- 1School of Dentistry, University of Utah Health, Salt Lake City, UT, United States
- 2Department of Biomedical Engineering, College of Engineering, The University of Utah, Salt Lake City, UT, United States
- 3Dental School, Faculty of Health and Medical Sciences, The University of Western Australia, Perth, WA, Australia
- 4College of Pharmacy, University of Utah Health, Salt Lake City, UT, United States
- 5Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
- 6Adams School of Dentistry, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 7Department of Neurobiology and Anatomy, Pathology, and Surgery, The University of Utah, Salt Lake City, UT, United States
In these studies, we explored for the first time the molecular relationship between the paired-domain-containing transcription factor, Pax9, and the ectodysplasin (Eda) signaling pathway during mouse incisor formation. Mice that were deficient in both Pax9 and Eda were generated, and the status of dentition analyzed in all progeny using gross evaluation and histomorphometric means. When compared to wildtype controls, Pax9+/–Eda–/– mice lack mandibular incisors. Interestingly, Fgf and Shh signaling are down-regulated while Bmp4 and Lef1 appear unaffected. These findings suggest that Pax9-dependent signaling involves the Eda pathway and that this genetic relationship is important for mandibular incisor development. Studies of records of humans affected by mutations in PAX9 lead to the congenital absence of posterior dentition but interestingly involve agenesis of mandibular central incisors. The latter phenotype is exhibited by individuals with EDA or EDAR mutations. Thus, it is likely that PAX9, in addition to playing a role in the formation of more complex dentition, is also involved with EDA signaling in the initiation of odontogenesis within the incisal domain.
Introduction
The formation of mammalian dentition is a remarkable developmental process and a valuable model for studying epithelial–mesenchymal signaling interactions that control patterning morphogenesis. Much of our understanding about the patterning of dentition comes from mouse studies. The use of transgenesis, gene targeting, expression analyses, functional tooth recombination, as well as bead implantation assays have advanced our knowledge about the patterning of the murine dentition. What has emerged is the realization that tooth development involves a complex series of genetic interactions between growth factors, transcription factors, signal receptors, and diffusible morphogens that interact within five critical pathways, namely, bone morphogenetic protein (Bmp), wingless-integrated site (Wnt), fibroblast growth factor (Fgf), sonic hedgehog (Shh), and ectodysplasin (Eda) (Lan et al., 2014; Balic and Thesleff, 2015; Huang et al., 2019).
That the patterning of dentition is under strict genetic control is best underscored by the condition of human tooth agenesis, a common inherited disorder that affects over 20% of the population. Classified as genetically and phenotypically heterogeneous, tooth agenesis most commonly affects third molars, mandibular second premolars, maxillary lateral incisors, and maxillary second premolars (Kapadia et al., 2007; Nieminen, 2009; Ye and Attaie, 2016; Williams and Letra, 2018). These commonly missing teeth represent the most distal members of each tooth family and fail to develop due to a disruption in normal signaling. This suggests that distinct distal-proximal morphogenetic gradients are involved in guiding the patterning of human dentition.
The appearance of tooth placodes marks the onset of odontogenesis and the formation of incisiform and molariform fields that develop when the inductive potential from dental epithelium is transferred to underlying mesenchyme (Jarvinen et al., 2018; Balic, 2019). This was proven in elegant recombination experiments where bud-staged molar mesenchyme was shown to fully transform non-dental epithelium to form teeth (Mina and Kollar, 1987; Lumsden, 1988). Systems biology approaches that integrated data from genome-wide expression profiling, bioinformatics, and in vivo genetic models show that canonical Wnt signals are the primary drivers of tooth signaling interactions (O’Connell et al., 2012). Lef1, a nuclear mediator of Wnt signaling, associates with β-catenin and activates Wnt-responsive genes. The molecular relationship between Wnt and Eda signaling pathways is well established in tooth development as Eda is down-regulated in Lef1–/– tooth organs that are arrested at the bud stage and Wnt6 can induce Eda expression in a Lef1-dependent manner (Laurikkala et al., 2001). Multiple lines of evidence further converge to support the hypothesis that the interactions between signaling pathways, rather than the intrinsic functions of transcription factors alone, dictate how the patterning of dentition is orchestrated.
Several studies point to the importance of the homeodomain-containing transcription factor, Pax9, as a key mediator of the odontogenic potential in the mesenchyme (Neubuser et al., 1997; Peters et al., 1998; Jernvall and Thesleff, 2000; Kapadia et al., 2006; Ogawa et al., 2006; Chen et al., 2009). Although Pax9-dependent signaling in tooth mesenchyme involves a partnership with Msx1 and the up-regulation of Bmp4 expression, its relationship with other key pathways is not well understood. These studies explored for the first time the relationship of Pax9 with the Eda signaling pathway during the formation of mouse dentition. Through the use of mouse genetics, we demonstrate that Pax9-dependent signaling is functionally integrated with Eda signaling during mandibular incisor development. Disruptions in this molecular relationship lead to downregulation of other signaling molecules. We also report on the findings of our long-term human genetic studies that show that mutations in PAX9, while dominantly affecting posterior dentition, often involve mandibular central incisors. The latter, we observe is a common occurrence in individuals with mutations in EDA or EDAR. Taken together, our results indicate that Pax9-mediated signaling involves the Eda pathway, impacting the initiation of odontogenesis within the mandibular incisiform field.
Materials and Methods
Mouse Strains
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Utah (Protocol #19-12012). Pax9+/– mice were provided by Dr. Rulang Jiang (Cincinnati Children’s Hospital), and Eda+/– mice (# 000314) were purchased from Jackson laboratory. Mouse colonies were maintained in the C57BL/6 background, and 2–8-month-old females were used for intercross mating.
Histology Analysis
For the whole-mount overview of mandibles and maxillae, 2-month-old mouse heads were removed using fine forceps under a stereomicroscope. Mandibles were carefully dissected after removal of the tongue, and images were taken through a stereomicroscope (Zeiss Stemi 508).
For H&E staining, whole heads were fixed in 4% paraformaldehyde (PFA) in PBS overnight and processed through serial gradients of ethanol and xylene for paraffin embedding. 7 μm-thick sagittal sections were stained with H&E and evaluated under a digital microscope (EVOS).
In situ Hybridizations
Sagittal paraffin sections of E13.5 and E14 embryos using digoxigenin-labeled RNA probes (1 μg/ml) as described previously (Jia et al., 2017). Embryo heads were fixed in 4% PFA in PBS overnight then processed through serial ethanol and xylene for paraffin embedding. 7 μm-thick sagittal sections were hybridized with digoxigenin-labeled antisense RNA probes (1 μg/ml) to Bmp4, Fgf3, Lef1, and Shh as described previously. An anti-digoxigenin-AP antibody (11093274910, ROCHE, 1:1000) was used to detect the labeled probe. Comparable images were taken with a digital microscope (EVOS). At least three biological replicates were used to establish the reproducibility of results.
Tooth Germ Dissections and Real-Time Reverse Transcription (RT)-PCR Analyses
The embryos heads were harvested at E13.5 in cold PBS. After the lower jaw and brain were removed under a dissecting microscope, the incisors were carefully dissected using fine forceps and stored individually at −80°C for total RNA extraction. After genotyping, 3 pairs of incisors were pooled and total RNA was extracted using the RNeasy Micro Kit (Qiagen). First-strand cDNA was synthesized using the SuperScript First-Strand Synthesis System IV (Thermo Fisher Scientific). Quantitative reverse transcription (RT)-PCRs were performed using the SYBR GreenER qPCR Supermix (Thermo Fisher Scientific). A list of gene-specific primers is provided in Supplementary Table S1. For each sample, the relative levels of target mRNAs were normalized to Gapdh using the standard curve method (Zhou et al., 2013). Three sets of biological replicates were analyzed for each gene.
Clinical Findings From Human Genetic Studies
Patients were identified and evaluated after approval was obtained from the Committee for the Protection of Human Subjects, University of Texas Health Science Center at Houston. Consent to participate (including a release of dental records) was obtained from a parental guardian, in the case of minors. Patient records are secured at the University of Utah and analyzed for these studies under HIPAA policies and without any personal identifiers. Medical and dental records of patients diagnosed with mutations in PAX9 and EDAR were studied along with radiographs and photographs. Patterns of tooth agenesis were noted, and records were selected to illustrate mandibular incisor agenesis in two individuals with mutations in PAX9 and EDAR, respectively.
Results
Pax9+/–Eda–/– Compound Mutant Mice Exhibited Fewer Molars and Missing Mandibular Incisors
Pax9+/– and Eda+/– mice exhibited normal dentition with no disruptions in the size, shape, and number of teeth. To begin to assess whether Pax9 and Eda genetically interact during tooth development, Pax9+/–Eda–/– mice were generated through a series of breeding. As shown in Figures 1A,D, Pax9+/–Eda+/+ mice had a full complement of six molars (3 + 3) and two incisors in each mandible and maxilla, respectively. The 2-month-old Pax9+/+Eda–/– mice (Figures 1B,E) had smaller molars with shallower cusps (as reported earlier) with six molars (3 + 3) and two incisors present in each mandible and maxilla analyzed. The compound mutant Pax9+/–Eda–/– mice exhibited missing third molars in both the mandible and maxilla, as well as missing mandibular incisors (Figures 1C,F,G). These data suggest that Pax9 and Eda-dependent signaling pathway genes share a genetic interaction in controlling the molar number and the formation of mandibular incisors.
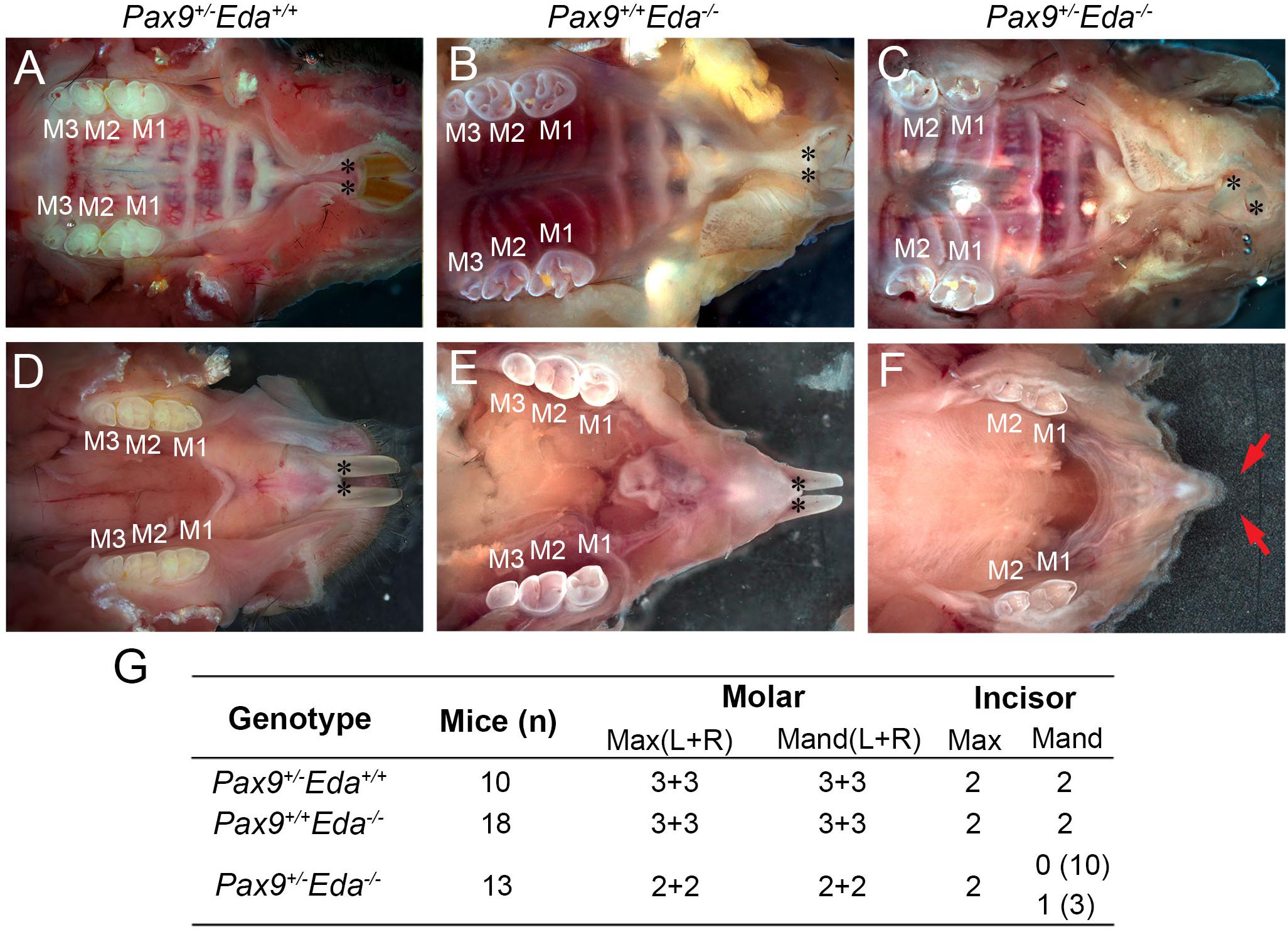
Figure 1. Missing teeth in Pax9+/–Eda–/– compound mutant mice. (A–C) Whole-mount view of maxillary teeth in 2-month-old mice. (A) Pax9+/–Eda+/+ mice showed six molars (3 + 3) and two incisors. (B) Pax9+/+Eda–/– mice showed six molars (3 + 3) with fewer cusps and two incisors. (C) Pax9+/–Eda–/– mice showed four molars (2 + 2) and two incisors. (D–F) Whole-mount view of mandibular teeth in 2-month-old mice. (D) Pax9+/–Eda+/+ mice showed two incisors and six molars (3 + 3). (E) Pax9+/+Eda–/– mice showed two incisors and six molars (3 + 3) with smaller size and fewer cusps. (F) Pax9+/–Eda–/– mice showed missing incisors and four molars (2 + 2) with smaller size and fewer cusps. (G) Summary of teeth phenotype in Pax9+/–Eda–/– compound mutants. Red arrows pointed to the space of missing incisors. M1, M2, and M3 are first, second, and third molar, respectively. Black * presents incisors. Max, maxillary; Mand, mandible.
Mandibular Incisor Development in Pax9+/–Eda–/– Compound Mutants
We examined embryos from E13.5 to P0, and found that at E13.5, the mandibular incisors formed bud-shape tooth germs in Pax9+/–Eda+/+, Pax9+/+Eda–/–, and Pax9+/–Eda–/– compound mutant mice (Figures 2A,E,I). At E14.5, the cervical loops of Pax9+/–Eda+/+ and Pax9+/+Eda–/– mandibular incisors were well formed, whereas the cervical loops of Pax9+/–Eda–/– compound mutant mandibular incisors appeared under-developed (Figures 2B,F,J). At P0, mandibular incisors showed asymmetric cervical loops at E17.5 and well-differentiated tooth organ layers in Pax9+/–Eda+/+ and Pax9+/+Eda–/– mandible (Figures 2C,D,G,H). In contrast, the mandibular incisors in Pax9+/–Eda–/– compound mutant exhibited residue of retarded tooth germs (Figures 2K,L).
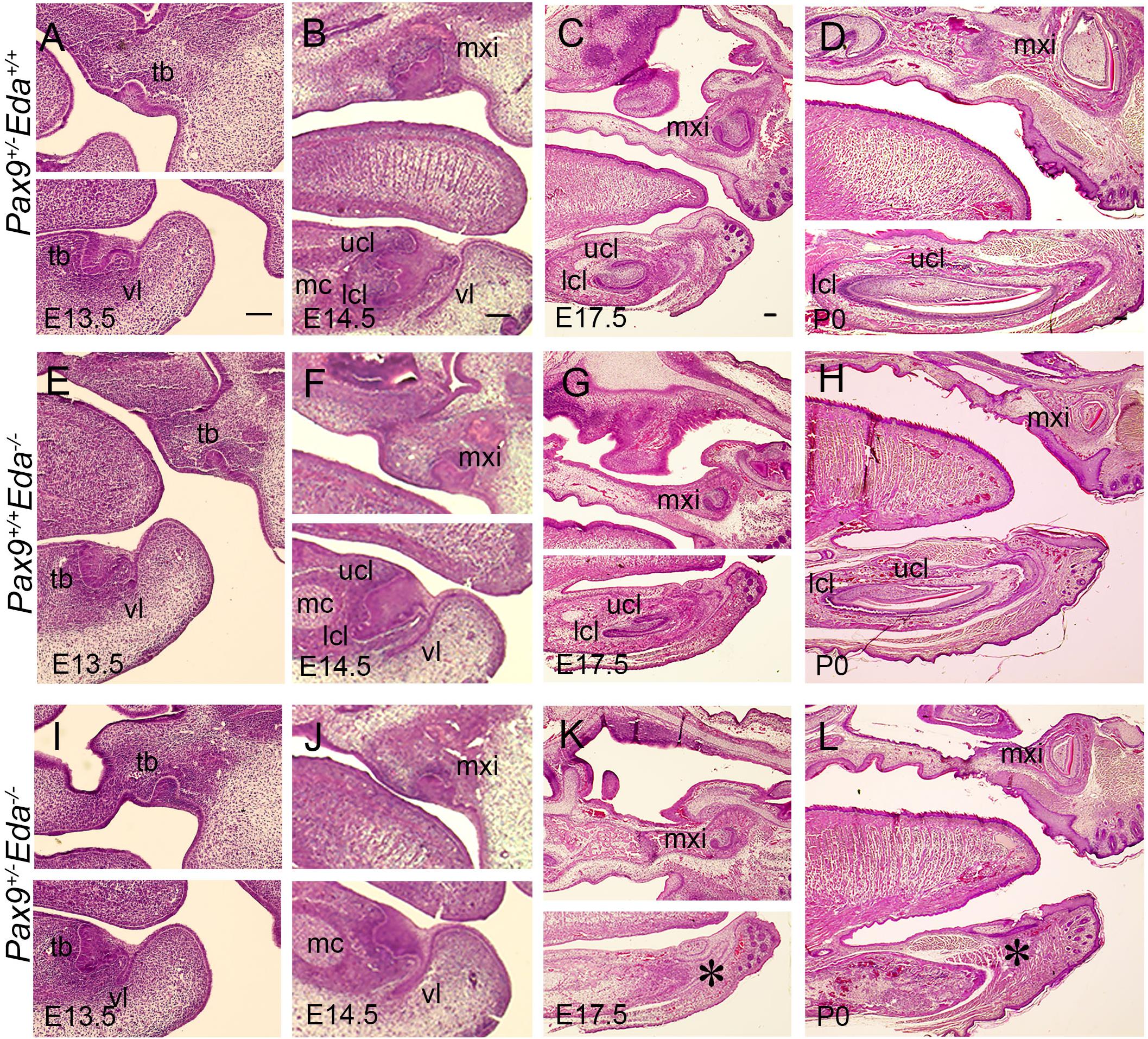
Figure 2. Incisor developmental defects in the Pax9+/–Eda–/– compound mutant embryos. Hematoxylin and eosin-stained (HE) sagittal sections through the developing incisor tooth germs. The Pax9+/–Eda+/+, Pax9+/+Eda–/–, and Pax9+/–Eda–/– compound mutants at E13.5 (A,E,I), E14.5 (B,F,J), E17.5 (C,G,K), and P0 (D,H,L), respectively, are shown. Scale bar presented 100 μm, * indicates the region of missing incisor. lcl, lower cervical loop; mc, Meckel’s cartilage; mxi, maxillary incisor; tb, tooth bud; ucl, upper cervical loop, vl, vestibular lamina.
The Activity of Fgf and Shh Signaling Was Reduced in Pax9+/–Eda–/– Mandibular Incisors
To investigate the potential interactions between Pax9 and the Eda/r signaling pathway, we analyzed the expression patterns of Bmp4, Fgf3, Lef1, and Shh in E13.5 and E14.0 Eda+/−, Eda–/–, and Pax9+/–Eda–/– incisor organs. Results of in situ hybridization indicate that the patterns of Bmp4 and Lef1 expression seen in both Eda+/– and Eda–/– incisor organs resembled that visible in Pax9+/–Eda–/– embryos (Figures 3A–L). The expression level of Bmp4 and Lef1 was not significantly reduced in the Pax9+/–Eda–/– mandibular incisors confirmed by quantitative RT-PCR (Figure 3M). In contrast, the expression pattern of Fgf3 and Shh appeared down-regulated in Pax9+/–Eda–/– incisors in comparison to that evident in both Eda+/– and Eda–/– samples (Figures 3N–Y). Quantitative RT-PCR revealed that the expression of Fgf3 in the Pax9+/–Eda–/– mandibular incisor was significantly reduced in comparison to that in the Eda+/– samples; and the expression of Shh in the Pax9+/–Eda–/– mandibular incisor was significantly reduced compared with that in both Eda+/– and Eda–/– samples (Figure 3Z).
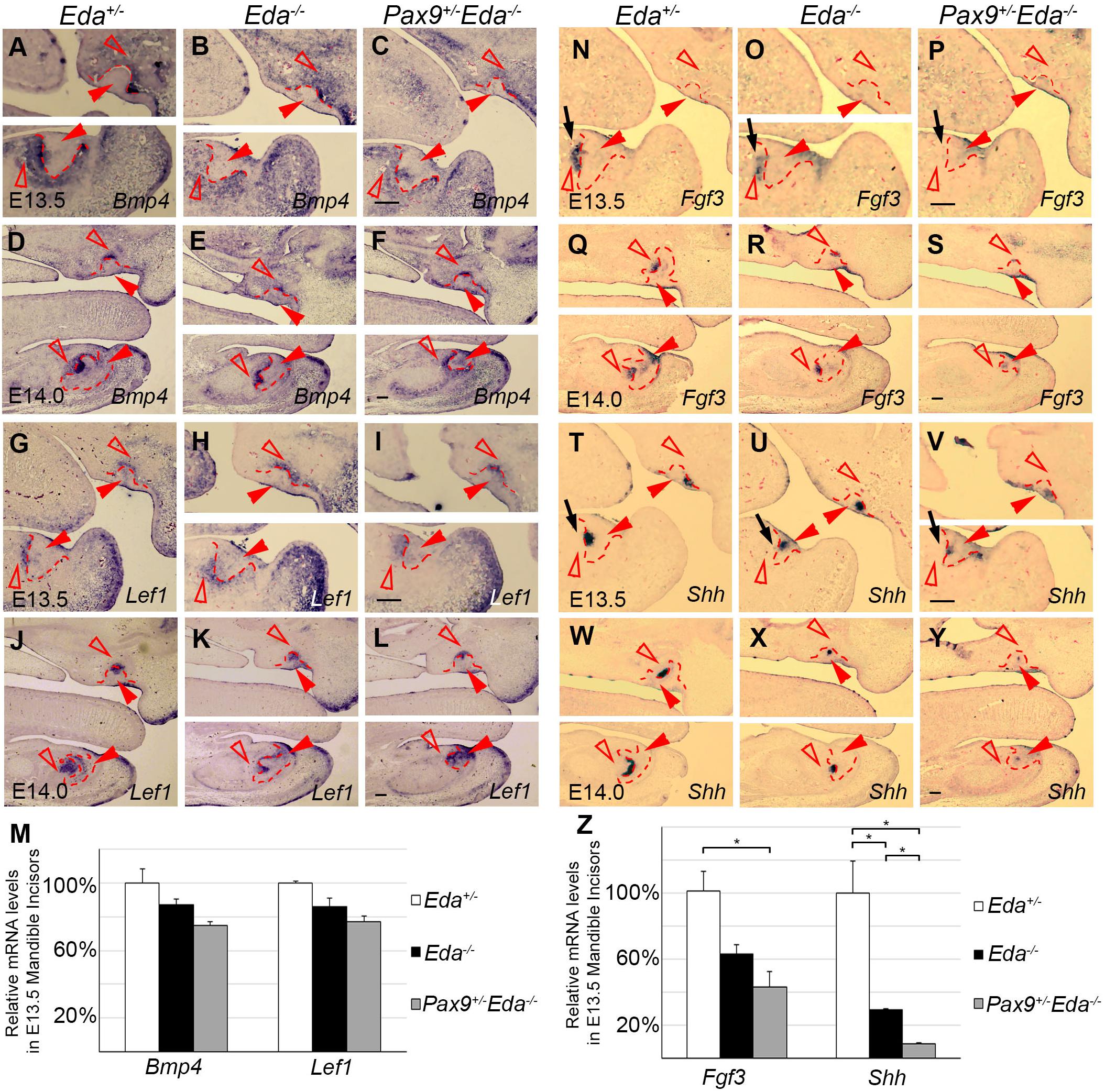
Figure 3. Comparison of incisor molecular marker expression in E13.5 and E14.0 tooth germs. The mRNA expression of Bmp4 and Lef1 were shown in Eda+/–, Eda–/–, and Pax9+/–Eda–/– mutants by in situ hybridization at E13.5 (A–C,G–H), as well as at E14.0 (D–F,G–L). The expression of Fgf3 and Shh was shown in Eda+/–, Eda–/–, and Pax9+/–Eda–/– mutants by in situ hybridization at E13.5 (N–P,T–V), as well as at E14.0 (Q–S,W–Y). Scale bar presented 100 μm, black arrows indicate mandibular incisor tooth germ. Red solid arrowheads indicate dental epithelia and red open arrowheads indicate dental mesenchyme. Red dashed lines indicate the boundary of incisor tooth germ mesenchyme and epithelia. (M,Z) The relative mRNA levels of Bmp4, Lef1, Fgf3, and Shh were analyzed by quantitative RT-PCR using the microdissected E13.5 mandibular incisors (n = 3). Error bars indicate SEM, *P < 0.01.
Patterns of Missing Teeth in Individuals With PAX9 and EDAR Mutations
Mutations in PAX9 result in a pattern of tooth agenesis that dominantly involves permanent maxillary and mandibular first, second, and third molars along with second premolars. However, agenesis of mandibular central incisors are often associated with missing molars and premolars, as shown in Figure 4B, and as previously reported by our group (Goldenberg et al., 2000; Stockton et al., 2000; Frazier-Bowers et al., 2002a,b). A sibling with an unaffected PAX9 gene shows a normal complement of permanent teeth (Figure 4A). For individuals with mutations in EDA or EDAR, tooth agenesis is more severe and mixed, typically including maxillary and central incisors while frequently involving mandibular central incisors (Figures 4C,D).
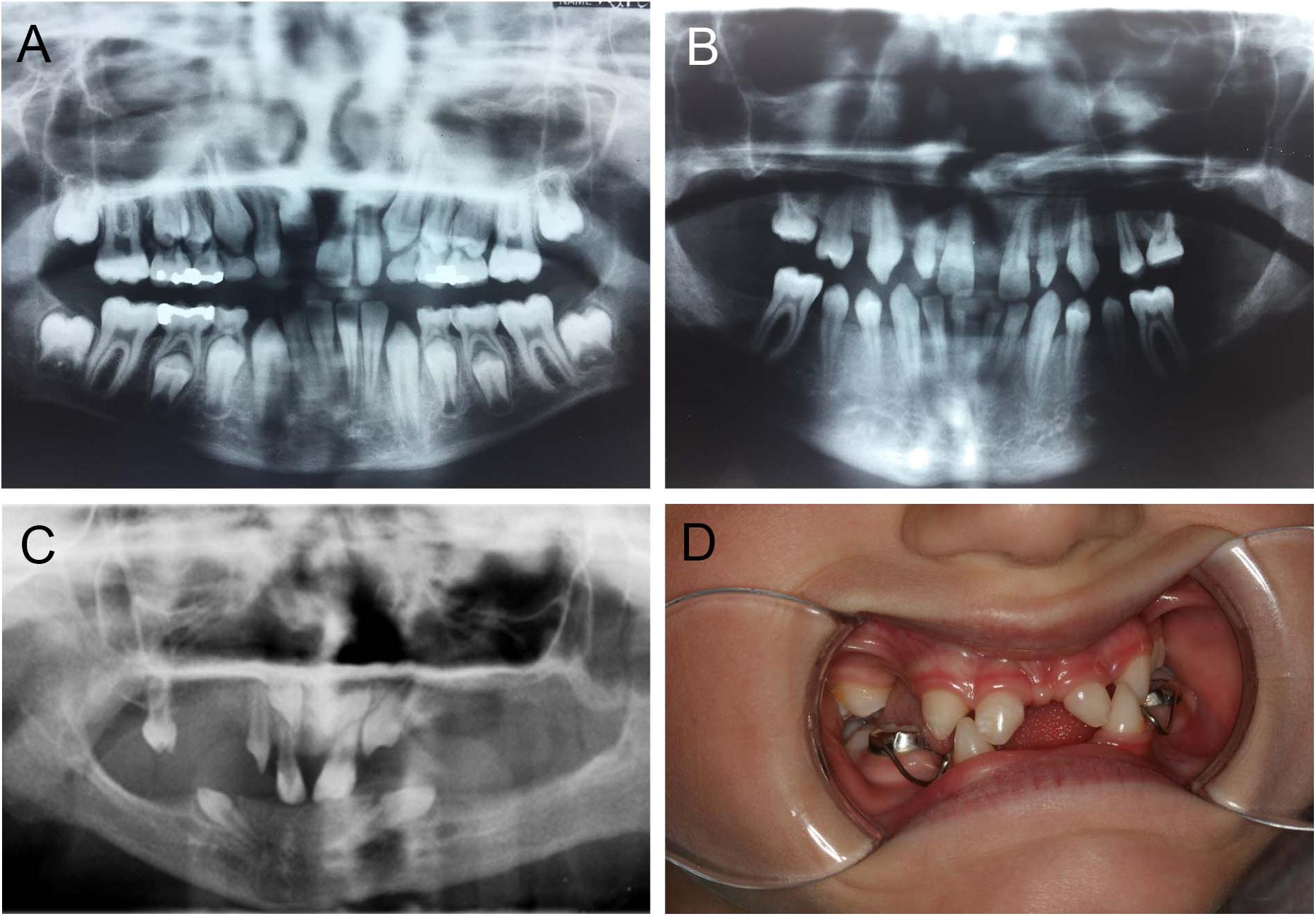
Figure 4. Abnormal tooth appearance, including agenesis of mandibular central incisors, premolars, and molars in a PAX9 mutant patient (B) compared to an unaffected sibling with normal permanent dentition (A); 5-year-old patient with a confirmed mutation of EDA gene (XLHED), no permanent teeth in the mandible and only primary canines (never erupted), conical incisors, canines, and one molar noted in the maxilla (C); female patient with autosomal recessive (AR) HED, missing mandibular incisors, and first primary molars, as well as maxillary lateral incisors and first primary molars (D).
Discussion
The patterning of dentition is a complex and unique developmental process involving multiple genes that control the precise shape, size, number, and position of teeth. While the past decades have advanced our understanding of the transcription factors that are involved, relatively little is known about their interactions with downstream effectors within the Bmp, Fgf, Wnt, Eda, and Shh pathways. Here, we sought to understand the relationship between Pax9, a key transcription factor in dental mesenchyme, and the Eda signaling pathway during the development of the dentition. Previous studies indicate that Pax9 regulates both Wnt and Bmp signaling pathways and Eda signaling has been reported linked with Wnt signaling (Peters et al., 1998; Laurikkala et al., 2001; Zhang et al., 2009; O’Connell et al., 2012; Voutilainen et al., 2015). Our studies show, for the first time, that Pax9-dependent signaling is functionally integrated with Eda signaling during odontogenesis. A mouse genetic model deficient in both Pax9 and Eda shows a consistent lack of maxillary and mandibular third molar development. Interestingly, mandibular incisor organs are arrested, a phenotypic change that suggests regionally specific interactions between Pax9-dependent signaling and the Eda pathway. In order to fully elucidate the nature of Pax9’s relationship with Eda signaling, further experiments such as chromatin immunoprecipitation (ChIP) or electrophoretic mobility shift assay (EMSA) are needed. Our mouse data align with observations from our human genetic analyses of individuals with mutations in PAX9 and EDA/R genes that show agenesis of mandibular central incisors. Taken together, these results underscore the need for further studies that will elucidate the relationship of Pax9 with other signaling molecules that direct key epithelial–mesenchymal interactions in tooth development.
As reported earlier, Eda–/– mice exhibit mild defects in dentition that include fewer and shallower cusps in molars, hypoplasia, and agenesis of maxillary and mandibular third molars in 20% of animals studied. These studies also showed that decreased levels of Eda signaling did not affect incisor development (Pispa et al., 1999; Parveen et al., 2019). Without exception, all of the 13 Pax9+/–Eda–/– mice studied showed maxillary and mandibular third molar agenesis with an overall reduction in size and cuspal morphology of residual first and second molars. Pax9+/– mice have a full complement of teeth while Pax9–/– mice consistently lack all molars (Kist et al., 2005; Mitsui et al., 2014). Furthermore, permanent molars are the dominant tooth group affected in individuals with mutations in PAX9 (Goldenberg et al., 2000; Stockton et al., 2000; Frazier-Bowers et al., 2002a). Therefore, it is likely that the combinatorial reduction of Pax9 and Eda signaling in Pax9+/–Eda–/– mice has an additive effect on maxillary and mandibular molar development.
Pax9+/–Eda–/– mice also exhibited a mandibular incisor phenotype as 10 of 13 embryos studied lacked lower incisors while in three of 13 embryos only a single mandibular incisor was present. In mouse genetic models disruptions in Pitx2, BCL11B and Wnt, Fgf, Bmp, and Wnt signaling pathway genes result in incisor organ agenesis or hypoplasia (Lin et al., 1999; Lu et al., 1999; Millar et al., 2003; Golonzhka et al., 2009; Yang et al., 2013; Yu et al., 2020). Furthermore, supernumerary incisors arise in mice lacking Spry2/4, Sostdc1, and Lrp4 (Ohazama et al., 2008; Munne et al., 2009; Charles et al., 2011). In these models, both maxillary and mandibular incisors were affected as compared to mice where the up-regulation of NF-κB activity and the reduction of Fam20B selectively affect mandibular vs. maxillary incisors, respectively (Wang et al., 2013; Tian et al., 2015). Our quantitative RT-PCR and in situ hybridization data revealed that the expression of Fgf3 in the Pax9+/–Eda–/– mandibular incisor was significantly reduced in comparison to that in the Eda+/– samples. Whereas, in comparison to the Eda+/– samples, the expression of Fgf3 was reduced in the Eda–/– incisors, but we found p > 0.05 with statistic analysis (Figure 3Z). Previous studies showed that Fgf3, Fgf10, and Fgf20 were related to tooth development, both Fgf3 and Fgf10 were down-regulated in Pax9+/–Msx1+/– embryos with missing mandibular incisors; and treatment with Fgf10 partially rescued the cusp defect in Eda–/– mouse (Pispa et al., 1999; Nakatomi et al., 2010). The expression of Fgf20 was reduced in the developing Eda–/– incisors, while the Fgf20 null mice had normal incisors (Häärä et al., 2012). Taken together, the Fgf signaling activity was reduced in Eda–/– tooth germs but appear adequate for regulating tooth formation; in Pax9+/–Eda–/– mandibles, the level of Fgf signaling was reduced to a certain level that appears inadequate for the normal development of mandibular incisors. Furthermore, the expression of Shh in the Pax9+/–Eda–/– mandibular incisor was significantly reduced compared with that in both Eda+/– and Eda–/– samples (Figure 3Z). The marker gene analysis indicates that both Fgf and Shh signaling pathways were involved in the genetic interaction between Pax9 and Eda during mandibular incisor formation. These data when viewed in the light of our findings suggest that a distinct cadre of genetic interactions are involved in the formation of mandibular incisors, whose initiation signifies the earliest zone of odontogenic induction in the mandibular arch.
As described by Nakatomi et al. (2010), Pax9 and Msx1 double heterozygous mutant mice show a consistent lack of mandibular incisors, a phenotype that is not evident in single mutant strains. Arrested Pax9+/–Msx1+/– mandibular incisor organs show a marked reduction in Fgf3 and Fgf10 expression within dental mesenchyme as well as reduced expression of Shh and Bmp2 in the dental epithelium. Our earlier biochemical analyses had first demonstrated that Pax9 interacts synergistically with Msx1 (Ogawa et al., 2006), so it is likely that this partnership plays a key role in driving key signaling interactions between dental mesenchyme and epithelium during the transition from the bud to the cap stage of development. Whether Eda signaling pathway genes are down-regulated in Pax9+/–Msx1+/– mandibular incisor organs offers a valuable direction for future studies. Our findings that signaling pathway genes are differentially affected in Pax9+/–Eda–/– mandibular incisor organs also warrant further investigation. In Pax9+/–Eda–/– maxillary incisors, Shh expression was slightly changed suggesting that the level of Shh was enough to induce the formation of maxillary incisors. Since the expression of Bmp4 remained unaffected in dental mesenchyme ofPax9+/–Eda–/– compound mandibular incisors, it is possible that the haploinsufficiency of Pax9 did not compromise Bmp4 expression in the developing incisor mesenchyme. Although it was reported that Wnt signaling pathway mediator Lef1 was down-regulated in the skin of Tabby mice (Durmowicz et al., 2002), our data showed that the expression of Lef1 was not affected in Pax9+/–Eda–/– compound mutant, which matched the regulatory hierarchy that Eda was downstream of Lef1 in the tooth organ (Laurikkala et al., 2001).
Patterns of tooth agenesis in humans provide valuable clues about the important roles that transcription factors in modulating epithelial–mesenchymal signaling during tooth development. As one of the best-studied genes, PAX9, is largely viewed as important for the patterning of human dentition. Our group’s clinical observations and that of others have documented that mutations in PAX9 consistently result in a pattern of tooth agenesis that involves posterior dentition, namely, molars and premolars. Intriguing is the finding that individuals with mutations in PAX9 often miss mandibular central incisors, the least patterned tooth in human dentition. While patients affected with mutations in EDA or EDAR lack posterior and anterior teeth in both arches, mandibular incisors are most often missing, a pattern of tooth agenesis that resembles that seen in individuals with mutations in WNT10A (Mues et al., 2014). Taken together, these data suggest that each tooth family (incisiform and cuspform/molariform) arises from distinct morphogenetic gradients or fields created by the differential actions of transcription factors and signaling pathways. However, the initiation of odontogenesis within each field is spatially regulated by unique molecular relationships such as for mandibular incisors where Pax9-dependent signaling and the Eda pathway appear to play an important role.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Committee for the Protection of Human Subjects, University of Texas Health Science Center at Houston. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Utah (Protocol #19-12012). Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
SHJ and RD’S contributed to the research design, data acquisition, and analyses, as well as the writing of the manuscript. JO contributed to the writing of the manuscript. ET and MR contributed to the mouse genetic data acquisition. MB contributed to the mouse genetic data analysis. JW contributed to the data acquisition for the medical and dental records of the patient. Each author gave final approval and agreed to be accountable for all aspects of the work, ensuring integrity and accuracy. All authors contributed to the article and approved the submitted version.
Funding
The following grants from the National Institutes of Health/National Institute of Dental and Craniofacial Research have supported this research: DE019471, DE019471-ARRA supplement, to RD’S, DE027255 to RD’S and SHJ, DE027355 supplement to MB. A fellowship was awarded to JO from the Ole and Marty Jensen Endowment Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Pascal Schneider for valuable feedback and advice throughout the project, the technical assistance of Greg Pratt.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.581843/full#supplementary-material
References
Balic, A. (2019). Concise review: cellular and molecular mechanisms regulation of tooth initiation. Stem Cells 37, 26–32. doi: 10.1002/stem.2917
Balic, A., and Thesleff, I. (2015). Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 115, 157–186. doi: 10.1016/bs.ctdb.2015.07.006
Charles, C., Hovorakova, M., Ahn, Y., Lyons, D. B., Marangoni, P., Churava, S., et al. (2011). Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development 138, 4063–4073. doi: 10.1242/dev.069195
Chen, J., Lan, Y., Baek, J. A., Gao, Y., and Jiang, R. (2009). Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev. Biol. 334, 174–185. doi: 10.1016/j.ydbio.2009.07.015
Durmowicz, M. C., Cui, C. Y., and Schlessinger, D. (2002). The EDA gene is a target of, but does not regulate Wnt signaling. Gene 285, 203–211. doi: 10.1016/s0378-1119(02)00407-9
Frazier-Bowers, S. A., Guo, D. C., Cavender, A., Xue, L., Evans, B., King, T., et al. (2002a). A novel mutation in human PAX9 causes molar oligodontia. J. Dent. Res. 81, 129–133. doi: 10.1177/154405910208100209
Frazier-Bowers, S. A., Scott, M. R., Cavender, A., Mensah, J., and D’Souza, R. N. (2002b). Mutational analysis of families affected with molar oligodontia. Connect. Tissue Res. 43, 296–300. doi: 10.1080/03008200290000961
Goldenberg, M., Das, P., Messersmith, M., Stockton, D. W., Patel, P. I., and D’Souza, R. N. (2000). Clinical, radiographic, and genetic evaluation of a novel form of autosomal-dominant oligodontia. J. Dent. Res. 79, 1469–1475. doi: 10.1177/00220345000790070701
Golonzhka, O., Metzger, D., Bornert, J. M., Bay, B. K., Gross, M. K., Kioussi, C., et al. (2009). Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 4278–4283. doi: 10.1073/pnas.0900568106
Häärä, O., Harjunmaa, E., Lindfors, P. H., Huh, S., FliniauxS, I., Åberg, T., et al. (2012). Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development 139, 3189–3199. doi: 10.1242/dev.079558
Huang, X., Wang, F., Zhao, C., Yang, S., Cheng, Q., Tang, Y., et al. (2019). Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cells Dev. 28, 683–694. doi: 10.1089/scd.2018.0230
Jarvinen, E., Shimomura-Kuroki, J., Balic, A., Jussila, M., and Thesleff, I. (2018). Mesenchymal Wnt/beta-catenin signaling limits tooth number. Development 145:dev158048. doi: 10.1242/dev.158048
Jernvall, J., and Thesleff, I. (2000). Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92, 19–29. doi: 10.1016/s0925-4773(99)00322-6
Jia, S., Zhou, J., Fanelli, C., Wee, Y., Bonds, J., Schneider, P., et al. (2017). Small-molecule Wnt agonists correct cleft palates in Pax9 mutant mice in utero. Development 144, 3819–3828. doi: 10.1242/dev.157750
Kapadia, H., Frazier-Bowers, S., Ogawa, T., and D’Souza, R. N. (2006). Molecular characterization of a novel PAX9 missense mutation causing posterior tooth agenesis. Eur. J. Hum. Genet. 14, 403–409. doi: 10.1038/sj.ejhg.5201574
Kapadia, H., Mues, G., and D’Souza, R. (2007). Genes affecting tooth morphogenesis. Orthod. Craniofac. Res. 10, 237–244. doi: 10.1111/j.1601-6343.2007.00407.x
Kist, R., Watson, M., Wang, X., Cairns, P., Miles, C., Reid, D. J., et al. (2005). Reduction of Pax9 gene dosage in an allelic series of mouse mutants causes hypodontia and oligodontia. Hum. Mol. Genet. 14, 3605–3617. doi: 10.1093/hmg/ddi388
Lan, Y., Jia, S., and Jiang, R. (2014). Molecular patterning of the mammalian dentition. Semin. Cell Dev. Biol. 2, 61–70. doi: 10.1016/j.semcdb.2013.12.003
Laurikkala, J., Mikkola, M., Mustonen, T., Aberg, T., Koppinen, P., Pispa, J., et al. (2001). TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev. Biol. 229, 443–455. doi: 10.1006/dbio.2000.9955
Lin, C. R., Kioussi, C., O’Connell, S., Briata, P., Szeto, D., Liu, F., et al. (1999). Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282. doi: 10.1038/45803
Lu, M. F., Pressman, C., Dyer, R., Johnson, R. L., and Martin, J. F. (1999). Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276–278. doi: 10.1038/45797
Lumsden, A. G. (1988). Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development 103(Suppl.) 155–169.
Millar, S. E., Koyama, E., Reddy, S. T., Andl, T., Gaddapara, T., Piddington, R., et al. (2003). Over- and ectopic expression of Wnt3 causes progressive loss of ameloblasts in postnatal mouse incisor teeth. Connect. Tissue Res. 44(Suppl. 1) 124–129. doi: 10.1080/713713645
Mina, M., and Kollar, E. J. (1987). The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch. Oral Biol. 32, 123–127. doi: 10.1016/0003-9969(87)90055-0
Mitsui, S. N., Yasue, A., Masuda, K., Watanabe, K., Horiuchi, S., Imoto, I., et al. (2014). Novel PAX9 mutations cause non-syndromic tooth agenesis. J. Dent. Res. 93, 245–249. doi: 10.1177/0022034513519801
Mues, G., Bonds, J., Xiang, L., Vieira, A. R., Seymen, F., Klein, O., et al. (2014). The WNT10A gene in ectodermal dysplasias and selective tooth agenesis. Am. J. Med. Genet. A 164A, 2455–2460. doi: 10.1002/ajmg.a.36520
Munne, P. M., Tummers, M., Jarvinen, E., Thesleff, I., and Jernvall, J. (2009). Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development 136, 393–402. doi: 10.1242/dev.025064
Nakatomi, M., Wang, X., Key, D., Lund, J. J., Turbe-Doan, A., Kist, R., et al. (2010). Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 340, 438–449. doi: 10.1016/j.ydbio.2010.01.031
Neubuser, A., Peters, H., Balling, R., and Martin, G. R. (1997). Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell 90, 247–255. doi: 10.1016/s0092-8674(00)80333-5
Nieminen, P. (2009). Genetic basis of tooth agenesis. J. Exp. Zool. B Mol. Dev. Evol. 312B, 320–342. doi: 10.1002/jez.b.21277
O’Connell, D. J., Ho, J. W., Mammoto, T., Turbe-Doan, A., O’Connell, J. T., Haseley, P. S., et al. (2012). A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci. Signal. 5:ra4. doi: 10.1126/scisignal.2002414
Ogawa, T., Kapadia, H., Feng, J. Q., Raghow, R., Peters, H., and D’Souza, R. N. (2006). Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J. Biol. Chem. 281, 18363–18369. doi: 10.1074/jbc.m601543200
Ohazama, A., Johnson, E. B., Ota, M. S., Choi, H. Y., Porntaveetus, T., Oommen, S., et al. (2008). Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One 3:e4092. doi: 10.1371/journal.pone.0004092
Parveen, A., Khan, S. A., Mirza, M. U., Bashir, H., Arshad, F., Iqbal, M., et al. (2019). Deleterious variants in WNT10A, EDAR, and EDA causing isolated and syndromic tooth agenesis: a structural perspective from molecular dynamics simulations. Int. J. Mol. Sci. 20:5282. doi: 10.3390/ijms20215282
Peters, H., Neubuser, A., Kratochwil, K., and Balling, R. (1998). Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12, 2735–2747. doi: 10.1101/gad.12.17.2735
Pispa, J., Jung, H. S., Jernvall, J., Kettunen, P., Mustonen, T., Tabata, M. J., et al. (1999). Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 216, 521–534. doi: 10.1006/dbio.1999.9514
Stockton, D. W., Das, P., Goldenberg, M., D’Souza, R. N., and Patel, P. I. (2000). Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19. doi: 10.1038/71634
Tian, Y., Ma, P., Liu, C., Yang, X., Crawford, D. M., Yan, W., et al. (2015). Inactivation of Fam20B in the dental epithelium of mice leads to supernumerary incisors. Eur. J. Oral Sci. 123, 396–402. doi: 10.1111/eos.12222
Voutilainen, M., Lindfors, P. H., Trela, E., Lonnblad, D., Shirokova, V., Elo, T., et al. (2015). Ectodysplasin/NF-κB promotes mammary cell fate via Wnt/β-catenin pathway. PLoS Genet. 11:e1005676. doi: 10.1371/journal.pgen.1005676
Wang, Y., Yan, M., Yu, Y., Wu, J., Yu, J., and Fan, Z. (2013). Estrogen deficiency inhibits the odonto/osteogenic differentiation of dental pulp stem cells via activation of the NF-kappaB pathway. Cell Tissue Res. 352, 551–559. doi: 10.1007/s00441-013-1604-z
Williams, M. A., and Letra, A. (2018). The changing landscape in the genetic etiology of human tooth agenesis. Genes 9:255. doi: 10.3390/genes9050255
Yang, Z., Hai, B., Qin, L., Ti, X., Shangguan, L., Zhao, Y., et al. (2013). Cessation of epithelial Bmp signaling switches the differentiation of crown epithelia to the root lineage in a beta-catenin-dependent manner. Mol. Cell. Biol. 33, 4732–4744. doi: 10.1128/mcb.00456-13
Ye, X., and Attaie, A. B. (2016). Genetic basis of nonsyndromic and syndromic tooth agenesis. J. Pediatr. Genet. 5, 198–208. doi: 10.1055/s-0036-1592421
Yu, W., Sun, Z., Sweat, Y., Sweat, M., Venugopalan, S. R., Eliason, S., et al. (2020). Pitx2-Sox2-Lef1 interactions specify progenitor oral/dental epithelial cell signaling centers. Development 147:dev186023. doi: 10.1242/dev.186023
Zhang, Y., Tomann, P., Andl, T., Gallant, N. M., Huelsken, J., Jerchow, B., et al. (2009). Reciprocal requirements for eda/edar/nf-kappab and wnt/beta-catenin signaling pathways in hair follicle induction. Dev. Cell 17, 49–61. doi: 10.1016/j.devcel.2009.05.011
Keywords: tooth development, signaling interaction, patterning, incisor development, Pax9, ectodysplasin pathway
Citation: Jia S, Oliver JD, Turner EC, Renouard M, Bei M, Wright JT and D’Souza RN (2020) Pax9’s Interaction With the Ectodysplasin Signaling Pathway During the Patterning of Dentition. Front. Physiol. 11:581843. doi: 10.3389/fphys.2020.581843
Received: 09 July 2020; Accepted: 05 November 2020;
Published: 26 November 2020.
Edited by:
Abigail Saffron Tucker, King’s College London, United KingdomReviewed by:
Anne Poliard, Université Paris Descartes, FranceBrad A. Amendt, The University of Iowa, United States
Copyright © 2020 Jia, Oliver, Turner, Renouard, Bei, Wright and D’Souza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rena N. D’Souza, UmVuYS5EU291emFAaHNjLnV0YWguZWR1
 Shihai Jia
Shihai Jia Jeremie D. Oliver
Jeremie D. Oliver Emma C. Turner3
Emma C. Turner3 Marianna Bei
Marianna Bei J. T. Wright
J. T. Wright Rena N. D’Souza
Rena N. D’Souza