- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Plant Protections, Agricultural University of Hebei, Baoding, China
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is an accurate and convenient technique for quantifying expression levels of the target genes. Selection of the appropriate reference gene is of the vital importance for RT-qPCR analysis. Hippodamia variegata is one of the most important predatory natural enemies of aphids. Recently, transcriptome and genome sequencings of H. variegata facilitate the gene functional studies. However, there has been rare investigation on the detection of stably expressed reference genes in H. variegata. In the current study, by using five analytical tools (Delta Ct, geNorm, NormFinder, BestKeeper, and RefFinder), eight candidate reference genes, namely, Actin, EF1α, RPL7, RPL18, RPS23, Tubulin-α, Tubulin-β, and TufA, were evaluated under four experimental conditions including developmental stages, tissues, temperatures, and diets. As a result, a specific set of reference genes were recommended for each experimental condition. These findings will help to improve the accuracy and reliability of RT-qPCR data, and lay a foundation for further exploration on the gene function of H. variegata.
Introduction
The reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is one of the most sensitive, accurate, and convenient methods for detecting quantitative analysis of gene expression in biological samples (Kubista et al., 2006; Provenzano and Mocellin, 2007; Derveaux et al., 2010). For RT-qPCR studies, it is necessary to select appropriate reference genes to correct and standardize the expression level of target genes. This will reduce the impact of RNA quality and cDNA synthesis efficiency on the results. Therefore, the validity of candidate internal reference gene needs to be evaluated before conducting RT-qPCR assay.
Reference genes, also called housekeeping genes, are supposed to be stably expressed in organisms under various biotic or abiotic conditions (Thellin et al., 1999; de Jonge et al., 2007; Zhu et al., 2014). The ideal internal reference gene should be stably expressed in different types of tissues and in different treatments of the same tissue, and its expression level is not affected by any endogenous or exogenous factors (Brym et al., 2013; Janská et al., 2013). Nevertheless more and more researches proved that the internal reference genes show significant various expression levels in different experimental treatments such as development stages, tissues, cells, and temperatures (Sun et al., 2008; Huis et al., 2010). Therefore, it is necessary to identify reference genes for their expression under different experimental conditions (Guo et al., 2016; Wan et al., 2017; Renard et al., 2018). Common reference genes include actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein S3 (RPS3), 18S ribosomal RNA (18S), 25S ribosomal RNA (25S), tubulin, translation elongation factor 1-alpha (EF1α), and others have been used extensively for RT-qPCR analysis (Lu et al., 2013, 2015; Yuan et al., 2014; Zhang et al., 2015; Ma et al., 2016; Yan et al., 2016; Gao et al., 2017; Hu et al., 2018; Yin et al., 2020).
Hippodamia variegata (Coccinellidae: Hippodamia) is one of the most important natural enemies of aphids infesting various crops in many countries (Natskova, 1973; Hameed et al., 1975; Belikova and Kosaev, 1985). The stably expressed reference genes in coccinellid insects such as Harmonia axyridis (Yang et al., 2018), Henosepilachna vigintioctomaculata (Lü et al., 2018a), Coccinella septempunctata (Yang et al., 2016), and Hippodamia convergens (Pan et al., 2015) have been screened under different experimental conditions. In this study, eight reference genes for insects, including actin, elongation factor-1α (EF1α), ribosomal protein L7 (RPL7), ribosomal protein L18 (RPL18), ribosomal protein L23 (RPL23), tubulin alpha (Tubulin-α), tubulin beta (Tubulin-β), and elongation factor Tu (TufA), have been selected as candidate reference genes for H. variegata. The expression stabilities of these genes were assessed with respect to different developmental stages, tissues, temperatures, and diet treatments using five different statistical algorithms. The five statistical algorithms are Delta Ct method (Silver et al., 2006), geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and RefFinder (Xie et al., 2012), which integrates four different statistical algorithms. Finally, the expression pattern of olfactory co-receptor (Orco) of H. variegata was assayed to verify the reference genes. The results will serve as a guide for gene expression studies in H. variegata.
Materials and Methods
Insect Rearing
The H. variegata were obtained from Langfang Experimental Station of Chinese Academy of Agricultural Sciences, Hebei Province, China. Larvae and adults were maintained in plastic containers (8 × 8 × 11 cm) with Myzus persicae as a food source. The colonies were reared under conditions of 25 ± 1°C, 65 ± 10% relative humidity, and a 14:10-h light/dark photoperiod.
Sample Treatment and Collection
Samples at developmental stages including eggs (50 eggs), first-instar larvae (10 individuals), second-instar larvae (10 individuals), third-instar larvae (2 individuals), fourth-instar larvae (1 individual), pupae (1 individual), and adults (1 male and 1 female individual) were selected as different treatment groups. Moreover, different tissues including head, thorax, abdomen, leg, and wing were dissected and harvested from adult females or males at 2 to 3 days (N = 30).
To evaluate the effects of temperature, third-instar larvae of H. variegata were exposed to 15, 25, and 35°C for 5 h. In diet treatment groups, 23-instar larvae of H. variegata were fed 3-day-old larvae of Spodoptera frugiperda (FAW), nymphs and adults of M. persicae, and canola pollen separately after 12 h of starvation (Schuldiner-Harpaz and Coll, 2017a, b).
Each experiment was replicated three times independently. Samples were preserved in 1.5-ml centrifuge tubes and snap frozen immediately in liquid nitrogen before storage at –80°C.
Total RNA Extraction and Reverse Transcription
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. RNA purity was checked on a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). Each sample of 2.0 μg total RNA was used to synthesize single-stranded cDNA using the FastQuant RT kit (Tiangen Biotech, Beijing Co., Ltd., Beijing, China).
Selection of Candidate Reference Genes
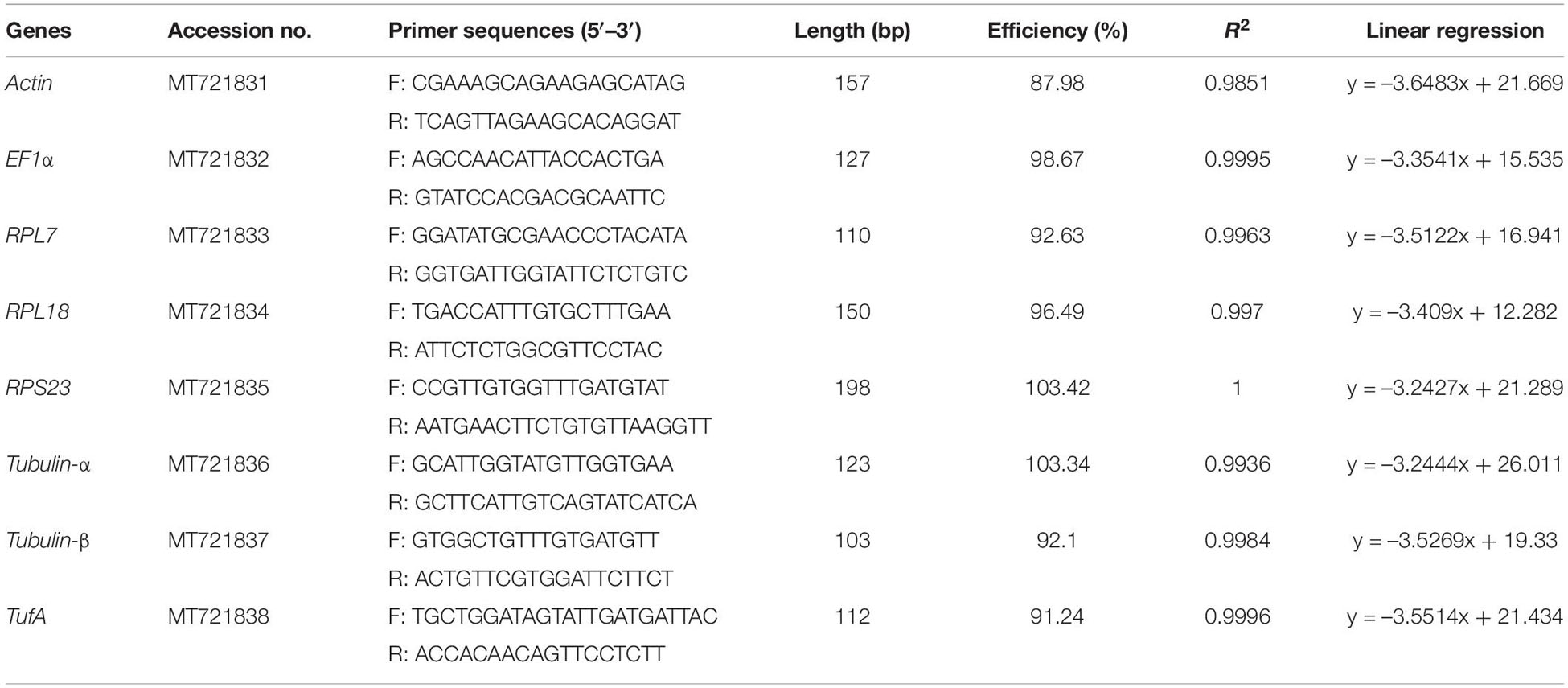
The reciprocal BLAST hits approach was used to screen reference genes from H. variegata transcriptome and genome data (unpublished data). The reference genes from other insect species in GenBank were downloaded from NCBI1 queried individually to H. variegata transcriptome and genome using the TBLASTN program with a permissive E-value cutoff of 10–5 to get the hits. And then, each of the queried hits was compared back against non-redundant database of NCBI by the BLASTX program (E < 10–5) to determine whether the original sequence was one of the hits. All the selected reference genes are listed in Table 1.
The primers of eight candidate reference genes were designed using Beacon Designer TM 7.9 software (Premier Biosoft International, CA, United States) according to sequences obtained from our recently sequenced transcriptomes and genome for H. variegata. Primer parameters were as follows: optimal temperature 60 ± 2°C, GC content 40–60%, and 18–24 bp in length. The amplification size was between 100 and 200 bp (Table 1).
Gene-specific primers (Supplementary Table 1) were designed using Primer Premier 6.0 software (Premier Biosoft International, CA, United States) to amplify ORF of Orco gene. PCR amplifications were performed in 50-μl reactions containing 25 μl 2 × Phanta Max Master Mix, 2 μl of each primer (10 μM each), 4 μl first-strand cDNA, and 17 μl ddH2O. The PCR parameters were as follows: one cycle of 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; a final cycle of 72°C for 10 min. PCR products were purified and cloned into the pcloneEZ-Blunt TOPO vector (Clone Smarter, United States) for sequencing confirmation.
RT-qPCR Analysis
The RT-qPCR measurements were performed on an ABI Prism 7500 system (Applied Biosystems, Carlsbad, CA, United States). The reaction system contained a mixture of 12.5 μl 2 × SuperReal PreMix Plus (Tiangen Biotech, Beijing Co., Ltd.), 0.75 μl of each primer (10 μM), 1 μg sample cDNA, 0.6 μl 50 × ROX Reference Dye, and proper volume of RNase-free water. PCR cycling parameters were as follows: 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, and cooled to 60°C for 32 s. Then, the PCR products were heated to 95°C for 15 s, cooled to 60°C for 1 min, heated to 95°C for 30 s, and cooled to 60°C for 15 s to measure the dissociation curves. To determine the amplification efficiency (E) and the correlation coefficients (R2), fivefold dilution series of cDNA (1:5, 1:25, 1:125, 1:625, and 1:3,125) were used as templates to construct a standard curve of each gene. Also, the homologous RT-qPCR efficiencies (E) were calculated according to the equation: E = (10[–1/slope] - 1) × 100.
Analysis of the Stability of Reference Gene Expression
Five tools (Delta Ct, geNorm, NormFinder, BestKeeper, RefFinder) were used to evaluate the stability of each reference gene. The Delta Ct method was used to select the optimal reference gene (Silver et al., 2006). geNorm was used to calculate the normalization factor to determine the optimal number of reference genes (Vandesompele et al., 2002); the proposed Vn/Vn + 1 ratio below V ≤ 0.15 does not significantly affect the normalization. The NormFinder algorithm, a model-based method, was used to rank the candidate reference genes by the stability value, and the gene with the lowest value was considered as the most stable reference gene (Andersen et al., 2004). The BestKeeper was used to rank the candidate reference genes based on the SDs and the coefficients of variation (CV) values (Pfaffl et al., 2004). Finally, RefFinder assigned an appropriate weight of the four methods to a single gene and calculated the geometric average of their weights for the overall final ranking (Xie et al., 2012).
Validation of the Selected Reference Genes
The Orco is highly conserved among insect species and is essential for localization of ORs in olfactory sensory neuron (ORN) dendrites and reception of odor signals (Leal, 2013). Thus, Orco was used as the target gene to assess the stability of candidate reference genes. Based on the selected candidate reference gene set, Orco expression levels in different tissues were calculated. The primers are shown in Supplementary Table 1. The gene expression was measured in various tissues and normalized by the optimal reference genes (TufA and RPS23) and the least stable reference genes (Tubulin-α and Tubulin-β). RT-qPCR analysis was performed as described previously. Data were calculated using the 2–△△Ct method (Schmittgen and Livak, 2008) and the expression level of target gene (Orco) has been normalized with the CT-value average of two best and least stable reference genes. All the experiments were performed in three replications, and the results are expressed as means ± SD.
Results
RT-qPCR Analysis
Before evaluating the applicability of the reference genes, the specificity and efficiency of PCR amplification should be first ascertained. PCR amplifications for each primer pair showed a single peak in melt curves (Supplementary Figure 1). The primer efficiency (E) ranged between 87.98 and 103.42% with linear regression coefficient (R2) values of 0.9851–1 (Table 1). Combining the three aforementioned factors, the results showed that the designed primers can precisely amplify candidate reference genes. Ct-values of the eight candidate reference genes ranged from 13.2 to 29.6, covering all the experimental conditions (Figure 1). EF1α was the most abundant reference gene with the lowest Ct-value, followed by RPL18, RPL7, Tubulin-β, RPS23, TufA, Actin, and Tubulin-α.
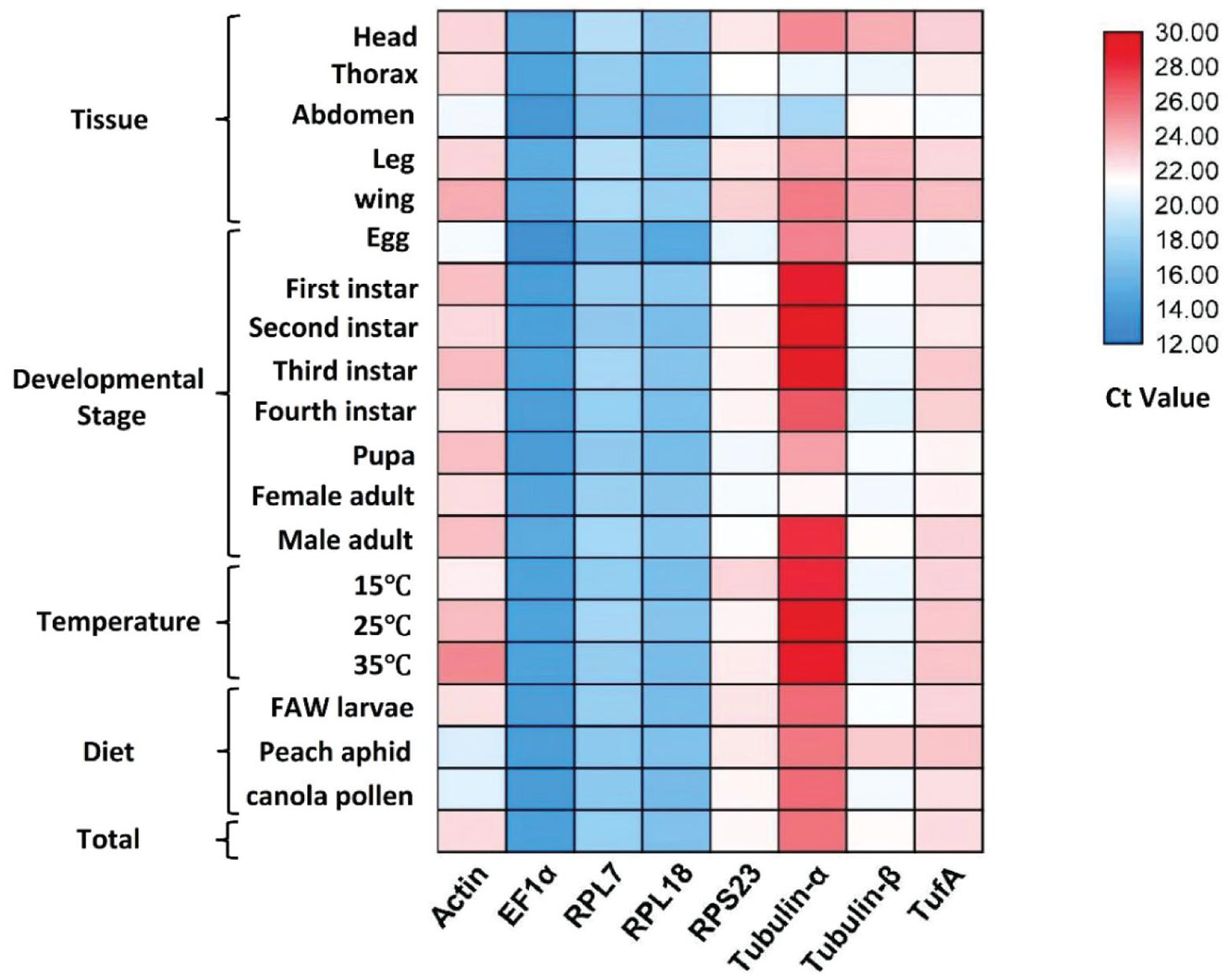
Figure 1. Ct-value of eight candidate reference genes under different experimental conditions. Numbers represent Ct-values.
Expression Stability of Candidate Reference Genes Under Biotic Conditions
For different developmental stages, geNorm ranked the stability from high to low as RPL7 = RPL18, EF1α, TufA, RPS23, Actin, Tubulin-β, and Tubulin-α. NormFinder provided a ranking as RPS23, TufA, RPL7, EF1α, RPL18, Actin, Tubulin-β, and Tubulin-α. BestKeeper offered a list as follows: EF1α, RPS23, RPL7, RPL18, Tubulin-β, TufA, Actin, and Tubulin-α. Delta Ct provided a ranking as RPL7, EF1α, TufA, RPL18, RPS23, Actin, Tubulin-β, and Tubulin-α. Combining the results of all four programs, RefFinder identified the two candidates RPL7 and EF1α with the highest consensus at different developmental stages. RPL7 was the most stable gene, whereas Tubulin-α was the most unstable candidate (Figure 2A and Table 2). The geNorm results demonstrated that the initial V-value at V2/3 < 0.015, indicating that only two reference genes were required to normalize the target gene data (Figure 3).
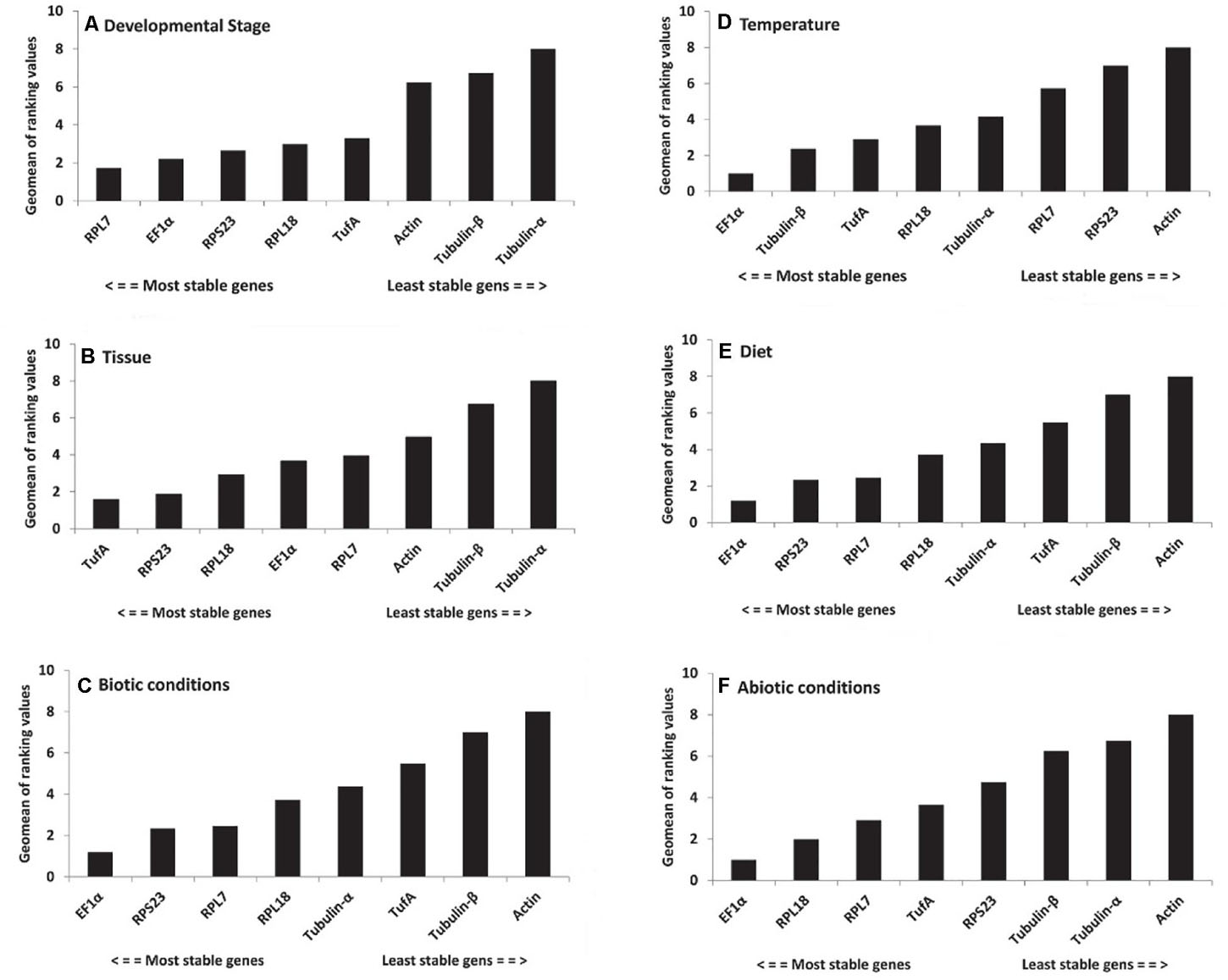
Figure 2. Expression stability of candidate reference genes under biotic and abiotic experimental conditions. (A) Development stages, (B) tissues, (C) biotic conditions, (D) temperatures, (E) diets, and (F) abiotic conditions. A lower Geomean value suggests stable expression.
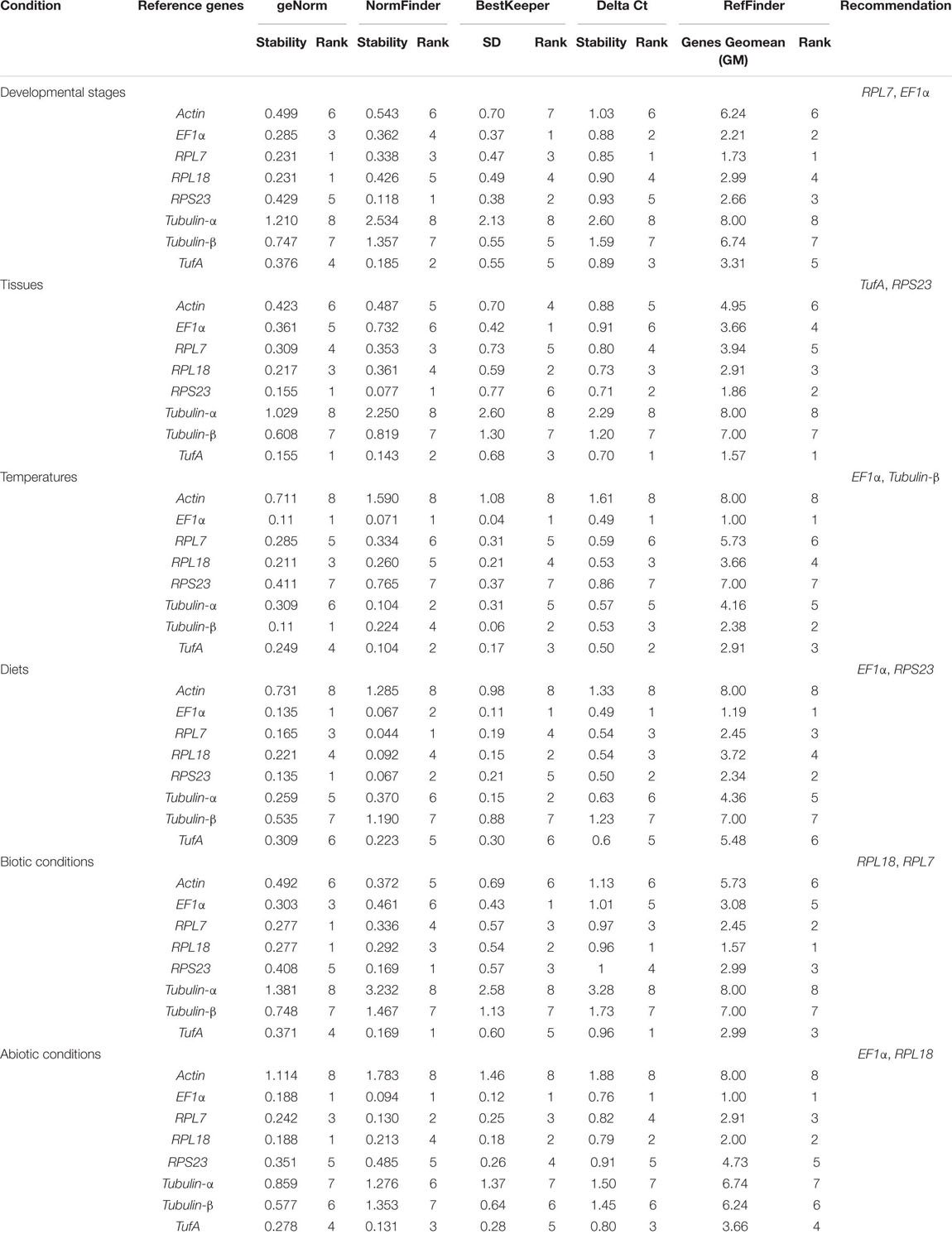
Table 2. Expression stability of eight candidate reference genes in Hippodamia variegata under different experimental conditions using five programs.
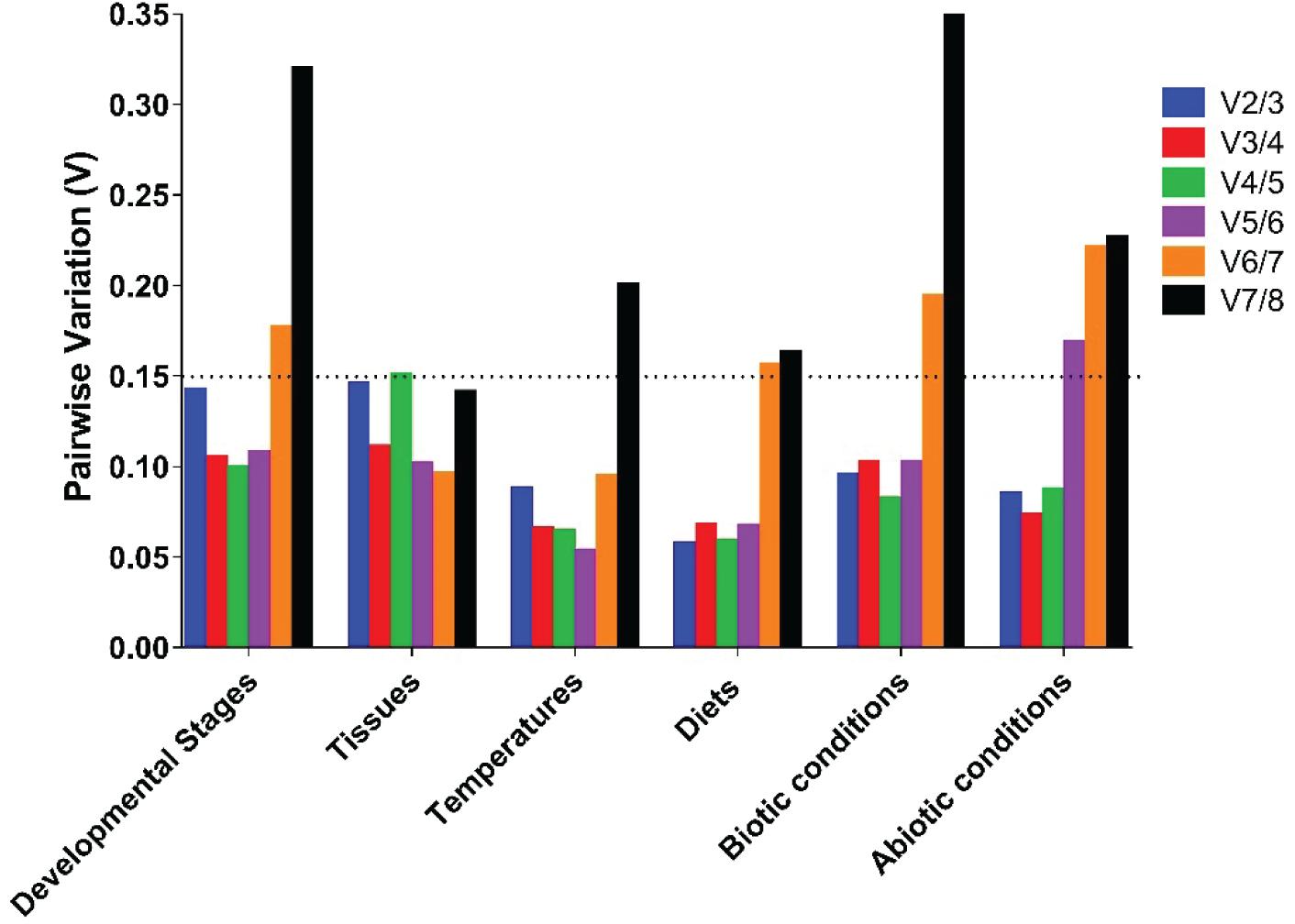
Figure 3. Optimal number of reference genes required for accurate normalization of gene expression by geNorm. Based on geNorm analysis, average pairwise variations were calculated between the normalization factors NFn and NFn + 1. Values <0.15 indicate that n + 1 genes were not required for the normalization of gene expression.
For the tissue-specific experiment, geNorm, NormFinder, and Delta Ct identified TufA and RPS23 as the most suitable reference genes, and BestKeeper identified EF1α and RPL18 as the most suitable reference genes (Table 2). Through the calculation of RefFinder, the overall ranking of the eight candidate internal reference genes was as follows: TufA > RPS23 > RPL18 > EF1α > RPL7 > Actin > Tubulin-β > Tubulin-α (Figure 2B and Table 2). The geNorm pairwise variation analysis also revealed that the first V < 0.15 emerged at V2/3 (Figure 3). Thus, the best combination of reference genes for tissue-specific samples of H. variegata was TufA and RPS23.
According to the comprehensive ranking of RefFinder, under biological conditions, the most stable to the least stable candidate reference genes were RPL18, RPL7, TufA, RPS23, EF1α, Actin, Tubulin-β, and Tubulin-α (Figure 2C and Table 2). The geNorm results identified an initial V < 0.015 at V2/3, indicating that only two references genes were needed to normalize the target gene data (Figure 3). Thus, the best combination of reference genes for tissue-specific samples of H. variegata was RPL18 and RPL7.
Stability of Candidate Reference Genes Under Abiotic Conditions
Under different temperature conditions, combining the results of four software analysis, the RefFinder rankings were as follows: EF1α, Tubulin-β, TufA, RPL18, Tubulin-α, RPL7, RPS23, and Actin (Figure 2D and Table 2). Through geNorm pairwise variation analysis, the results showed that the two reference genes were sufficient to normalize the target genes in different temperatures (Figure 3). Therefore, for H. variegata at different temperatures, the best combination of reference genes is EF1α and Tubulin-β.
For the diet treatment, geNorm and Delta Ct identified EF1α and RPS23 as the most suitable reference genes, NormFinder identified EF1α and RPL7, whereas BestKeeper identified EF1α and RPL18 as the most suitable reference genes (Table 2). Based on RefFinder analysis, the comprehensive reference genes are ranked from the most stable to the most unstable as follows: EF1α, RPS23, RPL7, RPL18, Tubulin-α, TufA, Tubulin-β, and Actin (Figure 2E and Table 2). In addition, geNorm pairwise variation analysis indicated that two reference genes were sufficient to normalize the target gene in dietary treatments (Figure 3). Therefore, we suggest that the best combination of reference genes in dietary treatments is EF1α and RPS23.
According to comprehensive ranking of RefFinder, under abiotic conditions, the order of the most stable to unstable candidate reference genes was as follows: EF1α, RPL18, RPL7, TufA, RPS23, Tubulin-β, Tubulin-α, and Actin (Figure 2F and Table 2). The geNorm results identified an initial V < 0.015 at V2/3, indicating that only two references genes were required to standardize the target gene data (Figure 3). Therefore, under different abiotic status, the best combination of reference genes of H. variegata was EF1α and RPL18.
Validation of Selected Reference Genes
When standardized with the two most stable reference genes (TufA and RPL23), the expression pattern of Orco in various tissues were similar. The expression level of Orco was as follows: head > leg > wing > abdomen > thorax. However, the expression level of Orco in the head was almost 600-fold higher than in the leg when the two most stable reference genes were used for normalization (Figure 4A). In contrast, when the two most unstable reference genes (Tubulin-α and Tubulin-β) were used for normalization, the Orco expression in the head was tremendously high. The calculated relative expression of Orco in head with two unstable genes was almost 10 times higher than that of normalized with two stable genes (Figure 4B).
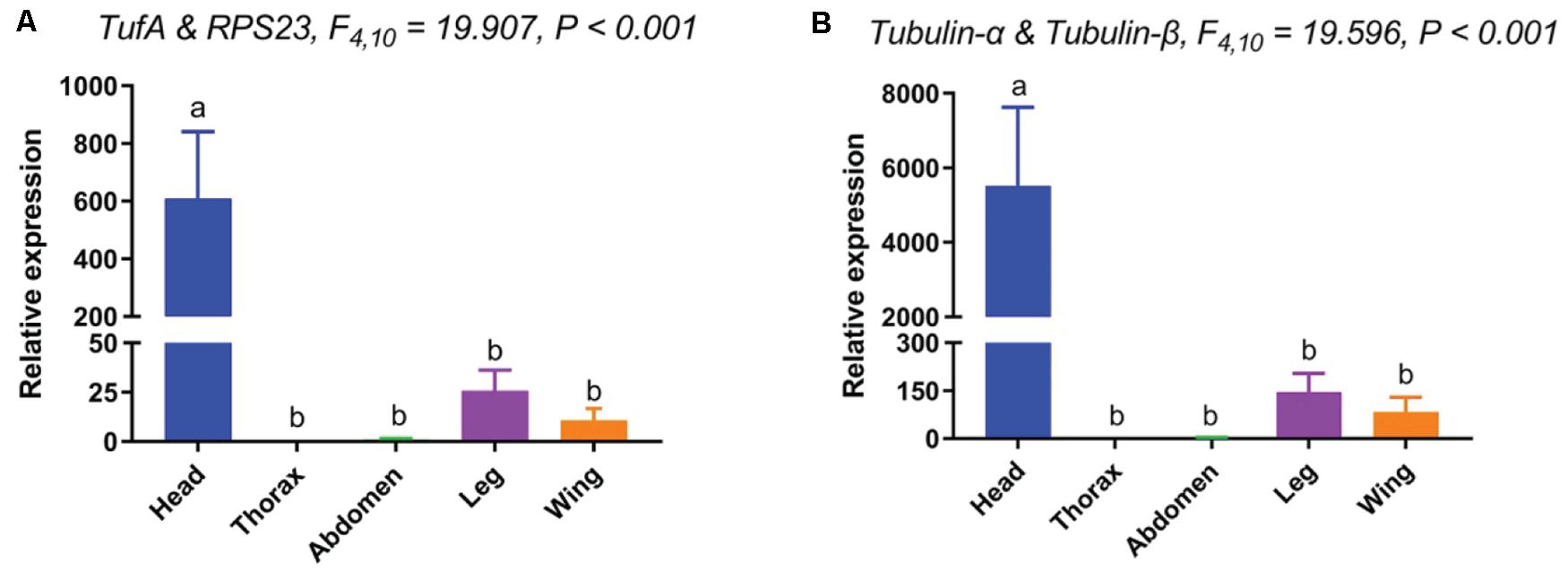
Figure 4. Relative expression levels of Orco in different tissues. The relative mRNA expression levels of Orco were normalized to the most suited [(A). TufA and RPL23) and the least suited [(B). Tubulin-α and Tubulin-β) reference genes. Values are means ± SEM. Different letters indicate significant differences (P < 0.05, two-way ANOVA followed by Tukey’s HSD multiple comparison).
Discussion
H. variegata is one of the most important predatory natural enemies of aphids in the Xinjiang Uygur Autonomous Region, the main cotton-producing area of China, which shows a strong control effect on cotton aphids (Feng et al., 2000). The genome and transcriptomes of both sexes of H. variegata have been sequenced recently (unpublished data). To further investigate the biological roles of any specific gene in H. variegata, quantitative gene expression through RT-qPCR is essential. Following the “Minimum Information for Publication of Quantitative Real-Time PCR Experiments” (MIQE) guideline, reference genes with high stability are necessary (Bustin et al., 2009). However, to our knowledge, the best or combination of best reference genes under various treatment conditions have not been identified in H. variegata so far.
In this study, BestKeeper (Pfaffl et al., 2004), geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and Delta values (Silver et al., 2006) were used to evaluate the expression stability of the candidate internal reference genes. The overall stability of selected reference genes were also evaluated using RefFinder (Xie et al., 2012), which is a web-based analysis tool that integrates all four major calculation programs. Because of the different algorithms, the stability levels derived from the four analysis tools may be different. Optimal number of reference genes could be determined using geNorm analysis by calculating the paired mutation value (Vn/n + 1). When Vn/n + 1 is <0.15, there is no need to add additional reference genes to improve accuracy (Vandesompele et al., 2002).
The expression stability of eight candidate reference genes showed that the most suitable candidate combinations of reference genes were as follows: RPL7 and EF1α for developmental stages, TufA and RPS23 for tissues, EF1α and Tubulin-β for temperature treatment, EF1α and RPS23 for diet treatment, RPL18 and RPL7 for biotic conditions, and EF1α and RPL18 for abiotic conditions. The EF1α, encoding a protein associated with translation elongation, is the most abundant protein in the cell and highly conserved in different species (Yin et al., 2020). Several research works have been reported that EF1α is the most stable reference gene in different group of insect species such as Lymantria dispar (Lepidoptera: Lymantriidae) (Yin et al., 2020), Cydia pommnella (Lepidoptera: Tortricidae) (Wei et al., 2020), Aphis gossypii (Hemiptera: Aphididae) (Ma et al., 2016), Locusta migratoria manilensis (Orthoptera: Acrididae) (Yang et al., 2014), and H. convergens (Pan et al., 2015). The ICG website2 counts the top 10 reported internal reference genes, among which, EF1α is in the first rank (Sang et al., 2017).
The RPL18 encodes a ribosomal protein that is a component of the 60S subunit and belongs to the L18E family of ribosomal proteins (Fagerberg et al., 2014). It has been demonstrated that under different experimental conditions, RPL18 is also one of the most stable reference genes in different group of insects such as Solenopsis invicta (Hymenoptera, Formicidae) (Cheng et al., 2013), Anastrepha obliqua (Diptera, Tephritidae) (Nakamura et al., 2016), and Cimex lectularius (Hemiptera, Cimicidae) (Mamidala et al., 2011). Consistently, RPL18 is stably expressed under different biotic and abiotic conditions and could be considered as a proper reference gene for normalization of RT-qPCR data in H. variegata. Similar to EF1α, RPL family genes are also ranked among the top five stable reference genes for RT-qPCR analysis by ICG website (Sang et al., 2017).
Ribosomal protein S family including RPS23 has also been reported as the most appropriate reference gene in insects (Fu et al., 2013) and other organisms (Yadav et al., 2012) and has been ranked seventh by the ICG website. In contrast, TufA is not a commonly used reference gene in insects (Lü et al., 2018b). However, based on our preliminary evaluation and main experimental data, it has been shown that TufA was stably expressed in different tissues and could be considered as a reference gene in H. variegata. Previous studies demonstrated that in different experimental conditions, TufA is one of the most stable reference genes in different species of bacteria (Savard and Roy, 2009; Jiang et al., 2019).
The Orco is highly conserved in all insects and is widely expressed in the most olfactory receptor neurons (ORNs) of olfactory sensilla, distributed on the head (mainly expressed in antennae and mouthparts), legs, wings, and abdomen (Jacquin-Joly and Merlin, 2004). To validate our obtained results, the two most stable reference genes and the two most unstable reference genes were applied for normalization of Orco. Using the two most stable internal reference genes to examine the expression pattern of Orco, the results were stable and almost similar in different tissues. On the other hand, by using the two most unstable internal reference genes, the relative expression level of Orco normalized with Tubulin-β was approximately double lower in the head than when normalized with Tubulin-α. A similar phenomenon was also observed when OBP20 was used as the target gene to verify the stability of the internal reference gene of Apolygus lucorum (Hemiptera, Miridae). The relative expression level of OBP20, normalized with the stable reference genes (RPL32 and RPL27), was approximately threefold higher in the head than when normalized with two unstable reference genes (ACT and βACT) (Luo et al., 2019). Under two normalization conditions, the expression of target genes in same tissue (head) was different, indicating that the inappropriate use of reference genes in any experimental condition may lead to inaccurate results. Therefore, selection and validation of the best reference genes are crucial to determine the accuracy of the expression pattern of different genes. Taken together, our findings provide a more rigorous method to normalize RT-qPCR data in H. variegata, which lays a foundation for the functional genomics research in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JX and AK conceived and designed the research. JX and CY conducted the experiments. TL and XL analyzed the data. JX and YZ wrote the article. All authors have read and agreed to the published version of the article.
Funding
This work was supported by the National Natural Science Foundation of China (31972338, 31672038, and 31621064) and the National Key Research and Development Program of China (2017YFD0201900 and 2019YFD0300100).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.669510/full#supplementary-material
Footnotes
References
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64:5245. doi: 10.1158/0008-5472.CAN-04-0496
Belikova, E. V., and Kosaev, E. M. (1985). The biology of the most important species of Coccinellidae and their role of controlling aphids in a cotton-lucerne rotation. Izvestiya Turkmeniskoi Akademii Nauk. Ser. Biol. Nauk 5, 61–63.
Brym, P., Ruść, A., and Kamiński, S. (2013). Evaluation of reference genes for qRT-PCR gene expression studies in whole blood samples from healthy and leukemia-virus infected cattle. Vet. Immunol. Immunop. 153, 302–307. doi: 10.1016/j.vetimm.2013.03.004
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cheng, D., Zhang, Z., and He, X. (2013). Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS One 8:e57718. doi: 10.1371/journal.pone.0057718
de Jonge, H. J. M., Fehrmann, R. S. N., Bont, E. S. J. M., Hofstra, R. M. W., Frans, G., Kamps, W. A., et al. (2007). Evidence based selection of housekeeping genes. PLoS One 2:e898. doi: 10.1371/journal.pone.0000898
Derveaux, S., Vandesompele, J., and Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. doi: 10.1016/j.ymeth.2009.11.001
Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Prteomics 13:397. doi: 10.1074/mcp.M113.035600
Feng, H., Wang, L., and Xiong, R. (2000). A study on the population dynamics and predacious function of Hippodamia (Adonia) variegate (Goeze). Entomol. Knowl. 4, 223–226, (in Chinese).
Fu, W., Xie, W., Zhang, Z., Wang, S., Wu, Q., Liu, Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9, 792–802. doi: 10.7150/ijbs.5862
Gao, X., Zhang, S., Luo, J., Wang, C., Lv, L., Zhang, L., et al. (2017). Identification and validation of reference genes for gene expression analysis in Aphidius gifuensis (Hymenoptera: Aphidiidae). PLoS One 12:e0188477. doi: 10.1371/journal.pone.0188477
Guo, H., Jiang, L., and Xia, Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genomics 291, 999–1004. doi: 10.1007/s00438-015-1125-4
Hameed, S. F., Sud, V. K., and Kashyap, N. P. (1975). Adonia variegata (Goeze) (Coccinellidae: Coleoptera), an important predator of the Indian grain aphid, Macrosiphum (Sitobion) miscanthi Tak in Kulu Valley (Himachal Pradesh). Indian J. Entomol. 37, 209–210.
Hu, Y., Fu, H., Qiao, H., Sun, S., Zhang, W., Jin, S., et al. (2018). Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 19:2258. doi: 10.3390/ijms19082258
Huis, R., Hawkins, S., and Neutelings, G. (2010). Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 10:71. doi: 10.1186/1471-2229-10-71
Jacquin-Joly, E., and Merlin, C. (2004). Insect olfactory receptors: contributions of molecular biology to chemical ecology. J. Chem. Ecol. 30, 2359–2397. doi: 10.1007/s10886-004-7941-3
Janská, A., Hodek, J., Svoboda, P., Zámečník, J., Prášil, I. T., Vlasáková, E., et al. (2013). The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol. Genet. Genomics 288, 639–649. doi: 10.1007/s00438-013-0774-4
Jiang, N., Lyu, Q., Han, S., Xu, X., Walcott, R. R., Li, J., et al. (2019). Evaluation of suitable reference genes for normalization of quantitative reverse transcription PCR analyses in Clavibacter michiganensis. MicrobiologyOpen 8:e928. doi: 10.1002/mbo3.928
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27, 95–125. doi: 10.1016/j.mam.2005.12.007
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Ann. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Lü, J., Chen, S., Guo, M., Ye, C., Qiu, B., Wu, J., et al. (2018a). Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9:1614. doi: 10.3389/fphys.2018.01614
Lü, J., Yang, C., Zhang, Y., and Pan, H. (2018b). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Lu, Y., Yuan, M., Gao, X., Kang, T., Zhan, S., Wan, H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8:e68059.
Lu, Y., Zheng, X., Qi, L., Xu, H., Yang, Y., Tian, J., et al. (2015). Evaluation and validation of reference genes for SYBR Green qRT-PCR normalization in Sesamia inferens (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 18, 669–675. doi: 10.1016/j.aspen.2015.08.002
Luo, J., Wang, A., Cheng, Y., Rong, H., Guo, L., Peng, Y., et al. (2019). Selection and validation of suitable reference genes for RT-qPCR analysis in Apolygus lucorum (Hemiptera: Miridae). J. Econ. Entomol. 113, 451–460. doi: 10.1093/jee/toz301
Ma, K., Li, F., Liang, P., Chen, X., Liu, Y., and Gao, X. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003
Mamidala, P., Rajarapu, S. P., Jones, S. C., and Mittapalli, O. (2011). Identification and validation of reference genes for quantitative real-time polymerase chain reaction in Cimex lectularius. J. Med. Entomol. 48, 947–951. doi: 10.1603/ME10262
Nakamura, A. M., Chahad-Ehlers, S., Lima, A. L. A., Taniguti, C. H., Sobrinho, I. Jr., Torres, F. R., et al. (2016). Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. U. K. 6:17480. doi: 10.1038/srep17480
Natskova, V. (1973). The effect of aphid predators on the abundance of aphids on peppers. Rastit Zashch 21, 20–22.
Pan, H., Yang, X., Siegfried, B. D., and Zhou, X. (2015). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10:e0125868. doi: 10.1371/journal.pone.0125868
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Provenzano, M., and Mocellin, S. (2007). Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv. Exp. Med. Biol. 593, 66–73. doi: 10.1007/978-0-387-39978-2_7
Renard, M., Vanhauwaert, S., Vanhomwegen, M., Rihani, A., and Willaert, A. (2018). Expressed repetitive elements are broadly applicable reference targets for normalization of reverse transcription-qPCR data in mice. Sci. Rep. U. K. 8:7642. doi: 10.1038/s41598-018-25389-6
Sang, J., Wang, Z., Li, M., Cao, J., Niu, G., Xia, L., et al. (2017). ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 46, D121–D126. doi: 10.1093/nar/gkx875
Savard, P., and Roy, D. (2009). Determination of differentially expressed genes involved in arabinoxylan degradation by Bifidobacterium longum NCC2705 using real-time RT-PCR. Probiotics Antimicro. 1, 121–129. doi: 10.1007/s12602-009-9015-x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schuldiner-Harpaz, T., and Coll, M. (2017a). Effect of diet history on prey and pollen food choice by two lady beetle species. J. Insect Behav. 30, 432–438. doi: 10.1007/s10905-017-9630-4
Schuldiner-Harpaz, T., and Coll, M. (2017b). Estimating the effect of plant-provided food supplements on pest consumption by omnivorous predators: lessons from two coccinellid beetles. Pest Manag. Sci. 73, 976–983. doi: 10.1002/ps.4410
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Sun, H., Meng, Y., Cui, G., Cao, Q., Li, J., and Liang, A. (2008). Selection of housekeeping genes for gene expression studies on the development of fruit bearing shoots in Chinese jujube (Ziziphus jujube Mill.). Mol. Biol. Rep. 36:2183. doi: 10.1007/s11033-008-9433-y
Thellin, O., Zorzi, W., Lakaye, B., Borman, B., Coumans, B., Hennen, G., et al. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295. doi: 10.1016/S0168-1656(99)00163-7
Vandesompele, J., Preter, K., Pattyn, F., Poppe, B., Roy, N., Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Wan, Q., Chen, S., Shan, Z., Yang, Z., and Zhou, X. (2017). Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS One 12:e0189405. doi: 10.1371/journal.pone.0189405
Wei, Z., Liu, M., Hu, C., and Yang, X. (2020). Overexpression of glutathione S-transferase genes in field λ-cyhalothrin-resistant population of Cydia pomonella: reference gene selection and expression analysis. J. Agr. Food Chem. 68, 5825–5834. doi: 10.1021/acs.jafc.0c01367
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2
Yadav, P., Deepak, S. D., Mukesh, M., Kataria, R. S., Yadav, A., Mohanty, A. K., et al. (2012). Identification of suitable housekeeping genes for expression analysis in mammary epithelial cells of buffalo (Bubalus bubalis) during lactation cycle. Livest. Sci. 147, 72–76. doi: 10.1016/j.livsci.2012.04.004
Yan, Z., Gao, J., Lv, X., Yang, W., Wen, S., Tong, H., et al. (2016). Quantitative evaluation and selection of reference genes for quantitative RT-PCR in mouse Acute Pancreatitis. Biomed Res. Int. 2016:8367063. doi: 10.1155/2016/8367063
Yang, C., Preisser, E., Zhang, H., Liu, Y., Dai, L., Pan, H., et al. (2016). Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 7:1672. doi: 10.3389/fpls.2016.01672
Yang, Q., Li, Z., Cao, J., Zhang, S., Zhang, H., Wu, X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS One 9:e98164. doi: 10.1371/journal.pone.0098164
Yang, X., Pan, H., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. U. K. 8:2689. doi: 10.1038/s41598-018-20612-w
Yin, J., Sun, L., Zhang, Q., and Cao, C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 749:144712. doi: 10.1016/j.gene.2020.144712
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 9:e86503. doi: 10.1371/journal.pone.0086503
Zhang, S., An, S., Li, Z., Wu, F., Yang, Q., Liu, Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402. doi: 10.1016/j.gene.2014.11.038
Keywords: RT-qPCR analysis, Hippodamia variegata, reference gene, expression stability, biotic conditions, abiotic conditions
Citation: Xie J, Liu T, Khashaveh A, Yi C, Liu X and Zhang Y (2021) Identification and Evaluation of Suitable Reference Genes for RT-qPCR Analysis in Hippodamia variegata (Coleoptera: Coccinellidae) Under Different Biotic and Abiotic Conditions. Front. Physiol. 12:669510. doi: 10.3389/fphys.2021.669510
Received: 19 February 2021; Accepted: 06 April 2021;
Published: 17 May 2021.
Edited by:
Patrizia Falabella, University of Basilicata, ItalyReviewed by:
Huipeng Pan, South China Agricultural University, ChinaZhi-Wei Kang, Shandong Agricultural University, China
Copyright © 2021 Xie, Liu, Khashaveh, Yi, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjun Zhang, eWp6aGFuZ0BpcHBjYWFzLmNu
 Jiaoxin Xie
Jiaoxin Xie Tinghui Liu2
Tinghui Liu2 Adel Khashaveh
Adel Khashaveh Yongjun Zhang
Yongjun Zhang