
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 16 July 2019
Sec. Invertebrate Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.00793
This article is part of the Research TopicTick and Tick-Borne Pathogens: Molecular and Immune Targets for Control StrategiesView all 25 articles
 Abid Ali1*
Abid Ali1* Munsif Ali Khan1
Munsif Ali Khan1 Hafsa Zahid1
Hafsa Zahid1 Pir Muhammad Yaseen1
Pir Muhammad Yaseen1 Muhammad Qayash Khan1
Muhammad Qayash Khan1 Javed Nawab2
Javed Nawab2 Zia Ur Rehman3
Zia Ur Rehman3 Muhammad Ateeq4
Muhammad Ateeq4 Sardar Khan5
Sardar Khan5 Mohammad Ibrahim4
Mohammad Ibrahim4Although ticks prevalent in various agro-systems of Pakistan are associated with economic losses, information is still missing about the tick’s diversity, hosts they infest, seasonal dynamics and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa (KP) Pakistan. This study for the first time enlisted ticks infesting diverse hosts including humans in various regions of KP. A total of 8,641 ticks were collected across the northern, southern and central regions of KP and were morpho-taxonomically categorized into six genera comprising 17 species, R. microplus (n = 3,584, 42%), Hyalomma anatolicum (n = 2,253, 27%), Argas persicus (n = 1,342, 16%), Hya. impeltatum (n = 586, 7%), R. turanicus (n = 161, 2%), R. haemaphysaloides (n = 142, 2%), R. annulatus (n = 132, 2%), Hae. montgomeryi (n = 123, 1.4%), Hya. marginatum (n = 110, 1.3%), R. sanguineus (n = 34, 0.4%), and Hae. longicornis (n = 31, 0.4%). Ticks infesting wild animals included Amblyomma gervaisi, Amb. exornatum, Amb. latum, Dermacentor marginatus, and Hae. indica, while ticks collected from humans included R. microplus, R. annulatus, Hya. anatolicum, Hya. marginatum, and Hae. punctata. The overall prevalence of ticks infesting domestic animals was 69.4% (536/772). Among animal hosts, cattle were found highly infested (87.2%, 157/180) followed by buffalos (79%, 91/114), domestic fowls (74.7%, 112/150), goats (68.3%, 82/120), dogs (66.7%, 32/48), horses (61.3%, 49/80), and sheep (16.3%, 13/80). Analysis revealed that the tick burden significantly differed among domestic animals and was found to be high in cattle, followed by buffalos, goats, sheep, domestic fowl, dogs, and horses. Seasonal patterns of ticks distribution showed highest prevalance in July, August, and September due to the prevailing high temperature and humidity during these months. The phylogenetic analysis of cattle tick R. microplus based on partial mitochondrial cytochrome oxidase subunit I (COX1), 16S ribosomal RNA (16S rRNA) and internal transcribed spacer 2 (ITS2) sequences, revealed that R. microplus prevalent in this region belongs to clade C which include ticks originating from Bangladesh, Malaysia, and India. Further large scale studies across the country are necessary to explore the molecular and cross breeding aspects at the geographical overlapping of various tick species and their associated pathogens to facilitate designing control strategies as well as awareness against tick infestation in the region.
Ticks are hematophagous arthropods, notorious vectors of human and animal pathogens and constitute anemerging economic and health problem in tropical and sub-tropical regions (de la Fuente et al., 2017). As a major challenge to public health and veterinary sector, ticks can harbor several pathogens that cause numerous infectious diseases. The emergence and resurgence of several tick-borne diseases pose increasing public health concerns (Gratz, 1999; Paddock and Childs, 2003; Jones et al., 2008). As a vector reservoir for several emerging and re-emerging infectious pathogens of medical and veterinary importance, ticks transmit a wide variety of arboviruses (Crimean-Congo hemorrhagic fever virus, tick-borne encephalitis virus), bacteria (Rickettsia spp.), spirochetes (Anaplasma spp. Borrelia spp. and Ehrlichia spp.) and protozoans (Babesia spp. and Theileria spp.), more than any other blood feeding arthropod (Labruna et al., 2004; Gondard et al., 2017; Schorderet-Weber et al., 2017). These voracious ectoparasites can infest wild, terrestrial, semi-aquatic vertebrates and humans, and are globally distributed from the Arctic to tropical regions (Sherrard-Smith et al., 2012; Yu et al., 2015). Ten percent of the total hard and soft tick species are known to transmit diseases to humans and other animals (Parola and Raoult, 2001; Lew-Tabor and Valle, 2016). Ticks and tick-borne diseases affect approximately 80% of the world’s cattle population, particularly in tropical and subtropical countries. Infestation with cattle tick Rhipicephalus microplus economically impact the livestock industry in different regions and annual losses due to only this tick have been estimated 22–30 billion US$ (Jabbar et al., 2015; Lew-Tabor and Valle, 2016).
The prevalence of ticks in different regions is mostly associated with various environmental factors that affect tick distribution and adaptation (Estrada-Peña, 2008). Like other hematophagous arthropods, ticks spend their life cycle in such an environment where they depend on host availability, host lifestyle, host interaction with other animals, vegetation coverage, habitat type and geo-climatic conditions (temperature and humidity) for their survival and development (Estrada-Peña, 2008; Gondard et al., 2017). Climatic changes influence tick distribution, shape the biodiversity of ticks and tick-borne pathogens and increase the risks of transmission of pathogens to humans and other hosts (Leger et al., 2013; Dantas-Torres, 2015). The dispersal of ticks in previously tick free areas is consociated with humans and other animals movements due to environmental changes (Estrada-Peña, 2008; Dantas-Torres, 2015) which has resulted in the emergence and re-emergence of tick-borne diseases.
Ticks that belong to the genus Rhipicephalus are responsible for severe economic losses in tropical and subtropical agro-systems. Previous studies have enlisted intraspecific variations in genus Rhipicephalus based on morpho-taxonomic complications in identification at species level (Lempereur et al., 2010; Barker and Walker, 2014). Morphological variations in Rhipicephaline ticks make it difficult to distinguish these tick species morphologically and to date, several valid species of Rhipicephalus (Boophilus) have been confirmed (Estrada-Peña et al., 2012; Guglielmone et al., 2014; Ali et al., 2016; Coimbra-Dores et al., 2018; Roy et al., 2018). The morpho-taxonomy of R. microplus (cattle tick) has been challenged in the last few years due to a hypothesis suggesting that biogeographical and ecological separations have occurred in Boophilid ticks across continents based on morphological and genetic variations (García-García et al., 1999; de la Fuente et al., 2000; Ali et al., 2016; Lew-Tabor et al., 2017). The genetic assemblage study based on mitogenome (12S rRNA and 16S rRNA) and microsatellites markers of R. microplus from America, Asia, Australia and Africa have confirmed that R. microplus consist of at least two different species (Labruna et al., 2009; Low et al., 2015; Ali et al., 2016; Baron et al., 2018). Mating experiments among these ticks evidenced that reproductive crosses between R. microplus ticks from Australia and Argentina or Mozambique are infertile while crosses between Boophilid ticks from Argentina and Mozambique are fertile. These findings based on genetic divergence and reproductive isolation put forward that the Australian R. microplus strain is different from the American, Asian and African strain. Phylogenetic analysis of the R. microplus mitogenome (COX1 and 16S rRNA) and internal transcribed spacer (ITS) gene revealed that R. microplus is a complex of species. The cryptic diversity of R. microplus complex includes R. annulatus, R. australis and two clades of R. microplus, clade A and B. Clade A R. microplus includes ticks from America and Africa and clade B includes ticks from China (Labruna et al., 2009; Burger et al., 2014; Low et al., 2015; Roy et al., 2018). Recent mitogenome approaches based on COX1 and 16S rRNA haplotypes suggested a distinct genetic assemblage of R. microplus from Malaysia resulting in novel clade C which includes ticks from Pakistan, Bangladesh, Myanmar, India and Malaysia (Low et al., 2015; Baron et al., 2018; Roy et al., 2018).
In Pakistan, valid studies have shown the prevalence of five tick genera, including Hyalomma, Haemaphysalis, Rhipicephalus, Ornithodoros, and Argus (Karim et al., 2017; Rehman et al., 2017). The notable species compositions of genus Hyalomma are Hya. anatolicum, Hya. scupense, Hya. kumari, Hya. isacci, Hya. turanicum and Hya. dromedarii; genus Haemaphysalis are Hae. bispinosa, Hae. montgomeryi, Hae. cornupunctata, Hae. kashmirensis, and Hae. sulcata; genus Rhipicephalus species compositions include R. microplus, R. annulatus, R. haemaphysaloides, R. turanicus, and R. sanguineus; the genus Argus is composed of Argas persicus and genus Ornithodoros is represented by Ornithodoros tolozani (Hoogstraal and Varma, 1962; Kaiser and Hoogstraal, 1964; Robertson et al., 1970; Karim et al., 2017; Rehman et al., 2017).
Pakistan is an agricultural country where livestock plays a crucialrole in the national economy. In Pakistan, climatic conditions are extremely favorable for the growth and survival of diverse tick species. These ticks cause severe problems for livestock holders which mainly include low income farmers. The majority of these farmers consider all tick species as one species and are mostly unaware of the capability of pathogens transmission to humans as well as other associated health risks. Tick and tick-borne diseases are prevalent in the province Khyber Pakhtunkhwa (KP) of Pakistan, causing economic loss to livestock holders (Jabbar et al., 2015; Karim et al., 2017; Rehman et al., 2017). However, there is a paucity of documented information about ticks spatial distribution and diversity, their diverse hosts and seasonal dynamics from this region. Therefore, the present endeavor was focused on tick diversity to investigate the present status of tick species infesting various animal hosts, their seasonal dynamics and to obtain the evolutionary information of cattle tick R. microplus which is prevalent in the study area.
During the present study five districts namely Charsadda (34°09′49.4′′N 71°44′53.4′′E), Mardan (34°11′54.6′′N 72°01′37.4′′E), Peshawar (34°01′36.2′′N 71°31′47.4′′E), Lower Kohistan (35°19′48.2′′N 73°13′50.7′′E), and Karak (33°06′37.5′′N 71°05′29.0′′E), in the northern, southern and central KP (Northwestern geographic state of Pakistan previously known as North-West Frontier Province) were selected to assess the diversity of ticks, their hosts and seasonal dynamics. Nine regions of each aforementioned districts were selected for ticks sampling (Figure 1). A Global Positioning System (GPS) was used to obtain geographic coordinates data and loaded onto a Microsoft Excel sheet to develop a distribution map for the study areas using ArcGIS 10.3.1.
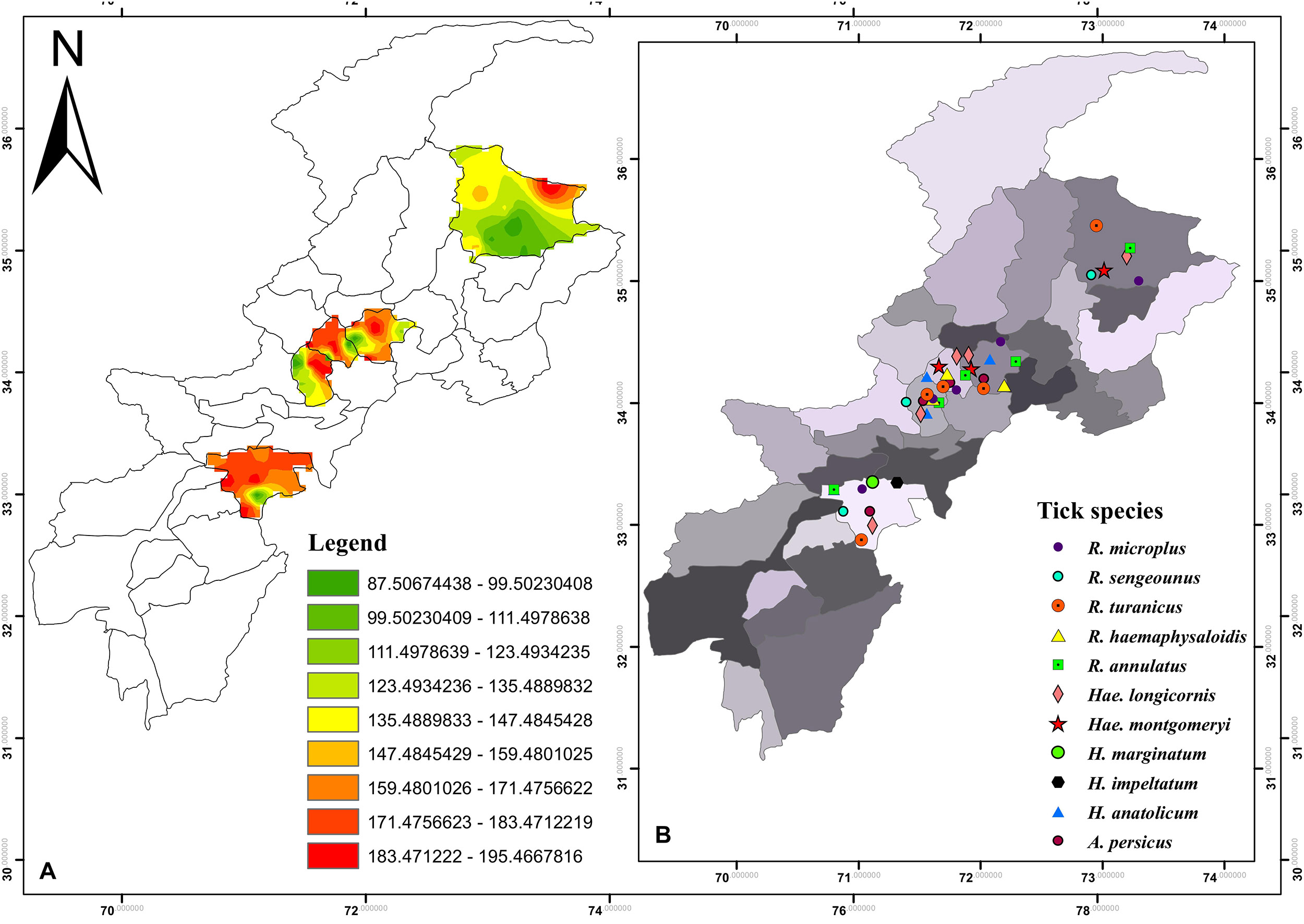
Figure 1. (A) Density map of tick burden in the selected districts (B) and spatial distribution of tick species in KP.
The protocol for the present study was approved by the ethical committee and board of study members of the Department of Zoology Abdul Wali Khan University Mardan. Written informed consent was obtained from the owners of animals and individuals infested by ticks prior to tick collection.
Tick samples were collected during April 2017 to March 2018, from different domestic animals (cattle, buffalo, horse, sheep, goat, dogs, and domestic fowl) at different localities (villages and towns) in various tehsils (an administrative division within the district) of the aforementioned districts (Figure 1). For ticks collection, a regular visit was made three times a month and sampling was done randomly with the help of fine tweezers from sampling animals reared at animal farms, houses or grazing in the field. Collected specimens were rinsed with distilled water followed by absolute ethanol to remove any surface confined microbes and host contaminating tissues and stored in properly labeled separate plastic tubes until further analyses. The relevant information including age of host, physical status, previous acaricides or any drugs used against ticks, the body region from where ticks were isolated, date and place of collection were recorded and all the samples were shipped to the Parasitology laboratory of the department of Zoology, Abdul Wali Khan University Mardan. Ticks were immediately identified or preserved (100% ethanol or in a mixture of 95% ethanol, 4% distilled water and 1% glycerol) (Walker et al., 2003) for further study.
During collection, ticks were accidentally found infesting humans and wild goats (locally known as markhor, a national animal of Pakistan mostly found on the Himalaya Mountains). Wild rodents (Indian gray mongoose), wild boars (wild pig), and wild reptiles (monitor lizard and Indian python) were captured by local farmers or found dead on highways. In this collection, ticks were also included from districts other than the above-mentioned districts such as Chitral (35°53′40.9′′N 71°41′31.1′′E), Haripur (33°59′41.0′′N 72°54′35.3′′E), Swabi (34°07′29.1′′N 72°27′25.7′′E), and Nowshera (34°00′27.8′′N 71°59′09.8′′E) (Figure 2).
Tick samples were identified morphologically using tick morpho-taxonomic characters under stereozoom microscope (HT Stereozoom) according to the standard relevant identification keys (Walker et al., 2003; Apanaskevich and Horak, 2005, 2008, 2009; Ghosh and Misra, 2012; Nowak-Chmura, 2012; Madder et al., 2014; Caetano et al., 2017).
A total of 225 (45 per district) morphologically identified R. microplus from nine locations in each of the five districts were selected for genomic DNA extraction. Tick samples from each district were separately pooled into five groups, each pool containing ticks of a different location within the district. Ticks were cleaned with distilled water, dried and cut into small pieces using a sterile scalpel and homogenized by a sterile micro pestle in separate 1.5 ml Eppendorf tubes. Genomic DNA was separately extracted from each pool of tick samples using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific) according to the standard DNA extraction protocol of the manufacturer. DNA concentration in the samples was quantified using a Nanodrop ND-100 (Thermo Fisher Scientific) and stored at −20°C until further processing. COX1, 16S rRNA and ITS2 gene fragments of R. microplus were amplified by PCR in a thermocycler (HT, ILF, UK) using specific primers (Table 1). PCR reactions were performed in a total volume of 25 μL reaction mixture with a composition of 1 μL of each forward and reverse primer, 2 μL of template DNA (50 ng), 7.5 μL of deionized water and 13.5 μL of master mix (DreamTaq PCR Master Mix [2X] (Thermo Fisher Scientific). The thermocycling conditions for COX1, 16S rRNA and ITS2 were optimized separately for each reaction. Standardized PCR conditions was, an initial denaturation of 5 min at 95°C followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 53.8°C for COX1, 55.9°C for 16S rRNA and 58°C for ITS2 for 30 s, an extension at 72°C for 30 s and a final extension of 10 min at 72°C. A negative control (distilled water) was included in each run of the amplification reaction for validation. PCR products were confirmed by running the amplified DNA in 2% ethidium bromide stained agarose gel with a 50 bp DNA marker (Thermo Fisher Scientific). The results were visualized using the GeneDoc (UVP BioDoc-It Imaging System).
The amplified PCR products were purified using the GeneClean II kit (Qbiogene) according to the manufacturer’s instructions. For the sequencing of PCR based amplified product, 45 DNA samples of each district were separately grouped containing fifteen samples for each gene fragment. The purified PCR amplicons (45 PCR product samples, 15 samples for each marker) were sequenced unidirectionally by Macrogen Inc. (Seoul, South Korea). The obtained sequences (nucleotide sequences) were analyzed using BioEdit V. 7.0.5 (Hall et al., 2011) and NCBI BLAST1 (Altschul et al., 1990). All the relevant library sequences of closely related species and of R. microplus complex available in GenBank were downloaded and saved for downstream analysis to construct a phylogenetic tree. The obtained sequences were trimmed for the removal of unnecessary nucleotides and those with 100% similarity with each other were discarded. Finally, 12 sequences were uploaded to NCBI GenBank including COX1 (4), 16S rRNA (3) and ITS2 (5).
The obtained trimmed sequences were aligned using ClustalW in BioEdit Sequence Alignment Editor V 7.0.5 (Hall et al., 2011). To test molecular phylogeny of the cattle tick R. microplus a multi locus analysis was performed for COX1 (Figure 3), 16S rRNA (Figure 4), and ITS2 (Figure 5) partial sequences using the Maximum likelihood method in MEGA v. X (Kumar et al., 2018) with bootstrapping at 1000 replications (Tamura et al., 2013).
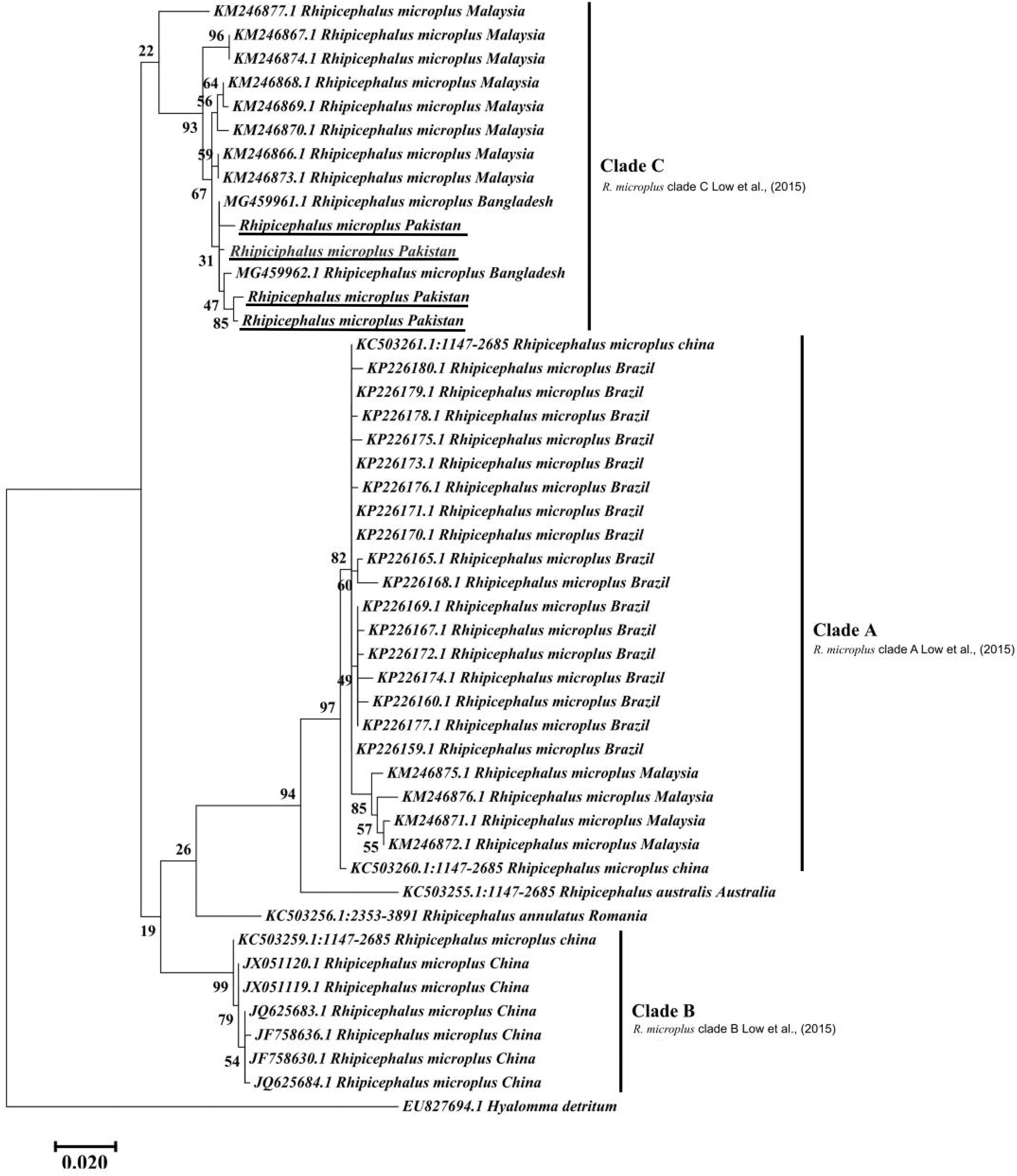
Figure 3. Maximum likelihood tree inferred from COX1 sequences of the genus Rhipicephalus and using Hyalomma sequence as outgrowth. GenBank accession numbers are followed by species name and location of collection. Clade A includes R. microplus ticks from America, Malaysia, and China, clade B includes tick from China and clade C includes ticks from Pakistan, India, Bangladesh, Myanmar, and Malaysia. Support values (Bootstrapping values) were indicated at each node. The bar represents 0.020 substitutions per site. Sequences obtained in the present study were underlined.
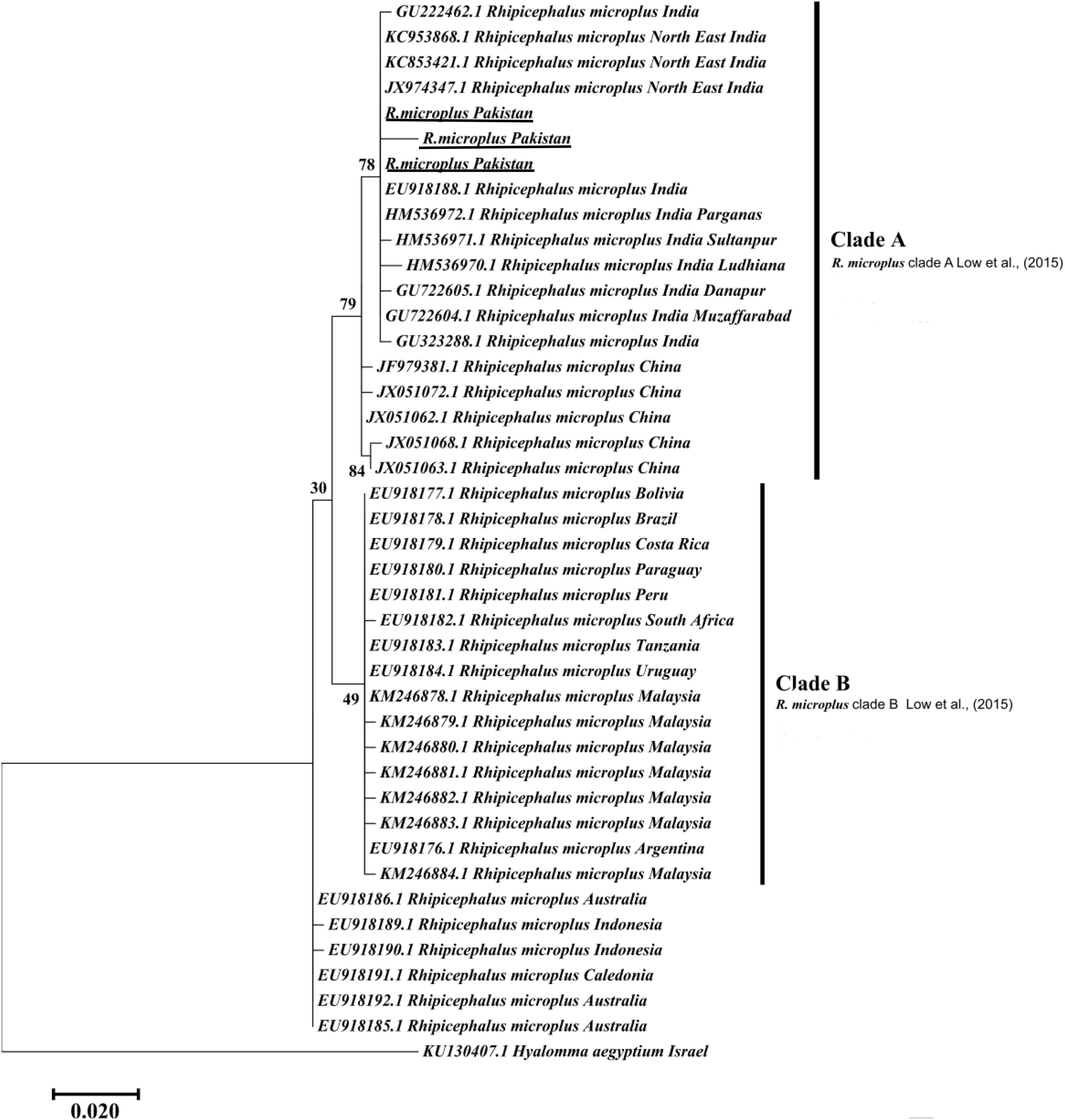
Figure 4. Maximum likelihood tree inferred from 16S rRNA sequences of the genus Rhipicephalus and using Hyalomma sequence as outgrowth. GenBank accession numbers are followed by species name and location of collection. The clade A includes R. microplus from Pakistan, India, and China while Clade B contains R. microplus from Africa, Malaysia and South America. Support values (Bootstrapping values) were indicated at each node. The bar represents 0.020 substitutions per site. Sequences obtained in the present study were underlined.
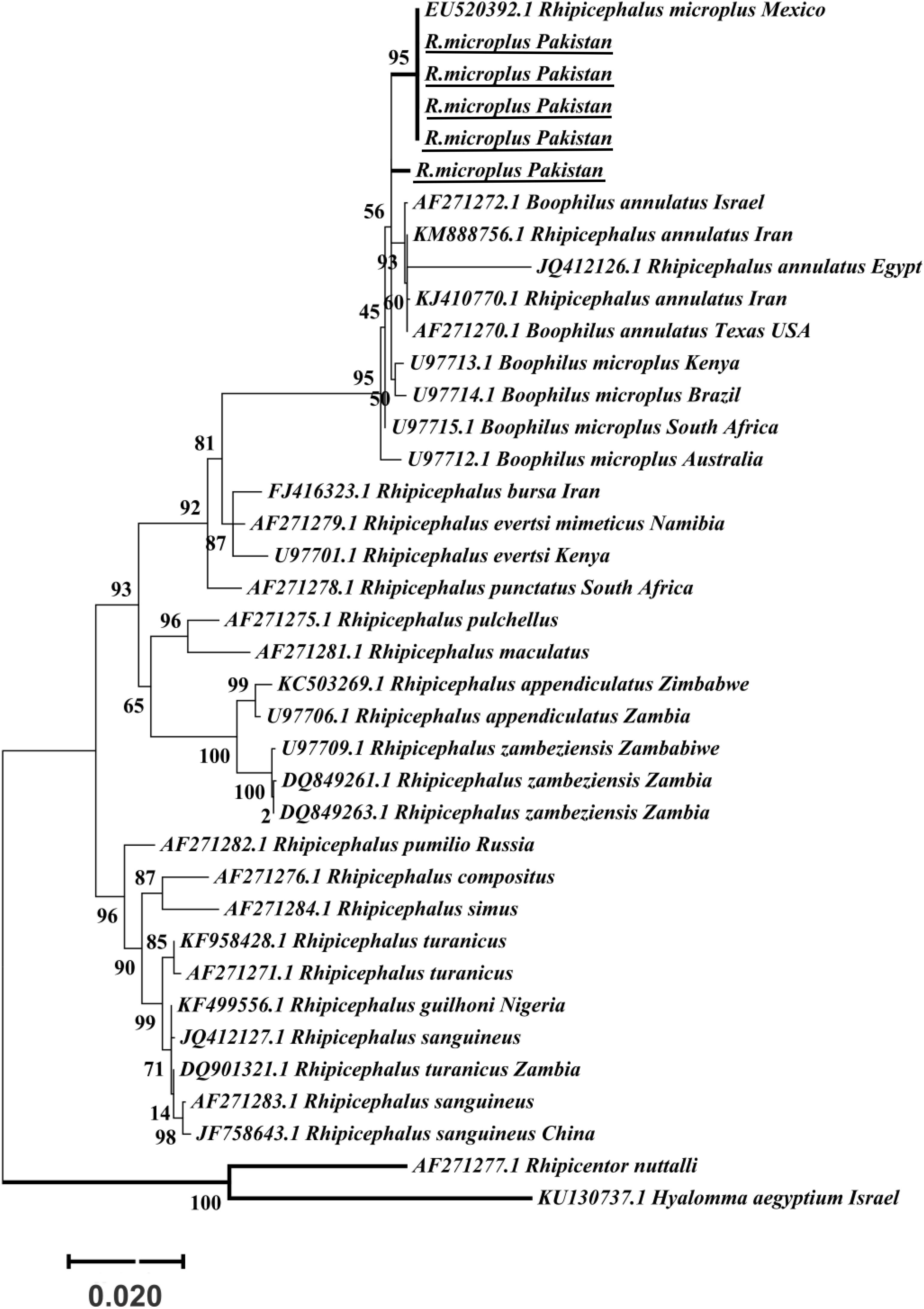
Figure 5. Maximum likelihood tree inferred from ITS2 sequences of the genus Rhipicephalus and using Rhipicentor and Hyalomma sequences as outgrowth. GenBank accession numbers are followed by species name and collection location. Support values (Bootstrapping values) were indicated at each node. The bar represents 0.020 substitutions per site. Sequences obtained in the present study were underlined.
The overall tick prevalence was calculated by the total ticks divided by total hosts, host wise infestation was calculated by the infested hosts divided by total examined. The mean percent of collected ticks and binomial confidence intervals for proportion were calculated (Table 2). The median tick burden was analyzed on hosts by applying the Kruskal–Wallis test (P < 0.001) among different hosts and the Pearson correlation test was applied for the temporal distribution of ticks. The results were considered statistically significant at p < 0.05.
Morpho-taxonomic analysis categorized collected ticks into six genera including 17 species (Figure 1). The highest number of collected tick species was R. microplus (n = 3584, 42%) followed by Hya. anatolicum (n = 2,253, 27%), A. persicus (n = 1,342, 16%), Hya. impeltatum (n = 586, 7%), R. turanicus (n = 161, 2%), R. haemaphysaloides (n = 142, 2%), R. annulatus (n = 132, 2%), Hae. montgomeryi (n = 123, 1.4%), Hya. marginatum (n = 110, 1.3%), R. sanguineus (n = 34, 0.4%), and Hae. longicornis (n = 31, 0.4%) (Table 2 and Figure 6A). Cattle were found infested separately by R. microplus, R. haemaphysaloides, Hya. anatolicum, Hya. impeltatum, Hya. marginatum and Hae. montgomeryi and buffalos were found infested separately by R. microplus, R. haemaphysaloides, Hya. anatolicum, and Hae. montgomeryi. On the other hand, goats were found infested by Hae. montgomeryi, Hae. longicornis, and Hya. impeltatum, and sheep were infested separately by two species Hae. longicornis and Hya. impeltatum. R. annulatus and R. sanguineus were collected from dogs. Horses and domestic fowls were found to be infested by a single species of R. turanicus and A. persicus, respectively. Hosts infested by multiple tick species were also recorded. For instance, R. microplus and Hya. anatolicum were found infesting both cattle and buffaloes (Table 3).
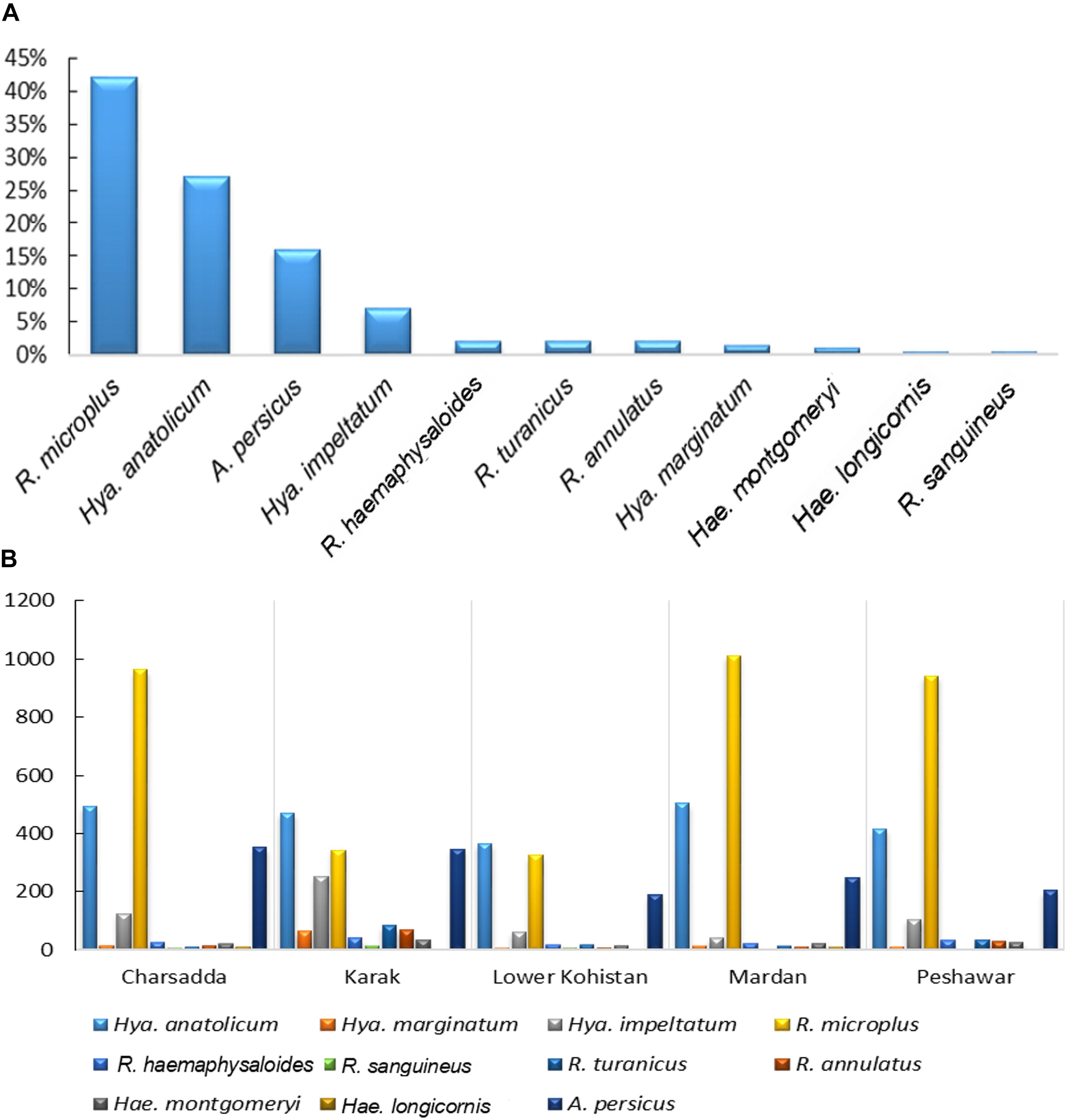
Figure 6. (A) shows percent composition of collected tick species (B) and spatial distribution of ticks in various districts of KP, Pakistan.
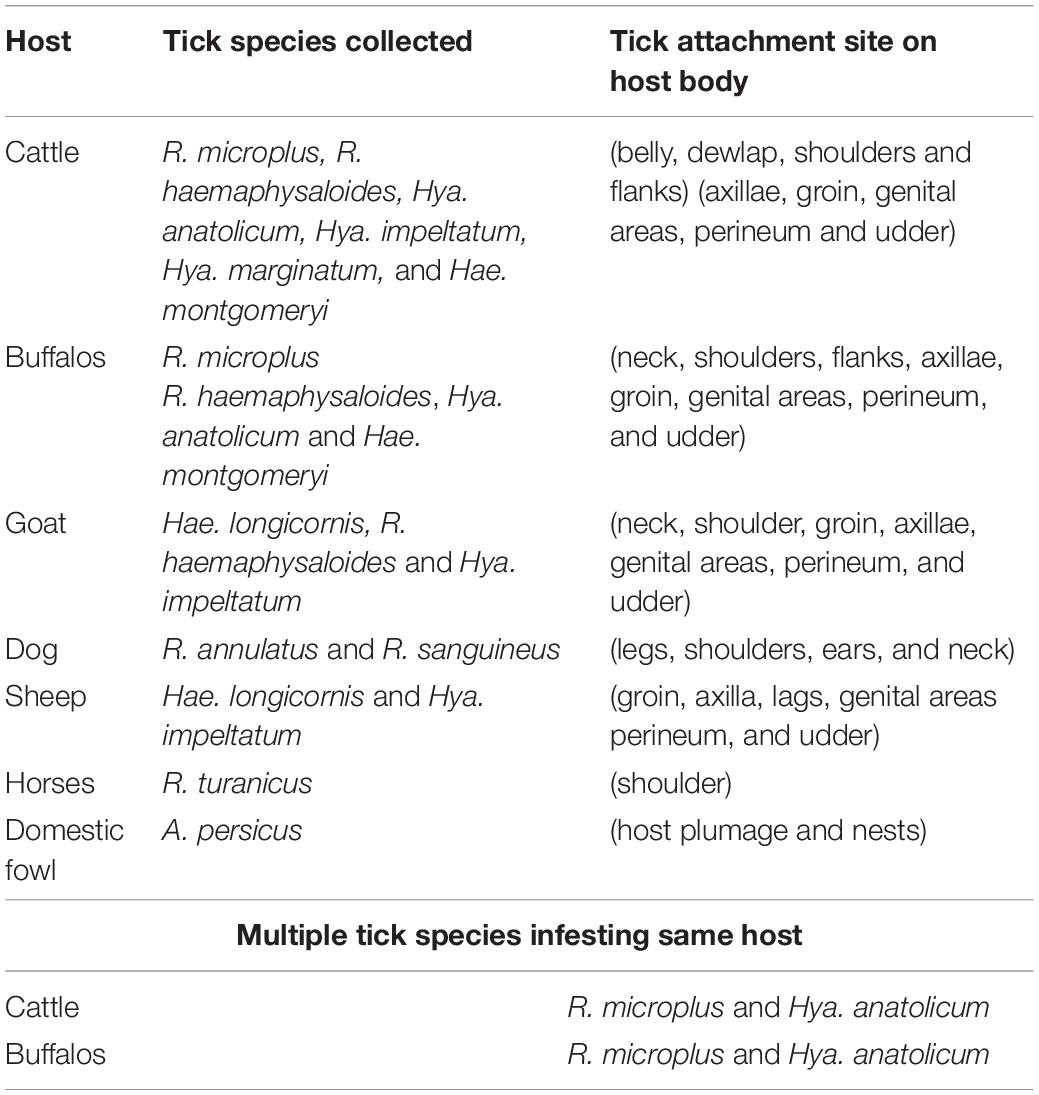
Table 3. Collected ticks, their hosts and preferred attachment sites on different hosts observed during present study.
A total of 772 animals were observed throughout the study period (April 2017 to March 2018) which included bovine (cattle, 180; buffalos, 114), ovine (sheep, 80), caprine (goats, 120), avian (domestic fowl, 150) equine (horse, 80) and carnivorous (dogs, 48) hosts, from which 8,498 ticks were collected. The overall prevalence of tick infested animals was 69.4% (536/772). Among the tick infested hosts; cattle were found highly infested (87.2%, 157/180) followed by buffalos (79.8%, 91/114), domestic fowl (74.7%, 112/150), goats (68.3%, 82/120), dogs (66.7%, 32/48), horses (61.3%, 49/80), and sheep (16.3%, 13/80).
Median tick burden recorded on host animals was 16 (median 16, Q1-Q3: 9-31) and showed a statistically significant difference among the different host groups (χ2 = 157.83, df = 6, p < 0.001). Median tick burden was high in cattle (median 29), followed by buffalos (median 21.5), goats (median 21), sheep (13), domestic fowl (12), dogs (8.5), and horses (6).
The tick species included in the seasonal dynamics analysis were; Hya. anatolicum, Hya. marginatum, Hya. impeltatum, R. microplus, R. haemaphysaloides, R. sanguineus, R. turanicus, R. annulatus, Hae. montgomeryi, Hae. longicornis, and A. persicus. Tick species showed spatio-temporal variations in their distribution. Specifically, species of the genus Hyalomma were mainly reported in dry regions while species belonging to the genus Rhipicephalus were reported in semi-arid regions of the study area (Figure 6B). During ticks collection, adult females (partially fed or nymphs) were highly prevalent followed by males and larvae (Table 2). Other tick species were not included in the temporal analysis because they were collected by chance from novel hosts (these hosts were not the focus of the present study) either from the study area or from other districts within KP Pakistan, during some parts of the year. This included tick species such as Amb. gervaisi, Amb. exornatum, Amb. latum, D. marginatus, Hae. indica, and R. sanguineus from monitor lizards, Indian pythons, markhor, mongoose and wild boars, respectively.
The highest number of ticks were collected in July followed by August and September as an increase in the infestation rate of ticks were recorded during ticks collection and these finding significantly correlated with an increase in mean temperature [r(10) = 0.929, p < 0.01] and relative humidity [r(10) = 0.831, p < 0.01] during these months in the region. As the mean temperature fell and relative humidity decreased, the infestation rate also decreased (Figure 7).
A total of 25 ticks were collected from humans (i.e., three genera and five species including R. microplus, R. annulatus, Hae. punctata, Hya. marginatum, and Hya. anatolicum). Ticks collected from wild animals included Hae. indica (wild rodent, n = 17), Amb. gervaisi and Amb. exornatum (monitor lizard, n = 21 and n = 14), Amb. latum (Indian python, n = 23), D. marginatus (wild goat, n = 31) and R. sanguineus (wild boar, n = 12) (Table 4 and Figure 8).
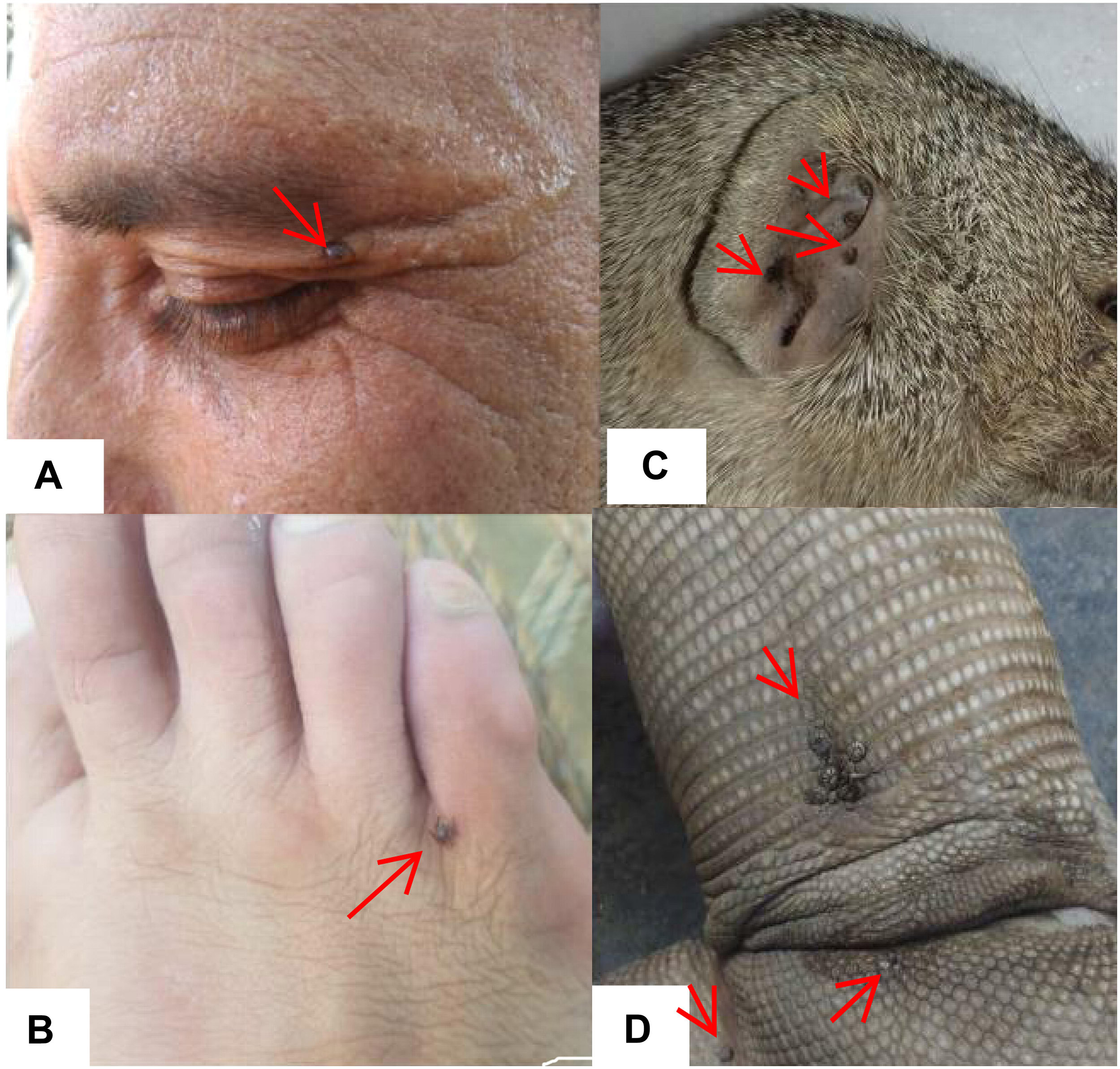
Figure 8. Figure showing ticks infesting humans (A,B) (written informed consent was obtained from the individuals for the publication of images), Hae. indica collected from mongoose (Herpestes edwardsi) (C), and Amb. gervaisi collected from monitor lizards (Varanus varanus) (D).
Genomic DNA extracted from morphologically identified R. microplus tick samples were amplified by PCR using COX1, 16S rRNA and ITS2 specific primers (Table 1). Results were observed on agarose gel electrophoresis and the resultant amplicons (COX1, 16S rRNA and ITS2) were 620, 376, and 267 bp, respectively. After sequencing, BLAST analysis of the COX1 nucleotide partial sequences showed 95–100% identity with the same sequence reported from Pakistan (accession no. MG459963), Bangladesh (accession no. MG459961, MG459962) Myanmar (accession no. MG459964) and China (accession no. KC503259). In the case of 16S rRNA nucleotide partial sequences, BLAST analysis revealed that it shares a 98–100% identity with previously reported sequences for R. microplus from India (accession no. KY458969; MG811555) and China (accession no. KU664521; MH208600). ITS2 partial nucleotide sequences showed that it shares 99% identity with the same sequences reported for the same tick species from India (accession no. JX974346; MG978179), China (accession no. JQ737125; JQ737124; KX450290; MG721034), Laos (accession no. KC503276), Colombia (accession no. MF353138), and Cambodia (accession no. KC503272). Similarly, the sequences of COX1 (accession no. MK812968, MK836289, MK858219, MK858220), 16S rRNA (accession no. MK463980, MK463981, MK463982) and ITS 2 (accession no. MK480725, MK524212, MK531135, MK578158, MK577644) obtained in the present study were deposited to the NCBI databank.
Maximum likelihood (ML) trees were inferred from COX1, 16S rRNA and ITS2 partial sequences of R. microplus to establish its phylogenetic relationship. ML analysis of COX1 nucleotide sequences revealed three clades, clade A includes ticks from America, Malaysia, and China, clade B was comprised of ticks originating from China (Burger et al., 2014; Low et al., 2015) and clade C includes ticks from Pakistan, India, Bangladesh, Myanmar, and Malaysia (Figure 3) (Roy et al., 2018). The COX1 phylogenetic analysis provided better support to resolve the evolutionary relationships of R. microplus. On the other hand, 16S rRNA partial sequences provided little phylogenetic structure of R. microplus complex and divided into two phylogenetic clades, the clade A comprised of the R. microplus tick species from China, India and Pakistan. Clade B includes R. microplus ticks originating from Malaysia, Africa and America and the R. australis (formerly recognized as R. microplus) from Australia, Indonesia and New Caledonia formed a separate subclade along with the aforementioned ticks (Figure 4). Based on ITS2 analysis, all species of the R. microplus complex were clustered together and the ITS2 tree provided support to the monophyly of R. microplus complex (Figure 5).
Climatic change and global warming have a vital impact on the distribution of ticks and tick-borne pathogens, as each tick species selects a set of ecological conditions and biotopes that determine its dispersal and outline risk areas for their associated pathogens transmission (Leger et al., 2013). Tick habitat expansion and novel pathogens (protozoans, bacteria, rickettsia, and viruses) are re-emerging in new geographic areas, posing a serious threat to public and veterinary health (Estrada-Peña et al., 2013; Jore et al., 2014). In KP Pakistan, the majority of farmers lack basic knowledge on ticks, their disease causing potential and the wide variety of hosts they infest, mostly resulting in a failure of controlling tick infestations. The present study encompasses the identification of diverse tick species and novel hosts, including humans, along with the seasonal dynamics and molecular phylogeny of R. microplus from north-western regions of Pakistan. The collected ticks were categorized into six genera including 17 species of medical and veterinary concern. We reported Amb. latum, Amb. gervaisi, Amb. exornatum, A. persicus, D. marginatus, Hae. indica, Hae. longicornis, Hae. punctata, Hae. montgomeryi, Hya. impeltatum, H. anatolicum, Hya. marginatum, R. annulatus, R. haemaphysaloides, R. microplus, R. sanguineus, and R. turanicus from the KP province. The majority of these ticks are responsible for the transmission of Babesia bovis, Theileria annulata, Anaplasma marginale, and Anaplasma centrale in the region (Jabbar et al., 2015). Our findings on the tick species prevalent in Pakistan correspond with previously reported studies (Manan et al., 2007; Karim et al., 2017; Rehman et al., 2017). Additionally, these ticks have been shown to be responsible for considerable losses in the livestock industry, causing severe threats to animal hosts and public health (Tadesse et al., 2012; Rehman et al., 2017).
Although ticks infestation on humans has been recorded from other regions of the world (Guglielmone and Robbins, 2018), ticks infesting humans and wild animals have not been previously reported in KP, Pakistan. During this study we found R. microplus, R. annulatus, Hya. anatolicum, Hya. marginatum and Hae. punctata (three genera and five species) infesting humans (Figure 8). These ticks, for instance, R. microplus (Labruna et al., 2005; Okino et al., 2010; Serra-Freire, 2010), R. annulatus (Horak et al., 2005; Bakirci et al., 2014; Kar et al., 2017) Hya. anatolicum (Estrada-Peña and Jongejan, 1999; Apanaskevich and Horak, 2005; Papa et al., 2011), Hya. marginatum (Santos-Silva et al., 2011), and Hae. punctata (Fernández-Soto et al., 2006; Briciu et al., 2011) parasitizing humans have already been reported in other parts of the world. Hae. indica from Indian gray mongoose are reported in this study, tick infestation on mongoose have been reported previously (Hoogstraal, 1970; Hoogstraal and Wassef, 1977; Horak et al., 1999; Cheng et al., 2018). In the present study Amb. gervaisi and Amb. exornatum parasitizing monitor lizards, and Amb. latum infesting Indian pythons were also reported. Amblyomma ticks are vectors of several pathogens known to transmit disease causing agents to humans and has never been investigated in Pakistan. Amblyomma species infesting reptiles are vectors of Aeromonas hydrophila that cause bacterial stomatitis, paralysis and pneumonia in snakes (Tenderio, 1953; Stephen and Achyutha, 1979; Marcus, 1981; Stenos et al., 2005; Hanson et al., 2007). D. marginatus were found to infest wild goats (locally known as markhor), which demands further studies to investigate the negative impact of this tick species in markhor which is considered as endangered species. The genus Dermacentor is of public and veterinary health concern as a vector reservoir for several pathogens with unknown potential risks in Pakistan. R. sanguineus, a vector of microorganisms of medical and veterinary health concern such as Hepatozoon canis and Ehrlichia canis (Dantas-Torres et al., 2018; Cabezas-Cruz et al., 2019) was collected for the first time from wild boars in the region.
Studies showing the seasonal pattern of tick distributions have never been reported from KP, Pakistan. During this study, information about the tick burden associated with relative humidity and temperature was recorded which is essential for the timing of potential control measures in tick infestation season. The target study region has high temperature and humidity during July, August and September therefore, high tick infestations were recorded during these months. On the other hand, due to the low temperatures during December, January, and February, several tick species were found to exhibit low infestation rates.
Several studies have used COX1, 16S rRNA and ITS2 as genetic markers for the accurate molecular identification and phylogenetic relationship of various organisms, including hard ticks such as R. microplus (Burger et al., 2014; Lv et al., 2014; Low et al., 2015; Coimbra-Dores et al., 2018). COX1 sequences have been used to show that cryptic species of R. microplus contain and display different population structures in different geographical regions (Burger et al., 2014; Low et al., 2015). Thus, R. microplus appears to consist of a complex of distinct genetic assemblages, namely R. australis, R. annulatus, R. microplus clade A (Burger et al., 2014), R. microplus clade B (Burger et al., 2014), and R. microplus clade C (Low et al., 2015). Burger et al. (2014) showed that R. microplus from clade B (Southern China and Northern India) is more closely related to R. annulatus than to R. microplus from clade A (Asia, South America, and Africa). In accordance with previous studies, phylogenetic trees based on COX1, 16S rRNA and ITS2 of R. microplus were obtained to establish the phylogenetic relationship of this tick (Lv et al., 2014; Low et al., 2015; Csordas et al., 2016; Rehman et al., 2017; Roy et al., 2018). COX1 was found to be an established marker for the phylogenetic analyses and informative for R. microplus phylogeny as compared to 16S rRNA and ITS2 as the latter provides monophyletic support to Boophild ticks (Burger et al., 2014). The COX1 phylogenetic analysis provided support to resolve the evolutionary relationship of R. microplus. Our findings about obtained COX1 sequences and phylogenetic analysis are parallel with previous reports from the region (Roy et al., 2018). ML analysis of COX1 nucleotide sequences revealed that these ticks are closely related to clade C (Low et al., 2015; Roy et al., 2018) (Figure 3). On the other hand, 16S rRNA partial sequences provided support to the monophyly of R. microplus complex and our tick sequences were clustered with clade A (Figure 4) (Brahma et al., 2014; Lv et al., 2014; Low et al., 2015). Based on ITS2 analysis, all species of the R. microplus complex were clustered together on the tree and provided support to the monophyly of R. microplus complex. The phylogenetic relationship in the R. microplus species complex was poor and could not accurately categorized as a sister species since ITS2 is highly conserved and is insufficient in distinguishing these closely related species (Figure 5).
This study for the first time explored the tick diversity infesting various hosts in KP, Pakistan. The important tick species of domestic animal hosts are R. microplus, R. annulatus, R. haemaphysaloides, R. sanguineus, R. turanicus, Hae. montgomeryi, Hae. longicornis, Hya. marginatum. Hya. anatolicum, Hya. impeltatum, and A. persicus. The seasonal pattern of several tick species showed high infestation during July, August and September in which high temperature and humidity persists in the region. The collection of tick species parasitizing humans in the study area may facilitate further studies to identify the functional role of these parasites in the transmission of pathogens that cause human diseases. Altogether, tick species such as Hae. indica, Amb. gervaisi, Amb. exornatum, Amb. latum, D. marginatus, and R. sanguineus were reported for the first time infesting wild animals in the region. The phylogenetic relationship of cattle tick R. microplus explored in this study will further help in defining control strategies. These findings will facilitate awareness among the local population about tick species diversity and their hosts and future control strategies against ticks and tick-borne diseases in Pakistan.
The datasets generated for this study can be found in NCBI, MK463980, MK463981, MK463982, MK480725, MK524212, MK531135, MK578158, MK577644, MK812968, MK836289, MK858219, MK858220.
AA designed the study and acquired the budget. AA, MAK, HZ, SK, PY, JN, ZUR, MA, and MI collected the samples. AA, HZ, SK, MQK, MAK, and JN carried out the statistical analysis. MAK, HZ, PY, ZUR, AA, MA, and MI conducted the experiments. All authors carried out critical revisions and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge the financial support provided by the Pakistan Science Foundation and the Higher Education Commission of Pakistan. We would like to thank Dr. Dmitry Apanaskevich, United States National Tick Collection (USNTC) for providing his expertise and facilities in the identification of ticks.
Ali, A., Parizi, L. F., Ferreira, B. R., Junior, V., and da Silva, I. (2016). A revision of two distinct species of Rhipicephalus: R. microplus and R. australis. Cienc. Rural 46, 1240–1248.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1006/jmbi.1990.9999
Apanaskevich, D. A., and Horak, I. G. (2005). The genus Hyalomma. II The taxonomic status of H. (Euhyalomma) anatolicum Koch 1844 and H. (Euhyalomma) excavatum Koch 1844 with the redescription of all stages. Acarina 13, 181–197.
Apanaskevich, D. A., and Horak, I. G. (2008). The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H. (Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int. J. Acarol. 34, 13–42. doi: 10.1080/01647950808683704
Apanaskevich, D. A., and Horak, I. G. (2009). The genus Hyalomma Koch, 1844. IX. Redescription of all parasitic stages of H. (Euhyalomma) impeltatum Schulze & Schlottke, 1930 and H. (E.) somalicum Tonelli Rondelli, 1935 (Acari: Ixodidae). Syst. Parasitol. 73, 199–218. doi: 10.1007/s11230-009-9190-x
Bakirci, S., Aysul, N., Eren, H., Unlu, A. H., and Karagenc, T. (2014). Diversity of ticks biting humans in Aydın province of Turkey. Ank Univ Vet Fak Derg. 61, 93–98. doi: 10.1501/vetfak_0000002611
Barker, S. C., and Walker, A. R. (2014). Ticks of Australia. The species that infest domestic animals and humans. Zootaxa. 3816, 1–144. doi: 10.11646/zootaxa.3816.1.1
Baron, S., van der Merwe, N. A., and Maritz-Olivier, C. (2018). The genetic relationship between R. microplus and R. decoloratus ticks in South Africa and their population structure. Mol. Phylogenet. Evol. 129, 60–69. doi: 10.1016/j.ympev.2018.08.003
Brahma, R. K., Dixit, V., Sangwan, A. K., and Doley, R. (2014). Identification and characterization of Rhipicephalus (Boophilus) microplus and Haemaphysalis bispinosa ticks (Acari: Ixodidae) of North East India by ITS2 and 16S rDNA sequences and morphological analysis. Exp. Appl. Acarol. 62, 253–265. doi: 10.1007/s10493-013-9732-4
Briciu, V. T., Titilincu, A., Ţãţulescu, D. F., Cârstina, D., Lefkaditis, M., and Mihalca, A. D. (2011). First survey on hard ticks (Ixodidae) collected from humans in Romania: possible risks for tick-borne diseases. Exp. Appl. Acarol. 54, 199–204. doi: 10.1007/s10493-010-9418-0
Burger, T. D., Shao, R., and Barker, S. C. (2014). Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick Rhipicephalus (Boophilus) microplus contains a cryptic species. Mol. Phylogenet. Evol. 76, 241–253. doi: 10.1016/j.ympev.2014.03.017
Cabezas-Cruz, A., Allain, E., Ahmad, A. S., Saeed, M. A., Rashid, I., Ashraf, K., et al. (2019). Low genetic diversity of Ehrlichia canis associated with high co-infection rates in Rhipicephalus sanguineus (s.l.). Parasit. Vectors 12:12. doi: 10.1186/s13071-018-3194-9
Caetano, R. L., Vizzoni, V. F., Bitencourth, K., Carrico, C., Sato, T. P., Pinto, Z. T., et al. (2017). Ultrastructural morphology and molecular analyses of tropical and temperate “species” of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) in Brazil. J. Med. Entomol. 54, 1201–1212. doi: 10.1093/jme/tjx066
Cheng, T., Halper, B., Siebert, J., Cruz-Martinez, L., Chapwanya, A., Kelly, P., et al. (2018). Parasites of small Indian mongoose, Herpestes auropunctatus, on St. Kitts, West Indies. Parasitol. Res. 117, 989–994. doi: 10.1007/s00436-018-5773-2
Coimbra-Dores, M. J., Maia-Silva, M., Marques, W., Oliveira, A. C., Rosa, F., and Dias, D. (2018). Phylogenetic insights on mediterranean and afrotropical Rhipicephalus species (Acari: Ixodida) based on mitochondrial DNA. Exp. Appl. Acarol. 75, 107–128. doi: 10.1007/s10493-018-0254-y
Csordas, B. G., Garcia, M. V., Cunha, R. C., Giachetto, P. F., Blecha, I. M. Z., and Andreotti, R. (2016). New insights from molecular characterization of the tick Rhipicephalus (Boophilus) microplus in Brazil. Rev. Bras. Parasitol. Vet. 25, 317–326. doi: 10.1590/S1984-29612016053
Dantas-Torres, F. (2015). Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int. J. Parasitol. Parasites Wildl. 4, 452–461. doi: 10.1016/j.ijppaw.2015.07.001
Dantas-Torres, F., Latrofa, M. S., Ramos, R. A. N., Lia, R. P., Capelli, G., Parisi, A., et al. (2018). Biological compatibility between two temperate lineages of brown dog ticks. Rhipicephalus sanguineus (sensu lato). Parasit. Vectors 11:398. doi: 10.1186/s13071-018-2941-2
de la Fuente, J., Antunes, S., Bonnet, S., Cabezas-Cruz, A., Domingos, A. G., Estrada-Peña, A., et al. (2017). Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front. Cell Infect. Microbiol. 7:114. doi: 10.3389/fcimb.2017.00114
de la Fuente, J., Rodriguez, M., and Garcia-Gari, J. C. (2000). Immunological control of ticks through vaccination with Boophilus microplus gut antigens. Ann. N Y Acad Sci. 916, 617–621. doi: 10.1111/j.1749-6632.2000.tb05347.x
Estrada-Peña, A. (2008). Climate, niche, ticks, and models: what they are and how we should interpret them. Parasitol. Res. 103, 87–95. doi: 10.1007/s00436-008-1056-7
Estrada-Peña, A., Gray, J. S., Kahl, O., Lane, R. S., and Nijhoff, A. M. (2013). Research on the ecology of ticks and tick-borne pathogens—methodological principles and caveats. Front. Cell Infect. Microbiol. 3:29. doi: 10.3389/fcimb.2013.00029
Estrada-Peña, A., and Jongejan, F. (1999). Ticks feeding on humans: a review of records on human-biting Ixodoidea with special reference to pathogen transmission. Exp. Appl. Acarol. 23, 685–715.
Estrada-Peña, A., Venzal, J. M., Nava, S., Mangold, A., Guglielmone, A. A., Labruna, M. B., et al. (2012). Reinstatement of Rhipicephalus (Boophilus) australis (Acari: Ixodidae) with redescription of the adult and larval stages. J. Med. Entomol. 49, 794–802. doi: 10.1603/me11223
Fernández-Soto, P., Pérez-Sánchez, R., Álamo-Sanz, R., and Encinas-Grandes, A. (2006). Spotted fever group rickettsiae in ticks feeding on humans in northwestern Spain: is Rickettsia conorii vanishing? Ann. N Y Acad Sci. 1078, 331–333. doi: 10.1196/annals.1374.063
García-García, J. C., Gonzalez, I. L., González, D. M., Valdés, M., Méndez, L., Lamberti, J., et al. (1999). Sequence variations in the Boophilus microplus Bm86 locus and implications for immunoprotection in cattle vaccinated with this antigen. Exp. Appl. Acarol. 23, 883–895.
Ghosh, H. S., and Misra, K. K. (2012). Scanning electron microscope study of a snake tick, Amblyomma gervaisi (Acari: Ixodidae). J. Parasit. Dis. 36, 239–250. doi: 10.1007/s12639-012-0117-0
Gondard, M., Cabezas-Cruz, A., Charles, R. A., Vayssier-Taussat, M., Albina, E., and Moutailler, S. (2017). Ticks and tick-borne pathogens of the Caribbean: current understanding and future directions for more comprehensive surveillance. Front. Cell Infect. Microbiol. 7:490. doi: 10.3389/fcimb.2017.00490
Gratz, N. G. (1999). Emerging and resurging vector-borne diseases. Annu. Rev. Entomol. 44, 51–75. doi: 10.1146/annurev.ento.44.1.51
Guglielmone, A. A. and Robbins, R. G., (2018). Hard Ticks (Acari: Ixodida: Ixodidae) Parasitizing Humans, (New York, NY: Springer), 314.
Guglielmone, A. A., Robbins, R. G., Apanaskevich, D. A., Petney, T. N., Estrada-Peña, A., and Horak, I. G. (2014). The hard ticks of the world. Dordrecht: Springer.
Hall, T., Biosciences, I., and Carlsbad, C. (2011). BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2, 60–61. doi: 10.1016/j.compbiolchem.2019.02.002
Hanson, B. A., Frank, P. A., Mertins, J. W., and Corn, J. L. (2007). Tick paralysis of a snake caused by Amblyomma rotundatum (Acari: Ixodidae). J. Med. Entomol. 44, 155–157. doi: 10.1093/jmedent/41.5.155
Hoogstraal, H. (1970). Identity, distribution, and hosts of Haemaphysalis (Rhipistoma) indica Warburton (resurrected) (Ixodoidea: Ixodidae), a carnivore parasite of the Indian subregion. J. Parasitol. 56, 1013–1022.
Hoogstraal, H., and Varma, M. G. R. (1962). Haemaphysalis cornupunctata sp. n. and H. kashmirensis sp. n. from Kashmir, with Notes on H. sundrai Sharif and H. sewelli Sharif of India and Pakistan (Ixodoidea, Ixodidae). J. Parasitol. 48, 185–194.
Hoogstraal, H., and Wassef, H. Y. (1977). Haemaphysalis (Ornithophysalis) kadarsani sp. n. (Ixodoidea: Ixodidae), a rodent parasite of virgin lowland forests in Sulawesi (Celebes). J. Parasitol. 63, 1103–1109.
Horak, I. G., Chaparro, F., Beaucournu, J. C., and Louw, J. P. (1999). Parasites of domestic and wild animals in South Africa. XXXVI. Arthropod parasites of yellow mongooses, Cynictis penicillata (G. Cuvier, 1829). Onderstepoort J. Vet. Res. 66, 33–38.
Horak, I. G., Fourie, L. J., and Braack, L. E. O. (2005). Small mammals as hosts of immature ixodid ticks. Onderstepoort J. Vet. Res. 72, 255–261.
Jabbar, A., Abbas, T., Saddiqi, H. A., Qamar, M. F., and Gasser, R. B. (2015). Tick-borne diseases of bovines in Pakistan: major scope for future research and improved control. Parasit. Vectors 8:283. doi: 10.1186/s13071-015-0894-2
Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., et al. (2008). Global trends in emerging infectious diseases. Nature 451, 990–993. doi: 10.1038/nature06536
Jore, S., Vanwambeke, S. O., Viljugrein, H., Isaksen, K., Kristoffersen, A. B., Woldehiwet, Z., et al. (2014). Climate and environmental change drives Ixodes ricinus geographical expansion at the northern range margin. Parasit. Vectors 7:11. doi: 10.1186/1756-3305-7-11
Kaiser, M. N., and Hoogstraal, H. (1964). The Hyalomma ticks (Ixodoidea, Ixodidae) of Pakistan, India, and Ceylon, with keys to subgenera and species. Acarologia 6, 257–286.
Kar, S., Yilmazer, N., Akyildiz, G., and Gargili, A. (2017). The human infesting ticks in the city of Istanbul and its vicinity with reference to a new species for Turkey. Syst. Appl. Acarol. 22, 2245–2256.
Karim, S., Budachetri, K., Mukherjee, N., Williams, J., Kausar, A., Hassan, M. J., et al. (2017). A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl. Trop Dis. 11:e0005681. doi: 10.1371/journal.pntd.0005681
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Labruna, M. B., Jorge, R. S., Sana, D. A., Jácomo, A. T., Kashivakura, C. K., Furtado, M. M., et al. (2005). Ticks (Acari: Ixodida) on wild carnivores in Brazil. Exp. Appl. Acarol. 36, 149–163.
Labruna, M. B., McBride, J. W., Bouyer, D. H., Camargo, L. M. A., Camargo, E. P., and Walker, D. H. (2004). Molecular evidence for a spotted fever group Rickettsia species in the tick Amblyomma longirostre in Brazil. J. Med. Entomol. 41, 533–537.
Labruna, M. B., Naranjo, V., Mangold, A. J., Thompson, C., Estrada-Peña, A., Guglielmone, A. A., et al. (2009). Allopatric speciation in ticks: genetic and reproductive divergence between geographic strains of Rhipicephalus (Boophilus) microplus. BMC Evol. Biol. 9:46. doi: 10.1186/1471-2148-9-46
Leger, E., Vourc’h, G., Vial, L., Chevillon, C., and McCoy, K. D. (2013). Changing distributions of ticks: causes and consequences. Exp. Appl. Acarol. 59, 219–244. doi: 10.1007/s10493-012-9615-0
Lempereur, L., Geysen, D., and Madder, M. (2010). Development and validation of a PCR–RFLP test to identify African Rhipicephalus (Boophilus) ticks. Acta Trop. 114, 55–58. doi: 10.1016/j.actatropica.2010.01.004
Lew-Tabor, A. E., Ali, A., Rehman, G., Rocha-Garcia, G., Zangirolamo, A. F., Malardo, T., et al. (2017). Cattle Tick Rhipicephalus microplus-host interface: a review of resistant and susceptible host responses. Front. Cell Infect Microbiol. 7:506. doi: 10.3389/fcimb.2017.00506
Lew-Tabor, A. E., and Valle, M. R. (2016). A review of reverse vaccinology approaches for the development of vaccines against ticks and tick borne diseases. Ticks Tick Borne Dis. 7, 573–585. doi: 10.1016/j.ttbdis.2015.12.012
Low, V. L., Tay, S. T., Kho, K. L., Koh, F. X., Tan, T. K., Lim, Y. A. L., et al. (2015). Molecular characterisation of the tick Rhipicephalus microplus in Malaysia: new insights into the cryptic diversity and distinct genetic assemblages throughout the world. Parasit. Vectors 8:341. doi: 10.1186/s13071-015-0956-5
Lv, J., Wu, S., Zhang, Y., Zhang, T., Feng, C., Jia, G., et al. (2014). Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA 25, 142–149. doi: 10.3109/19401736.2013.792052
Madder, M., Horak, I., and Stoltsz, H. (2014). Tick Identification. Pretoria: Faculty of veterinary Science University of Pretoria, 58.
Manan, A., Khan, Z., and Ahmad, B. (2007). Prevalence and identification of ixodid tick genera in frontier region Peshawar. J. Agric. Biol. Sci. 37, 173–176.
Marcus, L. C. (1981). Veterinary Biology and Medicine of Captive Amphibians and Reptiles. Philadelphia, PA: Lea and Febiger.
Nowak-Chmura, M. (2012). Teratological changes in tick morphology in ticks feeding on exotic reptiles. J. Nat. Hist. 46, 911–921. doi: 10.1080/00222933.2011.651635
Okino, T., Ushirogawa, H., Matoba, K., and Hatsushika, R. (2010). Bibliographical studies on human cases of hard tick (Acarina: Ixodidae) bites in Japan (5) cases of Ixodes acutitarsus and I. turdus infestation. Kawasaki Med. J. 36, 115–120.
Paddock, C. D., and Childs, J. E. (2003). Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64. doi: 10.1128/cmr.16.1.37-64.2003
Papa, A., Karabaxoglou, D., and Kansouzidou, A. (2011). Acute West Nile virus neuroinvasive infections: cross-reactivity with dengue virus and tick-borne encephalitis virus. J. Med. Virol. 83, 1861–1865. doi: 10.1002/jmv.22180
Parola, P., and Raoult, D. (2001). Ticks and tick-borne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32, 897–928. doi: 10.1086/319347
Rehman, A., Nijhof, A. M., Sauter-Louis, C., Schauer, B., Staubach, C., and Conraths, F. J. (2017). Distribution of ticks infesting ruminants and risk factors associated with high tick prevalence in livestock farms in the semi-arid and arid agro-ecological zones of Pakistan. Parasit. Vectors 10:190. doi: 10.1186/s13071-017-2138-0
Robertson, R. G., Wisseman, C. L., and Traub, R. (1970). Tick-Borne Rickettsiae of the Spotted Fever Group in West Pakistan: I. isolation of strains from ticks in different habitats. Am J Epidemiol. 92, 382–394. doi: 10.1093/oxfordjournals.aje.a121221
Roy, B. C., Krücken, J., Ahmed, J. S., Majumder, S., Baumann, M. P., Clausen, P. H., et al. (2018). Molecular identification of tick-borne pathogens infecting cattle in Mymensingh district of Bangladesh reveals emerging species of Anaplasma and Babesia. Transbound. Emerg. Dis. 65, 231–242. doi: 10.1111/tbed.12745
Santos-Silva, M. M., Beati, L., Santos, A. S., De Sousa, R., Núncio, M. S., Melo, P., et al. (2011). The hard-tick fauna of mainland Portugal (Acari: Ixodidae): an update on geographical distribution and known associations with hosts and pathogens. Exp. Appl. Acarol. 55, 85–121. doi: 10.1007/s10493-011-9440-x
Schorderet-Weber, S., Noack, S., Selzer, P. M., and Kaminsky, R. (2017). Blocking transmission of vector-borne diseases. Int. J. Parasitol. Drugs Drug Resist. 7, 90–109. doi: 10.1016/j.ijpddr.2017.01.004
Serra-Freire, N. M. (2010). Occurrence of ticks (Acari: Ixodidae) on human hosts, in three municipalities in the State of Pará, Brazil. Rev. Bras. Parasitol. Vet. 19, 141–147. doi: 10.1590/s1984-29612010000300003
Sherrard-Smith, E., Chadwick, E., and Cable, J. (2012). Abiotic and biotic factors associated with tick population dynamics on a mammalian host: Ixodes hexagonus infesting otters. Lutra. PloS One 7:e47131. doi: 10.1371/journal.pone.0047131
Stenos, J., Graves, S. R., and Unsworth, N. B. (2005). A highly sensitive and specific real-time PCR assay for the detection of spotted fever and typhus group Rickettsiae. Am. J. Trop Med. Hyg. 73, 1083–1085. doi: 10.4269/ajtmh.2005.73.1083
Stephen, S., and Achyutha, R. (1979). Q fever in South Kanara district: natural occurrence of Coxiella burneti in the tick (Aponomma gervaisi) preliminary report. Indian J. Med. Res. 69, 244–246.
Tadesse, F., Abadfaji, G., Girma, S., and Jibat, T. (2012). Identification of tick species and their preferred site on cattles body in and around Mizan Teferi, Southwestern Ethiopia. Vet. Med. Anim. Health 4, 1–5.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA 6 molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tenderio, J. (1953). Alguns dados sobre as stirpes de Coxiella burneti isoladas na Guine Portugusea. Bol. Cult. Guine Port. 8, 69–87.
Walker, A. R., Bouattour, A., Camicas, J. L., Estrada-Peña, A., Horak, I. G., Latif, A. A., et al. (2003). Ticks of Domestic Animals in Africa: A Guide to Identification of Species. Edinburgh: Bioscience Reports.
Keywords: ticks, hosts, R. microplus, Khyber Pakhtunkhwa, Pakistan
Citation: Ali A, Khan MA, Zahid H, Yaseen PM, Qayash Khan M, Nawab J, Ur Rehman Z, Ateeq M, Khan S and Ibrahim M (2019) Seasonal Dynamics, Record of Ticks Infesting Humans, Wild and Domestic Animals and Molecular Phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 10:793. doi: 10.3389/fphys.2019.00793
Received: 18 March 2019; Accepted: 06 June 2019;
Published: 16 July 2019.
Edited by:
Zhijun Yu, Hebei Normal University, ChinaReviewed by:
Nkululeko Nyangiwe, Dohne Agricultural Development Institute, South AfricaCopyright © 2019 Ali, Khan, Zahid, Yaseen, Qayash Khan, Nawab, Ur Rehman, Ateeq, Khan and Ibrahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, dW9wX2FsaUB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.