
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 21 October 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.753914
This article is part of the Research TopicPhysiological Response to Environmental Stressors in InvertebratesView all 5 articles
 Jian-hao Ding1
Jian-hao Ding1 Lu-xin Zheng1
Lu-xin Zheng1 Jie Chu1
Jie Chu1 Xin-hao Liang1
Xin-hao Liang1 Jun Wang1,2
Jun Wang1,2 Xiao-wen Gao3
Xiao-wen Gao3 Fu-an Wu1,2*
Fu-an Wu1,2* Sheng Sheng1,2*
Sheng Sheng1,2*Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) is a destructive mulberry pest, causing great damage to mulberry in China. Heat shock proteins (Hsps) are involved in various signal pathways and regulate lots of physiological processes in insects. The function of Hsps in G. pyloalis, however, has still received less attention. Here, we identified five Hsp genes from G. pyloalis transcriptome dataset including two Hsp70 family genes (GpHsp71.3 and GpHsp74.9) and three Hsp90 family genes (GpHsp82.4, GpHsp89, and GpHsp93.4). Quantitative Real-time PCR validation revealed that all Hsps of G. pyloalis have significant expression in pupal and diapause stage, at which the larvae arrest the development. Expressions of GpHsp71.3 and GpHsp82.4 were increased significantly after thermal treatment at 40°C, and this upregulation depended on heat treatment duration. Furthermore, silencing GpHsp82.4 by RNA interference led to a significant increase in mortality of G. pyloalis larvae under the heat stress compared to the control group. After starvation stress, the expression levels of GpHsp82.4 and GpHsp93.4 were significantly increased. At last, after being parasitized by the parasitoid wasp Aulacocentrum confusum, Hsp70 and Hsp90 genes of G. pyloalis were decreased significantly in the early stage of parasitization and this moderation was affected by time post-parasitization. This study highlights the function of G. pyloalis Hsps in response to environmental stress and provides a perspective for the control of this pest.
Heat shock protein (Hsp) is a highly conservative protein induced by extreme temperature, heavy metals, hunger, and other stresses in organisms, which can help the protein fold correctly, degrade the denatured protein, and exercise the function of molecular chaperone in the biological resistance to adverse environments (Tammariello et al., 1999). In insects, four major families of Hsps, along with several co-chaperones, are recognised, including the small heat shock proteins (sHsps), Hsp60, Hsp70, and Hsp90 (Kim et al., 1998). Hsp70 (72–80 kDa) and Hsp90 (81–99 kDa) are the two most widely studied Hsp families in insects (Feng et al., 2010). Due to the development of genomics, transcriptomics, and other sequencing technologies, an increasing number of Hsps have been reported in insects. For examples, Five CsHsp70 and CsHsp90 genes in Chilo suppressalis were identified (Lu et al., 2014), as well as two PxHsp90 and eight PxHsp70 genes in the Plutella xylostella genome (Xia et al., 2013). Hsp70 and Hsp90 function as molecular chaperones in response to a variety of stress factors, promote proper protein folding, and prevent the aggregation of denatured protein (Feder and Hofmann, 1999). Hsps have been found to be involved in many developmental processes, including insect diapause, metamorphosis, embryogenesis, apoptosis, and autophagy (Chu et al., 2020). Hsp synthesis in insects is differently regulated not only at series of developmental stages but also during the diapause process (Sasibhushan et al., 2013). The expression levels of Hsp70 were greatly increased in diapausing pupae of the blueberry maggot, Rhagoletis mendax (Teixeira and Polavarapu, 2005). In Omphisa fuscidentalis, OfHsp90 expression increased transiently during the termination of diapause (Tungjitwitayakul et al., 2008).
An important determinant of insect abundance and distribution is temperature, with extremes of heat and cold eliciting adaptive induction of Hsp gene expression required for survival (King and MacRae, 2015). When insects are threatened by extreme temperatures and other stresses, the synthesis of most proteins decreases but the synthesis of highly sensitive proteins increases, binding abnormal proteins and helping them to refold (Kim et al., 1998). Temperature is one of the important environmental factors. The heat resistance of insects is closely related to Hsp synthesis. Hsps in organisms can help to obtain the adaptability of high-temperature environments, improving thermal tolerance (Wang et al., 2018). Studies demonstrate that the regulation of Hsp genes varies with the intensity of thermal stress and the insect’s physiological state. P. xylostella showed a significant increase in the expression of PxHsp90 after heat stress (37°C) (Sonoda et al., 2006). While in Delia antiqua, low amounts of Hsp70 mRNA are exhibited, which builds up on exposure to heat and cold, followed by a gradual decline if cold persists (Chen et al., 2006). High and low temperatures boost Hsp90 mRNA in diapausing pupae of D. antiqua (Chen et al., 2005). In Ectomyelois ceratoniae, the mortality rates significantly increased in dsHsp90 injection larvae after thermal treatment, suggesting that the HSP90 plays an important role in conferring thermotolerance (Farahani et al., 2020). According to Rinehart et al. (2007), the deletion of Hsp70 mRNA reduces heat hardiness and cold hardiness in Sarcophaga crassipalpis, showing the role of Hsp in heat stress tolerance. In addition, Silence of PaHsp70 mRNA hinders heat shock recovery and repair of chilling injuries in adult Pyrrhocoris apterus (Koštál and Tollarová-Borovanská, 2009). Intermittent food shortages are often encountered by insects. During winter or under starvation stress, insects enter diapause stage as eggs, mature larvae, pupae, or adults (Zhang et al., 2019). In face of starvation, insect responses include changes in behavior and/or maintenance of low metabolic rates through physiological adaptation or regulation. Hsp are involved in these processes that maintain lower levels of physiological metabolic activity (King and MacRae, 2015). Silencing Hsp70 resulted in lower survivorship after food deprivation in Rhodnius prolixus (Paim et al., 2016). Starvation of 24 h can induce the expression of genes encoding Hsp90 in the endoparasitoid wasp, Pteromalus puparum (Wang et al., 2012).
Parasitoid wasps can be the biotic stressor for their host insects and can manipulate the development of the host in order to create a safe environment for their offspring (Rinehart et al., 2002). It has been suggested that there is a functional connection between host response and heat shock regulatory systems (Tutar and Tutar, 2010). For example, upregulation of Hsps in the flesh fly, S. crassipalpis in response to venom injected by the ectoparasitoid wasp Nasonia vitripennis (Rinehart et al., 2002). Hsp75 and Hsp90 of Pieris rapae were downregulated at 12 h post-parasitization and the minimum level was reached at 48 h during the time interval observed in response to parasitization by P. puparum (Zhu et al., 2013). Despite this, the function of Hsp in the response of host insects to parasitization stress still needs more exploration and verification.
The mulberry pyralid caterpillar, Glyphodes pyloalis, is a major pest of mulberry, causing serious losses to the production of mulberry leaves in China and threatens the development of the sericultural industry every year. G. pyloalis occurred four to seven generations in the mulberry-producing areas of eastern and southern China every year. It mainly outbreaks in summer or early autumn when the temperature gets as high as 42°C and it has strong adaptability to this thermal stress (Chu et al., 2020). The high temperature can alter the expression of many genes in G. pyloalis (Liu et al., 2017). G. pyloalis larvae can enter diapause stage when lacking of food in environment (Chen et al., 2015). Previous studies showed that sHsp are involved in response to high temperature stress in G. pyloalis (Chu et al., 2020), however, whether other Hsp families are involved in the response to these genes at high temperatures remains unknown. Meanwhile, it is urgent to elucidate the roles of Hsps when G. pyloalis encounter thermal stress as well as other environmental stresses, such as food deprivation and parasitization by their natural enemies.
In this study, we characterized Hsp70 and Hsp90 genes of G. pyloalis from transcriptome database and described their structural properties. The expressions of GpHsps in response to thermal and starvation stress were analyzed by RT-qPCR The impact of Aulacocentrum confusum parasitization on GpHsps expression was also examined. In addition, RNAi was used to study the functions of temperature-induced related Hsp genes. Thus, these results will further illustrate the molecular mechanism of insect Hsps in response to abiotic and biotic environmental stress. This study may also provide new targets and approaches for the control of G. pyloalis.
Glyphodes pyloalis larvae were collected from the leaves of a mulberry field at Sericulture Research Institute of the Chinese Academy of Agricultural Sciences in Zhen Jiang, China (32.20°N, 119.45°E). Collected larvae of G. pyloalis were reared in laboratory at 26 ± 1°C and 50 ± 5% relative humidity with a 16:8 h light/dark photoperiod and supplied with fresh mulberry leaves in boxes (15 cm × 10 cm × 8 cm) till pupation. After emergence, the male and female adults of G. pyloalis were transferred into a transparent glass jar (30 cm × 20 cm × 10 cm) for mating and spawning. A. confusum was obtained from rearing parasitized G. pyloalis larvae in the mulberry field. A. confusum were maintained using the larvae of G. pyloalis as the hosts. Adult wasps were kept in glass tube and fed with a 10% (w/v) honey solution.
The Hsp70 and Hsp90 genes were identified from the previously constructed transcriptome dataset (ACC.SRR9959773, Sheng et al., 2021). ORFfinder1 was used to predict the open reading frames (ORFs) of GpHsps and CLUSTALX analysis was conducted to obtain aligned amino acid sequences. ExPASy2 was used to predict the theoretical isoelectric point (pI) and molecular weight of each enzyme. Conserved motifs and domains were predicted using PFAM3 and SMART.4 The phylogenetic tree of Hsp70 and Hsp90 in G. pyloalis Walker and other insects were generated using neighbor-joining for 1000 times bootstrap replicates by MEGA 7 software.
The sample design for the initial experiment has been described previously. Two-day old 1st instar, 3rd instar, and 5th instar larvae, pupae, and male and female adults were collected and stored at −80°C until RNA extraction. The fifth instar larvae were collected and reared at 16°C for 7–10 days. Once the larvae molted into the pre-pupal stage to enter into the diapause, they were collected and kept for half a month without pupation to obtain diapause larvae. In the thermal treatment, 2-day old 5th instar larvae were selected for heat and cold shock, and three temperature levels (40, 25, and 0°C) were imposed on the larvae. To evaluate the effect of exposure time on gene expression, two levels of exposure duration (1 and 4 h) were examined for their short- and long-term effects. In the starvation treatment, 2-day old 4th instar G. pyloalis larvae were deprived for food for various times (0, 2, 4, and 6 days) to obtain the cohorts of starvation larvae. Control groups were provided with mulberry leaves. After starvation treatment, the surviving larvae were frozen and stored at −80°C as described above. Each treatment included at least three surviving larvae. The parasitization experiment was conducted following the methods described by Zheng et al. (2017). The parasitic conditions were as follows: twenty 4th larvae of G. pyloalis were placed into a transparent circular plastic box (90 × 90 × 100 cm) with a single mulberry leaf at the bottom, under which the mulberry leaves were moisturized with agar layer. One hour later, a mated female parasitoid was introduced into the box. After parasitism for 6 h, the female wasps were removed, and the larvae were collected separately and fed in plastic cups (5 × 5 × 2.5 cm). After the parasitoid cocoons formed, the parasitoids were taken out and placed in glass tubes separately till the adults emerged. These larvae at 0, 2nd, 4th, or 6th day after parasitization were collected and stored at −80°C until RNA extraction.
Total RNA samples were extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacture’s protocol, and the residual DNA was removed by RNase-Free DNase I (Promega, Madison, WI, United States). The integrity of total RNA was detected by 1% agarose gel electrophoresis, and the concentration and purity of total RNA from G. pyloalis were determined by Nanodrop 2000 spectrometer (Thermo Scientific, Waltham, MA, United States). All RNA samples were reverse transcribed using the PrimeScript RT reagent kit with gDNA Eraser (Takara Biotechnology Co. Ltd., Dalian, China). The qualified cDNA was stored at −20°C for later use.
Real-Time Quantitative PCR (RT-qPCR) was performed using QuantStudioTM Real-Time PCR system (Applied Biosystems, Foster, CA, United States). Each reaction was conducted in a 20 μL volume that consisted of the following: 10 μL SYBR Premix Ex Tap II (Takara Biotechnology Co. Ltd., Dalian, China), 0.4 μL each of the forward and reverse primer, 0.4 μL ROX Reference Dye (50×), 1.5 μL cDNA template, and 7.3 μL ddH2O. The beta-actin gene (MK450335.1) was used as the reference gene to normalize the expression of target genes and correct variation. Following RT-qPCR, the reaction process was as follows: 5 min at 95°C, followed by 45 cycles of 15 s at 95°C, 60°C for 31 s, and dissociation. A fluorescence melting curve from 55 to 95°C was used to ensure a single gene-specific peak and the absence of primer-dimer peaks. Each PCR reaction was replicated three times. The primers used in RT-qPCR analysis were designed by Primer-BLAST5 and are listed in Supplementary Table 1. The relative expression levels of GpHsps among the different samples were measured using 2–ΔΔCt method.
Primers (Supplementary Table 2) of dsRNA of GpHsp71.3 and GpHsp82.4 were designed with BLOCK-iTM RNAi Designer.6 Green fluorescent protein (GFP) was selected as a negative treatment. According to the manufacturer’s instructions, the dsRNA was synthesized using an in vitro Transcription T7 Kit (for siRNA Synthesis) (Takara Biotechnology Co. Ltd., Dalian, China). The integrity of the dsRNA was determined by 1% agarose gel electrophoresis. The quality and concentration of purified dsRNA were measured using NanoDrop 2000 spectrophotometer. To determine the RNAi efficiency, dsGpHsp71.3 and dsGpHsp82.4 were, respectively, dissolved to a final concentration of 1.0 μg/μL and 5th instar 2-day old larvae were selected for treatment and injection of 0.5, 1, 1.5, and 2 μL of dsGFP/dsRNA, respectively, using a Nanoject II microinjector (Drummond Scientific, Broomall, PA, United States). Triple biological replicates were done for each group. After 24 h of injection, both dsGFP and dsHsp larvae were collected and stored at −80°C as described above for RT-qPCR. RT-qPCR was used to check the efficiency of RNAi. Mortality rates of larvae of G. Pyloalis after RNAi of GFP or GpHsp were determined under heat shock treatment. After 24 h of injection, thirty 2-day old 5th instar larvae of G. pyloalis were exposed to 40°C for 1 h for larvae of dsGFP/dsGpHsp82.4 and 4 h for larvae of dsGFP/dsGpHsp71.3 to determine the effect of heat temperatures on survival, respectively. Mortality of larvae after heat shock treatment was recorded for 48 h separately.
A one-way analysis of variance (ANOVA) (Systat, Inc., Evanston, IL, United States) with Tukey’s post hoc test (p < 0.05) was used to check for significant differences in the expression levels of each target gene among the treatments. A two-way ANOVA is used to estimate significant differences in the expression levels affected by parasitization and durations post-parasitization. Chi-square teat was used to analyses mortality in bioassay. The data analysis was carried out in R 4.0.4 software (R Core Team, 2013).
Two Hsp70 genes (GpHsp71.3 and GpHsp74.9) and three Hsp90 genes (GpHsp82.4, GpHsp89, and GpHsp93.4) were identified, and the GenBank accession numbers were MK225533–MK225537. All GpHsp genes contained full-length Open Reading Frame, encoding 652–837 amino acid residues with predicted isoelectric point (pI) of 4.97–5.96 and molecular weight ranges from 71.3 to 93.4 kDa (Table 1). Alignment of the deduced amino acid sequences of ten Hsp70 in G. pyloalis and other lepidopterous insects showed that GpHsp70s have great similarities with the Hsp70 family of other lepidopteran insects (Figure 1). Both GpHsp70 have three signature motifs of the Hsp70 family: IDLGTTYS (amino acids 64–71), IFDLGGGTFDVSLL (amino acids 253–266), and IVLVGGSTRIPKVQQ (amino acids 395–409). Besides, they have a unique ATP/GTP binding site (AEAYLGKN) at amino acids 187–194 and a conservative amino acid sequence of EEVD motif at C-terminus. Multiple sequence alignments of Hsp90 showed a high similarity between species as shown in Figure 2. GpHsp90 has five signature sequences of the Hsp90 protein family: YSNKEIFLRELISNSSDALDKIR (residues 98–120), LGTIAKSGT (residues 167–175 residues), IGQFGVGFYSAYLVAD (residues 197–212), IKLYVRRVFI (residues 430–439), and GVVDSEDLPLNISRE (residues 456–470) and possess a conserved sequence MEEVD motif (residues 852–856) at the C-terminus. GpHsps displayed high amino acid identity with the Hsps of Mythimna separata (88%), Ostrinia furnacalis (88%), M. separata (87%), Spodoptera exigua (81%), and Amyelois transitella (79%) (Table 1).
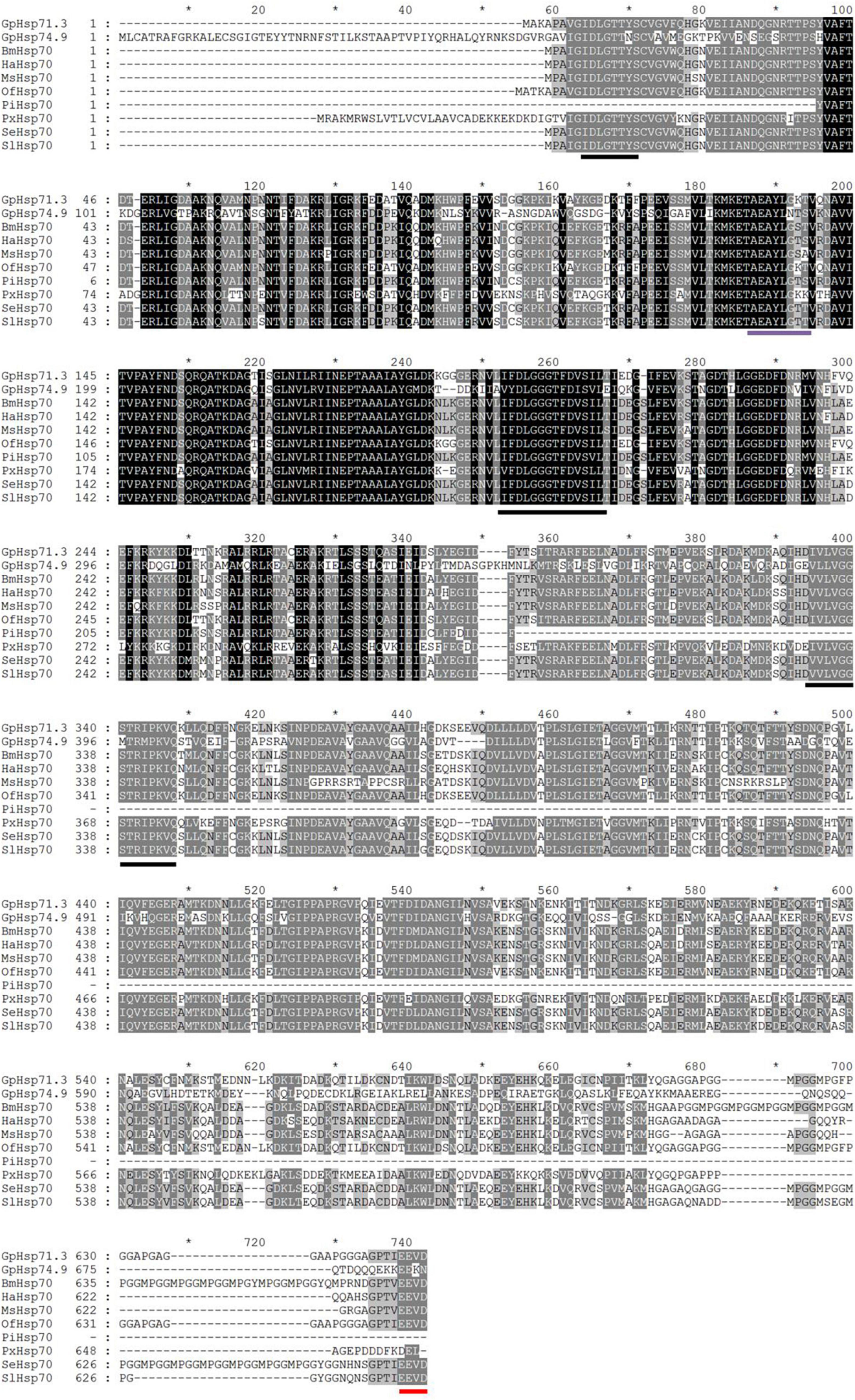
Figure 1. Alignment of the deduced amino acid sequences of Hsp70 genes in G. pyloalis and other Lepidoptera insect. The conserved domains of Hsp70 gene family is underlined in Black solid line. Red solid line indicates a conserved amino acid sequence of EEVD motif at C-terminus and purple solid line indicates a ATP/GTP binding site motif. Bombyx mori (Bm), Helicoverpa armigera (Ha), Manduca sexta (Ms), Ostrinia furnacalis (Of), Plodia interpunctella (Pi), Plutella xylostella (Px), Spodoptera exigua (Se), Spodoptera litura (Sl).

Figure 2. Alignment of the deduced amino acid sequences of Hsp90 genes in G. pyloalis and other Lepidoptera insect. The conserved domains of Hsp90 gene family is underlined in Black solid line. Blue solid line indicates a conserved amino sequence of MEEVD motif at C-terminus. Amyelois transitella (At), Mythimna separata (Ms), Trichoplusia ni (Tn), Bombyx mori (Bm), Spodoptera litura (Sl), Spodoptera exigua (Se), Grapholita molesta (Gm), Pieris melete (Pm), Pieris canidia (Pc), Pieris rapae (Pr).
Results of the phylogenetic tree showed that GpHsp71.3 and GpHsp74.9 were clustered into separate clusters and were adjacent with OfHsp71.4 and HaHsp74.9, respectively (Figure 3). Among three GpHsp90 (GpHsp82.4, GpHsp89, and GpHsp93.4), GpHsp82.4, and OfHsp90 were grouped into one cluster, while G. pylHsp89 and GpHsp93.4 was adjacent with GmHsp97 (Figure 4).
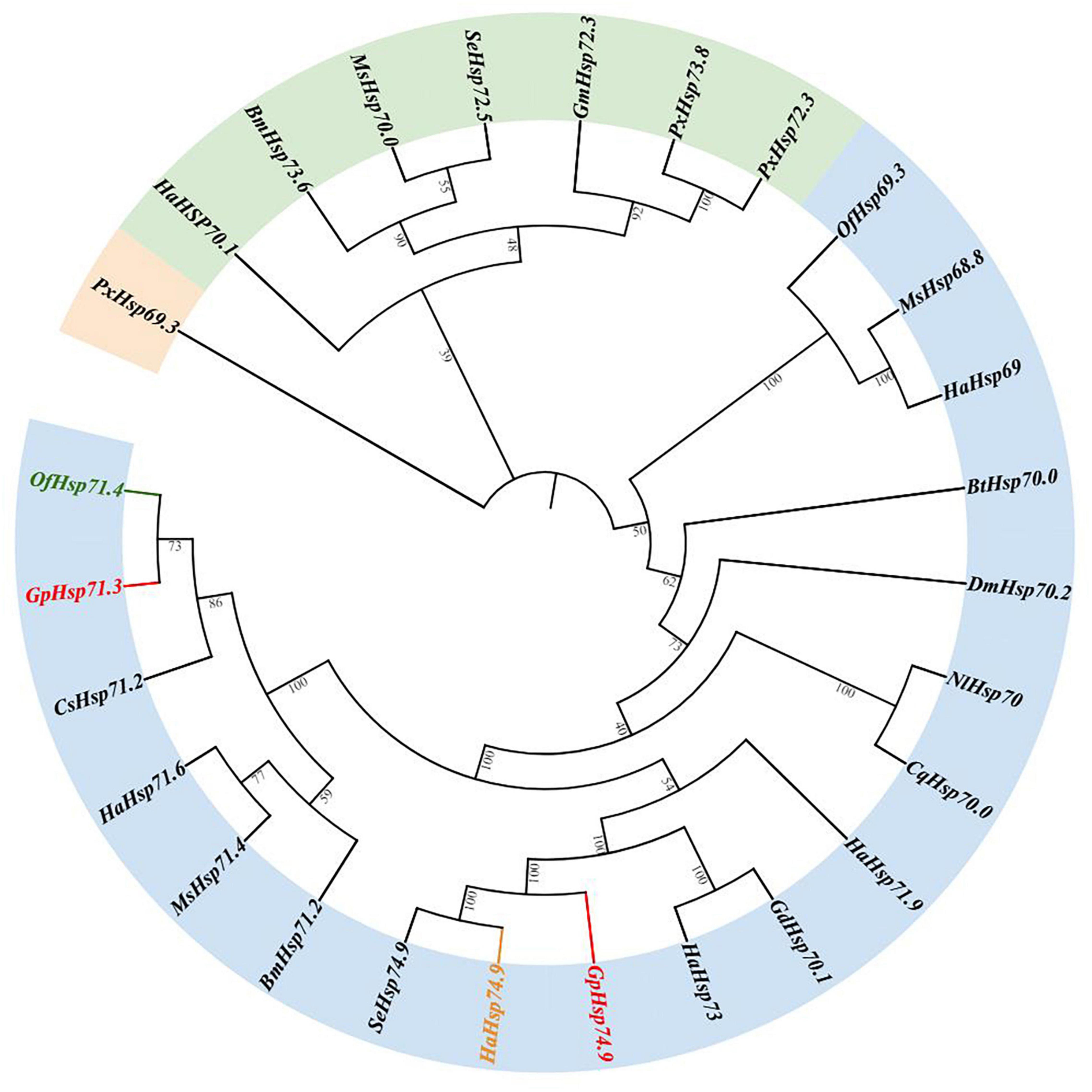
Figure 3. The neighbor-joining tree of GpHsp70s deduced from the coding sequence (CDS) between G. pyloalis and other insect species. GpHsp71.3 and GpHsp74.9 in red, OfHsp71.4 in green, and HaHsp74.9 in orange. Bombyx mori (Bm), Helicoverpa armigera (Ha), Manduca sexta (Ms), Ostrinia furnacalis (Of), Plutella xylostella (Px), Spodoptera exigua (Se), Bemisia tabaci (Bt), Nilaparvata lugens (Nl), Harmonia axyridis (Ha), Galeruca daurica (Gd), Drosophila melanogaster (Dm), Culex quinquefasciatus (Cq), Chilo suppressalis (Cs), Grapholita molesta (Gm).
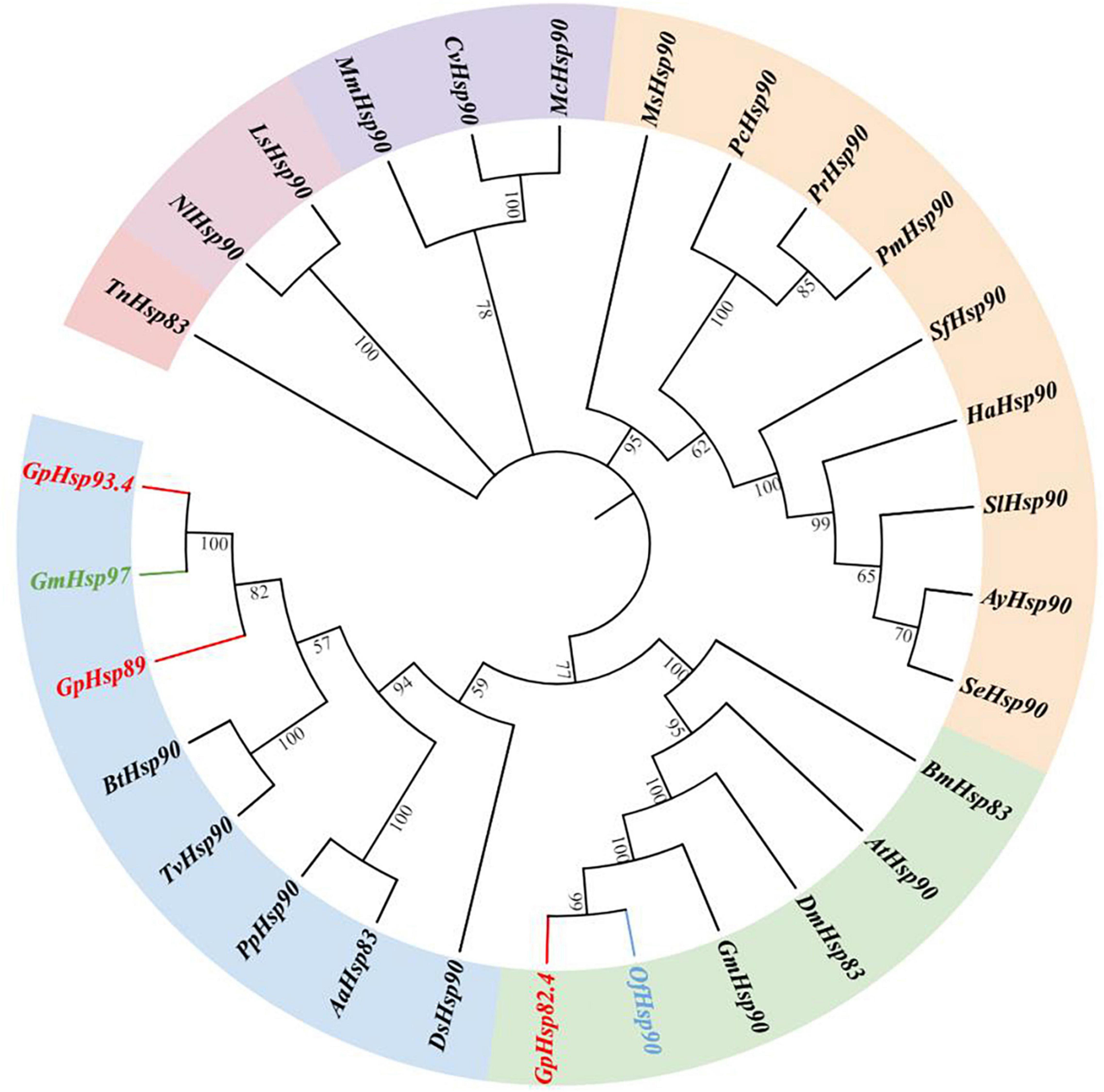
Figure 4. The neighbor-joining tree of GpHsp90s deduced from the coding sequence (CDS) between G. pyloalis and other insect species. GpHsp82.4, GpHsp89, and GpHsp93.4 in red. OfHsp90 in blue and GmHsp97 in green. Trichoplusia ni (Tn), Amyelois transitella (At), Helicoverpa armigera (Ha), Ostrinia furnacalis (Of), Microplitis mediator (Mm), Spodoptera litura (Sl), Plutella xylostella (Px), Spodoptera exigua (Se), Antheraea yamamai (Ay), Pieris canidia (Pc), Spodoptera frugiperda (Sf), Pieris rapae (Pr), Dendrolimus superans (Ds), Manduca sexta (Ms), Bombyx mori (Bm), Grapholita molesta (Gm), Pieris melete (Pm), Aedes aegypti (Aa), Laodelphax striatellus (Ls), Bemisia tabaci (Bt), Trialeurodes vaporariorum (Tv), Cotesia vestalis (Cv), Pteromalus puparum (Pp), Nilaparvata lugens (Nl), Macrocentrus cingulum (Mc), Harmonia axyridis (Ha), Drosophila melanogaster (Dm).
The results showed significant differences in the expression levels of GpHsps at different developmental stages. GpHsp71.3 was highly expressed in diapause larvae and pupae, and the expression of GpHsp74.9 was the highest at pupal stage. GpHsp82.4 and GpHsp89 were highly transcribed in pre-pupae and pupae. The expression levels of GpHsp93.4 were exhibited more significantly in pre-pupae, pupae, and diapause stages than the other developmental stages (Figure 5).
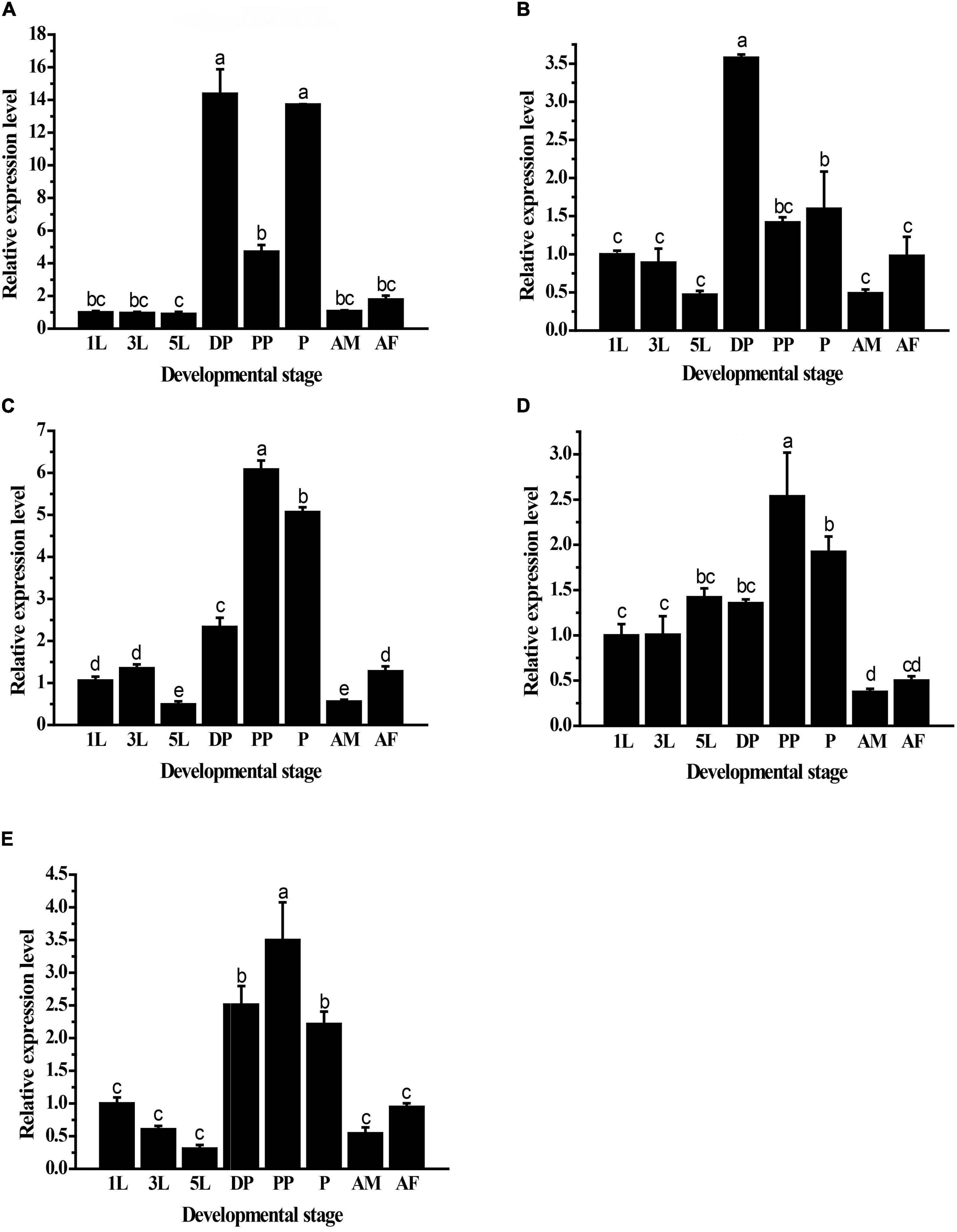
Figure 5. Relative mRNA expression levels of five GpHsps in different developmental stages. (A) GpHsp71.3; (B) GpHsp74.9; (C) GpHsp82.4; (D) GpHsp89; (E) GpHsp93.4. 1st instar larvae (1L), 3rd instar larvae (3L), 5th instar larvae (5L), diapause larvae (DP), prepupae (PP), pupa (P), male adults (AM), and female adults (AF) of G. pyloalis. The relative expression levels of the GpHsps were determined by comparison with the first instar larvae. Data are shown as the mean ± SE of three biological replicates. Differences among triple repeats were analyzed using one-way analysis of variance (Systat, Inc., Evanston, IL) with Tukey’s post hoc test. The different letters above the bars indicate significant differences (p < 0.05).
The expression patterns of GpHsp70s and GpHsp90s after exposure to cold (0°C) and heat (40°C) stress were analyzed. The expression of GpHsp74.9, GpHsp82.4, and G.pHsp93.4 were upregulated significantly after 1 h of high temperature stress at 40°C. GpHsp71.3 and GpHsp89 expression increased after 4 h at 40°C. The expressions of Hsp genes did not show significance under cold stress treatment, except for GpHsp74.9, which were upregulated compared to the control group after 4 h of treatment. These results indicate that GpHsps respond to temperature stress and are especially responsive to extreme temperatures but more susceptible to heat temperature induction (Figure 6).
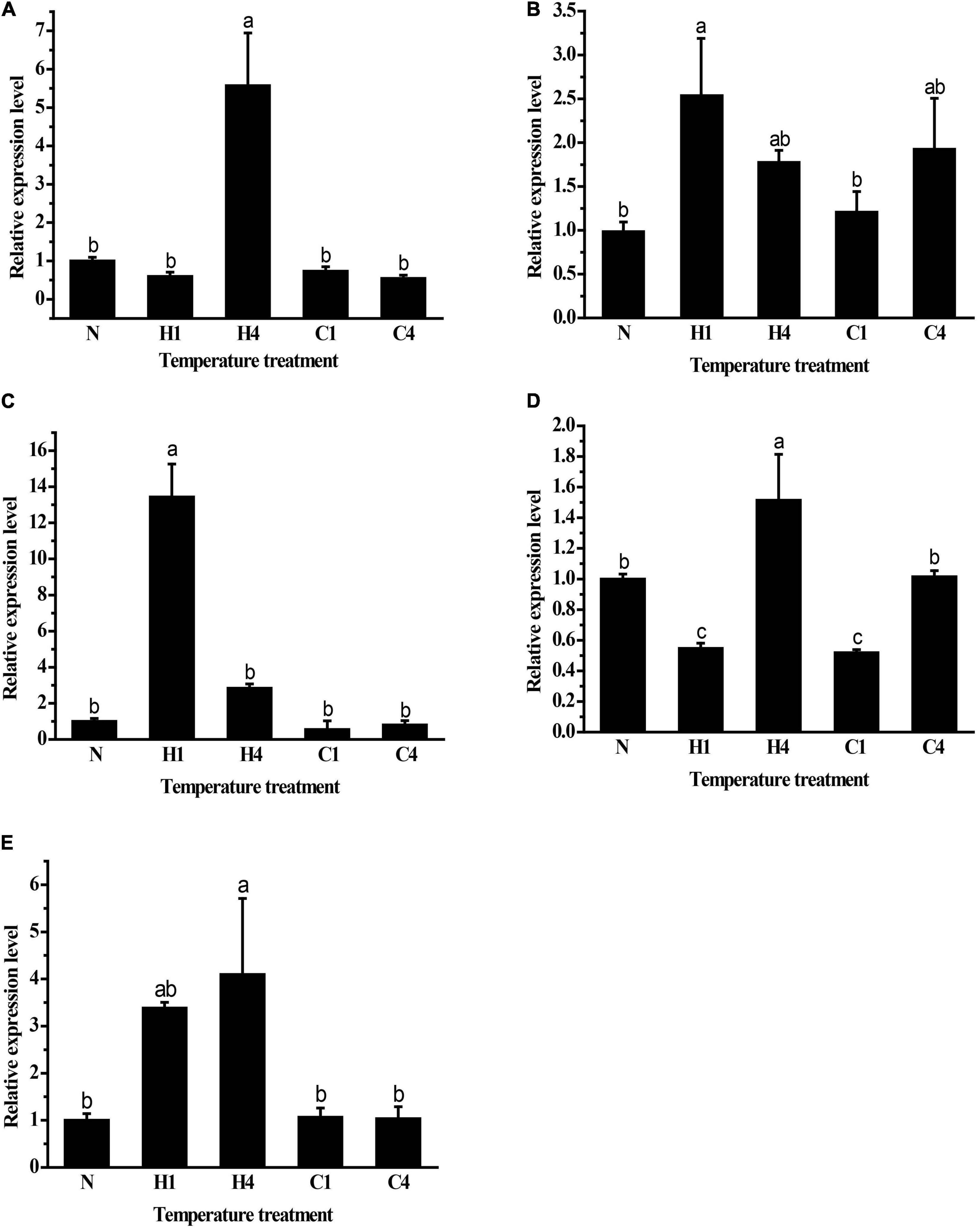
Figure 6. Relative mRNA expression levels of GpHsps at extreme cold and heat temperature stress. (A) GpHsp71.3; (B) GpHsp74.9; (C) GpHsp82.4; (D) GpHsp89; (E) GpHsp93.4.N: normal temperature control (25°C); H1: heat shock (40°C) for 1 h, H4: heat shock (40°C) for 4 h, C1: cold shock (0°C) for 1 h, C4: cold shock (0°C) for 4 h. Data are shown as the mean ± SE of three biological replicates. Differences among triple repeats were analyzed using one-way analysis of variance (Systat, Inc., Evanston, IL) with Tukey’s post hoc test. Different lowercase letters indicate significant differences (p < 0.05).
Both GpHsp82.4 and GpHsp93.4 showed significantly upregulated expression after 6 days of food deprivation, and the expression levels of GpHsp74.9 were downregulated at second day and fourth day after food deprivation while GpHsp71.3 is unaffected by starvation treatment. In contrast, the expression of GpHsp89 showed an opposite pattern with the increase in time (Figure 7).
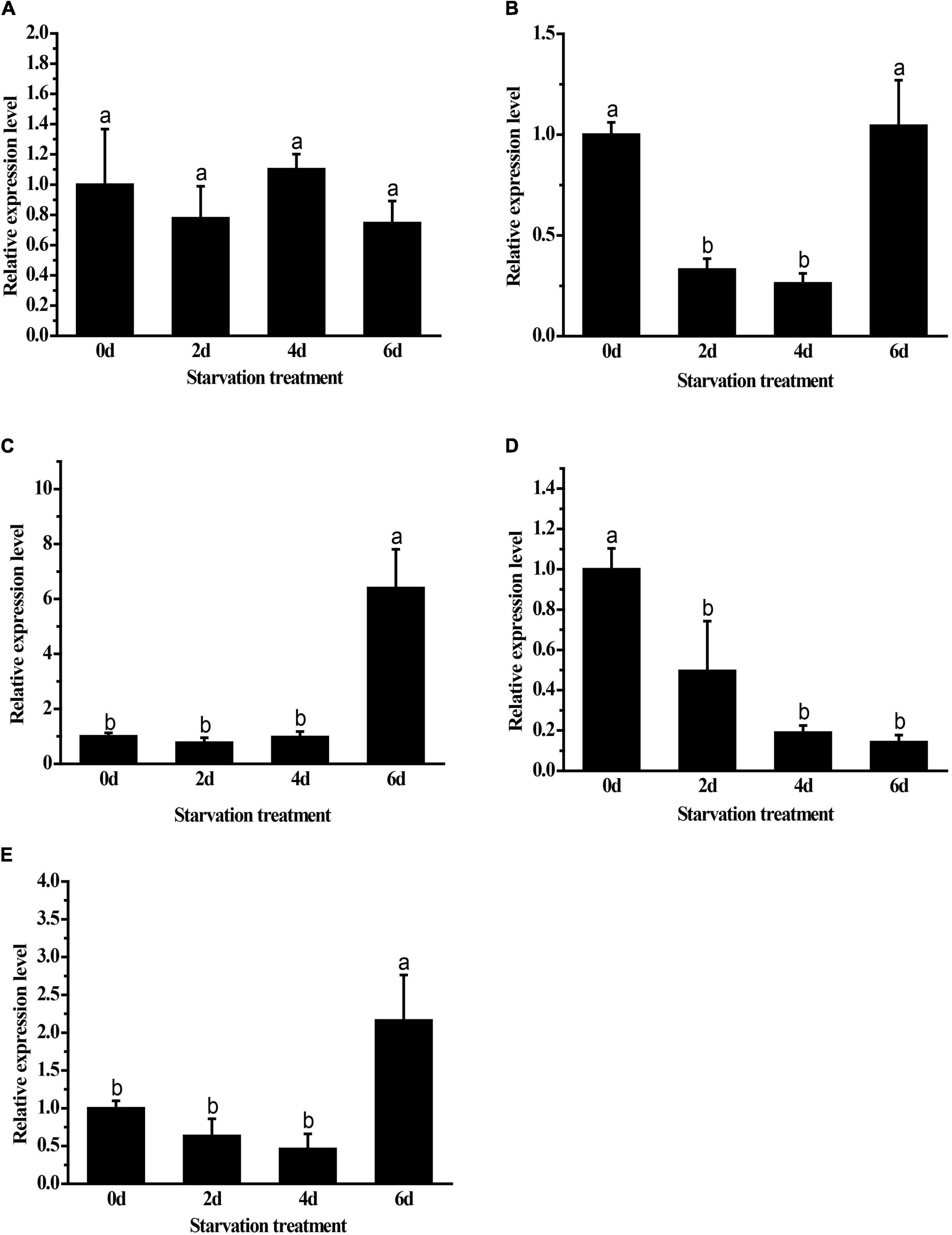
Figure 7. Expression levels of GpHsps after starvation stress. (A) GpHsp71.3; (B) GpHsp74.9; (C) GpHsp82.4; (D) GpHsp89; (E) GpHsp93.4. Data are shown as the mean ± SE of three biological replicates. Differences among triple repeats were analyzed using one-way analysis of variance (Systat, Inc., Evanston, IL) with Tukey’s post hoc test. Different lowercase letters indicate significant differences (p < 0.05).
The results showed that the parasitization by A. confusum and significantly regulated the expression levels of all candidate Hsps in G. pyloalis larvae and this moderation were affected by durations post-parasitization (The Two-way ANOVA: for GpHsp71.3, Fparasitization = 68.82, p < 0.001, Ftime = 6.82, p < 0.01, Fparasitization × time = 7.15, p < 0.01; for GpHsp74.9, Fparasitization = 52.62, p < 0.001, Ftime = 11.60, p < 0.001, Fparasitization × time = 29.45, p < 0.001; for GpHsp82.4, Fparasitization = 8.09, p < 0.01, Ftime = 9.81, p < 0.001, Fparasitization × time = 25.82, p < 0.001; for GpHsp89, Fparasitization = 31.56, p < 0.001, Ftime = 7.49, p < 0.01, Fparasitization × time = 22.72, p < 0.001; for GpHsp93.4, Fparasitization = 83.81, p < 0.001, Ftime = 55.43, p < 0.001, Fparasitization × time = 83.28, p < 0.001). All GpHsps expressed significantly lower on the first day after parasitization than on other time points. The expression level of GpHsp71.3 significantly decreased compared to the control group except for the second day after parasitization. The expression levels of GpHsp74.9 GpHsp82.4, and GpHsp89 increased significantly at sixth day after parasitism (Figure 8).
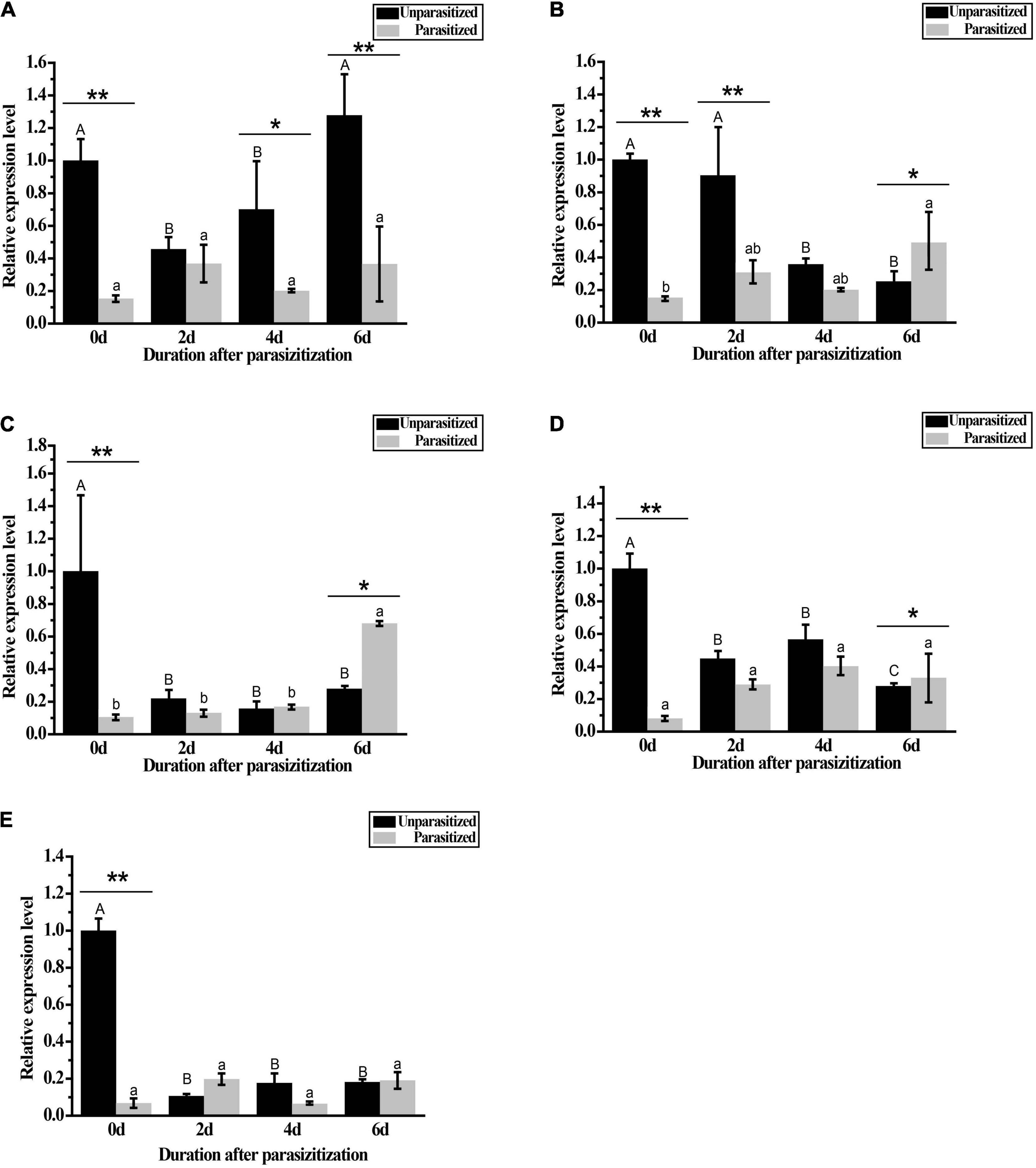
Figure 8. Expressions of GpHsps after parasitization by A. confusum. (A) GpHsp71.3; (B) GpHsp74.9; (C) GpHsp82.4; (D) GpHsp89; (E) GpHsp93.4. The black bar represents the healthy groups and the gray bar represents the parasitization groups. Data are shown as the mean ± SE of three biological replicates. Different upper and lower case letters indicate significant differences between healthy and parasitization groups within the different days (p < 0.05). Single asterisks (p < 0.05) and double asterisks (p < 0.01) indicate significant differences between the healthy and parasitization groups within the same day.
To further investigate the function of GpHsps in the thermal tolerance, GpHsp71.3 and GpHsp82.4 were selected for RNAi. The results showed that expression levels of both GpHsp71.3 and GpHsp82.4 were significantly decreased at 1.5–2 μL injection volume. In particular, the GpHsp71.3 mRNA level was significantly reduced at 2 μL injection volume compared to the control group. The GpHsp82.4 mRNA level was significantly downregulated at 1.5 μL injection volume compared to the control group (Figure 9).
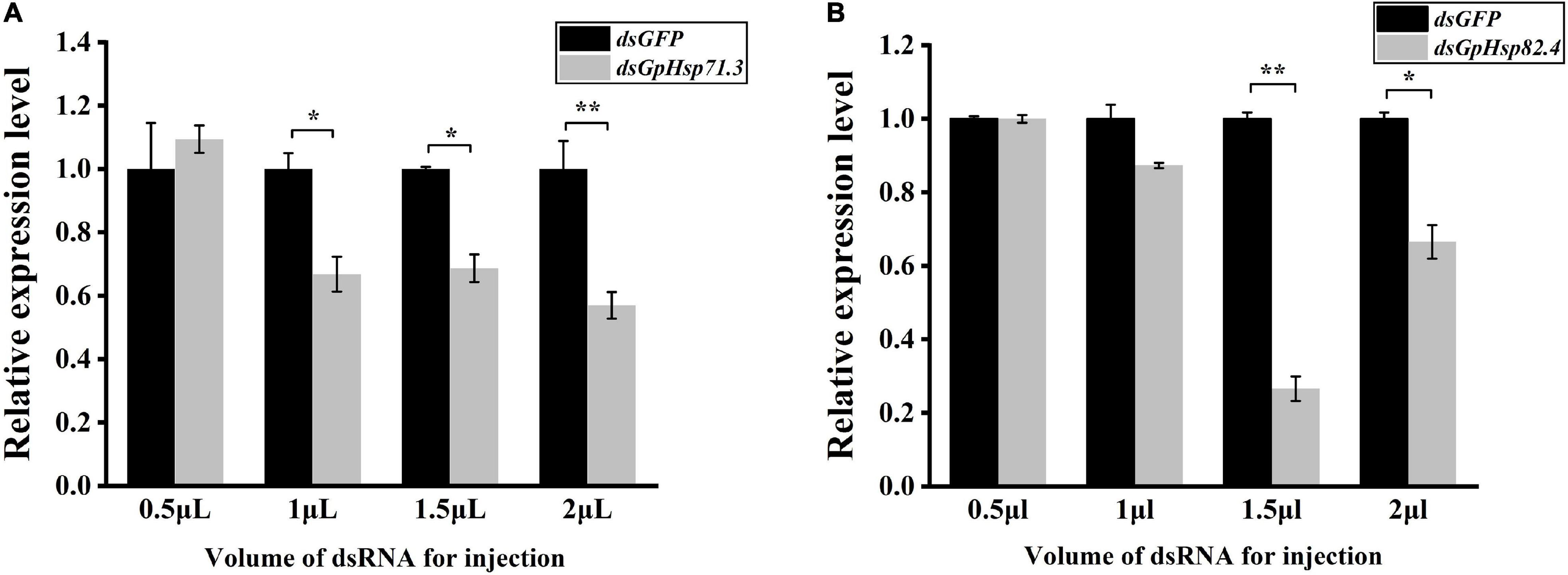
Figure 9. The analysis of the expression of two selected GpHsp genes after the knockdown at different injection volumes. (A) GpHsp71.3; (B) GpHsp82.4. Data are shown as the mean ± SE of three biological replicates. Differences in the expression levels of each target were compared using a one-way analysis of variance (Systat, Inc., Evanston, IL, United States) with Tukey’s post hoc test using R version 4.0.4. Significant differences are indicated by asterisks (p < 0.05).
In heat shock stress bioassay, we injected 2 μL dsHsp71.3 and 1.5 μL dsHsp82.4 into the individual larva, respectively, according to the optimal injection volume mentioned above. Compared to dsGFP control, knockdown of GpHsp71.3 and GpHsp82.4 significantly decreased the survivability of G. pyloalis larvae to high temperature stress for both 24 and 48 h (Figure 10). The knockdown of GpHsp71.3 and GpHsp82.4 resulted in greater mortalities after heat stress, suggesting that these genes are associated with improved protection from high temperature stress in G. pyloalis larvae.
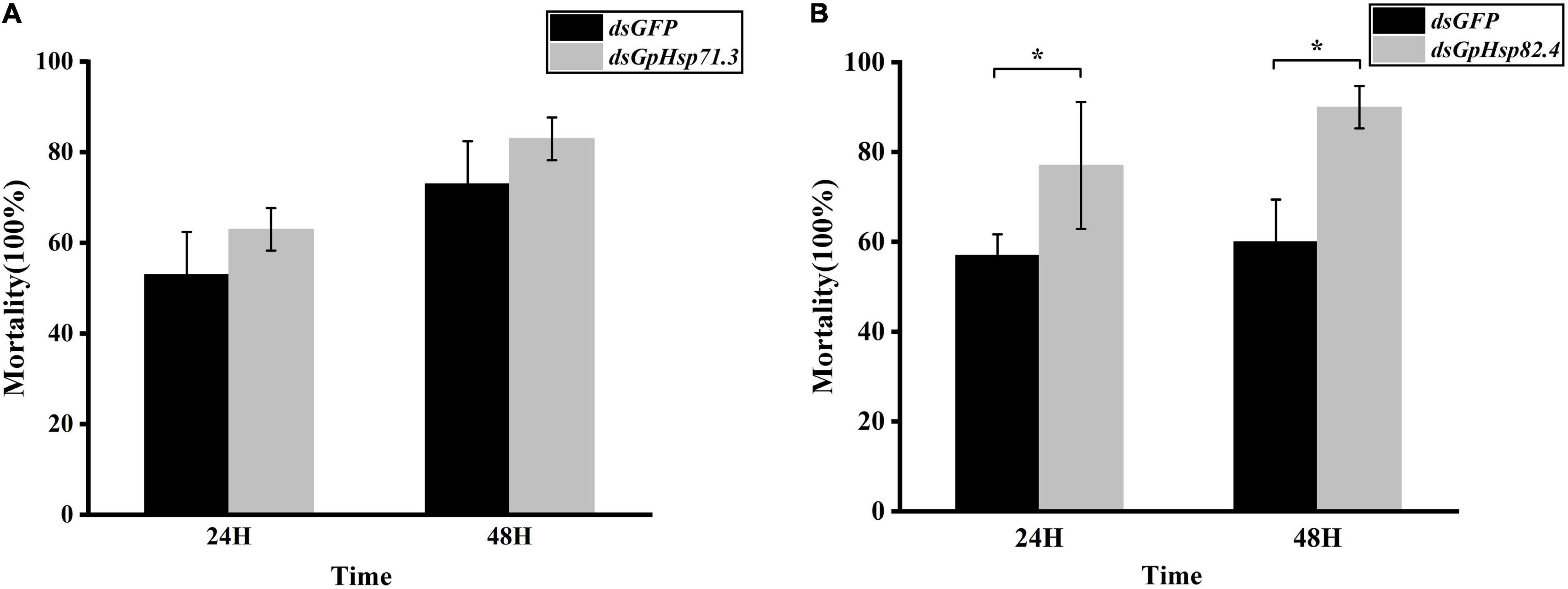
Figure 10. Mortality of G. pyloalis larvae to temperature stress after RNAi for 24/48 h. (A) GpHsp71.3; (B) GpHsp82.4. Reduction in heat tolerance of G. pyloalis injected with dsRNA and then exposed to 40°C (high temperature stress). Data are shown as the mean ± SE of three biological replicates and each replicate consists of ten individuals. Single asterisks (p < 0.05) indicate significant differences between dsGFP and dsGpHsp group.
Five Hsp genes from the G. pyloalis transcriptome dataset were identified, including two Hsp70 family genes (GpHsp71.3 and GpHsp74.9) and three Hsp90 family genes (GpHsp82.4, GpHsp89, and GpHsp93.4). The deduced protein, GpHsp71.3, and GpHsp74.9 have extremely high homology with Pyrausta nubilalis and S. exigua Hsp70 genes, indicating that insect Hsp70 has a high degree of interspecies conservation (Choi et al., 2014). Furthermore, the C-terminal end of these Hsp70s contains the highly conserved cytoplasm-specific EEVD motif, indicating that GpHsp70s functions in the cytoplasm (Gunter and Degnan, 2007; Zhang and Denlinger, 2010). This result is consistent with Galeruca daurica, Anaphothrips obscurus, Nilaparvata lugens, and Agrotis cnigrum in the previous studies (Wang et al., 2015; Lu et al., 2017; Tan et al., 2017; Guo and Feng, 2018).
Studies have found that Hsp play a crucial role in regulating insect growth, development, and metamorphosis (Gunter and Degnan, 2007). The expression level of Hsps can be expressed at various developmental stages. For example, the expression level of LsHsp70 in Laodelphax striatellus is higher in the first instar larvae (Zhang et al., 2014). The expression level of GdHsp70 is higher in eggs and first instar larvae of G. daurica (Tan et al., 2017). In Dastarcus helophoroides, DhHsp69.09 has the highest expression in the first instar larvae, and DhHsp70.11 and DhHsp71.88 are expressed highest in male adults (Zhang Z. et al., 2016). In the present study, GpHsp71.3 and GpHsp74.9 were highly expressed at diapause stage. The larvae of G. pyloalis will construct pupal chambers to molt into the diapause stage to successfully overwinter (Chen et al., 2015). The high expression of GpHsp71.3 and GpHsp74.9 in diapausing larvae indicates that the Hsp70 may play an important role in the successful overwintering process of G. pyloalis. Besides, the expression level of GpHsp71.3 in pupal stage was higher, and the expression level was significantly increased compared with that in pre-pupal stage, suggesting that GpHsp71.3 may play a crucial role in the metamorphosis process. This result is similar to Zhang B. et al. (2016) in Grapholitha molesta. In G. molesta, GmHsp70 can be highly expressed in diapausing larvae and can also be significantly expressed during the pupal stage. Similarly, the expression levels of Hsp90 genes in different insects are inconsistent with the changes in different developmental stages of insects. In L. striatellus, LsHsp90 has the high expression level in the third fourth instar larvae, and male adults (Zhang et al., 2014). In this study, the three GpHsp90 genes were highly expressed at prepupal stage followed by pupal stage. A similar scenario is also found in the Sitophilus zeamais. The SzHsp90 is expressed highest at the pupal stage (Tungjitwitayakul et al., 2015).
In many insects, the expression of the Hsp70 and Hsp90 genes is altered in response to temperature stress. The current study shows that heat/cold stress tends to induce high expression of Hsp70 and Hsp90 in insects. In C. pomonella, the expression of CpHsp70-1, CpHsp70-2, and C. pomHsp90 were all significantly increased in the heat-stressed state (40°C) (Yin et al., 2006). P. xylostella showed a significant increase in the expression of PxHsp70 and PxHsp90 after heat stress (37°C) (Zhang et al., 2015), and R. prolixus showed a significant rapid increase in the expression of R.proHsp70 under both cold stress (0°C) and heat stress (40°C) (Paim et al., 2016). The trend in the expression of NlHsp70 at different temperatures showed a continuous increase from 0 to 26°C, while the expression of NlHsp70 did not change at temperatures from 26 to 40°C (Lu et al., 2017). To evaluate the effect of exposure time on gene expression, two times of the exposure (1 and 4 h) were examined for their short- and long-term effects. The expression of all five Hsps in this study was elevated under heat stress, with GpHsp71.3 and GpHsp89 being significantly upregulated after 4 h of heat stress. GpHsp74.9, GpHsp82.4, and GpHsp93.4 were all upregulated after 1 h of heat stress. The divergence of expression levels of Hsps which depending on exposure time is also reported in other insects. For example, in Helicoverpa zea, Hsp90 is expressed at a higher level after a 30 min heat shock, while Hsp70 reaches its expression peak within 15 min heat exposure, which indicating that Hsp70 is more sensitive than Hsp90 to thermal stress in H. zea (Zhang and Denlinger, 2010). After 4 h of low-temperature stress, the expression of both GpHsp74.9 and GpHsp89 increased, and prolonged low-temperature induced the expression of GpHsp74.9 and GpHsp89 genes, presumably playing an essential role in the resistance of G. pyloalis to cold stress. Detail mechanism of the effects of long term cold stress on the expression levels of Hsps needs to be elucidated in the future study.
When faced with starvation stress, insects change their behavior and physiology to regulate the maintenance of a low metabolic state. The Hsp70 family plays role in response to starvation stress in insects. For example, In Harmonia axyridis, HaraxHSP68, HaraxHSP70A, and HaraxHSP70B reached maximal levels after starvation (Shen et al., 2015). In P. puparum expression of two Hsp genes PpHsp90 increased after 24 h starvation (Wang et al., 2012). In the starvation-treated Aphaenogaster picea, the expression level of ApHsp40 was twofold change higher than that of the control (Nguyen et al., 2017). We investigated the changes in the expression of GpHsp70s and GpHsp90s of G. pyloalis at different times after starvation and found that GpHsp74.9 expression was downregulated after 2 and 4 days of starvation. It has been shown that short-term starvation can affect protein homeostasis and nutrient balance in an organism’s cells, leading to a reduction in protein synthesis and nutrient content in the organism (Han et al., 2012). The synthesis and activity of Hsp70 require high energy consumption (Jahangirizadeh et al., 2018). We observed that in the absence of nutrients, the synthesis capacity of GpHsp74.9 was reduced, leading to a decrease in the expression of GpHsp74.9. The expression of GpHsp82.4 and GpHsp93.4 followed the same trend, with no significant in expression at the beginning of starvation (2 and 4 days) and a significant increase in expression after 6 days of starvation. It can be speculated that Hsp90 acts as a stress-protective protein and high expression of these proteins are effective in protecting G. pyloalis during starvation.
The G. pyloalis encounters many natural enemies in its natural environment. Clarifying the molecular response mechanism of the G. pyloalis after being parasitized by parasitic wasps can provide an effective, green, and safe strategy and a new method for the biological control of G. pyloalis. In this study, the G. pyloalis was parasitized by a parasitic wasp, A. confusum, and its Hsp70 family and Hsp90 family genes expression levels were measured. The result showed all Hsps of G. pyloalis downgraded significantly on the first day after parasitization, indicating that the parasitic behavior by A. confusum had a significant inhibitory effect on the expression of Hsp70 and Hsp90 families in G. pyloalis. Among them, the expression level of GpHsp71.3 was also significantly lower than that of the control after 4 and 6 days of parasitism, and the expression of GpHsp74.9 was also significantly lower than that of the control group after 2 days of parasitism. Compared to unparasitized larvae of first day, the expression of GpHsp74.9, GpHsp82.4, and GpHsp89 was significantly higher in the parasitized group after 6 days of parasitization. Our results showed that the expression level of the six GpHsps were significantly influenced by parasitism, and these effects depended on the developmental duration after parasitization. This result is similar to the results found in S. crassipalpis (Rinehart et al., 2002). Comparably, after P. rapae was parasitized by P. puparum, the expression of PrHsp90 continued to decrease within 48 h, which was significantly different from the control (Zhu et al., 2013). Downregulation of transcripts encoding Hsp90 in the flesh fly S. crassipalpis in response to venom stung by the ectoparasitoid wasp N. vitripennis have observed (Rinehart et al., 2002), and this result is similar to the present study. Besides, time duration post-parasitization is needed to be considered since the expression of GpHsp74.9, GpHsp82.4, and GpHsp89 was significantly higher after 6 days of parasitization. The possible reason for the downregulation of gene expression in the early stages of parasitism is that the parasitic wasp effectively suppresses the development of the host after injection of parasitic factors while disrupting the host’s immune system, which is largely suppressed by Hsps as an effective immune pathway for the host. Parasitic wasps inject venom and other parasitic factors into the host body while parasitizing (Moreau and Asgari, 2015). These factors cause infection of host blood cells, morphological and ultrastructural changes, apoptosis, inhibition of coagulation and encapsulation reactions, and alteration of actin cytoskeleton structure (Stettler et al., 1998; Carton et al., 2008). To date, parasitic wasp larvae have been shown to use a variety of strategies, such as inhibiting host development, to provide a suitable environment for their growth (Beckage and Gelman, 2004). Studies have shown that there is a functional relationship between hormones and the heat shock regulation system (Lezzi, 1996). Thus, parasitism-induced changes in the expression pattern of the host Hsp70 and Hsp90 genes of G. pyloalis suggest that these genes play a vital role in the parasitic process. It is implied that parasitic wasps may use maternal secretions to regulate these genes to create a suitable environment for parasitism. This may be one of the mechanisms by which parasitic wasps manipulate the development of their hosts. Further research is needed on the use of natural enemies such as parasitic wasps in controlling G. pyloalis.
In this study, two Hsp70 genes (GpHsp71.3 and GpHsp74.9) and three Hsp90 genes (GpHsp82.4, GpHsp89, and GpHsp93.4) were identified. We found five GpHsp genes that may be associated with diapause and pupation. Moreover, high-temperature environments can induce the upregulation of gene expression in different degrees, and parasitoid wasps can alter the expression of Hsp of G. pyloalis. This study presents a new idea for the comprehensive management and biological prevention and control of G. pyloalis. In conclusion, GpHsps is induced in response to both environmental and biotic stress and plays an important role in the physiological adaptation of G. pyloalis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR9959773.
SS and F-aW conceived and designed the experiments. J-hD, L-xZ, JC, and X-hL performed the experiments. J-hD, X-wG, and SS analyzed the data. J-hD, X-wG, and JW contributed reagents, materials, and analysis tools. J-hD and SS wrote the manuscript. All authors have read and approved the submitted manuscript.
This research was funded by the Key Research and Development Program (Modern Agriculture) of Zhenjiang City (NY2019021), the National Natural Science Foundation of China (31500312), the Jiangsu Agricultural Science and Technology Innovation Fund [CX(20)2029], and the Qing Lan Project of Jiangsu Province (Year of 2021).
X-wG is employed by Zhenjiang Runyu Biological Science and Technology Development Co., LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Sheng Qin from Jiangsu University of Science and Technology (Zhenjiang, PR China) for technical support in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.753914/full#supplementary-material
Beckage, N. E., and Gelman, D. B. (2004). Wasp parasitoid disruption of host development: implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 49, 299–330. doi: 10.1146/annurev.ento.49.061802.123324
Carton, Y., Poirié, M., and Nappi, A. J. (2008). Insect immune resistance to parasitoids. Insect Sci. 15, 67–87. doi: 10.1111/j.1744-7917.2008.00188.x
Chen, B., Kayukawa, T., Monteiro, A., and Ishikawa, Y. (2005). The expression of the HSP90 gene in response to winter and summer diapauses and thermal-stress in the onion maggot, Delia antiqua. Insect Mol. Biol. 14, 697–702. doi: 10.1111/j.1365-2583.2005.00602.x
Chen, B., Kayukawa, T., Monteiro, A., and Ishikawa, Y. (2006). Cloning and characterization of the HSP70 gene, and its expression in response to diapauses and thermal stress in the onion maggot, Delia antiqua. BMB Rep. 39, 749–758. doi: 10.5483/BMBRep.2006.39.6.749
Chen, W., Yang, Y., Zhang, F., and Dai, J. (2015). Preliminary study on diapause release and developmental period of overwintering larvae of Diaphania pyloalis. Jiangsu Sericul. 5, 9–13.
Choi, B.-G., Hepat, R., and Kim, Y. (2014). RNA interference of a heat shock protein, Hsp70, loses its protection role in indirect chilling injury to the beet armyworm, Spodoptera exigua. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 168, 90–95. doi: 10.1016/j.cbpa.2013.11.011
Chu, J., Jiang, D.-L., Yan, M.-W., Li, Y.-J., Wang, J., Wu, F.-A., et al. (2020). Identifications, characteristics, and expression patterns of small heat shock protein Genes in a Major Mulberry Pest, Glyphodes pyloalis (Lepidoptera: Pyralidae). J. Insect. Sci. 20:2. doi: 10.1093/jisesa/ieaa029
Farahani, S., Bandani, A. R., Alizadeh, H., Goldansaz, S. H., and Whyard, S. (2020). Differential expression of heat shock proteins and antioxidant enzymes in response to temperature, starvation, and parasitism in the Carob moth larvae, Ectomyelois ceratoniae (Lepidoptera: Pyralidae). PLoS One 15:e0228104. doi: 10.1371/journal.pone.0228104
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Feng, H., Wang, L., Liu, Y., He, L., Li, M., Lu, W., et al. (2010). Molecular characterization and expression of a heat shock protein gene (HSP90) from the carmine spider mite, Tetranychus cinnabarinus (Boisduval). J. Insect. Sci. 10:112. doi: 10.1673/031.010.11201
Gunter, H. M., and Degnan, B. M. (2007). Developmental expression of Hsp90, Hsp70 and HSF during morphogenesis in the vetigastropod Haliotis asinina. Dev. Genes 217, 603–612. doi: 10.1007/s00427-007-0171-2
Guo, X.-J., and Feng, J.-N. (2018). Comparisons of expression levels of heat shock proteins (hsp70 and hsp90) from Anaphothrips obscurus (Thysanoptera: Thripidae) in polymorphic adults exposed to different heat shock treatments. J. Insect. Sci. 18:15. doi: 10.1093/jisesa/iey059
Han, D., Huang, S. S., Wang, W.-F., Deng, D.-F., and Hung, S. S. (2012). Starvation reduces the heat shock protein responses in white sturgeon larvae. Environ. Biol. Fish. 93, 333–342.
Jahangirizadeh, Z., Ghafouri, H., Sajedi, R. H., Sarikhan, S., Taghdir, M., and Sariri, R. (2018). Molecular cloning, prokaryotic expression, purification, structural studies and functional implications of Heat Shock Protein 70 (Hsp70) from Rutilus frisii kutum. Int. J. Biol. Macromol. 108, 798–807. doi: 10.1016/j.ijbiomac.2017.10.174
Kim, K. K., Kim, R., and Kim, S.-H. (1998). Crystal structure of a small heat-shock protein. Nature 394, 595–599. doi: 10.1038/29106
King, A. M., and MacRae, T. H. (2015). Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 60, 59–75.
Koštál, V., and Tollarová-Borovanská, M. (2009). The 70 kDa heat shock protein assists during the repair of chilling injury in the insect, Pyrrhocoris apterus. PLoS One 4:e4546. doi: 10.1371/journal.pone.0004546
Lezzi, M. (1996). “Chromosome puffing: supramolecular aspects of ecdysone action,” in Metamorphosis, eds L. I. Gilbert, J. R. Tata, and B. G. Atkinson (San Diego, CA: Academic Press), 145–173.
Liu, Y., Su, H., Li, R., Li, X., Xu, Y., Dai, X., et al. (2017). Comparative transcriptome analysis of Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) reveals novel insights into heat stress tolerance in insects. BMC Genomics 18:974. doi: 10.1186/s12864-017-4355-5
Lu, K., Chen, X., Liu, W., Zhang, Z., Wang, Y., You, K., et al. (2017). Characterization of heat shock protein 70 transcript from Nilaparvata lugens (Stål): its response to temperature and insecticide stresses. Pestic. Biochem. Physiol. 142, 102–110. doi: 10.1016/j.pestbp.2017.01.011
Lu, M.-X., Liu, Z.-X., Cui, Y.-D., and Du, Y.-Z. (2014). Expression patterns of three heat shock proteins in Chilo suppressalis (Lepidoptera: Pyralidae). Ann. Entomol. Soc. Am. 107, 667–673. doi: 10.1603/AN13092
Moreau, S. J., and Asgari, S. (2015). Venom proteins from parasitoid wasps and their biological functions. Toxins 7, 2385–2412. doi: 10.3390/toxins7072385
Nguyen, A. D., DeNovellis, K., Resendez, S., Pustilnik, J. D., Gotelli, N. J., Parker, J. D., et al. (2017). Effects of desiccation and starvation on thermal tolerance and the heat-shock response in forest ants. J. Comp. Physiol. B 187, 1107–1116. doi: 10.1007/s00360-017-1101-x
Paim, R. M., Araujo, R. N., Leis, M., Sant’Anna, M. R., Gontijo, N. F., Lazzari, C. R., et al. (2016). Functional evaluation of Heat Shock Proteins 70 (HSP70/HSC70) on Rhodnius prolixus (Hemiptera, Reduviidae) physiological responses associated with feeding and starvation. Insect Biochem. Mol. Biol. 77, 10–20. doi: 10.1016/j.ibmb.2016.07.011
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rinehart, J. P., Denlinger, D. L., and Rivers, D. B. (2002). Upregulation of transcripts encoding select heat shock proteins in the flesh fly Sarcophaga crassipalpis in response to venom from the ectoparasitoid wasp Nasonia vitripennis. J. Invertebr. Pathol. 79, 62–63. doi: 10.1016/S0022-2011(02)00002-2
Rinehart, J. P., Li, A., Yocum, G. D., Robich, R. M., Hayward, S. A., and Denlinger, D. L. (2007). Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. U.S.A. 104, 11130–11137. doi: 10.1073/pnas.0703538104
Sasibhushan, S., Ponnuvel, K. M., and Vijayaprakash, N. B. (2013). Changes in diapause related gene expression pattern during early embryonic development in HCl-treated eggs of bivoltine silkworm Bombyx mori (Lepidoptera: Bombycidae). Braz. Arch. Biol. Technol. 56, 1–10. doi: 10.1590/S1516-89132013000100001
Shen, Q., Zhao, L., Xie, G., Wei, P., Yang, M., Wang, S., et al. (2015). Cloning three Harmonia axyridis (Coleoptera: Coccinellidae) heat shock protein 70 family genes: regulatory function related to heat and starvation stress. J. Entomol. Sci. 50, 168–185. doi: 10.18474/JES14-30.1
Sheng, S., Wang, J., Chu, J., Ding, J., Liu, Z. X., Jiang, D., et al. (2021). Analysis of the Glyphodes pyloalis larvae immune transcriptome in response to parasitization by its endoparasitoid, Aulacococentrum confusum. Comp. Biochem. Physiol. Part D Genomics Proteomics 38:100803. doi: 10.1016/j.cbd.2021.100803
Sonoda, S., Ashfaq, M., and Tsumuki, H. (2006). Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19. 5) from the diamondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch. Insect Biochem. Physiol. 62, 80–90. doi: 10.1002/arch.20124
Stettler, P., Trenczek, T., Wyler, T., Pfister-Wilhelm, R., and Lanzrein, B. (1998). Overview of parasitism associated effects on host haemocytes in larval parasitoids and comparison with effects of the egg-larval parasitoid Chelonus inanitus on its host Spodoptera littoralis. J. Insect Physiol. 44, 817–831.
Tammariello, S. P., Rinehart, J. P., and Denlinger, D. L. (1999). Desiccation elicits heat shock protein transcription in the flesh fly, Sarcophaga crassipalpis, but does not enhance tolerance to high or low temperatures. J. Insect Physiol. 45, 933–938. doi: 10.1016/S0022-1910(99)00073-6
Tan, Y., Zhang, Y., Huo, Z., Zhou, X., Shan, Y., and Pang, B. (2017). Molecular cloning and expression profiling of the heat shock protein gene GdHsp70 in Galeruca daurica (Coleoptera: Chrysomelidae). Acta Entomol. Sin. 60, 865–875. doi: 10.1017/S0007485317001079
Teixeira, L. A., and Polavarapu, S. (2005). Expression of heat shock protein 70 after heat stress during pupal diapause in Rhagoletis mendax (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 98, 966–972.
Tungjitwitayakul, J., Tatun, N., Singtripop, T., and Sakurai, S. (2008). Characteristic expression of three heat shock-responsive genes during larval diapause in the bamboo borer Omphisa fuscidentalis. Zool. Sci. 25, 321–333. doi: 10.2108/zsj.25.321
Tungjitwitayakul, J., Tatun, N., Vajarasathira, B., and Sakurai, S. (2015). Expression of heat shock protein genes in different developmental stages and after temperature stress in the maize weevil (Coleoptera: Curculionidae). J. Econ. Entomol. 108, 1313–1323. doi: 10.1093/jee/tov051
Tutar, L., and Tutar, Y. (2010). Heat shock proteins; an overview. Curr. Pharm. Biotechnol. 11, 216–222. doi: 10.2174/138920110790909632
Wang, H., Li, K., Zhu, J. Y., Fang, Q., Ye, G. Y., Wang, H., et al. (2012). cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch. Insect. Biochem. Physiol. 79, 247–263. doi: 10.1002/arch.21013
Wang, L., Yang, S., Zhao, K., and Han, L. (2015). Expression profiles of the heat shock protein 70 gene in response to heat stress in Agrotis c-nigrum (Lepidoptera: Noctuidae). J. Insect. Sci. 15:9. doi: 10.1093/jisesa/ieu169
Wang, Q., Liu, Y., Zheng, Z., Deng, Y., Jiao, Y., and Du, X. (2018). Adaptive response of pearl oyster Pinctada fucata martensii to low water temperature stress. Fish Shellfish Immunol. 78, 310–315. doi: 10.1016/j.fsi.2018.04.049
Xia, X., Lin, H., Zheng, D., Yang, G., and You, M. (2013). Identification and expression patterns of heat shock protein genes in the diamondback moth, Plutella xylostella (Lepidoptera: Yponomeutidae). Acta Entomol. Sin. 56, 457–464. doi: 10.16380/j.kcxb.2013.05.001
Yin, X., Wang, S., Tang, J., Hansen, J., and Lurie, S. (2006). Thermal conditioning of fifth-instar Cydia pomonella (Lepidoptera: Tortricidae) affects HSP70 accumulation and insect mortality. Physiol. Entomol. 31, 241–247. doi: 10.1111/j.1365-3032.2006.00512.x
Zhang, B., Peng, Y., Zheng, J., Liang, L., Hoffmann, A. A., and Ma, C.-S. (2016). Response of heat shock protein genes of the oriental fruit moth under diapause and thermal stress reveals multiple patterns dependent on the nature of stress exposure. Cell Stress Chaperones 21, 653–663. doi: 10.1007/s12192-016-0690-8
Zhang, D.-W., Xiao, Z.-J., Zeng, B.-P., Li, K., and Tang, Y.-L. (2019). Insect behavior and physiological adaptation mechanisms under starvation stress. Front. Physiol. 10:163. doi: 10.3389/fphys.2019.00163
Zhang, L. J., Wang, K. F., Jing, Y. P., Zhuang, H. M., and Wu, G. (2015). Identification of heat shock protein genes hsp70s and hsc70 and their associated mRNA expression under heat stress in insecticide-resistant and susceptible diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Eur. J. Entomol. 112, 215–226. doi: 10.14411/eje.2015.039
Zhang, Q., and Denlinger, D. L. (2010). Molecular characterization of heat shock protein 90, 70 and 70 cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J. Insect Physiol. 56, 138–150. doi: 10.1016/j.jinsphys.2009.09.013
Zhang, Q., Lu, M., and Zhu, S. (2014). Cloning, sequence analysis and expression profiling of heat shock protein 90 gene in the small brown plant hopper, Laodelphax striatellus (Hemiptera: Delphacidae). Acta Entomol. Sin. 57, 777–786. doi: 10.16380/j.kcxb.2014.07.003
Zhang, Z., Wang, H., Hao, C., Zhang, W., Yang, M., Chang, Y., et al. (2016). Identification, characterization and expression of methuselah-like genes in Dastarcus helophoroides (Coleoptera: Bothrideridae). Genes 7:91. doi: 10.3390/genes7100091
Zheng, Y., Liao, C., Yu, Z., Zhang, X., Jiao, W., Wu, S., et al. (2017). Determination of body weight and nutrient content in haemolymph of Diaphania pyloalis larvae after parasitism by Aulacococentrum confusum. Sci. Sericul. 43, 744–749.
Keywords: Glyphodes pyloalis, heat shock protein, environmental stress, RNA interference, parasitisation
Citation: Ding J-h, Zheng L-x, Chu J, Liang X-h, Wang J, Gao X-w, Wu F-a and Sheng S (2021) Characterization, and Functional Analysis of Hsp70 and Hsp90 Gene Families in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). Front. Physiol. 12:753914. doi: 10.3389/fphys.2021.753914
Received: 05 August 2021; Accepted: 28 September 2021;
Published: 21 October 2021.
Edited by:
Nikoletta Ntalli, Benaki Phytopathological Institute, GreeceReviewed by:
Jalal Jalali Sendi, University of Guilan, IranCopyright © 2021 Ding, Zheng, Chu, Liang, Wang, Gao, Wu and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fu-an Wu, ZnV3b3JkQDE2My5jb20=; Sheng Sheng, c2hlbmdzaGVuZ0BqdXN0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.