- Center for Craniofacial Molecular Biology, Herman Ostrow School of Dentistry, University of Southern California, Los Angeles, CA, United States
Macromolecular assembly of extracellular enamel matrix proteins (EMPs) is intimately associated with the nucleation, growth, and maturation of highly organized hydroxyapatite crystals giving rise to healthy dental enamel. Although the colocalization of two of the most abundant EMPs amelogenin (Amel) and ameloblastin (Ambn) in molar enamel has been established, the evidence toward their interaction is scarce. We used co-immunoprecipitation (co-IP) to show evidence of direct molecular interactions between recombinant and native Amel and Ambn. Ambn fragments containing Y/F-x-x-Y/L/F-x-Y/F self-assembly motif were isolated from the co-IP column and characterized by mass spectroscopy. We used recombinant Ambn (rAmbn) mutants with deletion of exons 5 and 6 as well as Ambn derived synthetic peptides to demonstrate that Ambn binds to Amel via its previously identified Y/F-x-x-Y/L/F-x-Y/F self-assembly motif at the N-terminus of its exon 5 encoded region. Using an N-terminal specific anti-Ambn antibody, we showed that Ambn N-terminal fragments colocalized with Amel from secretory to maturation stages of enamel formation in a single section of developing mouse incisor, and closely followed mineral patterns in enamel rod interrod architecture. We conclude that Ambn self-assembly motif is involved in its interaction with Amel in solution and that colocalization between the two proteins persists from secretory to maturation stages of amelogenesis. Our in vitro and in situ data support the notion that Amel and Ambn may form heteromolecular assemblies that may perform important physiological roles during enamel formation.
Introduction
Dental enamel formation involves a precisely orchestrated series of events in which assembly of extracellular enamel matrix proteins (EMPs) guides the formation of organized hydroxyapatite crystals (Margolis et al., 2006). Amelogenin (Amel) and ameloblastin (Ambn) are integral constituents of the forming enamel extracellular matrix. In vivo and in vitro studies reported that Amel is involved in regulating enamel thickness, controlling calcium phosphate mineral phase, and maintaining hydroxyapatite crystal growth and organization (Gibson et al., 2001; Fang et al., 2011; Bai et al., 2020), while Ambn was suggested to be involved in maintaining rod-interrod architecture (Paine et al., 2003), and ameloblast cell function (Fukumoto et al., 2004). Both proteins are intrinsically disordered and have strong tendency to self-assemble in vitro under a variety of solution conditions (Moradian-Oldak, 2012; Wald et al., 2013). Self-assembly domains within Amel and Ambn sequences have been previously identified and mutations in those domains affect enamel formation in mutant mouse models (Paine et al., 2002; Wald et al., 2017). Amel self-assembly is driven by two domains at its N‐ and C-termini, called as “domain A” and “domain B” respectively (Paine and Snead, 1997; Moradian-Oldak et al., 2000), whereas a Y/F-x-x-Y/L/F-x-Y/F motif located at the N-terminus of its exon 5 encoded region was identified for the self-assembly of Ambn (Wald et al., 2017).
Previous in vitro and in situ investigations have suggested molecular interactions and cooperative functions between EMPs during enamel crystal nucleation and maturation (Fan et al., 2011; Gallon et al., 2013). Specifically, it has been proposed that Amel and Ambn may function cooperatively to control some of the critical steps in the formation of enamel prismatic structure (Hatakeyama et al., 2009; Mazumder et al., 2016). Preliminary evidence that hints at interaction between Amel and Ambn begins at the secretory stage of enamel formation when these proteins are co-secreted through the same vesicles (Zalzal et al., 2008), and continues into maturation stage when their N-terminal fragments colocalize around molar enamel rods (Mazumder et al., 2014, 2016). Using surface plasmon resonance, Wald et al. (2017) suggested that Ambn self-assembly motif (YSRLGF motif in the mouse Ambn sequence) might play a role in binding to Amel but direct interactions between these two motifs were not demonstrated in solution.
Here, we provide direct evidence of the involvement of this conserved self-assembly domain in Amel-Ambn interaction using co-immunoprecipitation (co-IP). Recombinant Ambn (rAmbn) mutants with deletion of the sequences encoded by exons 5 and 6, Ambn-derived synthetic peptides representing the sequences encoded by exons 5 and 6, as well as an Ambn exon 5 peptide with a mutations in the YSRLGF self-assembly motif were used to confirm that this domain is indeed essential for binding of Ambn with Amel. Using mass spectrometry (MS) after co-IP of these two proteins from native porcine extract, we isolated Ambn fragments containing Y/F-x-x-Y/L/F-x-Y/F, confirming our findings from recombinant protein co-IP. In situ co-localization of Amel and N-terminal fragments of Ambn was analyzed from secretory to maturation stages in post-natal-day 8 (P8) wild-type mouse incisor enamel using immunohistochemistry. Longitudinal and transverse sectioning orientations were used to analyze the co-localization within the rod-interrod architecture. Our data confirm that the highly conserved self-assembly motif of Ambn can directly interact with Amel. We further demonstrate that colocalization of both proteins starts at the secretory stage and persists throughout the maturation stage.
Materials and Methods
Recombinant Protein Expression and Purification
Recombinant mouse Amel (rAmel) was expressed in BL21 Escherichia coli following published protocols and was precipitated by using saturated ammonium sulfate. The pallet was dissolved in 0.1% trifluoro acetic acid, and rAmel was purified by reversed phase HPLC (Simmer et al., 1994). Recombinant mouse Ambn (rAmbn) and rAmbn mutants with deletion of exons 5 or 6 (rAmbnΔ5 and rAmbnΔ6) were similarly expressed in BL21 E. coli. Protein from lysed E. coli was first concentrated by Ni-NTA column (QIAgen), and then dialyzed through a 10,000 Da dialysis membrane against ice cold phosphate buffer. Thioredoxin tag, S-tag, and histidine tag were cleaved by enzyme Enterokinase (light chain, New England Biolabs) at 37°C and rAmbn was purified by reversed phase HPLC (Su et al., 2019a,b). Purified rAmel and rAmbn proteins were characterized by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry (Supplementary Figure 1). Proteins were lyophilized and stored at −20°C until further use.
Porcine Enamel Matrix Protein Extraction
To obtain porcine EMPs, 6 month-old pig mandibles were purchased from Sierra for Medical Science (Whittier, CA, United States). Un-erupted second molars were extracted from their bony cavities, and EMPs were extracted from the crowns of the developing molars using previously published protocol (Uchida et al., 1995). Briefly, molar crowns were cleaned with ice cold phosphate buffered saline (PBS, pH 7.4) and newly formed enamel was scraped with a sharp razor blade on clean glass plates. Enamel scrapings were stirred overnight at 4°C in 0.5 M acetic acid. The slurry was desalted using Amicon Ultra 15 centrifugal filters to remove calcium phosphate. The supernatant was lyophilized and proteins were characterized using Western blots to confirm the presence of Amel and Ambn fragments (Supplementary Figure 2).
Ambn-Derived Synthetic Peptides
Synthetic peptides derived from mouse Ambn protein regions encoded by exons 5 and 6 are referred here as Ambn-exon 5 derived peptide (AB2) and Ambn-exon 6 derived peptide (AB4), respectively (Su et al., 2016). AB2 was further divided into N‐ and C-terminal peptides peptide derived from N-terminus of AB2 (AB2N) and peptide derived from C-terminus of AB2 (AB2C; Chempeptide Ltd., China). A mutant of AB2N was designed by replacing the key amino acids in the YSRLGF motif with glycine, making it GSRGGG (Wald et al., 2017), which we refer to as peptide AB2N with mutation p.Y67G_L70G_F72G (AB2N-GGG; Biomertech, United States; Supplementary Figures 3, 4; Supplementary Table 1).
Co-Immunoprecipitation
Co-immunoprecipitation protocol (Supplementary Scheme 1) was modified from Fan et al. (2009) and performed using a Pierce co-IP kit (Thermofisher Scientific). Manufacturer’s protocol was followed to bind either 10 μg anti-Amel (gift from Dr. Malcolm Snead) or 10 μg anti-Ambn antibody (AF3026, R&D systems) to the antibody coupling resin. Ten micrograms bait protein was added to the respective antibody bound column and incubated overnight at 4°C with gentle shaking. Columns were washed 5–6 times with Dulbecco’s PBS and 10 μg prey protein was added to the columns and bound for 4 h at 4°C. In case of native porcine EMP extract, the bait and prey proteins were loaded in the column as a mixture of all enamel matrix proteins. The bait-prey complex was eluted using 60 μl elution buffer provided with the kit. Control co-IP experiments were conducted in the absence of an antibody to visualize nonspecific binding to the column. Elution fractions were lyophilized and analyzed using SDS-PAGE, Western blots, and mass spectrometry.
Mass Spectrometry (MS)
Elution fractions of native porcine EMP co-IP were analyzed by mass spectrometry at Scripps Center for Metabolomics and Mass Spectrometry (San Diego, CA, United States). Detailed protocol is described in Supplementary Material. Briefly, MS analysis was performed using nanoelectrospray capillary column packed with Zorbax SB-C18 stationary phase (Agilent). MS/MS data were obtained with an LTQ linear ion trap mass spectrometer using a nanoelectrospray source at 2 kV at the tip. All MS/MS data were searched against the NCBI Mammalia (mammals) database using Mascot (version 2.3.02; Matrix Science, London, United Kingdom). Proteins with a p < 0.05 (corresponding to a Mascot ion score greater than 57) were identified with two or more peptides and considered at 95% confidence level.
Immunohistochemical Labeling of Post-Natal-Day 8 Mouse Incisors
Post-natal-day 8 wild-type C57BL/6 mice were euthanized following Institutional Animal Care and Use Committee (IACUC) protocols of the University of Southern California. Their mandibles were dissected, fixed (4% paraformaldehyde), decalcified (10% EDTA with 0.1% glutaraldehyde), paraffin embedded, and sectioned into 7 μm thick sections along the sagittal plane and anteroposteriorly along the transverse plane, maintaining the integrity of the developing incisor. Sections were immunohistochemically labeled with anti-Amel (gift from Dr. Malcolm Snead) or anti-Ambn (N-18, sc-33100, Santa Cruz Biotech, discontinued) primary antibodies (dilutions in Supplementary Table 2) and corresponding secondary antibodies conjugated with FITC or Alexa 488 for Amel and TRITC for Ambn using previously published protocol (Bapat and Moradian-Oldak, 2019). The slides were examined using a Leica SP-8 confocal microscope and data were analyzed with Leica Application Suite LAS-X version 1.8.1.13759. Manders’ colocalization coefficients (MCCs; Manders et al., 1993; Gallon et al., 2013) were utilized to quantify the colocalization between Amel and Ambn and calculated using Microsoft Excel. The change in colocalization coefficients between secretory and transition stage was calculated using a two-sample t-test, whereas the change in MCC values between secretory and maturation stages was calculated by Mann Whitney U-test in OriginPro data analysis software.
Alizarin Red S Staining
Anteroposterior sections of P8 mouse incisor were deparaffinized using decreasing concentrations of xylene and ethanol, stained in freshly prepared 2% (w/v) Alizarin red S solution (pH 4.1–4.3) for 2–3 min, dehydrated in acetone followed by acetone-xylene solution (1:1), cleared in xylene, and mounted with a synthetic mounting medium (Dahl, 1952). Sections were observed using a Keyence BZX-810 microscope in bright-field mode.
Results
Recombinant Amel and Ambn Bind in vitro
Direct binding between rAmel and rAmbn was confirmed by co-IP using anti-Amel and anti-Ambn antibodies. rAmbn bound to rAmel bait (Figure 1A, elution), and rAmel bound to rAmbn bait (Figure 1B, elution) to co-elute from the co-IP columns. Control experiment in the absence of anti-Amel antibody showed ~50% less rAmbn and rAmbn bound to the column (Figure 1A, control) and in the absence of anti-Ambn antibody no significant non-specific binding to the co-IP resin was observed (Figure 1B, control).
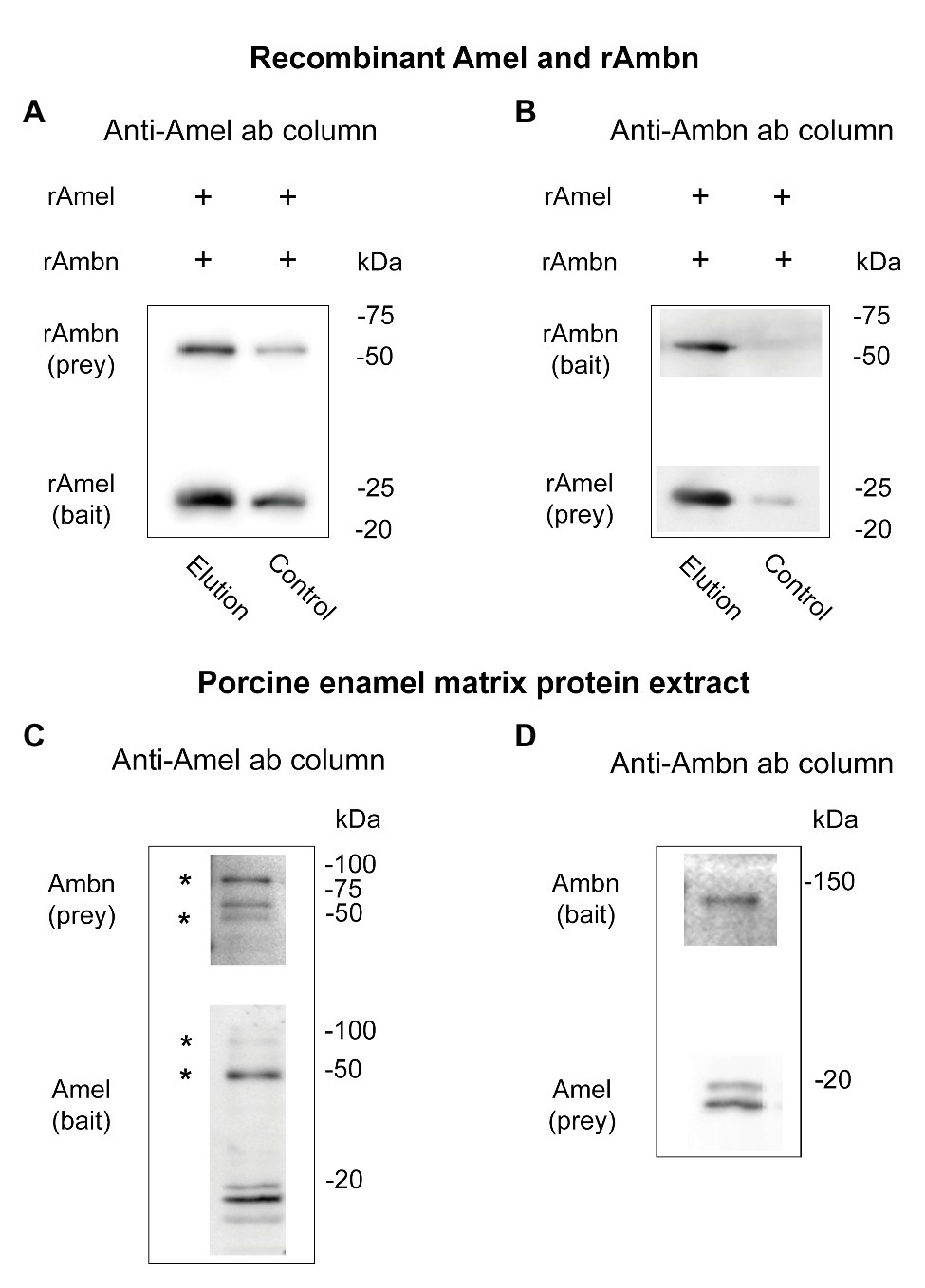
Figure 1. (A–D) Western blots showing (A,B) co-immunoprecipitation (co-IP) of recombinant Amel and Ambn; (A) lane 1 (elution) – co-IP of recombinant Ambn (rAmbn) with recombinant mouse Amel (rAmel) using rAmel as bait, lane 2 (control) – non-specific binding in the absence of an antibody is at least 50% less than elution; (B) lane 1 (elution) – co-IP of rAmel with rAmbn using rAmbn as bait, lane 2 (control) – no significant non-specific binding observed in the absence of antibody; (C,D) co-IP of native Amel and Ambn from porcine enamel matrix protein extract using (C) anti-Amel antibody column, (D) using anti-Ambn antibody column; asterisks (*) in (C) mark putative Amel-Ambn complexes.
Native Amel-Ambn Complexes Co-Elute From Porcine EMP Extract
Support for direct binding between native Amel and Ambn proteins was obtained by their co-IP from porcine second molar EMP extract (Figures 1C,D). Three bands at ~85, ~65, and ~45 kDa were labeled by the anti-Ambn antibody in the elution fractions of native EMP co-IP when anti-Amel antibody was used for the co-IP column (Figure 1C). Interestingly, the same 85 and 45 kDa bands were also labeled in anti-Amel Western blot (Figure 1C, asterisks). In the same blot, native Amel fragments were identified as previously described “18,” “20,” and central “13 k” fragments (Ryu et al., 1999; Figure 1C). Mass spectra from the elution fractions of this co-IP revealed Ambn peptides from exons 4 and 5 encoded region containing a partial Y/F-x-x-Y/L/F-x-Y/F motif with a protein score > 57, corresponding to 95% confidence level (Supplementary Table 3). The interaction was confirmed by reversing the experiment with anti-Ambn antibody in the co-IP column, which revealed Ambn in a single band at ~130 kDa, and Amel as 18 and 20 kDa bands (Figure 1D). Mass spectrometric analysis of this co-IP identified Ambn peptides matching the exons 3–5 encoded fragments, containing the entire Y/F-x-x-Y/L/F-x-Y/F motif (95% confidence level). Mass spectrometric analysis of co-IP elution fractions from both experiments also identified Amel peptides located within tyrosine rich amelogenin polypeptide (TRAP) containing the Y/F-x-x-Y/L/F-x-Y/F self-assembly motif but with confidence level < 95% (Supplementary Table 3).
YSRLGF Motif of Ambn Is Essential for Binding With Amel
In order to confirm the region of rAmbn that interacts with rAmel, co-IP experiments were repeated using mutants rAmbnΔ5 and rAmbnΔ6. Co-IP between rAmel and rAmbn was used as a positive control (Figure 2A, lane 1). Mutant rAmbnΔ6 retained its ability to bind to Amel and co-eluted with Amel (Figure 2A, lane 2); however, rAmbnΔ5 lost its ability to bind to Amel and did not co-elute (Figure 2A, lane 3); suggesting that exon 5 encoded region of Ambn is essential for its interaction with Amel. Co-IP of rAmel with Ambn exon 5 derived synthetic peptides AB2 and AB2N containing the YSRLGF motif confirmed the direct binding of this region to rAmel (Figure 2B, lanes 2 and 3, respectively). Peptide AB2C encoding the C-terminus of exon 5 lacking the YSRLGF motif did not bind to rAmel (Figure 2B, lane 4). Peptide AB4 representing Ambn exon 6 region also failed to bind to rAmel (Figure 2B, lane 1). Ambn peptide AB2N-GGG with YSRLGF motif mutated to GSRGGG also lost its ability to bind to rAmel (Figure 2B, lane 5). The sequences of Ambn synthetic peptides and mutant peptide AB2N-GGG are shown in Figure 2C. Masses of peptides are listed in Supplementary Table 1.
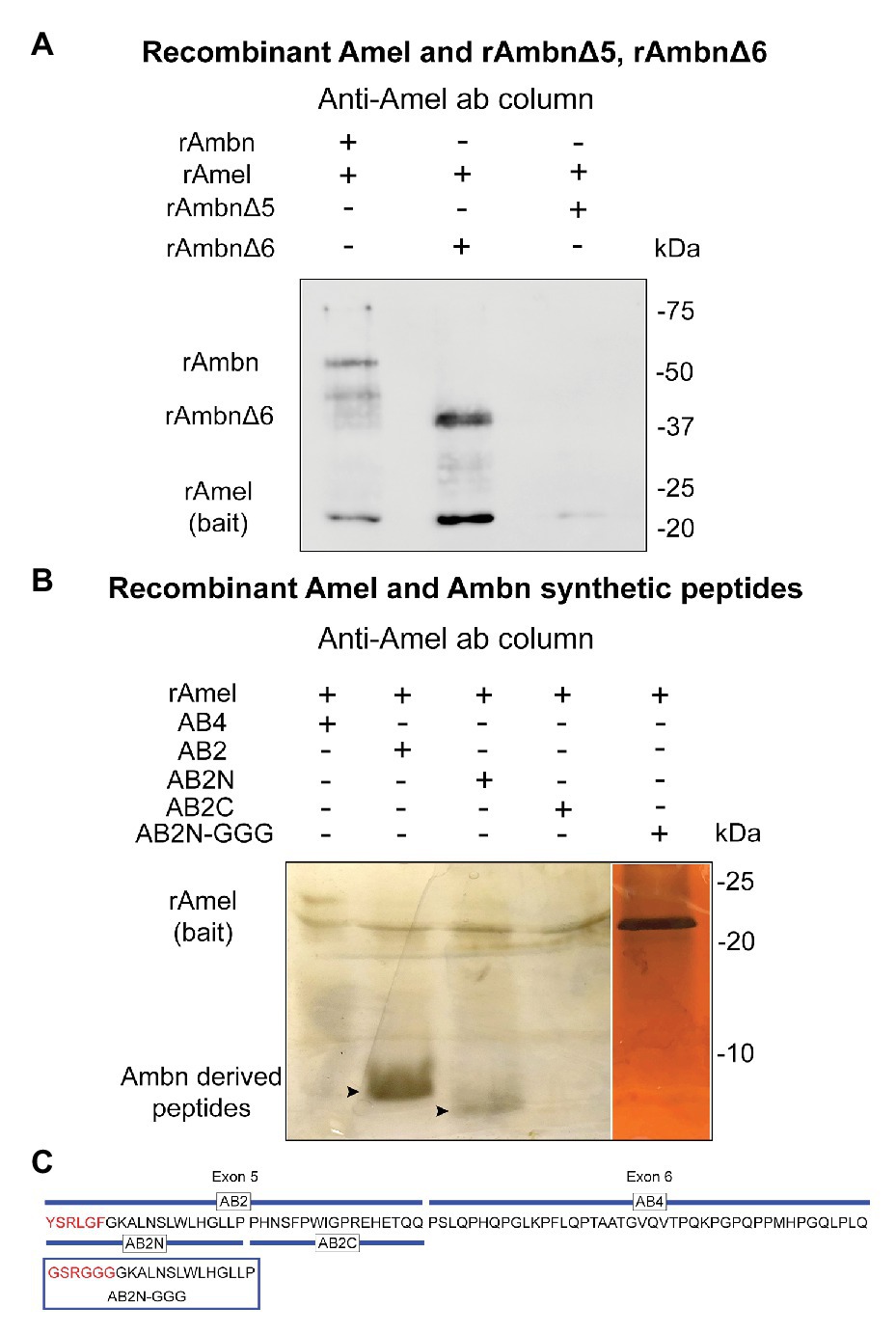
Figure 2. (A) Western blot showing lane 1 – rAmel and rAmbn co-IP positive control, lane 2 – rAmbnΔ6 co-elutes with rAmel, and lane 3 – rAmbnΔ5 fails to bind to rAmel; (B) 16% sodium dodecyl sulfate (SDS) gels stained with silver stain showing lane 1 – Ambn-exon 6 derived peptide (AB4) did not bind to rAmel, lane 2 – Ambn-exon 5 derived peptide (AB2) co-elutes with rAmel (arrowhead), lane 3 – peptide derived from N-terminus of AB2 (AB2N) containing the self-assembly motif YSRLGF co-elutes with rAmel (arrowhead), lane 4 – peptide derived from C-terminus of AB2 (AB2C) did not bind to rAmel, and lane 5 – peptide AB2N (AB2N-GGG) with mutated YSRLGF motif (p.Y67G_L70G_F72G) did not bind to rAmel; (C) schematic representation of mouse Ambn sequence encoded by exons 5 and 6 showing Ambn derived synthetic peptides AB2, AB2N, AB2C, AB4, and mutant peptide AB2N-GGG.
Amel-Ambn Co-Localization at Different Stages of Amelogenesis in Developing Mouse Incisor
Tile-scan image of P8 incisor showed the presence of Amel and N-18 antibody labeled Ambn within ameloblasts (Am), at the secretory fronts of ameloblasts, and throughout the entire bulk of the enamel thickness from secretory to maturation stage of enamel formation (Figure 3A). At a higher resolution, the maximum intensity projections of Z-stacks of secretory, transition, and maturation stages (Figure 3B-D) of enamel formation clearly depicted Amel-Ambn colocalization. In secretory stage (Figure 3B), the MCC for Amel was 0.95 and for Ambn 0.92 within ameloblasts. The values were similar to the MCC within secretory stage Tomes’ processes (TP), 0.96 for Amel, and 0.94 for Ambn (Figure 3E). In transition stage (Figure 3C), the MCC for both Amel and Ambn within ameloblasts increased to 0.98 (Figure 3F) but decreased as maturation progressed to 0.87 for Amel and 0.84 for Ambn (Figures 3D,G). Within the enamel matrix in secretory stage (Figure 3B), the MCC for Amel was 0.90 and for Ambn 0.85 (Figure 3E), meaning about 90% of labeled Amel colocalized with 85% of labeled Ambn fragments. In the transition stage enamel matrix (Figure 3C), the MCC values for Amel and Ambn increased to 0.94 and 0.91, respectively (Figure 3F), finally culminating at almost 0.99 for both Amel and Ambn in maturation stage enamel (Figures 3D,G). The Amel and Ambn colocalization within enamel matrix increased significantly from secretory to maturation stages (p < 0.001). The N-18 antibody labeled full-length Ambn, along with proteolytic cleavage products of Ambn. Based on N-18 antibody epitope, these fragments potentially contained regions in close proximity to the exon 5 encoded region (Supplementary Figure 3). For comparison, M300 antibody detected Ambn only within ameloblasts and at the Tomes’ processes but did not label Ambn proteolytic cleavage fragments within the bulk of the enamel matrix (Supplementary Figure 5).
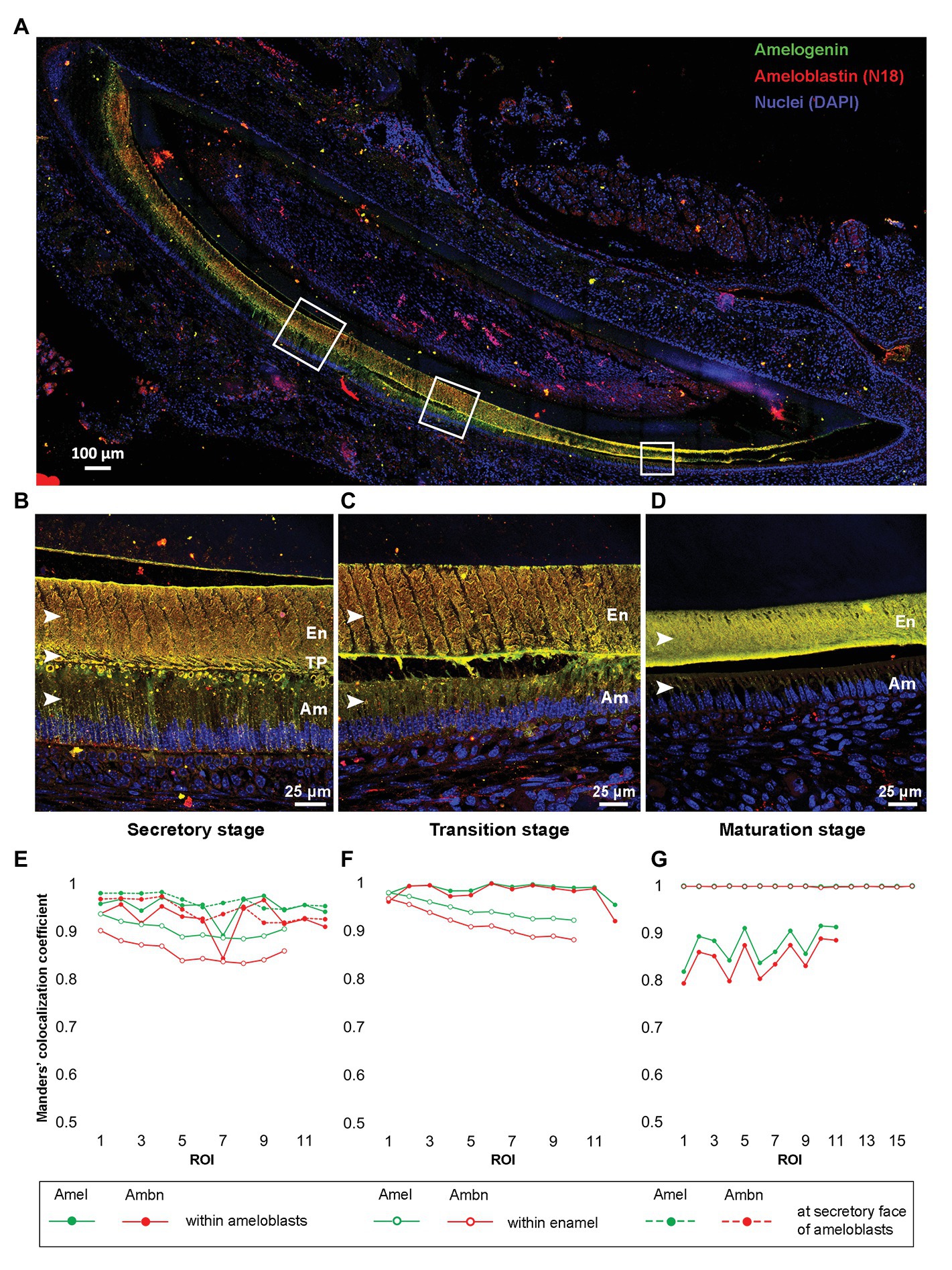
Figure 3. (A) Merged tile-scan confocal image of a longitudinal section from post-natal-day 8 (P8) mouse incisor co-labeled with anti-Amel (green) and anti-Ambn N-18 (red) antibodies. (B–D) Maximum intensity projections of Z-stacks of secretory, transition, and maturation stage areas marked by white squares in (A); (E–G) Manders’ colocalization coefficients (MCCs) for Amel and Ambn at different regions within secretory, transition, and maturation stages of enamel formation, respectively. (B,E) secretory stage, MCC calculated within enamel matrix, at secretory face of ameloblasts, and within ameloblasts (white arrowheads in B); (C,F) transition stage, MCC calculated within enamel matrix and inside ameloblasts (white arrowheads in C) and (D,G) maturation stage, MCC calculated within enamel matrix and within ameloblasts (white arrowhead in D). En, enamel; TP, Tomes’ processes; and Am, ameloblasts.
To confirm that Amel-Ambn co-localization persists during the maturation stage and to provide better visualization, anteroposterior sections through the tip of the incisor were used. Clearly demarcated enamel rod architecture was visible due to Amel co-localizing with N-terminal-containing fragments of Ambn in the maturing enamel (Figure 4A). Distal to the tip of the incisor, Amel-Ambn colocalization could be differentiated into surface enamel (SE), outer enamel (OE), and bulk enamel (BE) layers based on the colocalization pattern (Figure 4B). Alizarin Red S staining of demineralized sections revealed that there was residual calcium present in the maturation stage enamel even after EDTA demineralization (Figures 4C,D). The pattern of Amel-Ambn co-assembly in the anteroposterior sections closely mimicked the pattern of the mineral remnants and the rod-interrod structure observed previously in SEM images of non-demineralized mouse enamel (Lacruz et al., 2012).
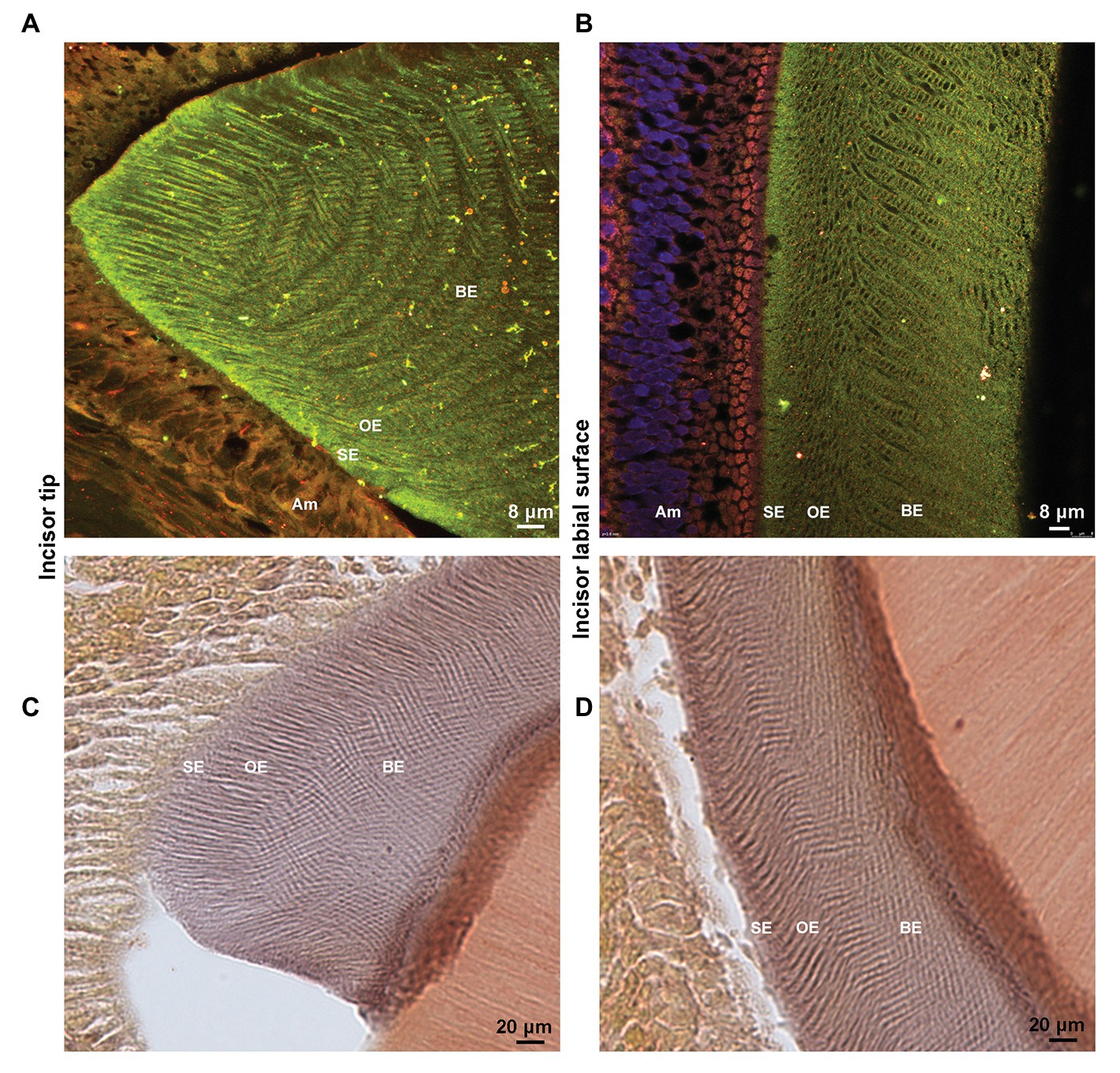
Figure 4. (A) Maximum projections of Z-stack confocal images and (B) a single confocal image of an anteroposterior section from maturation-stage P8 enamel showing co-localization of Amel (green) and Ambn (red, N18 antibody) and the rod-interrod architecture. (C,D) Bright field image of Alizarin red S stained P8 incisor sections depicting residual mineral in maturation stage enamel rod-interrod architecture. (A,C) tip of the incisor (B,D) labial surface of the incisor. Am, ameloblasts; SE, surface enamel; OE, outer enamel; and BE, bulk enamel.
Discussion
The essential physiological functions of both Amel and Ambn proteins in the formation of normal enamel have been documented in knockout and mutant animal models (Gibson et al., 2001; Paine et al., 2002; Fukumoto et al., 2004; Liang et al., 2019). Mutations in AMELX or AMBN genes in humans disrupt enamel formation and cause Amelogenesis Imperfecta (AI) with a range of clinical presentations including but not limited to very thin mineralized enamel or soft friable enamel (hypoplastic or hypomineralized, respectively; Smith et al., 2017).
In vitro and in vivo studies have suggested that protein assembly is the principal mechanism for controlling nucleation, growth, and organization of mineral crystals during enamel biomineralization (Margolis et al., 2006; Moradian-Oldak, 2012). Enamel extracellular proteins may function in a cooperative or synergic manner and direct interactions or co-assembly may be a requirement for their role (Fan et al., 2011; Wald et al., 2017; Tao et al., 2018).
The critical role of the Y/F-x-x-Y/L/F-x-Y/F motif at the N-terminus of Ambn exon 5-encoded region was recently identified in Ambn self-assembly (Wald et al., 2013, 2017). A mouse model having key residues of this domain mutated to glycine showed a lack of patterning in secreted Amel matrix leading to disorganized enamel crystals (Wald et al., 2017). The authors suggested that the Y/F-x-x-Y/L/F-x-Y/F motif may be involved in Amel-Ambn co-assembly along with Ambn self-assembly. The results of our current study show direct evidence of such interactions in solution, and our findings support that the abnormal enamel formation observed in the mutant animals by Wald et al. (2017) could be a result of interruptions in Ambn-Amel interactions.
Using co-IP, we confirmed that rAmbn directly binds to rAmel in solution. Here, the occurrence of this interaction in in vivo extracts is established by co-IP of native Amel and Ambn proteins extracted from porcine enamel in developing second molars. This confirmed that the interaction is occurring via a region devoid of any post translational modifications (Kobayashi et al., 2007) as it remained consistent in recombinant (E. coli derived) and native proteins. The 85 and 45 kDa bands detected in native porcine co-IP experiments with anti-Ambn antibody in the column could not be ascribed to either native Amel or Ambn fragments described in literature (Ryu et al., 1999; Iwata et al., 2007). These could be putative Amel and Ambn complexes.
As documented in mutant animal models, Ambn exons 5 and 6 are key in Ambn protein function and are involved in maintaining ameloblast cells (Fukumoto et al., 2004; Wazen et al., 2009). We identified that Ambn mutants with deletion of exons 5 (rAmbnΔ5) did not bind to Amel but rAmbnΔ6 retained its ability to bind to Amel. This implied that the region encoded by exon 5 of Ambn is essential for its interaction with Amel. AB2, the synthetic peptide derived from sequence encoded by exon 5 bound to Amel. To narrow down the location of the binding domain within AB2, we employed peptides AB2N, AB2C, and AB2N-GGG representing the N‐ and C-termini of AB2 and a peptide with mutated Y/F-x-x-Y/L/F-x-Y/F motif, respectively. AB2 and AB2N bound to Amel but AB2C and AB2N-GGG did not, thus confirming that the Y/F-x-x-Y/L/F-x-Y/F motif of Ambn is involved in its interaction with Amel. An AI case with a mutation within exon 5 of AMBN has not yet been identified, but deletion of exon 6 encoded region of AMBN which is adjacent to Ambn self-assembly Y/F-x-x-Y/L/F-x-Y/F motif does cause AI in humans (Poulter et al., 2014).
Moreover, a proline to threonine mutation adjacent to one of the self-assembly motifs of Amel within the TRAP region has also been identified which leads to hypomaturation type of AI (Collier et al., 1997). These mutations could potentially affect Amel-Ambn interactions intensifying the AI phenotype. The two evolutionary conserved Y/F-x-x-Y/L/F-x-Y/F motifs are also present within the TRAP peptide of Amel which overlaps with it’s self-assembly “A domain” (Paine and Snead, 1997; Wald et al., 2017). Mutations in the “A domain” lead to disruption of protein-protein interaction and ultimately to enamel malformation (Moradian-Oldak et al., 2000; Paine et al., 2002). It has been previously shown that TRAP accumulates in maturation stage enamel (Fincham et al., 1981; Mazumder et al., 2016) and can directly interact with Ambn in vitro (Su et al., 2016). In this study, mass spectrometry identified Amel peptides from the TRAP region, including one containing the self-assembly motif in co-IP elution fractions of porcine enamel matrix protein extract. Our current observation together with previously published data of Amel self-assembly supports the conclusion that the self-assembly domains on both Ambn as well as Amel may function as the interacting domains between the two proteins.
We previously reported colocalization of Amel and Ambn N-terminal fragments within the sheath space of maturation stage molar enamel (Mazumder et al., 2016). In this study, the rationale behind demonstrating Amel-Ambn colocalization in the mouse incisor model was to compare and identify possible changes in their colocalization as enamel maturation progressed. The persistent significant increase in colocalization coefficient observed from secretory to maturation stage enamel in a single incisor suggested continued interaction between not only nascent Amel and Ambn but also their proteolytic cleavage products.
The function of Amel-Ambn interactions and their biological significance are important subjects of our current and future investigations. One such function maybe related to maintaining the ameloblast-enamel matrix interface. We recently showed that Ambn localizes at the ameloblast-enamel matrix interface and interacts with ameloblast cell membrane (Su et al., 2019b, 2020). Considering that Amel is the main component of the matrix, binding of Ambn with Amel and with ameloblast cell membrane may together maintain the ameloblast-enamel matrix interface. This could also explain the severe separation of ameloblasts from underlying matrix in mice with deleted Ambn exons 5 and 6 (Fukumoto et al., 2004). Another aspect of such interactions may be related to the synergic function of these proteins in controlling mineral nucleation and growth (Fan et al., 2011; Tao et al., 2018). The ability of Ambn C-terminal region to bind calcium has been well-documented (Yamakoshi et al., 2001; Tarasevich et al., 2007; Zhang et al., 2011) suggesting Ambn’s involvement in mineralization. A recent report of AI with a point mutation in the calcium-binding region of Ambn described enamel with low mineral density (Lu et al., 2018).
In conclusion, we provide direct evidence for Ambn and Amel protein interactions in solution and demonstrate their colocalization in situ in mouse incisor, starting at the secretory stage and persisting until maturation stage. Dissecting Amel-Ambn co-assemblies and exploring the details of Amel-Ambn molecular interactions in the presence of calcium are the subjects for future investigations.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by University of Southern California IACUC.
Author Contributions
RAB designed and performed the experiments, analyzed the data, and wrote the manuscript. JS helped with protein synthesis and peptide design. JM-O designed experiments, analyzed data, wrote and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by NIH-NIDCR grants R01DE013414 and DE027632 to JM-O and USC Research Enhancement Fellowship to RAB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. M. Snead and M. Paine for providing Amel and Ambn antibodies, Dr. D. Eremin at the Agilent Center for Excellence in Biomolecular Characterization USC and Linh Hoang at the Scripps Center for Metabolomics and Mass Spectrometry for help with mass spectrometry; Gayathri Visakan for assistance in statistical analysis, and Drs. Natalie Kegulian and Bridget Samuels for proofreading the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.622086/full#supplementary-material
Abbreviations
AB2, Ambn-exon 5 derived peptide; AB2C, Peptide derived from C-terminus of AB2; AB2N, Peptide derived from N-terminus of AB2; AB2N, GGG, Peptide AB2N with mutation p.Y67G_L70G_F72G; AB4, Ambn-exon 6 derived peptide; AI, Amelogenesis imperfecta; Ambn/rAmbn, Ameloblastin/recombinant ameloblastin; AmbnΔ5, Ambn p.Y67_Q103del; AmbnΔ6, Ambn p.P104_V168del; Amel/rAmel, Amelogenin/recombinant amelogenin; Co-IP, Co-immunoprecipitation; EMP, Enamel matrix protein; MCC, Manders’ colocalization coefficient; SDS-PAGE, Sodium dodecyl sulfate polyacrylamide gel electrophoresis; TRAP, Tyrosine rich amelogenin polypeptide.
References
Bai, Y., Yu, Z., Ackerman, L., Zhang, Y., Bonde, J., Li, W., et al. (2020). Protein nanoribbons template enamel mineralization. Proc. Natl. Acad. Sci. 117, 19201–19208. doi: 10.1073/pnas.2007838117
Bapat, R. A., and Moradian-Oldak, J. (2019). “Immunohistochemical co-localization of amelogenin and ameloblastin in developing enamel matrix” in Odontogenesis. ed. P. Papagerakis (New York, NY: Humana Press), 219–228.
Collier, P., Sauk, J., Rosenbloom, J., Yuan, Z., and Gibson, C. (1997). An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch. Oral Biol. 42, 235–242. doi: 10.1016/S0003-9969(96)00099-4
Dahl, L. K. (1952). A simple and sensitive histochemical method for calcium. Proc. Soc. Exp. Biol. Med. 80, 474–479. doi: 10.3181/00379727-80-19661
Fan, D., Du, C., Sun, Z., Lakshminarayanan, R., and Moradian-Oldak, J. (2009). In vitro study on the interaction between the 32kDa enamelin and amelogenin. J. Struct. Biol. 166, 88–94. doi: 10.1016/j.jsb.2009.01.003
Fan, D., Iijima, M., Bromley, K. M., Yang, X., Mathew, S., and Moradian-Oldak, J. (2011). The cooperation of enamelin and amelogenin in controlling octacalcium phosphate crystal morphology. Cells Tissues Organs 194, 194–198. doi: 10.1159/000324208
Fang, P. -A., Conway, J. F., Margolis, H. C., Simmer, J. P., and Beniash, E. (2011). Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. 108, 14097–14102. doi: 10.1073/pnas.1106228108
Fincham, A., Belcourt, A., Termine, J., Butler, W., and Cothran, W. (1981). Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci. Rep. 1, 771–778. doi: 10.1007/BF01114799
Fukumoto, S., Kiba, T., Hall, B., Iehara, N., Nakamura, T., Longenecker, G., et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167, 973–983. doi: 10.1083/jcb.200409077
Gallon, V., Chen, L., Yang, X., and Moradian-Oldak, J. (2013). Localization and quantitative co-localization of enamelin with amelogenin. J. Struct. Biol. 183, 239–249. doi: 10.1016/j.jsb.2013.03.014
Gibson, C. W., Yuan, Z. A., Hall, B., Longenecker, G., Chen, E., Thyagarajan, T., et al. (2001). Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 276, 31871–31875. doi: 10.1074/jbc.M104624200
Hatakeyama, J., Fukumoto, S., Nakamura, T., Haruyama, N., Suzuki, S., Hatakeyama, Y., et al. (2009). Synergistic roles of amelogenin and ameloblastin. J. Dent. Res. 88, 318–322. doi: 10.1177/0022034509334749
Iwata, T., Yamakoshi, Y., Hu, J. C., Ishikawa, I., Bartlett, J. D., Krebsbach, P. H., et al. (2007). Processing of ameloblastin by MMP-20. J. Dent. Res. 86, 153–157. doi: 10.1177/154405910708600209
Kobayashi, K., Yamakoshi, Y., Hu, J. -C., Gomi, K., Arai, T., Fukae, M., et al. (2007). Splicing determines the glycosylation state of ameloblastin. J. Dent. Res. 86, 962–967. doi: 10.1177/154405910708601009
Lacruz, R. S., Nakayama, Y., Holcroft, J., Nguyen, V., Somogyi-Ganss, E., Snead, M. L., et al. (2012). Targeted overexpression of amelotin disrupts the microstructure of dental enamel. PLoS One 7:e35200. doi: 10.1371/journal.pone.0035200
Liang, T., Hu, Y., Smith, C. E., Richardson, A. S., Zhang, H., Yang, J., et al. (2019). AMBN mutations causing hypoplastic amelogenesis imperfecta and Ambn knockout-NLS-lacZ knockin mice exhibiting failed amelogenesis and Ambn tissue-specificity. Mol. Genet. Genomic Med. 7:e929. doi: 10.1002/mgg3.929
Lu, T., Li, M., Xu, X., Xiong, J., Huang, C., Zhang, X., et al. (2018). Whole exome sequencing identifies an AMBN missense mutation causing severe autosomal-dominant amelogenesis imperfecta and dentin disorders. Int. J. Oral Sci. 10:26. doi: 10.1038/s41368-018-0027-9
Manders, E., Verbeek, F., and Aten, J. (1993). Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x
Margolis, H. C., Beniash, E., and Fowler, C. E. (2006). Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 85, 775–793. doi: 10.1177/154405910608500902
Mazumder, P., Prajapati, S., Bapat, R., and Moradian-Oldak, J. (2016). Amelogenin-ameloblastin spatial interaction around maturing enamel rods. J. Dent. Res. 95, 1042–1048. doi: 10.1177/0022034516645389
Mazumder, P., Prajapati, S., Lokappa, S. B., Gallon, V., and Moradian-Oldak, J. (2014). Analysis of co-assembly and co-localization of ameloblastin and amelogenin. Front. Physiol. 5:274. doi: 10.3389/fphys.2014.00274
Moradian-Oldak, J. (2012). Protein-mediated enamel mineralization. Front. Biosci. 17, 1996–2023. doi: 10.2741/4034
Moradian-Oldak, J., Paine, M. L., Lei, Y. P., Fincham, A. G., and Snead, M. L. (2000). Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J. Struct. Biol. 131, 27–37. doi: 10.1006/jsbi.2000.4237
Paine, M. L., Lei, Y. -P., Dickerson, K., and Snead, M. L. (2002). Altered amelogenin self-assembly based on mutations observed in human X-linked amelogenesis imperfecta (AIH1). J. Biol. Chem. 277, 17112–17116. doi: 10.1074/jbc.M110473200
Paine, M. L., and Snead, M. L. (1997). Protein interactions during assembly of the enamel organic extracellular matrix. J. Bone Miner. Res. 12, 221–227. doi: 10.1359/jbmr.1997.12.2.221
Paine, M. L., Wang, H. -J., Luo, W., Krebsbach, P. H., and Snead, M. L. (2003). A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J. Biol. Chem. 278, 19447–19452. doi: 10.1074/jbc.M300445200
Poulter, J. A., Murillo, G., Brookes, S. J., Smith, C. E. L., Parry, D. A., Silva, S., et al. (2014). Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum. Mol. Genet. 23, 5317–5324. doi: 10.1093/hmg/ddu247
Ryu, O. H., Fincham, A. G., Hu, C. -C., Zhang, C., Qian, Q., Bartlett, J. D., et al. (1999). Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J. Dent. Res. 78, 743–750. doi: 10.1177/00220345990780030601
Simmer, J. P., Lau, E. C., Hu, C. C., Aoba, T., Lacey, M., Nelson, D., et al. (1994). Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif. Tissue Int. 54, 312–319. doi: 10.1007/BF00295956
Smith, C. E., Poulter, J. A., Antanaviciute, A., Kirkham, J., Brookes, S. J., Inglehearn, C. F., et al. (2017). Amelogenesis imperfecta; genes, proteins, and pathways. Front. Physiol. 8:435. doi: 10.3389/fphys.2017.00435
Su, J., Bapat, R. A., and Moradian-Oldak, J. (2019). “The expression and purification of recombinant mouse ameloblastin in E. coli” in Odontogenesis. ed. P. Papagerakis (New York, NY: Humana Press), 229–236.
Su, J., Bapat, R. A., Visakan, G., and Moradian-Oldak, J. (2020). An evolutionarily conserved helix mediates ameloblastin-cell interaction. J. Dent. Res. 99, 1072–1081. doi: 10.1177/0022034520918521
Su, J., Chandrababu, K. B., and Moradian-Oldak, J. (2016). Ameloblastin peptide encoded by exon 5 interacts with amelogenin N-terminus. Biochem. Biophys. Rep. 7, 26–32. doi: 10.1016/j.bbrep.2016.05.007
Su, J., Kegulian, N. C., Bapat, R. A., and Moradian-Oldak, J. (2019b). Ameloblastin binds to phospholipid bilayers via a helix-forming motif within the sequence encoded by exon 5. ACS Omega 4, 4405–4416. doi: 10.1021/acsomega.8b03582
Tao, J., Fijneman, A., Wan, J., Prajapati, S., Mukherjee, K., Fernandez-Martinez, A., et al. (2018). Control of calcium phosphate nucleation and transformation through interactions of enamelin and amelogenin exhibits the “goldilocks effect”. Cryst. Growth Des. 18, 7391–7400. doi: 10.1021/acs.cgd.8b01066
Tarasevich, B. J., Howard, C. J., Larson, J. L., Snead, M. L., Simmer, J. P., Paine, M., et al. (2007). The nucleation and growth of calcium phosphate by amelogenin. J. Cryst. Growth 304, 407–415. doi: 10.1016/j.jcrysgro.2007.02.035
Uchida, T., Fukae, M., Tanabe, T., Yamakoshi, Y., Satoda, T., Murakami, C., et al. (1995). Immunochemical and immunocytochemical study of a 15 kDa non-amelogenin and related proteins in the porcine immature enamel: proposal of a new group of enamel proteins ‘sheath proteins’. Biomed. Res. 16, 131–140. doi: 10.2220/biomedres.16.131
Wald, T., Osickova, A., Sulc, M., Benada, O., Semeradtova, A., Rezabkova, L., et al. (2013). Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J. Biol. Chem. 288, 22333–22345. doi: 10.1074/jbc.M113.456012
Wald, T., Spoutil, F., Osickova, A., Prochazkova, M., Benada, O., Kasparek, P., et al. (2017). Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc. Natl. Acad. Sci. 114, E1641–E1650. doi: 10.1073/pnas.1615334114
Wazen, R. M., Moffatt, P., Zalzal, S. F., Yamada, Y., and Nanci, A. (2009). A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 28, 292–303. doi: 10.1016/j.matbio.2009.04.004
Yamakoshi, Y., Tanabe, T., Oida, S., Hu, C. -C., Simmer, J., and Fukae, M. (2001). Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch. Oral Biol. 46, 1005–1014. doi: 10.1016/s0003-9969(01)00070-x
Zalzal, S. F., Smith, C. E., and Nanci, A. (2008). Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 27, 352–359. doi: 10.1016/j.matbio.2008.01.003
Keywords: enamel biomineralization, amelogenin (Amel), ameloblastin (Ambn), protein co-assembly, co-immunoprecipitation
Citation: Bapat RA, Su J and Moradian-Oldak J (2020) Co-Immunoprecipitation Reveals Interactions Between Amelogenin and Ameloblastin via Their Self-Assembly Domains. Front. Physiol. 11:622086. doi: 10.3389/fphys.2020.622086
Edited by:
Catherine Chaussain, Université de Paris, FranceReviewed by:
Thomas G. H. Diekwisch, Texas A&M University, United StatesVictor E. Arana-Chavez, University of São Paulo, Brazil
Copyright © 2020 Bapat, Su and Moradian-Oldak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janet Moradian-Oldak, am9sZGFrQHVzYy5lZHU=
 Rucha Arun Bapat
Rucha Arun Bapat Jingtan Su
Jingtan Su Janet Moradian-Oldak
Janet Moradian-Oldak