
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 19 November 2020
Sec. Craniofacial Biology and Dental Research
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.573214
This article is part of the Research Topic Contemporary Models in Ectodermal Organ Development, Maintenance and Regeneration View all 15 articles
 Charinya Kanchanasevee1
Charinya Kanchanasevee1 Kanokwan Sriwattanapong2
Kanokwan Sriwattanapong2 Thanakorn Theerapanon2
Thanakorn Theerapanon2 Sermporn Thaweesapphithak3
Sermporn Thaweesapphithak3 Wanna Chetruengchai4,5
Wanna Chetruengchai4,5 Thantrira Porntaveetus2*
Thantrira Porntaveetus2* Vorasuk Shotelersuk4,5
Vorasuk Shotelersuk4,5Tooth agenesis is one of the most common orodental anomalies that demonstrate phenotypic and genotypic heterogeneity with a prevalence of 2.5%–7%. Mutations in WNT10A have been proposed to be the most common cause of nonsyndromic tooth agenesis (NSTA). The aim of this study was to characterize the dental features and genetic variants of NSTA in a Thai population. We recruited 13 unrelated patients with NSTA who attended the Faculty of Dentistry, Chulalongkorn University, Thailand, from 2017 to 2019. All 13 underwent whole exome sequencing that identified likely pathogenic genetic variants, all in WNT10A, in five patients. All five patients had second premolar agenesis, while three also had absent or peg-shaped upper lateral incisors. Patient 1 possessed a novel heterozygous duplication, c.916_918dupAAC (p.Asn306dup) in WNT10A. Patients 2 and 3 harbored a heterozygous and homozygous c.637G > A (p.Gly213Ser) in WNT10A, respectively. Patients 4 possessed a heterozygous c.511C > T (p.Arg171Cys) in WNT10A. Patient 5 harbored a homozygous c.511C > T (p.Arg171Cys) in WNT10A and a novel heterozygous c.413A > T (p.Asn138Ile) in EDARADD, suggesting digenic inheritance. We recruited another 18 family members of these five patients. Out of 23 participants, homozygous WNT10A variants were identified in 2 patients and heterozygous variants in 17 individuals. Both homozygous patients had NSTA. Eight out of 17 heterozygous individuals (8/17) had NSTA or a peg-shaped lateral incisor, indicating a 47% penetrance of the heterozygous variants or 53% (10/19) penetrance of either homozygous or heterozygous variants in WNT10A. The frequencies of the c.511C > T in our in-house 1,876 Thai exome database, Asian populations, and non-Asian populations were 0.016, 0.005–0.033, and 0.001, respectively; while those of the c.637G > A were 0.016, 0.004–0.029, and 0.000, respectively. In conclusion, our study reports two novel variants with one each in WNT10A and EDARADD, expanding the genotypic spectra of NSTA. Second premolar agenesis is a common phenotype in affected individuals with variants in WNT10A; however, its penetrance is incomplete. Lastly, the different frequencies of WNT10A variants, c.511C > T and c.637G > A, in diverse populations might contribute to the prevalence range of NSTA between continents.
Tooth agenesis is the most common developmental dental anomaly in humans with a prevalence between 2.5 and 7% (Polder et al., 2004; Khalaf et al., 2014). Missing less than 6 teeth is called hypodontia, agenesis of 6 or more teeth is called oligodontia, and the complete absence of teeth is called anodontia. Multiple signaling pathways, including bone morphogenic protein, fibroblast growth factor, sonic hedgehog, and wingless (WNT), play important roles in the epithelial-mesenchymal interactions during tooth development. WNT/β-catenin signaling is involved from the early to late stages of tooth formation (Liu et al., 2008; Porntaveetus et al., 2012; Intarak et al., 2018). Among WNT family members, mutations in WNT10A (OMIM ∗606268) are predominantly related to tooth agenesis (TA) involving both nonsyndromic/isolated/selective tooth agenesis (NSTA/STHAG4, MIM #150400) and syndromic tooth agenesis, such as odontoonychodermal dysplasia (MIM #257980) and Schopf-Schulz-Passarge syndrome (MIM #224750).
Population-based studies have revealed that 28%–62% of tooth agenesis patients have with WNT10A variants (van den Boogaard et al., 2012; Mostowska et al., 2013; Arzoo et al., 2014). Heterozygous, homozygous, and compound heterozygous forms of WNT10A were associated with NSTA with phenotypic heterogeneity. Using WNT10A target sequencing, significantly elevated frequencies of WNT10A variants were observed in the tooth agenesis group compared with the control group (Song et al., 2014; Machida et al., 2017). Biallelic WNT10A variants were proposed to be a pathogenic factor for tooth agenesis with complete penetrance, while a single allelic variant, presenting in a significantly higher frequency in tooth agenesis patients, was considered to be a predisposing factor for tooth agenesis with reduced penetrance (Mues et al., 2014; Guazzarotti et al., 2018). These findings prompted us to investigate the dental phenotype and genotype in Thai patients with NSTA and determine the allele frequencies of WNT10A in Thais compared with Asian and non-Asian populations.
The study protocol was approved by the Institutional Review Board (HREC-DCU 2018-091), Faculty of Dentistry, Chulalongkorn University and complied with the Declaration of Helsinki. Written informed consents were obtained prior to the patients’ participation in this study. Thirteen unrelated patients with NSTA who attended the Faculty of Dentistry, Chulalongkorn University, Thailand between January 2017 and March 2019 and their family member were recruited. Clinical and radiographic examinations, dental history, and intraoral photographs of the probands were used to assess tooth agenesis. The size and shape of the remaining teeth were also observed. The patients did not have any signs or symptoms related to ectodermal organ defects, e.g. intolerance to heat, dry skin, abnormal sweating, sparse hair, or brittle nails (Bergendal et al., 2006). The dental phenotypes of the probands’ family members were obtained from clinical and radiographic examinations, dental records, or participant interviews.
Genomic DNA extracted from peripheral blood leukocytes was subjected to mutation analysis using whole exome sequencing (WES) (Porntaveetus et al., 2018). Briefly, genomic DNA was captured using a SureSelect Human All Exon version 4 kit (Agilent Technologies, Santa Clara, CA, United States) and sequenced using Hiseq2000 (Macrogen, Seoul, South Korea). The sequences were aligned to the human genome reference sequence1 using the Burrows-Wheeler Aligner2. Downstream processing was performed with SAMtools3 and annotated against the dbSNP and 1000 Genomes. After quality filtering, the variants were screened using the genes listed in HP: 0009804 (reduced number of teeth) in Human Phenotype Ontology (Köhler et al., 2018). All calls with a coverage < 10×, minor allele frequency > 5% in the 1000 Genomes Project, Genome Aggregation Database (gnomAD4), and our in-house database of 1,876 Thai exomes; non-coding variants; and synonymous exonic variants were filtered out. The identified variants were confirmed by Sanger sequencing (Supplementary Table 1). The alignment and conservation of amino acid residues were generated by Clustal Omega (Madeira et al., 2019).
Allele frequencies of WNT10A variants were screened with multiple variant databases comprising the Genome Aggregation Database (gnomAD), GenomeAsia100K consortium, Northeast Asian Reference Database (NARD), Han Chinese genome project (PGG.Han), 4.7K Japanese individual genome variation (4.7KJPN), Human Genetic Variation Database (HGVD), Korean Variant Archive (KOVA), and our in-house database, last access on June 3, 2020.
Bioinformatics tools consisting of PolyPhen-2 (Adzhubei et al., 2010), SIFT (Kumar et al., 2009), MutationTaster (Schwarz et al., 2014), and CADD (Rentzsch et al., 2018) were used to predict each variant’s pathogenicity.
We performed WES for 13 unrelated patients with NSTA during 2017–2019 and detected variants related to tooth agenesis (HP: 0009804) in five patients. All five patients (Patients 1–5) possessed variants in WNT10A (NM_025216.3). We then recruited 18 additional family members of these 5 index patients, characterized their dental phenotype, and performed Sanger sequencing (Figure 1 and Supplementary Figure 1).

Figure 1. Family pedigrees and genetic variants. Arrow indicates the proband. Tooth phenotypes of the participants were determined either by the dentist or reported by the participants. W+/+, homozygous WNT10A variant; W+/-, heterozygous WNT10A variant; W+/+, wild-type WNT10A variant; E+/+, homozygous EDARADD variant; E+/-, heterozygous EDARADD variant; E+/+, wild-type EDARADD variant.
Patient 1, a 16-year-old female, lacked nine permanent teeth, all premolars and the lower right first molar. The lower left second molar was extracted due to pulp necrosis. Oral examination revealed that she had peg-shaped upper lateral incisors, severely hypoplastic edentulous ridges, anterior deep bite, and malocclusion. WES identified that the patient possessed a novel heterozygous duplication, c.916_918dupAAC (p.Asn306dup), in WNT10A (ClinVar SCV001335264). This variant was detected in the patient’s mother and grandmother who had NSTA, and in the unaffected half aunt.
Patient 2, a 27-year-old female, had 6 missing teeth, 4 upper premolars, and 2 lower third molars. The known heterozygous missense mutation, c.637G > A (p.Gly213Ser), in WNT10A was identified in the patient, and in the patient’s father and two sisters who reported no missing teeth. The mothers had biallelic wild-type alleles.
Patient 3, a 28-year-old male, had 12 missing teeth comprising an upper right canine, 3 first premolars, 4 second premolars, and 4 third molars. The homozygous mutation, c.637G > A (p.Gly213Ser), in WNT10A was identified in the patient. The WNT10A heterozygous c.637G > A (p.Gly213Ser) variant was detected in the patient’s older sister who was missing her 4 second premolars, and the younger sister, father, and mother who did not have any teeth missing.
Patient 4, a 31-year-old female, was missing her 2 lower second premolars and peg-shaped upper lateral incisors. Her deciduous lower right and left second molars were retained. Four third molars were previously extracted. The known heterozygous missense variant, c.511C > T (p.Arg171Cys), in WNT10A was detected in the patient, and in the patient’s older sister and mother who did not have tooth agenesis. The father possessed wild-type alleles.
Patient 5, a 34-year-old male, had agenesis of 8 permanent teeth, 2 upper lateral incisors, 2 right second premolars, and 4 third molars. He had implant replacements at the upper right lateral incisor and lower right second premolar. The homozygous missense variant, c.511C > T (p.Arg171Cys) in WNT10A and a novel heterozygous missense variant, c.413A > T (p.Asn138Ile), in EDARADD (Clinvar SCV001335265) was identified in the patient. The heterozygous WNT10A c.511C > T variant was also detected in the patient’s older sister who was missing two upper lateral incisors and two lower third molars and the mother who had peg-shaped upper lateral incisors. The heterozygous EDARADD c.413A > T variant was present in the patient’s older brother, but not in the sister and mother.
In Patients 1–5, the number of missing teeth was between 2 and 12 or between 2 and 9 teeth (excluding the third molars). An absent second premolar or peg-shaped upper lateral incisor was frequently observed in the patients (Table 1).
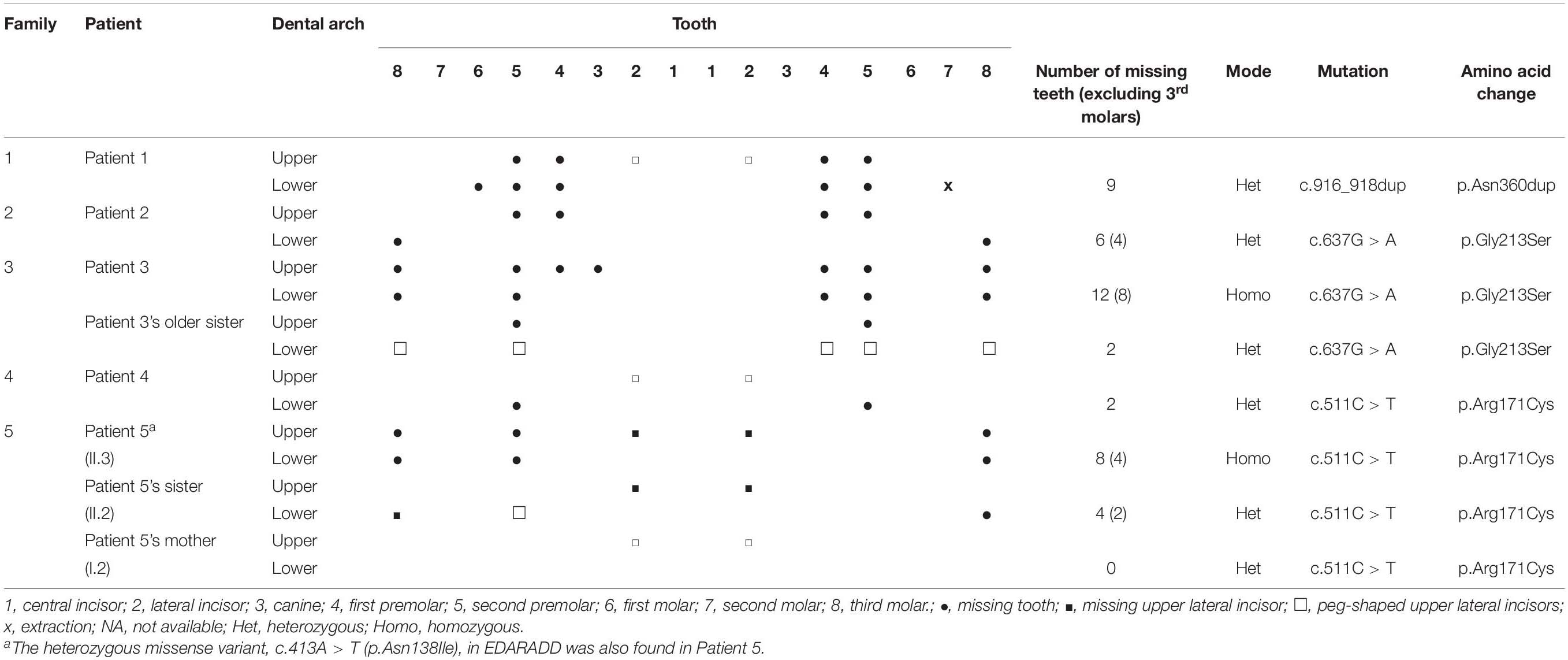
Table 1. The identified WNT10A variants in subjects with tooth agenesis or peg-shaped upper lateral incisors.
We used several bioinformatic tools to predict the pathogenicity of the variants. We found that the WNT10A c.511C > T (p.Arg171Cys) was predicted to be deleterious (CADD: 25.6), deleterious (SIFT: 0.0), possibly damaging (PolyPhen-2: 0.93), and disease causing (MutationTaster). The WNT10A c.637G > A (p.Gly213Ser) was predicted to be deleterious (CADD: 27.2), deleterious (SIFT: 0.0), probably damaging (PolyPhen-2: 1.0), and disease causing (MutationTaster). Furthermore, the EDARADD c.413A > T (p.Asn138Ile) was predicted to be deleterious (CADD: 27.3), deleterious (SIFT: 0.0), possibly damaging (PolyPhen: 1.0), and disease causing (MutationTaster).
The amino acid residues; p.Arg171, p.Gly213, and p.Asn306 in WNT10A, and p.Asn128 in EDARADD are conserved among multiple species (Supplementary Figure 2). According to the ACMG standards and guidelines, the WNT10A c.511C > T and c.637G > A and the EDARADD c.413A > T variants are considered to be likely pathogenic, while the WNT10A c.916_918dupAAC variant is considered as uncertain significance (Richards et al., 2015).
Eighteen members of the five index patients’ families (23 total) were included. Out of the 23 participants, the homozygous WNT10A variants were identified in 2 patients (3.II.2 and 5.II.3) and the heterozygous variants in 17 individuals. Both homozygous patients had nonsyndromic tooth agenesis (NSTA). Eight (1.I.2, 1.II.2, 1.III.1, 2.II.1, 3.II.1, 4.II.2, 5.I.2, and 5.II.2) out of 17 heterozygous individuals (8/17) had NSTA or a peg-shaped lateral incisor, indicating a 47% penetrance of the heterozygous variants, or a 53% (10/19) penetrance of either homozygous or heterozygous variants in WNT10A.
We screened the frequencies of the WNT10A c.511C > T and c.637G > A variants in multiple variant databases comprising the Genome Aggregation Database (gnomAD), GenomeAsia100K consortium, Northeast Asian Reference Database (NARD), Han Chinese genome project (PGG.Han), 4.7K Japanese individual genome variation (4.7KJPN), Human Genetic Variation Database (HGVD), Korean Variant Archive (KOVA), and our in-house database. The allele frequencies of WNT10A c.511C > T and c.637G > A in Asian populations was between 0.005 and 0.033 and between 0.004 and 0.029, respectively. In our in-house database (ThWES) of 1,876 Thai exomes, the frequency of c.511C > T variant was 0.016 and that of c.637G > A variant was 0.016, which were in the Asian population ranges. In contrast, the frequency of the c.511C > T variant was 0.001 and that of the c.637G > A variant was and 0.000 in non-Asian populations (Table 2). These results indicate that the WNT10A c.511C > T and c.637G > A variants are common and concentrated in Asian populations compared with non-Asian populations.
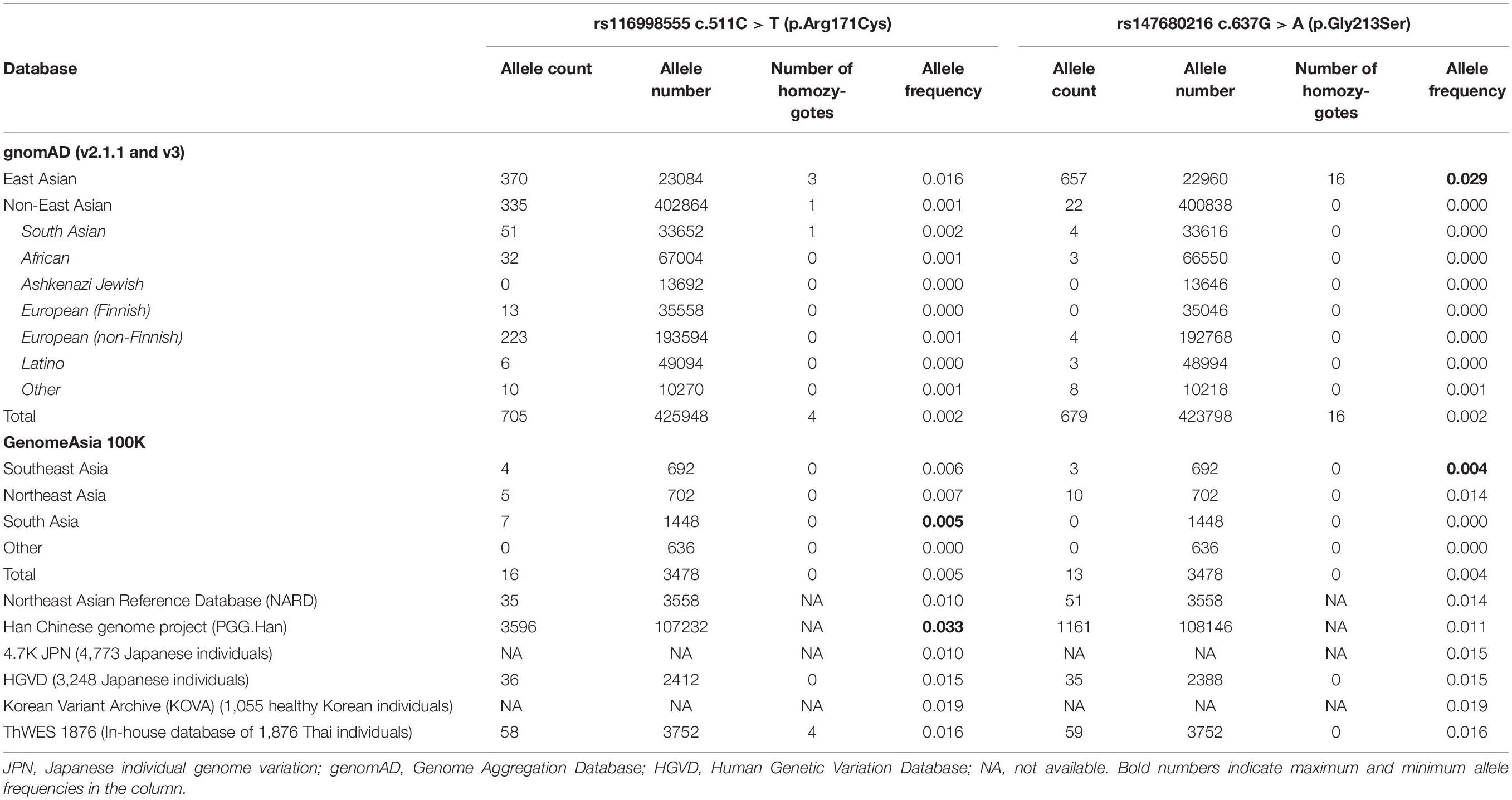
Table 2. Allele frequencies and details of WNT10A c.511C > T (p.Arg171Cys) and c.637G > A (p.Gly213Ser) variants.
In this study, we identified five index patients having NSTA and variants in WNT10A. Eighteen more family members were included. The number of missing teeth observed ranged from 2 to 12 teeth or 2 to 9 teeth, excluding third molars. WNT10A is the most common variant associated with NSTA (van den Boogaard et al., 2012; Mostowska et al., 2013; Arzoo et al., 2014). Here, we identified that all five patients possessed WNT10A variants. The novel heterozygous duplication, c.916_918dupAAC (p.Asn306dup), was identified in Patient 1. The heterozygous state of c.637G > A (p.Gly213Ser) variant was detected in Patient 2 and its homozygous state in Patient 3. The heterozygous state of c.511C > T (p.Arg171Cys) variant was detected in Patient 4 and its homozygous state was detected in Patient 5. The novel heterozygous c.413A > T (p.Asn138Ile) in EDARADD was observed in Patient 5.
The relationship between the heterozygous WNT10A variants (c.511C > T and c.637G > A) and NSTA was characterized in large well-phenotyped population cohorts (Song et al., 2014; Machida et al., 2017). Both variants were significantly associated with tooth agenesis compared with healthy control individuals. In other studies, the variants were shown to cause tooth agenesis with incomplete penetrance (He et al., 2013; Plaisancié et al., 2013; Kantaputra et al., 2014). Biallelic WNT10A variants were proposed to be the pathogenic factor for tooth agenesis with complete penetrance, while single allelic variants, presenting with a significantly higher frequency in tooth agenesis patients, were considered to be a predisposing factor for tooth agenesis with reduced penetrance (Mues et al., 2014; Guazzarotti et al., 2018). In addition, the phenotypic spectrum of WNT10A mutations was shown to be dose-dependent with variable expressivity, including within the same family (Park et al., 2019). Patients with biallelic WNT10A mutations had severe tooth agenesis, while heterozygous patients were either unaffected or had a mild tooth phenotype.
In our study, Patient 3 (3.II.2) and Patient 5 (5.II.3), who harbor the homozygous WNT10A c.637G > A and c.511C > T variants, respectively, have tooth agenesis and their phenotypes are more severe than the heterozygous individuals with the same variant. In our cohort, the heterozygous c.511C > T and c.637G > A variants demonstrated incomplete penetrance, which is consistent with previous reports. The evidence mentioned above suggests that the heterozygous WNT10A c.511C > T or c.637G > A allele can be a contributing factor for NSTA with low penetrance, while biallelic variants are associated with greater clinical severity. However, there might be other genetic or environmental factors influencing the phenotypic expression of NSTA patients.
Mutations in EDARADD (OMIM∗ 606603) are associated with autosomal dominant ectodermal dysplasia 11A (MIM# 614940) (Bal et al., 2007; Cluzeau et al., 2011) and autosomal recessive ectodermal dysplasia 11B (MIM# 614941) (Chaudhary et al., 2016). The EDARADD c.413A > T (p.Asn138Ile) variant identified in Patient 5 is located in the death domain that interacts with EDAR. The homozygous and heterozygous mutations in the EDAR death domain cause hypohidrotic ectodermal dysplasia (MIM #614940, #614941) with low penetrance (Bal et al., 2007; Cluzeau et al., 2011). To the best of our knowledge, only one heterozygous EDARADD variant, c.308C > T, p.Ser103Phe, was identified in patients with NSTA. The allelic frequency of the EDARRADD c.308C > T variant is found in up to 2% of a healthy population according to the dbSNP database. However, this variant has been associated with NSTA, but with low penetrance and variable expressivity. (Bergendal et al., 2011; Arte et al., 2013; Barbato et al., 2018; Martínez-Romero et al., 2019). Although the heterozygous EDARADD c.413A > T mutation was detected in the patient and his unaffected brother, it was not in his affected sister or mother.
Interestingly, the digenic heterozygous variants of WNT10A and other genes in the EDA pathway, including EDA, EDAR, and EDARADD, have been found in several patients with NSTA (Arte et al., 2013; He et al., 2013; Barbato et al., 2018; Martínez-Romero et al., 2019). The WNT and EDA pathways are suggested to play complementary roles during tooth development (Yu et al., 2019). In Family 5, the WNT10A c.511C > T variant was present in the homozygous state in the patient (8 missing teeth), in the heterozygous state in the sister (4 missing teeth) and proband’s mother (peg-shaped lateral incisors), and not detected in the proband’s brother (unaffected). Therefore, the role of the WNT10A variant in this family may be because its homozygous state is associated with more severe NSTA than those with the heterozygous variant, and the heterozygous variant shows incomplete penetrance. Moreover, the coexistence and variable penetrance of WNT10A and EDARADD variants may modulate the final phenotype of Patient 5.
Tooth agenesis is one of the most common anomalies in human development. Its prevalence in the general population is 2.5–7% (Polder et al., 2004). Mutations in WNT10A are the most frequently found variants associated with NSTA in several populations studied to date (van den Boogaard et al., 2012; Mostowska et al., 2013; Song et al., 2014; Tardieu et al., 2017). In particular, the WNT10A c.511C > T and c.637G > A variants are predominant in Asian populations compared with Europeans (Song et al., 2014; Machida et al., 2017). According to multiple genetic databases, we observed the allele frequencies of WNT10A c.511C > T and c.637G > A variants in Asian populations up to 0.033 and 0.029, respectively, compared with those in non-Asians, which are 0.000–0.001. The frequencies of the c.511C > T and c.637G > A variants in our in-house database of 1,876 Thai exomes are 0.016, which are in the range of Asian populations, including Japanese, Chinese, and Koreans. These results indicate that these two variants are relatively common in Asian populations. The difference in WNT10A allele frequencies among different ethnic groups may also partly explain the diverse prevalence of tooth agenesis on different continents (Khalaf et al., 2014).
The number and location of missing teeth are associated with mutations in specific genes (Al-Ani et al., 2017). A genotype-phenotype correlation study revealed that the second premolars were the most common missing teeth found in patient with WNT10A variants (Arzoo et al., 2014). All five patients in our study had agenesis of the second premolars, suggesting that WNT10A variants might be responsible for the absence of second premolars with high penetrance. Mutations in WNT10A have also been proposed to cause agenesis or microdontia of the upper lateral incisors (Kantaputra et al., 2014; Mostowska et al., 2015). Absent upper lateral incisors was observed in Patient 5 and his sister, and peg-shaped upper lateral incisors were found in Patient 1, Patient 4, and Patient 5’s mother who had the heterozygous WNT10A variants.
In conclusion, this study reports a novel in-frame duplication, c.916_918dupAAC (p.Asn306dup), in WNT10A and a novel heterozygous missense variant, c.413A > T (p.Asn138Ile), in EDARADD, expanding the genotypic spectrum related to NSTA. The heterozygous WNT10A c.511C > T and c.637G > A variants demonstrate incomplete penetrance. Both variants are more common in Asian populations compared with non-Asians, which might explain the diverse prevalence of NSTA in various continents.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Institutional Review Board (HREC-DCU 2018-091), Faculty of Dentistry, Chulalongkorn University and were complied with the Declaration of Helsinki. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was also obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
CK and TT contributed to the data acquisition and drafting of the manuscript. KS, ST, and WC contributed to the analysis and interpretation of the results. TP contributed to the study conception and design and drafting of the manuscript. VS contributed to the conception and data analysis. All authors revised and approved the submitted version.
This project was funded by the National Research Council of Thailand, Global partnership CU-C16F630029, Health Systems Research Institute, Thailand Research Fund (MRG6280001 and DPG6180001), and Faculty Research Grant (DRF64013) Faculty of Dentistry, Chulalongkorn University. KS is supported by Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University. ST is supported by the 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarship and The 90th Anniversary Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MA declared a past co-authorship with the authors TT and TP to the handling editor.
We thank Nut Pusuntisumpun, Adisa Suthirathikul, and Athipong Boonchanawiwat for their kind assistance with specimen collection, and Dr. Kevin A. Tompkins for the language revision of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.573214/full#supplementary-material
Supplementary Figure 1 | Clinical and radiographic features of the patients. ∗indicates missing tooth.
Supplementary Figure 2 | Schematic diagrams of the WNT10A and EDARADD genes and proteins showing the identified WNT10A variants c.916_918dupAAC (p.Asn306dup), c.637G > A (p.Gly213Ser), and c.511C > T (p.Arg171Cys).
Supplementary Table 1 | Primers for Sanger sequencing.
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. doi: 10.1038/nmeth0410-248
Al-Ani, A. H., Antoun, J. S., Thomson, W. M., Merriman, T. R., and Farella, M. (2017). Hypodontia: an update on its etiology, classification, and clinical management. BioMed Res. Int. 2017:9378325. doi: 10.1155/2017/9378325
Arte, S., Parmanen, S., Pirinen, S., Alaluusua, S., and Nieminen, P. (2013). Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS One 8:e73705. doi: 10.1371/journal.pone.0073705
Arzoo, P. S., Klar, J., Bergendal, B., Norderyd, J., and Dahl, N. (2014). WNT10A mutations account for of population-based isolated oligodontia and show phenotypic correlations. Am. J. Med. Genet. A 164a, 353–359. doi: 10.1002/ajmg.a.36243
Bal, E., Baala, L., Cluzeau, C., El Kerch, F., Ouldim, K., Hadj-Rabia, S., et al. (2007). Autosomal dominant anhidrotic ectodermal dysplasias at the EDARADD locus. Hum. Mut. 28, 703–709. doi: 10.1002/humu.20500
Barbato, E., Traversa, A., Guarnieri, R., Giovannetti, A., Genovesi, M. L., Magliozzi, M. R., et al. (2018). Whole exome sequencing in an Italian family with isolated maxillary canine agenesis and canine eruption anomalies. Arch. Oral. Biol. 91, 96–102. doi: 10.1016/j.archoralbio.2018.04.011
Bergendal, B., Klar, J., Stecksén-Blicks, C., Norderyd, J., and Dahl, N. (2011). Isolated oligodontia associated with mutations in EDARADD. AXIN2, MSX1, and PAX9 genes. Am. J. Med. Genet. A 155, 1616–1622. doi: 10.1002/ajmg.a.34045
Bergendal, B., Norderyd, J., Bågesund, M., and Holst, A. (2006). Signs and symptoms from ectodermal organs in young Swedish individuals with oligodontia. Int. J. Paediatr. Dent. 16, 320–326. doi: 10.1111/j.1365-263X.2006.00741.x
Chaudhary, A. K., Girisha, K. M., and Bashyam, M. D. (2016). A novel EDARADD 5’-splice site mutation resulting in activation of two alternate cryptic 5’-splice sites causes autosomal recessive hypohidrotic ectodermal dysplasia. Am. J. Med. Genet. A 170, 1639–1641. doi: 10.1002/ajmg.a.37607
Cluzeau, C., Hadj-Rabia, S., Jambou, M., Mansour, S., Guigue, P., Masmoudi, S., et al. (2011). Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum. Mut. 32, 70–72. doi: 10.1002/humu.21384
Guazzarotti, L., Tadini, G., Mancini, G. E., Sani, I., Pisanelli, S., Galderisi, F., et al. (2018). WNT10A gene is the second molecular candidate in a cohort of young Italian subjects with ectodermal derivative impairment (EDI). Clin. Genet. 93, 693–698. doi: 10.1111/cge.13147
He, H., Han, D., Feng, H., Qu, H., Song, S., Bai, B., et al. (2013). Involvement of and interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS One 8:e80393. doi: 10.1371/journal.pone.0080393
Intarak, N., Theerapanon, T., Srijunbarl, A., Suphapeetiporn, K., Porntaveetus, T., and Shotelersuk, V. (2018). Novel compound heterozygous mutations in KREMEN1 confirm it as a disease gene for ectodermal dysplasia. Br. J. Dermatol. 179, 758–760. doi: 10.1111/bjd.16541
Kantaputra, P., Kaewgahya, M., and Kantaputra, W. (2014). WNT10A mutations also associated with agenesis of the maxillary permanent canines, a separate entity. Am. J. Med. Genet. A 164, 360–363. doi: 10.1002/ajmg.a.36280
Khalaf, K., Miskelly, J., Voge, E., and Macfarlane, T. V. (2014). Prevalence of hypodontia and associated factors: a systematic review and meta-analysis. J. Orthod. 41, 299–316.
Köhler, S., Carmody, L., Vasilevsky, N., Jacobsen, J. O. B., Danis, D., Gourdine, J.-P., et al. (2018). Expansion of the human phenotype ontology (HPO) knowledge base and resources. Nucleic Acids Res. 47, D1018–D1027. doi: 10.1093/nar/gky1105
Kumar, P., Henikoff, S., and Ng, P. C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081. doi: 10.1038/nprot.2009.86
Liu, F., Chu, E. Y., Watt, B., Zhang, Y., Gallant, N. M., Andl, T., et al. (2008). Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 313, 210–224. doi: 10.1016/j.ydbio.2007.10.016
Machida, J., Goto, H., Tatematsu, T., Shibata, A., Miyachi, H., Takahashi, K., et al. (2017). WNT10A variants isolated from Japanese patients with congenital tooth agenesis. Hum. Genome Var. 4:17047. doi: 10.1038/hgv.2017.47
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Martínez-Romero, M. C., Ballesta-Martínez, M. J., López-González, V., Sánchez-Soler, M. J., Serrano-Antón, A. T., Barreda-Sánchez, M., et al. (2019). EDA, EDAR, EDARADD and WNT10A allelic variants in patients with ectodermal derivative impairment in the Spanish population. Orphanet J. Rare Dis. 14:281. doi: 10.1186/s13023-019-1251-x
Mostowska, A., Biedziak, B., Zadurska, M., Dunin-Wilczynska, I., Lianeri, M., and Jagodzinski, P. P. (2013). Nucleotide variants of genes encoding components of the Wnt signalling pathway and the risk of non-syndromic tooth agenesis. Clin. Genet. 84, 429–440. doi: 10.1111/cge.12061
Mostowska, A., Biedziak, B., Zadurska, M., Matuszewska-Trojan, S., and Jagodziński, P. P. (2015). WNT10A coding variants and maxillary lateral incisor agenesis with associated dental anomalies. Eur. J. Oral Sci. 123, 1–8. doi: 10.1111/eos.12165
Mues, G., Bonds, J., Xiang, L., Vieira, A. R., Seymen, F., Klein, O., et al. (2014). The WNT10A gene in ectodermal dysplasias and selective tooth agenesis. Am. J. Med. Genet. A 164A, 2455–2460. doi: 10.1002/ajmg.a.36520
Park, H., Song, J.-S., Shin, T. J., Hyun, H.-K., Kim, Y.-J., and Kim, J.-W. (2019). WNT10A mutations causing oligodontia. Arch. Oral. Biol. 103, 8–11. doi: 10.1016/j.archoralbio.2019.05.007
Plaisancié, J., Bailleul-Forestier, I., Gaston, V., Vaysse, F., Lacombe, D., Holder-Espinasse, M., et al. (2013). Mutations in WNT10A are frequently involved in oligodontia associated with minor signs of ectodermal dysplasia. Am. J. Med. Genet. A 161, 671–678. doi: 10.1002/ajmg.a.35747
Polder, B. J., Van’t Hof, M. A., Van der Linden, F. P. G. M., and Kuijpers-Jagtman, A. M. (2004). A meta-analysis of the prevalence of dental agenesis of permanent teeth. Am. J. Med. Genet. A 32, 217–226. doi: 10.1111/j.1600-0528.2004.00158.x
Porntaveetus, T., Abid, M. F., Theerapanon, T., Srichomthong, C., Ohazama, A., Kawasaki, K., et al. (2018). Expanding the oro-dental and mutational spectra of kabuki syndrome and expression of KMT2D and KDM6A in human tooth germs. Int. J. Biol. Sci. 14, 381–389. doi: 10.7150/ijbs.23517
Porntaveetus, T., Ohazama, A., Choi, H. Y., Herz, J., and Sharpe, P. T. (2012). Wnt signaling in the murine diastema. Eur. J. Orthod. 34, 518–524. doi: 10.1093/ejo/cjr049
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J., and Kircher, M. (2018). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894. doi: 10.1093/nar/gky1016
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405–423. doi: 10.1038/gim.2015.30
Schwarz, J. M., Cooper, D. N., Schuelke, M., and Seelow, D. (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362. doi: 10.1038/nmeth.2890
Song, S., Zhao, R., He, H., Zhang, J., Feng, H., and Lin, L. (2014). WNT10A variants are associated with non-syndromic tooth agenesis in the general population. Hum. Genet. 133, 117–124. doi: 10.1007/s00439-013-1360-x
Tardieu, C., Jung, S., Niederreither, K., Prasad, M., Hadj-Rabia, S., Philip, N., et al. (2017). Dental and extra-oral clinical features in 41 patients with WNT10A gene mutations: a multicentric genotype–phenotype study. Clin. Genet. 92, 477–486. doi: 10.1111/cge.12972
van den Boogaard, M. J., Créton, M., Bronkhorst, Y., van der Hout, A., Hennekam, E., Lindhout, D., et al. (2012). Mutations in WNT10A are present in more than half of isolated hypodontia cases. J. Med. Genet. 49, 327–331. doi: 10.1136/jmedgenet-2012-100750
Keywords: hypodontia, oligodontia, nonsynonymous, homozygous, heterozygous, ectoderm
Citation: Kanchanasevee C, Sriwattanapong K, Theerapanon T, Thaweesapphithak S, Chetruengchai W, Porntaveetus T and Shotelersuk V (2020) Phenotypic and Genotypic Features of Thai Patients With Nonsyndromic Tooth Agenesis and WNT10A Variants. Front. Physiol. 11:573214. doi: 10.3389/fphys.2020.573214
Received: 16 June 2020; Accepted: 27 October 2020;
Published: 19 November 2020.
Edited by:
Maisa Hanna-Maija Seppala, King’s College London, United KingdomReviewed by:
Dong Han, Peking University Hospital of Stomatology, ChinaCopyright © 2020 Kanchanasevee, Sriwattanapong, Theerapanon, Thaweesapphithak, Chetruengchai, Porntaveetus and Shotelersuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thantrira Porntaveetus, dGhhbnRyaXJhLnBAY2h1bGEuYWMudGg=; orcid.org/0000-0003-0145-9801
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.