
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 05 January 2023
Sec. Red Blood Cell Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1063294
This article is part of the Research TopicStress ErythropoiesisView all 6 articles
Steady state erythropoiesis produces new erythrocytes at a constant rate to replace the senescent cells that are removed by macrophages in the liver and spleen. However, infection and tissue damage disrupt the production of erythrocytes by steady state erythropoiesis. During these times, stress erythropoiesis is induced to compensate for the loss of erythroid output. The strategy of stress erythropoiesis is different than steady state erythropoiesis. Stress erythropoiesis generates a wave of new erythrocytes to maintain homeostasis until steady state conditions are resumed. Stress erythropoiesis relies on the rapid proliferation of immature progenitor cells that do not differentiate until the increase in serum Erythropoietin (Epo) promotes the transition to committed progenitors that enables their synchronous differentiation. Emerging evidence has revealed a central role for cell metabolism in regulating the proliferation and differentiation of stress erythroid progenitors. During the initial expansion stage, the immature progenitors are supported by extensive metabolic changes which are designed to direct the use of glucose and glutamine to increase the biosynthesis of macromolecules necessary for cell growth and division. At the same time, these metabolic changes act to suppress the expression of genes involved in erythroid differentiation. In the subsequent transition stage, changes in niche signals alter progenitor metabolism which in turn removes the inhibition of erythroid differentiation generating a bolus of new erythrocytes to alleviate anemia. This review summarizes what is known about the metabolic regulation of stress erythropoiesis and discusses potential mechanisms for metabolic regulation of proliferation and differentiation.
Steady state erythropoiesis maintains erythroid homeostasis, which requires precise control over the production and turnover of erythrocytes. The bone marrow has an enormous capacity for erythropoiesis and produces 2.5 × 10 ^ 6 erythrocytes per second in humans (Palis, 2014; Seu et al., 2017). This production is offset by a similar rate of turnover of erythrocytes in the spleen and liver such that the concentration of erythrocytes maintained in circulation optimizes oxygen delivery and minimizes problems due to increased blood viscosity (Klei et al., 2017). Situations that compromise oxygen delivery require a response that will increase tissue oxygenation. Blood loss and hypoxia can be compensated by increasing Epo levels which increases differentiation of committed erythroid progenitors (Tusi et al., 2018). In addition, sustained increases in serum Epo levels can skew hematopoiesis shunting progenitor cells down the CMP-MEP erythroid progenitor pathway at the expense of myelopoiesis (Grover et al., 2014; Singh et al., 2018). This phenomenon is enhanced steady state erythropoiesis. In contrast, inflammation caused by tissue damage or infection affects erythropoiesis at many levels. Pro-inflammatory cytokines inhibit steady state erythropoiesis (Rusten and Jacobsen, 1995; Zamai et al., 2000; Xiao et al., 2002; Tsopra et al., 2009). These factors also skew hematopoiesis towards myelopoiesis which increases the production of myeloid effector cells at the expense of steady state erythropoiesis (Oduro et al., 2012; Pietras et al., 2016; Pietras, 2017; Caiado et al., 2021). Pro-inflammatory cytokines like IL-6 increase the levels of hepcidin, which leads to sequestration of iron, making it unavailable for hemoglobin synthesis (Jurado, 1997; Cassat and Skaar, 2013; Soares and Weiss, 2015). Furthermore, inflammatory signals increase erythrocyte turnover, which further exacerbates the anemia (Libregts et al., 2011; Akilesh et al., 2019). To compensate for this loss in production, inflammation induces stress erythropoiesis (Paulson et al., 2020a). Unlike steady state erythropoiesis, the pro-inflammatory cytokines initiate stress erythropoiesis and promote the proliferation of immature progenitor cells (Bennett et al., 2019). The expansion of this progenitor population leads to increased production of erythroferrone (Erfe), which inhibits hepcidin expression so that once these progenitors start to differentiate, iron will be available for hemoglobin biosynthesis (Kautz et al., 2014a; Kautz et al., 2014b; Arezes et al., 2018). Furthermore, increased erythroid turnover by macrophages leads to heme dependent signaling, which increases production by macrophages of key factors like GDF15 and BMP4 that are required for stress erythropoiesis (Bennett et al., 2019). These observations support the idea that stress erythropoiesis is part of a coordinated inflammatory response, which allows bone marrow hematopoiesis to concentrate on the production of immune effector cells while erythroid homeostasis is maintained by extra-medullary stress erythropoiesis (Paulson et al., 2020a). In mice, this response is primarily in the spleen and liver. Stress erythropoiesis is highly conserved in humans (Xiang et al., 2015; Paulson et al., 2020b; Chen et al., 2020). Although the location of stress erythropoiesis in humans has not be definitively identified, there are many reports of extra-medullary stress erythropoiesis in anemia patients (for review see (Paulson et al., 2020b)).
Because of the stress response nature of stress erythropoiesis, the strategy for erythroid production is different from steady state erythropoiesis. As described above, steady state erythropoiesis constantly produces new erythrocytes. Stress erythropoiesis generates a bolus of new erythrocytes that maintain homeostasis until the source of the inflammation can be resolved (Perry et al., 2009; Paulson et al., 2020b). This response is more similar to stem cell-based tissue regeneration systems like those observed in muscle, lung and the intestinal epithelium (Asfaha, 2015; Tidball, 2017; Shen et al., 2022). These systems rely on tissue specific stem cells that are distinct from the stem cells that maintain tissue homeostasis. A common theme in tissue regeneration is that these stem cells respond to inflammatory signals which leads to proliferation of a transient amplifying population of progenitors that differentiates into mature cells that repair the damaged tissue. One of the better characterized regeneration systems is muscle regeneration, where satellite cells resident in the muscle are activated by inflammatory signals expressed by macrophages and infiltrating monocytes (Burzyn et al., 2013; Tidball, 2017; Scaramozza et al., 2019; Theret et al., 2019; Theret et al., 2022). These cells proliferate and differentiate to form new myotubes that repair the damaged muscle. The signals that drive the differentiation of myogenic precursors coincide with a switch from an inflammatory environment to a pro-resolving response. This example underscores the key role for inflammatory signals in initiating tissue regeneration, but also shows that resolving inflammatory signals plays a role in the transition to differentiation. Stress erythropoiesis shows a similar developmental trajectory. The initiation of stress erythropoiesis relies on inflammatory signals. Macrophage expression of TNFα, Interferon γ (Ifnγ) and IL-1β is transiently increased in the spleen following treatment with LPS and zymosan, which induces inflammatory anemia. In vitro, TNFα increases the proliferation of immature stress erythroid progenitors (SEPs) suggesting that transient TNFα signaling in SEPs drives their proliferation (Bennett et al., 2019). The mechanism by which TNFα promotes proliferation is not understood. At this stage, SEPs maintain stem cell characteristics, are capable of self-renewal, but do not differentiate (Harandi et al., 2010; Xiang et al., 2015). Analysis of SEPs showed that this transient amplifying population is heterogenous, made up of three distinct populations, with CD34+CD133+Kit + Sca1+ cells being the most immature, CD34negCD133+Kit + Sca1+ being the intermediate, and CD34negCD133negKit + Sca1+ being the most mature (Xiang et al., 2015). The transition from a proliferating SEP to a SEP committed to erythroid differentiation relies on erythropoietin (Epo) signaling in splenic macrophages, which alters the signals made by the niche. Epo signaling leads to a loss of pro-proliferative signals like canonical Wnts and an increase in signals that promote differentiation like Prostaglandin E2 (PGE2) (Chen et al., 2020). This change in signals is part of a change in the niche from a pro-inflammatory niche dominated by M1 macrophage type signals to a pro-resolving niche dominated by M2 macrophage like signals (Bennett et al., 2019). After this transition, SEPs lose their ability to self-renew and commit to differentiation, characterized by continued proliferation and subsequent entry into the terminal differentiation pathway (Harandi et al., 2010; Xiang et al., 2015). Although many of the signals that regulate stress erythropoiesis have been identified, the role of cellular metabolism and how these signals affect metabolism is largely unknown. In this review, we discuss what is known about metabolic regulation of stress erythropoiesis. We will identify open questions and discuss examples from other systems that illustrate potential paradigms for how changes in metabolism can regulate proliferation and differentiation of SEPs.
For efficient stress erythropoiesis, immature progenitor populations must be expanded prior to the commitment to differentiation. This process requires the translation of proliferative signals, like Wnt2b and 8a and GDF15, into changes in metabolism that support proliferation and prevent changes in chromatin structure associated with the activation of the erythroid gene program.
Our previous work identified signaling pathways that are required for the expansion of immature SEP populations. The questions that remain are how these signals regulate metabolism and how changes in metabolism result in the proliferating, self-renewing, uncommitted SEP populations we observe in vivo. To address these questions, we must first acknowledge that metabolism regulates all cellular processes. It is often discussed as though the different metabolic processes are independent pathways, however, that is not how we should think about metabolism. As articulated by Murphy and O’Neill, a more correct way to think about metabolism is that metabolic pathways are interconnected such that changes in one metabolic pathway affect the flux of metabolites through other pathways (Murphy and O'Neill, 2020). In stress erythropoiesis, the initial stage requires the expansion of immature SEP populations. Signals that drive the proliferation of SEPs establish a metabolism that favors anabolic processes required to generate sufficient amino acids, lipids, nucleotides and other macromolecules to generate biomass for cell division. In cancer cells, this type of metabolism is referred to as aerobic glycolysis or the “Warburg effect”, which is dominated by glycolysis and is present in rapidly proliferating cell populations (Warburg, 1956; Lunt and Vander Heiden, 2011). Although glycolysis is higher than oxidative phosphorylation at these times, this type of metabolism can really be thought of as a highly efficient anabolic metabolism where glycolytic metabolites can be shunted into anabolic pathways like the Pentose phosphate pathway (PPP) and the serine glycine (Ser/Gly) pathway, which generate nucleotides, amino acids, metabolites used in 1-carbon metabolism and regenerate NADPH levels (Wise et al., 2008; Lunt and Vander Heiden, 2011; Le et al., 2012). A role for these pathways in stress erythropoiesis was shown by Oburoglu et al. who demonstrated that commitment of human CD34+ cells to the erythroid lineage required glutamine and the PPP. Inhibition of glutamine metabolism with the glutamine analog 6-diazo-5-oxo-L-norleucine (DON) blocked erythroid commitment (Oburoglu et al., 2014; Oburoglu et al., 2016). This effect could be rescued with exogenous nucleotides further demonstrating the need for nucleotide biogenesis in erythropoiesis. Although this work used in vitro human CD34+ cell cultures, the same group showed that in vivo stress erythropoiesis induced by phenylhydrazine treatment in mice was blocked by DON, while myelopoiesis increased in the spleen (Oburoglu et al., 2014; Oburoglu et al., 2016). In cancer cells, highly efficient usage of the PPP and Ser/Gly pathways relies on the establishment of metabolons, like the purinosome, which is a complex of at least 10 enzymes that uses metabolic channeling to drive de novo purine biosynthesis (An et al., 2008; Zhao et al., 2015; Pedley et al., 2022). It remains to be demonstrated whether signals that promote the proliferation of SEPs also drive the formation of the purinosome and other metabolons to increase metabolic efficiency. The ability of the purinosome to increase purine biosynthesis has the potential to profoundly impact overall cell metabolism and cell signaling as increased levels of guanine and adenine activate the mTorC1 pathway (Ben-Sahra et al., 2016; Emmanuel et al., 2017; Hoxhaj et al., 2017). mTorC1 is a central regulator of metabolism. Blocking mTorC1 activity compromises stress erythropoiesis and limits SEP proliferation (Knight et al., 2014). Furthermore, through the downstream activation of ATF4, mTorC1 can feed back to increase purine biosynthesis by increasing the expression of Mthfd2, an enzyme involved in the mitochondrial tetrahydrofolate cycle (Ben-Sahra et al., 2016). Like mTorC1, mutations in ATF4 severely compromise stress erythropoiesis (Masuoka and Townes, 2002). In addition to purine biosynthesis, mTorC1 also increases pyrimidine biosynthesis by phosphorylating CAD, the initial enzyme in the de novo pyrimidine biosynthesis pathway (Ben-Sahra et al., 2013; Robitaille et al., 2013). This modification also leads to increased rRNA and ribosomal biogenesis, which are needed to increase protein biomass during cell division (Claiborne et al., 2022). Although data support the role of mTorC1/ATF4 dependent control of metabolism in proliferating SEPs, further work is needed to understand how pro-inflammatory signals establish this anabolic metabolism and how other key stress erythropoiesis signals like canonical Wnts, GDF15 and BMP4 impact these metabolic pathways.
Proliferating SEPs are able to self-renew and maintain an immature phenotype characterized by the expression of stem cell genes and low-level expression of erythroid genes (Xiang et al., 2015). How does metabolic regulation maintain this immature cell state? In macrophages, inflammatory signals alter the TCA cycle. This alteration is referred to as a “broken TCA cycle” (Lampropoulou et al., 2016; O'Neill, 2015; Ryan and O'Neill, 2020). Metabolites like citrate and succinate are exported from the mitochondria and act in other pathways. Glutamine is converted to alpha-ketoglutarate (αKG), which can generate both citrate and succinate through oxidative and reductive pathways (Mullen et al., 2014). When citrate and succinate are exported from the mitochondria, they contribute both to the anabolic metabolism and affect gene expression (Mills and O'Neill, 2014; Ryan et al., 2019; Tannahill et al., 2013; Williams and O'Neill, 2018). Exported citrate is converted by ATP Citrate lyase (Acly) to oxaloacetate and acetyl-CoA. In cancer cells, the latter is used by histone acetyltransferases to maintain expression of glycolytic enzymes (Wellen et al., 2009). Acetyl-CoA is also used in lipogenesis, which supports cell division. Succinate on the other hand acts as an inhibitor of αKG dependent dioxygenases, which includes the family of proline hydroxylases (PHDs) that regulate the stability of hypoxia inducible transcription factors (Hifs) (Tannahill et al., 2013). The inhibition of PHDs increases hypoxia dependent glycolysis. Other enzymes that are inhibited by succinate include jumonji-domain histone deacetylases and the TET proteins that demethylate DNA. These data suggest that increased succinate plays a role in inhibiting differentiation of SEPs by preventing changes in DNA and histone methylation. Further work is needed to establish that metabolic control of epigenetics marks inhibits erythroid differentiation, while at the same time increasing the expression of genes that promote proliferation.
GDF15 and the Hippo-Yap signaling pathways play an essential role in stress erythropoiesis (Hao et al., 2019a; Hao et al., 2019b). Mutation of these pathways significantly impairs proliferation of SEPs. We observed that both pathways regulate the expression of genes involved in glycolysis and the TCA cycle (Hao et al., 2019b). GDF15 signaling increases the expression of PDK1 and PDK3, two enzymes that limit the flow of pyruvate into the TCA cycle, which is consistent with an altered TCA cycle in SEPs. GDF15 also increases the expression of Hif1α and Glut1, which increases glycolysis. Similarly, the expression of Gls1 is increased with GDF15 treatment. Gls1 is an enzyme that catalyzes the breakdown of glutamine, which channels glutamine into anabolic pathways. Like GDF15, our analysis of Yap1, a transcriptional co-activator whose activity is regulated by the Hippo pathway and Wnt signaling, showed that it maintains the proliferative anabolic metabolism in immature SEPs(Azzolin et al., 2014; Enzo et al., 2015; Koo and Guan, 2018; Hao et al., 2019a; Ibar and Irvine, 2020). Mutation of Yap1 severely impairs the expansion of immature SEP populations. Yap1 regulates the expression of Psat1, an enzyme in the Ser/Gly pathway which generates Serine from the glycolytic metabolite 3-phosphoglycerate that is used in one carbon metabolism and the production of S-adenosyl-methionine. Serine is also needed for the production of folate, which feeds into purine and pyrimidine biosynthesis. Yap1 also regulates the expression of Got1, a transaminase that generates aspartate from αKG. Inhibition of Got1 blocks erythroid commitment to the erythroid lineage (Hao et al., 2019a). In addition to our data, work in several experimental systems has shown thatYap1 regulates the expression of glycolytic enzymes and other enzymes involved in glutamine metabolism (Enzo et al., 2015; Cox et al., 2016; Cox et al., 2018; Koo and Guan, 2018; Ibar and Irvine, 2020). These data suggest that the mechanisms that regulate the proliferation of SEPs rely on a complex integration of signaling pathways which shape the anabolic metabolism needed to support the expansion of these progenitor populations. Further work is needed to understand how this integration of cell signaling and metabolism is accomplished. (Figure 1).
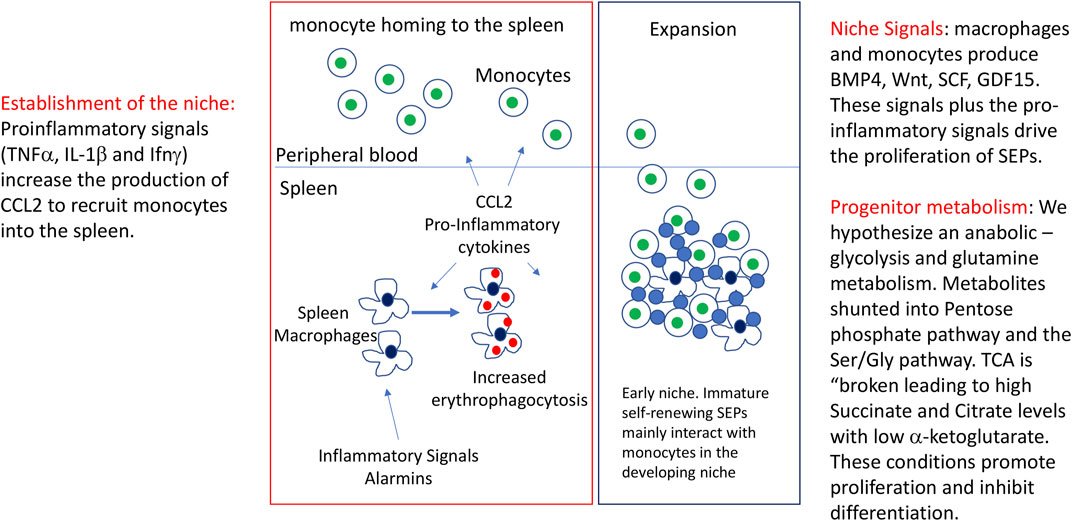
FIGURE 1. Initiation of stress erythropoiesis and the expansion of immature SEPs. Figure depicts the inflammatory signals that initiate stress erythropoiesis, the formation of the stress erythropoiesis niche and the expansion of a transient amplifying population of immature stress progenitors. Included in the figure are observed signals and potential changes that would be predicted to occur as discussed in the review.
The transition from uncommitted proliferating SEPs to SEPs committed to differentiation requires changes in the signals made by the niche. Epo dependent signaling leads to a loss of Wnt expression and an increase in PGE2 production, however, GDF15, BMP4 and Yap1 dependent signaling pathways remain in SEPs, but now these pathways promote differentiation (Chen et al., 2020). In addition, the niche also switches from one dominated by pro-inflammatory signals to one dominated by pro-resolving anti-inflammatory signals (Liao et al., 2018). This change in signals and microenvironment will have a profound impact on the differentiation of SEPs. Although we know little about the metabolic changes that drive differentiation, data from others have identified some paradigms that may also play a role in stress erythropoiesis. (Figure 2). Analysis of hematopoietic stem cells (HSCs) showed that quiescent HSCs prefer glycolysis, but as they become mobilized and differentiate, oxidative phosphorylation (OXPHOS) increases (Simsek et al., 2010; Suda et al., 2011; Harris et al., 2013; Takubo et al., 2013; Ito and Suda, 2014; Maryanovich et al., 2015). Again, this switch in metabolism does not mean that glycolysis is off and OXPHOS is on, but rather the relative amounts of metabolites that flux through these pathways change. The increase of OXPHOS and the TCA cycle leads to changes in metabolites that affect other signaling pathways. As mentioned above, high levels of succinate generated when the TCA cycle is broken can inhibit jumonji-domain histone demethylases and TET DNA demethylases, but when mitochondrial respiration is increased and the levels of αKG available for enzymes increases. During muscle regeneration, the H3K27 demethylases JmjD3 and UTX (KDM6A), which require αKG, increase their activity at different times during the recovery from muscle injury (Liu and Rando, 2016; Gabellini, 2022). JmjD3 activity is needed to increase the expression of Has2, an enzyme that initiates the production of hyaluronic acid. The expansion of the extracellular matrix leads to an exit from quiescence allowing for the proliferation of muscle stem cells (Nakka et al., 2022). UTX has no role at this stage, however, later UTX is required for the commitment of myocytes to differentiation. This step is characterized by the UTX dependent removal of H3K27me3 at the myogenin locus and at other loci associated with muscle development. Mutation of JmjD3 or UTX leads to increased levels of H3K27me, but they show distinct defects in muscle regeneration (Faralli et al., 2016). These two examples demonstrate how tight regulation of histone H3K27 demethylase activity can control the proliferation and differentiation of muscle stem cells during muscle regenration. Changes in niche signaling can also induce metabolic reprogramming and induce histone demethylase activity. Germinal center B cell differentiation is induced by IL-4 signaling, which increases mitochondrial respiration and αKG production in activated B cells. By increasing this metabolite, UTX activity increases and removes H3K27me3 marks at the Bcl6 enhancer and promoter. In addition to changing metabolism, IL-4 signaling activates Stat6 which recruits UTX to the enhancer and promoter in Bcl6. These data show that IL-4 signaling establishes a metabolism that increases UTX activity, but also targets UTX to key genes required for germinal center B cell development (Haniuda et al., 2020). These examples show how changes in metabolism lead to mobilization of chromatin remodeling enzymes that drive the changes in gene expression programs necessary for the differentiation of progenitor cells. The specificity of these epigenetic changes comes from the integration of cell signaling with metabolic reprogramming. Future work on SEP differentiation will be needed to determine how pro-resolving signals lead to changes in metabolism that de-repress the erythroid gene expression program and promote differentiation.
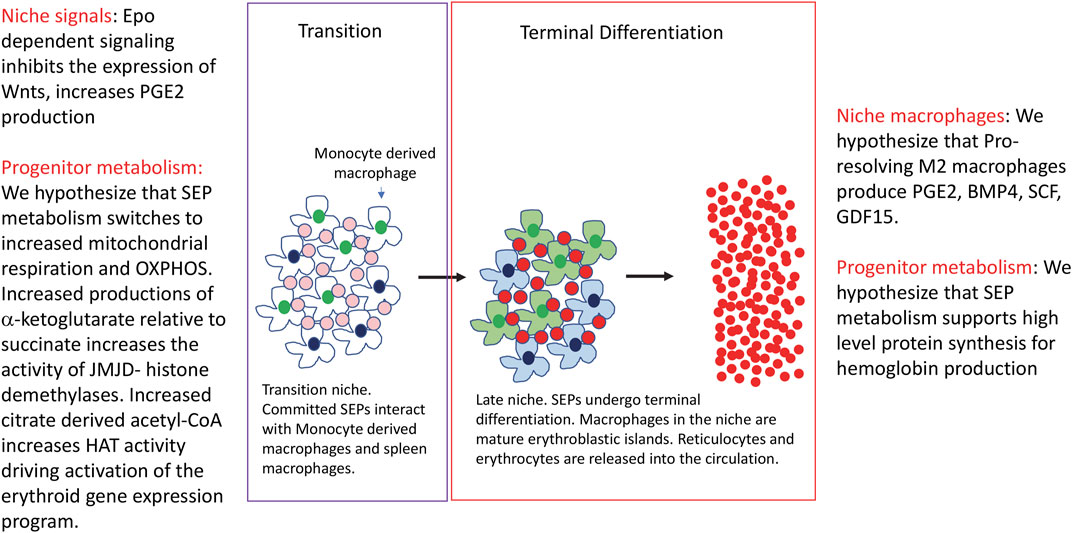
FIGURE 2. The transition to differentiation and the terminal differentiation of stress erythroid progenitors. The figure depicts the transition to differentiation and terminal differentiation. Changes in signals and metabolism in the niche and progenitors are indicated.
The increase in αKG coupled with a relative decrease in succinate and fumarate will also lead to the activation of the Ten-eleven translocation (TET) enzymes. Like the histone demethylases these enzymes require iron, oxygen and αKG (Laukka et al., 2016; Koivunen and Laukka, 2018). They catalyze the oxidation of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5hmC), which can be subsequently converted to un-methylated cytosine (Cimmino et al., 2011; Wu and Zhang, 2011). Changes in DNA methylation have been previously shown to regulate globin gene expression (Ley et al., 1982; Ginder et al., 1998; Siegfried et al., 1999). In addition, analysis of human, mouse and zebrafish showed that Tet2 plays a role in lineage commitment. Analysis of human CD34+ bone marrow cell cultures, showed that there is an increase in 5hmC during the erythroid commitment stage of the culture, which was associated with increased binding of erythroid transcription factors and increased chromatin accessibility (Madzo et al., 2014). Cultures of cord blood cells showed that inhibiting Tet2 leads to increased myeloid differentiation, while decreasing erythroid differentiation (Pronier et al., 2011). ShRNA knockdown Tet2 in CD34+ cell cultures led to increased proliferation of immature erythroid progenitors and impaired their differentiation (Yan et al., 2017). These observations are of interest as mutations in TET2 are often observed in myelodysplastic syndromes, which exhibit impaired erythroid differentiation and is associated with clonal hematopoiesis (Fujishima et al., 2021; Florez et al., 2022; Gurnari et al., 2022). These data are similar to what is observed when Tet2 is knocked down in zebrafish embryos (Ge et al., 2014). These data suggest a role for TET2 dependent changes in DNA methylation in the commitment of SEPs to differentiation. Further work will be needed to determine how changes in DNA methylation affect stress erythropoiesis.
This small set of examples illustrate some of the areas where metabolism could regulate stress erythropoiesis. The challenge for future research will be to integrate transcriptomic and metabolomics data with cell signaling. These analyses will address how metabolism impacts gene expression at each of the developmental stages of stress erythropoiesis, how changes in metabolism drive the transition from proliferating SEPs to differentiating SEPs and how signals from the niche impact and change metabolism so that sufficient erythrocytes are produced to maintain erythroid homeostasis.
RP and BR wrote and edited the manuscript.
Work in the Paulson Lab is supported by NIH grant HL146528 (RP), NIFA-USDA Hatch Project PEN04771 accession #0000005 (RP).
We would like to thank Sandeep Prabhu, Andrew Patterson, Imhoi Koo and Molly Hall for their help with the metabolomic studies and helpful insights and the Paulson lab for comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akilesh H. M., Buechler M. B., Duggan J. M., Hahn W. O., Matta B., Sun X., et al. (2019). Chronic TLR7 and TLR9 signaling drives anemia via differentiation of specialized hemophagocytes. Science 363 (6423), eaao5213. Epub 2019/01/12PubMed PMID: 30630901; PMCID: PMC6413693. doi:10.1126/science.aao5213
An S., Kumar R., Sheets E. D., Benkovic S. J. (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320 (5872), 103–106. Epub 2008/04/05PubMed PMID: 18388293. doi:10.1126/science.1152241
Arezes J., Foy N., McHugh K., Sawant A., Quinkert D., Terraube V., et al. (2018). Erythroferrone inhibits the induction of hepcidin by BMP6. Blood 132 (14), 1473–1477. Epub 2018/08/12PubMed PMID: 30097509; PMCID: PMC6238155 funding from Pfizer. N.F., A.S., V.T., A.B., M.T., E.R.L., O.C., M.L., and R.J. are Pfizer employees. N.F., O.C., R.J., J.A., K.M., S.J.D., and H.D. are named inventors on a patent application currently under evaluation. The remaining authors declare no competing financial interests. doi:10.1182/blood-2018-06-857995
Asfaha S. (2015). Intestinal stem cells and inflammation. Curr. Opin. Pharmacol. 25, 62–66. Epub 2015/12/15PubMed PMID: 26654865. doi:10.1016/j.coph.2015.11.008
Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., et al. (2014). YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158 (1), 157–170. Epub 2014/07/01PubMed PMID: 24976009. doi:10.1016/j.cell.2014.06.013
Ben-Sahra I., Howell J. J., Asara J. M., Manning B. D. (2013). Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339 (6125), 1323–1328. Epub 2013/02/23PubMed PMID: 23429703; PMCID: PMC3753690. doi:10.1126/science.1228792
Ben-Sahra I., Hoxhaj G., Ricoult S. J. H., Asara J. M., Manning B. D. (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351 (6274), 728–733. Epub 2016/02/26PubMed PMID: 26912861; PMCID: PMC4786372. doi:10.1126/science.aad0489
Bennett L. F., Liao C., Quickel M. D., Yeoh B. S., Vijay-Kumar M., Hankey-Giblin P., et al. (2019). Inflammation induces stress erythropoiesis through heme-dependent activation of SPI-C. Sci. Signal 12 (598), eaap7336. Epub 2019/09/12PubMed PMID: 31506384. doi:10.1126/scisignal.aap7336
Burzyn D., Kuswanto W., Kolodin D., Shadrach J. L., Cerletti M., Jang Y., et al. (2013). A special population of regulatory T cells potentiates muscle repair. Cell 155 (6), 1282–1295. Epub 2013/12/10PubMed PMID: 24315098; PMCID: PMC3894749. doi:10.1016/j.cell.2013.10.054
Caiado F., Pietras E. M., Manz M. G. (2021). Inflammation as a regulator of hematopoietic stem cell function in disease, aging, and clonal selection. J. Exp. Med. 218 (7), e20201541. Epub 2021/06/16PubMed PMID: 34129016; PMCID: PMC8210622. doi:10.1084/jem.20201541
Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13 (5), 509–519. Epub 2013/05/21PubMed PMID: 23684303; PMCID: 3676888. doi:10.1016/j.chom.2013.04.010
Chen Y., Xiang J., Qian F., Diwakar B. T., Ruan B., Hao S., et al. (2020). Epo receptor signaling in macrophages alters the splenic niche to promote erythroid differentiation. Blood 136 (2), 235–246. Epub 2020/05/01PubMed PMID: 32350523; PMCID: PMC7357191 Therapeutics. The remaining authors declare no competing financial interests. doi:10.1182/blood.2019003480
Cimmino L., Abdel-Wahab O., Levine R. L., Aifantis I. (2011). TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9 (3), 193–204. Epub 2011/09/03PubMed PMID: 21885017; PMCID: PMC3244690. doi:10.1016/j.stem.2011.08.007
Claiborne M. D., Sengupta S., Zhao L., Arwood M. L., Sun I. M., Wen J., et al. (2022). Persistent CAD activity in memory CD8(+) T cells supports rRNA synthesis and ribosomal biogenesis required at rechallenge. Sci. Immunol. 7 (71), eabh4271. Epub 2022/05/28PubMed PMID: 35622902. doi:10.1126/sciimmunol.abh4271
Cox A. G., Hwang K. L., Brown K. K., Evason K., Beltz S., Tsomides A., et al. (2016). Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 18 (8), 886–896. Epub 2016/07/19PubMed PMID: 27428308; PMCID: PMC4990146. doi:10.1038/ncb3389
Cox A. G., Tsomides A., Yimlamai D., Hwang K. L., Miesfeld J., Galli G. G., et al. (2018). Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth. Embo J. 37 (22), e100294. Epub 2018/10/24PubMed PMID: 30348863; PMCID: PMC6236334. doi:10.15252/embj.2018100294
Emmanuel N., Ragunathan S., Shan Q., Wang F., Giannakou A., Huser N., et al. (2017). Purine nucleotide availability regulates mTORC1 activity through the rheb GTPase. Cell Rep. 19 (13), 2665–2680. Epub 2017/06/29PubMed PMID: 28658616. doi:10.1016/j.celrep.2017.05.043
Enzo E., Santinon G., Pocaterra A., Aragona M., Bresolin S., Forcato M., et al. (2015). Aerobic glycolysis tunes YAP/TAZ transcriptional activity. Embo J. 34 (10), 1349–1370. Epub 2015/03/23PubMed PMID: 25796446; PMCID: PMC4491996. doi:10.15252/embj.201490379
Faralli H., Wang C., Nakka K., Benyoucef A., Sebastian S., Zhuang L., et al. (2016). UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 126 (4), 1555–1565. Epub 2016/03/22PubMed PMID: 26999603; PMCID: PMC4811158. doi:10.1172/jci83239
Florez M. A., Tran B. T., Wathan T. K., DeGregori J., Pietras E. M., King K. Y. (2022). Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem Cell 29 (6), 882–904. Epub 2022/06/07PubMed PMID: 35659875; PMCID: PMC9202417. doi:10.1016/j.stem.2022.05.006
Fujishima N., Kohmaru J., Koyota S., Kuba K., Saga T., Omokawa A., et al. (2021). Clonal hematopoiesis in adult pure red cell aplasia. Sci. Rep. 11 (1), 2253. Epub 2021/01/28PubMed PMID: 33500526; PMCID: PMC7838416. doi:10.1038/s41598-021-81890-5
Gabellini D. (2022). A regenerative niche for stem cells. Science 377 (6606), 578–579. Epub 2022/08/05PubMed PMID: 35926040. doi:10.1126/science.add6804
Ge L., Zhang R. P., Wan F., Guo D. Y., Wang P., Xiang L. X., et al. (2014). TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol. Cell Biol. 34 (6), 989–1002. Epub 2014/01/08PubMed PMID: 24396069; PMCID: PMC3958037. doi:10.1128/mcb.01061-13
Ginder G. D., Singal R., Little J. A., Dempsey N., Ferris R., Wang S. Z. (1998). Silencing and activation of embryonic globin gene expression. Ann. N. Y. Acad. Sci. 850, 70–79. Epub 1998/07/21PubMed PMID: 9668529. doi:10.1111/j.1749-6632.1998.tb10464.x
Grover A., Mancini E., Moore S., Mead A. J., Atkinson D., Rasmussen K. D., et al. (2014). Erythropoietin guides multipotent hematopoietic progenitor cells toward an erythroid fate. J. Exp. Med. 211 (2), 181–188. Epub 2014/02/05PubMed PMID: 24493804; PMCID: 3920567. doi:10.1084/jem.20131189
Gurnari C., Pagliuca S., Guan Y., Adema V., Hershberger C. E., Ni Y., et al. (2022). TET2 mutations as a part of DNA dioxygenase deficiency in myelodysplastic syndromes. Blood Adv. 6 (1), 100–107. Epub 2021/11/13PubMed PMID: 34768283; PMCID: PMC8753204. doi:10.1182/bloodadvances.2021005418
Haniuda K., Fukao S., Kitamura D. (2020). Metabolic reprogramming induces germinal center B cell differentiation through Bcl6 locus remodeling. Cell Rep. 33 (5), 108333. Epub 2020/11/05PubMed PMID: 33147467. doi:10.1016/j.celrep.2020.108333
Hao S., Matsui Y., Lai Z. C., Paulson R. F. (2019). Yap1 promotes proliferation of transiently amplifying stress erythroid progenitors during erythroid regeneration. Exp. Hematol. 80, 42–54. e4Epub 2019/11/23PubMed PMID: 31756359; PMCID: PMC6952558. doi:10.1016/j.exphem.2019.11.002
Hao S., Xiang J., Wu D. C., Fraser J. W., Ruan B., Cai J., et al. (2019). Gdf15 regulates murine stress erythroid progenitor proliferation and the development of the stress erythropoiesis niche. Blood Adv. 3 (14), 2205–2217. Epub 2019/07/22PubMed PMID: 31324641; PMCID: PMC6650738 interests. doi:10.1182/bloodadvances.2019000375
Harandi O. F., Hedge S., Wu D. C., McKeone D., Paulson R. F. (2010). Murine erythroid short-term radioprotection requires a BMP4-dependent, self-renewing population of stress erythroid progenitors. J. Clin. Invest. 120 (12), 4507–4519. Epub 2010/11/10PubMed PMID: 21060151; PMCID: 2993581. doi:10.1172/JCI41291
Harris J. M., Esain V., Frechette G. M., Harris L. J., Cox A. G., Cortes M., et al. (2013). Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood 121 (13), 2483–2493. Epub 2013/01/24PubMed PMID: 23341543; PMCID: PMC3612858. doi:10.1182/blood-2012-12-471201
Hoxhaj G., Hughes-Hallett J., Timson R. C., Ilagan E., Yuan M., Asara J. M., et al. (2017). The mTORC1 signaling network senses changes in cellular purine nucleotide levels. Cell Rep. 21 (5), 1331–1346. Epub 2017/11/02PubMed PMID: 29091770; PMCID: PMC5689476. doi:10.1016/j.celrep.2017.10.029
Ibar C., Irvine K. D. (2020). Integration of hippo-YAP signaling with metabolism. Dev. Cell 54 (2), 256–267. Epub 2020/07/22PubMed PMID: 32693058; PMCID: PMC7373816. doi:10.1016/j.devcel.2020.06.025
Ito K., Suda T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15 (4), 243–256. Epub 2014/03/22PubMed PMID: 24651542; PMCID: PMC4095859. doi:10.1038/nrm3772
Jurado R. L. (1997). Iron, infections, and anemia of inflammation. Clin. Infect. Dis. 25 (4), 888–895. Epub 1997/11/14. PubMed PMID: 9356804. doi:10.1086/515549
Kautz L., Jung G., Nemeth E., Ganz T. (2014). Erythroferrone contributes to recovery from anemia of inflammation. Blood 124 (16), 2569–2574. Epub 2014/09/07PubMed PMID: 25193872; PMCID: PMC4199959. doi:10.1182/blood-2014-06-584607
Kautz L., Jung G., Valore E. V., Rivella S., Nemeth E., Ganz T. (2014). Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat. Genet. 46 (7), 678–684. Epub 2014/06/02PubMed PMID: 24880340; PMCID: PMC4104984. doi:10.1038/ng.2996
Klei T. R., Meinderts S. M., van den Berg T. K., van Bruggen R. (2017). From the cradle to the grave: The role of macrophages in erythropoiesis and erythrophagocytosis. Front. Immunol. 8, 73. Epub 2017/02/18PubMed PMID: 28210260; PMCID: 5288342. doi:10.3389/fimmu.2017.00073
Knight Z. A., Schmidt S. F., Birsoy K., Tan K., Friedman J. M. (2014). A critical role for mTORC1 in erythropoiesis and anemia. Elife 3, e01913. Epub 2014/09/10PubMed PMID: 25201874; PMCID: PMC4179304. doi:10.7554/eLife.01913
Koivunen P., Laukka T. (2018). The TET enzymes. Cell Mol. Life Sci. 75 (8), 1339–1348. Epub 2017/12/01PubMed PMID: 29184981. doi:10.1007/s00018-017-2721-8
Koo J. H., Guan K. L. (2018). Interplay between YAP/TAZ and metabolism. Cell Metab. 28 (2), 196–206. Epub 2018/08/09PubMed PMID: 30089241. doi:10.1016/j.cmet.2018.07.010
Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E. E., Loginicheva E., et al. (2016). Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24 (1), 158–166. Epub 2016/07/05PubMed PMID: 27374498; PMCID: PMC5108454. doi:10.1016/j.cmet.2016.06.004
Laukka T., Mariani C. J., Ihantola T., Cao J. Z., Hokkanen J., Kaelin W. G., et al. (2016). Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 291 (8), 4256–4265. Epub 2015/12/26PubMed PMID: 26703470; PMCID: PMC4759199. doi:10.1074/jbc.M115.688762
Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., et al. (2012). Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15 (1), 110–121. Epub 2012/01/10PubMed PMID: 22225880; PMCID: PMC3345194. doi:10.1016/j.cmet.2011.12.009
Ley T. J., DeSimone J., Anagnou N. P., Keller G. H., Humphries R. K., Turner P. H., et al. (1982). 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N. Engl. J. Med. 307 (24), 1469–1475. Epub 1982/12/09PubMed PMID: 6183586. doi:10.1056/nejm198212093072401
Liao C., Prabhu K. S., Paulson R. F. (2018). Monocyte-derived macrophages expand the murine stress erythropoietic niche during the recovery from anemia. Blood 132 (24), 2580–2593. Epub 2018/10/17PubMed PMID: 30322871; PMCID: PMC6293871 interests. doi:10.1182/blood-2018-06-856831
Libregts S. F., Gutierrez L., de Bruin A. M., Wensveen F. M., Papadopoulos P., van Ijcken W., et al. (2011). Chronic IFN-gamma production in mice induces anemia by reducing erythrocyte life span and inhibiting erythropoiesis through an IRF-1/PU.1 axis. Blood 118 (9), 2578–2588. Epub 2011/07/05PubMed PMID: 21725055. doi:10.1182/blood-2010-10-315218
Liu L., Rando T. A. (2016). UTX in muscle regeneration--the right dose and the right time. J. Clin. Invest. 126 (4), 1233–1235. Epub 2016/03/22PubMed PMID: 26999609; PMCID: PMC4811162. doi:10.1172/jci86798
Lunt S. Y., Vander Heiden M. G. (2011). Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. Epub 2011/10/12PubMed PMID: 21985671. doi:10.1146/annurev-cellbio-092910-154237
Madzo J., Liu H., Rodriguez A., Vasanthakumar A., Sundaravel S., Caces D. B. D., et al. (2014). Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep. 6 (1), 231–244. Epub 2014/01/01PubMed PMID: 24373966; PMCID: PMC3976649. doi:10.1016/j.celrep.2013.11.044
Maryanovich M., Zaltsman Y., Ruggiero A., Goldman A., Shachnai L., Zaidman S. L., et al. (2015). An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat. Commun. 6, 7901. Epub 2015/07/30PubMed PMID: 26219591. doi:10.1038/ncomms8901
Masuoka H. C., Townes T. M. (2002). Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99 (3), 736–745. Epub 2002/01/25PubMed PMID: 11806972. doi:10.1182/blood.v99.3.736
Mills E., O'Neill L. A. (2014). Succinate: A metabolic signal in inflammation. Trends Cell Biol. 24 (5), 313–320. Epub 2013/12/24PubMed PMID: 24361092. doi:10.1016/j.tcb.2013.11.008
Mullen A. R., Hu Z., Shi X., Jiang L., Boroughs L. K., Kovacs Z., et al. (2014). Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 7 (5), 1679–1690. Epub 2014/05/27PubMed PMID: 24857658; PMCID: PMC4057960. doi:10.1016/j.celrep.2014.04.037
Murphy M. P., O'Neill L. A. J. (2020). How should we talk about metabolism? Nat. Immunol. 21 (7), 713–715. Epub 2020/05/16PubMed PMID: 32409776. doi:10.1038/s41590-020-0691-8
Nakka K., Hachmer S., Mokhtari Z., Kovac R., Bandukwala H., Bernard C., et al. (2022). JMJD3 activated hyaluronan synthesis drives muscle regeneration in an inflammatory environment. Science 377 (6606), 666–669. Epub 2022/08/05PubMed PMID: 35926054. doi:10.1126/science.abm9735
O'Neill L. A. (2015). A broken krebs cycle in macrophages. Immunity 42 (3), 393–394. Epub 2015/03/19PubMed PMID: 25786167. doi:10.1016/j.immuni.2015.02.017
Oburoglu L., Romano M., Taylor N., Kinet S. (2016). Metabolic regulation of hematopoietic stem cell commitment and erythroid differentiation. Curr. Opin. Hematol. 23 (3), 198–205. Epub 2016/02/13PubMed PMID: 26871253. doi:10.1097/moh.0000000000000234
Oburoglu L., Tardito S., Fritz V., de Barros S. C., Merida P., Craveiro M., et al. (2014). Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 15 (2), 169–184. Epub 2014/06/24PubMed PMID: 24953180. doi:10.1016/j.stem.2014.06.002
Oduro K. A., Liu F., Tan Q., Kim C. K., Lubman O., Fremont D., et al. (2012). Myeloid skewing in murine autoimmune arthritis occurs in hematopoietic stem and primitive progenitor cells. Blood 120 (11), 2203–2213. Epub 2012/08/03PubMed PMID: 22855602; PMCID: 3447779. doi:10.1182/blood-2011-11-391342
Palis J. (2014). Primitive and definitive erythropoiesis in mammals. Front. Physiol. 5, 3. Epub 2014/01/31PubMed PMID: 24478716; PMCID: 3904103. doi:10.3389/fphys.2014.00003
Paulson R. F., Hariharan S., Little J. A. (2020). Stress erythropoiesis: Definitions and models for its study. Exp. Hematol. 89, 43–54. e2. Epub 2020/08/05PubMed PMID: 32750404; PMCID: PMC7508762. doi:10.1016/j.exphem.2020.07.011
Paulson R. F., Ruan B., Hao S., Chen Y. (2020). Stress erythropoiesis is a key inflammatory response. Cells 9 (3), 634. Epub 2020/03/12PubMed PMID: 32155728; PMCID: PMC7140438. doi:10.3390/cells9030634
Pedley A. M., Pareek V., Benkovic S. J. (2022). The purinosome: A case study for a mammalian metabolon. Annu. Rev. Biochem. 91, 89–106. Epub 2022/03/24PubMed PMID: 35320684. doi:10.1146/annurev-biochem-032620-105728
Perry J. M., Harandi O. F., Porayette P., Hegde S., Kannan A. K., Paulson R. F. (2009). Maintenance of the BMP4-dependent stress erythropoiesis pathway in the murine spleen requires hedgehog signaling. Blood 113 (4), 911–918. Epub 2008/10/18PubMed PMID: 18927434; PMCID: 2630276. doi:10.1182/blood-2008-03-147892
Pietras E. M. (2017). Inflammation: A key regulator of hematopoietic stem cell fate in health and disease. Blood 130 (15), 1693–1698. Epub 2017/09/07PubMed PMID: 28874349; PMCID: 5639485. doi:10.1182/blood-2017-06-780882
Pietras E. M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L. V., Zhang S., et al. (2016). Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18 (6), 607–618. Epub 2016/04/26PubMed PMID: 27111842; PMCID: 4884136. doi:10.1038/ncb3346
Pronier E., Almire C., Mokrani H., Vasanthakumar A., Simon A., da Costa Reis Monte Mor B., et al. (2011). Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood 118 (9), 2551–2555. Epub 2011/07/08PubMed PMID: 21734233; PMCID: PMC3292425. doi:10.1182/blood-2010-12-324707
Robitaille A. M., Christen S., Shimobayashi M., Cornu M., Fava L. L., Moes S., et al. (2013). Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339 (6125), 1320–1323. Epub 2013/02/23PubMed PMID: 23429704. doi:10.1126/science.1228771
Rusten L. S., Jacobsen S. E. (1995). Tumor necrosis factor (TNF)-alpha directly inhibits human erythropoiesis in vitro: Role of p55 and p75 TNF receptors. Blood 85 (4), 989–996. Epub 1995/02/15. PubMed PMID: 7849320. doi:10.1182/blood.v85.4.989.bloodjournal854989
Ryan D. G., Murphy M. P., Frezza C., Prag H. A., Chouchani E. T., O'Neill L. A., et al. (2019). Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 1, 16–33. Epub 2019/04/30PubMed PMID: 31032474; PMCID: PMC6485344. doi:10.1038/s42255-018-0014-7
Ryan D. G., O'Neill L. A. J. (2020). Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol. 38, 289–313. Epub 2020/01/28PubMed PMID: 31986069. doi:10.1146/annurev-immunol-081619-104850
Scaramozza A., Park D., Kollu S., Beerman I., Sun X., Rossi D. J., et al. (2019). Lineage tracing reveals a subset of reserve muscle stem cells capable of clonal expansion under stress. Cell Stem Cell 24 (6), 944–957. e5. Epub 2019/04/23PubMed PMID: 31006621; PMCID: PMC6597014. doi:10.1016/j.stem.2019.03.020
Seu K. G., Papoin J., Fessler R., Hom J., Huang G., Mohandas N., et al. (2017). Unraveling macrophage heterogeneity in erythroblastic islands. Front. Immunol. 8, 1140. Epub 2017/10/06PubMed PMID: 28979259; PMCID: 5611421. doi:10.3389/fimmu.2017.01140
Shen M., Luo Z., Zhou Y. (2022). Regeneration-associated transitional state cells in pulmonary fibrosis. Int. J. Mol. Sci. 23 (12), 6757. Epub 2022/06/25PubMed PMID: 35743199; PMCID: PMC9223485. doi:10.3390/ijms23126757
Siegfried Z., Eden S., Mendelsohn M., Feng X., Tsuberi B. Z., Cedar H. (1999). DNA methylation represses transcription in vivo. Nat. Genet. 22 (2), 203–206. Epub 1999/06/16PubMed PMID: 10369268. doi:10.1038/9727
Simsek T., Kocabas F., Zheng J., Deberardinis R. J., Mahmoud A. I., Olson E. N., et al. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7 (3), 380–390. Epub 2010/09/02PubMed PMID: 20804973; PMCID: PMC4159713. doi:10.1016/j.stem.2010.07.011
Singh R. P., Grinenko T., Ramasz B., Franke K., Lesche M., Dahl A., et al. (2018). Hematopoietic stem cells but not multipotent progenitors drive erythropoiesis during chronic erythroid stress in EPO transgenic mice. Stem Cell Rep. 10 (6), 1908–1919. Epub 2018/05/15PubMed PMID: 29754961; PMCID: 5989815. doi:10.1016/j.stemcr.2018.04.012
Soares M. P., Weiss G. (2015). The Iron age of host-microbe interactions. EMBO Rep. 16 (11), 1482–1500. Epub 2015/10/18PubMed PMID: 26474900; PMCID: 4641501. doi:10.15252/embr.201540558
Suda T., Takubo K., Semenza G. L. (2011). Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9 (4), 298–310. Epub 2011/10/11PubMed PMID: 21982230. doi:10.1016/j.stem.2011.09.010
Takubo K., Nagamatsu G., Kobayashi C. I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12 (1), 49–61. Epub 2013/01/08PubMed PMID: 23290136; PMCID: PMC6592822. doi:10.1016/j.stem.2012.10.011
Tannahill G. M., Curtis A. M., Adamik J., Palsson-McDermott E. M., McGettrick A. F., Goel G., et al. (2013). Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496 (7444), 238–242. Epub 2013/03/29PubMed PMID: 23535595; PMCID: PMC4031686. doi:10.1038/nature11986
Theret M., Mounier R., Rossi F. (2019). The origins and non-canonical functions of macrophages in development and regeneration. Development 146 (9), dev156000. Epub 2019/05/03PubMed PMID: 31048317. doi:10.1242/dev.156000
Theret M., Saclier M., Messina G., Rossi F. M. V. (2022). Macrophages in skeletal muscle dystrophies, an entangled partner. J. Neuromuscul. Dis. 9 (1), 1–23. Epub 2021/09/21PubMed PMID: 34542080; PMCID: PMC8842758. doi:10.3233/jnd-210737
Tidball J. G. (2017). Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 17 (3), 165–178. Epub 2017/02/07PubMed PMID: 28163303; PMCID: PMC5452982. doi:10.1038/nri.2016.150
Tsopra O. A., Ziros P. G., Lagadinou E. D., Symeonidis A., Kouraklis-Symeonidis A., Thanopoulou E., et al. (2009). Disease-related anemia in chronic lymphocytic leukemia is not due to intrinsic defects of erythroid precursors: A possible pathogenetic role for tumor necrosis factor-alpha. Acta Haematol. 121 (4), 187–195. Epub 2009/05/27PubMed PMID: 19468203. doi:10.1159/000220331
Tusi B. K., Wolock S. L., Weinreb C., Hwang Y., Hidalgo D., Zilionis R., et al. (2018). Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555 (7694), 54–60. Epub 2018/02/22PubMed PMID: 29466336; PMCID: 5899604. doi:10.1038/nature25741
Warburg O. (1956). On the origin of cancer cells. Science 123 (3191), 309–314. Epub 1956/02/24PubMed PMID: 13298683. doi:10.1126/science.123.3191.309
Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324 (5930), 1076–1080. Epub 2009/05/23PubMed PMID: 19461003; PMCID: PMC2746744. doi:10.1126/science.1164097
Williams N. C., O'Neill L. A. J. (2018). A role for the krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 9, 141. Epub 2018/02/21PubMed PMID: 29459863; PMCID: PMC5807345. doi:10.3389/fimmu.2018.00141
Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., et al. (2008). Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U. S. A. 105 (48), 18782–18787. Epub 2008/11/27PubMed PMID: 19033189; PMCID: PMC2596212. doi:10.1073/pnas.0810199105
Wu H., Zhang Y. (2011). Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25 (23), 2436–2452. Epub 2011/12/14PubMed PMID: 22156206; PMCID: PMC3243055. doi:10.1101/gad.179184.111
Xiang J., Wu D. C., Chen Y., Paulson R. F. (2015). In vitro culture of stress erythroid progenitors identifies distinct progenitor populations and analogous human progenitors. Blood 125 (11), 1803–1812. Epub 2015/01/23PubMed PMID: 25608563; PMCID: 4357585. doi:10.1182/blood-2014-07-591453
Xiao W., Koizumi K., Nishio M., Endo T., Osawa M., Fujimoto K., et al. (2002). Tumor necrosis factor-alpha inhibits generation of glycophorin A+ cells by CD34+ cells. Exp. Hematol. 30 (11), 1238–1247. Epub 2002/11/09. PubMed PMID: 12423676. doi:10.1016/s0301-472x(02)00930-x
Yan H., Wang Y., Qu X., Li J., Hale J., Huang Y., et al. (2017). Distinct roles for TET family proteins in regulating human erythropoiesis. Blood 129 (14), 2002–2012. Epub 2017/02/09PubMed PMID: 28167661; PMCID: PMC5383871. doi:10.1182/blood-2016-08-736587
Zamai L., Secchiero P., Pierpaoli S., Bassini A., Papa S., Alnemri E. S., et al. (2000). TNF-related apoptosis-inducing ligand (TRAIL) as a negative regulator of normal human erythropoiesis. Blood 95 (12), 3716–3724. Epub 2000/06/14. PubMed PMID: 10845902.
Zhao H., Chiaro C. R., Zhang L., Smith P. B., Chan C. Y., Pedley A. M., et al. (2015). Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem. 290 (11), 6705–6713. Epub 2015/01/22PubMed PMID: 25605736; PMCID: PMC4358094. doi:10.1074/jbc.M114.628701
Keywords: stress erythropoiesis, glycolysis, anabolic metabolism, epigenetic regulation, tissue regeneration
Citation: Ruan B and Paulson RF (2023) Metabolic regulation of stress erythropoiesis, outstanding questions, and possible paradigms. Front. Physiol. 13:1063294. doi: 10.3389/fphys.2022.1063294
Received: 06 October 2022; Accepted: 21 December 2022;
Published: 05 January 2023.
Edited by:
Nermi Parrow, Saint Louis University, United StatesReviewed by:
Xiuli An, New York Blood Center, United StatesCopyright © 2023 Ruan and Paulson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert F. Paulson, cmZwNUBwc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.