
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 23 December 2021
Sec. Cardiac Electrophysiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.748570
This article is part of the Research Topic Cardiac Optogenetics: Using Light to Observe and Excite the Heart View all 13 articles
Optogenetics, involving the optical measurement and manipulation of cellular activity with genetically encoded light-sensitive proteins (“reporters” and “actuators”), is a powerful experimental technique for probing (patho-)physiological function. Originally developed as a tool for neuroscience, it has now been utilized in cardiac research for over a decade, providing novel insight into the electrophysiology of the healthy and diseased heart. Among the pioneering cardiac applications of optogenetic actuators were studies in zebrafish, which first demonstrated their use for precise spatiotemporal control of cardiac activity. Zebrafish were also adopted early as an experimental model for the use of optogenetic reporters, including genetically encoded voltage- and calcium-sensitive indicators. Beyond optogenetic studies, zebrafish are becoming an increasingly important tool for cardiac research, as they combine many of the advantages of integrative and reduced experimental models. The zebrafish has striking genetic and functional cardiac similarities to that of mammals, its genome is fully sequenced and can be modified using standard techniques, it has been used to recapitulate a variety of cardiac diseases, and it allows for high-throughput investigations. For optogenetic studies, zebrafish provide additional advantages, as the whole zebrafish heart can be visualized and interrogated in vivo in the transparent, externally developing embryo, and the relatively small adult heart allows for in situ cell-specific observation and control not possible in mammals. With the advent of increasingly sophisticated fluorescence imaging approaches and methods for spatially-resolved light stimulation in the heart, the zebrafish represents an experimental model with unrealized potential for cardiac optogenetic studies. In this review we summarize the use of zebrafish for optogenetic investigations in the heart, highlighting their specific advantages and limitations, and their potential for future cardiac research.
Optogenetics involves the measurement and manipulation of cellular activity using genetically encoded light-sensitive proteins (Deisseroth et al., 2006; Miesenböck, 2009). Originally developed as a set of tools for neuroscience to activate or silence neuronal circuits and observe neuronal activity (Li et al., 2005; Nagel et al., 2005), optogenetic “reporters” (for measurement of membrane potential (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002) or intracellular calcium [Ca2+] (Miyawaki et al., 1997, 1999; Baird et al., 1999)) and ‘actuators' [for modulation of membrane potential (Nagel et al., 2002, 2003; Boyden et al., 2005)] have now been utilized in cardiac research for over a decade (Entcheva and Kay, 2021). Cardiac optogenetics has had a wide-range of applications, including: (i) all-optical studies of cardiac electrophysiology and high-throughput drug screening; (ii) cell-specific measurement or control to investigate cardiac sub-populations (e.g., myocytes, Purkinje cells, fibroblasts, neurons, and immune cells); (iii) manipulation of cardiac ion channels, G protein- coupled receptor signaling, and energetics; (iv) control of action potential morphology or excitation waves; and (v) cardiac pacing, cardioversion/defibrillation, or arrhythmia termination/ablation. Some of the pioneering studies that applied optogenetics to the heart were performed in zebrafish (Danio rerio). Here we provide an overview of the use of zebrafish for cardiac optogenetic studies, highlighting their advantages, limitations, and future potential [for a more general consideration of cardiac optogenetics, please see the recent review by Entcheva and Kay (Entcheva and Kay, 2021)].
The zebrafish has become an important integrative animal model for cardiac research, based on its particular advantages as an experimental tool (Table 1) (Gut et al., 2017; Stoyek and Quinn, 2018). The zebrafish offers a fully sequenced genome, which can be easily altered using standard genetic techniques at relatively low cost (in terms of time, effort, and money) (Rafferty and Quinn, 2018; Stoyek et al., in press), and almost every cardiac gene has a human ortholog with analogous function (Howe et al., 2013). This high degree of genetic similarity has permitted researchers to recapitulate a variety of human cardiac diseases in the zebrafish (Bowley et al., in press), which can be studied in a high throughput manner (Kithcart and MacRae, 2018). Functionally, the zebrafish heart has comparable heart rate, action potential morphologies, ion channels (Ravens, 2018), and Ca2+-handling proteins (van Opbergen et al., 2018b) to human. Furthermore, it has been shown that cardiac regulatory pathways and mechanisms of both intracardiac (MacDonald et al., 2017) and extracardiac (Stoyek et al., 2016) origin are similar to human, and like the cardiac electrophysiology of the zebrafish (Nemtsas et al., 2010), are often more so than rodents.
Of course, as with any experimental model, there are also limitations to the zebrafish's use (Table 1). The zebrafish heart is small, has only two chambers (one atrium and one ventricle, rather than the four chambers found in human), and generates relatively low pressures (Hu et al., 2001). While over 70% of human genes have at least one zebrafish ortholog, 24% of genes have more than one ortholog (due to a duplication of the zebrafish genome), which can confer redundancy in gene function and confound results of genetic manipulations (Howe et al., 2013). Functionally, while cardiac electrophysiology appears strikingly similar to humans (Vornanen and Hassinen, 2016; Ravens, 2018), there are important differences in cellular calcium cycling (Genge et al., 2016; van Opbergen et al., 2018b). Zebrafish cardiomyocytes have a lack of transverse tubules (Brette et al., 2008), and even though sarcoplasmic reticulum Ca2+ levels are much higher in the zebrafish, release of Ca2+ from the sarcoplasmic reticulum following excitation (Ca2+-induced Ca2+ release) appears to be limited (due in part to a low sensitivity of ryanodine receptors to Ca2+) (Bovo et al., 2013). As a result, sarcolemmal Ca2+ influx is responsible for ~80% of the Ca2+ transient in zebrafish cardiomyocytes (compared to 25% in human) (Bovo et al., 2013), although this remains somewhat controversial, as others have shown a strong dependence of contractile force on sarcoplasmic reticulum Ca2+ release (Haustein et al., 2015) and the existence of Ca2+ sparks with characteristics similar to mammals (Llach et al., 2011). Zebrafish also have a higher sodium-Ca2+ exchanger current than in mammals, such that its reverse-mode has been shown to trigger sarcoplasmic reticulum Ca2+ release (Zhang et al., 2011).
Considering its use specifically for cardiac optogenetic studies, the zebrafish has a further advantage over other animal models, in that the entire zebrafish heart can be optically accessed in vivo in the transparent, externally developing embryo (van Opbergen et al., 2018a) or in situ in the relatively small, isolated adult heart (Stoyek et al., 2018), in a manner not possible in mammals. While other non-mammalian models may have a similar advantage (e.g., Drosophila melanogaster [Wolf et al., 2006) and Xenopus laevis (Warkman and Krieg, 2007)], they are limited in other ways. For instance, while Drosophila have been highly utilized for studies of cardiac genetics (Wolf et al., 2006), it is an invertebrate, and differences in the morphology of its heart—which is a tube—limits its applicability for functional studies (Rotstein and Paululat, 2016). The heart of Xenopus, on the other hand, is in some ways more anatomically similar to humans than zebrafish—for instance, it has a pulmonary circulation—but there is a limited genetic tool box for their transgenesis (Ishibashi et al., 2008).
Ultimately, the similarities of zebrafish to human, and its particular experimental advantages, have resulted in it being a popular experimental model for optogenetic investigations, both for neuroscience [the brain and nervous system can also be optically accessed in the whole animal (Del Bene and Wyart, 2012; Simmich et al., 2012; Portugues et al., 2013)] and for cardiovascular research (Table 2), which is the focus of this review.
The electrical activity of the heart has been optically monitored for decades, well before the emergence of modern optogenetics and the use of functional fluorescent proteins. In the 1970s, Salama and Morad published the first reports of the use of voltage sensitive fluorescent dyes to record cardiac action potentials (Salama and Morad, 1976; Morad and Salama, 1979). Since that time, optical mapping of membrane potential and intracellular Ca2+ in the whole heart or isolated tissue and cells has become a “standard” technique in many research labs (Herron et al., 2012; Jaimes et al., 2016; Berenfeld and Efimov, 2019), including studies using zebrafish (Sabeh et al., 2012; Lin et al., 2020). The use of optogenetic reporters (genetically encoded voltage and Ca2+ indicators, GEVIs and GECIs, respectively) have additional advantages as they allow for organ-, organelle-, and cell-specific measurements and for in vitro and in vivo longitudinal studies. There is now a wide array of GEVIs and GECIs available for use in the heart, with a range of excitation and emission spectra, light sensitivity and signal intensity, temporal dynamics, and other properties that dictate their specific use (Figure 1) (Kaestner et al., 2014, 2015; Koopman et al., 2017).
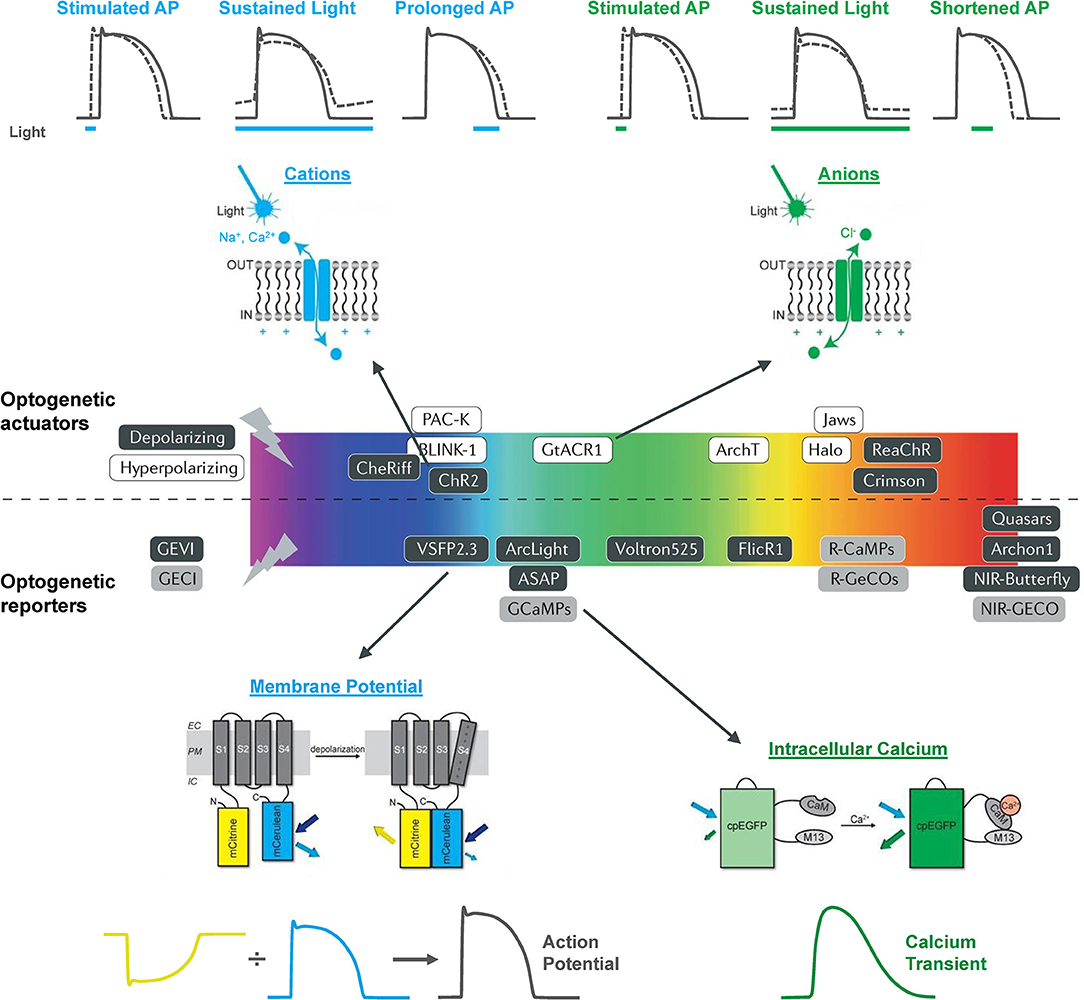
Figure 1. The optogenetic toolbox for measurement and manipulation of cardiac activity. There is a large array of optogenetic actuators and reporters with a broad range of activation spectra available for use in the heart. These comprise light-activated depolarizing (excitatory) and hyperpolarizing (inhibitory) opsins that pass cations, anions, and protons and genetically encoded voltage (GEVI) and calcium (GECI) indicators that can be used to measure membrane potential and intracellular calcium. Opsin schematic from Ferenczi et al. (2019), middle panel from Entcheva and Kay (2021), and GEVI and GECI schematics from van Opbergen et al. (2018a).
The first use of an optogenetic reporter in the heart was described in the early 2000s. This involved GCaMP2, which includes a circularly permutated EGFP within an M13/Calmodulin fusion protein that fluoresces when it binds Ca2+, to record Ca2+ waves in the isolated mouse heart and in open chest animals (Tallini et al., 2006). This was followed by a study that used GCaMP2 to demonstrate successful engraftment and electrical coupling of embryonic cardiomyocytes with surrounding myocardium in the infarcted mouse heart (Roell et al., 2007). Around the same time, the potential for using zebrafish to image intracellular Ca2+ in the intact animal was also being realized. The first published report involved the use of a transgenic zebrafish line with cardiac-specific expression of gGCaMP driven by the cardiac myosin light chain 2 (cmlc2) gene promoter (Tg(cmlc2:gCaMP)s878). It was used to investigate a transgenic zebrafish model of inherited long QT syndrome in which a loss of rapid delayed-rectifier potassium current (IKr) due to a mutation in the s290 allele of the kcnh2 gene (kcnh2s290) results in mechanical ventricular asystole (Arnaout et al., 2007). Using selective plane illumination microscopy (SPIM) and excitation-contraction uncoupling with a silent heart cardiac troponin (tnnt2) morpholino (to eliminate optical mapping motion artifact associated with contraction), Ca2+ transients were measured in vivo at various regions of the zebrafish atrium and ventricle in 2 days post-fertilization (dpf) embryos. In wild-type tnnt2 morpholino-injected zebrafish, repetitive fluorescent waves representing an increase in cytosolic Ca2+ during systole were visualized, spreading from the atrium through the atrioventricular junction and into the ventricle (Figure 2A). In contrast, in the kcnh2s290 homozygous mutants, Ca2+ waves were visible in the atrium but not in the ventricle (Figure 2A), implying impaired ventricular Ca2+ cycling.
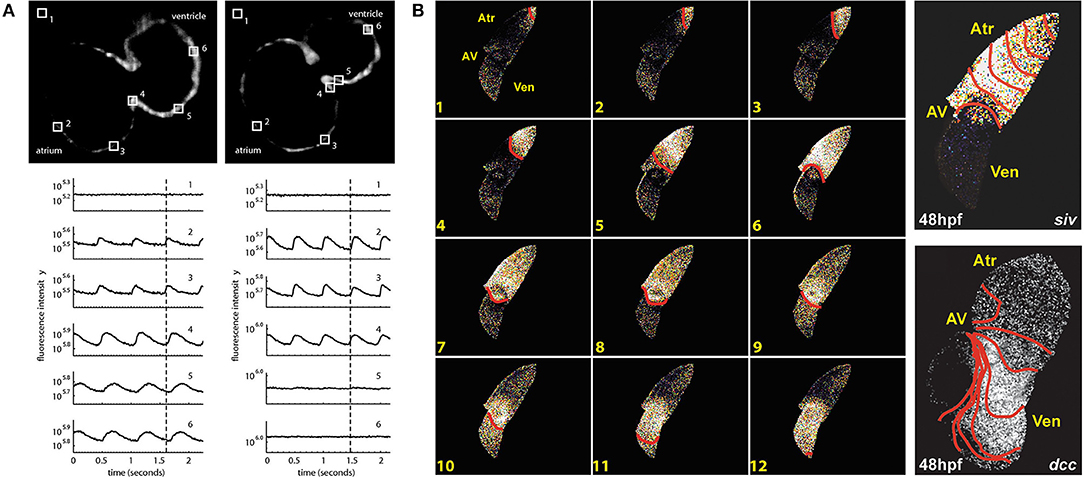
Figure 2. In vivo imaging of intracellular calcium using genetically encoded calcium indicators (GECIs) in intact zebrafish embryos. (A) Hearts in 48 h post-fertilization (hpf) wild-type embryos with cardiac-specific expression of gGCaMP exhibit atrial and ventricular calcium release (left), while hearts in kcnh2s290 mutants exhibit atrial but no ventricular release (right). Each selected region in the images has a corresponding fluorescence signal plotted below. The dotted lines mark an arbitrary point in time to facilitate comparison across the different signals. From Arnaout et al. (2007). (B) In hearts from 48 hpf wild-type embryos, calcium activation travels from the sinus venosus across the atrium (Atr) and ventricle (Ven), with a delay at the atrioventricular (AV) junction (left), while ventricular conduction is absent in silent ventricle (siv) mutants and disorganized in dococs215, 226 (dcc) mutants (right). Isochronal lines represent 60 ms. From Chi et al. (2008).
The above investigation was followed by a study using the same zebrafish line to map Ca2+ waves across the whole heart in intact embryos at various ages to investigate the development of the vertebrate cardiac conduction system (Figure 2B) (Chi et al., 2008). Four distinct stages of conduction development were identified, which corresponded to specific cellular and anatomical changes in the developing heart and were dependent on epigenetic mechanical factors such as hemodynamic flow and contraction. An in vivo optical mapping technique was then used as a phenotypic assay to perform a forward genetic screen, which identified 17 conduction-specific mutations (Figure 2B), thought to represent novel genetic regulators of the cardiac conduction system. A similar approach has been applied to other genes whose mutation is known to affect cardiac conduction, such as the Popeye domain containing (Popdc) gene family, with morpholino knock down of popdc2 in 5–6 dpf embryos causing sinoatrial node conduction failure, irregular atrial and ventricular activity, and varying degrees of atrioventricular block (Kirchmaier et al., 2012).
More recently, zebrafish have been used to help in the assessment of novel GECIs for cardiac-specific applications, such as those with a ratiometric readout, which is useful for assessing absolute changes in Ca2+ and to help correct for the motion artifact that occurs with optical mapping in the beating heart. Four available ratiometric Förster resonance energy transfer (FRET)-based GECIs with varying Ca2+-binding affinity (TN-XXL, Twitch-1, Twitch-2B, and Twitch-4) were transiently expressed in the hearts of zebrafish embryos (driven by the cmlc2 promoter) and kinetic parameters of atrial and ventricular Ca2+ transients were measured at 3 dpf under various conditions. Ultimately, this revealed that Twitch-1 and Twitch-4 are the most promising for use in the heart, based on their greater sensitivity, faster kinetics, and higher affinity for Ca2+ (Figure 3) (Salgado-Almario et al., 2020).
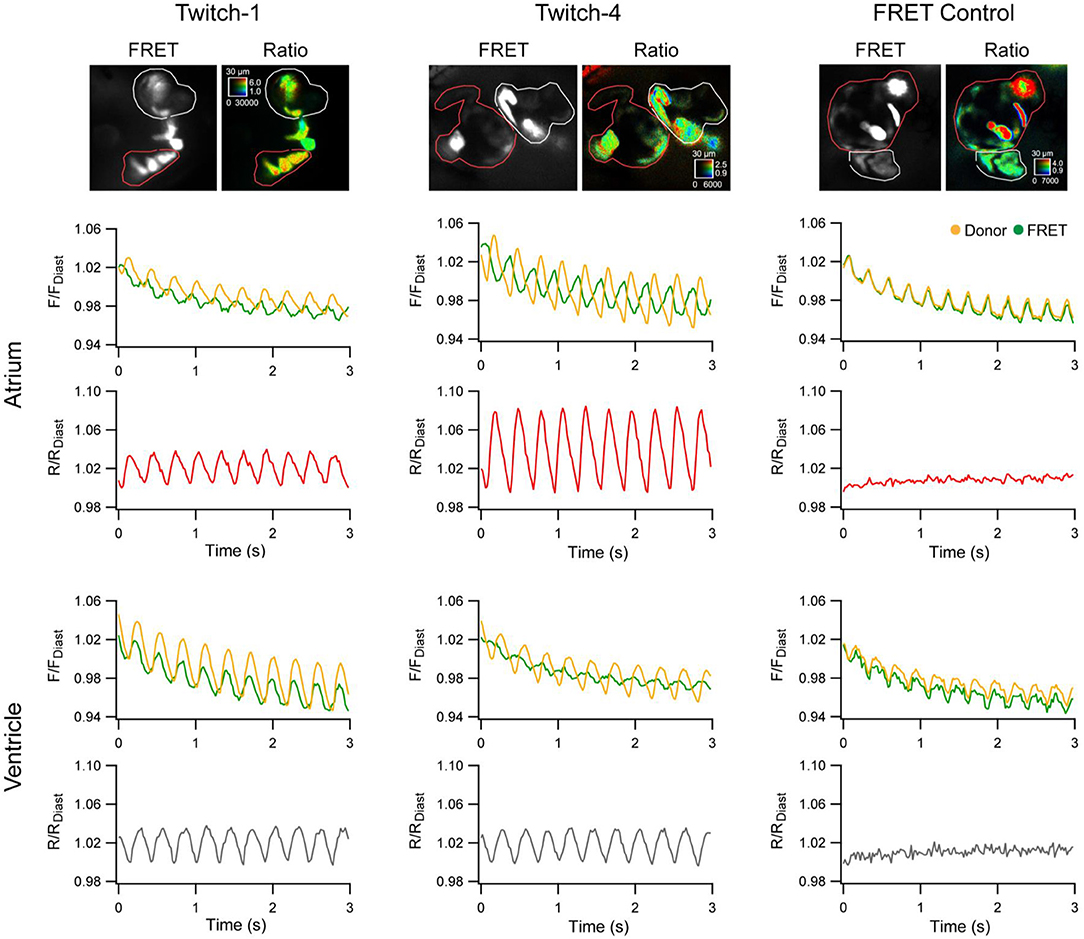
Figure 3. In vivo ratiometric intracellular calcium measurements with a genetically encoded calcium indicator (GECI) in intact zebrafish embryos. Ratiometric intracellular calcium signals were acquired with Twitch-1, Twitch-4, or a Förster resonance energy transfer (FRET) construct insensitive to calcium (ECFP-16aa-EYFP) from the atrium (red region-of-interest) and ventricle (white region-of-interest) of 3 days post-fertilization embryos. The change in fluorescence in the donor and FRET channels normalized to the first diastolic period (F/FDiast; upper graphs in atrium and ventricle) and their ratio (R/RDiast; lower graphs in atrium and ventricle) are shown. From Salgado-Almario et al. (2020).
The development of effective GEVIs has been slower than GECIs, owing to difficulties in achieving sufficiently fast kinetics and avoiding electrophysiological interference. Recently, significant progress has been made, which includes their application for cardiac research. The first reported use of GEVIs in the heart was in fact in zebrafish (Tsutsui et al., 2010), which utilized a FRET-based voltage-sensitive fluorescent protein (VSFP) called Mermaid (Tsutsui et al., 2008). The Mermaid construct consists of a green-emitting fluorescent donor (mUKG; Umi-Kinoko from Sarcophyton) and an orange-emitting fluorescent acceptor (mKOκ; Kusabira from Fungia concinna) fused to a voltage sensing phosphatase from Ciona intestinalis (Ci-VSP) with a transmembrane domain homologous to the S1–S4 segments of voltage-gated potassium (Kv) channels (Murata et al., 2005). The Mermaid reporter was expressed specifically in the zebrafish heart under the cmcl2 promotor [Tg(cmlc2:Mermaid)] and used for in vivo voltage mapping in 2–3 dpf embryos under normal conditions and after application of the histamine H1 receptor blocker astemizole (known to also block IKr). Measurements showed that astemizole disrupted the normal sequence of cardiac excitation, causing retrograde propagation from the atrioventricular boundary to the atrium (Figure 4A) (Tsutsui et al., 2010).
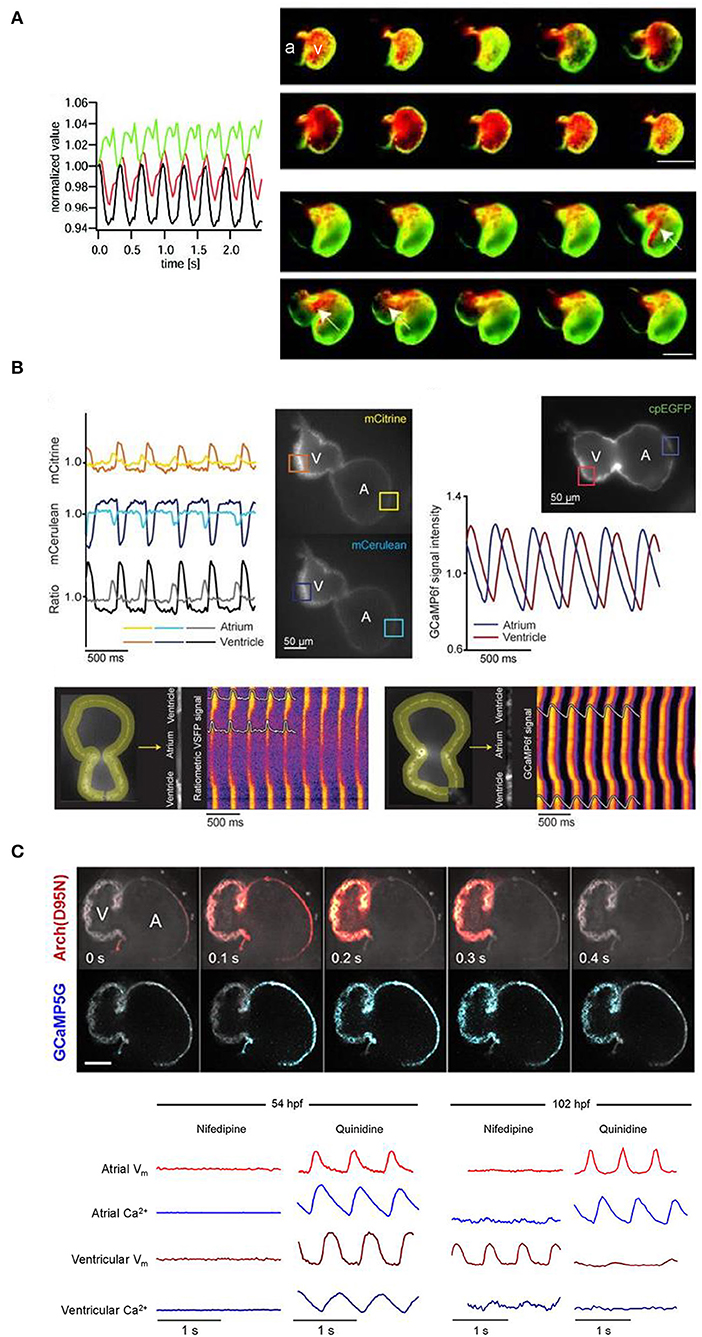
Figure 4. In vivo imaging of membrane potential using genetically encoded voltage indicators (GEVIs), combined with genetically encoded calcium indicators (GECIs) in intact zebrafish embryos. (A) Signals (left) from a donor (mUKG, green) and acceptor (mKOκ, red) Förster resonance energy transfer (FRET) pair of fluorescent proteins and their ratio (mKOκ/mUKG, black) acquired from the ventricle of a 3 days post-fertilization (dpf) zebrafish embryo with cardiac-specific expression of the GEVI Mermaid. Pseudo-colored ratio images (right) representing a single cardiac cycle in wild-type, Mermaid-expressing zebrafish (upper) showing propagation of excitation from the sinus venosus in the atrium (a) to the ventricle (v), and in astemizole-treated (5 μM, 15 min) zebrafish (lower) showing retrograde propagation from the ventricle to the atrium (highlighted with arrows). Scale bar, 100 μm. From Tsutsui et al. (2010). (B) Signals (upper left) from a donor (mCerulean, blue) and acceptor (mCitrine, yellow) Förster resonance energy transfer (FRET) pair of fluorescent proteins and their ratio (mCitrine/mCerulean, black) acquired from the regions of interest (boxes in fluorescent images) indicated on the atrium (A) and ventricle (V) of a 3 dpf zebrafish embryo with cardiac-specific expression of GEVI VSFP-butterfly CY. Signals (upper right) acquired from the atrium (blue) and ventricle (red) of a 3 dpf zebrafish embryo with cardiac-specific expression of the GECI GCaMP6f. Line plots of chimeric VSFP-butterfly CY (lower left) and GCaMP6f (lower right) background-corrected fluorescence intensities averaged across the width of the myocardial wall superimposed on heat maps of trajectory vs. time illustrating electrical impulse and Ca2+ propagation throughout the heart. cpEGFP, circularly permutated enhanced green fluorescent protein. From van Opbergen et al. (2018a). (C) Single optical sections of a 4 dpf zebrafish heart with cardiac-specific expression of the GEVI-GECI construct CaViar showing GEVI Arch(D95N) (top) and GECI GCaMP5G (middle) fluorescence as excitation propagates from the atrium (A) to ventricle (V). Voltage (Vm, red) and calcium (Ca2+, blue) signals acquired from the atrium and ventricle of 50 (upper left) and 102 (lower right) hours post-fertilization (hpf) zebrafish embryos exposed to nifedipine (L-type Ca2+ channel blocker) and quinidine (fast sodium channel blocker). At 52 hpf, nifedipine reversibly suppressed voltage and Ca2+ dynamics in both chambers, while quinidine had no effect. At 102 hpf, nifedipine largely suppressed calcium transients in both chambers but only suppressed atrial voltage, while quinidine largely suppressed voltage and calcium transients in the ventricle but did not affect either transient in the atrium. Scale bar, 50 μm. From Hou et al. (2014).
Since that time, a variety of new GEVIs have been developed with improved sensitivity and kinetics. In a recent study using zebrafish (van Opbergen et al., 2018a), the novel GEVI chimeric VSFP-butterfly CY (cyan-yellow, mCitrine/mCerulean) (Mishina et al., 2014) or the updated GECI GCaMP6f (Chen et al., 2013) were expressed in the heart of pigment-deficient, optically-transparent casper mutant zebrafish (White et al., 2008), with the myosin light chain 7 (myl7) promotor [Tg(myl7:chimeric VSFP-butterfly CY) and Tg(myl7:Gal4FF; UAS:GCaMP6f)]. The hearts of 3 dpf and 14 dpf zebrafish were imaged after administration of drugs targeting the sympathetic nervous system or various cardiac ion channels to assess effects on electrical activation, action potential morphology, and intracellular Ca2+ dynamics (van Opbergen et al., 2018a) (Figure 4B). It was shown that: (i) β adrenergic receptor stimulation (with isoproterenol) or blockade (with propranolol) increased or decreased diastolic Ca2+ levels and Ca2+ transient amplitudes, respectively; (ii) IKr block (with E-4031) increased action potential duration; (iii) L-type calcium current (ICa, L) block (with nifedipine) prevented Ca2+ transients, increased ventricular action potential duration, and disrupted atrioventricular conduction; and (iv) differences exist in atrial and ventricular Ca2+ recovery dynamics between 3 and 14 dpf zebrafish (but not in the Ca2+ upstroke).
Functional fluorescent dyes can be combined for simultaneous mapping of voltage and Ca2+ in the whole heart (Herron et al., 2012). There is similar interest in combining GEVIs and GECIs for dual voltage-Ca2+ imaging, however this is generally prevented by spectral overlap of the relevant fluorescent proteins. The first successful study using a GEVI-GECI construct in the heart was performed in the zebrafish, using a genetically encoded dual-function voltage-Ca2+ reporter (“CaViar,” created by fusing the GEVI Arch(D95N) with the GECI GCaMP5) under control of the heart-specific cmlc2 promoter [Tg(cmlc2:Arch(D95N)-GCaMP5G)] (Hou et al., 2014). Hearts of 1.5–4.5 dpf embryos were imaged during application of the ICa, L blocker nifedipine or fast sodium channel blocker quinidine, which showed that early in development the zebrafish cardiac AP is initiated by Ca2+, but by 4 dpf the ventricular AP becomes driven by sodium, while the atrial AP remains Ca2+ dependent (Figure 4C).
Optogenetic actuators are light-activated proteins that generate a transmembrane ion flux. The discovery and cloning of the cation-selective ion channel channelrhodopsin-2 (ChR2) from the green alga Chlamydomonas reinhardtii in 2003 (Nagel et al., 2003) has led to the development of an extensive toolkit that includes depolarizing (excitatory) and hyperpolarizing (inhibitory) opsins, which are activated across a wide spectrum of wavelengths, and may be used for manipulation of cardiac membrane potential (Figure 1) (Schneider-Warme, 2018; Ferenczi et al., 2019; Entcheva and Kay, 2021). As for the cardiac application of GEVIs and GECIs, one of the first reports of the use of optogenetic actuators in the heart was in zebrafish (Arrenberg et al., 2010). This involved the use of both ChR2 [Tg(E1b:Gal4s1101t; UAS:ChR2(H134R)-eYFPs1990t)] and the chloride-specific ion channel halorhodopsin from Natronomonas pharaonis (NpHR) (Zhang et al., 2007) [Tg(E1b:Gal4-VP16s1101t; UAS:NpHR-mCherrys1989t)], to locate and control cardiac pacemaker cells in intact 1–5 dpf zebrafish embryos. In NpHR-expressing zebrafish, maps were generated at each day post-fertilization by sequentially illuminating small, overlapping regions of the heart, and measuring the heart rate response, or the incidence of cardiac arrest or arrhythmia (Figure 5A). It was found that: (i) at 1 dpf, the heart stopped beating when a region at the venous pole was illuminated, indicating the location of the pacemaker; (ii) at 2 dpf, the pacemaker region was more confined to the sinoatrial ring, with illumination of large adjacent areas having no effect, and atrioventricular block (of varying degree, depending on light intensity) occurred with illumination of the atrioventricular canal; and (iii) at 3 dpf, the pacemaker region was more defined, being confined to the dorsal right quadrant of the sinoatrial ring. It was further found that in 4 dpf embryos, pulsed photo-stimulation of the sinoatrial ring at a frequency of 2.7–4.7 Hz with ChR2 was able to control heart rate.
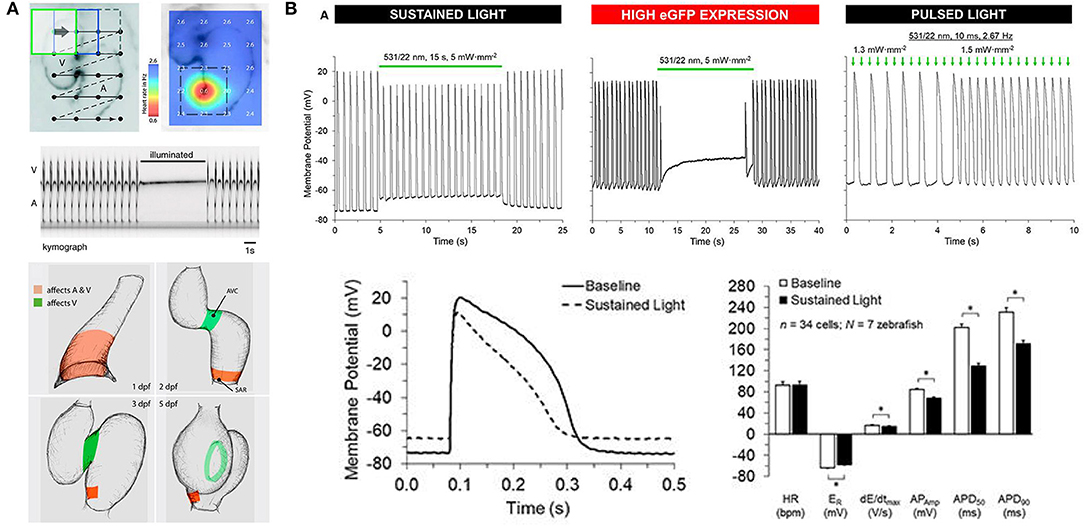
Figure 5. Optogenetic manipulation of membrane potential in zebrafish hearts. (A) in vivo stimulation of halorhodopsin by patterned illumination of 3 days post-fertilization (dpf) embryonic hearts (upper left) reveals the location of the pacemaker by a reduction in heart rate (upper right). Illumination of the whole heart could stop atrial (A) and ventricular (V) beating (middle). Areas that control atrial and ventricular (red) or only ventricular (green) contractions were revealed throughout embryogenesis (1–5 dpf, bottom). From Arrenberg et al. (2010). (B) Sustained stimulation of Guillardia theta anion channelrhodopsins 1 (GtACR1) by spot illumination on the ventricle of 3 months post-fertilization zebrafish isolated hearts caused an immediate increase in resting membrane potential (ER) and a decrease in the maximum rate of membrane depolarization (dE/dtmax), AP amplitude (APAmp), and APD at 50% and 90% repolarization (APD50 and APD90, upper left and bottom). In the case of particularly high GtACR1 expression (represented by high eGFP expression), ventricular excitation could be silenced (upper middle). Pulsed light, on the other hand, could be used to stimulate the heart (upper right). *Indicates p < 0.0001 by two-tailed paired Student's t-test. From Kopton et al. (2018).
Since the time of that pioneering study, another prominent family of chloride-specific light-activated ion channels has been developed, the Guillardia theta anion channelrhodopsin 1 and 2 (GtACR1 and GtACR2) (Govorunova et al., 2015), which have been shown to silence neuronal AP generation (including in zebrafish) (Malyshev et al., 2017; Mauss et al., 2017; Mohamed et al., 2017; Forli et al., 2018). The first cardiac application of GtACR1 involved zebrafish [using hearts isolated from 3 months post-fertilization adults with cardiac-specific GtACR1 expression, Tg(cmlc2:GtACR1-eGFP)], which in combination with experiments in genetically transfected single rabbit ventricular myocytes demonstrated that GtACR1 activation causes depolarization of ventricular myocytes when applied during the resting (diastolic) phase of the AP (and if suprathreshold, results in excitation), but causes repolarization when applied during the (systolic) plateau (resulting in shortening of the AP). This biphasic response relates to the reversal potential of chloride in ventricular myocytes, which is somewhere between −40 and −33 mV (Clemo et al., 1999), so that the flow of negative ions switches from outward (causing depolarization) to inward (causing repolarization) as cells are excited. As a result, pulsed illumination can be used to pace the heart, while sustained illumination can arrest the heart in a depolarized state (Figure 5B) (Kopton et al., 2018). This indicates that while GtACR1 does not address the need for optogenetic silencing through a physiological means (i.e., hyperpolarization), it is a potentially attractive tool for exciting cardiomyocytes by transient light-induced depolarization.
Unlike the prevalent use of optogenetics in zebrafish to study the nervous system (Del Bene and Wyart, 2012; Simmich et al., 2012; Portugues et al., 2013), there have been relatively few cardiac optogenetic studies performed in zebrafish (summarized in Table 2), yet those studies have been fundamental in progressing the application of optogenetic technologies to the heart. With the continual improvement of optogenetic techniques (Entcheva and Kay, 2021), the use of zebrafish for integrative (patho-)physiological cardiac structure-function studies holds great promise. Future research will be driven by technological advances such as high-speed, cell-accurate, three-dimensional mapping (Mickoleit et al., 2014; Weber et al., 2017; Sacconi et al., in press), more effective methods for cell-specific spatial and temporal gene expression (Reade et al., 2017; LaBelle et al., 2021), and novel optogenetic actuators and reporters with enhanced expression, fluorescence, and kinetics, combined with improved light delivery (Entcheva and Kay, 2021). As the field continues to develop, the zebrafish may be invaluable for cardiac optogenetic studies directly related to its strengths as an experimental model (Table 1; i.e., development, genetic screening, drug discovery, cardiotoxicity testing, disease modeling, all-optical studies of electrophysiology and cell signaling, and anti-arrhythmic strategy development). It may also be a powerful tool for fundamental investigations of the hetero-cellular heart (e.g., structure-function interactions of myocytes, fibroblasts, intracardiac neurons, and immune and endothelial cells) and in helping to overcome hindrances related to the clinical translation of optogenetic techniques (e.g., genetic transfection, immune responses, phototoxicity) (Richter and Bruegmann, 2020). This will be enhanced by the large number of currently available transgenic lines, genetic material, and tools (easily found through online resources and central repositories), facilitated by the open zebrafish community willing to share them (Rafferty and Quinn, 2018; Stoyek et al., in press).
While a majority of cardiac optogenetic studies in the zebrafish have been performed in the early stages of development, technological advances in fluorescence imaging approaches and methods for spatially-resolved light stimulation have the promise to enable studies to be performed in the adult isolated whole heart and in vivo. This will be aided by the continuing development of transgenic lines that lack pigment and are thus largely transparent throughout their lifespan (i.e., “casper” [White et al., 2008) and “crystal” (Antinucci and Hindges, 2016)], and can be used as a background on which to express optogenetic reporters and actuators along with mutations of interest.
Optogenetics is a powerful and highly successful (Deisseroth, 2010) set of techniques that has been instrumental in recent developments in neuroscience research (Deisseroth, 2015; Kim et al., 2017), and more recently also for cardiac research (Entcheva and Kay, 2021). Zebrafish provide specific advantages as an experimental model for optogenetic cardiac investigations (Gut et al., 2017; Stoyek and Quinn, 2018), and have been instrumental in its early development, suggesting a bright future for this little fish. With the increasing sophistication of optogenetic methods, the zebrafish represents an experimental model with great potential for cardiac optogenetic studies. Hopefully more cardiac researchers will soon begin to see the light.
JB wrote the manuscript. MS and TQ revised the manuscript. All authors approved the final version.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-04879 to TQ), the Heart and Stroke Foundation of Canada (G-18-0022185 to TQ), and the Canadian Institutes of Health Research (MOP 342562 to TQ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Antinucci, P., and Hindges, R. (2016). A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 6, 1–10. doi: 10.1038/srep29490
Arnaout, R., Ferrer, T., Huisken, J., Spitzer, K., Stainier, D. Y. R., Tristani-Firouzi, M., et al. (2007). Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. U. S. A. 104, 11316–11321. doi: 10.1073/pnas.0702724104
Arrenberg, A. B., Stainier, D. Y. R., Baier, H., and Huisken, J. (2010). Optogenetic control of cardiac function. Science 330, 971–974. doi: 10.1126/science.1195929
Ataka, K., and Pieribone, V. A. (2002). A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys. J. 82, 509–516. doi: 10.1016/S0006-3495(02)75415-5
Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (1999). Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 96, 11241–11246. doi: 10.1073/pnas.96.20.11241
Berenfeld, O., and Efimov, I. (2019). Optical mapping. Card. Electrophysiol. Clin. 11, 495–510. doi: 10.1016/j.ccep.2019.04.004
Bovo, E., Dvornikov, A. V., Mazurek, S. R., de Tombe, P. P., and Zima, A. V. (2013). Mechanisms of Ca2+ handling in zebrafish ventricular myocytes. Pflügers Arch. 465, 1775–1784. doi: 10.1007/s00424-013-1312-2
Bowley, G., Kugler, E., Wilkinson, R., Lawrie, A., Eeden, F., van Chico, T. J. A., and (in press). Zebrafish as a tractable model of human cardiovascular disease. Br. J. Pharmacol. doi: 10.1111/bph.15473.
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., and Deisseroth, K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. doi: 10.1038/nn1525
Brette, F., Luxan, G., Cros, C., Dixey, H., Wilson, C., and Shiels, H. A. (2008). Characterization of isolated ventricular myocytes from adult zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 374, 143–146. doi: 10.1016/j.bbrc.2008.06.109
Chen, T.-W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Chi, N. C., Shaw, R. M., Jungblut, B., Huisken, J., Ferrer, T., Arnaout, R., et al. (2008). Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6:e109. doi: 10.1371/journal.pbio.0060109
Clemo, H. F., Stambler, B. S., and Baumgarten, C. M. (1999). Swelling-activated chloride current is persistently activated in ventricular myocytes from dogs with tachycardia-induced congestive heart failure. Circ. Res. 84, 157–165. doi: 10.1161/01.RES.84.2.157
Deisseroth, K.. (2015). Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225. doi: 10.1038/nn.4091
Deisseroth, K., Feng, G., Majewska, A. K., Miesenböck, G., Ting, A., and Schnitzer, M. J. (2006). Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 26, 10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006
Del Bene, F., and Wyart, C. (2012). Optogenetics: a new enlightenment age for zebrafish neurobiology. Dev. Neurobiol. 72, 404–414. doi: 10.1002/dneu.20914
Entcheva, E., and Kay, M. W. (2021). Cardiac optogenetics: a decade of enlightenment. Nat. Rev. Cardiol. 18:349. doi: 10.1038/s41569-020-00478-0
Ferenczi, E. A., Tan, X., and Huang, C. L. H. (2019). Principles of optogenetic methods and their application to cardiac experimental systems. Front. Physiol. 10:1096. doi: 10.3389/fphys.2019.01096
Forli, A., Vecchia, D., Binini, N., Succol, F., Bovetti, S., Moretti, C., et al. (2018). Two-photon bidirectional control and imaging of neuronal excitability with high spatial resolution in vivo. Cell Rep. 22, 3087–3098. doi: 10.1016/j.celrep.2018.02.063
Genge, C. E., Lin, E., Lee, L., Sheng, X., Rayani, K., Gunawan, M., et al. (2016). The zebrafish heart as a model of mammalian cardiac function. Rev. Physiol. Biochem. Pharmacol. 171, 99–136. doi: 10.1007/112_2016_5
Govorunova, E. G., Sineshchekov, O. A., Janz, R., Liu, X., and Spudich, J. L. (2015). Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650. doi: 10.1126/science.aaa7484
Gut, P., Reischauer, S., Stainier, D. Y. R., and Arnaout, R. (2017). Little fish, big data: zebrafish as a model for cardiovascular and metabolic disease. Physiol. Rev. 97, 889–938. doi: 10.1152/physrev.00038.2016
Haustein, M., Hannes, T., Trieschmann, J., Verhaegh, R., Köster, A., Hescheler, J., et al. (2015). Excitation-contraction coupling in zebrafish ventricular myocardium is regulated by trans-sarcolemmal Ca2+ influx and sarcoplasmic reticulum Ca2+ release. PLoS ONE 10:e0125654. doi: 10.1371/journal.pone.0125654
Herron, T. J., Lee, P., and Jalife, J. (2012). Optical imaging of voltage and calcium in cardiac cells and tissues. Circ. Res. 110, 609–623. doi: 10.1161/CIRCRESAHA.111.247494
Hou, J. H., Kralj, J. M., Douglass, A. D., Engert, F., and Cohen, A. E. (2014). Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front. Physiol. 5:344. doi: 10.3389/fphys.2014.00344
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Hu, N., Yost, H. J., and Clark, E. B. (2001). Cardiac morphology and blood pressure in the adult zebrafish. Anat. Rec. 264, 1–12. doi: 10.1002/ar.1111
Ishibashi, S., Kroll, K. L., and Amaya, E. (2008). A method for generating transgenic frog embryos. Methods Mol. Biol. 461, 447–466. doi: 10.1007/978-1-60327-483-8_31
Jaimes, R. III., Walton, R. D., Pasdois, P., Bernus, O., Efimov, I. R., et al. (2016). Arrhythmias, electrophysiology, and optical mapping: a technical review of optical mapping of intracellular calcium within myocardial tissue. Am. J. Physiol. Heart Circ. Physiol. 310, H1388–H401. doi: 10.1152/ajpheart.00665.2015
Kaestner, L., Scholz, A., Tian, Q., Ruppenthal, S., Tabellion, W., Wiesen, K., et al. (2014). Genetically encoded Ca2+ indicators in cardiac myocytes. Circ. Res. 114, 1623–1639. doi: 10.1161/CIRCRESAHA.114.303475
Kaestner, L., Tian, Q., Kaiser, E., Xian, W., Müller, A., Oberhofer, M., et al. (2015). Genetically encoded voltage indicators in circulation research. Int. J. Mol. Sci. 16, 21626–21642. doi: 10.3390/ijms160921626
Kim, C. K., Adhikari, A., and Deisseroth, K. (2017). Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci. 18, 222–235. doi: 10.1038/nrn.2017.15
Kirchmaier, B. C., Poon, K. L., Schwerte, T., Huisken, J., Winkler, C., Jungblut, B., et al. (2012). The Popeye domain containing 2 (popdc2) gene in zebrafish is required for heart and skeletal muscle development. Dev. Biol. 363, 438–450. doi: 10.1016/j.ydbio.2012.01.015
Kithcart, A. P., and MacRae, C. A. (2018). Zebrafish assay development for cardiovascular disease mechanism and drug discovery. Prog. Biophys. Mol. Biol. 138, 126–131. doi: 10.1016/j.pbiomolbio.2018.07.002
Koopman, C. D., Zimmermann, W. H., Knöpfel, T., and de Boer, T. P. (2017). Cardiac optogenetics: using light to monitor cardiac physiology. Basic Res. Cardiol. 112, 1–13. doi: 10.1007/s00395-017-0645-y
Kopton, R. A., Baillie, J. S., Rafferty, S. A., Moss, R., Zgierski-Johnston, C. M., Prykhozhij, S. V., et al. (2018). Cardiac electrophysiological effects of light-activated chloride channels. Front. Physiol. 9:1806. doi: 10.3389/fphys.2018.01806
LaBelle, J., Ramos-Martinez, A., Shen, K., Motta-Mena, L. B., Gardner, K. H., Materna, S. C., et al. (2021). TAEL 2.0: an improved optogenetic expression system for zebrafish. Zebrafish 20–28. doi: 10.1089/zeb.2020.1951
Li, X., Gutierrez, D. V., Hanson, M. G., Han, J., Mark, M. D., Chiel, H., et al. (2005). Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U. S. A. 102, 17816–17821. doi: 10.1073/pnas.0509030102
Lin, E., Shafaattalab, S., Gill, J., Al-Zeer, B., Craig, C., Lamothe, M., et al. (2020). Physiological phenotyping of the adult zebrafish heart. Mar. Genomics 49:100701. doi: 10.1016/j.margen.2019.100701
Llach, A., Molina, C. E., Alvarez-Lacalle, E., Tort, L., Benítez, R., and Hove-Madsen, L. (2011). Detection, properties, and frequency of local calcium release from the sarcoplasmic reticulum in teleost cardiomyocytes. PLoS ONE 6:e23708. doi: 10.1371/journal.pone.0023708
MacDonald, E. A., Stoyek, M. R., Rose, R. A., and Quinn, T. A. (2017). Intrinsic regulation of sinoatrial node function and the zebrafish as a model of stretch effects on pacemaking. Prog. Biophys. Mol. Biol. 130, 198–211. doi: 10.1016/j.pbiomolbio.2017.07.012
Malyshev, A. Y., Roshchin, M. V., Smirnova, G. R., Dolgikh, D. A., Balaban, P. M., and Ostrovsky, M. A. (2017). Chloride conducting light activated channel GtACR2 can produce both cessation of firing and generation of action potentials in cortical neurons in response to light. Neurosci. Lett. 640, 76–80. doi: 10.1016/j.neulet.2017.01.026
Mauss, A. S., Busch, C., and Borst, A. (2017). Optogenetic neuronal silencing in drosophila during visual processing. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-14076-7
Mickoleit, M., Schmid, B., Weber, M., Fahrbach, F. O., Hombach, S., Reischauer, S., et al. (2014). High-resolution reconstruction of the beating zebrafish heart. Nat. Methods 11, 919–922. doi: 10.1038/nmeth.3037
Miesenböck, G.. (2009). The optogenetic catechism. Science 326, 395–399. doi: 10.1126/science.1174520
Mishina, Y., Mutoh, H., Song, C., and Knöpfel, T. (2014). Exploration of genetically encoded voltage indicators based on a chimeric voltage sensing domain. Front. Mol. Neurosci. 7:78. doi: 10.3389/fnmol.2014.00078
Miyawaki, A., Griesbeck, O., Heim, R., and Tsien, R. Y. (1999). Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc. Natl. Acad. Sci. U. S. A. 96, 2135–2140. doi.org/10.1073/pnas.96.5.2135 doi: 10.1073/pnas.96.5.2135
Miyawaki, A., Llopis, J., Heim, R., Michael McCaffery, J., Adams, J. A., Ikura, M., et al. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. doi: 10.1038/42264
Mohamed, G. A., Cheng, R.-K., Ho, J., Krishnan, S., Mohammad, F., Claridge-Chang, A., et al. (2017). Optical inhibition of larval zebrafish behaviour with anion channelrhodopsins. BMC Biol. 15:103. doi: 10.1186/s12915-017-0430-2
Morad, M., and Salama, G. (1979). Optical probes of membrane potential in heart muscle. J. Physiol. 292, 267–295. doi: 10.1113/jphysiol.1979.sp012850
Murata, Y., Iwasaki, H., Sasaki, M., Inaba, K., and Okamura, Y. (2005). Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243. doi: 10.1038/nature03650
Nagel, G., Brauner, M., Liewald, J. F., Adeishvili, N., Bamberg, E., and Gottschalk, A. (2005). Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284. doi: 10.1016/j.cub.2005.11.032
Nagel, G., Ollig, D., Fuhrmann, M., Kateriya, S., Musti, A. M., Bamberg, E., et al. (2002). Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296, 2395–2398. doi: 10.1126/science.1072068
Nagel, G., Szellas, T., Huhn, W., Kateriya, S., Adeishvili, N., Berthold, P., et al. (2003). Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U. S. A. 100, 13940–13945. doi: 10.1073/pnas.1936192100
Nemtsas, P., Wettwer, E., Christ, T., Weidinger, G., and Ravens, U. (2010). Adult zebrafish heart as a model for human heart? An electrophysiological study. J. Mol. Cell. Cardiol. 48, 161–171. doi: 10.1016/j.yjmcc.2009.08.034
Portugues, R., Severi, K. E., Wyart, C., and Ahrens, M. B. (2013). Optogenetics in a transparent animal: circuit function in the larval zebrafish. Curr. Opin. Neurobiol. 23, 119–126. doi: 10.1016/j.conb.2012.11.001
Rafferty, S. A., and Quinn, T. A. (2018). A beginner's guide to understanding and implementing the genetic modification of zebrafish. Prog. Biophys. Mol. Biol. 138, 3–19. doi: 10.1016/j.pbiomolbio.2018.07.005
Ravens, U.. (2018). Ionic basis of cardiac electrophysiology in zebrafish compared to human hearts. Prog. Biophys. Mol. Biol. 138, 38–44. doi: 10.1016/j.pbiomolbio.2018.06.008
Reade, A., Motta-Mena, L. B., Gardner, K. H., Stainier, D. Y., Weiner, O. D., and Woo, S. (2017). TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development 144, 345–355. doi: 10.1242/dev.139238
Richter, C., and Bruegmann, T. (2020). No light without the dark: perspectives and hindrances for translation of cardiac optogenetics. Prog. Biophys. Mol. Biol. 154, 39–50. doi: 10.1016/j.pbiomolbio.2019.08.013
Roell, W., Lewalter, T., Sasse, P., Tallini, Y. N., Choi, B.-R., Breitbach, M., et al. (2007). Engraftment of connexin 43-expressing cells prevents post-infarct arrhythmia. Nature 450, 819–824. doi: 10.1038/nature06321
Rotstein, B., and Paululat, A. (2016). On the morphology of the Drosophila heart. J. Cardiovasc. Dev. Dis. 3:15. doi: 10.3390/jcdd3020015
Sabeh, M. K., Kekhia, H., and MacRae, C. A. (2012). Optical mapping in the developing zebrafish heart. Pediatr. Cardiol. 33, 916–922. doi: 10.1007/s00246-012-0300-1
Sacconi, L., Silvestri, L., Rodríguez, E., Armstrong, G. A. B., Pavone, F., Shrier, A., and (in press). KHz-rate volumetric voltage imaging of the whole zebrafish heart. Cell Rep. doi: 10.2139/ssrn.3783403.
Sakai, R., Repunte-Canonigo, V., Raj, C. D., and Knöpfel, T. (2001). Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 13, 2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x
Salama, G., and Morad, M. (1976). Merocyanine 540 as an optical probe of transmembrane electrical activity in the heart. Science 191, 485–487. doi: 10.1126/science.191.4226.485
Salgado-Almario, J., Vicente, M., Vincent, P., Domingo, B., and Llopis, J. (2020). Mapping calcium dynamics in the heart of zebrafish embryos with ratiometric genetically encoded calcium indicators. Int. J. Mol. Sci. 21:6610. doi: 10.3390/ijms21186610
Schneider-Warme, F.. (2018). The power of optogenetics. Herzschrittmacherther. Elektrophysiol. 29, 24–29. doi: 10.1007/s00399-017-0545-8
Siegel, M. S., and Isacoff, E. Y. (1997). A genetically encoded optical probe of membrane voltage. Neuron 19, 735–741. doi: 10.1016/S0896-6273(00)80955-1
Simmich, J., Staykov, E., and Scott, E. (2012). Zebrafish as an appealing model for optogenetic studies. Prog. Brain Res. 196, 145–162. doi: 10.1016/B978-0-444-59426-6.00008-2
Stoyek, M. R., and Quinn, T. A. (2018). One fish, two fish, red fish, blue fish*: zebrafish as a model for cardiac research. Prog. Biophys. Mol. Biol. 138, 1–2. doi: 10.1016/j.pbiomolbio.2018.11.003
Stoyek, M. R., Quinn, T. A., Croll, R. P., and Smith, F. M. (2016). Zebrafish heart as a model to study the integrative autonomic control of pacemaker function. Am. J. Physiol. Heart Circ. Physiol. 311, H676–H688. doi: 10.1152/ajpheart.00330.2016
Stoyek, M. R., Rafferty, S. A., Quinn, T. A., and (in press). “Genetically modified zebrafish as experimental model,” in Handjournal of Molecular Biotechnology, ed D. Liu (Boca Raton, FL: CRC Press).
Stoyek, M. R., Rog-Zielinska, E. A., and Quinn, T. A. (2018). Age-associated changes in electrical function of the zebrafish heart. Prog. Biophys. Mol. Biol. 138, 91–104. doi: 10.1016/j.pbiomolbio.2018.07.014
Tallini, Y. N., Ohkura, M., Choi, B.-R., Ji, G., Imoto, K., Doran, R., et al. (2006). Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. U. S. A. 103, 4753–4758. doi: 10.1073/pnas.0509378103
Tsutsui, H., Higashijima, S., Miyawaki, A., and Okamura, Y. (2010). Visualizing voltage dynamics in zebrafish heart. J. Physiol. 588, 2017–2021. doi: 10.1113/jphysiol.2010.189126
Tsutsui, H., Karasawa, S., Okamura, Y., and Miyawaki, A. (2008). Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods 5, 683–685. doi: 10.1038/nmeth.1235
van Opbergen, C. J. M., Koopman, C. D., Kok, B. J. M., Knöpfel, T., Renninger, S. L., Orger, M. B., et al. (2018a). Optogenetic sensors in the zebrafish heart: a novel in vivo electrophysiological tool to study cardiac arrhythmogenesis. Theranostics 8, 4750–4764. doi: 10.7150/thno.26108
van Opbergen, C. J. M., van der Voorn, S. M., Vos, M. A., de Boer, T. P., and van Veen, T. A. B. (2018b). Cardiac Ca2+ signalling in zebrafish: translation of findings to man. Prog. Biophys. Mol. Biol. 138, 45–58. doi: 10.1016/j.pbiomolbio.2018.05.002
Vornanen, M., and Hassinen, M. (2016). Zebrafish heart as a model for human cardiac electrophysiology. Channels 10, 101–110. doi: 10.1080/19336950.2015.1121335
Warkman, A. S., and Krieg, P. A. (2007). Xenopus as a model system for vertebrate heart development. Semin. Cell Dev. Biol. 18, 46–53. doi: 10.1016/j.semcdb.2006.11.010
Weber, M., Scherf, N., Meyer, A. M., Panáková, D., Kohl, P., and Huisken, J. (2017). Cell-accurate optical mapping across the entire developing heart. Elife 6:e28307. doi: 10.7554/eLife.28307.025
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Wolf, M. J., Amrein, H., Izatt, J. A., Choma, M. A., Reedy, M. C., and Rockman, H. A. (2006). Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. U. S. A. 103, 1394–1399. doi: 10.1073/pnas.0507359103
Zhang, F., Wang, L.-P., Brauner, M., Liewald, J. F., Kay, K., Watzke, N., et al. (2007). Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639. doi: 10.1038/nature05744
Keywords: cardiac electrophysiology, opsins, membrane potential, intracellular calcium, genetically encoded voltage indicators (GEVIs), genetically encoded calcium indicators (GECIs)
Citation: Baillie JS, Stoyek MR and Quinn TA (2021) Seeing the Light: The Use of Zebrafish for Optogenetic Studies of the Heart. Front. Physiol. 12:748570. doi: 10.3389/fphys.2021.748570
Received: 28 July 2021; Accepted: 19 October 2021;
Published: 23 December 2021.
Edited by:
Stephan E. Lehnart, University Medical Center Göttingen, GermanyReviewed by:
Alexandra Zahradnikova, Slovak Academy of Sciences, SlovakiaCopyright © 2021 Baillie, Stoyek and Quinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: T. Alexander Quinn, YWxleC5xdWlubkBkYWwuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.