krueger E. W., Weller S. G., Johnson K. M., Casey C. A., Schott M. B., et al. (2020). Direct Lysosome-Based Autophagy of Lipid Droplets in Hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 117, 32443–32452. doi:10.1073/pnas.2011442117
Shi J., Wang L., Lu Y., Ji Y., Wang Y., Dong K., et al. (2017). Protective Effects of Seabuckthorn Pulp and Seed Oils against Radiation-Induced Acute Intestinal Injury. J. Radiat. Res. 58, 24–32. doi:10.1093/jrr/rrw069
Tranah T. H., Edwards L. A., Schnabl B., Shawcross D. L. (2021). Targeting the Gut-Liver-Immune axis to Treat Cirrhosis. Gut 70, 982–994. doi:10.1136/gutjnl-2020-320786
Wang X., Liu J., Zhang X., Zhao S., Zou K., Xie J., et al. (2018). Seabuckthorn berry Polysaccharide Extracts Protect against Acetaminophen Induced Hepatotoxicity in Mice via Activating the Nrf-2/ho-1-SOD-2 Signaling Pathway. Phytomedicine 38, 90–97. doi:10.1016/j.phymed.2017.11.007
Wang X., Yang H., Zhang L., Han L., Di S., Wei X., et al. (2021). Network Pharmacology-Based Prediction of Mechanism of Shenzhuo Formula for Application to DKD. Evidence-Based Complement. Altern. Med. 2021, 1–13. doi:10.1155/2021/6623010
Wei X., Huang L., Li J., Ni Y. (2019). “Probiotic Characteristics of Intestinal flora and Lactic Acid Bacteria of Cold Water Fish in High Latitudes of China,” in The 16th Annual Meeting of CFST Wuhan.
Wu N., Song Y.-L., Wang B., Zhang X.-Y., Zhang X.-J., Wang Y.-L., et al. (2016). Fish Gut-Liver Immunity during Homeostasis or Inflammation Revealed by Integrative Transcriptome and Proteome Studies. Sci. Rep. 6, 36048. doi:10.1038/srep36048
Wu N., Wang B., Cui Z.-W., Zhang X.-Y., Cheng Y.-Y., Xu X., et al. (2018). Integrative Transcriptomic and microRNAomic Profiling Reveals Immune Mechanism for the Resilience to Soybean Meal Stress in Fish Gut and Liver. Front. Physiol. 9, 1154. doi:10.3389/fphys.2018.01154
Wu N., Xu X., Wang B., Li X.-M., Cheng Y.-Y., Li M., et al. (2020). Anti-foodborne Enteritis Effect of Galantamine Potentially via Acetylcholine Anti-inflammatory Pathway in Fish. Fish Shellfish Immunol. 97, 204–215. doi:10.1016/j.fsi.2019.12.028
Xie J., Li M., Ye W., Shan J., Zhao X., Duan Y., et al. (2021). Sinomenine Hydrochloride Ameliorates Fish Foodborne Enteritis via α7nAchR-Mediated Anti-inflammatory Effect whilst Altering Microbiota Composition. Front. Immunol. 12, 766845. doi:10.3389/fimmu.2021.766845
Xiong S., Wu J., Jing J., Huang P., Li Z., Mei J., et al. (2017). Loss of Stat3 Function Leads to Spine Malformation and Immune Disorder in Zebrafish. Sci. Bull. 62, 185–196. doi:10.1016/j.scib.2017.01.008
Yang X., Lu D., Zhuo J., Lin Z., Yang M., Xu X. (2020). The Gut-Liver axis in Immune Remodeling: New Insight into Liver Diseases. Int. J. Biol. Sci. 16, 2357–2366. doi:10.7150/ijbs.46405
Ye Q., Feng Y., Wang Z., Zhou A., Xie S., Fan L., et al. (2019). Effects of Dietary Gelsemium Elegans Alkaloids on Intestinal Morphology, Antioxidant Status, Immune Responses and Microbiota of Megalobrama amblycephala. Fish Shellfish Immunol. 94, 464–478. doi:10.1016/j.fsi.2019.09.048
Ye X., Wu J., Zhang D., Lan Z., Yang S., Zhu J., et al. (2021). How Aconiti Radix Cocta Can Treat Gouty Arthritis Based on Systematic Pharmacology and UPLC-QTOF-MS/MS. Front. Pharmacol. 12, 618844. doi:10.3389/fphar.2021.618844
Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16, 284–287. doi:10.1089/omi.2011.0118
Yu T., Li Z., Xu L., Yang M., Zhou X. (2021). Anti-inflammation Effect of Qingchang Suppository in Ulcerative Colitis through JAK2/STAT3 Signaling Pathway In Vitro and In Vivo. J. Ethnopharmacology 266, 113442. doi:10.1016/j.jep.2020.113442
Zang L., Maddison L. A., Chen W. (2018). Zebrafish as a Model for Obesity and Diabetes. Front. Cel Dev. Biol. 6, 91. doi:10.3389/fcell.2018.00091
Zhang D., Zhang Y., Gao Y., Chai X., Pi R., Chan G., et al. (2020). Translating Traditional Herbal Formulas into Modern Drugs: a Network-Based Analysis of Xiaoyao Decoction. Chin. Med. 15, 25. doi:10.1186/s13020-020-00302-4
Zhang G., Gong S., Yu D., Yuan H. (2009). Propolis and Herba Epimedii Extracts Enhance the Non-specific Immune Response and Disease Resistance of Chinese Sucker, Myxocyprinus asiaticus. Fish Shellfish Immunol. 26, 467–472. doi:10.1016/j.fsi.2009.01.011
Zhang X., Zhu J., Yan J., Xiao Y., Yang R., Huang R., et al. (2020). Systems Pharmacology Unravels the Synergic Target Space and Therapeutic Potential of Rhodiola Rosea L. For Non-small Cell Lung Cancer. Phytomedicine 79, 153326. doi:10.1016/j.phymed.2020.153326
Zhang Z., Liu W., Zhao L., Huang Z., Chen X., Ma N., et al. (2018). Retinoblastoma 1 Protects T Cell Maturation from Premature Apoptosis by Inhibiting E2F1. Development 145, 158139. doi:10.1242/dev.158139
Zhang Z., Taylor L., Shommu N., Ghosh S., Reimer R., Panaccione R., et al. (2020). A Diversified Dietary Pattern Is Associated with a Balanced Gut Microbial Composition of Faecalibacterium and Escherichia/Shigella in Patients with Crohn's Disease in Remission. J. Crohns Colitis 14, 1547–1557. doi:10.1093/ecco-jcc/jjaa084
Zhao S., Sun H., Liu Q., Shen Y., Jiang Y., Li Y., et al. (2020). Protective Effect of Seabuckthorn berry Juice against Acrylamide‐induced Oxidative Damage in Rats. J. Food Sci. 85, 2245–2254. doi:10.1111/1750-3841.15313
Zheng P., Yu B., He J., Yu J., Mao X., Luo Y., et al. (2017). Arginine Metabolism and its Protective Effects on Intestinal Health and Functions in Weaned Piglets under Oxidative Stress Induced by Diquat. Br. J. Nutr. 117, 1495–1502. doi:10.1017/S0007114517001519
Keywords: herb therapy, anti-inflammation, intestinal microbiota, fatty acid metabolism, immune crosstalk
Citation: Li M, Zhao X, Xie J, Tong X, Shan J, Shi M, Wang G, Ye W, Liu Y, Unger BH, Cheng Y, Zhang W, Wu N and Xia X-Q (2022) Dietary Inclusion of Seabuckthorn (Hippophae rhamnoides) Mitigates Foodborne Enteritis in Zebrafish Through the Gut-Liver Immune Axis. Front. Physiol. 13:831226. doi: 10.3389/fphys.2022.831226
Received: 08 December 2021; Accepted: 21 March 2022;
Published: 06 April 2022.
Edited by:
Javier Santander, Memorial University of Newfoundland, CanadaReviewed by:
Patricia Diaz-Rosales, Centro de Investigación en Sanidad Animal (INIA-CSIC), SpainCopyright © 2022 Li, Zhao, Xie, Tong, Shan, Shi, Wang, Ye, Liu, Unger, Cheng, Zhang, Wu and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Wu, d3VuYW5AaWhiLmFjLmNu; Xiao-Qin Xia, eHF4aWFAaWhiLmFjLmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.