- 1Beijing Key Laboratory of Environment Friendly Management on Fruit Diseases and Pests in North China, Institute of Plant and Environment Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Chemosensory proteins (CSPs) have been identified in the sensory tissues of various insect species and are believed to be involved in chemical communication in insects. However, the physiological roles of CSPs in Halyomorpha halys, a highly invasive insect species, are rarely reported. Here, we focused on one of the antennal CSPs (HhalCSP15) and determined whether it was involved in olfactory perception. Reverse transcription PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR) analysis showed that HhalCSP15 was enriched in nymph and male and female adult antennae, indicating its possible involvement in the chemosensory process. Fluorescence competitive binding assays revealed that three of 43 natural compounds showed binding abilities with HhalCSP15, including β-ionone (Ki=11.9±0.6μM), cis-3-hexen-1-yl benzoate (Ki=10.5±0.4μM), and methyl (2E,4E,6Z)-decatrienoate (EEZ-MDT; Ki=9.6±0.8μM). Docking analysis supported the experimental affinity for the three ligands. Additionally, the electrophysiological activities of the three ligands were further confirmed using electroantennography (EAG). EEZ-MDT is particularly interesting, as it serves as a kairomone when H. halys forages for host plants. We therefore conclude that HhalCSP15 might be involved in the detection of host-related volatiles. Our data provide a basis for further investigation of the physiological roles of CSPs in H. halys, and extend the olfactory function of CSPs in stink bugs.
Introduction
Many insects rely on their sense of smell to locate food sources, search for mating partners, select oviposition sites, and avoid predators (Metcalf and Kogan, 1987; Takken, 1991; Bruce et al., 2005; Ebrahim et al., 2015). The detection of olfactory signals in insects is performed by olfactory receptor neurons located in olfactory sensilla, which are present on the antennae and other head appendices. Olfactory sensilla are perforated by numerous pores, forming a hollow structure filled with aqueous lymph that harbors the dendritic branches of olfactory receptor neurons and contains abundant small soluble binding proteins (Maida et al., 1993; Steinbrecht, 1997). In the initial stage of olfactory reception, odorants enter the olfactory sensillum cavity through pore canals, and are transported by soluble binding proteins to the olfactory receptor on dendrite membranes (Vogt and Riddiford, 1981; Kaissling, 2009; Leal, 2013).
Odorant binding proteins (OBPs) and chemosensory proteins (CSPs) are the two main types of soluble binding proteins. A large body of evidence from different approaches has extensively documented that OBPs bind to pheromones and odorants, with different degrees of affinity and selectivity for different OBPs (Pelosi et al., 2014; Zhang et al., 2017; Huang et al., 2018; Wang et al., 2020; Rihani et al., 2021). In several instances, it was demonstrated that OBPs are involved not just in detecting olfactory stimuli, but also in modulating stimulus sensitivity in the olfactory system (Xu et al., 2005; Larter et al., 2016; Gonzalez et al., 2020). CSPs may perform functions similar to OBPs in the olfactory system.
Insect CSPs are known as olfactory segment D (OS-D) or A-10 before being named as CSPs because of their high expression in chemosensory organs (McKenna et al., 1994; Pikielny et al., 1994; Angeli et al., 1999). CSPs are smaller than OBPs (100–120 residues) and bear no sequence similarity to OBPs. They present a motif of four conserved cysteines linked by disulfide bridges (Angeli et al., 1999). The three-dimensional (3D) structure of CSP protein is composed of six α-helices that define a hydrophobic cavity (Lartigue et al., 2002). As an olfactory protein, CSPs have been studied in Lepidoptera, Hemiptera, Hymenoptera, and Coleoptera (Gu et al., 2012; Sun et al., 2014; Peng et al., 2017; Fu et al., 2020). CSPs in the alfalfa plant bug Adelphocoris lineolatus have been implicated in mediating host recognition (Gu et al., 2012; Sun et al., 2015). However, numerous studies have shown that the expression of CSPs is not restricted to the antennae, and many CSP genes are expressed in other parts of the insect body, with functions different from olfaction. In non-olfactory tissues, they are believed to be involved in development, pheromone delivery, nutrient absorption, insecticide resistance, vision, and immune response (Nomura et al., 1992; Forêt et al., 2007; Maleszka et al., 2007; Bos et al., 2010; Xuan et al., 2015; Pelosi et al., 2017).
The brown marmorated stink bug Halyomorpha halys (Stål; Hemiptera: Pentatomidae), which is native to Asia, is an invasive pest that in the last few decades has rapidly spread globally, including in the United States, Canada, and Europe (Hoebeke and Carter, 2003; Wermelinger et al., 2008; Leskey and Nielsen, 2017). In its native and introduced range, H. halys feeds on more than 100 crops, and it has become a destructive pest of many crops in the world (Lee et al., 2013; Haye et al., 2015; Kriticos et al., 2017; Leskey and Nielsen, 2017). Being an important invasive pest worldwide, there are many studies examining the chemical ecology of H. halys (Khrimian et al., 2014; Harris et al., 2015; Leskey et al., 2015; Weber et al., 2017). In H. halys, antennal transcriptomic approaches have already led to the identification of 17 CSP genes (Sun et al., 2020), which now await functional characterization.
In this study, to examine the potential role of one antennal CSP (HhalCSP15) in olfaction perception, extensive expression profiling of HhalCSP15 transcripts was performed using semi-quantitative reverse transcription PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR) methods among different tissues in nymph and male and female adult stages. We further expressed HhalCSP15 in vitro and determined its binding affinities for 43 volatiles in fluorescence binding assays. Further, homology modeling and molecular docking were applied for predicting the key amino acids of HhalCSP15 that bind candidate ligands. In addition, electrophysiological activities of HhalCSP15 ligands were confirmed using electroantennography (EAG) recordings.
Materials and Methods
Insect Culture, Tissue Collection, Total RNA Isolation, and cDNA Synthesis
Overwintering H. halys adults were collected from Beijing Xishan National Forest Park, Beijing, China. The laboratory colony was established in plastic containers (20cm×13cm×8cm), which were maintained at 25±1°C, 60±10% relative humidity, and a 16L:8D photoperiod. The adults and nymphs were reared on green beans. Different tissues from third instar nymphs (antennae, mouthparts, heads, thoraxes, abdomens, and legs) and 1- to 3-day-old female and male adults (antennae, mouthparts, heads, thoraxes, abdomens, legs, and wings) were collected. All tissues were immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation.
Total RNA was extracted from different tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. The integrity and quantity of RNA samples were checked using 1.2% agarose gel electrophoresis and a NanoPhotometer N60 (Implen, München, Germany), respectively. cDNA from different tissues was synthesized from 2μg of RNA using the Fast Quant RT kit with gDNase (Tiangen, Beijing, China) for gene cloning and tissue expression pattern analyses.
Verification of the HhalCSP15 Sequence by Cloning and Sequencing
Gene-specific primers were designed to clone the open reading frame (ORF) of HhalCSP15. PCR was performed using one unit of KOD DNA polymerase (Taihe, Beijing, China) and 200ng cDNA under the following conditions: denaturation at 94°C for 2min, followed by 30cycles of denaturation at 94°C for 20s, annealing at 55°C for 30s, and extension at 68°C for 1min. The final extension step was at 68°C for 5min. The PCR products were cloned into a pCloneEZ-Blunt vector (Taihe, Beijing, China), and cloned products were sequenced using the M13 primer.
Reverse Transcription PCR
The expression of HhalCSP15 in different tissues of nymphs and male and female adults was analyzed by RT-PCR using Taq DNA polymerase (Biomed, Beijing, China). Each PCR volume (25μl) contained 200ng of cDNA from different tissues and was used as a template. The following cycling conditions were applied: 94°C for 4min, and for the subsequent 30cycles: 94°C for 30s, 55°C for 30s, and 72°C for 45s. The final extension step was at 72°C for 5min. The elongation factor 1-α (EF-1α, XM_014414739.2) was employed to assess the cDNA integrity for all samples. The amplification products were checked on 1.2% agarose gels. For each gene, one amplification product was sequenced to confirm its identity. The gene-specific primers were designed using Primer 31 and are listed in Supplementary Table S1.
Quantitative Real-Time PCR
The relative transcript abundance of HhalCSP15 in the antennae, mouthparts, and legs of nymphs and male and female adults was determined by qRT-PCR. qRT-PCR was conducted using an ABI Prism 7500 System (Applied Biosystems, Carlsbad, CA, United States) and SYBR Green SuperReal PreMix Plus (TianGen, Beijing, China). Each qRT-PCR reaction was conducted in a 20μl reaction mixture containing 10μl of 2× SuperReal PreMix Plus, 1μl (200ng) of sample cDNA, 0.4μl of 50× ROX Reference Dye, and 6.1μl of sterilized ultrapure water. Each qRT-PCR experiment was performed using three biological replicates, and each biological replicate was assessed three times. The ubiquitin conjugation factor E4 A (Ubiquitin, XM_014429239.2) and EF-1α were used as endogenous controls to normalize the target gene expression and correct for any sample-to-sample variation.
The comparative 2−∆∆CT method (Livak and Schmittgen, 2001) was used to calculate the relative transcript levels in each tissue. The primers of the target and reference genes are listed in Supplementary Table S1. The specificity of each primer set was validated by melting curve analysis, and the efficiency was calculated by analyzing the standard curves with a 5-fold cDNA dilution series. The comparative analyses of HhalCSP15 expression among different tissues and developmental stages were conducted with one-way ANOVA, followed by Tukey’s honestly significant difference (HSD) test using SPSS Statistics 18.0 (SPSS Inc., Chicago, IL, United States).
Expression and Purification of Recombinant HhalCSP15
HhalCSP15 was PCR-amplified using gene-specific primers (Supplementary Table S1). The PCR products were first subcloned into a T vector (Taihe, Beijing, China) and then into the bacterial expression vector pET30a (+; Novagen, Madison, WI, United States) between the NdeI and XhoI sites, and were verified by sequencing. The plasmids containing the correct insert were transformed into BL21 (DE3) competent cells. The protein was expressed in LB at 18°C for 16h through induction with 1mM of isopropyl-β-D-thiogalactopyranoside (IPTG). The cultures were harvested by centrifugation and resuspended in a 50mM Tris buffer (pH 7.4). After sonication and centrifugation, the recombinant proteins, which were mainly present in the supernatant, were purified by a standard Ni column (GE Healthcare, Waukesha, WI, United States). The His-tag was removed using a recombinant enterokinase (Novagen) following the manufacturer’s protocol. Purified HhalCSP15 was dialyzed in the Tris buffer, and its concentration was determined by the Bradford method (Bradford, 1976).
Fluorescence Competitive Binding Assays
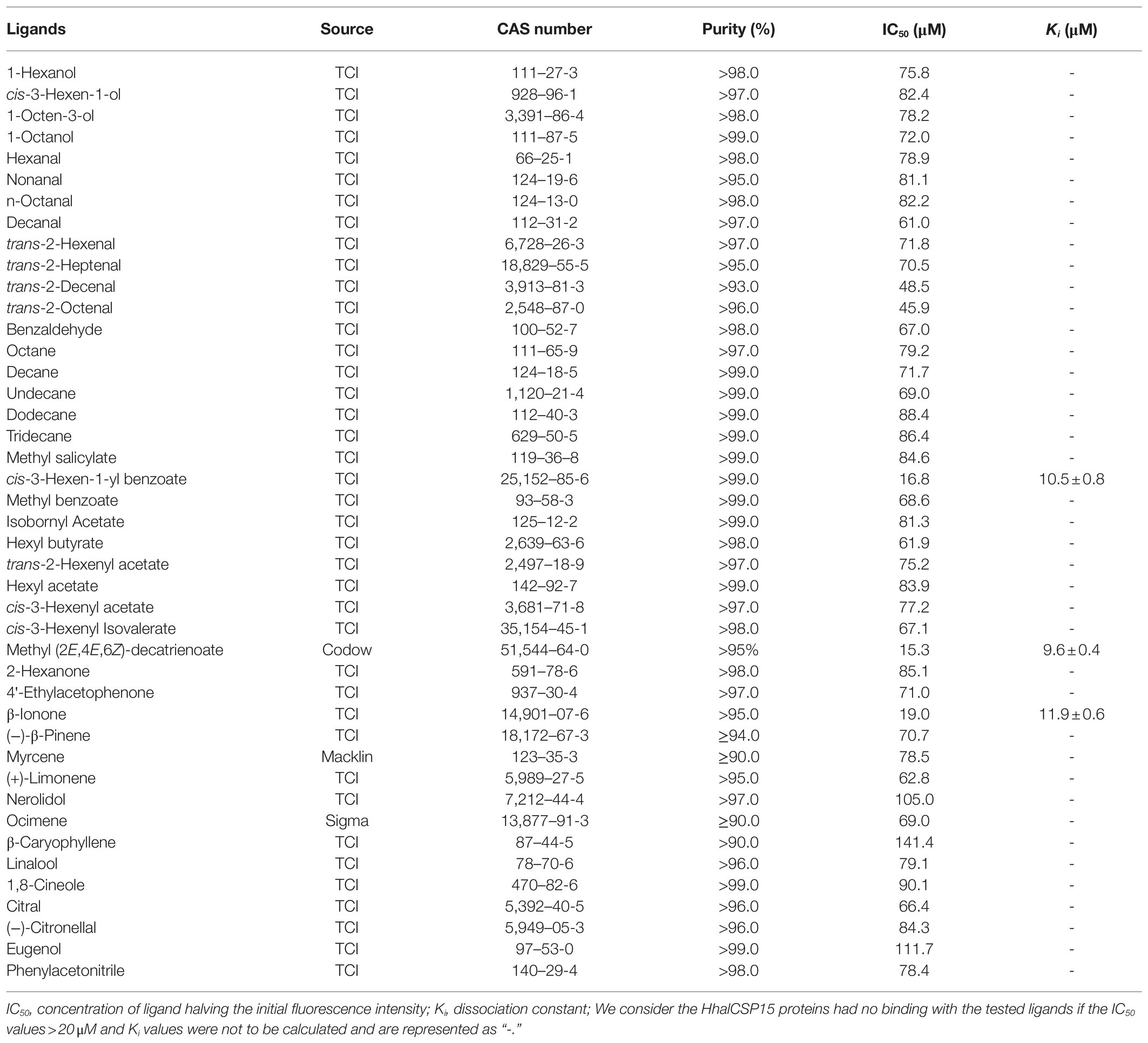
The binding abilities of HhalCSP15 to 43 volatiles were measured on an F-380 fluorescence spectrophotometer (Tianjin, China) using 10-nm slits and a 1-cm light path. As the fluorescent probe, N-phenyl-1-naphthylamine (1-NPN) was excited at the wavelength of 337nm, and emission spectra were recorded between 390 and 530nm. To measure the affinity of 1-NPN to the HhalCSP15 protein, a 2-μM solution of purified protein in 50-mM Tris-HCl at pH 7.4 was titrated with aliquots of 1-mM 1-NPN dissolved in methanol to final concentrations ranging from 2 to 16μM.
Competitive binding was measured by titration of the solution of both HhalCSP15 protein and 1-NPN at a concentration of 2μM by adding aliquots of 1-mM methanol solution of ligand to final concentrations of 2–20μM. Dissociation constants of the competitors were calculated by the equation Ki=IC50/(1+[1-NPN]/K1-NPN), where IC50 is the concentration of ligands halving the initial fluorescence value of 1-NPN, [1-NPN] is the free concentration of 1-NPN, and K1-NPN is the dissociation constant of the HhalCSP15/1-NPN complex. The experiments were performed in triplicate, excepting the ligands that did not show significant binding, which were analyzed in single experiments.
3D Structure Modeling and Molecular Docking
A suitable template for 3D modeling was identified using a sequence of similar searches at PSI-BLAST against sequences from the Protein Data Bank.2 Because of high sequence similarity with the HhalCSP15 protein, the high-resolution structure of a CSP from Mamestra brassicae (PDB: 1N8V) was selected for homology modeling using MODELLER 9.25.3 The structure refinement of the protein model was achieved by energy minimization via molecular dynamic simulation (MD) using GROMACSv5.0.7 with AMBER99SB force field (Abraham et al., 2015). A Ramachandran plot was performed to evaluate the rationality of the established 3D model using the online tool PROCHECK.4 The ligands were 3D optimized in ChemDraw 3D (PerkinElmer, Waltham, MA, United States) and refined with energy minimization. Ligands were docked to the model of HhalCSP15 using Autodock Vina.5 The best binding modes were selected according to the lowest free binding energy (kcalmol−1). The docked models of proteins interacting with ligands were displayed using PYMOL.6
Electrophysiological Recordings
The antennal responses of H. halys to the three ligands of HhalCSP15 were evaluated using conventional EAG methods as described in previous study (Wang et al., 2020). Briefly, antennae of the third instar nymphs and 1- to 3-day old male and female adults were dissected, and a few terminal segments at the distal end were excised. The treated antennae were attached to electrode holders with electrode gel. Ten microliters of tested chemicals (100μg/μl, diluted in paraffin oil) were applied to filter paper strips (1.0cm×2.0cm) and inserted into a glass Pasteur pipette as a cartridge. The test cartridge was connected to a stimulus controller (CS-55; Syntech, Kirchzarten, Germany) that generated a 0.5s stimulus at 30s intervals, with a constant flow of 10ml/s. The signals generated by the antennae were recorded using EAG Pro (Syntech). A blank stimulus (solvent control) was presented before testing the compound. For each compound, EAG responses were recorded from eight antennae of different insects. The EAG responses elicited by the test odor stimuli were corrected by subtracting EAG response from the solvent control. The corrected EAG data were statistically analyzed using ANOVA followed by Tukey’s HSD test.
Results
Sequence Analysis of HhalCSP15
The nucleotide sequence of HhalCSP15 was verified by molecular cloning and sequencing. Analysis of the HhalCSP15 sequence revealed full-length ORFs consisting of 366 nucleotides that encode 121 amino acid residues. At its N-terminus, HhalCSP15 is predicted to contain a signal peptide consisting of 19 amino acid residues (Supplementary Figure S1). The predicted molecular weight of HhalCSP15 protein was 11.91kDa, and the isoelectric point was 7.76. HhalCSP15 had the typical four-cysteine signature and fit the motif pattern of C1-X6-8-C2-X16-21-C3-X2-C4 of insect CSPs (Zhou et al., 2006).
Expression Profiles of HhalCSP15
We used RT-PCR to analyze the tissue-specific expression of the HhalCSP15 transcript in different nymph and adult tissues. The EF-1α gene was constitutively expressed in all tissues, thereby providing a stable control for the integrity of the cDNA templates (Figure 1). HhalCSP15 was detected specifically in the antennae of nymph and male and female adult, although minor bands were detected in other tissues, such as mouthpart and leg (Figure 1).
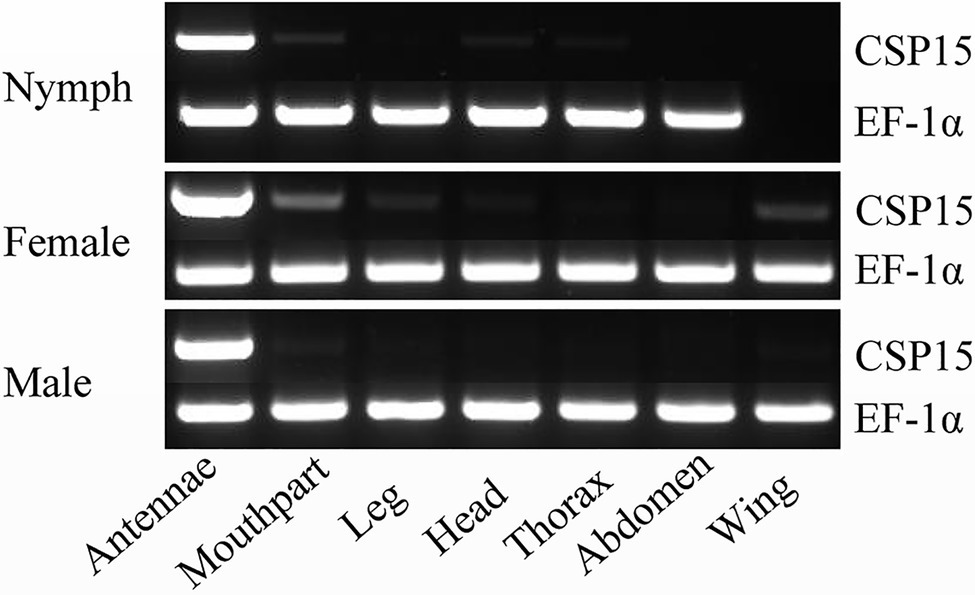
Figure 1. Tissue-specific expression of HhalCSP15 in different stages (nymph and male and female adult) measured by reverse transcription PCR (RT-PCR). Elongation factor 1-α (EF-1α) was used as a control gene.
Quantitative real-time PCR was used to measure the HhalCSP15 transcript levels in different tissues. The HhalCSP15 transcript was expressed significantly higher in the antennae than in other tissues; it was approximately 1,803, 3,023, and 2,130 times higher in the antennae than in the leg of nymphs and female and male adults, respectively. The expression of the HhalCSP15 gene was also detected in the mouthpart at 11.6-, 7.6-, and 2.4-fold higher than the leg of nymphs and female and male adults, respectively (Figure 2A). Furthermore, the expression levels were approximately 1.7- and 1.3-fold higher in the female and male adult antennae than in the nymph antennae (Figure 2B).
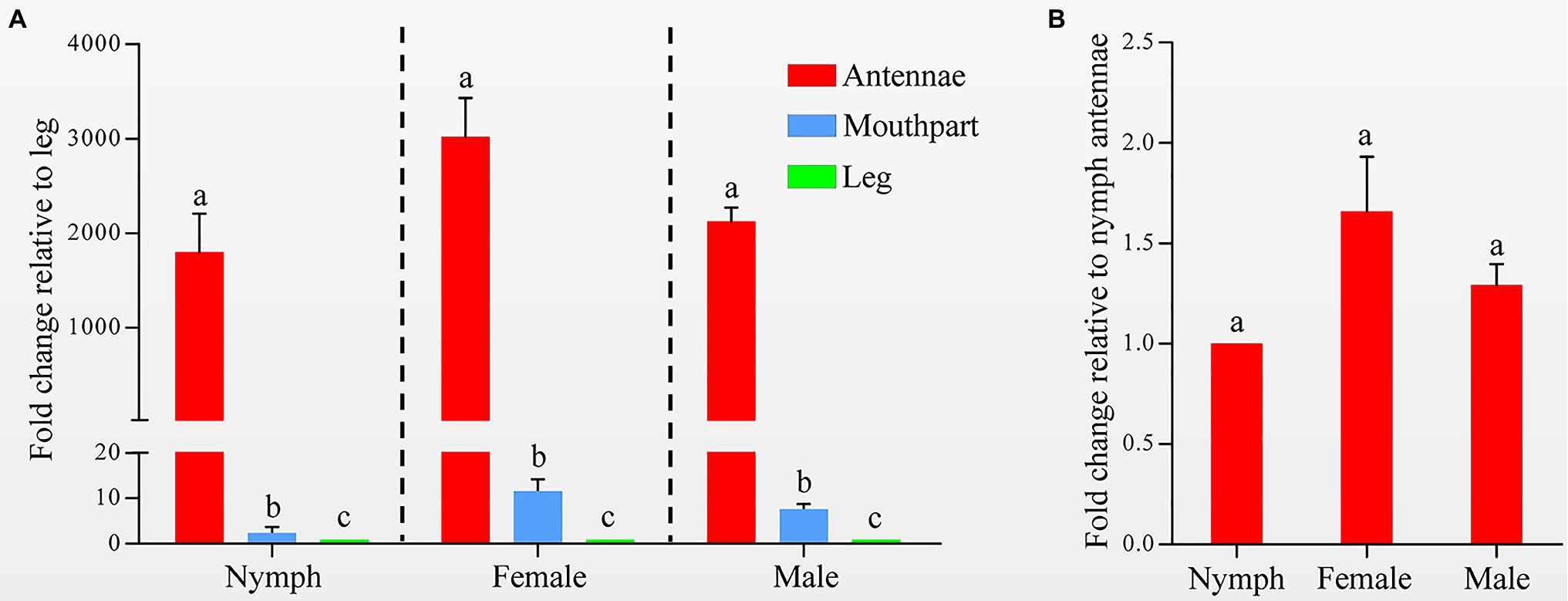
Figure 2. (A) Expression profiles of the HhalCSP15 gene in different tissues (antennae, mouthpart, and leg) measured by quantitative real-time PCR (qRT-PCR). The fold changes are relative to the transcript levels in the leg. (B) Relative transcript levels of the HhalCSP15 gene in the antennae at different stages. The fold changes are relative to the transcript levels in the antennae of nymph. Reference genes: EF-1α and Ubiquitin. The error bar and different letters represent the SE and significant differences, respectively (p<0.05).
Binding Characteristic of Recombinant HhalCSP15
The specific expression of HhalCSP15 in the antennae of nymphs and adults suggests that HhalCSP15 is potentially involved in peripheral olfactory reception for H. halys. To screen the putative ligands for HhalCSP15, we first expressed HhalCSP15 in a bacterial system. The protein was purified by affinity chromatography on Ni columns and was then used for ligand-binding experiments. The size and purity of the recombinant protein were examined by SDS–PAGE (Figure 3).
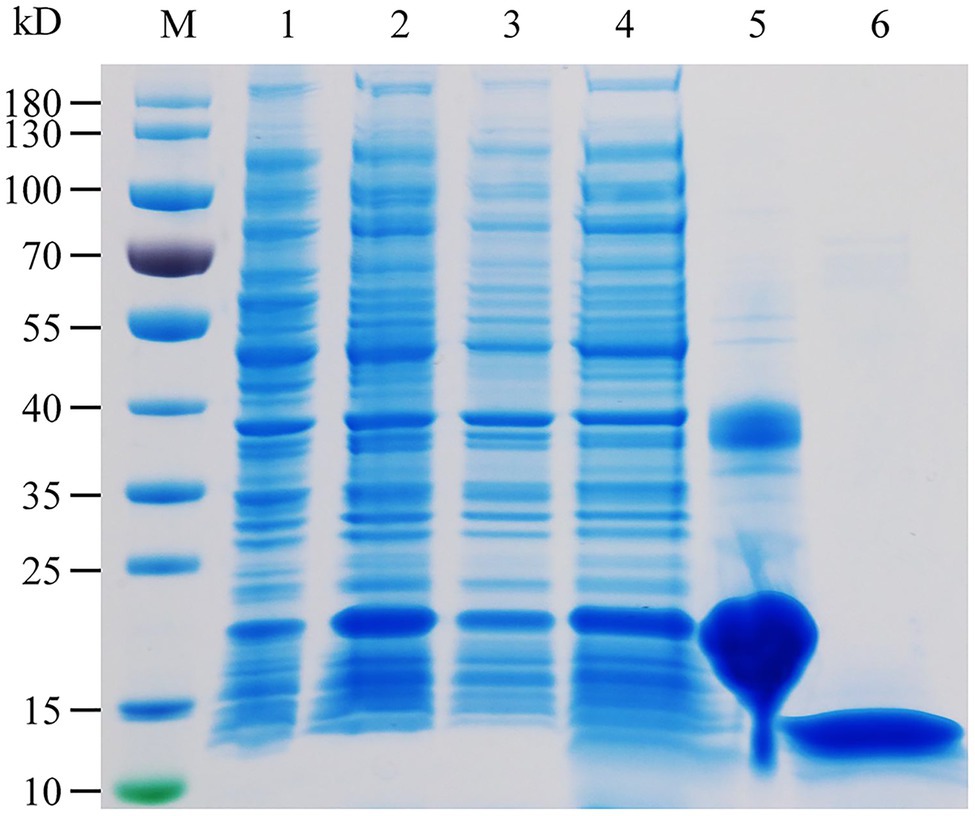
Figure 3. SDS-PAGE analysis of the recombinant HhalCSP15. M: molecular weight markers, 1: cell pellet before induction with IPTG, 2: cell pellet after induction, 3: pellet after sonication, 4: supernatant after sonication, 5: protein purified by affinity chromatography, and 6: purified protein after digestion with enterokinase.
We measured the protein affinity to 43 volatile compounds in competitive binding experiments using 1-NPN as a fluorescent probe. First, the affinity constant was measured for HhalCSP15 to 1-NPN (Figure 4A). HhalCSP15 binds reversibly to 1-NPN with a dissociation constant of 9.36μM, which indicates that 1-NPN is a suitable fluorescent reporter. HhalCSP15 displayed a relatively specific binding spectrum; of the 43 tested odorants, only three compounds showed binding affinities for HhalCSP15: β-ionone, cis-3-hexen-1-yl benzoate, and methyl (2E,4E,6Z)-decatrienoate (EEZ-MDT), which had binding affinities of 11.9±0.6, 10.5±0.4, and 9.6±0.8μM, respectively (Figure 4B; Table 1).
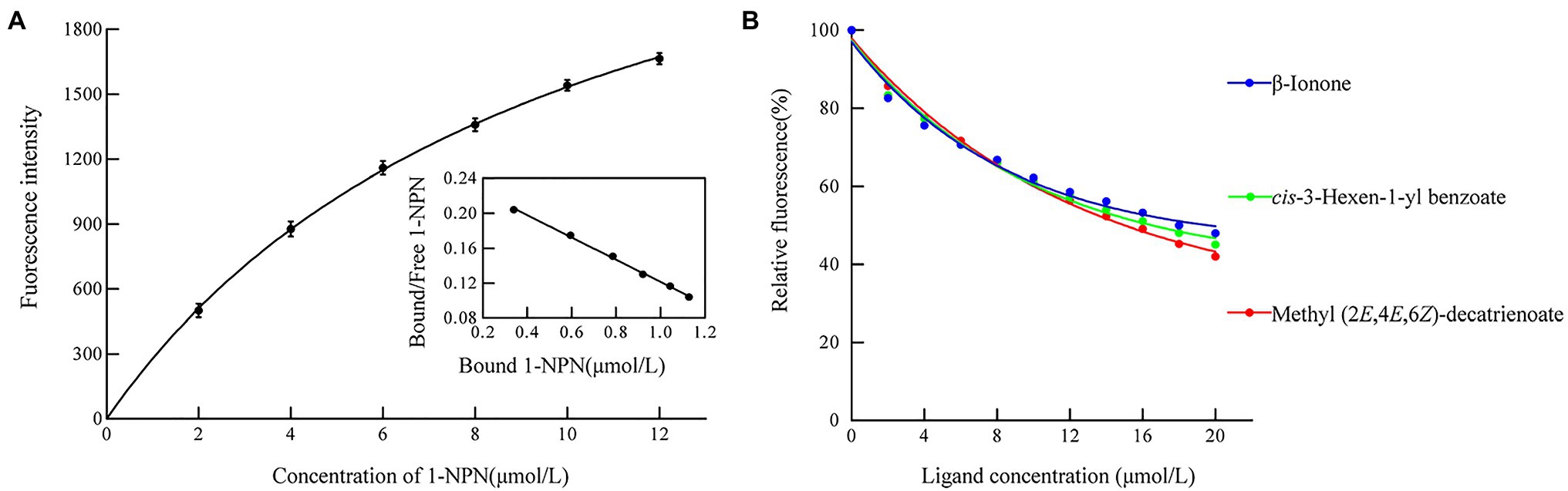
Figure 4. Binding properties of recombinant HhalCSP15. (A) Binding curve and Scatchard plot of N-phenyl-1-naphthylamine (1-NPN) to HhalCSP15. (B) Binding curves of HhalCSP15 to candidate ligands.
Protein Structure Prediction and Molecular Docking
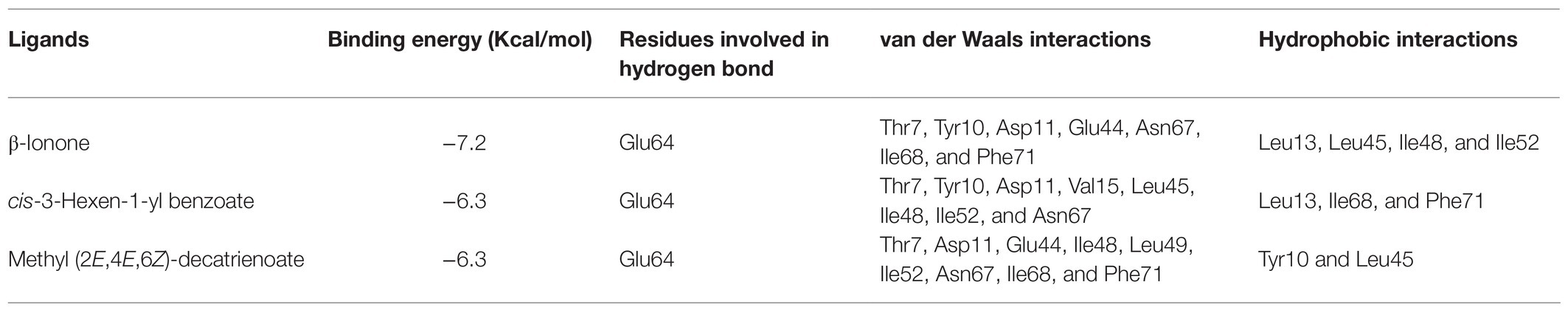
To support the results of our ligand binding assay and provide insight into the mechanism of HhalCSP15 interaction with ligands, molecular docking of the three ligands with HhalCSP15 was performed. The best model for HhalCSP15 (Supplementary Figure S2) was obtained using the crystal structure of the CSP from M. brassicae (PDB: 1N8V, 42% identity) as a template. The protein model was subjected to a 50ns MD simulation to energy minimize and stabilize the protein. The structural stability of the protein was measured by evaluating root mean square deviation (RMSD) and root mean square fluctuation (RMSF; Supplementary Figure S3). A Ramachandran plot was employed to estimate the rationality of the predicted protein structure. It revealed that 93.5% of the residues were in the most favored allowed region, 6.5% of the residues were in the additional allowed region, and none were in the disallowed region (Supplementary Figure S4), suggesting that the predicted model of HhalCSP15 is reasonable and reliable.
The docking results showed that the ligands tightly bind to the HhalCSP15 pocket with negative energy values (Table 2). The 2D and 3D ligand interaction diagram is shown in Figure 5. The results indicated that different residues from the binding pocket participated in the recognition of distinct ligands. The HhalCSP15 amino acid residue Glu64 forms a hydrogen bond with all three compounds. Apart from hydrogen bond formation, the compounds exhibited van der Waals as well as hydrophobic interactions with HhalCSP15 (Figure 5; Table 2).
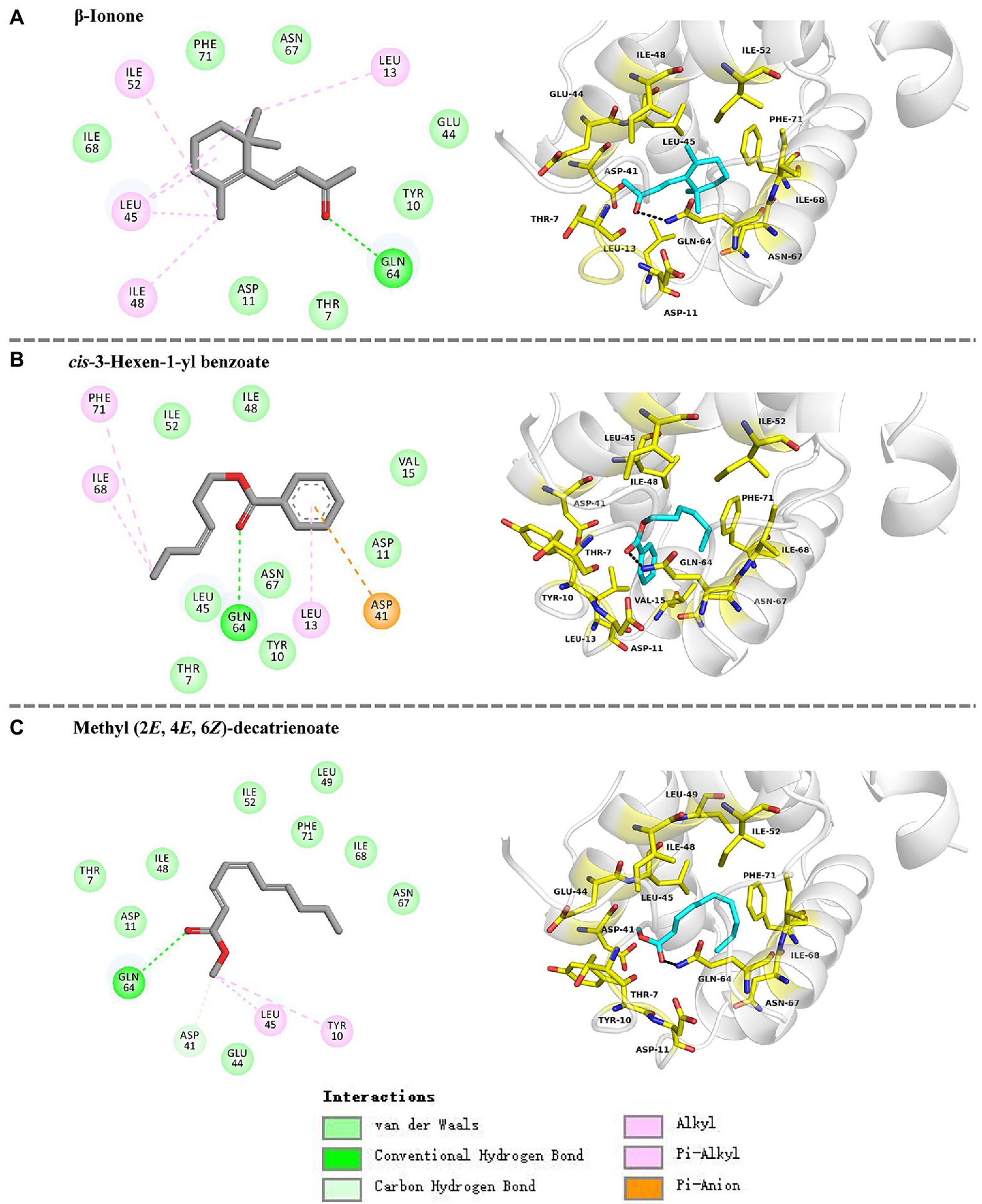
Figure 5. Molecular docking of HhalCSP15 with (A) β-ionone, (B) cis-3-hexen-1-yl benzoate, and (C) methyl (2E,4E,6Z)-decatrienoate.
Electrophysiological Activities of Putative Ligands of HhalCSP15
To determine whether the HhalCSP15 ligands have biological activity, we measured the electrophysiological responses of nymph and adult H. halys to these three volatiles using EAG recordings. The results indicated that all three volatiles elicited electrophysiological responses in the antennae of both nymphs and adults (Figure 6). Adult H. halys showed significantly greater responses than nymphs. Cis-3-hexen-1-yl benzoate and β-ionone elicited significantly greater responses in females compared with males and nymphs. EEZ-MDT stimulated significantly greater EAG responses in adults compared with nymphs, but there was no significant difference between the sexes of adults. Of note, cis-3-hexen-1-yl benzoate elicited the highest EAG response among all ligands in both nymphs and adults (Figure 6).
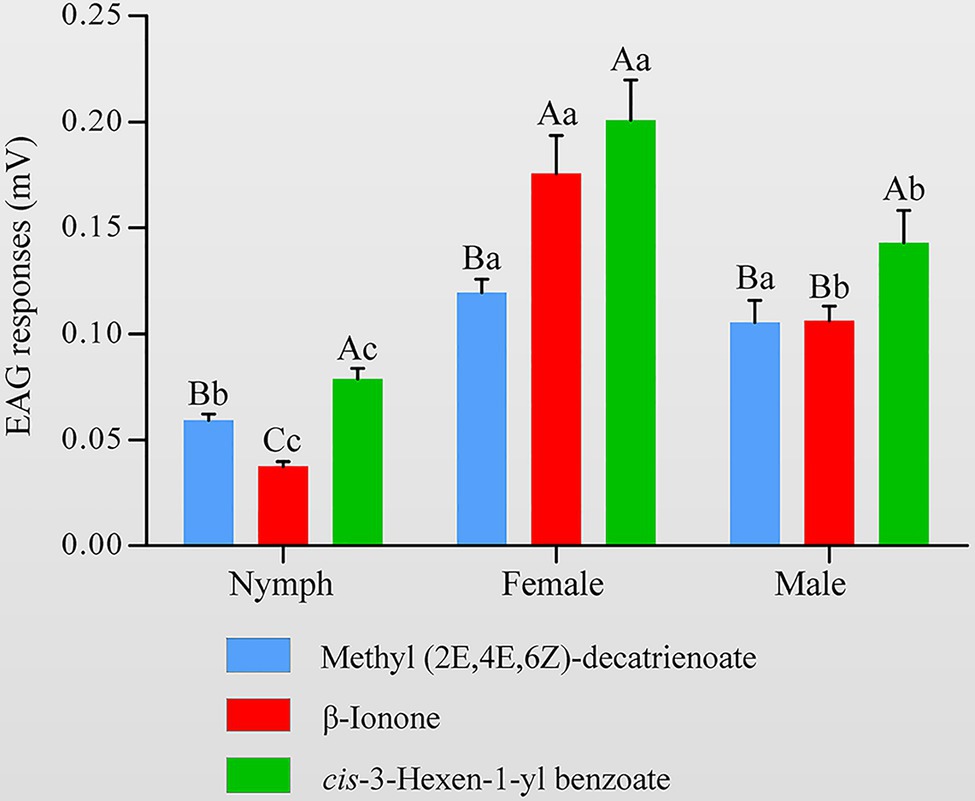
Figure 6. Electroantennography (EAG) activity of Halyomorpha halys antennae to different ligands of HhalCSP15. Different uppercase letters indicate significant differences among different chemicals, and different lowercase letters indicate significant differences among nymphs, females, and males (p<0.05).
Discussion
Insect CSPs exhibit broad expression profiles both in chemosensory organs and non-chemosensory organs. In H. halys antennal transcriptome, 17 CSPs were identified and half of them had diverse expression patterns (Sun et al., 2020). In southern green stink bug Nezara viridula, 13 CSP genes were identified from antennae and mouthpart transcriptome, and only four genes were primarily expressed in antennae (Wu et al., 2019). The broad and diverse expression patterns of stink bug CSPs are consistent with their possible multiple roles in chemoreception, development, and other processes. HhalCSP15 is orthologous with NvirCSP4 (GenBank: QCZ25118.1, 81% identity) and both of them are specifically expressed in antennae, which suggests their conserved roles in olfactory perception. NvirCSP4 was expressed roughly equally in both male and female antennae (Wu et al., 2019). Our data also reveal that no obvious difference was observed in expression levels of HhalCSP15 between male and female antennae. In addition, HhalCSP15 was also specifically expressed in nymph antennae. Thus, it is conceivable that HhalCSP15 is involved in the detection of odorants eliciting common stink bug behaviors, such as host location or intraspecific communication.
In fluorescent binding assays, 43 volatile compounds, including plant volatiles and H. halys volatiles, were selected as candidate ligands. Ligand-binding experiments demonstrated that HhalCSP15 has highly selective binding to volatile compounds. Some specific amino acids located in the hydrophobic cavities may be involved in the process of ligand binding in HhalCSP15 (Tomaselli et al., 2006). For example, in CSPsg4, I76 and W83 are involved in oleamide binding (Tomaselli et al., 2006); in CSP2, Y11 plays a key role in the binding of (E)-3,8-dimethyl-1,4,7-nonatriene (Li et al., 2021). Molecular docking analyses indicate favorable interactions between HhalCSP15 and its ligands. Glu64 forms a hydrogen bond with all ligands and could actively participate in forming the binding site of HhalCSP15. However, the specific binding sites that mediate the interactions between HhalCSP15 and ligands need to be investigated in future site-directed mutagenesis experiments.
β-Ionone and cis-3-hexen-1-yl benzoate are widely distributed in plants (Fraser et al., 2003; Simkin et al., 2004; Wei et al., 2011; Baldermann et al., 2012; Suckling et al., 2012). The binding experiments showed that the two compounds have strong binding abilities with HhalCSP15 and elicit an EAG response in both nymph and adult H. halys antennae. EEZ-MDT, which was identified as a binding ligand of HhalCSP15, is the aggregation pheromone of Plautia stali, which attracts stink bugs and is used as a lure in traps to monitor H. halys (Sugie et al., 1996; Aldrich et al., 2007; Morrison et al., 2017). H. halys does not emit EEZ-MDT though it may use EEZ-MDT as an indirect clue when searching for food plants (Funayama, 2008; Weber et al., 2017). These data further support a potential role of HhalCSP15 in H. halys host location. It was also found that HhalCSP15 could not bind with selected H. halys-derived volatiles, such as tridecane and (E)-2-decenal. (E)-2-decenal is an alarm pheromone in H. halys (Harris et al., 2015), and at least five H. halys OBPs showed high binding activities to it (Zhong et al., 2018). Thus, these findings also indicate that HhalCSP15 is not a pheromone binding protein of the stink bug and unlikely participates in the intraspecific communication for H. halys. However, gene editing and behavioral assays need to be further performed to verify the roles of this protein in the olfactory system of H. halys.
In conclusion, we report the antenna-specific expression as well as ligand binding capability of the CSP HhalCSP15 from H. halys, providing evidence for the possible olfactory roles of CSPs in the host-finding behavior of stink bugs. Although our results indicate that β-ionone and cis-3-hexen-1-yl benzoate are potential bioactive volatiles, further studies are necessary to confirm their behavioral activity as well as their possible applications for regulating the olfactory behavior of H. halys.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SW conceived and designed the research and wrote the manuscript. SW and ZW conducted all the experiments. ZW, AS, and FY analyzed the data. SS and YZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31801740) and the Promotion and Innovation of Beijing Academy of Agriculture and Forestry Sciences (KJCX201910).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.721247/full#supplementary-material
Footnotes
References
Abraham, M. J., Murtola, T., Schulz, R., Pall, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. doi: 10.1016/j.softx.2015.06.001
Aldrich, J. R., Khrimian, A., and Camp, M. J. (2007). Methyl 2,4,6-decatrienoates attract stink bugs and tachinid parasitoids. J. Chem. Ecol. 33:801. doi: 10.1007/s10886-007-9270-9
Angeli, S., Ceron, F., Scaloni, A., Monti, M., Monteforti, G., Minnocci, A., et al. (1999). Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 262, 745–754. doi: 10.1046/j.1432-1327.1999.00438.x
Baldermann, S., Kato, M., Fleischmann, P., and Watanabe, N. (2012). Biosynthesis of α- and β-ionone, prominent scent compounds, in flowers of Osmanthus fragrans. Acta Biochim. Pol. 59, 79–81. doi: 10.18388/abp.2012_2176
Bos, J. I., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216. doi: 10.1371/journal.pgen.1001216
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bruce, T. J., Wadhams, L. J., and Woodcock, C. M. (2005). Insect host location: a volatile situation. Trends Plant Sci. 10, 269–274. doi: 10.1016/j.tplants.2005.04.003
Ebrahim, S. A., Dweck, H. K., Stökl, J., Hofferberth, J. E., Trona, F., Weniger, K., et al. (2015). Drosophila avoids parasitoids by sensing their semiochemicals via a dedicated olfactory circuit. PLoS Biol. 13:e1002318. doi: 10.1371/journal.pbio.1002318
Forêt, S., Wanner, K. W., and Maleszka, R. (2007). Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Mol. Biol. 37, 19–28. doi: 10.1016/j.ibmb.2006.09.009
Fraser, A. M., Mechaber, W. L., and Hildebrand, J. G. (2003). Electroantennographic and behavioral responses of the sphinx moth Manduca sexta to host plant headspace volatiles. J. Chem. Ecol. 29, 1813–1833. doi: 10.1023/A:1024898127549
Fu, S., Li, F., Yan, X., and Hao, C. (2020). Expression profiles and binding properties of the chemosensory protein PxylCSP11 from the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J. Insect Sci. 20:17. doi: 10.1093/jisesa/ieaa107
Funayama, K. (2008). Seasonal fluctuations and physiological status of Halyomorpha halys (Stål) (Heteroptera: Pentatomidae) adults captured in traps baited with synthetic aggregation pheromone of Plautia crossota stali Scott (Heteroptera: Pentatomidae). Jpn. J. Appl. Entomol. Zool. 52, 69–75. doi: 10.1303/jjaez.2008.69
Gonzalez, D., Rihani, K., Neiers, F., Poirier, N., Fraichard, S., Gotthard, G., et al. (2020). The Drosophila odorant-binding protein 28a is involved in the detection of the floral odour ß-ionone. Cell. Mol. Life Sci. 77, 2565–2577. doi: 10.1007/s00018-019-03300-4
Gu, S. H., Wang, S. Y., Zhang, X. Y., Ji, P., Liu, J. T., Wang, G. R., et al. (2012). Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition. PLoS One 7:e42871. doi: 10.1371/journal.pone.0042871
Harris, C., Abubeker, S., Yu, M., Leskey, T., and Zhang, A. (2015). Semiochemical production and laboratory behavior response of the brown marmorated stink bug, Halyomorpha Halys. PLoS One 10:e0140876. doi: 10.1371/journal.pone.0140876
Haye, T., Gariepy, T., Hoelmer, K., Rossi, J. P., Streito, J. C., Tassus, X., et al. (2015). Range expansion of the invasive brown marmorated stinkbug, Halyomorpha halys: an increasing threat to field, fruit and vegetable crops worldwide. J. Pest. Sci. 88, 665–673. doi: 10.1007/s10340-015-0670-2
Hoebeke, E., and Carter, M. E. (2003). Halyomorpha halys (Stål) (Heteroptera: Pentatomidae): a polyphagous plant pest from Asia newly detected in North America. Proc. Entomol. Soc. Wash. 105, 225–237.
Huang, G. Z., Liu, J. T., Zhou, J. J., Wang, Q., Dong, J. Z., Zhang, Y. J., et al. (2018). Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 98, 34–47. doi: 10.1016/j.ibmb.2018.05.003
Kaissling, K. E. (2009). Olfactory perireceptor and receptor events in moths: a kinetic model revised. J. Comp. Physiol. A. 195, 895–922. doi: 10.1007/s00359-009-0461-4
Khrimian, A., Zhang, A., Weber, D. C., Ho, H. Y., Aldrich, J. R., Vermillion, K. E., et al. (2014). Discovery of the aggregation pheromone of the brown marmorated stink bug (Halyomorpha halys) through the creation of stereoisomeric libraries of 1-bisabolen-3-ols. J. Nat. Prod. 77, 1708–1717. doi: 10.1021/np5003753
Kriticos, D. J., Kean, J. M., Phillips, C. B., Senay, S. D., Acosta, H., and Haye, T. (2017). The potential global distribution of the brown marmorated stink bug, Halyomorpha halys, a critical threat to plant biosecurity. J. Pest. Sci. 90, 1033–1043. doi: 10.1007/s10340-017-0869-5
Larter, N. K., Sun, J. S., and Carlson, J. R. (2016). Organization and function of Drosophila odorant binding proteins. eLife 15:e20242. doi: 10.7554/eLife.20242
Lartigue, A., Campanacci, V., Roussel, A., Larsson, A. M., Jones, T. A., Tegoni, M., et al. (2002). X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 277, 32094–32098. doi: 10.1074/jbc.M204371200
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Lee, D. H., Short, B. D., Joseph, S. V., Bergh, J. C., and Leskey, T. C. (2013). Review of the biology, ecology, and management of Halyomorpha halys (Hemiptera: Pentatomidae) in China, Japan, and the Republic of Korea. Environ. Entomol. 42, 627–641. doi: 10.1603/EN13006
Leskey, T. C., Agnello, A., Bergh, J. C., Dively, G. P., Hamilton, G. C., Jentsch, P., et al. (2015). Attraction of the invasive Halyomorpha halys (Hemiptera: Pentatomidae) to traps baited with semiochemical stimuli across the United States. Environ. Entomol. 44, 746–756. doi: 10.1093/ee/nvv049
Leskey, T., and Nielsen, A. (2017). Impact of the invasive brown marmorated stink bug in North America and Europe: history, biology, ecology, and management. Annu. Rev. Entomol. 7, 599–618. doi: 10.1146/annurev-ento-020117-043226
Li, F., Dewer, Y., Li, D., Qu, C., and Luo, C. (2021). Functional and evolutionary characterization of chemosensory protein CSP2 in the whitefly, Bemisia tabaci. Pest Manag. Sci. 77, 378–388. doi: 10.1002/ps.6027
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maida, R., Steinbrecht, A., Ziegelberger, G., and Pelosi, P. (1993). The pheromone binding protein of Bombyx mori: purification, characterization and immunocytochemical localization. Insect Biochem. Mol. Biol. 23, 243–253. doi: 10.1016/0965-1748(93)90005-D
Maleszka, J., Forêt, S., Saint, R., and Maleszka, R. (2007). RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 217, 189–196. doi: 10.1007/s00427-006-0127-y
McKenna, M. P., Hekmat-Scafe, D. S., Gaines, P., and Carlson, J. R. (1994). Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 269, 16340–16347. doi: 10.1016/S0021-9258(17)34013-9
Metcalf, R. L., and Kogan, M. (1987). Plant volatiles as insect attractants. Crit. Rev. Plant Sci. 5, 251–301. doi: 10.1080/07352688709382242
Morrison, W. R., Park, C. G., Seo, B. Y., Park, Y. L., Kim, H. G., Rice, K. B., et al. (2017). Attraction of the invasive Halyomorpha halys in its native Asian range to traps baited with semiochemical stimuli. J. Pest. Sci. 90, 1205–1217. doi: 10.1007/s10340-016-0816-x
Nomura, A., Kawasaki, K., Kubo, T., and Natori, S. (1992). Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int. J. Dev. Biol. 36, 391–398.
Pelosi, P., Iovinella, I., Felicioli, A., and Dani, F. R. (2014). Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5:320. doi: 10.3389/fphys.2014.00320
Pelosi, P., Iovinella, I., Zhu, J., Wang, G., and Dani, F. R. (2017). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 93, 184–200. doi: 10.1111/brv.12339
Peng, Y., Wang, S. N., Li, K. M., Liu, J. T., Zheng, Y., Shan, S., et al. (2017). Identification of odorant binding proteins and chemosensory proteins in Microplitis mediator as well as functional characterization of chemosensory protein 3. PLoS One 12:e0180775. doi: 10.1371/journal.pone.0180775
Pikielny, C. W., Hasan, G., Rouyer, F., and Rosbash, M. (1994). Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 12, 35–49. doi: 10.1016/0896-6273(94)90150-3
Rihani, K., Ferveur, J. F., and Briand, L. (2021). The 40-year mystery of insect odorant-binding proteins. Biomol. Ther. 11:509. doi: 10.3390/biom11040509
Simkin, A. J., Schwartz, S. H., Auldridge, M., Taylor, M. G., and Klee, H. J. (2004). The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 40, 882–892. doi: 10.1111/j.1365-313X.2004.02263.x
Steinbrecht, R. A. (1997). Pore structures in insect olfactory sensilla: a review of data and concepts. Int. J. Insect Morphol. Embryol. 26, 229–245. doi: 10.1016/S0020-7322(97)00024-X
Suckling, D. M., Twidle, A. M., Gibb, A. R., Manning, L. M., Mitchell, V. J., Sullivan, T. E., et al. (2012). Volatiles from apple trees infested with light brown apple moth larvae attract the parasitoid Dolichogenidia tasmanica. J. Agric. Food Chem. 60, 9562–9566. doi: 10.1021/jf302874g
Sugie, H., Yoshida, M., Kawasaki, K., Noguchi, H., Moriya, S., Takagi, K., et al. (1996). Identification of the aggregation pheromone of the brown-winged green bug, Plautia stali Scott (Heteroptera: Pentatomidae). Appl. Entomol. Zool. 31, 427–431. doi: 10.1303/aez.31.427
Sun, H., Guan, L., Feng, H., Yin, J., Cao, Y., Xi, J., et al. (2014). Functional characterization of chemosensory proteins in the scarab beetle, Holotrichia oblita Faldermann (Coleoptera: Scarabaeida). PLoS One 9:e107059. doi: 10.1371/journal.pone.0107059
Sun, D., Huang, Y., Qin, Z., Zhan, H., Zhang, J., Liu, Y., et al. (2020). Identification of candidate olfactory genes in the antennal transcriptome of the stink bug Halyomorpha halys. Front. Physiol. 11:876. doi: 10.3389/fphys.2020.00876
Sun, L., Zhou, J. J., Gu, S. H., Xiao, H. J., Guo, Y. Y., Liu, Z. W., et al. (2015). Chemosensillum immunolocalization and ligand specificity of chemosensory proteins in the alfalfa plant bug Adelphocoris lineolatus (Goeze). Sci. Rep. 5:8073. doi: 10.1038/srep08073
Takken, W. (1991). The role of olfaction in host-seeking of mosquitoes: a review. Int. J. Trop. Insect Sci. 12, 287–295. doi: 10.1017/S1742758400020816
Tomaselli, S., Crescenzi, O., Sanfelice, D., Ab, E., Wechselberger, R., Angeli, S., et al. (2006). Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 45, 10606–10613. doi: 10.1021/bi060998w
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Wang, S. N., Shan, S., Yu, G. Y., Wang, H., Dhiloo, K. H., Khashaveh, A., et al. (2020). Identification of odorant-binding proteins and functional analysis of antenna-specific AplaOBP1 in the emerald ash borer, Agrilus planipennis. J. Pest. Sci. 93, 853–865. doi: 10.1007/s10340-019-01188-4
Weber, D. C., Morrison, W. R., Khrimian, A., Rice, K. B., Leskey, T. C., Rodriguez-Saona, C., et al. (2017). Chemical ecology of Halyomorpha halys: discoveries and applications. J. Pest. Sci. 90, 989–1008. doi: 10.1007/s10340-017-0876-6
Wei, S., Hannoufa, A., Soroka, J., Xu, N., Li, X., Zebarjadi, A., et al. (2011). Enhanced β-ionone emission in Arabidopsis over-expressing AtCCD1 reduces feeding damage in vivo by the crucifer flea beetle. Environ. Entomol. 40, 1622–1630. doi: 10.1603/EN11088
Wermelinger, B., Wyniger, D., and Forster, B. (2008). First records of an invasive bug in Europe: Halyomorpha halys Stål (Heteroptera: Pentatomidae), a new pest on woody ornamentals and fruit trees? Mitt. Schweiz. Entomol. Ges. 81, 1–8.
Wu, Z. Z., Cui, Y., Qu, M. Q., Lin, J. H., Chen, M. S., Bin, S. Y., et al. (2019). Candidate genes coding for odorant binding proteins and chemosensory proteins identified from dissected antennae and mouthparts of the southern green stink bug Nezara viridula. Comp. Biochem. Physiol. Part D Genomics Proteomics 31:100594. doi: 10.1016/j.cbd.2019.100594
Xu, P., Atkinson, R., Jones, D. N., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. doi: 10.1016/j.neuron.2004.12.031
Xuan, N., Guo, X., Xie, H.-Y., Lou, Q.-N., Lu, X.-B., Liu, G. X., et al. (2015). Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 22, 203–219. doi: 10.1111/1744-7917.12116
Zhang, R., Wang, B., Grossi, G., Falabella, P., Liu, Y., Yan, S., et al. (2017). Molecular basis of alarm pheromone detection in Aphids. Curr. Biol. 27, 55–61. doi: 10.1016/j.cub.2016.10.013
Zhong, Y. Z., Tang, R., Zhang, J. P., Yang, S. Y., Chen, G. H., He, K. L., et al. (2018). Behavioral evidence and olfactory reception of a single alarm pheromone component in Halyomorpha halys. Front. Physiol. 9:1610. doi: 10.3389/fphys.2018.01610
Keywords: Halyomorpha halys, chemosensory protein, expression profile, fluorescence binding assay, molecular docking, electroantennography
Citation: Wang Z, Yang F, Sun A, Shan S, Zhang Y and Wang S (2021) Expression Profiles and Functional Characterization of Chemosensory Protein 15 (HhalCSP15) in the Brown Marmorated Stink Bug Halyomorpha halys. Front. Physiol. 12:721247. doi: 10.3389/fphys.2021.721247
Edited by:
Jin Zhang, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Herbert Venthur, University of La Frontera, ChileYoussef Dewer, Agricultural Research Center, Egypt
Copyright © 2021 Wang, Yang, Sun, Shan, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanning Wang, d2FuZ3NoYW5uaW5nQHllYWgubmV0
 Zehua Wang
Zehua Wang Fan Yang
Fan Yang Ang Sun
Ang Sun Shuang Shan
Shuang Shan Yongjun Zhang
Yongjun Zhang Shanning Wang
Shanning Wang