- 1Engineering Research Center of Agricultural Microbiology Technology, Ministry of Education, Heilongjiang University, Harbin, China
- 2Heilongjiang Provincial Key Laboratory of Ecological Restoration and Resource Utilization for Cold Region, School of Life Sciences, Heilongjiang University, Harbin, China
- 3College of Life Science, Northeast Forestry University, Harbin, China
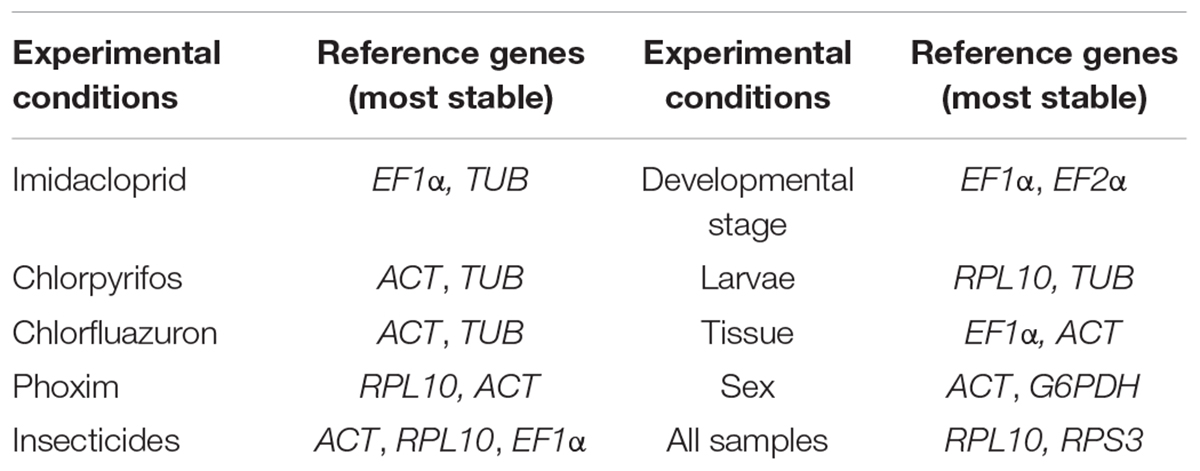
Bradysia odoriphaga (Diptera: Sciaridae) is the most serious root maggot pest which causes substantial damage to the Chinese chive. Organophosphate (OP) and neonicotinoid insecticides are widely used chemical pesticides and play important roles in controlling B. odoriphaga. However, a strong selection pressure following repeated pesticide applications has led to the development of resistant populations of this insect. To understand the insecticide resistance mechanism in B. odoriphaga, gene expression analysis might be required. Appropriate reference gene selection is a critical prerequisite for gene expression studies, as the expression stability of reference genes can be affected by experimental conditions, resulting in biased or erroneous results. The present study shows the expression profile of nine commonly used reference genes [elongation factor 1α (EF-1α), actin2 (ACT), elongation factor 2α (EF-2α), glucose-6-phosphate dehydrogenase (G6PDH), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L10 (RPL10), ribosomal protein S3 (RPS3), ubiquitin-conjugating enzyme (UBC), and α-tubulin (TUB)] was systematically analyzed under insecticide stress. Moreover, we also evaluated their expression stability in other experimental conditions, including developmental stages, sexes, and tissues. Five programs (NormFinder, geNorm, BestKeeper, RefFinder, and ΔCt) were used to validate the suitability of candidate reference genes. The results revealed that the most appropriate sets of reference genes were RPL10 and ACT across phoxim; ACT and TUB across chlorpyrifos and chlorfluazuron; EF1α and TUB across imidacloprid; EF1α and EF2α across developmental stages; RPL10 and TUB across larvae; EF1α and ACT across tissues, and ACT and G6PDH across sex. These results will facilitate the standardization of RT-qPCR and contribute to further research on B. odoriphaga gene function under insecticides stress.
Introduction
Bradysia odoriphaga Yang et Zhang (Diptera: Sciaridae) is a serious soil pest in China that feeds on 7 plant families and more than 30 plant species, including Chinese chive (Liliaceae), onion (Liliaceae), Chinese cabbage (Cruciferae), lettuce (Asteraceae), and so on (Li W. X. et al., 2015; Yang Y. T. et al., 2015). The main host plant of B. odoriphaga is Chinese chive (Allium tuberosum Rottle ex Spreng). Chinese chive is a perennial vegetable with a high economic value and is grown over a vast geographic area from Asia through the Middle East, to Europe and North America, and is widely cultivated in China. B. odoriphaga larvae usually gather in the roots, bulbs, and even in immature stems of Chinese chives, making the pest hard to control and allowing it to cause significant production losses of Chinese chives (Zhang et al., 2013; Chen et al., 2018). Yield loss of Chinese chive caused by B. odoriphaga has been reported to vary from 40 to 60% (Li et al., 2007). So far, the control efforts against B. odoriphaga still largely rely on the application of chemical insecticides, such as organophosphate (OP) and neonicotinoid insecticides (Chen et al., 2017). Phoxim, chlorpyrifos, imidacloprid, and chlorfluazuron are very popular insecticides that are used extensively for the purpose of B. odoriphaga control. Unfortunately, B. odoriphaga has developed increased resistance to insecticides because of heavy reliance on chemical insecticides (Chen et al., 2017). To investigate insecticide resistance mechanisms and promote integrated pest management (IPM) strategies, researchers have studied several pests over the past few decades and achieved important progress in several areas, including genomics (Xiao et al., 2021), transcriptomics (Cheng et al., 2020; Nazar et al., 2020; Nor Muhammad et al., 2020; Wang et al., 2020; Fu et al., 2021; Zou et al., 2021), proteomics (Prajapati et al., 2020; Chen et al., 2021), insecticide resistance (Wang et al., 2020; Gong et al., 2021; Ullah et al., 2021), RNA interference (Koo et al., 2020; Silver et al., 2021), and gene functions (Yu et al., 2020; Li L. L. et al., 2021; Luo et al., 2021). However, further studies on the mechanism of insecticide resistance are required to clarify the genes directly involved in resistance and regulatory mechanisms associated with those genes.
At present, real-time quantitative PCR is considered a reliable method to determine minor deviations in mRNA expression levels of a target gene due to its speed, accuracy, sensibility, throughput, cost, and reproducibility. The results of RT-qPCR must be normalized using reference genes because the threshold cycle (Ct) values are influenced by RNA quality and quantity, primer characteristics, PCR conditions, and variable transcriptional efficiencies. Since the validity and accuracy of RT-qPCR are highly dependent on the reference genes, it is imperative to identify the ideal candidate reference genes. An ideal reference gene should be constitutively and equally expressed in different cell types and tissues, regardless of internal and external factors or physiological cycles (Castanera et al., 2015). Many housekeeping genes that are necessary for regular cell functions have been universally used to normalize gene expression (Adeyinka et al., 2019; Chen et al., 2020). The top 10 most frequently used reference genes are Actin, RPL, Tubulin, GAPDH, RPS, 18S, EF1α, TATA, HSP, and SDHA (Lü et al., 2018). Several methods and programs have been developed to evaluate the stability of reference genes, including the ΔCt method (Silver et al., 2006), BestKeeper (Pfaffl et al., 2004), NormFinder (Andersen et al., 2004), geNorm (Vandesompele et al., 2002), and a web-based tool RefFinder (Xie et al., 2012). However, accumulating data of reference genes studies showed that an ideal reference gene that can keep stability in various experimental conditions does not exist (Yuan et al., 2014). Therefore, the reference genes should be selected cautiously, and their stability be validated before they are used under specific experimental conditions.
Consideration of the significance and diverse specificity of reference genes, many reference gene sets have been validated in various insect species, such as Spodoptera frugiperda (Zhou et al., 2021), Rhopalosiphum padi (Li M. et al., 2021), Aquatica leii (Fu and Meyer-Rochow, 2021), Tuta absoluta (Yan et al., 2021), Dichelops melacanthus (Pinheiro et al., 2020), Thermobia domestica (Bai et al., 2020), Apolygus lucorum (Luo et al., 2020), Lymantria dispar (Yin et al., 2020), Drosophila melanogaster (Kim et al., 2020), Phenacoccus solenopsis (Zheng et al., 2019), Chilo partellus (Adeyinka et al., 2019), Harmonia axyridis (Yang et al., 2018), Liriomyza trifolii (Chang et al., 2017), Myzus persicae (Kang et al., 2017), and B. odoriphaga (Shi et al., 2016) under various experimental conditions. However, a universal reference gene has not yet been identified. Therefore, the lack of a single universal reference for B. odoriphaga is not surprising. In this case, it’s important to choose reliable reference genes for gene expression analysis under various experimental conditions. Though appropriate references genes have been identified in B. odoriphaga under different biotic and abiotic conditions (Shi et al., 2016; Tang et al., 2019), a piece of comprehensive information is lacking for B. odoriphaga stressed by different groups of insecticides. Therefore, in this study, nine commonly used reference genes elongation factor 1α (EF-1α), actin2 (ACT), elongation factor 2α (EF-2α), glucose-6-phosphate dehydrogenase (G6PDH), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L10 (RPL10), ribosomal protein S3 (RPS3), ubiquitin-conjugating enzyme (UBC), and α-tubulin (TUB) were analyzed to assess their suitability for normalizing RT-qPCR data for B. odoriphaga under the stress of insecticides (phoxim, chlorpyrifos, imidacloprid, and chlorfluazuron). Additionally, the effects of developmental stages, tissues, and sexes were also evaluated. The objective of the present work was to identify different sets of suitable reference genes for further studies of toxicology-related target genes in B. odoriphaga.
Materials and Methods
Insects
B. odoriphaga was originally collected from The Institute of Plant Protection, Academy of Agricultural Sciences, Tianjin, China (39°10′36″N, 117°05′86″E) in 2018. The individuals were reared on scallions in an incubator at 20 ± 1°C, and 65 ± 5% relative humidity with a 12-h light:12-h dark photoperiod in culture dishes (Φ = 90 mm) filled with 2.5% agar solution at the liquid level of 0.5 mm and covered with filter paper.
Chemicals
Formulated insecticides, 50% phoxim EC, 40% chlorpyrifos EC, 20% imidacloprid SE, and 5% chlorfluazuron SE were manufactured by Xuzhou Shennong Chemical Co., Ltd., JiangSu, China, and kept in a refrigerator.
Analyzed Factors
The effects of the following factors on candidate reference genes mRNA were measured: insecticides (phoxim, chlorpyrifos, imidacloprid, and chlorfluazuron), developmental stages, tissues, and sexes. The samples processed by each factor were flash-frozen in liquid nitrogen and then stored at −80°C until analyzed by RT-qPCR. Each factor was assessed in four independent experiments.
Determination of LC50 Value of Insecticides
Groups of 15, third instar larvae were sprayed in a culture dish with 600 μL phoxim, chlorpyrifos, imidacloprid or chlorfluazuron, half on the body, half around, and fed scallion stained with pesticide (Li Z. N. et al., 2015). The control group was sprayed with distilled water. The number of dead individuals was checked after 24 h at 20°C and RH: 60–70%. LC50 was calculated for all the samples by survival analysis using SPSS 19.0 software for Windows (SPSS Inc., Chicago, IL, United States).
Insecticides Stress
The treatment groups of third instar larvae were sprayed with the LC50 value of phoxim, chlorpyrifos, imidacloprid, or chlorfluazuron. The control group was sprayed with distilled water. After 24, 48, and 72 h, 23 larvae in total were collected, flash-frozen, and stored.
Developmental Stages
B. odoriphaga samples were collected in a dish at each of the six developmental stages: first instar larvae, second instar larvae, third instar larvae, fourth instar larvae, pupa, and adult. Each dish contained 100 samples.
Tissues
The head, thorax, and abdomen from the fourth instar larvae were dissected by a dissection needle and a tweezer under a stereomicroscope. For each tissue, four replicates of 100 samples were collected.
Sexes
Hundred male and 100 female wingless B. odoriphaga adults were collected, flash-frozen in liquid nitrogen, and stored at −80°C until analyzed by RT-qPCR.
Primer Design
A set of nine candidate reference genes included EF1α, EF2α, ACT, GAPDH, G6PDH, RPL10, RPS3, TUB, and UBC. All of these genes are commonly used as reference genes in RT-qPCR analysis of other insects (Lü et al., 2018). The sequences of genes were obtained from B. odoriphaga transcriptome data (Chen et al., 2019). Primers were designed by NCBI Primer-BLAST1. The secondary structure of DNA template was predicted by the mfold web server2. Parameters were set as PCR products size 80–200 bp and size of primer 18–25 bp. Primers were synthetized by JINKAIRUI company, Wuhan, China. The details regarding the RT-qPCR primers are provided in Table 1.
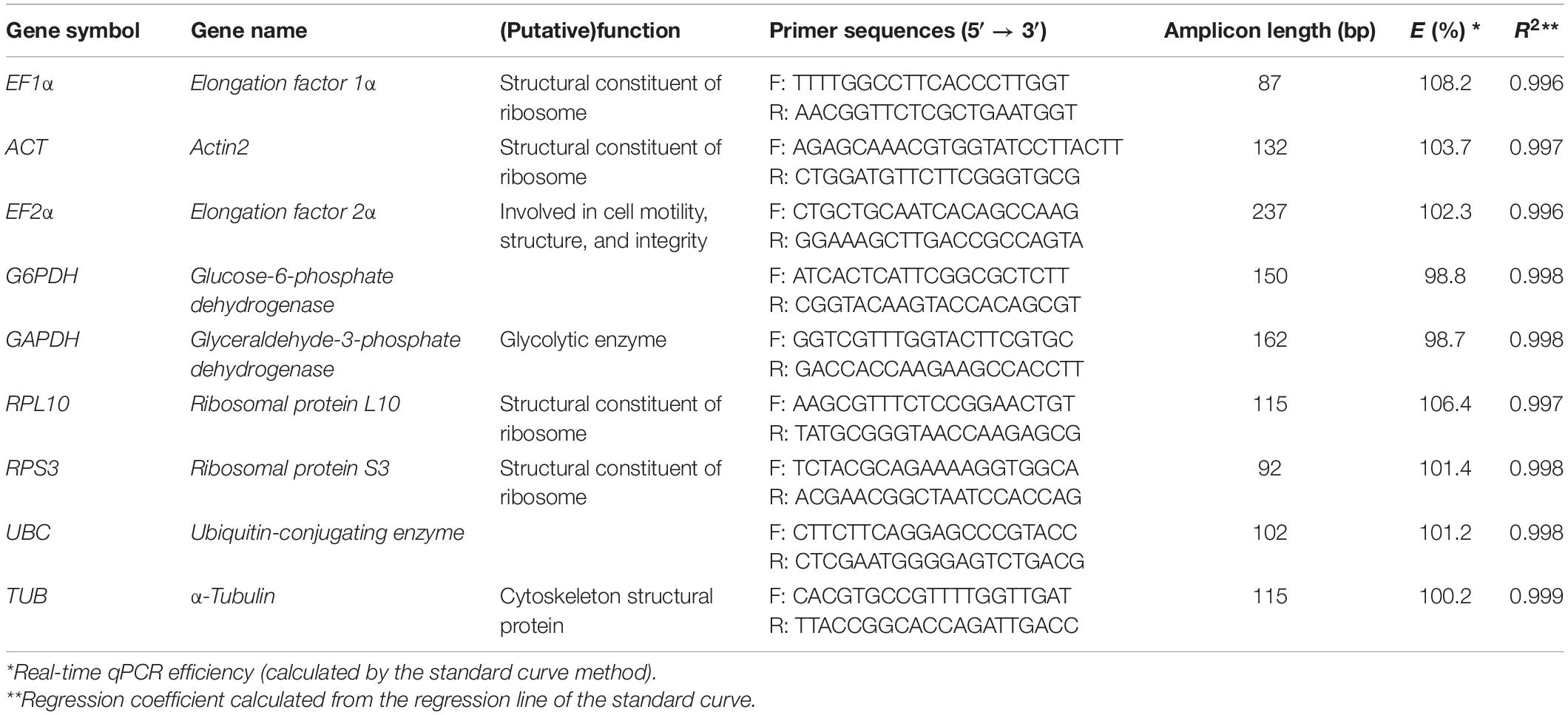
Table 1. Primer sequences and amplicon characteristics of the nine reference genes in B. odoriphaga samples.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted using the TRNzol Universal Reagent as described by the manufacturer (TaKaRa Bio, Dalian, China). The quantity and quality of RNA samples were assessed with a spectrophotometer 2000 (Thermo Scientific, Wilmington, DE, United States). RNA samples with OD ratio (A260/A280) ranging between 1.9 and 2.12 were selected for reverse transcription. Following the manufacturer’s instructions, the cDNA was synthesized using the Prime script TMRT reagent kit (TaKaRa Bio, Dalian, China). The synthesized cDNA was stored at −20°C.
RT-qPCR
The PCR reaction system was structured by SYBR Premix Ex Taq II kit (TaKaRa, Dalian, China). Each reaction was operated in a 20-μL solution including 2 μL mixture, 10 μL SYBR Premix Ex Taq II, 0.8 μL forward primer, 0.8 μL reverse primer, and 6.4 μL distilled water. The mixture was cDNA synthesized in different reverse transcription conditions. The amplification conditions for the RT-qPCR were set as following: 95°C for 30 s; followed by 40 cycles of 95°C for 5 s, 60°C for 34 s. The corresponding RT-qPCR efficiencies (E) were counted employing the equation: E = (10[–1/slope] − 1) × 100, with cDNA gradient dilution (1, 1/5, 1/25, 1/125, 1/625, and 1/3125) set as abscissa and Ct value as ordinate (Pfaffl, 2001). All samples were set three biological replicates and three technical replicates. The Ct values were obtained by analyzing the result from RT-qPCR using the SDS software of ABI 7500 (version 1.4).
Data Analysis
Data from RT-qPCR were analyzed by software SDS Shell.exe for ABI7500. The values were given as cycle threshold (Ct) numbers. All the Ct values were the average means of three biological replicates. The ΔCt method and three analysis applets NormFinder version 0.9533, GeNorm version 3.54, and BestKeeper5 were used to validate the stability of candidate reference genes. The comprehensive rank and a suitable number of reference genes were calculated by RefFinder6 and GeNorm, respectively.
Results
Verification of PCR Amplicons and PCR Amplification Efficiencies
The specific amplification of all primer pairs of candidate reference genes was confirmed with regular PCR and RT-qPCR. The PCR amplifications were identified by sequencing clones of the open reading frame (ORF). The results were consistent with the results of transcriptome sequencing. A single amplification peak for each candidate reference gene was observed in the melting curve (Figure 1). The size of amplicons ranged from 87 to 237 bp. The amplification efficiencies (E) for these genes varied from 98.7% for GAPDH to 108.2% for EF1α, and the correlation coefficients (R2) varied from 0.999 to 0.996 (Table 1).
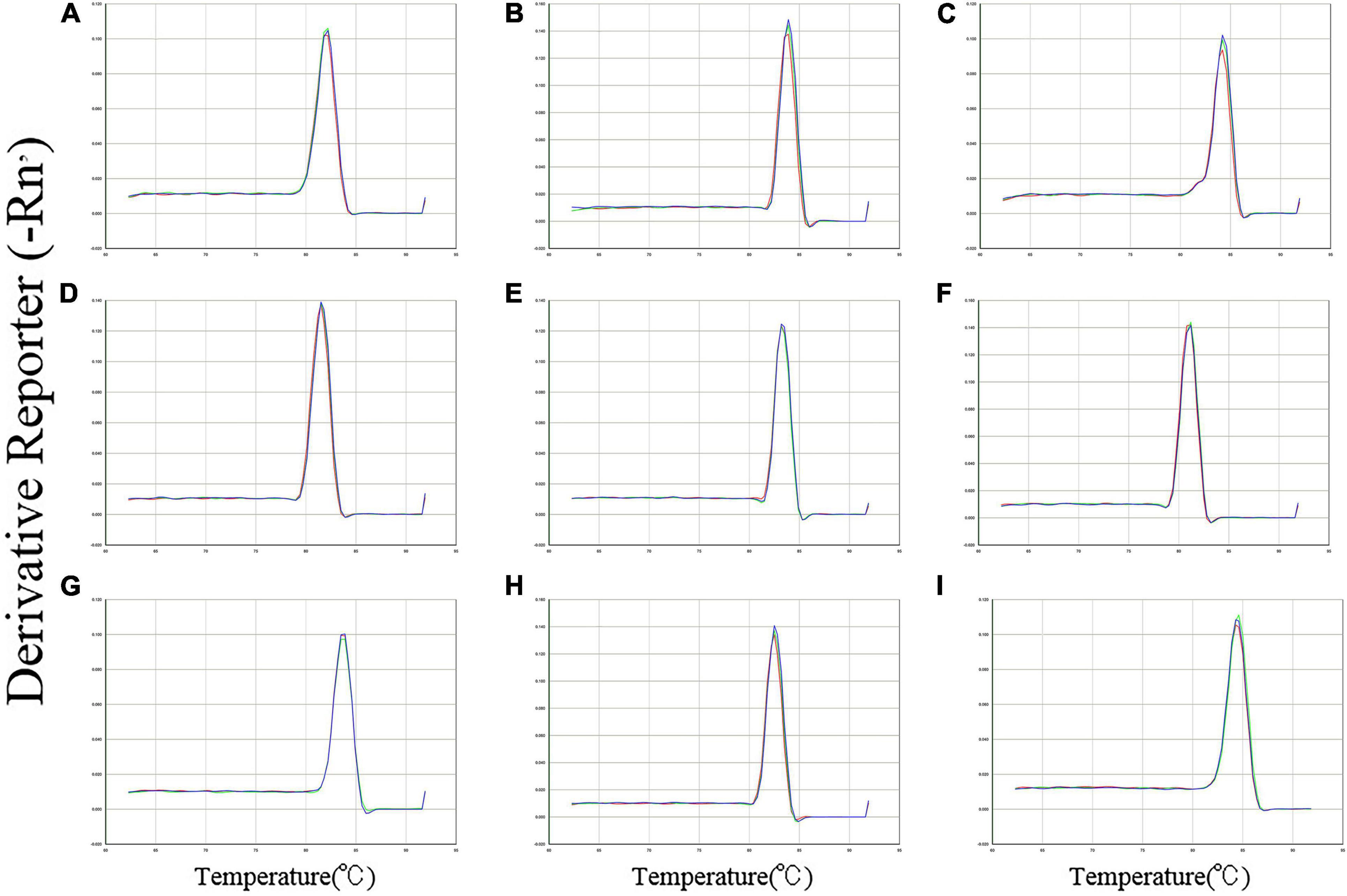
Figure 1. Specificity of primer pairs for RT-qPCR amplification in B. odoriphaga. Melting curves with single peaks were produced for all amplicons. (A) EF1α; (B) EF2α; (C) ACT; (D) GAPDH; (E) G6PDH; (F) RPL10; (G) RPS3; (H) TUB; and (I) UBC.
Expression Profiles of Candidate Reference Genes
The raw Ct values of the nine candidate reference genes for RT-qPCR were collected and are shown in Figure 2. The Ct values varied from 15.02 (EF-1α) to 37.26 (GAPDH), and the average Ct values ranged from 17.30 (EF-1α) to 22.10 (UBC), which indicates that noticeable differences exist in the expression profiles. Low Ct values correspond to high expression levels. Therefore, EF-1α exhibited the highest expression abundance, and UBC expressed the lowest level. Moreover, Ct values have also shown the differential expression variability, and EF-1α and TUB had a relatively narrow Ct range than other genes, indicating that these two genes might be expressed more stably.
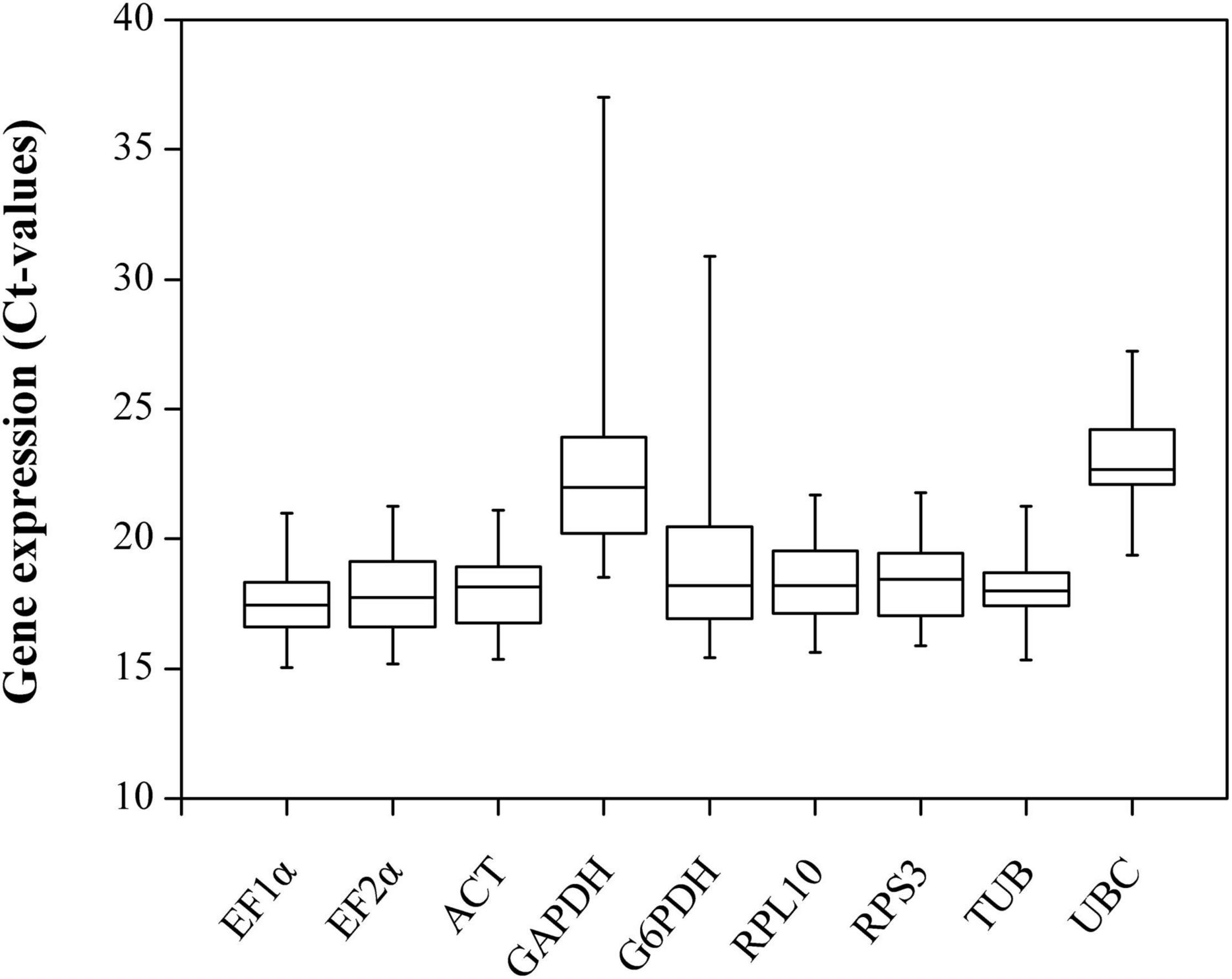
Figure 2. Candidate reference genes expression profiles in B. odoriphaga. The expression data are presented as mean Ct values for duplicate samples. Whiskers represent the maximum and minimum values. The lower and upper borders of boxes represent the 25th and 75th percentiles, respectively. The line across the box indicates the median Ct value.
Stability of Candidate Reference Genes
Imidacloprid
Based on ΔCt and the BestKeeper analyses, TUB and EF1α were the most stable genes (Table 2). However, the NormFinder analysis indicated RPL10 and ACT as the most stable genes (Table 2). All four analyses revealed G6PDH and GAPDH as the least stable genes (Table 2). The rank order for gene stability in the imidacloprid determined using RefFinder was as follows (most to least stable): EF1α, TUB, UBC, EF2α, RPL10, ACT, RPS3, G6PDH, and GAPDH (Figure 3A). The geNorm data indicated that the pairwise variation value for V2/3 was less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis suggested that EF1α and TUB are required to normalize target gene expression levels under imidacloprid stress (Table 3).
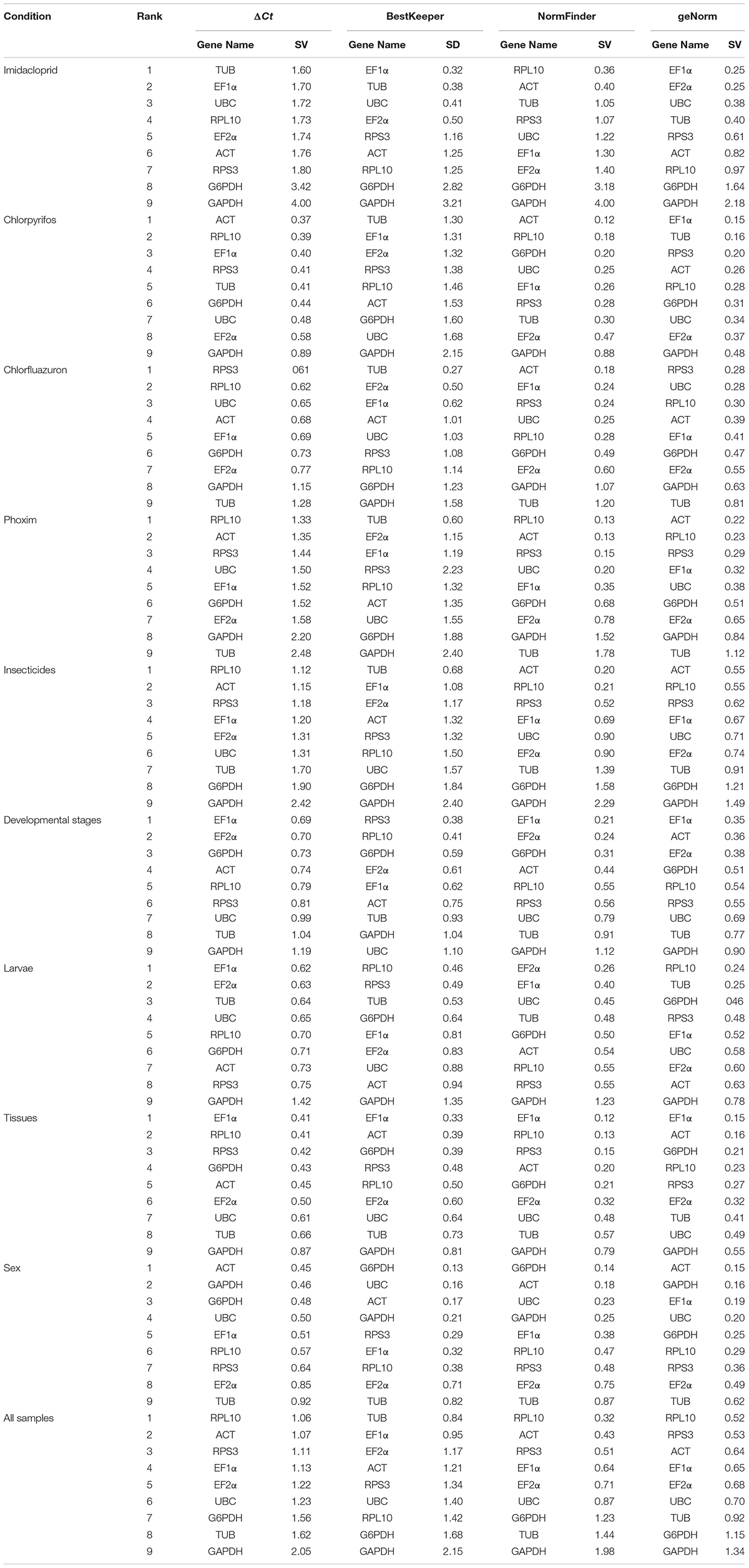
Table 2. Expression stability of the nine candidate reference genes in B. odoriphaga under various experimental conditions.
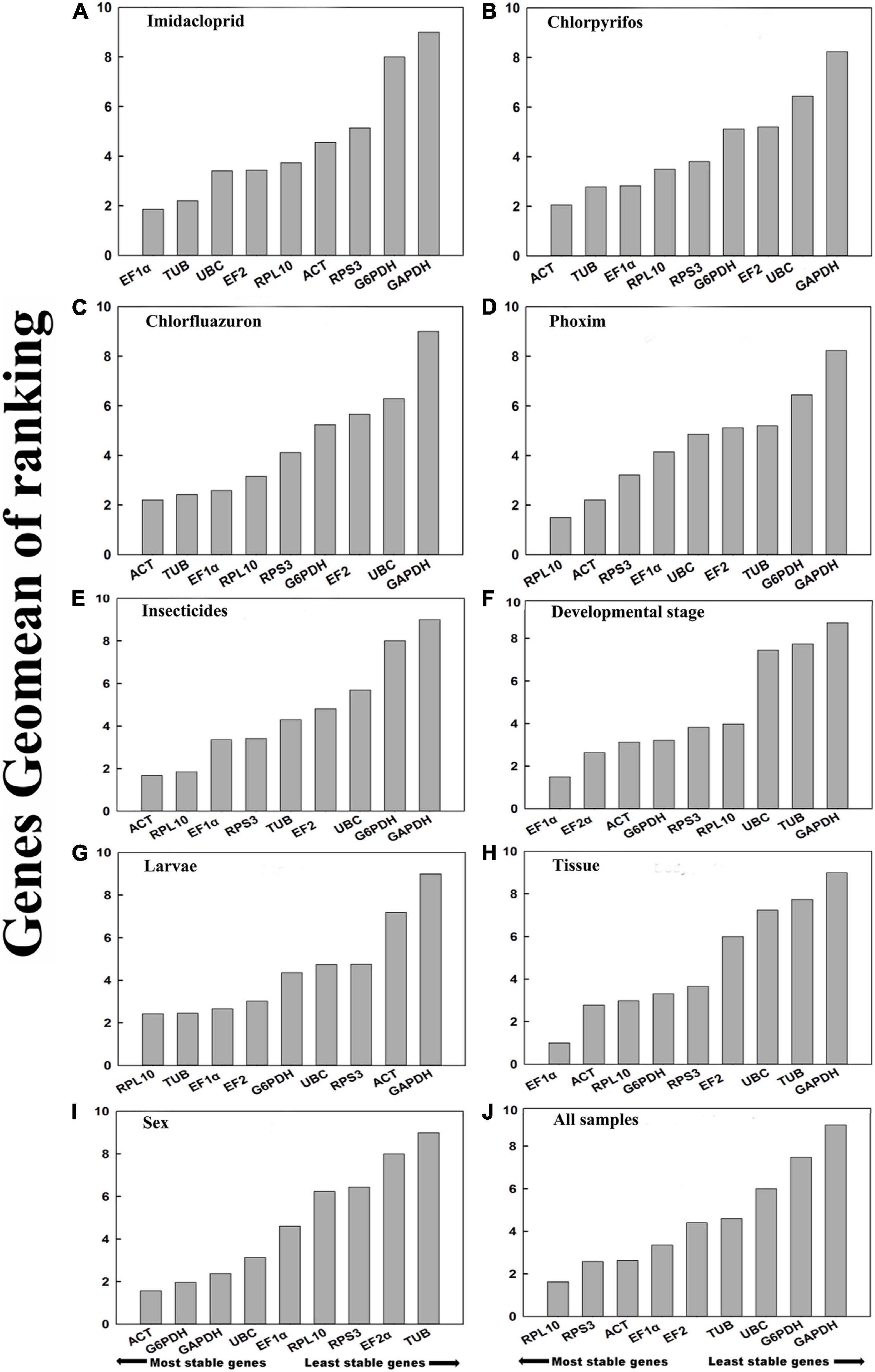
Figure 3. Stability of candidate reference genes in B. odoriphaga under various experimental conditions. In a RefFinder analysis, increasing Geomean values correspond to decreasing gene expression stability. The Geomean values for the following B. odoriphaga samples are presented: (A) imidacloprid: samples treated with imidacloprid; (B) chlorpyrifos: samples treated with chlorpyrifos; (C) chlorfluazuron: samples treated with chlorfluazuron; (D) phoxim:samples treated with phoxim; (E) insecticide treatment: adult samples treated with different insecticides; (F) developmental stage: samples for all developmental stages; (G) larvae: samples for larvae; (H) tissue: samples for different tissues; (I) adult samples for different sex; and (J) all samples: all samples for all treatments. The candidate reference genes are as follows: EF-1α, elongation factor 1α; ACT, actin2; EF-2α, elongation factor 2α; G6PDH, glucose-6-phosphate dehydrogenase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RPL10, ribosomal protein L10; RPS3, ribosomal protein S3; UBC, ubiquitin-conjugating enzyme; TUB, α-tubulin.
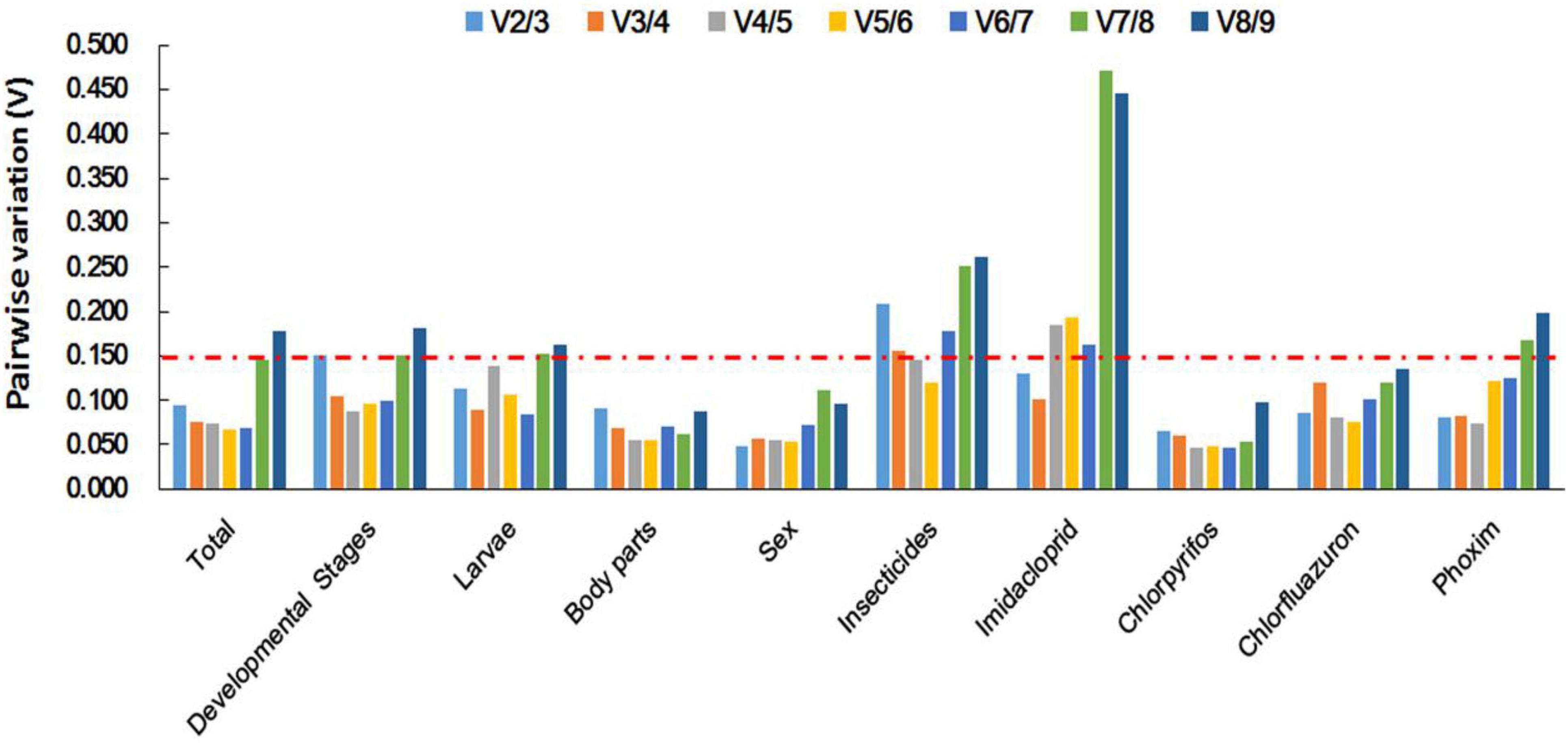
Figure 4. Optimal number of reference genes for accurate normalization as determined by geNorm. The Vn/n + 1 value indicates the pairwise variation (Y-axis) between two sequential normalization factors and was used to determine the optimal number of reference genes for an accurate data normalization. A-value < 0.15 indicates that an additional reference gene will not significantly improve the normalization.
Chlorpyrifos
Both ΔCt and NormFinder identified ACT and RPL10 as the most stable genes across chlorpyrifos samples (Table 2). In contrast, BestKeeper and geNorm detected TUB and EF1α as the most stable genes (Table 2). All analyses indicated that EF2α and GAPDH were the least stable genes. The RefFinder results for chlorpyrifos indicated the rank order for gene stability was as follows (most to least stable): ACT, TUB, EF1α, RPL10, RPS3, G6PDH, EF2α, UBC, and GAPDH (Figure 3B). The geNorm data indicated that all pairwise values were less than the proposed 0.15 cut-off (Figure 4). Based on the RefFinder analysis, ACT and TUB are required to normalize target gene expression levels across chlorpyrifos (Table 3).
Chlorfluazuron
The ΔCt analyses identified RPS3, RPL10, and UBC as the most stable genes across chlorfluazuron samples (Table 2). Similar results were obtained by geNorm (Table 2). However, BestKeeper identified TUB as the most stable gene (Table 2). The least stable gene was identified as TUB according to ΔCt, NormFinder, and geNorm. The RefFinder data indicated that the rank order for gene stability among chlorfluazuron samples was as follows (most to least stable): ACT, TUB, EF1α, RPL10, RPS3, G6PDH, EF2α, UBC, and GAPDH (Figure 3C). The geNorm analysis revealed that all pairwise variation values were less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis suggested that ACT and TUB are required to normalize target gene expression levels in chlorfluazuron-treated B. odoriphaga (Table 3).
Phoxim
All analyses except the BestKeeper indicated that RPL10, ACT, and RPS3 were the most stable genes, while GAPDH and TUB were the least stable genes (Table 2). In contrast, BestKeeper identified TUB as the most stable gene (Table 2). The rank order for gene stability determined using RefFinder was as follows (most to least stable): RPL10, ACT, RPS3, EF1α, UBC, EF2α, TUB, G6PDH, and GAPDH (Figure 3D). The geNorm data indicated that the pairwise variation value for V2/3 was less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis showed that target gene expression levels under phoxim stress conditions should be normalized against the expression of RPL10 and ACT (Table 3).
Integrative Analysis of Reference Genes Under Insecticides’ Stress
Regarding the insecticides’ stress effects, the ΔCt, geNorm, and NormFinder analyses indicated that the most stable genes were RPL10, ACT, and RPS3, whereas BestKeeper identified TUB, EF1α, and EF2α as the most stable genes (Table 2). All four analyses identified GAPDH and G6PDH as the least stable genes (Table 2). The RefFinder data indicated the rank order for gene stability was as follows (most to least stable): ACT, RPL10, EF1α, RPS3, TUB, EF2α, UBC, G6PDH, and GAPDH (Figure 3E). The geNorm analysis revealed that the pairwise variation value of V4/5 was less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis suggested that EF1α, TUB, and UBC are required to normalize target gene expression levels in B. odoriphaga under insecticides stress (Table 3).
Developmental Stages
Regarding the analyzed developmental stages, the ΔCt method, NormFinder, and geNorm, but not BestKeeper, indicated that EF-1α was the most stable gene and GAPDH and TUB were the least stable genes (Table 2). The BestKeeper analysis identified RPS3 and RPL10 as the most stable genes. In contrast, UBC was the least stable gene. The RefFinder analysis indicated the rank order for reference gene stability as follows (most to least stable): EF1α, EF2α, ACT, G6PDH, RPS3, RPL10, UBC, TUB, and GAPDH (Figure 3F). The geNorm analysis revealed that all pairwise variation values were less than the proposed 0.15 cut-off, except for V8/9 (Figure 4). A value less than 0.15 indicates that adding another reference gene will not change the normalization. The RefFinder analysis revealed that EF1α and EF2α are required for normalizing target gene expression levels in different B. odoriphaga developmental stages (Table 3).
Larvae
Both ΔCt and NormFinder identified EF1α and EF2α as the most stable genes and RPL10 as a moderately stable gene in larval samples (Table 2). However, the BestKeeper and geNorm analysis identified RPL10 as the most stable gene. According to the four algorithms, GAPDH was considered the least stable gene (Table 2). The rank order for gene stability based on the RefFinder results was as follows (most to least stable): RPL10, TUB, EF1α, EF2α, G6PDH, UBC, RPS3, ACT, and GAPDH (Figure 3G). The geNorm analysis indicated that the pairwise value of V2/3 was less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis suggested that RPL10 and TUB are required to normalize target gene expression levels in B. odoriphaga larval samples (Table 3).
Tissues
According to the four algorithms, the most stable gene was EF1α, and the least stable genes were GAPDH, UBC, and TUB across the tissues (Table 2). According to RefFinder, the reference gene stability rank order across tissues was as follows (most to least stable): EF1α, ACT, RPL10, G6PDH, RPS3, EF2α, UBC, TUB, and GAPDH (Figure 3H). The geNorm analysis results showed that all pair-wise variation values were less than the proposed 0.15 cut-off. The RefFinder analysis indicated EF1α and ACT are required for normalizing target gene expression levels in different B. odoriphaga tissues (Table 3).
Sex
Both ΔCt and geNorm identified ACT and GAPDH as the most stable genes across sex samples (Table 2). The BestKeeper and NormFinder analysis also identified G6PDH as the most stable gene, while UBC and ACT were the second and third stable genes, respectively (Table 2). According to the four algorithms, EF2α and TUB were identified as the least stable genes (Table 2). The rank order for gene stability among the examined sex samples based on the RefFinder results was as follows (most to least stable): ACT, G6PDH, GAPDH, UBC, EF1α, RPL10, RPS3, EF2α, and TUB (Figure 3I). The geNorm analysis indicated that the pairwise value of V2/3 was less than the proposed 0.15 cut-off (Figure 4). The RefFinder analysis suggested that ACT and G6PDH are required to normalize target gene expression levels in B. odoriphaga sex samples (Table 3).
Overall Ranking of Bradysia odoriphaga Reference Genes
Based on the RefFinder analysis, the overall rank order for the stability of B. odoriphaga genes was as follows (most to least stable): RPL10, RPS3, ACT, EF1α, EF2α, TUB, UBC, G6PDH, and GAPDH (Figure 3J). The geNorm analysis indicated that all pairwise variation values were less than the proposed 0.15 cut-off, except for V8/9 (Figure 4). The RefFinder data suggested that RPL10 and RPS3 are suitable internal reference genes for normalizing target gene expression levels in B. odoriphaga (Table 3).
Discussion
It is unquestionably true that gene expression quantification has never been easier than it is now, thanks to RT-qPCR technology. However, extreme care must be taken to avoid erroneous results (Liang et al., 2014). One of the most common strategies for correcting experimental errors introduced during the steps of RT-qPCR analysis is the normalization of RT-qPCR data with reference genes (Pinheiro et al., 2020). Inappropriate reference gene selection can obscure or magnify real biological changes caused by changes in reference gene expression (Zhu et al., 2014). Therefore, a reference gene with low expression variation must be chosen to ensure accurate normalization and avoid inaccurate quantification of gene expression (Huggett et al., 2005).
Earlier studies on reference genes evaluation and validation in insects under insecticide stress reported that the expression of reference genes varies under different insecticide stress even if they belong to the same group of insecticides (Liang et al., 2014). These findings further demonstrate that there is no single universal reference available under different conditions. Therefore, identifying suitable reference genes is critical for obtaining a reliable estimate for gene expression levels under different conditions.
The present study evaluated the expression stability of nine candidate reference genes in B. odoriphaga under four insecticides commonly applied for controlling this pest. Moreover, the stability of these selected candidate genes was also assessed in developmental stages, sexes, and different tissues of B. odoriphaga.
The assessment of RNA integrity and amplification efficiency must be conducted prior to RT-qPCR based analysis of genes expression. In the present work, RNA integrity results showed that the OD ratio (A260/A280) of all RNA samples varied between 1.8 and 2.0, and the amplification efficiency of the nine candidates ranged from 90 to 110% (R2 > 0.996) (Table 1). Thus, RNA quality and amplification were of sufficient quality to be used in RT-qPCR. Our RNA quality and amplification results agree with other reference gene validation studies conducted on the other insects (Shakeel et al., 2015; Pinheiro et al., 2020).
Our results of reference genes expression stability offered by five algorithms (geNorm, NormFinder, BestKeeper, Delta Ct, and RefFinder) indicated that the ranking order was different, such as TUB and EF1α were ranked as the most stable reference genes by ΔCt. In contrast, NormFinder ranked RPL10 and ACT as the most stable reference genes under imidacloprid stress. Similarly, geNorm indicated RPS3 and UBC as the most stable reference genes, whereas BestKeeper ranked TUB and EF2α as the most stable reference genes under chlorfluazuron stress. These discrepancies in the ranking order by different algorithms within the same tested insecticide might be because of the various analytical methods used (Shakeel et al., 2018). On the other hand, the difference in ranking of the reference genes under the stress of different insecticides in this study demonstrates the importance of evaluating their use under different sets of insecticides. Our findings provide more comprehensive information regarding reference genes selection under insecticide stress compared to the previous studies on B. odoriphaga (Shi et al., 2016; Tang et al., 2019).
The ACT gene, which is most frequently used as a reference gene, encodes a major structural protein that maintains organisms’ life activity and exhibits conservative structure during evolution. In the present study, our results demonstrated that ACT expression was highly stable under insecticide stress (chlorpyrifos, chlorfluazuron) and other experimental conditions, including tissues and both sexes, and developmental stages. Coincidentally, the results are consistent with the earlier reports. For example, ACT was identified as one of the most stable reference genes for normalizing target gene expression in Spodoptera litura treated with insecticides (Lu et al., 2013). Additionally, ACT expression was revealed to be most stable in Locusta migratoria under different insecticides stress (Yang et al., 2014). ACT also showed high stability in other insects under different experimental sets, such as in Plutella xylostella and Chilo suppressalis under different development stages (Teng et al., 2012), D. melanogaster after heat-stress (Ponton et al., 2011), Schistocerca gregaria in fifth instar nymphs (Van Hiel et al., 2009), and Orchesella cincta overall treatments (de Boer et al., 2009). Quite the contrary, ACT was a less stable reference gene for gene expression analyses in Bombyx mori, Spodoptera exigua (Teng et al., 2012), Coleomegilla maculata (Yang C. et al., 2015), Coccinella septempunctata (Yang et al., 2016), and Hippodamia convergens (Pan et al., 2015). In this study, ACT was not an ideal reference gene for the larval stage in B. odoriphaga. Thus, there is no single universal reference gene suitable for all insects and under all conditions, even the most commonly used housekeeping gene responds differently to various experimental conditions.
The TUB gene is assigned to the Eukaryotic structural gene family, and encodes cytoskeletal structure proteins that involve in the regulation of cell division, shape, motility, and intracellular activity. In previous studies, TUB exhibited a stable expression, for example, Nilaparvata lugens for geographic population (Yuan et al., 2014), Sogatella furcifera at different developmental stages and under different temperature stress (An et al., 2016), Thitarodes armoricanus for the fungal infections (Liu et al., 2016), and Bemisia tabaci MED across all sample sets (Dai et al., 2017). In this study, the stability of TUB was variable under different treatments in B. odoriphaga. It exhibited a stable expression under chlorpyrifos, imidacloprid, and chlorfluazuron stress, whereas its expression was unstable across different developmental stages and tissues. Similar results have also been noted in C. maculata (Yang C. et al., 2015). The above results clearly suggest that determining candidates and evaluating their suitability is required for each experimental condition.
In the present study, the EF1α gene expression levels was stable across different developmental stages, tissues, and under the treatment of imidacloprid. Indeed, EF1α has been commonly picked as reference genes across different developmental stages and temperature in many other insect species, such as Sesamia inferens (Sun et al., 2015), L. migratoria (Yang et al., 2014), Frankliniella occidentalis (Zheng et al., 2014), and H. convergens (Pan et al., 2015). However, EF1α was considered unstable in developmental stages and tissues, again, in B. odoriphaga (Shi et al., 2016). This discrepancy between our study and previous study might be caused by different candidate reference genes, diet, population, temperature, and photoperiods.
Notably, the GAPDH gene, which encodes a key enzyme involved in the energy metabolism and ranked as the fourth most widely used reference gene, showed poor stability among almost all experimental conditions in this study. There are also some reports suggesting that the GAPDH was not suitable to be used as reference gene under the specific condition in some species, for example, Bactrocera dorsalis in difference tissues (Shen et al., 2010), Musca domestica (Zheng et al., 2014), and Lucilia cuprina (Bagnall and Kotze, 2010) in difference developmental stages. On the other hand, GADPH was used as the most stable reference gene, such as S. litura in developmental stage and under temperature stress (Lu et al., 2013), P. xylostella in mechanical injury (Fu et al., 2013), Euscelidius variegatus, and Macrosteles quadripunctulatus by phytoplasma infection (Galetto et al., 2013). The results showed that the expression of candidate reference genes was not stable in all the tested conditions. Thus it is necessary to select different genes to normalize expression under different experimental conditions.
Conclusion
In summary, there was no single universal reference gene that could be used in all situations. It is indispensable to validate the expression of candidate genes before using them as the internal controls in qPCR. A suite of reference genes was specifically recommended for each experimental condition in this study. The suitable reference genes in different experimental conditions were EF1α and EF2α in development stages; EF1α and ACT in tissues; ACT and G6PDH in sex; RPL10 and ACT in phoxim treatment; ACT and TUB in chlorpyrifos treatment; EF1α and TUB in imidacloprid treatment; and ACT and TUB in chlorfluazuron treatment. The results of our experiment can be used for the further studies in B. odoriphaga.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Heilongjiang University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
FY and HF conceived and designed the research. TH, ZX, ChaoL, and CY conducted the experiments. ChunL and TW analyzed the data. HF wrote the manuscript. FS, FF, and FY revised the manuscript. All authors have read and approved the manuscript.
Funding
This study was supported by the Heilongjiang University Postdoctoral Research Fund (194359), National Natural Science Foundation of China (31672060), and Nature Scientific Foundation of Heilongjiang Province (ZD2017004). This work was supported by the Key Laboratory of Molecular Biology, College of Heilongjiang Province, and Heilongjiang Vegetable Collaborative Innovation and Extension System of Modern Agricultural Industry Technology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Researchersview, Shakeel M. for editing the English text of the draft of this manuscript.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome
- ^ http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form
- ^ http://www.mdl.dk/publications NormFinder.htm
- ^ http://GeNorm.cmgg.be
- ^ http://www.wzw.tum.de/gene-quantification/bestkeeper.html
- ^ https://www.heartcure.com.au/for-researchers
References
Adeyinka, O. S., Tabassum, B., Nasir, I. A., Yousaf, I., Sajid, I. A., Shehzad, K., et al. (2019). Identification and validation of potential reference gene for effective dsRNA knockdown analysis in Chilo partellus. Sci Rep. 9:13629. doi: 10.1038/s41598-019-49810-w
An, X. K., Hou, M. L., and Liu, Y. D. (2016). Reference Gene Selection and Evaluation for Gene Expression Studies Using qRT-PCR in the White-Backed Plant hopper, Sogatella furcifera (Hemiptera: delphacidae). J. Econ. Entomol. 109, 879–886. doi: 10.1093/jee/tov333
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Bagnall, N. H., and Kotze, A. C. (2010). Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med. Vet. Entomol. 24, 176–181. doi: 10.1111/j.1365-2915.2010.00866.x
Bai, Y., Lv, Y. N., Zeng, M., Jia, P. Y., Lu, H. N., Zhu, Y. B., et al. (2020). Selection of Reference Genes for Normalization of Gene Expression in Thermobia domestica (Insecta: zygentoma: lepismatidae). Genes 12:21. doi: 10.3390/genes12010021
Castanera, R., López-Varas, L., Pisabarro, A. G., and Ramírez, L. (2015). Validation of reference genes for transcriptional analyses in pleurotus ostreatus by using reverse transcription-quantitative PCR. Appl. Environ. Microbiol. 81, 4120–4129. doi: 10.1128/AEM.00402-15
Chang, Y., Chen, J., Lu, M., Gao, Y., Tian, Z., Gong, W., et al. (2017). Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: agromyzidae). PLoS One 12:e0181862. doi: 10.1371/journal.pone.0181862
Chen, C., Li, S., Zhu, H., Fan, B., Wang, Y., and Hao, D. (2020). Identification and evaluation of reference genes for gene expression analysis in the weevil pest Pagiophloeus tsushimanus using RT-qPCR. J. Asia Pacif. Entomol. 23, 336–344. doi: 10.1016/j.aspen.2020.01.010
Chen, C., Shi, X., Desneux, N., Han, P., and Gao, X. (2017). Detection of insecticide resistance in Bradysia odoriphaga Yang et Zhang (Diptera: sciaridae) in China. Ecotoxicology 26, 868–875. doi: 10.1007/s10646-017-1817-0
Chen, C., Wang, C., Liu, Y., Shi, X., and Gao, X. (2018). Transcriptome analysis and identification of P450 genes relevant to imidacloprid detoxification in Bradysia odoriphaga. Sci Rep. 8:2564. doi: 10.1038/s41598-018-20981-2
Chen, H., Lin, L., Xie, M., Zhong, Y., Zhang, G., and Su, W. (2019). Survey of the Bradysia odoriphaga Transcriptome Using PacBio Single-Molecule Long-Read Sequencing. Genes 10:481. doi: 10.3390/genes10060481
Chen, J., Guo, Y., Huang, S., Zhan, H., Zhang, M., Wang, J., et al. (2021). Integration of transcriptome and proteome reveals molecular mechanisms underlying stress responses of the cutworm, Spodoptera litura, exposed to different levels of lead (Pb). Chemosphere 283:131205. doi: 10.1016/j.chemosphere.2021.131205
Cheng, J., Su, Q., Xia, J., Yang, Z., Shi, C., Wang, S., et al. (2020). Comparative transcriptome analysis of differentially expressed genes in Bradysia odoriphaga Yang et Zhang (Diptera: sciaridae) at different acute stress temperatures. Genomics 112, 3739–3750. doi: 10.1016/j.ygeno.2020.04.019
Dai, T. M., Lü, Z. C., Liu, W. X., and Wan, F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS One 12:e0173821. doi: 10.1371/journal.pone.0173821
de Boer, M. E., de Boer, T. E., Mariën, J., Timmermans, M. J., Nota, B., van Straalen, N. M., et al. (2009). Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). BMC Mol. Biol. 10:54. doi: 10.1186/1471-2199-10-54
Fu, S., Duan, Y., Wang, S., Ren, Y., and Bu, W. (2021). Comparative transcriptomic analysis of Riptortus pedestris (Hemiptera: alydidae) to characterize wing formation across all developmental stages. Insects 12:226. doi: 10.3390/insects12030226
Fu, W., Xie, W., Zhang, Z., Wang, S., Wu, Q., Liu, Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: plutellidae). Int. J. Biol. Sci. 9, 792–802. doi: 10.7150/ijbs.5862
Fu, X., and Meyer-Rochow, V. B. (2021). Selection and validation of suitable reference genes for RT-qPCR analysis in the rare aquatic firefly Aquatica leii (Coleoptera: lampyridae). Insects 12:359. doi: 10.3390/insects12040359
Galetto, L., Bosco, D., and Marzachì, C. (2013). Selection of reference genes from two leafhopper species challenged by phytoplasma infection, for gene expression studies by RT-qPCR. BMC Res. Notes 6:409. doi: 10.1186/1756-0500-6-409
Gong, P., Li, X., Gao, H., Wang, C., Li, M., Zhang, Y., et al. (2021). Field evolved resistance to pyrethroids, neonicotinoids, organophosphates and macrolides in Rhopalosiphum padi (Linnaeus) and Sitobion avenae (Fabricius) from China. Chemosphere 269:128747. doi: 10.1016/j.chemosphere.2020.128747
Huggett, J., Dheda, K., Bustin, S., and Zumla, A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284. doi: 10.1038/sj.gene.6364190
Kang, Z., Liu, F., Tian, H., Zhang, M., Guo, S., and Liu, T. (2017). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid. Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Kim, Y., Kim, Y., and Kim, Y. H. (2020). Evaluation of reference genes for gene expression studies using quantitative real-time PCR in Drosophila melanogaster after chemical exposures. J. Asia Pacif. Entomol. 23, 385–394.
Koo, J., Chereddy, S. C. R. R., and Palli, S. R. (2020). RNA interference-mediated control of cigarette beetle, Lasioderma serricorne. Arch Insect Biochem Physiol. 104:e21680. doi: 10.1002/arch.21680
Li, H. J., He, X. K., Zeng, A. J., Liu, Y. J., and Jiang, S. R. (2007). Bradysia odoriphaga copulatory behavior and evidence of a female sex pheromone. J. Agric. Urban Entomol. 24, 27–34. doi: 10.3954/1523-5475-24.1.27
Li, L. L., Xu, J. W., Yao, W. C., Yang, H. H., Dewer, Y., Zhang, F., et al. (2021). Chemosensory genes in the head of Spodoptera litura larvae. Bull. Entomol. Res. 111, 454–463. doi: 10.1017/S0007485321000109
Li, M., Li, X., Wang, C., Li, Q., Zhu, S., Zhang, Y., et al. (2021). Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: aphididae). Front. Physiol. 12:663338. doi: 10.3389/fphys.2021.663338
Li, W. X., Yang, Y. T., Xie, W., Wu, Q. J., Xu, B. Y., Wang, S. L., et al. (2015). Effects of temperature on the age-stage, two-sex life table of Bradysia odoriphaga (Diptera: sciaridae). J. Econ. Entomol. 108, 126–134. doi: 10.1093/jee/tou011
Li, Z. N., Gu, X. S., Xu, W. H., Xu, J. Y., and Chen, J. R. (2015). Study on toxic effect of 8 kinds of insecticides to Bradysia odoriphaga. Shandong Agric. Sci. 47, 107–108.
Liang, P., Guo, Y., Zhou, X., and Gao, X. (2014). Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLoS One 9:e87514. doi: 10.1371/journal.pone.0087514
Liu, G., Qiu, X., Cao, L., Zhang, Y., Zhan, Z., and Han, R. (2016). Evaluation of reference genes for reverse transcription quantitative PCR Studies of Physiological Responses in the Ghost Moth, Thitarodes armoricanus (Lepidoptera, Hepialidae). PLoS One 11:e0159060. doi: 10.1371/journal.pone.0159060
Lü, J., Yang, C., Zhang, Y., and Pan, H. (2018). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Lu, Y., Yuan, M., Gao, X., Kang, T., Zhan, S., Wan, H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: noctuidae). PLoS One 8:e68059. doi: 10.1371/journal.pone.0068059
Luo, J., Wang, A., Cheng, Y., Rong, H., Guo, L., Peng, Y., et al. (2020). Selection and validation of suitable reference genes for RT-qPCR analysis in Apolygus lucorum (Hemiptera: miridae). J. Econ. Entomol. 113, 451–460. doi: 10.1093/jee/toz301
Luo, J., Zhang, Z., Li, D., Liu, J., Li, K., Sun, X., et al. (2021). Identification and functional analysis of SlitOBP11 from Spodoptera litura. Front. Physiol. 12:619816. doi: 10.3389/fphys.2021.619816
Nazar, M. Z., Freed, S., Hussain, S., Sumra, M. W., Shah, M. S., and Naeem, A. (2020). Characteristics of biochemical resistance mechanism of novel insecticides in Phenacoccus solenopsis Tinsley (Hemiptera: pseudococcidae). Crop Protect. 138:105320. doi: 10.1016/j.cropro.2020.105320
Nor Muhammad, N. A., Ramlee, I. A., Mohd Nor, D., Satyavenathan, M. V., Rahmat, N. L., Awang, A., et al. (2020). Data on RNA-seq analysis of the cocoa pod borer pest Conopomorpha cramerella (Snellen) (Lepidoptera: gracillariidae). Data Brief. 34:106638. doi: 10.1016/j.dib.2020.106638
Pan, H., Yang, X., Siegfried, B. D., and Zhou, X. (2015). A Comprehensive Selection of Reference Genes for RT-qPCR Analysis in a Predatory Lady Beetle, Hippodamia convergens (Coleoptera: coccinellidae). PLoS One 10:e0125868. doi: 10.1371/journal.pone.0125868
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Pinheiro, D. H., Moreira, R. O., Leite, N. A., Redoan, A. C., Xavier, A. D. S., Barros, B. A., et al. (2020). Suitable reference genes for RT-qPCR analysis in Dichelops melacanthus (Hemiptera: pentatomidae). Mol. Biol. Rep. 47, 4989–5000. doi: 10.1007/s11033-020-05550-z
Ponton, F., Chapuis, M. P., Pernice, M., Sword, G. A., and Simpson, S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850. doi: 10.1016/j.jinsphys.2011.03.014
Prajapati, V. K., Varma, M., and Vadassery, J. (2020). In silico identifification of effector proteins from generalist herbivore Spodoptera litura. BMC Genom. 21:819. doi: 10.1186/s12864-020-07196-4
Shakeel, M., Rodriguez, A., Tahir, U. B., and Jin, F. (2018). Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 40, 227–236. doi: 10.1007/s10529-017-2465-4
Shakeel, M., Zhu, X., Kang, T., Wan, H., and Li, J. (2015). Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Asia Pacif. Entomol. 18, 123–130. doi: 10.1016/j.aspen.2015.01.001
Shen, G. M., Jiang, H. B., Wang, X. N., and Wang, J. J. (2010). Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: tephritidae). BMC Mol. Biol. 11:76. doi: 10.1186/1471-2199-11-76
Shi, C., Yang, F., Zhu, X., Du, E., Yang, Y., Wang, S., et al. (2016). Evaluation of housekeeping genes for Quantitative Real-Time PCR analysis of Bradysia odoriphaga (Diptera: sciaridae). Int. J. Mol. Sci. 17:1034. doi: 10.3390/ijms17071034
Silver, K., Cooper, A. M., and Zhu, K. Y. (2021). Strategies for enhancing the efficiency of RNA interference in insects. Pest. Manag. Sci. 77, 2645–2658. doi: 10.1002/ps.6277
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Sun, M., Lu, M. X., Tang, X. T., and Du, Y. Z. (2015). Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: noctuidae). PLoS One 10:e0115979. doi: 10.1371/journal.pone.0115979
Tang, B., Dai, W., and Zhang, C. (2019). Selection of reference genes for quantitative real-time polymerase chain reaction normalization in Bradysia odoriphaga (Diptera: sciaridae). Entomol. Sci. 22, 422–436. doi: 10.1111/ens.12383
Teng, X., Zhang, Z., He, G., Yang, L., and Li, F. (2012). Validation of reference genes for quantitative expression analysis by real-time rt-PCR in four lepidopteran insects. J. Insect Sci. 12:60. doi: 10.1673/031.012.6001
Ullah, I., Wazir, S., Abbas, N., Naeem, M., Abdullah, K., Mahmood, Z., et al. (2021). Monitoring of field-evolved resistance to flonicamid, neonicotinoid, and conventional insecticides in the Oxycarenus hyalinipennis costa. Environ. Monit. Assess. 193:382. doi: 10.1007/s10661-021-09158-z
Vandesompele, J., Preter, K. D., Pattyn, F., Poppe, B., Roy, N. V., Paepe, A. D., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Van Hiel, M. B., Van Wielendaele, P., Temmerman, L., Van Soest, S., Vuerinckx, K., Huybrechts, R., et al. (2009). Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 10:56. doi: 10.1186/1471-2199-10-56
Wang, R., Che, W., Wang, J., and Luo, C. (2020). Monitoring insecticide resistance and diagnostics of resistance mechanisms in Bemisia tabaci Mediterranean (Q biotype) in China. Pestic. Biochem. Physiol. 163, 117–122.
Xiao, H. Y., Li, G. C., Wang, Z. Q., Guo, Y. R., and Liu, N. Y. (2021). Combined transcriptomic, proteomic and genomic analysis identifies reproductive-related proteins and potential modulators of female behaviors in Spodoptera litura. Genomics 113, 1876–1894. doi: 10.1016/j.ygeno.2021.04.006
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2
Yan, X., Zhang, Y., Xu, K., Wang, Y., and Yang, W. (2021). Selection and validation of reference genes for gene expression analysis in Tuta absoluta Meyrick (Lepidoptera: gelechiidae). Insects 12:589. doi: 10.3390/insects12070589
Yang, C., Pan, H., Noland, J. E., Zhang, D., Zhang, Z., Liu, Y., et al. (2015). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: coccinellidae). Sci Rep. 5:18201. doi: 10.1038/srep18201
Yang, C., Preisser, E. L., Zhang, H., Liu, Y., Dai, L., Pan, H., et al. (2016). Selection of Reference Genes for RT-qPCR Analysis in Coccinella septempunctata to Assess Un-intended Effects of RNAi Transgenic Plants. Front Plant Sci. 7:1672. doi: 10.3389/fpls.2016.01672
Yang, Q., Li, Z., Cao, J., Zhang, S., Zhang, H., Wu, X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: acrididae). PLoS One 9:e98164. doi: 10.1371/journal.pone.0098164
Yang, X., Pan, H., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. doi: 10.1038/s41598-018-20612-w
Yang, Y. T., Li, W. X., Xie, W., Wu, Q. J., Xu, B. Y., Wang, S. L., et al. (2015). Development of Bradysia odoriphaga (Diptera: sciaridae) as affected by humidity: an age–stage, two-sex, life-table study. Appl. Entomol. Zool. 50, 3–10. doi: 10.1007/s13355-014-0295-6
Yin, J., Sun, L., Zhang, Q., and Cao, C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: erebidae) using qRT-PCR. Gene 749:144712. doi: 10.1016/j.gene.2020.144712
Yu, H. Z., Li, N. Y., Xie, Y. X., Zhang, Q., Wang, Y., and Lu, Z. J. (2020). Identification and functional analysis of two chitin synthase genes in the common cutworm, Spodoptera litura. Insects 11:253. doi: 10.3390/insects11040253
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: delphacidae) using reverse-transcription quantitative PCR. PLoS One 9:e86503. doi: 10.1371/journal.pone.0086503
Zhang, H., Mallik, A., and Zeng, R. S. (2013). Control of Panama disease of banana by rotating and intercropping with Chinese chive (Allium tuberosum Rottler): role of plant volatiles. J. Chem. Ecol. 39, 243–252. doi: 10.1007/s10886-013-0243-x
Zheng, L., Zhang, Z., Zhang, J., Li, X., Huang, J., Lin, W., et al. (2019). Selection of reference genes for RT-qPCR analysis of Phenacoccus solenopsis (Hemiptera: pseudococcidae) sex-dimorphic development. J. Integr. Agric. 18, 854–864. doi: 10.1016/s2095-3119(18)61973-2
Zheng, Y. T., Li, H. B., Lu, M. X., and Du, Y. Z. (2014). Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: thripidae). PLoS One 9:e111369. doi: 10.1371/journal.pone.0111369
Zhou, L., Meng, J. Y., Ruan, H. Y., Yang, C. L., and Zhang, C. Y. (2021). Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: noctuidae). Arch. Insect. Biochem. Physiol. 108:e21831. doi: 10.1002/arch.21831
Zhu, X., Yuan, M., Shakeel, M., Zhang, Y., Wang, S., Wang, X., et al. (2014). Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS One 9:e84730. doi: 10.1371/journal.pone.0084730
Keywords: Bradysia odoriphaga, RT-qPCR, reference gene, insecticides stress, normalization
Citation: Fu H, Huang T, Yin C, Xu Z, Li C, Liu C, Wu T, Song F, Feng F and Yang F (2022) Selection and Validation of Reference Genes for RT-qPCR Normalization in Bradysia odoriphaga (Diptera: Sciaridae) Under Insecticides Stress. Front. Physiol. 12:818210. doi: 10.3389/fphys.2021.818210
Received: 19 November 2021; Accepted: 08 December 2021;
Published: 11 January 2022.
Edited by:
Ran Wang, Institute of Plant Protection and Environmental Protection, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Xingliang Wang, Nanjing Agricultural University, ChinaHuipeng Pan, South China Agricultural University, China
Haipeng Zhao, Shandong Agricultural University, China
Copyright © 2022 Fu, Huang, Yin, Xu, Li, Liu, Wu, Song, Feng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengshan Yang, MjAwNDA2NEBobGp1LmVkdS5jbg==
 Haiyan Fu
Haiyan Fu Tubiao Huang
Tubiao Huang Cheng Yin
Cheng Yin Zhenhua Xu1,2
Zhenhua Xu1,2 Chao Li
Chao Li Fuqiang Song
Fuqiang Song Fujuan Feng
Fujuan Feng Fengshan Yang
Fengshan Yang