
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 03 August 2021
Sec. Computational Physiology and Medicine
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.709230
This article is part of the Research TopicArtificial Intelligence in Heart ModellingView all 24 articles
Segmentation of cardiac fibrosis and scars is essential for clinical diagnosis and can provide invaluable guidance for the treatment of cardiac diseases. Late Gadolinium enhancement (LGE) cardiovascular magnetic resonance (CMR) has been successful in guiding the clinical diagnosis and treatment reliably. For LGE CMR, many methods have demonstrated success in accurately segmenting scarring regions. Co-registration with other non-contrast-agent (non-CA) modalities [e.g., balanced steady-state free precession (bSSFP) cine magnetic resonance imaging (MRI)] can further enhance the efficacy of automated segmentation of cardiac anatomies. Many conventional methods have been proposed to provide automated or semi-automated segmentation of scars. With the development of deep learning in recent years, we can also see more advanced methods that are more efficient in providing more accurate segmentations. This paper conducts a state-of-the-art review of conventional and current state-of-the-art approaches utilizing different modalities for accurate cardiac fibrosis and scar segmentation.
Necrosis regions found in the heart (including left atrium (LA) pre-ablation fibrosis, LA post-ablation scar and left ventricle (LV) infarction), depending on the location and size, can have various implications on the cardiac conditions of the patients. For example, ventricular scars can be signs of earlier episodes of myocardial infarction (MI) (Choi et al., 2001; Krittayaphong et al., 2008; Wu et al., 2008; Larose et al., 2010). Locating and quantifying the fibrosis and scars have also been demonstrated as a valuable tool for the treatment stratification of patients with atrial fibrillation (AF) (Allessie, 2002; Boldt, 2004) or ventricular tachycardia (Ukwatta et al., 2015) and provide guidance information for the surgical or ablation based procedures (Vergara and Marrouche, 2011). Imaging of post-ablation scars may also give valuable information on treatment outcomes (Peters et al., 2007; Badger et al., 2010).
Cardiovascular magnetic resonance (CMR) has been one of the modern imaging techniques, which is widely used for qualitative and quantitative evaluation of cardiac conditions and to support diagnosis, monitoring disease progression and treatment planning (Kim et al., 2009). In particular, Late Gadolinium enhancement (LGE) CMR has been an emerging technique for locating and quantifying regions of fibrosis and scars across the LA and the LV (Peters et al., 2007; McGann et al., 2008; Oakes et al., 2009; Akkaya et al., 2013; Bisbal et al., 2014). LGE CMR has also been shown to improve ablation strategy planning, treatment stratification and prognosis by pre-ablation fibrosis quantification via clinical validations (Akoum et al., 2011). It also enabled computationally guided and personalized targeted ablation in treating AF in clinical practices (Boyle et al., 2019).
Many algorithms have been developed for the segmentation of cardiac scarring regions, and a few challenges have benchmarked some of the high-performing methodologies (Table 1). Among these, 2-SD (standard deviation) has been advocated by the official guidelines (Kramer et al., 2013), while the full width at half maximum (FWHM) technique has been advocated as the most reproducible method to segment ventricular scars (Flett et al., 2011) (see Section 3.2 for descriptions of 2-SD and FWHM methods). As these algorithms are usually based on successful segmentation of the corresponding anatomical regions beforehand as an accurate initialization, there has also been rising attention to the automated segmentation of LA and LV anatomy from the LGE CMR images (Table 1).
With the development of artificial intelligence techniques, we can observe a rising number of various deep learning models using convolutional neural networks [e.g., fully connected neural network (FCNN) (Szegedy et al., 2016) and U-Net (Ronneberger et al., 2015)], which have demonstrated encouraging results in segmentations of cardiac substructures in recent years (Chen C. et al., 2020). It has also been found that deep learning can be directly applied to scar segmentation as a fully automated end-to-end solution for the input LGE CMR images. With co-registration of different modalities together and deep learning based transfer learning, the combination of LGE CMR with other CMR imaging modalities [e.g., balanced steady-state free precession (bSSFP)] may further improve the efficacy and efficiency of the segmentation results.
The use of Gadolinium-based contrast agent (GBCA) has led to concerns over the patient’s safety, particularly for the patient with renal impairments (Ledneva et al., 2009). With deep learning based methods, cardiac scarring regions can now be localized and quantified in non-Gadolinium enhanced CMR images without GBCA injections (Zhang et al., 2019).
As all pre-2016 and pre-2013 cardiac scarring segmentation have been carefully benchmarked and summarized by Karim et al. (2013, 2016), this paper instead focuses on the survey of all post-2016 methodologies in fibrosis and scars delineation and segmentation of the LA and LV anatomy from LGE CMR images. This study also discusses the potential use of the modalities other than LGE CMR in locating and quantifying the scars.
To identify related contributions, search engines like Scopus and Google Scholar were queried for papers on or after 01 Jan 2016 containing (“atrial” OR “ventricular”) and (“cardiac”) and (“segmentation”) with or without (“scar”) in their titles or abstracts. Papers that do not primarily focus on the segmentation of cardiac scar or scar-related cardiac anatomy were excluded. Each paper was reviewed and agreed upon by at least two of us (Y.W., Z.T., B.L.) before inclusion. We found 4,384 papers from the search engines and shortlisted 110 of them following the criterion above (Figure 1). After full-text screening for their relevances to the topic, we eventually included 47 of them into this study. The last update to the included papers was on 13 May 2021.
Fibrosis found in LA are signs of atrial structural remodeling and can be considered as a major risk factor in the progression of the atrial fibrillation (AF) (Allessie, 2002; Boldt, 2004), where the identification of scarring and fibrosis regions in LA has been crucial for diagnosis, prognosis and treatment planning. Native pre-ablation fibrosis can be a sign of AF recurrence (Oakes et al., 2009), and post-ablation detection of ablation induced scars can facilitate the identification of post-ablation ablation line gaps, which is the main reason of ablation failures (Peters et al., 2007; Badger et al., 2010). In contrast to the traditional method of the electro-anatomical mapping (EAM) system, which is an invasive technique in localization of the atrial scar and the fibrosis with suboptimal accuracy (Zhong et al., 2007; Schmidt et al., 2009), LGE CMR enables the atrial scarring and fibrosis regions to be localized and quantified non-invasively without ionizing radiation. LGE CMR employs the slow washout kinetics of Gadolinium in these regions to highlight these scarring and fibrosis regions (Peters et al., 2007; McGann et al., 2008; Oakes et al., 2009; Akkaya et al., 2013; Bisbal et al., 2014).
In addition to the atrium, LGE CMR has also been considered as a gold-standard modality for the assessment and quantification of the scarring regions in the left ventricle (Simonetti et al., 2001; Wu et al., 2001; Wagner et al., 2003a; Hendel et al., 2006), where fibrotic and scarring regions found can be considered as a sign of earlier or current episodes of the MI (Choi et al., 2001; Krittayaphong et al., 2008; Wu et al., 2008; Larose et al., 2010). In addition to MI, with growing prognostic evidence, LGE has been successful in the identification of scarring regions in cardiomyopathy, inflammatory and infiltrative conditions (Wagner et al., 2003b; Maceira et al., 2005; Smedema et al., 2005; Flett et al., 2009).
However, the LGE CMR modality often suffers from poor image qualities, which may be due to residual respiratory motions, variabilities in the heart rate and gadolinium wash-out during the currently long acquisition time (Yang et al., 2017). Particularly, the spatial resolution of the left atrium in the LGE CMR image is limited (To et al., 2011), considering the thin transmural thickness of the atrial wall [mean = 2.2–2.5 mm (Saìnchez-Quintana et al., 2005)] (Figure 2). The variable anatomical morphological shapes of the LA and pulmonary veins (PV) also impose an additional challenge to the LGE CMR segmentations. To improve the visualization of these scar regions, we can see a successful attempt by maximum intensity projection (MIP) to enhance intensities on post-ablation LA LGE CMR (Knowles et al., 2010). Moreover, some irrelevant cardiac substructures may be highlighted in LGE CMR images as well, in addition to the scarring and fibrosis regions. These may be due to, for example, the navigator beam artifact, which is often seen near the right PV, Gadolinium uptake by the aortic wall and valves, and confounded enhancement in the spine, esophagus, etc. (Karim et al., 2013; Yang et al., 2017). As a result, these can lead to a poor result in the delineation of LA and LV scar or fibrosis regions and even a significant amount of false positives in segmentations of these structures and regions.
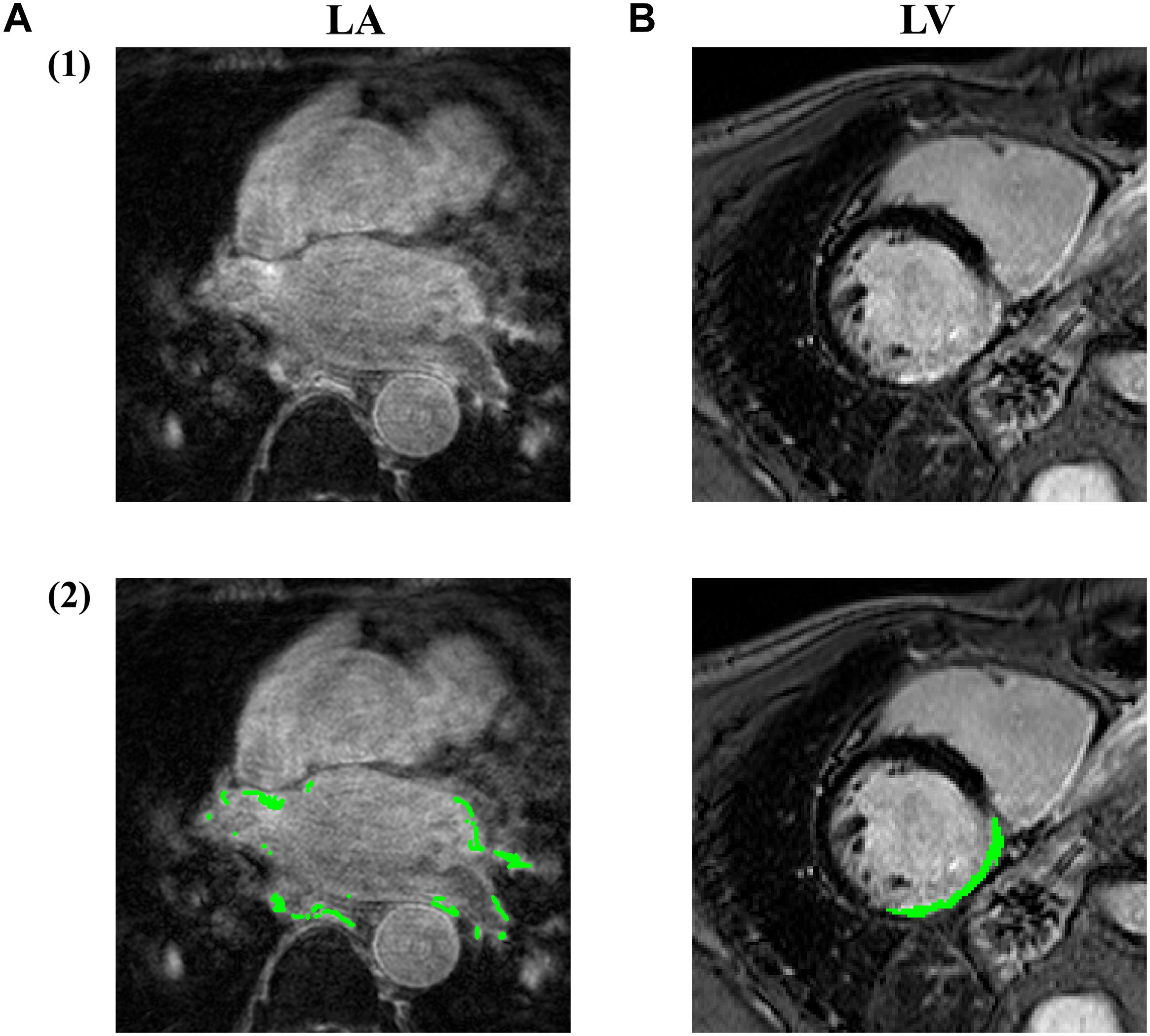
Figure 2. Examples of LGE CMR images acquired at (A) LA and (B) LV, with the fibrosis/infarction regions highlighted in green. By comparing (A2) and (B2), we can see the fibrosis region in LA is rather more discrete and thinner compared to LV infarction, making LA fibrosis regions more difficult to be accurately fully localized and quantified. Image source: (A) was extracted from pre-ablation CMR images in ISBI 2013 cDERMIS dataset (http://www.cardiacatlas.org/challenges/left-atrium-fibrosis-and-scar-segmentation-challenge/). (B) was extracted from MICCAI 2012 Ventricular Infarct Segmentation challenge dataset (http://www.cardiacatlas.org/challenges/ventricular-infarct -segmentation/).
In addition, although LGE CMR has been successful in being the gold standard reference technique for AF and MI, including LGE in MRI significantly extends the scanning time. There have been also increasingly growing concerns regarding the safety of the Gadolinium based contrast agent used, particularly for the patient with renal impairments (Ledneva et al., 2009).
In addition to LGE MRI, which could highlight the scarring regions, segmentation of the anatomy and scarring regions can also utilize other modalities (Figure 3) to further improve the accuracy if applied with LGR CMR by co-registering different modalities together (Zhuang, 2019).
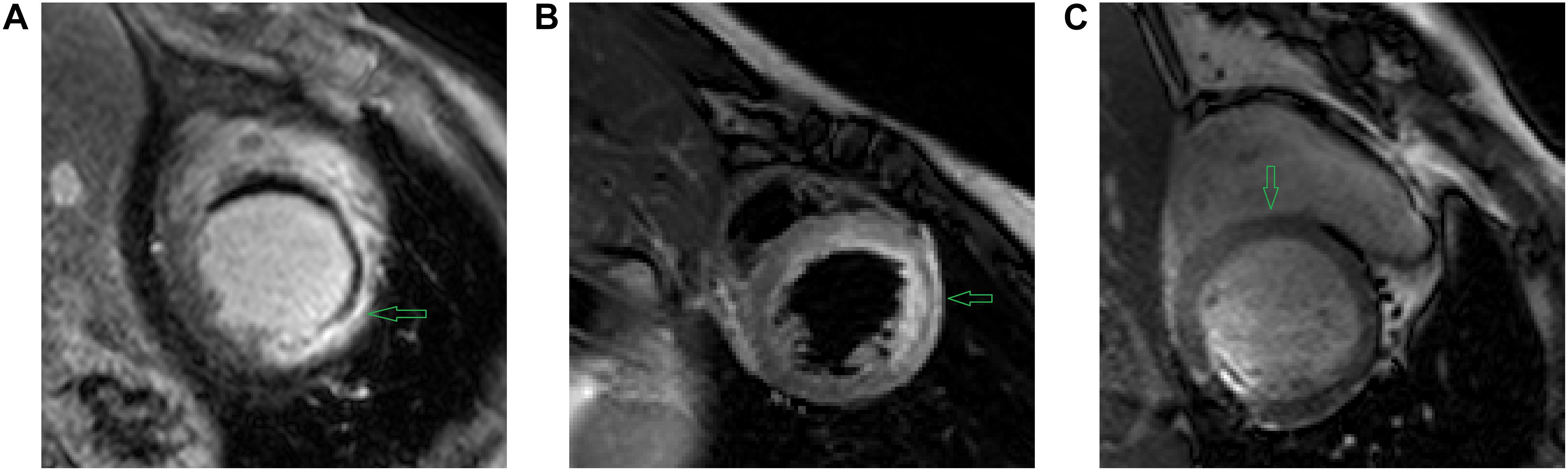
Figure 3. Example images using different CMR sequences acquired by (A) LGE CMR (B) T2 CMR (C) bSSFP CMR. As denoted by the green arrows, we can see (A) LGE CMR accentuates the scar tissue by high intensities on the images; (B) T2 CMR accentuates myocardial oedema by high intensities on the image; and (C) bSSFP CMR shows the distinct endo- and epi-cardial boundary of the myocardium clearly on the image. Image source: (A–C) extracted from the MS-CMR open challenge dataset (MS-CMR Challenge, 2019).
There have been challenges benchmarking a range of algorithms for the cross-modality fusion based segmentation of anatomy, scar and oedema.
(1) MS-CMR challenge (MS-CMR Challenge, 2019; Pop et al., 2020) presented a range of algorithms taking multiple modalities in to further improve the segmentation accuracy of LV myocardium, LV blood cavity and RV.
(2) MyoPS challenge (MyoPS Challenge, 2020; Zhuang and Li, 2020) presented algorithms to delineate LV myocardium with scarring and oedema.
Other modalities and sequences can include:
(1) Magnetic resonance angiography (MRA) sequence – to image LA and PV with high contrasts, which has been demonstrated by Tao et al. (2016) to improve the error distance in segmenting LA anatomy to within 1.5 mm. However, MRA is usually ungated and usually acquired in an inspiratory breath-hold, making anatomy delineated from MRA significantly distorted from LGE CMR.
(2) Balanced steady-state free precession (bSSFP) – provides a clear boundary between the myocardium and blood cavity under movements, which is usually respiratory and cardiac gated. It can offer cine CMR with a uniform texture.
(3) T2 – high intensities in T2 presents myocardial oedema with high specificity and sensitivity (Gannon et al., 2019), T2 could be helpful in segmenting myocardial oedema and scar simultaneously if incorporated with LGE-CMR and bSSFP (Zhu et al., 2017). Identification of oedema on CMR can help clinicians to differentiate between acute and remote myocardial infarction (Friedrich, 2017). The presence of oedema in patients without extensive irreversible injury (e.g., scar) can serve as a marker for clinicians to predict the recovery of LV systolic functions (Vermes et al., 2014).
Conventionally, a two-stage approach is adopted in the identification and evaluation of fibrotic and scarring tissue – (1) segmentation of the relevant anatomical structure (LA and PV in the case of LA fibrosis/scar segmentation and LV in the case of LV infarction segmentation) and (2) then segmentation of the fibrotic and scarring regions. This two-stage approach is particularly beneficial for LA and PV, as LA and PV are highly morphological variables and relatively small in size. We shall then elaborate on the recent developments of methodologies for each of them.
The delineation of anatomical structures, e.g., LA and LV wall, from others can be difficult in LGE CMR images. In LGE scarring tissues are significantly enhanced while the signals from the healthy tissues are attenuated (Keegan et al., 2015), making the segmentation of LA, PV and LV anatomical structures very challenging.
Accurate segmentation of the anatomy (LA or LV wall) is essential as it gives an accurate initialization for the scar segmentation. Therefore, traditionally, the segmentation of these structures were all done manually.
We could see in the cDEMRIS challenge in ISBI 2012 (Karim et al., 2013) algorithms with manually initialized LA segmentation showed significantly better performance than Others. It demonstrated the need for an accurate anatomy segmentation ahead of the scar segmentation along with Rajchl et al. (2015). Moccia et al. (2018) also demonstrated that manual and accurate segmentation of the LV wall could improve the deep learning based segmentation of the LV infarction.
In the early 21st century, radiologists looked between LGE CMR and cine CMR back and forth to delineate the myocardium region. To mimic that, we can see methods in the first decade and early second decade of this century utilizing both LGE and cine modalities by, for example, non-rigid registration to achieve high accuracy in segmentation of myocardium over LGE CMR (Dikici et al., 2004; Ciofolo et al., 2008; Wei et al., 2011, 2013). However, by doing so, the result may suffer from registration misalignment between LGE and cine modalities and the model may be computationally demanding. As such, from 2014 we can see methods that are less computationally demanding and using LGE modality only (Albà et al., 2014; Kurzendorfer et al., 2017a,b,c).
Conventional methods in medical image segmentation usually have limited efficacy. Representative methods are summarized in Table 2, which mainly include the following methodologies.
(1) Random forest (Kurzendorfer et al., 2017b).
(2) Image registration (Kurzendorfer et al., 2017c).
(3) Markov random field (MRF) model (Albà et al., 2014).
(4) Atlas-based modeling with active contour model (Kurzendorfer et al., 2017a).
(5) Principal component analysis (PCA) technique (Kurzendorfer et al., 2017c).
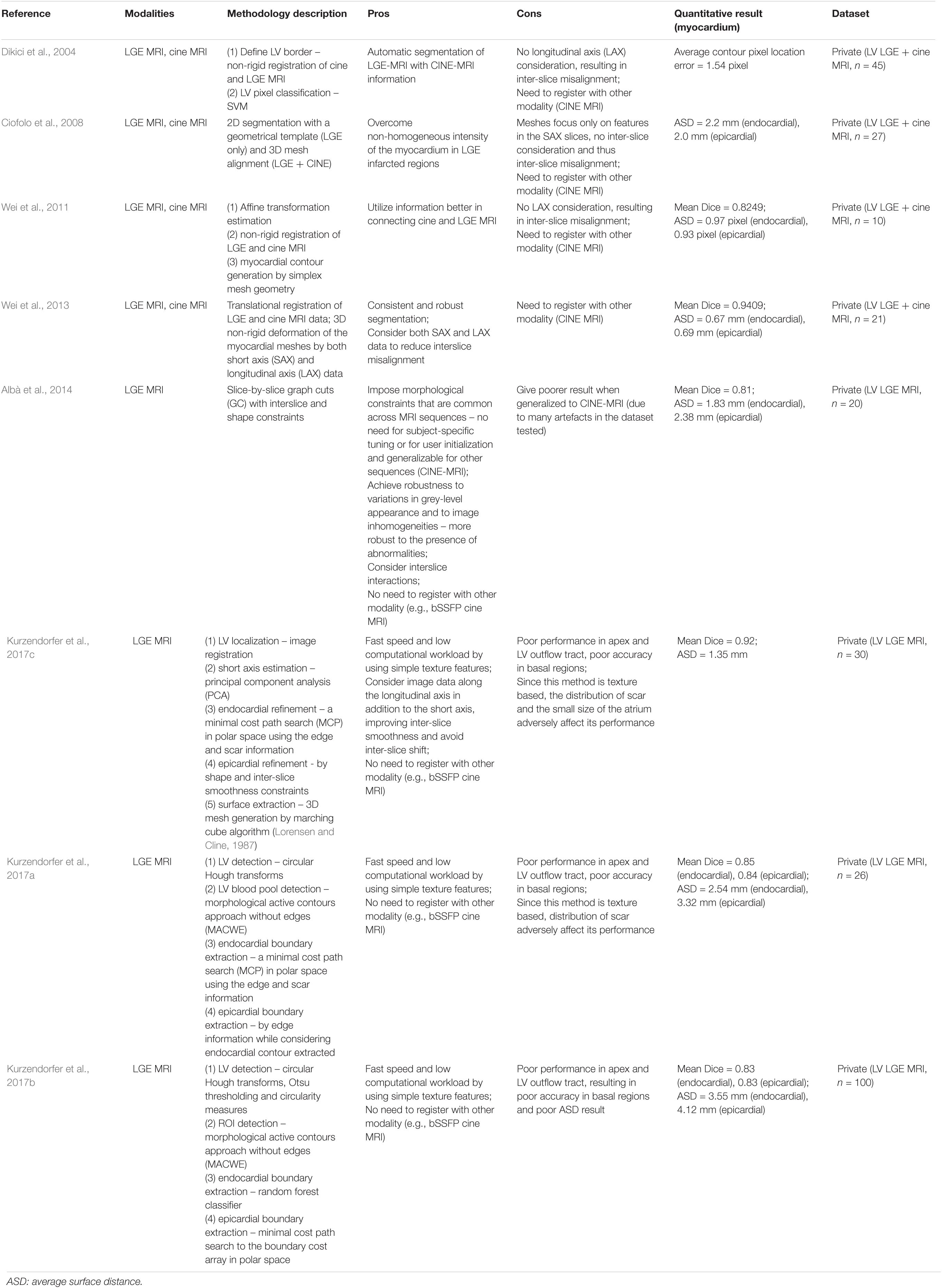
Table 2. Summary of representative conventional methodologies for segmentation of the myocardium on LGE-MRI.
For LA, in particular, the methods involving pre-defined shape priors (Zhu et al., 2013; Veni et al., 2017) often suffer from relatively poor error distance, which is more than 1–2 mm required (Xiong et al., 2021) under the clinical setting considering the thin LA wall (Zhao et al., 2017). However, one of them reported a relatively high Dice score (79%) (Zhu et al., 2013).
Upon successful segmentation of the anatomy, the scarring regions can be identified by a range of approaches. These approaches can be mainly divided into the following categories: threshold based methods, classification methods, or the combination of both.
Traditionally, the scarring regions can be detected as they are accentuated in LGE CMR. Among a range of conventional techniques, 2-SD has been advocated by official guidelines (Kramer et al., 2013), while the full width at half maximum (FWHM) technique has been advocated as the most reproducible method to segment ventricular scars (Flett et al., 2011).
2-SD and FWHM are both fixed threshold methods in segmenting the scarring region, where pixels with intensities above a fixed threshold would be labeled as the scar. 2-SD or even n-SD methods define such threshold as the sum of the mean and two or n standard deviations of signal intensities in a remote reference region, whereas FWHM defines such threshold as the half of the maximum signal intensity within the scar.
Karim et al. (2016) evaluated 2, 3, 4, 5, 6 -SD and FWHM methods on a public human LV infarct dataset and showed that FWHM superseded all n-SD methods tested by its Dice Scores and that the Dice Scores went slightly higher with the threshold rising from 2 to 6 -SD. However, it is not the case when Karim et al. (2013) evaluated 2, 3, 4 -SD and FWHM on a public human LA fibrosis/scar dataset. For pre-ablation LA fibrosis, FWHM performed much worse than all n-SD methods tested. For post-ablation LA scar, FWHM gave similar Dice Scores as 2-SD’s with 3, 4, 6 -SD methods’ Dice Scores much lower than these two.
However, these fixed-threshold techniques, including n-SD and FWHM, are unlikely to handle variations well (Oakes et al., 2009). The variations can come from two sources – scar itself and external circumstances. Scars are highly variable in their morphology and their brightness distribution on LGE CMR. Varied external factors including resolution, contrast, signal-to-noise ratio (SNR), inversion time and surface coil intensity variation can also adversely impact the accuracy of the segmentation. This is particularly the case for pulmonary veins, which are highly morphological variables.
An LV scar segmentation challenge (Karim et al., 2016) organized in MICCAI 2012 and LA scar segmentation (Karim et al., 2013) challenge organized in ISBI 2013 carefully benchmarked and summarized the majority of the pre-2013 conventional methods. In the LV segmentation challenge in 2012, it showed all of the algorithms benchmarked did not exhibit superiority against FWHM, although they did perform better than n-SD methods.
Conventional threshold based approaches are summarized in Table 3A, which mainly include the following methodologies.
(1) Otsu thresholding (Otsu, 1979; Tao et al., 2010).
(2) Histogram analysis (Karim et al., 2013).
(3) Hysteresis thresholding (Karim et al., 2013).
(4) Constrained watershed segmentation (Hennemuth et al., 2008).
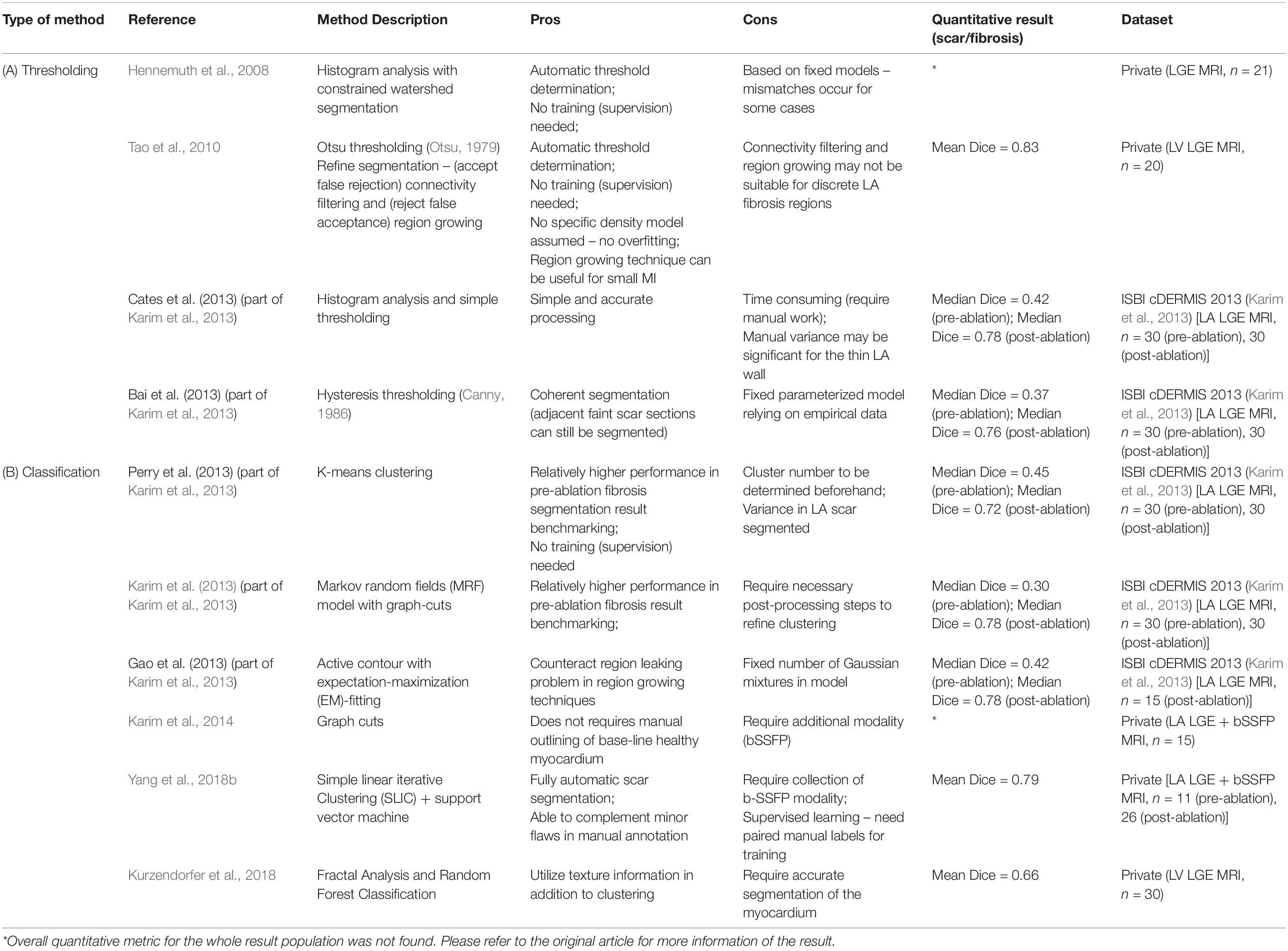
Table 3. Summary of representative conventional methodologies for segmentation of cardiac scar and fibrosis regions on LGE-MRI.
In addition, conventional classification approaches are summarized in Table 3B, which mainly include the following methodologies.
(1) K-means clustering (Karim et al., 2013).
(2) Graph cuts (Karim et al., 2013, 2014).
(3) Active contour with EM-fitting (Karim et al., 2013).
(4) Simple linear iterative clustering (SLIC) and support vector machine (Yang et al., 2018b).
(5) Random forest classification (Kurzendorfer et al., 2018).
Deep learning based methods are constructed from deep artificial neural networks. In this section, we will briefly introduce the common types of artificial neural networks (ANNs) and then focus on their variants targeting cardiac anatomy and scar segmentations. The authors would also like to recommend interested readers to refer to Goodfellow et al. (2016) for more detailed explanations and mathematical illustrations of these networks and Chen C. et al. (2020) for more thorough demonstrations of these networks in general cardiac imaging analysis.
Convoluted neural networks (CNNs), particularly fully convoluted neural networks (FCNNs), have demonstrated success in delineating anatomical structures in medical images (Shelhamer et al., 2017), especially in cardiac MR (Chen C. et al., 2020). Successful examples include ResNet (Szegedy et al., 2016), U-Net (Ronneberger et al., 2015), and etc. U-Net (Ronneberger et al., 2015), in particular, has been known for its ability to gather latent information in medical image analysis and thus to gain better performance in segmentation, which has become the most popular CNN backbone architecture, especially after demonstrating success in the ISBI cell tracking challenge in 2015.
The recurrent neural network (RNN) is another type of ANNs. The RNN is rather more useful in processing sequential data, as it could ‘memorize’ past data and utilize its ‘memory’ to assist with its current prediction. Widely used structures of RNNs include long-short-term memory (LSTM) (Hochreiter and Schmidhuber, 1997) and gated recurrent unit (GRU) (Cho et al., 2014).
Autoencoders (AEs) are also a type of ANNs, which are able to learn latent features of imaging data. Unlike CNNs and RNNs, AEs learn these features without supervision. With latent features gathered by AEs, it could be used to guide the segmentation of medical images (Oktay et al., 2016; Yue et al., 2019).
Generative Adversarial Networks (GANs) was initially proposed for image synthesis (Goodfellow et al., 2014). With its two-player model structure (a generator network to give a synthesized image and a discriminator network to try to differentiate that synthesized image from a true image), the model can enhance the resolution of the synthesized image by adversarial training. The GAN could also be used for segmentation, where its discriminator network would rather attempt to see if the output label is in an anatomically plausible shape (Luc et al., 2016).
There are a few challenges recently organized to benchmark the new methodologies proposed for the cardiac anatomy segmentation – 2018 LA Segmentation Challenge in MICCAI 2018 (LASC’18) (Xiong et al., 2021) for LA, MS-CMR (MS-CMR Challenge, 2019; Pop et al., 2020) in MICCAI 2019, and MyoPS 2020 (MyoPS Challenge, 2020; Zhuang and Li, 2020) in MICCAI 2020 for LV. With the recent development in deep learning, we can observe a range of methodologies developed for LA and LV segmentation in LGE CMR (Jamart et al., 2020).
In particular, in LASC’18, all deep learning methods had their mean surface distance in LA wall segmentation below 1.7 mm, with the minimum mean value of 0.748 mm. This demonstrated the efficacy of the deep learning based methods by the surface distance, which is required to be less than 1–2 mm under the clinical setting (Xiong et al., 2021).
Successful networks demonstrating success in delineating anatomical structures include VGG-net (Simonyan and Zisserman, 2014), U-Net (Zabihollahy et al., 2019b), and V-Net (Milletari et al., 2016). To further exploit the information on the z-axis, LSTM and its variants (Yang et al., 2018a; Zhang et al., 2020) and dilated residual learning blocks (Yang et al., 2018a) can be introduced to the widely used U-Net.
On top of the U-Net, Xiong et al. (2019) proposed a dual path U-Net variant, which is demonstrated to have the best Dice Score (0.942) followed by VGGNet (0.864) in their benchmarking of a range of popular CNNs including the original U-Net and one non-deep-learning based method (Zhu et al., 2013) in LA segmentation. Multi-view learning, incorporating axial, sagittal and coronal views together, gave superior performance compared to models based on one view only (Xiao et al., 2020).
On the contrary, further research showed that structural variations in U-Net are unlikely to cause a significant improvement of its performance in LA segmentation from LGE CMR (Wang et al., 2019), and that deep supervision and attention blocks are unlikely to further improve LA segmentation performance either (Borra et al., 2020b).
In addition to these supervised learning based methods, Chen J. et al. (2019) proposed a feature-matching based semi-supervised learning technique to further improve the segmentation efficacy.
All the methods discussed above are summarized in Table 4.
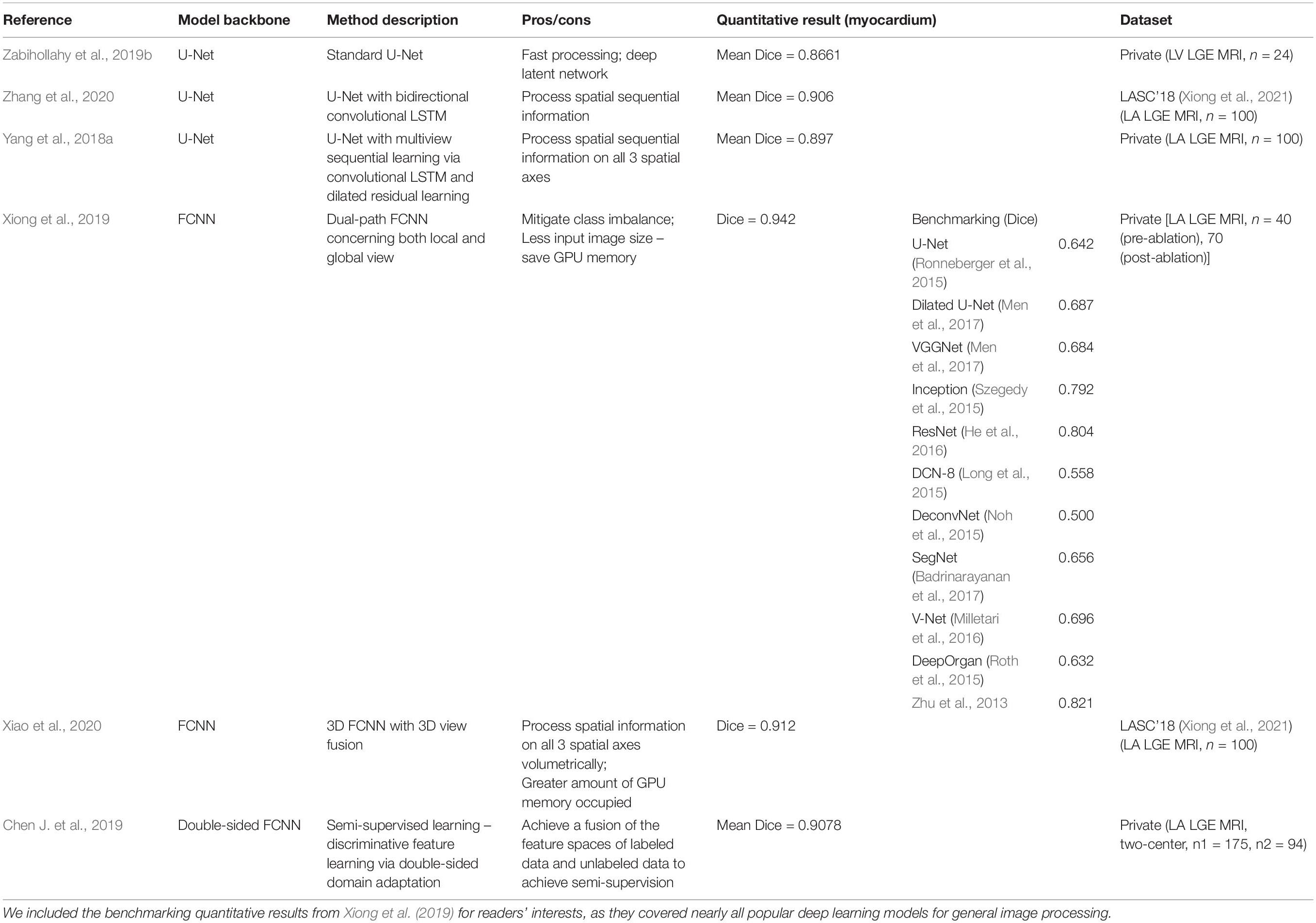
Table 4. Summary of representative deep learning based methodologies for segmentation of the myocardium on LGE-MRI.
We can observe a range of deep learning based methods in segmenting scars (Table 5).
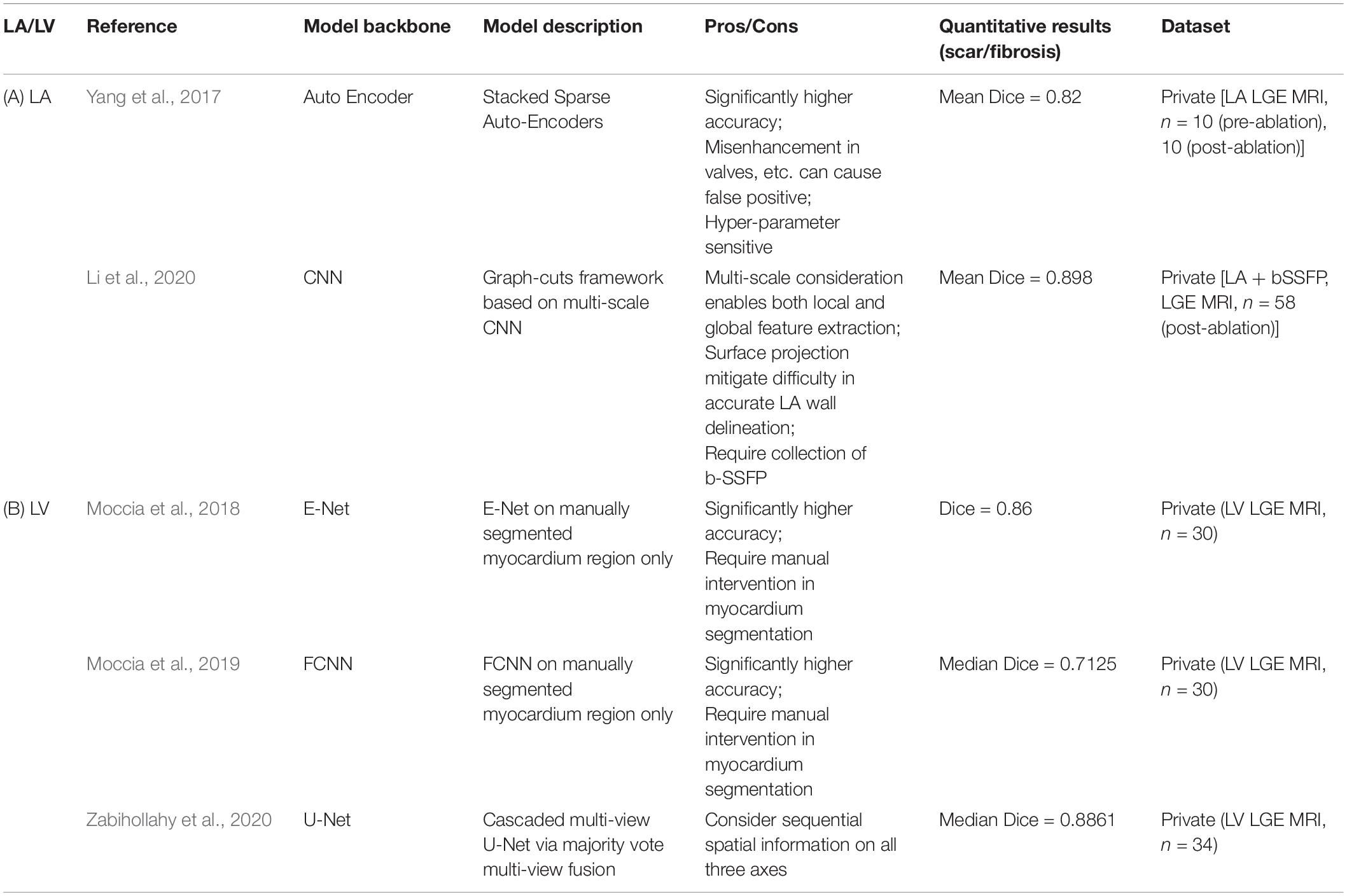
Table 5. Summary of representative deep learning based methodologies for segmentation of cardiac scar and fibrosis regions on LGE-MRI.
For LA (Table 5A), Yang et al. (2017) proposed a deep learning based method using Stacked Sparse Auto-Encoders to delineate the LA fibrosis region, which is based on accurate anatomical structure delineation. Li et al. (2020) proposed a graph-cuts framework based on multi-scale CNN to further incorporate local and global texture information of the images.
For LV (Table 5B), E-Net (Moccia et al., 2018) and FCNN (Moccia et al., 2019) were demonstrated for its high accuracy if with manually segmented LV walls. Then, multi-view U-Net has also been developed in segmenting the scar in a cascaded way (Zabihollahy et al., 2020).
With more recent developments of deep learning, the models can extract further latent information from the LGE CMR images and segment the scar directly from LGE CMR images without acquiring accurate segmentation of the relevant cardiac anatomical structures (e.g., LA wall) in advance while maintaining the accuracy. There has also been a range of methods (Table 6) that can complete the segmentation of both the anatomy of cardiac chambers and the scar simultaneously (referred to as “two tasks” below). This is particularly the case for LV, where there is much less variability in its anatomical shape.
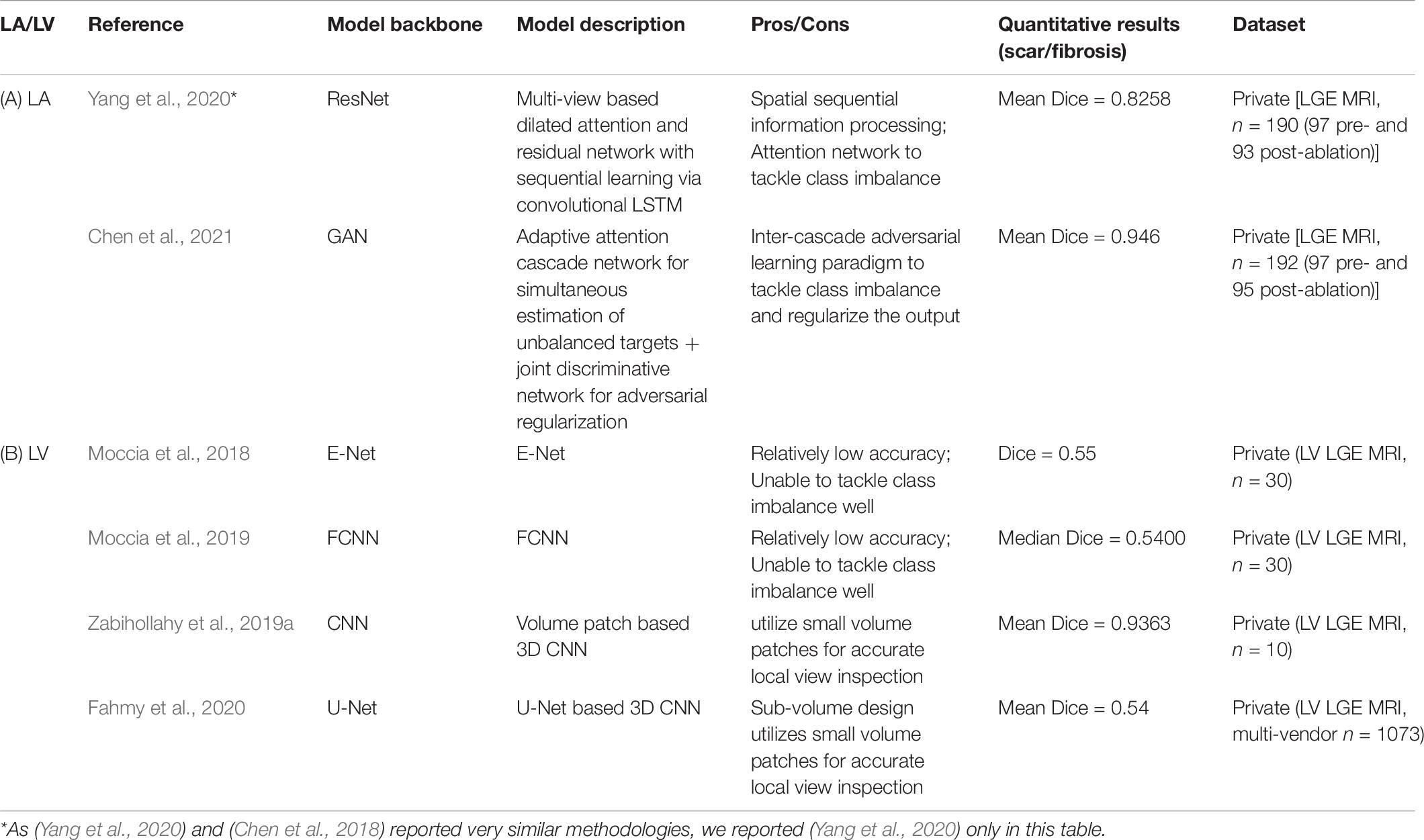
Table 6. Summary of representative end-to-end deep learning based methodologies for segmentation of cardiac scar and fibrosis regions on LGE-MRI.
For LA (Table 6A), due to the thin LA wall, it is particularly difficult to achieve an end-to-end segmentation of scar directly from LGE CMR. A multi-view two task (MVTT) deep learning based method with dilated attention network was proposed to complete the two tasks simultaneously (Chen et al., 2018; Yang et al., 2020). This study also benchmarked a range of popular deep learning networks such as U-Net and V-Net on each of the two tasks. It compared the performance of its network with conventional methods such as 2-SD and k-means to demonstrate the superiority of its network in completing both of the two tasks accurately on both pre-ablation and post-ablation datasets (Yang et al., 2020). This study also suggested that 2-SD, k-means and fuzzy c-means methods clearly over-estimated the enhanced LA scar region (Yang et al., 2020).
Later, with a joint GAN discriminator, Chen et al. were able to further improve the segmentation accuracy by dealing with the significantly unbalanced two LA targets (LA wall and scar) (Chen et al., 2021; Table 7). In their method, cascaded learning, a widely applied technique in learning labels with unbalanced classes in natural image segmentation (Dai et al., 2016; Murthy et al., 2016; Li et al., 2017; Lin et al., 2017; Ouyang et al., 2017; Cai and Vasconcelos, 2018; Chen K. et al., 2019), demonstrated superiority in learning.
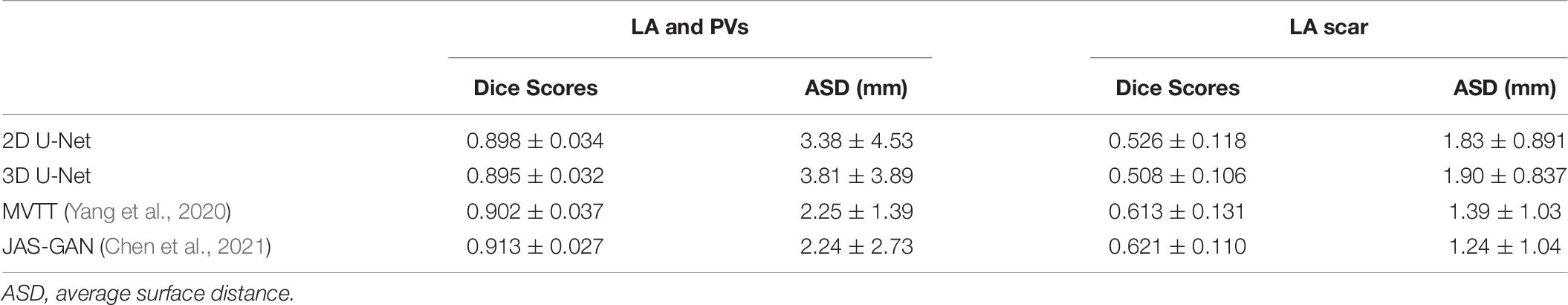
Table 7. Result of a private benchmarking (Chen et al., 2021) of different algorithms on the LASC’18 dataset, reported in their mean ± SD.
As LV has less variant morphology and greater size, there have been more successful methods demonstrating their efficacies andefficiencies in LV scar segmentation (Table 6B). E-Net (Moccia et al., 2018) and FCNN (Moccia et al., 2019) were the first few networks that demonstrated the ability to segment scar directly from LGE CMR. Although with relatively low Dice scores, they demonstrated that with an accurately segmented myocardium label it could perform better.
Recently, many deep learning methods have been proposed and demonstrated significantly higher efficacy compared to traditional threshold based methods. Zabihollahy et al. developed a CNN based network to classify each pixel by considering small volume patches around that pixel to greatly improve the mean segmentation accuracy in terms of its mean Dice score to 93.63, compared to the mean Dice scores of K-nearest neighbor (KNN) (77.85), FWHM (61.77), and 2SD (48.33) in their private benchmarking (Zabihollahy et al., 2019a).
In addition, Fahmy et al. (2020) proved that a 3D CNN deep learning based approach could be applied for LV scar segmentation for patients with hypertrophic cardiomyopathy (HCM) via a multicenter multivendor study.
Inspired by the two-stage approach, a multi-view cascaded U-Net driving for even higher efficacy in segmentation was developed to cascade the two tasks sequentially while considering sagittal, axial and coronal views (Moccia et al., 2019).
As explained in Section 3.1.2, traditionally, clinicians check both bSSFP cine and LGE MRI modalities to ensure accurate segmentation of the myocardium and then the scar. Therefore, many methods suggested the use of both bSSFP cine and LGE modalities in delineating anatomical structures and scar to mimic that. For LA, it is also known that MRA gives a clear boundary in PV to help with LA wall segmentation. We can see many methods taking MRA as an extra modality into their models to enhance their segmentation accuracy. However, many studies chose bSSFP over MRA, as bSSFP can be acquired in the same phase as LGE CMR by cardiac gating. Although MRA provides better resolution, MRA is not cardiac gated and can be difficult and error-prone in co-registration with LGE CMR, causing misalignments in registered images. Additionally, as explained in Section 2.2, integration with other modality (e.g., T2) may enable more findings from the CMR (e.g., oedema) in addition to scars.
There are few challenges benchmarking a range of algorithms for the cross-modality fusion based segmentation of anatomy, scar and oedema. MS-CMR challenge (MS-CMR Challenge, 2019; Pop et al., 2020) presented a range of algorithms taking multiple modalities in to further improve the segmentation accuracy of LV myocardium, LV blood cavity and RV. MyoPS challenge (MyoPS Challenge, 2020; Zhuang and Li, 2020) presented algorithms to delineate LV myocardium with scarring and oedema.
Common methods to segment anatomy and scar from multiple modalities include:
(1) Cross-modality style and feature propagation (typically from bSSFP to LGE-MRI) [e.g., multi-atlas label fusion (MAS) (Zhu et al., 2017)].
(2) Combination of multiple paired sequences and modalities for segmentation by either cross-modality image style transfer [e.g., Cycle-GAN (Zhu et al., 2017) and UNIT style transfer (Huang et al., 2018; Chen J. et al., 2020)] or multi-input models [e.g., Multi-variable mixture model (MvMM) (Zhuang, 2019)].
(3) A two-stage approach to firstly co-registering anatomical segmentation from one modality to another (typically from bSSFP segmentation to LGE-MRI) and then segment scars based on the co-registered anatomy segmentation (Leong et al., 2019).
However, respiratory and/or cardiac motion complications between acquisitions of different modalities can still cause errors in registration and possible misalignments.
Although LGE CMR has been very successful in being the gold standard reference technique for AF and MI, including LGE in an MRI scanning significantly extends the scanning Time. Moreover, there have been increasingly growing concerns regarding the safety of the Gadolinium based contrast agent used, particularly for the patient with renal impairments (Ledneva et al., 2009). There has been a rising attention in exploring methods to segment scars without injecting contrast agents to the patients on non-CA modalities. Non-CA modality based cardiac scar segmentation methods have been widely demonstrated for LV scar delineations but has not been realized for LA scar delineations.
Dastidar et al. (2019) and Liu et al. (2018) demonstrated the potential of pre-contrast scar segmentation by comparing the inter-modality manual observations of myocardial infarction regions on LGE CMR and native-T1 mapping without the Gadolinium contrast agents.
T1 and T2 (Messroghli et al., 2017) are modalities that are not enhanced by any contrast materials, where relaxation times in MI is longer compared to the healthy myocardium and could be referenced for MI region segmentation reproducibly (Abdel-Aty et al., 2004; Kali et al., 2014; Smulders et al., 2015). However, the relaxation time is field strength specific (Raman et al., 2013; Haaf et al., 2017) and requires the acquisition of images for additional breath holds, which significantly extends the CMR acquisition time.
Magnetic resonance imaging feature tracking is also an approach to differentiate MI induced cardiac wall abnormalities from normal myocardium, which can be acquired as part of a standard CMR scanning examination (Muser et al., 2017; Ogawa et al., 2017). However, this technique can only detect and locate the position of MI without quantifying it.
To further improve scar segmentation on non-contrast enhanced CMR, trained by co-registered LGE and cine MRI modalities, SVM based texture analysis in pre-contrast cine MRI only can discriminate between nonviable, viable and remote segments (Larroza et al., 2018). Non-contrasted enhanced CMR scar segmentation has also been demonstrated via neighborhood approximation forests (Bleton et al., 2016), Simple Linear Iterative Clustering (SLIC) (Achanta et al., 2012) based supervoxels (Popescu et al., 2017).
With the development of deep learning, a method based on a combination of Long short-term memory (LSTM), recurrent neural network (RNN) and fully convoluted neural network (FCNN) (Xu et al., 2017) and a GAN based method (Xu et al., 2018) have been demonstrated accuracy in detecting, locating and quantifying LV scarring regions from non-contrast enhanced CMR images. Zhang et al. proposed a deep learning based framework to greatly improve the efficacy of the segmentation of LV scar on cine MRI (with its stages consisting of (1) ROI localization, (2) RNN based motion pattern extraction, and (3) pixel classification by FCNN) and assess their network extensively under a clinical setting (Zhang et al., 2019). Xu et al. (2020) on top of the deep learning based workflow, proposed a progressive sequential causal generative adversarial network (GAN) to simultaneously synthesize LGE-equivalent images and multi-class tissue segmentation (including LV blood cavity, LV myocardium and scar region) from cine CMR images. A detailed summary and results of a private benchmarking of all these algorithms can be found in Table 8.
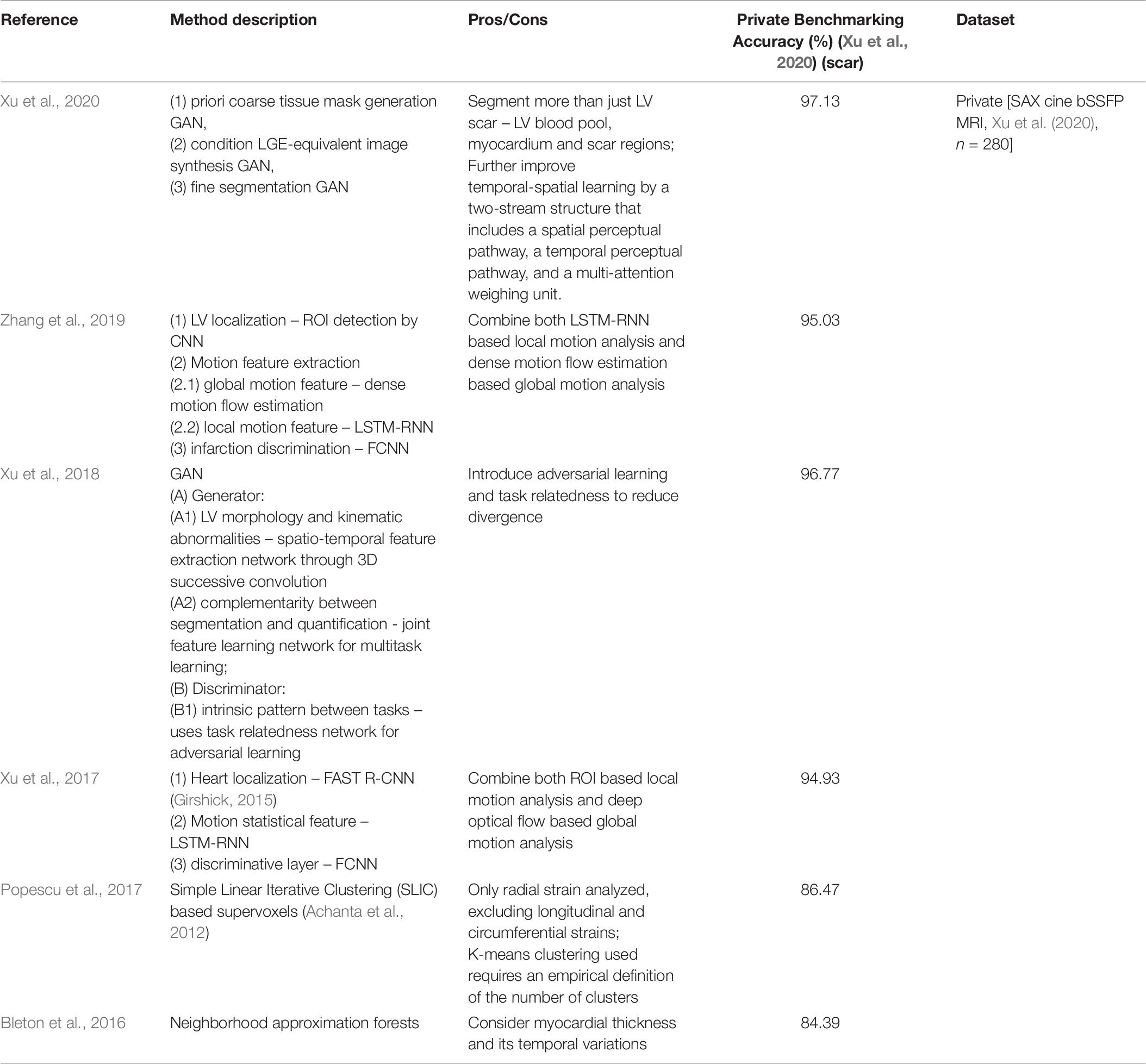
Table 8. Summary of representative machine learning/deep learning based scar segmentation in cine MRI for segmentation of cardiac scar regions on cine bSSFP MRI.
A range of evaluation metrics can be employed for assessing the results of the segmentation of the anatomy. These include Dice score, sensitivity, specificity, Hausdorff distance (HD) and surface-to-surface distance (STSD).
The Dice Score coefficient, DICE, is one of the most widely used evaluation metrics in segmentation accuracy evaluations. It is particularly sensitive to the difference between the ground truth label and the result label.
Given a 3D prediction label tensor, A, and 3D ground truth label tensor, B, the Dice score can be defined as:
Sensitivity score, also known as True Positive Rate, can be adapted to reflect the success of the algorithm for segmenting the foreground (cardiac anatomy) as:
where TP stands for true positive and FN stands for false negative.
Sensitivity score, also known as True Negative Rate, reflects the success of the algorithm for segmenting the background as:
where TN stands for true negative and FP stands for false positive.
Hausdorff distance, HD, is an important parameter in evaluating the geometrical characteristics which measures the maximum local distance between the surfaces of the predicted LA volume label tensor, A, and the ground truth label tensor, B, given by:
where a and b are all pixels locations within A and B.
In practice, the HD is not generally recommended to use it directly since it has a great sensitivity to outliers, and as noises and outliers are quite common in medical image segmentation (Gerig et al., 2001; Zhang and Lu, 2004). However, Huttenlocher et al. (1993) proposed a way to handle outliers by defining the HD as the qth quantile of distance instead of the maximum to exclude the outliers.
Surface-to-surface distance, STSD, measures the average distance error between the surfaces of the predicted LA volume and the ground truth.
where nA and nB are the numbers of pixels in A and B, respectively. Variables p and p′ describe all point between A and B.
The maximum error distance acceptable in the LA wall segmentation should be 1–2 mm under the clinical setting considering the thin LA wall (Xiong et al., 2021).
The anterior-posterior diameters of LA and LV are widely used as an essential clinical measure in clinical diagnosis and treatments.
The diameters can be estimated by finding the maximum Euclidean distance along the anterior-posterior axis of each CMR scan (Xiong et al., 2021).
The anatomical volumes of LA and LV are widely used as an essential clinical measure in clinical diagnosis and treatments.
The volume of the structure can be found as the sum of positively labeled voxels. Given the volume of the predicted anatomical structure, VA, and the volume of the ground truth, VA, the total volume error can be defined as:
In addition to the ones mentioned above, scar segmentation also employs a scar volume based metric in assessing the segmentation result, which is much more widely used as the quantification of scar is important for clinical use. They calculate the volumetric percentages of the scarring regions and compare them across the predicted and the ground truth labels.
The scar percentage is defined as the percentage of the volume of the scarring region, Vscar, relative to the volume of the relevant anatomical wall, Vwall (e.g., LA wall) (Tao et al., 2010).
For validation and benchmarking of different methods and training of deep learning based methods, accurate, consistent and reproducible acquisition of ground truth labels is essential.
Validation by employing labels from a single clinician may not be ideal as these labels may exhibit bias and intra-observer variances when the same clinician is asked to repeat their labeling. Thus, it is recommended that we take observations from multiple clinicians and fuse them together.
However, we can see significant inter-observer variances, particularly for LA anatomical segmentation in LGE-MRI where the boundaries of the LA walls are very blurred. Kurzendorfer et al. (2017c) attempted to compensate for inter-observer variances by additional smoothing but ended up with slight improvement in Dice Scores (+0.04).
It is recommended that the data source reports the inter- and intra- observer variances by employing evaluation metrics such as the Dice Score coefficient. The currently widely used method of label fusing is obtaining a 70% consensus label among multiple annotations, which can be low in their consistency levels. The level of each observer’s expertise (novice, medical student, trainee, junior clinician or senior clinician) must also be clearly noted, particularly when multiple observers are involved. It may be also recommended that the observers should all be experienced senior clinicians to maintain the high accuracy and low variance in the manual annotation.
Many methods use single-vendor single-center datasets to validate their methods, which may not demonstrate the ability to generalize the accurate segmentation methodology to centers with CMR machines of different settings and compositions.
There have been some trials assessing the performance of models based on multi-vendor and multi-center data (Engblom et al., 2016; Fahmy et al., 2020). However, evaluation based on multi-vendor and multi-center data with a more significant patient population should be introduced for a more comprehensive unbiased validation, comparison of performances of different methods and assessment for their scalability.
We would like to alert readers that nearly all studies summarized in this study used their own distinct private datasets when reporting results. Biased conclusion may be derived when directly comparing these quantitative metrics across studies. The authors would like to ask all readers to refer to the experimental settings in acquisitions of the datasets stated in their original articles when comparing quantitative results across different studies, instead of only looking at these numbers stated. We would also look forward to a public benchmarking of all these methods as a fairer review of their performances.
Also, the authors would like to ask readers to be cautious when directly comparing Dice Scores reported for the segmentation of the LV infarction than the LA necrosis’. As the LA is much smaller than the LV, an equivalent volume of discrepancy may trigger a more significant reduction in the LA necrosis’ Dice Score ratios than the LV infarction’s. Additionally, the LA necrosis tends to be more challenging to be accurately segmented than the LV infarction explained above.
In addition, the image quality, contrast, class imbalance and other factors of the image data can directly impact the result generated from it and thus the accuracy reported. In particular, the authors would advocate future literature to report (1) scar to blood pool contrast ratios (SC-BP) (Karim et al., 2014) to show the scar contrast, (2) signal-to-noise ratio (SNR) to show the noise variation along with evaluation metrics in results, so the readers can have a better understanding of the experimental settings before interpreting all the metrics reported quantitatively. These two additional metrics are essential, particularly when it comes to LA scar segmentation, where the scar segmentation is more difficult and where higher SC-BP can give higher Dice Scores in the results generated (Karim et al., 2014).
Obviously, as conventional methods are less demanding on the composition of the computing device, they can be deployed for wider clinical uses more easily. This is an advantage when it comes to the scalability and generalizability of the product, where a standard computer is enough for its deployment.
Conventional methods are also more explainable than deep learning. The explainability also guarantees easier acceptance from the clinicians, as the product may appear more trustworthy and more reliable.
Fixed threshold methods may not fit some LGE CMR images, as they are unlikely to handle variations well (Oakes et al., 2009).
Scars are highly variable in their morphology and their brightness distribution on LGE CMR. Some severe LV cardiac scar may appear bright in its surroundings and very dark in its center, as the center of the scar is so severely infarcted that very little GBCA carrying perfusion arrives there. N-SD and FWHM, which require the pixel intensity to be more than a certain threshold for that pixel to be recognized as a scar, may not label these dark centers as the scar. Additionally, due to the partial volume effect, fibrotic regions containing both intermingling bundles of fibrotic and viable myocytes will be darker than the complete necrosis region. The low intensity exhibited from such fibrotic regions may be below the fixed threshold set and make these fibrotic regions be falsely recognized as healthy myocardium.
Varied external factors including resolution, contrast, signal-to-noise ratio (SNR), inversion time and surface coil intensity variation can also adversely affect the accuracy of the scar segmentation. LGE CMR modality often suffers from poor image quality, which may be due to residual respiratory motion, variability in the heart rate and gadolinium wash-out during the currently long acquisition time (Yang et al., 2017). Considering the thin transmural thickness of the atrial wall [mean = 2.2–2.5 mm (Saìnchez-Quintana et al., 2005)] (Figure 2), the spatial resolution of LGE CMR images is relatively limited, particularly for the left atrium (To et al., 2011). The variable anatomical morphological shapes of pulmonary veins (PV) also impose an additional challenge to the LGE CMR segmentations. In addition, some uninterested cardiac substructures may be highlighted in LGE CMR images as well in addition to the scarring and fibrosis regions. These may be due to the navigator beam artifact (which is often seen near the right PV), Gadolinium uptake by the aortic wall and valves and confounded enhancement in the spine, esophagus, etc. (Karim et al., 2013; Yang et al., 2017).
Although adaptive conventional methods may mitigate adverse impacts from variable scar shapes and varied external factors, adaptive conventional methods can also be affected by sizes, variances and artifacts in testing image data as they utilize prior information learned. Kurzendorfer et al. (2017c) showed that a particular scar distribution over the myocardium could adversely affect their methods in segmenting endocardial contours. Such vulnerability may be more problematic when it comes to LA anatomical structures, as PV is a very morphological variable and LA walls are much smaller and thinner.
For detailed designs of the deep learning networks, LASC’18 benchmarked (Xiong et al., 2021) a range of U-Net variants in LA wall segmentation from LGE CMR. This challenge, along with other literature for cardiac scar segmentation, demonstrated the following.
(1) Image Sources
(a) Higher image qualities (as in signal-to-noise ratio) would result in a higher Dice Score, although not statistically significantly linearly related.
(b) In addition, models with contrast normalization as a pre-processing technique performed significantly better than the ones without using normalization.
(2) Model Backbone
(a) CNN based methods delivered better results compared to the atlas based methods.
(b) U-Net based methods outperformed other networks using VGGNet, ResNet, etc.
(c) There was no statistical difference between the segmentation performances of the models based on 2D CNNs and models based on 3D CNNs. However, further research showed that 3D CNNs greatly outperformed 2D CNNs with the same model architecture in terms of the Dice Scores of their segmentation results (Borra et al., 2020a).
(3) Segment on ROI or the Whole Image?
(a) Centring LA on ROI as an input to the second sequential model would make the model perform significantly better compared to the model with non-centered ROIs.
(b) Class imbalance induced by significantly big or small ROI size could lead to an adverse effect on the segmentation results in terms of Dice scores.
(c) Double sequential CNNs (Li et al., 2019; Xia et al., 2019; Yang et al., 2019; Xiong et al., 2021) (one detecting the region of interest first and then the second model performing regional segmentation within the region of interests (ROI) detected) achieved much better results compared to the methods with only one single CNN.
(d) Double sequential 3D CNN outperformed single 2D CNN and single 3D CNN models regarding its Dice scores, surface distance, LA diameter error and LA volume error.
(4) Model Architecture
(a) Models with residual connections performed significantly better compared to the ones without residual connections.
(b) The use of dropout blocks did not perform significantly better than the one without using dropout.
(c) Rectified Linear Unit (ReLU) trained models did not perform significantly better than the Parametric Rectified Linear Unit (PReLU) trained models.
(5) Loss Functions
(a) Dice loss trained models performed significantly better than the cross-entropy trained models.
Although we are able to observe much better results generated from deep learning based methods, we can also observe a rise in computational demand from deep learning networks. For deep learning based methods, high-end computer graphics processing units (GPUs) become a necessity when deploying these models, whereas standard computers with CPUs only are sufficient for most of the conventional methods to run. Under a clinical setting, hiring a GPU is not always possible, as it is not part of a standard clinical computing workstation. The requirement of a high-end computer with GPU in deploying a deep learning based method may significantly limit the ability of these methods to scale.
However, if a standard computer was only used to infer a deep learning model, its runtime may be a bit long but still falls within the maximum time limit that clinicians can accept (usually a few minutes per slice for models that are not extra complex). Therefore, we can see these models can be deployed and scaled only if they are sophisticatedly trained, as training on the clinician’s side, where unlikely they have a GPU, is not usually possible. As the inference time may vary significantly across different models over CPUs and depend on their architectures and complexities, reporting of inference time per slice on a standard computer without a GPU should also be mandatory in addition to the inference time over a GPU.
Training datasets with abundant paired labels are essential to the success of deep learning model training. However, there has been a scarcity of labels due to the tedious process of manually annotating the ground truths in medical imaging. In order to mitigate such scarcity in ground truth labels, several methods can be adopted, including the following.
(1) Data augmentation,
(2) Transfer learning with fine-tuning (Bai et al., 2018; Chen S. et al., 2019; Khened et al., 2019),
(3) Weak and semi-supervised learning (Bai et al., 2017, 2018; Can et al., 2018; Chartsias et al., 2018, 2019; Kervadec et al., 2019),
(4) Self-supervised learning (Bai et al., 2019) and,
(5) Unsupervised learning (Joyce et al., 2018).
In addition, to mitigate the challenging training process brought by the great data size required to train a scalable network, active learning (Mahapatra et al., 2018) has been introduced to reduce manual annotation workloads as well as the computational loads.
Although there has been a wide range of evidence demonstrating the efficacy of deep learning in medical image analysis, the deep learning networks behave more like a ‘black box,’ where its interpretability is poor. It has been shown that these deep learning networks can be attacked by adversarial noises or even just rotation in medical images (Finlayson et al., 2019), questioning the reliability and scalability of these deep learning models in assisting diagnosis. For alerting users of these possible failures, segmentation quality scores (Robinson et al., 2019) and confidence maps [e.g., uncertainty maps (Sander et al., 2019) and attention maps (Heo et al., 2018)] should be provided to highlight uncertainties in the model prediction.
Although many methods can accurately segment scars on non-CA cine MRI, the impact from different numbers of cardiac phases on cine MRI has not been assessed.
In addition, the binary class of either normal or scar may be too simplistic. Quantification of the so-called “gray-zone,” which has been proposed for the clinical implication of ventricular arrhythmia (Jablonowski et al., 2017), immediately surrounding the ventricular scar may be useful clinically.
Also, gadolinium based contrast agent is not only applied for scar imaging but also for assessing myocardial perfusion, which is usually assessed together in LGE CMR, for which additional classification and differentiation of ischemic and remote regions of myocardium would be useful (Leiner, 2019). To achieve that, Liu et al. (2016) demonstrated non-Gadolinium contrast adenosine stress and rest T1 Mapping for identification and classification of normal, infarcted, ischemic and remote regions in LV myocardium.
We are glad to see a range of algorithms demonstrated for LV scar segmentation in non-contrast enhanced CMR. However, this has not been realized for CMR images of LA, which is more difficult as the LA scarring regions in CMR suffers from greater variances in morphology and relatively lower resolution of CMR. Moreover, LA scars can appear in discrete regions (Figure 2), which imposes further challenges to the LA scar segmentation from non-CA modalities.
This study summarizes the recent developments in cardiac scar segmentation, covering a wide range of conventional and deep learning techniques. In particular, we presented and discussed the usefulness of non-LGE modalities in cardiac anatomy and scar segmentation. We then further discussed the recent progress in segmenting the cardiac scarring region from non-contrast-enhanced images. We hope this review can provide a comprehensive understanding of the segmentation methodologies for cardiac scar and fibrosis and increase the awareness of common challenges in these fields that can call for future research and contributions.
YW and GY contributed to study design and writing – original draft preparation. ZT, BL, YW, and GY contributed to data collection. YW, ZT, BL, and GY contributed to data visualization. YW, ZT, BL, DF, and GY contributed to writing – review and editing. DF and GY contributed to supervision and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This work was supported in part by the British Heart Foundation (Project Number: TG/18/5/34111, PG/16/78/32402), Heart Research UK (RG2584), the Hangzhou Economic and Technological Development Area Strategical Grant (Imperial Institute of Advanced Technology), the European Research Council Innovative Medicines Initiative on Development of Therapeutics and Diagnostics Combatting Coronavirus Infections Award “DRAGON: rapiD and secuRe AI imaging based diaGnosis, stratification, fOllow-up, and preparedness for coronavirus paNdemics” (H2020-JTI-IMI2 101005122), the AI for Health Imaging Award “CHAIMELEON: Accelerating the Lab to Market Transition of AI Tools for Cancer Management” (H2020-SC1-FA-DTS-2019-1 952172), and the UK Research and Innovation (MR/V023799/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Aty, H., Zagrosek, A., Schulz-Menger, J., Taylor, A. J., Messroghli, D., Kumar, A., et al. (2004). Delayed enhancement and T2-weighted cardiovascular magnetic resonance imaging differentiate acute from chronic myocardial infarction. Circulation 109, 2411–2416. doi: 10.1161/01.CIR.0000127428.10985.C6
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., and Süsstrunk, S. (2012). SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Patt. Anal. Mach. Intell. 34, 2274–2282. doi: 10.1109/TPAMI.2012.120
Akkaya, M., Higuchi, K., Koopmann, M., Burgon, N., Erdogan, E., Damal, K., et al. (2013). Relationship between left atrial tissue structural remodelling detected using late gadolinium enhancement MRI and left ventricular hypertrophy in patients with atrial fibrillation. Europace 15, 1725–1732. doi: 10.1093/europace/eut147
Akoum, N., Daccarett, M., Mcgann, C., Segerson, N., Vergara, G., Kuppahally, S., et al. (2011). Atrial fibrosis helps select the appropriate patient and strategy in catheter ablation of atrial fibrillation: a DE-MRI guided approach. J. Cardiovasc. Electrophysiol. 22, 16–22. doi: 10.1111/j.1540-8167.2010.01876.x
Albà, X., Figueras i Ventura, R. M., Lekadir, K., Tobon-Gomez, C., Hoogendoorn, C., and Frangi, A. F. (2014). Automatic cardiac LV segmentation in MRI using modified graph cuts with smoothness and interslice constraints. Magn. Reson. Med. 72, 1775–1784. doi: 10.1002/mrm.25079
Allessie, M. (2002). Electrical, contractile and structural remodeling during atrial fibrillation. Cardiovasc. Res. 54, 230–246. doi: 10.1016/S0008-6363(02)00258-4
Badger, T. J., Daccarett, M., Akoum, N. W., Adjei-Poku, Y. A., Burgon, N. S., Haslam, T. S., et al. (2010). Evaluation of left atrial lesions after initial and repeat atrial fibrillation ablation. Circ. Arrhythmia Electrophysiol. 3, 249–259. doi: 10.1161/CIRCEP.109.868356
Badrinarayanan, V., Kendall, A., and Cipolla, R. (2017). SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495. doi: 10.1109/TPAMI.2016.2644615
Bai, W., Chen, C., Tarroni, G., Duan, J., Guitton, F., Petersen, S. E., et al. (2019). “Self-supervised learning for cardiac MR image segmentation by anatomical position prediction,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019: 22nd International Conference, Proceedings, Part II. Lecture Notes in Computer Science, eds D. Shen, T. Liu, T. M. Peters, L. H. Staib, C. Essert, S. Zhou, et al. (Berlin: Springer), 541–549. doi: 10.1007/978-3-030-32245-8_60
Bai, W., Oktay, O., Sinclair, M., Suzuki, H., Rajchl, M., Tarroni, G., et al. (2017). “Semi-supervised learning for network-based cardiac MR image segmentation,” in Medical Image Computing and Computer Assisted Intervention - MICCAI 2017 - 20th International Conference, Proceedings, eds P. Jannin, S. Duchesne, M. Descoteaux, A. Franz, D. L. Collins, and L. Maier-Hein (Berlin: Springer), 253–260. doi: 10.1007/978-3-319-66185-8_29
Bai, W., Sinclair, M., Tarroni, G., Oktay, O., Rajchl, M., Vaillant, G., et al. (2018). Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 20:65. doi: 10.1186/s12968-018-0471-x
Bisbal, F., Guiu, E., Cabanas-Grandío, P., Berruezo, A., Prat-Gonzalez, S., Vidal, B., et al. (2014). CMR-guided approach to localize and ablate gaps in repeat AF ablation procedure. JACC Cardiovasc. Imaging 7, 653–663. doi: 10.1016/j.jcmg.2014.01.014
Bleton, H., Margeta, J., Lombaert, H., Delingette, H., and Ayache, N. (2016). “Myocardial infarct localization using neighbourhood approximation forests,” in Proceedings of the 6th International Workshop on Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges, Vol. 9534, eds O. Camara, T. Mansi, M. Pop, K. Rhode, M. Sermesant, and A. Young (Cham: Springer), 108–116. doi: 10.1007/978-3-319-28712-6_12
Boldt, A. (2004). Fibrosis in left atrial tissue of patients with atrial fibrillation with and without underlying mitral valve disease. Heart 90, 400–405. doi: 10.1136/hrt.2003.015347
Borra, D., Andalò, A., Paci, M., Fabbri, C., and Corsi, C. (2020a). A fully automated left atrium segmentation approach from late gadolinium enhanced magnetic resonance imaging based on a convolutional neural network. Quant. Imaging Med. Surg. 10, 1894–1907. doi: 10.21037/qims-20-168
Borra, D., Portas, D., Andalò, A., Fabbri, C., and Corsi, C. (2020b). “Performance comparison of deep learning approaches for left atrium segmentation from LGE-MRI Data,” in Proceedings of the 2020 Computing in Cardiology, Rimini. doi: 10.22489/CinC.2020.306
Boyle, P. M., Zghaib, T., Zahid, S., Ali, R. L., Deng, D., Franceschi, W. H., et al. (2019). Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat. Biomed. Eng. 3, 870–879. doi: 10.1038/s41551-019-0437-9
Cai, Z., and Vasconcelos, N. (2018). “Cascade R-CNN: delving into high quality object detection,” in Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, 6154–6162. doi: 10.1109/CVPR.2018.00644
Can, Y. B., Chaitanya, K., Mustafa, B., Koch, L. M., Konukoglu, E., and Baumgartner, C. F. (2018). “Learning to segment medical images with scribble-supervision alone,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2018, ML-CDS 2018. Lecture Notes in Computer Science, Vol. 11045, eds D. Stoyanov et al. (Cham: Springer), 236–244. doi: 10.1007/978-3-030-00889-5_27
Canny, J. (1986). A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698. doi: 10.1109/TPAMI.1986.4767851
Chartsias, A., Joyce, T., Papanastasiou, G., Semple, S., Williams, M., Newby, D., et al. (2018). “Factorised spatial representation learning: application in semi-supervised myocardial segmentation,” in Medical Image Computing and Computer Assisted Intervention, eds A. L. Martel, P. Abolmaesumi, D. Stoyanov, D. Mateus, M. A. Zuluaga and S. K. Zhou (Cham: Springer), 490–498. doi: 10.1007/978-3-030-00934-2_55
Chartsias, A., Joyce, T., Papanastasiou, G., Semple, S., Williams, M., Newby, D. E., et al. (2019). Disentangled representation learning in cardiac image analysis. Med. Image Anal. 58:101535. doi: 10.1016/j.media.2019.101535
Chen, C., Qin, C., Qiu, H., Tarroni, G., Duan, J., Bai, W., et al. (2020). Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med 7:25. doi: 10.3389/fcvm.2020.00025
Chen, J., Li, H., Zhang, J., and Menze, B. (2020). “Adversarial convolutional networks with weak domain-transfer for multi-sequence cardiac MR images segmentation,” in Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges: 10th International Workshop, STACOM 2019, eds M. Pop, M. Sermesant, O. Camara, X. Zhuang, S. Li, A. Young, et al. (Cham: Springer), 317–325. doi: 10.1007/978-3-030-39074-7_34
Chen, J., Yang, G., Gao, Z., Ni, H., Angelini, E., Mohiaddin, R., et al. (2018). “Multiview two-task recursive attention model for left atrium and atrial scars segmentation,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, Vol. 11071, eds A. Frangi, J. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer), 455–463. doi: 10.1007/978-3-030-00934-2_51
Chen, J., Yang, G., Khan, H., Zhang, H., Zhang, Y., Zhao, S., et al. (2021). JAS-GAN generative adversarial network based joint atrium and scar segmentation on unbalanced atrial targets. IEEE J. Biomed. Heal. Informatics. doi: 10.1109/JBHI.2021.3077469 (accessed July 11, 2021)
Chen, J., Zhang, H., Zhang, Y., Zhao, S., Mohiaddin, R., Wong, T., et al. (2019). “Discriminative consistent domain generation for semi-supervised learning,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 Lecture Notes in Computer Science, eds D. Shen, T. Liu, T. M. Peters, L. H. Staib, C. Essert, S. Zhou, et al. (Cham: Springer), 595–604. doi: 10.1007/978-3-030-32245-8_66
Chen, K., Ouyang, W., Loy, C. C., Lin, D., Pang, J., Wang, J., et al. (2019). “Hybrid task cascade for instance segmentation,” in Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, 4969–4978. doi: 10.1109/CVPR.2019.00511
Chen, S., Ma, K., and Zheng, Y. (2019). Med3D: transfer learning for 3D medical image analysis. arXiv [Preprint]. Available online at: http://arxiv.org/abs/1904.00625 (accessed May 13, 2021).
Cho, K., van Merrienboer, B., Gulcehre, C., Bahdanau, D., Bougares, F., Schwenk, H., et al. (2014). “Learning phrase representations using RNN encoder–decoder for statistical machine translation,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), (Stroudsburg, PA: Association for Computational Linguistics), 1724–1734. doi: 10.3115/v1/D14-1179
Choi, K. M., Kim, R. J., Gubernikoff, G., Vargas, J. D., Parker, M., and Judd, R. M. (2001). Transmural extent of acute myocardial infarction predicts long-term improvement in contractile function. Circulation 104, 1101–1107. doi: 10.1161/hc3501.096798
Ciofolo, C., Fradkin, M., Mory, B., Hautvast, G., and Breeuwer, M. (2008). “Automatic myocardium segmentation in late-enhancement MRI,” in Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Piscataway, NJ, 225–228. doi: 10.1109/ISBI.2008.4540973
Dai, J., He, K., and Sun, J. (2016). “Instance-aware semantic segmentation via multi-task network cascades,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, 3150–3158. doi: 10.1109/CVPR.2016.343
Dastidar, A. G., Harries, I., Pontecorboli, G., Bruno, V. D., De Garate, E., Moret, C., et al. (2019). Native T1 mapping to detect extent of acute and chronic myocardial infarction: comparison with late gadolinium enhancement technique. Int. J. Cardiovasc. Imaging 35, 517–527. doi: 10.1007/s10554-018-1467-1
Dikici, E., O’Donnell, T., Setser, R., and White, R. D. (2004). “Quantification of delayed enhancement MR images,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2004. MICCAI 2004. Lecture Notes in Computer Science, Vol. 3216, eds C. Barillot, D. R. Haynor, and P. Hellier (Berlin: Springer), 250–257. doi: 10.1007/978-3-540-30135-6_31
Engblom, H., Tufvesson, J., Jablonowski, R., Carlsson, M., Aletras, A. H., Hoffmann, P., et al. (2016). A new automatic algorithm for quantification of myocardial infarction imaged by late gadolinium enhancement cardiovascular magnetic resonance: experimental validation and comparison to expert delineations in multi-center, multi-vendor patient data. J. Cardiovasc. Magn. Reson. 18:27. doi: 10.1186/s12968-016-0242-5
Fahmy, A. S., Neisius, U., Chan, R. H., Rowin, E. J., Manning, W. J., Maron, M. S., et al. (2020). Three-dimensional Deep Convolutional neural networks for automated myocardial scar quantification in hypertrophic cardiomyopathy: a multicenter multivendor study. Radiology 294, 52–60. doi: 10.1148/radiol.2019190737
Finlayson, S. G., Bowers, J. D., Ito, J., Zittrain, J. L., Beam, A. L., and Kohane, I. S. (2019). Adversarial attacks on medical machine learning. Science 363, 1287–1289. doi: 10.1126/science.aaw4399
Flett, A. S., Hasleton, J., Cook, C., Hausenloy, D., Quarta, G., Ariti, C., et al. (2011). Evaluation of techniques for the quantification of myocardial scar of differing etiology using cardiac magnetic resonance. JACC Cardiovasc. Imaging 4, 150–156. doi: 10.1016/j.jcmg.2010.11.015
Flett, A. S., Westwood, M. A., Davies, L. C., Mathur, A., and Moon, J. C. (2009). The prognostic implications of cardiovascular magnetic resonance. Circ. Cardiovasc. Imaging 2, 243–250. doi: 10.1161/CIRCIMAGING.108.840975
Friedrich, M. G. (2017). Why edema is a matter of the heart. Circ. Cardiovasc. Imaging 10:e006062. doi: 10.1161/CIRCIMAGING.117.006062
Gannon, M. P., Schaub, E., Grines, C. L., and Saba, S. G. (2019). State of the art: evaluation and prognostication of myocarditis using cardiac MRI. J. Magn. Reson. Imaging 49, e122–e131. doi: 10.1002/jmri.26611
Gerig, G., Jomier, M., and Chakos, M. (2001). “Valmet: a new validation tool for assessing and improving 3D object segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2001. MICCAI 2001. Lecture Notes in Computer Science, Vol. 2208, eds W. J. Niessen and M. A. Viergever (Berlin: Springer), 516–523. doi: 10.1007/3-540-45468-3_62
Girshick, R. (2015). “Fast R-CNN,” in Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, 1440–1448. doi: 10.1109/ICCV.2015.169
Goodfellow, I. J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). Generative adversarial nets. Adv. Neural Inform. Process. Syst. 11, 2672–2680. doi: 10.3156/jsoft.29.5_177_2
Haaf, P., Garg, P., Messroghli, D. R., Broadbent, D. A., Greenwood, J. P., and Plein, S. (2017). Cardiac T1 Mapping and Extracellular Volume (ECV) in clinical practice: a comprehensive review. J. Cardiovasc. Magn. Reson. 18:89. doi: 10.1186/s12968-016-0308-4
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, 770–778. doi: 10.1109/CVPR.2016.90
Hendel, R. C., Patel, M. R., Kramer, C. M., Poon, M., Hendel, R. C., Carr, J. C., et al. (2006). ACCF/ACR/SCCT/SCMR/ASNC/NASCI/SCAI/SIR 2006 appropriateness criteria for cardiac computed tomography and cardiac magnetic resonance imaging: a report of the American college of cardiology foundation quality strategic directions committee appropriateness criteria working group, American college of radiology, society of cardiovascular computed tomography, society for cardiovascular magnetic resonance, American society of nuclear cardiology, north American society for cardiac imaging, society for cardiovascular angiography and interventions, and society of interventional radiology. J. Am. Coll. Cardiol. 48, 1475–1497. doi: 10.1016/j.jacc.2006.07.003
Hennemuth, A., Seeger, A., Friman, O., Miller, S., Klumpp, B., Oeltze, S., et al. (2008). A comprehensive approach to the analysis of contrast enhanced cardiac MR images. IEEE Trans. Med. Imaging 27, 1592–1610. doi: 10.1109/TMI.2008.2006512
Heo, J., Lee, H. B., Kim, S., Lee, J., Kim, K. J., Yang, E., et al. (2018). “Uncertainty-aware attention for reliable interpretation and prediction,” in Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS 2018), Montréal.
Hochreiter, S., and Schmidhuber, J. (1997). Long short-term memory. Neural Comput. 9, 1735–1780. doi: 10.1162/neco.1997.9.8.1735
Huang, X., Liu, M.-Y., Belongie, S., and Kautz, J. (2018). “Multimodal unsupervised image-to-image translation,” in ECCV 2018. LNCS, Vol. vol. 11207, eds V. Ferrari, M. Hebert, C. Sminchisescu, and Y. Weiss (Cham: Springer), 179–196. doi: 10.1007/978-3-030-01219-9_11
Huttenlocher, D. P., Klanderman, G. A., and Rucklidge, W. J. (1993). Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 15, 850–863. doi: 10.1109/34.232073
Jablonowski, R., Chaudhry, U., van der Pals, J., Engblom, H., Arheden, H., Heiberg, E., et al. (2017). Cardiovascular magnetic resonance to predict appropriate implantable cardioverter defibrillator therapy in ischemic and nonischemic cardiomyopathy patients using late gadolinium enhancement border zone. Circ. Cardiovasc. Imaging 10:e006105. doi: 10.1161/CIRCIMAGING.116.006105
Jamart, K., Xiong, Z., Maso Talou, G. D., Stiles, M. K., and Zhao, J. (2020). Mini review: deep learning for atrial segmentation from late gadolinium-enhanced MRIs. Front. Cardiovasc. Med. 7:86. doi: 10.3389/fcvm.2020.00086
Joyce, T., Chartsias, A., and Tsaftaris, S. A. (2018). “Deep multi-class segmentation without ground-truth labels,” in Proceedings of the 9th International Conference on Medical Imaging With Deep Learning, Amsterdam.
Kali, A., Cokic, I., Tang, R. L. Q., Yang, H.-J., Sharif, B., Marbán, E., et al. (2014). Determination of location, size, and transmurality of chronic myocardial infarction without exogenous contrast media by using cardiac magnetic resonance imaging at 3 T. Circ. Cardiovasc. Imaging 7, 471–481. doi: 10.1161/CIRCIMAGING.113.001541
Karim, R., Arujuna, A., Housden, R. J., Gill, J., Cliffe, H., Matharu, K., et al. (2014). A method to standardize quantification of left atrial scar from delayed-enhancement MR images. IEEE J. Transl. Eng. Heal. Med. 2, 1–15. doi: 10.1109/JTEHM.2014.2312191
Karim, R., Bhagirath, P., Claus, P., James Housden, R., Chen, Z., Karimaghaloo, Z., et al. (2016). Evaluation of state-of-the-art segmentation algorithms for left ventricle infarct from late Gadolinium enhancement MR images. Med. Image Anal. 30, 95–107. doi: 10.1016/j.media.2016.01.004
Karim, R., Housden, R. J., Balasubramaniam, M., Chen, Z., Perry, D., Uddin, A., et al. (2013). Evaluation of current algorithms for segmentation of scar tissue from late Gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. J. Cardiovasc. Magn. Reson. 15:105. doi: 10.1186/1532-429X-15-105
Keegan, J., Gatehouse, P. D., Haldar, S., Wage, R., Babu-Narayan, S. V., and Firmin, D. N. (2015). Dynamic inversion time for improved 3D late gadolinium enhancement imaging in patients with atrial fibrillation. Magn. Reson. Med. 73, 646–654. doi: 10.1002/mrm.25190
Kervadec, H., Dolz, J., Tang, M., Granger, E., Boykov, Y., and Ben Ayed, I. (2019). Constrained-CNN losses for weakly supervised segmentation. Med. Image Anal. 54, 88–99. doi: 10.1016/j.media.2019.02.009
Khened, M., Kollerathu, V. A., and Krishnamurthi, G. (2019). Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 51, 21–45. doi: 10.1016/j.media.2018.10.004
Kim, H. W., Farzaneh-Far, A., and Kim, R. J. (2009). Cardiovascular magnetic resonance in patients with myocardial infarction. J. Am. Coll. Cardiol. 55, 1–16. doi: 10.1016/j.jacc.2009.06.059
Knowles, B. R., Caulfield, D., Cooklin, M., Rinaldi, C. A., Gill, J., Bostock, J., et al. (2010). 3-D visualization of acute rf ablation lesions using MRI for the simultaneous determination of the patterns of necrosis and edema. IEEE Trans. Biomed. Eng. 57, 1467–1475. doi: 10.1109/TBME.2009.2038791
Kramer, C. M., Barkhausen, J., Flamm, S. D., Kim, R. J., and Nagel, E. (2013). Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update. J. Cardiovasc. Magn. Reson. 15:91. doi: 10.1186/1532-429X-15-91
Krittayaphong, R., Laksanabunsong, P., Maneesai, A., Saiviroonporn, P., Udompunturak, S., and Chaithiraphan, V. (2008). Comparison of cardiovascular magnetic resonance of late gadolinium enhancement and diastolic wall thickness to predict recovery of left ventricular function after coronary artery bypass surgery. J. Cardiovasc. Magn. Reson. 10:41. doi: 10.1186/1532-429X-10-41
Kurzendorfer, T., Breininger, K., Steidl, S., Brost, A., Forman, C., and Maier, A. (2018). “Myocardial scar segmentation in LGE-MRI using fractal analysis and random forest classification,” in Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, 3168–3173. doi: 10.1109/ICPR.2018.8545636
Kurzendorfer, T., Brost, A., Forman, C., and Maier, A. (2017a). “Automated left ventricle segmentation in 2-D LGE-MRI,” in Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, 831–834. doi: 10.1109/ISBI.2017.7950646
Kurzendorfer, T., Forman, C., Brost, A., and Maier, A. (2017b). “Random forest based left ventricle segmentation in LGE-MRI,” in Functional Imaging and Modelling of the Heart. FIMH 2017. Lecture Notes in Computer Science, Vol. 10263, eds M. Pop and G. Wright (Cham: Springer), doi: 10.1007/978-3-319-59448-4_15
Kurzendorfer, T., Forman, C., Schmidt, M., Tillmanns, C., Maier, A., and Brost, A. (2017c). Fully automatic segmentation of left ventricular anatomy in 3-D LGE-MRI. Comput. Med. Imaging Graph. 59, 13–27. doi: 10.1016/j.compmedimag.2017.05.001
Larose, E., Rodés-Cabau, J., Pibarot, P., Rinfret, S., Proulx, G., Nguyen, C. M., et al. (2010). Predicting late myocardial recovery and outcomes in the early hours of ST-segment elevation myocardial infarction. J. Am. Coll. Cardiol. 55, 2459–2469. doi: 10.1016/j.jacc.2010.02.033
Larroza, A., López-Lereu, M. P., Monmeneu, J. V., Gavara, J., Chorro, F. J., Bodí, V., et al. (2018). Texture analysis of cardiac cine magnetic resonance imaging to detect nonviable segments in patients with chronic myocardial infarction. Med. Phys. 45, 1471–1480. doi: 10.1002/mp.12783
Ledneva, E., Karie, S., Launay-Vacher, V., Janus, N., and Deray, G. (2009). Renal safety of gadolinium-based contrast media in patients with chronic renal insufficiency. Radiology 250, 618–628. doi: 10.1148/radiol.2503080253
Leiner, T. (2019). Deep learning for detection of myocardial scar tissue: Goodbye to gadolinium? Radiology 291, 618–619. doi: 10.1148/radiol.2019190783
Leong, C. O., Lim, E., Tan, L. K., Abdul Aziz, Y. F., Sridhar, G. S., Socrates, D., et al. (2019). Segmentation of left ventricle in late gadolinium enhanced MRI through 2D-4D registration for infarct localization in 3D patient-specific left ventricular model. Magn. Reson. Med. 81, 1385–1398. doi: 10.1002/mrm.27486
Li, C., Tong, Q., Liao, X., Si, W., Sun, Y., Wang, Q., et al. (eds) (2019). Attention Based Hierarchical Aggregation Network for 3D Left Atrial Segmentation, Vol. 11395, eds M. Pop et al. (Cham: Springer), 255–264. doi: 10.1007/978-3-030-12029-0_28
Li, L., Wu, F., Yang, G., Xu, L., Wong, T., Mohiaddin, R., et al. (2020). Atrial scar quantification via multi-scale CNN in the graph-cuts framework. Med. Image Anal. 60, 101595. doi: 10.1016/j.media.2019.101595
Li, X., Liu, Z., Luo, P., Loy, C. C., and Tang, X. (2017). “Not All pixels are equal: difficulty-aware semantic segmentation via deep layer cascade,” in Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, 6459–6468. doi: 10.1109/CVPR.2017.684
Lin, D., Chen, G., Cohen-Or, D., Heng, P.-A., and Huang, H. (2017). “Cascaded feature network for semantic segmentation of RGB-D images,” in Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, 1320–1328. doi: 10.1109/ICCV.2017.147
Liu, A., Wijesurendra, R. S., Francis, J. M., Robson, M. D., Neubauer, S., Piechnik, S. K., et al. (2016). Adenosine stress and rest T1 mapping can differentiate between ischemic, infarcted, remote, and normal myocardium without the need for gadolinium contrast agents. JACC Cardiovasc. Imaging 9, 27–36. doi: 10.1016/j.jcmg.2015.08.018
Liu, X., Hou, J., Yang, Z., Xia, C., Xie, L., Ye, P., et al. (2018). Native T 1 mapping for characterization of acute and chronic myocardial infarction in swine: comparison with contrast-enhanced MRI. J. Magn. Reson. Imaging 47, 1406–1414. doi: 10.1002/jmri.25871
Long, J., Shelhamer, E., and Darrell, T. (2015). “Fully convolutional networks for semantic segmentation,” in Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, 3431–3440. doi: 10.1109/CVPR.2015.7298965
Lorensen, W. E., and Cline, H. E. (1987). “Marching cubes: a high resolution 3D surface construction algorithm,” in Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques - SIGGRAPH ’87, (New York, NY: ACM Press), 163–169. doi: 10.1145/37401.37422
Luc, P., Couprie, C., Chintala, S., and Verbeek, J. (2016). “Semantic segmentation using adversarial networks,” in Proceedings of the NIPS Workshop on Adversarial Training, Barcelona.
Maceira, A. M., Joshi, J., Prasad, S. K., Moon, J. C., Perugini, E., Harding, I., et al. (2005). Cardiovascular magnetic resonance in cardiac amyloidosis. Circulation 111, 186–193. doi: 10.1161/01.CIR.0000152819.97857.9D
Mahapatra, D., Bozorgtabar, B., Thiran, J.-P., and Reyes, M. (2018). “Efficient active learning for image classification and segmentation using a sample selection and conditional generative adversarial network,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, Vol. 11071, eds A. Frangi, J. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer), 580–588. doi: 10.1007/978-3-030-00934-2_65
McGann, C. J., Kholmovski, E. G., Oakes, R. S., Blauer, J. J. E., Daccarett, M., Segerson, N., et al. (2008). New magnetic resonance imaging-based method for defining the extent of left atrial wall injury after the ablation of atrial fibrillation. J. Am. Coll. Cardiol. 52, 1263–1271. doi: 10.1016/j.jacc.2008.05.062
Men, K., Dai, J., and Li, Y. (2017). Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med. Phys. 44, 6377–6389. doi: 10.1002/mp.12602
Messroghli, D. R., Moon, J. C., Ferreira, V. M., Grosse-Wortmann, L., He, T., Kellman, P., et al. (2017). Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2∗ and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imagi. J. Cardiovasc. Magn. Reson. 19:75. doi: 10.1186/s12968-017-0389-8
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). “V-net: fully convolutional neural networks for volumetric medical image segmentation,” in Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, 565–571. doi: 10.1109/3DV.2016.79
Moccia, S., Banali, R., Martini, C., Muscogiuri, G., Pontone, G., Pepi, M., et al. (2018). “Automated scar segmentation from cardiac magnetic resonance-late gadolinium enhancement images using a deep-learning approach,” in Proceedings of the 2018 Computing in Cardiology Conference (CinC), Maastricht. doi: 10.22489/CinC.2018.278
Moccia, S., Banali, R., Martini, C., Muscogiuri, G., Pontone, G., Pepi, M., et al. (2019). Development and testing of a deep learning-based strategy for scar segmentation on CMR-LGE images. Magn. Reson. Mater. Phys, Biol. Med. 32, 187–195. doi: 10.1007/s10334-018-0718-4
MS-CMR Challenge (2019). MICCAI2019. Available online at: www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mscmrseg/ (accessed May 13, 2021)
Murthy, V. N., Singh, V., Chen, T., Manmatha, R., and Comaniciu, D. (2016). “Deep decision network for multi-class image classification,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, 2240–2248. doi: 10.1109/CVPR.2016.246
Muser, D., Tioni, C., Shah, R., Selvanayagam, J. B., and Nucifora, G. (2017). Prevalence, correlates, and prognostic relevance of myocardial mechanical dispersion as assessed by feature-tracking cardiac magnetic resonance after a first ST-segment elevation myocardial infarction. Am. J. Cardiol. 120, 527–533. doi: 10.1016/j.amjcard.2017.05.019
MyoPS Challenge (2020). MICCAI 2020. Available online at: https://zmiclab.github.io/projects/myops20/index.html (accessed May 13, 2021)
Noh, H., Hong, S., and Han, B. (2015). “Learning deconvolution network for semantic segmentation,” in Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, 1520–1528. doi: 10.1109/ICCV.2015.178
Oakes, R. S., Badger, T. J., Kholmovski, E. G., Akoum, N., Burgon, N. S., Fish, E. N., et al. (2009). Detection and quantification of left atrial structural remodeling with delayed-enhancement magnetic resonance imaging in patients with atrial fibrillation. Circulation 119, 1758–1767. doi: 10.1161/CIRCULATIONAHA.108.811877
Ogawa, R., Kido, T., Nakamura, M., Kido, T., Kurata, A., Miyagawa, M., et al. (2017). Diagnostic capability of feature-tracking cardiovascular magnetic resonance to detect infarcted segments: a comparison with tagged magnetic resonance and wall thickening analysis. Clin. Radiol. 72, 828–834. doi: 10.1016/j.crad.2017.05.010
Oktay, O., Bai, W., Lee, M., Guerrero, R., Kamnitsas, K., Caballero, J., et al. (2016). “Multi-input cardiac image super-resolution using convolutional neural networks,” in Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, Vol. 9902, eds S. Ourselin, L. Joskowicz, M. Sabuncu, G. Unal, and W. Wells (Cham: Springer), 246–254. doi: 10.1007/978-3-319-46726-9_29
Ouyang, W., Wang, K., Zhu, X., and Wang, X. (2017). “Chained cascade network for object detection,” in Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, 1956–1964. doi: 10.1109/ICCV.2017.214
Peters, D. C., Wylie, J. V., Hauser, T. H., Kissinger, K. V., Botnar, R. M., Essebag, V., et al. (2007). Detection of pulmonary vein and left atrial scar after catheter ablation with three-dimensional navigator-gated delayed enhancement MR imaging: initial experience 1. Radiology 243, 690–695. doi: 10.1148/radiol.2433060417
Pop, M., Sermesant, M., Camara, O., Zhuang, X., Li, S., Young, A., et al. (eds) (2020). “Statistical atlases and computational models of the heart,” in Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges, (Cham: Springer). doi: 10.1007/978-3-030-39074-7
Popescu, I. A., Irving, B., Borlotti, A., Dall’Armellina, E., and Grau, V. (2017). “Myocardial scar quantification using SLIC Supervoxels - parcellation based on tissue characteristic strains,” in Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges. STACOM 2016. Lecture Notes in Computer Science, Vol. 10124, eds T. Mansi, K. McLeod, M. Pop, K. Rhode, M. Sermesant, and A. Young (Cham: Springer), 182–190. doi: 10.1007/978-3-319-52718-5_20
Rajchl, M., Stirrat, J., Goubran, M., Yu, J., Scholl, D., Peters, T. M., et al. (2015). Comparison of semi-automated scar quantification techniques using high-resolution, 3-dimensional late-gadolinium-enhancement magnetic resonance imaging. Int. J. Cardiovasc. Imaging 31, 349–357. doi: 10.1007/s10554-014-0553-2
Raman, F. S., Kawel-Boehm, N., Gai, N., Freed, M., Han, J., Liu, C.-Y., et al. (2013). Modified look-locker inversion recovery T1 mapping indices: assessment of accuracy and reproducibility between magnetic resonance scanners. J. Cardiovasc. Magn. Reson. 15:64. doi: 10.1186/1532-429X-15-64
Robinson, R., Valindria, V. V., Bai, W., Oktay, O., Kainz, B., Suzuki, H., et al. (2019). Automated quality control in image segmentation: application to the UK Biobank cardiovascular magnetic resonance imaging study. J. Cardiovasc. Magn. Reson. 21:18. doi: 10.1186/s12968-019-0523-x
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, Vol. 9351, eds N. Navab, J. Hornegger, W. Wells and A. Frangi (Cham: Springer), 234–241. doi: 10.1007/978-3-319-24574-4_28
Roth, H. R., Lu, L., Farag, A., Shin, H.-C., Liu, J., Turkbey, E. B., et al. (2015). “DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, Vol. 9349, eds N. Navab, J. Hornegger, W. Wells, and A. Frangi (Cham: Springer), 556–564. doi: 10.1007/978-3-319-24553-9_68
Saìnchez-Quintana, D., Cabrera, J. A., Climent, V., Farreì, J., de Mendonc̨a, M. C., and Ho, S. Y. (2005). Anatomic relations between the esophagus and left atrium and relevance for ablation of atrial fibrillation. Circulation 112, 1400–1405. doi: 10.1161/CIRCULATIONAHA.105.551291
Sander, J., de Vos, B. D., Wolterink, J. M., and Išgum, I. (2019). “Towards increased trustworthiness of deep learning segmentation methods on cardiac MRI,” in Medical Imaging 2019: Image Processing, eds E. D. Angelini and B. A. Landman ((Bellingham, WA: SPIE), 44. doi: 10.1117/12.2511699
Schmidt, E. J., Mallozzi, R. P., Thiagalingam, A., Holmvang, G., D’Avila, A., Guhde, R., et al. (2009). Electroanatomic mapping and radiofrequency ablation of porcine left atria and atrioventricular nodes using magnetic resonance catheter tracking. Circ. Arrhythmia Electrophysiol. 2, 695–704. doi: 10.1161/CIRCEP.109.882472
Shelhamer, E., Long, J., and Darrell, T. (2017). Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 640–651. doi: 10.1109/TPAMI.2016.2572683
Simonetti, O. P., Kim, R. J., Fieno, D. S., Hillenbrand, H. B., Wu, E., Bundy, J. M., et al. (2001). An improved MR imaging technique for the visualization of myocardial infarction. Radiology 218, 215–223. doi: 10.1148/radiology.218.1.r01ja50215
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. Available online at: https://arxiv.org/abs/1409.1556 (accessed May 13, 2021)
Smedema, J.-P., Snoep, G., van Kroonenburgh, M. P. G., van Geuns, R.-J., Dassen, W. R. M., Gorgels, A. P. M., et al. (2005). Evaluation of the Accuracy of Gadolinium-Enhanced Cardiovascular Magnetic Resonance in the Diagnosis of Cardiac Sarcoidosis. J. Am. Coll. Cardiol. 45, 1683–1690. doi: 10.1016/j.jacc.2005.01.047
Smulders, M. W., Bekkers, S. C. A. M., Kim, H. W., Van Assche, L. M. R., Parker, M. A., and Kim, R. J. (2015). Performance of CMR methods for differentiating acute from chronic MI. JACC Cardiovasc. Imaging 8, 669–679. doi: 10.1016/j.jcmg.2014.12.030
Szegedy, C., Ioffe, S., Vanhoucke, V., and Alemi, A. (2016). Inception-v4, inception-ResNet and the impact of residual connections on learning. arXiv [Preprint]. Available online at: https://arxiv.org/abs/1602.07261 (accessed May 13, 2021)
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. (2015). “Going deeper with convolutions,” in Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, 1–9. doi: 10.1109/CVPR.2015.7298594
Tao, Q., Ipek, E. G., Shahzad, R., Berendsen, F. F., Nazarian, S., and van der Geest, R. J. (2016). Fully automatic segmentation of left atrium and pulmonary veins in late gadolinium-enhanced MRI: towards objective atrial scar assessment. J. Magn. Reson. Imaging 44, 346–354. doi: 10.1002/jmri.25148
Tao, Q., Milles, J., Zeppenfeld, K., Lamb, H. J., Bax, J. J., Reiber, J. H. C., et al. (2010). Automated segmentation of myocardial scar in late enhancement MRI using combined intensity and spatial information. Magn. Reson. Med. 64, 586–594. doi: 10.1002/mrm.22422
To, A. C. Y., Flamm, S. D., Marwick, T. H., and Klein, A. L. (2011). Clinical utility of multimodality LA imaging. JACC Cardiovasc. Imaging 4, 788–798. doi: 10.1016/j.jcmg.2011.02.018
Ukwatta, E., Arevalo, H., Rajchl, M., White, J., Pashakhanloo, F., Prakosa, A., et al. (2015). Image-based reconstruction of three-dimensional myocardial infarct geometry for patient-specific modeling of cardiac electrophysiology. Med. Phys. 42, 4579–4590. doi: 10.1118/1.4926428
Veni, G., Elhabian, S. Y., and Whitaker, R. T. (2017). ShapeCut: bayesian surface estimation using shape-driven graph. Med. Image Anal. 40, 11–29. doi: 10.1016/j.media.2017.04.005
Vergara, G. R., and Marrouche, N. F. (2011). Tailored management of atrial fibrillation using a LGE-MRI based model: from the clinic to the electrophysiology laboratory. J. Cardiovasc. Electrophysiol. 22, 481–487. doi: 10.1111/j.1540-8167.2010.01941.x
Vermes, E., Childs, H., Faris, P., and Friedrich, M. G. (2014). Predictive value of CMR criteria for LV functional improvement in patients with acute myocarditis. Eur. Hear. J. Cardiovasc. Imaging 15, 1140–1144. doi: 10.1093/ehjci/jeu099
Wagner, A., Mahrholdt, H., Holly, T. A., Elliott, M. D., Regenfus, M., Parker, M., et al. (2003a). Contrast-enhanced MRI and routine single photon emission computed tomography (SPECT) perfusion imaging for detection of subendocardial myocardial infarcts: an imaging study. Lancet 361, 374–379. doi: 10.1016/S0140-6736(03)12389-6
Wagner, A., Schulz-Menger, J., Dietz, R., and Friedrich, M. G. (2003b). Long-term follow-up of patients with acute myocarditis by magnetic resonance imaging. MAGMA Magn. Reson. Mater. Phys. Biol. Med. 16, 17–20. doi: 10.1007/s10334-003-0007-7
Wang, C., Ukwatta, E., Rajchl, M., and Chan, A. (2019). “An ensemble of U-Net architecture variants for left atrial segmentation,” in Proceedings of the SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, San Diego, CA, 21. doi: 10.1117/12.2512905
Wei, D., Sun, Y., Chai, P., Low, A., and Ong, S. H. (2011). “Myocardial segmentation of late gadolinium enhanced MR images by propagation of contours from cine MR images,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2011. MICCAI 2011. Lecture Notes in Computer Science, Vol. 6893, eds G. Fichtinger, A. Martel, and T. Peters (Berlin: Springer), 428–435. doi: 10.1007/978-3-642-23626-6_53
Wei, D., Sun, Y., Ong, S.-H., Chai, P., Teo, L. L., and Low, A. F. (2013). Three-dimensional segmentation of the left ventricle in late gadolinium enhanced MR images of chronic infarction combining long- and short-axis information. Med. Image Anal. 17, 685–697. doi: 10.1016/j.media.2013.03.001
Wu, E., Judd, R. M., Vargas, J. D., Klocke, F. J., Bonow, R. O., and Kim, R. J. (2001). Visualisation of presence, location, and transmural extent of healed Q-wave and non-Q-wave myocardial infarction. Lancet 357, 21–28. doi: 10.1016/S0140-6736(00)03567-4
Wu, E., Ortiz, J. T., Tejedor, P., Lee, D. C., Bucciarelli-Ducci, C., Kansal, P., et al. (2008). Infarct size by contrast enhanced cardiac magnetic resonance is a stronger predictor of outcomes than left ventricular ejection fraction or end-systolic volume index: prospective cohort study. Heart 94, 730–736. doi: 10.1136/hrt.2007.122622
Xia, Q., Yao, Y., Hu, Z., and Hao, A. (2019). “Automatic 3D atrial segmentation from GE-MRIs using volumetric fully convolutional networks,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), eds M. Pop, M. Sermesant, J. Zhao, S. Li, K. McLeod, and A. Young (Cham: Springer), 211–220. doi: 10.1007/978-3-030-12029-0_23
Xiao, J., Si, D., Wu, Y., Li, M., Yin, J., and Ding, H. (2020). “Multi-view learning for 3D LGE-MRI left atrial cavity segmentation,” in Proceedings of the 2020 3rd International Conference on Artificial Intelligence and Pattern Recognition, (New York, NY: ACM), 84–87. doi: 10.1145/3430199.3430203
Xiong, Z., Fedorov, V. V., Fu, X., Cheng, E., Macleod, R., and Zhao, J. (2019). Fully automatic left atrium segmentation from late gadolinium enhanced magnetic resonance imaging using a dual fully convolutional neural network. IEEE Trans. Med. Imaging 38, 515–524. doi: 10.1109/TMI.2018.2866845
Xiong, Z., Xia, Q., Hu, Z., Huang, N., Bian, C., Zheng, Y., et al. (2021). A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. Image Anal. 67, 101832. doi: 10.1016/j.media.2020.101832
Xu, C., Xu, L., Brahm, G., Zhang, H., and Li, S. (2018). “MuTGAN: simultaneous segmentation and quantification of myocardial infarction without contrast agents via joint adversarial learning,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, Vol. 11071, eds A. Frangi, J. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer), 525–534. doi: 10.1007/978-3-030-00934-2_59
Xu, C., Xu, L., Gao, Z., Zhao, S. S., Zhang, H., Zhang, Y., et al. (2017). “Direct detection of pixel-level myocardial infarction areas via a deep-learning algorithm,” in Medical Image Computing and Computer Assisted Intervention - MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science, Vol. 10435, eds M. Descoteaux, L. Maier-Hein, A. Franz, P. Jannin, D. Collins, and S. Duchesne (Cham: Springer), 240–249. doi: 10.1007/978-3-319-66179-7_28
Xu, C., Xu, L., Ohorodnyk, P., Roth, M., Chen, B., and Li, S. (2020). Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs. Med. Image Anal. 62:101668. doi: 10.1016/j.media.2020.101668
Yang, G., Chen, J., Gao, Z., Li, S., Ni, H., Angelini, E., et al. (2020). Simultaneous left atrium anatomy and scar segmentations via deep learning in multiview information with attention. Futur. Gener. Comput. Syst. 107, 215–228. doi: 10.1016/j.future.2020.02.005
Yang, G., Chen, J., Gao, Z., Zhang, H., Ni, H., Angelini, E., et al. (2018a). “Multiview sequential learning and dilated residual learning for a fully automatic delineation of the left atrium and pulmonary veins from late gadolinium-enhanced cardiac MRI Images,” in Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, 1123–1127. doi: 10.1109/EMBC.2018.8512550
Yang, G., Zhuang, X., Khan, H., Haldar, S., Nyktari, E., Li, L., et al. (2018b). Fully automatic segmentation and objective assessment of atrial scars for long-standing persistent atrial fibrillation patients using late gadolinium-enhanced MRI. Med. Phys. 45, 1562–1576. doi: 10.1002/mp.12832
Yang, G., Zhuang, X., Khan, H., Haldar, S., Nyktari, E., Ye, X., et al. (2017). “Segmenting atrial fibrosis from late gadolinium-enhanced cardiac MRI by deep-learned features with stacked sparse auto-encoders,” in MIUA2017 Communications in Computer and Information Science, eds M. Valdés Hernández and V. González-Castro (Cham: Springer), 195–206. doi: 10.1007/978-3-319-60964-5_17
Yang, X., Wang, N., Wang, Y., Wang, X., Nezafat, R., Ni, D., et al. (2019). “Combating uncertainty with novel losses for automatic left atrium segmentation,” in Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science, Vol. 11395, eds M. Pop et al. (Cham: Springer), 246–254. doi: 10.1007/978-3-030-12029-0_27
Yue, Q., Luo, X., Ye, Q., Xu, L., and Zhuang, X. (2019). “Cardiac segmentation from LGE MRI using deep neural network incorporating shape and spatial priors,” in MICCAI 2019, Part II. LNCS, Vol. 11765, eds D. Shen et al. (Cham: Springer), 559–567. doi: 10.1007/978-3-030-32245-8_62
Zabihollahy, F., Rajchl, M., White, J. A., and Ukwatta, E. (2020). Fully automated segmentation of left ventricular scar from 3D late gadolinium enhancement magnetic resonance imaging using a cascaded multi-planar U-Net (CMPU-Net). Med. Phys. 47, 1645–1655. doi: 10.1002/mp.14022
Zabihollahy, F., White, J. A., and Ukwatta, E. (2019a). Convolutional neural network-based approach for segmentation of left ventricle myocardial scar from 3D late gadolinium enhancement MR images. Med. Phys. 46, 1740–1751. doi: 10.1002/mp.13436
Zabihollahy, F., White, J. A., and Ukwatta, E. (2019). “Fully automated segmentation of left ventricular myocardium from 3D late gadolinium enhancement magnetic resonance images using a U-net convolutional neural network-based model,” in Proceedings of the SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 109503C (13 March 2019), San Diego, CA. doi: 10.1117/12.2512146
Zhang, D., and Lu, G. (2004). Review of shape representation and description techniques. Pattern Recognit. 37, 1–19. doi: 10.1016/j.patcog.2003.07.008
Zhang, N., Yang, G., Gao, Z., Xu, C., Zhang, Y., Shi, R., et al. (2019). Deep learning for diagnosis of chronic myocardial infarction on nonenhanced cardiac Cine MRI. Radiology 291, 606–617. doi: 10.1148/radiol.2019182304
Zhang, Z., Wang, K., Li, Q., Liu, Y., Yuan, Y., Li, Y., et al. (2020). “Automatic segmentation of the left atrium from LGE-MRI based on U-net and bidirectional convolutional LSTM,” in Proceedings of the 2020 Computing in Cardiology, Rimini, 1–4. doi: 10.22489/CinC.2020.288
Zhao, J., Hansen, B. J., Wang, Y., Csepe, T. A., Sul, L. V., Tang, A., et al. (2017). Three-dimensional integrated functional, structural, and computational mapping to define the structural “fingerprints” of heart-specific atrial fibrillation drivers in human heart ex vivo. J. Am. Heart Assoc. 6:e005922. doi: 10.1161/JAHA.117.005922
Zhong, H., Lacomis, J. M., and Schwartzman, D. (2007). On the accuracy of CartoMerge for guiding posterior left atrial ablation in man. Hear. Rhythm 4, 595–602. doi: 10.1016/j.hrthm.2007.01.033
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017). “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV0), Venice, 2242–2251. doi: 10.1109/ICCV.2017.244
Zhu, L., Gao, Y., Yezzi, A., and Tannenbaum, A. (2013). Automatic segmentation of the left atrium from MR images via variational region growing with a moments-based shape prior. IEEE Trans. Image Process. 22, 5111–5122. doi: 10.1109/TIP.2013.2282049
Zhuang, X. (2019). Multivariate mixture model for myocardial segmentation combining multi-source images. IEEE Trans. Pattern Anal. Mach. Intell. 41, 2933–2946. doi: 10.1109/TPAMI.2018.2869576
Keywords: cardiac magnetic resonance, late gadolinium enhancement, scar segmentation, deep learning, atrial fibrillation, myocardial infarction
Citation: Wu Y, Tang Z, Li B, Firmin D and Yang G (2021) Recent Advances in Fibrosis and Scar Segmentation From Cardiac MRI: A State-of-the-Art Review and Future Perspectives. Front. Physiol. 12:709230. doi: 10.3389/fphys.2021.709230
Received: 13 May 2021; Accepted: 28 June 2021;
Published: 03 August 2021.
Edited by:
Linwei Wang, Rochester Institute of Technology, United StatesCopyright © 2021 Wu, Tang, Li, Firmin and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinzhe Wu, eWluemhlLnd1MThAaW1wZXJpYWwuYWMudWs=; Guang Yang, Zy55YW5nQGltcGVyaWFsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.