
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 01 December 2020
Sec. Invertebrate Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.593962
Insect ecdysis triggering hormone (ETH) receptors (ETHRs) are rhodopsin-like G protein-coupled receptors. Upon binding its ligand ETH, ETHR initiates a precisely programed ecdysis behavior series and physiological events. In Drosophila melanogaster, the ethr gene produces two functionally distinct splicing isoforms, ethra and ethrb. ETH/ETHRA activates eclosion hormone (EH), kinin, crustacean cardioactive peptide (CCAP), and bursicon (burs and pburs) neurons, among others, in a rigid order, to elicit the behavioral sequences and physiological actions for ecdysis at all developmental stages, whereas ETH/ETHRB is required at both pupal and adult ecdysis. However, the role of ETHRB in regulation of molting has not been clarified in any non-drosophila insects. In the present paper, we found that 20-hydroxyecdysone (20E) signaling triggers the expression of both ethra and ethrb in a Coleopteran insect pest, the Colorado potato beetle Leptinotarsa decemlineata. RNA interference (RNAi) was performed using double-stranded RNAs (dsRNAs) targeting the common (dsethr) or isoform-specific (dsethra, dsethrb) regions of ethr. RNAi of dsethr, dsethra, or dsethrb by the final-instar larvae arrested larva development. The arrest was not rescued by feeding 20E. All the ethra depleted larvae stopped development at prepupae stage; the body cavity was expanded by a large amount of liquid. Comparably, more than 80% of the ethrb RNAi larvae developmentally halted at the prepupae stage. The remaining Ldethrb hypomorphs became pupae, with blackened wings and highly-expressed burs, pburs and four melanin biosynthesis genes. Therefore, ETHRA and ETHRB play isoform-specific roles in regulation of ecdysis during larva-pupa transition in L. decemlineata.
Insects are enclosed in a sclerotized exoskeleton. For a remarkable increase in size, the integument must be shed and replaced periodically during ecdysis, a precisely timed behavior sequence that is further separated into pre-ecdysis, ecdysis, and post-ecdysis (Krüger et al., 2015; de Oliveira et al., 2019). Each of these stages correlates with major behavioral, molecular, and cellular changes (Truman, 2005). At the endocrine level, ecdysis is initiated by a near-complete discharge of ecdysis triggering hormone (ETH) from Inka cells, fueled by an endocrine positive feedback with centrally produced eclosion hormone (EH; Ewer et al., 1997; Kingan et al., 1997). ETH directly targets neurons in the central nervous system that express its receptors ETHRA and ETHRB, two functionally distinct splice variants found in many insect species (Kim et al., 2006b; Arakane et al., 2008; Dai and Adams, 2009; Roller et al., 2010; Jiang et al., 2014; Shi et al., 2017) to regulate ecdysis behavior series and ecdysis-related physiological preparations (Clark, 2004; de Oliveira et al., 2019).
In Drosophila melanogaster, ETHRA and ETHRB splice isoforms are known to differ both in their expression patterns within the nervous system (Kim et al., 2006a,b, 2015; Diao et al., 2016) and their affinities for the ETH peptides (Iversen et al., 2002; Park et al., 2003). The ETHRA-expressing neurons include almost all peptidergic ones, such as kinin, diuretic hormone (DH), eclosion hormone (EH), FMRFamide, crustacean cardioactive peptide (CCAP), myoinhibitory peptide (MIP), bursicon (burs/pburs heterodimer), neuropeptide F (NPF), and short neuropeptide F (sNPF; Kim et al., 2006a,b, 2015; Diao et al., 2016). In contrast, ETHRB-expressing neurons are mainly located in the dorsolateral and dorsomedial sites in each ganglion along ventral nerve cord, with a few in the brain (Kim et al., 2006b; Diao et al., 2016). Moreover, ETHRB has an around 450-fold higher affinity for ETH than ETHRA (Iversen et al., 2002; Park et al., 2003). Recently, it is discovered that the two ETHR variants functionally diverge in D. melanogaster. Ethra-expressing neurons are required for ecdysis at all developmental stages. At the time of the pupal molt, the functionally distinct Ethra-expressing subsets regulate not only ecdysis behavior but also fluid balance. In contrast, ethrb-expressing neurons are required at both pupal and adult ecdysis (Kim et al., 2015; Diao et al., 2016, 2017; Mena et al., 2016).
However, the specific functions of ETHR isoforms have only been functionally characterized by RNA interference (RNAi) in two non-drosophila insect species (Diao et al., 2016; Meiselman et al., 2017, 2018; Shi et al., 2017, 2019; Kim et al., 2018). In Bactrocera dorsalis, selective RNAi of ethra, but not ethrb, causes developmental failure of ecdysis at early larval stages. The dsRNA-treated larvae cannot shed the old cuticle followed by death (Shi et al., 2017). Similarly, knockdown of ethra rather than ethrb at pharate pupae in Tribolium castaneum results in developmental arrest at the pharate adult stage before initiation of ecdysis (Arakane et al., 2008). Given that both ethr transcripts are widely expressed throughout larval development stages (Meiselman et al., 2017), which role does ETHRB play during larva-pupa-adult transformation in non-drosophila insects?
In the present paper, two ETHR isoforms were identified in the Colorado potato beetle Leptinotarsa decemlineata. Since this beetle is sensitive to RNAi (Deng et al., 2018; Meng et al., 2018, 2019; Xu et al., 2018), we intended to clarify the isoform-specific functions of ETHR in larval development by RNAi-aided knockdown of all ethr variants, or each of them, and comparison of the defective phenotypes.
The Leptinotarsa decemlineata beetles were kept in an insectary according to a previously described method (Shi et al., 2013), with potato foliage at the vegetative growth or young tuber stages in order to assure sufficient nutrition. At this feeding protocol, the larvae progressed the first-, second-, penultimate-, and final-instar stages with approximate periods of 2, 2, 2, and 4 days, respectively. Upon reaching full size, the final larval instars stopped feeding and entered the wandering stage. The wandering larvae (prepupae) dropped to the ground and finally burrowed to the soil. The prepupae spent an approximately 3 days to pupate. The pupae lasted about 5 days and the adults emerged.
The putative Ldethr isoforms were obtained from the genome (Schoville et al., 2018) and transcriptome data (Shi et al., 2013) of L. decemlineata. The correctness of the sequences was substantiated by polymerase chain reaction (PCR) using primers in Supplementary Table S1 and Supplementary Figure S1.
Three dsRNAs against two Ldethr isoforms (dsethr), either Ldethra (dsethra) or Ldethrb (dsethrb), were designed (Supplementary Figure S1). Other dsRNAs including dsSHD, dsEcR, dsFTZ-F1 and dsegfp were derived from LdSHD, LdEcR, and LdFTZ-F1 genes from L. decemlineata, and enhanced green fluorescent protein (egfp) gene from Aequorea victoria. These dsRNA fragments were cloned using specific primers listed in Supplementary Table S1. All dsRNAs were individually expressed using Escherichia coli HT115 (DE3) competent cells lacking RNase III following the established method (Meng et al., 2019). Individual colonies were inoculated and grown until cultures reached an OD600 value of 1.0. The colonies were then induced to express dsRNA by addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.1 mM. The expressed dsRNA was extracted and confirmed by electrophoresis on 1% agarose gel. Bacteria cells were centrifuged at 5000 × g for 10 min and resuspended in an equal original culture volume of 0.05 M phosphate buffered saline (PBS, pH 7.4). The bacterial solutions (at a dsRNA concentration of about 0.5 μg/ml) were used for experiment.
An ecdysteroid agonist halofenozide (Hal) and 20-hydroxyecdysone (20E) were respectively purchased from ChemService (West Chester, United States) and Sigma-Aldrich (United States). Chemicals were dissolved in distilled water with added surfactant (Tween 20, 1 g/L) to give solution at the concentration of 100 ng/ml. It was further serially diluted 10-folds with distilled water if necessary.
Four independent bioassays were carried out using newly-ecdysed fourth-instar larvae. The first bioassay was to test the influence of 20E and Hal on the expression of Ldethr isoforms and had three treatments: (1) water (control), (2) 100 ng/ml 20E, or (3) 100 ng/ml Hal. Potato leaves were immersed for 5 s, removed, and dried for 2 h under airflow on filter paper. Five treated leaves were then placed in a Petri dish (9 cm diameter and 1.5 cm height). Ten fourth-instar larvae were confined in petri dishes containing treated leaves for 1 day as a repeat. Each treatment was replicated three times. The second to fourth bioassays were to knock down LdSHD, LdEcR, and LdFTZ-F1 and had three treatments: (1) PBS-, (2) dsegfp-, (3) dsSHD-, dsEcR-, or dsFTZ-F1-immerged leaves. Each treatment (10 fourth-instar larvae) was replicated three times and fed for 3 days (replaced with freshly treated ones each day). The resultant larvae were used to analyze the mRNA levels of target gene and Ldethr variants.
The same method as previously reported was used to introduce dsRNA into larvae (Meng et al., 2019). Potato leaves were immersed with a bacterial suspension containing a dsRNA. The PBS‐ and dsegfp-dipped leaves were used as controls. The newly-ecdysed final-instar larvae were starved for at least 4 h prior to the experiment. Then, 10 larvae were transferred to each dish as a repeat. For each treatment, nine repeats were set and randomly allocated to three groups, each containing three replicates. The first and second groups were allowed to feed on dsRNA-dipped foliage for 3 days (replaced with freshly treated ones each day) and on untreated foliage until reaching the wandering stage. Three replicates were used to observe the defective phenotypes, pupation and adult emergence and the other three were used to extract body fluid. The third group was collected after continuously fed on treated foliage 3 days to extract total RNA.
Two bioassays were carried out as previously described (Meng et al., 2019) using newly-ecdysed final-instar larvae, to test the rescuing effect of 20E at the concentration of 10 ng/ml. Ten larvae in a repeat were firstly allowed to ingest potato foliage immersed with PBS, dsegfp or dsethra (dsethrb) for 2 days, and then to consume leaves dipped with PBS, dsegfp, dsethra (dsethrb), dsethra (dsethrb) + 20E, for an additional day. The larvae were then transferred to untreated foliage if necessary. For each treatment, nine (for the bioassay to test rescuing effect to dsethra ingestion) or six (for the bioassay to test rescuing effect to dsethrb ingestion) repeats were set. Three replicates were randomly sampled for total RNA extraction 3 days after initiation of the bioassay. The other three repeats were used to observe the defective phenotypes, pupation and adult emergence. An addition of three repeats in the bioassay to test rescuing effect to dsethra ingestion was used to extract body liquid (see section “Assay of Body Fluid Volume” for detail).
For temporal expression analysis, RNA templates were derived from eggs, the larvae from the first through final instars, wandering larvae,pupae (5 days after burring into soil) and adults (5 days after emerging). For analysis of the tissue expression patterns, templates were from the brain-corpora cardiaca-corpora allata complex, ventral nerve cord, foregut, midgut, hindgut, Malpighian tubules, fat body, epidermis, and trachea of the day 4 fourth-instar larvae. For analysis of the effects of treatments, total RNA was extracted from treated larvae. Each sample contained at least 5–10 individuals and repeated three times. The RNA was extracted using SV Total RNA Isolation System Kit (Promega). Purified RNA was subjected to DNase I to remove any residual DNA according to the manufacturer’s instructions. Quantitative mRNA measurements were performed by real-time quantitative PCR (qRT-PCR) in technical triplicate, using four internal control genes (LdRP4, LdRP18, LdARF1, and LdARF4, the primers listed in Supplementary Table S1) according to our published results (Shi et al., 2013). An RT negative control (without reverse transcriptase) and a non-template negative control were included for each primer set to confirm the absence of genomic DNA and to check for primer-dimer or contamination in the reactions, respectively.
According to a previously described method (Bustin et al., 2009), the generation of specific PCR products was confirmed by gel electrophoresis. The primer pair for each gene was tested with a 10-fold logarithmic dilution of a cDNA mixture to generate a linear standard curve [crossing point (CP) plotted vs. log of template concentration], which was used to calculate the primer pair efficiency. All primer pairs amplified a single PCR product with the expected sizes, showed a slope less than −3.0, and exhibited efficiency values ranging from 2.3 to 2.4. Data were analyzed by the 2−ΔΔCT method using the geometric mean of the four internal control genes for normalization.
Twelve day after initiation of experiment, extractable body fluid volume was measured in the resultant beetles having ingested PBS, dsegfp, dsethra, and dsethrb as larvae, according to a previously described method (Diao et al., 2016), with some modifications. Briefly, the collected animals were weighed and then crushed in a pre-chilled mortar using a pellet pestle and centrifuged at 3,000 rpm in an Eppendorf micro centrifuge. The volume of fluid collected after centrifugation was measured and the ratio of this volume to the starting weight was calculated for each treatment.
We used SPSS for Windows (Chicago, IL, United States) for statistical analyses. The averages (±SE) were submitted to ANOVA with the Tukey-Kramer test.
By mining the genome (Schoville et al., 2018) and transcriptome data (Shi et al., 2013), two Ldethr transcripts were identified. Using the TMHMM server, we predicted that both proteins contained seven TMs (Figure 1A), similar to their partners from other insect species (Kim et al., 2015; de Oliveira et al., 2019). The gene structure of Ldethr was analyzed. Ldethr included four exons. The first two exons, which encode the first four of the seven TMs, were shared by both isoforms. A specific exon encoded the later three TMs specific in each Ldethr isoform (Figure 1B). Comparable gene structures of ethr genes have been documented in other insects such as D. melanogaster (Diao et al., 2016).
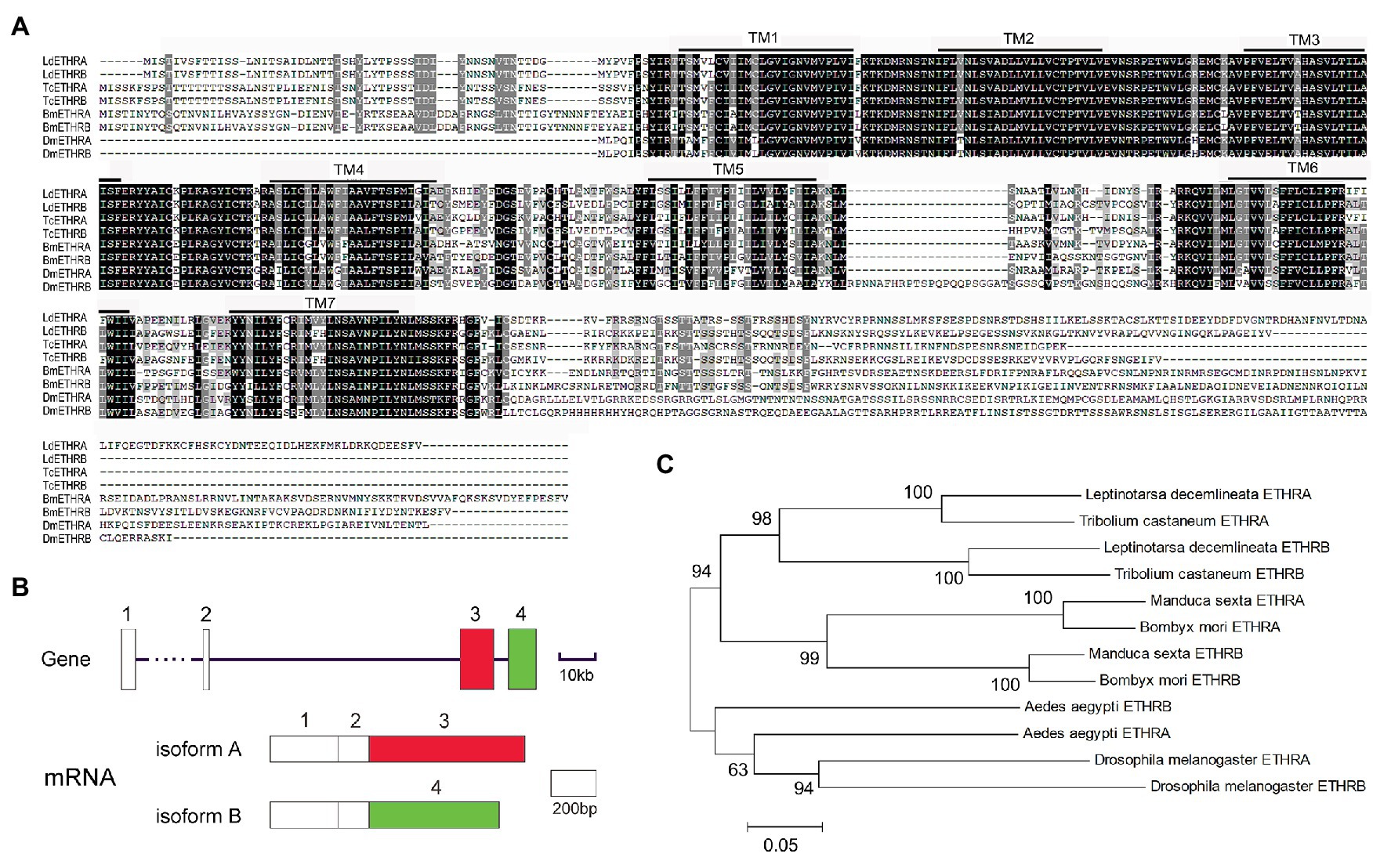
Figure 1. Alignment (A), exon/intron structure (B), and phylogenetic analysis (C) of ecdysis triggering hormone receptor (ETHR) derived from Leptinotarsa decemlineata. (A) Sequence alignment of LdETHR. Increasing background intensity (from light to dark) indicates an increase in sequence similarity. Gaps have been introduced to permit alignment. The transmembrane domains (TMs) are highlighted. (B) Boxes indicate exons. Black lines mark introns. (C) ETHR proteins are from two coleopteran L. decemlineata (ETHRA, QBH70335.1; ETHRB, QBH70336.1) and Tribolium castaneum (ETHRA, ABN79653; ETHRB, ABN79654), two lepidopteran Bombyx mori (ETHRA, AB330426; ETHRB, AB330427), Manduca sexta (ETHRA, AAX19163; ETHRB, AAX19164), two dipteran Drosophila melanogaster (ETHRA, NP_650960; ETHRB, NP_996255), and Aedes aegypti (ETHRA, ABI93273; ETHRB, ABI93274). The tree is constructed using the neighbor-joining method based on the full-length protein sequence alignments. Bootstrap analyses of 1,000 replications are carried out and bootstrap values >50% are shown on the tree.
An unrooted tree was constructed to examine the phylogenetic relationships of LdETHR variants with homologous proteins from representative species across three insect orders (Coleoptera, Diptera, and Lepidoptera). In the tree, the proteins were order-specific. The two ETHR isoforms sorted with ETHR splicing variants from another coleopteran T. castaneum. The two ETHR variants were accordingly named as ETHRA and ETHRB (Figure 1C).
To identify stages at which Ldethr isoforms are expressed and potentially required, cDNA samples prepared from the whole bodies of larvae, prepupae, pupae and sexually mature adults were analyzed by qRT-PCR. Ldethra and Ldethrb transcripts were detectable from embryo (egg) to adult by qRT-PCR. Ldethra was abundantly expressed at the adult and embryo; Ldethrb was highly transcribed at the embryo and wandering larval stages. In contrast, both Ldethra and Ldethrb were lowly expressed in the fourth-instar larval stage. Within the larval stage, copious mRNA levels of Ldethra and Ldethrb were found in the late stage of the first-, second-, third-, and fourth-instar larvae (Figures 2A,B).
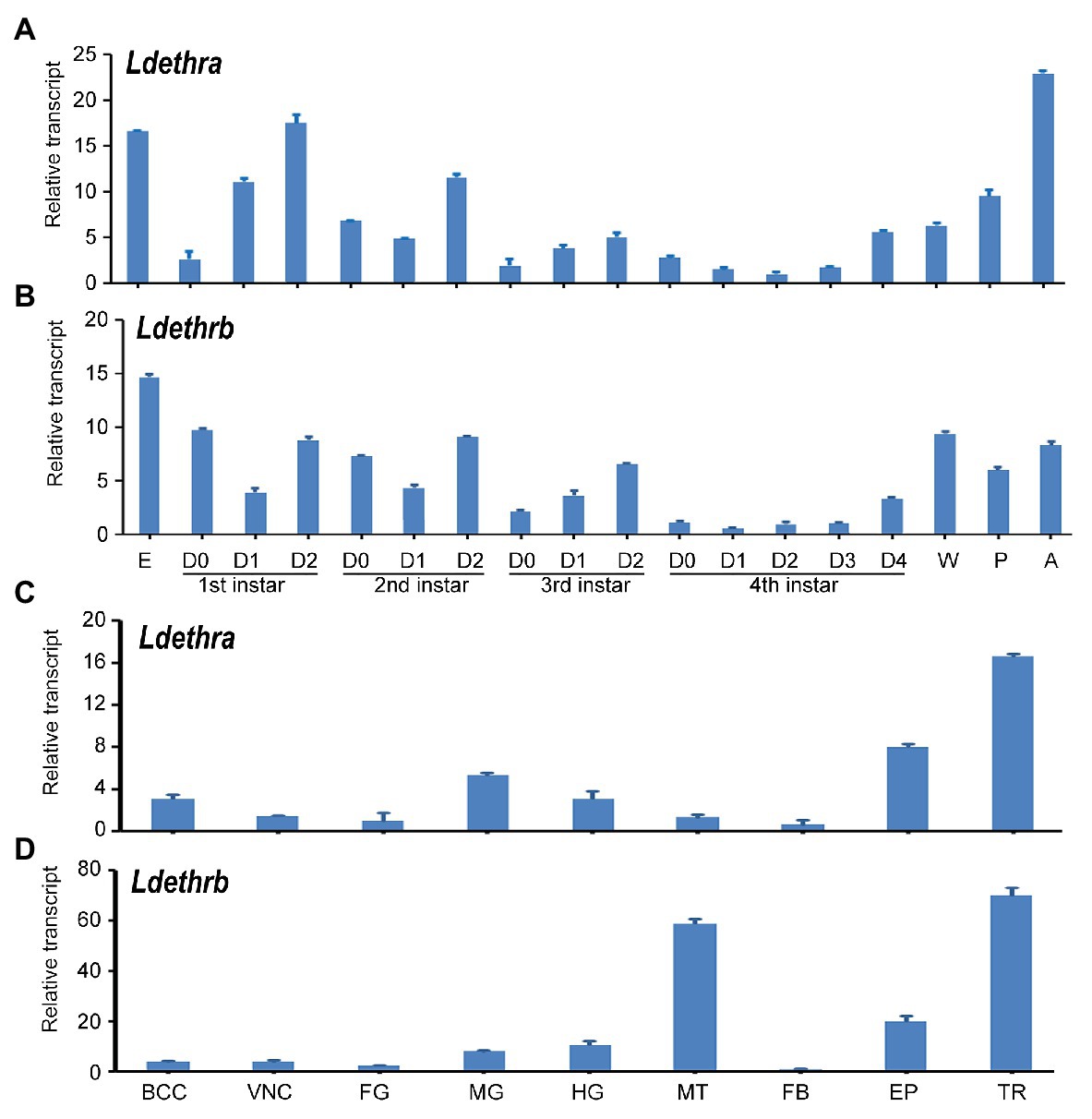
Figure 2. Temporal and tissue transcription patterns of ecdysis triggering hormone receptor (Ldethr) isoforms in L. decemlineata. For analysis of the temporal expression patterns of Ldethra (A) and Ldethrb (B), complementary DNA templates were derived from the eggs, first, second, third, and fourth larval instars at an interval of 1 day (D0 indicated newly ecdysed larvae), wandering larvae, pupae (5 days after burring into soil) and adults (5 days after emerging). For analysis of the tissue expression patterns of Ldethra (C) and Ldethrb (D), RNA templates were derived from the brain-corpora cardiaca-corpora allata complex (BCC), ventral nerve cord (VNC), foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), fat body (FB), epidermis (EP) and trachea (TR) of the day 4 fourth-instar larvae. For each sample, three independent pools of 5–10 individuals were measured in technical triplicate using real-time quantitative PCR (qRT-PCR). The values were calculated using the 2−ΔΔCT method. The lowest transcript levels are set as 1. The columns represent averages with vertical lines indicating SE.
Analysis of tissue expression patterns uncovered that Ldethra and Ldethrb were widely transcribed in the brain-corpora cardiaca-corpora allata complex, ventral nerve cord, foregut, midgut, hindgut, Malpighian tubules, fat body, epidermis, and trachea of the day 4 fourth-instar larvae. The levels of Ldethra were high in the trachea, epidermis, midgut, hindgut and brain-corpora cardiaca-corpora allata complex; while the levels of Ldethrb were high in the trachea, Malpighian tubules, epidermis, midgut and hindgut (Figures 2C,D).
The expression patterns uncover that the surges of mRNA levels of Ldethra and Ldethrb are correlated with the pulses of hemolymph 20E. To check whether 20E activates the transcription of Ldethra and Ldethrb in vivo, the newly-ecdysed fourth-instar larvae were allowed to feed 20E or Hal. After ingestion of 20E or Hal for 1 day, the mRNA levels of the early ecdysone-response genes LdHR3 and LdE75 were significantly elevated (Supplementary Figure S2). This finding indicates that in vivo ecdysteroid introduction activates 20E signaling. Consequently, the mRNA levels of Ldethra and Ldethrb were dramatically increased in the larvae having ingested Hal or 20E, compared with control specimens (Figure 3A).
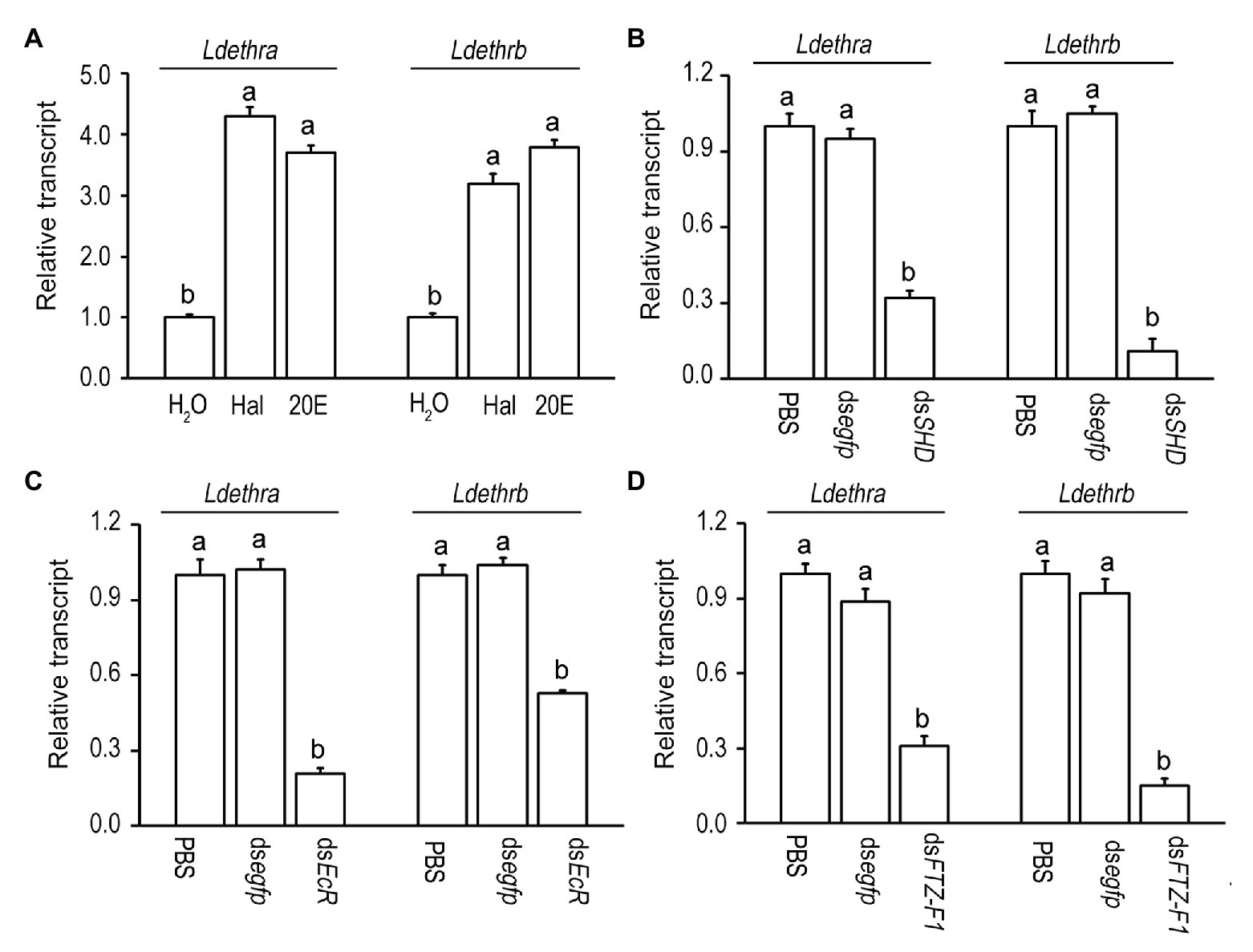
Figure 3. Induction of the expression of Ldethr isoforms by 20-hydroxyecdysone (20E) in L. decemlineata. For in vivo induction of 20E (A), the newly-ecdysed fourth-instar larvae have ingested potato foliage treated with water (control), 100 ng/ml halofenozide (Hal) or 20E for 1 day. For knockdown of the genes encoding Halloween protein SHADE (SHD) (B), the heterodimer of ecdysone receptor (EcR) (C) and a late ecdysone response gene fushi tarazu factor-1 (FTZ-F1) (D), the newly-molted fourth-instar larvae had ingested dsSHD, dsEcR, and dsFTZ-F1 dipped leaves for 3 days. The larvae having ingested PBS and dsegfp immersed foliage were used as controls. The values were calculated using the 2−ΔΔCT method. The transcript level at control is set as 1. Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test.
Moreover, silencing an ecdysteroidogenesis gene Shade (LdSHD; Kong et al., 2014; Supplementary Figure S3A) significantly decreased the mRNA levels of Ldethra and Ldethrb (Figure 3B), compared with control specimens. In addition, depleting each of two 20E signaling genes (LdEcR or LdFTZ-F1; Liu et al., 2014; Xu et al., 2019, 2020; Supplementary Figures S3B,C) significantly reduced the mRNA levels of Ldethra and Ldethrb (Figures 3C,D).
To characterize ETHR function, we took advantage of RNAi by immersing the potato foliage with dsethr targeting the common sequence of both Ldethr variants. The results were examined according to the experimental procedure timing listed in Supplementary Figure S4. Ingesting dsethr significantly declined the target mRNA level (Figure 4A). Larval growth was inhibited in the dsethr fed beetles (Figures 4B,F vs. G). Defective phenotypes were noted during larva-pupa-adult transition. The most severe defect occurred in around 22% of the Ldethr RNAi larvae. These animals remained on the soil surface, gradually dried and finally died within 10 days (Figure 4C).
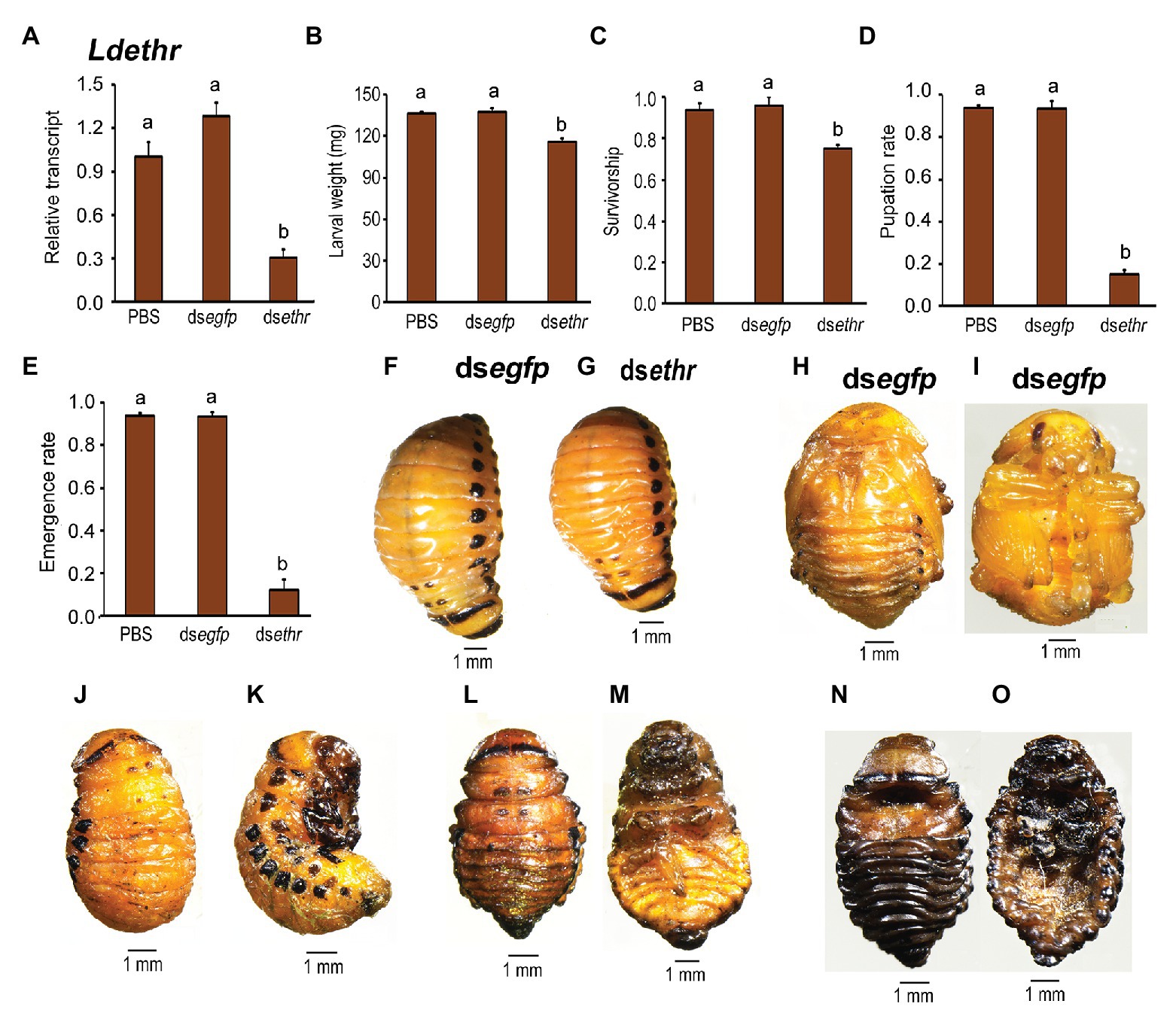
Figure 4. Ingestion of dsethr by the final instar larvae affects larval performance in L. decemlineata. The newly-ecdysed fourth instar larvae had ingested PBS-, dsegfp-, and dsethr-dipped leaves for 3 days, and untreated foliage for an addition of 1 day. Timing of the experimental procedures was shown. The expression level of Ldethr was measured 3 days after the initiation of the bioassay (A). Relative transcripts are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which is set as 1. The larvae were weighed 4 days after the initiation of the bioassay (B,F vs. G). The survivorship was observed through the fourth-instar larvae and prepupae. The pupation and emergence rates were recorded during a 4-week trial period (C–E). The bars represent values (±SE). Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test. While the PBS‐ and dsegfp-fed larvae pupated 8 days (H,I) after initiation of bioassay, more than 80% of the Ldethr RNAi larvae remain as prepupae (D) 10 (J,K) and 15 (L,M) days after initiation of bioassay, completely wrapped in the old larval exuviae. These prepupae become withered, dried and darkened gradually, and finally die in soil (N,O).
The remaining 78% of the dsethr-treated larvae showed moderate or slight defective phenotypes. They displayed similar wandering behavior and dug into soil normally after developing a similar period to those in the dsegfp‐ and PBS-fed larvae. More than 80% of these Ldethr RNAi prepupae mined into soil exhibited a modest defect. They failed to pupate (Figure 4D) 10 (Figures 4J,K) and 15 (Figures 4L,M) days after initiation of bioassay, wrapped completely in the old larval exuviae, unlike the dsegfp‐ and PBS-fed larvae pupating 8 days after initiation of bioassay (Figures 4D,H,I). These prepupae gradually darkened and withered, and finally died (Figures 4N,O).
Lastly, less than 20% of these Ldethr RNAi prepupae that mined into soil displayed a minor defect. They normally pupated and emerged as adults (Figures 4D,E). However, all the Ldethr RNAi adults eventually died in a week after emergence.
In the larvae having fed for 3 days on foliage immersed with an isoform-specific dsRNA (dsethra or dsethrb), the mRNA level of target transcript was significantly decreased. In contrast, ingestion either dsethra or dsethrb did not affect the transcript level of non-target isoform (Figures 5A,B).
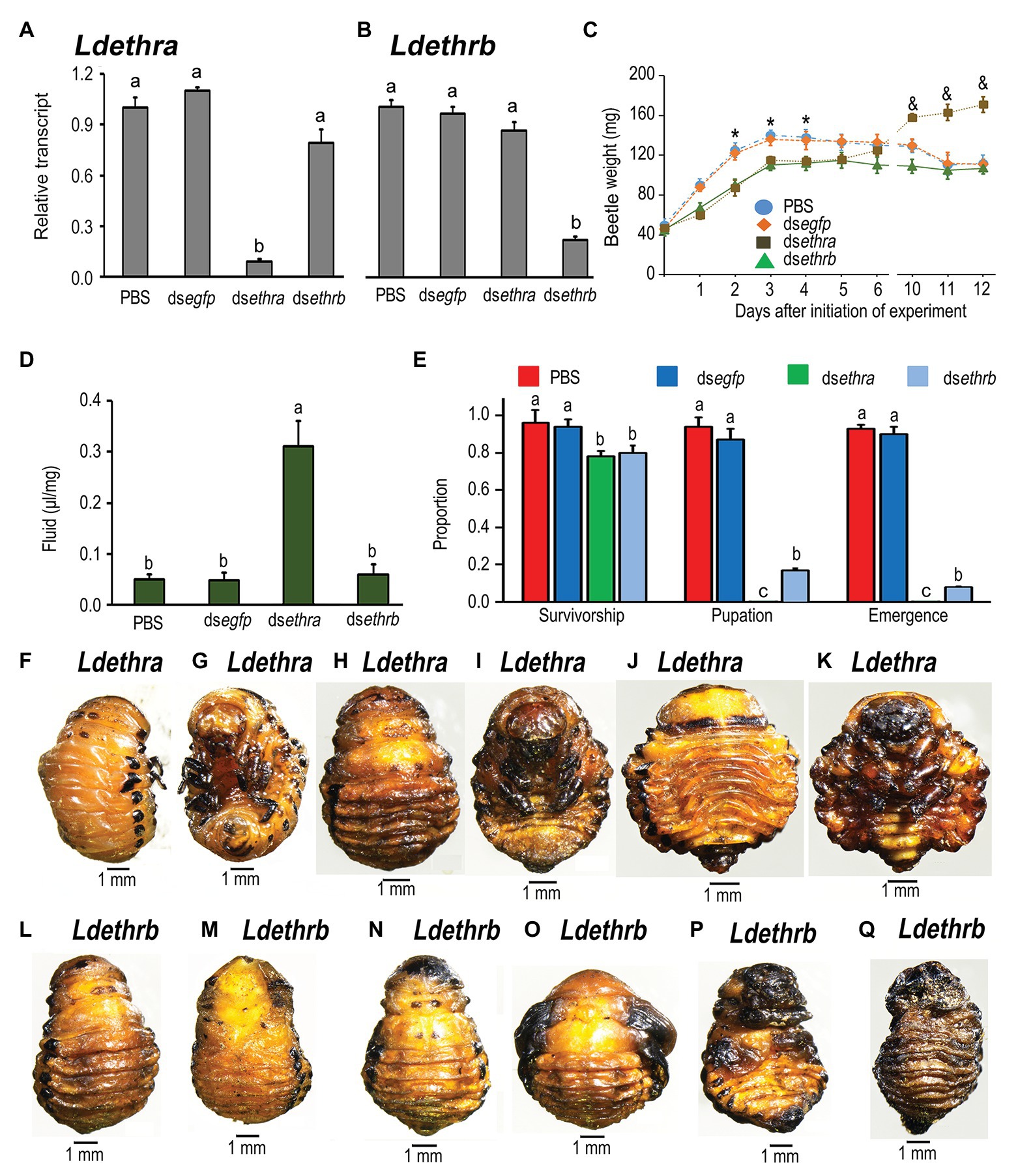
Figure 5. Two splice ETHR isoforms have distinct roles in larva-pupa transition in L. decemlineata. The newly-ecdysed fourth instar larvae had ingested PBS-, dsegfp-, dsethra- and dsethrb-immersed potato foliage for 3 days, and untreated foliage for an addition of 1 day. The expression levels of Ldethra and Ldethrb were tested 3 days after the initiation of the bioassay (A,B). Relative transcripts are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which are set as 1. The larvae were weighed 1–6 and 10–12 days after the initiation of the bioassay (C). The extracellular fluid volumes were determined 12 days after the initiation of the bioassay (D). The survivorship, pupation and emergence rates were recorded during a 4-week trial period (E). The bars represent values (±SE). Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test. The defective phenotypes in prepupae or pupae having ingested dsethra (F–K) or dsethrb (L–Q) as larvae are shown.
Ingestion of dsethra or dsethrb suppressed larval growth (Figure 5C). The fresh weights of the resulting larvae having fed on either dsethra or dsethrb were significantly lighter than those of PBS‐ and dsegfp-fed larvae 2, 3, and 4 days after initiation of bioassay. When the larvae entered prepupae stages (5 and 6 days after experiment), the difference in body weights became smaller (Figure 5C). Similar to knockdown of two ethr isoforms, consumption of either dsethra or dsethrb caused various defective phenotypes, varying in degree. Firstly, around 20% larval mortalities were noted before entering the soil (Figure 5E). Secondly, although the remaining approximately 80% dsethra‐ or dsethrb-treated larvae displayed similar wandering behavior and dug into soil normally after a similar developing period to those in the dsegfp‐ and PBS-fed larva, feeding dsethra or dsethrb arrested development.
In contrast to the dsegfp‐ and PBS-fed larvae that pupated 8 days after initiation of bioassay (Figure 5E), all Ldethra depleted larvae remained as prepupae (Figures 5E–I), completely wrapped in the old larval exuviae. The fresh weights of the resultant prepupae were greatly increased 10, 11, and 12 days after initiation of bioassay (Figure 5C). At the same time, the body cavity was expanded. A large amount of liquid was found in the cavity (Figures 5J,K). We assayed the accumulation of extracellular fluid by directly measuring the extractable volume of liquid normalized to body weight [liquid volume (μl) per mg fresh weight]. We found that fluid accumulation was significantly elevated 12 days after the initiation of bioassay (Figure 5D).
As for RNAi of Ldethrb, approximately 80% of the resulting larvae were developmentally halted at the prepupae stage (Figures 5L–N), completely or partially wrapped in the old larval cuticles. These prepupae gradually darkened and dried, and finally died (Figures 5P,Q). Around 20% of the Ldethrb hypomorphs shed their larval exuvia and became pupae, with blackened wings (Figures 5E,O), in contrast to control pupae (Figures 4H,I).
The darkened wings indicate the deposition of dark black and brown melanins in wings, a process induced by bursicon heterodimer (burs/pburs; Kim et al., 2006a,b, 2015; Diao et al., 2016). The biosynthesis of melanin pigments from tyrosine is catalyzed by tyrosine hydroxylase (TH), dopa decarboxylase (DDC), and prophenol oxidase (PPO), among others (Radovic et al., 2002; Protas and Patel, 2008; Wittkopp et al., 2009; Wittkopp and Beldade, 2009; Kronforst et al., 2012; Takahashi, 2013; Massey and Wittkopp, 2016). We accordingly tested the expression level of Ldbur, Ldpbur, Ldth, Ldddc, Ldppox1, Ldppox2, and Ldppox3 in these wing-darkened pupae. As expected, the expression levels of Ldbur, Ldpbur, Ldth, Ldppox1, Ldppox2, and Ldppox3 were increased compared with those in the control pupae (Figure 6).
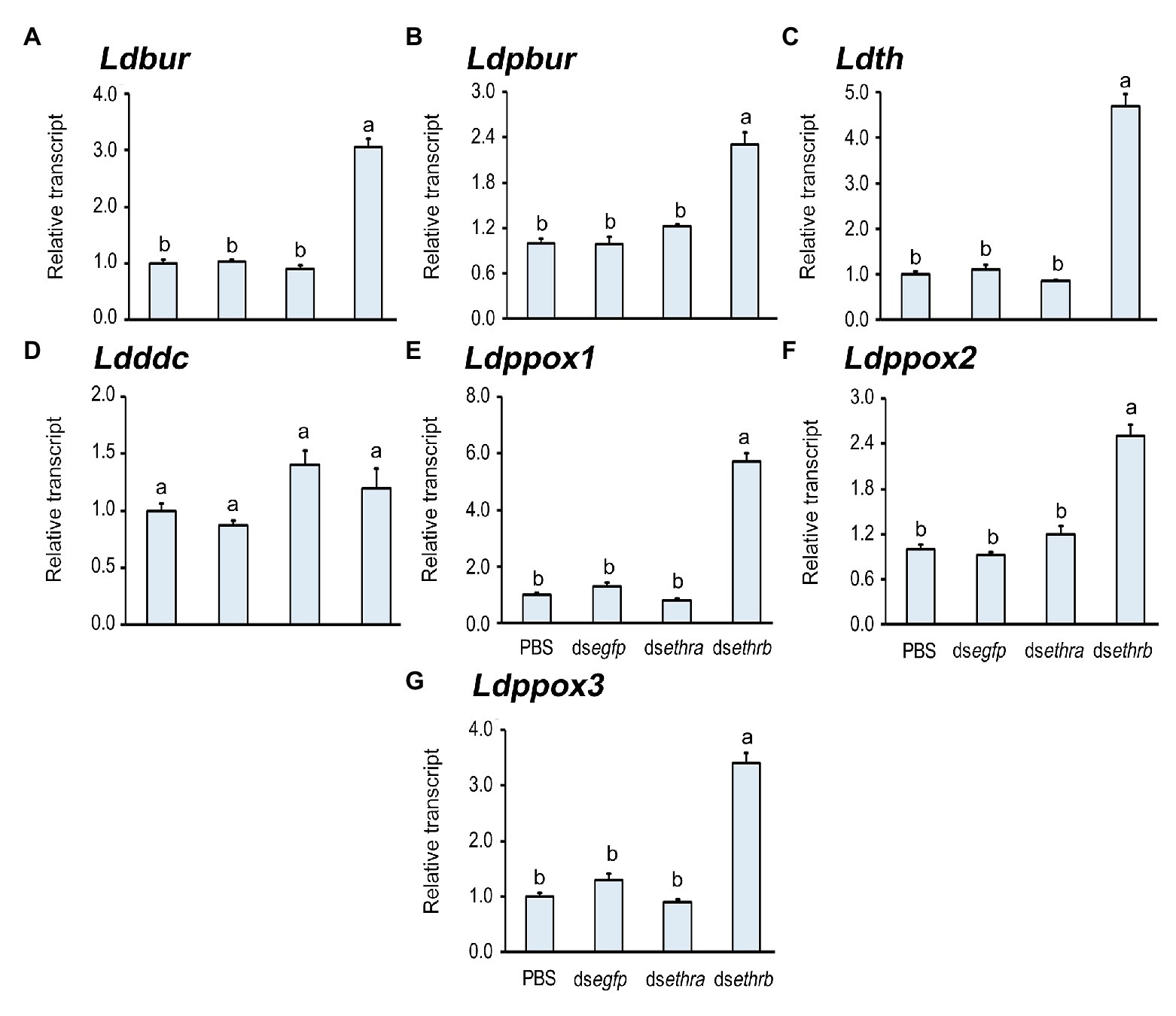
Figure 6. Knockdown of ethrb isoform prematurely activates melanin biosynthesis in L. decemlineata. The newly-ecdysed fourth instar larvae had ingested PBS-, dsegfp-, dsethra-, and dsethrb-immersed potato foliage for 3 days, and untreated foliage for an addition of 1 day. The expression levels of Ldbur, Ldpbur, Ldth, Ldddc, Ldppox1, Ldppox2, and Ldppox3 were tested 12 days after the initiation of the bioassay (A–G). Relative transcripts are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which are set as 1. Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test.
A few Ldethrb RNAi pupae with darkened wings finally emerged as adults (Figure 5E). All these Ldethrb RNAi adults appeared normal in shape, but eventually died within 1 week after eclosion.
20E ingestion by Ldethra RNAi larvae did not rescue the decreased expression levels of Ldethra (Figure 7A). Moreover, 20E introduction neither lessened the accumulated extracellular fluid (Figure 7B) nor elevated the low pupation rate (Figure 7C). While the dsegfp‐ and PBS-fed larvae pupated 8 days after initiation of bioassay (Figures 7D,E), almost all of the resultant larvae ingested dsethra and dsethra + 20E developmentally arrested at prepupae stage (Figure 7F). Furthermore, 10, 11, and 12 days after initiation of bioassay, the body cavity of the Ldethra hypomorph was expanded, especially at the abdomen (Figure 7G).
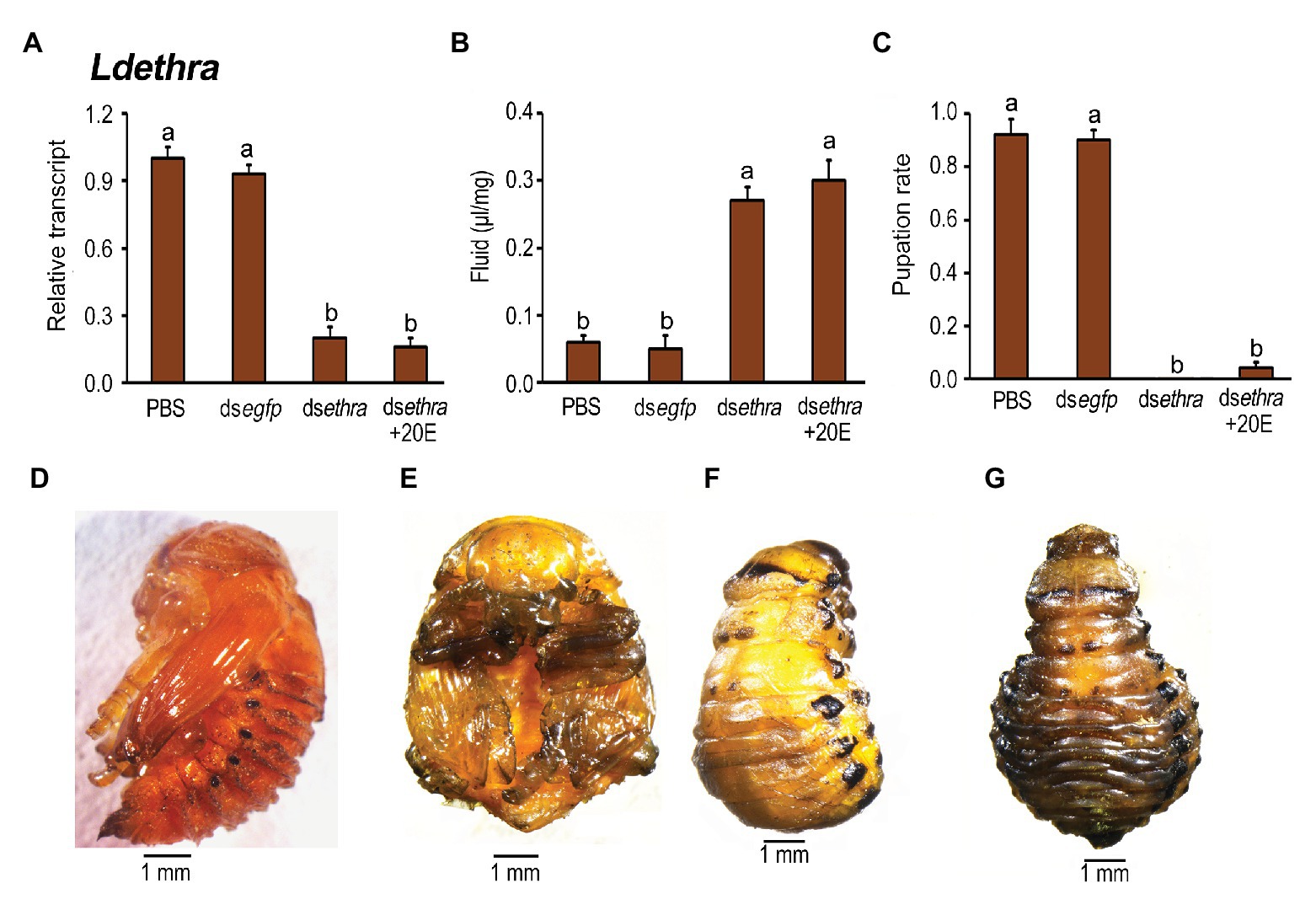
Figure 7. The rescuing effects of 20E on pupation in the Ldethra RNAi larvae in L. decemlineata. The newly-ecdysed final instar larvae had ingested PBS, dsegfp, dsethra-, and dsethra + 20E-dipped leaves. The expression level of Ldethra was tested (A). Relative transcript levels are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which is set as 1. The extracellular fluid volumes were determined 12 days after the initiation of the bioassay (B). The pupation rate was recorded during a 4-week trial period (C). The bars represent values (±SE). Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test. The prepupa defective phenotypes that have fed on dsethra + 20E as larvae are shown (F,G), compared with that in control (D,E).
We repeated the rescuing experiment in Ldethrb RNAi larvae using 20E and found that ingestion of 20E did not restore the lowed expression level of Ldethrb (Figure 8A), nor alleviated the impairment of pupation and adult eclosion (Figures 8B,C). Around 80% of the resultant larvae ingested dsethrb and dsethrb + 20E remained developmentally stationary in the prepupal stage (Figures 8D,E).
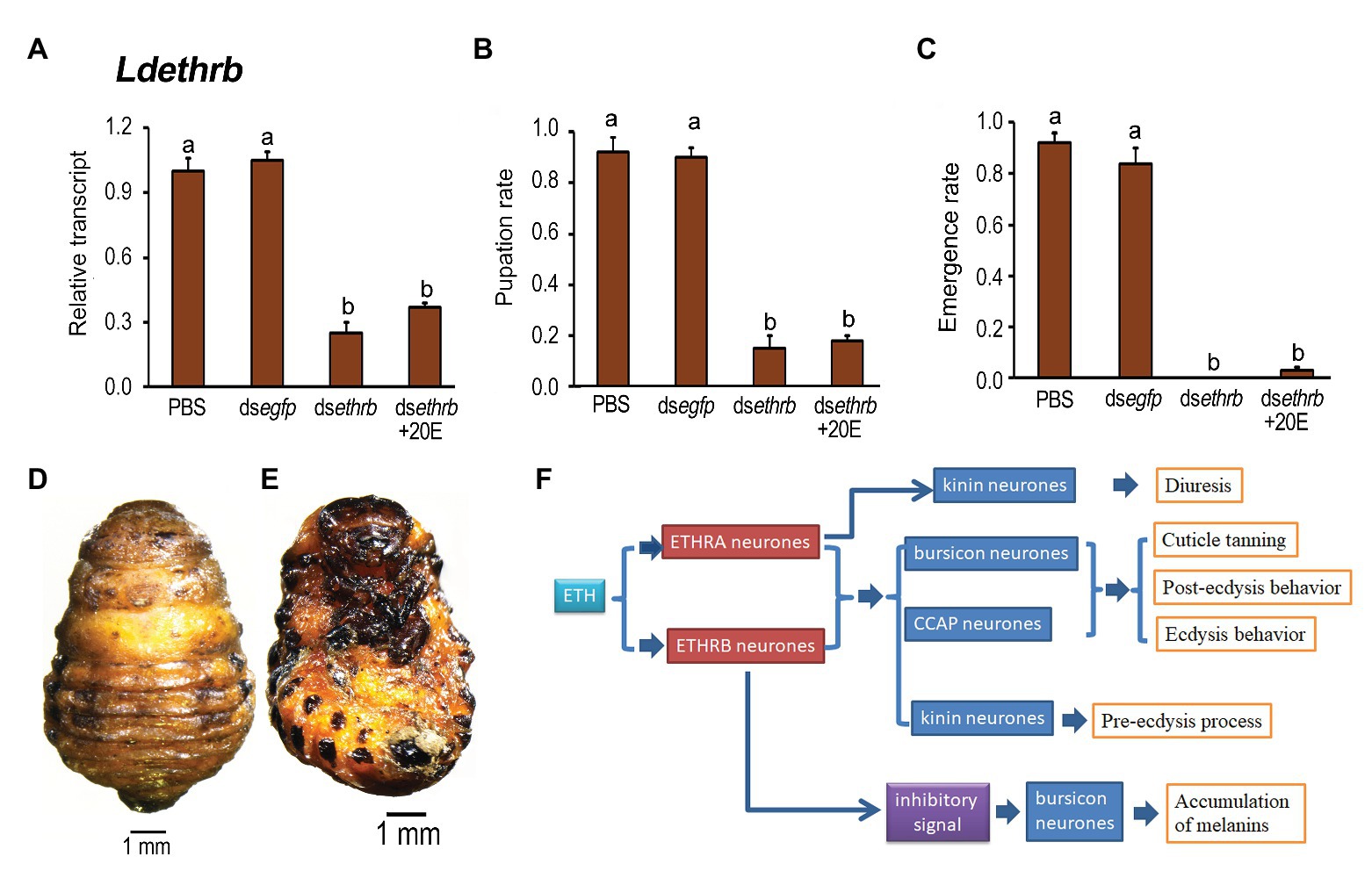
Figure 8. The rescuing effects of 20-hydroxyecdysone (20E) on pupation in the Ldethrb RNAi larvae in L. decemlineata. The newly-ecdysed final instar larvae had ingested PBS, dsegfp, dsethrb-, and dsethrb + 20E-immersed potato foliage. The expression level of Ldethrb was determined (A). Relative transcript levels are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which is set as 1. The pupation and emergence rates were observed during a 4-week trial period (B,C). The bars represent values (±SE). Different letters indicate significant difference at p < 0.05 compared with ANOVA and the Tukey-Kramer test. The prepupa defective phenotypes that have fed on dsethrb + 20E as larvae are given (D,E). A model depicts the combinatory role of ETHRA and ETHRB on regulation of ecdysis in L. decemlineata (see section “Discussion” for details) (F).
The rescuing findings reveal that both ETHRA and ETHRB act downstream of 20E biosynthesis.
Ecdysis process consists of behavioral routines and physiological events that are expressed in a specific sequence. In D. melanogaster, the sequential nature of ecdysis is based on the progressive activation of different ETH targets: the ETHRB‐ and ETHRA-expressing neurons (Kim et al., 2015; Diao et al., 2016, 2017; Mena et al., 2016). In this survey, we found that alternative use of exon causes different ETHR variants, ETHRA and ETHRB, in L. decemlineata (Figure 1). RNAi-aided knockdown of both Ldethr transcripts in L. decemlineata (Figure 4) phenocopied ETH depletion insects such as D. melanogaster (Park et al., 2002; Cho et al., 2014), B. dorsalis (Shi et al., 2017), and Schistocerca gregaria (Lenaerts et al., 2017), indicating that the ethr gene encodes the sole receptor for ETH in mediating ecdysis in L. decemlineata. In this survey, we focused on the isoform-specific roles of the two splicing variants in larva-pupa transition in L. decemlineata.
In this study, we found that the expression peaks of either Ldethra or Ldethrb in first through fourth larval instars exhibited a clear correlation with 20E pulses just before ecdysis (Figure 2). Consistent with our result, the peaks in ethr transcript levels are observed together with the peaks in ecdysteroid titers in M. sexta, A. aegypti, and S. gregaria (Kim et al., 2006b; Dai and Adams, 2009; Lenaerts et al., 2017). Based on the above researches in both holometabolans and hemimetabolans, the temporal expression profiles of both Ldethra and Ldethrb suggest that they may regulate by an ecdysteroid pulse.
Consistent with the suggestion, we found that ingestion of either Hal or 20E augmented the expression of both Ldethra and Ldethrb, whereas knockdown of SHD decreased their transcription in L. decemlineata. Furthermore, transcript levels of both Ldethra and Ldethrb were significantly reduced in LdEcR or LdFTZ-F1 knockdown fourth larval instars, compared to control beetles (Figure 3). In agreement with our results, 20E regulates ethr expression in the larval stages of moths, mosquitoes, desert locust and D. melanogaster adults (Žitňan et al., 1999, 2007; Dai and Adams, 2009; Lenaerts et al., 2017; Meiselman et al., 2017).
The findings that the transcript levels of both Ldethra and Ldethrb are highly dependent on functional 20E signaling indicate that both variants are essential for larva metamorphoses in L. decemlineata. Conversely, in B. dorsalis, only ethra displays a clear correlation with 20E titer. As expected, only ETHRA is responsible for the regulation of larva-pupa metamorphosis; whereas ETHRB control adult ovary growth (Shi et al., 2017, 2019).
In this survey, we uncovered that feeding ethra at the final instar stage impaired larva-pupa ecdysis. The defects could not be rescued by 20E. All the ethra hypomorphs were developmentally halted at the prepupae stage in L. decemlineata (Figures 5, 7). Consistent with our results, suppression of ETHRA-expressing neurons leads to 100% larval lethality in D. melanogaster, with ecdysis deficits; restricting suppression of ETHRA-expressing neurons at the pupal and pharate adult stages blocks adult ecdysis and results in 100% lethality (Diao et al., 2016). Similar results have been documented using RNAi in holometabolans including B. dorsalis (Shi et al., 2017) and T. castaneum (Arakane et al., 2008), hemimetabolans such as S. gregaria (Lenaerts et al., 2017) and other arthropods for instance Macrobrachium nipponense (Liang et al., 2017).
An obvious defective phenotype was the accumulation of extracellular liquid in the ethra depletion prepupae in L. decemlineata, which resulted in the expanded bodies 10, 11, and 12 days after initiation of bioassay (Figure 5). It is well known that ETHRA-expressing neurons include kinin-, EH-, CCAP-, and bursicon-producing ones that regulate different phases of the ecdysis sequence (Kim et al., 2006a,b, 2015; Diao et al., 2016). In D. melanogaster, kinin regulates fluid balance by targeting Malpighian tubules (Terhzaz et al., 1999). Suppression of ETHRA within all kinin-expressing neurons accumulates a larger amount of water in the resultant pupae (Diao et al., 2016).
The similar defective phenotypes demonstrate that the critical role of ETHRA during larva-pupa transition is conserved among dipterans such as D. melanogaster (Kim et al., 2006a,b, 2015; Diao et al., 2016) and B. dorsalis (Shi et al., 2017), and coleopterans such as T. castaneum (Arakane et al., 2008) and L. decemlineata (this study).
In the present paper, our results revealed that ETHRB acts downstream of 20E biosynthesis to regulate larva-pupa molting in L. decemlineata (Figures 5, 8). Similar role of ETHRB has been reported in D. melanogaster (Diao et al., 2016), but not in B. dorsalis (Shi et al., 2017) and T. castaneum (Arakane et al., 2008).
In this study, we established that some Ldethrb RNAi pupae had blackened wings (Figure 5), a phenotype caused by the accumulation of melanins (Radovic et al., 2002; Protas and Patel, 2008; Wittkopp et al., 2009; Wittkopp and Beldade, 2009; Kronforst et al., 2012; Takahashi, 2013; Massey and Wittkopp, 2016). Moreover, we uncovered that the expression levels of Ldbur, Ldpbur, Ldth, Ldppox1, Ldppox2, and Ldppox3 were upraised (Figure 6). Our results provide, for the first time, a compelling piece of experimental evidence that the inhibitory inputs from ETHRB-expressing neurons indirectly act on bursicon neurons to regulate the production and secretion of burs and pburs. During wandering stage, Ldethrb was highly expressed. The resultant ETHRB acts as an indirect inhibitor of bursicon-producing neurons. Bursicon does not release until after adult emergence. In the present paper, depletion of ETHRB by RNAi lessens these inhibitory inputs, burs and pburs that are prematurely expressed and released, the melanins are precociously biosynthesized and accumulated in the pupal wings in L. decemlineata. This issue deserves further research to clarify.
In summary, our results establish the isoform-specific roles of ETHRA and ETHRB on regulation of ecdysis in L. decemlineata. Accordingly, we propose a model summarizing these findings (Figure 8F). During larva-pupa transition, ETH regulates ecdysis behavior series and cuticle tanning by triggering ETHRA and ETHRB to coordinately activate kinin, CCAP, and bursicon neurons. Moreover, the ETH/ETHRA way specifically controls kinin signal for fluid balance by targeting Malpighian tubules; whereas ETH/ETHRB inhibitory signal is critical for suppression of the premature expression and release of burs and pburs in bursicon neurons.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
G-QL, LJ, C-HS, and Q-YX conceived the study. LJ, C-HS, Q-YX, and G-QL participated in the design of the experiments and the interpretation of the results. C-HS, Q-YX, K-YF, W-CG, and LJ performed the experiments. G-QL, C-HS, and LJ wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key R&D Program of China (2017YFD0200900) and China Agriculture Research System (CARS-09-P22).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.593962/full#supplementary-material
Arakane, Y., Li, B., Muthukrishnan, S., Beeman, R. W., Kramer, K. J., and Park, Y. (2008). Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech. Dev. 125, 984–995. doi: 10.1016/j.mod.2008.09.002
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cho, K. H., Daubnerová, I., Park, Y., Žitnan, D., and Adams, M. E. (2014). Secretory competence in a gateway endocrine cell conferred by the nuclear receptor βFTZ-F1 enables stage-specific ecdysone responses throughout development in Drosophila. Dev. Biol. 385, 253–262. doi: 10.1016/j.ydbio.2013.11.003
Clark, A. C. (2004). Neuroendocrine control of larval ecdysis behavior in Drosophila: complex regulation by partially redundant neuropeptides. J. Neurosci. 24, 4283–4292. doi: 10.1523/JNEUROSCI.4938-03.2004
Dai, L., and Adams, M. E. (2009). Ecdysis triggering hormone signaling in the yellow fever mosquito Aedes aegypti. Gen. Comp. Endocrinol. 162, 43–51. doi: 10.1016/j.ygcen.2009.03.004
de Oliveira, A. L., Calcino, A., and Wanninger, A. (2019). Ancient origins of arthropod moulting pathway components. eLife 8:e46113. doi: 10.7554/eLife.46113
Deng, P., Xu, Q. -Y., Fu, K. -Y., Guo, W. -C., and Li, G. -Q. (2018). RNA interference against the putative insulin receptor substrate gene chico affects metamorphosis in Leptinotarsa decemlineata. Insect Biochem. Mol. Biol. 103, 1–11. doi: 10.1016/j.ibmb.2018.10.001
Diao, F., Elliott, A. D., Diao, F., Shah, S., and White, B. H. (2017). Neuromodulatory connectivity defines the structure of a behavioral neural network. eLife 6:e29797. doi: 10.7554/eLife.29797
Diao, F., Mena, W., Shi, J., Park, D., Diao, F., Taghert, P., et al. (2016). The splice isoforms of the Drosophila ecdysis triggering hormone receptor have developmentally distinct roles. Genetics 202, 175–189. doi: 10.1534/genetics.115.182121
Ewer, J., Gammie, S. C., and Truman, J. W. (1997). Control of insect ecdysis by a positive-feedback endocrine system: roles of eclosion hormone and ecdysis triggering hormone. J. Exp. Biol. 200, 869–881.
Iversen, A., Cazzamali, G., Williamson, M., Hauser, F., and Grimmelikhuijzen, C. J. P. (2002). Molecular identification of the first insect ecdysis triggering hormone receptors. Biol. Biophys. Res. Commun. 299, 924–931. doi: 10.1016/S0006-291X(02)02798-5
Jiang, H., Wei, Z., Nachman, R. J., Adams, M. E., and Park, Y. (2014). Functional phylogenetics reveals contributions of pleiotropic peptide action to ligand-receptor coevolution. Sci. Rep. 4:6800. doi: 10.1038/srep06800
Kim, D. H., Han, M. R., Lee, G., Lee, S. S., Kim, Y. J., and Adams, M. E. (2015). Rescheduling behavioral subunits of a fixed action pattern by genetic manipulation of peptidergic signaling. PLoS Genet. 11:e1005513. doi: 10.1371/journal.pgen.1005513
Kim, D. H., Kim, Y. J., and Adams, M. E. (2018). Endocrine regulation of airway clearance in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 115, 1535–1540. doi: 10.1073/pnas.1717257115
Kim, Y. J., Zitnan, D., Cho, K. H., Schooley, D. A., Mizoguchi, A., and Adams, M. E. (2006b). Central peptidergic ensembles associated with organization of an innate behavior. Proc. Natl. Acad. Sci. U. S. A. 103, 14211–14216. doi: 10.1073/pnas.0603459103
Kim, Y. J., Zitnan, D., Galizia, C. G., Cho, K. H., and Adams, M. E. (2006a). A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Curr. Biol. 16, 1395–1407. doi: 10.1016/j.cub.2006.06.027
Kingan, T. G., Gray, W., Žitnan, D., and Adams, M. E. (1997). Regulation of ecdysis-triggering hormone release by eclosion hormone. J. Exp. Biol. 200, 3245–3256.
Kong, Y., Liu, X. -P., Wan, P. -J., Shi, X. -Q., Guo, W. -C., and Li, G. -Q. (2014). The P450 enzyme shade mediates the hydroxylation of ecdysone to 20-hydroxyecdysone in the colorado potato beetle, Leptinotarsa decemlineata. Insect Mol. Biol. 23, 632–643. doi: 10.1111/imb.12115
Kronforst, M. R., Barsh, G. S., Kopp, A., Mallet, J., Monteiro, A., Mullen, S. P., et al. (2012). Unraveling the thread of nature’s tapestry: the genetics of diversity and convergence in animal pigmentation. Pigment Cell Melanoma Res. 25, 411–433. doi: 10.1111/j.1755-148X.2012.01014.x
Krüger, E., Mena, W., Lahr, E. C., Johnson, E. C., and Ewer, J. (2015). Genetic analysis of eclosion hormone action during Drosophila larval ecdysis. Development 142, 4279–4287. doi: 10.1242/dev.126995
Lenaerts, C., Cools, D., Verdonck, R., Verbakel, L., Vanden Broeck, J., and Marchal, E. (2017). The ecdysis triggering hormone system is essential for successful moulting of a major hemimetabolous pest insect, Schistocerca gregaria. Sci. Rep. 7:46502. doi: 10.1038/srep46502
Liang, G. X., Fu, H. T., Qiao, H., Sun, S. M., Zhang, W. Y., Jin, S. B., et al. (2017). Molecular cloning and characterization of a putative ecdysis-triggering hormone receptor (ETHR) gene from Macrobrachium nipponense. J. World Aquacult. Soc. 49, 1081–1094. doi: 10.1111/jwas.12451
Liu, X. -P., Fu, K. -Y., Lü, F. -G., Meng, Q. -W., Guo, W. -C., and Li, G. -Q. (2014). Involvement of FTZ-F1 in the regulation of pupation in Leptinotarsa decemlineata (say). Insect Biochem. Mol. Biol. 55, 51–60. doi: 10.1016/j.ibmb.2014.10.008
Massey, J. H., and Wittkopp, P. J. (2016). The genetic basis of pigmentation differences within and between Drosophila species. Curr. Top. Dev. Biol. 119, 27–61. doi: 10.1016/bs.ctdb.2016.03.004
Meiselman, M. R., Kingan, T. G., and Adams, M. E. (2018). Stress-induced reproductive arrest in Drosophila occurs through ETH deficiency-mediated suppression of oogenesis and ovulation. BMC Biol. 16:18. doi: 10.1186/s12915-018-0484-9
Meiselman, M., Lee, S. S., Tran, R. T., Dai, H., Ding, Y., Rivera-Perez, C., et al. (2017). Endocrine network essential for reproductive success in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 114, E3849–E3858. doi: 10.1073/pnas.1620760114
Mena, W., Diegelmann, S., Wegener, C., and Ewer, J. (2016). Stereotyped responses of Drosophila peptidergic neuronal ensemble depend on downstream neuromodulators. eLife 5:e19686. doi: 10.7554/eLife.19686
Meng, Q. -W., Xu, Q. -Y., Deng, P., Fu, K. -Y., Guo, W. -C., and Li, G. -Q. (2018). Involvement of methoprene-tolerant (Met) in the determination of the final body size in Leptinotarsa decemlineata (Say) larvae. Insect Biochem. Mol. Biol. 97, 1–9. doi: 10.1016/j.ibmb.2018.04.003
Meng, Q. -W., Xu, Q. -Y., Zhu, T. -T., Jin, L., Fu, K. -Y., Guo, W. -C., et al. (2019). Hormonal signaling cascades required for phototaxis switch in wandering Leptinotarsa decemlineata larvae. PLoS Genet. 15:e1007423. doi: 10.1371/journal.pgen.1007423
Park, Y., Kim, Y. J., Dupriez, V., and Adams, M. E. (2003). Two subtypes of ecdysistriggering hormone receptor in Drosophila melanogaster. J. Biol. Chem. 278, 17710–17715. doi: 10.1074/jbc.M301119200
Park, Y., Valery, F., Gill, S. S., and Adams, M. E. (2002). Deletion of the ecdysis triggering hormone gene leads to lethal ecdysis deficiency. Development 129, 493–503.
Protas, M. E., and Patel, N. H. (2008). Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24, 425–446. doi: 10.1146/annurev.cellbio.24.110707.175302
Radovic, A., Wittkopp, P. J., Long, A. D., and Drapeau, M. D. (2002). Immunohistochemical colocalization of yellow and male-specific fruitless in Drosophila melanogaster neuroblasts. Biochem. Biophys. Res. Commun. 293, 1262–1264. doi: 10.1016/S0006-291X(02)00366-2
Roller, L., Zitnanová, I., Dai, L., Simo, L., Park, Y., Satake, H., et al. (2010). Ecdysis triggering hormone signaling in arthropods. Peptides 31, 429–441. doi: 10.1016/j.peptides.2009.11.022
Schoville, S. D., Chen, Y. H., Andersson, M. N., Benoit, J. B., Bhandari, A., Bowsher, J. H., et al. (2018). A model species for agricultural pest genomics: the genome of the colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 8:1931. doi: 10.1038/s41598-018-20154-1
Shi, X. -Q., Guo, W. -C., Wan, P. -J., Zhou, L. -T., Ren, X. -L., Ahmat, T., et al. (2013). Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (say). BMC. Res. Notes 6:93. doi: 10.1186/1756-0500-6-93
Shi, Y., Jiang, H. B., Gui, S. H., Liu, X. Q., Pei, Y. X., Xu, L., et al. (2017). Ecdysis triggering hormone signaling (ETH/ETHR-A) is required for the larva-larva ecdysis in Bactrocera dorsalis (Diptera: Tephritidae). Front. Physiol. 8:587. doi: 10.3389/fphys.2017.00587
Shi, Y., Liu, T. Y., Jiang, H. B., Liu, X. Q., Dou, W., Park, Y., et al. (2019). The ecdysis triggering hormone system, via ETH/ETHR-B, is essential for successful reproduction of a major pest insect, Bactrocera dorsalis (Hendel). Front. Physiol. 10:151. doi: 10.3389/fphys.2019.00151
Takahashi, A. (2013). Pigmentation and behavior: potential association through pleiotropic genes in Drosophila. Genes Genet. Syst. 88, 165–174. doi: 10.1266/ggs.88.165
Terhzaz, S., O’Connell, F. C., Pollock, V. P., Kean, L., Davies, S. A., Veenstra, J. A., et al. (1999). Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J. Exp. Biol. 202, 3667–3676.
Truman, J. W. (2005). Hormonal control of insect ecdysis: endocrine cascades for coordinating behavior with physiology. Vitam. Horm. 73, 1–30. doi: 10.1016/S0083-6729(05)73001-6
Wittkopp, P. J., and Beldade, P. (2009). Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin. Cell Dev. Biol. 20, 65–71. doi: 10.1016/j.semcdb.2008.10.002
Wittkopp, P. J., Stewart, E. E., Arnold, L. L., Neidert, A. H., Haerum, B. K., Thompson, E. M., et al. (2009). Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 326, 540–544. doi: 10.1126/science.1176980
Xu, Q. -Y., Deng, P., Li, A., Zhang, Q., Mu, L. -L., Fu, K. -Y., et al. (2019). Functional characterization of ultraspiracle in Leptinotarsa decemlineata using RNA interference assay. Insect Mol. Biol. 28, 676–688. doi: 10.1111/imb.12580
Xu, Q. -Y., Deng, P., Zhang, Q., Li, A., Fu, K. -Y., Guo, W. -C., et al. (2020). Ecdysone receptor isoforms play distinct roles in larval-pupal-adult transition in Leptinotarsa decemlineata. Insect Sci. 27, 487–499. doi: 10.1111/1744-7917.12662
Xu, Q. -Y., Meng, Q. -W., Deng, P., Guo, W. -C., and Li, G. -Q. (2018). Leptinotarsa hormone receptor 4 (HR4) tunes ecdysteroidogenesis and mediates 20-hydroxyecdysone signaling during larval-pupal metamorphosis. Insect Biochem. Mol. Biol. 94, 50–60. doi: 10.1016/j.ibmb.2017.09.012
Žitňan, D., Kim, Y. J., Žitňanová, I., Roller, L., and Adams, M. E. (2007). Complex steroid-peptide-receptor cascade controls insect ecdysis. Gen. Comp. Endocrinol. 153, 88–96. doi: 10.1016/j.ygcen.2007.04.002
Keywords: Leptinotarsa decemlineata, ecdysis triggering hormone receptor, isoform specificity, ecdysis behavior, larva-pupa transition
Citation: Shen C-H, Xu Q-Y, Fu K-Y, Guo W-C, Jin L and Li G-Q Two Splice Isoforms of Leptinotarsa Ecdysis Triggering Hormone Receptor Have Distinct Roles in Larva-Pupa Transition. Front. Physiol. 11:593962. doi: 10.3389/fphys.2020.593962
Received: 12 August 2020; Accepted: 11 November 2020;
Published: 01 December 2020.
Edited by:
Guy Smagghe, Ghent University, BelgiumReviewed by:
Shi Yan, Qingdao Agricultural University, ChinaCopyright © 2020 Shen, Xu, Fu, Guo, Jin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Jin, amlubGluQG5qYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.