
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 17 November 2021
Sec. Chronobiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.776826
This article is part of the Research TopicBiological Rhythms: Evolution, Population Biology, and AdaptationView all 9 articles
With approximately 160,000 identified species of butterflies and moths, Lepidoptera are among the most species-rich and diverse insect orders. Lepidopteran insects have fundamental ecosystem functions as pollinators and valuable food sources for countless animals. Furthermore, Lepidoptera have a significant impact on the economy and global food security because many species in their larval stage are harmful pests of staple food crops. Moreover, domesticated species such as the silkworm Bombyx mori produce silk and silk byproducts that are utilized by the luxury textile, biomedical, and cosmetics sectors. Several Lepidoptera have been fundamental as model organisms for basic biological research, from formal genetics to evolutionary studies. Regarding chronobiology, in the 1970s, Truman’s seminal transplantation experiments on different lepidopteran species were the first to show that the circadian clock resides in the brain. With the implementation of molecular genetics, subsequent studies identified key differences in core components of the molecular circadian clock of Lepidoptera compared to the dipteran Drosophila melanogaster, the dominant insect species in chronobiological research. More recently, studies on the butterfly Danaus plexippus have been fundamental in characterizing the interplay between the circadian clock and navigation during the seasonal migration of this species. Moreover, the advent of Next Generation Omic technologies has resulted in the production of many publicly available datasets regarding circadian clocks in pest and beneficial Lepidoptera. This review presents an updated overview of the molecular and anatomical organization of the circadian clock in Lepidoptera. We report different behavioral circadian rhythms currently identified, focusing on the importance of the circadian clock in controlling developmental, mating and migration phenotypes. We then describe the ecological importance of circadian clocks detailing the complex interplay between the feeding behavior of these organisms and plants. Finally, we discuss how the characterization of these features could be useful in both pest control, and in optimizing rearing of beneficial Lepidoptera.
The daily rotation of the Earth causes predictable 24-h environmental cycles that have resulted in the evolution of circadian clocks, endogenously maintained timing mechanisms in almost all organisms, including bacteria, fungi, plants, and metazoan (Paranjpe and Sharma, 2005; Rosbash, 2009; Eelderink-Chen et al., 2021). These internal clocks are synchronized (entrained) by environmental stimuli, such as daylight or temperature, enabling organisms to adapt to environmental changes in phase with the 24-h day. Moreover, they regulate molecular, cellular, physiological, and behavioral rhythms, which show a periodicity of ̴ 24h in the absence of any external cue (free-running condition). In insects, circadian clocks control the rhythmicity of several behaviors and physiological features, including egg-hatching, pupation, adult eclosion, locomotion, mating, feeding, and metabolism (Saunders, 2002; Tomioka and Matsumoto, 2010; Franco et al., 2018). The first circadian clock gene (period) was identified in Drosophila melanogaster (Konopka and Benzer, 1971); since then, the species has been the dominant model to elucidate the molecular genetic mechanisms underlying circadian rhythms in insects. However, studies have also been conducted in other insect species, many including Lepidoptera.
The order of Lepidoptera comprises butterflies and moths and is among the most species-rich and diverse insect groups. It includes approximately 160,000 identified species, subdivided into ̴ 43–45 superfamilies (Mitter et al., 2017; Kawahara et al., 2019; Figure 1). Lepidoptera are holometabolous insects, showing juvenile stages (larvae or caterpillars) morphologically distinct from the mature adult form (moths and butterflies). The majority of Lepidoptera are nocturnal (about 75–85% of the identified species), while the remaining 15–25% of diurnal species also includes butterflies (Papilionoidea, ̴19,000 described species; Kawahara et al., 2018). The overwhelming majority of Lepidoptera (>99%) are herbivores with over 90% having a host plant range of three or fewer species (Pierce, 1995); they are found in almost all terrestrial ecosystems and have fundamental ecosystem roles as pollinators and prey. They also have a significant impact on the human economy, as many species represent serious food crop pests, while others such as the domesticated silkworm Bombyx mori (Goldsmith et al., 2005) and Antheraea pernyi (Li et al., 2017) produce silk that is utilized by the textile industry. Moreover, silk is employed in innovative applications of both the biomedical and cosmetics sectors.
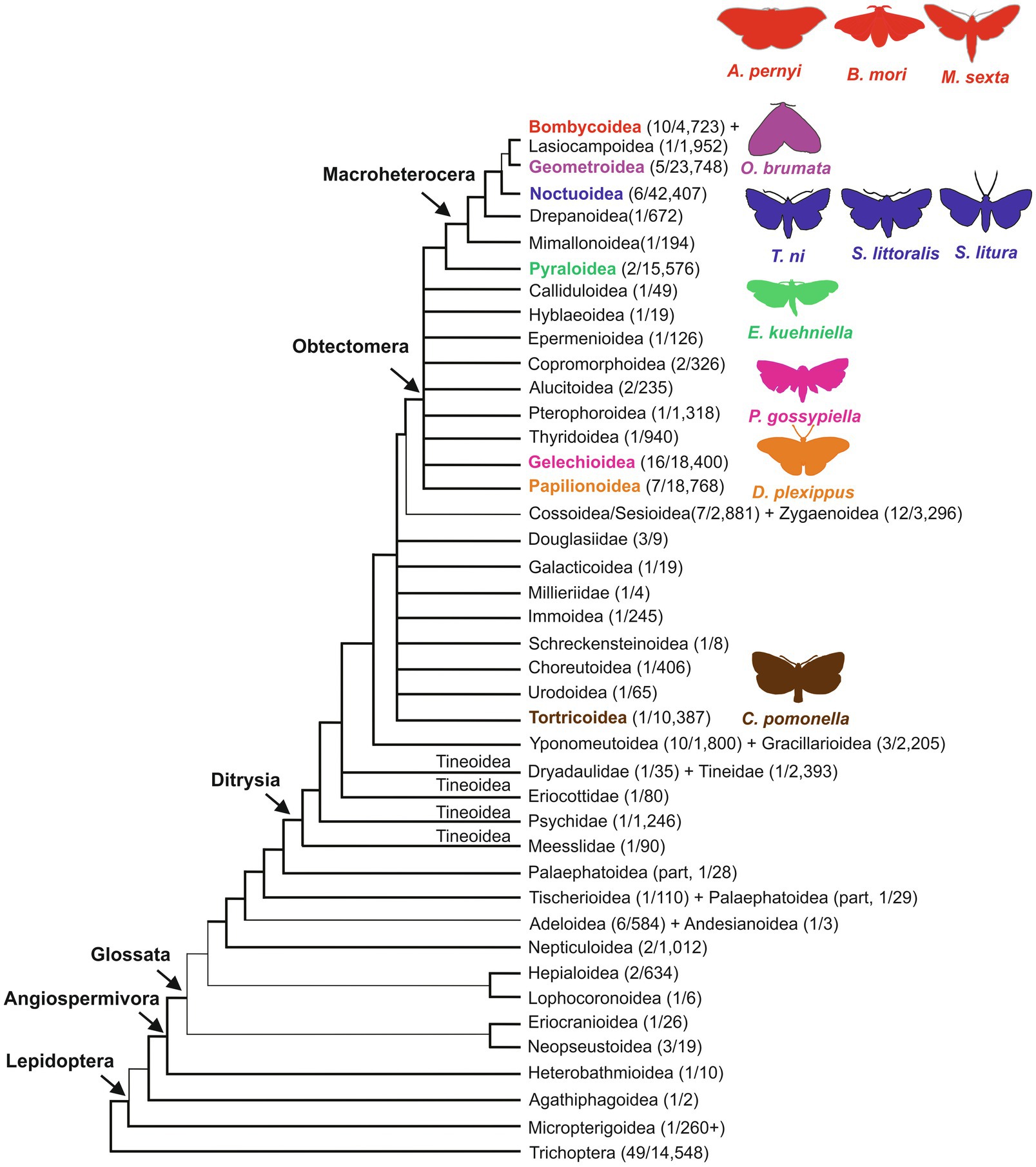
Figure 1. Phylogeny of Lepidoptera superfamilies, including Trichoptera, sister group to Lepidoptera (Mitter et al., 2017). Superfamilies, including species with at least one circadian phenotype described in this review, are reported with different colors: brown: Tortricoidea; orange: Papilionoidea; magenta: Gelechioidea; green: Pyraloidea; blue: Noctuoidea; purple: Geometroidea; and red: Bombycoidea. Representative species of each superfamily are shown using the same color code. As in Mitter et al. (2017), thicker lines indicate more strongly recognized groupings; numbers within parenthesis indicate the number of families/species. On the left, the following higher-level classification groups are shown: Angiospermivora, angiosperm feeding; Glossata, acquisition of the proboscis in adults; Ditrysia, a clade containing 98% of lepidopteran species; Obtectomera, a clade comprising butterflies and macro-moths; Macroheterocera, a clade of moths including superfamilies previously placed in the Macrolepidoptera group, which is currently not considered a monophyletic group (Mimallonoidea, Drepanoidea, Noctuoidea, Geometroidea, Lasiocampoidea, and Bombycoidea; Mitter et al., 2017). Modified from Mitter et al. (2017).
A recent phylogenomic analysis dated the most recent common ancestor of Lepidoptera in the late Carboniferous, about 300million years ago. Evolutionary studies support the hypothesis that Lepidoptera developed initially as internal feeders of non-vascular plants, then differentiated and coevolved in parallel with angiosperms (Figure 1). In this process, the larval stages became external plant feeders, while adults evolved a tube-like proboscis, used for the efficient acquisition of nutritious nectar from flowering plants (Mitter et al., 2017; Kawahara et al., 2019; Figure 1). These adaptations along with an increased ability to fly and colonize new plants are believed to be associated with the large body size observed in many moths and butterflies (Powell et al., 1998; Kawahara et al., 2019).
Several of the larger lepidopteran species have been model organisms in the study of different biological phenomena, including development, physiology, ecology, and evolution (Roe et al., 2009). Pioneering Lepidoptera-based investigations have been fundamental in the study of circadian biology, showing for the first time that the brain is the site of a light-entrainable circadian clock controlling behavioral rhythms (Williams, 1963; Truman and Riddiford, 1970).
This review presents an updated overview of the circadian clock in Lepidoptera. After a general description of the different behavioral circadian rhythms currently identified, we report the molecular and anatomical organization of circadian clocks in the adult stage and during development. We then describe the ecological importance of circadian clocks detailing the complex interplay between lepidopteran feeding behavior and plants. Finally, we discuss how the characterization of these features could be useful in both pest control and rearing improvements of beneficial Lepidoptera.
From embryo to adulthood, both nocturnal and diurnal Lepidoptera exhibit multiple daily behavioral rhythms restricted to species-specific daytimes. Furthermore, for some species, these rhythmic activities have been demonstrated to be maintained under free-running conditions, indicating they are controlled by an endogenous circadian clock (Merlin and Reppert, 2009; Groot, 2014).
One of the first described circadian events occurring during lepidopteran development is egg-hatching, i.e., the emergence of the larva from the protective envelopes of the egg. In several moth species, egg-hatching occurs in the first hours of the day with a peak anticipating morning light-on, when maintained in light/dark (LD) entraining conditions (Minis and Pittendrigh, 1968; Sakamoto and Shimizu, 1994; Sauman and Reppert, 1998). This egg-hatching rhythm was demonstrated to be truly circadian as it was entrainable by environmental stimuli (such as light and/or temperature) from mid embryogenesis and was maintained in constant darkness (DD) regimes in the pink bollworm Pectinophora gossypiella (superfamily Gelechioidea; Minis and Pittendrigh, 1968) and the Chinese oak silk moth Antheraea pernyi (superfamily Bombycoidea; Sauman et al., 1996; Sauman and Reppert, 1998). Circadian egg-hatching rhythms have been described in other lepidopteran species, including the domesticated silkworm B. mori (superfamily Bombycoidea; Sakamoto and Shimizu, 1994; Table 1).
During post-embryonic development, lepidopteran larvae are characterized by intense feeding and recent studies demonstrated that for some Lepidoptera, feeding behavior in the last larval instar occurs with a daily rhythm in entraining conditions (Goodspeed et al., 2012; Suszczynska et al., 2017; Niepoth et al., 2018; Zhang et al., 2021). Studies on different pest species belonging to the Noctuoidea superfamily, including the cabbage looper Trichoplusia ni, as well as the cotton leafworms Spodoptera littoralis and Spodoptera litura, analyzed feeding behavior in both LD and DD conditions. They indicated that in these species larval feeding is under circadian control (Goodspeed et al., 2012; Suszczynska et al., 2017; Zhang et al., 2021). Moreover, S. litura larvae showed rhythmic circadian locomotor and defecation activities, respectively peaking during the night and the day, in both LD and DD conditions (Zhang et al., 2021; Table 1). Additionally, gut purging, which occurs at the end of larval development to empty the gut before entering the pupal stage, was shown to be under circadian control in the silkmoth Samia cynthia var. ricini (superfamily Bombycoidea; Mizoguchi and Ishizaki, 1982; Table 1).
Adult eclosion, i.e., adult emergence from the pupal exuvia, is one of the most vulnerable events during an insect’s life cycle, and in many species is controlled by the circadian clock. Eclosion occurs at species-specific times of the day in different lepidopteran species when maintained in LD conditions and likely restricts the timing of this behavior to the most advantageous moments of the day (Merlin and Reppert, 2009). In constant conditions, adult eclosion maintains a 24-h rhythmic profile, as demonstrated for species belonging to the superfamilies of Bombycoidea (the giant silkmoth Hyalophora cecropia; Truman and Riddiford, 1970; B. mori; Truman, 1972; Shimizu and Matsui, 1983; A. pernyi; Truman and Riddiford, 1970), Noctuoidea (the fall webworm Hyphantria cunea; Morris and Takeda, 1994), Pyraloidea (the southwestern corn borer Diatraea grandiosella; Takeda, 1983), and Papilionoidea (the monarch butterfly Danaus plexippus; Froy et al., 2003; Table 1).
Adult flight is related to vital activities for both moths and butterflies, including nectar feeding and mating. To date, the circadian control of flight behavior has been evaluated in relatively few Lepidoptera. Nevertheless, a flight circadian rhythmicity has been demonstrated in species belonging to the superfamily Bombycoidea, initially in Hyalophora cecropia, S. cynthia var. ricini, A. pernyi (Truman, 1974), and more recently in the hawkmoths Manduca sexta and Hyles lineata (Broadhead et al., 2017). In LD conditions, the nocturnal moths H. cecropia, S. cynthia var. ricini, A. pernyi, and M. sexta display rhythmic flight restricted to the dark period of the day, while the crepuscular H. lineata shows a bimodal activity, with a first peak after light-on and a second after light-off. In DD, all these species maintain 24h rhythmic flight, although with some modifications in their profiles (Truman, 1974; Broadhead et al., 2017; Table 1).
In moths, several behavioral activities related to reproduction have been shown to have a rhythmic daily species-specific pattern (reviewed in Groot, 2014). In these species, reproduction is characterized by stereotypical behaviors anticipating actual mating, associated with the production and/or release of sex pheromones by females (reviewed in Groot, 2014; Stepien et al., 2020). During sex pheromone emission, females adopt typical postures (named calling), characterized by a modification of the abdomen position and the extrusion of the pheromone gland to better disperse sex pheromones in the environment (Stepien et al., 2020). Males perceive sex pheromones through the antennae and fly toward the female for mating (Stepien et al., 2020). In several moths, female calling, male response to female sex pheromones, and mating have been shown to have a rhythmic daily species-specific pattern in LD conditions (Groot, 2014). Some studies examined these behaviors under constant conditions, demonstrating a circadian control of these daily activities. For example, female calling behavior was shown to maintain its rhythmicity in DD in species of the superfamily Noctuoidea (the true armyworm moth Pseudaletia unipuncta, the oriental tobacco budworm moth Helicoverpa assulta, and S. littoralis; Turgeon and McNeil, 1982; Delisle and McNeil, 1987; Kamimura and Tatsuki, 1993; Silvegren et al., 2005; Sadek et al., 2012), in some Tortricoidea (the oriental fruit moth Grapholita molesta and the codling moth Cydia pomonella; Baker and Cardé, 1979; Castrovillo and Cardé, 1979), and in the Mediterranean flour moth Ephestia kuehniella (superfamily Pyraloidea; Závodská et al., 2012; Table 1). A 24h rhythmic male response to female sex pheromones was demonstrated in constant conditions in some Noctuoidea moths (the turnip moth Agrotis segetum, T. ni, Heliothis virescens, Spodoptera exigua, and S. littoralis; Shorey and Gaston, 1965; Castrovillo and Cardé, 1979; Linn et al., 1996; Rosén et al., 2003; Silvegren et al., 2005) and in the Tortricoidea C. pomonella (Castrovillo and Cardé, 1979; Table 1). A 24h rhythm was detected in mating frequency of S. littoralis in both 12:12 LD and DD conditions (Silvegren et al., 2005). Additionally, in S. littoralis, optimized mating efficiency requires synchronized circadian clocks in phase between the two sexes, as a decreased frequency in mating was detected when males and females possessed out of phase circadian clocks (Silvegren et al., 2005).
Various lepidopteran species show a migratory behavior due to seasonally changing environments (Williams, 1957; Walker, 1980). Migration has been extensively studied in the eastern North American populations of the monarch butterfly D. plexippus, which are characterized by oriented flight and each Autumn can travel up to 4,500km to reach wintering sites in central Mexico. Migrants traveling southward are characterized by a reproductive diapause and enhanced longevity. In Spring, adults exit from reproductive diapause, mate, and remigrate to the north, with females laying fertilized eggs in the southern United States. The progeny of these butterflies continues migrating North, reaching the original sites in North America, in a mostly multigenerational migratory phenomenon (Perez et al., 1997; recently reviewed in Reppert et al., 2016; Reppert and de Roode, 2018; Merlin et al., 2020). To generate accurate orientation during migration, monarchs possess well-characterized sun and magnetic compasses, which incorporate daylight cues and the Earth’s magnetic field information, respectively (Mouritsen and Frost, 2002; Froy et al., 2003; Guerra et al., 2014). The sun compass requires the circadian clock to compensate for the apparent movement of the sun in the sky during the day (Mouritsen and Frost, 2002; Froy et al., 2003; Merlin et al., 2009). Additionally, a specific molecular clock element with a clock-independent role (DpCRYPTOCHROME1, DpCRY1) is involved in the proper functioning of the magnetic compass (Gegear et al., 2008, 2010; Wan et al., 2021). As the role of DpCRY1 in D. plexippus magnetoreception is not related to its function in the circadian clock, this aspect is not further discussed in this review (for interested readers: Reppert et al., 2016; Reppert and de Roode, 2018; Merlin et al., 2020; Wan et al., 2021).
The involvement of the circadian clock for time-compensated navigation during migration has been hypothesized in other butterflies such as Aphrissa statira and Phoebis argante (Oliveira et al., 1998; Table 1). Migration has great relevance for the functionality of an ecosystem since newly arrived migrants can have a role as pollinators, representing a food source for insectivores and in turn for the other components of a trophic system. As migration is typical of several butterflies and moths (Williams, 1957), it would be interesting to understand whether the circadian clock is involved in orientation during navigation of other lepidopteran species.
The core of the molecular circadian clock is driven by positive and negative interlocking transcription–translation feedback loops (TTLs), which sustain a rhythmic production of mRNA and proteins of key clock factors (recently reviewed in Patke et al., 2020). In D. melanogaster, the positive limb of the major TTL includes the basic helix–loop–helix (bHLH), Per-Arnt-Sim (PAS), transcriptional factors dCLOCK (dCLK) and dCYCLE (dCYC, an ortholog of BMAL1), which acting as a heterodimer (dCLK:dCYC), bind conserved sequences (E-boxes) in the promoters of dperiod (dper) and dtimeless (dtim), activating their transcription. dPER and dTIM proteins belong to the negative limb of the major TTL and act in the nucleus as inhibitors of the dCLK:dCYC transcriptional regulators (Allada et al., 1998; Bae et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Glossop et al., 1999; Lee et al., 1999; Figure 2A). However, at both post-transcriptional and post-translational levels dper and dtim mRNAs and proteins are subjected to several modifications which control dPER and dTIM synthesis, stability, accumulation, and nuclear translocation, generating fundamental temporal delays in the TTL, and in turn determining the precise timing of the circadian clock (Curtin et al., 1995; Saez and Young, 1996; Price et al., 1998; Rothenfluh et al., 2000; Martinek et al., 2001; Patke et al., 2020). In a second TTL, dCLK:dCYC dimer induces the expression of dVRILLE (dVRI) and dPAR DOMAIN PROTEIN1ε (dPDP1ε), which inhibits and promotes dClk expression, respectively (Cyran et al., 2003; Figure 2A). A third TTL helps sustain a robust rhythmic oscillation and includes the transcriptional repressor dclockwork orange (dcwo; Patke et al., 2020). dcwo transcription is promoted by dCLK:dCYC, and dCWO modulates dCLK:dCYC activity. In particular, dCWO represses dCLK:dCYC-mediated transcription by binding to E-box elements of clock genes, including dper and dtim (Kadener et al., 2007; Lim et al., 2007). Recent data indicate that CWO also exerts an indirect promotion of dCLK:dCYC activity, by repressing transcription of the Drosophila ortholog of mouse Clock Interacting Protein Circadian (Cipc), an inhibitor of dCLK:dCYC activity (Rivas et al., 2021; Figure 2A).
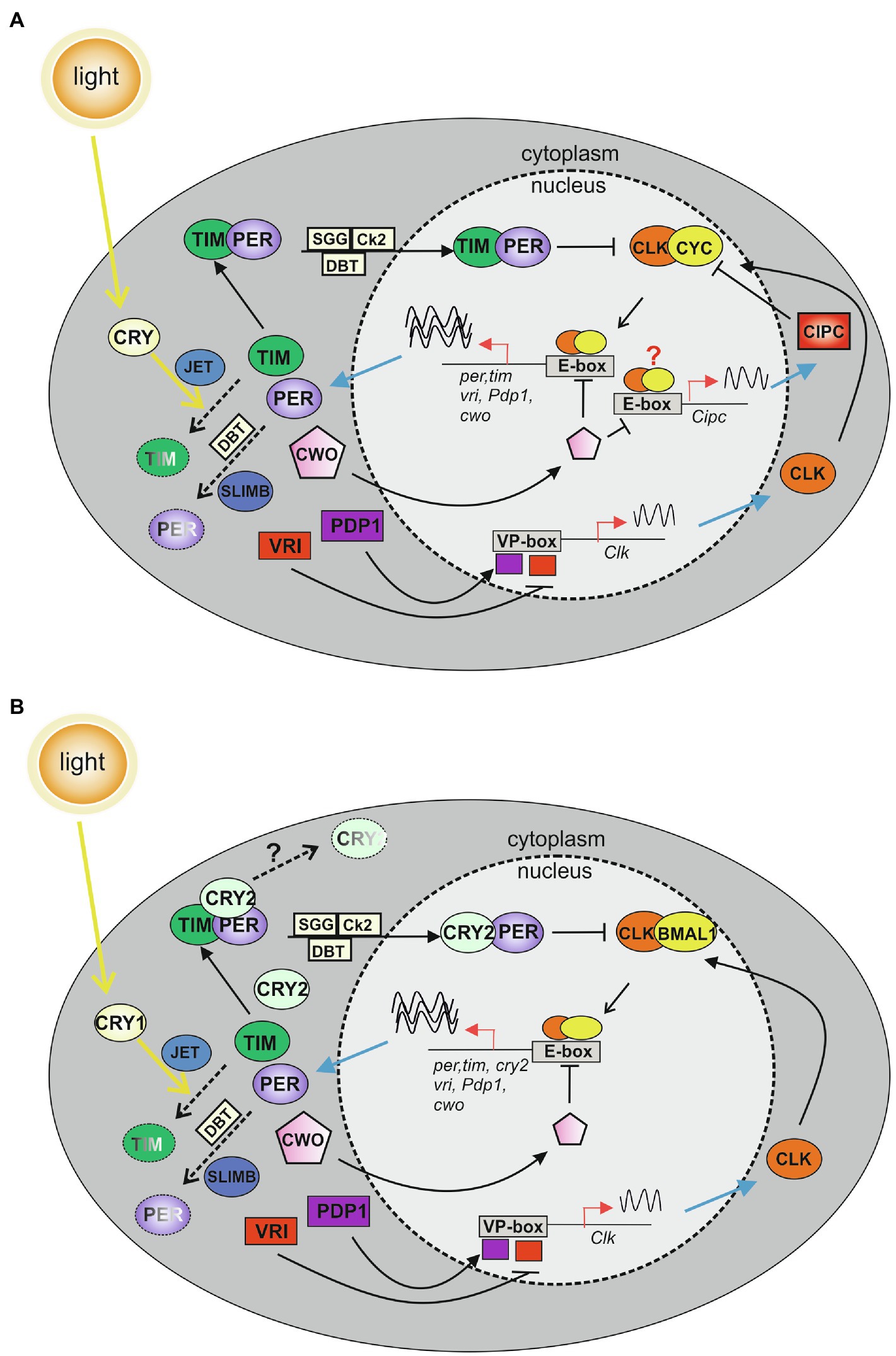
Figure 2. Model of the major transcription–translation feedback loops (TTLs) in the circadian molecular clock of Drosophila melanogaster (A) and Lepidoptera (B). (A) In the first TTL of D. melanogaster, CLOCK (CLK) and CYCLE (CYC) dimerize and bind to E-boxes in the promoters of per and tim clock genes. PERIOD (PER) and TIMELESS (TIM) enter the nucleus as a complex, and inhibit CLK-CYC activity (Allada et al., 1998; Bae et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Glossop et al., 1999; Lee et al., 1999). A second TTL modulates Clk expression: CLK:CYC dimers induce the transcription of vrille (vri) and par domain protein 1 (Pdp1) genes. VRI and PDP1 compete for the same element (VP-box) in the Clk promoter, controlling Clk transcription (Cyran et al., 2003). In the third TTL, CLK:CYC dimers activate the transcription of clockwork orange (cwo). CWO enters the nucleus and inhibits CLK:CYC activity by binding to E-box elements of clock genes (Kadener et al., 2007; Lim et al., 2007; Richier et al., 2008). CWO acts also indirectly promoting CLK:CYC-mediated transcription, by repressing the production of the CLK:CYC inhibitor CLOCK INTERACTING PROTEIN CIRCADIAN (CIPC; Rivas et al., 2021). Cipc transcription modulation might occur at the level of the E-boxes detected in the Cipc locus (red question mark; Rivas et al., 2021). Phosphorylation mediated by DOUBLETIME (DBT), CASEIN KINASE 2 (CK2), and SHAGGY (SGG) modulate clock protein activities, regulating protein–protein interactions, nuclear translocation, and degradation (Curtin et al., 1995; Saez and Young, 1996; Price et al., 1998; Rothenfluh et al., 2000; Martinek et al., 2001). Light activates the photoreceptor CRYPTOCHROME (CRY), which interacts with TIM and mediates its degradation in combination with JETLAG (JET). SUPERNUMERARY LIMBS (SLIMB) signals PER degradation (Emery et al., 1998; Stanewsky et al., 1998; Lin et al., 2001; Koh et al., 2006). (B) The first lepidopteran TTL includes CLK and BMAL1, which dimerize and bind to the E-boxes in the promoters of per, tim, and cry2 genes. In the cytoplasm, PER, TIM, and CRY2 form a complex. CRY2, stabilized by PER, enters into the nucleus to inhibit CLK:BMAL1 activity (Zhu et al., 2005, 2008; Yuan et al., 2007). The second and third TTLs are speculative and include lepidopteran orthologs of Drosophila core and regulatory clock components identified in lepidopteran assembled genomes (Zhan et al., 2011; Derks et al., 2015). In the second TTL, the CLK:BMAL1 complex induces the transcription of vri and Pdp1 genes, which, once translated, compete for the same element (VP-box) in the Clk promoter, controlling Clk transcription. In the third TTL, a CLK:BMAL1 dimer activates the transcription of cwo. CWO enters the nucleus and inhibits CLK:BMAL1 activity by binding to E-box elements of clock genes. Phosphorylation mediated by DBT, CK2, and SGG modulate clock protein activities, regulating protein–protein interactions, nuclear translocation, and degradation. Light is responsible for TIM degradation in a process mediated by CRY1 and JET. SLIMB is involved in PER degradation. Pathways triggering to CRY2 degradation are still unknown (black question mark). Dashed arrows indicate degradation pathways; sinusoidal lines show transcription activity.
For virtually all organisms, light is the main environmental stimulus that entrains the circadian clock with the 24-h LD cycle. In Drosophila, light synchronization is mainly determined by the blue light photoreceptor dCRY (Emery et al., 1998; Stanewsky et al., 1998). Once activated by light, dCRY interacts with dTIM inducing its degradation which in turn impacts dPER stability, thereby resetting the molecular clock (Lin et al., 2001; Koh et al., 2006; Figure 2A).
The molecular elements among insect circadian clocks are homologous, although, some important differences have been identified between the circadian factors of Drosophila and Lepidoptera (Figure 2B). PER, TIM, CLOCK, and BMAL1 clock components from A. pernyi have been demonstrated to act in the positive (ApCLOCK, ApBMAL1) and negative (ApPER, ApTIM, the latter with a modulatory activity) limbs of the major TTL, using Drosophila embryonic Schneider (S2) cells (Chang et al., 2003). However, a bioinformatic analysis mapped the main transactivation domain essential for the ApCLK:ApBMAL1 function in the C-terminal region of ApBMAL1 protein, and not at the level of ApCLK, as previously shown for dCLK in Drosophila (Allada et al., 1998; Darlington et al., 1998; Chang et al., 2003). This appears a common feature in Lepidoptera, as the presence of a BMAL1 transactivation domain was reported in other species belonging to this order, including B. mori and D. plexippus (Markova et al., 2003; Zhang et al., 2017). Interestingly, with the exception of Drosophila, the localization of the transactivation domain in the BMAL1 C-terminal region is highly conserved in other non-lepidopteran insects and vertebrates (Chang et al., 2003). These observations suggested an evolutionary model, in which the common ancestor of insects and vertebrates possessed a BMAL homolog characterized by a C-terminal transactivation domain. The latter was subsequently lost in the Drosophila lineage, likely because of the acquisition of a functionally related transactivation domain within dCLK (Chang et al., 2003). For these reasons, in this review, we use “BMAL1” to identify the lepidopteran ortholog of Drosophila dCYC, although, both BMAL1 and CYC are currently employed in the lepidopteran circadian clock literature.
The most informative experiments elucidating the structure and functioning of the lepidopteran molecular clock have been performed in D. plexippus, in a huge series of studies initiated by Reppert and colleagues to understand the role of the circadian clock in the seasonal migration typical of this butterfly (Mouritsen and Frost, 2002; Froy et al., 2003; Zhu et al., 2005, 2008; Zhan et al., 2011; Table 1). These investigations demonstrated that D. plexippus possesses two CRY proteins, one ortholog to the light-sensitive Drosophila dCRY (henceforth DpCRY1) and a second vertebrate-like light-insensitive CRY, named DpCRY2 (Figure 2B). DpCRY1 was able to mediate the light-sensitive degradation of DpTIM in monarch butterfly DpN1 embryonic cell lines (Palomares et al., 2003; Zhu et al., 2008). On the other hand, DpCRY2 coimmunoprecipitated with DpPER and DpTIM in both DpN1 cells and D. plexippus brains and showed an inhibitory activity of the DpCLK:DpBMAL1-mediated transcription in Drosophila and monarch cell lines (Zhu et al., 2005, 2008; Yuan et al., 2007). In addition, DpCRY2 was the only D. plexippus clock factor showing circadian translocation into the nuclei of the putative Danaus brain clock neurons (see below), while the role of DpPER was associated with a DpCRY2 stabilization rather than with an active transcriptional repression (Zhu et al., 2008). As a consequence, a Danaus molecular clock model in which DpCRY2 represents the major inhibitor of the DpCLK:DpBMAL1 clock-controlled transcription in the first TTL was developed (Zhu et al., 2008; Zhan et al., 2011; Figure 2B). This model was further confirmed by the generation of D. plexippus Dpcry2 and DpClk knock-out (KO) mutants, which indicated DpCRY2 and DpCLK, respectively, as a transcriptional repressor and a transcriptional activator of the monarch circadian clock in vivo (Merlin et al., 2013; Markert et al., 2016). In addition, the analysis of CRISPR/Cas9-mediated D. plexippus mutants with different modifications in the DpBMAL1 C-terminal region and luciferase assays in Drosophila cells mapped the amino acid portions of DpBMAL1 and DpCLK important for the DpCRY2 repression activity and regulation of circadian rhythmicity (Zhang et al., 2017). It was demonstrated that the DpCRY2 inhibitory activity on DpCLK:DpBMAL1 occurs with a dual mechanism. In particular, one mechanism which appears to be independent from the DpBMAL1 transactivation domain, involves a specific residue of DpCLK (E333 in the PAS-B domain) and it is important for the generation of circadian rhythmicity. The second mechanism implies the interaction with both the DpBMAL1 transactivation domain and DpCLK (at the residue W328) and plays a role in the regulation of the circadian phase or period (Zhang et al., 2017).
The D. plexippus molecular clockwork, with CRY2 acting as main transcriptional repressor of the CLK:BMAL1 dimer in the first TTL, is currently considered the model of the molecular circadian clock in Lepidoptera, as two functional cry genes have been detected in several species, including those belonging to the superfamilies of Bombycoidea (B. mori and A. pernyi; Zhu et al., 2005; Kawamoto et al., 2019), Noctuoidea (S. littoralis, S. litura, A. segetum, the pink stalk borer Sesamia nonagrioides, and the cotton bollworm Helicoverpa armigera; Merlin et al., 2007; Tomioka and Matsumoto, 2010; Yan et al., 2013; Chang et al., 2019; Zhang et al., 2021) and Geometroidea (the winter moth Operophtera brumata; Derks et al., 2015).
Finally, the orthologs of other Drosophila core and regulatory circadian clock components have been identified with the assembly of several Lepidoptera genomes, including those of D. plexippus and O. brumata, resulting in a lepidopteran molecular circadian clock model incorporating three main TTLs (Zhan et al., 2011; Derks et al., 2015; Figure 2B). Further work will be fundamental to functionally demonstrate the role of all these elements in the molecular clock of this order.
Currently, the molecular circadian clock has been deeply characterized in a relatively small number of Lepidoptera. Nevertheless, the knowledge derived from these studies has given a fundamental contribution to the comprehension of circadian clock evolution in insects. In particular, the identification of two functionally different cry genes in Lepidoptera led to the detection of both cry1 and cry2 in other dipteran non-Drosophilid species such as the mosquitoes Anopheles gambiae and Aedes aegypti (Zhu et al., 2005; Yuan et al., 2007). On the contrary, species such as the hymenopteran Apis mellifera and Bombus impatiens as well as the coleopteran Tribolium castaneum have been demonstrated to possess only cry2 (Rubin et al., 2006; Yuan et al., 2007). When tested in Drosophila cell cultures, CRY2s belonging to these species were not light-sensitive and behaved as repressors of a CLK:BMAL1-mediated transcription (Yuan et al., 2007). A clock bearing only light-insensitive cry2 has also been detected in the fire ant Solenopsis invicta, belonging to the order Hymenoptera (Ingram et al., 2012). These findings led to hypothesize that the Lepidoptera-like molecular clock, with both a light-sensitive CRY1 and a transcriptional repressor CRY2, represents an ancestral clock, while those characterized by only a functional CRY1 (as in Drosophila) or only a functional CRY2 (as in bees, beetles, and ants) are derived clocks, in which cry2 or cry1 have been lost, respectively (Reppert, 2007; Yuan et al., 2007; Merlin and Reppert, 2009).
In Lepidoptera, the central clock controlling the rhythmicity of several behavioral phenotypes resides in the brain. This was first demonstrated in A. pernyi and H. cecropia by transplantation and ablation experiments, which showed that the dorsolateral brain region houses a master clock controlling circadian behavioral phenotypes such as adult eclosion and flight (Truman, 1972, 1974).
Most of the studies aimed at identifying the brain master clock neurons in Lepidoptera have been performed in four species: A. pernyi (Sauman and Reppert, 1996; Mohamed et al., 2014), M. sexta (Wise et al., 2002), B. mori (Sehadová et al., 2004), and D. plexippus (Sauman et al., 2005; Zhu et al., 2008). To map the clock neurons within the brains of these organisms, in situ mRNA hybridization analyses and/or immunolocalization studies using specific antibodies against different clock proteins have been performed. Several species-specific differences in brain expression patterns have been detected, with M. sexta and A. pernyi showing the highest and the lowest number of clock gene expressing neurons, respectively (Sauman and Reppert, 1996; Wise et al., 2002; Mohamed et al., 2014). However, a common feature characterizing the four species was the expression of the analyzed clock factors in two clusters of large type Ia1 neurosecretory cells, symmetrically located in each Pars Lateralis (PL) of the brain, which are considered to represent the circadian clock neurons in the brain of these Lepidoptera (Wise et al., 2002; Sehadová et al., 2004; Sauman et al., 2005; Zhu et al., 2008; Figure 3A).
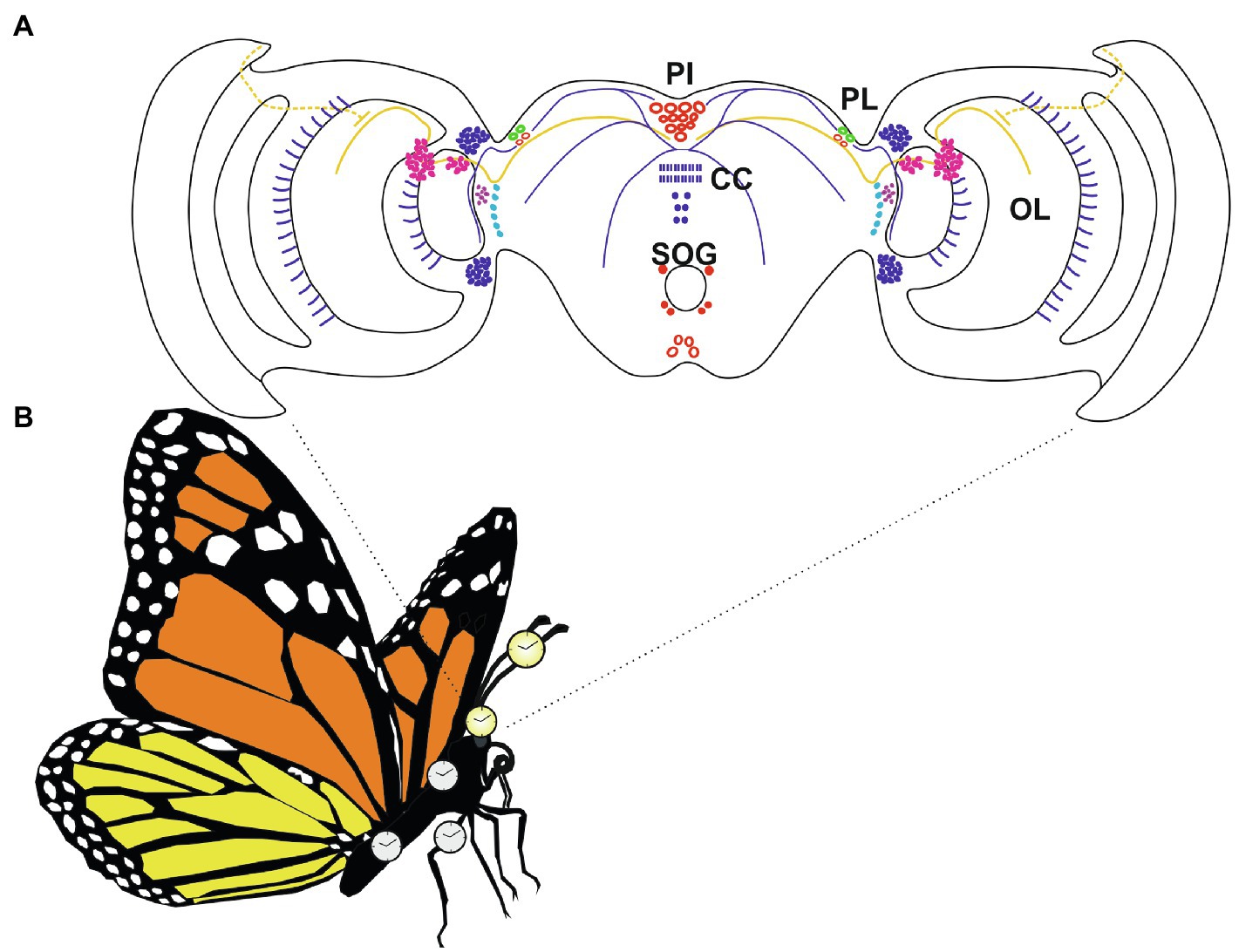
Figure 3. The circadian system in Danaus plexippus adults, with multiple oscillators located in the brain and body. (A) Relative positions of neurons and fibers expressing clock factors in D. plexippus brain. In each brain hemisphere, two Pars Lateralis (PL; green) neurons co-express DpTIM, DpPER, DpCRY1, and DpCRY2. These PL cells are believed the putative circadian clock neurons of the D. plexippus brain. Neurons expressing DpTIM, DpPER, DpCRY1, and/or DpCRY2 are depicted in red. In these cells, the four clock molecules partially colocalize. Regions expressing DpTIM or DpCRY1 are indicated in magenta. In these areas, TIM and CRY1 do not colocalize. Cells exclusively expressing TIM or PER are represented in purple and light blue, respectively. CRY2-positive neurons and fibers are shown in blue. Continuous yellow lines indicate fibers expressing CRY1. Dotted yellow lines indicate projections of the dorsal rim area. PL, Pars lateralis; PI, Pars intercerebralis; CC, central body; SOG, Subesophageal ganglion; and OL, optic lobe (modified from Sandrelli et al., 2008; see text for explanation). (B) Schematic representation of an adult monarch butterfly. Yellow clocks represented autonomous oscillators located in the brain and antennae (Sauman et al., 2005; Zhu et al., 2008; Merlin et al., 2009). White clocks indicate monarch butterfly tissues (thorax, legs, and abdomen) which showed a rhythmic expression of different clock factors in LD conditions (Merlin et al., 2009) and are likely sites of peripheral oscillators.
In the A pernyi central brain, four PL neurons per hemisphere have been reported as the only cells expressing ApPER and ApTIM (Sauman and Reppert, 1996). More recently, ApPER-/ApTIM-positive PL neurons have been shown to co-express ApBMAL1 and ApCLK proteins (Mohamed et al., 2014). In LD conditions, ApPER and ApTIM were rhythmically expressed in both cytoplasm and axonal projections, with higher levels during the night and lower during the day. However, the expression profiles of ApPER and ApTIM proteins oscillated with the same phase of Apper and Aptim mRNAs, without any delay between mRNA and protein accumulation (Sauman and Reppert, 1996). In addition, the staining was restricted to cytoplasmic and axonal subcellular compartments, while no ApPER and/or ApTIM signals were detected within the nuclei (Sauman and Reppert, 1996). These data provided the first indication that the functioning of these elements in the putative lepidopteran adult clock neurons could be different from that of Drosophila, as later shown in the monarch butterfly (Sauman et al., 2005; Zhu et al., 2008).
In M. sexta, only the distribution of Msper mRNA and its corresponding protein was analyzed in the adult brain (Wise et al., 2002). In PL neurons, MsPER protein was localized in both the cytoplasm and nucleus, but no evident 24h cycling expression was detected (Wise et al., 2002).
In B. mori adult brains, PL neurons expressed different clock factors, including BmPER, BmCYC, and BmCRY-like proteins. At this level, the staining of all the proteins was cytoplasmatic, with only BmPER signal showing a cycling intensity in LD (Sehadová et al., 2004).
In D. plexippus, the four PL neurons of each brain hemisphere expressed DpTIM and DpCRY2. In each PL cluster, two neurosecretory cells also co-expressed DpPER and DpCRY1, and are considered the circadian clock neurons in the monarch brain (Sauman et al., 2005; Zhu et al., 2008; Figure 3A). In these neurons DpTIM and DpPER showed a cycling expression in both LD and DD, with a peak during the night. However, DpTIM, DpPER, and DpCRY1 were localized in the cytoplasm. On the contrary, DpCRY2 showed a cycling expression and a localization at the nuclear level at the beginning of the day, when the maximal transcriptional repression of Dpper mRNA is recorded in the monarch brain (Zhu et al., 2008).
In all these species, apart from A. pernyi, additional expression of the examined clock factors have been reported in other regions of the brain, including the Pars Intercerebralis (PI), a region known to have a role in insect insulin signaling, aging and diapause, as well as the optic lobe (OL), and subesophageal ganglion (SOG; Wise et al., 2002; Sehadová et al., 2004; Sauman et al., 2005; Zhu et al., 2008). In B. mori, PI neurons expressing BmPER appeared at the identical position to those expressing BmCYC (Sehadová et al., 2004). In the monarch butterfly, several PI neurons expressed DpTIM, DpPER, DpCRY1, and DpCRY2, and a cluster of neurons clearly co-expressed DpTIM, DpCRY1, and DpPER (Sauman et al., 2005; Zhu et al., 2008; Figure 3A). Moreover, both DpCRY1 and DpCRY2 have been detected in multiple neuronal projections connecting PL neurons to brain regions important for migration (Figure 3A). DpCRY1-positive fibers appeared to connect PL neurons to the dorsal rim area (DRA), an eye region involved in the perception of the polarized light input (Sauman et al., 2005; see below). DpCRY2-positive projections appeared to link PL and PI neurons, and cycling DpCRY2 expression was found in the fibers of the central complex, a conserved high-order neuronal structure of the insect brain, which in the monarch is considered the central site of the sun-compass (Zhu et al., 2008; reviewed in Reppert et al., 2016; Reppert and de Roode, 2018). Interestingly, a recent analysis mapped the brain expression of per and tim genes in E. kuehniella (superfamily Pyraloidea), a pest moth which in the evolution of Lepidoptera represents a less divergent species compared to A. pernyi, B. mori, and M. sexta, which belong to the Macroheterocera clade (Kobelková et al., 2015; Mitter et al., 2017; Figure 1). Immunolocalization studies detected a wide expression of EkPER throughout E. kuehniella brain, with hundreds of EkPER-positive neurons (Kobelková et al., 2015). In most of the E. kuehniella neurons, intense but non-cycling EkPER signals were detected at the level of both cytoplasm and nuclei. However, in the PI a group of 4–5 large neurosecretory cells showed EkPER localization restricted to the cytoplasm and neuronal projections, with only the latter displaying a daily rhythm in staining intensity, and higher signals during the night in LD conditions (Kobelková et al., 2015). To date, the localization of the master clock neurons in this species is still unknown. However, the high number of EkPER-positive neurons in its brain has been hypothesized to be a characteristic of an ancient clock, as already proposed for more ancestral Apterygota and Exopterygota, compared to holometabolous insects (Závodská et al., 2003; Kobelková et al., 2015).
The presence of circadian clocks outside the brain has been demonstrated in adults of different lepidopteran species, including A. pernyi, M. sexta, B. mori, S. littoralis, D. plexippus, E. kuehniella, and H. armigera (Sauman and Reppert, 1996; Wise et al., 2002; Syrova et al., 2003; Iwai et al., 2006; Schuckel et al., 2007; Kotwica et al., 2009; Merlin et al., 2009; Kobelková et al., 2015; Yan et al., 2019). In insects, these circadian clocks appear to be important for the daily regulation of the physiological activities typical of peripheral tissues or organs (Tomioka et al., 2012). Some of these peripheral clocks depend on the clock located in the brain, others are self-sustained oscillators, autonomously ticking with respect to the master clock, as nicely demonstrated in the dipteran D. melanogaster (Plautz et al., 1997). Throughout expression studies and/or physiological analyses, peripheral clocks have been detected in different adult structures, such as eyes, antennae, thorax, abdomen, flight muscles, midgut, testis, and legs (Sauman and Reppert, 1996; Wise et al., 2002; Iwai et al., 2006; Schuckel et al., 2007; Merlin et al., 2009; Yan et al., 2014, 2019; Kobelková et al., 2015; Figure 3B). Here, we will focus on peripheral clocks located in the eye, antenna, and testis, for which most of the data are available.
Like all insects, Lepidoptera possess a compound eye, composed of units known as ommatidia, each formed by a variable number of photoreceptor cells (e.g., nine in Papilionoidea and Sphingidae; eight in Noctuoidea; Friedrich et al., 2006). Most of the studies aimed at characterizing lepidopteran eye clocks were conducted by mapping per mRNA and/or protein expression in A. pernyi, M. sexta, and E. kuehniella (Sauman and Reppert, 1996; Wise et al., 2002; Kobelková et al., 2015). In M. sexta, both Msper mRNA and MsPER protein were detected in photoreceptor nuclei without any clear rhythmic expression (Wise et al., 2002). Conversely, in A. pernyi and E. kuehniella a rhythmic per mRNA accumulation was observed in the nuclei of these cells, with a maximum signal in the dark phase of the LD cycle. Moreover, several hours following the mRNA peak, intense PER protein staining was detected in the photoreceptor nuclei (Sauman and Reppert, 1996; Kobelková et al., 2015). Both the temporal delay between mRNA and protein accumulation and protein nuclear translocation are considered among the principal characteristics of the Drosophila circadian clock. However, they have not been reported for per in the A. pernyi and E. kuehniella brains (see above). These observations suggest that in A. pernyi and E. kuehniella the molecular mechanisms governing the circadian clock in the eye and central brain are distinct. This is not surprising since dissimilarities between the central and peripheral clocks have already been reported in D. melanogaster. In particular, dCRY which is fundamental for light synchronization in both fly brain and peripheral clocks, in antennae and Malpighian tubules has an additional role for maintaining a persistent oscillation, possibly acting as a transcriptional repressor in the core molecular clock (Ivanchenko et al., 2001; Krishnan et al., 2001; Tomioka et al., 2012). Finally, in the photoreceptors of the D. plexippus compound eye, only the expression of Dpcry1 mRNA and its corresponding protein has been reported to date (Wan et al., 2021).
The compound eye perceives light cues and forms visual images (Sondhi et al., 2021), playing a key role for several insect behaviors, such as flight, foraging, mating, and predator avoidance. Additionally, within the compound eye, the dorsal rim area (DRA) is involved in detecting polarized light (Labhart and Meyer, 1999; Reppert et al., 2004). In the study of D. plexippus migration, sun and polarized light detected by the main retina and DRA of the compound eye have been demonstrated to represent the sensing information transmitted to the sun compass, that is likely located in the central complex of the brain (Froy et al., 2003; Reppert et al., 2004; Sauman et al., 2005; Heinze and Reppert, 2011, 2012).
At a molecular level, light is perceived by opsins, G-coupled seven-transmembrane proteins, which combined with a chromophore are key light-sensing factors for color vision. In Lepidoptera and other insects, five distinct opsins have been detected in both nocturnal and diurnal species (Feuda et al., 2016). Among these, the most studied are the long wavelength sensitive opsin (peak absorbance 500–600nm), the blue sensitive opsin (peak absorbance 400–500nm) and the UV sensitive opsin (peak absorbance 300–400nm; Sondhi et al., 2021). mRNA in situ hybridization studies in the monarch showed that the three opsins are expressed in the compound eye, except for the DRA in which only the UV opsin mRNA was detected (Sauman et al., 2005).
A quantitative PCR study on H. armigera compound eyes showed a daily rhythmic oscillation in the transcription levels of the three opsins, with a peak of expression during daytime, in LD conditions and the first day of DD (Yan et al., 2014). However, the daily mRNA oscillations dumped from the second day of DD and in constant light (LL), indicating an interaction between the circadian clock and environmental light in the control of opsin expression in this organism (Yan et al., 2014). The molecular mechanism connecting opsin daily expression and circadian clocks is still unknown. However, cycling mRNA levels of different core clock genes (Haper, Hatim, Hacry1, and Hacry2) have been detected in H. armigera eyes, suggesting the presence of a peripheral clock (Yan et al., 2014, 2019).
A better described clock is located in the antennae, appendages of the adult lepidopteran head involved in detecting odor, taste, temperature, humidity variations, and mechanoreception. Antennae are long and slight structures, composed of two basal segments and a flagellum, formed of multiple units (Hansson and Stensmyr, 2011). Antennae play a fundamental role in reproduction as they are involved in pheromone detection, which occurs in a specific time window of the day and is under circadian control in several species (Groot, 2014; Table 1).
The presence of a peripheral clock in the lepidopteran antenna has been suggested by electrophysiological studies demonstrating in M. sexta and S. littoralis that the electroantennogram (EAG) response after olfactory stimuli shows a daily variation in both LD and DD conditions (Hoballah et al., 2005; Merlin et al., 2007; Fenske et al., 2018). Additionally, cycling mRNA and/or protein expression of different circadian clock factors was detected in the antennae of several lepidopteran species, including B. mori (Bmper; Iwai et al., 2006), M. sexta (Msper; Schuckel et al., 2007), and D. plexippus (Dpper, Dptim, and Dpcry2; Merlin et al., 2009). Importantly, in the monarch the daily oscillation in DpTIM was maintained in explanted antennae, cultured in both LD and DD conditions (Merlin et al., 2009). Moreover, DpTIM abundance was consistently lower during the light phase of the day compared to the night, independently from the phase of the 24h LD cycle, indicating that the D. plexippus antenna possesses an autonomous light-entrainable circadian clock (Merlin et al., 2009; Figure 3B).
In the monarch butterfly, the antennal peripheral clock has been implicated in the circadian compensation of the sun compass, as migratory D. plexippus with an intact brain but without any functional antennae lost the time-compensated feature of the sun compass when analyzed in a flight simulator (Merlin et al., 2009; Guerra et al., 2012). However, butterflies with a single functional antenna showed an operating time-compensated sun compass, indicating that each antennal structure can provide the timing information to the sun compass brain center, in a neuronal circuit that has still to be fully characterized (Guerra et al., 2012).
A peripheral circadian clock controlling the timing of sperm release from the male testis has been suggested by studies performed in C. pomonella (superfamily Tortricoidea; Giebultowicz and Brooks, 1998), as well as in the gypsy moth Lymantria dispar and S. littoralis (superfamily Noctuoidea; Giebultowicz et al., 1997; Syrova et al., 2003). Expression analyses have indicated a rhythmic expression of per mRNA and/or the corresponding protein at a testis level in C. pomonella, B. mori, and S. littoralis (Gvakharia et al., 2000; Iwai et al., 2006; Kotwica et al., 2009). Additionally, in a S. littoralis functional study, an in vitro Spper RNA interference (RNAi) caused a delay in sperm release, indicating that a molecular oscillator involving Spper plays an essential role in regulating rhythmic sperm release in this species (Kotwica et al., 2009).
Several studies aimed to characterize the molecular circadian clock during development in different lepidopteran species. Although in most of the cases, such investigations are at an early stage, compelling evidence indicates that their results are both important for understanding the regulation of the different circadian rhythms occurring in embryonic and larval stages and can provide useful information for the management of both pest and beneficial Lepidoptera, as discussed below.
A circadian clock driving egg-hatching rhythmicity has been shown in several species from mid-embryogenesis (Table 1). Antheraea pernyi transplantation experiments demonstrated that the circadian clock controlling egg-hatching rhythm resides in the brain (Sauman and Reppert, 1998). Moreover, a fundamental role of TIM and PER in this circadian output has been recently confirmed in B. mori Bmtim and Bmper KO mutants (Ikeda et al., 2019; Nartey et al., 2020). In particular, Bmtim KO egg-hatching activity was significantly different compared to that of the wild-type, when monitored every 4h in 12:12 LD and DD conditions, suggesting an egg-hatching rhythm alteration in Bmtim KO individuals (Nartey et al., 2020). Additionally, an accurate analysis was performed on Bmper KO mutants, recording hatching every 30min, in both 12:12 LD and DD regimes (Ikeda et al., 2019). In DD conditions, Bmper KO mutants lost egg-hatching rhythm, indicating a disruption of the circadian clock (Ikeda et al., 2019). In 12:12 LD, Bmper KO animals displayed a main hatching peak 1h after light-on, likely produced by a direct response to light. However, differently from controls, these mutants showed additional hatching activities also at other times of the 24hday (Ikeda et al., 2019). These data were also consistent with the wider “egg-hatching gate” characterizing Bmper knockdown (KD) B. mori lines when compared to controls under LD regimes, a result indicating that a Bmper KD moderately affected egg-hatching rhythms (Sandrelli et al., 2007).
The presence of a maturing circadian clock in the brain of developing embryos was first shown in A. pernyi, in which ApTIM and ApPER proteins were found in the cytoplasm of the PL neurons in 4- and 6-day-old embryos, respectively (Sauman et al., 1996). In these neurons, ApTIM and ApPER signals did not show any cycling variation or nuclear localization; however, during embryonic maturation and in the first instar larvae initially ApTIM and, subsequently, ApPER were also observed in the axon projections of PL neurons (Sauman et al., 1996).
More recently, a cycling expression of several clock genes, including Bmper, Bmtim, Bmcry1, Bmcry2, and BmClk was demonstrated in B. mori embryos under both LD and DD conditions (Xu et al., 2011; Tao et al., 2017; Ikeda et al., 2019; Nartey et al., 2020). Additionally, immunolocalization studies showed BmPER nuclear translocation after a transient light pulse in late-stage embryonic heads, suggesting that a molecular feedback loop including Bmper controls egg-hatching rhythm in B. mori (Tao et al., 2017). Interestingly, in Bmper KO embryos the levels of Bmper mRNAs did not vary during the day and were extremely low when compared to those of wild-type individuals (Ikeda et al., 2019). As the lack of an operating PER/TIM-negative feedback loop would predict constitutively higher Bmper expression levels, Ikeda et al. (2019) hypothesized that a nonsense-mediated mRNA decay (NMD) phenomenon affecting the stability of the aberrant Bmper mRNA was active in Bmper KO mutants. NMD is a protecting mechanism acting as mRNA surveillance, which induces the degradation of mRNAs carrying non-sense mutations in all eukaryotes, including B. mori (Houseley and Tollervey, 2009; Komoto et al., 2009). NMD might also explain the low Bmtim expression levels detected in Bmtim KO embryos in both LD and DD regimes (Nartey et al., 2020).
Besides the brain, other tissues express clock factors during embryonic development. In A. pernyi, for example, non-cycling ApPER staining was detected in the cytoplasm of fat body cells (Sauman et al., 1996). Additionally, in the midgut epithelium of A. pernyi embryos and first instar larvae, ApPER showed a cycling profile in LD and DD and a rhythmic nuclear translocation occurring late in the night in LD conditions (Sauman et al., 1996; Sauman and Reppert, 1998). However, head ligation experiments in first instar larvae disrupted ApPER nuclear translocation demonstrating that the gut peripheral clock depends on the brain master clock (Sauman and Reppert, 1998). Although extensive and standardized studies have not always been performed, several tissues have been shown to express multiple clock factors during larval development in species such as B. mori, S. litura, and S. littoralis. Specifically, last (5th) instar larvae of B. mori actively transcribe the clock genes Bmtim, Bmper, BmClk, and BmBmal1 in the head, midgut, fat body, and silk glands (Markova et al., 2003; Iwai et al., 2006; Xu et al., 2011; Nobata et al., 2012). In the midgut, a significant cycling expression of Bmtim was demonstrated in both LD and DD, while Bmper mRNA levels tended to be higher during the night compared to the day (Nobata et al., 2012). Among the multiple cell types composing the silkworm midgut, BmPER staining was limited to goblet cells, known for their role in the active transport of potassium from hemolymph into the gut lumen and in the release of digestive enzymes (Daimon et al., 2008; Park et al., 2014). Future work employing the recently generated Bmper KO mutants (Ikeda et al., 2019) might functionally clarify the meaning of the goblet cell-restricted BmPER expression in the silkworm midgut.
Transcriptomic and quantitative PCR experiments performed on the head, midgut, and fat body of S. litura larvae at the last (6th) developmental stage showed a daily cycling expression in several clock genes in LD conditions (Slituper, Slitutim, Slitucry1, Slitucry2, SlituCwo, and Slituvri with unimodal expression profiles; SlituBmal1 and SlituClk displaying bimodal expression profiles; Zhang et al., 2021). To date, only S. litura larval heads have been analyzed in DD conditions, showing a rhythmic expression of SlituCwo, SlituBmal1, and SlituClk mRNAs, while Slituper and Slitutim mRNA levels did not display any cycling profile (Zhang et al., 2021). Insect heads are composed of different structures, and a detailed clock gene expression map of the larval brain is still missing. However, these data indicate that a fully mature circadian clock bearing a self-sustained oscillation of per and tim components is absent in S. litura larvae. Nevertheless, it has been suggested that other molecular clock mechanisms, independent from Slituper and/or Slitutim mRNA oscillation(s), can guarantee an endogenous timekeeping system at this developmental stage, since S. litura larvae show circadian rhythmicity in feeding, defecation, and locomotor behaviors from the 5th instar onward, in both LD and DD conditions (Zhang et al., 2021; Table 1). It will be interesting to evaluate whether these behavioral rhythms are in some ways associated with circadian variations occurring at a protein level, including activity, post transcriptional modification/s, and nuclear translocation of the different elements composing the larval molecular clock.
Similar expression studies have been conducted in S. littoralis, a closely related species to S. litura, showing a cycling expression in different circadian genes in both midgut and fat body of last instar larvae, in LD and DD conditions (midgut: Sptim; Spcry1, and Spcry2; fat body: Spper; Sptim, Spcry2, and Spvri; Suszczynska et al., 2017). However, the absence of a Spper mRNA cycling transcription in DD at the midgut level prompted authors to suggest that this organ lacks a canonical molecular circadian clock. On the contrary, in fat bodies Spper and Sptim rhythmic expressions persisted in DD and a rhythmic SpPER protein nuclear translocation occurred in both LD and DD, suggesting that a mature circadian clock is operating in fat body of S. littoralis larvae (Suszczynska et al., 2017).
In nature, adult moths and butterflies are crucial pollinators feeding on nectar, and in some cases, on pollen. However, almost all lepidopteran larvae are voraciously phytophagous, feeding on leaves and, less frequently, on stems, flowers, fruits, seeds, or wood. Complex plant-Lepidoptera interactions have evolved, with plants having developed defense strategies to protect from caterpillar attacks (Howe and Jander, 2008), while also attracting adult lepidopteran pollinators (Raguso, 2008; Rausher, 2008). Conversely, caterpillars have developed mechanisms to avoid plant defense systems (e.g., expansion of detoxification genes; Cheng et al., 2017; Gouin et al., 2017; Pearce et al., 2017) and adult moths and butterflies have evolved structures to efficiently reach their food sources, working at a same time as pollinators (Powell et al., 1998; Mitter et al., 2017; Kawahara et al., 2019; Figure 1). In this survival game, circadian systems of both plants and Lepidoptera appear to have played a fundamental role in controlling the timing of different activities/physiological phenomena, such as the emission (plants)/perception (moths/butterflies) of attraction signals, or the production of toxic substances (plants) and detoxification enzyme (caterpillars) at the proper time of the day.
A series of studies aimed at exploring the relationship between M. sexta moth and the plant Petunia axillaris indicate that an interaction between their respective circadian-controlled phenotypes is ecologically and evolutionary important for an efficient mutualistic relationship in which a plant requiring out-crossing provides nectar as a source of food to adult Lepidoptera which act as pollinators (Hoballah et al., 2005; Broadhead et al., 2017; Fenske et al., 2018; Figure 4A). Although, the M. sexta larval form is a pest for Solanaceae crops, adult moths are generally known as crepuscular/nocturnal nectivores of different plants species, including P. axillaris (Metcalf et al., 1993; Hoballah et al., 2005; Riffell and Hildebrand, 2016). Petunia axillaris flowers show clear rhythmicity in scent emission, with a peak during the night, in a mechanism under circadian control (Hoballah et al., 2005; Fenske and Imaizumi, 2016). Manduca sexta shows circadian rhythmicity in flight behavior, with higher activity during the night in both LD and DD (Broadhead et al., 2017; Fenske et al., 2018). Manipulating the circadian system of both moths and plants, Fenske et al. (2018) demonstrated that the moth and petunia circadian clocks must be in phase to ensure an efficient level of flower visits by the moths (Figure 4A). In fact, when the moth clocks were out of phase compared to that of the plant and vice versa, the number of flower visits was reduced compared to a condition in which the plant and moth circadian systems oscillated in phase. The importance of the circadian clock was emphasized by the fact that moth flight activity increased significantly when exposed to flower scent during the subjective night compared to when the same odor signal was administered during the subjective day. It has been demonstrated that the ability to perceive odors resides in the antenna, which also contains a peripheral clock in M. sexta moths (Schuckel et al., 2007). Additionally, in response to synthetic flower scent, M. sexta antennae show circadian rhythmicity in electrophysiological activity, with a peak during the night when the moths display greater foraging behavior (Hoballah et al., 2005; Fenske et al., 2018). Therefore, it can be hypothesized that in the moth the central and the peripheral clocks interact to coordinate the organismal locomotor and odor perception circadian activities.
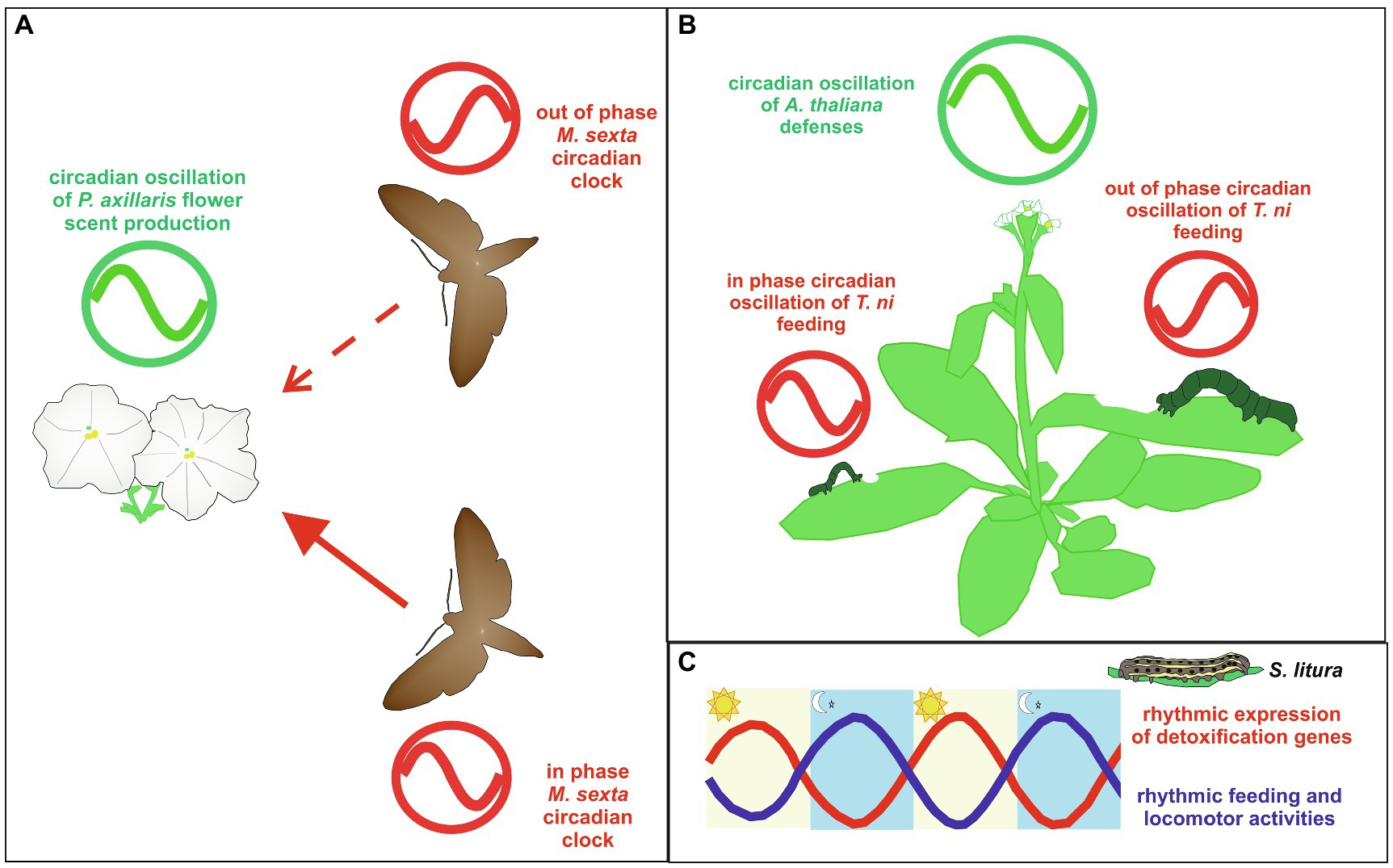
Figure 4. Examples of circadian clock-controlled interactions between Lepidoptera and plants. (A) The interplay between Manduca sexta and Petunia axillaris circadian clocks. Petunia axillaris flower scent production displays a circadian oscillation with a peak during the night (green clock). When M. sexta and P. axillaris clocks are synchronized (in phase), moths show higher levels of flower visits regarding moths with a circadian clock shifted by 12h compared to the plant clock (out of phase; Fenske et al., 2018). Manduca sexta circadian clocks are indicated in red. Solid and dashed red lines represent the higher and lower number of flower visits, respectively. (B) Trichoplusia ni caterpillars feeding on Arabidopsis thaliana. Arabidopsis thaliana jasmonate-mediated defenses show a circadian oscillation, which peaks during the day (green clock). When T. ni and A. thaliana clocks are synchronized (in phase), caterpillars feed less and, in turn, grow less rapidly compared to caterpillars with an internal clock shifted by 12h compared to the plant clock (out of phase; Goodspeed et al., 2012). Trichoplusia ni circadian clocks are indicated in red. (C) Schematic representations of the daily oscillations in locomotor and feeding activities (blue line) and expression of detoxification genes (red line) in Spodoptera litura caterpillars. In light/dark (LD) conditions, midguts and fat bodies show a 24h rhythmic transcription of detoxification genes with a peak during the daytime, when larvae are inactive, do not feed, and show a digestion activity at the highest levels (Zhang et al., 2021; see text for explanation).
In the study of the interactions between plants and Lepidoptera there is also evidence indicating that an in phase synchronization between the two organismal circadian clocks have likely promoted a clock-controlled mechanism enhancing the plants resistance to caterpillar diurnal attacks. One such example derived from a study analyzing the relationship between the circadian clocks of the generalist herbivore T. ni and the model plant Arabidopsis thaliana (Goodspeed et al., 2012; Figure 4B). Trichoplusia ni is commonly known as the cabbage looper since larvae preferentially feed on crucifers but successfully feed on over 160 plant hosts. Arabidopsis thaliana possesses a well-characterized circadian system (Barak et al., 2000; Robertson, 2000), and can host cabbage looper larvae. When fed on A. thaliana, T. ni larvae showed a circadian feeding behavior, with a higher feeding activity during the end of the day and lower during the night, in both LD and DD conditions (Goodspeed et al., 2012; Table 1). However, it was demonstrated that T. ni growth was enhanced when the larval feeding rhythm was entrained to free-run out of phase with respect to A. thaliana circadian rhythmicity, and reduced when the caterpillar free-running rhythm was in phase with that of A. thaliana’s clock (Goodspeed et al., 2012; Figure 4B). The variable larval growth rate was linked to the interaction between the clock-controlled larval feeding behavior and the plant clock-mediated production of jasmonic acid, a hormone triggering plant herbivore defense (Reymond et al., 2000; Jander et al., 2001; Kessler and Baldwin, 2002; Howe and Jander, 2008), which shows a peak of accumulation in the middle of the day (Goodspeed et al., 2012). In fact, when caterpillars were fed on arrhythmic or jasmonate-defective A. thaliana mutants, their weight increment was independent of the phase of circadian feeding behavior (Goodspeed et al., 2012). These data suggest a protective effect of the circadian system in A. thaliana, as the circadian clock controlled phase patterns of jasmonate accumulation are consistent with an anticipated preparation to the peak of the circadian caterpillar feeding behavior, occurring at the end of the day (Goodspeed et al., 2012).
On the other hand, different works have indicated that the lepidopteran counterparts might have evolved circadian-controlled strategies for avoiding plant defense mechanisms. For example, the absence of any rhythmic daily profile in feeding behavior detected in the tobacco hornworm M. sexta when reared on an artificial diet has been suggested as a strategy to limit the negative effects of the diurnal defense mechanisms typical of its natural host Nicotiana attenuata (Reynolds et al., 1986; Kim et al., 2011; Jander, 2012). However, further experiments are required, since in some species, the rhythmicity in larval feeding behavior can be influenced by several environmental factors, including the type of diet. This is the case of the tobacco budworm Heliothis virescens (superfamily Noctuoidea), whose feeding behavior was arrhythmic when maintained on an artificial diet in LD laboratory conditions but rhythmic when reared on a corn-based diet in a greenhouse, with a LD and temperature cycling environment (Niepoth et al., 2018).
A large body of evidence indicating the presence of circadian-controlled mechanisms for escaping plant defense strategies derived from a series of studies performed on nocturnal Spodoptera spp., pest insects with a wide host range, including several plant species of economic importance. In particular, S. exigua, S. littoralis, and S. litura have shown rhythmic feeding behavior, with a peak at night, when maintained on artificial diet in LD conditions (Kim and Hong, 2015; Suszczynska et al., 2017; Zhang et al., 2021). In S. littoralis and S. litura this rhythmicity was shown to persist in DD, suggesting that feeding behavior is a true circadian rhythm in the larvae of both species (Suszczynska et al., 2017; Zhang et al., 2021; Table 1). Spodoptera litura larvae showed a similar circadian profile in their locomotor behavior, with a peak of activity during the night in both LD and DD conditions (Zhang et al., 2021; Table 1; Figure 4C). Interestingly, transcriptomic analyses on S. litura larval midguts and fat bodies in LD conditions showed daily rhythmicity in the transcriptional levels of detoxification genes with a peak during the daytime, when larvae were most inactive and their digestion activity was at its highest levels (Zhang et al., 2021; Figure 4C). Although, the same analyses have not been performed in DD conditions, the importance of the circadian clock in the control of detoxification processes was suggested by transient KD experiments indicating that different core clock elements (SlituClk, SlituBmal1, and SlituCwo) are involved in modulating the transcription of several detoxification genes, with SlituCLK and SlituBMAL1 acting as promoting factors and SlituCWO as an inhibitor (Zhang et al., 2021). A molecular link between the S. litura circadian system and detoxification rhythm was further suggested by in vitro luciferase assays in S. litura cell lines, indicating that intact E-boxes in the promoters of representative detoxification genes are important for the regulation of their expression, with SlituCLK, SlituBMAL1 acting as transcriptional promoters and SlituPER and SlituCWO as inhibitors (Zhang et al., 2021). Therefore, it might be speculated that in S. litura larvae the circadian system exerts a protective effect by maintaining a clear temporal separation between night feeding and daytime digestion/detoxification processes (Zhang et al., 2021).
It is important to note that the S. litura studies open the way for possible applications in agricultural pest control, as Zhang et al. (2021) demonstrated in S. litura larvae that topical treatments with insecticides were more efficient in inducing mortality when applied during nighttime than daytime. These findings are particularly relevant considering that crop management is evolving rapidly towards “precision agriculture” that supports management decisions for profitable and sustainable agricultural production. Automated systems and drones may also permit the distribution of pesticides when they are most effective against their target species. However, as species-specific differences are expected, preliminary studies of the larval circadian system in target pest Lepidoptera will enable specific and effective “chronobiological treatments,” limiting the spread of toxic insecticides in the environment.
To date, the circadian clock has been examined in a relatively small number of species compared to the multitude of extant Lepidoptera. Nevertheless, the studies performed so far indicate the ecological relevance of the lepidopteran circadian system, as it influences the fitness of a single organism and in turn impacts the biodiversity of an ecosystem. It is easy to predict that the technological advances in Next Generation Omic technologies, as well as genome editing methodologies will facilitate the study and manipulation of model and non-model species. When applied to lepidopteran chronobiology, these methodologies will expand our current knowledge on the role of the circadian clocks in these organisms when considered in their natural habitat. Similar approaches will also be useful for the systematic study of species with economic importance such as B. mori, to understand whether manipulating the circadian clock can improve both rearing conditions and/or silk production efficiency. It is worthy to mention that Bmper KD lines exhibited a reduction in developmental time which did not affect silk productivity parameters (Sandrelli et al., 2007). Thus, the employment of genome editing techniques which generated Bmper and Bmtim KO B. mori mutants (Ikeda et al., 2019; Nartey et al., 2020) for producing a battery of silkworm lines carrying mutations in different clock genes might permit an efficient evaluation of these aspects in B. mori.
FS and DB: manuscript organization. FS, SC, DB, and AS: writing and editing. All authors contributed to the article and approved the submitted version.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 765937 (CINCHRON).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allada, R., White, N. E., So, W. V., Hall, J. C., and Rosbash, M. (1998). A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. doi: 10.1016/s0092-8674(00)81440-3
Bae, K., Lee, C., Sidote, D., Chuang, K. Y., and Edery, I. (1998). Circadian regulation of a Drosophila homolog of the mammalian clock gene: per and tim function as positive regulators. Mol. Cell. Biol. 18, 6142–6151. doi: 10.1128/mcb.18.10.6142
Baker, T. C., and Cardé, R. T. (1979). Endogenous and exogenous factors affecting periodicities of female calling and male sex pheromone response in Grapholitha molesta (Busck). J. Insect Physiol. 25, 943–950. doi: 10.1016/0022-1910(79)90107-0
Barak, S., Tobin, E. M., Andronis, C., Sugano, S., and Green, R. M. (2000). All in good time: the Arabidopsis circadian clock. Trends Plant Sci. 5, 517–522. doi: 10.1016/s1360-1385(00)01785-4
Broadhead, G. T., Basu, T., von Arx, M., and Raguso, R. A. (2017). Diel rhythms and sex differences in the locomotor activity of hawkmoths. J. Exp. Biol. 220, 1472–1480. doi: 10.1242/jeb.143966
Castrovillo, P. J., and Cardé, R. T. (1979). Environmental regulation of female calling and male pheromone response periodicities in the codling moth (Laspeyresia pomonella). J. Insect Physiol. 25, 659–667. doi: 10.1016/0022-1910(79)90116-1
Chang, H., Guo, J. L., Fu, X. W., Wang, M. L., Hou, Y. M., and Wu, K. M. (2019). Molecular characterization and expression profiles of cryptochrome genes in a long-distance migrant, agrotis segetum (Lepidoptera: Noctuidae). J. Insect Sci. 19:8. doi: 10.1093/jisesa/iey127
Chang, D. C., McWatters, H. G., Williams, J. A., Gotter, A. L., Levine, J. D., and Reppert, S. M. (2003). Constructing a feedback loop with circadian clock molecules from the silkmoth, Antheraea pernyi. J. Biol. Chem. 278, 38149–38158. doi: 10.1074/jbc.M306937200
Cheng, T., Wu, J., Wu, Y., Chilukuri, R. V., Huang, L., Yamamoto, K., et al. (2017). Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756. doi: 10.1038/s41559-017-0314-4
Curtin, K. D., Huang, Z. J., and Rosbash, M. (1995). Temporally regulated nuclear entry of the drosophila period protein contributes to the circadian clock. Neuron 14, 365–372. doi: 10.1016/0896-6273(95)90292-9
Cyran, S. A., Buchsbaum, A. M., Reddy, K. L., Lin, M. C., Glossop, N. R., Hardin, P. E., et al. (2003). Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112, 329–341. doi: 10.1016/s0092-8674(03)00074-6
Daimon, T., Taguchi, T., Meng, Y., Katsuma, S., Mita, K., and Shimada, T. (2008). β-fructofuranosidase genes of the silkworm, Bombyx mori: insights into enzymatic adaptation of B. mori to toxic alkaloids in mulberry latex. J. Biol. Chem. 283, 15271–15279. doi: 10.1074/jbc.M709350200
Darlington, T. K., Wager-Smith, K., Ceriani, M. F., Staknis, D., Gekakis, N., Steeves, T. D., et al. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603. doi: 10.1126/science.280.5369.1599
Delisle, J., and McNeil, J. N. (1987). Calling behavior and pheromone titer of the true armyworm pseudaletia-unipuncta (Haw; Lepidoptera, Noctuidae) under different temperature and photoperiodic conditions. J. Insect Physiol. 33, 315–324. doi: 10.1016/0022-1910(87)90119-3
Derks, M. F., Smit, S., Salis, L., Schijlen, E., Bossers, A., Mateman, C., et al. (2015). The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol. Evol. 7, 2321–2332. doi: 10.1093/gbe/evv145
Eelderink-Chen, Z., Bosman, J., Sartor, F., Dodd, A. N., Kovács, Á. T., and Merrow, M. (2021). A circadian clock in a nonphotosynthetic prokaryote. Sci. Adv. 7:eabe2086. doi: 10.1126/sciadv.abe2086
Emery, P., So, W. V., Kaneko, M., Hall, J. C., and Rosbash, M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. doi: 10.1016/s0092-8674(00)81637-2
Fenske, M. P., and Imaizumi, T. (2016). Circadian rhythms in floral scent emission. Front. Plant Sci. 7:462. doi: 10.3389/fpls.2016.00462
Fenske, M. P., Nguyen, L. P., Horn, E. K., Riffell, J. A., and Imaizumi, T. (2018). Circadian clocks of both plants and pollinators influence flower seeking behavior of the pollinator hawkmoth Manduca sexta. Sci. Rep. 8:2842. doi: 10.1038/s41598-018-21251-x
Feuda, R., Marlétaz, F., Bentley, M. A., and Holland, P. W. (2016). Conservation, duplication, and divergence of five opsin genes in insect evolution. Genome Biol. Evol. 8, 579–587. doi: 10.1093/gbe/evw015
Franco, D. L., Frenkel, L., and Ceriani, M. F. (2018). The underlying genetics of Drosophila circadian behaviors. Physiology 33, 50–62. doi: 10.1152/physiol.00020.2017
Friedrich, M., Dong, Y., and Jackowska, M. (2006). Insect interordinal relationships: evidence from the visual system. Arthropod. Syst. Phyl. 64, 133–148.
Froy, O., Gotter, A. L., Casselman, A. L., and Reppert, S. M. (2003). Illuminating the circadian clock in monarch butterfly migration. Science 300, 1303–1305. doi: 10.1126/science.1084874
Gegear, R. J., Casselman, A., Waddell, S., and Reppert, S. M. (2008). Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018. doi: 10.1038/nature07183
Gegear, R. J., Foley, L. E., Casselman, A., and Reppert, S. M. (2010). Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463, 804–807. doi: 10.1038/nature08719
Giebultowicz, J. M., and Brooks, N. (1998). The circadian rhythm of sperm release in the codling moth, Cydia pomonella. Entomol. Exp. Appl. 88, 229–234. doi: 10.1046/j.1570-7458.1998.00367.x
Giebultowicz, J. M., Weyda, F., Erbe, E. F., and Wergin, W. P. (1997). Circadian rhythm of sperm release in the gypsy moth, Lymantria dispar: ultrastructural study of transepithelial penetration of sperm bundles. J. Insect Physiol. 43, 1133–1147. doi: 10.1016/s0022-1910(97)00061-9
Glossop, N. R., Lyons, L. C., and Hardin, P. E. (1999). Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768. doi: 10.1126/science.286.5440.766
Goldsmith, M. R., Shimada, T., and Abe, H. (2005). The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 50, 71–100. doi: 10.1146/annurev.ento.50.071803.130456
Goodspeed, D., Chehab, E. W., Min-Venditti, A., Braam, J., and Covington, M. F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U. S. A. 109, 4674–4677. doi: 10.1073/pnas.1116368109
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J. M., Duvic, B., et al. (2017). Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7:11816. doi: 10.1038/s41598-017-10461-4
Groot, A. T. (2014). Circadian rhythms of sexual activities in moths: a review. Front. Ecol. Evol. 2:43. doi: 10.3389/fevo.2014.00043
Guerra, P. A., Gegear, R. J., and Reppert, S. M. (2014). A magnetic compass aids monarch butterfly migration. Nat. Commun. 5:4164. doi: 10.1038/ncomms5164
Guerra, P. A., Merlin, C., Gegear, R. J., and Reppert, S. M. (2012). Discordant timing between antennae disrupts sun compass orientation in migratory monarch butterflies. Nat. Commun. 3:958. doi: 10.1038/ncomms1965
Gvakharia, B. O., Kilgore, J. A., Bebas, P., and Giebultowicz, J. M. (2000). Temporal and spatial expression of the period gene in the reproductive system of the codling moth. J. Biol. Rhythms 15, 4–12. doi: 10.1177/074873040001500102
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Heinze, S., and Reppert, S. M. (2011). Sun compass integration of skylight cues in migratory monarch butterflies. Neuron 69, 345–358. doi: 10.1016/j.neuron.2010.12.025
Heinze, S., and Reppert, S. M. (2012). Anatomical basis of sun compass navigation I: the general layout of the monarch butterfly brain. J. Comp. Neurol. 520, 1599–1628. doi: 10.1002/cne.23054
Hoballah, M. E., Stuurman, J., Turlings, T. C., Guerin, P. M., Connétable, S., and Kuhlemeier, C. (2005). The composition and timing of flower odour emission by wild Petunia axillaris coincide with the antennal perception and nocturnal activity of the pollinator Manduca sexta. Planta 222, 141–150. doi: 10.1007/s00425-005-1506-8
Houseley, J., and Tollervey, D. (2009). The many pathways of RNA degradation. Cell 136, 763–776. doi: 10.1016/j.cell.2009.01.019
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Ikeda, K., Daimon, T., Sezutsu, H., Udaka, H., and Numata, H. (2019). Involvement of the clock gene period in the circadian rhythm of the silkmoth Bombyx mori. J. Biol. Rhythms 34, 283–292. doi: 10.1177/0748730419841185
Ingram, K. K., Kutowoi, A., Wurm, Y., Shoemaker, D., Meier, R., and Bloch, G. (2012). The molecular clockwork of the fire ant Solenopsis invicta. PLoS One 7:e45715. doi: 10.1371/journal.pone.0045715
Ivanchenko, M., Stanewsky, R., and Giebultowicz, J. M. (2001). Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms 16, 205–215. doi: 10.1177/074873040101600303
Iwai, S., Fukui, Y., Fujiwara, Y., and Takeda, M. (2006). Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori. J. Insect Physiol. 52, 625–637. doi: 10.1016/j.jinsphys.2006.03.001
Jander, G. (2012). Timely plant defenses protect against caterpillar herbivory. Proc. Natl. Acad. Sci. U. S. A. 109, 4343–4344. doi: 10.1073/pnas.1201443109
Jander, G., Cui, J., Nhan, B., Pierce, N. E., and Ausubel, F. M. (2001). The TASTY locus on chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni. Plant Physiol. 126, 890–898. doi: 10.1104/pp.126.2.890
Kadener, S., Stoleru, D., McDonald, M., Nawathean, P., and Rosbash, M. (2007). Clockwork orange is a transcriptional repressor and a new drosophila circadian pacemaker component. Genes Dev. 21, 1675–1686. doi: 10.1101/gad.1552607
Kamimura, M., and Tatsuki, S. (1993). Diel rhythms of calling behavior and pheromone production of oriental tobacco budworm moth, Helicoverpa assulta (Lepidoptera, Noctuidae). J. Chem. Ecol. 19, 2953–2963. doi: 10.1007/BF00980595
Kawahara, A. Y., Plotkin, D., Espeland, M., Meusemann, K., Toussaint, E. F. A., Donath, A., et al. (2019). Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. U. S. A. 116, 22657–22663. doi: 10.1073/pnas.1907847116
Kawahara, A. Y., Plotkin, D., Hamilton, C. A., Gough, H., St Laurent, R., Owens, H. L., et al. (2018). Diel behavior in moths and butterflies: a synthesis of data illuminates the evolution of temporal activity. Org. Divers. Evol. 18, 13–27. doi: 10.1007/s13127-017-0350-6
Kawamoto, M., Jouraku, A., Toyoda, A., Yokoi, K., Minakuchi, Y., Katsuma, S., et al. (2019). High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 107, 53–62. doi: 10.1016/j.ibmb.2019.02.002
Kessler, A., and Baldwin, I. T. (2002). Plant responses to insect herbivory: the emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. doi: 10.1146/annurev.arplant.53.100301.135207
Kim, Y., and Hong, Y. (2015). Regulation of hemolymph trehalose level by an insulin-like peptide through diel feeding rhythm of the beet armyworm, Spodoptera exigua. Peptides 68, 91–98. doi: 10.1016/j.peptides.2015.02.003
Kim, S. G., Yon, F., Gaquerel, E., Gulati, J., and Baldwin, I. T. (2011). Tissue specific diurnal rhythms of metabolites and their regulation during herbivore attack in a native tobacco, Nicotiana attenuata. PLoS One 6:e26214. doi: 10.1371/journal.pone.0026214
Kobelková, A., Závodská, R., Sauman, I., Bazalová, O., and Dolezel, D. (2015). Expression of clock genes period and timeless in the central nervous system of the Mediterranean flour moth, Ephestia kuehniella. J. Biol. Rhythms 30, 104–116. doi: 10.1177/0748730414568430
Koh, K., Zheng, X., and Sehgal, A. (2006). JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312, 1809–1812. doi: 10.1126/science.1124951
Komoto, N., Quan, G. X., Sezutsu, H., and Tamura, T. (2009). A single-base deletion in an ABC transporter gene causes white eyes, white eggs and translucent larval skin in the silkworm w-3oe mutant. Insect Biochem. Mol. Biol. 39, 152–156. doi: 10.1016/j.ibmb.2008.10.003
Konopka, R. J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 68, 2112–2116. doi: 10.1073/pnas.68.9.2112
Kotwica, J., Bebas, P., Gvakharia, B. O., and Giebultowicz, J. M. (2009). RNA interference of the period gene affects the rhythm of sperm release in moths. J. Biol. Rhythms 24, 25–34. doi: 10.1177/0748730408329109
Krishnan, B., Levine, J. D., Lynch, M. K., Dowse, H. B., Funes, P., Hall, J. C., et al. (2001). A new role for cryptochrome in a Drosophila circadian oscillator. Nature 411, 313–317. doi: 10.1038/35077094
Labhart, T., and Meyer, E. P. (1999). Detectors for polarized skylight in insects: a survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc. Res. Tech. 47, 368–379. doi: 10.1002/(SICI)1097-0029(19991215)47:6<368::AID-JEMT2>3.0.CO;2-Q
Lee, C., Bae, K., and Edery, I. (1999). PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19, 5316–5325. doi: 10.1128/mcb.19.8.5316
Li, W., Zhang, Z., Lin, L., and Terenius, O. (2017). Antheraea pernyi (Lepidoptera: Saturniidae) and its importance in sericulture, food consumption, and traditional chinese medicine. J. Econ. Entomol. 110, 1404–1411. doi: 10.1093/jee/tox140
Lim, C., Chung, B. Y., Pitman, J. L., McGill, J. J., Pradhan, S., Lee, J., et al. (2007). Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr. Biol. 17, 1082–1089. doi: 10.1016/j.cub.2007.05.039
Lin, F. J., Song, W., Meyer-Bernstein, E., Naidoo, N., and Sehgal, A. (2001). Photic signaling by cryptochrome in the Drosophila circadian system. Mol. Cell Biol. 21, 7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001
Linn, C. E., Campbell, M. G., Poole, K. R., Wu, W. Q., and Roelofs, W. L. (1996). Effects of photoperiod on the circadian timing of pheromone response in male Trichoplusia ni: relationship to the modulatory action of octopamine. J. Insect Physiol. 42, 881–891. doi: 10.1016/0022-1910(96)00034-0
Markert, M. J., Zhang, Y., Enuameh, M. S., Reppert, S. M., Wolfe, S. A., and Merlin, C. (2016). Genomic access to monarch migration using TALEN and CRISPR/Cas9-mediated targeted mutagenesis. G3 6, 905–915. doi: 10.1534/g3.116.027029
Markova, E. P., Ueda, H., Sakamoto, K., Oishi, K., Shimada, T., and Takeda, M. (2003). Cloning of Cyc (Bmal1) homolog in Bombyx mori: structural analysis and tissue specific distributions. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 535–542. doi: 10.1016/s1096-4959(03)00004-6
Martinek, S., Inonog, S., Manoukian, A. S., and Young, M. W. (2001). A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779. doi: 10.1016/s0092-8674(01)00383-x
Merlin, C., Beaver, L. E., Taylor, O. R., Wolfe, S. A., and Reppert, S. M. (2013). Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 23, 159–168. doi: 10.1101/gr.145599.112
Merlin, C., Gegear, R. J., and Reppert, S. M. (2009). Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325, 1700–1704. doi: 10.1126/science.1176221
Merlin, C., Iiams, S. E., and Lugena, A. B. (2020). Monarch butterfly migration moving into the genetic era. Trends Genet. 36, 689–701. doi: 10.1016/j.tig.2020.06.011
Merlin, C., Lucas, P., Rochat, D., François, M. C., Maïbèche-Coisne, M., and Jacquin-Joly, E. (2007). An antennal circadian clock and circadian rhythms in peripheral pheromone reception in the moth Spodoptera littoralis. J. Biol. Rhythms. 22, 502–514. doi: 10.1177/0748730407307737
Merlin, C., and Reppert, S. M. (2009). “Lepidopteran circadian clocks: From molecules to behavior,” in Genetics and Molecular Biology of Lepidoptera. eds. M. R. Goldsmith and F. Marec (Boca Raton, FL: CRC Press), 137–152.
Metcalf, R. L., Metcalf, R. A., and Metcalf, C. L. (1993). Destructive and Useful Insects: Their Habits and Control. 5th Edn. New York, USA: McGraw-Hill.
Minis, D. H., and Pittendrigh, C. S. (1968). Circadian oscillation controlling hatching: its ontogeny during embryogenesis of a moth. Science 159, 534–536. doi: 10.1126/science.159.3814.534
Mitter, C., Davis, D. R., and Cummings, M. P. (2017). Phylogeny and evolution of Lepidoptera. Annu. Rev. Entomol. 62, 265–283. doi: 10.1146/annurev-ento-031616-035125
Mizoguchi, A., and Ishizaki, H. (1982). Prothoracic glands of the saturniid moth Samia cynthia ricini possess a circadian clock controlling gut purge timing. Proc. Natl. Acad. Sci. U. S. A. 79, 2726–2730. doi: 10.1073/pnas.79.8.2726
Mohamed, A. A., Wang, Q., Bembenek, J., Ichihara, N., Hiragaki, S., Suzuki, T., et al. (2014). N-acetyltransferase (nat) is a critical conjunct of photoperiodism between the circadian system and endocrine axis in Antheraea pernyi. PLoS One 9:e92680. doi: 10.1371/journal.pone.0092680
Morris, M. C., and Takeda, S. (1994). The adult eclosion rhythm in Hyphantria cunea (Lepidoptera: Arctiidae): endogenous and exogenous light effects. Biol. Rhythm. Res. 25, 464–476. doi: 10.1080/09291019409360316
Mouritsen, H., and Frost, B. J. (2002). Virtual migration in tethered flying monarch butterflies reveals their orientation mechanisms. Proc. Natl. Acad. Sci. U. S. A. 99, 10162–10166. doi: 10.1073/pnas.152137299
Nartey, M. A., Sun, X., Qin, S., Hou, C. X., and Li, M. W. (2020). CRISPR/Cas9-based knockout reveals that the clock gene timeless is indispensable for regulating circadian behavioral rhythms in Bombyx mori. Insect Sci. 28, 1414–1425. doi: 10.1111/1744-7917.12864
Niepoth, N., Ke, G., de Roode, J. C., and Groot, A. T. (2018). Comparing behavior and clock gene expression between caterpillars, butterflies, and moths. J. Biol. Rhythms 33, 52–64. doi: 10.1177/0748730417746458
Nobata, H., Oishi, K., Takeda, M., and Sakamoto, K. (2012). Photoreception in decapitated larvae of silkworm Bombyx mori. Entomol. Sci. 15, 392–399. doi: 10.1111/j.1479-8298.2012.00526.x
Oliveira, E. G., Srygley, R. B., and Dudley, R. (1998). Do neotropical migrant butterflies navigate using a solar compass? J. Exp. Biol. 201, 3317–3331. doi: 10.1242/jeb.201.24.3317
Palomares, L. A., Joosten, C. E., Hughes, P. R., Granados, R. R., and Shuler, M. L. (2003). Novel insect cell line capable of complex N-glycosylation and sialylation of recombinant proteins. Biotechnol. Prog. 19, 185–192. doi: 10.1021/bp025598o
Paranjpe, D. A., and Sharma, V. K. (2005). Evolution of temporal order in living organisms. J. Circadian Rhythms 3:7. doi: 10.1186/1740-3391-3-7
Park, M. S., Egi, Y., Takeda, M., and Sakamoto, K. (2014). The clock protein PERIOD is expressed in goblet cells of the larval midgut in the silkworm, Bombyx mori. Biol. Rhythm. Res. 45, 369–374. doi: 10.1080/09291016.2013.830506
Patke, A., Young, M. W., and Axelrod, S. (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2, 67–84. doi: 10.1038/s41580-019-0179-2
Pearce, S. L., Clarke, D. F., East, P. D., Elfekih, S., Gordon, K. H. J., Jermiin, L. S., et al. (2017). Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 15:63. doi: 10.1186/s12915-017-0402-6
Perez, S., Taylor, O., and Jander, R. (1997). A sun compass in monarch butterflies. Nature 387:29. doi: 10.1038/387029a0
Pierce, N. E. (1995). Predatory and parasitic Lepidoptera: carnivores living on plants. J. Lepid. Soc. 49, 412–453.
Plautz, J. D., Kaneko, M., Hall, J. C., and Kay, S. A. (1997). Independent photoreceptive circadian clocks throughout Drosophila. Science 278, 1632–1635. doi: 10.1126/science.278.5343.1632
Powell, J. A., Mitter, C., and Farrell, B. D. (1998). “Evolution of larval food preferences in Lepidoptera,” in Evolution, Systematics, and Biogeography Handbook of Zoology. ed. N. P. Kristensen (Berlin: de Gruyter), 403–422.
Price, J. L., Blau, J., Rothenfluh, A., Abodeely, M., Kloss, B., and Young, M. W. (1998). Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. doi: 10.1016/s0092-8674(00)81224-6
Raguso, R. A. (2008). Wake up and smell the roses: the ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 39, 549–569. doi: 10.1146/annurev.ecolsys.38.091206.095601
Rausher, M. D. (2008). Evolutionary transitions in floral color. Int. J. Plant Sci. 169, 7–21. doi: 10.1086/523358
Reppert, S. M. (2007). The ancestral circadian clock of monarch butterflies: role in time-compensated sun compass orientation. Cold Spring Harb. Symp. Quant. Biol. 72, 113–118. doi: 10.1101/sqb.2007.72.056
Reppert, S. M., and de Roode, J. C. (2018). Demystifying monarch butterfly migration. Curr. Biol. 28, R1009–R1022. doi: 10.1016/j.cub.2018.02.067
Reppert, S. M., Guerra, P. A., and Merlin, C. (2016). Neurobiology of monarch butterfly migration. Annu. Rev. Entomol. 61, 25–42. doi: 10.1146/annurev-ento-010814-020855
Reppert, S. M., Zhu, H., and White, R. H. (2004). Polarized light helps monarch butterflies navigate. Curr. Biol. 14, 155–158. doi: 10.1016/j.cub.2003.12.034
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. doi: 10.1105/tpc.12.5.707
Reynolds, S. E., Yeomans, M. R., and Timmins, W. A. (1986). The feeding behaviour of caterpillars (Manduca sexta) on tobacco and on artificial diet. Physiol. Entomol. 11, 39–51. doi: 10.1111/j.1365-3032.1986.tb00389.x
Richier, B., Michard-Vanhee, C., Lamouroux, A., Papin, C., and Rouyer, F. (2008). The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J. Biol. Rhythm. 23, 103–116. doi: 10.1177/0748730407313817
Riffell, J. A., and Hildebrand, J. G. (2016). “Adaptive processing in the insect olfactory system,” in The Ecology of Animal Senses: Matched Filters for Economical Sensing. eds. G. von der Emde and E. Warrant (Cham, Swizterland: Springer International Publishing), 3–24.
Rivas, G. B., Zhou, J., Merlin, C., and Hardin, P. E. (2021). Clockwork orange promotes CLOCK-CYCLE activation via the putative Drosophila ortholog of clock interacting protein circadian. Curr. Biol. 31, 4207.e4–4218.e4. doi: 10.1016/j.cub.2021.07.017
Robertson, M. C. (2000). Circadian rhythms in plants: a millennial view. Physiol. Plant. 109, 359–371. doi: 10.1034/j.1399-3054.2000.100401.x
Roe, A., Weller, S., Baixeras, J., Brown, J. W., Cummings, M. P., Davis, D. R., et al. (2009). “Evolutionary framework for Lepidoptera model systems,” in Genetics and Molecular Biology of Lepidoptera. eds. M. R. Goldsmith and F. Marec (Boca Raton, FL: CRC Press) 1–24.
Rosbash, M. (2009). The implications of multiple circadian clock origins. PLoS Biol. 7:e62. doi: 10.1371/journal.pbio.1000062
Rosén, W. Q., Han, G. B., and Löfstedt, C. (2003). The circadian rhythm of the sex-pheromone-mediated behavioral response in the turnip moth, Agrotis segetum, is not controlled at the peripheral level. J. Biol. Rhythms 18, 402–408. doi: 10.1177/0748730403256869
Rothenfluh, A., Young, M. W., and Saez, L. (2000). A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26, 505–514. doi: 10.1016/s0896-6273(00)81182-4
Rubin, E. B., Shemesh, Y., Cohen, M., Elgavish, S., Robertson, H. M., and Bloch, G. (2006). Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16, 1352–1365. doi: 10.1101/gr.5094806
Rutila, J. E., Suri, V., Le, M., So, W. V., Rosbash, M., and Hall, J. C. (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. doi: 10.1016/s0092-8674(00)81441-5
Sadek, M. M., von Wowern, G., Lofstedt, C., Rosén, W. Q., and Anderson, P. (2012). Modulation of the temporal pattern of calling behavior of female Spodoptera littoralis by exposure to sex pheromone. J. Insect Physiol. 58, 61–66. doi: 10.1016/j.jinsphys.2011.09.016
Saez, L., and Young, M. W. (1996). Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17, 911–920. doi: 10.1016/s0896-6273(00)80222-6
Sakamoto, K., and Shimizu, I. (1994). Photosensitivity in the circadian hatching rhythm of the carotenoid-depleted silkworm, Bombyx mori. J. Biol. Rhythms 9, 61–70. doi: 10.1177/074873049400900106
Sandrelli, F., Cappellozza, S., Benna, C., Saviane, A., Mastella, A., Mazzotta, G. M., et al. (2007). Phenotypic effects induced by knock-down of the period clock gene in Bombyx mori. Genet. Res. 89, 73–84. doi: 10.1017/S0016672307008713
Sandrelli, F., Costa, R., Kyriacou, C. P., and Rosato, E. (2008). Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 17, 447–463. doi: 10.1111/j.1365-2583.2008.00832.x
Sauman, I., Briscoe, A. D., Zhu, H., Shi, D., Froy, O., Stalleicken, J., et al. (2005). Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron 46, 457–467. doi: 10.1016/j.neuron.2005.03.014
Sauman, I., and Reppert, S. M. (1996). Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of period protein regulation. Neuron 17, 889–900. doi: 10.1016/s0896-6273(00)80220-2
Sauman, I., and Reppert, S. M. (1998). Brain control of embryonic circadian rhythms in the silkmoth Antheraea pernyi. Neuron 20, 741–748. doi: 10.1016/s0896-6273(00)81012-0
Sauman, I., Tsai, T., Roca, A. L., and Reppert, S. M. (1996). Period protein is necessary for circadian control of egg hatching behavior in the silkmoth Antheraea pernyi. Neuron 17, 901–909. doi: 10.1016/s0896-6273(00)80221-4
Schuckel, J., Siwicki, K. K., and Stengl, M. (2007). Putative circadian pacemaker cells in the antenna of the hawkmoth Manduca sexta. Cell Tissue Res. 330, 271–278. doi: 10.1007/s00441-007-0471-x
Sehadová, H., Markova, E. P., Sehnal, F., and Takeda, M. (2004). Distribution of circadian clock-related proteins in the cephalic nervous system of the silkworm, Bombyx mori. J. Biol. Rhythms 19, 466–482. doi: 10.1177/0748730404269153
Shimizu, I., and Matsui, K. (1983). Photoreceptions in the eclosion of silkworm, Bombyx mori. Photochem. Photobiol. 37, 409–413. doi: 10.1111/j.1751-1097.1983.tb04493.x
Shorey, H. H., and Gaston, L. K. (1965). Sex pheromones of noctuid moths. V. Circadian rhythm of pheromone-responsiveness in males of Autographa californica, Heliothis virescens, Spodoptera exigua, and Trichoplusia ni (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 58, 597–600. doi: 10.1093/aesa/58.5.597
Silvegren, G., Lofstedt, C., and Rosén, W. Q. (2005). Circadian mating activity and effect of pheromone pre-exposure on pheromone response rhythms in the moth Spodoptera littoralis. J. Insect Physiol. 51, 277–286. doi: 10.1016/j.jinsphys.2004.11.013
Sondhi, Y., Ellis, E. A., Bybee, S. M., Theobald, J. C., and Kawahara, A. Y. (2021). Light environment drives evolution of color vision genes in butterflies and moths. Commun. Biol. 4:177. doi: 10.1038/s42003-021-01688-z
Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., et al. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. doi: 10.1016/s0092-8674(00)81638-4
Stepien, T. L., Zmurchok, C., Hengenius, J. B., Rivera, R. M. C., D’Orsogna, M. R., and Lindsay, A. E. (2020). Moth mating: modeling female pheromone calling and male navigational strategies to optimize reproductive success. Appl. Sci. 10:6543. doi: 10.3390/app10186543
Suszczynska, A., Kaniewska, M. M., Bebas, P., Giebultowicz, J. M., and Kotwica-Rolinska, J. (2017). Circadian regulation of caterpillar feeding and growth. J. Insect Physiol. 101, 113–122. doi: 10.1016/j.jinsphys.2017.07.009
Syrova, Z., Sauman, I., and Giebultowicz, J. M. (2003). Effects of light and temperature on the circadian system controlling sperm release in moth Spodoptera littoralis. Chronobiol. Int. 20, 809–821. doi: 10.1081/cbi-120024217
Takeda, M. (1983). Ontogeny of the circadian system governing ecdysial rhythms in a holometabolous insect, Diatraea grandiosella (Pyralidae). Physiol. Entomol. 8, 321–331. doi: 10.1111/j.1365-3032.1983.tb00364.x
Tao, H., Li, X., Qiu, J. F., Liu, H. J., Zhang, D. Y., Chu, F., et al. (2017). The light cycle controls the hatching rhythm in Bombyx mori via negative feedback loop of the circadian oscillator. Arch. Insect Biochem. Physiol. 96:e21408. doi: 10.1002/arch.21408
Tomioka, K., and Matsumoto, A. (2010). A comparative view of insect circadian clock systems. Cell. Mol. Life Sci. 67, 1397–1406. doi: 10.1007/s00018-009-0232-y
Tomioka, K., Uryu, O., Kamae, Y., Umezaki, Y., and Yoshii, T. (2012). Peripheral circadian rhythms and their regulatory mechanism in insects and some other arthropods: a review. J. Comp. Physiol. B. 182, 729–740. doi: 10.1007/s00360-012-0651-1
Truman, J. W. (1972). Physiology of insect rhythms II. The silkmoth brain as the location of the biological clock controlling eclosion. J. Comp. Physiol. 81, 99–114. doi: 10.1007/BF00693553
Truman, J. W. (1974). Physiology of insect rhythms IV. Role of the brain in the regulation of flight rhythm of the giant silkmoths. J. Comp. Physiol. 95, 281–296. doi: 10.1007/BF00609702
Truman, J. W., and Riddiford, L. M. (1970). Neuroendocrine control of ecdysis in silkmoths. Science 167, 1624–1626. doi: 10.1126/science.167.3925.1624
Turgeon, J., and McNeil, J. (1982). Calling behavior of the army worm, Pseudaletia unipuncta. Entomol. Exp. Appl. 31, 402–408. doi: 10.1111/j.1570-7458.1982.tb03168.x
Walker, T. J. (1980). Migrating Lepidoptera: are butterflies better than moths? Florida Entomol. 63, 79–98. doi: 10.2307/3494658
Wan, G., Hayden, A. N., Iiams, S. E., and Merlin, C. (2021). Cryptochrome 1 mediates light-dependent inclination magnetosensing in monarch butterflies. Nat. Commun. 12:771. doi: 10.1038/s41467-021-21002-z
Williams, C. B. (1957). Insect migration. Annu. Rev. Entomol. 2, 163–180. doi: 10.1146/annurev.en.02.010157.001115
Williams, C. M. (1963). Control of pupal diapause by the direct action of light on the insect brain. Science 140:386. doi: 10.1126/science.140.3565.386
Wise, S., Davis, N. T., Tyndale, E., Noveral, J., Folwell, M. G., Bedian, V., et al. (2002). Neuroanatomical studies of period gene expression in the hawkmoth, Manduca sexta. J. Comp. Neurol. 447, 366–380. doi: 10.1002/cne.10242
Xu, L., Liang, H., Sima, Y. H., and Xu, S. Q. (2011). Timeless is a critical gene in the diapause of silkworm, Bombyx mori. Afr. J. Biotechnol. 10, 16594–16601. doi: 10.5897/AJB10.2522
Yan, S., Liu, Y. J., Zhu, J. L., Cui, W. N., Zhang, X. F., Yang, Y. H., et al. (2019). Daily expression of two circadian clock genes in compound eyes of Helicoverpa armigera: evidence for peripheral tissue circadian timing. Insect Sci. 26, 217–228. doi: 10.1111/1744-7917.12541
Yan, S., Ni, H., Li, H., Zhang, J., Liu, X., and Zhang, Q. (2013). Molecular cloning, characterization, and mRNA expression of two cryptochrome genes in Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 106, 450–462. doi: 10.1603/ec12290
Yan, S., Zhu, J., Zhu, W., Zhang, X., Li, Z., Liu, X., et al. (2014). The expression of three opsin genes from the compound eye of Helicoverpa armigera (Lepidoptera: Noctuidae) is regulated by a circadian clock, light conditions and nutritional status. PLoS One 9:e111683. doi: 10.1371/journal.pone.0111683
Yuan, Q., Metterville, D., Briscoe, A. D., and Reppert, S. M. (2007). Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955. doi: 10.1093/molbev/msm011
Závodská, R., Fexová, S., von Wowern, G., Han, G. B., Dolezel, D., and Sauman, I. (2012). Is the sex communication of two pyralid moths, Plodia interpunctella and Ephestia kuehniella, under circadian clock regulation? J. Biol. Rhythms. 27, 206–216. doi: 10.1177/0748730412440689
Závodská, R., Sauman, I., and Sehnal, F. (2003). Distribution of PER protein, pigment-dispersing hormone, prothoracicotropic hormone, and eclosion hormone in the cephalic nervous system of insects. J. Biol. Rhythms 18, 106–122. doi: 10.1177/0748730403251711
Zhan, S., Merlin, C., Boore, J. L., and Reppert, S. M. (2011). The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185. doi: 10.1016/j.cell.2011.09.052
Zhang, J., Li, S., Li, W., Chen, Z., Guo, H., Liu, J., et al. (2021). Circadian regulation of night feeding and daytime detoxification in a formidable Asian pest Spodoptera litura. Commun. Biol. 4:286. doi: 10.1038/s42003-021-01816-9
Zhang, Y., Markert, M. J., Groves, S. C., Hardin, P. E., and Merlin, C. (2017). Vertebrate-like CRYPTOCHROME 2 from monarch regulates circadian transcription via independent repression of CLOCK and BMAL1 activity. Proc. Natl. Acad. Sci. U. S. A. 114, E7516–E7525. doi: 10.1073/pnas.1702014114
Zhu, H., Sauman, I., Yuan, Q., Casselman, A., Emery-Le, M., Emery, P., et al. (2008). Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6:e4. doi: 10.1371/journal.pbio.0060004
Keywords: Lepidoptera, circadian clock, circadian behaviors, larval circadian clock, pest insects, beneficial insects
Citation: Brady D, Saviane A, Cappellozza S and Sandrelli F (2021) The Circadian Clock in Lepidoptera. Front. Physiol. 12:776826. doi: 10.3389/fphys.2021.776826
Received: 14 September 2021; Accepted: 25 October 2021;
Published: 17 November 2021.
Edited by:
Pamela Menegazzi, University of Wuerzburg, GermanyReviewed by:
Joanna Kotwica-Rolinska, Academy of Sciences of the Czech Republic (ASCR), CzechiaCopyright © 2021 Brady, Saviane, Cappellozza and Sandrelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federica Sandrelli, ZmVkZXJpY2Euc2FuZHJlbGxpQHVuaXBkLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.