
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 03 November 2022
Sec. Aquatic Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1049776
This article is part of the Research Topic Aquatic Biotechnology and Its Application: Physiological Foundations of Germplasm Improvement View all 6 articles
 Xingxing Lai1
Xingxing Lai1 Zhongxuan Zhong1
Zhongxuan Zhong1 Bing Lin1
Bing Lin1 Yuxin Wu1
Yuxin Wu1 Yonghao Ma1
Yonghao Ma1 Cuiping Zhang1
Cuiping Zhang1 Yang Yang2
Yang Yang2 Mingqing Zhang1
Mingqing Zhang1 Weijian Qin1
Weijian Qin1 Xiaoqin Fu1*
Xiaoqin Fu1* Hu Shu1*
Hu Shu1*Hypoxia is a critical problem in intensive Epinephelus coioides aquaculture systems. In the present study, the physiological responses of E. coioides muscle to acute hypoxic stress (DO = 0.6 ± 0.1 mg/L) and reoxygenation (DO = 6.0 ± 0.1 mg/L) were analyzed by transcriptome sequencing (RNA-seq) and quantitative real-time PCR (qRT–PCR). RNA-seq was conducted on the muscle tissues of E. coioides in the hypoxia-tolerant (EMS), hypoxia-sensitive (EMW), and normoxic (CM) groups. Among the three groups, a total of 277 differentially expressed genes (DEGs) were identified. KEGG analysis revealed that the pathways significantly enriched after hypoxic stress are involved in the immune response, glycolysis/gluconeogenesis, energy metabolism, vasodilation and proliferation, cell proliferation, and apoptosis. qRT‒PCR verified that the differentially expressed genes FIH-1, PHD-2, PPARα, BCL-XL, LDH-A, and Flt-1 were significantly upregulated after hypoxic stress and returned to normal levels after reoxygenation, suggesting that these DEGs play important roles in responding to hypoxia treatment. In addition, the HIF-1 signaling pathway was also activated under hypoxic stress, and qRT‒PCR confirmed that the expression level of HIF-1α was significantly elevated under acute hypoxic stress, indicating that the HIF-1 signaling pathway is the central pathway in the E. coioides hypoxic response mechanism and activates other related pathways to adapt to hypoxic stress. These pathways jointly regulate energy metabolism, substance synthesis, blood vessel proliferation, cell proliferation, and differentiation and prolong survival time. These results provide ideas for understanding physiological regulation after hypoxic stress and reoxygenation and provide basic insights for the future breeding of hypoxia-tolerant E. coioides.
All organisms must deal with a variety of pressures that may affect their growth, health, and reproduction (Wilson, 2020). Oxygen is an important substrate for the normal biological processes of most organisms and is an essential factor to maintain metabolism and life activities (Childress and Seibel, 1998). Unlike that in the terrestrial environment, the dissolved oxygen (DO) level in the aquatic environment has a wide range of temporal and spatial variations (Ma et al., 2013). Hypoxia occurs when DO levels in aquatic environments drop below 2.0 mg/L and is accelerated by various factors, such as human activities, water pollution, and intensive fish farming (Vajner et al., 2006; Saetan et al., 2020; Sula et al., 2020). It has been well documented that hypoxia not only affects the survival and development of many aquatic species, but is also an important ecological factor leading to diseases (Cai et al., 2010; Wingfield, 2013). Aquaculture production declines sharply when there is no effective method to relieve or offset hypoxic stress (Dawood et al., 2021), which may also lead to huge economic losses and increasing declines in biodiversity and ecosystem function (Riedel et al., 2012; Villnäs et al., 2012).
Hypoxic stress is one of the most critical aquatic environmental factors that adversely affects the physiological processes of fish behavior, growth and development, reproduction, metabolism, and survival (Timmerman and Chapman, 2004; Papandreou et al., 2005; Pollock et al., 2007; Wang et al., 2020; Liu et al., 2021; Zheng et al., 2022). For example, hypoxia can reduce the fecundity of Fundulus grandis (Landry et al., 2007) and Cyprinus carpio (Wu et al., 2003) and inhibit the embryonic development of Danio rerio (Wang et al., 2020). In Oreochromis niloticus, acute hypoxia decreases the feeding activity and time, and growth rate (Zhou et al., 2001) and impairs the immune system (Gallage et al., 2016). Hypoxia adaptation is a complex process involving various physiological and biochemical processes, and molecular responses, such as hypoxia can increase erythrocyte number, reduce protein synthesis, enhance anaerobic metabolism regulate red blood cell proliferation, inhibit apoptosis, stimulate angiogenesis, and upregulate HIF-1a and other hypoxia-related gene expression (Carmeliet et al., 1998; Michiels, 2004; Terova et al., 2008; Muz et al., 2015; Lee et al., 2020). Under hypoxic conditions, glycogen content and lactate concentration decrease, and phosphoenolpyruvate carboxylate kinase expression increases in the liver of Lateolabrax maculatus (Yan et al., 2021).
Epinephelus coioides is the most economically important marine fish in China and other Asian countries (Føre et al., 2018; Yu et al., 2018). As this cultured fish is favored by consumers, its aquaculture industry is developing toward intensive modes such as cage farming and industrial farming to meet people’s demand for a higher yield. However, due to the natural environment and high nutrient input, high-density aquaculture usually leads to hypoxia, which restricts the cultivation of E. coioides (Wang et al., 2013; Huang et al., 2016). To date, few studies have uncovered the mechanism of the response to hypoxic stress in E. coioides.
As an important part of the fish body, muscle can maintain organism movement, anabolism, and catabolism, but its role can be affected by hypoxic stress (Liao et al., 2013). However, the hypoxia acclimation mechanism of muscle in E. coioides remains unknown. In this study, transcriptome sequencing (RNA-seq) was used to investigate the transcriptome profiles of the E. coioides muscle tissues among the hypoxia-tolerant (EMS), hypoxia-sensitive (EMW), and normoxic (CM) groups. Quantitative real-time PCR (qRT‒PCR) was used to analyze the expression sequences of related genes under hypoxic stress and reoxygenation according to the RNA-seq-screened hypoxia-related genes. Our results provide new insights for understanding the adaptation of fish to environmental stress and provide a theoretical basis for solving the problems of modern intensive aquaculture, especially high-density aquaculture.
Healthy E. coioides (weight, 150 ± 5 g; length 15 ± 5 cm) were achieved from the Huizhou base of the Guangdong Marine Fisheries Experiment Center. These fish were acclimated in cylindrical tanks with constant ventilation for 2 weeks before prescription. The tanks were equipped with artificially disinfected self-circulating seawater with a salinity of 20–30 ppt and a temperature of 30 ± 1°C. The DO of the seawater was 6.0 ± 0.1 mg/L, as measured by a water quality analyzer (YSI, United States, ProQuatro). During acclimation, the fish were fed a commercial diet of 3% of their body weight daily. After adaptation, the fish were used for the hypoxic stress and reoxygenation experiment.
Fifty-four individuals were kept in a breeding pool to conduct the pre-experiment. Briefly, the tank was sealed with a transparent plastic film, and nitrogen was continuously pumped through the nitrogen plastic ventilator until the fish were unbalanced (Pichavant et al., 2000; Li et al., 2018). At this moment, the water’s DO was found to be 0.6 ± 0.1 mg/L, which served as the benchmark for the next formal hypoxic stress test on E. coioides.
The experimental fish were split into two groups, the hypoxic group (H group) and the normoxic control group (CM group), with three replicates each and 54 fish for each replication. In the H group, the fish tanks were membrane-sealed and continuously nitrogen-injected to keep the DO level at 0.6 ± 0.1 mg/L. After 8 h of hypoxic stress, the reoxygenation (R) experiment was performed on the H group for 72 h, and the DO in the water was restored to 6.0 ± 0.1 mg/L. Throughout the hypoxia and reoxygenation experiment, the fish in the CM group tanks were continuously aerated to keep the DO at 6.0 ± 0.1 mg/L.
Sampling for RNA-seq began when half of the fish died after the hypoxic condition (DO 0.6 ± 0.1 mg/L) lasted for an hour in the H group. Following that, depending on their physiological states, the live fish were divided into the EMS and EMW group. Fish from each of the three groups (CM, EMS, EMW) were taken and dissected (three fish per replicate for a total of nine fish per group), and muscle tissues were taken and kept in liquid nitrogen.
Sampling for qRT‒PCR was performed when half of the fish were floating and unbalanced in the H group. Muscle tissues of the fish from the CM group and H group were collected at 0 h, 2 h, 4 h, 6 h, and 8 h after hypoxic stress. In the R group and CM group, muscle samples for qRT‒PCR were obtained at 0 h, 4 h, 8 h, 12 h, 24 h, 48 h, and 72 h after reoxygenation treatment. Muscle tissues were taken from 3 fish in each parallel group at each interval, and placed at −80°C for further analysis.
Following the manufacturer’s instructions, total RNA from the muscle was extracted using TRIzol kits (Promega, Madison, WI, United States). The quantity and quality of the RNA were determined using a Bioanalyzer 2100 (Agilent Technologies, United States), ND-2000 (NanoDrop Technologies, United States), and RNase-free agarose gel electrophoresis, respectively. High-quality RNA (RIN> 7) was applied to create the sequencing libraries. Three biological replicate samples were set up in each group. Finally, nine cDNA libraries (CM-3, EMS-3, EMW-3) were scanned using paired-end technology (Guangzhou Gene Jirui Gene Technology Co., Ltd., Guangzhou, China) around an Illumina HiSeq 2500 sequencing (Illumina, San Diego, CA, United States).
After CASAVA base calling, the raw data from the Illumina NovaSeq 6000 sequencing machine were transformed into text data and stored in FASTQ format as raw reads. Fastp (v 0.12) was used to remove low-quality sequences from the raw reads (Altschul et al., 1997). The effectiveness of RNA-sequencing was assessed by the quality scores of the Q20, Q30, and GC contents. The clean data were aligned to the E. coioides genome utilizing TOPHAT2 (v.2.0.4). The relative expression abundance of unigenes was determined as the reads per kb per million reads (RPKM).
DESeq2 was applied to discover differentially expressed genes (DEGs) among the three groups. Absolute log2 fold change |log2FC| > 1 and false discovery rate (FDR) < 0.05 were defined as DEGs. The DEGs were subjected to GO functional annotation and pathway enrichment analysis via GOATOOLS (v. 0.6.5) (Ye et al., 2018) and KOBAS v.2.1 (Wu et al., 2006), respectively. Enrichment analysis of the KEGG pathway provided functional annotations of the DEGs. An FDR-adjusted p value ≤0.05 was used to determine the significantly enriched GO terms or pathways. DEGs implicated in metabolism, cellular processes, environmental information processing, organismal systems, human diseases, and genetic information processing were screened for further investigation to clarify the physiological changes in E. coioides under hypoxia.
qRT‒PCR of the selected DEGs was performed to evaluate the reliability of the RNA-seq results. The specific primers shown in Supplementary Table S1 for the chosen DEGs were created through Primer v.6.0. qRT‒PCR was carried out on a 384-well plate qRT‒PCR device (ABI 7500; Applied Biosystems, Waltham, MA, United States) with a final volume of 12.5 μL including 6.25 μL of SYBR qRT‒PCR Master Mix (Novizan, China), 4.25 μL of ddH2O, 0.5 μL of each primer (the primers used for qRT‒PCR are displayed in Supplementary Table S1), and 1 μL of template DNA (50 ng/μL). The qRT‒PCR conditions were as follows: 95°C for 5 min; 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 10 min for data collection. Each sample was examined in triplicate for this experiment. The gene expression levels were determined by the 2−ΔΔCT method with β-actin as the internal reference. Statistical analyses of the experimental data were carried out by SPSS (v 20.0). The results are presented as the mean ± SE and were graphed using Origin 9.0 software (Zhong et al., 2021).
Total RNA from each sample was extracted by RNA isolater Total RNA Extraction Reagent following the manufacturer’s instructions. RNA concentration and purity were measured on a spectrophotometer. First-strand cDNA was synthesized according to the instructions of the HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) reverse transcription kit and then stored at -80°C for further study. The primers for qRT‒PCR detection of hypoxic stress- and reoxygenation-related genes in muscle tissue of E. coioides are shown in Supplementary Table S2. The qRT‒PCR conditions and protocol were the same as those described in Section 2.6.
Transcriptomic sequencing was performed from the muscle of E. coioides. After mass filtration, a total of 45,597,702 raw reads (68.83 Gb) were obtained, of which 45,313,712 passed the quality control process. The size of the clean reads in each library ranged from 6.50 Gb to 8.99 Gb. The clean reads were compared with the reference genome, and the statistical parameters of clean reads in the CM, EMS, and EMW groups were as follows: Q20, 97.46–97.92%; Q30, 93.41–94.36%; GC content, 51.01–51.89%; and error rate, 0.025–0.026%. Clean reads from each library were sequentially mapped with reference genomic information from Epinephelus spp., with a total ratio of 87.60%–89.71% per sample, and more than 5.75 Gb of data for each library was obtained (Table 1).
As shown in Supplementary Figure S1, 277 DEGs were found in the CM vs. EMS, CM vs. EMW, and EMS vs. EMW groups (Supplementary Figure S1). In the CM vs. EMS groups, a total of 172 DEGs were identified, with 133 upregulated DEGs and 39 downregulated DEGs (Figures 1A,B). The comparison of CM and EMW displayed 165 DEGs, of which 50 were upregulated and 115 were downregulated (Figures 1A,C). There were only 25 DEGs between the EMS and EMW groups, with 4 DEGs upregulated and 21 DEGs downregulated (Figures 1A,D).
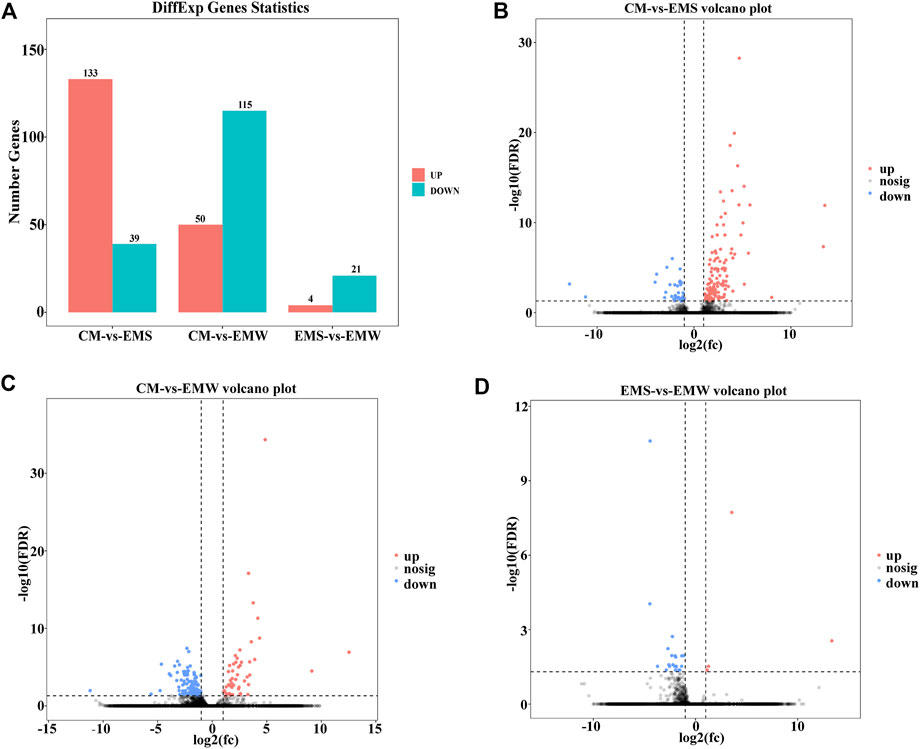
FIGURE 1. Differences in gene number in CM vs. EMS, CM vs. EMW, and EMS vs. EMW groups. (A) The bar chart shows the number of genes in the CM vs. EMS, CM vs. EMW, and EMS vs. EMW groups. (B) Volcano plot of DEGs in CM vs. EMS. (C) Volcano plot of DEGs in CM vs. EMW. (D) Volcano plot of EMS vs. EMW.
In the CM vs. EMS comparison, 172 DEGs were screened for GO enrichment analysis and functional classification, and the results showed that they were divided into three main functional categories, namely, the biological process (BP), cellular component (CC), and molecular function (MF) categories. The DEGs were mainly associated with BP terms such as the cellular process, tissue process, biological regulation, stimulus-response and metabolic process terms (Figure 2A). To further elucidate the biological events of DEGs primarily involved in the hypoxia response, the top 20 most enriched GO terms were listed (Figure 2B). The DEGs were mainly enriched in energy metabolism terms (GO:0034098 VCP-NPL4-UFD1 AAA ATPase complex), cilium-related terms (GO:0072372 primary cilium, GO:0031513 nonmotile primary cilium), and other BP terms (Figure 2B).
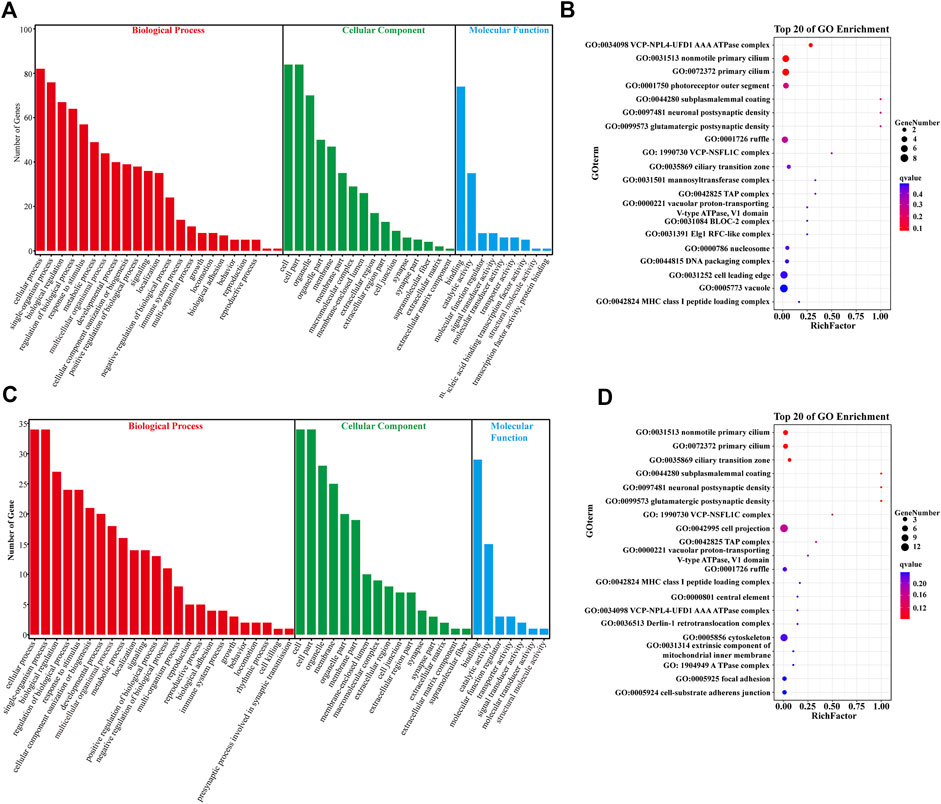
FIGURE 2. Gene ontology (GO) analysis of DEGs. (A) GO pathway analysis of DEGs in the CM vs. EMS groups. (B) Top 20 enriched terms by DEGs in the CM vs. EMS groups. (C) GO pathway analysis of DEGs in the CM vs. EMW groups. (D) Top 20 enriched terms by DEGs in the CM vs. EMW groups. The color of each dot represents the corrected p-value of the corresponding item.
In the CM vs. EMW comparison, GO enrichment analysis revealed that the165 screened DEGs were also divided into BP, CC, and MF groups, and the DEGs were mainly related to BP terms (Figure 2C). To further elucidate the biological events of DEGs primarily participated in the hypoxia response, the 20 most enriched GO terms were listed, which were mainly associated with BP terms such as cytoskeleton movement (GO:0005856 cytoskeleton), cilia movement (GO:0072372 primary cilium), and synthesis of ATPase complex (Figure 2D).
KEGG analysis showed that a total of 43 pathways were enriched between the CM group and EMS group, classified into six functional categories: metabolism, environmental information processing, organic systems, human diseases, and cellular processes (Figure 3A). Signal transduction, global and overview maps, carbohydrate metabolism, and cell growth and death were the dominant pathways of enrichment (Figure 3A). All DEGs obtained in CM vs. EMS were mapped to KEGG to identify specific biological pathways of the hypoxia response. The top 20 most abundant KEGG pathways associated with the hypoxia mechanism were the HIF-1 signaling pathway, apoptosis, NF-kappa B signaling pathway, FOXO signaling pathway, MAPK signaling pathway, C-Type Lectin receptor signaling pathway, and glycolysis/gluconeogenesis (Figure 3B). The above-enriched pathways were mainly involved in the immune response, cell proliferation, apoptosis, and energy metabolism.
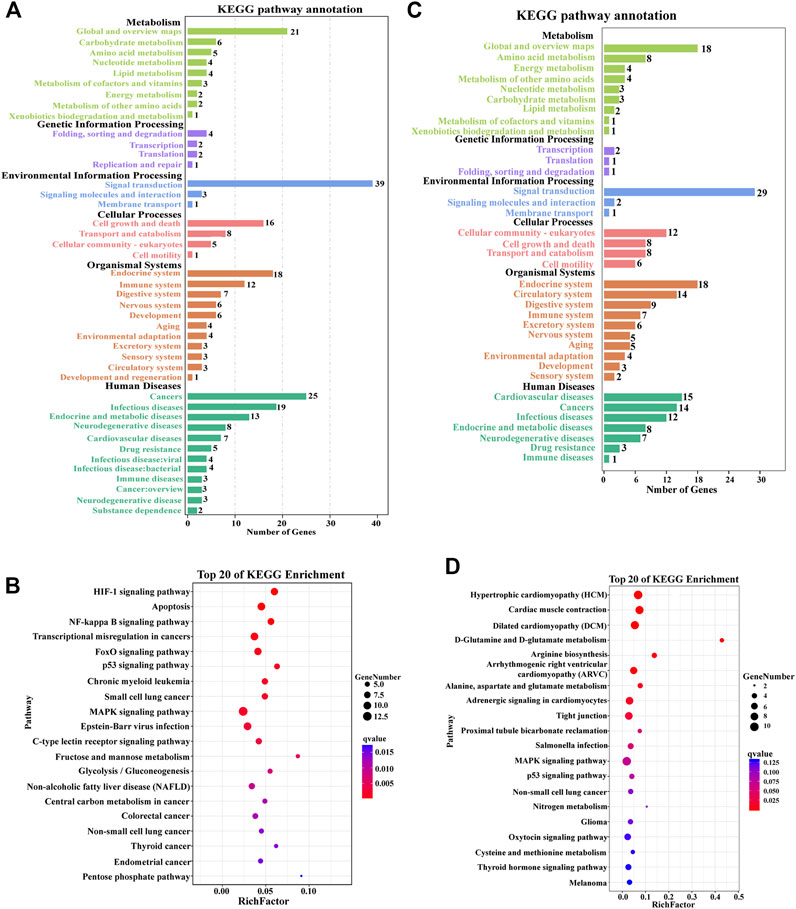
FIGURE 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs in the muscle of E. coioides. (A) KEGG pathway enrichment analysis of DEGs in the CM vs. EMS groups. (B) Top 20 enriched pathways by DEGs in the CM vs. EMS groups. (C) KEGG pathway enrichment analysis of DEGs in the CM vs. EMW groups. (D) Top 20 enriched pathways by DEGs in the CM vs. EMW groups.
In the comparison of the CM and EMW groups, 36 significantly enriched KEGG annotations were found, consisting of 6 different functional categories. These pathways were mainly related to signal transduction, fatty acid metabolism, cell growth and death, the endocrine system, and the circulatory system (Figure 3C). Among the 20 most enriched KEGG pathways, the MAPK signaling pathway, adrenergic signaling in cardiomyocytes, and cardiac muscle contraction were significantly enriched and involved in the hypoxia response (Figure 3D).
PPI network analysis showed that in the EMS group, the DEGs were mainly involved in the HIF-1 signaling pathway, FOXO signaling pathway, AMPK signaling pathway, MAPK signaling pathway, NF-κB signaling pathway, vascular endothelial growth factor (VEGF) signaling pathway, cAMP signaling pathway, and glycolysis/gluconeogenesis signaling pathway (Figures 4A,B), while there was no significant difference in the EMW group.
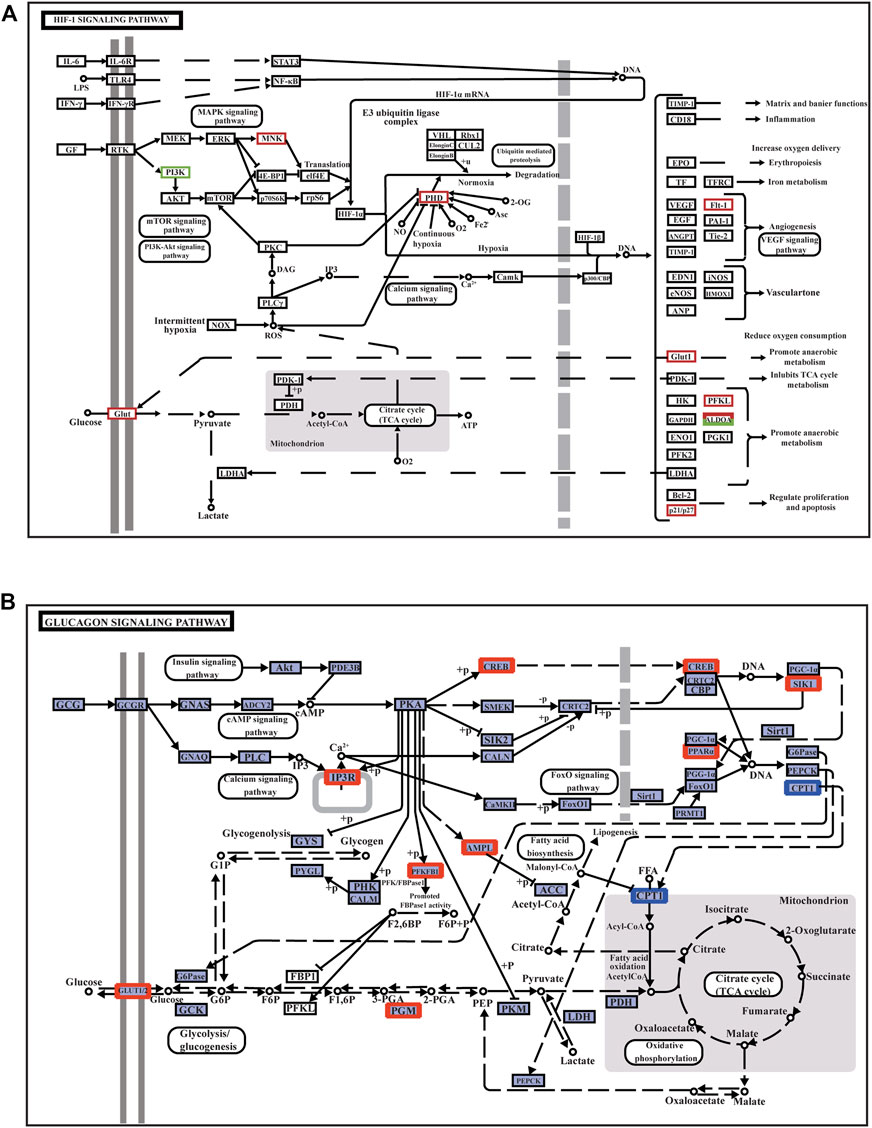
FIGURE 4. Annotation diagram of muscle signaling pathway and its related DEGs. (A) HIF-1 signaling pathway. (B) Glucagon signaling pathway. The red box indicates the upregulated genes in the pathways; the green box represents the downregulated genes in the pathway.
The HIF-1 signaling pathway is the main pathway activated when muscle cells are damaged by hypoxic stress. In the HIF-1 signaling pathway, the MNK factor in the MAPK signaling pathway is upregulated; thus, HIF-1α mRNA expression is upregulated. The related gene VEGF receptor-1 (Flt-1) is upregulated to trigger the VEGF signaling pathway, promote angiogenesis and proliferation, and accelerate blood oxygen delivery to adjust to a hypoxic environment. Upregulation of cyclin p21/p27 regulates cell proliferation and reduces apoptosis (Figure 4A). The upregulation of phosphofructokinase-1 (PFKL) and recombinant human fructose-bisphosphate aldolase A (ALDOA) promotes anaerobic metabolism and reduces oxygen consumption. Glucose transporter 1 (Glut-1) is involved in glycolysis/gluconeogenesis in response to hypoxia to enhance glucose transport and utilization (Figure 4B). The glycolysis/gluconeogenesis pathway is regulated by the FOXO signaling pathway and the calcium signaling pathway (Supplementary Figure S2). In the FOXO signaling pathway, cyclin p27 is an important member of the cyclin-dependent inhibitor CIP/KIP family. P27 and growth arrest- and DNA damage-inducible 45 (Gadd45), which function to regulate the cell cycle, were also upregulated when muscle tissue was damaged under hypoxic stress. Gadd45 can also regulate antioxidant stress and DNA repair in the hypoxia response (Supplementary Figure S2).
To comprehend the mechanism underlying E. coioides’s adaptive reaction to hypoxia, the DEGs associated with hypoxia were further screened. As shown in Figure 5, 26 DEGs were discovered to be significantly upregulated in the EMS group, while in the EMW group, only 2 DEGs, proline hydroxylase-1 (PHD-1) and superoxide dismutase (SOD), were detected as significantly upregulated. As expected, none of these 28 DEGs were found in the CM group (Figure 5). The screened DEGs in the EMS group were divided into two categories. One group included upstream pathway DEGs, including HIF-1α, factor inhibiting HIF-1 (FIH-1), PHD-2, protein kinase (AKT), MAPK-activated kinase (MNK) and von Hipel-Lindau protein (VHL). Another group of downstream pathway DEGs included genes associated with energy metabolism, such as pyruvate dehydrogenase kinase (PDK), lactate dehydrogenase A (LDH-A, GLUT-1, ALDOA, and hexokinase (HK), and genes involved in regulating cell proliferation and apoptosis, including poly (rC) binding protein 2 (PCBP2), DNA damage-inducible transcript 4 (DDIT4), CCAAT/enhancer-binding protein delta (CEBPD), Ras-related protein Rab-13 (RAB13), purine nucleoside phosphorylase (PNP), C-X-C motif chemokine 10 (CXCL10), peroxisome proliferator-activated receptor-α (PPARα), B-cell lymphoma-extra-large (BCL-XL) and GADD45. In addition, the downstream pathway DEGs also included VEGF-A, Flt-1, and enolase (EN O 1), which were related to the VEGF-A pathway, proliferation, and anaerobic metabolism. The activation of these genes is involved in the major biological processes that participate in the response to hypoxic stress (Figure 5).
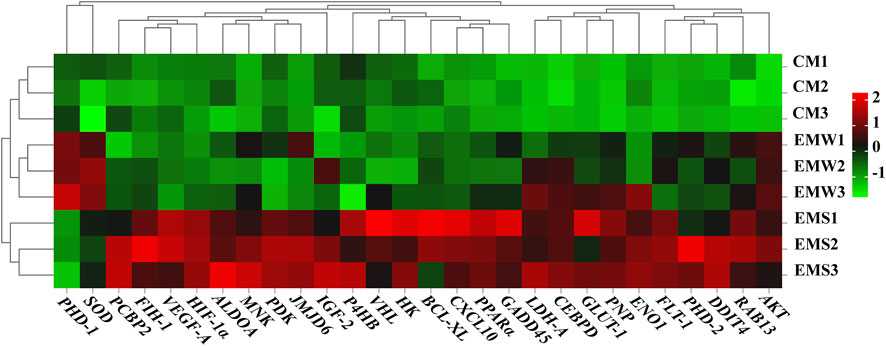
FIGURE 5. Heat map of DEGs in the control and experimental groups. Red represents gene up-regulation, green represents gene down-regulation. CM indicates the normoxic control group of muscle tissue; EMS represents hypoxia-tolerant group of muscle tissue; EMW represents hypoxia-sensitive group of muscle tissue.
To confirm the accuracy of the RNA-seq data, the transcript levels of HIF-1α, PHD-2, LDH-A, VHL, VEGF-A, FIH-1, insulin-like growth factor-2 (IGF-2), SOD, HK, and pyruvate kinase (PK) in the EMS group and those of HIF-1α, PHD-2, LDH-A, VHL, VEGF-A, PHD-1, FIH-1, IGF-2, SOD, catalase (CAT), HK and PK in the EMW group were analyzed and verified by qRT‒PCR. Correlation analysis was performed on the obtained results using the log2FC values. The fitting curve equation of the EMS group was y = 0.01269 + 0.98838x, R2 = 0.99018 (Figure 6A), while that of the EMW group was y = 0.02831 + 1.08217x, R2 = 0.99514 (Figure 6B). The EMS and EMW groups’ R2 values were both greater than 0.7, demonstrating the high accuracy of the data from transcriptome sequencing.
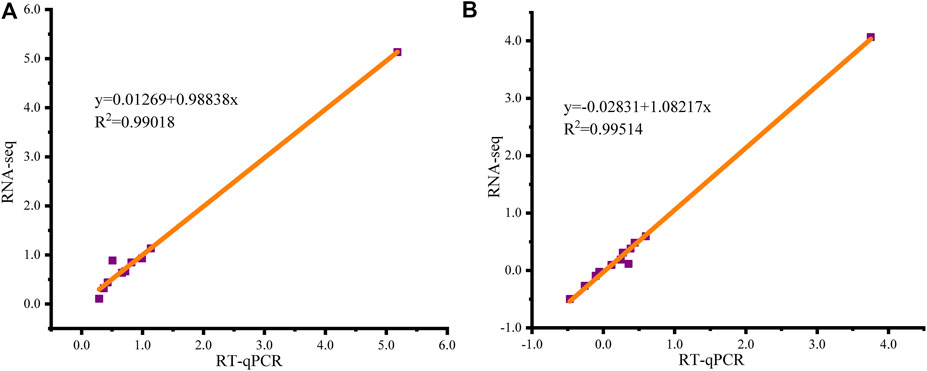
FIGURE 6. Fitting curve of correlation between qRT‒PCR and RNA-seq gene expression results. X-axis is the log2FC value of qRT‒PCR, Y-axis is the log2FC value of RNA-seq, and the red line is the fitting curve. (A) Correlation analysis of EMS group. (B) Correlation analysis of EMW group.
qRT‒PCR was carried out to illustrate the mRNA expression levels of seven significant DEGs in the muscle after hypoxia and reoxygenation treatment. As shown in Figure 7, the expression levels of HIF-1α, FIH-1, PPARα, BCL-XL, LDH-A and Flt-1 were remarkably upregulated at all time points after acute hypoxic stress compared with normoxic controls, peaking at 4 h of hypoxia (H4), H2, H4, H6, H6, and H4, respectively. However, the expression level of PHD-2 was significantly increased only at H8. After the restoration of dissolved oxygen, the expression of these genes successively returned to levels similar to those of normoxic controls. The expression of both HIF-1α and LDH-A recovered to normal control levels at 24 h of reoxygenation (R24), Flt-1, PPARα and BCL-XL all returned to normal levels at R12, while PHD-2 and FIH-1 returned to normal expression levels at R48 and R72, respectively (Figure 7).
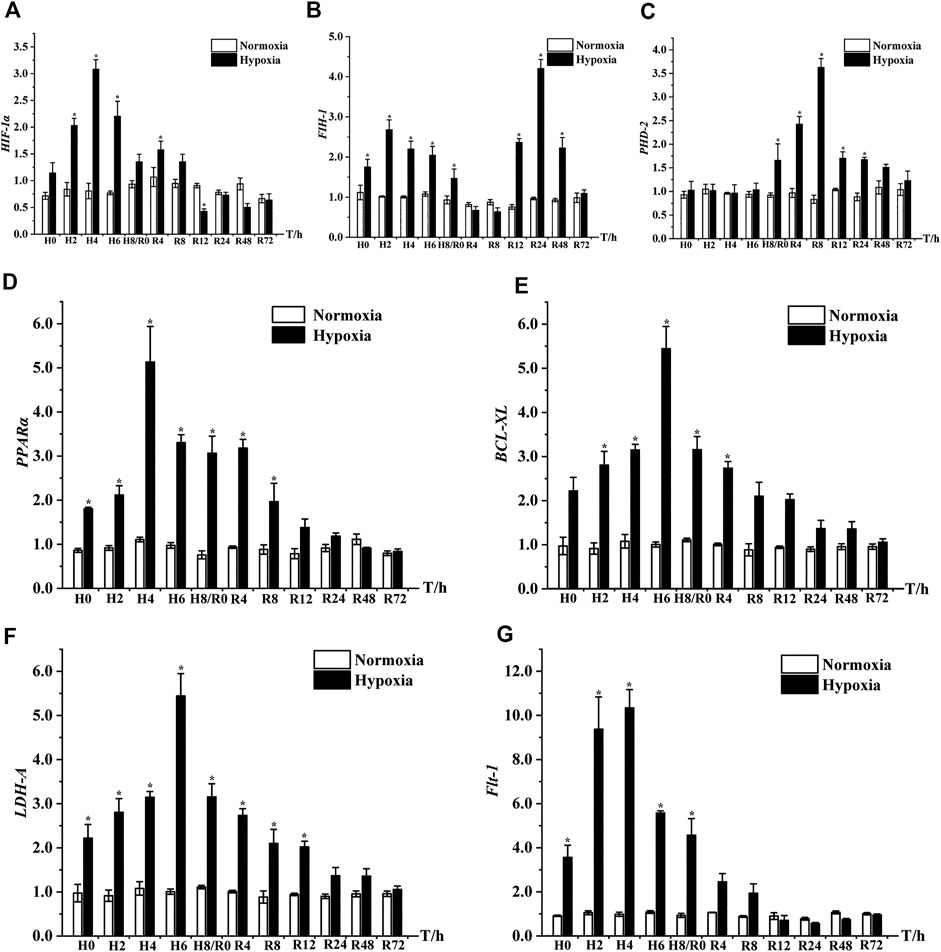
FIGURE 7. Expression patterns of seven key DEGs in muscle of E. coioides after hypoxic stress and reoxygenation, including HIF-1α (A), FIH-1 (B), PHD-2 (C), PPARα (D), BCL-XL (E), LDH-A (F), Flt-1 (G). H: Hypoxia; R: Reoxygenation. The results were represented by mean and standard error, and the difference of labeled lowercase letters indicated significant difference (p < 0.05).
As we all know, hypoxia is a major problem in high-density fish aquaculture. Therefore, screening for candidate genes associated with hypoxia tolerance traits is extremely important for the selection and breeding of hypoxia-tolerant species in intensive fish farming. To provide a theoretical basis for solving the problems of modern intensive aquaculture of E. coioides, the main aim of this paper is to identify underlying candidate genes related to hypoxia tolerance. In the present study, RNA-seq was performed on the EMS, EMW and CM individuals, and qRT‒PCR was adopted to analyze the expression level of DEGs screened from the RNA-seq based on the samples of hypoxic stress and reoxygenation. Fortunately, some significantly enriched pathways and DEGs were identified from the hypoxia-tolerant group by RNA-seq and qRT‒PCR analyses.
In the current study, KEGG analysis of DEGs showed that the upstream HIF-1 signaling pathway was enriched in the EMS group, while HIF-1α or HIF-1α mRNA expression was upregulated in the HIF-1 signaling pathway annotation. Previous studies have shown that the HIF-1 signaling pathway is the central pathway in the response to hypoxia (Sheng et al., 2017) and that it can be activated quickly after hypoxic stress to enable adaptation to a hypoxic environment. As a critical regulator in the HIF-1 signaling pathway, the HIF-1α gene plays an important role in the homeostatic regulation of organisms (Minet et al., 1999). Our results showed that HIF-1α expression was dramatically elevated under acute hypoxic stress but returned to normal after reoxygenation, which is similar to findings in Lateolabrax japonicus (Terova et al., 2008), implying that HIF-1α plays a key role in the response to hypoxic stress. These results suggest that the HIF-1 signaling pathway is the core of the molecular regulatory pathway that is activated during hypoxia and reoxygenation of E. coioides and triggers a series of metabolic changes, including changes in energy metabolism, the immune response, and other metabolic pathways.
Flt-1 can stimulate the VEGF signaling pathway and promote vascular dilation and proliferation (Jiang et al., 1997; Shan et al., 2012). qRT‒PCR analysis on Flt-1 showed significant upregulation under hypoxic stress, which was consistent with the transcriptome sequencing results. Both the PHD-2 and FIH-1 genes can theoretically inhibit HIF-1α expression (Grandcourt et al., 2009; Jelkmann, 2011; Wang et al., 2015). In this study, similar to that of HIF-1α, the expression of FIH-1 was significantly increased under hypoxic stress in the H0-H8 stage, unlike the expression of PHD-2, which did not differ significantly from that of the control group. After 12 h of reoxygenation, PHD-2 and FIH-1 expression levels were significantly increased compared with those of the normoxic group, while the HIF-1α level decreased significantly until the R24 period (Figure 7). These results suggest that PHD-2 and FIH-1 are inhibitory factors of HIF-1α expression within 8–24 h after reoxygenation. Unexpectedly, PHD-2 expression peaked at R8 point in the reoxygenation stage, we speculated that there were two possible reasons for this. On the one hand, species variability leads to a different expression pattern of E. coioides PHD-2 in the reoxygenation stage than that in other fish, on the other hand, PHD-2 may participate in down-regulating the expression of HIF-1α and other genes during the reoxygenation phase, which needs further study in the future.
During hypoxic stress, extracellular signaling factors specifically bind to intracellular or cell membrane receptors to induce intracellular signals and trigger cascade reactions, thus affecting cell functions (Chen et al., 2001; Sauer et al., 2001). The HIF-1 signaling pathway coordinates the repair of inflammation by activating the NF-κB signaling pathway (Spirina et al., 2017; Mohamed et al., 2019). The AMPK and FOXO signaling pathways complete cell proliferation and differentiation, complete the distribution and reorganization of intracellular energy metabolism, and repair the inflammation caused by metabolic regulation (Nagata et al., 2003; Lee et al., 2013; Dengler, 2020). Our results showed that oxidative phosphorylation, the NF-κB signaling pathway, the FOXO signaling pathway, the p53 signaling pathway, the MAPK signaling pathway, and apoptosis were highly enriched in the EMS group and that DEGs in these pathways were significantly upregulated. For example, PPARα and BCL-XL are involved in the immune response process to repair inflammatory damage, slow apoptosis, and prolong survival time. The qRT‒PCR results showed that PPARα and BCL-XL were also significantly upregulated under hypoxic stress (Figure 7), indicating that PPARα and BCL-XL can repair muscle tissue inflammatory damage caused by hypoxic stress.
Activation of anaerobic metabolism is related to hypoxic stress. Under hypoxic stress, fish respiration changes from aerobic to anaerobic respiration (Maccormack et al., 2006). Anaerobic glycolysis/glucose metabolism is the cause of the high energy demand of fish under hypoxic stress. Glycolysis is an important energy-providing pathway during hypoxic stress (Connett et al., 1990; Semenza et al., 1994; Xie et al., 2019; Kierans and Taylor, 2021). Here, the glycolysis/glucose metabolism signaling pathway was also highly enriched in the EMS group. As a crucial REDOX enzyme in the glycolysis pathway, LDH-A can reversely catalyze the conversion of lactic acid to pyruvate and provide ATP and prevent acidosis caused by excess lactic acid (Shim et al., 1997; Fantin et al., 2006). We found that LDH-A expression was also significantly elevated under acute hypoxia treatment but returned to normal under reoxygenation, in line with the outcomes shown in Oncorhynchus mykiss (Davie et al., 1986). These results suggest that glycolysis and glucose metabolism are of great significance in physiological adaptation and the response to acute hypoxic stress and reoxygenation.
The mechanism of the E. coioides muscle response to acute hypoxia and reoxygenation was investigated by RNA-seq and qRT‒PCR in this study. 277 DEGs were identified, and several key DEGs were verified by qRT‒PCR to be significantly upregulated after hypoxic stress but restored to normal levels after reoxygenation. The significantly enriched pathways after hypoxia induction were involved in immune processes, energy metabolism, apoptosis and so on. These results are beneficial for understanding adaptability to hypoxic environments and provide valuable information for the breeding of E. coioides and other fish with low oxygen tolerance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) are below: NCBI (accession: PRJNA895010).
The animal study was reviewed and approved by the Experimental Animal Ethics Committee of the Guangzhou University of China.
XL: Data curation, visualization, methodology, software, writing original draft. ZZ: Methodology, data curation, software. BL: Data curation, software, formal analysis. YW: Methodology, Formal analysis. YM: Data curation, methodology. CZ: Data curation, methodology, formal analysis. YY: Methodology, investigation, data curation. MZ: Methodology, validation. WQ: Methodology, validation. XF: Conceptualization, methodology, software, resources, data curation, visualization, writing. HS: Conceptualization, methodology, software, resources, data curation, visualization, writing-review and editing, supervision, project administration.
This research was supported by grants from Science and Technology Program of Guangdong Province (2019B030316022), Agriculture and Rural Affairs Department of Guangdong Province (SDYY-2018-10), and the Key Research Project of Grouper Breeding and Ecological regulation in Pond culture of Guangdong Province (No. 68112).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1049776/full#supplementary-material
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-blast: A new generation of protein database search programs. Nucleic Acids Res. 25 (17), 3389–3402. doi:10.1093/nar/25.17.3389
Cai H., Zhao S., Wu C., Zhu A., Yu J., Zhang X. (2010). “Environmental pollution and marine aquaculture ecosystem health assessment,” in 2010 4th international conference on bioinformatics and biomedical engineering (IEEE), 1–4.
Carmeliet P., Dor Y., Herbert J.-M., Fukumura D., Brusselmans K., Dewerchin M., et al. (1998). Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394 (6692), 485–490. doi:10.1038/28867
Chen E. Y., Mazure N. M., Cooper J. A., Giaccia A. J. (2001). Hypoxia activates a platelet-derived growth factor receptor/phosphatidylinositol 3-kinase/Akt pathway that results in glycogen synthase kinase-3 inactivation. Cancer Res. 6, 2429–2433.
Childress J. J., Seibel B. A. (1998). Life at stable low oxygen levels: Adaptations of animals to oceanic oxygen minimum layers. J. Exp. Biol. 201 (8), 1223–1232. doi:10.1242/jeb.201.8.1223
Connett R., Honig C., Gayeski T., Brooks G. (1990). Defining hypoxia: A systems view of VO2, glycolysis, energetics, and intracellular PO2. J. Appl. Physiol. 68 (3), 833–842. doi:10.1152/jappl.1990.68.3.833
Davie P. S., Wells R. M., Tetens V. (1986). Effects of sustained swimming on rainbow trout muscle structure, blood oxygen transport, and lactate dehydrogenase isozymes: Evidence for increased aerobic capacity of white muscle. J. Exp. Zool. 237 (2), 159–171. doi:10.1002/jez.1402370203
Dawood M. A., Noreldin A. E., Sewilam H. (2021). Long term salinity disrupts the hepatic function, intestinal health, and gills antioxidative status in Nile tilapia stressed with hypoxia. Ecotoxicol. Environ. Saf. 220, 112412–112424. doi:10.1016/j.ecoenv.2021.112412
Dengler F. (2020). Activation of AMPK under hypoxia: Many roads leading to rome. Int. J. Mol. Sci. 21 (7), 2428–2443. doi:10.3390/ijms21072428
Fantin V. R., St-Pierre J., Leder P. (2006). Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 9 (6), 425–434. doi:10.1016/j.ccr.2006.04.023
Føre M., Frank K., Norton T., Svendsen E., Alfredsen J. A., Dempster T., et al. (2018). Precision fish farming: A new framework to improve production in aquaculture. Biosyst. Eng. 173, 176–193. doi:10.1016/j.biosystemseng.2017.10.014
Gallage S., Katagiri T., Endo M., Futami K., Endo M., Maita M. (2016). Influence of moderate hypoxia on vaccine efficacy against Vibrio anguillarum in Oreochromis niloticus (Nile tilapia). Fish. Shellfish Immunol. 51, 271–281. doi:10.1016/j.fsi.2016.02.024
Grandcourt E., Al Abdessalaam T., Francis F., Al Shamsi A., Hartmann S. (2009). Reproductive biology and implications for management of the orange‐spotted grouper Epinephelus coioides in the southern Arabian Gulf. J. Fish. Biol. 74 (4), 820–841. doi:10.1111/j.1095-8649.2008.02163.x
Huang W., Liu Q., Xie J., Wang W., Xiao J., Li S., et al. (2016). Characterization of triploid hybrid groupers from interspecies hybridization (Epinephelus coioides♀× Epinephelus lanceolatus♂). Aquac. Res. 47 (7), 2195–2204. doi:10.1111/are.12672
Jelkmann W. (2011). Regulation of erythropoietin production. J. Physiol. 589 (6), 1251–1258. doi:10.1113/jphysiol.2010.195057
Jiang B.-H., Agani F., Passaniti A., Semenza G. L. (1997). V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: Involvement of HIF-1 in tumor progression. Cancer Res. 57 (23), 5328–5335.
Kierans S. J., Taylor C. T. (2021). Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 599 (1), 23–37. doi:10.1113/JP280572
Landry C., Steele S., Manning S., Cheek A. (2007). Long term hypoxia suppresses reproductive capacity in the estuarine fish, Fundulus grandis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 148 (2), 317–323. doi:10.1016/j.cbpa.2007.04.023
Lee H.-H., Chang C.-C., Shieh M.-J., Wang J.-P., Chen Y.-T., Young T.-H., et al. (2013). Hypoxia enhances chondrogenesis and prevents terminal differentiation through PI3K/Akt/FoxO dependent anti-apoptotic effect. Sci. Rep. 3 (1), 2683–2712. doi:10.1038/srep02683
Lee P., Chandel N. S., Simon M. C. (2020). Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 21 (5), 268–283. doi:10.1038/s41580-020-0227-y
Li M., Wang X., Qi C., Li E., Du Z., Qin J. G., et al. (2018). Metabolic response of Nile tilapia (Oreochromis niloticus) to acute and chronic hypoxia stress. Aquaculture 495, 187–195. doi:10.1016/j.aquaculture.2018.05.031
Liao X., Cheng L., Xu P., Lu G., Wachholtz M., Sun X., et al. (2013). Transcriptome analysis of crucian carp (Carassius auratus), an important aquaculture and hypoxia-tolerant species. PloS one 8 (4), e62308. doi:10.1371/journal.pone.0062308
Liu B., Wen H., Li X., Yang J., Li G., Zhang M., et al. (2021). Acute hypoxia effects on Keap1/Nrf2 (Mafs)-GST pathway related oxidative metabolism in muscle of Japanese flounder (Paralichthys olivaceus). Sci. Total Environ. 795, 148646. doi:10.1016/j.scitotenv.2021.148646
Ma Q., Li R., Feng J., Wang L. (2013). Relationships between total dissolved gas and dissolved oxygen in water. Fresenius Environ. Bull. 22 (11), 3243–3250.
Maccormack T. J., Lewis J. M., Almeida‐Val V. M. F., Val A. L., Driedzic W. R. (2006). Carbohydrate management, anaerobic metabolism, and adenosine levels in the armoured catfish, Liposarcus pardalis (Castelnau), during hypoxia. J. Exp. Zool. A Comp. Exp. Biol. 305 (4), 363–375. doi:10.1002/jez.a.274
Michiels C. (2004). Physiological and pathological responses to hypoxia. Am. J. Pathol. 164 (6), 1875–1882. doi:10.1016/S0002-9440(10)63747-9
Minet E., Mottet D., Michel G., Roland I., Raes M., Remacle J., et al. (1999). Hypoxia-induced activation of HIF-1: Role of HIF-1alpha-Hsp90 interaction. FEBS Lett. 460 (2), 251–256. doi:10.1016/s0014-5793(99)01359-9
Mohamed A. A.-R., Metwally M. M., Khalil S. R., Salem G. A., Ali H. A. (2019). Moringa oleifera extract attenuates the CoCl2 induced hypoxia of rat's brain: Expression pattern of HIF-1α, NF-kB, MAO and EPO. Biomed. Pharmacother. 109, 1688–1697. doi:10.1016/j.biopha.2018.11.019
Muz B., de la Puente P., Azab F., Azab A. K. (2015). The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 3, 83–92. doi:10.2147/HP.S93413
Nagata D., Mogi M., Walsh K. (2003). AMP-activated protein kinase (AMPK) signaling in endothelial cells is essential for angiogenesis in response to hypoxic stress. J. Biol. Chem. 278 (33), 31000–31006. doi:10.1074/jbc.M300643200
Papandreou I., Powell A., Lim A. L., Denko N. (2005). Cellular reaction to hypoxia: Sensing and responding to an adverse environment. Mutat. Res. 569 (1-2), 87–100. doi:10.1016/j.mrfmmm.2004.06.054
Pichavant K., Person-Le-Ruyet J., Le Bayon N., Severe A., Le Roux A., Quéméner L., et al. (2000). Effects of hypoxia on growth and metabolism of juvenile turbot. Aquaculture 188 (1-2), 103–114. doi:10.1016/s0044-8486(00)00316-1
Pollock M., Clarke L., Dubé M. (2007). The effects of hypoxia on fishes: From ecological relevance to physiological effects. Environ. Rev. 15, 1–14. doi:10.1139/a06-006
Riedel B., Zuschin M., Stachowitsch M. (2012). Tolerance of benthic macrofauna to hypoxia and anoxia in shallow coastal seas: A realistic scenario. Mar. Ecol. Prog. Ser. 458, 39–52. doi:10.3354/meps09724
Saetan W., Tian C., Yu J., Lin X., He F., Huang Y., et al. (2020). Comparative transcriptome analysis of gill tissue in response to hypoxia in silver Sillago (Sillago sihama). Animals. 10 (4), E628–E958. doi:10.3390/ani10040628
Sauer H., Wartenberg M., Hescheler J. (2001). Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell. Physiol. biochem. 11 (4), 173–186. doi:10.1159/000047804
Semenza G. L., Roth P. H., Fang H.-M., Wang G. L. (1994). Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269 (38), 23757–23763. doi:10.1016/s0021-9258(17)31580-6
Shan B., Gerez J., Haedo M., Fuertes M., Theodoropoulou M., Buchfelder M., et al. (2012). RSUME is implicated in HIF-1-induced VEGF-A production in pituitary tumour cells. Endocr. Relat. Cancer 19 (1), 13–27. doi:10.1530/ERC-11-0211
Sheng L., Mao X., Yu Q., Yu D. (2017). Effect of the PI3K/AKT signaling pathway on hypoxia-induced proliferation and differentiation of bone marrow-derived mesenchymal stem cells. Exp. Ther. Med. 13 (1), 55–62. doi:10.3892/etm.2016.3917
Shim H., Dolde C., Lewis B. C., Wu C.-S., Dang G., Jungmann R. A., et al. (1997). c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U. S. A. 94 (13), 6658–6663. doi:10.1073/pnas.94.13.6658
Spirina L. V., Usynin Y. A., Yurmazov Z. A., Slonimskaya E. M., Kolegova E. S., Kondakova I. V. (2017). Transcription factors NF-kB, HIF-1, HIF-2, growth factor VEGF, VEGFR2 and carboanhydrase IX mRNA and protein level in the development of kidney cancer metastasis. Mol. Biol. 51 (2), 372–377. doi:10.7868/S0026898417020197
Sula E., Aliko V., Barceló D., Faggio C. (2020). Combined effects of moderate hypoxia, pesticides and PCBs upon crucian carp fish, Carassius carassius, from a freshwater lake- in situ ecophysiological approach. Aquat. Toxicol. 228, 105644–111027. doi:10.1016/j.aquatox.2020.105644
Terova G., Rimoldi S., Corà S., Bernardini G., Gornati R., Saroglia M. (2008). Acute and chronic hypoxia affects HIF-1α mRNA levels in sea bass (Dicentrarchus labrax). Aquaculture 279 (1-4), 150–159. doi:10.1016/j.aquaculture.2008.03.041
Timmerman C. M., Chapman L. J. (2004). Behavioral and physiological compensation for chronic hypoxia in the sailfin molly (Poecilia latipinna). Physiol. Biochem. Zool. 77 (4), 601–610. doi:10.1086/421754
Vajner L., Vytášek R., Lachmanová V., Uhlík J., Konrádová V., Novotná J., et al. (2006). Acute and chronic hypoxia as well as 7‐day recovery from chronic hypoxia affects the distribution of pulmonary mast cells and their MMP‐13 expression in rats. Int. J. Exp. Pathol. 87 (5), 383–391. doi:10.1111/j.1365-2613.2006.00493.x
Villnäs A., Norkko J., Lukkari K., Hewitt J., Norkko A. (2012). Consequences of increasing hypoxic disturbance on benthic communities and ecosystem functioning. PLOS ONE 7 (10), 44920–44932. doi:10.1371/journal.pone.0044920
Wang H., Huang C., Chen N., Zhu K., Chen B., Wang W., et al. (2015). Molecular characterization and mRNA expression of HIF-prolyl hydroxylase-2 (phd2) in hypoxia-sensing pathways from Megalobrama amblycephala. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 186, 28–35. doi:10.1016/j.cbpb.2015.04.001
Wang T., Cheng Y., Liu Z., Yan S., Long X. (2013). Effects of light intensity on growth, immune response, plasma cortisol and fatty acid composition of juvenile Epinephelus coioides reared in artificial seawater. Aquaculture 414, 135–139. doi:10.1016/j.aquaculture.2013.08.004
Wang Y., Liu X., Xie B., Yuan H., Zhang Y., Zhu J. (2020). The NOTCH1-dependent HIF1α/VGLL4/IRF2BP2 oxygen sensing pathway triggers erythropoiesis terminal differentiation. Redox Biol. 28, 101313. doi:10.1016/j.redox.2019.101313
Wilson R., Hepgul N., Higginson I. J., Gao W. (2020). End-of-life care and place of death in adults with serious mental illness: A systematic review and narrative synthesis. Palliat. Med. 40 (2), 49–68. doi:10.1177/0269216319867847
Wingfield J. C. (2013). Ecological processes and the ecology of stress: The impacts of abiotic environmental factors. Funct. Ecol. 27 (1), 37–44. doi:10.1111/1365-2435.12039
Wu J., Mao X., Cai T., Luo J., Wei L. (2006). KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34 (2), W720–W724. doi:10.1093/nar/gkl167
Wu R. S., Zhou B. S., Randall D. J., Woo N. Y., Lam P. K. (2003). Aquatic hypoxia is an disrupter and impairs fish reproduction. Environ. Sci. Technol. 37 (6), 1137–1141. doi:10.1021/es0258327
Xie Y., Shi X., Sheng K., Han G., Li W., Zhao Q., et al. (2019). PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 19 (2), 783–791. doi:10.3892/mmr.2018.9713
Yan L., Wang P., Zhao C., Fan S., Lin H., Guo Y., et al. (2021). Toxic responses of liver in Lateolabrax maculatus during hypoxia and re-oxygenation. Aquat. Toxicol. 236, 105841–105851. doi:10.1016/j.aquatox.2021.105841
Ye J., Zhang Y., Cui H., Liu J., Wu Y., Cheng Y., et al. (2018). Wego 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46 (W1), W71–W75. doi:10.1093/nar/gky400
Yu H., You X., Li J., Zhang X., Zhang S., Jiang S., et al. (2018). A genome-wide association study on growth traits in orange-spotted grouper (Epinephelus coioides) with RAD-seq genotyping. Sci. China. Life Sci. 61 (8), 934–946. doi:10.1007/s11427-017-9161-4
Zheng C., Zhao Q., Li E., Zhao D., Sun S. (2022). Role of hypoxia in the behaviour, physiology, immunity and response mechanisms of crustaceans: A review. Rev. Aquac. 14 (2), 676–687. doi:10.1111/raq.12618
Zhong D., Zhang M., Lan X., Li S., Shu H. (2021). Molecular cloning and functional characterization of growth hormone–releasing hormone in Mastacembelus armatus. Fish. Physiol. Biochem. 47 (1), 69–78. doi:10.1007/s10695-020-00895-5
Keywords: Epinephelus coioides, hypoxia stress, HIF-1 signaling pathway, transcriptome, qRT-PCR, HIF-1α
Citation: Lai X, Zhong Z, Lin B, Wu Y, Ma Y, Zhang C, Yang Y, Zhang M, Qin W, Fu X and Shu H (2022) RNA-seq and qRT-PCR analyses reveal the physiological response to acute hypoxia and reoxygenation in Epinephelus coioides. Front. Physiol. 13:1049776. doi: 10.3389/fphys.2022.1049776
Received: 21 September 2022; Accepted: 21 October 2022;
Published: 03 November 2022.
Edited by:
Hongyu Ma, Shantou University, ChinaReviewed by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaCopyright © 2022 Lai, Zhong, Lin, Wu, Ma, Zhang, Yang, Zhang, Qin, Fu and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqin Fu, cWlueGlhb3FpbnhpYW9AMTYzLmNvbQ==; Hu Shu, c2h1aHUwMDFAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.