
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 15 April 2020
Sec. Invertebrate Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.00320
Invading pathogens are recognized by peptidoglycan recognition proteins (PGRPs) that induce translocation of NF-κB transcription proteins and expression of robust antimicrobial peptides (AMPs). Tenebrio molitor PGRP-LE (TmPGRP-LE) has been previously identified as a key sensor of Listeria monocytogenes infection. Here, we present that TmPGRP-LE is highly expressed in the gut of T. molitor larvae and 5-day-old adults in the absence of microbial infection. In response to Escherichia coli and Candida albicans infections, TmPGRP-LE mRNA levels are significantly upregulated in both the fat body and gut. Silencing of TmPGRP-LE by RNAi rendered T. molitor significantly more susceptible to challenge by E. coli infection and, to a lesser extent, Staphylococcus aureus and C. albicans infections. Reduction of TmPGRP-LE levels in the larval gut resulted in downregulation of eight AMP genes following exposure to E. coli, S. aureus, and C. albicans. However, the transcriptional levels of AMPs more rapidly reached a higher level in the dsEGFP-treated larval gut after challenge with E. coli, which may suggest that AMPs induction were more sensitive to E. coli than S. aureus and C. albicans. In addition, TmPGRP-LE RNAi following E. coli and C. albicans challenges had notable effects on TmRelish, TmDorsal X1 isoform (TmDorX1), and TmDorX2 expression level in the fat body and gut. Taken together, TmPGRP-LE acts as an important gut microbial sensor that induces AMPs via Imd activation in response to E. coli, whereas involvement of TmPGRP-LE in AMPs synthesize is barely perceptible in the hemocytes and fat body.
Invading infectious agents stimulate the innate immune system through complex intracellular pathways. To successfully combat invading pathogens, host immune cells express germline-encoded pathogen recognition receptors (PRRs) that recognize evolutionarily conserved molecular targets, known as pathogen-associated molecular patterns (PAMPs) (Kumar et al., 2011).
There is compelling evidence that innate immunity has ancient evolutionary origins in vertebrates and invertebrates. Phylogenetic studies of innate immune-related genes have revealed that mammals and insects share highly conserved generalized signaling components, which allows insects to be used as models for host-pathogen interaction studies (Ausubel, 2005). However, there are some considerable differences in the innate immune systems of vertebrates and invertebrates, including the mechanism of interaction between terminal molecules and receptors, the signal transduction pathways involved, and the effect of signaling pathway activation on the secretion or production of specific molecules (Sheehan et al., 2018). Mammalian Toll-like receptor (TLR) and tumor necrosis factor alpha (TNFα) signaling pathways are activated in response to invading microorganisms and this results in the translocation of NF-κB transcription factors to the nucleus, which stimulate the production of pro-inflammatory cytokines and chemokines, induce apoptosis, and connect innate and adaptive immunities (Ganesan et al., 2011; Ting and Bertrand, 2016). Insect NF-κB, Toll, and immune deficiency (Imd) signaling pathways, however, induce production of antimicrobial peptides (AMPs) (Lemaitre and Hoffmann, 2007).
The Drosophila Imd pathway (a homolog of the mammalian TNF pathway) is predominantly activated through the recognition of meso-diaminopimelic acid-containing peptidoglycans (DAP-type PGN) of gram-negative bacteria and some gram-positive bacteria (Bacillus and Listeria spp.) by peptidoglycan-recognition proteins (PGRPs) (Lemaitre and Hoffmann, 2007). The periplasmic murein (PGN) sacculus, which is composed of long glycan chains cross-linked by peptides, is the only cell surface component unique to bacteria, and is common to both gram-positive and gram-negative bacteria (Vollmer and Bertsche, 2008).
PGRP family members are evolutionarily conserved components of the innate immune system, with variants in insects and mammals sharing one or more C-terminal PGRP domain (around 165 amino acids) that is homologous to bacteriophage type 2 amidases and T7 lysozymes (Kang et al., 1998; Dziarski, 2004). However, some PGRPs do not have amidase activity, seemingly due to a reduction in number of amino acid residues necessary for cleavage and degradation of PGN in the presence of zinc (Wang et al., 2019).
The 19-kDa PGRP protein was first discovered in the hemolymph and cuticle of the silkworm, Bombyx mori (Yoshida et al., 1996). Since then, more than 100 PGRP orthologs have been identified in vertebrate and invertebrate species, including Trichoplusia ni (moth) (Kang et al., 1998), Mus musculus (mouse) (Kang et al., 1998), Rattus norvegicus (Rat) (Rehman et al., 2001), Drosophila melanogaster (Werner et al., 2000, 2003), Anopheles gambiae (mosquito) (Christophides et al., 2002), different fishes (Jang et al., 2013; Sun et al., 2014; Sun and Sun, 2015; Zhang et al., 2016), and humans (Liu et al., 2001). However, PGRP genes are not present in plants and nematodes (Dziarski and Gupta, 2006).
PGRPs can be separated into three categories based on general structure and transcript size: short extracellular PGRPs (PGRP-S, molecular weight of 20–25 kDa), intermediate PGRPs (PGRP-I, 40–45 kDa), and long intracellular, extracellular, or transmembrane PGRPs (PGRP-L, up to 90 kDa) (Dziarski, 2004; Mao et al., 2019). Previous studies have revealed a relatively high number of PGRP genes in insects (e.g., 13 genes in Drosophila) compared to mammals (only four PGRP genes). PGRP members can be further categorized into two groups: catalytic PGRPs and sensor PGRPs (non-catalytic). The former shares a conserved three-amino-acid structure critical for hydrolyzing PGN through cleavage of the amide bond between MurNAc and L-Ala (Mellroth et al., 2003; Guan et al., 2004). The latter does not cleave PGN due to a missing cysteine residue, but members of this group can bind to PNG, inducing Toll and Imd signaling pathways that lead to the production of AMPs (Table 1; Guan and Mariuzza, 2007; Wang et al., 2019).
In D. melanogaster, PGRP-LE (DmPGRP-LE) is a long, secreted PGRP that acts as an intracellular and extracellular microbial sensor. DmPGRP-LE recognizes the polymeric DAP-type PGN and tracheal cytotoxin (TCT, a monomeric DAP-type PGN) from gram-negative bacteria and Listeria monocytogenes. Binding of the ligand triggers independent activation of the Imd pathway and induces autophagy (Kaneko et al., 2006; Yano et al., 2008). Following L. monocytogenes challenge, stimulation of the Janus kinase-signal transducers and activators of transcription (JAK-STAT) pathway results in Listericin induction, which is cooperatively regulated by DmPGRP-LE (Goto et al., 2010). After PGRP-LE binds to DAP-type PGN in the cytoplasm of immune cells, it interacts synergistically with PGRP-LC, which in-turn activates the receptor multimerization that is required for signal transduction in the Imd pathway. Moreover, overexpression of PGRP-LE and PGRP-LC leads to activation of the prophenoloxidase cascade (Takehana et al., 2004; Kurata, 2014). Exploration of the D. melanogaster-Photorhabdus model has highlighted the importance of DmPGRP-LE in the hemolymph of flies during immune response to Photorhabdus luminescens and Photorhabdus asymbiotica, but not to non-pathogenic Escherichia coli (Chevée et al., 2019). Interestingly, PGRP-LE knockdown in the pupae of the red flour beetle, Tribolium castaneum, has been shown to decrease the expression of Defensin3 following E. coli challenge, suggesting that TcPGRP-LE has independent activity against gram-negative bacteria (Koyama et al., 2015).
In a related study, the immune significance of Tenebrio molitor PGRP-SA (TmPGRP-SA) and TmPGRP-SC2 proteins was demonstrated (Table 1; Yu et al., 2010). Our team has previously identified the TmPGRP-LE protein (37.3 kDa) as a non-catalytic PGRP that is essential for larval survival upon L. monocytogenes infection (Tindwa et al., 2013). Taking into account the known biological differences in immune responses between Tenebrio and Drosophila, the focus of this study was to investigate the role of TmPGRP-LE as a multifunctional component of the Imd pathway, specifically as a fine regulator of key immune-related genes (including AMP genes) in the fat body, gut, and hemocytes of Tenebrio molitor following challenges with E. coli, Staphylococcus aureus, or Candida albicans.
Stocks were raised in an insectarium on an artificial diet at 27 ± 1°C and 60 ± 5% relative humidity in the dark, in accordance with a previous study (Keshavarz et al., 2019). Healthy, normal larvae at the 10th–12th instar (around 2.4 cm in length) were used for experiments (n = 20 per group).
Evidence of the constitutive expression of TmPGRP-LE during the developmental stages (Tindwa et al., 2013) prompted us to elucidate the tissue-specific pattern of TmPGRP-LE expression in the integument, fat body, hemocytes, gut, Malpighian tubules, ovary, and testes of larvae and adults.
Total RNA was extracted from all tissue samples using the LogSpin RNA isolation method with minor modifications (Yaffe et al., 2012). Briefly, the samples were homogenized in guanidine thiocyanate-based RNA lysis buffer mixed with 99% ethanol and transferred to silica spin columns (Bioneer, Korea, KA-0133-1). The resultant RNA was treated with DNase (Promega, M6101, United States) to eliminate the genomic DNA contamination for 15 min at 37°C. Preceding all steps each silica column was then washed twice with the supplied wash buffers using 3 M sodium acetate buffer and 80% ethanol and dried for 1 min. Total RNA was eluted with 30 μL of distilled water (Sigma, W4502-1L, United States). Next, 2 μg of total RNAs were converted to cDNA using the AccuPower® RT PreMix (Bioneer, Korea) kit with an oligo-(dT) 12–18 primer according to the protocol recommended by the manufacturer. The synthesized cDNAs (1:20 dilution with DNase/RNase free water) processed for quantitative reverse-transcription PCR (qRT-PCR) using AccuPower® 2X GreenStar qPCR Master Mix (Bioneer, Korea). Data was normalized to T. molitor 60 S ribosomal protein L27a (TmL27a) transcripts and values were calculated using the comparative CT method (2–ΔΔCT method) (Schmittgen and Livak, 2008). Specific primers for qRT-PCR were designed using Primer 3.0 plus1 and are listed in Table 2. For detailed information, including PCR conditions, see our previous paper (Keshavarz et al., 2019).
The gram-negative bacterium E. coli (starin K12) and gram-positive bacterium S. aureus (strain RN4220) were cultured in Luria-Bertani (LB) broth, and the fungus C. albicans was cultivated in Sabouraud dextrose broth at 37°C overnight. Cells from overnight microbial cultures were concentrated by centrifugation at 3,500 rpm for 15 min at room temperature (∼25°C). The supernatant was discarded, after which the pellet was washed (three times) and resuspended in phosphate-buffered saline (PBS; pH 7.0). The cell concentration was determined by optical density (OD) measurement at 600 nm. Based on OD600 values, the microorganism suspensions were adjusted to 106 cells/μL for E. coli and S. aureus, and 5 × 105 cells/μL for C. albicans.
Microbial challenges were conducted on T. molitor larvae (10th–12th instar) by injecting 1 μL of E. coli (1 × 106 cells/μL), S. aureus (1 × 106 cells/μL), C. albicans (5 × 104 cells/μL), or PBS control between the 3rd and 4th abdominal segment. For quantification of TmPGRP-LE mRNA expression, whole insects or dissected immune-related tissues (fat body, gut, and hemocytes) were collected at various time points (at 3, 6, 9, 12, and 24 h post-infection). Subsequently, total RNA extraction, cDNA synthesis, and qRT-PCR were performed as previously described.
Double-stranded RNA for use in RNAi experiments were synthesized as previously described (Tindwa et al., 2013). Concisely, the PCR product (636 bp sequence) tailed with T7 promotor sequence of TmPGRP-LE was amplified using AccuPower® Pfu PCR PreMix with forward (dsTmPGRP-LE_Fw) and reverse (dsTmPGRP-LE_Rv) primers under the following conditions: denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 30 s, and extension at 72°C for 5 min (Table 2). The synthesized dsTmPGRP-LE was purified using an AccuPrep® PCR Purification Kit (Bioneer, Korea). Subsequently, the purified product was used as a template to synthesize dsTmPGRP-LE in vitro using an EZTM T7 High Yield in vitro Transcription Kit (Enzynomics, Korea) as per the manufacturer’s instructions. Then it was precipitated with 5 M ammonium acetate and washed with 70, 80, and 99.9% ethanol sequentially. After drying at room temperature, the precipitate was resuspended in 30 μL distilled water (Sigma, W4502-1L, United States) to obtain the final product.
An additional dsRNA stock of EGFP (dsEGFP) was generated using the PCR product (546 bp sequence) of the enhanced green fluorescent protein (EGFP) gene derived from the plasmid EGFP-C1. dsEGFP was used as a negative control in subsequent RNAi experiments.
A key question concerning TmPGRP-LE is its role in combatting larval infection; to address this question, we examined the viability of larvae exposed to microbes after silencing TmPGRP-LE expression. However, it is crucial to verify that merely TmPGRP-LE depletion do not affect the survival percent. To quantitatively address this question, we injected 1 μL (1 μg) of dsTmPGRP-LE into 10th–12th instar T. molitor larvae. The knockdown efficiency for the target gene (TmPGRP-LE) was measured on the 6th day post-treatment. After confirmation of silencing, dsRNA-injected larvae (n = 10 per group) were challenged with E. coli (1 × 106 cells/μL), S. aureus (1 × 106 cells/μL), or C. albicans (5 × 104 cells/μL). PBS were injected into dsRNA-injected larvae as a control. Survivors were counted daily for a duration of 10 days. The experiments were repeated three times, with 10 larvae per group for each experiment.
To understand the function of TmPGRP-LE in regulating AMP genes, we examined the gene expression profiles of 14 AMPs, namely TmTenecin-1 (TmTene1), TmTenecin-2 (TmTene2), TmTenecin-3 (TmTene3), TmTenecin-4 (TmTene4), TmAttacin-1a (TmAtt1a), TmAttacin-1b (TmAtt1b), TmAttacin-2 (TmAtt2), TmDefensin-1 (TmDef1), TmDefensin-2 (TmDef2), TmColeoptericin-1 (TmCole1), TmColeoptericin-2 (TmCole2), TmCecropin-2 (TmCec2), TmThaumatin-like protein-1 (TmTLP1), and TmThaumatin-like protein-2 (TmTLP2) in the TmPGRP-LE-silenced larvae after microbial challenges. In addition to AMP gene expression profiling, the mRNA levels of three previously identified transcription factors composed of TmRelish, TmDorsal X1 isoform (TmDorX1), and TmDorsal X2 isoform (TmDorX2) were also measured. dsEGFP was used as a negative control, and PBS served as a wound control. Knowledge of the immunological role of TmPGRP-LE in different tissues is valuable; thus, we dissected the fat body, gut, and hemocytes of experimental samples 24 h post-injection. Each experiment was independently repeated trice (n = 20 per group). Samples were processed for cDNA synthesis, and qRT-PCR analysis was conducted using AMP-specific primers (Table 2).
Three independent biological replicates were used for all experiments. Values were reported as mean ± SE. Differences between groups were analyzed using one-way statistical analysis of variance (ANOVA) and Tukey’s test; p < 0.05 were considered significant. The results for the mortality assay were analyzed using the Kaplan-Meier plot (log-rank Chi-square test) in Excel.2
As an intracellular receptor, cytoplasmic DmPGRP-LE was previously detected in hemocytes and Malpighian tubules. However, as an extracellular PGN receptor, it was previously detected in the fat body and hemocytes; in these tissues, DmPGRP-LE can enhance DmPGRP-LC-mediated recognition of PGN and activate the Imd signaling pathway (Takehana et al., 2004; Kaneko et al., 2006). Similar to other long PGRPs (excluding PGRP-LB), TmPGRP-LE lacks a signal peptide. Thus, it is constitutively expressed in the cytoplasm during all developmental stages and acts as an intracellular scavenger of PGN (Tindwa et al., 2013).
In this study, we employed qRT-PCR to investigate the tissue distribution of TmPGRP-LE transcripts (Figure 1). TmPGRP-LE mRNA was detected in all T. molitor larval tissues, with the highest expression level observed in the gut, followed by that in hemocytes and Malpighian tubules, while the lowest mRNA quantities were found in the integument and fat body (Figure 1A). Similarly, there was high expression of TmPGRP-LE in the gut and hemocytes of 5-day-old adults, with lower expression found in the integuments. The transcription of TmPGRP-LE was weakly detected in the fat body, Malpighian tubules, and ovary. The lowest expression of TmPGRP-LE was found in the testis (Figure 1B).
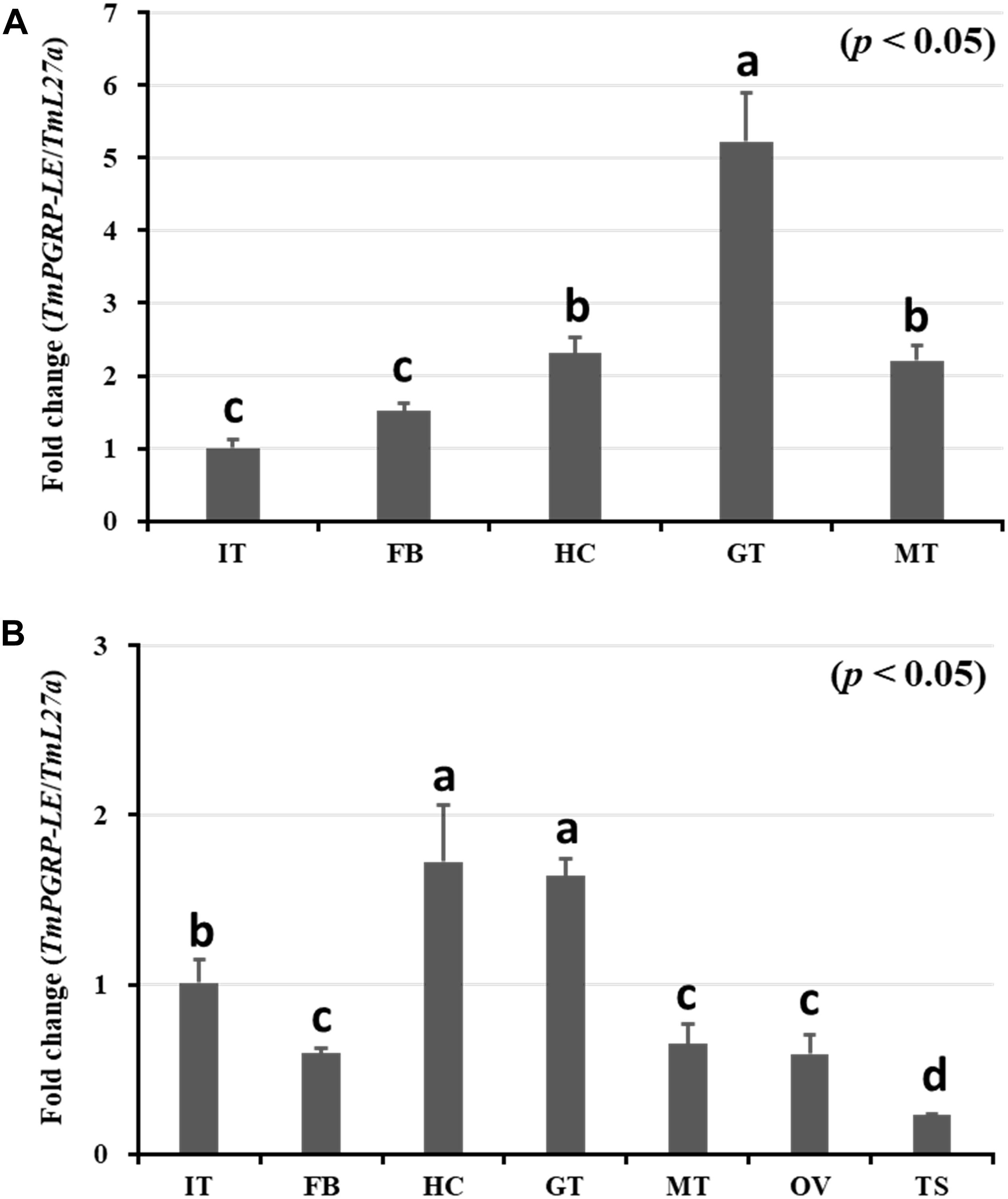
Figure 1. Quantitative determination of TmPGRP-LE expression in different tissues of late-instar T. molitor larvae (A) and 5-day-old adults (B) measured by qRT-PCR. The mRNA levels of TmPGRP-LE in the integument (IT), fat body (FB), hemocytes (HC), gut (GT), and Malpighian tubules (MT) are shown for late-instar larvae (A). TmPGRP-LE transcripts in these tissues, as well as in the ovary (OV) and testis (TS), are shown for 5-day-old adults (B). All measurements are depicted relative to the expression levels of T. molitor 60S ribosomal protein L27a (TmL27a) as an endogenous control. Each vertical bar represents mean ± SE (n = 20 per group). The significant differences between groups were determined using One-way ANOVA and Tukey’s multiple range test at 95% confidence level (p < 0.05). Bars in each graph with the same letter are not significantly different from each other.
The Drosophila PGRP-LE protein serves as a master bacterial peptidoglycan-sensing molecule in the gut that mediates NF-κB-induced responses to infectious pathogens (Bosco-Drayon et al., 2012). Thus, to elucidate how whole organs and tissues of T. molitor (including fat body, gut, and hemocytes) respond to various microbes, we evaluated changes in the transcriptional abundance of TmPGRP-LE after challenging hosts with E. coli, S. aureus, and C. albicans at various time points (3, 6, 9, 12, and 24 h post-infection) (Figure 2). In the whole body, fat body, and gut tissues, E. coli infection led to a gradual but significant upregulation in transcription of TmPGRP-LE, resulting in an up to threefold increase in expression over the PBS-injected controls by 9 h post-infection (p < 0.05). Levels of TmPGRP-LE in the whole body and fat body increased at early time points (3, 6, and 9 h) but did not persist at 12 and 24 h post-infection. Similarly, TmPGRP-LE expression levels were significantly increased in the gut, but there was a slight decrease (p < 0.05) in expression after 9 h (Figure 2A). These results indicate that the T. molitor larval gut responds to infection by E. coli and that the responses involve TmPGRP-LE.
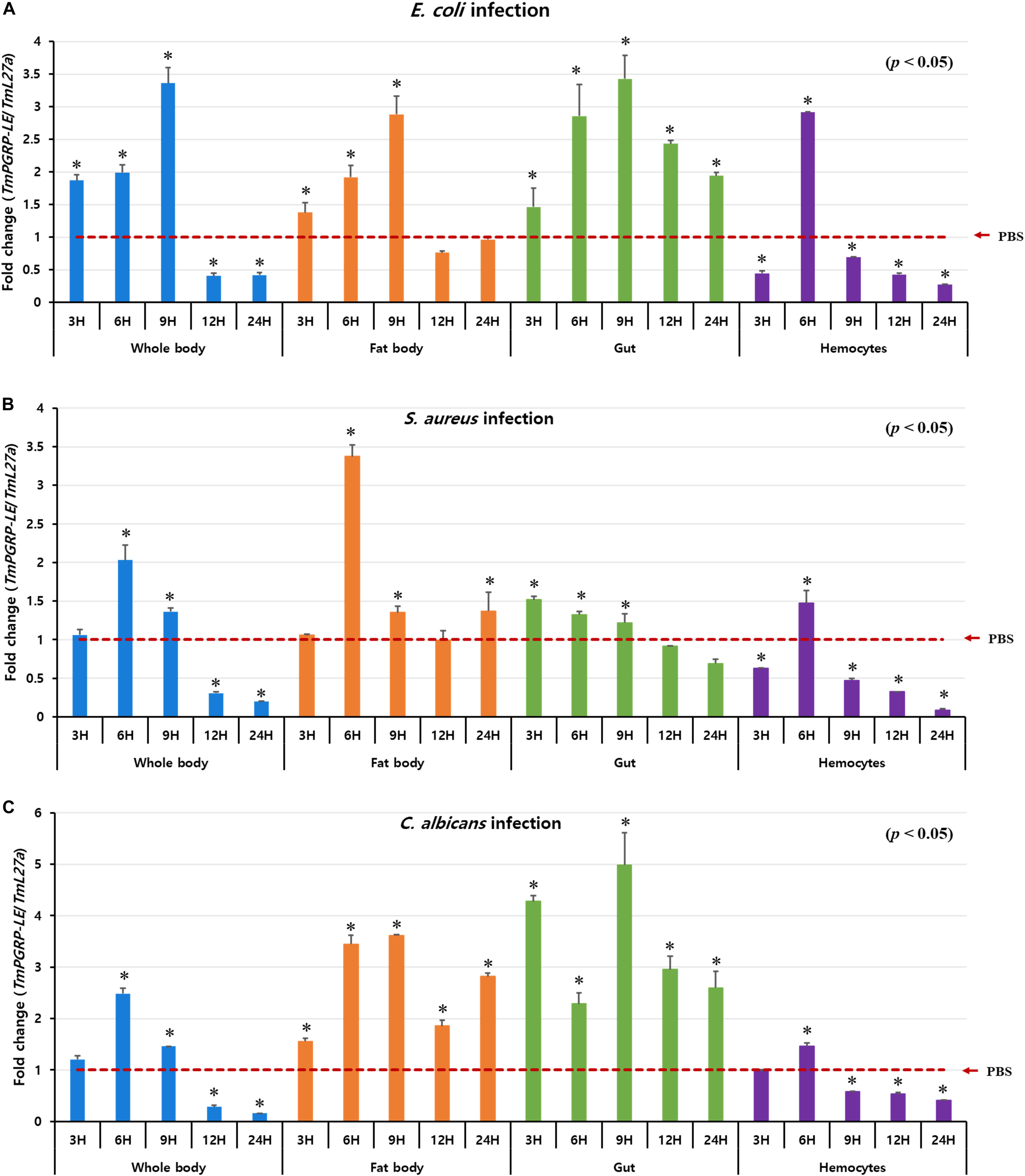
Figure 2. Relative expression profiles of TmPGRP-LE in the whole body, fat body, gut, and hemocytes of T. molitor young (10th–12th instar) larvae, after experimental challenge with E. coli (A), S. aureus (B), and C. albicans (C) (n = 20 per treatment group per time point) at 3, 6, 9, 12, and 24 h post-infection were examined by qRT-PCR. TmPGRP-LE mRNA levels were normalized against the internal control, T. molitor 60S ribosomal protein L27a (TmL27a), followed by normalization against the average relative expression of PBS-challenged controls. Each vertical bar represents mean ± SE. Statistical differences are denoted by asterisks (*) (p < 0.05).
As shown in Figure 2B, S. aureus challenge primarily induced TmPGRP-LE expression at 6 h post-infection in the whole body and fat body, whereas its expression dropped noticeably at later time points. In the gut, S. aureus exposure resulted in weak expression of TmPGRP-LE and its expression remained mostly unchanged at 3, 6, and 9 h post-infection (p < 0.05) (Figure 2B). Similar results were observed for TmPGRP-LE transcription following exposure to C. albicans as were found after infection with E. coli. The fold-increase in TmPGRP-LE mRNA levels was highest at 9 h post-infection with C. albicans in the fat body and gut in comparison to mock controls (p < 0.05) (Figure 2C).
Although microbial infection (E. coli, S. aureus, and C. albicans) within hemocytes was different in T. molitor larvae, the mRNA expression patterns of TmPGRP-LE were extremely similar. These results show a significant increase in TmPGRP-LE expression 6 h after the onset of infection followed by a dramatic decline in expression during later time points (p < 0.05) (Figures 2A–C). In hemocytes, the expression level of TmPGRP-LE after E. coli infection was more potent than it was after infection with S. aureus and C. albicans (Figure 2A).
In insects, the conserved RNAi pathway has been widely described, and RNAi techniques using dsRNA injection have been applied for gene function studies for over a decade (Brown et al., 1999; Amdam et al., 2003). To assess the response of the T. molitor young larvae (10th–12th instar) to dsTmPGRP-LE injection compared to dsEGFP control injection, the knockdown efficiency was measured by qRT-PCR. The transcription of TmPGRP-LE was significantly reduced (80%) 6 days after dsRNA injection (p < 0.05) (Figure 3A). As expected, silencing of TmPGRP-LE had no effect on PBS-injected larvae mortality (Supplementary Figure S1). Furthermore, in order to determine the necessity of TmPGRP-LE in larval survival, we challenged dsRNA-treated larvae with E. coli, S. aureus, and C. albicans and assessed insect mortality over 10 days. The survival rate of TmPGRP-LE-silenced larvae that were infected with E. coli dropped dramatically to 30% compared to >60% survival of the controls (p < 0.05) (Figure 3B). The percent survival resulting from S. aureus (50%, Figure 3C) and C. albicans (30%, Figure 3D) challenges in the TmPGRP-LE-silenced groups were significantly lower than that in the dsEGFP-injected controls (p < 0.05).
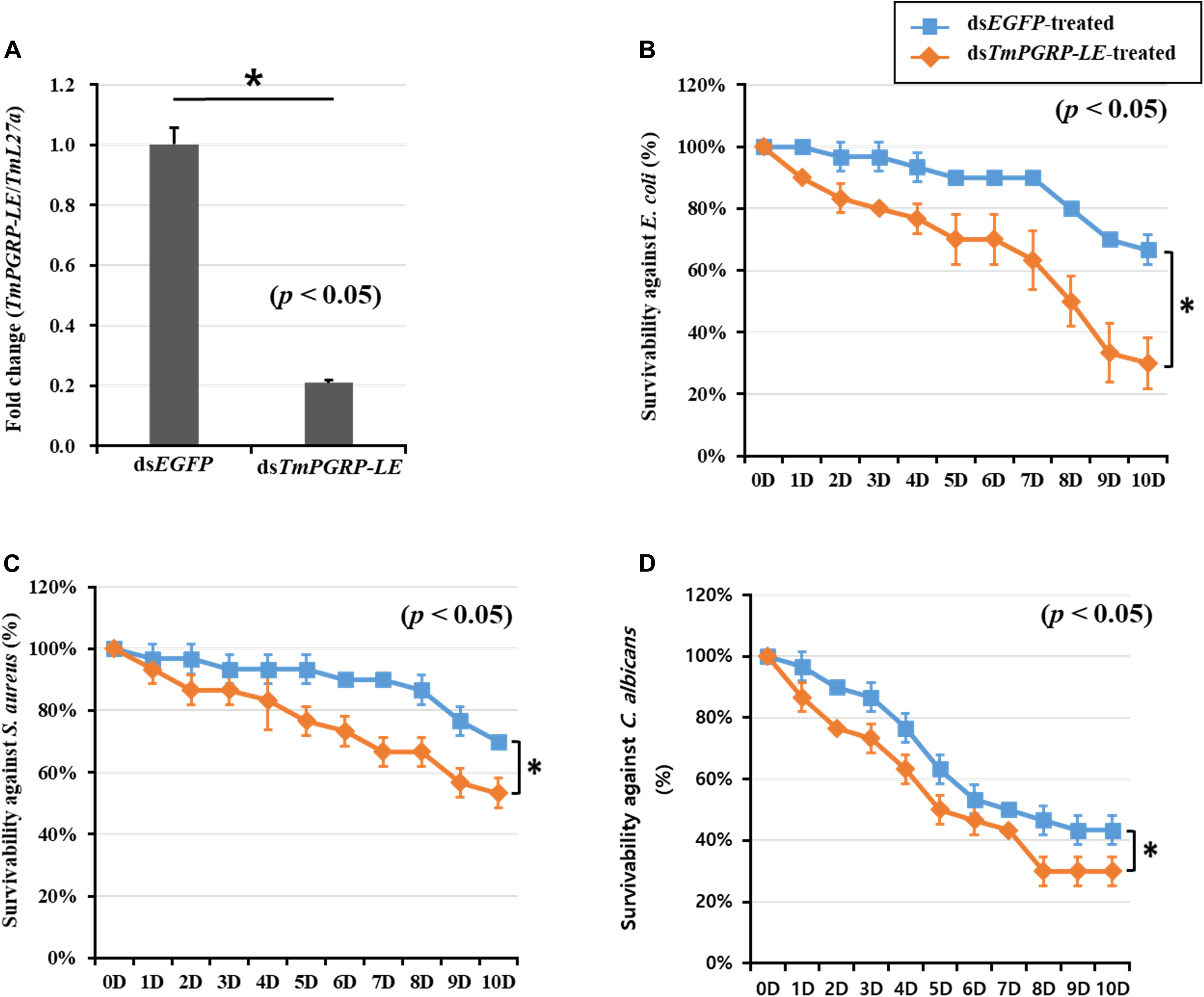
Figure 3. Effect of TmPGRP-LE silencing on the larval survival of T. molitor (n = 10 per treatment group), monitored for 10 days after microbial challenge. Knockdown efficiency of TmPGRP-LE in T. molitor larvae injected with target gene-specific dsRNA (A). Young larvae (n = 3 per group) were injected with 1 μL (1 μg) of TmPGRP-LE dsRNA and, after 6 days, the TmPGRP-LE transcript was determined by qRT-PCR. Survival of dsTmPGRP-LE-injected larvae during immune challenge with E. coli (B), S. aureus (C), and C. albicans (D). Larvae injected with dsRNA targeting enhanced green fluorescent protein (dsEGFP) was used as a negative control. Results are an average of three independent biological replicates. Asterisks (*) denote significant differences between EGFP- and TmPGRP-LE-silenced larvae (p < 0.05).
Among the previously described receptors of the Imd signaling pathway, DmPGRP-LE senses bacteria-derived PGN either independently or synergistically with DmPGRP-LC depending on its localization. It eventually conveys the signal transduction to the NF-κB transcription proteins and in-turn regulates immunity effector genes, i.e., AMPs (Takehana et al., 2004). Considering the significant mortality ratios observed in TmPGRP-LE-silenced larvae groups after microbial invasion, we reasoned that these phenotypes could be due to TmPGRP-LE knockdown which would subsequently impair the antimicrobial responses. These observations prompted us to investigate the expression levels of 14 AMP genes in larvae with depleted levels of TmPGRP-LE. Therefore, we injected 1 μL (1 μg) of dsTmPGRP-LE and dsEGFP into two sets of T. molitor larvae and confirmed the knockdown efficiency (80%) of the target genes after 6 days. Given that immune tissues might be variably tolerant to infection (Neyen et al., 2012), we examined the larval fat body, gut, and hemocytes of dsTmPGRP-LE- and dsEGFP-treated cohorts 24 h after challenge with E. coli, S. aureus, and C. albicans.
In dsEGFP-injected controls, E. coli infection induced the expression of TmTene1 (Figures 4A, 5A), TmTene2 (Figures 4B, 5B), TmTene4 (Figures 4D, 5D), TmAtt1a (Figures 4E, 5E), TmAtt1b (Figures 4F, 5F), TmAtt2 (Figures 4G, 5G), TmCole1 (Figures 4H, 5H), TmCole2 (Figures 4I, 5I), TmDef1 (Figures 4J, 5J), TmDef2 (Figures 4K, 5K), and TmCec2 (Figures 4L, 5L) in both the fat body and gut (Figures 4, 5). In dsTmPGRP-LE-injected groups, E. coli-induced gut expression of all 11 AMP genes was significantly downregulated (Figure 5), while fat body expression of TmTene1 (Figure 4A), TmTene2 (Figure 4B), TmAtt1b (Figure 4F), TmCole1 (Figure 4H), and TmDef1 (Figure 4J) was upregulated.
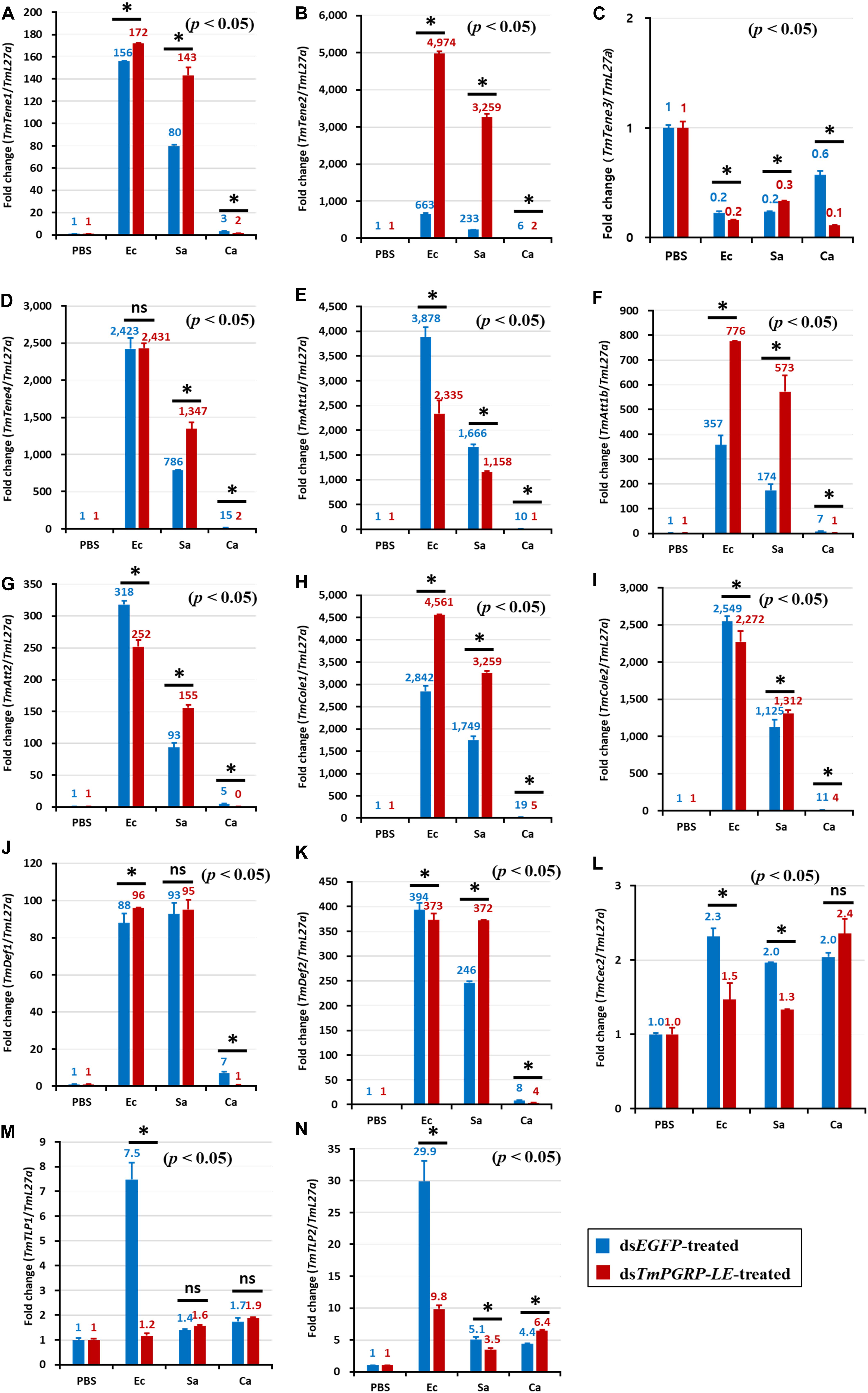
Figure 4. Antimicrobial peptide (AMP) induction profiles in the TmPGRP-LE-silenced T. molitor larval fat body in response to E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) challenges. Double-stranded RNA specific to TmPGRP-LE was injected (1 μg) into young larvae. Six days after dsRNA treatment, TmPGRP-LE mRNA levels were reduced by 80% in the dsTmPGRP-LE-injected groups compared to dsEGFP-injected groups. The larvae were then infected with E. coli, S. aureus, and C. albicans (n = 20 per group). The expression levels of 14 AMP genes were evaluated by qRT-PCR: TmTenecin-1 (TmTene1, A); TmTenecin-2 (TmTene2, B); TmTenecin-3 (TmTene3, C); TmTenecin-4 (TmTene4, D); TmAttacin-1a (TmAtt1a, E); TmAttacin-1b (TmAtt1b, F); TmAttacin-2 (TmAtt2, G); TmColeptericin-1 (TmCole1, H); TmColeptericin-2 (TmCole2, I); TmDefensin-1 (TmDef1, J); TmDefensin-2 (TmDef2, K); TmCecropin-2 (TmCec2, L); TmTLP-1 (TmTLP1, M); and TmTLP-2 (TmTLP2, N). EGFP dsRNA injection served as a negative control, and the mRNA levels of the respective AMP genes are presented relative to those for TmL27a as an internal control. Each vertical bar represents mean ± SE of three independent biological replicates and the numbers above the bars show AMP transcription levels. Significant differences between dsEGFP- and dsTmPGRP-LE-treated cohorts are shown by asterisks (*) (p < 0.05) and ns, not significant.
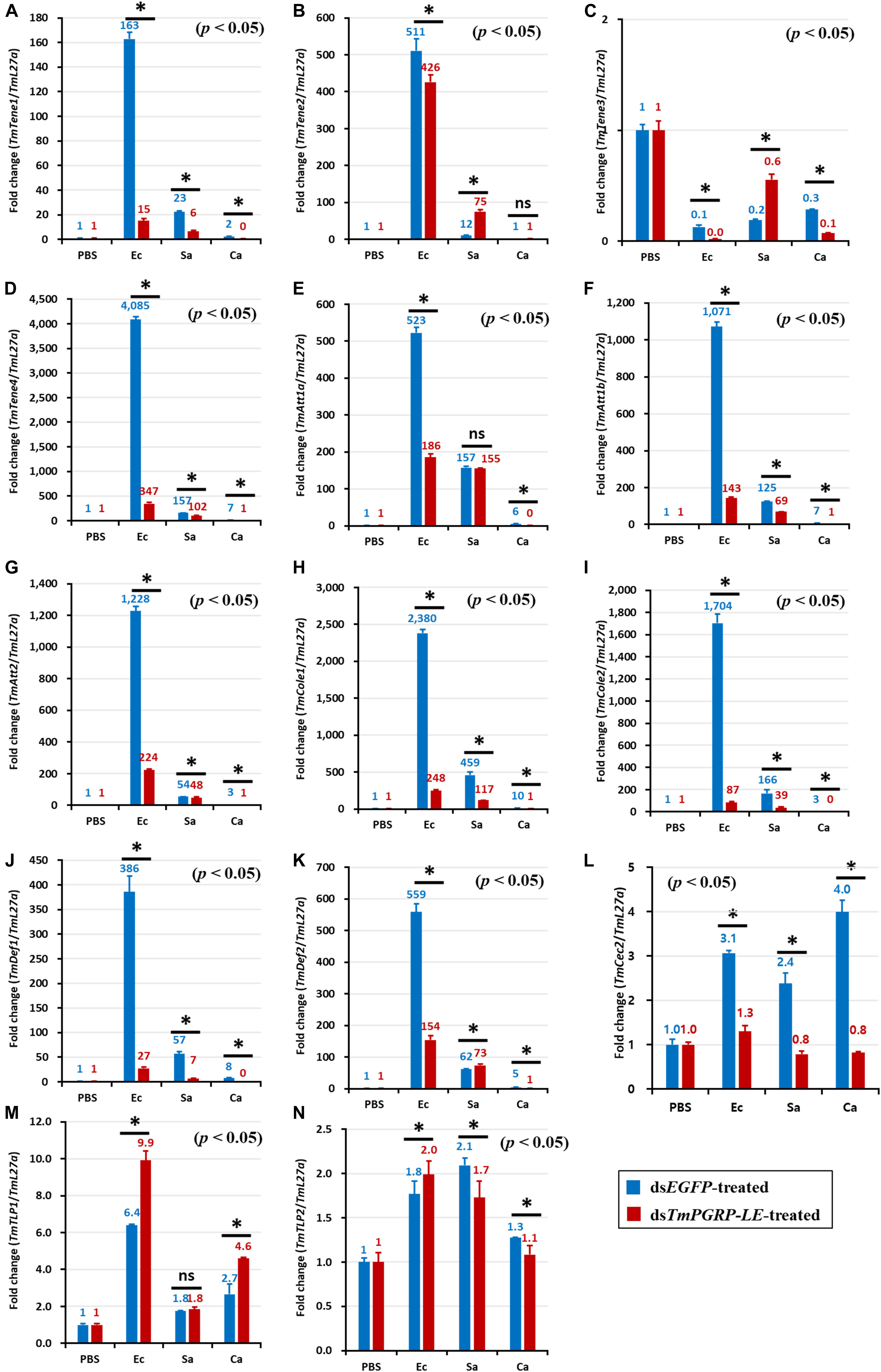
Figure 5. The effect of TmPGRP-LE silencing on induction of antimicrobial peptides (AMPs) in the gut of T. molitor larvae in response to pathogenic microbial stimuli of E. coli (Ec), S. aureus (Sa), and C. albicans (Ca). Young T. molitor larvae were treated with 1 μL (1 μg) of dsRNA of TmPGRP-LE. Six days later, the dsRNA-treated larvae were challenged with E. coli, S. aureus, and C. albicans (n = 20 per group). The mRNA quantity of 14 AMP genes, namely TmTenecin-1 (TmTene1, A), TmTenecin-2 (TmTene2, B), TmTenecin-3 (TmTene3, C), TmTenecin-4 (TmTene4, D), TmAttacin-1a (TmAtt1a, E), TmAttacin-1b (TmAtt1b, F), TmAttacin-2 (TmAtt2, G), TmColeptericin-1 (TmCole1, H), TmColeptericin-2 (TmCole2, I), TmDefensin-1 (TmDef1, J), TmDefensin-2 (TmDef2, K), TmCecropin-2 (TmCec2, L), TmTLP-1 (TmTLP1, M), and TmTLP-2 (TmTLP2, N), were measured in relation to L27a by qRT-PCR 24 h post-infection. The other details were the same as in Figure 4.
In the fat body, C. albicans-induced expression of almost all AMPs was decreased in TmPGRP-LE-silenced larvae with the exception of four AMPs: TmTene3 (Figure 4C), TmCec2 (Figure 4L), TmTLP1 (Figure 4M), and TmTLP2 (Figure 4N). This suggests that TmPGRP-LE is crucial for the production of AMPs after C. albicans infection (Figure 4). In contrast, S. aureus-dependent expression of AMPs was dramatically increased in TmPGRP-LE-silenced larvae with the exception of TmAtt1a (Figure 4E), TmCec2 (Figure 4L), and TmTLP2 (Figure 4N). Furthermore, TmPGRP-LE knockdown did not affect the mRNA levels of TmDef1 (Figure 4J) and TmTLP1 (Figure 4M) after S. aureus infection and of TmCec2 (Figure 4L) and TmTLP1 (Figure 4M) following C. albicans challenge.
Notably, AMP genes were upregulated dramatically in the gut of dsEGFP-injected larvae infected with E. coli (Figure 5). The upregulation of TmTene1 (Figure 5A), TmTene2 (Figure 5B), TmTene4 (Figure 5D), TmAtt1a (Figure 5E), TmAtt1b (Figure 5F), TmAtt2 (Figure 5G), TmCole1 (Figure 5H), TmCole2 (Figure 5I), TmDef1 (Figure 5J), and TmDef2 (Figure 5K) was notably suppressed in dsTmPGRP-LE-treated larvae. This result highlights the critical role of TmPGRP-LE in the humoral immune response of the gut.
However, in contrast to the strong AMP induction observed in the gut following E. coli challenge, AMP expression was comparatively mild in S. aureus and C. albicans infections.
Consistently, silencing of PGRP-LE decreased the expression of S. aureus-induced AMP genes, including TmTene1 (Figure 5A), TmTene4 (Figure 5D), TmAtt1b (Figure 5F), TmAtt2 (Figure 5G), TmCole1 (Figure 5H), TmCole2 (Figure 5I), TmDef1 (Figure 5J), and TmCec2 (Figure 5L) in comparison with their levels in dsEGFP-injected larvae. Compared with the prominent effect of TmPGRP-LE on AMPs expression upon E. coli challenge, TmPGRP-LE acts as a positive regulator during S. aureus infection, where it conveys the signal to produce AMPs. As shown Figure 5, silencing TmPGRP-LE had surprising effects on the induction of gut AMPs by C. albicans. More precisely, the mRNA levels of 11 AMP genes, namely TmTene1 (Figure 5A), TmTene4 (Figure 5D), TmAtt1a (Figure 5E), TmAtt1b (Figure 5F), TmAtt2 (Figure 5G), TmCole1 (Figure 5H), TmCole2 (Figure 5I), TmDef1 (Figure 5J), TmDef2 (Figure 5K), and TmCec2 (Figure 5L), were significantly decreased.
The antimicrobial responses to fungal and bacterial challenges in hemocytes of dsTmPGRP-LE-treated larvae were weaker compared to the other tissues studied (Figure 6). Similar to fat body and gut responses, when dsTmPGRP-LE-injected larvae were challenged with E. coli, the transcription levels of TmTene1 (Figure 6A), TmTene4 (Figure 6D), TmAtt1a (Figure 6E), TmAtt1b (Figure 6F), TmAtt2 (Figure 6G), TmCole1 (Figure 6H), TmCole2 (Figure 6I), TmDef1 (Figure 6J), and TmDef2 (Figure 6K) were downregulated compared to dsEGFP-treated controls. In contrast, however, TmTene1 (Figure 6A), TmTene2 (Figure 6B), TmTene4 (Figure 6D), TmAtt1a (Figure 6E), TmCole2 (Figure 6I), TmDef1 (Figure 6J), and TmDef2 (Figure 6K) gene expression following infection with S. aureus remained inducible in dsTmPGRP-LE-injected larvae, suggesting that TmPGRP-LE acts as a negative signal transducer in hemocytes following S. aureus challenge. Note that levels of expression of AMP genes in larval hemocytes after challenge with C. albicans were not significantly different in TmPGRP-LE-silenced larvae compared to dsEGFP controls (Figure 6).
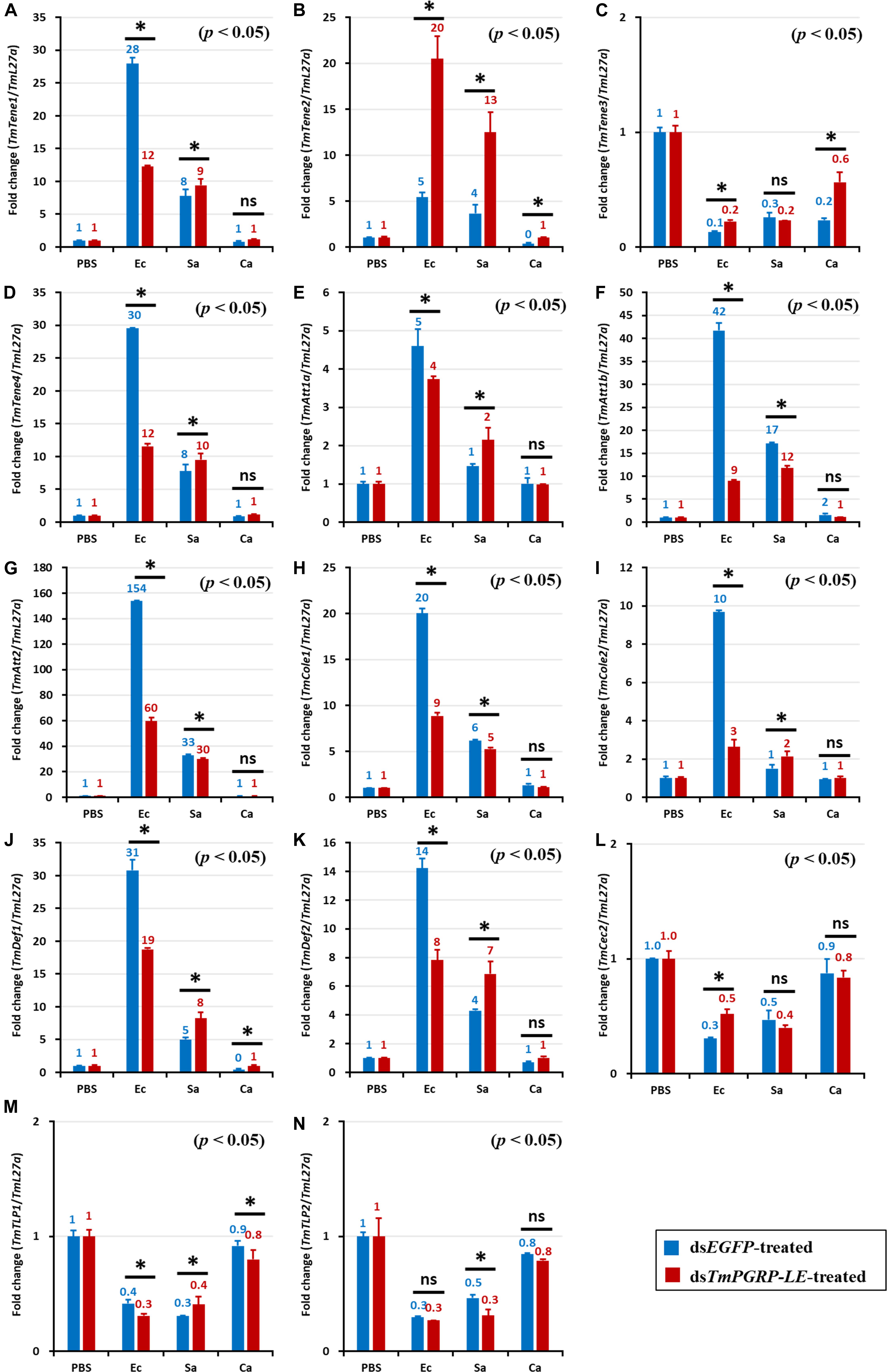
Figure 6. Effect of TmPGRP-LE knockdown on induction of microbial AMP genes in the hemocytes of T. molitor larvae in response to E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) infections. On the sixth day after dsTmPGRP-LE injection (knockdown efficiency of 80%), the larvae were experimentally exposed to E. coli, S. aureus, or C. albicans (n = 20 per group). Twenty-four hours post-infection; the larvae were dissected. qRT-PCR was used to evaluate the expression profile of 14 AMP genes: TmTenecin-1 (TmTene1, A), TmTenecin-2 (TmTene2, B), TmTenecin-3 (TmTene3, C), TmTenecin-4 (TmTene4, D), TmAttacin-1a (TmAtt1a, E), TmAttacin-1b (TmAtt1b, F), TmAttacin-2 (TmAtt2, G), TmColeptericin-1 (TmCole1, H), TmColeptericin-2 (TmCole2, I), TmDefensin-1 (TmDef1, J), TmDefensin-2 (TmDef2, K), TmCecropin-2 (TmCec2, L), TmTLP-1 (TmTLP1, M), and TmTLP-2 (TmTLP2, N). The other details were the same as in Figure 4.
Altogether, these results indicate that reducing TmPGRP-LE levels in the fat body, gut, and hemocytes led to a significant downregulation of AMP genes after E. coli infection. Similarly, TmPGRP-LE depletion was sufficient to suppress the expression of several AMP genes in the fat body and gut following C. albicans challenge, highlighting the critical role of TmPGRP-LE as a receptor of the NF-κB signaling pathway. In contrast to the responses to E. coli and C. albicans infection, no drastic reduction in the transcription of AMP genes was observed during S. aureus infections in the fat body of TmPGRP-LE-silenced larvae.
Previous studies in insect immunity have revealed that different transcription factors regulate the Toll and Imd immunity pathways. Relish and Dorsal, key downstream proteins in the Imd and Toll pathways, respectively, from dimers to participate in the precise control of the production of AMPs (Sagisaka et al., 2004; Shin et al., 2005; Schlüns and Crozier, 2007; Tanaka et al., 2007; Oeckinghaus and Ghosh, 2009; Keshavarz et al., 2019). We wanted to find out whether the reduction of TmPGRP-LE, which is a sensor of PGN (Tindwa et al., 2013), would have an effect on the expression levels of T. molitor transcription proteins. To address this question, we assessed the mRNA expression of TmRelish, TmDorsal X1 isoform (TmDorX1), and TmDorsal X2 isoform (TmDorX2) in the fat body, gut, and hemocytes of TmPGRP-LE-silenced larvae following infection with E. coli, S. aureus, and C. albicans.
After E. coli and C. albicans infections, depletion of TmPGRP-LE appeared to have a significant effect in downregulating TmRelish and TmDorX1 transcription in the larval fat body and gut (Figures 7A, B). TmPGRP-LE knockdown larvae showed slightly decreased TmDorX1 expression in the fat body and hemocytes after S. aureus infection (Figure 7B).
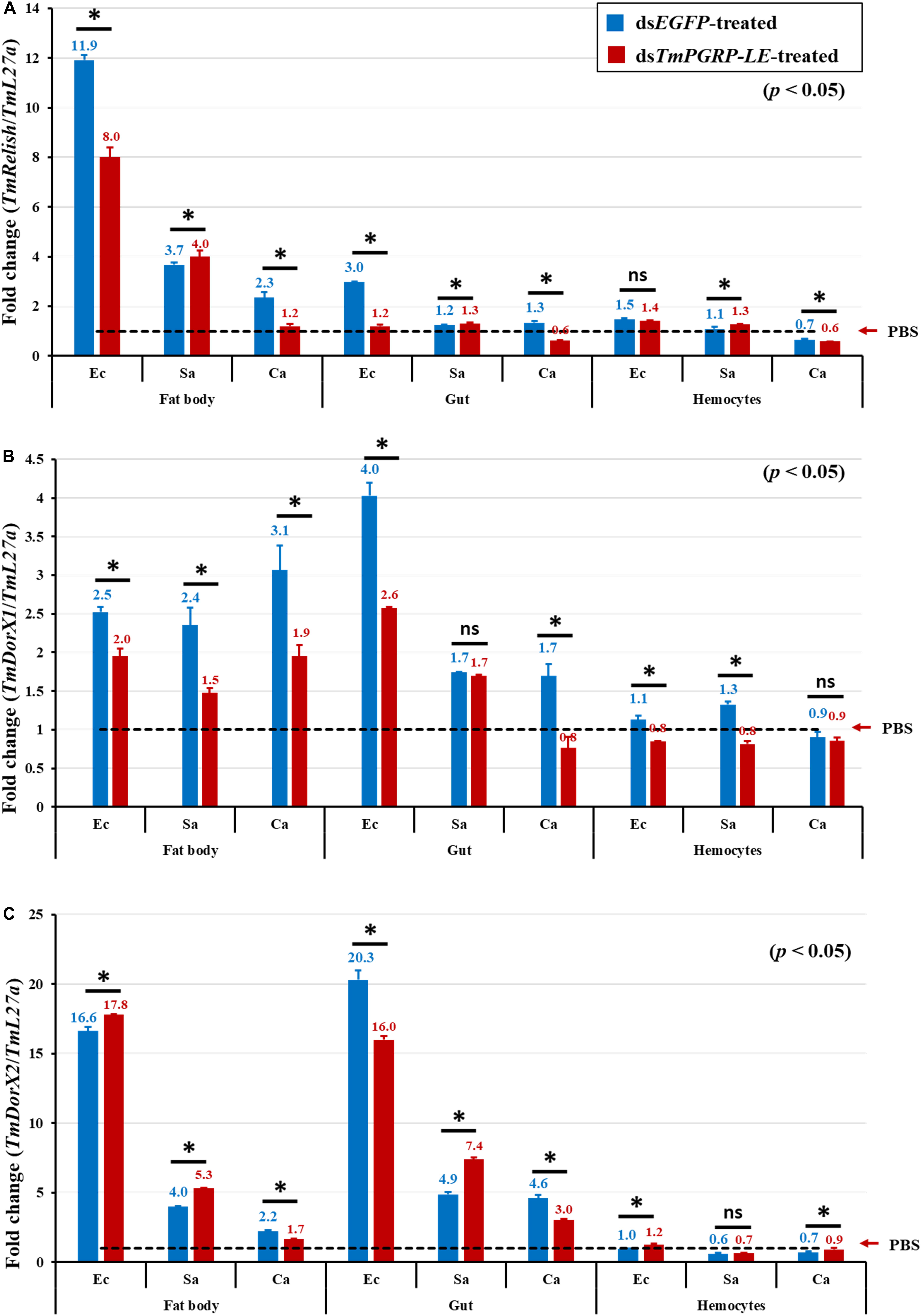
Figure 7. Transcriptional activation of different NF-κB genes in the fat body, hemocytes, and gut of TmPGRP-LE dsRNA-injected T. molitor larvae after inoculation with E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) (n = 20 per group). mRNA quantities of TmRelish (A), TmDorX1 (B), and TmDorX2 (C) in TmPGRP-LE knockdown larvae were measured relative to those for L27a at 24 h post-challenge by qRT-PCR. EGFP RNAi was used as a negative control. Bars represent mean ± SE of three independent experiments and the numbers above the bars indicate the transcription levels of NF-κB genes. Significant differences between dsEGFP- and dsTmPGRP-LE-treated groups are presented by asterisks (*) (p < 0.05) and ns, not significant.
In dsTmPGRP-LE-injected larvae, expression of TmDorX2 was significantly decreased in both the fat body and gut following C. albicans challenge (Figure 7C). In response to E. coli infection, there was a moderate decline in the expression of TmDorX2 in the gut (Figure 7C) and of TmDorX1 in hemocytes (Figure 7B).
Collectively, all NF-κB genes were significantly downregulated in the larval gut of TmPGRP-LE knockdown groups after infection with either E. coli or C. albicans. Of note, a similar result was observed in the fat body of dsTmPGRP-LE-injected larvae following C. albicans challenge. It is plausible that the Toll and Imd pathways synergistically activate through TmPGRP-LE (Tanji et al., 2007). This conclusion is in agreement with our findings regarding the expression of AMP genes in the fat body and gut.
The recognition of foreign microbes is a crucial aspect of the host defense mechanism that relies on the activation of PGRPs as central sensors and regulators of innate immune response. DmPGRP-LC is a transmembrane protein in the Imd pathway, which is fundamental to the production of an array of potent AMPs following E. coli challenge (Rämet et al., 2002). It is not, however, the only upstream factor that activates the Imd pathway (Gottar et al., 2002). The multifunctional protein PGRP-LE, a constitutive hemolymph protein, triggers both the activation of the prophenoloxidase (pro-PO) cascade and the Imd pathway (Takehana et al., 2002). Importantly, DmPGRP-LE is expressed in many cells and tissues, including the fat body, hemocytes, hemolymph, gut, trachea, Malpighian tubules, and cuticle (Takehana et al., 2002, 2004; Kaneko et al., 2006). In this study, we found that TmPGRP-LE was expressed in the integument, fat body, hemocytes, gut, and Malpighian tubules, which is in agreement with the results of previous studies. Interestingly, we observed that TmPGRP-LE expression was comparatively higher in the gut and hemocytes of T. molitor both larvae and adults. The results of our study, along with previously published results (Bosco-Drayon et al., 2012; Neyen et al., 2012), may indicate that PGRP-LE is necessary and sufficient to react promptly to PGN in both tissues. Additionally, in the case of hemocytes, considerable expression of TmPGRP-LE is related to the activation of the proPO cascade as a secondary humoral response (Takehana et al., 2002). Our results revealed a TmPGRP-LE-mediated induction of the gut immune response following E. coli infection. Likewise, T. castaneum and Armigeres subalbatus showed modest expression of PGRP-LE after E. coli infection (Wang and Beerntsen, 2013; Koyama et al., 2015).
In Drosophila, a combination of multiple receptors, namely PGRP-SA, PGRP-SD, and gram-negative binding protein 1 (GNBP1), is required to trigger an adequate immune response against gram-positive bacteria (S. aureus) (Bischoff et al., 2004). However, further investigation underlined the main role of PGRP-SD in presenting the PGN of gram-negative bacteria to PGRP-LC, which in-turn activates the Imd pathway (Iatsenko et al., 2016). The interaction between PGRP-SD and PGRP-LE remains to be elucidated. Similar to the Drosophila model, the Tenebrio PGRP-SA and GNBP1 complex is critical for the activation of the Toll and proPO pathways by Lys-type PGN of S. aureus (Park et al., 2007). Here we reported that TmPGRP-LE was slightly, but significantly, expressed following S. aureus challenge. Unexpectedly, we found that TmPGRP-LE is markedly induced in C. albicans-infected larvae in both the fat body and gut. Previous genomic analysis of T. castaneum revealed a poor activation in TcPGRP-LE expression in C. albicans-challenged beetles (Zou et al., 2007).
T. molitor larvae with decreased levels of TmPGRP-LE (silenced with RNAi) have reduced viability following infection. We propose that the most plausible cause of TmPGRP-LE-silenced larval death is related to the transcription of AMP genes, which are regulated by two intracellular signaling pathways: the Toll and the Imd pathways. Under this framework, if expression levels of AMPs decline as a result of TmPGRP-LE depletion, TmPGRP-LE can be considered a positive regulator of AMPs in larvae, and TmPGRP-LE-silenced larvae are more susceptible to microbial infections.
In a previous study, expression levels of TmTene1, TmTene2, TmTene4, TmAtt1a, TmAtt1b, TmAtt2, TmCole1, TmCole2, and TmDef2 were significantly reduced in E. coli-challenged T. molitor following silencing of immune deficiency (TmIMD) expression (Jo et al., 2019). These findings are consistent with our own, in which the expression levels of these genes were downregulated in the gut of E. coli-challenged T. molitor larvae following silencing of TmPGRP-LE. This raises the question of whether expression of NF-κB genes are affected by TmPGRP-LE RNAi in the gut. In this manuscript, we report that silencing of TmPGRP-LE reduced the expression levels of larval gut TmRelish, TmDorX1, and TmDorX2 after E. coli challenge. This is consistent with our recent report demonstrating that all the aforementioned AMPs were positively regulated by TmDorX2 (Keshavarz et al., 2019) and TmRelish (Keshavarz et al., 2020). These results leave open the possibility that detection of invading E. coli by TmPGRP-LE in the gut of T. molitor larvae results in signal transduction to both TmDorX2 and TmRelish. Although, more work is needed to examine whether translocation of TmDorX2 and TmRelish occurs in dsTmPGRP-LE larvae. Furthermore, following E. coli infection, the expression of seven AMP genes, namely TmTene1, TmTene4, TmAtt1b, TmAtt2, TmCole1, TmCole2, and TmDef2, showed a dramatic reduction in the hemocytes of the dsTmPGRP-LE-injected group. We also know that the same AMPs were downregulated in the dsTmIMD, dsTmRelish, and dsTmDorX2-injected cohorts (Jo et al., 2019; Keshavarz et al., 2019). Moreover, in the fat body of dsTmPGRP-LE-injected insects, TmAtt1a, TmCole2, TmDef2, and TmCec2 were significantly suppressed following exposure to E. coli. Similarly, depletion of TmIMD, TmRelish, and TmDorX2 led to a reduction in the same AMPs in the larval fat body after E. coli infection (Jo et al., 2019; Keshavarz et al., 2019). Mortality assays and AMP expression analyses from our present work and those of others suggest that, in comparison to hemocytes and the fat body, the T. molitor gut plays a pivotal role in the response to E. coli infection via activation of the Imd pathway. Additionally, these findings showed that there was robust TmPGRP-LE-dependent AMPs induction following E. coli in the larval gut, while there was slight TmPGRP-LE-dependent transcription of AMP genes following S. aureus and C. albicans. Collectively, the presently-identified signaling components of the Imd pathway (comprising TmPGRP-LE, TmIMD, and TmRelish) positively regulate TmAtt1a, TmCole2, and TmDef2 gene expression following E. coli infection in immune-related tissues (fat body, gut, and hemocytes).
Drosophila gut immune defense mechanisms function independently from Imd-induced AMPs following infection by the gram-positive bacteria, S. aureus (Hori et al., 2018). As opposed to Drosophila, S. aureus-infected T. molitor larvae exhibited increased mortality following TmPGRP-LE silencing, and the transcription of nine AMPs decreased in TmPGRP-LE knockdown larval gut. It should also be mentioned that TmRelish is a positive regulator of TmTene1, TmTene4, TmAtt1b, TmAtt2, TmCole1, and TmCole2 in the gut of S. aureus-infected larvae (Keshavarz et al., 2020) suggesting that the Imd pathway is critical for combatting infection of the larval gut infected with S. aureus via AMP production. We further show that the involvement of TmPGRP-LE in the control of the Imd pathway is limited to only some AMPs in the fat body and hemocytes of S. aureus-challenged larvae. TmAttacin-2, an anti-gram positive bacterial protein (Jo et al., 2018), is regulated by TmPGRP-LE in the fat body, gut, and hemocytes during S. aureus infection.
While there is a scarcity of information available regarding the recognition of fungi by PGRP-LE in insects, our current study suggests that C. albicans is sensed in the gut and fat body by TmPGRP-LE, which leads to the expression of immune-related AMP-coding genes. We observed a notable effect of TmPGRP-LE silencing on the mRNA expression of transcription proteins for both Imd (TmRelish) and Toll (TmDorX1 and 2) signaling pathways in response to C. albicans infection in the fat body and gut. In this context, following C. albicans challenge, downregulation of TmPGRP-LE-induced genes (i.e., AMPs) is mediated by TmDorX2 in the gut (Keshavarz et al., 2019), whereas the AMP transcription levels are mainly regulated by TmRelish in the fat body (Keshavarz et al., 2020).
Finally, this recent finding uncovers the pivotal role of TmPGRP-LE in the immune response of the gut by regulating the production of eight AMP genes, namely TmTene1, TmTene4, TmAtt1b, TmAtt2, TmCole1, TmCole2, TmDef1, and TmCec2, following infection by E. coli, S. aureus, and C. albicans (Figure 8). Our study provides valuable information about the critical involvement of TmPGRP-LE in the regulation of AMP genes in response to microbial infections. More work needs to be done in order to elucidate the cooperation between TmPGRP-LE and T. molitor NF-κB genes using double-knockdown and immunocytochemistry methods.
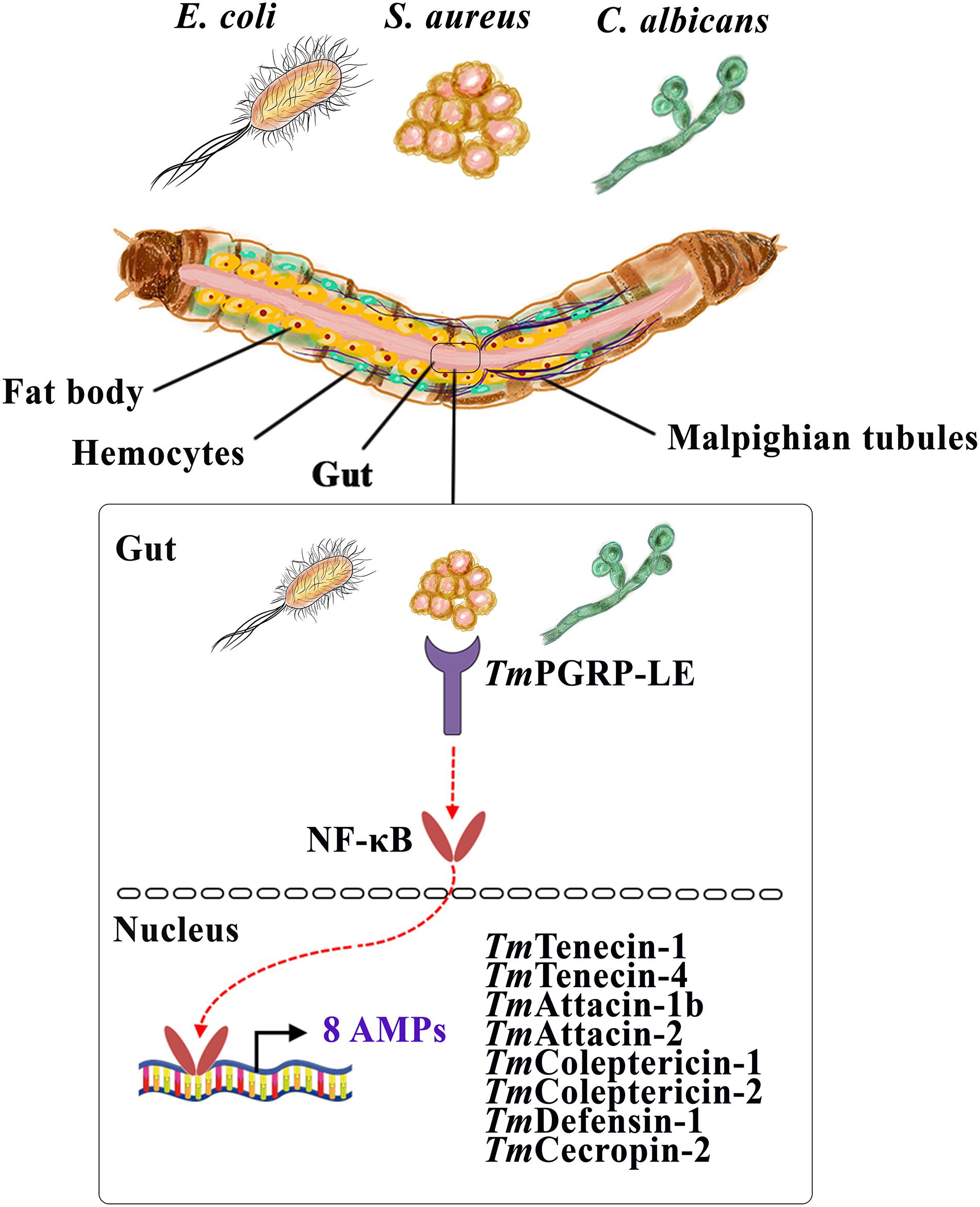
Figure 8. A schematic representation of the role of a central sensor of intracellular immune pathways, TmPGRP-LE, in regulating antimicrobial peptides (AMPs) in the larval gut against E. coli, S. aureus, and C. albicans.
Previous analysis of T. molitor PGRP-LE provides insight into the functional role of this non-catalytic PGRP in detecting DAP-type PGN and inducing autophagy, which is the basis of T. molitor response to L. monocytogenes (Tindwa et al., 2013). However, further studies were needed to explore the exact role of TmPGRP-LE in response to gram-negative and gram-positive bacteria and fungi in other innate immune signaling pathways. In this study, we indicate that TmPGRP-LE in the T. molitor gut induces 10 AMP genes in response to E. coli infection. Moreover, S. aureus- and C. albicans-dependent induction of 9 and 11 AMPs, respectively, are regulated by TmPGRP-LE in the larval gut. In conclusion, TmPGRP-LE is required for the detection of gram-negative bacteria (E. coli) in the gut of T. molitor, which subsequently transduces the signal to NF-κB transcription proteins to induce AMPs.
The datasets generated for this study are available on request to the corresponding author.
YH and YJ conceived and designed the experiments and revised the manuscript. MK and TE performed the experiments. MK analyzed the data and wrote the manuscript. YH procured reagents, materials, and analysis tools.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (Grant No. 2019R1I1A3A01057848).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00320/full#supplementary-material
Amdam, G. V., Simões, Z. L., Guidugli, K. R., Norberg, K., and Omholt, S. W. (2003). Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol. 3:1. doi: 10.1186/1472-6750-3-1
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979.
Basbous, N., Coste, F., Leone, P., Vincentelli, R., Royet, J., Kellenberger, C., et al. (2011). The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 12, 327–333. doi: 10.1038/embor.2011.19
Bischoff, V., Vignal, C., Boneca, I. G., Michel, T., Hoffmann, J. A., and Royet, J. (2004). Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 5, 1175–1180.
Bosco-Drayon, V., Poidevin, M., Boneca, I. G., Narbonne-Reveau, K., Royet, J., and Charroux, B. (2012). Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165. doi: 10.1016/j.chom.2012.06.002
Brown, S. J., Mahaffey, J. P., Lorenzen, M. D., Denell, R. E., and Mahaffey, J. W. (1999). Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1, 11–15.
Chevée, V., Sachar, U., Yadav, S., Heryanto, C., and Eleftherianos, I. (2019). The peptidoglycan recognition protein PGRP-LE regulates the Drosophila immune response against the pathogen Photorhabdus. Microb. Pathog. 136:103664. doi: 10.1016/j.micpath.2019.103664
Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., et al. (2002). Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165.
Costechareyre, D., Capo, F., Fabre, A., Chaduli, D., Kellenberger, C., Roussel, A., et al. (2016). Tissue-specific regulation of Drosophila NF-κB pathway activation by peptidoglycan recognition protein SC. J. Innate Immun. 8, 67–80.
Dziarski, R., and Gupta, D. (2006). Mammalian PGRPs: novel antibacterial proteins. Cell. Microbiol. 8, 1059–1069.
Dziarski, R., and Gupta, D. (2018). A balancing act: PGRPs preserve and protect. Cell Host Microbe 23, 149–151. doi: 10.1016/j.chom.2018.01.010
Ganesan, S., Aggarwal, K., Paquette, N., and Silverman, N. (2011). NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 349, 25–60.
Gendrin, M., Zaidman-Rémy, A., Broderick, N. A., Paredes, J., Poidevin, M., Roussel, A., et al. (2013). Functional analysis of PGRP-LA in Drosophila immunity. PLoS One 8:e69742. doi: 10.1371/journal.pone.0069742
Goto, A., Yano, T., Terashima, J., Iwashita, S., Oshima, Y., and Kurata, S. (2010). Cooperative regulation of the induction of the novel antibacterial Listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway. J. Biol. Chem. 285, 15731–15738. doi: 10.1074/jbc.M109.082115
Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., et al. (2002). The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644.
Guan, R., and Mariuzza, R. A. (2007). Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 15, 127–134.
Guan, R., Roychowdhury, A., Ember, B., Kumar, S., Boons, G.-J., and Mariuzza, R. A. (2004). Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17168–17173.
Hori, A., Kurata, S., and Kuraishi, T. (2018). Unexpected role of the IMD pathway in Drosophila gut defense against Staphylococcus aureus. Biochem. Biophys. Res. Commun. 495, 395–400. doi: 10.1016/j.bbrc.2017.11.004
Iatsenko, I., Kondo, S., Mengin-Lecreulx, D., and Lemaitre, B. (2016). PGRP-SD, an extracellular pattern-recognition receptor, enhances peptidoglycan-mediated activation of the Drosophila Imd pathway. Immunity 45, 1013–1023. doi: 10.1016/j.immuni.2016.10.029
Jang, J. H., Kim, H., and Cho, J. H. (2013). Rainbow trout peptidoglycan recognition protein has an anti-inflammatory function in liver cells. Fish Shellfish Immunol. 35, 1838–1847.
Jo, Y. H., Park, S., Park, K. B., Noh, M. Y., Cho, J. H., Ko, H. J., et al. (2018). In silico identification, characterization and expression analysis of attacin gene family in response to bacterial and fungal pathogens in Tenebrio molitor. Entomol. Res. 48, 45–54.
Jo, Y. H., Patnaik, B. B., Hwang, J., Park, K. B., Ko, H. J., Kim, C. E., et al. (2019). Regulation of the expression of nine antimicrobial peptide genes by Tm IMD confers resistance against Gram-negative bacteria. Sci. Rep. 9:10138.
Kaneko, T., Yano, T., Aggarwal, K., Lim, J.-H., Ueda, K., Oshima, Y., et al. (2006). PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723.
Kang, D., Liu, G., Lundström, A., Gelius, E., and Steiner, H. (1998). A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. U.S.A. 95, 10078–10082.
Keshavarz, M., Jo, Y. H., Park, K. B., Ko, H. J., Edosa, T. T., Lee, Y. S., et al. (2019). Tm DorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 9:16878.
Keshavarz, M., Jo, Y. H., Patnaik, B. B., Park, K. B., Ko, H. J., Kim, C. E., et al. (2020). TmRelish is required for regulating the antimicrobial responses to Escherichia coli and Staphylococcus aureus in Tenebrio molitor. Sci. Rep. 10, 1–18. doi: 10.1038/s41598-020-61157-1
Koyama, H., Kato, D., Minakuchi, C., Tanaka, T., Yokoi, K., and Miura, K. (2015). Peptidoglycan recognition protein genes and their roles in the innate immune pathways of the red flour beetle, Tribolium castaneum. J. Invertebr. Pathol. 132, 86–100. doi: 10.1016/j.jip.2015.09.003
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Kurata, S. (2010). Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int. Immunol. 22, 143–148. doi: 10.1093/intimm/dxp128
Kurata, S. (2014). Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 42, 36–41.
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743.
Liu, C., Xu, Z., Gupta, D., and Dziarski, R. (2001). Peptidoglycan recognition proteins a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276, 34686–34694.
Lu, X., Wang, M., Qi, J., Wang, H., Li, X., Gupta, D., et al. (2006). Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 281, 5895–5907.
Mao, X., Xu, X., Yang, X., Li, Z., Yang, J., and Liu, Z. (2019). Molecular and functional characterization of ApPGRP from Anatolica polita in the immune response to Escherichia coli. Gene 690, 21–29. doi: 10.1016/j.gene.2018.12.036
Mellroth, P., Karlsson, J., and Steiner, H. (2003). A scavenger function for a DrosophilaPeptidoglycan recognition protein. J. Biol. Chem. 278, 7059–7064.
Michel, T., Reichhart, J.-M., Hoffmann, J. A., and Royet, J. (2001). Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759.
Neyen, C., Poidevin, M., Roussel, A., and Lemaitre, B. (2012). Tissue-and ligand-specific sensing of gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J. Immunol. 189, 1886–1897. doi: 10.4049/jimmunol.1201022
Oeckinghaus, A., and Ghosh, S. (2009). The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 1:a000034. doi: 10.1101/cshperspect.a000034
Park, J. W., Je, B.-R., Piao, S., Inamura, S., Fujimoto, Y., Fukase, K., et al. (2006). A synthetic peptidoglycan fragment as a competitive inhibitor of the melanization cascade. J. Biol. Chem. 281, 7747–7755.
Park, J.-W., Kim, C.-H., Kim, J.-H., Je, B.-R., Roh, K.-B., Kim, S.-J., et al. (2007). Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. U.S.A. 104, 6602–6607.
Rämet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B., and Ezekowitz, R. A. B. (2002). Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648.
Rehman, A., Taishi, P., Fang, J., Majde, J. A., and Krueger, J. M. (2001). The cloning of a rat peptidoglycan recognition protein (PGRP) and its induction in brain by sleep deprivation. Cytokine 13, 8–17.
Sagisaka, A., Tanaka, H., Furukawa, S., and Yamakawa, M. (2004). Characterization of a homologue of the Rel/NF-κB transcription factor from a beetle, Allomyrina dichotoma. Biochim. Biophys. Acta 1678, 85–93.
Schlüns, H., and Crozier, R. (2007). Relish regulates expression of antimicrobial peptide genes in the honeybee, Apis mellifera, shown by RNA interference. Insect Mol. Biol. 16, 753–759.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 3, 1101–1108.
Sheehan, G., Garvey, A., Croke, M., and Kavanagh, K. (2018). Innate humoral immune defences in mammals and insects: the same, with differences? Virulence 9, 1625–1639. doi: 10.1080/21505594.2018.1526531
Shin, S. W., Kokoza, V., Bian, G., Cheon, H.-M., Kim, Y. J., and Raikhel, A. S. (2005). REL1, a homologue of Drosophila dorsal, regulates toll antifungal immune pathway in the female mosquito Aedes aegypti. J. Biol. Chem. 280, 16499–16507.
Sun, L., Liu, S., Wang, R., Li, C., Zhang, J., and Liu, Z. (2014). Pathogen recognition receptors in channel catfish: IV. Identification, phylogeny and expression analysis of peptidoglycan recognition proteins. Dev. Comp. Immunol. 46, 291–299. doi: 10.1016/j.dci.2014.04.018
Sun, Q.-L., and Sun, L. (2015). A short-type peptidoglycan recognition protein from tongue sole (Cynoglossus semilaevis) promotes phagocytosis and defense against bacterial infection. Fish Shellfish Immunol. 47, 313–320. doi: 10.1016/j.fsi.2015.09.021
Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T., et al. (2002). Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 99, 13705–13710.
Takehana, A., Yano, T., Mita, S., Kotani, A., Oshima, Y., and Kurata, S. (2004). Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23, 4690–4700.
Tanaka, H., Matsuki, H., Furukawa, S., Sagisaka, A., Kotani, E., Mori, H., et al. (2007). Identification and functional analysis of Relish homologs in the silkworm, Bombyx mori. Biochim. Biophys. Acta 1769, 559–568.
Tanji, T., Hu, X., Weber, A. N., and Ip, Y. T. (2007). Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 27, 4578–4588.
Tindwa, H., Patnaik, B., Kim, D., Mun, S., Jo, Y., Lee, B., et al. (2013). Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection. Int. J. Mol. Sci. 14, 22462–22482. doi: 10.3390/ijms141122462
Ting, A. T., and Bertrand, M. J. (2016). More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545. doi: 10.1016/j.it.2016.06.002
Vollmer, W., and Bertsche, U. (2008). Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778, 1714–1734.
Wang, M., Liu, L.-H., Wang, S., Li, X., Lu, X., Gupta, D., et al. (2007). Human peptidoglycan recognition proteins require zinc to kill both gram-positive and gram-negative bacteria and are synergistic with antibacterial peptides. J. Immunol. 178, 3116–3125.
Wang, Q., Ren, M., Liu, X., Xia, H., and Chen, K. (2019). Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 106, 69–76. doi: 10.1016/j.molimm.2018.12.021
Wang, S., and Beerntsen, B. T. (2013). Insights into the different functions of multiple peptidoglycan recognition proteins in the immune response against bacteria in the mosquito, Armigeres subalbatus. Insect Biochem. Mol. Biol. 43, 533–543. doi: 10.1016/j.ibmb.2013.03.004
Werner, T., Borge-Renberg, K., Mellroth, P., Steiner, H., and Hultmark, D. (2003). Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J. Biol. Chem. 278, 26319–26322.
Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H., and Hultmark, D. (2000). A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 13772–13777.
Yaffe, H., Buxdorf, K., Shapira, I., Ein-Gedi, S., Zvi, M. M.-B., Fridman, E., et al. (2012). LogSpin: a simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes 5:45. doi: 10.1186/1756-0500-5-45
Yano, T., Mita, S., Ohmori, H., Oshima, Y., Fujimoto, Y., Ueda, R., et al. (2008). Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916. doi: 10.1038/ni.1634
Yoshida, H., Kinoshita, K., and Ashida, M. (1996). Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 271, 13854–13860.
Yu, Y., Park, J.-W., Kwon, H.-M., Hwang, H.-O., Jang, I.-H., Masuda, A., et al. (2010). Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem. 285, 32937–32945. doi: 10.1074/jbc.M110.144014
Zaidman-Rémy, A., Poidevin, M., Hervé, M., Welchman, D. P., Paredes, J. C., Fahlander, C., et al. (2011). Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One 6:e17231. doi: 10.1371/journal.pone.0017231
Zhang, L., Gao, C., Liu, F., Song, L., Su, B., and Li, C. (2016). Characterization and expression analysis of a peptidoglycan recognition protein gene, SmPGRP2 in mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 56, 367–373. doi: 10.1016/j.fsi.2016.07.029
Zhang, Y., Van Der Fits, L., Voerman, J. S., Melief, M.-J., Laman, J. D., Wang, M., et al. (2005). Identification of serum N-acetylmuramoyl-l-alanine amidase as liver peptidoglycan recognition protein 2. Biochim. Biophys. Acta 1752, 34–46.
Keywords: Tenebrio molitor, TmPGRP-LE, nuclear factor κB, expression patterns, antimicrobial peptides
Citation: Keshavarz M, Jo YH, Edosa TT and Han YS (2020) Tenebrio molitor PGRP-LE Plays a Critical Role in Gut Antimicrobial Peptide Production in Response to Escherichia coli. Front. Physiol. 11:320. doi: 10.3389/fphys.2020.00320
Received: 20 December 2019; Accepted: 20 March 2020;
Published: 15 April 2020.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Roman Dziarski, Indiana University, United StatesCopyright © 2020 Keshavarz, Jo, Edosa and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Hun Jo, eWh1bjEyMjhAam51LmFjLmty; Yeon Soo Han, aGFueXNAam51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.