
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 27 September 2021
Sec. Gastrointestinal Sciences
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.715506
This article is part of the Research Topic The Microbiome in Hepatobiliary and Intestinal Disease View all 18 articles
 Salma Sultan1
Salma Sultan1 Mohammed El-Mowafy2
Mohammed El-Mowafy2 Abdelaziz Elgaml2,3
Abdelaziz Elgaml2,3 Tamer A. E. Ahmed1,4
Tamer A. E. Ahmed1,4 Hebatoallah Hassan1,5
Hebatoallah Hassan1,5 Walid Mottawea1,2*
Walid Mottawea1,2*Inflammatory bowel diseases (IBD) are chronic medical disorders characterized by recurrent gastrointestinal inflammation. While the etiology of IBD is still unknown, the pathogenesis of the disease results from perturbations in both gut microbiota and the host immune system. Gut microbiota dysbiosis in IBD is characterized by depleted diversity, reduced abundance of short chain fatty acids (SCFAs) producers and enriched proinflammatory microbes such as adherent/invasive E. coli and H2S producers. This dysbiosis may contribute to the inflammation through affecting either the immune system or a metabolic pathway. The immune responses to gut microbiota in IBD are extensively discussed. In this review, we highlight the main metabolic pathways that regulate the host-microbiota interaction. We also discuss the reported findings indicating that the microbial dysbiosis during IBD has a potential metabolic impact on colonocytes and this may underlie the disease progression. Moreover, we present the host metabolic defectiveness that adds to the impact of symbiont dysbiosis on the disease progression. This will raise the possibility that gut microbiota dysbiosis associated with IBD results in functional perturbations of host-microbiota interactions, and consequently modulates the disease development. Finally, we shed light on the possible therapeutic approaches of IBD through targeting gut microbiome.
Inflammatory Bowel Disease (IBD) is a chronic and remitting disorder characterized by relapsing episodes of gastrointestinal inflammation (Ramos and Papadakis, 2019). IBD is comprised of two major classes; ulcerative colitis (UC) and Crohn’s disease (CD). The inflammation in UC is restricted to the mucosal layer, leading to superficial damage of the bowel wall, while CD is associated with inflammation of all layers of the bowel wall (Kobayashi et al., 2020). Epidemiological studies showed that IBD is spreading widely throughout the world leading to a public health challenge worldwide (Zheng H. et al., 2017; Windsor and Kaplan, 2019; Kaplan and Windsor, 2021). It is widely accepted that the IBD pathogenesis results from an interplay between gut microbiota and the host immune system with the predisposition of genetic susceptibility and environmental factors (Ma et al., 2019; O’Donnell et al., 2019; Ramos and Papadakis, 2019; Pang et al., 2021). The microbiome-immune system interaction leads to improper immune activation responsible for the clinical and endoscopic observations in IBD patients (Ramos and Papadakis, 2019; Zheng et al., 2020).
The human gastrointestinal tract comprises very rich and diverse microbial community that includes more than 100 trillion microorganisms (Thursby and Juge, 2017). The gut microbiome plays an important role in human health and in the development of several chronic diseases. The altered composition of the gut microbiota (dysbiosis), in addition to the disturbance of the metabolic harmony of such microbial community plays a crucial role in the pathogenesis of IBD (Nishida et al., 2018; Zuo and Ng, 2018; Khan et al., 2019). Indeed, recent advances in molecular biology techniques together with improving the usability of microbial databases led to comprehensive characterization of microbial communities and revealing the association between gut microbiota dysbiosis and IBD (Wright et al., 2015; Lloyd-Price et al., 2019; Loeffler et al., 2020; Ryan et al., 2020). However, the actual contributions of this dysbiosis to the inflammation and the cause/effect relationship between gut microbe and IBD are still unclear.
In this review, we outline the recent findings that correlate gut microbiota dysbiosis and metabolic dysfunctionality to IBD. We highlight the metabolic pathways by which microbial dysbiosis could contribute to the inflammation seen in IBD. We also consider the connection among different metabolic pathways in relation to disease progression. In addition, we clarify the strategies of manipulating gut microbiota to promote gut health in IBD.
Our gut is populated by a complex and dynamic microbial ensemble, which is considered an additional organ of the human body and collectively known as the gut microbiota. This cohort consists mainly of bacteria with low percentage of archeae, eukaryotic and viral members (Qin et al., 2010). Using fecal samples from 124 Europeans and Illumina-based metagenomic sequencing, the gut microbial gene catalog was estimated to be 100–150-fold that of human genes with 99.1% of bacterial origin. The bacterial species that compose the entire community were calculated to range from 1,000 to 1,150 prevalent species with at least 160 species per individual (Qin et al., 2010).
Studying mucosal and luminal microbiota structure via sequencing 16S rDNA clones has revealed that approximately 90% of gut microbes are related to main two phyla; Firmicutes (51%) and Bacteroidetes (48%), with the remaining 10% distributed among Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia (Eckburg et al., 2005). At lower levels, the majority of the detected Firmicutes were related to the Clostridia class, with the vast majority classified under clostridial clusters IV, XIVa, and XVI. Bacteroidetes, on the other hand, has showed less diversity than Firmicutes with 67% of its sequences classified under 3 main phylotypesB. vulgatus, Prevotellaceae, and B. thetaiotamicron. B. thetaiotamicron was the only common Bacteroidetes among all tested individuals. Other low abundant species related to other phyla included Proteobacteria species (Sutterellawadsworthensis, Desulfomonaspigra, and Bilophilawadsworthia), Actinobacteria genera (e.g., Actinomyces, Collinsella, and Bifidobacterium) and the mucin-degrading Verrucomicrobia species, Akkermansiamuciniphila (Eckburg et al., 2005). Along with bacteria, the most common coexisting fungal genera in the gut are Saccharomyces, Candida, Galactomyces, Pleospora, Bullera, Aspergillus, Trametes, Sclerotinia, Penicillium, and Rhodotorula, while virome mainly exists in the form of bacteriophages (Ungaro et al., 2019a; Beheshti-Maal et al., 2021).
The diversity and the composition of gut bacteria vary along the length of the gastrointestinal tract (GIT) with a gradual increase in bacterial load and diversity from the stomach and duodenum (102 CFU/gm content) to the colon (1011–1012 CFU/gm content) (Eckburg et al., 2005; Eckburg and Relman, 2007; Frank et al., 2007; Sartor, 2008). The composition also varies at different intestinal locations starting with aerobic Streptococcus and Lactobacillus species in the duodenum and ending with strict anaerobes such as Bacteroides, Bifidobacterium and clostridial clusters in the colon (Frank et al., 2007). This difference in composition and diversity may be attributed to nutritional substrate availability, oxygen content, luminal acidity, and other physiological and immunological conditions at different parts of the GIT.
In addition to the stable differences in the composition at different intestinal locations, the gut bacteria are known to, significantly, vary among individuals (Eckburg et al., 2005). Furthermore, gut bacteria have been shown to be in continuous temporal variation within the same individual as revealed by monitoring the gut microbiota structure of two subjects for 15 months (Caporaso et al., 2011). These temporal variations could be attributed to small perturbations in some environmental factor, such as a short-term change in diet, a gastrointestinal infection, or due to a reversible change in the immunological state of the host (Caporaso et al., 2011; Lozupone et al., 2012). However, the gut microbiota tolerates these temporal changes and returns to its original structure once all physical and physiological conditions of the gut return to the normal state, which is known as resilience of the gut microbial ecosystem (Lozupone et al., 2012). Contrary to this, gut microbiota can lose this resilience by exposure to certain permanent factors, such as broad spectrum antibiotics (Lozupone et al., 2012), chronic diseases [e.g., Diabetes (Qin et al., 2012), IBD (Sartor, 2008)], obesity (Ray, 2012), or with age (Claesson et al., 2011). With regards to age, gut microbiota begins to colonize the intestine immediately after birth with a few aerobes (Enterobacteria, Staphylococcus, and Streptococcus), which gradually are replaced by anaerobic bacteria to reach the same complexity as mentioned above for adults by the first year of life (Palmer et al., 2007). However, differences between children and adults are still significant even after the first year of life. For example, children 1–7 years of age have lower fecal microbiota diversity with higher abundance of Enterobacteria than adults (Hopkins et al., 2001). Also, a more recent comprehensive study has indicated a substantial difference in elderly people when compared to the established adult pattern (Claesson et al., 2011), with Bacteroidetes as the predominant phyla and lower abundance of Firmicutes. This age-related pattern has been confirmed previously by comparative assessment of Firmicutes/Bacteroidetes ratio at different ages by quantitative PCR (Mariat et al., 2009). Also, individual bacterial species such as Escherichia coli, Enterococcispp., Bacteroidesspp., Bifidobacterium spp., and lactobacilli have been demonstrated to exhibit specific age-related profiles in adults and elderly subjects (Woodmansey, 2007; Enck et al., 2009). Yatsunenko et al. (2012) linked the microbial diversity to both age and geographical location. They surveyed the gut microbiome structure in the stool of 314 Americans, 114 Malawians, and 100 Amerindians at different ages. They reported that the inter-subject variability is higher in the early stages of life compared to the adult microbiome with a progressive increase in microbial diversity with age (Yatsunenko et al., 2012). The authors also illustrated that the infant microbiome is dominated by Bifidobacteria and reaches an adult-like composition by the age of three (Yatsunenko et al., 2012). Moreover, the composition of the gut microbiota was more similar between Malawians and Amerindians and its diversity was higher than the American’s reflecting the association between geography and gut microbiota diversity.
Many reports have described humans as a superorganism that live in symbiosis with different microbes within various parts of the body. This ecosystem offers many benefits to the human host that are essential for good health. The gut harbors the greatest human microbial assembly (Sultan et al., 2020; El-Mowafy et al., 2021). The first role of the gut bacteria involves its metabolic capacity to process undigestible food particles such as complex carbohydrates, plant glycans, choline and bile acids (Tremaroli and Backhed, 2012). The microbial processing of indigestible polysaccharides generates beneficial short chain fatty acids (SCFAs) such as butyrate, acetate and propionate, which represent around 90% of SCFAs produced in the human gut (Cummings and Macfarlane, 1991). Butyrate is considered the major source of energy to the colonocytes. For example, the colonocytes of germ-free mice develop impaired mitochondrial respiration and increased autophagy in comparison with conventionally raised mice, and these findings have been reversed by adding butyrate to germ free colonocytes (Donohoe et al., 2011). On the other hand, acetate and propionate exert extra intestinal roles, where they act as metabolic substrates for lipogenesis and gluconeogenesis (Bergman, 1990; Tremaroli and Backhed, 2012). The major producers of SCFAs include the genus Bacteroides, Clostridium clusters IV and XIVa and Bifidobacterium (Cummings and Macfarlane, 1991; Louis et al., 2010; Martens et al., 2011). While Eubacterium rectale, Roseburiafaecis, Eubacterium hallii, and Faecalibacterium prausnitzii are the major butyrate producers in the gut as revealed by investigating the butyryl-CoA:acetate CoA-transferase gene (Louis et al., 2010), B. thetaiotamicron and B. ovatus showed a high genomic content of carbohydrate active enzymes (CAZymes) that enable them to metabolize indigestible plant and host glycans (Martens et al., 2011). This might explain the predominance of Bacteroidetes in the gut of rural Africans who have a mainly plant-based diet (De Filippo et al., 2010). The highest percentage of SCFAs in the large bowel is seen in the cecum and proximal colon, and decreases gradually toward the distal colon (Cummings and Macfarlane, 1991). This gradient may be explained by the higher prevalence of substrates in the proximal colon, which decreases progressively toward the rectum (Cummings and Macfarlane, 1991). The majority of the SCFAs produced are absorbed by the gut or delivered to peripheral tissues such as the liver and muscles (Bergman, 1990; Cummings and Macfarlane, 1991). Hence using feces to measure the gut metabolites may be inappropriate.
Gut microbiota, via various mechanisms, contributes to intestinal epithelial integrity. The first of these factors is the production of SCFAs, notably butyrate. Butyrate is a main modulator of mucin release, which acts as the first barrier against gut microbial invasion (Barcelo et al., 2000). In addition, butyrate controls gene expression of the colonocytes either via inhibition of histone deacetylase (HDAC) or through binding to G-protein coupled receptors (GPR41 or GPR43) (Tremaroli and Backhed, 2012). For example, sodium butyrate up-regulates the expression of the tight junction proteins and their mRNA via the inhibition of HDAC (Bordin et al., 2004). Furthermore, intra-rectal delivery of C. tyrobutyricum to immunocompetent and immunodeficient pathogen free mice showed a protective effect against dextran sulfate sodium (DSS)-induced colitis. This is mediated by inducing the expression of zonula occludens (ZO)-1 tight junction proteins, as well as MUC-2 mucin both of which are directly related to the luminal level of butyrate (Hudcovic et al., 2012). In addition to SCFAs, other microbial components contribute to the epithelial integrity. Germ free mice showed a thinner mucus layer relative to microbiota colonized mice, and this was corrected following exposure to LPS or peptidoglycan (Petersson et al., 2011). Recently, it has been shown that gut microbiota, also, induces intestinal mucosal endothelial and mesenchymal cells via stimulation of toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) like receptor pathways (Schirbel et al., 2013). Together, this emphasizes the critical role of gut microbiota or their metabolites in maintaining the integrity of the intestinal barrier.
In addition to the development of tissues and cells, the gut microbiota is responsible for the shaping and maturation of the immune system. Germ free mice showed defective gut associated lymphoid tissues (GALT), fewer and smaller Peyer’s patches, less cellular mesenteric lymph nodes, less cellular lamina propria, lower expression of TLRs and class II major histocompatibility complex (MHC II) molecules, and finally reduced intraepithelial lymphocytes and CD4+ T cells in comparison to conventional mice. Additionally, gut colonization with microbes corrected some of these deficiencies (Lee and Mazmanian, 2010). In relation to specific pathogen free mice, germ free mice also showed accumulation of invariant natural killer T cells (iNKT) in the lamina propria of the colon and lung, which resulted in higher mortality rates (Olszak et al., 2012). However, the exposure of neonatal germ free mice, but not adults, to commensal bacteria was protective against iNKT-accumulation and its undesirable consequences (Olszak et al., 2012). Likewise, mice who received CD4+CD62L+ lymphocytes from germ free mice developed colitis faster than mice who received the same regulatory T cells (Tregs) from conventionally housed mice (Strauch et al., 2005). This suggests a critical role of the gut microbiota in the development of the intestinal immune system. This gut immune maturation is dependent on the host-specific microbiota. Colonization of germ free mice with either human or rat microbiota resulted in fewer intestinal T cells and dendritic cells, and lower antimicrobial peptide expression relative to germ free mice colonized with a murine microbiota, and humanized mice were more susceptible to salmonella infection (Chung et al., 2012). Both humanized mice and the mice colonized with a murine microbiota showed similar bacterial diversity at higher taxonomic levels, but they harbored different species (Chung et al., 2012). This indicates that each host selects a specific microbial consortium that shapes its immune system and maintains intestinal health.
As mentioned above, our gut is populated with a complex and dynamic ecosystem, which under normal circumstances is characterized by a stable structure at various intestinal segments in everyone. Any alteration of this consortium may disrupt its functionality and eventually, a diseased state will appear. The dysbiosis of the intestinal microbiota is well reported in different diseases such as irritable bowel syndrome (IBS), obesity, diabetes, and IBD (Shanahan, 2007; Saulnier et al., 2011; Hansen et al., 2012; Qin et al., 2012; Everard et al., 2013). The relationship between IBD and gut microbiota dysbiosis was first established by studying animal models of colitis. Germ free IL 10–/– mice do not develop colitis unless they are colonized by enteric bacteria (Schwerbrock et al., 2004). Garrett et al. (2007) were able to clearly demonstrate that the alteration of the gut microbiota composition could induce colitis in immunocompetent mice. They reported that deficiency of T-bet, a transcriptional factor that is important for gut homeostasis, resulted in microbial population shifts into a colitogenic community. This colitogenic microbiota was able to drive the intestinal inflammation in genetically intact mice (Garrett et al., 2007). Many molecular studies have illustrated the changes in gut microbial composition of IBD patients in comparison with non-IBD controls. The gut microbiota of individuals with IBD is characterized by low microbial diversity (Ott et al., 2004; Andoh et al., 2012), a reduced abundance of Bifidobacterium spp. (Joossens et al., 2011; Andoh et al., 2012), Lactobacillus spp. (Ott et al., 2004), and F. prausnitzii (Sokol et al., 2009; Joossens et al., 2011; Andoh et al., 2012) and a higher abundance of pathobionts such as adherent/invasive E. coli (Darfeuille-Michaud et al., 2004; Sokol et al., 2006a) and C. difficile (Rodemann et al., 2007), resulting in lower SCFA concentrations compared with healthy individuals (Huda-Faujan et al., 2010).
Studies using 16S rDNA sequencing have shown a decrease in the diversity of gut biota in IBD mucosal specimens (Baumgart et al., 2007; Frank et al., 2007). Using single strand confirmation polymorphism (SSCP) fingerprint, based on 16S rRNA showed that the diversity decreased by 50 and 30% in CD and UC, respectively (Ott et al., 2004). Frank et al. (2007) illustrated this imbalance by sequencing SS-rRNA clones from 190 biopsies. In their study, the Lachnospiraceae family of Firmicutes and Bacteroidetes were depleted in IBD subjects, with a relative increase of Proteobacteria, Actinobacteria, and Bacillus subgroups of Firmicutes. FISH analysis, on the other hand, has illustrated an increase in the relative abundance of Bacteroidetes and a low abundance of some butyrate producing bacteria such as F. prausnitziiin mucosal IBD specimens (Swidsinski et al., 2005; Sokol et al., 2008b, 2009). Using 454 pyrosequencing of the 16S rRNA V5 and V6 regions extracted from the fecal materials of concordant and discordant twins, ileal CD, colonic CD and healthy individuals were differentiated from each other according to their microbial profile (Willing et al., 2010). However, the authors were not able to discriminate between UC and healthy subjects by following the same approach (Willing et al., 2010). Colonic CD was characterized by higher Firmicutes (mainly Faecalibacterium, and Ruminococsulus), Bifidobacteriaceae (Bifidobacterium), Coriobacteriaceae (Collinsella), and Aneroplasmataceae. Conversely, ileal CD showed depletion of Ruminococaceae family, especially Faecalibacterium, and Collinsella with higher abundance of Proteobacteria due to the increase of the Enterobacteriaceae family (Willing et al., 2010). For UC, Willing et al. (2010) were able to identify few differences such as depletion of Prevotella, Streptococcus, and Asteroleplasma. In contrast, other study was able to discriminate between UC and healthy individuals by calculating the mean of 16S rDNA-clone libraries taken from sigmoid colon biopsies of 62 individuals (Lepage et al., 2011). The microbiota dysbiosis of UC was characterized by less diversity, fewer Lachnospiraceae and Ruminococcacea families with higher abundance of Proteobacteria and Actinobacteria (Lepage et al., 2011). These two studies confirm the association between IBD and gut microbiota dysbiosis. However, the discrepancy between them, regarding the UC microbiota structure, underlines the importance of the sampling approach in these types of studies (Mottawea et al., 2019).
For pediatric IBD, some studies have monitored the gut microbiota in IBD compared to healthy controls. Conte et al. (2006) described the alteration in microbiota composition along the length of the intestine in pediatric IBD patients in comparison to control subjects. They used the conventional culture-based techniques and 16S rRNA-based real time PCR for quantifying the mucosa associated bacteria at the ileum, caecum, and rectum of 42 subjects. One important observation of this study is that, in contrast to adult IBD, B. vulgatus was found at lower abundance in patients with IBD compared to controls. This supports the idea that pediatric IBD is a unique form of IBD. Moreover, a study investigating the relative abundance of 9 bacterial groups using real-time PCR showed higher number of E. coli and lower number of F. prausnitzii in children with CD in comparison to control subjects (Schwiertz et al., 2010). Furthermore, microbiota diversity in pediatric IBD patients was shown to differ from that of adults (Cucchiara et al., 2009). However, these studies solely examined the dominant bacterial groups by applying conventional culture-based techniques and simple molecular methodology. Since it is widely accepted that 80% of gut microbiota are unculturable (Eckburg et al., 2005), a comprehensive molecular survey of the gut microbiota in pediatric IBD is necessary. Two studies have applied a high throughput molecular approach to characterize gut microbiota in pediatric IBD (Hansen et al., 2012; Papa et al., 2012). The first study applied synthetic learning in microbial ecology (SLiME) analytic approach of 454 pyrosequencing data obtained from fecal samples of 91 individuals and other published datasets. They were able to differentiate between children with IBD and healthy individuals or those with other diseases based on their microbiota composition (Papa et al., 2012). The drawback of this study is that they relayed upon stool samples for their analysis, and it is known that fecal materials contain different microbiota when compared to mucosa associated bacteria. Moreover, Hansen et al. (2012) reported that the microbiota diversity is lower in CD but not in UC relative to control subjects. Pairwise comparison among the 3 groups identified only 7 differentially abundant taxa. One important result is that F. prausnitzii was highly abundant in CD compared to controls, which is the reverse of what has been previously documented in adults with IBD (Sokol et al., 2008b; Hansen et al., 2012), indicating once again the unique microbiota composition of pediatric IBD. Using intestinal mucosal colonoscopic washes, Mottawea et al. (2016) have reported dysbiosis of gut microbiota in new onset pediatric IBD with enrichment of H2S producing bacteria. Overall, these studies support the association between intestinal microbiota imbalance and pediatric IBD at different ages. Nevertheless, the cause/effect relationship between these conditions and gut microbiota is still unclear.
Generally, microbiota dysbiosis is significantly greater in patients with CD than with UC (Pascal et al., 2017), where the microbial community stability and diversity is significantly lower in case of CD than in UC (Ryan et al., 2020). In case of CD a specific microbial signature that is comprised of eight groups is well reported. Eight groups of microorganisms involving Anaerostipes, Methanobrevibacter, Faecalibacterium, an unknown Peptostreptococcaceae, Collinsella, an unknown Christensenellaceae, and Escherichia, Fusobacterium could be utilized to differentiate between CD from non-CD, where, the first six groups are relatively low and the latest two groups are relatively high in case of CD (Pascal et al., 2017; Ryan et al., 2020). Moreover, it is noteworthy fecal (stool) and mucosal sampling is critical during the determination of the microbial dysbiosis in IBD (Lo Presti et al., 2019). The IBD stool samples are characterized by reduced diversity of microbiota in comparison to IBS and healthy population (Mei et al., 2021). IBD cases are characterized by decreased Verrucomicrobia and Bacteroidetes than healthy population (Lo Presti et al., 2019; Mei et al., 2021). On the other hand, in case of IBS Bacteroidetes are increased in comparison to healthy population. Moreover, IBD are reported to harbor less population of Bacteroidetes and Verrucomicrobia, and higher abundance of Actinobacteria in comparison to IBS (Lo Presti et al., 2019; Ryan et al., 2020; Mei et al., 2021). Lactobacillus, Ruminococcus, and Streptococcus are significantly higher in IBD than healthy population, while Oscillospira, Lachnospiraceae, Ruminococcaceae, and Rikenellaceaeas well as Akkermansia muciniphila are diminished in IBD (Lo Presti et al., 2019; Ryan et al., 2020; Mei et al., 2021). In case of IBS, Pseudomonas and Lactococcus are decreased in comparison to healthy population, while Parabacteroides distasonisis relatively increased (Forbes et al., 2016; Lo Presti et al., 2019). By comparing IBS and IBD, Rikenellaceae, Bacteroides, Butyricimonas, Oscillospira, Mogibacteriaceae, Anaerostipes, Barnesiellaceae, Roseburia, Parabacteroides, P. distasonis, and A. muciniphila were more abundant in IBS than in IBD, while Granulicatella was relatively decreased (Dziarski et al., 2016; Lo Presti et al., 2019; Cuffaro et al., 2020). Similarly, to fecal samples, mucosal samples exhibited reduced microbiota diversity passing from healthy population to IBS to IBD (Lo Presti et al., 2019). Generally, there is no significant difference in microbiota population between inflamed and not-inflamed tissue samples of IBD. On contrary, the microbiota of inflamed mucosa of IBD patients exhibited low abundance of Firmicutes and Bacteroidetes and higher abundance of Proteobacteria in comparison to healthy population (Zuo and Ng, 2018). The abundance level of Enterobacteriaceae was significantly increased and the abundance levels of Lachnospiraceae, Ruminococcaceae, Rikenellaceae, Bacteroides, Coprococcus, F. prausnitzii, and P. distasonis were diminished in IBD inflamed mucosa compared to healthy population (Lo Presti et al., 2019; Aldars-García et al., 2021).
In addition to dysbiosis of bacteria, the dramatic changes that occur in the fungal community named as “mycobiome” is relatively important during IBD (Liu et al., 2020). Mechanistically, it appears that fungi may paly crucial role in the progression of IBD through either affecting the gut microbiota composition or increasing the production of pro-inflammatory cytokines (Iliev and Leonardi, 2017). In addition, mycobiome dysbiosis is a well-reported case in IBD (Beheshti-Maal et al., 2021). It is reported that fungal diversity is higher in CD patients than in healthy controls (Ott et al., 2008).
An increased ratio of Basidiomycota/Ascomycota is a characteristic feature of IBD (Sokol et al., 2017). Qiu et al. (2017) reported higher abundances of Aspergillus, Wickerhamomyces, Candida, and Sterigmatomyces and lower abundances of Alternaria, Penicillium, Exophiala, Emericella, Acremonium, Epicoccum, and Trametes in patients suffering from UC in comparison to healthy controls. However, there was no significant association between Basidiomycota/Ascomycota ratio and the increased levels of pro-inflammatory cytokines. Moreover, Candida spp., particularly C. albicans, is significantly increased in patients suffering from CD or general IBD (Li et al., 2014; Chehoud et al., 2015; Sokol et al., 2017). It is reported that specific-pathogen-free (SPF) Clec7a–/– mice exhibit more severe colitis symptoms when colonized with the pathogenic fungus, C. tropicalis compared to uncolonized Clec7a–/– mice or colonized wild type mice (Iliev et al., 2012; Tang et al., 2015). These studies confirm the crucial link between mycobiome dysbiosis, especially Candida spp. and IBD (Li et al., 2019). Li et al. (2014) illustrated higher levels of Candida spp. in the inflamed mucosa of IBD patients than in healthy population. In addition, Standaert-Vitse et al. (2009) illustrated higher colonization of familial CD patients by C. albicans. Moreover, other reports reported high levels of both C. albicans and C. glabrata in CD patients (Liguori et al., 2016; Sokol et al., 2017). In addition, Kowalska-Duplaga et al. (2019) documented the high abundance of Candida in CD patients, which was decreased by therapeutic intervention, especially with anti-TNF-α treatment.
Besides Candida spp., Malasseziarestricta, which is a skin normal fungus, significantly increases in patients suffering from CD (Limon et al., 2019). This fungus was found to aggravate the colitis in mouse models via mechanisms demanding on a protein included in antifungal immunity; CARD9 (Limon et al., 2019). Moreover, depletion of Saccharomyces cerevisiae is reported in IBD patients (Sokol et al., 2017). Tiago et al. (2015) depicted the protective effects of S. cerevisiae UFMG A-905 in mice suffering from UC. In addition, Sivignon et al. (2015) reported that adherent-invasive E. coli (AIEC)-induced ileal colitis can be reduced by S. cerevisiae CNCM I-3856 in a murine model. However, on the other hand, Chiaro et al., 2017 demonstrated that S. cerevisiae worsens the disease condition in a murine model of colitis (Chiaro et al., 2017).
The crosstalk between fungi and bacteria may be a pivotal concern in IBD patients. However, in pediatric IBD patients there was no significant correlation between leading fungal species with particular bacterial taxa and it has been hypothesized that fungal dysbiosis may be a result of or a cause for the gut bacterial dysbiosis (Chehoud et al., 2015). On the other hand, previous report showed a positive correlation between C. tropicalis and both E. coli and Serratia marcescens (Hoarau et al., 2016). It is noteworthy that the correlation between mycobiota-bacterial dysbiosis with the disease severity in IBD patients may afford evidence for the crucial role for gut mycobiome during IBD.
In addition to microbiota and mycobiota, gut virome, which is comprised of viruses infecting both prokaryotes and eukaryotes, constitute a large portion of gut microbiome (Manrique et al., 2016; Lin and Lin, 2019). Bacteriophages are the major components of the enteric virome (Clooney et al., 2019). Generally, in patients suffering from IBD, alteration of the virome reflects microbiota dysbiosis (Clooney et al., 2019). One of the first studies, which reported virome dysbiosis in IBD patients revealed higher abundance of bacteriophages infecting Alteromonadales, Clostridiales, and Clostridium acetobutylicum as well as Retroviridae family in IBD patients in comparison to healthy population (Pérez-Brocal et al., 2015). It is well reported that Caudovirales phage families, including Siphoviridae, Myoviridae, and Podoviridae, are significantly enriched in IBD subjects (Wagner et al., 2013; Norman et al., 2015; Duerkop et al., 2018). A previous study illustrated the high abundance of Caudovirales phage in children suffering from IBD (Fernandes et al., 2019). On the other hand, another study reported a decreased diversity and consistency of Caudovirales that is directly connected to the inflammation degree of the intestine in UC patients (Zuo et al., 2019). Moreover, phages infecting Enterobacteria and Escherichia were reported to be abundant in UC patients (Zuo et al., 2019). A recent study established in germ-free mice showed that certain phages including Escherichia, Lactobacillus, and Bacteroides infecting phages as well as phage DNA exacerbate gut inflammation and contribute to IBD pathogenesis through increased production of IFN-γ via a TLR9-dependent pathway (Gogokhia et al., 2019).
In addition to bacteriophages, phages infecting and incorporating into eukaryotic cells are relatively important and have been related to IBD pathogenesis due to their ability of integration in human genome and affecting physiological state of the intestinal cells (Gloor et al., 2010; Beller and Matthijnssens, 2019; Santiago-Rodriguez and Hollister, 2019). Previous reports conducted on patients suffering from UC revealed higher abundance of Pneumoviridae and lower abundance of Anelloviridae compared to healthy population (Zuo et al., 2019). On the other hand, a study that was established on a small cohort of patients suffering from UC and CD depicted the higher abundance of Herpesviridae compared to healthy control (Wang et al., 2015). Other studies demonstrated that infection with Norovirus contributes to intestinal inflammation and can increase the rate of colitis incidence (Cadwell et al., 2010; Basic et al., 2014). Moreover, recent studies depicted the high abundance of Hepeviridae and Hepadnaviridae in the intestinal mucosa of patients suffering from CD and UC, respectively (Ungaro et al., 2019b). Despite all these studies, the actual role of virome dysbiosis during IBD has not been fulfilled yet, even with some reports revealing that intestinal inflammation may be initiated by eukaryotic viruses. By detecting and identifying viruses that infect IBD patients during the early stages of intestinal inflammation, it will be promising to establish a complete correlation between virome and disease progression.
Major dysbiosis of gut microbiota, mycobiome, and virome during IBD from different studies is summarized in Table 1. Moreover, the contribution of this dysbiosis to worsening the condition and progression of the disease is illustrated in Figure 1.
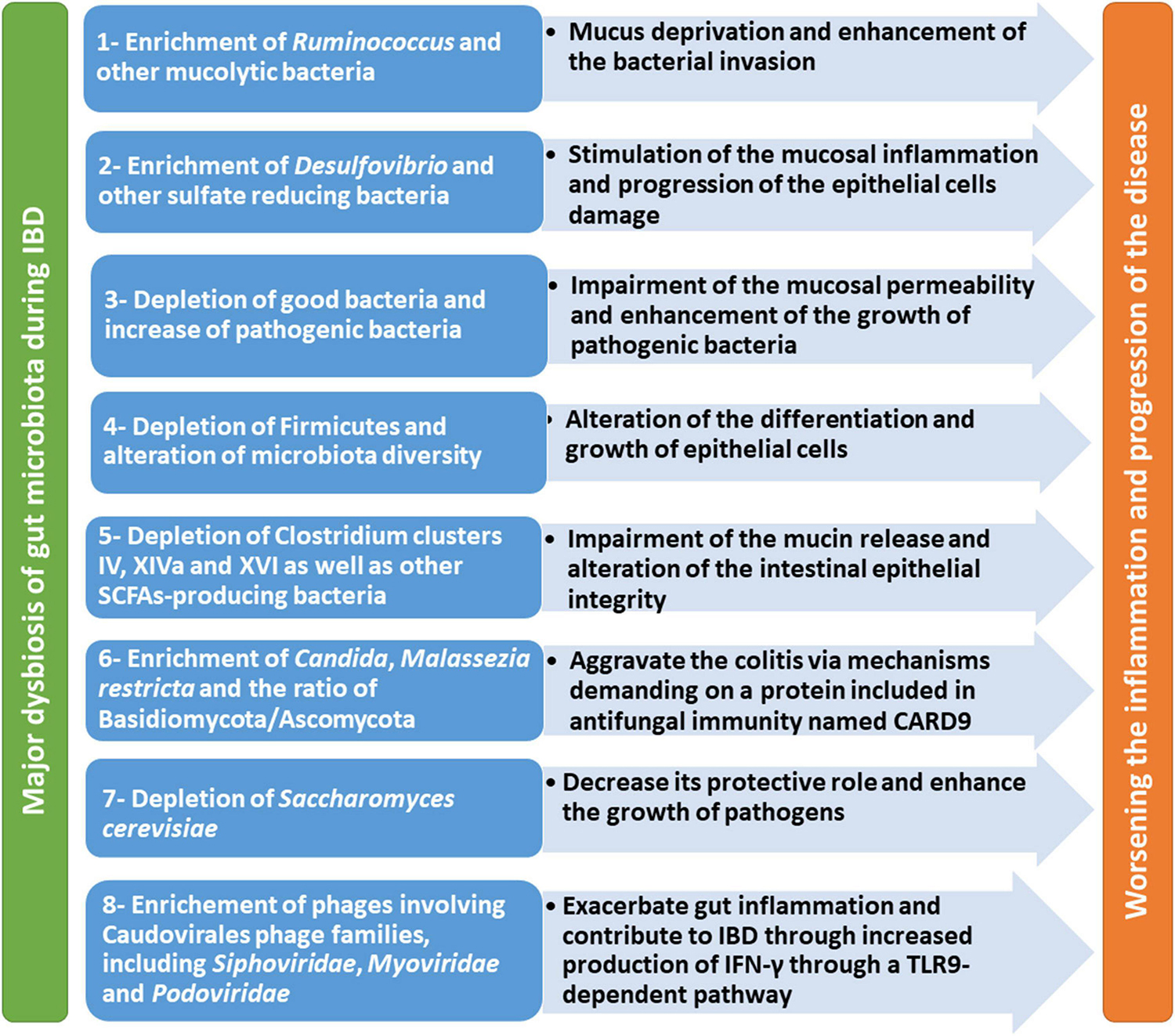
Figure 1. Major dysbiosis of gut microbiome during IBD and its impact on worsening the condition and progression of the disease.
Two metagenomic studies tried to link the dysfunctionality of the gut microbiome and IBD via two different approaches (Greenblum et al., 2012; Morgan et al., 2012). First, Greenblum et al. (2012) treated the gut microbiota as one single organism and constructed the metabolic network of the entire community and compared the odd ratios, of the topology of different metabolic pathways, genes and biological processes of obese and IBD cases against healthy individuals. This approach yielded an association between IBD and genes involved in NO2 production and metabolism of choline and p-cresol (Greenblum et al., 2012). In contrast to the metagenomic analysis of the community as a single supra-organism, Morgan et al. (2012) identified the microbiota composition of each disease phenotype. Then, they extracted the genome of the microbiota composing each community and constructed a gene catalog for each phenotype, followed by genes and pathway assessment using sparse multivariate analysis (Morgan et al., 2012). They also confirmed their findings by shotgun metagenomic analysis of 4 CD and 7 healthy fecal microbiota (Morgan et al., 2012). They found an IBD-associated enrichment of genes involved in glutathione transport and metabolism, sulfur amino acid metabolism, redox homeostasis, mucin degradation, secretion systems, adhesion, and invasion as well as a depletion of genes involved in the biosynthesis of SCFAs, some nucleotides and amino acids (Morgan et al., 2012). Recently, an interesting study identified a network of bacterial metabolite interactions including sulfur metabolism as an important player correlated to CD activity (Metwaly et al., 2020). To better understand the dysfunctionality of gut microbiota in IBD, we will discuss the IBD-associated metabolic shift in more detail.
Gut microbiota generate hydrogen sulfide (H2S) via 2 different biochemical pathways. First, sulfate reducing bacteria (SRB) couple the reduction of sulfate as a terminal electron acceptor to the oxidation of H2 or other organic compounds such as lactate as an electron donor during their anaerobic respiration (Carbonero et al., 2012). The second pathway is followed by bacteria that ferment sulfur containing amino acids. These bacteria might depend on desulfhydrases such as cysteine desulfhydrase or other enzymes, which have the ability to metabolize cysteine (Awano et al., 2005). Sulfate reducing bacteria mainly include members of Deltaproteobacteria in addition to Desulfotomaculum, Desulfosporosinus, Thermodesulfobacterium, and Thermodesulfovibrio genera (Blachier et al., 2010). Using multiplex PCR to identify fecal-SRB isolates, luminal SRB are dominated by Desulfovibriopiger, D. fairfieldensis, and D. desulfuricans (Loubinoux et al., 2002). A diversity analysis of Desulfovibrio using the dissimilatory sulfite reductase (dsrAB) gene confirmed the predominance of D. piger with the detection of a new unclassified SRB (Scanlan et al., 2009). Another study identified 8 sulfate and sulphite reducing bacteria in addition to the sulphite reducing bacterium, Bilophilawadsworthia via 454 pyrosequencing of dsrAB gene fragment (Jia et al., 2012). The identified bacteria include four known species; D. piger, D. vulgaris, Desulfovibrio sp. NY682, and D. desulfuricans F28-1, and four new species highly similar to D. desulfuricans F28-1 (93% dsrABsequence similarity); D. oxamicus (84% identity), Desulfotomaculum sp. Lac2 (80% identity), and D. simplex (88% identity) (Jia et al., 2012). H2S producing microbiota through fermentation of amino acids include, for example, Fusobacterium nucleatum, Atopobium spp., Gemella sanguinis, Micromonas micros, Streptococcus spp., Actinomyces spp., Eubacterium spp., Veillonella spp., Bulleidiamoorei., Prevotella spp., Campylobacter spp., and Selenomonas spp. (Washio et al., 2005). Mottawea et al. (2016) have reported the enrichment of H2S producers in children with CD with one H2S generating organism; A. parvulum, has proinflammatory characteristics in IL10–/– mice. Recently, an interesting study identified a network of bacterial metabolite interactions including sulfur metabolism as an important player correlated to CD activity (Metwaly et al., 2020). The authors showed that CD patients with active disease are enriched in members belonging to Enterococcus, Fusobacterium, Haemophilus, Megasphaera, and Campylobacter, while Roseburia, Christensenellaceae, Oscillibacter, and Odoribacter are enriched in CD patients with inactive disease (Metwaly et al., 2020). Another report detected two dominating sulfate-reducing bacteria morphotypes that differ in colonial size and quantitate in the feces of healthy and patients with colitis. In the feces of healthy individuals, 93% of sulfate-reducing bacteria of morphotype I prevailed (Desulfovibrio) while morphotype II included only 7% (Desulfomicrobium); in the feces of patients with colitis, the ratio of these morphotypes was 99:1, respectively (Kushkevych et al., 2021). In addition to microbiota-released H2S, it can also be synthesized endogenously by intestinal colonocytes from L-cysteine via two main enzymes; cystathionine β-synthase and cystathionine γ-lyase (Shibuya et al., 2013). Two additional pathways (3-mercaptopyruvate sulfurtransferase in combination with cysteine aminotransferase and 3-mercaptopyruvate sulfurtransferase coupled with D-amino acid oxidase) were identified in H2S production, peripherally (Shibuya et al., 2013; Guo et al., 2016). Normally, the luminal H2S concentration of the human large intestine is 1.0–2.4 mmol/L (Macfarlane et al., 1992), while the concentration in the fecal contents ranges from 0.17 to 3.38 mmol/kg (Florin, 1991; Magee et al., 2000). Taking into consideration the lipid solubility and the passive diffusion of H2S through the intestinal mucosa, these concentrations are underestimated.
At lower concentrations (<1 mM), H2S is considered a cytoprotective metabolite that induces some cellular anti-inflammatory responses. These cellular responses include, for example, prevention of caspase activation and apoptotic cell death (Rose et al., 2005), inhibition of leukocyte adhesion to vascular endothelium, which decreases infiltration of neutrophils and lymphocytes (Zanardo et al., 2006), induction of cyclooxygenase-2 (COX-2) expression (Wallace et al., 2009), and promotion of neutrophil apoptosis (Mariggio et al., 1998). The later was contradicted by Rinaldi et al. (2006) who concluded that H2S accelerates the resolution of the inflammation process via inhibition of polymorphonucelar (PMN) apoptosis. In another in vitro study, H2S at normal colonic concentration lowered the proliferation of different colonic cancerous and normal cells and induced autophagy through the AMP-activated protein kinase (AMPK) pathway (Wu et al., 2012). At higher concentrations, H2S acts as a genotoxic and/or cytotoxic transmitter to the colonocytes by affecting genes responsible for cell cycle progression, DNA repair and inflammatory responses (Attene-Ramos et al., 2010). The main cytotoxic effect of H2S is the inhibition of cytochrome c oxidase activity, which is the terminal oxidase of mitochondrial respiration (Roediger et al., 1993; Leschelle et al., 2005). This leads to the prevention of the oxidation of essential metabolites such as n-butyrate, L-glutamine and acetate, which eventually decreases the bioenergetic performance of the cell (Roediger et al., 1993; Leschelle et al., 2005). Sulfide has also been shown to inhibit butyrate oxidation in rat colonocytes through inhibition of short chain acyl dehydrogenation of activated fatty acids (Moore et al., 1997). The antagonistic effect of H2S on butyrate may induce hyperproliferation of the colonic mucosa (Christl et al., 1996). An indirect cytotoxicity of H2S was reported, where increased sulfide production induces the conversion of nitrite to nitric oxide, which had a damaging effect on colonocytes (Vermeiren et al., 2012).
To keep the local concentration of H2S at a harmless level, the colonic mucosa expresses a special H2S oxidation system that degrades H2S to sulfate and thiosulfate (Furne et al., 2001). This oxidation system consists of sulfide quinone reductase (SQR), dioxygenase ethylmalonic encephalopathy protein 1 (ETHE1), and thiosulfate sulfur transferase (TST that is also known as rhodanese) (Mimoun et al., 2012). This mitochondrial oxidation of H2S is not essential for cell respiration, but instead, its main purpose is the detoxification of excess H2S (Mimoun et al., 2012). In contrast, the respiratory capacity of the cell is an important parameter that affects the efficiency of H2S detoxification independent of the mitochondrial oxidation system (Mimoun et al., 2012).
A higher abundance of H2S generated by gut microbiota is considered one of the strongest models associated with the pathogenesis of IBD. Higher luminal H2S concentration in IBD could arise from either increased abundance of H2S-producing bacteria or a deficient H2S-detoxification pathway. The association between H2S and IBD was first reported in UC. In Roediger et al. (1997) reviewed the role of H2S in the pathogenesis of UC. They stated that the level of colonic sulfide and the relative abundance of sulfate reducing bacteria was higher in UC patients compared to healthy subjects (Roediger et al., 1997). In addition, the bacteria isolated from UC patients showed higher generation of H2S than those separated from control cases (Roediger et al., 1997). Treatment of those patients with 5-aminosalicylic acid containing drugs lowered the production of H2S as indicated by the stool sulfide level and this was proposed to contribute to the therapeutic activities of these drugs (Edmond et al., 2003). Sulfate reducing bacteria were exclusive to patients with UC, where they were isolated from 80% of UC pouches but not form patients with familial adenomatous polyposis (Duffy et al., 2002). Other indirect evidences for the role of H2S in the pathogenesis of UC are also available. The first comes from studies of diet consumption, where high protein intake is associated with a higher risk of IBD (Ng et al., 2013). Higher protein means increased sulfur containing amino acids and subsequently, elevated H2S levels. The second is the higher activity of fecal mucin sulphatase in UC patients (Tsai et al., 1995). Mucin sulphatase releases sulfate from the mucosal sulfomucin and this endogenous sulfate provides the source for H2S biosynthesis by SRB (Tsai et al., 1995). Regarding CD, few reports have linked H2S production with disease activity. Jia et al. (2012) have reported no difference in the general abundance of SRB between CD and healthy subjects. Mottawea et al. (2016) have illustrated the increased abundance of H2S producing bacteria along with downregulation of mitochondrial proteins implicated in H2S detoxification in children with CD. Some indirect links are also available. For example, increased metabolism of sulfur containing amino acids such as methionine and cysteine associated with a decrease in the metabolism of sulfur lacking amino acids such as lysine and glutamine are characteristics of ileal CD (Morgan et al., 2012). Also, the same study reported an over representation of sulfate transport genes in CD patients. Both sulfate and sulfur containing amino acids are the main precursors for H2S biosynthesis as mentioned above. The second factor that contributes to higher colonic sulfide concentration is the dysfunctionality of H2S detoxification. The dysfunctionality of H2S detoxification genes in IBD is still up for debate. Pitcher et al. (1998) showed that the activity of thiol-methyl transferase (TMT) is higher in the peripheral blood of UC patients. This was then confirmed in 2007, when the activity of TMT and rhodanese were found to be higher in the erythrocytes of UC patients than controls (Picton et al., 2007). However, no difference in TMT and rhodanese activity was detected in the rectal biopsies of the same individuals. In patients with CD neither the erythrocytes nor the rectal biopsies of patients showed a change in that enzymatic activity (Picton et al., 2007). In 2009, the role of H2S detoxification in IBD emerged again when the activity and the expression of rhodanese were shown to decrease in parallel to the development of dextran sodium sulfate-induced colitis in mice (Taniguchi et al., 2009). In concordance with formerly mentioned studies, this loss of activity is followed by an increase in its activity in red blood cells (Taniguchi et al., 2009). Impaired detoxification of H2S has been confirmed for UC patients via the assessment of TST expression level and activity in colonic mucosal biopsies (De Preter et al., 2012). As well, it has been confirmed for CD where metaproteomic and expression analyses reported the decreased abundance of H2S detoxification proteins and transcripts in children with new onset CD and UC (Mottawea et al., 2016). These includes the sulfur dioxygenase (ETHE1), the thiosulfate sulfurtransferase (TST) and the components of complexes III and IV of the mitochondrial respiratory chain, and tst, cytochrome c oxidase subunit IV (hcox41) and the sulfide dehydrogenase genes (SQRDL) transcripts (Mottawea et al., 2016). All in all, the association between higher H2S generation and IBD is well established. Additionally, the impaired intestinal H2S detoxification in IBD is evident.
Butyrate is known as the salt or ester of butanoic (butyric) acid, which is a weak acid with a pKa of 4.8. By considering the pH of the intestine, which is approximately neutral, most of the butyrate in the intestine will be in the anionic form rather than the free acid form. Butyrate biosynthesis by the gut microbiota starts by condensation of two molecules of acetyl coA to generate one molecule of butyrylcoA. Next, butyrate are generated from butyrylcoA via two main pathways; the enzymes butyrate kinase and phosphotransbutyrylase or butyryl-CoA: acetate-CoA-transferase (Louis and Flint, 2009). The second pathway has been shown to be predominant among butyrate producing microbiota in human colon (Duncan et al., 2002). The major butyrate producing bacteria that inhabit the human gut are related to Clostridium clusters XIVa and IV with few percentage of Clostridium clusters I, XV, and XVI (Louis and Flint, 2009), while acetate and propionate are mainly produced by Bacteroides (Martens et al., 2011). It was indicated that the terminal ileum and proximal colon are the main sites of butyrate production, while acetate and propionate are generated in the distal colon, where Bacteroidesis the predominant bacteria (Walker et al., 2005). Particularly, the role of butyrate in preventing IBD is illustrated in several studies as mentioned below.
A large body of evidence has revealed the association between IBD, perturbations of butyrate metabolism and/or depleted butyrate producing bacteria in the gut. Initially, a raised luminal butyrate level, as a result of impaired oxidation by colonocytes, was correlated with the severity of mucosal inflammation and was considered as a biomarker in UC patients (Roediger, 1980; Roediger et al., 1982). On the other hand, butyrate intake was shown to have a protective effect against development of colitis independent of restored butyrate oxidation in UC patients (De Preter et al., 2011). For example, administration of butyrate either orally or via local enema to mice or rats with chemically induced colitis alleviated the mucosal inflammation (Butzner et al., 1996; Vieira et al., 2012). Indeed, butyrate exhibits several anti-inflammatory activities. First, butyrate inhibits NFκB activation, which results in suppression of proinflammatory cytokines in UC patients (Luhrs et al., 2002) and rats with trinitrobenzene sulphonic acid (TNBS) induced colitis (Segain et al., 2000). This inhibitory action is impaired in IBD individuals as revealed by assessment of butyrate effect on cytokines production by peripheral blood mononuclear cells (PBMC) isolated from IBD and healthy subjects in response to TLR-2 activation (Kovarik et al., 2011). Secondly, butyrate was reported to induce Fas-mediated apoptosis of T cells via inhibition of HDAC-1 in mice (Zimmerman et al., 2012). This in turn inhibits IFN-γ-induced STAT1 activation, which results in reduced colonocyte expression of inflammatory mediators such as nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX2) (Zimmerman et al., 2012). Finally, Butyrate is known to reduce the inflammation through contributions to intestinal barrier integrity. For example, intra-rectal inoculation of C. tyrobutyricum, the potent butyrate producer, has restored MUC-2 secretion, upregulated expression of (ZO)-1 tight junction protein and reduced cytokine release in DSS-treated mice (Hudcovic et al., 2012). Recently, Magnusson et al. (2020) compared impacts of butyrate on the intestinal immune profile of UC patients with active disease and non-inflamed controls. They found that butyrate exhibits different impact on gene regulation and more strongly leads to down-regulation of +expressed genes of inflammatory pathways in non-inflamed controls than in inflamed tissue of UC patients (Magnusson et al., 2020). According to the authors, such discrepancies can partly elucidate why expected anti-inflammatory impacts of local butyrate stimulation or supplementation are not always obtained.
Regarding butyrate producing bacteria, screening of fecal microbiota of 6 healthy and 6 CD individuals via 16S rDNA clone sequencing and FISH analysis revealed a depletion of C. leptum cluster in CD cases compared to the healthy group (Manichanh et al., 2006). In agreement with this, Sokol et al. (2008b) illustrated that F. prausnitzii was depleted in CD patients mucosal microbiota and this reduction in abundance was associated with a higher risk of recurrence of ileal CD. Though, they have shown that the anti-inflammatory activity of this bacterium is independent of butyrate production using in vitro cellular models and in vivo colitis model of mice. Recently, depletion of F. prausnitziiand other butyrate producers was confirmed in the fecal microbiota of adults with CD compared to their unaffected relatives and other healthy controls via denaturing gradient gel electrophoresis and real time PCR (Joossens et al., 2011). This is not the case in pediatric CD, where some studies reported that F. prausnitzii had higher abundance in CD in comparison to controls (Hansen et al., 2012; Mottawea et al., 2016). An important difference in the pediatric studies is that the samples were collected at the time of diagnosis by colonoscopy, in contrast to Sokol et al. (2008b) where specimens were taken at surgical resection. This indicates the importance of sample collection time for these kinds of microbiota studies. The age groups screened by different studies were also different confirming the different phenotypes of the disease at different ages.
Geirnaert et al. (2017) reported the therapeutic effect of butyrate-producing bacteria in CD patients in addition to its role in enhanced intestinal epithelial barrier integrity. The four-strain probiotic supplement (containing Lactobacillus acidophilus NCIMB 30175, Lactobacillus plantarum NCIMB 30173, Lactobacillus rhamnosus NCIMB 30174, and Enterococcus faecium NCIMB 30176) was found to positively influence the immune response via colonic butyrate production in vitro and facilitating modulation of the gut microbiota composition and metabolic enhancing (Bjarnason et al., 2019; Moens et al., 2019).
Bile acids are self signaling molecules that regulate their own biosynthesis through a negative feedback mechanism (Sayin et al., 2013). Primary bile acids are the product of cholesterol breakdown in the liver, while secondary bile acids are the products of gut microbiota metabolism. primary and secondary bile acids have been found to act as signaling players on a group of cell membrane and nuclear receptors together named “bile acid-activated receptors.” These receptors are highly expressed all over the gastrointestinal tract and control the bilateral communications of the intestinal microbiota with the host immune system (Fiorucci et al., 2021). Bile acid absorption is affected in models of inflammatory bowel disease (Fitzpatrick and Jenabzadeh, 2020). The expression of bile acid transporter apical sodium dependent bile acid transporter was inhibited in rats with colitis in addition to murine, canine and rabbit models of intestinal inflammation (Fitzpatrick and Jenabzadeh, 2020).
Bile acids mainly act as digestive aids to facilitate digestion of cholesterol, fat-soluble vitamins and triglycerides into water soluble products so they can be absorbed from the small intestine (Russell, 2003). They are also nutrient regulatory molecules that mediate some endocrine functions (Houten et al., 2006; Hylemon et al., 2009). They activate some nuclear receptors and cell signaling pathways to regulate lipid and glucose metabolism, energy expenditure and triglyceride homeostasis (Watanabe et al., 2006; Hylemon et al., 2009; Trauner et al., 2010). In addition, they have been shown to exert antimicrobial activity either directly through their detergent characteristics on bacterial membranes or via indirect physiological function (Hofmann and Eckmann, 2006; Duboc et al., 2013). Their indirect antimicrobial activity mainly occurs in the distal small intestine, where they promote the activation of nuclear farnesoid X receptor (FXR) (Inagaki et al., 2006). Activation of FXR induces the expression of several host genes involved in mucosal defense including genes involved in oxidative stress, and antibacterial peptide biosynthesis (Bernstein et al., 1999; Hofmann and Eckmann, 2006; Inagaki et al., 2006). The expression and function of bile acid-activated receptors including FXR in addition to other receptors; G-Protein bile acid-activated receptor, pregnane-X-receptor, vitamin D receptor, and related orphan receptor gamma are strongly linked to the composition of the intestinal microbiota and negatively regulated by intestinal inflammation (Fiorucci et al., 2021).
The enterohepatic circulation of bile acids starts at the liver, where 14 enzymes are required to synthesize the two primary forms of bile acids namely cholic acid and chenodeoxycholic acid from cholesterol (Russell, 2003). Thereafter, they are conjugated to glycine or taurine to form bile salts (also termed conjugated bile acids), which are stored in the gallbladder during interdigestive periods. After meals, bile salts are released into the duodenum via the biliary duct to facilitate lipid digestion and absorption (Ridlon et al., 2006). In the gut lumen, the microbiota induce biotransformation of the primary forms of bile acids into secondary bile acids products via deconjugation, oxidation of hydroxyl groups and dehydoroxylation (Ridlon et al., 2006). This increases the hydrophobicity of bile acids so that they can passively diffuse through the small intestine and proximal colon in addition to their active transport from the small intestine to the blood stream and back to the liver (Ridlon et al., 2006). Approximately 95% of bile acids are recycled and transported through the circulation to return to the liver while 5% (400–600 mg) enter the colon, where they are dehydroxylated by colonic bacteria into secondary bile acid products [Deoxycholic acid (DCA) and Lithocholic acid (LCA); Ridlon and Hylemon, 2012].
Gut microbiota start the metabolism of bile salts via the deconjugation step through bile salt hydrolases (BSH) that generate free primary bile acids and amino acids (Ridlon et al., 2006). The distribution of BSH activity among gut microbiota was estimated via metagenomic analysis of 89,856 fecal clones (Jones et al., 2008). Firmicutes were the most predominant BSH-expressing bacteria (30%) followed by Bacteroidetes (14.4%) and Actinobacteria (8.9%) (Jones et al., 2008). Firmicutes with BSH activity include the genera Eubacterium, Coprococcus, Clostridium, Ruminococcus, Dorea, Lactobacillus, Enterococcus, Listeria, and Lactococcus while Bifidobacterium and Collinsella are the two Actinobacteria genera that express BSH (Jones et al., 2008). The same study revealed that Firmicutes and Actinobacteria are capable of deconjugating both glyco- and tauro-conjugated bile acids while Bacteroidetes are only able to hydrolyse tauro-conjugated bile acids. Also 9 BSH open reading frames have been identified among the gut microbiota community, one of them shares 56% homology with a protein encoded by the gut, archeon Methanobrevibacter smithii, which has been confirmed to have BSH activity against both glyco- and tauro-conjugated bile acids (Jones et al., 2008). The second step of bile acid metabolism by intestinal bacteria is the oxidation of the hydroxyl group at carbon 3, 7, or 12 of deconjugated bile acids via hydroxysteroid dehydrogenases to generate the oxo-derivative of bile acids (Ridlon et al., 2006). Gut microbiota expressing hydroxysteroid dehydrogenases include, but are not limited to Clostridium spp., Eggerthella lentum, Ruminococcus spp., Bacteroides spp., and E. coli (Ridlon et al., 2006; Fukiya et al., 2009). Simultaneously, some microbes promote 7α-/β-dehydroxylation of free cholic acid and chenodeoxycholic acid to generate deoxycholic acid (DCA) and lithocholic acid (LCA), respectively (Ridlon and Hylemon, 2012; Kakiyama et al., 2013). This dehydroxylation activity is common among the order Clostridiales of gut bacteria including Ruminococcaceae, Lachnospiraceae, and Blautia (Kakiyama et al., 2013). Heinken et al. (2019) performed an interesting study that included a systematic workflow to computationally model bile acid metabolism by gut microbes and microbial communities. The authors found that, each microbe could produce maximally 6 secondary bile acids in silico, while microbial pairs could produce up to 12 bile acids, suggesting bile acid biotransformation being a microbial community task (Heinken et al., 2019). Das et al. (2019) performed a shotgun metagenomic analysis of the bile salt biotransformation genes and their distribution at the phyla level. They reported that IBD patients harbored a significantly lower abundance of these genes in comparison with healthy individuals; many of these genes originated from Firmicutes (Das et al., 2019).
Bile acids metabolism by gut microbes has a controversial contribution to both the host and the microbe. The deconjugation step has been suggested to benefit gut microbiota colonization by increasing their resistance to bile salts, where the antimicrobial effect of bile acids benefit the host via alleviating the bacterial overgrowth (Desmet et al., 1995; Ridlon et al., 2006; Jones et al., 2008). Also, bile acid modification results in reducing cholesterol level and controls lipid metabolism, which is considered a protective factor against metabolic disorders such as obesity, cardiac diseases and diabetes (Tonack et al., 2013; Wu et al., 2013). Another link between bile acids and metabolic diseases arises from its effect on gut microbiota composition. Using 16S rDNA clones sequencing of rats’ cecal microbiota has revealed that cholic acid is a host modifier of the gut microbiota structure that is equivalent to high fat diet (Islam et al., 2011). One more benefit to the host from bile acid metabolism is the protection from pathogen colonization such as vegetative C. difficile (Sorg and Sonenshein, 2008). However, high levels of secondary bile acids are associated with some diseases such as gastrointestinal cancer and gallstones (Berr et al., 1996; Bernstein et al., 2005).
Four decades of research have established the association between IBD and bile acid dysmetabolism (Andersson et al., 1978; Heuman et al., 1983; Nyhlin et al., 1994; Duboc et al., 2013; Iwamoto et al., 2013). Different studies have linked the dysmetabolism of bile acids to gut microbiota dysbiosis in IBD (Wohlgemuth et al., 2011; Duboc et al., 2013). It is clear now that cholic acid or a high fat diet act as a regulatory host factor that selects for gut microbes that have the machinery to detoxify bile acid metabolites or utilize their conjugates, glycine and taurine as metabolic substrates (De Filippo et al., 2010; Islam et al., 2011; Devkota et al., 2012). Devkota et al. (2012) reported that diets high in saturated fat induce the expansion of the H2S producing pathobiont, Bilophila wadsworthia, which in turn promotes colitis in IL10–/– mice. This proinflammatory effect of high saturated fat diet was attributed to taurocholic acid (Devkota et al., 2012). Taurine, the conjugate in taurocholic acid, is known to have a sulfonic acid moiety that could be dissimilated by gut microbiota into H2S as a microbial by-product (Laue et al., 2001). Diets high in fat or meat lead to more conjugation of taurine to bile acids and consequently more H2S production, which is considered as a potential risk factor for IBD (Magee et al., 2000; Devkota et al., 2012). Alternatively, microbial dysbiosis associated with IBD may result in bile acid dysmetabolism and this in turn may affect the anti-inflammatory characteristics of bile acids (Duboc et al., 2013). Duboc et al. (2013) has reported the dysfunctionality of bacterial metabolism of bile acids because of microbial dysbiosis. Moreover, they illustrated that this intestinal dysmetabolism disturbed the intestinal bile acid pool, which in turn impacted the anti-inflammatory characteristics of bile acids (Duboc et al., 2013). Indeed, bile acids are known as anti-inflammatory mediators that inhibit NFκB activation and consequently reduce cytokine production by macrophages (Wang et al., 2011). This anti-inflammatory effect is characteristic of secondary bile acids but not the conjugated forms, which stresses the importance of deconjugation by gut microbiota (Duboc et al., 2013). Another anti-inflammatory effect of bile acids may be attributed to the activation of FXR, the bile acid receptors, which has been revealed as a protective factor against chemically induced colitis in mice (Gadaleta et al., 2011). Mice lacking FXR receptors have developed compromised intestinal barrier and antimicrobial defense in small intestine (Inagaki et al., 2006). The role of bile acids in protection against inflammation has been confirmed recently after identifying the association between a genetic variation of NR1H4, the gene encoding FXR receptor, and IBD (Attinkara et al., 2012). A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease demonstrated that bile acid profiles were in general alike among patients with IBD and healthy controls (Vaughn et al., 2019). Oral administration of secondary bile acids in mice was reported to reduce the severity of colitis and ameliorate colitis-associated fecal dysbiosis at the phylum level (Van den Bossche et al., 2017). In accordance with the last study, dysbiosis of gut microbiota was found to induce deficiency in secondary bile acids in inflammatory-prone UC patients, which in turn leads to pro-inflammatory status in the intestine that may be treated via restoring secondary bile acids (Sinha et al., 2020). Like bile acid dysmetabolism, bile acid malabsorption was reported to be common reason of diarrhea in CD and colitis patients (Hou et al., 2018; Mena Bares et al., 2019).
Oxidative stress is thought to be one of the key players in the tissue damage associated with IBD. Oxidative stress is defined as an imbalance between reactive oxygen species and intracellular antioxidants. Hence, oxidative stress arises from either a higher production of oxidative free radicals (Keshavarzian et al., 2003) and/or deficient antioxidant machinery (Koutroubakis et al., 2004). The inflammatory cascade starts by the infiltration of proinflammatory cells to the intestinal mucosa, which release reactive oxygen species (ROS) and/or reactive nitrogen metabolites. For example, chemiluminescence analysis of ROS has elucidated a higher release of these free radicals by monocytes and polymorphonuclear cells extracted from both CD and UC biopsies (Kitahora et al., 1988; Keshavarzian et al., 1992). These free radicals induce more infiltration of proinflammatory cells and so this cycle is sustained and eventually causes tissue damage. The disruption of the intestinal epithelium exposes the immune system to the gut microbiota or other antigenic luminal components, which exacerbate the inflammation resulting in the active phenotype of the disease (Keshavarzian et al., 2003). For the cells to protect themselves against oxidative stress, they have to induce the production of antioxidant metabolites, which is not the case in IBD. It has been reported that IBD is associated with a depleted total antioxidant capacity of the cells or individual antioxidants. For example, IBD patients are characterized by a depletion of copper/zinc containing protein (superoxide dismutase and metallothionein), lower glutathione transferase activity with higher glutathione peroxidase in UC, and lower colonic ascorbate (Koutroubakis et al., 2004). Indeed, it is well reported that increased oxidative stress level in IBD patients and the detection of oxidative stress index rate could be used as predictors for the pathogenesis of IBD (Yuksel et al., 2017; Luceri et al., 2019). Bourgonje et al. (2019) showed that plasma free thiols are reduced in patients with CD, reflection of systemic oxidative stress, in clinical remission. The authors recommended that systemic oxidative stress and plasma free thiols may be a relevant therapeutic target and biomarker to monitor disease activity in CD (Bourgonje et al., 2019). Oral administration of probiotics in IBD patients were reported to effectiveness via the decrease of oxidative stress values (Ballini et al., 2019).
For the gut microbiota to maintain their homeostasis at this oxidative stress, they must develop oxidative stress resistance machinery. Morgan et al. (2012) demonstrated a shift of IBD microbiota toward microbes that possess the glutathione generation and reduction capability, which enables them to compensate for the oxidative stress. This machinery includes higher cysteine biosynthesis, which is a precursor of glutathione, riboflavin and NADPH, which are cofactors of glutathione reduction reaction and glutathione transfer gene (Morgan et al., 2012). Glutathione is a tripeptide (γ-glutamylcysteinylglycine) thiol that is produced by the majority of living cells (Anderson, 1998). The reduced form of glutathione protects the cells from toxic oxygen metabolites and other electrophiles via keeping the cell in a reduced state (Anderson, 1998). It also has other protective functions via regulation of gene expression, cell apoptosis and transport of organic solutes (Hammond et al., 2001). With regards to gut microbiota, glutathione has been reported to be biosynthesized by Proteobacteria members and a limited number of gram positive bacteria such as some Streptococcus spp. and Staphylococcus aureus, but not by Clostridium, Bacillus or Micrococcus members (Fahey et al., 1978). Also, glutathione sulfur transferase encoding genes that possess peroxidase activity are expressed by Proteobacteria members (Bartels et al., 1999). It is well documented now that IBD microbiota is dominated by Proteobacteria, which is known to produce some proinflammatory metabolites such as enterotoxins or LPS. This might generate a testable hypothesis that the inflammation associated stresses in the gut, such as oxidative stress, might constitute a selective pressure that induces a microbial shift toward the stress-resistant microbes. Furthermore, this shift might develop a colitogenic microbiota that could maintain the active chronic phenotype of the inflammation.
Medications for IBD can be classified into five classes: aminosalicylates (sulfasalazine, olsalazine, and mesalamine), Corticosteroids (cortisone, prednisone, prednisolone, hydrocortisone, methylprednisolone, beclometasone, and budesonide), immunosuppressive agents (6-Mercaptopurine, azathioprine, methotrexate, and tacrolimus), antibody agents (Anti-TNF agents (infliximab, adalimumab, certolizumab pegol), and antibiotics (metronidazole, ciprofloxacin and rifaximin) (Gade et al., 2020; Targownik et al., 2020). The choice of the treatment strategy depends on the severity of the disease and the response to previous therapy. The effect of some of the above-mentioned drugs on gut microbiota are summarized in Table 2. Interestingly some drugs that are used for treatment of IBD requires metabolic activation via the gut microbiota e.g., sulfasalazine, balsalazide (mesalamine prodrug), olsalazine, and methotrexates (Crouwel et al., 2020).
It should be mentioned that some IBD medications are reported to affect either the metabolism of gut microbiota or the metabolic status of intestinal cells by altering the intestinal biota. Sulfasalazine was reported to enhance carbohydrate metabolism, citrate cycle and decrease the oxidative stress (riboflavin, sulfur, cysteine) (Zheng H. et al., 2017). Dahl et al. (2017) showed that, mesalamine was able to decrease polyphosphate levels in bacteria, including members of the human gut microbiota. This reduction leads to bacterial sensitization to oxidative stress and decreases bacterial colonization (Arthur et al., 2012). Effenberger et al. (2021) performed an in-silico metabolic prediction analysis by including azathioprine or anti-TNF antibodies-treated IBD groups and assessed the effect of gut microbiota function on remission status. They found that the predicted butyrate synthesis was significantly enriched in patients achieving clinical remission. The use of oral steroids in IBD patients was demonstrated to affect two biosynthetic pathways of methanogenesis and one pathway in the biosynthesis of vitamin B2 and nucleosides (Vich Vila et al., 2020). The oral administration of metronidazole was found to reduce basal oxidative stress in colonic tissue of healthy rat (Pelissier et al., 2007) and increase the thickness of colonic mucosal layer by about twofolds (Pelissier et al., 2010). Moreover, the metronidazole-treated microbiota in murine fecal donors retained its ability to control inflammation co-occurring with enrichment of Lactobacillus and innate immune responses including invariant natural killer T cells in experimental colitis (Pelissier et al., 2010).
Probiotics are living organisms that when are given in appropriate amount to the host result in health benefit (Hill et al., 2014). Prebiotics are substances, which are utilized by probiotics leading to enhancing the health (Gibson et al., 2017). Postbiotics are non-living microorganisms with or without their cell components and metabolites that confer health benefits (Salminen et al., 2021). Although probiotics have the ability to modulate microbiome composition resulting in enhancing the growth of good species and inhibiting the growth of pathogenic ones, their use in IBD treatment is recommended only in the context of clinical trials in adults and children (Su et al., 2020). Probiotics have anti-inflammatory impact and enhance intestinal barrier functions (Abraham and Quigley, 2017). In adults and children with pouchitis, the use of eight-strain combination of Lactobacillus plantarum, Lactobacillus paracasei subspparacasei, Lactobacillus delbrueckii subsp bulgaricus, Lactobacillus acidophilus, Bifidobacterium breve, Bifidobacterium longum subsp. longum, Bifidobacterium longum subsp. infantis, and Streptococcus salivarius subsp. thermophilus is recommended (Su et al., 2020). It is well documented that the probiotic cocktail VSL#3, which is composed of 3 Bifidobacteria strains, 4 lactobacilli strains, and 1 Streptococcus strain, is promising for treatment of patients suffering from IBD (Bibiloni et al., 2005; Miele et al., 2009; Fedorak et al., 2015). Also, the probiotic Lactobacillus reuteri ATCC 55730 was speculated to be helpful during the treatment of IBD cases (Oliva et al., 2012). In addition, the probiotic cocktail composed of Lactobacillus acidophilus NCIMB 30175, Lactobacillus plantarum NCIMB 30173, Lactobacillus rhamnosus NCIMB 30174, and Enterococcus faecium NCIMB 30176 has a well-documented effect in the treatment of IBD patients especially UC (Bjarnason et al., 2019; Moens et al., 2019). More recently, next generation probiotics such as A. muciniphila and F. prausnitzii and their supernatants (postbiotics) are reported to exhibit beneficial effects during IBD treatment (Sokol et al., 2008b; O’Toole et al., 2017). Fermented foods are good sources of probiotics including miso, tempeh, kefir, kimchi, pickled vegetables, yogurt with live active cultures, kombucha tea and sauerkraut (Sultan et al., 2020).
Also, prebiotics such as inulin, resistant starch, gums, pectins, and fructo-oligosaccharides are reported to be beneficial in the treatment of IBD patients through enhancing functions of the intestinal barrier and protecting vs. invasion and translocation of pathogens (Akram et al., 2019). Although there are many commercial products of these prebiotic fibers, healthy diets are considered their main source such as bananas, beans, onions, raw version of leeks, oats, dandelion greens, wheat, garlic, asparagus, artichokes, barley, seaweed, and other fruits and vegetables that are rich in fibers and indigestible carbohydrates (Sultan et al., 2020). Such prebiotics aid the growth of normal gut microbiota and the production of SCFAs, which results in enhancing the activity of immune cells, maintaining the levels of glucose and cholesterol as well as decreasing the pH of the colon, which results in enhancing the condition (Akram et al., 2019; Sultan et al., 2020).
Phages are the most ubiquitous organisms worldwide and they are characterized by their selectivity and specificity to bind their target host (Abd El-Aziz et al., 2019). Phages can bind and lyse specific bacterial strains within certain species (El-Mowafy et al., 2021). This capability gives the phage the advantage to be safer during the treatment of bacterial infections than commonly used antibiotics and to have limited effect on microbiota of the host (Abd El-Aziz et al., 2019; El-Mowafy et al., 2021).
The use of Russian coliphage or oral T4-like coliphages in children with bacterial infection-induced diarrhea does not induce any side effect, however, it was unsuccessful to enhance the conditions (Sarker et al., 2016). Recently, it is recommended to use phages against AIEC in patients suffering from IBD (Galtier et al., 2017). AIEC is an abnormal pathogen, which is commonly found in the ileal mucosa of IBD patients (Barnich et al., 2007). The use of bacteriophages to lyse AIEC significantly reduced the symptoms of DSS-induced colitis in transgenic mice expressing the human receptor for AIEC named CEACAM6 (Galtier et al., 2017). Therefore, phages targeting AIEC may be a promising therapeutic approach for the treatment of IBD patients.
FMT is defined as the transplantation of healthy donor fecal microbiota into the gut of a patient as a trial to reverse the dysbiosis, restore homeostasis of the gut microbiota and improve the condition (Burrello et al., 2018; Hvas et al., 2019). This approach was reported to be successful during the treatment of recurrent C. difficile infection (CDI) (Khoruts and Sadowsky, 2016). The successful treatment of IBD patients using FMT depends on three main factors including; early treatment, content of fecal matter of the donor and usage of multiple FMTs (Moayyedi et al., 2015). Treatment of IBD patients using FMT is based on the idea that regaining the gut microbiota homeostasis will alter the growth of pathogens like C. difficile that are responsible for the disease (Khoruts et al., 2016). This is achieved through competition between the regained microbiota and pathogens for nutrients and colonization, production of antimicrobials by regained microbiota that directly affect pathogens, and vegetative growth and spore germination inhibition through bile acid mediated mechanism (Khoruts and Sadowsky, 2016). However, it is noteworthy that the fungal and viral content of the stool of donors may affect the outcome of IBD treatment using FMT (Draper et al., 2018; Zuo et al., 2018). It is reported that using stool rich in C. albicans significantly decreases the treatment efficacy (Zuo et al., 2018). Moreover, it was depicted that following FMT, different patients who received fecal matter from the same donor exhibited extremely individualized virus colonization patterns (Draper et al., 2018).
Costello et al. (2019) showed that 1 week treatment with anaerobically prepared fecal material, in mild to moderate UC patients, led to a higher probability of remission after 8 weeks. Another interesting work followed a randomized controlled study to investigate the role of FMT to maintain remission in CD patients (Sokol et al., 2020). The authors found a low similarity index between donor and recipient microbiota in some patients suggesting that a single FMT might not be enough to stimulate significant changes in these patients. An interesting study aimed to investigate the optimum timing of FMT for maintaining the long-term clinical benefits in UC (Li et al., 2020). The authors demonstrated that UC patients could take the 2nd course of FMT within 4 months after the initial course of FMT, which will allow clinicians to consider sequential FMTs as a long-term treatment strategy for UC. Moreover, they showed that the relative abundance of Eubacterium, Ruminococcus, Eggerthella, and Lactobacillus in UC patients can be used as predictors for the long-term efficacy of FMT (Li et al., 2020). Similarly, the species Eubacterium hallii, Ruminococcus bromii, and Roseburia inulivorans were recommended to predict the success of FMT therapy in UC patients (Paramsothy et al., 2019). However, the application of FMT has many disadvantages. First, different samples represent different bacterial ensembles which means variable efficacy and the outcome will rely on the sample (Choi and Cho, 2016). Additionally, safety risks may arise from the presence of undesirable strain/functionality within the donated stool sample such as transferable antibiotic resistance elements, virulence factors or pathogenic strains enteropathogenic E. coli (Nataro and Kaper, 1998; Choi and Cho, 2016).
There is a tight bond between diet, gut microbiota, colonocytes, and immune cells during both the intestinal healthy conditions (homeostasis) and inflammation (Celiberto et al., 2018). Healthy diet patterns including diets rich in fruits, vegetables, probiotics, fermentable food, fibers, prebiotics, and adequate concentrations of vitamin D are good for intestinal health and homeostasis (Rinninella et al., 2019). Healthy diets generally result in more microbial diversity and an increase of good bacteria including F. prausnitzii, Bifidobacterium spp., and Lactobacillus spp. that result in increased production of SCFAs, especially butyrate (Celiberto et al., 2018; Rinninella et al., 2019). Butyrate has strong anti-inflammatory characters as it improves the function of intestinal barrier and endorses the proliferation of regulatory T cells (Treg) (Kespohl et al., 2017). Moreover, butyrate promotes the dendritic cells (DC) within the lamina propria and commensal microbiota antigens within the intestinal lumen. Dendritic cells mainly trigger Treg cells to produce transforming growth factor-β (TGF-β) and interleukin-10 (IL-10) through the production of TGF-β resulting in tolerant immune response and intestinal homeostasis (Zheng L. et al., 2017). These modulations result in more thick mucus layer, which is an important property of intestinal homeostasis, giving a protective shield between epithelial cells and luminal bacteria (Butzner et al., 1996; Segain et al., 2000; Geirnaert et al., 2017). Moreover, healthy diets represent a good source of aryl hydrocarbon receptor (AhR) which induce the production of IL-22, via the innate lymphoid cell 3 (ILC3) that maintains intestinal barrier function (Song et al., 2020).
Healthy diets harboring adequate concentrations of vitamin D as well as vitamin D supplements are reported to be helpful during IBD cases (Celiberto et al., 2018). It is reported that vitamin D is crucial for maintaining the composition of gut normal microbiota, where vitamin D receptor (VDR) deficient mice exhibited increased abundance of Bacteroidetes and Proteobacteria as well as diminished Lactobacillus spp. resembling the dysbiotic cases of IBD patients (Winter et al., 2017; Windsor and Kaplan, 2019). Vitamin D enhances the epithelial barrier function and promotes the production of antimicrobial peptides (Celiberto et al., 2018). It also promotes the proliferation of dendritic cells, which in turn promotes the production of the immunosuppressive IL-10 through provoking Treg cells (Sokol et al., 2006b; Jin et al., 2015). Vitamin D also decreases the production of pro-inflammatory cytokines including TNF-γ, IL-17, and IL-21 through its inhibitory effect on IL-12 and IL-23 that are responsible for the responses of Th-1 and Th-17, respectively (Santos-Antunes et al., 2016; Winter et al., 2017).
On the other hand, unhealthy diets such as Western dietary patterns those are low in fruits, vegetables, probiotics, fermentable food, fibers, prebiotics, and vitamin D are directly related to dysbiosis of the microbiota and diminished microbial diversity, decreasing SCFAs production, reducing good bacteria and increasing the pathobionts such as E. coli and Clostridium difficile (Sugihara and Kamada, 2021). This results in more patchy and thinner mucus layer, which as a result provides less protection between colonocytes and luminal bacteria (Sugihara and Kamada, 2021). In addition, the compromised intestinal barrier function, due to decreased expression of cellular tight junctions, leads to escape of harmful bacterial products including lipopolysaccharides (LPS) from the intestinal lumen to the lamina propria (Ghosh et al., 2020). LPS trigger the macrophages via binding to toll-like receptors (TLR) leading to the production of TNF-α. TNF-α endorses the proliferation of T helper cell type 1 (Th1) that produce the proinflammatory cytokines including TNF-α and TNF-γ resulting in inflammation and dysfunctionality of the intestinal barrier (Grassin-Delyle et al., 2020). Moreover, the condition may worsen due to decreasing Treg cells responsible for the production of IL-10, which mitigates the intestinal inflammation (Iyer and Cheng, 2012).
From what has been discussed above, gut microbiota exerts many beneficial roles to human health, where any permanent disturbance of this ensemble could metabolically lead to a disease state. Gut microbiota of IBD patients is characterized by depletion of Firmicutes and Bacteroidetes with increased abundance of Proteobacteria and Actinobacteria. These changes in microbial composition result in perturbations of microbial functions that could be summarized in less short chain fatty acid production, less bile acid hydrolysis, higher redox potential and increased H2S production (Figure 2). This pattern of microbial metabolic perturbations is associated with defectiveness of some human cellular pathways such as defective H2S detoxification, SCFAs transport and oxidation, and high luminal redox potential. The interaction between both sides of dysfunctionality is considered pathogenic to the host and might modulate the chronicity of the disease. However, the knowledge of the cause/effect relationship between these metabolic perturbations and the inflammation has so far been limited. In addition, the actual molecular mechanisms that regulate the interaction between both sides of the equation still need to be identified. Still, different strategies are being developed to manipulate gut microbiota to reduce the intestinal inflammation and improve the disease outcomes. These include live biotherapeutics such as FMT and probiotics. Taking in account the disadvantages of FMT and enrichment-based approaches in general, new approaches may rely on bottom-up rational to design identified bacterial consortia that are metabolically interdependent and exert a variety of protective functions to the host and the gut microbiota. These functions include generation of beneficial metabolites such as SCFAs and indole, deconjugation and conversion of bile acids, competition for critical nutrients and synthesis of antimicrobial molecules against opportunistic pathogens.
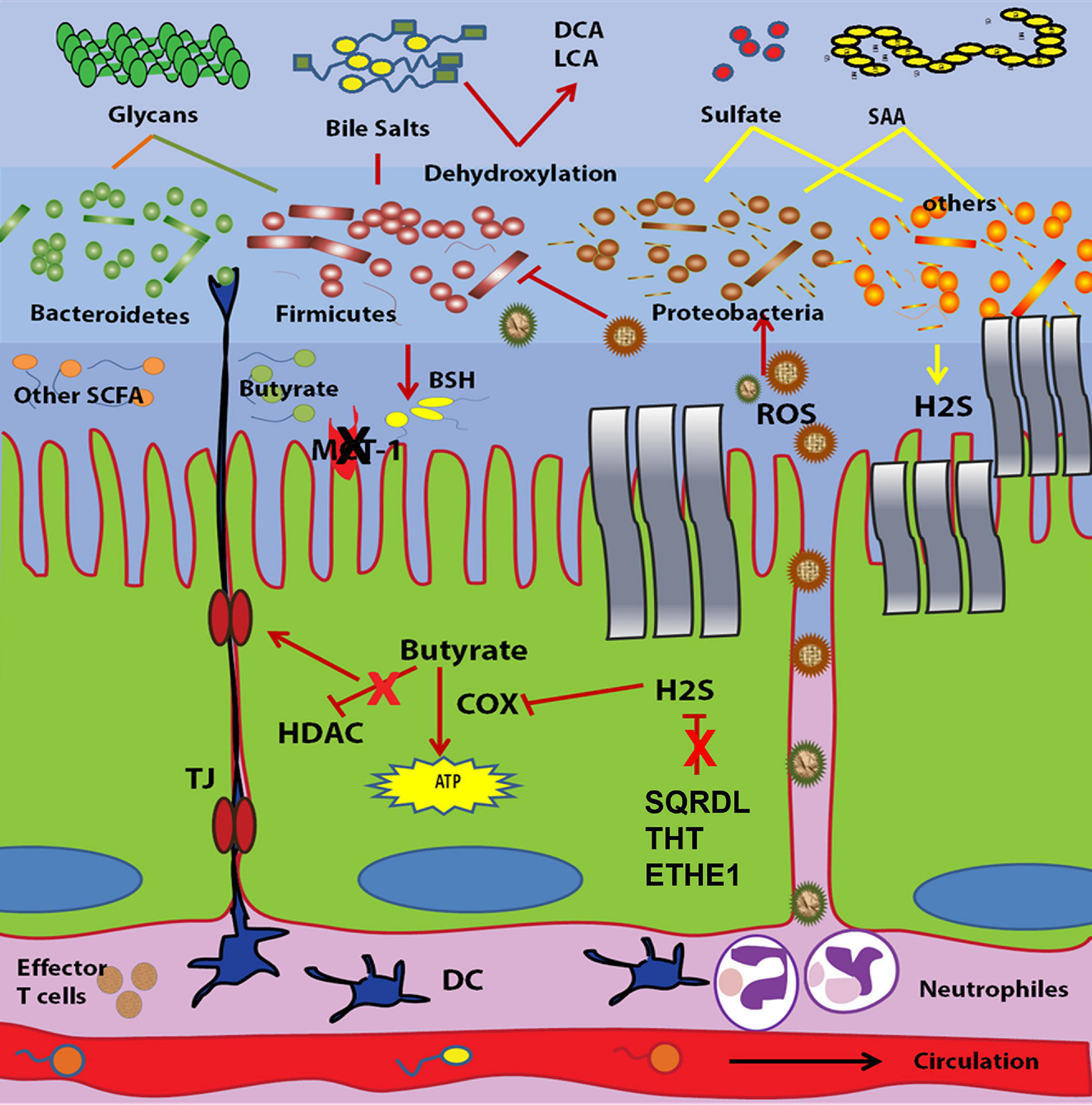
Figure 2. Proposed Host-microbe metabolic dysfunctionalities in inflammatory bowel disease. The oxidative stress environment created by inflammation acts as a selective pressure. This pressure favors the microbes that are able to resist that stress such as Proteobacteria members but not Firmicutes members. This in turn leads to less short chain fatty acids (SCFAs) production, less bile salts hydrolases (BSHs) in association with higher H2S release. From the host side, impaired butyrate transport and oxidation, less mucin secretion, low tight junction (TJ) expression, and defective H2S detoxification are the major host metabolic perturbations associated with IBD.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd El-Aziz, A. M., Elgaml, A., and Ali, Y. M. (2019). Bacteriophage therapy increases complement-mediated lysis of bacteria and enhances bacterial clearance after acute lung infection with multidrug-resistant Pseudomonas aeruginosa. J. Infect. Dis. 219, 1439–1447. doi: 10.1093/infdis/jiy678
Abraham, B. P., and Quigley, E. M. M. (2017). Probiotics in inflammatory bowel disease. Gastroenterol. Clin. North. Am. 46, 769–782.
Akram, W., Garud, N., and Joshi, R. (2019). Role of inulin as prebiotics on inflammatory bowel disease. Drug Discov. Ther. 13, 1–8. doi: 10.5582/ddt.2019.01000
Aldars-García, L., Chaparro, M., and Gisbert, J. P. (2021). Systematic review: the gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 9:977. doi: 10.3390/microorganisms9050977
Anderson, M. E. (1998). Glutathione: an overview of biosynthesis and modulation. Chem. Biol. Interact. 111–112, 1–14. doi: 10.1016/s0009-2797(97)00146-4
Andersson, H., Filipsson, S., and Hulten, L. (1978). Determination of the faecal excretion of labelled bile salts after i.v. administration of 14C-cholic acid. An evaluation of the bile salt malabsorption before and after surgery in patients with Crohn’s disease. Scand. J. Gastroenterol. 13, 249–255. doi: 10.3109/00365527809181756
Andoh, A., Kuzuoka, H., Tsujikawa, T., Nakamura, S., Hirai, F., Suzuki, Y., et al. (2012). Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J. Gastroenterol. 47, 1298–1307.
Arthur, J. C., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T. J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123.
Attene-Ramos, M. S., Nava, G. M., Muellner, M. G., Wagner, E. D., Plewa, M. J., and Gaskins, H. R. (2010). DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 int cells. Environ. Mol. Mutagenesis 51, 304–314.
Attinkara, R., Mwinyi, J., Truninger, K., Regula, J., Gaj, P., Rogler, G., et al. (2012). Association of genetic variation in the NR1H4 gene, encoding the nuclear bile acid receptor FXR, with inflammatory bowel disease. BMC Res. Notes 5:461. doi: 10.1186/1756-0500-5-461
Awano, N., Wada, M., Mori, H., Nakamori, S., and Takagi, H. (2005). Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 71, 4149–4152. doi: 10.1128/aem.71.7.4149-4152.2005
Ballini, A., Santacroce, L., Cantore, S., Bottalico, L., Dipalma, G., Topi, S., et al. (2019). Probiotics efficacy on oxidative stress values in inflammatory bowel disease: a randomized double-blinded placebo-controlled pilot study. Endocr. Metab. Immune Disord Drug Targets 19, 373–381. doi: 10.2174/1871530319666181221150352
Barcelo, A., Claustre, J., Moro, F., Chayvialle, J. A., Cuber, J. C., and Plaisancie, P. (2000). Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut 46, 218–224. doi: 10.1136/gut.46.2.218
Barnich, N., Carvalho, F. A., Glasser, A. L., Darcha, C., Jantscheff, P., Allez, M., et al. (2007). CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574. doi: 10.1172/jci30504
Bartels, F., Backhaus, S., Moore, E. R., Timmis, K. N., and Hofer, B. (1999). Occurrence and expression of glutathione-S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiology 145(Pt 10), 2821–2834. doi: 10.1099/00221287-145-10-2821
Basic, M., Keubler, L. M., Buettner, M., Achard, M., Breves, G., Schröder, B., et al. (2014). Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm. Bowel Dis. 20, 431–443. doi: 10.1097/01.mib.0000441346.86827.ed
Baumgart, M., Dogan, B., Rishniw, M., Weitzman, G., Bosworth, B., Yantiss, R., et al. (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 1, 403–418. doi: 10.1038/ismej.2007.52
Beheshti-Maal, A., Shahrokh, S., Ansari, S., Mirsamadi, E. S., Yadegar, A., Mirjalali, H., et al. (2021). Gut mycobiome: The probable determinative role of fungi in IBD patients. Mycoses 64, 468–476. doi: 10.1111/myc.13238
Beller, L., and Matthijnssens, J. (2019). What is (not) known about the dynamics of the human gut virome in health and disease. Curr. Opin. Virol. 37, 52–57. doi: 10.1016/j.coviro.2019.05.013
Bergman, E. N. (1990). Energy contributions of volatile fatty-acids from the gastrointestinal-tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Bernstein, C., Bernstein, H., Payne, C. M., Beard, S. E., and Schneider, J. (1999). Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39, 68–72. doi: 10.1007/s002849900420
Bernstein, H., Bernstein, C., Payne, C. M., Dvorakova, K., and Garewal, H. (2005). Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. Rev. Mutat. Res. 589, 47–65. doi: 10.1016/j.mrrev.2004.08.001
Berr, F., Kullak-Ublick, G. A., Paumgartner, G., Munzing, W., and Hylemon, P. B. (1996). 7 alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology 111, 1611–1620. doi: 10.1016/s0016-5085(96)70024-0
Bibiloni, R., Fedorak, R. N., Tannock, G. W., Madsen, K. L., Gionchetti, P., Campieri, M., et al. (2005). VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 100, 1539–1546.
Bjarnason, I., Sission, G., and Hayee, B. (2019). A randomised, double-blind, placebo-controlled trial of a multi-strain probiotic in patients with asymptomatic ulcerative colitis and Crohn’s disease. Inflammopharmacology 27, 465–473.
Blachier, F., Davila, A. M., Mimoun, S., Benetti, P. H., Atanasiu, C., Andriamihaja, M., et al. (2010). Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids 39, 335–347. doi: 10.1007/s00726-009-0445-2
Bordin, M., D’Atri, F., Guillemot, L., and Citi, S. (2004). Histone deacetylase inhibitors up-regulate the expression of tight junction proteins. Mol. Cancer Res. 2, 692–701.
Bourgonje, A. R., von Martels, J. Z. H., Bulthuis, M. L. C., van Londen, M., Faber, K. N., Dijkstra, G., et al. (2019). Crohn’s disease in clinical remission is marked by systemic oxidative stress. Front. Physiol. 10:499.
Burrello, C., Garavaglia, F., Cribiù, F. M., Ercoli, G., Lopez, G., Troisi, J., et al. (2018). Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat. Commun. 9:5184.
Butzner, J. D., Parmar, R., Bell, C. J., and Dalal, V. (1996). Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut 38, 568–573. doi: 10.1136/gut.38.4.568
Cadwell, K., Patel, K. K., Maloney, N. S., Liu, T. C., Ng, A. C., Storer, C. E., et al. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145. doi: 10.1016/j.cell.2010.05.009
Caporaso, J. G., Lauber, C. L., Costello, E. K., Berg-Lyons, D., Gonzalez, A., Stombaugh, J., et al. (2011). Moving pictures of the human microbiome. Genome Biol. 12:R50.
Carbonero, F., Benefiel, A. C., and Gaskins, H. R. (2012). Contributions of the microbial hydrogen economy to colonic homeostasis. Nat. Rev. Gastroenterol. Hepatol. 9, 504–518. doi: 10.1038/nrgastro.2012.85
Celiberto, L. S., Graef, F. A., Healey, G. R., Bosman, E. S., Jacobson, K., Sly, L. M., et al. (2018). Inflammatory bowel disease and immunonutrition: novel therapeutic approaches through modulation of diet and the gut microbiome. Immunology 155, 36–52. doi: 10.1111/imm.12939
Chehoud, C., Albenberg, L. G., Judge, C., Hoffmann, C., Grunberg, S., Bittinger, K., et al. (2015). Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 21, 1948–1956. doi: 10.1097/mib.0000000000000454
Chiaro, T. R., Soto, R., Zac Stephens, W., Kubinak, J. L., Petersen, C., Gogokhia, L., et al. (2017). A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044
Choi, H. H., and Cho, Y.-S. (2016). Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin. Endosc. 49, 257–265. doi: 10.5946/ce.2015.117
Christl, S. U., Eisner, H. D., Dusel, G., Kasper, H., and Scheppach, W. (1996). Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa – A potential role for these agents in the pathogenesis of ulcerative colitis. Digestive Dis. Sci. 41, 2477–2481. doi: 10.1007/bf02100146
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. doi: 10.1016/j.cell.2012.04.037
Claesson, M. J., Cusack, S., O’Sullivan, O., Greene-Diniz, R., de Weerd, H., Flannery, E., et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4586–4591.
Clooney, A. G., Sutton, T. D. S., Shkoporov, A. N., Holohan, R. K., Daly, K. M., O’Regan, O., et al. (2019). Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778.e5.
Conte, M. P., Schippa, S., Zamboni, I., Penta, M., Chiarini, F., Seganti, L., et al. (2006). Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55, 1760–1767. doi: 10.1136/gut.2005.078824
Costello, S. P., Hughes, P. A., Waters, O., Bryant, R. V., Vincent, A. D., Blatchford, P., et al. (2019). Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 321, 156–164. doi: 10.1001/jama.2018.20046
Crouwel, F., Buiter, H. J. C., and de Boer, N. K. (2020). Gut microbiota-driven drug metabolism in inflammatory bowel disease. J. Crohns Colitis 15, 307–315. doi: 10.1093/ecco-jcc/jjaa143
Cucchiara, S., Iebba, V., Conte, M. P., and Schippa, S. (2009). The microbiota in inflammatory bowel disease in different age groups. Dig. Dis. 27, 252–258. doi: 10.1159/000228558
Cuffaro, B., Assohoun, A. L. W., Boutillier, D., Súkeníková, L., Desramaut, J., Boudebbouze, S., et al. (2020). In vitro characterization of gut microbiota-derived commensal strains: selection of Parabacteroides distasonis strains alleviating TNBS-induced colitis in mice. Cells 9:2104. doi: 10.3390/cells9092104
Cummings, J. H., and Macfarlane, G. T. (1991). The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x
Dahl, J., Gray, M. J., Bazopoulou, D., Beaufay, F., Lempart, J., Koenigsknecht, M. J., et al. (2017). The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat. Microbiol. 2:16267. doi: 10.1038/nmicrobiol.2016.267
Dai, L., Tang, Y., Zhou, W., Dang, Y., Sun, Q., Tang, Z., et al. (2020). Gut microbiota and related metabolites were disturbed in ulcerative colitis and partly restored after mesalamine treatment. Front. Pharmacol. 11:620724.
Dalmasso, G., Cottrez, F., Imbert, V., Lagadec, P., Peyron, J. F., Rampal, P., et al. (2006). Saccharomyces boulardii inhibits inflammatory bowel disease by trapping T cells in mesenteric lymph nodes. Gastroenterology 131, 1812–1825. doi: 10.1053/j.gastro.2006.10.001
Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A. L., Barnich, N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421. doi: 10.1053/j.gastro.2004.04.061
Das, P., Marcisauskas, S., Ji, B., and Nielsen, J. (2019). Metagenomic analysis of bile salt biotransformation in the human gut microbiome. BMC Genomics 20:517. doi: 10.1186/s12864-019-5899-3
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
De Preter, V., Arijs, I., Windey, K., Vanhove, W., Vermeire, S., Schuit, F., et al. (2012). Decreased mucosal sulfide detoxification is related to an impaired butyrate oxidation in ulcerative colitis. Inflamm. Bowel Dis. 18, 2371–2380. doi: 10.1002/ibd.22949
De Preter, V., Geboes, K. P., Bulteel, V., Vandermeulen, G., Suenaert, P., Rutgeerts, P., et al. (2011). Kinetics of butyrate metabolism in the normal colon and in ulcerative colitis: the effects of substrate concentration and carnitine on the beta-oxidation pathway. Aliment. Pharmacol. Therap. 34, 526–532. doi: 10.1111/j.1365-2036.2011.04757.x
Dennis, K. L., Wang, Y., Blatner, N. R., Wang, S., Saadalla, A., Trudeau, E., et al. (2013). Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res. 73, 5905–5913. doi: 10.1158/0008-5472.can-13-1511
Desmet, I., Vanhoorde, L., Woestyne, M. V., Christiaens, H., and Verstraete, W. (1995). Significance of bile-salt hydrolytic activities of Lactobacilli. J. Appl. Bacteriol. 79, 292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x
Devkota, S., Wang, Y. W., Musch, M. W., Leone, V., Fehlner-Peach, H., Nadimpalli, A., et al. (2012). Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10(-/-) mice. Nature 487, 104–108. doi: 10.1038/nature11225
Donohoe, D. R., Garge, N., Zhang, X., Sun, W., O’Connell, T. M., Bunger, M. K., et al. (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526. doi: 10.1016/j.cmet.2011.02.018
Draper, L. A., Ryan, F. J., Smith, M. K., Jalanka, J., Mattila, E., Arkkila, P. A., et al. (2018). Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome 6:220.
Duboc, H., Rajca, S., Rainteau, D., Benarous, D., Maubert, M. A., Quervain, E., et al. (2013). Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62, 531–539. doi: 10.1136/gutjnl-2012-302578
Duerkop, B. A., Kleiner, M., Paez-Espino, D., Zhu, W., Bushnell, B., Hassell, B., et al. (2018). Murine colitis reveals a disease-associated bacteriophage community. Nat. Microbiol. 3, 1023–1031. doi: 10.1038/s41564-018-0210-y
Duffy, M., O’Mahony, L., Coffey, J. C., Collins, J. K., Shanahan, F., Redmond, H. P., et al. (2002). Sulfate-reducing bacteria colonize pouches formed for ulcerative colitis but not for familial adenomatous polyposis. Dis. Colon Rectum 45, 384–388. doi: 10.1007/s10350-004-6187-z
Duncan, S. H., Barcenilla, A., Stewart, C. S., Pryde, S. E., and Flint, H. J. (2002). Acetate utilization and butyryl coenzyme A (CoA): acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. App. Environ. Microbiol. 68, 5186–5190. doi: 10.1128/aem.68.10.5186-5190.2002
Dziarski, R., Park, S. Y., Kashyap, D. R., Dowd, S. E., and Gupta, D. (2016). Pglyrp-regulated gut microflora prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One 11:e0146162. doi: 10.1371/journal.pone.0146162
Eckburg, P. B., and Relman, D. A. (2007). The role of microbes in Crohn’s disease. Clin. Infect. Dis. 44, 256–262.
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Edmond, L. M., Hopkins, M. J., Magee, E. A., and Cummings, J. H. (2003). The effect of 5-aminosalicylic acid-containing drugs on sulfide production by sulfate-reducing and amino acid-fermenting bacteria. Inflamm. Bowel Dis. 9, 10–17. doi: 10.1097/00054725-200301000-00002
Effenberger, M., Reider, S., Waschina, S., Bronowski, C., Enrich, B., Adolph, T. E., et al. (2021). Microbial butyrate synthesis indicates therapeutic efficacy of azathioprine in IBD patients. J. Crohns Colitis 15, 88–98. doi: 10.1093/ecco-jcc/jjaa152
El-Mowafy, M., Elgaml, A., El-Mesery, M., Sultan, S., Ahmed, T. A. E., Gomaa, A. I., et al. (2021). Changes of gut-microbiota-liver axis in hepatitis c virus infection. Biology (Basel) 10:55. doi: 10.3390/biology10010055
Enck, P., Zimmermann, K., Rusch, K., Schwiertz, A., Klosterhalfen, S., and Frick, J. S. (2009). The effects of ageing on the colonic bacterial microflora in adults. Zool. Gastroenterol. 47, 653–658. doi: 10.1055/s-0028-1109055
Ettreiki, C., Gadonna-Widehem, P., Mangin, I., Coëffier, M., Delayre-Orthez, C., and Anton, P. M. (2012). Juvenile ferric iron prevents microbiota dysbiosis and colitis in adult rodents. World J. Gastroenterol. 18, 2619–2629. doi: 10.3748/wjg.v18.i21.2619
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Fahey, R. C., Brown, W. C., Adams, W. B., and Worsham, M. B. (1978). Occurrence of glutathione in bacteria. J. Bacteriol. 133, 1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978
Fava, F., and Danese, S. (2011). Intestinal microbiota in inflammatory bowel disease: friend of foe? World J. Gastroenterol. 17, 557–566. doi: 10.3748/wjg.v17.i5.557
Favier, C., Neut, C., Mizon, C., Cortot, A., Colombel, J. F., and Mizon, J. (1997). Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig. Dis. Sci. 42, 817–822.
Fedorak, R. N., Feagan, B. G., Hotte, N., Leddin, D., Dieleman, L. A., Petrunia, D. M., et al. (2015). The probiotic VSL#3 has anti-inflammatory effects and could reduce endoscopic recurrence after surgery for Crohn’s disease. Clin. Gastroenterol. Hepatol. 13, 928–935.e2.
Fernandes, M. A., Verstraete, S. G., Phan, T. G., Deng, X., Stekol, E., LaMere, B., et al. (2019). Enteric virome and bacterial microbiota in children with ulcerative colitis and crohn disease. J. Pediatr. Gastroenterol. Nutr. 68, 30–36. doi: 10.1097/mpg.0000000000002140
Fiorucci, S., Carino, A., Baldoni, M., Santucci, L., Costanzi, E., Graziosi, L., et al. (2021). Bile acid signaling in inflammatory bowel diseases. Dig. Dis. Sci. 66, 674–693. doi: 10.1007/s10620-020-06715-3
Fitzpatrick, L. R., and Jenabzadeh, P. (2020). IBD and bile acid absorption: focus on pre-clinical and clinical observations. Front. Physiol. 11:564.
Florin, T. H. (1991). Hydrogen sulphide and total acid-volatile sulphide in faeces, determined with a direct spectrophotometric method. Clin. Chim. Acta 196, 127–134. doi: 10.1016/0009-8981(91)90065-k
Forbes, J. D., Van Domselaar, G., and Bernstein, C. N. (2016). The gut microbiota in immune-mediated inflammatory diseases. Front. Microbiol. 7:1081.
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Fujimoto, T., Imaeda, H., Takahashi, K., Kasumi, E., Bamba, S., Fujiyama, Y., et al. (2013). Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J. Gastroenterol. Hepatol. 28, 613–619. doi: 10.1111/jgh.12073
Fukiya, S., Arata, M., Kawashima, H., Yoshida, D., Kaneko, M., Minamida, K., et al. (2009). Conversion of cholic acid and chenodeoxycholic acid into their 7-oxo derivatives by Bacteroides intestinalis AM-1 isolated from human feces. FEMS Microbiol. Lett. 293, 263–270. doi: 10.1111/j.1574-6968.2009.01531.x
Furne, J., Springfield, J., Koenig, T., DeMaster, E., and Levitt, M. D. (2001). Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem. Pharmacol. 62, 255–259. doi: 10.1016/s0006-2952(01)00657-8
Gadaleta, R. M., van Erpecum, K. J., Oldenburg, B., Willemsen, E. C. L., Renooij, W., Murzilli, S., et al. (2011). Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60, 463–472. doi: 10.1136/gut.2010.212159
Gade, A. K., Douthit, N. T., and Townsley, E. (2020). Medical management of Crohn’s disease. Cureus 12:e8351.
Galtier, M., De Sordi, L., Sivignon, A., de Vallée, A., Maura, D., Neut, C., et al. (2017). Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohns Colitis 11, 840–847.
Garrett, W. S., Lord, G. M., Punit, S., Lugo-Villarino, G., Mazmanian, S. K., Ito, S., et al. (2007). Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45. doi: 10.1016/j.cell.2007.08.017
Geirnaert, A., Calatayud, M., Grootaert, C., Laukens, D., Devriese, S., Smagghe, G., et al. (2017). Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 7:11450.
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392.
Ghosh, S. S., Wang, J., Yannie, P. J., and Ghosh, S. (2020). Intestinal barrier dysfunction, LPS translocation, and disease development. J. Endocrine Soc. 4:bvz039.
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Gloor, G. B., Hummelen, R., Macklaim, J. M., Dickson, R. J., Fernandes, A. D., MacPhee, R., et al. (2010). Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5:e15406. doi: 10.1371/journal.pone.0015406
Gogokhia, L., Buhrke, K., Bell, R., Hoffman, B., Brown, D. G., Hanke-Gogokhia, C., et al. (2019). Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25, 285–299.e8.
Grassin-Delyle, S., Abrial, C., Salvator, H., Brollo, M., Naline, E., and Devillier, P. (2020). The role of toll-like receptors in the production of cytokines by human lung macrophages. J. Innate Immun. 12, 63–73. doi: 10.1159/000494463
Greenblum, S., Turnbaugh, P. J., and Borenstein, E. (2012). Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl. Acad. Sci. U.S.A. 109, 594–599. doi: 10.1073/pnas.1116053109
Guo, F. F., Yu, T. C., Hong, J., and Fang, J. Y. (2016). Emerging roles of hydrogen sulfide in inflammatory and neoplastic colonic diseases. Front. Physiol. 7:156.
Håkansson, Å, Tormo-Badia, N., Baridi, A., Xu, J., Molin, G., Hagslätt, M. L., et al. (2015). Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin. Exp. Med. 15, 107–120. doi: 10.1007/s10238-013-0270-5
Halfvarson, J., Brislawn, C. J., Lamendella, R., Vázquez-Baeza, Y., Walters, W. A., Bramer, L. M., et al. (2017). Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2:17004.
Hammond, C. L., Lee, T. K., and Ballatori, N. (2001). Novel roles for glutathione in gene expression, cell death, and membrane transport of organic solutes. J. Hepatol. 34, 946–954. doi: 10.1016/s0168-8278(01)00037-x
Hansen, R., Russell, R. K., Reiff, C., Louis, P., McIntosh, F., Berry, S. H., et al. (2012). Microbiota of de-novo pediatric IBD: increased Faecalibacterium Prausnitzii and reduced bacterial diversity in Crohn’s but not in ulcerative colitis. Am. J. Gastroenterol. 107, 1913–1922. doi: 10.1038/ajg.2012.335
Heinken, A., Ravcheev, D. A., Baldini, F., Heirendt, L., Fleming, R. M. T., and Thiele, I. (2019). Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 7:75.
Heuman, R., Sjodahl, R., Tobiasson, P., and Tagesson, C. (1983). Decreased absorption of ingested unconjugated chenodeoxycholic acid in patients with Crohn’s disease. Scand. J. Gastroenterol. 18, 23–26. doi: 10.3109/00365528309181553
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document, the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hoarau, G., Mukherjee, P. K., Gower-Rousseau, C., Hager, C., Chandra, J., Retuerto, M. A., et al. (2016). Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7, e01250–16.
Hofmann, A. F., and Eckmann, L. (2006). How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 4333–4334. doi: 10.1073/pnas.0600780103
Hopkins, M. J., Sharp, R., and Macfarlane, G. T. (2001). Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48, 198–205. doi: 10.1136/gut.48.2.198
Hou, R.-G., Fan, L., Liu, J.-J., Cheng, Y., Chang, Z.-P., Wu, B., et al. (2018). Bile acid malabsorption is associated with diarrhea in acute phase of colitis. Can. J. Physiol. Pharmacol. 96, 1328–1336. doi: 10.1139/cjpp-2018-0017
Houten, S. M., Watanabe, M., and Auwerx, J. (2006). Endocrine functions of bile acids. EMBO J. 25, 1419–1425. doi: 10.1038/sj.emboj.7601049
Huda-Faujan, N., Abdulamir, A. S., Fatimah, A. B., Anas, O. M., Shuhaimi, M., Yazid, A. M., et al. (2010). The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 4, 53–58. doi: 10.2174/1874091x01004010053
Hudcovic, T., Kolinska, J., Klepetar, J., Stepankova, R., Rezanka, T., Srutkova, D., et al. (2012). Protective effect of Clostridium tyrobutyricum in acute dextran sodium sulphate-induced colitis: differential regulation of tumour necrosis factor-alpha and interleukin-18 in BALB/c and severe combined immunodeficiency mice. Clin. Exp. Immunol. 167, 356–365. doi: 10.1111/j.1365-2249.2011.04498.x
Hvas, C. L., Dahl Jørgensen, S. M., Jørgensen, S. P., Storgaard, M., Lemming, L., Hansen, M. M., et al. (2019). Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent clostridium difficile infection. Gastroenterology 156, 1324–1332.e3.
Hylemon, P. B., Zhou, H. P., Pandak, W. M., Ren, S. L., Gil, G., and Dent, P. (2009). Bile acids as regulatory molecules. J. Lipid Res. 50, 1509–1520. doi: 10.1194/jlr.r900007-jlr200
Iliev, I. D., and Leonardi, I. (2017). Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 17, 635–646. doi: 10.1038/nri.2017.55
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. doi: 10.1126/science.1221789
Inagaki, T., Moschetta, A., Lee, Y. K., Peng, L., Zhao, G., Downes, M., et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 3920–3925. doi: 10.1073/pnas.0509592103
Islam, K. B. M. S., Fukiya, S., Hagio, M., Fujii, N., Ishizuka, S., Ooka, T., et al. (2011). Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781. doi: 10.1053/j.gastro.2011.07.046
Iwamoto, J., Saito, Y., Honda, A., Miyazaki, T., Ikegami, T., and Matsuzaki, Y. (2013). Bile Acid malabsorption deactivates pregnane x receptor in patients with Crohn’s disease. Inflamm. Bowel Dis. 19, 1278–1284. doi: 10.1097/mib.0b013e318281f423
Iyer, S. S., and Cheng, G. (2012). Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 32, 23–63. doi: 10.1615/critrevimmunol.v32.i1.30
Jacobs, J. P., Goudarzi, M., Singh, N., Tong, M., McHardy, I. H., Ruegger, P., et al. (2016). A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol. Gastroenterol. Hepatol. 2, 750–766. doi: 10.1016/j.jcmgh.2016.06.004
Jia, W., Whitehead, R. N., Griffiths, L., Dawson, C., Bai, H., Waring, R. H., et al. (2012). Diversity and distribution of sulphate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS Immunol. Med. Microbiol. 65, 55–68. doi: 10.1111/j.1574-695x.2012.00935.x
Jin, D., Wu, S., Zhang, Y. G., Lu, R., Xia, Y., Dong, H., et al. (2015). Lack of Vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin. Ther. 37, 996–1009.e3.
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585. doi: 10.1073/pnas.0804437105
Joossens, M., Huys, G., Cnockaert, M., De Preter, V., Verbeke, K., Rutgeerts, P., et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637. doi: 10.1136/gut.2010.223263
Kakiyama, G., Pandak, W. M., Gillevet, P. M., Hylemon, P. B., Heuman, D. M., Daita, K., et al. (2013). Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58, 949–955. doi: 10.1016/j.jhep.2013.01.003
Kaplan, G. G., and Windsor, J. W. (2021). The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 18, 56–66. doi: 10.1038/s41575-020-00360-x
Kernbauer, E., Ding, Y., and Cadwell, K. (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. doi: 10.1038/nature13960
Keshavarzian, A., Banan, A., Farhadi, A., Komanduri, S., Mutlu, E., Zhang, Y., et al. (2003). Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut 52, 720–728. doi: 10.1136/gut.52.5.720
Keshavarzian, A., Sedghi, S., Kanofsky, J., List, T., Robinson, C., Ibrahim, C., et al. (1992). Excessive production of reactive oxygen metabolites by inflamed colon: analysis by chemiluminescence probe. Gastroenterology 103, 177–185. doi: 10.1016/0016-5085(92)91111-g
Kespohl, M., Vachharajani, N., Luu, M., Harb, H., Pautz, S., Wolff, S., et al. (2017). The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front. Immunol. 8:1036.
Khan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., et al. (2019). Alteration of gut microbiota in Inflammatory Bowel Disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8:126. doi: 10.3390/pathogens8030126
Khoruts, A., and Sadowsky, M. J. (2016). Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 13, 508–516. doi: 10.1038/nrgastro.2016.98
Khoruts, A., Rank, K. M., Newman, K. M., Viskocil, K., Vaughn, B. P., Hamilton, M. J., et al. (2016). Inflammatory bowel disease affects the outcome of fecal microbiota transplantation for recurrent clostridium difficile infection. Clin. Gastroenterol. Hepatol. 14, 1433–1438. doi: 10.1016/j.cgh.2016.02.018
Kitahora, T., Suzuki, K., Asakura, H., Yoshida, T., Suematsu, M., Watanabe, M., et al. (1988). Active oxygen species generated by monocytes and polymorphonuclear cells in Crohn’s disease. Dig. Dis. Sci. 33, 951–955. doi: 10.1007/bf01535990
Kobayashi, T., Siegmund, B., Le Berre, C., Wei, S. C., Ferrante, M., Shen, B., et al. (2020). Ulcerative colitis. Nat. Rev. Dis. Primers 6:74.
Koutroubakis, I. E., Malliaraki, N., Dimoulios, P. D., Karmiris, K., Castanas, E., and Kouroumalis, E. A. (2004). Decreased total and corrected antioxidant capacity in patients with inflammatory bowel disease. Dig. Dis. Sci. 49, 1433–1437. doi: 10.1023/b:ddas.0000042242.22898.d9
Kovarik, J. J., Tillinger, W., Hofer, J., Holzl, M. A., Heinzl, H., Saemann, M. D., et al. (2011). Impaired anti-inflammatory efficacy of n-butyrate in patients with IBD. Eur. J. Clin. Invest. 41, 291–298. doi: 10.1111/j.1365-2362.2010.02407.x
Kowalska-Duplaga, K., Krawczyk, A., Sroka-Oleksiak, A., Salamon, D., Wêdrychowicz, A., Fyderek, K., et al. (2019). Dependence of colonization of the large intestine by candida on the treatment of Crohn’s disease. Pol. J. Microbiol. 68, 121–126. doi: 10.21307/pjm-2019-014
Kushkevych, I., Dordevic, D., and Vitezova, M. (2021). Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J. Adv. Res. 27, 71–78. doi: 10.1016/j.jare.2020.03.007
Laue, H., Friedrich, M., Ruff, J., and Cook, A. M. (2001). Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183, 1727–1733. doi: 10.1128/jb.183.5.1727-1733.2001
Lee, Y. K., and Mazmanian, S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773. doi: 10.1126/science.1195568
Lepage, P., Hasler, R., Spehlmann, M. E., Rehman, A., Zvirbliene, A., Begun, A., et al. (2011). Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236. doi: 10.1053/j.gastro.2011.04.011
Leschelle, X., Goubern, M., Andriamihaja, M., Blottiere, H. M., Couplan, E., Gonzalez-Barroso, M. D., et al. (2005). Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim. Biophys. Acta 1725, 201–212. doi: 10.1016/j.bbagen.2005.06.002
Li, Q., Ding, X., Liu, K., Marcella, C., Liu, X., Zhang, T., et al. (2020). fecal microbiota transplantation for ulcerative colitis: the optimum timing and gut microbiota as predictors for long-term clinical outcomes. Clin. Transl. Gastroenterol. 11:e00224. doi: 10.14309/ctg.0000000000000224
Li, Q., Wang, C., Tang, C., He, Q., Li, N., and Li, J. (2014). Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J. Clin. Gastroenterol. 48, 513–523. doi: 10.1097/mcg.0000000000000035
Li, X. V., Leonardi, I., and Iliev, I. D. (2019). Gut mycobiota in immunity and inflammatory disease. Immunity 50, 1365–1379. doi: 10.1016/j.immuni.2019.05.023
Liguori, G., Lamas, B., Richard, M. L., Brandi, G., da Costa, G., Hoffmann, T. W., et al. (2016). Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohns Colitis 10, 296–305. doi: 10.1093/ecco-jcc/jjv209
Lim, E. S., Zhou, Y., Zhao, G., Bauer, I. K., Droit, L., Ndao, I. M., et al. (2015). Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234. doi: 10.1038/nm.3950
Limon, J. J., Tang, J., Li, D., Wolf, A. J., Michelsen, K. S., Funari, V., et al. (2019). Malassezia Is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388.e6.
Lin, D. M., and Lin, H. C. (2019). A theoretical model of temperate phages as mediators of gut microbiome dysbiosis. F1000Res 8:F1000FacultyRev–997.
Liu, F., Ma, R., Riordan, S. M., Grimm, M. C., Liu, L., Wang, Y., et al. (2017). Azathioprine, mercaptopurine, and 5-aminosalicylic acid affect the growth of IBD-associated campylobacter species and other enteric microbes. Front. Microbiol. 8:527.
Liu, S., Zhao, W., Lan, P., and Mou, X. (2020). The microbiome in inflammatory bowel diseases: from pathogenesis to therapy. Protein Cell 15, 331–345. doi: 10.1007/s13238-020-00745-3
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662.
Lo Presti, A., Zorzi, F., Del Chierico, F., Altomare, A., Cocca, S., Avola, A., et al. (2019). Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front. Microbiol. 10:1655.
Loeffler, C., Karlsberg, A., Martin, L. S., Eskin, E., Koslicki, D., and Mangul, S. (2020). Correction to: Improving the usability and comprehensiveness of microbial databases. BMC Biol. 18:92. doi: 10.1186/s12915-020-00831-2
Loubinoux, J., Bronowicki, J. P., Pereira, I. A. C., Mougenel, J. L., and Le Faou, A. E. (2002). Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40, 107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Louis, P., Young, P., Holtrop, G., and Flint, H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314. doi: 10.1111/j.1462-2920.2009.02066.x
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Luceri, C., Bigagli, E., Agostiniani, S., Giudici, F., Zambonin, D., Scaringi, S., et al. (2019). Analysis of oxidative stress-related markers in Crohn’s disease patients at surgery and correlations with clinical findings. Antioxidants (Basel) 8:378. doi: 10.3390/antiox8090378
Luhrs, H., Gerke, T., Muller, J. G., Melcher, R., Schauber, J., Boxberge, F., et al. (2002). Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 37, 458–466. doi: 10.1080/003655202317316105
Ma, Y., Xu, X., Li, M., Cai, J., Wei, Q., and Niu, H. (2019). Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol. Med. 25:35.
Macfarlane, G. T., Gibson, G. R., and Cummings, J. H. (1992). Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72, 57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x
Machiels, K., Joossens, M., Sabino, J., De Preter, V., Arijs, I., Eeckhaut, V., et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. doi: 10.1136/gutjnl-2013-304833
Magee, E. A., Richardson, C. J., Hughes, R., and Cummings, J. H. (2000). Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 72, 1488–1494. doi: 10.1093/ajcn/72.6.1488
Magnusson, M. K., Isaksson, S., and Ohman, L. (2020). The anti-inflammatory immune regulation induced by butyrate is impaired in inflamed intestinal mucosa from patients with ulcerative colitis. Inflammation 43, 507–517. doi: 10.1007/s10753-019-01133-8
Manichanh, C., Rigottier-Gois, L., Bonnaud, E., Gloux, K., Pelletier, E., Frangeul, L., et al. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211. doi: 10.1136/gut.2005.073817
Manrique, P., Bolduc, B., Walk, S. T., van der Oost, J., de Vos, W. M., and Young, M. J. (2016). Healthy human gut phageome. Proc. Natl. Acad. Sci. U.S.A. 113, 10400–10405. doi: 10.1073/pnas.1601060113
Mariat, D., Firmesse, O., Levenez, F., Guimaraes, V., Sokol, H., Dore, J., et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9:123. doi: 10.1186/1471-2180-9-123
Mariggio, M. A., Minunno, V., Riccardi, S., Santacroce, R., De Rinaldis, P., and Fumarulo, R. (1998). Sulfide enhancement of PMN apoptosis. Immunopharmacol. Immunotoxicol. 20, 399–408. doi: 10.3109/08923979809034822
Martens, E. C., Lowe, E. C., Chiang, H., Pudlo, N. A., Wu, M., McNulty, N. P., et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. doi: 10.1371/journal.pbio.1001221
Martin, H. M., Campbell, B. J., Hart, C. A., Mpofu, C., Nayar, M., Singh, R., et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127, 80–93.
Mei, L., Zhou, J., Su, Y., Mao, K., Wu, J., Zhu, C., et al. (2021). Gut microbiota composition and functional prediction in diarrhea-predominant irritable bowel syndrome. BMC Gastroenterol. 21:105. doi: 10.1186/s12876-021-01693-w
Mena Bares, L. M. F., Benitez Cantero, J. M., Iglesias Flores, E., Gros Alcalde, B., Moreno Ortega, E., Maza Muret, F. R., et al. (2019). Bile acid malabsorption in patients with chronic diarrhea and Crohn’s disease. Rev. Esp. Enferm. Dig. 111, 40–45.
Metwaly, A., Dunkel, A., Waldschmitt, N., Raj, A. C. D., Lagkouvardos, I., Corraliza, A. M., et al. (2020). Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 11:4322.
Miele, E., Pascarella, F., Giannetti, E., Quaglietta, L., Baldassano, R. N., and Staiano, A. (2009). Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am. J. Gastroenterol. 104, 437–443.
Mimoun, S., Andriamihaja, M., Chaumontet, C., Atanasiu, C., Benamouzig, R., Blouin, J. M., et al. (2012). Detoxification of H2S by differentiated colonic epithelial cells: implication of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxidants Redox Signal. 17, 1–10. doi: 10.1089/ars.2011.4186
Moayyedi, P., Surette, M. G., Kim, P. T., Libertucci, J., Wolfe, M., Onischi, C., et al. (2015). Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6.
Moens, F., Van den Abbeele, P., Basit, A. W., Dodoo, C., Chatterjee, R., Smith, B., et al. (2019). A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int. J. Pharm. 555, 1–10. doi: 10.1016/j.ijpharm.2018.11.020
Moore, J. W., Babidge, W., Millard, S., and Roediger, W. E. (1997). Effect of sulphide on short chain acyl-CoA metabolism in rat colonocytes. Gut 41, 77–81. doi: 10.1136/gut.41.1.77
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79.
Mottawea, W., Butcher, J., Li, J., Abujamel, T., Manoogian, J., Mack, D., et al. (2019). The mucosal-luminal interface: an ideal sample to study the mucosa-associated microbiota and the intestinal microbial biogeography. Pediatr. Res. 85, 895–903. doi: 10.1038/s41390-019-0326-7
Mottawea, W., Chiang, C. K., Muhlbauer, M., Starr, A. E., Butcher, J., Abujamel, T., et al. (2016). Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 7:13419.
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. doi: 10.2174/978160805192211001010142
Ng, S. C., Bernstein, C. N., Vatn, M. H., Lakatos, P. L., Loftus, E. V. Jr., Tysk, C., et al. (2013). Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 62, 630–649. doi: 10.1136/gutjnl-2012-303661
Nishida, A., Inoue, R., Inatomi, O., Bamba, S., Naito, Y., and Andoh, A. (2018). Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11, 1–10.
Norman, J. M., Handley, S. A., Baldridge, M. T., Droit, L., Liu, C. Y., Keller, B. C., et al. (2015). Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. doi: 10.1016/j.cell.2015.01.002
Nyhlin, H., Merrick, M. V., and Eastwood, M. A. (1994). Bile acid malabsorption in Crohn’s disease and indications for its assessment using SeHCAT. Gut 35, 90–93. doi: 10.1136/gut.35.1.90
O’Donnell, S., Borowski, K., Espin-Garcia, O., Milgrom, R., Kabakchiev, B., Stempak, J., et al. (2019). The unsolved link of genetic markers and crohn’s disease progression: a north american cohort experience. Inflamm. Bowel Dis. 25, 1541–1549. doi: 10.1093/ibd/izz016
Oliva, S., Di Nardo, G., Ferrari, F., Mallardo, S., Rossi, P., Patrizi, G., et al. (2012). Randomised clinical trial: the effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment Pharmacol. Ther. 35, 327–334. doi: 10.1111/j.1365-2036.2011.04939.x
Olszak, T., An, D. D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
O’Toole, P. W., Marchesi, J. R., and Hill, C. (2017). Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2:17057.
Ott, S. J., Kühbacher, T., Musfeldt, M., Rosenstiel, P., Hellmig, S., Rehman, A., et al. (2008). Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43, 831–841. doi: 10.1080/00365520801935434
Ott, S. J., Musfeldt, M., Wenderoth, D. F., Hampe, J., Brant, O., Folsch, U. R., et al. (2004). Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53, 685–693. doi: 10.1136/gut.2003.025403
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Pang, J. X. Q., Kheirkhahrahimabadi, H., Bindra, S., Bindra, G., Panaccione, R., Eksteen, B., et al. (2021). Differential effect of genetic burden on disease phenotypes in Crohn’s disease and ulcerative colitis in a Canadian cohort. J. Can. Assoc. Gastroenterol. 4, 65–72. doi: 10.1093/jcag/gwaa002
Papa, E., Docktor, M., Smillie, C., Weber, S., Preheim, S. P., Gevers, D., et al. (2012). Non-Invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7:e39242. doi: 10.1371/journal.pone.0039242
Paramsothy, S., Nielsen, S., Kamm, M. A., Deshpande, N. P., Faith, J. J., Clemente, J. C., et al. (2019). Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology 156, 1440–1454 e2.
Pascal, V., Pozuelo, M., Borruel, N., Casellas, F., Campos, D., Santiago, A., et al. (2017). A microbial signature for Crohn’s disease. Gut 66, 813–822. doi: 10.1136/gutjnl-2016-313235
Pelissier, M. A., Marteau, P., and Pochart, P. (2007). Antioxidant effects of metronidazole in colonic tissue. Dig. Dis. Sci. 52, 40–44. doi: 10.1007/s10620-006-9231-0
Pelissier, M. A., Vasquez, N., Balamurugan, R., Pereira, E., Dossou-Yovo, F., Suau, A., et al. (2010). Metronidazole effects on microbiota and mucus layer thickness in the rat gut. FEMS Microbiol. Ecol. 73, 601–610.
Pérez-Brocal, V., García-López, R., Nos, P., Beltrán, B., Moret, I., and Moya, A. (2015). Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm. Bowel Dis. 21, 2515–2532. doi: 10.1097/mib.0000000000000549
Petersson, J., Schreiber, O., Hansson, G. C., Gendler, S. J., Velcich, A., Lundberg, J. O., et al. (2011). Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G327–G333.
Picton, R., Eggo, M. C., Langman, M. J., and Singh, S. (2007). Impaired detoxication of hydrogen sulfide in ulcerative colitis? Dig. Dis. Sci. 52, 373–378. doi: 10.1007/s10620-006-9529-y
Pitcher, M. C., Beatty, E. R., Harris, R. M., Waring, R. H., and Cummings, J. H. (1998). Sulfur metabolism in ulcerative colitis: investigation of detoxification enzymes in peripheral blood. Dig. Dis. Sci. 43, 2080–2085.
Ponziani, F. R., Zocco, M. A., D’Aversa, F., Pompili, M., and Gasbarrini, A. (2017). Eubiotic properties of rifaximin: disruption of the traditional concepts in gut microbiota modulation. World J. Gastroenterol 23, 4491–4499. doi: 10.3748/wjg.v23.i25.4491
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65.
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60.
Qiu, X., Ma, J., Jiao, C., Mao, X., Zhao, X., Lu, M., et al. (2017). Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget 8, 107577–107588. doi: 10.18632/oncotarget.22534
Ramos, G. P., and Papadakis, K. A. (2019). Mechanisms of disease: inflammatory bowel diseases. Mayo Clin. Proc. 94, 155–165.
Ray, K. (2012). Gut microbiota: adding weight to the microbiota’s role in obesity–exposure to antibiotics early in life can lead to increased adiposity. Nat. Rev. Gastroenterol. Hepatol. 9:615. doi: 10.1038/nrgastro.2012.175
Ridlon, J. M., and Hylemon, P. B. (2012). Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7alpha-dehydroxylating intestinal bacterium. J. Lipid Res. 53, 66–76. doi: 10.1194/jlr.m020313
Ridlon, J. M., Kang, D. J., and Hylemon, P. B. (2006). Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259. doi: 10.1194/jlr.r500013-jlr200
Rinaldi, L., Gobbi, G., Pambianco, M., Micheloni, C., Mirandola, P., and Vitale, M. (2006). Hydrogen sulfide prevents apoptosis of human PMN via inhibition of p38 and caspase 3. Lab. Invest. 86, 391–397. doi: 10.1038/labinvest.3700391
Rinninella, E., Cintoni, M., Raoul, P., Lopetuso, L. R., Scaldaferri, F., Pulcini, G., et al. (2019). Food components and dietary habits: keys for a healthy gut microbiota composition. Nutrients 11:2393. doi: 10.3390/nu11102393
Rodemann, J. F., Dubberke, E. R., Reske, K. A., Seo, D. H., and Stone, C. D. (2007). Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 5, 339–344.
Rodríguez-Nogales, A., Algieri, F., Garrido-Mesa, J., Vezza, T., Utrilla, M. P., Chueca, N., et al. (2018). Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: Impact on microRNAs expression and gut microbiota composition. J. Nutr. Biochem. 61, 129–139. doi: 10.1016/j.jnutbio.2018.08.005
Roediger, W. E. (1980). The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet 2, 712–715. doi: 10.1016/s0140-6736(80)91934-0
Roediger, W. E. W., Heyworth, M., Willoughby, P., Piris, J., Moore, A., and Truelove, S. C. (1982). Luminal ions and short chain fatty-acids as markers of functional-activity of the mucosa in ulcerative-colitis. J. Clin. Pathol. 35, 323–326. doi: 10.1136/jcp.35.3.323
Roediger, W. E., Duncan, A., Kapaniris, O., and Millard, S. (1993). Sulphide impairment of substrate oxidation in rat colonocytes: a biochemical basis for ulcerative colitis? Clin. Sci. (Lond) 85, 623–627. doi: 10.1042/cs0850623
Roediger, W. E., Moore, J., and Babidge, W. (1997). Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig. Dis. Sci. 42, 1571–1579.
Rooks, M. G., Veiga, P., Wardwell-Scott, L. H., Tickle, T., Segata, N., Michaud, M., et al. (2014). Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 8, 1403–1417.
Rose, P., Moore, P. K., Ming, S. H., Nam, O. C., Armstrong, J. S., and Whiteman, M. (2005). Hydrogen sulfide protects colon cancer cells from chemopreventative agent beta-phenylethyl isothiocyanate induced apoptosis. World J. Gastroenterol. 11, 3990–3997. doi: 10.3748/wjg.v11.i26.3990
Russell, D. W. (2003). The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72, 137–174. doi: 10.1146/annurev.biochem.72.121801.161712
Ryan, F. J., Ahern, A. M., Fitzgerald, R. S., Laserna-Mendieta, E. J., Power, E. M., Clooney, A. G., et al. (2020). Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 11:1512.
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al. (2021). The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Santiago-Rodriguez, T. M., and Hollister, E. B. (2019). Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 11:656. doi: 10.3390/v11070656
Santoru, M. L., Piras, C., Murgia, A., Palmas, V., Camboni, T., Liggi, S., et al. (2017). Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 7:9523.
Santos-Antunes, J., Nunes, A. C., Lopes, S., and Macedo, G. (2016). The relevance of vitamin d and antinuclear antibodies in patients with inflammatory bowel disease under anti-tnf treatment: a prospective study. Inflamm. Bowel Dis. 22, 1101–1106. doi: 10.1097/mib.0000000000000697
Sarker, S. A., Sultana, S., Reuteler, G., Moine, D., Descombes, P., Charton, F., et al. (2016). Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137. doi: 10.1016/j.ebiom.2015.12.023
Sartor, R. B. (2008). Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594. doi: 10.1053/j.gastro.2007.11.059
Saulnier, D. M., Riehle, K., Mistretta, T. A., Diaz, M. A., Mandal, D., Raza, S., et al. (2011). Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141, 1782–1791. doi: 10.1053/j.gastro.2011.06.072
Sayin, S. I., Wahlstrom, A., Felin, J., Jantti, S., Marschall, H. U., Bamberg, K., et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235. doi: 10.1016/j.cmet.2013.01.003
Scanlan, P. D., Shanahan, F., and Marchesi, J. R. (2009). Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol. Ecol. 69, 213–221. doi: 10.1111/j.1574-6941.2009.00709.x
Schirbel, A., Kessler, S., Rieder, F., West, G., Rebert, N., Asosingh, K., et al. (2013). Pro-Angiogenic activity of TLRs and NLRs: a novel link between gut microbiota and intestinal angiogenesis. Gastroenterology 144, 613–623.e9.
Schwerbrock, N. M. J., Makkink, M. K., van der Sluis, M., Buller, H. A., Einerhand, A. W. C., Sartor, R. B., et al. (2004). Interleukin 10-deficient mice exhibit defective colonic Muc2 synthesis before and after induction of colitis by commensal bacteria. Inflamm. Bowel Dis. 10, 811–823. doi: 10.1097/00054725-200411000-00016
Schwiertz, A., Jacobi, M., Frick, J. S., Richter, M., Rusch, K., and Kohler, H. (2010). Microbiota in pediatric inflammatory bowel disease. J. Pediatr. 157, 240–244 e1.
Segain, J. P., de la Bletiere, D. R., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NF kappa B inhibition: implications for Crohn’s disease. Gut 47, 397–403. doi: 10.1136/gut.47.3.397
Seksik, P., Rigottier-Gois, L., Gramet, G., Sutren, M., Pochart, P., Marteau, P., et al. (2003). Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 52, 237–242. doi: 10.1136/gut.52.2.237
Shanahan, F. (2007). Irritable bowel syndrome: Shifting the focus toward the gut microbiota. Gastroenterology 133, 340–342. doi: 10.1053/j.gastro.2007.05.030
Shibuya, N., Koike, S., Tanaka, M., Ishigami-Yuasa, M., Kimura, Y., Ogasawara, Y., et al. (2013). A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat. Commun. 4:1366.
Sinha, S. R., Haileselassie, Y., Nguyen, L. P., Tropini, C., Wang, M., Becker, L. S., et al. (2020). Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 27, 659–670.e5.
Sivignon, A., de Vallée, A., Barnich, N., Denizot, J., Darcha, C., Pignède, G., et al. (2015). Saccharomyces cerevisiae CNCM I-3856 prevents colitis induced by AIEC bacteria in the transgenic mouse model mimicking Crohn’s disease. Inflamm. Bowel Dis. 21, 276–286. doi: 10.1097/mib.0000000000000280
Sokol, H., Landman, C., Seksik, P., Berard, L., Montil, M., Nion-Larmurier, I., et al. (2020). Fecal microbiota transplantation to maintain remission in Crohn’s disease: a pilot randomized controlled study. Microbiome 8:12.
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., et al. (2008b). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad Sci. U.S.A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Sokol, H., Lay, C., Seksik, P., and Tannock, G. W. (2008a). Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm. Bowel Dis. 14, 858–867. doi: 10.1002/ibd.20392
Sokol, H., Leducq, V., Aschard, H., Pham, H. P., Jegou, S., Landman, C., et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Sokol, H., Lepage, P., Seksik, P., Doré, J., and Marteau, P. (2006a). Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J. Clin. Microbiol. 44, 3172–3177. doi: 10.1128/jcm.02600-05
Sokol, H., Seksik, P., Furet, J. P., Firmesse, O., Nion-Larmurier, I., Beaugerie, L., et al. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15, 1183–1189. doi: 10.1002/ibd.20903
Sokol, H., Seksik, P., Rigottier-Gois, L., Lay, C., Lepage, P., Podglajen, I., et al. (2006b). Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 12, 106–111. doi: 10.1097/01.mib.0000200323.38139.c6
Song, D., Lai, L., and Ran, Z. (2020). Metabolic regulation of group 3 innate lymphoid cells and their role in inflammatory bowel disease. Front. Immunol. 11:580467.
Sorg, J. A., and Sonenshein, A. L. (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512. doi: 10.1128/jb.01765-07
Standaert-Vitse, A., Sendid, B., Joossens, M., François, N., Vandewalle-El Khoury, P., Branche, J., et al. (2009). Candida albicans colonization and ASCA in familial Crohn’s disease. Am. J. Gastroenterol. 104, 1745–1753. doi: 10.1038/ajg.2009.225
Stewardson, A. J., Gaia, N., Francois, P., Malhotra-Kumar, S., Delemont, C., Martinez de Tejada, B., et al. (2015). Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin. Microbiol. Infect. 21, 344.e1–e11.
Strati, F., Pujolassos, M., Burrello, C., Giuffre, M. R., Lattanzi, G., Caprioli, F., et al. (2021). Antibiotic-associated dysbiosis affects the ability of the gut microbiota to control intestinal inflammation upon fecal microbiota transplantation in experimental colitis models. Microbiome 9:39.
Strauch, U. G., Obermeier, F., Grunwald, N., Gurster, S., Dunger, N., Schultz, M., et al. (2005). Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut 54, 1546–1552. doi: 10.1136/gut.2004.059451
Su, G. L., Ko, C. W., Bercik, P., Falck-Ytter, Y., Sultan, S., Weizman, A. V., et al. (2020). AGA clinical practice guidelines on the role of probiotics in the management of gastrointestinal disorders. Gastroenterology 159, 697–705. doi: 10.1053/j.gastro.2020.05.059
Sugihara, K., and Kamada, N. (2021). Diet-Microbiota Interactions in Inflammatory Bowel Disease. Nutrients 13, 1533. doi: 10.3390/nu13051533
Sultan, S., El-Mowafy, M., Elgaml, A., El-Mesery, M., El Shabrawi, A., Elegezy, M., et al. (2020). Alterations of the treatment-naive gut microbiome in newly diagnosed hepatitis C virus infection. ACS Infect. Dis. 7, 1059–1068. doi: 10.1021/acsinfecdis.0c00432
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P., and Lochs, H. (2005). Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389. doi: 10.1128/jcm.43.7.3380-3389.2005
Tang, C., Kamiya, T., Liu, Y., Kadoki, M., Kakuta, S., Oshima, K., et al. (2015). Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T Cell expansion in the intestine. Cell Host Microbe 18, 183–197. doi: 10.1016/j.chom.2015.07.003
Taniguchi, E., Matsunami, M., Kimura, T., Yonezawa, D., Ishiki, T., Sekiguchi, F., et al. (2009). Rhodanese, but not cystathionine-gamma-lyase, is associated with dextran sulfate sodium-evoked colitis in mice: a sign of impaired colonic sulfide detoxification? Toxicology 264, 96–103. doi: 10.1016/j.tox.2009.07.018
Targownik, L. E., Benchimol, E. I., Bernstein, C. N., Singh, H., Tennakoon, A., Zubieta, A. A., et al. (2020). Combined biologic and immunomodulatory therapy is superior to monotherapy for decreasing the risk of inflammatory bowel disease-related complications. J. Crohns Colitis 14, 1354–1363. doi: 10.1093/ecco-jcc/jjaa050
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/bcj20160510
Tiago, F. C., Porto, B. A., Ribeiro, N. S., Moreira, L. M., Arantes, R. M., Vieira, A. T., et al. (2015). Effect of Saccharomyces cerevisiae strain UFMG A-905 in experimental model of inflammatory bowel disease. Benef. Microbes 6, 807–815. doi: 10.3920/bm2015.0018
Tonack, S., Tang, C., and Offermanns, S. (2013). Endogenous metabolites as ligands for G protein-coupled receptors modulating risk factors for metabolic and cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 304, H501–H513.
Tong, M., Li, X., Wegener Parfrey, L., Roth, B., Ippoliti, A., Wei, B., et al. (2013). A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One 8:e80702. doi: 10.1371/journal.pone.0080702
Trauner, M., Claudel, T., Fickert, P., Moustafa, T., and Wagner, M. (2010). Bile acids as regulators of hepatic lipid and glucose metabolism. Dig. Dis. 28, 220–224. doi: 10.1159/000282091
Tremaroli, V., and Backhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. doi: 10.1038/nature11552
Tsai, H. H., Dwarakanath, A. D., Hart, C. A., Milton, J. D., and Rhodes, J. M. (1995). Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut 36, 570–576. doi: 10.1136/gut.36.4.570
Ungaro, F., Massimino, L., D’Alessio, S., and Danese, S. (2019a). The gut virome in inflammatory bowel disease pathogenesis: from metagenomics to novel therapeutic approaches. United European Gastroenterol. J. 7, 999–1007. doi: 10.1177/2050640619876787
Ungaro, F., Massimino, L., Furfaro, F., Rimoldi, V., Peyrin-Biroulet, L., D’Alessio, S., et al. (2019b). Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early-diagnosed inflammatory bowel disease. Gut Microbes 10, 149–158. doi: 10.1080/19490976.2018.1511664
Van den Bossche, L., Hindryckx, P., Devisscher, L., Devriese, S., Van Welden, S., Holvoet, T., et al. (2017). Ursodeoxycholic acid and its taurine- or glycine-conjugated species reduce colitogenic dysbiosis and equally suppress experimental colitis in mice. Appl. Environ. Microbiol. 83, e02766–16.
Vaughn, B. P., Kaiser, T., Staley, C., Hamilton, M. J., Reich, J., Graiziger, C., et al. (2019). A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin. Exp. Gastroenterol. 12, 9–19. doi: 10.2147/ceg.s186097
Verma, R., Verma, A. K., Ahuja, V., and Paul, J. (2010). Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J. Clin. Microbiol. 48, 4279–4282. doi: 10.1128/jcm.01360-10
Vermeiren, J., Van de Wiele, T., Van Nieuwenhuyse, G., Boeckx, P., Verstraete, W., and Boon, N. (2012). Sulfide- and nitrite-dependent nitric oxide production in the intestinal tract. Microb. Biotechnol. 5, 379–387. doi: 10.1111/j.1751-7915.2011.00320.x
Vich Vila, A., Collij, V., Sanna, S., Sinha, T., Imhann, F., Bourgonje, A. R., et al. (2020). Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 11:362.
Vieira, E. L. M., Leonel, A. J., Sad, A. P., Beltrao, N. R. M., Costa, T. F., Ferreira, T. M. R., et al. (2012). Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J. Nutr. Biochem. 23, 430–436. doi: 10.1016/j.jnutbio.2011.01.007
Virgin, H. W. (2014). The virome in mammalian physiology and disease. Cell 157, 142–150. doi: 10.1016/j.cell.2014.02.032
Vrakas, S., Mountzouris, K. C., Michalopoulos, G., Karamanolis, G., Papatheodoridis, G., Tzathas, C., et al. (2017). Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS One 12:e0170034. doi: 10.1371/journal.pone.0170034
Wagner, J., Maksimovic, J., Farries, G., Sim, W. H., Bishop, R. F., Cameron, D. J., et al. (2013). Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19, 1598–1608. doi: 10.1097/mib.0b013e318292477c
Walker, A. W., Duncan, S. H., Leitch, E. C. M., Child, M. W., and Flint, H. J. (2005). pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 71, 3692–3700. doi: 10.1128/aem.71.7.3692-3700.2005
Walker, A. W., Sanderson, J. D., Churcher, C., Parkes, G. C., Hudspith, B. N., Rayment, N., et al. (2011). High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 11:7. doi: 10.1186/1471-2180-11-7
Wallace, J. L., Vong, L., McKnight, W., Dicay, M., and Martin, G. R. (2009). Endogenous and exogenous hydrogen sulfide promotes resolution of colitis in rats. Gastroenterology 137, 569–78, 578.e1.
Walters, S. S., Quiros, A., Rolston, M., Grishina, I., Li, J., Fenton, A., et al. (2014). Analysis of gut microbiome and diet modification in patients with Crohn’s disease. SOJ Microbiol. Infect. Dis. 2, 1–13.
Wang, W., Chen, L., Zhou, R., Wang, X., Song, L., Huang, S., et al. (2014). Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 52, 398–406. doi: 10.1128/jcm.01500-13
Wang, W., Jovel, J., Halloran, B., Wine, E., Patterson, J., Ford, G., et al. (2015). Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm. Bowel Dis. 21, 1419–1427.
Wang, Y. D., Chen, W. D., Yu, D. N., Forman, B. M., and Huang, W. D. (2011). The G-Protein-coupled bile acid receptor, gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated b cells (NF-kappa B) in mice. Hepatology 54, 1421–1432. doi: 10.1002/hep.24525
Washio, J., Sato, T., Koseki, T., and Takahashi, N. (2005). Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour. J. Med. Microbiol. 54, 889–895. doi: 10.1099/jmm.0.46118-0
Watanabe, M., Houten, S. M., Mataki, C., Christoffolete, M. A., Kim, B. W., Sato, H., et al. (2006). Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489. doi: 10.1038/nature04330
Willing, B. P., Dicksved, J., Halfvarson, J., Andersson, A. F., Lucio, M., Zheng, Z., et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–U105.
Windsor, J. W., and Kaplan, G. G. (2019). Evolving epidemiology of IBD. Curr. Gastroenterol. Rep. 21:40.
Winter, R. W., Collins, E., Cao, B., Carrellas, M., Crowell, A. M., and Korzenik, J. R. (2017). Higher 25-hydroxyvitamin D levels are associated with greater odds of remission with anti-tumour necrosis factor-α medications among patients with inflammatory bowel diseases. Aliment Pharmacol. Ther. 45, 653–659. doi: 10.1111/apt.13936
Wohlgemuth, S., Keller, S., Kertscher, R., Stadion, M., Haller, D., Kisling, S., et al. (2011). Intestinal steroid profiles and microbiota composition in colitic mice. Gut Microbes 2, 159–166. doi: 10.4161/gmic.2.3.16104
Woodmansey, E. J. (2007). Intestinal bacteria and ageing. J. Appl. Microbiol. 102, 1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x
Wright, E. K., Kamm, M. A., Teo, S. M., Inouye, M., Wagner, J., and Kirkwood, C. D. (2015). Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm. Bowel Dis. 21, 1219–1228.
Wu, T. Z., Bound, M. J., Standfield, S. D., Jones, K. L., Horowitz, M., and Rayner, C. K. (2013). Effects of taurocholic acid on glycemic, glucagon-like peptide-1, and insulin responses to small intestinal glucose infusion in healthy humans. J. Clin. Endocrinol. Metab. 98, E718–E722.
Wu, Y. C., Wang, X. J., Yu, L., Chan, F. K., Cheng, A. S., Yu, J., et al. (2012). Hydrogen sulfide lowers proliferation and induces protective autophagy in colon epithelial cells. PLoS One 7:e37572. doi: 10.1371/journal.pone.0037572
Xu, J., Chen, N., Wu, Z., Song, Y., Zhang, Y., Wu, N., et al. (2018). 5-aminosalicylic acid alters the gut bacterial microbiota in patients with ulcerative colitis. Front. Microbiol. 9:1274.
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Yuksel, M., Ates, I., Kaplan, M., Arikan, M. F., Ozin, Y. O., Kilic, Z. M. Y., et al. (2017). is oxidative stress associated with activation and pathogenesis of inflammatory bowel disease? J. Med. Biochem. 36, 341–348. doi: 10.1515/jomb-2017-0013
Zanardo, R. C. O., Brancaleone, V., Distrutti, E., Fiorucci, S., Cirino, G., and Wallace, J. L. (2006). Hydrogen sulfide is an endogenous modulator of leukocyte-mediated inflammation. FASEB J. 20, 2118–2120. doi: 10.1096/fj.06-6270fje
Zhang, L., Lee, H., Grimm, M. C., Riordan, S. M., Day, A. S., and Lemberg, D. A. (2014). Campylobacter concisus and inflammatory bowel disease. World J. Gastroenterol. 20, 1259–1267. doi: 10.3748/wjg.v20.i5.1259
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Zheng, H., Chen, M., Li, Y., Wang, Y., Wei, L., Liao, Z., et al. (2017). Modulation of gut microbiome composition and function in experimental colitis treated with sulfasalazine. Front. Microbiol. 8:1703.
Zheng, L., Kelly, C. J., Battista, K. D., Schaefer, R., Lanis, J. M., Alexeev, E. E., et al. (2017). Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of Claudin-2. J. Immunol. (Baltimore, Md. : 1950) 199, 2976–2984. doi: 10.4049/jimmunol.1700105
Zheng, N. X., Wang, Y., Hu, D. D., Yan, L., and Jiang, Y. Y. (2015). The role of pattern recognition receptors in the innate recognition of Candida albicans. Virulence 6, 347–361. doi: 10.1080/21505594.2015.1014270
Zimmerman, M. A., Singh, N., Martin, P. M., Thangaraju, M., Ganapathy, V., Waller, J. L., et al. (2012). Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G1405–G1415.
Zuo, T., and Ng, S. C. (2018). The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front. Microbiol. 9:2247.
Zuo, T., Lu, X. J., Zhang, Y., Cheung, C. P., Lam, S., Zhang, F., et al. (2019). Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179. doi: 10.1136/gutjnl-2018-318131
Keywords: IBD, gut microbiota, dysbiosis, SCFAs (short chain fatty acids), metabolome
Citation: Sultan S, El-Mowafy M, Elgaml A, Ahmed TAE, Hassan H and Mottawea W (2021) Metabolic Influences of Gut Microbiota Dysbiosis on Inflammatory Bowel Disease. Front. Physiol. 12:715506. doi: 10.3389/fphys.2021.715506
Received: 27 May 2021; Accepted: 18 August 2021;
Published: 27 September 2021.
Edited by:
Phillipp Hartmann, University of California, San Diego, United StatesReviewed by:
Gabriela Fonseca-Camarillo, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (INCMNSZ), MexicoCopyright © 2021 Sultan, El-Mowafy, Elgaml, Ahmed, Hassan and Mottawea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walid Mottawea, d21vdHRhd2VAdW90dGF3YS5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.