
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 26 July 2019
Sec. Invertebrate Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.00977
This article is part of the Research TopicTick and Tick-Borne Pathogens: Molecular and Immune Targets for Control StrategiesView all 25 articles
Ticks and tick-borne pathogens affect health and welfare of companion animals worldwide, and some human tick-borne diseases are associated with exposure to domestic animals. Vaccines are the most environmentally friendly alternative to acaracides for the control of tick infestations, and to reduce the risk for tick-borne diseases affecting human and animal health. However, vaccines have not been developed or successfully implemented for most vector-borne diseases. The main limitation for the development of effective vaccines is the identification of protective antigens. To address this limitation, in this study we used an experimental approach combining vaccinomics based on transcriptomics and proteomics data with vaccination trials for the identification of tick protective antigens. The study was focused on Ixodes ricinus and Dermacentor reticulatus that infest humans, companion animals and other domestic and wild animals, and transmit disease-causing pathogens. Tick larvae and adult salivary glands were selected for analysis to target tick organs and developmental stages playing a key role during tick life cycle and pathogen infection and transmission. Two I. ricinus (heme lipoprotein and uncharacterized secreted protein) and five D. reticulatus (glypican-like protein, secreted protein involved in homophilic cell adhesion, sulfate/anion exchanger, signal peptidase complex subunit 3, and uncharacterized secreted protein) proteins were identified as the most effective protective antigens based on the criteria of vaccine E > 80%. The putative function of selected protective antigens, which are involved in different biological processes, resulted in vaccines affecting multiple tick developmental stages. These results suggested that the combination of some of these antigens might be considered to increase vaccine efficacy through antigen synergy for the control of tick infestations and potentially affecting pathogen infection and transmission. These antigens were proposed for commercial vaccine development for the control of tick infestations in companion animals, and potentially in other hosts for these tick species.
Tick-borne pathogens cause medically important infections affecting dogs and other pet species worldwide (Liu et al., 2018; Skotarczak, 2018), and some human tick-borne diseases are associated with exposure to domestic animals (Estrada-Peña et al., 2008; Kwit et al., 2018; Escárcega-Ávila et al., 2019). The tick species (Acari: Ixodidae), Ixodes ricinus (Linnaeus, 1758) and Dermacentor reticulatus (Fabricius, 1794) infest humans, pets and other domestic and wild animals. I. ricinus transmits disease-causing pathogens such as Borrelia spp. (Lyme disease and various borreliosis), tick-borne encephalitis virus (TBEV; tick-borne encephalitis) and Anaplasma phagocytophilum (human and animal anaplasmosis) while D. reticulatus is a vector for Francisella tularensis (tularemia), Rickettsia spp. (human and animal rickettsiosis), Omsk hemorrhagic fever virus (OHFV; Omsk hemorrhagic fever), and Babesia canis (canine babesiosis) (Glickman et al., 2006; de la Fuente et al., 2008, 2015; Beugnet and Marié, 2009).
Vaccines have not been developed or successfully implemented for most vector-borne diseases (VBD) affecting humans and animals (de la Fuente et al., 2017b). Therefore, reduction of arthropod vector infestations is important for the control of VBD (de la Fuente and Kocan, 2003; Sperança and Capurro, 2007; Karunamoorthi, 2011; Coller et al., 2012; de la Fuente and Contreras, 2015; de la Fuente et al., 2017b; de la Fuente, 2018). Traditional control methods for arthropod vector infestations are based on the use of chemical acaricides with associated drawbacks such as selection of arthropod-resistant strains and contamination of both the environment and animal products (de la Fuente and Kocan, 2003; de la Fuente et al., 2017b). Vaccination is an environmentally friendly alternative for the control of vector infestations and pathogen infections that allows control of several VBD by targeting their common vector (de la Fuente et al., 2007, 2011, 2017b; de la Fuente and Contreras, 2015; de la Fuente, 2018). Vaccines could be developed to target different tick developmental stages and functions on various hosts with the advantage of avoiding environmental contamination and selection of pesticide resistant arthropod vectors while improving animal welfare and production (de la Fuente et al., 2017b; de la Fuente, 2018). The experience with the only commercial vaccines available for the control of ectoparasite infestations, TickGard and Gavac, demonstrated that these vaccines contribute to the control of cattle tick populations while reducing acaricide applications, but were difficult to introduce into the market because of the absence of immediate effect on tick infestations and the application in combination with other control measures (de la Fuente and Kocan, 2003, 2006; Willadsen, 2004; de la Fuente et al., 2007). The hypothesis behind tick vaccine protective capacity is that ticks feeding on immunized hosts ingest antibodies specific for the target antigen that could reduce its levels and biological activity and/or interact with conserved epitopes in other proteins resulting in reduced tick feeding, development and reproduction (de la Fuente et al., 2011, 2017b; Moreno-Cid et al., 2011; de la Fuente, 2018).
The limiting step in developing tick vaccines is the identification of protective antigens (de la Fuente and Kocan, 2003; de la Fuente et al., 2018). Recent developments in omics analyses of both ticks and tick-borne pathogen and the application of systems biology to the study of tick-host-pathogen molecular interactions have advanced our understanding of the genetic factors and molecular pathways involved at the tick-host, tick-pathogen and host-pathogen interface (de la Fuente, 2012; de la Fuente et al., 2017a). These technologies are generating extensive information, but algorithms are needed to use these data for advancing knowledge on basic biological questions and the discovery of candidate protective antigens for the development of improved vaccines for the control of ticks and VBD (de la Fuente and Merino, 2013; de la Fuente et al., 2016, 2018; Contreras et al., 2018).
Acaricides have been effectively used to reduce tick infestations in companion animals (Otranto, 2018). However, ticks and transmitted pathogens continue to be a major health problem in dogs and other companion animals that require the development of effective vaccines (Otranto, 2018). Previous experiments have shown the possibility of using vaccines with tick gut protein extracts, Subolesin/Akirin and Bm96 for the control of Rhipicephalus sanguineus infestations in dogs (de la Fuente et al., 2015), and with pathogen-derived antigens for the control of VBD (e.g., Alhassan et al., 2018; Grosenbaugh et al., 2018). Nevertheless, vaccines for the control of tick infestations and transmitted diseases in companion animals have not been registered and commercialized.
In this study, we used a vaccinomics approach based on transcriptomics and proteomics data (de la Fuente and Merino, 2013; de la Fuente et al., 2016) in combination with vaccination trials for the discovery of tick protective antigens for the control of I. ricinus and D. reticulatus infestations in companion animals. Dogs and rabbits were used as model animals. Tick larvae and adult salivary glands were selected for analysis. Tick larvae are the first developmental stage to infest hosts and acquire infection or transmit pathogens that are transovarially transmitted. Salivary glands produce a cocktail of anti-clotting, anti-platelet, anti-inflammatory, vasodilatatory and immunomodulatory proteins that interfere with host defenses to aid in tick blood feeding and pathogen infection and transmission (Ayllón et al., 2015; Chmelaø et al., 2016; Nuttall, 2018). The experiments resulted in the identification of new antigens that showed a protective efficacy of vaccination against I. ricinus and D. reticulatus infestations in rabbits and dogs.
White rabbits (Oryctolagus cuniculus) and Beagle dogs (Canis lupus familiaris) were used in the experiments. Animal experiments were conducted in strict accordance with the recommendations of the European Guide for the Care and Use of Laboratory Animals. Animals were housed and experiments conducted at LLC ACRO Vet Lab (Pylipovichi village, Kiev region, Ukraine) with the approval and supervision of the Ukrainian Commission for Bioethics and Biosafety for animals under the studies “Tick vaccine experiment on rabbits” number 000369 and “Tick vaccine experiments on dogs” numbers 000576, 000577 and 000761.
Ixodes ricinus ticks were originally obtained from the reference laboratory colony maintained at the tick rearing facility of the Institute of Parasitology of the Biology Centre of the Academy of Sciences of the Czech Republic (Genomic Resources Development Consortium, Contreras et al., 2014). D. reticulatus ticks were obtained from a Dutch colony maintained at the Utrecht Centre for Tick-borne Diseases (UCTD), Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands (Villar et al., 2014). Tick colonies were maintained at the LLC ACRO Vet Lab (Pylipovichi village, Kiev region, Ukraine) and used for analysis and vaccination trials. I. ricinus unfed larvae whole internal tissues (IL) and unfed adult salivary glands (ISG), and D. reticulatus unfed larvae whole internal tissues (DL) and unfed adult salivary glands (DSG) were processed. RNA and proteins were extracted from approximately 500 larvae using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA, United States) and from 50 salivary glands (25 females and 25 males) using Tri Reagent (Sigma-Aldrich, St. Louis, MO, United States) according to manufacturer instructions. RNA was purified with the RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, United States) and characterized using the Agilent 2100 Bioanalyzer (Santa Clara, CA, United States) in order to evaluate the quality and integrity of RNA preparations. RNA concentration was determined using the Nanodrop ND-1000 (NanoDrop Technologies Wilmington, DE, United States). Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, United States) with BSA as standard. RNA yield was 10 μg (IL), 4 μg (ISG), 3.5 μg (DL), and 6 μg (DSG). Protein yield was 8 mg (IL and DL), 1.4 mg (ISG), and 3 mg (DSG).
Purified RNAs were used for library preparation using the TruSeq RNA sample preparation kit v.1 and the standard low throughput procedure (Illumina, San Diego, CA, United States) as previously reported (Genomic Resources Development Consortium, Contreras et al., 2014; Villar et al., 2014). Briefly, 1 μg total RNA was used as starting material for library preparation with the exception of the DL sample for which 0.7 μg were used. Messenger RNA was captured using poly-dT magnetic beads and purified polyA+ RNA was chemically fragmented and reverse-transcribed. Remaining RNA was enzymatically removed, and the second strand generated following an end repair procedure and preparation of double-stranded cDNA for adaptor ligation. Adaptor oligonucleotides containing the signals for subsequent amplification and sequencing were ligated to both ends and cDNA samples were washed using AMPure SPRI-based magnetic beads (Beckman Coulter, IZASA, Barcelona, Spain). Adapters contained identifiers, which allow multiplexing in the sequencing run. An enrichment procedure based on PCR was then performed to ensure that all molecules in the library conserved the adapters at both ends. The number of PCR cycles was adjusted to 10 for all samples except for DL which needed up to 15 cycles. The final amplified library was checked again on a BioAnalyzer 2100 (Agilent, Santa Clara, CA, United States). Libraries were titrated by quantitative PCR using a reference standard to characterize molecule concentration per library (IL, 5.51 nM; ISG, 62.16 nM; DL, 12.44 nM; DSG, 100.95 nM). Libraries were denatured and seeded on the respective lanes of the flowcells at a final concentration after re-naturalization of 10–14 pM. IL and ISG samples were run in single-nucleotide flowcells while DL and DSG samples were run in double-nucleotide flowcells to allow for pair-end sequencing. After binding, clusters were formed in the flowcells by local amplification using a Cluster Station apparatus (Illumina). Following sequencing primer annealing, flowcells were loaded into a GAIIx equipment (Illumina) to perform sequencing using the TruSeq® system (Illumina). IL and ISG samples were run under a 1 × 75 bp single-end read protocol while DL and DSG samples were run under a pair-end 2 × 100 bp protocol for de novo RNA sequencing (RNAseq). After sequencing and quality filtering, reads were split according to their different identifiers and fastq files were generated to proceed to quality analysis and gene expression analysis and/or de novo transcript assembly. Transcriptomics data was submitted to Dryad Digital Repository1 or published here (Supplementary Dataset 1) and by Villar et al. (2014) for I. ricinus and D. reticulatus samples, respectively.
The pipeline used for bioinformatics analysis of I. ricinus transcriptomics data was similar to that described by Twine et al. (2011) as previously reported (Genomic Resources Development Consortium, Contreras et al., 2014). The reads from both IL and ISG samples were mapped to the reference I. scapularis genome (assembly JCVI_ISG_i3_1.02) using a TopHat-Cufflinks-Cuffdiff pipeline (Langmead et al., 2009; Trapnell et al., 2009). Transcript abundance was estimated using Cufflinks by analyzing the number of reads mapped to each transcript and handling the deviations due to the sample preparation (Langmead et al., 2009). Finally, differential expression between IL and ISG was characterized using Cuffdiff, integrated into the Cufflinks package (Langmead et al., 2009). For analysis of genes coding for secreted proteins, 6 subcellular localization predictors were chosen: WoLF PSORT3, TargetP4, Protein Prowler5, SLP-Local6, PredSL7 and CELLO8. All these tools have web services that enable batch protein sequence submissions. Results for genes encoding for predicted secreted proteins in IL and ISG were included in Supplementary Dataset 2.
For D. reticulatus transcriptomics data, the 21,838,507 reads (∼2.2 Gb) in DSG were compared against the I. ricinus selected reference gene set (Table 1) using PROmer (Kurtz et al., 2004). The query sequences were considered in their 3 reading frames and the reference sequences only in the frame 1. PROmer results were filtered by selecting the hits with a minimum length of 60 bp for the extended similarity region and a minimum identity of 80% in this region. Reads with hits fulfilling these requirements were clustered to the selected gene with which they have PROmer hits. We selected the read with hits and always also its paired read. The reads belonging to each cluster were then assembled with Velvet (Zerbino and Birney, 2008; Zerbino et al., 2009) to produce a set of contigs clustered to the reference genes. A BLASTX was then done against all proteins from Ixodidae included in the nr database. For each cluster of contigs, we chose the most similar protein to which the contigs aligned with more similarity, and substituted that protein as the new reference protein for that cluster. As expected, in many cases the most similar protein was identical to the initial I. ricinus reference protein. Differential gene expression between DL and DSG samples was determined using χ2 test statistics with Bonferroni correction (P = 0.05) in the IDEG6 software9.
Proteins were resuspended in 5% SDS and frozen at −20°C until use. Two hundred μg of protein extracts from the four samples, IL, ISG, DL and DSG, were mixed with the same quantity of Laemmli sample buffer x2 and applied using a 5-well comb on a conventional SDS-PAGE gel (1.5 mm-thick, 4% stacking, 10% resolving). The electrophoretic run was stopped as soon as the front entered 3 mm into the resolving gel, so that the whole proteome became concentrated in the stacking/resolving gel interface. The unseparated protein bands were visualized by Coomassie Brilliant Blue R-250 staining, excised, cut into cubes (2 × 2 mm) and treated with 0.1% RapiGest SF surfactant (Waters, Milford. MA, United States) according to manufacturer’s protocol in order to enhance the subsequent in-gel trypsin digestion. After RapiGest treatment, samples were digested overnight at 37°C with 60 ng/μl trypsin (Promega, Madison, WI, United States) at 5:1 protein:trypsin (w/w) ratio in 50 mM ammonium bicarbonate, pH 8.8 containing 10% (v/v) acetonitrile. The resulting tryptic peptides from each proteome were extracted by 1 h-incubation in 12 mM ammonium bicarbonate, pH 8.8. Trifluoroacetic acid (TFA) was added to a final concentration of 1% and the peptides were finally desalted onto C18 OASIS HLB extraction cartridges (Waters, Milford, MA, United States) and dried-down prior to reverse phase high performance liquid chromatography (RP-HPLC) method coupled with mass spectrometry (RP-HPLC-MS/MS) analysis using a Surveyor LC system coupled to an ion trap mass spectrometer model LCQ Fleet (Thermo-Finnigan, San Jose, CA, United States). Peptides were eluted using a 180-min gradient from 5 to 40% solvent B in solvent A (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile). The LCQ Fleet was programmed to perform a data-dependent MS/MS scan on the 3 most intense precursors detected in a full scan from 400 to 1600 amu (1 μscan, 200 ms injection time). Protein identification was conducted using SEQUEST algorithm (Proteome Discoverer 1.1 package, Thermo Finnigan), allowing optional modifications in Methionine (oxidation) and Cysteine (carboxamidomethylation). Proteomics data was included in Supplementary Dataset 3 or published by Villar et al. (2014) for I. ricinus and D. reticulatus samples, respectively.
The MS/MS raw files were searched against the Ixodidae Uniprot database (28,771 entries in February 2012) with the following constraints: tryptic cleavage after Arginine and Lysine, up to two missed cleavage sites, and tolerances of 0.5 Da for precursor ions and 0.8 Da for MS/MS fragment ions. Protein assignations were first filtered with a SEQUEST XCorr > 2, but only proteins with false discovery rate (FDR) ≤ 0.01 and identified with at least 4 peptides were selected for further analysis from the Ixodidae database. The MS/MS data was also used to search against the database of predicted secreted proteins encoded by the selected top 1,000 expressed genes in both IL and ISG. In this case, proteins with sequence span > 2% were selected for further analysis. Differential protein representation between L and SG samples was determined using χ2 test statistics with Bonferroni correction (P = 0.001) in the IDEG6 software (see footnote 9).
Gene ontology (GO) analysis for Molecular Function (MF), Cellular Component (CC) and/or Biological Process (BP) was conducted using the cDNA Annotation System software (dCAS; Bioinformatics and Scientific IT Program (BSIP), Office of Technology Information Systems (OTIS), National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD, United States10), AmiGO11 and UniProt12 using non-redundant sequence database (nr) and databases of tick-specific sequences13, 14. The GO annotations for selected candidate tick protective antigens were included in Supplementary Dataset 4.
First, selected I. ricinus orthologs and D. reticulatus de novo generated contigs were amplified by RT-PCR with I. scapularis or D. reticulatus sequence-specific primers and conditions (Supplementary Table 1) using IL, ISG, DL and DSG RNA and the iScript One-Step RT-PCR Kit with SYBR Green and the iQ5 thermal cycler (Bio-Rad, Hercules, CA, United States) following manufacturer’s recommendations. The genes that did not produce an amplicon after RT-PCR were eliminated from further analyses (Tables 1, 2). Remaining coding regions for I. scapularis sequences corresponding to selected I. ricinus orthologous sequences and D. reticulatus de novo generated contig sequences were synthesized (GenScript, Hong Kong) with optimized codon usage for Escherichia coli (Supplementary Dataset 5).
For gene expression and the production of the recombinant proteins, E. coli BL21 Star (DE3) One Shot cells (Invitrogen-Life Technolgies, Inc., Grand Island, NY, United States) were transformed with the target gene cloned into the pET101/D-TOPO expression vector (Invitrogen-Life Technolgies). Recombinant E. coli were inoculated into 10 ml of Luria-Bertani (LB) containing 50 μg/ml ampicillin (Sigma-Aldrich, St Louis, MO, United States) and 0.4% glucose (Laboratorios CONDA S.A., Madrid, Spain) and kept growing overnight at 37°C with shaking. Two ml of the overnight culture was propagated into 250 ml flasks containing 50 ml LB, 50 μg/ml ampicillin and 0.4% glucose for 2 h at 37°C and 200 rpm, and then for 4 h after addition of 0.5 mM final concentration of isopropyl-β-d-thiogalactopyranoside (IPTG, Sigma-Aldrich) for induction of gene expression. The cells were harvested by centrifugation at 3,900 × g for 15 min at 4°C and stored at −80°C for protein purification. One g of the cells harvested after induction of gene expression were resuspended in 5 ml of lysis buffer (100 mM Tris-HCl, pH 7.5, 250 mM NaCl, 7 M Urea, 10 mM imidazole) containing protease inhibitor (Ref. 04693132001, Roche, San Cugat del Vallés, Barcelona, Spain) and disrupted using a cell sonicator (Model MS73; Bandelin Sonopuls, Berlin, Germany). After disruption, the insoluble protein fraction containing the recombinant antigens as inclusion bodies were collected by centrifugation at 15,000 × g for 15 min at 4°C and stored at −20°C before purification. The purification of the recombinant protein was conducted using the automated Maxwell 16 Polyhistidine Protein Purification Kit (Promega, Madison, WI, United States). The eluted fraction containing the purified denatured proteins was dialyzed against 1000 volumes of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), pH 7.4 for 12 h at 4°C. Recombinant proteins were then concentrated using an Amicon Ultra-15 ultrafiltration device (cut off 10 kDa) (Millipore-Merck, Darmstadt, Germany), and adjusted to 0.5 mg/ml. Protein concentration was determined using bicinchoninic acid (Pierce BCA Protein Assay Kit, Thermo Scientific, Rockford, IL, United States). For vaccine formulation, recombinant proteins or saline control were adjuvated in Montanide ISA 50 V2 (Seppic, Paris, France) for rabbit trials or Montanide PET GEL A (Seppic), a ready-to-disperse polymeric adjuvant designed to improve the safety and efficacy of vaccines for companion animals (Parker et al., 2009) for dog trials to a final protein concentration of 250 μg/ml (Merino et al., 2013; Contreras and de la Fuente, 2016, 2017).
Three two-year-old rabbits per group were assigned to I. ricinus or D. reticulatus trial injected subcutaneously (dorsum between shoulders) at days 0 (T1) and 14 (T2) with 0.2 ml (50 μg) doses of recombinant protein vaccine formulations or adjuvant/saline using a syringe with removable needle (0.45 × 13 mm). Two weeks after the last immunization (day 28; T3), rabbits in vaccinated and control groups were infested with 200 I. ricinus or D. reticulatus tick larvae of approximately 21 days after hatching placed on bags located on each rabbit’s shaved ear as described previously (Contreras and de la Fuente, 2016, 2017). Immunizations, tick larval infestations, collections and evaluations were done blinded and the key to the experimental groups was not disclosed until the end of the experiment. Engorged tick larvae were collected, counted and weighted as they dropped off between days 31 to 34. Fed larvae were incubated at 21°C, 80–82% humidity with 7 h dark and 17 h light photoperiod until molting. The tick larvae successfully molting to nymphal stage were collected and counted between days 51 to 65. The number of engorged larvae, weight/larvae, and percent of larvae molting to nymphs were evaluated. Data were analyzed statistically to compare results for each tick species between individuals fed on vaccinated and adjuvant/saline injected control rabbits by Student’s t-test with unequal variance (P = 0.05). Vaccine efficacy (E) was calculated as E (%) = 100 [1−(DL × DMN)], where DL is the reduction in the number of engorged larvae and DMN is the reduction in the percent of larvae molting to nymphs in ticks fed on vaccinated rabbits when compared to the controls fed on adjuvant/saline injected rabbits (Contreras and de la Fuente, 2016, 2017). Only parameters with statistically significant differences were included in vaccine E calculation. Results were included in Supplementary Dataset 6.
Three randomly mixed breed male and female dogs (age > 6 months; body weight > 2 Kg) per group were assigned to I. ricinus or D. reticulatus trial and injected subcutaneously (loose skin on the side of the chest) at days 0 (T1) and 21 (T2) with 0.2 ml (50 μg) doses of recombinant protein vaccine formulations or adjuvant/saline using a syringe with removable needle (0.45 × 13 mm). Two weeks after the last immunization (day 36; T3), dogs in vaccinated and control groups were infested with I. ricinus or D. reticulatus 25:30 (female:male ratio) adults of at least 10 days after molting and 100 nymphs of approximately 15 days after molting placed on feeding chambers (one chamber per tick stage) glued on each dog’s shaved flank. Immunizations, tick infestations, collections and evaluations were done blinded and the key to the experimental groups was not disclosed until the end of the experiment. Engorged tick females were collected, counted and weighted as they dropped off between days 41 and 48. Fed females were incubated at 21°C, 80–82% humidity with 24 h dark photoperiod for approximately 1 month until oviposition. Egg mass per tick was weighted and incubated for fertility determined by counting the number of larvae per tick. Engorged tick nymphs were collected and counted as they dropped off between days 40 to 43. Detached engorged nymphs were incubated at 21°C, 80–82% humidity with 8 h dark and 16 h light photoperiod until molting. The tick nymphs successfully molting to adults were collected and counted approximately 1 month after collection. The number of engorged nymphs and adult females, weight/female, oviposition (egg mass/tick), fertility (No. hatching larvae), and number of nymphs molting to adults were evaluated. Data were analyzed statistically to compare results for each tick species between individuals fed on vaccinated and adjuvant/saline injected control rabbits by Chi2-test (P = 0.05). Vaccine efficacy (E) was calculated as E (%) = 100 [1−(DN × DMA × DT × DO × DF)], where DN is the reduction in the number of engorged nymphs, DMA is the reduction in the number of nymphs molting to adults, DT is the reduction in the number of engorged female ticks, DO is the reduction in oviposition, and DF is the reduction in fertility in ticks fed on vaccinated rabbits when compared to the controls fed on adjuvant/saline injected dogs. Only parameters with statistically significant differences were included in vaccine E calculation. Results were included in Supplementary Dataset 6.
In rabbits, blood samples were collected from each animal before each immunization (T1 and T2) and tick infestation (T3). In dogs, blood samples were collected from each animal before each immunization (T1 and T2), tick infestation (T3) and at day 75 (T4) to better evaluate the duration of the antibody-mediated immune response in the major vaccine target species. Blood samples were collected into sterile tubes and maintained at 4 °C until arrival at the laboratory. Serum was then separated by centrifugation and stored at −20°C. An indirect ELISA test was performed to detect IgG antibodies against recombinant antigens in serum samples from vaccinated and control animals collected at T1–T3 for rabbits or T1–T4 for dogs as described previously (Merino et al., 2013; Contreras and de la Fuente, 2016, 2017). High absorption capacity polystyrene microtiter plates were coated with 50 μl (0.02 μg/ml solution of purified recombinant proteins) per well in carbonate-bicarbonate buffer (Sigma-Aldrich). After an overnight incubation at 4°C, coated plates were blocked with 200 μl/well of blocking solution (5% skim milk in PBS). Serum samples or PBS as negative control were diluted (1:1000, 1:10,000, 1:100,000 v/v; optimal dilution, 1:10,000) in blocking solution and 50 μl/well were added into duplicate wells of the antigen-coated plates. After an overnight incubation at 4°C, the plates were washed three times with a washing solution (PBS containing 0.05% Tween 20). A goat anti-rabbit or anti-dog IgG-peroxidase conjugate (Sigma-Aldrich) was added (diluted 1:3000 v/v in blocking solution) and incubated at RT for 1 h. After three washes with washing solution, 200 μl/well of substrate solution (Fast OPD, Sigma-Aldrich) was added. Finally, the reaction was stopped with 50 μl/well of 3N H2SO4 and the optical density (OD) was measured in an ELISA plate reader at 450 nm. Antibody levels in vaccinated and control animals were expressed as the OD450nm (ODanimal sera−ODPBS control) and compared between vaccinated and control groups by ANOVA test (P = 0.05).
For Western blot analysis, 10 μg of selected purified recombinant proteins and Rhipicephalus microplus Subolesin (Merino et al., 2013) as negative control were separated by electrophoresis in an SDS-12% polyacrylamide gel (Life Science, Hercules, CA, United States) and transferred to a nitrocellulose membrane. The membrane was blocked with 5% BSA (Sigma-Aldrich) for 2 h at room temperature (RT) and washed three times with TBS (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.05% Tween 20). Pooled sera collected at T3 from vaccinated dogs were used as primary antibodies. Primary antibodies were used at a 1:300 dilution in TBS, and the membrane was incubated overnight at 4°C and washed three times with TBS. The membrane was then incubated with an anti-dog IgG-horseradish peroxidase (HRP) conjugate (Sigma-Aldrich) diluted 1:15000 in TBS with 2% BSA. The membrane was washed four times with TBS and finally developed with TMB (3,3′, 5,5′- tetramethylbenzidine) stabilized substrate for HRP (Promega, Madrid, Spain) according to the manufacturer recommendations.
The expression of selected genes was characterized using total RNA extracted from three separate pools of adult tick salivary glands (25 females and 25 males each pool) and larvae (500 larvae each pool) using Tri Reagent (Sigma-Aldrich) according to manufacturer instructions. Real-time RT-PCR was performed on RNA samples using gene-specific oligonucleotide primers (Supplementary Table 1) and the iScript One-Step RT-PCR Kit with SYBR Green and the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). A dissociation curve was run at the end of the reaction to ensure that only one amplicon was formed and that the amplicons denatured consistently in the same temperature range for every sample. The mRNA levels were normalized against tick ribosomal protein S4 (rps4) as described previously using the genNorm method (ddCT method as implemented by Bio-Rad iQ5 Standard Edition, Version 2.0) (Ayllón et al., 2013). Normalized Ct values were compared between salivary glands and larvae by Student’s t-test with unequal variance (P = 0.05).
To characterize the subcellular localization of proteins encoded by selected genes, the proteomes of IL and DL were obtained as described above but purifying membrane proteins prior to analysis. Approximately 200 D. reticulatus and I. ricinus larvae were pulverized in liquid nitrogen and homogenized with a glass homogenizer (20 strokes) in 4 ml buffer (0.25 M sucrose, 1 mM MgCl2, 10 mM Tris-HCl, pH 7.4) supplemented with complete mini protease inhibitor cocktail (Roche, Basel, Switzerland). Samples were sonicated for 1 min in an ultrasonic cooled bath followed by 10 sec of vortex. After 3 cycles of sonication-vortex, the homogenate was centrifuged at 200 × g for 5 min at RT to remove cellular debris. The supernatant was then centrifuged at 12000 × g for 30 min at 4°C and the pellet fraction enriched in crude plasma membranes was collected, resuspended in 100 μl Laemmli sample buffer and applied onto 1.2-cm wide wells on a 12% SDS-PAGE gel. The MS/MS raw files were searched against the Ixodida (40,849 entries in June 2013) Uniprot database and the protein database of candidate protective antigens (Tables 1, 2) using the SEQUEST algorithm (Proteome Discoverer 1.3, Thermo Scientific) with the following constraints: tryptic cleavage after Arginine and Lysine, up to two missed cleavage sites, and tolerances of 1 Da for precursor ions and 0.8 Da for MS/MS fragment ions and the searches were performed allowing optional Met oxidation and Cys carbamidomethylation. A FDR < 0.01 was considered as condition for successful peptide assignments. Results are shown in Tables 1, 2.
According to Elvin and Kemp (1994) the conditions necessary and sufficient for an effective tick protective antigen include that (a) host antibodies should be able to gain access to the tick antigen, (b) sufficient antibodies must gain access to the target antigen, and (c) the formation of the antibody-antigen complex should disrupt the normal function of the tick protein. In our study, the hypothesis was that the genes encoding for functionally important secreted proteins that are highly represented in both L and SG would result in good candidate tick protective antigens. These antigens should fulfill the above conditions (a–c) as abundant functionally important secreted proteins would be highly exposed to host antibodies and their interaction should affect protein function, and thus tick feeding and/or development and pathogen infection and transmission.
To fulfill these criteria, an algorithm was developed using a vaccinomics approach to tick protective antigen discovery (de la Fuente and Merino, 2013: de la Fuente et al., 2016). The algorithm included the generation of transcriptomics and proteomics data in tick L and SG, and selection of gene transcripts and proteins highly represented in both samples and encoding for predicted functionally important secreted proteins. The initial dataset was obtained from I. ricinus because of the availability of the I. scapularis genome sequence for data mining (Gulia-Nuss et al., 2016). Genes selected in I. ricinus as encoding for tick candidate protective antigens (Table 1) were then used to find orthologous genes in D. reticulatus (Table 2). Tick IL and ISG were selected to cover both initial (larvae) and final (adult) developmental stages and tissues that are essential for pathogen acquisition and transmission, respectively (Ayllón et al., 2015; Chmelaø et al., 2016; Nuttall, 2018). I. ricinus and D. reticulatus acquire infection when larvae feed on infected hosts and after transtadial transmission, pathogen is secreted from salivary glands during feeding of nymph and adult ticks. Therefore, affecting these two developmental stages should reduce both tick infestations and pathogen infection/transmission.
After RNAseq, the number of reads/sample corresponded to 27.7 and 27.2 millions for IL and ISG, respectively. Of them, 5,731,608 and 7,024,484 corresponded to pass filter high quality reads aligned to IL and ISG samples, respectively. Of the 23,297 identified gene transcripts, 49% were included in the analysis and not discarded due to not enough alignment, high complexity or shallowly sequenced (Figure 1A and Supplementary Dataset 1). Of them, 13% were expressed in IL or ISG only and 18% were overexpressed on either tissue, but most of the identified genes (70%) did not show differential expression between IL and ISG (Figure 1A and Supplementary Dataset 1).
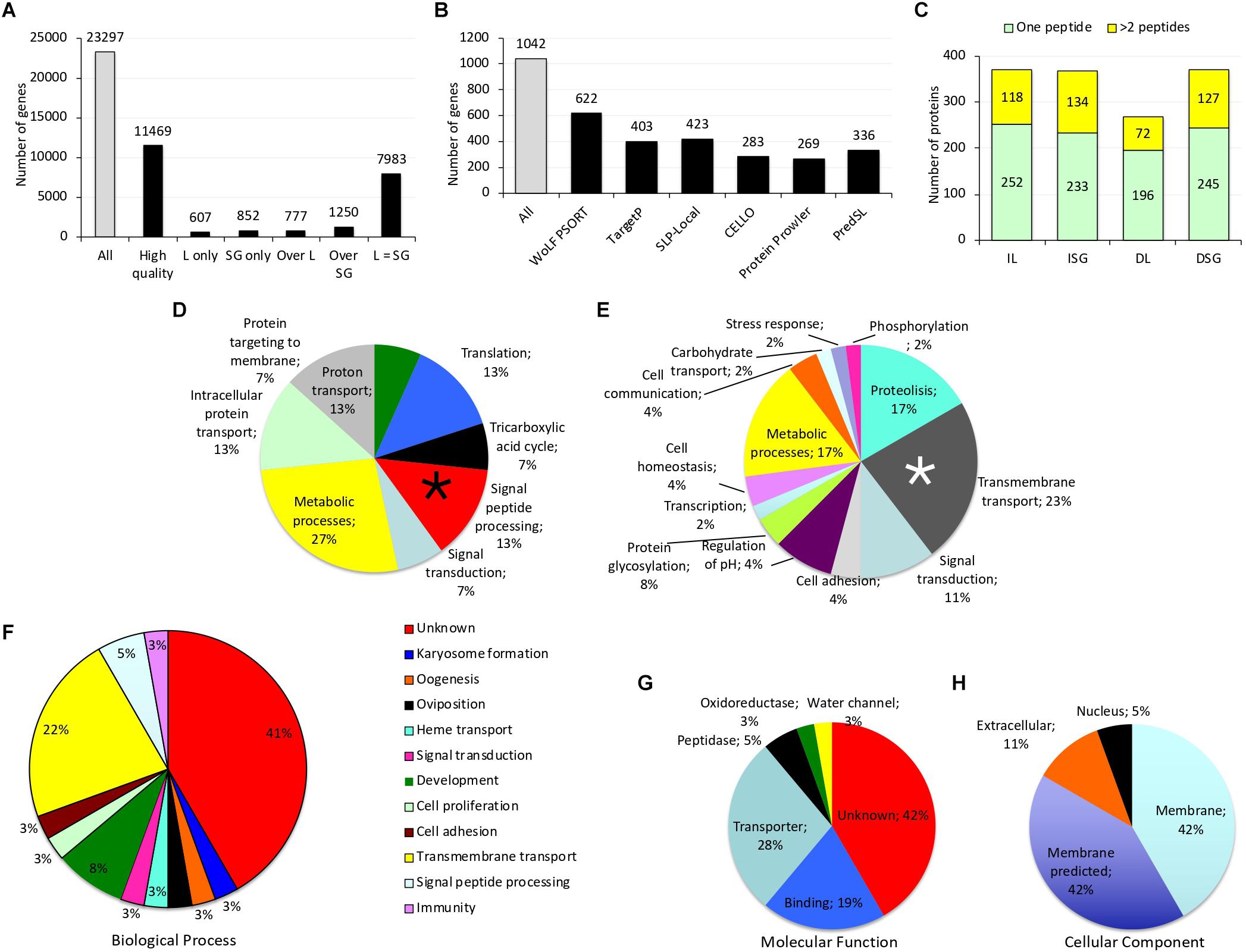
Figure 1. Tick transcriptomics and proteomics data. (A) Differential gene expression in I. ricinus larvae (L) and salivary gland (SG) samples. (B) Genes predicted by different algorithms to encode for secreted proteins in I. ricinus larvae (IL) and salivary glands (ISG) samples. (C) Proteins identified with XCorr > 2 in the Ixodidae database in IL, ISG, D. reticulatus larvae (DL) and salivary gland (DSG) samples. (D,E) Biological process (BP) GO for genes predicted as coding for secreted proteins that were either (D) among the top 1,000 expressed genes in both IL and ISG or (E) overexpressed in ISG. Genes coding for hypothetical proteins or with unknown function were excluded from the graph. The BPs selected for further analysis are marked with asterisks. (F–H) GO analysis for (F) biological process (BP), (G) molecular function (MF) and (H) cell component (CC) of genes selected as coding for tick protective antigens. Genes coding for nuclear DNA-binding proteins were removed from further analysis. Complete datasets are on Supplementary Datasets 1–4.
Genes encoding for secreted proteins were predicted using different algorithms. Predictor’s performance was different with a total of 1,042 genes predicted as encoding for secreted proteins (Figure 1B). Protein Prowler performed rather poor (269 predicted proteins) and almost all of them (259 proteins) were identical to those predicted by TargetP. CELLO predicted a low number of secreted proteins. Moreover, it predicted as many as twelve different subcellular localizations, which made the results difficult to analyze. Therefore, the results from Protein Prowler and CELLO were discarded. Five hundred and one genes were selected as encoding for secreted proteins (Supplementary Dataset 2). These genes were predicted by at least two predictors. Of them, 208 (41%) were overexpressed in ISG and 97 (19%) were among the top 1,000 expressed genes in both IL and ISG with 6 of them overexpressed in IL (Supplementary Dataset 2). These 6 genes were selected as encoding for candidate tick protective antigens (Figure 2 and Table 1).
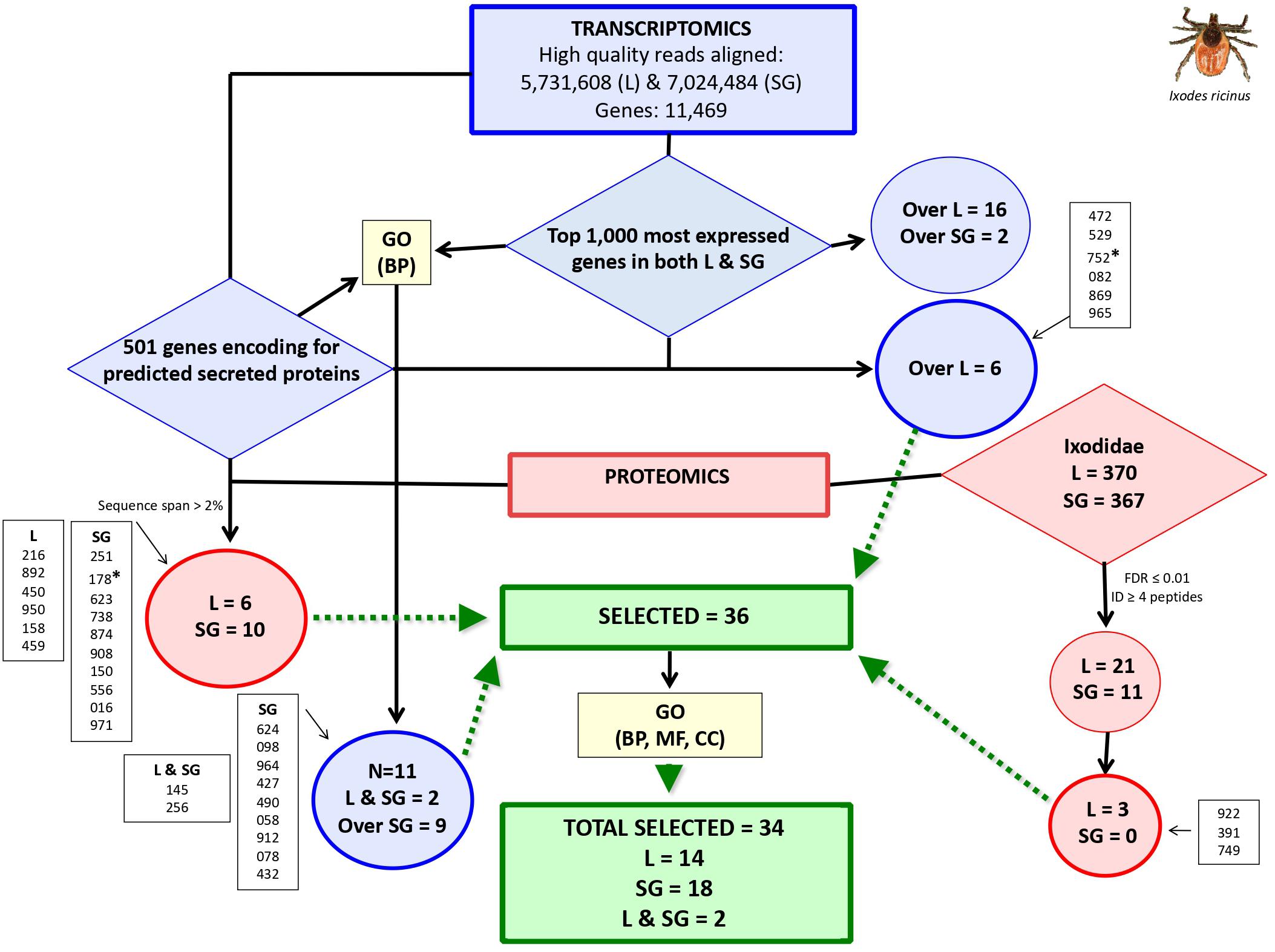
Figure 2. Selection of genes coding for candidate I. ricinus tick protective antigens. Trascriptomics data were obtained from I. ricinus larvae (L) and salivary gland (SG) samples followed by selection of top 1,000 most expressed genes in both samples and genes coding for secreted proteins. GO analysis for biological process (BP) was then used to select for functionally relevant genes. Proteomics data was used for mining databases of Ixodidae and predicted secreted proteins. Selected genes (N = 36; IDs. shown inside boxes) expressed or overexpressed (Over) in I. ricinus larvae (IL) and salivary glands (ISG) samples were finally subjected to GO analysis for BP, molecular function (MF) and cell component (CC) to remove two genes coding for nuclear DNA-binding proteins (marked with asterisks), resulting in a final set of 34 selected genes coding for candidate protective antigens. Complete datasets are on Supplementary Datasets 1–4.
A total of 3,454 and 2,339 proteins were identified in the Ixodidae database in IL and ISG, respectively. Of them, after applying XCorr > 2, a total of 370 and 367 proteins were identified in IL and ISG, respectively (Figure 1C and Supplementary Dataset 3). An additional selection criterion using FDR ≤ 0.01 and protein identification with at least 4 peptides resulted in the selection of 21 and 11 proteins in IL and ISG, respectively (Figure 2). Finally, after filtering for redundant biological function, 3 genes were selected from IL as encoding for candidate tick protective antigens (Figure 2 and Table 1). The search against the database of predicted secreted proteins encoded by the selected top 1,000 expressed genes in both IL and ISG (Supplementary Dataset 4) using XCorr > 2 and sequence span > 2% resulted in the identification of 6 and 10 proteins in IL and ISG, respectively that were selected as candidate tick protective antigens (Figure 2 and Table 1).
The GO analysis of transcriptomics data resulted in different BP for genes predicted as encoding for secreted proteins that were either overexpressed in ISG or among the top 1,000 expressed genes in both IL and ISG (Figures 1D,E). In genes highly expressed in both IL and ISG, metabolic processes was the most represented (27%) BP (Figure 1D). In genes overexpressed in ISG, transmembrane transport (23%) and metabolic processes (17%) were the most represented BP (Figure 1E). Genes in the signal peptide processing BP (Figure 1D) and transmembrane transport (Figure 1E) were filtered for highly similar sequences and resulted in 11 genes (9 overexpressed in ISG and 2 highly expressed in both IL and ISG) that were selected as encoding for candidate tick protective antigens (Figure 2 and Table 1).
Finally, GO analysis for BP, MF and CC was conducted for the selected 36 genes encoding for candidate protective antigens (Figure 2 and Table 1). The results showed that approx. 40% of genes had unknown BP and MF (Figures 1F,G). As predicted, most of the genes (95%) encoded for membrane and secreted proteins, but two genes (ID Nos. 752 and 178) encoded for predicted nuclear DNA-binding proteins (Figure 1H). Although these two genes were initially predicted as encoding for secreted proteins, they were removed from further analysis due to the GO CC prediction (Figure 2 and Table 1). The most represented BP and MF corresponded to transmembrane transport (22%; Figure 1F) and transporter activity (28%; Figure 1G), respectively. Additionally, BP involved in development (8%), reproduction (oogenesis and oviposition; 6%), protein secretion (signal peptide processing; 3%) and immunity (3%) were also represented (Figure 1F).
The final set of genes coding for I. ricinus tick candidate protective antigens included 34 candidates (Figures 2, 3, Table 1, and Supplementary Dataset 4).
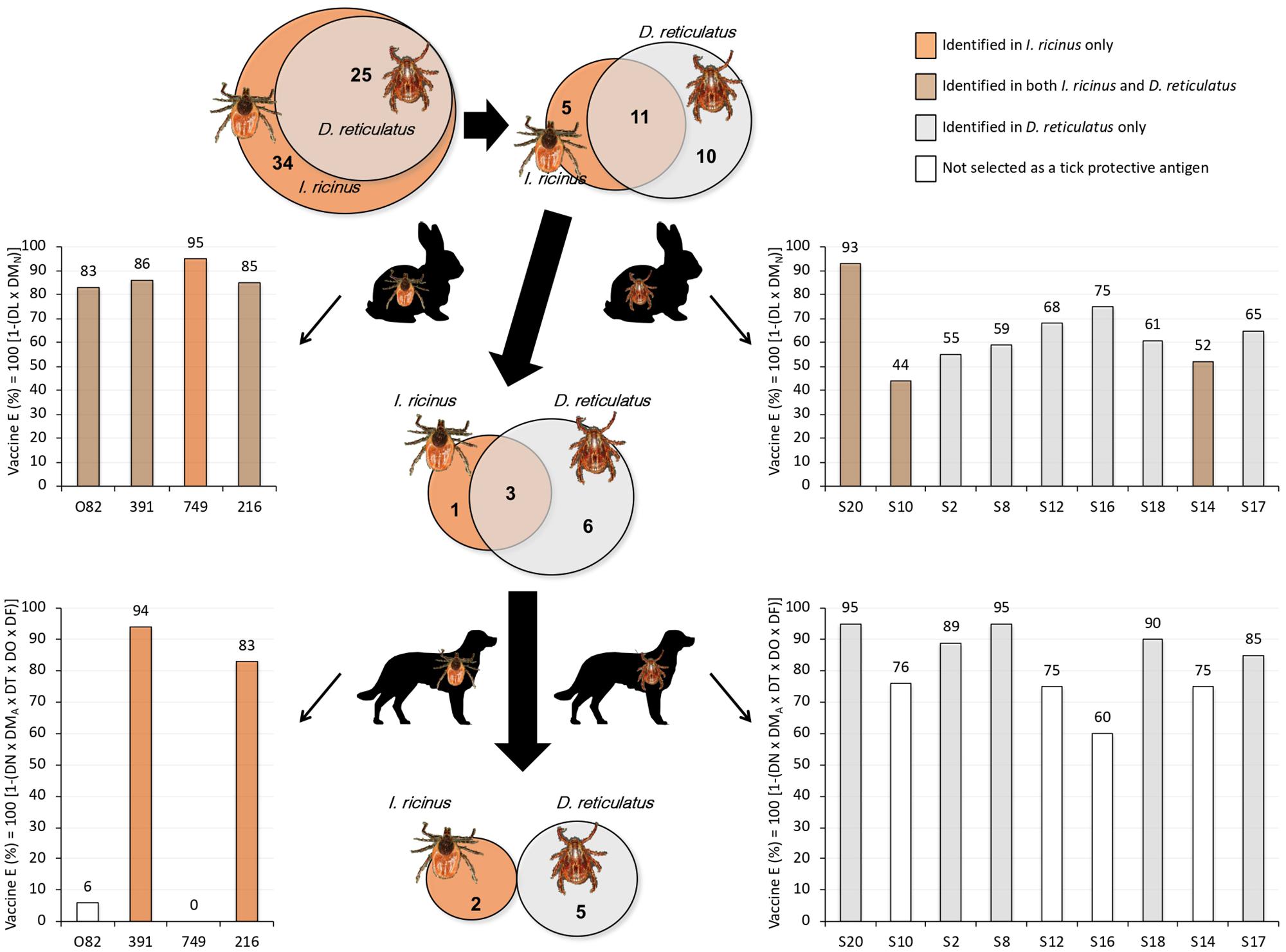
Figure 3. Pipeline for the identification of tick protective antigens. The Venn diagrams depict the set of genes selected for further analysis as coding for candidate tick protective antigens in I. ricinus and D. reticulatus. The graphs represent the vaccine E after vaccination trials in rabbits and dogs. Of the selected genes coding for I. ricinus (N = 34) and D. reticulatus (N = 25) tick protective antigens, only 16 and 21, respectively, were produced as recombinant proteins for initial vaccination screening in rabbits. After vaccination trial in rabbits, the criteria for the final selection of tick protective antigens for vaccination in dogs was based on vaccine E > 40%, resulting in 4 I. ricinus and 9 D. reticulatus antigens. The vaccination trial in dogs resulted in an overall vaccine E > 80% for 216 and 391 gene antigens in I. ricinus, and S2, S8, S17, S18, and S20 gene antigens in D. reticulatus. Full gene description and results are on Tables 1–3 and Supplementary Dataset 6.
The number of reads/sample corresponded to 21.7 millions for DL and 21.8 millions for DSG. The 21,838,507 reads (∼2.2 Gb) in DSG were compared against the selected set of I. ricinus genes encoding for candidate protective antigens (Table 1), clustered and assembled to produce a set of contigs with homology to selected reference genes. These contigs were then analyzed for differential expression between DL and DSG samples and compared to results in I. ricinus (Table 2). The results showed that 16/27 (59%) of D. reticulatus genes had an expression pattern similar to I. ricinus (Table 2).
A total of 268 and 372 proteins were identified in the Ixodidae database in DL and DSG, respectively, and all remained after data filtering (Figure 1C and Supplementary Dataset 3). The additional selection criteria using FDR ≤ 0.01 and protein identification with at least 4 peptides did not produce new candidate tick protective antigens.
As previously described for I. ricinus, two genes (IDs S3 and S4) were removed from further analysis due to the GO CC prediction (Figure 2 and Tables 1, 2). Therefore, the final set of genes coding for D. reticulatus tick candidate protective antigens included 25 candidates (Figure 3, Table 2, and Supplementary Dataset 4).
Of the selected genes encoding for I. ricinus (N = 34) and D. reticulatus (N = 25) tick protective antigens, only 16 and 21 respectively were produced as recombinant proteins for initial vaccination screening in rabbits (Figure 3 and Tables 1, 2). Of them, 11 were identified in both tick species (Figure 3). After rabbit vaccination and infestation with I. ricinus or D. reticulatus tick larvae, the criteria for the final selection of tick protective antigens for vaccination trials in dogs was based on vaccine E > 40%. Selection criteria need to be applied in order to move forward in the vaccinomics pipeline of selecting candidate protective antigens, and E > 40% was set as the minimum vaccine E for tick larvae. The selection of tick protective antigens resulted in 4 I. ricinus and 9 D. reticulatus antigens (Figure 3, Table 3, and Supplementary Dataset 6). Of them, 3 were identified in both tick species (Figure 3). The results in dogs showed that vaccination with I. ricinus antigens affected different tick developmental stages (Supplementary Dataset 6) and resulted in an overall vaccine E > 80% for gene antigens 216 and 391 (Figure 3 and Table 3). Vaccination with D. reticulatus antigens mainly affected nymph feeding and molting and oviposition (Supplementary Dataset 6) with an overall vaccine E ranging from 75 to 95% (Figure 3 and Table 3). Finally, based on the criteria of vaccine E > 80%, which is considered a good vaccine E for selection of candidate protective antigens, the I. ricinus 216 and 391 and the D. reticulatus S2, S8, S17, S18, and S20 were identified as the most effective tick protective antigens (Figure 3 and Table 3).
The candidate protective antigens were produced in E. coli and showed a molecular weight similar to that predicted by the amino acid sequence composition (Figure 4A and Supplementary Dataset 5). The mRNA levels of selected genes coding for the most effective protective antigens were determined by real-time RT-PCR and confirmed the results of the transcriptomics analysis (Figure 4B and Tables 1, 2).
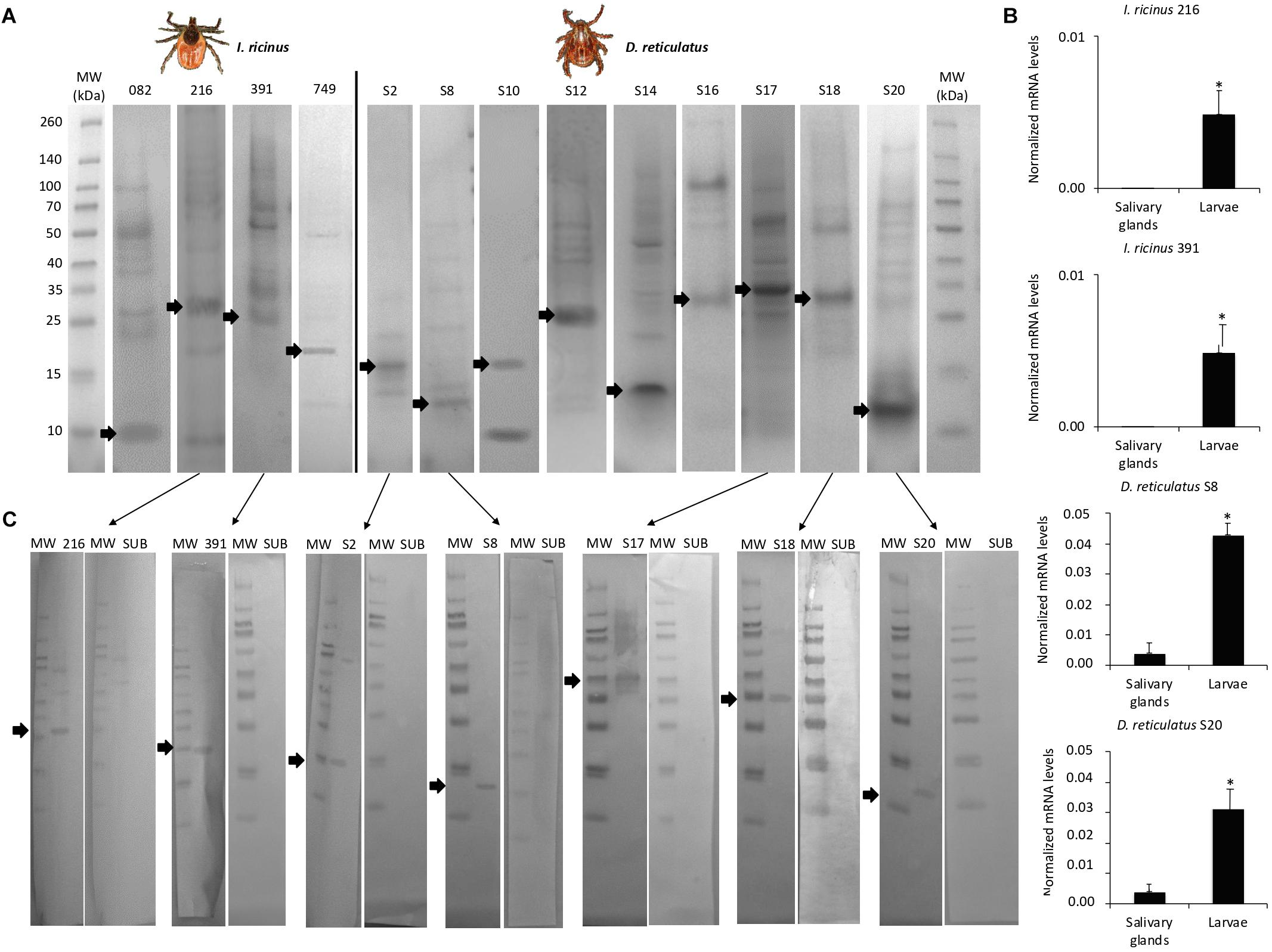
Figure 4. Characterization of candidate tick protective antigens. The 4 I. ricinus and 9 D. reticulatus candidate protective antigens were selected after vaccination trial in rabbits with vaccine E > 40%. (A) The histidine-tagged recombinant proteins were produced using the pET101/D-TOPO expression vector in E. coli BL21 Star (DE3) cells. After induction of gene expression with IPTG, collected cells were disrupted and the insoluble protein fraction containing the recombinant antigens as inclusion bodies were collected by centrifugation and purified by affinity to 16 polyhistidine. The eluted fraction containing the purified denatured proteins was dialyzed and concentrated by ultrafiltration to 0.5 mg/ml. Ten μg of purified recombinant antigens were separated by SDS-12% polyacrylamide gel electrophoresis and stained with Coomassie blue. Arrows indicate the position of recombinant antigens based on the predicted molecular weight. (B) The mRNA levels of selected genes coding for most effective vaccine antigens in dogs were determined by real-time RT-PCR in tick salivary glands and larvae (IDs refer to annotations in Tables 1–3). The mRNA levels were normalized against tick rps4 and normalized Ct values (Ave+SD) were compared between salivary glands and larvae by Student’s t-test with unequal variance (*P < 0.05; N = 3 biological replicates). (C) Western blot analysis of selected recombinant proteins and Subolesin as negative control using pooled sera from vaccinated dogs collected at T3. Selected proteins were those corresponding to the most effective protective antigens with vaccine E > 80%. Arrows indicate the position of recombinant antigens. MW, molecular weight marker.
The immunogenicity of candidate protective antigens (Table 3) was characterized by Western blot (Figure 4C) and indirect ELISA (Figures 5A,B) tests. The results supported the antigen-specific antibody response in vaccinated animals, and except for S2 and S8 antigens the background IgG levels were low in rabbits (Figure 5A). However, in dogs the background IgG levels before vaccination at T1 were higher than in rabbits (Figure 5B), suggesting host-specific differences in IgG affinity for tick antigens, and/or possible previous tick infestations at least in some of these dogs. Nevertheless, the IgG antibody response increased in response to vaccination in both rabbits and dogs, and for most antigens remained higher than in control animals until the end of the experiment (Figures 5A,B).
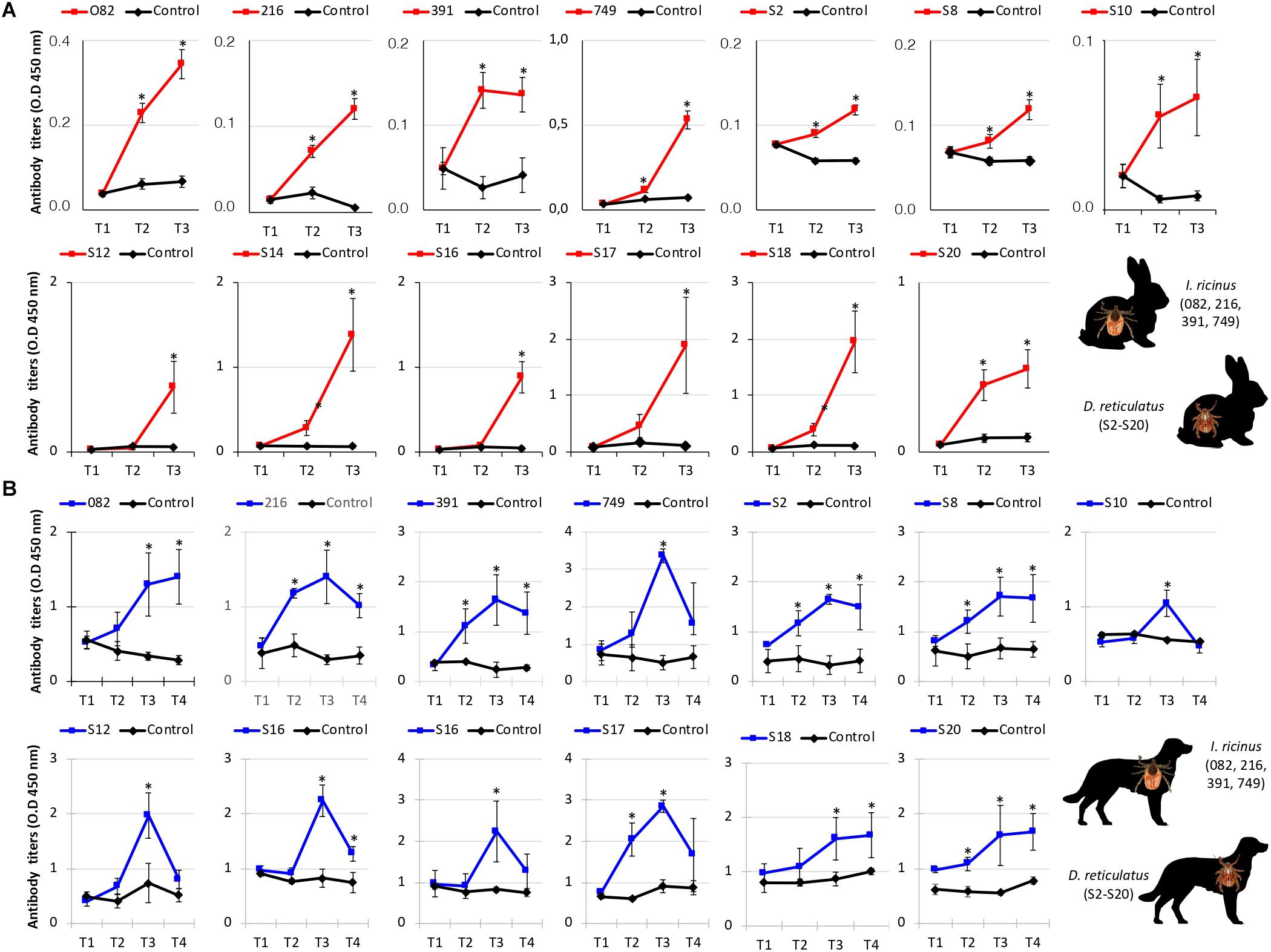
Figure 5. Immune IgG antibody response in vaccinated rabbits and dogs. An indirect ELISA test was performed to detect IgG antibodies against recombinant antigens in serum samples from vaccinated and control (A) rabbits and (B) dogs. Blood samples were collected from each animal before immunization (T1 and T2) and tick infestation (T3), and at day 75 (T4) in dogs. Serum samples or PBS as negative control were diluted at the optimal dilution of 1:10,000 (v/v). Antibody levels in vaccinated and control animals were expressed as the OD450nm (ODanimal sera–ODPBS control) and compared between vaccinated and control groups by ANOVA test (*P < 0.05). IDs refer to annotations in Tables 1–3.
Focusing on the selected tick protective antigens after vaccination trials in dogs (Table 3), the I. ricinus antigens 216 and 391, and the D. reticulatus antigens S8, S17, S18 and S20 were identified in the membrane fraction derived from D. reticulatus and I. ricinus larvae (Figure 6 and Tables 1, 2), thus confirming the prediction of the algorithms used for selection of membrane/secreted antigens. These antigens showed an effect on different tick developmental stages (Figure 6).
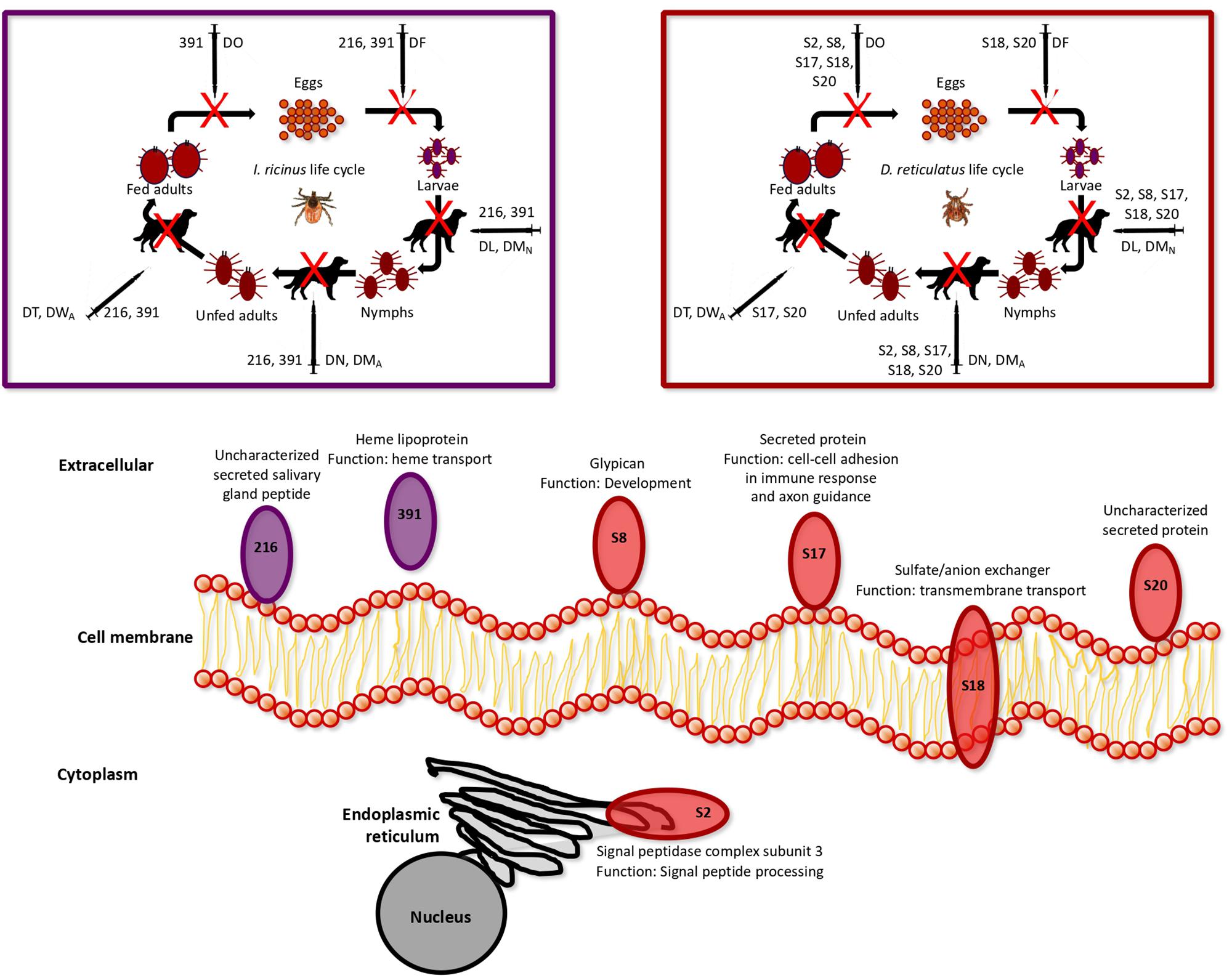
Figure 6. Summary representation of subcellular localization, function and vaccine efficacy of the selected protective antigens after vaccination trial in dogs. Complete GO annotations are in Supplementary Dataset 4, and the results of the vaccination trial in rabbits and dogs are in Table 3 and Supplementary Dataset 6. The I. ricinus antigens 216 and 391, and the D. reticulatus antigens S8, S17, S18 and S20 were identified in the membrane fraction derived from D. reticulatus and I. ricinus larvae. The effect of vaccination on tick life cycle was based on the parameters used to calculate vaccine E in rabbits and dogs, where DL is the reduction in the number of engorged larvae, DMN is the reduction in the percent of larvae molting to nymphs, DN is the reduction in the number of engorged nymphs, DMA is the reduction in the number of nymphs molting to adults, DT is the reduction in the number of engorged female ticks, DO is the reduction in oviposition, and DF is the reduction in fertility in ticks fed on vaccinated animals when compared to the controls fed on adjuvant/saline injected animals. Only parameters with statistically significant differences were included in vaccine E calculation.
Among these proteins, I. ricinus 216 and D. reticulatus S20 are uncharacterized secreted proteins (Figure 6). However, heme lipoproteins such as I. ricinus 391 has been proposed as candidate vaccine protective antigens as being among the most abundant proteins involved in heme transport from hemolymph to tissues to complement the loss of the heme synthesis pathway in ticks (Braz et al., 1999; Maya-Monteiro et al., 2000; Lara et al., 2003, 2005; Hajdusek et al., 2009; Kluck et al., 2018). In agreement with this function, vaccination with 391 affected all the developmental stages during I. ricinus life cycle (Figure 6).
In D. reticulatus, a glypican-like protein S8 was identified (Figure 6). Glypicans have been used for cancer vaccines against hepatocellular carcinoma (Sun et al., 2016; Li et al., 2018), but not as protective antigens against ectoparasites or vector-borne pathogens. Glypicans are a group of cell-surface glycoproteins involved in the regulation of cellular morphology, growth, motility and differentiation (Li et al., 2018). The results of the vaccination trials suggested that these proteins might play a role in oviposition and development of immature tick stages (Figure 6).
The D. reticulatus secreted protein S17 was annotated as involved in homophilic cell adhesion via plasma membrane adhesion molecules and axon guidance (Figure 6). Proteins with cell-cell adhesion have a role in immune response and neuronal wiring, and are highly conserved across evolution (Samanta and Almo, 2015; Jin and Li, 2019). This protein family has been recently identified in ticks (Jin and Li, 2019), and our results suggested a role for this protein in all tick developmental stages except egg fertility (Figure 6).
Sulfate/anion exchanger molecules such as D. reticulatus S18 function as transmembrane transporters and are involved in different biological processes including regulation of blood pressure (Wall, 2016). Vaccination with this antigen affected larval and nymphal developmental stages, oviposition and fertility, suggesting a still uncharacterized function of this protein during these developmental processes (Figure 6).
The only protein with subcelllular localization in the endoplasmic reticulum (ER) was the D. reticulatus S2 antigen (Figure 6). This protein was annotated as a signal peptidase complex subunit 3, which is involved in protein targeting to ER through signal peptide processing. The disruption of the catalytic subunit of these conserved proteins leads to cell death in yeast (Bohni et al., 1988; Fang et al., 1997), Plasmodium falciparum (Tuteja et al., 2008), and Leishmania manilensis major (Taheri et al., 2010). Furthermore, knockdown by RNA interference of the gene coding for this enzyme in Locusta migratoria (LmSPC1) affected insect molting, feeding, reproduction and embryonic development (Zhang and Xia, 2014). Vaccination with this antigen reduced development of tick immature stages and oviposition, supporting the role of this protein in tick survival (Figure 6).
The experimental approach used in this study resulted in the identification of tick protective antigens by using a vaccinomics approach in combination with vaccination trials. Based on the criteria of vaccine E > 80%, two I. ricinus and five D. reticulatus proteins were identified as the most effective protective antigens (Figure 6). Consequently, these antigens were proposed for commercial vaccine development for the control of tick infestations in companion animals, and other hosts for these tick species.
The putative function of selected protective antigens, which are involved in different biological processes, resulted in vaccines affecting multiple tick developmental stages (Figure 6). These results suggested that the combination of some of these antigens might be considered to increase vaccine efficacy through antigen synergy for the control of tick infestations and potentially affecting pathogen infection and transmission.
Finally, additional facts should be considered for vaccine development including that (a) a protective antigen is necessary but not sufficient for an effective vaccine as vaccine formulation, adjuvant and antigen composition including tick-derived and pathogen-derived antigens highly influence the final vaccine efficacy, (b) tick vaccines cannot be designed to prevent tick attachment and feeding because the immune response to vaccination (e.g., antibodies) needs to interact with target antigens in order to have an effect on different biological processes affecting life cycle of ticks and transmitted pathogens, (c) tick vaccines for wild animals are difficult to deliver, but appropriate vaccine formulations (i.e., virus-based) may address this problem, and (d) vaccines for companion animals may be acceptable if they increase animal welfare and health by reducing the duration of tick feeding and pathogen transmission.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
White rabbits (Oryctolagus cuniculus) and Beagle dogs (Canis lupus familiaris) were used in the experiments. Animal experiments were conducted in strict accordance with the recommendations of the European Guide for the Care and Use of Laboratory Animals. Animals were housed and the experiments were conducted at LLC ACRO Vet Lab (Pylipovichi Village, Kyiv Region, Ukraine) with the approval and supervision of the Ukrainian Commission for Bioethics and Biosafety for animals under the studies “Tick vaccine experiment on rabbits” number 000369 and “Tick vaccine experiments on dogs” numbers 000576, 000577, and 000761.
MV and JdlF conceived the study and designed the experiments. MC and MV performed the experiments. All authors performed the data analysis, wrote the manuscript, and contributed to the final version of the manuscript and approved it for publication.
This research was financially supported by Beaphar B.V. (Raalte, Netherlands). MC was funded by the Consejo Superior de Investigaciones Científicas (CSIC), Spain. MV was funded by the Universidad de Castilla–La Mancha (UCLM), Spain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Francisco Ruiz-Fons (SaBio, IREC, Spain) for the tick pictures included in the figures.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00977/full#supplementary-material
Alhassan, A., Liu, H., McGill, J., Cerezo, A., Jakkula, L. U. M. R., Nair, A. D. S., et al. (2018). Rickettsia rickettsii whole cell antigens offer protection against rocky mountain spotted fever in the canine host. Infect. Immun. 87:e00628-18. doi: 10.1128/IAI.00628-18
Ayllón, N., Villar, M., Busby, A. T., Kocan, K. M., Blouin, E. F., Bonzón-Kulichenko, E., et al. (2013). Anaplasma phagocytophilum inhibits apoptosis and promotes cytoskeleton rearrangement for infection of tick cells. Infect. Immun. 81, 2415–2425. doi: 10.1128/IAI.00194-13
Ayllón, N., Villar, V., Galindo, R. C., Kocan, K. M., Šíma, R., López, J. A., et al. (2015). Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet. 11:e1005120. doi: 10.1371/journal.pgen.1005120
Beugnet, F., and Marié, J. L. (2009). Emerging arthropod-borne diseases of companion animals in europe. Vet. Parasitol. 163, 298–305. doi: 10.1016/j.vetpar.2009.03.028
Bohni, P. C., Deshaies, R. J., and Schekman, R. W. (1988). SEC11 is required for signal peptide processing and yeast cell growth. J. Cell. Biol. 106, 1035–1042. doi: 10.1083/jcb.106.4.1035
Braz, G. R., Coelho, H. S., Masuda, H., and Oliveira, P. L. (1999). A missing metabolic pathway in the cattle tick Boophilus microplus. Curr. Biol. 9, 703–706. doi: 10.1016/s0960-9822(99)80312-1
Chmelaø, J., Kotál, J., Karim, S., Kopacek, P., Francischetti, I. M. B., Pedra, J. H. F., et al. (2016). Sialomes and mialomes: a systems-biology view of tick tissues and tick-host interaction. Trends Parasitol. 32, 242–254. doi: 10.1016/j.pt.2015.10.002
Coller, B. A., Clements, D. E., Martyak, T., Yelmene, M., Thorne, M., and Parks, D. E. (2012). Advances in flavivirus vaccine development. IDrugs 13, 880–884.
Contreras, M., and de la Fuente, J. (2016). Control of ixodes ricinus and dermacentor reticulatus tick infestations in rabbits vaccinated with the Q38 subolesin/akirin chimera. Vaccine 34, 3010–3013. doi: 10.1016/j.vaccine.2016.04.092
Contreras, M., and de la Fuente, J. (2017). Control of infestations by ixodes ricinus tick larvae in rabbits vaccinated with aquaporin recombinant antigens. Vaccine 35, 1323–1328. doi: 10.1016/j.vaccine.2017.01.052
Contreras, M., Villar, M., Artigas-Jerónimo, S., Kornieieva, L., Mótrofanov, S., and de la Fuente, J. (2018). A reverse vaccinology approach to the identification and characterization of Ctenocephalides felis candidate protective antigens for the control of cat flea infestations. Parasite. Vectors 11:43. doi: 10.1186/s13071-018-2618-x
de la Fuente, J. (2012). Vaccines for vector control: exciting possibilities for the future. Vet. J. 194, 139–140. doi: 10.1016/j.tvjl.2012.07.029
de la Fuente, J. (2018). Controlling ticks and tick-borne diseases looking forward. Ticks Tick Borne Dis. 9, 1354–1357. doi: 10.1016/j.ttbdis.2018.04.001
de la Fuente, J., Almazán, C., Canales, M., Pérez de la Lastra, J. M., Kocan, K. M., and Willadsen, P. (2007). A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 8, 23–28. doi: 10.1017/s1466252307001193
de la Fuente, J., Antunes, S., Bonnet, S., Cabezas-Cruz, A., Domingos, A., Estrada-Peña, A., et al. (2017a). Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front. Cell. Infect. Microbiol. 7:114. doi: 10.3389/fcimb.2017.00114
de la Fuente, J., and Contreras, M. (2015). Tick vaccines: current status and future directions. Expert Rev. Vaccines 14, 1367–1376. doi: 10.1586/14760584.2015.1076339
de la Fuente, J., Contreras, M., Estrada-Peña, A., and Cabezas-Cruz, A. (2017b). Targeting a global health problem: vaccine design and challenges for the control of tick-borne diseases. Vaccine 35, 5089–5094. doi: 10.1016/j.vaccine.2017.07.097
de la Fuente, J., Estrada-Peña, A., Venzal, J. M., Kocan, K. M., and Sonenshine, D. E. (2008). Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 13, 6938–6946. doi: 10.2741/3200
de la Fuente, J., and Kocan, K. M. (2003). Advances in the identification and characterization of protective antigens for development of recombinant vaccines against tick infestations. Expert Rev. Vaccines 2, 583–593. doi: 10.1586/14760584.2.4.583
de la Fuente, J., and Kocan, K. M. (2006). Strategies for development of vaccines for control of ixodid tick species. Parasite Immunol. 28, 275–283. doi: 10.1111/j.1365-3024.2006.00828.x
de la Fuente, J., Kopáèek, P., Lew-Tabor, A., and Maritz-Olivier, C. (2016). Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol 38, 754–769. doi: 10.1111/pim.12339
de la Fuente, J., and Merino, O. (2013). Vaccinomics, the new road to tick vaccines. Vaccine 31, 5923–5929. doi: 10.1016/j.vaccine.2013.10.049
de la Fuente, J., Moreno-Cid, J. A., Canales, M., Villar, M., Pérez de la Lastra, J. M., Kocan, K. M., et al. (2011). Targeting arthropod subolesin/akirin for the development of a universal vaccine for control of vector infestations and pathogen transmission. Vet. Parasitol. 181, 17–22. doi: 10.1016/j.vetpar.2011.04.018
de la Fuente, J., Villar, M., Contreras, M., Moreno-Cid, J. A., Merino, O., Pérez de la Lastra, J. M., et al. (2015). Prospects for vaccination against the ticks of pets and the potential impact on pathogen transmission. Vet. Parasitol. 208, 26–29. doi: 10.1016/j.vetpar.2014.12.015
de la Fuente, J., Villar, M., Estrada-Peña, A., and Olivas, J. A. (2018). High throughput discovery and characterization of tick and pathogen vaccine protective antigens using vaccinomics with intelligent big data analytic techniques. Expert Rev. Vaccines 17, 569–576. doi: 10.1080/14760584.2018.1493928
Elvin, C. M., and Kemp, D. H. (1994). Generic approaches to obtaining efficacious antigens from vector arthropods. Int. J. Parasitol. 24, 67–79. doi: 10.1016/0020-7519(94)90060-4
Escárcega-Ávila, A. M., de la Mora-Covarrubias, A., Quezada-Casasola, A., and Jiménez-Vega, F. (2019). Occupational risk for personnel working in veterinary clinics through exposure to vectors of rickettsial pathogens. Ticks Tick Borne Dis. 10, 299–304. doi: 10.1016/j.ttbdis.2018.10.012
Estrada-Peña, A., Venzal, J. M., Kocan, K. M., and Sonenshine, D. E. (2008). Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 13:6938–6946.
Fang, H., Mullins, C., and Green, N. (1997). In addition to SEC11, a newly identified gene, SPC3, is essential for signal peptidase activity in the yeast endoplasmic reticulum. J. Biol. Chem. 272, 13152–13158. doi: 10.1074/jbc.272.20.13152
Genomic Resources Development Consortium, Contreras, M., de la Fuente, J., Estrada-Peña, A., Grubhoffer, L., and Tobes, R. (2014). Genomic Resources Notes accepted 1 April 2014 – 31 May 2014. Mol. Ecol. Resour. 14:1095. doi: 10.1111/1755-0998.12298
Glickman, L. T., Moore, G. E., Glickman, N. W., Caldanaro, R. J., Aucoin, D., and Lewis, H. B. (2006). Purdue University-Banfield National Companion Animal Surveillance Program for emerging and zoonotic diseases. Vector Borne Zoonotic Dis. 6, 14–23. doi: 10.1089/vbz.2006.6.14
Grosenbaugh, D. A., De Luca, K., Durand, P. Y., Feilmeier, B., DeWitt, K., Sigoillot-Claude, C., et al. (2018). Characterization of recombinant OspA in two different borrelia vaccines with respect to immunological response and its relationship to functional parameters. BMC Vet. Res. 14:312. doi: 10.1186/s12917-018-1625-7
Gulia-Nuss, M., Nuss, A. B., Meyer, J. M., Sonenshine, D. E., Roe, R. M., Waterhouse, R. M., et al. (2016). Genomic insights into the ixodes scapularis tick vector of lyme disease. Nat. Commun. 7:10507. doi: 10.1038/ncomms10507
Hajdusek, O., Sojka, D., Kopacek, P., Buresova, V., Franta, Z., Sauman, I., et al. (2009). Knockdown of proteins involved in iron metabolism limits tick reproduction and development. Proc. Natl. Acad. Sci. U.S.A. 106, 1033–1038. doi: 10.1073/pnas.0807961106
Jin, Y., and Li, H. (2019). Revisiting DSCAM diversity: lessons from clustered protocadherins. Cell. Mol. Life Sci. 76, 667–680. doi: 10.1007/s00018-018-2951-4
Karunamoorthi, K. (2011). Vector control: a cornerstone in the malaria elimination campaign. Clin. Microbiol. Infect. 17, 1608–1616. doi: 10.1111/j.1469-0691.2011.03664.x
Kluck, G. E. G., Silva Cardoso, L., De Cicco, N. N. T., Lima, M. S., Folly, E., and Atella, G. C. (2018). A new lipid carrier protein in the cattle tick Rhipicephalus microplus. Ticks Tick Borne Dis. 9, 850–859. doi: 10.1016/j.ttbdis.2018.03.010
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
Kwit, N. A., Schwartz, A., Kugeler, K. J., Mead, P. S., and Nelson, C. A. (2018). Human tularaemia associated with exposure to domestic dogs-United States, 2006-2016. Zoonoses Public Health 2018, 1–5. doi: 10.1111/zph.12552
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lara, F. A., Lins, U., Bechara, G. H., and Oliveira, P. L. (2005). Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J. Exp. Biol. 208, 3093–3101. doi: 10.1242/jeb.01749
Lara, F. A., Lins, U., Paiva-Silva, G., Almeida, I. C., Braga, C. M., Miguens, F. C., et al. (2003). A new intracellular pathway of haem detoxification in the midgut of the cattle tick Boophilus microplus: aggregation inside a specialized organelle, the hemosome. J. Exp. Biol. 206, 1707–1715. doi: 10.1242/jeb.00334
Li, N., Gao, W., Zhang, Y. F., and Ho, M. (2018). Glypicans as cancer therapeutic targets. Trends Cancer. 4, 741–754. doi: 10.1016/j.trecan.2018.09.004
Liu, J., Drexel, J., Andrews, B., Eberts, M., Breitschwerdt, E., and Chandrashekar, R. (2018). Comparative evaluation of 2 in-clinic assays for vector-borne disease testing in dogs. Top. Companion Anim. Med. 33, 114–118. doi: 10.1053/j.tcam.2018.09.003
Maya-Monteiro, C. M., Daffre, S., Logullo, C., Lara, F. A., Alves, E. W., Capurro, M. L., et al. (2000). HeLp, a heme lipoprotein from the hemolymph of the cattle tick. Boophilus microplus. J. Biol. Chem. 275, 36584–36589. doi: 10.1074/jbc.m007344200
Merino, M., Antunes, S., Mosqueda, J., Moreno-Cid, J. A., Pérez de la Lastra, J. M., Rosario-Cruz, R., et al. (2013). Vaccination with proteins involved in tick-pathogen interactions reduces vector infestations and pathogen infection. Vaccine 31, 5889–5896. doi: 10.1016/j.vaccine.2013.09.037
Moreno-Cid, J. A., Jiménez, M., Cornelie, S., Molina, R., Alarcón, P., Lacroix, M. N., et al. (2011). Characterization of Aedes albopictus akirin for the control of mosquito and sand fly infestations. Vaccine 29, 77–82. doi: 10.1016/j.vaccine.2010.10.011
Nuttall, P. A. (2018). Wonders of tick saliva. Ticks Tick Borne Dis. 10, 470–481. doi: 10.1016/j.ttbdis.2018.11.005
Otranto, D. (2018). Arthropod-borne pathogens of dogs and cats: from pathways and times of transmission to disease control. Vet. Parasitol. 251, 68–77. doi: 10.1016/j.vetpar.2017.12.021
Parker, R., Deville, S., Dupuis, L., Bertrand, F., and Aucouturier, J. (2009). Adjuvant formulation for veterinary vaccines: montanidetm gel safety profile. Procedia Vaccinol. 1, 140–147. doi: 10.1016/j.provac.2009.07.026
Samanta, D., and Almo, S. C. (2015). Nectin family of cell-adhesion molecules: structural and molecular aspects of function and specificity. Cell. Mol. Life Sci. 72, 645–658. doi: 10.1007/s00018-014-1763-4
Skotarczak, B. (2018). The role of companion animals in the environmental circulation of tick-borne bacterial pathogens. Ann. Agric. Environ. Med. 25, 473–480. doi: 10.26444/aaem/93381
Sperança, M. A., and Capurro, M. L. (2007). Perspectives in the control of infectious diseases by transgenic mosquitoes in the post-genomic era-a review. Mem. Inst. Oswaldo Cruz 102, 425–433. doi: 10.1590/s0074-02762007005000054
Sun, Z., Zhu, Y., Xia, J., Sawakami, T., Kokudo, N., and Zhang, N. (2016). Status of and prospects for cancer vaccines against hepatocellular carcinoma in clinical trials. Biosci. Trends 10, 85–91. doi: 10.5582/bst.2015.01128
Taheri, T., Salmanian, A.-H., Gholami, E., Doustdari, F., Zahedifard, F., and Rafati, S. (2010). Leishmania major: disruption of signal peptidase type I and its consequences on survival, growth and infectivity. Exp. Parasitol. 126, 135–145. doi: 10.1016/j.exppara.2010.04.009
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tuteja, R., Pradhan, A., and Sharma, S. (2008). Plasmodium falciparum signal peptidase is regulated by phosphorylation and required for intra-erythrocytic growth. Mol. Biochem. Parasitol. 157, 137–147. doi: 10.1016/j.molbiopara.2007.10.007
Twine, N. A., Janitz, K., Wilkins, M. R., and Janitz, M. (2011). Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by alzheimer’s disease. PLoS One 6:e16266. doi: 10.1371/journal.pone.0016266
Villar, M., Popara, M., Ayllón, N., Fernández de Mera, I. G., Mateos-Hernández, L., Galindo, R. C., et al. (2014). A systems biology approach to the characterization of stress response in Dermacentor reticulatus tick unfed larvae. PLoS One 9:e89564. doi: 10.1371/journal.pone.0089564
Wall, S. M. (2016). The role of pendrin in blood pressure regulation. Am. J. Physiol. Renal Physiol. 310, F193–F203. doi: 10.1152/ajprenal.00400.2015
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Zerbino, D. R., McEwen, G. K., Margulies, E. H., and Birney, E. (2009). Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS One 4:e8407. doi: 10.1371/journal.pone.0008407
Keywords: dog, rabbit, vaccine, tick, Ixodes, Dermacentor, transcriptomics, proteomics
Citation: Contreras M, Villar M and de la Fuente J (2019) A Vaccinomics Approach for the Identification of Tick Protective Antigens for the Control of Ixodes ricinus and Dermacentor reticulatus Infestations in Companion Animals. Front. Physiol. 10:977. doi: 10.3389/fphys.2019.00977
Received: 13 May 2019; Accepted: 11 July 2019;
Published: 26 July 2019.
Edited by:
Itabajara Silva Vaz Jr., Federal University of Rio Grande do Sul, BrazilReviewed by:
Maria Kazimirova, Institute of Zoology, Slovak Academy of Sciences, SlovakiaCopyright © 2019 Contreras, Villar and de la Fuente. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: José de la Fuente, am9zZV9kZWxhZnVlbnRlQHlhaG9vLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.