
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Physiol., 20 September 2019
Sec. Developmental Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.01197
This article is part of the Research TopicXenopus Models of Organogenesis and DiseaseView all 20 articles
Congenital heart disease (CHD) is a significant cause of mortality in infants and adults. Currently human genomic analysis has identified a number of candidate genes in these patients. These genes span diverse categories of gene function suggesting that despite the similarity in cardiac lesion, the underlying pathophysiology may be different. In fact, patients with similar CHDs can have drastically different outcomes, including a dramatic decrease in myocardial function. To test these human candidate genes for their impact on myocardial function, we need efficient animals models of disease. For this purpose, we paired Xenopus tropicalis with our microangiography technique, hemoglobin contrast subtraction angiography (HCSA). To demonstrate the gene-teratogen-physiology relationship, we modeled human cardiomyopathy in tadpoles. First we depleted the sarcomeric protein myosin heavy chain 6 (myh6) expression using morpholino oligos. Next, we exposed developing embryos to the teratogen ethanol and in both conditions showed varying degrees of cardiac dysfunction. Our results demonstrate that HCSA can distinguish biomechanical phenotypes in the context of gene dysfunction or teratogen. This approach can be used to screen numerous candidate CHD genes or suspected teratogens for their effect on cardiac function.
Congenital heart disease (CHD) occurs in ~8 out of 1,000 live births and effects 1.3 million newborns per year worldwide (Marelli et al., 2007; van der Linde et al., 2011). Currently the prevalence of adults with CHD exceeds the pediatric population, shifting the burden of disease to adulthood, and enlarging the cohort of CHD patients. Despite the early corrective measures and close, multidisciplinary care, adults with CHD still have a significantly higher mortality rate (Pierpont et al., 2007). Sudden death and early onset congestive heart failure remains the most common causes of mortality suggesting that premature myocardial failure may be associated with adult CHD (Sabatine et al., 2006; O'Donnell and Nabel, 2011; Kathiresan and Srivastava, 2012). Currently very little is known about the interactions between the malformed heart and biomechanical function which ultimately affects life expectancy and the quality of life.
Human genomics technologies are now enabling genetic analyses of patients with CHD (Fakhro et al., 2011; Sanders et al., 2012; Zaidi and Brueckner, 2017; Pierpont et al., 2018). Based on the diversity of the genes identified in similar phenotypic patients, we suggest that genetic heterogeneity may explain the differences seen in patient outcomes. To better understand this relationship between a candidate gene and the biomechanical phenotype, we need to develop efficient animal models to evaluate the effect of candidate CHD genes on cardiac function. This would be the first step to investigate the functional variability seen in CHD patients.
Imaging small animal cardiovascular system remains to be a challenge because of the small heart sizes as well as the fast heart rates requiring high resolution, high speed imaging. Cardiac MRI with gating has been used for imaging anesthetized mice and rat cardiovascular system yet image acquisition time reaching to hours making it difficult to use for any screening purposes (Ramirez and Bankson, 2007; Ramirez et al., 2007; Esparza-Coss et al., 2008). Alternatively micro-CT systems are designed to provide fast (1–2 s) imaging. Combined with the cardio-respiratory gating and laborious segmentation, 4D data sets can be formed to measure cardiac function (Yushkevich et al., 2006; Clark et al., 2013). Both of these modalities have limited utilization for rapid, high-throughput screening since they require extensive image processing. As an alternative modality our group and the others have implemented Optical Coherence Tomography (OCT) imaging for small animal cardiovascular imaging (Tearney et al., 1996; Luo et al., 2006; Männer et al., 2009; Yoo et al., 2011; Wang et al., 2016; Deniz et al., 2017; Grishina et al., 2017). Principally OCT imaging is similar to the ultrasound but light is utilized instead of sound and provides cross-sectional images. With scan rates of ~100 2D frames/s and implemented Doppler technology, OCT imaging has been evolving in functional analysis of cardiovascular system in small animal models (Davis et al., 2008; Jenkins et al., 2010). The main disadvantages of OCT are the expense of the commercially available products and the limitations of Doppler application. For accurate quantification of blood flow by doppler application requires high speed systems to prevent phase-wrapping artifacts and also relies on laminar flow. This is very difficult to obtain from motile, trabeculated embryonic ventricle, limiting Doppler OCT application at its current state for accurately quantifying cardiac ventricle function.
We previously demonstrated that hemoglobin contrast subtraction angiography (HCSA) can exploit the wavelength-sensitive absorption of hemoglobin as a molecular-specific source of endogenous flow contrast and can be applied to Xenopus tadpole hearts in a straightforward manner (Deniz et al., 2012) using an EOS camera. HCSA can quantify changes in the embryonic cardiac function after perturbation with a calcium channel blocker. In this brief report, we demonstrate that Xenopus can be a fast gene-teratogen-function assay and when coupled with HCSA, can be used to quantify physiological cardiac phenotypes over a short period of time. Xenopus, as a model of cardiac development, has a favorable balance between human modeling and cost/efficiency that is required for high throughput screening (Warkman and Krieg, 2007; Khokha, 2012). The tadpole cardiovascular system develops within 72 h and remains optically accessible throughout the early stages of development. Importantly, during this period of development, tadpoles can survive with severely malformed hearts since nutrient and O2 delivery is not dependent on blood circulation. Here, we capitalized on these advantages to model human cardiomyopathy in Xenopus in two ways, first by reducing the expression of the sarcomeric protein myosin heavy chain 6 (MYH6) using morpholino oligonucleotides (MO) and, in a separate set of experiments, by exposing them to the known teratogen ethanol (EtOH).
Cardiac muscle myosin consists of two heavy chain subunits, two light chain subunits, and two regulatory subunits. We used MYH6 morpholino to block the translation start site of the mRNA encoding the alpha heavy chain. It has been shown that mutations in MYH6 gene cause cardiomyopathy both in humans (Hershberger et al., 2010) and Xenopus suggesting a well-conserved disease mechanism (Abu-Daya et al., 2009). One substantial advantage of MOs is that gene dosage can be titrated based on the amount of MO injected providing fine dosage control. This is important since most cardiomyopathy patients are heterozygous (Zaidi and Brueckner, 2017) and therefore gene dosage may play a critical role.
In parallel to the knockdown approach, we exposed the developing embryo to EtOH, a known cardiac teratogen, affecting one in eight pregnant women (Floyd and Sidhu, 2004) and leading to fetal alcohol syndrome (FAS). Worldwide FAS affects many children with a prevalence of 23 per 1,000 (Roozen et al., 2016). Patients with FAS also present with structural birth defects including cardiac defects (Jones et al., 2010). To investigate the effects of EtOH on the developing cardiovascular system, studies in zebrafish found increased heart volumes, decreased thickness of the ventricular wall, and decreased basal heart rates (Dlugos and Rabin, 2010). Similarly, in Xenopus embryos exposed to EtOH, a reduction in trabeculae formation was found (Yelin et al., 2007). More elaborate work in chick suggested that ethanol-induced alterations in early cardiac function may potentially lead to late-stage valve and septal defects (Karunamuni et al., 2014). Together, EtOH appears to alter myocardial formation and pump function.
In this report we show that HCSA imaging can detect changes in embryonic cardiac function using several different anatomic and physiological metrics, establishing a rapid, non-destructive, low-cost method to investigate cardiac gene-function relationship.
Xenopus tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by the Yale Institutional Animal Care and Use Committee (IACUC) in accordance with NIH guidelines. Female and male mature Xenopus tropicalis were obtained from National Xenopus Resource.
In vitro fertilization and microinjections were done as previously described (del Viso and Khokha, 2012). In order to deplete myh6, we injected fertilized eggs with either 1 or 2 ng of MYH6-MOs and the tracer dye Alexa488 (Invitrogen) at one cell stage. Controls were injected with only tracer dye Alexa488. The advantage of MOs is that “gene dosage” can be titrated based on amount of MO injected providing fine dosage control. We used a translational-blocking start site morpholino oligo (MO) (Morcos, 2007) to deplete the alpha heavy chain, MYH6 (sequence 5′ AGTCTGCCATCAGGGCATCACCCAT-3′–Gene-Tools, LLC) which was previously utilized and verified in Xenopus (Abu-Daya et al., 2009). Embryos were injected at the one cell stage with borosilicate glass needles. Post-injections, embryos were incubated in 1/9 Modified Ringer's solution supplemented with 50 μg/ml of gentamycin at 25°C. Injections were confirmed by fluorescent lineage tracing with a Zeiss Lumar fluorescence stereomicroscope at stage 28 and tadpoles further raised at until stage 45 (Nieuwkoop—Faber staging. ~ post-fertilization day 4 at 25°C Nieuwkoop and Faber, 1994) for imaging.
We incubated Xenopus embryos in 1% EtOH in 1/9 Modified Ringer's solution at 25°C following mid-blastula stage (stage 9, Nieuwkoop—Faber staging Nieuwkoop and Faber, 1994) as described previously (Yelin et al., 2007) until stage 45 to determine the effects on the myocardium using HCSA imaging. EtOH solution refreshed daily for 4–5 days until tadpoles reached stage 45.
We used a Nikon SMZ800 stereomicroscope equipped with Canon EOS 5D Mark II digital camera and Sugar CUBE—LED (4,000 Lumens) with ring illuminator. Image acquisition setup: Zoom 6.2/Gain: 0/Depth 14 bit/LED 4,000 Lumens. Fifty RAW images acquired with 0.2 s intervals. High Speed color movie obtained with Motion Xtra N4-IDT, Inc. N4 Camera. For cardiac functional analysis, we embedded Stage 44–45 (post fertilization day 3 at 28°C) tadpoles in low melt agarose and positioned ventral surface exposed to the imaging plane (Supplemental Figure 1A). First, we obtained a scout image to record the tadpole stage, tadpole position and overall development. Next, we imaged the cardiac sac and confirmed that ventricle borders were clearly delineated. We obtained 50 still images (14-bit frames in RAW format) in the time lapse mode with a 0.2 s interframe interval using a Canon EOS 5D Mark II digital camera (Supplemental Figure 1B). Acquiring 50 images effectively captured end-systolic and end-diastolic stages of the cardiac cycle. Next, we processed the images and calculated various metrics using our custom image processing and analysis software. The custom software was developed in Matlab (The Mathworks, Natick, USA) Version 2012B. The application allows the user to load a high-bit depth image series in PNG format, sort, and select images of systole or diastole, segment the outline of the ventricle and store results as comma-separated value files. The software is available open-source1.
The graphical user interface allows the user to browse through the acquired images and select the frames corresponding to the maximal end-systole and end-diastole (Supplemental Figure 1C—Right Panel). Then on the selected images the ventricle is manually segmented (Supplemental Figure 1C—white dots). Using an Otsu-based thresholding function in MATLAB, we classified pixels into those that contain hemoglobin and those that do not. Finally, we quantified the number of hemoglobin-containing pixels (referred as Hb blush) within a manually segmented ventricle at the end of systole and diastole (Supplemental Figure 1D).
Based on hemoglobin pixel counts and the total ventricular area delineated by manual segmentation, we estimated the total ventricular end-diastolic area (tEDA), the total ventricular end-systolic area (tESA), percent change in the total ventricular area (Δ%tA = 100*(tEDA-tESA)/tEDA), end-diastolic blood area (EDBA), end-systolic blood area (ESBA), the stroke area (SA = EDBA-ESBA), and ejection fraction (EF = 100*SA/EDBA). We also derived a “myocardial fraction” metric where we estimated the myocardial area and referred to that as the myocardial mass index (MMI). To derive MMI, we assumed that during peak systole, where the myocardium is at its maximal contraction, most of the blood is ejected minimizing this confounding volume. We then subtracted the end-systolic blood area from the total ventricular end-systolic area assuming that the remainder of the segmented area would be a rough estimate of the myocardial mass at maximal contraction.
Tadpole raised and positioned in 1% agar as described above. Using microinjector (Harvard Apparatus PLI-100 Pico-Injector Microinjection System) Brilliant Blue FCF (Sigma-Aldrich) dye injected to the cardinal vein. Movie obtained with Redlake/IDT Y4 Lite grayscale camera.
For comparison we utilized the Mann-Whitney test (non-parametric and not paired) and used a scatter plot with the bar showing the median and 95% confidence interval. We also included before-after graph including standard error of the mean. All significant differences between groups indicated in the figure legends. Significance was determined when the p-value is lower than 0.05.
We first analyzed cardiac function in MYH6 morphant group. We analyzed HCSA images from 29 control embryos, 39 embryos injected with 1 ng MYH6 morpholino, and 13 embryos injected with 2 ng MYH6 morpholino. Overall, the 2 ng-morphant hearts were smaller in size as ascertained using total ventricular end-diastolic area (tEDA) (Figures 1A,B), whereas 1 ng-morphants had similar end diastolic area but slightly larger end systolic area when compared to the controls. The percent change in the ventricle area during contraction (Δ%tA) was significantly reduced in both morphants suggesting myocardial dysfunction (Figure 1C). We derived these measurements with manual segmentation as described in methods. Then we applied HCSA, where we were able to quantify end-diastolic and end-systolic Hb blush (EDBA: end-diastolic blood area; ESBA: end-systolic blood area), which is a surrogate for blood area. In end-diastole, due to their overall smaller cardiac size, 2 ng morphants filled with less blood whereas 1 ng morphants, given their normal cardiac size, filled with a similar amount of blood when compared to the controls (Figures 1D,E). Importantly, at the end of systole, they both showed a significant failure in emptying the ventricle leading to an increase in end systolic blood area (Figures 1D,E). Therefore, the ejection fraction and the stroke area diminished, so we concluded that the magnitude of the dysfunction was worse in 2 ng morphants (Figures 1F,G). Then we estimated MMI at maximal ventricular contraction. As shown in Figure 1H, 1 ng myh6 morphants did not have a significant reduction in myocardial mass in contrast to 2 ng morphants where MMI was significantly reduced. In conclusion, 2 ng morphants showed both structural loss of myocardium size and functional loss of contractility. Nevertheless, even without a change in MMI in 1 ng myh6 morphants, HCSA imaging could still delineate changes in myocardial function.
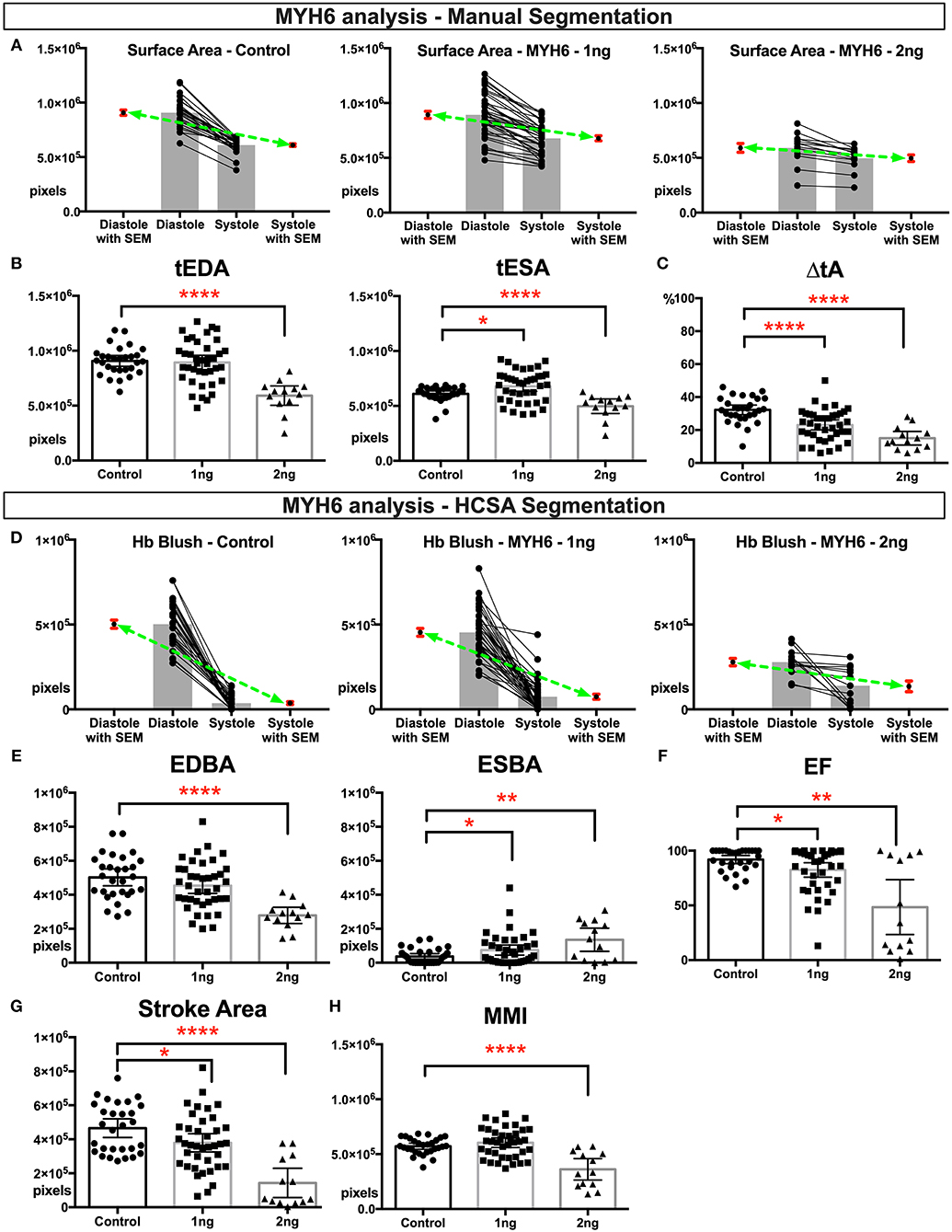
Figure 1. Physiological quantification of embryo heart function in MYH6 morphants. For this experiment 29 control, 39 morphants-−1 ng morpholino injected and 13 morphants-−2 ng morpholino injected analyzed. Measurements derived from manual segmentation include surface area at the end of diastole and systole and the change in ventricle area. (A) Before-after graph presented with extra columns pointing mean with SEM in red. Green line between means flattens as change in surface area diminishes with increasing MO dose. (B,C) As the MO dose is increased, the change in ventricle area is reduced, and the ventricle sizes are smaller. (D) Measurements derived from HCSA application include Hb blush at the end of diastole and systole and the change in blush area. Before-after graph presented with extra columns pointing mean with SEM in red. Green line between means flattens as change in blush area diminishes with increasing MO dose. (E,F) Following HCSA application, Hb blush is quantified at the end of diastole and systole. Ejection Fraction is derived from these measurments. Morphants demonstrated a dose dependent diminished ejection fraction and (G) stroke area. (H) Myocardial mass index estimates the amount of cardiac mass within the manually segmented heart. Myocardial mass index is affected in 2 ng morphants but remains within normal limits in 1 ng morphants. SEM, standard error of the mean; Hb Blush, Hemoglobin-containing pixels; tEDA, total end-diastolic area; tESA, total end-systolic area; ΔtA, change in total area; EDBA, end diastolic blood area; ESBA, end-systolic blood area; EF, ejection fraction; MMI, myocardiac mass index; HCSA, Hb contrast subtraction angiography. ****p < 0.0001, **p < 0.01, and *p < 0.05.
Next we analyzed the ethanol exposed tadpoles. In the ethanol group, we analyzed HCSA images from 30 control and 30 exposed tadpoles. Grossly dysmorphic embryos were excluded. In this group, ventricles were smaller than the control group (Figures 2A,B) and the change in ventricle area was diminished (Figure 2C), suggesting that EtOH exposed ventricle can‘t shorten appropriately. Aslo ESBA, a surrogate for blood volume at the end of systole, was significantly higher than controls (Figures 2D,E) which is also likely due to the reduced ability to generate adequate ventricular force, shown as reduced ejection fraction and stroke area (Figures 2F,G). Therefore, together these tadpoles had slightly smaller looking ventricles with reduced myocardial pump function. Furthermore, “myocardial fraction” in systole shows a significant reduction suggesting a loss of myocardial mass (Figure 2H).
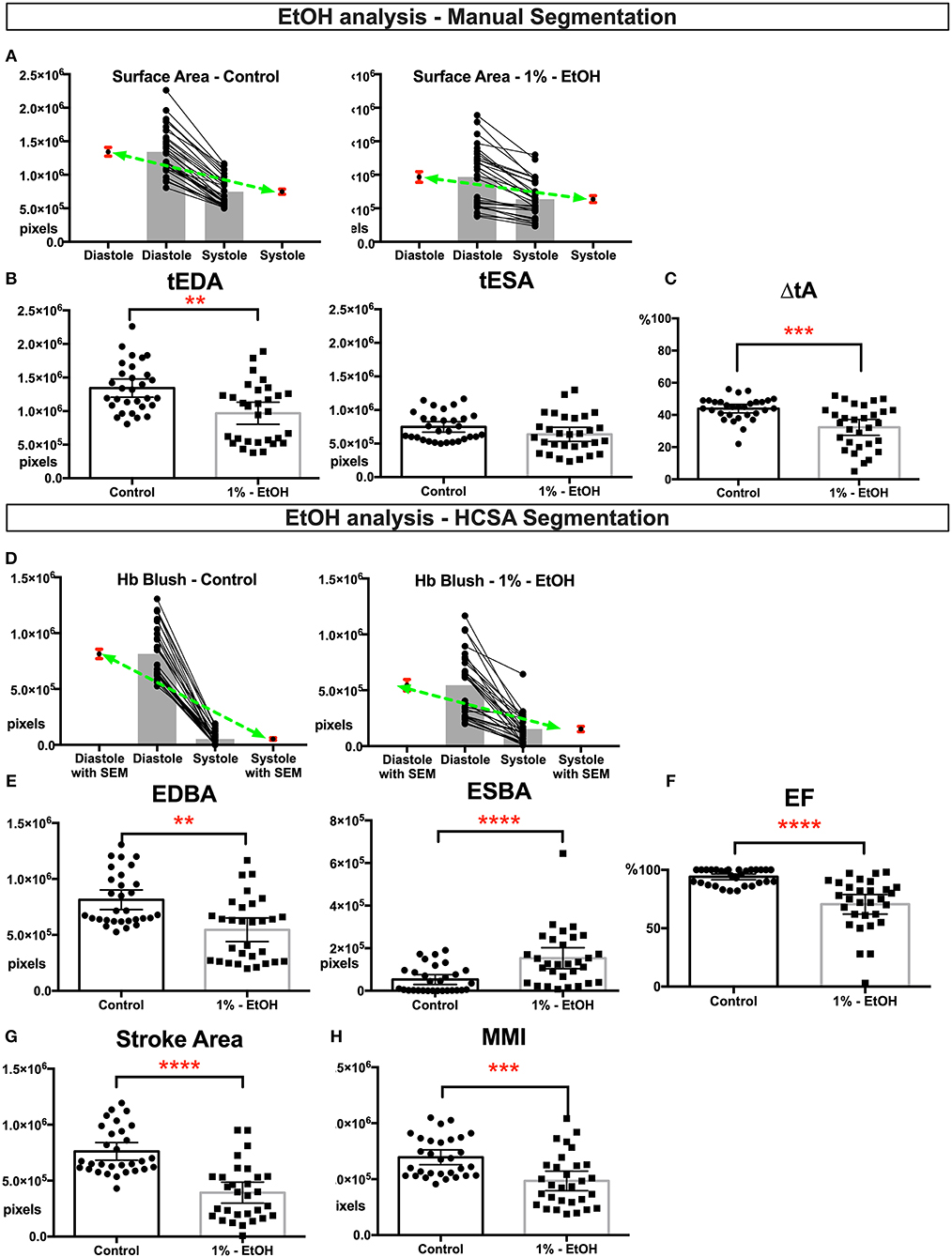
Figure 2. Physiological quantification of embryo heart function in EtOH treated tadpoles. For this experiment 30 controls and 30 EtOH exposed tadpoles analyzed. Measurements derived from manual segmentation includes surface area at the end of diastole and systole and the change in ventricle area. (A) Before-after graph presented with extra columns pointing mean with SEM in red. Green line between means flattens as change in surface area diminishes in EtOH group. (B,C) Ventricles of the ethanol exposed tadpoles were slightly smaller in size but had diminished change in the ventricle area. Following HCSA application, Hb blush quantified at the end of diastole and systole. Ejection fraction is derived from these measurements. (D) Before-after graph presented with extra columns pointing mean with SEM in red. Green line between means flattens as change in blush area diminishes in EtOH group. (E,F) Ethanol exposed tadpoles had less blood at the end of diastole and blood is not propelled efficiently leading an increase in end systolic Hb blush, diminished ejection fraction, and diminished (G) stroke area. (H) Myocardial Mass Index. In ethanol exposed tadpoles myocardial mass is diminished. SEM, standard error of the mean; Hb Blush, Hemoglobin-containing pixels; tEDA, total end-diastolic area; tESA, total end-systolic area; ΔtA, change in total area; EDBA, end diastolic blood area; ESBA, end-systolic blood area; EF, ejection fraction; MMI, myocardiac mass index; HCSA, Hb contrast subtraction angiography. ****p < 0.0001, ***p < 0.001, and **p < 0.01.
The incidence of CHD is staggering: ~1% of live births and 10% of aborted fetuses have CHD worldwide (Bernier et al., 2010; van der Linde et al., 2011; Jorgensen et al., 2014). New genomics technologies are enabling genetic analyses of CHD patients and identifying sequence variations in patients with CHD (Fakhro et al., 2011; Zaidi et al., 2013; Glessner et al., 2014; Homsy et al., 2015; McKean et al., 2016; Priest et al., 2016; Jin et al., 2017; Hoang et al., 2018; Manheimer et al., 2018). Genes identified in genomic studies are many but their role in cardiac development and function is uncertain; therefore, functional assays must be relatively rapid and inexpensive. We have previously shown that Xenopus tropicalis with the CRISPR/Cas9 system enables the analysis of hundreds of different genes in cardiac development (Deniz et al., 2018) and in this brief report we demonstrate that HSCA imaging can be used to screen for types of CHDs that present with sarcomeric dysfunction, for example cardiomyopathies.
The advantage of our HCSA method is that we can quantitate different metrics across multiple embryos to identify statistically significant differences between groups rather than relying on a qualitative assessment of function. In myh6 morphants with increasing doses of morpholino we showed worsening cardiac function, yet 1 ng morphants didn't show differences in EDA, EDBA, and MMI suggesting that even subtle changes in sarcomeric function can be demonstrated with HCSA imaging. Similarly EtOH treated hearts were smaller overall and failed to eject adequate blood, again suggesting sarcomeric dysfunction that was easily quantified with HCSA imaging.
As a major advantage HCSA imaging is non-destructive, reproducible, faster, and most importantly non-lethal to the tadpole enabling multiple viewing session at various stages. This is important in analyzing numerous candidate genes. Fluoroscopy-based angiography requires microinjection in tadpoles, is fatal preventing subsequent imaging, and is difficult to reproduce limiting its utility for screening purposes (Movie 1). As the high-speed color cameras (>250 fps) are more available and cheaper, more detailed functional analysis including simultaneous assessment for cardiac and peripheral vasculature structures will be more feasible (Movie 2) providing structural details comparable to angiography images.
Clinical cardiology has demonstrated that imaging modalities (e.g., echocardiography and cardiac angiography) are essential to identify abnormal heart structures and heart function (Cua and Feltes, 2012; Roest and de Roos, 2012). Current advances in optical imaging are enabling comprehensive, high-resolution cardiac imaging in small animal models, which facilitates studying gene-phenotype and teratogen-phenotype relationships (Choma et al., 2010). Xenopus is ideal for these studies given the ease of in vivo optical access to the cardiac structures as well as the optimal balance between human modeling and cost/efficiency (Warkman and Krieg, 2007). In this report we showed the effects of a human cardiomyopathy gene (MYH6) and a known teratogen, EtOH (known to cause Fetal Alcohol Syndrome) on early cardiac development using HCSA imaging. Broadly our results support the utility of HCSA imaging, when paired with Xenopus, to screen the impact of novel candidate CHD genes or potential teratogens on cardiac function.
All datasets generated for this study are included in the manuscript/Supplementary Files.
Xenopus tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by the Yale Institutional Animal Care and Use Committee (IACUC) in accordance with NIH guidelines. Female and male mature Xenopus tropicalis were obtained from National Xenopus Resource.
ED, SJ, MK, and MC designed the study. ED and SJ acquired and analyzed the data. ED wrote the first draft of the article. SJ, MK, and MC contributed to the interpretation of the results and the writing of the manuscript. All authors have approved the final manuscript.
This work was supported by NIH 5R33HL120783 to MK. MK is a Mallinckrodt scholar.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank all the members of the Khokha Lab and Choma Lab their support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01197/full#supplementary-material
Supplemental Figure 1. Experimental and quantitative analysis workflow. (A) For myh6 experiments, one cell Xenopus embryos are injected with either MOs and the tracer dye (Alexa488) or only the tracer dye (control group). In ethanol exposure experiments, we incubated post-midblastula stage embryos in1% EtOH. On day five, NF Stage 44 tadpoles are embedded in low melt agarose (1%) for non-pharmacological immobilization to facilitate imaging. (B) Scout Image and Magnified Images acquired. (C) Custom software interface. Right panel allows the user navigate through the acquired images and adjust the brightness and the contrast. Once maximal end-diastolic and maximal-end-systolic frames are identified, the ventricle is manually segmented. (D) Images processed and Hb blush is quantified within the segmented area using the HCSA software interface. IVF, in vitro fertilization; lta, left truncus arteriosus; rta, right truncus arteriosus; v, ventricle; oft, out flow tract; Hb, hemoglobin.
Movie 1. Grayscale movie demonstrating injection of Brilliant Blue dye via cardinal tail vein and tracking of flow from the venous system sequentially to the atrium, ventricle, and outflow tract that eventually propagates to the systemic circulation.
Movie 2. High speed (250 fps—color) color movie and processed HCSA movie.
1. ^http://mhealthgroup.net/software (Note to editors and reviewers: the software will be made available upon publication of the manuscript).
Abu-Daya, A., Sater, A. K., Wells, D. E., Mohun, T. J., and Zimmerman, L. B. (2009). Absence of heartbeat in the Xenopus tropicalis mutation muzak is caused by a nonsense mutation in cardiac myosin myh6. Dev. Biol. 336, 20–29. doi: 10.1016/j.ydbio.2009.09.019
Bernier, P. L., Stefanescu, A., Samoukovic, G., and Tchervenkov, C. I. (2010). The challenge of congenital heart disease worldwide: epidemiologic and demographic facts. Semin. Thorac. Cardiovasc. Surg. Pediatr. Card. Surg. Annu. 13, 26–34. doi: 10.1053/j.pcsu.2010.02.005
Choma, M. A., Suter, M. J., Vakoc, B. J., Bouma, B. E., and Tearney, G. J. (2010). Heart wall velocimetry and exogenous contrast-based cardiac flow imaging in Drosophila melanogaster using Doppler optical coherence tomography. J. Biomed. Opt. 15:056020. doi: 10.1117/1.3503418
Clark, D., Badea, A., Johnson, G. A., and Badea, C. T. (2013). Constructing a 4D murine cardiac micro-CT atlas for automated segmentation and phenotyping applications. Med. Imag. 2013 Image Process. 8669:86691P. doi: 10.1117/12.2007043
Cua, C. L., and Feltes, T. F. (2012). Echocardiographic evaluation of the single right ventricle in congenital heart disease. Circ. J. 76, 22–31. doi: 10.1253/circj.CJ-11-1267
Davis, A. M., Rothenberg, F. G., Shepherd, N., and Izatt, J. A. (2008). In vivo spectral domain optical coherence tomography volumetric imaging and spectral Doppler velocimetry of early stage embryonic chicken heart development. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 25, 3134–3143. doi: 10.1364/JOSAA.25.003134
del Viso, F., and Khokha, M. (2012). Generating diploid embryos from Xenopus tropicalis. Methods Mol. Biol. 917:33–41. doi: 10.1007/978-1-61779-992-1_3
Deniz, E., Jonas, S., Hooper, M., Griffin, N. J., Choma, M. A., and Khokha, M. K. (2017). Analysis of craniocardiac malformations in Xenopus using optical coherence tomography. Sci. Rep. 7:42506. doi: 10.1038/srep42506
Deniz, E., Jonas, S., Khokha, M., and Choma, M. A. (2012). Endogenous contrast blood flow imaging in embryonic hearts using hemoglobin contrast subtraction angiography. Opt. Lett. 37, 2979–2981. doi: 10.1364/OL.37.002979
Deniz, E., Mis, E. K., Lane, M., and Khokha, M. K. (2018). CRISPR/Cas9 F0 screening of congenital heart disease genes in Xenopus tropicalis. Methods Mol. Biol. 1865, 163–174. doi: 10.1007/978-1-4939-8784-9_12
Dlugos, C. A., and Rabin, R. A. (2010). Structural and functional effects of developmental exposure to ethanol on the zebrafish heart. Alcohol. Clin. Exp. Res. 34, 1013–1021. doi: 10.1111/j.1530-0277.2010.01176.x
Esparza-Coss, E., Ramirez, M. S., and Bankson, J. A. (2008). Wireless self-gated multiple-mouse cardiac cine MRI. Magn. Reson. Med. 59, 1203–1206. doi: 10.1002/mrm.21562
Fakhro, K. A., Choi, M., Ware, S. M., Belmont, J. W., Towbin, J. A., Lifton, R. P., et al. (2011). Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc. Natl. Acad. Sci. U.S.A. 108, 2915–2920. doi: 10.1073/pnas.1019645108
Floyd, R. L., and Sidhu, J. S. (2004). Monitoring prenatal alcohol exposure. Am. J. Med. Genet. C Semin. Med. Genet. 127C, 3–9. doi: 10.1002/ajmg.c.30010
Glessner, J. T., Bick, A. G., Ito, K., Homsy, J., Rodriguez-Murillo, L., Fromer, M., et al. (2014). Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ. Res. 115, 884–896. doi: 10.1161/CIRCRESAHA.115.304458
Grishina, O. A., Wang, S., and Larina, I. V. (2017). Speckle variance optical coherence tomography of blood flow in the beating mouse embryonic heart. J. Biophotonics 10, 735–743. doi: 10.1002/jbio.201600293
Hershberger, R. E., Norton, N., Morales, A., Li, D., Siegfried, J. D., and Gonzalez-Quintana, J. (2010). Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ. Cardiovasc. Genet. 3, 155–161. doi: 10.1161/CIRCGENETICS.109.912345
Hoang, T. T., Goldmuntz, E., Roberts, A. E., Chung, W. K., Kline, J. K., Deanfield, J. E., et al. (2018). The congenital heart disease genetic network study: cohort description. PLoS ONE 13:e0191319. doi: 10.1371/journal.pone.0191319
Homsy, J., Zaidi, S., Shen, Y., Ware, J. S., Samocha, K. E., Karczewski, K. J., et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. doi: 10.1126/science.aac9396
Jenkins, M. W., Peterson, L., Gu, S., Gargesha, M., Wilson, D. L., Watanabe, M., et al. (2010). Measuring hemodynamics in the developing heart tube with four-dimensional gated Doppler optical coherence tomography. J. Biomed. Opt. 15:066022. doi: 10.1117/1.3509382
Jin, S. C., Homsy, J., Zaidi, S., Lu, Q., Morton, S., DePalma, S. R., et al. (2017). Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601. doi: 10.1038/ng.3970
Jones, K. L., Hoyme, H. E., Robinson, L. K., Del Campo, M., Manning, M. A., Prewitt, L. M., et al. (2010). Fetal alcohol spectrum disorders: Extending the range of structural defects. Am. J. Med. Genet. A 152A, 2731–2735. doi: 10.1002/ajmg.a.33675
Jorgensen, M., McPherson, E., Zaleski, C., Shivaram, P., and Cold, C. (2014). Stillbirth: the heart of the matter. Am. J. Med. Genet. A. 164A, 691–699. doi: 10.1002/ajmg.a.36366
Karunamuni, G., Gu, S., Doughman, Y. Q., Peterson, L. M., Mai, K., McHale, Q., et al. (2014). Ethanol exposure alters early cardiac function in the looping heart: a mechanism for congenital heart defects? Am. J. Physiol. Heart Circ. Physiol. 306, H414–H421. doi: 10.1152/ajpheart.00600.2013
Kathiresan, S., and Srivastava, D. (2012). Genetics of human cardiovascular disease. Cell 148, 1242–1257. doi: 10.1016/j.cell.2012.03.001
Khokha, M. K. (2012). Xenopus white papers and resources: folding functional genomics and genetics into the frog. Genesis 50, 133–142. doi: 10.1002/dvg.22015
Luo, W., Marks, D. L., Ralston, T. S., and Boppart, S. A. (2006). Three-dimensional optical coherence tomography of the embryonic murine cardiovascular system. J. Biomed. Opt. 11:021014. doi: 10.1117/1.2193465
Manheimer, K. B., Richter, F., Edelmann, L. J., D'Souza, S. L., Shi, L., Shen, Y., et al. (2018). Robust identification of mosaic variants in congenital heart disease. Hum. Genet. 137, 183–193. doi: 10.1007/s00439-018-1871-6
Männer, J., Thrane, L., Norozi, K., and Yelbuz, T. M. (2009). In vivo imaging of the cyclic changes in cross-sectional shape of the ventricular segment of pulsating embryonic chick hearts at stages 14 to 17: a contribution to the understanding of the ontogenesis of cardiac pumping function. Dev. Dyn. 238, 3273–3284. doi: 10.1002/dvdy.22159
Marelli, A. J., Mackie, A. S., Ionescu-Ittu, R., Rahme, E., and Pilote, L. (2007). Congenital heart disease in the general population: changing prevalence and age distribution. Circulation 115, 163–172. doi: 10.1161/CIRCULATIONAHA.106.627224
McKean, D. M., Homsy, J., Wakimoto, H., Patel, N., Gorham, J., DePalma, S. R., et al. (2016). Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat. Commun. 7:12824. doi: 10.1038/ncomms12824
Morcos, P. A. (2007). Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem. Biophys. Res. Commun. 358, 521–527. doi: 10.1016/j.bbrc.2007.04.172
Nieuwkoop, P. D., and Faber, J. (1994). Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development From the Fertilized Egg Till the End of Metamorphosis (New York, NY: Garland Publisher), 252, 10 leaves of plates.
O'Donnell, C. J., and Nabel, E. G. (2011). Genomics of cardiovascular disease. N. Engl. J. Med. 365, 2098–2109. doi: 10.1056/NEJMra1105239
Pierpont, M. E., Basson, C. T., Benson, D. W., Gelb, B. D., Giglia, T. M., Goldmuntz, E., et al. (2007). Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115, 3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056
Pierpont, M. E., Brueckner, M., Chung, W. K., Garg, V., Lacro, R. V., McGuire, A. L., et al. (2018). Genetic basis for congenital heart disease: revisited: a scientific statement from the American Heart Association. Circulation 138, e653–711. doi: 10.1161/CIR.0000000000000631
Priest, J. R., Osoegawa, K., Mohammed, N., Nanda, V., Kundu, R., Schultz, K., et al. (2016). De novo and rare variants at multiple loci support the oligogenic origins of atrioventricular septal heart defects. PLoS Genet. 12:e1005963. doi: 10.1371/journal.pgen.1005963
Ramirez, M. S., and Bankson, J. A. (2007). A practical method for 2D multiple-animal MRI. J. Magn. Reson. Imaging. 26, 1162–1166. doi: 10.1002/jmri.21112
Ramirez, M. S., Ragan, D. K., Kundra, V., and Bankson, J. A. (2007). Feasibility of multiple-mouse dynamic contrast-enhanced MRI. Magn. Reson. Med. 58, 610–615. doi: 10.1002/mrm.21348
Roest, A. A., and de Roos, A. (2012). Imaging of patients with congenital heart disease. Nat. Rev. Cardiol. (2012). 9, 101–115. doi: 10.1038/nrcardio.2011.162
Roozen, S., Peters, G. J., Kok, G., Townend, D., Nijhuis, J., and Curfs, L. (2016). Worldwide prevalence of fetal alcohol spectrum disorders: a systematic literature review including meta-analysis. Alcohol. Clin. Exp. Res. 40, 18–32. doi: 10.1111/acer.12939
Sabatine, M. S., Seidman, J. G., and Seidman, C. E. (2006). Cardiovascular genomics. Circulation 113, e450–e455. doi: 10.1161/CIRCULATIONAHA.105.560151
Sanders, S. J., Murtha, M. T., Gupta, A. R., Murdoch, J. D., Raubeson, M. J., Willsey, A. J., et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. doi: 10.1038/nature10945
Tearney, G. J., Bouma, B. E., Boppart, S. A., Golubovic, B., Swanson, E. A., and Fujimoto, J. G. (1996). Rapid acquisition of in vivo biological images by use of optical coherence tomography. Opt. Lett. 21, 1408–1410. doi: 10.1364/OL.21.001408
van der Linde, D., Konings, E. E., Slager, M. A., Witsenburg, M., Helbing, W. A., Takkenberg, J. J., et al. (2011). Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241–2247. doi: 10.1016/j.jacc.2011.08.025
Wang, S., Lakomy, D. S., Garcia, M. D., Lopez, A. L., Larin, K. V., and Larina, I. V. (2016). Four-dimensional live imaging of hemodynamics in mammalian embryonic heart with Doppler optical coherence tomography. J. Biophotonics 9, 837–847. doi: 10.1002/jbio.201500314
Warkman, A. S., and Krieg, P. A. (2007). Xenopus as a model system for vertebrate heart development. Semin. Cell Dev. Biol. 18, 46–53. doi: 10.1016/j.semcdb.2006.11.010
Yelin, R., Yelin, D., Oh, W. Y., Yun, S. H., Boudoux, C., Vakoc, B. J., et al. (2007). Multimodality optical imaging of embryonic heart microstructure. J. Biomed. Opt. 12:064021. doi: 10.1117/1.2822904
Yoo, J., Larina, I. V., Larin, K. V., Dickinson, M. E., and Liebling, M. (2011). Increasing the field-of-view of dynamic cardiac OCT via post-acquisition mosaicing without affecting frame-rate or spatial resolution. Biomed. Opt. Express 2, 2614–2622. doi: 10.1364/BOE.2.002614
Yushkevich, P. A., Piven, J., Hazlett, H. C., Smith, R. G., Ho, S., Gee, J. C., et al. (2006). User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. doi: 10.1016/j.neuroimage.2006.01.015
Zaidi, S., and Brueckner, M. (2017). Genetics and genomics of congenital heart disease. Circ. Res. 120, 923–940. doi: 10.1161/CIRCRESAHA.116.309140
Keywords: Xenopus tadpole, hemoglobin subtraction angiography, human cardiomyopathy, animal model cardiovascular system, videomicroscopy
Citation: Deniz E, Jonas S, Khokha MK and Choma MA (2019) Quantitative Phenotyping of Xenopus Embryonic Heart Pathophysiology Using Hemoglobin Contrast Subtraction Angiography to Screen Human Cardiomyopathies. Front. Physiol. 10:1197. doi: 10.3389/fphys.2019.01197
Received: 28 November 2018; Accepted: 03 September 2019;
Published: 20 September 2019.
Edited by:
Theresia Kraft, Hannover Medical School, GermanyReviewed by:
Rebecca Maree Dyson, University of Otago, New ZealandCopyright © 2019 Deniz, Jonas, Khokha and Choma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Engin Deniz, ZW5naW4uZGVuaXpAeWFsZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.