
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 18 June 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.662651
Neuropeptide F (NPF) is an important signaling molecule that acts as a neuromodulator to regulate a diversity of physiological and behavioral processes from vertebrates to invertebrates by interaction with NPF receptors, which are G protein-coupled receptors (GPCR). However, nothing is known about NPF in Chinese white pine beetle, Dendroctonus armandi, a destructive pest of natural and coniferous forests in the middle Qinling Mountains of China. We have cloned and characterized cDNAs encoding one NPF precursor and two NPF receptors in D. armandi and made bioinformatics predictions according to the deduced amino acid sequences. They were highly similar to that of Dendroctonus ponderosa. The transcription levels of these genes were different between larvae and adults of sexes, and there were significant differences among the different developmental stages and tissues and between beetles under starvation and following re-feeding states. Additionally, downregulation of NPF and NPFR by injecting dsRNA into beetles reduced their food intake, caused increases of mortality and decreases of body weight, and also resulted in a decrease of glycogen and free fatty acid and an increase of trehalose. These results indicate that the NPF signaling pathway plays a significant positive role in the regulation of food intake and provides a potential target for the sustainable management of this pest.
Neuropeptides play significant regulatory roles in both vertebrates and invertebrates. Neuropeptide Y (NPY) is not only one of the most widespread neuropeptides in the central nervous system (CNS) of vertebrates (Allen, 1990) but also an important molecule to regulate diverse physiological and behavioral processes (Cerdá-Reverter and Larhammar, 2000). Neuropeptide F (NPF) is a homolog of NPY found in invertebrates, which is highly similar to NPY in structure and function but differs from NPY in having a phenylalanine (F) instead of a tyrosine (Y) at the C-terminus (Maule et al., 1991; de Jong-Brink et al., 2001; Roller et al., 2008; Cui and Zhao, 2020; Yeoh et al., 2017). NPFs have been identified in 30 Coleopteran species (Pandit et al., 2019), which are evolutionarily well conserved. The NPFs of most insects contain more than 28 residues (generally 28–45 amino acids) and are characterized by a RxRFamide consensus sequence at the carboxy terminus. However, some short peptides of 8–10 amino acids in length were found to be encoded by another gene, the short NPF (sNPF), characterized by an M/T/L/FRF amide carboxyterminal motif (Nässel and Wegener, 2011). NPFs operate through interacting with the NPF receptors, which belong to members of the G protein-coupled receptor (GPCR) superfamily. The NPF/NPFR signaling system provides a new paradigm for exploring the central regulation of cooperative and behavioral processes (Wu et al., 2003). NPFRs have been identified in Drosophila and Anopheles (Garczynski et al., 2002, 2005) and are also predicted in various other insect species (Caers et al., 2012). NPFs have a significant role in regulating feeding and foraging behaviors (Shen and Cai, 2001; Wu et al., 2003; Fadda et al., 2019). As a matter of fact, regulation of feeding was the first observed role of NPF, and most functional insights have been mainly obtained in studies with Drosophila melanogaster (Brown et al., 1999; Wu et al., 2005a, b; Wang et al., 2013). For example, the NPF signaling system has an effect on consuming noxious food in larvae of D. melanogaster (Wu et al., 2005b). Overexpression of NPFR in fly larvae causes an augmentation of noxious food uptake, while RNAi-mediated knockdown of NPFR shows the opposite phenotype (Wu et al., 2005b). Similarly, NPF signaling also stimulates feeding behavior under cold conditions in Drosophila larvae (Lingo et al., 2007).
Additionally, NPFs are also involved in many other functions, such as sexual and male characteristic courtship behavior (Lee et al., 2006; Kim et al., 2013), ethanol sensitivity (Wen et al., 2005), learning and memory (Krashes et al., 2009), aggression (Dierick and Greenspan, 2007), locomotor activity, and circadian rhythm (Hermann et al., 2012; Erion et al., 2016). However, the role of NPF in other insects has been rarely explored and has mainly focused on the functions related to feeding behavior. The association between NPF signaling and feeding-related processes, as documented in Drosophila, has also been observed in Locusta migratoria, with starvation causing an increase of LmiNPF1 expression level, and downregulation of LmiNPF1 notably reduced food intake (Tan et al., 2019). Furthermore, the NPF expression levels in Acyrthosiphon pisum were significantly higher in starved aphids than in satiated aphids, and RNAi-mediated knockdown of NPF in adult aphids markedly inhibited feeding behavior (Li et al., 2018). Moreover, NPF transcript levels of Schistocerca gregaria were significantly upregulated in starved animals compared with feeding animals. Injection of trNPF in locust adults caused an increase in food intake, while RNAi knockdown showed the opposite effect (Van Wielendaele et al., 2013). The NPF signaling system has also been shown to regulate feeding and growth development in Bombyx mori, where dsRNA-mediated knock-down of BomNPFR leads to a significant reduction in food intake and body weight (Deng et al., 2014).
Dendroctonus armandi Tsai and Li (Coleoptera: Curculionidae: Scolytinae) is a destructive pest of natural and coniferous forests in the middle Qinling Mountains of China, which only attacks healthy Pinus armandi, devastating the forest ecological system and incurring heavy economic losses (Chen and Tang, 2007; Hu et al., 2013). Although the NPFs have been explored in some insect species, there are no reports on the functional roles of NPF in bark beetles. In this study, to investigate whether NPF signaling in D. armandi is involved in food intake or not, we identified and cloned cDNAs encoding a NPF precursor and two NPFRs from D. armandi, which were used for further investigations of related functions. These information will serve as a considerable step forward to provide a potential molecular target for the sustainable management of bark beetles.
D. armandi was collected from infested Pinus armandi trees at the Huoditang Experimental Forest Station, which is located on the southern slope of the middle Qinling Mountains, in Shaanxi, China (33°18′N, 108°21′E). The newly infected P. armandi was transported to the laboratory from the sampling point after felling, where emerged adults and larvae were collected. They were reared in glass dishes (90 × 15 mm) at 20 ± 1°C, 70% relative humidity, and in the dark in an artificial climate cabinet of the laboratory using the meridic diet as described previously. Adults were sexed by the shape of external genitals and other male-characteristic auditory cues (Dai et al., 2014; Zhao et al., 2017).
Total RNA was isolated from three development stages (larvae, pupae, and adults) using the UNlQ-10 Column Trizol Total RNA Isolation Kit (Sangon Biotech, Shanghai, China). Total mixed RNA for cDNA was synthesized using the Fast King RT reagent Kit (with gDNase) (Tiangen, China) and then stored at −20°C until use.
The synthesized cDNA obtained from the samples was used as a template for PCR. Each pair of specific primers (Supplementary Table 1) was designed in Primer Premier 5.0, based on NPF and NPFR sequences of other related insect species from NCBI1 GenBank. All the PCR amplifications were performed with a C1000 thermocycler (Bio-Rad Laboratories, Hercules, CA, United States) in a final reaction mixture of 20 μl, containing 1 μl cDNA (1:4 dilution), 0.25 μl of each primer, and 2 × EcoTaq PCR SuperMix (TransGen, Beijing, China). The sequenced PCR products were manually edited using the DNAMAN software.
cDNA-specific primers for 5′ and 3′ RACE (Supplementary Table 1) were designed according to the obtained sequence. Touchdown PCR (annealing temperatures: 65–55°C) was used to improve the amplification specificity of the 5′-UTR and 3′-UTR sequences. The amplified products were cloned and sequenced as followed by previous description “cDNA amplification and cloning.” To obtain the full-length sequences, we designed specific primers containing the putative initiation and terminator codons (Supplementary Table 1).
The three cDNA sequences obtained were submitted in the GenBank, and accession numbers are listed in Table 1. The open reading frames (ORFs) of full-length cDNA sequences were obtained using ORF Finder2. Multiple sequence alignment of sequences was carried out in DNAMAN6.0. Molecular mass (kDa) and isoelectric points were determined in the ProtParam tool (Gasteiger et al., 2005). The putative signal peptide was predicted using Signal P 4.1 Server3. All putative D. armandi NPF and NPFR proteins were predicted for subcellular localization using the TARGETP tool4 (Emanuelsson et al., 2000) with the default parameters. TMHMM v. 2.05 was used to visually display predictions of topological and transmembrane domains. The phylogenetic trees were constructed by software MEGA 6.0 (Tamura et al., 2013) using the maximum likelihood method with a Whelan and Goldman (WAG) model and a gamma parameter value. The support for each node of bootstrap was estimated by a bootstrap program after 500 replicates.
D. armandi larvae were separated into two substages: larvae (eating on host phloem for development) and mature larvae (cease feeding). Pupae were separated into two substages: early pupae (newly transformed from larvae) and late pupae (approach to become adults). We separated the adults into three substages: teneral adults (light body color), emerged adults, and feeding adults (invading the bark) (Dai et al., 2014). The heads, thoraxes, foreguts, midguts, hindguts, pheromone glands, and body fat of emerged adults, and heads, thoraxes, foreguts, midguts, hindguts, and body fat of larvae were dissected for measurement of expression level in tissues. Samples were collected in triplicate, killed by submerging beetles in liquid nitrogen, and then stored at −80°C until use.
The males and females of emerged adults were divided into eight groups, and larvae were divided into seven groups. One group of collected insects were killed at 0 h of feeding, which were used as the control. Each of the other groups of emerged adults was immediately placed in glass dishes (90 × 15 mm) with normal food for 24 and 48 h in the artificial climate cabinet. After 48 h of normal feeding, the adults and larvae were reared without food and starved for 72 and 48 h, respectively. Then, the alive insects were subsequently re-fed for 24 h after starvation treatment. The experiments of each treatment for real-time PCR analyses were explored in three biological replicates independently, with 15 adults of each sex and 20 larvae at each repeat.
The frozen samples were used for RNA extraction and cDNA synthesis following the above description “Total RNA Extraction and cDNA Synthesis.” All samples were placed in the CFX-96TM real-time PCR Detection System (Bio-Rad, CA, United States) for RT-qPCR. The β-actin (GenBank accession no. KJ507199.1) and CYP4G55 (GenBank accession no. JQ855658.1) sequence of D. armandi was used as the internal control (Vandesompele et al., 2002; Dai et al., 2015). RT-qPCR primers were designed using Primer Premier 5.0 according to the obtained sequences (Supplementary Table 1). The amplification efficiency of each transcript was analyzed with relative standard curves at a different dilution series (1.0, 10–1, 10–2, 10–3, and 10–4) of cDNAs, and the efficiency values of the primers were analyzed by 100 ± 5%. The reaction mixture (20 μl) contained 1 μl of each primer, 2.5 μl of cDNA (diluted four times), 5.5 μl of ddH2O, and 10 μl of 2 × SYBR Premix Ex Taq (Roche Diagnostics GmbH, Mannheim, Germany). The specificity of qPCR primers was estimated by the melting curve analysis. Three technical replicates of each treatment contained three biological replicates, which were performed to verify reproducibility. The relative expression level of each gene was analyzed by the 2–ΔΔCt method (Livak and Schmittgen, 2001, 2008).
The synthesis of dsRNA was prepared using the T7 Ribo-MAXTM Express RNAi System (Promega, Madison, MI, United States). The primers (Supplementary Table 1) used for RNAi were designed according to the obtained sequences. The final dsRNA products were diluted to 1,000 ng/μl with DEPC water. Before injection, emerged adults and larvae were anesthetized by placing on a tray in an ice bath for 20 min. Afterward, each of emerged adults was injected with 0.2 μl dsRNA solution and larvae were injected with 0.l μl visa Hamilton MicroliterTM syringes (700 series, RN) with 32 G sharp-point needles (Hamilton, Bonaduz, Switzerland). Non-injected insects and insects injected with DEPC-treated water were used as controls in all experiments. Then, beetles were kept in an artificial climate cabinet under the feeding or starvation condition. Each treatment group contained 40 individuals, and 6 individuals of each treatment and after 72 h were collected and then stored at −80°C until qRT-PCR. Each treatment group contained three biological replicates.
After 72 h of dsRNA injection, the beetles were kept at room temperature for 1 h, and animals that did not move were considered to be dead (Fu et al., 2019). Adults and larvae mortality were quantified under the different treatments and control conditions to determine their effect at different time points (12, 24, 36, 48, 60, and 72 h). The body weight of each alive sample was immediately measured by an electronic balance (d = 0.0001 g, Tianjin, AL204; Mettler-Toledo Ltd., China). The measurement included three replicates.
For emerged adults and larvae at 72 h after injection, we measured three physiological indices in each treatment group, including the content of glycogen, trehalose, and free fatty acid, via relevant biochemical methods. Three biological replicates (six beetles for one replicate) were performed for each measurement. Whole-body homogenates of each group were used to measure glycogen, trehalose, and free fatty acid, respectively. The three content levels were measured with a spectrophotometer (UV-1800PC, Shanghai Mapada Instrument Co., Ltd., Shanghai, China) using the relevant kit (TY-2-Y for glycogen, FFA-1-W for free fatty acid, and HT-2-Y for trehalose, respectively; SuzhouComin Biotechnology Co. Ltd., Jiangsu, China).
All data were statistically analyzed with SPSS Statistics 19.0 (IBM, Chicago, IL, United States). Significant differences between different treatments were determined using post hoc Tukey tests through one-way ANOVA. Student’s t-test was employed to perform the two-sample analyses. Graphs were plotted with Prism 6.0 (GraphPad Software, CA, United States).
We successfully sequenced cDNAs encoding NPF and NRFRs from D. armandi with full-length sequences, and NPF cDNA encoded 133 amino acids with a predicted molecular mass of 15.55 kDa and isoelectric point of 9.97 (Table 1). Two splicing variants encoded by the NPFR gene were identified, with NPFR1 and NPFR2 comprising 418 and 400 amino acids, respectively. The cellular localization of these proteins shows that NPF is a secretory protein, whereas NPFR1 and NPFR2 are membrane proteins (Table 1).
The NPF precursor of D. armandi consisted of 133 amino acids, and the first 25 amino acids were predicted to be an N-terminal signal peptide, followed by the mature NPF (Supplementary Figure 1). The NPF prepropeptide contained processing sites at the C-terminal F residue (RPRFGKR), including an amidation site (G) and followed by a dibasic cleavage site (KR) (Supplementary Figure 1). The protein sequence of NPFR was predicted to have seven transmembrane domains and showed the typical characteristic of the rhodopsin-like GPCR family (Supplementary Figure 2). Further identities showed that DaNPF and DaNPFRs had the highest degree similarity with that of fellow Coleoptera member Dendroctonus ponderosa (Table 2). The phylogenetic tree of NPF (Supplementary Figure 3) and NPFRs (Supplementary Figure 4) revealed that these proteins were clustered with the Coleoptera group. Although only a few NPF sequences were listed in this study, we could also find more relevant information of protein sequences with other Coleoptera species (Veenstra, 2019).
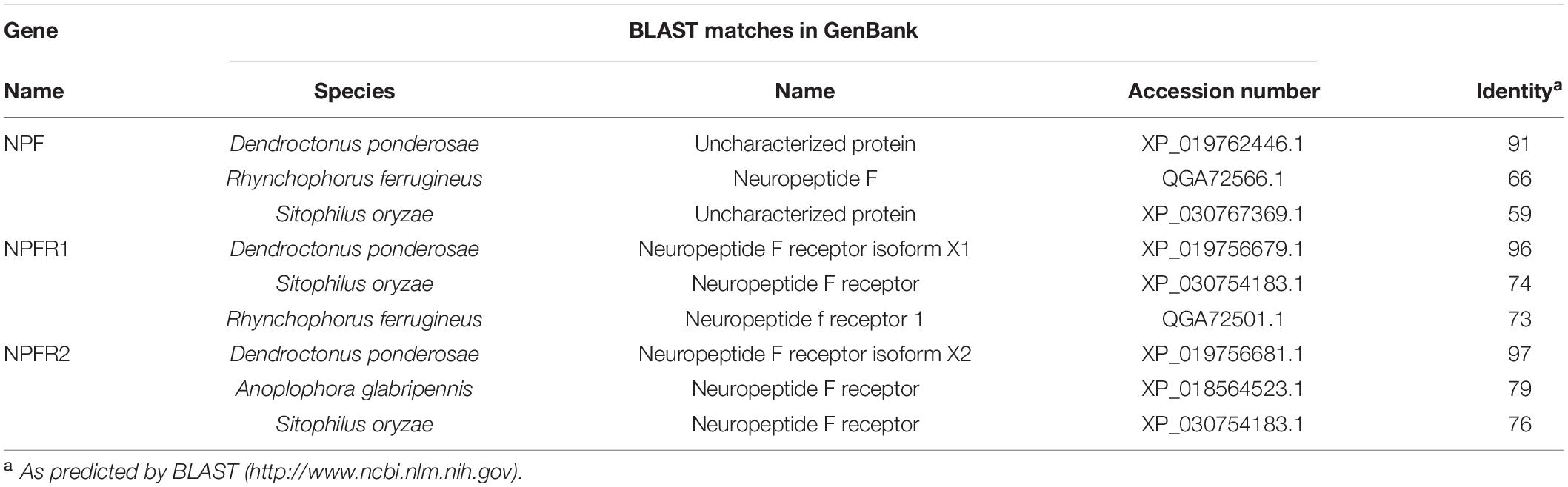
Table 2. Identity of NPF and NPFR genes from D. armandi with relevant gene sequences in other insect species.
NPF, NPFR1, and NPFR2 were broadly expressed in all developmental stages of D. armandi, but with different patterns. All of them were highly expressed in emerged adults, followed by pupae, and the lowest in mature larvae (Figure 1). Compared with NPFR1 and NPFR2, the expression levels of NPF in the larval stage and two pupal stages were not significantly different (Figure 1A). In the adult stage, the expression levels of NPF and NPFR2 in males were higher than in females (Figures 1A,C), while NPFR1 showed the opposite result (Figure 1B).
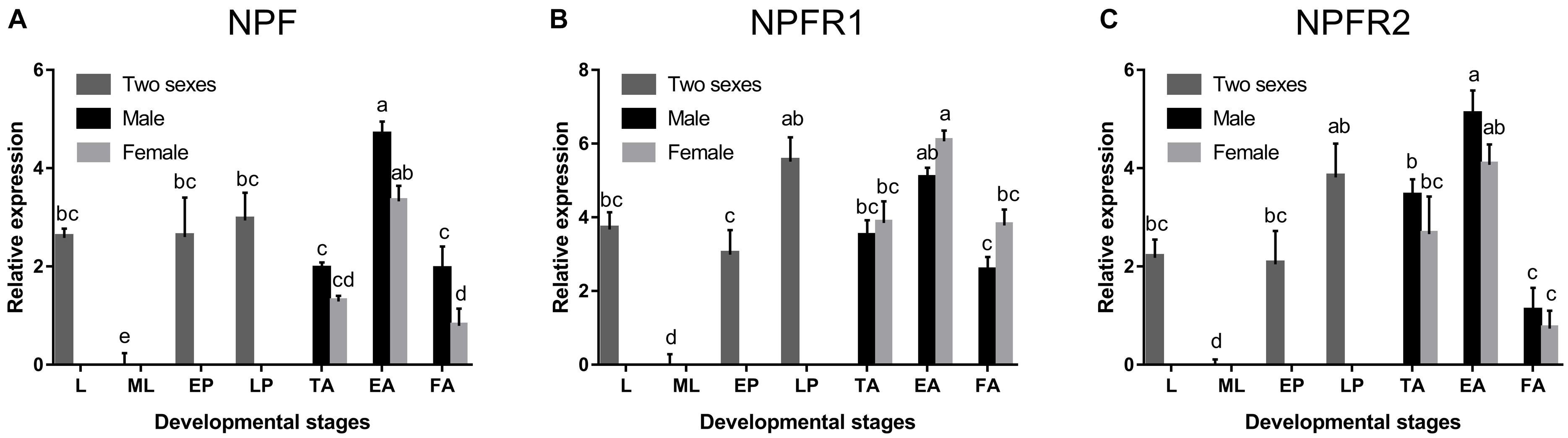
Figure 1. Relative mRNA expression levels of NPF (A), NPFR1 (B), and NPFR2 (C) in different developmental stages of D. armandi. The relative expression levels were normalized with β-actin and CYP4G55 using the expression levels in the mature larvae for calibration. Different lowercase letters indicate significant differences at the 0.05 level. All values are mean ± SE, n = 3. L, larvae; ML, mature larvae; EP, early stage pupae; LP, late stage puage; TA, teneral adults; EA, emerged adults; FA, feeding adults.
NPF, NPFR1, and NPFR2 were expressed at different levels and with occasional sex differences among the different tissues (Figure 2). NPF was highly expressed only in the head and midgut of adults (Figure 2A) and larvae (Figure 2D), while NPFR1 (Figures 2B,E) and NPFR2 (Figures 2C,F) were expressed in different tissues. All of them were highly expressed in midgut, followed by head in adults and larvae. NPF was more highly expressed in females than in males in head, thorax, and midgut (Figures 2A,D). NPFR1 and NPFR2 were highly expressed in head, foregut, and midgut, with a strikingly higher expression in females than in males among the three tissues. Specifically, the expression of NPFR2 in the hindgut of males was significantly higher than that of females (Figure 2C).
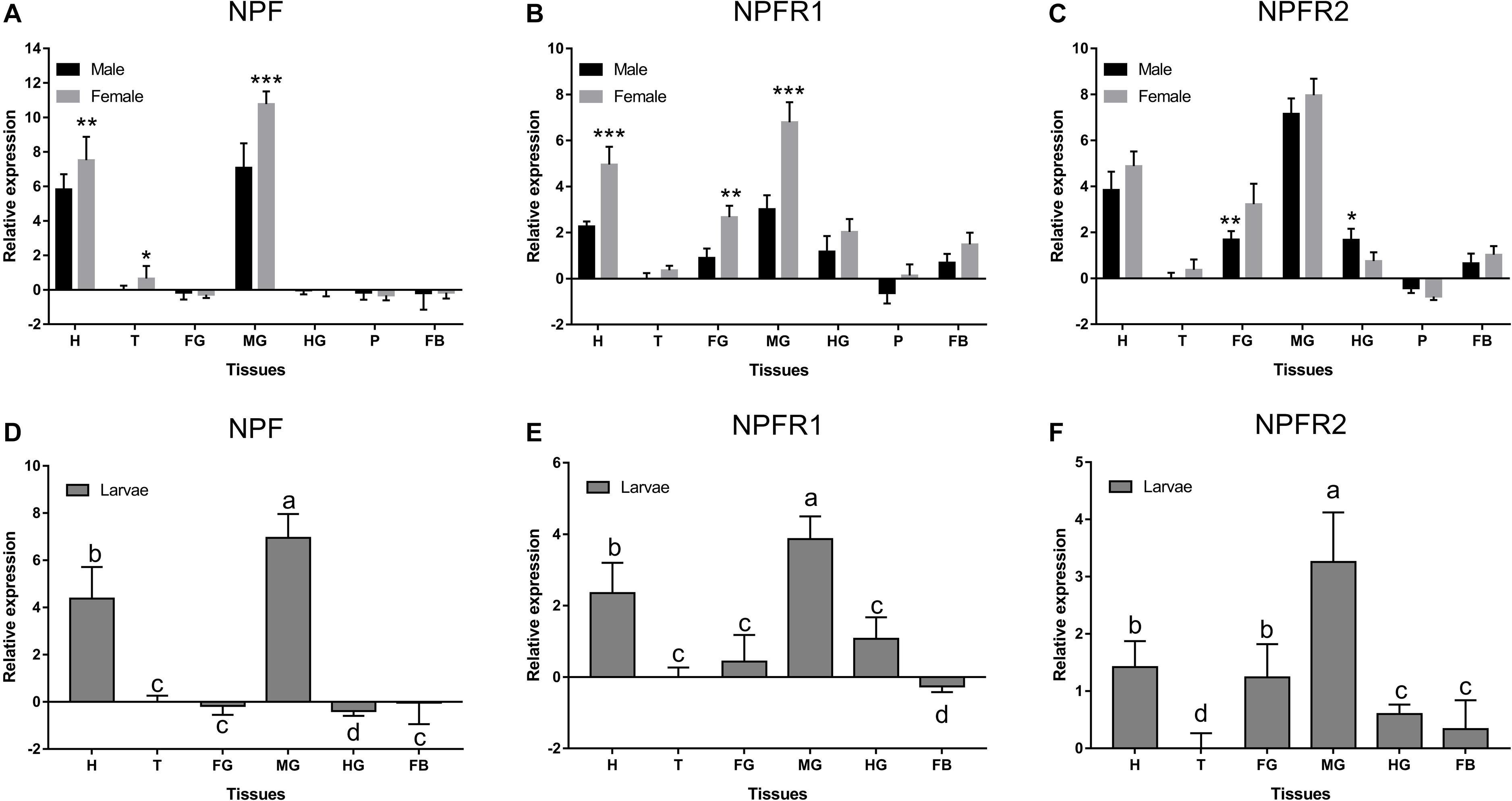
Figure 2. Relative expression levels of emerged adults of NPF (A), NPFR1 (B), NPFR2 (C), and larvae of NPF (D), NPFR1 (E), and NPFR2 (F) in different tissues of D. armandi. The relative expression levels were normalized with β-actin and CYP4G55 using the expression levels in the thorax for calibration. Different lowercase letters indicate significant differences at the 0.05 level. The asterisk indicates a significant difference between female and male expression levels (*P < 0.05, **P < 0.01, ***P < 0.001, independent Student’s t-test). All values are mean ± SE, n = 3. H, head; T, thorax; FG, foregut; MG, midgut; HG, hindgut; P, pheromone gland; FB, fat body.
Expression of three genes analyzed in adults and larvae showed a different response to starvation stress (Figure 3). Compared with the feeding groups, the NPF and NPFR1 expression levels in adults (Figures 3A,B) and larvae (Figures 3D,E) were highly upregulated in the starvation groups and reached the highest level at 72 h. Moreover, during the refeeding experiment after food deprivation, the NPF and NPFR1 expression levels showed a steady decline and then returned to the original level after refeeding for 24 h. The expression levels of NPF (Figure 3A) and NPFR1 (Figure 3B) in male adults were lower than those in female adults at 48 h after feeding, and the highest at 72 h after starvation. However, the expression level of NPFR2 was not affected in either the starvation or the following refeeding assay (Figures 3C,F).

Figure 3. Relative expression levels of emerged adults of NPF (A), NPFR1 (B), NPFR2 (C), and mature larvae of NPF (D), NPFR1 (E), and NPFR2 (F) in D. armandi after starvation and subsequent re-feeding treatment. The relative expression levels were normalized with β-actin and CYP4G55 using the expression levels in 0 h for calibration. Different letters indicate significant differences at the 0.05 level (uppercase for males, lowercase for females and uppercase for males and larvae, no letter means no significant difference among all time points). All values are mean ± SE, n = 3.
In both the starvation and the feeding groups, compared with the other two control groups, the expression level of NPF (Figures 4A–C) and NPFR1 (Figures 4D–F) in adults and larvae was significantly downregulated at 72 h after dsRNA injection, respectively, and the expression level of NPF and NPFR1 in the starvation group was decreased more than the feeding group; after 72 h of dsNPFR2 injection, there was no obvious change in the male (Figure 4G) and female (Figure 4H) adults, but the expression level of the larvae was downregulated in both the starvation group and the feeding group (Figure 4I).
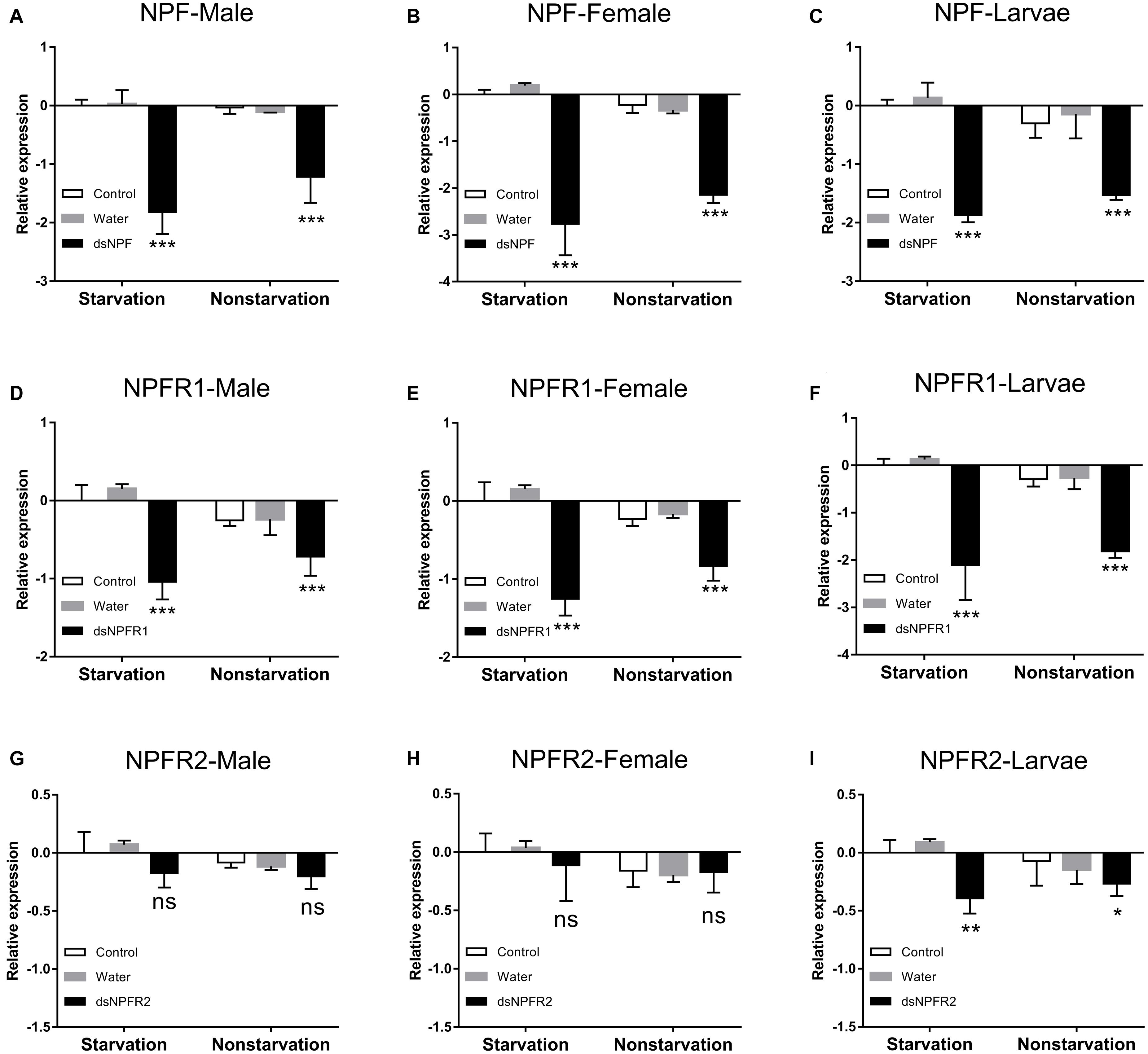
Figure 4. Knockdown of NPF-Male (A), NPF-Female (B), NPF-Larvae (C), NPFR1-Male (D), NPFR1-Female (E), NPFR1-Larvae (F), NPFR2-Male (G), NPFR2-Female (H), and NPFR2-Larvae (I) expression levels in D. armandi after injecting dsRNA for 72 h in the state of starvation and non-starvation. The asterisk indicates a significant difference between control and RNAi treatments according to Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant). All values are mean ± SE, n = 3.
In both the feeding and starvation groups, the mortality of the dsRNA injection in adults and larvae was higher than that of the non-injected and water-injected controls (Figure 5). The mortality significantly increased when the adults and larvae were injected with dsNPF and dsNPFR1 from 0 to 72 h. However, in the dsNPFR2-injected group, there was no significant change. The highest mortality was found after injection of dsNPF at 72 h. Moreover, the mortality of larvae was the highest and female adults was the lowest, 89.2 and 58.3%, respectively (Figures 5C,E). However, the mortality of the starvation group was significantly higher than that of the feeding group (Figure 5).
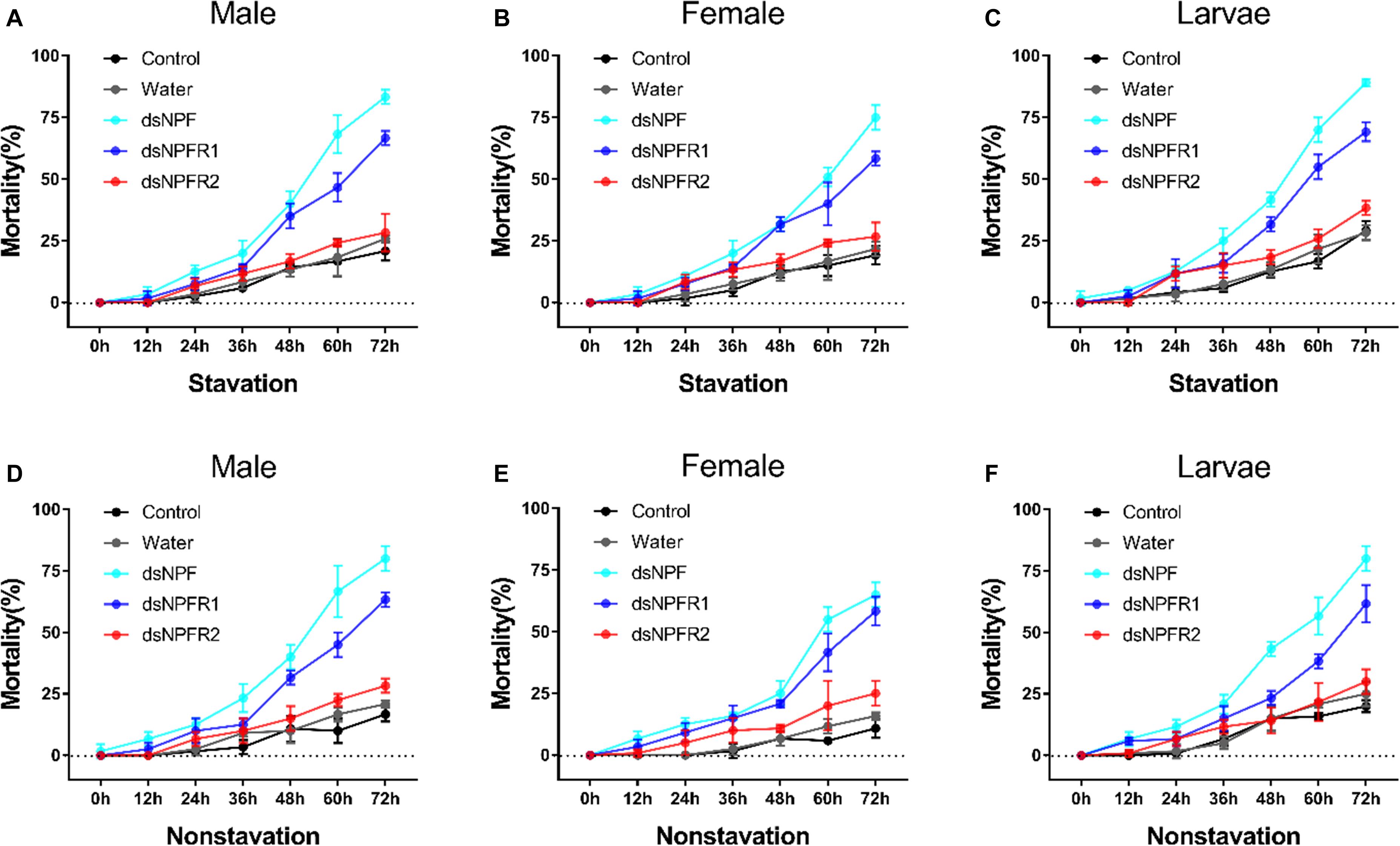
Figure 5. Mortality responses of RNAi D. armandi male (A), female (B), and larvae (C) in starvation and male (D), female (E), and larvae (F) in non-starvation to different time points. Mortality responses of dsRNA-treated, water-injected, and non-injected in D. armandi to different time points (0, 12, 24, 36, 48, 60, and 72 h). All experiments were analyzed by Student’s t-test, and in (A–F), experimental beetles significantly differed (P < 0.01) from two controls.
Compared with the two control groups, the average weight of adults and larvae significantly decreased after 72 h of injection of dsNPF and dsNPFR1 in the starvation and feeding groups; among them, the male and the female adults had the most and the least weight loss, with 54.2 and 28.9%, respectively (Figures 6A,B). In particular, the average body weight of animals injected with dsNPFR2 did not change significantly, except for the larvae in the starvation group. Compared with the feeding group, the average weight of adults and larvae in the starvation group showed more reductions after dsRNA injection (Figure 6). These results indicate that the RNAi mediated downregulation of NPF inhibited food intake of beetles.
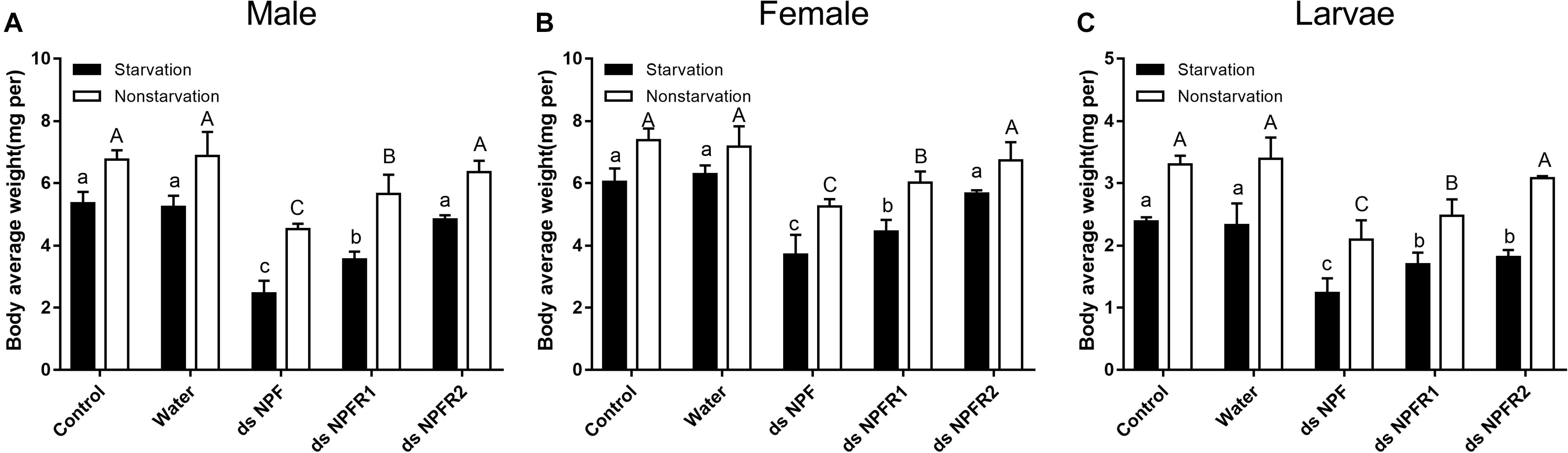
Figure 6. Effect of dsNPF, dsNPFR1, and dsNPFR2 on body average weight of D. armandi. Samples were collected and assayed at 72 h after injection. In the RNAi experiments, body average weight of male (A), female (B), and larvae (C) in the state of starvation and non-starvation was analyzed. Different letters indicate significant differences at the 0.05 level (uppercase for non-starvation, lowercase for starvation). All values are mean ± SE, n = 3.
Compared with the two control groups, the glycogen and free fatty acid contents of adults and larvae were significantly decreased after injection of dsNPF and dsNPFR1 for 72 h in the starvation group and the feeding group, and the largest decline was in the dsNPF injection group (Figure 7). In particular, there was no significant change in the glycogen content of the larvae injected with dsNPFR2, but the free fatty acid content of the larvae injected with dsNPFR2 was decreased (Figure 7F). More interestingly, compared with the two control groups, the trehalose content of adults and larvae increased significantly at 72 h after injection of dsNPF and dsNPFR1, respectively, but there was no obvious change in the trehalose content of male and female adults injected with dsNPFR2 (Figure 7). Compared with the feeding group, the content of glycogen and free fatty acid decreased in the starvation group after injection of dsRNA. On the contrary, the content of trehalose increased (Figure 7).
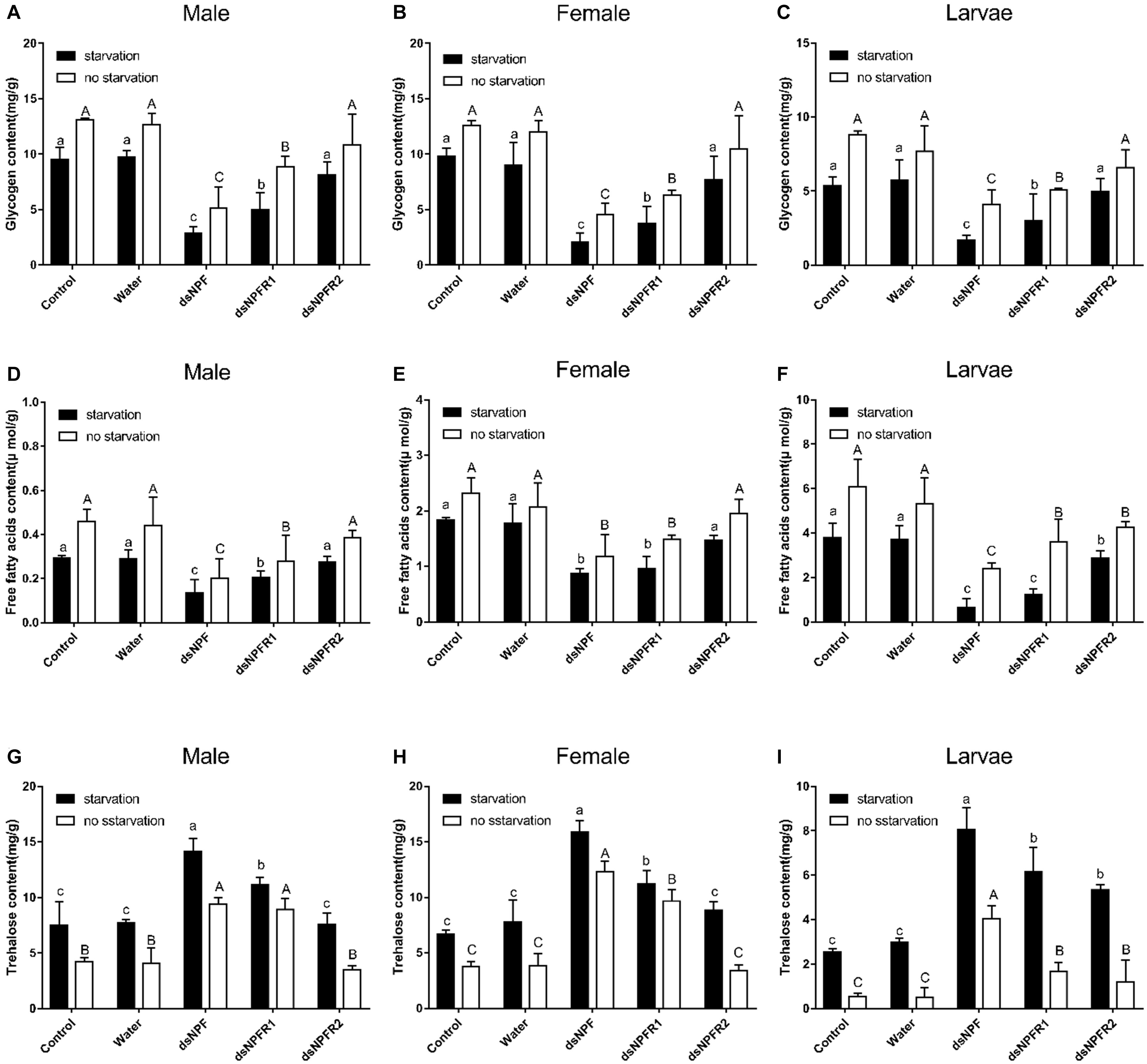
Figure 7. Effect of dsNPF, dsNPFR1, and dsNPFR2 on energy metabolism of D. armandi. Samples were collected and assayed at 72 h after injection. Whole-body homogenates were used to measure glycogen of male (A), female (B), and larvae (C), Free fatty acids of male (D), female (E), and larvae (F) and trehalose contents of male (G), female (H), and larvae (I) in the state of starvation and non-starvation. Different letters indicate significant differences at the 0.05 level (uppercase for non-starvation, lowercase for starvation). All values are mean ± SE, n = 3.
Most NPFs are encoded by one gene copy per species, based on genomic sequence data from invertebrates (Nässel and Wegener, 2011). However, some species have two homologous genes encoding NPF1 and NPF2, among which have an RYamide C-terminus (Wu et al., 2005a; Roller et al., 2008; Huang et al., 2011; Liu et al., 2013), and NPF1 even produces two splicing variants, NPF1a and NPF1b, which contain RPRFamide at the C-terminus (Garczynski et al., 2005; Roller et al., 2008; Yue et al., 2016). In general, NPF1b is coded by optional intron within the NPF1a coding region. In the present study, one NPF-encoding gene was successfully cloned and identified in D. armandi, which is in consonance with the results of most insect studies so far. The cDNA sequence of NPF confirms the presence of 133 amino acids, and mature NPF is produced with an amidated RPRFamide C-terminus, which has a typical characteristic of the NPF1 precursor family. All NPF sequences of most insects contain the conserved C-terminal ending RxRFamide (Nässel and Wegener, 2011), which display the form of ancestors in invertebrates from an evolutionary point of view. We also cloned and identified cDNAs encoding NPFR1 and NPFR2 from D. armandi, which code a length of 418 and 400 amino acids, respectively. They contained some characteristic residues in the seven transmembrane domains including GN in helix 1, NLX3DX8P in helix 2, SX6IX2DRY in helix 3, WX8P in helix 4, PX7Y in helix 5, FX3WXP in helix 6, and NPX2YX6F in helix 7. This typical pattern shows that the NPFRs belong to the rhodopsin-like GPCR superfamily (Costanzi, 2012). Amino acid sequence identity analysis showed that NPF and NPFRs of D. armandi were very similar to that of fellow Coleoptera member D. ponderosa and clustered with the Coleoptera group.
We found that both the NPF and NPFRs transcripts were expressed throughout various ages of D. armandi, suggesting that the NPF signal system might be involved in regulating some physiological processes in the growth and development of beetles. The distribution and expression level of D. melanogaster NPF have indicated that NPF is mainly expressed in the CNS and midgut endocrine cells of larvae and adults (Brown et al., 1999). Furthermore, endocrine cells that produce and secrete NPF were found in some other analyzed insect species, such as Helicoverpa zea, Aedes aegypti, Rhodnius prolixus, Reticulitermes flavipes, and Chilo suppressalis (Stanek et al., 2002; Gonzalez and Orchard, 2008; Nuss et al., 2010; Huang et al., 2011; Xu et al., 2016). Here, the expression level of D. armandi NPF in the midgut is higher than that in the brain of adults and larvae. This is consistent with the situation in Helicoverpa assulta and Helicoverpa armigera (Liu et al., 2013; Yue et al., 2016). On the contrary, in nymphs of L. migratoria, the NPF expression level in the midgut is significantly lower than that in the CNS (Tan et al., 2019). Additionally, the expression of NPF is similar in the midgut and CNS in Anopheles gambiae (Garczynski et al., 2005). However, the midgut is not the only part of NPF expression in the digestive system; some previous studies showed the existence of NPF expression in the foregut of the R. flavipes and H. assulta (Nuss et al., 2010; Liu et al., 2013). Furthermore, significantly higher expression levels of D. armandi NPF were observed in female midguts and brains compared to male’s, but this contrasts with the situation in S. gregaria, which showed that the expression level of male brains was higher than the female brains (Van Wielendaele et al., 2013). Interestingly, immunocytochemistry indicated that the male adults of D. melanogaster showed additional male characteristic NPF expressing neurons, which may be involved in the regulation of circadian rhythm and courtship behavior (Lee et al., 2006). The expression levels of D. armandi NPFRs in the head and midgut were in line with obtained data in other insect species (Garczynski et al., 2002; Nässel and Wegener, 2011; Deng et al., 2014; Zhang et al., 2020). For examples, the NPFR expression level was significantly high in the midgut, brain, and accessory nerve of Drosophila larvae through in situ hybridization experiments (Garczynski et al., 2002). NPFR was highly expressed in the brain, mid-gut, and fat body of B. mori larvae, and it was also detected in such parts as the silk gland, malpighian tubule, ovary, and testis (Yamanaka et al., 2008; Deng et al., 2014). In Rhynchophorus ferrugineus, NPF corresponding receptor gene was also highly expressed in gut and CNS of eighth instar larvae (Zhang et al., 2020).
In this study, we found that with the prolongation of starvation time, the expression level of NPF in D. armandi increased, but the following refeeding caused a continuous drop to the original level. Starvation and re-feeding experiments provided direct evidence that NPF signaling system has an effect on feeding behavior. The result was similar to those observed in other insect species previously. For example, NPF expression levels of D. melanogaster larvae were higher in the wandering period in comparison to the following feeding stage period (Wu et al., 2003). Additionally, NPF increased expression in male Nicrophorus vespilloides when food-deprived, and the decreased expression of NPFR was detected during parenting (Cunningham et al., 2016). Similarly, in Apis mellifera, NPF expression levels were shown to be higher in foragers than nurses, but did not appear to be regulated by nutritional status in the workers (Ament et al., 2011). Furthermore, NPF levels were also demonstrated to be higher in prior to feeding conditions compared to the post-feeding conditions in the hemolymph of A. aegypti (Stanek et al., 2002). The expression pattern of NPFR1 in the starvation and re-feeding experiments was consistent with NPF. It is obvious that the expression level of NPFR2 was not significantly changed in starved or refed beetles. These results show that the state of feeding has an influence on the expression levels of NPF and NPFR1 in D. armandi, while NPFR2 is more stable under different states. The need for nutrients will increase when beetles are starved. As a result of this, the feeding behavior and food intake might be stimulated in the starvation state. The higher NPF expression level was observed in starved beetles that is likely related to a promotion of appetite in response to starvation.
NPF is a critical factor to manage the selection of food, which regulates the transition between acceptance and rejection of harmful food (Wu et al., 2005a). With overexpression of the Drosophila NPF in the CNS of larvae, the feeding time was prolonged, while RNAi-mediated knockdown of the NPF gene caused refusal of food (Wu et al., 2003). Furthermore, this is also consistent with the observation that knockdown of NPF or NPFR of Drosophila neurons suppresses food attractiveness (Beshel and Zhong, 2013). In this study, we knocked NPF down in adults and larvae by injecting dsNPF and the results showed that this method could effectively inhibit the expression of NPF, with smaller body weight. It has caused the beetles to eat less, leading to a delay in growth and development, resulting in body weight loss. This indicates that silencing of NPF expression in D. armandi suppresses their appetite, likely leading to a change in feeding behavior.
The mortality was significantly higher in dsRNA-treated adults and larvae than in the two controls (water-injected and non-injected). Thus, silencing the target gene NPF not only inhibited their expression levels but also resulted in augmentation of mortality in the state of both starvation and feeding. The result was consistent with previous studies on H. armigera; the NPF RNAi led to high mortality of this species, in which all animals could not pupate or emerge due to abnormal growth and development (Yue et al., 2017).
A previous study reported in D. melanogaster that the NPF signal system is regulated by the insulin through the InR/PI3K/S6K pathway (Wu et al., 2005b). In this study, we observed that the D. armandi NPF regulates feeding behavior through the effect on energy metabolism, in which downregulation of NPF leads to a decrease of glycogen and an increase of trehalose. Presumably, with less food or even starvation, NPF not only promotes biosynthesis or energy storage but also inhibits metabolism or energy utilization. Feeding behavior provides more nutrients to reduce the metabolic demands of stored glycogen and free fatty acid. This pattern was consistent with H. armigera and Ostrinia furnacalis, and in contrast to O. furnacalis, downregulated NPF causes a decrease of free fatty acid in D. armandi instead of total lipid (Yue et al., 2016, 2017), suggesting that the NPF system is involved in insulin signal to regulate energy metabolism, which is a hypothesis that needs to be further investigated.
It should be noted that the regulation of feeding behavior not only is related to NPF but also involves the expression changes of its receptors in invertebrates. In this study, we found that the effect of NPFR1-knockdown experiment was consistent with the NPF-knockdown experiment, while there was no significant change in silencing NPFR2 because of the low silence efficiency of dsNPFR2. These results suggest that NPFR has similar functions in the regulation of feeding behavior. However, it is not clear how NPF/NPFR signaling system regulates to modify feeding behavior at the neural circuit, molecular, and cellular levels in D. armandi. Further studies will be needed to clarify a more clear explanation of the mechanism involved in the regulation of feeding behavior.
The generation of insecticide resistance is increasingly becoming the primary problem of pest management, and the regulation of the NPF/NPFR signal system is a potential control method. NPF not only is involved in the regulation of many important insect behaviors but also has the characteristics of wide distribution and high conservation, which might become a potential target of new insecticides. In the future research, it is still necessary to further clarify the upstream and downstream action elements of NPF/NPFR, so as to screen out stable and highly active repressors by pharmacology, and finally achieve specific pest control.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
BL, MT, and HC designed the experiments and revised the manuscript. BL, DF, HG, HN, and YS preformed the experiments. BL analyzed data and drafted manuscript. All authors read and approved the manuscript for final submission.
This research was funded by the National Natural Science Foundation of China (31870636) and the National Key Research and Development Program of China (2017YFD0600104).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.662651/full#supplementary-material
Allen, J. M. (1990). Molecular structure of neuropeptide Y. Ann. N. Y. Acad. Sci. 611, 86–98. doi: 10.1111/j.1749-6632.1990.tb48924.x
Ament, S. A., Velarde, R. A., Kolodkin, M. H., Moyse, D., and Robinson, G. E. (2011). Neuropeptide Y-like signalling and nutritionally mediated gene expression and behaviour in the honey bee. Insect Mol. Biol. 20, 335–345. doi: 10.1111/j.1365-2583.2011.01068.x
Beshel, J., and Zhong, Y. (2013). Graded encoding of food odor value in the Drosophila brain. J. Neurosci. 33, 15693–15704. doi: 10.1523/JNEUROSCI.2605-13.2013
Brown, M. R., Crim, J. W., Arata, R. C., Cai, H. N., Chun, C., and Shen, P. (1999). Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides 20, 1035–1042. doi: 10.1016/S0196-9781(99)00097-2
Caers, J., Verlinden, H., Zels, S., Vandersmissen, H. P., Vuerinckx, K., and Schoofs, L. (2012). More than two decades of research on insect neuropeptide GPCRs: an overview. Front. Endocrinol. (Lausanne) 3:151. doi: 10.3389/fendo.2012.00151
Cerdá-Reverter, J. M., and Larhammar, D. (2000). Neuropeptide Y family of peptides: structure, anatomical expression, function, and molecular evolution. Biochem. Cell Biol. 78, 371–392. doi: 10.1139/o00-004
Chen, H., and Tang, M. (2007). Spatial and temporal dynamics of bark beetles in Chinese white pine in Qinling Mountains of Shaanxi province. China. Environ. Entomol. 36, 1124–1130. doi: 10.1093/ee/36.5.1124
Costanzi, S. (2012). Homology modeling of class a G protein-coupled receptors. Methods Mol. Biol. 857, 259–279. doi: 10.1007/978-1-61779-588-6_11
Cui, H., and Zhao, Z. (2020). Structure and function of neuropeptide F in insects. J. Integ. Agri. 19, 1429–1438. doi: 10.1016/S2095-3119(19)62804-2
Cunningham, C. B., Vandenheuvel, K., Khana, D. B., McKinney, E. C., and Moore, A. J. (2016). The role of neuropeptide F in a transition to parental care. Biol. Lett. 12:20160158. doi: 10.1098/rsbl.2016.0158
Dai, L., Ma, M., Wang, C., Shi, Q., Zhang, R., and Chen, H. (2015). Cytochrome P450s from the Chinese white pine beetle, Dendroctonus armandi (Curculionidae: Scolytinae): expression profiles of different stages and responses to host allelochemicals. Insect Biochem. Mol. Biol. 65, 35–46. doi: 10.1016/j.ibmb.2015.08.004
Dai, L., Wang, C., Zhang, X., Yu, J., Zhang, R., and Chen, H. (2014). Two CYP4 genes of the Chinese white pine beetle, Dendroctonus armandi (Curculionidae: Scolytinae), and their transcript levels under different development stages and treatments. Insect Mol. Biol. 23, 598–610. doi: 10.1111/imb.12108
de Jong-Brink, M., ter Maat, A., and Tensen, C. P. (2001). NPY in invertebrates: molecular answers to altered functions during evolution. Peptides 22, 309–315. doi: 10.1016/S0196-9781(01)00332-1
Deng, X., Yang, H., He, X., Liao, Y., Zheng, C., Zhou, Q., et al. (2014). Activation of Bombyx neuropeptide G protein-coupled receptor A4 via a Gαi-dependent signaling pathway by direct interaction with neuropeptide F from silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 45, 77–88. doi: 10.1016/j.ibmb.2013.12.007
Dierick, H. A., and Greenspan, R. J. (2007). Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat. Genet. 39, 678–682. doi: 10.1038/ng2029
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Erion, R., King, A. N., Wu, G., Hogenesch, J. B., and Sehgal, A. (2016). Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. eLife 5:e13552. doi: 10.7554/eLife.13552
Fadda, M., Hasakiogullari, I., Temmerman, L., Beets, I., Zels, S., and Schoofs, L. (2019). Regulation of feeding and metabolism by neuropeptide F and short neuropeptide F in invertebrates. Front. Endocrinol. (Lausanne) 10:64. doi: 10.3389/fendo.2019.00064
Fu, D., Dai, L., Gao, H., Sun, Y., Liu, B., and Chen, H. (2019). Identification, expression patterns and RNA interference of aquaporins in Dendroctonus armandi (Coleoptera: Scolytinae) larvae during overwintering. Front. Physiol. 10:967. doi: 10.3389/fphys.2019.00967
Garczynski, S. F., Brown, M. R., Shen, P., Murray, T. F., and Crim, J. W. (2002). Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides 23, 773–780. doi: 10.1016/s0196-9781(01)00647-647
Garczynski, S. F., Crim, J. W., and Brown, M. R. (2005). Characterization of neuropeptide F and its receptor from the African malaria mosquito, Anopheles gambiae. Peptides 26, 99–107. doi: 10.1016/j.peptides.2004.07.014
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Gonzalez, R., and Orchard, I. (2008). Characterization of neuropeptide F-like immunoreactivity in the blood-feeding hemipteran. Rhodnius prolixus. Peptides 29, 545–558. doi: 10.1016/j.peptides.2007.11.023
Hermann, C., Yoshii, T., Dusik, V., and Helfrich-Forster, C. (2012). Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J. Comp. Neurol. 520, 970–987. doi: 10.1002/cne.22742
Hu, X., Wang, C., Chen, H., and Ma, J. (2013). Differences in the structure of the gut bacteria communities in development stages of the Chinese white pine beetle (Dendroctonus armandi). Int. J. Mol. Sci. 14, 21006–21020. doi: 10.3390/ijms141021006
Huang, Y., Crim, J. W., Nuss, A. B., and Brown, M. R. (2011). Neuropeptide F and the corn earworm, Helicoverpa zea: a midgut peptide revisited. Peptides 32, 483–492. doi: 10.1016/j.peptides.2010.09.014
Kim, W. J., Jan, L. Y., and Jan, Y. N. (2013). A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron 80, 1190–1205. doi: 10.1016/j.neuron.2013.09.034
Krashes, M. J., DasGupta, S., Vreede, A., White, B., Armstrong, J. D., and Waddell, S. (2009). A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139, 416–427. doi: 10.1016/j.cell.2009.08.035
Lee, G., Bahn, J. H., and Park, J. H. (2006). Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc. Natl. Acad. Sci. U S A. 103, 12580–12585. doi: 10.1073/pnas.0601171103
Li, X., Qu, M. J., Zhang, Y., Li, J. W., and Liu, T. X. (2018). Expression of neuropeptide F gene and its regulation of feeding behavior in the Pea Aphid, Acyrthosiphon pisum. Front. Physiol. 9:87. doi: 10.3389/fphys.2018.00087
Lingo, P. R., Zhao, Z., and Shen, P. (2007). Co-regulation of coldresistant food acquisition by insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience 148, 371–374. doi: 10.1016/j.neuroscience.2007.06.010
Liu, X., Zhang, Y., Zhou, Z., Zhao, Z., and Liu, X. (2013). Cloning and sequence analysis of neuropeptide F from the oriental tobacco budworm Helicoverpa assulta (Guenee). Arch. Insect Biochem. Physiol. 84, 115–129. doi: 10.1002/arch.21119
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livak, K. J., and Schmittgen, T. D. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Maule, A. G., Shaw, C., Halton, D. W., Thim, L., Johnston, C. F., Fairweather, I., et al. (1991). Neuropeptide F: a novel parasitic flatworm regulatory peptide from Moniezia expansa (Cestoda: Cyclophyllidea). Parasitology 102, 309–316. doi: 10.1017/S0031182000062648
Nässel, D. R., and Wegener, C. (2011). A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides 32, 1335–1355. doi: 10.1016/j.peptides.2011.03.013
Nuss, A. B., Forschler, B. T., Crim, J. W., TeBrugge, V., Pohl, J., and Brown, M. R. (2010). Molecular characterization of neuropeptide F from the eastern subterranean termite Reticulitermes flavipes (Kollar) (Isoptera: Rhinotermitidae). Peptides 31, 419–428. doi: 10.1673/031.008.6801
Pandit, A. A., Davies, S. A., Smagghe, G., and Dow, J. A. T. (2019). Evolutionary trends of neuropeptide signaling in beetles - A comparative analysis of Coleopteran transcriptomic and genomic data. Insect Biochem. Mol. Biol. 114:103227. doi: 10.1016/j.ibmb.2019.103227
Roller, L., Yamanaka, N., Watanabe, K., Daubnerova, I., Zitnan, D., Kataoka, H., et al. (2008). The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1147–1157. doi: 10.1016/j.ibmb.2008.04.009
Shen, P., and Cai, H. N. (2001). Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J. Neurobiol. 47, 16–25. doi: 10.1002/neu.1012
Stanek, D. M., Pohl, J., Crim, J. W., and Brown, M. R. (2002). Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides 23, 1367–1378. doi: 10.1016/S0196-9781(02)00074-8
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6:molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, S., Li, A., Wang, Y., and Shi, W. (2019). Role of the neuropeptide F 1 in regulating the appetite for food in Locusta migratoria. Pest Manag. Sci. 75, 1304–1309. doi: 10.1002/ps.5244
Van Wielendaele, P., Dillen, S., Zels, S., Badisco, L., and Vanden Broeck, J. (2013). Regulation of feeding by Neuropeptide F in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 43, 102–114. doi: 10.1016/j.ibmb.2012.10.002
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Veenstra, J. A. (2019). Coleoptera genome and transcriptome sequences reveal numerous differences in neuropeptide signaling between species. Peerj 7:e7144. doi: 10.7717/peerj.7144
Wang, Y., Pu, Y., and Shen, P. (2013). Neuropeptide-gated perception of appetitive olfactory inputs in Drosophila larvae. Cell Rep. 3, 820–830. doi: 10.1016/j.celrep.2013.02.003
Wen, T., Parrish, C. A., Xu, D., Wu, Q., and Shen, P. (2005). Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc. Natl. Acad. Sci. U S A. 102, 2141–2146. doi: 10.1073/pnas.0406814102
Wu, Q., Wen, T., Lee, G., Park, J. H., Cai, H. N., and Shen, P. (2003). Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147–161. doi: 10.1016/S0896-6273(03)00396-9
Wu, Q., Zhang, Y., Xu, J., and Shen, P. (2005a). Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. U S A. 102, 13289–13294. doi: 10.1073/pnas.0501914102
Wu, Q., Zhao, Z., and Shen, P. (2005b). Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat. Neurosci. 8, 1350–1355. doi: 10.1038/nn1540
Xu, G., Gu, G., Teng, Z., Wu, S., Huang, J., Song, Q., et al. (2016). Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci. Rep. 6:28976. doi: 10.1038/srep28976
Yamanaka, N., Yamamoto, S., Zitnan, D., Watanabe, K., Kawada, T., Satake, H., et al. (2008). Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One 3:e3048. doi: 10.1371/journal.pone.0003048
Yeoh, J. G. C., Pandit, A. A., Zandawala, M., Nässel, D. R., Davies, S. A., and Dow, J. A. T. (2017). DINeR: database for insect neuropeptide research. Insect Biochem. Mol. Biol. 86, 9–19. doi: 10.1016/j.ibmb.2017.05.001
Yue, Z., Li, X., Zhang, E., Liu, X., and Zhao, Z. (2017). A potential and novel type transgenic corn plant for control of the corn borer. Sci. Rep. 7:44105. doi: 10.1038/srep44105
Yue, Z., Liu, X., Zhou, Z., Hou, G., Hua, J., and Zhao, Z. (2016). Development of a novel-type transgenic cotton plant for control of cotton bollworm. Plant Biotechnol. J. 14, 1747–1755. doi: 10.1111/pbi.12534
Zhang, H., Bai, J., Huang, S., Liu, H., Lin, J., and Hou, Y. (2020). Neuropeptides and G-protein coupled receptors (GPCRs) in the red palm weevil Rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae). Front. Physiol. 11:159. doi: 10.3389/fphys.2020.00159
Keywords: Dendroctonus armandi, Neuropeptide F, Neuropeptide F receptor, food intake, energy metabolism, RNA interference
Citation: Liu B, Fu D, Gao H, Ning H, Sun Y, Chen H and Tang M (2021) Cloning and Expression of the Neuropeptide F and Neuropeptide F Receptor Genes and Their Regulation of Food Intake in the Chinese White Pine Beetle Dendroctonus armandi. Front. Physiol. 12:662651. doi: 10.3389/fphys.2021.662651
Received: 01 February 2021; Accepted: 12 May 2021;
Published: 18 June 2021.
Edited by:
Robert Huber, Bowling Green State University, United StatesReviewed by:
Gong-yin Ye, Zhejiang University, ChinaCopyright © 2021 Liu, Fu, Gao, Ning, Sun, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Chen, Y2hlbmh1aUBud3N1YWYuZWR1LmNu; Ming Tang, dGFuZ21AbndzdWFmLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.