
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 17 March 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.648481
This article is part of the Research Topic Invertebrate UDP-Glycosyltransferases: Nomenclature, Diversity and Functions View all 5 articles
UDP-glycosyltransferases (UGTs) are important conjugation enzymes found in all kingdoms of life, catalyzing a sugar conjugation with small lipophilic compounds and playing a crucial role in detoxification and homeostasis. The UGT gene family is defined by a signature motif in the C-terminal domain where the uridine diphosphate (UDP)-sugar donor binds. UGTs have been identified in a number of insect genomes over the last decade and much progress has been achieved in characterizing their expression patterns and molecular functions. Here, we present an update of the complete repertoire of UGT genes in Drosophila melanogaster and provide a brief overview of the latest research in this model insect. A total of 35 UGT genes are found in the D. melanogaster genome, localized to chromosomes 2 and 3 with a high degree of gene duplications on the chromosome arm 3R. All D. melanogaster UGT genes have now been named in FlyBase according to the unified UGT nomenclature guidelines. A phylogenetic analysis of UGT genes shows lineage-specific gene duplications. Analysis of anatomical and induced gene expression patterns demonstrate that some UGT genes are differentially expressed in various tissues or after environmental treatments. Extended searches of UGT orthologs from 18 additional Drosophila species reveal a diversity of UGT gene numbers and composition. The roles of Drosophila UGTs identified to date are briefly reviewed, and include xenobiotic metabolism, nicotine resistance, olfaction, cold tolerance, sclerotization, pigmentation, and immunity. Together, the updated genomic information and research overview provided herein will aid further research in this developing field.
UDP-glycosyltransferases (UGTs) are a superfamily of enzymes found in all kingdoms of life, including animals, plants, fungi, bacteria, and some viruses (Bock, 2016). UGTs catalyze the covalent addition of sugars from uridine diphosphate (UDP) sugar donors to a broad range of lipophilic small molecules, playing a crucial role in conjugation, detoxification and elimination of exogenous and endogenous toxic compounds, as well as in regulation and distribution of endogenous signal molecules and metabolites (Meech et al., 2019). Mammalian UGTs were previously called “UDP-glucuronosyltransferases” as most research articles in drug metabolism dealt with enzymes that mainly use UDP-glucuronic acid as the sugar donor; however, the UGT Nomenclature Committee recommended the use of “UDP-glycosyltransferase” in order to include enzymes that do not use UDP-glucuronic acid (Mackenzie et al., 2005). The same notion has been adopted for non-mammalian UGTs (Meech et al., 2012), including insects as they predominantly use UDP-glucose as the sugar donor (Myers and Smith, 1954; Dutton and Ko, 1964; Ahmad and Forgash, 1976; Kramer and Hopkins, 1987; Rausell et al., 1997; Wang et al., 1999).
The first evidence of UGT activity in insects was obtained by a chromatographic analysis of m-aminophenyl glucoside from feces of a locust, Locusta migratoria, suggesting insects conjugate the hydroxyl compounds with glucose, instead of glucuronic acid (Myers and Smith, 1954). Biochemical studies in a variety of insect species indicated that the glucose conjugation plays an important role in diverse physiological processes in insects, such as detoxification (Smith, 1955; Wilkinson, 1986; Ahn et al., 2011), sclerotization (Kramer and Hopkins, 1987; Hopkins, 1992), pigmentation (Hopkins and Ahmad, 1991; Wiesen et al., 1994), and insecticide resistance (Lee et al., 2005). Molecular studies revealed that a UGT is responsible for the glycosylation of flavonoids in the silkworm cocoon (Daimon et al., 2010). Antenna-specific UGTs were detected by gene expression analysis in a moth, Spodoptera littoralis, suggesting specific roles in olfaction (Bozzolan et al., 2014). It was revealed that benzoxazinoids, the indole-derived plant defense compounds, are stereoselectively inactivated by UGT enzymes in the fall armyworm, Spodoptera frugiperda (Israni et al., 2020). Also, some UGTs were shown to be associated with insecticide resistance (Li et al., 2017; Chen et al., 2019, 2020; Zhou et al., 2019; Pan et al., 2020). Several UGTs have been identified and characterized in the Drosophila genus, with a focus on the model organism D. melanogaster. Drosophila UGTs have been shown to function in diverse processes including xenobiotic metabolism, nicotine resistance, olfaction, cold tolerance, sclerotization, pigmentation, and immunity (summarized in Table 1). Among non-insect arthropods, the two-spotted spider mite, Tetranychus urticae, has been intensively studied for the substrate specificity of its UGTs (Snoeck et al., 2019), which are most likely acquired from bacteria via horizontal gene transfer (Ahn et al., 2014).
During the last two decades, genome and transcriptome sequencing of insects has generated genome-wide analyses of UGT genes in a variety of insects (Luque and O’Reilly, 2002; Huang et al., 2008; Ahn et al., 2012; Hu B. et al., 2019), revealing that the UGT gene family comprises multiple genes in each species, ranging from 12 (honeybee) to 58 (aphid) (Ahn et al., 2012). Given these and similar studies of non-insect genomes, the UGT Nomenclature Committee was formed to assign systematic names to the large number of UGTs, defining the families (e.g., UGT36) and subfamilies (e.g., UGT36A) at >45% and >60% amino acid sequence identity, respectively1. Originally, families 1–50 are reserved for animals, 51–70 for fungi and yeasts, 71–100 for plants, and 101–200 for bacteria; if these number assignments become depleted, the family number increases by 10-fold (Mackenzie et al., 1997). For insects and insect viruses, the UGT family numbers have been assigned from 31 to 50, resuming in the range 301–500 (Ahn et al., 2012).
As a model insect, it is particularly important that the UGT genes of D. melanogaster are identified and named in accordance with the UGT Nomenclature Committee guidelines; these genes define the range of insect UGT family numbers, and also provide a consensus standard to study UGT genes from other insects that will be annotated in the future. For this purpose, we report here the complete repertoire of D. melanogaster UGT genes with updated nomenclature, genomic architecture and gene expression data. We also identify orthologous genes from 18 additional Drosophila species in order to view the D. melanogaster UGTs from an evolutionary perspective.
The first Drosophila melanogaster UGT gene to be identified, Dorothy (currently Ugt36A1), was named after a character of The Wizard of Oz (Rodriguez et al., 1996). A little later, five other D. melanogaster UGT genes, Ugt35a, Ugt35b, Ugt37a1, Ugt37b1, and Ugt37c1 (lowercase letters were initially used to indicate subfamily membership), were among the first UGT genes to be named in consultation with the UGT Nomenclature Committee (Wang et al., 1999). Subsequently, several other D. melanogaster UGTs were directly named in FlyBase according to their cytogenetic locations (e.g., Ugt36Ba – Ugt36Bc, Ugt58Fa, and Ugt86Da – Ugt86Dj) (Table 2), which is evidently confusing given the superficial resemblance between this notation and the UGT Committee nomenclature. Ahn et al. (2012) revised and curated the D. melanogaster UGTs, employing the systematic names to maintain consistency with the universal nomenclature and the five previously assigned official names. In the current study, we have completed the list of D. melanogaster UGT genes and have updated the gene symbols and names within FlyBase to adopt the systematic nomenclature. Furthermore, we have added a UGT “gene group” page to FlyBase that conveniently lists all these genes in a single report to facilitate further analysis and download of associated data2.
Wang et al. (1999) identified 9–10 putative UGT gene sequences, including the five named ones (see above), from cDNA libraries and the incomplete genome databases available at the time. Upon completion of the D. melanogaster genome (Adams et al., 2000), the first genome-wide annotation of multiple UGT genes was conducted and a total of 33 putative UGT genes were reported together with a phylogenetic and genomic analysis (Luque and O’Reilly, 2002). Ahn et al. (2012) revised the sequences in detail and identified an additional gene (Ugt50B3). The current study has added one further gene (Ugt305A1), resulting in a complete repertoire of 35 UGT genes in D. melanogaster (Table 2). They are grouped into 13 families according to the nomenclature system: UGT35 (6 genes), UGT36 (4 genes), UGT37 (8 genes), UGT49 (3 genes), UGT50 (1 gene), UGT301 (1 gene), UGT302 (3 genes), UGT303 (4 genes), and 1 gene in each of UGT304, UGT305, UGT307, UGT316, and UGT317 (Table 2 and Figure 1).
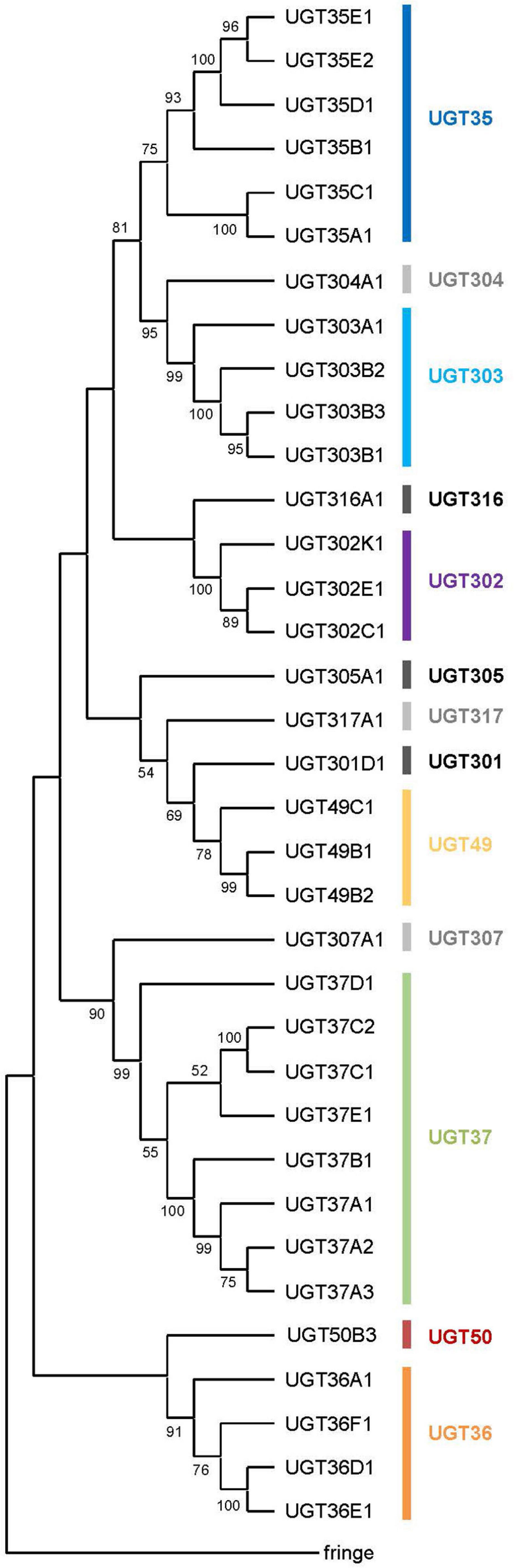
Figure 1. A phylogenetic tree of the UDP-glycosyltransferases from Drosophila melanogaster. All the 35 UGT protein sequences and the fringe protein sequence (as an outgroup) were aligned using ClustalW and a consensus phylogenetic tree was constructed using the Maximum Likelihood method and JTT matrix-based model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Those less than 50% are omitted). Evolutionary analyses were conducted in MEGA X.
All 35 UGT genes are found on the two major autosomes (chromosome 2 with 16 genes and chromosome 3 with 19 genes); none are located on the minor autosome (chromosome 4) or the sex chromosomes (Table 2 and Supplementary Figure 1). Among different chromosomal arms, about half (17 UGT genes) lie on 3R (the right arm of chromosome 3), followed by 2L (11 UGT genes), 2R (5 genes) and 3L (2 genes). A large cluster of UGT genes is found on 3R at the cytogenetic location of 86D4 – 86D6, where ten closely related UGT genes are positioned in tandem. The other multiplied gene families are found in one or two genomic locations in close proximity, whereas the members of another large family, UGT37, are spread across three different chromosomal arms (five in 2L, one in 2R, and two in 3R) (Table 2 and Supplementary Figure 1). It is noteworthy that 3L harbors only two UGT genes (Ugt305A1 and Ugt316A1), both of which seem to be unique in their sequences, and are unusually long (Table 2).
All 35 UGT genes are interrupted by intron(s) except for Ugt37C1 and Ugt37C2 (Table 2). These two intron-less genes do not seem to originate from bacterial UGT genes due to their sequence similarity to animal UGTs (see Ahn et al., 2014). D. melanogaster UGT genes are composed of one to six exons: a majority of genes (19 genes; 54%) comprise 2 exons and the rest of genes have 1, 3, 4 or 5 exons, except one gene (Ugt50B3) has 6 exons in its coding sequence (Table 2 and Supplementary Figure 2). The lengths of intron sequences are mostly within the range of 48–85 bp (41 introns) or 108–584 bp (14 introns). Exceptionally, Ugt50B3 is interrupted by three long introns (1,389, 1,0432, and 8,198 bp) followed by two short ones (63 and 52 bp) (Supplementary Table 1 and Supplementary Figure 3). This, together with the fact it is phylogenetically distinguished from the others (Figure 1) and highly conserved in insects in general (Ahn et al., 2012), suggests Ugt50B3 is one of the oldest UGT genes.
Splicing variants are found in two UGT genes, Ugt50B3 and Ugt303B1, where two alternative transcripts have been reported (Table 2). The Ugt50B3 variant is annotated to have an alternative start codon in the middle of what is otherwise the third exon, producing a protein that is 89 amino acids (aa) shorter than the normal one. The Ugt303B1 variants seem to be derived from alternative splicing sites at the 3’-end of the first exon, resulting in a difference of only 9 nucleotides (3 aa) (Table 2).
The average length of D. melanogaster UGT proteins is 532 aa with two outliers, Ugt305A1 (583 aa) and Ugt316A1 (636 aa), which, as noted above, are phylogenetically unique and located in different genomic positions from the other UGT genes. All the UGTs contain an N-terminal signal peptide and a C-terminal transmembrane (TM) domain (Table 2 and Supplementary Figure 4), indicating that the D. melanogaster UGTs are located in the endoplasmic reticulum (ER) with their catalytic domains facing the ER lumen, as shown in other animals (Meech et al., 2012). The UGT-defining 44-aa signature sequence in the C-terminal domain, which is predicted to be intimately involved in the binding of UDP-sugar (Meech et al., 2019), is well conserved across the 35 UGTs (Supplementary Figure 5). However, variations shown in some residues in the signature sequence imply different specificity to different sugar donors other than UDP-glucose.
A consensus Maximum-likelihood tree constructed with deduced amino acid sequences revealed lineage-specific gene amplifications in several families such as UGT35, UGT36, UGT37, UGT49, UGT302, and UGT303 (Figure 1). For example, upon divergence from a common ancestor with Ugt307A1, UGT37 seems to have diversified into the largest gene family in D. melanogaster UGTs. It is noteworthy that the UGT37 members are spread across five different genomic locations. On the other hand, other multiplied UGTs are most likely diversified by tandem gene duplications, as they are found in the same genomic scaffolds in close proximity (Supplementary Figure 1).
Tissue-specific expression patterns of D. melanogaster UGT genes were analyzed previously by Ahn et al. (2012) using microarray data present in FlyAtlas (Chintapalli et al., 2007). Here, we have revisited this analysis using the higher quality RNAseq data available from the FlyAtlas2 database (Leader et al., 2018) – full data for adult males, adult females and larvae are included in Supplementary Table 2; representative data for adult males and larvae are in Figure 2. UGTs from each family are expressed in every adult and larval tissue at some level. Some UGT genes belonging to multi-gene families (Ugt35D1 and Ugt37E1) are undetectable in any tissue, while several others are expressed only in restricted patterns. In contrast, many UGT genes appear to be expressed ubiquitously, with high expression levels often seen within the digestive and excretory systems, particularly for members of the UGT35 and UGT37 families. Across all UGTs, the highest expression is seen within the adult midgut and larval Malpighian tubules. Of note, Ugt50B3, the sole representative of the UGT50 family, shows unusually high expression within the male accessory gland and the female spermatheca, whereas Ugt305A1 is only expressed at appreciable levels in the testis. Such restricted expression patterns suggest particularly important roles of Ugt50B3 and Ugt305A1 within these tissues.
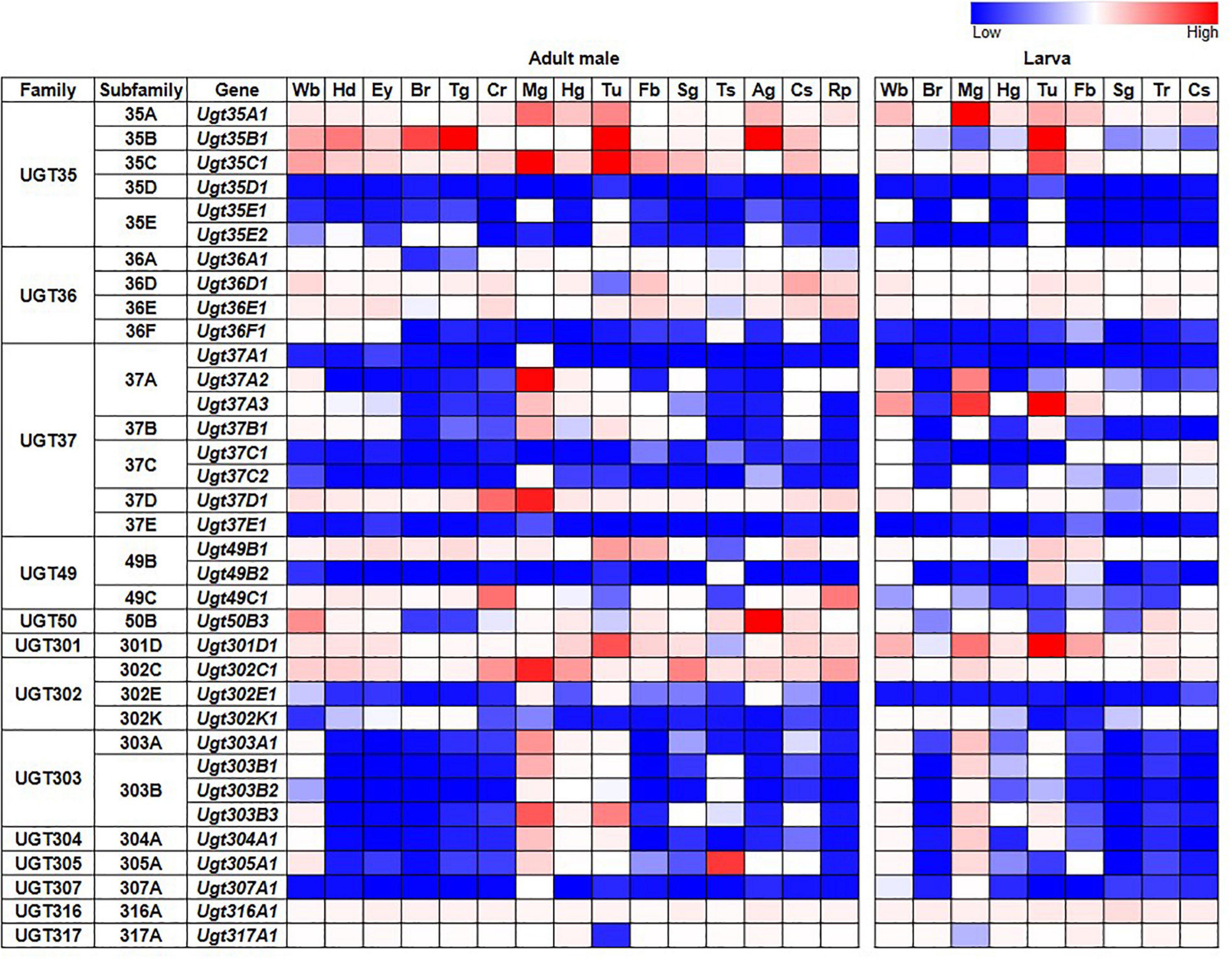
Figure 2. Expression of D. melanogaster UGT genes in different tissues of adult males and larvae (FlyAtlas2, Leader et al., 2018). Wb: whole body; Hd: head; Ey: eye; Br: brain/CNS; Tg: thoracicoabdominal ganglion; Cr: crop; Mg: midgut; Hg: hindgut; Tu: Malpighian tubules; Fb: fat body; Sg: salivary gland; Ts: testis; Ag: accessory glands; Cs: carcass; Rp: rectal pad; Tr: trachea. See Supplementary Table 2 for details and equivalent data for adult females.
Given the documented role of some UGTs in detoxification, we also examined whether D. melanogaster UGT gene expression is induced after exposure to various environmental and chemical treatments by examining RNAseq data generated by the modENCODE project (Brown et al., 2014) – the full dataset is in Supplementary Table 3; representative subsets are in Figure 3. The expression of most UGT genes is not upregulated in response to the majority of treatments. However, six genes from four different UGT families (Ugt35A1, Ugt37A2, Ugt37A3, Ugt37D1, Ugt49B1, and Ugt302C1) clearly show upregulated expression in response to the addition of caffeine, rotenone or ethanol to the diet, or exposure to Sindbis virus. On the other hand, certain treatments, including cold exposure and increased dietary copper or zinc, have no/little effect on the expression of any UGT gene.
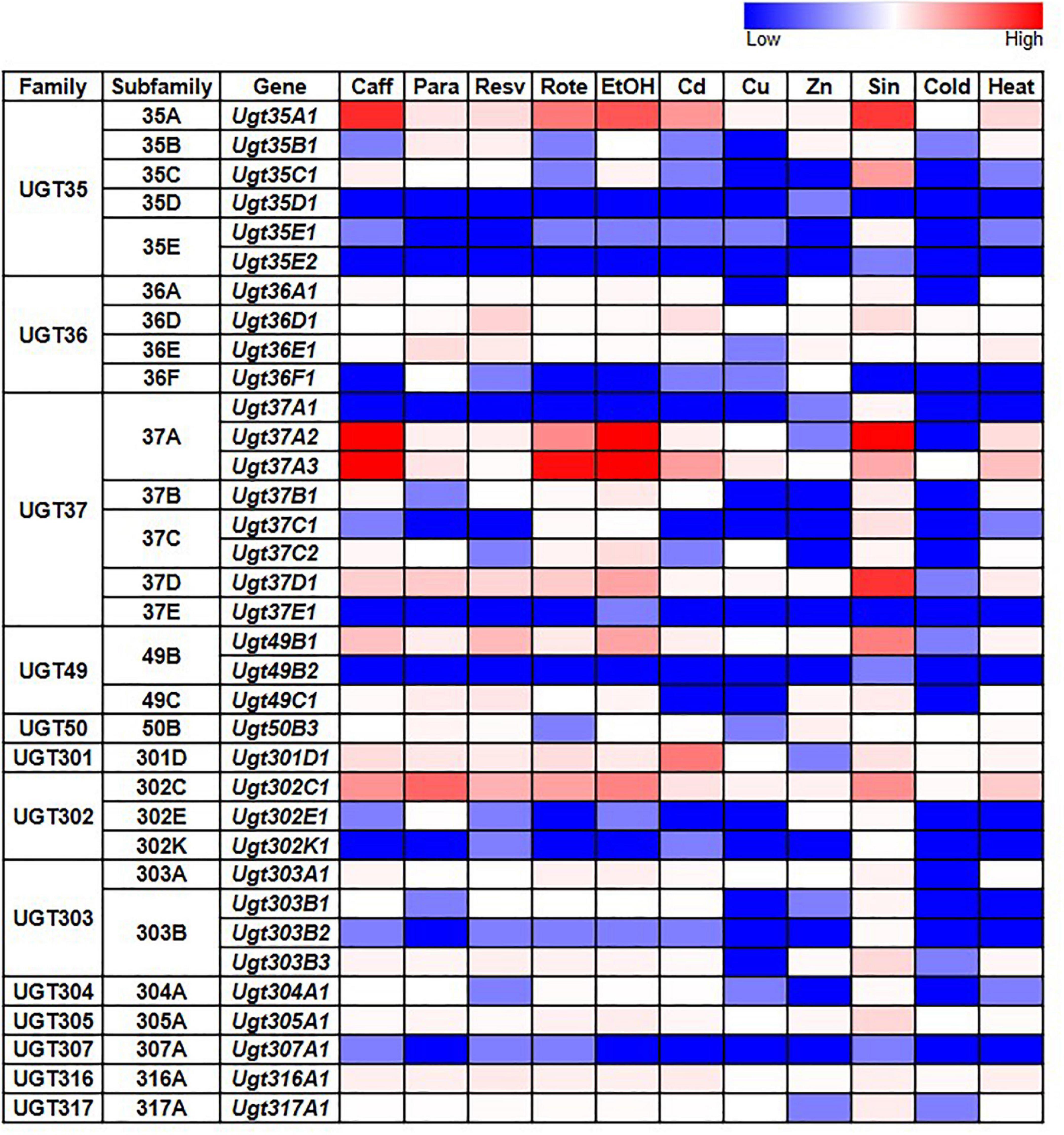
Figure 3. Expression of D. melanogaster UGT genes in wild type larvae/adults after various treatments (modENCODE; Brown et al., 2014). Caff: starved L3 larvae were fed 5 mg/ml caffeine for 4 h; Para: 3-day-old adults were fed 10 mM paraquat for 24 h; Resv: 2-day-old adults were fed 100 μM resveratrol continuously for 10 days; Rote: Feeding L3 larvae were fed 2 μg/ml rotenone for 6 h; EtOH: L3 larvae were treated with 5% ethanol; Cd: starved L3 larvae were fed 0.05 mM CdCl2 for 12 h; Cu: starved L3 larvae were fed 0.5 mM CuSO4 for 12 h; Zn: 2-day-old adults were fed 4.5 mM ZnCl2 for 48 h; Sin: L3 larvae were exposed to Sindbis virus; Cold: 4-day-old adults were kept at 0°C for 9 h, followed by 2 h of recovery at 25°C; Heat: 4-day-old adults were kept at 36°C for 1 h followed by a 30-min recovery at 25°C. See Supplementary Table 3 for details.
We identified UGT genes in 18 additional Drosophila species and deduced their orthologous relationships to the D. melanogaster genes (Figure 4; see section “Materials and Methods”). The total number of UGT genes per genome varies from 29 in D. elegans, D. pseudoobscura, and D. mojavensis, to 50 in D. takahashii. Some UGT families have been preserved, whereas others have been multiplied or lost through evolution (Figure 4 and Supplementary Table 4). The conserved UGT families are mostly single-member families, such as UGT50, UGT301, UGT304, UGT305, UGT307, UGT316, and UGT317, and show little or no gene additions/losses. The other UGT families comprising multiple genes show variable gene additions or losses in the different species (Supplementary Table 4). One of the most fluctuating families is UGT37: there are 8 gene members in D. melanogaster, but the number increases up to double (16 genes) in D. rhopaloa followed by D. willistoni (15 genes), and decreases down to half (4 genes) in D. erecta and D. grimshawi. The UGT49 family also shows a high degree of species difference: there are 3 gene members in D. melanogaster, but the number increases up to 11 in D. bipectinata followed by 8 in D. ananassae.
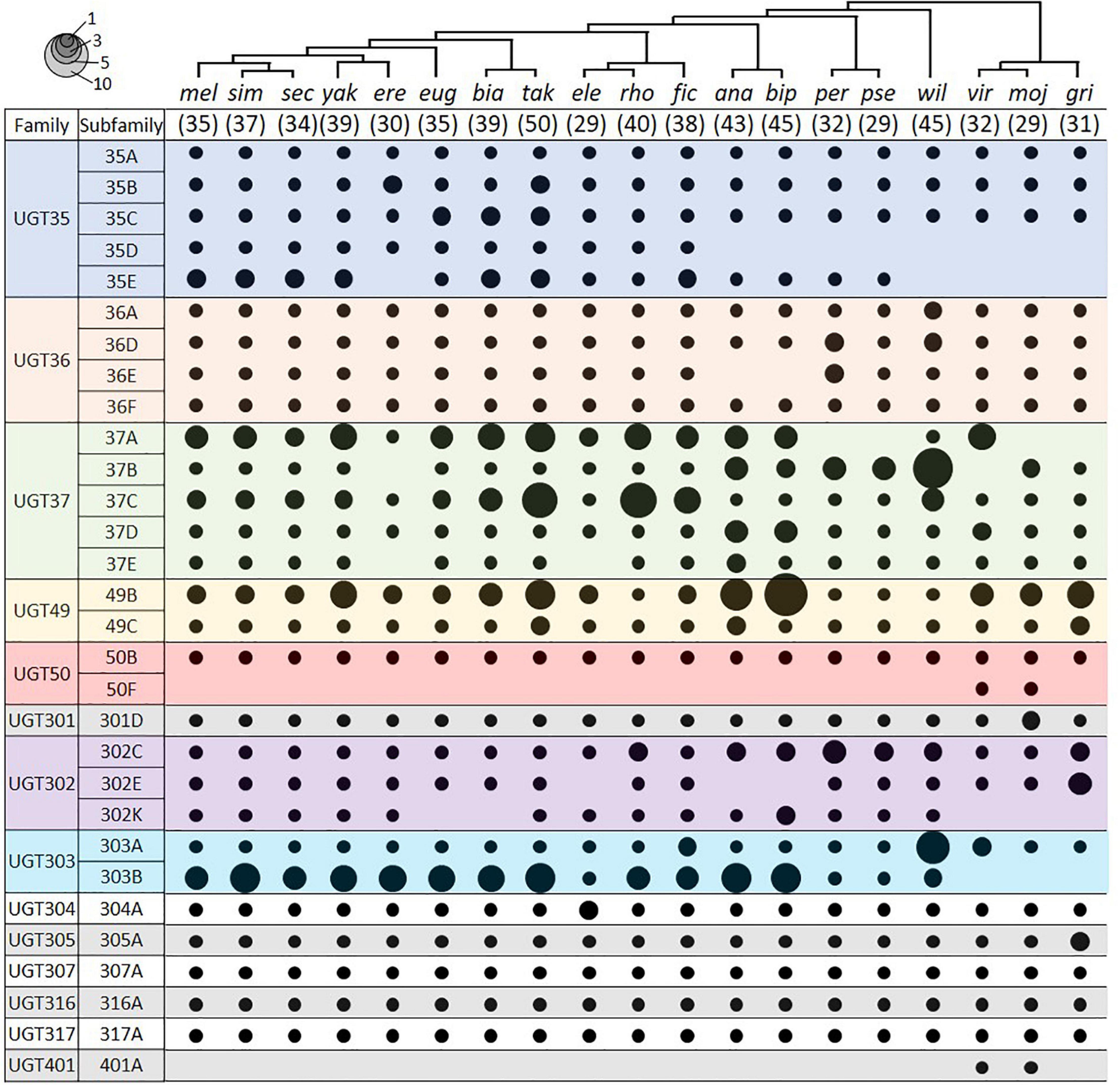
Figure 4. UGT orthologs in 19 Drosophila species. Circle size represents the number of genes in the indicated group. The species tree is adapted from Seetharam and Stuart (2013). The number in parenthesis under the tree represents the total number of UGT genes in the given species. Species names refer to D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. eugracilis, D. biarmipes, D. takahashii, D. elegans, D. rhopaloa, D. ficusphila, D. ananassae, D. bipectinata, D. persimilis, D. pseudoobscura, D. willistoni, D. virilis, D. mojavensis, and D. grimshawi. See Supplementary Table 4 for details.
Two UGTs that are not orthologous with any D. melanogaster UGTs were detected in both D. virilis and D. mojavensis. One pair is an additional member of the UGT50 family, named as the UGT50F subfamily in this study. The other pair defines a new UGT family, named here as Ugt401A. By BLAST search in NCBI, additional UGT50F members were found in three other species not included in this study (D. arizonae, D. navojoa, and D. hydei), whereas orthologs of UGT401A were present in seven other species (D. arizonae, D. navojoa, D. hydei, D. novamexicana, D. albomicans, D. innubila, and D. busckii). As all of these species form a distant group (“repleta-virilis” group) from D. melanogaster, the UGT401A genes might have been lost after divergence of two sub-genera, Sophophora and Drosophila, or newly emerged in this group, probably playing a unique role.
Further comparative analyses amongst Drosophila and related species will become possible as additional genomes are sequenced and annotation pipelines are improved. This will likely reveal other interesting evolutionary patterns. For example, our preliminary analysis of the genome (Gloss et al., 2019) and transcriptome (Whiteman et al., 2012) of Scaptomyza flava, a herbivorous leaf-mining species belonging to the Drosophilidae family (Whiteman et al., 2011), reveals that this species has only 23 UGT genes (data not shown), the smallest number among the species surveyed in this study.
The UGT gene family is one of the largest in the glycosyltransferase (GT) superfamily (EC:2.4.x.y). Since the pioneering work by Myers and Smith (1954), a large body of research outcomes on insect UGTs has been accumulated (Nagare et al., 2020). However, their molecular characteristics are less defined compared to the other detoxification enzymes, such as cytochrome P450s, glutathione S-transferases, and carboxylesterases. One of the reasons is that UGT genes have been incorrectly annotated in many genome sequencing projects. The nomenclature updates and genome-wide analyses of the D. melanogaster UGTs in this study will facilitate future work and communication in this growing research domain.
Conjugation with sugar residues changes the properties of aglycone substrate molecules by decreasing the reactivity of functional groups and by increasing solubility, thereby combating toxic xenobiotics (Heckel, 2018). The six genes (Ugt35A1, Ugt37A2, Ugt37A3, Ugt37D1, Ugt49B1, and Ugt302C1) upregulated upon noxious treatments would be the most promising elements potentially responsible for metabolic detoxification of xenobiotics. On the other hand, UGT genes that are highly expressed in specific tissues (e.g., Ugt35B1, Ugt50B3, and Ugt305A1) are likely to play important physiological roles by conjugating endogenous molecules. Two olfactory UGTs (Ugt35B1 and Ugt36E1) may give a new insight on management of the congeneric pest species, D. suzukii. Much more remains to be discovered in relation to the molecular functions of UGTs in sclerotization, pigmentation, immunity and other processes.
Genomic data for D. melanogaster UGTs were obtained from FlyBase (flybase.org; Thurmond et al., 2019) using release FB2020_05, which includes D. melanogaster genome annotation R6.36. Genomic data for other Drosophila species were obtained from NCBI – sequence assemblies and annotation versions are given in Supplementary Table 4. Supplementary Data File 1 contains all Drosophila UGT protein sequences in fasta format. The signal peptides and transmembrane domains shown in Supplementary Table 1 were predicted by SignalP-5.0 Server3 and TMHMM Server v. 2.04, respectively.
Deduced amino acid sequences of 35 D. melanogaster UGT sequences were aligned by ClustalW and a consensus phylogenetic tree was constructed using the Maximum Likelihood method and JTT matrix-based model with 1,000 bootstrappings. As an outgroup, fringe (CG10580), an N-acetylglucosaminyltransferase, was used. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). The species phylogenetic tree of Drosophila used in Figure 4 was adapted from that in (Seetharam and Stuart, 2013).
Tissue expression (RNAseq) data were downloaded from FlyAtlas2 (5 Leader et al., 2018). Gene FPKM (Fragments Per Kilobase of transcript per Million mapped reads) and Enrichment (measuring the abundance of a gene in a particular tissue relative to that in the whole fly) data for adult males, adult females and larvae were downloaded as TSV files and processed in Excel (Supplementary Table 2). FPKM data for adult males and larvae are presented in Figure 2.
modENCODE treatment expression (RNAseq) data (Brown et al., 2014) for were obtained from FlyBase (6 Thurmond et al., 2019) using the Batch Download tool operated on the gene_rpkm_report precomputed file. Data were processed in Excel (Supplementary Table 3) and a subset of representative data are presented in Figure 3.
UDP-glycosyltransferases genes in 18 non-melanogaster species were additionally identified, which are D. ananassae (taxID: 7217), D. biarmipes (taxID: 125945), D. bipectinata (taxID: 42026), D. elegans (taxID: 30023), D. erecta (taxID: 7220), D. eugracilis (taxID: 29029), D. ficusphila (taxID: 30025), D. grimshawi (taxID: 7222), D. mojavensis (taxID: 7230), D. persimilis (taxID: 7234), D. pseudoobscura (taxID: 7237), D. rhopaloa (taxID: 1041015), D. sechellia (taxID: 7238), D. simulans (taxID: 7240), D. takahashii (taxID: 29030), D. virilis (taxID: 7244), D. willistoni (taxID: 7260), and D. yakuba (taxID: 7245), in alphabetic order. All the UGTs were classified into families/subfamilies using three complementary approaches. First, D. melanogaster UGT gene/protein sequences were used as queries of other Drosophila genomes available at NCBI using NCBI BLAST. In case of multiple genes in a same gene family, genomic locations were further compared with those of D. melanogaster to confirm the orthologous families/subfamilies they belong. Second, the InterPro database (release 82.0;7 Mitchell et al., 2019) was queried using the InterPro signature “UDP-glucuronosyl/UDP-glucosyltransferase” (IPR002213), which is diagnostic of UGT proteins, within the Drosophila genus (taxon ID 7215). Third, the OrthoDB v10.1 database (8 Kriventseva et al., 2019) was also queried using the IPR002213 signature within the Drosophila genus (taxon ID 7215) to identify orthologous groups comprising UGT genes. In addition, OrthoDB v9.1 data were obtained via D. melanogaster orthology data present in FlyBase (FB2020_05), primarily to obtain OrthoDB groupings for genes in Drosophila species absent from v10.1 (D. simulans, D. sechellia, D. persimilis). Data were cross-referenced using the NCBI gene IDs, FlyBase gene IDs and/or UniProt accessions present in each database, and the integrated data are shown in Supplementary Table 4. There is a large (mainly 1:1) agreement between the UGT subfamilies defined by the UGT Nomenclature Committee and the orthologous groups defined by OrthoDB (see Supplementary Table 5 for details). Note that several UGT gene models are incorrectly annotated at FlyBase/NCBI, e.g., some gene models need to be split, others need to be merged, others require extending (see Supplementary Table 4 for details). Also note that all non-melanogaster gene models and IDs have been retired from FlyBase and are now annotated and maintained by the NCBI (see the FB2018_06 and FB2020_03 release notes9). However, since archived non-melanogaster data are still present in FlyBase, and FlyBase IDs/symbols are still present in many databases, FlyBase gene IDs for the non-melanogaster species are included in Supplementary Table 4.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
S-JA and SM designed the research, performed the analyses, evaluated the data, interpreted the results, and wrote the manuscript.
This material is based upon work that is supported by the National Institute of Food and Agriculture, United States Department of Agriculture, Hatch-Multistate project under accession number MIS-311360 and by the Mississippi Agricultural and Forestry Experiment Station to S-JA. SM was funded by a grant from the National Human Genome Research Institute of the National Institutes of Health (U41HG000739) to Norbert Perrimon (PI) and Nicholas Brown (co-PI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Michael Court in the UGT Nomenclature Committee for consultations and many contributors to the bioinformatic databases used in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.648481/full#supplementary-material
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Ahmad, S., and Forgash, A. J. (1976). Nonoxidative enzymes in the metabolism of insecticides. Drug Metab. Rev. 5, 141–164. doi: 10.3109/03602537608995840
Ahn, S.-J., Badenes-Pérez, F. R., Reichelt, M., Svatoš, A., Schneider, B., Gershenzon, J., et al. (2011). Metabolic detoxification of capsaicin by UDP-glycosyltransferase in three Helicoverpa species. Arch. Insect Biochem. Physiol. 78, 104–118. doi: 10.1002/arch.20444
Ahn, S.-J., Dermauw, W., Wybouw, N., Heckel, D. G., and van Leeuwen, T. (2014). Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem. Mol. Biol. 50, 43–57. doi: 10.1016/j.ibmb.2014.04.003
Ahn, S.-J., Vogel, H., and Heckel, D. G. (2012). Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42, 133–147. doi: 10.1016/j.ibmb.2011.11.006
Bock, K. W. (2016). The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: animal-plant arms-race and co-evolution. Biochem. Pharmacol. 99, 11–17. doi: 10.1016/j.bcp.2015.10.001
Bozzolan, F., Siaussat, D., Maria, A., Durand, N., Pottier, M. A., Chertemps, T., et al. (2014). Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 25, 539–549. doi: 10.1111/imb.12100
Brown, J. B., Boley, N., Eisman, R., May, G. E., Stoiber, M. H., Duff, M. O., et al. (2014). Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–399. doi: 10.1038/nature12962
Chen, P. S., Mitchell, H. K., and Neuweg, M. (1978). Tyrosine glucoside in Drosophila busckii. Insect Biochem. 8, 279–286. doi: 10.1016/0020-1790(78)90038-0
Chen, X., Tang, C., Ma, K., Xia, J., Song, D., and Gao, X. W. (2020). Overexpression of UDP-glycosyltransferase potentially involved in insecticide resistance in Aphis gossypii Glover collected from Bt cotton fields in China. Pest Manag. Sci. 76, 1371–1377. doi: 10.1002/ps.5648
Chen, X., Xia, J., Shang, Q., Song, D., and Gao, X. (2019). UDP-glucosyltransferases potentially contribute to imidacloprid resistance in Aphis gossypii glover based on transcriptomic and proteomic analyses. Pestic. Biochem. Physiol. 159, 98–106. doi: 10.1016/j.pestbp.2019.06.002
Chintapalli, V. R., Wang, J., and Dow, J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720. doi: 10.1038/ng2049
Daimon, T., Hirayama, C., Kanai, M., Ruike, Y., Meng, Y., Kosegawa, E., et al. (2010). The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proc. Natl. Acad. Sci. U.S.A. 107, 11471–11476. doi: 10.1073/pnas.1000479107
Dutton, G. J., and Ko, V. (1964). The apparent absence of uridine diphosphate glucuronyltransferase for detoxication in Musca domestica. Comp. Biochem. Physiol. 11, 269–272. doi: 10.1016/0010-406X(62)90025-7
Ferré, J., Real, M. D., Mensua, J. L., and Jacobson, K. B. (1985). Xanthurenic acid 8-O-β-D-glucoside, a novel tryptophan metabolite in eye-color mutants of Drosophila melanogaster. J. Biol. Chem. 260, 7509–7514. doi: 10.1016/S0021-9258(17)39636-9
Fraichard, S., Legendre, A., Lucas, P., Chauvel, I., Faure, P., Neiers, F., et al. (2020). Modulation of sex pheromone discrimination by a UDP-glycosyltransferase in Drosophila melanogaster. Genes 11:237. doi: 10.3390/genes11030237
Gloss, A. D., Nelson Dittrich, A. C., Lapoint, R. T., Goldman-Huertas, B., Verster, K. I., Pelaez, J. L., et al. (2019). Evolution of herbivory remodels a Drosophila genome. bioRxiv [Preprint], 767160. doi: 10.1101/767160 bioRxiv:767160
Heckel, D. G. (2018). “Insect detoxification and sequestration strategies,” in Annual Plant Reviews; Plant Insect Interactions, eds C. Voelckel, and G. Jander (Chichester: Wiley-Blackwell) 77–114. doi: 10.1002/9781119312994.apr0507
Highfill, C. A., Tran, J. H., Nguyen, S. K. T., Moldenhauer, T. R., Wang, X., and Macdonald, S. J. (2017). Naturally segregating variation at Ugt86Dd contributes to nicotine resistance in Drosophila melanogaster. Genetics 207, 311–325. doi: 10.1534/genetics.117.300058/-/DC1.1
Hopkins, T. (1992). Insect cuticle sclerotization. Annu. Rev. Entomol. 37, 273–302. doi: 10.1146/annurev.ento.37.1.273
Hopkins, T. L., and Ahmad, S. A. (1991). Flavonoid wing pigments in grasshoppers. Experientia 47, 1089–1091. doi: 10.1007/BF01923349
Hu, B., Zhang, S. H., Ren, M. M., Tian, X. R., Wei, Q., Mburu, D. K., et al. (2019). The expression of Spodoptera exigua P450 and UGT genes: tissue specificity and response to insecticides. Insect Sci. 26, 199–216. doi: 10.1111/1744-7917.12538
Huang, F.-F., Chai, C.-L., Zhang, Z., Liu, Z.-H., Dai, F.-Y., Lu, C., et al. (2008). The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9:563. doi: 10.1186/1471-2164-9-563
Israni, B., Wouters, F. C., Luck, K., Seibel, E., Ahn, S., Paetz, C., et al. (2020). The fall armyworm Spodoptera frugiperda utilizes specific UDP-glycosyltransferases to inactivate maize defensive benzoxazinoids. Front. Physiol. 11:604754. doi: 10.3389/fphys.2020.604754
Kimbrell, D. A., Hice, C., Bolduc, C., Kleinhesselink, K., and Beckingham, K. (2002). The Dorothy enhancer has tinman binding sites and drives hopscotch-induced tumor formation. Genesis 34, 23–28. doi: 10.1002/gene.10134
Königer, A., and Grath, S. (2018). Transcriptome analysis reveals candidate genes for cold tolerance in Drosophila ananassae. Genes 9:624. doi: 10.3390/genes9120624
Kramer, K. J., and Hopkins, T. L. (1987). Tyrosine metabolism for insect cuticle tanning. Arch. Insect Biochem. Physiol. 6, 279–301. doi: 10.1002/arch.940060406
Kriventseva, E. V., Kuznetsov, D., Tegenfeldt, F., Manni, M., Dias, R., Simão, F. A., et al. (2019). OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47, D807–D811. doi: 10.1093/nar/gky1053
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Leader, D. P., Krause, S. A., Pandit, A., Davies, S. A., and Dow, J. A. T. (2018). FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46, D809–D815. doi: 10.1093/nar/gkx976
Lee, S.-W., Shono, T., Tashiro, S., and Ohta, K. (2005). Metabolism of pyraclofos in housefly, Musca domestica. J. Asia Pac. Entomol. 8, 387–392. doi: 10.1016/S1226-8615(08)60261-7
Li, X., Zhu, B., Gao, X., and Liang, P. (2017). Over-expression of UDP–glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 73, 1402–1409. doi: 10.1002/ps.4469
Luque, T., and O’Reilly, D. R. (2002). Functional and phylogenetic analyses of a putative Drosophila melanogaster UDP-glycosyltransferase gene. Insect Biochem. Mol. Biol. 32, 1597–1604. doi: 10.1016/S0965-1748(02)00080-2
Macdonald, S. J., and Highfill, C. A. (2020). A naturally-occurring 22-bp coding deletion in Ugt86Dd reduces nicotine resistance in Drosophila melanogaster. BMC Res. Notes 13:188. doi: 10.1186/s13104-020-05035-z
Mackenzie, P. I., Bock, K. W., Burchell, B., Guillemette, C., Ikushiro, S., Iyanagi, T., et al. (2005). Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics 15, 677–685. doi: 10.1097/01.fpc.0000173483.13689.56
Mackenzie, P. I., Owens, I. S., Burchell, B., Bock, K. W., Bairoch, A., Belanger, A., et al. (1997). The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7, 255–269. doi: 10.1097/00008571-199708000-00001
Marriage, T. N., King, E. G., Long, A. D., and Macdonald, S. J. (2014). Fine-mapping nicotine resistance loci in Drosophila using a multiparent advanced generation inter-cross population. Genetics 198, 45–57. doi: 10.1534/genetics.114.162107
Meech, R., Hu, D. G., McKinnon, R. A., Mubarokah, S. N., Haines, A. Z., Nair, P. C., et al. (2019). The UDP-glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms. Physiol. Rev. 99, 1153–1222. doi: 10.1152/physrev.00058.2017
Meech, R., Miners, J. O., Lewis, B. C., and MacKenzie, P. I. (2012). The glycosidation of xenobiotics and endogenous compounds: versatility and redundancy in the UDP glycosyltransferase superfamily. Pharmacol. Ther. 134, 200–218. doi: 10.1016/j.pharmthera.2012.01.009
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Myers, C. M., and Smith, J. N. (1954). Comparative detoxication. 2. Glucoside formation from phenols in locusts. Biochem. J. 56, 498–503. doi: 10.1042/bj0560498
Nagare, M., Ayachit, M., Agnihotri, A., Schwab, W., and Joshi, R. (2020). Glycosyltransferases: the multifaceted enzymatic regulator in insects. Insect Mol. Biol. doi: 10.1111/imb.12686
Okubo, S. (1958). Occurrence of N-acetylhydroxytryamine glucoside in Drosophila melanogaster. Med. J. Osaka Univ. 9, 321–331.
Pan, Y., Wen, S., Chen, X., Gao, X., Zeng, X., Liu, X., et al. (2020). UDP-glycosyltransferases contribute to spirotetramat resistance in Aphis gossypii Glover. Pestic. Biochem. Physiol. 166:104565. doi: 10.1016/j.pestbp.2020.104565
Rausell, C., Llorca, J., and Dolores Real, M. (1997). Separation by FPLC chromatofocusing of UDP-glucosyltransferases from three developmental stages of Drosophila melanogaster. Arch. Insect Biochem. Physiol. 34, 347–358. doi: 10.1002/(SICI)1520-6327199734:3<347::AID-ARCH8<3.0.CO;2-R
Real, M. D., and Ferré, J. (1989). Distribution of xanthurenic acid glucoside in species of the genus Drosophila. Insect Biochem. 19, 111–115. doi: 10.1016/0020-1790(89)90080-2
Real, M. D., and Ferré, J. (1990). Biosynthesis of xanthurenic acid 8-O-β-D-glucoside in Drosophila. Characterization of the xanthurenic acid:UDP-glucosyltransferase activity. J. Biol. Chem. 265, 7407–7412. doi: 10.1016/S0021-9258(19)39128-8
Real, M. D., Ferré, J., and Chapa, F. J. (1991). UDP-glucosyltransferase activity toward exogenous substrates in Drosophila melanogaster. Anal. Biochem. 194, 349–352. doi: 10.1016/0003-2697(91)90239-P
Rodriguez, A., Zhou, Z., Tang, M. L., Meller, S., Chen, J., Bellen, H., et al. (1996). Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics 143, 929–940.
Seetharam, A. S., and Stuart, G. W. (2013). Whole genome phylogeny for 21 Drosophila species using predicted 2b-RAD fragments. PeerJ 1:e226. doi: 10.7717/peerj.226
Smith, J. N. (1955). Detoxification mechanisms in insects. Biol. Rev. 30, 455–475. doi: 10.1111/j.1469-185X.1955.tb01548.x
Snoeck, S., Pavlidi, N., Pipini, D., Vontas, J., Dermauw, W., and Van Leeuwen, T. (2019). Substrate specificity and promiscuity of horizontally transferred UDP-glycosyltransferases in the generalist herbivore Tetranychus urticae. Insect Biochem. Mol. Biol. 109, 116–127. doi: 10.1016/j.ibmb.2019.04.010
Thurmond, J., Goodman, J. L., Strelets, V. B., Attrill, H., Gramates, L. S., Marygold, S. J., et al. (2019). FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759–D765. doi: 10.1093/nar/gky1003
Wang, Q., Hasan, G., and Pikielny, C. W. (1999). Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 274, 10309–10315. doi: 10.1074/jbc.274.15.10309
Whiteman, N. K., Gloss, A. D., Sackton, T. B., Groen, S. C., Humphrey, P. T., Lapoint, R. T., et al. (2012). Genes involved in the evolution of herbivory by a leaf-mining, Drosophilid fly. Genome Biol. Evol. 4, 900–916. doi: 10.1093/gbe/evs063
Whiteman, N. K., Groen, S. C., Chevasco, D., Bear, A., Beckwith, N., Gregory, T. R., et al. (2011). Mining the plant–herbivore interface with a leafmining Drosophila of Arabidopsis. Mol. Ecol. 20, 995–1014. doi: 10.1111/j.1365-294X.2010.04901.x
Wiesen, B., Krug, E., Fiedler, K., Wray, V., and Proksch, P. (1994). Sequestration of host-plant-derived flavonoids by Lycaenid butterfly Polyommatus icarus. J. Chem. Ecol. 20, 2523–2538. doi: 10.1016/s0305-1978(01)00036-9
Wilkinson, C. F. (1986). “Xenobiotic conjugation in insects,” in Xenobiotic Conjugation Chemistry ACS Symposium Series, eds G. D. Paulson, J. Caldwell, D. H. Hutson, and J. J. Menn (Washington, DC: American Chemical Society), 3–48. doi: 10.1021/bk-1986-0299.ch003
Younus, F., Chertemps, T., Pearce, S. L., Pandey, G., Bozzolan, F., Coppin, C. W., et al. (2014). Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 53, 30–43. doi: 10.1016/j.ibmb.2014.07.003
Zhou, Y., Fu, W. B., Si, F. L., Yan, Z. T., Zhang, Y. J., He, Q. Y., et al. (2019). UDP-glycosyltransferase genes and their association and mutations associated with pyrethroid resistance in Anopheles sinensis (Diptera: Culicidae). Malar. J. 18:62. doi: 10.1186/s12936-019-2705-2
Keywords: Drosophila melanogaster, UDP-glycosyltransferase, UGT, nomenclature, detoxification, conjugation
Citation: Ahn S-J and Marygold SJ (2021) The UDP-Glycosyltransferase Family in Drosophila melanogaster: Nomenclature Update, Gene Expression and Phylogenetic Analysis. Front. Physiol. 12:648481. doi: 10.3389/fphys.2021.648481
Received: 31 December 2020; Accepted: 22 February 2021;
Published: 17 March 2021.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Markus Friedrich, Wayne State University, United StatesCopyright © 2021 Ahn and Marygold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seung-Joon Ahn, c2V1bmdqb29uLmFobkBtc3N0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.