
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 09 November 2022
Sec. Cell Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1008364
This article is part of the Research Topic Interplay of Environmental Factors, Genetic, and Epigenetic in Aging-related Diseases View all 7 articles
Circular RNAs (circRNAs) are a new type of non-coding RNAs originating from precursor messenger RNAs. Recent research has confirmed that circRNAs play a significant role in various biological and pathological processes, including cell viability, migration, and apoptosis. Emerging studies have demonstrated that the deregulated circRNA–miRNA–mRNA interaction network plays a key role in the development of many diseases. Increasing evidence has highlighted the role of ncRNAs (mainly miRNAs and lncRNAs) in the pathogenesis of keloids. Recently, several publications also indicated that circRNAs contribute to keloid development. The discovery of circRNAs changed the current understanding of the biology of keloids It is crucial to elucidate a circRNA–miRNA–mRNA network to understand the pathological mechanism of keloids. In the present review, we summarize the aberrant expression of regulatory roles of circRNAs in keloids. We discuss the potential clinical application of circRNAs in the diagnosis and treatment of keloids.
Keloids are pathological scars caused by skin injury and irritation. These are benign skin tumors characterized by hyperproliferation of fibroblasts and excessive deposition of collagen fibers (Limandjaja et al., 2020). Keloids can extend beyond the area of injury and invade the adjacent normal skin (Tan et al., 2019). Keloids significantly reduce people’s life quality by causing esthetic deformity, pruritus, hyperesthesia, and pain (Olaitan, 2009). Conventional treatment ways, consisting of surgical excision, cryotherapy, topical steroids, and laser therapy, remain unsatisfactory, and relapse is common (Park et al., 2014; Berman et al., 2017; Ekstein et al., 2021). It is urgent to understand the mechanism underlying keloid formation for better treatment approaches.
Circular RNAs (circRNAs) are a new type of non-coding RNAs originating from precursor messenger RNAs. Different from linear RNA, circRNAs are single-stranded RNAs that form a closed loop. CircRNAs lack 5′ and 3’ ends, which makes them more stable (Jeck et al., 2013; Kristensen et al., 2019). Since circRNAs were first discovered in plant viroids in 1976, thousands of circRNAs across species have been identified (Sanger et al., 1976; Vo et al., 2019). CircRNAs were considered the byproduct of wrong splicing for a long time. Recent research has confirmed that circRNAs play a significant role in various biological and pathological processes, including cell viability, migration, and apoptosis (Memczak et al., 2013; Di Timoteo et al., 2020). CircRNAs exert their biological functions at the post-transcriptional level, including in transcription and splicing, interfering with miRNA activities or signaling pathways, and serving as translation templates (Chen, 2020). In particular, circRNAs contain various binding sites for miRNAs and absorb miRNAs like a sponge, serving as competitive endogenous RNAs (ceRNAs) or miRNA sponges (Memczak et al., 2013). Emerging studies have demonstrated that the deregulated circRNA–miRNA–mRNA interaction network plays a key role in the development of many diseases (Liang et al., 2020; Sakshi et al., 2021).
Increasing evidence has highlighted the role of ncRNAs (mainly miRNAs and lncRNAs) in the pathogenesis of keloids (Babalola et al., 2013). Recently, several publications also indicated that circRNAs contribute to keloid development. The discovery of circRNAs changed the current understanding of the biology of keloids. It is crucial to elucidate a circRNA–miRNA–mRNA network to understand the pathological mechanism of keloids. In the present review, we summarize the aberrant expression of regulatory roles of circRNAs in keloids. We discuss the potential clinical application of circRNAs in the diagnosis and treatment of keloid.
Genetic, epigenetic, and environmental factors are crucial in molecular pathogenesis underlying keloid formation (He et al., 2017). The inheritance pattern of keloids may follow autosomal dominance with incomplete penetrance. In addition, keloids tend to be polygenic, not following a simple Mendelian monogenic manner. Individuals with dark skin color show an increased prevalence of keloids with an estimated incidence of 4%–16%, 15 times higher than in Caucasian populations. NEDD4 was proven to be a candidate gene in Chinese Han and Japanese populations (Fujita et al., 2019). Epigenetic modifications are composed of DNA methylations, histone modifications, and non-coding RNA regulations. Non-coding RNAs are mainly composed of three types, microRNAs—miRNAs, long non-coding RNAs—lncRNAs, and circular RNAs—circRNAs. Emerging research showed that epigenetics plays a crucial role in the molecular pathogenesis of keloids. Epigenetic modification is considered to be an important regulator in the initial and sustained activation of keloid fibroblasts (Lv et al., 2020).
High-throughput sequencing and gene microarray have demonstrated aberrant expression profiles of circRNAs in keloid tissue and fibroblasts (Table 1). The altered expression profiles of circRNAs in keloid tissue may contribute to the etiology and pathophysiology of keloids by impacting signaling pathways relevant to the scaring process.
In a high-throughput sequencing research, Zhang et al. identified 411 differentially expressed (DE) circRNAs performed in three HKFs and normal dermal fibroblasts, with 206 circRNAs upregulated and 205 circRNAs downregulated. Bioinformatics analyses showed that 411 DE circRNAs mainly participated in cell apoptosis and focal adhesion processes, as well as PI3K-Akt, Rap1, and metabolic signaling pathways (Zhang et al., 2020a). Shi et al. used a circRNA microarray assay to determine circRNA expression in keloid tissue compared with paired normal skin tissue. They showed that 52 circRNAs were upregulated and 24 downregulated in keloids (Shi et al., 2020). In addition, further analysis found that circRNAs could interact with miR-29a, miR-23a-5p, and miR-1976 (Shi et al., 2020). Wang et al. performed high-throughput sequencing research in keloid tissue compared with normal skin tissue. Among 154 DE circRNAs, 81 circRNAs were upregulated and 73 circRNAs were downregulated (Wang et al., 2019). Li et al. performed high-throughput sequencing and showed that circRNAs might act as ceRNAs in the development of human hypertrophic scars (Li et al., 2018). Pang et al. performed microarray technology in four patient-derived keloid dermal fibroblasts (KDFs) compared with normal dermal fibroblasts (NDFs) (Pang et al., 2022). They detected a total of 327 DE circRNAs, with 195 upregulated and 132 downregulated circRNAs. The DE circRNAs were mainly enriched in cell function of the cytoskeleton, tight junctions, axonal guidance, and morphogenesis of the epithelium.
Myofibroblasts derived from quiescent resident skin fibroblasts are the principal cell type responsible for extracellular matrix (ECM) accumulation. The imbalance between fibroblast proliferation and apoptosis is the cytological basis for the continuous proliferation of keloids, and it highlights the epigenetic contribution to keloid formation by modulating the balance between fibroblast proliferation and apoptosis.
Lv et al. showed that circCOL5A1 was upregulated in keloid tissues and HKFs. Silencing of circCOL5A1 inhibited HKF proliferation, migration, and ECM deposition and promoted the rate of apoptosis. Moreover, circCOL5A1 sponges miR-7-5p to release Epac1 through the PI3K/Akt signaling pathway (Lv et al., 2021). Similarly, Jiao et al. showed that circCOL5A1 expression is obviously higher in keloid tissues and HKFs. CircCOL5A1 knockdown hindered HKF proliferation, invasion migration, and ECM deposition, while promoting the rate of cell apoptosis. Moreover, circCOL5A1 could upregulate the expression level of EGR1 via sponging miR-877–5p (Jiao et al., 2022). Liu et al. found increased hsa_circ_0043688 and FGF2 and decreased miR-145-5p in human keloid samples and HKFs using RT-qPCR. Functional analysis showed that silencing of hsa_circ_0043688 repressed HKF proliferation, invasion, and migration and promoted apoptosis. Collectively, hsa_circ_0043688 modulated keloid progression via miR-145-5p/FGF2 (Liu et al., 2022a). CircPDE7B was highly expressed in keloid samples and HKFs. High circPDE7B accelerates HKF proliferation, migration, and invasion and hindered the rate of apoptosis. Moreover, circPDE7B functioned as a ceRNA for miR-661. The circPDE7B/miR-661/FGF2 ceRNA regulatory axis plays crucial roles in the pathogenesis of keloids (Wu et al., 2022). YANG et al. showed that circ_101,238 was significantly increased in keloid samples. Circ_101,238 was proven to sponge miR-138-5p, with CDK6 as a target. Transfection with sh-circ_101,238 inhibited HKF proliferation, while promoting apoptosis via regulating the miR-138-5p/CDK6 pathway (Yang et al., 2020). Runt-related transcription factors (Runx) play critical roles in the development and cancers (Date and Ito, 2020). Knockdown of circ_0008450 may reduce cell proliferation, migration, and EMT process of human keratinized epithelial cells and promoted apoptosis through increasing Runx3 and repressing the TGF-β/Smad signal pathway (Chen et al., 2020). The TGF-β/Smad signaling pathway is the most crucial pathway involved in the excessive production of collagen in the fibroblasts and myofibroblasts (Zhang et al., 2020b). WANG et al. demonstrated circNRIP1 was higher in keloid tissue than in adjacent skin tissue. Absence of circNRIP1 inhibited the proliferation and ECM-associated protein production while increasing apoptosis in HKFs. CircNRIP1 maintained FXR1 stability by inhibiting ubiquitination and degradation of FXR1, which increased the expression of miR-503-3p and miR-503-5p. In summary, circNRIP1 contributes to keloid development via FXR1-mediated upregulation of miR-503-3p and miR-503-5p (Wang et al., 2021). Gao et al. demonstrated that hsa_circ_0057452 and AFF4 are remarkably higher in keloids than in matched normal skin tissues. Hsa_circ_0057452 knockdown suppressed cell proliferation, viability, and migration, while accelerating the rate of apoptosis of HKFs. MiR-1225-3p is downregulated and showed a reverse effect on HKF function. Collectively, hsa_circ_0057452 regulates AFF4 and promotes keloid formation by sponging miR-1225-3p (Gao et al., 2022). Zhu et al. showed that circ_005745 induced keloid progression via upregulating GAB1 (Zhu et al., 2022).
Yuan et al. showed that the level of circSLC8A1 declined in keloid tissues and HKFs. Overexpression of circSLC8A1 inhibited cell proliferation, migration, and ECM production and elevated cell apoptosis of HKFs. MiR-181a-5p is a direct sponging target of circSLC8A1, and HIF1AN was the downstream effect factor of miR-181a-5p. Taken together, circSLC8A1 inhibited keloid progression by regulating the miR-181a-5p/HIF1AN axis (Yuan et al., 2022). CircPTPN12 expression was downregulated in keloid tissue compared with the adjacent normal skin. Silencing of circPTPN12 accelerated HKF proliferation, migration, and invasion and suppressed apoptosis. CircPTPN12 could sponge miR-21-5p, while SMAD7 was the downstream effect factor of miR-21-5p. MiR-21-5p was a direct target of circPTPN12. In summary, silencing of circPTPN12 promotes keloid formation by activating the Wnt pathway sponging miR-21-5p (Liu et al., 2022b) (Figure 1 and Table 1).
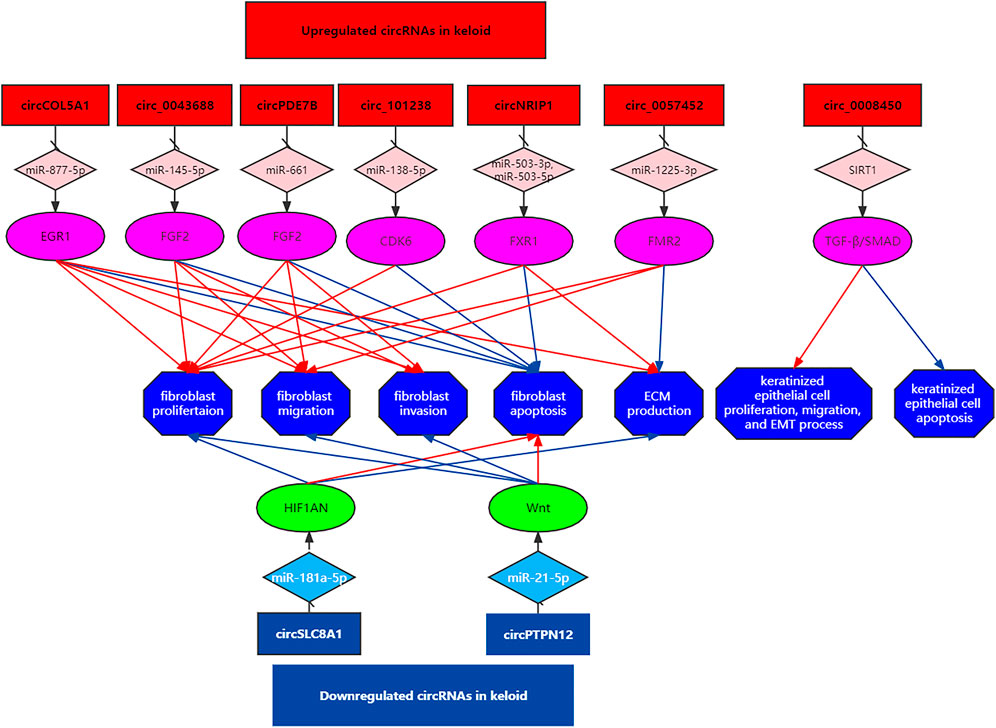
FIGURE 1. CircRNAs regulated gene expression via sponging miRNAs and played crucial roles in keloid development.
Some circRNAs are profibrotic and their upregulation contributed to the development of keloids. Those profibrotic circRNAs include circCOL5A1、circ_0043688、circPDE7B、circ_101,238、circ_0008450、circNRIP1, and circ_0057452. However, some circRNAs are anti-fibrotic and their reduction inhibits the development of keloids. Those anti-fibrotic circRNAs include circSLC8A1 and circPTPN12. Gain- and loss-of-function studies have proven that deregulated circRNAs may regulate the processes underlying keloid formation and development.
CircRNAs are expected to be a potential diagnostic and therapeutic target in the management of keloids. For example, si-circCOL5A1 inhibited the growth and ECM deposition of keloids in the skin of nude mice (Lv et al., 2021). Further investigation into keloid-related circRNAs is needed to identify more effective prophylactic and clinical treatment strategies for keloids.
XY, XZ, LL, and GG conceptualized the study and analyzed the data. XY has carried out the bioinformatics assay. XY and LL revised the manuscript.
This work is supported by National Natural Science Foundation of China (82270051), Reform and Development Program of Beijing Institute of Respiratory Medicine (ysrh2022006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Babalola O., Mamalis A., Lev-Tov H., Jagdeo J. (2013). The role of microRNAs in skin fibrosis. Arch. Dermatol. Res. 305 (9), 763–776. Epub 2013/09/12. doi:10.1007/s00403-013-1410-1
Berman B., Maderal A., Raphael B. (2017). Keloids and hypertrophic scars: Pathophysiology, classification, and treatment. Dermatol. Surg. 43, S3. doi:10.1097/DSS.0000000000000819
Chen H., Xu X., Lai L., Huo R., Chen M. (2020). Circ_0008450 downregulates Runx3 to promote the proliferation and epithelial-mesenchymal transition of human keratinized epithelial cells. Cell Cycle 19 (23), 3303–3316. Epub 2020/11/03. doi:10.1080/15384101.2020.1842665
Chen L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21 (8), 475–490. Epub 2020/05/06. doi:10.1038/s41580-020-0243-y
Date Y., Ito K. (2020). Oncogenic RUNX3: A link between p53 deficiency and myc dysregulation. Mol. Cells 43 (2), 176–181. Epub 2020/01/30. doi:10.14348/molcells.2019.0285
Di Timoteo G., Rossi F., Bozzoni I. (2020). Circular RNAs in cell differentiation and development. Development 147 (16), dev182725. Epub 2020/08/26. doi:10.1242/dev.182725
Ekstein S. F., Wyles S. P., Moran S. L., Meves A. (2021). Keloids: A review of therapeutic management. Int. J. Dermatol. 60 (6), 661–671. Epub 2020/09/10. doi:10.1111/ijd.15159
Fujita M., Yamamoto Y., Jiang J. J., Atsumi T., Tanaka Y., Ohki T., et al. (2019). NEDD4 is involved in inflammation development during keloid formation. J. Invest. Dermatol. 139 (2), 333–341. Epub 2018/10/03. doi:10.1016/j.jid.2018.07.044
Gao H., Hu Z., Zhang X. (2022). Circular RNA hsa_circ_0057452 facilitates keloid progression by targeting the microRNA-1225-3p/AF4/FMR2 family member 4 axis. Bioengineered 13 (5), 13815–13828. Epub 2022/06/17. doi:10.1080/21655979.2022.2084460
He Y., Deng Z., Alghamdi M., Lu L., Fear M. W., He L. (2017). From genetics to epigenetics: New insights into keloid scarring. Cell Prolif. 50 (2), e12326. Epub 2017/01/06. doi:10.1111/cpr.12326
Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 (2), 141–157. Epub 2012/12/20. doi:10.1261/rna.035667.112
Jiao H., Ji G., Luo B., Chen C. (2022). CircCOL5A1 inhibits proliferation, migration, invasion, and extracellular matrix production of keloid fibroblasts by regulating the miR-877-5p/EGR1 axis. Burns 2022, 35184918. doi:10.1016/j.burns.2021.12.013
Kristensen L. S., Andersen M. S., Stagsted L. V. W., Ebbesen K. K., Hansen T. B., Kjems J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. Epub 2019/08/10. doi:10.1038/s41576-019-0158-7
Li M., Wang J., Liu D., Huang H. (2018). Highthroughput sequencing reveals differentially expressed lncRNAs and circRNAs, and their associated functional network, in human hypertrophic scars. Mol. Med. Rep. 18 (6), 5669–5682. Epub 2018/10/16. doi:10.3892/mmr.2018.9557
Liang Z. Z., Guo C., Zou M. M., Meng P., Zhang T. T. (2020). circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 20, 173. Epub 2020/05/30. doi:10.1186/s12935-020-01245-4
Limandjaja G. C., Niessen F. B., Scheper R. J., Gibbs S. (2020). The keloid disorder: Heterogeneity, histopathology, mechanisms and models. Front. Cell Dev. Biol. 8, 360. Epub 2020/06/13. doi:10.3389/fcell.2020.00360
Liu F., Li T., Zhan X. (2022). Silencing circular RNAPTPN12 promoted the growth of keloid fibroblasts by activating Wnt signaling pathway via targeting microRNA-21-5p. Bioengineered 13 (2), 3503–3515. Epub 2022/01/25. doi:10.1080/21655979.2022.2029108
Liu Y., Wang X., Ni Z., Li Y., Song J., Zhu F., et al. (2022). Circular RNA hsa_circ_0043688 serves as a competing endogenous RNA for microRNA-145-5p to promote the progression of Keloids via Fibroblast growth factor-2. J. Clin. Lab. Anal. 36, e24528. Epub 2022/06/28. doi:10.1002/jcla.24528
Lv W., Liu S., Zhang Q., Hu W., Wu Y., Ren Y. (2021). Circular RNA CircCOL5A1 sponges the MiR-7-5p/epac1 Axis to promote the progression of keloids through regulating PI3K/akt signaling pathway. Front. Cell Dev. Biol. 9, 626027. Epub 2021/02/09. doi:10.3389/fcell.2021.626027
Lv W., Ren Y., Hou K., Hu W., Yi Y., Xiong M., et al. (2020). Epigenetic modification mechanisms involved in keloid: Current status and prospect. Clin. Epigenetics 12 (1), 183. Epub 2020/11/28. doi:10.1186/s13148-020-00981-8
Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–338. Epub 2013/03/01PubMed PMID: 23446348. doi:10.1038/nature11928
Olaitan P. B. (2009). Keloids: Assessment of effects and psychosocial-impacts on subjects in a black african population. Indian J. dermatol. Venereol. Leprol. 75 (4), 368–372. Epub 2009/07/09. doi:10.4103/0378-6323.53132
Pang Q., Lin X., Sun J., Hu J., Dai S., Shen Y., et al. (2022). Comprehensive analysis of circular RNA expression in ceRNA networks and identification of the effects of hsa_circ_0006867 in keloid dermal fibroblasts. Front. Mol. Biosci. 9, 800122. doi:10.3389/fmolb.2022.800122
Park T. H., Park J. H., Chang C. H. (2014). Are auricular keloids and persistent hypertrophic scars resectable? The role of intrascar excision. Ann. Plast. Surg. 72 (3), 369. Epub 2013/05/10. doi:10.1097/SAP.0b013e31827f5131
Sakshi S., Jayasuriya R., Ganesan K., Xu B., Ramkumar K. M. (2021). Role of circRNA-miRNA-mRNA interaction network in diabetes and its associated complications. Mol. Ther. Nucleic Acids 26, 1291–1302. Epub 2021/12/03. doi:10.1016/j.omtn.2021.11.007
Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. Epub 1976/11/01. doi:10.1073/pnas.73.11.3852
Shi J., Yao S., Chen P., Yang Y., Qian M., Han Y., et al. (2020). The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol. Biol. Rep. 47 (1), 201–209. Epub 2019/10/16. doi:10.1007/s11033-019-05120-y
Tan S., Khumalo N., Bayat A. (2019). Understanding keloid pathobiology from a quasi-neoplastic perspective: Less of a scar and more of a chronic inflammatory disease with cancer-like tendencies. Front. Immunol. 10, 1810. Epub 2019/08/24. doi:10.3389/fimmu.2019.01810
Vo J. N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176 (4), 869–881. e13Epub 2019/02/09. doi:10.1016/j.cell.2018.12.021
Wang B., Yin H., Zhang H., Wang T. (2021). circNRIP1 facilitates keloid progression via FXR1mediated upregulation of miR5033p and miR5035p. Int. J. Mol. Med. 47 (5), 70. Epub 2021/03/03. doi:10.3892/ijmm.2021.4903
Wang J., Wu H., Xiao Z., Dong X. (2019). Expression profiles of lncRNAs and circRNAs in keloid. Plast. Reconstr. Surg. Glob. Open 7 (6), e2265. Epub 2019/10/19. doi:10.1097/GOX.0000000000002265
Wu F., He H., Chen Y., Zhu D., Jiang T., Wang J. (2022). CircPDE7B/miR-661 axis accelerates the progression of human keloid fibroblasts by upregulating fibroblast growth factor 2 (FGF2). Mol. Cell. Biochem. 477 (4), 1113–1126. Epub 2022/01/27. doi:10.1007/s11010-021-04345-5
Yang D., Li M., Du N. (2020). Effects of the circ_101238/miR-138-5p/CDK6 axis on proliferation and apoptosis keloid fibroblasts. Exp. Ther. Med. 20 (3), 1995–2002. Epub 2020/08/13. doi:10.3892/etm.2020.8917
Yuan X., Chen B., Wang X. (2022). CircSLC8A1 targets miR-181a-5p/HIF1AN pathway to inhibit the growth, migration and extracellular matrix deposition of human keloid fibroblasts. Burns 2022, 35610079. doi:10.1016/j.burns.2022.04.009
Zhang T., Wang X. F., Wang Z. C., Lou D., Fang Q. Q., Hu Y. Y., et al. (2020). Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed. Pharmacother. = Biomedecine Pharmacother. 129, 110287. Epub 2020/06/17. doi:10.1016/j.biopha.2020.110287
Zhang Z., Yu K., Liu O., Xiong Y., Yang X., Wang S., et al. (2020). Expression profile and bioinformatics analyses of circular RNAs in keloid and normal dermal fibroblasts. Exp. Cell Res. 388 (1), 111799. Epub 2020/01/07. doi:10.1016/j.yexcr.2019.111799
Keywords: circular RNAs, circRNAs, keloid, miRNA, mRNA
Citation: Yu X, Zhu X, Li L and Gao G (2022) Circular RNAs: Emerging players in the pathogenesis of keloid. Front. Physiol. 13:1008364. doi: 10.3389/fphys.2022.1008364
Received: 31 July 2022; Accepted: 15 September 2022;
Published: 09 November 2022.
Edited by:
Lifeng Lao, Shanghai Jiao Tong University, ChinaReviewed by:
Cheng Huang, China-Japan Friendship Hospital, ChinaCopyright © 2022 Yu, Zhu, Li and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linfeng Li, ZmdyM3I0QHllYWgubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.