- 1Key Laboratory of Green Prevention and Control on Fruits and Vegetables in South China Ministry of Agriculture and Rural Affairs, Guangdong Provincial Key Laboratory of High Technology for Plant Protection, Institute of Plant Protection, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2College of Plant Protection, South China Agricultural University, Guangzhou, China
RT-qPCR remains a vital approach for molecular biology studies aimed at quantifying gene expression in a range of physiological or pathological settings. However, the use of appropriate reference genes is essential to attain meaningful RT-qPCR results. Anastatus japonicus Ashmead (Hymenoptera: Helicopteridae) is an important egg parasitoid wasp and natural enemy of fruit bugs and forest caterpillars. While recent transcriptomic studies have analyzed gene expression profiles in A. japonicus specimens, offering a robust foundation for functional research focused on this parasitoid, no validated A. japonicus reference genes have yet been established, hampering further research efforts. Accordingly, this study sought to address this issue by screening for the most stable internal reference genes in A. japonicus samples to permit reliable RT-qPCR analyses. The utility of eight candidate reference genes (ACTIN, TATA, GAPDH, TUB, RPL13, RPS6, EF1α, RPS3a) was assessed under four different conditions by comparing developmental stages (larvae, pupae, adults), tissues (abdomen, chest, head), sex (male or female adults), or diapause states (diapause induction for 25, 35, 45, or 55 days, or diapause termination). RefFinder was used to calculate gene stability based on the integration of four algorithms (BestKeeper, Normfinder, geNorm, and ΔCt method) to determine the optimal RT-qPCR reference gene. Based on this approach, RPS6 and RPL13 were found to be the most reliable reference genes when assessing different stages of development, while ACTIN and EF1α were optimal when comparing adults of different sexes, RPL13 and EF1α were optimal when analyzing different tissues, and TATA and ACTIN were optimal for different diapause states. These results provide a valuable foundation for future RT-qPCR analyses of A. japonicus gene expression and function under a range of experimental conditions.
Introduction
Anastatus japonicus Ashmead (Hymenoptera: Helicopteridae) is an important egg parasitoid wasp that preys on a range of fruit bugs and forest caterpillars including Halyomorpha halys Stal, Tessaratoma papillosa Drury, and Riptortus pedestris Fabricius (Zhao et al., 2019a; Chen et al., 2019; Mi et al., 2020; Chen et al., 2021; Zhao et al., 2021; Wei et al., 2022). In commercial settings, A. japonicus is used as a form of biological control agent that is reared within Antheraea pernyi Guerin-Meneville eggs (Lepidoptera: Saturniidae) (Zhao et al., 2021). Moderate springtime temperatures induce diapause in A. japonicus, resulting in delayed adult emergence and interfering with the optimal timing for pest control, thus limiting the biocontrol value of this wasp species (Zhao et al., 2019b; Zhao et al., 2021). Relative to A. japonicus individuals not in diapause, those in diapause exhibit higher survival rates following storage at 10°C for 180 days (Zhao et al., 2021). Our group recently conducted transcriptomic sequencing for non-diapause and diapause A. japonicus individuals (unpublished data), revealing the presence of many genes associated with energy metabolism, development, and oxidation-reduction reactions. Further studies of these genes, however, will be vital to clarify the molecular basis for diapause in A. japonicus. No published studies to date have sought to analyze A. japonicus gene expression profiles, highlighting the need for additional foundational research to support further study of this potentially economically valuable biocontrol species.
Quantitative real-time PCR (RT-qPCR) analyses offer a rapid, sensitive, efficient, accurate, and reproducible means of measuring gene expression under a range of analytical conditions while also permitting single nucleotide polymorphism and restriction fragment length polymorphism analyses (Wang et al., 2019). However, the reliability of RT-qPCR analyses is strongly dependent on factors including the quality and yield of utilized templates, the amplification efficiency of the selected primers, and underlying biological signal (Jiang et al., 2015; Shu et al., 2018; Hu et al., 2019). In the process of RT-qPCR, in order to ensure the accuracy of quantitative results and achieve normalization and correction of different samples, the introduction of internal reference genes is indispensable (Lu et al., 2013; Taylor et al., 2016). Housekeeping genes often used for this purpose across species and treatment conditions include 18S rRNA and the genes encoding tubulin, ACTIN, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). However, a growing number of studies have revealed that these commonly utilized housekeeping genes do not exhibit stable expression patterns across all experimental conditions, potentially leading to skewed, inaccurate data. It is thus vital that the most appropriate reference genes for particular RT-qPCR applications be selected in a systematic manner to maximize the reliability and accuracy of associated experimental results.
Here, eight commonly utilized reference genes were evaluated to establish their value when normalizing RT-qPCR data for A. japonicus specimens. These genes included ribosomal protein S3a (RPS3a), GAPDH, ACTIN, elongation factor 1α (EF1α), TATA-box binding protein (TATA), ribosomal protein S6 (RPS6), β -tubulin (TUB), and ribosomal protein L13 (RPL13). All of which have been widely used as reference genes in different organisms because they are considered to have a uniform expression.The EF1α and TUB were thought to be good reference genes for most animals (Zhao et al., 2019a; Wang et al., 2021). However, the absence of poly-A in rRNA makes it impossible to isolate it using oligo (dT)-based techniques, which is a significant disadvantage when employing rRNA as a normalizer in RT-qPCR experiments (Xue et al., 2010). Thus, we focused on genes transcribed by RNA polymerase II whose transcripts can be isolated using oligo (dT), like the genes linked to GAPDH, ACTIN and ribosomal proteins (e.g., RPS3a, RPS6, and RPL13) (Galetto et al., 2013; Xu et al., 2021; Li et al., 2021; Shen et al., 2022). And the TATA gene is required for initiating the RNA polymerase (I, II, and III)-mediated transcription of genes with promoters with or without a TATA box (Pugh, 2000). So, Those eight reference genes were chosen. Expression dynamics for these candidate reference genes were compared with RefFinder, which integrates results from four different algorithms (BestKeeper, NormFinder, geNorm, and the ΔCt method). The stability of these genes was compared across three biotic conditions (sex, tissue type, or developmental stage) and one abiotic condition (diapause state). The goal of this study was to define the A. japonicus reference genes that are most appropriate for use across a range of experimental conditions. These data may offer value as a foundation for future more reliably RT-qPCR-based studies of A. japonicus.
Materials and methods
Insect rearing
A. japonicus Ashmead specimens were obtained from the Institute of Plant Protection of the Chinese Academy of Agricultural Sciences, Ministry of Agriculture and Rural Affairs-CABI Biosafety Joint Laboratory. The initial A. japonicus colony was established from brown marmorated stink bug (Halyomorpha halys Stal) eggs that had been parasitized under field conditions and were collected from the suburbs of Beijing. A. japonicus colonies were maintained for 1 year under laboratory conditions (24°C, 16 h light/8 h dark cycle) using Antheraea pernyi eggs. After newly emerging, a solution of 10% honey and water was used to feed A. japonicus individuals in a 32 cm × 25 cm × 9 cm plastic box under these same laboratory conditions.
Sample preparation
Biotic factors
All A. japonicus developmental stages were analyzed for this study (including mature larvae (n = 8), pupae (n = 10), and male (n = 20) and female (n = 10) adult individuals sampled on day 1 of the corresponding stage. In addition, samples from the head, chest, and abdomen of ∼40 adult female individuals were collected for analysis.
Abiotic factors
To induce diapause, Antheraea pernyi eggs were parasitized by A. japonicus for 2 days under standard conditions (24°C ± 0.5°C, 70% relative humidity, 16L:8D photoperiod), after which they were transferred to diapause-inducing conditions (17°C, 10L:14D photoperiod) for 25, 35, 45, or 55 days. Diapause was terminated after 55 days by transferring A. japonicus individuals back to normal developmental conditions (24°C, 16L:8D) for 12 days.
Each treatment was repeated in triplicate (n = 15 individuals/treatment). Samples were stored in 1.5 ml RNA-free tubes, snap-frozen with liquid nitrogen, and stored at −80°C.
Total RNA extraction and cDNA synthesis
The MiniBEST Universal RNA Extraction Kit (TaKaRa, China) was used to isolate RNA based on provided directions. RNA concentration levels were measured with a NanoDrop ND-2000C instrument (Thermo Fisher Scientific, MA, United States). Total RNA was prepared in a 50–100 µl volume of ddH2O at the following concentrations (mean ± SEM): 651.8 ± 35.6 ng/μl for mature larvae, 1,013.7 ± 180.3 ng/μl for pupae, 317.9 ± 162.8 ng/μl for female adults, 275.9 ± 119.8 ng/μl for male adults, 335.3 ± 50.7 ng/μl for head samples, 429.8 ± 86.5 ng/μl for chest samples, 596.6 ± 233.2 ng/μl for abdomen samples, 1,180.9 ± 74.7, 328.1 ± 184.9, 278.1 ± 80.4 and 266.9 ± 83.5 ng/μl for 25, 35, 45, and 55 days under diapause-inducing conditions, and 258.9 ± 30.7 ng/μl for diapause termination.
All samples exhibited OD260/280 values from 1.9 to 2.1. First-strand cDNA synthesis was performed using a PrimeScript RT kit containing gDNA Eraser (Perfect Real Time, TaKaRa, China). All cDNA samples were diluted 10-fold for RT-qPCR analyses.
Primer design
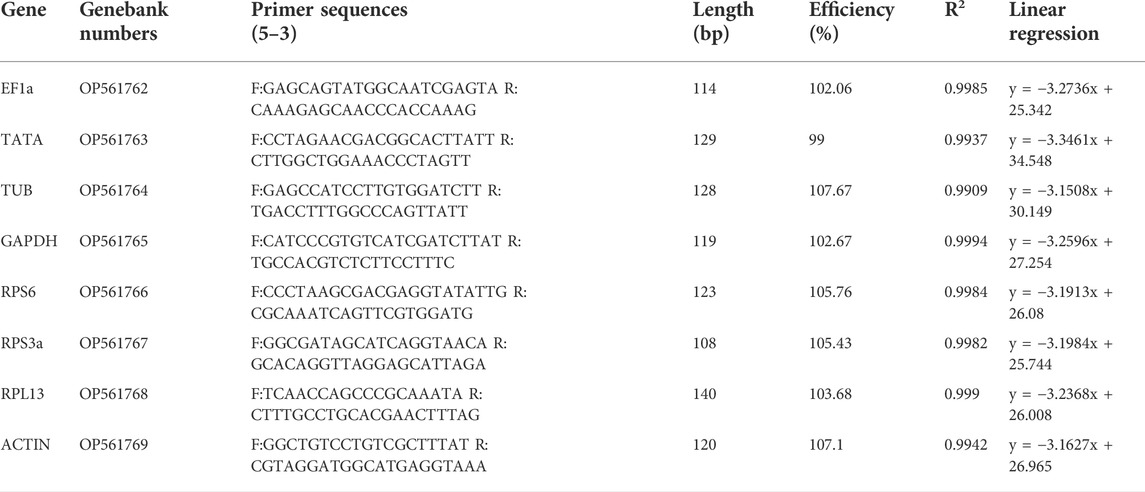
For this study, RT-qPCR was used to examine the expression of eight commonly utilized reference gene candidates (Table 1), using primers designed with the PrimerQuest Tool (https://sg.idtdna.com/PrimerQuest/Home/Index) based on sequences derived from our recent transcriptomic analysis of A. japonicus (unpublished data).
Quantitative real-time PCR analysis
TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara) and a CFX Connect Real-Time Instrument (Bio-Rad, CA, United States) were used for all RT-qPCR analyses, which were conducted in a total volume of 50 µl containing 25 µl of TB green, 2.5 µl of each primer (F + R; 100 µM), 2.5 µl of diluted cDNA, and 17.5 µl of RNase-free ddH2O. Amplification settings were as follows: 95°C for 30 s; 39 cycles of 95°C for 5 s and 55°C for 30 s; Melting curves for the product were determined by raising the temperature from 55°C to 90°C in sequential steps of 0.5°C for 1 s. For each primer, standard curves were produced using a two-fold dilution series of cDNA as the template according to the linear regression model. All analyses were conducted using three biological and three technical replicates. RT-qPCR amplification efficiency (E) were measured as follows: E = [10 (−1/slope)−1] × 100 (Pfaffl, 2001).
Data analysis
The number of cycle (Ct) values required for amplification levels to reach a fixed exponential phase threshold (set to 500 for all genes) during PCR analyses was used to compute gene expression. RefFinder (http://blooge.cn/RefFinder/), which is a tool that integrates results from the NormFinder (Andersen et al., 2004), geNorm (Vandesompele et al., 2002), ΔCt method (Silver, et al., 2006) and BestKeeper (Pfaffl et al., 2004)analytical tools, was utilized to assess the stability of these different A. japonicus reference gene candidates. In addition, the geNorm tool was utilized to establish the optimal number of reference genes based on Vn/Vn+1 values, with a Vn/Vn+1 > 0.15 necessitating the addition of an additional reference gene whereas no further reference genes are required when Vn/Vn+1 < 0.15, with 0.15 thus serving as the threshold for additional reference genes gene incorporation (Vandesompele et al., 2002; Liang et al., 2014).
Results
Amplification efficiencies
All eight of the tested reference gene candidates were expressed at detectable levels in the analyzed A. japonicus samples, with 1% agarose gel electrophoresis results for each transcript exhibiting a single amplicon of the expected size. Amplification efficiency for each gene was estimated using two-point standard curves with known RNA concentrations, while melt curves were used to confirm the specificity of these amplification results based on the presence of a single peak without any evidence of primer-dimer peaks (Figure 1). Amplification efficiency (E) values for these genes ranged from 99% to 107.67%, with an R2 > 0.9909 (Table 1).
Expression profiles of candidate reference genes
Expression levels for these candidate reference genes varied widely across the tested experimental conditions with respect to the measured Ct values (Figure 2), ranging from a Ct value of 17.80 for GAPDH to 28.69 for TATA. Mean respective Ct values for TATA and TUB were 27.65 and 24.28, with these values being notably higher than those for other tested genes. Moderate expression was observed for the other candidate reference genes included in this study, with respective mean Ct values for EF1α, GAPDH, RPS6, RPS3a, RPL13, and ACTIN of 19.53, 20.07, 20.40, 19.88, 19.90, and 20.56.

FIGURE 2. Expression profiles of the eight candidate reference genes in all four experiments for A. japonicus. The expression levels of the reference genes are shown in terms of the Cq-value for each experimental condition. (A) Development stage, (B) Tissue, (C) Sex, (D) Diapause treatment.
Analyses of the stability of A. japonicus reference genes under different experimental conditions and optimum number of genes for normalization
Developmental stage analysis
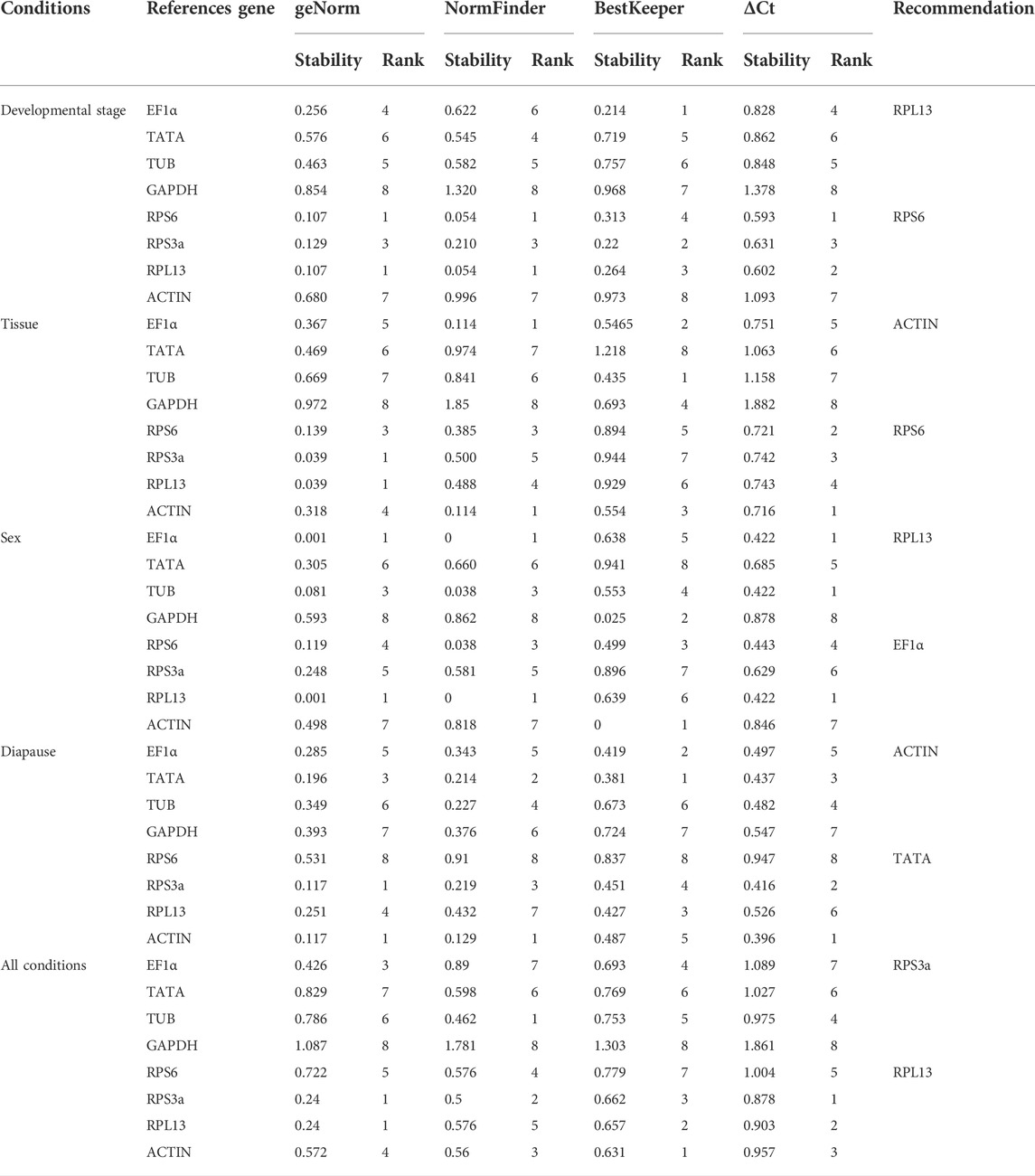
When analyzing A. japonicus samples collected during different stages of development, the geNorm, NormFinder, and ΔCt analytical approaches revealed RPS6 and RPL13 as the most stable reference genes (Table 2). In contrast, BestKeeper analyses suggested that Ef1α and RPS3a were instead the most stable, with ACTIN and GAPDH as the least stable genes. Per RefFinder, the overall rank order for the stability of these reference genes was: RPL13, RPS6, RPS3a, EF1α, TATA, TUB, ACTIN, GAPDH (Figure 3A). In geNorm analyses, the pairwise V2/3 value was below the established 0.15 cut-off threshold (Figure 4). Overall, RefFinder thus suggested that RPL13 and RPS6 are the most appropriate target genes when analyzing A. japonicus samples across different stages of development (Table 2).
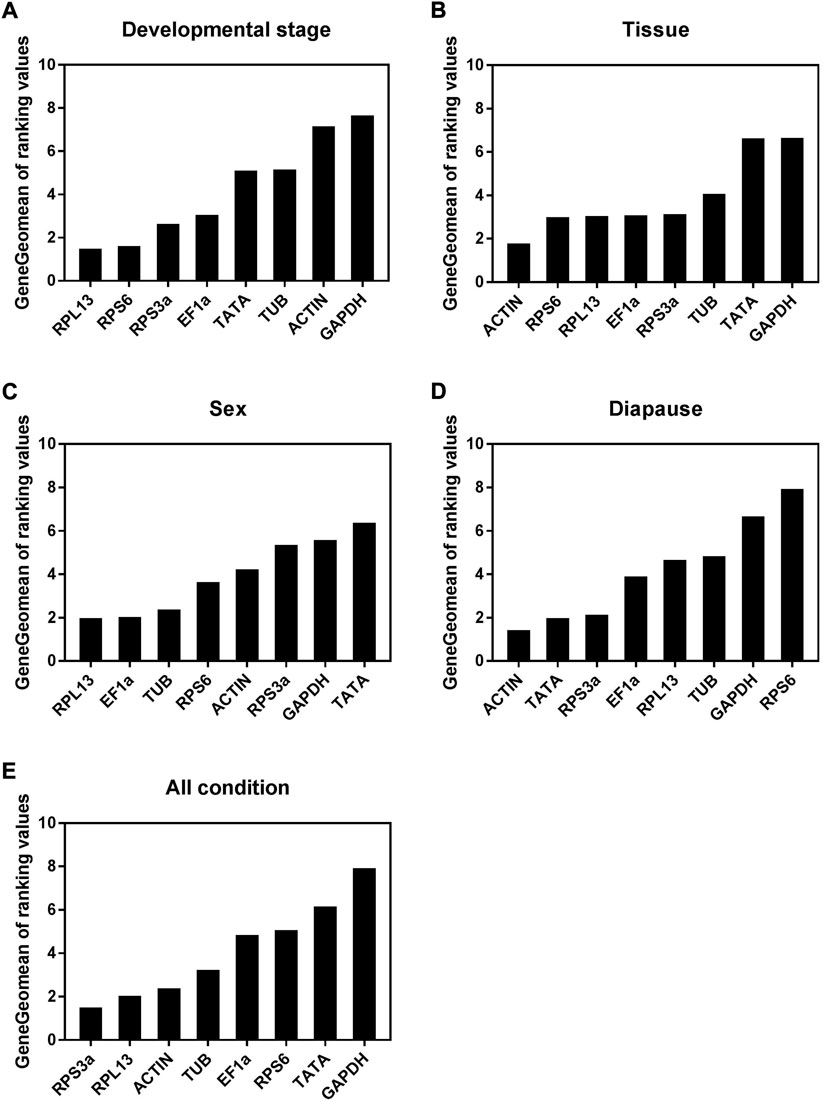
FIGURE 3. Stability of the eight candidate reference gene expressions in A. japonicus under different treatment conditions analyzed using RefFinder. (A) Development stage, (B) Tissue, (C) Sex, (D) Diapause treatment, (E)All condition.
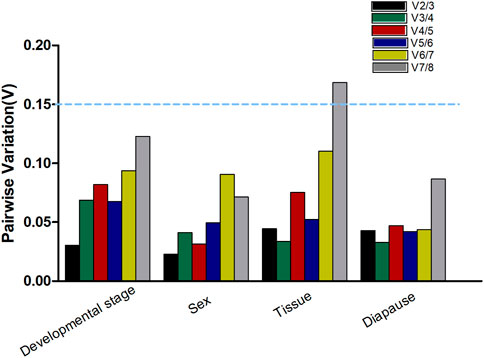
FIGURE 4. Optimum number of reference genes required for accurate normalization of gene expression. Pairwise variation (V) values in the four groups using geNorm.
Tissue-based analyses
Different reference genes were found to be the most stable when performing tissue-based analyses of A. japonicus samples using these different software programs (Table 2). Specifically, RPS3a and RPL13 were identified as the most stable reference genes in these different tissue types according to geNorm, while EF1α and ACTIN were the most stable according to NormFinder, EF1α and TUB were the most stable according to BestKeeper, and the ΔCt identified ACTIN and RPS6 as the most stable reference gene candidates. GAPDH was identified as being the least stable reference gene in the geNorm, NormFinder, and BestKeeper analyses. Per RefFinder, the overall rank order for the stability of these reference genes was: ACTIN, RPS6, RPL13, EF1α, RPS3a, TUB, TATA, GAPDH (Figure 3B). The pairwise V2/V3 value identified based on geNorm data was below the established 0.15 cut-off threshold, although the V7/8 value was above this threshold (Figure 4). Overall, the RefFinder analyses indicated that ACTIN and RPS6 are the most appropriate reference genes for use when normalizing A. japonicus gene expression across different tissue types (Table 2).
Sex-based analyses
When comparing gene expression results for male and female A. japonicus specimens, the geNorm, NormFinder, and ΔCt methods identified EF1α and RPL13 as the most stable candidate reference genes (Table 2). While BestKeeper found ACTIN and GAPDH to be the most stable genes, the other three analytical tools suggested that GAPDH was instead the least stable reference gene. Per RefFinder, the overall rank order for the stability of these reference genes was: RPL13, EF1α, TUB, RPS6, ACTIN, RPS3a, GAPDH, TATA (Figure 3C). In the geNorm analysis, the V2/V3 pairwise value was below the established 0.15 cut-off threshold (Figure 4). Overall, these RefFinder results suggest that RPL13 and EF1α are the most appropriate reference genes for use when normalizing gene expression levels in A. japonicus specimens of different sexes (Table 2).
Diapause treatment analyses
When evaluating the different tested diapause treatments, ACTIN and TATA were identified by geNorm analyses as the most stable reference genes, whereas RPS6 was the least stable. The ΔCt and NormFinder analyses also yielded similar results (Table 2). BestKeeper, in contrast, identified RPL13 and EF1α as being the most stable. Per RefFinder, the overall rank order for the stability of these reference genes was: TATA, ACTIN, RPL13, RPS3a, EF1α, TUB, GAPDH, and RPS6 (Figure 3D). All pairwise variation values were under the 0.15 cut-off threshold in geNorm analyses (Figure 4). According to the RefFinder results, ACTIN and TATA were thus the most reliable reference genes when normalizing target gene expression over a range of diapause treatment conditions.
All conditions
When evaluating all treatment conditions, the ΔCt and geNorm analyses revealed that RPL13 and RPS3a were the most stable reference genes (Table 2). From most to least stable, the RefFinder analysis ranked the stability of these candidate A. japonicus reference genes as follows: RPS3a, RPL13, ACTIN, TUB, EF1α, RPS6, TATA, GADPH (Figure 3E). In geNorm analyses, all pairwise variation values were under the selected cut-off value of 0.15 (Figure 4). Overall, this RefFinder analysis indicated that RPS3a and RPL13 are the most appropriate genes for internal reference use when normalizing A. japonicus gene expression in RT-qPCR analyses (Table 2).
Reference gene validation
FD (Farnesol dehydrogenase) is an important enzyme in the diapause process of insects and can oxidize farnesol to farnesol (the precursor of JH) (Mayoral et al., 2009). Relative FD expression levels were next used for the validation of these reference genes among the tested diapause treatment conditions. In general, expression patterns were similar across conditions, although these expression levels were 54- and 85-fold higher on diapause day 25 relative to non-diapause treatment when respectively normalized to the most stable and least stable reference genes (Figure 5). When using ACTIN and TATA, which were the most stable reference genes, significant increases in FD expression were observed in D55 relative to D25 samples, whereas no significant differences were evident between these two time points when using unstable reference genes such as GAPDH and RPS6.
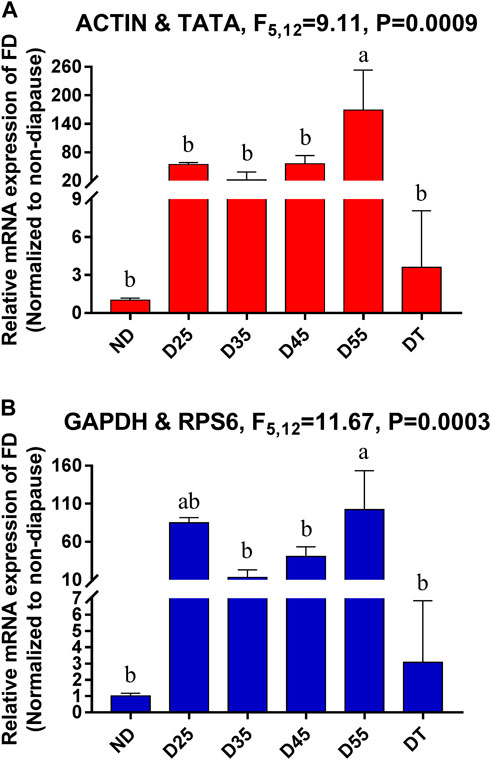
FIGURE 5. Relative gene expression of FD in different tissues of A. japonicus. The relative abundance of FD in the ND, D25, D35, D45, D55 and DT were normalized to the best stable (A, ACTIN and TATA) and least stable (B, GAPDH and RPS6) reference genes, respectively. ND, non-diapause mature larvae; D25, D35, D45, D55, diapausing mature larvae which incubated in diapause-inducing conditions for 25, 35, 45, 55 days; DT, diapause termination mature larvae. The values are means + SE. Different letters indicate significant differences in gene expression among different tissues of A. japonicus (p < 0.05).
Discussion
Appropriate reference gene selection is dependent on species- and development stage-specific variability. For example, EF1α and Arm have been identified as reliable targets for normalizing RT-qPCR data derived from Australian plague locusts under varying rearing density conditions (Chapuis et al., 2011), while GAPDH and UCCR have served as Spodoptera litura reference genes (Lu et al., 2013). Indeed, no one reference gene or set of reference genes is universally appropriate across insect species. Zhang et al. (2015) demonstrated that while RPS15 and RPL13 were the most stable reference genes when comparing a range of Helicoverpa armigera larval tissue types, RPL27 and EF were more appropriate when evaluating adult samples. Five studies have explored reference gene selection in Bemisia tabaci, yet these studies have yielded conflicting results (Li et al., 2013; Su et al., 2013; Collins et al., 2014; Liang et al., 2014; Dai et al., 2017).
To date, reliable reference genes have been identified for many Hymenoptera species (Table 3), including Cotesia chilonis (Li et al., 2019), Apis mellifera (Lourenço et al., 2008), Lysiphlebia japonica (Gao et al., 2017a), Aphidius gifuensis (Gao et al., 2017b), Tamarixia radiate (Guo et al., 2020), and Solenopsis invicta (Cheng et al., 2013). A. japonicus is an important natural predator of a range of agricultural pests, and further molecular analyses of this species may thus support further biocontrol efforts. However, no prior studies have selected or validated reference genes in this parasitic wasp species. As such, eight commonly utilized reference genes were herein analyzed for their stability under four different sets of experimental conditions using a panel of software programs.
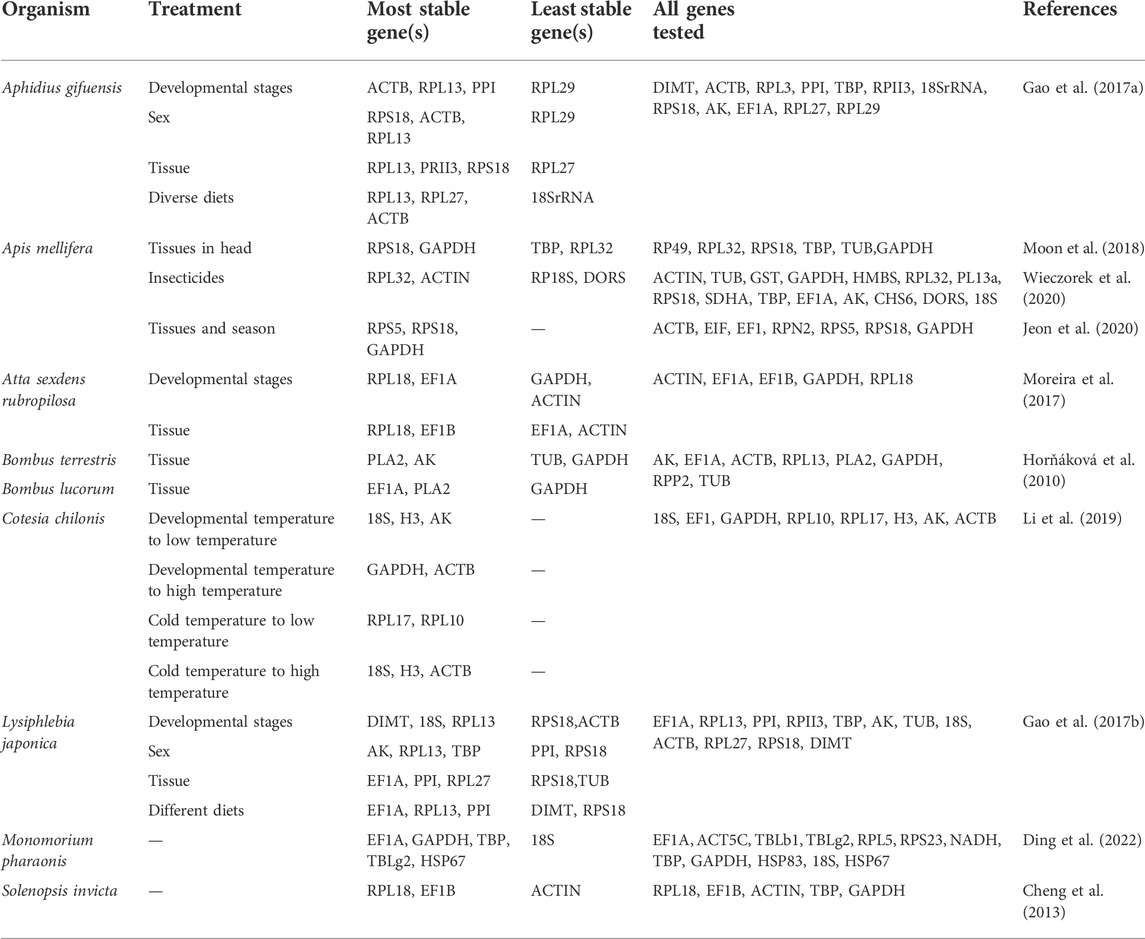
TABLE 3. Summary of the stability rankings of reference genes from studies conducted in Hymenoptera.
Ribosomal proteins are highly conserved across different species (Petibon et al., 2021), and many ribosomal genes have successfully been leveraged as stable reference genes when conducting transcriptomic analyses (Liu et al., 2016; Gao et al., 2017a). Consistently, RPS3a and RPL13 were herein identified as the most stable A. japonicus reference genes across all conditions. In a prior study of Agasicles hygrophilao, these two genes were similarly found to exhibit stable expression in most tested experimental samples (Wang et al., 2019).In the present study, RPS6 was the second most stable reference gene across different tissue types and developmental stages in A. japonicus, while RPL13 was the most effective reference target when analyzing samples from different developmental stages or adults of different sexes. In Rhopalosiphum padi, RPS18 and RPL13 have been reported as the most stable reference genes across tissue types, while RPS6 was the second most stable such reference gene in response to temperature or antibiotic treatment conditions (Li et al., 2021).
ACTIN is an integral component of the cytoskeleton, controlling the structural integrity of cells such that it can be readily leveraged as a reference gene (Ponton et al., 2011). Indeed, ACTIN was found to be the most stable tested reference gene in A. japonicus under different tested diapause treatment conditions and in various tissues, in line with prior evidence regarding insecticide stress conditions in Locusta migratoria (Yang et al., 2014) and Spodoptera litura (Lu et al., 2013). Similarly, ACTIN showed high stability in other insects under different experimental sets, for the Orchesella cincta, ACTIN was the dominant most stable gene under temperature, desiccation and starvation treatments (de Boer et al., 2009), while high levels of ACTIN stability were evident in Bradysia odoriphaga specimens exposed to insecticides including chlorfluazuron or chlorpyrifos, in addition to remaining stable across developmental stages and tissue types (Fu et al., 2022). However, these results are not universal, with ACTIN having been established as the least stable gene in Henosepilachna vigintioctopunctata across different tissues, host plants, and stages of development (Lü et al., 2018). These findings emphasize the importance of individually evaluating reference gene stability under experimental conditions of interest.
Here reference gene stability was tested with the geNorm, NormFinder, BestKeeper, and ΔCt programs, and these different software applications each yielded differing results with respect to target gene stability. Under the developmental stages, the analysis results of NormFinder, geNorm and ΔCt are relatively similar, and the analysis results of BestKeeper are quite different from the other three analysis results. For example, RPS6/RPL13 ranks as the most stable reference gene in NormFinder, geNorm and ΔCt, but ranks 4th and 3rd in BestKeeper, respectively. Inconsistent results were obtained for these four software programs across tissue types, with RPS3a/RPL13, EF1α/Actin, TUB, and ACTIN ultimately being the most stable reference genes. When comparing specimens of different sexes, NormFinder and geNorm yielded similar results with EF1α and RPL13 as the two top-ranked genes, whereas TATA, Actin, and GAPDH were the bottom-ranked genes. In contrast, BestKeeper identified Actin and GAPDH as the two top-ranked genes. Under the tested diapause conditions, TATA and ACTIN were the top-ranked reference genes according to geNorm, NormFinder, and ΔCt, with Actin and TATA respectively being ranked first in the NormFinder and were TATA and ΔCt analyses. However, BestKeeper ranked RPL13 and EF1α as the two top genes. These inconsistent results across programs are attributable to the differences in the calculation strategies that they employ, with NormFinder and geNorm yielding more stable results for each reference gene. geNorm calculates M values for each reference gene to determine the optimal reference gene, with NormFinder using a similar approach while also analyzing each independent internal reference gene (Fu et al., 2013). BestKeeper produces correlation coefficients, standard deviations, and coefficients of variation for each gene pairing, enabling the simultaneous analysis of up to 10 genes (Chen and Lu, 2014). The inconsistent ranked lists produced by these programs can make the selection of optimal reference genes challenging. As such, RefFinder was used in this study owing to its ability to integrate these algorithms and to thereby better clarify overall candidate gene stability.
These results revealed marked differences in the optimal internal reference genes for use when comparing different sample types. This aligns well with growing interest in recent years to utilize more than one reference gene when normalizing gene expression data (Guo et al., 2021). The geNorm software offers the ability to establish the optimal number of reference genes for use through the calculation of pairwise variation (Vn/n + 1) based on normalization factors NFn and NFn + 1, where n is greater than or equal to 2. Using this algorithm, n is defined as the optimal number of reference genes when Vn/n + 1 is under 0.15. Accordingly, the optimal number of reference gene combinations identified herein when comparing A. japonicus samples corresponding to different development stages, tissue types, sexes, and diapause treatments were RPL13 and RPS6, ACTIN and RPS6, RPL13 and EF1α, and ACTIN and TATA, respectively.
Conclusion
In conclusion, these results underscore the importance of selecting application-specific reference genes when conducting RT-qPCR analyses of A. japonicus, as no one reference gene is optimal under all conditions. In this study, the most stable sets of reference genes were compared, leading to the identification of a combination of RPS3a and RPL13 as the most appropriate reference genes on average across the tested experimental conditions. However, the ideal reference genes ultimately varied among experiments, with RPL13 and RPS6 being the most reliable for developmental stage-based analyses, ACTIN and RPS6 as the most reliable when comparing different tissue types, ACTIN and TATA as the most stable when comparing different diapause treatments, and RPL13 and EF1α as the most stable when analyzing female and male A. japonicus specimens (de Boer et al., 2009; Chen and Lu, 2014; Lü et al., 2018; Wang et al., 2020).
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
CZ and ZL conceived and designed research. ZL and JX conducted the experiments. ZL analyzed the data and wrote the manuscript. YX and QW contributed material. CZ and DL revised the manuscript. All authors have read and approved the manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (32001968); the Science and Technology Program of Guangzhou, China (202102020211); the earmarked fund for CARS-32-10; and the Science and Technology Project of Maoming City (2021S0063).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 (15), 5245–5250. doi:10.1158/0008-5472.can-04-0496
Chapuis M. P., Tohidi-Esfahani D., Dodgson T., Blondin L., Ponton F., Cullen D., et al. (2011). Assessment and validation of a suite of reverse transcription-quantitative PCR reference genes for analyses of density-dependent behavioural plasticity in the Australian plague locust. BMC Mol. Biol. 12, 7. doi:10.1186/1471-2199-12-7
Chen F., Lu Y. (2014). Selection of reference genes in Phenacoccus solenopsis (Hemiptera: Pseudococcidae) under heat stress. Acta Entomol. Sin. 57 (10), 1146–1154. doi:10.16380/j.kcxb.2014.10.008
Chen Y. M., Gibson G. A. P., Peng L. F., Iqbal A., Zang L. S. (2019). Anastatus motschulsky (Hymenoptera, eupelmidae): Egg parasitoids of Caligula japonica moore (Lepidoptera, Saturniidae) in China. Zookeys 17 (881), 109–134. doi:10.3897/zookeys.881.34646
Chen Y. M., Qu X. R., Li T. H., Iqbal A., Wang X. G., Ren Z. Y., et al. (2021). Performances of six eupelmid egg parasitoids from China on Japanese giant silkworm Caligula japonica with different host age regimes. J. Pest Sci. (2004). 94, 309–319. doi:10.1007/s10340-020-01271-1
Cheng D., Zhang Z., He X., Liang G. (2013). Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS ONE 8 (2), e57718. doi:10.1371/journal.pone.0057718
Collins C., Patel M. V., Colvin J., Bailey D., Seal S. (2014). Identification and evaluation of suitable reference genes for gene expression studies in the whitefly Bemisia tabaci (Asia I) by reverse transcription quantitative realtime PCR. J Insect Sci 14 (63). doi:10.1093/jis/14.1.63
Dai T., Lü Z., Liu W., Wan F. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PLoS One 12, e0173821. doi:10.1371/journal.pone.0173821
de Boer M. E., de Boer T. E., Mariën J., Timmermans M. J., Nota B., van Straalen N. M., et al. (2009). Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). BMC Mol. Biol. 10, 54. doi:10.1186/1471-2199-10-54
Ding G., Gao Q., Chen J., Zhao J., Zhang G., Liu W. (2022). Validation of potential reference genes for real-time qPCR analysis in pharaoh ant, Monomorium pharaonis (Hymenoptera: Formicidae). Front. Physiol. 13, 852357. doi:10.3389/fphys.2022.852357
Fu H., Huang T., Yin C., Xu Z., Li C., Liu C., et al. (2022). Selection and Validation of reference genes for RT-qPCR normalization in Bradysia odoriphaga (Diptera: Sciaridae) under insecticides stress. Front. Physiol. 12, 818210. doi:10.3389/fphys.2021.818210
Fu W., Xie W., Zhang Z., Wang S., Wu Q., Liu Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9 (8), 792–802. doi:10.7150/ijbs.5862
Galetto L., Bosco D., Marzachì C. (2013). Selection of reference genes from two leafhopper species challenged by phytoplasma infection, for gene expression studies by RT-qPCR. BMC Res. Notes 6, 409. doi:10.1186/1756-0500-6-409
Gao X., Zhang S., Luo J., Wang C., Lü L., Zhang L., et al. (2017a). Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene 637, 211–218. doi:10.1016/j.gene.2017.09.057
Gao X., Zhang S., Luo J., Wang C., Lü L., Zhang L., et al. (2017b). Identification and validation of reference genes for gene expression analysis in Aphidius gifuensis (Hymenoptera: Aphidiidae). PLoS One 12 (11), e0188477. doi:10.1371/journal.pone.0188477
Guo C., Pan H., Zhang L., Ou D., Lu Z., Khan M. M., et al. (2020). Comprehensive assessment of candidate reference genes for gene expression studies using RT-qPCR in Tamarixia radiata, a predominant parasitoid of Diaphorina citri. Genes 11 (10), 1178. doi:10.3390/genes11101178
Guo Y., Yang Y., Chai Y., Gao L., Ma R. (2021). Identification and evaluation of reference genes for quantitative PCR normalization in Alligator Weed Flea Beetle (Coleoptera: Chrysomelidae). J. Insect Sci. 1 (5), 9. doi:10.1093/jisesa/ieab067
Horňáková D., Matoušková P., Kindl J., Valterová I., Pichová I. (2010). Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397 (1), 118–120. doi:10.1016/j.ab.2009.09.019
Hu Y., Fu H., Qiao H., Sun S., Zhang W., Jin S., et al. (2019). Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 19 (8), 2258–2273. doi:10.3390/ijms19082258
Jeon J. H., Moon K., Kim Y., Kim Y. H. (2020). Reference gene selection for qRT-PCR analysis of season- and tissue-specific gene expression profiles in the honey bee Apis mellifera. Sci. Rep. 10 (1), 13935. doi:10.1038/s41598-020-70965-4
Jiang H., Qian Z., Lu W., Ding H., Yu H., Wang H., et al. (2015). Identification and characterization of reference genes for normalizing expression data from red swamp crawfish Procambarus clarkia. Int. J. Mol. Sci. 16 (9), 21591–21605. doi:10.3390/ijms160921591
Li R., Xie W., Wang S., Wu Q., Yang N., Yang X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8, e53006. doi:10.1371/journal.pone.0053006
Li Q. Y., Li Z. L., Lu M. X., Cao S. S., Du Y. Z. (2019). Selection of valid reference genes for quantitative real-time PCR in Cotesia chilonis (Hymenoptera: Braconidae) exposed to different temperatures. PLOS ONE 14 (12), e0226139. doi:10.1371/journal.pone.0226139
Li M., Li X., Wang C., Li Q., Zhu S., Zhang Y., et al. (2021). Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 12, 663338. doi:10.3389/fphys.2021.663338
Liang P., Guo Y., Zhou X., Gao X. (2014). Expression profiling in Bemisia tabaci under insecticide treatment: Indicating the necessity for custom reference gene selection. PLoS ONE 9 (1), e87514. doi:10.1371/journal.pone.0087514
Liu G., Qiu X., Cao L., Zhang Y., Zhan Z., Han R. (2016). Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth, Thitarodes armoricanus (Lepidoptera, Hepialidae). PLoS One 11 (7), e0159060. doi:10.1371/journal.pone.0159060
Lourenço A. P., Mackert A., Cristino A. d. S., Paulino Sim˜oes Z. L. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. PLoS One 11 (5), e0155640. doi:10.1371/journal.pone.0155640
Lü J., Chen S., Guo M., Ye C., Qiu B., Wu J., et al. (2018). Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9, 1614. doi:10.3389/fphys.2018.01614
Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS ONE 8 (7), e68059. doi:10.1371/journal.pone.0068059
Mayoral J. G., Nouzova M., Navare A., Noriega F. G. (2009). NADP+-dependent farnesol dehydrogenase, a corporaallata enzyme involved in juvenile hormone synthesis. Proc. Natl. Acad. Sci. U. S. A. 106 (50), 21091–21096. doi:10.1073/pnas.0909938106
Mi Q., Zhang J., Haye T., Zhang B., Zhao C., Lei Y., et al. (2020). Fitness and interspecific competition of Trissolcus japonicus and Anastatus japonicus, egg parasitoids of Halyomorpha halys. Biol. Control 152, 104461. doi:10.1016/j.biocontrol.2020.104461
Moon K., Lee S. H., Kim Y. H. (2018). Validation of quantitative real-time PCR reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of Western honey bee, Apis mellifera. PLOS ONE 13 (7), e0200369. doi:10.1371/journal.pone.0200369
Moreira A., Adriana M., Carneiro R., Bueno O., Souza D. (2017). Validation of reference genes in leaf-cutting ant Atta sexdens rubropilosa in different developmental stages and tissues. IJEAB 2 (2), 743–755. doi:10.22161/ijeab/2.2.23
Petibon C., Malik Ghulam M., Catala M., Abou Elela S. (2021). Regulation of ribosomal protein genes: An ordered anarchy. Wiley Interdiscip. Rev. RNA 12 (3), e1632. doi:10.1002/wrna.1632
Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi:10.1093/nar/29.9.e45
Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 (6), 509–515. doi:10.1023/b:bile.0000019559.84305.47
Ponton F., Chapuis M-P., Pernice M., Sword G. A., Simpson S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57 (6), 840–850. doi:10.1016/j.jinsphys.2011.03.014
Pugh B. F. (2000). Control of gene expression through regulation of the TATA-binding protein. Gene 255, 1–14. doi:10.1016/s0378-1119(00)00288-2
Shen C, H., Peng L, J., Zhang Y, X., Zeng H, R., Yu H, F., Jin L., et al. (2022). Reference genes for expression analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects 13 (2), 140. doi:10.3390/insects13020140
Shu B., Zhang J., Cui G., Sun R., Sethuraman V., Yi X., et al. (2018). Evaluation of reference genes for real-time quantitative PCR analysis in larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 9, 372. doi:10.3389/fphys.2018.00372
Silver N., Best S., Jiang J., Thein S. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7 (1), 33. doi:10.1186/1471-2199-7-33
Su Y., He W., Wang J., Li J., Liu S., Wang X. (2013). Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 106 (3), 1446–1455. doi:10.1603/EC12459
Taylor C. M., Jost R., Erskine W., Nelson M. N. (2016). Identifying stable reference genes for qRT-PCR normalisation in gene expression studies of narrow-leafed lupin (Lupinus angustifolius L.). PLoS One 11 (2), e0148300. doi:10.1371/journal.pone.0148300
Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., de Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (7), RESEARCH0034. doi:10.1186/gb-2002-3-7-research0034
Wang Z., Meng Q., Zhu X., Sun S., Gao S., Gou Y., et al. (2019). Evaluation and validation of reference genes for quantitative real-time PCR in Helopeltis theivora waterhouse (Hemiptera: Miridae). Sci. Rep. 9 (1), 13291. doi:10.1038/s41598-019-49479-1
Wang G., Cheng H., Li M., Zhang C., Deng W., Li T. (2020). Selection and validation of reliable reference genes for Tolypocladium guangdongense gene expression analysis under differentially developmental stages and temperature stresses. Gene 734, 144380. doi:10.1016/j.gene.2020.144380
Wang W., Hu S., Cao Y., Chen R., Wang Z., Cao X. (2021). Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol. Biol. Rep. 48 (2), 1115–1126. doi:10.1007/s11033-021-06153-y
Wei X. Y., Chen Y. M., Wang X., Lv R. E., Zang L. S. (2022). Demography and fitness of Anastatus japonicus reared from Antheraea pernyi as a biological control agent of Caligula japonica. Insects 13 (4), 349. doi:10.3390/insects13040349
Wieczorek P., Frąckowiak P., Obrępalska-Stęplowska A. (2020). Evaluation of the expression stability of reference genes in Apis mellifera under pyrethroid treatment. Sci. Rep. 10 (1), 16140. doi:10.1038/s41598-020-73125-w
Xu J., Welker D, L., James R, R. (2021). Variation in expression of reference genes across life stages of a bee, Megachile rotundata. Insects 12 (1), 36. doi:10.3390/insects12010036
Xue J. L., Salem T. Z., Turney C. M ., Cheng X. W. (2010). Strategy of the use of 28S rRNA as a housekeeping gene in real-time quantitative PCR analysis of gene transcription in insect cells infected by viruses. J. Virol. Methods 163 (2), 210–215. doi:10.1016/j.jviromet.2009.09.019
Yang Q., Li Z., Cao J., Zhang S., Zhang H., Wu X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (orthoptera: Acrididae). PLoS One 9, e98164. doi:10.1371/journal.pone.0098164
Zhang S., An S., Li Z., Wu F., Yang Q., Liu Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555 (2), 393–402. doi:10.1016/j.gene.2014.11.038
Zhao C., Xia Y., Guo Y., Li M., Zhang B., Li D. (2019a). The study on diapause induction in Anastatus japonicus Ashmead. Chin. J. Biol. Control 35 (02), 282–287. doi:10.16409/j.cnki.2095-039x.2019.02.017
Zhao D., Wang X., Chen J., Huang Z., Huo H., Jiang C., et al. (2019b). Selection of reference genes for qPCR normalization in buffalobur (Solanum rostratum Dunal). Sci. Rep. 9 (1), 6948. doi:10.1038/s41598-019-43438-6
Keywords: Anastatus japonicus, RT-qPCR, reference gene, diapause, normalization
Citation: Liu Z, Xiao J, Xia Y, Wu Q, Zhao C and Li D (2022) Selection and validation of reference genes for RT-qPCR-based analyses of Anastatus japonicus Ashmead (Hymenoptera: Helicopteridae). Front. Physiol. 13:1046204. doi: 10.3389/fphys.2022.1046204
Received: 16 September 2022; Accepted: 05 October 2022;
Published: 19 October 2022.
Edited by:
Lian-Sheng Zang, Guizhou University, ChinaCopyright © 2022 Liu, Xiao, Xia, Wu, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Can Zhao, emhhb2NhbkBnZGFhcy5jbg== Dunsong Li, ZHNsaUBnZHBwcmkuY24=
†This author share first authorship
 Zixin Liu
Zixin Liu Junjiang Xiao1,2
Junjiang Xiao1,2