
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 25 January 2019
Sec. Integrative Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01897
Background: We obtained conflicting results regarding the relationship between the genetic role of the rs1138272 C/T polymorphism of the GSTP1 (Glutathione S-Transferase pi) gene and the risk of various cancers.
Methods: Using the presently available data, a meta-analysis was conducted to comprehensively evaluate the genetic relationship between the GSTP1 rs1138272 polymorphism and cancer susceptibility.
Results: A total of 43 studies including 15,688 cases and 17,143 controls were recruited into our quantitative synthesis. In the overall population, we observed an increased risk of overall cancer cases, compared with unrelated controls, in the genetic models of allele T vs. allele C (P-association = 0.007, OR = 1.17), carrier T vs. carrier C (P-association = 0.035, OR = 1.11), TT vs. CC (P-association = 0.002, OR = 1.45), TT vs. CC+CT (P-association = 0.009, OR = 1.42), and CT+TT vs. CC (P-association = 0.027, OR = 1.13). We detected similar positive results within the Asian population. Additionally, there was a significant increase in the incidence of cancer for Africans under all genetic models (all P-association < 0.05, OR > 1). When targeting the Caucasian population, we detected a positive association with the TT vs. CC and TT vs. CC+CT models in the “Colorectal cancer” (P-association < 0.05, OR < 1) and “Head and neck cancer” (P-association < 0.05, OR > 1) subgroups. For the “Lung cancer” subgroup, we observed a slightly increased risk in Caucasians under the models of allele T vs. allele C, carrier T vs. carrier C, CT vs. CC, and CT+TT vs. CC (P-association < 0.05, OR > 1).
Conclusion: The TT genotype of the GSTP1 rs1138272 polymorphism is likely related to the susceptibility to overall cancer in the Asian and African populations and, specifically, “Colorectal” and “Head and neck” cancers in the Caucasian population. In addition, the CT genotype of the GSTP1 rs1138272 polymorphism may be linked to the risk of lung cancer in Caucasians. Additional evidence is required to confirm this conclusion.
The human GSTP1 gene is located on chromosome 11 (11q13.2) (Sharma et al., 2017), and the GSTP1 (Glutathione S-Transferase pi) protein participates in the drug resistance process of cancer cells (Singh, 2015). Two commonly occurring polymorphisms within the exon 5/6 region of the GSTP1 gene, namely, rs1695 (A313G, IIe105Val) and rs1138272 (C341T, Ala114Val), may be related to the occurrence and development of certain diseases (Huang et al., 2013; Xie et al., 2014; Tan and Chen, 2015; Zhou et al., 2015; Wang et al., 2016). For instance, the GSTP1 rs1695 polymorphism is likely associated with the risk of Alzheimer’s disease, based on a previous meta-analysis (Wang et al., 2016). There have been several comprehensive analyses concerning the potential role of the GSTP1 rs1695 polymorphism in the susceptibility to cancer. However, the results varied between cancer types. For example, GSTP1 rs1695 was reported to be associated with the risk of esophageal cancer and malignant melanoma in the Caucasian population (Tan and Chen, 2015; Zhou et al., 2015), but not childhood acute lymphoblastic leukemia (ALL) (Zhao et al., 2018) or bladder cancer (Yu et al., 2016). To the best of our knowledge, very limited comprehensive analyses on the relationship between GSTP1 rs1138272 and overall cancer risk have been reported.
Huang et al. (2013), one relevant meta-analysis containing 28 case-control studies was reported, assessing the potential effect of the GSTP1 rs1138272 C/T polymorphism on the risk of overall cancer. In view of the publication of new relevant articles in the last 5 years, we performed an updated meta-analysis to gain insight into the genetic association between the rs1138272 C/T polymorphism of the GSTP1 gene and the risk of cancer. Altogether, 43 eligible case-control studies were recruited into our statistical analysis.
Five online databases extending until September 2018, including PubMed, Embase, Cochrane, Scopus, and WOS (web of science), were utilized for the article identification. Referring Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Moher et al., 2009) were considered. The search terms are shown in Supplementary Table S1.
First, duplicate articles or articles with overlapping data were removed. In addition, review articles, meta-analyses, meeting abstracts and case reports were excluded. Articles that lacked normal control data or the complete genotype data on the CC, CT, TT status of GSTP1 rs1138272 in the cases/controls were also removed. The basic information was then collected and summarized, and P-HWE (P-value for Hardy–Weinberg equilibrium) was calculated. The quality appraisal of each study was also performed using the Newcastle-Ottawa Scale (NOS) system. Articles with P-HWE > 0.05 for the controls and an NOS score > = 5 were included. Eligible case-control studies were finally considered.
A fixed-effects model was applied in the Mantel–Haenszel statistics of association test when the P-heterogeneity of Cochran’s Q statistic was larger than 0.1 or the I2 value was less than 50%. When those criteria were not met, a random effects model was used in the DerSimonian and Laird statistics of association test. For the assessment of the pooled effect size, we obtained the odds ratio (OR), 95% confidence interval (CI) and P-Association (P-value of association test) from each meta-analysis and subsequent subgroup analysis by ethnicity, control source, or cancer type.
We used the Begg’s and Egger’s tests to assess the potential publication bias when the number of enrolled case-control studies was larger than 10. We also performed a sensitivity analysis to evaluate the data stability and possible sources of heterogeneity. The STATA software (version 12.0, StataCorp, United States) was used to analyze the following genetic models in the association test, Begg’s test, Egger’s test, and sensitivity analysis: the allele model (allele T vs. allele C), homozygote model (TT vs. CC), heterozygote model (CT vs. CC), dominant model (CT+TT vs. CC), recessive model (TT vs. CC+CT), and carrier model (carrier T vs. carrier C).
A flow chart illustrating the process of study selection is presented in Figure 1. Briefly, we initially obtained a total of 2,804 records by searching five databases, including 736 records from PubMed, 484 records from Embase, 60 records from Cochrane, 723 records from Scopus and 801 records from WOS. Then, we removed 1,202 duplicate records and excluded the following 1,506 records: 158 reviews; 70 meta-analyses; 42 case reports; 160 meeting abstracts; 62 articles with data on mouse, rat or dog models; 111 articles with in vitro data on cell lines; 792 articles focusing on other diseases, other genes or other variants of the GSTP1 gene; and 111 articles containing data on methylation or gene expression. Next, we assessed the eligibility of the remaining 96 full-text articles. An additional 56 articles were excluded, including 56 articles with unavailable data on the genotype frequency of CC, CT, and TT within GSTP1 rs1138272, and 4 articles in which the data were not in HWE. After a quality evaluation, 40 articles (Harris et al., 1998; Saarikoski et al., 1998; Park et al., 1999; Wadelius et al., 1999; Welfare et al., 1999; Marshall et al., 2000; Stanulla et al., 2000; Krajinovic et al., 2002; Wang et al., 2003, 2011; Sorensen et al., 2004; Yang et al., 2004; Garcia-Closas et al., 2005; Landi et al., 2005, 2007; Moore et al., 2005; De Roos et al., 2006; Lira et al., 2006; Marciniak et al., 2006; Jiao et al., 2007; Li et al., 2007, 2010; Murphy et al., 2007; Al-Dayel et al., 2008; Kury et al., 2008; Siraj et al., 2008; Van Emburgh et al., 2008; Zienolddiny et al., 2008; Canova et al., 2009; Northwood et al., 2010; Sainz et al., 2011; Ebrahimkhani et al., 2012; Garcia-Gonzalez et al., 2012; Ibarrola-Villava et al., 2012; Dura et al., 2013; Oskina et al., 2014; Ghatak et al., 2016; De Mattia et al., 2017; Minina et al., 2017; Rajesh et al., 2018) of good quality were included. Finally, we included a total of 43 case-control studies for our quantitative synthesis. All of the data in these articles were in HWE. The detailed characteristics of these articles are provided in Table 1.
First, we analyzed the relationship between the GSTP1 rs1138272 polymorphism and the risk of cancer through a meta-analysis of the overall population. As shown in Table 2, a total of 43 case-control studies with 15,688 cases and 17,143 controls were enrolled for the models of allele T vs. allele C, carrier T vs. carrier C, CT vs. CC, CT+TT vs. CC; in addition, 40 studies with 15,479 cases and 16,765 controls were enrolled for the models of TT vs. CC and TT vs. CC+CT. Because there was not a high degree of heterogeneity observed in the homozygote and recessive genetic models, a fixed-effects model was used in the Mantel–Haenszel statistics of association test for those genetic models. For the other genetic models, a random effects model was used in the DerSimonian and Laird statistics of association test. The quantitative synthesis results (Table 2) revealed an increased risk for cancer, compared with the control group, for the genetic models of allele T vs. allele C (P-association = 0.007, OR = 1.17), carrier T vs. carrier C (P-association = 0.035, OR = 1.11), TT vs. CC (P-association = 0.002, OR = 1.45), TT vs. CC+CT (P-association = 0.009, OR = 1.42), and CT+TT vs. CC (P-association = 0.027, OR = 1.13). Nevertheless, no significant effect on cancer risk was observed for the model of CT vs. CC (Table 2, P-association = 0.106). Supplementary Figures S1–S4 presents the forest plot data under the allele, carrier, heterozygote and dominant models. In summary, the TT genotype of the GSTP1 rs1138272 polymorphism may be associated with an increased susceptibility to cancer.
Next, we performed three subgroup analyses based upon ethnicity (Table 3), control source (Supplementary Table S2) and cancer type (Supplementary Table S3) in the overall population. As shown in Table 3, similar positive results were detected in the subgroup “Asian” under the allele, homozygote, recessive and dominant models (Table 3, all P-association < 0.05, OR > 1). As shown in Supplementary Table S2, we also assessed the difference between cancer cases and PB-based controls under the TT vs. CC (P-association = 0.006, OR = 1.45) and TT vs. CC+CT (P-association = 0.007, OR = 1.44) models. Figures 2, 3 present the relative forest plot of the subgroup analysis by ethnicity under the TT vs. CC and TT vs. CC+CT models, while Supplementary Figure S5 shows the forest plot of subgroup analysis by control source under the TT vs. CC model. Moreover, compared with the controls, an increased cancer risk was observed in the “African” subgroup under all of the genetic models (Table 3, all P-association < 0.05, OR > 1), but this was not the case for the “Caucasian” (Table 3) and “Hospital-based, HB” (Supplementary Table S2) subgroups (all P-association > or = 0.05). In addition, no significant association was found based on cancer type under most of the genetic models, except for the TT vs. CC (Supplementary Table S3, P-association = 0.001, OR = 3.11) and TT vs. CC+CT (P-association = 0.001, OR = 3.07) models of the “Head and neck cancer” subgroup. Supplementary Figure S6 presents the forest plot of subgroup analysis by cancer type in the overall population under the allele T vs. allele C model, and Supplementary Table S3 shows the pooled data of the “Colorectal cancer” subgroup with nine case-control studies (4,858 cases and 4,998 controls), the “Lung cancer” subgroup with seven case-control studies (2,123 cases and 2,266 controls) and the “Head and neck cancer” subgroup with six case-control studies (1,190 cases and 1,827 controls). Therefore, the rs1138272 polymorphism of the GSTP1 gene appears to be correlated with an increased risk of cancer in the Asian and African populations. Moreover, the TT genotype of GSTP1 rs1138272 may be associated with the risk of head and neck cancer in the overall population.
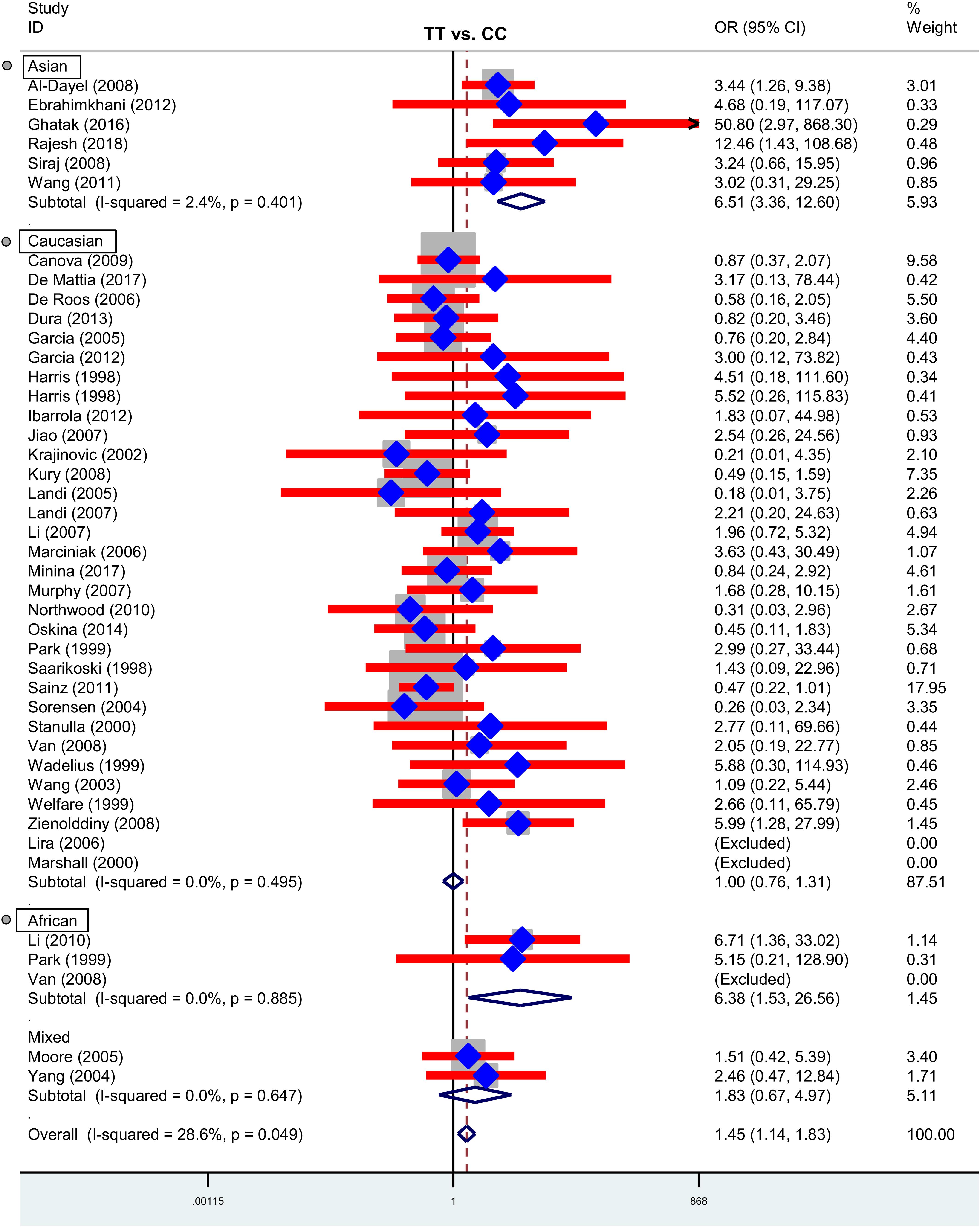
Figure 2. Forest plot of the subgroup analysis by ethnicity in the overall population (TT vs. CC model).
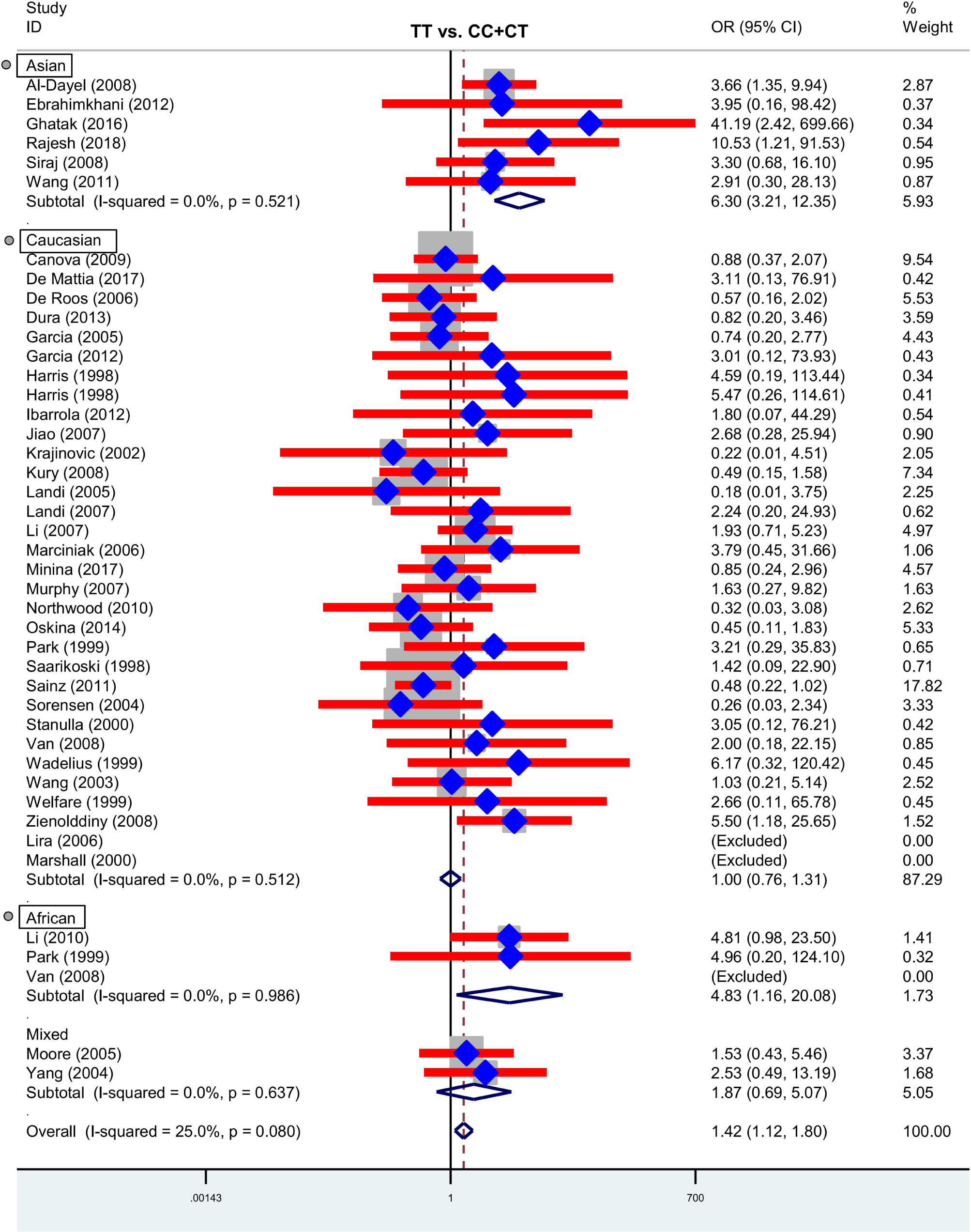
Figure 3. Forest plot of the subgroup analysis by ethnicity in the overall population (TT vs. CC+CT model).
Next, we performed subgroup analyses based upon control source (Supplementary Table S4) and cancer type (Supplementary Table S5 and Supplementary Figures S7–S10), targeting the Caucasian population. Similar positive results were detected in the “Head and neck cancer” subgroup analysis (Supplementary Table S5). Even though no significant associations were found in the “PB” or “HB” subgroup analyses (Supplementary Table S4, all P-association > 0.05), there was a positive association between GSTP1 rs1138272 and the risk of colorectal cancer in the Caucasian population for the models of TT vs. CC (Supplementary Table S5, P-association = 0.52, OR = 1.21) and TT vs. CC+CT (P-association = 0.023, OR = 0.52). With regard to “Lung cancer,” we observed a slightly increased risk in the Caucasian population under the models of allele T vs. allele C (P-association = 0.015, OR = 1.21), carrier T vs. carrier C (P-association = 0.044, OR = 1.18), CT vs. CC (P-association = 0.032, OR = 1.20), and CT+TT vs. CC (P-association = 0.020, OR = 1.22). Further, to eliminate the effects of the HB controls in the results of the Caucasian population, we also performed another subgroup analysis based upon cancer type using the Caucasian cases and population-based negative controls. Similar results were detected for colorectal and lung cancer (Supplementary Table S6). These data revealed that the TT genotype of the GSTP1 rs1138272 polymorphism may decrease susceptibility to “Colorectal” cancers but increase susceptibility to “Head and neck” cancers, while the CT genotype may be associated with lung cancer risk in the Caucasian population.
In the meta-analysis of the overall population, the Begg’s and Egger’s tests revealed (Supplementary Table S7) a presence of potential publication bias for the allele model in the Begg’s test (P-Begg’s test = 0.028) or the allele (P-Egger = 0.013), carrier (P-Egger = 0.035), homozygote (P-Egger = 0.013), recessive (P-Egger = 0.013), and dominant (P-Egger = 0.046) models in the Egger’s test. For the analysis targeting the Caucasian population (Supplementary Table S8), we only detected potential publication bias for the homozygote and recessive models (P-Egger < 0.05; P-Egger < 0.05). However, this slight publication bias only existed for the homozygote (P-Egger = 0.049) and recessive (P-Egger = 0.044) models using the Caucasian cases and population-based negative controls in the Egger’s test (Supplementary Table S9). Figures 4A,B and Supplementary Figures S11A,B, S12A,B present the relative publication bias plots according to the Begg’s tests as examples.
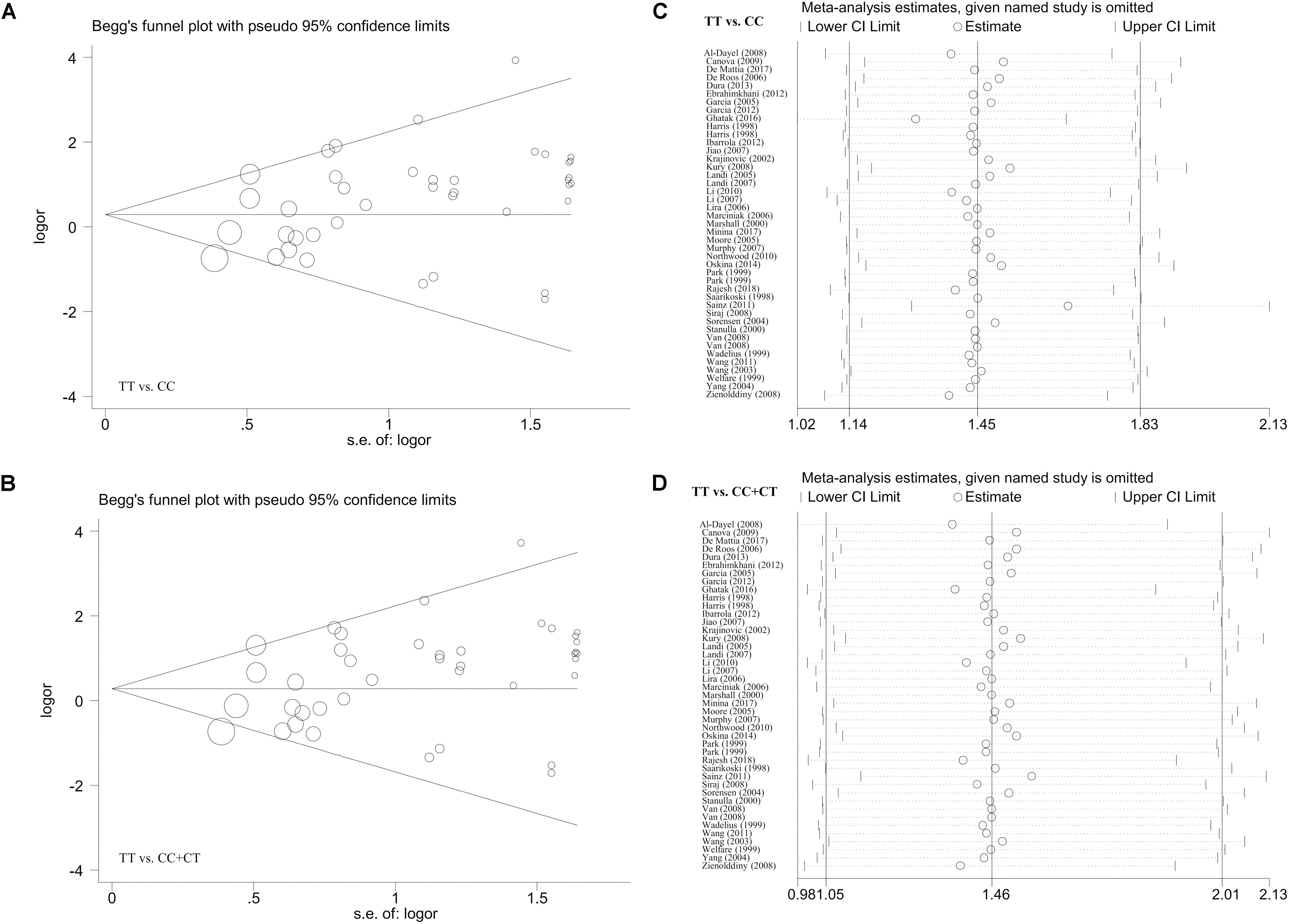
Figure 4. The publication bias plot according to the Begg’s test and the sensitivity analysis data for the overall population under the models of TT vs. CC and TT vs. CC+CT. (A,B) Begg’s test; (C,D) sensitivity analysis.
Additionally, we did not observe any remarkable alteration of the summary OR and corresponding 95% CI value when the individual case-control studies were removed one by one in our sensitivity analysis, confirming the abovementioned stability of the results. Some of the sensitivity analysis data (Figures 4C,D and Supplementary Figures S11C,D, S12C,D) are shown as examples.
The GSTP1 rs1138272 polymorphism may be related to the risk of non-small cell lung cancer in the Norwegian population (Zienolddiny et al., 2008) and lung cancer in the Caucasian population of the United States (Wang et al., 2003). Nevertheless, no association was found between this polymorphism and lung cancer in Denmark (Sorensen et al., 2004) or with lung cancer in individuals in Russia who smoke (Minina et al., 2017). Hence, comprehensive analyses via the meta-analysis approach are meaningful.
In 2013, 28 case-control studies including 26 articles (Harris et al., 1998; Saarikoski et al., 1998; Wadelius et al., 1999; Wang et al., 2003, 2011; Barnette et al., 2004; Sorensen et al., 2004; Yang et al., 2004; Garcia-Closas et al., 2005; Landi et al., 2005, 2007; Moore et al., 2005; Marciniak et al., 2006; Jiao et al., 2007; Kim et al., 2007; Murphy et al., 2007; Al-Dayel et al., 2008; Kury et al., 2008; Siraj et al., 2008; Van Emburgh et al., 2008; Zienolddiny et al., 2008; Canova et al., 2009; Marie-Genica Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk, 2010; Northwood et al., 2010; Ebrahimkhani et al., 2012; Ibarrola-Villava et al., 2012) were recruited into a meta-analysis performed by Huang et al. (2013). The results indicated that the GSTP1 rs1138272 polymorphism appears to be associated with an increased risk of cancer, particularly lung cancer in the Asian population (Huang et al., 2013). In our analysis, we collected the available published articles as thoroughly as possible through a systematic search of five online electronic databases. The included case-control studies that were selected using our strict inclusion and exclusion criteria. We removed one case-control study in which the data were not in HWE (Marie-Genica Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk, 2010), and we removed two additional studies (Barnette et al., 2004) because they failed to meet the requirement of reporting the genotype frequency in both the case and control group. Moreover, 17 new articles (Park et al., 1999; Welfare et al., 1999; Marshall et al., 2000; Stanulla et al., 2000; Krajinovic et al., 2002; De Roos et al., 2006; Lira et al., 2006; Li et al., 2007, 2010; Sainz et al., 2011; Garcia-Gonzalez et al., 2012; Dura et al., 2013; Oskina et al., 2014; Ghatak et al., 2016; De Mattia et al., 2017; Minina et al., 2017; Rajesh et al., 2018) were added. Finally, a total of 40 articles were included in our updated meta-analysis. After the data extraction, 43 case-control studies were enrolled in the meta-analysis under the allele, heterozygote, dominant, and carrier genetic models. All of the studies follow Hardy–Weinberg equilibrium and exhibit high quality. Three studies (Marshall et al., 2000; Lira et al., 2006; Van Emburgh et al., 2008) were excluded in the homozygote and recessive models because the CC genotype frequency in both the case and control group was equal to zero. We detected a potential correlation between the TT genotype of GSTP1 rs1138272 and cancer susceptibility in the Asian population, which is partly in agreement with the previously reported data (Huang et al., 2013). In addition, we found that the GSTP1 rs1138272 polymorphism may be associated with an increased risk of cancer in the African population.
Ye et al. (2006) recruited four case-control studies (Harris et al., 1998; Wang et al., 2003; Sorensen et al., 2004; Yang et al., 2004) to conduct a meta-analysis on the association between GSTP1 rs1138272 and lung cancer risk (Ye et al., 2006). This group did not provide evidence for a strong association between GSTP1 rs1138272 and lung cancer susceptibility (Ye et al., 2006). Yan et al. (2016) included five case-control studies (Harris et al., 1998; Wang et al., 2003; Yang et al., 2004; Zienolddiny et al., 2008; Vural et al., 2012) to perform another relative meta-analysis (Yan et al., 2016) wherein an association between GSTP1 rs1138272 and increased lung cancer risk was detected (Yan et al., 2016). Here, in our subgroup analysis of lung cancer, we removed one study that was not in HWE (Vural et al., 2012) and added two case-control studies (Saarikoski et al., 1998; Minina et al., 2017) for the pooled analysis. Based on the available data within seven articles (Harris et al., 1998; Saarikoski et al., 1998; Wang et al., 2003; Sorensen et al., 2004; Yang et al., 2004; Zienolddiny et al., 2008; Minina et al., 2017), we failed to detect a relationship between GSTP1 rs1138272 and lung cancer risk in the overall population. However, when we enrolled the Caucasian cases and population-based negative controls in six studies (Harris et al., 1998; Saarikoski et al., 1998; Wang et al., 2003; Sorensen et al., 2004; Zienolddiny et al., 2008; Minina et al., 2017) to perform another subgroup analysis by cancer type, we found that the CT genotype of GSTP1 rs1138272 may confer the highest susceptibility to the lung cancer in the Caucasian population.
Previously, three meta-analyses of data on brain tumors were published (Lai et al., 2005; Fan et al., 2013; Geng et al., 2016). Each of these meta-analyses included four case-control studies (Ezer et al., 2002; De Roos et al., 2003; Wrensch et al., 2004; Schwartzbaum et al., 2007). Unfortunately, based on our screening strategy, these studies could not be enrolled in our comprehensive analyses. One of the studies was not in HWE (Ezer et al., 2002), and the others failed to provide the complete genotype frequency of CT and CT within GSTP1 rs1138272 (De Roos et al., 2003; Wrensch et al., 2004; Schwartzbaum et al., 2007).
With regard to colorectal cancer, Li et al. (2013) performed a relevant meta-analysis including seven case-control studies (3,173 cases/3,323 controls) (Welfare et al., 1999; Sachse et al., 2002; Landi et al., 2005; Kury et al., 2008; Sainz et al., 2011; Wang et al., 2011; Ebrahimkhani et al., 2012) in 2013 and reported a negative association between GSTP1 rs1138272 and colorectal cancer risk (Li et al., 2013). Herein, we ruled out one the studies included by Li et al. (2013) because it deviated from Hardy–Weinberg equilibrium (Sachse et al., 2002), and we included three new eligible studies (Harris et al., 1998; Moore et al., 2005; Northwood et al., 2010) to perform an updated analysis. Compared with the “colorectal cancer” subgroup of Huang et al. (2013), two case-control studies (Welfare et al., 1999; Sainz et al., 2011) were added. Despite the additional studies, a similar negative conclusion in the overall population was observed in our updated meta-analysis. However, when targeting the Caucasian population, we found that the TT genotype of GSTP1 rs1138272 may be positively linked to a decreased risk of colorectal cancer in Caucasians. Some environmental factors, such as nutrition and other exposures, may serve as the potential contributory reasons for the observed differences of susceptibility in different populations or cancers.
Although the results of the sensitivity analysis indicated the stability of the data, our study is not without several limitations. The issue of small sample sizes should be considered fully when interpreting certain results. For example, an elevated cancer risk was observed for the “African” subgroup under all of the genetic models; however, only two case-control studies were included for the homozygote and recessive models. Due to the lack of data, we had to consider all cancers together for the Asian and African populations. Despite the positive conclusions obtained, more case-control studies in the Asian and African population are warranted to enable more accurate cancer type-specific subgroup analyses.
Although we observed a potential relationship between GSTP1 rs1138272 and the risk of colorectal, lung, head and neck cancers within Caucasians, no more than 10 case-control studies were enrolled, and more detailed head and neck cancer types were not evaluated due to the lack of sufficient data. Furthermore, the role of GSTP1 rs1138272 in other cancer types has not yet been investigated. Only one case-control study was available for the stratified analysis of bladder, liver, or pancreatic cancer.
In addition, a high degree of inter-study heterogeneity and potential publication bias were observed in certain comparisons. The level of heterogeneity and publication bias was reduced in the analyses of the Caucasian population, suggesting that the “ethnicity” factor is essential for the assessment of the distinct role of GSTP1 rs1138272 in cancer risk.
Considering the role of possible linkage disequilibrium in the genetic susceptibility to different cancers, we tried to extract the data of GSTP1 haplotypes in the enrolled case-control studies. Nevertheless, not enough relevant data supported the performance of pooling analysis. In addition, the GSTP1 rs1138272 polymorphism together with the GSTM1 (glutathione S-transferase M1) null genotype was reported to be associated with the risk of colon or rectal cancer in the Indian population (Wang et al., 2011). The limited availability of useable data also prevented us from exploring the genetic effects of the GSTP1 polymorphism combined with variants of other genes in specific cancer types. The factors, such as the age of onset, sex, lifestyle, environmental exposure, cancer source, linkage disequilibrium, synergistic interaction between genes, etiologies, relapses, and other patient clinical characterizations should be considered carefully when more data is available.
Above all, our pooled analysis consisting of the currently available eligible case-control studies demonstrated that the GSTP1 rs1138272 polymorphism is associated with the susceptibility to overall cancer in the Asian and African populations and, moreover, this polymorphism may be linked to the risk of colorectal, lung or head and neck cancers in the Caucasian population. More eligible case-control studies containing cases with distinct cancers in various ethnic backgrounds are necessary for a more precise and relatively objective estimation.
FD, J-PL, and YZ were conducted by database searching and screening process. FD, G-HQ, and Z-CS was performed by statistical analysis. FD and J-PL wrote the manuscript. Y-HY reviewed the manuscript. All authors approved the final version of the manuscript.
This work was supported by grants from Natural Science Foundation of Shandong Province (Grant No. ZR2015HM041).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01897/full#supplementary-material
FIGURE S1 | Forest plot of the meta-analysis in the overall population (allele T vs. allele C model).
FIGURE S2 | Forest plot of the meta-analysis in the overall population (carrier T vs. carrier C model).
FIGURE S3 | Forest plot of the meta-analysis in the overall population (CT vs. CC model).
FIGURE S4 | Forest plot of the meta-analysis in the overall population (CT+TT vs. CC model).
FIGURE S5 | Forest plot of the subgroup analysis by control source in the overall population (TT vs. CC model).
FIGURE S6 | Forest plot of the subgroup analysis by cancer type in the overall population (the allele T vs. allele C model).
FIGURE S7 | Forest plot of the subgroup analysis by cancer type in the Caucasian population (TT vs. CC model).
FIGURE S8 | Forest plot of the subgroup analysis by cancer type in the Caucasian population (TT vs. CC+CT model).
FIGURE S9 | Forest plot of the subgroup analysis by cancer type in the Caucasian population (CT vs. CC model).
FIGURE S10 | Forest plot of the subgroup analysis by cancer type in the Caucasian population (CT+TT vs. CC model).
FIGURE S11 | The publication bias plot according to the Begg’s test and sensitivity analysis data for the Caucasian population under the models of TT vs. CC and TT vs. CC+CT. (A,B) Begg’s test; (C,D) sensitivity analysis.
FIGURE S12 | The publication bias plot according to the Begg’s test and sensitivity analysis data for the Caucasian population under the models of CT vs. CC and CT+TT vs. CC. (A,B) Begg’s test; (C,D) sensitivity analysis.
TABLE S1 | Database searching terms (up to September 2018).
TABLE S2 | Subgroup analysis by control source in the overall population.
TABLE S3 | Subgroup analysis by cancer type in the overall population.
TABLE S4 | Subgroup analysis by control source in the Caucasian population.
TABLE S5 | Subgroup analysis by cancer type in the Caucasian population.
TABLE S6 | Subgroup analysis by cancer type using the Caucasian cases and population-based negative controls.
TABLE S7 | Publication bias analysis in the overall population.
TABLE S8 | Publication bias analysis in the Caucasian population.
TABLE S9 | Publication bias analysis in the Caucasian cases and population-based negative controls.
Al-Dayel, F., Al-Rasheed, M., Ibrahim, M., Bu, R., Bavi, P., Abubaker, J., et al. (2008). Polymorphisms of drug-metabolizing enzymes CYP1A1, GSTT and GSTP contribute to the development of diffuse large B-cell lymphoma risk in the Saudi Arabian population. Leuk. Lymphoma 49, 122–129. doi: 10.1080/10428190701704605
Barnette, P., Scholl, R., Blandford, M., Ballard, L., Tsodikov, A., Magee, J., et al. (2004). High-throughput detection of glutathione s-transferase polymorphic alleles in a pediatric cancer population. Cancer Epidemiol. Biomarkers Prev. 13, 304–313. doi: 10.1158/1055-9965.EPI-03-0178
Canova, C., Hashibe, M., Simonato, L., Nelis, M., Metspalu, A., Lagiou, P., et al. (2009). Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res. 69, 2956–2965. doi: 10.1158/0008-5472.can-08-2604
De Mattia, E., Cecchin, E., Polesel, J., Bignucolo, A., Roncato, R., Lupo, F., et al. (2017). Genetic biomarkers for hepatocellular cancer risk in a caucasian population. World J. Gastroenterol. 23, 6674–6684. doi: 10.3748/wjg.v23.i36.6674
De Roos, A. J., Gold, L. S., Wang, S., Hartge, P., Cerhan, J. R., Cozen, W., et al. (2006). Metabolic gene variants and risk of non-Hodgkin’s lymphoma. Cancer Epidemiol. Biomarkers Prev. 15, 1647–1653. doi: 10.1158/1055-9965.epi-06-0193
De Roos, A. J., Rothman, N., Inskip, P. D., Linet, M. S., Shapiro, W. R., Selker, R. G., et al. (2003). Genetic polymorphisms in GSTM1, -P1, -T1, and CYP2E1 and the risk of adult brain tumors. Cancer Epidemiol. Biomarkers Prev. 12, 14–22.
Dura, P., Salomon, J., Te Morsche, R. H., Roelofs, H. M., Kristinsson, J. O., Wobbes, T., et al. (2013). No role for glutathione S-transferase genotypes in Caucasian esophageal squamous cell or adenocarcinoma etiology: an European case-control study. BMC Gastroenterol. 13:97. doi: 10.1186/1471-230x-13-97
Ebrahimkhani, S., Asgharian, A. M., Nourinaier, B., Ebrahimkhani, K., Vali, N., Abbasi, F., et al. (2012). Association of GSTM1, GSTT1, GSTP1 and CYP2E1 single nucleotide polymorphisms with colorectal cancer in Iran. Pathol. Oncol. Res. 18, 651–656. doi: 10.1007/s12253-011-9490-8
Ezer, R., Alonso, M., Pereira, E., Kim, M., Allen, J. C., Miller, D. C., et al. (2002). Identification of glutathione S-transferase (GST) polymorphisms in brain tumors and association with susceptibility to pediatric astrocytomas. J. Neurooncol. 59, 123–134. doi: 10.1023/A:1019601305210
Fan, Z., Wu, Y., Shen, J., and Zhan, R. (2013). Glutathione S-transferase M1, T1, and P1 polymorphisms and risk of glioma: a meta-analysis. Mol. Biol. Rep. 40, 1641–1650. doi: 10.1007/s11033-012-2213-8
Garcia-Closas, M., Malats, N., Silverman, D., Dosemeci, M., Kogevinas, M., Hein, D. W., et al. (2005). NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish bladder cancer study and meta-analyses. Lancet 366, 649–659. doi: 10.1016/s0140-6736(05)67137-1
Garcia-Gonzalez, M. A., Quintero, E., Bujanda, L., Nicolas, D., Benito, R., Strunk, M., et al. (2012). Relevance of GSTM1, GSTT1, and GSTP1 gene polymorphisms to gastric cancer susceptibility and phenotype. Mutagenesis 27, 771–777. doi: 10.1093/mutage/ges049
Geng, P., Li, J., Wang, N., Ou, J., Xie, G., Sa, R., et al. (2016). Genetic contribution of polymorphisms in glutathione S-transferases to brain tumor risk. Mol. Neurobiol. 53, 1730–1740. doi: 10.1007/s12035-015-9097-2
Ghatak, S., Yadav, R. P., Lalrohlui, F., Chakraborty, P., Ghosh, S., Ghosh, S., et al. (2016). Xenobiotic pathway gene polymorphisms associated with gastric cancer in high risk mizo-mongoloid population, Northeast India. Helicobacter 21, 523–535. doi: 10.1111/hel.12308
Harris, M. J., Coggan, M., Langton, L., Wilson, S. R., and Board, P. G. (1998). Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics 8, 27–31. doi: 10.1097/00008571-199802000-00004
Huang, S. X., Wu, F. X., Luo, M., Ma, L., Gao, K. F., Li, J., et al. (2013). The glutathione S-transferase P1 341C > T polymorphism and cancer risk: a meta-analysis of 28 case-control studies. PLoS One 8:e56722. doi: 10.1371/journal.pone.0056722
Ibarrola-Villava, M., Martin-Gonzalez, M., Lazaro, P., Pizarro, A., Lluch, A., and Ribas, G. (2012). Role of glutathione S-transferases in melanoma susceptibility: association with GSTP1 rs1695 polymorphism. Br. J. Dermatol. 166, 1176–1183. doi: 10.1111/j.1365-2133.2012.10831.x
Jiao, L., Bondy, M. L., Hassan, M. M., Chang, D. Z., Abbruzzese, J. L., Evans, D. B., et al. (2007). Glutathione S-transferase gene polymorphisms and risk and survival of pancreatic cancer. Cancer 109, 840–848. doi: 10.1002/cncr.22468
Kim, J. Y., Kim, S. T., Jeon, S. H., and Lee, W. Y. (2007). Midbrain transcranial sonography in Korean patients with Parkinson’s disease. Mov. Disord. 22, 1922–1926. doi: 10.1002/mds.21628
Krajinovic, M., Labuda, D., and Sinnett, D. (2002). Glutathione S-transferase P1 genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukaemia. Pharmacogenetics 12, 655–658. doi: 10.1097/00008571-200211000-00010
Kury, S., Buecher, B., Robiou-du-Pont, S., Scoul, C., Colman, H., Le Neel, T., et al. (2008). Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer 8:326. doi: 10.1186/1471-2407-8-326
Lai, R., Crevier, L., and Thabane, L. (2005). Genetic polymorphisms of glutathione S-transferases and the risk of adult brain tumors: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 14, 1784–1790. doi: 10.1158/1055-9965.epi-05-0105
Landi, S., Gemignani, F., Moreno, V., Gioia-Patricola, L., Chabrier, A., Guino, E., et al. (2005). A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet. Genomics 15, 535–546. doi: 10.1097/01.fpc.0000165904.48994.3d
Landi, S., Gemignani, F., Neri, M., Barale, R., Bonassi, S., Bottari, F., et al. (2007). Polymorphisms of glutathione-S-transferase M1 and manganese superoxide dismutase are associated with the risk of malignant pleural mesothelioma. Int. J. Cancer 120, 2739–2743. doi: 10.1002/ijc.22590
Li, D., Dandara, C., and Parker, M. I. (2010). The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet. 11:47. doi: 10.1186/1471-2156-11-47
Li, D., Wang, L. E., Chang, P., El-Naggar, A. K., Sturgis, E. M., and Wei, Q. (2007). In vitro benzo[a]pyrene diol epoxide-induced DNA adducts and risk of squamous cell carcinoma of head and neck. Cancer Res. 67, 5628–5634. doi: 10.1158/0008-5472.CAN-07-0983
Li, F., Xu, B., Yang, Z., Wu, Y., Dong, S., and Qian, J. (2013). GSTP1 Ala114Val polymorphism and colorectal cancer risk: a meta-analysis. Tumour Biol. 34, 1825–1831. doi: 10.1007/s13277-013-0723-1
Lira, M. G., Provezza, L., Malerba, G., Naldi, L., Remuzzi, G., Boschiero, L., et al. (2006). Glutathione S-transferase and CYP1A1 gene polymorphisms and non-melanoma skin cancer risk in Italian transplanted patients. Exp. Dermatol. 15, 958–965. doi: 10.1111/j.1600-0625.2006.00500.x
Marciniak, P., Drobnik, K., Ziemnicka, K., Gut, P., Slomski, R., and Sowinski, J. (2006). [Transferase S-glutathione class pi gene (GSTP1) polymorphism in thyroid cancer patients]. Endokrynol. Pol. 57, 509–515.
Marie-Genica Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. (2010). Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res. Treat. 119, 463–474. doi: 10.1007/s10549-009-0407-0
Marshall, S. E., Bordea, C., Haldar, N. A., Mullighan, C. G., Wojnarowska, F., Morris, P. J., et al. (2000). Glutathione S-transferase polymorphisms and skin cancer after renal transplantation. Kidney Int. 58, 2186–2193. doi: 10.1111/j.1523-1755.2000.00392.x
Minina, V. I., Soboleva, O. A., Glushkov, A. N., Voronina, E. N., Sokolova, E. A., Bakanova, M. L., et al. (2017). Polymorphisms of GSTM1, GSTT1, GSTP1 genes and chromosomal aberrations in lung cancer patients. J. Cancer Res. Clin. Oncol. 143, 2235–2243. doi: 10.1007/s00432-017-2486-3
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Moore, L. E., Huang, W. Y., Chatterjee, N., Gunter, M., Chanock, S., Yeager, M., et al. (2005). GSTM1, GSTT1, and GSTP1 polymorphisms and risk of advanced colorectal adenoma. Cancer Epidemiol. Biomarkers Prev. 14, 1823–1827. doi: 10.1158/1055-9965.epi-05-0037
Murphy, S. J., Hughes, A. E., Patterson, C. C., Anderson, L. A., Watson, R. G., Johnston, B. T., et al. (2007). A population-based association study of SNPs of GSTP1, MnSOD, GPX2 and Barrett’s esophagus and esophageal adenocarcinoma. Carcinogenesis 28, 1323–1328. doi: 10.1093/carcin/bgm007
Northwood, E. L., Elliott, F., Forman, D., Barrett, J. H., Wilkie, M. J., Carey, F. A., et al. (2010). Polymorphisms in xenobiotic metabolizing enzymes and diet influence colorectal adenoma risk. Pharmacogenet. Genomics 20, 315–326. doi: 10.1097/FPC.0b013e3283395c6a
Oskina, N. A., Årmolenko, N. A., Boyarskih, U. A., Lazarev, A., Petrova, V. D., Ganov, D. I., et al. (2014). Associations between SNPs within antioxidant genes and the risk of prostate cancer in the Siberian region of Russia. Pathol. Oncol. Res. 20, 635–640. doi: 10.1007/s12253-014-9742-5
Park, J. Y., Schantz, S. P., Stern, J. C., Kaur, T., and Lazarus, P. (1999). Association between glutathione S-transferase pi genetic polymorphisms and oral cancer risk. Pharmacogenetics 9, 497–504.
Rajesh, D., Balakrishna, S., Azeem Mohiyuddin, S. M., Suresh, T. N., and Moideen Kutty, A. V. (2018). GSTP1 c.341C > T gene polymorphism increases the risk of oral squamous cell carcinoma. Mutat. Res. 831, 45–49. doi: 10.1016/j.mrgentox.2018.04.006
Saarikoski, S. T., Voho, A., Reinikainen, M., Anttila, S., Karjalainen, A., Malaveille, C., et al. (1998). Combined effect of polymorphic GST genes on individual susceptibility to lung cancer. Int. J. Cancer 77, 516–521. doi: 10.1002/(SICI)1097-0215(19980812)77:4<516::AID-IJC7>3.0.CO;2-X
Sachse, C., Smith, G., Wilkie, M. J., Barrett, J. H., Waxman, R., Sullivan, F., et al. (2002). A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis 23, 1839–1849. doi: 10.1093/carcin/23.11.1839
Sainz, J., Rudolph, A., Hein, R., Hoffmeister, M., Buch, S., von Schonfels, W., et al. (2011). Association of genetic polymorphisms in ESR2, HSD17B1, ABCB1, and SHBG genes with colorectal cancer risk. Endocr. Relat. Cancer 18, 265–276. doi: 10.1530/erc-10-0264
Schwartzbaum, J. A., Ahlbom, A., Lonn, S., Warholm, M., Rannug, A., Auvinen, A., et al. (2007). An international case-control study of glutathione transferase and functionally related polymorphisms and risk of primary adult brain tumors. Cancer Epidemiol. Biomarkers Prev. 16, 559–565. doi: 10.1158/1055-9965.epi-06-0918
Sharma, V., Nandan, A., Sharma, A. K., Singh, H., Bharadwaj, M., Sinha, D. N., et al. (2017). Signature of genetic associations in oral cancer. Tumour Biol. 39:1010428317725923. doi: 10.1177/1010428317725923
Singh, S. (2015). Cytoprotective and regulatory functions of glutathione S-transferases in cancer cell proliferation and cell death. Cancer Chemother. Pharmacol. 75, 1–15. doi: 10.1007/s00280-014-2566-x
Siraj, A. K., Ibrahim, M., Al-Rasheed, M., Abubaker, J., Bu, R., Siddiqui, S. U., et al. (2008). Polymorphisms of selected xenobiotic genes contribute to the development of papillary thyroid cancer susceptibility in Middle Eastern population. BMC Med. Genet. 9:61. doi: 10.1186/1471-2350-9-61
Sorensen, M., Autrup, H., Tjonneland, A., Overvad, K., and Raaschou-Nielsen, O. (2004). Glutathione S-transferase T1 null-genotype is associated with an increased risk of lung cancer. Int. J. Cancer 110, 219–224. doi: 10.1002/ijc.20075
Stanulla, M., Schrappe, M., Brechlin, A. M., Zimmermann, M., and Welte, K. (2000). Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a case-control study. Blood 95, 1222–1228.
Tan, X., and Chen, M. (2015). Association between glutathione S-transferases P1 Ile105Val polymorphism and susceptibility to esophageal cancer: evidence from 20 case-control studies. Mol. Biol. Rep. 42, 399–408. doi: 10.1007/s11033-014-3781-6
Van Emburgh, B. O., Hu, J. J., Levine, E. A., Mosley, L. J., Perrier, N. D., Freimanis, R. I., et al. (2008). Polymorphisms in CYP1B1, GSTM1, GSTT1 and GSTP1, and susceptibility to breast cancer. Oncol. Rep. 19, 1311–1321. doi: 10.3892/or.19.5.1311
Vural, B., Yakar, F., Derin, D., Saip, P., Yakar, A., Demirkan, A., et al. (2012). Evaluation of glutathione S-transferase P1 polymorphisms (Ile105Val and Ala114Val) in patients with small cell lung cancer. Genet. Test Mol. Biomarkers 16, 701–706. doi: 10.1089/gtmb.2011.0315
Wadelius, M., Autrup, J. L., Stubbins, M. J., Andersson, S. O., Johansson, J. E., Wadelius, C., et al. (1999). Polymorphisms in NAT2, CYP2D6, CYP2C19 and GSTP1 and their association with prostate cancer. Pharmacogenetics 9, 333–340. doi: 10.1097/00008571-199906000-00008
Wang, J., Jiang, J., Zhao, Y., Gajalakshmi, V., Kuriki, K., Suzuki, S., et al. (2011). Genetic polymorphisms of glutathione S-transferase genes and susceptibility to colorectal cancer: a case-control study in an Indian population. Cancer Epidemiol. 35, 66–72. doi: 10.1016/j.canep.2010.07.003
Wang, M., Li, Y., Lin, L., Song, G., and Deng, T. (2016). GSTM1 null genotype and GSTP1 Ile105Val polymorphism are associated with Alzheimer’s disease: a meta-analysis. Mol. Neurobiol. 53, 1355–1364. doi: 10.1007/s12035-015-9092-7
Wang, Y., Spitz, M. R., Schabath, M. B., Ali-Osman, F., Mata, H., and Wu, X. (2003). Association between glutathione S-transferase p1 polymorphisms and lung cancer risk in Caucasians: a case-control study. Lung Cancer 40, 25–32. doi: 10.1016/S0169-5002(02)00537-8
Welfare, M., Monesola Adeokun, A., Bassendine, M. F., and Daly, A. K. (1999). Polymorphisms in GSTP1, GSTM1, and GSTT1 and susceptibility to colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 8(4 Pt 1), 289–292.
Wrensch, M., Kelsey, K. T., Liu, M., Miike, R., Moghadassi, M., Aldape, K., et al. (2004). Glutathione-S-transferase variants and adult glioma. Cancer Epidemiol. Biomarkers Prev. 13, 461–467.
Xie, P., Liang, Y., Liang, G., and Liu, B. (2014). Association between GSTP1 Ile105Val polymorphism and glioma risk: a systematic review and meta-analysis. Tumour Biol. 35, 493–499. doi: 10.1007/s13277-013-1069-4
Yan, F., Wang, R., and Geng, L. (2016). The 341C/T polymorphism in the GSTP1 gene and lung cancer risk: a meta-analysis. Genet. Mol. Res. 15:gmr.15038172. doi: 10.4238/gmr.15038172
Yang, P., Bamlet, W. R., Ebbert, J. O., Taylor, W. R., and de Andrade, M. (2004). Glutathione pathway genes and lung cancer risk in young and old populations. Carcinogenesis 25, 1935–1944. doi: 10.1093/carcin/bgh203
Ye, Z., Song, H., Higgins, J. P., Pharoah, P., and Danesh, J. (2006). Five glutathione s-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med. 3:e91. doi: 10.1371/journal.pmed.0030091
Yu, Y., Li, X., Liang, C., Tang, J., Qin, Z., Wang, C., et al. (2016). The relationship between GSTA1, GSTM1, GSTP1, and GSTT1 genetic polymorphisms and bladder cancer susceptibility: a meta-analysis. Medicine 95:e4900. doi: 10.1097/md.0000000000004900
Zhao, T., Ma, F., and Yin, F. (2018). Role of polymorphisms of GSTM1, GSTT1 and GSTP1 Ile105Val in childhood acute lymphoblastic leukemia risk: an updated meta-analysis. Minerva Pediatr. 70, 185–196. doi: 10.23736/s0026-4946.17.04657-6
Zhou, C. F., Ma, T., Zhou, D. C., Shen, T., and Zhu, Q. X. (2015). Association of glutathione S-transferase pi (GSTP1) Ile105Val polymorphism with the risk of skin cancer: a meta-analysis. Arch. Dermatol. Res. 307, 505–513. doi: 10.1007/s00403-015-1576-9
Keywords: GSTP1, polymorphism, cancer, meta-analysis, risk
Citation: Ding F, Li J-P, Zhang Y, Qi G-H, Song Z-C and Yu Y-H (2019) Comprehensive Analysis of the Association Between the rs1138272 Polymorphism of the GSTP1 Gene and Cancer Susceptibility. Front. Physiol. 9:1897. doi: 10.3389/fphys.2018.01897
Received: 14 May 2018; Accepted: 18 December 2018;
Published: 25 January 2019.
Edited by:
Brian James Morris, The University of Sydney, AustraliaReviewed by:
George P. Chrousos, National and Kapodistrian University of Athens, GreeceCopyright © 2019 Ding, Li, Zhang, Qi, Song and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Hua Yu, c2R5b25naHVheXVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.