
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 22 December 2022
Sec. Invertebrate Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1107045
This article is part of the Research TopicThe Side Effects of Insecticides on Insects and the Adaptation Mechanisms of Insects to InsecticidesView all 15 articles
In recent years, the leaf beetle Galeruca daurica has broken out in the northern grasslands of Inner Mongolia, its management still mainly depends on chemical control using traditional insecticides or with novel action. The study was aim to identify mutation locus associated with resistance to diamide insecticides in field population of G. daurica, to provide a reference for rational selection of insecticides and to avoid the rapid resistance development to diamide insecticides. We cloned the full length of the ryanodine receptor gene of G. daurica (GdRyR), constructed 3D model and transmembrane regions by homologous modeling based on deduced amino acid sequence. Two potential mutation loci (Gly4911Glu and Ile4754Met) and allelic mutation frequencies were detected in individuals of G. daurica. In addition, their binding patterns to two diamide insecticides (chlorantraniliprole, cyantraniliprole) were analyzed separately using a molecular docking method. The full-length cDNA sequence of GdRyR (GenBank accession number: OP828593) was obtained by splicing and assembling, which is 15,399 bp in length and encodes 5,133 amino acids. The amino acid similarity of GdRyR with that of other Coleopteran insects were 86.70%–91.33%, which possessed the typical structural characteristics. An individual resistance allelic mutation frequency test on fifty field leaf beetles has identified 12% and 32% heterozygous individuals at two potential mutation loci Gly4911Glu and Ile4754Met, respectively. The affinity of the I4754M mutant model of GdRyR for chlorantraniliprole and cyantraniliprole was not significantly different from that of the wild type, and all had non-covalent interactions such as hydrogen bonding, hydrophobic interactions and π-cation interactions. However, the G4911E mutant model showed reduced affinity and reduced mode of action with two diamide insecticides, thus affecting the binding stability of the ryanodine receptor to the diamide insecticides. In conclusion, the G4911E mutation in GdRyR may be a potential mechanism for the development of resistance to diamide insecticides on G. daurica and should be a key concern for resistance risk assessment and reasonable applications of diamide insecticides for control in future. Moreover, this study could provide a reference for ryanodine receptor structure-based insecticides design.
Galeruca daurica (Coleoptera: Chrysomelidae), a serious grassland pest since its sudden outbreaks on the Inner Mongolia grasslands in 2009 (Zhou et al., 2016, Zhou et al., 2019), has continually spread and caused great losses to pasture on the Inner Mongolia grasslands (Ma et al., 2019, Ma et al., 2021), and levels of damage has increased year by year (Duan et al., 2022). The application of traditional chemical insecticides to control outbreaking pests in desert steppes has the advantages of a good insecticidal effect with rapid results, especially organophosphorus, pyrethroids, chloronicotinyl insecticide (Dong et al., 2016; Gao et al., 2022). Currently, the management of G. daurica is dependent on above-mentioned insecticides (Chang et al., 2015; Zhang et al., 2022). Besides, plant-derived insecticides have been reported to provide good protection against the key pest G. daurica, such as: neem, matrine and nicotine analogues (Chang et al., 2015; Du et al., 2016). In recent years, the chemical insecticides with novel action, high efficiency, high selectivity, low toxicity and low residue are being chosen to control this outbreaking insect in desert steppe. Among them, chlorantraniliprole has been reported to have strong insecticidal activity against the G. daurica and recommended for pest management (Zhang et al., 2022).
Diamide insecticides are a new class of highly effective, broad-spectrum, low toxicity, good systemic absorption and highly selective insecticides with O-amido benzamide chemical structure, mainly with stomach toxicity and high effectiveness against Coleoptera, Diptera and Lepidoptera insects (Tohnishi et al., 2005; Temple et al., 2009; Hughes et al., 2013; Huang et al., 2021). The main products currently on the market include: Chlorantraniliprole, Fluorobenzamide, Cyantraniliprole, Cyclaniliprole, Tetrachlorantraniliprole, Broflanilide, and Tetraniliprole (Sparks and Nauen, 2014). These chemical ligands can open calcium channels in insects by binding to the Ryanodine receptor (RyR), causing the freeing of large amounts of Ca2+ from the sarcoplasmic reticulum and endoplasmic reticulum, therefore, a significant reduction in the Ca2+ content triggers an imbalance in the insect’s internal environment, resulting in the cessation of feeding, functional muscle disorders, neurological disorders, and even death (Sattelle et al., 2008; Jeanguenat, 2013; Guo et al., 2021). RyR is a calcium channel protein distributed in the endoplasmic reticulum and sarcoplasmic reticulum of animal myocytes, and consists of a homotetrameric structure with four subunits containing 5,000 amino acid residues (Amador et al., 2009). Currently, diamide insecticides have been widely used against key agricultural insects, such as: diamond back moth Plutella xylostella (Guo et al., 2014, Guo et al., 2021), beet armyworm Spodoptera exigua (Zuo et al., 2017), Spodoptera frugiperda (Lv et al., 2021), rice stem borer Chilo suppressalis (Gao et al., 2013; Huang et al., 2020) and so on. Among them, chlorantraniliprole and flubendiamide were reported the most widely-used diamide insecticides with high insecticidal activity, low toxicity and safety to mammals and humans (Troczka et al., 2015).
In recent years, it has been continuously reported that the overuse of diamide insecticides led to the occurrence of resistance, with the resistance mechanism of the diamond back moth P. xylostella as a lepidopteran model pest being more intensively studied. Troczka et al. (2015) reported that the G4946E mutation in RyR was detected in resistant P. xylostella populations in Thailand and the Philippines, and was confirmed to affect the binding of diamide insecticides to RyR by in vitro expression in Sf9 cells. More recent studies have demonstrated that the I4790M/K mutation in PxRyR is associated with resistance to diamide insecticides in field populations of diamond back moth (Guo et al., 2014; Jouraku et al., 2020; Wang et al., 2020; Jiang et al., 2021; Richardson et al., 2021). It was reported that the high frequencies of mutations were found among six Chinese field populations collected from 2016 to 2019 resulting in RyR I4743M substitutions on Spodoptera exigua, however, no significant correlation was found between chlorantraniliprole resistance level and SeRyR I4743M allele frequency but CYP9A186 F116V substitutions (Teng et al., 2022). Boaventura et al. (2019) reported that a I4734M mutation of the FAW ryanodine receptor (RyR) C-terminal domains II to VI has recently been described to confer target-site resistance to diamides in Lepidopteran insect Spodoptera frugiperda. Among the reported mutant loci of the insect ryanodine receptor involving resistance to diamide insecticides, G4946E and I4790M on P. xylostella are the classical allele substitutions with the highest frequency of resistance, corresponding to G4911 and I4754 of RyR in G. daurica, respectively. It has not been reported that resistance occurrence, resistance levels of diamide insecticides on G. daurica, whether there are amino acid changes in GdRyR in field populations and whether they may contribute to resistance to diamide insecticides.
With the development of bioinformatics and computer simulation technology, protein homology modeling, molecular docking and other technical tools are becoming increasingly sophisticated. Novel ideas for studying pest resistance mechanisms to insecticides are proposed by predicting the binding pattern of receptors and ligands (Chen et al., 2021). Homology modeling uses the crystal structure of a protein molecule with high homology to its amino acid sequence as a template for sequence alignment, and predicts the structure of an unknown, homologous protein from the conserved domain of known crystal protein molecule (Boris and Ke, 2022). At present, the crystal structure of the GdRyR protein has not been identified, but the structure analysis of some mammalian ryanodine receptors is more mature and can be used as a reference approach for structural simulation and molecular docking (Ma et al., 2020). The deduced amino acid sequence was obtained from the cloned GdRyR gene sequence, and homology modeling and molecular docking were applied to construct the 3D model and analyze its affinity conformation, respectively.
In the study, we cloned the full-length GdRyR gene, examined the two classic mutation allele frequency of GdRyR associated with resistance to diamide insecticides in individuals of field populations of G. daurica, and applied homology modeling to construct a three-dimensional model of GdRyR and its transmembrane region. The binding patterns of GdRyR and diamide insecticides were studied by molecular docking, and the relationship between potential mutation sites of GdRyR and resistance was analyzed. The aim of this study was to explore the mechanism of resistance to diamide insecticides from the perspective of structural biology, and to lay a theoretical foundation for further analysis of the resistance mechanism of diamide insecticides.
The overwintering eggs of G. daurica were collected from the Xianghuang Banner grasslands (44°62′N, 115°80′E) of Inner Mongolia, China on September 2021, were brought back to the Research Center for Grassland Entomology (40°48′N, 111°42′E), Inner Mongolia Agricultural University (Hohhot, Inner Mongolia, China), and were maintained in intelligent light artificial incubator (PRX-350C, Ningbo HaiShuSaiFu experimental instrument Co., Ltd.) (25°C ± 1°C, RH = 70 ± 10%, L14:D10). After hatching out, the larvae were fed on Allium mongolicum in a incubator under the same conditions just as described by Tan et al. (2018). The healthy third instar larvae in consistent physiological state were collected for further experiments.
Bioassays were conducted using a leaf-dip method based on methods of He et al. (2012) and Zhang et al. (2022) with minor modifications. 95% chlorantraniliprole (DuPont) was dissolved in acetone solution, and then diluted into five to six concentrations with distilled water containing .05% Triton X-100 at a triple gradient dose. The .05% Triton X-100 solution was used as blank control. The fresh leek cultured in the laboratory measuring 5 cm × .5 cm were immersed in the prepared various concentrations of chlorantraniliprole for 15 s, and taken out in ventilated cupboard to air dry and then placed into a Petri dish lined with filter paper. Twenty healthy 2-day third instar larvae were placed into each Petri dish with six pieces of leek leaves with four replications for toxicity assessment bioassays. Mortality was assessed after 48 h of exposure, larvae that did not move when gently pushed with a fine hair brush were considered dead. The control groups was below 5%, mortality was corrected using Abbott’s formula (Abbott, 1925). The LC50 value and slope were calculated by regression probit analysis conducted with the POLO-Plus software (2002).
Sequences annotated as ryanodine receptor genes were screened from the transcriptome data published in Genbank (Bioproject No. 785282), and specific primers listed in Table 1 were designed for segmental cloning. The Total RNA was extracted from the whole body using RNAiso reagent (TaKaRa, Dalian, China) based on the manufacturer’s instructions, cDNA was synthesized by reverse transcription using the PrimeScript® RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) and as the PCR templates. Total PCR reaction system (50 μl) included cDNA template (2 μl), forward and reverse primers (1 μl, 0.2 μmol/L), amplification enzyme mix (25 μl), RNAse Free ddH2O (21 μl). 2 × Vazyme LAmp® Master Mix was purchased online from Vazyme Co., Ltd. and used for high fidelity and rapid PCR, the annealing velocity is high to 1 kb/s. PCR amplification conditions were performed as follows: 98°C for 3 min, followed by 35 cycles of 98°C for 10 s, 60°C for X s (X: each primer used a different annealing time), and 72°C for 3 min. Finally, it was extended for 5–10 min at 72°C (Table 1). The expected-size fragments were purified, ligated into pMD19-T vector (TaKaRa, code: D102A), and positive transformants were selected for plasmid isolation using MiniBEST Plasmid Purification Kit (TaKaRa, Dalian, China) and sent to Sangon Biotech company for sequencing. The fragments were overlapped and aligned with the annotated GdRyR gene identified from the transcriptome database using DNAMAN software, and the spliced complete CDS sequence of the GdRyR gene was submitted to the NCBI database, the Genbank accession number is OP828593.
ORF Finder was applied to search the open reading frame of the GdRyR gene, and sequence integrity was verified by NCBI’s Blastx tool to predict the gene isoelectric point, molecular mass, atomic composition, protein structural domain, signal peptide, and transmembrane region. Phylogenetic tree was constructed using the BLAST tool and DNAMAN V6.0 software to align amino acid sequence homology, using the neighbor-joining method in MEGA software and repeatedly running 1,000 times.
The total RNA was isolated from individuals of G. daurica population collected from the Xianghuang Banner grasslands for cDNA preparation and PCR. The two pairs of specific primers containing the potential mutation sites G4911 and I4754 were designed based on the sequence of the C-terminal transmembrane region (Table 1). The specific bands obtained by PCR amplification were purified and sent to company for sequencing, and the sequences from different individuals were compared in the point mutation region to detect the frequency of allelic mutation sites.
The GdRyR amino acid sequence was submitted to the SWISS-MODEL server, and the predicted rabbit RyR cryo-electron microscopy structure (PDB: 7CF9) with 61.8% amino acid sequence homology to GdRyR reported by Ma et al. (2020) was used as a template to construct a model of GdRyR using Modeller 10.2. The Align2D module was used to compare GdRyR with the template sequence, and then the Model-single module was used to build 100 candidate models. The stability of the models was measured by the Dope parameter, and the model with the lowest energy was selected as the optimal model for homology modeling, which was named as WT GdRyR. To obtain more accurate 3D structure for molecular docking, the amino acid sequence of the C-terminus of GdRyR was submitted on the AlphaFold2 platform on the Wemol online website, and the AlphaFold2 tool was used to construct a protein model of the C-terminus of GdRyR (WT GdRyR-C). The 3D structures of protein crystals obtained from Modeller and AlphaFold2 simulations were verified for stereochemical plausibility and energetic stability using the Molprobity tool. The rationality of the Pis and Phi angles between residues was demonstrated by the Ramachandran Plot, and the rationality of the protein model was evaluated based on the percentage of optimal regions, generally allowed regions, etc. in the Ramachandran Plot (Williams et al., 2018).
The amino acid sequence of GdRyR was compared with domains containing mutation loci for P. xylostella RyR (G4946E, I4790M), S. frugiperda for (G4900E, I4743M) and S. exigua (G4891E, I4743M) (Zou et al., 2019), which correspond to G4900E, I4754M mutation sites of GdRyR. Mutation of “Ile” was replaced by “Met” in I4754 in GdRyR using AlphaFold2, the mutation model GdRyR-C-M4754 was constructed. Similarly, “Gly” was substituted into “Glu” in G4900 in GdRyR, and the mutation model GdRyR-C-E4900 was constructed. The models WT GdRyR-C, GdRyR-C-M4754 and GdRyR-C-E4911were processed separately using Autodock Tool-1.5.7 for energy minimization and dehydrogenation as molecular docking receptors.
The 3D structures of chlorantraniliprole and cyanobacteriamide were downloaded from the PubChem website, and converted into a suitable format for molecular docking using Open Babel-2.4.1, and used as ligands for molecular docking using Autodock Tool-1.5.6 for energy minimization etc.
Based on the reported binding sites of the rabbit RyR complex crystal structure with chlorantraniliprole, the active pockets of the constructed 3D model were predicted and the docking-box was constructed. The box parameters used for the docking of GdRyRs were as follows: center-x = 8.18, center-y = −1.56, center-z = .877; size-x = 99.75, size-y = 99.75, size-z = 99.75. The processed protein receptors were docked to the ligands using Autodock vina1.1.2 software. Computing platform was as follows: Microsoft-PC: Intel(R) Core(TM) i5-1035G4CPU@1.10GHz 1.50 GHz.
By molecular docking, the two ligands were selected for optimal conformation to construct complexes with the ryanodine receptor, and the binding modes were analyzed using the PLIP online tool, including: hydrogen bonding, hydrophobic interactions, π-π, π-cation, halogen bonding and non-covalent interaction forces such as interactions. Pymol-2.1.0 was used to visualize the binding modes.
Homologous sequence alignment: https://blast.ncbi.nlm.nih.gov/Blast.cgi; Predicting protein properties such as gene isoelectric point, molecular mass and atomic composition: ProtParam online tool; Protein domain prediction: https://www.ebi.ac.uk/interpro/result/InterProScan/(InterProscan); Signal peptide prediction: http://www.cbs.dtu.dk/services/SigRyRlP/; Homologous modeling: https://swissmodel.expasy.org/(SWISS-MODEL) (Waterhouse et al., 2018); Protein quality evaluation: https://Main page-MolProbity (duke.edu) (Molprobity) (Williams et al., 2018); Protein crystal structure PDB data: http://www1.rcsb.org/; NCBI databse: https://www.ncbi.nlm.nih.gov/; Transmembrane region prediction (TMHMM): http://www.cbs.dtu.dk/services/TMHMM/(Möller et al., 2001); Action force analysis tools (PLIP): https://projects.biotec.tu-dresden.de/plip-web/plip (Melissa et al., 2021); Small molecular crystal structure: PubChem (nih.gov).
MEGA X was applied to construct phylogenetic tree (Kumar et al., 2018); Open Babel-2.4.1 was used to convert the file format of the chemical structure type (O’Boyle et al., 2011); The 3D protein crystal structure was visualized by Pymol-2.1.0 (Delano, 2002); Modeller (Version-10.2) was used for homologous modeling (Chen et al., 2021); Autodock Tool-1.5.7 (Morris et al., 2009) and Autodock vina 1.1.2 (Trott and Olson., 2010) were used for molecular docking; AlphaFold2 was applied for deep learning modeling (Jumper et al., 2021).
Linear regression of the dose–mortality relationship (Y = −2.557 ± 2.291 X) was fitted to the observed data (i.e. no significant deviation between the observed and the expected data; χ2 = 1.81, df = 3) and LC50 was considered valid (inf lim < LC50 < sup lim: 21.25 < 44.10 mg/L < 80.33).
Based on the GdRyR sequence information in the transcriptome, a total of four target fragments were amplified. After overlapped, a complete open reading frame (ORF) of 15,399 bp, encoding 5,133 amino acids, was obtained by BlastX alignment and sequence splicing. The molecular weight of the protein is 582.53 kDa, and the isoelectric point (pI) is 5.51. The GdRyR consists of five elements: carbon (C), hydrogen (H), Nitrogen (N), Oxygen (O) and Sulphur (S), and the total number of atoms is 81,490, the chemical formula is C258925H40538N6992O7799S236, it has six transmembrane structure regions and no signal peptide. As shown in Figure 1, the N-terminal part of the gene contains one MIR domain (Mannosyltransferase, IP3R and RyR) located between amino acid sites 220-397, two RIH domains (RyR and IP3R Homology) located at amino acid sites 447-642 and 2254-2484, respectively, and three SPRY domains (SPla and RyR) located at amino acid sites 664-802, 1091-1214 and 1563-1706, respectively. The gene also has four RyR domains located at amino acid sites 854-944, 967-1056, 2861-2952, 2979-3062, and one highly conserved RIH-associated domain located at amino acid sites 4021–4137 before the transmembrane helix; The C-terminus has six transmembrane helices (TM1 to TM6) located at amino acid sites 4474-4496, 4658-4680, 4739-4761, 4881-4903, 4929-4951, and 5009-5028, respectively.
The amino acid sequences encoded by the GdRyR gene were blasted with RyR of other insect species researched in the NCBI database, and a phylogenetic tree was constructed using the neighbour-joining (NJ) method. Figure 2 showed that GdRyR has the highest homology with RyR of potato beetle Leptinotarsa decemlineata (GenBank Accession No: QZZ63290.1), and amino acid sequence identity is 91.33%. The amino acid homology with Hypothenemus hampei RyR (QEE14187.1) and Tribolium castaneum RyR (NP 001308588.1) were 88.05% and 86.70%, respectively, and the RyR of the four Coleopteran species clustered as one clade. The clustering results indicated that the GdRyR gene is highly similar to taxonomically similar insect taxa.
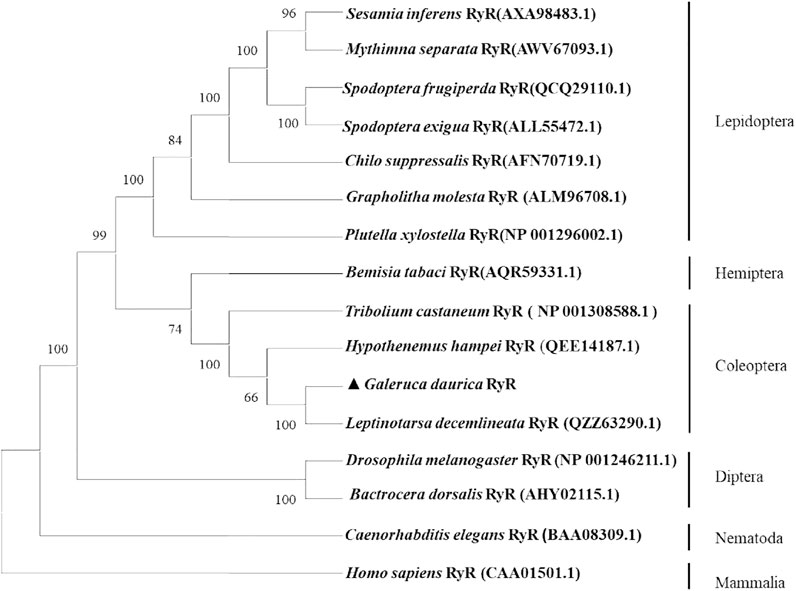
FIGURE 2. Phylogenetic tree of ryanodine receptor amino acid sequences from G. daurica and other insects. Note: The tree was generated by MEGA X using the Neighbor-Joning (NJ) method, using model p-distance. The topology was tested using bootstrap analyses (1,000 replicates). Numbers at nodes are bootstrap values.
The two potential allelic mutation sites and the occurrence frequency of Gly4911Glu and Ile4754Met were examined by individual extraction of RNA, using reverse-transcribed cDNA as a template, respectively. Fifty beetles collected from the grasslands of Xilingol Xianghuang Banner, Inner Mongolia, were conducted to gene sequence testing, the mutation loci and allelic frequencies of the GdRyR are summarized in Table 2. The results showed that only the heterozygous genotype G4911E was present in the test population, accounting for approximately 12% of the population; the heterozygous and homozygous genotypes of I4754M were present in 32% and 2% of the population, respectively.
The Modeller and AlphaFold2 are applied to construct the GdRyR 3D crystal structure (WT GdRyR), the C-terminal wild-type 3D structure of GdRyR (WT GdRyR-C) and the mutant structure (GdRyR-C-M4754) (GdRyR-C-E4911), respectively. Figure 3A showed the 3D structure of GdRyR consists of multiple α-helices, β-folds, β-turns, irregular curls, and some extended extension structures. In lateral view, the protein crystal structure resembles the letter “Y,” with a wide N-terminus and a narrow C-terminus, separated by a small angle at the C-terminus. As it can be seen from Figures 3C, E, G that the 3D structure of the C-terminal region of GdRyR is mainly composed of several parallel sets of α-helices, exhibiting obvious transmembrane protein properties. The 3D structural model of Ramachandran Plot diagram and the test results are shown in Figure 3 and Table 3, respectively. The percentages of residues in the optimal regions for WT GdRyR, WT GdRyR-C and GdRyR-C-M4754 and GdRyR-C-E4911 were 86.9%, 88.3%, 87.8%, and 88.1%, respectively. The proportions of amino acid residues in the acceptable region were 95.8%, 94.6%, 94.9%, and 94.6% respectively, all are greater than 90%, indicating that the constructed models were all reasonable.
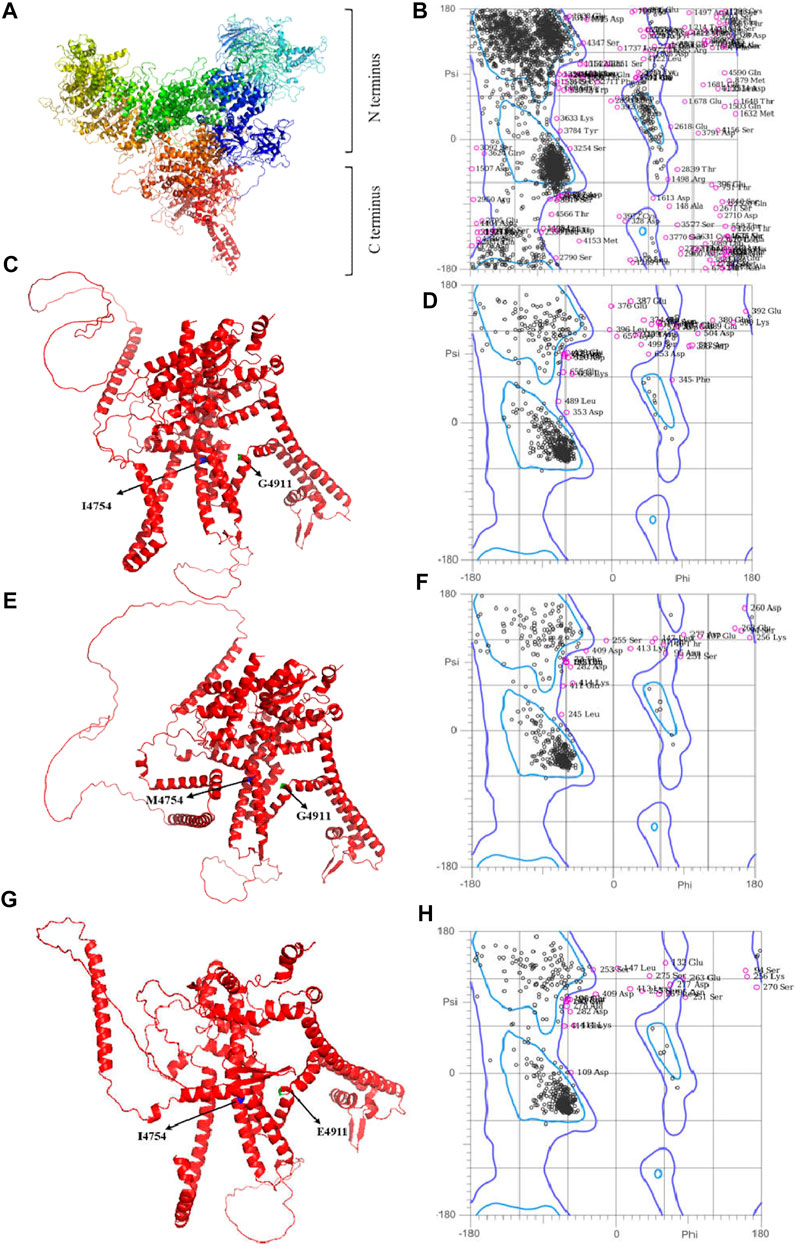
FIGURE 3. The models and evaluation results of WT GdRyR, WT GdRyR-C, GdRyR-C-M4754, and GdRyR-C-E4911. Note: (A) Three-dimension RyR model of WT GdRyR; (B) The result of Molprobity evaluation of WT GdRyR; (C) Three-dimension RyR model of the C-terminal transmembrane region of WT GdRyR-C; (D) The result of Molprobity evaluation of WT GdRyR-C; (E) Three-dimension RyR model of GdRyR-C-M4954; (F) The results of Molprobity evaluation of GdRyR-C-M4954. (G) Three-dimension RyR model of GdRyR-C-E4911; (H) The results of Molprobity evaluation of GdRyR-C-E4911.
The binding affinities of WT GdRyR-C to the optimal conformation of chlorantraniliprole and cyantraniliprole were −31.38, −33. 89 kJ/mol, respectively. The binding pattern was analyzed using PLIP and drawn by Pymol. As Figure 4 shows: Y4660 (Tyr-220), K4663 (Lys-223) form a hydrophobic interaction with the benzene and pyrazole rings of chlorantraniliprole, K4663 forms a hydrogen bond with the oxygen atom on the carbonyl group, K4762 (Lys-322) forms a hydrogen bond with the nitrogen atom of the pyrazole ring, G4911 (Gly-471) forms a hydrogen bond with the NH of the methylamino structure on the benzene ring, R4916 (Arg-476) forms a hydrogen bond with the nitrogen atom of the pyridine ring (Figure 4A). Y4660 forms a hydrophobic interaction with the methyl group on the benzene ring of cyantraniliprole, K4762 forms a hydrogen bond with the nitrogen atom of the pyrazole ring, G4911 forms a hydrogen bond with the NH of the amide structure on the side of the pyrazole ring, the nitrogen atom of R4916 forms a π-cation with the pyrazole ring of cyantraniliprole (Figure 4B).
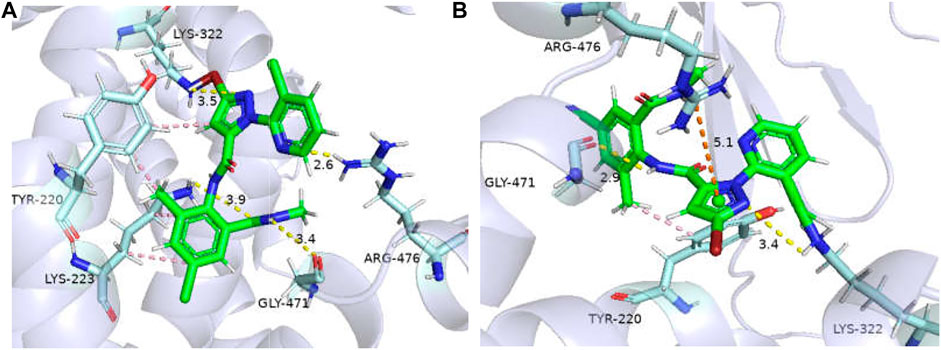
FIGURE 4. Binding modes of WT GdRyR and chlorantraniliprole (A) and cyanobacteriamide (B). Note: Hydrophobic effect: pink; Hydrogen bond: yellow; π-cation: orange; Hydrogen: white; Oxygen: red; Nitrogen: blue; Selphur: dark orange.
The binding affinities of GdRyR-M4754 to the optimal conformation of chlorantraniliprole and cyantraniliprole were −32.23, −33. 91 kJ/mol, respectively. Combined pattern analysis diagram was seen in Figure 5: Y4660 (Tyr-220) and K4663 (Lys-223) form hydrophobic interactions with the pyridine ring of chlorantraniliprole, K4913 (Lys-473) forms hydrophobic interactions with the benzene ring as well as the methyl group, Y4660 forms a hydrogen bond with the oxygen atom on the carbonyl group of the benzene ring, K4663 forms a hydrogen bond with the nitrogen atom of the pyrazole ring, R4916 (Arg-476) forms a hydrogen bond with the NH of the amide structure on the pyrazole side (Figure 5A). Y4660, V4910 (Val-470) and K4663 form hydrophobic interactions with the pyridine ring, pyrazole ring and methyl group of cyantraniliprole, respectively. K4663 forms a hydrogen bond with nitrogen on the pyridine ring, K4762 (Lys-332) forms a hydrogen bond with the nitrogen atom of the pyrazole ring, G4911 (Gly-471) forms a hydrogen bond with NH of the amide structure on the benzene ring side, R4916 forms a hydrogen bond with the oxygen atom of the carbonyl group on the pyrazole ring side, the nitrogen atom of K4663 forms a π-cation interaction with the pyridine ring of cyantraniliprole (Figure 5B).
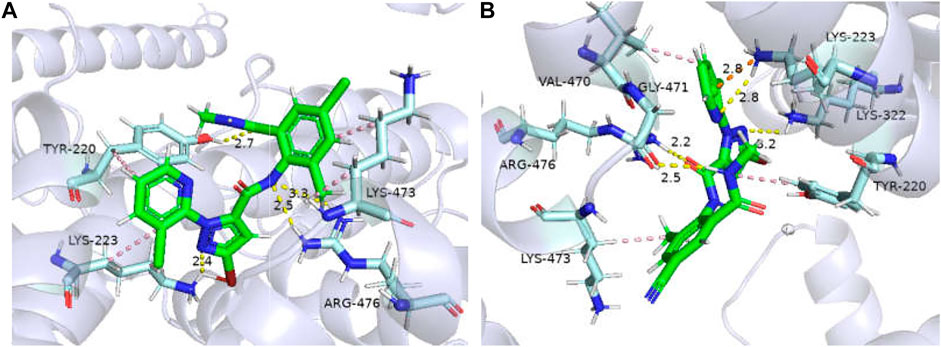
FIGURE 5. Binding modes of GdRyR-M4754 and chlorantraniliprole (A) and cyanobacteriamide (B). Note: Hydrophobic effect: pink; Hydrogen bond: yellow; π-cation: orange; Hydrogen: white; Oxygen: red; Nitrogen: blue; Selphur: dark orange.
The binding affinities of GdRyR-E4911 to the optimal conformation of chlorantraniliprole and cyantraniliprole were −25.89, −27.54 kJ/mol, respectively. Combined pattern analysis diagram was seen in Figure 6: K4762 (Lys-329) forms hydrophobic interactions with methyl of chlorantraniliprole, K4769 (Lys-322) and R4770 (Arg-330) form hydrophobic interactions with the pyridine ring and the chlorine ion on the pyridine ring, respectively. D4878 (Asp-438) forms hydrophobic interactions and hydrogen bonds with the pyrazole ring and the NH of the pyrazole side amide structure, respectively (Figure 6A). L4497 (Leu-60) and L4500 (Leu-57) form hydrophobic interactions with the methyl and benzene rings of cyantraniliprole, respectively. M4501 (Met-61) and Q4868 (Gln-428) form hydrophobic interactions with the pyridine ring, Y4759 (Tyr-341) and F4881 (Phe-441) form hydrophobic interactions with the benzene ring, Y4759 forms a hydrogen bond with the NH of the amide structure on the side of the benzene ring, Q4868 (Gln-428) forms a hydrogen bond with the nitrogen atom of the pyrazole ring (Figure 6B).
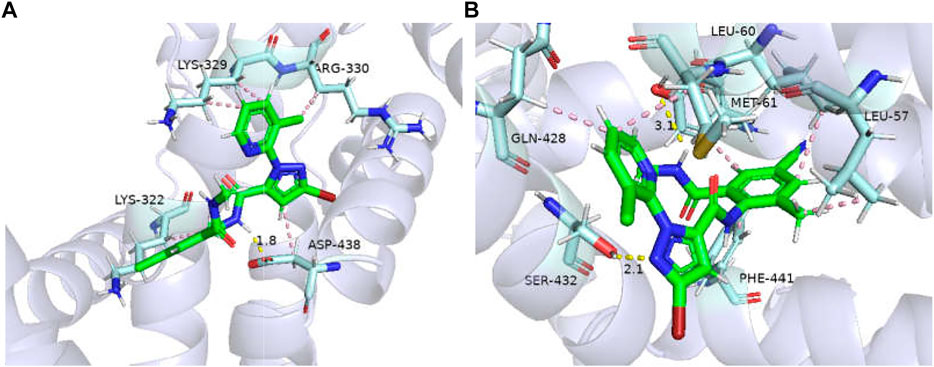
FIGURE 6. Binding modes of GdRyR-E4911 and chlorantraniliprole (A) and cyanobacteriamide (B). Note: Hydrophobic effect: pink; Hydrogen bond: yellow; π-cation: orange; Hydrogen: white; Oxygen: red; Nitrogen: blue; Selphur: dark orange.
As shown in Table 4: The mutation of isoleucine (Ile) to methionine (Met) at position 4754 in GdRyR showed no decrease in binding affinity to diamide insecticides; the mutation of glycine (Gly) to glutamic acid (Glu) at position 4911 in GdRyR showed a significant decrease in binding affinity to diamide insecticides. The binding modes of two diamide insecticides with WT GdRyR, GDRYR-M4754 and GDRYR-E4911 are shown in Figures 4–6: After mutating isoleucine (Ile, position 4754) of GdRyR to methionine (Met), the mode of interaction with diamide insecticides, the residues involved in the formation of the action force and WT GdRyR are essentially the same, mainly because the isoleucine (Ile) at position 4754 of WT GdRyR is not involved in the formation of the force action between ligands and receptors. The mutation to glutamic acid (Glu) in WT GdRyR, which has a hydrogen bonding interaction between glycine (Gly) at position 4911 and the diamide insecticide, prevents the formation of hydrogen bonds with the diamide insecticide, resulting in a reduced receptor-ligand interaction and decreased affinity. The mutations of amino acid from Gly to Glu at position 4911 in WT GdRyR may lead to resistance to diamide insecticides on G. daurica, while further studies are needed to determine whether the I4734M mutation is associated with resistance.
The insect ryanodine receptor, one of the largest Ca2+ releasing channels, has an important role in muscle excitation-contraction coupling and is an important action target of diamide insecticides. Currently, in addition to the model insect Drosophila, the RyR genes of P. xylostella (Sun et al., 2012), Cnaphalocrocis medinalis (Wang et al., 2012), Ostrinia furnacalis (Cui et al., 2013), S. exigua (Zuo et al., 2017), C. suppressalis (Gao et al., 2013; Huang et al., 2021) and S. frugiperda (Li et al., 2021) have been successfully cloned, and different mutant sites have been reported to be associated with diamide insecticide resistance. In this study, the full-length cDNA sequence of the GdRyR gene was obtained by segmental cloning technique, which was assembled by splicing. Sequence homology analysis showed that GdRyR had 86.70%, 88.05%, and 91.33% amino acid similarity with the ryanodine receptors of the other Coleopteran insects, T. castaneum (Herbst), H. hampei (Ferrari) and L. decemlineata (Say), respectively. The structural domains were predicted to have one MIR structural domain, two RIH structural domains, three SPRY structural domains, four RyR structural domains and one RIH-associated structural domain at the N-terminal part of GdRyR, and six hydrophobic transmembrane motifs at the C-terminal end of GdRyR, this result is highly similar to the reported positions of the functional structural domains of insects such as Carposina sasakii (Sun et al., 2012), C. suppressalis (Peng et al., 2017). The MIR structural domain functions as a transferring ligand in mammals, and the RIH and MIR structural domains are involved in the formation of IP3 binding sites in IP3Rs (Ponting, 2000). The SPRY structural domain is often considered to have a function in regulating protein interactions (Woo et al., 2006; Cui et al., 2009). These structural features are similar to those of Drosophila DmRyR, suggesting that the GdRyR gene can encode a functional Ca2+ channel protein (Xu et al., 2000; Bouakaz et al., 2002).
Diamide insecticides, a class of chemical insecticides acting on insect ryanodine receptors, have good efficacy against a wide range of pests and are widely used in the chemical control on Lepidoptera, Coleoptera and Diptera. In recent years, with the widespread use of diamide insecticides, many insects have developed a high level of resistance (Lai et al., 2011; Guo et al., 2014; Wang et al., 2018). Resistance mechanism studies suggested that the diamide insecticide resistance is associated with target mutation site of the ryanodine receptor (Troczka et al., 2015; Nauen and Steinbach, 2016). Field populations of S. exigua with a homozygous mutation in the ryanodine receptor gene I4743M have developed up to 154-fold higher levels of resistance to chlorantraniliprole (Zuo et al., 2019). Teng et al. (2022) found that the key mutation was I4743M in the resistant population of S. exigua in six regions of eastern China by sequencing the transmembrane region of RyR, and the mutation frequency was determined to be as high as 70%–100%. In this study, two potential mutation loci (G4911 and I4754) of the ryanodine receptor were examined in individuals from field populations of G. daurica, there were 12% of the heterozygous mutants G4911E in the population, while 32% of the heterozygous and 2% of heterozygous mutants were I4754M. The study showed that the G4946E and I4790M/K mutations in RyR were detected in resistant populations of P. xylostella (Guo et al., 2014; Troczka et al., 2015; Jouraku et al., 2020). It was confirmed that the G4946E and I4790M/K mutations could affect the binding of diamide insecticides to RyR by in vitro expression in Sf9 cells and construction of UAS-PxRyR transgenic Drosophila strain (Jouraku et al., 2020; Jiang et al., 2021; Richardson et al., 2021). Resistance to diamide insecticides in S. frugiperda is associated with mutations in the I4743M and G4891E loci of RyR (Boaventura et al., 2019). Therefore, the two amino acid sites G4911 and I4754 in GdRyR may be potential target binding sites for diamide insecticides, and mutations at the two sites could also be a potential resistance mechanism to diamide insecticides. In order to verify the inference, we analyzed the binding pattern of diamide insecticides to GdRyR and its mutants through protein homology modeling and molecular docking techniques.
The binding pattern analysis of insect RyR with diamide insecticides has been a hot topic of research in structural biology. Steinbach et al. (2015) had constructed a homology model of PxRyR, on which the G4946E and I4790M mutations were introduced, and the binding mode of diamide insecticides to PxRyR were analyzed, suggesting that the binding site of RyR to diamide insecticides may be close to the two mutation sites, but the exact binding mode, interaction force and residues forming the force are not clear. Studies on the mechanisms of resistance to diamide insecticides in Coleoptera are poorly reported. Based on the previous study, the rabbit ryanodine receptor (PDB: 7CF9) was selected as the template for constructing a homologous 3D model of GdRyR. In addition, GdRyR-E49111 and GdRyR-M4754 mutant models were constructed based on potential resistance allelic frequency detection by introducing the G4911E and I4754M mutations, and analyzed for their binding patterns to diamide insecticides. The results showed that mutating Gly of GdRyR to Glu resulted in reduced affinity to diamide insecticides. The current study showed that both of the G4946E mutation in PxRyR and the G4900E mutation in SeRyR can lead to resistance to diamide insecticides. Therefore, a substitution from Gly to Glu at 4911 position of GdRyR may lead to resistance to diamide insecticides on G. daurica. The frequency of mutations in GdRyR at G4911E should be a key concern for resistance risk assessment and reasonable applications of diamide insecticides for G. daurica control in future. The I4790M mutation of PxRyR and I4734M of SfRyR are also involved in insect resistance to diamide insecticides (Guo et al., 2014; Boaventura et al., 2019). The amino acid at position I4754 of GdRyR was not involved in its binding to diamide insecticides, and the substitution from Ile to Met may not reduce the affinity of GdRyR for diamide insecticides. Our result is similar with that of Teng et al. (2022), who reported that no significant correlation was found between chlorantraniliprole resistance level and RyR I4743M allele frequency in the six field populations of S. exigua. Therefore, the I4754M mutation may not be involved in the resistance to diamide insecticides on G. daurica, but whether the I4754M mutation can lead to resistance needs to be further explored inin vitro functional experiments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP828593. http://www.wwpdb.org/, PDB: 7CF9.
The animal subject used in this study is a kind of leaf beetle in northern steppe, which is an invertebrate and exempt from this requirement. No specific permits were required for the collection from the field and for maintenance in laboratory. This study did not involve any endangered species, protected species, or protected areas.
YTA and HR conceived research; HR, HZ, RN, WW, and YTI conducted experiment; HR, RN, YL, and LL analyzed data and conducted statistical analyses; YTA secured funding; HR, B-PP, and YTA wrote the manuscript; All authors read and approved the Manuscript.
This work was supported by the Inner Mongolian Science and Technology Key Project-2021 grant funded by the Inner Mongolian Chamber of Science and Technology (2021GG0108), Higher Education Young Scientific and Technology Talent Plan Fund (NJYT-19-B22), and the Project of Central Support for Local Development (2022-184).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbott W. S. (1925). A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18, 265–267. doi:10.1093/jee/18.2.265a
Amador F. J., Liu S., Ishiyam N., Plevin M. J., Wilson A., MacLennan D. H., et al. (2009). Crystal structure of type I ryanodine receptor amino-terminal -trefoil domain reveals a disease-associated mutation "Hot Spot" Loop. P Natl. Acad. Sci. U. S. A. 106, 11040–11044. doi:10.1073/pnas.0905186106
Boaventura D., Bolzan A., Padovez F., Okuma D., Omoto C., Nauen R. (2019). Detection of a ryanodine receptor target-site mutation in diamide insecticide resistant fall armyworm, Spodoptera frugiperda. Pest Manag. Sci. 76, 47–54. doi:10.1002/ps.5505
Boris S. Z., Ke D. (2022). Pyrethroids in an AlphaFold2 model of the insect sodium channel. Insects 13, 745. doi:10.3390/insects13080745
Bouakaz A., De J. N., Gerfault L., Cachard C. (2002). Regulation of mammalian ryanodine receptors. Front. Biosci-landmrk. 35, d2072–d2080. doi:10.2741/A899
Chang J., Zhou X. R., Li H. P., Pang B. P. (2015). Synergistic effects of metarhizium anisopliae mixed with three pesticides against Galeruca daurica. Chin. J. Pest. Sci. 17, 54–59.
Chen X. Y., Wang Y., Ma Y., Wang R. N., Zhao D. X. (2021). To explore the Radix Paeoniae Rubra-Flos Carthami herb pair's potential mechanism in the treatment of ischemic stroke by network pharmacology and molecular docking technology. Medicine 100, 27752. doi:10.1097/MD.0000000000027752
Cui L., Yang D. B., Yan X. J., Rui C., Wang Z. Y., Yuan H. Z. (2013). Molecular cloning, characterization and expression profiling of a ryanodine receptor gene in Asian corn borer, Ostrinia furnacalis (Guenee). PLOS One 8, e75825. doi:10.1371/journal.pone.0075825
Cui Y. F., Shen H. T., Norris N. C., Karunasekara Y., Pouliquin P., Board P. G., et al. (2009). A dihydropyridine receptor α 1s loop region critical for skeletal muscle contraction is intrinsically unstructured and binds to a SPRY domain of the type 1 ryanodine receptor. Int. J. Biochem. Cell. B 41, 677–686. doi:10.1016/j.biocel.2008.08.004
Dong W., Zhang X. B., Zhang X. Y., Wu H. H., Zhang M., Ma E. B., et al. (2016). Susceptibility and potential biochemical mechanism of Oedaleus asiaticus to beta-cypermethrin and deltamethrin in the Inner Mongolia, China. Pestic. Biochem. Phys. 132, 47–52. doi:10.1016/j.pestbp.2015.11.011
Du G. L., Ma C. Y., Hong J., Zhang Z. R., Li X. Y., Yu C. (2016). Occurrence trend of Galeruca daurica Joannis in grassland and control efficiency of botanical pesticides. Plant Prot. Sci. 42, 253–256.
Duan T. F., Gao S. J., Wang H. C., Li L., Li Y. Y., Tan Y., et al. (2022). MicroRNA let-7-5p targets the juvenile hormone primary response gene Krüppel homolog 1 and regulates reproductive diapause in Galeruca daurica. Insect Biochem. Molec 142, 103727. doi:10.1016/j.ibmb.2022.103727
Gao C. F., Yao R., Zhang Z. Z., Wu M., Zhang S., Su J. Y. (2013). Susceptibility baseline and chlorantraniliprole resistance monitoring in Chilo suppressalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 106, 2190–2194. doi:10.1603/ec13058
Gao S. J., Tan Y., Han H. B., Guo N., Gao H. Y., Wang N., et al. (2022). Resistance to beta-cypermethrin, azadirachtin, and matrine, and detoxification enzyme activity in Oedaleus asiaticus (Bey-Bienko) in Inner Mongolia, northern China. J. Insect Sci. 22, 1–8. doi:10.1093/jisesa/ieac063
Guo L., Li C., Coupland G., Liang P., Chu D. (2021). Up-regulation of calmodulin involved in the stress response to cyantraniliprole in the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Sci. 28, 1745–1755. doi:10.1111/1744-7917.12887
Guo L., Wang Y., Zhou X. G., Li Z. Y., Liu S. Z., Liang P., et al. (2014). Functional analysis of a point mutation in the ryanodine receptor of Plutella xylostella (L.) associated with resistance to chlorantraniliprole. Pest Manag. Sci. 70, 1083–1089. doi:10.1002/ps.3651
He Y. X., Zhao J. W., Zheng Y., Desneux N., Wu K. M. (2012). Lethal effect of imidacloprid on the coccinellid predator Serangium japonicum and sublethal effects on predator voracity and on functional response to the whitefly Bemisia taboci. Ecotoxicol. 21, 1291e1300.
Huang J. M., Rao C., Wang S., He F. L., Zhao S. Q., Zhou L. Q., et al. (2020). Multiple target-site mutations occurring in Lepidopterans confer resistance to diamide insecticides. Insect Biochem. Molec 121, 103367. doi:10.1016/j.ibmb.2020.103367
Huang J. M., Sun H., He L. F., Liu C., Ge W. c., Ni H., et al. (2021). Double ryanodine receptor mutations confer higher diamide resistance in rice stem borer, Chilo suppressalis. Chilo suppressalis 77, 4971–4979. doi:10.1002/ps.6539
Hughes K. A., Lahm G. P., Selby T. P., Stevenson T. M. (2013). Cyano anthranilamide insecticides. Patent: USPTO TOSUS8475819B2.
Jeanguenat A. (2013). The story of a new insecticidal chemistry class: The diamides. Pest Manag. Sci. 69, 7–14. doi:10.1002/ps.3406
Jiang D., Qian C., Wang D. H., Wang F. L., Zhao S., Yang Y. H., et al. (2021). Varying contributions of three ryanodine receptor point mutations to diamide insecticide resistance in Plutella xylostella. Pest Manag. Sci. 77, 4874–4883. doi:10.1002/ps.6534
Jouraku A., Kuwazaki S., Miyamoto K., Uchiyama M., Kurokawa T., Mori E., et al. (2020). Ryanodine receptor mutations (G4946E and I4790K) differentially responsible for diamide insecticide resistance in diamondback moth, Plutella xylostella L. Insect Biochem. Molec 118, 103308. doi:10.1016/j.ibmb.2019.103308
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lai T. C., Li J., Su J. Y. (2011). Monitoring of beet armyworm Spodoptera exigua (Lepidoptera: Noctuidae) resistance to chlorantraniliprole in China. Pestic. Biochem. Phys. 101, 198–205. doi:10.1016/j.pestbp.2011.09.006
Li K., Wang K., Tao L., Li L., Li J., Wu W. W., et al. (2021). Analysis of binding modes and resistance risk between ryanodine receptor of Spodoptera frugiperda and diamide insecticides. Chin. J. Pest Sci. 23, 856–868.
Lv S. L., Shi Y., Zhang J. C., Liang P., Zhang L., Gao X. W. (2021). Detection of ryanodine receptor target-site mutations in diamide insecticide-resistant Spodoptera frugiperda in chiRyR. Insect Sci. 28, 639–648. doi:10.1111/1744-7917.12896
Ma H. Y., Li Y. Y., Li L., Tan Y., Pang B. P. (2021). Juvenile hormone regulates the reproductive diapause through Methoprene-tolerant gene in Galeruca daurica. Insect Mol. Biol. 30, 446–458. doi:10.1111/imb.12710
Ma H. Y., Zhou X. R., Tan Y., Pang B. P. (2019). Proteomic analysis of adult Galeruca daurica (Coleoptera: Chrysomelidae) at different stages during summer diapause. Comp. Biochem. Phys. D. 29, 351–357. doi:10.1016/j.cbd.2019.01.007
Ma R. F., Haji-Ghassemi O., Ma D., Jiang H., Lin L. Y., Yao L., et al. (2020). Structural basis for diamide modulation of ryanodine receptor. Nat. Chem. Biol. 16, 1246–1254. doi:10.1038/s41589-020-0627-5
Melissa F. A., Katja L. L., Sarah N. B., Florian K., Sebastian S., Joachim V. H., et al. (2021). Plip 2021: Expanding the scope of the protein-ligand interaction profiler to DRYR and RRYR. Nucleic Acids Res. 49, 530–534.
Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi:10.1002/jcc.21256
Nauen R., Steinbach D. (2016). “Resistance to diamide insecticides in lepidopteran pests,” in Advances in insect control and resistance management. Editors A. R. Horowitz, and I. Ishaaya (Basel: Springer Nature Switzerland AG), 219–240.
O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R. (2011). Open Babel: An open chemical toolbox. J. Cheminformatics 3, 33. doi:10.1186/1758-2946-3-33
Peng Y. C., Sheng C. W., Casida J. E., Zhao C. Q., Han Z. J. (2017). Ryanodine receptor genes of the rice stem borer, Chilo suppressalis: Molecular cloning, alternative splicing and expression profiling. Pestic. Biochem. Phys. 135, 69–77. doi:10.1016/j.pestbp.2016.06.002
Ponting C. P. (2000). Novel repeats in ryanodine and IP 3 receptors and protein O-mannosyltransferases. Trends Biochem. Sci. 25, 48–50. doi:10.1016/s0968-0004(99)01513-3
Richardson W., Homem R. A., Troczka B. J., George C. H., Ebbinghaus-Kintscher U., Williamson M. S., et al. (2021). Diamide insecticide resistance in transgenic Drosophila and Sf9-cells expressing a full-length diamondback moth ryanodine receptor carrying an I4790M mutation. Pest Manag. Sci. 78, 869–880. doi:10.1002/ps.6730
Sattelle D. B., Cordova D., Cheek T. R. (2008). Insect ryanodine receptors: Molecular targets for novel pest control chemicals. Invertebr. Neurosci. 8, 107–119. doi:10.1007/s10158-008-0076-4
Sparks T. C., Nauen R. (2014). Irac: Mode of action classification and insecticide resistance management. Pestic. Biochem. Phys. 121, 122–128. doi:10.1016/j.pestbp.2014.11.014
Steinbach D., Gutbrod O., Lummen P., Matthiesen S., Schorn C., Nauen R. (2015). Geographics spread, genetics and functional characteristics of ryanodine based targer-site resistance to diamide insecticides in diamondback moth, Plutella xylostella. Insect Biochem. Mol. Biol. 63, 14–22.
Sun L. N., Cui L., Rui C. H., Yan X. J., Yang D. B., Yuan H. Z. (2012). Modulation of the expression of ryanodine receptor from Plutella xylostella as a result of diamide insecticide application. Gene 511, 265–273. doi:10.1016/j.gene.2012.09.027
Tan Y., Zhang Y., Huo Z. J., Zhou X. R., Pang B. P. (2018). Molecular cloning of heat shock protein 10(Hsp10) and 60 (Hsp60) cDNAs from Galeruca daurica (Coleoptera: Chrysomelidae) and their expression analysis. B Entomol. Res. 108, 510–522. doi:10.1017/S0007485317001079
Temple J. H., Pommireddy P. L., Cokk D. R., Leonard B. R. (2009). Arthropod M AnAgeMent susceptibility of selected Lepidopteran pests to rynaxypyr®, a novel insecticide. J. Cotton Sci. 13, 23–31.
Teng H. Y., Zuo Y. Y., Yuan J., Fabrick J. A., Wu Y. D., Yang Y. H. (2022). High frequency of ryanodine receptor and cytochrome P450 CYP9A186 mutations in insecticide-resistant field populations of Spodoptera exigua from China. Pestic. Biochem. Phys. 186, 105153. doi:10.1016/j.pestbp.2022.105153
Tohnishi M., Nakao H., Furuya T., Seo A., Kodama H., Tsubata K., et al. (2005). Flubendiamide, a novel insecticide highly active against lepidopterous insect pests. J. Pestic. Sci. 30, 354–360. doi:10.1584/jpestics.30.354
Troczka B. J., Williams A. J., Williams M. A., Field L. M., Luemmen P., Davies T. G. E. (2015). Stable expression and functional characterisation of the diamondback moth ryanodine receptor G4946E variant conferring resistance to diamide insecticides. Sci. Rep-uk. 5, 14680. doi:10.1038/srep14680
Trott O., Olson A. J. (2010). AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Wang J. J., Li Y. Q., Han Z. J., Zhu Y. L., Xie Z. J., Wang J., et al. (2012). Molecular characterization of a ryanodine receptor gene in the rice leaffolder, Cnaphalocrocis medinalis (Guenée). PLOS One 7, e36623. doi:10.1371/journal.pone.0036623
Wang R., Wang J. D., Che W. N., Luo C. (2018). First report of field resistance to cyantraniliprole, a new anthranilic diamide insecticide, on Bemisia tabaci MED in China. J. Integr. Agric. 17, 158–163. doi:10.1016/s2095-3119(16)61613-1
Wang X. L., Cao X. W., Jiang D., Yang Y. M., Wu Y. D. (2020). CRISPR/Cas9 mediated ryanodine receptor I4790M knock-in confers unequal resistance to diamides in Plutella xylostella. Insect Biochem. Molec 125, 103453. doi:10.1016/j.ibmb.2020.103453
Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296-W303–W303. doi:10.1093/nar/gky427
Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. doi:10.1002/pro.3330
Woo J. S., Suh H. Y., Park S. Y., Oh B. H. (2006). Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell. 24, 967–976. doi:10.1016/j.molcel.2006.11.009
Xu X., Bhat M. B., Nishi M., Takeshima H., Ma J. (2000). Molecular Cloning of cDNA encoding a Drosophila ryanodine receptor and functional studies of the Carboxyl-terminal calcium release channel. Biophys. J. 78, 1270–1281. doi:10.1016/S0006-3495(00)76683-5
Zhang W. B., Ren H., Sun F. L., Shen T. T., Yuan S., Gao X. W., et al. (2022). Evaluation of the toxicity of chemical and biogenic insecticides to three outbreaking insects in desert steppes of northern China. Toxins 14, 546. doi:10.3390/toxins14080546
Zhou X. R., Gao J. C., Pang B. P. (2016). Effects of temperature on the termination of egg diapause and post-diapause embryonic development of Galeruca daurica (Coleoptera: Chrysomelidae). Environ. Entomol. 45, 1076–1080. doi:10.1093/ee/nvw074
Zhou X. R., Shan Y. M., Tan Y., Zhang Z. R., Pang B. P. (2019). Comparative analysis of transcriptome responses to cold stress in Galeruca daurica (Coleoptera: Chrysomelidae). J. Insect Sci. 19, 8–6. doi:10.1093/jisesa/iez109
Zuo Y. Y., Ma H. H., Lu W. J., Wang X. L., Wu S. W., Nauen R., et al. (2019). Identification of the ryanodine receptor mutation I4743M and its contribution to diamide insecticide resistance in Spodoptera exigua (Lepidoptera: Noctuidae). Insect Sci. 27, 791–800. doi:10.1111/1744-7917.12695
Keywords: Galeruca daurica, ryanodine receptor, diamide insecticide, homologous modeling, molecular docking, binding modes
Citation: Ren H, Zhang H, Ni R, Li Y, Li L, Wang W, Tian Y, Pang B and Tan Y (2022) Detection of ryanodine receptor G4911E and I4754M mutation sites and analysis of binding modes of diamide insecticides with RyR on Galeruca daurica (Coleoptera: Chrysomelidae). Front. Physiol. 13:1107045. doi: 10.3389/fphys.2022.1107045
Received: 24 November 2022; Accepted: 12 December 2022;
Published: 22 December 2022.
Edited by:
Xiaoming Xia, Shandong Agricultural University, ChinaReviewed by:
Hu Wan, Huazhong Agricultural University, ChinaCopyright © 2022 Ren, Zhang, Ni, Li, Li, Wang, Tian, Pang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao Tan, ODUwMzEwLnRhbmh1YWY0QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.