
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 18 January 2022
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.785637
This article is part of the Research Topic Hormone Regulation of Insect Development View all 7 articles
 Jinxin Wu1,2
Jinxin Wu1,2 Guanwang Shen1,2
Guanwang Shen1,2 Die Liu3
Die Liu3 Haoran Xu3
Haoran Xu3 Mengyao Jiao1,2
Mengyao Jiao1,2 Yungui Zhang1,2
Yungui Zhang1,2 Ying Lin1,2*
Ying Lin1,2* Ping Zhao1,2*
Ping Zhao1,2*
Estrogen-related receptor (ERR) is an orphan nuclear receptor that was first discovered in animals, and play an important role in metabolism, development, and reproduction. Despite extensive research on the function of ERR, its transcriptional regulation mechanism remains unclear. In this study, we obtained the upstream region of Bombyx mori ERR (BmERR) and confirmed the promoter activity of this region. Interestingly, we found that 10 and 50 nM 20-hydroxyecdysone (20E) up-regulated the transcriptional activity of BmERR promoter. In addition, eight putative ecdysone response elements (EcREs) were predicted in the upstream sequence of BmERR. Based on their positions, the upstream sequence of BmERR was truncated into different fragments. Finally, an EcRE-like sequence (5′-AGTGCAGTAAACTGT-3′) was identified. Electrophoretic mobility shift assay (EMSA) and cell transfection experiments confirmed that this motif specifically binds to the complex formed between ecdysone receptor (BmEcR) and the ultraspiracle (BmUSP), a key complex in the 20E signaling pathway. Interference of BmERR or BmEcR mRNA in the embryonic cells of Bombyx mori significantly affected the expression of BmEcR and BmUSP. Overall, these results suggested that an EcRE element was identified from BmERR, and this will help understanding the detailed regulatory mechanism of ERR in insects.
Nuclear receptors are a large family of eukaryotic transcription factors (TFs) with important functions such as regulation of ligand-mediated gene expression and signaling pathways (Beato et al., 1995). Estrogen-related receptors (ERRs) are the orphan nuclear receptors and belong to the third nuclear receptor subfamily (NR3). Due to the high similarity in the ligand binding domain and DNA binding domain between ERR and estrogen receptor (ER), ERR can participating in the ER signaling pathway, sharing target genes, co-regulatory proteins, ligands and sites of action with the ER (Giguere, 2002; Greschik et al., 2002; Horard and Vanacker, 2003).
In mammals, there are three subtypes of ERRs, ERRα, ERRβ, and ERRγ, which play important roles in physiological and pathological functions (Deblois and Giguere, 2011). In particular, ERRs are closely associated with tumorigenesis. ERRα and ERRγ are two potential biological targets for the detection of breast cancer in women (Ariazi et al., 2002). ERRβ is shown to inhibit the growth of prostate cancer (Misawa and Inoue, 2015). In addition, ERRs play vital roles in energy metabolism, mitochondrial biogenesis, oxidative phosphorylation, fat metabolism, and cell growth (Carrier et al., 2004; Tremblay and Giguere, 2007; Deblois and Giguere, 2011; Eichner and Giguere, 2011; Sailland et al., 2014).
Most studies on ERR have focused on mammals. A homologous receptor of ERR has only been identified as a single subtype in insects. In Drosophila, ERR controls energy metabolism by regulating genes expression in the glycolytic pathway. ERR-knockout in Drosophila strains resulted in glycogen accumulation, and the glycolysis process was blocked, leading to death of the flies (Tennessen et al., 2011; Beebe et al., 2020). Genetic knockdown the expression of ERR in the testis of Drosophila inhibited the development of the testis and sperm production (Misra et al., 2017). ERR expression in Apis cerana and Chironomus riparius could be induced by external stress such as exposure to insecticides, ultraviolet rays, and fungicides (Park and Kwak, 2010; Zhang et al., 2016). Decreasing the expression ERR in male Agrotis ipsilon affected its sexual behavior (Bozzolan et al., 2017). To date, most studies on ERRs in insects focus on their physiological functions, whereas the mechanisms driving the regulation of ERRs remains unclear.
Since the completion of the genome sequence, the silkworm (Bombyx mori) has become a widely used model insect for lepidopteran research, and a general model organism for life sciences, toxicology, and fungal infections (International Silkworm Genome, 2008; Suetsugu et al., 2013; Meng et al., 2017; Abdelli et al., 2018; Nakajima et al., 2018; Matsumoto and Sekimizu, 2019). In previous research, we have demonstrated that the Bombyx mori ERR (BmERR) is involved in the growth and development of silkworm embryos and larvae by regulating the expression of glucose metabolism-related genes (Long et al., 2020; Shen et al., 2021). and vitellogenin via the 20-hydroxyecdysone (20E) -EcR pathway in silkworm (Shen et al., 2018). Therefore, we hypothesized that the activation of the 20E signaling pathway may be a key event in the transcriptional regulatory of ERR. To evaluate this possibility, we cloned the upstream sequence of BmERR including the transcription start site (TSS) using 5′-rapid amplification of cDNA ends (5′RACE), analyzed its transcriptional activity and relationship with 20E, and explored the molecular mechanism of BmERR response to 20E.
The silkworm strain of dazao was provided by the silkworm gene bank in the Southwest University, China.1 The silkworms were fed with mulberry leaves at 25°C and grown with a photoperiod of 12 h light/12 h dark and 55 ± 5% relative humidity.
Previous studies found that the fat body of silkworms has the expression of BmERR (Shen et al., 2018). So, in this article, the fat body was used for the extraction of total RNA. The total RNA was extracted using the Trizol extraction kit (Invitrogen, United States), then digested carefully with DNase I (TAKARA, Japan) to avoid genomic DNA contamination. For race polymerase chain reaction (PCR), the first-stand cDNA of fat body was synthesized with the SMARTer RACE 5′/3′ kit (TAKARA, Japan) as the manufacturer instructions and then stored at −20°C. For the quantitative real time-PCR (qRT-PCR), M-MLV reverse transcriptase (Promega, United States) was used to generate BmE cells’ first-strand cDNA. Genomic DNA was extracted from the whole silkworm using the Tissue DNA kit (OMEGA, United States) as the manufacturer’s instruction.
Quantitative real time-PCR was performed to evaluate the expression levels of BmERR (GenBank: KT268294), BmEcR (GenBank: L35266), and BmUSP (GenBank: U06073.1) using the SYBR Premix Ex TaqTM (TAKARA Biotech, Japan) and an ABI StepOne v2.1 Sequence Detection System (Applied Biosystems, United States). The relative mRNA expression levels of target genes were calculated with the 2–ΔΔCT method and the silkworm translation initiation factor 4A (BmTIF4A, NM_001043911.1) was used as an endogenous control. The primers used for PCR were listed in Table 1.
To obtain the complete BmERR 5′-UTR sequence and the transcription start site (TSS), RACE was employed to clone the 5′-untranslated region of BmERR. The universal primer mix (UPM) from the 5′-Full RACE Kit (TAKARA, Japan) was used as the forward primer, and the specific reverse primer BmERR-R (Table 1) was designed based on the partial BmERR 5′-UTR sequence in our previous fat body transcriptome sequencing results (date not shown). The 5′-UTR was amplified by nested PCR using the synthesized cDNA as a template under the following program: 30 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 30 s. The PCR products were cloned into the pMD19-T simple clone vector (TAKARA, Japan) and then sequenced.
The upstream sequences of BmERR were obtained by 5′RACE-PCR and transcriptome sequencing of silkworm fat body. The cis-acting regulatory elements (CREs) in the upstream sequences of BmERR were predicted using JASPAR.2
Using the high-fidelity DNA polymerase (TransGen Biotech, China), different lengths of BmERR promoter fragments were cloned from the silkworm genomic DNA with different primers (shown in Table 1) under the following program: 95°C for 5 min; 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 2 min; and 72°C for 10 min, and then inserted into the firefly luciferase reporter vector pGL3-Basic (Promega, United States) between SacI and XhoI (TAKARA, Japan) restriction sites.
The psl 1180-HR3-A4-DsRed-SV40 (Liu et al., 2019) vector was stored in our laboratory and used for overexpression in B. mori cells. Using the ligation free cloning kit (abm, China), the open reading frame (ORF) of BmEcR and BmUSP were amplified from the cDNA and cloned into psl 1180-HR3-A4 SV40 vector restricted with BamHI and NotI in the manner of homologous recombination. The primers were given in Table 1.
The luminescent reporter assay was performed according to the manufacturer’s instruction. The B. mori embryonic cell line (BmE) was cultured in a 24-wells plate at 27°C in Grace (Gibco, United States) insect cell culture medium supplemented with 10% fetal bovine serum (FBS) (Gibco, United States). After 12 h, every well was transfected with a mixture of 1 μg recombinant plasmid, 0.1 μg internal control plasmid pRL-78ML (Liu et al., 2018; Shen et al., 2018), and 3 μL of Lipofectamine 2000 (Invitrogen, United States) in the insect medium without FBS. After 6 h, the transfection mixture was replaced with 500 μL fresh insect medium containing 10% FBS.
Twenty-hydroxyecdysone (Sigma, United States) were dissolved in DMSO (Sigma, United States) at a stock concentration of 5 mg/mL and stored at −20°C. After 6 h of cell transfection, different concentrations of 20E were added to the 24-well cell culture plate, and the equal amount of DMSO was added as a control. The cells were harvested after 48 h of transfection and assayed with the Dual-Luciferase Reporter System (Promega, United States).
Oligonucleotide sequences of four EcRE-like motifs 1/2/3/8 (EcRE-like 1/2/3/8) predicted at positions −1,325 to −1,311 (5′-AGTGCAGTAAACTGT-3′), −1,258 to −1,244 (5′-ACCTTTATTAAATT-3′), −1,199 to −1,185 (5′-TAGTGGTACTTTAGA-3′), and −28 to −14 (5′-GCGTTAGCTGATATT-3′) in the BmERR promoter were synthesized as probes for electrophoretic mobility shift assay (EMSA) (Sangon Biotech, China). The single-stranded sequences were labeled with biotin at the 3′-end and annealed to produce a double-stranded probe. To evaluate interactions between the regulatory elements and prokaryotic expressed proteins BmEcR and BmUSP (Shen et al., 2018), EMSA was performed as previously described using a Chemiluminescent EMSA Kit (Beyotime, China) as the manufacturer’s instructions. After incubation at 25°C for 25 min, the reaction mixtures were loaded to 5% native polyacrylamide gels and electrophoresis was conducted in Tris–borate—EDTA buffer (1 mM EDTA and 45 mM Tris–borate, pH 8.3). The proteins were transferred to the nylon membrane (Roche, United States). and then imaged with the enhanced chemiluminescence using a Clinx ChemiScope 3400 Mini system (Science Instruments, China) after incubation with Streptavidin-horseradish peroxidase.
A double-stranded RNA interference (dsRNAi) approach was performed to evaluate the relationships among BmERR, BmECR, and BmUSP. The 584- and 504-bp fragments of BmERR and BmECR were, respectively, selected to synthesize double-stranded RNA (dsRNA) (Jin et al., 2020; Long et al., 2020). The fragments containing the bacteriophage T7 promoter sequence were obtained through PCR and then cloned into the pMD19-T simple vector (TAKARA, Japan). The dsRNA was generated using the T7 RiboMAX Express RNAi System (Promega, United States). A fragment of enhanced green fluorescence protein (EGFP) (458 bp) was used as a negative control. After 20 min of incubation, the mixture (5 μg dsRNA and 10 μL Lipofectamine 2000) (Invitrogen, United States) was added into the BmE cells. The primer sequences were shown in Table 1.
The results are presented as the mean ± SD from three independent experiments. Statistical analyses were performed using Microsoft excel (Microsoft, United States). Differences between groups were analyzed with Student’s t-tests, and *P < 0.05, **P < 0.01, and ***P < 0.001 were accepted as statistically significant.
To identify the key sequence of BmERR promoter, we analyzed the upstream region of BmERR from the silkworm genome database.3 The TSS and complete (5′-UTR) sequence were successfully identified through 5′-RACE PCR (Figure 1). The TSS was located 458 bp upstream of the translation initiation site and the 5′-UTR sequence was transcribed and spliced by two exon regions. Using the online website4, we predicted a typical TATA BOX in the region from −64 to −57 bp and eight putative ecdysone response elements (EcREs, shown in the Table 2) in −1,333 bp upstream of the TSS.

Figure 1. Sequence analysis of the 5′-flanking region of Bombyx mori ERR (BmERR). The nucleotides are numbered relative to the translation start site indicated by + 1, with upstream sequences preceded by “–.” The transcription start site (TSS) and translation start site are indicated in red. The putative transcription factor (TF)-binding sites are labeled. The light gray area represents the first exon sequence and the dark box represents the second exon sequence. Eight putative EcRE motif are predicted on the BmERR promoter.
To verify the transcriptional activity of the BmERR promoter, the 1,333 bp promoter fragment was amplified with a specific primer (shown in the Table 1) from silkworm genomic DNA. A cell expression vector based on the dual luciferase reporter system was constructed and then constructed vector was transfected into BmE cells. The promoter activity was measured post 48 h of transfection using a dual luciferase reporter system. Compared to the control, the BmERR promoter showed higher transcriptional activity (Figure 2A).
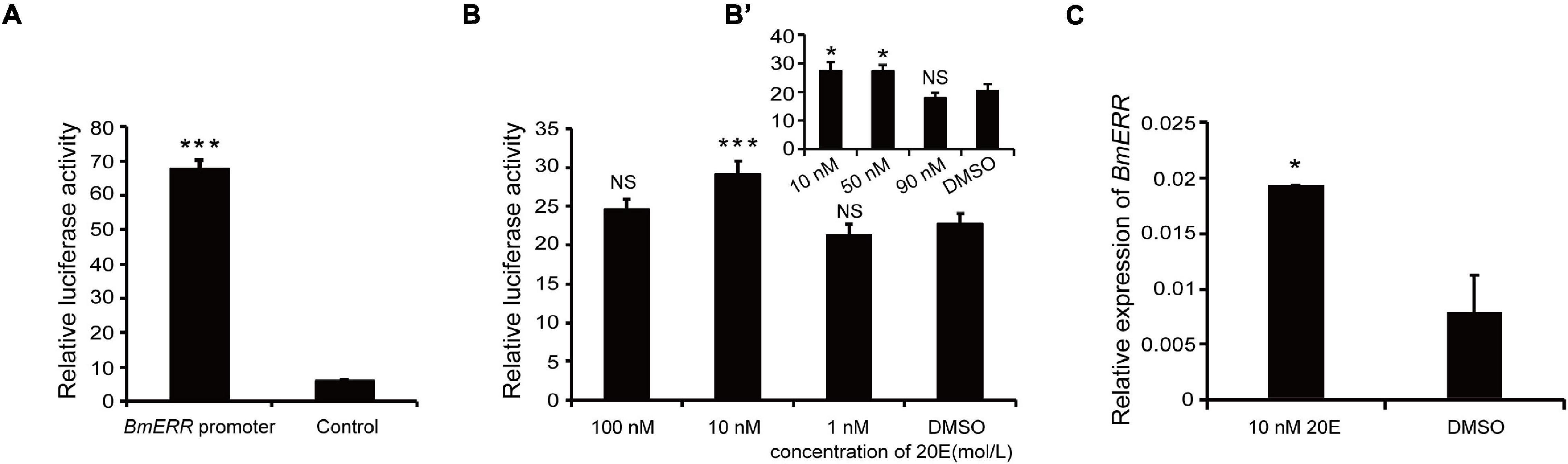
Figure 2. Effect of 20-hydroxyecdysone (20E) on Bombyx mori ERR (BmERR) promoter activity. (A) Comparison of transcriptional activity between the BmERR promoter and control pGL3-basic plasmids. The luciferase activity was corrected for transfection efficiency with the Renilla luciferase pRL-78ML plasmid and is presented as ratios to the activity of the BmERR promoter. (B,B’) Changes of BmERR promoter activity with different concentrations of 20E. (C) Quantitative real time-PCR (qRT-PCR) detection of BmERR expression in BmE cells treated with 10 nM 20E. Dimethyl sulfoxide was used as the control treatment. *P < 0.05, ***P < 0.001, and t-test.
To validate the effect of 20E on BmERR promoter activity, BmE cells were transfected with BmERR promoter in the presence of different concentrations (100, 10, and 1 nM) 20E. After 48 h, BmERR promoter activity was up-regulated only by 10 nM (Figure 2B) and 50 nM 20E (Figure 2B’). In addition, exposure of BmE cells to 10 nM 20E for 48 h resulted in a similar effect that the transcriptional level of BmERR significantly increased (Figure 2C).
Eight EcRE-like elements were predicted at −1,329 to −1,315 bp, −1,262 to −1,248 bp, −1,216 to −1,202 bp, −836 to −822 bp, −686 to −672 bp, −550 to −536 bp, −250 to −236 bp, and −28 to −14 bp on the BmERR promoter, and designated EcRE-like 1–8, respectively. We constructed six vectors for cell transfection with different lengths of BmERR promoter fragments as the position of these elements on the promoter. The effects of EcRE-like elements on promoter activity were evaluated by overexpression of BmEcR and BmUSP and Red fluorescent protein (RFP) was transfected as a control in BmE cells. The luciferase activity assay showed that BmERR promoter activity was significantly increased compared with that of the control when BmEcR and BmUSP proteins were overexpressed. However, compared with the control, when the promoter fragments containing EcRE-like 1/2/3 were removed, there was no significant change in BmERR promoter activity, indicating that these elements may play a positive regulatory role. After the promoter fragment containing the EcRE-like elements 4/5/6/7 were truncated, respectively, the promoter activity did not show significant difference under the case of overexpression of BmEcR and BmUSP. But compared with the control, the promoter still showed obviously higher transcriptional activity (Figure 3). These findings suggested that EcRE-like elements 1–3 and 8 are likely involved in the regulation of BmERR promoter activity mediated by EcR/USP.

Figure 3. Regulation of EcRE on the Bombyx mori ERR (BmERR) promoter. Comparison of the activity of different truncated forms of BmERR promoter with the overexpression of BmEcR and BmUSP and RFP through the dual luciferase reporter system. 1–8, EcRE like 1–8, respectively. EcR and USP, the overexpression of BmEcR and BmUSP protein. RFP, the overexpression of red fluorescent protein (RFP). *P < 0.05. NS, not significant; t-test.
Electrophoretic mobility shift assay was performed to further identify these EcRE-like elements. Each biotin-labeled probe was incubated with BmEcR and BmUSP protein. Only an obvious band shift was evident after incubation with the EcRE-like 1 labeled probe (Figures 4A,B). The concentrations of the labeled probes were 50, 500, and 5 μM. The concentrations of the competing probes were 1, 5, and 50 μM, respectively. No band appeared when the sequence of EcRE-like 1 probe 5′- ATTAAGTAGCA-GTAAACTGTGACCT-3′ was mutated to 5′- ATTAGACGATGACGGGTCATG ACCT -3′ (Figure 4C).
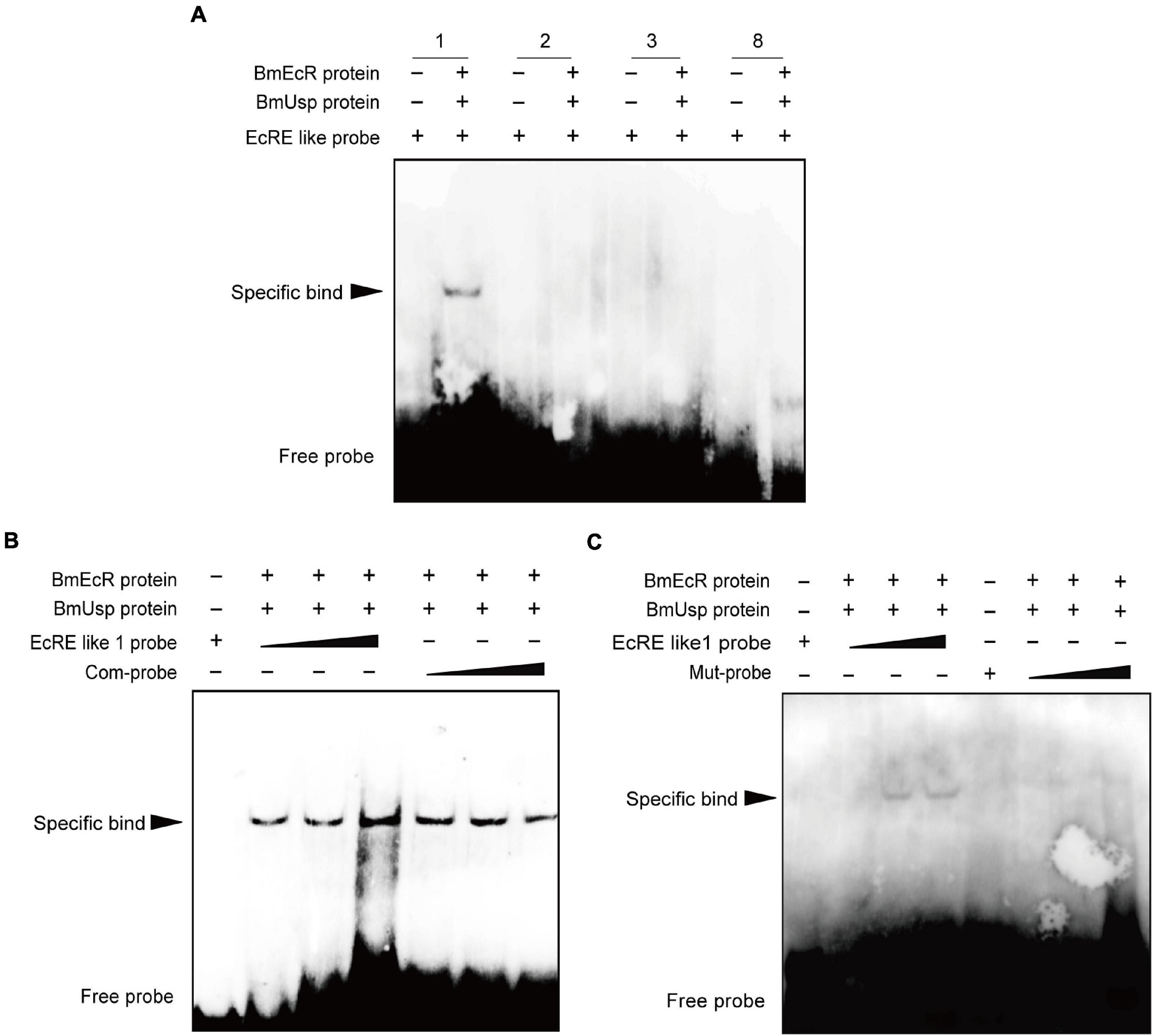
Figure 4. Identification of EcRE on the Bombyx mori ERR (BmERR) promoter. (A–C) DNA-binding activity of EcRE-like 1/2/3/8 probes with purified BmEcR and BmUSP protein verified by electrophoretic mobility shift assay (EMSA). 1/2/3/8, biotin-labeled EcRE-like 1/2/3/8 probe. Com-probe is the biotin-unlabeled competition probe which is of the same sequence as EcRE-like 1. Mut-probe is the mutated biotin-labeled EcRE-like 1 probe. The final concentration of EcRE probe was 5 μM, the concentration of BmEcR protein is 0.3 μg/μL and the concentration of BmUSP protein is 0.45 μg/μL. The ratio of EcRE probe to the competing probe is 1:10.
To further study whether the EcRE motif can respond to 20E, recombinant vectors were constructed by inserting EcRE-like 1 motif and the mutated EcRE-like 1 motif (Mut-EcRE 1) into a basal vector pGL3- VgP78ML (Liu et al., 2019), which does not respond to 20E, designated EcRE 1-P and Mut-EcRE 1-P, respectively. These two vectors were transfected into BmE cells followed by treatment with 10 nM 20E. The luciferase assay showed that only the activity of EcRE-1-P was significantly up-regulated after 20E treatment (Figure 5), suggesting that EcRE1 motif responds to 20E to up-regulate the basic promoter activity.
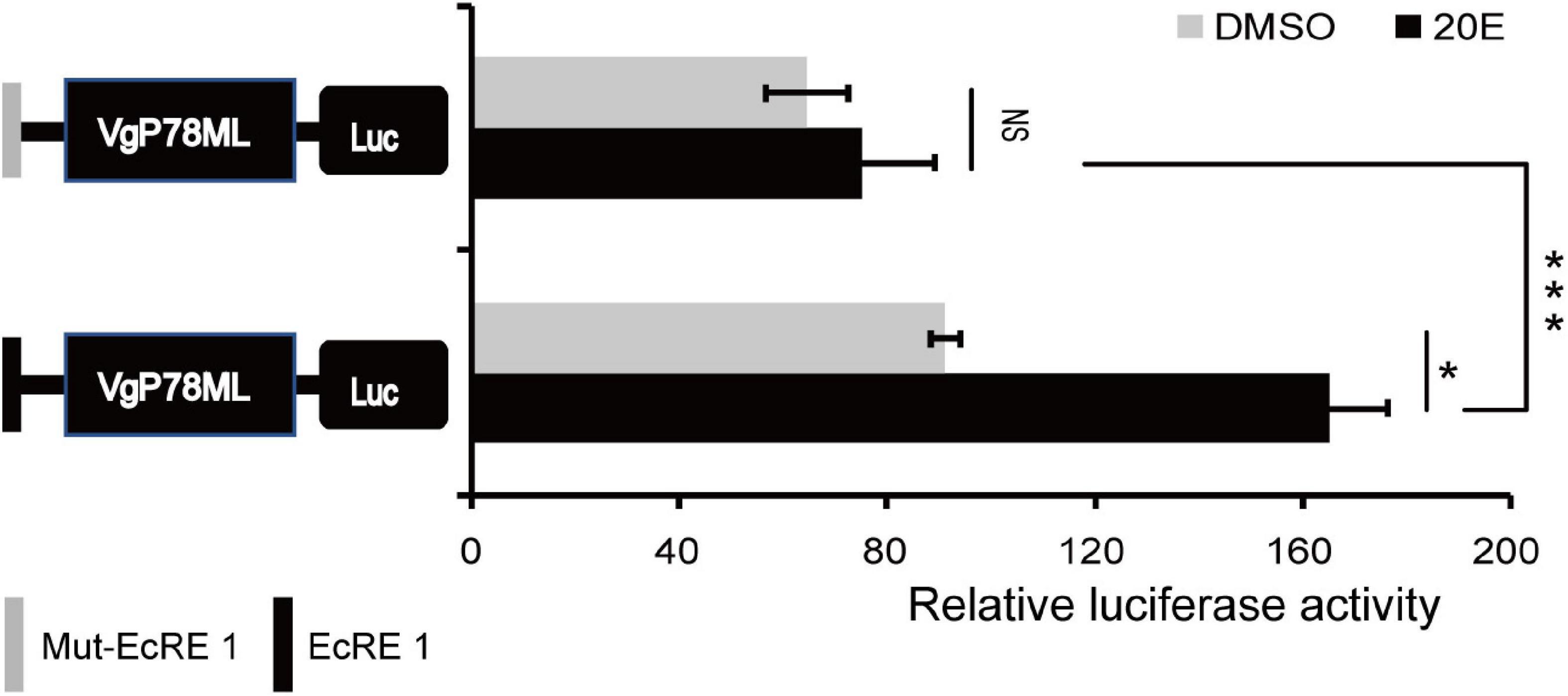
Figure 5. Effect of 10 nM 20-hydroxyecdysone (20E) on the EcRE-1. Luciferase activity of the basic promoters containing EcRE-1 and mut-EcRE 1 after treatment with 10 nM 20E and DMSO, respectively. Mut, mutated, NS, not significant. *P < 0.05, ***P < 0.001; t-test.
In order to further explore the mechanism of 20E regulating the transcription activity of BmERR, the relationship between BmERR, BmEcR, and BmUSP was evaluated by RNA interference in BmE cells. qRT-PCR showed that the expression of BmEcR was significantly decreased after the dsBmEcR fragment was transfected into BmE cells (Figure 6A1), and the expression of BmERR and BmUSP were significantly reduced compared with the control (Figures 6A2,A3). RNAi of BmERR caused a significant reduction of BmERR expression (Figure 6B1). Contrast with the effects of dsBmEcR, the expression of BmEcR and BmUSP were significantly increased (Figures 6B2,B3). These results suggested a complicated network of cross-talking between BmERR, BmEcR, and BmUSP.
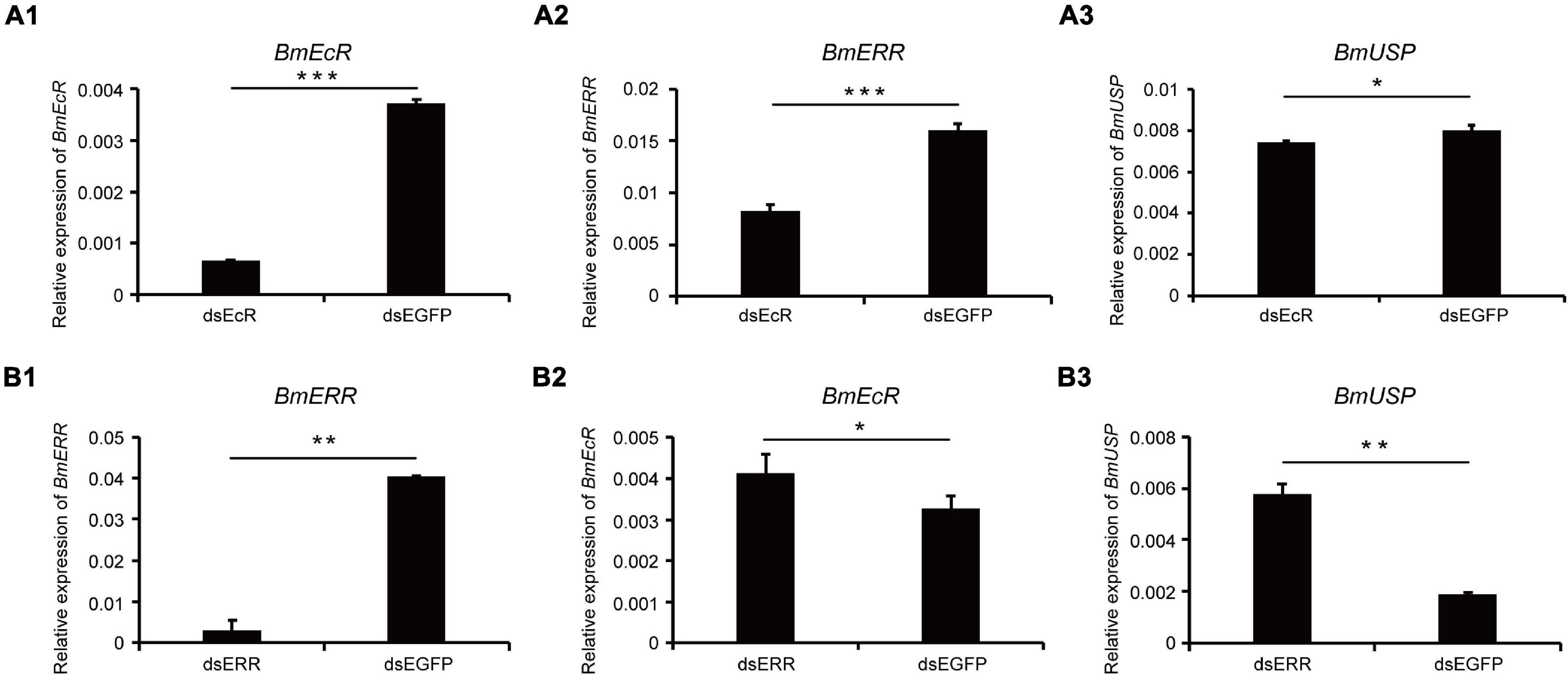
Figure 6. Expression levels of Bombyx mori ERR (BmERR), BmEcR, and BmUSP in BmE cells after dsRNA interference. (A1–A3) Expression levels of BmERR, BmEcR, and BmUSP after treatment with dsEcR. (B1–B3) Expression levels of BmERR, BmEcR, and BmUSP after treatment with dsERR. *P < 0.05, **P < 0.01, and ***P < 0.001; t-test.
Twenty-hydroxyecdysone is an important hormone that regulates the growth, development, metabolism, and apoptosis of insects (Thummel and Chory, 2002; Yang et al., 2014). High 20E titer inhibited the expression of ERR in Drosophila, and the expression of genes related to carbohydrate metabolism were also significantly reduced (Kovalenko et al., 2019). Here, we cloned the nucleotide sequence of the BmERR promoter and confirmed that BmERR promoter can respond to 20E. EMSA showed BmEcR and BmUSP likely bind to the EcRE-like 1 motif on the BmERR promoter. After mutation of EcRE-like 1 motif, the basal promoter will not respond to 20E. This indicated that 20E up-regulated the transcriptional activity by activating the BmEcR and BmUSP complex to bind to the EcRE motif on the BmERR promoter.
Bombyx mori estrogen-related receptor and BmEcR have functional cross-talking in the BmE cells. Decreasing the expression of BmEcR by dsRNAi reduced the mRNA levels of both BmERR and BmUSP, whereas the expression of BmEcR and BmUSP were increased when the expression of BmERR was down-regulated. This is similar to the research on other insects. In the Teleogryllus emma, ERR and EcR both affected the development of testes. The expression of TeEcR and TeERR are regulated by each other (Jin et al., 2017). In Drosophila, EcR and ERR jointly regulated carbohydrate metabolism (Kovalenko et al., 2019). These implied that ERR may function in insects by participating in the 20E signaling pathway.
Twenty-hydroxyecdysone regulated the expression of glycolysis-related genes in the fat body of the silkworm through the ecdysone receptor EcR-USP (Tian et al., 2013; Keshan et al., 2017). In Antheraea pernyi, 20E participated in trehalose catabolism by regulating the expression of trehalase gene (Li et al., 2020). These indicated that 20E was closely related to the energy metabolism of insects. In addition, ERR was involved in carbohydrate metabolism, hypoxic metabolism and energy metabolism in Drosophila (Tennessen et al., 2011; Li et al., 2013; Kovalenko et al., 2019). Our previous research also found that BmERR regulated the expression of glycolysis-related genes to participate in the development of silkworm embryos, and affected the glucose concentration in the midgut by regulating the expression of trehalase (Long et al., 2020; Shen et al., 2021). These further implied that ERR might regulate 20E signaling by mediating nutritional metabolism, then affects insulin pathway and finally exerts its influence on 20E signaling.
So far, the research on ERR mainly focused on the function and the role in the 20E signaling pathway in insects. Previous report showed that 1 μM 20E inhibited the expression of ERR in Drosophila larvae, but there was no significant difference in S2 cells treated with 0.3 μM 20E (Kovalenko et al., 2019). It showed that the expression of ERR was very sensitive to the concentration of 20E. Our research found that 10 and 50 nM 20E could up-regulate the activity of BmERR promoter. Although eight EcRE motifs were predicted on the 1,333 bp BmERR promoter region, there was only one EcRE motif response to 20E. In addition, the 1,333 bp BmERR promoter region had significant transcriptional activity, but it might not contain all hormone response elements completely. It was reported that the distal sequence of the promoter also contained the motifs which respond to 20E (Nishita, 2014). These results supplied that the expression of BmERR was not only very sensitive to the dose of 20E, but also depended on length of the BmERR promoter region. In summary, our study analyzed how 20E regulate the expression of ERR and provided a perspective in the regulation of ERR expression in insects.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
JW and GS contributed to the conception and design of the study. JW performed the statistical analysis and wrote the first draft of the manuscript. All authors contributed to experiment, manuscript revision, read, and approved the submitted version.
This work was supported by the Doctoral Fund Project of Southwest University, China (Grant No. 7110300034) and the Natural Science Foundation of Chongqing, China (Grant No. cstc2020jcyj-cxttX0001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Huawei He of Southwest University, Chongqing, China for his careful and professional revision of English grammar, and revising of this manuscript.
Abdelli, N., Peng, L., and Keping, C. (2018). Silkworm, Bombyx mori, as an alternative model organism in toxicological research. Environ. Sci. Pollut. Res. Int. 25, 35048–35054. doi: 10.1007/s11356-018-3442-8
Ariazi, E. A., Clark, G. M., and Mertz, J. E. (2002). Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 62, 6510–6518.
Beato, M., Herrlich, P., and Schutz, G. (1995). Steroid hormone receptors: many actors in search of a plot. Cell 83, 851–857. doi: 10.1016/0092-8674(95)90201-5
Beebe, K., Robins, M. M., Hernandez, E. J., Lam, G., Horner, M. A., and Thummel, C. S. (2020). Drosophila estrogen-related receptor directs a transcriptional switch that supports adult glycolysis and lipogenesis. Genes Dev. 34, 701–714. doi: 10.1101/gad.335281.119
Bozzolan, F., Durand, N., Demondion, E., Bourgeois, T., Gassias, E., and Debernard, S. (2017). Evidence for a role of oestrogen receptor-related receptor in the regulation of male sexual behaviour in the moth Agrotis ipsilon. Insect. Mol. Biol. 26, 403–413. doi: 10.1111/imb.12303
Carrier, J. C., Deblois, G., Champigny, C., Levy, E., and Giguere, V. (2004). Estrogen-related receptor alpha (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J. Biol. Chem. 279, 52052–52058. doi: 10.1074/jbc.M410337200
Deblois, G., and Giguere, V. (2011). Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim. Biophys. Acta 1812, 1032–1040. doi: 10.1016/j.bbadis.2010.12.009
Eichner, L. J., and Giguere, V. (2011). Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion 11, 544–552. doi: 10.1016/j.mito.2011.03.121
Giguere, V. (2002). To ERR in the estrogen pathway. Trends Endocrinol. Metab. 13, 220–225. doi: 10.1016/s1043-2760(02)00592-1
Greschik, H., Wurtz, J. M., Sanglier, S., Bourguet, W., van Dorsselaer, A., Moras, D., et al. (2002). Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell 9, 303–313. doi: 10.1016/s1097-2765(02)00444-6
Horard, B., and Vanacker, J. M. (2003). Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J. Mol. Endocrinol. 31, 349–357. doi: 10.1677/jme.0.0310349
International Silkworm Genome, C. (2008). The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect. Biochem. Mol. Biol. 38, 1036–1045. doi: 10.1016/j.ibmb.2008.11.004
Jin, W. J., Jia, Y. S., Tan, E., and Xi, G. S. (2017). Relevance of estrogen-related receptor gene and ecdysone receptor gene in adult testis of the cricket Teleogryllus emma (Orthoptera: Gryllidae). Sci. Nat. 104, 11–12. doi: 10.1007/s00114-017-1518-9
Jin, X. L., Wu, X. Y., Zhou, L. T., He, T., Yin, Q., and Liu, S. P. (2020). 20-Hydroxyecdysone-responsive microRNAs of insects. Rna Biol. 17, 1454–1471. doi: 10.1080/15476286.2020.1775395
Keshan, B., Thounaojam, B., and Kh, S. D. (2017). Insulin and 20-hydroxyecdysone action in Bombyx mori: Glycogen content and expression pattern of insulin and ecdysone receptors in fat body. Gen. Comp. Endocrinol. 241, 108–117. doi: 10.1016/j.ygcen.2016.06.022
Kovalenko, E. V., Mazina, M. Y., Krasnov, A. N., and Vorobyeva, N. E. (2019). The Drosophila nuclear receptors EcR and ERR jointly regulate the expression of genes involved in carbohydrate metabolism. Insect Biochem. Mol. Biol. 112:103184. doi: 10.1016/j.ibmb.2019.103184
Li, Y., Padmanabha, D., Gentile, L. B., Dumur, C. I., Beckstead, R. B., and Baker, K. D. (2013). HIF- and non-HIF-regulated hypoxic responses require the estrogen-related receptor in Drosophila melanogaster. PLoS Genet. 9:e1003230. doi: 10.1371/journal.pgen.1003230
Li, Y. N., Liu, Y. B., Xie, X. Q., Zhang, J. N., and Li, W. L. (2020). The Modulation of Trehalose Metabolism by 20-Hydroxyecdysone in Antheraea pernyi (Lepidoptera: Saturniidae) During its Diapause Termination and Post-Termination Period. J. Insect Sci. 20:5. doi: 10.1093/jisesa/ieaa108
Liu, H., Lin, Y., Shen, G., Gu, J., Ruan, Y., Wu, J., et al. (2019). A novel GATA transcription factor GATAbeta4 promotes vitellogenin transcription and egg formation in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 107, 10–18. doi: 10.1016/j.ibmb.2019.01.004
Liu, H., Lin, Y., Shen, G., Gu, J., Zhang, H., Wu, J., et al. (2018). [Establishment of a suitable control reporter plasmid of a dual luciferase reporter gene system for hormone research in silkworm cell lines]. Sheng Wu Gong Cheng Xue Bao 34, 1631–1641. doi: 10.13345/j.cjb.180007
Long, W., Wu, J., Shen, G., Zhang, H., Liu, H., Xu, Y., et al. (2020). Estrogen-related receptor participates in regulating glycolysis and influences embryonic development in silkworm Bombyx mori. Insect Mol. Biol. 29, 160–169.
Matsumoto, Y., and Sekimizu, K. (2019). Silkworm as an experimental animal for research on fungal infections. Microbiol. Immunol. 63, 41–50. doi: 10.1111/1348-0421.12668
Meng, X., Zhu, F., and Chen, K. (2017). Silkworm: A Promising Model Organism in Life Science. J. Insect Sci. 17:5. doi: 10.1093/jisesa/iex064
Misawa, A., and Inoue, S. (2015). Estrogen-Related Receptors in Breast Cancer and Prostate Cancer. Front. Endocrinol. 6:83. doi: 10.3389/fendo.2015.00083
Misra, S., Pandey, A. K., Gupta, S., Kumar, A., Khanna, P., Shankar, J., et al. (2017). Estrogen related receptor is required for the testicular development and for the normal sperm axoneme/mitochondrial derivatives in Drosophila males. Sci. Rep. 7:40372. doi: 10.1038/srep40372
Nakajima, H., Matsumoto, Y., and Sekimizu, K. (2018). Establishment of a gnotobiotic silkworm model. Drug Discov. Ther. 12, 291–294. doi: 10.5582/ddt.2018.01048
Nishita, Y. (2014). Ecdysone response elements in the distal promoter of the Bombyx Broad-Complex gene. BmBR-C. Insect Mol. Biol. 23, 341–356. doi: 10.1111/imb.12085
Park, K., and Kwak, I. S. (2010). Molecular effects of endocrine-disrupting chemicals on the Chironomus riparius estrogen-related receptor gene. Chemosphere 79, 934–941. doi: 10.1016/j.chemosphere.2010.03.002
Sailland, J., Tribollet, V., Forcet, C., Billon, C., Barenton, B., Carnesecchi, J., et al. (2014). Estrogen-related receptor alpha decreases RHOA stability to induce orientated cell migration. Proc. Natl. Acad. Sci. U S A 111, 15108–15113. doi: 10.1073/pnas.1402094111
Shen, G., Wu, J., Han, C., Liu, H., Xu, Y., Zhang, H., et al. (2018). Oestrogen-related receptor reduces vitellogenin expression by crosstalk with the ecdysone receptor pathway in female silkworm, Bombyx mori. Insect Mol. Biol. 27, 454–463. doi: 10.1111/imb.12385
Shen, G., Wu, J., Lin, Y., Hua, X., Xia, Q., and Zhao, P. (2021). Estrogen-Related Receptor Influences the Hemolymph Glucose Content by Regulating Midgut Trehalase Gene Expression in the Last Instar Larvae of Bombyx mori. Int. J. Mol. Sci. 22:9. doi: 10.3390/ijms22094343
Suetsugu, Y., Futahashi, R., Kanamori, H., Kadono-Okuda, K., Sasanuma, S., Narukawa, J., et al. (2013). Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3 3, 1481–1492. doi: 10.1534/g3.113.006239
Tennessen, J. M., Baker, K. D., Lam, G., Evans, J., and Thummel, C. S. (2011). The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13, 139–148. doi: 10.1016/j.cmet.2011.01.005
Thummel, C. S., and Chory, J. (2002). Steroid signaling in plants and insects–common themes, different pathways. Genes Dev. 16, 3113–3129. doi: 10.1101/gad.1042102
Tian, L., Ma, L., Guo, E., Deng, X., Ma, S., Xia, Q., et al. (2013). 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 9, 1172–1187. doi: 10.4161/auto.24731
Tremblay, A. M., and Giguere, V. (2007). The NR3B subgroup: an ovERRview. Nucl. Recept. Signal 5:e009. doi: 10.1621/nrs.05009
Yang, C., Lin, Y., Liu, H., Shen, G., Luo, J., Zhang, H., et al. (2014). The Broad Complex isoform 2 (BrC-Z2) transcriptional factor plays a critical role in vitellogenin transcription in the silkworm Bombyx mori. Biochim. Biophys. Acta 1840, 2674–2684. doi: 10.1016/j.bbagen.2014.05.013
Keywords: silkworm, estrogen-related receptor, 20-hydroxyecdysone, ecdysone response element, transcriptional activity, 20E signal pathway
Citation: Wu J, Shen G, Liu D, Xu H, Jiao M, Zhang Y, Lin Y and Zhao P (2022) The Response of the Estrogen-Related Receptor to 20-Hydroxyecdysone in Bombyx mori: Insight Into the Function of Estrogen-Related Receptor in Insect 20-Hydroxyecdysone Signaling Pathway. Front. Physiol. 12:785637. doi: 10.3389/fphys.2021.785637
Received: 29 September 2021; Accepted: 29 December 2021;
Published: 18 January 2022.
Edited by:
Sheng Li, South China Normal University, ChinaReviewed by:
Erik C. Johnson, Wake Forest University, United StatesCopyright © 2022 Wu, Shen, Liu, Xu, Jiao, Zhang, Lin and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Lin, bHk5MDhAc3d1LmVkdS5jbg==; Ping Zhao, emhhb3BAc3d1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.