
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 08 February 2022
Sec. Invertebrate Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.823418
This article is part of the Research Topic Hormone Regulation of Insect Development View all 7 articles
 Takumi Kamiyama1
Takumi Kamiyama1 Ryusuke Niwa2*
Ryusuke Niwa2*Steroid hormones are responsible for coordinating many aspects of biological processes in most multicellular organisms, including insects. Ecdysteroid, the principal insect steroid hormone, is biosynthesized from dietary cholesterol or plant sterols. In the last 20 years, a number of ecdysteroidogenic enzymes, including Noppera-bo, Neverland, Shroud, Spook/Spookier, Cyp6t3, Phantom, Disembodied, Shadow, and Shade, have been identified and characterized in molecular genetic studies using the fruit fly Drosophila melanogaster. These enzymes are encoded by genes collectively called the Halloween genes. The transcriptional regulatory network, governed by multiple regulators of transcription, chromatin remodeling, and endoreplication, has been shown to be essential for the spatiotemporal expression control of Halloween genes in D. melanogaster. In this review, we summarize the latest information on transcriptional regulators that are crucial for controlling the expression of ecdysteroid biosynthetic enzymes and their roles in insect development.
Steroid hormones are bioactive small molecules that induce many physiological events in both vertebrates and invertebrates. Steroid hormones are received by their specific nuclear receptors and activate downstream gene transcription. In arthropods, including insects, the principal steroid hormones, ecdysteroids, including the most biologically active 20-hydroxyecdysone (20E), play essential roles in their development, such as embryogenesis, molting, metamorphosis, oogenesis, diapause, immune response, stress resistance, seasonal plasticity, learning, memory acquisition, and longevity (Ishimoto and Kitamoto, 2011; Uryu et al., 2015; Belles, 2020; Yamanaka and Okamoto, 2020; Kamiyama and Niwa, 2021a,b; Kurogi et al., 2021; Nunes et al., 2021; van der Burg and Reed, 2021).
During larval and pupal development, ecdysteroids are biosynthesized from dietary cholesterol and plant sterols in a special endocrine organ called the prothoracic gland (PG) (Figure 1). Studies during the 2000s and early 2010s successfully identified and characterized a number of essential ecdysteroid biosynthetic enzymes (Niwa and Niwa, 2014a). These enzyme-encoding genes are collectively called the Halloween genes (Gilbert, 2004). After their discovery, it was found that the levels of expression of Halloween genes in the PG correlated well with the levels of hemolymph ecdysteroid titers. This finding was reminiscent of the expression of mammalian steroidogenic genes, which reflects the production of steroid hormones (Mizutani et al., 2015). In mammals, the strict control of the expression of steroidogenic genes is regulated by several key steroidogenic transcription factors (TFs), including Ad4BP/steroidogenic factor 1 (SF-1) and liver receptor homolog-1 (Parker et al., 2002). By contrast, elucidation of ecdysteroidogenic TFs and their regulatory roles in insects has lagged far behind that of mammalian steroidogenic transcriptional regulation.
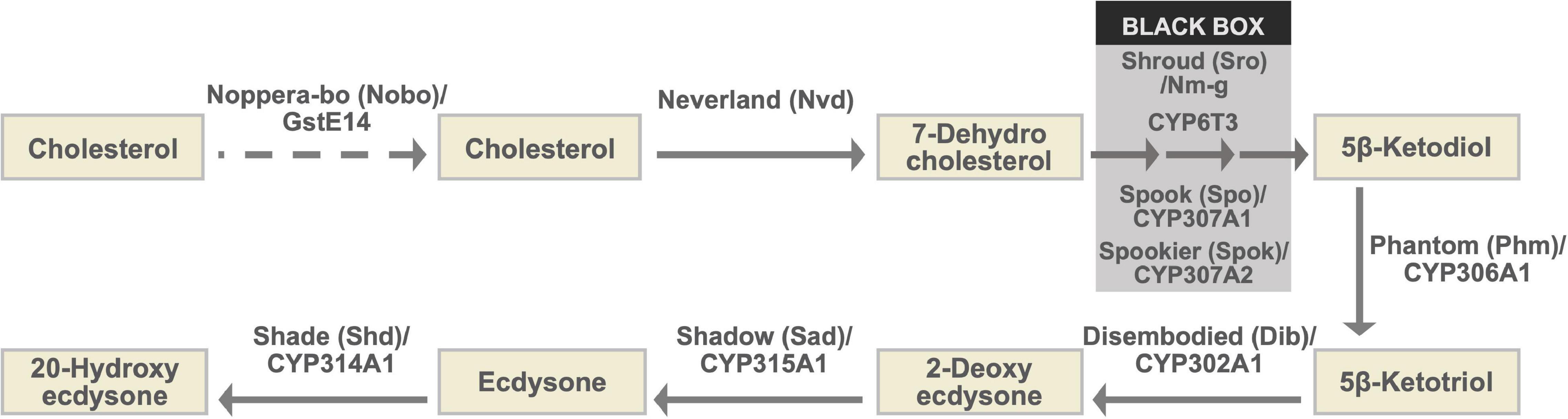
Figure 1. The ecdysteroid biosynthesis pathway and Halloween enzymes. As the conversion steps between 7-dehydrocholesterol and 5β-ketodiol have not been chemically characterized, they are collectively called the “Black Box.” Notably, spo and spok are paralogous to each other (Ono et al., 2006; Komura-Kawa et al., 2015). Although genetic and physiological analyses have suggested that Nobo plays a role in cholesterol intake or trafficking in the PG, endogenous substrates of Nobo have not been identified. Therefore, the role of Nobo is indicated by the dashed arrow in this illustration.
Here, we describe the recent findings regarding the transcriptional regulation and chromatin remodeling in the expression of Halloween genes. Particularly, we aim to update our knowledge on the role of ecdysteroidogenic TFs in the control of ecdysteroid biosynthesis in the PG during development.
Some of the genes encoding the ecdysteroidogenic enzymes were first identified in a series of Drosohila melanogaster classical genetic mutants that caused embryonic lethality and a defect in cuticle structure characterized by naked and polished cuticles (Chávez et al., 2000; Gilbert, 2004). Because naked cuticle embryos were superficially characterized, these mutants were referred to as ghosts and specters, including disembodied (dib), noppera-bo (nobo), phantom (phm), shadow (sad), shade (shd), spook (spo), and shroud (sro) (Chávez et al., 2000; Enya et al., 2014). D. melanogaster embryonic lethal mutants showing the naked cuticle phenotype and defects related to ecdysteroid biosynthesis were designated and classified as “Halloween mutants” (Gilbert, 2004). In addition, the genes responsible for Halloween mutants and their products were named “Halloween genes” and “Halloween enzymes,” respectively. However, embryonic lethal phenotypes identical to Halloween mutants have not been reported for a part of ecdysteroid biosynthetic enzymes (Niwa and Niwa, 2014a). Despite that, researchers usually collectively utilize the term “Halloween genes” and “Halloween enzymes” to refer to all identified ecdysteroid biosynthetic genes and enzymes, respectively. In this review article, we follow this customary scientific terminology.
The ecdysteroid biosynthetic pathway and ecdysteroid biosynthetic enzymes encoded by Halloween genes are illustrated in Figure 1 and Table 1 (Gilbert et al., 2002; Niwa and Niwa, 2014a; Kamiyama and Niwa, 2021b, and references therein). All Halloween enzymes, except for Shd, act as catalytic enzymes in the PG. Endogenous substrates of Neverland (Nvd), Phm, Dib, Sad, and Shd have been identified (Figure 1 and Table 1), while the precise biochemical roles of other enzymes have not been elucidated. Nevertheless, Sro, Spo, Spok, and CYP6T3 are thought to function in the “Black Box” because the larval arrest phenotype of knockdown of sro, spok, or Cyp6t3 can be rescued by oral administration of 5β-ketodiol, but not 7-dehydrocholesterol (Figure 1; Ono et al., 2006; Niwa et al., 2010; Ou et al., 2011). After release from the PG to the hemolymph, ecdysone is converted to 20E by Shd in the peripheral tissues (Figure 1).
Among the Halloween enzymes, Nobo, also known as GstE14 in D. melanogaster and GSTE7 in the silkworm Bombyx mori, is the most recently identified one known to contribute to ecdysteroid biosynthesis. Nobo belongs to the family of glutathione S-transferase (GST). Therefore, we briefly introduce Nobo in this review article. Nobo is predominantly expressed in the embryonic PG precursor, larval PG, and adult ovarian follicle cells (Chanut-Delalande et al., 2014; Enya et al., 2014, 2015). D. melanogaster genetic null mutants of nobo show embryonic lethality and typical cuticular patterning defects, which are identical to the phenotypes of other Halloween embryonic mutants (Chávez et al., 2000). In addition, the larval-arrest phenotype of PG-specific RNAi against nobo was almost fully rescued by oral administration of 20E or cholesterol. As cholesterol is thought to be the most upstream precursor, it was hypothesized that Nobo does not convert ecdysteroidal intermediates, but rather plays a crucial role in cholesterol intake and transport in the PG (Chanut-Delalande et al., 2014; Enya et al., 2014). The primary reaction catalyzed by GSTs is the conjugation of the reduced form of L-γ-glutamyl-L-cysteinylglycine or glutathione (GSH) to substrates. Consistent with the fact that Nobo is a GST, gclc, encoding an enzyme required for the de novo synthesis of GSH, was shown to partly phenocopy nobo mutants in D. melanogaster (Enya et al., 2017). Although the endogenous substrates of Nobo remain unclear, a high-throughput chemical screen and X-ray crystallography revealed that several small chemical compounds can be inserted into the putative ligand-binding pocket of Nobo, including the mammalian female sex hormone, 17β-estradiol (Fujikawa et al., 2015; Koiwai et al., 2020, 2021). This implies that a steroid could also be an endogenous substrate of Nobo, but future research is needed to clarify this point.
Ecdysteroids play a pivotal role in the development of arthropods. Consistent with this fact, ecdysteroid biosynthetic enzyme genes are mostly well conserved in the genomes of insects as well as other arthropods (Markov et al., 2009; Schumann et al., 2018). However, some enzymes are curiously missing from specific taxonomic groups of arthropods. For example, the genomes of Myriapoda and Chelicerata lack phm (Schumann et al., 2018) and thus the ability to perform carbon-25 hydroxylation of ecdysteroids. Consequently, it is thought that the main ecdysteroid is ponasterone A instead of 20E in these species. Likewise, no clear orthologs of nvd have been identified in any coleopteran species (R.N., unpublished observation). This is paradoxical, as the red flour beetle Tribolium castaneum requires sterols for larval development (Fraenkel and Blewett, 1943a,b). Orthologs of nobo are well conserved in dipteran and lepidopteran species, but are not found in other insect species (Enya et al., 2014; Koiwai et al., 2020). More strikingly, spok and Cyp6t3 are found only in Drosophilidae species (Ono et al., 2006; Sztal et al., 2007; Ou et al., 2011). These facts have suggested that some ecdysteroid biosynthetic enzymes have evolved and are maintained in very limited clades, as reported in a recent comprehensive phylogenetic analysis (Schumann et al., 2018).
The temporal profiles of Halloween genes have been found to correlate well with the changes in the 20E titer during larval development (Niwa and Niwa, 2014b; Yamanaka and Okamoto, 2020). In addition, all known Halloween genes, except for shd, display high tissue specificity, as they are predominantly expressed in the PG and ovaries (Niwa and Niwa, 2014a; Kamiyama and Niwa, 2021b). Such temporally dynamic and spatially restricted expression profiles of the Halloween genes imply a tight transcriptional control network. To date, many transcription factors (TFs) have been reported to be responsible for the expression of Halloween genes (Niwa and Niwa, 2016b). In addition, posttranscriptional and epigenetic regulation are crucial for the expression of Halloween genes. All known validated and putative ecdysteroidogenic TFs required in PG are listed in Table 2. The modes of action of some ecdysteroidogenic TFs are illustrated in Figure 2.
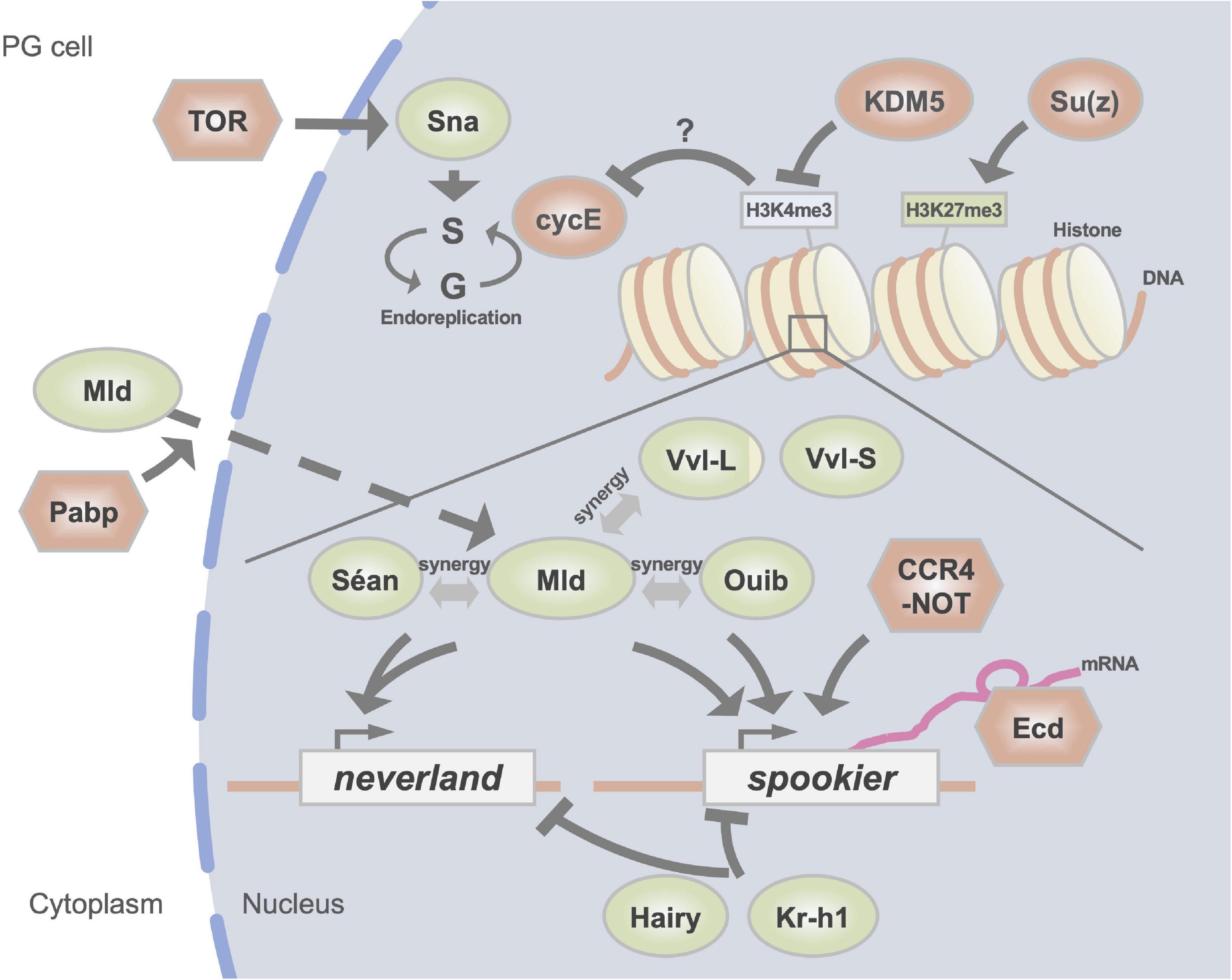
Figure 2. A schematic illustration of regulators of the expression of Halloween genes in the PG. DNA-binding transcription factors are indicated in green circles. Red circles represent histone modification enzymes and cell cycle regulators. Other factors are indicated in red hexagons. H3K4me3 and H3K27me3 indicate trimethylation of the fourth and 27th lysine residues of histone H3, respectively.
The first identified TF that influences the levels of expression of validated Halloween genes is βFtz-f1, a critical regulator of insect metamorphosis in many tissues (Parvy et al., 2005). A study on D. melanogaster showed that the protein levels of Phm and Dib were significantly reduced following the loss of βFtz-f1 function in PG cells (Parvy et al., 2005). It was also revealed that the activity of βFtz-f1 in PG is modulated by SUMOylation (Talamillol et al., 2008; Talamillo et al., 2013). Notably, βFtz-f1 is a nuclear receptor (NR) homologous to Ad4BP/SF-1, a key TF for steroidogenic genes in mammalian cells (Morohashi et al., 2013). Moreover, mammalian Ad4BP/SF-1 is also SUMOylated, resulting in the modulation of the expression of steroidogenic genes, thus influencing adrenocortical steroidogenesis (Suda et al., 2011). These data imply that the role of βFtz-f1/Ad4BP/SF-1 in steroidogenesis and the posttranslational modification of them are evolutionarily conserved between insects and vertebrates, at least in part.
βFtz-f1 is well known to be transcriptionally induced by a surge of 20E during development (Yamada et al., 2000). Given the fact that 20E signaling plays a pivotal role in the sequential induction of not only βFtz-f1 but also other NR genes (Thummel, 2002), it is plausible that other NRs are also involved in the transcriptional regulation of Halloween genes. Indeed, this is the case, as 20E-responsive NR genes other than βFtz-f1 also regulate the expression of Halloween genes in the PG.
One such 20E-responsive NR is Ecdysone receptor (EcR) itself (Moeller et al., 2013; Parvy et al., 2014). Dynamic feedback regulation of 20E-EcR signaling in the PG is important for ecdysteroid biosynthesis. During the larval stages, reduced 20E-EcR signaling in the PG decreases the expression of Halloween genes, leading to a delay in the larva-to-prepupa transition (Moeller et al., 2013). This observation indicates that 20E has a positive feedback effect on PG, rapidly amplifying its own synthesis to trigger pupariation. In contrast, after pupariation, reduced 20E-EcR signaling increases the expression of Halloween genes, leading to metamorphosis. These findings indicate that 20E conversely has a negative feedback signal that ensures a decline in the level of 20E in the prepupa-to-pupa transition. These opposite effects are partly regulated by the 20E-responsive Broad (Br) gene, as described in the next section.
Downstream of 20E-EcR signaling, other 20E-responsive NRs, E75, βFtz-f1, and DHR3, regulate ecdysteroid biosynthesis in the PG (Bialecki et al., 2002; Cáceres et al., 2011; Moeller et al., 2013; Parvy et al., 2014). E75 and βFtz-f1 enhance ecdysteroid biosynthesis, whereas DHR3 suppresses it. In addition, loss of function of these TFs influences the expression of a part of Halloween genes (Table 1). Moreover, the expression of E75 is affected by EcR, whereas the expression of DHR3 is affected by E75 and βFtz-f1. These findings suggest that the NR-mediated 20E-responsive gene expression cascade is involved in the control of the expression of Halloween genes.
DHR4, which is known as a 20E-responsive gene (King-Jones et al., 2005), negatively regulates ecdysteroid biosynthesis in the PG (Ou et al., 2011). PG-specific DHR4 knockdown in the larval stage caused a higher ecdysteroid titer and precocious puparium formation. Interestingly, the subcellular localization of DHR4 is temporally regulated by prothoracicotropic hormone (PTTH), the important and most classical humoral factor that stimulate the biosynthesis and secretion of ecdysteroids in the PG (Kawakami et al., 1990; McBrayer et al., 2007; Rewitz et al., 2009b; Ou et al., 2011). When PTTH signaling is active in the PG, DHR4 is exported from the nucleus to the cytoplasm, leading to the suppression of DHR4-mediated gene expression. One of the essential targets of DHR4 is Cyp6t3.
Another 20E-responsive NR, Ultraspiracle (Usp), which is a heterodimer partner of EcR, is also involved in the expression of Halloween genes (Koyama et al., 2014). This point is described in more detail later.
The Broad-complex (Br-C) gene belongs to the Br-C, Tramtrack, Bric-a-brac/Poxvirus, and zinc finger (BTB/POZ) family of C2H2-zinc finger TFs. It is well known that Br-C is crucial for the appropriate regulation of many 20E-inducible genes as larvae enter metamorphosis (Belles, 2020). In D. melanogaster, Br-C expresses 4 different isoforms, designated Br-Z1 to Br-Z4. All Br-C isoforms are expressed in the PG (Zhou et al., 2004). In addition, overexpression of any of the isoforms in the PG prevents larval molting or pupariation to varying degrees, which is consecutively rescued by administration of 20E (Zhou et al., 2004). These results suggest an inhibitory effect of Br-C on ecdysteroid biosynthesis in the PG. However, in reality, this situation is not very simple.
PG-specific RNAi experiments revealed that, among these isoforms, Br-Z1 and Br-Z4, not Br-Z2 or Br-Z3, are involved in the regulation of ecdysteroid biosynthesis in the PG (Xiang et al., 2010; Moeller et al., 2013). Interestingly, Br-Z4 regulates the expression of Halloween genes positively, whereas Br-Z1 negatively. The functional differences between Br-Z4 and Br-Z1 are also reflected by the different responses to 20E, as the expression of Br-Z4 is more acutely induced by 20E than Br-Z1 (Moeller et al., 2013). In fact, the 20E-EcR-mediated positive and negative feedback regulation of ecdysteroid biosynthesis in the PG, was demonstrated to be dependent on the differential functions of Br-Z4 and Br-Z1. The positive feedback regulatory mechanism requires Br-Z4 to induce the expression of Halloween genes in the late third larval instar, whereas the negative feedback regulatory mechanism requires Br-Z1 to suppress the expression of Halloween genes in the prepupal and pupal stages (Moeller et al., 2013). Taken together, Br-C plays a crucial role in the EcR-dependent switching from positive to negative feedback regulation, shaping the pulses of the 20E titer during the larval-to-pupal transition.
Besides Br-C, other C2H2-zinc finger TFs, namely Molting defective (Mld), Séance (Séan), and Ouija board (Ouib) are also involved in ecdysteroid biosynthesis in the PG. All 3 TFs belong to the zinc finger associated domain (ZAD)-C2H2 zinc finger family.
Mld was identified from a mutant with a larval molting defect and low 20E titer (Neubueser et al., 2005). In addition, a classical temperature-sensitive mutant lethal(3)dts3, which shows ecdysteroid deficiency at the restrictive temperature (Walker et al., 1987), has been recognized as an allele of mld (Simon et al., 2003; Ishimoto et al., 2013). Studies have also shown that loss of mld function in the PG specifically impairs the expression of nvd and spok, but not that of other Halloween genes (Neubueser et al., 2005; Ono et al., 2006; Danielsen et al., 2014). Importantly, the lethal phenotype of loss-of-mld function mutants is rescued by the simultaneous expression of both nvd and spo (the paralog of spok) (Uryu et al., 2018). These data indicate that, at least during larval development, the essential downstream targets of Mld are only nvd and spok.
Two other TFs, Séan (also known as Numerous disordered muscles; Dobi et al., 2014) and Ouib, are also essential for the transcriptional regulation of nvd and spok (Komura-Kawa et al., 2015; Niwa and Niwa, 2016a; Uryu et al., 2018). Strikingly, loss-of-function mutations in séan or ouib result in a larval-arrest phenotype with a dramatic reduction in the expression of nvd and spok, respectively, but not in that of other Halloween genes. More importantly, this lethal phenotype is rescued by expression of nvd or spo alone, respectively (Komura-Kawa et al., 2015; Uryu et al., 2018). These data indicate that, at least during larval development, only nvd and spok are essential downstream targets of Séan and Ouib, respectively. It should also be noted that among all identified ecdysteroidogenic TFs, Ouib is the only ecdysteroidogenic TF that is specifically localized in the PG and not in other tissues (Komura-Kawa et al., 2015).
Further analyses using a cell culture system have revealed that these 3 TFs cooperatively control the expression of nvd and spok. Coexistence of both Mld and Séan in cultured cells results in a drastic induction of nvd promoter-driven reporter gene expression compared with that of cells treated with Mld or Séan alone (Uryu et al., 2018). Similarly, coexistence of both Mld and Ouib dramatically induces spok promoter-driven reporter gene expression compared with Mld or Ouib alone (Komura-Kawa et al., 2015). These studies have revealed the unique feature of the cooperative role of Mld, Séan, and Ouib in regulating a very small subset of genes, that is, nvd and spok. As the ZAD domain is known to be protein-protein interaction domain (Jauch et al., 2003), this suggest that a physical interaction between Mld and Séan or between Mld and Ouib is possible. However, such protein-protein interactions have not been reported to date. Therefore, it remains unclear how this cooperativity is achieved (Uryu et al., 2018).
Of note, the gene loci of nvd and spok, but not that of other Halloween genes, are located in the pericentric constitutive heterochromatin region in the D. melanogaster genome (Ono et al., 2006; Yoshiyama et al., 2006; Fitzpatrick et al., 2011). Although constitutive heterochromatin has long been thought to be transcriptionally inert in cells, a number of studies have revealed that it contains active genes essential for viability (Dimitri et al., 2009). To date, Mld, Séan, and Ouib represent the first example of a set of TFs that regulate genes located in constitutive heterochromatin in a spatiotemporal-specific manner.
From an evolutionary point of view, mld, séan, and ouib are also unique because their orthologs are found only in genomes of Drosophilidae species (Neubueser et al., 2005; Ono et al., 2006; Komura-Kawa et al., 2015; Uryu et al., 2018). This trend is partly consistent with the fact that spok (but not nvd) is only conserved in genomes of Drosophilidae species (Ono et al., 2006). Therefore, it has been suggested that some essential ecdysteroidogenic TFs might have rapidly evolved only in very limited insect clade(s).
Ventral vein lacking (Vvl), also known as Ventral veinless or Drifter (Drf), belongs to the POU homeobox TF family and plays versatile roles in cell fate specification, organ patterning, and immune defense. As a TF that regulates the expression of Halloween genes, Vvl was independently identified in studies using D. melanogaster (Danielsen et al., 2014) and the red flour beetle Tribolium castaneum (Cheng et al., 2014). The role of vvl orthologs in ecdysteroid biosynthesis has also been confirmed in other insects, such as the silkworm Bombyx mori (Meng et al., 2015) and the milkweed bug Oncopeltus fasciatus (Sarwar et al., 2020). In D. melanogaster, loss of vvl function results in the reduced expression of most Halloween genes (Danielsen et al., 2014). Conversely, overexpression of the B. mori ortholog of vvl, known as POU-M2, can induce the expression of some Halloween genes in B. mori culture cells (Meng et al., 2015). In addition, POU-M2 was demonstrated to directly bind to potential cis-regulatory elements in the phm promoter region. These data suggest that the function of Vvl in regulating the expression of Halloween genes is conserved in insects.
A recent study demonstrated that the vvl gene produces long and short isoforms through a stop codon readthrough process in the PG during larval development (Zhao et al., 2021). Stop codon readthrough is a phenomenon in which near-cognate tRNAs are able to decode stop codons as sense codons, resulting in a read-frame extension even in a conventional 3′ untranslated region. While the error rate of translational termination should be less than 0.1%, surprisingly, the stop codon readthrough of vvl mRNA reaches a rate of 50% in the PG (Zhao et al., 2021). Consequently, the C-terminal extension of Vvl produces a long isoform, which possesses an extra 286 amino acid residues compared with the short isoform (427 amino acid length) encoded by the conventional vvl coding sequence. Loss-of-function mutations that lead to the specific lack of the long isoform have been associated with pupariation delay and decreased expression of nvd, spok, dib, and sad, even in the presence of the short isoform.
A chromosomal deletion of a human homolog of vvl, known as POU3F2, has been associated with hypogonadotropic hypogonadism and adrenal insufficiency (Bonaglia et al., 2008; Izumi et al., 2013). Therefore, it is suggested that the Vvl/POU3F2 family participates in the conserved regulatory mechanisms of steroidogenesis between insects and vertebrates.
Cap-n-collar (CncC), the D. melanogaster ortholog of mammalian nuclear factor erythroid 2-related factor 2 (Nrf2), and its cofactor Kelch-like ECH associated protein 1 (Keap1) are evolutionary conserved key TFs that regulate xenobiotic responses and redox homeostasis (Ulasov et al., 2021). Although CncC-Keap1 signaling is indispensable for these biological processes, it has also been found to be involved in ecdysteroid biosynthesis (Deng and Kerppola, 2013). Studies have shown that PG-specific knockdown of CncC or Keap1 leads to decreased expression of a wide variety of Halloween genes in the PG. In addition, accelerated pupariation induced by the overexpression of a constitutively activated oncogenic RAS is suppressed by knocking down CncC. Considering that RAS is necessary for PTTH signaling (Rewitz et al., 2009b; Pan et al., 2020; Kannangara et al., 2021), CncC seems to be downstream of the PTTH-RAS pathway. In addition, a more recent study has also found that loss of function of Keap1 results in the reduction in the expression of several Halloween genes in the ovary, another ecdysteroid biosynthetic organ (Carlson et al., 2022).
Forkhead box transcription factor class O (FoxO) regulates the metabolism, cellular proliferation, stress tolerance, and lifespan in most animals, including insects (Calnan and Brunet, 2008). In D. melanogaster PG, FoxO functions as a negative regulator of the expression of Halloween genes just before the early generation of a small peak of 20E in the third (final) larval instar of D. melanogaster (Koyama et al., 2014). This small peak of 20E has been proposed to induce a developmental transition known as the critical weight (Mirth and Shingleton, 2012). Critical weight is the minimum body weight of larvae that is essential for the transition to pupa. Before critical weight, FoxO is localized in the nuclei of PG, suppressing the expression of Halloween genes, and thus of ecdysteroid biosynthesis. In contrast, after critical weight, FoxO is exported to the cytoplasm and excluded from the nuclei, resulting in the generation of a small 20E peak. Interestingly, FoxO has been found to be localized in the nuclei of larvae that are reared under starved conditions at the same time point. As they cannot generate this small 20E peak, they consequently cannot grow beyond the critical weight (Koyama et al., 2014). These data suggest that nuclear localization of FoxO determines the critical weight state attainment by suppressing the expression of the Halloween genes before this time point. Moreover, FoxO is known to physically interact with the nuclear hormone receptor Usp, which is also required for the suppression of the expression of Halloween genes (Koyama et al., 2014).
Besides D. melanogaster, the role of FoxO in ecdysteroid biosynthesis in the PG was also demonstrated in T. castaneum. RNAi of FoxO results in a delay of ecdysteroid biosynthesis (Lin et al., 2018). In the RNAi animals, expression of spo is significantly delayed, whereas that of phm, dib, sad, or shd is not.
It is well known that the nuclear localization of FoxO is negatively regulated by the activity of insulin/insulin-like growth factor signaling (IIS) (Calnan and Brunet, 2008). Moreover, IIS has been demonstrated to enhance ecdysteroid biosynthesis in D. melanogaster PG (Colombani et al., 2005). Therefore, FoxO, and possibly Usp, might be crucial for the suppression of the expression of Halloween genes before the critical weight when the activity of IIS is low (Koyama et al., 2014). As the critical weight 20E peak responds to environmental cues, including nutritional status (Mirth and Shingleton, 2012), both IIS and FoxO-Usp seem to play an essential role in the developmental transition influenced by nutritional availability.
Homeotic (Hox) transcription factors confer segmental identity along the anteroposterior axis of insect embryos (Mallo and Alonso, 2013). Sex comb reduced (Scr), a homeotic transcription factor, is required for the differentiation and morphogenesis of PG (Sánchez-Higueras et al., 2014). Of note, PG has a homologous origin with the respiratory tracheal system in D. melanogaster and is derived from the primordium in the labial segment of the embryo. Scr cooperates with signal transducer and activator of transcription (STAT), inducing the expression of vvl in the labial patch. Subsequently, a subgroup of vvl-expressing cells activates the Zn-finger gene Snail (Sna), a key regulator of the epithelial-mesenchymal transition. The differentiated PG cells eventually express the specific marker TF gene spalt. These TF associations appear to be essential for establishing the cellular status of PG cells (Sánchez-Higueras et al., 2014).
In addition to shaping the segment-specific establishment of the PG, some homeotic transcription factors also regulate the expression of Halloween genes in differentiated PG. In particular, 2 homeotic transcription factors, Antennapedia (Antp) and Scr, regulate the expression of Halloween genes in B. mori (Meng et al., 2015; Daimon et al., 2020). Antp is highly expressed in the B. mori PG, and its temporal expression pattern in the PG is positively correlated with that of Halloween genes. In addition, Antp directly binds to potential cis-regulatory elements in the phm promoter region (Meng et al., 2015).
Scr was identified as a gene responsible for Moltinism B. mori strains (Daimon et al., 2020). Moltinism strains molt 3, 4, or 5 times, which is determined by allelic variations at a single autosomal locus. Positional cloning has revealed that this locus overlaps with the Scr gene region. While Moltinism strains do not exhibit any abnormalities in segmental identity, mutations in these strains induce abnormal expression of Scr in the PG. Inhibiting the function of Scr in the PG upregulates the expression of nvd, hence increasing the basal ecdysteroid titer during the early fifth larval instar. This result suggest that increased basal ecdysteroid titer leads to the extra molt to the sixth instar larvae, but not pupae (Daimon et al., 2020). This interpretation resembles the “leaky prothoracic glands” hypothesis, which proposes that the effects of low levels of ecdysone over a long period trigger the molt, similar to high levels of ecdysone over a brief period (Nijhout, 1976; Callier and Nijhout, 2011). Taken together, these results suggest that Scr plays a role in controlling the number of proper molts through the suppression of the expression of nvd in the PG.
Period (Per) and Timeless (Tim) are indispensable TFs that consist of the central components of the circadian pacemaker clock gene regulatory network in insects (Dubowy and Sehgal, 2017). Both Per and Tim oscillate in the PG of larvae (Di Cara and King-Jones, 2016) and pupae (Myers et al., 2003; Morioka et al., 2012). PG-specific knockdown of either per, tim, or other clock component genes results in the larval-arrest phenotype (Di Cara and King-Jones, 2016). In addition, PG-specific overexpression of tim affects eclosion timing (Myers et al., 2003). Moreover, at least in the larval stage, both Per and Tim are required for the transcriptional upregulation of Halloween genes (Di Cara and King-Jones, 2016). These data suggest that Per and Tim are part of the PG circadian clock that influences developmental progress via the transcriptional regulation of Halloween genes.
In some insects, molting is determined by circadian “gating,” where a ecdysteroid pulse is delayed by 24 h if the appropriate time window is missed (Truman and Riddiford, 1974; Mizoguchi and Ishizaki, 1982). However, not only molting in D. melanogaster is not dependent on such gating, but also its genetic null mutants of per and tim are viable (Zerr et al., 1994; Myers et al., 1995). Nevertheless, paradoxically, PG-specific knockdown of either per or tim results in a larval-arrest phenotype. This discrepancy might be explained by the hypothesis that PG-specific knockdown against circadian components causes dis-synchronization between the PG and external timing signals, leading to catastrophic effects on ecdysteroid biosynthesis (Di Cara and King-Jones, 2016). Indeed, Tim is known to couple the circadian machinery directly to IIS and PTTH signaling in the PG. Moreover, the active IIS directly modulates the function of Tim, suggesting that the local clock in the PG is normally synchronized with systemic insulin cues (Di Cara and King-Jones, 2016). Because both PTTH and systemic IIS are themselves under circadian control, the desynchronization of the local endocrine clock in the PG with external circadian cues appears to be the primary cause for the failure in ecdysteroid biosynthesis.
In some insects, including the eri silk moth Samia cynthia ricini, the PG is thought to contain a photoreceptor that regulates timed developmental events (Mizoguchi and Ishizaki, 1982). A recent RNA-seq study has revealed that cryptochrome (cry), which encodes the circadian transcriptional repressor essential for photoentrainment, is expressed in B. mori PG (Bian et al., 2019). Moreover, the oscillation in the expression of tim in the PG has been shown to be diminished or weakened in the cry mutant background in D. melanogaster (Myers et al., 2003; Morioka et al., 2012). However, PG-specific knockdown of cry is not associated with any developmental timing/arrest phenotype in D. melanogaster (Di Cara and King-Jones, 2016). Therefore, the role of cry in the PG circadian clock remains unclear.
Sna is another TF exhibiting versatile functions during insect development (Nieto, 2002). As described in the section “Homeotic/Hox transcription factors,” Sna is necessary for epithelial-mesenchymal transition in the PG. Moreover, Sna also plays an essential role in endoreplication, which is the replication of genomic DNA that is not followed by cell division, in the PG (Zeng et al., 2020). Sna shows a temporally fluctuating expression pattern in the PG of the third instar in D. melanogaster larvae. Interestingly, knocking down sna leads to PG cells failing to undergo the endoreplication cycle, thus resulting in a significant reduction in their nuclear size. In addition, sna knockdown significantly suppress the expression of almost all Halloween genes (Zeng et al., 2020).
Of note, PG cells grow continuously by endoreplication throughout the larval stage. Inhibition of endoreplication, specifically in the PG, causes developmental arrest in the larval stage (Ohhara et al., 2017). Endoreplication in PG cells is essential for the progression of the third instar development in D. melanogaster after critical weight. Blocking endoreplication in the PG causes a significant reduction in the 20E titer because of the suppression in the expression of most Halloween genes. Moreover, progression of endoreplication in PG cells requires the upregulation in the expression of cyclin E by the nutrient sensor target of rapamycin (TOR) signaling. Therefore, nutrient-dependent endoreplication is essential for critical weight attainment, resulting in the strong expression of Halloween genes timely before pupariation for the initiation of metamorphosis (Ohhara et al., 2017, 2019). Conclusively, Sna is considered to regulate the expression of Halloween genes by coordinating endoreplication in a TOR-dependent manner in the PG; however, the essential downstream targets of Sna remain unclear (Zeng et al., 2020).
A basic helix-loop-helix (bHLH) transcription factor, Hairy, is known as one of the signal transducers of the sesquiterpenoid insect hormone, juvenile hormone (JH). A recent study has revealed that Hairy acts as a negative regulator of the expression of Halloween genes in a JH-dependent manner in D. melanogaster and B. mori (Yang et al., 2021). Curiously, the expression of hairy is suppressed by trimethylation of histone H3 on lysine 27 (H3K27me3), which is catalyzed by Polycomb repressive complex 2 (PRC2) with histone methyltransferase activity. H3K27me3 level dynamically increases during the final larval instar of D. melanogaster and B. mori, at which 20E titer is elevated. Consistent with the correlation between H3K27me3 level and 20E titer, PG-specific knockdown of suppressor of zeste 12 (Su(z)12), encoding a component of PRC2, results in reduction in H3K27me3, diminishing ecdysteroid biosynthesis and disturbs the larval–pupal transition in D. melanogaster (Yang et al., 2021). Mechanistically, H3K27me3 targets hairy to repress its transcription in the PG, leading to an upregulation in the expression of Halloween genes. Moreover, the application of a JH mimic to PG decreases both the levels of H3K27me3 and the expression of Su(z)12. An attenuation in the expression of some Halloween genes has also been observed in B. mori injected with the chemical inhibitor of PRC2 (Yang et al., 2021). Taken together, these data suggest that PRC2-mediated H3K27me3 at the hairy locus in the PG is negatively regulated by JH and required for ecdysteroid biosynthesis during the larval–pupal transition.
The suppressive function of JH on ecdysteroid biosynthesis in the PG is reminiscent of the downregulation of Halloween genes by another JH signal transducer, Krüppel-homolog 1 (Kr-h1) (Zhang et al., 2018). These studies provide valuable insights into JH-mediated ecdysteroid biosynthesis.
A number of other TFs are also involved in ecdysteroid biosynthesis in the PG.
Knirps (Kni) is a well-known gap gene product essential for the segmentation process during early embryogenesis (Jaeger, 2011). Kni was identified through in silico analyses of cis-regulatory elements as a candidate TF that can bind to the enhancer regions of spok, phm, and dib (Danielsen et al., 2014). kni is strongly expressed in the larval PG cells, and PG-specific knockdown of kni results in a larval-arrest phenotype.
CCCTC-binding factor (CTCF), a highly conserved insulator protein, has also been reported to regulate ecdysteroid biosynthesis (Fresán et al., 2015). PG-specific knockdown of CTCF causes low transcriptional levels of nobo, spok, and sad. Curiously, puparium formation delay caused by PG-specific CTCF knockdown is rescued by administration of both cholesterol and 20E, but not by cholesterol or 20E alone. Therefore, CTCF might play a role not only in ecdysteroid biosynthesis, but also in cholesterol homeostasis in the PG.
Without children (woc) is a C2H2-type zinc finger protein. Homozygous mutations of woc results in larval lethality with ecdysteroid deficiency (Wismar et al., 2000). Because administration of 7-dehydrocholesterol partially rescues this mutant (Warren et al., 2001), it has been hypothesized that Woc activates the transcription of a gene involved in 7,8-dehydrogenation of cholesterol to produce 7dC. However, it is unlikely that Woc regulates the expression of nvd, encoding the cholesterol 7,8-dehydrogenase, in PG (Yoshiyama et al., 2006). The actual downstream target of Woc is currently unknown.
HLH54F is a basic helix-loop-helix TF gene that is predominantly expressed in the PG of both D. melanogaster and B. mori (Namiki et al., 2009). However, no validated targets of HLH54F in PG have been reported.
The chromatin remodeling factor lysine demethylase 5 (KDM5) is also known to be involved in ecdysteroid biosynthesis (Drelon et al., 2019). KDM5 is a trimethylated histone H3 on lysine 4 (H3K4me3) demethylase. A loss–of-function KDM5 genetic mutant exhibits severe pupariation delay (Drelon et al., 2019). Consistent with this phenotype, the 20E titer and the level of expression of Halloween genes are decreased in this mutant. Unexpectedly, although KDM5 is believed to participate in the H3K4me3 demethylation process in almost all cells, the PG-specific forced expression of KDM5 rescued the pupariation delay and decreased ecdysteroid titer in the KDM5 genetic mutant. In addition, the pupariation delay of KDM5 genetic mutants is also partially rescued by the PG-specific forced expression of the PTTH receptor gene torso or cycE. These data indicate that the KDM5-mediated demethylation of H3K4me3 is required for proper gene expression in PG cells, at least sustaining PTTH signaling and endoreplication (Drelon et al., 2019).
Recent RNA-seq analyses have reported the predominant expression of other TFs in the ring gland, an endocrine tissue complex containing PG in higher dipteran species; however, their functions in the PG have not been elucidated (Ou et al., 2016; Christesen et al., 2017; Nakaoka et al., 2017; Moulos et al., 2018; Bian et al., 2019). These uncharacterized factors include CG6163 (MADF-BESS domain), CG33557 (bHLH), Escargot (C2H2 zinc finger), Hand (bHLH), Mesoderm-expressed 2 (MADF-BESS domain), Pou domain motif 3 (Pdm3) (POU-Hox), and Silk gland factor 3 (POU-Hox). It would be intriguing to examine whether any of these TFs are produced in the PG and influence the expression of Halloween genes.
Some studies have revealed that posttranscriptional regulation, such as splicing and polyadenylated [poly(A)]-tail modification, are also important in modulating the expression of Halloween genes.
The role of splicing in the expression of Halloween genes has mostly been elucidated in a study on spok and ecdysoneless (ecd) in D. melanogaster. Ecd was originally discovered as a genetic mutant resulting in ecdysteroid deficiency (Garen et al., 1977; Gaziova et al., 2004). It encodes an evolutionarily conserved protein that acts as a member of the spliceosome complex. As Ecd seems to regulate only a part of the splicing process, its dysfunction was reported to impact the splicing of spok but not that of phm pre-mRNA among Halloween genes in the PG (Claudius et al., 2014). Notably, the larval-arrest phenotype of ecd loss-of-function mutants is rescued by overexpression of the human ortholog of ecd (Claudius et al., 2014), suggesting that the function of Ecd as a regulator of splicing is conserved in humans as well.
Poly(A)-tail modifiers are also involved in the regulation of the expression of spok. Loss-of-function of a key subunit of the CCR4-NOT complex Pop2 and poly(A)-binding protein (Pabp) decreases in the expression of spok (Zeng et al., 2018; Kamiyama et al., 2020). CCR4-NOT is an mRNA deadenylation complex. Generally, it is thought that a longer poly(A)-tail contributes to mRNA stability. However, loss of the function of Pop2 in larvae leads to a larval-arrested phenotype, extended poly(A)-tail of spok mRNA, and decreased levels of spok mRNA (Zeng et al., 2018). Therefore, the CCR4-NOT complex might regulate the transcription of spok directly in the PG. Loss-of-function of Pabp has also been reported to lead to a decrease in the expression of spok (Kamiyama et al., 2020). In PG-specific pabp-knockdown animals, Mld, which is indispensable for the expression of spok, is abnormally accumulated in the cytoplasm of the PG. Such abnormal accumulation is not observed in the case of Vvl and Pdm3, which are also expressed in the PG. These data imply that Pabp contributes specifically to the nuclear localization of Mld, while it is unclear whether it regulates the subcellular localization of Mld directly or indirectly. Of note, PG-specific knockdown of the nuclear poly(A) polymerase gene hiiragi also causes developmental arrest in the larval stage (Danielsen et al., 2016). Taken together, these data suggest that poly(A)-related proteins are specifically involved in the regulation of the expression of Halloween genes.
In the last 16 years, since βFtz-F1 was identified as an ecdysteroidogenic TF in the PG in 2005 (Parvy et al., 2005), many ecdysteroidogenic TFs (Table 2) have been identified and characterized, particularly in D. melanogaster. However, there are still many important issues to be solved for a better understanding of the transcriptional regulation of ecdysteroid biosynthesis. Below, we list some remaining questions that must be addressed in the future.
To date, 20 types of TFs (Table 2) have been identified and characterized as ecdysteroidogenic TFs. However, it remains largely unclear whether and how these TFs interact or coordinate with each other to regulate the expression of Halloween genes. For example, in D. melanogaster, the expression of phm in the PG is transcriptionally influenced by at least 14 TFs, including Br-C, CncC, DHR3, Keap1, E75, EcR, FoxO, βFtz-F1, Hairy, Kni, Kr-h1, Sna, Usp, and Vvl (Table 2). The involvement of multiple TFs is also the case regarding other Halloween genes (Table 2). Currently, we do not understand why multiple TFs are required for the expression of a single Halloween gene. Does each ecdysteroidogenic TF differentially regulate Halloween genes in a context-dependent manner, such as nutritional availability? Does each ecdysteroidogenic TF cooperate with other TFs to induce the expression of Halloween genes? If so, do any of these ecdysteroidogenic TFs physically interact with each other? These questions must be answered for a comprehensive understanding of the role of TFs in the transcriptional regulation of Halloween genes. Notably, a recent study successfully conducted immunoprecipitation followed by mass spectrometry to identify physically bound proteins to a protein of interest from the PG samples (Huynh et al., 2019). This technology might be applicable for revealing the TF-mediated network to regulate Halloween genes.
It is also intriguing to examine which TF or combination of TFs is sufficient for the induction of the expression of Halloween genes. In mammals, Ad4BP/SF-1 is both necessary and sufficient for the induction and maintenance of the expression of steroidogenic genes. For example, overexpression of Ad4BP/SF-1 induces the differentiation of cultured stem cells into steroidogenic cells (Miyamoto et al., 2011). Moreover, transgenic expression of Ad4BP/SF-1 in mice leads to ectopic adrenal formation (Zubair et al., 2009). Among ecdysteroidogenic TFs, simultaneous forced expression of mld and ouib induces the expression of spok in D. melanogaster S2 cultured cells (Uryu et al., 2018). However, no studies have examined whether any other ecdysteroidogenic TFs, including βFtz-F1, are sufficient for inducing the expression of Halloween genes in non-steroidogenic cells in insects.
Ecdysteroid biosynthesis in the PG is tightly regulated by endocrine and autocrine humoral factors. Endocrine factors include PTTH (Kawakami et al., 1990; McBrayer et al., 2007), Drosophila insulin-like peptides (Dilps) (Colombani et al., 2005), serotonin (Shimada-Niwa and Niwa, 2014), Hedgehog (Rodenfels et al., 2014), Decapentaplegic (Setiawan et al., 2018), PDGF- and VEGF-related factors 2 and 3, and Jelly belly (Pan and O’Connor, 2021). Autocrine factors include octopamine (Ohhara et al., 2015) and epidermal growth factor (Cruz et al., 2020). While receptors for activin (Gibbens et al., 2011) and neuropeptide F (Kannangara et al., 2020) are also involved in ecdysteroid biosynthesis and the expression of Halloween genes in the PG, the source of these ligands is unclear. Most of these humoral factors affect the levels of expression of Halloween genes (Niwa and Niwa, 2016b; Pan et al., 2020; Texada et al., 2020; Kannangara et al., 2021). Therefore, the important issue in drawing a signaling network for controlling ecdysteroid biosynthesis is to understand which ecdysteroidogenic TFs act downstream of the extracellular stimuli-triggered signaling pathway. However, the mechanism by which these receptors regulate the transcription of Halloween genes remains largely unclear.
To date, DHR4 is the only good example of an ecdysteroidogenic TF acting downstream of PTTH signaling (Ou et al., 2011). A crucial function of DHR4 is to negatively regulate the expression of Cyp6t3 when PTTH signaling is suppressed in the PG. However, DHR4 does not seem to be extensively involved in regulating the expression of any other Halloween gene (Ou et al., 2011), indicating that PTTH signaling must target other ecdysteroidogenic TFs. The PTTH receptor Torso is known to activate the Ras-ERK signaling pathway (Rewitz et al., 2009b). Therefore, it is likely that ERK might phosphorylate some ecdysteroidogenic TFs in the PG to regulate their transcriptional activity; however, a proteomic approach for the identification of phosphorylated proteins in the PG failed to detect the phosphorylation of identified ecdysteroidogenic TFs (Rewitz et al., 2009a). Alternatively, the Ras-ERK signaling pathway might affect epigenetic regulation in the PG, as ERK signaling is known to regulate gene expression by activating histone modification enzymes in mammals (Vicent et al., 2010).
While Sna is an indispensable TF that regulates endoreplication and the expression of Halloween genes (Zeng et al., 2020), it remains unclear the mechanism by which Sna is activated at a proper time point to evoke endoreplication in the PG. It has been suggested that the Warts-Yorkie signaling, IIS, and Ras-ERK signaling are regulators of endoreplication in D. melanogaster (Costa et al., 2021). These signaling pathways are active and essential for ecdysteroid biosynthesis in the PG (Colombani et al., 2005; Rewitz et al., 2009b; Moeller et al., 2017). Although TOR-dependent endoreplication in the PG requires Sna (Zeng et al., 2020), the roles of Warts-Yorkie and Ras-ERK have not been examined. Interestingly, an interaction has been suggested between chromatin remodeling by KDM5 and endoreplication (Drelon et al., 2019). Potentially, the progress of endoreplication in the PG might depend on the developmental timing-specific chromatin status during larval development.
The means by which the expression of Halloween genes is induced in PG cells with high ploidy is also unclear. A recent study has reported the use of a genetic screen for the identification of essential genes involved in ecdysteroid biosynthesis, but not the endoreplication process in the PG (Ohhara et al., 2019). The TFs and epigenetic regulators identified in this screen might help to understand the molecular mechanism of the expression of Halloween genes downstream of the endoreplication event.
Chromatin status might be temporally changed during larval development in the PG, as the levels of H3K27me3 are gradually increased during the third larval instar (Yang et al., 2021). In fact, a large-scale RNAi screen was performed to knock down genes specifically in the PG and identified only the Su(z)12 gene among 31 genes involved in methylation, demethylation, acetylation, deacetylation, and phosphorylation of histones (Danielsen et al., 2016). Although oversights might occur in such large-scale screens, the study implies that Su(z)12 might be a key factor in regulating the temporal dynamics of histone modification in the PG. Recent advances in the Assay for Transposase-Accessible Chromatin with RNA-seq, known as ATAC-seq (Cusanovich et al., 2018), and chromatin immunoprecipitation-RNA-seq technology using samples with a low number of cells (Akhtar et al., 2019) will allow us to examine a genome-wide chromatin modification in the PG in the future.
To date, studies on ecdysteroidogenic TFs have focused on their functions in PG cells. However, ecdysteroid biosynthesis also occurs in other types of cells during embryonic and adult stages.
Studies on D. melanogaster and B. mori have demonstrated that ecdysteroids are essential for cell differentiation and morphogenesis during embryogenesis (Kozlova and Thummel, 2003; Wang et al., 2018; Fujinaga et al., 2020; Gu et al., 2021; Taira et al., 2021; Yoo et al., 2021). In D. melanogaster embryos, ecdysteroids are actively biosynthesized during mid-embryogenesis before the formation of PG primordial cells (Maróy et al., 1988). At this stage, ecdysteroid biosynthesis occurs in the epidermal and amnioserosa cells, in which Halloween genes are strongly expressed (Chávez et al., 2000; Petryk et al., 2003; Niwa et al., 2004, 2010; Warren et al., 2004; Namiki et al., 2005; Ono et al., 2006; Enya et al., 2014). Accordingly, the temporal fluctuation in the expression of Halloween genes during embryogenesis has been shown to correlate with that of the embryonic ecdysteroid titer (Niwa et al., 2010; Enya et al., 2014).
In addition to PG, ovaries are known sites of ecdysteroid biosynthesis. Ovarian ecdysteroids are required for many aspects of oogenesis, such as the regulation of germline stem cell numbers, oocyte maturation, and follicle rupture (Sieber and Spradling, 2015; Uryu et al., 2015; Ables and Drummond-Barbosa, 2017; Knapp and Sun, 2017; Yoshinari et al., 2019; Hoshino and Niwa, 2021) as well as for ovary-gut, and ovary-to-neuron communication (Ahmed et al., 2020; Hadjieconomou et al., 2020; Zipper et al., 2020). Indeed, Halloween genes are expressed in ovarian follicle or nurse cells (Chávez et al., 2000; Petryk et al., 2003; Niwa et al., 2004, 2010; Warren et al., 2004; Namiki et al., 2005; Ono et al., 2006; Domanitskaya et al., 2014; Enya et al., 2014; Ameku and Niwa, 2016; Ameku et al., 2017). However, it is unclear which TFs regulate the ovarian expression of Halloween genes, except for βFtz-f1, which appears to regulate the expression of dib in follicle cells (Talamillo et al., 2013).
As shown in Table 2, most studies on ecdysteroidogenic TFs have been conducted using D. melanogaster. Currently, Vvl and FoxO are the exception of an ecdysteroidogenic TF shown to act in other insects. In particular, a knockdown of vvl results in defects in ecdysteroid biosynthesis in the T. castaneum and O. fasciatus coleopteran insects (Cheng et al., 2014; Sarwar et al., 2020). However, besides Vvl, the involvement of other ecdysteroidogenic TFs identified in D. melanogaster in the regulation of ecdysteroid biosynthesis has not been explored in other insects. In fact, a comparative gene expression study has reported that, among 13 TFs identified as specifically expressed in the D. melanogaster ring glands (Ou et al., 2016), only 2 of their orthologs, tim and vvl, are expressed in B. mori PGs (Moulos et al., 2018). More strikingly, mld, séan, and ouib are only found in Drosophilidae species but not in any other insect species (Ono et al., 2006; Komura-Kawa et al., 2015; Uryu et al., 2018). This situation is quite paradoxical, especially for mld and séan, because nvd, the Halloween gene that is transcriptionally induced by the cooperation of Mld and Séan, is widely conserved in most insect species (Yoshiyama et al., 2006; Yoshiyama-Yanagawa et al., 2011; Schumann et al., 2018). These data imply that, during evolution, each phylogenetic class of insects might have acquired or lost essential ecdysteroidogenic TFs that are not conserved in D. melanogaster. Therefore, future studies using non-D. melanogaster species are extremely important. To search for ecdysteroidogenic TFs in species other than D. melanogaster, a large-scale, unbiased RNAi screen system in T. castaneum might be useful (Schmitt-Engel et al., 2015; Schultheis et al., 2019). Future challenges with a wide range of insect species will allow us to understand the common and diverse mechanisms of transcriptional regulation of ecdysteroid biosynthesis.
TK and RN wrote, revised the manuscript, and approved the submitted version. Both authors contributed to the article and approved the submitted version.
This work was supported by a grant from JSPS KAKENHI 21J10894 to TK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
TK was a recipient of a fellowship from the Japan Society for the Promotion of Science. We thank Yuya Ohhara for carefully reading and providing critical comments on this manuscript. We would also like to thank Editage (www.editage.com) for English language editing.
Ables, E. T., and Drummond-Barbosa, D. (2017). Steroid hormones and the physiological regulation of tissue-resident stem cells: lessons from the Drosophila ovary. Curr. Stem Cell Rep. 3, 9–18. doi: 10.1007/S40778-017-0070-Z
Ahmed, S. M. H., Maldera, J. A., Krunic, D., Paiva-Silva, G. O., Pénalva, C., Teleman, A. A., et al. (2020). Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature 584, 415–419. doi: 10.1038/s41586-020-2462-y
Akhtar, J., More, P., Albrecht, S., Marini, F., Kaiser, W., Kulkarni, A., et al. (2019). TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay. Life Sci. Alliance 2:e201900318. doi: 10.26508/LSA.201900318
Ameku, T., and Niwa, R. (2016). Mating-induced increase in germline stem cells via the neuroendocrine system in female Drosophila. PLoS Genet. 12:e1006123. doi: 10.1371/journal.pgen.1006123
Ameku, T., Yoshinari, Y., Fukuda, R., and Niwa, R. (2017). Ovarian ecdysteroid biosynthesis and female germline stem cells. Fly 11, 185–193. doi: 10.1080/19336934.2017.1291472
Belles, X. (2020). Insect Metamorphosis: From Natural History to Regulation of Development and Evolution. 1st ed. Cambridge: Academic Press.
Bialecki, M., Shilton, A., Fichtenberg, C., Segraves, W. A., and Thummel, C. S. (2002). Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell 3, 209–220. doi: 10.1016/s1534-5807(02)00204-6
Bian, H. X., Chen, D. B., Zheng, X. X., Ma, H. F., Li, Y. P., Li, Q., et al. (2019). Transcriptomic analysis of the prothoracic gland from two lepidopteran insects, domesticated silkmoth Bombyx mori and wild silkmoth Antheraea pernyi. Sci. Rep. 9:5313. doi: 10.1038/s41598-019-41864-0
Bonaglia, M. C., Ciccone, R., Gimelli, G., Gimelli, S., Marelli, S., Verheij, J., et al. (2008). Detailed phenotype–genotype study in five patients with chromosome 6q16 deletion: narrowing the critical region for Prader–Willi-like phenotype. Eur. J. Hum. Genet. 16, 1443–1449. doi: 10.1038/ejhg.2008.119
Cáceres, L., Necakov, A. S., Schwartz, C., Kimber, S., Roberts, I. J. H., and Krause, H. M. (2011). Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 25, 1476–1485. doi: 10.1101/gad.2064111
Callier, V., and Nijhout, H. F. (2011). Control of body size by oxygen supply reveals size-dependent and size-independent mechanisms of molting and metamorphosis. Proc. Natl. Acad. Sci. U. S. A. 108, 14664–14669. doi: 10.1073/PNAS.1106556108
Calnan, D. R., and Brunet, A. (2008). The FoxO code. Oncogene 27, 2276–2288. doi: 10.1038/onc.2008.21
Carlson, J., Price, L., Cook, I., and Deng, H. (2022). Drosophila Keap1 xenobiotic response factor regulates developmental transcription through binding to chromatin. Dev. Biol. 481, 139–147. doi: 10.1016/J.YDBIO.2021.10.003
Chanut-Delalande, H., Hashimoto, Y., Pelissier-Monier, A., Spokony, R., Dib, A., Kondo, T., et al. (2014). Pri peptides are mediators of ecdysone for the temporal control of development. Nat. Cell Biol. 16, 1035–1044. doi: 10.1038/ncb3052
Chávez, V. M., Marqués, G., Delbecque, J. P., Kobayashi, K., Hollingsworth, M., Burr, J., et al. (2000). The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127, 4115–4126.
Cheng, C., Ko, A., Chaieb, L., Koyama, T., Sarwar, P., Mirth, C. K., et al. (2014). The POU factor ventral veins lacking/drifter directs the timing of metamorphosis through ecdysteroid and juvenile hormone signaling. PLoS Genet. 10:e1004425. doi: 10.1371/JOURNAL.PGEN.1004425
Christesen, D., Yang, Y. T., Somers, J., Robin, C., Sztal, T., Batterham, P., et al. (2017). Transcriptome analysis of Drosophila melanogaster third instar larval ring glands points to novel functions and uncovers a cytochrome p450 required for development. G3 (Bethesda) 3, 467–479. doi: 10.1534/G3.116.037333
Claudius, A.-K., Romani, P., Lamkemeyer, T., Jindra, M., and Uhlirova, M. (2014). Unexpected role of the steroid-deficiency protein ecdysoneless in Pre-mRNA splicing. PLoS Genet. 10:e1004287. doi: 10.1371/journal.pgen.1004287
Colombani, J., Bianchini, L., Layalle, S., Pondeville, E., Dauphin-Villemant, C., Antoniewski, C., et al. (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310, 667–670. doi: 10.1126/SCIENCE.1119432
Costa, C. A. M., Wang, X.-F., Ellsworth, C., and Deng, W.-M. (2021). Polyploidy in development and tumor models in Drosophila. Semin. Cancer Biol. doi: 10.1016/J.SEMCANCER.2021.09.011 [Epub Online ahead of Print].
Cruz, J., Martín, D., and Franch-Marro, X. (2020). Egfr signaling is a major regulator of ecdysone biosynthesis in the Drosophila prothoracic gland. Curr. Biol. 30, 1547–1554.e4. doi: 10.1016/j.cub.2020.01.092
Cusanovich, D. A., Reddington, J. P., Garfield, D. A., Daza, R. M., Aghamirzaie, D., Marco-Ferreres, R., et al. (2018). The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542. doi: 10.1038/nature25981
Daimon, T., Koyama, T., Yamamoto, G., Sezutsu, H., Mirth, C. K., and Shinoda, T. (2020). The number of larval molts is controlled by hox in caterpillars. Curr. Biol. 31, 884–891.e3. doi: 10.1016/j.cub.2020.11.017
Danielsen, E., Moeller, M. E., Yamanaka, N., King-Jones, K., O’connor, M. B., Rewitz Correspondence, K. F., et al. (2016). A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Dev. Cell 37, 558–570. doi: 10.1016/j.devcel.2016.05.015
Danielsen, E. T., Moeller, M. E., Dorry, E., Komura-Kawa, T., Fujimoto, Y., Troelsen, J. T., et al. (2014). Transcriptional control of steroid biosynthesis genes in the Drosophila prothoracic gland by ventral veins lacking and knirps. PLoS Genet. 10:e1004343. doi: 10.1371/journal.pgen.1004343
Deng, H., and Kerppola, T. K. (2013). Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS Genet. 9:e1003263. doi: 10.1371/journal.pgen.1003263
Di Cara, F., and King-Jones, K. (2016). The circadian clock is a key driver of steroid hormone production in Drosophila. Curr. Biol. 26, 2469–2477. doi: 10.1016/J.CUB.2016.07.004
Dimitri, P., Caizzi, R., Giordano, E., Carmela Accardo, M., Lattanzi, G., and Biamonti, G. (2009). Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma 118, 419–435. doi: 10.1007/S00412-009-0211-Y/FIGURES/4
Dobi, K. C., Halfon, M. S., and Baylies, M. K. (2014). Whole-genome analysis of muscle founder cells implicates the chromatin regulator Sin3A in muscle identity. Cell Rep. 8, 858–870. doi: 10.1016/J.CELREP.2014.07.005
Domanitskaya, E., Anllo, L., and Schüpbach, T. (2014). Phantom, a cytochrome P450 enzyme essential for ecdysone biosynthesis, plays a critical role in the control of border cell migration in Drosophila. Dev. Biol. 386, 408–418. doi: 10.1016/J.YDBIO.2013.12.013
Drelon, C., Rogers, M. F., Belalcazar, H. M., and Secombe, J. (2019). The histone demethylase KDM5 controls developmental timing in Drosophila by promoting prothoracic gland endocycles. Development 146:dev182568. doi: 10.1242/dev.182568
Dubowy, C., and Sehgal, A. (2017). Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205, 1373–1397. doi: 10.1534/GENETICS.115.185157
Enya, S., Ameku, T., Igarashi, F., Iga, M., Kataoka, H., Shinoda, T., et al. (2014). A Halloween gene noppera-bo encodes a glutathione S-transferase essential for ecdysteroid biosynthesis via regulating the behaviour of cholesterol in Drosophila. Sci. Rep. 4:6586. doi: 10.1038/srep06586
Enya, S., Daimon, T., Igarashi, F., Kataoka, H., Uchibori, M., Sezutsu, H., et al. (2015). The silkworm glutathione S-transferase gene noppera-bo is required for ecdysteroid biosynthesis and larval development. Insect Biochem. Mol. Biol. 61, 1–7. doi: 10.1016/J.IBMB.2015.04.001
Enya, S., Yamamoto, C., Mizuno, H., Esaki, T., Lin, H.-K., Iga, M., et al. (2017). Dual roles of glutathione in ecdysone biosynthesis and antioxidant function during the larval development in Drosophila. Genetics 207, 1519–1532. doi: 10.1534/genetics.117.300391
Fitzpatrick, K. A., Sinclair, D. A., Schulze, S. R., Syrzycka, M., and Honda, B. M. (2011). A genetic and molecular profile of third chromosome centric heterochromatin in Drosophila melanogaster. Genome 48, 571–584. doi: 10.1139/G05-025
Fraenkel, G., and Blewett, M. (1943a). The basic food requirements of several insects. J. Exp. Biol. 20, 28–34. doi: 10.1242/JEB.20.1.28
Fraenkel, G., and Blewett, M. (1943b). The sterol requirements of several insects. Biochem. J. 37, 692–695. doi: 10.1042/BJ0370692
Fresán, U., Cuartero, S., O’Connor, M. B., and Espinàs, M. L. (2015). The insulator protein CTCF regulates Drosophila steroidogenesis. Biol. Open 4, 852–857. doi: 10.1242/bio.012344
Fujikawa, Y., Morisaki, F., Ogura, A., Morohashi, K., Enya, S., Niwa, R., et al. (2015). A practical fluorogenic substrate for high-throughput screening of glutathione S-transferase inhibitors. Chem. Commun. 51, 11459–11462. doi: 10.1039/C5CC02067K
Fujinaga, D., Gu, J., Kawahara, H., Ogihara, M. H., Kojima, I., Takeshima, M., et al. (2020). Twenty-hydroxyecdysone produced by dephosphorylation and ecdysteroidogenesis regulates early embryonic development in the silkmoth, Bombyx mori. Insect Biochem. Mol. Biol. 127:103491. doi: 10.1016/J.IBMB.2020.103491
Garen, A., Kauvar, L., and Lepesant, J. A. (1977). Roles of ecdysone in Drosophila development. Proc. Natl. Acad. Sci. U. S. A. 74, 5099–5103. doi: 10.1073/PNAS.74.11.5099
Gaziova, I., Bonnette, P. C., Henrich, V. C., and Jindra, M. (2004). Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development 131, 2715–2725. doi: 10.1242/DEV.01143
Gibbens, Y. Y., Warren, J. T., Gilbert, L. I., and O’Connor, M. B. (2011). Neuroendocrine regulation of Drosophila metamorphosis requires TGFβ/Activin signaling. Development 138, 2693–2703. doi: 10.1242/DEV.063412
Gilbert, L. I. (2004). Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol. Cell. Endocrinol. 215, 1–10. doi: 10.1016/J.MCE.2003.11.003
Gilbert, L. I., Rybczynski, R., and Warren, J. T. (2002). Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 47, 883–916. doi: 10.1146/ANNUREV.ENTO.47.091201.145302
Gu, S. H., Chen, C. H., and Lin, P. L. (2021). Changes in expressions of ecdysteroidogenic enzyme and ecdysteroid signaling genes in relation to Bombyx embryonic development. J. Exp. Zool. Part A Ecol. Integr. Physiol. 335, 477–488. doi: 10.1002/JEZ.2466
Hadjieconomou, D., King, G., Gaspar, P., Mineo, A., Blackie, L., Ameku, T., et al. (2020). Enteric neurons increase maternal food intake during reproduction. Nature 587, 455–459. doi: 10.1038/s41586-020-2866-8
Hoshino, R., and Niwa, R. (2021). Regulation of mating-induced increase in female germline stem cells in the fruit fly Drosophila melanogaster. Front. Physiol. 12:785435. doi: 10.3389/fphys.2021.785435
Huynh, N., Ou, Q., Cox, P., Lill, R., and King-Jones, K. (2019). Glycogen branching enzyme controls cellular iron homeostasis via Iron Regulatory Protein 1 and mitoNEET. Nat. Commun. 10:5463. doi: 10.1038/s41467-019-13237-8
Ishimoto, H., and Kitamoto, T. (2011). Beyond molting-roles of the steroid molting hormone ecdysone in regulation of memory and sleep in adult Drosophila. Fly 5, 215–220. doi: 10.4161/fly.5.3.15477
Ishimoto, H., Wang, Z., Rao, Y., Wu, C. F., and Kitamoto, T. (2013). A novel role for ecdysone in Drosophila conditioned behavior: linking GPCR-mediated non-canonical steroid action to cAMP signaling in the adult brain. PLoS Genet. 9:e1003843. doi: 10.1371/JOURNAL.PGEN.1003843
Izumi, K., Housam, R., Kapadia, C., Stallings, V. A., Medne, L., Shaikh, T. H., et al. (2013). Endocrine phenotype of 6q16.1–q21 deletion involving SIM1 and Prader–Willi syndrome-like features. Am. J. Med. Genet. Part A 161, 3137–3143. doi: 10.1002/AJMG.A.36149
Jaeger, J. (2011). The gap gene network. Cell. Mol. Life Sci. 68, 243–274. doi: 10.1007/S00018-010-0536-Y/FIGURES/9
Jauch, R., Bourenkov, G. P., Chung, H. R., Urlaub, H., Reidt, U., Jäckle, H., et al. (2003). The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure 11, 1393–1402. doi: 10.1016/J.STR.2003.09.015
Kamiyama, T., and Niwa, R. (2021a). “20-Hydroxyecdysone,” in Handbook of Hormones, eds H. Ando, K. Ukena, and S. Nagata (Cambridge: Academic Press), 983–986.
Kamiyama, T., and Niwa, R. (2021b). “Ecdysteroid,” in Handbook of Hormones, eds H. Ando, K. Ukena, and S. Nagata (Cambridge: Academic Press), 981–982.
Kamiyama, T., Sun, W., Tani, N., Nakamura, A., and Niwa, R. (2020). Poly(A) binding protein is required for nuclear localization of the ecdysteroidogenic transcription factor molting defective in the prothoracic gland of Drosophila melanogaster. Front. Genet. 11:636. doi: 10.3389/fgene.2020.00636
Kannangara, J. R., Henstridge, M. A., Parsons, L. M., Kondo, S., Mirth, C. K., and Warr, C. G. (2020). A new role for neuropeptide F signaling in controlling developmental timing and body size in Drosophila melanogaster. Genetics 216, 135–144. doi: 10.1534/GENETICS.120.303475
Kannangara, J. R., Mirth, C. K., and Warr, C. G. (2021). Regulation of ecdysone production in Drosophila by neuropeptides and peptide hormones. Open Biol. 11:200373. doi: 10.1098/RSOB.200373
Kawakami, A., Kataoka, H., Oka, T., Mizoguchi, A., Kimura-Kawakami, M., Adachi, T., et al. (1990). Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science 247, 1333–1335. doi: 10.1126/SCIENCE.2315701
King-Jones, K., Charles, J. P., Lam, G., and Thummel, C. S. (2005). The ecdysone-induced dhr4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell 121, 773–784. doi: 10.1016/J.CELL.2005.03.030
Knapp, E., and Sun, J. (2017). Steroid signaling in mature follicles is important for Drosophila ovulation. Proc. Natl. Acad. Sci. U. S. A. 114, 699–704. doi: 10.1073/PNAS.1614383114
Koiwai, K., Inaba, K., Morohashi, K., Enya, S., Arai, R., Kojima, H., et al. (2020). An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo. J. Biol. Chem. 295, 7154–7167. doi: 10.1074/jbc.RA119.011463
Koiwai, K., Morohashi, K., Inaba, K., Ebihara, K., Kojima, H., Okabe, T., et al. (2021). Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S-transferase Noppera-bo. J. Pestic. Sci. 46, 75–87. doi: 10.1584/jpestics.D20-072
Komura-Kawa, T., Hirota, K., Shimada-Niwa, Y., Yamauchi, R., Shimell, M. J., Shinoda, T., et al. (2015). The Drosophila Zinc finger transcription factor Ouija board controls ecdysteroid biosynthesis through specific regulation of spookier. PLoS Genet. 11:e1005712. doi: 10.1371/journal.pgen.1005712
Koyama, T., Rodrigues, M. A., Athanasiadis, A., Shingleton, A. W., and Mirth, C. K. (2014). Nutritional control of body size through FoxO-Ultraspiracle mediated ecdysone biosynthesis. Elife 3:e03091. doi: 10.7554/eLife.03091
Kozlova, T., and Thummel, C. S. (2003). Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science 301, 1911–1914. doi: 10.1126/SCIENCE.1087419
Kurogi, Y., Mizuno, Y., Imura, E., and Niwa, R. (2021). Neuroendocrine regulation of reproductive dormancy in the fruit fly Drosophila melanogaster: a review of juvenile hormone-dependent regulation. Front. Ecol. Evol. 9:715029. doi: 10.3389/FEVO.2021.715029
Lang, M., Murat, S., Clark, A., Gouppil, G., Blais, C., Matzkin, L. M., et al. (2012). Mutations in the neverland gene turned Drosophila pachea into an obligate specialist species. Science 337, 1658–1661. doi: 10.1126/science.1224829
Lin, X., Yu, N., and Smagghe, G. (2018). FoxO mediates the timing of pupation through regulating ecdysteroid biosynthesis in the red flour beetle, Tribolium castaneum. Gen. Comp. Endocrinol. 258, 149–156. doi: 10.1016/J.YGCEN.2017.05.012
Mallo, M., and Alonso, C. R. (2013). The regulation of Hox gene expression during animal development. Development 140, 3951–3963. doi: 10.1242/DEV.068346
Markov, G. V., Tavares, R., Dauphin-Villemant, C., Demeneix, B. A., Baker, M. E., and Laudet, V. (2009). Independent elaboration of steroid hormone signaling pathways in metazoans. Proc. Natl. Acad. Sci. U. S. A. 106, 11913–11918. doi: 10.1073/PNAS.0812138106
Maróy, P., Kaufmann, G., and Dübendorfer, A. (1988). Embryonic ecdysteroids of Drosophila melanogaster. J. Insect Physiol. 34, 633–637. doi: 10.1016/0022-1910(88)90071-6
McBrayer, Z., Ono, H., Shimell, M. J., Parvy, J. P., Beckstead, R. B., Warren, J. T., et al. (2007). Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857–871. doi: 10.1016/J.DEVCEL.2007.11.003
Meng, M., Cheng, D. J., Peng, J., Qian, W. L., Li, J. R., Dai, D. D., et al. (2015). The Homeodomain transcription factors antennapedia and POU-M2 regulate the transcription of the steroidogenic enzyme gene phantom in the silkworm. J. Biol. Chem. 290, 24438–24452. doi: 10.1074/JBC.M115.651810
Mirth, C. K., and Shingleton, A. W. (2012). Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front. Endocrinol. 3:49. doi: 10.3389/FENDO.2012.00049
Miyamoto, K., Yazawa, T., Mizutani, T., Imamichi, Y., Kawabe, S., Kanno, M., et al. (2011). Stem cell differentiation into steroidogenic cell lineages by NR5A family. Mol. Cell. Endocrinol. 336, 123–126. doi: 10.1016/J.MCE.2010.11.031
Mizoguchi, A., and Ishizaki, H. (1982). Prothoracic glands of the saturniid moth Samia cynthia ricini possess a circadian clock controlling gut purge timing. Proc. Natl. Acad. Sci. U. S. A. 79, 2726–2730. doi: 10.1073/PNAS.79.8.2726
Mizutani, T., Ishikane, S., Kawabe, S., Umezawa, A., and Miyamoto, K. (2015). Transcriptional regulation of genes related to progesterone production. Endocr. J. 62, 757–763. doi: 10.1507/ENDOCRJ.EJ15-0260
Moeller, M. E., Danielsen, E. T., Herder, R., O’Connor, M. B., and Rewitz, K. F. (2013). Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development 140, 4730–4739. doi: 10.1242/dev.099739
Moeller, M. E., Nagy, S., Gerlach, S. U., Soegaard, K. C., Danielsen, E. T., Texada, M. J., et al. (2017). Warts signaling controls organ and body growth through regulation of ecdysone. Curr. Biol. 27, 1652–1659.e4. doi: 10.1016/j.cub.2017.04.048
Morioka, E., Matsumoto, A., and Ikeda, M. (2012). Neuronal influence on peripheral circadian oscillators in pupal Drosophila prothoracic glands. Nat. Commun. 3:909. doi: 10.1038/ncomms1922
Morohashi, K., Baba, T., and Tanaka, M. (2013). Steroid hormones and the development of reproductive organs. Sex. Dev. 7, 61–79. doi: 10.1159/000342272
Moulos, P., Alexandratos, A., Nellas, I., and Dedos, S. G. (2018). Refining a steroidogenic model: an analysis of RNA-seq datasets from insect prothoracic glands. BMC Genomics 19:537. doi: 10.1186/S12864-018-4896-2
Myers, E. M., Yu, J., and Sehgal, A. (2003). Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr. Biol. 13, 526–533. doi: 10.1016/S0960-9822(03)00167-2
Myers, M. P., Wager-Smith, K., Wesley, C. S., Young, M. W., and Sehgal, A. (1995). Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270, 805–808. doi: 10.1126/SCIENCE.270.5237.805
Nakaoka, T., Iga, M., Yamada, T., Koujima, I., Takeshima, M., Zhou, X., et al. (2017). Deep sequencing of the prothoracic gland transcriptome reveals new players in insect ecdysteroidogenesis. PLoS One 12:e0172951. doi: 10.1371/JOURNAL.PONE.0172951
Namiki, T., Niwa, R., Higuchi, A., Yoshiyama, T., Mita, K., and Kataoka, H. (2009). A Basic-HLH transcription factor, HLH54F, is highly expressed in the prothoracic gland in the Silkworm Bombyx mori and the fruit fly Drosophila melanogaster. Biosci. Biotechnol. Biochem. 73, 762–765. doi: 10.1271/BBB.80737
Namiki, T., Niwa, R., Sakudoh, T., Shirai, K., Takeuchi, H., and Kataoka, H. (2005). Cytochrome P450 CYP307A1/Spook: a regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 337, 367–374. doi: 10.1016/J.BBRC.2005.09.043
Neubueser, D., Warren, J. T., Gilbert, L. I., and Cohen, S. M. (2005). Molting defective is required for ecdysone biosynthesis. Dev. Biol. 280, 362–372. doi: 10.1016/j.ydbio.2005.01.023
Nieto, M. A. (2002). The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3, 155–166. doi: 10.1038/nrm757
Nijhout, H. F. (1976). The rôle of ecdysone in pupation of Manduca sexta. J. Insect Physiol. 22, 453–463. doi: 10.1016/0022-1910(76)90017-2
Niwa, R., Matsuda, T., Yoshiyama, T., Namiki, T., Mita, K., Fujimoto, Y., et al. (2004). CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 279, 35942–35949. doi: 10.1074/jbc.M404514200
Niwa, R., Namiki, T., Ito, K., Shimada-Niwa, Y., Kiuchi, M., Kawaoka, S., et al. (2010). Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the ‘Black Box’ of the ecdysteroid biosynthesis pathway. Development 137, 1991–1999. doi: 10.1242/dev.045641
Niwa, R., and Niwa, Y. S. (2014a). Enzymes for ecdysteroid biosynthesis: their biological functions in insects and beyond. Biosci. Biotechnol. Biochem. 78, 1283–1292. doi: 10.1080/09168451.2014.942250
Niwa, Y. S., and Niwa, R. (2014b). Neural control of steroid hormone biosynthesis during development in the fruit fly Drosophila melanogaster. Genes Genet. Syst. 89, 27–34. doi: 10.1266/GGS.89.27
Niwa, Y. S., and Niwa, R. (2016b). Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Dev. Growth Differ. 58, 94–105. doi: 10.1111/dgd.12248
Niwa, Y. S., and Niwa, R. (2016a). Ouija board: a transcription factor evolved for only one target in steroid hormone biosynthesis in the fruit fly Drosophila melanogaster. Transcription 7, 196–202. doi: 10.1080/21541264.2016.1210370
Nunes, C., Sucena, É, and Koyama, T. (2021). Endocrine regulation of immunity in insects. FEBS J. 288, 3928–3947. doi: 10.1111/FEBS.15581
Ohhara, Y., Kobayashi, S., and Yamanaka, N. (2017). Nutrient-dependent endocycling in steroidogenic tissue dictates timing of metamorphosis in Drosophila melanogaster. PLoS Genet. 13:e1006583. doi: 10.1371/journal.pgen.1006583
Ohhara, Y., Nakamura, A., Kato, Y., and Yamakawa-Kobayashi, K. (2019). Chaperonin TRiC/CCT supports mitotic exit and entry into endocycle in Drosophila. PLoS Genet. 15:e1008121. doi: 10.1371/journal.pgen.1008121
Ohhara, Y., Shimada-Niwa, Y., Niwa, R., Kayashima, Y., Hayashi, Y., Akagi, K., et al. (2015). Autocrine regulation of ecdysone synthesis by β3-octopamine receptor in the prothoracic gland is essential for Drosophila metamorphosis. Proc. Natl. Acad. Sci. U. S. A. 112, 1452–1457. doi: 10.1073/pnas.1414966112
Ono, H., Rewitz, K. F., Shinoda, T., Itoyama, K., Petryk, A., Rybczynski, R., et al. (2006). Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 298, 555–570. doi: 10.1016/j.ydbio.2006.07.023
Ou, Q., Magico, A., and King-Jones, K. (2011). Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol. 9:e1001160. doi: 10.1371/journal.pbio.1001160
Ou, Q., Zeng, J., Yamanaka, N., Brakken-Thal, C., O’Connor, M. B., and King-Jones, K. (2016). The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep. 16, 247–262. doi: 10.1016/J.CELREP.2016.05.053
Pan, X., Connacher, R. P., and O’Connor, M. B. (2020). Control of the insect metamorphic transition by ecdysteroid production and secretion. Curr. Opin. Insect Sci. 43, 11–20. doi: 10.1016/j.cois.2020.09.004
Pan, X., and O’Connor, M. B. (2021). Coordination among multiple receptor tyrosine kinase signals controls Drosophila developmental timing and body size. Cell Rep. 36:109644. doi: 10.1016/J.CELREP.2021.109644
Parker, K. L., Rice, D. A., Lala, D. S., Ikeda, Y., Luo, X., Wong, M., et al. (2002). Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 57, 19–36. doi: 10.1210/rp.57.1.19
Parvy, J.-P., Blais, C., Bernard, F., Warren, J. T., Petryk, A., Gilbert, L. I., et al. (2005). A role for βFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev. Biol. 282, 84–94. doi: 10.1016/j.ydbio.2005.02.028
Parvy, J.-P., Wang, P., Garrido, D., Maria, A., Blais, C., Poidevin, M., et al. (2014). Forward and feedback regulation of cyclic steroid production in Drosophila melanogaster. Development 141, 3955–3965. doi: 10.1242/dev.102020
Petryk, A., Warren, J. T., Marqués, G., Jarcho, M. P., Gilbert, L. I., Kahler, J., et al. (2003). Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. U. S. A. 100, 13773–13778. doi: 10.1073/PNAS.2336088100
Rewitz, K. F., Yamanaka, N., Gilbert, L. I., and O’Connor, M. B. (2009b). The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326, 1403–1405. doi: 10.1126/SCIENCE.1176450
Rewitz, K. F., Larsen, M. R., Lobner-Olesen, A., Rybczynski, R., O’Connor, M. B., and Gilbert, L. I. (2009a). A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem. Mol. Biol. 39, 475–483. doi: 10.1016/J.IBMB.2009.04.005
Rodenfels, J., Lavrynenko, O., Ayciriex, S., Sampaio, J. L., Carvalho, M., Shevchenko, A., et al. (2014). Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev. 28, 2636–2651. doi: 10.1101/gad.249763.114
Sánchez-Higueras, C., Sotillos, S., and Castelli-Gair Hombría, J. (2014). Common origin of insect trachea and endocrine organs from a segmentally repeated precursor. Curr. Biol. 24, 76–81. doi: 10.1016/J.CUB.2013.11.010
Sarwar, P. F., McDonald, I. R., Wang, V. R., and Suzuki, Y. (2020). The POU factor Ventral veins lacking regulates ecdysone and juvenile hormone biosynthesis during development and reproduction of the milkweed bug, Oncopeltus fasciatus. Dev. Biol. 459, 181–193. doi: 10.1016/j.ydbio.2019.12.001
Schmitt-Engel, C., Schultheis, D., Schwirz, J., Ströhlein, N., Troelenberg, N., Majumdar, U., et al. (2015). The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat. Commun. 6:7822. doi: 10.1038/ncomms8822
Schultheis, D., Weißkopf, M., Schaub, C., Ansari, S., Dao, V. A., Grossmann, D., et al. (2019). A Large Scale Systemic RNAi screen in the red flour beetle Tribolium castaneum identifies novel genes involved in insect muscle development. G3 (Bethesda) 3, 1009–1026. doi: 10.1534/G3.118.200995
Schumann, I., Kenny, N., Hui, J., Hering, L., and Mayer, G. (2018). Halloween genes in panarthropods and the evolution of the early moulting pathway in Ecdysozoa. R. Soc. Open Sci. 5:180888. doi: 10.1098/RSOS.180888
Setiawan, L., Pan, X., Woods, A. L., O’Connor, M. B., and Hariharan, I. K. (2018). The BMP2/4 ortholog Dpp can function as an inter-organ signal that regulates developmental timing. Life Sci. Alliance 1:e201800216. doi: 10.26508/lsa.201800216
Shimada-Niwa, Y., and Niwa, R. (2014). Serotonergic neurons respond to nutrients and regulate the timing of steroid hormone biosynthesis in Drosophila. Nat. Commun. 5:5778. doi: 10.1038/ncomms6778
Sieber, M. H., and Spradling, A. C. (2015). Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr. Biol. 25, 993–1004. doi: 10.1016/J.CUB.2015.02.019
Simon, A. F., Shih, C., Mack, A., and Benzer, S. (2003). Steroid control of longevity in Drosophila melanogaster. Science 299, 1407–1410. doi: 10.1126/SCIENCE.1080539
Suda, N., Shibata, H., Kurihara, I., Ikeda, Y., Kobayashi, S., Yokota, K., et al. (2011). Coactivation of SF-1-Mediated transcription of steroidogenic enzymes by Ubc9 and PIAS1. Endocrinology 152, 2266–2277. doi: 10.1210/EN.2010-1232
Sztal, T., Chung, H., Gramzow, L., Daborn, P. J., Batterham, P., and Robin, C. (2007). Two independent duplications forming the Cyp307a genes in Drosophila. Insect Biochem. Mol. Biol. 37, 1044–1053. doi: 10.1016/J.IBMB.2007.05.017
Taira, Y., Wada, H., Hayashi, S., and Kageyama, Y. (2021). polished rice mediates ecdysone-dependent control of Drosophila embryonic organogenesis. Genes Cells 26, 269–281. doi: 10.1111/GTC.12841
Talamillo, A., Herboso, L., Pirone, L., Pérez, C., González, M., Sánchez, J., et al. (2013). Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila Steroidogenesis. PLoS Genet. 9:e1003473. doi: 10.1371/JOURNAL.PGEN.1003473
Talamillol, A., Sánchez, J., Cantera, R., Pérez, C., Martín, D., Caminero, E., et al. (2008). Smt3 is required for Drosophila melanogaster metamorphosis. Development 135, 1659–1668. doi: 10.1242/DEV.020685
Texada, M. J., Koyama, T., and Rewitz, K. (2020). Regulation of body size and growth control. Genetics 216, 269–313. doi: 10.1534/GENETICS.120.303095
Thummel, C. S. (2002). Ecdysone-regulated puff genes 2000. Insect Biochem. Mol. Biol. 32, 113–120. doi: 10.1016/S0965-1748(01)00112-6
Truman, J. W., and Riddiford, L. M. (1974). Physiology of insect rhythms III. The temporal organization of the endocrine events underlying pupation of the tobacco hornworm. J. Exp. Biol. 60, 371–382. doi: 10.1242/JEB.60.2.371
Ulasov, A. V., Rosenkranz, A. A., Georgiev, G. P., and Sobolev, A. S. (2021). Nrf2/Keap1/ARE signaling: towards specific regulation. Life Sci. 291:120111. doi: 10.1016/J.LFS.2021.120111
Uryu, O., Ameku, T., and Niwa, R. (2015). Recent progress in understanding the role of ecdysteroids in adult insects: germline development and circadian clock in the fruit fly Drosophila melanogaster. Zool. Lett. 1:32. doi: 10.1186/s40851-015-0031-2
Uryu, O., Ou, Q., Komura-Kawa, T., Kamiyama, T., Iga, M., Syrzycka, M., et al. (2018). Cooperative control of ecdysone biosynthesis in Drosophila by transcription factors séance, ouija board, and molting defective. Genetics 208, 605–622. doi: 10.1534/genetics.117.300268
van der Burg, K. R., and Reed, R. D. (2021). Seasonal plasticity: how do butterfly wing pattern traits evolve environmental responsiveness? Curr. Opin. Genet. Dev. 69, 82–87. doi: 10.1016/J.GDE.2021.02.009
Vicent, G. P., Nacht, A. S., Zaurín, R., Ballaré, C., Clausell, J., and Beato, M. (2010). Minireview: role of kinases and chromatin remodeling in progesterone signaling to chromatin. Mol. Endocrinol. 24, 2088–2098. doi: 10.1210/ME.2010-0027
Walker, V. K., Watson, K. L., Holden, J. J. A., and Steel, C. G. H. (1987). Vitellogenesis and fertility in Drosophila females with low ecdysteroid titres; the L(3)3DTS mutation. J. Insect Physiol. 33, 137–142. doi: 10.1016/0022-1910(87)90139-9
Wang, C. F., Zhang, Z., and Sun, W. (2018). Ecdysone oxidase and 3-dehydroecdysone-3β-reductase contribute to the synthesis of ecdysone during early embryonic development of the silkworm. Int. J. Biol. Sci. 14, 1472–1482. doi: 10.7150/IJBS.26227
Warren, J. T., Petryk, A., Marques, G., Jarcho, M., Parvy, J.-P., Dauphin-Villemant, C., et al. (2002). Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 99, 11043–11048. doi: 10.1073/pnas.162375799
Warren, J. T., Petryk, A., Marqués, G., Parvy, J.-P., Shinoda, T., Itoyama, K., et al. (2004). Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 34, 991–1010. doi: 10.1016/J.IBMB.2004.06.009
Warren, J. T., Wismar, J., Subrahmanyam, B., and Gilbert, L. I. (2001). Woc (without children) gene control of ecdysone biosynthesis in Drosophila melanogaster. Mol. Cell. Endocrinol. 181, 1–14. doi: 10.1016/S0303-7207(01)00404-X
Wismar, J., Habtemichael, N., Warren, J. T., Dai, J. D., Gilbert, L. I., and Gateff, E. (2000). The Mutation without children(rgl) Causes Ecdysteroid Deficiency in Third-Instar Larvae of Drosophila melanogaster. Dev. Biol. 226, 1–17. doi: 10.1006/DBIO.2000.9811
Xiang, Y., Liu, Z., and Huang, X. (2010). br regulates the expression of the ecdysone biosynthesis gene npc1. Dev. Biol. 344, 800–808. doi: 10.1016/j.ydbio.2010.05.510
Yamada, M., Murata, T., Hirose, S., Lavorgna, G., Suzuki, E., and Ueda, H. (2000). Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development 127, 5083–5092. doi: 10.1242/dev.127.23.5083
Yamanaka, N., and Okamoto, N. (2020). “Molecular functions of ecdysteroids in insects,” in Advances in Invertebrate (NEURO)Endocrinology, eds S. Saleuddin, A. B. Lange, and I. Orchard (Palm Bay: Apple Academic Press), 77–127.
Yang, Y., Zhao, T., Li, Z., Qian, W., Peng, J., Wei, L., et al. (2021). Histone H3K27 methylation–mediated repression of Hairy regulates insect developmental transition by modulating ecdysone biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 118:e2101442118. doi: 10.1073/PNAS.2101442118
Yoo, B., Kim, H. Y., Chen, X., Shen, W., Jang, J. S., Stein, S. N., et al. (2021). 20-hydroxyecdysone (20E) signaling regulates amnioserosa morphogenesis during Drosophila dorsal closure: EcR modulates gene expression in a complex with the AP-1 subunit, Jun. Biol. Open 10:bio058605. doi: 10.1242/BIO.058605
Yoshinari, Y., Kurogi, Y., Ameku, T., and Niwa, R. (2019). Endocrine regulation of female germline stem cells in the fruit fly Drosophila melanogaster. Curr. Opin. Insect Sci. 31, 14–19. doi: 10.1016/J.COIS.2018.07.001
Yoshiyama, T., Namiki, T., Mita, K., Kataoka, H., and Niwa, R. (2006). Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133, 2565–2574. doi: 10.1242/dev.02428
Yoshiyama-Yanagawa, T., Enya, S., Shimada-Niwa, Y., Yaguchi, S., Haramoto, Y., Matsuya, T., et al. (2011). The conserved rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J. Biol. Chem. 286, 25756–25762. doi: 10.1074/JBC.M111.244384
Zeng, J., Huynh, N., Phelps, B., and King-Jones, K. (2020). Snail synchronizes endocycling in a TOR-dependent manner to coordinate entry and escape from endoreplication pausing during the Drosophila critical weight checkpoint. PLoS Biol. 18:e3000609. doi: 10.1371/journal.pbio.3000609
Zeng, J., Kamiyama, T., Niwa, R., and King-Jones, K. (2018). The Drosophila CCR4-NOT complex is required for cholesterol homeostasis and steroid hormone synthesis. Dev. Biol. 443, 10–18. doi: 10.1016/J.YDBIO.2018.08.012
Zerr, D. M., Hall, J. C., Rosbash, M., Siwicki, K. K., Neurosci, J., Wheeler, D. A., et al. (1994). Block in nuclear localization of period protein by a second clock mutation, timeless. Science 263, 1606–1609. doi: 10.1126/SCIENCE.8128247
Zhang, T., Song, W., Li, Z., Qian, W., Wei, L., Yang, Y., et al. (2018). Krüppel homolog 1 represses insect ecdysone biosynthesis by directly inhibiting the transcription of steroidogenic enzymes. Proc. Natl. Acad. Sci. U. S. A. 115, 3960–3965. doi: 10.1073/pnas.1800435115
Zhao, Y., Lindberg, B. G., Esfahani, S. S., Tang, X., Piazza, S., and Engström, Y. (2021). Stop codon readthrough alters the activity of a POU/Oct transcription factor during Drosophila development. BMC Biol. 19:185. doi: 10.1186/S12915-021-01106-0
Zhou, X., Zhou, B., Truman, J. W., and Riddiford, L. M. (2004). Overexpression of broad: a new insight into its role in the Drosophila prothoracic gland cells. J. Exp. Biol. 207, 1151–1161. doi: 10.1242/JEB.00855
Zipper, L., Jassmann, D., Burgmer, S., Görlich, B., and Reiff, T. (2020). Ecdysone steroid hormone remote controls intestinal stem cell fate decisions via the PPARγ-homolog Eip75B in Drosophila. Elife 9:e55795. doi: 10.7554/eLife.55795
Keywords: ecdysteroids, ecdysone, steroidogenic enzyme, transcription factors (TFs), Drosophila, Halloween genes
Citation: Kamiyama T and Niwa R (2022) Transcriptional Regulators of Ecdysteroid Biosynthetic Enzymes and Their Roles in Insect Development. Front. Physiol. 13:823418. doi: 10.3389/fphys.2022.823418
Received: 27 November 2021; Accepted: 12 January 2022;
Published: 08 February 2022.
Edited by:
Sheng Li, South China Normal University, ChinaReviewed by:
Naoki Yamanaka, University of California, Riverside, United StatesCopyright © 2022 Kamiyama and Niwa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryusuke Niwa, cnl1c3VrZS1uaXdhQHRhcmEudHN1a3ViYS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.