
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 03 September 2021
Sec. Mitochondrial Research
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.729452
This article is part of the Research TopicRole of Mitochondria-Associated Non-Coding RNAs in Intracellular CommunicationView all 11 articles
Mutations in human mitochondrial tRNAs (mt-tRNAs) are responsible for several and sometimes severe clinical phenotypes, classified among mitochondrial diseases. In addition, post-transcriptional modifications of mt-tRNAs in correlation with several stress signals can affect their stability similarly to what has been described for their nuclear-encoded counterparts. Many of the perturbations related to either point mutations or aberrant modifications of mt-tRNAs can lead to specific cleavage and the production of mitochondrial tRNA-derived fragments (mt-tRFs). Although mt-tRFs have been detected in several studies, the exact biogenesis steps and biological role remain, to a great extent, unexplored. Several mt-tRFs are produced because of the excessive oxidative stress which predominantly affects mitochondrial DNA integrity. In addition, mt-tRFs have been detected in various diseases with possible detrimental consequences, but also their production may represent a response mechanism to external stimuli, including infections from pathogens. Finally, specific point mutations on mt-tRNAs have been reported to impact the pool of the produced mt-tRFs and there is growing evidence suggesting that mt-tRFs can be exported and act in the cytoplasm. In this review, we summarize current knowledge on mitochondrial tRNA-deriving fragments and their possible contribution to gene expression regulation.
Mitochondria have evolved as the powerhouse of cells and the central nexus for major metabolic pathways that utilize oxygen for ATP production in eukaryotic cells. As such, they represent a major hub of the intracellular communication that define the cell’s fate. Mitochondria-related networking and the bidirectional pathways that rule it have long been identified to have profound effects on cell’s metabolism, bioenergetics, and homeostasis. However, several details regarding the molecular mechanisms and gene networks involved still remain unknown. Human mitochondrial DNA exists in multiple copies in each organelle and consists of 37 genes including 2 mt-rRNAs, 22 mutations in human mitochondrial tRNAs (mt-tRNAs), and 13 coding genes which are translated through specific decoding by mitochondrial ribosomes to the essential subunits of respiratory complexes (Anderson et al., 1981). The expression of the mitochondrial genes provides, among others, energy supply and antioxidant defense depending also on several sets of devoted nuclear-encoded genes that act either in the mitochondrial milieu or at the mitochondrial membrane. Nuclear-encoded non-coding RNAs are also known to indirectly affect mitochondrial function either through regulating the expression and post-translational modifications of the approximately 1,500 proteins targeted to mitochondria or through their direct import and interaction with proteins and RNAs inside mitochondria (Gusic and Prokisch, 2020). Several miRNAs or long non-coding RNAs, like SAMMSON, modulate mitochondrial function by affecting the import of proteins (Leucci et al., 2016; Vendramin et al., 2017; Jeandard et al., 2019; Kaliatsi et al., 2020). It should be noted that in human, although RNA import mechanisms into mitochondria have been proposed and involve PNPase (polynucleotide phosphorylase) activity located in the intermembrane space (reviewed in Vendramin et al., 2017 and Jeandard et al., 2019), an export mechanism is still elusive.
Current knowledge suggests that several perturbations in the regulation of the mitochondrial gene expression can lead to dysfunction of mitochondria. In turn, defective mitochondria are characterized by impaired energy production and depending on the broadness of the mutations, can lead to disorders with severe clinical symptoms that mainly affect the highly energy-demanding tissues, like brain, heart, and muscles. More specifically, mitochondrial DNA mutations, especially on mt-tRNAs, have been linked initially to syndromes like MERRF (myoclonic epilepsy with ragged red fibers), MELAS (mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) and cardiomyopathies (Gorman et al., 2016), and nowadays to the emergence of specific types of cancer (Stewart et al., 2015). The fact that many of the known mitochondrial diseases have their molecular basis on mutations in mt-tRNA genes indicates the central role of mitochondrial translation in the pathophysiology of the cells and the tissues. Point mutations in mt-tRNAs can affect the efficiency of the 5' and 3' processing, the epigenetic changes deriving from specific post-transcriptional modifications, the accuracy of tRNA aminoacylation and decoding during translation, and overall the mt-tRNA stability, resulting in many cases in severe inherited mitochondrial diseases (Yan et al., 2006; Zifa et al., 2007; Xue et al., 2019; Chujo and Tomizawa, 2021; Ji et al., 2021; Karasik et al., 2021). It should be noted that several mitochondrial tRNAs appear truncated in their mature form when compared to their cytosolic counterparts and in the case of tRNASer(AGY), the D-stem and loop are missing. In addition, different point mutations in the same mt-tRNA molecule can result in different diseases, like in the case of mt-tRNAGlu where specific mutations have been linked with maternally inherited diabetes and deafness, while others in the same gene with infantile transient mitochondrial myopathy (Webb et al., 2020). Moreover, mutations in mt-tRNAs can co-exist with mutations in genes encoding mitochondrial enzymes, an observation that highlights the phenotypic diversity among mitochondrial mutations with possible clinical significance (Zifa et al., 2008). The accurate transcription and maturation of mt-tRNAs are essential to successfully decode several codon deviations from the conventional universal genetic code, like the UGA stop codon which encodes for tryptophan, the isoleucine AUA codon which encodes for methionine, and the two arginine codons AGA and AGG which in mitochondria designate termination (Kummer and Ban, 2021). As a result, defects that affect the mt-tRNA stability and utilization as substrate can impact not only the mitochondrial translation rate but also the pool of available molecules leading to specific tRNA fragmentation that could affect the mitochondrial gene expression and potentially the crosstalk with the cytoplasm and the nucleus.
Although fragments of tRNAs were sporadically observed many years ago, the recent advance of sequencing technologies and bioinformatics tools brought tRNA-derived fragments (tRFs) to the spotlight, categorizing them in groups depending on their site of origin on tRNA molecules (Kumar et al., 2016). More specifically, tRFs are classified as tRF-1s, tRF-5s, tRF-3s, 5'-tRNA halves, 3' tRNA halves, or itRFs (internal tRFs) and are produced depending also on the external stimulus. More interestingly, each distinct class has been linked to specific biological function; global translation inhibition has been attributed mainly to 5' tRNA halves (but also to tRF-5s), translation repression of specific mRNAs mainly to tRF-3s, inhibition of specific gene transcription to tRF-5s, inhibition of viral RNA translation to tRF-1s, inhibition of apoptosis to 5' tRNA halves, and regulation of retrotransposon expression both to tRF-3s and tRF-5s (Schimmel, 2018; Su et al., 2020). In general, although tRFs production can be induced by stress, or the lack of specific post-transcriptional tRNA modifications, a certain number of tRFs has been proposed to act like miRNAs and to mediate gene silencing (Kumar et al., 2014; Skeparnias et al., 2020). Finally, although several known ribonucleases seem to contribute in the production of specific tRFs (see below), it is also likely that additional ribonucleolytic activities and/or biogenesis steps may mediate tRFs biogenesis (Su et al., 2020).
The advance of high throughput methodologies, such as next-generation sequencing and meta-analysis of existing data, has lately revealed the existence of mitochondrial tRNA-derived fragments (mt-tRFs) with possible correlation to pathological phenotypes. Strikingly, mt-tRFs appear as contributors to the synchronization of a series of essential cellular and mitochondrial biological processes, acting as “couriers” not only between nucleus and mitochondrion, but also between different cells of the same or different species (Saikia et al., 2014; Meseguer, 2021). Herein, the putative production pathways of mt-tRFs and their effect on gene expression regulation, along with the role of tRNA processing factors that may contribute to differential mt-tRF expression profiles in pathogenic phenotypes, are summarized.
Nowadays, the powerful methodological tools can provide an unprecedented opportunity to approach the nuclear-mitochondrial communication and the delicate coordination of nuclear- and mitochondrial-encoded factors, enzymes, and cofactors aiming to delineate the mechanisms that drive to specific mitochondrial diseases. Although at the beginning, tRFs were first considered tRNA degradation byproducts, later studies showed they are abundant in all species and are generated following a specific endonucleolytic cleavage of precursor or mature tRNAs (Grafanaki et al., 2019; Su et al., 2020). Today, tRFs represent important modulators of various processes, including translation, apoptosis, anti-viral defense, and response to nutrient starvation or oxidative stress. Interestingly, with the accumulation of sequencing data and the development of highly specialized bioinformatic platforms, it becomes evident that not only nuclear-encoded tRNAs but also mt-tRNAs can produce tRFs (Kumar et al., 2015; Zheng et al., 2016; Pliatsika et al., 2018; Looney et al., 2020). Of note, differential expression of mt-tRFs has been observed in cancer, as a result of mtDNA mutations and as a response to external stimuli, including infections from pathogens (Meseguer et al., 2019; Nätt et al., 2019; Karousi et al., 2020; Looney et al., 2020). Subsequently, mt-tRFs appear to actively participate in the intracellular communication and the mitochondrial pathophysiology, affecting also the cytoplasmic translation machinery (Loher et al., 2017; Meseguer et al., 2019; Looney et al., 2020). Finally, there is growing interest regarding the possible transportation of mt-tRFs across the mitochondrial membrane which raises further questions on which and how many ribonucleases and modifiers could be implicated to mt-tRF biogenesis.
So far, specific endoribonucleases have been identified that participate in tRF production. The first endoribonucleases identified were RNase ZL/ELAC2 and Angiogenin (ANG). RNase ZL/ELAC2 is responsible for the maturation of the 3' end of tRNAs through pre-tRNA cleavage downstream of the discriminator base and the only activity which is localized both in the nucleus and mitochondria. ANG, on the other hand, cleaves predominantly in the anticodon loop and is mainly responsible for stress-induced tRFs production (Lee et al., 2009; Yamasaki et al., 2009; Siira et al., 2018). Additional enzymes that have been identified to participate in the generation of tRFs include SLFN11 (Schlafen 11), SLFN13, and RNase L (Li et al., 2012, 2018; Donovan et al., 2017; Yang et al., 2018), but interestingly, none of these activities have been detected in mitochondria. Although, the cleavage site of SLFN11 is unknown, SLFN13 cleaves in the junction of the T and anticodon stems of tRNAs, while RNase L cleaves at the anticodon loop (Donovan et al., 2017; Li et al., 2018; Yang et al., 2018). Dicer is also implicated to the generation of tRFs but only in specific cases (Hasler et al., 2016; Kuscu et al., 2018). For example, precursor tRNAs that escape the La-dependent maturation pathway are folded into structures that are recognized by exportin 5 (Xpo5) as miRNA precursors and exported in the cytoplasm, where they are cleaved by DICER producing tRFs, which are then loaded to AGO proteins (Hasler et al., 2016). Aside from La that protects tRNAs from being cleaved, it was found recently that SLFN2 binds mature tRNAs upon T-cell activation and inhibits oxidative stress-induced tRF biogenesis by ANG, thus preventing global translation inhibition (Yue et al., 2021). However, in most cases, the mechanism for producing tRNA halves, tRF-5s, tRF-3s, and itRFs is still elusive and the question whether specific tRNA-halves production is tightly regulated remains open (Su et al., 2019, 2020). Given the current knowledge regarding the activities that mediate tRFs production from nuclear-encoded tRNAs, the respective activities that are responsible for mt-tRFs production are still unknown (Figure 1A). It should also be noted that it remains debatable whether and which mt-tRNAs are processed either in the mitochondrion or the cytoplasm to generate mt-tRFs. Enzymes that are targeted into mitochondria could affect mt-tRF biogenesis, like RNase ZL/ELAC2, which has been identified as a hotspot for mutations linked with mitochondrial diseases, such as hypertrophic cardiomyopathy but also with prostate cancer (Siira et al., 2018; Saoura et al., 2019). Although most of these mutations impair the efficient maturation of the 3' end of both the nuclear and mitochondrial tRNAs, the effect in mitochondria is more intense. For example, the R781H substitution in RNase ZL/ELAC2, which has been linked with prostate cancer and hypertrophic cardiomyopathy, mainly affects mt-tRNA processing while has no major differences on the nuclear pre-tRNA processing (Minagawa et al., 2005; Saoura et al., 2019). This effect could potentially lead to production of differential mt-tRFs levels and a broader effect in gene expression regulation, both inside and outside mitochondria. Another candidate involved in the mt-tRF generation is LACTB2 (Lactamase B2), a mitochondrially targeted ribonuclease that is able to cleave single-stranded RNAs in vitro (Levy et al., 2016). Although LACTB2 is mainly involved in mt-mRNA turnover, in vitro cleavage assays demonstrated that in the presence of a hairpin-structured RNA substrate, LACTB2 cleaves only in the loop region and may also target the T, D, or anticodon loops of mt-tRNAs, generating mt-tRFs. Given the putative existence that a mechanism that actively shuffles mitochondrial RNAs across the mitochondrial membrane to be processed in the cytoplasm by known endoribonucleases, the possibility of mt-tRFs production by cytoplasmic enzymes cannot be excluded (Bruni et al., 2017; Jády et al., 2018; Figure 1A). Interestingly, it was shown recently that silencing of either DICER or AGO2 downregulated the expression of mt-itRFGlu(UUC), mt-5'tRFLeu(UUA) carrying the A3243G mutation and mt-3'tRFVal(UAC) (Meseguer et al., 2019). This observation suggests that DICER could potentially cleave mt-tRNAs as well, however, whether this occurs in the cytoplasm or inside mitochondria is unclear (Figure 1A; Meseguer, 2021). The reason why AGO2 silencing leads to decreased expression of mt-tRFs is also not clear. Considering the possibility that AGO2-bound mt-tRFs exhibit increased half-lives, the absence of AGO2 could increase the susceptibility of mt-tRFs to degradation. Although the defined mitochondrial export mechanism is still elusive, it is apparent that apoptosis and autophagic turnover of mitochondria cause mt-tRNAs to translocate in the cytoplasm (Marnef et al., 2016; Jády et al., 2018). Under these conditions, several RNA-binding proteins, including AGO2, Y-box-binding proteins (YBX1 and YBX3), mRNA-binding proteins (SRSF1, SRSF2, SRSF3, hnRNP A1, and hnRNP H), and polypyrimidine-binding proteins, have been found to interact with a subset of mt-tRNAs in the cytoplasm. This interaction may indicate a possible mechanism via which, specific mt-tRNAs are processed by cytoplasmic ribonucleases, like ANG and DICER to produce mt-tRFs (Maniataki and Mourelatos, 2005; Marnef et al., 2016; Jády et al., 2018). However, as discussed above, additional ribonucleases may also be involved in cytosolic mt-tRF biogenesis.
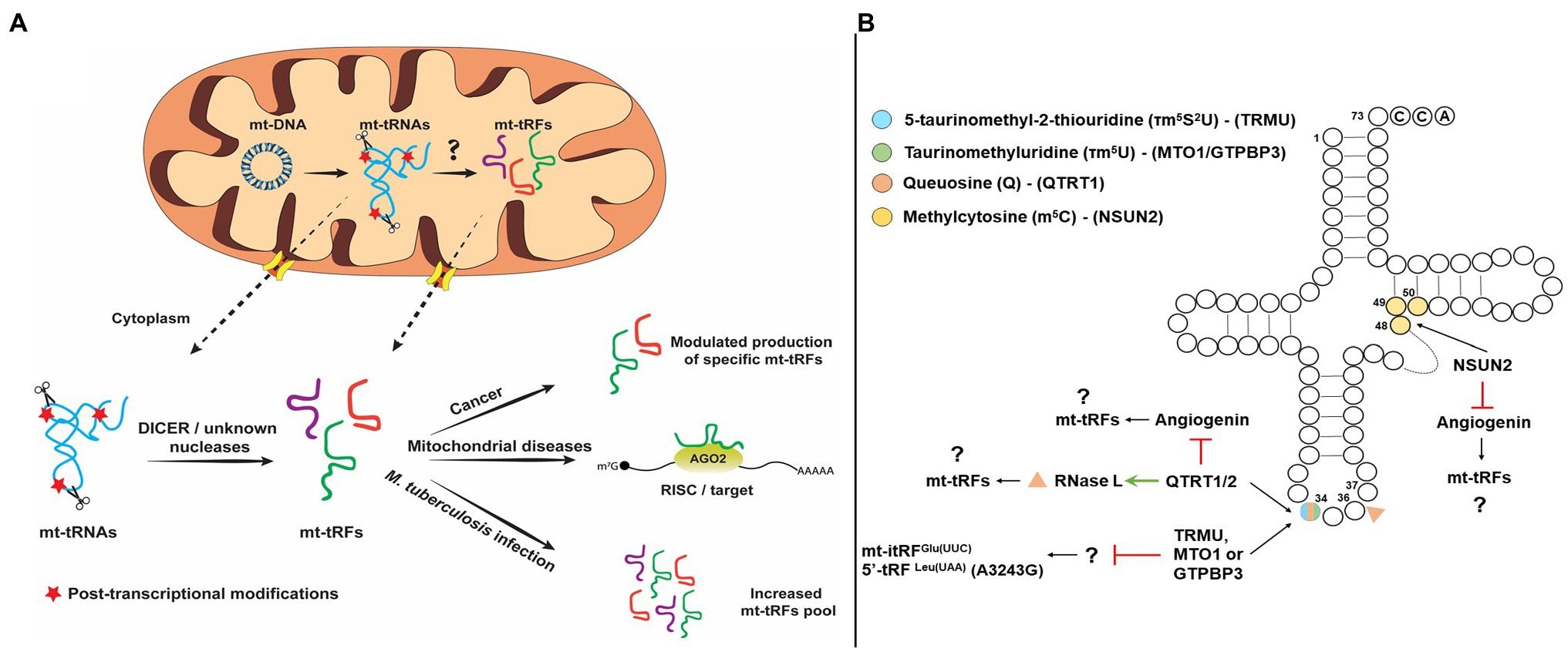
Figure 1. (Α) Schematic representation of putative pathways of mitochondrial tRNA-derived fragments (mt-tRFs) biogenesis and biological function and (Β) Hypothetic model of mt-tRNA cleavage regulation by post-transcriptional modifications.
Apart from the previously mentioned mechanisms, specific modification patterns on mt-tRNAs are key factors and not only contribute to structural stability and recognition by enzymes and proteins, but also act protectively against aberrant cleavage and fragmentation. More interestingly, lack of specific modifications has been linked with the production of specific tRFs under different conditions. Queuosine (Q) is an important modification of G34 of GUN anticodons and responsible for the efficient decoding of NAY codons in the cytosol and mitochondria (where N corresponds to any base and Y to pyrimidine) and lack of this modification affects the cleavage of both RNase L and ANG (Suzuki, 2021). More specifically, RNase L cleaves the in vitro-transcribed tRNAHis in three different sites; however, isolation of endogenous and modified with Q34 tRNAHis is only cleaved at position 36 (Donovan et al., 2017). On the other hand, Q34 inhibits ANG cleavage altogether (Wang et al., 2018). Thus, considering that Q34 modification status besides the decoding fidelity and efficiency affects also the cleavage specificity of mt-tRNAs and it may modulate mt-tRF production. Interestingly, the absence of an identified biosynthetic pathway for the micronutrient queuine in human, which is required for queuosine formation, suggests that the microbiome is mainly responsible for the existence of this modification (Müller et al., 2019). This observation provides a possible link between the role of the microbiome in tRF biogenesis. In the same line, a recent report that analyzed in detail the human tRNA modification landscape underlined the fact that several modifications occur near sites that are susceptible to potential pathogenic mutations. The presence of non-canonical nucleotides near modification sites could affect the binding affinity of modifying enzymes and/or affect their enzymatic efficiency, resulting in hypomodified and unstable mt-tRNAs, which are unable to participate efficiently in mitochondrial translation and are prone to cleavage (Suzuki et al., 2020). Fruit flies and mice lacking NSUN2 (NOP2/Sun methyltransferase 2) or DNMT2 (DNA methyltransferase 2), the methyltransferases that modify cytosines at position 5 (m5C), were found to bear increased levels of stress-induced tRFs that are produced by ANG (Figure 1B). The correlation of NSUN2 with other modification enzymes that modify mt-tRNAs regulates their stability and functional properties (Suzuki, 2021). NSUN2 has recently been detected in mitochondria and is responsible for introduction of m5 at C48, C49, or C50 of mt-tRNAs (Shinoda et al., 2019; Van Haute et al., 2019). Taking into account that NSUN2 expression is dynamically modulated in various physiological conditions, such as cancer, neurodegenerative diseases, and during differentiation, it is possible that differential methyltransferase activity in mitochondria may also drives the production of specific mt-tRFs (Blanco et al., 2014; Chellamuthu and Gray, 2020). Of note, queuosine hypomodification has been reported in several cancers and in multiple sclerosis via reduced activity of tRNA guanine transglycosylase (TRT) or via promoter methylation of the genes coding for the TRT subunits (QTRT1 and QTRT2) (Baranowski et al., 1994; Pathak et al., 2005; Varghese et al., 2017; Hayes et al., 2020).
Several tRNA modifications that are mitochondria specific-like 5-taurinomethyluridine (τm5U), 5-taurinomethyl-2-thiouridine (τm5S2U), and 5-formylcytidine (f5C) that could play an important role in the regulation of mt-tRNA cleavage. Silencing of either TRMU, GTPBP3, or MTO1 genes that code for enzymes that modify U34 of mt-tRNAs led to increased expression of mt-itRFGlu(UUC), indicating that the modification state of mt-tRNAs can modulate mt-tRF biogenesis (Figure 1B; Schaefer et al., 2010; Tuorto et al., 2012; Meseguer et al., 2019). Also, the mt-tRNALeu(UUR) harboring the MELAS mutation A3243G which is known to prevent the τm5 modification of U34, also leads to the upregulation of a mt-tRF-5 derived from mt-tRNALeu(UUR). Based on these observations, it has been proposed that lack of τm5U34 could drive the generation of specific mt-tRFs (Meseguer et al., 2019). Finally, τm5U frequency in cultured cell lines and in animal models is highly affected by dietary uptake of taurine (Asano et al., 2018). Upon taurine starvation in cultured cells and animal tissues, τm5U is replaced by cmnm5U (5-carboxymethylaminomethyluridine), which could potentially alter the cleavage patterns of mt-tRNAs by specific ribonucleases, leading to the production of differential mt-tRF profiles.
The role of tRFs in gene expression regulation has gradually drawn the interest, with specific tRFs deriving from nuclear-encoded tRNA species to be considered as novel diagnostic and prognostic biomarkers of several types of disorders, such as cancer, as well as possible druggable targets for cancer treatment (Srinivasan et al., 2019; Zhu et al., 2019). Although, the biological function of mt-tRFs has not been clearly defined, they could bear similar value (Table 1). A previous extensive analysis of sRNA-seq data from mitoplasts derived from 143B osteosarcoma cells revealed the existence of several sRNA (small RNA) species that map to mitochondrial tRNAs (Mercer et al., 2011). Interestingly, most of the mt-tRFs detected in that study correspond to 5' mt-tRFs but also to the region downstream of tRNA 3' cleavage side, which correspond to tRF-1s (Mercer et al., 2011). Moreover, the 5' mt-tRFs harbored several sequencing mismatches at positions known for being modified, indicating their origin from mature mt-tRNAs. Several studies have attempted to correlate the differential mt-tRFs patterns with various pathophysiological conditions. A reported correlation between differential abundance and length distribution of mt-tRFs between datasets from breast invasive carcinoma (BRCA; derived from The Cancer Genome Atlas project) and patient-derived lymphoblastoid cell lines (LCL) highlighted the importance of mt-tRFs patterns as biomarkers (Telonis et al., 2015). It is likely that specific mt-tRNAs produce only a specific type of mt-tRF (i.e., either 5'-tRF or 3'-tRF or i-tRF) while others can be fragmented to all types. Moreover, LCL and BRCA datasets contained not only different mt-tRF pools, but also different types of mt-tRFs generated from the same type of mt-tRNA, suggesting that the type of mt-tRF produced by each individual mt-tRNA is cancer type specific. A recent report interconnected the low overall survival of patients suffering from chronic lymphocytic leukemia (CLL) with the presence of an internal mt-tRF (Karousi et al., 2020). In this study, non-coding RNAs were quantified via bioinformatic analysis in CLL patients. Interestingly, a novel mt-tRF was identified derived from the mt-tRNAPhe bearing the anticodon GAA and termed i-tRFPhe(GAA). Specifically, i-tRFPhe(GAA) could serve as a potential prognostic biomarker in CLL, since its high expression level was linked with poor outcome of the disease (Karousi et al., 2020). Although these observations await experimental validation and more in-depth investigation to attribute its biological significance, they illustrate the dynamic nature of mt-tRF biogenesis, which may reflect to their distinct biological functions under different conditions or pathophysiologies. Moreover, apart from the apparent correlation of mt-tRFs with several cancer types, their role has been also emerged in infections from pathogens. Although most studies have focused on viral infections, it was recently reported that infection with Listeria monocytogenes and Mycobacterium tuberculosis affected the tRF production in macrophages and more importantly, infection by M. tuberculosis significantly affected the abundance of the mt-tRFs pool (Looney et al., 2020). The study suggested that this effect could be driven by the hypoxic conditions that usually accompany M. tuberculosis infections which, in turn, alter the mitochondrial physiology and subsequently the mt-tRF production.
Very recently, the role of diet as factor that induces mt-tRFs production was described in relation to human sperm motility. Human sperm responds rapidly to changes in a sugar-rich diet exhibiting increased motility and upregulation of specific nuclear and mt-tRFs. Strikingly, although both nuclear and mt-tRFs production are sugar sensitive, the high percentage of mt-tRFs could correlate to the improved sperm motility (Nätt et al., 2019). In addition, considering that the modification state of tRNAs can affect the tRFs biogenesis, it seems that the mt-tRNA epitranscriptome can also give rise to the mt-tRFome (Martínez-Zamora et al., 2015). For example, the prevalent mutation A3243G, which leads to the MELAS disease, can cause differential expression of several mt-tRFs. This mutation resides inside the mt-tRNALeu(UUR) and prevents the writing of the τm5U34 modification that stabilizes the U-G wobble pair and makes the mutated mt-tRNA unable to decode the UUG codon efficiently (Kirino et al., 2004). Notably, a 5' mt-tRF derived from the mutated mt-tRNALeu(UUR) was upregulated in MELAS cybrid cells, indicating that the loss of τm5 modification at U34 favors the production of this mt-tRF. Finally, specific mt-tRFs and particularly, mt-itRFGlu(UUC), mt-5'tRFLeu(UUA) carrying the A3243G mutation and another mt-5'tRFLeu(UAA), apart from being accumulated inside the mitochondria, have been detected also in the cytosol.
Similar to their cytoplasmic counterparts, mt-tRFs presumably can act like miRNAs to induce gene silencing and, in some cases, may share common mRNA targets (Meseguer et al., 2019; Skeparnias et al., 2020). Interestingly, the differential expressed mt-tRFs in MELAS cybrid cells are predicted to target several mRNAs involved in the regulation of striated and cardiac muscle contraction but also in the development of these tissues (Meseguer et al., 2019). Since myopathy and muscle spasms are common symptoms in MELAS, these mt-tRFs may facilitate disease progression by affecting muscle tissue function, on top of the mitochondrial dysfunction that results from reduced mitochondrial translation. In this context, a mt-itRF derived from mt-tRNAGlu(UUC) was found upregulated in MELAS cybrid cells. This specific tRF directly interacts to and downregulates MPC1 mRNA which encodes for the mitochondrial pyruvate carrier protein, leading to extracellular accumulation of lactate. Finally, mt-tRNASer(UCN) is a hotspot for mutations (T7510C, T7511C, and A7445G) that can lead to non-syndromic hearing loss, meaning that hearing impairment is the main, and usually the only symptom. It is known that inner-ear cells have structurally unique mitochondria which may be why this is usually the main symptom in individuals carrying this mtDNA mutation (Lesus et al., 2019). The pathogenesis of these mutations has been attributed to the reduced steady state levels of mt-tRNASer(UCN) because RNase ZL/ELAC2 cannot process these transcripts efficiently (Yan et al., 2006). One possibility that should be further explored is whether these mutations could result in the production of specific mt-tRFs deriving from mt-tRNASer(UCN) and whether can be related to the progression of non-syndromic hearing loss.
Collectively, current knowledge suggests that among the main factors that lead to detection of differential levels of mt-tRFs in cancer includes, among others, rewiring of mitochondrial metabolism through translation deregulation. The latter indicates that several types of mitochondrial dysfunction, such as loss of mitochondrial membrane potential and mitochondrial calcium overload, can affect mt-tRF production, via their impact on mtDNA transcription and translation and possibly via impaired efficiency of non-coding RNAs and proteins import. Aberrant translation could increase the availability of mt-tRNAs for cleavage thus leading to production of mt-tRFs. On the other hand, reduced mtDNA transcription could lead to a reduction of the mt-tRNA pool and subsequently to overall availability for mt-tRFs production. Moreover, alterations during import of important proteins that are targeted into mitochondria can also potentially lead to differential mt-tRFs levels and the modulation of the genes that are potential targets.
Mitochondria, although rather simple organelles, facilitate essential metabolic cascades and sense environmental cues to regulate energy production, manage oxidative stress, and serve an overall normal cellular function. Due to the dynamic crosstalk with other subcellular compartments and their profound effects on pathophysiology, mitochondria represent a unique field for studying several regulatory circuits that affect gene expression. During recent years, tRFs have been established as a novel class of small non-coding RNAs that predominantly have protective effect but can also play role in the deregulation of translation. Based on the central role of mitochondrial tRNAs during mitochondrial translation, the corresponding mt-tRFs could modulate gene expression regulation in a similar way to achieve fine-tuning of the communication with the nucleus.
It must be underlined that almost half the mutations in mtDNA occur on tRNA genes with various effects on mt-tRNA biogenesis, aminoacylation efficiency, and mRNA decoding (Zifa et al., 2007; Yarham et al., 2010; Suzuki, 2021). The increased instability of such mutated mt-tRNAs as well as of mt-tRNAs bearing aberrant modifications could be the leading cause for the differential mt-tRF production related to disease onset or progression. In addition to known point mutations on mt-tRNA genes or to deficiencies of ribonucleases and modifiers, external stimuli spanning from bacterial infections to specific nutrients can affect the homeostasis of mitochondria and therefore can directly affect mt-tRFs production. For instance, taurine administration has been shown to ameliorate stroke episodes in MELAS patients and is now used as a treatment regimen in Japan (Rikimaru et al., 2012; Ohsawa et al., 2019; Suzuki, 2021). Therefore, it is worth studying the effect of taurine intake on mt-tRF production in afflicted tissues of MELAS patients. It is evident that exploring the mt-tRF biogenesis and post-transcriptional modifications of both nuclear and mitochondrial tRNAs consist a critical step to unlock new RNA-mediated nuclear-mitochondrial circuits implicated in the cell metabolism and pathological conditions.
Apart from the undisputed existence of mt-tRFs and their correlation to specific human disorders, several questions regarding their biogenesis and role remain open. It is important to decipher how and which mt-tRNAs or mt-tRFs are exported from mitochondria but also in which subcellular compartment production of the majority of mt-tRF occurs. Import and export through the mitochondrial membrane currently remain a mystery. For this reason, experimental tools, such as specific RNA-aptamers, antisense oligonucleotides, or even delivery of free RNA, could be developed to carefully and reliably determine mt-tRFs spatiotemporal localization (Dovydenko et al., 2021). The putative mechanisms which control import and export of mt-tRNAs and mt-tRFs could be targeted to treat mitochondrial diseases or to ameliorate secondary effects. The identification of specific enzymatic activities that are involved in mt-tRF production will provide valuable knowledge and will allow the better understanding of the possible role of mt-tRFs during intracellular communication. Given that the majority of mt-tRFs is accumulated inside mitochondria, it should be delineated which ribonucleases get involved, whether they are transported inside mitochondria and which is the cleavage pattern of their mt-tRNA substrates. Finally, considering that mitochondria originated from endosymbiotic events with prokaryotes, and regarding the similarities between mitochondria and bacteria, ranging from genome organization, membrane structure, and protein synthesis to overall structure, elucidation of the mechanism of mt-tRF biogenesis could provide new knowledge on the potential roles and biogenesis pathways of bacterial tRFs and vice versa.
A-NS, EK, VS, and CS contributed to the manuscript writing and the figure design and approved the review for publication. All authors contributed to the article and approved the submitted version.
This research was funded by the “INSPIRED-The National Research Infrastructures on Integrated Structural Biology, Drug Screening Efforts and Drug target functional characterization” (MIS 5002550), which is implemented under the action “Reinforcement of the Research and Innovation Infrastructure,” funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund). A-NS is recipient of a fellowship from the “RIBOMAP” grant implemented under the Operational Program: Human Resources Development, Education and Lifelong Learning which is co-financed by the European Social Fund (ESF) and National Resources (MIS 5047175 to CS). EK is a recipient of a fellowship from the State Scholarships Foundation (IKY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H. L., Coulson, A. R., Drouin, J., et al. (1981). Sequence and organization of the human mitochondrial genome. Nature 290, 457–465. doi: 10.1038/290457a0
Asano, K., Suzuki, T., Saito, A., Wei, F., Ikeuchi, Y., Numata, T., et al. (2018). Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 46, 1565–1583. doi: 10.1093/nar/gky068
Baranowski, W., Dirheimer, G., Jakowicki, J. A., and Keith, G. (1994). Deficiency of queuine, a highly modified purine base, in transfer RNAs from primary and metastatic ovarian malignant tumors in women. Cancer Res. 54, 4468–4471.
Blanco, S., Dietmann, S., Flores, J. V., Hussain, S., Kutter, C., Humphreys, P., et al. (2014). Aberrant methylation of t RNA s links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039. doi: 10.15252/embj.201489282
Bruni, F., Lightowlers, R. N., and Chrzanowska-Lightowlers, Z. M. (2017). Human mitochondrial nucleases. FEBS J. 284, 1767–1777. doi: 10.1111/febs.13981
Chellamuthu, A., and Gray, S. G. (2020). The RNA Methyltransferase NSUN2 and its potential roles in cancer. Cell 9:1758. doi: 10.3390/cells9081758
Chujo, T., and Tomizawa, K. (2021). Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 1–27. doi: 10.1111/febs.15736 [Epub ahead of print]
Donovan, J., Rath, S., Kolet-Mandrikov, D., and Korennykh, A. (2017). Rapid RNase L–driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671. doi: 10.1261/rna.062000.117
Dovydenko, I., Meschaninova, M., Heckel, A. M., Tarassov, I., Venyaminova, A., and Entelis, N. (2021). Lipophilic conjugates for carrier-free delivery of RNA importable into human mitochondria. Methods Mol. Biol. 2277, 49–67. doi: 10.1007/978-1-0716-1270-5_4
Gorman, G. S., Chinnery, P. F., DiMauro, S., Hirano, M., Koga, Y., McFarland, R., et al. (2016). Mitochondrial diseases. Nat. Rev. Dis. Prim. 2:16080. doi: 10.1038/nrdp.2016.80
Grafanaki, K., Anastasakis, D., Kyriakopoulos, G., Skeparnias, I., Georgiou, S., and Stathopoulos, C. (2019). Translation regulation in skin cancer from a tRNA point of view. Epigenomics 11, 215–245. doi: 10.2217/epi-2018-0176
Gusic, M., and Prokisch, H. (2020). ncRNAs: new players in mitochondrial health and disease? Front. Genet. 11:95. doi: 10.3389/fgene.2020.00095
Hasler, D., Lehmann, G., Murakawa, Y., Klironomos, F., Jakob, L., Grässer, F. A., et al. (2016). The lupus autoantigen La prevents Mis-channeling of tRNA fragments into the human MicroRNA pathway. Mol. Cell 63, 110–124. doi: 10.1016/j.molcel.2016.05.026
Hayes, P., Fergus, C., Ghanim, M., Cirzi, C., Burtnyak, L., McGrenaghan, C. J., et al. (2020). Queuine micronutrient deficiency promotes Warburg metabolism and reversal of the mitochondrial ATP synthase in hela cells. Nutrients 12, 1–18. doi: 10.3390/nu12030871
Jády, B. E., Ketele, A., and Kiss, T. (2018). Dynamic association of human mRNP proteins with mitochondrial tRNAs in the cytosol. RNA 24, 1706–1720. doi: 10.1261/rna.066738.118
Jeandard, D., Smirnova, A., Tarassov, I., Barrey, E., Smirnov, A., and Entelis, N. (2019). Import of non-coding RNAs into human mitochondria: a critical review and emerging approaches. Cell 8:286. doi: 10.3390/cells8030286
Ji, Y., Nie, Z., Meng, F., Hu, C., Chen, H., Jin, L., et al. (2021). Mechanistic insights into mitochondrial tRNAAla 3'-end metabolism deficiency. J. Biol. Chem. 297:100816. doi: 10.1016/j.jbc.2021.100816
Kaliatsi, E. G., Giarimoglou, N., Stathopoulos, C., and Stamatopoulou, V. (2020). Non-coding RNA-driven regulation of rRNA biogenesis. Int. J. Mol. Sci. 21:9738. doi: 10.3390/ijms21249738
Karasik, A., Wilhelm, C. A., Fierke, C. A., and Koutmos, M. (2021). Disease-associated mutations in mitochondrial precursor tRNAs affect binding, m1R9 methylation, and tRNA processing by mtRNase P. RNA 27, 420–432. doi: 10.1261/rna.077198.120
Karousi, P., Adamopoulos, P. G., Papageorgiou, S. G., Pappa, V., Scorilas, A., and Kontos, C. K. (2020). A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clin. Biochem. 85, 20–26. doi: 10.1016/j.clinbiochem.2020.07.005
Kirino, Y., Yasukawa, T., Ohta, S., Akira, S., Ishihara, K., Watanabe, K., et al. (2004). Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. 101, 15070–15075. doi: 10.1073/pnas.0405173101
Kumar, P., Anaya, J., Mudunuri, S. B., and Dutta, A. (2014). Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12:78. doi: 10.1186/s12915-014-0078-0
Kumar, P., Kuscu, C., and Dutta, A. (2016). Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 41, 679–689. doi: 10.1016/j.tibs.2016.05.004
Kumar, P., Mudunuri, S. B., Anaya, J., and Dutta, A. (2015). tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145. doi: 10.1093/nar/gku1138
Kummer, E., and Ban, N. (2021). Mechanisms and regulation of protein synthesis in mitochondria. Nat. Rev. Mol. Cell Biol. 22, 307–325. doi: 10.1038/s41580-021-00332-2
Kuscu, C., Kumar, P., Kiran, M., Su, Z., Malik, A., and Dutta, A. (2018). tRNA fragments (tRFs) guide ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105. doi: 10.1261/rna.066126.118
Lee, Y. S., Shibata, Y., Malhotra, A., and Dutta, A. (2009). A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649. doi: 10.1101/gad.1837609
Lesus, J., Arias, K., Kulaga, J., Sobkiv, S., Patel, A., Babu, V., et al. (2019). Why study inner ear hair cell mitochondria? HNO 67, 429–433. doi: 10.1007/s00106-019-0662-2
Leucci, E., Vendramin, R., Spinazzi, M., Laurette, P., Fiers, M., Wouters, J., et al. (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522. doi: 10.1038/nature17161
Levy, S., Allerston, C. K., Liveanu, V., Habib, M. R., Gileadi, O., and Schuster, G. (2016). Identification of LACTB2, a metallo-β-lactamase protein, as a human mitochondrial endoribonuclease. Nucleic Acids Res. 44, 1813–1832. doi: 10.1093/nar/gkw050
Li, M., Kao, E., Gao, X., Sandig, H., Limmer, K., Pavon-Eternod, M., et al. (2012). Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491, 125–128. doi: 10.1038/nature11433
Li, M., Kao, E., Malone, D., Gao, X., Wang, J. Y. J., and David, M. (2018). DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 25, 1047–1058. doi: 10.1038/s41594-018-0142-5
Loher, P., Telonis, A. G., and Rigoutsos, I. (2017). MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 7:41184. doi: 10.1038/srep41184
Looney, M. M., Lu, Y., Karakousis, P. C., and Halushka, M. K. (2020). Mycobacterium tuberculosis infection drives mitochondria-biased dysregulation of host transfer RNA–derived fragments. J. Infect. Dis. 223, 1796–1805. doi: 10.1093/infdis/jiaa596
Maniataki, E., and Mourelatos, Z. (2005). Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11, 849–852. doi: 10.1261/rna.2210805
Marnef, A., Jády, B. E., and Kiss, T. (2016). Human polypyrimidine tract-binding protein interacts with mitochondrial tRNA(Thr) in the cytosol. Nucleic Acids Res. 44, 1342–1353. doi: 10.1093/nar/gkv1355
Martínez-Zamora, A., Meseguer, S., Esteve, J. M., Villarroya, M., Aguado, C., Enríquez, J. A., et al. (2015). Defective expression of the mitochondrial-tRNA modifying enzyme GTPBP3 triggers AMPK-mediated adaptive responses involving complex I assembly factors, uncoupling protein 2, and the mitochondrial pyruvate carrier. PLoS One 10:e0144273. doi: 10.1371/journal.pone.0144273
Mercer, T. R., Neph, S., Dinger, M. E., Crawford, J., Smith, M. A., Shearwood, A. M. J., et al. (2011). The human mitochondrial transcriptome. Cell 146, 645–658. doi: 10.1016/j.cell.2011.06.051
Meseguer, S. (2021). MicroRNAs and tRNA-derived small fragments: key messengers in nuclear–mitochondrial communication. Front. Mol. Biosci. 8:643575. doi: 10.3389/fmolb.2021.643575
Meseguer, S., Navarro-González, C., Panadero, J., Villarroya, M., Boutoual, R., Sánchez-Alcázar, J. A., et al. (2019). The MELAS mutation m.3243A>G alters the expression of mitochondrial tRNA fragments. Biochim. Biophys. Acta, Mol. Cell Res. 1866, 1433–1449. doi: 10.1016/j.bbamcr.2019.06.004
Minagawa, A., Takaku, H., Takagi, M., and Nashimoto, M. (2005). The missense mutations in the candidate prostate cancer gene ELAC2 do not alter enzymatic properties of its product. Cancer Lett. 222, 211–215. doi: 10.1016/j.canlet.2004.09.013
Müller, M., Legrand, C., Tuorto, F., Kelly, V. P., Atlasi, Y., Lyko, F., et al. (2019). Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 47, 3711–3727. doi: 10.1093/nar/gkz063
Nätt, D., Kugelberg, U., Casas, E., Nedstrand, E., Zalavary, S., Henriksson, P., et al. (2019). Human sperm displays rapid responses to diet. PLoS Biol. 17:e3000559. doi: 10.1371/journal.pbio.3000559
Ohsawa, Y., Hagiwara, H., Nishimatsu, S., Hirakawa, A., Kamimura, N., Ohtsubo, H., et al. (2019). Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicentre, open-label, 52-week phase III trial. J. Neurol. Neurosurg. Psychiatry 90, 529–536. doi: 10.1136/jnnp-2018-317964
Pathak, C., Jaiswal, Y. K., and Vinayak, M. (2005). Hypomodification of transfer RNA in cancer with respect to queuosine. RNA Biol. 2, 143–148. doi: 10.4161/rna.2.4.2417
Pliatsika, V., Loher, P., Magee, R., Telonis, A. G., Londin, E., Shigematsu, M., et al. (2018). MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the cancer genome atlas projects. Nucleic Acids Res. 46, D152–D159. doi: 10.1093/nar/gkx1075
Rikimaru, M., Ohsawa, Y., Wolf, A. M., Nishimaki, K., Ichimiya, H., Kamimura, N., et al. (2012). Taurine ameliorates impaired the mitochondrial function and prevents stroke-like episodes in patients with MELAS. Intern. Med. 51, 3351–3357. doi: 10.2169/internalmedicine.51.7529
Saikia, M., Jobava, R., Parisien, M., Putnam, A., Krokowski, D., Gao, X.-H., et al. (2014). Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 34, 2450–2463. doi: 10.1128/MCB.00136-14
Saoura, M., Powell, C. A., Kopajtich, R., Alahmad, A., Al-Balool, H. H., Albash, B., et al. (2019). Mutations in ELAC2 associated with hypertrophic cardiomyopathy impair mitochondrial tRNA 3'-end processing. Hum. Mutat. 40, 1731–1748. doi: 10.1002/humu.23777
Schaefer, M., Pollex, T., Hanna, K., Tuorto, F., Meusburger, M., Helm, M., et al. (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595. doi: 10.1101/gad.586710
Schimmel, P. (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58. doi: 10.1038/nrm.2017.77
Shinoda, S., Kitagawa, S., Nakagawa, S., Wei, F.-Y., Tomizawa, K., Araki, K., et al. (2019). Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 47, 8734–8745. doi: 10.1093/nar/gkz575
Siira, S. J., Rossetti, G., Richman, T. R., Perks, K., Ermer, J. A., Kuznetsova, I., et al. (2018). Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep. 19:e46198. doi: 10.15252/embr.201846198
Skeparnias, I., Anastasakis, D., Grafanaki, K., Kyriakopoulos, G., Alexopoulos, P., Dougenis, D., et al. (2020). Contribution of miRNAs, tRNAs and tRFs to aberrant Signaling and translation deregulation in lung cancer. Cancers 12:3056. doi: 10.3390/cancers12103056
Srinivasan, S., Yeri, A., Cheah, P. S., Chung, A., Danielson, K., De Hoff, P., et al. (2019). Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462. doi: 10.1016/j.cell.2019.03.024
Stewart, J. B., Alaei-Mahabadi, B., Sabarinathan, R., Samuelsson, T., Gorodkin, J., Gustafsson, C. M., et al. (2015). Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 11:e1005333. doi: 10.1371/journal.pgen.1005333
Su, Z., Kuscu, C., Malik, A., Shibata, E., and Dutta, A. (2019). Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3–mediated gene silencing. J. Biol. Chem. 294, 16930–16941. doi: 10.1074/jbc.RA119.009272
Su, Z., Wilson, B., Kumar, P., and Dutta, A. (2020). Noncanonical roles of tRNAs: tRNA fragments and Beyond. Annu. Rev. Genet. 54, 47–69. doi: 10.1146/annurev-genet-022620-101840
Suzuki, T. (2021). The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 11, 375–392. doi: 10.1038/s41580-021-00342-0
Suzuki, T., Yashiro, Y., Kikuchi, I., Ishigami, Y., Saito, H., Matsuzawa, I., et al. (2020). Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11:4269. doi: 10.1038/s41467-020-18068-6
Telonis, A. G., Loher, P., Honda, S., Jing, Y., Palazzo, J., Kirino, Y., et al. (2015). Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 6, 24797–24822. doi: 10.18632/oncotarget.4695
Tuorto, F., Liebers, R., Musch, T., Schaefer, M., Hofmann, S., Kellner, S., et al. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905. doi: 10.1038/nsmb.2357
Van Haute, L., Lee, S.-Y., McCann, B. J., Powell, C. A., Bansal, D., Vasiliauskaitė, L., et al. (2019). NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 47, 8720–8733. doi: 10.1093/nar/gkz559
Varghese, S., Cotter, M., Chevot, F., Fergus, C., Cunningham, C., Mills, K. H., et al. (2017). In vivo modification of tRNA with an artificial nucleobase leads to full disease remission in an animal model of multiple sclerosis. Nucleic Acids Res. 45, 2029–2039. doi: 10.1093/nar/gkw847
Vendramin, R., Marine, J., and Leucci, E. (2017). Non-coding RNA s: the dark side of nuclear–mitochondrial communication. EMBO J. 36, 1123–1133. doi: 10.15252/embj.201695546
Wang, X., Matuszek, Z., Huang, Y., Parisien, M., Dai, Q., Clark, W., et al. (2018). Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313. doi: 10.1261/rna.067033.118
Webb, B. D., Diaz, G. A., and Prasun, P. (2020). Mitochondrial translation defects and human disease. J. Transl. Genet. Genomics 4, 71–80. doi: 10.20517/jtgg.2020.11
Xue, L., Chen, Y., Tang, X., Yao, J., Huang, H., Wang, M., et al. (2019). A deafness-associated mitochondrial DNA mutation altered the tRNASer(UCN) metabolism and mitochondrial function. Mitochondrion 46, 370–379. doi: 10.1016/j.mito.2018.10.001
Yamasaki, S., Ivanov, P., Hu, G., and Anderson, P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42. doi: 10.1083/jcb.200811106
Yan, H., Zareen, N., and Levinger, L. (2006). Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the transfer ribonuclease Z cleavage site, processing kinetics, and substrate secondary structure. J. Biol. Chem. 281, 3926–3935. doi: 10.1074/jbc.M509822200
Yang, J.-Y., Deng, X.-Y., Li, Y.-S., Ma, X.-C., Feng, J.-X., Yu, B., et al. (2018). Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 9:1165. doi: 10.1038/s41467-018-03544-x
Yarham, J. W., Elson, J. L., Blakely, E. L., Mcfarland, R., and Taylor, R. W. (2010). Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA 1, 304–324. doi: 10.1002/wrna.27
Yue, T., Zhan, X., Zhang, D., Jain, R., Wang, K., Choi, J. H., et al. (2021). SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell–mediated immunity. Science 80:eaba4220. doi: 10.1126/science.aba4220
Zheng, L.-L., Xu, W.-L., Liu, S., Sun, W.-J., Li, J.-H., Wu, J., et al. (2016). tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 44, W185–W193. doi: 10.1093/nar/gkw414
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., et al. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18:74. doi: 10.1186/s12943-019-1000-8
Zifa, E., Giannouli, S., Theotokis, P., Stamatis, C., Mamuris, Z., and Stathopoulos, C. (2007). Mitochondrial tRNA mutations: clinical and functional perturbations. RNA Biol. 4, 38–66. doi: 10.4161/rna.4.1.4548
Keywords: mitochondrial tRNA-derived fragments, mitochondrial tRNAs, tRNA-derived fragments, ncRNAs, mitochondria
Citation: Shaukat A-N, Kaliatsi EG, Stamatopoulou V and Stathopoulos C (2021) Mitochondrial tRNA-Derived Fragments and Their Contribution to Gene Expression Regulation. Front. Physiol. 12:729452. doi: 10.3389/fphys.2021.729452
Received: 23 June 2021; Accepted: 09 August 2021;
Published: 03 September 2021.
Edited by:
Nina Entelis, Génomique et microbiologie (GMGM), FranceReviewed by:
Pooja Jadiya, Temple University, United StatesCopyright © 2021 Shaukat, Kaliatsi, Stamatopoulou and Stathopoulos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constantinos Stathopoulos, Y3N0YXRoQG1lZC51cGF0cmFzLmdy; Vassiliki Stamatopoulou, di5zdGFtQHVwYXRyYXMuZ3I=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.