- Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
Obesity, a global public health issue, is characterized by excessive adiposity and is strongly related to some chronic diseases including cardiovascular diseases and diabetes. Extra energy intake-induced adipogenesis involves various transcription factors and long noncoding RNAs (lncRNAs) that control lipogenic mRNA expression. Currently, lncRNAs draw much attention for their contribution to adipogenesis and adipose tissue function. Increasing evidence also manifests the pivotal role of lncRNAs in modulating white, brown, and beige adipose tissue development and affecting the progression of the diseases induced by adipose dysfunction. The aim of this review is to summarize the roles of lncRNAs in adipose tissue development and obesity-caused diseases to provide novel drug targets for the treatment of obesity and metabolic diseases.
Introduction
Emerging data show that excessive body fat, particularly obesity, is a major risk factor of mortality worldwide (Peeters et al., 2003; Blüher, 2019; Chooi et al., 2019). As the main lipid storage organ, excess adipose tissue is closely related to the occurrence and development of obesity (Sun et al., 2011). When obesity occurs, pathological changes in the morphology, composition, and function of adipose tissues can lead to the occurrence of various metabolic diseases, such as insulin resistance, fatty liver, diabetes, and cardiovascular diseases (Lavie et al., 2009; Blüher, 2019; Ghaben and Scherer, 2019; Schetz et al., 2019). It is of great significance to identify new therapeutic targets for obesity and its related metabolic diseases.
Adipose tissues are physiologically classified into white adipose tissue (WAT) and brown adipose tissue (BAT). WAT is mainly responsible for unnecessary energy storage, whereas BAT functions as fuel oxidation and energy expenditure because of containing abundant mitochondria. With the drug treatments or thermogenic stimuli, WAT possesses the potential to convert into “brown-like” cells. These brite adipocytes can also dissipate energy. Thus, promoting WAT browning might be an effective strategy to prevent obesity.
Recently, many studies have focused on the roles of nonconding RNAs in regulation of adipose tissue activities. Thereinto, long noncoding RNAs (lncRNAs) are defined as long RNA transcripts (>200 bp) not encoding proteins and these lncRNAs are a class of RNA observed to play modulatory roles in many biological processes consistent with their tissue-specificity. LncRNAs are crucial to the regulatory network of adipocyte biology, generating both positive and negative control in lipogenesis and adipogenesis. They also affect adipose tissue functions like white fat browning and brown fat thermogenesis.
This wide range in regulatory roles may make lncRNAs, a promising new therapeutic area in the fight against obesity and related metabolic diseases. However, reviews on the roles of lncRNAs in adipose tissue dysplasia, abnormal lipid metabolism, and associated diseases are very limited. Thus, it is necessary to summarize the latest research progress into the regulation of lncRNAs in lipid metabolism and adipocyte biology. The present review focuses on summarizing the potential of lncRNAs as therapeutic targets for obesity and related diseases caused by lipid metabolism disorders and adipose tissue dysfunction.
LncRNAs: the Emerging Regulators
Discovery and Definition of LncRNAs
The development of new technologies, including genome tiling arrays, Global Nuclear Run-On sequencing (GRO-Seq), and Chromatin Isolation by RNA Purification (ChIRP-Seq), helped to identify a mass of new RNAs. LncRNAs are defined as RNA molecules longer than 200 nucleotides (Core et al., 2008; Guttman et al., 2009; Chu et al., 2011). H19, reported in 1990, may be the first identified lncRNA. After transcription by RNA polymerase II, H19 is spliced and polyadenylated like an mRNA, but it encodes no almost protein (Brannan et al., 1990; Bartolomei et al., 1991).
Since the discovery of H19, advances in genome sequencing have identified many more lncRNAs. They generally possess little potential to encode protein for the lack of open reading frames, 3′ untranslated regions, and typical terminal regions (Jarroux et al., 2017), but they play critical roles in a diversity of cellular processes, such as translation, transcription, and epigenetic modification.
Structure and Function of LncRNAs
A vast number of lncRNAs have been identified, but they urgently need to be annotated to be useful in therapeutic applications. Annotation begins by dividing newly discovered lncRNAs into one of five categories based on their relative location in the genome to protein-coding genes (Mattick and Rinn, 2015): sense lncRNAs overlap the nearest protein coding gene along the sense direction; antisense lncRNAs are those whose transcription overlaps an mRNA in any portion; intronic lncRNAs are located in an intron of a protein-coding gene; intergenic lncRNAs that are found between two protein-coding genes; and bidirectional lncRNAs are those whose transcription start site is within 1,000 base pairs (bp) from the neighboring exon and is transcribed in the opposite direction (Figure 1A).
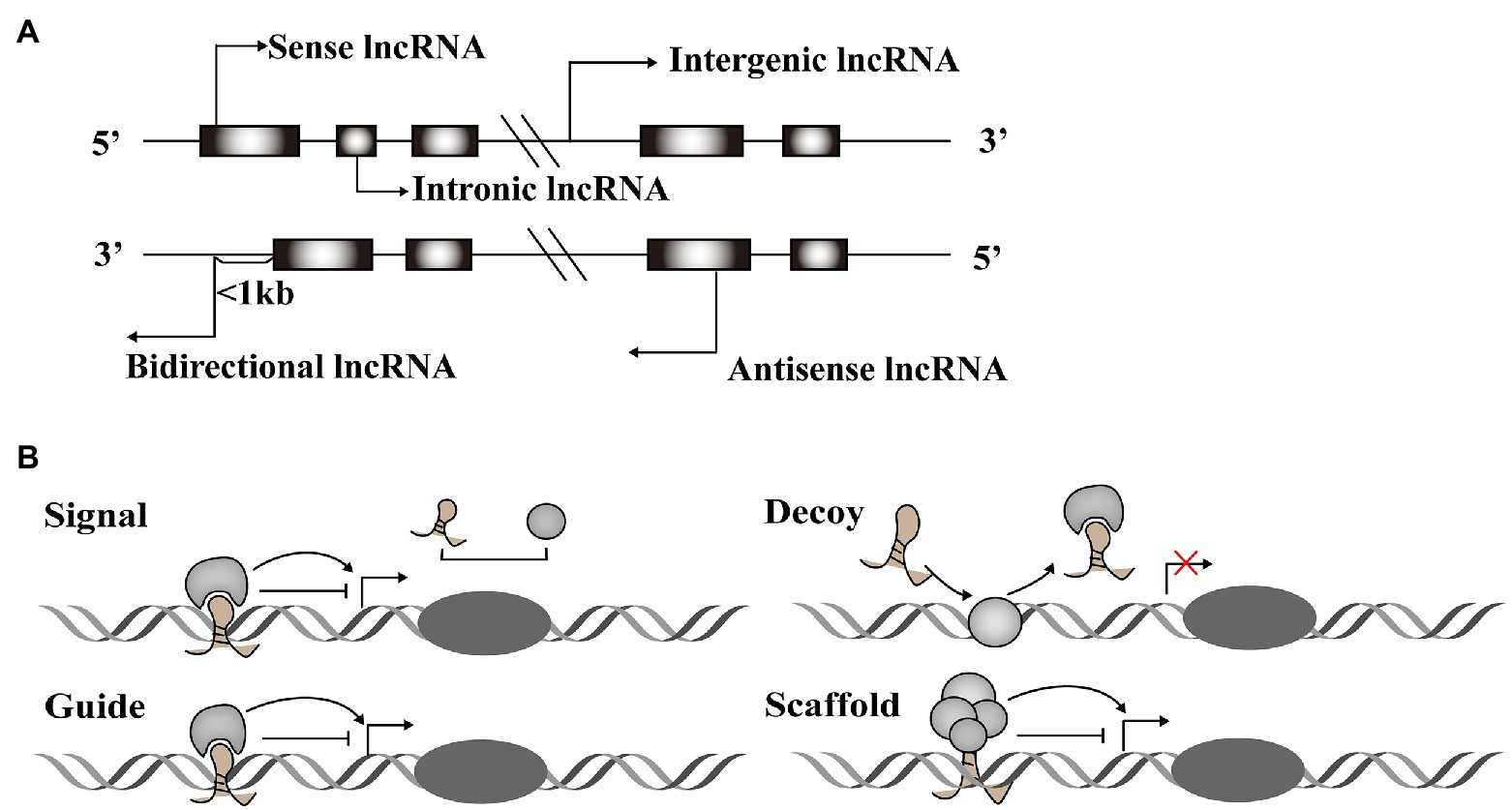
Figure 1. Classification and function modes of Long noncoding RNAs (lncRNAs). (A) LncRNAs can be classified into five categories, including sense, antisense, intergenic, intronic, and bidirectional groups according to their relative location with the protein-coding genes. (B) LncRNAs function in four archetypes. Archetype I: As signals, lncRNAs can take part in signaling pathways to regulate gene expression. Archetype II: As decoys, lncRNAs combine with transcription factors to block the pathways. Archetype III: As guides, lncRNAs direct protein complexes to specific genome sites. Archetype IV: As scaffolds, lncRNAs recruit several proteins to form ribonucleoprotein complexes.
Long noncoding RNAs have diverse structural motifs, including pseudoknots, stem-loops, G-quadruplexes, and triplexes. They mediate gene expression by interacting with DNA and mRNA in the nucleus, or miRNA and protein in the cytoplasm (Willingham et al., 2005; Ulitsky and Bartel, 2013; Sun and Kraus, 2015; Qian et al., 2019). Some lncRNAs act as molecular signals to promote transcription in various metabolic pathways. Others play modulatory roles like decoy, blocking pathways by binding transcription factors. LncRNAs also function as guides, joining with protein complexes and directing them to specific genome sites. These regulatory RNAs can also form scaffolds that recruit modifying enzymes to integrate different signaling pathways (Figure 1B). These four roles are interconnected, and a single lncRNA may exhibit different functions depending on cellular conditions (Wang and Chang, 2011). Taken together, the regulatory role of lncRNAs acts across the whole process of gene expression variability.
LncRNAs and Diseases
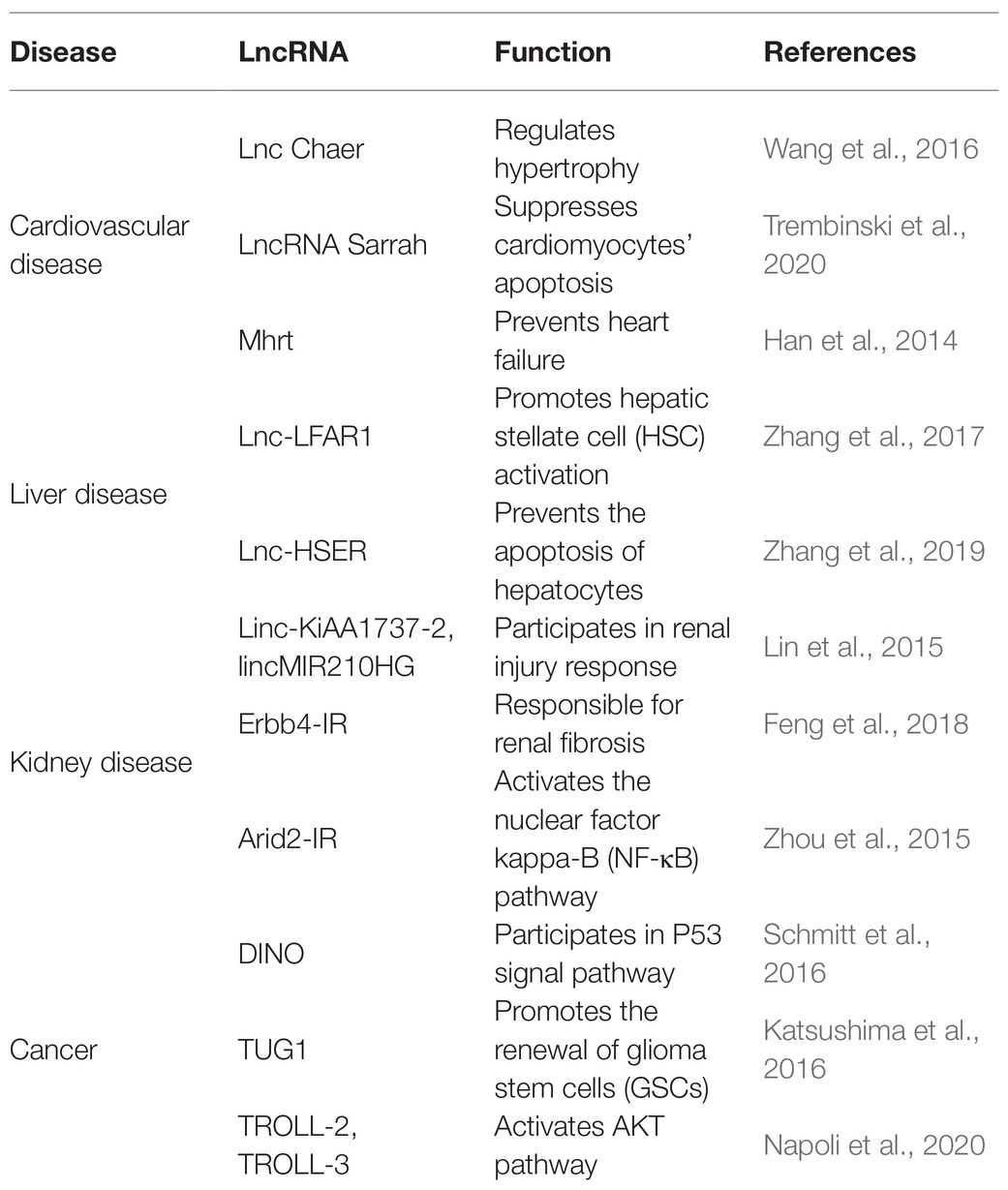
Genome-wide association studies (GWAS) have recognized thousands of single nucleotide polymorphisms (SNPs) from noncoding regions associated with clinical phenotypes, and this intimately links lncRNAs to cardiovascular, liver, and kidney disease as well as some cancers (Figure 2; Table 1; Gao and Wei, 2017; Gong et al., 2018; Hu et al., 2019).
Figure 2. Long noncoding RNAs play critical roles in the pathogenesis of cardiovascular, liver, and kidney disease as well as cancers.
Long noncoding RNAs are variably expressed in the cardiovascular system under different physiological and pathological conditions. For instance, some lncRNAs regulate apoptosis of cardiomyocytes, like lncRNA Sarrah, and hypertrophy, like lncRNA cardiac-hypertrophy-associated epigenetic regulator (Chaer). Some lncRNAs can also reduce the risk of heart failure and acute myocardial infarction. For example, Mhrt prevents Brg1, a chromatin remodeling factor, from binding its DNA targets to prevent heart failure (Han et al., 2014). Another lncRNA, ZFAS1, is a marker of acute myocardial infarction in cardiac systolic function by inhibiting the activity of SERCA2a protein (Zhang Y. et al., 2018). These functions indicate that the roles of lncRNAs are critical to cardiovascular diseases.
High-throughput technologies have also characterized some lncRNAs in liver fibrosis (Bian et al., 2019). Evidence strongly support that lncRNAs are involved in regulating protein-encoding genes in liver fibrosis. For example, Zhang et al. have demonstrated that lncLFAR1 could activate TGFβ and Notch signaling pathways to promote hepatic stellate cell (HSC) activation and liver fibrosis (Zhang et al., 2017). Lnc-HSER, specifically expressed in hepatocytes (HCs), was reported to prevent the apoptosis of hepatocytes through C5AR1-Hippo-YAP pathway and alleviate hepatic fibrosis by inhibiting the HCs epithelial-mesenchymal transition mediated by Notch signaling pathway (Zhang et al., 2019).
Long noncoding RNAs also play vital roles in kidney pathogenesis (Moghaddas Sani et al., 2018). Whole transcriptome profiling analyses identified some lncRNAs associated with acute and chronic renal injury in human proximal renal tubular epithelial cells. Among them, two highlighted lncRNAs, lnc-KiAA1737-2 and lnc-MIR210HG, might participate in renal injury response (Lin et al., 2015). Furthermore, Feng et al. (2018) reported a novel lncRNA Erbb4-IR mediated by transforming growth factor/(TGFβ)/Smad3 responsible for renal fibrosis. Zhou et al. (2015) found Arid2-ir stimulated the nuclear factor kappa-B (NF-κB)- dependent renal inflammation pathway to function in in vitro and in vivo fibrotic models.
Long noncoding RNAs also participate in the emergence and progression of cancers (Iyengar et al., 2016; Peng et al., 2017). For instance, lncRNA DINO forms a positive feed-back loop with p53 protein to amplify DNA damage signals in the nucleus (Schmitt et al., 2016). Also, activation of the Notch1 signal pathway in glioma stem cells (GSCs) specifically induced expression of the lncRNA TUG1. TUG1 functions to sponge miR-145 in cytoplasm and recruit polycomb in the nucleus to promote the renewal of GSCs (Katsushima et al., 2016). Two TAp63-regulated lncRNAs, TROLL-2, and TROLL-3, can form a trimer complex with the effector WDR26 in cytoplasm to activate AKT pathway (Napoli et al., 2020).
In summary, lncRNAs participate in different disease processes, hinting at their key roles in maintaining homeostasis of human bodies.
Key Roles of LncRNAs in Controlling Lipid Metabolism and Adipocyte Development
The Roles of LncRNAs in Controlling White Adipogenesis
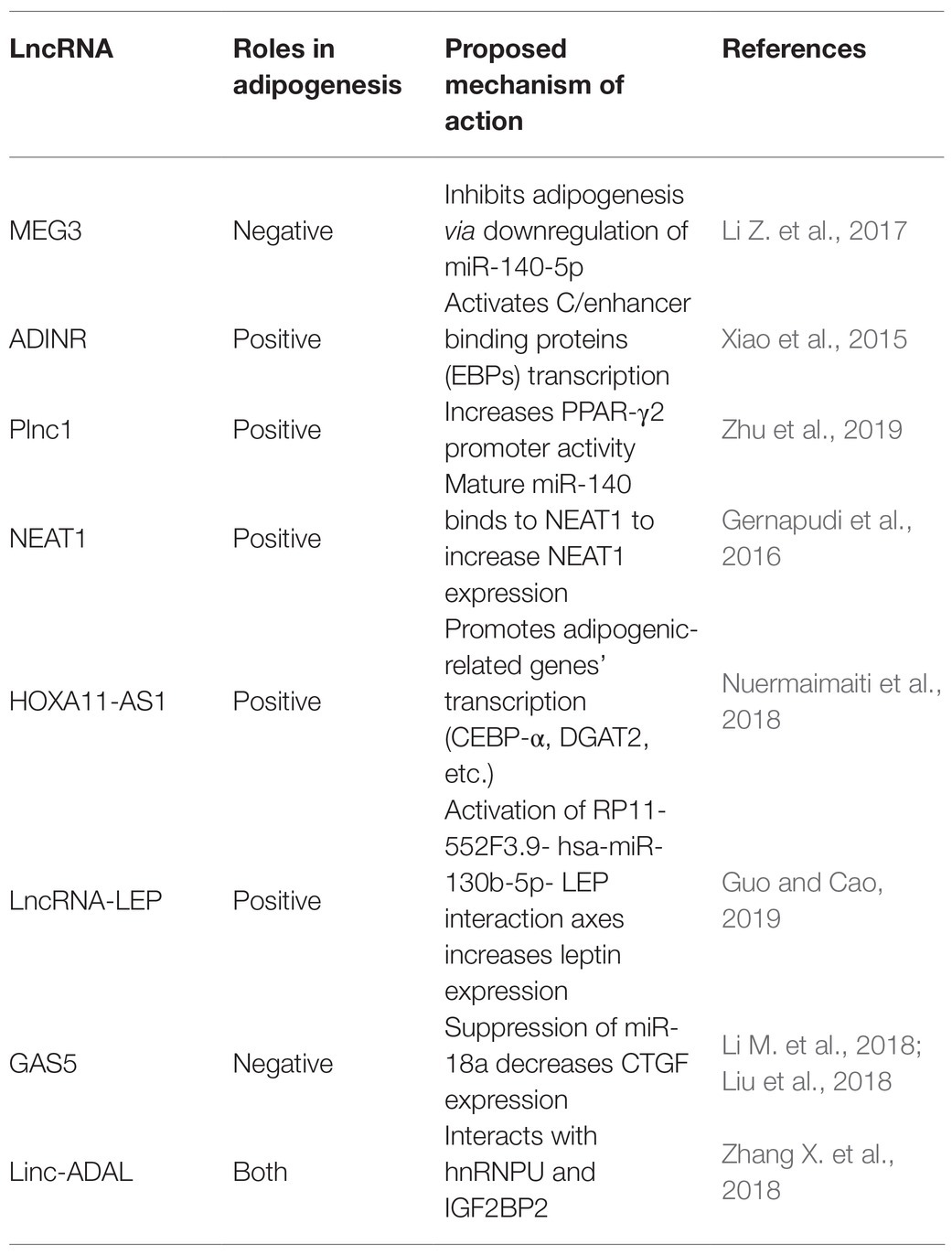
Adipogenesis mainly includes two stages. The first stage occurs when embryonic stem cells or mesenchymal stem cells in adipose tissue differentiate into adipose progenitor cells and then to preadipocytes. In the second stage, preadipocytes differentiate into mature adipocytes (Figure 3; Ali et al., 2013; Ambele et al., 2020). The whole process is accompanied by the temporal expression of many crucial adipogenesis-related genes and key transcriptional factors such as lipoprotein lipase (LPL) and sterol regulatory element binding proteins-1c (SREBP-1C). Peroxisome proliferators-activated receptor γ (PPARγ) and CCAAT/enhancer binding proteins (C/EBPs) are common markers of mature adipocytes and the major drivers of adipocyte differentiation. An increasing body of research has found that lncRNAs can regulate these pivotal genes and exert key roles in adipogenesis (Cipolletta et al., 2012; Ghaben and Scherer, 2019).
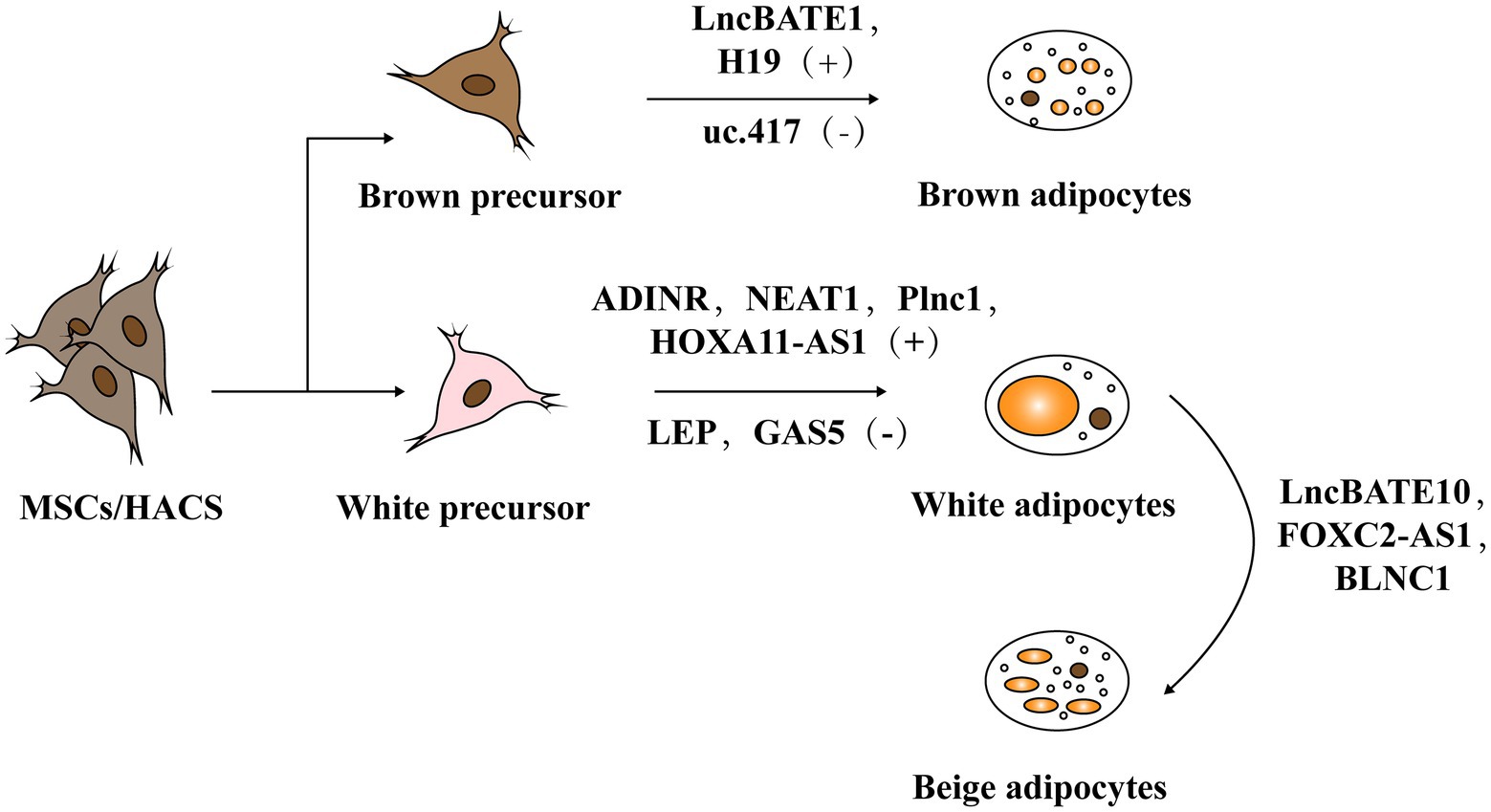
Figure 3. The role of lncRNA in adipogenesis. Adipogenesis mainly includes two stages: mesenchymal stem cells (MSCs) or human adipose tissue-derived stem cells (hASCs) in adipose tissue differentiate into adipose progenitor cells and further into preadipocytes. In the second stage, preadipocytes differentiate into mature adipocytes including white adipose tissue (WAT) and brown adipose tissue (BAT).
Human adipose tissue-derived stem cells (hASCs) have the ability to differentiate into both osteocytes and adipocytes. LncRNA MEG3 is upregulated in osteocyte differentiation and downregulated in adipocyte differentiation. Accordingly, MEG3 knockouts promote adipogenic differentiation but suppress osteogenic differentiation, suggesting that MEG3 may serve as a switch for hASCs’ adipogenic or osteogenic differentiation (Li Z. et al., 2017).
Long noncoding RNA ADINR, transcribed from a position about 450 bp upstream of the C/EBPα gene, is induced during adipogenic differentiation. Xiao et al. (2015) revealed that ADINR increases the H3K4me3 methylation but impairs the H3K27me3 histone modification of C/EBPα during adipogenic differentiation to coordinate the transcription of C/EBPα, and this finally promoted adipogenesis in hMSCs. Plnc1 was an lncRNA transcribed from a position 25,000 bp upstream of PPARγ2. In vitro studies showed that a plnc1 knockout decreases the expression of PPARγ, C/EBPα, and Fatty Acid Binding Protein 4 (AP2), which subsequently suppresses the differentiation of ST2 cells and BMSCs into mature adipocytes. However, overexpression of Plnc1 has the opposite effect. Mechanistically, plnc1 enhances the transcriptional activity of PPARγ2 by decreasing the methylation level of CPG in PPARγ2 promoter (Zhu et al., 2019).
Long noncoding RNA MIR-140 knockout decreases the expression of key transcription factors (C/EBP and PPARγ), which directly impairs the mice’s adipogenic capabilities. It suggestd that MIR-140 is a necessary regulatory factor for adipocyte differentiation. Non-coding RNA qPCR array showed that NEAT1 is highly conserved between humans and mice and is upregulated in hASC differentiation. Using RNA hybrid, Gernapudi et al. (2016) identified the miR140 binding site in NEAT1 and they found that mature miR-140 can physically interact with NEAT1 in the nucleus. Further experimental data indicated that the binding of MIR-140 to NEAT1 in the nucleus increases NEAT1 stability, thus promoting adipogenesis.
Nuermaimaiti et al. (2018) found HOXA11-AS1 knockdown inhibits the transcription of adipogenesis-related genes in hASCs differentiation model, leading to the suppression of adipogenic differentiation and alleviation of lipid accumulation. Moreover, HOXA11-AS1 is highly expressed during the process of adipocyte differentiation in obese patients, indicating that it might be a potential target for the treatment of obesity.
In addition, bioinformatics analyses were used to identify a series of key mRNAs, miRNAs, and lncRNAs in hASCs adipogenesis. Several interaction axes were observed to regulate the adipogenic differentiation of hASCs, among which the leptin (LEP)-related axis was particularly important by analyzing the region upstream and downstream of leptin gene (Guo and Cao, 2019).
Some lncRNAs are reported to be potential targets that inhibit adipogenesis. The investigation based on a cDNA chip to analyze the adipogenesis regulatory genes found that the expression of lnc-U90926 was negatively correlated with the differentiation of 3 T3-L1 preadipocyte. Then, using RNA fluorescence in situ hybridization (FISH), researchers confirmed lnc-U90926 mainly localized to the cytoplasm of mice’s preadipocytes. Gain- and loss-of-function experiments showed that the overexpression of lnc-U90926 blocked adipocyte differentiation in 3 T3-L1 as evidenced by reductions in lipid accumulation and specific protein expression, like that of PPARγ and AP2. Additionally, lnc-U90926 had lower expression levels in obese mice, which indicates it can inhibit adipogenesis by suppressing the transcriptional activity of key genes (Chen et al., 2017).
In a study on adipogenic differentiation, Li M. et al. (2018) found lncRNA GAS5 is negatively associated with adipogenesis of mesenchymal stromal cells (MSCs). Using luciferase reporter assays they further discovered that GAS5 inhibits MSCs’ adipogenic differentiation by competitively sponging miR-18a. Likewise, Liu et al. (2018) determined that GAS5 has a negative role in 3 T3 cells’ adipogenesis by repressing miR-21a-5p. Thus, GAS5 is an important regulator in the adipogenic differentiation.
LncHCG11 is another target for inhibiting adipogenesis. Li et al. (2020) found that the expression of lncHCG11 declines during the adipogenic differentiation in an in vitro hADMSCs differentiation model. Specifically, both the activity of related lipogenesis enzymes and the expressions of adipogenic proteins increase in HCG11 knockdowns, while the reverse response is observed when HCG11 is overexpressed. Bioinformatics analyses of the HCG11/miR204-5p/SIRT1 axis, in addition to experimental evidence, show that when co-transfected with a miR-204-5p mimic and pcDNA-HCG11, the miR-204-5p mimic reduced SIRT1’s inhibitory effects on the expression of lipogenesis enzymes and adipogenic marker proteins to reverse pcDNA-HCG11’s depression effects on adipogenesis.
The majority of lncRNAs are poorly conserved among mammals, many therapeutic applications necessitate that more attention is placed on identifying and characterizing lncRNAs in human adipose tissue. Zhang X. et al. (2018) performed RNA-seq on subcutaneous biopsy samples from healthy, lean humans and detected 1,001 adipose-enriched lncRNAs, among which lnc-ADAL is the most highly expressed. Lnc-ADAL is a non-conserved lncRNA closely tied to obesity. ShRNA-mediated knockouts suppressed the expression of lipid synthesis genes, while ASO-mediated knockouts not only impaired the expression of lipid synthesis genes in mature adipocytes but also damaged the preadipocyte differentiation. Researchers then verified that lnc-ADAL interacted with both the nuclear protein hnRNPU and cytoplasmic protein IGF2BP2 to control preadipocyte differentiation and de novo lipogenesis. These studies collectively support that lncRNAs emerge as important regulatory players in the process of white adipogenesis (Table 2).
The Roles of LncRNAs in Brown/Beige Fat Development and Their Function
Brown adipocytes (BAT) possess abundant mitochondria in the cytoplasm and high levels of uncoupling protein 1 (UCP1; Cannon et al., 1982). Accordingly, they can generate heat through uncoupling the lipid oxidative phosphorylation, facilitating the burning of fatty acid and glucose. Earlier studies supported that BAT is present and active in newborns to maintain their body temperature through non-shivering thermogenesis (Cannon and Nedergaard, 2004). Positron emission tomography (PET) detected considerable amounts of BAT in adult males, suggesting that BAT also plays an essential role in adult metabolism (Nedergaard et al., 2007). To date, a series of studies have indicated that lncRNAs are indispensable regulators in brown adipogenesis and thermogenesis (You et al., 2015).
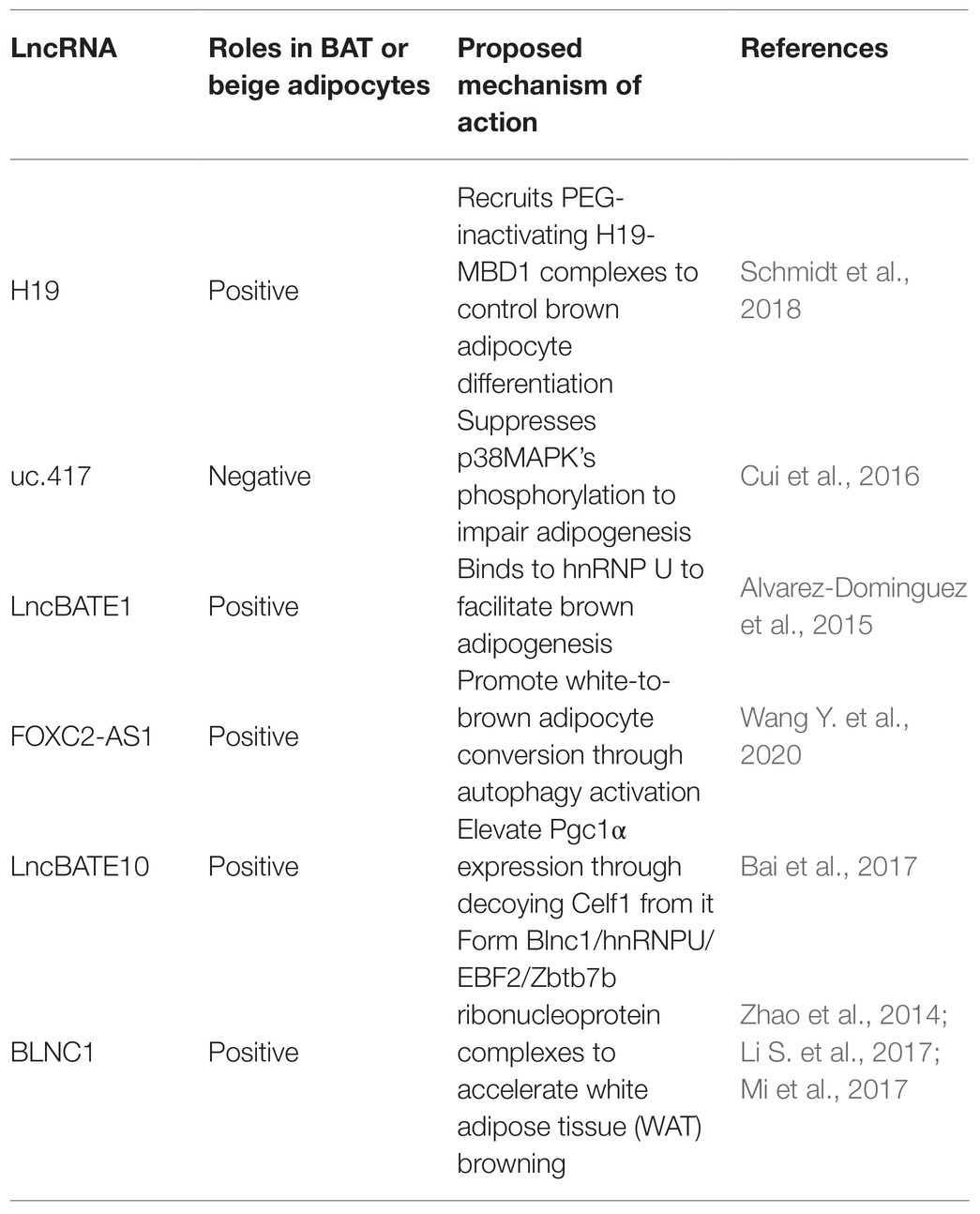
For example, H19, a maternally inherited lncRNA, is inversely correlated with Body Mass Index (BMI) in humans. Schmidt et al. (2018) reported that H19 overexpression promotes adipogenesis and mitochondrial respiration in BAT by recruiting PEG-inactivating H19-MBD1 complexes. This study illustrated the function of H19 in regulating the BAT thermogenic gene program and metabolism.
Cui et al. (2016) identified uc.417, an ultra-conserved lncRNA that is abundant in differentiated brown adipocytes. They found that overexpression of uc.417 inhibits the differentiation of brown fat cells. They also analyzed oxygen consumption of brown adipocytes with uc.417 overexpressed and evaluated the negative roles of uc.417 overexpression in BAT’s thermogenesis progress. However, knockouts of uc.417 had no significant impact on the differentiation and thermogenesis of brown adipocytes. Another lncRNA, lncBATE1, has been found to interact with hnRNP U that is necessary for brown adipogenesis and maintaining its thermogenic capacity (Alvarez-Dominguez et al., 2015). These data strongly support lncRNAs’ roles in driving brown fat formation and maintaining energy balance.
Beige fat is usually stored in white fat warehouses and can be differentiated into specific beige precursor cells in WAT or derived directly from the browning of mature white fat cells under exposure to cold or other stimuli. Because they can highly express UCP1 protein and function as BAT, induction of beige fat adipogenesis helps resist obesity and is assumed to be a promising strategy to covert unhealthy WAT into metabolically active BAT (Guerra et al., 1998; Bartelt and Heeren, 2014; Wang and Seale, 2016).
Wang Y. et al. (2020) found high expression of FOXC2-AS1 in the forskolin (Fsk)-stimulating adipocytes with high levels of UCP1 and peroxisome proliferator-activated receptorγcoactivator-1α (PGC1α). They found that FOXC2-AS1 may promote WAT browning and thermogenic program through the autophagy signaling pathway. The result showed that lncRNAs also play crucial roles in the development and functional activation of beige adipose.
In fact, some lncRNAs regulate through the co-expression network. For instance, Bai et al. (2017) found a large number of lncRNAs embedded into metabolic pathways by establishing an mRNA-lncRNA co-expression network. Through this network, they identified lncBATE10 that is rich in BAT and can decoy Celf1 from Pgc1α, activating Pgc1α expression and promoting thermogenesis and WAT browning.
Additionally, AK079912 is another BAT-enriched lncRNA. Knockdown of AK079912 decreases the expression of vital adipogenic and thermogenic genes; while overexpression upregulates the thermogenic program by increasing protein expressions of UCP1 and mitochondria electron transport chain (ETC) complexes. Moreover, Xiong et al. (2018) found expression of AK079912 in inguinal WAT could induce their browning.
Blnc1 is rich in both the brown and beige adipocytes. Over-expression of BLNC1 in brown adipocytes increases the expression of thermogenic genes through the formation of the Blnc1/hnRNPU/EBF2 ribonucleoprotein complex (Mi et al., 2017). The effects of Blnc1 on beige adipocytes were also evaluated. During brown fat whitening induced by a high-fat diet (HFD), specific deactivation of Blnc1 in the fat tissue not only accelerated the BAT to bleach, but also exacerbated the inflammation. However, fat-specific Blnc1 transgenic mice have the opposite effects. The molecular mechanism is that Blnc1, as a target of EBF2, built a feedforward regulatory loop to promote browning of WAT (Zhao et al., 2014). Additionally, Lin et al. (2015) found that BTB domain-containing 7b (Zbtb7b) could recruit the lncRNA Blnc1 through hnRNP U to increase thermogenic genes expression, and the function of Blnc1 is conserved in mice and humans (Li S. et al., 2017).
Thus, lncRNAs are important regulators for activating brown/beige adipocytes to function with the benefit of decreasing serum triglycerides and fighting against obesity (Table 3).
LncRNA and Lipid Homeostasis in the Liver
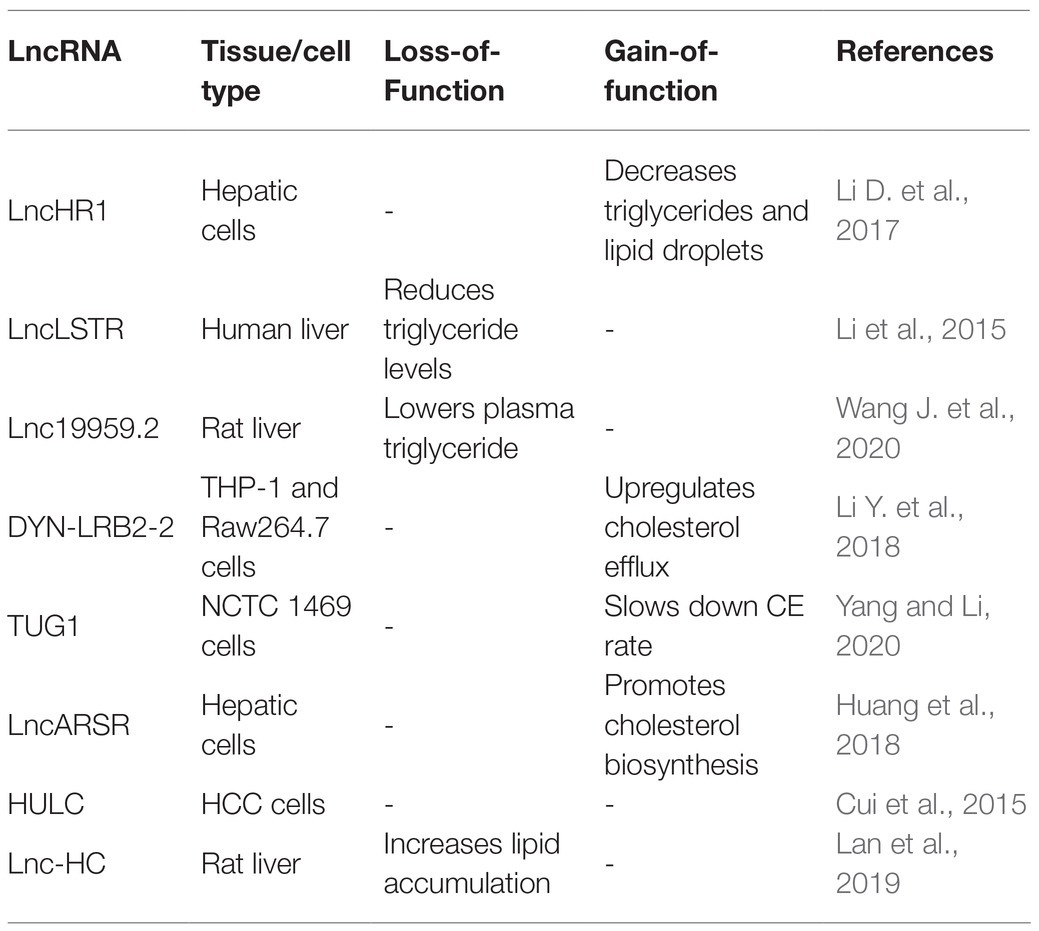
Besides the known roles in adipocytes, a series of studies have shown that lncRNAs regulate lipid metabolism in the liver by targeting several crucial transcription factors, such as liver X receptor (LXRs), sterol-regulatory element binding proteins (SREBPs), and peroxisome proliferation-activated receptor α (PPARα). These transcription factors are regulators of gene expression for the synthesis and uptake of cholesterol, fatty acids, and phospholipids in the liver (Sun and Lin, 2019). Here, we summarize the regulatory mechanisms of lncRNAs in liver lipid homeostasis (Table 4).
LncHR1, identified in human hepatoma cells infected with HCV, negatively regulates the expression of SREBP-1c and fatty acid synthase. Overexpression of lncHR1 inhibits the accumulation of triglyceride and lipid droplets in liver cells (Li D. et al., 2017). Li et al. (2015) found liver-enriched lncLSTR could decrease the plasma triglyceride levels (TDP43/FXR/APOC2) by competitively binding with TDP-43 to regulate the expression of Cyp8b1, leading to the activation of Apoc2 via the nuclear receptor farnesoid-X-receptor (FXR) pathway.
Additionally, Wang J. et al. (2020) found that Lnc19959.2 was upregulated in high fat-induced hepatocytes. Mechanically, overexpressed lnc19959.2 promotes the expression of ApoA4 by interacting with nuclear protein Purb. Lnc19959.2 specifically binds to hnRNPA2B1 to negatively regulate the expression of genes related to TG metabolism. Taken together, the investigations of lncHR1 and lnc19959.2 indicate that lncRNAs specifically expressed in liver are emerging as key players in the regulation of triglycerides.
Long noncoding RNAs are also closely related to the cholesterol metabolism. Hu et al. (2019) found that ox-LDL could significantly upregulate the expression of lincRNA-DYN-LRB2-2, directly leading to an increased expression of ATP-binding cassette transporter A1 (ABCA1). Elevated expression of ABCA1 mediated cholesterol efflux (CE) in foam cells, thus reducing cholesterol levels (Li Y. et al., 2018). Another study found that lncTUG1 inhibited CE by inhibiting the expression of APOM in an miR-92a/FXR1 dependent manner (Yang and Li, 2020). Additionally, Huang et al. (2018) found overexpression of lncARSR can activate PI3K/Akt signal pathway, promoting the expression of transcription factor SREBP2. This transcription factor, in turn, increased the expression level of the rate-liming enzyme in cholesterol, HMG-CoA reductase (HMGCR), and accelerated cholesterol biosynthesis in liver.
Another group of lncRNAs can influence both the cholesterol and triglyceride level. HULC was reported to mediate abnormal lipid metabolism in hepatocellular carcinoma and elevate the levels of intracellular triglycerides and cholesterol by activating ACSL1/miR-9/PPARA signaling pathway (Cui et al., 2015). Moreover, Lan et al. (2019) identified a novel noncoding RNA lnc-HC that not only reduced cholesterol efflux by inhibiting the expression of cholesterol 7α-hydroxylase (Cyp7a1) and ABCA1, but also promoted hepatic triglyceride metabolism by negatively regulating PPARγ expression. Overall, lncRNAs play vital roles in maintaining lipid homeostasis in the liver and emerge as important targets to alleviate liver diseases caused by fat metabolic disorders, such as nonalcoholic fatty liver disease.
The Potential of Lncrnas As Therapeutic Targets of Related Diseases Induced By Adipocyte Dysfunction
Obesity is becoming a global pandemic and usually leads to some metabolic diseases, including nonalcoholic fatty liver disease (NAFLD), diabetes, and other diabetic complications (Kusminski et al., 2009). However, therapeutic targets and methods for the treatment of obesity and related metabolic diseases remain limited in the clinic (Kakkar and Dahiya, 2015). The existing approved drugs mainly function to combat obesity by reducing intestinal fat absorption or suppressing appetite (Tsilingiris et al., 2020). Treatments to improve related metabolic disease are mainly through the use of drugs with hypoglycemic, anti-hypertensive or lipid-lowering activity. However, these agents usually have larger side effects on human body and their protective effect is limited. For examples, statins commonly cause muscular adverse reactions such as fatal rhabdomyolysis (Bouitbir et al., 2020). Chronic administration of niacin can decrease glucose tolerance and increase uric acid level, potentially induce gouty attacks and impaired liver function (Kei et al., 2011). Thus, there is an urgent need to identify novel targets and develop new effective and safe drug candidates. Functional studies of lncRNAs provide new insight for the establishment of related drugs.
The Roles of LncRNAs in Insulin Resistance
Low-grade chronic inflammation, as a marker of obesity, has been identified as a vital risk factor for the occurrence of insulin resistance (Glass and Olefsky, 2012). The increased free fatty acids in the obese can promote NF-κB signaling, which upregulates the expression of pro-inflammatory cytokines, such as TNF-α and IL-6 (Tangvarasittichai, 2015). The pro-inflammatory signals then inhibit the function of insulin in metabolic tissue, thereby mediating insulin resistance.
Because the inflammation reaction is a phenotype caused by macrophages responding to excessive lipid accumulation, researchers have focused on macrophages to reduce this inflammation. These studies have found several novel lncRNAs enriched in macrophages and differentially expressed in diet-induced mice models with obesity and early diabetes. For example, Stapleton et al. (2020) found that MIST was associated with a macrophage anti-inflammatory phenotype during gain- and loss-of-function experiments. When transcription or interaction with RNAs of MIST was disrupted, expression of inflammatory genes heavily increased. They then discovered that MIST interacted with poly ADP-ribose polymerase (PARP1) in the nucleus. It may act as a protective lncRNA by interfering with the formation of pro-inflammatory cytokines that are closely correlated with insulin sensitivity index. In addition, Zhang et al. (2020) identified lncRNA uc.333 that improved obesity-induced insulin resistance by binding to miR-223. Moreover, Liu S. et al. (2014) reported that knocking out lncRNA SRA protected mice against high fat diet-induced obesity and improved their glucose tolerance. All the above indicate that lncRNAs are potential therapeutic targets for improving insulin resistance.
The Roles of LncRNAs in Hepatic Steatosis
Metabolic disorders often cause non-alcoholic fatty liver disease (NAFLD) and hepatic steatosis (NASH) that are characterized by the accumulation of liver lipids (Kawano and Cohen, 2013). If NAFLD is not treated in time, it can gradually develop into NASH and then into hepatocellular cancer or other malignant diseases (Kanwal et al., 2018; Lindenmeyer and McCullough, 2018). LncRNAs may be checkpoints to enable unhealthy hepatic lipogenesis and impair liver lipid homeostasis. In an animal model with NAFLD, Wang (2018) found that lncRNA-NEAT1 knockdown may alleviate the NAFLD via regulating the mTOR/S6K1 signaling pathway. Another lncRNA, MALAT1, is highly expressed in livers of ob/ob mice. Mechanism research demonstrated that inhibiting MALAT1 suppresses hepatic lipid accumulation and attenuates hepatic steatosis by reducing the stability of nuclear SREBP-1c protein in hepatocytes (Yan et al., 2016). Considering that hepatic expression of BLNC1 is evidently elevated in the obesity and NAFLD in mice, researchers studied the effects of BLNC1 on HFD – induced hepatic steatosis. They found that BlNC1 deficiency greatly inhibits both the plasma TAG levels and the induction of SREBP1 protein expression by LXR agonists. Additionally, liver-specific BLNC1 knockout mice exhibited resistance to HFD-induced hepatic steatosis, manifested as reduced hepatic damage and fibrosis. These responses indicated that BLNC1 may work cooperatively with LXR to control hepatic lipid metabolism, which may serve as a therapeutic target for the treatment of NAFLD patients (Zhao et al., 2018).
The Roles of LncRNAs in Atherosclerosis
The pathogenesis of atherosclerosis is complex, although current research as established that dyslipidemia (abnormal plasma cholesterol and lipoprotein levels) is one of the crucial risk factors (Gisterå and Ketelhuth, 2018). Since many studies have confirmed lncRNAs’ regulatory roles in lipid homeostasis, some researchers think they may take part in the development of atherosclerosis. LncRNA KCNQ1OT1 could inhibit cholesterol efflux and promote lipid accumulation in macrophages via the miR-452-3p/HDAC3/ABCA1 pathway, and, thus, contribute to the development of atherosclerosis (Yu et al., 2020). A key node in atherosclerosis is when macrophages uptake lipoproteins and form foam cells (Tabas and Bornfeldt, 2020). Kanwal et al. (2018) found lncRNA E330013P06 upregulated the expression of CD36 in macrophages to promote foam cells formation (Reddy et al., 2014). Another lncRNA, AT102202, controls the expression of mRNA-3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) to affect the accumulation of cholesterol in macrophages (Liu et al., 2015). Although the function and mechanism of lncRNAs in atherosclerosis still need further exploration, lncRNAs are apparently vital regulatory factors implicated in the pathological process of atherosclerosis. LncRNAs have may have future clinical applications as biomarkers and potential therapeutic targets of atherosclerosis.
The Roles of LncRNAs in Diabetic Complications
Epidemiological studies have reported a large increase in the prevalence of diabetes, which mainly happens among people with long-term abdominal obesity (Ampofo and Boateng, 2020). The main harm of diabetes lies in its severe complications, such as diabetic retinopathy, nephropathy, cardiomyopathy, etc.; however, the therapeutic targets and drugs still remain limited (Harding et al., 2019). LncRNAs recently gained attention for their regulatory roles in diabetic complications (Figure 4).
Diabetic retinopathy is a common complication caused by hyperglycemia and dyslipidemia. At present, retinal gene expression profiles have identified more than 300 differentially expressed lncRNAs associated with diabetic retinopathy (Yan et al., 2014). One study showed that an lncRNA MALAT1 knockdown can inhibit the proliferation, migration, and tube formation of retinal endothelial cells (Liu J.-Y. et al., 2014). Radhakrishnan and Kowluru (2021) verified MALAT1 suppression attenuates oxidative damage via the Keap1-Nrf2 pathway to improve retinal vascular function and slow diabetic retinopathy. Additionally, Yan et al. found lncRNA MIAT can form a feedback loop with VEGF and miR-150-5p to regulate endothelial cell function and improve the microvascular dysfunction induced by diabetes (Yan et al., 2015). Thus, lncRNAs are involved in the development of diabetic retinopathy and may be potential therapeutic targets for the disease.
Chronic hyperglycemia and dyslipidemia are also the main causes of diabetic nephropathy, a microvascular complication characterized by the damage of glomerular capillaries (Opazo-Ríos et al., 2020). Accumulating evidence supports lncRNAs’ involvement in the occurrence and development of this disease.
For example, LncRNA Tug1 was found to interact with PGC-1 to regulate its expression and affect the mitochondrial bioenergetics in podocytes (Long et al., 2016). Additionally, in type II diabetic patients with end-stage renal disease, lncPVT1 controlled the accumulation of the extracellular matrix and the progression of renal cells fibrosis, thereby mediating the development of diabetic kidney disease (Alvarez and DiStefano, 2011).
Diabetic cardiomyopathy is mainly caused by cardiac lipotoxicity (Nakamura and Sadoshima, 2020). LncRNA GAS5 regulates the miR-34b-3p/AHR pathway to repress the pyroptosis induced by NLRP3 inflammasome activation, making GAS5 a potential therapeutic target (Xu et al., 2020). Wang et al. (2021) found that silencing lncRNA MALAT1 could inhibit EZH2 expression via the EZH2/miR-22/ABCA1 signaling axis, which prevents cardiomyocyte apoptosis and attenuates cardiac dysfunction. Above all, many studies have demonstrated that lncRNAs play functional roles in the pathological processes of diabetic complications and have potential therapeutic significance for the diseases.
Conclusion
This review mainly summarized the studies on the regulation of lncRNAs in lipid metabolism in the liver as well as the development and function of adipose tissue. Meanwhile, numerous examples have been provided of a series of lncRNAs involved in adipocyte dysfunction-induced diseases, such as insulin resistance, hepatic steatosis, and diabetic complications. Currently, most of the drugs used to fight obesity target proteins, but these drugs have side effects because of the unintended regulation of non-target protein. Thus, the development of new drugs that target nucleic acids might provide a novel therapeutic strategy for the treatment of obesity and its related diseases.
Some targeted nucleic acid therapies, such as antibacterial and anticancer therapy are gradually being applied to treat some diseases (Aradi et al., 2020; Javanmard et al., 2020). Nucleic acid targeting methods are considered the third generation of therapeutic drugs, and three main strategies have been reported thus far: (i) small interfering RNA (siRNA), targeting cytoplasmic lncRNAs, like Givlaari (Givosiran) which was approved to treat adult acute hepatic porphyria; (ii) antisense oligonucleotides (ASO), targeting nuclear lncRNAs. LNA gapmeR ASO-targeting lncRNA MALAT1 possesses anti-multiple myeloma activity (Amodio et al., 2018); (iii) CRISPR/Cas9 technology is suitable for dual-located lncRNAs, and it has been widely used in the discovery and annotation of lncRNAs but, as of yet, is not ideal for systematic drug delivery. These three therapies have been approved for clinical application, but all face off-target problems (Caffrey et al., 2011; Fu et al., 2014; Rinaldi and Wood, 2018).
Some lncRNAs positively regulate white adipogenesis and are upregulated in the obese patients, and they might be suitable ASO targets. However, no lncRNA targeting drugs have entered clinical trials, and many therapies are still in the preclinical stage. Because many lncRNAs are poorly conserved, researchers often struggle to transfer successful mouse model experiments to human treatments. Thus, more humanized lncRNAs remain to be probed and more applicable preclinical study models need to be established. On the other hand, the regulation network of lncRNAs is complex and it is not easy to achieve accurate regulation in vivo. Accordingly, it is necessary to establish a highly organized lncRNA research database, D-LNC platform is such an attempt to query and analyze the modification effects of drugs on the expression of lncRNAs (Jiang et al., 2019).
To sum up, the study of lncRNAs in adipose metabolism and obesity-caused diseases, as well as the therapeutic strategies presented above, may provide novel medication for the treatment of obesity and related metabolic diseases.
Author Contributions
BZ, YX, and SuX contributed to the conception of the review. BZ and SaX contributed significantly to the complete manuscript preparation. SaX and JL contributed to constructive discussions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Sciences Foundation for Young Scientists of China (grant no. 81803803) and the CAMS Innovation Fund for Medical Sciences (CIFMS; grant no. 2019-I2M-1-005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ali, A. T., Hochfeld, W. E., Myburgh, R., and Pepper, M. S. (2013). Adipocyte and adipogenesis. Eur. J. Cell Biol. 92, 229–236. doi: 10.1016/j.ejcb.2013.06.001
Alvarez, M. L., and DiStefano, J. K. (2011). Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 6:e18671. doi: 10.1371/journal.pone.0018671
Alvarez-Dominguez, J. R., Bai, Z., Xu, D., Yuan, B., Lo, K. A., Yoon, M. J., et al. (2015). De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. doi: 10.1016/j.cmet.2015.04.003
Ambele, M. A., Dhanraj, P., Giles, R., and Pepper, M. S. (2020). Adipogenesis: a complex interplay of multiple molecular determinants and pathways. Int. J. Mol. Sci. 21:4283. doi: 10.3390/ijms21124283
Amodio, N., Stamato, M. A., Juli, G., Morelli, E., Fulciniti, M., Manzoni, M., et al. (2018). Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 32, 1948–1957. doi: 10.1038/s41375-018-0067-3
Ampofo, A. G., and Boateng, E. B. (2020). Beyond 2020: modelling obesity and diabetes prevalence. Diabetes Res. Clin. Pract. 167:108362. doi: 10.1016/j.diabres.2020.108362
Aradi, K., Di Giorgio, A., and Duca, M. (2020). Aminoglycoside conjugation for RNA targeting: antimicrobials and beyond. Chemistry 26, 12273–12309. doi: 10.1002/chem.202002258
Bai, Z., Chai, X. R., Yoon, M. J., Kim, H. J., Lo, K. A., Zhang, Z. C., et al. (2017). Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 15:e2002176. doi: 10.1371/journal.pbio.2002176
Bartelt, A., and Heeren, J. (2014). Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 10, 24–36. doi: 10.1038/nrendo.2013.204
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155. doi: 10.1038/351153a0
Bian, E. B., Xiong, Z. G., and Li, J. (2019). New advances of lncRNAs in liver fibrosis, with specific focus on lncRNA-miRNA interactions. J. Cell. Physiol. 234, 2194–2203. doi: 10.1002/jcp.27069
Blüher, M. (2019). Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298. doi: 10.1038/s41574-019-0176-8
Bouitbir, J., Sanvee, G. M., Panajatovic, M. V., Singh, F., and Krähenbühl, S. (2020). Mechanisms of statin-associated skeletal muscle-associated symptoms. Pharmacol. Res. 154:104201. doi: 10.1016/j.phrs.2019.03.010
Brannan, C. I., Dees, E. C., Ingram, R. S., and Tilghman, S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36. doi: 10.1128/mcb.10.1.28
Caffrey, D. R., Zhao, J., Song, Z., Schaffer, M. E., Haney, S. A., Subramanian, R. R., et al. (2011). siRNA off-target effects can be reduced at concentrations that match their individual potency. PLoS One 6:e21503. doi: 10.1371/journal.pone.0021503
Cannon, B., Hedin, A., and Nedergaard, J. (1982). Exclusive occurrence of thermogenin antigen in brown adipose tissue. FEBS Lett. 150, 129–132. doi: 10.1016/0014-5793(82)81319-7
Cannon, B., and Nedergaard, J. (2004). Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359. doi: 10.1152/physrev.00015.2003
Chen, J., Liu, Y., Lu, S., Yin, L., Zong, C., Cui, S., et al. (2017). The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int. J. Obes. 41, 299–308. doi: 10.1038/ijo.2016.189
Chooi, Y. C., Ding, C., and Magkos, F. (2019). The epidemiology of obesity. Metabolism 92, 6–10. doi: 10.1016/j.metabol.2018.09.005
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E., and Chang, H. Y. (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. doi: 10.1016/j.molcel.2011.08.027
Cipolletta, D., Feuerer, M., Li, A., Kamei, N., Lee, J., Shoelson, S. E., et al. (2012). PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553. doi: 10.1038/nature11132
Core, L. J., Waterfall, J. J., and Lis, J. T. (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. doi: 10.1126/science.1162228
Cui, M., Xiao, Z., Wang, Y., Zheng, M., Song, T., Cai, X., et al. (2015). Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857. doi: 10.1158/0008-5472.CAN-14-1192
Cui, X., You, L., Li, Y., Zhu, L., Zhang, F., Xie, K., et al. (2016). A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. 30, 4301–4312. doi: 10.1096/fj.201600694R
Feng, M., Tang, P. M., Huang, X. R., Sun, S. F., You, Y. K., Xiao, J., et al. (2018). TGF-β mediates renal fibrosis via the Smad3-Erbb4-IR long noncoding RNA Axis. Mol. Ther. 26, 148–161. doi: 10.1016/j.ymthe.2017.09.024
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808
Gao, P., and Wei, G. H. (2017). Genomic insight into the role of lncRNA in cancer susceptibility. Int. J. Mol. Sci. 18:1239. doi: 10.3390/ijms18061239
Gernapudi, R., Wolfson, B., Zhang, Y., Yao, Y., Yang, P., Asahara, H., et al. (2016). MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol. Cell. Biol. 36, 30–38. doi: 10.1128/MCB.00702-15
Ghaben, A. L., and Scherer, P. E. (2019). Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 20, 242–258. doi: 10.1038/s41580-018-0093-z
Gisterå, A., and Ketelhuth, D. F. J. (2018). Lipid-driven immunometabolic responses in atherosclerosis. Curr. Opin. Lipidol. 29, 375–380. doi: 10.1097/MOL.0000000000000540
Glass, C. K., and Olefsky, J. M. (2012). Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 15, 635–645. doi: 10.1016/j.cmet.2012.04.001
Gong, Z., Tang, J., Xiang, T., Lin, J., Deng, C., Peng, Y., et al. (2018). Genome-wide identification of long noncoding RNAs in CCl4-induced liver fibrosis via RNA sequencing. Mol. Med. Rep. 18, 299–307. doi: 10.3892/mmr.2018.8986
Guerra, C., Koza, R. A., Yamashita, H., Walsh, K., and Kozak, L. P. (1998). Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 102, 412–420. doi: 10.1172/JCI3155
Guo, Z., and Cao, Y. (2019). An lncRNA-miRNA-mRNA ceRNA network for adipocyte differentiation from human adipose-derived stem cells. Mol. Med. Rep. 19, 4271–4287. doi: 10.3892/mmr.2019.10067
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Han, P., Li, W., Lin, C. H., Yang, J., Shang, C., Nuernberg, S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. doi: 10.1038/nature13596
Harding, J. L., Pavkov, M. E., Magliano, D. J., Shaw, J. E., and Gregg, E. W. (2019). Global trends in diabetes complications: a review of current evidence. Diabetologia 62, 3–16. doi: 10.1007/s00125-018-4711-2
Hu, W., Ding, H., Ouyang, A., Zhang, X., Xu, Q., Han, Y., et al. (2019). LncRNA MALAT1 gene polymorphisms in coronary artery disease: a case-control study in a Chinese population. Biosci. Rep. 39:BSR20182213. doi: 10.1042/bsr20182213
Huang, J., Chen, S., Cai, D., Bian, D., and Wang, F. (2018). Long noncoding RNA lncARSR promotes hepatic cholesterol biosynthesis via modulating Akt/SREBP-2/HMGCR pathway. Life Sci. 203, 48–53. doi: 10.1016/j.lfs.2018.04.028
Iyengar, N. M., Gucalp, A., Dannenberg, A. J., and Hudis, C. A. (2016). Obesity and cancer mechanisms: tumor microenvironment and inflammation. J. Clin. Oncol. 34, 4270–4276. doi: 10.1200/JCO.2016.67.4283
Jarroux, J., Morillon, A., and Pinskaya, M. (2017). History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 1008, 1–46. doi: 10.1007/978-981-10-5203-3_1
Javanmard, S. H., Vaseghi, G., Ghasemi, A., Rafiee, L., Ferns, G. A., Esfahani, H. N., et al. (2020). Therapeutic inhibition of microRNA-21 (miR-21) using locked-nucleic acid (LNA)-anti-miR and its effects on the biological behaviors of melanoma cancer cells in preclinical studies. Cancer Cell Int. 20:384. doi: 10.1186/s12935-020-01394-6
Jiang, W., Qu, Y., Yang, Q., Ma, X., Meng, Q., Xu, J., et al. (2019). D-lnc: a comprehensive database and analytical platform to dissect the modification of drugs on lncRNA expression. RNA Biol. 16, 1586–1591. doi: 10.1080/15476286.2019.1649584
Kakkar, A. K., and Dahiya, N. (2015). Drug treatment of obesity: current status and future prospects. Eur. J. Intern. Med. 26, 89–94. doi: 10.1016/j.ejim.2015.01.005
Kanwal, F., Kramer, J. R., Mapakshi, S., Natarajan, Y., Chayanupatkul, M., Richardson, P. A., et al. (2018). Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology 155, 1828.e1822–1837.e1822. doi: 10.1053/j.gastro.2018.08.024
Katsushima, K., Natsume, A., Ohka, F., Shinjo, K., Hatanaka, A., Ichimura, N., et al. (2016). Targeting the notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 7:13616. doi: 10.1038/ncomms13616
Kawano, Y., and Cohen, D. E. (2013). Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 48, 434–441. doi: 10.1007/s00535-013-0758-5
Kei, A., Liberopoulos, E. N., and Elisaf, M. S. (2011). What restricts the clinical use of nicotinic acid? Curr. Vasc. Pharmacol. 9, 521–530. doi: 10.2174/157016111796197215
Kusminski, C. M., Shetty, S., Orci, L., Unger, R. H., and Scherer, P. E. (2009). Diabetes and apoptosis: lipotoxicity. Apoptosis 14, 1484–1495. doi: 10.1007/s10495-009-0352-8
Lan, X., Wu, L., Wu, N., Chen, Q., Li, Y., Du, X., et al. (2019). Long noncoding RNA lnc-HC regulates PPARγ-mediated hepatic lipid metabolism through miR-130b-3p. Mol. Ther. Nucleic Acids 18, 954–965. doi: 10.1016/j.omtn.2019.10.018
Lavie, C. J., Milani, R. V., and Ventura, H. O. (2009). Obesity and cardiovascular disease: risk factor, paradox, and impact of weight loss. J. Am. Coll. Cardiol. 53, 1925–1932. doi: 10.1016/j.jacc.2008.12.068
Li, D., Cheng, M., Niu, Y., Chi, X., Liu, X., Fan, J., et al. (2017). Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int. J. Biol. Sci. 13, 349–357. doi: 10.7150/ijbs.16635
Li, Z., Jin, C., Chen, S., Zheng, Y., Huang, Y., Jia, L., et al. (2017). Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol. Cell. Biochem. 433, 51–60. doi: 10.1007/s11010-017-3015-z
Li, D., Liu, Y., Gao, W., Han, J., Yuan, R., Zhang, M., et al. (2020). LncRNA HCG11 inhibits adipocyte differentiation in human adipose-derived mesenchymal stem cells by sponging miR-204-5p to upregulate SIRT1. Cell Transplant. 29:963689720968090. doi: 10.1177/0963689720968090
Li, S., Mi, L., Yu, L., Yu, Q., Liu, T., Wang, G. X., et al. (2017). Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc. Natl. Acad. Sci. U. S. A. 114, E7111–E7120. doi: 10.1073/pnas.1703494114
Li, P., Ruan, X., Yang, L., Kiesewetter, K., Zhao, Y., Luo, H., et al. (2015). A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 21, 455–467. doi: 10.1016/j.cmet.2015.02.004
Li, Y., Shen, S., Ding, S., and Wang, L. (2018). LincRNA DYN-LRB2-2 upregulates cholesterol efflux by decreasing TLR2 expression in macrophages. J. Cell. Biochem. 119, 1911–1921. doi: 10.1002/jcb.26352
Li, M., Xie, Z., Wang, P., Li, J., Liu, W., Tang, S., et al. (2018). The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 9:554. doi: 10.1038/s41419-018-0627-5
Lin, J., Zhang, X., Xue, C., Zhang, H., Shashaty, M. G., Gosai, S. J., et al. (2015). The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am. J. Physiol. Ren. Physiol. 309, F901–F913. doi: 10.1152/ajprenal.00290.2015
Lindenmeyer, C. C., and McCullough, A. J. (2018). The natural history of nonalcoholic fatty liver disease-an evolving view. Clin. Liver Dis. 22, 11–21. doi: 10.1016/j.cld.2017.08.003
Liu, H., Li, H., Jin, L., Li, G., Hu, S., Ning, C., et al. (2018). Long noncoding RNA GAS5 suppresses 3T3-L1 cells adipogenesis through miR-21a-5p/PTEN signal pathway. DNA Cell Biol. 37, 767–777. doi: 10.1089/dna.2018.4264
Liu, S., Sheng, L., Miao, H., Saunders, T. L., MacDougald, O. A., Koenig, R. J., et al. (2014). SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 289, 13000–13009. doi: 10.1074/jbc.M114.564658
Liu, J.-Y., Yao, J., Li, X. M., Song, Y. C., Wang, X. Q., Li, Y. J., et al. (2014). Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 5:e1506. doi: 10.1038/cddis.2014.466
Liu, G., Zheng, X., Xu, Y., Lu, J., Chen, J., and Huang, X. (2015). Long non-coding RNAs expression profile in HepG2 cells reveals the potential role of long non-coding RNAs in the cholesterol metabolism. Chin. Med. J. 128, 91–97. doi: 10.4103/0366-6999.147824
Long, J., Badal, S. S., Ye, Z., Wang, Y., Ayanga, B. A., Galvan, D. L., et al. (2016). Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 126, 4205–4218. doi: 10.1172/JCI87927
Mattick, J. S., and Rinn, J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7. doi: 10.1038/nsmb.2942
Mi, L., Zhao, X. Y., Li, S., Yang, G., and Lin, J. D. (2017). Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 6, 101–110. doi: 10.1016/j.molmet.2016.10.010
Moghaddas Sani, H., Hejazian, M., Hosseinian Khatibi, S. M., Ardalan, M., and Zununi Vahed, S. (2018). Long non-coding RNAs: an essential emerging field in kidney pathogenesis. Biomed. Pharmacother. 99, 755–765. doi: 10.1016/j.biopha.2018.01.122
Nakamura, M., and Sadoshima, J. (2020). Cardiomyopathy in obesity, insulin resistance and diabetes. J. Physiol. 598, 2977–2993. doi: 10.1113/JP276747
Napoli, M., Li, X., Ackerman, H. D., Deshpande, A. A., Barannikov, I., Pisegna, M. A., et al. (2020). Pan-cancer analysis reveals TAp63-regulated oncogenic lncRNAs that promote cancer progression through AKT activation. Nat. Commun. 11:5156. doi: 10.1038/s41467-020-18973-w
Nedergaard, J., Bengtsson, T., and Cannon, B. (2007). Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293, E444–E452. doi: 10.1152/ajpendo.00691.2006
Nuermaimaiti, N., Liu, J., Liang, X., Jiao, Y., Zhang, D., Liu, L., et al. (2018). Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem. Biophys. Res. Commun. 495, 1878–1884. doi: 10.1016/j.bbrc.2017.12.006
Opazo-Ríos, L., Mas, S., Marín-Royo, G., Mezzano, S., Gómez-Guerrero, C., Moreno, J. A., et al. (2020). Lipotoxicity and diabetic nephropathy: novel mechanistic insights and therapeutic opportunities. Int. J. Mol. Sci. 21:2632. doi: 10.3390/ijms21072632
Peeters, A., Barendregt, J. J., Willekens, F., Mackenbach, J. P., Al Mamun, A., and Bonneux, L. (2003). Obesity in adulthood and its consequences for life expectancy: a life-table analysis. Ann. Intern. Med. 138, 24–32. doi: 10.7326/0003-4819-138-1-200301070-00008
Peng, W. X., Koirala, P., and Mo, Y. Y. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667. doi: 10.1038/onc.2017.184
Qian, X., Zhao, J., Yeung, P. Y., Zhang, Q. C., and Kwok, C. K. (2019). Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem. Sci. 44, 33–52. doi: 10.1016/j.tibs.2018.09.012
Radhakrishnan, R., and Kowluru, R. A. (2021). Long noncoding RNA MALAT1 and regulation of the antioxidant defense system in diabetic retinopathy. Diabetes 70, 227–239. doi: 10.2337/db20-0375
Reddy, M. A., Chen, Z., Park, J. T., Wang, M., Lanting, L., Zhang, Q., et al. (2014). Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 63, 4249–4261. doi: 10.2337/db14-0298
Rinaldi, C., and Wood, M. J. A. (2018). Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 14, 9–21. doi: 10.1038/nrneurol.2017.148
Schetz, M., De Jong, A., Deane, A. M., Druml, W., Hemelaar, P., Pelosi, P., et al. (2019). Obesity in the critically ill: a narrative review. Intensive Care Med. 45, 757–769. doi: 10.1007/s00134-019-05594-1
Schmidt, E., Dhaouadi, I., Gaziano, I., Oliverio, M., Klemm, P., Awazawa, M., et al. (2018). LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 9:3622. doi: 10.1038/s41467-018-05933-8
Schmitt, A. M., Garcia, J. T., Hung, T., Flynn, R. A., Shen, Y., Qu, K., et al. (2016). An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 48, 1370–1376. doi: 10.1038/ng.3673
Stapleton, K., Das, S., Reddy, M. A., Leung, A., Amaram, V., Lanting, L., et al. (2020). Novel long noncoding RNA, macrophage inflammation-suppressing transcript (MIST), regulates macrophage activation during obesity. Arterioscler. Thromb. Vasc. Biol. 40, 914–928. doi: 10.1161/ATVBAHA.119.313359
Sun, M., and Kraus, W. L. (2015). From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 36, 25–64. doi: 10.1210/er.2014-1034
Sun, K., Kusminski, C. M., and Scherer, P. E. (2011). Adipose tissue remodeling and obesity. J. Clin. Invest. 121, 2094–2101. doi: 10.1172/JCI45887
Sun, L., and Lin, J. D. (2019). Function and mechanism of long noncoding RNAs in adipocyte biology. Diabetes 68, 887–896. doi: 10.2337/dbi18-0009
Tabas, I., and Bornfeldt, K. E. (2020). Intracellular and intercellular aspects of macrophage immunometabolism in atherosclerosis. Circ. Res. 126, 1209–1227. doi: 10.1161/CIRCRESAHA.119.315939
Tangvarasittichai, S. (2015). Oxidative stress, insulin resistance, dyslipidemia and type 2 diabetes mellitus. World J. Diabetes 6, 456–480. doi: 10.4239/wjd.v6.i3.456
Trembinski, D. J., Bink, D. I., Theodorou, K., Sommer, J., Fischer, A., van Bergen, A., et al. (2020). Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat. Commun. 11:2039. doi: 10.1038/s41467-020-15995-2
Tsilingiris, D., Liatis, S., Dalamaga, M., and Kokkinos, A. (2020). The fight against obesity escalates: new drugs on the horizon and metabolic implications. Curr. Obes. Rep. 9, 136–149. doi: 10.1007/s13679-020-00378-x
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
Wang, X. (2018). Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell. Biochem. 119, 1567–1574. doi: 10.1002/jcb.26317
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, Y., Hua, S., Cui, X., Cao, Y., Wen, J., Chi, X., et al. (2020). The effect of FOXC2-AS1 on white adipocyte browning and the possible regulatory mechanism. Front. Endocrinol. 11:565483. doi: 10.3389/fendo.2020.565483
Wang, C., Liu, G., Yang, H., Guo, S., Wang, H., Dong, Z., et al. (2021). MALAT1-mediated recruitment of the histone methyltransferase EZH2 to the microRNA-22 promoter leads to cardiomyocyte apoptosis in diabetic cardiomyopathy. Sci. Total Environ. 766:142191. doi: 10.1016/j.scitotenv.2020.142191
Wang, W., and Seale, P. (2016). Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 17, 691–702. doi: 10.1038/nrm.2016.96
Wang, J., Xiang, D., Mei, S., Jin, Y., Sun, D., Chen, C., et al. (2020). The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1. Mol. Metab. 37:100996. doi: 10.1016/j.molmet.2020.100996
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wang, Z., Zhang, X. J., Ji, Y. X., Zhang, P., Deng, K. Q., Gong, J., et al. (2016). The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139. doi: 10.1038/nm.4179
Willingham, A. T., Orth, A. P., Batalov, S., Peters, E. C., Wen, B. G., Aza-Blanc, P., et al. (2005). A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573. doi: 10.1126/science.1115901
Xiao, T., Liu, L., Li, H., Sun, Y., Luo, H., Li, T., et al. (2015). Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Rep. 5, 856–865. doi: 10.1016/j.stemcr.2015.09.007
Xiong, Y., Yue, F., Jia, Z., Gao, Y., Jin, W., Hu, K., et al. (2018). A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 409–419. doi: 10.1016/j.bbalip.2018.01.008
Xu, Y., Fang, H., Xu, Q., Xu, C., Yang, L., and Huang, C. (2020). LncRNA GAS5 inhibits NLRP3 inflammasome activation-mediated pyroptosis in diabetic cardiomyopathy by targeting miR-34b-3p/AHR. Cell Cycle 19, 3054–3065. doi: 10.1080/15384101.2020.1831245
Yan, C., Chen, J., and Chen, N. (2016). Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 6:22640. doi: 10.1038/srep22640
Yan, B., Tao, Z. F., Li, X. M., Zhang, H., Yao, J., and Jiang, Q. (2014). Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 55, 941–951. doi: 10.1167/iovs.13-13221
Yan, B., Yao, J., Liu, J. Y., Li, X. M., Wang, X. Q., Li, Y. J., et al. (2015). lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 116, 1143–1156. doi: 10.1161/CIRCRESAHA.116.305510
Yang, L., and Li, T. (2020). LncRNA TUG1 regulates ApoM to promote atherosclerosis progression through miR-92a/FXR1 axis. J. Cell. Mol. Med. 24, 8836–8848. doi: 10.1111/jcmm.15521
You, L. H., Zhu, L. J., Yang, L., Shi, C. M., Pang, L. X., Zhang, J., et al. (2015). Transcriptome analysis reveals the potential contribution of long noncoding RNAs to brown adipocyte differentiation. Mol. Gen. Genomics. 290, 1659–1671. doi: 10.1007/s00438-015-1026-6
Yu, X. H., Deng, W. Y., Chen, J. J., Xu, X. D., Liu, X. X., Chen, L., et al. (2020). LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. 11:1043. doi: 10.1038/s41419-020-03263-6
Zhang, K., Han, X., Zhang, Z., Zheng, L., Hu, Z., Yao, Q., et al. (2017). The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and notch pathways. Nat. Commun. 8:144. doi: 10.1038/s41467-017-00204-4
Zhang, Y., Jiao, L., Sun, L., Li, Y., Gao, Y., Xu, C., et al. (2018). LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ. Res. 122, 1354–1368. doi: 10.1161/circresaha.117.312117
Zhang, Y., Sun, J., Yao, H., Lin, Y., Wei, J., Hu, G., et al. (2020). Ultraconserved element uc.333 increases insulin sensitivity by binding to miR-223. Aging 12, 6667–6679. doi: 10.18632/aging.103020
Zhang, X., Xue, C., Lin, J., Ferguson, J. F., Weiner, A., Liu, W., et al. (2018). Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci. Transl. Med. 10:eaar5987. doi: 10.1126/scitranslmed.aar5987
Zhang, K., Zhang, M., Yao, Q., Han, X., Zhao, Y., Zheng, L., et al. (2019). The hepatocyte-specifically expressed lnc-HSER alleviates hepatic fibrosis by inhibiting hepatocyte apoptosis and epithelial-mesenchymal transition. Theranostics 9, 7566–7582. doi: 10.7150/thno.36942
Zhao, X. Y., Li, S., Wang, G. X., Yu, Q., and Lin, J. D. (2014). A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 55, 372–382. doi: 10.1016/j.molcel.2014.06.004
Zhao, X. Y., Xiong, X., Liu, T., Mi, L., Peng, X., Rui, C., et al. (2018). Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat. Commun. 9:2986. doi: 10.1038/s41467-018-05383-2
Zhou, Q., Huang, X. R., Yu, J., Yu, X., and Lan, H. Y. (2015). Long noncoding RNA Arid2-IR is a novel therapeutic target for renal inflammation. Mol. Ther. 23, 1034–1043. doi: 10.1038/mt.2015.31
Keywords: long noncoding RNAs, lipid metabolism, adipogenesis, brown/beige adipose, fat, insulin resistance
Citation: Zhang B, Xu S, Liu J, Xie Y and Xiaobo S (2021) Long Noncoding RNAs: Novel Important Players in Adipocyte Lipid Metabolism and Derivative Diseases. Front. Physiol. 12:691824. doi: 10.3389/fphys.2021.691824
Edited by:
Tizhong Shan, Zhejiang University, ChinaReviewed by:
Weiqin Chen, Augusta University, United StatesYan Xiong, Southwest Minzu University, China
Tongxing Song, Huazhong Agricultural University, China
Copyright © 2021 Zhang, Xu, Liu, Xie and Xiaobo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Xie, eXhpZUBpbXBsYWQuYWMuY24=; Sun Xiaobo, c3VuX3hpYW9ibzE2M0AxNjMuY29t
†These authors have contributed equally to this work
 Bin Zhang
Bin Zhang Saijun Xu
Saijun Xu Jinyan Liu
Jinyan Liu Yong Xie*
Yong Xie* Sun Xiaobo
Sun Xiaobo