- 1Center for Molecular Genetics, Institute of Translational Medicine, The Affiliated Hospital of Qingdao University, College of Medicine, Qingdao University, Qingdao, China
- 2School of Basic Medicine, Qingdao University, Qingdao, China
- 3The Affiliated Cardiovascular Hospital of Qingdao University, Qingdao, China
- 4Jiangsu Provincial Engineering Research Center for Biomedical Materials and Advanced Medical Device, Huaiyin Institute of Technology, Huaian, China
- 5Max-Delbrück-Center for Molecular Medicine (MDC), Berlin, Germany
- 6School of Bioengineering, Suqian University, Suqian, China
Excessive production of free radicals can induce cellular damage, which is associated with many diseases. RNA is more susceptible to oxidative damage than DNA due to its single-stranded structure, and lack of protective proteins. Yet, oxidative damage to RNAs received little attention. Accumulating evidence reveals that oxidized RNAs may be dysfunctional and play fundamental role in the occurrence and development of type 2 diabetes (T2D) and its complications. Oxidized guanine nucleoside, 8-oxo-7, 8-dihydroguanine (8-oxoGuo) is a biomarker of RNA oxidation that could be associated with prognosis in patients with T2D. Nowadays, some clinical trials used antioxidants for the treatment of T2D, though the pharmacological effects remained unclear. In this review, we overview the cellular handling mechanisms and the consequences of the oxidative RNA damage for the better understanding of pathogenesis of T2D and may provide new insights to better therapeutic strategy.
Introduction
Free radicals are the normal cellular products of energy metabolism (Kong and Lin, 2010). In living organisms, radicals are mostly derived from reactive oxygen species (ROS) and reactive nitrogen species (RNS). However, excessive production of free radicals may cause mitochondria dysfunction, which could lead to the changing of energy metabolism and affect the normal equilibrium of cells (Valko et al., 2007). Free radicals can damage the DNAs, RNAs and proteins as well as other cell components (Haigis and Yankner, 2010; Majzunova et al., 2013; McArdle and Jackson, 2019). Oxidative stress occurs when free radicals are over-produced or due to the decrease in the ability of the antioxidant defense system. Indeed, oxidative damage exists widely in the body, which is mainly manifested by the damage to the structure and function of biological macromolecules (such as DNA, proteins, lipids, etc.), which leads to gene mutation, cell carcinogenesis, and aging. The pathogenesis of some age-related diseases such as neurological disorders (i.e., Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s disease) and atherosclerosis have also been attributed to the oxidation of the chemicals induced by free radicals (Poulsen et al., 2012; Fimognari, 2015).
Nowadays, noncommunicable diseases (NCDs), including diabetes, cancer, and cardiovascular diseases, are the major cause of death globally. NCDs account for 63 percent of all deaths worldwide every year. Diabetes is one of the most essential NCDs that threatening human health in the world, the rapid growth of diabetes has brought an alarming global burden to the social and economic development (Jaacks et al., 2016). The international diabetes federation (IDF) reported that the number of diabetes patients could reach over six hundred million by 2040 (Ogurtsova et al., 2017; Luo et al., 2019). In diabetes, Type 2 diabetes (T2D) constitutes the main type (Ogurtsova et al., 2017). T2D could cause a series of concomitant diseases, such as coronary heart disease, lower limb arteriopathy, retinopathy, diabetic nephropathy (Sung et al., 2018).
And now, there are many strategies for the treatment of T2D, mainly includes surgery, drug therapy, exercise therapy, dietary nutrition, and multifactorial treatment (Durrer Schutz et al., 2019; Koch and Shope, 2021). However, due to the diversity of disease factors in T2D, these methods cannot effectively reduce the increase in morbidity and mortality. In addition, the level of venous blood glucose and biomarkers like glycated hemoglobin (HbA1c) also have limitations to be used as prognostic and predictive biomarkers for mortality, especially for elders or patients with hyperlipidemia (Durrer Schutz et al., 2019). Therefore, much work remains to be done to understand the underlying mechanism involved in the regulation of glucose metabolism and insulin sensitivity, which will unveil a better understanding of blood glucose homeostasis and find more potential therapy targets.
In this review, we discuss the consequences and the mechanism of intracellular response to RNA oxidation. Furthermore, we review the relationship between the ROS and T2D which may provide new intervention strategy for T2D.
The History of RNA Functions and Structure
Although F. Miescher discovered the nuclein in 1868, it did not attract much attention for more than half a century (Sankaran, 2016). In the 1920s and 1930s, it was confirmed that there were two kinds of nucleic acids DNA and RNA in nature, and the composition of nucleotides was clarified. In the decades that followed, life sciences focused on the application and analysis of the genomic DNA. In the 1980s, W. Gilbert et al. discovered that RNA had the activity of catalytic enzymes, which opened a window for the study of RNA (Sankaran, 2016). In addition to transferring information from DNA to protein, RNA molecules also play vital role in maintaining, regulating, and protecting the genomes of all organisms and have more structural and functional diversity than DNA (Sharp, 2009; Cech and Steitz, 2014).
Biomolecules have unique molecular structure, which is essential for their function. However, we know very little about the structure of RNA. The exploration of RNA structure started from the Click coined the hypothesis that RNA played an intermediary role in the transmission of genetic information to proteins in 1953. And in 1965, Holly and his colleagues used nuclease to sequence yeast tRNA (Holley et al., 1965). Noller and Chaires found that kethoxal modifies affected the function of rRNA in 1972 (Noller and Chaires, 1972). Subsequently, the studies focus on the secondary and tertiary structures of RNA (Peattie and Gilbert, 1980). High-throughput sequencing and new computational methods to interrogate RNA structure are beginning to provide new insights into RNA structures (Kertesz et al., 2010; Underwood et al., 2010; Lucks et al., 2011). Driven by technology and cognition, more and more structural changes, modifications, and types of RNA have been gradually discovered. At the same time, the remarkable diversity of RNA structure and function revealed an enormous potential of RNA in future clinical applications is gradually emerging.
The Generation and Damage of ROS in T2D
The theory of ROS in organisms was put forward in the 1950s (Commoner et al., 1954; Newsholme et al., 2016). ROS can be produced by exogenous and endogenous sources (Kohen and Gati, 2000). Exogenous sources may include ultraviolet radiation (Liu et al., 2020), ultrasound, drugs, radiation and exposure to pollutants and toxic chemicals. Endogenous sources may include neutrophils, cytokines (Obata et al., 2001), enzymes that directly produce ROS. These enzymes mainly come from mitochondria, such as dihydroorotate dehydrogenase, succinate dehydrogenase and NADPH oxidases (NOX) (Le Bras et al., 2005; Ahmad et al., 2017). Among those enzymes, NOX is an enzyme that localizes on cell membrane and catalyzes the reduction of oxygen molecules to superoxide anion (Dubois-Deruy et al., 2020; Masselli et al., 2020). It is the main source of ROS generation. As a coenzyme, NOX participates in cellular electron transfer and servers as a second messenger in many signaling pathways (Bae et al., 2011).
ROS have been reported to contributed the pathology of T2D in many ways (Figure 1). High glucose intake in the body increases mitochondrial production of ATP, which produces large amounts of free radicals and increases ROS levels. And excess sugar can bind to proteins to form advanced glycation products (AGEs).
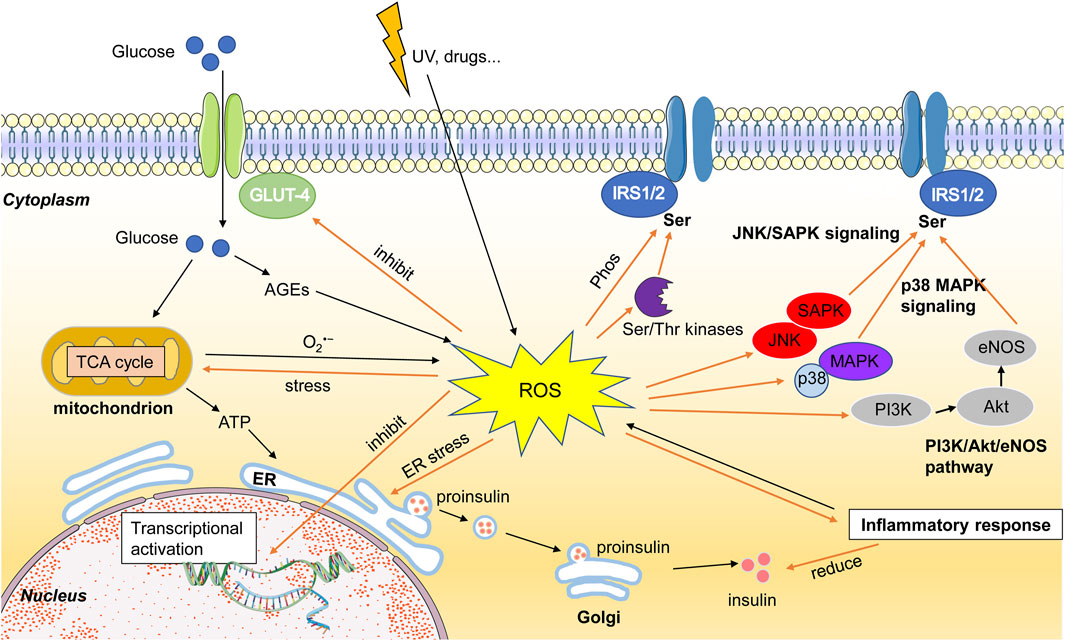
FIGURE 1. Schematic diagram of the effect of ROS toxicity on insulin secretion and insulin resistance signaling pathways. High level of ROS can modulate insulin resistance by altering the insulin-signaling ways, such as Akt, P13K, and JNK. ROS also can inhibit the function of cell organelles, and then reduce the energy supply, induce the apoptosis.
Accumulating studies demonstrated that ROS plays a vital role in the occurrence and development of T2D by inducing insulin resistance and beta cell dysfunction (Rochette et al., 2014; Liu et al., 2016). A classical mechanism of insulin abnormality is dysregulation of the insulin-regulated pathway, such as inappropriate phosphorylation or inhibition of the insulin receptor-associated proteins. The insulin binds to insulin receptors will lead to phosphorylation of residues, resulting in changes in receptor structure and triggering downstream signal transduction. Excess free radicals can disturb beta cell function, inhibiting proliferation and regeneration (Porte and Kahn, 2001; Liang et al., 2016; White et al., 2016; Wang and Wang, 2017).
The insulin receptor substrate (IRS) proteins are the cytoplasmic proteins that regulate growth and metabolism, which response to some stimuli including hormones and cytokines (Lee and White, 2004). The family of IRS was compound of six type named IRS-1, IRS-2, IRS-3, IRS-4, IRS-5, and IRS-6. Among them, IRS-1 and IRS-2 are the key control of IR (Copps and White, 2012). ROS could induce IR by leading the phosphorylation of the serine at serin 307 of IRS-1. Excess free radicals could also reduce the migration and relocation of insulin-dependent IRS (Yaribeygi et al., 2019). The high level of ROS can also impair the insulin sensitivity by activating the IKKβ/NF-κB and JNK pathway, resulting the degradation of IRS-1/2 (Nishikawa et al., 2000; Evans et al., 2005). ROS-mediated activation of JNK signaling leads to decreased insulin secretion via nucleocytoplasmic translocation of pancreatic and duodenal homeobox-1 (PDX-1), a key transcription factor that binds to the insulin promoter and induces insulin expression (Kajimoto and Kaneto, 2004). High levels of intracellular oxidation can also inhibit p38 MAPK pathway, PI3K/Akt/eNOS pathway, PI3K/JUK pathway, JNK/SAPK pathway, modulate serine/threonine kinases such as Akt (or PKB), GSK-3, AMPK, and mTOR, which cause IR (Bloch-Damti and Bashan, 2005; Rains and Jain, 2011; Yaribeygi et al., 2020).
Glucose transporter-4 (GLUT-4) have the function of transporting the glucose into cells, any factor that reduces the expression of GLUT-4 have a significant effect on the insulin signal (Klip et al., 2019). Clinal researches showed the expression of GLUT-4 was lower than the control in the patients with insulin resistance (Hussey et al., 2012; Reno et al., 2017). ROS could suppress the expression and relocation of GLUT-4 by inducing the point mutation (Hurrle and Hsu, 2017). And some studies reported that ROS also could suppress the GLUT-4 expression by effecting the expression of its transcriptional factors such as PPAR-γ, CEB/Ps, nuclear factor-1, p85, HIF-1α, MEF2, and NF-κB (Cooke and Lane, 1999; Pessler et al., 2001; She and Mao, 2011).
ROS is mainly derived from mitochondria, but can also in turn lead to mitochondrial stress, mitochondrial dysfunction, mitochondrial division, and cell apoptosis (Medda et al., 2021). As noted above that the mitochondrial dysfunction can affect the energy supply and indirectly affect insulin secretion.
Endoplasmic reticulum (ER) is the main organelle for intracellular modification and synthesis. The stimuli of oxidative stress could disturb the function of ER and induce the ER stress (Lemmer et al., 2021). ER stress could disrupt proper protein folding leading to accumulation of misfolded proteins, which can lead to reduced insulin synthesis and decreased insulin stability in beta cells (Han et al., 2013). ER stress can suppress the expression of GLUT-4 and relocation on the cell membrane (Yang et al., 2010). Ozcan et al. (2004) reported the ER stress could induce IR by directly suppressing the insulin pathway through hyperactivation of c-Jun N-terminal kinase (JNK) and subsequent serine phosphorylation of IRS-1 (Nakatani et al., 2005). The accumulation of misfolded proteins in ER stress may cause the unfolded protein response (UPR), which will promote the cell death during the prolong stress (Fernández et al., 2015).
Oxidative stress stimulates the inflammatory response, which in turn worsens oxidative levels, and numerous studies have shown that oxidative stress and inflammation are key factors in the development of insulin resistance and the establishment of T2D (Keane et al., 2015). ROS could increase inflammatory responses which in turn induces insulin resistance in several pathways, such as IKK, JNK and NF-κB signaling pathway (Cai et al., 2005), IL-6, TNF-a/JNK/IRS-1 pathway (Akash et al., 2018).
The Indicator of RNA Oxidation: 8-oxoGuo
Oxidation of RNA could cause strand breaks, lose the expression of RNA, and then induce modified nucleobases, lipid, and sugar (Li et al., 2006; Jacobs et al., 2010; Resendiz et al., 2012). The oxidation products of RNA mainly include 5-hydroxycytidine, 5-hydroxyuridine, 8-hydroxyadenosine and 8-oxoGuo. Among various oxidation formations, the oxidized nucleobase 8-oxoGuo in RNA is currently used as an essential indicator of RNA oxidation because it appears to be abundant and mutagenic (Li et al., 2006).
8-oxoGuo is an oxidative adduct produced by reactive oxygen radicals such as hydroxyl radicals and singlet oxygen that attack the carbon 8 of the guanine base in RNA molecules (Figure 2). The degree of oxidative damage and repair in vivo can be evaluated by the detection of 8-oxoGuo, and the relationship between oxidative stress and RNA damage (Schöttker et al., 2020). It is of great significance for the study of degenerative diseases, aging mechanism, carcinogenesis mechanism, the relationship between environmental toxins and oxidative stress. It can also be used to evaluate the efficacy of antioxidants in the treatment of RNA oxidative damage.
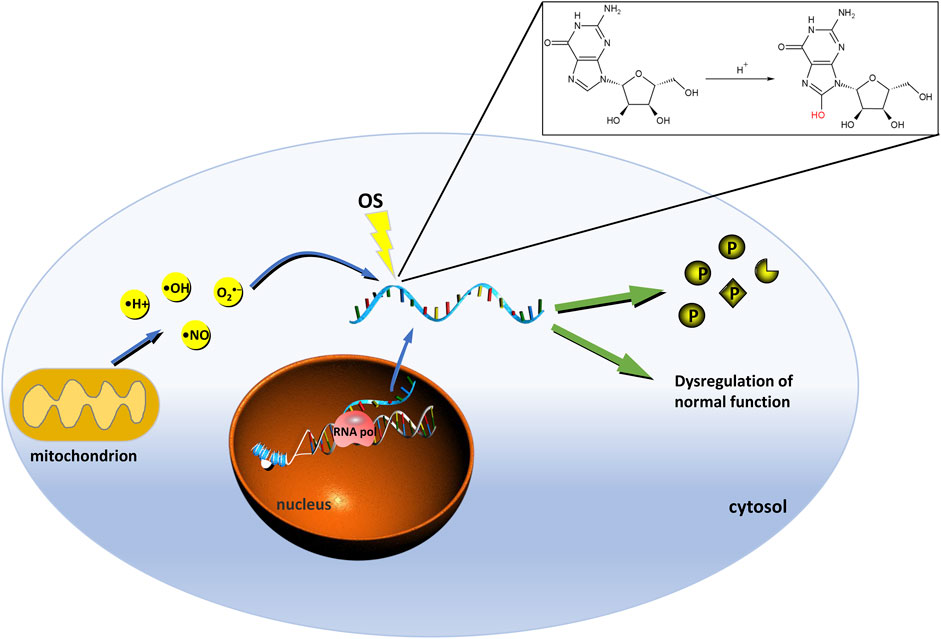
FIGURE 2. Mode of the occurrence and consequence of RNA oxidation. Free radicals attacked the eighth carbon atom of RNA to turn G into 8-oxoGuo. Once RNA is oxidized, the original function would become abnormal. Oxidized coding RNAs lead to decrease in the level and activity of protein synthesis and increase the non-functional, incomplete protein, and mutated proteins. Oxidation of non-coding RNA could affect their regulatory functions. The green marked P ball represents a normal protein, the notched sphere indicates non-functional or truncated protein. Rhombohedral sphere indicates mutated protein. RNA pol, RNA polymerase; P, protein; OS, oxidative stress.
As mentioned above, DNA was discovered and studied first, and most previous studies were focus on the DNA oxidation. The first report to assessed the level of 8-hydroxydeoxyguanosine in urine was in 1962 (Loft et al., 1992). Franzke et al. (2018) found that exercise, nutrition and cognitive training increased antioxidant activity by measuring oxidative markers in blood plasma. Larsen et al. (2019) pointed that clinical study must be considered the study population and size, as many factors such as environment can contribute to differences.
Several methods have been reported to detect RNA oxidation levels. However, due to the lack in the RNA oxidation studies, the methods need to be developed and updated. Nowadays, the measure of oxidized RNA products is based on the chemical structure or immune properties of oxidized nucleotides. Nucleosides and bases could be detected by high-performance liquid phase chromatography coupled with tandem mass spectrometry (HPLC–MS/MS) or ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS) (Fiala et al., 1989; Shen et al., 2000; Hofer et al., 2006). Another detection method relies on the interaction of antigens and antibodies, including dot blot, ELISA, Northwestern blot, immunoprecipitation, and immunohistochemistry (Nunomura et al., 1999; Honda et al., 2005; Shan and Lin, 2006; Looi et al., 2008; McKinlay et al., 2012). Elisa methodology is one of the earliest, most widely used and relatively mature detection methods, which benefit from the reaction between antibodies (Cooke et al., 2001; Chiou et al., 2003). At present, there are a lot of kits on the market to detect oxidized nucleic acid are based on this principle (Chao et al., 2021). HPLC/UPLC-MS are relatively mature methods for the detection of RNA oxidation products (Wang et al., 2017; Jorgensen et al., 2018; Jørs et al., 2020). These methods have high accuracy while requiring many samples. ELISA and other biochemical reaction methods can detect components in body fluids, but the operation is complex and time-consuming. Since there is only one oxygen atom difference between 8-oxoGuo and G in structure, it is a challenge for the specificity of antibodies (Barregard et al., 2013). It is impossible for DNA or RNA specific recognition antibodies to reach all oxidative damage sites in nucleic acids, which could affect the quality of experimental data.
Even though there are many methods to detect the content of 8-oxoGuo in different samples, it is still quite challenging to measure it accurately. Because it will generate the uncontrollable new oxidation reactions during nucleic acid extraction and subsequent operations (Barregard et al., 2013). In order to increase the accuracy of the results as much as possible, metal chelation or antioxidants were added during the experiment to decrease the false new RNA oxidative damage during the experiment (Hofer et al., 2005). In addition, because the RNA oxidation could impede reverse transcription, the rate of reverse transcription is also used to assess the level of RNA oxidation (Gong et al., 2006).
Further studies reported that oxidation did not affect the association of RNA and polysome in the process of transcription, but affect the properties of the product (Shan et al., 2003; Shan et al., 2007). Oxidized coding RNAs lead to a decrease in the level and activity of protein synthesis and increase the non-functional, incomplete protein, and mutated proteins (Figure 2). Oxidized non-coding RNAs can inhibit their regulatory functions in the process of protein synthesis, leading to cell apoptosis and cell death. For instance, the microRNAs (miRNAs) are crucial member of non-coding RNAs and have been recognized as the vital regulator in the gene expression (Saliminejad et al., 2019; Kozak et al., 2020). MiR-184 upon oxidative modification could mismatch the 3′UTR of Bcl-xL and Bcl-w resulting in their reduction and this mismatch is involved in the initiation of apoptosis in vitro and vivo (Wang et al., 2015). Oxidized miR-1 could redirect the new targets in heart hypertrophy pathways that influence the redox mediated gene expression (Seok et al., 2020).
Coping With the RNA Oxidation
Living organisms need to reduce the risk of damage mediated by RNA oxidation. The cells must have some protective mechanisms to maintain the normal function and to survive under stress conditions, such as monitoring and inhibiting the process of RNA oxidation or keeping and reducing the excess oxidation molecules. Unfortunately, little is known about any of the mechanisms. Therefore, we discuss the progress and hypothesize potential ways in which oxidized RNAs may be involved in regulation (Figure 3).
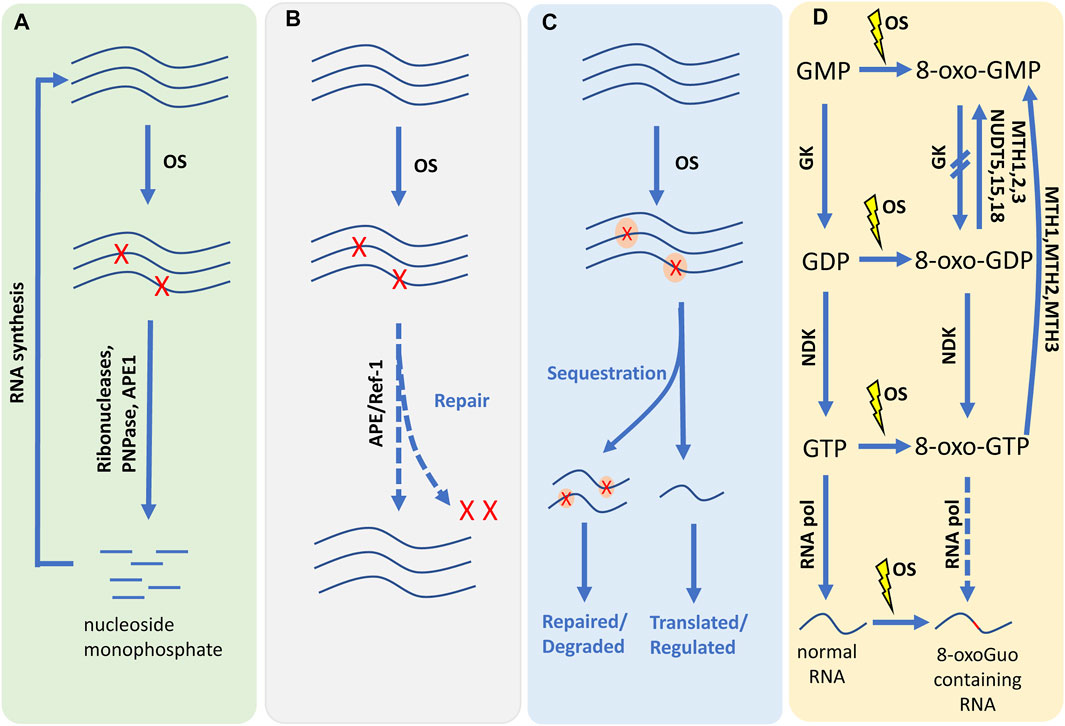
FIGURE 3. The schematic diagram of coping with RNA oxidation. (A) The oxidized RNA may lose its normal function, it would be degraded into nucleoside monophosphate by ribonucleases, PNPase or APE1. The nucleoside monophosphate can be reused in RNA synthesis. PNPase: Polynucleotide phosphorylase; APE1: Apurinic/apyrimidinic endonuclease 1. (B) The pathway of RNA repair has not been reported yet, but there are many studies speculating that it exists. Under the action of some substances, the oxidative damage of RNA will be repaired to perform its normal function. (C) Oxidized RNA molecules can be labeled with specific binding proteins. Once identified, oxidized RNA can be separated from normal RNA. Sequestration may help recruit repair/degradation activities that will eventually eliminate oxidized RNA. The orange ovals indicate binding proteins. (D) 8-oxoGuo containing RNA can be generated by oxidizing RNA and a few can be synthesis by 8-oxo-GTP. 8-oxo-GTP can be generated by oxidation of GTP as well as by phosphorylation of 8-oxo-GDP by NDK. 8-oxo-GDP can be generated by oxidation of GDP as well as by phosphorylation of 8-oxo-GMP by GK. MTH1, MTH2, MTH3 could hydrolyze 8-oxo-GDP and 8-oxo-GTP to 8-oxo-GMP. NUDT5, NUDT15, NUDT18 could hydrolyze 8-oxo-GDP to 8-oxo-GMP. GK: Guanylate kinase; NDK, nucleotide diphosphate kinase; MTH1, MutT homologue 1; MTH2, MutT homologue 2; MTH3, MutT homologue 3; NUDT5, Nudix type 5; NUDT15, Nudix type 15; NUDT18, Nudix type 18. OS, oxidized stress; The red blot in the 8-oxoG containing RNA indicates the oxidized nucleotide. Blue lines represent RNA molecules. Red “X” represents oxidized residues in RNA.
Degradation
Degradation can play an important role in RNA metabolism and eliminating oxidized RNA. This irreversibly eliminating pathway is probably the major system dealing with oxidized RNA. Ribonucleases have the degradation activity for oxidized RNA, and it can eradicate aberrant RNAs. However, further studies show that RNase is usually assisted by polyadenylation and RNA helicase activity (Deutscher, 2006). Polynucleotide phosphorylase (PNPase) is widely distributed across organisms of all kingdoms (Das et al., 2011). In E.coli, PNPase could regulate many aspects of RNA metabolism, including degradation of defective rRNA and tRNA, etc. (Kushner, 2002; Basturea et al., 2011). Human PNPase (hPNPase) is mainly localized in mitochondrial intermembrane space (Piwowarski et al., 2003; Chen et al., 2006). In human mitochondria, hPNPase and RNA helicase Hsuv3 form a complex, which maintains the homeostasis of some mt-mRNAs. And in cell culture, the hPNPase plays a vital role in the degradation of mitochondrial mRNA, c-myc RNA, and miRNAs. Several studies reported Apurinic/apyrimidinic endonuclease 1 (APE1) could control the RNA quality and degrade the oxidized RNA (Li et al., 2014).
Repair
At present, very little is known about the repair mechanisms of oxidative RNA damage. It is very costly for cells to synthesize RNAs because those pathways require energy. If the damage to RNA is sublethal and can be repaired and reused, it could save cells energy and be evolutionarily beneficial. Although RNA is like DNA, RNA lacks a complementary strand; RNA-repair pathways may be different from the DNA repair mechanisms. Indeed, some studies have confirmed that RNA damage can be repaired. For instance, the methyl-guanine-methyl transferases (MGMT), which repair O6-mG (Kaina et al., 2007), and the oxidative demethylases, such as A1KB, repair m1A and m3C (Dinglay et al., 1998; Aas et al., 2003; Fu et al., 2010). The DNA-repair enzyme APE/Ref-1 has rRNA quality control (Fu et al., 2010).
Sequestration
The oxidized RNA may lose its original biological function, some specific factors could discriminate and mark oxidized nucleotides during RNA synthesis or translation (such as RNA polymerase, etc.). And then, it will activate the recruitment of repair enzymes or degradation reactions (Deutscher, 2006). Several proteins have been demonstrated to recognize and bind oxidized RNAs. For instance, PNPase protein or hPNPase, which has a higher binding affinity to 8-oxoGuo than other normal oligonucleotides (Hayakawa et al., 2001; Hayakawa and Sekiguchi, 2006; Wu and Li, 2008). Mammalian Y box-binding protein 1 (YB-1), heterogeneous nuclear ribonucleoprotein D0 (HNRNPD), splicing factor 3B subunit 4 (SF3B4), heterogeneous nuclear ribonucleoprotein C1/C2 (HNRNPC) and splicing isoform 1 of DAZ-associated protein 1 (DAZAP1) also show affinity comparable to hPNPase protein (Hayakawa et al., 2002; Hayakawa et al., 2010).
Interdicting Incorporation of Oxidized Nucleotides Into RNA
Oxidized nucleotides triphosphates from the nucleotide pools can be metabolically incorporated into RNA through the synthesis pathway, and oxidized nucleotides can be produced by degrading oxidized RNAs (Li et al., 2006). Under oxidative stress, nucleosides and nucleotides may be oxidized. RNA polymerase could discriminate guanosine-5′-triphosphate (GTP) and 8-oxo-7,8-dihydroguanosine-5′-triphosphate (8-oxo-GTP) during the process of RNA synthesis. The rate of misincorporation of 8-oxo-GTP into RNA was only 2% that of guanine (Hayakawa et al., 1999). 8-oxo-7,8-dihydroguanosine 5′-monophosphate (8-oxo-GMP) can be transformed into 8-oxo-7,8-dihydro-guanosine 5′-diphosphate (8-oxo-GDP) under the catalysis of guanylate kinase (GK), similarly, 8-oxo-GDP can be phosphorylated to 8-oxo-GTP through the nucleotide diphosphate kinase (NDK) (Ishibashi et al., 2005; Sekiguchi et al., 2013). Several enzymes can dephosphorylate oxidized nucleoside triphosphates, thus preventing their incorporation into RNA, have been identified. MutT homologue 1 (MTH1), MTH2, and MTH3 can hydrolyze 8-oxo-GDP or 8-oxo-GTP to 8-oxo-GMP (Hayakawa et al., 1999; Ishibashi et al., 2005; Takagi et al., 2012). Nudix type 5 (NUDT5), NUDT15, NUDT18 can hydrolyze 8-oxo-GDP to 8-oxo-GMP (Ishibashi et al., 2005). And GK could also block the phosphorylation of 8-oxo-GMP to 8-oxo-GDP (Li et al., 2014).
RNA Oxidation and T2D
Many studies reported that T2D is associated with 8-oxoGuo. The first research about the relationship of RNA oxidation marker 8-oxoGuo and T2D was reported in 2011. Broedbaek and his colleagues analyzed approximately fourteen hundred diagnosed T2D patients and found that the level of 8-oxoGuo was positively correlated with the mortality of patients (Broedbaek et al., 2011). Moreover, Broedbaek confirmed the association between 8-oxoGuo and the mortality of T2D in the same cohort (Broedbaek et al., 2013). Besides, the levels of 8-oxoGuo were independent of other risk factors. Those findings suggest that the 8-oxoGuo could serve as a new clinical biomarker in diabetes.
Later, some studies reported that high RNA oxidation is associated with the risk of death in patients with all-cause, cardiovascular and microalbuminuria in patients with T2D (Kjær et al., 2017; Kjaer et al., 2018), and the imbalanced redox system could be a molecular mechanism contributing to the progression of T2D (Poulsen et al., 2019; Schöttker et al., 2020). RNA oxidation occurs earlier than DNA oxidation and is more closely associated with diabetic nephropathy. 8-oxoGuo may represent a new and easily detectable biomarker for diabetic nephropathy (Poulsen et al., 2019; Schöttker et al., 2020). Interestingly, this association of 8-oxoGuo in T2D was weak in younger patients, likely due to the higher tolerance to oxidative stress in youth (Schöttker et al., 2020).
The etiology of T2D is complicated. Currently, the main drugs include glipizide, biguanide, thiazolidinedione, and other oral drugs, as well as insulin and insulin-like injection preparations, were reported to decrease the level of oxidation. Long-term clinical follow-up studies have found that taking anti-hypertensive drugs such as losartan, valsrtan, and olmesartan could reduce DNA oxidation markers compared with the placebo (Kuboki et al., 2007; Ogawa et al., 2009; Nakamura et al., 2013; Pan et al., 2015).
Interestingly, in the therapeutic aspect of RNA oxidation, Broedbaek and his colleagues found that the usage of lipid-lowering drugs was associated with lower RNA oxidation, which could be the potential therapeutic drugs (Broedbaek et al., 2011). Another study reported that in T2D patients, a decrease of 8-oxoGuo was observed after treatment with sevelamer, and the inflammatory factors IL-2 and IL-6 also tended to decrease (Brønden et al., 2020). Biguanides and sulfonylureas have been used to lower blood sugar, like metformin, glimepiride, pioglitazone and dapagliflozin. In clinical studies, hypoglycemic drugs can reduce the level of oxidation in vivo while lowering blood glucose (Sova et al., 2013; Wang et al., 2013; Shigiyama et al., 2017).
Environmental factors are also important influencing factors for oxidation of organisms. Previous studies have found that Mediterranean diet and carbohydrate-reduced high-protein dietary intervention can also reduce the level of oxidation in the body (Gutierrez-Mariscal et al., 2012; Skytte et al., 2020). Consumption of Green tea catechin, watermelon powder, and supplementation of paricalcitol and l-arginine also reduced oxidative damage levels (Oyama et al., 2010; Glenn et al., 2018; Fan et al., 2019). Although exercise can increase the body’s energy metabolism, produce more potential oxidation factors. Clinical and animal experiments have shown that physical exercise can increase the activity of antioxidant enzymes in cells and improve oxidative resistance (Radak et al., 2007; Tweedie et al., 2011; Villaño et al., 2015; Franzke et al., 2018; Larsen et al., 2020).
As we all know, T2D can cause many complications, and the mortality rate of cardiovascular disease is very high. Shokri suggested that increasing PON1 expression can reduce plasma oxidized-LDL level, reduce the ability of macrophages to absorb oxidized-LDL, and reduce cardiovascular complications in T2D patients. Therefore, the strategy of raising or restoring PON1 level is useful for reducing or preventing cardiovascular complications (Shokri et al., 2020).
Conclusion
Oxidative stress has been advocated as an essential pathological factor for many diseases, especially in aging and neurodegenerative disorders. As noted above that pancreatic islets and pancreatic B cells have a high metabolic reaction and a low expression of antioxidant substances, so they are vulnerable to oxidative free radicals (Shah et al., 2007). As noted above that the oxidative stress in pancreatic B cells could reduce insulin synthesis, even make the cell prone to apoptosis (Keane et al., 2015; Poulsen et al., 2019). Furthermore, oxidative stress can lead to insulin resistance by disturbing the insulin receptor signaling pathway. Some researchers reported that oxidative stress can activate the proinflammatory signaling pathways of NF-kB and c-Jun N-terminal protein kinase, which can cause serine hyperphosphorylation in insulin receptor substrates (such as IRS1 and IRS2) (Evans et al., 2005; Shah et al., 2007; Xie et al., 2019). This may also suppress the function of GLUT-4 (Hurrle and Hsu, 2017).
Watson proposed T2D as a redox disease (Watson, 2014). In the redox research, DNA oxidation has been attracted more attention in the past few decades whereas a less focus on RNA oxidation. However, RNA oxidation has become the focus of current research because of its role in many diseases. RNA are more susceptible to oxidation than DNA (Hofer et al., 2006), which may be inferred from the following observations. First, its single-stranded nature and widely distributed around the mitochondria that produce reactive oxygen species, making it vulnerable to free radical (Hofer et al., 2006; Nunomura et al., 2012; Poulsen et al., 2012). Second, the repair mechanism of RNA activity is not fully understood, degradation and elimination are the main ways that are coping with RNA oxidative damage. Third, DNA is protected by proteins such as histones, and few proteins bind to RNA (Hofer et al., 2006; Nunomura et al., 2012; Poulsen et al., 2012).
8-oxoGuo is one of the products of RNA oxidation, so the actual oxidation levels in the body would be higher than measured. In the previous studies, 8-oxoGuo has the potential role of diagnostic detection, prognosis, and treatment target. Although there is strong evidence that oxidative stress has pathophysiological effects and the antioxidant effects of existing diabetes-related treatments have been proposed, however, the clinical outcomes of antioxidant trials have been disappointing. Many drugs cannot cure T2D fundamentally, they can only relieve symptoms. Recently studies have found some drugs can reduce the oxidation of T2D and reduce the incidence of complications, but its mechanism is unclear and further studies are still required to determine causality in T2D patients. Increasing intracellular antioxidant defense and controlling the production of free radicals may be helpful to the treatment of T2D, like developing the new compounds that inhibiting the reactive oxygen-producing enzymes. In this review, we summarize the consequences and cellular handling mechanisms of the oxidative RNA damage that may provide new insights to the pathogenesis of T2D and lead to a better therapeutic strategy.
Author Contributions
XC, LA, and PL generated the idea, and edited the manuscript. XC prepared the manuscript. HY, ZhL, ZiL, JG, YW, XL, LZ, NA, and MB read and revise the manuscript.
Funding
This project was supported by the National Natural Science Foundation of China Research Fund for International Young Scientists (82050410450), Natural Science Foundation of Shandong Province (ZR2019BH014), and the Research Program of the National Natural Science Foundation of China (91849209, 51775221, and 81850410551).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aas P. A., Otterlei M., Falnes P. Ø., Vågbø C. B., Skorpen F., Akbari M., et al. (2003). Human and Bacterial Oxidative Demethylases Repair Alkylation Damage in Both RNA and DNA. Nature 421 (6925), 859–863. doi:10.1038/nature01363
Ahmad W., Ijaz B., Shabbiri K., Ahmed F., Rehman S. (2017). Oxidative Toxicity in Diabetes and Alzheimer's Disease: Mechanisms behind ROS/RNS Generation. J. Biomed. Sci. 24 (1), 76. doi:10.1186/s12929-017-0379-z
Akash M. S. H., Rehman K., Liaqat A. (2018). Tumor Necrosis Factor‐Alpha: Role in Development of Insulin Resistance and Pathogenesis of Type 2 Diabetes Mellitus. J. Cel. Biochem. 119 (1), 105–110. doi:10.1002/jcb.26174
Bae Y. S., Oh H., Rhee S. G., Yoo Y. D. (2011). Regulation of Reactive Oxygen Species Generation in Cell Signaling. Mol. Cell 32 (6), 491–509. doi:10.1007/s10059-011-0276-3
Barregard L., Møller P., Henriksen T., Mistry V., Koppen G., Rossner P., et al. (2013). Human and Methodological Sources of Variability in the Measurement of Urinary 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine. Antioxid. Redox Signal. 18 (18), 2377–2391. doi:10.1089/ars.2012.4714
Basturea G. N., Zundel M. A., Deutscher M. P. (2011). Degradation of Ribosomal RNA during Starvation: Comparison to Quality Control during Steady-State Growth and a Role for RNase PH. RNA 17 (2), 338–345. doi:10.1261/rna.2448911
Bloch-Damti A., Bashan N. (2005). Proposed Mechanisms for the Induction of Insulin Resistance by Oxidative Stress. Antioxid. Redox Signal. 7 (11-12), 1553–1567. doi:10.1089/ars.2005.7.1553
Broedbaek K., Siersma V., Henriksen T., Weimann A., Petersen M., Andersen J. T., et al. (2013). Association between Urinary Markers of Nucleic Acid Oxidation and Mortality in Type 2 Diabetes. Diabetes Care 36 (3), 669–676. doi:10.2337/dc12-0998
Broedbaek K., Siersma V., Henriksen T., Weimann A., Petersen M., Andersen J. T., et al. (2011). Urinary Markers of Nucleic Acid Oxidation and Long-Term Mortality of Newly Diagnosed Type 2 Diabetic Patients. Diabetes Care 34 (12), 2594–2596. doi:10.2337/dc11-1620
Brønden A., Larsen E. L., Karstoft K., Henriksen T., Vilsbøll T., Poulsen H. E., et al. (2020). Changes in Oxidative Nucleic Acid Modifications and Inflammation Following One-Week Treatment with the Bile Acid Sequestrant Sevelamer: Two Randomised, Placebo-Controlled Trials. J. Diabetes its Complications 34 (2), 107446. doi:10.1016/j.jdiacomp.2019.107446
Cai D., Yuan M., Frantz D. F., Melendez P. A., Hansen L., Lee J., et al. (2005). Local and Systemic Insulin Resistance Resulting from Hepatic Activation of IKK-β and NF-κB. Nat. Med. 11 (2), 183–190. doi:10.1038/nm1166
Cech T. R., Steitz J. A. (2014). The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 157 (1), 77–94. doi:10.1016/j.cell.2014.03.008
Chao M.-R., Evans M. D., Hu C.-W., Ji Y., Møller P., Rossner P., et al. (2021). Biomarkers of Nucleic Acid Oxidation - A Summary State-Of-The-Art. Redox Biol. 42, 101872. doi:10.1016/j.redox.2021.101872
Chen H.-W., Rainey R. N., Balatoni C. E., Dawson D. W., Troke J. J., Wasiak S., et al. (2006). Mammalian Polynucleotide Phosphorylase Is an Intermembrane Space RNase that Maintains Mitochondrial Homeostasis. Mol. Cel Biol. 26 (22), 8475–8487. doi:10.1128/mcb.01002-06
Chiou C. C., Chang P. Y., Chan E. C., Wu T. L., Tsao K. C., Wu J. T. (2003). Urinary 8-Hydroxydeoxyguanosine and its Analogs as DNA Marker of Oxidative Stress: Development of an ELISA and Measurement in Both Bladder and Prostate Cancers. Clin. Chim. Acta 334 (1-2), 87–94. doi:10.1016/s0009-8981(03)00191-8
Commoner B., Townsend J., Pake G. E. (1954). Free Radicals in Biological Materials. Nature 174 (4432), 689–691. doi:10.1038/174689a0
Cooke D. W., Lane M. D. (1999). The Transcription Factor Nuclear Factor I Mediates Repression of the GLUT4 Promoter by Insulin. J. Biol. Chem. 274 (18), 12917–12924. doi:10.1074/jbc.274.18.12917
Cooke M. S., Patel K., Ahmad J., Holloway K., Evans M. D., Lunec J. (2001). Monoclonal Antibody to Single-Stranded DNA: A Potential Tool for DNA Repair Studies. Biochem. Biophysical Res. Commun. 284 (1), 232–238. doi:10.1006/bbrc.2001.4954
Copps K. D., White M. F. (2012). Regulation of Insulin Sensitivity by Serine/Threonine Phosphorylation of Insulin Receptor Substrate Proteins IRS1 and IRS2. Diabetologia 55 (10), 2565–2582. doi:10.1007/s00125-012-2644-8
Das S. K., Bhutia S. K., Sokhi U. K., Dash R., Azab B., Sarkar D., et al. (2011). Human Polynucleotide Phosphorylase (hPNPaseold-35): An Evolutionary Conserved Gene with an Expanding Repertoire of RNA Degradation Functions. Oncogene 30 (15), 1733–1743. doi:10.1038/onc.2010.572
Deutscher M. P. (2006). Degradation of RNA in Bacteria: Comparison of mRNA and Stable RNA. Nucleic Acids Res. 34 (2), 659–666. doi:10.1093/nar/gkj472
Dinglay S., Gold B., Sedgwick B. (1998). Repair in Escherichia C alkB Mutants of Abasic Sites and 3-Methyladenine Residues in DNA. Mutat. Research/DNA Repair 407 (2), 109–116. doi:10.1016/s0921-8777(97)00065-7
Dubois-Deruy E., Peugnet V., Turkieh A., Pinet F. (2020). Oxidative Stress in Cardiovascular Diseases. Antioxidants 9 (9), 864. doi:10.3390/antiox9090864
Durrer Schutz D., Busetto L., Dicker D., Farpour-Lambert N., Pryke R., Toplak H., et al. (2019). European Practical and Patient-Centred Guidelines for Adult Obesity Management in Primary Care. Obes. Facts 12 (1), 40–66. doi:10.1159/000496183
Evans J. L., Maddux B. A., Goldfine I. D. (2005). The Molecular Basis for Oxidative Stress-Induced Insulin Resistance. Antioxid. Redox Signal. 7 (7-8), 1040–1052. doi:10.1089/ars.2005.7.1040
Fan Y.-G., Guo T., Han X.-R., Liu J.-L., Cai Y.-T., Xue H., et al. (2019). Paricalcitol Accelerates BACE1 Lysosomal Degradation and Inhibits Calpain-1 Dependent Neuronal Loss in APP/PS1 Transgenic Mice. EBioMedicine 45, 393–407. doi:10.1016/j.ebiom.2019.07.014
Fernández A., Ordóñez R., Reiter R. J., González‐Gallego J., Mauriz J. L. (2015). Melatonin and Endoplasmic Reticulum Stress: Relation to Autophagy and Apoptosis. J. Pineal Res. 59 (3), 292–307. doi:10.1111/jpi.12264
Fiala E. S., Conaway C. C., Mathis J. E. (1989). Oxidative DNA and RNA Damage in the Livers of Sprague-Dawley Rats Treated with the Hepatocarcinogen 2-Nitropropane. Cancer Res. 49 (20), 5518–5522.
Fimognari C. (2015). Role of Oxidative RNA Damage in Chronic-Degenerative Diseases. Oxidative Med. Cell Longev. 2015, 358713. doi:10.1155/2015/358713
Franzke B., Schober-Halper B., Hofmann M., Oesen S., Tosevska A., Henriksen T., et al. (2018). Age and the Effect of Exercise, Nutrition and Cognitive Training on Oxidative Stress - the Vienna Active Aging Study (VAAS), a Randomized Controlled Trial. Free Radic. Biol. Med. 121, 69–77. doi:10.1016/j.freeradbiomed.2018.04.565
Fu D., Brophy J. A. N., Chan C. T. Y., Atmore K. A., Begley U., Paules R. S., et al. (2010). Human AlkB Homolog ABH8 Is a tRNA Methyltransferase Required for Wobble Uridine Modification and DNA Damage Survival. Mol. Cel Biol. 30 (10), 2449–2459. doi:10.1128/mcb.01604-09
Glenn K., Klarich D. S., Kalaba M., Figueroa A., Hooshmand S., Kern M., et al. (2018). Effects of Watermelon Powder and L-Arginine Supplementation on Azoxymethane-Induced Colon Carcinogenesis in Rats. Nutr. Cancer 70 (6), 938–945. doi:10.1080/01635581.2018.1490782
Gong X., Tao R., Li Z. (2006). Quantification of RNA Damage by Reverse Transcription Polymerase Chain Reactions. Anal. Biochem. 357 (1), 58–67. doi:10.1016/j.ab.2006.06.025
Gutierrez-Mariscal F. M., Perez-Martinez P., Delgado-Lista J., Yubero-Serrano E. M., Camargo A., Delgado-Casado N., et al. (2012). Mediterranean Diet Supplemented with Coenzyme Q10 Induces Postprandial Changes in P53 in Response to Oxidative DNA Damage in Elderly Subjects. Age 34 (2), 389–403. doi:10.1007/s11357-011-9229-1
Haigis M. C., Yankner B. A. (2010). The Aging Stress Response. Mol. Cel 40 (2), 333–344. doi:10.1016/j.molcel.2010.10.002
Han J., Back S. H., Hur J., Lin Y.-H., Gildersleeve R., Shan J., et al. (2013). ER-Stress-Induced Transcriptional Regulation Increases Protein Synthesis Leading to Cell Death. Nat. Cel Biol. 15 (5), 481–490. doi:10.1038/ncb2738
Hayakawa H., Fujikane A., Ito R., Matsumoto M., Nakayama K. I., Sekiguchi M. (2010). Human Proteins that Specifically Bind to 8-Oxoguanine-Containing RNA and Their Responses to Oxidative Stress. Biochem. Biophysical Res. Commun. 403 (2), 220–224. doi:10.1016/j.bbrc.2010.11.011
Hayakawa H., Hofer A., Thelander L., Kitajima S., Cai Y., Oshiro S., et al. (1999). Metabolic Fate of Oxidized Guanine Ribonucleotides in Mammalian Cells. Biochemistry 38 (12), 3610–3614. doi:10.1021/bi982361l
Hayakawa H., Kuwano M., Sekiguchi M. (2001). Specific Binding of 8-Oxoguanine-Containing RNA to Polynucleotide Phosphorylase Protein. Biochemistry 40 (33), 9977–9982. doi:10.1021/bi010595q
Hayakawa H., Sekiguchi M. (2006). Human Polynucleotide Phosphorylase Protein in Response to Oxidative Stress. Biochemistry 45 (21), 6749–6755. doi:10.1021/bi052585l
Hayakawa H., Uchiumi T., Fukuda T., Ashizuka M., Kohno K., Kuwano M., et al. (2002). Binding Capacity of Human YB-1 Protein for RNA Containing 8-Oxoguanine. Biochemistry 41 (42), 12739–12744. doi:10.1021/bi0201872
Hofer T., Badouard C., Bajak E., Ravanat J.-L., Mattsson Å., Cotgreave I. A. (2005). Hydrogen Peroxide Causes Greater Oxidation in Cellular RNA Than in DNA. Biol. Chem. 386 (4), 333–337. doi:10.1515/bc.2005.040
Hofer T., Seo A. Y., Prudencio M., Leeuwenburgh C. (2006). A Method to Determine RNA and DNA Oxidation Simultaneously by HPLC-ECD: Greater RNA Than DNA Oxidation in Rat Liver after Doxorubicin Administration. Biol. Chem. 387 (1), 103–111. doi:10.1515/bc.2006.014
Holley R. W., Apgar J., Everett G. A., Madison J. T., Marquisee M., Merrill S. H., et al. (1965). Structure of a Ribonucleic Acid. Science 147 (3664), 1462–1465. doi:10.1126/science.147.3664.1462
Honda K., Smith M. A., Zhu X., Baus D., Merrick W. C., Tartakoff A. M., et al. (2005). Ribosomal RNA in Alzheimer Disease Is Oxidized by Bound Redox-Active Iron. J. Biol. Chem. 280 (22), 20978–20986. doi:10.1074/jbc.M500526200
Hurrle S., Hsu W. H. (2017). The Etiology of Oxidative Stress in Insulin Resistance. Biomed. J. 40 (5), 257–262. doi:10.1016/j.bj.2017.06.007
Hussey S. E., McGee S. L., Garnham A., McConell G. K., Hargreaves M. (2012). Exercise Increases Skeletal Muscle GLUT4 Gene Expression in Patients with Type 2 Diabetes. Diabetes Obes. Metab. 14 (8), 768–771. doi:10.1111/j.1463-1326.2012.01585.x
Ishibashi T., Hayakawa H., Ito R., Miyazawa M., Yamagata Y., Sekiguchi M. (2005). Mammalian Enzymes for Preventing Transcriptional Errors Caused by Oxidative Damage. Nucleic Acids Res. 33 (12), 3779–3784. doi:10.1093/nar/gki682
Jaacks L. M., Siegel K. R., Gujral U. P., Narayan K. M. V. (2016). Type 2 Diabetes: A 21st Century Epidemic. Best Pract. Res. Clin. Endocrinol. Metab. 30 (3), 331–343. doi:10.1016/j.beem.2016.05.003
Jacobs A. C., Resendiz M. J. E., Greenberg M. M. (2010). Direct Strand Scission from a Nucleobase Radical in RNA. J. Am. Chem. Soc. 132 (11), 3668–3669. doi:10.1021/ja100281x
Jorgensen A., Siersma V., Davidsen A. S., Weimann A., Henriksen T., Poulsen H. E., et al. (2018). Markers of DNA/RNA Damage from Oxidation as Predictors of a Registry-Based Diagnosis of Psychiatric Illness in Type 2 Diabetic Patients. Psychiatry Res. 259, 370–376. doi:10.1016/j.psychres.2017.11.017
Jørs A., Lund M. A. V., Jespersen T., Hansen T., Poulsen H. E., Holm J.-C. (2020). Urinary Markers of Nucleic Acid Oxidation Increase with Age, Obesity and Insulin Resistance in Danish Children and Adolescents. Free Radic. Biol. Med. 155, 81–86. doi:10.1016/j.freeradbiomed.2020.05.009
Kaina B., Christmann M., Naumann S., Roos W. P. (2007). MGMT: Key Node in the Battle against Genotoxicity, Carcinogenicity and Apoptosis Induced by Alkylating Agents. DNA Repair 6 (8), 1079–1099. doi:10.1016/j.dnarep.2007.03.008
Kajimoto Y., Kaneto H. (2004). Role of Oxidative Stress in Pancreatic β-Cell Dysfunction. Ann. N. Y Acad. Sci. 1011, 168–176. doi:10.1007/978-3-662-41088-2_17
Keane K. N., Cruzat V. F., Carlessi R., de Bittencourt P. I. H., Newsholme P. (2015). Molecular Events Linking Oxidative Stress and Inflammation to Insulin Resistance and β-Cell Dysfunction. Oxidative Med. Cell Longevity 2015, 1–15. doi:10.1155/2015/181643
Kertesz M., Wan Y., Mazor E., Rinn J. L., Nutter R. C., Chang H. Y., et al. (2010). Genome-Wide Measurement of RNA Secondary Structure in Yeast. Nature 467 (7311), 103–107. doi:10.1038/nature09322
Kjaer L. K., Oellgaard J., Henriksen T., Gaede P., Pedersen O., Poulsen H. E. (2018). Indicator of RNA Oxidation in Urine for the Prediction of Mortality in Patients with Type 2 Diabetes and Microalbuminuria: A Post-Hoc Analysis of the Steno-2 Trial. Free Radic. Biol. Med. 129, 247–255. doi:10.1016/j.freeradbiomed.2018.09.030
Kjær L. K., Cejvanovic V., Henriksen T., Petersen K. M., Hansen T., Pedersen O., et al. (2017). Cardiovascular and All-Cause Mortality Risk Associated with Urinary Excretion of 8-oxoGuo, a Biomarker for RNA Oxidation, in Patients with Type 2 Diabetes: A Prospective Cohort Study. Diabetes Care 40 (12), 1771–1778. doi:10.2337/dc17-1150
Klip A., McGraw T. E., James D. E. (2019). Thirty Sweet Years of GLUT4. J. Biol. Chem. 294 (30), 11369–11381. doi:10.1074/jbc.REV119.008351
Koch T. R., Shope T. R. (2021). Laparoscopic Vertical Sleeve Gastrectomy as a Treatment Option for Adults with Diabetes Mellitus. Adv. Exp. Med. Biol. 1307, 299–320. doi:10.1007/5584_2020_487
Kohen R., Gati I. (2000). Skin Low Molecular Weight Antioxidants and Their Role in Aging and in Oxidative Stress. Toxicology 148 (2-3), 149–157. doi:10.1016/s0300-483x(00)00206-7
Kong Q., Lin C.-l. G. (2010). Oxidative Damage to RNA: Mechanisms, Consequences, and Diseases. Cell. Mol. Life Sci. 67 (11), 1817–1829. doi:10.1007/s00018-010-0277-y
Kozak J., Jonak K., Maciejewski R. (2020). The Function of miR-200 Family in Oxidative Stress Response Evoked in Cancer Chemotherapy and Radiotherapy. Biomed. Pharmacother. 125, 110037. doi:10.1016/j.biopha.2020.110037
Kuboki K., Iso K., Murakami E., Abe S., Araki E., Ueshiba H., et al. (2007). Effects of Valsartan on Inflammatory and Oxidative Stress Markers in Hypertensive, Hyperglycemic Patients: An Open-Label, Prospective Study. Curr. Ther. Res. 68 (5), 338–348. doi:10.1016/j.curtheres.2007.10.008
Kushner S. R. (2002). mRNA Decay in Escherichia C Comes of Age. J. Bacteriol. 184 (17), 4658–4665. doi:10.1128/jb.184.17.4658-4665.2002
Larsen E. L., Poulsen H. E., Michaelsen C., Kjær L. K., Lyngbæk M., Andersen E. S., et al. (2020). Differential Time Responses in Inflammatory and Oxidative Stress Markers after a marathon: An Observational Study. J. Sports Sci. 38 (18), 2080–2091. doi:10.1080/02640414.2020.1770918
Larsen E. L., Weimann A., Poulsen H. E. (2019). Interventions Targeted at Oxidatively Generated Modifications of Nucleic Acids Focused on Urine and Plasma Markers. Free Radic. Biol. Med. 145, 256–283. doi:10.1016/j.freeradbiomed.2019.09.030
Le Bras M., Clément M. V., Pervaiz S., Brenner C. (2005). Reactive Oxygen Species and the Mitochondrial Signaling Pathway of Cell Death. Histol. Histopathol 20 (1), 205–219. doi:10.14670/hh-20.205
Lee Y. H., White M. F. (2004). Insulin Receptor Substrate Proteins and Diabetes. Arch. Pharm. Res. 27 (4), 361–370. doi:10.1007/bf02980074
Lemmer I. L., Willemsen N., Hilal N., Bartelt A. (2021). A Guide to Understanding Endoplasmic Reticulum Stress in Metabolic Disorders. Mol. Metab. 47, 101169. doi:10.1016/j.molmet.2021.101169
Li Z., Malla S., Shin B., Li J. M. (2014). Battle against RNA Oxidation: Molecular Mechanisms for Reducing Oxidized RNA to Protect Cells. WIREs RNA 5 (3), 335–346. doi:10.1002/wrna.1214
Li Z., Wu J., Deleo C. (2006). RNA Damage and Surveillance under Oxidative Stress. IUBMB Life 58 (10), 581–588. doi:10.1080/15216540600946456
Liang J., Wu S. Y., Zhang D., Wang L., Leung K. K., Leung P. S. (2016). NADPH Oxidase-Dependent Reactive Oxygen Species Stimulate β-Cell Regeneration through Differentiation of Endocrine Progenitors in Murine Pancreas. Antioxid. Redox Signal. 24 (8), 419–433. doi:10.1089/ars.2014.6135
Liu X., Gan W., Zou Y., Yang B., Su Z., Deng J., et al. (2016). Elevated Levels of Urinary Markers of Oxidative DNA and RNA Damage in Type 2 Diabetes with Complications. Oxidative Med. Cell Longevity 2016, 4323198. doi:10.1155/2016/4323198
Liu Z., Chen X., Li Z., Ye W., Ding H., Li P., et al. (2020). Role of RNA Oxidation in Neurodegenerative Diseases. Int. J. Mol. Sci. 21 (14), 5022. doi:10.3390/ijms21145022
Loft S., Vistisen K., Ewertz M., Tjønneland A., Overvad K., Poulsen H. E. (1992). Oxidative DNA Damage Estimated by 8-Hydroxydeoxyguanosine Excretion in Humans: Influence of Smoking, Gender and Body Mass index. Carcinogenesis 13 (12), 2241–2247. doi:10.1093/carcin/13.12.2241
Looi M.-L., Mohd Dali A. Z. H., Md Ali S. A., Wan Ngah W. Z., Mohd Yusof Y. A. (2008). Oxidative Damage and Antioxidant Status in Patients with Cervical Intraepithelial Neoplasia and Carcinoma of the Cervix. Eur. J. Cancer Prev. 17 (6), 555–560. doi:10.1097/CEJ.0b013e328305a10b
Lucks J. B., Mortimer S. A., Trapnell C., Luo S., Aviran S., Schroth G. P., et al. (2011). Multiplexed RNA Structure Characterization with Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension Sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. U.S.A. 108 (27), 11063–11068. doi:10.1073/pnas.1106501108
Luo J., Gonsalves G., Greene J. (2019). Insulin for All: Treatment Activism and the Global Diabetes Crisis. Lancet 393 (10186), 2116–2117. doi:10.1016/s0140-6736(19)31090-6
Majzunova M., Dovinova I., Barancik M., Chan J. Y. (2013). Redox Signaling in Pathophysiology of Hypertension. J. Biomed. Sci. 20 (1), 69. doi:10.1186/1423-0127-20-69
Masselli E., Pozzi G., Vaccarezza M., Mirandola P., Galli D., Vitale M., et al. (2020). ROS in Platelet Biology: Functional Aspects and Methodological Insights. Int. J. Mol. Sci. 21 (14), 4866. doi:10.3390/ijms21144866
McArdle A., Jackson M. J. (2019). An Introduction to a Special Issue of Free Radical Biology and Medicine - "Reactive Oxygen Species and Musculoskeletal Aging". Free Radic. Biol. Med. 132, 1–2. doi:10.1016/j.freeradbiomed.2018.12.038
McKinlay A., Gerard W., Fields S. (2012). Global Analysis of RNA Oxidation in Saccharomyces C. Biotechniques 52 (2), 109–111. doi:10.2144/000113801
Medda N., De S. K., Maiti S. (2021). Different Mechanisms of Arsenic Related Signaling in Cellular Proliferation, Apoptosis and Neo-Plastic Transformation. Ecotoxicology Environ. Saf. 208, 111752. doi:10.1016/j.ecoenv.2020.111752
Nakamura A., Shikata K., Nakatou T., Kitamura T., Kajitani N., Ogawa D., et al. (2013). Combination Therapy with an Angiotensin-Converting-Enzyme Inhibitor and an Angiotensin II Receptor Antagonist Ameliorates Microinflammation and Oxidative Stress in Patients with Diabetic Nephropathy. J. Diabetes Invest. 4 (2), 195–201. doi:10.1111/jdi.12004
Nakatani Y., Kaneto H., Kawamori D., Yoshiuchi K., Hatazaki M., Matsuoka T.-A., et al. (2005). Involvement of Endoplasmic Reticulum Stress in Insulin Resistance and Diabetes. J. Biol. Chem. 280 (1), 847–851. doi:10.1074/jbc.M411860200
Newsholme P., Cruzat V. F., Keane K. N., Carlessi R., de Bittencourt P. I. H. (2016). Molecular Mechanisms of ROS Production and Oxidative Stress in Diabetes. Biochem. J. 473 (24), 4527–4550. doi:10.1042/bcj20160503c
Nishikawa T., Edelstein D., Du X. L., Yamagishi S.-I., Matsumura T., Kaneda Y., et al. (2000). Normalizing Mitochondrial Superoxide Production Blocks Three Pathways of Hyperglycaemic Damage. Nature 404 (6779), 787–790. doi:10.1038/35008121
Noller H. F., Chaires J. B. (1972). Functional Modification of 16S Ribosomal RNA by Kethoxal. Proc. Natl. Acad. Sci. U.S.A. 69 (11), 3115–3118. doi:10.1073/pnas.69.11.3115
Nunomura A., Moreira P. I., Castellani R. J., Lee H.-g., Zhu X., Smith M. A., et al. (2012). Oxidative Damage to RNA in Aging and Neurodegenerative Disorders. Neurotox Res. 22 (3), 231–248. doi:10.1007/s12640-012-9331-x
Nunomura A., Perry G., Hirai K., Aliev G., Takeda A., Chiba S., et al. (1999). Neuronal RNA Oxidation in Alzheimer's Disease and Down's Syndrome. Ann. NY Acad Sci 893, 362–364. doi:10.1111/j.1749-6632.1999.tb07855.x
Obata T., Yamanaka Y., Kinemuchi H., Oreland L. (2001). Release of Dopamine by Perfusion with 1-Methyl-4-Phenylpyridinium Ion (MPP(+)) into the Striatum Is Associated with Hydroxyl Free Radical Generation. Brain Res. 906 (1-2), 170–175. doi:10.1016/s0006-8993(01)02238-7
Ogawa S., Kobori H., Ohashi N., Urushihara M., Nishiyama A., Mori T., et al. (2009). Angiotensin II Type 1 Receptor Blockers Reduce Urinary Angiotensinogen Excretion and the Levels of Urinary Markers of Oxidative Stress and Inflammation in Patients with Type 2 Diabetic Nephropathy. Biomark Insights 4, 97–102. doi:10.4137/bmi.s2733
Ogurtsova K., da Rocha Fernandes J. D., Huang Y., Linnenkamp U., Guariguata L., Cho N. H., et al. (2017). IDF Diabetes Atlas: Global Estimates for the Prevalence of Diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 128, 40–50. doi:10.1016/j.diabres.2017.03.024
Oyama J.-I., Maeda T., Sasaki M., Kozuma K., Ochiai R., Tokimitsu I., et al. (2010). Green tea Catechins Improve Human Forearm Vascular Function and Have Potent Anti-Inflammatory and Anti-Apoptotic Effects in Smokers. Intern. Med. 49 (23), 2553–2559. doi:10.2169/internalmedicine.49.4048
Özcan U., Cao Q., Yilmaz E., Lee A.-H., Iwakoshi N. N., Özdelen E., et al. (2004). Endoplasmic Reticulum Stress Links Obesity, Insulin Action, and Type 2 Diabetes. Science 306 (5695), 457–461. doi:10.1126/science.1103160
Pan Y., Qiao Q., Pan L., Zhou D., Hu C., Gu H., et al. (2015). Losartan Reduces Insulin Resistance by Inhibiting Oxidative Stress and Enhancing Insulin Signaling Transduction. Exp. Clin. Endocrinol. Diabetes 123 (3), 170–177. doi:10.1055/s-0034-1395658
Peattie D. A., Gilbert W. (1980). Chemical Probes for Higher-Order Structure in RNA. Proc. Natl. Acad. Sci. U.S.A. 77 (8), 4679–4682. doi:10.1073/pnas.77.8.4679
Pessler D., Rudich A., Bashan N. (2001). Oxidative Stress Impairs Nuclear Proteins Binding to the Insulin Responsive Element in the GLUT4 Promoter. Diabetologia 44 (12), 2156–2164. doi:10.1007/s001250100024
Piwowarski J., Grzechnik P., Dziembowski A., Dmochowska A., Minczuk M., Stepien P. P. (2003). Human Polynucleotide Phosphorylase, hPNPase, Is Localized in Mitochondria. J. Mol. Biol. 329 (5), 853–857. doi:10.1016/s0022-2836(03)00528-x
Porte D., Kahn S. E. (2001). Beta-Cell Dysfunction and Failure in Type 2 Diabetes: Potential Mechanisms. Diabetes 50 (Suppl. 1), S160–S163. doi:10.2337/diabetes.50.2007.s160
Poulsen H. E., Specht E., Broedbaek K., Henriksen T., Ellervik C., Mandrup-Poulsen T., et al. (2012). RNA Modifications by Oxidation: A Novel Disease Mechanism? Free Radic. Biol. Med. 52 (8), 1353–1361. doi:10.1016/j.freeradbiomed.2012.01.009
Poulsen H. E., Weimann A., Henriksen T., Kjær L. K., Larsen E. L., Carlsson E. R., et al. (2019). Oxidatively Generated Modifications to Nucleic Acids In Vivo: Measurement in Urine and Plasma. Free Radic. Biol. Med. 145, 336–341. doi:10.1016/j.freeradbiomed.2019.10.001
Radak Z., Kumagai S., Nakamoto H., Goto S. (2007). 8-Oxoguanosine and Uracil Repair of Nuclear and Mitochondrial DNA in Red and white Skeletal Muscle of Exercise-Trained Old Rats. J. Appl. Physiol. 102 (4), 1696–1701. doi:10.1152/japplphysiol.01051.2006
Rains J. L., Jain S. K. (2011). Oxidative Stress, Insulin Signaling, and Diabetes. Free Radic. Biol. Med. 50 (5), 567–575. doi:10.1016/j.freeradbiomed.2010.12.006
Reno C. M., Puente E. C., Sheng Z., Daphna-Iken D., Bree A. J., Routh V. H., et al. (2017). Brain GLUT4 Knockout Mice Have Impaired Glucose Tolerance, Decreased Insulin Sensitivity, and Impaired Hypoglycemic Counterregulation. Diabetes 66 (3), 587–597. doi:10.2337/db16-0917
Resendiz M. J. E., Pottiboyina V., Sevilla M. D., Greenberg M. M. (2012). Direct Strand Scission in Double Stranded RNA via a C5-Pyrimidine Radical. J. Am. Chem. Soc. 134 (8), 3917–3924. doi:10.1021/ja300044e
Rochette L., Zeller M., Cottin Y., Vergely C. (2014). Diabetes, Oxidative Stress and Therapeutic Strategies. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1840 (9), 2709–2729. doi:10.1016/j.bbagen.2014.05.017
Saliminejad K., Khorram Khorshid H. R., Soleymani Fard S., Ghaffari S. H. (2019). An Overview of microRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cel Physiol 234 (5), 5451–5465. doi:10.1002/jcp.27486
Sankaran N. (2016). The RNA World at Thirty: A Look Back with its Author. J. Mol. Evol. 83 (5-6), 169–175. doi:10.1007/s00239-016-9767-3
Schöttker B., Xuan Y., Gào X., Anusruti A., Brenner H. (2020). Oxidatively Damaged DNA/RNA and 8-Isoprostane Levels Are Associated with the Development of Type 2 Diabetes at Older Age: Results from a Large Cohort Study. Diabetes Care 43 (1), 130–136. doi:10.2337/dc19-1379
Sekiguchi T., Ito R., Hayakawa H., Sekiguchi M. (2013). Elimination and Utilization of Oxidized Guanine Nucleotides in the Synthesis of RNA and its Precursors. J. Biol. Chem. 288 (12), 8128–8135. doi:10.1074/jbc.M112.418723
Seok H., Lee H., Lee S., Ahn S. H., Lee H.-S., Kim G.-W. D., et al. (2020). Position-Specific Oxidation of miR-1 Encodes Cardiac Hypertrophy. Nature 584 (7820), 279–285. doi:10.1038/s41586-020-2586-0
Shah S., Iqbal M., Karam J., Salifu M., McFarlane S. I. (2007). Oxidative Stress, Glucose Metabolism, and the Prevention of Type 2 Diabetes: Pathophysiological Insights. Antioxid. Redox Signal. 9 (7), 911–929. doi:10.1089/ars.2007.1629
Shan X., Chang Y., Glenn Lin C. L. (2007). Messenger RNA Oxidation Is an Early Event Preceding Cell Death and Causes Reduced Protein Expression. FASEB j. 21 (11), 2753–2764. doi:10.1096/fj.07-8200com
Shan X., Lin C.-L. G. (2006). Quantification of Oxidized RNAs in Alzheimer's Disease. Neurobiol. Aging 27 (5), 657–662. doi:10.1016/j.neurobiolaging.2005.03.022
Shan X., Tashiro H., Lin C.-l. G. (2003). The Identification and Characterization of Oxidized RNAs in Alzheimer's Disease. J. Neurosci. 23 (12), 4913–4921. doi:10.1523/jneurosci.23-12-04913.2003
She H., Mao Z. (2011). Regulation of Myocyte Enhancer Factor-2 Transcription Factors by Neurotoxins. Neurotoxicology 32 (5), 563–566. doi:10.1016/j.neuro.2011.05.019
Shen Z., Wu W., Hazen S. L. (2000). Activated Leukocytes Oxidatively Damage DNA, RNA, and the Nucleotide Pool through Halide-Dependent Formation of Hydroxyl Radical. Biochemistry 39 (18), 5474–5482. doi:10.1021/bi992809y
Shigiyama F., Kumashiro N., Miyagi M., Ikehara K., Kanda E., Uchino H., et al. (2017). Effectiveness of Dapagliflozin on Vascular Endothelial Function and Glycemic Control in Patients with Early-Stage Type 2 Diabetes Mellitus: DEFENCE Study. Cardiovasc. Diabetol. 16 (1), 84. doi:10.1186/s12933-017-0564-0
Shokri Y., Variji A., Nosrati M., Khonakdar-Tarsi A., Kianmehr A., Kashi Z., et al. (2020). Importance of Paraoxonase 1 (PON1) as an Antioxidant and Antiatherogenic Enzyme in the Cardiovascular Complications of Type 2 Diabetes: Genotypic and Phenotypic Evaluation. Diabetes Res. Clin. Pract. 161, 108067. doi:10.1016/j.diabres.2020.108067
Skytte M. J., Samkani A., Astrup A., Larsen T. M., Frystyk J., Poulsen H. E., et al. (2020). Effects of a Highly Controlled Carbohydrate-Reduced High-Protein Diet on Markers of Oxidatively Generated Nucleic Acid Modifications and Inflammation in Weight Stable Participants with Type 2 Diabetes; a Randomized Controlled Trial. Scand. J. Clin. Lab. Invest. 80 (5), 401–407. doi:10.1080/00365513.2020.1759137
Sova H., Puistola U., Morin-Papunen L., Karihtala P. (2013). Metformin Decreases Serum 8-Hydroxy-2′-Deoxyguanosine Levels in Polycystic Ovary Syndrome. Fertil. Sterility 99 (2), 593–598. doi:10.1016/j.fertnstert.2012.10.013
Sung K.-C., Lee M. Y., Kim Y.-H., Huh J. H., Kim J.-Y., Wild S. H., et al. (2018). Obesity and Incidence of Diabetes: Effect of Absence of Metabolic Syndrome, Insulin Resistance, Inflammation and Fatty Liver. Atherosclerosis 275, 50–57. doi:10.1016/j.atherosclerosis.2018.05.042
Takagi Y., Setoyama D., Ito R., Kamiya H., Yamagata Y., Sekiguchi M. (2012). Human MTH3 (NUDT18) Protein Hydrolyzes Oxidized Forms of Guanosine and Deoxyguanosine Diphosphates. J. Biol. Chem. 287 (25), 21541–21549. doi:10.1074/jbc.M112.363010
Tweedie C., Romestaing C., Burelle Y., Safdar A., Tarnopolsky M. A., Seadon S., et al. (2011). Lower Oxidative DNA Damage Despite Greater ROS Production in Muscles from Rats Selectively Bred for High Running Capacity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300 (3), R544–R553. doi:10.1152/ajpregu.00250.2010
Underwood J. G., Uzilov A. V., Katzman S., Onodera C. S., Mainzer J. E., Mathews D. H., et al. (2010). FragSeq: Transcriptome-wide RNA Structure Probing Using High-Throughput Sequencing. Nat. Methods 7 (12), 995–1001. doi:10.1038/nmeth.1529
Valko M., Leibfritz D., Moncol J., Cronin M. T. D., Mazur M., Telser J. (2007). Free Radicals and Antioxidants in normal Physiological Functions and Human Disease. Int. J. Biochem. Cel Biol. 39 (1), 44–84. doi:10.1016/j.biocel.2006.07.001
Villaño D., Vilaplana C., Medina S., Cejuela-Anta R., Martínez-Sanz J. M., Gil P., et al. (2015). Effect of Elite Physical Exercise by Triathletes on Seven Catabolites of DNA Oxidation. Free Radic. Res. 49 (8), 973–983. doi:10.3109/10715762.2015.1025388
Wang J., Wang H. (2017). Oxidative Stress in Pancreatic Beta Cell Regeneration. Oxid Med. Cel Longev 2017, 1930261. doi:10.1155/2017/1930261
Wang W.-X., Luo S.-B., Jiang P., Xia M.-M., Hei A.-l., Mao Y.-H., et al. (2017). Increased Oxidative Damage of RNA in Early-Stage Nephropathy in Db/db Mice. Oxidative Med. Cell Longevity 2017, 2353729. doi:10.1155/2017/2353729
Wang J. X., Gao J., Ding S. L., Wang K., Jiao J. Q., Wang Y., et al. (2015). Oxidative Modification of miR-184 Enables It to Target Bcl-xL and Bcl-w. Mol. Cell 59 (1), 50–61. doi:10.1016/j.molcel.2015.05.003
Wang Y., Ye S., Hu Y., Zhao L., Zheng M. (2013). The Effect of Hydrochloride Pioglitazone on Urinary 8-hydroxy -deoxyguanosine Excretion in Type 2 Diabetics. J. Diabetes its Complications 27 (1), 75–77. doi:10.1016/j.jdiacomp.2012.08.004
Watson J. D. (2014). Type 2 Diabetes as a Redox Disease. The Lancet 383 (9919), 841–843. doi:10.1016/s0140-6736(13)62365-x
White M. G., Shaw J. A. M., Taylor R. (2016). Type 2 Diabetes: The Pathologic Basis of Reversible β-Cell Dysfunction. Diabetes Care 39 (11), 2080–2088. doi:10.2337/dc16-0619
Wu J., Li Z. (2008). Human Polynucleotide Phosphorylase Reduces Oxidative RNA Damage and Protects HeLa Cell against Oxidative Stress. Biochem. Biophysical Res. Commun. 372 (2), 288–292. doi:10.1016/j.bbrc.2008.05.058
Xie W., Ma L. L., Xu Y. Q., Wang B. H., Li S. M. (2019). METTL3 Inhibits Hepatic Insulin Sensitivity via N6-Methyladenosine Modification of Fasn mRNA and Promoting Fatty Acid Metabolism. Biochem. Biophysical Res. Commun. 518 (1), 120–126. doi:10.1016/j.bbrc.2019.08.018
Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S. (2010). Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance. Cel Metab. 11 (6), 467–478. doi:10.1016/j.cmet.2010.04.005
Yaribeygi H., Farrokhi F. R., Butler A. E., Sahebkar A. (2019). Insulin Resistance: Review of the Underlying Molecular Mechanisms. J. Cell Physiol. 234 (6), 8152–8161. doi:10.1002/jcp.27603
Keywords: oxidative damage, RNA oxidation, 8-oxoGuo, type 2 diabetes, therapeutic strategy
Citation: Chen X, Yu H, Li Z, Ye W, Liu Z, Gao J, Wang Y, Li X, Zhang L, Alenina N, Bader M, Ding H, Li P and Aung LHH (2022) Oxidative RNA Damage in the Pathogenesis and Treatment of Type 2 Diabetes. Front. Physiol. 13:725919. doi: 10.3389/fphys.2022.725919
Received: 16 June 2021; Accepted: 11 March 2022;
Published: 28 March 2022.
Edited by:
Murugesan Velayutham, West Virginia University, United StatesReviewed by:
Vittorio Calabrese, University of Catania, ItalyLucia Coppo, Karolinska Institutet (KI), Sweden
Copyright © 2022 Chen, Yu, Li, Ye, Liu, Gao, Wang, Li, Zhang, Alenina, Bader, Ding, Li and Aung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peifeng Li, cGVpZmxpQHFkdS5lZHUuY24=; Lynn Htet Htet Aung, bHlubmF1bmdAcWR1LmVkdS5jbg==
 Xiatian Chen
Xiatian Chen Hua Yu3
Hua Yu3 Zhe Li
Zhe Li Ziqian Liu
Ziqian Liu Natalia Alenina
Natalia Alenina Michael Bader
Michael Bader Peifeng Li
Peifeng Li Lynn Htet Htet Aung
Lynn Htet Htet Aung