- 1Ganzhou Key Laboratory of Nanling Insect Biology/Ganzhou Key Laboratory of Greenhouse Vegetables/National Navel Orange Engineering Research Center, College of Life Sciences, Gannan Normal University, Ganzhou, Jiangxi, China
- 2Entomology and Nematology Department, University of Florida, Gainesville, FL, United States
The Asian citrus psyllid, Diaphorina citri, is a notorious pest that is an efficient vector for Candidatus Liberibacter asiaticus (CLas), the causal agent of citrus huanglongbing (HLB). The olfactory system of insects is crucial for foraging and mating behavior. Antennae-abundant odorant degrading enzymes (ODEs), including cytochrome P450 (CYPs), are important in degrading redundant odorant molecules to recover the insect olfactory. In this study, to isolate the antennal CYP genes of D. citri, we generated four transcriptomes from female/male antennae and body through deep sequencing of RNA libraries. Seven DcCYP genes preferentially expressed in antennae were first identified by comparing the antennal and body transcriptomes. Phylogenetic analysis grouped four DcCYPs (DcCYP6a13, DcCYP6j1, DcCYP6k1, and DcCYP6a2) into the CYP3 class, whereas DcCYP4d2, DcCYP4c62, and DcCYP4d8 were clustered in the CYP4 clade. qRT-PCR analyses across developmental stages and tissues showed they were antennae-abundant in both genders and constantly expressed from the first instar nymph to the adult. The results presented here highlight the isolation and expression of CYP genes in D. citri antennae, providing valuable insights into their putative role in odorant degradation.
Introduction
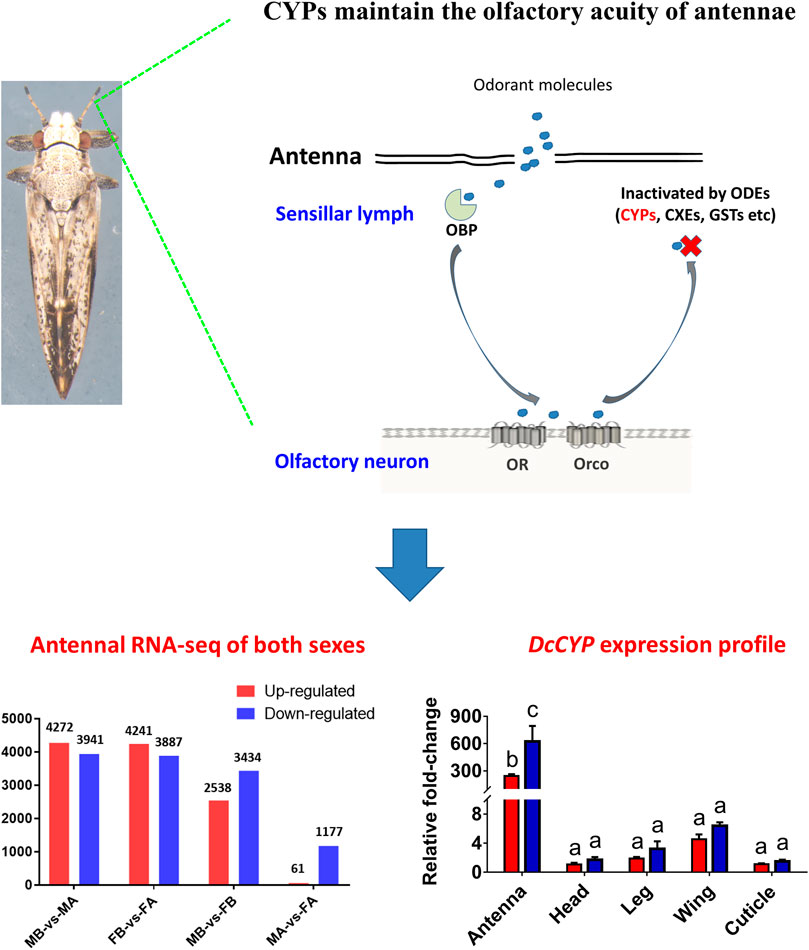
Insect antennae are complex sensory organs that detect various volatile compounds for securing food, mating, and oviposition. The antennal system in insects has been intensively studied using various electrophysiological, molecular, and omics techniques, providing a broad range of biological insights into olfactory perception (Oh et al., 2019; Ahn et al., 2020). The biologic processes of olfactory perception involve interactions between exogenous odor molecules and various families of chemoreception-related proteins, including the odorant-binding proteins (OBPs), chemosensory proteins (CSPs), olfactory receptors (ORs), and odorant-degrading enzymes (ODEs) (Pelosi, 1996; Brito et al., 2016; Fleischer et al., 2018). OBPs and CSPs are small soluble proteins proposed to selectively trap and transport hydrophobic odorant molecules through the sensillum lymph towards the dendrites of olfactory sensory neurons, where ORs are located on the membrane surface (Brito et al., 2016; Cheema et al., 2021; Zhou and Jander, 2022). ORs act as a biotransducer to convert chemical signals into electrophysiological signals. Upon ORs are successfully activated, the odor molecules must be rapidly removed by ODEs, allowing the insect olfactory system to recover and maintain the olfactory acuity (Fleischer et al., 2018; Li F et al., 2018).
ODEs play an integral role in insect chemoreception and they prevent the accumulation of stimulants and subsequent sensory adaptation (Vogt and Riddiford 1981; Younus et al., 2014). Insect ODEs have been mostly known for their involvement in the metabolism of endogenous hormones and steroids, as well as exogenous xenobiotics and allelochemicals (Younus et al., 2014; Blomquist et al., 2021). They include a few antenna-specific or antenna-enriched cytochrome P450 (CYPs), glutathione S-transferases (GSTs), carboxylesterases (CXEs), carboxyl/cholinesterases (CCEs) and UDP-glycosyltransferases (UGTs) (Rogers et al., 1999; Maïbèche-Coisne et al., 2002; Durand et al., 2010a; Durand et al., 2010b; Keeling et al., 2013; Bozzolan et al., 2014; Tan et al., 2014; Younus et al., 2014; He et al., 2015; Li F et al., 2018; Blomquist et al., 2021; Wang et al., 2021; Wei et al., 2021). The olfactory-specific GSTs protect the olfactory system from harmful xenobiotics and inactivate the components of sex pheromone in the Manduca sexta and Bombyx mori (Rogers et al., 1999; Tan et al., 2014). In the Oriental fruit moth Grapholita molesta, four antenna-enriched CXEs are found to modulate foraging and mating behaviors, by hydrolyzing the acetate sex pheromone components (Z/E)-8-dodecenyl and the ester host plant volatiles (Wei et al., 2021).
CYPs represent an important supergene family of detoxification enzymes widely occurring in vertebrates and invertebrates (Nauen et al., 2022). The first antennae-specific CYP, CYP345E2, is functionally characterized in the pine beetle Dendroctonus ponderosae. This CYP enzyme catalyzes the oxidation of monoterpene pine host volatiles (Keeling et al., 2013). Subsequent studies in D. ponderosae document that antennae-abundant CYP6DE1, CYP6DJ1, CYP6BW1 and CYP6BW3, could oxidize and remove terpenoids from antennae, as well as detoxify host terpenoids to overcome plant defenses, in which some of the terpenoid detoxification products are used as pheromones by both sexes (Chiu et al., 2019a; Chiu et al., 2019b; Chiu et al., 2019c; Blomquist et al., 2021). Examples include antennal CYP6DE1 catalyzes the conversion of α-pinene into trans-verbenol, an aggregation pheromone released by female D. ponderosae; CYP6DJ1 oxidizes terpinolene and limonene to alcohols and an epoxide, CYP6BW1 and CYP6BW3 oxidize several diterpene resin acids to cope with host defenses (Chiu et al., 2019a; Chiu et al., 2019b; Chiu et al., 2019c). Recent years, with antennal transcriptome analysis, some antennal CYPs have been identified in Locusta migratoria (Wu H et al., 2020) and Drosophila (Younus et al., 2014; Baldwin et al., 2021).
The Asian citrus psyllid Diaphorina citri Kuwayama serves as an efficient insect vector of huanglongbing (HLB) in citrus production. This disease presents an unprecedented challenge to citrus production by reducing fruit yield and quality (Yu and Killiny, 2020; Zhou, 2020). Foliar application of chemical pesticides is the major strategy for controlling D. citri. This is challenged by the fact that resistance development has been reported in many citrus-growing areas (Chen et al., 2018). A potential alternative and effective strategy for insect pest management via RNA interfere (RNAi) technique requires the selection of optimal target genes (Yu et al., 2016; Yu and Killiny, 2020). Antennae-abundant ODEs such as CYPs hold a great promise for the potential application of RNAi, in which DcCYP-silenced insects are expected to exhibit decreased or disordered mating and foraging behaviors.
The availability of genomic and transcriptomic data facilitated the identification of OBPs and CSPs genes in D. citri (Wu et al., 2016; Zhang et al., 2020; Liu et al., 2021). However, to our knowledge, no DcCYPs have been reported to contribute to odorant degradation. To identify DcCYPs correlated with odorant processing in this study, we performed as follows: 1) a comparative transcriptome analysis of antennae and body from both female and male; 2) isolation and in silico analysis of DcCYPs abundant in antennae; 3) identification of the expression profile of candidate DcCYP genes among tissues and developmental stages.
Materials and methods
Insect rearing
A colony of D. citri was collected in 2015 using field populations from Nankang District, Jiangxi (Yu et al., 2022). The culture was continuously maintained on Murraya paniculata seedlings in insect rearing cages kept at 27–28°C and relative humidity of 60–65% with a 14:10 h light: dark photoperiod.
Sample preparation and RNA extraction
For antennae collection, adult D. citri were collected using an aspirator and anesthetized with CO2 for sex separation based on the appearance of their abdomen (Yu and Killiny, 2018). Using fine forceps, a pool of approximately 500 antennae was carefully dissected from males and females, respectively. The whole body of twenty males and females without antennae was prepared as the control. Insect samples were dissected on ice under a stereomicroscope and stored at −80°C until RNA extraction. Total RNAs were isolated from the D. citri tissues using Trizol (Sigma, St. Louis, MO, United States), and quantified using a NanoDrop OneC spectrophotometer (Thermo Fisher Scientific, MA, United States). The integrity of RNA was assessed on 1% agarose gels.
cDNA library construction and sequencing
cDNA library construction and Illumina sequencing of D. citri samples were performed at Huada Gene Sequencing Center, Wuhan, China. Briefly, Oligo (dT)-attached magnetic beads were used to purify mRNA. Purified mRNA was fragmented into small pieces to synthesize first-strand cDNA using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis. After end-repair and ligation of adaptors, the products were amplified by PCR. The double-stranded PCR products were heated, denatured and circularized by the splint oligo sequence to create a cDNA library. Pair-end sequencing of the cDNA library was performed on a BGIseq500 platform (BGI-Wuhan, China).
De novo transcriptome data processing
RNA-seq was carried out on pools of antennae and bodies from both sexes with three biological replicates. The sequencing data was filtered to obtain clean reads with SOAPnuke (v1.5.2) by removing low-quality reads (i.e., low-quality base ratio > 20%) and reads with adaptor sequences and/or unknown nucleotides (>5%). Clean reads were aligned to the reference genome [Diaci version 3.0 (Hosmani et al., 2019; Yu et al., 2022)] using HISAT2 (version 2.0.4). Transcriptome de novo assembly and functional annotation were conducted in BGI Company as described by Yi et al. (2021). Transcript abundances were calculated by RSEM (version 1.2.12), and differentially expressed genes among samples were determined using DESeq2 (version 1.4.5) with Q value ≤0.05.
Identification and phylogenetic analysis of antennal DcCYP genes
The functional annotation of differentially expressed unigenes or contigs was performed by BLAST algorithm with a cut-off E-value of 10–5 in public databases, including NCBI-Nr (http://ftp.ncbi.nlm.nih.gov/blast/db) and Swiss-Prot (http://ftp.ebi.ac.uk/pub/databases/swissprot). The open reading frames (ORFs) of putative antennal DcCYP genes were predicted using the ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). DcCYP sequences were submitted to Pfam database (https://pfam.xfam.org/) and SMART (http://smart.embl.de/) to predict the conserved domain. The number of amino acids, molecular weights (MWs), and theoretical isoelectric points (pIs) of DcCYPs were calculated on ExPASy (http://web.expasy.org/protparam/). The putative N-terminal signal peptides (SPs) of deduced DcCYP proteins were predicted using the SignalP 6.0 server (https://services.healthtech.dtu.dk/service.php?SignalP-6.0). The DcCYP genomic sequences were retrieved from citrusgreening.org. The exon-intron structure was determined on the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) by comparing the full-length ORF sequence with the corresponding genomic DNA sequence. The antennal DcCYPs and CYPs of Acyrthosiphon pisum were used for the phylogenetic analysis (Ramsey et al., 2010; Supplementary Table S1). The alignment of CYP protein sequences was performed using CLUSTAL_X (version 1.83). The joint unrooted phylogenetic tree was constructed with MEGA11 using the maximum likelihood method with 1,000 bootstrap replicates (Tamura et al., 2021).
Quantitative real-time PCR analysis
To examine the expression of DcCYP genes, a variety of D. citri tissues (antenna, head, leg, wing and cuticle) and samples of D. citri at six stages, including the first instar nymph, second instar nymph, third instar nymph, fourth instar nymph, fifth instar nymph, as well as adult male and female were collected separately. First-strand cDNA synthesis was initiated with 500–1,000 ng of purified RNA using the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). The synthesized cDNA was stored at −20 C until use.
qRT-PCR reactions were carried out in a 20 μl volume: 4 μl of diluted cDNA, 0.4 μM of each primer and 10 μl of PerfectStart® Green qPCR SuperMix (TransGen Biotech, Beijing, China). All the samples were placed in a Roche LightCycler 96® system (Roche Diagnostics, Mannheim, Germany). Two reference genes, actin (GenBank accession number DQ675553) and GAPDH (GenBank accession number XM_017447140), were used to normalize the amount of cDNA added to the PCR reactions. The relative value of DcCYP gene expression was analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001). Specific primers for each gene were listed in Supplementary Table S2. Three replicates were performed for each treatment.
Statistical analysis
To compare the expression level of DcCYP gene among different tissues and developmental stages, the ANOVA with Tukey’s multiple comparisons test was performed using GRAPHPAD PRISM version 6.0 (GraphPad Software Inc., La Jolla, CA, United States). A value of p < 0.05 was considered statistically significant.
Results
Transcriptome sequencing and analysis
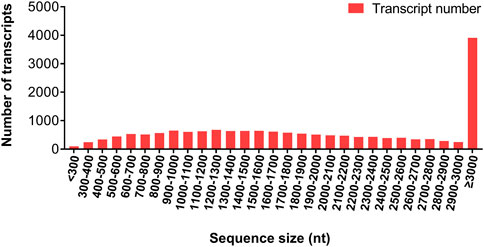
Twelve RNA samples from female antennae (FA), female body (FB), male antennae (MA), and male body (MB) were sequenced using a DNBSEQ platform, with total raw reads of 45.44 M per sample. After data filtering, a total base of 6.46 ± 0.02 Gb per FA, 6.47 ± 0.03 Gb per FB, 6.45 ± 0.02 Gb per MA, and 6.48 ± 0.02 Gb per MB were generated, respectively (Supplementary Table S3). The clean data were subjected to de novo transcriptome assembly using Trinity software. A total of 20,520 unigenes were generated, which refer to a uniquely assembled transcript or a cluster of genes that perform a particular function. Among them, 13,827 unigenes (67.38%) were longer than 1,000 bp, and 7,747 (37.75%) were longer than 2000 bp (Figure 1). Additionally, 17,212 (83.88%) unigenes were annotated in at least one public database (e.g. GO, KEGG, KOG, NR, NT, Pfam, and SwissProt databases), while 3,308 (16.12%) unigenes had no matching sequences in any of these databases. All RNA-seq data generated for this study were deposited in the GenBank under Bio-Project accession number PRJNA857870.
Analysis of differentially expressed genes
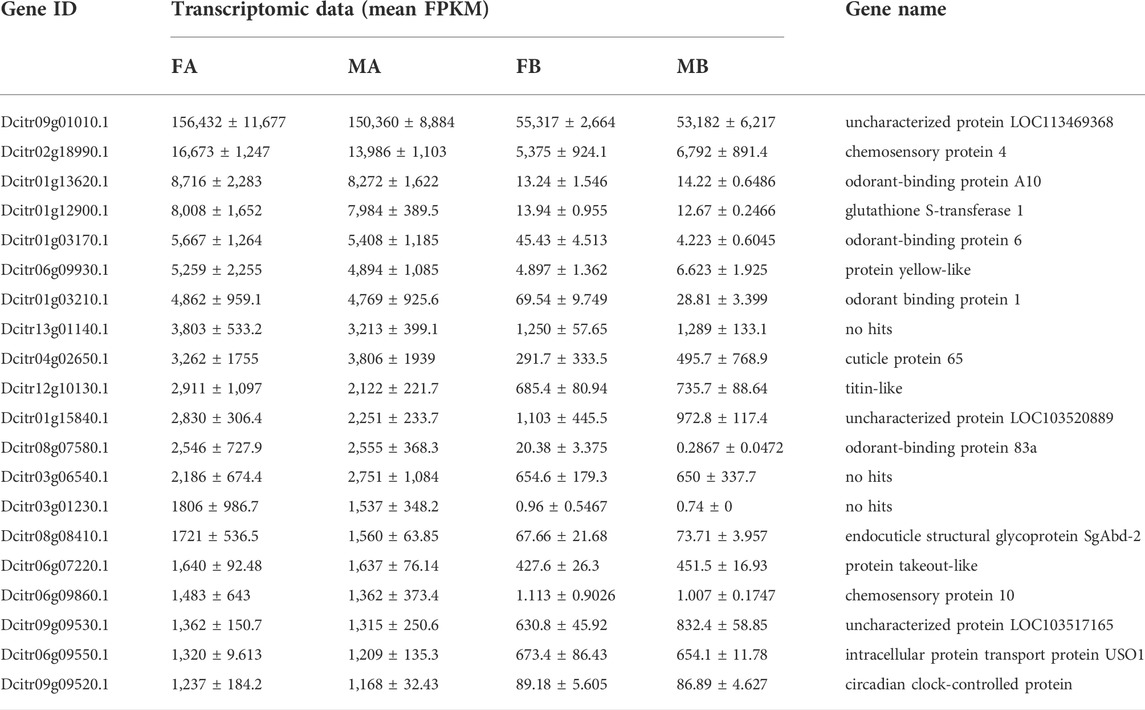
Comparative analyses of the antennal and body transcriptomes from both sexes in this study provide useful information to identify the antennae-specific and sex-biased antennal genes. A total of 665 unigenes were detected only in the antenna of females or males; 192 unigenes were unique for MA and 157 were expressed only in FA (Figure 2A). DEGs with a Q value ≤ 0.05 were further identified in antennae, by comparing with the transcriptomes of body tissues. Pairwise comparisons of transcript abundance revealed that 4,241 and 4,272 unigenes were significantly upregulated in FA and MA, respectively (Figure 2B). Twenty DEGs that were most abundant in antennae were listed in Table 1. These included transcripts of odorant binding protein (DcOBP1, DcOBP6, DcOBPA10, and DcOBP83a), chemosensory protein (DcCSP4 and DcCSP10), etc. Notably, DcOBP1, DcOBP6, DcOBPA10, DcOBP83a, and DcCSP10 exhibited an antennae-specific expression pattern, in which their antennal FPKM values were >100-fold higher than in the body (Table 1).
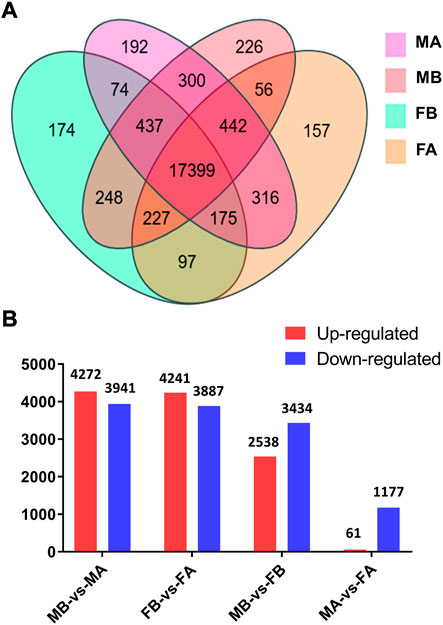
FIGURE 2. Expression of unigenes in D. citri samples. (A) Venn diagram of unigenes in the FA, FB, MA and MB of D. citri. (B) Number of DEGs identified by a pairwise comparison of the four samples. FA, female antennae; FB, female body; MA, male antennae; MB, male body.
Identification of candidate DcCYP genes
Seven DEGs abundant in antennae were identified to be DcCYPs by blasting against the Nr database. Their redundancy levels were found to be 15.86–167.70 FPKM in antennae, which were >3-fold higher compared to body group. The predicted Mw was from 28.45 to 60.30 kDa with the predicted pI of 6.14–9.04. All DcCYP proteins harbored a conserved P450 domain (Pfam ID: PF00067), and were predicted to have no signal peptide, indicating their functional location inside the cell (Table 2). Gene structure analysis showed that the length of genomic sequence (1,116–14,366 nt) and the number of introns (2–7) varied dramatically among DcCYP genes (Figure 3). A maximum likelihood phylogenetic tree was constructed using the amino-acid sequence of the P450 domain from candidate DcCYPs and A. pisum CYPs. Four subclasses (CYP2, CYP3, CYP4, and mitochondrial CYP) were well clustered in relevant phylogenetic branches. DcCYP6a13, DcCYP6j1, DcCYP6k1, and DcCYP6a2 were categorized in the CYP3 clan; the other three DcCYP genes, DcCYP4d2, DcCYP4c62 and DcCYP4d8 were clustered in the CYP4 class (Figure 4).
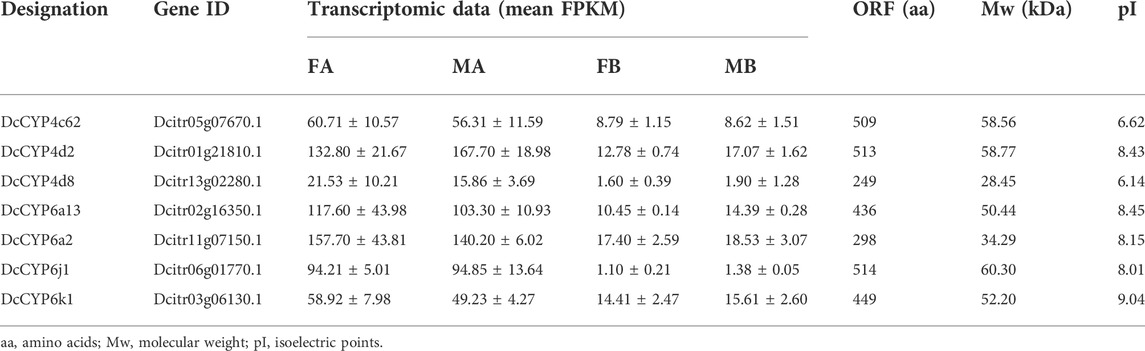
TABLE 2. Identification of candidate DcCYP genes in different expression values (FPKM) in the D. citri antennal and body transcriptomes.
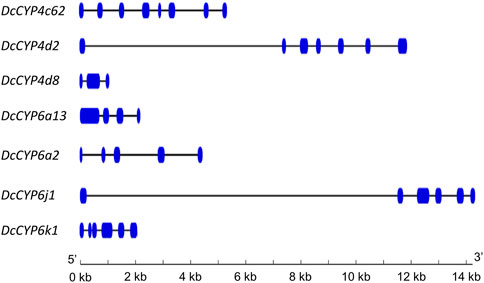
FIGURE 3. Exon-intron structure of DcCYP genes. Exons and introns are indicated by blue boxes and lines, respectively. The length of genes can be inferred by the scale at the bottom.
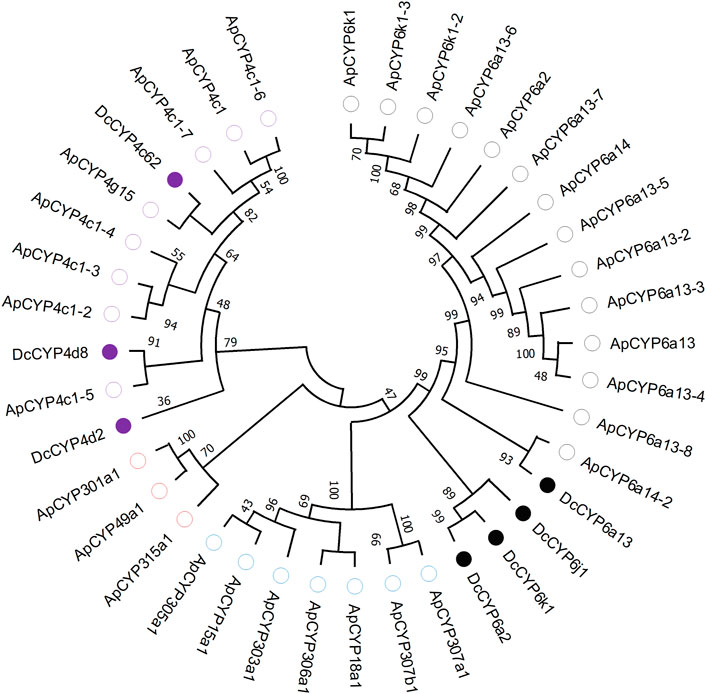
FIGURE 4. Phylogenetic relationship of 39 CYP proteins from D. citri (Dc, 7) and A. pisum (Ap, 32). Amino acid sequences were completely aligned using ClustalX 1.83, and a maximum likelihood tree was constructed using MEGA 11. Bootstrap values are shown at the branch points and indicate support as a percentage of 1,000 replicates. Seven D. citri CYPs are marked with filled circles. The reference CYPs from A. pisum are marked with empty circles. CYPs assigned to CYP2-, CYP3- and CYP4-subclass are labelled by blue, black and purple circles, respectively. Three mitochondrial CYPs are marked by red circles. GenBank accession numbers for the CYP used in phylogenetic analysis are provided in Supplementary Table S1.
Developmental and tissue expression analysis for DcCYP genes
To identify the DcCYPs that correlated with odorant degradation in the antennae, DcCYP transcripts abundant in the antennae basing on the FKPM methods were selected for developmental and tissue expression analysis. The selected DcCYP genes were constantly expressed from the first instar nymph to the adult, along with the low or undetectable levels detected in eggs. However, DcCYP4d2 transcripts accumulated at a higher level in the last nymphal instar (fourth–fifth), and DcCYP4k1 exhibited relatively higher expression in the first–fourth nymphal instar (Figure 5). Tissue expression analysis revealed that DcCYP4c62, DcCYP4d2, DcCYP6a13, DcCYP6a2, DcCYP6j1 and DcCYP6k1 were expressed higher in both male and female antennae than in other non-olfactory tissues, such as heads, legs, wings, and cuticles; DcCYP4d8 had higher expression levels in the antennae and legs of both sexes (Figure 6). Interestingly, the expression of DcCYP6j1 calculated by qRT-PCR were a bit higher in the male antennae than in the female antennae, which was inconsistent with the RNA-Seq data (Table 2; Figure 6); this possibly attributed to the difference in sensitivity of two methods.
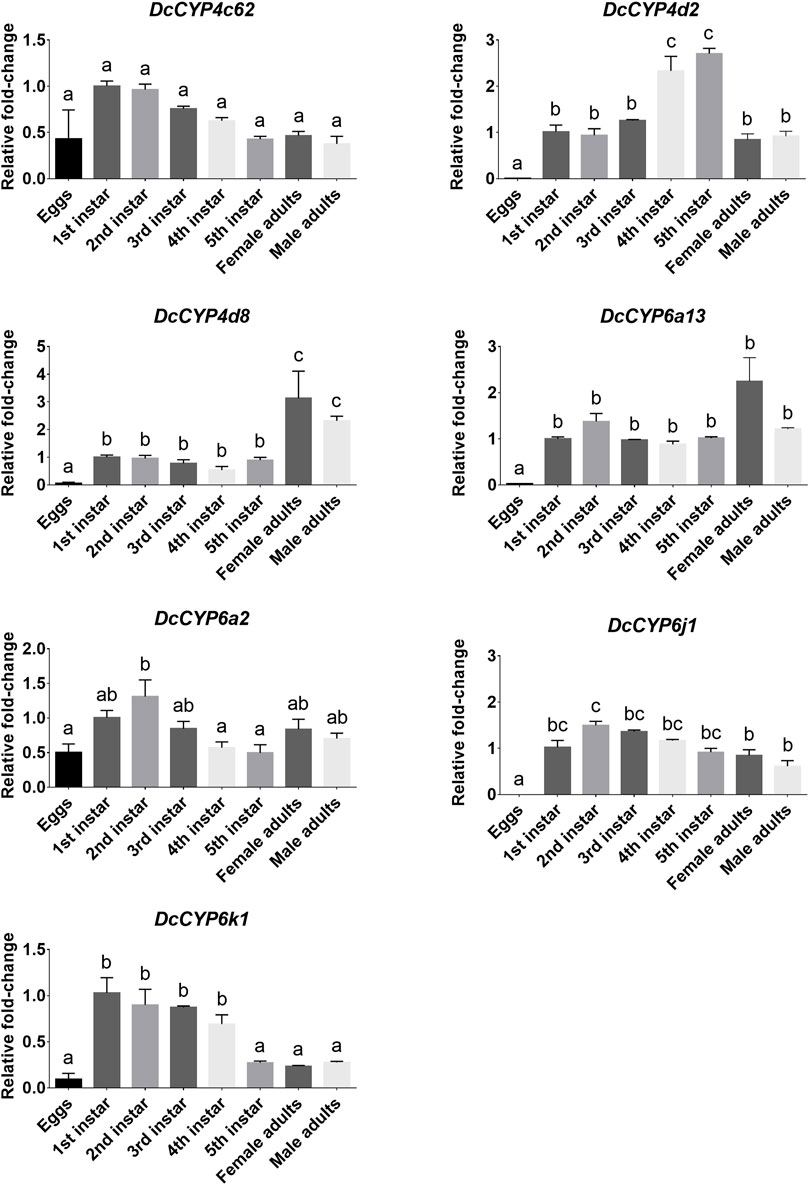
FIGURE 5. DcCYP expression at different life stages of D. citri. The expression level of DcCYP in first instar nymph was assigned an arbitrary value of 1. The expression levels in other samples are presented relative to the average first instar levels. Error bars represent standard error. Different letters above the error bar indicate significant differences among developmental stages (p < 0.05; one-way ANOVA, Tukey’s multiple comparisons test).
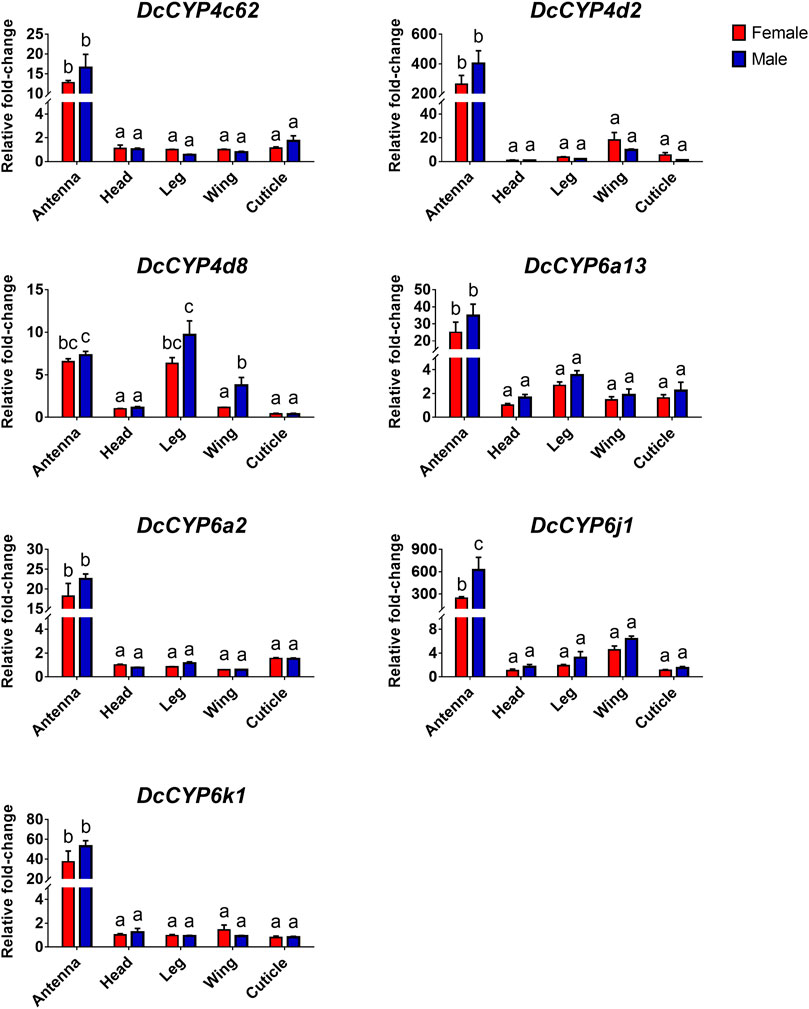
FIGURE 6. The relative abundance of DcCYP transcripts between different sexes and tissues. The expression level of DcCYP in female head was assigned an arbitrary value of 1. The expression levels in other samples are presented relative to the average female head levels. Different letters above the error bar indicate significant differences among female and male tissues (p < 0.05; two-way ANOVA, Tukey’s multiple comparisons test).
Discussion
Chemical control of D. citri is threatened by the increased insecticide resistance (Chen et al., 2018). Insect antennae are the main structures responsible for odorant reception via sensilla (Ahn et al., 2020; Zhou and Jander, 2022). Identification of antennal ODE such as CYP genes, could provide insights into the odorant recognition mechanism of D. citri and further help us to better control this agricultural pest insect. In this study, a total of seven antennae-enriched DcCYP genes were first identified by comparing the antennal and body transcriptome data from both sexes. qRT-PCR analyses showed they were antenna-biased and constantly expressed from the first instar nymph to the adult, with DcCYP6j1 expressed in male antennae at relatively higher levels.
Transcriptome analyses in this study identified several antennae-specific and/or antennae-enriched genes in D. citri. As expected, chemosensory gene OBPs and CSPs were highly expressed in antennae. Among the top twenty DEGs that were abundant in antennae, four transcripts were identified as OBP genes (DcOBP1, DcOBP6, DcOBPA10 and DcOBP83a), and two DEGs were CSP genes (DcCSP4 and DcCSP10), this was consistent with the putative olfactory role of insect antennae. Antennal OBP and CSP genes have been reported in many other insect species including spotted-wing drosophila suzukii (Ahn et al., 2020), ladybird Aphidius gifuensis (Kang et al., 2021), as well as hemipteran aphid (Zhou et al., 2010; Wang et al., 2019), hawthorn lace bug Corythucha ciliata (Li G. W et al., 2018) and brown plant hopper Nilaparvata lugens (Zhou et al., 2014). In green peach aphid Myzus persicae, three OBP genes (MpOBP6/7/10) were specifically expressed in antennae, and five OBP genes (MpOBP2/4/5/8/9) were expressed antennae enriched (Wang et al., 2019).
Insect CYPs perform a variety of important physiological functions, and evidence for their metabolic clearance of plant volatiles or host phytochemical detoxification is accumulating (Blomquist et al., 2021; Vandenhole et al., 2021; Nauen et al., 2022). Insect CYPs could be categorized into CYP2, CYP3, CYP4 and mitochondrial CYP clans (Feyereisen, 2012). Many members of CYP3 and CYP4 clan are currently known or suspected to participate in herbivore adaptation to host plant (Feyereisen, 2012; Blomquist et al., 2021; Vandenhole et al., 2021). For example, the CYP3 P450 genes in Dendroctonus armandi and Oedaleus asiaticus were shown to be induced by the host terpenoids (pinene and 3-carene) or flavonoid rutin (Dai et al., 2016; Huang et al., 2017); in another study, larval exposure to the plant volatiles induced by Spodoptera litura herbivory enhanced transcript levels of CYP3 genes (Sun et al., 2021). From the four clades of P450s commonly found in insects, we found four transcripts (DcCYP6a13, DcCYP6j1, DcCYP6k1 and DcCYP6a2) were clustered in CYP3 family, and three genes (DcCYP4d2, DcCYP4c62 and DcCYP4d8) were grouped in CYP4 clan.
Antennae-abundant CYPs have been functionally demonstrated as ODEs involved in terpenoid detoxification and odorant processing (Keeling et al., 2013; Chiu et al., 2019a; Chiu et al., 2019b; Chiu et al., 2019c; Blomquist et al., 2021). A combined transcriptome and qRT-PCR analysis in this study revealed that seven DcCYP genes (DcCYP4d2, DcCYP4c62, DcCYP4d8, DcCYP6a13, DcCYP6j1, DcCYP6k1 and DcCYP6a2) were preferentially expressed in antennae. Unexpectedly, six totally different DcCYP genes (CYP3175B1, CYP3178A1, CYP3640A2, CYP4C67, CYP380C15 and CYP3167A7) were reported to be antennae-abundant in the previous study (Wu Z et al., 2020), by comparing antennal transcriptome with that of gut tissues, reproductive organs, Malpighia tubules, brain, and fat body. However, the selected small tissues and organs are not representatives of non-olfactory tissues, this may contribute to the conflicting results of previous studies. In addition, the expression of DcCYP6j1 was observed to be a bit higher in male antennae. Our results, together with previous findings that male-biased CYPs were usually implicated in the recognition processes of sex pheromone (Maïbèche-Coisne et al., 2004; Feng et al., 2017), suggesting a putative role of these DcCYPs in inactivation of plant volatile semiochemicals and/or sex pheromone. Future research elucidating their physiological role will help understand the olfactory mechanism of hemipteran insect, and offer new targets for developing behavioral interference control strategy against D. citri.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA857870.
Author contributions
XY conceived the study; YK and YX reared the insects and conducted the laboratory work; XY and YK carried out the analyses; XC helped to modify the manuscript; XY wrote the manuscript.
Funding
This work is funded by the National Natural Science Foundation of China (grant no. 32160634), the Natural Science Foundation for Distinguished Young Scholars of Jiangxi province (grant no. 20212ACB215001), the Double Thousand Plan of Jiangxi Province (grant no. jxsq2019101058), and the Major Science and Technology R&D Program of Jiangxi Province (grant no. 20194ABC28007).
Acknowledgments
The authors wish to acknowledge the assistance of Yan Liu, Zhidan Hu and Fang Liang with the psyllid samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1004192/full#supplementary-material
References
Ahn S. J., Oh H. W., Corcoran J., Kim J. A., Park K. C., Park C. G., et al. (2020). Sex-biased gene expression in antennae of Drosophila suzukii. Arch. Insect Biochem. Physiol. 104, e21660. doi:10.1002/arch.21660
Baldwin S. R., Mohapatra P., Nagalla M., Sindvani R., Amaya D., Dickson H. A., et al. (2021). Identification and characterization of CYPs induced in the Drosophila antenna by exposure to a plant odorant. Sci. Rep. 11, 20530–20614. doi:10.1038/s41598-021-99910-9
Blomquist G. J., Tittiger C., MacLean M., Keeling C. I. (2021). Cytochromes P450: terpene detoxification and pheromone production in bark beetles. Curr. Opin. Insect Sci. 43, 97–102. doi:10.1016/j.cois.2020.11.010
Bozzolan F., Siaussat D., Maria A., Durand N., Pottier M. A., Chertemps T., et al. (2014). Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 23, 539–549. doi:10.1111/imb.12100
Brito N. F., Moreira M. F., Melo A. C. (2016). A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 95, 51–65. doi:10.1016/j.jinsphys.2016.09.008
Cheema J. A., Carraher C., Plank N. O., Travas-Sejdic J., Kralicek A. (2021). Insect odorant receptor-based biosensors: Current status and prospects. Biotechnol. Adv. 53, 107840. doi:10.1016/j.biotechadv.2021.107840
Chen X. D., Gill T. A., Ashfaq M., Pelz-Stelinski K. S., Stelinski L. L. (2018). Resistance to commonly used insecticides in asian citrus psyllid: stability and relationship to gene expression. J. Appl. Entomol. 142, 967–977. doi:10.1111/jen.12561
Chiu C. C., Keeling C. I., Bohlmann J. (2019a). Cytochromes P450 preferentially expressed in antennae of the mountain pine beetle. J. Chem. Ecol. 45, 178–186. doi:10.1007/s10886-018-0999-0
Chiu C. C., Keeling C. I., Bohlmann J. (2019b). The cytochrome P450 CYP6DE1 catalyzes the conversion of α-pinene into the mountain pine beetle aggregation pheromone trans-verbenol. Sci. Rep. 9, 1477–1510. doi:10.1038/s41598-018-38047-8
Chiu C. C., Keeling C. I., Henderson H. M., Bohlmann J. (2019c). Functions of mountain pine beetle cytochromes P450 CYP6DJ1, CYP6BW1 and CYP6BW3 in the oxidation of pine monoterpenes and diterpene resin acids. PLoS One 14, e0216753. doi:10.1371/journal.pone.0216753
Dai L., Ma M., Gao G., Chen H. (2016). Dendroctonus armandi (Curculionidae: Scolytinae) cytochrome P450s display tissue specificity and responses to host terpenoids. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 201, 1–11. doi:10.1016/j.cbpb.2016.06.006
Durand N., Carot-Sans G., Chertemps T., Bozzolan F., Party V., Renou M., et al. (2010a). Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS One 5, e15026. doi:10.1371/journal.pone.0015026
Durand N., Carot-Sans G., Chertemps T., Montagné N., Jacquin-Joly E., Debernard S., et al. (2010b). A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol. Biol. 19, 87–97. doi:10.1111/j.1365-2583.2009.00939.x
Feng B., Zheng K., Li C., Guo Q., Du Y. (2017). A cytochrome P450 gene plays a role in the recognition of sex pheromones in the tobacco cutworm, Spodoptera litura. Insect Mol. Biol. 26, 369–382. doi:10.1111/imb.12307
Feyereisen R. (2012). “Insect CYP genes and P450 enzymes,” in Insect molecular biology and biochemistry. Editor L. I. Gilbert (Amsterdam: Elsevier BV), 236–316.
Fleischer J., Pregitzer P., Breer H., Krieger J. (2018). Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 75, 485–508. doi:10.1007/s00018-017-2627-5
He P., Zhang Y. N., Yang K., Li Z. Q., Dong S. L. (2015). An antenna-biased carboxylesterase is specifically active to plant volatiles in Spodoptera exigua. Pestic. Biochem. Physiol. 123, 93–100. doi:10.1016/j.pestbp.2015.03.009
Hosmani P. S., Flores-Gonzalez M., Shippy T., Vosburg C., Massimino C., Tank W., et al. (2019). Chromosomal length reference assembly for Diaphorina citri using single-molecule sequencing and Hi-C proximity ligation with manually curated genes in developmental, structural and immune pathways. bioRxiv [Preprint]. doi:10.1101/869685
Huang X., Whitman D. W., Ma J., McNeill M. R., Zhang Z. (2017). Diet alters performance and transcription patterns in Oedaleus asiaticus (Orthoptera: Acrididae) grasshoppers. PLoS One 12, e0186397. doi:10.1371/journal.pone.0186397
Kang Z. W., Liu F. H., Xu Y. Y., Cheng J. H., Lin X. L., Jing X. F., et al. (2021). Identification of candidate odorant-degrading enzyme genes in the antennal transcriptome of Aphidius gifuensis. Entomol. Res. 51, 36–54. doi:10.1111/1748-5967.12489
Keeling C. I., Henderson H., Li M., Dullat H. K., Ohnishi T., Bohlmann J. (2013). CYP345E2, an antenna-specific cytochrome P450 from the mountain pine beetle, Dendroctonus ponderosae Hopkins, catalyses the oxidation of pine host monoterpene volatiles. Insect Biochem. Mol. Biol. 43, 1142–1151. doi:10.1016/j.ibmb.2013.10.001
Li F., Fu N., Li D., Chang H., Qu C., Wang R., et al. (2018). Identification of an alarm pheromone-binding chemosensory protein from the invasive sycamore lace bug Corythucha ciliata (Say). Front. Physiol. 9, 354. doi:10.3389/fphys.2018.00354
Li G. W., Chen X. L., Xu X. L., Wu J. X. (2018). Degradation of sex pheromone and plant volatile components by an antennal glutathione S‐transferase in the oriental fruit moth, Grapholita molesta Busck (Lepidoptera: Tortricidae). Arch. Insect Biochem. Physiol. 99, e21512. doi:10.1002/arch.21512
Liu X. Q., Jiang H. B., Fan J. Y., Liu T. Y., Meng L. W., Liu Y., et al. (2021). An odorant-binding protein of Asian citrus psyllid, Diaphorina citri, participates in the response of host plant volatiles. Pest Manag. Sci. 77, 3068–3079. doi:10.1002/ps.6352
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Maïbèche-Coisne M., Jacquin-Joly E., François M. C., Nagnan-Le Meillour P. (2002). cDNA cloning of biotransformation enzymes belonging to the cytochrome P450 family in the antennae of the noctuid moth Mamestra brassicae. Insect Mol. Biol. 11, 273–281. doi:10.1046/j.1365-2583.2002.00335.x
Maïbèche-Coisne M., Nikonov A. A., Ishida Y., Jacquin-Joly E., Leal W. S. (2004). Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. U. S. A. 101, 11459–11464. doi:10.1073/pnas.0403537101
Nauen R., Bass C., Feyereisen R., Vontas J. (2022). The role of cytochrome P450s in insect toxicology and resistance. Annu. Rev. Entomol. 67, 105–124. doi:10.1146/annurev-ento-070621-061328
Oh H. W., Jeong S. A., Kim J., Park K. C. (2019). Morphological and functional heterogeneity in olfactory perception between antennae and maxillary palps in the pumpkin fruit fly, Bactrocera depressa. Arch. Insect Biochem. Physiol. 101, e21560. doi:10.1002/arch.21560
Pelosi P. (1996). Perireceptor events in olfaction. J. Neurobiol. 30, 3–19. doi:10.1002/(SICI)1097-4695(199605)30:1<3::AID-NEU2>3.0.CO;2-A
Ramsey J. S., Rider D. S., Walsh T. K., De Vos M., Gordon K., Ponnala L., et al. (2010). Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol. Biol. 19, 155–164. doi:10.1111/j.1365-2583.2009.00973.x
Rogers M. E., Jani M. K., Vogt R. G. (1999). An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta. J. Exp. Biol. 202, 1625–1637. doi:10.1242/jeb.202.12.1625
Sun Z., Lin Y., Wang R., Li Q., Shi Q., Baerson S. R., et al. (2021). Olfactory perception of herbivore-induced plant volatiles elicits counter‐defences in larvae of the tobacco cutworm. Funct. Ecol. 35, 384–397. doi:10.1111/1365-2435.13716
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Tan X., Hu X. M., Zhong X. W., Chen Q. M., Xia Q. Y., Zhao P. (2014). Antenna-specific glutathione S-transferase in male silkmoth Bombyx mori. Int. J. Mol. Sci. 15, 7429–7443. doi:10.3390/ijms15057429
Vandenhole M., Dermauw W., Van Leeuwen T. (2021). Short term transcriptional responses of P450s to phytochemicals in insects and mites. Curr. Opin. Insect Sci. 43, 117–127. doi:10.1016/j.cois.2020.12.002
Vogt G. R., Riddiford L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi:10.1038/293161a0
Wang Q., Zhou J. J., Liu J. T., Huang G. Z., Xu W. Y., Zhang Q., et al. (2019). Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids. Insect Mol. Biol. 28, 1–22. doi:10.1111/imb.12513
Wang M. M., Long G. J., Guo H., Liu X. Z., Wang H., Dewer Y., et al. (2021). Two carboxylesterase genes in Plutella xylostella associated with sex pheromones and plant volatiles degradation. Pest Manag. Sci. 77, 2737–2746. doi:10.1002/ps.6302
Wei H., Tan S., Li Z., Li J., Moural T. W., Zhu F., et al. (2021). Odorant degrading carboxylesterases modulate foraging and mating behaviors of Grapholita molesta. Chemosphere 270, 128647. doi:10.1016/j.chemosphere.2020.128647
Wu Z. Z., Zhang H., Bin S. Y., Chen L., Han Q. X., Lin J. T. (2016). Antennal and abdominal transcriptomes reveal chemosensory genes in the Asian citrus psyllid, Diaphorina citri. PLoS One 11, e0159372. doi:10.1371/journal.pone.0159372
Wu H., Liu Y., Shi X., Zhang X., Ye C., Zhu K. Y., et al. (2020). Transcriptome analysis of antennal cytochrome P450s and their transcriptional responses to plant and locust volatiles in Locusta migratoria. Int. J. Biol. Macromol. 149, 741–753. doi:10.1016/j.ijbiomac.2020.01.309
Wu Z., Pu X., Shu B., Bin S., Lin J. (2020). Transcriptome analysis of putative detoxification genes in the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 76, 3857–3870. doi:10.1002/ps.5937
Yi J., Wang S., Wang Z., Wang X., Li G., Zhang X., et al. (2021). Identification of candidate carboxylesterases associated with odorant degradation in Holotrichia parallela antennae based on transcriptome analysis. Front. Physiol. 12, 674023. doi:10.3389/fphys.2021.674023
Younus F., Chertemps T., Pearce S. L., Pandey G., Bozzolan F., Coppin C. W., et al. (2014). Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect biochem. Mol. Biol. 53, 30–43. doi:10.1016/j.ibmb.2014.07.003
Yu X., Killiny N. (2018). Effect of parental RNA interference of a transformer-2 homologue on female reproduction and offspring sex determination in Asian citrus psyllid. Physiol. Entomol. 43, 42–50. doi:10.1111/phen.12223
Yu X., Killiny N. (2020). RNA interference-mediated control of Asian citrus psyllid, the vector of the huanglongbing bacterial pathogen. Trop. Plant Pathol. 45, 298–305. doi:10.1007/s40858-020-00356-7
Yu X. D., Liu Z. C., Huang S. L., Chen Z. Q., Sun Y. W., Duan P. F., et al. (2016). RNAi-mediated plant protection against aphids. Pest Manag. Sci. 72, 1090–1098. doi:10.1002/ps.4258
Yu X., Marshall H., Liu Y., Xiong Y., Zeng X., Yu H., et al. (2022) Sex-specific transcription and DNA methylation landscapes of the Asian citrus psyllid, a vector of huanglongbing pathogens. bioRxiv [Preprint]. doi:10.1101/2022.01.28.478167
Zhang H., Chen J. L., Lin J. H., Lin J. T., Wu Z. Z. (2020). Odorant-binding proteins and chemosensory proteins potentially involved in host plant recognition in the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 76, 2609–2618. doi:10.1002/ps.5799
Zhou S., Jander G. (2022). Molecular ecology of plant volatiles in interactions with insect herbivores. J. Exp. Bot. 73, 449–462. doi:10.1093/jxb/erab413
Zhou J. J., Vieira F. G., He X. L., Smadja C., Liu R., Rozas J., et al. (2010). Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 19, 113–122. doi:10.1111/j.1365-2583.2009.00919.x
Zhou S. S., Sun Z., Ma W., Chen W., Wang M. Q. (2014). De novo analysis of the Nilaparvata lugens (Stål) antenna transcriptome and expression patterns of olfactory genes. Comp. Biochem. Physiol. Part D. Genomics Proteomics 9, 31–39. doi:10.1016/j.cbd.2013.12.002
Keywords: asian citrus psyllid, antennal transcriptome, odorant degrading enzyme, cytochrome P450, gene expression
Citation: Kuang Y, Xiong Y, Chen XD and Yu X (2022) Antennae-abundant expression of candidate cytochrome P450 genes associated with odorant degradation in the asian citrus psyllid, Diaphorina citri. Front. Physiol. 13:1004192. doi: 10.3389/fphys.2022.1004192
Received: 27 July 2022; Accepted: 22 August 2022;
Published: 13 September 2022.
Edited by:
Wei Zhang, Guizhou University, ChinaReviewed by:
Sabir Hussain, Mir Chakar Khan Rind University, PakistanGhulam Sarwar Solangi, Sindh Agriculture University, Pakistan
Copyright © 2022 Kuang, Xiong, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiudao Yu, eXV4aXVkYW9AMTYzLmNvbQ==
 Yinhui Kuang1
Yinhui Kuang1 Xue Dong Chen
Xue Dong Chen Xiudao Yu
Xiudao Yu