
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 09 April 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.571826
 John E. Ramos1
John E. Ramos1 Ritesh G. Jain2
Ritesh G. Jain2 Charles A. Powell2
Charles A. Powell2 William O. Dawson3
William O. Dawson3 Siddarame Gowda3
Siddarame Gowda3 Dov Borovsky4*
Dov Borovsky4* Robert G. Shatters Jr.1
Robert G. Shatters Jr.1Citrus Greening or Huanglongbing (HLB) is a disease of citrus, causing high reduction in citrus production and is transmitted by the Asian citrus psyllid Diaphorina citri Kuwayama vectoring a phloem-limited bacterium Candidatus Liberibacter sp. We report research results using crowdsourcing challenge strategy identifying potential gene targets in D. citri to control the insect using RNA interference (RNAi). From 63 submitted sequences, 43 were selected and tested by feeding them to D. citri using artificial diet assays. After feeding on artificial diet, the three most effective dsRNAs causing 30% mortality above control silenced genes expressing iron-sulfur cluster subunit of the mitochondrial electron transport chain complex (Rieske), heme iron-binding terminal oxidase enzyme (Cytochrome P450) and tetrahydrobiopterin (BH4) pathway enzyme (Pterin 4α-Carbinolamine Dehydratase). These sequences were cloned into a citrus phloem-limited virus (Citrus tristeza virus, CTV T36) expressing dsRNA against these target genes in citrus. The use of a viral mediated “para-transgenic” citrus plant system caused higher mortality to adult D. citri than what was observed using artificial diet, reaching 100% when detached citrus leaves with the engineered CTV expressing dsRNA were fed to adult D. citri. Using this approach, a virus-induced gene silencing (VIGS) can be used to test future transgenic cultivars before genetically engineering citrus. RNA Seq analysis after feeding D. citri CTV-RIE on infected leaves identified transcriptionally modified genes located upstream and downstream of the targeted RIE gene. These genes were annotated showing that many are associated with the primary function of the Rieske gene that was targeted by VIGS.
Citrus Greening or Huanglongbing (HLB) is a disease of citrus, first discovered in the state of Florida in 2005 (Stokstad, 2012), that is responsible for a greater than fifty percent reduction in citrus production and is now present in other citrus producing states (National Agricultural and Statistics Service, 2005; Putnman et al., 2011). D. citri (Diaphorina citri Kuwayama) is the only known insect vector of the HLB associated phloem-limited bacterium Candidatus Liberibacter sp. In spite of aggressive approach to control D. citri, the disease has continued to spread in Florida and there is concern over the loss of pesticide effectiveness and residue accumulation in the fruit and in the environment (Ichinose et al., 2010; Tiwari et al., 2011). In an effort to develop a biological pest control strategy a global crowd sourcing challenge was issued by the United States citrus industry to identify the best D. citri transcript target(s) for RNA interference (RNAi) control strategy. RNAi is a eukaryotic genetic defense strategy that evolved in order to confer primitive immunity to infection (Covey et al., 1997; Qiu et al., 2005; Ben Berkhout, 2006; Marques et al., 2006; Wang et al., 2006; Maillard et al., 2013). dsRNAs from Viral RNA genomes and foreign transcripts are recognized by RNase III (an endoribonuclease) that clips dsRNA molecules into small (20–25 base) fragments, or small interfering RNAs (siRNAs). A single siRNA strand is then incorporated into the RNA Induced Silencing Complex (RISC), which uses the siRNA to bind (by hybridization) mRNAs in the cytosol by sequence complementary recognition. The catalytic subunit of the RISC (a member of the Argonaut protein family with endonuclease activity) then cleaves the mRNA rendering it non-functional (Bernstein et al., 2001; Lipardi et al., 2001). This effect can be redirected against the host cell by the exogenous introduction of dsRNAs having sequence identity for host genes, resulting in the reduction of host cell mRNA abundance and subsequent loss of function (Fire et al., 1998; Gregory et al., 2005). RNAi silencing of host genes has previously been demonstrated in eukaryotic organisms (including insects) following oral uptake of synthetic dsRNA molecules (Velásquez et al., 2009; Borgio, 2010; Gong et al., 2011; Pitino et al., 2011; Wuriyanghan et al., 2011; Galdeano et al., 2017; Yu et al., 2017; Yu and Killiny, 2018, 2020; Lu et al., 2019).
Preliminary reports suggest that D. citri and Bactericera cockerelli (potato/tomato psyllid) are vulnerable to dsRNA induced RNAi toxicity (Wuriyanghan et al., 2011; El-Shesheny et al., 2013; Galdeano et al., 2017; Yu et al., 2017; Yu and Killiny, 2018; Lu et al., 2019). These observations led to the support of the Florida Citrus industry through the Florida Research and Development Foundation (CDRF) to fund a web-based challenge through InnoCentive, Inc. Sequences that induce an RNAi-based toxicity in D. citri were identified by “solvers” (scientists and experts) as potential D. citri gene targets for silencing by oral uptake of synthetic dsRNAs using artificial diet (Hall et al., 2010). Forty-three genes were ultimately targeted and screened for susceptibility to gene silencing causing D. citri mortality. Of these, the Rieske gene, that expresses an iron sulfur cluster, a soluble domain of Complex III of the electron transport chain (ETC), is known to cause mortality in other insects. Therefore, proteins involved in ETC aerobic metabolism could be used as potential targets for pest control using RNA interference (Wuriyanghan et al., 2011). In addition, RNAi targeting was observed on G-protein coupled receptors with transmembrane sequences like chemokine receptors inhibiting HIV-1 replication (Martinez et al., 2002), as well as soluble proteins like juvenile hormone acid methyl transferase, causing inhibition of pupal to adult development in mosquitoes (Van Ekert et al., 2014).
Delivery of dsRNA to target insects is usually done by injection or by oral feeding using artificial diets. The latter is preferred because it does not cause injury to the target insect and was successfully demonstrated with Aedes aegypti larvae, Triatomine Bug (Rhodnius prolixus), light brown apple moth (Epiphyas postvittana), tsetse fly (Glossina morsitans morsitans), and diamondback moth (Plutella xylostella) (Velásquez et al., 2009; Borgio, 2010; Gong et al., 2011; Wuriyanghan et al., 2011; Van Ekert et al., 2014). Initially, dsRNA was mixed with the artificial diet that was used to feed insects. Later, long hair pin dsRNA was expressed in specific target plants to control insects (Martinez et al., 2002; Hall et al., 2010; Wuriyanghan et al., 2011; El-Shesheny et al., 2013) or in yeast cells (Van Ekert et al., 2014) to control mosquito larvae. A novel new technique using Citrus tristeza virus (CTV) expressing dsRNA in citrus has been recently introduced. Using this technique an abnormal wing disk (Awd) dsRNA has been expressed in citrus causing D. citri nymphs that fed on CTV infected trees to develop into adults with malformed wings (Hajeri et al., 2014). Bioassays using in planta approach was used by Taning et al. (2016) showed that D. citri is sensitive to ingested dsRNA showing a strong RNAi response prompted us to explore the potential of RNAi strategy to control the Asian citrus psyllid.
This report describes the biological activity of dsRNA molecules initially tested by feeding adult C. citri using artificial diet. Although many dsRNA molecules caused mortalities at a high dose, we selected only dsRNA molecules that exhibited dose response RNA interference. Because CTV is the most abundant virus in Florida citrus and is found where D. citri populations affect citrus (Britt et al., 2020) we engineered these molecules into attenuated CTV and expressed them in citrus leaves to protect against D. citri. We also report that targeting ETC membrane proteins by dsRNA in artificial diet is not as effective as targeting the soluble ETC subunit Rieske. These results suggest that soluble extra membranous domains such as Rieske or those that share similar functionality (iron-sulfur binding), are more sensitive to RNA interference (Boykin et al., 2012).
Diaphorina citri were reared on Citrus macrophylla plants in screen cages in an insectary at 25°C with 13:11 h light: dark cycle. Every week, 200 D. citri were removed to freshly flushing plants, allowed to lay eggs for 7 days and then removed from the plant leaves by aspiration. Nymphs and eggs were collected by brushing them off the flush with a small paint brush or by gently removing them with the tip of a pin. Insects that were used for RNA isolation were stored at −80°C.
Bioassays were performed using 4-week-old D. citri (10–20 male and female adults) in a 35 mm × 10 mm Greiner Bio-One petri dishes. D. citri, were contained in dishes with stretched parafilm and diet containing dsRNA (200 μL) (Supplementary Table 1) was applied to the exterior-top surface of the stretched parafilm and sandwiched by a second sheet of stretched parafilm. Adult D. citri (10–20 per group) 1 week after adult eclosion were fed on four different concentrations of dsRNAs for 6 days. Control groups were fed artificial diets without dsRNA (Hall et al., 2010) and each feeding experiment was repeated six times. Forty-three different dsRNA (48 ng/μL) were fed to D. citri for 6 days and adult mortality expressed as percentage was recorded. Results were corrected for mortalities caused by diet without dsRNA and expressed as means of 6 determinations ± S.E.M. (Figure 1A). Student’s t-tests using Graphpad Prism 6.0 were used for statistical analyses of the data for all the 43 dsRNA. Bioassays were performed weekly for 6 days and three gene targets were tested using dsRNA (275–325 bp) enriched diets at concentrations of 48, 24, 12, 6, and 3 ng/μL and each assay was repeated six-times. Homologous D. citri sequences were identified using existing genomic and transcriptomic data mining, primers were designed to amplify a short region (275–325 bp) of each gene which served as a template for dsRNAs synthesis using the MEGAscript RNAi kit (Life Technologies) (Supplementary Tables 1, 2). Diet without dsRNA was used to measure background mortality (Hall et al., 2010) that were subtracted from the mortality in each chamber. Since dsRNA of gfp a gene from Aequorea victoria that is not found in psyllids it has a very low effect on psyllid mortality (Figure 1C) testing psyllids without the addition of dsRNA to the artificial media is a good control to check for background mortality as was shown earlier by Hall et al. (2010). Feeding was stopped at day 6 because mortalities beyond day 6 often exceeded 40% when D. citri were fed on diet alone without dsRNA. Each experiment was repeated at least six times.
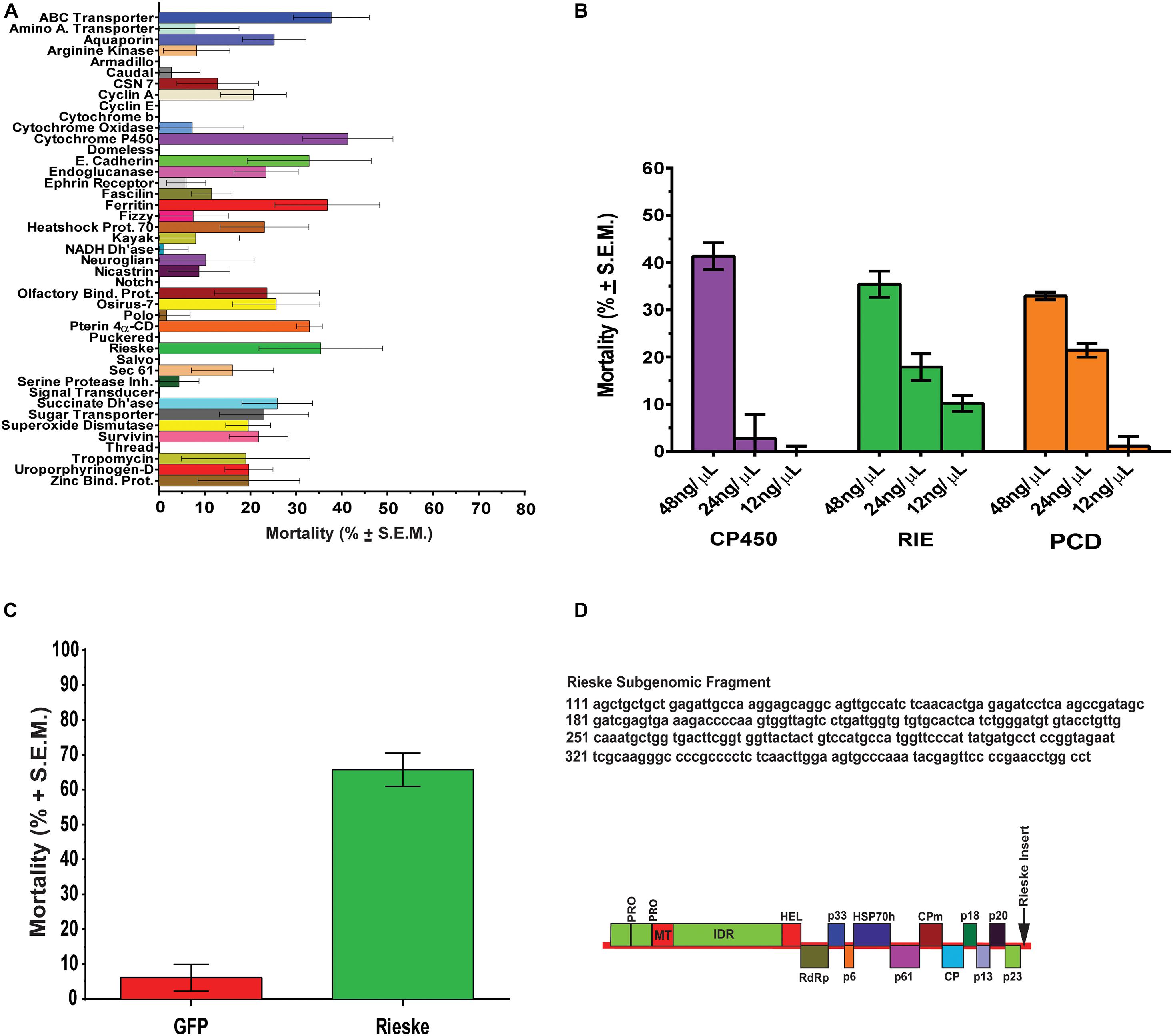
Figure 1. RNA interference studies. (A) Mortality of D. citri after feeding on dsRNA (48 ng/μL) of different target genes. The results are expressed as mean mortalities (%) corrected with mortalities observed by feeding on diet without dsRNA (control) of 6 determinations ± S.E.M. Twenty dsRNAs out of 43 tested caused significant mortalities above control diets. (B) Dose response of genes (CP450, RIE, and PCD) that were most significantly down regulated by a single concentration of dsRNA (48 ng/μL) in (A) using concentrations of 48, 24, and 12 ng/μl, respectively. (C) Effect of engineered CTV-RIE and CTV-gfp on D. citri feeding on C. macrophylla infected leaves using single leaf bioassays. The infected C. macrophylla leaves expressed RIE (Rieske) and gfp subgenomic dsRNAs of Green Fluorescent Protein of A. victoria GFP gene sequence that does not share sequence identity with D. citri genes. Mortalities are expressed as means of 6 determination ± SEM and were corrected with mortalities observed by feeding D. citri on leaves infected with CTV-wt. (D) CTV viral amplicon used to infect C. macrophylla is shown including the RIE subgenomic sequence that was cloned at the p23 locus in the 3′-NTR (non-translated region, arrow).
Citrus tristeza virus is a single-stranded (+ sense) RNA virus that forms a dsRNA replicative genome form at p23, a 3′ replicon, and expresses variable lengths of subgenomic dsRNAs (sgRNA) (Figure 1D). The 283 bp subgenomic Rieske sequence (RIE) was cloned into pCTV9, a cDNA clone of CTV T36 (GenBank Accession number AY170468), between the p23 locus and the 3′NTR (Figure 1D) using PacI and StuI restriction sites (Ayllón et al., 2003; Tatineni et al., 2008).
The engineered virus was amplified by PCR and transfected into Nicotiana benthamiana using polyethylene glycol (Mehta et al., 2009; Sztal et al., 2012) and virions extracted from snap frozen protoplasts in 40 mM Na2HPO4 buffer pH 8.2 (Navas-Castillo et al., 1998; Satyanarayana et al., 1999). After centrifugation for 4 min at 150 g, the supernatant was screened for CTV by Northern blot analysis using DIG Northern Starter Kit (Roche) and Northern Max Kit (Life Technologies) and inoculated into 1 year old C. macrophylla trees and allowed to incubate for 6 months. After incubation, leaves were extracted and screened by RT-qPCR using primers against RIE dsRNA sequence expressed by CTV. Relative CTV titer was determined using a ratio of CTV amplification efficiency (ACTV) to citrus dehydrin (CD) gene (a ubiquitous citrus reference gene) amplification efficiency (ACD), before inoculating the genetically engineered CTV into C. macrophylla for virus-induced gene silencing (VIGS). An average relative CTV titer of 5.3 for this study was calculated using the equation: (ACTV–Ct)/(ACD–Ct) (Livak and Schmittgen, 2001; Yuan et al., 2006).
Single and fully expanded young leaves were removed from citrus trees that were inoculated with genetically engineered CTV and the petiole placed in Eppendorf tube filled with water (1.2 mL). The tubes were sealed with parafilm and transferred into a conical tube (50 mL) containing adult D. citri (15–20) and the tube capped with a screened lid (El-Desouky et al., 2013). Adult D. citri were fed on leaves infected with engineered CTV-RIE and compared with adult D. citri that were fed on leaves infected with CTV-GFP.
RNA was extracted from 10 whole adult D. citri or surgically removed 20 guts with TRIzol® (Life Technologies, Carlsbad, CA, United States) following a modified manufacturer protocol (Chomczynski and Sacchi, 1987). Briefly, adults and guts were separately transferred into tubes containing acid-washed glass beads (180 μm) (Sigma). TRIzol was added and the samples were incubated for 5 min at room temperature. Tissues were broken in a Fast Prep instrument for 40 s at speed 6 m/s, chloroform added, and the samples incubated for 10 min at room temperature. Tissues were pelleted by centrifugation (12,000 rpm for 10 min at 4°C) and the aqueous phase transferred into a fresh tube. RNA was precipitated with isopropanol and samples were incubated at room temperature for 10 min, centrifuged (12,000 rpm for 8 min at 4°C). The pellet was washed with 75% ethanol, samples centrifuged (8,500 rpm for 5 min at 4°C) and ethanol was removed, and the RNA pellet air-dried for 10 min. The pellets were dissolved in water, and RQ1 DNase and DNase buffer (Promega, Madison, WI, United States) were added and samples incubated at 37°C for 1 h. After incubation, water, and phenol (pH 4.3) were added and the samples centrifuged (12,000 rpm for 6 min at 4°C), the upper phase transferred into a fresh tube and an equal volume of phenol/chloroform was added, samples centrifuged, and the upper phase transferred into a fresh tube and precipitated in sodium acetate (3M, pH 5.2) and 100% ethanol at −20°C overnight. The pellets were centrifuged (12,000 rpm for 15 min at 4°C), washed in 75% ice cold ethanol, re-centrifuged (12,000 rpm for 8 min at 4°C) and the ethanol removed and the pellet, air dried and suspended in water. RNA concentrations were determined at A260 and A280 in a NanoDrop 1000 instrument (Thermo Fisher Scientific, Waltham, MA, United States), and stored at −80°C. Extracted RNA was amplified using SMARTTM mRNA amplification kit (Clonetech) using SMARTTM MMLV Reverse Transcriptase. Poly G SMARTTM T7 extension primers, Advantage 2 polymerase mix, and RNAse H were used to create a sense cDNA template with T7 promoter. SMARTTM RNA polymerase was used to amplify a sense mRNA with linear amplification maintaining a relative constant cDNA copy number (Clonetech). The cDNA and mRNA were purified using NucleoSpin Gel and PCR Clean-up and NucleoSpin RNA II Purification kit (Macherey-Nagel), respectively. The concentration, nucleotide size distribution, and RNA integrity number (RIN) of the purified mRNA was determined using a 2100 Bioanalyzer and RNA 6000 Pico Reagents Kit and 2100 Expert software (Agilent Technologies). RNA was synthesized in a thermocycler at 37°C for 7 h using MEGAscript kit (Life Technologies) and linear dsDNA primers (1–2 μg) with T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′).
Linear dsDNA was synthesized using real-time reverse transcriptase (RT) PCR in RotorGene 6000 thermocycler using Rotor-Gene SYBR Green Two-Step RT-PCR Kit (Qiagen). Adult D. citri extracted mRNA (250 ng) was used as template in 12.5 μL reactions. Primers were designed using Generunner software1 with optimal tm in 50 nM NaCl of 60°C and product length of 300 bp. NCBI Blast, Flybase.org, and sohomopter.org databases were used to obtain D. Citri gene sequences. Thermocycler settings for the PCR were: 10 min at 95°C for the reverse transcription step followed with 40 cycles of 95°C for 5 s, 55°C for 15 s, and 72°C for 30 s. Melt was performed from 60 to 95°C with 0.2°C increments. Following PCR, the amplified DNA was purified using NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). T7 dsDNA was amplified with Platinum PCR SuperMix (Life Technologies) and purified using NucleoSpin PCR Clean-up kit (Macherey-Nagel).
Quantitative Real-time Reverse Transcriptase (RT-qPCR) was run as was described for RT-PCR (above). The following primer set amplifying a region of the D. citri Ribosomal S20 (ew) psyllid gene (GenBank Accession # number DQ673424) was used as an internal reference to quantify psyllid cell number in each reaction: using primers: forward (5′ GCCCAAGGGCCCAATCA 3′, tm 69°C) and reverse (5′ GGAGTCTTACGGGTGGTTATTCTG 3′, tm 67°C) (Ammar et al., 2016). Analysis was performed using RotorGene 6000 software and the results were normalized using a D. citri ubiquitous ribosomal protein [RPS20 as a reference gene and ΔΔCt (Livak and Schmittgen, 2001; Wilkening and Bader, 2004; Yuan et al., 2006)]. Comparative quantitation was used to acquire representative Ct take-off points (TOP) and representative amplification efficiencies (A) for target (target) and reference genes (ref) in experimental (exp) and control (cntrl) conditions. The following formula for relative mRNA abundance was used: (Aexp–targetTOP/Acntrl–targetTOP)/(Aexp–refTOP/Acntrl–refTOP). A primer set flanking the RIE dsRNA subgenomic sequence, not to detect synthetic or viral RIE dsRNA, was used to determine the relative as compared with controls of Rieske mRNA abundance in total RNA of tissue extracts. These results were normalized with a ubiquitous ribosomal protein (RPS20) of D. citri reference gene using ΔΔCt as follows: (ARIE–RIECt/Acntrl–RIECt)/(ARIE–RPSCt/Acntrl–RPSCt), where ARIE is the amplification of the dsRNA RIE treated sample, Acntrl is the amplification of the diet alone sample, RIE Ct is the cycle threshold (Ct) using Rieske primers forward (5′ GGCTGATGTGTTGGCAATGG 3′, tm 63°C) and reverse (5′ CTAAGTGTCTCGCGGCTGGTGTG 3′, tm 52°C) and RPSCt is the Ct using RPS20 primers (Livak and Schmittgen, 2001; Wilkening and Bader, 2004; Yuan et al., 2006).
Total RNA was extracted from D. citri guts that were fed on CTV-wt and CTV-RIE infected leaves using TRIzol reagent (GIBCO BRL, Gaithersburg, MD, United States) following modified manufacturer protocol (Lee et al., 2002; Griebler et al., 2008). Denatured gut mRNA (5 μg per lane) was run by electrophoresis on 1.5% denaturing gel using Northern MAX kit (Ambion Life Technology, Austin, TX, United States), transferred to BrightStar+ nylon membrane (Ambion) and hybridized with a DIG-labeled 273 bp cDNA probe (Roche, Life Sciences) that was amplified by PCR from D. citri Rieske by using primers RIE (forward) (5′-AGCTGCTGCTGAGATTGCCAAGG-3′) and RIE (reverse) (5′-GCTCAAGGGGCTTGGACCGGA-3′) using a melt cycle at 94°C for 60 s following with 94°C for 20 s, 55°C for 20 s, and 72°C for 60 s for 30 cycles and an extension cycle of 72°C for 10 min. The blot was washed according to the manufacturer’s instructions and exposed to X-ray film for 24 h. The Rieske mRNA transfer of CTV-RIE and CTV-wt fed D. citri guts was compared with Ribosomal RNA of D. citri guts that were similarly treated.
Whole transcriptome of D. citri gut cDNA library was constructed from adult D. citri gut mRNA (500 ng) (see RNA preparation above) that were fed on citrus leaves that were infected or not infected with engineered CTV. The purified transcript was treated with RNase III hydrolyzing dsRNA and stem loop structures and creating uniformly sized mRNAs (100–200 nt). The transcripts were hybridized for 1 h at 30°C with an adapter at the 5′PO4 and 3′OH sites following by reverse transcription with SuperScript® III enzyme mix for 30 min at 42°C. The cDNA was then amplified by PCR using Platinum® PCR SuperMix (Life Technologies) and barcode primers that are computer-identifiable and sequence specific and the PCR amplification conditions followed the manufacturer specifications (Life Technologies). Each cDNA library was constructed three times and amplified with different bar code primers. Each reverse transcription and PCR reaction was treated with magnetic bead cleanup kit (Life Technologies) and cDNA size selection (200 bp or higher) was done using 2% E-Gel (Life Technologies). The concentration and size distribution of the cDNA library was quantified using Bioanalyzer HS DNA Reagent Kit (Agilent Technologies). Libraries with average length of 150–300 bp and at concentration of 20 pM were clonal amplified and fixed on ion sphere particles (ISP) with ion one touch 2 instrument using ion PGM template OT2 200 kit (Life Technologies). Template positive ISPs were enriched with Ion ES, Ion PGM Template OT2 Solutions 200 Kit, and Dynabeads® MyOneTM Streptavidin C1 Beads (Life Technologies). The Dynabeads® ISPs complexes were immobilized with a magnet, washed thoroughly and afterward the ISPs were melted off releasing the ISPs- cDNAs conjugates. The percentage of template positive ISPs was quantified using Qubit 2.0 Fluorimeter and Ion SphereTM quality control kit by measuring the ratio of fluorescence emission from Alexa-Fluor 488 probe-hybridized ISPs and Alexa-Fluor 647 probe-hybridized template. Samples with a ratio of relative fluorescence emission (647 nm/488 nm) of 4–50% were used following manufacturer guidelines. The amplified ISP-cDNAs were annealed with sequencing primers by melting the cDNA at 95°C for 2 min and then annealing the primers at 37°C for 2 min following by the addition of DNA sequencing polymerase and the mixture added to Ion 318 chip and sequenced using PGM sequencer (Life Technologies).
cDNA libraries of adult D. citri guts that were isolated after feeding for 4 days on engineered CTV infected leaves were transcribed, deposited on Ion 318 chip (Ion Torrent, Life Technologies) and sequenced. More than 4 × 106 reads (length 130–200 bp) were obtained after feeding D. citri on CTV-RIE, CTV-gfp and CTV-free leaves. Only high quality (>90% and containing <40% polyclonal) sequencing runs (2 – 6 × 106 reads) were used. A de novo D. citri reference assembly was created in Geneious 7.0 (Biomatters) using sequences from www.sohomoptera.org (Reese et al., 2014). A comparative RNA Seq analysis using QSeq and Array Star (Lasergene 11.0 suite) generated 100,517 combined reference contig hits for CTV-RIE fed and control (CTV-free) gut sequence. Moderated t-tests and 99% confidence intervals were used to analyze the reference hits that contained 149 contigs that were 8-fold differentially expressed showing increase or decrease between citrus leaves fed to D. citri expressing CTV-RIE and CTV-free leaves. No differentially expressed genes were found in gut transcriptomes of CTV-gfp fed and control D. citri.
Diaphorina citri contigs were analyzed by blastx against UniProt (SwissProt), NCBI (nr), and refseq_protein, databases in Blast2Go and in flybase.org using 1E–6 cut off. From 149 significant genes 25 were reliably annotated with GO terminology, showing contigs that are over and under expressed in D. citri gut transcriptome fed CTV-RIE (Supplementary Figure 2 and Supplementary Table 3). Blast2Go was used to generate GO ontology maps for the two GO term categories, biological process, and molecular function (Figure 2). The ontology map denotes under or over expressed contigs and shared processes or functions between related annotations. Similar annotations were grouped by node score (NS) [NS = Σ#Seqs × α (α = dist to node)] (Conesa et al., 2005; Götz et al., 2008; Figure 2).
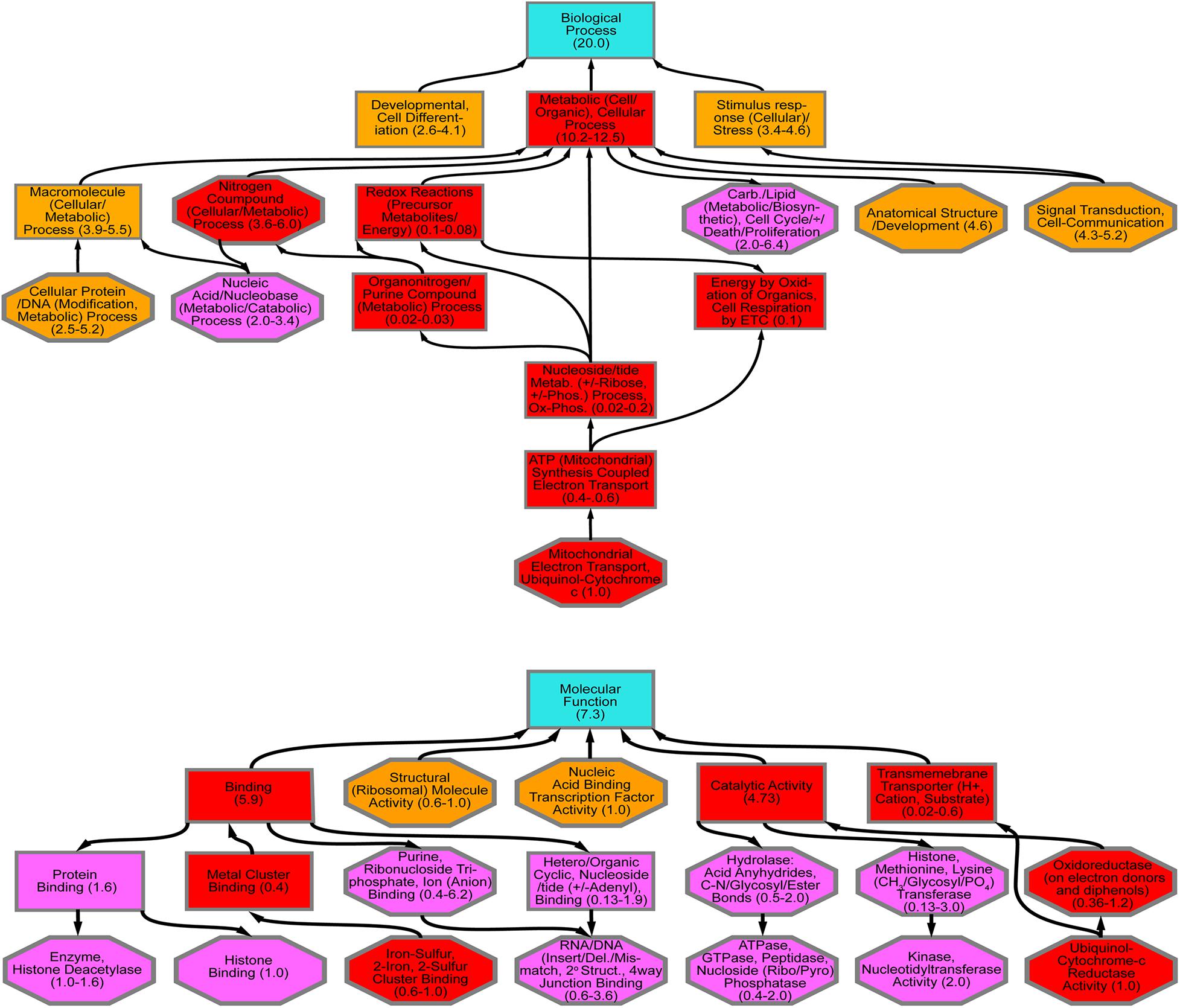
Figure 2. GO Annotation of comparative RNA Seq Transcriptome analysis. Contigs with the most significant variations showing up or down transcriptional abundance were identified in D. citri gut transcriptome after feeding on C. macrophylla leaves engineered with CTV-RIE. Using two major categories of GO Ontology (Biological Process and Molecular Function upper and lower hierarchical maps, respectively). The hierarchical maps show groups of contig specific annotations (in octagons) linked by shared functions or processes (in rectangles) each with a range of node scores in parenthesis. Each term in an octagon or rectangle is associated with a node score [Node Score = (no. of sequences) × αdist, with α = constant = 0.6, dist = a measure of the relative association to the node term]. Annotations of the contig identified as the Rieske gene are contained in the lowest octagons in each map above. Upward arrows connect to other red rectangles and octagons and represent “downstream” effects. Downward arrows (from red shapes) connect to magenta shapes that represent “upstream effects.” Orange shapes contain terms that are not (or have not yet been) associated with downstream effects of dsRNA-RIE.
Data were analyzed by Two-Way ANOVA to identify diets significant in mortality. Diets targeting genes with more than four significant bioassays were tested for normality by D’Agostino and Pearson’s Test followed by Two-Way ANOVA for mortality and dose response. Dunnett’s Multiple Comparison Test and One-Way ANOVA with F-test for equal variance, as well as Unpaired Student t-tests with Bartlett’s Test for variance were also used. All statistical analyses were performed using Graph pad Prism 6.0. Results were considered statistically significant when p < 0.05 and results are expressed as means of 6 determinations ± S.E.M., except where otherwise stated.
Using crowd-sourcing challenge 43 unique dsRNA sequences targeting specific D. citri genes were identified (Supplementary Table 1). The proposed dsRNA molecules targeted diverse groups of key biological processes in D. citri. In order to synthesize dsRNAs for each target homologous D. citri sequences were identified using genomic and transcriptomic data mining and primers were designed to amplify a 200–300 bp dsRNAs. Feeding of different concentrations of the dsRNAs (Supplementary Table 1) (48, 24, and 12 ng/μL) to psyllids were statistically analyzed. Lower dsRNA concentrations (6 and 3 ng/μL) were not significantly effective and the lowest effective dsRNA concentration is 12 ng/μL. Twenty-one diets of 43 tested dsRNAs, showed significant mortality when fed 48 ng/μL, however, only 3 dsRNA (Rieske, Cytochrome P450 4g15-like2 and Pterin 4α-Carbinolamine Dehydratase) showed significant mortalities at 48 ng/mL greater than 30% mortality above diet alone and significant mortalities at concentrations of 24 ng/μL and 12 ng/μL (Tables 1, 2). To find out if the mean mortalities of the three tested dsRNA concentrations that were fed to psyllids are significantly different in causing mortalities, and to find out if the mean mortalities between different pairs (48–24 ng/mL and 24–12 ng/mL) are also statistically significant. Data were analyzed by ANOVA multiple comparison tests with Fisher’s LSD (Tables 2, 3). The three most effective dsRNA targeted genes express (a) an iron-sulfur cluster subunit of the mitochondrial electron transport chain complex (Rieske), (b) a heme iron-binding terminal oxidase enzyme (Cytochrome P450), and (c) a tetrahydrobiopterin (BH4) pathway enzyme (Pterin 4α-Carbinolamine Dehydratase) (Tables 2, 3). In addition, three other dsRNA caused mortalities greater than 30%, only at a concentration of 48 ng/μL, they include: (d) a small molecule-ion ATP Binding Cassette Transporter subunit (ABC Transporter), (e) a ubiquitous iron binding storage protein (Ferritin), and (f) a cell adhesion calcium binding protein (E-Cadherin). Five of these six proteins (a, b, d, e, and f above) are important in cellular maintenance of iron levels (Iwata et al., 1996; Bilello et al., 2003; Mehta et al., 2009; Sztal et al., 2012; Yan et al., 2012). Sequence repeats of 3–9 base pairs that were reported in the literature in successful RNA interference studies were also identified in the dsRNAs that were used in this study. Three of these sequences, GCCC, ATGC, and TTCG were identified in the subgenomic Rieske sequence (three, two, and three times, respectively) whereas ATGC in Pterin 4α-CD once (Table 4; Elbashir et al., 2001; Yoshinari et al., 2004). The lengths of the three most effective dsRNA sequences (Rieske, Pterin 4α-CD, and Cytochrome P450) are smaller than 284 bps, and they are 10% shorter than the average length of the other 40 genes that were screened (Table 4; Lee et al., 2002).
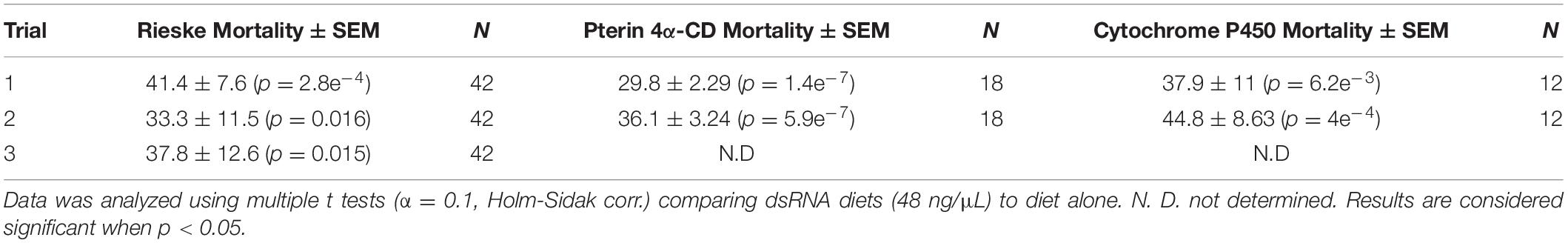
Table 1. Statistical analysis of D. citri mortality of the top three dsRNAs fed by artificial diet showing dose response (Figure 1B).
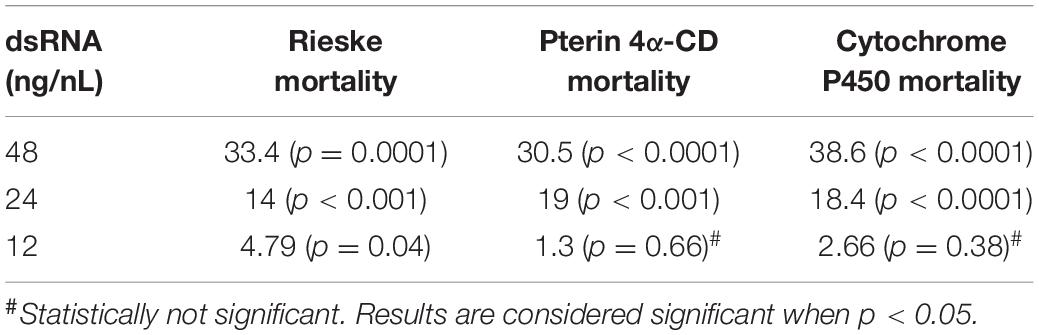
Table 2. Statistical analysis of D. citri mortalities using 2-way ANOVA with Fisher’s LSD test and different concentrations the best three dsRNA diets showing dose response (Figure 1B) as compared with diet alone.
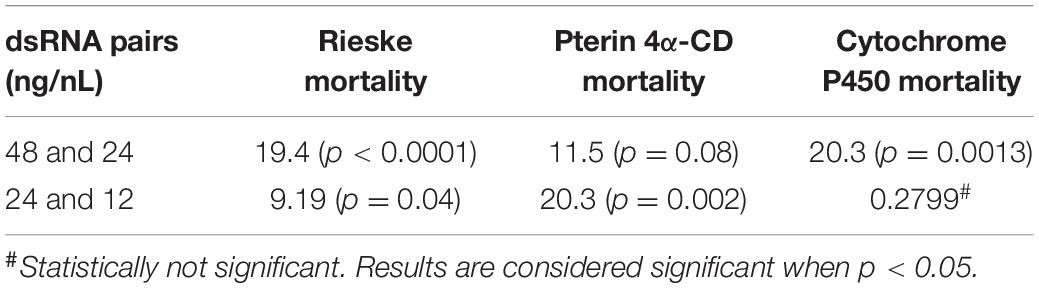
Table 3. Comparison of different dsRNA concentrations by 2-way ANOVA with Fisher’s LSD test for the three best dsRNA diets showing dose response (Figure 1B).
The artificial diet bioassays identified genes that are affected by oral uptake of dsRNA by adult D. citri. Mortalities, however, never exceeded 40% after subtracting the mortalities of controls that were fed on diet without dsRNAs. Mortalities of adult D. citri fed on best performing dsRNAs targeting Rieske, Pterin 4α-CD and Cytochrome P450 genes, were 34%, 30.5%, and 38.6%, respectively, after subtracting mortalities caused by feeding on artificial diet alone (Figures 1A,B). Because artificial diet feeding causes mortalities of 15.7–39.1% whereas feeding on citrus leaves that expressed dsRNA-gfp (Supplementary Figure 1) causes very low mortalities (6%) (Figure 1C), a non-pathogenic strain of the Citrus tristeza virus (CTV) isolate (T36) was therefore engineered to express dsRNA in citrus leaves. The engineered virus was inoculated into Citrus macrophylla (C. mac), producing paratransgenic trees (Figure 3B) with leaves that express (in phloem ducts) subgenomic viral dsRNAs targeting D. citri genes that were shown to be effective in artificial diet screens.
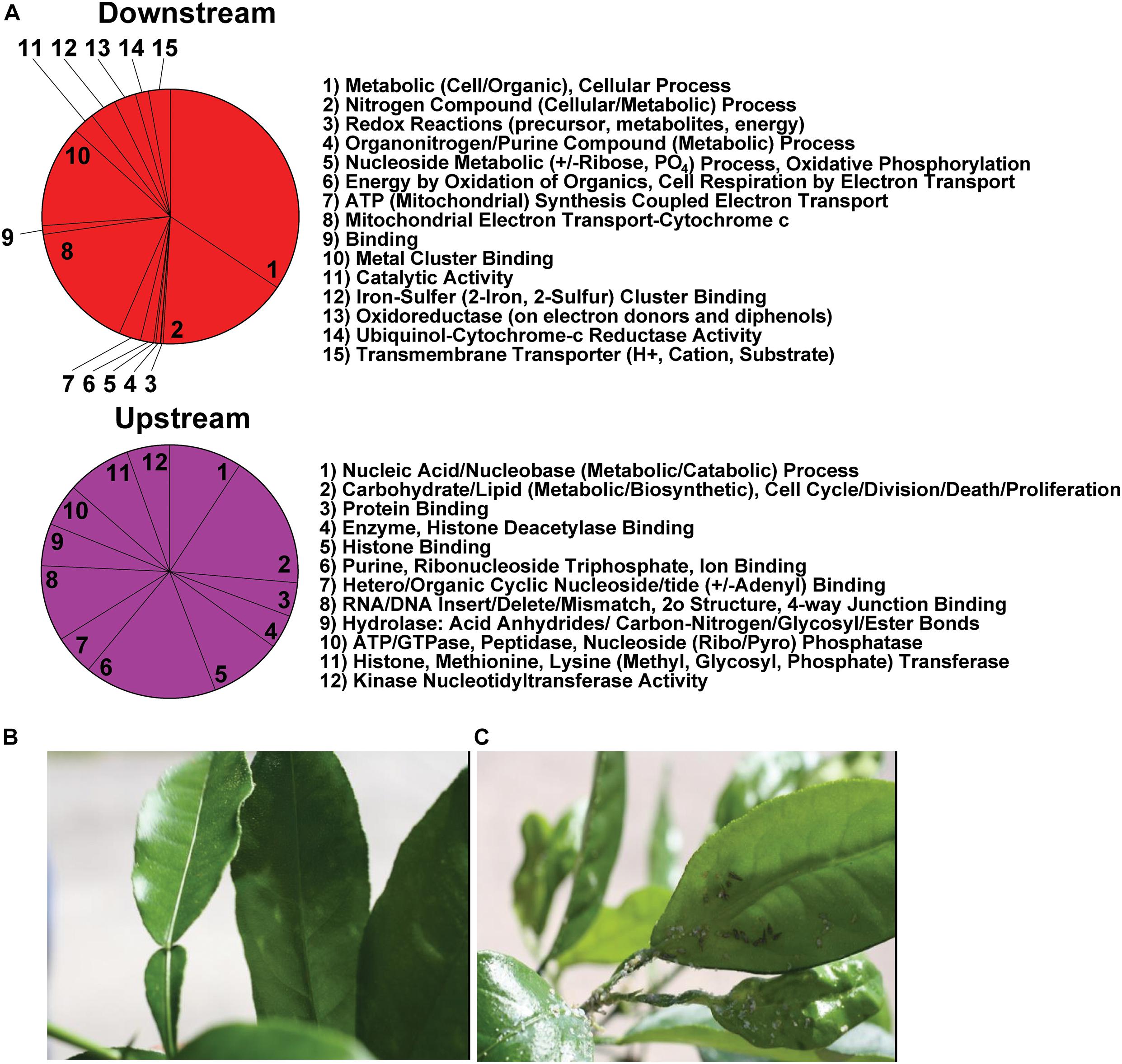
Figure 3. (A) Ontological processes arranged by upstream and downstream biological processes and unrelated effects after dsRNA-RIE targeting. Downstream and upstream effects that are not related specifically to down regulation of Rieske on the red and magenta pie charts include for downstream events: Nitrogen, Organonitrogen/Purine, Nucleoside and cellular and metabolic processes, and for upstream effects: RNA/DNA, and Nucleotide (Organo/Hetero) binding, metabolism, catabolism, hydrolysis, modification and repair as well as carbohydrate and lipid metabolism. (B) Citrus leaves infected with CTV-RIE are resistant to D. citri infestation, whereas (C) non-infected citrus leaves are not resistant to D. citri infestation.
Paratransgenic trees were created by inoculating CTV infected leaves expressing dsRNA (Satyanarayana et al., 1999). Single-detached-leaves infected with CTV expressing D. citri Rieske dsRNA in virus induced gene silencing (VIGS) experiments were compared with C. Macrophylla leaves expressing A. victoria gfp dsRNA (control). Because GFP is produced by CTV earlier than the gfp dsRNA the protein is visible in the leaves by fluorescence microscopy confirming that the plants have been successfully transfected expressing dsRNA-gfp (Supplementary Figure 1). Single and fully expanded young leaves from CTV engineered citrus trees were removed and the petiole placed in water filled microfuge tube (1.2 mL), sealed with parafilm, and transferred into a conical tube (50 mL) containing 15–20 adult D. citri then the tube was capped with a screened lid (El-Desouky et al., 2013). Adult Asian Citrus Psyllids (ACP) feeding on leaves infected with engineered CTV-RIE showed an average mortality of 76.0% whereas adults feeding on leaves that were infected with CTV-GFP showed an average mortality of 6.2% (Figure 1C). In contrast with artificial diets that causes high mortality in the absence of dsRNA only 9.5% of adult D. citri feeding on CTV-free leaves died (results not shown).
RT-qPCR separately quantified the Rieske mRNA abundance in adult D. citri and its digestive tract. Total RNA was extracted from insects that fed on RIE artificial diets and CTV-RIE paratransgenic plants for 4 days. Analyzing adult D. citri in which guts were removed prior to analysis, showed that the abundance of the Rieske transcript was not affected by feeding RIE dsRNA (Figure 4A). On the other hand, gut extract transcripts that were analyzed by RT-qPCR show that feeding D. citri RIE dsRNA expressed in citrus leaves by engineered CTV or in artificial diet causes significant reduction in the level of Rieske transcript of 71.3 ± 3.79% and 29.8 ± 3.61%, respectively (Figure 4A). A large reduction in the RIE transcript was also observed by Northern blot analysis when gut RNA was analyzed after feeding D. citri on leaves infected with engineered CTV expressing RIE dsRNA as compared with insects that were fed on control leaves that were not infected (Figure 4B) or infected with gfp-CTV (not shown). No effect was found by Northern blot analysis when RIE dsRNA was fed and D. citri gut RS20 ribosomal RNA subunits were analyzed (Figure 4B). These results indicate that the Rieske transcript is affected only in the gut of the insect by feeding on artificial diet mixed with the dsRNA. If, on the other hand, the dsRNA is expressed in the leaf phloem by engineered CTV the effect is more pronounced.
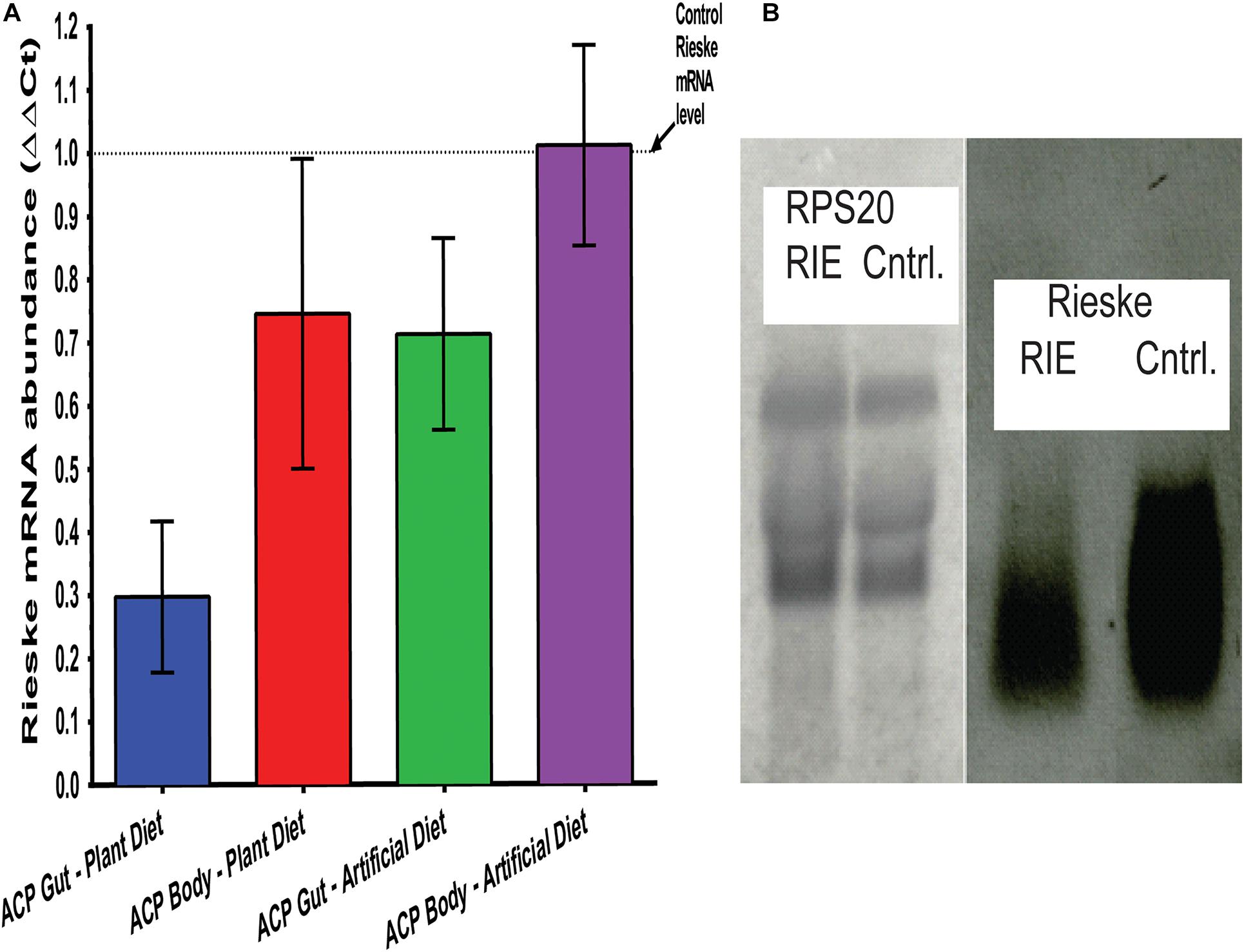
Figure 4. Transcriptional modification of RIE in D. citri. (A) mRNA-RIE abundance (ΔΔCt) after feeding D. citri on CTV-RIE infected citrus leaf or artificial diet containing dsRNA-RIE analyzed by qRT-PCR of gut and body extracts. (B) Northern blot analyses of control C. macrophylla leaf infected with CTV-RIE and CTV-wt (control), respectively, probed with RPS20 ribosomal proteins probe (left two lanes) and RIE probe (right two lanes).
Transcriptome analysis was performed on amplified mRNA extracted from the gut tissue of D. citri fed on CTV transformed detached leaves and sequences were assembled onto reference contigs. Of the 149 contigs showing under or overexpression of 8-fold or greater, with 99% confidence interval, after feeding D. citri dsRNAs expressed by CTV-RIE or CTV-gfp, 25 contigs were reliably annotated using GO terminology (Supplementary Table 1). Blast2Go generated gene ontology maps for the biological processes and the molecular functions (Figure 2). The annotated genes that were directly affected by the targeting of Rieske transcripts in these assays are found downstream (downstream effect) of the target (Rieske) gene and are colored red (Figure 2). They consist mainly of metabolic processes, cellular processes, nitrogen compound processes, mitochondria and electron transport processes and metal cluster binding (Figure 3A). Upstream effects that occur by targeting the Rieske gene or are the consequences of downstream effects were also annotated and are colored magenta (Figure 2). They consist mainly of nucleic acid processes, carbohydrate and lipid metabolism, histone binding, purine, ribonucleotide triphosphate and ion binding and RNA and DNA processes. In general, these biological processes are involved in DNA, RNA Cell Cycle, and nutrient regulation (Figure 3A). Annotated processes that were not directly attributable (given the limitations of the GO annotation database) for targeting the Rieske transcript are colored orange (Figure 2). A ranking of upstream and downstream effects on cell processes was analyzed by using the previously calculated node scores (Figure 2) allowed us to quantitate and correlate the impact of these processes with upstream and downstream effects of Rieske on D. citri (Figure 3A, Supplementary Figure 2 and Supplementary Table 3).
The gene ontology studies of Rieske dsRNA with CTV-RIE infected citrus leaves prompted us to test the possibility that citrus trees can be protected against D. citri. One year old C. macrophylla seedlings (approximately two feet tall and stem of a pencil thickness) were infected with either CTV-RIE or CTV-wt control and were exposed to 100 D. citri adults caged in insect rearing cages (30 × 15.5 × 15 inches) and kept in growth rooms. After 3 weeks of exposure trees were observed for nymph development, adults, laid eggs, and honey-dew secretions. C. macrophylla plant that was infected with CTV-RIE is devoid of adults, nymphs, eggs or honey-dew secretions, whereas C. macrophylla that was infected with CTV-wt shows dense population of adults, nymphs, eggs and copious secretion of honeydew (Figure 3B) indicating that expression of Rieske dsRNA confers resistance against D. citri.
We report an innovative approach to control D. citri an agricultural pest insect that threatens the citrus industry in the United States and beyond. Employing a crowdsourcing strategy, we identified potentially target genes in D. citri that cause mortality when silenced by RNA interference. Using this information, we mined and used the D. citri genomics, Citrus genetics, viral engineering, molecular biology and computational biology to screen potential genes that are suitable candidates for a biological control strategy. Using gene silencing, a technique that has been shown to successfully combat insect of medical importance, as well as agricultural pest insects (Baum et al., 2007; Van Ekert et al., 2014) we rapidly (within weeks) screened potential genes for RNA interference studies using artificial diet containing dsRNA to feed adult D. citri. The artificial diet allowed us to rapidly monitor psyllids mortality and physiological effects. Using artificial diets to test dsRNA interference by feeding was reported for Coleoptera (Diabrotica virgifera virgifera and Phylloreta striolata) (Baum et al., 2007; Zhao et al., 2008), Diptera (Glossina morsitans morsitans) (Walshe et al., 2009), Hemiptera (Acrthosiphon pisum and Rhodnius prolixus) (Araujo et al., 2006; Shakesby et al., 2009), Hymenopterans (Apis mellifera) (Aronstein et al., 2006; Nunes and Simões, 2009), Lepidoptera (Helicoverpa armigera, Pluttela xylostella, Spodoptera frugiperda), and Gubernatora (Mao et al., 2007; Griebler et al., 2008; Bautista et al., 2009). The artificial diet screening identified 6 gene targets causing significant mortalities (>30%) after 6 days of feeding. Five of these genes express proteins that are directly involved or associated with proteins that maintain iron in the cell (Dhur et al., 1989; Köster, 2001; Gubernatora et al., 2003; Ana et al., 2005). These proteins include an iron-sulfur cluster domain of the mitochondrial electron transport chain (Rieske), a heme containing iron binding terminal oxidase (CP450), an iron storage protein (Ferritin), an enzyme involved in the BH4 biosynthetic pathway (PCD), and an ATP binding cassette transporter (ABC Transporter) indicating that cellular iron is important for D. citri survival.
Rieske mRNA, which is translated into a soluble domain of Complex III of the Electron Transport Chain (Figure 5), significantly affected mortality by dsRNA-RIE (Figure 1A). Feeding D. citri artificial diet containing different concentrations (12, 24, and 48 ng/μL) of dsRNA-RIE caused a significant dose response mortality (Tables 1–3 and Figure 1B). Several reports suggest that, for every target gene an optimal concentration has to be empirically determined in order to induce optimal silencing and exceeding that optimal concentration does not necessarily result in more silencing (Liu et al., 2000; El-Desouky et al., 2013). Indeed, poor dose response relationship was found when dsRNA against CP450 was fed to D. citri using the same concentrations that produced good linear relationship in feeding dsRNA-RIE and dsRNA-PCD (Figure 1B). Although other genes were targeted in the electron transport chain (ETC) such as NADH Dehydrogenase, Succinate Dehydrogenase, Cytochrome b Reductase, and Cytochrome Oxidase (components of Complexes I, II, III, and IV, respectively) (Figure 5) and several showed significant mortalities. The effect was not as pronounced as observed when dsRNA-RIE diet was used (El-Desouky et al., 2013; Figures 1A,B). Of the five ETC transcripts screened, only the Rieske transcript is expressed in the mitochondrial intermembrane space soluble protein (ISP) of cytochrome bc1 complex. The other ETC proteins that were targeted are either membrane domains of NADH dehydrogenase, cytochrome b subunit of cytochrome bc1 complex, cytochrome c oxidase subunit VIa, or subunits located in the mitochondrial matrix like the δ subunit of ATP synthase and the Flavoprotein subunit of succinate dehydrogenase (Figure 5). The failure to affect the mitochondrial components that are part of the mitochondrial membrane or are found inside the mitochondrial matrix is perhaps due to a lower turnover of these proteins as compared with the ISP subunit (Rieske) of cytochrome bc1 complex located in the inter-membrane space. It is interesting to note that the turnover of mitochondrial proteins is generally low (6–12%) (Augustin et al., 2007). Mitochondrial proteases that are involved in mitochondrial proteins turnover are found in both the mitochondrial inter-membrane space, inner membrane and matrix, however, their activities and roles may be differently regulated (Koppen and Langer, 2007). Thus, subunits that exhibit a higher turnover are better targets for RNA silencing. Indeed, when dsRNA against Cytb, a subunit of cytochrome bc1 complex found in the inner mitochondrial membrane (Herrman and Neupert, 2003) was fed to D. citri no effect was observed (Figure 1A) indicating that perhaps the location of the ETC subunits in the mitochondria may determine their turnover and vulnerability to RNA silencing. Our feeding experiments did not test mixtures of dsRNAs targeting several components of the ETC. In future experiments, however, down regulating multiple components of this pathway may prove to be more effective than targeting only one transcript (Figure 1). RNA interference studies continue to highlight the complexity of insect orally induced mortality using exogenous dsRNAs. Thus, it is important to use gene internal sequences that correlate well with published and experimentally tested RNAi effects recognizing that secondary structure and base-pair length of the selected dsRNA are important in optimizing RNAi silencing effect (Elbashir et al., 2001; Lee et al., 2002; Yoshinari et al., 2004). Therefore, it is possible that designing alternate dsRNA constructs against domain genes that reside in the inter membrane space of the mitochondria like Rieske, and combinations of dsRNA targeting different genes may cause higher mortalities by oral uptake as was shown in lung cancer by reversing drug resistance (Ganesh et al., 2013) or in corneal fibroblast in which a combination of triple dsRNAs were used to target three different scarring genes (Sriram et al., 2013). In Tribolium castaneum 5,000 randomly screened genes were tested as RNAi potential targets and 11 genes were identified as highly efficient RNAi targets showing effective inhibition however no qPCR data or Northern blot analyses to correlate the effect of these genes with mortalities is provided. The authors similarly to our approach use GO term combinations to show the efficiency of the RNAi target genes (Ulrich et al., 2015). These genes, however, were not tested by us and in future studies we may test them even though there are not known reports that they ever were effective on the Asian Citrus psyllids.
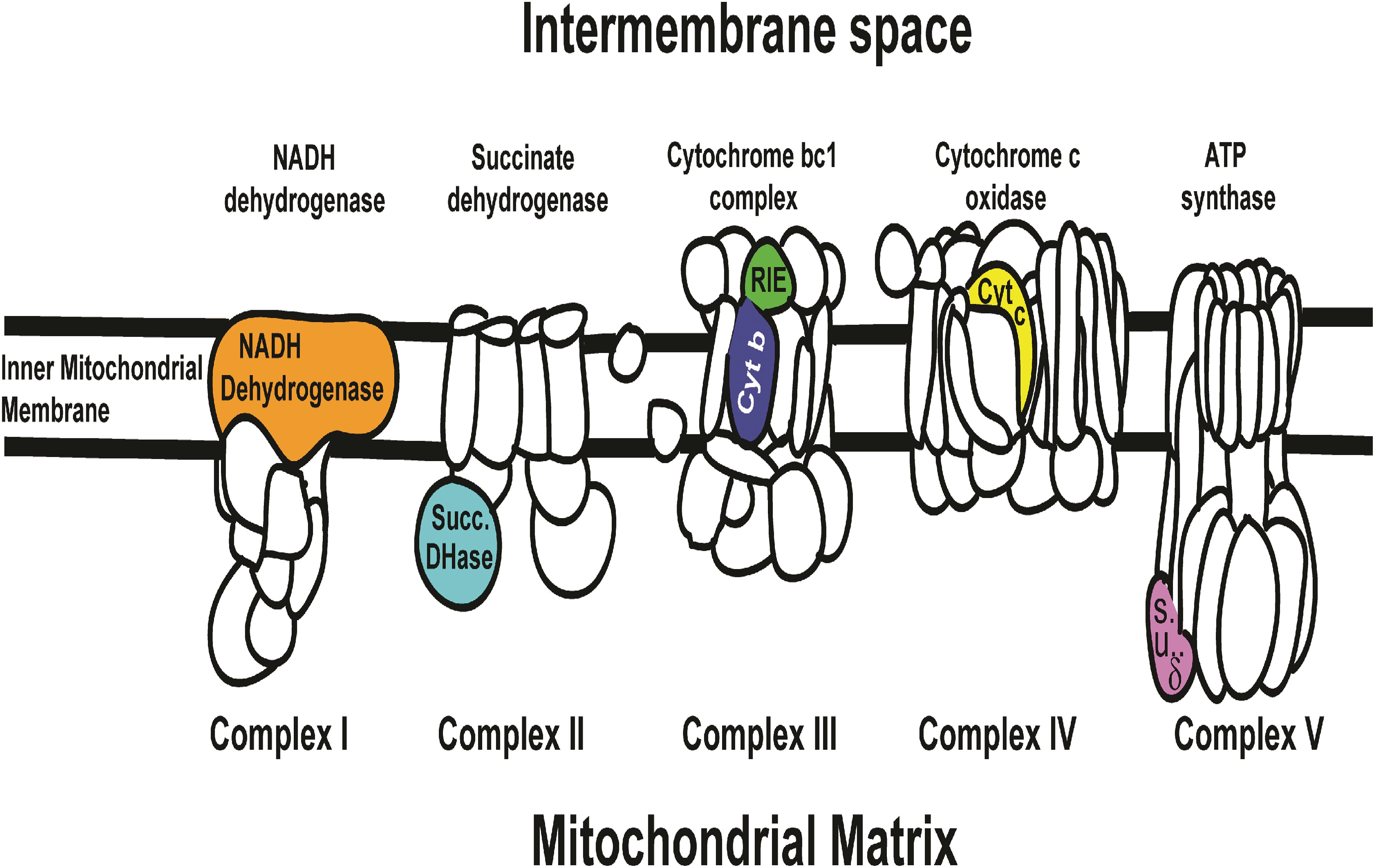
Figure 5. Six ETC (Electron Transport Chain complexes) genes that were screened in artificial diet bioassay (Figure 1A) in relation to the KEGG map cartoon of the oxidative phosphorylation chain in complexes I, II, III, IV, and V (NADH dehydrogenase membrane domain, Succinate dehydrogenase flavoprotein, Cytochrome bc1 complex ISP and Cytb, Cytochrome c oxidase VIa and ATP synthase δ subunit, respectively) and their relative position in relation to the intermembrane space, inter mitochondrial membrane and matrix is shown. Diets targeting Rieske (green color) and Succinate Dehydrogenase (blue color) transcripts resulted in significant mortality (see Figure 1). Cognate proteins of these two gene correspond to soluble domains of the ETC. None of the membranous domains targeted showed significant mortality (see Figure 1). Succinate Dehydrogenase (blue color) is oriented toward the mitochondrial matrix while Rieske (green color) is oriented toward the intermembrane space with greater access to the cytosolic environment. The ETC cartoon is based on available models of NADH dehydrogenase (Thermus thermophilus), Succinate dehydrogenase (E. coli), Cytochrome bc1 complex (bovine), Cytochrome c oxidase (bovine), and ATP synthase (E. coli). http://www.genome.jp/kegg-bin/show_pathway?map00190.
Expressing dsRNAs in citrus using engineered CTV-RIE and testing feeding D. citri on single leaves caused 2-fold higher mortality than feeding on artificial diet containing dsRNA (65.7% and 35.4%, respectively) (Figures 1A–C), and in several cases 100% mortality was observed. These results indicate that artificial diet is inferior to feeding on engineered leaves that contain all the nutrients that are not found in artificial diet specifically when controls give very high mortality and thus these artificial diets cannot be used in long duration studies. We used artificial diets to rapidly screen several hundred dsRNA candidates that were generated by crowd sourcing to select few dsRNA candidates that were then cloned and express by engineered CTV in citrus and allowed us to test D. citri by feeding on engineered CTV in citrus leaves. Gong et al. (2011) reported that coating cabbage leaves with three siRNAs-RIE of Plutella xylostella caused significant mortalities lowering the Rieske transcript. Similarly, citrus leaves that were infected with CTV-RIE and fed to D. citri showed significant reduction in Rieske transcript abundance in D. citri gut tissue as compared with the whole insect tissue devoid of its gut. These observations support our hypothesis that oral delivery of dsRNA produces a predominantly localized response in the gut tissue and that adult D. citri do not amplify or transport dsRNA templates systemically as nematodes and Tribolium (Fire et al., 1998; Miller et al., 2012). The most economical way to deliver dsRNA to insects is to express dsRNA as a long hairpin (LHP) RNA in plants or cells that are eaten by insects. Baum et al. (2007) expressed LHP RNA of vacuolar ATPase (V-ATPase) in corn as an alternative to Bacillus thuringiensis corn expressing the Bt bacterial toxin and showed significant reduction in corn root damage after exposing the transgenic plants to western corn rootworm (D. virgifera virgifera). Following this report, two model plants Nicotiana tabacum and Arabidopsis thaliana were genetically engineered to express LHP RNA of cytochrome P450 of the cotton bollworm (H. armigera). Feeding this insect on leaves of the genetically transformed plants reduced cytochrome P450 transcript and larval growth was affected. Van Ekert et al. (2014) transformed yeast cells (Pichia pastoris) and expressed LHP-RNA against Aedes aegypti juvenile hormone acid methyltransferase transcript (jmtA). Mosquito larvae that were fed on these recombinant cells stopped larval development and the few adults that emerged died within 3 days, indicating that this technique is useful against agricultural pests, as well as disease transmitting insects. It is possible that the potency of our dsRNA-RIE could be attributed to resistance to nuclease digestion. In Tribolium castaneum Cao et al. (2018) reported that nucleases play a role in the stability and effectiveness of RNAi. In psyllids it was shown that dsRNA was not degraded by insect saliva that was secreted into an artificial medium during feeding. The tested dsRNA was not degraded from saliva that is excreted into the medium after feeding for 120 h (Galdeano et al., 2017). Killiny and Kishk (2017) reported that the target of the ingested dsRNA in D. citri is found in the gut by mixing dsRNA with a dye and microscopically showed that the dsRNA- dye was found in the psyllid gut. However, the presence of nucleases in D. citri gut was not tested and it is unknown what role nucleases play in digesting dsRNA in D. citri gut. Thus, it is important in future studies to find whether nucleases degrade dsRNA-RIE in D. citri gut. It is also possible that some of the primers that we have used were not optimal or were amplifying shorter regions that are optimal and may have to be retested in future studies.
An RNA Seq analysis of the genes affected by the dsRNA targeting the aerobic metabolic Rieske gene was performed to find out the cause of death of D. citri after Rieske silencing. Transcriptome analysis showed multiple potential pathways that are vulnerable after transcriptional modification in D. citri that could be used in future RNAi strategies (Figures 2, 3). The “downstream effects” that are not the result of Rieske or ETC malfunction include nitrogen, organonitrogen, nucleoside and nucleotide metabolic processes. Conversely, nucleic acid, nucleotide, and nucleoside binding, catalysis, phosphorylation, hydrolysis, and metabolism were identified as being indirectly affected as a consequence of the metabolic pathways of aerobic metabolism (ETC) being stressed, or “upstream effects” (Figures 2, 3). These pathways may also be sensitive to a decrease in the products of the ETC metabolism such as the oxidative state of the cell, cellular energy production, and phosphorylation. These processes are associated with cell-cycle regulation, cell-death, cell-division, cell-proliferation, DNA/RNA repair, nutrient metabolism (including carbon-nitrogen, glycosyl, peptide hydrolysis, lipid and carbohydrate biosynthesis.
Recent RNAi and enzymatic activity studies in Drosophila implicate an energy (or ATP) deficiency role in protein folding within the endoplasmic reticulum leading to cell-cycle arrest/delay at the G1 and G2/M checkpoints (Thomas et al., 2013). The role of cell-cycle regulation in cell-division/proliferation is well established (Zhou et al., 2002), recent studies showed that when the ER (Endoplasmic reticulum) is under stress impairment in the expression of mediators will cause cell-death, energy and nutritional deficiencies (Miller et al., 2012). The role of p53, an activator of DNA repair enzymes and mediator of cell-death (Cho et al., 1994; Xie et al., 2001), and a CHK1, a serine/threonine-protein kinase involved in cell-cycle checkpoint regulation by phosphorylation (Liu et al., 2000) in cell-death is correlated with ER stress. Thus, our virus-induced gene silencing (VIGS) is an important intermediate step in the engineering of transgenic D. citri resistant cultivars. Using RNA Seq analyses and next generation sequencing we identified genes (other than those targeted) that were transcriptional modified (Ratcliff et al., 1997; Ma et al., 2006; Morilla et al., 2006; Folimonov et al., 2007; Hunter et al., 2008; Velásquez et al., 2009; Gan et al., 2010; Jiang et al., 2012; Liu et al., 2013). These genes were functionally annotated, and the majority (either upstream or downstream) were found to be functionally associated with the primary function performed by the Rieske (ETC) gene targeted by VIGS (Dhur et al., 1989; Baum et al., 2007; Wuriyanghan et al., 2011; Figures 2, 3).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JR performed the experiments, involved in the experimental design, analyzed the data, and wrote the manuscript. RJ performed the experiments. CP and WD analyzed the data and involved in the experimental design. SG performed the experiments, involved in the experimental design, and analyzed the data. DB and RS analyzed the data, involved in the experimental design, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
DB is an established scientist fellow at the Oak Ridge Institute for Science and Education (ORISE). This research was supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the United States Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-AC05-06OR23100. All opinions expressed in the manuscript are the author’s and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE. This work was partially supported with grants from the Florida citrus industry to DB, RS, WD, SG, and CP.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.571826/full#supplementary-material
Supplementary Figure 1 | Fluorescence image (4X microscopic magnification) of leaf sample inoculated with CTV-gfp.
Supplementary Figure 2 | Right side, Scatter plot of contigs matching mRNA sequenced from psyllid guts post paratransgenic CTV-RIE and CTV-gfp (control) oral uptake. Yellow contigs are equally expressed in guts from psyllid CTV-RIE or CTV-gfp paratransgenic leaves, red contigs overexpressed in CTV-RIE ACP, blue contigs overexpressed in CTV-gfp fed ACP (or under expressed in CTV-RIE fed adult Asian psyllids). The most significant (99% of contigs expression >8-fold) of under and over expression is represented by white contigs (on axes). Left side, 25 of 149 of these contigs have been annotated and quantified by relative RNA abundancies as read per million (RPM) showing underexpressed and overexpressed contigs (negative and positive fold changes, respectively) in CTV-RIE paratransgenic citrus fed insects.
Supplementary Table 3 | Differential Gene Expression of up and down regulated genes after feeding D. citri CTV-RIE paratransgenic citrus.
CP450, the subgenomic fragment of the Cytochrome P450 gene; CTV, Citrus tristeza virus; ETC, electron transport chain; ISP, iron sulfur protein; PCD, the subgenomic fragment of the Pterin 4- α Carbinolamine Dehydratase; RIE, the subgenomic fragment of the Rieske gene; RIN, RNA integrity number; VIGS, virus induced gene silencing; wt, wild type.
Ammar, E.-D., Ramos, J. E., Hall, D. G., Dawson, W. O., and Shatters, R. G. Jr. (2016). Acquisition, replication and inoculation of Candidatus Liberibacter asiaticus following various acquisition periods on Huanglongbing infected citrus by nymphs and adults of the asian citrus psyllid. PLoS One 11:e0159595. doi: 10.371/journal.pone.0159594
Ana, M. P., Melo, A. M. P., Loboa, S. A. L., Sousaa, F. L., Fernandesa, A. S., Pereira, M. M., et al. (2005). AnhaD Na+/H + antiporter and a pcd homologues are among the Rhodothermus marinus complex I genes. Biochim. Biophys. Acta 1709, 95–103. doi: 10.1016/j.bbabio.2005.06.003
Araujo, R. N., Santos, A., Pinto, F. S., Gontijo, N. F., Lehane, M. J., and Pereira, M. H. (2006). RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 36, 683–693. doi: 10.1016/j.ibmb.2006.05.012
Aronstein, K., Pankiw, T., and Saldivar, E. (2006). SID-1 is implicated in systemic gene silencing in the honeybee. J. Apic. Res. 45, 20–24. doi: 10.1080/00218839.2006.11101307
Augustin, S., Nolden, M., Muller, S., Hardt, O., Arnold, I., and Langer, T. (2007). Characterization of peptides released from mitochondria evidence for constant proteolysis and peptide efflux. J. Biol. Chem. 280, 2691–2699. doi: 10.1074/jbc.m410609200
Ayllón, M. A., Gowda, S., Satyanarayana, T., Karasev, A. V., Adkins, S., Mawassi, M., et al. (2003). Effects of modification of the transcription initiation site context on Citrus Tristeza Virus Subgenomic RNA synthesis. J. Virol. 77, 9232–9243. doi: 10.1128/jvi.77.17.9232-9243.2003
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Bautista, M. A. M., Miyata, T., Miura, K., and Tanaka, T. (2009). RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem. Mol. Biol. 39, 38–46. doi: 10.1016/j.ibmb.2008.09.005
Ben Berkhout, J. H. (2006). The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 580, 2896–2902. doi: 10.1016/j.febslet.2006.02.070
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. (2001). Role for bidentate ribonuclease in the initiation step in RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Bilello, J. P., Cable, E. E., and Isom, H. C. (2003). Expression of E-cadherin and othe paracellular junction genes is decreased in iron loaded hepatocytes. Am. J. Phatol. 162, 1322–1338.
Borgio, J. F. (2010). RNAi mediated gene knockdown in sucking and chewing insect pests. J. Biopestic. 3, 386–393.
Boykin, L. M., De Barro, P., Hall, D. G., Hunter, W. B., McKenzie, C. L., Powell, C. A., et al. (2012). Overview of worldwide diversity of Diaphorina citri Kuwayama mitochondrial cytochrome oxidase 1 haplotypes: two Old World lineages and a NewWorld invasion. Bull. Entomol. Res. 102, 573–582. doi: 10.1017/s0007485312000181
Britt, K., Gebben, S., Levy, A., Rwahnih, M. A., and Batuman, O. (2020). The detection and surveillance of Asian Citrus Psyllid (Diaphorina citri)- associated virus in Florida citrus groves. Front. Plant Sci 10:1687. doi: 10.3389/fpl.2019.01687
Cao, M., Gatehouse, J. A., and Fitches, E. C. (2018). A systemic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosphon pisum following injection and ingestion of analogous dsRNA. Int. J. Med. Sci. 19:1079. doi: 10.3390/ijms19041079
Cho, Y., Gorina, S., Jeffrey, P. D., and Pavletich, N. P. (1994). Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265, 346–355. doi: 10.1126/science.8023157
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. doi: 10.1006/abio.1987.9999
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Covey, S. N., Al-Kaff, N. S., Langara, A., and Turner, D. S. (1997). Plants combat infection by gene silencing. Nature 385, 781–782. doi: 10.1038/385781a0
Dhur, A., Galan, P., and Hercberg, S. (1989). Effects of different degrees of iron deficiency on cytochrome P450 complex and pentose phosphate pathway dehydrogenases in the rat. J. Nutr. 119, 40–47. doi: 10.1093/jn/119.1.40
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. (2001). Funtional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888. doi: 10.1093/emboj/20.23.6877
El-Desouky, A., Walter, A. J., and Hall, D. G. (2013). New excised-leaf assay method to test inoculativity of asian citrus psyllid (Hemiptera: syllidae) with Candidatus liberibacter asiaticus associated with citrus huanglongbing disease. J. Econ. Entomol. 106, 25–35. doi: 10.1603/ec12245
El-Shesheny, I., Hajeri, S., El-Hawary, I., Gowda, S., and Killiny, N. (2013). Silencing abnormal wing disc gene of the Asian Citrus Psyllid, Diaphornia citri disrupts adult wing development and increases nymph mortality. PLoS One. 8:e65392. doi: 10.1371/journal.pone.0065392
Fire, A., Xue, S.-Q., Montgomery, M. K., Kostas, S. A., Driver, S. A., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditiselegans. Nature 391, 806–811. doi: 10.1038/35888
Folimonov, A. S., Folimonova, S. Y., Bar-Joseph, M., and Dawson, W. O. (2007). A stable RNA virus-based vector for citrus trees. Virology 368, 205–216. doi: 10.1016/j.virol.2007.06.038
Galdeano, D. M., Breton, M. C., Lopes, J. R. S., Falk, B. W., and Machado, M. A. (2017). Oral delivery of double-strandedRNAs inducesmortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri. PLoSOne 12:e0171847. doi: 10.1371/journal.pone.0171847
Gan, D., Zheng, J., Jiang, H., Jiang, T., Zhu, S., and Cheng, B. (2010). Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 29, 1261–1268. doi: 10.1007/s00299-010-0911-z
Ganesh, S., Iyer, A. K., Weiler, J., Morrissey, D. V., and Amiji, M. M. (2013). Combination of siRNA-directed gene silencing with cisplatin reverses drug resistance in human non-small cell lung cancer. Mol. Ther. Nucleic Acids 2:e110. doi: 10.1038/mtna.2013.29
Gong, L., Yang, X., Zhang, B., Zhonga, G., and Hua, M.-Y. (2011). Silencing of Rieske iron–sulfur protein using chemically synthesised siRNA as a potential biopesticide against Plutella xylostella. Pest Manag. Sci. 67, 514–520. doi: 10.1002/ps.2086
Götz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Gregory, R. I., Chendrimada, T. P., Cooch, N., and Shiekhattar, R. (2005). Human RISC couples MicroRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640. doi: 10.1016/j.cell.2005.10.022
Griebler, M., Westerlund, S. A., Hoffmann, K. H., and Meyering-Vos, M. (2008). RNA interference with the allatoregulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the hemolymph. J. Insect Physiol. 54, 997–1007. doi: 10.1016/j.jinsphys.2008.04.019
Gubernatora, B., Seidlerb, A., Rögnerb, M., and Szczepaniaka, A. (2003). Overexpression and reconstitution of a Rieske iron–sulfur protein from the higher plant. Protein Expr. Purif. 29, 8–14. doi: 10.1016/s1046-5928(03)00016-0
Hajeri, S., Killiny, N., Ei-Mohtar, C., Dawson, W. O., and Gowda, S. (2014). Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphornia citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing). J. Biotechnol. 176, 42–49. doi: 10.1016/j.jbiotec.2014.02.010
Hall, D. G., Shatters, R. G., Carpenter, J. E., and Shapiro, J. P. (2010). Research toward an artificial diet for adult asian citrus psyllid. Ann. Entomol. Soc. Am. 103, 611–617. doi: 10.1603/an10004
Herrman, J. M., and Neupert, W. (2003). Protein insertion into the inner membrane of mitochondria. IUBMA Life 55, 219–225.
Hunter, W. B., Shelby, K., Dowd, S., McKenzie, C., and Shatters, R. Jr. (2008). “Gene expression in asian citrus psyllid adults feeding from florida citrus: application to biology and vector control,” in Proceedings of the InternationalResearch Conference on Huanglongbing, (Orlando, FL), 233–237.
Ichinose, K., Miyazi, K., Matsuhira, K., Yasuda, K., Sadoyama, Y., Tuan, D.-H., et al. (2010). Unreliable pesticide control of the vector psyllid Diaphorina citri (Hemiptera: Psyllidae) for the reduction of microorganism disease transmission. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 45, 466–472. doi: 10.1080/03601231003800263
Iwata, S., Saynovits, M., Link, A. T., and Hartmut, M. (1996). Structure of water soluble fragment of the Rieske iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution. Structure 4, 567–579. doi: 10.1016/s0969-2126(96)00062-7
Jiang, F., Yang, M., Guo, W., Wang, X., and Kang, L. (2012). Large-scale transcriptome analysis of retroelements in the migratory locust, Locusta migratoria. PLoS One 7:e40532. doi: 10.1371/journal.pone.0040532
Killiny, N., and Kishk, A. (2017). Delivery of dsRNA through topical feeding for RNA interference in the citrus sap piercing-sucking hemipteran, Diaphornia citri. Arch. Insect Biochem. Physiol. 2017:e21394. doi: 10.1002/arch.21394
Koppen, M., and Langer, T. (2007). Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 42, 221–242. doi: 10.1080/10409230701380452
Köster, W. (2001). ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152, 291–301. doi: 10.1016/s0923-2508(01)01200-1
Lee, N. S., Dohjima, T., Bauer, G., Li, H., Li, M.-J., Ehsani, A., et al. (2002). Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20, 500–505. doi: 10.1038/nbt0502-500
Lipardi, C., Wei, Q., and Paterson, B. M. (2001). RNAi as Random Degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107, 297–307. doi: 10.1016/s0092-8674(01)00537-2
Liu, J., Smagghe, G., and Swevers, L. (2013). Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J. Insect Physiol. 59, 646–654. doi: 10.1016/j.jinsphys.2013.03.013
Liu, Q., Guntuku, S., Cui, X.-S., Matsuoka, S., Cortez, D., Tamai, K., et al. (2000). Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Z. J., Huang, Y. L., Yu, H. Z., Li, N. Y., Xie, Y. X., Zhang, Q., et al. (2019). Silencing of the Chitin Synthase Gene Is Lethal to the Asian Citrus Psyllid, Diaphorina citri. Int. J. Mol. Sci. 20:3734. doi: 10.3390/ijms20153734
Ma, Y., Creanga, A., Lum, L., and Beachy, P. A. (2006). Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359–363. doi: 10.1038/nature05179
Maillard, P. V., Ciaudo, C., Marchais, A., Li, Y., Jay, F., Ding, S. W., et al. (2013). Antiviral RNA interference in mammalian cells. Science 342, 235–238. doi: 10.1126/science.1241930
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Marques, J. T., Devosse, T., Wang, D., Zamanian-Daryoush, M., Serbinowski, P., Hartmann, R., et al. (2006). A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24, 559–565. doi: 10.1038/nbt1205
Martinez, M. A., Gutierrez, A., Armand-Ugon, M., Blanco, J., Gomez, J., Clotet, B., et al. (2002). Suppression of chemokine receptor expression by RNA interference for inhibition of HIV-1 replication. AIDS 16, 2385–2390. doi: 10.1097/00002030-200212060-00002
Mehta, A., Deshpande, L., Bettedi, L., and Missirlis, F. (2009). Ferritin accumulation under iron scarcity in Drosophila iron cells. Biochimie 91, 1331–1334. doi: 10.1016/j.biochi.2009.05.003
Miller, S. C., Miyata, K., Brown, S. J., and Tomoyasu, Y. (2012). Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One. 7:e47431. doi: 10.1371/journal.pone.0047431
Morilla, G., Castillo, A. G., Preiss, W., Jeske, H., and Bejarano, E. R. (2006). A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication. J. Virol. 80, 3624–3633. doi: 10.1128/jvi.80.7.3624-3633.2006
National Agricultural and Statistics Service. (2005). Agricultural Chemical Usage Postharvest Applications Oranges Summary. Washington, DC: United States Department of Agriculture.
Navas-Castillo, J., Diaz, J. A., Sanchez-Campos, S., and Moriones, E. (1998). Improvement of the print-capture polymerase chain reaction procedure for efficient amplification of DNA virus genomes from plants and insect vectors. J. Virol. Methods 75, 195–198. doi: 10.1016/s0166-0934(98)00110-4
Nunes, F. M. F., and Simões, Z. L. (2009). A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem. Mol. Biol. 39, 157–160. doi: 10.1016/j.ibmb.2008.10.011
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J., and Hogenhout, S. A. (2011). Silencing of aphid genes by dsRNA feeding from plants. PLoS One 6:e25709. doi: 10.1371/journal.pone.0025709
Putnman, A. H., Geuder, J. K., Mongiovi, N. L., and Dixon, W. N. (2011). Florida Citrus Statistics 2009-2010. Tallahassee, FL: Florida department of agriculture and consumer services.
Qiu, S., Adema, C. M., and Lane, T. (2005). A computational study of off-target effects of RNA interference. Nucleic Acids Res. 33, 1834–1847. doi: 10.1093/nar/gki324
Ratcliff, F., Harrison, B. D., and Baulcombe, D. C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. doi: 10.1126/science.276.5318.1558
Reese, J., Christenson, M. K., Leng, N., Saha, S., Cantarel, B., Lindeberg, M., et al. (2014). Characterization of the asian citrus psyllid transcriptome. J. Genomics 2, 54–58.
Satyanarayana, T., Gowda, S., Boyko, V. P., Albiach-Marit, M. R., Mawassi, M., Navas-Castillo, J., et al. (1999). An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. U.S.A. 96, 7433–7438. doi: 10.1073/pnas.96.13.7433
Shakesby, A. J., Wallace, I. S., Isaacs, H. V., Pritchard, J., Roberts, D. M., and Douglas, A. E. (2009). A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem. Mol. Biol. 39, 1–10. doi: 10.1016/j.ibmb.2008.08.008
Sriram, S., Robinson, P., Pi, L., Lewin, A. S., and Schultz, G. (2013). Triple combination of siRNAs targeting TGFb1,TGFbR2, and CTGF enhances reduction of collagen I and smooth muscle actin in corneal fibroblasts. Invest Ophthalmol. Vis. Sci. 54, 8214–8223. doi: 10.1167/iovs.13-12758
Stokstad, E. (2012). Dread citrus disease turns up in California. Texas Sci. 336, 283–284. doi: 10.1126/science.336.6079.283
Sztal, T., Chung, H., Berger, S., Currie, P. D., Batterham, P., and Daborn, P. J. (2012). A cytochrome P450 conserved in insects is involved in cuticle formation. PLoS One 7:e36544. doi: 10.1371/journal.pone.0036544
Taning, N. T., Andrade, E. C., Hunter, W. B., Christiaens, O., and Smagghe, G. (2016). Asian citrus psyllid RNAi pathway-RNAi evidence. Sci. Rep. 6:38082. doi: 10.1038/srep38082
Tatineni, S., Robertson, C. J., Garnsey, S. M., Bar-Joseph, M., Gowda, S., and Dawson, W. O. (2008). Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 376, 297–307. doi: 10.1016/j.virol.2007.12.038
Thomas, S. E., Malzer, E., Ordóñez, A., Dalton, L. E., van T Wout, E. F. A., Liniker, E., et al. (2013). p53 and translation attenuation regulate distinct cell cycle checkpoints during endoplasmic reticulum (ER) stress. J. Biol. Chem. 288, 7606–7617. doi: 10.1074/jbc.m112.424655
Tiwari, S., Rogers, M. E., and Stelinski, L. L. (2011). Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest. Manag. Sci. 67, 1258–1268. doi: 10.1002/ps.2181
Ulrich, J., Dao, V. A., Majumdar, U., Schmitt-Engel, C., Schwirz, J., Schultheis, D., et al. (2015). Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteosome as prime target. BMC Genomics 16:674.
Van Ekert, E., Powell, C. A., Shatters, R. G. Jr., and Borovsky, D. (2014). Control of larval and egg development in Aedes aegypti with RNA interference against juvenile hormone acid methyl transferase. J. Insect Physiol. 70, 143–150. doi: 10.1016/j.jinsphys.2014.08.001
Velásquez, A. C., Chakravarthy, S., Gregory, B., and Martin, G. B. (2009). Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 28:1292.
Walshe, D. P., Lehane, S. M., Lehane, M. J., and Haines, L. R. (2009). Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 18, 11–19. doi: 10.1111/j.1365-2583.2008.00839.x
Wang, X.-H., Aliyari, R., Li, W.-X., Li, H.-W., Kim, K., Carthew, R., et al. (2006). RNA interferance directs innate immunity against viruses in adult Drosophila. Science 312, 452–454. doi: 10.1126/science.1125694
Wilkening, S., and Bader, A. (2004). Quantitative real-time polymerase chain reaction: methodical analysis and mathematical model. J. Biomol. Tech.. 15, 107–111.
Wuriyanghan, H., Rosa, C., and Falk, B. W. (2011). Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato Psyllid, Bactericerca cockerelli. PLoS One 6:e27736. doi: 10.1371/journal.pone.0027736
Xie, S., Wu, H., Wang, Q., Cogswell, J. P., Husain, I., Conn, C., et al. (2001). Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J. Biol. Chem. 276, 43305–43312. doi: 10.1074/jbc.m106050200
Yan, R., Urdaneta-Marquez, L., Keller, K., James, C. E., Davey, M. W., and Prichard, R. K. (2012). The role of several ABC transporter genes in ivermectin resistance in Caenorhabditis elegans. Vet. Parasitol. 190, 519–529. doi: 10.1016/j.vetpar.2012.06.038
Yoshinari, K., Miyagishi, M., and Taira, K. (2004). Effects on RNAi of the tight structure, sequence and position of the target region. Nucleic Acids Res. 32, 691–699. doi: 10.1093/nar/gkh221
Yu, X., Gowda, S., and Killiny, N. (2017). Double stranded RNA delivery through soaking, mediates silencing of the muscle protein 20 and increases mortality to the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 73, 1846–1853. doi: 10.1002/ps.4549
Yu, X., and Killiny, N. (2018). Effect of parental RNA interference of a transformer-2 homolog on female reproduction and offspring sex determination in Asian citrus psyllid. Physiol. Entomol. 43, 42–50. doi: 10.1111/phen.12223
Yu, X., and Killiny, N. (2020). RNA interferance-mediated control of Asian citru psyllid, the vector of huanglongbing bacterial pathogen. Trop. Plant Pathol. 45, 298–305. doi: 10.1007/s40858-020-00356-7
Yuan, J. S., Reed, A., Chen, F., and Stewart, C. N. Jr. (2006). Statistical analysis of real-time PCR data. BMC Bioinform. 7:85.
Zhao, Y. Y., Yang, G., Wang-Pruski, G., and You, M. S. (2008). Phyllotreta striolata (Coleoptera: Chrysomelidae): arginine kinase cloning and RNAi-based pest control. Eur. J. Entomol. 105, 815–822. doi: 10.14411/eje.2008.108
Keywords: Diaphorina citri, RNA interference, Citrus tristeza virus engineering, gene silencing with dsRNA, Rieske down regulation
Citation: Ramos JE, Jain RG, Powell CA, Dawson WO, Gowda S, Borovsky D and Shatters RG Jr (2021) Crowdsourced Identification of Potential Target Genes for CTV Induced Gene Silencing for Controlling the Citrus Greening Vector Diaphorina citri. Front. Physiol. 12:571826. doi: 10.3389/fphys.2021.571826
Received: 11 June 2020; Accepted: 19 March 2021;
Published: 09 April 2021.
Edited by:
Guy Smagghe, Ghent University, BelgiumReviewed by:
Mohammad Mehrabadi, Tarbiat Modares University, IranCopyright © 2021 Ramos, Jain, Powell, Dawson, Gowda, Borovsky and Shatters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dov Borovsky, ZG92Ym9yb3Zza3lAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.