- 1Centro de Ensino Superior de Maringá (CESUMAR), Curitiba, Brazil
- 2Programa de Pós-graduação em Biotecnologia Aplicada à Saúde da Criança e do Adolescente, Faculdades Pequeno Príncipe, Curitiba, Brazil
- 3Instituto de Pesquisas Pelé Pequeno Príncipe, Curitiba, Brazil
Nutrigenomics is the study of the gene-nutrient interaction and it indicates that some nutrients, called bioactive compounds, can mold the genetic expression or change the nucleotide chain. Polyphenols are secondary metabolites found in plants that are regularly consumed in functional foods and help prevent or delay the onset of type 2 diabetes mellitus (T2DM) and its complications. This article objected to review studies about the interaction of diet with polyphenols and Mediterranean diet in the expression of human genes related to T2DM. Resveratrol acts as an antioxidant, anti-inflammatory, and increases mitochondrial function. Regular consumption of quercetin resulted in improvement of hypertension and suppression of diabetes-induced vasoconstriction. Genistein also showed positive results in T2DM, such as increased cell mass and improved glucose tolerance and insulin levels. Catechins showed efficiency in inducing genes in triacylglycerol biosynthesis, inhibition of fatty acids and cholesterol, and resulting in their participation in mitigating complications of diabetes. Lastly, curcumin was demonstrated to be a protector of the pancreatic islets against streptozotocin-induced oxidative stress. Growing evidence suggest that bioactive compounds such as polyphenols have an important role in T2DM and the prevention and treatment of its complication, as they cause activation or inhibition of related genes.
Introduction
Diabetes mellitus (DM) is a syndrome of multiple etiologies, characterized mainly by chronic hyperglycemia with dysfunctions related to the metabolism of proteins and lipids. The increase in glucose concentrations in the bloodstream (hyperglycemia) may be associated with the inability to produce, secrete, or fail to absorb insulin, or even with a set of all these abnormalities (Kuzuya et al., 2002).
Among the types of diabetes, type 2 diabetes mellitus (T2DM) represents 90% of cases, and it occurs when the body does not properly use the insulin produced or does not produce the required hormone capable of controlling blood glucose (Holman et al., 2015). Some of the patients do not present, at the beginning of metabolic alterations, symptoms, such as thirst, increased diuresis, leg pains, and visual alterations, however, these can manifest late, becoming aggravating factors, and when the diagnosis is not made early, the complications generated by the disease can be greater. The treatment usually consists of changes in eating habits, physical exercise, and pharmacological therapy (Zheng et al., 2018).
According to Steemburgo et al. (2009), several chronic diseases, such as T2DM, have their pathogenesis associated with genetic and environmental aspects. Among the latter, the diet has the power to contribute to the incidence and the severity of these pathologies. Nevertheless, the diet can have a modulating action on phenotypes linked to genetic changes, and this action is related to gene and nutrient interaction.
A variety of habits and environmental factors, including food, can influence the expression of genes involved in T2DM which could be beneficial or harmful in relation to disease. Great progress has been made in the study of these interactions after the Human Genome Project and with the emergence of genetic nutrition, a field of nutrition that studies the relationship between genome and eating habits (Billings and Florez, 2010; Cole and Florez, 2020).
In this perspective, genetic nutrition highlights what is most recent in the science of nutrition. The concepts of nutrigenomics and nutrigenetics are related but follow a different approach to the understanding of the association between genes and diet. Nutrigenomics studies the nutrients and food structures capable of acting on the expression of genes, in contrast, nutrigenetics studies the variables of the personal genome in relation to how we respond to foods or compounds consumed in a diet (Mickelson et al., 2019). Polyphenols are among these compounds. They are secondary metabolites produced by plants that are part of the human diet. They have the potential to interact with genetic material and may alter the expression of important genes. In addition, they act as antioxidants, anti-inflammatories have been studied in the prevention and treatment of type 2 diabetes (Nunes et al., 2018; Li et al., 2019).
Thus, this study reviewed the relationship between polyphenols and gene expression in T2DM identifying major genes and scientific evidence.
Diabetes Mellitus
Diabetes mellitus is a metabolic disorder characterized by persistent hyperglycemia in the bloodstream as a result of the disabled action and/or failure in production of the hormone insulin, which has as function to promote glucose entry into cells (Kuzuya et al., 2002; Holman et al., 2015). When insulin is absent or its function is impaired, cells are unable to absorb glucose, which remains in the bloodstream causing hyperglycemia (Asmat et al., 2016).
There are three main types of diabetes: type 1 diabetes mellitus, type 2 diabetes, and gestational diabetes. T2DM stands out among them by being present in about 90% of cases (Cole and Florez, 2020). T2DM is a multifactorial polygenic disease, which is believed to be a result of interaction between multiple genes and environmental factors (Rheinheimer et al., 2017).
According to Mahler and Adler (1999), the pathophysiology of T2DM includes peripheral resistance to insulin, increased hepatic glucose production, and functional impairment of pancreatic cells. In the initial stage of the disease, a decrease in insulin sensitivity known as insulin resistance is observed and, to compensate, pancreatic cells increase insulin secretion resulting in a state of hyperinsulinemia. As the disease progresses, these cells lose the ability to secrete large amounts of insulin to maintain balance and the individual develops a deficiency of this hormone (Asmat et al., 2016; American Diabetes Association, 2020). The main characteristic of T2DM is the development and persistence of hyperglycemia, which occurs in conjunction with hyperglucagonemia and increased hepatic glucose production (García-Chapa et al., 2017; Furmli et al., 2018). Multiple metabolic disorders, such as impaired lipid and lipoprotein metabolism, oxidative stress, subclinical inflammation, vascular endothelial dysfunction, and hypertension accompany T2DM (Spranger et al., 2003; Gadi and Samaha, 2007). These disorders have long-term consequences, such as micro and macrovascular complications, neuropathy, retinopathy, nephropathy, and therefore increased mortality rate (Lloyd et al., 2001; Constantino et al., 2013).
Oxidative Stress and Diabetes
Free radicals are highly reactive molecules that contain oxygen (or nitrogen) and are naturally generated in small amounts during metabolic reactions. Oxidative stress is an imbalance that occurs when the production of free radicals exceeds the antioxidant defenses resulting in damage to vital biomolecules to membranes and DNA, proteins, and lipids (Wu and Cederbaum, 2003).
The cellular damage caused by these reactive oxygen species (ROS) is related to the pathological process of diseases such as cancer and T2DM (Dandona et al., 1996). Hyperglycemia in DM induces an increase in oxidative stress, favoring the progression, and the appearance of complications of the disease (Nishikawa et al., 2000; Rajendran et al., 2011). The reduction of oxidative stress can happen due to antioxidants, molecules that play an important role against free radicals, acting in order to eliminate them or transform them into less toxic products for the cell (Sies, 1993).
Insulin resistance and pancreatic beta-cell dysfunction are associated with oxidative stress. Diabetic patients showed lower enzyme and antioxidants levels, low markers of oxidative stress, and increased production of ROS, which can contribute to vascular complications in DM (Dandona et al., 1996; Weyer et al., 1999; Kaneto et al., 2007; Jiménez-Osorio et al., 2014).
Nutrigenomics and Dietary Factors
Nutrigenomics studies how nutrients affect gene expression (Marcum, 2020), bringing the perspective of designing and prescribe customized diets according to the individual genetic makeup and expanding strategies for prevention and treatment of non-communicable diseases (NCDs), such as obesity, T2DM, inflammatory bowel disease (IBD), and cancer (Fialho et al., 2008). It seeks to observe the variations of genetic polymorphisms, responsible for the absorption, metabolism, and excretion of nutrients and bioactive compounds, acting in conjunction with other sub-areas of studies, including metabolomics, transcriptomics, and proteomics (Dimitrov et al., 2016) that together, allow the discovery of the influences of nutrients in the epigenome or genome and how each individual can be affected (Rist et al., 2006).
The diet alone or in conjunction with other environmental factors may cause epigenetic changes (Fenech et al., 2011), and these changes in the genes have great influences on cellular processes associated with health and disease, hormonal balance, cell signaling, carcinogen metabolism, apoptosis, cell cycle control, changes in energy levels, and angiogenesis (Ferguson, 2006). In addition, offspring can also be affected through embryonic development and long-term health (Trujillo et al., 2006). Therefore, it is necessary to understand the health status and correlate it with the individual nutritional needs (Picó et al., 2019).
Functional foods are then able to interact with the genome, being defined as foods that contain physiologically active components that perform a beneficial function to health in addition to the basic nutritional function (Henry, 2010). These components are called bioactive compounds that, even when present in small amounts, their frequent intake has the ability to reduce the risk of chronic diseases. It is recommended that these compounds can be obtained in their natural form. As examples of bioactive compounds, we can mention the polyphenols, such as resveratrol, quercetin, curcumin, and genistein (Karasawa and Mohan, 2018).
The genes may change during intrauterine life, when the nutrients and other food compounds can modulate gene expressions or even change the nucleotide sequence and modify the response of the organism in the presence of toxic and infectious compounds, in addition, the inherited individual genetic sequence can also influence diet, leading to the suppression of nutrients and risks for NCDs. The knowledge of these interactions between the genome and food contributes to the promotion of health and reduces the risks for NCDs through personalized diets (Paparo et al., 2014; Raiten and Bremer, 2020).
Polyphenols and Diabetes Genes
The Mediterranean diet, rich in polyphenol and others nutrients, consists of a balanced intake of fruits, vegetables, fish, cereals, and polyunsaturated fats, combined with a reduction in the consumption of meat and dairy products and a moderate intake of alcohol, mainly red wine (Di Daniele et al., 2017). This diet has been working to prevent different metabolic disorders such as cardiovascular disease and T2DM, and has been shown to decrease the incidence of neurodegenerative diseases and cancer (Bach-Faig et al., 2011). The application of the Mediterranean diet resulted in a reduction of the rate of diabetes incidence by 52% (Salas-Salvadó et al., 2011).
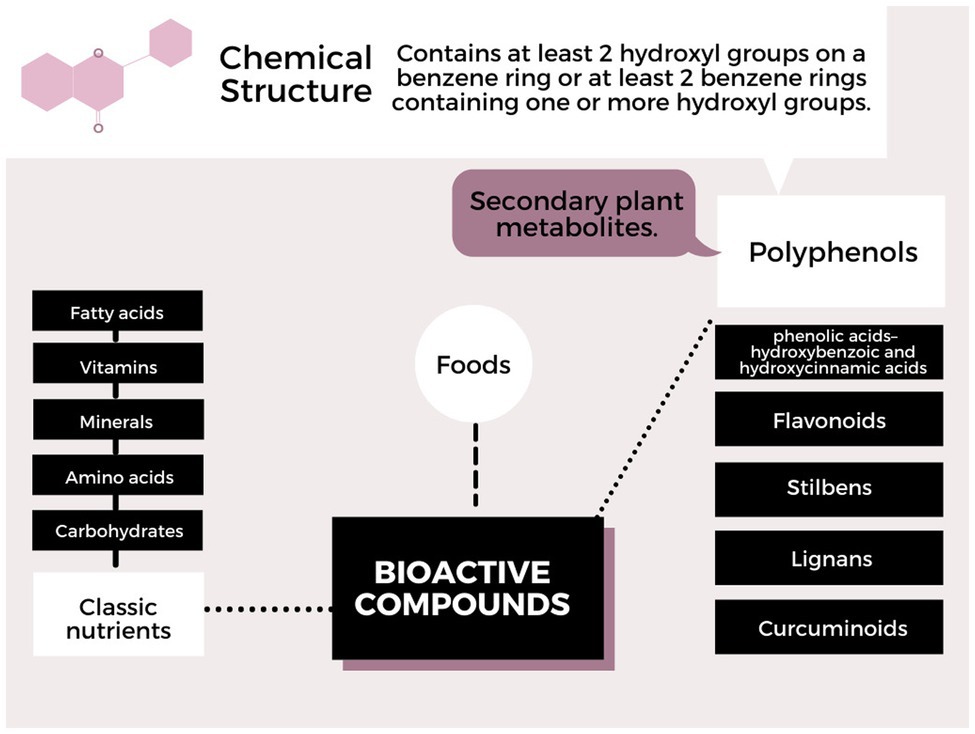
Polyphenols can interact with the DNA molecule, RNA, or with proteins involved in the activation cascade, changing number, function, and structure. Generally epigenetic mechanisms, such as methylation, DNA demethylation, and histone modifications, whether by phosphorylation, acetylation, or others, may arise from the interaction between the compounds found in food and the genes involved. These modifications are reproduced in the phenotype that can change the state of health and disease. But it is still very complex, due to genetic variability, interaction complexity, and variation in the mode of action of polyphenols. The classification of polyphenols according to the chemical structure is shown in Figure 1 (Nunes et al., 2018; Papuc et al., 2020). Furthermore, the absorption and metabolism of polyphenols in the human body (stomach, intestine, and liver) may have a different impact on human health, and factors, such as bioavailability, intestinal microbiota, and transport proteins, and the type of polyphenol may affect the bioactivity of the consumed polyphenol (Scalbert et al., 2002; Manach et al., 2004; Hoda et al., 2019).
Polyphenols can interact with the epigenome in different ways, which can alter gene expression, causing inhibition or activation. Curcumin, for example, can cause demethylation and interact with transcription factors; catechins such as epigallocatechin-3 gallate (EGCG) can also reduce the methylation mechanism by inhibiting the DNA methyltransferase enzyme and cause phosphorylation of serine and tyrosine residues of histone proteins. Flavonoids, such as Luteolin and genistein can cause acetylation of histone H3 and cause hypermethylation of genes and cause inhibition, as well as resveratrol and folic acid (Han, 2003; Collins et al., 2007; Berner et al., 2011; Vetterli et al., 2011; Goh et al., 2014; Boyanapalli and Kong, 2015; Li et al., 2015).
Genes Related to Type 2 Diabetes Mellitus
Oxidative stress is associated with T2DM (Jiménez-Osorio et al., 2014), therefore, genes as NFE2 and NFE2L2 with a regulatory role in the expression of antioxidant proteins can be targeted for protection against oxidative stress (Fu et al., 2017). Animal models showed that Nrf2 agonists improved insulin resistance and obesity, and prevented pancreatic beta-cell apoptosis (Zhao et al., 2011; Bhakkiyalakshmi et al., 2014; Matzinger et al., 2018).
The PRKAA2 gene (AMPK) is responsible for preventing the production of glucose, cholesterol, and triglycerides by promoting the oxidation of fatty acids. This gene has a relationship with the SIRT1 gene because its functioning results in the activation of the SIRT1 gene, and this causes an increase in the substrate NAD+. The SIRT1 gene is responsible for deacetylation processes and modulation of several other genes and therefore can control hepatic glucose production, lipid metabolism, and sensitivity and insulin production. It may, for example, regulate the activity of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1) causing its deacetylation, which has the function to suppress the production of ROS and regulate mitochondrial biogenesis. And lastly, it is able to reduce the production of hepatic glucose by deacetylation and activation of adenosine monophosphate-activated protein kinase (AMPK; Price et al., 2012; Rato et al., 2014). Therefore, there is a reciprocal activation between AMPK and SIRT1, which is suspended by hyperglycemia, decreasing the expression of AMPK and consequently decreasing the expression of SIRT1 (Cantó et al., 2009; Clarke et al., 2014; Li et al., 2019).
Other genes are related to insulin signaling, activation, and production, such as the PI3KR1, IRS1, FFAR1, HNF4A, and ENPP1 genes. The PI3KR1 gene encodes a phosphoinositide-3-kinase regulatory subunit 1 enzyme with direct function in the insulin signaling pathway (Karadoğan et al., 2018). The IRS1 gene encodes the insulin receptor substrate-1, which after phosphorylate regulates growth cascades, metabolism, and glucose transporter (Keshavarzi and Golsheh, 2019). The FFAR1 gene carries the code for the formation of the Free fatty acid receptor 1 protein (Ffar1). This protein and agonists (substances capable of activating Ffar1) can amplify insulin secretion in pancreatic beta cells and control blood glucose (Kohara et al., 2019). The HNF4A gene acts by maintaining glucose homeostasis (Azizi et al., 2019) as it directly activates the expression of the insulin gene. In addition, SNPs in the promoter region of the HNF4A gene were correlated with predisposition to T2DM (Bartoov-Shifman et al., 2002). Finally, the ENPP1 gene inhibits insulin receptor signaling, which is why it is related to the development of insulin resistance when overexpressed (Bacci et al., 2007; Neamati et al., 2017).
Some genes are more directly related to pancreatic β cells. The IGF2BP2 gene plays an important role in regulating the function of pancreatic cells (Huang et al., 2010) and its deregulation is associated with insulin resistance (Cao et al., 2018). In studies with rats, total ablation of IGF2BP2 results in increased insulin sensitivity and glucose tolerance (Yang et al., 2020). The overexpression of the PARP1 gene is associated with tissue damage and destruction of β cells, being a highly relevant factor in endothelial dysfunction in diabetes (Garcia Soriano et al., 2001; Pacher and Szabo, 2005).
Glucose transport and production also stand out as important processes in T2DM. The genes SLC2A1 and SLC2A2, for example, encode proteins that transport glucose into the cells, which reduces blood sugar and prevents disease (Kilpeläinen et al., 2007; Fu et al., 2017). The PCK1 and PCK2 genes encode proteins related to the production of glucose and have increased expression in people with diabetes (Cao et al., 2004). The reduction in the expression of the TCF7L2 gene was associated with an increase in β cell apoptosis (Shu et al., 2008) and an increase in hepatic glucose production and a reduction in insulin secretion (Lyssenko et al., 2007). The overexpression of the G6PC gene was observed in glucose intolerance and hyperinsulinemia (Im et al., 2011). Estrogens can also regulate the transport and control the levels of glucose in the adipose tissue and muscle, for example, so the gene ESR1 can also be targeted in the treatment or prevention of T2DM (Barreto-Andrade et al., 2018).
Finally, we can also mention genes related to inflammatory processes and oxidative stress. Hyperglycemia promotes the formation of advanced glycation end-products (AGEs) that induce inflammation and oxidative stress, so polymorphisms in the AGER gene have been associated with the risk of type 2 diabetes (Kang et al., 2012; Lin et al., 2012). In addition, the inhibition of the NFKB1 and NFKB2 genes shows a decrease in the inflammatory process, consequently improving hypertension and suppressing vasoconstriction induced by diabetes (Gautam et al., 2017; Behera et al., 2020). In patients with T2DM, we can also observe an increased expression of the FTO gene, which may be involved in oxidative metabolism, lipogenesis, and oxidative stress (Bravard et al., 2011).
The functions associated with T2DM of the commented genes, in addition to the respective bioactive compounds found in foods that show some type of interaction are described in Table 1.
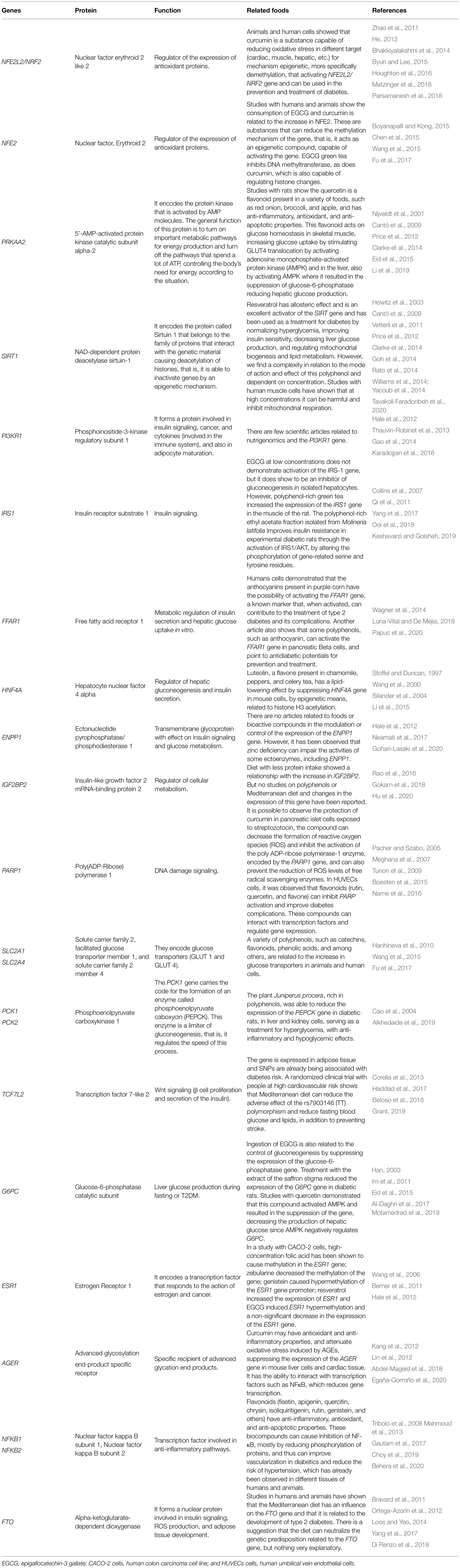
Table 1. Genes associated with type 2 diabetes mellitus (T2DM) and respective bioactive compounds found in foods that are capable to change in expression.
Future Perspectives
One way to prevent T2DM is to know the related genes and define the foods that can interact with them in a positive way. The results demonstrate that the active ingredients found in some foods, such as resveratrol, quercetin, genistein, catechins, curcumin, and anthocyanins, interact with DNA and show protective effects in relation to T2DM. These compounds interact with related genes mainly in the control of insulin secretion and signaling, oxidative stress, inflammatory processes, cellular apoptosis, and glucose and lipid metabolism. With that, we can say that the bioactive compounds present in functional foods have established functions in the prevention and treatment of T2DM and its complications. Many of the described genes need further studies to become more important in the prevention and treatment of T2DM and other diseases, like the IRS1, TCF7L2, IGF2BP2, PI3KR1, PCK1, PCK2, and FTO genes that are related to the etiology or control of the disease but there are not many studies on compounds that can modulate their expression. The PRKAA and SIRT1 genes, on the other hand, are very well-studied and, therefore, their manipulation could be used to benefit patients with T2DM or prevent the disease, since both genes, besides being associated with each other, interfere in the expression of other genes. Most studies only seek to know whether there is a change in the expression level of genes or not, there is a lack of information about the mode of action of these bioactives. Finally, most genes are directly related to insulin or glucose metabolism, but there is a great need to study genes involved in other important metabolic processes, such as inflammation, apoptosis, and oxidative stress, which may be linked to the prevention and treatment of the disease in an indirect way, it is also important to recognize that the genetic variants of each gene may respond differently to the compounds. It is noteworthy that most genes are also associated with other chronic diseases, which could encourage further studies on the subject.
Author Contributions
KF, JG, LB-S, and IG contributed to the conception and design and drafted and critically revised the manuscript. All authors gave final approval and contributed to the article and approved the submitted version.
Funding
This work was supported by Instituto de Pesquisas Pelé Pequeno Príncipe. KF is a Ph.D. student receiving a grant from Instituto de Pesquisa Pelé Pequeno Príncipe.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdel-Mageid, A. D., Abou-Salem, M. E. S., Salaam, N. M. H. A., and El-Garhy, H. A. S. (2018). The potential effect of garlic extract and curcumin nanoparticles against complication accompanied with experimentally induced diabetes in rats. Phytomedicine 43, 126–134. doi: 10.1016/j.phymed.2018.04.039
Al-Daghri, N. M., Pontremoli, C., Cagliani, R., Forni, D., Alokail, M. S., Al-Attas, O. S., et al. (2017). Susceptibility to type 2 diabetes may be modulated by haplotypes in G6PC2, a target of positive selection. BMC Evol. Biol. 17:43. doi: 10.1186/s12862-017-0897-z
Alkhedaide, A., Abdo Nassan, M., Ahmed Ismail, T., Soliman, M. M., Hassan Mohamed, E., Hassan Amer, H., et al. (2019). Hypoglycemic and antioxidant effect of Juniperus procera extract on rats with streptozotocin-induced diabetes. Pathophysiology 26, 361–368. doi: 10.1016/j.pathophys.2019.11.001
American Diabetes Association (2020). Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care 43, S14–S31. doi: 10.2337/dc20-S002
Asmat, U., Abad, K., and Ismail, K. (2016). Diabetes mellitus and oxidative stress—A concise review. Saudi Pharm. J. 24, 547–553. doi: 10.1016/j.jsps.2015.03.013
Azizi, S. M., Sarhangi, N., Afshari, M., Abbasi, D., Meybodi, H. R. A., and Hasanzad, M. (2019). Association analysis of the HNF4A common genetic variants with type 2 diabetes mellitus risk. Int. J. Mol. Cell. Med. 8, 56–62. doi: 10.22088/IIJMCM.BUMS.8.2.56
Bacci, S., De Cosmo, S., Prudente, S., and Trischitta, V. (2007). ENPP1 gene, insulin resistance and related clinical outcomes. Curr. Opin. Clin. Nutr. Metab. Care 10, 403–409. doi: 10.1097/MCO.0b013e3281e386c9
Bach-Faig, A., Berry, E. M., Lairon, D., Reguant, J., Trichopoulou, A., Dernini, S., et al. (2011). Mediterranean diet pyramid today. Science and cultural updates. Public Health Nutr. 14, 2274–2284. doi: 10.1017/S1368980011002515
Barreto-Andrade, J. N., de Fátima, L. A., Campello, R. S., Guedes, J. A. C., de Freitas, H. S., Okamoto, M. M., et al. (2018). Estrogen receptor 1 (ESR1) enhances Slc2a4/GLUT4 expression by a SP1 cooperative mechanism. Int. J. Med. Sci. 15, 1320–1328. doi: 10.7150/ijms.26774
Bartoov-Shifman, R., Hertz, R., Wang, H., Wollheim, C. B., Bar-Tana, J., and Walker, M. D. (2002). Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4α. J. Biol. Chem. 277, 25914–25919. doi: 10.1074/jbc.M201582200
Behera, S., Lamare, A. A., Rattan, R., Patnaik, B., and Das, S. (2020). Association of NFkB1 gene polymorphism with inflammatory markers in patients of type 2 diabetes mellitus with or without renal involvement in eastern India. J. Diabetes Mellit. 10, 169–181. doi: 10.4236/jdm.2020.103014
Beloso, C., Souto, J., Fábregat, M., Romanelli, G., Javiel, G., and Mimbacas, A. (2018). Association of TCF7L2 mutation and atypical diabetes in a Uruguayan population. World J. Diabetes 9, 157–164. doi: 10.4239/wjd.v9.i9.157
Berner, C., Aumüller, E., Gnauck, A., Nestelberger, M., Just, A., and Haslberger, A. G. (2011). Epigenetic control of estrogen receptor expression and tumor suppressor genes is modulated by bioactive food compounds. Ann. Nutr. Metab. 57, 183–189. doi: 10.1159/000321514
Bhakkiyalakshmi, E., Shalini, D., Sekar, T. V., Rajaguru, P., Paulmurugan, R., and Ramkumar, K. M. (2014). Therapeutic potential of pterostilbene against pancreatic beta-cell apoptosis mediated through Nrf2. Br. J. Pharmacol. 171, 1747–1757. doi: 10.1111/bph.12577
Billings, L. K., and Florez, J. C. (2010). The genetics of type 2 diabetes: what have we learned from GWAS? Ann. N. Y. Acad. Sci. 1212, 59–77. doi: 10.1111/j.1749-6632.2010.05838.x
Boesten, D. M. P. H. J., Von Ungern-Sternberg, S. N. I., Den Hartog, G. J. M., and Bast, A. (2015). Protective pleiotropic effect of flavonoids on NAD+ levels in endothelial cells exposed to high glucose. Oxidative Med. Cell. Longev. 2015:894597. doi: 10.1155/2015/894597
Boyanapalli, S. S. S., and Kong, A. N. T. (2015). “Curcumin, the king of spices”: epigenetic regulatory mechanisms in the prevention of cancer, neurological, and inflammatory diseases. Curr. Pharmacol. Rep. 1, 129–139. doi: 10.1007/s40495-015-0018-x
Bravard, A., Lefai, E., Meugnier, E., Pesenti, S., Disse, E., Vouillarmet, J., et al. (2011). FTO is increased in muscle during type 2 diabetes, and its overexpression in myotubes alters insulin signaling, enhances lipogenesis and ROS production, and induces mitochondrial dysfunction. Diabetes 60, 258–268. doi: 10.2337/db10-0281
Byun, H. G., and Lee, J. K. (2015). Chlorella ethanol extract induced phase II enzyme through NFE2L2 (nuclear factor [Erythroid-derived] 2-Like 2, NRF2) activation and protected ethanol-induced hepatoxicity. J. Med. Food 18, 182–189. doi: 10.1089/jmf.2014.3159
Cantó, C., Gerhart-Hines, Z., Feige, J. N., Lagouge, M., Noriega, L., Milne, J. C., et al. (2009). AMPK regulates energy expenditure by modulating NAD + metabolism and SIRT1 activity. Nature 458, 1056–1060. doi: 10.1038/nature07813
Cao, J., Mu, Q., and Huang, H. (2018). The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int. 2018:4217259. doi: 10.1155/2018/4217259
Cao, H., Van Der Veer, E., Ban, M. R., Hanley, A. J. G., Zinman, B., Harris, S. B., et al. (2004). Promoter polymorphism in pck1 (phosphoenolpyruvate carboxykinase gene) associated with type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 89, 898–903. doi: 10.1210/jc.2003-031361
Chen, B., Lu, Y., Chen, Y., and Cheng, J. (2015). The role of Nrf2 in oxidative stress-induced endothelial injuries. J. Endocrinol. 225, R83–R99. doi: 10.1530/JOE-14-0662
Choy, K. W., Murugan, D., Leong, X. F., Abas, R., Alias, A., and Mustafa, M. R. (2019). Flavonoids as natural anti-inflammatory agents targeting nuclear factor-kappa B (NFκB) signaling in cardiovascular diseases: a mini review. Front. Pharmacol. 10:1295. doi: 10.3389/fphar.2019.01295
Clarke, N. E., Belyaev, N. D., Lambert, D. W., and Turner, A. J. (2014). Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 126, 507–516. doi: 10.1042/CS20130291
Cole, J. B., and Florez, J. C. (2020). Genetics of diabetes mellitus and diabetes complications. Nat. Rev. Nephrol. 16, 377–390. doi: 10.1038/s41581-020-0278-5
Collins, Q. F., Liu, H. Y., Pi, J., Liu, Z., Quon, M. J., and Cao, W. (2007). Epigallocatechin-3-gallate (EGCG), a green tea polyphenol, suppresses hepatic gluconeogenesis through 5′-AMP-activated protein kinase. J. Biol. Chem. 282, 30143–30149. doi: 10.1074/jbc.M702390200
Constantino, M. I., Molyneaux, L., Limacher-Gisler, F., Al-Saeed, A., Luo, C., Wu, T., et al. (2013). Long-term complications and mortality in young-onset diabetes: type 2 diabetes is more hazardous and lethal than type 1 diabetes. Diabetes Care 36, 3863–3869. doi: 10.2337/dc12-2455
Corella, D., Carrasco, P., Sorlí, J. V., Estruch, R., Rico-Sanz, J., Martínez-González, M. Á., et al. (2013). Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: A randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care 36, 3803–3811. doi: 10.2337/dc13-0955
Dandona, P., Thusu, K., Cook, S., Snyder, B., Makowski, J., Armstrong, D., et al. (1996). Oxidative damage to DNA in diabetes mellitus. Lancet 347, 444–445. doi: 10.1016/S0140-6736(96)90013-6
Di Daniele, N. D., Noce, A., Vidiri, M. F., Moriconi, E., Marrone, G., Annicchiarico-Petruzzelli, M., et al. (2017). Impact of Mediterranean diet on metabolic syndrome, cancer and longevity. Oncotarget 8, 8947–8979. doi: 10.18632/oncotarget.13553
Di Renzo, L., Cioccoloni, G., Falco, S., Abenavoli, L., Moia, A., Sinibaldi Salimei, P., et al. (2018). Influence of FTO rs9939609 and Mediterranean diet on body composition and weight loss: a randomized clinical trial NCT01890070 NCT. J. Transl. Med. 16:308. doi: 10.1186/s12967-018-1680-7
Dimitrov, D., Thiele, I., and Ferguson, L. R. (2016). Editorial: the human gutome: nutrigenomics of host-microbiome interactions. Front. Genet. 7:158. doi: 10.3389/fgene.2016.00158
Egaña-Gorroño, L., López-Díez, R., Yepuri, G., Ramirez, L. S., Reverdatto, S., Gugger, P. F., et al. (2020). Receptor for advanced glycation end products (Rage) and mechanisms and therapeutic opportunities in diabetes and cardiovascular disease: insights from human subjects and animal models. Front. Cardiovasc. Med. 7:37. doi: 10.3389/fcvm.2020.00037
Eid, H. M., Nachar, A., Thong, F., Sweeney, G., and Haddad, P. S. (2015). The molecular basis of the antidiabetic action of quercetin in cultured skeletal muscle cells and hepatocytes. Pharmacogn. Mag. 11, 74–81. doi: 10.4103/0973-1296.149708
Fenech, M., El-Sohemy, A., Cahill, L., Ferguson, L. R., French, T. A. C., Tai, E. S., et al. (2011). Nutrigenetics and nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. J. Nutrigenet. Nutrigenomics 4, 69–89. doi: 10.1159/000327772
Ferguson, L. R. (2006). Nutrigenomics: integrating genomic approaches into nutrition research. Mol. Diagnosis Ther. 10, 101–108. doi: 10.1007/BF03256449
Fialho, E., Moreno, F. S., and Ong, T. P. (2008). Nutrition in the post-genome era: “Omic” tools basics and applications. Rev. Nutr. 21, 757–766. doi: 10.1590/S1415-52732008000600014
Fu, Q. Y., Li, Q. S., Lin, X. M., Qiao, R. Y., Yang, R., Li, X. M., et al. (2017). Antidiabetic effects of tea. Molecules 22:849. doi: 10.3390/molecules22050849
Furmli, S., Elmasry, R., Ramos, M., and Fung, J. (2018). Therapeutic use of intermittent fasting for people with type 2 diabetes as an alternative to insulin. BMJ Case Rep. 2018:bcr2017221854. doi: 10.1136/bcr-2017-221854
Gadi, R., and Samaha, F. F. (2007). Dyslipidemia in type 2 diabetes mellitus. Curr. Diab. Rep. 7, 228–234. doi: 10.1007/s11892-007-0036-0
Gao, X., Liu, X., Xu, J., Xue, C., Xue, Y., and Wang, Y. (2014). Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 118, 476–481. doi: 10.1016/j.jbiosc.2014.03.001
Garcia Soriano, F., Virág, L., Jagtap, P., Szabó, É., Mabley, J. G., Liaudet, L., et al. (2001). Diabetic endothelial dysfunction: the role of poly(ADP-ribose) polymerase activation. Nat. Med. 7, 108–113. doi: 10.1038/83241
García-Chapa, E. G., Leal-Ugarte, E., Peralta-Leal, V., Durán-González, J., and Meza-Espinoza, J. P. (2017). Genetic epidemiology of type 2 diabetes in mexican mestizos. Biomed. Res. Int. 2017:3937893. doi: 10.1155/2017/3937893
Gautam, A., Gupta, S., Mehndiratta, M., Sharma, M., Singh, K., Kalra, O. P., et al. (2017). Association of NFKB1 gene polymorphism (rs28362491) with levels of inflammatory biomarkers and susceptibility to diabetic nephropathy in Asian Indians. World J. Diabetes 8:66. doi: 10.4239/wjd.v8.i2.66
Goh, K. P., Lee, H. Y., Lau, D. P., Supaat, W., Chan, Y. H., and Koh, A. F. Y. (2014). Effects of resveratrol in patients with type 2 diabetes mellitus on skeletal muscle SIRT1 expression and energy expenditure. Int. J. Sport Nutr. Exerc. Metab. 24, 2–13. doi: 10.1123/ijsnem.2013-0045
Gohari-Lasaki, S., Sharafshah, A., Abbaspour, S., and Keshavarz, P. (2020). Single locus and haplotype association of ENPP1 gene variants with the development of retinopathy among type 2 diabetic patients. Int. Ophthalmol. 40, 639–647. doi: 10.1007/s10792-019-01224-3
Gokarn, R., Solon-Biet, S. M., Cogger, V. C., Cooney, G. J., Wahl, D., McMahon, A. C., et al. (2018). Long-term dietary macronutrients and hepatic gene expression in aging mice. J. Gerontol. A Biol. Sci. Med. Sci. 73, 1618–1625. doi: 10.1093/gerona/gly065
Grant, S. F. A. (2019). The TCF7L2 locus: a genetic window into the pathogenesis of type 1 and type 2 diabetes. Diabetes Care 42, 1624–1629. doi: 10.2337/dci19-0001
Haddad, S. A., Palmer, J. R., Lunetta, K. L., Ng, M. C. Y., Ruiz-Narváez, E. A., Shriner, D., et al. (2017). A novel TCF7L2 type 2 diabetes SNP identified from fine mapping in African American women. PLoS One 12:e172577. doi: 10.1371/journal.pone.0172577
Hale, P. J., López-Yunez, A. M., and Chen, J. Y. (2012). Genome-wide meta-analysis of genetic susceptible genes for type 2 diabetes. BMC Syst. Biol. 6:S16. doi: 10.1186/1752-0509-6-S3-S16
Han, M. K. (2003). Epigallocatechin gallate, a constituent of green tea, suppresses cytokine-induced pancreatic β-cell damage. Exp. Mol. Med. 35, 136–139. doi: 10.1038/emm.2003.19
Hanhineva, K., Törrönen, R., Bondia-Pons, I., Pekkinen, J., Kolehmainen, M., Mykkänen, H., et al. (2010). Impact of dietary polyphenols on carbohydrate metabolism. Int. J. Mol. Sci. 11, 1365–1402. doi: 10.3390/ijms11041365
He, H.-J. (2012). Curcumin attenuates Nrf2 signaling defect, oxidative stress in muscle and glucose intolerance in high fat diet-fed mice. World J. Diabetes 3:94. doi: 10.4239/wjd.v3.i5.94
Hoda, M., Hemaiswarya, S., and Doble, M. (2019). “Pharmacokinetics and pharmacodynamics of polyphenols,” in Role of phenolic phytochemicals in diabetes management. (Singapore: Springer), 159–173.
Holman, N., Young, B., and Gadsby, R. (2015). Current prevalence of type 1 and type 2 diabetes in adults and children in the UK. Diabet. Med. 32, 1119–1120. doi: 10.1111/dme.12791
Houghton, C. A., Fassett, R. G., and Coombes, J. S. (2016). Sulforaphane and other nutrigenomic Nrf2 activators: can the clinician’s expectation be matched by the reality? Oxidative Med. Cell. Longev. 2016:7857186. doi: 10.1155/2016/7857186
Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., et al. (2003). Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nat. Publ. 425, 191–196. doi: 10.1038/nature01960
Hu, X., Peng, W. X., Zhou, H., Jiang, J., Zhou, X., Huang, D., et al. (2020). IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 27, 1782–1794. doi: 10.1038/s41418-019-0461-z
Huang, Q., Yin, J. Y., Dai, X. P., Pei, Q., Dong, M., Zhou, Z. G., et al. (2010). IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol. Sin. 31, 709–717. doi: 10.1038/aps.2010.47
Im, S. S., Kim, M. Y., Kwon, S. K., Kim, T. H., Bae, J. S., Kim, H., et al. (2011). Peroxisome proliferator-activated receptor α is responsible for the up-regulation of hepatic glucose-6-phosphatase gene expression in fasting and db/db mice. J. Biol. Chem. 286, 1157–1164. doi: 10.1074/jbc.M110.157875
Jiménez-Osorio, A. S., Picazo, A., González-Reyes, S., Barrera-Oviedo, D., Rodríguez-Arellano, M. E., and Pedraza-Chaverri, J. (2014). Nrf2 and redox status in prediabetic and diabetic patients. Int. J. Mol. Sci. 15, 20290–20305. doi: 10.3390/ijms151120290
Kaneto, H., Katakami, N., Kawamori, D., Miyatsuka, T., Sakamoto, K., Matsuoka, T. A., et al. (2007). Involvement of oxidative stress in the pathogenesis of diabetes. Antioxid. Redox Signal. 9, 355–366. doi: 10.1089/ars.2006.1465
Kang, P., Tian, C., and Jia, C. (2012). Association of RAGE gene polymorphisms with type 2 diabetes mellitus, diabetic retinopathy and diabetic nephropathy. Gene 500, 1–9. doi: 10.1016/j.gene.2012.03.056
Karadoğan, A. H., Arikoglu, H., Göktürk, F., İşçioğlu, F., and İpekçi, S. H. (2018). PIK3R1 gene polymorphisms are associated with type 2 diabetes and related features in the Turkish population. Adv. Clin. Exp. Med. 27, 921–927. doi: 10.17219/acem/68985
Karasawa, M. M. G., and Mohan, C. (2018). Fruits as prospective reserves of bioactive compounds: a review. Nat. Prod. Bioprospect. 8, 335–346. doi: 10.1007/s13659-018-0186-6
Keshavarzi, F., and Golsheh, S. (2019). IRS1-rs10498210 G/A and CCR5-59029 A/G polymorphisms in patients with type 2 diabetes in Kurdistan. Mol. Genet. Genomic Med. 7:e631. doi: 10.1002/mgg3.631
Kilpeläinen, T. O., Lakka, T. A., Laaksonen, D. E., Laukkanen, O., Lindström, J., Eriksson, J. G., et al. (2007). Physical activity modifies the effect of SNPs in the SLC2A2 (GLUT2) and ABCC8 (SUR1) genes on the risk of developing type 2 diabetes. Physiol. Genomics 31, 264–272. doi: 10.1152/physiolgenomics.00036.2007
Kohara, K., Obata, A., Kimura, T., Shimoda, M., Moriuchi, S., Okauchi, S., et al. (2019). Suppression of free fatty acid receptor 1 expression in pancreatic β-cells in obese type 2 diabetic db/db mice: a potential role of pancreatic and duodenal homeobox factor 1. Endocr. J. 66, 43–50. doi: 10.1507/endocrj.EJ18-0203
Kuzuya, T., Nakagawa, S., Satoh, J., Kanazawa, Y., Iwamoto, Y., Kobayashi, M., et al. (2002). Report of the committee on the classification and diagnostic criteria of diabetes mellitus. Diabetes Res. Clin. Pract. 55, 65–85. doi: 10.1016/S0168-8227(01)00365-5
Li, J., Inoue, J., Choi, J. M., Nakamura, S., Yan, Z., Fushinobu, S., et al. (2015). Identification of the flavonoid luteolin as a repressor of the transcription factor hepatocyte nuclear factor 4α. J. Biol. Chem. 290, 24021–24035. doi: 10.1074/jbc.M115.645200
Li, R., Zhang, Y., Rasool, S., Geetha, T., and Babu, J. R. (2019). Effects and underlying mechanisms of bioactive compounds on type 2 diabetes mellitus and Alzheimer’s disease. Oxidative Med. Cell. Longev. 2019:8165707. doi: 10.1155/2019/8165707
Lin, J., Tang, Y., Kang, Q., Feng, Y., and Chen, A. (2012). Curcumin inhibits gene expression of receptor for advanced glycation end-products (RAGE) in hepatic stellate cells in vitro by elevating PPARγ activity and attenuating oxidative stress. Br. J. Pharmacol. 166, 2212–2227. doi: 10.1111/j.1476-5381.2012.01910.x
Lloyd, A., Sawyer, W., and Hopkinson, P. (2001). Impact of long-term complications on quality of life in patients with type 2 diabetes not using insulin. Value Health 4, 392–400. doi: 10.1046/j.1524-4733.2001.45029.x
Loos, R. J. F., and Yeo, G. S. H. (2014). The bigger picture of FTO—the first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 10, 51–61. doi: 10.1038/nrendo.2013.227
Luna-Vital, D. A., and De Mejia, E. G. (2018). Anthocyanins from purple corn activate free fatty acid-receptor 1 and glucokinase enhancing in vitro insulin secretion and hepatic glucose uptake. PLoS One 13:e0200449. doi: 10.1371/journal.pone.0200449
Lyssenko, V., Lupi, R., Marchetti, P., Del Guerra, S., Orho-Melander, M., Almgren, P., et al. (2007). Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 117, 2155–2163. doi: 10.1172/JCI30706
Mahler, R. J., and Adler, M. L. (1999). Clinical review 102: type 2 diabetes mellitus: update on diagnosis, pathophysiology, and treatment. J. Clin. Endocrinol. Metab. 84, 1165–1171. doi: 10.1210/jcem.84.4.5612
Mahmoud, M. F., Hassan, N. A., El Bassossy, H. M., and Fahmy, A. (2013). Quercetin protects against diabetes-induced exaggerated vasoconstriction in rats: effect on low grade inflammation. PLoS One 8:e63784. doi: 10.1371/journal.pone.0063784
Manach, C., Scalbert, A., Morand, C., Rémésy, C., and Jiménez, L. (2004). Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 79, 727–747. doi: 10.1093/ajcn/79.5.727
Marcum, J. A. (2020). Nutrigenetics/nutrigenomics, personalized nutrition, and precision healthcare. Curr. Nutr. Rep. 9, 338–345. doi: 10.1007/s13668-020-00327-z
Matzinger, M., Fischhuber, K., and Heiss, E. H. (2018). Activation of Nrf2 signaling by natural products-can it alleviate diabetes? Biotechnol. Adv. 36, 1738–1767. doi: 10.1016/j.biotechadv.2017.12.015
Meghana, K., Sanjeev, G., and Ramesh, B. (2007). Curcumin prevents streptozotocin-induced islet damage by scavenging free radicals: a prophylactic and protective role. Eur. J. Pharmacol. 577, 183–191. doi: 10.1016/j.ejphar.2007.09.002
Mickelson, B., Herfel, T. M., Booth, J., and Wilson, R. P. (2019). “Nutrition,” in The laboratory rat 3rd Edn. (United States: Academic Press), 243–347.
Motamedrad, M., Shokouhifar, A., Hemmati, M., and Moossavi, M. (2019). The regulatory effect of saffron stigma on the gene expression of the glucose metabolism key enzymes and stress proteins in streptozotocin-induced diabetic rats. Res. Pharm. Sci. 14, 255–262. doi: 10.4103/1735-5362.258494
Narne, P., Ponnaluri, K. C., Siraj, M., and Ishaq, M. (2016). Association analysis of polymorphisms in genes related to oxidative stress in south Indian type 2 diabetic patients with retinopathy. Ophthalmic Genet. 37, 1–8. doi: 10.3109/13816810.2014.895015
Neamati, N., Hosseini, S. R., Hajiahmadi, M., Halalkhor, S., Nooreddini, H., Niaki, H. A., et al. (2017). The ENPP1 K121Q polymorphism modulates developing of bone disorders in type 2 diabetes: a cross sectional study. Gene 637, 100–107. doi: 10.1016/j.gene.2017.09.042
Nijveldt, R. J., Van Nood, E., Van Hoorn, D. E. C., Boelens, P. G., Van Norren, K., and Van Leeuwen, P. A. M. (2001). Flavonoids: a review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 74, 418–425. doi: 10.1093/ajcn/74.4.418
Nishikawa, T., Edelstein, D., Du, X. L., Yamagishi, S. I., Matsumura, T., Kaneda, Y., et al. (2000). Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 404, 787–790. doi: 10.1038/35008121
Nunes, M. A., Rodrigues, F., Vinha, A. F., Alves, R. C., and Oliveira, M. B. P. P. (2018). Nutrigenomics and polyphenols. Cambridge: Woodhead Publishing, Elsevier Inc.
Ooi, D. J., Adamu, H. A., Imam, M. U., Ithnin, H., and Ismail, M. (2018). Polyphenol-rich ethyl acetate fraction isolated from Molineria latifolia ameliorates insulin resistance in experimental diabetic rats via IRS1/AKT activation. Biomed. Pharmacother. 98, 125–133. doi: 10.1016/j.biopha.2017.12.002
Ortega-Azorín, C., Sorlí, J. V., Asensio, E. M., Coltell, O., Martínez-González, M. T., Salas-Salvadó, J., et al. (2012). Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc. Diabetol. 11:137. doi: 10.1186/1475-2840-11-137
Pacher, P., and Szabo, C. (2005). Role of poly (ADP-ribose) Polymerase-1 activation in the pathogenesis of diabetic complications. Antioxid. Redox Signal. 7, 1568–1580. doi: 10.1089/ars.2005.7.1568
Paparo, L., Di Costanzo, M., Di Scala, C., Cosenza, L., Leone, L., Nocerino, R., et al. (2014). The influence of early life nutrition on epigenetic regulatory mechanisms of the immune system. Nutrients 6, 4706–4719. doi: 10.3390/nu6114706
Papuc, C., Goran, G. V., Predescu, C. N., Tudoreanu, L., and Ștefan, G. (2020). Plant polyphenols mechanisms of action on insulin resistance and against the loss of pancreatic beta cells. Crit. Rev. Food Sci. Nutr. 1–28. doi: 10.1080/10408398.2020.1815644 [Epub ahead of print].
Parsamanesh, N., Moossavi, M., Bahrami, A., Butler, A. E., and Sahebkar, A. (2018). Therapeutic potential of curcumin in diabetic complications. Pharmacol. Res. 136, 181–193. doi: 10.1016/j.phrs.2018.09.012
Picó, C., Serra, F., Rodríguez, A. M., Keijer, J., and Palou, A. (2019). Biomarkers of nutrition and health: new tools for new approaches. Nutrients 11:1092. doi: 10.3390/nu11051092
Price, N. L., Gomes, A. P., Ling, A. J. Y., Duarte, F. V., Martin-Montalvo, A., North, B. J., et al. (2012). SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15, 675–690. doi: 10.1016/j.cmet.2012.04.003
Qi, Q., Bray, G. A., Smith, S. R., Hu, F. B., Sacks, F. M., and Qi, L. (2011). Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial the preventing overweight using novel dietary strategies (POUNDS LOST) trial. Circulation 124, 563–571. doi: 10.1161/CIRCULATIONAHA.111.025767
Raiten, D. J., and Bremer, A. A. (2020). Exploring the nutritional ecology of stunting: new approaches to an old problem. Nutrients 12:371. doi: 10.3390/nu12020371
Rajendran, R., Garva, R., Krstic-Demonacos, M., and Demonacos, C. (2011). Sirtuins: molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J. Biomed. Biotechnol. 2011:368276. doi: 10.1155/2011/368276
Rao, P., Wang, H., Fang, H., Gao, Q., Zhang, J., Song, M., et al. (2016). Association between IGF2BP2 polymorphisms and type 2 diabetes mellitus: a case-control study and meta-analysis. Int. J. Environ. Res. Public Health 13:574. doi: 10.3390/ijerph13060574
Rato, L., Duarte, A. I., Tomás, G. D., Santos, M. S., Moreira, P. I., Socorro, S., et al. (2014). Pre-diabetes alters testicular PGC1-α/SIRT3 axis modulating mitochondrial bioenergetics and oxidative stress. Biochim. Biophys. Acta Bioenerg. 1837, 335–344. doi: 10.1016/j.bbabio.2013.12.008
Rheinheimer, J., de Souza, B. M., Cardoso, N. S., Bauer, A. C., and Crispim, D. (2017). Current role of the NLRP3 inflammasome on obesity and insulin resistance: a systematic review. Metabolism 74, 1–9. doi: 10.1016/j.metabol.2017.06.002
Rist, M. J., Wenzel, U., and Daniel, H. (2006). Nutrition and food science go genomic. Trends Biotechnol. 24, 172–178. doi: 10.1016/j.tibtech.2006.02.001
Salas-Salvadó, J., Bulló, M., Babio, N., Martínez-González, M. Á., Ibarrola-Jurado, N., Basora, J., et al. (2011). Reduction in the incidence of type 2 diabetes with the mediterranean diet: results of the PREDIMED-Reus nutrition intervention randomized trial. Diabetes Care 34, 14–19. doi: 10.2337/dc10-1288
Scalbert, A., Mor, C., Manach, C., and Rémésy, C. (2002). Absorption and metabolism of polyphenols in the gut and impact on health. Biomed. Pharmacother. 56, 276–282. doi: 10.1016/S0753-3322(02)00205-6
Shu, L., Sauter, N. S., Schulthess, F. T., and Matveyenko, A. V. (2008). Transcription factor 7-like 2 regulates-cell survival and function in human pancreatic islets. Diabetes 57, 645–653. doi: 10.2337/db07-0847.N.S.S
Sies, H. (1993). Strategies of antioxidant defense. Eur. J. Biochem. 215, 213–219. doi: 10.1111/j.1432-1033.1993.tb18025.x
Silander, K., Mohlke, K. L., Scott, L. J., Peck, E. C., Hollstein, P., Skol, A. D., et al. (2004). Genetic variation near the hepatocyte nuclear factor-4α gene predicts susceptibility to type 2 diabetes. Diabetes 53, 1141–1149. doi: 10.2337/diabetes.53.4.1141
Spranger, J., Kroke, A., Möhlig, M., Hoffmann, K., Bergmann, M. M., Ristow, M., et al. (2003). Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based european prospective investigation into cancer and nutrition (EPIC)-Potsdam study. Diabetes 52, 812–817. doi: 10.2337/diabetes.52.3.812
Steemburgo, T., de Azevedo, M. J., and Martínez, J. A. (2009). Gene-nutrient interaction and its association with obesity and diabetes mellitus. Arq. Bras. Endocrinol. Metabol. 53, 497–508. doi: 10.1590/S0004-27302009000500003
Stoffel, M., and Duncan, S. A. (1997). The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc. Natl. Acad. Sci. U. S. A. 94, 13209–13214. doi: 10.1073/pnas.94.24.13209
Tavakoli Faradonbeh, R., Zakerkish, M., Karimi Akhormeh, A., Mohammadtaghvaei, N., Jalali, M. T., and Yaghooti, H. (2020). Association of the rs3758391 polymorphism in the SIRT1 gene with diabetic nephropathy and decreased estimated glomerular filtration rate (GFR) in a population from Southwest Iran. Int. J. Diabetes Dev. Ctries. 40, 99–105. doi: 10.1007/s13410-019-00742-1
Thauvin-Robinet, C., Auclair, M., Duplomb, L., Caron-Debarle, M., Avila, M., St-Onge, J., et al. (2013). PIK3R1 mutations cause syndromic insulin resistance with lipoatrophy. Am. J. Hum. Genet. 93, 141–149. doi: 10.1016/j.ajhg.2013.05.019
Tribolo, S., Lodi, F., Connor, C., Suri, S., Wilson, V. G., Taylor, M. A., et al. (2008). Comparative effects of quercetin and its predominant human metabolites on adhesion molecule expression in activated human vascular endothelial cells. Atherosclerosis 197, 50–56. doi: 10.1016/j.atherosclerosis.2007.07.040
Trujillo, E., Davis, C., and Milner, J. (2006). Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J. Am. Diet. Assoc. 106, 403–413. doi: 10.1016/j.jada.2005.12.002
Tunon, M., Garcia-Mediavilla, M., Sanchez-Campos, S., and Gonzalez-Gallego, J. (2009). Potential of flavonoids as anti-inflammatory agents: modulation of pro-inflammatory gene expression and signal transduction pathways. Curr. Drug Metab. 10, 256–271. doi: 10.2174/138920009787846369
Vetterli, L., Brun, T., Giovannoni, L., Bosco, D., and Maechler, P. (2011). Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E β-cells and human islets through a SIRT1-dependent mechanism. J. Biol. Chem. 286, 6049–6060. doi: 10.1074/jbc.M110.176842
Wagner, R., Staiger, H., Ullrich, S., Stefan, N., Fritsche, A., and Häring, H. U. (2014). Untangling the interplay of genetic and metabolic influences on beta-cell function: examples of potential therapeutic implications involving TCF7L2 and FFAR1. Mol. Metab. 3, 261–267. doi: 10.1016/j.molmet.2014.01.001
Wang, X., Chen, H., Liu, J., Ouyang, Y., Wang, D., Bao, W., et al. (2015). Association between the NF-E2 related factor 2 gene polymorphism and oxidative stress, anti-oxidative status, and newly-diagnosed type 2 diabetes mellitus in a Chinese population. Int. J. Mol. Sci. 16, 16483–16496. doi: 10.3390/ijms160716483
Wang, J. Y., Grabacka, M., Marcinkiewicz, C., Staniszewska, I., Peruzzi, F., Khalili, K., et al. (2006). Involvement of α1β1 integrin in insulin-like growth factor-1-mediated protection of PC12 neuronal processes from tumor necrosis factor-α-induced injury. J. Neurosci. Res. 83, 7–18. doi: 10.1002/jnr.20712
Wang, H., Maechler, P., Antinozzi, P. A., Hagenfeldt, K. A., and Wollheim, C. B. (2000). Hepatocyte nuclear factor 4α regulates the expression of pancreatic β-cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J. Biol. Chem. 275, 35953–35959. doi: 10.1074/jbc.M006612200
Weyer, C., Bogardus, C., Mott, D. M., and Pratley, R. E. (1999). The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J. Clin. Invest. 104, 787–794. doi: 10.1172/JCI7231
Williams, C. B., Hughes, M. C., Edgett, B. A., Scribbans, T. D., Simpson, C. A., Perry, C. G. R., et al. (2014). An examination of resveratrol’s mechanisms of action in human tissue: impact of a single dose in vivo and dose responses in skeletal muscle ex vivo. PLoS One 9:e102406. doi: 10.1371/journal.pone.0102406
Wu, D., and Cederbaum, A. I. (2003). Alcohol, oxidative stress, and free radical damage. Alcohol Res. Health 65, 278–290. doi: 10.1079/pns2006496
Yacoub, R., Lee, K., and He, J. C. (2014). The role of SIRT1 in diabetic kidney disease. Front. Endocrinol. 5:166. doi: 10.3389/fendo.2014.00166
Yang, M., Gallo-ebert, C., Hayward, M., Liu, W., Mcdonough, V., and Nickels, J. T. (2020). Human insulin growth factor 2 mRNA binding protein 2 increases microRNA 33a/b inhibition of liver ABCA1 expression and alters low-density apolipoprotein levels in mice. Mol. Cell. Biol. 40, e00058–e00020. doi: 10.1128/MCB.00058-20
Yang, Q., Xiao, T., Guo, J., and Su, Z. (2017). Complex relationship between obesity and the fat mass and obesity locus. Int. J. Biol. Sci. 13, 615–629. doi: 10.7150/ijbs.17051
Zhao, S. G., Li, Q., Liu, Z. X., Wang, J. J., Wang, X. X., Qin, M., et al. (2011). Curcumin attenuates insulin resistance in hepatocytes by inducing Nrf2 nuclear translocation. Hepato-Gastroenterology 58, 2106–2111. doi: 10.5754/hge11219
Keywords: nutrigenomics, type 2 diabetes mellitus, chronic disease, bioactive compounds, nutrients, gene-nutrient interactions, polyphenols
Citation: Felisbino K, Granzotti JG, Bello-Santos L and Guiloski IC (2021) Nutrigenomics in Regulating the Expression of Genes Related to Type 2 Diabetes Mellitus. Front. Physiol. 12:699220. doi: 10.3389/fphys.2021.699220
Edited by:
Atsushi Masamune, Tohoku University, JapanReviewed by:
Kei Takahashi, Tohoku University, JapanNorihisa Nishimura, Nara Medical University, Japan
Yoichi Kakuta, Tohoku University Hospital, Japan
Copyright © 2021 Felisbino, Granzotti, Bello-Santos and Guiloski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Izonete Cristina Guiloski, aXpvbmV0ZS5jcmlzdGluYUBwZWxlcGVxdWVub3ByaW5jaXBlLm9yZy5icg==; aWd1aWxvc2tpQGdtYWlsLmNvbQ==
 Karoline Felisbino1,2,3
Karoline Felisbino1,2,3 Izonete Cristina Guiloski
Izonete Cristina Guiloski