- 1School of Pharmacy, Binzhou Medical University, Yantai, China
- 2Yantai Affiliated Hospital of Binzhou Medical University, Yantai, China
Background: Coronary artery disease (CAD) remains the leading cause of mortality worldwide, and its susceptibility is closely associated with genetic modifications. The association between inflammation and CAD has been investigated in detail. This meta-analysis was conducted based on the PRISMA guidelines to evaluate the association between the tumor necrosis factor superfamily member 4 (TNFSF4) gene polymorphisms (rs3861950 T > C and rs1234313 A > G) and the risk of CAD.
Methods: The selected criteria included 11 eligible articles containing 18 studies (nine studies included 7,395 cases and 5,296 controls for rs3861950 and nine studies with 6,951 cases and 4,959 controls for rs1234313). Correlations between the two polymorphisms and CAD were estimated by pooling the odds ratios (ORs) with 95% confidence interval (95% CI) in allelic, dominant, recessive, heterozygous, and homozygous models.
Results: The pooled analyses demonstrated that the rs3861950 T > C polymorphism was significantly associated with an increased risk of CAD in the Asian population in the allelic model, dominant model, and homozygous model. Furthermore, subgroup analysis based on disease type showed that TNFSF4 rs3861950 T > C had a robust correlation with increased risk of cerebral infarction (CI) in the allelic model, dominant model, heterozygous model, and homozygous model. However, the rs1234313 A > G polymorphism mostly tended to decrease the risk of CAD in the Asian and Caucasian populations in the allelic and dominant model. This single nucleotide polymorphism (SNP) had a close relation to myocardial infarction (MI) susceptibility in the allelic model, dominant model, and heterozygous model.
Conclusion: This meta-analysis identified two novel SNPs in TNFSF4 significantly associated with CAD susceptibility.
Introduction
Coronary artery disease (CAD) is a severe condition caused by the accumulation of plaque in the coronary arteries that leads to dysfunction of coronary arteries endothelial cells at an early stage (Marenberg et al., 1995; Kessler et al., 2016). Epidemiological and translational medical research has improved our understanding the mechanism underlying CAD (Khera and Kathiresan, 2017). However, CAD remains the leading cause of mortality worldwide. An increasing number of studies demonstrated that age, race, gender, smoking status, blood pressure, diabetes, obesity, and an unhealthy lifestyle are associated with an increased risk of CAD (Kessler et al., 2016). High-throughput sequencing and functional studies have confirmed that the susceptibility of CAD is regulated by a combination of genetic and environmental factors. Moreover, family-based studies indicated that the heritable variation was closely associated with CAD susceptibility (Marenberg et al., 1995). Therefore, there is a current need to identify robust genetic markers for the precise diagnosis of CAD in a specific population.
The close association between atherosclerosis and inflammation is well-known as the pathological basis of CAD(Christodoulidis et al., 2014). Furthermore, many well-established inflammatory mediators were thought to be potential clinical biomarkers of CAD. For example, TNFSF4, also called OX40L, belongs to the tumor necrosis factor superfamily and it is a membrane-bound protein predominantly expressed on antigen-presenting cells, CD4/8-positive T cells, and vascular endothelial cells (Baum et al., 1994). TNFSF4 has been suggested to bind to its receptor TNFRSF4 and improves the proliferation and development of memory T cells (Van Wanrooij et al., 2007; Kaur and Brightling, 2012). Previous studies have found that a high expression of TNFSF4 may play a vital role in atherosclerotic lesions and the accumulation of plagues in the arteries (Van Wanrooij et al., 2007). Therefore, there is a current need to improve our understand of the association between TNFSF4 variations and CAD risks. Single nucleotide polymorphisms (SNPs) are associated with disease susceptibility by altering gene expression or function. Human TNFSF4 gene maps to chromosome 1q25 and many SNPs have been found in the TNFSF4 gene (Baum et al., 1994). Furthermore, case-control studies have already shown a robust association between some TNFSF4 SNPs and CAD, such as rs17568, rs3861950, rs1234314, rs1234313, and rs3850641(Wang et al., 2019). Recently, a meta-analysis has reported the SNPs of TNFSF4 and evaluated the associations between several SNPs and CAD risk(Wang et al., 2019). However, conclusions were drawn from insufficient data, and this caught our attention. Considering that no significant association was found between TNFSF4 rs3861950 T > C and rs1234313 A > G polymorphisms and CAD ris, there is a need to re-evaluate the potential risks of CAD based on additional case-control studies.
Methods
Protocol and Registration
The implementation of this meta-analysis was completed in accordance with the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) guidelines (Moher et al., 2009). The PRISMA checklist is provided in Supplementary Table 1.
Literature Search Strategy
A systematic and comprehensive literature search based on the Participant, Intervention, Comparison, Outcomes, and Studies (PICOS) framework was performed in PubMed, EBSCO, Web of Science, and CNKI databases to collect the relevant articles published prior to February 2020 without language restrictions using the following terms and keywords: (tumor necrosis factor superfamily member 4” OR “TNFSF4” OR “OX40L”) AND (“polymorphism*” OR “variant*”) AND [“coronary artery disease” OR “CAD” OR “coronary heart disease” OR “CHD” OR “ischemic heart disease” OR “IHD” OR “angina pectoris” OR “myocardial infarction (MI)” OR “MI” OR (“coronary” AND “atherosclerosis”) OR “acute coronary syndrome”]. Moreover, the references cited by these articles were also evaluated to include additional relevant articles. Two investigators (S Liu and X Wang) independently read and checked the associated articles and retrieved articles separately.
Eligibility Criteria
PICOS inclusion criteria were used for this meta-analysis (Supplementary Table 2). Briefly, studies were included if they met the following criteria: (1) association of TNFSF4 polymorphisms (rs3861950 T > C and rs1234313 A > G) with susceptibility to CAD; (2) CAD diagnosis by using coronary angiography and international validated criteria; (3) case-control or cohort studies; (4) availablility of original data and sufficient genotype data provided; (5) human studies; and (6) studies subjected to the Hardy-Weinberg equilibrium (HWE), (p > 0.05). Additionally, the exclusion criteria were as follows: (1) duplicate studies and repetitive data; (2) reviews, meeting abstracts, and meta-analysis; (3) studies on the association between other gene polymorphisms and CAD risks; and (4) family-based and case-only studies. The χ2-test was used to determine whether the observed genotype frequencies conformed to HWE, and p < 0.05 was considered statistically significant (Thakkinstian et al., 2005; Bowden et al., 2011).
Literature Quality Assessment
The Newcastle-Ottawa scale (NOS) was applied to assess article quality, which consists of three perspectives: selection, comparability, and exposure. A total score of five or above was considered a high-quality study, while scores lower than five were further excluded (Stang, 2010). The scores of NOS range from 0 to 9 and the scores of each study are listed in Supplementary Table 3.
Data Extraction
The following variables for the rs3861950 T > C and rs1234313 A > G polymorphisms were extracted from the eligible studies by two investigators (S Liu and X Wang) independently: first author, publication year, type of diseases, country and ethnicity of the study population, sample size of cases and controls, genotyping methods, percentage of males and mean age of the study population, number of genotypes and alleles in both groups, and study design. All investigators participated in the discussion to reach a consensus.
Statistical Analysis
The allele frequencies of the polymorphism from each study were calculated from genotype distributions or from the minor allele frequencies (MAF) provided in the studies. The association between the two TNFSF4 polymorphisms and susceptibility to CAD was assessed from the odds ratios (ORs) and their corresponding 95% confidence intervals (95% CIs) for each study in five genetic models: allelic, dominant, recessive, heterozygous, and homozygous models. Complete and subgroup analyses based on ethnicity and diseases were performed. A p < 0.05 was considered to be statistically significant. The Cochran's Q statistical test and I2-values were used to evaluate the heterogeneity of the eligible studies, and the pooled OR and its 95% CI were obtained by using either the fixed-effect model (p > 0.1 and I2 ≤ 50%) or the random-effect model (p ≤ 0.1 or I2 > 50%)(Bowden et al., 2011). Considering the large heterogeneity, meta-regression analyses were carried out. The following variables were considered to determine the potential source: publication year, population ethnicity (Asian or Caucasian), disease type, genotype methods, source of controls, and the case sample size (defined as >500 or <500 cases);
p < 0.05 was considered to be statistically significant. Begg's funnel plot and the Egger's test were used to determine the publication bias with p < 0.05 being statistically significant (Begg and Mazumdar, 1994; Egger, 1997). The robustness of results was tested by using sensitivity analysis. STATA version 12.0 was used to perform all statistical analyses (Thakkinstian et al., 2005).
Results
Characteristics of the Included Studies
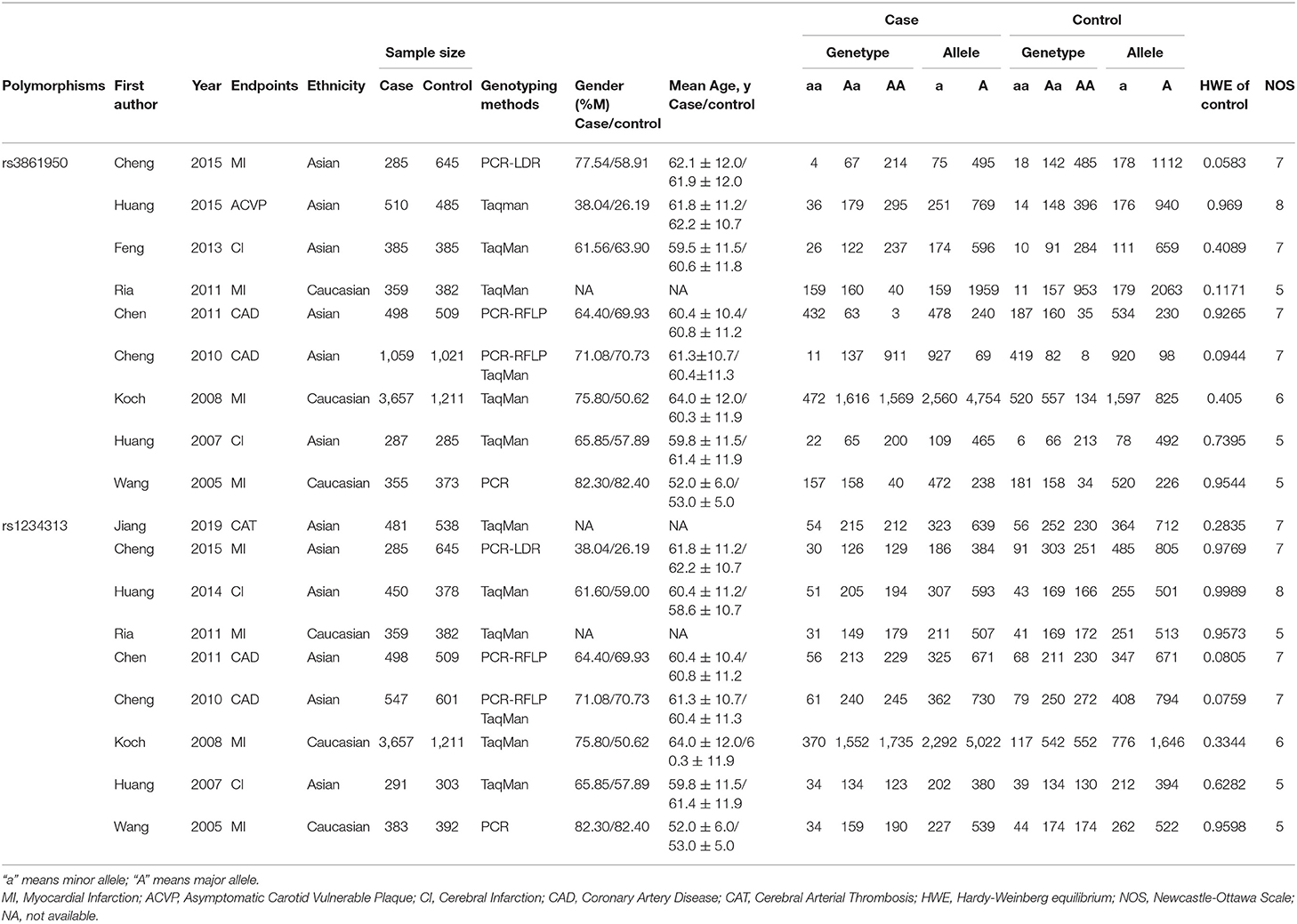
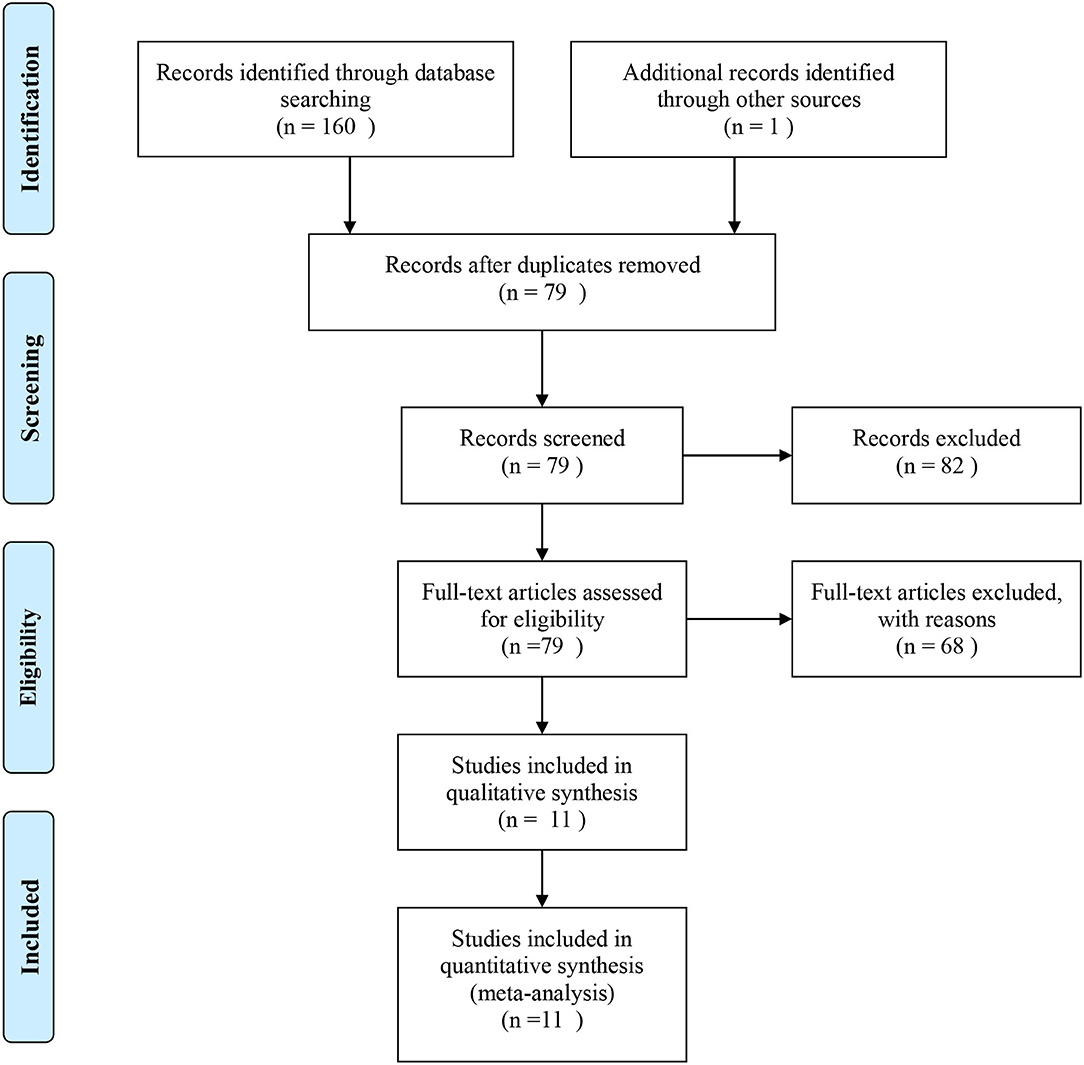
A total of 161 papers were initially obtained from PubMed, Web of Science, EBSCO, CNKI, and Google Scholar using the keywords search before February 2020. Among these studies, 150 articles were further excluded using the selection criteria described above and 11 of the articles (Wang et al., 2005; Koch et al., 2008; Chen et al., 2011; Cheng et al., 2011, 2015; Ria et al., 2011; Feng et al., 2013; Huang et al., 2014, 2016; Jiang et al., 2019; Huang) were included for quality assessment and evaluation of the Hardy-Weinberg equilibrium (HWE) (Thakkinstian et al., 2005). Finally, the 11 eligible articles containing nine studies of rs3861950 T > C and nine studies of rs1234313 A > G were included based on NOS scores >5 (Supplementary Table 3). The overview of the study selection and exclusion process based on the PRISMA diagram is shown in Figure 1, and the main characteristics of the eligible studies are shown in Table 1.
Overall and Subgroup Meta-Analyses
Nine studies with 7,395 cases and 5,296 controls for rs3861950 T > C and nine studies with 6,951 cases and 4,959 controls for rs1234313 A > G were analyzed. The overall and subgroup meta-analyses based on ethnicity and type of disease of the association between the two TNFSF4 SNPs and CAD risk were performed in allelic, dominant, recessive, heterozygous, and homozygous models. The results were summarized in Table 3.
Association Between TNFSF4 rs3861950 T > C Polymorphism and CAD Risk
Six and three studies in the Asian and Caucasian populations, respectively, were included for rs3861950 T > C. Our overall analysis showed no significant correlation between the TNFSF4 rs3861950 T > C polymorphism and CAD risk under the five genetic models (Table 2). However, we observed an increased risk of CAD risk in the Asian population in the allelic model (C vs. T, OR = 1.33, 95% CI: 1.04–1.70, p = 0.021), dominant model (CCCT vs. TT, OR = 1.34, 95% CI: 1.00–1.78, p = 0.046) and homozygous model (CC vs. TT, OR = 2.07, 95% CI: 1.12–3.81, p = 0.020), but not in the Caucasian population (Figure 2A and Table 2). Furthermore, subgroup analysis based on types of disease demonstrated that in TNFSF4 rs3861950 T > C polymorphism there is a robust correlation with increased risk of cerebral infarction (CI) in the allelic model (C vs. T, OR = 1.63, 95% CI: 1.33–1.99, p = 0.000), dominant model (CCCT vs. TT, OR = 1.55, 95% CI: 1.22–1.96, p = 0.000), heterozygous model (CT vs. TT, OR = 1.35, 95% CI: 1.06–1.74, p = 0.017) and homozygous model (CC vs. TT, OR = 3.42, 95% CI: 1.91–6.12, p = 0.000) (Figure 2B and Table 2). These results indicated that TNFSF4 rs3861950 T > C polymorphism was a risk factor for CAD, specifically in the Asian population, and this SNP increases cerebral infarction susceptibility.
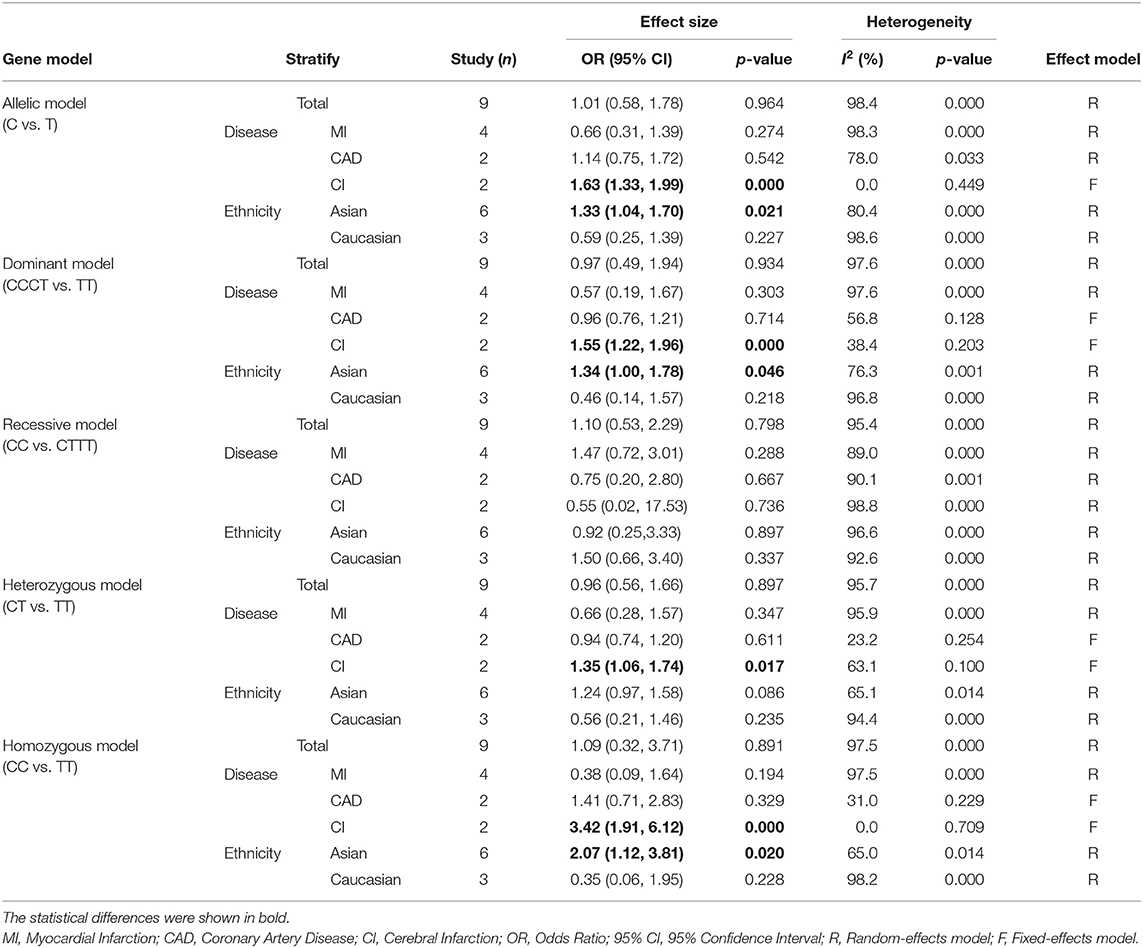
Table 2. Overall and stratified analyses of the association between TNFSF4 rs3861950 T > C and coronary artery disease risk.
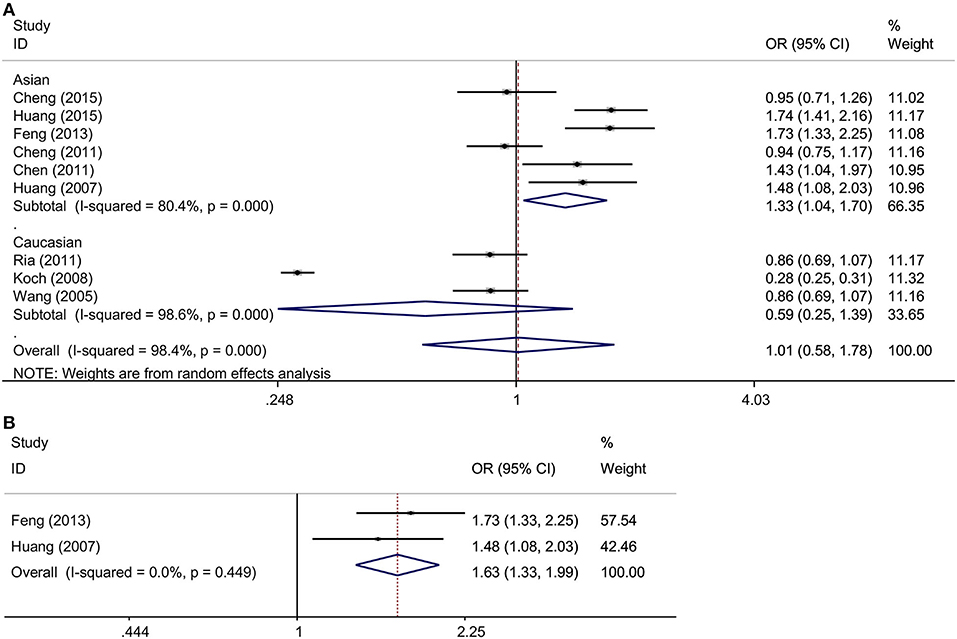
Figure 2. (A) ORs with their 95% CI for the association between TNFSF4 rs3861950 T>C polymorphism and the susceptibility to CAD in the overall population stratified by ethnicity in the random-effects model (allelic model: C vs. T); (B) ORs with their 95% CI for the association between TNFSF4 rs3861950 T>C polymorphism and the susceptibility to CI in the fixed-effects model (allelic model: C vs. T).
Association Between TNFSF4 rs1234313 A > G Polymorphism and CAD Risk
Overall, this meta-analysis revealed a significant association between TNFSF4 rs1234313 A > G polymorphism and decreased risk of CAD in the allelic model (G vs. A, OR = 0.94, 95% CI: 0.89–1.00, p = 0.034) and dominant model (GGGA vs. AA, OR = 0.93, 95% CI: 0.86–1.00, p = 0.049) (Figure 3A and Table 3). Additionally, it showed that this SNP was a protective factor in the Caucasian population in the dominant model (GGGA vs. AA, OR = 0.89, 95% CI: 0.80–1.00, p = 0.045) and heterozygous model (GA vs. AA, OR = 0.89, 95% CI: 0.79–1.00, p = 0.048), but not in the Asian model (Figure 3B and Table 3). Subgroup analysis based on types of disease showed that this SNP was also a favorable marker for decreased risk of myocardial infarction (MI) in the allelic model (G vs. A, OR = 0.91, 95% CI: 0.84–0.98, p = 0.018), dominant model (GGGA vs. AA, OR = 0.88, 95% CI: 0.79–0.97, p = 0.012) and heterozygous model (GA vs. AA, OR = 0.88, 95% CI: 0.79–0.98, p = 0.019) (Figure 3A and Table 3). These results indicated that TNFSF4 rs1234313 A > G polymorphism was a protective factor for CAD risk.
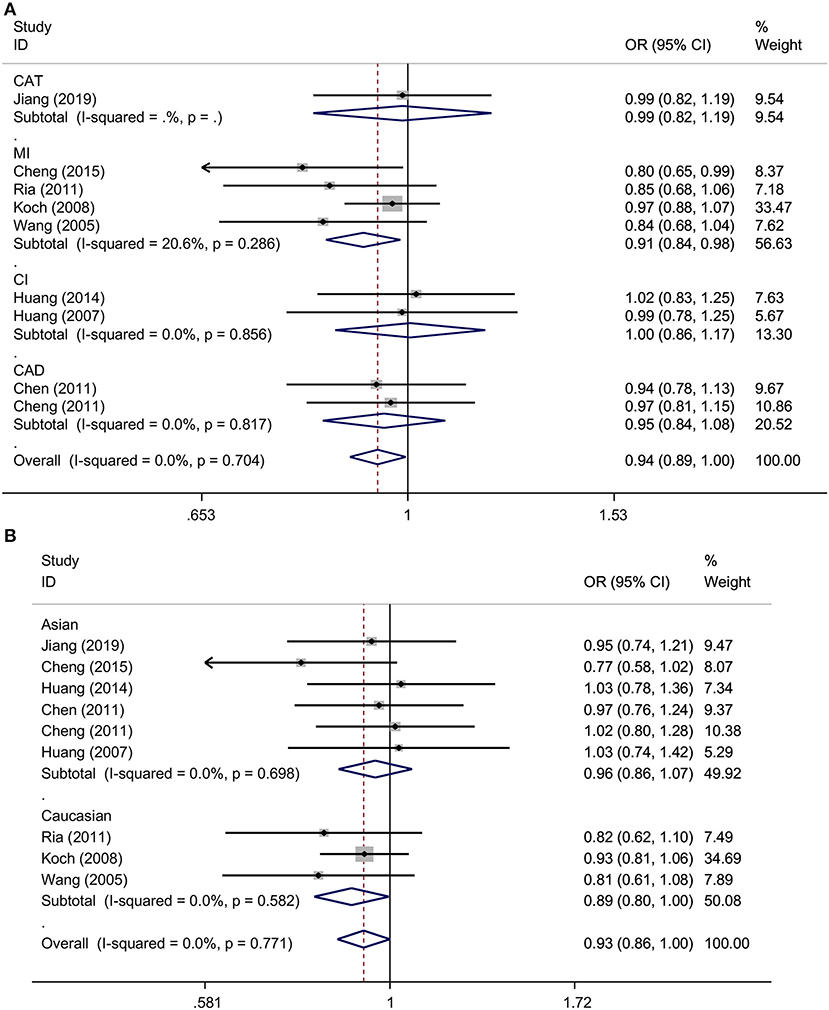
Figure 3. ORs with their 95% CI for the association between TNFSF4 rs1234313 A>G polymorphism and the susceptibility to CAD in the overall population stratified by disease (allelic model: G vs. A) (A) and ethnicity (dominant model: GGGA vs. AA) (B) both in the fixed-effects model.
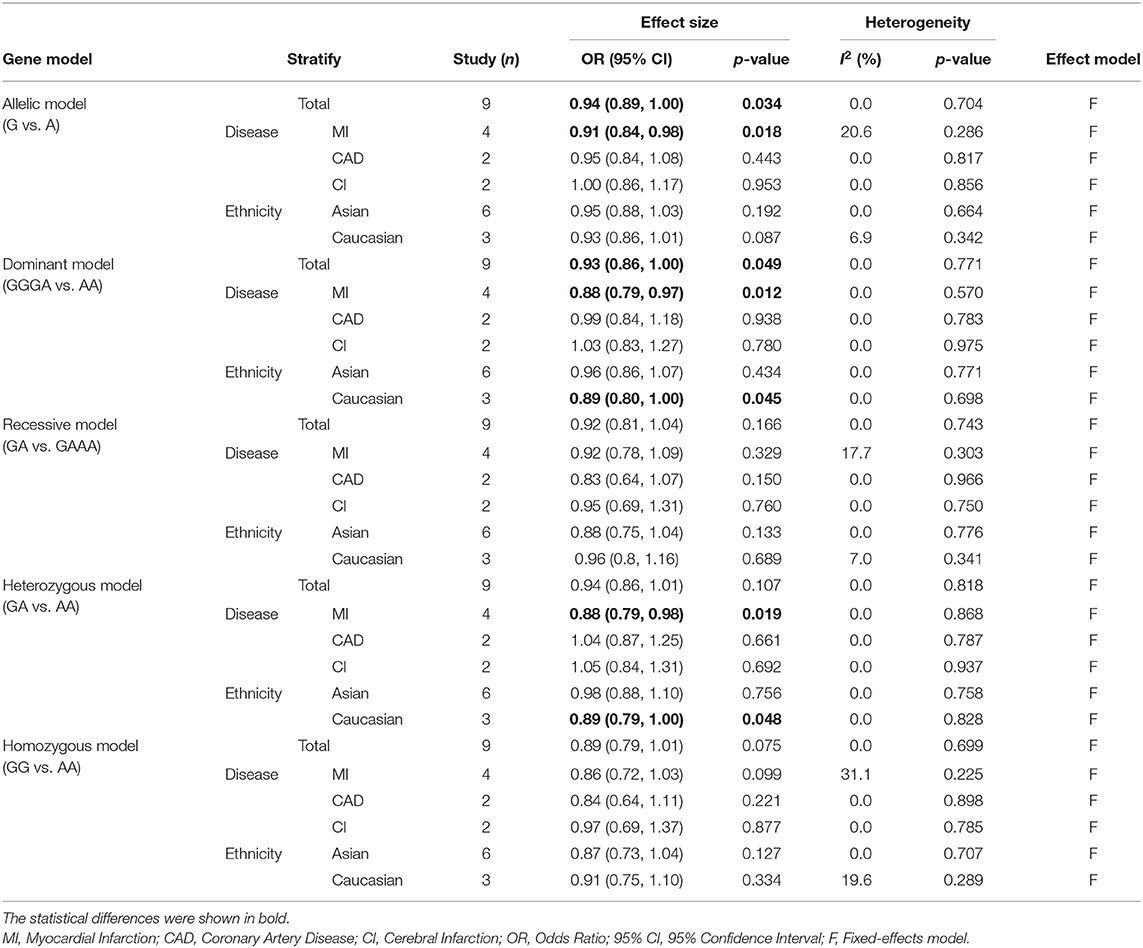
Table 3. Overall and stratified analyses of the association between TNFSF4 rs1234313 A > G and coronary artery disease risk.
Evaluation of Heterogeneity and Sensitivity Analysis
No heterogeneity was observed for rs1234313 A > G polymorphism in the five genetic models analyzed in our meta-analysis (Table 3). However, although a significant association between rs3861950 T > C polymorphism and CAD risk was established under the allelic, dominant, and homozygote models in the Asian population, a large heterogeneity among studies was also observed (Table 2). Therefore, a meta-regression analysis was carried out to observe the source of heterogeneity in the general variables using data from the allelic model under the random-effects model. However, our results showed that the year of publication, population ethnicity, type of disease, genotype methods, source of controls, and the case sample size were not all associated with the large heterogeneity (p > 0.05). The sensitivity analysis was next conducted to determine the stability of the results. This process involved omitting one study at a time to obtain a further pooled ORs. Our results showed a significant association between rs3861950 T > C polymorphism and CAD risk remained constant in the Asian population under the homozygote model in the random-effects model (Figure 4A), which demonstrated the robustness of the results. The stability of the results was established from the sensitivity analysis of the rs1234313 A > G polymorphism in the overall population under the allelic model in the fixed-effects model (Figure 4B).
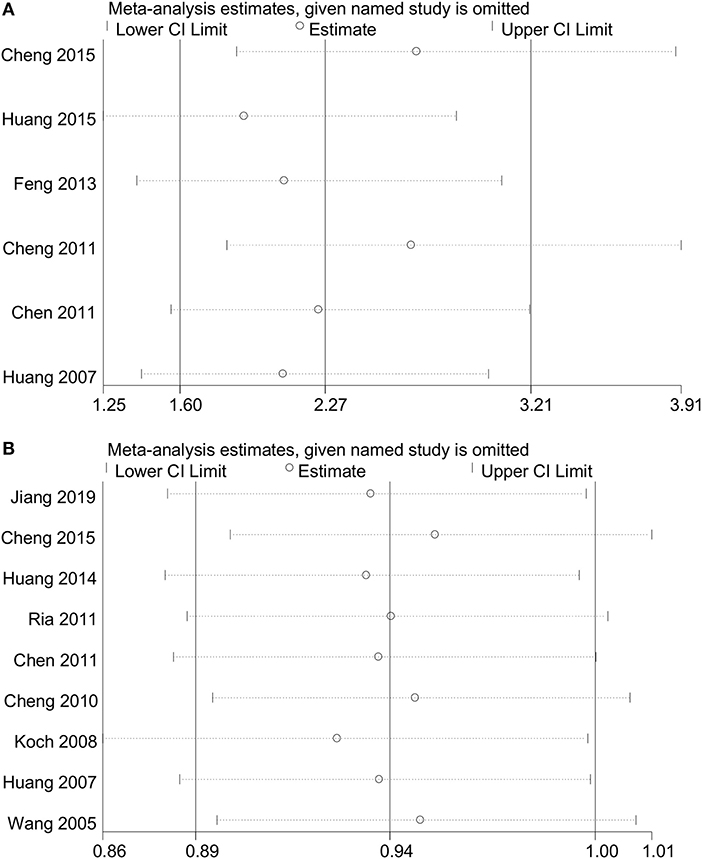
Figure 4. Sensitivity analyses for TNFSF4 rs3861950 T>C (A) and rs1234313 A>G (B) polymorphisms respectively, in the Asian population (homozygous model: CC vs. TT) in random-effects model and overall (allelic model: G vs. A) population in the fixed-effects model.
Publication Bias
The Egger's test and Begg's funnel plot were used to assess the publication bias of the studies involved in this meta-analysis for rs3861950 T > C and rs1234313 A > G polymorphisms (Begg and Mazumdar, 1994; Egger, 1997). The results showed that there was no statistically significant evidence of publication bias for rs3861950 T > C (Egger's test: t = −0.00, p = 0.996>0.05; Begg's test: z = 0.38; p = 0.707>0.05) (Figures 5A,B) in the Asian population and rs1234313 A > G (Egger's test: t = −1.16, p = 0.285>0.05; Begg's test: z = 0.94; p = 0.348>0.05; Figures 5C,D) in the overall population.
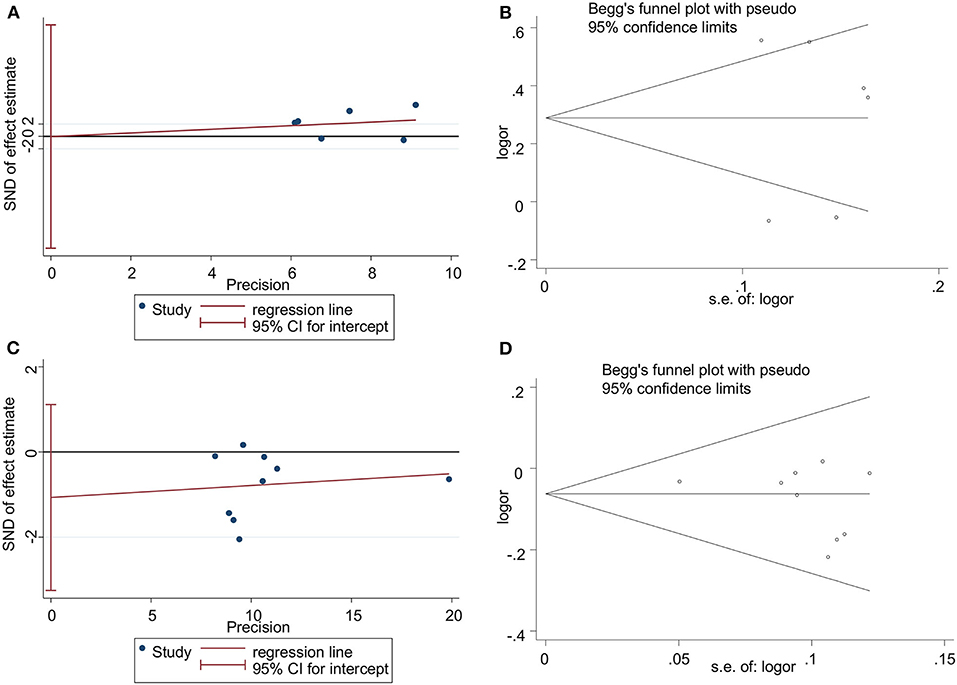
Figure 5. Publication bias analyzed by Egger's regression and Begg's funnel plot for TNFSF4 rs3861950 T>C (A,B) and rs1234313 A>G (C,D) polymorphisms, respectively, in the Asian population (allelic model: C vs. T) in random-effects model and overall population (allelic model: G vs. A) in fixed-effects model.
Discussion
Because CAD is a complex metabolism-related disease, multiple separate factors should be considered for the treatment of CAD in different geographical regions. Gene sequencing studies have identified many promising biomarkers for CAD. For example, the inactivating mutant of LDLR (Do et al., 2015), LPL (Nioi et al., 2016), and APOA5 (Do et al., 2015) has been conferred to increase the risk of CAD. In contrast, PCSK9 (Cohen et al., 2006), NPC1L1, APOC3 (Jørgensen et al., 2014), and ANGPTL4 (Dewey et al., 2016) inactivating mutants have been reported to decreased CAD susceptibility. Drug therapy for specific gene mimics such as Alirocumab, Evolucumab (Cohen et al., 2006), and Ezetimibe (Nioi et al., 2016) have been approved by the Food Drug Administration (FDA) and the European Medicine Agency (EMA), but these drugs may have side effects and have shown partial toxicity in animal models (Desai et al., 2007; Dewey et al., 2016). Therefore, additional systematic analyses of the genetic function are needed to identify alternative biomarkers for diagnosis and drug development of CAD.
Previous studies revealed that TNFSF4 expression was higher in atherosclerotic lesions, and both in vivo and in vitro studies found that SIRT6 decreased TNFSF4 gene expression by binding to and deacetylating the H3K9 at TNFSF4 gene promoter and had a protective effect against endothelial dysfunction and atherosclerosis (Van Wanrooij et al., 2007; Xu et al., 2016), which provoked us to consider whether TNFSF4 expression and the SNPs in TNFSF4 are closely correlated with CAD risk. TNFSF4 rs3861950 T > C is located in the 2nd intron prior to the 3rd exon and rs1234313 A > G is located in the region of 1st intron between the 2nd and 3rd exon (Baum et al., 1994). Even though the location of the two SNPs are in the non-coding intron region, the bioinformatics analysis performed by mining the GTEx (https://gtexportal.org/home/) and HaploReg v4 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) databases showed that rs3861950 had little effect on TNFSF4 expression. However, eQTL studies showed rs1234313 may have the potential to promoted the cis expression of TNFSF4 via enhancer epigenetic regulation, but the mechanistic details need further experimental validation (Ward and Kellis, 2012). Wang et al. have assessed the associations between rs3861950 T > C and rs1234313 A > G polymorphisms with CAD risk (Wang et al., 2019). Six studies were evaluated in Wang's meta-analysis for rs3861950 T > C (Wang et al., 2005; Koch et al., 2008; Chen et al., 2011; Cheng et al., 2011, 2015; Feng et al., 2013) and another six for rs1234313 A > G (Wang et al., 2005; Koch et al., 2008; Chen et al., 2011; Cheng et al., 2011, 2015; Huang et al., 2014, 2016). However, sufficient data was only available in 4 studies, both for rs3861950 T > C (Koch et al., 2008; Chen et al., 2011; Cheng et al., 2011; Feng et al., 2013) and rs1234313 A > G (Koch et al., 2008; Chen et al., 2011; Cheng et al., 2011; Huang et al., 2014). Therefore, it was necessary to reassess their associations using a greater number of studies. The conclusions in our meta-analysis based on additional available studies revealed that rs3861950 T > C increased the risk of CAD in the Asian population under allelic, dominant, and homozygous models (Figure 2A and Table 2). In addition, analyses according to disease type showed that it could also contribute to CI risk (Figure 2B and Table 2). These results indicated that rs3861950 T>C polymorphism might be a specific risk factor in the Asian population. The genetic function of rs3861950 and the difference between Asians and other ethnicities is worth investigating further. The meta-analysis of rs1234313 A>G polymorphism in this study revealed that this SNP is significantly associated with a decreased risk of CAD. This result indicated that the rs1234313 A>G polymorphism might serve as a novel clinical biomarker for CAD diagnosis.
Furthermore, the rs1234313 A>G polymorphism was sensitive to myocardial infarction (MI) susceptibility, and ethnic subgroup analysis obtained from present data showed that this SNP might be a more promising biomarker in Caucasians than in Asians. In summary, this meta-analysis revealed that two novel SNPs in TNFSF4 have a robust association with CAD susceptibility in different races, which may lay a foundation for precise CAD diagnosis, and provided theoretical support for personalized CAD treatment. Nevertheless, there were still some limitations that need to be addressed in this meta-analysis. First, the number of studies for each SNP in the different regions included was not abundant, and more independent studies were needed to evaluate our conclusion. The difference between ethnicities was smaller due to the limited number of studies included. Second, the number of studies for certain types of disease was not enough to perform further impartial analysis. Third, the effects of gender, age, smoking status, lifestyle, and other environmental factors on CAD were not considered in this meta-analysis due to the limitations or unavailable data. Moreover, the genotyping methods for SNPs identification were also inconsistent; all methods were based on PCR, such as PCR-RFLP, Taqman, and PCR-LDR, and each method had its own merits and limitations, so more appropriate methods and guidelines for SNPs identification needed to be discussed. Fourth, significant heterogeneity among studies on the association between rs3861950 and CAD risk in the Asian population was observed. However, the source of heterogeneity was not found in the meta-regression analysis, but the sensitivity analysis confirmed a robust association between the two. Therefore, more case-control studies with larger sample sizes and more available data are necessary to obtain a more defined conclusion in the future.
Conclusion
Herein, this meta-analysis identified two novel gene polymorphisms of TNFSF4, rs3861950, and rs1234313, which may affect the predisposition to CAD in different genetic models, suggesting that these two SNPs of TNFSF4 may potentially be used as biomarkers for CAD diagnosis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
SL and XW: data curation. SL, XW, MY, and YP: formal analysis. ZX: funding acquisition. SY: methodology. GZ and ZX: supervision. MY and YP: validation. SL: writing—original draft. SY, GZ, and ZX: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants (81902710 to ZX) from National Natural Science Foundation of China and grants (2019KYQD11 to ZX) from Binzhou Medical University and we also are supported from Major Basic Research Project of Shandong Provincial Natural Science Foundation (ZR2019ZD27), Shandong Province Higher Educational Youth Innovation Science and Technology Program (2019KJE013), and Yantai Development Project of University and Government Integration (2019XDRHXMPT14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.539288/full#supplementary-material
References
Baum, P. R., Ramsdell, F., Srinivasan, S., Sorensen, R. A., Watson, M. L., Seldin, M. F., et al. (1994). Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 13, 3992–4001. doi: 10.1002/j.1460-2075.1994.tb06715.x
Begg, C. B., and Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101. doi: 10.2307/2533446
Bowden, J., Tierney, J. F., Copas, A. J., and Burdett, S. (2011). Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med. Res. Methodol. 11:41. doi: 10.1186/1471-2288-11-41
Chen, M. Z., Cheng, G. H., Ma, L., Wang, H., Qiu, R. F., Xue, F. Z., et al. (2011). Association study between TNFSF4 and coronary heart disease. Yi Chuan 33, 239–245. doi: 10.3724/SP.J.1005.2011.00239
Cheng, G., Wang, H., Chen, M., Li, L., Gong, Y., and Liu, Q. (2011). Lack of evidence to support the association of polymorphisms within the TNFSF4 gene and coronary heart disease in a Chinese Han population. Exp. Ther. Med. 2, 275–280. doi: 10.3892/etm.2010.188
Cheng, J., Cen, J. M., Cai, M. Y., Xu, S., Li, L., Li, Z. C., et al. (2015). Association between TNFSF4 tagSNPs and myocardial infarction in a Chinese Han population. Genet. Mol. Res. 14, 6136–6145. doi: 10.4238/2015.June.8.11
Christodoulidis, G., Vittorio, T. J., Fudim, M., Lerakis, S., and Kosmas, C. E. (2014). Inflammation in coronary artery disease. Cardiol. Rev. 22, 279–288. doi: 10.1097/CRD.0000000000000006
Cohen, J. C., Boerwinkle, E., Mosley, T. H., and Hobbs, H. H. (2006). Sequence variations in PCSK9, Low LDL and protection against coronary heart disease. N Engl. J. Med. 354, 1264–1272. doi: 10.1056/NEJMoa054013
Desai, U., Lee, E.-C., Chung, K., Gao, C., Gay, J., Key, B., et al. (2007). Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 104, 11766–11771. doi: 10.1073/pnas.0705041104
Dewey, F. E., Gusarova, V., O'Dushlaine, C., Gottesman, O., Trejos, J., Hunt, C., et al. (2016). Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl. J. Med. 374, 1123–1133. doi: 10.1056/NEJMoa1510926
Do, R., Stitziel, N. O., Won, H. H., Jorgensen, A. B., Duga, S., Merlini, P. A., et al. (2015). Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 518, 102–106. doi: 10.1038/nature13917
Egger, M. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634. doi: 10.1136/bmj.315.7109.629
Feng, J., Liu, Y. H., Yang, Q. D., Zhu, Z. H., Xia, K., Tan, X. L., et al. (2013). TNFSF4 gene polymorphism rs3861950 but not rs3850641 is associated with the risk of cerebral infarction in a Chinese population. J. Thromb. Thrombolysis 36, 307–313. doi: 10.1007/s11239-012-0849-9
Huang, Q., Liu, X., Feng, J., Wen, Y., and Liu, Y. (2016). Association between tumor necrosis factor superfamily member 4 gene polymorphism and risk of asymptomatic carotid vulnerable plaque in a Chinese population. Zhonghua Liu Xing Bing Xue Za Zhi 36, 998–1001.
Huang, Q., Yang, Q. D., Tan, X. L., Feng, J., Tang, T., Xia, J., et al. (2014). Absence of association between atherosclerotic cerebral infarction and TNFSF4/TNFRSF4 single nucleotide polymorphisms rs1234313, rs1234314 and rs17568 in a Chinese population. J. Int. Med. Res. 42, 436–443. doi: 10.1177/0300060514521154
Huang, Q. The study on the association between TNFSF4/TNFRSF4 gene and cerebral infarction (doctoral dissertation), Central South University, Changsha, China.
Jiang, Y., Liu, X., Du, Y., and Zhou, S. (2019). rs1234313 and rs45454293 are risk factors of cerebral arterial thrombosis, large artery atherosclerosis, and carotid plaque in the Han Chinese population: a case-control study. BMC Neurol. 19:31. doi: 10.1186/s12883-019-1259-9
Jørgensen, A. B., Frikke-Schmidt, R., Nordestgaard, B. G., and Tybjærg-Hansen, A. (2014). Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. J. Vasc. Surg. 371:1096. doi: 10.1016/j.jvs.2014.08.087
Kaur, D., and Brightling, C. (2012). OX40/OX40 ligand interactions in T-cell regulation and asthma. Chest 141, 494–499. doi: 10.1378/chest.11-1730
Kessler, T., Vilne, B., and Schunkert, H. (2016). The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol. Med. 8, 688–701. doi: 10.15252/emmm.201506174
Khera, A. V., and Kathiresan, S. (2017). Genetics of coronary artery disease: discovery, biology and clinical translation. Nat. Rev. Genet. 18, 331–344. doi: 10.1038/nrg.2016.160
Koch, W., Hoppmann, P., Mueller, J. C., Schomig, A., and Kastrati, A. (2008). Lack of support for association between common variation in TNFSF4 and myocardial infarction in a German population. Nat. Genet. 40, 1386–1387. doi: 10.1038/ng1208-1386
Marenberg, M. E., Risch, N., Berkman, L. F., Floderus, B., and de Faire, U. (1995). Genetic susceptibility to death from coronary heart disease in a study of twins. J. Occupat. Environ. Med. 330, 1041–1046. doi: 10.1056/NEJM199404143301503
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Nioi, P., Sigurdsson, A., Thorleifsson, G., Helgason, H., and Stefansson, K. (2016). Variant ASGR1 associated with a reduced risk of coronary artery disease. N Engl. J. Med. 374:2131. doi: 10.1056/NEJMoa1508419
Ria, M., Lagercrantz, J., Samnegård, A., Boquist, S., Hamsten, A., and Eriksson, P. (2011). A common polymorphism in the promoter region of the TNFSF4 gene is associated with lower allele-specific expression and risk of myocardial infarction. PLoS ONE 6:e17652. doi: 10.1371/journal.pone.0017652
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605. doi: 10.1007/s10654-010-9491-z
Thakkinstian, A., McElduff, P., D'Este, C., Duffy, D., and Attia, J. (2005). A method for meta-analysis of molecular association studies. Stat. Med. 24, 1291–1306. doi: 10.1002/sim.2010
Van Wanrooij, E. J. A., van Puijvelde, G. H. M., de Vos, P., Yagita, H., va Berkel, T. J. C., and Kuiper, J. (2007). Interruption of the Tnfrsf4/Tnfsf4 (OX40/OX40L) pathway attenuates atherogenesis in low-density lipoprotein receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 27, 204–210. doi: 10.1161/01.ATV.0000251007.07648.81
Wang, X., Luan, Y., and Zhang, C. (2019). A meta-analysis on correlations of OX40L variants with atherosclerotic disorders. J. Cell. Biochem. 120, 9624–9630. doi: 10.1002/jcb.28240
Wang, X., Ria, M., Kelmenson, P. M., Eriksson, P., Higgins, D. C., Samnegård, A., et al. (2005). Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 37, 365–372. doi: 10.1038/ng1524
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. doi: 10.1093/nar/gkr917
Keywords: polymorphism, coronary artery disease, meta-analysis, susceptibility, TNFSF4
Citation: Liu S, Wang X, Yu S, Yan M, Peng Y, Zhang G and Xu Z (2020) A Meta-Analysis on the Association Between TNFSF4 Polymorphisms (rs3861950 T > C and rs1234313 A > G) and Susceptibility to Coronary Artery Disease. Front. Physiol. 11:539288. doi: 10.3389/fphys.2020.539288
Received: 10 March 2020; Accepted: 17 September 2020;
Published: 26 November 2020.
Edited by:
John D. Imig, Medical College of Wisconsin, United StatesReviewed by:
Suowen Xu, University of Science and Technology of China, ChinaXiang Xie, Xinjiang Medical University, China
Jian Xu, Nanjing Medical University, China
Copyright © 2020 Liu, Wang, Yu, Yan, Peng, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guilong Zhang, Z2x6aGFuZ0Biem1jLmVkdS5jbg==; Zhaowei Xu, emhhb3dlaXh2QGJ6bWMuZWR1LmNu
†These authors have contributed equally to this work
 Shuyan Liu1†
Shuyan Liu1† Zhaowei Xu
Zhaowei Xu