- 1College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, Guangdong, China
- 2Department of Animal Genetics Breeding and Reproduction, College of Animal Science, South China Agricultural University, Guangzhou, China
The birth weight of chickens does not significantly affect the weight at slaughter, while the different growth rate after birth was one of the important reasons for the difference in slaughter weight. Also, the increase in chickens’ postnatal skeletal muscle weight is the main cause of the slaughter weight gain, but which genes are involved in this biological process is still unclear. In this study, by integrating four transcriptome datasets containing chicken muscles at different developmental times or different chicken tissues in public databases, a total of nine candidate genes that may be related to postnatal muscle development in chickens were obtained, including RPL3L, FBP2, ASB4, ASB15, CKMT2, PGAM1, YIPF7, PFKM, and LDHA. One of these candidate genes is RPL3L, whose 42 bp insertion/deletion (indel) mutation significantly correlated with multiple carcass traits in the F2 resource population from Xinghua chickens crossing with White Recessive Rock (WRR) chickens, including live weight, carcass weight, half eviscerated weight, eviscerated weight, breast meat weight, wing weight, leg muscle shear force, and breast muscle shear force. Also, there was a very significant difference between different genotypes of the RPL3L 42 bp indel mutation in these trains. Further experiments showed that RPL3L was highly expressed in chicken skeletal muscle, and its overexpression could promote the proliferation and inhibit the differentiation of chicken myoblasts by regulating ASB4 and ASB15 expression. Our findings demonstrated that the RPL3L 42 bp indel may be one of the molecular markers of chicken weight-related traits.
Introduction
Growth traits are one of the most important economic traits of chickens, and individual body weight is one of the most concerned economic indicators. About 40% of the body weight of a chicken comes from its skeletal muscles (Schiaffino and Reggiani, 2011). The development of skeletal muscle is closely related to meat yield and quality. Skeletal muscle composed of breast and leg muscles is one of the important protein sources for humans. Chicken skeletal muscle weight is determined by the number of muscle fibers and volume of muscle fibers, and postnatal skeletal muscle growth occurs primarily by increasing muscle fiber volume rather than muscle fiber number (Smith, 1963). The increase in muscle fiber volume is essentially the result of cellular protein accumulation, a net increase in protein synthesis minus protein degradation (Pereira et al., 2017). The rate of protein synthesis depends on two factors: translation efficiency and translation capacity, corresponding to protein synthesis per unit of RNA and the total amount of ribosomes (Figueiredo and McCarthy, 2019). Although much progress has been made in the past in exploring the molecular mechanisms underlying chicken skeletal muscle growth and development, most studies have focused on the embryonic stage (Davis et al., 2015; Taylor, 2017; Li Y. et al., 2019). It has been reported that the birth weight of chickens does not significantly affect the weight at slaughter (7–8 W) (Lara et al., 2005). However, chickens of different breeds but with similar birth weights can have large differences in the weight of 7-day-olds and 41-day-olds (Tona et al., 2017) or at slaughter (Dou et al., 2017). In other words, the growth rate of different chicken breeds may not differ much during the embryonic period. The difference in growth rate after birth is one of the important reasons for the difference in slaughter weight. However, the current research studies have not been able to fully explain this phenomenon.
The development of chicken skeletal muscle can be divided into two stages: the embryonic stage and postnatal stage. Chicken embryonic muscle grows mainly through the proliferation of myoblasts. At the late embryonic stage, muscle satellite cells attach to the lower basal layer of mature muscle fibers, and their main function is to participate in postnatal muscle damage repair and hypertrophy (Gros et al., 2005). Therefore, the number of myoblasts affects not only the number of muscle fibers but also the number of satellite cells present during postnatal growth. The development of chicken skeletal muscle during the entire embryonic period is mainly divided into the following stages: formatting the primary muscle fibers (primary fibers, also known as primary myotubes) at about 7 days of incubation, completing the secondary muscle fibers (secondary fibers, also known as the formation of secondary myotubes) at about 12 days of incubation, and basically completing the differentiation of myotubes and fixing the number of muscle fibers at about 18 days of incubation (Picard et al., 2010; Chal and Pourquie, 2017). The metabolism and contraction functions of muscle fibers mature within 3 days after birth, after which the main changes in muscle fibers are thickening and muscle fiber type switching (Smith, 1963; Chal and Pourquie, 2017). Due to the fusion of muscle satellite cells with muscle fibers during the late embryonic stage of chickens, the volume of muscle fibers increases, thereby limiting the increase in muscle weight at birth (Egner et al., 2016). After hatching, the rapidly proliferating muscle satellite cells fuse with the muscle fiber, thereby providing a large number of nuclei for the muscle fiber. More nuclei can enhance the transcription efficiency of the muscle fiber and improve the protein synthesis ability and the muscle growth ability (Dumont et al., 2015; Murach et al., 2018). It has been reported that the content of muscle satellite cells in fast-growing large broiler breeds was significantly higher than that in common breeds (Velleman et al., 2000). Although muscle satellite cells are in a resting state under normal conditions, they will be activated by the action of some specific factors. For example, muscle satellite cells will be activated when muscles are injured, and the cells will re-enter the process of proliferation and differentiation. In addition, the muscle-enhancing effect achieved by exercise often utilizes this feature of muscle satellite cells (Murach et al., 2018; Dornen and Dittmar, 2021).
Making full use of bioinformatics methods to study the regulation mechanism of poultry genetic information on phenotypic traits can effectively improve the speed of poultry breeding. Chickens with similar birth weights of different breeds can vary widely in slaughter weight. Previous research studies have not fully elucidated the phenomenon. Are there some specifically expressed genes in regulating chicken muscle growth and development after they are born? Therefore, this study aims to obtain the key genes and their signaling pathways that may affect the muscle growth and development of chickens after birth by integrating the chicken muscle-related RNA-seq data in the public database and validating one of the candidate genes, RPL3L, in chicken muscle growth and development. The results of this study will provide a theoretical reference for the study of chicken muscle growth and development.
Materials and methods
Data sources
In this study, four datasets were downloaded from three databases, including the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) (Harrison et al., 2021), and the National Genomics Data Center, China National Center for Bioinformation (CNCB-NGDC, https://ngdc.cncb.ac.cn/) (CNCB-NGDC Members and Partners, 2022).
The PRJCA001556 dataset in the original study (Li et al., 2020) contained transcriptome data of Shouguang chickens’ breast muscle tissues from 17 individuals at 6-time points, including the embryo day (E) 12, E17, the age of 1 day (D), 2 weeks (W), 8 W, and 14 W. In our study, the transcriptome data of 11 individuals were selected from 4-time points for analysis, including E12, E17, D1, and 8 W. The PRJCA001192 dataset in the original study (Xing et al., 2020) contained transcriptome data of Jingxin Yellow chickens’ breast muscle and abdominal adipose tissues from 27 individuals at 9-time points of E12, E17, D1, 1 W, 3 W, 8 W, 14 W, 20 W, and 26 W. In the current study, the data of breast muscle tissues from 12 individuals at 4-time points of E12, E17, D1, and 8 W were selected for analysis. In the original study (Jin et al., 2021), the GSE162148 dataset had the transcriptome data of seven tissues from 21 Tibetan chicken individuals, including skeletal muscle, liver, heart, spleen, lung, kidney, and fat. In the present study, the data from six tissues from 18 individuals were selected for analysis, including breast muscle, liver, spleen, lung, kidney, and fat. The PRJNA665193 dataset in the original study (Kern et al., 2021) contained transcriptome data of eight tissues from 16 White Leghorn chickens, including skeletal muscle, liver, spleen, lung, fat, cerebellum, cerebral cortex, and hypothalamus. All data from this dataset were selected for analysis in our study. The fastq format files of the datasets PRJCA001556 (Li et al., 2020), PRJCA001192 (Xing et al., 2020), and PRJNA665193 (Kern et al., 2021) were directly downloaded through FileZilla FTP Client (FileZilla 64-bit installation version, https://www.filezilla.cn/) and IBM Aspera Connect (ibm-aspera-connect-3.8.1.161,274-linux). The SRA file of GSE162148 (Jin et al., 2021) was downloaded through IBM Aspera Connect and used the convert SRA to fastq files plugin of TBtools (TBtools_windows-x64_1_09867, Chen et al., 2020) to convert it into a fastq format file. The information on all the datasets used in this study is listed in Supplementary Table S1.
Quality control and gene expression quantification
Moreover, fastp (version 0.23.2, Chen et al., 2018) software was used to perform quality control on raw data, including removing adapter sequences and trimming low-quality reads. Also, Kallisto (Kallisto 0.44.0, Bray et al., 2016) software was used to perform unmatched direct quantification of quality-controlled sequencing data. The result of the Kallisto analysis was the expression level at the transcript level, which was converted into gene expression level using the tran value sum plugin of TBtools software (Kern et al., 2021).
Principal component analysis
Principal component analysis (PCA) was performed on the data using the basic PCA analysis plugin of TBtools software (Chen et al., 2020).
Differentially expressed gene analysis and functional enrichment analysis
Differentially expressed gene (DEG) analysis was performed on the read count obtained by Kallisto using the differential gene expression analysis (DESeq2, Love et al., 2014) plugin of TBtools software (Chen et al., 2020), and the upregulated genes in each group were screened with log2|FC| > 1 and FDR < 0.05 as thresholds. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs were performed via the website https://biit.cs.ut.ee/gprofiler/gost. The GO database classified DEGs into three categories according to their molecular functions (MF), cellular components (CC), and biological processes (BP) involved.
Protein–protein interaction network analysis
The protein–protein interaction (PPI) network was constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) (version 10.0, https://cn.string-db.org/) online database. Cytoscape (version 3.6.1, https://cytoscape.org/) was used to visualize the PPI network, and its cytoHubba plugin was used to screen the top 10 as hub genes (key genes) according to the degrees of nodes.
Insertion/deletion (indel) typing
The genomic DNA samples of 350 individuals’ blood from the F2 resource population of Xinghua chickens crossed with WRR chickens were extracted following the instructions of the HiPure Blood DNA Mini Kit (Guangzhou Magen Biotechnology Co., Ltd., Guangzhou, China). All of them were amplified by PCR with RPL3L-INDEL primers (Supplementary Table S2), and the products were analyzed by electrophoresis on agarose gel at a concentration of 2%.
Animals and cells
All animal experiments performed in this study were approved by the Institutional Animal Protection and Utilization Committee of South China Agricultural University (approval ID: SCAU#0014). Furthermore, the care and use of animals complied with the local animal welfare laws, guidelines, and policies. The chicken embryos used in this study were as previously described (Luo et al., 2014). The experimental animals were purchased from Zhaoqing Fengkai Zhicheng Poultry Breeding Co., Ltd. During hatch, the temperature was controlled at 38.5°C, 38°C, 37.9°C, and 37.3°C–37.5°C in the first 6 days, 7–14 days, 15th day, and 16–21 days of hatching, respectively. The relative humidity was maintained at 60%–70%, 50%–55%, and 65%–70% for the 1–7 days, 8–16 days, and after the 17th day of incubation, respectively. The chicks after hatching drank freely and were fed with chick feed which was provided by Zhaoqing Fengkai Zhicheng Poultry Breeding Co., Ltd. The breast muscle samples were collected from embryonic day 10 (E10) to E15 and post-hatch day 1 chicks, and the breast muscle, leg muscle, heart, liver, spleen, lung, brain, cerebellum, pituitary, abdominal fat, and cartilage samples were collected from 7-week-old Xinghua chickens. The information of 100 individuals from 15 chicken populations were as previously described (Luo et al., 2020). The 15 chicken populations were composed of nine Chinese nationwide indigenous chicken breeds, including six Huiyang Bearded chickens (BC), 20 Hetian chickens (HT), six Baier Yellow chickens (BEH), six Xianju chickens (XJ), six Liyang chickens (LY), six Jining Bairi chickens (BR), six Yunyang Da chickens (YY), six Lindian chickens (LD), and six Tulufan gamecock chickens (TLF); four typical commercial populations, including six White Leghorn chickens (LH), six White Recessive Rocks (WRR), six Cobb RS308 chickens (RS), and six Rhode Island Reds (RIR); one Red jungle fowl population from Guangxi Province (five individuals, RJF) and one gamecock population from Laos (three individuals, Laos).
The primary myoblasts of chicken were isolated from the leg muscles of E11 chicks and cultured in DMEM (Invitrogen Trading Co. Ltd., Shanghai, China) as previously characterized (Luo et al., 2014). Myoblast differentiation and DF-1 cell culturing were as previously reported (Luo et al., 2015). The chicken satellite cells were isolated from the breast muscle of 5-day-old Xinghua chicks and cultured in DMEM/F12 (Invitrogen) were the same as previously described (Han et al., 2019).
Plasmid construction and cell transfection
The overexpression vector of the chicken RPL3L gene coding sequence (CDS) region (pcDNA3.1-RPL3L) and RPL3L gene indel firefly luciferase reporters (PGL3-RPL3L-II and PGL3-RPL3L-DD) were synthesized by Wuhan GeneCreate Biological Engineering Co., Ltd. The cell transfection was performed using LipofectamineTM 3000 Transfection Reagent following the manufacturer’s instructions (Invitrogen).
Quantitative real-time PCR
Total RNA of breast muscle tissues of Xinghua chickens before or after birth, different tissues of 7 W Xinghua chickens, and the primary myoblasts and satellite cells of chicken that transfected with or without overexpression of RPL3L was extracted according to the instructions of the HiPure Total RNA Mini Kit (Guangzhou Magen Biotechnology Co., Ltd., Guangzhou, China). Then, following the instructions of the MonScript™ RTIII All-in-One Mix with dsDNase kit (Monad Biotech Co. Ltd., Wuhan, China) to synthesize cDNA. Quantitative real-time PCR (qRT-PCR) was carried out according to the instructions of the MonAmp™ ChemoHS qPCR Mix kit (Monad Biotech Co. Ltd., Wuhan, China). Fluorescence quantitative results were calculated by Excel software to calculate the 2-△△Ct value to compare the relative gene (β-actin) expression. All primers used for qRT-PCR were listed in Supplementary Table S2.
Cell proliferation assay
A total of 3 × 103 primary myoblasts of chicken per well were seeded in 96-well plates, after 0, 6, 12, 24, and 48 h transfection with pcDNA3.1-RPL3L or pcDNA3.1, the Cell Counting Kit-8 (CCK-8) experiment was performed according to the instructions of the CCK-8 kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). A total of 5 × 105 primary myoblasts of chicken per well were seeded in 12-well plates, after 48 h transfection with pcDNA3.1-RPL3L or pcDNA3.1, the cell cycle assay was performed as previously described (Luo et al., 2015), and the EdU experimental was carried out according to the protocol of the EdU Cell Proliferation Detection kit (Ribobio Co., Ltd., Guangzhou, China). All experiments were repeated at least three times.
Western blotting
A total of 1.2 × 106 primary myoblasts of chicken per well were seeded into 6-well plates, cultured in DMEM, and transfected with pcDNA3.1-RPL3L or pcDNA3.1 plasmids. After 48 h of transfection, the whole-cell lysate was extracted by the lysate (a 10:1 ratio of RIPA lysate and PMSF) and their concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). A total of 5 µg proteins were separated by 8%–12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA) that was pre-soaked in methanol. The membrane was then blocked with Quick Blocking Buffer (Beyotime Biotechnology Co., Ltd., Shanghai, China) for 15 min at room temperature before incubation overnight at 4°C with primary detection antibodies, including the primary antibodies specific for anti-RPL3L (Abcepta Biotech Ltd., Co., Suzhou, China; 1:500), anti-Myhc, anti-Myog, anti-Myod (Thermo Fisher Scientific; 1:500 for each of them), and anti-β-actin (Beyotime Biotechnology Co., Ltd., Shanghai, China; 1:1000). The PVDF membrane was washed three times with western wash buffer (Beyotime Biotechnology Co., Ltd., Shanghai, China), followed by incubation for 2 h at room temperature with the secondary antibody, horseradish peroxidase (HRP)-labeled anti-mouse and anti-rabbit immunoglobulin G (IgG) (Beyotime Biotechnology Co., Ltd., Shanghai, China; 1:2000). Bands were detected using the BeyoECL Star chemiluminescence substrate (Beyotime Biotechnology Co., Ltd., Shanghai, China).
Dual-luciferase reporter assay
A total of 5 × 104 DF-1 cells/well were seeded in 96-well plates and transfected with a 1:20 ratio of PGL3-RPL3L-II or PGL3-RPL3L-DD to renella luciferase reporter (pRL-TK) plasmid. After 24 h of transfection, the cells were lysed and centrifuged at 12,000 rpm for 1 min to collect the supernatant. The dual-luciferase reporter assay was performed according to the instructions of the Dual-Glo® Luciferase Assay System (Promega Biotech Co., Ltd., Beijing, China).
Transcription factor prediction
The online website http://gene-regulation.com/pub/programs/alibaba2/index.html was used to predict the transcription factor.
Association analysis and statistical analysis
As previously described (Li W. et al., 2019; Lin et al., 2021; Wei et al., 2022), the mixed linear model in SPSS software (version 24.0) was used to analyze the association between different genotypes of indel and the different phenotypic traits of F2 resource population from Xinghua chickens crossing with WRR chickens. The analysis model was as follows,
Among them, Yijklm represents the trait phenotype value of the individual, μ represents the overall population mean, Gi is the fixed effect of the marker genotype (i = 3), Sj represents the fixed effect of sex, Hk represents the fixed effect of the batch, fl is the fixed effect of the family, and eijklm represents the random error. A least-squares analysis was used to investigate the effect of the polymorphic genotypes on the target traits. p < 0.05 indicated a significant level, and Bonferroni’s adjustment was performed to control for multiple comparisons.
Data from the other experiments were shown as the mean values with the standard error of the mean. Statistical differences were determined using Student’s t-test or one-way ANOVA. p < 0.05 was considered to indicate statistical significance.
Results
Results of quality control and principal component analysis
After filtering the PRJCA001556 dataset, the average size of single-sample sequencing data was 11.4 Gb, the average number of reads was 76.36 M, and the Q20 and Q30 were 98.12% and 94.64%, respectively (Supplementary Table S3). From the filtered results of the PRJCA001192 dataset, the average size of single-sample sequencing data was 7.45 Gb, and the average number of reads was 60.93 M. Q20 and Q30 were 95.16% and 89.83%, respectively (Supplementary Table S3). After filtering the GSE162148 dataset, the average size of single-sample sequencing data was 14.26 Gb, and the average number of reads was 95.46 M. Q20 and Q30 were 97.78% and 94.06%, respectively (Supplementary Table S3). As a result of the filtered PRJNA665193 dataset, the average size of single-sample sequencing data was 13.00 Gb, the average number of reads was 132.90 M, and the Q20 and Q30 rates were 98.80% and 96.12%, respectively (Supplementary Table S3). These results showed that the overall quality of the clean reads of the sequencing data fulfilled the requirements of subsequent data analysis.
The four datasets PRJCA001556, PRJCA001192, GSE162148, and PRJNA665193 were found to be clustered together after performing PCA (Figure 1), suggesting that the samples were well grouped and could be used for further analysis.
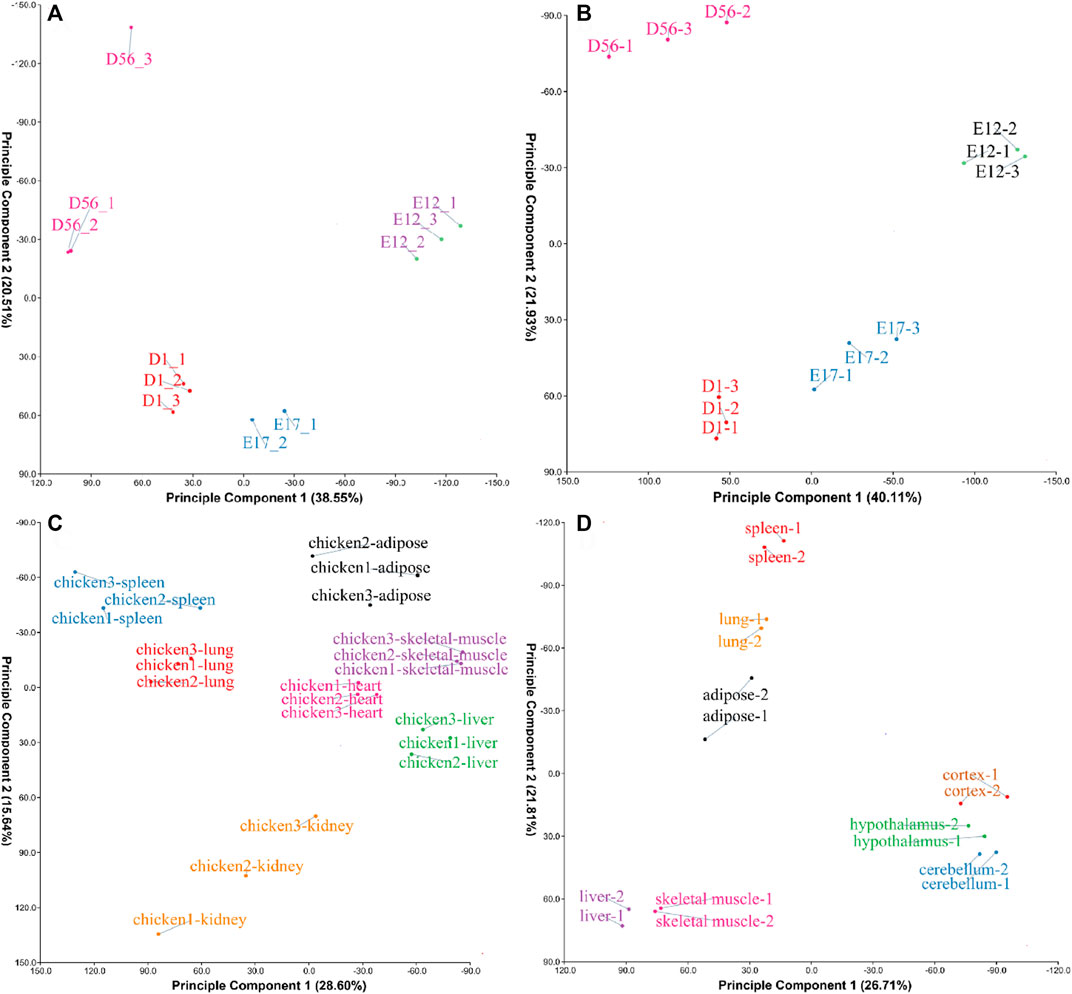
FIGURE 1. Principal component analysis (PCA) of samples from different datasets. (A) The PCA result of the PRJCA001556 dataset; (B) PCA result of the PRJCA001192 dataset; (C) PCA result of the GSE162148 dataset; and (D) PCA result of the PRJNA665193 dataset.
Differentially expressed genes and functional enrichment analysis
In order to screen the genes that are specifically highly expressed in chickens after birth, the D1, 8 W muscle transcriptome data were compared with those of E12 and E17 from the PRJCA001556 and PRJCA001192 datasets, respectively. As a result, compared with E12 and E17, a total of 531 DEGs were upregulated in both D1 and 8 W in the PRJCA001556 dataset (Figure 2A), and 341 differentially expressed genes (DEGs) were upregulated in both D1 and 8 W in the PRJCA001192 dataset (Figure 2B). Additionally, a total of 200 postnatal upregulated DEGs were overlapped in these two datasets (Figure 2C). According to the top five GO terms with the smallest p-values in the three categories of MF, CC, and BP of the 200 DEGs between the embryonic stage and after hatching, the GO enrichment results of postnatal-specific highly expressed genes included NAD binding, anion transmembrane transporter activity, transmembrane transporter activity, transporter activity, and active transmembrane transporter activity (Figure 2D). As a result of KEGG pathway enrichment analysis, the significantly enriched pathways included the insulin signaling pathway, ABC transporter, and 2-oxocarboxylate metabolism(Figure 2E).
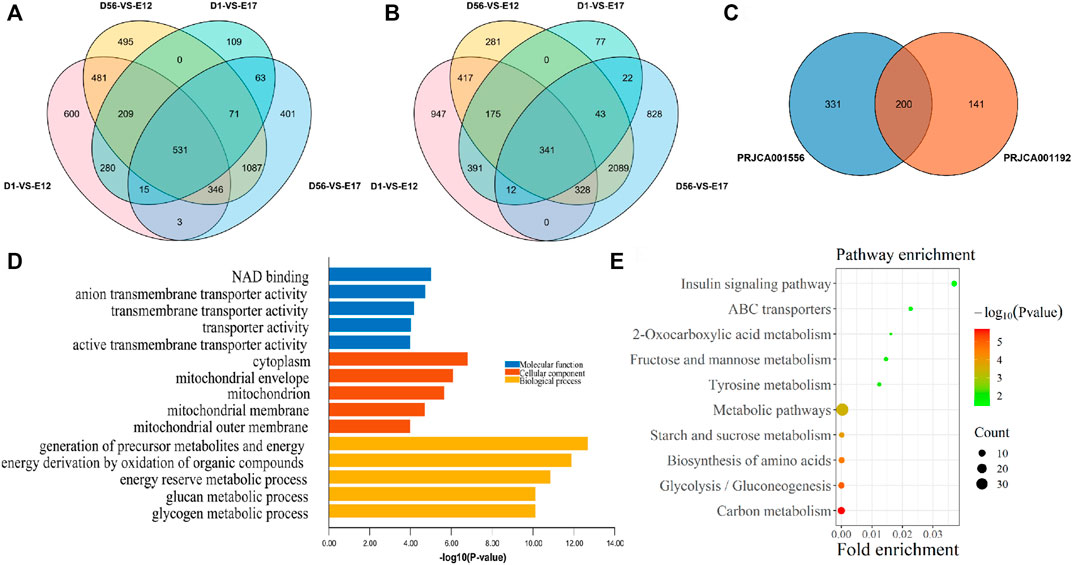
FIGURE 2. Functional enrichment analysis of postnatal-specific highly expressed genes. (A) Venn diagram of DEGs from the PRJCA001556 dataset; (B) Venn diagram of DEGs from the PRJCA001192 dataset; (C) overlapping DEGs between PRJCA001556 and PRJCA001192 datasets; (D) GO enrichment analysis results of postnatal-specific highly expressed genes; and (E) KEGG enrichment analysis results of postnatal-specific highly expressed genes.
For exploring genes that were specifically highly expressed in muscle, the muscle tissues and non-muscle tissues transcriptome data of GSE162148 and PRJNA665193 datasets were analyzed, respectively. As a result, compared with non-muscle tissues, there were 921 upregulated DEGs in muscle tissues in the GSE162148 dataset (Figure 3A), and 1673 upregulated DEGs in muscle tissue in the PRJNA665193 dataset (Figure 3B). In addition, a total of 560 muscle upregulated DEGs were overlapped in these two datasets (Figure 3C). According to the top five GO terms with the smallest p-values in the three categories of MF, CC, and BP of the 560 DEGs upregulated in muscle tissues, the GO enrichment results of muscle-specific highly expressed genes included actin binding, cytoskeletal protein binding, and protein serine/threonine phosphatase activity (Figure 3D). As muscle-specific were highly expressed genes, they were significantly enriched in various GO terms related to muscle composition and muscle development. Also, the result of the KEGG pathway enrichment analysis found that they were significantly enriched in the pathways of carbon metabolism, glycolysis/gluconeogenesis, oxidative phosphorylation, and myocardial contraction (Figure 3E).
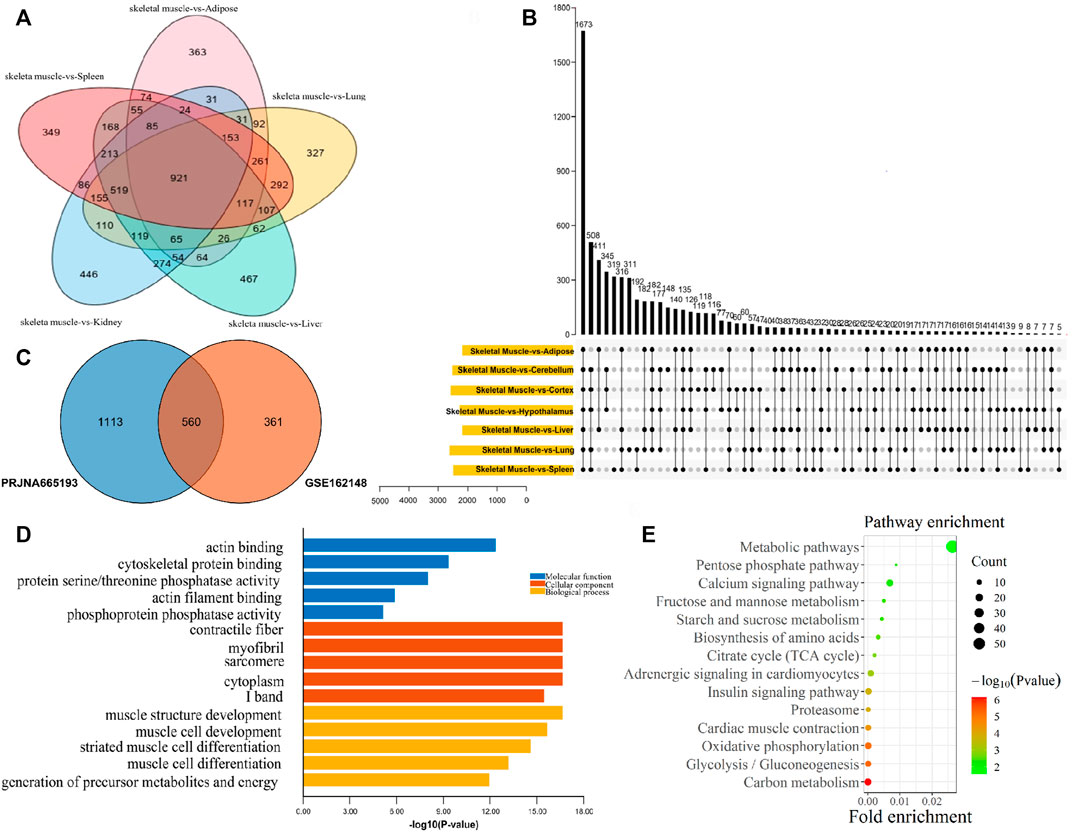
FIGURE 3. Functional enrichment analysis of muscle-specific highly expressed genes. (A) Venn diagram of DEGs from the GSE162148 dataset; (B) Venn diagram of DEGs from the PRJNA665193 dataset; (C) overlapping DEGs between the postnatal-specific high-expressed gene set and the muscle-specific high-expressed gene set; (D) GO enrichment analysis results of muscle-specific highly expressed genes; and (E) KEGG enrichment analysis results of muscle-specific highly expressed genes.
In order to find the postnatal and muscle-specific high-expression genes, the postnatal-specific high-expression gene set and the muscle-specific high-expression gene set obtained earlier were intersected. A total of 79 postnatal and muscle-specific highly expressed genes were screened (Figure 4A and Supplementary Table S4). From the results of GO and KEGG enrichment analysis on these 79 genes, it was found that the GO MF enrichment mainly included anion channel activity and anion transmembrane transporter activity (Figure 4B). In addition, the GO CC enrichment mainly included the mitochondrial envelope, mitochondrial membrane, cytoplasm, sarcomere, and organelle envelope, and the GO BP enrichment mainly contained the production of precursor metabolites and energy, the process of carbohydrate metabolism, and the oxidation of organic compounds to generate energy (Figure 4B). Moreover, as a result of KEGG pathway enrichment, they were significantly enriched in the pathways of glycolysis/gluconeogenesis, carbon metabolism, and amino acid biosynthesis (Figure 4C).
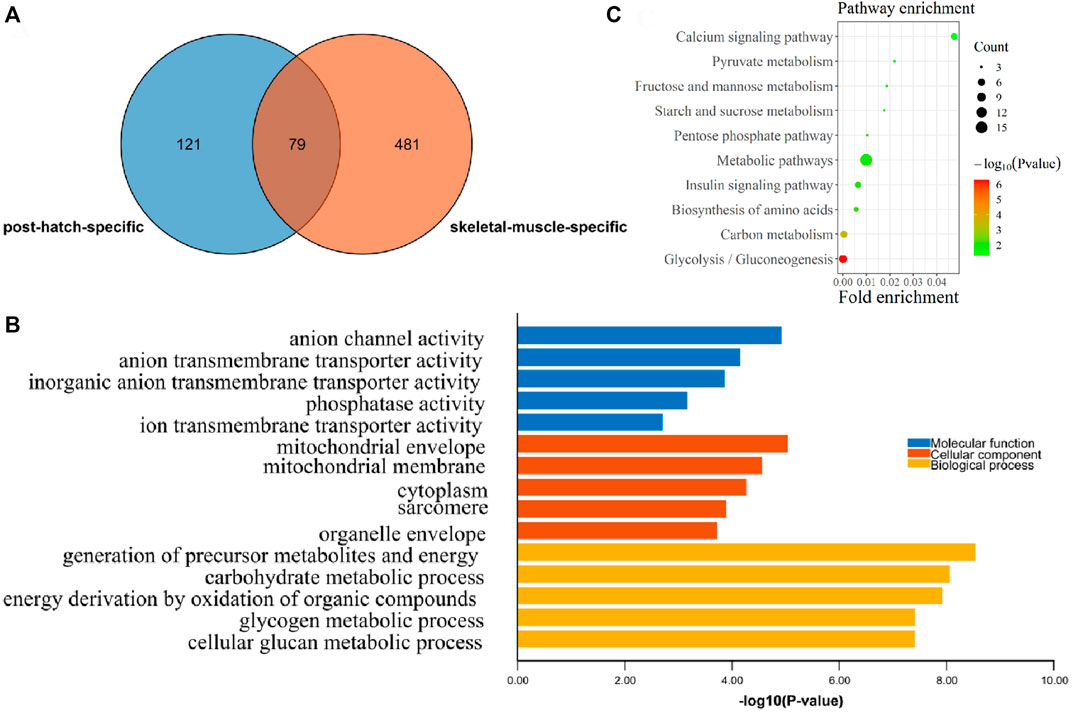
FIGURE 4. Functional enrichment analysis of postnatal muscle-specific highly expressed genes. (A) Overlapping DEGs between the postnatal-specific high-expressed gene set and the muscle-specific high-expressed gene set; (B) GO enrichment analysis results of postnatal muscle-specific highly expressed genes; and (C) KEGG enrichment analysis results of postnatal muscle-specific highly expressed genes.
Protein–protein interaction network analysis results
To reveal the interactions between the 79 DEGs, we analyzed and constructed a PPI network, and screened the hub genes. As a result, there was a protein–protein interaction (PPI) network, which consisted of 50 nodes and 123 edges in total (Figure 5A). Also, 10 hub genes were screened out, including RPL3L, FBP2, ASB4, ASB15, CKMT2, PGAM1, YIPF7, PFKM, LDHA, and GAPDH (Figure 5B). Among them, except for YIPF7 and GAPDH, they all have been reported to play a certain regulatory role in muscle growth and development.
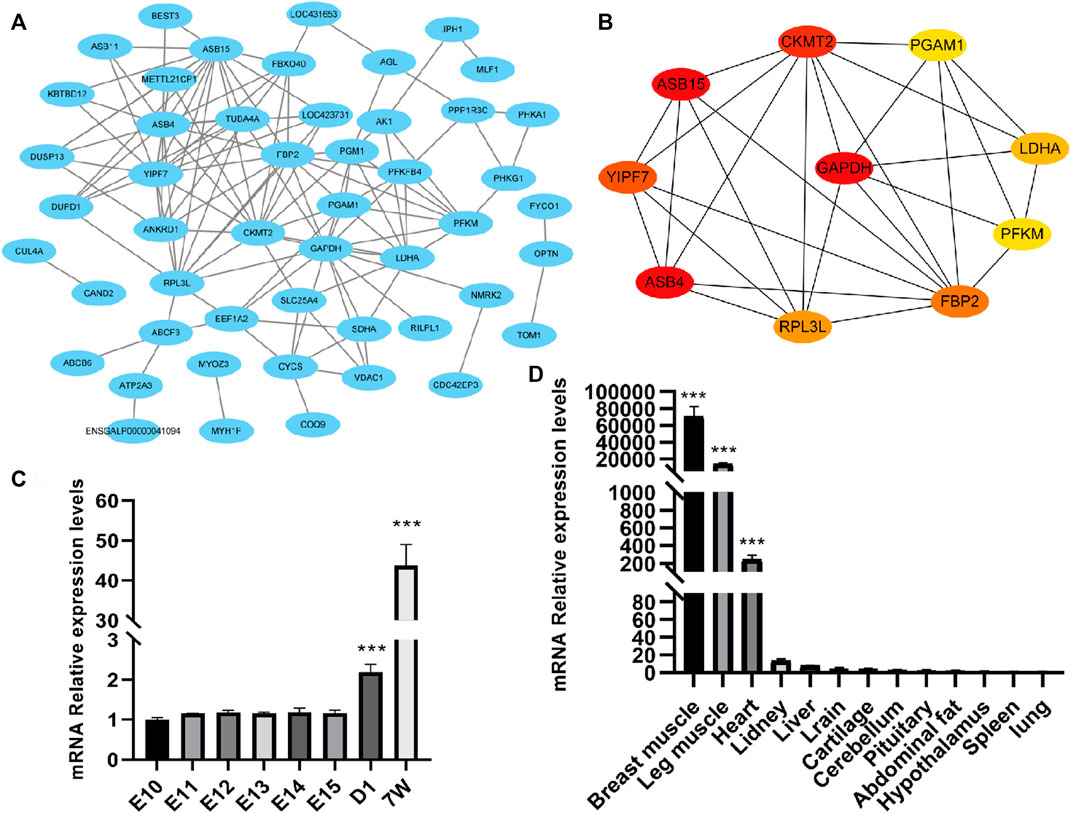
FIGURE 5. Protein–protein interaction (PPI) network analysis and validation of one of the hub genes RPL3L. (A) PPI network composed of differential genes; (B) top 10 hub genes according to the degree score; (C) expressed level of RPL3L in different developmental stages of the breast muscle of Xinghua chicken; and (D) expression level of RPL3L in different tissues of Xinghua chicken. ***p < 0.001.
Considering that the RPL3L has not been reported in livestock and poultry and its fold change (after birth/embryonic stage) was higher than that of the other genes, we chose RPL3L from the 10 hub genes for further study. From the results of qRT-PCR, it was shown that the expression level of RPL3L was significantly higher after birth than the embryonic stage in breast muscle tissues of Xinghua chickens (Figure 5C), and its expression level in muscle tissues of 7-week-old Xinghua chickens was also significantly higher than that in non-muscle tissues, among which breast muscle had the highest expression level (Figure 5D).
RPL3L promotes chicken myoblast proliferation and inhibits myoblast differentiation
In order to preliminarily study the effect of RPL3L on chicken muscle development, we detected its expression level in different proliferation and differentiation stages of primary myoblasts and muscle satellite cells. It was found that RPL3L first decreased and then increased during the proliferation and differentiation stage of myoblasts (Supplementary Figure S1A), while in muscle satellite cells, it continued to rise along with the cell differentiation (Supplementary Figure S1B). These results suggest that RPL3L may be involved in the regulation of muscle growth and development.
Then, the overexpression vector of RPL3L was transfected into chicken primary myoblasts. The CCK-8 results showed that overexpression of RPL3L could significantly enhance cell viability (Figure 6A). From the result of the cell cycle assay, it was found that cells in the G1 phase were significantly reduced, and cells in the S phase were significantly increased after overexpression of RPL3L compared with the control group (Figure 6B). The EDU results also showed that overexpressing RPL3L could promote the proliferation of chicken primary myoblasts (Figure 6C). These results suggest that RPL3L can significantly promote the proliferation of primary myoblasts. Moreover, overexpression of RPL3L in the differentiation stage of myoblasts significantly inhibited the transcription levels of differentiation-related marker genes, including Myog, Myod, Myomaker, Myhc, Mef2c, and Myf5 (Figure 6D), and the protein levels of Myhc, Myog, Myod, and Myomaker (Figure 6E), implying that RPL3L may inhibit primary myoblast differentiation by downregulating the expression of these differentiation-related proteins.
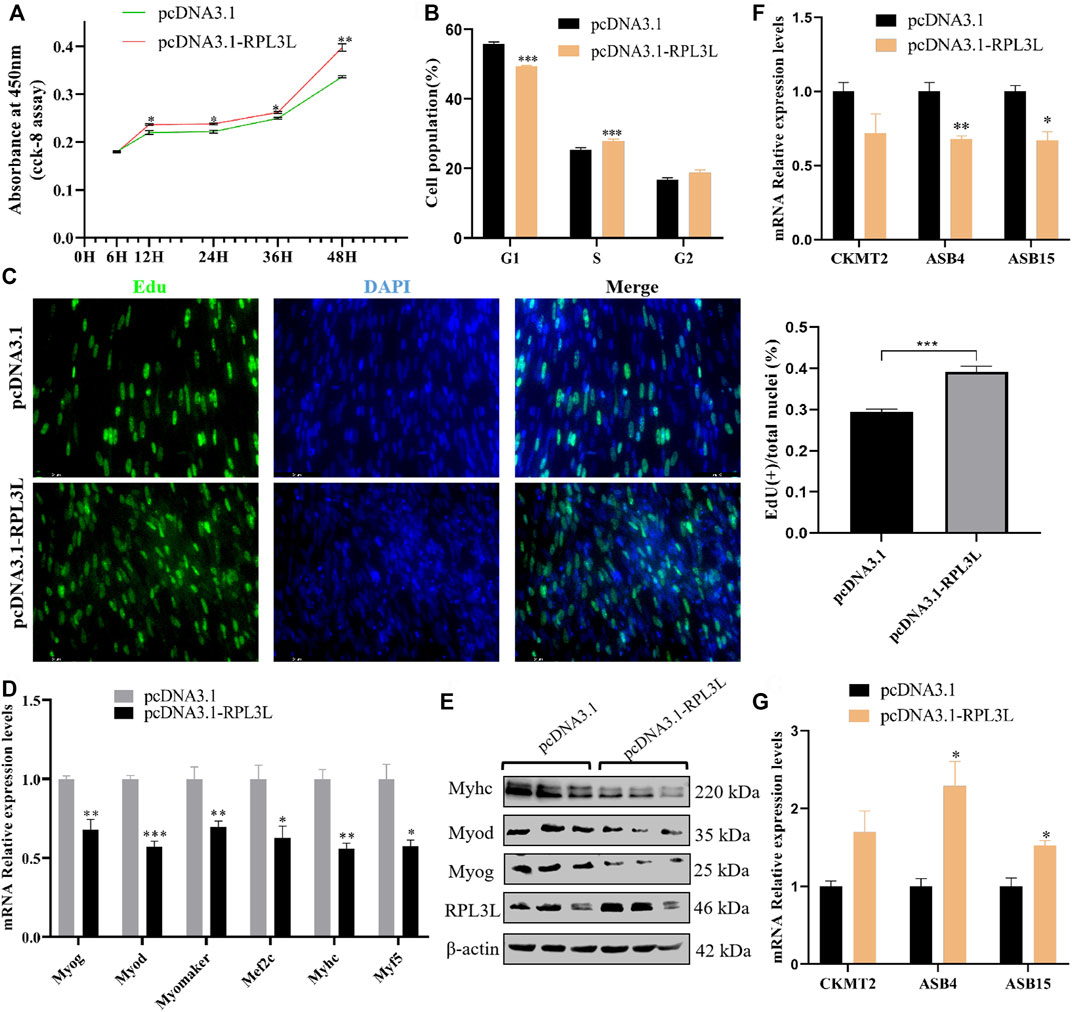
FIGURE 6. RPL3L promotes cell proliferation and inhibits cell differentiation of chicken primary myoblast. (A) CCK-8 result with the overexpressed chicken RPL3L and its negative control; (B) cell cycle assay result after transfecting with the overexpressed chicken RPL3L and its negative control; (C) EdU result with the overexpressed chicken RPL3L and its negative control; (D) mRNA expression levels of differentiation-related marker genes after overexpressing chicken RPL3L; (E) protein expression levels of differentiation-related marker genes after overexpressing chicken RPL3L; (F) expression levels of CKMT2, ASB4, and ASB15 detected by overexpression of RPL3L in myoblast proliferation stage, respectively; and (G) expression levels of CKMT2, ASB4, and ASB15 detected by the overexpression of RPL3L in the myoblast differentiation stage, respectively. ns, not significant, *p < 0.05, **p < 0.01, and ***p < 0.001.
According to the previously mentioned PPI results, there was a certain interaction between RPL3L and several genes known to affect the growth and development of chicken skeletal muscle, such as CKMT2, ASB4, and ASB15. In this study, RPL3L was overexpressed in the primary myoblast proliferation phase (Supplementary Figure S1A), and it was found that RPL3L could inhibit the transcription of ASB4 and ASB15 genes in the cell proliferation phase (Figure 6F). Overexpression of RPL3L during differentiation could significantly promote the transcription levels of these two genes (Figure 6G). This result demonstrated that chicken RPL3L may regulate muscle growth and development by affecting the expression of ASB4 or ASB15.
RPL3L polymorphism affects chicken growth and development
From the RPL3L gene structure and conservation among different species, we found that the chicken RPL3L gene was in the length of 7359 bp, located on chicken chromosome 14, and consists of 10 exons and nine introns. The closest distance between chicken RPL3L and duck was found by constructing nucleotide evolutionary trees of 10 different species (data not shown). Comparing the conservation of the protein encoded by RPL3L in 10 different animals, including chicken, human, pig, and mouse, it was found that the RPL3L protein was very conserved, indicating that the function of RPL3L may be similar among animals (data not shown).
Based on a 42 bp indel (Figure 7A) and the SNP sites of the RPL3L gene, an evolutionary tree was constructed. It was found that the RPL3L gene was strongly selected in Huiyang Bearded chicken, Rhode Island Reds chicken, and WRR chicken (Figure 7B). Also, the frequency of this 42 bp mutation in fast-growing chicken breeds such as WRR chickens was significantly higher than that in other Chinese landraces with slow-growing breeds (Figure 7C). These results imply that the mutation may be one of the important molecular markers of chicken growth traits.
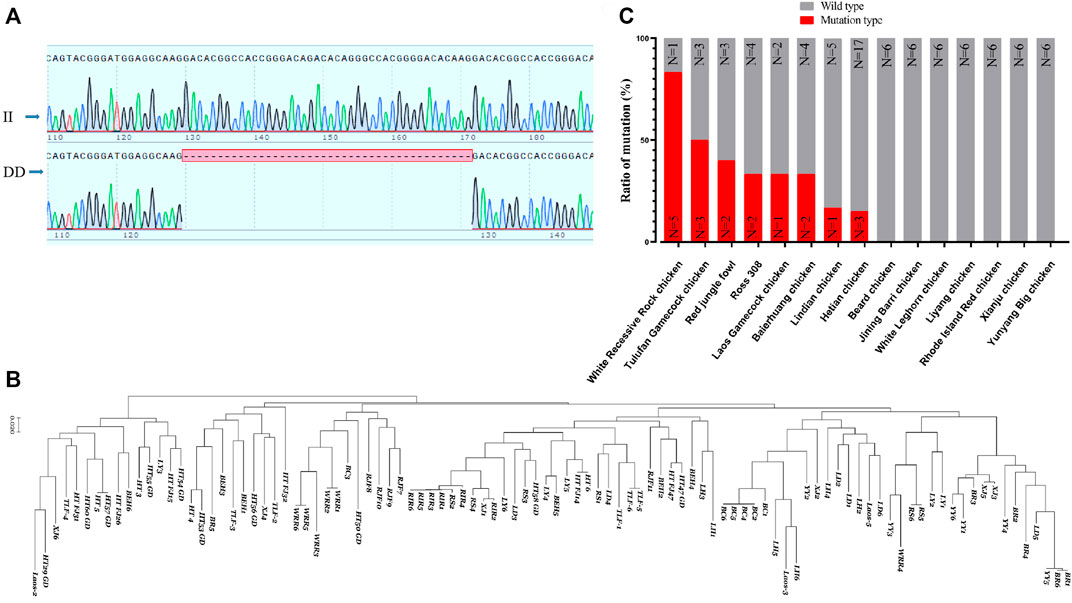
FIGURE 7. Polymorphism of Chicken RPL3L. (A) The 42 bp indel of chicken RPL3L; (B) evolutionary tree diagram of RPL3L in different chicken breeds; (C) mutation ratio of RPL3L 42 bp indel in different chicken breeds. BC, Huiyang Bearded Chicken; WRR, White Recessive Rock Chicken; HT, Hetian chicken; BEH, Baier Yellow chicken; XJ, Xianju chicken; LY, Liyang chicken; BR, Jining Bairi chicken; YY, Yunyang Da chicken; LD, Lindian chicken; TLF, Tulufan gamecock chicken; LH, White Leghorn chicken; RS, Cobb RS308 chicken; RIR, Rhode Island Reds; RJF, Red jungle fowl chicken; and Laos, one gamecock population from Laos.
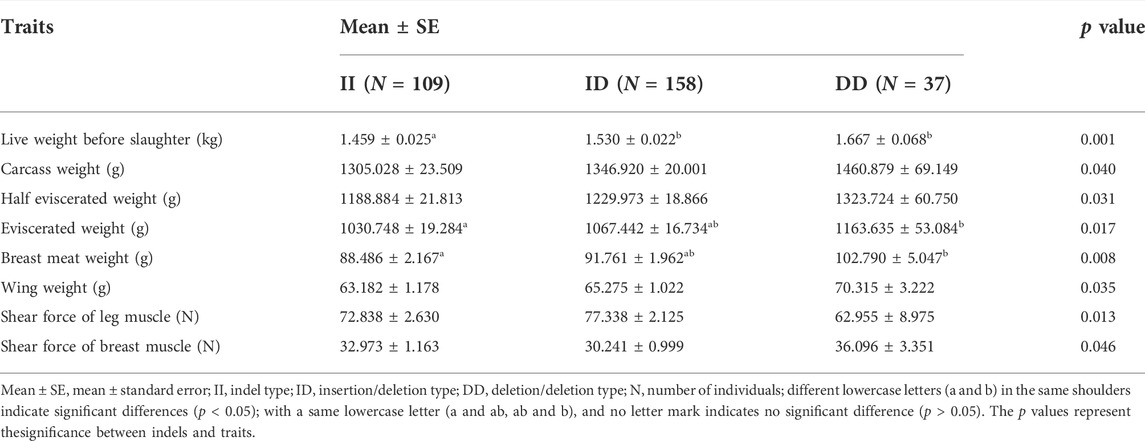
Using gel electrophoresis, we classified the indels into insertion/insertion (II) type, insertion/deletion (ID) type, and deletion/deletion (DD) type (Supplementary Figure S2). The SPSS software was used to analyze the association between the indel and the growth traits of the F2 resource population from Xinghua chickens crossing with WRR chickens. It was shown that the different genotypes of this mutation were significantly correlated with multiple growth traits of chickens, including live weight, carcass weight, half eviscerated weight, eviscerated weight, breast meat weight, wing weight, leg muscle shearing force, and chest muscle shear force (p < 0.05) (Table 1). The results indicated that individuals with the DD genotype had significant advantages in multiple economic traits.
Indels of RPL3L influence its transcriptional activity
In order to analyze the effect of this 42 bp indel on the expression level of the RPL3L gene, the PGL3-RPL3L-II and PGL3-RPL3L-DD vectors were transfected into DF-1 cells for dual-luciferase reporter experiments to detect the transcriptional activity of these two genotypes of the RPL3L promoter. It was found that the transcriptional activity of the II genotype was significantly higher than that of the DD genotype (p < 0.0001) (Supplementary Figure S3A), suggesting that the 42 bp deletion could inhibit the transcriptional activity of RPL3L. Transcription factor prediction through online tools found that genotype DD may lose binding sites for a total of eight transcription factors, including CPE-bind, C/EBP, c-Myc, Max1, Sp1, AP-2alph, NF-1, and GATA-1 (Supplementary Figure S3B,C.
Discussion
With the continuous development of sequencing technology, bioinformatics has played a pivotal role in the field of poultry breeding. Different biological information databases contain a large amount of poultry genetic information, and there could be new valuable information by integrated analysis of these public data again. In this study, by integrating the transcriptome sequencing data of different chicken tissues and chicken muscles at different developmental times, it was found that the GO BP terms of the differentially highly expressed genes after birth were mainly the production and metabolism of organic matter and energy. The KEGG enrichment results mainly included pathways of amino acid biosynthesis and carbon metabolism. Just like in previous studies, muscle growth and development are often accompanied by the production and metabolism of a large amount of organic matter. Skeletal muscle can absorb and utilize glucose and fatty acids, and a lack of energy can trigger muscle atrophy. Protein synthesis consumes energy, and a key sensor of cellular energy levels (AMPK) is sensitive to the nutritional status of the body (Kimura et al., 2003). The protein synthesis of skeletal muscle decreases when the body lacks energy and nutrients (Zhu et al., 2004).
From the result of the PPI network analysis of these 79 DEGs, it was found that PFKM, LDHA, PGAM1, FBP2, CKMT2, ASB4, ASB15, and RPL3L were the hug genes, which were reported to play a certain regulatory role in muscle growth and development. For example, muscle phosphofructo-1 kinase (PFKM) is a key regulatory enzyme of glycolysis (Garcia et al., 2009), and regulates the insulin-stimulated glycolytic pathway in chicken skeletal muscle (Seki et al., 2006). Lactate dehydrogenase A (LDHA) inhibits the proliferation and promotes differentiation of chicken primary myoblasts through being targeted by miR-29b-1-5p (Li et al., 2022). Studies have pointed out that white streaks and lignified chicken breasts in broiler breasts may be related to LDHA, but no causal relationship has been established (Malila et al., 2019). Phosphoglycerate mutase 1 (PGAM1) may be a potential molecular marker of chicken tenderness, and its expression is positively correlated with broiler breast muscle shear stress (Kubota et al., 2021). Fructose bisphosphatase 2 (FBP2) can promote fetal birth weight, and is positively correlated with the development of middle and late skeletal muscle in sheep (Xu et al., 2014). In addition, it can increase glucose uptake in skeletal muscle (Bakshi et al., 2018; Takahashi et al., 2020). Mitochondrial creatine kinase 2 (CKMT2) has been shown to be an important protein related to energy metabolism during muscle development in tissue in swine (Voillet et al., 2018), bovine (Cassar-Malek et al., 2017), chicken (Bottje et al., 2017), and other species. Multiple members of the ankyrin repeat and suppressor (ASB) family of cytokine signaling cassettes encoding E3 ubiquitination ligases are thought to be important genes during skeletal muscle growth and development (Ehrlich et al., 2020). It has been reported that ASB4 can promote vascular differentiation and myogenesis in mice by degrading protein ID2 (DNA-binding inhibitor 2), a protein that negatively regulates myogenesis and vascular differentiation (Townley-Tilson et al., 2014). ASB15 has been shown to be involved in muscle cell development by promoting protein synthesis and regulating muscle cell differentiation (McDaneld et al., 2006). Also, multiple SNP loci of ASB15 were significantly associated with chicken muscle-related carcass traits (Wang et al., 2015; Wang et al., 2017).
Ribosomal proteins are the main components involved in the composition of ribosomes. The ribosomal proteins are composed of the large subunit ribosomal protein (RPL) and the small subunit ribosomal protein (Wool, 1979). Many studies have shown that ribosomal proteins are involved in cell proliferation (Lindström and Zhang, 2008; Li et al., 2012), cell differentiation (Schneider et al., 2016), apoptosis (Khanna et al., 2003), regulation of cell development (Komili et al., 2007; Zhang et al., 2013), malignant transformation of normal cells (Challagundla et al., 2011; Pourdehnad et al., 2013) and extraribosomal functions (Warner and McIntosh, 2009). Due to the large number of ribosomal protein genes and limited current research methods, the expression patterns of ribosomal protein genes are still unclear (Uprety et al., 2018; Derenzini et al., 2019). Previous studies have demonstrated that reduced levels of ribosomal protein genes in organisms can adversely affect growth, development, and physiological functions. Most ribosomal protein genes exhibit specific expression patterns at specific developmental stages or specific tissues (Bortoluzzi et al., 2001), and haploinsufficiency of individual ribosomal proteins can lead to non-lethal phenotypic malformations and abnormal translation efficiency (Coelho et al., 2005; Marygold et al., 2005). There are 80 different ribosomal proteins in eukaryotic cells, among them, the ribosomal protein L3-like (RPL3L) is a member of the large subunit ribosomal protein. RPL3L was found to be a muscle-specifically expressed protein in humans and mice. Previous studies have shown that it was specific highly expressed in human muscle (Gupta and Warner, 2014) and a missense mutation in the human RPL3L gene could cause neonatal dilated cardiomyopathy (Ganapathi et al., 2020). The expression of RPL3L could significantly inhibit myotube formation in mouse myoblast C2C12 cells (Chaillou et al., 2016), and its knockdown in mice increases muscle fiber length (Kao et al., 2021). However, whether it plays a role in the growth and development of chicken skeletal muscle is still unknown. From the results of this study, we found that RPL3L has a certain interaction with several genes known to affect the growth and development of chicken skeletal muscle, such as CKMT2, ASB4, and ASB15. In this study, RPL3L was highly expressed in the proliferation and differentiation stages of primary myoblasts, respectively. It was found that RPL3L could inhibit the transcription of ASB4 and ASB15 genes in the cell proliferation stage but promote the transcription of them in the cell differentiation stage. Hence, we speculated that the chicken RPL3L may regulate muscle growth and development by affecting the expression of ASB4 or ASB15.
The number of skeletal muscle fibers is mainly determined during the embryonic stage, and the increase in meat production is mainly due to the increase in muscle cell volume (Egner et al., 2016). Under the regulation of extracellular factors, neonatal skeletal muscle is mainly generated by the activation, proliferation, and differentiation of satellite cells (Morgan and Partridge., 2003; Dumont et al., 2015). Our study found that the RPL3L gene was significantly highly expressed in chicken skeletal muscle, and its expression was significantly increased after birth, which was consistent with the results obtained by previous studies of RPL3L in mammals. By studying its role in muscle satellite cells and myoblasts, it was found that RPL3L can significantly promote cell proliferation and inhibit cell differentiation. This result suggested that the molecular function of chicken RPL3L in skeletal muscle was the same as that of mammals. It shows that RPL3L was indeed a negative regulator gene involved in muscle growth and development. However, the specific signaling pathways through which this gene regulates muscle growth and development remain to be further explored.
As a new generation of molecular genetic markers, indel plays an important role in animal genetics and breeding research (Wang et al., 2013; Zang et al., 2016; Li et al., 2017). Previous studies have been applied to poultry genetics and breeding by mining effective indels of different muscle growth and development-related genes (Ren et al., 2020). We found an interesting indel of 42 bp by screening the genetic sequences of the chicken RPL3L gene in different breeds, and its DD genotype has a significant mutation rate in large chicken breeds. We speculated that it may be somewhat associated with weight-related traits in chickens. Therefore, using the F2 resource population of Xinghua chicken crossing with WRR chicken to conduct an association analysis of multiple economic traits and genotypes with different mutation sites, it was found that the DD genotype had significantly higher performance in weight-related traits, which may be an important weight-related molecular genetic marker. In this mutated region, we identified a loss of at least eight important transcription factor binding sites, which may contribute to the decreased transcriptional activity of RPL3L during transcription in individuals with the DD genotype. Lower transcriptional activity increased muscle mass production due to the negative regulatory role of RPL3L during muscle growth and development. The transcription factor c-Myc has previously been reported to inhibit the differentiation of primary myoblasts (Luo et al., 2019). It is speculated that individuals with the DD genotype have stronger muscle growth and development ability due to the failure of c-Mycbinding because of base deletion, thereby reducing the effect of c-Myc on inhibiting the differentiation of primary myoblasts.
Conclusion
It was found from multiple datasets that RPL3L, FBP2, ASB4, ASB15, CKMT2, PGAM1, YIPF7, PFKM, and LDHA may be key genes affecting chicken muscle development. Among them, 42 bp indel in the RPL3L gene may be one of the molecular markers of chicken weight-related traits. In addition, the RPL3L could promote chicken myoblast proliferation and inhibit chicken myoblast differentiation to participate in chicken muscle development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Protection and Utilization Committee of South China Agricultural University.
Author contributions
XZ, MX, and SL participated in the study design. MX performed bioinformatics analysis and the experiments. SL assisted in bioinformatics analysis and wrote and revised the manuscript. LZ and XZ revised the manuscript. XZ finally agreed to publish it. The remaining authors gave necessary support to the experiments. All authors read and approved the final manuscript.
Funding
This work was supported by the Science and Technology Program of Guangzhou, China (201804020088), the China Agriculture Research System of MOF and MARA (CARS-41), and the Local Innovative and Research Teams Project of Guangdong Province (2019BT02N630).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1033075/full#supplementary-material
Abbreviations
DD, deletion/deletion mutation; DEG, differentially expressed genes; GO, Gene Ontology; II, insertion/insertion mutation; Indel, insertion/deletion mutation; KEGG, Kyoto Encyclopedia of Genes and Genomes; RPL3L, ribosomal protein large 3-like; WRR, White Recessive Rock chickens.
References
Bakshi I., Suryana E., Small L., Quek L. E., Brandon A. E., Turner N., et al. (2018). Fructose bisphosphatase 2 overexpression increases glucose uptake in skeletal muscle. J. Endocrinol. 237, 101–111. doi:10.1530/JOE-17-0555
Bortoluzzi S., d'Alessi F., Romualdi C., Danieli G. A. (2001). Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17, 1152–1157. doi:10.1093/bioinformatics/17.12.1152
Bottje W. G., Lassiter K., Dridi S., Hudson N., Kong B. W. (2017). Enhanced expression of proteins involved in energy production and transfer in breast muscle of pedigree male broilers exhibiting high feed efficiency. Poult. Sci. 96, 2454–2458. doi:10.3382/ps/pew453
Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi:10.1038/nbt.3519
Cassar-Malek I., Boby C., Picard B., Reverter A., Hudson N. J. (2017). Molecular regulation of high muscle mass in developing Blonde d'Aquitaine cattle foetuses. Biol. Open 6, 1483–1492. doi:10.1242/bio.024950
Chaillou T., Zhang X., McCarthy J. J. (2016). Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J. Cell. Physiol. 231, 1894–1902. doi:10.1002/jcp.25294
Chal J., Pourquié O. (2017). Making muscle: Skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122. doi:10.1242/dev.151035
Challagundla K. B., Sun X. X., Zhang X., DeVine T., Zhang Q., Sears R. C., et al. (2011). Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol. Cell. Biol. 31, 4007–4021. doi:10.1128/MCB.05810-11
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi:10.1093/bioinformatics/bty560
CNCB-NGDC Members, and Partners (2022). Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 50, D27–D38. doi:10.1093/nar/gkab951
Coelho C. M., Kolevski B., Walker C. D., Lavagi I., Shaw T., Ebert A., et al. (2005). A genetic screen for dominant modifiers of a small-wing phenotype in Drosophila melanogaster identifies proteins involved in splicing and translation. Genetics 171, 597–614. doi:10.1534/genetics.105.045021
Davis R. V., Lamont S. J., Rothschild M. F., Persia M. E., Ashwell C. M., Schmidt C. J. (2015). Transcriptome analysis of post-hatch breast muscle in legacy and modern broiler chickens reveals enrichment of several regulators of myogenic growth. PLoS One 10, e0122525. doi:10.1371/journal.pone.0122525
Derenzini E., Agostinelli C., Rossi A., Rossi M., Scellato F., Melle F., et al. (2019). Genomic alterations of ribosomal protein genes in diffuse large B cell lymphoma. Br. J. Haematol. 185, 330–334. doi:10.1111/bjh.15442
Dörnen J., Dittmar T. (2021). The role of MSCs and cell fusion in tissue regeneration. Int. J. Mol. Sci. 22, 10980. doi:10.3390/ijms222010980
Dou T., Zhao S., Rong H., Gu D., Li Q., Huang Y., et al. (2017). Biological mechanisms discriminating growth rate and adult body weight phenotypes in two Chinese indigenous chicken breeds. BMC Genomics 18, 469. doi:10.1186/s12864-017-3845-9
Dumont N. A., Bentzinger C. F., Sincennes M. C., Rudnicki M. A. (2015). Satellite cells and skeletal muscle regeneration. Compr. Physiol. 5, 1027–1059. doi:10.1002/cphy.c140068
Egner I. M., Bruusgaard J. C., Gundersen K. (2016). Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development 143, 2898–2906. doi:10.1242/dev.134411
Ehrlich K. C., Lacey M., Ehrlich M. (2020). Epigenetics of skeletal muscle-associated genes in the ASB, LRRC, TMEM, and OSBPL gene families. Epigenomes 4, 1. doi:10.3390/epigenomes4010001
Figueiredo V. C., McCarthy J. J. (2019). Regulation of ribosome biogenesis in skeletal muscle hypertrophy. Physiol. (Bethesda) 34, 30–42. doi:10.1152/physiol.00034.2018
Ganapathi M., Argyriou L., Martínez-Azorín F., Morlot S., Yigit G., Lee T. M., et al. (2020). Bi-allelic missense disease-causing variants in RPL3L associate neonatal dilated cardiomyopathy with muscle-specific ribosome biogenesis. Hum. Genet. 139, 1443–1454. doi:10.1007/s00439-020-02188-6
García M., Pujol A., Ruzo A., Riu E., Ruberte J., Arbós A., et al. (2009). Phosphofructo-1-kinase deficiency leads to a severe cardiac and hematological disorder in addition to skeletal muscle glycogenosis. PLoS Genet. 5, e1000615. doi:10.1371/journal.pgen.1000615
Gros J., Manceau M., Thomé V., Marcelle C. (2005). A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435, 954–958. doi:10.1038/nature03572
Gupta V., Warner J. R. (2014). Ribosome-omics of the human ribosome. RNA 20, 1004–1013. doi:10.1261/rna.043653.113
Han S., Cui C., Wang Y., He H., Liu Z., Shen X., et al. (2019). Knockdown of CSRP3 inhibits differentiation of chicken satellite cells by promoting TGF-β/Smad3 signaling. Gene 707, 36–43. doi:10.1016/j.gene.2019.03.064
Harrison P. W., Ahamed A., Aslam R., Alako B. T. F., Burgin J., Buso N., et al. (2021). The European nucleotide archive in 2020. Nucleic Acids Res. 49, D82–D85. doi:10.1093/nar/gkaa1028
Jin L., Tang Q., Hu S., Chen Z., Zhou X., Zeng B., et al. (2021). A pig BodyMap transcriptome reveals diverse tissue physiologies and evolutionary dynamics of transcription. Nat. Commun. 12, 3715. doi:10.1038/s41467-021-23560-8
Kao B. R., Malerba A., Lu-Nguyen N. B., Harish P., McCarthy J. J., Dickson G., et al. (2021). Knockdown of muscle-specific ribosomal protein L3-like enhances muscle function in healthy and dystrophic mice. Nucleic Acid. Ther. 31, 457–464. doi:10.1089/nat.2020.0928
Kern C., Wang Y., Xu X., Pan Z., Halstead M., Chanthavixay G., et al. (2021). Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 12, 1821. doi:10.1038/s41467-021-22100-8
Khanna N., Sen S., Sharma H., Singh N. (2003). S29 ribosomal protein induces apoptosis in H520 cells and sensitizes them to chemotherapy. Biochem. Biophys. Res. Commun. 304, 26–35. doi:10.1016/s0006-291x(03)00532-1
Kimura N., Tokunaga C., Dalal S., Richardson C., Yoshino K., Hara K., et al. (2003). A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes. cells. 8, 65–79. doi:10.1046/j.1365-2443.2003.00615.x
Komili S., Farny N. G., Roth F. P., Silver P. A. (2007). Functional specificity among ribosomal proteins regulates gene expression. Cell. 131, 557–571. doi:10.1016/j.cell.2007.08.037
Kubota S., Promkhun K., Sinpru P., Suwanvichanee C., Molee W., Molee A. (2021). RNA profiles of the korat chicken breast muscle with increased carnosine content produced through dietary supplementation with β-alanine or L-histidine. Animals. 11, 2596. doi:10.3390/ani11092596
Lara L. J. C., Baião N. C., Cançado S. V., et al. (2005). Influência do peso inicial sobre o desempenho e o rendimento de carcaça e cortes de frangos de corte. Arq. Bras. Med. Veterinária Zootec. 57, 799. doi:10.1590/S0102-09352005000600015
Li W., Liu D., Tang S., Li D., Han R., Tian Y., et al. (2019). A multiallelic indel in the promoter region of the Cyclin-dependent kinase inhibitor 3 gene is significantly associated with body weight and carcass traits in chickens. Poult. Sci. 98, 556–565. doi:10.3382/ps/pey404
Li Y., Chen Y., Jin W., Fu S., Li D., Zhang Y., et al. (2019). Analyses of MicroRNA and mRNA expression profiles reveal the crucial interaction networks and pathways for regulation of chicken breast muscle development. Front. Genet. 10, 197. doi:10.3389/fgene.2019.00197
Li C., Ge M., Yin Y., Luo M., Chen D. (2012). Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol. Cell. Biochem. 370, 127–139. doi:10.1007/s11010-012-1404-x
Li J., Zhu X., Ma L., Xu H., Cao X., Luo R., et al. (2017). Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion 11, 143–150. doi:10.1080/19336896.2017.1300740
Li Y., Zhai B., Yuan P., Fan S., Jin W., Li W., et al. (2022). MiR-29b-1-5p regulates the proliferation and differentiation of chicken primary myoblasts and analysis of its effective targets. Poult. Sci. 101, 101557. doi:10.1016/j.psj.2021.101557
Lin W., Ren T., Li W., Liu M., He D., Liang S., et al. (2021). Novel 61-bp indel of RIN2 is associated with fat and hatching weight traits in chickens. Front. Genet. 12, 672888. doi:10.3389/fgene.2021.672888
Lindström M. S., Zhang Y. (2008). Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 283, 15568–15576. doi:10.1074/jbc.M801151200
Liu J., Lei Q., Li F., Zhou Y., Gao J., Liu W., et al. (2020). Dynamic transcriptomic analysis of breast muscle development from the embryonic to post-hatching periods in chickens. Front. Genet. 10, 1308. doi:10.3389/fgene.2019.01308
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Luo W., Chen J., Li L., Ren X., Cheng T., Lu S., et al. (2019). c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell. Death Differ. 26, 426–442. doi:10.1038/s41418-018-0129-0
Luo W., Li E., Nie Q., Zhang X. (2015). Myomaker, regulated by MYOD, MYOG and miR-140-3p, promotes chicken myoblast fusion. Int. J. Mol. Sci. 16, 26186–26201. doi:10.3390/ijms161125946
Luo W., Luo C., Wang M., Guo L., Chen X., Li Z., et al. (2020). Genome diversity of Chinese indigenous chicken and the selective signatures in Chinese gamecock chicken. Sci. Rep. 10, 14532. doi:10.1038/s41598-020-71421-z
Luo W., Wu H., Ye Y., Li Z., Hao S., Kong L., et al. (2014). The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell. Death Dis. 5, e1347. doi:10.1038/cddis.2014.289
Malila Y., Thanatsang K., Arayamethakorn S., Uengwetwanit T., Srimarut Y., Petracci M., et al. (2019). Absolute expressions of hypoxia-inducible factor-1 alpha (HIF1A) transcript and the associated genes in chicken skeletal muscle with white striping and wooden breast myopathies. PLoS One 14, e0220904. doi:10.1371/journal.pone.0220904
Marygold S. J., Coelho C. M., Leevers S. J. (2005). Genetic analysis of RpL38 and RpL5, two minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics 169, 683–695. doi:10.1534/genetics.104.034124
McDaneld T. G., Hannon K., Moody D. E. (2006). Ankyrin repeat and SOCS box protein 15 regulates protein synthesis in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1672–R1682. doi:10.1152/ajpregu.00239.2005
Morgan J. E., Partridge T. A. (2003). Muscle satellite cells. Int. J. Biochem. Cell. Biol. 35, 1151–1156. doi:10.1016/s1357-2725(03)00042-6
Murach K. A., Englund D. A., Dupont-Versteegden E. E., McCarthy J. J., Peterson C. A. (2018). Myonuclear domain flexibility challenges rigid assumptions on satellite cell contribution to skeletal muscle fiber hypertrophy. Front. Physiol. 9, 635. doi:10.3389/fphys.2018.00635
Pereira M. G., Dyar K. A., Nogara L., Solagna F., Marabita M., Baraldo M., et al. (2017). Comparative analysis of muscle hypertrophy models reveals divergent gene transcription profiles and points to translational regulation of muscle growth through increased mTOR signaling. Front. Physiol. 8, 968. doi:10.3389/fphys.2017.00968
Picard B., Berri C., Lefaucheur L., Molette C., Sayd T., Terlouw C. (2010). Skeletal muscle proteomics in livestock production. Brief. Funct. Genomics 9, 259–278. doi:10.1093/bfgp/elq005
Pourdehnad M., Truitt M. L., Siddiqi I. N., Ducker G. S., Shokat K. M., Ruggero D. (2013). Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. U. S. A. 110, 11988–11993. doi:10.1073/pnas.1310230110
Ren T., Yang Y., Lin W., Li W., Xian M., Fu R., et al. (2020). A 31-bp indel in the 5' UTR region of GNB1L is significantly associated with chicken body weight and carcass traits. BMC Genet. 21, 91. doi:10.1186/s12863-020-00900-z
Schiaffino S., Reggiani C. (2011). Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531. doi:10.1152/physrev.00031.2010
Schneider R. K., Schenone M., Ferreira M. V., Kramann R., Joyce C. E., Hartigan C., et al. (2016). Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 22, 288–297. doi:10.1038/nm.4047
Seki Y., Sato K., Kono T., Akiba Y. (2006). Two types of phosphofructokinase-1 differentially regulate the glycolytic pathway in insulin-stimulated chicken skeletal muscle. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 143, 344–350. doi:10.1016/j.cbpb.2005.12.006
Smith J. H . (1963). Relation of body size to muscle cell size and number in the chicken. Poult. Sci. 42, 283–290. doi:10.3382/ps.0420283
Takahashi K., Kitaoka Y., Matsunaga Y., Hatta H. (2020). Effect of post-exercise lactate administration on glycogen repletion and signaling activation in different types of mouse skeletal muscle. Curr. Res. Physiol. 3, 34–43. doi:10.1016/j.crphys.2020.07.002
Taylor M. V. (2017). Skeletal muscle development on the 30th anniversary of MyoD. Semin. Cell. Dev. Biol. 72, 1–2. doi:10.1016/j.semcdb.2017.11.019
Tona K., Onagbesan O. M., Jego Y., Kamers B., Decuypere E., Bruggeman V. (2004). Comparison of embryo physiological parameters during incubation, chick quality, and growth performance of three lines of broiler breeders differing in genetic composition and growth rate. Poult. Sci. 83, 507–513. doi:10.1093/ps/83.3.507
Townley-Tilson W. H., Wu Y., Ferguson J. E., Patterson C. (2014). The ubiquitin ligase ASB4 promotes trophoblast differentiation through the degradation of ID2. PLoS One 9, e89451. doi:10.1371/journal.pone.0089451
Uprety B., Kaja A., Bhaumik S. R. (2018). TOR facilitates the targeting of the 19S proteasome subcomplex to enhance transcription complex assembly at the promoters of the ribosomal protein genes. Mol. Cell. Biol. 38, 004699-17–e517. doi:10.1128/MCB.00469-17
Velleman S. G., Liu X., Nestor K. E., McFarland D. C. (2000). Heterogeneity in growth and differentiation characteristics in male and female satellite cells isolated from Turkey lines with different growth rates. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 125, 503–509. doi:10.1016/s1095-6433(00)00178-1
Voillet V., San Cristobal M., Père M. C., Billon Y., Canario L., Liaubet L., et al. (2018). Integrated analysis of proteomic and transcriptomic data highlights late fetal muscle maturation process. Mol. Cell. Proteomics 17, 672–693. doi:10.1074/mcp.M116.066357
Wang Y. C., Han R. L., Li Z. J., Geng J., Tian Y. D., Jiang R. R., et al. (2017). Polymorphisms of flanking region of the ASB15 gene and their associations with performance traits in chicken. Anim. Biotechnol. 28, 53–60. doi:10.1080/10495398.2016.1200986
Wang Y. C., Jiang R. R., Kang X. T., Li Z. J., Han R. L., Geng J., et al. (2015). Identification of single nucleotide polymorphisms in the ASB15 gene and their associations with chicken growth and carcass traits. Genet. Mol. Res. 14, 11377–11388. doi:10.4238/2015.September.25.5
Wang Z., Qu L., Yao J., Yang X., Li G., Zhang Y., et al. (2013). An EAV-HP insertion in 5' Flanking region of SLCO1B3 causes blue eggshell in the chicken. PLoS Genet. 9, e1003183. doi:10.1371/journal.pgen.1003183
Warner J. R., McIntosh K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 34, 3–11. doi:10.1016/j.molcel.2009.03.006
Wei C., Niu Y., Chen B., Qin P., Wang Y., Hou D., et al. (2022). Genetic effect of an InDel in the promoter region of the NUDT15 and its effect on myoblast proliferation in chickens. BMC genomics 23, 138. doi:10.1186/s12864-022-08362-6
Wool I. G. (1979). The structure and function of eukaryotic ribosomes. Annu. Rev. Biochem. 48, 719–754. doi:10.1146/annurev.bi.48.070179.003443
Xing S., Liu R., Zhao G., Liu L., Groenen M., Madsen O., et al. (2020). RNA-seq analysis reveals Hub genes involved in chicken intramuscular fat and abdominal fat deposition during development. Front. Genet. 11, 1009. doi:10.3389/fgene.2020.01009
Xu L., Zhao F., Ren H., Li L., Lu J., Liu J., et al. (2014). Co-expression analysis of fetal weight-related genes in ovine skeletal muscle during mid and late fetal development stages. Int. J. Biol. Sci. 10, 1039–1050. doi:10.7150/ijbs.9737
Zang L., Wang Y., Sun B., Zhang X., Yang C., Kang L., et al. (2016). Identification of a 13 bp indel polymorphism in the 3'-UTR of DGAT2 gene associated with backfat thickness and lean percentage in pigs. Gene 576, 729–733. doi:10.1016/j.gene.2015.09.047
Zhang Y., Duc A. C., Rao S., Sun X. L., Bilbee A. N., Rhodes M., et al. (2013). Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev. Cell. 24, 411–425. doi:10.1016/j.devcel.2013.01.018
Keywords: chicken, muscle development, transcriptome, data mining, indel, ribosomal protein L3-like (RPL3L)
Citation: Lin S, Xian M, Ren T, Mo G, Zhang L and Zhang X (2022) Mining of chicken muscle growth genes and the function of important candidate gene RPL3L in muscle development. Front. Physiol. 13:1033075. doi: 10.3389/fphys.2022.1033075
Received: 31 August 2022; Accepted: 14 October 2022;
Published: 03 November 2022.
Edited by:
Takeshi Ohkubo, Ibaraki University, JapanReviewed by:
Yanfa Sun, Longyan University, ChinaZhigang Song, Shandong Agricultural University, China
Monika Proszkowiec-Weglarz, Agricultural Research Service (USDA), United States
Copyright © 2022 Lin, Xian, Ren, Mo, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiquan Zhang, eHF6aGFuZ0BzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Shudai Lin
Shudai Lin Mingjian Xian
Mingjian Xian Tuanhui Ren
Tuanhui Ren Guodong Mo
Guodong Mo Li Zhang
Li Zhang Xiquan Zhang
Xiquan Zhang