
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 12 January 2022
Sec. Membrane Physiology and Membrane Biophysics
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.802057
This article is part of the Research TopicTargeting Signals in Protein Trafficking and TransportView all 15 articles
In higher plants, chloroplasts are essential semi-autonomous organelles with complex compartments. As part of these sub-organellar compartments, the sheet-like thylakoid membranes contain abundant light-absorbing chlorophylls bound to the light-harvesting proteins and to some of the reaction center proteins. About half of the thylakoid membrane proteins are encoded by nuclear genes and synthesized in the cytosol as precursors before being imported into the chloroplast. After translocation across the chloroplast envelope by the Toc/Tic system, these proteins are subsequently inserted into or translocated across the thylakoid membranes through distinct pathways. The other half of thylakoid proteins are encoded by the chloroplast genome, synthesized in the stroma and integrated into the thylakoid through a cotranslational process. Much progress has been made in identification and functional characterization of new factors involved in protein targeting into the thylakoids, and new insights into this process have been gained. In this review, we introduce the distinct transport systems mediating the translocation of substrate proteins from chloroplast stroma to the thylakoid membrane, and present the recent advances in the identification of novel components mediating these pathways. Finally, we raise some unanswered questions involved in the targeting of chloroplast proteins into the thylakoid membrane, along with perspectives for future research.
Chloroplasts of higher plants, the semi-autonomous organelles with internal sheet-like membrane-bound structure, are not only the sites for photosynthesis, but also for the synthesis of many essential metabolites, such as fatty acids, amino acids, vitamins, tetrapyrroles, and some phytohormones (Neuhaus and Emes, 2000; Wicke et al., 2011). Chloroplasts are believed to have evolved from an ancient cyanobacteria-like endosymbiont. Following this endosymbiosis most of the endosymbiont genes were transferred to the host nuclear genome. As a result, ∼3,000 chloroplast proteins are encoded by nuclear genes, while only ∼100 proteins are encoded by the chloroplast genome and synthesized in the stroma (Lee et al., 2017; Sjuts et al., 2017; Nakai, 2018; Richardson and Schnell, 2020). Chloroplasts contain three distinct membranes: outer envelope, inner envelope, and thylakoid membranes, which separate the chloroplast into three compartments: the intermembrane space, chloroplast stroma and thylakoid lumen (Staehelin, 2003; Kirchhoff, 2019). The thylakoid contains several photosynthetic complexes including photosystem I (PSI), photosystem II (PSII), the cytochrome b6f complex (Cyt b6f), and ATP synthase (ATPase), which are responsible for the primary reactions of photosynthesis. About half of the subunits of these complexes are encoded by the nuclear genome, synthesized in the cytosol, imported into chloroplasts and transported to the thylakoids. However, the complex structure of chloroplasts makes translocation of these proteins across the membrane system for correct targeting to their specific sub-compartment highly challenging.
The nuclear-encoded thylakoid proteins with an N-terminal transit peptide (TP) are first imported into the stroma by the Toc/Tic (translocon at the outer/inner envelope membrane of chloroplasts) complexes, and then the TP is removed by the stromal processing peptidase, and the proteins are targeted into or across the thylakoid membranes through sophisticated targeting pathways (Martin et al., 2002; Leister, 2003). To date, several independent pathways have been reported, of which the chloroplast signal recognition particle (cpSRP) pathway, the spontaneous insertion pathway and the recently discovered chloroplast Guided Entry of TA proteins (cpGET) pathway are responsible for inserting proteins into the thylakoid membrane (Figure 1), while the chloroplast secretory (cpSec) and the chloroplast twin-arginine translocase (cpTat) pathways are needed for translocating proteins into the thylakoid lumen (Schünemann, 2007; Ouyang et al., 2020; Anderson et al., 2021; Xu et al., 2021a,b). In contrast, proteins encoded by the chloroplast genome are cotranslationally transported from the stroma to thylakoids (Figure 1). Although several translocation events can be distinguished through their components and energy requirements, how the imported chloroplast proteins are recognized and sorted exactly to their distinct pathways is still largely unknown. In this review, we describe the distinct transport mechanisms for targeting nuclear- and chloroplast-encoded proteins from the stroma to the thylakoid membrane. Moreover, we review recent advances and the discovery of novel components of these transport systems. Finally, we discuss current challenges and present new perspectives in this promising field.
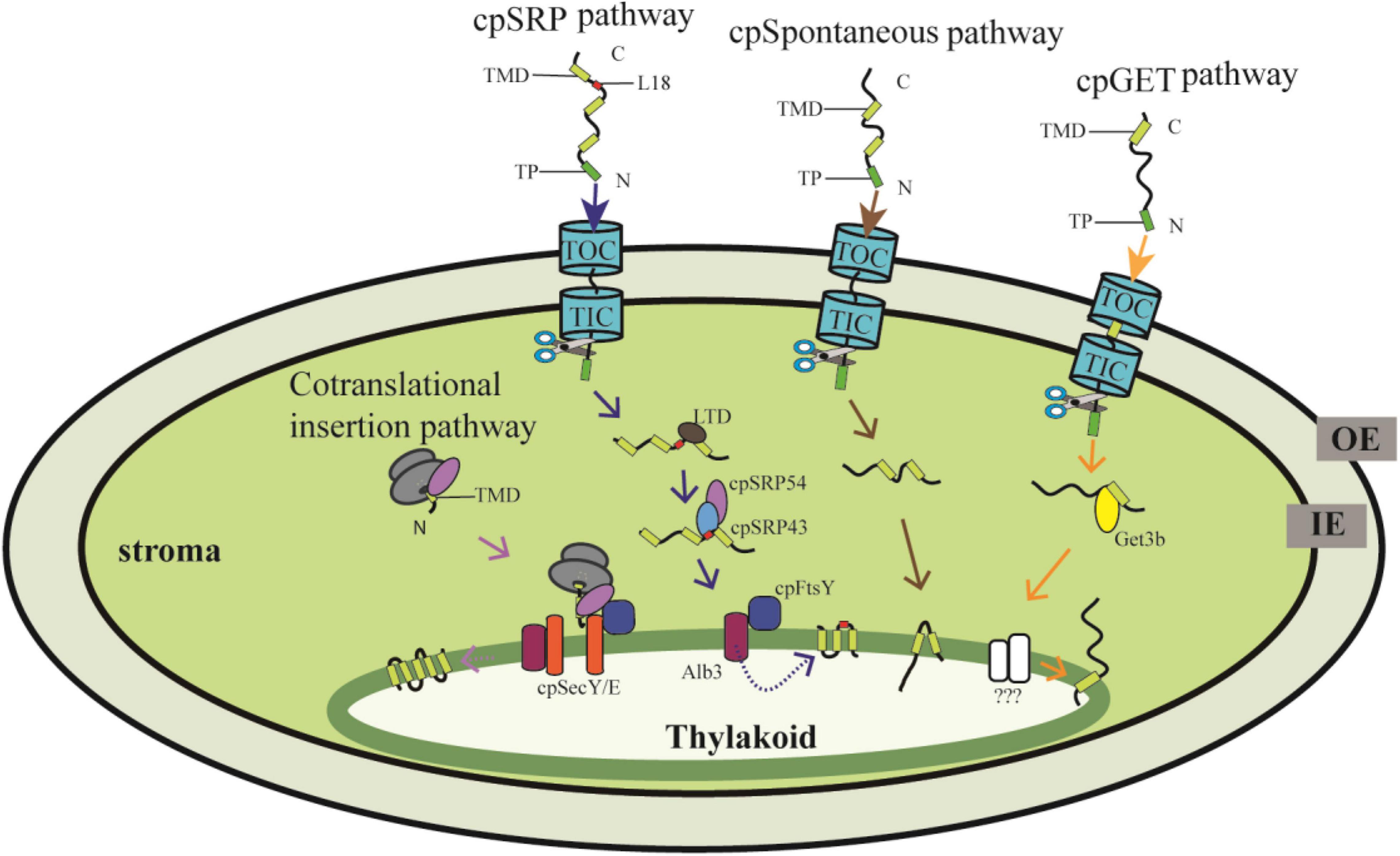
Figure 1. Proteins targeting into thylakoid membranes through distinct pathways. The nuclear-encoded preprotein with an N-terminal transit peptide (TP) is translocated into the chloroplast stroma through the Toc/Tic system, and then the TP is cleaved by stromal processing peptidase. The thylakoid membrane proteins are further targeted mainly through the cpSRP pathway, cpGET pathway and the spontaneous pathway. In the cpSRP-dependent pathway, LHCP is transferred from the Toc/Tic system to the SRP43/SRP54 complex with the help of LTD., cpSRP43 interacts with the L18 region between the second and third TMD of LHCP. Then the cpSRP/LHCP complex interacts with cpFtsY and finally LHCP is insert into the thylakoid membrane by the integrase Alb3. In the cpGET pathway, imported TA proteins are transferred to the targeting factor Get3b, followed by transferr to the unknown integrases. Some proteins are translocated into thylakoid membranes spontaneously. On the other hand, the chloroplast-encoded proteins are synthesized in the stroma, and cotranslationally integrated into the thylakoid membrane with the requirement of cpSec and cpSRP components. OE, outer chloroplast envelope; IE, inner chloroplast envelope. TP, transit peptide. TMD, transmembrane domain.
The thylakoid membrane harbors the photosynthetic apparatus composed of PSI, PSII, Cyt b6f, and ATP synthase. Besides, several proteins involved in thylakoid biogenesis and homeostasis are also anchored in the thylakoid membrane. These proteins are encoded by both the nuclear and chloroplast genomes, of which the nuclear-encoded proteins are translocated into thylakoid membranes through the cpSRP pathway, the spontaneous insertion pathway, and the cpGET pathway, whereas chloroplast-encoded proteins are inserted into the thylakoid membrane cotranslationally (Figure 1).
The classical SRP system is responsible for cotranslationally delivering newly synthesized proteins to their correct cellular membranes. The functional core of cytosolic SRP is composed of the SRP54 GTPase and SRP RNA, which is tightly bound to SRP54. Notably, the cpSRP system in green plants is different from all other SRP systems because the cpSRP lacks the associated RNA and instead consists of a conserved cpSRP54 and a unique chloroplast-specific cpSRP43 subunit. Contrary to the universally conserved cytosolic SRP system which only binds to the short nascent polypeptide chains and functions cotranslationally, the cpSRP system evolved posttranslational activity to target the light-harvesting chlorophyll a/b binding proteins (LHCPs) from the stroma to the thylakoid membrane (Ziehe et al., 2017, 2018; Costa et al., 2018). LHCP membrane integration requires the cpSRP43/cpSRP54 complex and its receptor cpFtsY, the integral translocase Alb3, and GTP, which is hydrolyzed by the cpSRP54 and cpFtsY GTPases (Moore et al., 2003; Ziehe et al., 2018; Figure 1).
It was first confirmed that the L18 region between the second and third transmembrane domain (TMD) of LHCP is the binding site for cpSRP43 (DeLille et al., 2000; Tu et al., 2000; Stengel et al., 2008). Recently, however, McAvoy et al. (2018) found that cpSRP43 makes more extensive interactions with all the TMDs within LHCP, thus protecting the hydrophobic LHCP from aggregation in the stroma. These new interaction sites are important for the chaperone activity of cpSRP43 based on the result that a class of cpSRP43 mutants are specifically deficient in their ability of chaperoning full-length LHCP but are not affected in their association activity with the L18 region. NMR analysis showed three conformations of cpSRP43. Interaction with cpSRP54 produces the active state through structural rearrangement, thereby improving the substrate binding efficiency of cpSRP43 (Gao et al., 2015; Liang et al., 2016). The assembly of cpSRP54 to the cpSRP43/LHCP complex leads to a much smaller LHCP/cpSRP transit complex (∼170 kDa) than the cpSRP43/LHCP complex (∼450 kDa), most likely to enable efficient LHCP insertion (Dünschede et al., 2015).
Upon formation of the cpSRP/LHCP transit complex, it is guided to the thylakoid membrane and docks to the membrane-bound cpSRP receptor cpFtsY, and then transfers LHCP to the integrase Alb3 for insertion into the thylakoid membrane. cpFtsY and cpSRP54 form a complex by association of their homologous NG domains (the N-terminal four-helix bundle and the GTPase domain), and stabilization of the cpSRP54/cpFtsY complex is dramatically activated by anionic phospholipids and the Alb3 translocase (Chandrasekar and Shan, 2017). Moreover, a new interaction site between a positively charged cluster within the cpFtsY G-domain (the GTPase domain) and a negatively charged cluster in the cpSRP54 M-domain has been identified, in which the M domain of cpSRP54 plays a similar role as the classical SRP RNA to enhance the cpSRP54/cpFtsY complex assembly (Chandrasekar et al., 2017; Figure 2).
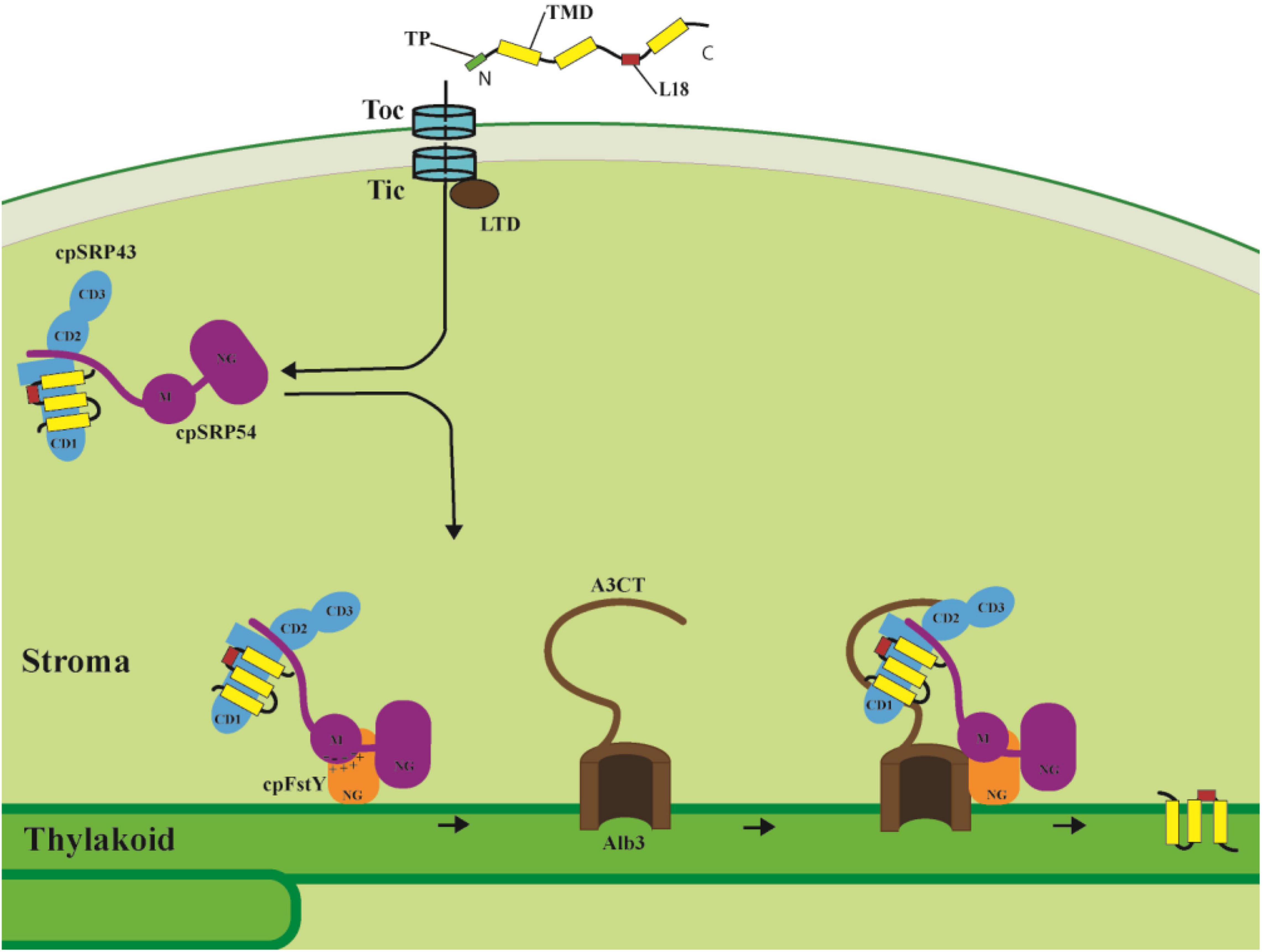
Figure 2. LHCPs targeting into thylakoid membranes through the cpSRP pathway. After import into the stroma through the Toc/Tic system, the LHCPs are transferred to the cpSRP43/cpSRP54 complex with the help of LTD. The L18 region between the second and third TMD of LHCP is the binding site for cpSRP43. Then cpSRP54 interacts with cpFtsY through their homologous NG domains, while the negatively charged cluster of cpSRP54 M domain interacts with the positively charged cluster of cpFtsY G domain and enhances the cpSRP54/cpFtsY complex assembly. Finally, Alb3 C-terminal tail (A3CT) interacts with the C-terminal chromodomains CD2 and CD3 of cpSRP43, and thus facilitates LHCP delivery to the Alb3 translocase and insertion into the thylakoid membrane. TP, transit peptide. TMD, transmembrane domain.
The recruitment of the transit complex to the Alb3 translocase for LHCP insertion is mediated through the cpSRP43/Alb3 interaction. The linear motif in the Alb3 C-terminal tail (A3CT) interacts with the C-terminal chromodomains CD2 and CD3 of cpSRP43, which enables the efficient delivery of LHCP to the Alb3 integrase (Falk et al., 2010; Horn et al., 2015; Figure 2). A novel model has been proposed in which the conformational dynamics of cpSRP43 enables LHCP capture and release. Distinct conformations of cpSRP43 allow it to be activated by cpSRP54 assembly in the stroma to capture LHCP and inactivated by association with Alb3 translocase in the thylakoid membrane to release LHCP (Liang et al., 2016). Interestingly, cpSRP43 also directly interacts with the N-terminal of glutamyl-tRNA reductase (GluTR), a rate-limiting enzyme in tetrapyrrole biosynthesis (TBS), chaperones and stabilizes GluTR, and consequently optimizes chlorophyll biosynthesis. This interaction between cpSRP43 and GluTR reveals a posttranslational coordination for LHCP insertion with chlorophyll biosynthesis (Wang et al., 2018). More strikingly, high temperature drives the dissociation of the cpSRP43/54 complex, thus freeing cpSRP43 to interact with GluTR, CHLH (Mg-chelatase H subunit) and GUN4 (Genomes uncoupled 4) to protect them from heat-induced aggregation (Ji et al., 2021). Taken together, cpSRP43 not only functions as the hub for LHCP membrane insertion since it recruits cpSRP54, chaperones LHCP, and provides the docking site for Alb3 translocase, but also chaperones proteins for chlorophyll biosynthesis and thus coordinates the assembly of chlorophyll and LHCP into the light-harvesting complex.
Several new components of the cpSRP pathway have also been identified. An Arabidopsis ankyrin protein, LTD., which is located in the stroma and interacts with the Tic complex, LHCP and cpSRP43, was demonstrated to handover LHCP from the Tic translocon to the cpSRP43/cpSRP54 complex (Ouyang et al., 2011). In addition, STIC1 (suppressor of tic40) and STIC2 were revealed to act in the cpSRP54/cpFtsY-involved transport pathway. The STIC1 gene encodes the Alb3 paralog protein, Alb4, while STIC2 encodes a novel stromal protein that associates with Alb3 and Alb4. Loss of STIC1/Alb4 and STIC2 proteins may mitigate the defect of protein import caused by the TIC40 mutation, thus revealing a potential link between the envelope and thylakoid protein transport systems (Bédard et al., 2017).
Many proteins such as the photosystem subunit PsaG, PsaK, PsbW, PsbX, and PsbY, the ATPase subunit CFo-II, and the cpTat translocase subunits Tha4 and Hcf106, are integrated into the thylakoid membrane via the spontaneous insertion pathway which does not need any energy or proteinaceous components. This direct insertion pathway was first proposed for CFo-II, a nuclear-encoded single-membrane-spanning component of the ATP synthase. Treatments that disrupt the cpSRP, cpSec and cpTat dependent pathways do not affect the integration of CFo-II, suggesting a direct interaction of CFo-II with the lipid components of the membrane (Michl et al., 1994; Robinson et al., 1996). CFo-II, PsbW, and PsbX all span the thylakoid membrane with a single hydrophobic domain, whereas PsaG and PsaK insert into the membrane with two transmembrane spans (Lorkovic et al., 1995; Kim et al., 1998; Thompson et al., 1998; Mant et al., 2001; Zygadlo et al., 2006; Aldridge et al., 2009). For PsbY, the two transmembrane spans partition into the lipid bilayer of the thylakoid membrane and provide the driving force required for the translocation of the intervening charged region through hydrophobic interactions (Thompson et al., 1999).
The mechanism of insertion of the M13 procoat protein into the cytoplasmic membrane of E. coli was also originally proposed to be by the spontaneous pathway. However, it was subsequently found that YidC, the homolog of the chloroplast Alb3 insertase, is actually required for the insertion of the M13 procoat protein, which raised the question if there exists a truly spontaneous insertion pathway (Kuhn et al., 1986; Samuelson et al., 2000). PsbW and PsbX have remarkable structural similarities with the M13 procoat protein. However, inactivation of Alb3 did not affect PsbW and PsbX insertion into the thylakoid membrane, suggesting a direct insertion pathway for these proteins (Woolhead et al., 2001).
In a typical eukaryote cell, 3%∼5% of the membrane proteins are tail-anchored (TA) proteins with the feature of a cytosolic-facing N-terminal domain and a single C-terminal TMD followed by a tail within 30 amino acids. In these proteins TMD acts both as a membrane anchor and a targeting signal (Shigemitsu et al., 2016; Mateja and Keenan, 2018). TA proteins are found in all cellular membranes and play key roles in protein translocation, membrane fusion, vesicular trafficking, apoptosis, organelle biogenesis, and other essential cellular processes (Denic et al., 2013).
The posttranslational insertion of TA proteins from the cytosol to cellular membranes depends on the GET pathway in yeast and the TRC (Transmembrane Recognition Complex) pathway in mammals (Farkas et al., 2019; Farkas and Bohnsack, 2021). The yeast cytosolic GET machinery includes a pretargeting complex consisting of Sgt2, a small glutamine-rich tetratricopeptide repeat (TPR)-containing protein (SGTA in mammals), Get4 (TRC35 in mammals), and Get5 (UBL4 in mammals), of which Sgt2 captures newly synthesized TA proteins from the ribosomal exit tunnel and is considered the most upstream factor in the GET pathway (Wang et al., 2010; Shao et al., 2017; Zhang et al., 2021). Subsequently Get4 and Get5 build up a “scaffolding complex” by recruiting Get3 and the Sgt2-TA complex, respectively. This scaffolding complex prevents aggregation and promotes loading of TA proteins onto Get3 (Bat3 in mammals), the central targeting factor that forms a closed structure after binding ATP. The resulting hydrophobic groove recognizes the TMD of TA proteins. At the endoplasmic reticulum (ER) membrane, Get2 (CAML in mammals) aids Get1 (WRB in mammals) to form a tight complex with nucleotide-free Get3, resulting in the release of the TA protein for insertion into the membranes. Finally, Get3 is recycled to the cytosol to initiate a new round of targeting (Schuldiner et al., 2008; Gristick et al., 2014; Rome et al., 2014; Wang et al., 2014; Rao et al., 2016; Figure 3A). A recent study showed that the cytoplasmic helix α3′ of Get2/CAML forms a “gating” interaction with Get3/TRC40, which is important for guiding TA proteins insertion into membranes (McDowell et al., 2020).
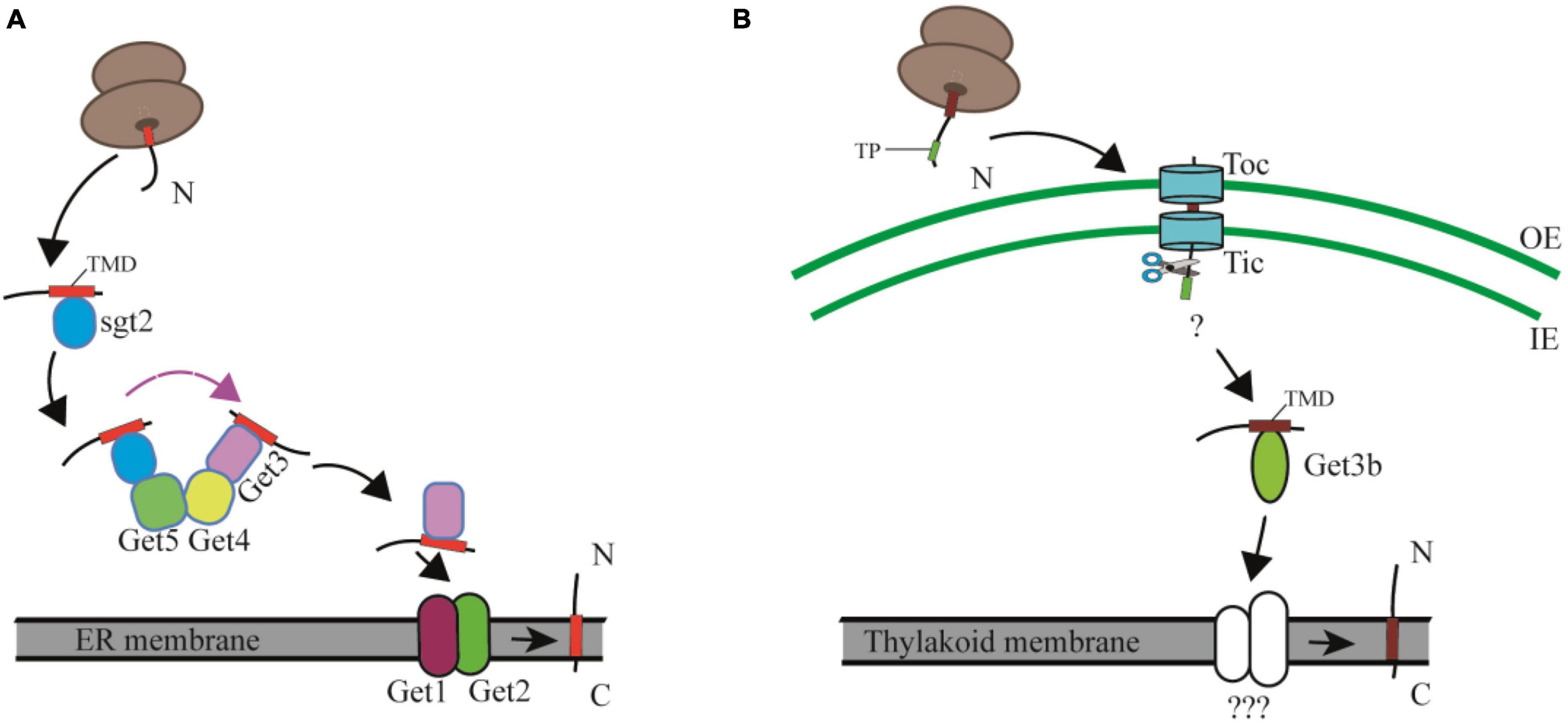
Figure 3. Comparison of the cytosolic and chloroplast GET pathway. (A) In the cytosolic GET pathway, the newly synthesized TA proteins are firstly captured by Sgt2, and then transferred to the targeting factor Get3 by the Get4/Get5 scaffold complex, and finally integrated into ER membranes with the facilitation of Get1 and Get2. TMD, transmembrane domain. (B) In the cpGET pathway, imported TA proteins are transferred to the targeting factor Get3b, and then integrated into the thylakoid membrane with the aid of unknown integrases. OE, outer chloroplast envelope; IE, inner chloroplast envelope. TP, transit peptide. TMD, transmembrane domain.
While the GET pathway was originally identified in mammals and yeasts, it was recently found to be partially conserved in higher plants as well. Orthologs of Get1, Get3, Get4, Get5, and Sgt2, but not Get2 have been identified in plant genomes through bioinformatic approaches Get1, Get3, Get4, Get5, and Sgt2, Get2 (Paul et al., 2013; Srivastava et al., 2017). Further phylogenetic analysis of 18 species revealed two Get3 clades termed Get3a and Get3bc, of which AtGet3a, AtGet3b, and AtGet3c are localized in the cytosol, chloroplast and mitochondria, respectively. Loss of AtGet1 or AtGet3a results in a significant reduction of the root hair-specific protein SYP123, and thus leads to root hair growth defects in these Atget mutant lines (Xing et al., 2017). Most surprisingly, overexpression of AtGet3a in the Atget1 mutant causes a more severe phenotype in root, silique and seed development compared with the parental lines. The absence of GET pathway components may trigger alternative insertion pathways. However, overexpression of AtGet3a in the receptor mutant Atget1 might cause cytosolic AtGet3a/TA protein aggregates and a consequent TA protein insertion block through the alternative insertion pathway (Xing et al., 2017).
Recently, Arabidopsis Get2 was also discovered by an immunoprecipitation-mass spectrometry (IP-MS) method. Using the AtGet1-GFP transgenic plants, G1IP (AtGet1-interacting protein) was identified. G1IP exhibits low sequence similarity but high structural similarity to Get2/CAML (Asseck et al., 2021). Both G1IP and AtGet1 show a subcellular ER localization and share the same expression profile. The g1ip mutant shows reduced root hair elongation, which is similar to other Atget lines. Moreover, G1IP interacts with AtGet1 and AtGet3a, and expression of G1IP and AtGet1 together can complement the yeast GET receptor mutant Δget1get2 strain. Besides, the G1IP N terminus contains a conserved cluster of positively charged amino acids which is essential for the binding of Get3/TRC40, and alteration of this motif in G1IP is sufficient to inhibit TA protein transport. These results strongly suggest that Arabidopsis G1IP encodes the functional ortholog of Get2, although only a small part of its sequence is conserved between yeasts, plants and mammals (Asseck et al., 2021).
Based on the facts that only Get3b and Get3c, but no other Get system components are found in organelles, and that Get3bc cannot rescue the growth defect of the Atget3a mutant, it was previously considered to be unlikely that the organellar Get3 homologs were involved in TA protein insertion (Xing et al., 2017). However, Anderson et al. recently demonstrated that AtGet3b is structurally similar to the cytosolic Get3 protein and serves as a targeting factor to deliver TA proteins from the chloroplast stroma to the thylakoid membrane (Anderson et al., 2021; Figure 3B). Chloroplast membranes contain several TA proteins, such as cpSecE1, a component of the thylakoid Sec1 translocase and cpSecE2, a component of the chloroplast inner envelope Sec2 translocase (Schüenemann et al., 1999; Li et al., 2015). Both in vitro and in vivo assays indicated that AtGet3b interacts with cpSecE1 rather than cpSecE2 to form a homodimer or oligomer in the chloroplast stroma, mediated by the C-X-X-C zinc coordination motif of AtGet3b (Anderson et al., 2021). A domain-swapping strategy used between cpSecE1 and cpSecE2 suggests that the TMDs and C-terminal tails of TA proteins are key factors that specify the interaction between AtGet3b and its client TA proteins (Anderson et al., 2019). The physicochemical features of the TMD and C-terminal regions of cpSecE2 make it incompatible with the hydrophobic groove formed by AtGet3b oligomerization (Anderson et al., 2021).
Loss of get3b in Arabidopsis displays a visually indistinguishable phenotype from the wild type. Similar observations have been made with the yeast get3 mutant, suggesting that the Get system may operate in parallel or at least be partially redundant with other insertion pathways (Anderson et al., 2021). Consistent with this notion, the get3bsrp54 double mutant and the get3bsrp43srp54 triple mutant show much more severe defects in growth, including pronounced dwarfism, obvious chlorosis and dramatically reduced PSI and PSII activities. These results demonstrate synergistic interactions between cpGET and the cpSRP pathways in delivering proteins into thylakoid membranes (Anderson et al., 2021).
Chloroplast genes encode ∼37 thylakoid membrane proteins such as the PSII reaction center protein D1 and D2, and the PSI subunits PsaA and PsaB, which are synthesized in the stroma and integrated into thylakoid membranes through a cotranslational process (Zoschke and Barkan, 2015). However, little is known about the mechanisms that target these proteins into the thylakoid membrane. The cpSRP54 (ffc) mutant contains much lower levels of D1, D2, PsaA, and PsaB proteins, suggesting the involvement of cpSRP54 in the cotranslational pathway (Amin et al., 1999). Cross-linking experiments revealed an interaction between the D1-ribosome nascent chain complex and cpSRP54, but not cpSRP43 (Nilsson et al., 1999). cpSRP54 is present in two stroma pools. In one of them it is recruited by cpSRP43 to function in the posttranslational targeting for LHCPs, and in the other pool it is associated with 70S ribosomes to function in the cotranslational import of proteins such as D1 (Franklin and Hoffman, 1993; Schüenemann et al., 1998). The interaction between cpSRP54 and D1 involves the first TMD of D1, and only occurs when the D1 nascent chain is still attached to ribosomes, suggesting that cpSRP54 functions in the early steps of D1 biogenesis (Nilsson and van Wijk, 2002).
Besides cpSRP54, the SRP receptor cpFtsY is also considered to function in the cotranslational integration process of chloroplast-encoded thylakoid membrane proteins, based on the observation of a strong reduction of D1 in the cpftsy mutant (Tzvetkova-Chevolleau et al., 2007; Asakura et al., 2008). Zhang et al. (2001) demonstrated a direct interaction between D1 elongation intermediates and the chloroplast Sec translocon component cpSecY, suggesting that the cpSec pathway not only functions in posttranslational targeting of nuclear-encoded proteins, but also in cotranslational insertion of chloroplast-encoded proteins. The Alb3 integrase of the cpSRP pathway is also reported to act in a later insertion process, facilitate release of D1 from the cpSecY translocase, and promote the assembly of D1 into PSII (Ossenbühl et al., 2004; Walter et al., 2015). Thus, the integration of D1 into the thylakoid membrane suggests a functional link between the cpSRP and the cpSec translocation machinery. Further evidence for this speculation comes from the integration of Cytochrome f. Cytochrome f insertion into the thylakoid membrane was initially thought to be mediated through the cpSec translocon. However, further analysis showed a severely decreased level of Cytochrome f in the cpftsy mutant, indicating a coordinated mechanism between the cpSRP and cpSec translocation pathways (Nohara et al., 1996; Asakura et al., 2008).
Recently, Zoschke and Barkan performed a comprehensive analysis of chloroplast ribosome profiling with separated soluble and membrane fractions in maize. They found that about half of the chloroplast-encoded thylakoid membrane proteins are targeted cotranslationally while the other half is targeted posttranslationally. Synthesis of these cotranslationally integrated proteins are initiated on stromal ribosomes, and then transferred to the thylakoid-attached ribosomes in a nuclease-resistant fashion shortly after the emergence of the first transmembrane segment of the nascent peptide. In contrast, membrane proteins whose translation terminates before a transmembrane segment emerge from the ribosome’s exit channel will be integrated into thylakoid membranes posttranslationally. These results suggest a key role of the first transmembrane segment in linking ribosomes to the thylakoid membrane for cotranslational targeting (Zoschke and Barkan, 2015).
In the past decades, much progress has been made in understanding the molecular details of targeting both nuclear- and chloroplast-encoded proteins into the thylakoid membranes. However, several central issues still need to be further investigated in the future. One avenue of study will be to explore novel components of these translocation pathways. During evolution, ancient translocation systems acquired and developed new proteins and mechanisms to facilitate the correct targeting of chloroplast preproteins. Thus, identification of these novel components may provide new insights into how plant cells have adapted prokaryotic mechanisms to the eukaryotic environment. Another area yet to be examined is the overall structure of these distinct translocases, a challenging task given their highly dynamic and transient nature. Furthermore, the recent discovered cpGET pathway has broadened our understanding of protein targeting into thylakoid membranes. However, only Get3b in this system has been identified, thus the search for the remaining components of the cpGET pathway constitutes an important task for the future.
DZ and XX designed and wrote the manuscript. DZ, JW, CZ, and DL collected and analyzed the data. LZ and XX revised the manuscript. HX produced the figures. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (32170258), CAS Interdisciplinary Innovation Team (JCTD-2020-06), the 111 Project of China (D16014), and Program for Innovative Research Team (in Science and Technology) in University of Henan Province (22IRTSTHN024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Jean-David Rochaix (University of Geneva) and Leonard Krall (Yunnan University) for critical reading of this manuscript.
Aldridge, C., Cain, P., and Robinson, C. (2009). Protein transport in organelles: protein transport into and across the thylakoid membrane. FEBS J. 276, 1177–1186.
Amin, P., Sy, D. A., Pilgrim, M. L., Parry, D. H., Nussaume, L., and Hoffman, N. E. (1999). Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121, 61–70. doi: 10.1104/pp.121.1.61
Anderson, S. A., Satyanarayan, M. B., Wessendorf, R. L., Lu, Y., and Fernandez, D. E. (2021). A homolog of GuidedEntry of Tail-anchored proteins3 functions in membrane-specific protein targeting in chloroplasts of Arabidopsis. Plant Cell 33, 2812–2833. doi: 10.1093/plcell/koab145
Anderson, S. A., Singhal, R., and Fernandez, D. E. (2019). Membrane specific targeting of tail-anchored proteins SECE1 and SECE2 within chloroplasts. Front. Plant Sci. 10:1401. doi: 10.3389/fpls.2019.01401
Asakura, Y., Kikuchi, S., and Nakai, M. (2008). Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J. 56, 1007–1017. doi: 10.1111/j.1365-313X.2008.03659.x
Asseck, L. Y., Mehlhorn, D. G., Monroy, J. R., Ricardi, M. M., Breuninger, H., Wallmeroth, N., et al. (2021). Endoplasmic reticulum membrane receptors of the GET pathway are conserved throughout eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 118:e2017636118. doi: 10.1073/pnas.2017636118
Bédard, J., Trösch, R., Wu, F., Ling, Q., Flores-Pérez, Ú, and Töpel, M. (2017). Suppressors of the chloroplast protein import mutant tic40 reveal a genetic link between protein import and thylakoid biogenesis. Plant Cell 29, 1726–1747. doi: 10.1105/tpc.16.00962
Chandrasekar, S., and Shan, S. O. (2017). Anionic phospholipids and the albino3 translocase activate signal recognition particle-receptor interaction during light-harvesting chlorophyll a/b-binding protein targeting. J. Biol. Chem. 292, 397–406. doi: 10.1074/jbc.M116.752956
Chandrasekar, S., Sweredoski, M. J., Sohn, C. H., Hess, S., and Shan, S. (2017). Co-evolution of two GTPases enables efficient protein targeting in an RNA-less chloroplast signal recognition particle pathway. J. Biol. Chem. 292, 386–396. doi: 10.1074/jbc.M116.752931
Costa, E. A., Subramanian, K., Nunnari, J., and Weissman, J. S. (2018). Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692. doi: 10.1126/science.aar3607
DeLille, J., Peterson, E. C., Johnson, T., Moore, M., Kight, A., and Henry, R. (2000). A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 97, 1926–1931. doi: 10.1073/pnas.030395197
Denic, V., Dötsch, V., and Sinning, I. (2013). Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb. Perspect. Biol. 5:a013334. doi: 10.1101/cshperspect.a013334
Dünschede, B., Träger, C., Schröder, C. V., Ziehe, D., Walter, B., Funke, S., et al. (2015). Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J. Biol. Chem. 290, 13104–13114. doi: 10.1074/jbc.M114.597922
Falk, S., Ravaud, S., Koch, J., and Sinning, I. (2010). The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962. doi: 10.1074/jbc.M109.084996
Farkas, Á, and Bohnsack, K. E. (2021). Capture and delivery of tail-anchored proteins to the endoplasmic reticulum. J. Cell Biol. 220:e202105004. doi: 10.1083/jcb.202105004
Farkas, Á, De Laurentiis, E. I., and Schwappach, B. (2019). The natural history of Get3-like chaperones. Traffic 20, 311–324. doi: 10.1111/tra.12643
Franklin, A. E., and Hoffman, N. E. (1993). Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J. Biol. Chem. 268, 22175–22180. doi: 10.1016/s0021-9258(20)80664-4
Gao, F., Kight, A. D., Henderson, R., Jayanthi, S., Patel, P., Murchison, M., et al. (2015). Regulation of structural dynamics within a signal recognition particle promotes binding of protein targeting substrates. J. Biol. Chem. 290, 15462–15474. doi: 10.1074/jbc.M114.624346
Gristick, H. B., Rao, M., Chartron, J. W., Rome, M. E., Shan, S. O., and Clemons, W. M. (2014). Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat. Struct. Mol. Biol. 21, 437–442. doi: 10.1038/nsmb.2813
Horn, A., Hennig, J., Ahmed, Y. L., Stier, G., Wild, K., and Sattler, M. (2015). Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction. Nat. Commun. 6:8875. doi: 10.1038/ncomms9875
Ji, S., Siegel, A., Shan, S. O., Grimm, B., and Wang, P. (2021). Chloroplast SRP43 autonomously protects chlorophyll biosynthesis proteins against heat shock. Nat. Plants 7, 1420–1432. doi: 10.1038/s41477-021-00994-y
Kim, S. J., Robinson, C., and Mant, A. (1998). Sec/SRP-independent insertion of two thylakoid membrane proteins bearing cleavable signal peptides. FEBS Lett. 424, 105–108. doi: 10.1016/s0014-5793(98)00148-3
Kirchhoff, H. (2019). Chloroplast ultrastructure in plants. New Phytol. 223, 565–574. doi: 10.1111/nph.15730
Kuhn, A., Wickner, W., and Kreil, G. (1986). The cytoplasmic carboxy terminus of M13 procoat is required for the membrane insertion of its central domain. Nature 322, 335–339. doi: 10.1038/322335a0
Lee, D. W., Lee, J., and Hwang, I. (2017). Sorting of nuclear-encoded chloroplast membrane proteins. Curr. Opin. Plant Biol. 40, 1–7. doi: 10.1016/j.pbi.2017.06.011
Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19, 47–56. doi: 10.1016/s0168-9525(02)00003-3
Li, Y., Singhal, R., Taylor, I. W., McMinn, P. H., Chua, X. Y., Cline, K., et al. (2015). The Sec2 translocase of the chloroplast inner envelope contains a unique and dedicated SECE2 component. Plant J. 84, 647–685. doi: 10.1111/tpj.13028
Liang, F. C., Kroon, G., McAvoy, C. Z., Chi, C., Wright, P. E., and Shan, S. (2016). Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc. Natl. Acad. Sci. U.S.A. 22, 1615–1624. doi: 10.1073/pnas.1524777113
Lorkovic, Y. J., Schroder, W. P., Pakrasi, H. B., Irrgang, K. D., Herrmann, R. G., and Oelmuller, R. (1995). Molecular characterization of PsbW, a nuclear-encoded component of the photosystem II reaction center complex in spinach. Proc. Natl. Acad. Sci. U.S.A. 92, 8930–8934. doi: 10.1073/pnas.92.19.8930
Mant, A., Woolhead, C. A., Moore, M., Henry, R., and Robinson, C. (2001). Insertion of PsaK into the thylakoid membrane in a “Horseshoe” conformation occurs in the absence of signal recognition particle, nucleoside triphosphates, or functional albino3. J. Biol. Chem. 276, 36200–36206. doi: 10.1074/jbc.M102914200
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251. doi: 10.1073/pnas.182432999
Mateja, A., and Keenan, R. J. (2018). A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 51, 195–202. doi: 10.1016/j.sbi.2018.07.009
McAvoy, C. Z., Siegel, A., Piszkiewicz, S., Miaou, E., Yu, M. S., Nguyen, T., et al. (2018). Two distinct sites of client protein interaction with the chaperone cpSRP43. J. Biol. Chem. 293, 8861–8873. doi: 10.1074/jbc.RA118.002215
McDowell, M. A., Heimes, M., Fiorentino, F., Mehmood, S., Farkas, Á, Coy-Vergara, J., et al. (2020). Structural basis of tail-anchored membrane protein biogenesis by the GET Insertase complex. Mol. Cell 80, 72–86. doi: 10.1016/j.molcel.2020.08.012
Michl, D., Robinson, C., Shackleton, J. B., Herrmann, R. G., and Klösgen, R. B. (1994). Targeting of proteins to the thylakoids by bipartite presequences: CFoII is imported by a novel, third pathway. EMBO J. 13, 1310–1317.
Moore, M., Goforth, R. L., Mori, H., and Henry, R. (2003). Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J. Cell Biol. 162, 1245–1254. doi: 10.1083/jcb.200307067
Nakai, M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59, 1111–1119. doi: 10.1093/pcp/pcy083
Neuhaus, H. E., and Emes, M. J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140. doi: 10.1146/annurev.arplant.51.1.111
Nilsson, R., and van Wijk, K. J. (2002). Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett. 524, 127–133. doi: 10.1016/s0014-5793(02)03016-8
Nilsson, R., Brunner, J., Hoffman, N. E., and van Wijk, K. J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18, 733–742. doi: 10.1093/emboj/18.3.733
Nohara, T., Asai, T., Nakai, M., Sugiura, M., and Endo, T. (1996). Cytochrome f encoded by the chloroplast genome is imported into thylakoids via the SecA-dependent pathway. Biochem. Biophys. Res. Commun. 224, 2474–2478. doi: 10.1006/bbrc.1996.1051
Ossenbühl, F., Göhre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J. D., and Eichacker, L. A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16, 1790–1800. doi: 10.1105/tpc.023226
Ouyang, M., Li, X., Ma, J., Chi, W., Xiao, J., Zou, M., et al. (2011). LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat. Commun. 2:277. doi: 10.1038/ncomms1278
Ouyang, M., Li, X., Zhang, J., Feng, P., Pu, H., Kong, L., et al. (2020). Liquid-liquid phase transition drives intra-chloroplast cargo sorting. Cell 180, 1144–1159. doi: 10.1016/j.cell.2020.02.045
Paul, P., Simm, S., Blaumeiser, A., Scharf, K. D., Fragkostefanakis, S., Mirus, O., et al. (2013). The protein translocation systems in plants-composition and variability on the example of Solanum lycopersicum. BMC Genom. 14:189. doi: 10.1186/1471-2164-14-189
Rao, M., Okreglak, V., Chio, U. S., Cho, H., Walter, P., and Shan, S. O. (2016). Multiple selection filters ensure accurate tail-anchored membrane protein targeting. Elife. 5:e21301. doi: 10.7554/eLife.21301
Richardson, L. G. L., and Schnell, D. J. (2020). Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids. J. Exp. Bot. 71, 1226–1238. doi: 10.1093/jxb/erz517
Robinson, D., Karnauchov, I., Herrmann, R. G., Klosgen, R. B., and Robinson, C. (1996). Protease-sensitive thylakoidal import machinery for the Sec-, ΔpH- and signal recognition particle-dependent protein targeting pathways, but not for CFoII integration. Plant J. 10, 149–155. doi: 10.1046/j.1365-313x.1996.10010149.x
Rome, M. E., Chio, U. S., Rao, M., Gristick, H., and Shan, S. O. (2014). Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E4929–E4935. doi: 10.1073/pnas.1411284111
Samuelson, J. C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 575–577. doi: 10.1038/35020586
Schüenemann, D., Amin, P., Hartmann, E., and Hoffman, N. E. (1999). Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J. Biol. Chem. 274, 12177–12182. doi: 10.1074/jbc.274.17.12177
Schüenemann, D., Gupta, S., Persello-Cartieaux, F., Klimyuk, V. I., Jones, J. D. G., Nussaume, L., et al. (1998). A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 10312–10316. doi: 10.1073/pnas.95.17.10312
Schuldiner, M., Metz, J., Schmid, V., Denic, V., Rakwalska, M., Schmitt, H. D., et al. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645. doi: 10.1016/j.cell.2008.06.025
Schünemann, D. (2007). Mechanisms of protein import into thylakoids of chloroplasts. Biol. Chem. 388, 907–915. doi: 10.1515/BC.2007.111
Shao, S., Rodrigo-Brenni, M. C., Kivlen, M. H., and Hegde, R. S. (2017). Mechanistic basis for a molecular triage reaction. Science 355, 298–302. doi: 10.1126/science.aah6130
Shigemitsu, S., Cao, W., Terada, T., Shimizu, K., Kutay, U., Hartmann, E., et al. (2016). Development of a prediction system for tail-anchored proteins. BMC Bioinform. 17:378. doi: 10.1186/s12859-016-1202-7
Sjuts, I., Soll, J., and Bölter, B. (2017). Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front. Plant Sci. 8:168. doi: 10.3389/fpls.2017.00168
Srivastava, R., Zalisko, B. E., Keenan, R. J., and Howell, S. H. (2017). The GET system inserts the tailanchored protein. SYP72, into endoplasmic reticulum membranes. Plant Physiol. 173, 1137–1145. doi: 10.1104/pp.16.00928
Staehelin, L. A. (2003). Chloroplast structure: from chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth. Res. 76, 185–196. doi: 10.1023/A:1024994525586
Stengel, K. F., Holdermann, I., Cain, P., Robinson, C., Wild, K., and Sinning, I. (2008). Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321, 253–256. doi: 10.1126/science.1158640
Thompson, S. J., Kim, S. J., and Robinson, C. (1998). Sec-independent insertion of thylakoid membrane proteins. Analysis of insertion forces and identification of a loop intermediate involving the signal peptide. J. Biol. Chem. 273, 18979–18983. doi: 10.1074/jbc.273.30.18979
Thompson, S. J., Robinson, C., and Mant, A. (1999). Dual signal peptides mediate the signal recognition particle/Sec-independent insertion of a thylakoid membrane polyprotein, PsbY. J. Biol. Chem. 274, 4059–4066. doi: 10.1074/jbc.274.7.4059
Tu, C. J., Peterson, E. C., Henry, R., and Hoffman, N. E. (2000). The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J. Biol. Chem. 275, 13187–13190. doi: 10.1074/jbc.c000108200
Tzvetkova-Chevolleau, T., Hutin, C., Noel, L. D., Goforth, R., Carde, J. P., and Caffarri, S. (2007). Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19, 1635–1648. doi: 10.1105/tpc.106.048959
Walter, B., Hristou, A., Nowaczyk, M. M., and Schünemann, D. (2015). In vitro reconstitution of co-translational D1 insertion reveals a role of the cpSec-Alb3 translocase and Vipp1 in photosystem II biogenesis. Biochem. J. 468, 315–324. doi: 10.1042/BJ20141425
Wang, F., Brown, E. C., Mak, G., Zhuang, J., and Denic, V. (2010). A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40, 159–171. doi: 10.1016/j.molcel.2010.08.038
Wang, F., Chan, C., Weir, N. R., and Denic, V. (2014). The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 512, 441–444. doi: 10.1038/nature13471
Wang, P., Liang, F. C., Wittmann, D., Siegel, A., Shan, S. O., and Grimm, B. (2018). Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 115, E3588–E3596. doi: 10.1073/pnas.1719645115
Wicke, S., Schneeweiss, G. M., Pamphilis, C. W., Müller de, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Woolhead, C. A., Thompson, S. J., Moore, M., Tissier, C., Mant, A., Rodger, A., et al. (2001). Distinct Albino3-dependent and-independent pathways for thylakoid membrane protein insertion. J. Biol. Chem. 276, 40841–40846. doi: 10.1074/jbc.M106523200
Xing, S., Mehlhorn, D. G., Wallmeroth, N., Asseck, L. Y., Kar, R., Voss, A., et al. (2017). Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc. Natl. Acad. Sci. U.S.A. 114, E1544–E1553. doi: 10.1073/pnas.1619525114
Xu, X., Ouyang, M., Lu, D., Zheng, C., and Zhang, L. (2021a). Protein sorting within chloroplasts. Trends Cell Biol. 31, 9–16. doi: 10.1016/j.tcb.2020.09.011
Xu, X., Zheng, C., Lu, D., Song, C. P., and Zhang, L. (2021b). Phase separation in plants: new insights into cellular compartmentalization. J. Integr. Plant Biol. 63, 1835–1855. doi: 10.1111/jipb.13152
Zhang, L., Paakkarinen, V., Suorsa, M., and Aro, E. M. (2001). A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J. Biol. Chem. 276, 37809–37814. doi: 10.1074/jbc.M105522200
Zhang, Y., De Laurentiis, E., Bohnsack, K. E., Wahlig, M., Ranjan, N., Gruseck, S., et al. (2021). Ribosome-bound Get4/5 facilitates the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun. 12:782. doi: 10.1038/s41467-021-20981-3
Ziehe, D., Dünschede, B., and Schünemann, D. (2017). From bacteria to chloroplasts: evolution of the chloroplast SRP system. Biol. Chem. 398, 653–661. doi: 10.1515/hsz-2016-0292
Ziehe, D., Dünschede, B., and Schünemann, D. (2018). Molecular mechanism of SRP-dependent light-harvesting protein transport to the thylakoid membrane in plants. Photosynth. Res. 138, 303–313. doi: 10.1007/s11120-018-0544-6
Zoschke, R., and Barkan, A. (2015). Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. U.S.A. 112, E1678–E1687. doi: 10.1073/pnas.1424655112
Zygadlo, A., Robinson, C., Scheller, H. V., Mant, A., and Jensen, P. E. (2006). The properties of the positively charged loop region in PSI-G are essential for its ‘spontaneous’ insertion into thylakoids and rapid assembly into the photosystem I complex. J. Biol. Chem. 281, 10548–10554. doi: 10.1074/jbc.M512687200
Keywords: chloroplast protein import, cpSRP pathway, cpGet pathway, spontaneous pathway, cotranslational protein transport
Citation: Zhu D, Xiong H, Wu J, Zheng C, Lu D, Zhang L and Xu X (2022) Protein Targeting Into the Thylakoid Membrane Through Different Pathways. Front. Physiol. 12:802057. doi: 10.3389/fphys.2021.802057
Received: 26 October 2021; Accepted: 07 December 2021;
Published: 12 January 2022.
Edited by:
Inhwan Hwang, Pohang University of Science and Technology, South KoreaReviewed by:
Chiung-Chih Chu, Academia Sinica, TaiwanCopyright © 2022 Zhu, Xiong, Wu, Zheng, Lu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiumei Xu, eHV4aXVtZWlAdmlwLmhlbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.