
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 01 April 2021
Sec. Integrative Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.624950
This article is part of the Research TopicThe Integrative Physiology of Metabolic DownstatesView all 10 articles
Tissue-resident stem cells may enter a dormant state, also known as quiescence, which allows them to withstand metabolic stress and unfavorable conditions. Similarly, hibernating mammals can also enter a state of dormancy used to evade hostile circumstances, such as food shortage and low ambient temperatures. In hibernation, the dormant state of the individual and its cells is commonly known as torpor, and is characterized by metabolic suppression in individual cells. Given that both conditions represent cell survival strategies, we here compare the molecular aspects of cellular quiescence, particularly of well-studied hematopoietic stem cells, and torpor at the cellular level. Critical processes of dormancy are reviewed, including the suppression of the cell cycle, changes in metabolic characteristics, and cellular mechanisms of dealing with damage. Key factors shared by hematopoietic stem cell quiescence and torpor include a reversible activation of factors inhibiting the cell cycle, a shift in metabolism from glucose to fatty acid oxidation, downregulation of mitochondrial activity, key changes in hypoxia-inducible factor one alpha (HIF-1α), mTOR, reversible protein phosphorylation and autophagy, and increased radiation resistance. This similarity is remarkable in view of the difference in cell populations, as stem cell quiescence regards proliferating cells, while torpor mainly involves terminally differentiated cells. A future perspective is provided how to advance our understanding of the crucial pathways that allow stem cells and hibernating animals to engage in their ‘great slumbers.’
The difference between life and death of individual cells or animals depends on their ability to survive, particularly during periods of scarcity. When environmental conditions are unfavorable, or nutrients are scarce, individual cells may enter a dormant state (quiescence). Similarly, some mammals may hibernate to cope with such conditions by suppressing metabolism in a state called torpor. Given that they represent cell survival strategies triggered by external factors, cell quiescence and hibernation both deploy molecular adaptations to survive environmental stress such as low temperature and shortage of nutrients, and even increase their resistance to withstand periods of low oxygen supply. In both cases, phenotypic plasticity is of paramount importance to ensure survival, yet it is undocumented whether the mechanism governing entry or exit from cellular dormancy and torpidity are similar. Here, we summarize mechanisms used in cellular quiescence and mammalian hibernation and use the collective findings to establish their resemblance.
Cellular dormancy is the ability to enter a quiescence state (reversible cellular arrest) by withdrawing from the cell cycle and entering the so called G0 phase (Nakamura-Ishizu et al., 2014). The cell cycle is divided into four phases: G1 phase (interphase), S phase (DNA synthesis), G2 phase (interphase), and M phase (mitosis) (Yang and Sheridan, 2014). Cells that overcome the G1 checkpoint commit to divide and proceed to the S phase, culminating in cell division. In the early G1 phase, cells that are non-proliferating, undivided, senescent (permanent cell cycle arrest) or terminally differentiated, can withdraw from the cell cycle and enter a dormant or quiescent state (G0) (Figel and Fenstermaker, 2018). Quiescent cells are characterized by low mobility, low metabolic activity and rare division (Rocheteau et al., 2014). Once quiescent, cells may either re-enter the G1 phase in response to growth signals and commit to divide (reversible quiescence) or continue dormancy, which may or may not ultimately lead to senescence, i.e., a state of permanent cell cycle arrest with high metabolic activity and secretion of inflammatory factors (Sabbatinelli et al., 2019). In contrast, non-proliferating cells that are either terminally differentiated or senescent are irreversibly arrested.
In humans, reversible quiescence commonly occurs in many somatic cells including hematopoietic stem cells (Nakamura-Ishizu et al., 2014), muscle stem cells (Chakkalakal et al., 2012), epithelial stem cell (Coloff et al., 2016), neural stem cell (Kalamakis et al., 2019), and hair follicle stem cell (Wang et al., 2016). Although quiescence is a key characteristic of tissue-resident stem cells, which function as a dormant reserve to replenish the tissue loss throughout life, the discovery of highly proliferative stem cells in several tissues has challenged the concept that quiescence is an integral property of all stem cells (Clevers and Watt, 2018). Given the divergent mechanisms governing quiescence in different stem cells, this review will focus on the most extensively and well-characterized tissue-resident stem cells: hematopoietic stem cells (HSCs) (Richmond et al., 2016). Nevertheless, quiescence can also be found in non-stem cells such as endothelial cells (Sabbatinelli et al., 2019) and mature hepatocytes, the latter being considered long-term quiescent cells essential for liver regeneration (Zimmermann, 2004; Berasain and Avila, 2015). Although quiescence is not an inherent property that characterizes stem cells or distinguishes them from non-stem cells (e.g., consider mature hepatocytes), dysregulation and loss of quiescence affects homeostasis of many progenitor cell populations, ultimately leading to stem cell exhaustion, i.e., the depletion of stem cells with impact on health (Orford and Scadden, 2008). Stem cell exhaustion is particularly noticeable in HSCs, due to their multi-lineage capacity of differentiation and self-renewal potential (Pietras et al., 2011). HSCs give rise to progenitor cells that differentiate into all lineages of mature blood cells. However, continuous self-renewal of HSCs is insufficient for lifelong maintenance, as the inevitable accumulation of damage would result in dysfunctional hematopoiesis, leading to diseases such as leukemia (Riether et al., 2015). Hence quiescence is considered an essential feature to prevent HSCs exhaustion. To avoid this potential hazard, HSCs are kept quiescent in a unique microenvironment in the bone marrow. Quiescence is actively maintained in HSCs, in which the microenvironment plays a crucial role to assure their longevity and function. Furthermore, computational modeling of HSCs kinetics infers that human HSCs complete the cell cycle once every 18 years to self-renew and generate progenitor cells. Quiescence thus allows stem cells to prolong their lifespan to maintain critical physiological functions (Hao et al., 2016).
Dormancy in animals can be subdivided into four subclasses: hibernation (Lee, 2009), diapause (Renfree and Shaw, 2000), estivation (Masaki, 2009; Storey and Storey, 2012), and brumation (McEachern et al., 2015). Hibernation is often described as winter dormancy and is adopted by both warm and cold-blooded vertebrates. Furthermore, hibernation is characterized by alternating periods of low metabolic activity (torpor), and normalization of metabolism and physiology (arousals) (Carey et al., 2003). Diapause refers to a spontaneous interruption of the development, characterized by a reduction of metabolic activity and is mainly observed in insects and a few mammalian species (Renfree and Shaw, 2000; Denlinger, 2002). Estivation occurs in vertebrates and invertebrates and is characterized by reduced metabolic rate and inactivity to avoid desiccation during hot periods with soaring temperatures (Masaki, 2009; Storey and Storey, 2012). Brumation is mostly seen in reptiles and strictly induced by low ambient temperatures. It is characterized by long periods of inactivity with lowered respiration rate, intersected by brief periods of activity required to drink (McEachern et al., 2015). Despite having evolved different forms of dormancy, the end goal of these animals is the same: survival of periods with low energy supply.
Between these forms of animal dormancy, hibernation is the best explored regarding molecular changes, which is why we will almost exclusively focus on hibernation in mammals. Depending on the species and environmental challenges hibernation takes different forms, mostly consisting of seasonal hibernation with multi-day torpor bouts and brief arousals (12–18 h) versus daily torpor during which metabolism is suppressed for 6–10 h (Turbill et al., 2011). Likely, similar molecular mechanisms govern both, as some species switch between multi-day and daily torpor (Wilz and Heldmaier, 2000; Dausmann et al., 2004). Hibernation consists of (daily) torpor phases that are characterized by low metabolism, which are interspersed by brief periods of arousal with restoration of metabolic rate to levels of non-hibernating animals. Torpidity is a state in which physical activity, development, growth and metabolism are transitorily and profoundly reduced in response to harsh environmental conditions and to reduce energy dissipation (Heldmaier et al., 2000). In this review, we discriminate between the two different forms of torpidity by using ‘torpor’ and ‘daily torpor.’
During torpor, animals undergo profound physiological, morphological and behavioral changes. For example, body temperature of seasonal hibernators in cold environments sharply declines to as low as 0–4°C, heart rate and respiration decreases by 95%, and renal function is significantly reduced (Carey et al., 2003). Most hibernators synchronize their dormancy with environmental changes, with some animals entering dormancy only after the start of unfavorable conditions (consequential dormancy, such as facultative hibernation in the Syrian hamster), while others have a yearly rhythmicity allowing them to enter in advance of harsh conditions (predictive dormancy, such as seasonal hibernation in ground squirrels) (Lee, 2009; Masaki, 2009; Ruf and Geiser, 2015).
Cellular quiescence in HSC is associated with three key changes in cell physiology: (i) cell cycle arrest by inhibition of cyclin-dependent kinases (CDKs) upon an increase in expression of cyclin-dependent kinase inhibitors (CKIs), (ii) lowering of metabolism with a switch from carbohydrate to lipid-based metabolism and (iii) resistance to accumulating cellular damage conferred by differential expression of genes involved in apoptosis, proliferation and oxidative stress.
The exact signals that induce torpidity in mammals are still not known, yet reduction of metabolic rate is at its heart (Heldmaier et al., 2000; Storey and Storey, 2004). Torpor entry is achieved by active suppression of metabolism and by limiting ATP-expensive activities, ultimately leading to a change of physiology in cells, tissues and organs. In torpor, vital functions including respiratory and heart rate strongly decline secondary to the metabolic suppression, while temperature regulation is adjusted to accommodate lower body temperatures (Tsiouris, 2005; Ruf and Geiser, 2015). Moreover, reversible cell cycle arrest has been reported in hypoxic red-eared slider turtles and torpid 13-lined squirrels (Biggar and Storey, 2009; Wu and Storey, 2012a), in concert with the metabolic suppression and shift in energy source from carbohydrates to fatty acids (Storey et al., 2010).
Because of the similarity between the overall regulation of HSC quiescence and torpidity, it is conceivable that some form of reversible quiescence occurs during hibernation and might even be necessary for the induction of hibernation. The interaction between these mechanisms may set the stage for both reversible cellular quiescence and hibernation allowing them to withstand stress conditions and extend their life span. However, it must be underlined that the impact of mechanisms involved, particularly those regulating cell cycle, differ largely between quiescence in stem cells and torpidity. While in stem cells cell cycle regulation drives their proliferation and self-renewal, it is less clear what its role is in terminally differentiated cells, as studied in torpor.
While numerous intrinsic and extrinsic factors regulate the cell cycle, they generally converge on the cell cycle master regulators, cyclin-dependent kinases, CDKs (Fisher, 1997; Coller et al., 2006; Valcourt et al., 2012; Cheung and Rando, 2013; Lim and Kaldis, 2013). CDK activation requires binding to the proper cyclin regulatory subunit (A, B, D, and E) and together they drive cell cycle progress (Murray, 2004; Garcia and Su, 2008; Figel and Fenstermaker, 2018). Differential gene expression of cyclins during specific cell cycle phases drives cell cycle progression or arrest (Malumbres and Barbacid, 2005; Camins et al., 2013). Cyclin-CDK complex formation is antagonized by CKIs that competitively bind to the catalytic site of the cyclin-CDK complex (Dai and Grant, 2003; Sánchez-Martínez et al., 2015). Two families of CKIs exist, namely INK4 (p16INK4a, p15INK4b, p18INK4c, p19INK4d) and CIP/KIP (p21cip1, p27kip1, p57kip2). The balance between CDKs, cyclins and CKIs determines whether a cell will commit to proliferation or maintains cell cycle arrest. The major factors contributing to regulation of the cell cycle with relevancy to quiescence or torpor are depicted in Figure 1.
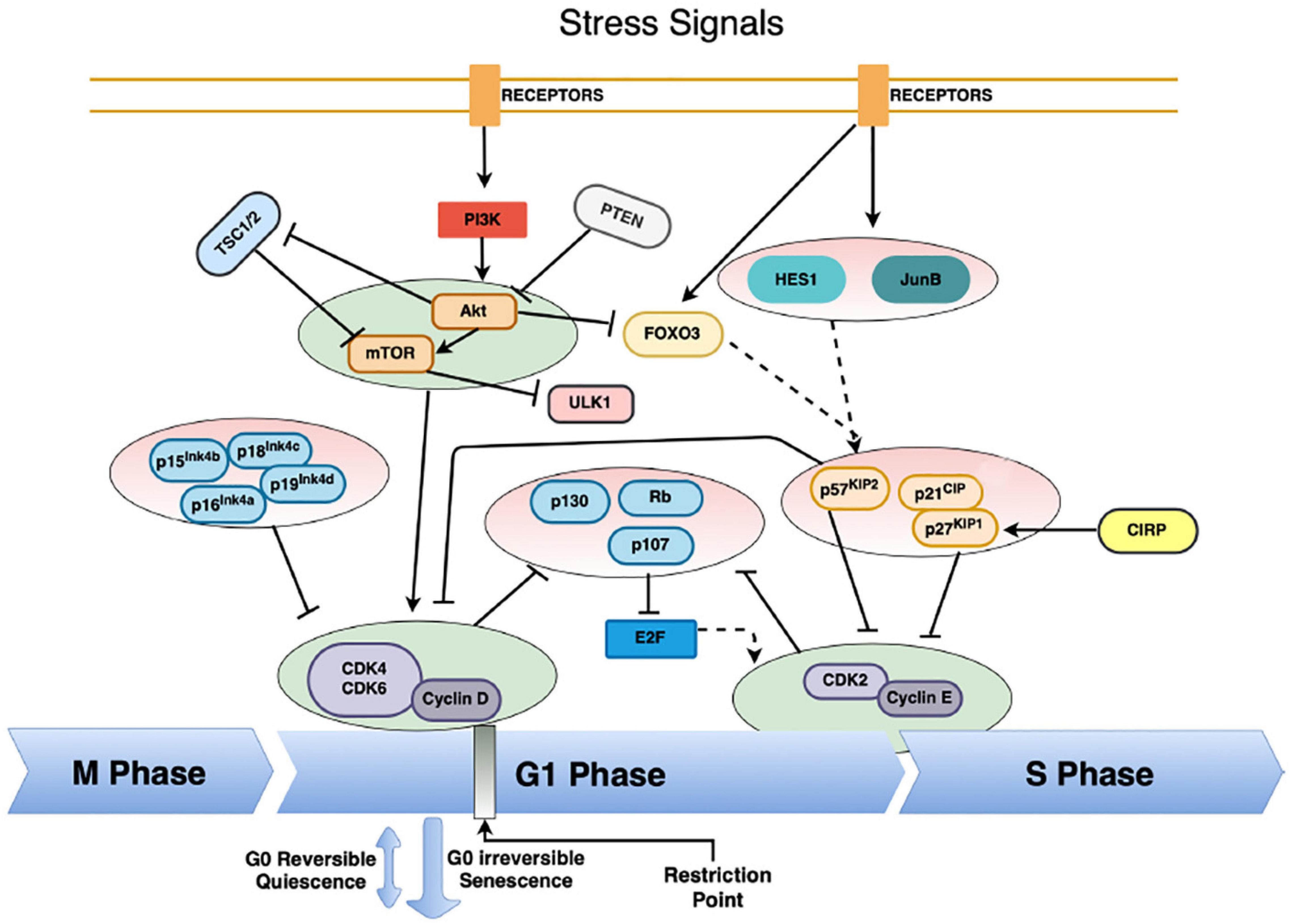
Figure 1. The complex regulation of HSC – and possibly hibernators – cell cycle entry. Cyclin-CDK complexes drive the progression of the cell cycle. Their inhibition forces entry into quiescence from G1 into the G0 phase of the cell, mainly by the action of cyclin-dependent kinase inhibitors (CKI) from the INK and CIP/KIP family. Various pathways converge on the CKIs, thus instituting quiescence. Low activity of the PI3K/Akt/mTOR pathway is vital to enter quiescence, mainly through the inhibition of cyclin-D-CDK4/6 complexes and suppression of the Rb-E2F pathway. This pathway is massively regulated, primarily through inhibition by the tumor suppressors phosphatase and tensin homolog (PTEN) and tuberous sclerosis proteins 1 and 2 (TSC1/2). Furthermore, the activity of the cyclin D-CDK4/6 complexes is inhibited both by the Ink4 and CIP/KIP CKIs. The Cyclin E-CDK2 complex regulates progression from the G1 to the S phase, and it is mainly regulated by the CIP/KIP family of CKIs, as well as by tumor suppressors from the retinoblastoma protein family (Rb, p107, p130). Extrinsic signals activate the transcription factors HES1, JunB and FOXO3a expression, which in turn regulate the transcription of the CIP/KIP family members. Moreover, entry into cellular quiescence is also subject to regulation via the tumor suppressor p53, as a response to cellular damage. Solid arrows indicate direct activation pathways, solid T-lines indicate inhibitory pathways and dashed arrows indicate transcriptional regulation activity.
Mitogen-induced signaling pathways tightly regulate cell growth and proliferation. In the presence of plentiful nutrients, growth factors activate transmembrane receptors, eliciting downstream signaling cascades, including the rat sarcoma oncogene (RAS) (Beauséjour et al., 2003), myelocytomatosis (Myc) (Rahl et al., 2010) and the serine/threonine-protein kinase B (PI3K/Akt) pathways. This is followed by sequential activation of the mitogen-activated protein kinases (MAPKs), which induce the transcription of cyclin D that binds to cyclin-dependent kinases (CDK) 4 and 6, forming activated complexes that initiate the downstream phosphorylation of DNA synthesis associated proteins (Seger and Krebs, 1995; Pimienta and Pascual, 2007; Risal et al., 2016; Seger and Wexler, 2016). The cyclin-D-CDK(4,6) complex phosphorylates and inactivates the tumor suppressor retinoblastoma protein (Rb) and its homologs p107 and p130. Rb inhibition releases its inhibiting of the E2F transcription factor, thus activating E2F-binding to DNA promoter regions (Infante et al., 2008; Kent and Leone, 2019), allowing the transcription of E2F-dependent genes such as cyclin A and cyclin E. These cyclins form a complex with CDK-2, activating its kinase activity. Cyclin(A,E)-CDK-2 complexes further phosphorylate Rb resulting in its complete inactivation (Dyson, 1998; Harbour and Dean, 2000; Bracken et al., 2004). The increase in cyclins and the activated cyclin(D,E)-CDKs(2/4/6) complexes are essential to drive cells from G1 to the S phase and commit the cell to proliferation.
On the other hand, the absence of growth factors reduces the activity of the RAS (MAPKs) and PI3K/Akt pathways, thus leading to the activation of the glycogen synthase kinase 3 beta (GSK3β), which halts the cell cycle by phosphorylation and subsequent degradation of cyclin-D (Dong et al., 2005; Theeuwes et al., 2017; Tessier et al., 2019). Degradation of cyclin D reduces cyclin(D)-CDK(4,6) complex formation followed by an increase in activated Rb, leading to strong suppression of the E2F transcription factor and downstream genes. Cyclin D thus comprises a rate-limiting factor of cell cycle progression through G1 (Trimarchi and Lees, 2002; Malumbres and Barbacid, 2009). Quiescent cells display low levels of activators of the cyclin-CDK-Rb-E2F pathway, such as cyclin D, CDK 2, 4, and 6, and high levels of the pathway repressors, including Rb protein and family homologs (p107 and p130) (Harbour and Dean, 2000; Bracken et al., 2004; Peng et al., 2015) and the CKIs p21 (Cheng et al., 2000) and p27 (Coats et al., 1996). However, when conditions turn favorable again, quiescent HSCs increase Cyclin D and E abundance, thus outcompeting CKIs and activating CDK2/4/6 (Murray, 2004; Choi and Anders, 2014; Figel and Fenstermaker, 2018).
Hematopoietic stem cells quiescence primarily results from cell cycle arrest through inhibition of CDKs by an increase in the abundance of CKIs. When conditions for HSCs survival are unfavorable, intrinsic and extrinsic signals upregulate the expression of CKIs, modulating the formation of cyclin-CDK complexes and allowing the formation of the Rb-E2F complex, thus effectively halting the cell cycle. Meanwhile, quiescent HSCs ensure reversibility by upregulation of the chromatin remodeler helix-loop-helix protein 1 (HES1), which promotes transcriptional repression through alteration of chromatin recruiting histone deacetylases (HDACs) (Sang and Coller, 2009), promoting tight packaging of DNA into heterochromatin and downregulation of p21 (Yu et al., 2006; Sang et al., 2008), p27 (Murata et al., 2005), and E2F-dependent proteins (Hartman et al., 2004). Quiescent HSCs deploy additional mechanisms to protect DNA integrity by raising defense mechanisms against excessive oxidative stress to protect cells from accumulating damage and apoptosis. Low levels of reactive oxygen species (ROS) are tolerated by an increased antioxidant defense, including the NADPH-dependent glutathione reductase system (Hosokawa et al., 2006; Jang and Sharkis, 2007), FOXO3a (Miyamoto et al., 2008; Rimmelé et al., 2015), and Sirtuin1 (Ezoe et al., 2008; Matsui et al., 2012). Further, DNA repair systems are enhanced in HSCs, including non-homologous end joining (NHEJ) and p53-mediated DNA damage response (Maynard et al., 2008; Mohrin et al., 2010; Dannenmann et al., 2015). In addition, increased expression of p53 serves to further enhance quiescence in HSCs by upregulation of downstream genes, including p21, Necdin, Gfi-1, BTG2, BAX, and PUMA (Lacorazza et al., 2006; Liu et al., 2009; Asai et al., 2012).
The ability to recommence the cell cycle following reversible arrest is crucial to the functionality of quiescent cells. Upon sufficient extrinsic growth stimulation, MAPKs activate the transcription factor Myc, which promotes the transcription of several cell-cycle promoting genes, including cyclin D, cyclin E, E2F2, and CDK4. Moreover, evidence suggests that the upregulation of Myc (Eilers et al., 1991; Kretzner et al., 1992), E2F (Johnson et al., 1993; Kowalik et al., 1995) and Cyclin E (Blomen and Boonstra, 2007; Fox et al., 2011) alone can drive a cell out of quiescence into cell cycle progression. Nevertheless, downregulation of CKIs activity, p21 (Cheng et al., 2000; Kippin et al., 2005) and p27 (Coats et al., 1996; Rivard et al., 1996), and downregulation of all three retinoblastoma protein family members (Rb, p107, p130) result in quiescence exit and cellular proliferation (Dannenberg et al., 2000; Sage et al., 2003; Viatour et al., 2008). Moreover, the upregulation of cell cycle progression genes leads to a shift in energy metabolism from lipid based oxidation (FAO) back to carbohydrate oxidation (glycolysis). This switch is essential to supply the demand of the ATP-expensive processes to meet the energy demand during proliferation (Valcourt et al., 2012; Takubo et al., 2013). It is of note that quiescent cells may resume cell proliferation only if they express a specific group of genes. One of the genes essential to the reversibility of quiescence is HES1. Hes1 is upregulated in quiescent cells and prevents premature senescence or terminal differentiation in response to specific signals (hypoxia, wnt signaling, Hedgehog pathways) (Baek et al., 2006; Sang et al., 2008). Although the exact mechanism by which Hes1 governs cell quiescence is still unknown, Hes1 can bind to the DNA enhancer site of the CKIs p21, p27, and p57 in HSCs (Yu et al., 2006), expectedly resulting in their inhibition of cyclins, thus arresting the cell cycle.
The majority of the pathways involved in stem cell quiescence have also been implicated in hibernators, as torpor phases feature the molecular signature of cell cycle arrest and a reduction of energy-consuming processes such as transcription and translation (Biggar and Storey, 2009; Ruf et al., 2012; Wu and Storey, 2012a; Schwartz et al., 2013; Blanco and Zehr, 2015; Al-attar and Storey, 2020). Modulation of key players in cell cycle arrest due to low nutrient levels is mainly effectuated through the reduction in cellular ATP-consuming processes. A low [ATP]:[AMP] ratio activates the energy-sensing AMP-activated protein kinase (AMPK), initiating a signaling cascade that minimizes energy expenditure. Many genes involved in the cell cycle arrest are upregulated by AMPK dependent phosphorylation, including the transcription factors Hes1, JunB, and FOXO3a. In turn, they induce transcription of CIP/KIP CKIs that strongly inhibit cyclin D and E, which in proliferating cells halts the cell cycle and initiates G0 cell arrest (Coller et al., 2006; Li and Bhatia, 2011; Valcourt et al., 2012). In line with these molecular changes, the long term torpor molecular profile matches suppression of cell cycle progression in the liver of thirteen-lined ground squirrel (Ictidomys tridecemlineatus) reflected in upregulation of CKIs (p15INK4b and p21CIP1) and downregulation of cyclins (D and E) and CDKs2/4/6 (Andrews, 2007). Moreover, the highly proliferative intestinal epithelial cells of thirteen-lined ground squirrel and Caenorhabditis elegans embryos (Nystul et al., 2003) halt their mitotic activity during deep torpor because of arrest in the G2 phase of the cell cycle (Matthews and Fisher, 1968; Kruman et al., 1988; Kruman, 1992). Yet, similar changes have been observed in terminally differentiated skeletal muscle cells of Brand’s bat (Myotis brandtii) (Wu and Storey, 2016) and brown adipose tissue of arctic ground squirrel (Urocitellus parryii) (Zhu et al., 2005; Yan et al., 2008). Furthermore, cellular stress such as low temperature, UV radiation and hypoxia, upregulates the expression of the cold-inducible RNA-binding protein (CIRP), which induces the translation of the CKI, p27KIP1. High activity of CIRP has been reported both in HSCs and during torpor in brain, which suggests that this protein plays a crucial role in inducing cell cycle arrest and in inhibiting proliferation of HSCs at low temperatures (Zhu et al., 2016; Roilo et al., 2018).
Changes observed in torpor are basically reversed during interbout arousals. While HES1 constitutes an essential factor to leave cell quiescence in HSC, there is currently limited information about HES1 activity in hibernators, warranting further studies to understand its role in hibernation-induced cellular dormancy.
With their survival depending on an inherent mechanism to maintain adequate metabolic function even during unfavorable environmental conditions, quiescent cells and torpid hibernators undergo a series of interconnected adjustments in metabolism. Key changes in relevant metabolic processes are depicted in Figure 2.
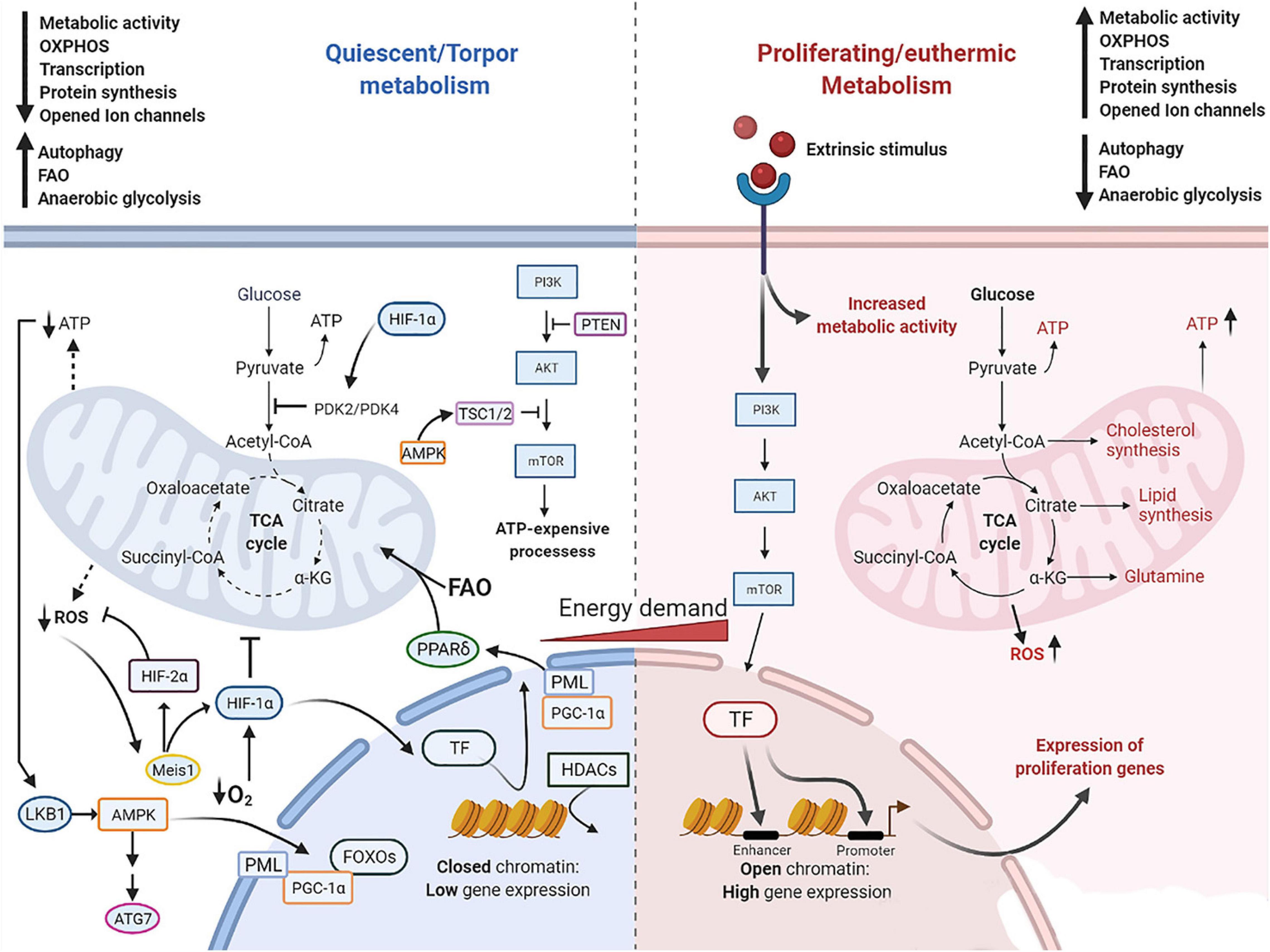
Figure 2. Molecular mechanism governing cell quiescence and torpidity in contrast with cell proliferation and euthermia. Quiescent and torpid cells feature a metabolic shift from glucose fueled OXPHOS to FAO. Low nutrient supply leads to depletion of ATP which is detected by LKB1 that in turn activates AMPK, eliciting a downstream cascade including the activation of TSC1/2 that suppresses PI3K/Akt pathway and downstream ATP-expensive processes. AMPK further shuts down gene expression via activation of HDACs which tightly pack histone-DNA complexes into heterochromatin. Also, AMPK promotes autophagy by activating ULK1/ATG7, lipolysis and fatty acid oxidation via activation of OXPHOS regulator FOXOs proteins and PGC-1α. In addition, PGC-1α, a transcriptional coregulator of PPAR target genes, promotes the upregulation of PPARδ. The PML-PPARδ-FAO further promotes β-oxidation of lipids and the metabolic shift toward FAO. HIF-1α promotes FAO via upregulation of PDK4 and inhibition of the first step in the TCA cycle, and promotes resistance to hypoxia stress and ROS damage. HIF-1α can be activated by its upstream regulator the Meis1 protein, which is activated by the elevation of intracellular ROS, and by low levels of O2. Dotted lines represent a restricted generation of molecules pointed at. Solid lines represent activated pathways. T lines indicate the suppression of the pathway. AMPK, AMP-activated protein kinase; ATG7, autophagy related 7; FOXOs, forkhead proteins; HDACs, histone deacetylases; HIF-1α, hypoxia-inducible factor 1α; HIF-2α, hypoxia-inducible factor 2α; LKB1, liver kinase B1; OXPHOS, oxidative phosphorylation; PDK4, pyruvate dehydrogenase kinases 4; PGC-1a, peroxisome proliferator-activated receptor-coactivator 1α; PML, promyelocyte leukemia protein; PML, promyelocyte leukemia protein; PPARs, peroxisome proliferator-activated receptors; PTEN, phosphatase and tensin homolog; TCA, tricarboxylic acid cycle; TF, transcription factors; TSC1/2, tuberous sclerosis proteins 1 and 2.
One of the remarkable adjustments is the phenotypic switch from glucose fueled mitochondrial OXPHOS to FAO, which involves the differential expression of genes and changes in metabolic processes in response to both intrinsic and extrinsic signals (Lee, 2009; Yao, 2014). Under normal physiological conditions, cellular proliferation requires high levels of energy in the form of ATP, fueled mainly by glucose through mitochondrial OXPHOS of the end product of glycolysis, pyruvate (Yang and Sheridan, 2014). Also, mitochondrial activity, especially oxidative phosphorylation, is the main cellular ROS generator, which can damage macromolecules and organelles, including DNA, proteins and mitochondria (Ito and Ito, 2016). Quiescent cells thus switch from glucose to FAO (β-oxidation), anaerobic glycolysis and autophagy, to secure a minimal energy supply, thereby protecting them from metabolic dysregulation and DNA-damaging ROS production (Valcourt et al., 2012; Ito and Suda, 2014; Yao, 2014). Hypoxia-inducible factor 1α (HIF-1α) is critically involved in this metabolic switch. HIF-1α is a transcription factor expressed in mammalian cells residing under hypoxic conditions, which activates transcription factors and confers post-translational modifications that lower oxygen consumption. HIF-1α promotes anaerobic glycolysis by upregulating glycolytic genes and repressing glucose fueled OXPHOS through transcription of pyruvate dehydrogenase kinases PDK2 and PDK4 (Takubo et al., 2010, 2013). In turn, these kinases inhibit pyruvate dehydrogenase (PDH) (Zhang et al., 2014), which is an essential enzyme to convert glucose derived pyruvate to acetyl-CoA. Inhibition of PDH lowers carbohydrate use by limiting the flow of glycolysis products toward the citric acid cycle and promoting β-oxidation of ketones and fatty acids. At the same time, the lipolytic protein triacylglycerol lipase (PTL), which stimulates lipolysis by breaking down adiposomes (fat droplets), liberates fatty acids for FAO. As HSCs reside in a hypoxic niche in the bone marrow, HIF-1α actively maintains the quiescent state relying on glycolysis and β-oxidation to support low levels of ATP generation. Consequently, inhibition of HIF-1α led to the depletion of mice HSCs, while HIF-1α overexpression induced their quiescence (Takubo et al., 2010). Simsek et al. (2010) showed that HIF-1α expression in HSCs is controlled by the DNA-binding transcription factor myeloid ecotropic viral integration site 1 homolog (Meis1). Further, HSCs upregulate PTL to secure the supply of fatty acids (Liu et al., 2018).
In concert with the metabolic rewiring, adaptations also constitute mechanisms that repress energy-consuming processes by inhibiting gene expression related to cell proliferation, anabolic processes and oxidative phosphorylation through the action of HDACs (Sang et al., 2010). Quiescent cells rapidly reduce their energy expenditure by downregulating ATP-demanding processes, such as DNA replication, macromolecular synthesis, macromolecular turnover and ion pumping activities (Cheung and Rando, 2013; Yao, 2014), which is a requirement for HSC quiescence. Signer et al. showed that chemical induction of protein synthesis leads to the loss of quiescence and promotion of proliferation of mice HCSs (Signer et al., 2014). The switch in energy metabolism back from FAO to glucose fueled OXPHOS is essential to meet the energy demand for differentiation. Indeed, inhibition of mitochondrial respiration blocks differentiation of HSCs (Yu et al., 2013), while inhibition of FAO led to cell proliferation (Ito et al., 2012).
Hypoxia also induces expression of the RNA binding motif protein 3 (RBM3), a critical translation facilitator (Wellmann et al., 2010; Zhu et al., 2019). Loss of RBM3 results in increased damage, mitotic dysfunction and apoptosis (Sureban et al., 2008), reduced neuronal structural plasticity (Peretti et al., 2015) and has been postulated to increase the translational efficacy in HSCs under hypoxic conditions. Other genes that are upregulated in quiescent cells may enhance resistance to apoptosis (NFKB2, MET) (Lin and Karin, 2003; Huh et al., 2004), suppress proliferation (MXI1, TP53, FAT) (Schreiber-Agus et al., 1998; Coller et al., 2006) and protect against accumulating oxidative damage (FOXO, HIF-1α, LKB1, SOD3, PRDX4, EPHX1) (Kops et al., 2002; Shen and Nathan, 2002; Serra et al., 2003; Shackelford and Shaw, 2009; Gurumurthy et al., 2010; Takubo et al., 2010).
The mammalian target of rapamycin (mTOR) pathway regulates growth and metabolism and embodies a crucial switch from high-expensive energy state (anabolic) to hypometabolism (catabolic). Upstream of mTOR is the highly conserved phosphatidylinositol-3 kinase/protein kinase B (PI3K/Akt) signaling pathway, which is activated by various signals from activated tyrosine kinase receptors. When activated, mTOR drives cell proliferation, growth and survival by activating cyclin-D-CDK4/6 complexes. The Akt/mTOR pathway is regulated by the tumor suppressors phosphatase and tensin homolog (PTEN) and tuberous sclerosis complex (TSC1/2). PTEN and TSC1/2 regulate reversible protein phosphorylation (RPP) of the mTOR pathway determining the “on” or “off” state of many energy-expensive processes. PTEN is considered a major regulator of metabolic reprogramming and has been shown to regulate PDK1, a critical activator of the insulin pathway. Normally, when carbohydrates and glucose are plentiful, insulin signaling activates the PI3K/Akt pathway, which induces glucose uptake and breakdown via glycolysis mechanisms (Rigano et al., 2017; Jansen et al., 2019). In the absence of nutrients, Akt/mTOR is inactivated by the action of PTEN (Yilmaz et al., 2006) and TSC 1/2 (Chen et al., 2008), reducing the energy-intensive processes and inducing catabolism. In a low nutrient environment, levels of glucose are reduced as well as insulin signaling (Martin, 2008; McCain et al., 2013), which reduces PI3K/Akt signaling, activating the forkhead protein (FOXO) family that lowers ROS production to protect from oxidative damage. Quiescent HSCs show the typical changes related to mTOR suppression, including the upregulation of PTEN (Zhang et al., 2006; Kalaitzidis et al., 2012; Porter et al., 2016). In HSCs, activation of the PI3K/Akt/mTOR pathway, reversing the upregulation of PTEN, occurs when cells migrate toward a more oxygen-rich microenvironment, which induces a switch from FAO to glucose fueled OXPHOS and promotes cellular respiration, in turn increasing the levels of ROS and cell cycle progression.
Mitochondrial respiration and production of ROS are strictly regulated by the activity of FOXOs and HIF-1α. Following their activation by growth-repressive signals in the PI3/Akt/mTOR pathway, FOXO proteins repress a large number of mitochondrial genes, inhibiting not only mitochondrial activity, but also biogenesis. Consistent with HSCs’ low dependency on mitochondrial respiration as a source of energy, HSCs are characterized by a low number of mitochondria, which are immature and display underdeveloped cristae with globular morphology (Piccoli et al., 2005). Because of their low metabolic activity, quiescent stem cells produce low levels of ROS (Eckers et al., 2014), yet deploy unique mechanisms protecting them against DNA-damaging ROS (Wanet et al., 2015). The role of Meis1 in counteracting oxidative stress is well established (Kocabas et al., 2012; Unnisa et al., 2012; Papa et al., 2019), by serving as an upstream regulator in response to high ROS levels through activation of HIF-1α and HIF-2α (Simsek et al., 2010; Kocabas et al., 2012; Simonetti et al., 2016). In addition to Meis1, FOXO proteins are essential regulators of oxidative stress and deemed essential to maintain quiescence in long term HSCs. FOXOs inhibit mitochondrial respiratory chain protein expression and transcriptionally activate antioxidant enzymes such as catalases, sestrins, and superoxide dismutase 2 (SOD2). FOXO proteins, notably FOXO3a but also FOXO1, FOXO4, and FOXO6, inhibit mitochondrial gene expression (Tothova et al., 2007; Rimmelé et al., 2015). FOXO3a is highly expressed in HSCs and is also a main transcriptional regulator of antioxidants enzymes (Yalcin et al., 2010; Liang et al., 2016). Conditional deletion of FOXO 1/2/3/4, especially FOXO3a, results in a reduction of the HSC pool (Tothova et al., 2007; Liang et al., 2016; Bigarella et al., 2017). Apart from regulating mitochondrial ROS production and biogenesis, FOXOs also induce the expression of CKIs of the CIP/KIP family (Kops et al., 2002; Camperio et al., 2012), thereby inducing HSC cell cycle arrest. FOXOs are activated by the tumor suppressor liver kinase B1 kinase (LKB1), which also activates AMPK (Shackelford and Shaw, 2009). LKB1 regulates AMPK activity and downstream promotion of ATP production. A relatively high AMP level is indicative of an energy-depleted state and leads to the activation of LKB1, a master kinase that in turn activates the downstream AMPK and 12 other related kinases (Lizcano et al., 2004; Shackelford and Shaw, 2009). Conditional deletion of LKB1 led to loss of quiescence and increased the number of hematopoietic progenitor cells, while depleting HSCs in mice (Gan et al., 2010; Nakada et al., 2010). In addition, LKB1 is an upstream regulator of the peroxisome proliferator-activated receptor-coactivator 1α (PGC-1α), a central regulator of mitochondrial biogenesis and oxidative metabolism, and the deletion of LKB1 in HSCs led to a downregulation of PGC-1α resulting in mitochondrial dysfunction (Gan et al., 2010). On the other hand, the knockout of PGC-1α in early subsets of HSCs showed that hematopoiesis is minimally affected in these cells. Nevertheless, knockout of PGC-1α lead to susceptibility to oxidative stress and modulation of long-term HSC re-population (Basu et al., 2013).
When HSCs commit to proliferation by moving to the high oxygen osteoblastic niche, there is a rapid increase in mitochondrial biogenesis and activity (Jang and Sharkis, 2007; Chen et al., 2008; Valcourt et al., 2012; Nakamura-Ishizu et al., 2014). Proliferating HSCs face high ROS levels and upregulate p38 mitogen-activated protein kinase (MAPK) and mTOR, compatible with the proliferative and differentiation phenotype (Kopp et al., 2005; Parmar et al., 2007). Recent investigation suggests that FOXO3a regulates mitochondrial biogenesis gene transcription, and loss of FOXO3a leads to a dysfunctional metabolic shift and impaired OXPHOS (Rimmelé et al., 2015; Menon and Ghaffari, 2018). However, the mechanism of this regulation remains unknown.
Inhibition of mTOR increases PPARs signaling in HSCs, which has the function of nutrient sensing and transcriptional control of metabolic pathways (Desvergne and Wahli, 1999), especially fatty acid transport and FAO (Braissant et al., 1996; Takahashi et al., 2007). HSC fate and self-renewal decisions are critically dependent on the PML-PPARδ pathway. Promyelocyte leukemia protein (PML) nuclear bodies maintain quiescence in HSC by activating peroxisome proliferator-activated receptors (PPARs), which in turn reprogram cellular metabolism by suppressing the Akt/mTOR pathway. The deletion of PML led to the loss of HSCs quiescence and subsequent exhaustion. Further investigation showed that PPAR signaling and FAO were significantly reduced during HSCs differentiation, while induction of PPARδ by a PML-targeting compound induced quiescence (Ito et al., 2012). The depletion of PML and inhibition of mitochondrial FAO in HSC resulted in symmetric division (two committed HSC daughter cells, i.e., cells with a progressive differentiation toward a particular type of red or white blood cell) both in vitro and in vivo. Conversely, pharmacological activation of PPARδ increased asymmetric division (one daughter committed, the other for self-renewal) (Ito et al., 2012; Ito and Ito, 2013). Hence, PML and PPARδ activation play an essential role in maintaining the stem cell pool (Lallemand-Breitenbach and de Thé, 2010; Ito and Suda, 2014).
Autophagy is a conserved mechanism by which cytoplasmic proteins and organelles are engulfed within autophagosomes and degraded in lysosomes, providing ATP and metabolites. Autophagy is governed by the activation of AMPK in response to nutritional deprivation, leading to phosphorylation of TSC1/2, which in turn inhibits mTORC1 (Yang and Klionsky, 2010). Low levels of mTORC1 activate Unc-51 like autophagy activating kinase, ULK1 (Kim et al., 2008; Mizushima, 2010), which recruits additional proteins to form a complex that promotes autophagosome formation and autophagy. Autophagy related 7 (Atg7) is consistently upregulated in quiescent cells, and deletion of Atg7 in mice decreased the number of HSCs and progenitors cells of various lineages and increased the number of abnormal mitochondria and ROS levels (Mortensen et al., 2011). Furthermore, a mutation in the autophagy gene Atg12 increased levels of ROS, protein synthesis and glucose fueled OXPHOS in mouse HSCs (Vessoni et al., 2012; Revuelta and Matheu, 2017). These results suggest that disruption of autophagy induces a loss of quiescence and a switch back to the proliferating phenotype in HSCs, consistent with the idea that autophagy influences cell fate decisions through metabolic reprogramming of HSC.
Hibernators have implemented metabolic adaptations very similar to those found in quiescence. Mammalian hibernators also switch from carbohydrate fueled OXPHOS to lipid-based metabolism (FAO) during torpor (Vermillion et al., 2015). This is reflected in the respiratory quotient (RQ: the quotient of CO2 production over O2 use), which reflects carbohydrate (RQ ∼ 1.0) versus lipid oxidation (RQ ∼ 0.7). During torpor, European hedgehog (Erinaceus europaeus) and Arctic ground squirrel show RQ values of 0.7 indicating an exclusive use of FAO. However, during arousals, RQ rises to values > 0.85 suggesting a partial return to glucose fueled OXPHOS metabolism (Tähti, 1978; Buck and Barnes, 2000). Hibernator cells also readily suppress most ATP-consuming processes, including transcription and translation responsible for an estimated 20–30% of cellular energy consumption (Frerichs et al., 1998; Wieser and Krumschnabel, 2001; Armitage et al., 2003). This is thought to be only one of the mechanisms contributing to the metabolic reduction in hibernation, since the metabolic rate reduction during hibernation is much larger (∼83%) (Armitage et al., 2003; Storey and Storey, 2004). Although global transcription inhibition during hibernation is still debated (Carey and Martin, 1996; Hittel D. and Storey, 2002), there is increasing evidence that transcription modulation is tissue-dependent during torpor (Soukri et al., 1996; Van Breukelen and Martin, 2002). For example, thirteen-lined ground squirrels suppress DNA transcription and replication by a twofold decrease in transcriptional initiation, reducing elongation during torpor (Storey, 2003; Tsiouris, 2005; Tessier and Storey, 2014). To avoid energy expenditure by transcription of genes unnecessary in torpor, histone deacetylases (HDACs) silence genes through chromatin remodeling. Protein levels of HDAC1 and HDAC4 are significantly upregulated, and RNA polymerase II activity is downregulated by 57% in thirteen-lined ground squirrels during torpor (Morin and Storey, 2006), suggesting a tight regulation of energy-consuming gene transcription by chromatin remodeling and protein synthesis due to low temperature and low levels of mRNA turnover in hibernation. Given that protein synthesis is even more energetically expensive than transcription, expectedly, most of protein synthesis is actively repressed during torpor. Global mRNA translation is inhibited in torpid golden-mantled ground squirrels (Zhegunov et al., 1988) and Syrian hamsters (Osborne et al., 2004), even though some proteins are tissue-specifically synthesized at low rate during torpor. Conversely, very high rates of protein synthesis and cell proliferation are observed shortly after interbout arousal compared to squirrels that had been active for 1–2 days after hibernation, suggesting an initial compensatory mechanism to replenish proteins lost during torpor (Zhegunov et al., 1988).
Although the mechanistic control of the switch from glucose to fat combustion during hibernation has not been fully elucidated, it seems to be similar to mechanisms observed in starvation (Pilegaard et al., 2003), diabetes (Wu et al., 1999; Kim et al., 2006), and caloric restriction (Lee C. K. et al., 2002). Animals tested in these conditions show upregulation of the transcriptional targets of HIF-1α, including PDK4, PTL, and repression of PDH. However, HIF-1α was not investigated in these studies. Similarly, hibernating ground squirrels showed upregulation of PDK4 and PTL in heart, skeletal muscle and white adipose tissue (Andrews et al., 1998; Buck et al., 2002; Brauch et al., 2005). PTL shows high lipolytic activity at low ambient temperatures as an intrinsic feature of the protein across all mammal lineages. Yet, during torpor, PTL is further upregulated, which is an unique feature observed only in hibernators (Squire et al., 2004). While regulation of PDK4 and PTL only suggests involvement of HIF-1α, direct evidence for its upregulation in torpor comes from two studies. Maistrovski et al. (2012) found increased HIF-1α protein levels in skeletal muscle of 13-lined squirrels and little brown bat (Myotis lucifugus), and in liver of little brown bat during hibernation. Previously, HIF-1α protein levels were shown to increase by 60–70% in brown adipose tissue in 13-lined squirrels (Morin and Storey, 2005). Moreover, a recent study using fasting-induced daily torpor in B6N and B6J mice showed that the promoters of the HIF-1α signaling pathway are highly activated during torpidity (Sunagawa et al., 2018). On the other hand, some species such as Arctic ground squirrels (AGS) show significantly higher levels of HIF-1α during late-arousal and euthermic conditions compared to torpor (Ma et al., 2005). These contrasting findings could be due to species differences and/or differences in timing of sample collection. For example, normalization of oxygen consumption rate (OCR) in the brain upon arousal differs greatly among species, taking ∼60 min in Horseshoe bats (Rhinolophus ferrumequinum) (Lee M. et al., 2002), while taking ∼4 h in AGS (Zhu et al., 2005). Since the expression and activity of HIF-1α is also regulated by oxygen-independent mechanisms such as ROS levels, mechanical stress, and growth factors (Chun et al., 2002), HIF-1α expression during hibernation might be tissue-specific with differential expression associated with species differences.
In addition to HIF-1α, the hypoxia related RBM3 provides cytoprotection in hibernators by maintaining protein homeostasis under low metabolic conditions (Fedorov et al., 2009). Moreover, several of the genes offering protection in quiescent cell are reported upregulated in hibernating 13-lined ground squirrels, including FOXOs, HIF-1α, SOD3, p21, and p27, as discussed above.
Hibernation also features changes in expression of the negative regulators of mTOR, i.e., PTEN and TSC1/2. PTEN levels are significantly elevated (1.4 fold) in late torpor in skeletal muscle of thirteen-lined squirrels compared to summer euthermic controls (Wu et al., 2013a, 2015). Activation and inhibition of the Akt/mTOR pathway are paramount for cell cycle arrest during torpor and proliferation during arousal (Huang and Manning, 2008; Wu and Storey, 2012b). Moreover, hibernators re-activate the mTOR pathway upon arousal by supporting the switch from FAO to glucose fueled OXPHOS, increasing oxygen consumption and mitochondrial biogenesis (Wilson et al., 2008; Foudi et al., 2009).
Hibernating thirteen-lined squirrels reduce their O2 consumption by 98% from basal levels and increase it by 300% during arousals (Boyer and Barnes, 1999). In addition, an increase in mitochondrial ROS production also activates HIF-1α via the oxidative stress-sensitive transcription factor nuclear factor erythroid 2-related factor 2 (NRF2) (Hawkins et al., 2016; Lacher et al., 2018), which inhibits mitochondrial respiration and sequentially activates LKB1/AMPK (Simsek et al., 2010). However, the underlying mechanism of these molecular processes are not fully understood (Hwang et al., 2014; Li et al., 2015). Recent reports showed a significant upregulation of HIF-1α in the heart and skeletal muscle tissue of thirteen-lined squirrels during hibernation, suggesting that it confers protection against mitochondrial hyperpolarization as a possible mechanism against cellular stress (Maistrovski et al., 2012; Wu et al., 2013b). Mitochondrial hyperpolarization results from the disruption of their electrochemical gradient by the blockade of ATP-synthase, which may ultimately lead to Fas-induced apoptosis (Gergely et al., 2002; Perl et al., 2004). Recently, Ou et al. (2018) reported that exposure of human induced pluripotent stem cell-derived neurons (iPSC-neurons) to low temperature (4°C) produced mitochondrial hyperpolarization and accumulation of ROS, while mitochondrial from iPSC-neurons from thirteen-lined squirrels were depolarized and produced significant less ROS (Ou et al., 2018). High levels of ROS also induced accumulation of HIF-1α in brown adipose tissue (BAT), while HIF-1α knockdown in mice BAT led to reduced levels of glucose consumption, lactate export and glycolytic capacity (Choudhry and Harris, 2018). This suggests that glycolysis is dependent on HIF-1α regulation under hypoxic conditions, which maximizes metabolism in BAT.
Mitochondrial numbers and activity are differentially regulated between torpor and interbout arousal (Martin et al., 1999; Staples, 2014). Mitochondrial activity during hibernation of the 13-lined ground squirrel is tissue-specifically regulated and significantly increases in BAT and brain cortex (Ballinger et al., 2016), while mitochondria number was reported unchanged in ground squirrel liver (Brown et al., 2012), skeletal muscle (Hittel D. S. and Storey, 2002), and heart muscle (Staples and Brown, 2008) during hibernation. Yet, mitochondrial respiration exhibited no apparent suppression in heart muscle, moderate suppression in skeletal muscle and significant suppression in liver. Suppression of the uptake, transport or synthesis of specific substrates of OXPHOS may be a possible mechanism conferring suppression of mitochondrial respiration in liver cells (Brown et al., 2012). This possibility is in line with the decreased succinate dehydrogenase levels during hibernation reported previously (Gehnrich and Aprille, 1988; Cho, 2018).
While LKB1 is considered a master regulator of cellular metabolism in quiescent cells by inhibiting mitochondrial function and biogenesis through activation of FOXO and PGC-1α proteins, its relevance in torpor is still unknown. Nevertheless, in thirteen-lined ground squirrels, AMPK levels in white adipose tissue (WAT) were three times higher than those of summer animals (Horman et al., 2005). The activity of AMPK and LKB1 might be regulated by sex hormones, as dihydrotestosterone (DHT) inhibits AMPK activation, while androgens and estrogens inhibit LKB1 activation (McInnes et al., 2012) and declines in steroid hormone production seem to be a precondition for males to enter hibernation. As such, high levels of testosterone inhibit entrance into torpor in hamster (Hall and Goldman, 1980), hedgehog (Webb and Ellison, 1998), and Belding’s ground squirrel (Lee et al., 1990; Boonstra et al., 2001). Also, transcription of mtDNA and mitochondrial proteins such as PGC-1α, uncoupling proteins (UCP1, UCP3) and AMPK is 4-fold higher in BAT than in other tissues during hibernation (Boyer et al., 1998; Xu et al., 2013; Ballinger et al., 2016). PGC-1α is a central regulator of mitochondrial biogenesis and respiration and it induces the transcription of nuclear respiratory factors (NRF1 and NRF2) that activate the replication of mtDNA.
The crucial role of PML in HSCs quiescence (Ishida, 2009; Ito et al., 2012) would suggest that a similar mechanism is present during mammalian torpor (Lee et al., 2007). However, to the best of our knowledge, there are no reports about the specific role of PML during hibernation, although increasing evidence suggests downstream PPARs to constitute the master transcriptional regulators of changes in lipid metabolism. For example, both protein and mRNA of PPARα were upregulated in WAT, heart, kidney and liver of six species of hibernating bats (Han et al., 2015), jerboa (Jaculus orientalis) (Kabine et al., 2004) and 13-lined squirrels (Eddy et al., 2005). PPARα not only induces the activation of genes involved in lipid metabolism, but also induces the expression of uncoupling proteins (UCPs), which pump protons back into the mitochondrial matrix generating heat without synthesizing ATP. The high uncoupled thermogenesis activity of mitochondrial respiration in BAT plays an essential role in thermoregulation and contributes significantly to the rewarming of the organism during arousal from hibernation. Further, PML was also reported to be highly active in thirteen-lined ground squirrels brain during torpor and is associated with massive SUMOylation and increased tolerance to brain ischemia (Lee et al., 2007; Lee and Hallenbeck, 2013).
Autophagy is poorly examined in hibernators. In the heart of hibernating Syrian hamster, autophagy seems already initiated during (late) torpor and executed during early arousal, reflected by the gradual increase in active autophagosomes during torpor followed by a peak at early arousal, returning to normal late in arousal (Wiersma et al., 2018). This might be due to the build-up of damaged or misfolded proteins being gradually formed during torpor, since there is an accumulation of cells in the G2 phase during torpor (Matthews and Fisher, 1968; Kruman et al., 1988). While autophagy is an essential mechanism to sustain quiescence in HSC, its role in maintaining cell dormancy in hibernators is poorly understood. More research into autophagy in hibernators may unclose relevant knowledge on the preservation of life under stress conditions.
Under normal physiological conditions, the body has stem cells in all phases of the cell cycle (Lyle and Moore, 2011). Radiation generates high levels of ROS, which increases cellular stress and causes irreversible cellular damage leading to senescence and apoptosis. Particular stem cells are resistant to radiation: cancer stem cells (CSC). Not surprisingly, HIF-1α is significantly upregulated in cancerous cells due to the hypoxic environment created by the rapidly proliferating cells (Majmundar et al., 2010). HIF-1α regulates the switch from glucose to fatty acid combustion, a characteristic of quiescent cells, indicating that HIF-1α might play an important role in conferring radiation resistance in dormant cells. Other mechanisms, including the reduction in histone acetylation because of increased activity of HDACs, lead to a tighter packing of DNA into heterochromatin, which plays a significant role in the radiation resistance of quiescent cells (Diehn and Clarke, 2006). Quiescent cells, especially human stem cells, significantly upregulate antioxidant gene expression, creating an environment where the cells can resist ROS production by radiation (Oberley et al., 1995). In cancer cells, this appears under the control of a particular gene from the FOXO family, FOXM1. FOXM1 downregulation in quiescent cells elevated expression of antioxidant genes such as manganese superoxide dismutase (MnSOD), catalase (CAT), and peroxiredoxin (PRDX3) (Eckers et al., 2014). Radiotherapy is usually administered when treating cancer cells, but quiescent CSCs are resistant to this therapy designed to eliminate proliferating cells (Luk et al., 1986). Knockdown of FOXM1 increases sensitivity to radiation therapy in quiescent and cancer stem cells. The amount of therapeutic radiation is still restricted by its toxicity to normal tissue. For example, multiple metastases cannot be treated without exceeding the tolerance of the healthy organ nearby. By putting specific tissues or organs of patients into dormancy, cells may potentially tolerate higher doses of radiation (Cerri et al., 2016).
Studies conducted in hibernators have shown that torpor limits radiation-induced DNA damage in squirrels, hamsters, and mice (Ghosh et al., 2017; Tinganelli et al., 2019), which has awakened the interest of its utility in cancer therapies and long-haul space missions. The mechanisms that mediate radioprotection during torpor are not known. Likely, they parallel those found in quiescent cells, as torpid animals upregulate antioxidant genes and activate HDACs. Recent studies indicate that radiation resistance in torpor may also relate to the hypothermia, as cell cooling limits DNA damage and leads to a different dynamics in DNA damage repair (Baird et al., 2011).
Quiescent (hematopoietic stem) cells and torpor share many similarities (Table 1). First, these include activation of molecular mechanisms that stall the cell cycle of proliferating cells including a major overlap in regulation of essential cell cycle genes and proteins associated with the maintenance of quiescence in HSCs and torpidity during hibernation. In both cases, the entry into dormancy is associated with differential gene expression of proteins that includes cyclins, CDKs and CKIs. Yet the molecular mechanism conferring cellular quiescence in stem cells are described in great detail, whereas this is not the case for hibernators. Moreover, it is evident that induction of HES1 during stem cell quiescence is a prerequisite to enable reversal from cell arrest. However, there is a marked paucity of data on Hes1 activity in hibernators, warranting further studies to understand its role in torpor. HSC may provide a blueprint to disclose mechanisms used by hibernators that govern activation of the molecular machinery of cell cycle arrest in response to environmental changes, even though hibernator cells largely represent terminally differentiated cells.
Secondly, cell quiescence and torpor share a similar metabolic rewiring. Both in quiescent and torpid cells, energy conservation is brought about by the reduction in metabolic rate and the switch from glucose fueled OXPHOS to FAO as the primary mechanism to supply ATP, supported by the radical suppression of anabolic processes, such as DNA replication, transcription and protein synthesis. HIF-1α coordinates the cellular adaptation to restore the balance between oxygen supply and metabolic demand, leading to a reduction in the consumption of oxygen. Under ATP-deprived conditions resulting from nutrient deprivation or hypoxia, activation of the energy-sensing liver kinase B1 (LKB1) and the downstream AMP-activated protein kinase (AMPK) precede the upregulation of HIF-1α (Hudson et al., 2002; Lee et al., 2003). Moreover, the activation of LKB1 and AMPK stimulates autophagy through phosphorylation of ULK1 and inhibits the mTOR pathway (Hudson et al., 2002; Li et al., 2015; Mohammad et al., 2019). The Akt/mTOR pathway regulates many energy-expensive processes and is inhibited in quiescence and hibernation by the action of PTEN and TSC1/2 coupled with downstream RPP signaling. High levels of PTEN and TSCs both in HSCs and during hibernation suggest that these proteins are essential to maintain quiescence and torpor. Several of these pathways need further study in hibernators to define their contribution to metabolic suppression, including HIF-1α and autophagy. Further, the PML-PPARδ-FAO pathway appears to play a vital role in the maintenance of HSC quiescence and plays a critical role in its cell fate and self-renewal decisions. However, very little is known about the function and activity of PML during hibernation. Also, quiescent cells and hibernators share the upregulation of cell protective, anti-apoptotic pathways, suggesting that a similar mechanisms is activated during quiescence and mammalian torpor (Heldmaier et al., 2000; Hefler et al., 2015; Zhang et al., 2016).
Thirdly, both quiescent cells and hibernators share radiation resistance, which seems conferred by the combination of upregulation of antioxidant defense and heterochromatin formation. In addition to the antioxidant environment, HIF-1α recruits HDACs to tightly pack the DNA into heterochromatin resulting in resistance to radiation. FOXM1 appears to be a master regulator of antioxidants, and its downregulation resulted in lower radiation sensitivity of cancer stem cells. While strict control over oxidation is crucial for hibernators to survive torpor/arousal switches, there is no literature of the activity and function of FOXM1 in hibernators available. Consequently, mechanisms of radiation resistance in both stem cells and torpor are still ill-defined. At present, it is unclear whether radiation resistance merely exists as a bystander effect of metabolic suppression and antioxidant defense, including a tighter packaging of DNA, or whether it is conferred by specific mechanisms; a question not so easily addressed.
As outlined, the three phenomena discussed above show substantial crosstalk, with the one activating or promoting the other. Although processes are quite similar in HSC and torpor, there may be a clear distinction in their order. For instance, in HSC the regulation of cell cycle and metabolism seems tightly integrated, as interventions in both drive cells out of quiescence. Whether this is true for hibernators is unclear. Possibly, the inhibition of the cell cycle of the differentiated cells in hibernators is merely a consequence of strong metabolic suppression, including the inhibition of DNA synthesis and transcription. To explore differences in orchestration, an accurate delineation of the critical factors and their sequence of events during entry into torpor is warranted.
Reversible protein phosphorylation (RPP) is a crucial post-translational modifier of proteins and regulator of cell homeostasis. In HSCs and hibernators, RPP is especially important to support the exit from and re-entering of the cell cycle without spending much energy on anabolic processes. While the LKB1-AMPK route plays a crucial role in inactivating many anabolic processes such as protein synthesis, RPP signaling seems of crucial importance to inactivate ion channels. The preponderance of cellular processes that expend energy is directly or indirectly affiliated with membranes preserving concentration gradients. Thus, suppression of membrane-associated (ion channels) proteins by RPP has a profound impact on metabolic reduction and ATP turnover reduction. For example, phosphorylation of sodium-potassium pump (Na+/K+-ATPase) led to a decrease in activity by up to 60% in golden-mantled squirrels (Storey and Storey, 2004). Therefore, RPP could be responsible for not only maintaining lipid-based metabolism but also to initiate the metabolic repression in hibernation preparation through AMPK signaling.
The large overlap between quiescent (hematopoietic stem) cells and hibernator adaptations may have some future implications. First, studies on (stem) cell quiescence have identified a number of crucial genes/pathways, of which the relevance in hibernation has been insufficiently explored, in particular LKB1, HES1, HIF-1α, and PML. One study explored the effects of LKB1 knockout in Caenorhabditis elegans (Narbonne and Roy, 2009). Interestingly, the LKB1 knockout worms entered Dauer state, a survival mechanism that arrests feeding while retaining activity, motility and acquiring stress-resistance, but rapidly consumed their stored energy leading to failure of vital organs and death. Although C. elegans is not a true hibernator nor a mammal, the phenotype is similar to LKB1 knockout in HSC. LKB1 might thus also be an essential protein in hibernators under energetically unfavorable conditions to maintain energy and oxidative stress homeostasis. PML-PPARδ regulation of FAO is essential to maintain HSC quiescence and acts as a negative regulator of Akt/mTOR, a crucial element in the metabolic shift. In hibernators, there is only a single study reporting increased PML-PPARδ activity in thirteen-lined squirrel. Given its critical role in FAO and metabolic reprogramming, PML is an interesting target to address in hibernation. Given the activation of the machinery governing cell cycle block during torpidity, it is of great importance to study HES-1, the factor that retains the option for stem cells to re-enter cell cycle, in particular in relation to arousals. Finally, although upregulation of HIF-1α has been documented in hibernators, details of its regulation and effects need a deeper understanding.
Nevertheless, exploring the relevance of these and other proteins in hibernation is not a trivial task. The main limitation to infer causality in factors contributing to hibernation, is the absence of (conditional) knock-out models in true hibernators. Generation of such models would require genetic modification of hibernator blastocyst stem cells to introduce LoxP sites into genes of interest and generation of Cre expressing lines, preferable harboring inducible promotors, like the tamoxifen-induced CRE-ERT2 system. Such developments may be accelerated by the recent advancements of CRISPR/Cas9 technology, but will still require considerable effort and funding, and specialized knowledge. An alternative approach may be to use inducible knockouts of genes in the house mouse (Mus musculus), a species long known to be capable of serial daily torpor (Hudson and Scott, 1979). Conditional knockout mouse lines of a number of factors discussed are readily available. Moreover, the present variety of Cre mouse lines and the superior toolbox to introduce LoxP sites, signify that making the appropriate knockout in mouse is far easier compared to true hibernators. However, it is unclear to date to what extent the molecular mechanisms of mouse daily torpor resemble those of torpor found in true hibernators. The first step should therefore consist of exploring the molecular footprint of cell cycle arrest and metabolic rewiring in mouse torpor. A third option to explore specific genes might be the use of induced pluripotent stem cells (iPSCs) from hibernators (Takahashi and Yamanaka, 2006). These iPSCs can be differentiated into any cell type to study molecular biology in vitro. Also, if the selected cell type does not depend on a heterotypic, complex environment, but represents an autonomous cell, it can be further maturated into engineered 3D tissue or organoids to mimic physiological behavior. A recent study showed that iPSC-derived neurons from thirteen-lined squirrel behaved differently than human neurons with higher resistance to cold (Ou et al., 2018). Knocking out genes in iPSC-derived hibernator cells may at least explore the important question whether some of the mechanism present in HSC quiescence may be induced in a cell autonomous way by starvation or hypoxia.
Most cells in a mammal, whether or not a hibernator, are capable of quiescence. The similarity between molecular mechanisms conferring quiescence in HSC and hibernation may signify that there is a unifying (epi)genetic-metabolic program governing the both states. If true, it may mean that all non-hibernator species could be capable of hibernation, provided that cellular quiescence is induced in high energy consuming organs.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This study was supported by a grant from the European Space Agency to RH (Research agreement collaboration 4000123556).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Al-attar, R., and Storey, K. B. (2020). Suspended in time: Molecular responses to hibernation also promote longevity. Exp. Gerontol. 134:110889. doi: 10.1016/j.exger.2020.110889
Andrews, M. T. (2007). Advances in molecular biology of hibernation in mammals. BioEssays 29, 431–440. doi: 10.1002/bies.20560
Andrews, M. T., Squire, T. L., Bowen, C. M., and Rollins, M. B. (1998). Low-temperature carbon utilization is regulated by novel gene activity in the heart of a hibernating mammal. Proc. Natl. Acad. Sci. U. S. A. 95, 8392–8397. doi: 10.1073/pnas.95.14.8392
Armitage, K. B., Blumstein, D. T., and Woods, B. C. (2003). Energetics of hibernating yellow-bellied marmots (Marmota flaviventris). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 134, 101–114. doi: 10.1016/S1095-6433(02)00219-2
Asai, T., Liu, Y., Di Giandomenico, S., Bae, N., Ndiaye-Lobry, D., Deblasio, A., et al. (2012). Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood 120, 1601–1612. doi: 10.1182/blood-2011-11-393983
Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T., and Kageyama, R. (2006). Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 133, 2467–2476. doi: 10.1242/dev.02403
Baird, B. J., Dickey, J. S., Nakamura, A. J., Redon, C. E., Parekh, P., Griko, Y. V., et al. (2011). Hypothermia postpones DNA damage repair in irradiated cells and protects against cell killing. Mutat. Res. Fundam. Mol. Mech. Mutagen. 711, 142–149. doi: 10.1016/j.mrfmmm.2010.12.006
Ballinger, M. A., Hess, C., Napolitano, M. W., Bjork, J. A., and Andrews, M. T. (2016). Seasonal changes in brown adipose tissue mitochondria in a mammalian hibernator: From gene expression to function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 311, R325–R336. doi: 10.1152/ajpregu.00463.2015
Basu, S., Broxmeyer, H. E., and Hangoc, G. (2013). Peroxisome proliferator-activated-γ coactivator-1α-mediated mitochondrial biogenesis is important for hematopoietic recovery in response to stress. Stem Cells Dev. 22, 1678–1692. doi: 10.1089/scd.2012.0466
Beauséjour, C. M., Krtolica, A., Galimi, F., Narita, M., Lowe, S. W., Yaswen, P., et al. (2003). Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 22, 4212–4222. doi: 10.1093/emboj/cdg417
Berasain, C., and Avila, M. A. (2015). Regulation of hepatocyte identity and quiescence. Cell. Mol. Life Sci. 72, 3831–3851. doi: 10.1007/s00018-015-1970-7
Bigarella, C. L., Li, J., Rimmelé, P., Liang, R., Sobol, R. W., and Ghaffari, S. (2017). FOXO3 transcription factor is essential for protecting hematopoietic stem and progenitor cells from oxidative DNA damage. J. Biol. Chem. 292, 3005–3015. doi: 10.1074/jbc.M116.769455
Biggar, K., and Storey, K. (2009). Perspectives in Cell Cycle Regulation: Lessons from an Anoxic Vertebrate. Curr. Genomics 10, 573–584. doi: 10.2174/138920209789503905
Blanco, M. B., and Zehr, S. M. (2015). Striking longevity in a hibernating lemur. J. Zool. 296, 177–188. doi: 10.1111/jzo.12230
Blomen, V. A., and Boonstra, J. (2007). Cell fate determination during G1 phase progression. Cell. Mol. Life Sci. 64, 3084–3104. doi: 10.1007/s00018-007-7271-z
Boonstra, R., Hubbs, A. H., Lacey, E. A., and McColl, C. J. (2001). Seasonal changes in glucocorticoid and testosterone concentrations in free-living arctic ground squirrels from the boreal forest of the Yukon. Can. J. Zool. 79, 49–58. doi: 10.1139/z00-175
Boyer, B. B., and Barnes, B. M. (1999). Molecular and metabolic aspects of mammalian hibernation. Bioscience 49, 713–724. doi: 10.2307/1313595
Boyer, B. B., Barnes, B. M., Lowell, B. B., and Grujic, D. (1998). Differential regulation of uncoupling protein gene homologues in multiple tissues of hibernating ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 275, R1232. doi: 10.1152/ajpregu.1998.275.4.r1232
Bracken, A. P., Ciro, M., Cocito, A., and Helin, K. (2004). E2F target genes: Unraveling the biology. Trends Biochem. Sci. 29, 409–417. doi: 10.1016/j.tibs.2004.06.006
Braissant, O., Foufelle, F., Scotto, C., Dauça, M., and Wahli, W. (1996). Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology 137, 354–366. doi: 10.1210/endo.137.1.8536636
Brauch, K. M., Dhruv, N. D., Hanse, E. A., and Andrews, M. T. (2005). Digital transcriptome analysis indicates adaptive mechanisms in the heart of a hibernating mammal. Physiol. Genomics 2005, 00076. doi: 10.1152/physiolgenomics.00076.2005
Brown, J. C. L., Chung, D. J., Belgrave, K. R., and Staples, J. F. (2012). Mitochondrial metabolic suppression and reactive oxygen species production in liver and skeletal muscle of hibernating thirteen-lined ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 00230. doi: 10.1152/ajpregu.00230.2011
Buck, C. L., and Barnes, B. M. (2000). Effects of ambient temperature on metabolic rate, respiratory quotient, and torpor in an arctic hibernator. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279, r255. doi: 10.1152/ajpregu.2000.279.1.r255
Buck, M. J., Squire, T. L., and Andrews, M. T. (2002). Coordinate expression of the PDK4 gene: A means of regulating fuel selection in a hibernating mammal. Physiol. Genomics 2002, 5–13. doi: 10.1152/physiolgenomics.00076.2001
Camins, A., Pizarro, J. G., and Folch, J. (2013). Cyclin-Dependent Kinases. Brenner’s Encyclop. Genet. 2013, 260–266. doi: 10.1016/B978-0-12-374984-0.00370-3
Camperio, C., Caristi, S., Fanelli, G., Soligo, M., De Porto, P., and Piccolella, E. (2012). Forkhead Transcription Factor FOXP3 Upregulates CD25 Expression through Cooperation with RelA/NF-κB. PLoS One 7:0048303. doi: 10.1371/journal.pone.0048303
Carey, H. V., and Martin, S. L. (1996). Preservation of intestinal gene expression during hibernation. Am. J. Physiol. Gastrointest. Liver Physiol. 271, g805. doi: 10.1152/ajpgi.1996.271.5.g805
Carey, H. V., Andrews, M. T., and Martin, S. L. (2003). Mammalian hibernation: Cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev. 83, 1153–1181. doi: 10.1152/physrev.00008.2003
Cerri, M., Tinganelli, W., Negrini, M., Helm, A., Scifoni, E., Tommasino, F., et al. (2016). Hibernation for space travel: Impact on radioprotection. Life Sci. Sp. Res. 11, 1–9. doi: 10.1016/j.lssr.2016.09.001
Chakkalakal, J. V., Jones, K. M., Basson, M. A., and Brack, A. S. (2012). The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360. doi: 10.1038/nature11438
Chen, C., Liu, Y., Liu, R., Ikenoue, T., Guan, K. L., Liu, Y., et al. (2008). TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205, 2397–2408. doi: 10.1084/jem.20081297
Cheng, T., Rodrigues, N., Shen, H., Yang, Y. G., Dombkowski, D., Sykes, M., et al. (2000). Hematopoietic stem cell quiescence maintained by p21(cip1/waf1). Science 287, 1804–1809. doi: 10.1126/science.287.5459.1804
Cheung, T. H., and Rando, T. A. (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340. doi: 10.1038/nrm3591
Cho, E. H. (2018). Succinate as a regulator of hepatic stellate cells in liver fibrosis. Front. Endocrinol. 9:00455. doi: 10.3389/fendo.2018.00455
Choi, Y. J., and Anders, L. (2014). Signaling through cyclin D-dependent kinases. Oncogene 33, 1890–1903. doi: 10.1038/onc.2013.137
Choudhry, H., and Harris, A. L. (2018). Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 27, 281–298. doi: 10.1016/j.cmet.2017.10.005
Chun, Y. S., Kim, M. S., and Park, J. W. (2002). Oxygen-dependent and -independent regulation of HIF-1alpha. J. Korean Med. Sci. 2002, 581. doi: 10.3346/jkms.2002.17.5.581
Clevers, H., and Watt, F. M. (2018). Defining Adult Stem Cells by Function, not by Phenotype. Annu. Rev. Biochem. 87, 1015–1027. doi: 10.1146/annurev-biochem-062917-012341
Coats, S., Flanagan, W. M., Nourse, J., and Roberts, J. M. (1996). Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science 272, 877–880. doi: 10.1126/science.272.5263.877
Coller, H. A., Sang, L., and Roberts, J. M. (2006). A new description of cellular quiescence. PLoS Biol. 4:0329–0349. doi: 10.1371/journal.pbio.0040083
Coloff, J. L., Murphy, J. P., Braun, C. R., Harris, I. S., Shelton, L. M., Kami, K., et al. (2016). Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 23, 867–880. doi: 10.1016/j.cmet.2016.03.016
Dai, Y., and Grant, S. (2003). Cyclin-dependent kinase inhibitors. Curr. Opin. Pharmacol. 3, 362–370. doi: 10.1016/S1471-4892(03)00079-1
Dannenberg, J. H., Van Rossum, A., Schuijff, L., and Te Riele, H. (2000). Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14, 3051–3064. doi: 10.1101/gad.847700
Dannenmann, B., Lehle, S., Hildebrand, D. G., Kübler, A., Grondona, P., Schmid, V., et al. (2015). High glutathione and glutathione peroxidase-2 levels mediate cell-type-specific DNA damage protection in human induced pluripotent stem cells. Stem Cell Reports 4, 886–898. doi: 10.1016/j.stemcr.2015.04.004
Dausmann, K. H., Glos, J., Ganzhorn, J. U., and Heldmaier, G. (2004). Physiology: Hibernation in a tropical primate. Nature 429, 825–826. doi: 10.1038/429825a
Denlinger, D. L. (2002). Regulation of diapause. Annu. Rev. Entomol. 47, 93–122. doi: 10.1146/annurev.ento.47.091201.145137
Desvergne, B., and Wahli, W. (1999). Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 20, 649–688. doi: 10.1210/er.20.5.649
Diehn, M., and Clarke, M. F. (2006). Cancer stem cells and radiotherapy: New insights into tumor radioresistance. J. Natl. Cancer Inst. 98, 1755–1757. doi: 10.1093/jnci/djj505
Dong, J. J., Peng, J., Zhang, H., Mondesire, W. H., Jian, W., Mills, G. B., et al. (2005). Role of glycogen synthase kinase 3β in rapamycin-mediated cell cycle regulation and chemosensitivity. Cancer Res. 65, 1961–1972. doi: 10.1158/0008-5472.CAN-04-2501
Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262. doi: 10.1101/gad.12.15.2245
Eckers, J. C., Kalen, A. L., Sarsour, E. H., Tompkins, V. S., Janz, S., Son, J. M., et al. (2014). Forkhead Box M1 Regulates Quiescence-Associated Radioresistance of Human Head and Neck Squamous Carcinoma Cells. Radiat. Res. 182, 420–429. doi: 10.1667/RR13726.1
Eddy, S. F., Morin, P., and Storey, K. B. (2005). Cloning and expression of PPARγ and PGC-1α from the hibernating ground squirrel, Spermophilus tridecemlineatus. Mol. Cell. Biochem. 269, 175–182. doi: 10.1007/s11010-005-3459-4
Eilers, M., Schirm, S., and Bishop, J. M. (1991). The MYC protein activates transcription of the α-prothymosin gene. EMBO J. 10, 133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x
Ezoe, S., Matsumura, I., Tanaka, H., Satoh, Y., Yokota, T., Oritani, K., et al. (2008). SIRT1 Deficiency Suppresses the Maintenance of Hematopoietic Stem Cell Pool. Blood 112, 1394–1394. doi: 10.1182/blood.v112.11.1394.1394
Fedorov, V. B., Goropashnaya, A. V., Tøien, O., Stewart, N. C., Gracey, A. Y., Chang, C., et al. (2009). Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus). Physiol. Genomics 37, 108–118. doi: 10.1152/physiolgenomics.90398.2008
Figel, S., and Fenstermaker, R. A. (2018). “Cell-Cycle Regulation,” in Handbook of Brain Tumor Chemotherapy, Molecular Therapeutics, and Immunotherap. H B. Newton (Elsevier: Amsterdam) 257–269. doi: 10.1016/B978-0-12-812100-9.00018-8
Fisher, R. P. (1997). CDKs and cyclins in transition(s). Curr. Opin. Genet. Dev. 7, 32–38. doi: 10.1016/S0959-437X(97)80106-2
Foudi, A., Hochedlinger, K., Van Buren, D., Schindler, J. W., Jaenisch, R., Carey, V., et al. (2009). Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90. doi: 10.1038/nbt.1517
Fox, P. M., Vought, V. E., Hanazawa, M., Lee, M. H., Maine, E. M., and Sched, T. (2011). Cyclin e and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138, 2223–2234. doi: 10.1242/dev.059535
Frerichs, K. U., Smith, C. B., Brenner, M., Degracia, D. J., Krause, G. S., Marrone, L., et al. (1998). Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc. Natl. Acad. Sci. U. S. A. 95, 14511–14516. doi: 10.1073/pnas.95.24.14511
Gan, B., Hu, J., Jiang, S., Liu, Y., Sahin, E., Zhuang, L., et al. (2010). Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704. doi: 10.1038/nature09595
Gehnrich, S. C., and Aprille, J. R. (1988). Hepatic gluconeogenesis and mitochondrial function during hibernation. Comp. Biochem. Physiol. Part B Biochem. 91, 11–16. doi: 10.1016/0305-0491(88)90107-1
Gergely, P., Grossman, C., Niland, B., Puskas, F., Neupane, H., Allam, F., et al. (2002). Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum 46, 175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015<3.0.CO;2-H
Ghosh, S., Indracanti, N., Joshi, J., Ray, J., and Indraganti, P. K. (2017). Pharmacologically induced reversible hypometabolic state mitigates radiation induced lethality in mice. Sci. Rep. 7, 15002–15007. doi: 10.1038/s41598-017-15002-7
Gurumurthy, S., Xie, S. Z., Alagesan, B., Kim, J., Yusuf, R. Z., Saez, B., et al. (2010). The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659–663. doi: 10.1038/nature09572
Hall, V., and Goldman, B. (1980). Effects of gonadal steroid hormones on hibernation in the Turkish hamster (Mesocricetus brandti). J. Comp. Physiol. B 135, 107–114. doi: 10.1007/BF00691200
Han, Y., Zheng, G., Yang, T., Zhang, S., Dong, D., and Pan, Y. H. (2015). Adaptation of peroxisome proliferator-activated receptor alpha to hibernation in bats Genome evolution and evolutionary systems biology. BMC Evol. Biol. 15:373–376. doi: 10.1186/s12862-015-0373-6
Hao, S., Chen, C., and Cheng, T. (2016). Cell cycle regulation of hematopoietic stem or progenitor cells. Int. J. Hematol. 103, 487–497. doi: 10.1007/s12185-016-1984-4
Harbour, J. W., and Dean, D. C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409. doi: 10.1101/gad.813200
Hartman, J., Müller, P., Foster, J. S., Wimalasena, J., and Gustafsson, J. Å, and Ström, A. (2004). HES-1 inhibits 17β-estradiol and heregulin-β1-mediated upregulation of E2F-1. Oncogene 23, 8826–8833. doi: 10.1038/sj.onc.1208139
Hawkins, K. E., Joy, S., Delhove, J. M. K. M., Kotiadis, V. N., Fernandez, E., Fitzpatrick, L. M., et al. (2016). NRF2 Orchestrates the Metabolic Shift during Induced Pluripotent Stem Cell Reprogramming. Cell Rep. 14, 1883–1891. doi: 10.1016/j.celrep.2016.02.003
Hefler, J., Wu, C. W., and Storey, K. B. (2015). Transcriptional activation of p53 during cold induced torpor in the 13-lined ground squirrel Ictidomys tridecemlineatus. Biochem. Res. Int. 2015, 731595. doi: 10.1155/2015/731595
Heldmaier, G., Klingenspor, M. F., and Klaus, S. (2000). Life in the Cold: Eleventh International Hibernation Symposium. New York, NY: Springer
Hittel, D. S., and Storey, K. B. (2002). Differential expression of mitochondria-encoded genes in a hibernating mammal. J. Exp. Biol. 205, 1625–1631.
Hittel, D., and Storey, K. B. (2002). The translation state of differentially expressed mRNAs in the hibernating 13-lined ground squirrel (Spermophilus tridecemlineatus). Arch. Biochem. Biophys. 401, 244–254. doi: 10.1016/S0003-9861(02)00048-6
Horman, S., Hussain, N., Dilworth, S. M., Storey, K. B., and Rider, M. H. (2005). Evaluation of the role of AMP-activated protein kinase and its downstream targets in mammalian hibernation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 142, 374–382. doi: 10.1016/j.cbpb.2005.08.010
Hosokawa, K., Arai, F., Yoshihara, H., Takubo, K., Ito, K., Matsuoka, S., et al. (2006). Reactive Oxygen Species Control Hematopoietic Stem Cell-Niche Interaction through the Regulation of N-Cadherin. Blood 108, 86–86. doi: 10.1182/blood.v108.11.86.86
Huang, J., and Manning, B. D. (2008). The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 412, 179–190. doi: 10.1042/BJ20080281
Hudson, C. C., Liu, M., Chiang, G. G., Otterness, D. M., Loomis, D. C., Kaper, F., et al. (2002). Regulation of Hypoxia-Inducible Factor 1α Expression and Function by the Mammalian Target of Rapamycin. Mol. Cell. Biol. 22, 7004–7014. doi: 10.1128/mcb.22.20.7004-7014.2002
Hudson, J. W., and Scott, I. M. (1979). Daily Torpor in the Laboratory Mouse, Mus musculus Var. Albino. Physiol. Zool. 1979, 30152564. doi: 10.1086/physzool.52.2.30152564
Huh, C. G., Factor, V. M., Sánchez, A., Uchida, K., Conner, E. A., and Thorgeirsson, S. S. (2004). Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. U. S. A. 101, 4477–4482. doi: 10.1073/pnas.0306068101
Hwang, A. B., Ryu, E. A., Artan, M., Chang, H. W., Kabir, M. H., Nam, H. J., et al. (2014). Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 111, E4458–E4467. doi: 10.1073/pnas.1411199111
Infante, A., Laresgoiti, U., Fernández-Rueda, J., Fullaondo, A., Galán, J., Díaz-Uriarte, R., et al. (2008). E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle 7, 3915–3927. doi: 10.4161/cc.7.24.7379
Ishida, N. (2009). Role of PPAR in the control of torpor through FGF21-NPY pathway: From circadian clock to seasonal change in mammals. PPAR Res. 2009:412949. doi: 10.1155/2009/412949
Ito, K., and Ito, K. (2013). Newly identified roles of PML in stem cell biology. Front. Oncol. 3:00050. doi: 10.3389/fonc.2013.00050
Ito, K., and Ito, K. (2016). Metabolism and the Control of Cell Fate Decisions and Stem Cell Renewal. Annu. Rev. Cell Dev. Biol. 32, 399–409. doi: 10.1146/annurev-cellbio-111315-125134
Ito, K., and Suda, T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15, 243–256. doi: 10.1038/nrm3772
Ito, K., Carracedo, A., Weiss, D., Arai, F., Ala, U., Avigan, D. E., et al. (2012). A PML-PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18, 1350–1358. doi: 10.1038/nm.2882
Jang, Y. Y., and Sharkis, S. J. (2007). A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110, 3056–3063. doi: 10.1182/blood-2007-05-087759
Jansen, H. T., Trojahn, S., Saxton, M. W., Quackenbush, C. R., Evans Hutzenbiler, B. D., Nelson, O. L., et al. (2019). Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun. Biol. 2, 574–574. doi: 10.1038/s42003-019-0574-4
Johnson, D. G., Schwarz, J. K., Cress, W. D., and Nevins, J. R. (1993). Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365, 349–352. doi: 10.1038/365349a0
Kabine, M., El Kebbaj, Z., Oaxaca-Castillo, D., Clémencet, M. C., El Kebbaj, M. S., Latruffe, N., et al. (2004). Peroxisome proliferator-activated receptors as regulators of lipid metabolism; Tissue differential expression in adipose tissues during cold acclimatization and hibernation of jerboa (Jaculus orientalis). Biochimie 86, 763–770. doi: 10.1016/j.biochi.2004.10.003
Kalaitzidis, D., Sykes, S. M., Wang, Z., Punt, N., Tang, Y., Ragu, C., et al. (2012). MTOR complex 1 plays critical roles in hematopoiesis and pten-loss-evoked leukemogenesis. Cell Stem Cell 11, 429–439. doi: 10.1016/j.stem.2012.06.009
Kalamakis, G., Brüne, D., Ravichandran, S., Bolz, J., Fan, W., Ziebell, F., et al. (2019). Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell 176, 1407.e–1419.e. doi: 10.1016/j.cell.2019.01.040
Kent, L. N., and Leone, G. (2019). The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338. doi: 10.1038/s41568-019-0143-7
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P., and Guan, K. L. (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945. doi: 10.1038/ncb1753
Kim, Y. I., Lee, F. N., Choi, W. S., Lee, S., and Youn, J. H. (2006). Insulin regulation of skeletal muscle PDK4 mRNA expression is impaired in acute insulin-resistant states. Diabetes 55, 2311–2317. doi: 10.2337/db05-1606
Kippin, T. E., Martens, D. J., and Van Der Kooy, D. (2005). P21 Loss Compromises the Relative Quiescence of Forebrain Stem Cell Proliferation Leading To Exhaustion of Their Proliferation Capacity. Genes Dev. 19, 756–767. doi: 10.1101/gad.1272305
Kocabas, F., Zheng, J., Thet, S., Copeland, N. G., Jenkins, N. A., DeBerardinis, R. J., et al. (2012). Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120, 4963–4972. doi: 10.1182/blood-2012-05-432260
Kopp, H. G., Avecilla, S. T., Hooper, A. T., and Rafii, S. (2005). The bone marrow vascular niche: Home of HSC differentiation and mobilization. Physiology 20, 349–356. doi: 10.1152/physiol.00025.2005
Kops, G. J. P. L., Dansen, T. B., Polderman, P. E., Saarloos, I., Wirtz, K. W. A., Coffer, P. J., et al. (2002). Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321. doi: 10.1038/nature01036
Kowalik, T. F., DeGregori, J., Schwarz, J. K., and Nevins, J. R. (1995). E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J. Virol. 69, 2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995
Kretzner, L., Blackwood, E. M., and Eisenman, R. N. (1992). Myc and Max proteins possess distinct transcriptional activities. Nature 359, 426–429. doi: 10.1038/359426a0
Kruman, I. I. (1992). Comparative analysis of cell replacement in hibernators. Comp. Biochem. Physiol. Part A Physiol. 1992, 90620–90626. doi: 10.1016/0300-9629(92)90620-6
Kruman, I. I., Ilyasova, E. N., Rudchenko, S. A., and Khurkhulu, Z. S. (1988). The intestinal epithelial cells of ground squirrel (Citellus undulatus) accumulate at G2 phase of the cell cycle throughout a bout of hibernation. Comp. Biochem. Physiol. Part A Physiol. 90, 233–236. doi: 10.1016/0300-9629(88)91109-7
Lacher, S. E., Levings, D. C., Freeman, S., and Slattery, M. (2018). Identification of a functional antioxidant response element at the HIF1A locus. Redox Biol. 19, 401–411. doi: 10.1016/j.redox.2018.08.014
Lacorazza, H. D., Yamada, T., Liu, Y., Miyata, Y., Sivina, M., Nunes, J., et al. (2006). The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9, 175–187. doi: 10.1016/j.ccr.2006.02.017
Lallemand-Breitenbach, V., and de Thé, H. (2010). PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000661. doi: 10.1101/cshperspect.a000661
Lee, C. K., Allison, D. B., Brand, J., Weindruch, R., and Prolla, T. A. (2002). Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc. Natl. Acad. Sci. U. S. A. 99, 14988–14993. doi: 10.1073/pnas.232308999
Lee, M., Choi, I., and Park, K. (2002). Activation of stress signaling molecules in bat brain during arousal from hibernation. J. Neurochem 2002:01022.x. doi: 10.1046/j.1471-4159.2002.01022.x
Lee, M., Hwang, J. T., Lee, H. J., Jung, S. N., Kang, I., Chi, S. G., et al. (2003). AMP-activated protein kinase activity is critical for hypoxia-inducible factor-1 transcriptional activity and its target gene expression under hypoxic conditions in DU145 cells. J. Biol. Chem. 278, 39653–39661. doi: 10.1074/jbc.M306104200
Lee, R. E. (2009). “Hibernation,” in Encyclopedia of Insects. V H. Resh and R T. Card (Cambridge, MA: Academic Press)449–449. doi: 10.1016/B978-0-12-374144-8.00127-2
Lee, T. M., Pelz, K., Licht, P., and Zucker, I. (1990). Testosterone influences hibernation in golden-mantled ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 259:r760. doi: 10.1152/ajpregu.1990.259.4.r760
Lee, Y. J., and Hallenbeck, J. M. (2013). SUMO and ischemic tolerance. NeuroMolecular Med. 15, 771–781. doi: 10.1007/s12017-013-8239-9
Lee, Y. J., Miyake, S. I., Wakita, H., McMullen, D. C., Azuma, Y., Auh, S., et al. (2007). Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J. Cereb. Blood Flow Metab. 27, 950–962. doi: 10.1038/sj.jcbfm.9600395
Li, H., Satriano, J., Thomas, J. L., Miyamoto, S., Sharma, K., Pastor-Soler, N. M., et al. (2015). Interactions between HIF-1α and AMPK in the regulation of cellular hypoxia adaptation in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 309, F414–F428. doi: 10.1152/ajprenal.00463.2014
Li, L., and Bhatia, R. (2011). Stem cell quiescence. Clin. Cancer Res. 17, 4936–4941. doi: 10.1158/1078-0432.CCR-10-1499
Liang, R., Rimmelé, P., Bigarella, C. L., Yalcin, S., and Ghaffari, S. (2016). Evidence for AKT-independent regulation of FOXO1 and FOXO3 in haematopoietic stem and progenitor cells. Cell Cycle 15, 861–867. doi: 10.1080/15384101.2015.1123355
Lim, S., and Kaldis, P. (2013). Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Dev. 140, 3079–3093. doi: 10.1242/dev.091744
Lin, A., and Karin, M. (2003). NF-κB in cancer: A marked target. Semin. Cancer Biol. 13, 107–114. doi: 10.1016/S1044-579X(02)00128-1
Liu, C., Han, T., Stachura, D. L., Wang, H., Vaisman, B. L., Kim, J., et al. (2018). Lipoprotein lipase regulates hematopoietic stem progenitor cell maintenance through DHA supply. Nat. Commun. 9, 03775–y. doi: 10.1038/s41467-018-03775-y
Liu, Y., Elf, S. E., Miyata, Y., Sashida, G., Liu, Y., Huang, G., et al. (2009). p53 Regulates Hematopoietic Stem Cell Quiescence. Cell Stem Cell 4, 37–48. doi: 10.1016/j.stem.2008.11.006
Lizcano, J. M., Göransson, O., Toth, R., Deak, M., Morrice, N. A., Boudeau, J., et al. (2004). LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843. doi: 10.1038/sj.emboj.7600110
Luk, C. K., Keng, P. C., and Sutherland, R. M. (1986). Radiation response of proliferating and quiescent subpopulations isolated from multicellular spheroids. Br. J. Cancer 54, 25–32. doi: 10.1038/bjc.1986.148
Lyle, S., and Moore, N. (2011). Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011:396076. doi: 10.1155/2011/396076
Ma, Y. L., Zhu, X., Rivera, P. M., Tøien, Ø, Barnes, B. M., LaManna, J. C., et al. (2005). Absence of cellular stress in brain after hypoxia induced by arousal from hibernation in Arctic ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005:00260. doi: 10.1152/ajpregu.00260.2005
Maistrovski, Y., Biggar, K. K., and Storey, K. B. (2012). HIF-1α regulation in mammalian hibernators: Role of non-coding RNA in HIF-1α control during torpor in ground squirrels and bats. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 182, 849–859. doi: 10.1007/s00360-012-0662-y
Majmundar, A. J., Wong, W. J., and Simon, M. C. (2010). Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol. Cell 40, 294–309. doi: 10.1016/j.molcel.2010.09.022
Malumbres, M., and Barbacid, M. (2005). Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30, 630–641. doi: 10.1016/j.tibs.2005.09.005
Malumbres, M., and Barbacid, M. (2009). Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 9, 153–166. doi: 10.1038/nrc2602
Martin, S. L. (2008). Mammalian hibernation: A naturally reversible model for insulin resistance in man? Diabetes Vasc. Dis. Res. 5, 76–81. doi: 10.3132/dvdr.2008.013
Martin, S. L., Maniero, G. D., Carey, C., and Hand, S. C. (1999). Reversible depression of oxygen consumption in isolated liver mitochondria during hibernation. Physiol. Biochem. Zool. 72, 255–264. doi: 10.1086/316667
Masaki, S. (2009). Aestivation. Encyclop. Insects 2009, 2–4. doi: 10.1016/B978-0-12-374144-8.00002-3
Matsui, K., Ezoe, S., Oritani, K., Shibata, M., Tokunaga, M., Fujita, N., et al. (2012). NAD-dependent histone deacetylase, SIRT1, plays essential roles in the maintenance of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 418, 811–817. doi: 10.1016/j.bbrc.2012.01.109
Matthews, L. H., and Fisher, K. C. (1968). Mammalian Hibernation III. J. Anim. Ecol. 37:724. doi: 10.2307/3087
Maynard, S., Swistowska, A. M., Lee, J. W., Liu, Y., Liu, S.-T., Da Cruz, A. B., et al. (2008). Human Embryonic Stem Cells Have Enhanced Repair of Multiple Forms of DNA Damage. Stem Cells 26, 2266–2274. doi: 10.1634/stemcells.2007-1041
McCain, S., Ramsay, E., and Kirk, C. (2013). The effects of hibernation and captivity on glucose metabolism and thyroid hormones in American black bear (Ursus Americanus). J. Zoo Wildl. Med. 44, 324–332. doi: 10.1638/2012-0146R1.1
McEachern, M. A., Yackel Adams, A. A., Klug, P. E., Fitzgerald, L. A., and Reed, R. N. (2015). Brumation of Introduced Black and White Tegus, Tupinambis merianae (Squamata: Teiidae), in Southern Florida. Southeast. Nat. 14, 319–328. doi: 10.1656/058.014.0207
McInnes, K. J., Brown, K. A., Hunger, N. I., and Simpson, E. R. (2012). Regulation of LKB1 expression by sex hormones in adipocytes. Int. J. Obes. 36, 982–985. doi: 10.1038/ijo.2011.172
Menon, V., and Ghaffari, S. (2018). Transcription factors FOXO in the regulation of homeostatic hematopoiesis. Curr. Opin. Hematol. 25, 290–298. doi: 10.1097/MOH.0000000000000441
Miyamoto, K., Miyamoto, T., Kato, R., Yoshimura, A., Motoyama, N., and Suda, T. (2008). FoxO3a regulates hematopoietic homeostasis through a negative feedback pathway in conditions of stress or aging. Blood 112, 4485–4493. doi: 10.1182/blood-2008-05-159848
Mizushima, N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139. doi: 10.1016/j.ceb.2009.12.004
Mohammad, K., Dakik, P., Medkour, Y., Mitrofanova, D., and Titorenko, V. I. (2019). Quiescence entry, maintenance, and exit in adult stem cells. Int. J. Mol. Sci. 20:20092158. doi: 10.3390/ijms20092158
Mohrin, M., Bourke, E., Alexander, D., Warr, M. R., Barry-Holson, K., Le Beau, M. M., et al. (2010). Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185. doi: 10.1016/j.stem.2010.06.014
Morin, P., and Storey, K. B. (2005). Cloning and expression of hypoxia-inducible factor 1α from the hibernating ground squirrel, Spermophilus tridecemlineatus. Biochim. Biophys. Acta Gene Struct. Expr. 2005:009. doi: 10.1016/j.bbaexp.2005.02.009
Morin, P., and Storey, K. B. (2006). Evidence for a reduced transcriptional state during hibernation in ground squirrels. Cryobiology 53, 310–318. doi: 10.1016/j.cryobiol.2006.08.002
Mortensen, M., Soilleux, E. J., Djordjevic, G., Tripp, R., Lutteropp, M., Sadighi-Akha, E., et al. (2011). The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208, 455–467. doi: 10.1084/jem.20101145
Murata, K., Hattori, M., Hirai, N., Shinozuka, Y., Hirata, H., Kageyama, R., et al. (2005). Hes1 Directly Controls Cell Proliferation through the Transcriptional Repression of p27Kip1. Mol. Cell. Biol. 25, 4262–4271. doi: 10.1128/mcb.25.10.4262-4271.2005
Murray, A. W. (2004). Recycling the Cell Cycle: Cyclins Revisited. Cell 116, 221–234. doi: 10.1016/S0092-8674(03)01080-8
Nakada, D., Saunders, T. L., and Morrison, S. J. (2010). Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658. doi: 10.1038/nature09571
Nakamura-Ishizu, A., Takizawa, H., and Suda, T. (2014). The analysis, roles and regulation of quiescence in hematopoietic stem cells. Dev. 141, 4656–4666. doi: 10.1242/dev.106575
Narbonne, P., and Roy, R. (2009). Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457, 210–214. doi: 10.1038/nature07536
Nystul, T. G., Goldmark, J. P., Padilla, P. A., and Roth, M. B. (2003). Suspended Animation in C. elegans Requires the Spindle Checkpoint. Science 302, 1038–1041. doi: 10.1126/science.1089705
Oberley, T. D., Schultz, J. L., Li, N., and Oberley, L. W. (1995). Antioxidant enzyme levels as a function of growth state in cell culture. Free Radic. Biol. Med. 19, 53–65. doi: 10.1016/0891-5849(95)00012-M
Orford, K. W., and Scadden, D. T. (2008). Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9, 115–128. doi: 10.1038/nrg2269
Osborne, P. G., Gao, B., and Hashimoto, M. (2004). Determination in vivo of newly synthesized gene expression in hamsters during phases of the hibernation cycle. Jpn. J. Physiol. 54, 295–305. doi: 10.2170/jjphysiol.54.295
Ou, J., Ball, J. M., Luan, Y., Zhao, T., Miyagishima, K. J., Xu, Y., et al. (2018). iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell 173, 851.e–863.e. doi: 10.1016/j.cell.2018.03.010
Papa, L., Djedaini, M., and Hoffman, R. (2019). Mitochondrial role in stemness and differentiation of hematopoietic stem cells. Stem Cells Int. 2019:4067162. doi: 10.1155/2019/4067162
Parmar, K., Mauch, P., Vergilio, J. A., Sackstein, R., and Down, J. D. (2007). Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc. Natl. Acad. Sci. U. S. A. 104, 5431–5436. doi: 10.1073/pnas.0701152104
Peng, H., Park, J. K., Katsnelson, J., Kaplan, N., Yang, W., Getsios, S., et al. (2015). MicroRNA-103/107 family regulates multiple epithelial stem cell characteristics. Stem Cells 33, 1642–1656. doi: 10.1002/stem.1962
Peretti, D., Bastide, A., Radford, H., Verity, N., Molloy, C., Martin, M. G., et al. (2015). RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature 518, 236–239. doi: 10.1038/nature14142
Perl, A., Gergely, P., Nagy, G., Koncz, A., and Banki, K. (2004). Mitochondrial hyperpolarization: A checkpoint of T-cell life, death and autoimmunity. Trends Immunol. 25, 360–367. doi: 10.1016/j.it.2004.05.001
Piccoli, C., Ria, R., Scrima, R., Cela, O., D’Aprile, A., Boffoli, D., et al. (2005). Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells: Novel evidence of the occurrence of NAD(P)H oxidase activity. J. Biol. Chem. 280, 26467–26476. doi: 10.1074/jbc.M500047200
Pietras, E. M., Warr, M. R., and Passegué, E. (2011). Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720. doi: 10.1083/jcb.201102131
Pilegaard, H., Saltin, B., and Neufer, P. D. (2003). Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes 52, 657–662. doi: 10.2337/diabetes.52.3.657
Pimienta, G., and Pascual, J. (2007). Canonical and alternative MAPK signaling. Cell Cycle 6, 2628–2632. doi: 10.4161/cc.6.21.4930
Porter, S. N., Cluster, A. S., Signer, R. A. J., Voigtmann, J., Monlish, D. A., Schuettpelz, L. G., et al. (2016). Pten cell autonomously modulates the hematopoietic stem cell response to inflammatory cytokines. Stem Cell Reports 6, 806–814. doi: 10.1016/j.stemcr.2016.04.008
Rahl, P. B., Lin, C. Y., Seila, A. C., Flynn, R. A., McCuine, S., Burge, C. B., et al. (2010). C-Myc regulates transcriptional pause release. Cell 141, 432–445. doi: 10.1016/j.cell.2010.03.030
Renfree, M. B., and Shaw, G. (2000). Diapause. Annu. Rev. Physiol. 62, 353–375. doi: 10.1146/annurev.physiol.62.1.353
Revuelta, M., and Matheu, A. (2017). Autophagy in stem cell aging. Aging Cell 16, 912–915. doi: 10.1111/acel.12655
Richmond, C. A., Shah, M. S., Carlone, D. L., and Breault, D. T. (2016). An enduring role for quiescent stem cells. Dev. Dyn. 245, 718–726. doi: 10.1002/dvdy.24416
Riether, C., Schürch, C. M., and Ochsenbein, A. F. (2015). Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 22, 187–198. doi: 10.1038/cdd.2014.89
Rigano, K. S., Gehring, J. L., Evans Hutzenbiler, B. D., Chen, A. V., Nelson, O. L., Vella, C. A., et al. (2017). Life in the fat lane: seasonal regulation of insulin sensitivity, food intake, and adipose biology in brown bears. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 187, 649–676. doi: 10.1007/s00360-016-1050-9
Rimmelé, P., Liang, R., Bigarella, C. L., Kocabas, F., Xie, J., Serasinghe, M. N., et al. (2015). Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO 3. EMBO Rep. 16, 1164–1176. doi: 10.15252/embr.201439704
Risal, S., Adhikari, D., and Liu, K. (2016). Animal models for studying the in vivo functions of cell cycle CDKs. Methods Mol. Biol. 1336, 155–166. doi: 10.1007/978-1-4939-2926-9_13
Rivard, N., L’Allemain, G., Bartek, J., and Pouysségur, J. (1996). Abrogation of p27(Kip1) by cDNA antisense suppresses quiescence (G0 state) in fibroblasts. J. Biol. Chem. 271, 18337–18341. doi: 10.1074/jbc.271.31.18337
Rocheteau, P., Vinet, M., and Chretien, F. (2014). Dormancy and quiescence of skeletal muscle stem cells. Results Probl. Cell Differ. 56, 215–235. doi: 10.1007/978-3-662-44608-9_10
Roilo, M., Kullmann, M. K., and Hengst, L. (2018). Cold-inducible RNA-binding protein (CIRP) induces translation of the cell-cycle inhibitor p27Kip1. Nucleic Acids Res. 46, 3198–3210. doi: 10.1093/nar/gkx1317
Ruf, T., and Geiser, F. (2015). Daily torpor and hibernation in birds and mammals. Biol. Rev. 90, 891–926. doi: 10.1111/brv.12137
Ruf, T., Bieber, C., and Turbill, C. (2012). “Survival, Aging, and Life-History Tactics in Mammalian Hibernators,” in Living in a Seasonal World. T. RufClaudia, B. Arnold, E. Millesi (New York, NY:Springer). doi: 10.1007/978-3-642-28678-0_11
Sabbatinelli, J., Prattichizzo, F., Olivieri, F., Procopio, A. D., Rippo, M. R., and Giuliani, A. (2019). Where Metabolism Meets Senescence: Focus on Endothelial Cells. Front. Physiol. 10:1523. doi: 10.3389/fphys.2019.01523
Sage, J., Miller, A. L., Pérez-Mancera, P. A., Wysocki, J. M., and Jacks, T. (2003). Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424, 223–228. doi: 10.1038/nature01764
Sánchez-Martínez, C., Gelbert, L. M., Lallena, M. J., and De Dios, A. (2015). Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorganic Med. Chem. Lett. 25, 3420–3435. doi: 10.1016/j.bmcl.2015.05.100
Sang, L., and Coller, H. A. (2009). Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle 8, 2161–2167. doi: 10.4161/cc.8.14.9104
Sang, L., Coller, H. A., and Roberts, J. M. (2008). Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100. doi: 10.1126/science.1155998
Sang, L., Roberts, J. M., and Coller, H. A. (2010). Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol. Med. 16, 17–26. doi: 10.1016/j.molmed.2009.11.001
Schreiber-Agus, N., Meng, Y., Hoang, T., Hou, H., Ghen, K., Greenberg, R., et al. (1998). Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393, 483–487. doi: 10.1038/31008
Schwartz, C., Hampton, M., and Andrews, M. T. (2013). Seasonal and Regional Differences in Gene Expression in the Brain of a Hibernating Mammal. PLoS One 8:0058427. doi: 10.1371/journal.pone.0058427
Seger, R., and Krebs, E. G. (1995). The MAPK signaling cascade. FASEB J. 9, 726–735. doi: 10.1096/fasebj.9.9.7601337
Seger, R., and Wexler, S. (2016). “The MAPK Signaling Cascades,” in Encyclopedia of Cell Biology. R. A. Bradshaw and P. D. Stahl (Amsterdam: Elsevier) doi: 10.1016/B978-0-12-394447-4.30014-1
Serra, V., Von Zglinicki, T., Lorenz, M., and Saretzki, G. (2003). Extracellular superoxide dismutase is a major antioxidant in human fibroblasts and slows telomere shortening. J. Biol. Chem. 278, 6824–6830. doi: 10.1074/jbc.M207939200
Shackelford, D. B., and Shaw, R. J. (2009). The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575. doi: 10.1038/nrc2676
Shen, C., and Nathan, C. (2002). Nonredundant antioxidant defense by multiple two-cysteine peroxiredoxins in human prostate cancer cells. Mol. Med. 8, 95–102. doi: 10.1007/bf03402079
Signer, R. A. J., Magee, J. A., Salic, A., and Morrison, S. J. (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 508, 49–54. doi: 10.1038/nature13035
Simonetti, G., Padella, A., Manfrini, M., do Valle, I. F., Papayannidis, C., Fontana, M. C., et al. (2016). Aggressive Aneuploid Acute Myeloid Leukemia Is Dependent on Alterations of P53, Gain of APC and PLK1 and Loss of RAD50. Blood 128, 1702–1702. doi: 10.1182/blood.v128.22.1702.1702
Simsek, T., Kocabas, F., Zheng, J., Deberardinis, R. J., Mahmoud, A. I., Olson, E. N., et al. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390. doi: 10.1016/j.stem.2010.07.011
Soukri, A., Valverde, F., Hafid, N., Elkebbaj, M. S., and Serrano, A. (1996). Occurrence of a differential expression of the glyceraldehyde-3-phosphate dehydrogenase gene in muscle and liver from euthermic and induced hibernating jerboa (Jaculus orientalis). Gene 181, 139–145. doi: 10.1016/S0378-1119(96)00494-5
Squire, T. L., Lowe, M. E., Bauer, V. W., and Andrews, M. T. (2004). Pancreatic triacylglycerol lipase in a hibernating mammal. II. Cold-adapted function and differential expression. Physiol. Genomics 16, 131–140. doi: 10.1152/physiolgenomics.00168.2002
Staples, J. F. (2014). Metabolic suppression in mammalian hibernation: The role of mitochondria. J. Exp. Biol. 217, 2032–2036. doi: 10.1242/jeb.092973
Staples, J. F., and Brown, J. C. L. (2008). Mitochondrial metabolism in hibernation and daily torpor: A review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 178, 811–827. doi: 10.1007/s00360-008-0282-8
Storey, K. B. (2003). Mammalian hibernation: Transcriptional and translational controls. Advances in Experimental Medicine and Biology 2003, 21–38. doi: 10.1007/978-1-4419-8997-0_3
Storey, K. B., and Storey, J. M. (2004). Metabolic rate depression in animals: Transcriptional and translational controls. Biol. Rev. Camb. Philos. Soc. 79, 207–233. doi: 10.1017/S1464793103006195
Storey, K. B., and Storey, J. M. (2012). Aestivation: Signaling and hypometabolism. J. Exp. Biol. 215, 1425–1433. doi: 10.1242/jeb.054403
Storey, K. B., Heldmaier, G., and Rider, M. H. (2010). Mammalian hibernation: Physiology, cell signaling, and gene controls on metabolic rate depression. Top. Curr. Genet. 21, 227–252. doi: 10.1007/978-3-642-12422-8_13
Sunagawa, G. A., Deviatiiarov, R., Ishikawa, K., Gazizova, G., Gusev, O., and Takahashi, M. (2018). Integrative Transcription Start Site Analysis and Physiological Phenotyping Reveal Torpor-specific Expressions in Mouse Skeletal Muscle. bioRxiv 2018:374975. doi: 10.1101/374975
Sureban, S. M., Ramalingam, S., Natarajan, G., May, R., Subramaniam, D., Bishnupuri, K. S., et al. (2008). Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene 27, 4544–4556. doi: 10.1038/onc.2008.97
Tähti, H. (1978). Seasonal differences in O2 consumption and respiratory quotient in a hibernator (Erinaceus europaeus L.). Ann. Zool. Fenn. 15, 69–75.
Takahashi, K., and Yamanaka, S. (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Takahashi, S., Tanaka, T., and Sakai, J. (2007). New therapeutic target for metabolic syndrome: PPARδ. Endocr. J. 54, 347–357. doi: 10.1507/endocrj.KR-99
Takubo, K., Goda, N., Yamada, W., Iriuchishima, H., Ikeda, E., Kubota, Y., et al. (2010). Regulation of the HIF-1α level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402. doi: 10.1016/j.stem.2010.06.020
Takubo, K., Nagamatsu, G., Kobayashi, C. I., Nakamura-Ishizu, A., Kobayashi, H., Ikeda, E., et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61. doi: 10.1016/j.stem.2012.10.011
Tessier, S. N., and Storey, K. B. (2014). To be or not to be: The regulation of mRNA fate as a survival strategy during mammalian hibernation. Cell Stress Chaperones 19:9. doi: 10.1007/s12192-014-0512-9
Tessier, S. N., Wu, C. W., and Storey, K. B. (2019). Molecular control of protein synthesis, glucose metabolism, and apoptosis in the brain of hibernating thirteen-lined ground squirrels. Biochem. Cell Biol. 97, 536–544. doi: 10.1139/bcb-2018-0256
Theeuwes, W. F., Gosker, H. R., Langen, R. C. J., Verhees, K. J. P., Pansters, N. A. M., Schols, A. M. W. J., et al. (2017). Inactivation of glycogen synthase kinase-3β (GSK-3β) enhances skeletal muscle oxidative metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 3075–3086. doi: 10.1016/j.bbadis.2017.09.018
Tinganelli, W., Hitrec, T., Romani, F., Simoniello, P., Squarcio, F., Stanzani, A., et al. (2019). Hibernation and radioprotection: Gene expression in the liver and testicle of rats irradiated under synthetic torpor. Int. J. Mol. Sci. 20:20020352. doi: 10.3390/ijms20020352
Tothova, Z., Kollipara, R., Huntly, B. J., Lee, B. H., Castrillon, D. H., Cullen, D. E., et al. (2007). FoxOs Are Critical Mediators of Hematopoietic Stem Cell Resistance to Physiologic Oxidative Stress. Cell 128, 325–339. doi: 10.1016/j.cell.2007.01.003
Trimarchi, J. M., and Lees, J. A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20. doi: 10.1038/nrm714
Tsiouris, J. A. (2005). Metabolic depression in hibernation and major depression: An explanatory theory and an animal model of depression. Med. Hypotheses 65, 829–840. doi: 10.1016/j.mehy.2005.05.044
Turbill, C., Bieber, C., and Ruf, T. (2011). Hibernation is associated with increased survival and the evolution of slow life histories among mammals. Proceedings.Biological Sci. R. Soc. 278, 3355–3363. doi: 10.1098/rspb.2011.0190
Unnisa, Z., Clark, J. P., Roychoudhury, J., Thomas, E., Tessarollo, L., Copeland, N. G., et al. (2012). Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood 120, 4973–4981. doi: 10.1182/blood-2012-06-435800
Valcourt, J. R., Lemons, J. M. S., Haley, E. M., Kojima, M., Demuren, O. O., and Coller, H. A. (2012). Staying alive: Metabolic adaptations to quiescence. Cell Cycle 11, 1680–1696. doi: 10.4161/cc.19879
Van Breukelen, F., and Martin, S. L. (2002). Reversible depression of transcription during hibernation. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 172, 355–361. doi: 10.1007/s00360-002-0256-1
Vermillion, K. L., Anderson, K. J., Marshall, H., and Andrews, M. T. (2015). Gene expression changes controlling distinct adaptations in the heart and skeletal muscle of a hibernating mammal. Physiol. Genomics 47, 58–74. doi: 10.1152/physiolgenomics.00108.2014
Vessoni, A. T., Muotri, A. R., and Okamoto, O. K. (2012). Autophagy in stem cell maintenance and differentiation. Stem Cells Dev. 21, 513–520. doi: 10.1089/scd.2011.0526
Viatour, P., Somervaille, T. C., Venkatasubrahmanyam, S., Kogan, S., McLaughlin, M. E., Weissman, I. L., et al. (2008). Hematopoietic Stem Cell Quiescence Is Maintained by Compound Contributions of the Retinoblastoma Gene Family. Cell Stem Cell 3, 416–428. doi: 10.1016/j.stem.2008.07.009
Wanet, A., Arnould, T., Najimi, M., and Renard, P. (2015). Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 24, 1957–1971. doi: 10.1089/scd.2015.0117
Wang, L., Siegenthaler, J. A., Dowell, R. D., and Yi, R. (2016). Stem cells: Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science 351, 613–617. doi: 10.1126/science.aad5440
Webb, P. I., and Ellison, J. (1998). Normothermy, torpor, and arousal in hedgehogs (erinaceus europaeus) from Dunedin. N. Zeal. J. Zool. 25, 85–90. doi: 10.1080/03014223.1998.9518139
Wellmann, S., Truss, M., Bruder, E., Tornillo, L., Zelmer, A., Seeger, K., et al. (2010). The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr. Res. 67, 35–41. doi: 10.1203/PDR.0b013e3181c13326
Wiersma, M., Beuren, T. M. A., de Vrij, E. L., Reitsema, V. A., Bruintjes, J. J., Bouma, H. R., et al. (2018). Torpor-arousal cycles in Syrian hamster heart are associated with transient activation of the protein quality control system. Comp. Biochem. Physiol. Part - B Biochem. Mol. Biol. 223, 23–28. doi: 10.1016/j.cbpb.2018.06.001
Wieser, W., and Krumschnabel, G. (2001). Hierarchies of ATP-consuming processes: Direct compared with indirect measurements, and comparative aspects. Biochem. J. 355, 389–395. doi: 10.1042/0264-6021:3550389
Wilson, A., Laurenti, E., Oser, G., van der Wath, R. C., Blanco-Bose, W., Jaworski, M., et al. (2008). Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 135, 1118–1129. doi: 10.1016/j.cell.2008.10.048
Wilz, M., and Heldmaier, G. (2000). Comparison of hibernation, estivation and daily torpor in the edible dormouse, Glis glis. J. Comp. Physiol. Biochem. Syst. Environ. Physiol. 170, 511–521.
Wu, C. W., and Storey, K. B. (2012a). Pattern of cellular quiescence over the hibernation cycle in liver of thirteen-lined ground squirrels. Cell Cycle 11, 1714–1726. doi: 10.4161/cc.19799
Wu, C. W., and Storey, K. B. (2012b). Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J. Exp. Biol. 215, 1720–1727. doi: 10.1242/jeb.066225
Wu, C. W., and Storey, K. B. (2016). Life in the cold: Links between mammalian hibernation and longevity. Biomol. Concepts 7, 41–52. doi: 10.1515/bmc-2015-0032
Wu, C. W., Bell, R. A., and Storey, K. B. (2015). Post-translational regulation of PTEN catalytic function and protein stability in the hibernating 13-lined ground squirrel. Biochim. Biophys. Acta Gen. Subj. 1850, 2196–2202. doi: 10.1016/j.bbagen.2015.07.004
Wu, C.-W., Bell, R., and Storey, K. B. (2013a). 49. Regulation of PTEN function and structural stability in hibernating thirteen-lined ground squirrels. Cryobiology 66, 355–356. doi: 10.1016/j.cryobiol.2013.02.055
Wu, J., Chen, P., Li, Y., Ardell, C., Der, T., Shohet, R., et al. (2013b). HIF-1α in heart: Protective mechanisms. Am. J. Physiol. Hear. Circ. Physiol. 305, H821–H828. doi: 10.1152/ajpheart.00140.2013
Wu, P., Inskeep, K., Bowker-Kinley, M. M., Popov, K. M., and Harris, R. A. (1999). Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes 48, 1593–1599. doi: 10.2337/diabetes.48.8.1593
Xu, R., Andres-Mateos, E., Mejias, R., MacDonald, E. M., Leinwand, L. A., Merriman, D. K., et al. (2013). Hibernating squirrel muscle activates the endurance exercise pathway despite prolonged immobilization. Exp. Neurol. 247, 392–401. doi: 10.1016/j.expneurol.2013.01.005
Yalcin, S., Marinkovic, D., Mungamuri, S. K., Zhang, X., Tong, W., Sellers, R., et al. (2010). ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3 -/- mice. EMBO J. 29, 4118–4131. doi: 10.1038/emboj.2010.292
Yan, J., Barnes, B. M., Kohl, F., and Marr, T. G. (2008). Modulation of gene expression in hibernating arctic ground squirrels. Physiol. Genomics 32, 170–181. doi: 10.1152/physiolgenomics.00075.2007
Yang, N., and Sheridan, A. M. (2014). “Cell Cycle,” in Encyclopedia of Toxicology.P. Wexler (Amsterdam:Elsevier) 753–758. doi: 10.1016/B978-0-12-386454-3.00273-6
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: A history of macroautophagy. Nat. Cell Biol. 12, 814–822. doi: 10.1038/ncb0910-814
Yao, G. (2014). Modelling mammalian cellular quiescence. Interface Focus 4:0074. doi: 10.1098/rsfs.2013.0074
Yilmaz, ÖH., Valdez, R., Theisen, B. K., Guo, W., Ferguson, D. O., Wu, H., et al. (2006). Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441, 475–482. doi: 10.1038/nature04703
Yu, W. M., Liu, X., Shen, J., Jovanovic, O., Pohl, E. E., Gerson, S. L., et al. (2013). Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 12, 62–74. doi: 10.1016/j.stem.2012.11.022
Yu, X., Alder, J. K., Chun, J. H., Friedman, A. D., Heimfeld, S., Cheng, L., et al. (2006). HES1 Inhibits Cycling of Hematopoietic Progenitor Cells via DNA Binding. Stem Cells 24, 876–888. doi: 10.1634/stemcells.2005-0598
Zhang, J., Grindley, J. C., Yin, T., Jayasinghe, S., He, X. C., Ross, J. T., et al. (2006). PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441, 518–522. doi: 10.1038/nature04747
Zhang, S., Hulver, M. W., McMillan, R. P., Cline, M. A., and Gilbert, E. R. (2014). The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 11:10. doi: 10.1186/1743-7075-11-10
Zhang, Y., Aguilar, O. A., and Storey, K. B. (2016). Transcriptional activation of muscle atrophy promotes cardiac muscle remodeling during mammalian hibernation. PeerJ 2016:2317. doi: 10.7717/peerj.2317
Zhegunov, G., Kudokotseva, E., and Mikulinsky, Y. (1988). Hyperactivation of protein synthesis in tissues of hibernating animals on arousal. Cryo-Left 9, 236–245.
Zhu, X., Bührer, C., and Wellmann, S. (2016). Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. 73, 3839–3859. doi: 10.1007/s00018-016-2253-7
Zhu, X., Smith, M. A., Perry, G., Wang, Y., Ross, A. P., Zhao, H. W., et al. (2005). MAPKs are differentially modulated in arctic ground squirrels during hibernation. J. Neurosci. Res. 80, 862–868. doi: 10.1002/jnr.20526
Zhu, X., Yan, J., Bregere, C., Zelmer, A., Goerne, T., Kapfhammer, J. P., et al. (2019). RBM3 promotes neurogenesis in a niche-dependent manner via IMP2-IGF2 signaling pathway after hypoxic-ischemic brain injury. Nat. Commun. 10:11870–x. doi: 10.1038/s41467-019-11870-x
Keywords: cell cycle, cell dormancy, hibernation, metabolism, torpor, quiescence
Citation: Dias IB, Bouma HR and Henning RH (2021) Unraveling the Big Sleep: Molecular Aspects of Stem Cell Dormancy and Hibernation. Front. Physiol. 12:624950. doi: 10.3389/fphys.2021.624950
Received: 01 November 2020; Accepted: 11 March 2021;
Published: 01 April 2021.
Edited by:
Alessandro Silvani, University of Bologna, ItalyReviewed by:
Kelly Drew, University of Alaska Fairbanks, United StatesCopyright © 2021 Dias, Bouma and Henning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert H. Henning, ci5oLmhlbm5pbmdAdW1jZy5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.