
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol. , 07 August 2019
Sec. Integrative Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.01014
This article is part of the Research Topic E3 Ubiquitin Ligases: From Structure to Physiology View all 13 articles
Homologous to the E6AP carboxyl terminus (HECT) and regulator of chromosome condensation 1 (RCC1)-like domain-containing proteins (HERCs) belong to the superfamily of ubiquitin ligases. HERC proteins are divided into two subfamilies, Large and Small HERCs. Despite their similarities in terms of both structure and domains, these subfamilies are evolutionarily very distant and result from a convergence phenomenon rather than from a common origin. Large HERC genes, HERC1 and HERC2, are present in most metazoan taxa. They encode very large proteins (approximately 5,000 amino acid residues in a single polypeptide chain) that contain more than one RCC1-like domain as a structural characteristic. Accumulating evidences show that these unusually large proteins play key roles in a wide range of cellular functions which include neurodevelopment, DNA damage repair, and cell proliferation. To better understand the origin, evolution, and function of the Large HERC family, this minireview provides with an integrated overview of their structure and function and details their physiological implications. This study also highlights and discusses how dysregulation of these proteins is associated with severe human diseases such as neurological disorders and cancer.
Proteins containing a HECT domain are ubiquitin ligases (E3). These enzymes participate in the ubiquitylation process accepting ubiquitin from a ubiquitin-conjugating enzymes (E2) and catalysing its transfer to the protein to be ubiquitylated (Buetow and Huang, 2016). In animals, HECT E3 ligases can be divided into 16 groups including the Large HERC family (Marín, 2010), which is the subject of the present minireview. This family is comprised by HERC1 and HERC2, two gigantic proteins of close to 5,000 amino acid residues in a single polypeptide chain. They are the largest HECT-containing proteins1.
Although traditionally classified together with the Small HERC proteins, Large and Small HERCs form two distant protein families (Marín, 2010). Large HERCs contain more than one RCC1-like domains (RLDs), differing from Small HERCs, which carry only one. Structural differences were observed between the RLDs in Large and Small HERCs (Hadjebi et al., 2008). The explanation for the differences among these two protein groups is that they result from convergent evolution of ancestors belonging to distant families (Marín, 2010).
While HERC2 appears in some choanoflagellates such as Monosiga brevicollis and Salpingoeca rosetta, the emergence of HERC1 occurred in Metazoa. Both proteins are already present in the placozoan Trichoplax adhaerens and in most metazoan phyla, with the absence of HERC1 in certain insect clades (Marín, 2010). Phylogenetic analysis of Large HERCs amino acid sequences segregates them in two clusters: one for HERC1 and one for HERC2; displaying higher similarity between orthologues (Figure 1). The phylogenetic relationships of the sequences within each cluster correlate with those in the evolution of the species. It is noteworthy that HERC2 from S. rosetta presents a SPRY domain, which is characteristic of HERC1 (Figure 2A; Garcia-Gonzalo et al., 2005). However, it cannot be considered homologous to that of T. adhaerens HERC1 [19.6% identity, e = 0.17; as shown by BLAST-p comparison (Altschul et al., 1997, 2005)]. Thus, this presence is likely due to convergence, a relatively frequent event in HECT proteins along evolution (Marín, 2010).
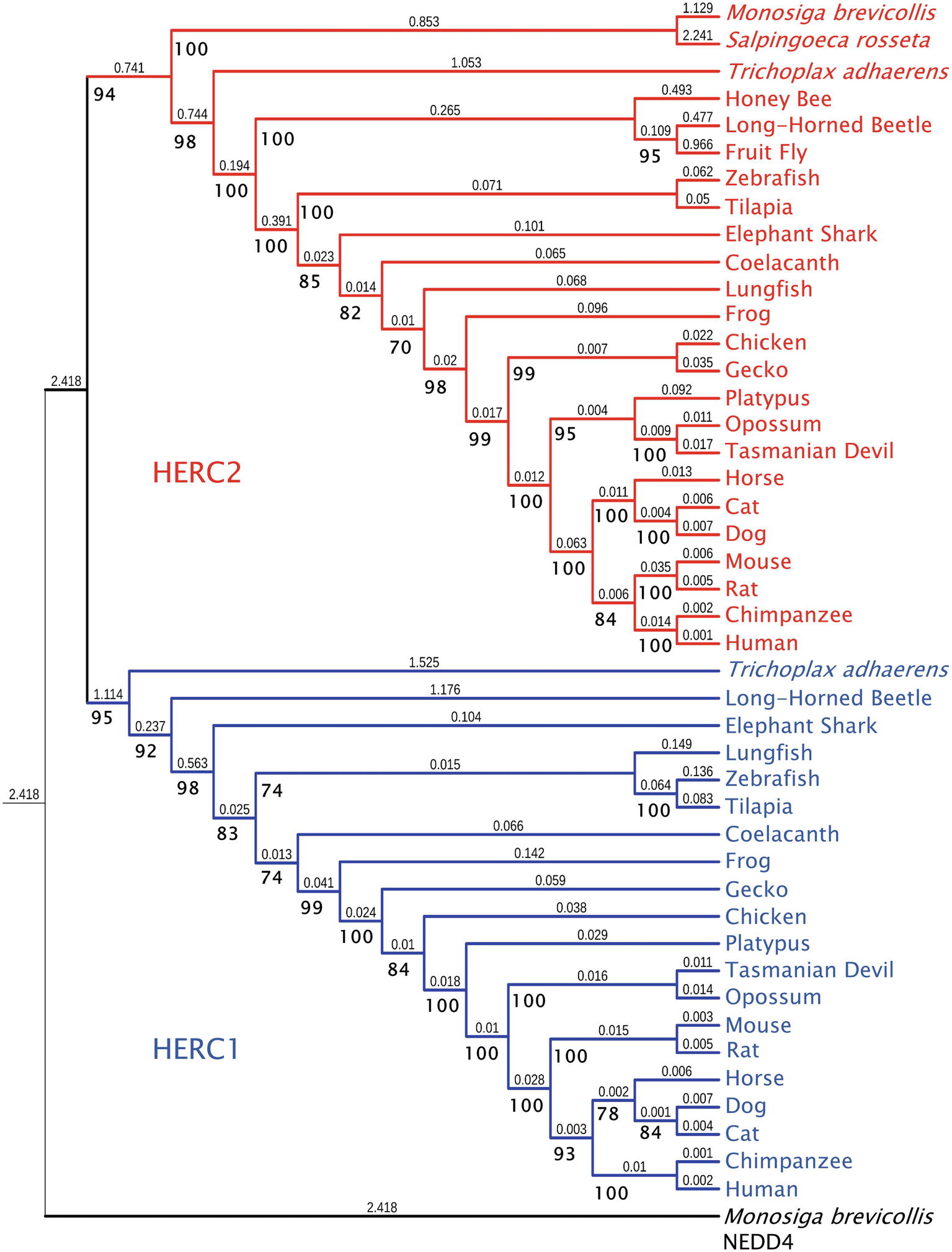
Figure 1. Molecular phylogeny of Large HERC proteins. The tree shows the clustering of HERC1 (blue) and HERC2 (red) amino acid sequences from significative choanoflagellate and metazoan species. Ultrafast bootstrap support (1,000 replicates) is shown next to the branches in bold. Smaller numbers indicate branch lengths. NEDD4 protein from Monosiga brevicollis was used as a outgroup.
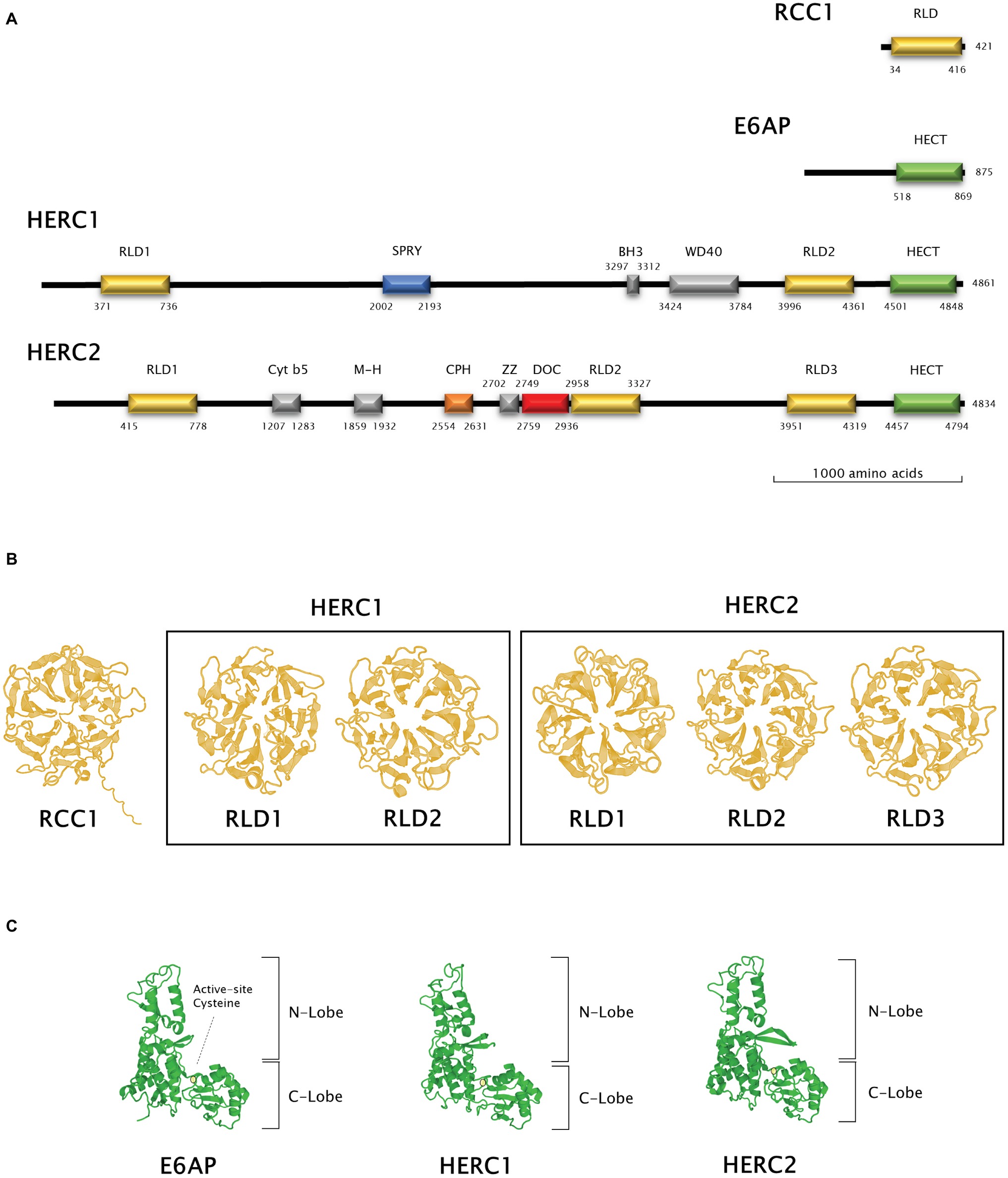
Figure 2. Structural features of Large HERC proteins. (A) Primary structure of human RCC-1, E6AP, HERC1, and HERC2 proteins. Scheme shows the approximate relative position of each significative protein domain as indicated by ScanProSite domain predictions with the amino acid sequences. (B,C) Three-dimensional structures of homologous RCC1-like and HECT domains in HERC1 and HERC2. Three-dimensional structures of RCC1 and E6AP HECT domains were modelled using the Swiss-Model online software and used as a template for HERC1 and HERC2 RCC1-like (B) and HECT (C) domains modelling. Active-site cysteine location in the HECT domains is indicated by a pale yellow circle.
The presence of RLDs is a structural feature of Large HERCs (Figure 2A). RCC1 is necessary for maintaining chromosomes decondensed during DNA replication. It is also a guanine exchange factor (GEF) for the GTPase Ran, a nuclear import protein (Nishimoto et al., 1978; Bischoff and Ponstingl, 1991). RCC1 tertiary structure is composed of seven β blades resembling the shape of a propeller (Figure 2B; first panel). Structure prediction models have been used since three-dimensional structure determination of Large HERCs has not been possible to date (Waterhouse et al., 2018). Large HERC RLDs structure is very similar to that of RCC1 (Figure 2B; framed panels).
HERC1 is implicated in intracellular vesicle trafficking by interacting through its RLD2 with ARF1 and Clathrin (Rosa et al., 1996; Rosa and Barbacid, 1997). HERC1 RLD1 may also function as a GDP releasing factor (GRF) for ARF proteins in the presence of phosphatidylinositol-4,5-bisphosphate (Garcia-Gonzalo et al., 2004, 2005). As a small GTPase regulator, HERC1 interacts, among others, with IQGAP1, which is a key interactor centre for such proteins (Jacquemet and Humphries, 2013). No GEF or GRF activities have been reported in HERC2. Of note, HERC2 forms a complex with and stimulates the E6AP ubiquitin ligase activity through its RLD2 (Kuhnle et al., 2011).
Large HERCs display a HECT domain at their carboxyl end. In silico predicted models show structural similarity with the HECT domain of E6AP (Figure 2C). They form a bilobed structure consisting of a helix-turn-helix motif packed with two and four antiparallel β sheets at the N and C ends, respectively. The lobes are joined to the hinge formed by a core of α helices. This bilobed structure facilitates transmission of the ubiquitin residue to its target protein. Thus, the N-terminal-facing lobe is able to bind the E2 enzyme from which the ubiquityl residue is transferred to the catalytic cysteine within the C-terminal-facing lobe of the domain (Figure 2C; circled). Following that, the ubiquitin is transferred to a lysyl residue or to the amino terminus in the target protein (Metzger et al., 2012; Streich and Lima, 2014).
Large HERCs play a role in protein stability. HERC1 regulates C-RAF stability through ubiquitylation leading to proteasomal degradation (Schneider et al., 2018) and is also implicated in the stability of TSC2/tuberin (Chong-Kopera et al., 2006). HERC2 ubiquitylates for proteasomal degradation proteins involved in DNA repair such as XPA and BRCA1, Ubiquitin Specific Proteases (USP) such as USP33 and USP20, and proteins involved in iron metabolism such as FBXl5 and NCOA4 (reviewed in Sánchez-Tena et al., 2016). HERC2 also promotes degradation of the LKB1 kinase when acetylated (Bai et al., 2016).
HERC2 is implicated in different types of cancer. In osteosarcoma, the increase of HERC2-binding protein SOX18 enhances cell proliferation correlating with a decrease in HERCs mRNA levels, especially those of HERC2 (Zhu et al., 2018). Certain HERC2 genetic variants are risk factors in cutaneous and uveal melanomas (Ibarrola-Villava et al., 2010; Amos et al., 2011; Kosiniak-Kamysz et al., 2014). Frameshift mutations in HERC2 have been described in gastric and colorectal carcinomas with microsatellite instability (Yoo et al., 2011).
HERC2 is also implicated in DNA damage repair (DDR). HERC2 induces BRCA1 degradation in breast cancer. This is inhibited either by binding of TUSC4 to HERC2 (Peng et al., 2015) or by BARD1 binding BRCA1 itself (Wu et al., 2010). Moreover, HERC2 targets XPA for degradation. ATR phosphorylates XPA thus preventing this ubiquitylation while WIP dephosphorylates XPA in a circadian manner (Kang et al., 2010, 2011; Lee et al., 2014). Besides, ATR phosphorylates and also stabilizes USP20 by unbinding it from HERC2. In turn, USP20 stabilizes Claspin, which increases the activity of the ATR-Chk1 axis (Yuan et al., 2014; Zhu et al., 2014). On the other hand, HERC2 binds Claspin among other proteins upon MCM2 phosphorylation, and facilitates replication origin firing (Izawa et al., 2011). In a different way, HERC2 promotes resistance to cisplatin and UV-induced DNA damage by stabilizing the Ubc13-RNF8 tandem and RNF168 and enhancing the recruitment of repair factors such as 53BP1, RAP80, and BRCA1 to the damaged sites (Bekker-Jensen et al., 2010; Danielsen et al., 2012; Bonanno et al., 2016; Mohiuddin et al., 2016). Ubc13-RNF8 binding is facilitated by HERC2 by interacting with the RNF8 FHA domain upon phosphorylation of threonine 4827 of HERC2 triggered by ionizing radiation (IR) (Bekker-Jensen et al., 2010) and by acting as a docking platform upon DNA damage, when HERC2 is SUMOylated by PIAS4 (Bekker-Jensen and Mailand, 2010; Danielsen et al., 2012). Both RNF8 and RNF168 are required for ubiquitin foci formation upon IR (Oestergaard et al., 2012), which are necessary for DDR maintenance. In addition to that, HERC2 stabilizes USP16 which, in turn, deubiquitylates histone 2A so as to terminate the DDR (Zhang et al., 2014). Moreover, during S phase, HERC2 is necessary for RPA ubiquitylation which plays a role in clearing G-quadruplex DNA structures by binding the RecQ DNA helicases BLM and WRN (Wu et al., 2018). Finally, HERC2 along with NEURL4 controls cell proliferation by regulating the transcriptional activity of the tumour suppressor protein p53 through regulation of its oligomerization (Cubillos-Rojas et al., 2014, 2017). All these data suggest that HERC2 functions to control the cell response to genotoxic insults and helps maintain genome stability.
HERC1 is also involved in DDR and cancer. HERC1 deletions affect MSH2 protein levels (Diouf et al., 2011). Besides, it also promotes degradation of BAK in the presence of the E6 protein from the HPV5β virus and prevents cell death in response to UV (Holloway et al., 2015). Furthermore, HERC1 is recurrently mutated in metastatic triple negative (Craig et al., 2013) and in invasive lobular cancer in the breast (Ping et al., 2016). In addition to that, an atypical HERC1-PML transcript fusion mRNA (Walz et al., 2010) and HERC1 mutations are also found in leukaemia (Diouf et al., 2011; Neumann et al., 2015; Johansson et al., 2018). HERC1 binds to the M2 isoform of pyruvate kinase, which is typically found in proliferating tissues and cancer cells although the physiological role for this association has not been elucidated to date (Garcia-Gonzalo et al., 2003; Mazurek, 2008). Moreover, as stated above, HERC1 regulates cell proliferation through C-RAF stability. In particular, accumulation of C-RAF upon HERC1 knockdown results in increased cell proliferation (Schneider et al., 2018). Finally, one singular clinical case of pulmonary sclerosing pneumocytoma revealed several somatic mutations on HERC1 along with other genes such as TSC1 and AKT1 (Fan et al., 2018).
In mice, recessive mutations of the Herc2 gene are associated with defects in growth, motor coordination and fertility (Lehman et al., 1998; Walkowicz et al., 1999). While Herc2 knockout animals are not viable, heterozygous mice have motor impairment (Cubillos-Rojas et al., 2016). In humans, the HERC2 gene locates to chromosome 15 among genes responsible for such disorders as Angelman and Prader-Willi syndromes and Autism spectrum disorders (Ji et al., 1999; Dimitropoulos and Schultz, 2007; Roberts et al., 2014). Recessive mutations in the HERC2 locus are related to symptoms ranging from cognitive delay, ataxia, speech disorders, microcephalia, seizures, facial dysmorphism, hypopigmentation, and other secondary signs such as infections and behavioural alterations (Puffenberger et al., 2012; Harlalka et al., 2013; Neubert et al., 2013; Han et al., 2016; Morice-Picard et al., 2016).
Little is known about the precise molecular mechanism by which HERC2 affects neuronal development and function. HERC2-NEURL4 complex binds RNF8 in neurons regulating synapse formation in vivo. Knockdown of HERC2 resembles the effects in RNF8 depletion and inhibition of RNF8-Ubc13 signalling – an increase in parallel fibres, synaptic boutons, and synapse formation with Purkinje cells (Valnegri et al., 2017). HERC2 could also be linked to other neurological diseases. Parkinson’s disease-associated kinase LRRK2 is known to bind to the HERC2-NEURL4 complex thus regulating endosomal vesicular trafficking and promoting the internalization of the Delta-like 1/Delta Notch ligand, affecting its signalling (Imai et al., 2015).
HERC1 has also major implications in neurodevelopment. Herc1 Gly483Glu mutant mice, termed tambaleante, present with delayed growth, short body, high juvenile mortality, and severe ataxia, the latter due to Purkinje cell loss beyond the age of 2 months, correlating with autophagy and decreased mTOR signalling (Mashimo et al., 2009). Tambaleante mice have impaired neurotransmitter release in neuromuscular junctions (Bachiller et al., 2015), defective ubiquitin-proteasome-driven protein aggregate clearance, and increased autophagic flux in neocortical pyramidal, CA3 hippocampal pyramidal and spinal cord motor neurons (Ruiz et al., 2016) and poor myelinisation and delayed action potential transmission (Bachiller et al., 2018). Furthermore, they have impaired associative learning (Perez-Villegas et al., 2018). In humans, patients with recessive mutations of HERC1 present with intellectual disability, macrocephalia, facial dysmorphism, feeding difficulties, kyphoscoliosis, thicker corpus callosum, seizures, and other clinical signs resembling autism (Ortega-Recalde et al., 2015; Aggarwal et al., 2016; Hashimoto et al., 2016; Nguyen et al., 2016; Kannan et al., 2017; Utine et al., 2017).
The human HERC2 gene locus is upstream of that of the OCA2 gene (mutated in oculocutaneous albinism) and certain HERC2 SNPs can interfere OCA2’s expression thus affecting eye, skin, and hair pigmentation (Eiberg et al., 2008; Kayser et al., 2008; Sturm et al., 2008; Branicki et al., 2009; Nan et al., 2009). Certain phenotypes are favoured by evolution with a traceable population gradient from Europe to Asia (Ulivi et al., 2013; Wilde et al., 2014). Moreover, polymorphisms in the HERC2 locus relate to rosacea (Aponte et al., 2018), macular degeneration (Klein et al., 2014), vitiligo (Jin et al., 2012), and skin photosensitivity (Hernando et al., 2018). As for HERC1, some of its catalogued polymorphisms are more likely to occur in the East Asian population than in the rest of the world (Xue et al., 2009; Yuasa et al., 2009).
HERC2 is also related to inflammation and autoimmune diseases such as Crohn’s disease, ulcerative colitis, type 1 diabetes mellitus, or sarcoidosis (Franke et al., 2008; Weersma et al., 2009; Wang et al., 2010; Fischer et al., 2011; Garcia-Heredia and Carnero, 2017). With regard to HERC1, a downregulation of its expression has been observed in an LPS-induced inflammation model following coenzyme Q10 treatment. (Schmelzer and Doring, 2010).
In addition to this, HERC2 is involved in iron metabolism and ferritinophagy by regulating stability of FBXL5, IRP2, and NCOA4 proteins (Moroishi et al., 2011, 2014; Galligan et al., 2015; Mancias et al., 2015; Ryu et al., 2018) and in toxicology to the response to diisocyanate, toluene, or alcohol intake (Sulovari et al., 2015; Yucesoy et al., 2015; Lim et al., 2017). In regard to toxicology, HERC1 may also play a role in the response of the nervous system to heroin (Drakenberg et al., 2006).
Large-HERC family members are staggeringly complex proteins that can intervene in a wide range of physiological processes, such as proliferation, DNA repair, neurodevelopment, inflammation, or ferritinophagy among others. HERC1 and HERC2 sequences are quite conserved through animal evolution, evolving linearly together with the increase of complexity in nervous, endocrine, and immune systems of organisms. Mutations or reduced expression of Large HERCs are associated with neurological disorders, DNA repair defects, and cancer pointing out the importance that Large HERC proteins have in the abovementioned physiological processes.
Amino acid sequences were aligned using the Mafft FFT-NS-i algorithm (Katoh, 2002). The phylogenetic tree in Figure 1 was inferred using the maximum likelihood method with IQ-Tree 1.6.9 software (Nguyen et al., 2015). The model used for the analysis was JTT + F + G4, determined using ModelFinder (Kalyaanamoorthy et al., 2017). The IQ-Tree search parameters were set to perturbation strength = 0.8 and 500 unsuccessful iterations to stop (numstop = 500). Ultrafast Bootstrap support (Hoang et al., 2018) was calculated from 1,000 replicates. The final tree obtained with IQ-Tree was visualized using Interactive Tree of Life v4 (Letunic and Bork, 2019). NEDD4 protein from M. brevicollis was used as an outgroup. The accession numbers of the sequences used are: for NEDD4: M. brevicollis (XP_001750085.1), for HERC1: T. adhaerens (XP_002116356.1), Anoplophora glabripennis (Asian long-horned beetle) (XP_018567016.1), Callorhinchus milii (Elephant shark) (XP_007906088.1), Latimeria chalumnae (Coelacanth) (XP_005987767.1), Danio rerio (Zebrafish) (XP_021333511.1), Oreochromis niloticus (Tilapia) (XP_013121789.1), Xenopus tropicalis (Frog) (XP_004916026.1), Gekko japonicus (Gecko) (XP_015275623.1), Gallus gallus (Chicken) (XP_004943789.1), Ornithorhynchus anatinus (Platypus) (XP_028921613), Monodelphis domestica (Opossum) (XP_016284147.1), Sarcophilus harrisii (Tasmanian devil) (XP_023353554.1), Equus caballus (Horse) (XP_023471806.1), Rattus norvegicus (Rat) (XP_017451567.1), Mus musculus (Mouse) (NP_663592.3), Homo sapiens (Human) (NP_003913.3), Pan troglodytes (Chimpanzee) (XP_001174017.1), Canis lupus familiaris (Dog) (XP_544717.3), Felis catus (Cat) (XP_023110993.1), and Lepidosiren paradoxa (Lungfish) (Translated from GEHZ01042047.1, NCBI TSA database). For HERC2: M. brevicollis (XP_001743304.1), S. rosetta (XP_004996155.1), T. adhaerens (XP_002112421), Asian long-horned beetle (XP_018562376.1), Drosophila melanogaster (Fruit fly) (NP_608388.2), Apis mellifera (Honey bee) (XP_01677273), Coelacanth (XP_014343905.1), Zebrafish (XP_021332870.1), Tilapia (XP_013130527.1), Frog (XP_012813156.1), Gecko (XP_015266462.1), Chicken (XP_015133197.1), Platypus (XP_016083014.1), Opossum (XP_007501467.1), Tasmanian devil (XP_023356163.1), Horse (XP_023507879.1), Mouse (NP_001347009.1), Rat (XP_006229362.1), Human (NP_004658.3), Chimpanzee (XP_024204823.1), Dog (XP_022272508.1), Cat (XP_023110801.1), Elephant shark (XP_007889299.1), and Lungfish (Translated from GEHZ01036559.1, NCBI TSA).
Human RCC1 (NCBI accession number P18754) and HECT domain from E6AP (NCBI accession number Q05086.4, amino acids from 518 to 869) three-dimensional structures were modelled using Swiss-Model online software (Waterhouse et al., 2018). RLDs and HECT domains from HERC1 and HERC2 were modelled using a User Template mode. The query sequences were the following: Q15751 for HERC1 (amino acids 371–736 and 3996–4361 for RLD1 and RLD2, respectively, and 4501–4848 for the HECT domain) and O95714 for HERC2 (amino acids 415–778, 2958–3327 and 3951–4319 for RLD1, RLD2, and RLD3, respectively, and 4457–4794 for the HECT domain). PDB files from RCC1 and E6AP HECT domain models obtained as described above were used as templates in these queries. The amino acid position of each domain identified was verified using ScanProSite domain predictions (Sigrist et al., 2012).
JG-C and JR conceived and designed the manuscript. JG-C, AM-M, and JR analysed the data and performed figures and tables. JG-C, AM-M, JS-G, LP, and JR wrote the manuscript.
This work was supported by the Spanish Ministerio de Economía, Industria y Competitividad (MINECO-AEI/FEDER, UE; BFU2016-80295-R; and SAF2017-90900-REDT). JG-C received a contract from the Universitat de Barcelona postdoctoral programme in collaboration with the “La Caixa” bank foundation. AM-M was awarded an “FI” PHD Fellowship by the Agència de Gestió dels Ajuts Universitaris i de Recerca (AGAUR) of the Generalitat de Catalunya regional government. JS-G was supported by an FPU PhD (FPU17/02413) grant from the Spanish Ministry of Science, Innovation and Universities and LP received a postdoctoral contract from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 203,528/2017-4), from the Ministry of Science, Technology and Innovation Education of Brazil.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Joan Martí-Carreras for technical assistance and commentaries.
1. https://www.uniprot.org/uniprot/?query=reviewed%3Ayes+AND+organism%3A%22Homo+sapiens+%28Human%29+%5B9606%5D%22&sort=mass; (Bateman et al., 2017).
Aggarwal, S., Bhowmik, A. D., Ramprasad, V. L., Murugan, S., and Dalal, A. (2016). A splice site mutation in HERC1 leads to syndromic intellectual disability with macrocephaly and facial dysmorphism: further delineation of the phenotypic spectrum. Am. J. Med. Genet. A 170, 1868–1873. doi: 10.1002/ajmg.a.37654
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., et al. (2005). Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x
Amos, C. I., Wang, L.-E., Lee, J. E., Gershenwald, J. E., Chen, W. V., Fang, S., et al. (2011). Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 20, 5012–5023. doi: 10.1093/hmg/ddr415
Aponte, J. L., Chiano, M. N., Yerges-Armstrong, L. M., Hinds, D. A., Tian, C., Gupta, A., et al. (2018). Assessment of rosacea symptom severity by genome-wide association study and expression analysis highlights immuno-inflammatory and skin pigmentation genes. Hum. Mol. Genet. 15, 2762–2772. doi: 10.1093/hmg/ddy184
Bachiller, S., Roca-Ceballos, M. A., Garcia-Dominguez, I., Perez-Villegas, E. M., Martos-Carmona, D., Perez-Castro, M. A., et al. (2018). HERC1 ubiquitin ligase is required for normal axonal myelination in the peripheral nervous system. Mol. Neurobiol. 55, 8856–8868. doi: 10.1007/s12035-018-1021-0
Bachiller, S., Rybkina, T., Porras-Garcia, E., Perez-Villegas, E., Tabares, L., Armengol, J. A., et al. (2015). The HERC1 E3 ubiquitin ligase is essential for normal development and for neurotransmission at the mouse neuromuscular junction. Cell. Mol. Life Sci. 72, 2961–2971. doi: 10.1007/s00018-015-1878-2
Bai, B., Man, A. W. C., Yang, K., Guo, Y., Xu, C., Tse, H.-F., et al. (2016). Endothelial SIRT1 prevents adverse arterial remodeling by facilitating HERC2-mediated degradation of acetylated LKB1. Oncotarget 7, 39065–39081. doi: 10.18632/oncotarget.9687
Bateman, A., Martin, M. J., O’Donovan, C., Magrane, M., Alpi, E., Antunes, R., et al. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Bekker-Jensen, S., and Mailand, N. (2010). Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9, 1219–1228. doi: 10.1016/j.dnarep.2010.09.010
Bekker-Jensen, S., Rendtlew Danielsen, J., Fugger, K., Gromova, I., Nerstedt, A., Lukas, C., et al. (2010). HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12, 12–80. doi: 10.1038/ncb2008
Bischoff, F. R., and Ponstingl, H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82. doi: 10.1038/354080a0
Bonanno, L., Costa, C., Majem, M., Sanchez, J.-J., Rodriguez, I., Gimenez-Capitan, A., et al. (2016). Combinatory effect of BRCA1 and HERC2 expression on outcome in advanced non-small-cell lung cancer. BMC Cancer 16:312. doi: 10.1186/s12885-016-2339-5
Branicki, W., Brudnik, U., and Wojas-Pelc, A. (2009). Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype. Ann. Hum. Genet. 73, 160–170. doi: 10.1111/j.1469-1809.2009.00504.x
Buetow, L., and Huang, D. T. (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642. doi: 10.1038/nrm.2016.91
Chong-Kopera, H., Inoki, K., Li, Y., Zhu, T., Garcia-Gonzalo, F. R., Rosa, J. L., et al. (2006). TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J. Biol. Chem. 281, 8313–8316. doi: 10.1074/jbc.C500451200
Craig, D. W., O’Shaughnessy, J. A., Kiefer, J. A., Aldrich, J., Sinari, S., Moses, T. M., et al. (2013). Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol. Cancer Ther. 12, 104–116. doi: 10.1158/1535-7163.MCT-12-0781
Cubillos-Rojas, M., Amair-Pinedo, F., Peiró-Jordán, R., Bartrons, R., Ventura, F., and Rosa, J. L. (2014). The E3 ubiquitin protein ligase HERC2 modulates the activity of tumor protein p53 by regulating its oligomerization. J. Biol. Chem. 289, 14782–14795. doi: 10.1074/jbc.M113.527978
Cubillos-Rojas, M., Schneider, T., Bartrons, R., Ventura, F., Rosa, J. L., Cubillos-Rojas, M., et al. (2017). NEURL4 regulates the transcriptional activity of tumor suppressor protein p53 by modulating its oligomerization. Oncotarget 8, 61824–61836. doi: 10.18632/oncotarget.18699
Cubillos-Rojas, M., Schneider, T., Hadjebi, O., Pedrazza, L., de Oliveira, J. R., Langa, F., et al. (2016). The HERC2 ubiquitin ligase is essential for embryonic development and regulates motor coordination. Oncotarget 7, 56083–56106. doi: 10.18632/oncotarget.11270
Danielsen, J. R., Povlsen, L. K., Villumsen, B. H., Streicher, W., Nilsson, J., Wikstrom, M., et al. (2012). DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197, 179–187. doi: 10.1083/jcb.201106152
Dimitropoulos, A., and Schultz, R. T. (2007). Autistic-like symptomatology in Prader-Willi syndrome: a review of recent findings. Curr. Psychiatry Rep. 9, 159–164. doi: 10.1007/s11920-007-0086-7
Diouf, B., Cheng, Q., Krynetskaia, N. F., Yang, W., Cheok, M., Pei, D., et al. (2011). Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat. Med. 17, 1298–1303. doi: 10.1038/nm.2430
Drakenberg, K., Nikoshkov, A., Horvath, M. C., Fagergren, P., Gharibyan, A., Saarelainen, K., et al. (2006). Opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc. Natl. Acad. Sci. USA 103, 7883–7888. doi: 10.1073/pnas.0600871103
Eiberg, H., Troelsen, J., Nielsen, M., Mikkelsen, A., Mengel-From, J., Kjaer, K. W., et al. (2008). Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 123, 177–187. doi: 10.1007/s00439-007-0460-x
Fan, X., Lin, L., Wang, J., Wang, Y., Feng, A., Nie, L., et al. (2018). Genome profile in a extremely rare case of pulmonary sclerosing pneumocytoma presenting with diffusely-scattered nodules in the right lung. Cancer Biol. Ther. 19, 13–19. doi: 10.1080/15384047.2017.1360443
Fischer, A., Nothnagel, M., Franke, A., Jacobs, G., Saadati, H. R., Gaede, K. I., et al. (2011). Association of inflammatory bowel disease risk loci with sarcoidosis, and its acute and chronic subphenotypes. Eur. Respir. J. 37, 610–616. doi: 10.1183/09031936.00049410
Franke, A., Balschun, T., Karlsen, T. H., Hedderich, J., May, S., Lu, T., et al. (2008). Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat. Genet. 40, 713–715. doi: 10.1038/ng.148
Galligan, J. T., Martinez-Noel, G., Arndt, V., Hayes, S., Chittenden, T. W., Harper, J. W., et al. (2015). Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J. Proteome Res. 14, 953–966. doi: 10.1021/pr501005v
Garcia-Gonzalo, F. R., Bartrons, R., Ventura, F., and Rosa, J. L. (2005). Requirement of phosphatidylinositol-4,5-bisphosphate for HERC1-mediated guanine nucleotide release from ARF proteins. FEBS Lett. 579, 343–348. doi: 10.1016/j.febslet.2004.11.095
Garcia-Gonzalo, F. R., Cruz, C., Munoz, P., Mazurek, S., Eigenbrodt, E., Ventura, F., et al. (2003). Interaction between HERC1 and M2-type pyruvate kinase. FEBS Lett. 539, 78–84. doi: 10.1016/S0014-5793(03)00205-9
Garcia-Gonzalo, F. R., Munoz, P., Gonzalez, E., Casaroli-Marano, R. P., Vilaro, S., Bartrons, R., et al. (2004). The giant protein HERC1 is recruited to aluminum fluoride-induced actin-rich surface protrusions in HeLa cells. FEBS Lett. 559, 77–83. doi: 10.1016/S0014-5793(04)00030-4
Garcia-Heredia, J. M., and Carnero, A. (2017). The cargo protein MAP17 (PDZK1IP1) regulates the immune microenvironment. Oncotarget 8, 98580–98597. doi: 10.18632/oncotarget.21651
Hadjebi, O., Casas-Terradellas, E., Garcia-Gonzalo, F. R., and Rosa, J. L. (2008). The RCC1 superfamily: from genes, to function, to disease. Biochim. Biophys. Acta, Mol. Cell Res. 1783, 1467–1479. doi: 10.1016/j.bbamcr.2008.03.015
Han, J. Y., Park, J., Jang, W., Chae, H., Kim, M., and Kim, Y. (2016). A twin sibling with Prader-Willi syndrome caused by type 2 microdeletion following assisted reproductive technology: a case report. Biomed. reports 5, 18–22. doi: 10.3892/br.2016.675
Harlalka, G. V., Baple, E. L., Cross, H., Kuhnle, S., Cubillos-Rojas, M., Matentzoglu, K., et al. (2013). Mutation of HERC2 causes developmental delay with Angelman-like features. J. Med. Genet. 50, 65–73. doi: 10.1136/jmedgenet-2012-101367
Hashimoto, R., Nakazawa, T., Tsurusaki, Y., Yasuda, Y., Nagayasu, K., Matsumura, K., et al. (2016). Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 61, 199–206. doi: 10.1038/jhg.2015.141
Hernando, B., Sanz-Page, E., Pitarch, G., Mahiques, L., Valcuende-Cavero, F., and Martinez-Cadenas, C. (2018). Genetic variants associated with skin photosensitivity in a southern European population from Spain. Photodermatol. Photoimmunol. Photomed. 34, 415–422. doi: 10.1111/phpp.12412
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Holloway, A., Simmonds, M., Azad, A., Fox, J. L., and Storey, A. (2015). Resistance to UV-induced apoptosis by beta-HPV5 E6 involves targeting of activated BAK for proteolysis by recruitment of the HERC1 ubiquitin ligase. Int. J. Cancer 136, 2831–2843. doi: 10.1002/ijc.29350
Ibarrola-Villava, M., Fernandez, L. P., Pita, G., Bravo, J., Floristan, U., Sendagorta, E., et al. (2010). Genetic analysis of three important genes in pigmentation and melanoma susceptibility: CDKN2A, MC1R and HERC2/OCA2. Exp. Dermatol. 19, 836–844. doi: 10.1111/j.1600-0625.2010.01115.x
Imai, Y., Kobayashi, Y., Inoshita, T., Meng, H., Arano, T., Uemura, K., et al. (2015). The Parkinson’s disease-associated protein kinase LRRK2 modulates notch signaling through the endosomal pathway. PLoS Genet. 11:e1005503. doi: 10.1371/journal.pgen.1005503
Izawa, N., Wu, W., Sato, K., Nishikawa, H., Kato, A., Boku, N., et al. (2011). HERC2 interacts with Claspin and regulates DNA origin firing and replication fork progression. Cancer Res. 71, 5621–5625. doi: 10.1158/0008-5472.CAN-11-0385
Jacquemet, G., and Humphries, M. J. (2013). IQGAP1 is a key node within the small GTPase network. Small GTPases 4, 199–207. doi: 10.4161/sgtp.27451
Ji, Y., Walkowicz, M. J., Buiting, K., Johnson, D. K., Tarvin, R. E., Rinchik, E. M., et al. (1999). The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum. Mol. Genet. 8, 533–542. doi: 10.1093/hmg/8.3.533
Jin, Y., Birlea, S. A., Fain, P. R., Ferrara, T. M., Ben, S., Riccardi, S. L., et al. (2012). Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680. doi: 10.1038/ng.2272
Johansson, P., Klein-Hitpass, L., Choidas, A., Habenberger, P., Mahboubi, B., Kim, B., et al. (2018). SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J. 8:11. doi: 10.1038/s41408-017-0036-5
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kang, T.-H., Lindsey-Boltz, L. A., Reardon, J. T., and Sancar, A. (2010). Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 107, 4890–4895. doi: 10.1073/pnas.0915085107
Kang, T.-H., Reardon, J. T., and Sancar, A. (2011). Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 39, 3176–3187. doi: 10.1093/nar/gkq1318
Kannan, M., Bayam, E., Wagner, C., Rinaldi, B., Kretz, P. F., Tilly, P., et al. (2017). WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc. Natl. Acad. Sci. USA 114, E9308–E9317. doi: 10.1073/pnas.1713625114
Katoh, K. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kayser, M., Liu, F., Janssens, A. C. J. W., Rivadeneira, F., Lao, O., van Duijn, K., et al. (2008). Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am. J. Hum. Genet. 82, 411–423. doi: 10.1016/j.ajhg.2007.10.003
Klein, B. E. K., Howard, K. P., Iyengar, S. K., Sivakumaran, T. A., Meyers, K. J., Cruickshanks, K. J., et al. (2014). Sunlight exposure, pigmentation, and incident age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 55, 5855–5861. doi: 10.1167/iovs.14-14602
Kosiniak-Kamysz, A., Marczakiewicz-Lustig, A., Marcinska, M., Skowron, M., Wojas-Pelc, A., Pospiech, E., et al. (2014). Increased risk of developing cutaneous malignant melanoma is associated with variation in pigmentation genes and VDR, and may involve epistatic effects. Melanoma Res. 24, 388–396. doi: 10.1097/CMR.0000000000000095
Kuhnle, S., Kogel, U., Glockzin, S., Marquardt, A., Ciechanover, A., Matentzoglu, K., et al. (2011). Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J. Biol. Chem. 286, 19410–19416. doi: 10.1074/jbc.M110.205211
Lee, T.-H., Park, J.-M., Leem, S.-H., and Kang, T.-H. (2014). Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair. Oncogene 33, 19–25. doi: 10.1038/onc.2012.539
Lehman, A. L., Nakatsu, Y., Ching, A., Bronson, R. T., Oakey, R. J., Keiper-Hrynko, N., et al. (1998). A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc. Natl. Acad. Sci. USA 95, 9436–9441. doi: 10.1073/pnas.95.16.9436
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Lim, J.-H., Song, M.-K., Cho, Y., Kim, W., Han, S. O., and Ryu, J.-C. (2017). Comparative analysis of microRNA and mRNA expression profiles in cells and exosomes under toluene exposure. Toxicol. In Vitro 41, 92–101. doi: 10.1016/j.tiv.2017.02.020
Mancias, J. D., Pontano Vaites, L., Nissim, S., Biancur, D. E., Kim, A. J., Wang, X., et al. (2015). Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. elife 4:e10308. doi: 10.7554/eLife.10308
Marín, I. (2010). Animal HECT ubiquitin ligases: evolution and functional implications. BMC Evol. Biol. 10:56. doi: 10.1186/1471-2148-10-56
Mashimo, T., Hadjebi, O., Amair-Pinedo, F., Tsurumi, T., Langa, F., Serikawa, T., et al. (2009). Progressive Purkinje cell degeneration in tambaleante mutant mice is a consequence of a missense mutation in HERC1 E3 ubiquitin ligase. PLoS Genet. 5:e1000784. doi: 10.1371/journal.pgen.1000784
Mazurek, S. (2008). “Pyruvate kinase type M2: a key regulator within the tumour metabolome and a tool for metabolic profiling of tumours” in Ernst Schering Foundation symposium proceedings (Germany), 99–124.
Metzger, M. B., Hristova, V. A., and Weissman, A. M. (2012). HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537. doi: 10.1242/jcs.091777
Mohiuddin, Kobayashi, S., Keka, I. S., Guilbaud, G., Sale, J., Narita, T., et al. (2016). The role of HERC2 and RNF8 ubiquitin E3 ligases in the promotion of translesion DNA synthesis in the chicken DT40 cell line. DNA Repair 40, 67–76. doi: 10.1016/j.dnarep.2016.02.002
Morice-Picard, F., Benard, G., Rezvani, H. R., Lasseaux, E., Simon, D., Moutton, S., et al. (2016). Complete loss of function of the ubiquitin ligase HERC2 causes a severe neurodevelopmental phenotype. Eur. J. Hum. Genet. 25, 52–58. doi: 10.1038/ejhg.2016.139
Moroishi, T., Nishiyama, M., Takeda, Y., Iwai, K., and Nakayama, K. I. (2011). The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab. 14, 339–351. doi: 10.1016/j.cmet.2011.07.011
Moroishi, T., Yamauchi, T., Nishiyama, M., and Nakayama, K. I. (2014). HERC2 targets the iron regulator FBXL5 for degradation and modulates iron metabolism. J. Biol. Chem. 289, 16430–16441. doi: 10.1074/jbc.M113.541490
Nan, H., Kraft, P., Qureshi, A. A., Guo, Q., Chen, C., Hankinson, S. E., et al. (2009). Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 129, 2250–2257. doi: 10.1038/jid.2009.62
Neubert, G., von Au, K., Drossel, K., Tzschach, A., Horn, D., Nickel, R., et al. (2013). Angelman syndrome and severe infections in a patient with de novo 15q11.2-q13.1 deletion and maternally inherited 2q21.3 microdeletion. Gene 512, 453–455. doi: 10.1016/j.gene.2012.10.061
Neumann, M., Vosberg, S., Schlee, C., Heesch, S., Schwartz, S., Gokbuget, N., et al. (2015). Mutational spectrum of adult T-ALL. Oncotarget 6, 2754–2766. doi: 10.18632/oncotarget.2218
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nguyen, L. S., Schneider, T., Rio, M., Moutton, S., Siquier-Pernet, K., Verny, F., et al. (2016). A nonsense variant in HERC1 is associated with intellectual disability, megalencephaly, thick corpus callosum and cerebellar atrophy. Eur. J. Hum. Genet. 24, 455–458. doi: 10.1038/ejhg.2015.140
Nishimoto, T., Eilen, E., and Basilico, C. (1978). Premature chromosome condensation in a ts DNA-mutant of BHK cells. Cell 15, 475–483. doi: 10.1016/0092-8674(78)90017-X
Oestergaard, V. H., Pentzold, C., Pedersen, R. T., Iosif, S., Alpi, A., Bekker-Jensen, S., et al. (2012). RNF8 and RNF168 but not HERC2 are required for DNA damage-induced ubiquitylation in chicken DT40 cells. DNA Repair 11, 892–905. doi: 10.1016/j.dnarep.2012.08.005
Ortega-Recalde, O., Beltran, O. I., Galvez, J. M., Palma-Montero, A., Restrepo, C. M., Mateus, H. E., et al. (2015). Biallelic HERC1 mutations in a syndromic form of overgrowth and intellectual disability. Clin. Genet. 88, e1–e3. doi: 10.1111/cge.12634
Peng, Y., Dai, H., Wang, E., Lin, C. C.-J., Mo, W., Peng, G., et al. (2015). TUSC4 functions as a tumor suppressor by regulating BRCA1 stability. Cancer Res. 75, 378–386. doi: 10.1158/0008-5472.CAN-14-2315
Perez-Villegas, E. M., Negrete-Diaz, J. V., Porras-Garcia, M. E., Ruiz, R., Carrion, A. M., Rodriguez-Moreno, A., et al. (2018). Mutation of the HERC 1 ubiquitin ligase impairs associative learning in the lateral amygdala. Mol. Neurobiol. 55, 1157–1168. doi: 10.1007/s12035-016-0371-8
Ping, Z., Siegal, G. P., Harada, S., Eltoum, I.-E., Youssef, M., Shen, T., et al. (2016). ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget 7, 80655–80663. doi: 10.18632/oncotarget.13019
Puffenberger, E. G., Jinks, R. N., Wang, H., Xin, B., Fiorentini, C., Sherman, E. A., et al. (2012). A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 33, 1639–1646. doi: 10.1002/humu.22237
Roberts, J. L., Hovanes, K., Dasouki, M., Manzardo, A. M., and Butler, M. G. (2014). Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene 535, 70–78. doi: 10.1016/j.gene.2013.10.020
Rosa, J. L., and Barbacid, M. (1997). A giant protein that stimulates guanine nucleotide exchange on ARF1 and Rab proteins forms a cytosolic ternary complex with clathrin and Hsp70. Oncogene 15, 1–6. doi: 10.1038/sj.onc.1201170
Rosa, J. L., Casaroli-Marano, R. P., Buckler, A. J., Vilaró, S., and Barbacid, M. (1996). p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. EMBO J. 15, 4262–4273. doi: 10.1002/j.1460-2075.1996.tb00801.x
Ruiz, R., Perez-Villegas, E. M., Bachiller, S., Rosa, J. L., and Armengol, J. A. (2016). HERC 1 ubiquitin ligase mutation affects neocortical, CA3 hippocampal and spinal cord projection neurons: an ultrastructural study. Front. Neuroanat. 10:42. doi: 10.3389/fnana.2016.00042
Ryu, M.-S., Duck, K. A., and Philpott, C. C. (2018). Ferritin iron regulators, PCBP1 and NCOA4, respond to cellular iron status in developing red cells. Blood Cells Mol. Dis. 69, 75–81. doi: 10.1016/j.bcmd.2017.09.009
Sánchez-Tena, S., Cubillos-Rojas, M., Schneider, T., and Rosa, J. L. (2016). Functional and pathological relevance of HERC family proteins: a decade later. Cell. Mol. Life Sci. 73, 1955–1968. doi: 10.1007/s00018-016-2139-8
Schmelzer, C., and Doring, F. (2010). Identification of LPS-inducible genes downregulated by ubiquinone in human THP-1 monocytes. Biofactors 36, 222–228. doi: 10.1002/biof.93
Schneider, T., Martinez-Martinez, A., Cubillos-Rojas, M., Bartrons, R., Ventura, F., Rosa, J. L., et al. (2018). The E3 ubiquitin ligase HERC1 controls the ERK signaling pathway targeting C-RAF for degradation. Oncotarget 9, 31531–31548. doi: 10.18632/oncotarget.25847
Sigrist, C. J. A., de Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2012). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Streich, F. C., and Lima, C. D. (2014). Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379. doi: 10.1146/annurev-biophys-051013-022958
Sturm, R. A., Duffy, D. L., Zhao, Z. Z., Leite, F. P. N., Stark, M. S., Hayward, N. K., et al. (2008). A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 82, 424–431. doi: 10.1016/j.ajhg.2007.11.005
Sulovari, A., Kranzler, H. R., Farrer, L. A., Gelernter, J., and Li, D. (2015). Eye color: a potential indicator of alcohol dependence risk in European Americans. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168B, 347–353. doi: 10.1002/ajmg.b.32316
Ulivi, S., Mezzavilla, M., and Gasparini, P. (2013). Genetics of eye colours in different rural populations on the Silk Road. Eur. J. Hum. Genet. 21, 1320–1323. doi: 10.1038/ejhg.2013.41
Utine, G. E., Taskiran, E. Z., Kosukcu, C., Karaosmanoglu, B., Guleray, N., Dogan, O. A., et al. (2017). HERC1 mutations in idiopathic intellectual disability. Eur. J. Med. Genet. 60, 279–283. doi: 10.1016/j.ejmg.2017.03.007
Valnegri, P., Huang, J., Yamada, T., Yang, Y., Mejia, L. A., Cho, H. Y., et al. (2017). RNF8/UBC13 ubiquitin signaling suppresses synapse formation in the mammalian brain. Nat. Commun. 8:1271. doi: 10.1038/s41467-017-01333-6
Walkowicz, M., Ji, Y., Ren, X., Horsthemke, B., Russell, L. B., Johnson, D., et al. (1999). Molecular characterization of radiation- and chemically induced mutations associated with neuromuscular tremors, runting, juvenile lethality, and sperm defects in jdf2 mice. Mamm. Genome 10, 870–878. doi: 10.1007/s003359901106
Walz, C., Grimwade, D., Saussele, S., Lengfelder, E., Haferlach, C., Schnittger, S., et al. (2010). Atypical mRNA fusions in PML-RARA positive, RARA-PML negative acute promyelocytic leukemia. Genes Chromosomes Cancer 49, 471–479. doi: 10.1002/gcc.20757
Wang, K., Baldassano, R., Zhang, H., Qu, H.-Q., Imielinski, M., Kugathasan, S., et al. (2010). Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 19, 2059–2067. doi: 10.1093/hmg/ddq078
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Weersma, R. K., Stokkers, P. C. F., Cleynen, I., Wolfkamp, S. C. S., Henckaerts, L., Schreiber, S., et al. (2009). Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch-Belgian cohort. Am. J. Gastroenterol. 104, 630–638. doi: 10.1038/ajg.2008.112
Wilde, S., Timpson, A., Kirsanow, K., Kaiser, E., Kayser, M., Unterlander, M., et al. (2014). Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc. Natl. Acad. Sci. USA 111, 4832–4837. doi: 10.1073/pnas.1316513111
Wu, W., Rokutanda, N., Takeuchi, J., Lai, Y., Maruyama, R., Togashi, Y., et al. (2018). HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-Quadruplex DNA. Cancer Res. 78, 6371–6385. doi: 10.1158/0008-5472.CAN-18-1877
Wu, W., Sato, K., Koike, A., Nishikawa, H., Koizumi, H., Venkitaraman, A. R., et al. (2010). HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 70, 6384–6392. doi: 10.1158/0008-5472.CAN-10-1304
Xue, Y., Zhang, X., Huang, N., Daly, A., Gillson, C. J., Macarthur, D. G., et al. (2009). Population differentiation as an indicator of recent positive selection in humans: an empirical evaluation. Genetics 183, 1065–1077. doi: 10.1534/genetics.109.107722
Yoo, N. J., Park, S. W., and Lee, S. H. (2011). Frameshift mutations of ubiquitination-related genes HERC2, HERC3, TRIP12, UBE2Q1 and UBE4B in gastric and colorectal carcinomas with microsatellite instability. Pathology 43, 753–755. doi: 10.1097/PAT.0b013e32834c7e78
Yuan, J., Luo, K., Deng, M., Li, Y., Yin, P., Gao, B., et al. (2014). HERC2-USP20 axis regulates DNA damage checkpoint through Claspin. Nucleic Acids Res. 42, 13110–13121. doi: 10.1093/nar/gku1034
Yuasa, I., Umetsu, K., Nishimukai, H., Fukumori, Y., Harihara, S., Saitou, N., et al. (2009). HERC1 polymorphisms: population-specific variations in haplotype composition. Cell Biochem. Funct. 27, 402–405. doi: 10.1002/cbf.1582
Yucesoy, B., Kaufman, K. M., Lummus, Z. L., Weirauch, M. T., Zhang, G., Cartier, A., et al. (2015). Genome-wide association study identifies novel loci associated with diisocyanate-induced occupational asthma. Toxicol. Sci. 146, 192–201. doi: 10.1093/toxsci/kfv084
Zhang, Z., Yang, H., and Wang, H. (2014). The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J. Biol. Chem. 289, 32883–32894. doi: 10.1074/jbc.M114.599605
Zhu, D., Yang, D., Li, X., and Feng, F. (2018). Heterogeneous expression and biological function of SOX18 in osteosaroma. J. Cell. Biochem. 119, 4184–4192. doi: 10.1002/jcb.26635
Keywords: ubiquitin, ligase, HERC, structure, function, cancer, neurobiology, evolution
Citation: García-Cano J, Martinez-Martinez A, Sala-Gaston J, Pedrazza L and Rosa JL (2019) HERCing: Structural and Functional Relevance of the Large HERC Ubiquitin Ligases. Front. Physiol. 10:1014. doi: 10.3389/fphys.2019.01014
Received: 26 November 2018; Accepted: 23 July 2019;
Published: 07 August 2019.
Edited by:
Julien Licchesi, University of Bath, United KingdomReviewed by:
Michael B. Morris, University of Sydney, AustraliaCopyright © 2019 García-Cano, Martinez-Martinez, Sala-Gaston, Pedrazza and Rosa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jose Luis Rosa, am9zZWx1aXNyb3NhQHViLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.