
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 07 July 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.690185
This article is part of the Research Topic The Morphology and Physiology of Insect Chemosensory Systems – Its Origin and Evolution View all 17 articles
 Cong Huang1†
Cong Huang1† Xue Zhang1,2†
Xue Zhang1,2† Dongfeng He1,3
Dongfeng He1,3 Qiang Wu1
Qiang Wu1 Rui Tang4
Rui Tang4 Longsheng Xing1
Longsheng Xing1 Wanxue Liu5
Wanxue Liu5 Wenkai Wang3
Wenkai Wang3 Bo Liu1
Bo Liu1 Yu Xi1
Yu Xi1 Nianwan Yang5*
Nianwan Yang5* Fanghao Wan1,2,5*
Fanghao Wan1,2,5* Wanqiang Qian1*
Wanqiang Qian1*Insect olfaction is vital for foraging, mating, host-seeking, and avoidance of predators/pathogens. In insects, odorant binding proteins (OBPs) are involved in transporting hydrophobic odor molecules from the external environment to receptor neurons. The codling moth, Cydia pomonella, one of the most destructive insect fruit pests, causes enormous economic losses. However, little is known about the number, variety, gains and losses, and evolution of OBP genes in C. pomonella. Here we report the identification of 40 OBPs in C. pomonella, most (75%) of which are classic OBPs, using genomic and transcriptomic analyses. Two OBP genes were lost in C. pomonella relative to possible distant ancestor in Lepidoptera lineage based on an analysis of gene gains and losses. The phylogenetic tree and chromosome location showed that the expansion of OBP genes mainly resulted from tandem duplications, as the CpomGOBP2 gene was duplicated twice along with loss of CpomPBPB. Two positive selection sites of the CpomGOBP1 gene were identified while other OBP genes evolved under purifying selection. Our results provide fundamental knowledge of OBP genes allowing further study of their function in C. pomonella.
Insects rely on their olfactory system to sense environmental odors related to behaviors such as foraging, host-seeking, mating, and oviposition, as well as avoiding predators and pathogens (Andersson et al., 2015). Odorant binding proteins (OBPs) are small water-soluble globular proteins with molecular masses of 10–30 kDa (Sun et al., 2018). OBPs are highly expressed in the hydrophilic lymph of insect olfactory sensilla (Pelosi et al., 2014). When lipophilic semiochemicals from the environment enter the lymph through micropores on the surface of olfactory sensilla, the OBPs bind, solubilize, and deliver the semiochemicals to the receptor proteins, e.g., odorant receptors (ORs) or ionotropic receptors (IRs), which are located on the membranes of olfactory sensory neurons. This delivery activates a series of downstream olfactory signal transductions accompanied by corresponding behavioral movements in insects (Zhang et al., 2020). OBPs are clearly essential in communications between insects and environmental semiochemicals including both pheromones and host volatiles.
OBPs are involved in the initial step of recognizing host volatiles or sex pheromones, suggesting that the functional divergence of OBPs is associated with speciation or host diversity. Considering the low sequence identities between orthologous/paralogous OBP genes, OBP genes have likely been evolving at a rapid rate through gene gains or losses (McKenzie et al., 2014) and positive selection (Campanini and de Brito, 2016). Most OBP genes are tandemly arranged in chromosomes, indicating that the occurrence of these genes arose from tandem duplication (Hekmat-Scafe et al., 2002; Gong D. P. et al., 2009; Manoharan et al., 2013; Dippel et al., 2014). The duplicate genes then gradually diverge in function through mutation or pseudogenization (Nei and Rooney, 2005; Vieira et al., 2007).
Studies on the origin, evolution, and structural variation of OBP genes provide insight into the functional differentiation of OBPs and host preference in insects. However, there is little knowledge of the numbers, structures, and evolution of the OBP gene family in important insect crop pests such as the codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae). C. pomonella is an economically threatening pest worldwide (Witzgall et al., 2008; Kumar et al., 2015; Zhu et al., 2017). It mainly destroys apples and pears as well as other seed and stone fruits. Some studies focused on the structures and functions of pheromone binding proteins (PBPs) (Liu et al., 2016; Tian et al., 2016a, b; Tian and Zhang, 2016) and identification of general odorant binding proteins (GOBPs) (Garczynski et al., 2013). In contrast, studies on OBPs are lacking in C. pomonella, with little information of their roles in recognizing hosts or locating mates.
To understand the evolution and function of OBPs in C. pomonella, we identified and annotated its OBP genes by combining transcriptome data with the high quality genome we released previously (Wan et al., 2019). The gene gains and losses of OBPs were estimated by CAFÉ 3.0 (Han et al., 2013) for seven moth species. Subsequently, a phylogenetic tree of OBP genes from three lepidopteran insects (C. pomonella, Bombyx mori, and Manduca sexta) was constructed to explore their evolutionary relationships. The collinearity and chromosome locations were used to compare the divergence of OBP genes between C. pomonella and B. mori. Finally, the positive selection of genes and structural homology model were analyzed to predict the functional divergence of selected OBPs. Our results provide insights into the evolution of OBP genes in C. pomonella, which will facilitate future functional studies.
The protein sequences of seven lepidopteran insect OBPs were collected from deposited data of published articles, which have been identified from their genomes, and these species included B. mori (Gong D. P. et al., 2009), M. sexta (Vogt et al., 2015), Plutella xylostella (Cai et al., 2020), Spodoptera litura (Cheng et al., 2017), Spodoptera frugiperda (Gouin et al., 2017), Helicoverpa armigera (Pearce et al., 2017), and Danaus plexippus (Zhan et al., 2011). These protein sequences were then used as queries in iterative BLASTP searches with parameter “-e-value 1e–5” against the C. pomonella genome (Wan et al., 2019) to find candidate OBP genes. A local command line HMMER (version 3.1b2) search was conducted for these candidate OBP genes against the Pfam-A database (El-Gebali et al., 2019) to find the PBP_GOBP (PF01395) HMM profile. The identified OBP genes were subsequently used as queries to align the C. pomonella genome using tBLASTn search with parameter “-e-value 1e-5” to identify the missing OBP genes during gene prediction for the genome. We used an in-house Perl script to extract DNA sequences of novel genes from the genome, followed by predicting the CDS using the online website FGENESH (Solovyev et al., 2006). Gene prediction was verified by comparing with the transcriptome data that we used in the C. pomonella genome paper to confirm the complete gene structure (Wan et al., 2019). Finally, we used GMAP (Wu and Watanabe, 2005) to rebuild gene structures of all OBP genes. For B. mori, we used the 44 OBPs of B. mori which were identified by Gong (Gong D. P. et al., 2009), to perform tBLASTn search against the newest version of the B. mori genome (Kawamoto et al., 2019) and rebuilt their gene structures by GMAP (Wu and Watanabe, 2005).
To check the conserved cysteine pattern, which is the predominant feature of OBP genes, we first performed multiple sequence alignment of OBP sequences using MAFFT v7 (Katoh et al., 2002) with default parameters. Then, the aligned sequences were trimmed by trimAl v1.2 (Capella-Gutierrez et al., 2009) to remove gaps and low-quality regions with the parameter ‘‘-automated1.’’ The trimmed sequences were subsequently submitted to ESPript 3.0 Server1 for visualization.
To explore gene gains and losses of OBPs in moths, seven moth species with available genomes and past investigations of the OBP gene family were selected, including S. litura, S. frugiperda, H. armigera, B. mori, M. sexta, C. pomonella, and P. xylostella. Orthologous and paralogous groups of these species were inferred by OrthoFinder v2.3.1 (Emms and Kelly, 2015) with default parameters. Orthologous groups including only single copy genes for each species were selected to construct the species tree. Protein sequences of each orthologous group were independently aligned using MAFFT v7 (Katoh et al., 2002), trimmed by trimAl v1.2 (Capella-Gutierrez et al., 2009), and then concatenated into one super-sequence. The phylogenetic tree was inferred using maximum likelihood (ML) in RAxML with the best-fit model (JTT + I + F) estimated by ProtTest3 v3.4.2 (Darriba et al., 2011). The Bayesian Relaxed Molecular Clock (BRMC) approach was adopted to estimate the neutral evolutionary rate and species divergence time using the program MCMCTree, implemented in PAML v4.9b package (Yang, 2007). The tree was calibrated with the following time frames adopted from TimeTree (Kumar et al., 2017) to constrain the age of the nodes between the species: 99–121 million years ago (Mya) for B. mori and H. armigera, and 80–243 Mya for C. pomonella and P. xylostella.
The OBP gene gains and losses were estimated by CAFÉ v3.0 (Han et al., 2013). Gene numbers of the OBP gene family in each insect were collected from published articles for S. litura (Cheng et al., 2017), S. frugiperda (Gouin et al., 2017), H. armigera (Pearce et al., 2017), M. sexta (Vogt et al., 2015), and P. xylostella (Cai et al., 2020), while the numbers of OBP genes in B. mori and C. pomonella were identified in this study (see section “Materials and Methods”). This gene number matrix together with the phylogenetic tree corrected by MCMCTree were used as input files for CAFÉ 3.0 (Han et al., 2013).
A total of 133 OBP genes from three species (C. pomonella, B. mori, and M. sexta) were used in the phylogenetic analysis. These gene sequences were aligned using MAFFT v7 (Katoh et al., 2002) with default parameters, then the alignments were trimmed by trimAl v1.2 (Capella-Gutierrez et al., 2009) with the parameter “-automated1.” RAxML (Stamatakis, 2014) was used to construct a maximum likelihood evolutionary tree with the best-fit model (LG) estimated by ProtTest3 v3.4.2 (Darriba et al., 2011). FigTree v1.4.32 and Adobe Illustrator CC 2017 were used to visualize and annotate the phylogenetic tree.
We mapped the 44 OBP genes of B. mori (Gong D. P. et al., 2009) to the chromosomes in the newest version of the B. mori genome (Kawamoto et al., 2019) and rebuilt their gene structure by GMAP (Wu and Watanabe, 2005). However, only 43 OBP genes were successfully mapped: BmorOBP24 is a pseudogene that was discarded. Subsequently, the best reciprocal BLAST hit was used to identify the orthologous OBP gene pairs in C. pomonella and B. mori genomes. MapGene2Chrom web v23 was used to draw the distribution map of OBP genes on the chromosomes of both species. Orthologous gene pairs or blocks were linked by lines.
To estimate whether natural selection acted on the evolution of OBP genes in C. pomonella, we inferred the ratio of the normalized non-synonymous rate (dN) to the synonymous rate (dS) of nucleotide substitutions (ω = dN/dS) by a maximum likelihood method using the Codeml program in PAML v4.9b (Yang, 2007), with ω > 1, ω = 1, ω < 1 indicating positive selection, neutral evolution, and purifying selection, respectively. We first aligned the protein sequences for each analysis in MAFFT v7 (Katoh et al., 2002), then these protein alignments were converted to CDS alignments by the PAL2NAL program4. Subsequently, the protein alignments were trimmed by trimAl v1.2 (Capella-Gutierrez et al., 2009) and were used in MEGA6 (Tamura et al., 2013) to build Neighbor-Joining (NJ) trees with the Jones-Taylor-Thornton (JTT) model and 1,000 bootstrap replications.
We used the site model for each group of OBP orthologous/paralogous genes clustered by the phylogenetic tree to test which genes or sites might have evolved under positive selection. In this site model, we performed a test of heterogeneity across sites by comparing the M0 and M3 models with K = 3 categories. Another test of positive selection on sites involved fitting a beta distribution of ω values across sites by comparing M7 and M8 models. Considering the evolutionary specificity of GOBP and PBP genes in lepidopteran insects (Yasukochi et al., 2018), we used the branch-site model to test the genes as well as their amino acid sites that evolved under positive selection in nine lepidopteran insects including B. mori, C. pomonella, D. plexippus, Heliconius melpomene, M. sexta, Operophtera brumata, Papilio xuthus, P. xylostella, and S. litura. In the phylogenetic tree of each gene (GOBP1, GOBP2, PBPA, PBPC, and PBPD, but not PBPB due to lack of gene numbers), we labeled the branch composed of genes from C. pomonella as the foreground branch and the remaining branches as background branches to test positive selection in C. pomonella GOBP and PBP genes. We compared model A (the alternative model), in which some sites on the foreground branch were allowed to change to a value of ω > 1, with the null model of neutral evolution.
The likelihood ratio tests (LRTs) statistic (2ΔL), which approximates a χ2 distribution, was used for comparisons between models, and significant results were determined using χ2-tests. If the LRT was significant, Bayes empirical Bayes (BEB) was used to identify sites of positive selection. The sites with posterior probabilities (PPs) of ≥ 0.95 were considered positively selected, thus they were defined as positively selected sites (PSS).
To further understand the functional significance of the identified positively selected sites, we labeled them on protein sequences and tertiary structures. The amino acid sequences of CpomGOBP1 gene which evolved under positive selection in C. pomonella were submitted to the SWWISS-MODEL Server5 to predict and refine 3D structures. The best template was BmorGOBP2 (PDB ID: 2WCK), which has 53.57% identity with CpomGOBP1. Subsequently, we used the SAVES server6 and RAMPAGE server to estimate the quality of the predicted 3D structure. SAVES assesses the quality of protein 3D structure based on the PROCHECK (Laskowski et al., 1993), ERRAT (Colovos and Yeates, 1993), and VERIFY 3D (Lüthy et al., 1992) program. RAMPAGE assesses the quality based on the Ramachandran plot (Wang et al., 2016). The generated model structures were rendered and visualized using Visual Molecular Dynamics (VMD) v1.9.3 (Humphrey et al., 1996).
We collected 48 odorant molecules including pheromones and host plant volatiles by mining literatures (Bengtsson et al., 2001; Ansebo et al., 2004; Bengtsson et al., 2014; Knight et al., 2019). Then, docking was carried out by AutoDock Vina (Trott and Olson, 2010). The binding patterns between CpomGOBP1 and the odorant molecule was visualized using VMD (Humphrey et al., 1996). The key binding site analysis was performed using LigPlus (Wallace et al., 1995).
We calculated the expression levels of 40 CpomOBPs in several sensory tissues based on transcriptome data. The sensory tissues were collected from female and male adults, including the antennae, head, leg, wing, labial palp, each sample with three biological replicates. The paired-end clean reads were mapped to the C. pomonella genome using HISAT2 (Kim et al., 2015). The FPKM (Fragments Per Kilobase of transcript per Million mapped reads) was calculated by StringTie software (Pertea et al., 2016). The heatmap.2 function of R package “gplots” to draw the heatmap of expression profiling based on the FPKM values.
A total of 40 OBP genes were identified in the C. pomonella genome. Complete CDS were determined by cross-checking with transcriptome assemblies. All gene information including gene names, CDS, amino acid sequences, chromosomes, and gene lengths and classification are provided in Supplementary Table 1. We compared the 40 CpomOBPs with those previous reported (Garczynski et al., 2013) and renamed them, and those that were not well matched were renamed by the sequence length. The amino acid sequences range from 133 to 339 amino acids (Supplementary Table 1). Multiple sequence alignments show that most have six typical conserved cysteine residues (Figure 1 and Supplementary Table 1). Based on the number and location of the conserved cysteines, the 40 CpomOBPs were classified into four subfamilies, including classic, minus-C, plus-C, and atypical. In total, 30 CpomOBPs belong to the classic subfamily, which had six typical conserved cysteine residues. Four CpomOBPs contained more cysteines than the classic OBPs, and were classified into the plus-C subfamily. Four CpomOBPs belong to the minus-C subfamily, which had fewer than six cysteines. The remaining two CpomOBPs, which exhibited none of the above characteristics, were classified into the atypical subfamily.
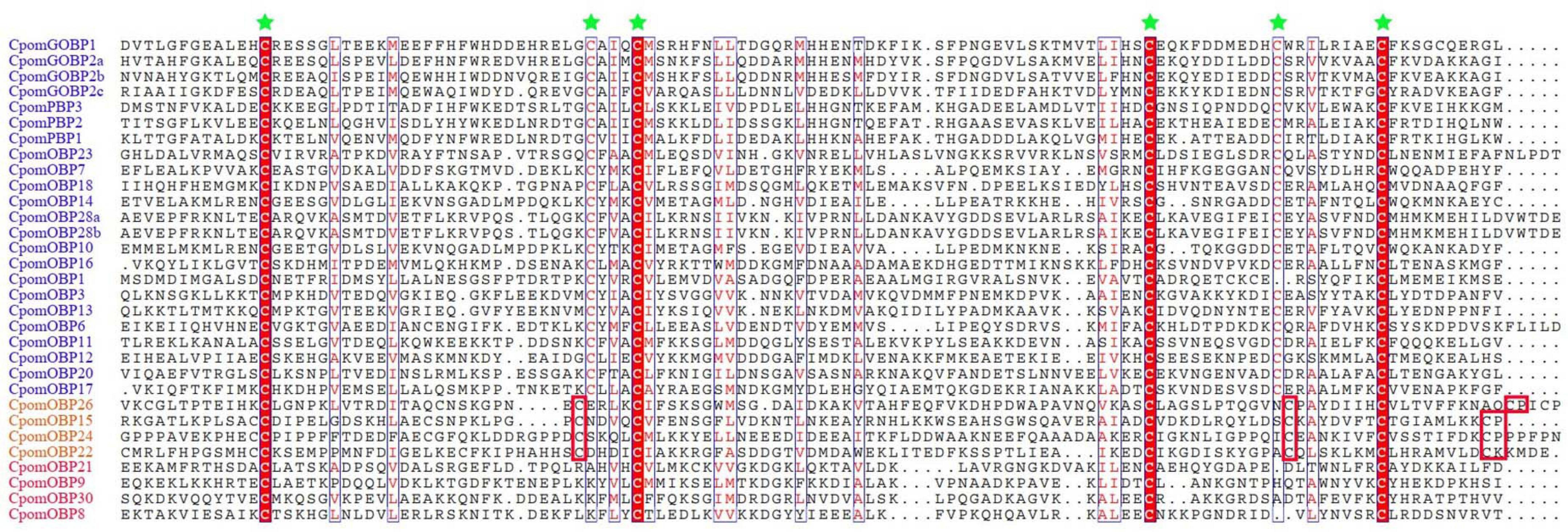
Figure 1. Amino acid alignment of various C. pomonella OBP family members. The alignment was performed by MAFFT v7, aligned sequences were depicted with ESPript 3.0 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Highly conserved cysteine residues are marked by green stars. The gene IDs of classic, plus-C, and minus-C OBPs are shown in blue, orange and red color, respectively. Only the cysteines which are conserved in all of alignment sequences are highlighted in red block. Seven classic and two atypical OBPs are not shown in this figure.
A statistical gene birth and death analysis for OBP genes from seven moth species (S. litura, S. frugiperda, H. armigera, B. mori, M. sexta, C. pomonella, and P. xylostella) was performed by CAFÉ (Figure 2). Forty-two OBP genes were inferred in the common ancestor node of moth species considered in this study at 162 Mya. The gene gains and losses range from -1 (lost one gene) to + 1 (gained one gene) between the adjacent ancestor nodes. However, different species have various gene gains or losses ranging from 1 to 7 compared with their adjacent ancestors. For example, gene gains occurred in S. frugiperda (+ 7) and M. sexta (+ 6), while gene losses occurred in S. litura (-7), H. armigera (-2), B. mori (-1), C. pomonella (-2), and P. xylostella (-3) compared with their adjacent ancestors. Our results suggested that the gains and losses of OBP genes may be associated with functional divergence which results from adaptation.
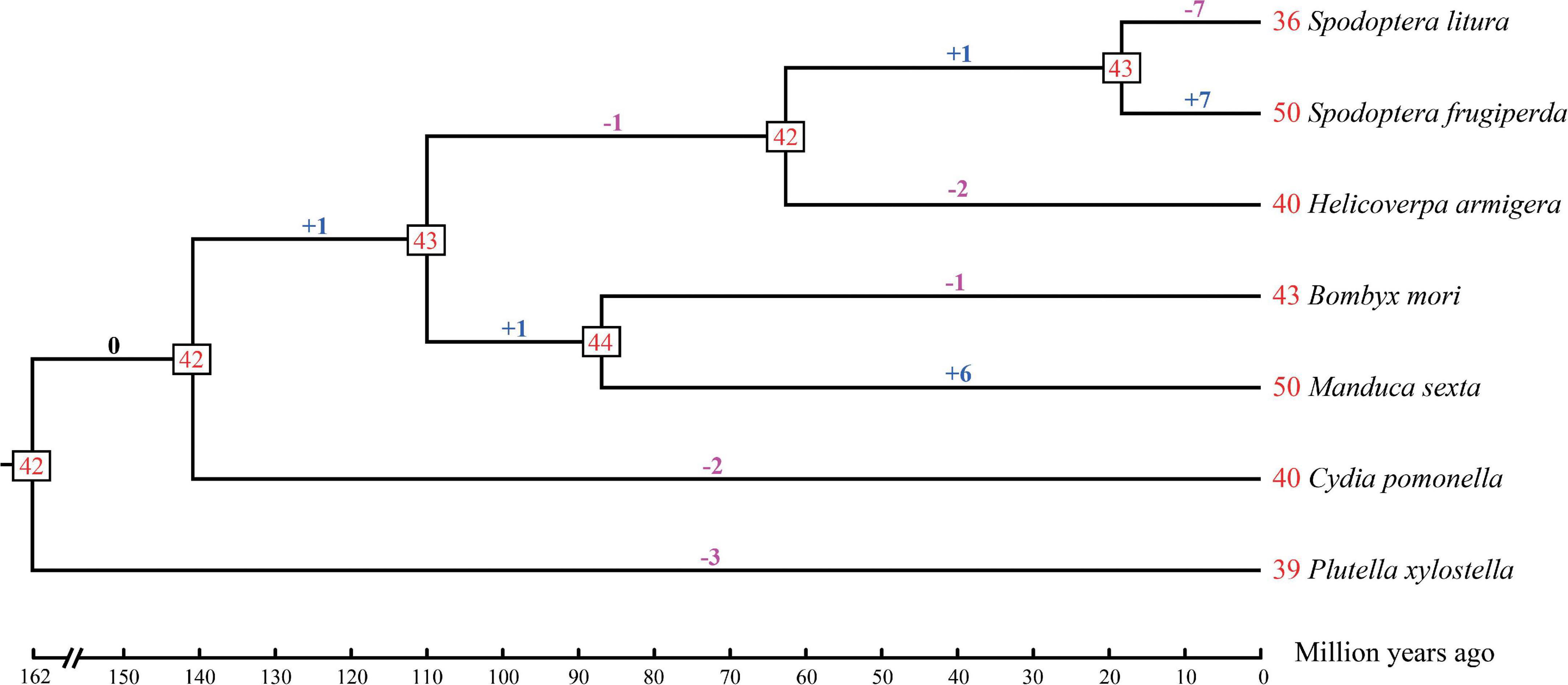
Figure 2. Estimation of OBP gene gains and losses during the evolution of seven moth species. The phylogenetic tree with estimated divergence times (million year ago) was inferred by MCMCTree in PAML v4.9b. The numbers at the tree termini are the numbers of genes in each species, which we found in the literature: Spodoptera litura, Spodoptera frugiperda, Helicoverpa armigera, Manduca sexta, and Plutella xylostella, the numbers of OBP genes in Cydia pomonella were identified in this study, while the number of OBP genes in Bombyx mori was corrected by this study (see section “Materials and Methods”). The numbers at the tree nodes are the numbers of genes in their most recent common ancestors. The numbers of gene gains and losses are shown above the branches, where the symbol “+” represents gene gains while “-” represents gene losses. The numbers before each species represent the OBP genes of the specific moth.
The phylogenetic tree was inferred using a total of 133 amino acid sequences of OBP genes, including 40 OBPs from C. pomonella, 43 OBPs from B. mori, and 50 OBPs from M. sexta (Figure 3). We classified these OBP genes into 12 groups (Groups 1–12) according to the clusters in the phylogenetic tree. Eleven of them were orthologous groups shared among these three species with nearly 1:1 orthologous genes in each group from each species, except that Group 2 has a lineage-specific expansion in M. sexta. The GOBP/PBP subfamily is a specific cluster in lepidopteran species (Vogt et al., 2015; Yasukochi et al., 2018), consisting of six typical tandem genes including GOBP1, GOBP2, and PBPA-PBPD. In our study, Group 1 was the conserved GOBP/PBP cluster, including six genes from B. mori (GOBP1, GOBP2, PBPA-1, PBPA-2, PBPC, and PBPD), six genes from M. sexta (GOBP1, GOBP2, PBPA, PBPB, PBPC, and PBPD), and seven genes from C. pomonella (GOBP1, GOBP2a, GOBP2b, GOBP2c, PBP3, PBP2, and PBP1). The PBPB gene was lost in C. pomonella, while the GOBP2 gene is duplicated twice, which suggested that GOBP2 may be under positive selection. In general, the OBP gene family is evolutionarily conserved in Lepidoptera insects.
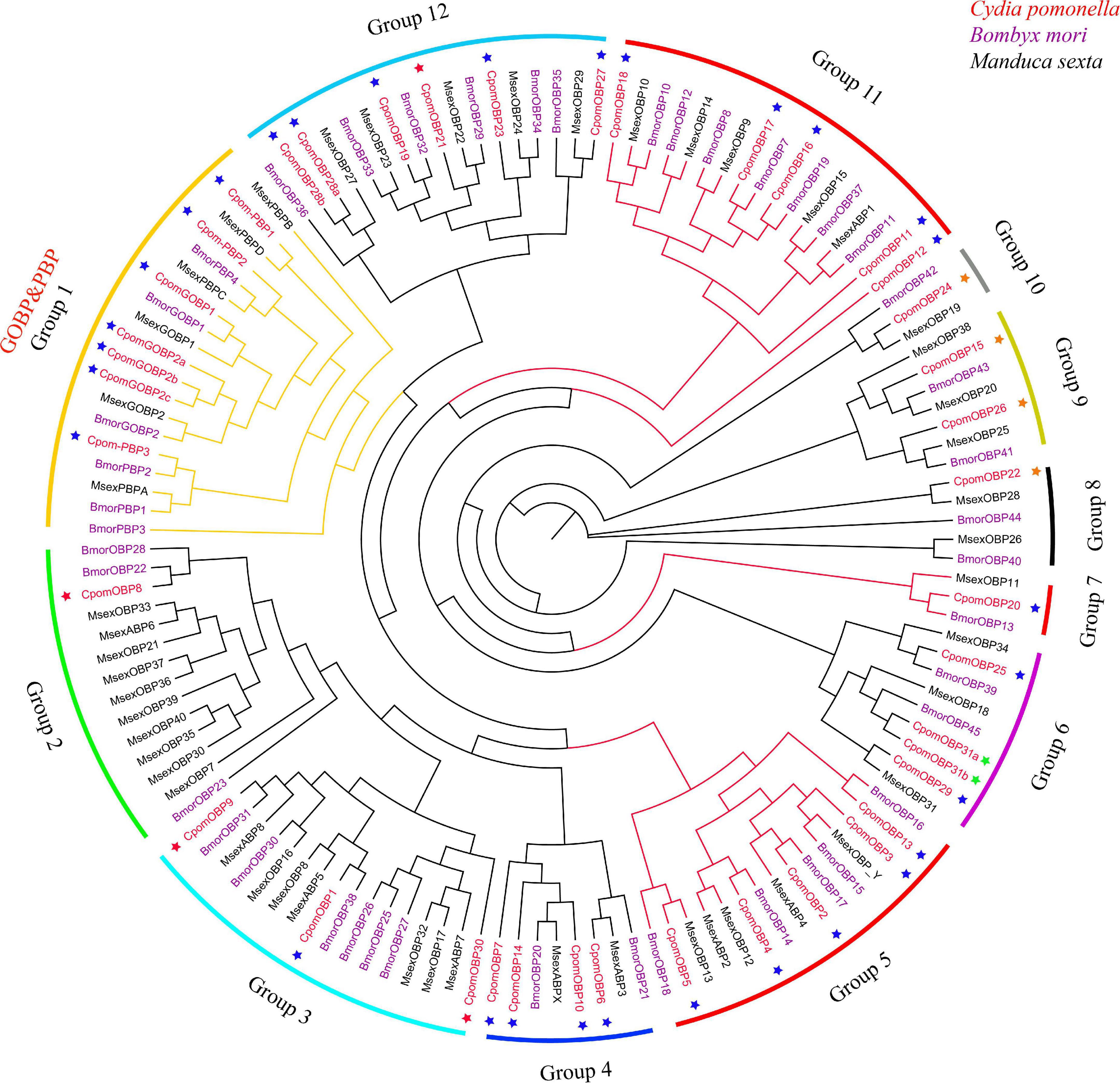
Figure 3. The phylogenetic tree of the OBP gene family in lepidopteran insects. C. pomonella OBPs are classified as Classic, Minus-C, Plus-C, and Atypical, which are represented by blue, red, orange, and green stars, respectively. The GOBP/PBP genes are shown in orange.
All 40 OBP genes were located on 11 C. pomonella chromosomes (Figure 4). These genes are organized into two major clusters on chromosomes 18 and 8, while the other chromosomes contain scattered and few OBP genes. The largest cluster contains 11 tandem OBP genes on chromosome 18, accounting for 27.5% of the total number of OBP genes. These genes have a collinearity block in chromosome 18 of B. mori that contains 12 tandem OBP genes. Furthermore, this collinearity block of OBP genes was clustered into Groups 5, 7, and 11 in the phylogenetic tree, which belong to antennal binding protein I (ABPI) and antennal binding protein II (ABPII) (Figure 3) (Gong D. P. et al., 2009). Another big cluster on chromosome 8 contains seven OBP genes, which account for 17.5% of the total number of OBP genes. Six of them were in a tandem GOBP/PBP gene cluster, including GOBP1, GOBP2a, PBP3, PBP2, and PBP1. There was also a tandem GOBP/PBP gene cluster on chromosome 19 in B. mori. There was a collinearity block of the GOBP/PBP gene cluster between C. pomonella and B. mori, and they were clustered into Group 1 in the phylogenetic tree (Figure 3). Seven OBP genes in C. pomonella have no collinearity compared with B. mori.
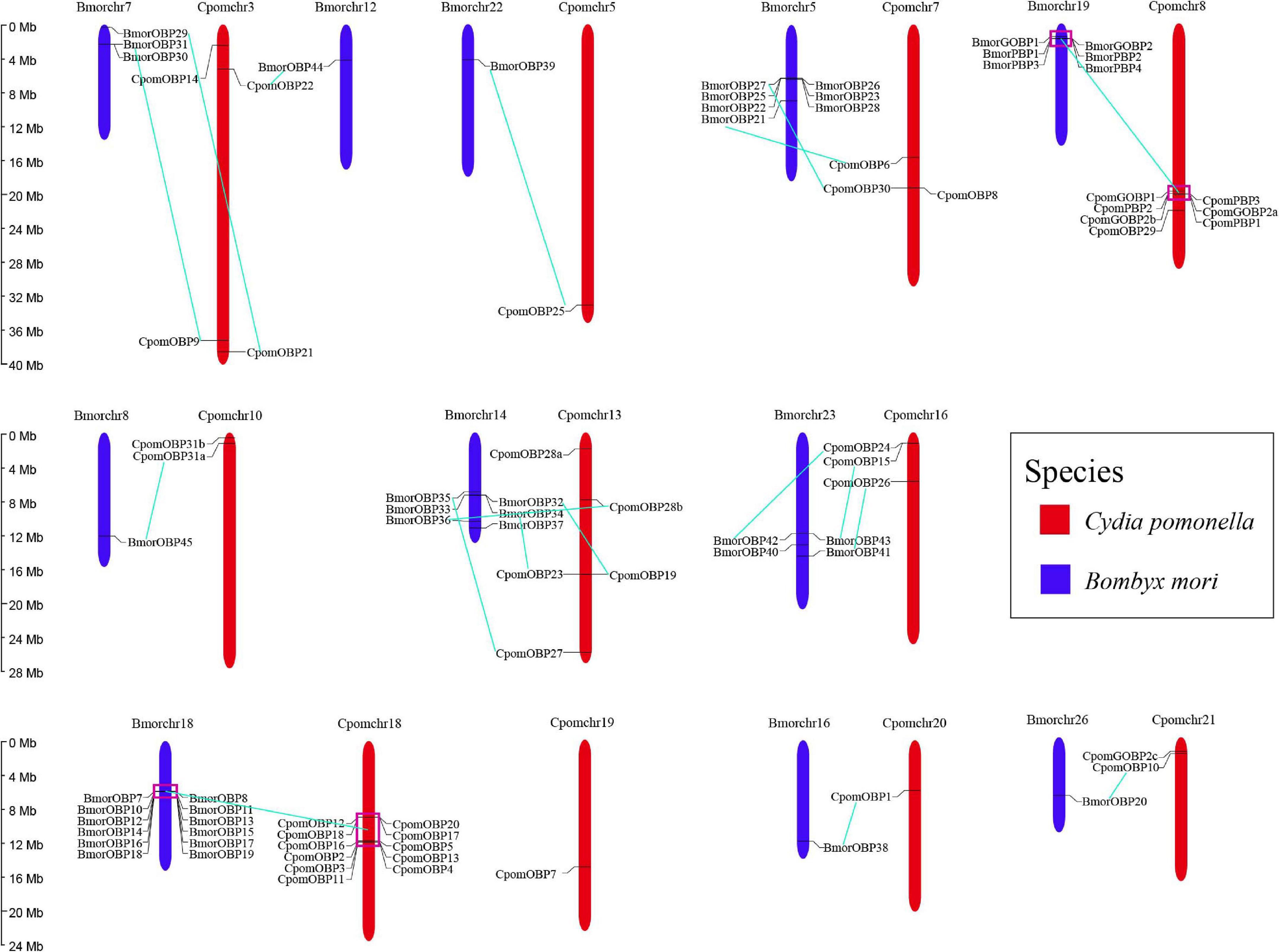
Figure 4. Chromosomal distribution of OBP genes in C. pomonella (red) and B. mori (blue). Cyan solid lines represent the correspondence between C. pomonella and B. mori OBP genes.
We selected eight clades (groups) from the phylogeny to test whether some orthologous/paralogous OBP genes of three moths (including B. mori, C. pomonella, and M. sexta) evolved under positive selection. Selected groups included Groups 2–6, 9, 11, and 12 (Table 1). Groups 7–8 and 10 were excluded since they had too few genes. Group 1 was later tested independently using the branch-site model.

Table 1. Tests of positive selection on the orthologous/paralogous OBP genes of moths by site model.
According to tests of the one-ratio model (M0), which assumes a single ω for all amino acid sites, the ω values of eight clades ranged from 0.00547 to 0.15846 (Table 1), suggesting the existence of strong purifying selection. However, the comparison between models M0 and M3 (discrete) provided strong evidence of variation in selective pressures at different amino acid sites in Groups 2–3, 5, 9, and 11 (P < 0.01, Table 1), indicating that purifying selection has been relaxed at some amino acid sites. We further compared models M7 and M8 for clades showing 0.5 < dS < 1 to investigate whether some amino acid sites actually evolved under positive selection. Only Group 2 presented evidence of positive selection (P = 0.0008) with one positively selected site (PSS). However, the Bayes empirical Bayes (BEB) analysis showed that the PSS only had a 93.4% posterior probabilities (PPs), which is not statistically significant.
We used the branch-site model to test the positive selection in each codon for different gene clades of GOBPs and PBPs from nine species (B. mori, C. pomonella, D. plexippus, Heliconius melpomene, M. sexta, Operophtera brumata, Papilio xuthus, P. xylostella, and S. litura) (Figure 5). Only GOBP1 was identified as being under positive selection for C. pomonella after the likelihood ratio test (P = 0.0318). We further used the BEB approach to detect the positive sites in GOBP1, which showed that sites 41S and 43G were significant signs of positive selection with PPs of 0.994 and 0.972, respectively (Table 2).
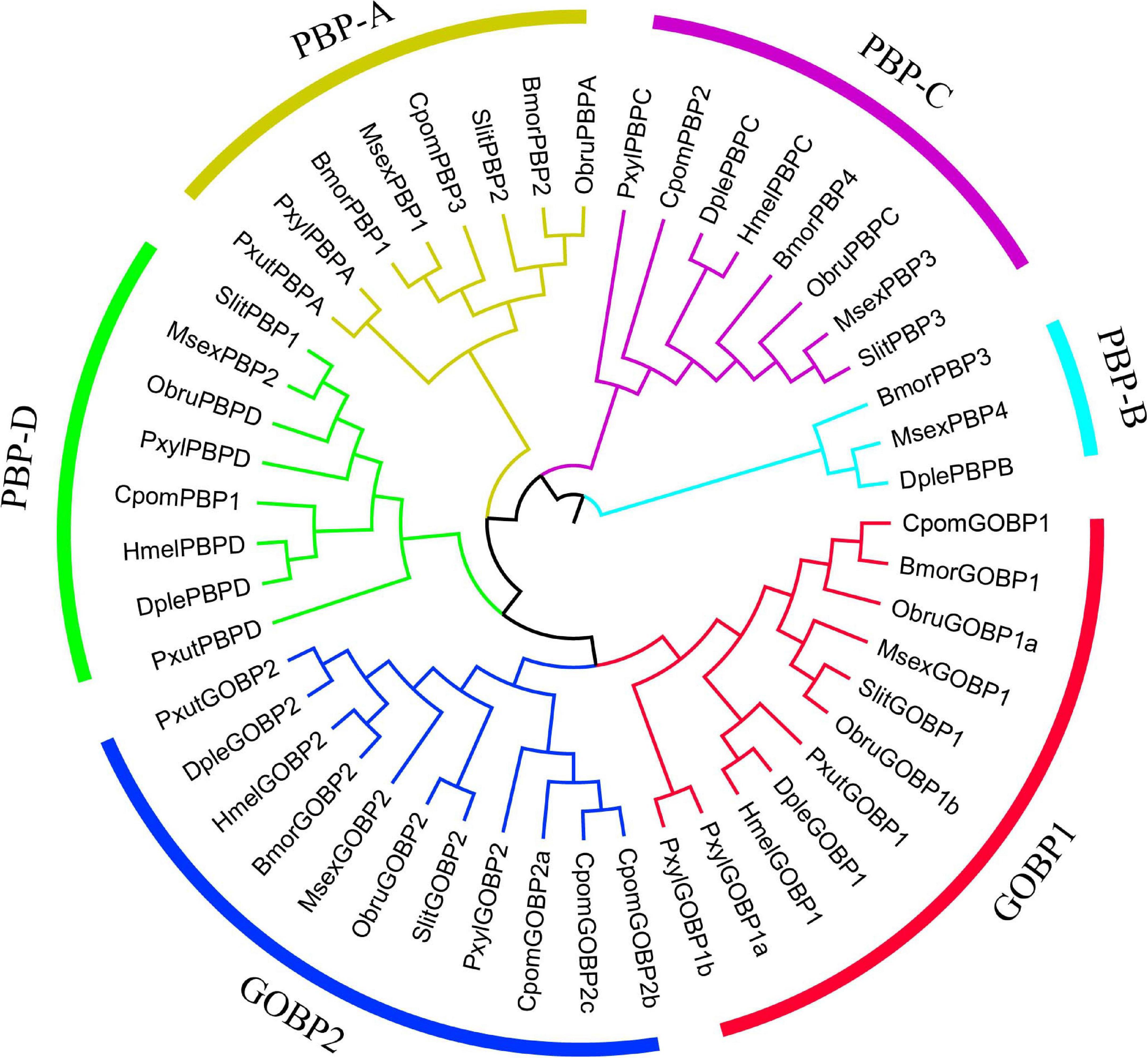
Figure 5. Phylogenetic relationships of GOBP and PBP genes in lepidopteran insects. The maximum likelihood tree was constructed based on the 49 GOBP and PBP genes from nine lepidopteran species. The amino acid sequences were aligned by MAFFT v7 with default parameters, and then the alignments were trimmed by trimAl with the parameter “-automated1.” (Bmor: Bombyx mori, Cpom: Cydia pomonella, Dple: Danaus plexippus, Hmel: Heliconius melpomene, Msex: Manduca sexta, Obru: Operophtera brumata, Pxut: Papilio xuthus, Pxyl: Plutella xylostella, Slit: Spodoptera litura). MEGA v6 was used to construct a Neighbor-Joining tree with the Jones-Taylor-Thornton (JTT) model and 1,000 bootstrap replications. FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and Adobe Illustrator CC 2017 were used to visualize and annotate the phylogenetic tree.
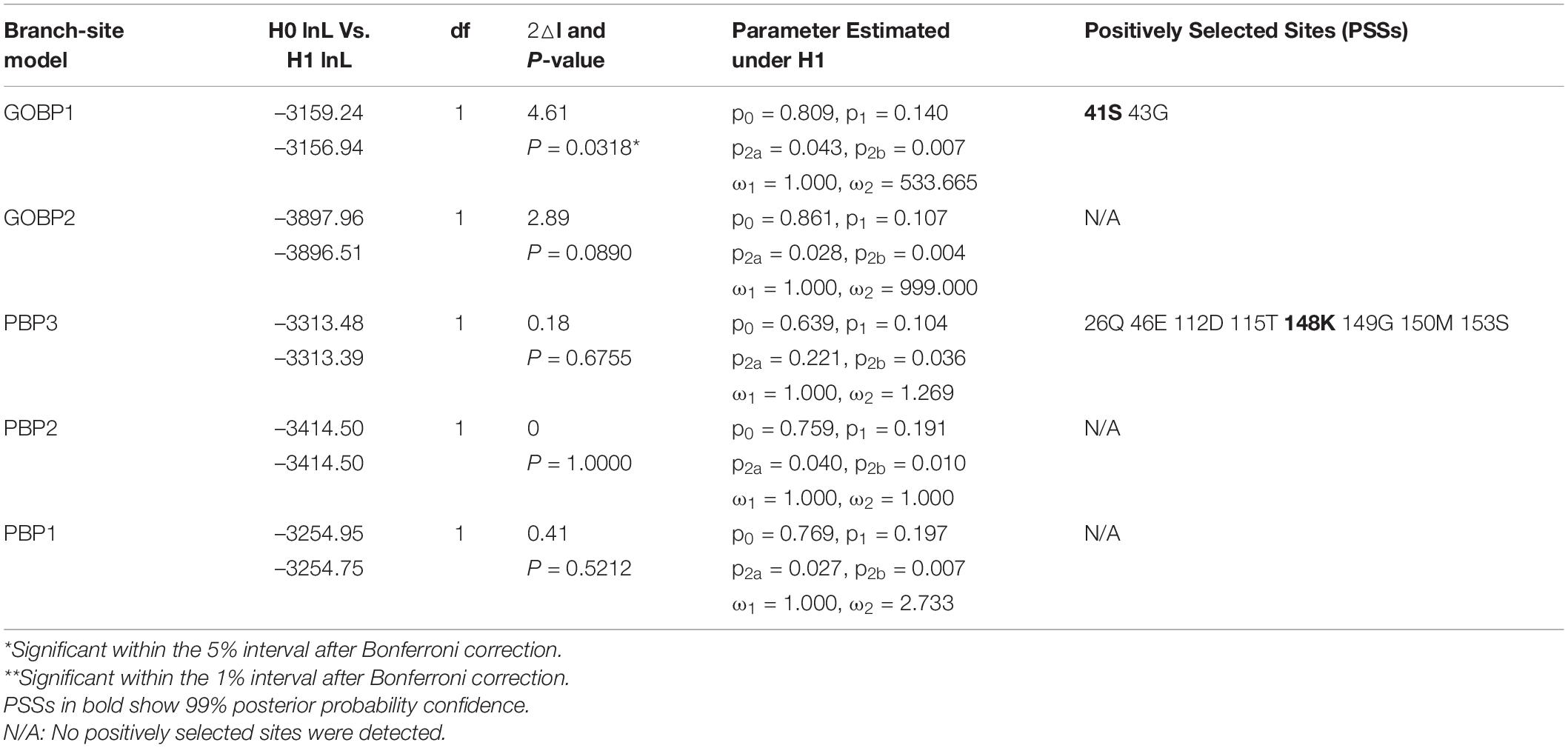
Table 2. Tests of positive selection on Lepidopteran GOBP and PBP genes by branch-site model (Branch labels referring to Figure 5).
To get additional insight into the functional significance of PSSs, we mapped the PSSs to the multiple sequence alignments of GOBP1 protein sequences from nine species, and labeled them on the structural homology model of C. pomonella GOBP1 (Figure 6A). Compared to the other eight species, the 41st amino acid Glutamic acid (E) was substituted by Serine (S), while the 43rd amino acid Glutamine (Q) was substituted by Glycine (G) (Figure 6B). The structural homology model of C. pomonella GOBP1 showed that both the 41st and 43rd amino acids were located in the loop near the first helix (Figure 6A).
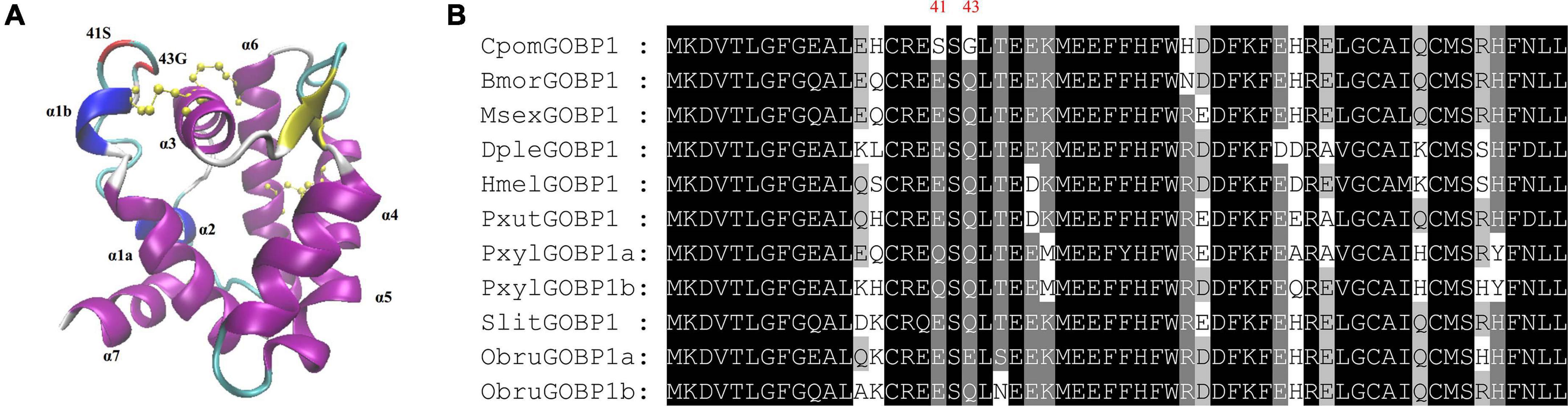
Figure 6. Structural homology model and positively selected sites of C. Pomonella GOBP1. (A) Structural homology model of C. pomonella GOBP1, positively selected sites are marked in red. (B) Multiple sequence alignments of GOBP1 genes from ten moth species.
The binding energies that CpomGOBP1 bind with 48 odorant molecules were assessed by AutoDock Vina (Trott and Olson, 2010). Among them, β-bourbonene had the lowest binding energy (-9.3 KJ/mol) with CpomGOBP1 (Supplementary Table 2). β-bourbonene was located in the binding cavity composed of 11 hydrophobic amino acid residues, including Phe12, Phe33, Phe36, Phe76, Phe118, Ile52, Ile94, Val8, Trp37, Met5, and Leu61 (Figures 7A,B).
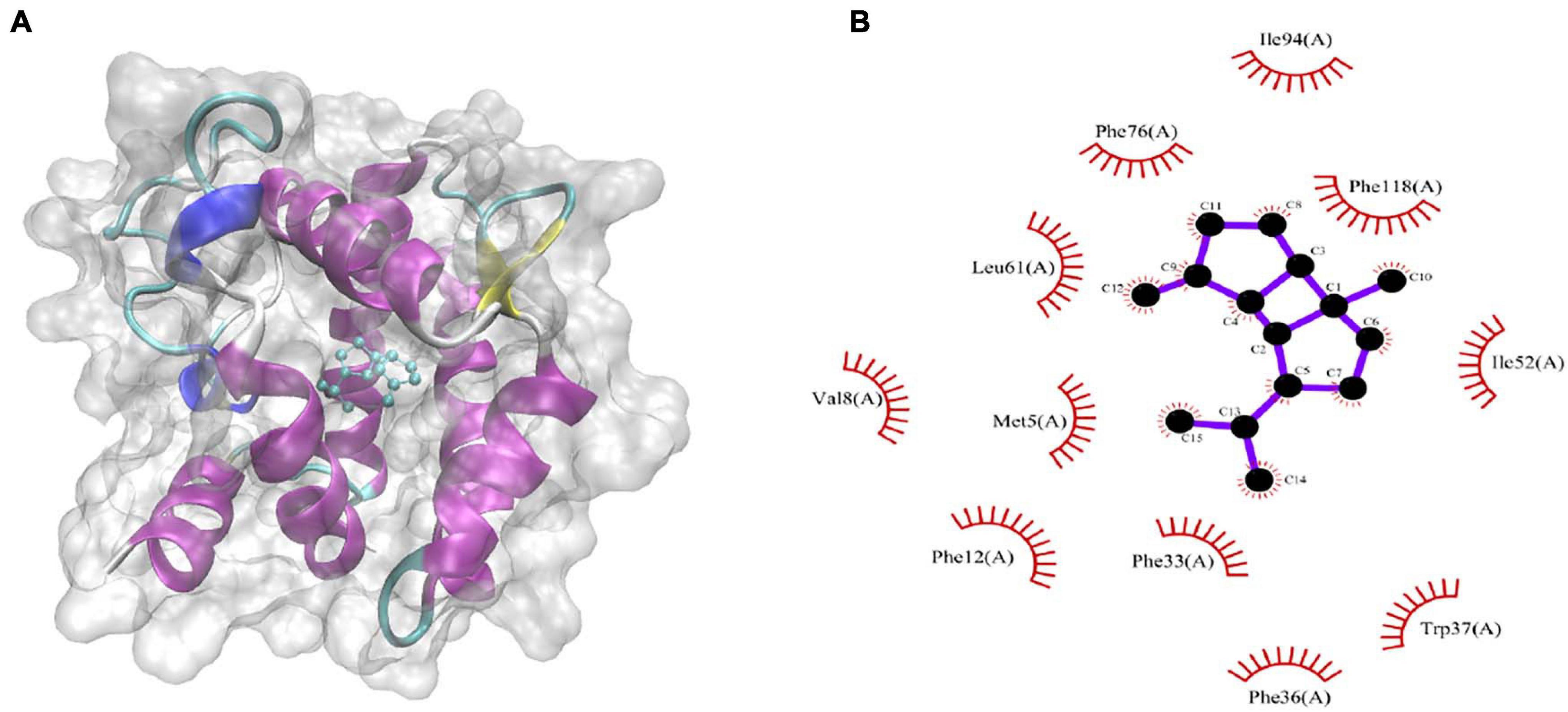
Figure 7. Molecular docking of CpomGOBP1 with β-bourbonene. (A) β-bourbonene located in the binding cavity of CpomGOBP1. (B) Key amino acid residues that interact with β-bourbonene.
The expression profiling of all 40 CpomOBPs were assessed using FPKM values based on transcriptome data (Figure 8). The result showed that 31 CpomOBPs expressed (FPKM ≥ 10) in the antennae, head, leg, wing, and labial palp, except CpomOBP23, CpomOBP2, CpomOBP18, CpomOBP28a, CpomOBP28b, CpomOBP19, CpomOBP21, CpomOBP27, and CpomOBP17. There were 25 and 26 CpomOBPs that are expressed in the antennae of female and male adults, respectively, 22 of them were classic OBPs. The other enriched expression tissue is the labial palp, in which there are 27 and 25 CpomOBPs expressed in female and male adults. Three CpomPBPs, CpomGOBP1, CpomGOBP2a, and CpomGOBP2b, were mainly expressed in the antennae and labial palp. It is interesting to note that CpomGOBP2c was specifically expressed in wing.
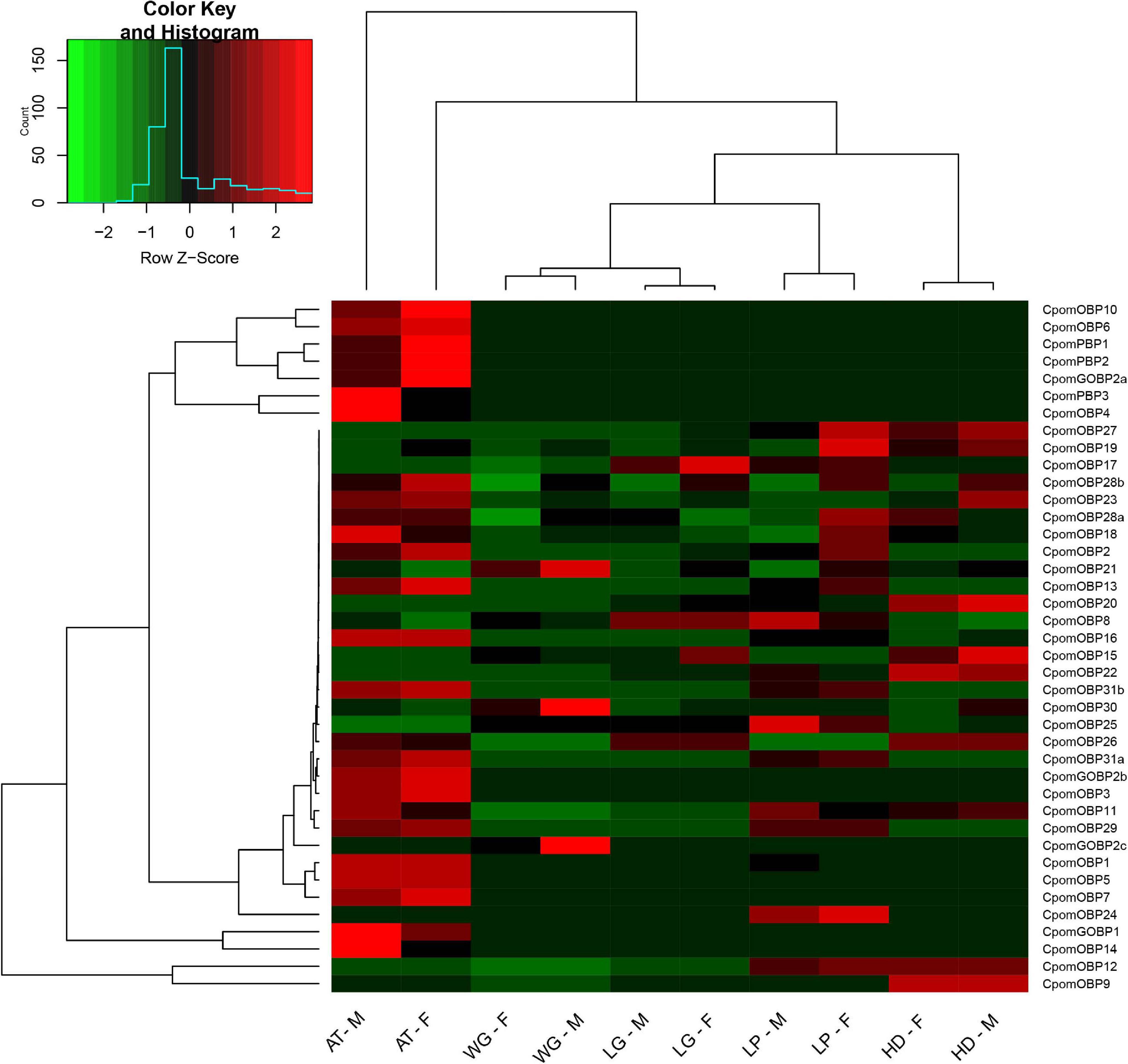
Figure 8. Expression profiling of 40 CpomOBPs in different tissues. F, female; M, male; AT, antenna; HD, head (antennae removed); WG, wing; LG, leg; LP, labial palp.
In this study, we identified 40 OBP genes in C. pomonella, which is similar to several lepidopteran moth species e.g., 43 OBPs in B. mori (Gong D. P. et al., 2009), 39 OBPs in P. xylostella (Cai et al., 2020), 40 OBPs in H. armigera (Pearce et al., 2017), and 36 OBPs in S. litura (Cheng et al., 2017). However, C. pomonella has fewer OBPs than M. sexta and S. frugiperda, both of which have 50 OBP genes (Gouin et al., 2017). Variation in the numbers of OBP genes among moth species suggests that the evolution of OBP genes occurred during the speciation adaptation process and functional requirements for each species. A study in B. mori found that classic OBPs were dominant in the OBP gene family: B. mori has 29 classic OBPs, five plus-C OBPs, and eight minus-C OBPs (Gong D. P. et al., 2009). Similarly, in our study the 40 OBP genes of C. pomonella were classified as 30 classic OBPs, four plus-C OBPs, four minus-C OBPs, and two atypical OBPs. A previous study suggested that most of the classic OBPs and all ABPIIs are likely involved in chemoreception, since they show increased chemosensory tissue expression (Dippel et al., 2014). The fact that 75% of OBPs in C. pomonella are classic OBPs indicated that these genes are essential in recognizing host plants or pheromones such as sex pheromones. The result of expression profiling indicates that 22 classic OBPs were expressed in the antennae, which is similar to the finding in Tribolium castaneum (Dippel et al., 2014).
We used the CAFÉ software to estimate gene gains and losses, rather than directly comparing the number of OBP genes, because it considers a birth-and-death model in the evolutionary process (Han et al., 2013). In the most recent common ancestor of moths considered in this study, approximately 162 Mya inferred by two time frames adopted from TimeTree (see section “Materials and Methods”), 42 OBP genes were shared. There were no more than two expanded and contracted genes in each ancestor node, which indicated that speciation may not be driven by the evolution of OBP genes. The gene gains or losses of each species compared to their distant ancestors range from 1 to 7. According to this result, we suggest that the functional divergence of OBP genes occurred mainly after speciation, as a result of adapting to a new diversity of environments such as new host plants or pheromones. As a result, the OBP genes may be under positive selection. However, the variation of OBPs in moths is smaller than the odorant receptors (ORs) or gustatory receptors (GRs): the expanded or contracted genes of these two gene families is as high as 54 (Engsontia et al., 2014). In general, we found that C. pomonella lost two OBP genes compared to its closest ancestor.
To further explore which OBP genes have expanded or contracted in C. pomonella, we built a phylogenetic tree and performed collinearity analysis in chromosome location by comparing with related moth species. The results showed that OBP genes were conserved except the genes in Group 2, which contains many expanded genes in M. sexta. We also noticed some gene gains and losses in the conserved clade Group 1, composed of GOBP and PBP genes. The GOBPs and PBPs were a specific conserved subfamily in butterflies and moths, including GOBP1-2 and PBPA-D, which are in a tandem array with a fixed order in the same chromosome. These genes were thought to be involved in the recognition of volatile organic compounds (VOC) and sex pheromones of insects (Liu et al., 2020). However, recent studies showed some variations in this subfamily, including gene gains, losses, inversions, and translocations (Yasukochi et al., 2018). Although GOBP1 and GOBP2 were regarded as conserved in lepidopteran species (Vogt et al., 2015), some studies found that gene gains occurred in the GOBP1 genes, such as duplication events of GOBP1 in P. xylostella (Yasukochi et al., 2018) and Operophtera brumata (Yasukochi et al., 2018). In our study, we found a duplication event of a GOBP2 gene that generated three GOBP2 (GOBP2a, GOBP2b, and GOBP2c), two of which have been reported by Garczynski (Garczynski et al., 2013). PBP gene gains and losses occurred more commonly; most Lepidoptera have lost the PBPB gene (Yasukochi et al., 2018), while PBPA was expanded in B. mori (Gong D. P. et al., 2009). Similarly, we found that the PBPB gene was also lost in C. pomonella, which suggests that this gene may be undergoing a gene fusion event (Yasukochi et al., 2018).
Most OBP genes result from tandem duplications in insects, such as Drosophila melanogaster (Hekmat-Scafe et al., 2002) and Anopheles gambiae (Xu et al., 2003) in Diptera; Tribolium castaneum (Dippel et al., 2014) in Coleoptera; and B. mori (Gong D. P. et al., 2009) and C. pomonella (this study) in Lepidoptera. However, in earlier diverging ancestor orders including Hemiptera and Hymenoptera, there are fewer OBP genes without large tandem duplications (Vieira and Rozas, 2011), as in Acyrthosiphon pisum (Zhou et al., 2010) and Bemisia tabaci (Zeng et al., 2019) in Hemiptera; and Apis mellifera (Foret and Maleszka, 2006) and Solenopsis invicta (Pracana et al., 2017) in Hymenoptera. We also found a very consistent collinearity between B. mori and C. pomonella. These findings strongly suggest that the expansion of most OBP genes is caused by tandem duplications, and the tandem duplications of OBP genes in Lepidoptera occurred before speciation, indicating the existence of mainly purifying selection in moth OBP genes. In addition, the duplicated CpomGOBP2c gene is located in chromosome 21, instead of the GOBP/PBP gene cluster in chromosome 8, which indicates functional differentiation.
Some studies showed that single-point mutation of an amino acid could cause functional differentiation (Leary et al., 2012; Yang et al., 2017). Therefore, we further tested whether there are some positive sites in the OBP genes in C. pomonella. The results of evolutionary analysis by site model showed that most OBP genes evolved under purifying selection with ω ranging from 0.00547 to 0.15846 estimated by the M0 model. Similarly, most OBP genes in B. mori also evolved under purifying selection (Gong D. P. et al., 2009). The purifying selection of OBP genes is potentially due to functional constraints (Gong D. P. et al., 2009). However, among the OBP genes of lepidopteran species, the major function of PBPs is mainly to sense pheromones (Gong Y. et al., 2009), while GOBPs mainly sense the volatiles of host plants (Vogt et al., 2002). We assumed that GOBP/PBP genes may evolve under positive selection due to the vast diversity of sex pheromones and host volatiles. The results of the branch-site model on GOBP/PBP genes suggested that the GOBP1 gene in C. pomonella evolved under positive selection. We detected two positively selected sites (41 S and 43 G) in CpomGOBP1, both located in the loop, close to the first disulfide bridge on helix 1. The mutations of these two amino acid residues may influence the fold shape of the binding cavity by modifying the disulfide bridge, which will cause functional differentiation (Sanchez-Gracia and Rozas, 2008). The docking result suggests that CpomGOBP1 may have the ability to bind with β-bourbonene, however this must be functionally validated.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
FW, WQ, and NY conceived, designed this study, and revised the manuscript. CH, XZ, DH, QW, RT, LX, WL, WW, BL, and YX collected the data and completed bioinformatics analysis. CH and XZ drafted the manuscript. All authors read and approved the final manuscript.
This research was supported by Shenzhen Science and Technology Program (KQTD20180411143628272), China Postdoctoral Science Foundation (2020M683001), the Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (caascx-2017-2022-IAS), the Youth Program of National Natural Science Foundation of China (31901949 and 31801803), and the Special Fund for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (PT202001-05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.690185/full#supplementary-material
Andersson, M. N., Löfstedt, C., and Newcomb, R. D. (2015). Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 3:53. doi: 10.3389/fevo.2015.00053
Ansebo, L., Coracini, M. D. A., Bengtsson, M., Liblikas, I., Ramírez, M., Borg-Karlson, A. K., et al. (2004). Antennal and behavioural response of codling moth Cydia pomonella to plant volatiles. J. Appl. Entomol. 128, 488–493. doi: 10.1111/j.1439-0418.2004.00878.x
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagne, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bengtsson, M., Bäckman, A. C., Liblikas, I., Ramirez, M. I., Borg-Karlson, A. K., Ansebo, L., et al. (2001). Plant odor analysis of apple: antennal response of codling moth females to apple volatiles during phenological development. J. Agri. Food Chem. 49, 3736–3741. doi: 10.1021/jf0100548
Cai, L. J., Zheng, L. S., Huang, Y. P., Xu, W., and You, M. S. (2020). Identification and characterization of odorant binding proteins in the diamondback moth, Plutella xylostella. Insect Sci. 29, 531–544. doi: 10.1111/imb.12664
Campanini, E. B., and de Brito, R. A. (2016). Molecular evolution of odorant-binding proteins gene family in two closely related Anastrepha fruit flies. BMC Evol. Biol. 16:198. doi: 10.1186/s12862-016-0775-0
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cheng, T. C., Wu, J. Q., Wu, Y. Q., Chilukuri, R. V., Huang, L. H., Yamamoto, K., et al. (2017). Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756. doi: 10.1038/s41559-017-0314-4
Colovos, C., and Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519. doi: 10.1002/pro.5560020916
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest-HPC: fast selection of best-fit models of protein evolution. Lect. Notes Comput. Sci. 6586, 177–184. doi: 10.1007/978-3-642-21878-1_22
Dippel, S., Oberhofer, G., Kahnt, J., Gerischer, L., Opitz, L., Schachtner, J., et al. (2014). Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15:1141. doi: 10.1186/1471-2164-15-1141
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432.
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.
Engsontia, P., Sangket, U., Chotigeat, W., and Satasook, C. (2014). Molecular evolution of the odorant and gustatory receptor genes in Lepidopteran insects: implications for their adaptation and speciation. J. Mol. Evol. 79, 21–39. doi: 10.1007/s00239-014-9633-0
Foret, S., and Maleszka, R. (2006). Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16, 1404–1413. doi: 10.1101/gr.5075706
Garczynski, S. F., Coates, B. S., Unruh, T. R., Schaeffer, S., Jiwan, D., Koepke, T., et al. (2013). Application of Cydia pomonella expressed sequence tags: identification and expression of three general odorant binding proteins in codling moth. Insect Sci. 20, 559–574. doi: 10.1111/j.1744-7917.2012.01560.x
Gong, D. P., Zhang, H. J., Zhao, P., Xia, Q. Y., and Xiang, Z. H. (2009). The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics 10:332. doi: 10.1186/1471-2164-10-332
Gong, Y., Pace, T. C., Castillo, C., Bohne, C., O’Neill, M. A., and Plettner, E. (2009). Ligand-interaction kinetics of the pheromone- binding protein from the gypsy moth. L. dispar: insights into the mechanism of binding and release. Chem. Biol. 16, 162–172. doi: 10.1016/j.chembiol.2009.01.005
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J.-M., Duvic, B., et al. (2017). Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7:11816.
Han, M. V., Thomas, G. W. C., Lugo-Martinez, J., and Hahn, M. W. (2013). Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997. doi: 10.1093/molbev/mst100
Hekmat-Scafe, D. S., Scafe, C. R., McKinney, A. J., and Tanouye, M. A. (2002). Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12, 1357–1369. doi: 10.1101/gr.239402
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Katoh, K., Misawa, K., Kuma, K-i., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kawamoto, M., Jouraku, A., Toyoda, A., Yokoi, K., Minakuchi, Y., Katsuma, S., et al. (2019). High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem. Mole. Biol. 107, 53–62. doi: 10.1016/j.ibmb.2019.02.002
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Knight, A. L., Mujica, V., Herrera, S. L., and Tasin, M. (2019). Addition of terpenoids to pear ester plus acetic acid increases catches of codling moth (Lepidoptera: Tortricidae). J. Appl. Entomol. 143, 813–821. doi: 10.1111/jen.12646
Kumar, S., Neven, L. G., Zhu, H., and Zhang, R. (2015). Assessing the global risk of establishment of Cydia pomonella (Lepidoptera: Tortricidae) using CLIMEX and MaxEnt niche models. J. Econ. Entomol. 108, 1708–1719. doi: 10.1093/jee/tov166
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291. doi: 10.1107/s0021889892009944
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Liu, J. Y., Tian, Z., and Zhang, Y. L. (2016). Structure-based discovery of potentially active semiochemicals for Cydia pomonella (L.). Sci. Rep. 6:34600.
Liu, Y., Hu, Y., Bi, J., Kong, X., Long, G., Zheng, Y., et al. (2020). Odorant-binding proteins involved in sex pheromone and host-plant recognition of the sugarcane borer Chilo infuscatellus (Lepidoptera: Crambidae). Pest Manag. Sci. 76, 4064–4076. doi: 10.1002/ps.5961
Lüthy, R., Bowie, J. U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356, 83–85. doi: 10.1038/356083a0
Manoharan, M., Ng Fuk Chong, M., Vaïtinadapoulé, A., Frumence, E., Sowdhamini, R., and Offmann, B. (2013). Comparative genomics of odorant binding proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 5, 163–180. doi: 10.1093/gbe/evs131
McKenzie, S. K., Oxley, P. R., and Kronauer, D. J. C. (2014). Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genomics 15:718. doi: 10.1186/1471-2164-15-718
Nei, M., and Rooney, A. P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. doi: 10.1146/annurev.genet.39.073003.112240
Pearce, S. L., Clarke, D. F., East, P. D., Elfekih, S., Gordon, K. H. J., Jermiin, L. S., et al. (2017). Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 15:63. doi: 10.1186/s12915-017-0402-6
Pelosi, P., Iovinella, I., Felicioli, A., and Dani, F. R. (2014). Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5:320. doi: 10.3389/fphys.2014.00320
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11:1650. doi: 10.1038/nprot.2016.095
Pracana, R., Levantis, I., Martinez-Ruiz, C., Stolle, E., Priyam, A., and Wurm, Y. (2017). Fire ant social chromosomes: differences in number, sequence and expression of odorant binding proteins. Evol. Lett. 1, 199–210. doi: 10.1002/evl3.22
Sanchez-Gracia, A., and Rozas, J. (2008). Divergent evolution and molecular adaptation in the Drosophila odorant-binding protein family: inferences from sequence variation at the OS-E and OS-F genes. BMC Evol. Biol. 8:323. doi: 10.1186/1471-2148-8-323
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7:S10.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, J. S., Xiao, S., and Carlson, J. R. (2018). The diverse small proteins called odorant-binding proteins. Open Biol. 8:180208. doi: 10.1098/rsob.180208
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tian, Z., Liu, J. Y., and Zhang, Y. L. (2016a). Key residues involved in the interaction between Cydia pomonella pheromone binding protein 1 (CpomPBP1) and codlemone. J. Agric. Food Chem. 64, 7994–8001. doi: 10.1021/acs.jafc.6b02843
Tian, Z., Liu, J. Y., and Zhang, Y. L. (2016b). Structural insights into Cydia pomonella pheromone binding protein 2 mediated prediction of potentially active semiochemicals. Sci. Rep. 6:22336.
Tian, Z., and Zhang, Y. (2016). Molecular characterization and functional analysis of pheromone binding protein 1 from Cydia pomonella (L.). Insect Mol. Biol. 25, 769–777. doi: 10.1111/imb.12261
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461.
Vieira, F. G., and Rozas, J. (2011). Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490. doi: 10.1093/gbe/evr033
Vieira, F. G., Sanchez-Gracia, A., and Rozas, J. (2007). Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 8:R235.
Vogt, R. G., Grosse-Wilde, E., and Zhou, J. J. (2015). The Lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62, 142–153. doi: 10.1016/j.ibmb.2015.03.003
Vogt, R. G., Rogers, M. E., Franco, M. D., and Sun, M. (2002). A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera). J. Exp. Biol. 205, 719–744. doi: 10.1242/jeb.205.6.719
Wallace, A. C., Laskowski, R. A., and Thornton, J. M. (1995). LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134. doi: 10.1093/protein/8.2.127
Wan, F., Yin, C., Tang, R., Chen, M., Wu, Q., Huang, C., et al. (2019). A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 10:4237.
Wang, W., Xia, M., Chen, J., Deng, F. N., Yuan, R., Zhang, X. P., et al. (2016). Data set for phylogenetic tree and RAMPAGE Ramachandran plot analysis of SODs in Gossypium raimondii and G. arboreum. Data. Brief 9, 345–348. doi: 10.1016/j.dib.2016.05.025
Witzgall, P., Stelinski, L., Gut, L., and Thomson, D. (2008). Codling moth management and chemical ecology. Annu. Rev. Entomol 53, 503–522. doi: 10.1146/annurev.ento.53.103106.093323
Wu, T. D., and Watanabe, C. K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. doi: 10.1093/bioinformatics/bti310
Xu, P. X., Zwiebel, L. J., and Smith, D. P. (2003). Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 12, 549–560. doi: 10.1046/j.1365-2583.2003.00440.x
Yang, K., Huang, L. Q., Ning, C., and Wang, C. Z. (2017). Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. Elife 6:e29100.
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yasukochi, Y., Yang, B., Fujimoto, T., Sahara, K., Matsuo, T., and Ishikawa, Y. (2018). Conservation and lineage-specific rearrangements in the GOBP/PBP gene complex of distantly related ditrysian Lepidoptera. PLoS One 13:e0192762. doi: 10.1371/journal.pone.0192762
Zeng, Y., Yang, Y. T., Wu, Q. J., Wang, S. L., Xie, W., and Zhang, Y. J. (2019). Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 26, 620–634. doi: 10.1111/1744-7917.12576
Zhan, S., Merlin, C., Boore, J. L., and Reppert, S. M. (2011). The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185. doi: 10.1016/j.cell.2011.09.052
Zhang, H., Chen, J. L., Lin, J. H., Lin, J. T., and Wu, Z. Z. (2020). Odorant-binding proteins and chemosensory proteins potentially involved in host plant recognition in the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 76, 2609–2618. doi: 10.1002/ps.5799
Zhou, J. J., Vieira, F. G., He, X. L., Smadja, C., Liu, R., Rozas, J., et al. (2010). Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 19, 113–122. doi: 10.1111/j.1365-2583.2009.00919.x
Keywords: odorant binding proteins, codling moth, Cydia pomonella, positive selection, comparative genomics, gene gains and losses
Citation: Huang C, Zhang X, He D, Wu Q, Tang R, Xing L, Liu W, Wang W, Liu B, Xi Y, Yang N, Wan F and Qian W (2021) Comparative Genomics Provide Insights Into Function and Evolution of Odorant Binding Proteins in Cydia pomonella. Front. Physiol. 12:690185. doi: 10.3389/fphys.2021.690185
Received: 02 April 2021; Accepted: 15 June 2021;
Published: 07 July 2021.
Edited by:
Paivi H. Torkkeli, Dalhousie University, CanadaReviewed by:
William Benjamin Walker III, United States Department of Agriculture (USDA-ARS), United StatesCopyright © 2021 Huang, Zhang, He, Wu, Tang, Xing, Liu, Wang, Liu, Xi, Yang, Wan and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nianwan Yang, eWFuZ25pYW53YW5AY2Fhcy5jbg==; Fanghao Wan, d2FuZmFuZ2hhb0BjYWFzLmNu; Wanqiang Qian, cWlhbndhbnFpYW5nQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.