
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Physiol. , 06 November 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01560
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a reliable technique for quantifying expression levels of targeted genes during various biological processes in numerous areas of clinical and biological research. Selection of appropriate reference genes for RT-qPCR normalization is an elementary prerequisite for reliable measurements of gene expression levels. Here, by analyzing datasets published between 2008 and 2017, we summarized the current trends in reference gene selection for insect gene expression studies that employed the most widely used SYBR Green method for RT-qPCR normalization. We curated 90 representative papers, mainly published in 2013–2017, in which a total of 78 insect species were investigated in 100 experiments. Furthermore, top five journals, top 10 frequently used reference genes, and top 10 experimental factors have been determined. The relationships between the numbers of the reference genes, experimental factors, analysis tools on the one hand and publication date (year) on the other hand was investigated by linear regression. We found that the more recently the paper was published, the more experimental factors it tended to explore, and more analysis tools it used. However, linear regression analysis did not reveal a significant correlation between the number of reference genes and the study publication date. Taken together, this meta-analysis will be of great help to researchers that plan gene expression studies in insects, especially the non-model ones, as it provides a summary of appropriate reference genes for expression studies, considers the optimal number of reference genes, and reviews the average number of experimental factors and analysis tools per study.
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a premier molecular biology tool and a powerful method for quantification of gene expression levels in real-time (Vandesompele et al., 2002). Although RT-qPCR is one of the most efficient, reliable, and reproducible techniques to quantify gene expression, multiple factors, including the quality and integrity of RNA samples, efficiency of cDNA synthesis, and PCR efficiency, can significantly influence signal normalization (Bustin et al., 2005; Strube et al., 2008). RT-qPCR generally involves normalization of expression levels of multiple genes to the expression levels of a suite of stable reference genes. Even though reference gene transcript levels should ideally be stable across a range of different conditions, previous studies have shown that expression of many commonly used reference genes differs dramatically under different treatment conditions (Kalushkov and Hodek, 2004; Bustin et al., 2013). It is clear that the expression level of many reference genes is condition-specific and accordingly, there is no universal gene that can be used for internal control for all application scenarios, strongly indicating the necessity of conducting custom reference gene selection for RT-qPCR analyses on a case-by-case basis, even for the same species.
Over the last 10 years, RT-qPCR has been increasingly used in genome/transcriptome expression studies in insect species. Furthermore, considerable advancements have been made for identification and validation of appropriate reference genes across various biotic and abiotic experimental conditions in many insect species (Table 1). In RT-qPCR experiments, SYBR Green and TaqMan probes have been the two most frequently used methodologies, with the SYBR Green method being utilized much more frequently. Here, we have summarized only the studies that used the SYBR Green method. It is well known that characterization of reference genes is an onerous task requiring well-designed molecular experiments followed by elaborate computational analyses (Andersen et al., 2004; Pfaffl et al., 2004). Therefore, a comprehensive summary of published sets of experimentally validated reference genes in conjunction with the description of relevant experimental conditions and analysis tools would be timely (Sang et al., 2017).
In order to fill this gap and provide molecular biologists with informative guidance on selecting the reference genes to customize their RT-qPCR experiments, this present review summarizes the current trends in reference gene selection for RT-qPCR normalization in gene expression studies performed on insects between 2008 and 2017 (Table 1). Specifically, the insect species, reference genes, experimental conditions, analysis tools, and publication year have been summarized. Furthermore, the relationships between the numbers of the reference genes, experimental factors, analysis tools, and publication date (year) were investigated by linear regression. We hoped that our meta-analysis would be of great help for researchers that plan gene expression studies in insects, especially the non-model ones, as it provides a summary of appropriate reference genes for expression studies, considers the optimal number of reference genes, and reviews average numbers of experimental factors and analysis tools per study.
The relevant publications that analyzed reference gene expression in insects in 2008–2017 are summarized in Table 1. All data were extracted from databases such as https://www.ncbi.nlm.nih.gov/pubmed, https://scholar.google.com/, https://link.springer.com/, http://onlinelibrary.wiley.com/, and https://www.sciencedirect.com/ using the following search terms: (“internal control genes” OR “reference genes” OR “housekeeping genes”) AND (“qPCR” OR “quantitative PCR” OR “qRT-PCR” OR “RT-qPCR”) occurring in the Title/Abstract. Additionally, we also curated relevant papers that came to our attention independently but were not uncovered by the above search algorithm. We found and curated 90 representative papers published in 36 journals. The top five journals by the number of published studies on gene expression in insects were PLoS One (26/90), Scientific Reports (9/90), Journal of Economic Entomology (6/90), Journal of Insect Science (5/90), and BMC Research Notes (4/90; Table 1). These papers were mainly published between 2013 and 2017 with an average of 14 papers published over the last 5 years (Figure 1A). We can clearly see that open access journals provide the main platform for publications on this topic.
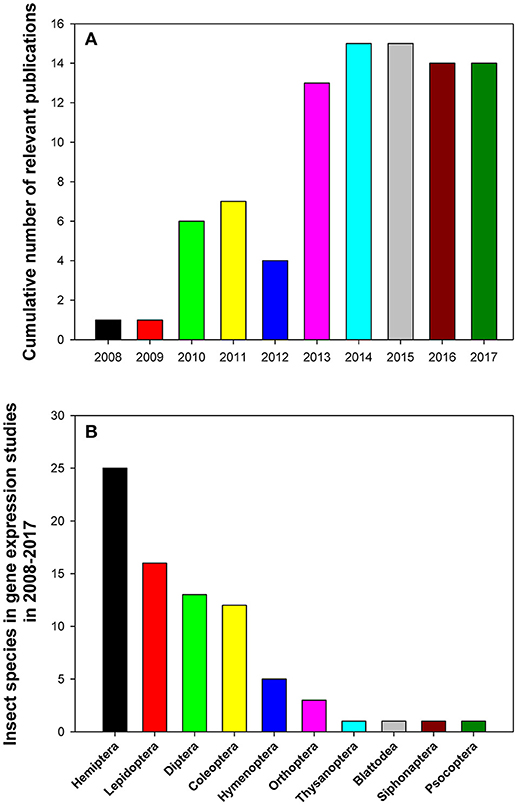
Figure 1. Cumulative numbers of relevant publications (A) and distribution of insect species belonging to different taxonomic orders (B) in relevant gene expression studies performed in 2008–2017 that utilized expression levels of reference genes to normalize RT-qPCR data.
The 90 reviewed papers reported results of gene expression studies in 78 insect species in 100 separate experiments (Table 1). These insects were from 10 insect orders (Figure 1B). They predominantly belonged to the following four insect orders: Hemiptera (25 insect species), Lepidoptera (16 insect species), Coleoptera (12 insect species), and Diptera (13 insect species; Figure 1B). Some insects, such as Bemisia tabaci (Li et al., 2013; Su et al., 2013; Collins et al., 2014; Liang et al., 2014; Dai et al., 2017; Lü et al., 2017) and Helicoverpa armigera (Chandra et al., 2014; Shakeel et al., 2015; Zhang et al., 2015), which cause serious damage to crops, were investigated extensively and frequently. There were six and three papers, respectively, for the above-mentioned species that analyzed expression levels of reference genes and were published during the last 5 years.
In the 90 papers, 3–21 reference genes were investigated per single study (Figure 2). In the majority of studies, the expression level of 5–10 reference genes was determined (Figure 2A). The breakdown of the papers that analyzed expression of multiple reference genes was as follows: five genes (10%), six genes (16%), seven genes (14%), eight genes (15%), nine genes (14%), and ten genes (10%). Recently, in some studies, more than 10 candidate reference genes were analyzed to provide more choices for expression level comparisons and normalization (Table 1). However, linear regression analysis did not reveal a significant correlation between the number of reference genes used in the study and its publication date (year; Figure 2B).
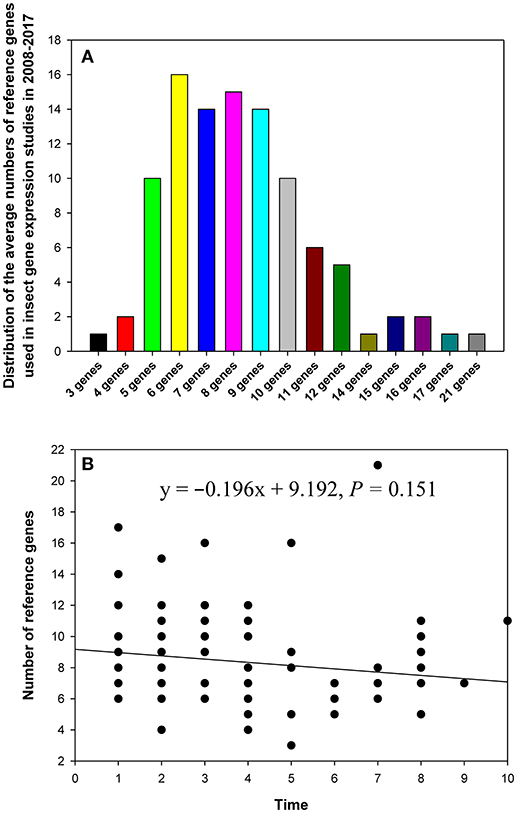
Figure 2. The distribution of the numbers of reference genes per study in relevant publications about gene expression in insects in 2008–2017 (A), and the relationship between the number of reference genes and study publication date (year) fitted by linear regression (B). The numbers 1–10 on the X-axis represent years from 2017 to 2008, respectively.
In the set of curated 90 papers, the expression level of reference genes was determined for 841 times. The number of experiments that utilized top 10 most frequently used reference genes, including Actin, RPL, Tubulin, GAPDH, RPS, 18S, EF1A, TATA, HSP, and SDHA, are shown in Figure 3. Actin, which encodes a major structural protein, is expressed at various levels in many cell types. It is considered the ideal reference gene for RT-qPCR analysis and has been investigated most frequently (Figure 3). For example, previous studies have shown that the expression of Actin was the most stable among other reference genes across different developmental stages of many insects, including Apis mellifera, Schistocerca gregaria, Drosophila melanogaster, Plutella xylostella, Chilo suppressalis, Chortoicetes terminifera, Liriomyza trifolii, and Diuraphis noxia (Scharlaken et al., 2008; Van Hiel et al., 2009; Chapuis et al., 2011; Ponton et al., 2011; Teng et al., 2012; Sinha and Smith, 2014; Chang et al., 2017). Nonetheless, the expression of Actin was less stable in several insects, including those of the species, Coleomegilla maculata, Coccinella septempunctata, and Hippodamia convergens of the family Coccinellidae (Pan et al., 2015b; Yang et al., 2015c, 2016).
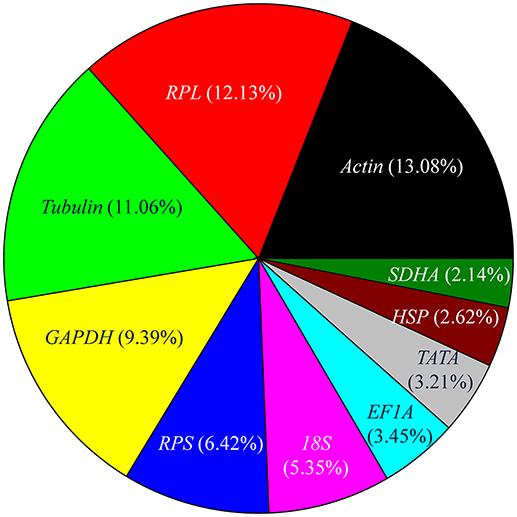
Figure 3. Frequency of the top 10 most popular reference genes in relevant insect gene expression studies performed during 2008–2017. RPL includes RPL3, RPL4, RPL5, RPL7, RPL8, RPL9, RPL10, RPL11, RPL12, RPL13, RPL14, RPL15, RPL17, RPL18, RPL19, RPL22, RPL23, RPL26, RPL27, RPL28, RPL29, RPL32, RPL40, and RPL50; RPS includes RPS2, RPS3, RPS5, RPS6, RPS7, RPS8, RPS9, RPS11, RPS13, RPS15, RPS17, RPS18, RPS20, RPS23, RPS24, RPS26, and RPS27, Tubulin includes α-tubulin, β-tubulin, and γ-tubulin; HSP includes HSP20, HSP22, HSP40, HSP60, HSP70, and HSP90.
Ribosomal protein (RP), a principal component of ribosomes, is among the most highly conserved proteins across all life forms. The fraction of studies in which the expression level of RPL and RPS family genes was used as reference was 18.55%. Together, these genes were the most widely selected reference genes for expression studies in insects during the past 10 years. In most of these studies, RP-encoding genes were stable reference genes. For example, RPS24 and RPS18 were stable reference genes across different developmental stages and sex treatments of C. maculata (Yang et al., 2016); RPS13 and RPS23 were stable reference genes across different developmental stages of P. xylostella (Fu et al., 2013); whereas RPL11, RPS8, and RPL14 were the three most stable reference genes across different developmental stages and under different temperature conditions of Aphis craccivora (Yang et al., 2015b). However, under some conditions, expression levels of RP-encoding genes may be unstable. For example, RPS20 was the least stable gene in P. xylostella strains that were collected in different fields, grown under different temperatures, exposed to different photoperiods, or presented different insecticide susceptibility (Fu et al., 2013).
Tubulin (α-tubulin, β-tubulin, and γ-tubulin), which encodes cytoskeletal structure proteins, was ranked as the third most widely investigated reference gene (Figure 3). In many studies, the stability of Tubulin was variable under different treatments for the same species. For example, a-tubulin exhibits a stable expression in different tissues and sexes of C. maculata, whereas its expression was unstable across different developmental stages and following dsRNA treatments (Yang et al., 2015c).
GAPDH is another commonly used reference gene, ranked as the fourth most widely utilized reference gene (Figure 3). Occasionally, the stability of GAPDH expression was variable under different treatments within the same species. For example, GAPDH expression was not affected by tissue type, sex, photoperiod, or dsRNA treatment in H. convergens, but it varied across different developmental stages and at different temperatures (Pan et al., 2015b). GAPDH was a stable reference gene whose expression was not appreciably altered under different temperatures or by mechanical injury in different strains of P. xylostella; however, its expression was unstable across different developmental stages and was affected by photoperiod (Fu et al., 2013).
18S ribosomal RNA, a part of the ribosomal RNA, was ranked as the sixth most widely investigated reference gene (Figure 3). It was stably expressed throughout the vast majority of biotic and abiotic conditions in most studies that employed its expression level as reference (Table 1). However, it is generally acknowledged that the use of rRNA for normalization of RT-qPCR signals is problematic as rRNA forms a significant proportion of the total RNA pool (>80%), whereas mRNA accounts for a mere 3–5%, so the subtle changes in target gene expression levels may be potentially masked. With this in mind, it is much better to use the mRNA species of the ribosomal machinery, such as RPL and RPS genes, instead of rRNA.
Altogether, the expression level of EF1A, TATA, HSP, and SDHA genes was used as a reference in 11.42% of the experiments. These four genes transiently exhibited variable expression under different treatments in different insect species. For example, EF1A was the least stable reference gene in A. craccivora across different developmental stages and at different temperatures (Yang et al., 2015b). In contrast, EF1A was one of the best reference genes in H. convergens with its expression level being unaffected by three biological factors (developmental stage, tissue type, and sex) and three abiotic conditions (temperature, photoperiod, and dietary RNAi; Pan et al., 2015b).
In the 90 papers, changes in the reference gene expression level were investigated under the influence of one to seven experimental factors. Most of these studies analyzed the influence of one (10%), two (16%), or three (14%) experimental factors (Figure 4A). The relationship between the number of experimental factors and study publication date (year) was investigated by linear regression. We found that the more recently the paper was published, the more experimental factors it tended to explore (Figure 4B).
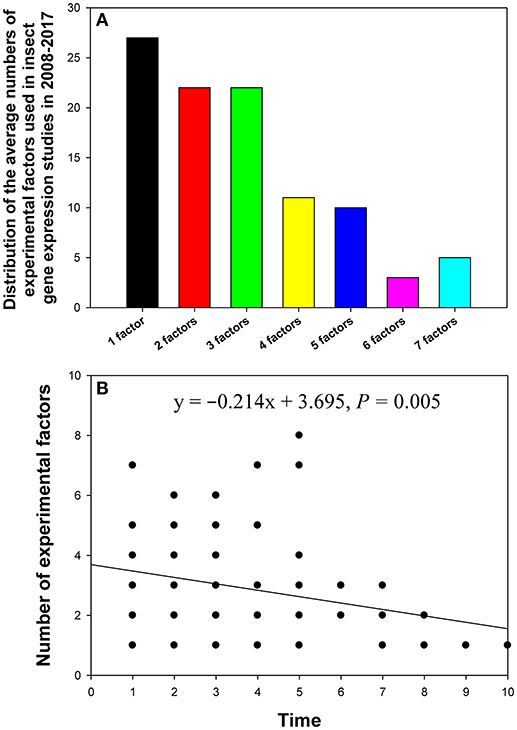
Figure 4. Distribution of the number of experimental factors in relevant insect gene expression studies performed during 2008–2017 (A), and the relationship between the number of experimental factors per study and study publication date (year) investigated by linear regression (B). The numbers 1–10 on the X-axis represent years from 2017 to 2008.
A total of 39 experimental factors were investigated in these 90 papers, with the top 10 experimental factors (in the descending order) being developmental stage, tissue, temperature, insecticide, diet, population, virus, sex, photoperiod, and starvation (Figure 5).
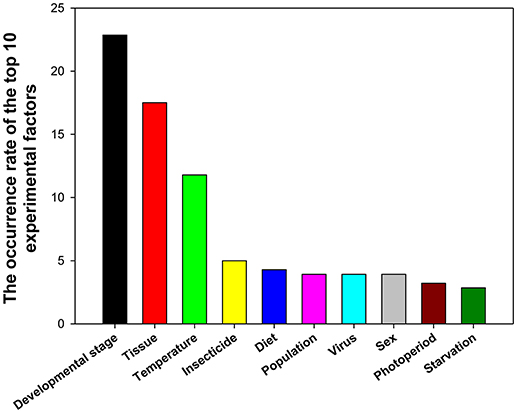
Figure 5. Frequency of top 10 experimental factors in relevant insect gene expression studies performed during 2008–2017.
RNA interference (RNAi) is a conserved mechanism whereby messenger RNA transcripts are targeted by small interfering RNAs in a sequence-specific manner, leading to downregulation of gene expression. During the past 20 years, RNAi has been widely used as a tool to investigate functions of insect genes (Zotti et al., 2018), whereas RT-qPCR is the method of choice to study gene expression in terms of its sensitivity and specificity. The genes that play important roles during insect metamorphosis and affect different tissues can serve as target genes for manipulations that kill the insect or retard its growth. This is why gene expression profiles are widely assessed at different developmental stages and in different tissues. The effect of these two factors on gene expression was investigated frequently with the use of reference gene expression levels in 22.86 and 17.50% of studies, respectively (Figure 5).
Insects are ectothermic organisms, and the body temperature of most insects is affected by changes in ambient temperature, ultimately influencing their growth, and development. Temperature was ranked as the third most widely investigated factor at 11.79% (Figure 5). We found that the numbers/kinds of reference genes under different temperatures varied in different insects. For instance, GAPDH, and EF1A were the best stable gene combinations in Spodoptera litura (Lu et al., 2013), while RPS15, β-tubulin, and EF1A were the most stable reference genes in Nilaparvata lugens (Yuan et al., 2014).
Many insects, including the 78 insect species summarized in this study have developed resistance to insecticides. Insecticide resistance presents as a major challenge for pest control. The molecular mechanisms underlying insecticide resistance are under intense scrutiny; RT-qPCR is an important technology for investigating the gene functions involved in insecticide resistance. Insecticides ranked as the fourth most widely investigated factor at 5.00% (Figure 5). We found that different reference genes were used in different insects to study the effect of various insecticide treatments. RPS15 and RPL32 were stably expressed reference genes in insecticide treatment experiments in H. armigera (Zhang et al., 2015); while RPS11, EF1A, and β-tubulin were the best choice in the insecticide-stressed N. lugens (Yuan et al., 2014). Different classes of insecticides have warranted different sets of reference genes to normalize target gene expression in B. tabaci (Liang et al., 2014).
Diet was ranked as the fifth most widely investigated factor at 4.29% (Figure 5). Different gene combinations were required for different diet conditions. For examples, RPL10 and GAPDH were the most stable reference genes in S. litura that were reared on different diets (Lu et al., 2013); whereas, Actin, RPS18, and RPS15 were the most stable reference genes among different diets in Bradysia odoriphaga (Shi et al., 2016), Actin and 18S were the best reference gene combination for feeding assay experiments with Aphis gossypii (Ma et al., 2016).
Population, virus, and sex were all ranked as the sixth most widely investigated factor at 3.93%(Figure 5). Different reference gene combinations were suggested for the studies of each factor. For example, RPL10 and EF1A were the most stable reference genes in S. litura collected from different locations (Lu et al., 2013), EF1A, Actin, and GAPDH were the more stable reference genes in P. xylostella (Fu et al., 2013). The combination of Actin and EF1A was very useful for experiments involving A. gossypii (Ma et al., 2016). In addition, in viral infection experiments, different reference gene combinations were recommended for different insects. For example, GAPDH, RPL27, and β-tubulin was the best reference gene combination for nuclear polyhedrosis virus infection (Zhang et al., 2015), HSP90 and RPL29 were the most stable reference genes in B. tabaci when the whitefly carried the tomato yellow leaf curl virus and when it did not (Li et al., 2013). Moreover, in females and males, different reference gene combinations were recommended for different insects. For instance, GAPDH and CypA were most stable reference genes for H. convergens (Pan et al., 2015b), HSP90 and RP49 were the most stable ones for Harmonia axyridis (Yang et al., 2018), and 18S, EF1A, and GAPDH were the best for gene expression normalization in Sesamia inferens (Sun et al., 2015).
Photoperiod and starvation ranked as the seventh and eighth most widely investigated factors at 3.21 and 2.86%, respectively (Figure 5). Different reference gene combinations were recommended for different insects for these two factors. For instance, under photoperiod stressed conditions, GAPDH and CypA were most stable reference genes in for H. convergens (Pan et al., 2015b), EF1A and V-ATPase A were the most stable ones for Danaus plexippus (Pan et al., 2015a), and HSP90 and β-tubulin were the best reference genes for H. armigera (Shakeel et al., 2015). Under starvation conditions, RPL28 and RPS15 were the most stable reference genes for H. armigera (Shakeel et al., 2015), RPS3 and Actin were the best reference genes for S. litura (Lu et al., 2013), and RPS11, ArgK, and EF1A were recommended for N. lugens (Yuan et al., 2014).
In the 90 papers, one to five analysis tools were used to evaluate gene expression stability, with one tool (4%) and three tools (34%) being the least and most frequently used variants in these studies, respectively (Figure 6A). Linear regression analysis showed that the more recently the paper was published, the more analysis tools it used (Figure 6B).
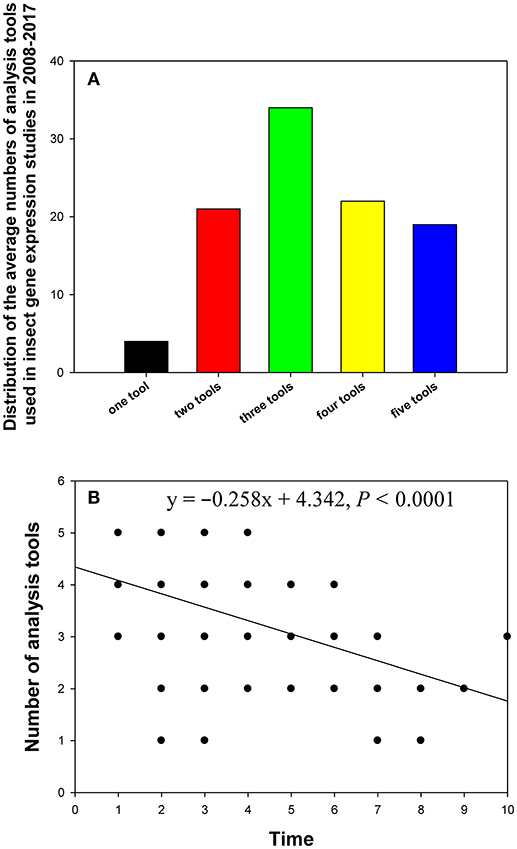
Figure 6. Distribution of the numbers of analysis tools in relevant insect gene expression studies performed during 2008–2017 (A), and the relationship between the number of analysis tools per study and study publication date (year) investigated by linear regression (B). The numbers 1–10 on the X-axis represent years from 2017 to 2008.
Our review clearly suggests that no reference gene is universally stably expressed because variable expression levels even for the most popular reference genes have been observed under different circumstances in the same insect species or under the same experimental condition among different insects. In order to obtain reliable experimental data for the target gene, it is necessary to perform internal reference gene screening under specific experimental conditions. Given that the best internal reference genes in different species under different conditions often have large differences in expression, it may result in a multi-fold difference of target gene expression, or even false conclusion, if used improperly. For instance, the expression of V-ATPase A in the gut ranged from 7.7- to 22.4-fold higher than that in the carcass of C. septempunctata when normalized to the most- and least-stable sets of reference genes, respectively (Yang et al., 2016). Furthermore, the relative hsp83 expression was noticeably variable when a less stable reference gene was used for RT-qPCR normalization in different tissues and developmental stages of S. inferens, whereas hsp83 was uniformly expressed when stable reference genes were used for normalization (Sun et al., 2015). Therefore, better accuracy in gene expression analysis can promote the investigation of gene function. We strongly recommend that prior to each RT-qPCR experiment, the reference gene expression stability must be validated. Furthermore, multiple reference genes should be used to achieve the best results. This review should help researchers select the best reference genes and optimize their experiments to examine gene expression levels in insects, especially the non-model ones, in terms of the number of reference genes chosen, experimental factors manipulated, and the analysis tools used.
HP and YZ conceived the topic of the review. HP, CY, and JL performed literature review analyzed the data. HP and CY wrote the manuscript.
This work was supported by the National Key R&D Program of China (grant No. 2017YFD0200900), project supported by GDUPS (2017), a start-up fund from the South China Agricultural University. The granting agencies have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
An, X. K., Hou, M. L., and Liu, Y. D. (2016). Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 109, 879–886. doi: 10.1093/jee/tov333
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Arun, A., Baumlé, V., Amelot, G., and Nieberding, C. M. (2015). Selection and validation of reference genes for qRT-PCR expression analysis of candidate genes involved in olfactory communication in the butterfly Bicyclus anynana. PLoS ONE 10:e0120401. doi: 10.1371/journal.pone.0120401
Arya, S. K., Jain, G., Upadhyay, S. K., Singh, H., Dixit, S., and Verma, P. C. (2017). Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 7:13520. doi: 10.1038/s41598-017-13925-9
Bagnall, N. H., and Kotze, A. C. (2010). Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med. Vet. Entomol. 24, 176–181. doi: 10.1111/j.1365-2915.2010.00866.x
Bansal, R., Mamidala, P., Mian, M. A., Mittapalli, O., and Michel, A. P. (2012). Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 105, 1432–1438. doi: 10.1603/EC12095
Bansal, R., Mittapelly, P., Chen, Y., Mamidala, P., Zhao, C., and Michel, A. (2016). Quantitative RT-PCR gene evaluation and RNA interference in the brown marmorated stink bug. PLoS ONE 11:e0152730. doi: 10.1371/journal.pone.0152730
Bassan, M. M., Angelotti-Mendonça, J., Alves, G. R., Yamamoto, P. T., and Mourão Filho, F. D. A. A. (2017). Selection of reference genes for expression studies in Diaphorina citri (Hemiptera: Liviidae). J. Econ. Entomol. 110, 2623–2629. doi: 10.1093/jee/tox253
Baumann, A., Lehmann, R., Beckert, A., Vilcinskas, A., and Franta, Z. (2015). Selection and evaluation of tissue specific reference genes in Lucilia sericata during an immune challenge. PLoS ONE 10:e0135093. doi: 10.1371/journal.pone.0135093
Bustin, S. A., Benes, V., Garson, J., Hellemans, J., Huggett, J., Kubista, M., et al. (2013). The need for transparency and good practices in the qPCR literature. Nat. Methods 10, 1063–1067. doi: 10.1038/nmeth.2697
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755
Cameron, R. C., Duncan, E. J., and Dearden, P. K. (2013). Stable reference genes for the measurement of transcript abundance during larval caste development in the honeybee. Apidologie 44, 357–366. doi: 10.1007/s13592-012-0187-0
Chandra, G. S., Asokan, R., Manamohan, M., Kumar, N. K., and Sita, T. (2014). Evaluation of reference genes for quantitative real-time PCR normalization in cotton bollworm, Helicoverna armigera. Mol. Biol. 48, 813–822. doi: 10.1134/S0026893314060156
Chang, Y. W., Chen, J. Y., Lu, M. X., Gao, Y., Tian, Z. H., Gong, W. R., et al. (2017). Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS ONE 12:e0181862. doi: 10.1371/journal.pone.0181862
Chapuis, M. P., Tohidiesfahani, D., Dodgson, T., Blondin, L., Ponton, F., Cullen, D., et al. (2011). Assessment and validation of a suite of reverse transcription-quantitative PCR reference genes for analyses of density-dependent behavioural plasticity in the Australian plague locust. BMC Mol. Biol. 12, 1–11. doi: 10.1186/1471-2199-12-7
Cheng, D., Zhang, Z., He, X., and Liang, G. (2013). Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS ONE 8:e57718. doi: 10.1371/journal.pone.0057718
Collins, C., Patel, M. V., Colvin, J., Bailey, D., Seal, S., and Wolfner, M. (2014). Identification and evaluation of suitable reference genes for gene expression studies in the whitefly Bemisia tabaci (Asia I) by reverse transcription quantitative real time PCR. J. Insect Sci. 14:63. doi: 10.1673/031.014.63
Cristiano, G., Grossi, G., Scala, A., Fanti, P., Zhou, J. J., Bufo, S. A., et al. (2016). Validation of reference genes for qRT-PCR analysis in Megoura viciae (Hemiptera Aphididae). B. Insectol. 69, 229–238.
Dai, T. M., Lü, Z. C., Liu, W. X., and Wan, F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS ONE 12:e0173821. doi: 10.1371/journal.pone.0173821
Dzaki, N., Ramli, K. N., Azlan, A., Ishak, I. H., and Azzam, G. (2017). Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci. Rep. 7:43618. doi: 10.1038/srep43618
Fu, W., Xie, W., Zhang, Z., Wang, S., Wu, Q., Liu, Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9:792. doi: 10.7150/ijbs.5862
Galetto, L., Bosco, D., and Marzach,ì, C. (2013). Selection of reference genes from two leafhopper species challenged by phytoplasma infection, for gene expression studies by RT-qPCR. BMC Res. Notes 6:409. doi: 10.1186/1756-0500-6-409
Gao, X. K., Zhang, S., Luo, J. Y., Wang, C. Y., Lü, L. M., Zhang, L. J., et al. (2017). Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene 637, 211–218 doi: 10.1016/j.gene.2017.09.057
Guo, H., Jiang, L., and Xia, Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genomics 291:999. doi: 10.1007/s00438-015-1125-4
Hornáková, D., Matousková, P., Kindl, J., Valterová, I., and Pichová, I. (2010). Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397, 118–120. doi: 10.1016/j.ab.2009.09.019
Ibanez, F., and Tamborindeguy, C. (2016). Selection of reference genes for expression analysis in the potato psyllid, Bactericera cockerelli. Insect Mol. Biol. 25, 227–238. doi: 10.1111/imb.12219
Jiang, H. B., Liu, Y. H., Tang, P. A., Zhou, A. W., and Wang, J. J. (2010). Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostsrychophila (Psocoptera: Liposcelididae). Mol. Biol. Rep. 37:1019. doi: 10.1007/s11033-009-9803-0
Kalushkov, P., and Hodek, I. (2004). The effects of thirteen species of aphids on some life history parameters of the ladybird Coccinella septempunctata. Biol. Control 49, 21–32. doi: 10.1023/B:BICO.0000009385.90333.b4
Kang, Z. W., Liu, F. H., Tian, H. G., Zhang, M., Guo, S. S., and Liu, T. X. (2017). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Koramutla, M. K., Aminedi, R., and Bhattacharya, R. (2016). Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 6:25883. doi: 10.1038/srep25883
Li, R., Xie, W., Wang, S., Wu, Q., Yang, N., Yang, X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 8:e53006. doi: 10.1371/journal.pone.0053006
Liang, P., Guo, Y., Zhou, X., and Gao, X. (2014). Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLoS ONE 9:e87514. doi: 10.1371/journal.pone.0087514
Ling, D., and Salvaterra, P. M. (2011). Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS ONE 6:e17762. doi: 10.1371/journal.pone.0017762
Liu, G., Qiu, X., Cao, L., Zhang, Y., Zhan, Z., and Han, R. (2016). Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth, Thitarodes armoricanus (Lepidoptera, Hepialidae). PLoS ONE 11:e0159060. doi: 10.1371/journal.pone.0159060
Lord, J. C., Hartzer, K., Toutges, M., and Oppert, B. (2010). Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Meth. 80, 219–221. doi: 10.1016/j.mimet.2009.12.007
Lu, Y., Yuan, M., Gao, X., Kang, T., Zhan, S., Wan, H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS ONE 8:e68059. doi: 10.1371/journal.pone.0068059
Lü, Z. C., Wang, L. H., Dai, R. L., Zhang, G. F., Guo, J. Y., and Wan, F. H. (2014). Evaluation of endogenous reference genes of Bactrocera (tetradacus) minax by gene expression profiling under various experimental conditions. Fla. Entomol. 97, 597–604. doi: 10.1653/024.097.0235
Lü, Z. H., Pan, H. P., Zhang, W., Ding, T. B., and Chu, D. (2017). Reference gene selection for RT-qPCR analysis in two invasive whiteflies after the acquisition of vectored or non-vectored viruses. J. Asia-Pac. Entomol. 21, 19–24. doi: 10.1016/j.aspen.2017.10.001
Ma, K. S., Li, F., Liang, P. Z., Chen, X. W., Liu, Y., and Gao, X. W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003
Majerowicz, D., Alves-Bezerra, M., Logullo, R., Fonseca-de-Souza, A. L., Meyer-Fernandes, J. R., Braz, G. R. C., et al. (2011). Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus, (Hemiptera: Reduviidae). Insect Mol. Biol. 20, 713–722. doi: 10.1111/j.1365-2583.2011.01101.x
Mamidala, P., Rajarapu, S. P., Jones, S. C., and Mittapalli, O. (2011). Identification and validation of reference genes for quantitative real-time polymerase chain reaction in Cimex lectularius. J. Med. Entomol. 48, 947–951. doi: 10.1603/ME10262
Marchal, E., Hult, E. F., Huang, J., and Tobe, S. S. (2013). Sequencing and validation of housekeeping genes for quantitative real-time PCR during the gonadotrophic cycle of Diploptera punctata. BMC Res. Notes 6:237. doi: 10.1186/1756-0500-6-237
Maroniche, G. A., Sagadín, M., Mongelli, V. C., Truol, G. A., and Del, V. M. (2011). Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. Virol. J. 8:308. doi: 10.1186/1743-422X-8-308
Matta, B. P., Bitner-Mathé, B. C., and Alves-Ferreira, M. (2011). Getting real with real-time qPCR: a case study of reference gene selection for morphological variation in Drosophila melanogaster, wings. Dev. Genes Evol. 221, 49–57. doi: 10.1007/s00427-011-0356-6
Mcintosh, C. H., Baird, J., Zinser, E., Woods, D. J., Campbell, E. M., and Bowman, A. S. (2016). Reference gene selection and RNA preservation protocol in the cat flea, Ctenocephalides felis, for gene expression studies. Parasitology 143, 1532–1542. doi: 10.1017/S0031182016001025
Nakamura, A. M., Chahadehlers, S., Lima, A. L., Taniguti, C. H., Sobrinho, I. J. Jr., Torres, F. R., et al. (2016). Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. 6:17480. doi: 10.1038/srep17480
Niu, J., Cappelle, K., de Miranda, J. R., Smagghe, G., and Meeus, I. (2014). Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. J. Invertebr. Pathol. 115:76. doi: 10.1016/j.jip.2013.10.011
Omondi, B. A., Latorre-Estivalis, J. M., Oliveira, I. H., Ignell, R., and Lorenzo, M. G. (2015). Evaluation of reference genes for insect olfaction studies. Parasite. Vector. 8:243. doi: 10.1186/s13071-015-0862-x
Paim, R. M., Pereira, M. H., Di Ponzio, R., Rodrigues, J. O., Guarneri, A. A., Gontijo, N. F., et al. (2012). Validation of reference genes for expression analysis in the salivary gland and the intestine of Rhodnius prolixus (Hemiptera, Reduviidae) under different experimental conditions by quantitative real-time PCR. BMC Res. Notes 5:128. doi: 10.1186/1756-0500-5-128
Pan, H., Yang, X., Bidne, K., Hellmich, R. L., Siegfried, B. D., and Zhou, X. (2015a). Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio-indicator. PLoS ONE 10:e0129482. doi: 10.1371/journal.pone.0129482
Pan, H., Yang, X., Siegfried, B. D., and Zhou, X. (2015b). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS ONE 10:e0125868. doi: 10.1371/journal.pone.0125868
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Piron Prunier, F., Chouteau, M., Whibley, A., Joron, M., and Llaurens, V. (2016). Selection of valid reference genes for reverse transcription quantitative PCR analysis in Heliconius numata (Lepidoptera: Nymphalidae). J. Insect Sci. 16:50. doi: 10.1093/jisesa/iew034
Ponton, F., Chapuis, M. P., Pernice, M., Sword, G. A., and Simpson, S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850. doi: 10.1016/j.jinsphys.2011.03.014
Rajarapu, S. P., Mamidala, P., and Mittapalli, O. (2012). Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis). Insect Sci. 19, 41–46. doi: 10.1111/j.1744-7917.2011.01447.x
Reim, T., Thamm, M., Rolke, D., Blenau, W., and Scheiner, R. (2013). Suitability of three common reference genes for quantitative real-time PCR in honey bees. Apidologie 44, 342–350. doi: 10.1007/s13592-012-0184-3
Ridgeway, J. A., and Timm, A. E. (2015). Reference gene selection for quantitative real-time PCR normalization in larvae of three species of Grapholitini (Lepidoptera: Tortricidae). PLoS ONE 10:e0129026. doi: 10.1371/journal.pone.0129026
Rodrigues, T. B., Dhandapani, R. K., Duan, J. J., and Palli, S. R. (2017). RNA interference in the Asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 7:8913. doi: 10.1038/s41598-017-08813-1
Rodrigues, T. B., Khajuria, C., Wang, H., Matz, N., Cunha, C. D., Valicente, F. H., et al. (2013). Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE 9:e109825. doi: 10.1371/journal.pone.0109825
Sagri, E., Koskinioti, P., Gregoriou, M. E., Tsoumani, K. T., Bassiakos, Y. C., and Mathiopoulos, K. D. (2017). Housekeeping in Tephritid insects: the best gene choice for expression analyses in the medfly and the olive fly. Sci. Rep. 7:45634. doi: 10.1038/srep45634
Sang, J., Wang, Z., Li, M., Cao, J., Niu, G., Xia, L., et al. (2017). ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 46, D121–D126. doi: 10.1093/nar/gkx875
Sang, W., He, L., Wang, X. P., Zhusalzman, K., and Lei, C. L. (2015). Evaluation of reference genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) under UVB stress. Environ. Entomol. 44:418. doi: 10.1093/ee/nvv010
Scharlaken, B., Graaf, D. C. D., Goossens, K., Brunain, M., Peelman, L. J., and Jacobs, F. J. (2008). Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 8, 1–10. doi: 10.1673/031.008.3301
Shakeel, M., Zhu, X., Kang, T., Wan, H., and Li, J. (2015). Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera, (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 18, 123–130. doi: 10.1016/j.aspen.2015.01.001
Shang, F., Wei, D. D., Jiang, X. Z., Wei, D., Shen, G. M., Feng, Y. C., et al. (2015). Reference gene validation for quantitative PCR under various biotic and abiotic stress conditions in Toxoptera citricida (Hemiptera, Aphidiae). J. Econ. Entomol. 108, 2040–2047. doi: 10.1093/jee/tov184
Shen, G. M., Huang, Y., Jiang, X. Z., Dou, W., and Wang, J. J. (2013). Effect of β-cypermethrin exposure on the stability of nine housekeeping genes in Bactrocera dorsalis (Diptera: Tephritidae). Fla. Entomol. 96, 442–450. doi: 10.1653/024.096.0208
Shen, G. M., Jiang, H. B., Wang, X. N., and Wang, J. J. (2010). Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Mol. Biol. 11:76. doi: 10.1186/1471-2199-11-76
Shi, C., Yang, F., Zhu, X., Du, E., Yang, Y., Wang, S., et al. (2016). Evaluation of housekeeping genes for quantitative real-time PCR analysis of Bradysia odoriphaga (Diptera: Sciaridae). Int. J. Mol. Sci. 17:1034. doi: 10.3390/ijms17071034
Shi, X. Q., Guo, W. C., Wan, P. J., Zhou, L. T., Ren, X. L., Ahmat, T., et al. (2013). Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (say). BMC Res. Notes 6:93. doi: 10.1186/1756-0500-6-93
Sinha, D. K., and Smith, C. M. (2014). Selection of reference genes for expression analysis in Diuraphis noxia (Hemiptera: Aphididae) fed on resistant and susceptible wheat plants. Sci. Rep. 4:5059. doi: 10.1038/srep05059
Strube, C., Buschbaum, S., Wolken, S., and Schnieder, T. (2008). Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus. Gene 425, 36–43. doi: 10.1016/j.gene.2008.08.001
Su, Y. L., He, W. B., Wang, J., Li, J. M., Liu, S. S., and Wang, X. W. (2013). Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 106, 1446–1455. doi: 10.1603/EC12459
Sun, M., Lu, M. X., Tang, X. T., and Du, Y. Z. (2015). Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS ONE 10:e0115979. doi: 10.1371/journal.pone.0115979
Tan, Q. Q., Zhu, L., Li, Y., Liu, W., Ma, W. H., Lei, C. L., et al. (2015). A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS ONE 10:e0118693. doi: 10.1371/journal.pone.0118693
Tan, Y., Zhou, X. R., and Pang, B. P. (2017). Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). B. Entomol. Res. 107, 359–368. doi: 10.1017/S0007485316000948
Tang, P. A., Duan, J. Y., Wu, H. J., Ju, X. R., and Yuan, M. L. (2017). Reference gene selection to determine differences in mitochondrial gene expressions in phosphine-susceptible and phosphine-resistant strains of Cryptolestes ferrugineus, using qRT-PCR. Sci. Rep. 7:7047. doi: 10.1038/s41598-017-07430-2
Teng, X., Zhang, Z., He, G., Yang, L., and Li, F. (2012). Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J. Insect Sci. 12, 1–17. doi: 10.1673/031.012.6001
Toutges, M. J., Hartzer, K., Lord, J., and Oppert, B. (2010). Evaluation of reference genes for quantitative polymerase chain reaction across life cycle stages and tissue types of Tribolium castaneum. J. Agr. Food Chem. 58:8948. doi: 10.1021/jf101603j
Van Hiel, M. B., Temmerman, L., Van, S. S., Vuerinckx, K., Huybrechts, R., et al. (2009). Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 10:56. doi: 10.1186/1471-2199-10-56
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034-1. doi: 10.1186/gb-2002-3-7-research0034
Wang, W. X., Lai, F. X., Li, K. L., and Fu, Q. (2014). Selection of reference genes for gene expression analysis in Nilaparvata lugens with different levels of virulence on rice by quantitative real-time PCR. Rice Sci. 21, 305–311. doi: 10.1016/S1672-6308(14)60272-9
Wang, X. Y., Xiong, M., Wang, J. L., Lei, C. L., and Zhu, F. (2015). Reference gene stability of a synanthropic fly, Chrysomya megacephala. Parasit. Vectors 8:565. doi: 10.1186/s13071-015-1175-9
Wang, Y., Wang, Z. K., Huang, Y., Liao, Y. F., and Yin, Y. P. (2014). Identification of suitable reference genes for gene expression studies by qRT-PCR in the blister beetle Mylabris cichorii. J. Insect Sci. 14:94. doi: 10.1093/jis/14.1.94
Wu, K., Liu, W., Mar, T., Liu, Y., Wu, Y., and Wang, X. (2014). Sequencing and validation of reference genes to analyze endogenous gene expression and quantify yellow dwarf viruses using RT-qPCR in viruliferous Rhopalosiphum padi. PLoS ONE 9:e97038. doi: 10.1371/journal.pone.0097038
Xu, J., Lu, M. X., Cui, Y. D., and Du, Y. Z. (2017). Selection and evaluation of reference genes for expression analysis using qRT-PCR in Chilo suppressalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 110, 683–691. doi: 10.1093/jee/tow297
Yang, C., Li, H., Pan, H., Ma, Y., Zhang, D., Liu, Y., et al. (2015a). Stable reference gene selection for RT-qPCR analysis in nonviruliferous and viruliferous Frankliniella occidentalis. PLoS ONE 10:e0135207. doi: 10.1371/journal.pone.0135207
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2014). Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS ONE 9:e110454. doi: 10.1371/journal.pone.0110454
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2015b). Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS ONE 10:e0130593. doi: 10.1371/journal.pone.0130593
Yang, C., Pan, H., Noland, J. E., Zhang, D., Zhang, Z., Liu, Y., et al. (2015c). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5:18201. doi: 10.1038/srep18201
Yang, C., Preisser, E. L., Zhang, H., Liu, Y., Dai, L., Pan, H., et al. (2016). Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 7:e53006. doi: 10.3389/fpls.2016.01672
Yang, Q., Li, Z., Cao, J., Zhang, S., Zhang, H., Wu, X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust, Locusta migratoria, (Orthoptera: Acrididae). PLoS ONE 9:e98164. doi: 10.1371/journal.pone.0098164
Yang, X., Pan, H., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. doi: 10.1038/s41598-018-20612-w
Yu, S. H., Pu, Y., Sun, T., Qi, Q., Wang, X. Q., Xu, D. L., et al. (2016). Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. Springerplus 5, 1–8. doi: 10.1186/s40064-016-2548-z
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 9:e86503. doi: 10.1371/journal.pone.0086503
Zhai, Y., Lin, Q., Zhou, X., Zhang, X., Liu, T., and Yu, Y. (2014). Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae). PLoS ONE 9:e106800. doi: 10.1371/journal.pone.0106800
Zhang, L., Zhang, Q. L., Wang, X. T., Yang, X. Z., Li, X. P., and Yuan, M. L. (2017). Selection of reference genes for qRT-PCR and expression analysis of high-altitude-related genes in grassland caterpillars (Lepidoptera: Erebidae: Gynaephora) along an altitude gradient. Ecol. Evol. 7, 9054–9065. doi: 10.1002/ece3.3431
Zhang, S., An, S., Li, Z., Wu, F., Yang, Q., Liu, Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402. doi: 10.1016/j.gene.2014.11.038
Zheng, Y. T., Li, H. B., Lu, M. X., and Du, Y. Z. (2014). Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLoS ONE 9:e111369. doi: 10.1371/journal.pone.0111369
Zhong, M., Wang, X., Wen, J., Cai, J., Wu, C., and Aly, S. M. (2013). Selection of reference genes for quantitative gene expression studies in the house fly (Musca domestica L.) using reverse transcription quantitative real-time PCR. Acta Bioch. Bioph. Sin. 45:1069. doi: 10.1093/abbs/gmt111
Zhu, X., Yuan, M., Shakeel, M., Zhang, Y., Wang, S., Wang, X., et al. (2014). Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS ONE 9:e84730. doi: 10.1371/journal.pone.0084730
Keywords: RT-qPCR, reference genes, SYBR green method, experimental factors, analysis tools
Citation: Lü J, Yang C, Zhang Y and Pan H (2018) Selection of Reference Genes for the Normalization of RT-qPCR Data in Gene Expression Studies in Insects: A Systematic Review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Received: 20 August 2018; Accepted: 18 October 2018;
Published: 06 November 2018.
Edited by:
Fernando Ariel Genta, Fundação Oswaldo Cruz (Fiocruz), BrazilReviewed by:
Mauro Mandrioli, Università degli Studi di Modena e Reggio Emilia, ItalyCopyright © 2018 Lü, Yang, Zhang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huipeng Pan, cGFuaHVpcGVuZ0BzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.