- 1State Key Laboratory for Ecological Pest Control of Fujian/Taiwan Crops and College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory of Integrated Pest Management for Fujian-Taiwan Crops, Ministry of Agriculture and Rural Affairs, Fuzhou, China
- 3International Joint Research Laboratory of Ecological Pest Control, Ministry of Education, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Comprehensive Technology Service Center of Quanzhou Customs, Quanzhou, China
Chemosensory proteins (CSPs) can bind and transport odorant molecules, which are believed to be involved in insect chemoreception. Here, we investigated three CSPs in perception of volatiles in Empoasca onukii. Expression profiles showed that although EonuCSP4, EonuCSP 6-1 and EonuCSP6-2 were ubiquitously expressed in heads, legs, thoraxes and abdomen, they were all highly expressed in the antennae of E. onukii. Further, fluorescence competitive binding assays revealed that EonuCSP4 and 6-1 had binding affinities for three plant volatiles, suggesting their possible involvement in the chemosensory process. Among them, EonuCSP6-1 showed relatively high binding affinities for benzaldehyde. Behavioral assays revealed that the adults of E. onukii showed a significant preference for two compounds including benzaldehyde. The predicted three-dimensional (3D) structures of these 3 CSP have the typical six α-helices, which form the hydrophobic ligand-binding pocket. We therefore suggest that Eoun6-1 might be involved in the chemoreception of the host-related volatiles for E. onukii. Our data may provide a chance of finding a suitable antagonist of alternative control strategies which block the perception of chemosensory signals in pest, preventing the food- orientation behaviors.
Introduction
Molecular sensing of chemicals is important for locating food, searching mating partners, selecting oviposition sites, socializing among groups, and avoiding predators of insects (Bruce et al., 2005; Ebrahim et al., 2015; Pelosi et al., 2018). The communication of the insect chemosensory system is characterized by stability, sensitivity and specificity (Carey et al., 2010; Mao et al., 2010; Leal 2013). Multiple olfactory proteins involved in the odorants perception were identified (Hua et al., 2021; Zhan et al., 2021; Zhang et al., 2021). Firstly, hydrophobic odorants enter the insects through the olfactory pores of cuticular chemosensilla (Stocker 1994; Wang et al., 2020). Secondly, odorant binding proteins (OBPs) and chemosensory proteins (CSPs) bind to hydrophobic odorants and transport them to the olfactory sensory neurons (OSNs) (Vogt and Riddiford 1981; Zhang et al., 2018; Zhang et al., 2021). Then, odorant receptors (ORs) as odorant-gated ion channels play roles in insect olfaction. They are formed by a heteromultimeric complex of the odorant receptor co-receptor (Orco) and a ligand-selective Or, which are activated when the odorant binds to the binding site (Laughlin et al., 2008; Sato et al., 2008; Wicher et al., 2008; Stengl and Funk 2013; Butterwick et al., 2018; Del Mármol et al., 2021) and convert the chemical signals into nerve impulses (Ha and Smith 2009). Next, odorants are degraded by odorant degrading enzymes (ODEs) (Ishida and Leal 2005; Wang M. M. et al., 2021). Subsequently, nerve impulses are transmitted to the central nervous system (CNS) to guide the insects’ behaviors (Leal et al., 2005).
CSPs are small water-soluble proteins and are major binding proteins in insects with small molecular weight of ∼12 kDa. CSPs possess a conserved cysteine CSP motif (C1-X6-8-C2-X16-21-C3-X2-C4) that form two disulfide bridges (C1-X6-8-C2, C3-X2-C4) (Zhou et al., 2006). The disulfide bonds in the CSP motif are inter-helical with two small loops, forming a rigid hydrophobic pocket involved in ligand binding (Lartigue et al., 2002). This folding conformation are different from those of insect OBPs (Tegoni et al., 2004). Some CSPs function in chemosensory signal transduction and solubilization of pheromone components (Zhan et al., 2021; Zhang et al., 2021). Other CSPs are involved in insect physiological processes and behavior, such as moulting, tissue formation, regeneration, reproduction and resistance (Li et al., 2015; Zhu et al., 2016; Pelosi et al., 2018). BodoCSP1 is involved in host plant volatiles perception in Bradysia odoriphaga (Zhang et al., 2021). CforCSP1, 5 and 6 are involved in the chemical communication between Cylas formicarius and host plant volatiles (Hua et al., 2021). Besides the chemosensory functions, CSPs are found to have various physiological functions. In Locusta migratoria, CSPs are involved in the physiological transition from solitary to the gregarious phase (Guo et al., 2011). CSP5 in Apis mellifera functions in embryo development (Maleszka et al., 2007). In Solenopsis invicta, CSP9 is responsible for cuticle development (Cheng et al., 2015). In Periplaneta Americana, CSP10 participates in leg regeneration (Kitabayashi et al., 1998). BmorCSPs of Bombyx mori are associated with insect resistance to insecticide abamectin (B1a and B1b avermectins) (Xuan et al., 2015).
The tea green leafhopper, Empoasca onukii (Hemiptera: Cicadellidae) is a serious pest of tea plants in East Asia. The nymphs and adults suck the sap from the fresh buds while female adults lay eggs in tender plant shoots, which leads to early symptoms of chlorosis and leaf curling, followed by browning, shriveling and necrosis that ultimately destroy the whole leaf (Jin et al., 2012). Outbreaks of E. onukii lead to significantly reduction of tea yields both in summer and autumn, two periods of tea harvest, with economic losses up to 15%–50% in China (Chen et al., 2019) and 33% in Japan (Zhang et al., 2019). For now, chemical insecticides remains the main control strategy for its management. However, the development of insecticide resistance/tolerance in the tea green hopper makes its control difficult. Meanwhile, the excessive use of insecticides damages the ecological environment and causes the residue problems of tea. Therefore, it is urgent to develop the safe strategies to control this pest. Understanding the mechanism and signals that are involved in the odorant reception of E. onukii may provide the clues of control methods, which enhance the tea safety.
Twenty-six species-expanded CSP genes have been recently identified in E. onukii based on genome data (Zhao et al., 2022). However, there is no information about the CSPs that can bind to the volatiles from tea, which can be further used to develop the alternative control strategies against E. onukii.
In the current study, we focused on analyzing the CSPs distributing in clusters on Chromosome 1. Results showed that there are 7 CSPs distributing in clusters on chromosome 1. However, only 3 CSPs were successfully expressed in the expression vector. Bases on phylogenetic analysis with aphid, plant bug, and plant hoppers, these 3 CSPs belonged to aphid CSP4 and aphid CSP6 cluster respectively. We named them as EonuCSP4, EonuCSP6-1, and EonuCSP6-2. To identify the roles of these CSPs, we analyzed their expression patterns. Besides, binding properties of the 3 CSPs were investigated by fluorescence binding assays. In addition, we use a Y-tube olfactometer to detect the behavioral responses of E. onukii to volatiles. These results provide clues for better understanding the chemosensory mechanisms of E. onukii, which lead to the novel way of pest control strategies.
Materials and methods
Sample collection, total RNA extraction and cDNA synthesis
E. onukii adult samples were collected in Fuzhou, Fijian province, southeastern China in May 2020 (on the tea cultivar of Fudingdabai), and maintained on tea plants in lab. The insectarium environment was set at 25°C ± 1°C and 60 ± 5% relative humidity (RH) with a photoperiod (light: dark = 14:10).
Total RNA was extracted using Eastep® Super Kit (Promega, Beijing, China) according to the protocol. The concentration of RNA was assessed by NanoDrop 2000 spectrophotometry (NanoDrop, DE, USA) by measuring the OD at 260 nm. Then, the cDNAs were synthesized using FastKing gDNA Dispelling RT SuperMix (Tiangen, Beijing, China) according to the protocol.
Phylogenetic analysis
CSP sequences from plant bugs, aphid and plant hoppers were from previous study (Wang Q. et al., 2019). Sequences alignments were conducted with the MUSCLE alignment program (http://www.drive5.com/muscle/manual/). We used Neighbor-Joining algorithm (NJ) to construct unrooted phylogenies of these sequences with MEGA7 (Kumar et al., 2016). Bootstrap values were calculated with 1000 replicates; bootstrap values < 50% were deleted from the branches.
3D modeling
We used the strategies to predict the 3D modeling of the CSPs, which was described before (Zhang et al., 2020). By homology searching, the CSPs that we identified have > 40% homology with the CSP templates in the Protein Data Bank (http://www/rcsb.org/pdb). Based on the high sequence similarity (Supplementary Table S2) with the 3 E. onukii CSPs, NMR solution structure of CSPsg4 (PDB ID: 2gvs.1.A) was used as the template to build the 3D structures of the 3 CSPs using the online program SWISS MODEL. Information of the template was shown in Supplementary Table S2. The final 3D model was assessed using Verification Server (http://services.mbi.ucla.edu/SAVES/).
Expression patterns analysis
Body parts were collected from both adult males and females including head, thorax, abdomen, leg. Different parts were dissected from 1 to 5 days old adults (300 males and 300 females). Five biological replicates were prepared. Total RNA was extracted following the protocols above (section of Total RNA extraction and cDNA synthesis). We performed the qPCR to analyze the expression patterns of the 3 CSP genes (primers are listed in Supplementary Table S1). qPCR was performed on ABI Prism 7500 Fast Detection System (Applied Biosystems, Carlsbad, CA, United States). Each reaction contained 10 μl of 2 × GoTaq qPCR Master Mix, 0.4 μl of each primer (10 μM), 7 μl of nuclease-free water, 0.2 μl of CXR References Dye and 2 μl of sample cDNA (500 ng μl−1). The thermocycler program had an initial 95 denaturation step followed by 40 cycles consisting of a 10 s denaturation at 95, a 40 s annealing at 60, and a 30 s extension step at 72. The relative expression levels of BodoCSP1 were analyzed using the 2−ΔΔCT method (Li et al., 2021). β-actin gene was used as a control to normalize target gene expression and correct for sample-to-sample variation.
Bacterial expression and purification of recombinant CSP proteins
Primers designed for constructs were shown in Supplementary Table S1. Purified PCR products were ligated into the expression vector pET32a (+) with TRX-6 His tag (∼17 kDa) and the resulting construct were transformed into Escherichia coli BL21 (DE3) competent cells. These recombinant plasmid CSPs were firstly confirmed by sequencing. Bacteria transformed with recombinant plasmids were cultured in 1000 ml Luria−Bertani (LB) medium containing 100 μg/ml of ampicillin. Then the recombinant protein was induced at 16°C for 24 h with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when OD600 reached 0.4–0.6. We performed the centrifugation to collect the cells at 8000g for 5 min at 4°C and then sonicated in ice. The recombinant protein was further purified by Ni-NTA resin (GE). The poly-histidine tag was not removed following studies before (Terpe. 2003; Li et al., 2016; Tian and Zhang. 2016; Cui et al., 2018; Zhang et al., 2020). Purified protein was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Finally, purified protein was dialyzed with Buffer B. The molecular weights of recombinant EonuCSP proteins are consist with the predicted molecular weight of EonuCSPs (predicted at https://web.expasy.org/protparam/) plus a TRX-6 His tag (∼ 17 kDa).
Competitive fluorescence binding assays
Based on the results of previous studies (Zhao et al., 2002; Zhang et al., 2012; Zhao et al., 2014; Cai et al., 2017; Xu et al., 2017), we chose 15 typical volatile components as ligands for the fluorescence competitive binding assays (Table 1). These volatile components are the volatile components in tea (Cai et al., 2017) with purity > 98% (Table 1). N-phenyl-1-naphthylamine (1-NPN) was used as the fluorescent probe. The excitation wavelength in the fluorescence spectrometer was set to 337 nm, which was the results of an optimization that we achieved in lab. The scanning wavelength was 420–600 nm.
The tested chemicals were dissolved in methanol in preparation for 1 mM stock solution. To measure the affinity of 1-NPN to the CSP proteins, 2 μM solution of purified protein in buffer B was titrated with aliquots of 1 mM 1-NPN dissolved in methanol to final concentrations ranging from 2 to 20 μM. Then the affinities of ligands were tested by competitive binding assays through titrating the chemical competitor from 2 to 20 μM into the 1-NPN and EonuCSPs mixed solution (both at 2 μM). We determined the binding constants of 1-NPN by Scatchard formula. The dissociation constant (Kd) for binding between CSPs and 1-NPN was determined with the Scatchard linear regressive equation in the software GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA). The binding affinities of the competitors were evaluated from the corresponding IC50 according to the equation: Ki = IC50/(1 + [1-NPN]/K1-NPN). [1 – NPN] is the free concentration of 1 – NPN, while K1-NPN is the dissociation constant of the protein/1-NPN complex (Zhang et al., 2021). Tested compounds with Ki < 20 μM shows relatively high binding affinities to EonuCSPs.
Y-tube olfactometer assay
We use a Y-tube olfactometer (1.5 cm in diameter, arms with 8 cm length, and a stem with 18 cm length) to detect the behavioral response of E. onukii to volatiles. The incoming air to the tube was firstly filtered using active carbon and then humidified with the ultrapure water. The air through the arms was blown at a constant flow (300 ML/min). A total of 20 μL tested volatile oil (10 μg/μL in methanol (HPLC)) pipetted onto 1 cm diameter filter paper. This filter paper was placed in the chamber, which connected to one arm of the Y-tube olfactometer. As control, the same filter paper treated with 20 μL of methanol was placed in the chamber connected to another arm of the Y-tube. This experiment was performed in the dark room, which employed a light-emitting diode as the light source. Each adult was introduced into the middle of the stem of the Y-tube and the individuals that made a choice within 8 min were recorded. Individuals moving toward the odorant source for half of the arm distance and stayed for 1 minute were recorded as odorant choice. For each volatile component, 90 adult were used. After 10 insects were tested, the Y-tube olfactometer was washed with 75% ethanol and air-dried, and tested volatiles were placed in another arm for the subsequent tests. Odor resource are shown in Table 1.
Result
Sequence and homology analysis
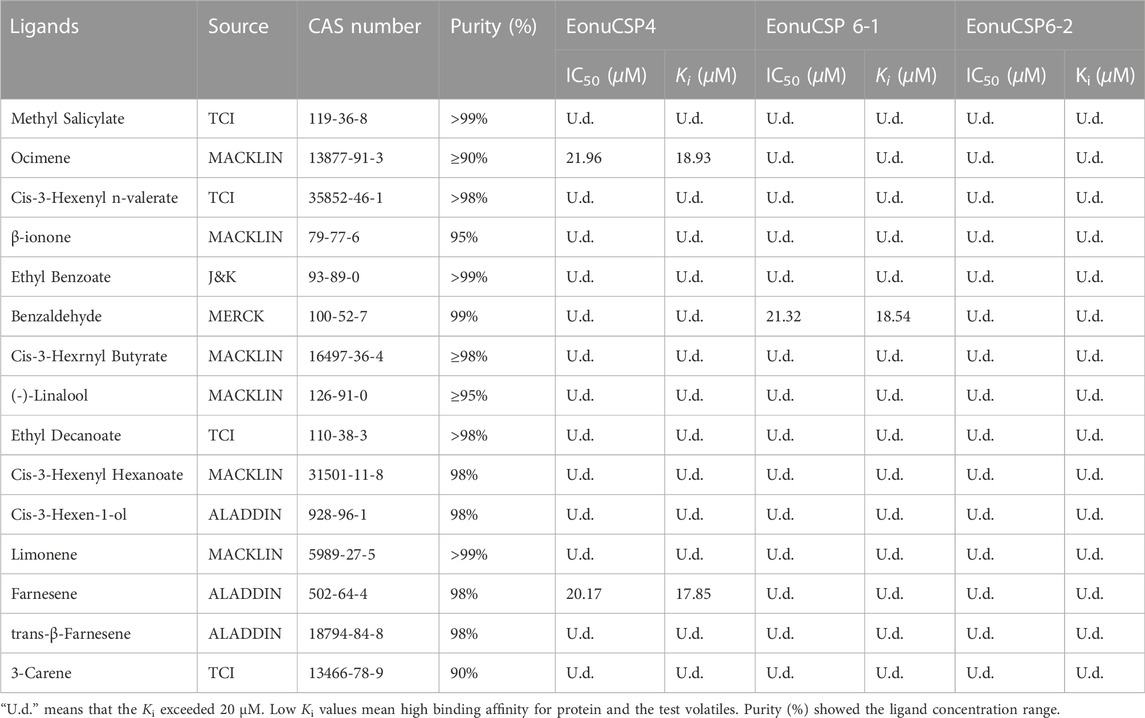
The full length cDNAs encoding EonuCSP4, 6-1, and 6-2 were amplified (Supplementary Figure S1), cloned and further verified by sequencing. The gene sequences were submitted to GenBank with accession number MF509603.1, MF509616.1 and MF509617.1. According to the genome annotation of E. onukii (Zhao et al., 2022), these 3 CSPs are located on Chromosome 1, with EonuCSP6-1 and 6-2 tandemly distributed (Supplementary Figure S2). Sequence analysis showed that their ORF sequences are 372 bp, 396 bp, and 399 bp respectively (Supplementary Table S3). Meanwhile, the EonuCSP 4, 6-1 and 6-2 proteins were predicted to contain the signal peptides of 18 amino acides at their N-terminus. Furthermore, the alignments of these cloned CSPs were done with homologues from other insects (Figure 1A). As the results showed, EonuCSP4, 6-1, and 6-2 were found to contain typical characteristics including 4 conserved cysteine residues with the following pattern: C1-X6-8-C2-X16-21-C3-X2-C4 (Figure 1A).
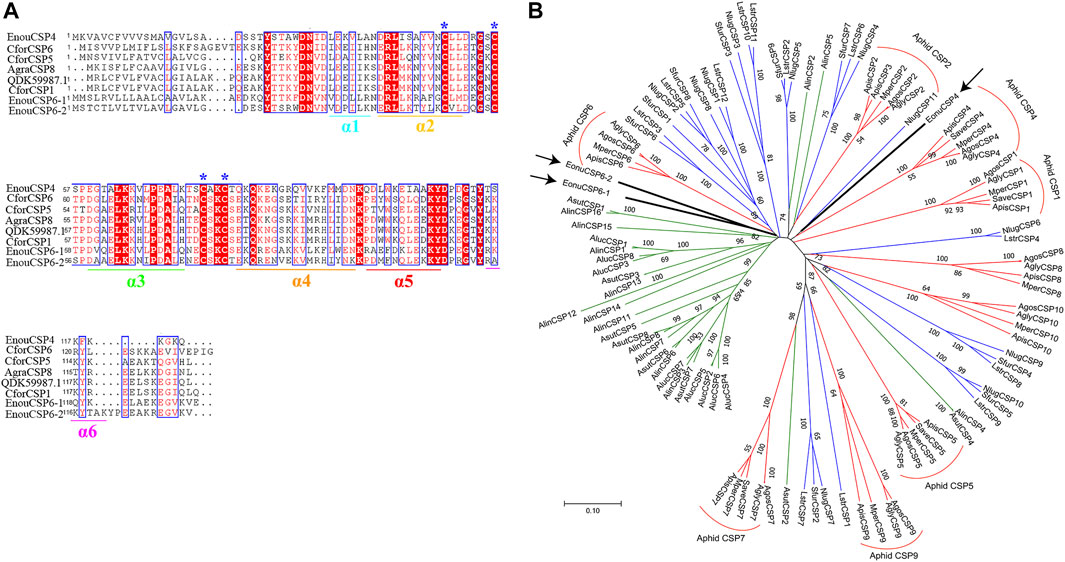
FIGURE 1. Homology analysis and phylogenetic relationship of EonuCSPs with CSPs from other insects. (A). Secondary structure of E. onukii chemosensory proteins (EonuCSP4, 6-1, and 6-2). α -helices are displayed as straight. Identical residues are highlighted in white letters with a red background. Residues with similar physicochemical properties are shown in red letters with a blue frame. The conserved cysteines are labeled with asterisk. (B). Phylogenetic relationships of the CSPs. Species includes hoppers (N. lugens (Nlug), S. furcifera (Sfur), and L. striatellus (Lstr)], aphids (A. pisum (Apis), M. persicae (Mper), A. gossypii (Agos), A. glycines (Agly), and S. avenae (Save)), and plant bugs (N. lugens (Alug), L. striatellus (Lstr), and S. furcifera (Sfur)). E. onukii CSPs was marked with arrowhead. CSPs are classified into 8 subfamilies in Aphid based on previous study (Wang Q. et al., 2019), which were marked in red. The 3 CSPs belonged to aphid CSP4 and aphid CSP6 cluster respectively. Bootstrap values were calculated with 1000 replicates; bootstrap values < 50% were deleted from the branches.
Moreover, we searched the CSPs in other Hemiptera species including aphid (Acyrthosiphon pisum (Api), Myzus persicae (Mpe), Aphis gossypii (Ago), Aphis glycines (Agl) and Sitobion avenae (Sav)), plant bugs (Adelphocoris lineolatus (Alin), Adelphocoris suturalis (Asu), Apolygus lucorum (Alu)), and plant hoppers [Empoasca onukii (Eon), Laodelphax striatellus (Lst), Nilaparvata lugens (Nlu) and Sogatella furcifera (Sfu)]. Based on the results, EonuCSPs are clearly clustered together to form 2 homologous subgroups named CSP4 and CSP6 supported by high bootstrap values (Figure 1B).
3D model of the 3 EonuCSP
The 3D structurally determined CSP, chemosensory protein CSP-sg4 (PBD ID: 2gvs.1.A) was found to share sequence similarities of more than 45% with the 3 CSPs (Supplementary Table S2). Thus, we selected this structurally determined CSP as template to build the 3D structural model of EonuCSP4, 6-1, and 6-2 using SWISS-MODEL (Figure 2). The results or Ramachandran plot showed that 87.9%, 85.1%, and 87.2% of the residues were in preferred regions, all the residues (100%) were in the allowed region (Supplementary Figure S3), suggesting that the predicted models of the 3 CSPs are generally reliable. The predicted 3D structures of these 3 CSPs consist of six α-helices (Figures 1A, 2).
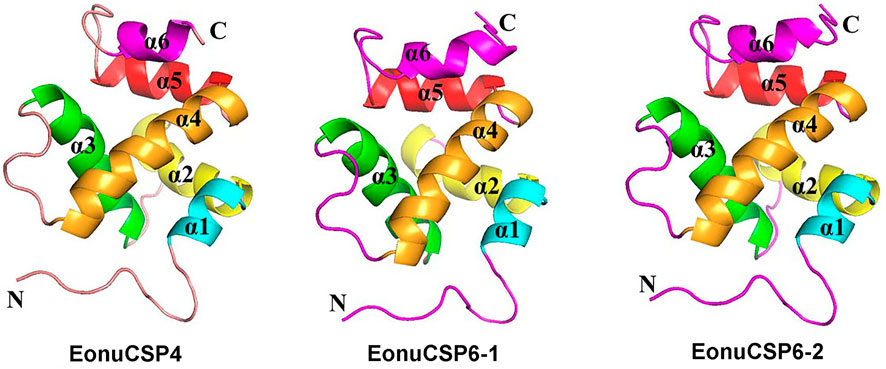
FIGURE 2. Three-dimensional structure of EonuCSPs. N-terminus, C-terminus, and helices are labeled. The six α-helices are labeled as α1- α6.
Expression profile of EonuCSP4, 6-1 and 6-2
Based on the previous assessment, EonuCSP4, 6-1, and 6-2 were predominantly expressed in antennae (Zhao et al., 2017). For more accurate estimation of the expression profiles, we analyzed the expression patterns of the CSPs in multiple body parts. Results showed that none of the 3 CSP genes were expressed in a specific body part (Figure 3). EonuCSP4 was expressed higher in the head and abdomen (p < 0.05); while EonuCSP6-1 and EonuCSP6-2 exhibited a higher expression in thorax than in other body parts (p < 0.05; Figure 3).
FIGURE 3. Expression profiles of the 3 CSPs in E. onukii. Transcript levels of the CSPs were normalized byβ-actin.
Bacterial expression and purification of the 3 EonuCSP
The high expression of the 3 CSPs in the antennae suggested that they were potentially involved in peripheral olfactory reception for E. onukii. In order to screen the putative ligands for these CSPs, we first expressed the 3 CSPs in a bacterial system. pET-32a (+)/EonuCSPs were successfully induced and expressed in BL21 (DE3) cells. All these 3 EonuCSPs were mainly present in the supernatant. Subsequently, expression and purification of EonuCSP4, EonuCSP 6-1 and EonuCSP6-2 recombinant proteins containing His-Tag were assessed by SDS-PAGE with the molecular weight of ∼30.61 kDa, ∼32.06 kDa, and ∼32.12 kDa respectively (Figure 4).
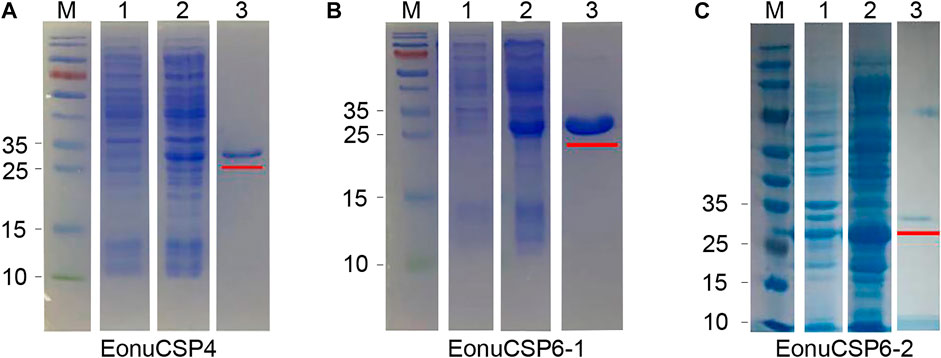
FIGURE 4. Recombinant protein analyzed by SDS-PAGE. (A). Expression and purification of recombinant protein EonuCSP4; (B). Expression and purification of recombinant protein EonuCSP6-1; (C). Expression and purification of recombinant protein EonuCSP6-2. Lane M: molecular marker. Lane 1: cell pellet before induction with IPTG. Lane 2: cell pellet after induction with IPTG. Lane 3: purified protein, which was also marked in red lines.
Fluorescence binding assay
For the ligand binding assay, the binding affinity of the fluorescent probe 1-NPN with the 3 purified CSP proteins were firstly measured (Figure 5A). Results revealed that the 3 purified CSP proteins (EonuCSP 4, 6-1 and 6-2) were capable of binding 1-NPN with the dissociation constants (Kd) were 11.02, 12.52 and 10.22 μM respectively. Then, the fluorescence competitive binding assay was performed to determine the binding affinities of EonuCSP4, 6-1 and 6-2 (Figure 5B). Results revealed that different CSP protein displayed various binding spectrum (Table 1; Figure 5B). And of the 15 tested compounds, each CSP showed a relatively narrow binding spectrum (Table 1; Figure 5B). For EonuCSP4, the binding test results indicated that this CSP showed much higher affinity to farnesene (Ki = 17.85) and ocimene (Ki = 18.93); whereas EonuCSP6-1 showed binding to benzaldehyde (Ki = 18.54). However, EonuCSP6-2 showed little binding to all the tested compounds, with Ki >20.
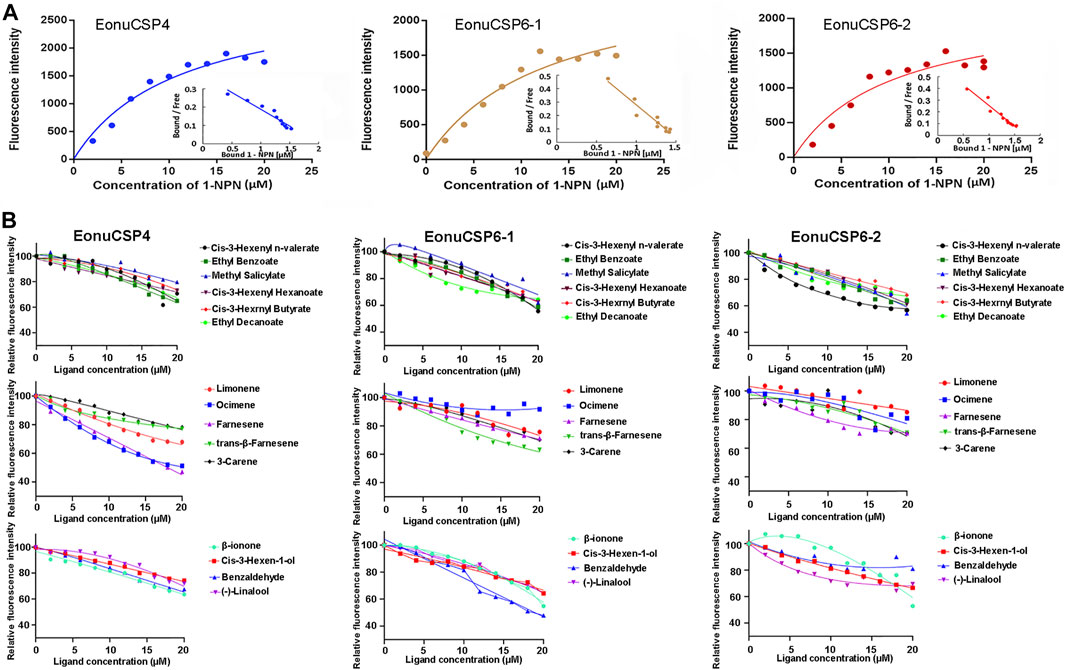
FIGURE 5. Fluorescence competitive ligand-binding assays of EonuCSPs. (A). Binding curve and the Scatchard plot of N-phenyl-1-naphthylamine (1-NPN) to EonuCSP4, 6-1, and 6-2 respectively. (B). Competitive binding curves of EonuCSP4, 6-1, and 6-2 with various ligands including ester substances (Cis-3-Hexenyl n-valerate, Ethyl Benzoate, Methyl Salicylate, Cis-3-Hexenyl Hexanoate, Cis-3-Hexrnyl Butyrate, and Ethyl Decanoate), alkene substances (Limonene, Ocimene, Farnesene, trans-β-Farnesene, and 3-Carene), and aldehydes, ketones and alcohols [(-)-Linalool, Cis-3-Hexen-1-ol, β-ionone, and Benzaldehyde]. Dissociation constants (Ki) of all examined ligands were evaluated. Low Ki values mean high binding affinity for protein and the plant volatiles.
Behavioral trials
The behavioral responses of E. onukii adults to the tested compounds were investigated in a Y-tube olfactometer. Results showed that the adults of E. onukii displayed a significant preference for benzaldehyde and cis-3-hexenyl n-valerate (Figure 6), suggesting that other volatiles had no significant effect on the behavior of these insects at concentration of 10 μg/μl.
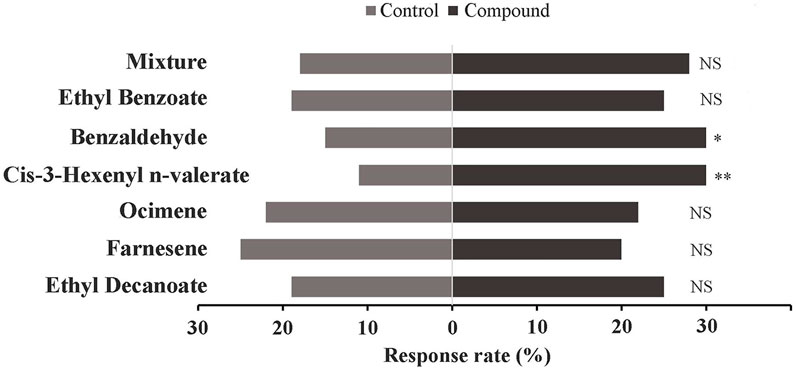
FIGURE 6. Response of E. onukii to 10 μg/μL compounds. Asterisks indicate significant differences; *p < 0.05, and **p < 0.01. NS indicates no significant difference (χ2 test). Two compounds, Benzaldehyde (chi-square: 5; p value: 0.026) and Cis-3-Hexenyl n-valerate (chi-square: 8.8; p value: 0.003), showed significant differences in this assay.
Discussion
CSPs of insects could recognize, bind, and transport the chemicals to odorant receptors, which play vital roles in chemoreception (Leal 2013; Pelosi et al., 2018; Zhang et al., 2021). Insect CSPs are also widely expressed in various tissues, which are proved to be involved in multiple physiological processes (Iovinella et al., 2013; Zhao et al., 2017). In this study, 3 CSPs of E. onukii were cloned from E. onukii, and the respective proteins they encoded were determined to have four typical conserved cysteine spacing motif, which showed a relative high identity with other insect CSPs (Zhang et al., 2020; Hua et al., 2021; Zhan et al., 2021). We investigated the biological binding characteristics of the 3 E. onukii CSPs to various plant volatiles. In addition, the 3 CSPs were functionally characterized using behavior bioassays to investigate the involvement of the examined CSPs in plant–volatile detection and plant host orientation behavior.
CSPs of insects exhibit broad expression profiles in various tissues (Wang Z. et al., 2021). qRT-PCR results also showed that the expression of EonuCSP 4, 6-1 and 6-2 were expressed in various body parts. Based on the transcriptomes of E. onukii, these three CSPs were also expressed in both antennae and body (Zhao et al., 2017). These broad and diverse expression profiles of EonuCSP 4, 6-1 and 6-2 may consistent with their potential multiple roles in chemoreception and contact chemoreception. Similar results were observed in Bradysia odoriphaga, B. mori, and Nilaparvata lugens, in which CSPs was expressed in antennae and the body (e.g. heads, wings, abdomen, and legs) (Gong et al., 2007; Waris et al., 2018; Younas et al., 2018; Zhang et al., 2021), suggesting the potential roles in contact chemoreception. Because EonuCSP 4, 6-1 and 6-2 expression were also found in heads, antennae and legs, we speculated that these 3 CSPs function in chemosensation.
The predicted 3D structure of the 3 CSPs exhibited conserved structural features, including six α-helices that form the hydrophobic ligand-biding pocket (Lartigue et al., 2002; Zhang et al., 2021). To further infer the potential roles of EonuCSP 4, 6-1 and 6-2, the fluorescence competitive binding assay were performed. Of the 15 tested compounds, only 3 compounds showed relatively high binding affinities (Ki < 20 μM) for EonuCSP4 (binding to farnesene and ocimene) and EonuCSP6-1 (benzaldehyde), whereas EonuCSP6-2 showed weak binding to all the tested compounds (Ki > 20 μM). These results indicated that EonuCSP4 and EonuCSP6-1 might have the appropriate binding sites for the compounds, which involved in chemoreception of E. onukii. Behavioral responses also showed that adult E. onukii exhibited a significant preference for benzaldehyde, which was confirmed by previous studies (Cai et al., 2017). Benzaldehyde is a prominent scent compound which greatly contribute to the fruity, floral smells of flowers, plants, and fruits. Also, benzaldehyde has a typical almond-like odor, which is one of the key volatile components in all types of tea, especially green tea, black tea, and oolong tea (Yang et al., 2013; Wang X. et al., 2019). Thus, these findings indicated that EonuCSP6-1 is a chemoreception protein, which may be involved in attraction of E. onukii adults to host. Behavioral responses also indicated that adult E. onukii exhibited a significant preference for Cis-3-Hexenyl n-valerate. However, we did not identified the potential CSPs that involved in binding this compound.
In summary, we report the expressions and ligand binding capabilities of 3 CSP of E. onukii, proving the potential olfactory roles of CSPs in host-location behavior of tea leafhopper. Totally, three volatiles exhibited strong binding abilities for CSPs in E. onukii, and one of them were able to elicit behavioral responses. These results provide clues into the mechanism of olfactory recognition of E. onukii. Besides, this study indicate that the CSPs in E. onukii are involved into the functions of host plant volatiles perception, and can be used as a molecular target for screening behaviorally active compounds for alternative control strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. Coding sequences of EonuCSP4, 6-1, and 6-2 are deposited in GenBank with accession number MF509603.1, MF509616.1 and MF509617.1, respectively.
Ethics statement
The pests were collected from the tea plants and maintained on tea plants in lab.
Author contributions
QZ and MY designed the research. QZ analyzed the sequence structure, constructed phylogenetic trees, and wrote the manuscript. HH, MZ, and FA collected the samples. LZ cloned the sequences and performed the fluorescence competitive binding assay. MZ and FA conducted the qPCR and analyzed the results. MZ performed the behavioral assays. MY edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by The National Key R & D Program of China (2019YFD1002100), Natural Science Foundation of Fujian Province, China (Grant No. 2020J01525), Fujian Agriculture and Forestry University Construction Project for Technological Innovation and Service System of Tea Industry Chain (Grant No. K1520005A03), and Quanzhou City Science & Technology Program of China (Grant No. 2020N008s).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1068543/full#supplementary-material
Supplementary Figure S1 | PCR amplification of 3 EonuCSPs. Line 1 represents another EonuCSP lacking two α-helices, which was exclude from further analysis.
Supplementary Figure S2 | Chromosomal distribution of EonuCSP4, 6-1, and 6-2.</b> All these three genes were located on chromosome 1 of E. onukii.
Supplementary Figure S3 | PROCHECK results from the predicted 3D model of EonuCSP4, 6-1, and 6-2. Ramachandran plot showed that > 85% of the residues were in preferred regions, all the residues (100%) were in the allowed region.
References
Bruce T. J., Wadhams L. J., Woodcock C. M. (2005). Insect host location: A volatile situation. Trends Plant Sci. 10, 269–274. doi:10.1016/j.tplants.2005.04.003
Butterwick J. A., Del Mármol J., Kim K. H., Kahlson M. A., Rogow J. A., Walz T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. doi:10.1038/s41586-018-0420-8
Cai X., Bian L., Xu X., Luo Z., Li Z., Chen Z. M. (2017). Field background odour should be taken into account when formulating a pest attractant based on plant volatiles. Sci. Rep. 7, 41818. doi:10.1038/srep41818
Carey A. F., Wang G., Su C. Y., Zwiebel L. J., Carlson J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Anopheles Gamb. Nat. 464, 66–71. doi:10.1038/nature08834
Chen L. L., Yuan P., Pozsgai G., Chen P., Zhu H., You M. S. (2019). The impact of cover crops on the predatory mite Anystis baccarum (Acari, Anystidae) and the leafhopper pest Empoasca onukii (Hemiptera, Cicadellidae) in a tea plantation. Pest Manag. Sci. 75, 3371–3380. doi:10.1002/ps.5489
Cheng D., Lu Y., Zeng L., Liang G., He X. (2015). Si-CSP9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 5, 9245. doi:10.1038/srep09245
Cui X., Liu D., Sun K., He Y., Shi X. (2018). Expression profiles and functional characterization of two odorant-binding proteins from the apple buprestid beetle Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 111 (3), 1420–1432. doi:10.1093/jee/toy066
Del Mármol J., Yedlin M. A., Ruta V. (2021). The structural basis of odorant recognition in insect olfactory receptors. Nature 597 (7874), 126–131. doi:10.1038/s41586-021-03794-8
Ebrahim S. A., Dweck H. K., Stokl J., Hofferberth J. E., Trona F., Weniger K., et al. (2015). Drosophila avoids parasitoids by sensing their semiochemicals via a dedicated olfactory circuit. PLoS Biol. 13, e1002318. doi:10.1371/journal.pbio.1002318
Gong D. P., Zhang H. J., Zhao P., Lin Y., Xia Q. Y., Xiang Z. H. (2007). Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 37, 266–277. doi:10.1016/j.ibmb.2006.11.012
Guo W., Wang X., Ma Z., Xue L., Han J., Yu D., et al. (2011). CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 7, e1001291. doi:10.1371/journal.pgen.1001291
Ha T. S., Smith D. P. (2009). Odorant and pheromone receptors in insects. Front. Cell Neurosci. 3, 10. doi:10.3389/neuro.03.010.2009
Hua J., Fu Y., Zhou Q., Huang Y., Li H., Chen T., et al. (2021). Three chemosensory proteins from the sweet potato weevil, Cylas formicarius, are involved in the perception of host plant volatiles. Pest Manag. Sci. 77, 4497–4509. doi:10.1002/ps.6484
Iovinella I., Bozza F., Caputo B., Della Torre A., Pelosi P. (2013). Ligand-binding study of Anopheles gambiae chemosensory proteins. Chem. Senses 38, 409–419. doi:10.1093/chemse/bjt012
Ishida Y., Leal W. S. (2005). Rapid inactivation of a moth pheromone. Proc. Natl. Acad. Sci. U. S. A. 102, 14075–14079. doi:10.1073/pnas.0505340102
Jin S., Chen Z. M., Backus E. A., Sun X. L., Xiao B. (2012). Characterization of EPG waveforms for the tea green leafhopper, Empoasca vitis Gothe (Hemiptera: Cicadellidae), on tea plants and their correlation with stylet activities. J. Insect Physiol. 58, 1235–1244. doi:10.1016/j.jinsphys.2012.06.008
Kitabayashi A. N., Arai T., Kubo T., Natori S. (1998). Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem. Mol. Biol. 28, 785–790. doi:10.1016/s0965-1748(98)00058-7
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lartigue A., Campanacci V., Roussel A., Larsson A. M., Jones T. A., Tegoni M., et al. (2002). X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 277, 32094–32098. doi:10.1074/jbc.M204371200
Laughlin J. D., Ha T. S., Jones D. N. M., Smith D. P. (2008). Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265. doi:10.1016/j.cell.2008.04.046
Leal W. S., Chen A. M., Ishida Y., Chiang V. P., Erickson M. L., Morgan T. I., et al. (2005). Kinetics and molecular properties of pheromone binding and release. Proc. Natl. Acad. Sci. U. S. A. 102, 5386–5391. doi:10.1073/pnas.0501447102
Leal W. S. (2013). Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Li G. W., Zhang Y., Y Li Y. P., Wu J. X., Xu X. L. (2016). Cloning, expression, and functional analysis of three odorant-binding proteins of the oriental fruit moth, Grapholita molesta (BUSCK) (Lepidoptera: Tortricidae). Arch. Insect Biochem. Physiol. 91, 67–87. doi:10.1002/arch.21309
Li T., Zhang L., Li Y., You M. S., Zhao Q. (2021). Functional analysis of the orphan genes Tssor-3 and Tssor-4 in male Plutella xylostella. J. Integr. Agr. 20, 1880–1888. doi:10.1016/s2095-3119(21)63655-9
Li Z. Q., Zhang S., Luo J. Y., Zhu J., Cui J. J., Dong S. L. (2015). Expression analysis and binding assays in the chemosensory protein gene family indicate multiple roles in helicoverpa armigera. J. Chem. Ecol. 41, 473–485. doi:10.1007/s10886-015-0574-x
Maleszka J., Foret S., Saint R., Maleszka R. (2007). RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 217, 189–196. doi:10.1007/s00427-006-0127-y
Mao Y., Xu X., Xu W., Ishida Y., Leal W. S., Ames J. B., et al. (2010). Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc. Natl. Acad. Sci. U. S. A. 107, 19102–19107. doi:10.1073/pnas.1012274107
Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R. (2018). Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb Philos. Soc. 93, 184–200. doi:10.1111/brv.12339
Sato K., Pellegrino M., Nakagawa T., Nakagawa T., Vosshall L. B., Touhara K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi:10.1038/nature06850
Stengl M., Funk N. W. (2013). The role of the coreceptor Orco in insect olfactory transduction. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 199, 897–909. doi:10.1007/s00359-013-0837-3
Stocker R. F. (1994). The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 275, 3–26. doi:10.1007/BF00305372
Tegoni M., Campanacci V., Cambillau C. (2004). Structural aspects of sexual attraction and chemical communication in insects. Trends Biochem. Sci. 29, 257–264. doi:10.1016/j.tibs.2004.03.003
Terpe K. (2003). Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60, 523–533. doi:10.1007/s00253-002-1158-6
Tian Z., Zhang Y. (2016). Molecular characterization and functional analysis of pheromone binding protein 1 from Cydia pomonella (L.) Insect Mol. Biol. 25, 769–777. doi:10.1111/imb.12261
Vogt R. G., Riddiford L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi:10.1038/293161a0
Wang H., Ma Y. F., Wang M. M., Chen G. L., Dewer Y., He M., et al. (2020). Expression, affinity, and functional characterization of the specific binding of two putative pheromone-binding proteins in the omnivorous German cockroach Blattella germanica. Blattella Ger. J. Agric. Food Chem. 68, 13573–13583. doi:10.1021/acs.jafc.0c02456
Wang M. M., Long G. J., Guo H., Liu X. Z., Wang H., Dewer Y., et al. (2021). Two carboxylesterase genes in Plutella xylostella associated with sex pheromones and plant volatiles degradation. Pest Manag. Sci. 77, 2737–2746. doi:10.1002/ps.6302
Wang Q., Zhou J. J., Liu J. T., Huang G. Z., Xu W. Y., Zhang Q., et al. (2019). Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids. Insect Mol. Biol. 28, 1–22. doi:10.1111/imb.12513
Wang X., Zeng L., Liao Y., Zhou Y., Xu X., Dong F., et al. (2019). An alternative pathway for the formation of aromatic aroma compounds derived from l-phenylalanine via phenylpyruvic acid in tea (Camellia sinensis (L.) O. Kuntze) leaves. Food Chem. 270, 17–24. doi:10.1016/j.foodchem.2018.07.056
Wang Z., Yang F., Sun A., Shan S., Zhang Y., Wang S. (2021). Expression profiles and functional characterization of chemosensory protein 15 (HhalCSP15) in the Brown marmorated stink bug Halyomorpha halys. Front. Physiol. 12, 721247. doi:10.3389/fphys.2021.721247
Waris M. I., Younas A., Ul Qamar M. T., Hao L., Ameen A., Ali S., et al. (2018). Silencing of chemosensory protein gene NlugCSP8 by RNAi induces declining behavioral responses of Nilaparvata lugens. Nil. lugens Front. Physiol. 9, 379. doi:10.3389/fphys.2018.00379
Wicher D., Schafer R., Bauernfeind R., Stensmyr M. C., Heller R., Heinemann S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi:10.1038/nature06861
Xu X., Cai X., Bian L., Luo Z., Li Z., Chen Z. (2017). Does background odor in tea gardens mask attractants? Screening and application of attractants for Empoasca onukii matsuda. J. Econ. Entomol. 110 (6), 2357–2363. doi:10.1093/jee/tox269
Xuan N., Guo X., Xie H. Y., Lou Q. N., Lu X. B., Liu G. X., et al. (2015). Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 22, 203–219. doi:10.1111/1744-7917.12116
Yang Z., Baldermann S., Watanabe N. (2013). Recent studies of the volatile compounds in tea. Food Res. Int. 53, 585–599. doi:10.1016/j.foodres.2013.02.011
Younas A., Waris M. I., Tahir Ul Qamar M., Shaaban M., Prager S. M., Wang M. Q. (2018). Functional analysis of the chemosensory protein MsepCSP8 from the oriental armyworm Mythimna separata. Front. Physiol. 9, 872. doi:10.3389/fphys.2018.00872
Zhan H., Dewer Y., Zhang J., Tian J., Li D., Qu C., et al. (2021). Odorant-binding protein 1 plays a crucial role in the olfactory response of Bemisia tabaci to R-curcumene. J. Agric. Food Chem. 69, 12785–12793. doi:10.1021/acs.jafc.1c03825
Zhang C., Tang B., Zhou T., Yu X., Hu M., Dai W. (2021). Involvement of chemosensory protein BodoCSP1 in perception of host plant volatiles in Bradysia odoriphaga. J. Agric. Food Chem. 69, 10797–10806. doi:10.1021/acs.jafc.1c02807
Zhang F. M., Merchant A., Zhao Z. B., Zhang Y. H., Zhang J., Zhang Q. W., et al. (2018). The odorant binding protein 6 expressed in sensilla chaetica displays preferential binding affinity to host plants volatiles in Ectropis obliqua. Front. Physiol. 9, 534–611. doi:10.3389/fphys.2018.00534
Zhang H., Chen J. L., Lin J. H., Lin J. T., Wu Z. Z. (2020). Odorant-binding proteins and chemosensory proteins potentially involved in host plant recognition in the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 76, 2609–2618. doi:10.1002/ps.5799
Zhang H., Liu F., Li H., Wang D. (2012). Advances of studies on tea plant volatiles. Tea Sci. Technol. 4, 4–8.
Zhang L., Wang F., Qiao L., Dietrich C. H., Matsumura M., Qin D. (2019). Population structure and genetic differentiation of tea green leafhopper, Empoasca (Matsumurasca) onukii, in China based on microsatellite markers. Sci. Rep. 9, 1202. doi:10.1038/s41598-018-37881-0
Zhao D., Gao J., Chen Z., Cheng J., Xu H. (2002). Orientation response of Empoasca vitis to tea shoots volatiles. J. South China Agric. Univ. Nat. Sci. Ed. 23, 27–29.
Zhao L., Cui H., Zhang L., Chen L., Yu J., Li H. (2014). Molecular binding characterization with tea plant volatiles of a general odorant-binding protein EoblGOBP2 in the tea geometrid, Ectropis oblique Prout (Lepidoptera: Geometridae). J. Tea Sci. 34, 165–171.
Zhao Q., Shi L., He W., lI J., You S., Chen S., et al. (2022). Genomic variation in the tea leafhopper reveals the basis of adaptive evolution. Genom Proteom Bioinf. doi:10.1016/j.gpb.2022.05.011
Zhao Y., Li H., Wang Q., Liu j., Zhang L., Mu W., et al. (2017). Identification and expression analysis of chemosensory genes in the tea green leafhopper, Empoasca onukii Matsuda. J. Appl. Entomol. 142, 828–846. doi:10.1111/jen.12530
Zhou J. J., Kan Y., Antoniw J., Pickett J. A., Field L. M. (2006). Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem. Senses 31, 453–465. doi:10.1093/chemse/bjj050
Keywords: chemosensory proteins, Empoasca onukii, insect chemoreception, plant volatiles, benzaldehyde
Citation: Zhang L, Zhao M, Aikeremu F, Huang H, You M and Zhao Q (2023) Involvement of three chemosensory proteins in perception of host plant volatiles in the tea green leafhopper, Empoasca onukii. Front. Physiol. 13:1068543. doi: 10.3389/fphys.2022.1068543
Received: 13 October 2022; Accepted: 12 December 2022;
Published: 04 January 2023.
Edited by:
Amr A. Mohamed, Cairo University, EgyptReviewed by:
Nathália F. Brito, National Cancer Institute (INCA), BrazilSebastien Lebreton, Institut de Recherche en Sémiochimie et Ethologie Appliquée (IRSEA), France
Alberto Maria Cattaneo, Swedish University of Agricultural Sciences, Sweden
Copyright © 2023 Zhang, Zhao, Aikeremu, Huang, You and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minsheng You, bXN5b3VAZmFmdS5lZHUuY24=; Qian Zhao, emhhb3FpYW45NzcyMjhAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Liwen Zhang
Liwen Zhang Mingxian Zhao1,2†
Mingxian Zhao1,2† Minsheng You
Minsheng You Qian Zhao
Qian Zhao