- 1Department of Agricultural Entomology, University of Agricultural Sciences, Raichur, India
- 2ICAR-National Bureau of Agricultural Insect Resources, Bengaluru, India
- 3ICAR-Indian Institute of Horticultural Research, Bengaluru, India
- 4Department of Genetics and Breeding, University of Agricultural Sciences, Raichur, India
- 5Department of Horticulture, University of Agricultural Sciences, Raichur, India
The shoot and fruit borer, Leucinodes orbonalis (Lepidoptera: Crambidae) is the major cause of low productivity in eggplant and insecticides being the mainstay of management of L. orbonalis. However, field control failures are widespread due to the evolution of insecticide resistance. Taking advantage of the whole genome sequence information, the present study investigated the level of insecticide resistance and the expression pattern of individual carboxylesterase (CE) and glutathione S-transferases (GSTs) genes in various field collected populations of L. orbonalis. Dose-mortality bioassays revealed a very high level of resistance development against fenvalerate (48.2–160-fold), phosalone (94-534.6-fold), emamectin benzoate (7.2–55-fold), thiodicarb (9.64–22.7-fold), flubendiamide (187.4–303.0-fold), and chlorantraniliprole (1.6–8.6-fold) in field populations as compared to laboratory-reared susceptible iso-female colony (Lo-S). Over-production of detoxification enzymes viz., CE and GST were evident upon enzyme assays. Mining of the draft genome of L. orbonalis yielded large number of genes potentially belonging to the CE and GST gene families with known history of insecticide resistance in other insects. Subsequent RT-qPCR studies on relative contribution of individual genes revealed over-expression of numerous GSTs and few CEs in field populations, indicating their possible involvement of metabolic enzymes in insecticide resistance. The genomic information will facilitate the development of novel resistance management strategies against this pest.
Highlights
- High levels of multiple insecticide resistance in field collected Leucinodes orbonalis populations.
- Enhanced midgut activities of carboxylesterase and glutathione S-transferase enzymes.
- Qualitative changes in esterase banding pattern in resistant populations.
- Mining and retrieval of 94 carboxylesterase and 33 glutathione S-transferase full length gene sequences from draft assembled genome and transcriptome of L. orbonalis.
- Over-expression of large number of glutathione S-transferase and few carboxylesterase genes in resistant field collected populations.
Introduction
Eggplant or aubergine (Solanum melongena), also called as brinjal, is one of the most popular vegetable crops in south-east Asia especially India, China, and Bangladesh. The most important limiting factor in brinjal cultivation is the damage caused by shoot and fruit borer, Leucinodes orbonalis Guenee (Lepidoptera: Crambidae). It was first described from India and now it is distributed all over Asia, Africa, and in few parts of Europe (Mally et al., 2015). In India and Bangladesh, it causes severe yield losses up to 93% despite best management practices (Kodandaram et al., 2017; Prodhan et al., 2018). Surveys in India and Bangladesh indicated that farmers spray chemical insecticides up to 84 times during a 6–7 month cropping season. A very high level of pesticide load at a concentration 40–450 times higher than the maximum residue limit (MRL) in marketed brinjal fruits poses major health concerns (Srinivasan, 2008; Latif et al., 2010; Kariyanna et al., 2020b). It is also considered as a pest of quarantine significance to the number of countries outside its native range. L. orbonalis has high reproductive potential with overlapping generations. Studies reporting high level of insecticide resistance development in L. orbonalis might be attributed to the long-term indiscriminate use of various insecticides on eggplant (Kodandaram et al., 2017; Shirale et al., 2017).
Resistant insects overcome the toxic effect of insecticide molecules by adopting one or more mechanisms ranging from cuticular thickening, nerve impenetration, altered production of metabolic enzymes, target site insensitivity, enhanced excretion through ABC transporters as well as by gut symbionts (Mohan et al., 2015). The enhanced detoxification of insecticides by metabolic enzymes viz., carboxylesterases (CE) and glutathione S-transferases (GSTs) are commonly reported in resistant populations of many insect species and mites (Ranson et al., 2002; Enayati et al., 2005; Oakeshott et al., 2005; Perry et al., 2011; Das and Dutta, 2014; Akıner and Ekşi, 2015; Benoit et al., 2015). The resistant individuals metabolize the insecticides faster due to higher catalytic rate or enhanced production of the enzyme as a consequence of increased transcription or gene duplication (Panini et al., 2016; Kariyanna et al., 2020a). The GSTs are phase II metabolic enzymes and are distributed in most of the animals (Hayes and Pulford, 1995). Among several subclasses of GST, the enzymes which are frequently associated with the insecticide resistance are Delta and Epsilon classes (Friedman, 2011; Lumjuan et al., 2011). Similarly, enhanced metabolism of organophosphates, carbamates, and pyrethroids are frequently associated with CEs through gene amplification, upregulation, mutations by coding sequence, or a combination of all these mechanisms (Zhang et al., 2012; Cui et al., 2015).
Considering the economic and social impact of L. orbonalis, genetically modified eggplant expressing insecticidal cry1Ac gene from Bacillus thuringiensis has been developed (Shelton et al., 2018), but, a moratorium on its commercialization was imposed by Indian Government due to public concern. Though Bt eggplant is commercialized in Bangladesh way back in 2014, yet the area under cultivation is <2500 Ha (Shelton et al., 2018) due to various reasons. Baring Bt brinjal, the insecticides remain the sole method of controlling L. orbonalis in all the eggplant growing countries. With the availability of genome sequence, the present study investigated the level of insecticide and the expression pattern of CE and GST genes from field collected insecticide resistant L. orbonalis populations, to pinpoint the key metabolic genes involved in insecticide degradation.
Materials and Methods
Insect Collection and Maintenance
The field populations of L. orbonalis larvae were collected during 2017–2018 from intensive eggplant growing regions of India viz., Raichur (16.2120° N, 77.3439° E), Dharmapuri (12.0933° N, 78.2020° E), Bhubaneshwar (20.2961° N, 85.8245° E), Pune (18.5204° N, 73.8567° E), and Varanasi (25.3176° N, 82.9739° E). All the field populations were reared under laboratory conditions at 27 ± 2°C, 60–70% relative humidity (RH), and a photoperiod of 14:10 h (L:D) on a natural diet and the F1 individuals were used for bioassay and biochemical studies. The insecticide susceptible iso-female line (National Accession number: NBAIR-IS-CRA-01A), designated as Lo-S (65th generation) was originally derived from L. orbonalis collected near Bengaluru (12.9716° N, 77.5946° E) and maintained at insect genomic resources laboratory at ICAR-NBAIR.
Insecticide Resistance Bioassays
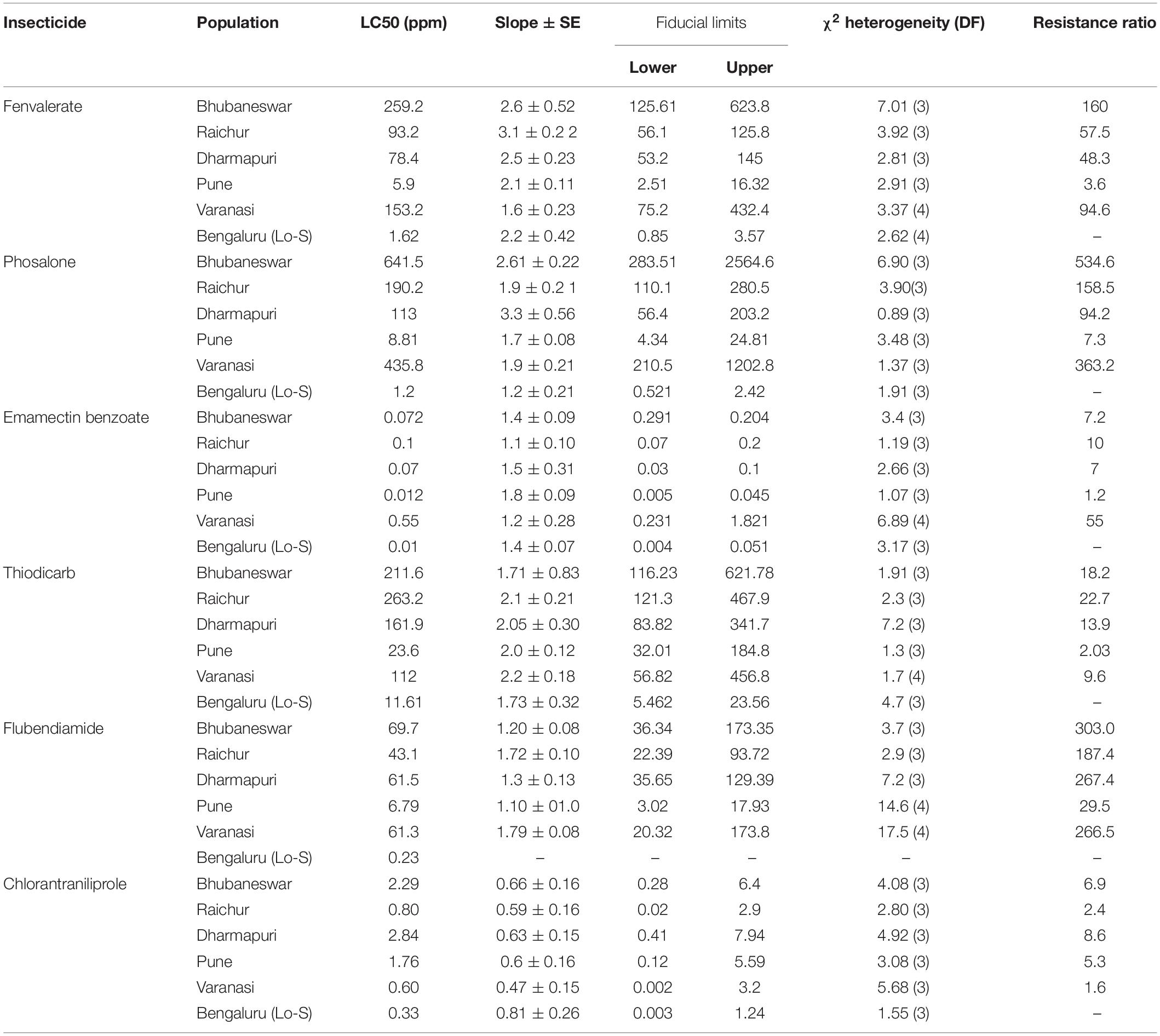
Insecticides fenvalarate (20% EC), emamectin benzoate (5% EC), phosalone (35%), thiodicarb (75%), flubendiamide (20%), and chlorantraniliprole (18.5%) were selected for conducting dose-mortality bioassays based on their usage history on brinjal by farmers. Filter paper residue assay (Cheng et al., 2010) with essential modifications was used for insecticide resistance bioassays on early second instar larvae of L. orbonalis. Five to seven appropriate concentrations of each of the selected insecticides were prepared based on the dose bracketing technique. The filter paper discs (4.5 cm diameter) were dipped in the appropriate dilutions and dried vertically under shade for 1 h. Then, the discs were placed individually in plastic containers (5 cm diameter) and 10 larvae were released in each container. Moistened filter paper discs without insecticide were used in control. After 24 h, the larvae were transferred to untreated natural diet. All the assays were replicated and repeated at least thrice on alternate days. The mortality of the larvae was assessed after 48 h. The pooled larval mortality data were subjected to probit analysis using the software POLO (LeOra, 1987) and the lethal concentration to kill 50% of the test larvae (LC50) was calculated for each population. Resistance ratios (RRs) were calculated with the following formula: LC50 of field population/LC50 of insecticide susceptible Lo-S population.
Preparation of Midgut Homogenate
The starved late second instar larvae were used for the preparation of midgut homogenate. Midguts were dissected and homogenized with homogenization buffer (0.1 M sodium phosphate buffer pH 7.8 containing 1 mM each of DTT, EDTA, PTU, and PMSF). The content was centrifuged at 12,000 rpm for 20 min and the clear supernatant was used as a enzyme source for estimating the titers of carboxylesterase and GST. The total protein content of the preparations was assessed by Coomassie brilliant blue G-250 dye-binding method using bovine serum albumin (BSA) as the standard (Bradford, 1976).
Assays on Carboxylesterase and Glutathione S-Transferase
The activity of carboxylesterase was determined using the method described by van Asperen (1962) with essential modifications and α-naphthyl acetate as a substrate. The assay mixture in a 96 well microplate (iMark, Biorad microplate reader) consisted of an appropriate amount of midgut homogenate, 0.1 M sodium phosphate buffer pH 7.8 and 800 μl of 3 mM α-naphthyl acetate containing 0.3 mM eserine. The mixture was incubated at 30°C for 30 min. Finally, 200 μl of 0.1% tetrazotized o-dianisidine (Fast blue B) in 3.5% sodium dodecyl sulfate was added and incubated for 20 min at room temperature in dark. The α-naphthol formation was measured at 590 nm and the enzyme activity was computed from α-naphthol standard curve.
Glutathione S-transferase activity was determined using 1-chloro-2,4-dinitrobenzene (CDNB) as a substrate (Kao et al., 1989). The assay mixture composed of 50 mM each of CDNB and L-glutathione reduced and 10 μl gut homogenate. The change in absorbance was measured at 340 nm for 5 min and the enzyme activity in terms of nmoles of CDNB conjugated per minute per mg protein using the molar extinction coefficient of 5.3 mM–1 (optimized for the path length: 0.55 cm).
Esterase Isozyme Studies
The native polyacrylamide gel electrophoresis (PAGE) with 10% resolving gel was performed as per Davis (1964) to visualize the esterase isozymes from the 2nd instar midgut larval homogenates of the L. orbonalis populations tested. Midgut homogenates with 20 μg protein were loaded onto the native PAGE and run at a constant voltage of 70 for 1.5 h. Gel was briefly stained with freshly prepared 0.05% (w/v) α-naphthyl acetate and 0.1% (w/v) fast blue B in 50 mM phosphate buffer pH 7.8.
Genome Mining and Phylogenetic Analysis
The draft assembled genome (NCBI Bioproject ID: PRJNA377400) of L. orbonalis was used as query for extracting the gene sequences of glutathione S-transferases (GSTs) and carboxylesterases (CEs). The hmm model for carboxylesterase and GST (CE-PF00135, PF02230, and GST-C PF00043, GST-N PF02798, GST-PF13417, PF14497, PF17171, PF17172) were obtained from PFAM database1. A total of 94 and 33 putative CE and GST sequences were mined using hmm search with HMMER3 software package2. Among them, the genes with known history of involvement in insecticide resistance in other insects were selected and further confirmed with NCBI BLASTX. The GST and CE protein sequences of Drosophila melanogaster was downloaded from its genome database (Marygold et al., 2013), GenBank3 and Uniprot4. We combined these putative GST protein sequences with D. melanogaster’s annotated protein sequences for each of the six subclasses of GST (Epsilon, Sigma, Omega, Delta, Zeta, and Theta). The sequences were multiple aligned using CLUSTALW and a maximum likelihood tree with 1000 bootstrap value was constructed using MEGA75 and TreeDyn online server6 was used to visualize the tree. Similarly, 94 CE protein sequences under seven subclasses (α-esterases, gliotactin, glutactin, neurolignin, juvenile hormone esterases, acetylcholinesterases, and cricklet) were combined, multiple aligned using MAFFT online server using conserved domain feature and phylogenetic tree of CE genes from L. orbonalis using D. melanogaster as a reference were constructed with the help of CLC workbench (QIAGEN Bioinformatics7) using maximum likelihood approach.
Quantitative Real-Time PCR
The late 2nd instar larvae were used for RNA extraction (ISOLATE II RNA mini kit) by following the manufacturer’s guidelines (Bioline). Purity and concentration of total RNA were measured in a spectrophotometer (NanoDrop Lite, Thermo Fisher Scientific) and denatured agarose gel (Masek et al., 2005). RNA samples with an A260/A280 ratio ranging from 1.8 to 2.0 and A260/A230 ratio >2.0 were used for cDNA preparation. First strand of complementary DNA was synthesized from 4 μg of total RNA using Revert AID first strand cDNA synthesis kit (Thermo Fisher ScientificTM, Lithuania) following the suppliers guidelines and stored at −80°C till further use.
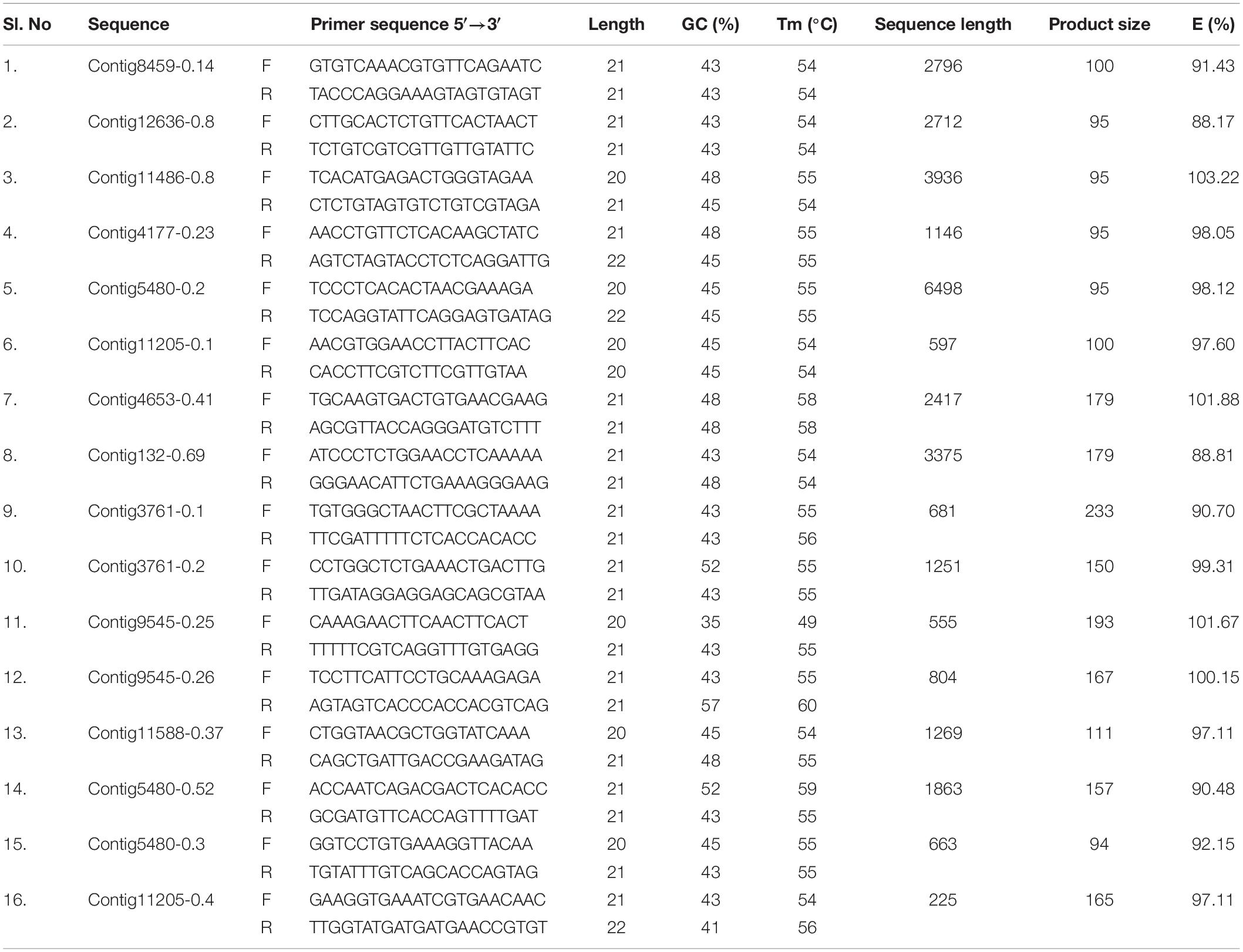
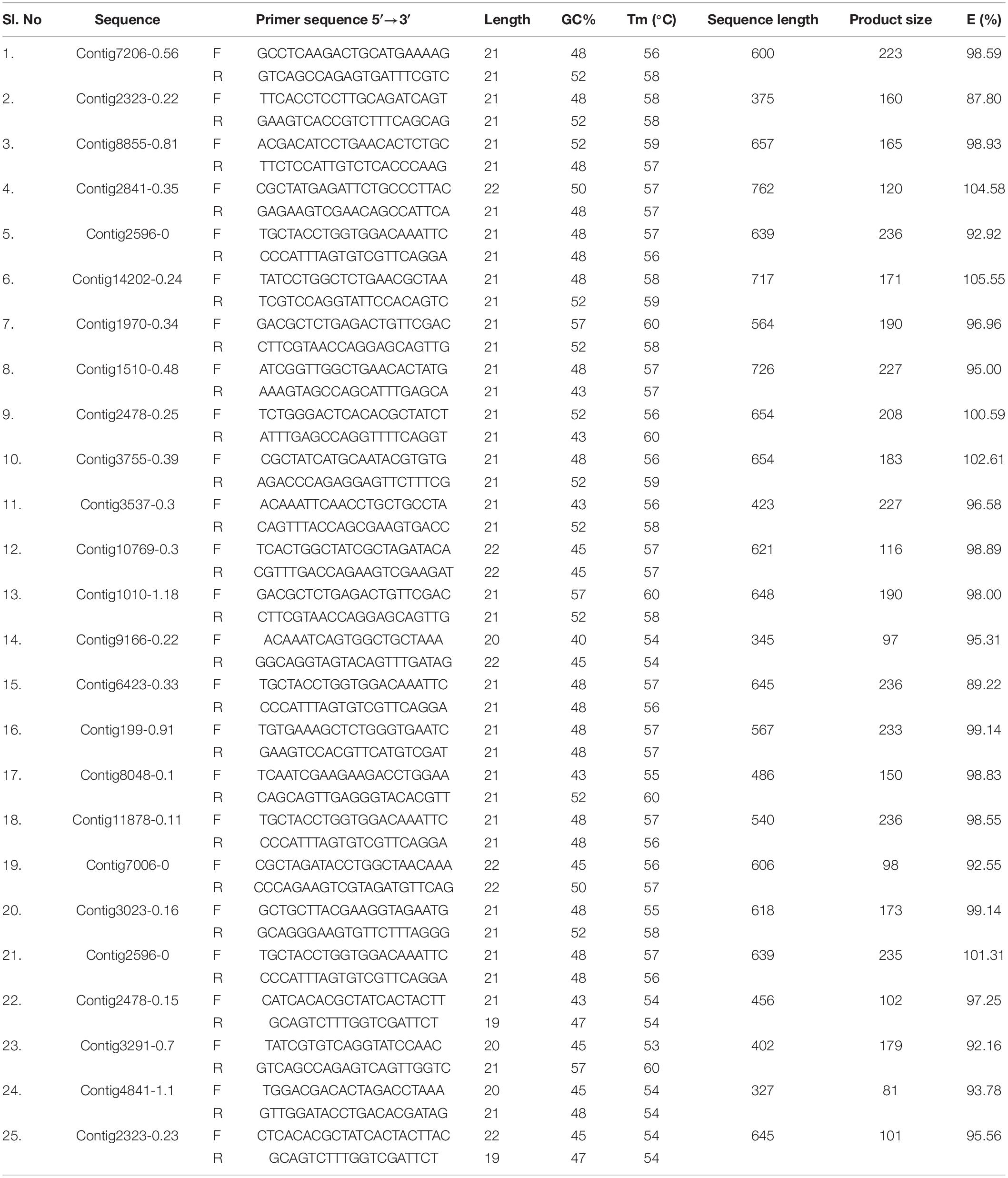
qPCR primers for 25 GST and 16 CE gene sequences were designed using Primer 3.0 software. The detailed information of the primers used in the current study is listed in Tables 1, 2. To remove the primer dimer and confirm the efficiency, OligoEvaluatorTM sequence analysis tool (accessed on July 20188) was used. Primer specificity analysis was observed by single peak in the melting curve and single band from the agarose gel of the all candidate genes of CE and GST ranged from 90.36 to 104.58% (Tables 1, 2). Quantitative real-time PCR (RT-qPCR) amplifications were performed in 20 μL reaction consisted of 10 μL 2×SYBR® Premix EX TaqTM II (Tli RNaseH Plus, TAKARA®, Japan), 1 μl cDNA and gene specific primer pair. The parameters used for RT-qPCR are: One cycle of 95°C for 3 min; 35 cycles of 95°C for 30 s, 53°C for 45 s, and 72°C for 1 min; a final cycle of 72°C for 10 min using Roche 480II machine. All the samples including control (no template) and internal control were performed in triplicates. The PCR products were electrophoresed on 2.0% agarose gel in a 1.0×TAE buffer. Relative expression levels for the CE and GST genes were calculated by 2–ΔΔCT method. The 28SR3 (28S ribosomal protein S3 mitochondrial) (Kariyanna et al., 2019) gene was used as an internal control to normalize the expression of target genes.
Results
Insecticide Resistance Monitoring
The LC50 values against fenvalerate, phosalone, thiodicarb, emamectin benzoate, flubendiamide and chlorantraniliprole indicated that there is a large shift in susceptibility of field-populations of L. orbonalis as compared to the laboratory reared susceptible iso-female colony (Lo-S). The field collected populations exhibited 3.6–160-fold resistance against fenvalerate, 7.3–534.6-fold resistance against phosalone, 7.0–55.0-fold resistance against emamectin benzoate, 2.0–22.7-fold resistance against thiodicarb, 29.5–303.0-fold resistance against flubendiamide, and 1.6–8.6-fold resistance against chlorantraniliprole. However, the susceptibility level of all the field populations and the Lo-S did not vary significantly against chlorantraniliprole based on the overlapping fiducial limit values. High level of resistance development against phosalone and flubendiamide was observed in L. orbonalis collected from Varanasi whereas other field populations, the RRs were non-significant as compared to Lo-S. L. orbonalis populations from Bhubaneswar showed a significantly very high levels of resistance against fenvalerate, phosalone and flubendiamide as compared to Lo-S. The population of L. orbonalis collected from high hills near Pune recorded less RR and the susceptibility status was almost on par with Lo-S population (Table 3).
Activities of Detoxification Enzymes
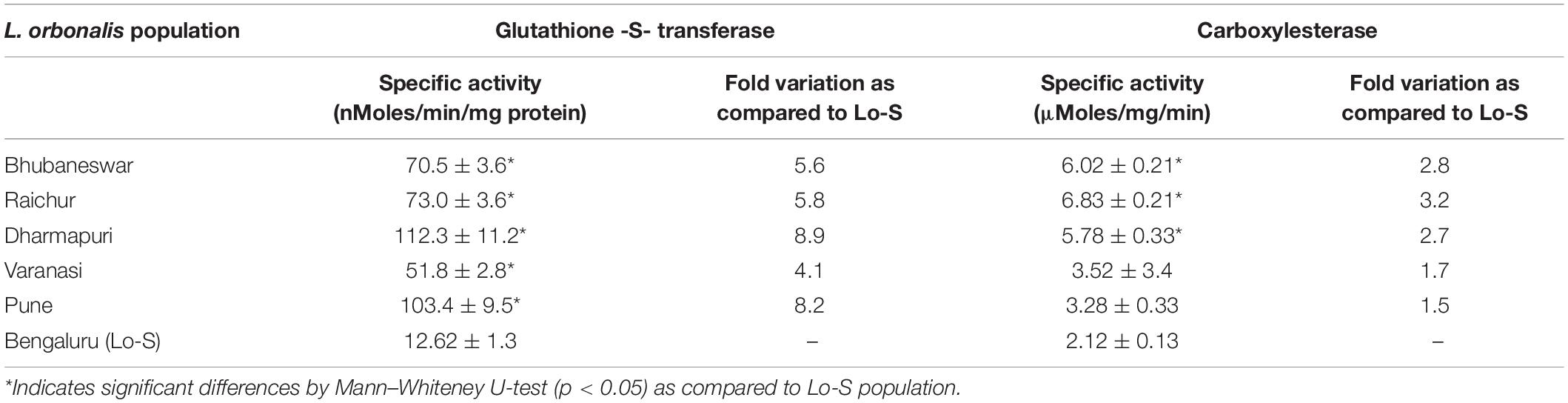
Quantitative differences in the titer of GST and CE were determined using α-naphthyl acetate and 1-chloro-2,4-dinitrobenzene (CDNB) as substrates, respectively. Significant level of elevated GST activities (4.1–8.9-fold) was observed in field collected L. orbonalis as compared to susceptible Lo-S population. However, the overproduction of CE has not pronounced much (1.5–3.2-fold), but there was a significant level of overproduction in two field populations based on Mann–Whiteney U-test. The results indicated that GST activity was altered more profoundly than the CE (Table 4).
The qualitative differences in carboxylesterase isozymes were visualized under native PAGE after incubation of the gels in the α-naphthyl acetate as a substrate. Among the four major esterase activity bands (E1 to E4), the high molecular weight E1 and E2 bands were absent in Lo-S and Pune populations and faint in case of Varanasi population. However, the E1 band was very prominent and intense in other field populations indicating its over-production nature (Figure 1).
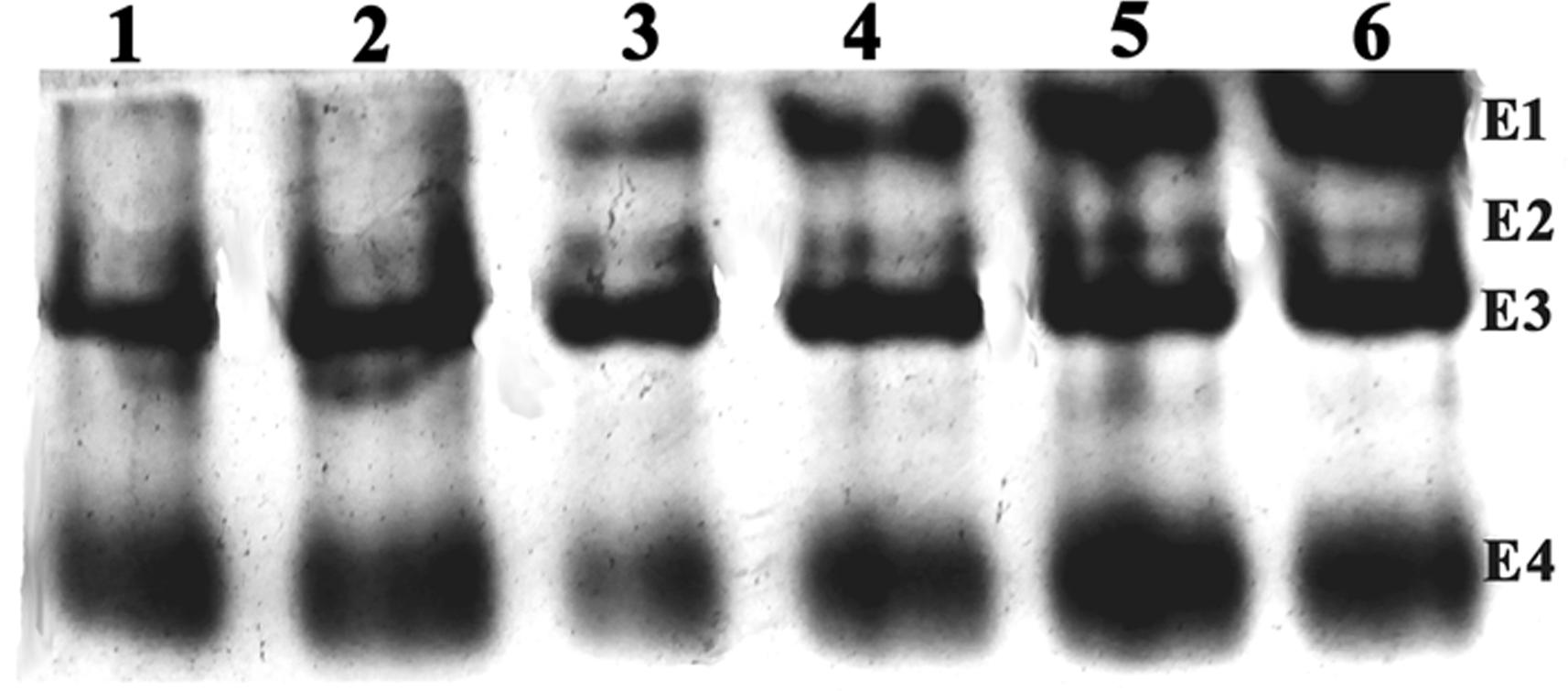
Figure 1. The Native PAGE of the esterase isozyme pattern of L. orbonalis (1. Lo-S, 2. Pune, 3. Varanasi, 4. Raichur, 5. Bhubaneswar, 6. Dharmapuri).
Differential Expression of CE Genes
Ninety-four CE genes were identified from the genome and transcriptome sequences of L. orbonalis. They were classified under seven subfamilies viz., α-esterases, gliotactin, glutactin, neurolignin, juvenile hormone esterases, acetylcholinesterases, and cricklet like orthologs (Figure 2). The length of the gene sequences ranged from 225 to 6498 bp. Expression profiling of 16 genes (homologous to resistant genes in other insects with ≥90% query coverage) was examined from the mRNA samples derived from late second instar larvae of five field-collected L. orbonalis populations by RT-qPCR. The CE genes contig11486-0.8 and contig4653-0.41 were over-expressed more than 10-fold as compared to susceptible Lo-S population (Figures 3, 4).
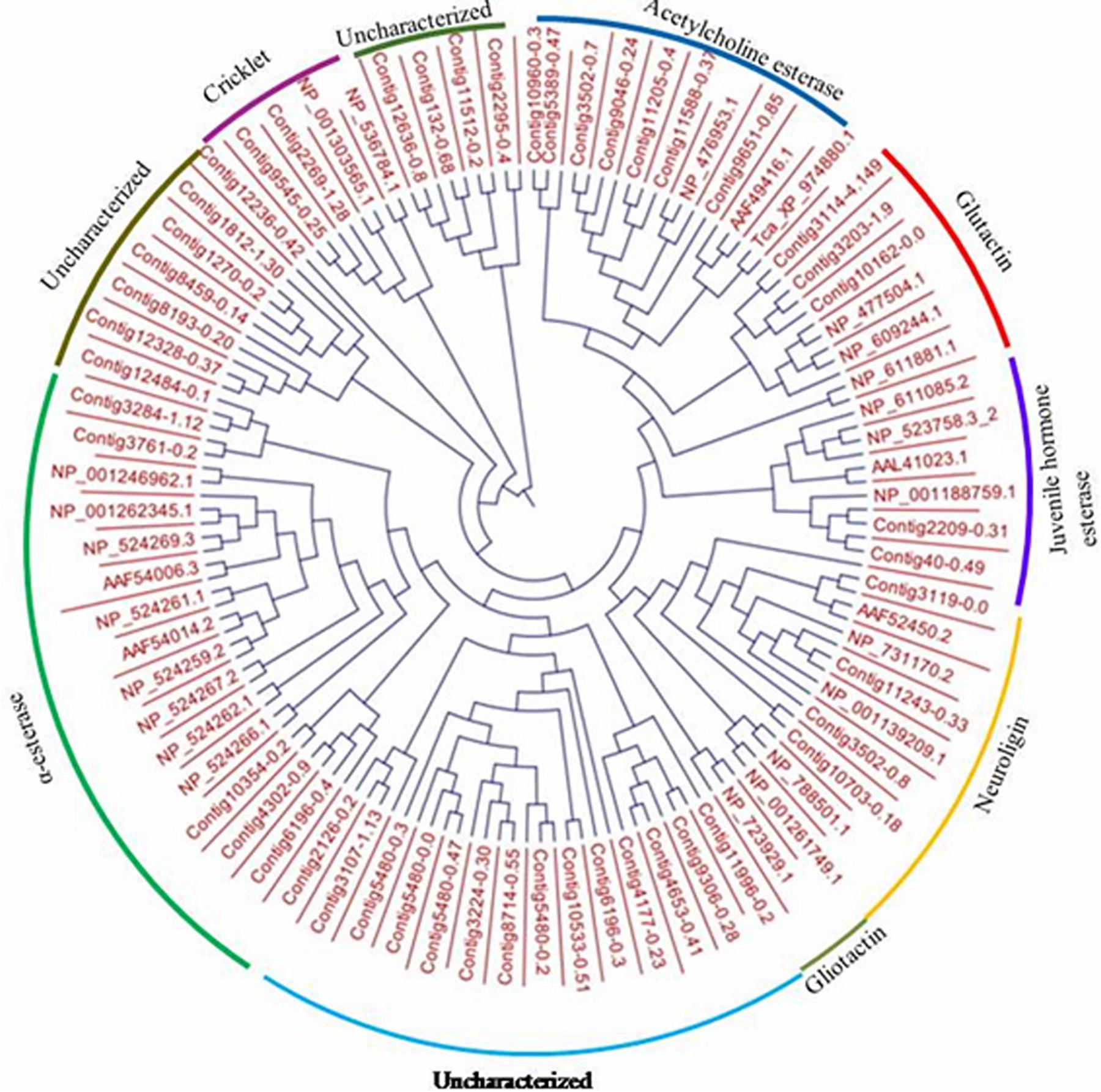
Figure 2. Phylogenetic relationships of 94 CE proteins of L. orbonalis with selected D. melanogaster protein representative of seven characterized carboxylesterase classes. Putative CE proteins in L. orbonalis were identified using hmmsearch against PFAM CE families (CE PF00135). The Maximum likelihood tree was constructed using CLC workbench.
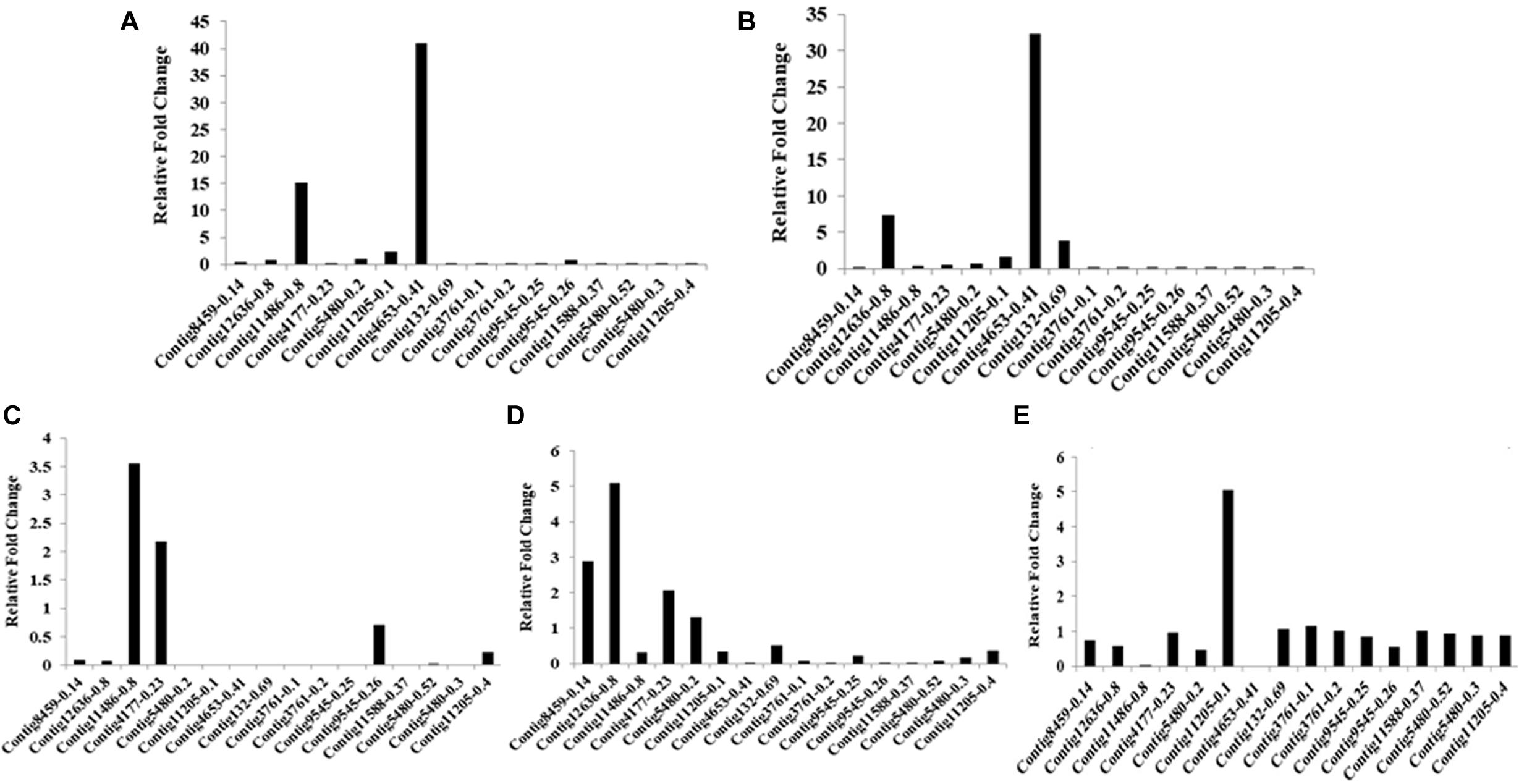
Figure 3. Transcription profiling of carboxylesterase genes in field collected populations of L. orbonalis depicted as fold change over the susceptible Lo-S colony. (A) Bhubaneswar, (B) Dharmapuri, (C) Pune, (D) Raichur, (E) Varanasi.
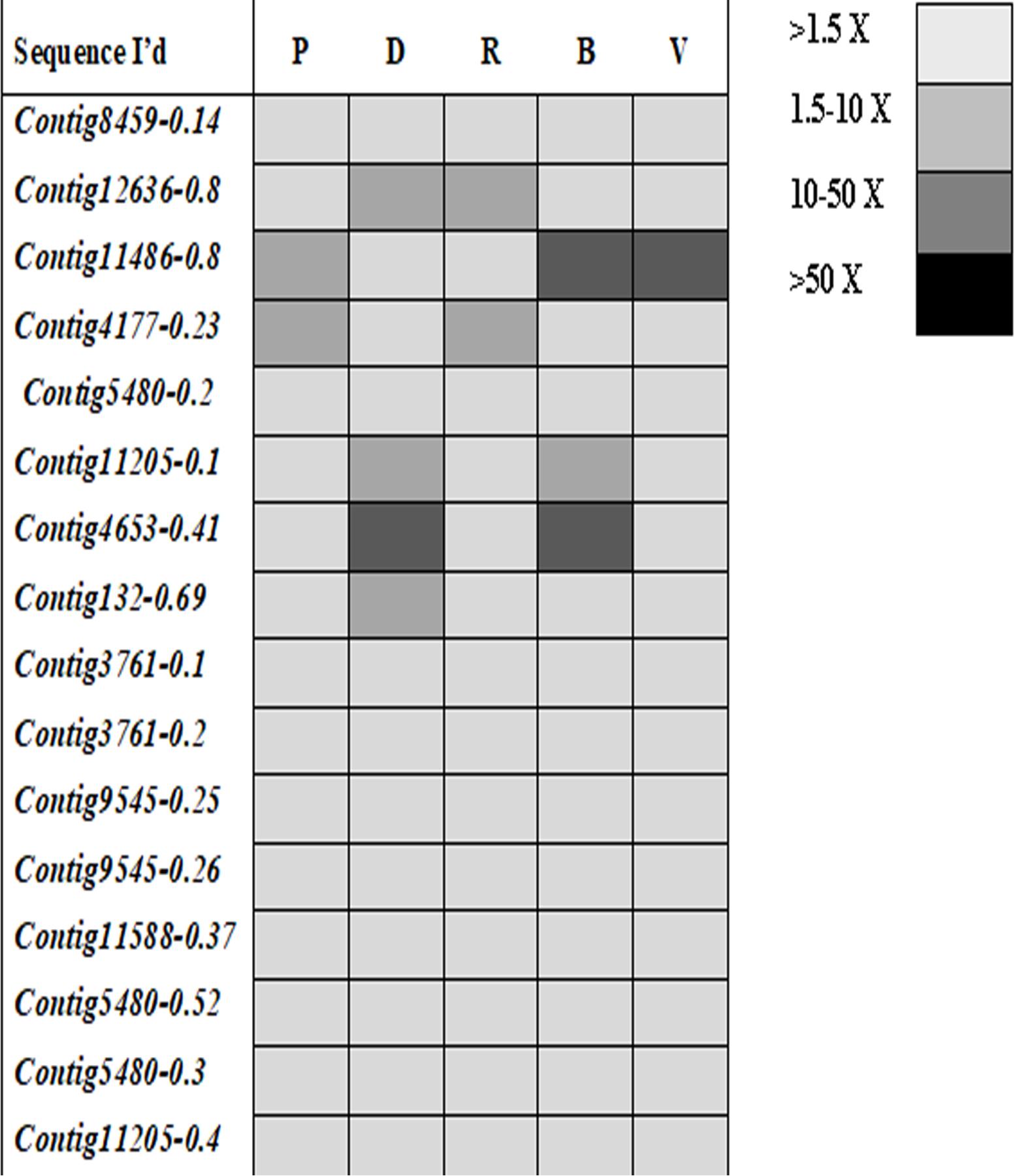
Figure 4. Expression profiles (fold changes over Lo-S population) of carboxylesterase genes across field collected L. orbonalis populations (P, Pune; D, Dharmapuri; R, Raichur; B, Bhubaneswar; V, Varanasi). The fold changes are indicated in different shades indicate significant difference as per Mann–Whiteney U-test p-value < 0.05.
Differential Expression of Glutathione S-Transferase Genes
Mining of genome and transcriptome data of L. orbonalis yielded 33 unigenes codings for GSTs. Lengths of the identified GST unigene sequences ranged from 370 to 5,048 bp. The phylogenetic analysis revealed that the predicted GST genes are represented under all six classes of GST viz., Epsilon (10 genes), Sigma (6 genes), Omega (5 genes), Delta (8 genes), Zeta (3 genes), and Theta (1 gene) (Figure 5). RT-qPCR analysis of 25 GST genes (homologous to other insects resistant genes) revealed that many of them were over-expressed in larvae collected from the field. The GST genes named contig2323-0.22, contig14202-0.24, contig3755-0.39, contig3537-0.3, contig199-0.91, contig3023-0.16, contig3291-0.7, contig4841-1.1, and contig2323-0.23 were very highly expressed (>50-fold) in one or more resistant field-collected L. orbonalis populations (Figures 6, 7) over the susceptible Lo-S population.
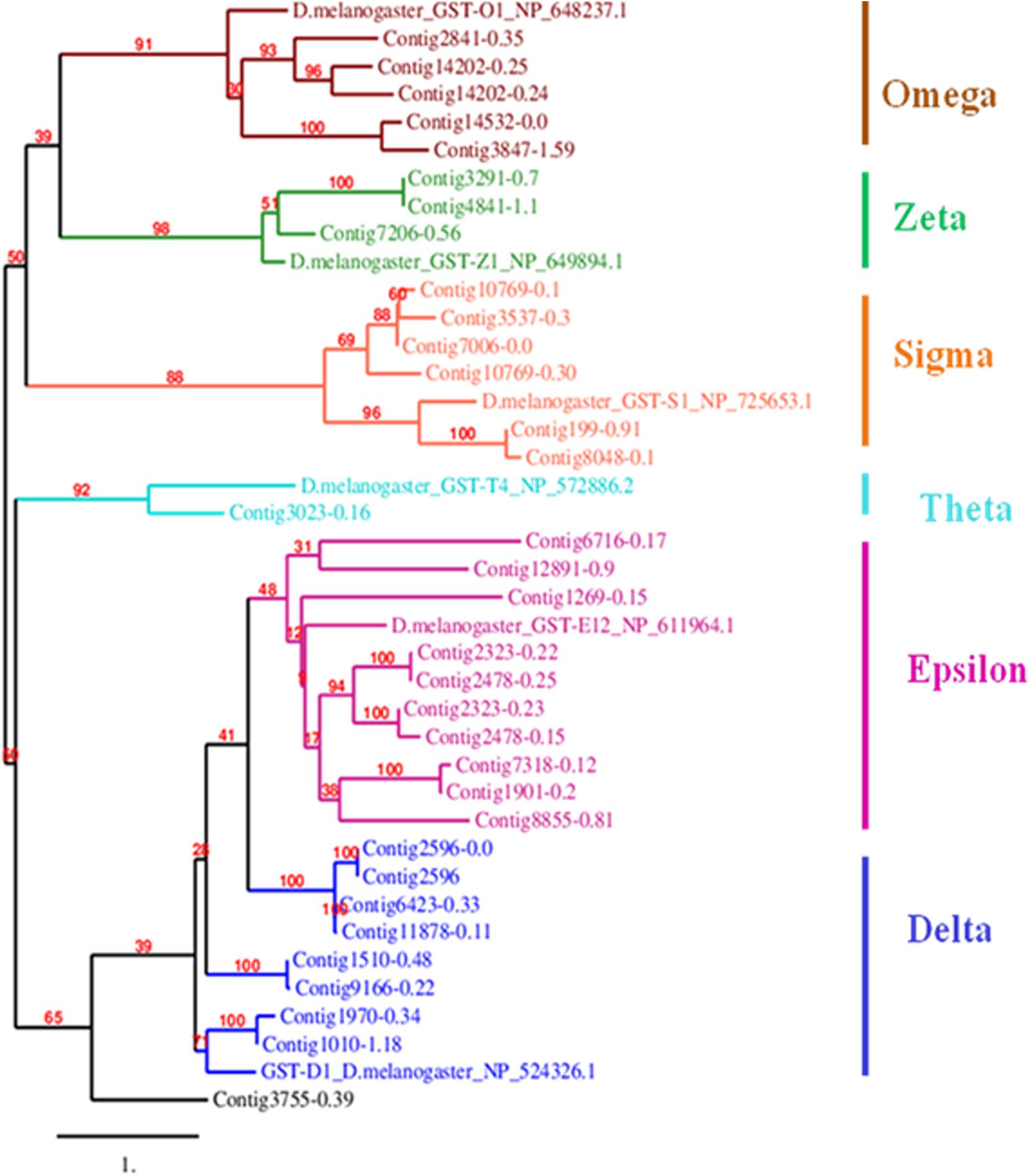
Figure 5. Phylogenetic tree of 34 predicted L. orbonalis glutathione S-transferase proteins with D. melanogaster genes representative of the six characterized GST classes. Putative GST proteins in L. orbonalis were identified using hmmsearch against PFAM GST families (GST-C PF00043, GST-N PF02798), the Maximum Likelihood tree with 1000 bootstraps was constructed using MEGA7 and TreeDyn online server was used to visualize the tree. Number on each node is bootstrap values in percent. Scale = 1 amino acid substitution per site.
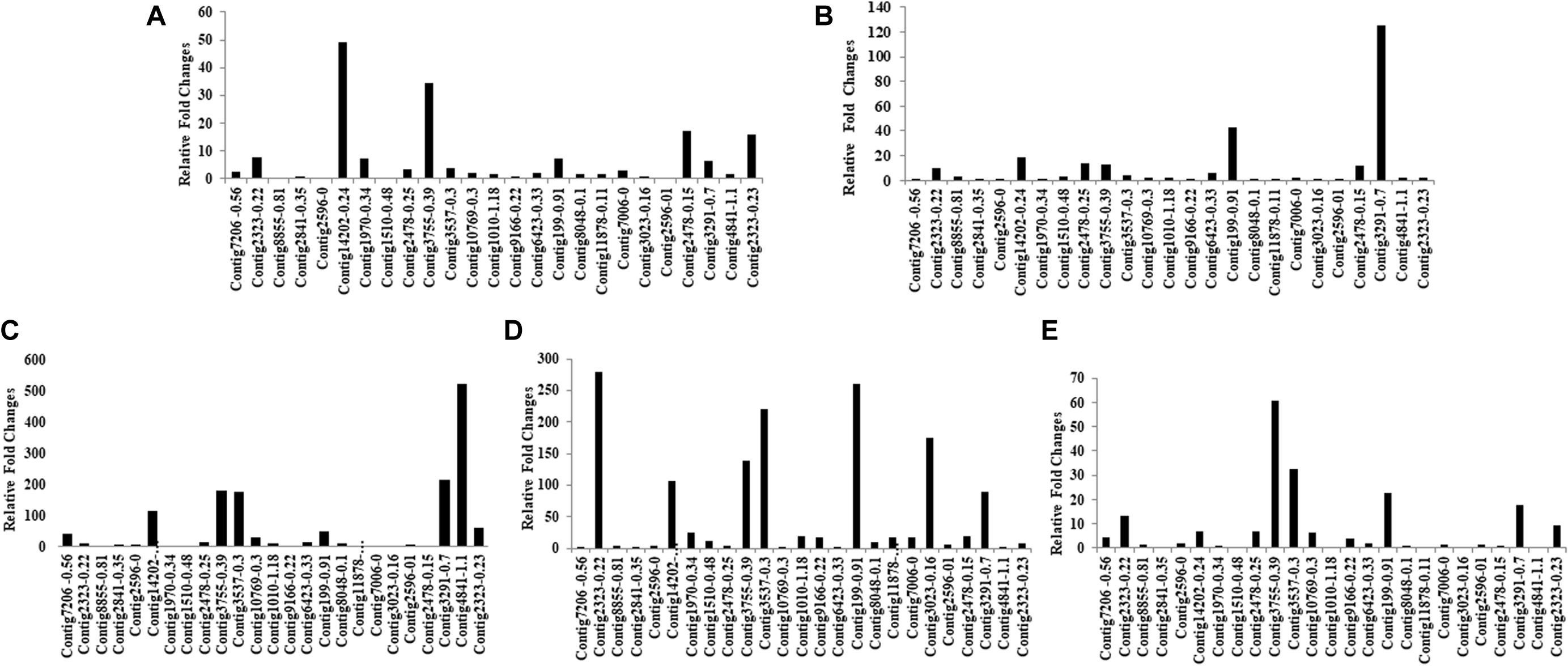
Figure 6. Transcription profiling of glutathione -S- transferase genes in field collected populations of L. orbonalis depicted as fold change over the susceptible Lo-S colony. (A) Bhubaneswar, (B) Dharmapuri, (C) Pune, (D) Raichur, (E) Varanasi.
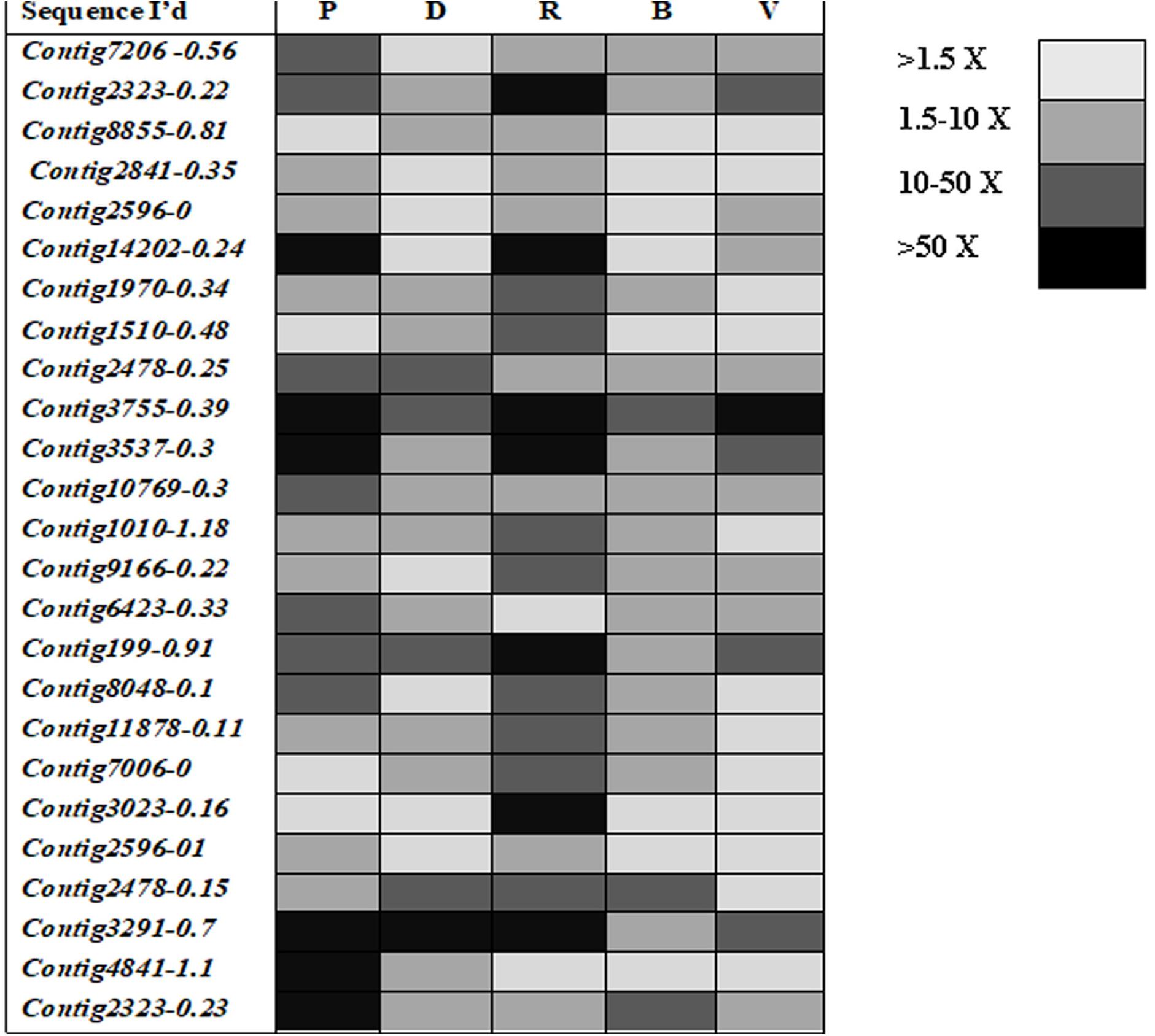
Figure 7. Expression profiles (fold changes over Lo-S population) of glutathione -S- transferase genes across field collected L. orbonalis populations (P, Pune; D, Dharmapuri; R, Raichur; B, Bhubaneswar; V, Varanasi). The fold changes are indicated in different shades indicate significant difference as per Mann–Whiteney U-test p-value < 0.05.
Discussion
Damage caused by L. orbonalis is the major limiting factor in realizing maximum productivity of eggplant in many Asian countries including India, China, and Bangladesh. The high reproductive potential and shorter life cycle of the pest coupled with the evolution of multiple insecticide resistance pose a major challenge in profitable cultivation of eggplant (Prodhan et al., 2018). The insecticide resistance levels detected in the field collected populations ranged from low (>10-fold) to moderate (10–100-fold) and high (>100-fold) against fenvalerate (48.2–160-fold), phosalone (94–534.58-fold), emamectin benzoate (7–55-fold), thiodicarb (9.64–22.67-fold), and flubendiamide (29.51–363.91-fold). The resistance levels reflect the occurrence of differential selection pressure associated with long term history of insecticide use against L. orbonalis. However, the response to chlorantraniliprole was similar among the resistant field collected populations with RRs below 10-fold. The efficacy of chlorantraniliprole has been reported earlier (Kodandaram et al., 2013; Munje et al., 2015). The chlorantraniliprole and flubendiamide are anthranilic and phthalic diamides, respectively, are the newer insecticides with little or no cross-resistance among them and also with other classes of insecticides. They are the activators of the ryanodine receptor.
Insect employs various mechanisms to nullify the toxic effect of xenobiotic compounds. Insect metabolic enzymes play a major role in detoxification of plant allelochemicals and pesticidal compounds in addition to other physiological roles. Insect pests have evolved large reservoirs of various detoxification enzymes. Glutathione S-transferases mediated detoxification can be direct or by the metabolism of secondary products generated from other detoxification enzymes. The insecticides are metabolized rapidly into readily excretable water-soluble metabolites by reductive dehydrochlorination or by conjugation with reduced glutathione (Pavlidi et al., 2018). The carboxylesterase is a phase-1 detoxification enzyme that mainly hydrolyses organophosphates, carbamates, and synthetic pyrethroids.
The quantitative mechanisms underlying metabolic resistance is characterized by over-production due to gene amplification or transcriptional upregulation (Mohan et al., 2015; Feyereisen, 2015; Pavlidi et al., 2018). A qualitative mechanism occurs as a result of changes in the enzymatic characteristics. The present investigation on the enhanced metabolism of insecticides in resistant populations of L. orbonalis resistance is very prominent as inferred by elevated GST (1.2–2.6-fold) and carboxylesterases (6.3–13.1-fold) titers. The involvement of CEs and GSTs in insecticide resistance is commonly reported in many other insect species (Mohan et al., 2015; Pavlidi et al., 2018).
With the advent of genome and transcriptome information, multiple GSTs and carboxylesterases encoding genes have been characterized from insects and mites such as Plutella xylostella, Culex spp., Tribolium castaneum, Bombyx mori, Leptinotarsa decemlineata, Tetranychus cinnabarinus, Musca domestica and in many other insect species (Yang and Liu, 2011; Wang et al., 2014; Cui et al., 2015; Meisel and Scott, 2018; Pavlidi et al., 2018). L. orbonalis genome has large expansion of CE and GST genes which is comparable to many other insects. However, the GSTs have prominent role in insecticide resistance as compared to CEs. mRNA level of at least nine GSTs were over-expressed >50-fold where as none of the CEs showed very high levels of expression in the field collected resistant populations. The over-expression of few CE genes in field populations, especially from Varanasi, Raichur, and Bhubaneswar, also aligned with additional high molecular weight esterase band (E1) probably contributed to overall resistance. A similar high molecular weight esterase band associated with B-esterase type was reported from diamondback moth (Mohan and Gujar, 2003). In addition to qualitative changes in esterase banding pattern, there is a significant increase in the production of the CEs in at least three field populations. The over-production and appearance of new esterase isozymes might be the consequence of increased transcription or gene duplication events (Malathi et al., 2015; Panini et al., 2016).
Most of the GST gene expansions are from epsilon and delta subclasses together accounting for 18 genes (54.5%) out of 33 genes identified from the draft genome of L. orbonalis. These two GST subclasses are insect-specific, widely expanded in many other insect species whereas other classes have wider taxonomic distribution (Friedman, 2011). There were at least nine GSTs over-expressed >50-fold in one or more of the field-collected resistant populations of L. orbonalis. In Anopheles gambiae, Apis mellifera, B. mori, D. melanogaster, Nasonia vitripennis, and T. castaneum respectively 28, 8, 23, 37, 19, 35 GSTs and 51, 24, 76, 35, 41, 49 CEs were identified (Yu et al., 2008, 2009; Oakeshott et al., 2010). Similarly, the gene expansion of cytochromoe p450 monooxygenases (CYP) and their role in insecticide resistance in L. orbonalis were recently documented (Kariyanna, 2019; Kariyanna et al., 2020a). Together with the results of present and earlier studies it is very much evident that multiple metabolic enzymes contribute toward overall insecticide resistance in L. orbonalis. However, in many insect species including L. orbonalis, it is difficult to distinguish whether the expansion of these metabolic genes and their involvement in insecticide detoxification are driven from ancestry or adaptation to stress or both.
Conclusion
In conclusion, the present study provides the first genomic level information on GST and CE gene families of L. orbonalis and their role in insecticide resistance. The information further supports insect resistance management through use of reduced risk pesticides including biologicals.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
BKr: conducted the experiment, analysis, manuscript writing, materials and methods, and hypothesis. AP: analysis, material methods, and corrections. RA: analysis and technical suggestion, manuscript correction, and resource supply. AA and PJ: bioinformatics and analysis. RG and BKl: resources supply and, technical guidance and suggestions. TV: resources supply, technical guidance and suggestions, and reviews and literatures. MB and JD: analysis, technical guidance and suggestions, and reviews and literatures. MM: designing experiment, resources supply, manuscript corrections, analysis, material and methods, technical guiding, and suggestions. YP: technical guidance and suggestions, and reviews and literatures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors profusely thank Indian Council of Agricultural Research for generous funding under ICAR-CRP Genomics project. The first author acknowledges the award of DST-Inspire fellowship (2016/IF160900) and UAS Raichur for necessary support. Authors are thankful to Director, ICAR-NBFGR for timely release of fund. The facilities extended by Director, ICAR-NBAIR, and Director, ICAR-IIHR, Bengaluru are gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.594845/full#supplementary-material
Supplementary Table 1 | Highlights of the research.
Supplementary Table 2 | Sequence information GST and CE from L. orbonalis.
Supplementary Table 3 | Resistance genes of GST and CE from L. orbonalis.
Supplementary Table 4 | Detailed sequence information of CE from L. orbonalis.
Supplementary Table 5 | Detailed sequence information of GST from L. orbonalis.
Supplementary Table 6 | NCBI ID and description for the Reference protein taken in GST and CE phylogeny.
Supplementary Table 7 | Comparative characterization L. orbonalis CE genes with other group based on the nr hit.
Supplementary Table 8 | Descriptive information of the L. orbonalis CE sequences.
Supplementary Presentation 1 | Graphical representation of the manuscript.
Footnotes
- ^ http://pfam.xfam.org
- ^ http://hmmer.janelia.org
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://www.uniprot.org/
- ^ http://www.megasoftware.net
- ^ www.treedyn.org
- ^ https://www.qiagenbioinformatics.comproductsclc-main-workbench
- ^ http://www.sigmaaldrich.com/life-science/custom-oligos/custom-dna/learning-center/calculator.html
References
Akıner, M. M., and Ekşi, E. (2015). Evaluation of insecticide resistance and biochemical mechanisms of Culex pipiens L. in four localities of east and middle mediterranean basin in Turkey. Int. J. Mosq. Res. 2, 39–44.
Benoit, J., Adelman, B. Z. N., Reinhardt, K., Dolan, A., Poelchau, M., Jennings, E. C., et al. (2015). Unique features of the bed bug, a global human ectoparasite, identified through genome sequencing. Nat. Commun. 7:10165. doi: 10.1038/ncomms10165
Bradford, M. M. (1976). Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cheng, X., Chang, C., and Daia, S.-M. (2010). Responses of striped stem borer, Chilo suppressalis (Lepidoptera: Pyralidae), from Taiwan to a range of insecticides. Pest Manag. Sci. 66, 762–766. doi: 10.1002/ps.1939
Cui, F., Li, M. X., Chang, H. J., Mao, Y., Zhang, H. Y., Lu, L. X., et al. (2015). Carboxylesterase-mediated insecticide resistance: quantitative increase induces broader metabolic resistance than qualitative change. Pestic. Biochem. Physiol. 121, 88–96. doi: 10.1016/j.pestbp.2014.12.016
Das, M., and Dutta, P. (2014). Status of insecticide resistance and detoxifying enzyme activity of Aedes albopictus population in Sonitpur district of Assam. India. Int. J. Mosq. Res. 1, 35–41.
Davis, B. J. (1964). Disc electrophoresis II. Method and application to human serum proteins. Ann. N.Y. Acad. Sci. 121, 404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Molec. Biol. 14, 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Feyereisen, R. (2015). Insect P450 inhibitors and insecticides: challenges and opportunities. Pest Manag. Sci. 71, 793–800. doi: 10.1002/ps.3895
Friedman, R. (2011). Genomic organization of the glutathione S-transferase family in insects. Molec. Phylogenet. Evol. 61, 924–932. doi: 10.1016/j.ympev.2011.08.027
Hayes, J. D., and Pulford, D. J. (1995). The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Molec. Biol. 30, 445–600. doi: 10.3109/10409239509083491
Kao, C. H., Hung, C. F., and Sun, C. N. (1989). Parathion and methyl parathion resistance in diamondback moth (Lepidoptera: Plutellidae) larvae. J. Econ. Entomol. 82, 1299–1304. doi: 10.1093/jee/82.5.1299
Kariyanna, B. (2019). Identification and Functional Analysis of Genes Involved in Insecticide Resistance in Leucinodes orbonalis Guenee (Lepidoptera: Crambidae). PhD thesis, University of Agricultural Sciences, Raichur, Karnataka, 1–199.
Kariyanna, B., Prabhuraj, A., Asokan, R., Babu, P., Jalali, S. K., Venkatesan, T., et al. (2019). Identification of suitable reference genes for normalization of RT-qPCR data in eggplant fruit and shoot borer (Leucinodes orbonalis Guenée). Biologia 75, 289–297. doi: 10.2478/s11756-019-00346-4
Kariyanna, B., Prabhuraj, A., Asokan, R., Ramkumar, G., Venkatesan, T., Gracy, R. G., et al. (2020a). Genomewide analysis and expression patterns of cytochrome P450 genes involved in insecticide resistance in Leucinodes orbonalis (Lepidoptera: Crambidae). Biotechnol. Appl. Biochem. 1–12. doi: 10.1002/bab.1997
Kariyanna, B., Prabhuraj, A., Bheemanna, M., Kalmat, B., Pamapanna, Y., Mohan, M., et al. (2020b). Insecticide usage pattern and evolution of resistance in brinjal shoot and fruit borer, Leucinodes orbonalis (Lepidoptera: Crambidae) in India. Plant Arch. 20, (Suppl. 2), 1255–1261.
Kodandaram, M. H., Rai, A. B., Sharma, S. K., and Singh, B. (2017). Shift in the level of susceptibility and relative resistance of brinjal shoot and fruit borer Leucinodes orbonalis (Guen) to diamide insecticides. Phytoparasitica 45, 151–154. doi: 10.1007/s12600-017-0584-z
Kodandaram, M. H., Sujoy Saha, A. B. R., and Prakash, S. N. (2013). Compendium on Pesticide use in Vegetables. Extension Bulletin No. 50. Varanasi: Indian Institute of Vegetable Research, 133.
Latif, M. A., Rahman, M. M., and Alam, M. Z. (2010). Efficacy of nine insecticides against shoot and fruit borer, Leucinodes orbonalis Guenee (Lepidoptera: Pyralidae) in eggplant. J. Pest Sci. 83, 391–397. doi: 10.1007/s10340-010-0309-2
Lumjuan, N., Rajatileka, S., Changsom, D., Wicheer, J., Leelapat, P., Prapanthadara, L. A., et al. (2011). The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem. Molec. Biol. 41, 203–209. doi: 10.1016/j.ibmb.2010.12.005
Malathi, V. M., Jalali, S. K., Gowda, D. K., Mohan, M., and Venkatesan, T. (2015). Establishing the role of detoxifying enzymes in field-evolved resistance to various insecticides in the brown planthopper (Nilaparvata lugens) in South India. Insect Sci. 24, 35–46. doi: 10.1111/1744-7917.12254
Mally, R., Korycinska, A., Agassiz, D. J. L., Hall, J., Hodgetts, J., and Nuss, M. (2015). Discovery of an unknown diversity of Leucinodes species damaging Solanaceae fruits in sub-Saharan Africa and moving in trade (Insecta. Lepidoptera, Pyraloidea). Zookeys 472, 117–162. doi: 10.3897/zookeys.472.8781
Marygold, S. J., Leyland, P. C., Seal, R. L., Goodman, J. L., Thurmond, J., Strelets, V. B., et al. (2013). FlyBase: improvements to the bibliography. Nucleic Acids Res. 41, D751–D757.
Masek, T., Vopalensky, V., Suchomelovam, P., and Pospisek, M. (2005). Denaturing RNA electrophoresis in TAE agarose gels. Anal. Biochem. 336, 46–50. doi: 10.1016/j.ab.2004.09.010
Meisel, R. P., and Scott, J. G. (2018). Using genomic data to study insecticide resistance in the house fly. Musca domestica. Pestic. Biochem. Physiol. 151, 76–81. doi: 10.1016/j.pestbp.2018.01.001
Mohan, M., and Gujar, G. T. (2003). Local variation in susceptibility of the diamondback moth, Plutella xylostella (Linnaeus) to insecticides and role of detoxification enzymes. Crop Prot. 22, 495–504. doi: 10.1016/s0261-2194(02)00201-6
Mohan, M., Venkatesan, T., Sivakumar, G., Yandigeri, M. S., and Verghese, A. (2015). Fighting Pesticide Resistance in Arthropods. New Delhi: Westville Publishers.
Munje, S. S., Salunke, P. B., and Botre, B. S. (2015). Toxicity of newer insecticides against Leucinodes orbonalis (Guen.). Asian J. Biol. Sci. 10, 106–109. doi: 10.15740/has/ajbs/10.1/106-109
Oakeshott, J. G., Claudianos, C., Campbell, P. M., Newcomb, R. D., and Russell, R. J. (2005). “Biochemical genetics and genomics of insect esterases,” in Comprehensive Molecular Insect Science-Pharmacology, Vol. 5, eds L. I. Gilbert, K. Latrou, and S. S. Gill (Oxford: Elsevier), 309–381. doi: 10.1016/b0-44-451924-6/00073-9
Oakeshott, J. G., Johnson, R. M., Berenbaum, M. R., Ranson, H., Cristino, A. S., and Claudianos, C. (2010). Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Biochem. Molec. Biol. 19, 147–163. doi: 10.1111/j.1365-2583.2009.00961.x
Panini, M., Manicardi, G. C., Moores, G., and Mazzoni, E. (2016). An overview of the main pathways of metabolic resistance in insects. Invertebrate Surviv. J. 13, 326–335.
Pavlidi, N., Vontas, J., and Leeuwen, T. V. (2018). The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opi. Insect Sci. 27, 97–102. doi: 10.1016/j.cois.2018.04.007
Perry, T., Batterham, P., and Daborn, P. J. (2011). The biology of insecticidal activity and resistance. Insect Biochem. Molec. Biol. 41, 411–422. doi: 10.1016/j.ibmb.2011.03.003
Prodhan, M. Z., Hasan, H. M. T., Chowdhury, M. M. I., Alam, M. S., Rahman, M. L., Azad, A. K., et al. (2018). Bt eggplant (Solanum melongena L.) in Bangladesh: fruit production and control of eggplant fruit and shoot borer (Leucinodes orbonalis Guenee), effects on non-target arthropods and economic returns. PLoS One 13:e0205713. doi: 10.1371/journal.pone.0205713
Ranson, H., Claudianos, C., Ortelli, F., Abgrall, C., Hemingway, J., Sharakhova, M. V., et al. (2002). Evolution of supergene families associated with insecticide resistance. Science 298, 179–181. doi: 10.1126/science.1076781
Shelton, A. M., Hossain, M. J., Paranjape, V., Azad, A. K., Rahman, M. L., Khan, A. S. M. M. R., et al. (2018). Bt eggplant project in bangladesh: history, present status, and future direction. Front. Bioeng. Biotechnol. 6:106. doi: 10.3389/fbioe.2018.00106
Shirale, D., Patil, M., and Parimi, S. (2017). Insecticide resistance in field populations of Leucinodes orbonalis (Lepidoptera: Crambidae) in India. Can. Entomol. 149, 399–407. doi: 10.4039/tce.2017.3
Srinivasan, R. (2008). Integrated pest management for eggplant fruit and shoot borer (Leucinodes orbonalis) in south and southeast Asia: past, present and future. J. Biopesticides. 1, 105–112.
van Asperen, K. (1962). A study of house fly esterase by means of a sensitive colorimetric method. J. Insect Physiol. 8, 401–416. doi: 10.1016/0022-1910(62)90074-4
Wang, Y., Zhao, S., Shi, L., Xu, Z., and He, L. (2014). Resistance selection and Biochemical mechanism of resistance against cyflumetofen in Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 111, 24–30. doi: 10.1016/j.pestbp.2014.04.004
Yang, T., and Liu, N. (2011). Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes. Culex quinquefasciatus. PLoS One 6:e29418. doi: 10.1371/journal.pone.0029418
Yu, Q., Lu, C., Li, B., Fang, S., Zuo, W., Dai, F., et al. (2008). Identification, genomic organization and expression pattern of glutathione S-transferase in the silkworm, Bombyx mori. Insect Biochem. Molec. Biol. 38, 1158–1164. doi: 10.1016/j.ibmb.2008.08.002
Yu, Q. Y., Lu, C., Li, W. L., Xiang, Z. H., and Zhang, Z. (2009). Annotation and expression of carboxylesterases in the silkworm, Bombyx mori. BMC Genomics 10:553. doi: 10.1186/1471-2164-10-553
Keywords: Leucinodes orbonalis, insecticide resistance, genome, arboxylesterase, gene expression, glutathione S-transferase, carboxylesterase.
Citation: Kariyanna B, Prabhuraj A, Asokan R, Agrawal A, Gandhi Gracy R, Jyoti P, Venkatesan T, Bheemanna M, Kalmath B, Diwan JR, Pampanna Y and Mohan M (2020) Genome Mining and Expression Analysis of Carboxylesterase and Glutathione S-Transferase Genes Involved in Insecticide Resistance in Eggplant Shoot and Fruit Borer, Leucinodes orbonalis (Lepidoptera: Crambidae). Front. Physiol. 11:594845. doi: 10.3389/fphys.2020.594845
Received: 14 August 2020; Accepted: 22 October 2020;
Published: 19 November 2020.
Edited by:
Kai Lu, Fujian Agriculture and Forestry University, ChinaReviewed by:
Wei Dou, Southwest University, ChinaYuji Yasukochi, National Agriculture and Food Research Organization (NARO), Japan
Copyright © 2020 Kariyanna, Prabhuraj, Asokan, Agrawal, Gandhi Gracy, Jyoti, Venkatesan, Bheemanna, Kalmath, Diwan, Pampanna and Mohan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Mohan, TW9oYW4uTUBpY2FyLmdvdi5pbg==
 B. Kariyanna
B. Kariyanna A. Prabhuraj1
A. Prabhuraj1