- Center for Craniofacial Molecular Biology, Herman Ostrow School of Dentistry, University of Southern California, Los Angeles, CA, United States
The distinct morphology adopted by ameloblasts during amelogenesis is highly stage specific and involved intimately with the development of a hierarchical enamel microstructure. The molecular mechanisms that govern the development of an elongated and polarized secretory ameloblast morphology and the potential roles played by the enamel matrix proteins in this process are not fully understood. Thus far, the in vitro models that have been developed to mimic these early cell-matrix interactions have either been unable to demonstrate direct morphological change or have failed to adapt across ameloblast cell lines. Here, we use a recently established 3D cell culture model to examine the interactions between HAT-7 cells and the major enamel matrix proteins, amelogenin and ameloblastin. We demonstrate that HAT-7 cells selectively respond to functional EMPs in culture by forming clusters of tall cells. Aspect ratio measurements from three-dimensional reconstructions reveal that cell elongation is 5-times greater in the presence of EMPs when compared with controls. Using confocal laser scanning microscopy, we observe that these clusters are polarized with asymmetrical distributions of Par-3 and claudin-1 proteins. The behavior of HAT-7 cells in 3D culture with EMPs is comparable with that of ALC and LS-8 cells. The fact that the 3D model presented here is tunable with respect to gel substrate composition and ameloblast cell type highlights the overall usefulness of this model in studying ameloblast cell morphology in vitro.
Introduction
Enamel’s unique mechanical strength stems from the hierarchical arrangement of hydroxyapatite crystals into prisms with interprismatic mineral (Boyde, 1997; Chai et al., 2009). Although mature enamel is acellular, the generation of a prismatic enamel microstructure has been linked to features of ameloblast morphology during the early stages of amelogenesis - like the Tomes’ processes. During enamel development, ameloblasts undergo a complicated set of movements with each one contributing to unique structural features in mature enamel’s final microstructure. Ameloblasts move away from the developing enamel matrix, and adjacent sheets of ameloblasts move relative to one another resulting in a decussating pattern of enamel rods in higher mammals (Boyde, 1964; Boyde, 1997). However, the mechanisms underlying the formation of this unique, highly specialized ameloblast morphology are not fully understood.
The roles of enamel matrix proteins in amelogenesis - either individually or corporately are constantly evolving (Hatakeyama et al., 2009; Moradian-Oldak, 2012; Mazumder et al., 2016; Lacruz et al., 2017; Bapat et al., 2020; Moradian-Oldak and George, 2021). Genetically engineered mouse models have shown that, in addition to affecting enamel mineralization, components of the extracellular milieu like matrix proteins (ameloblastin and enamelin) and proteinases (MMP20) may influence ameloblast cell behavior (Fukumoto et al., 2005; Guan and Bartlett, 2013; Hu et al., 2014; Shin et al., 2018). The truncation of ameloblastin (removal of exons 5 and 6) results in the loss of secretory ameloblast elongation and polarization with a resultant de-differentiation into an inner enamel epithelium-like stage (Fukumoto et al., 2004). MMP20 overexpression disrupts the coordinated ameloblast movements, resulting in an abnormal migration of the ameloblasts into the papillary layers with a concomitant loss of ameloblast cell polarity (Shin et al., 2018). Additionally, MMP20 has been shown to regulate ameloblast movement through Wnt- and beta-catenin-mediated pathways by catalyzing the cleavage of ameloblast cell surface cadherins (Bartlett et al., 2011; Guan et al., 2016).
Reliable in vitro 3D cell culture models have proven invaluable for examining the interactions between ameloblasts and the enamel extracellular matrix (He et al., 2010; Földes et al., 2021). In vitro and cell culture systems provide for precise control over experimental conditions and the ability to isolate and examine the role of individual components in the matrix. When primary human enamel epithelial cells are co-cultured with dental pulp cells in a 3D Matrigel, they form spheroidal structures, increasing both their size and the expression levels of integrins (Li et al., 2006; He et al., 2010). Since ameloblasts are lost after amelogenesis, several ameloblast cell lines have been developed that are immortalized, either spontaneously (Nakata et al., 2003) or by viral transfection (Chen et al., 1992). These ameloblast cell lines exhibit characteristic and distinct phenotypes in culture and can represent different ameloblast stages (Sarkar et al., 2014). Using bioreactor-based 3D embedded cultures of HAT-7 cells, it was shown that HAT-7 cells display immunoreactivity to amelogenin and actin when co-cultured with dental pulp cells and in the presence of enamel matrix proteins (Pandya et al., 2021). The differentiation of cells into an ameloblast-like state in 3D culture is often demonstrated with differential gene expression patterns (Jiang et al., 2018). Given the pivotal roles played by ameloblast morphology in patterning the overall microstructure of enamel, ameloblast cell responses need to be methodically characterized, particularly the morphological changes occurring in response to the extracellular matrix composition.
We recently established a 3D cell culture model that is adaptable across gel substrates and allows for the quantification of changes in cell morphology and polarization in response to the matrix composition (Visakan et al., 2022a; Visakan et al., 2022b). Here, our optimized 3D model will be used to culture HAT-7 cells, which are often the cell line of choice for examining functional cell polarization (Bori et al., 2016). Using a systematic protocol for assessing changes in cell morphology by means of cell aspect ratio measurements and polarization by confocal microscopy, we observed that the addition of recombinant amelogenin (AMEL) and ameloblastin (AMBN) to the gel resulted in the formation of elongated and polarized HAT-7 cell clusters. These changes in HAT-7 cells are comparable to those of ALC and LS-8 cells, which have been previously studied (Visakan et al., 2022a). Since the composition of the 3D gel matrix is tunable, this model also allowed us to tailor the cell microenvironment using a combination of proteins or their proteolytic cleavage fragments as shown here with the recombinant AMBN 17 kDa. The proteolytic processing of ameloblastin by MMP-20 results in the production of three N-terminal cleavage fragments that weigh 17, 15 and 13 kDa (Iwata et al., 2007; Chun et al., 2010). Among these N-terminal cleavage fragments, the 17 kDa fragment predominates and exhibits potential regenerative properties when used on artificial periodontal defects (Fukae et al., 2006). We also examined the effect of ameloblastin lacking a potential cell binding domain (Su et al., 2020) on HAT-7 cells, suggesting that this system can be expanded to model disorders of enamel development.
Methods
Recombinant protein expression and purification
Recombinant amelogenin and ameloblastin (AMEL, AMBN, AMBNΔ5) were expressed and purified based on published protocols and as described in the Supplementary Material (Su et al., 2019a; Bapat et al., 2020).
Recombinant 17 kDa ameloblastin cleavage fragment expression, purification and characterization
To purify the 17 kDa fragment of ameloblastin, the full-length Ambn pET32a plasmid was first purified using the Monarch Miniprep kit (New England Biolabs). The 17 kDa fragment within the plasmid was selectively amplified using forward (atatatggatccgtgccggcatttcctcaac) and reverse primers (ttttttctcgagtcaacgggcgatctggaac). The PCR product and new pET32a plasmids (EMD Millipore) were digested using BamHI and XhoI enzymes (New England Biolabs). Then, they were purified using a QIAgen gel extraction kit and ligated. The ligated plasmid was transferred into Dh5α competent E. coli using the heat shock technique. Bacterial colonies were screened for successful transformation using ampicillin agar plates. The 17 kDa Ambn plasmid from selected colonies was amplified, purified, and sequenced. 17kDa Ambn was expressed in BL21 E. coli, and the protein was purified using the previously published protocol for full-length Ambn and characterized as described in the Supplementary Material.
HAT-7 characterization in 2D cell culture
HAT-7 cells were obtained as a gift from Prof. Hidemitsu Harada (Tokyo). HAT-7 cells were cultured following standard protocols (Kawano et al., 2002). Briefly, cells were cultured in 13 mm culture dishes (Corning) with low glucose Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Thermo) supplemented with 10% FBS (Thermo) and 1% penicillin/streptomycin (Thermo). This supplemented DMEM is referred to as cell culture media. Cultures were maintained under 5% CO2 at 37 °C until they achieved 80% or greater confluence. HAT-7 cells were characterized using immunofluorescence staining for amelogenin and ameloblastin proteins.
3D cell culture and aspect ratio measurement
A previously developed, optimized 3D-on-top type culture technique for the culture of ameloblast cell lines was used (Lee et al., 2007; Visakan et al., 2022a; Visakan et al., 2022b). Briefly, pre-chilled glass-bottomed 96-well plates (Mattek) were coated with 20 ug/ml test (AMBN, AMEL, AMBN Δ5, AMBN 17 kDa) and control (heat denatured AMBN, BSA and PBS) proteins. Ice-cold, growth-factor-reduced Geltrex (GFR Geltrex; Thermo) was overlaid on coated plates and incubated at 37 °C for 30 min. HAT-7 cells were detached from the culture dishes at 80% or greater confluence and were centrifuged at 200 x g for 5 min to obtain a soft pellet. The cells were resuspended in cell culture media and inoculated atop set GFR Geltrex gels to create a 3D-on-top-type culture (Figure 1). These 3D gels were incubated at 37 °C in a standard cell culture incubator for 24–72 h. At the end of the experiment, the cells were fixed with 25% glutaraldehyde for 15 min at room temperature. Cells in the 3D gel were labeled with 1:1000 DiD and 1:1000 DAPI and were visualized using Keyence BZX810 with objective PlanApo λ NA 0.75. Sequential Z stacks were recorded with a pitch of 0.4 μm. Z stacks were reconstructed using the Keyence Image Viewer (software version 1.1.1.8), and the 3D measure-tool was used to record the cell width along the XY plane and cell height along the Z axis. These values were tabulated, and the aspect ratio was calculated using the following formula:
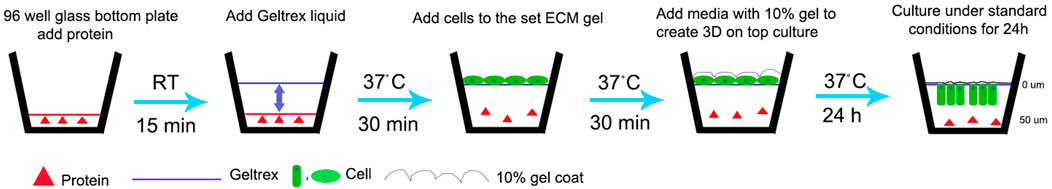
FIGURE 1. Schematic representation of modified 3D-on-top culture method (side view). Red triangle represents proteins-test (AMEL, AMBN, AMBN 17kDa, AMBN ∆5) and control (BSA, heat denatured AMBN). Green oval and rectangle represent HAT-7 cells. Solid line- GFR Geltrex after polymerization and scalloped line denotes 10% gel overlaid on cells. 0 and 50um markings are used to indicate the Z position on the surface (0 um) and within the gel (50 um) after 24 h resulting in elongation of the cells as shown in Figures 2D,E and Figure 4.
Immunofluorescence labelling
All steps were carried out in the dark at room temperature. Immunofluorescence labeling of whole cultures was carried out without the extraction of cells from the 3D gel (Lee et al., 2007). Cells in 3D that were fixed with glutaraldehyde were permeabilized with 0.1% Triton-X-100 for 5 min at room temperature. Then, the cells were incubated with 10% serum from the species of secondary antibody for 30 min at room temperature and were labelled with primary antibodies against Par-3, Actin and claudin-1. Fluorescently conjugated secondary antibodies (Jackson Immuno) were used for immunodetection for 1 h at room temperature. For characterization of HAT-7 cells, anti-amelogenin and anti-ameloblastin antibodies were used. A 2D monolayer of cells was fixed using 4% paraformaldehyde, was permeabilized using 0.1% Triton-X-100, and was labeled following standard protocols for immunofluorescence. Antibody dilutions for both 3D and 2D are provided in Supplementary Table S1. All images were created with confocal microscopy as described, except the image in Figure 2 and Supplementary Figure S2 which was made using an inverted fluorescence microscope (Keyence BZX810, Keyence, United States).
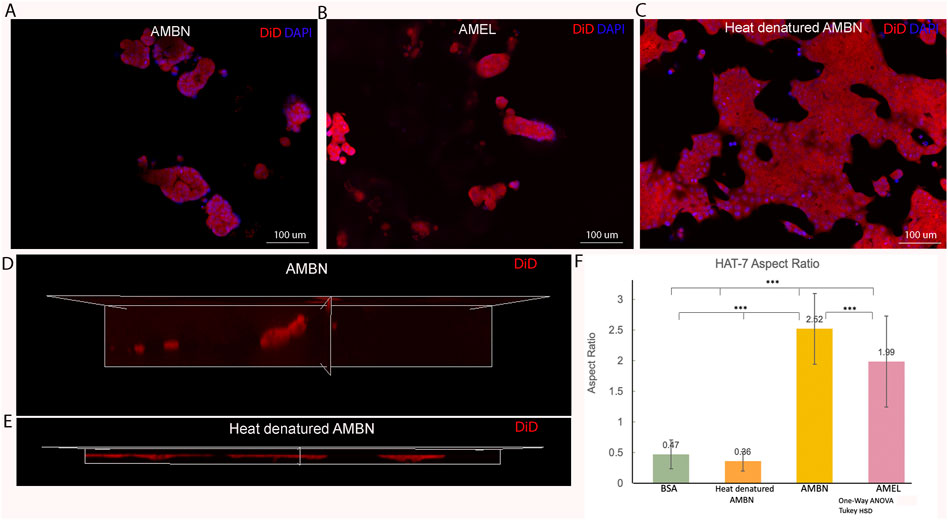
FIGURE 2. HAT-7 cells in modified 3D-on-top culture using growth-factor reduced Geltrex; 24h. (A–C). 2D surface images (top view, XY plane) of HAT-7 cells in the presence of AMBN, AMEL and heat denatured AMBN (negative control) respectively. Cell membrane labeled with DiD (red) and nucleus with DAPI (blue). HAT-7 cells organize into distinct cell clusters in (A) and (B) unlike in the control (C) where the cells are spread along the XY plane. (D, E). 3D reconstruction and axial view (side view, XZ plane) of representative samples from AMBN and heat denatured AMBN groups respectively. Cell outline as visualized by DiD reveals elongated, tall cells in (D) unlike the planar, flat cells with (E). (F) Aspect ratio measurements of HAT-7 cells from test and control groups. HAT-7 cells are statistically significantly elongated in the presence of AMEL and AMBN. *p < 0.05; **p < 0.01; ***p < 0.001.
Confocal imaging
Confocal imaging was carried out using the Lecia Stellaris 5 confocal microscope with an oil immersion objective HCX PL APO CS 63 x (NA 1.4). Sequential Z stacks were recorded with an optical pitch of 0.5 µm. Alexa Fluor 488 was detected at 502–552 nm (excitation at 488 nm), DiD was detected at 645–695 nm (excitation at 633 nm), and Alexa Fluor 647 was detected at 650–695 nm (excitation at 647 nm). Z stacks were 3D reconstructed and viewed using the LAS-X version 1.8.1.13759.
Statistical analysis
Data from aspect ratio measurements was analyzed with Microsoft Excel (version 16.66.1) using One-way ANOVA, independent Student’s t-test and Tukey HSD. p values less than 0.05 were considered as statistically significant. All experiments were performed in triplicates. For all experiments 90 cells in total were examined.
Results
3D culture to analyze morphology and aspect ratios in HAT-7 cells in response to AMEL and AMBN
Confluent 2D monolayers of HAT-7 cells were characterized by immunofluorescence staining for amelogenin and ameloblastin. We observed the expression of both proteins intracellularly using confocal microscopy, confirming the ameloblast characteristics of the HAT-7 cells (Supplementary Figure S1). HAT-7 cells that were cultured in the presence of enamel matrix proteins in 3D culture displayed characteristic differences in their morphology when compared with controls at the end of 24 h (Figure 2). HAT-7 cells organized into clusters in the presence of AMBN and AMEL as visualized with cell membrane labeling using DiD (Figures 2A,B). This morphology adopted by the HAT-7 cells contrasts strikingly with the controls (gel alone, BSA and heat denatured ameloblastin) where cells-maintained cell-cell contacts and were spread along the XY plane (Figure 2C). 3D reconstruction from sequential Z stacks revealed that the HAT-7 clusters in the presence of AMEL and AMBN were composed of tall cells that preferentially elongated along the Z axis (Figure 2D). Meanwhile, in the controls, the cells appeared planar without any significant height (Figure 2E). To quantify the cell elongation that occurs in the presence of enamel matrix proteins (EMPs), the aspect ratio of the individual cells was recorded. Repeated measurements of the cell aspect ratio revealed that, after 24 h of 3D culture, HAT-7 cells elongated 5.42 times greater with EMPs when compared to the controls (i.e., in the presence of BSA and heat denatured AMBN). This increase in the aspect ratio was statistically significantly different between all groups (n = 90; p < 0.001) with the greatest cell elongation achieved in the presence of AMBN (n = 90; p < 0.001) (Figure 2F). This pattern of cell behavior was consistent at the end of 72 h of culture (Supplementary Figure S2).
Morphology and aspect ratios in HAT-7 cells in response to 17 kDa AMBN and AMBN Δ5
Recent work has identified the presence of a highly conserved amphipathic helix (AH) forming, cell binding motif within the exon 5 region of AMBN (Su et al., 2019b; Su et al., 2020). To verify the function of the N-terminal domain identified within exon 5 of Ambn, the 3D culture model was used to examine the effect of 17 kDa AMBN (a proteolytic fragment contains the AH domain) and AMBN Δ5 (a mutant lacks the AH domain) on HAT-7 morphology. The theoretical molecular mass of AMBN 17 kDa was calculated using ProtParam and was found to be 17,990.53 Da. SDS-PAGE gels revealed that the protein migrated at an apparent molecular weight higher than the expected theoretical value (Supplementary Figure S3A). Mass spectra (ESI) of the band around 20 kDa in SDS-PAGE showed that the molecular mass of the protein was 17,991 Da, close to the theoretical value of 17,990.53 Da, suggesting that the purified protein was the AMBN 17 kDa fragment (Supplementary Figure S3B). The difference in the theoretical and experimental mass of the protein was 0.47 Da, suggesting that the protein identified in the peak was intact. The amino acid sequence of the 17 kDa cleavage fragment is shown in Supplementary Figure S3C. When cultured with the recombinant AMBN 17 kDa cleavage fragment, HAT-7 cells responded similarly to full-length ameloblastin. At comparable molar concentrations of both proteins (0.48 μM), the 17 kDa fragment was sufficient to recapitulate the cell clustering and elongation achieved with full-length AMBN (Figures 3A,B). Measurements of the aspect ratio confirmed this, where no statistically significant difference was observed between the 17 kDa fragment and the full-length AMBN (p > 0.05) (Figure 3C). The 3D model was then used to observe how the removal of highly conserved key domains from AMBN affected HAT-7 morphology. Culturing HAT-7 cells with AMBN Δ5 lacking the exon 5 encoded region resulted in a complete reversal of the cells’ behavior in 3D. HAT-7 cells failed to cluster and, instead, appeared distinct like in the negative controls (Figures 3D,E). Aspect ratio measurements confirmed the lack of cell elongation in the AMBN Δ5 mutant when compared with the wild-type AMBN (p < 0.001) (Figure 3F).
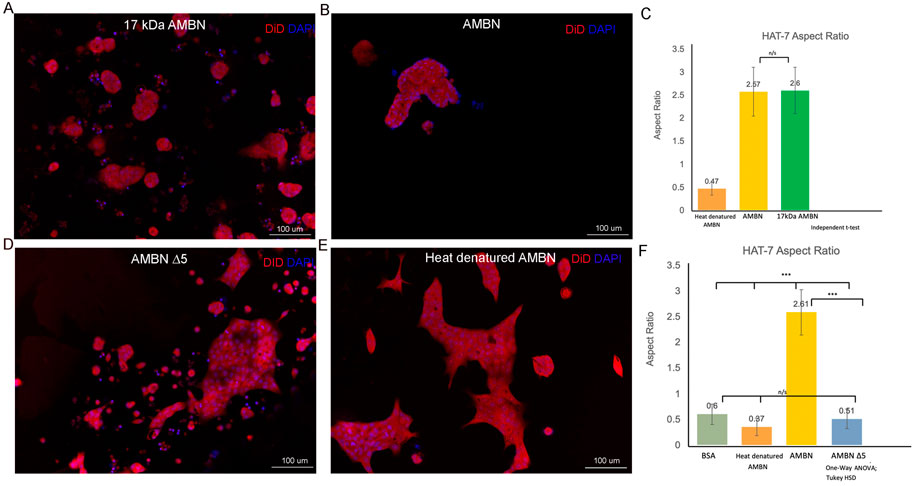
FIGURE 3. HAT-7 cells cultured with AMBN 17 kDa fragment and AMBN ∆5 mutant. (A,B). XY surface images of HAT-7 cells cultured in the presence of AMBN 17 kDa fragment and wild-type full-length AMBN respectively. HAT-7 cells organized into distinct clusters of cells in both groups as visualized using DiD (red) and DAPI (blue). (C). Aspect ratio measurements of HAT-7 cells cultured with 17 kDa fragment, positive and negative controls revealing statistically comparable HAT-7 cell elongation in AMBN 17 kDa cleavage fragment and positive control (full-length AMBN); p > 0.05. (D,E). XY surface images of HAT-7 cells cultured with mutant AMBN ∆5, and heat denatured AMBN respectively. (F). Aspect ratio measurements of HAT-7 cells in the presence of AMBN ∆5. Removal of exon 5 encoded sequence resulted in HAT-7 cells remaining planar with aspect ratio values statistically significantly lower than wild-type AMBN p < 0.001. AMBN ∆5 aspect ratios are comparable with those of negative controls p > 0.05. *p < 0.05; **p < 0.01; ***p < 0.001.
Asymmetric distribution of Par-3 and claudin-1 upon HAT-7 polarization with AMBN
3D clusters of HAT-7 cells that were elongated in the presence of AMBN were labeled with anti-actin, anti-Par-3, and anti-claudin-1 antibodies (Figure 4). They were then examined under a confocal laser scanning microscope with the 3D reconstruction of sequential Z stacks. Actin labeling (grey pseudo color) confirmed the cell elongation (Supplementary Figure S4A,B) observed earlier with DiD (Figure 2). Actin labeling patterns within individual cells in HAT-7 clusters (formed in the presence of AMBN) were uniform and symmetrical with signals being detected throughout the entire outline of the cells (Figure 4B). Cell polarity protein Par-3 (red) localization, however, was polarized with signals being restricted to the cell membrane basal to nucleus (Figure 4C). Additionally, colocalization of actin and Par-3 was restricted to one pole of the cell (white arrows in Figure 4D). XY plane images acquired at two different Z depths from the surface of the cells revealed that, at higher Z depths, only actin signals were detectable, which confirms the polarized distribution of Par-3 (Supplementary Figures S5A–F). Like Par-3, tight junction protein claudin-1 (green) was also asymmetrical in its distribution compared to that of cell membrane label DiD (grey pseudo color) with its localization being restricted basal to nucleus (Figures 4E–H). The planar cells in the control did not display any polarization in the distribution of Par-3 (Supplementary Figure S6). The HAT-7 clusters formed in the presence of 17 kDa AMBN were also polarized with the Par-3 localization patterns being comparable with that of the full-length AMBN (Figures 4I–L).
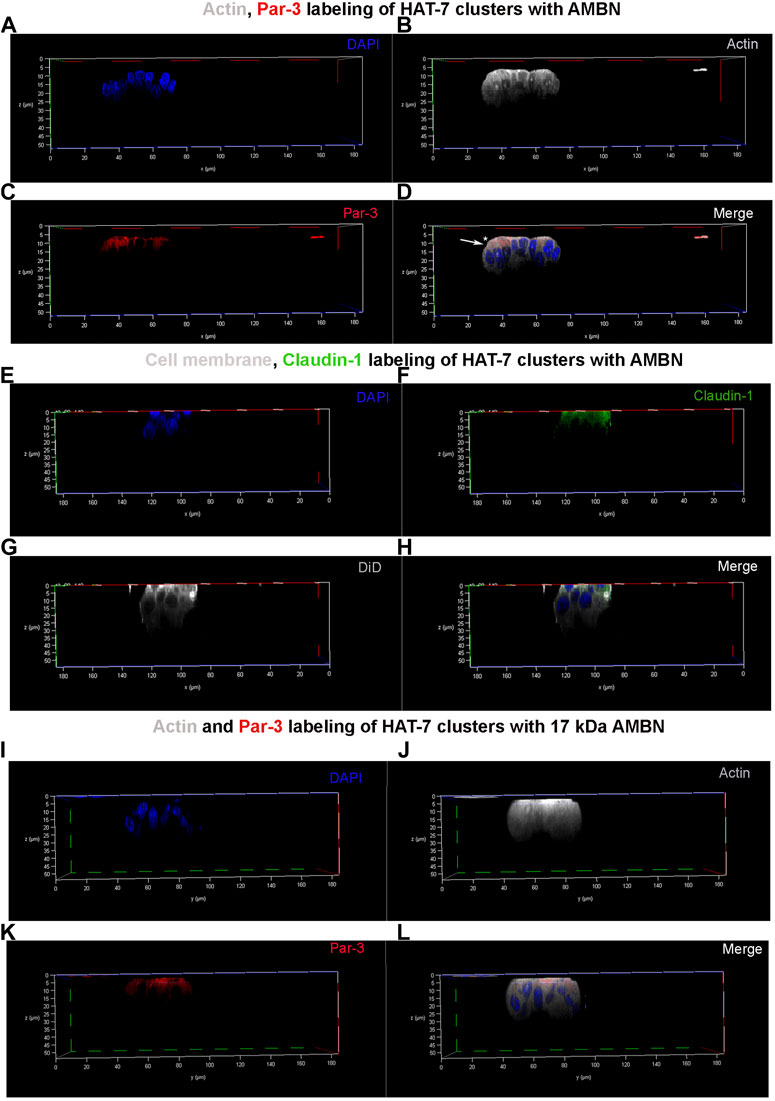
FIGURE 4. Par-3 and Claudin-1 polarization in HAT-7 clusters in a 3D axial view. (A–C) Individual channels for nucleus (DAPI), actin (grey pseudo color) and Par-3 (red) distribution respectively in HAT-7 cells cultured with AMBN. (D) Merged image of all three channels revealing a uniform symmetrical labeling of actin with an asymmetrical and polarized distribution of Par-3 (white arrow). (E–G) Individual channels for nucleus (blue), claudin-1 (green) and DiD (grey pseudo color) respectively in HAT-7 cells cultured with ameloblastin. (H) Merged images of all three channels revealing selective claudin-1 labeling within the pole of the cells basal to nucleus. DiD labeling is uniform throughout the entire outline of the cells. (I–K) Individual channels for nucleus (DAPI), actin (grey pseudo color) and Par-3 (red) distribution in HAT-7 cells cultured with AMBN 17 kDa cleavage fragment respectively. (L) Merged image of all three channels revealing a uniform symmetrical labeling of actin with an asymmetrical and polarized distribution of Par-3 like the observations in (D).
Discussion
It has been proposed that ameloblast morphology is directly or indirectly related to the development of an overall prismatic enamel architecture (Boyde, 1964, Boyde, 1997; Nanci, 2017). Ameloblast elongation and polarization were severely impacted in genetically engineered mouse models lacking functional ameloblastin or enamelin (Fukumoto et al., 2004; Hu et al., 2014), resulting in a lack of true enamel. The severe dysmorphology in genetically engineered animal models makes it challenging to observe and examine the molecular mechanisms that govern the formation of this highly specialized ameloblast morphology, thereby necessitating the need for in vitro models.
Here, we demonstrate that the changes in cell morphology and polarization upon addition of enamel matrix proteins can be modeled using a recently-established, in-vitro 3D cell culture (Visakan et al., 2022a). This model enabled such differential cell responses to be readily visualized and quantified in a systematic manner in three different ameloblast-like cell lines (i.e., ALC, LS-8 and HAT-7). Using immunofluorescent staining and aspect ratio measurements, we observed that HAT-7 cells exhibited a preferential elongation along the Z axis, resulting in the formation of clusters of tall cells when cultured in the presence of AMBN or AMEL. Formation of these tall cells in the presence of the 17 kDa AMBN suggests a potential in vivo significance of this response, considering that the 17 kDa cleavage fragment persists in the prism sheaths of the developing enamel matrix (Uchida et al., 1991).
Loss of function of AMBN in cell elongation and polarization was further demonstrated using recombinant mutant AMBN protein lacking the exon 5 encoded region (AMBN Δ5). This mutant lacked the ability to induce cell clustering or elongation in HAT-7 cells. This observation can be the result of loss of AMBN -cell binding, (Su et al., 2020), or AMBN self-assembly (Wald., et al., 2017). AMBN exon 5 encoded region contains the amphipathic helix (AH) cell binding domain that recently was found to be highly conserved among mammals and had a strong evolutionary relationship with enamel prismatic structure (Su et al., 2022). The same region contains the Y/F-x-x-Y/L/F-x-Y/F self-assembly motif, and the disruption of this motif resulted in a perturbed enamel prismatic architecture (Wald et al., 2017). The Y/F-x-x-Y/L/F-x-Y/F motif within exon 5 encoded region also is the site of co-assembly with AMEL (Bapat et al., 2020).
The model can also be used to examine and characterize the polarization status of cells in 3D culture as demonstrated using immunofluorescent labeling of Par-3, claudin-1, and actin proteins. We originally developed this 3D on top culture model to generate non-uniform contact surfaces for cells to mimic the enamel extracellular matrix during development (Visakan et al., 2022a). The developing ameloblasts are in contact with the ECM only along their functional apical membranes, and this is what was modeled in the modified 3D culture system. Culturing HAT-7 cells in 3D-on-top-type cultures reveal a differential cell behavior in the presence of AMEL and AMBN with the HAT-7 cells adopting distinctly different morphologies. Like previous observations with other ameloblast cell lines - ALC and LS-8 cells (Visakan et al., 2022a; Visakan et al., 2022b). HAT-7 cells exhibit preferential elongation along their Z axes with aspect ratios 5-times greater than control when amelogenin and ameloblastin were added to the 3D cell culture. In repeated experiments, AMBN was observed to exert a greater influence on HAT-7 elongation than amelogenin did. These morphological changes were specific to the presence of functional enamel matrix proteins, as this behavior could be reversed using heat denatured AMBN.
HAT-7 cells derived from rat molars are functional ameloblast-like cells expressing amelogenin and ameloblastin proteins (Kawano et al., 2002). They are often the cell line of choice for examining functional cell polarization (Bori et al., 2016), the epithelial-mesenchymal interactions during odontogenesis (Matsumoto et al., 2011), and for modeling disease (Földes et al., 2021). When cultured on permeable transwell filters, HAT-7 cells formed polarized epithelial layers as characterized by apical membrane permeability to CO2 and basolateral membrane uptake of bicarbonate (Bori et al., 2016). These functional polarized layers also express the tight junctional proteins claudins-1, 4 and 8. In the 3D-on-top culture, we observed that the HAT-7 cells cultured in the presence of AMBN and 17 kDa AMBN exhibited features of structural cell polarization (Figure 4).
Confocal laser scanning microscopy revealed a selective accumulation of polarity protein Par-3 and tight junctional protein claudin-1 along the HAT-7 membrane basal to nucleus position in 3D (Figure 4). Claudin-1 was chosen as a marker for polarization as it has been shown to be highly upregulated in the elongated secretory ameloblasts (Zhang et al., 2019). Furthermore, in fully polarized mouse ameloblasts, it has been observed that Par-3 localized along the functional base and claudin-1 along the functional apical membrane (Inai et al., 2008). 3D reconstruction of Z stacks permits the precise localization of claudin-1 and Par-3 within the overall outline of the cell visualized with actin. Par-3 and claudin-1 labeling patterns in 3D culture of ALC (Visakan et al., 2022a) and HAT-7 cells (Figure 4) contrasts with what is observed in vivo. In 3D cultures, both proteins were observed to be localizing along the same pole of the cell. This discrepancy could be due to the differences in the height of the secretory stage ameloblasts in fully developed enamel organs compared to the individual cells in 3D culture. We have recently reported that the addition of functional AMBN to LS-8 cells in culture resulted in an upregulation of planar cell polarity protein Vangl2 (2.8 times) and cell polarity protein Par-3 (3.8 times) when compared to controls (heat denatured AMBN and AMBN Δ5) (Su et al., 2022). Vangl2 planar cell polarity protein is highly expressed particularly in the Tomes’ processes of secretory stage ameloblasts (Nishikawa and Kawamoto, 2012).
Upon their secretion into the enamel matrix, amelogenin and ameloblastin are rapidly processed into N- and C-terminal cleavage fragments by MMP-20 (Bartlett and Simmer, 1999). An evolutionarily conserved novel amphipathic helix (AH) forming cell binding domain within the exon-5 encoded region of ameloblastin was recently identified (Su et al., 2019b). Using the immunofluorescent localization of peptides, it was observed that the AH domain could be specifically localized to ALC cell processes (Su et al., 2020). Furthermore, in ameloblast cell lines, the gene expression levels of cell polarity and planar cell polarity proteins were upregulated in the presence of ameloblastin (Su et al., 2022). In the present study, given the rapid in vivo processing of ameloblastin, the newly expressed recombinant 17 kDa ameloblastin containing the exon 5 encoded region was tested in 3D on HAT-7 cells to examine the potential in vivo significance of ameloblastin-cell interactions. All the cell effects that were observed with full-length ameloblastin were observed in the presence of the 17 kDa ameloblastin cleavage fragment as well. HAT-7 cells form clusters of elongated cells with polarized distributions of Par-3 (Figures 3, 4).
In conclusion, 3D cell culture models are increasingly replacing conventional 2D monolayer culture techniques, as they result in physiologically comparable cell responses (Kozlowski et al., 2009; Baker and Chen, 2012; Duval et al., 2017). However, the validity of the model lies in large part in being replicable across cell lines and substrate types. The 3D-on-top model presented here is adaptable across gel substrates (GFR Geltrex, type I collagen and gelatin) and can be used with multiple ameloblast-like cell lines (ALC, LS-8 and HAT-7) (Visakan et al., 2022a; Visakan et al., 2022b). Although the 3D system presented here permits visualization of the cells and the structures they adopt in 3D, they are limited in their localization of the EMPs within the entirety of the gel. Examination of the diffusion gradients within the gel and the effect of the gel’s rheological properties remain outside the scope of this study. The interactions of the ameloblast-like cells with enamel extracellular matrix proteins can be modeled using the in vitro 3D cell culture system. Such 3D culture models can be expanded to observe the concerted effects of the various bioactive molecules in the extracellular milieu on the cells, and the knowledge thus gained may be translated into future biomimetic attempts at enamel repair.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author contributions
GV designed and performed the experiments, analyzed the data, and wrote the manuscript. RB and JS helped with the recombinant protein design and synthesis. JM-O designed the experiments, analyzed the data, and both wrote and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the National Institutes of Health–National Institute of Dental and Craniofacial Research grants R01DE013414, DE-027529, and R01DE027632 to JM-O.
Acknowledgments
We would like to thank Hidemitsu Harada for providing HAT-7 cells; Salony Jassar for technical assistance with cell culture; and Emma Burris-Janssen for proofreading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1069519/full#supplementary-material
References
Baker B. M., Chen C. S. (2012). Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J. Cell Sci. 125 (13), 3015–3024. doi:10.1242/jcs.079509
Bapat R. A., Su J., Moradian-Oldak J. (2020). Co-immunoprecipitation reveals interactions between amelogenin and ameloblastin via their self-assembly domains. Front. Physiol. 11, 622086. doi:10.3389/fphys.2020.622086
Bartlett J. D., Yamakoshi Y., Simmer J. P., Nanci A., Smith C. E. (2011). MMP20 cleaves E-cadherin and influences ameloblast development. Cells Tissues Organs 194 (2-4), 222–226. doi:10.1159/000324205
Bartlett J., Simmer J. (1999). Proteinases in developing dental enamel. Crit. Rev. Oral Biol. Med. 10 (4), 425–441. doi:10.1177/10454411990100040101
Bori E., Guo J., Rácz R., Burghardt B., Földes A., Kerémi B., et al. (2016). Evidence for bicarbonate secretion by ameloblasts in a novel cellular model. J. Dent. Res. 95 (5), 588–596. doi:10.1177/0022034515625939
Boyde A. (1997). “Microstructure of enamel,” in Ciba foundation symposium (Wiley Online Library), 18–31.
Boyde A. (1964). The structure and development of mammalian enamel. Doctoral dissertation. Queen Mary University of London.
Chai H., Lee J. J.-W., Constantino P. J., Lucas P. W., Lawn B. R. (2009). Remarkable resilience of teeth. Proc. Natl. Acad. Sci. U. S. A. 106 (18), 7289–7293. doi:10.1073/pnas.0902466106
Chen L., Couwenhoven R., Hsu D., Luo W., Snead M. (1992). Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch. Oral Biol. 37 (10), 771–778. doi:10.1016/0003-9969(92)90110-t
Chun Y.-H., Yamakoshi Y., Yamakoshi F., Fukae M., Hu J.-C., Bartlett J. D., et al. (2010). Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J. Dent. Res. 89 (8), 785–790. doi:10.1177/0022034510366903
Duval K., Grover H., Han L.-H., Mou Y., Pegoraro A. F., Fredberg J., et al. (2017). Modeling physiological events in 2D vs. 3D cell culture. Physiology 32 (4), 266–277. doi:10.1152/physiol.00036.2016
Földes A., Sang-Ngoen T., Kádár K., Rácz R., Zsembery Á., DenBesten P., et al. (2021). Three-dimensional culture of ameloblast-originated HAT-7 cells for functional modeling of defective tooth enamel formation. Front. Pharmacol. 12, 682654. doi:10.3389/fphar.2021.682654
Fukae M., Kanazashi M., Nagano T., Tanabe T., Oida S., Gomi K. (2006). Porcine sheath proteins show periodontal ligament regeneration activity. Eur. J. Oral Sci. 114, 212–218. doi:10.1111/j.1600-0722.2006.00309.x
Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167 (5), 973–983. doi:10.1083/jcb.200409077
Fukumoto S., Yamada A., Nonaka K., Yamada Y. (2005). Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells Tissues Organs 181 (3-4), 189–195. doi:10.1159/000091380
Guan X., Bartlett J. (2013). MMP20 modulates cadherin expression in ameloblasts as enamel develops. J. Dent. Res. 92 (12), 1123–1128. doi:10.1177/0022034513506581
Guan X., Xu M., Millar S. E., Bartlett J. D. (2016). Beta-catenin is essential for ameloblast movement during enamel development. Eur. J. Oral Sci. 124 (3), 221–227. doi:10.1111/eos.12261
Hatakeyama J., Fukumoto S., Nakamura T., Haruyama N., Suzuki S., Hatakeyama Y., et al. (2009). Synergistic roles of amelogenin and ameloblastin. J. Dent. Res. 88 (4), 318–322. doi:10.1177/0022034509334749
He P., Zhang Y., Kim S. O., Radlanski R. J., Butcher K., Schneider R. A., et al. (2010). Ameloblast differentiation in the human developing tooth: Effects of extracellular matrices. Matrix Biol. 29 (5), 411–419. doi:10.1016/j.matbio.2010.03.001
Hu J. C.-C., Hu Y., Lu Y., Smith C. E., Lertlam R., Wright J. T., et al. (2014). Enamelin is critical for ameloblast integrity and enamel ultrastructure formation. PloS one 9 (3), e89303. doi:10.1371/journal.pone.0089303
Inai T., Sengoku A., Hirose E., Iida H., Shibata Y. (2008). Differential expression of the tight junction proteins, claudin-1, claudin-4, occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisors. Anat. Rec. 291 (5), 577–585. doi:10.1002/ar.20683
Iwata T., Yamakoshi Y., Hu J.-C., Ishikawa I., Bartlett J., Krebsbach P., et al. (2007). Processing of ameloblastin by MMP-20. J. Dent. Res. 86 (2), 153–157. doi:10.1177/154405910708600209
Jiang N., Chen L., Ma Q., Ruan J. (2018). Nanostructured Ti surfaces and retinoic acid/dexamethasone present a spatial framework for the maturation and amelogenesis of LS-8 cells. Int. J. Nanomedicine 13, 3949–3964. doi:10.2147/IJN.S167629
Kawano S., Morotomi T., Toyono T., Nakamura N., Uchida T., Ohishi M., et al. (2002). Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect. Tissue Res. 43 (2-3), 409–412. doi:10.1080/03008200290000637
Kozlowski M., Gajewska M., Majewska A., Jank M., Motyl T. (2009). Differences in growth and transcriptomic profile of bovine mammary epithelial monolayer and three-dimensional cell cultures. J. Physiol. Pharmacol. 60 (1), 5–14.
Lacruz R. S., Habelitz S., Wright J. T., Paine M. L. (2017). Dental enamel formation and implications for oral health and disease. Physiol. Rev. 97 (3), 939–993. doi:10.1152/physrev.00030.2016
Lee G. Y., Kenny P. A., Lee E. H., Bissell M. J. (2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4 (4), 359–365. doi:10.1038/nmeth1015
Li W., Machule D., Gao C., DenBesten P. K. (2006). Growth of ameloblast-lineage cells in a three-dimensional Matrigel environment. Eur. J. Oral Sci. 114, 159–163. doi:10.1111/j.1600-0722.2006.00308.x
Matsumoto A., Harada H., Saito M., Taniguchi A. (2011). Induction of enamel matrix protein expression in an ameloblast cell line co-cultured with a mesenchymal cell line in vitro. Vitro Cell. Dev. Biol. Anim. 47 (1), 39–44. doi:10.1007/s11626-010-9362-7
Mazumder P., Prajapati S., Bapat R., Moradian-Oldak J. (2016). Amelogenin-ameloblastin spatial interaction around maturing enamel rods. J. Dent. Res. 95 (9), 1042–1048. doi:10.1177/0022034516645389
Moradian-Oldak J., George A. (2021). Biomineralization of enamel and dentin mediated by matrix proteins. J. Dent. Res. 100 (10), 1020–1029. doi:10.1177/00220345211018405
Moradian-Oldak J. (2012). Protein-mediated enamel mineralization. Front. Biosci. 17, 1996–2023. doi:10.2741/4034
Nakata A., Kameda T., Nagai H., Ikegami K., Duan Y., Terada K., et al. (2003). Establishment and characterization of a spontaneously immortalized mouse ameloblast-lineage cell line. Biochem. Biophys. Res. Commun. 308 (4), 834–839. doi:10.1016/s0006-291x(03)01467-0
Nanci A. (2017). Ten cate's oral histology-e-book: Development, structure, and function. St.Louis, MO: Elsevier Health Sciences.
Nishikawa S., Kawamoto T. (2012). Planar cell polarity protein localization in the secretory ameloblasts of rat incisors. J. Histochem. Cytochem. 60 (5), 376–385. doi:10.1369/0022155412438887
Pandya M., Lyu H., Luan X., Diekwisch T. G. (2021). Polarized, amelogenin expressing ameloblast-like cells from cervical loop/dental pulp co-cultures in bioreactors. Stem Cells Dev. 30 (16), 797–805. doi:10.1089/scd.2021.0115
Sarkar J., Simanian E. J., Tuggy S. Y., Bartlett J. D., Snead M. L., Sugiyama T., et al. (2014). Comparison of two mouse ameloblast-like cell lines for enamel-specific gene expression. Front. Physiol. 5, 277. doi:10.3389/fphys.2014.00277
Shin M., Chavez M., Ikeda A., Foster B., Bartlett J. (2018). MMP20 overexpression disrupts molar ameloblast polarity and migration. J. Dent. Res. 97 (7), 820–827. doi:10.1177/0022034518758657
Su J., Arun Bapat R., Visakan G., Moradian-Oldak J. (2022). Co-emergence of the amphipathic helix on ameloblastin with mammalian prismatic enamel. Mol. Biol. Evol. 39, msac205. doi:10.1093/molbev/msac205
Su J., Bapat R. A., Moradian-Oldak J. (2019a). The expression and purification of recombinant mouse ameloblastin in E. coli. Methods Mol. Biol. 1922, 229–236. doi:10.1007/978-1-4939-9012-2_23
Su J., Bapat R., Visakan G., Moradian-Oldak J. (2020). An evolutionarily conserved helix mediates ameloblastin-cell interaction. J. Dent. Res. 99 (9), 1072–1081. doi:10.1177/0022034520918521
Su J., Kegulian N. C., Bapat R. A., Moradian-Oldak J. (2019b). Ameloblastin binds to phospholipid bilayers via a helix-forming motif within the sequence encoded by exon 5. ACS omega 4 (2), 4405–4416. doi:10.1021/acsomega.8b03582
Uchida T., Tanabe T., Fukae M., Shimizu M., Yamada M., Miake K., et al. (1991). Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry 96 (2), 129–138. doi:10.1007/BF00315983
Visakan G., Su J., Moradian-Oldak J. (2022a). Ameloblastin promotes polarization of ameloblast cell lines in a 3-D cell culture system. Matrix Biol. 105, 72–86. doi:10.1016/j.matbio.2021.11.002
Visakan G., Su J., Moradian-Oldak J. (2022b). Data from ameloblast cell lines cultured in 3D using various gel substrates in the presence of ameloblastin. Data Brief. 42, 108233. doi:10.1016/j.dib.2022.108233
Wald T., Spoutil F., Osickova A., Prochazkova M., Benada O., Kasparek P., et al. (2017). Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc. Natl. Acad. Sci. 114 (9), E1641–E1650. doi:10.1073/pnas.1615334114
Keywords: ameloblasts, ameloblastin, amelogenin, polarization, cell morphology, Geltrex®, 3D cell culture
Citation: Visakan G, Bapat RA, Su J and Moradian-Oldak J (2022) Modeling ameloblast-matrix interactions using 3D cell culture. Front. Physiol. 13:1069519. doi: 10.3389/fphys.2022.1069519
Received: 14 October 2022; Accepted: 21 November 2022;
Published: 01 December 2022.
Edited by:
Pamela Den Besten, University of California, San Francisco, United StatesReviewed by:
Gabor Varga, Semmelweis University, HungaryYan Zhang, University of California, San Francisco, United States
Copyright © 2022 Visakan, Bapat, Su and Moradian-Oldak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janet Moradian-Oldak, am9sZGFrQHVzYy5lZHU=
 Gayathri Visakan
Gayathri Visakan Rucha Arun Bapat
Rucha Arun Bapat Jingtan Su
Jingtan Su Janet Moradian-Oldak
Janet Moradian-Oldak