
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Physiol., 21 November 2018
Sec. Membrane Physiology and Membrane Biophysics
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01406
This article is part of the Research TopicThe evolution of ion channelsView all 5 articles
The appearance of voltage-gated, sodium-selective channels with rapid gating kinetics was a limiting factor in the evolution of nervous systems. Two rounds of domain duplications generated a common 24 transmembrane segment (4 × 6 TM) template that is shared amongst voltage-gated sodium (Nav1 and Nav2) and calcium channels (Cav1, Cav2, and Cav3) and leak channel (NALCN) plus homologs from yeast, different single-cell protists (heterokont and unikont) and algae (green and brown). A shared architecture in 4 × 6 TM channels include an asymmetrical arrangement of extended extracellular L5/L6 turrets containing a 4-0-2-2 pattern of cysteines, glycosylated residues, a universally short III-IV cytoplasmic linker and often a recognizable, C-terminal PDZ binding motif. Six intron splice junctions are conserved in the first domain, including a rare U12-type of the minor spliceosome provides support for a shared heritage for sodium and calcium channels, and a separate lineage for NALCN. The asymmetrically arranged pores of 4x6 TM channels allows for a changeable ion selectivity by means of a single lysine residue change in the high field strength site of the ion selectivity filter in Domains II or III. Multicellularity and the appearance of systems was an impetus for Nav1 channels to adapt to sodium ion selectivity and fast ion gating. A non-selective, and slowly gating Nav2 channel homolog in single cell eukaryotes, predate the diversification of Nav1 channels from a basal homolog in a common ancestor to extant cnidarians to the nine vertebrate Nav1.x channel genes plus Nax. A close kinship between Nav2 and Nav1 homologs is evident in the sharing of most (twenty) intron splice junctions. Different metazoan groups have lost their Nav1 channel genes altogether, while vertebrates rapidly expanded their gene numbers. The expansion in vertebrate Nav1 channel genes fills unique functional niches and generates overlapping properties contributing to redundancies. Specific nervous system adaptations include cytoplasmic linkers with phosphorylation sites and tethered elements to protein assemblies in First Initial Segments and nodes of Ranvier. Analogous accessory beta subunit appeared alongside Nav1 channels within different animal sub-phyla. Nav1 channels contribute to pace-making as persistent or resurgent currents, the former which is widespread across animals, while the latter is a likely vertebrate adaptation.
The superfamily of voltage-gated cation channels containing 24 transmembrane segments are classified as sodium-selective, calcium-selective, or non-selective channels that span the plasma membrane. Their gene numbers, their expression patterns and functions have been reported in differing body plans that range from single cell eukaryotes, to invertebrates, to the greatest gene complexity of ion channel isoforms in vertebrates. There are twenty-one mammalian genes : Ten are sodium channel (SCNxA) genes coding for Nav1.1 to Nav1.9 (Catterall et al., 2005a), expressing mostly sodium-selective currents and Nax (Noda and Hiyama, 2015); Ten are calcium channel (CACNA1x) genes, coding for Cav1.1 to Cav1.4, Cav2.1 to Cav2.3, and Cav3.1 to Cav3.3 (Catterall et al., 2005b), that generate mostly calcium-selective channel currents (Figure 1). NALCN is a unique, orphan, leak channel gene within the superfamily of voltage-gated cation channels (Cochet-Bissuel et al., 2014; Figure 1) but are poorly understood, because its ion channel characteristics have not been identified and validated by in vitro expression (Senatore et al., 2013; Senatore and Spafford, 2013; Boone et al., 2014). Here, we start by exploring the features in key extant life forms which provide insights into the evolutionary history of the voltage-gated, sodium-selective channels. The most basal life form for appearance of voltage-gated sodium channel classes is in the single cell eukaryotes (Zakon, 2012). Prokaryotes have representative ion channels for potassium- (Doyle et al., 1998) and sodium-selective (Yue et al., 2002) pores, which provides insights into the selectivity mechanisms of eukaryotic 4 × 6 TM channels.
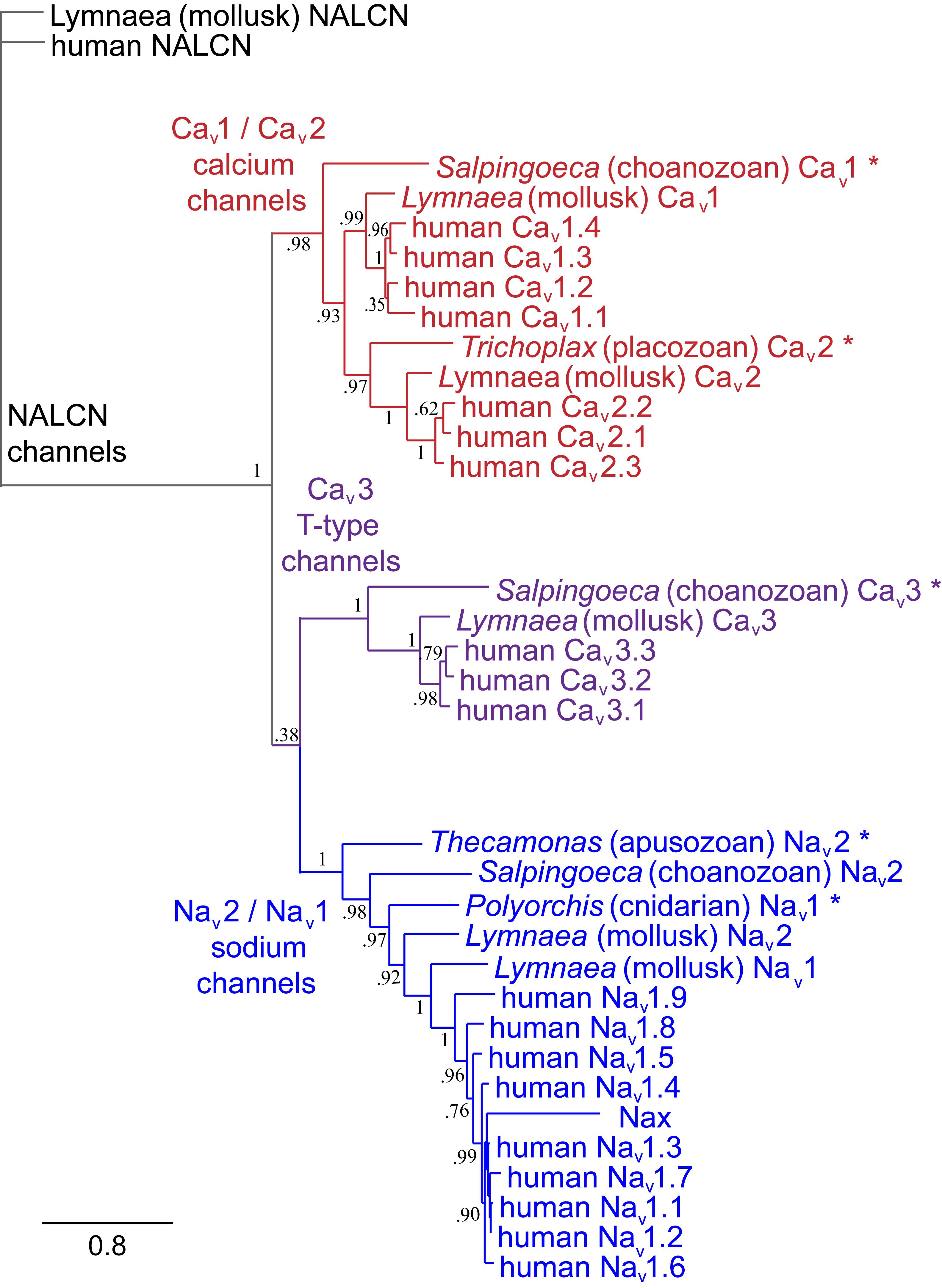
FIGURE 1. Gene tree of 4 × 6 TM (TM = transmembrane) voltage-gated sodium channel α subunits (Nav1 and Nav2), calcium channel α1 subunits (Cav1, Cav2, and Cav3) and NALCN. Included in the multiple alignment are the 21 human homologs and the 6 homologs from representative protostome invertebrate, pulmonate snail, Lymnaea stagnalis. Basal genes of each class (indicated in bold and ∗) including apusozoan, Thecamonas trahens (Nav2), single cell choanoflagellate, Salpingoeca rosetta (Cav1 and Cav3), placozoan, Trichoplax adhaerens (Cav2), and cnidarians (hydrozoan jellyfish), Polyorchis penicillatus (Nav1). More distant Na Leak Conductance channel (NALCN) serve as the out-group for the phylogenetic tree. Bootstrap values are shown at the nodes. Notably weak branch strength is in the positioning for Cav3 T-type channels which is relatively equidistant from sodium channels and other calcium channels. The gene tree illustrate the close kinship amongst Nav2 and Nav1 channels and between Cav1 and Cav2 channels. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017). Gene trees were constructed in PhyML 3.0 (Guindon et al., 2010) using Phylogeny.fr web interface (Dereeper et al., 2008).
The voltage-gated (K+, Ca2+, and Na+) channels consist of two semi-autonomous domains, a pore domain that provides an aqueous pathway for ion selection through the plasma membrane, and a voltage-sensor domain, that transduces pore gate movements (opening/closing events), based on changes to the state of the membrane electric field. There are two major templates for the pore domain that first appear in bacterial representatives, one that is a highly selective pore domain for potassium ions (Doyle et al., 1998) and a different pore domain that is selective for sodium ions (Payandeh et al., 2011). The overall structure of pore domains consist of two transmembrane helices (the so-called S5–S6 segments in voltage-gated channels), with an intervening Pore- (P-) loop, that forms a pore lining border that resembles an inverted teepee, including a descending pore helix from the end of S5 and an ascending loop to the end of S6 (Kim and Nimigean, 2016; Figure 2). At the base of the inverted teepee between the pore helices is a pore-selectivity filter that ascends to form the most constricted point of access of ions, between the outer vestibule facing the wide, extracellular milieu above, and the more expansive aqueous lake within the membrane channel below (Kim and Nimigean, 2016) (Figures 3A,B). The first of the high resolution structures of the potassium selective pore is KcSA, a pore only protein isolated from the bacteria, Streptomyces lividans (Doyle et al., 1998). The signature sequence of the potassium selective pore is the five amino acid residues: T(V/I)GYG contributed by each of four subunits forming identical quadrants contributing to the pore lining selectivity filter. The side chains residues face outwards, and the pore-lining backbone carbonyls form an octet of oxygens, serving to accommodate four dehydrated potassium ions, occupied in every other position (1 and 3 or 2 and 4) at one time in potassium channels (Doyle et al., 1998; Figure 3A). The K channel pore achieves an exclusive K ion selectivity because only the K channel will shed its hydration shell, for optimized, energetically favorable binding conditions within the pore selectivity filter, and passage through by electrochemical force generated by a transmembranal ion gradient (Doyle et al., 1998). Both prokaryotic and eukaryotic potassium-selective channels share the same signature, selectivity filter of T(V/I)GYG residues (Kim and Nimigean, 2016).
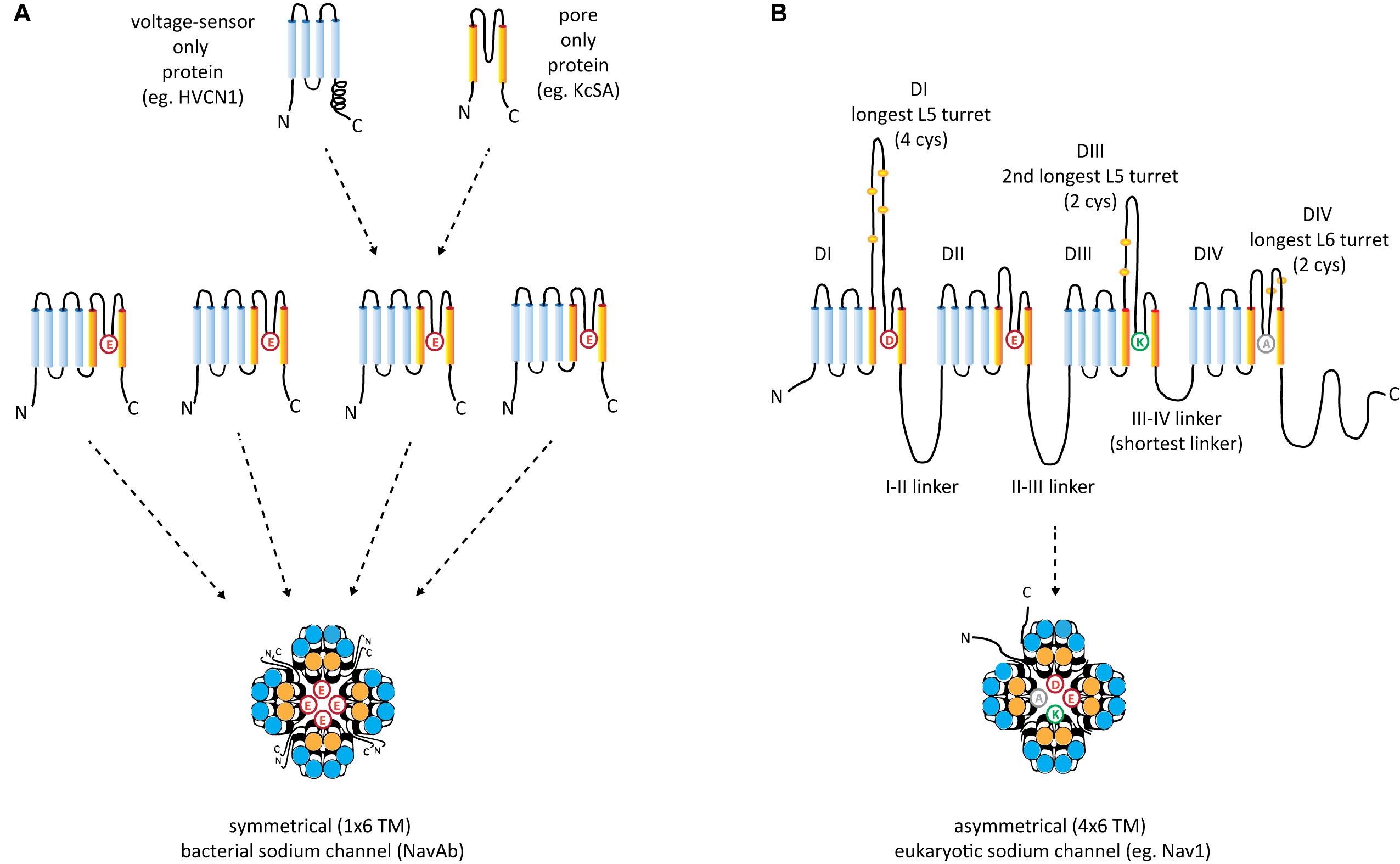
FIGURE 2. Structural comparisons between (A) symmetrical, 1 × 6 TM (TM = transmembrane) homotetrameric, bacterial Na channels consisting of four repeat subunit (B) asymmetrical, eukaryotic four domain calcium and sodium channels consisting of 24 transmembrane segments (4 × 6 TM). Each of the four sodium and calcium channel domains consist of a voltage-sensor domain of four transmembrane segments, S1–S4 (blue), akin to voltage sensor only protein (e.g., HVCN1) and a pore domain, S5–S6 (orange), resembling the size of the two membrane segments of the inward rectifying K channels (e.g., KcsA). A dramatic difference between the 1 × 6 TM bacterial sodium channels and 4 × 6 TM calcium and sodium channels are the larger sizes of extracellular turrets rising before the pore selectivity filter, especially L5I and L5III which range from 40 to 105 amino acids long. The longest extracellular turret in Domain IV in 4 × 6 TM channels is always the extracellular turret rising after the pore selectivity filter (L6IV), while L5IV (the extracellular loop just before the pore selectivity filter) is always short. Comparatively speaking, 1 × 6 TM channels all possess short extracellular turrets of 10–15 amino acids long. Eight core, conserved cysteines populate the L5I - L5II - L5III - L6IV extracellular turrets in a 4-0-2-2 configuration in all eukaryotic 4 × 6 TM channels. Also novel are singleton N- and C-termini, and cytoplasmic linkers, of which the III-IV linker is almost always 53–57 amino acids long in eukaryotic 4 × 6 TM channels.
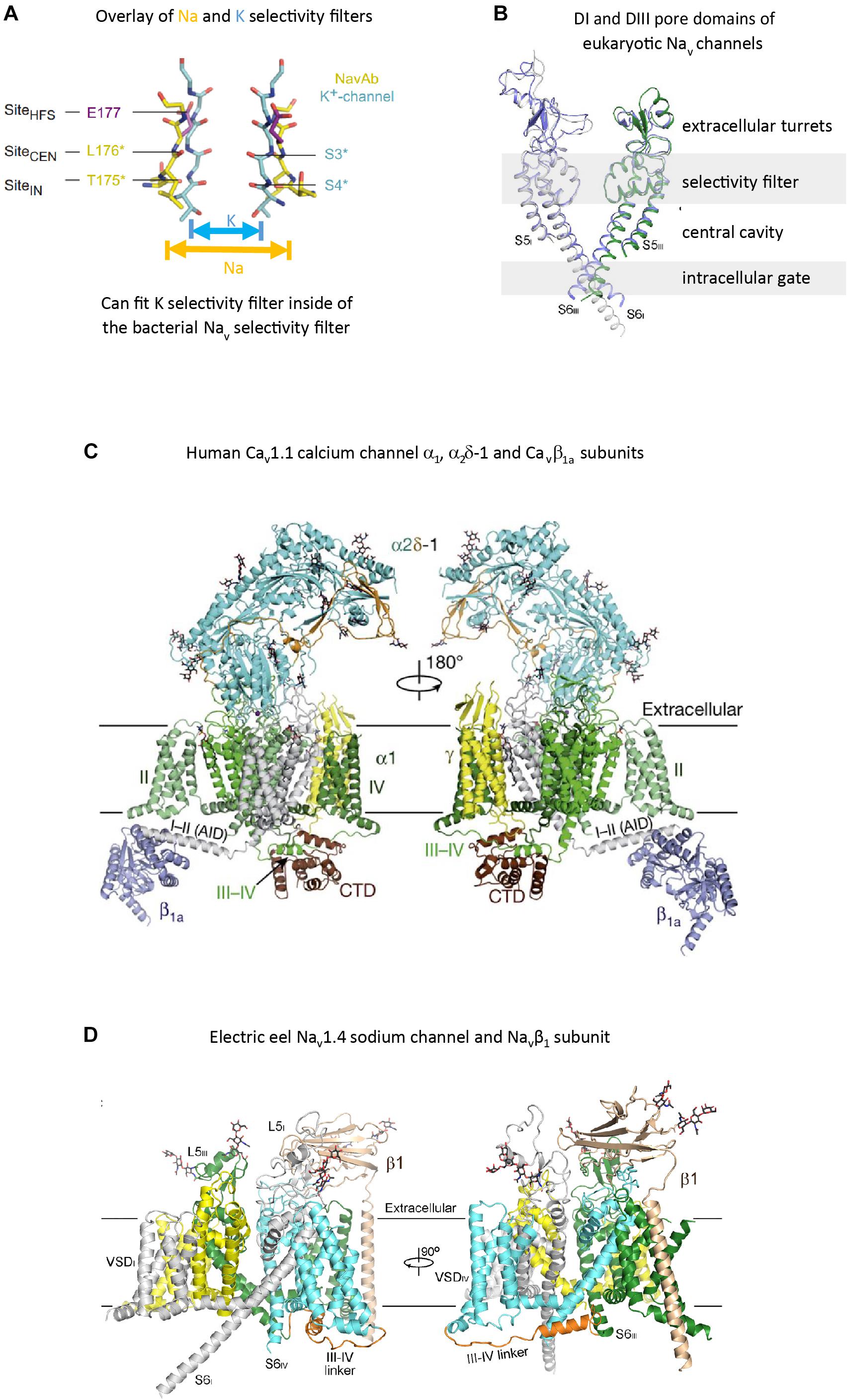
FIGURE 3. High resolution structures of pore selectivity filters, a pore domain and full length voltage gated calcium and sodium channels. (A) Overlay of the structure of the pore selectivity filter of bacterial KcsA potassium channel (Doyle et al., 1998) and bacterial sodium channel NavAb (Payandeh et al., 2012). Only two subunits are shown for clarity. Backbone carbonyls in the selectivity filter are red. The narrower pore selectivity filter for potassium ions fits within the broader selectivity filter for sodium (or calcium) ions. (B) Overlay of Domain I and III pores of arthropod Nav1 and electric eel Nav1.4. Key amino acid side chain residues of the selectivity filter project into the pore center to regulate ion passage through sodium and calcium channels pores. Long extensive extracellular turrets rising from segment 5 to the pore selectivity filter, and shorter extensions from the pore selectivity filter to segment 6 project rise above the membrane, serving as a first contact for incoming ions and drugs, before passing through the selectivity filter below. (C) α1 subunit of the human Cav1.1 calcium channel, illustrating the extended extracellular turret loops of L5I, L5III, L6IV and the intracellular globular domain formed by the III-IV linker and the proximal C-terminal Domain (CTD) containing the calmodulin binding IQ motif. The proximal C-terminus omitted in the right panel to better illustrate the III–IV linker. Accessory α2Δ-1 and β1A subunits are illustrated with the pore-forming α1 subunit. Potential Ca2+ ions in the selectivity filter vestibule are illustrated as green spheres. (D) The structure of the electric eel Nav1.4 (each domain individually colored) and β1(wheat colored) complex, with glycosyl moieties shown as black sticks. The short cytoplasmic III-IV linker (orange color) regulates fast inactivation of Nav1 channels. (A) reproduced from Payandeh, et al (2011). Nature, 10:475(7356):353-8 with permission. (B,D) reproduced from Yan et al. (2017). (C) reproduced from J Wu et al. (2016).
The overall structure for the sodium-selective pore in eukaryotes approximates the unique dimensions of a sodium-selective channel pore identified in bacteria, exemplified by NavAb, the first bacterial sodium channel resolved by X-ray crystallography, isolated from Arcobacter butzleri (Payandeh et al., 2011). The pore selectivity filter is a broader and shorter conduit for ion passage across the membrane, such that one can fit the K ion-selective pore between the dimensions of the bacterial or eukaryotic Na -selective pore (Payandeh et al., 2011; Figures 3A,B). There is an additional pore helix ascending from the ion selectivity filter (P2 helix) in both bacteria sodium channels (Payandeh et al., 2011) and eukaryotic sodium and calcium channels (Wu et al., 2016), in addition to the descending pore helix leading to the ion selectivity filter (P1 helix, also shared in potassium channels), generating a wider and more structured outer vestibule facing the channel exterior.
The equivalent of the potassium channel’s signature residues of the pore selectivity filter is TLESWSM in the bacteria sodium channel (Figure 3A), where the negatively charged carboxylates side chain of the glutamate residue (E) faces towards the pore center (rather than away from the pore center of the K selective pore), creating a highly electronegative, high field strength (HFS) site, contributed by the four glutamates (EEEE) in identical position (Payandeh et al., 2011). At this most constricted width of the Na ion selective pore, is a 4.6 × 4.6 angstrom square HFS site, that is large enough to accommodate a sodium ion with two planar waters of hydration (Payandeh et al., 2011; Figure 3A). This contrasts with the narrow ion traversing pathway where all waters are stripped from the potassium ion before entering the pore selectivity filter (Figure 3A).
Potassium-selective pores can be voltage-gated with addition of N-terminally attached voltage-sensor domains, consisting of four (S1–S4) segments (Groome, 2014). The voltage-sensor is optional in potassium channels, and can operate as a semi-autonomous unit. An example of voltage-sensor only proteins are the voltage-gated proton channels that allow proton transport into phagosomes via the voltage sensor (e.g., human HVCN1) (Takeshita et al., 2014; Figure 2A). All eukaryotic sodium and calcium channels possess voltage-sensor domains like the voltage-gated potassium channel, where the S4 segments have a variable number (4–8) positive charged residues (lysine or arginine) every third amino acid, forming a highly charged side of an alpha helix (Groome, 2014). S4 segments respond to membrane potential changes, by movement of their positive charged residues along negatively charged, counter-charges formed by S1–S3 segments, transducing the mechanical coupling of an amphipathic S4–S5 helix to pore gating movements that lead to iris-like, occlusion or widening of the pore helical bundle formed by the distal ends of S6 segments (Groome, 2014).
Sodium and calcium channels only exist in as voltage-gated channels subunits with voltage-sensors and pore domains (6TM, TM = TransMembrane) fused together, unlike potassium channels which can exist as pore-only proteins (Figure 2A). The 1 × 6 TM bacterial sodium channels have been identified in proteobacteria and actinobacteria, but also have spread to eukaryotes, including diatoms, which they likely have received by horizontal gene transfer from bacteria (Verret et al., 2010).
Typical eukaryotic calcium and sodium channels are always approximately four times the equivalency in size of 1 × 6 TM prokaryotic sodium channels and voltage-gated potassium channels (Figure 2B). The closest homolog to the prokaryotic 1 × 6 TM Na-selective channels are the eukaryotic transient receptor potential (TRP) family of channels, whose members are usually non-selective or sodium selective channels, with exceptions, such as Catsper1 with an electronegative HFS sites which are key for conferring a calcium-selectivity (Wu et al., 2010; Figure 4). Two domain TPC channels are sodium-selective in mammals, but TPC homologs in basal representatives such as Salpingoeca rosetta resemble calcium-selective channels with a glutamate or aspartate in Domains I or II of the HFS site of the pore selectivity filter (Figure 4). The example of differing ion selectivities in TPC channels is suggestive of experimentation between sodium and calcium selective pores in possible two domain channel ancestors to eukaryotic 4 × 6 TM channels (Peng et al., 2015). As expected from a two domain intermediate that duplicated into a four domain channel such as 4 × 6 TM channels (Strong et al., 1993), the first domain of TPC is more similar to Domain I and III of 4 × 6 TM channels and the second domain of TPC is more similar to Domain II and IV of 4 × 6 TM channels (Rahman et al., 2014).

FIGURE 4. Alignment of amino acid sequences contributing to the pore selectivity filters in representative calcium, sodium and NALCN channel homologs from single cell protists, yeast, algae, invertebrate and human, as well as 2 × 6 TM TPC channels, 1 × 6 TM channels (TRP, Catsper, and bacterial Na), and 1 × 2 TM channels (bacterial K). The pores of 4 × 6 TM channels from brown and green algae, yeast, single-cell choanoflagellates, cnidarian and mollusks are illustrated alongside human representatives, to illustrate the relationship of pore selectivity filters in different life forms. The selectivity filter is flanked by a descending P1 helix and ascending P2 helix. Residues contributing to the central (CEN) and inner (IN) sites of the selectivity filter are highlighted blue. Negative charged residues contributing to the selectivity filters (HFS sites), and to the ring of outer carboxylates are a red/brown color. A positively charged lysine residue (green color) populate the HFS site in Domain II or III of all Nav1 channels. The aspartate (D) residue in Domain II at the Ca site, next to the HFS site, is conserved in Cav1, Cav2 and Cav3 channels. Nav2 and Nav1 channels are shortened in the pore selectivity filter in Domain II and lack the Ca site altogether. Instead, a glutamate (E) residue is conserved at the Na site (HFS+4) in Domain II of Nav2 and Nav1 channels. Exceptionally conserved tryptophans (W site) form inter-repeat hydrogen bonds that stabilize the pore loop region. Outer ring carboxylates (Na site, especially in Domain IV) contribute to TTX sensitivity, which when altered lowers TTX insensitivity. Note the amino acid changes (yellow colored residues) in Domain IV of the TTX-sensitive hNav1.4 and TTX-low sensitive channels from garter snake (Thamnophis sirtalis) that adapts to feed on TTX-ladened newts by neutralizing a negative charge at the Na site. Bacterial Na channel, NavAb becomes calcium-selective with aspartate (D) substitutions at the HFS, Ca and Na sites. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
All the 4 × 6 TM channels of the superfamily of Cav and Nav channels appear to derive from the same stem eukaryotic channel with a fundamentally shared template. There is evidence in a shared genomic structure: Sodium channels (Nav2 and Nav1) and calcium channels (Cav1, Cav2, and Cav3) in animals possess five common splice site locations in the first of four domains, including each of the first four transmembrane segments (S1–S4) of Domain I of the voltage-sensor domain (D1S1, D1S2, D1S3, and D1S4) and two splice site locations in the pore loop (S5–S6) of Domain I (DIS5-D1S6) (Figure 5). The D1S1 splice site is conserved in almost all known calcium and sodium channels (Spafford et al., 1999) and is a highly rare, unconventional, U12- type (AT-AC) splice site that representing just 700-800 putative genes (or ∼4%) of total genes in mammalian genomes (Parada et al., 2014; Figure 6). The exception in the conservation of splice sites in Domain I is that the 6th splice site in the D1 pore loop is lacking in Cav3 channels (Figure 5). A shared genomic structure in the first of four domain may indicate constraints on the structural divergence of the first domain shared between four domain calcium and sodium channels. These six conserved splice sites are not present in NALCN channels (Figure 5), including the unconventional, U12- type splice site governed by the minor spliceosome shared in homologous position of D1S1 (Figure 5). The lack of conservation of intron splice sites reflects the weaker evolutionary link between NALCN from the cluster of more closely related families of eukaryotic voltage-gated calcium and sodium channels.
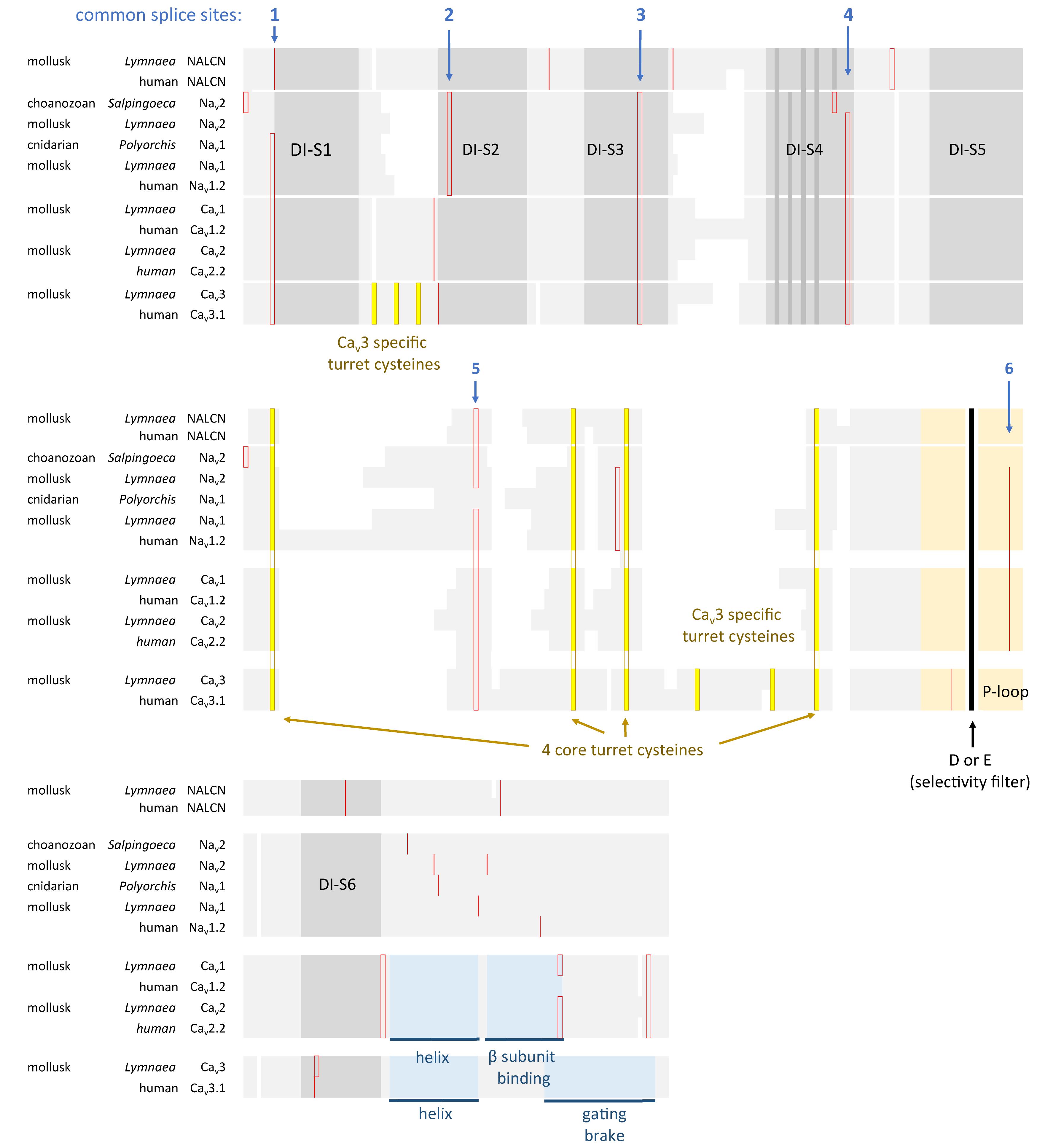
FIGURE 5. Amino acid alignment of the first domain of representative 4 × 6 TM channels illustrating the conservation of six intron splice sites (red vertical lines) in voltage-gated calcium (Cav1, Cav2, and Cav3) and sodium (Nav1 and Nav2) channels. Conserved placement of intron splice junctions (red vertical lines) span each transmembrane segment of the voltage-sensor domain (D1S1, D1S2, D1S3, and D1S4) and two in the pore loop of Domain I (DIS5-D1S6). The sixth splice site is lacking in Cav3 channels. NALCN lacks all of these splice sites, supporting a more distant relationship for NALCN compared to the eukaryotic calcium and sodium channels. The six membrane spanning helices S1–S6 are indicated in gray shading, with the High Field Strength (HFS) site (D or E) indicated by black vertical line surrounded by the rest of the pore loop region (light orange shading). Darker gray shading in S4 indicate the positions of positive charges serving the voltage-sensor domain. Four cysteine residues are conserved in the extracellular turret (L5) of all 4 × 6TM channels, and the location of two extra cysteines in L5 and S1–S2 are only found in Cav3 T-type channels. Cysteine residues are indicated in a bright yellow color. Illustrated in blue color is a rigid, helix in the proximal I-II cytoplasmic linker, upstream of an accessory CaVβ subunit binding site in Cav1 and Cav2 channels, and the “gating brake” of Cav3 channels which harbors a nano-molar affinity binding site for calmodulin. Genes in the alignment include NALCN, sodium channels (Nav2 and Nav1) and calcium channels (Cav1, Cav2, and Cav3) from human representatives, as well as protostome invertebrate, pond snail L. stagnalis, and basally branching species from single cell choanoflagellate, S. rosetta, placozoan, Trichoplax adhaerens, and cnidarians: Nematostella vectensis (sea-anemone) and Polyorchis penicillatus (hydrozoan jellyfish). Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
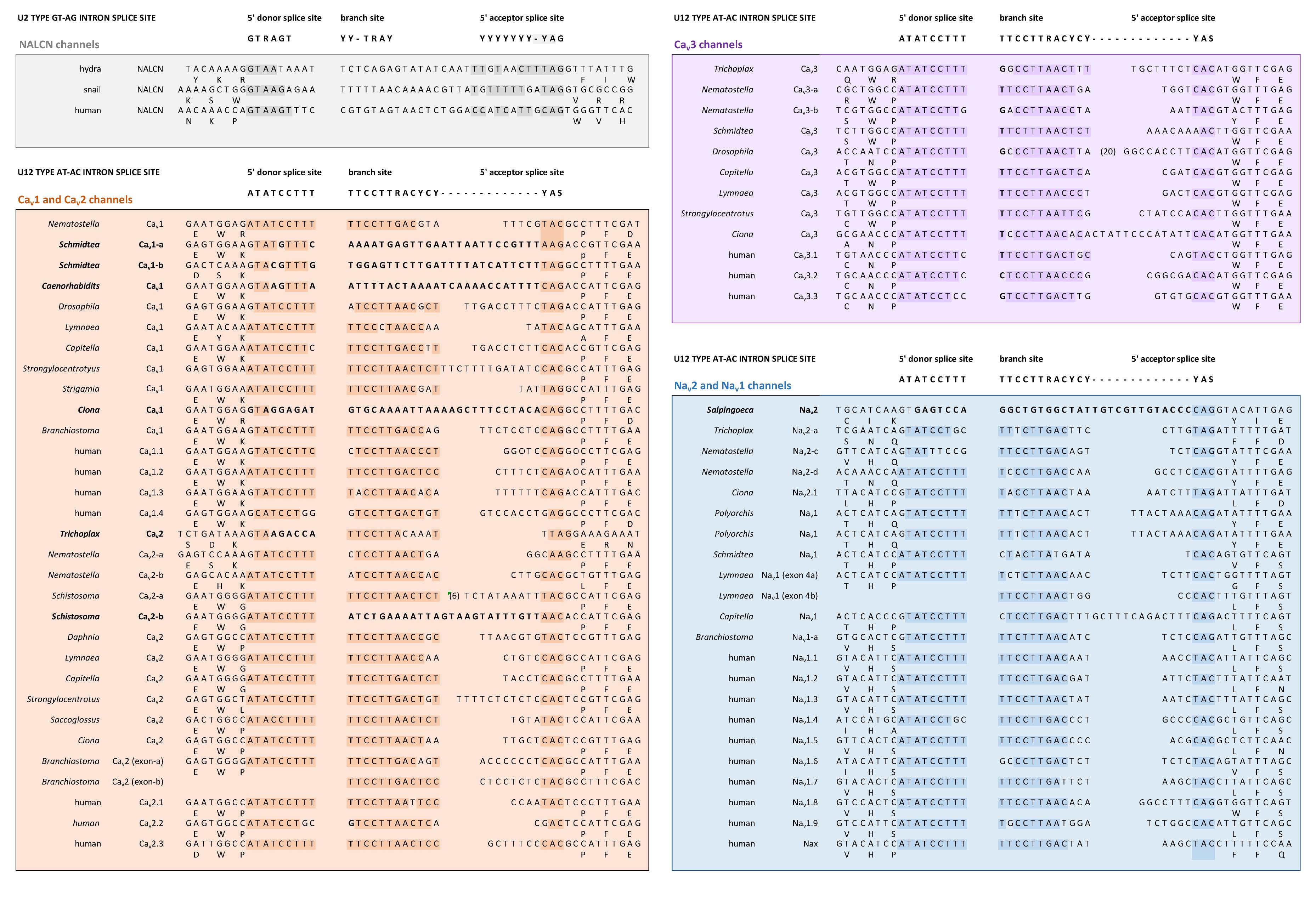
FIGURE 6. Conservation of a very rare U12 type intron splice site in Domain 1, segment 1 of Nav2, Nav1, Cav1, Cav2, and Cav3 channels, but not NALCN channels. Exceptionally rare U12-type splice site represent just 700–800 putative genes (or ∼4%) of total genes in mammalian genomes. Consensus 5′ donor splice site (ATATCCTTT), branch site (TTCCTTRACYCY) and 5′ acceptor splice site (YAS) recognition sequences of minor U12-type spliceosome varies from conventional major U2-type spliceosome recognition sequences with consensus 5′ donor splice site (GTRAGT), branch site (YY-TRAY) and 5′ acceptor splice site (YYYYYYY-YAG). NALCN has a variable intron splice junction in this region, with resemblances to the major U2-type spliceosome recognition sequence. DNA sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
Eukaryotic 4 × 6 TM channels adopted a particular asymmetrical, architecture followed domain duplication which generated the four domain channel. The signature, asymmetrical structure is common to all known cation channels including sodium, calcium and NALCN channels in animals, including Nav2, Cav1, and Cav3 homologs in the single cell choanoflagellate, S. rosetta, voltage-gated cation channels from other protists (e.g., dinoflagellates, ciliates, oomycetes, and diatoms), yeast calcium channel (e.g., Cch1p) (Locke et al., 2000), as well as species of brown and green algae (chlorophytes and prasinophytes) (Figure 7). Major eukaryotic groups lacking the 4 × 6 TM channel structure are the embryophytes (the land plants) and red algae (Verret et al., 2010). Ancestors to the land plants appeared to have purged the eukaryotic 4 × 6 TM channels, alongside key components of the animal toolkit for cation influx across the plasma membrane which includes the IP3 receptors, ATP-gated purinergic receptors, the cys-loop superfamily of ligand-gated ion channels and TRP channels (Edel et al., 2017). A single two-domain, TPC1 homolog is retained in land plants as it relates to comparable signaling across internal membrane compartments of plant vacuoles and animal organelles (Hedrich and Marten, 2011).
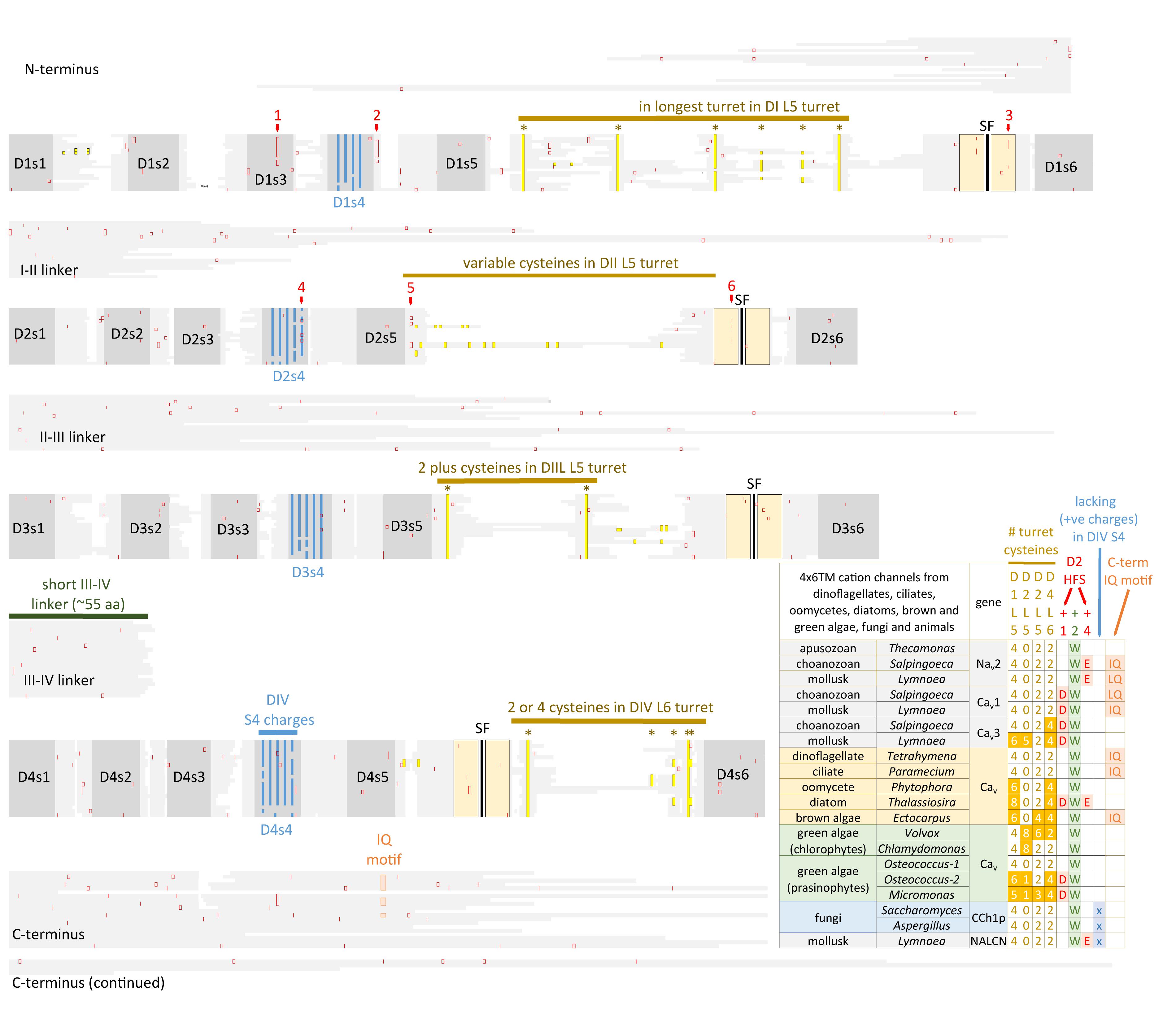
FIGURE 7. Alignment of 4 × 6 TM cation channel representatives illustrating a shared fundamental architecture of asymmetrically arranged extracellular and intracellular regions from single cell protists (choanoflagellates, dinoflagellates, oomycetes, and ciliates), green algae (chlorophytes and prasinophytes), brown algae, yeast, and Nav2, Nav1, Cav1, Cav2, and Cav3 channels from protostome invertebrate (pond snail, L. stagnalis). A concerted evolutionary change to a common asymmetrical architecture of 4 × 6 TM cation channels contrasts with the pseudo-symmetry of cyclic nucleotide-gated potassium channel (CNGK) channels which contain four nearly identical domains chained together. Long extensive extracellular turrets prior to the pore selectivity filter (SF) include L51, L5III and more rarely L5II and a longer turret after the SF in L6IV, as well as a 4-0-2-2 framework of core cysteines populating L5I-L5II-L5III-L6IV extracellular turrets in 4 × 6 TM channels. L5IV extracellular turret is always short in 4 × 6 TM channels. The cytoplasmic III-IV linkers are always short (∼53–57 amino acids) compared in contrast to more variable I-II linker, II-III linkers, and N- and C-termini in 4 × 6 TM cation channels. Intron splice sites are indicated by red vertical lines. Dinoflagellate, ciliate, oomycete, diatom, green and brown algae channels illustrated in the figure all possess a conserved HFS site resembling an animal calcium channel (EEEE). Notably lacking in most of these protist channels is the conservation in the pore selectivity filter of Domain II of a “calcium beacon” aspartate (D) (HFS+1) omni-present in animal calcium channels, or a “sodium beacon” glutamate (HFS+4) omni-present in animal sodium channels, and a C-terminal IQ motif. A tryptophan ring at HFS+2 is present in all four domains of all channels except Domain I of Cch1p. A notable absence of S4 charges is found for yeast Cch1p and NALCN. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
Generation of the four pores and voltage-sensor domains after domain duplication, provided opportunity for divergence and specialization of the four individual domains of 4 × 6 TM channels. This is evident in studies assessing the relative contribution of voltage-sensor domains in Nav1 channels for example, where the first three of four domains appear to be faster in gating charge movement, and are necessary and sufficient to regulate channel opening, while the fourth domain is slower to mobilize and contributes more to the refractory, inactivated state of the channel after prolonged channel opening (Capes et al., 2013).
Associated structural changes for the coordinated movements of individual domains for channel gating include a unique “domain swapping” topology found in 1x6TM (Doyle et al., 1998; Payandeh et al., 2011) and 4 × 6 TM (Wu et al., 2015; Shen et al., 2017) voltage-gated cation channels, where voltage sensor modules are rotated clockwise with respect to pore modules of each domain. The voltage sensor module of Domain I, for instance, surrounds the Domain II pore. The domain swapping arrangement is lacking in many potassium channel classes [e.g., Eag1 (Whicher and MacKinnon, 2016), CNG (Li et al., 2017), HCN (Lee and MacKinnon, 2017) Slo1 (Tao et al., 2017) and Slo2.2 (Hite and MacKinnon, 2017)], and lacking in some TRP channels like TRPV6 while present in other TRP channels (e.g., TRPV1 and TRPV2) (Singh et al., 2017). The absence of a domain swapping topology correlates with an S4-S5 linker that is too short to accommodate the swapping of voltage-sensor and pore domains. The longer S4-S5 linkers supports a rotary coupling of interlocked pore domains for more concerted actions in gated pore movements within voltage-gated cation channels (Bagneris et al., 2015).
There is a specific asymmetrical pattern of long (15–105 aa) extracellular turrets shared in the pore domains of all animal calcium, sodium and NALCN channels, and more broadly with most eukaryotic lifeforms (outside of land plants and red algae), including yeast, ciliates, dinoflagellates, oomycetes, diatoms and brown and green (chorophytes and prasinophytes) algae (Figures 2, 7). Bacterial sodium and potassium channels possess extracellular L5 and L6 turrets that are always short (10–15 aa) in length (Stephens et al., 2015; Figures 2, 7). 4 × 6 TM channels possess a rising extracellular turret from the S5 transmembrane helix (dubbed L5) before the pore selectivity filter, and a second shorter extracellular turret after the pore before S6 transmembrane helix (dubbed L6) (Figures 2, 3C,D, 7; Stephens et al., 2015). The longest L5 extracellular turret is in Domain I (ranging from 60 to 105 aa). L5 in Domain III possess the second longest turret (ranging from 40 to 60 aa), while Domain II and IV turrets are the shortest L5 extracellular turrets (15–30 aa) in size. The longest and most variable L6 extracellular turret amongst 4 × 6 TM channels is contained in Domain IV, with L6 extracellular turrets being shorter and less variable in Domains I, II and III. Populated within the four turret domains is a fundamental set of eight conserved cysteines that are present in all 4x6 TM channels: two in L5I, two in L5III and two in L6IV (Figure 7; Stephens et al., 2015). The extended extracellular turrets appear to form intra-loop disulphide bonds which stabilize the structure as a “windowed dome” of interlocked extracellular loops in the human Cav1.1 calcium channel (Figure 3C; Wu et al., 2016) or electric eel Nav1.4 sodium channel complex (Figure 3D; Yan et al., 2017), resolved in the cryo- electron microscopy of channel protein nanoparticles. L5 turrets of Domains I, II and III (i.e., L5I, L5II, and L5III) as well as L5 and L6 turrets of Domains III and IV, respectively, form highly negatively charged vestibules above the pore selectivity filter that serves as an electro-attractant for cation passage before reaching the selectivity filter below (Figures 3D,E). The eight conserved cysteines in extracellular turrets appear to be necessary for proper folding of 4 × 6 TM channels, as mutations of these conserved cysteines prevents their ion channel expression (Karmazinova et al., 2010). Varying from this conserved set of 8 extracellular turret cysteines in 4 × 6 TM channels are additional 1 to 3 cysteines in L5II of Nav1 channels in vertebrates only (Stephens et al., 2015). Cav3 T-type channels, but not Cav1 or Cav2 channels, possess extra cysteines – more than double the number beyond the core 8 cysteines in L5I (+2), L5II (from 0 to +5), L5IV (+2), L6IV (0 to +2), and S1-S2 in Domain I (+3) (Senatore et al., 2014). The positioning of numbers of cysteines and size of L5II and L6IV turrets is exploited to generate a variable ion selectivity from calcium to highly sodium selectivity, available in most invertebrates Cav3 (T-type) channels by alternative splicing (Senatore et al., 2014).
The external surface of voltage-gated channels are coated with (glycans) sugar groups, by an enzymatic, co- and post-translational process fundamentally critical for the proper folding and functional expression of voltage-gated ion channels (Baycin-Hizal et al., 2014). N-linked glycosylation involves sugars attached to the nitrogen atom of an asparagine amino-acid side chains (Mellquist et al., 1998) and is the most common form of glycosylation (Apweiler et al., 1999). O-linked glycosylation has a more promiscuous consensus binding site to different possible oxygen atoms of nascent proteins (Van den Steen et al., 1998), but has been reported for Nav1 channels (Ednie et al., 2015).
Glycosylation sites are enriched in the vicinity of the conserved cysteines on the longest L5I and second longest L5III extracellular loops of 4 × 6 TM voltage-gated cation channels. Electron microscopy structures of frozen sodium channel nano-particles resolves 20 and 16 sugars, incorporated into seven glycosylation sites on insect Nav1 (Shen et al., 2017) and electric eel Nav1.4 (Yan et al., 2017) sodium channels, with four and three glycosylation sites contained in L5I and L5III extracellular loops, respectively. While there are a conserved positioning of cysteines in extracellular loops, putative locations of glycosylation sites are not fundamentally shared between sodium and calcium channel families. Individual sodium (Nav2/Nav1), calcium channel (Cav1/Cav2) or T-type (Cav3) channel genes can highly vary in their density of glycosylation sites. An often cited example of enrichment is in mammalian Nav1.4 sodium channel, which possesses four extra glycosylation site harbored in a unique ∼36 amino acid fragment added to the L5I extracellular loop compared to non-mammalian, but vertebrate (e.g., eel) Nav1.4 (Bennett, 2002). Each sugar moiety contains a terminal sialic acid in N-glycosylation sites, contributing to the total electro-negativity of channel surface charge, with consequences to channel gating in biasing the voltage sensors by surface charge screening (Ednie and Bennett, 2012). The high electro-negativity due to glycosylation appears to reaches a saturation in mammalian Nav1.4, so that additional sialic acids contributed by the two known glycosylation sites of co-expressed mammalian beta subunit (Navβ1, 25 kDa), influence channel gating on the more weakly glycosylated mammalian sodium channels (Nav1.2, Nav1.5, and Nav1.7) but not on channel gating of the heavily, saturated Nav1.4 channel with glycosylation sites (Johnson et al., 2004). The longest extracellular loops (L5I and L5III) of the pore-forming α1 subunit (∼170 kDa) of Cav1 and Cav2 channels are preoccupied in cradling a large, mostly extracellular α2 subunit (∼150 kDa) (Leung et al., 1987). Almost all of the glycosylation sites (15 out of 16) in the resolved structure of the mammalian Cav1.1 channel complex are associated with the α2 subunit, with a single glycosylation site contained in L5I extracellular loop of the pore forming α1 subunit of Cav1.1 (Wu et al., 2016). Cav3 T-type channels resemble Nav sodium channels in lacking association with a large accessory subunit, and appear to possess more secondary structure with greater potential variations for glycosylation in their longer (L5I and L5III) as well as L6IV extracellular loops like the Nav channels (Lazniewska and Weiss, 2017).
A critical feature that emerges from the structural asymmetry in eukaryotic 4 × 6 TM channels is a highly changeable ion selectivity that ranges from highly calcium-selective channels (like Cav1/Cav2 channels) and highly sodium-selective channels (like Nav1 channels). The 4 × 6 TM calcium and sodium channels are at least a thousand-fold and ten-fold more selective for their particular cation, respectively, amongst competing native cations (e.g., Ca2+, Na+, and K+) (Hille, 1972; Hille, 2001; Finol-Urdaneta et al., 2014). The high calcium or sodium selectivity is largely converted by a singular, determinative residue at the most constricted point of the funnel-like pore known as the HFS site of the pore selectivity filter (Heinemann et al., 1992; Schlief et al., 1996). The HFS site is a negatively charged carboxylate, derived from a glutamate residue in the bacterial sodium channel pore of NavAb of TLESWSM (Yue et al., 2002). The equivalent HFS site in eukaryotic calcium-selective channels is contributed by different amino acids from Domains I, II, III and IV, forming a ring of electronegative, glutamate residues configured as “EEEE” in Cav1 and Cav2 channels (Figure 8). The HFS site of sodium-selective channels are always differentiated with a positively charged lysine (K) residue configured in the third domain as “DEKA” in most Nav1 channels (Noda et al., 1984).
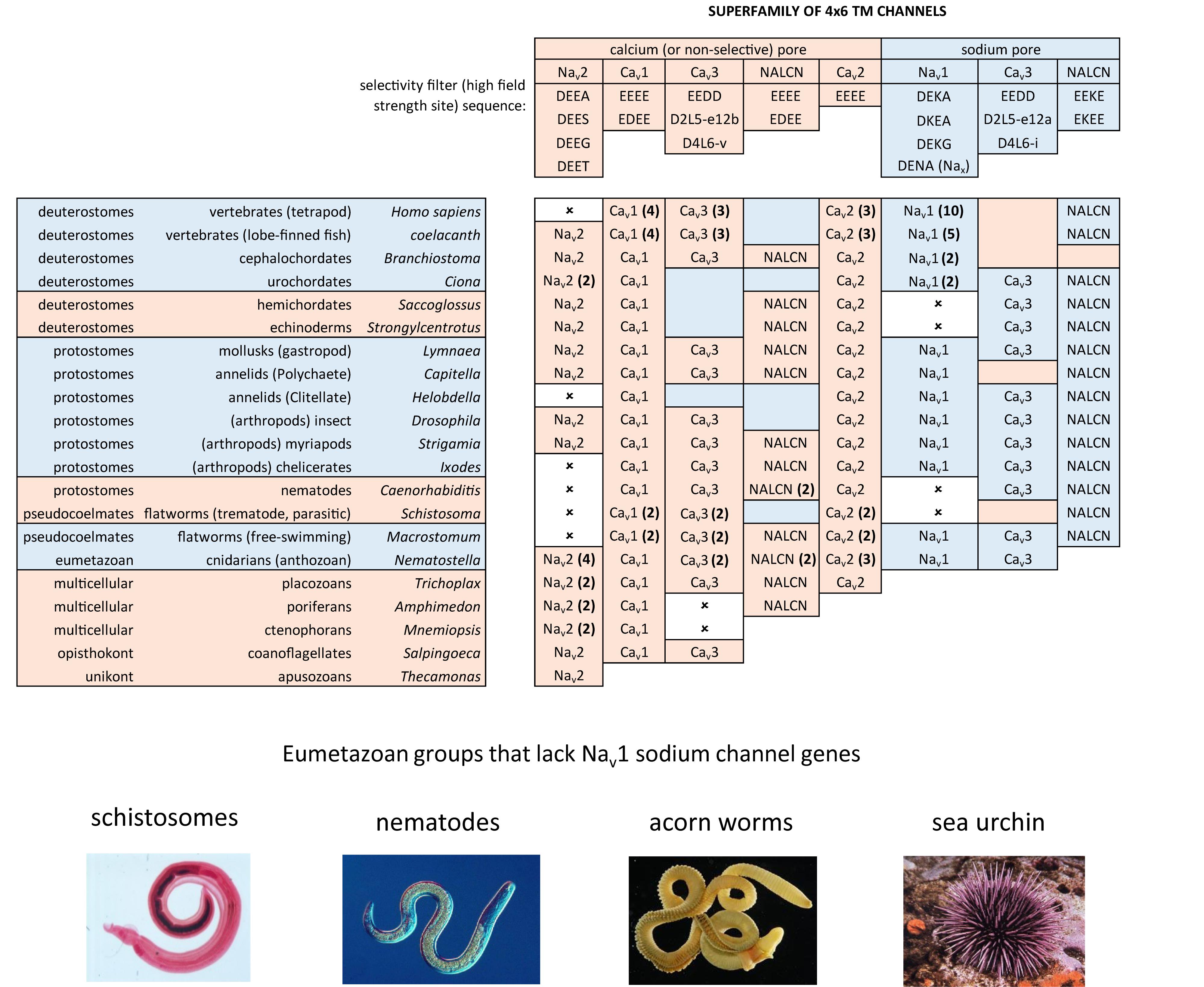
FIGURE 8. Illustration of the distribution of HFS site sequences in NALCN, sodium channels (Nav2 and Nav1) and calcium channels (Cav1, Cav2, and Cav3) in different animals groups, as well as single cell eukaryotes Thecamonas (apusozoan) and Salpingoeca (choanoflagellate) (Top panel). Illustrations of animals (images imported from Wikipedia) lacking Nav1 channel genes (Bottom panel). Lifeforms with calcium-selective pores (shaded orange color) include HFS site sequences mostly of negatively charged glutamate (E) or aspartate (D) residues: DEEA, DEES, DEEG, DEET, EEEE, EDEE, NEEE, while sodium-selective pores (shaded blue color) have a lysine (K) residue in Domains II (EKEE and DKEA) or a lysine residue in Domain III (EEKE, DEKA, and DEKG). Cav3 T-type calcium channels have a selectivity filter of EEDD and may be calcium or non-selective channels (shaded orange color), with high sodium selectivity engendered in invertebrate Cav3 channels by alternative splicing of exon 12a coding for a novel L5II turret (shaded blue color). Greater sodium selectivity also involves a D4L6i extracellular turret found in invertebrates instead of the D4L6v turret found in cnidarians and vertebrates. Numbers in brackets next to the gene indicate the number of gene isoforms (2, 3, 4, 5, or 10) that have been identified in a species, outside of one. An () indicates loss of gene in the indicated species. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
Native bacterial channels are not known to possess calcium-selective channels, but such a channel can be artificially created by engineering symmetrical rings of negatively charged carboxylates from aspartates (TLDDWSD from TLESWSM) at twelve amino acid positions in the pore, spanning the HFS site glutamate residue, and downstream amino acids dubbed “Ca”, (position +1) and “Na”, (position +4). (NavAb mutant, Figure 4; Tang et al., 2014). The “Ca” and “Na” positions contribute to the outermost vestibule of the re-entrant pore selectivity filter, where they appear to be engineered to serve as potential “beacons” for attracting “Ca” or “Na” ions to the eukaryotic pore.
Molecular dynamics modeling illustrate how sodium selectivity is conserved in eukaryotic 4 × 6 TM channels involving four key amino acid residues (Zhang et al., 2018). The “Na beacon” position in Domain II (HFS+4) is always a glutamate residue (E) in known Nav2 and Nav1 channels, where it is optimally positioned in the outer vestibule to attract incoming cations (Zhang et al., 2018). The “Na beacon” relays incoming cations to the aspartate residue (DEKA) of the HFS in Domain I and then to the glutamate residue (DEKA) of the HFS site of Domain II, in a set of likely side-chain swinging transitions between these three negatively charged carboxylate residues in the pore selectivity filter (Zhang et al., 2018). The omnipresent, positively charged lysine residue of Nav1 channels of the HFS site (DEKA) protrudes into the spacious central cavity, repulsing the positive cations to the opposite side wall of the pore, where the ions are funneled, sequentially to the negatively charged (D and E) side chains of the HFS site (DEKA and DEKA) of the pore selectivity filter (Zhang et al., 2018). A critical structural deviation only found in Nav2 and Nav1 channels is a shortening of the selectivity filter in the second domain (Tikhonov and Zhorov, 2012), bringing the “Na beacon” residue in closer proximity to the other carboxylate site chains of the HFS site (D and E) residues required for coordination and selective passage of Na+ ions through the Nav1 channel pore (Shen et al., 2017). Homologous positioning of the pore selectivity filters and the Na site of 4 × 6 TM sodium channels are not easy to translate into sequence alignments with calcium channels or bacterial sodium channels. A single gap in amino acid sequence is inserted into multiple sequence alignments to convey this discrepancy for the 4 × 6 TM sodium channel pores (see Figure 4; Tikhonov and Zhorov, 2018).
Known variations in the HFS site of the pore selectivity filter of Nav1 channels including substitution of the 4th position from alanine (DEKA) to glycine (DEKG) identified in flatworm Nav1 channels (Jeziorski et al., 1997) and many alternative isoforms of duplicated Nav1 genes in teleost fish (Jost et al., 2008). The 4th position of the HFS site is not involved in coordinating ion selectivity, as predicted by modeling (Shen et al., 2017) and experimental evidence (Wu et al., 2013). The HFS lysine residue is relocated from Domain III (DEKA) in most metazoans to Domain II (DKEA) in cnidarians. The HFS site variation likely reflects a mirror imaging of selectivity filters swapped between Domain III and Domain II in cnidarian Nav1 channels without major consequences to their sodium selectivity (Spafford et al., 1998; Figure 8). Indeed, native sodium currents in cnidarians highly resemble mammalian Nav1 currents with a similar profile of ion selectivity for monovalent and divalent ions, suggesting that the cnidarian pore containing a DKEA HFS likely confers a similarly high sodium selectivity as the DEKA configuration of vertebrate Nav1 channels (Grigoriev et al., 1996; Spafford et al., 1996). The molecular constituents necessary for these channels to generate a high sodium selectivity hasn’t been assessed in cnidarian Nav1 channels, because these channels do not generate expressible ion currents in vitro, like other non-pancrustacean, invertebrate Nav1 channels (Schlief et al., 1996; Gur Barzilai et al., 2012).
The replacement of a positively charged lysine residue in the HFS site of Domain II or III with a negatively charged glutamate residue as DEEA differentiate the Nav2 channels, which are always lacking in sodium ion selectivity (Zhou et al., 2004). Nav2 channels otherwise closely resemble the Nav1 channels in structure and are distributed in genomes from single cell eukaryotes and multicellular animals including vertebrates, but not including ray-finned fish or tetrapods, like mammals (Zakon et al., 2017; Figure 8). Molecular modeling suggest a much wider and accommodating “DEEA” pore for Na+, K+, and Ca2+ ions in the absence of a protruding positively charged lysine residue, funneling ions to the side wall containing the carboxylate (D and E) side chains of the HFS site in Nav2 channels (Shen et al., 2017). Substitution of small amino acids (glycine, DEEG; serine, DEES; threonine, DEET) for the alanine in the 4th position of the “DEEA” HFS site occurs in Nav2 channels (Moran et al., 2015) in a homologous position as Nav1 channel substitutions in the 4th position of the HFS site. The omni-presence of the “Na beacon” in Domain II and the first and second position of “DE” within the HFS, reflects a common mode of ion selectivity for Nav2 and Nav1 channels, outside the uniquely protruding, positively charged lysine residue in the HFS site, which is required for limiting ion selectivity to sodium ions for Nav1 channels.
A uniquely, more symmetrical, pore selectivity filter is shared amongst calcium channels, compared to 4 × 6 TM sodium channels. An aspartate (D) residue is ubiquitously located in the “Ca” position (HFS+1) of calcium channels (Cav1, Cav2, and Cav3) (Figure 8) and dubbed a “Ca beacon” for its position just above the HFS site, in an opportune position to attract incoming calcium ions to the pore selectivity filter below (Tikhonov and Zhorov, 2011). The “Ca beacon” is located in the vicinity of the outer pore of Domain II as the “Na beacon” of sodium channels (Figure 4; Tikhonov and Zhorov, 2011). 4 × 6 TM calcium channels vary from 4 × 6 TM sodium channels in possess a pseudo-symmetry of their pore selectivity filters, more resembling the four-fold symmetry of bacterial 1 × 6 TM sodium channels. Calcium channels lack the shortened pore selectivity filter for Domain II found in sodium channels, and bear a more symmetrical, electro-negative ring of glutamate residues in the HFS site, configured as “EEEE” in Cav1 and Cav2 channels (Figure 8). The only known deviation of the “EEEE” configuration of the HFS site is the basal Cav1 homolog isolated from S. rosetta, which retains a high calcium selective, even though the carboxylate side chain in Domain II is shortened by one carbon chain as “EDEE” (Mehta, 2016).
NALCN channels are usually a single representative in most animal groups, and possess variable HFS sites that resemble calcium channels “EEEE” (or rarely “EDEE”) or can resemble Nav1 sodium channels with a positively charged lysine (K) residue in Domain II or Domain III as EKEE or EEKE, respectively (Figure 8; Senatore et al., 2013). Many invertebrates groups possess a flexibility in generating alternative pores which resemble calcium- or sodium-selective NALCN channels specialized for different tissues (Senatore et al., 2013). A duplication of Exon 15 coding for region spanning the selectivity filter residue in Domain II of NALCN creates alternative calcium-selective (EEEE) and sodium-selective (EKEE) pores in the Platyhelminthes (flatworms) and protostomes of the lophotrochozoan lineage (mollusks and annelids), and non-chordate deuterostomes (echinoderms and hemichordates) (Figure 8; Senatore et al., 2013). A separate duplication event in Exon 31 of the Ecdysozoan lineage creates different alternative calcium-selective (EEEE) and sodium-selective (EEKE) NALCN pores that are retained in myriapods (includes centipedes, millipedes) and chelicerates (includes Arachnids like mites and ticks) (Figure 8; Senatore et al., 2013). NALCN channels are well described as a cation leak conductance that play roles in generating rhythmic behaviors in invertebrates and vertebrates (Cochet-Bissuel et al., 2014), while also highly curious channels in retaining both calcium selective and sodium selective pore structures in many non-vertebrates (Senatore et al., 2013).
Cav3 T-type channels have a HFS site in the selectivity filter that resembles calcium channels (EEEE), but notably different, and universally EEDD, shortened in carbon chains of the carboxylate side chain residues (glutamate (E) to aspartate (D) in Domains III and IV) compared to Cav1 and Cav2 channels (Perez-Reyes et al., 1998). The most calcium-selective, Cav3 T-type channel, Cav3.1, is more sodium permeable (∼1 Na+ per 5 Ca2+) (Shcheglovitov et al., 2007; Senatore et al., 2014) than Cav1 and Cav2 calcium channels (∼1 Na+ per 1,000 Ca2+), suggesting that the HFS site of EEDD and other contributing pore selectivity filter residues, support a less calcium selective pore in Cav3 T-type channels. Invertebrates generate a high sodium selectivity in their Cav3 T-type channels resembling Nav1 channels through alternative splicing of a novel cysteine-enriched L5II extracellular turret coded in exon 12 (Senatore et al., 2014).
Nav, Cav and NALCN channels possess a universal set of HFS site configurations with calcium- or non-selective pores with electronegative residues in Domains II and III (EEEE, EDEE, EEDD, DEEA, DEES, DEEG, and DEET) and sodium-selective pores with a lysine in Domains II (DKEA, DKEG, and EKEE) or Domain III (DEKA and EEKE). More variable HFS pores are found in the 4 × 6 TM representatives outside of the animal/fungi supergroup of Unikonts (lifeforms with one flagella), such as diatoms, brown algae, oomycetes, cilates and dinoflagellates, and within branches of the Bikonts (lifeforms with two flagella), such as the green algae (chlorophytes and prasinophytes) (Verret et al., 2010). Approximately half of these non-animal representatives possess a HFS site resembling a calcium-selective pore (NEEE, EEEE, DDDD, and EEDE) (e.g., see representatives in Figure 7) and the rest of the pores are characterized by a pattern that represents experimentation and divergence. These pores may resemble a sodium-selective channel DDKD or possess HFS sites that are unlike any animal representative (e.g., RSDD, RADD, SESE, TDEE, and TEND) (Verret et al., 2010).
The outermost position “Na” (HFS site +4) is populated to form a negative-charged ring in the uppermost pore in many of the pore domains of eukaryotic sodium and calcium channels (Shen et al., 2018; Figure 4). The negatively charged aspartate (D) of the “Na” site in Domain IV is a frequently altered residue in animals adapted for resistance to pore-blocking, Nav1 channel toxin tetrodotoxin (TTX) (see TTX insensitive Nav1.4, Figure 4; Feldman et al., 2012). TTX is a bacterial toxin produced commonly by Vibrio alginolyticus which exists symbiotically within many discovered hosts, including notably pufferfish and many other fish species, tubellarian flatworms, blue-ringed octopus, western newt, toads, sea stars, angelfish, polyclad flatworms, Chaetognath arrow worms, nemertean ribbon worms and xanthid horseshoe crabs (Chau et al., 2011). A striking example of adaptation in the sodium channel pore is illustrated in the TTX-resistance of garter snakes (Thamnophis sirtalis) in districts where toxic prey newts (Taricha granulosa) are present, but not in the same species of garter snake in neighboring districts where these newts are absent (Geffeney et al., 2005). Similar residue changes in the “Na” position also provides for resistance to the analog saxitoxin (STX), contributing to the paralytic shellfish toxin, produced in toxic algal blooms in marine environments (Terlau et al., 1991). The many marine invertebrates that are completely resistant to TTX or STX, can comes at a cost of a lowered sodium ion selectivity in their Nav1 sodium channels (Grigoriev et al., 1996). TTX is a primary tool for the discrimination of Nav1 channels currents in doses ranging from the nanomolar to micromolar concentrations (Toledo et al., 2016). Nonetheless, the outer pore-blocking mechanism of TTX or STX or the micro-conotoxins (μ-CTXs) from venomous cone snails, is more the exception than the rule to how animal toxins regulate Nav1 channels. The vast majority of animal toxins are gating modifiers which take advantage of the greater structural variance outside of the outer pore of different Nav1 channels (Kalia et al., 2015). Another form of blockade are the frequency-, use- or state-dependent drugs that inhabit the aqueous vestibule within the channel pore, at higher affinities in particular channel states, such as when the inactivation gate is closed (Hille, 2001). Nav1 channels possess a much smaller aqueous vestibule within the pore for harboring drugs (e.g., local anesthetics like lidocaine) (Shen et al., 2017) compared to Cav1 channels which possess a more accommodating pocket within the pore for harboring drugs (e.g., nifedipine, verapamil, diltiazem) (Wu et al., 2016). Both the bacterial Nav channels and eukaryotic Nav and Cav channels possess side-portals or “fenestrations” between domains, providing an access pathway of small hydrophobic drugs to the inside of the pore without required passage through the pore selectivity filter during channel pore opening (Catterall and Swanson, 2015).
Single cell eukaryotes, like diatoms generate an animal-like, rapid action potential spike of a few ms in length from a resting membrane potential of ∼-68 mV (Taylor, 2009). The action potential in diatoms is generated with a fast inward current carried by sodium and calcium ions, peaking at -∼ 20 mV, and recovering quickly from inactivation (τ = ∼7 ms) (Taylor, 2009). While diatoms functionally generate an animal-like sodium and calcium current (Figure 9A), structurally it can only be derived from a 1x6TM bacterial sodium channel gene or a 4 × 6 TM template with an architecture distantly resembling calcium channels from animals (Verret et al., 2010; Figure 7). The diatoms belong to the Heterokonts (multiple different shaped flagella), which are separate from the Unikonts (single or no flagella), which are the lineage which includes the animals. The most basal extant representative of a recognizable homolog to animal genes is a 4 × 6 TM Nav2 homolog found in an apusozoan, represented by Thecamonas trahens (Figures 1, 8; Cai, 2012). Apusozoans are Unikonts that lies outside the Opisthokonts (bearing a posterior flagella) that includes the choanoflagellates, animals and fungi (Cavalier-Smith and Chao, 2010). Choanoflagellates (posterior flagella; possessing ciliated collars like sponge choanocytes) are closer relatives to the animals and fungi than the apusozoans. Choanoflagellates are represented by S. rosetta which possess a Nav2 homolog, SroNav2 (Mehta, 2016), but also have expressible homologs for Cav1 and Cav3 channels too (Figure 1). Expression of SroNav2 channel from this extant representative of an earliest branching single-celled organism, reveals a non-selective ion channel, equally passing Na+, K+, or Ca2+ ions, and also possesses extremely slow gating kinetics (Figure 9A; Mehta, 2016). Gating of a typical sodium channel from the classical squid giant axon is akin to mammalian neurons is rapid, enabling action potential spikes of 1–2 ms in length (Figure 9A; Hodgkin and Huxley, 1952). SroNav2 is more than tenfold slower, taking more than 10 ms to fully activate, with a slow inactivation decay (τ of >50 ms), requiring ∼ more than 1/2 s to completely inactivate, and ∼12 s to fully recover from inactivation in a whole cell patch clamp recording of currents recorded in HEK293T cell lines (Mehta, 2016).
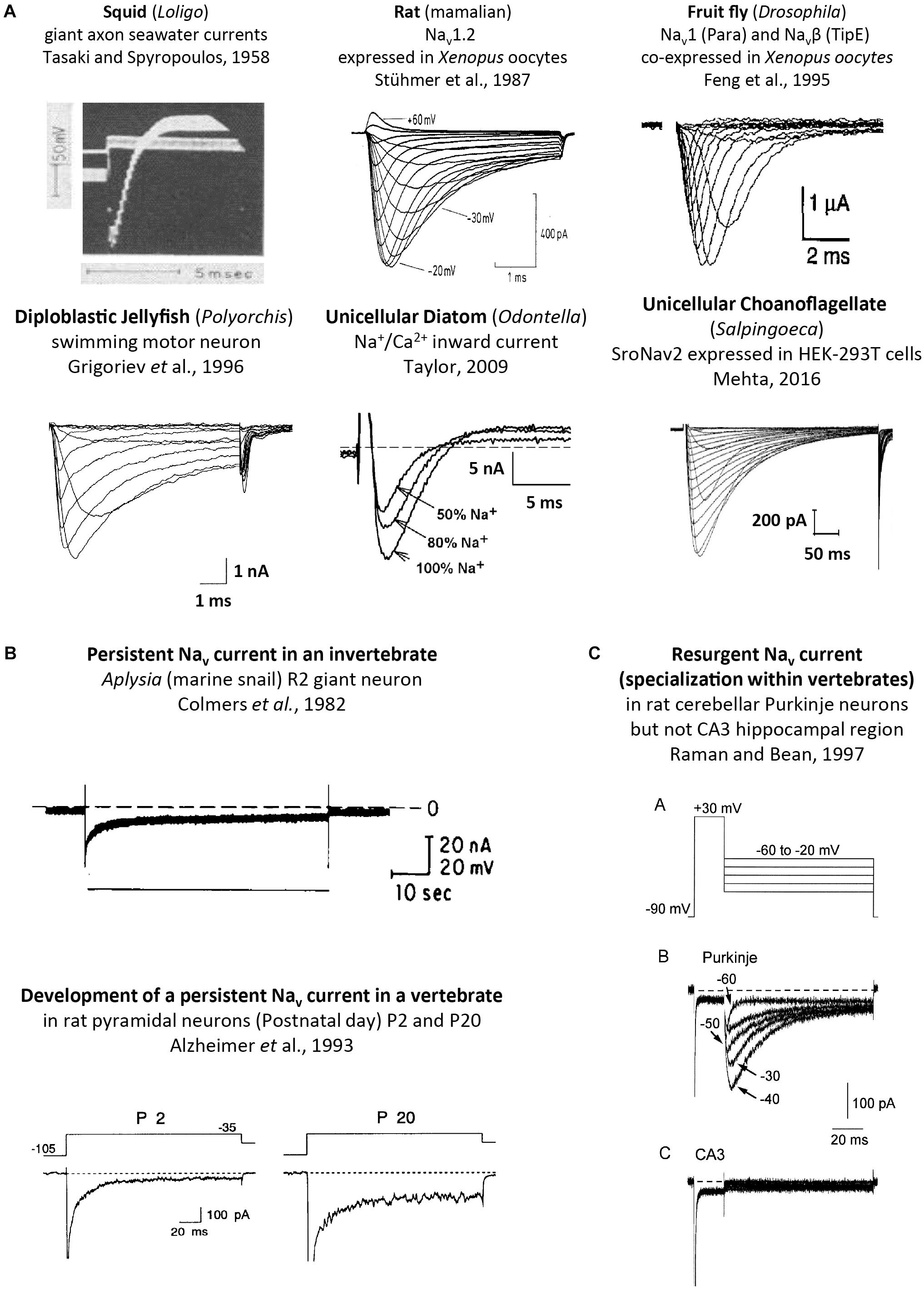
FIGURE 9. Sodium ion containing currents recorded in voltage-clamp studies of cells or expressed genes derived from single cell eukaryotes, invertebrates and vertebrates. (A) Squid, rat, fruit fly, jellyfish, diatom and choanoflagellates, reproduced with permission from Tasaki and Spyropoulos (1958), Stühmer et al. (1987), Feng et al. (1995), Grigoriev et al. (1996), Taylor (2009), Mehta (2016). (B) Persistent sodium currents in both invertebrates (top panel) and vertebrate neurons (bottom panel), reproduced with permission from Colmers et al. (1982) and Alzheimer et al. (1993), respectively. (C) Resurgent sodium currents, a property associated only with particular vertebrate neurons, reproduced with permission from Raman and Bean (1997).
Nav2 homologs have also been recorded from arthropods such as cockroach (Zhou et al., 2004), honeybee (Gosselin-Badaroudine et al., 2016) and fruit fly (Zhang et al., 2011) as well as sea anemone (a cnidarian) (Gur Barzilai et al., 2012), the latter of which has four Nav2 channel homologs. Single Nav2 channel homologs have been identified in vertebrates species, including cartilaginous fish (Australian ghostshark), lobed-finned fish (coelacanth), and jawless fish (lampreys) (Zakon et al., 2017). All expressed Nav2 homologs possess slow gating kinetics, and are non-selective channels passing calcium and sodium ions (Gosselin-Badaroudine et al., 2016). The Nav2 channel from honeybee is unique in generating mostly a calcium current, contributed by a smaller-sized sodium current (Gosselin-Badaroudine et al., 2016). The honeybee Nav2 channel may be mostly a calcium current but does not possess the nearly exclusive (1,000:1) calcium over sodium ion selectivity characteristic of Cav1 or Cav2 calcium channels (Hille, 2001).
Nav2 channels are represented from single cell choanoflagellates to vertebrates mostly as a single gene, but it is not a required gene in every species. Nav2 channel genes appear to be completely lacking in yeast, flatworms, nematodes, chelicerates (within the arthropods) and clitellates (within the annelids), and also lacking in many vertebrates, including ray-finned fishes and tetropods, including mammals (Figure 8). The absence of either sodium (Nav2 and Nav1) channel genes in different animal species is in contrast with calcium channels (Cav1, Cav2, and Cav3) which are omni-present, and not known to be secondarily lost in any animal group.
A possible function associated with Nav2 channels has been inferred from gene knockdowns in fruit fly (Drosophila), which possess a diminished ability to smell (Kulkarni et al., 2002), and a hyperkinetic mobility that is intensified by heat shock and starvation (Zhang et al., 2013). Nav2 channels have increased in gene number beyond a single homolog in urochordates such as Ciona intestinalis (two genes) ctenophores, sponges and placozoans (two genes), and as many as four genes in anthozoan or scyphozoan cnidarians (Figure 8). The diversification of Nav2 channels particularly within animals lacking true nervous systems (ctenophores, sponges and placozoans) or possessing vestigial nervous systems (tunicates), is suggestive of an expansion of roles for Nav2 filling of niches in the absence of animals with a mature nervous system. All vertebrate Nav2 channels identified in non-teleost fish, for example are only expressed outside of the nervous system (Zakon et al., 2017).
Unicellular organisms (yeast, protozoans) and the earliest diverging multicellular animals, such as ctenophores (comb jellies), sponges and placozoans (Trichoplax adhaerens) lack Nav1 channels (Figure 8). These early branching lifeforms such as sponges (Leys, 2015) or placozoans (Senatore et al., 2017) have a “toolkit” of many of the building blocks for nervous systems, without possessing eu-metazoan nervous tissue. Ctenophores have a nervous system, but are lacking most of the classical transmitter receptors and ion channels, and other neuronal elements common to the eu-metazoan nervous system template (Moroz and Kohn, 2016). Inward currents generating membrane excitability containing calcium ions as a charge carrier is limited because of competing roles of calcium ions as an exquisitely sensitive intracellular messenger. Calcium ions are toxic to cells when levels rise too high and calcium ions have a propensity to precipitate with phosphate (as bone matrix) (Hille, 2001). Free concentrations of intracellular calcium is necessarily kept at very low (nM) within cells by intracellular calcium buffering, compartmental storage of calcium in intracellular organelles, and extrusion to the cell exterior (Hille, 2001). Sodium ions are relatively inert, maintained at high (mM) concentrations within cells, and serves as a major ion in generating osmotic pressure (Hille, 2001) and as a workhorse in the secondary active transport of hydrogen ions or amino acids (Hille, 2001). A key step in the appearance of nervous systems, was the amino acid changes in the HFS site and surrounding residues, allowing for the sodium-selectivity of Nav1 channels, and an ability to exploit the steep, calcium ion free electro-chemical gradient, for generating rapid, mostly inert, action potential spikes along nervous tissue.
The cnidarians are the simplest eu-metazoans with Nav1 channel genes and they also are the simplest organism to possess a true nervous systems (Spafford et al., 1998). Cnidarians have a simple body plan of two germ layers (diploblastic), are lacking a coelom, and are radially symmetrical animals without cephalization (a brain localized in the anterior position) (Zapata et al., 2015). The Nav1 sodium channels in these basal species possess motor neurons which regulate the pulsating contractions of the jellyfish bell during swimming, are almost indistinguishable from those of the giant axons of the Atlantic squid or frog sciatic nerve, in their rapid millisecond gating and high sodium selectivity (Grigoriev et al., 1996). Sodium-selective Nav1 channels remain as a singleton gene in most invertebrates with functional currents limited in detection to nervous tissue, and lacking in functional protein expression in heart muscle or striated (skeletal-like) muscle or glands. mRNA coding for invertebrate Nav1 channels can be detected outside the nervous system, e.g., (Hong and Ganetzky, 1994; Gosselin-Badaroudine et al., 2015), but sodium currents have never been resolved outside the nervous system in known protostome invertebrates from the diverse lophotrochozoans to the ecdysozoans, e.g., (Hagiwara et al., 1964; Mounier and Vassort, 1975; Salkoff and Wyman, 1983; Brezina et al., 1994; Collet and Belzunces, 2007; Senatore et al., 2014). The primary expression of Nav1 sodium channel, “para” is in insect nervous systems, where Drosophila flies with Para locus mutations exhibit a temperature-sensitive paralyzes from nerve conduction blockade (Loughney et al., 1989). A niche left from a lack of Nav1 sodium channels outside of the nervous system can be filled with a sodium-selective Cav3 T-type channel isoform, generated by the unique alternative splicing in the extracellular turret upstream of the Domain II pore in most invertebrates (Senatore et al., 2014). Presence of an alternative 12a exon engenders Cav3 T-type channels with sodium selectivity (Figure 8), and is the only splice isoform found in the molluscan heart. The exon 12a containing T-type sodium channel isoform is almost ubiquitously present isoform in all eu-metazoans with organ-containing body cavities (coelomates), including some flatworms (pseudocoelomates) (Senatore et al., 2014).
Vertebrates underwent an expansion of Nav1 channel gene numbers to ten alongside the dramatic changes in body plan that include a greater sophistication of tissues, with different Nav1.x isoforms expressed at high density in particular tissues such as brain (e.g., Nav1.6), heart (e.g., Nav1.5) and skeletal muscle (Nav1.4) (Zakon et al., 2017).
A loss of Nav1 sodium channel genes is also common in non-vertebrate animals. For example, Nav1 sodium channels are present in free living flatworms (e.g., Schmidtea mediterranea) but are lacking in parasitic flatworms (Schistosoma). Nav1 channels are also lacking in nematodes, approximately half of animals in this phyla which are parasitic (Zhang, 2013; Figure 8). Nematodes lack both Nav2 and Nav1 channels, and possess simple, vestigial nervous systems that may related to their often obligate parasitism of host species (Zhang, 2013; Figure 8). All-or-none action potentials of body wall muscles requires inward Na+ influx in nematodes (Liu et al., 2011), but it is likely derived from other sources such as sodium-selective Cav3 T-type channels, in lieu of Nav2 and Nav1 channels (Senatore et al., 2014). Deuterostomes such as the echinoderms (e.g., Strongylocentrotus purpuratus) and their relatives the hemichordates (e.g., Saccoglossus kowalevskii) have sparse, radially symmetrical nervous systems and also lack Nav1 channel genes (Figure 8). In comparison, genes coding for all three calcium channels (Cav1, Cav2, Cav3) and NALCN is present in every known extant, eu-metazoan to date (Senatore et al., 2016), while the rise and fall of Nav1 channel genes (from zero to ten) appears to relate to a species’ necessity for sodium dependent action potentials, a vital feature of metazoan nervous systems (Figure 8).
The heritage of the first Nav1 channels emerging from Nav2 channel common ancestors is evident in the shared genomic structures of Nav1 and Nav2 channel genes. 20 intron splice sites are homologously shared in Nav1 and Nav2 channels, and these are all the intron splice sites spanning the conserved pore and voltage sensor domains (Figure 10). The same conservation of intron splice sites is paralleled within the calcium channel family, where Cav1 and Cav2 channels possess 31 shared intron splice sites. Both the Nav1 and Cav2 channels emerged from primordial Nav2 and Cav1 channel ancestors under a selection pressure for the evolution of specialized nervous tissue. Emergent roles of Nav1 and Cav2 channels appeared for fast conduction along nerve axons, and secretion of neurotransmitters between nerve synapses, respectively. The pairs of ion channel homologs in the most basal extant organisms known including Nav2/Nav1 in cnidarians (Gur Barzilai et al., 2012), or Cav1/Cav2 channels in placozoan, Trichoplax adhaerens (Senatore et al., 2016), are not easily distinguished from each other, as one would expect in extant organisms which are phylogenetically closer to the common ancestor containing the first Nav1 channels or Cav2 channels.
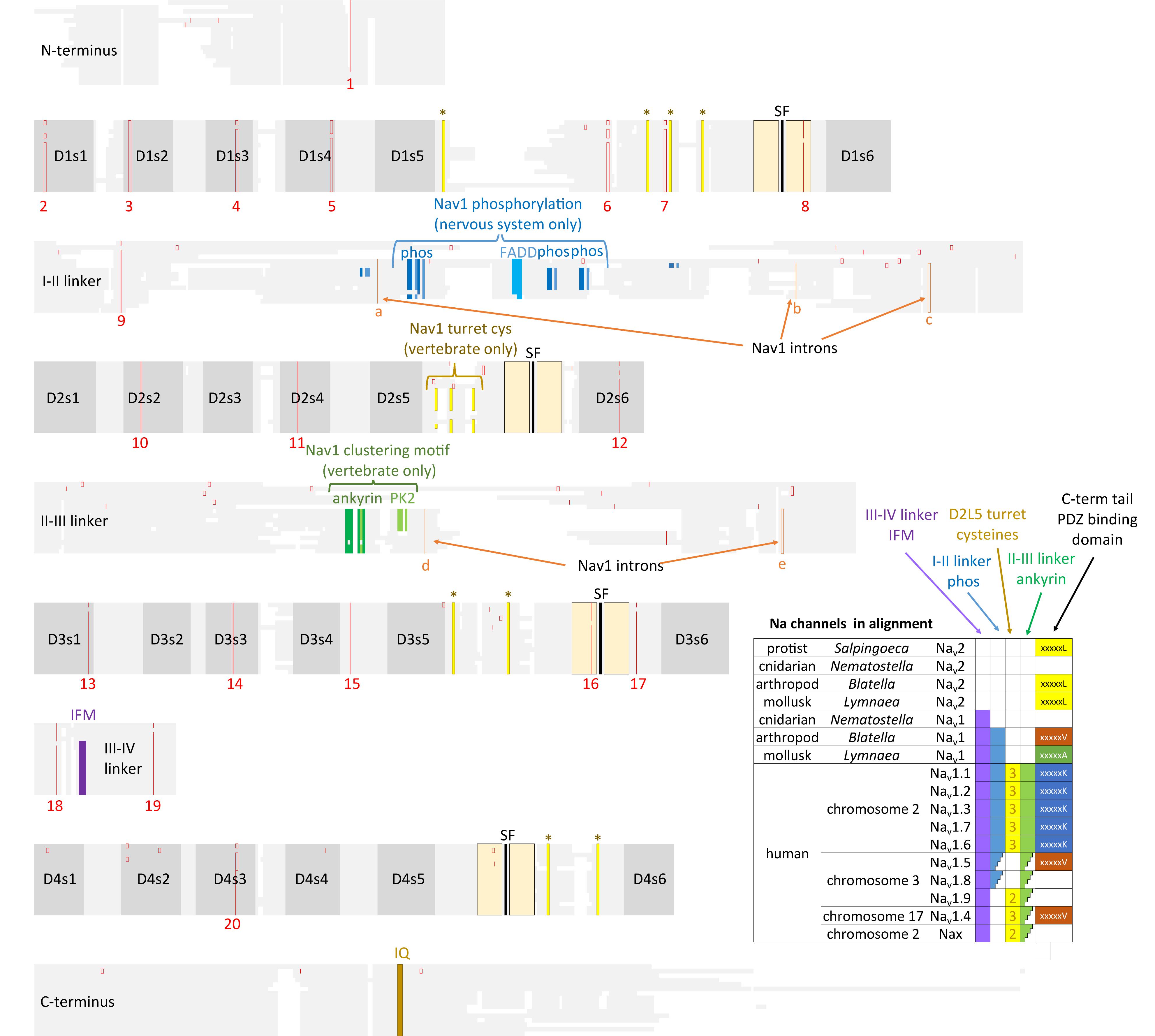
FIGURE 10. Amino acid alignments illustrating the conservation of twenty intron splice sites (red vertical lines) shared between Nav2 channels and Nav1 channels. The close kinship between the Nav2 channel class only found in non-vertebrates and the classical Nav1 channels is evident in the shared genomic structure. Nav1 differ primarily from Nav2 channels in containing three additional intron splice sites (a–c) in the I-II linker and two splice sites (d,e) in the II-III linker. Extra exons in the I-II linker provides exons coding for conserved phosphorylation sites within invertebrate Nav1 and vertebrate homologs (Nav1.1, 1.2, 1.3, 1.6, and 1.7) expressed in central nervous systems. Vertebrate, but not invertebrate Nav1 channel homologs contain three extra L5II turret cysteines, and an ankyrin binding site that contribute to the localized clustering of Nav1 channels in vertebrate Axon Initial Segments and nodes of Ranvier (Hill et al., 2008). Nav1 channels possess an IFM motif and surrounding residues within the III-IV linker which contribute to the fast inactivation (West et al., 1992). Nav2 and Nav1 channels possess a conserved proximal C-terminus containing an IQ motif required for calmodulin binding (Ben-Johny et al., 2014, 2015). Different C-terminal motifs containing PDZ binding domains are possessed in sodium channels. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
A defining feature that ubiquitously separates Nav1 and Nav2 channels is the lysine residue in Domain II (cnidarians) and III (all others metazoans) of the HFS site in the selectivity filter that confers the high sodium selectivity of Nav1 channels (Heinemann et al., 1992; Schlief et al., 1996). With substitution of the glutamate residue for a lysine residue DKEA or DEKA HFS site in Nav1 for the glutamate (E) in the DEEA selectivity filter of Nav2 channel of single-cell choanoflagellate, S. rosetta, the Nav2 channel transforms from being non-selective for calcium and sodium ions to exclusively selective for sodium ions (Mehta, 2016), resembling the Nav1 channels found in animals with nervous systems.
Sodium channels expanded from one gene (the condition in invertebrates) to ten vertebrate genes by gene duplication (Figure 1), and this heritage can be traced by their linkage to HOX (homeotic) developmental genes (Lopreato et al., 2001). Vertebrate sodium channels Nav1.1, Nav1.2, Nav1.3 and Nav1.7, are expressed primarily in central and peripheral nervous systems and are coded on human chromosome number 2 (linked to HOX D gene) (Lopreato et al., 2001). Nav1.6 is the primary sodium channel found in vertebrate myelinated axons at Nodes of Ranvier, is coded on chromosome number 12, and is linked to HOX C gene (Lopreato et al., 2001). Nav1.8 and Nav1.9 are expressed in dorsal root ganglia, and Nav1.5 is primarily expressed in heart muscles (Lopreato et al., 2001). Nav1.5, Nav1.8, and Nav1.9 are clustered on chromosome 3, linked to Hox A (Lopreato et al., 2001). Nav1.4 is specialized to vertebrate skeletal muscle, and is found on chromosome 17, linked to Hox B (Lopreato et al., 2001). A tenth vertebrate sodium channel, Nax, (located on chromosome 2) is the most diverged in sequence, is lacking voltage-sensitive gating and plays a likely role as a putative salt sensor in the subfornical organ (Noda and Hiyama, 2015).
A consequence of chaining of domains together in the four domain structure of the 4 × 6 TM channels is that it generates opportunities for divergence and specialization within the differing voltage-sensor and pore domains. It creates unique extensions of differing extracellular turrets or intracellular linkers linking the four domains, and a trimming down to singleton, asymmetrically arranged, amino- and carboxyl-termini in 4 × 6 TM channels compared to the four N- and C- terminal tails in the repeating four subunits composing the 1x6TM potassium channels (Long et al., 2005) or bacterial sodium channels (Payandeh et al., 2011) or (Figure 2). Cyclic nucleotide-gated potassium channel (CNGK) channels are a 4 × 6 TM channel representative amongst the potassium channel superfamily. CNGK channels lack the uniquely different extracellular turret extensions in differing domains or asymmetrical intracellular linkers between domains that are characteristic of 4 × 6 TM sodium or calcium or NALCN channels (Bonigk et al., 2009). CNGK channels possess a pseudo-symmetry of four nearly identically repeating domains, linked together by equally short and repeating, cytoplasmic linkers and extracellular regions. There is a similarly positioned cyclic nucleotide binding domain (CNBD) within each of their four domains [Bonigk et al., 2009; Fechner et al., 2015]. The CNGK channels appears to be ancient like the lineage of 4 × 6 TM calcium channels with representatives in single cell choanoflagellates (Fechner et al., 2015), in marine invertebrates (Bonigk et al., 2009) and in vertebrates (fish) (Fechner et al., 2015). There appears to be no obvious adaptations or specializations in the four domains of CNGK channel homologs, despite their apparent ancient history. CNGK channels contain four repeating subunits linked together, but only the sodium, calcium and NALCN channel lineage adopted a particular asymmetrical architecture.
The most constrained of the cytoplasmic linkers is the III-IV linker which is always shorter than the I-II and II-III linkers in 4 × 6 TM channels (Figure 2B), and limited in size to mostly 53 to 57 amino acids (Stephens et al., 2015). The exception is the unusual optional splicing of inserts (e.g. exon 25C) in invertebrate and vertebrate Cav3 T-type calcium channels (Senatore and Spafford, 2012). Also highly conserved is a calcium-calmodulin binding IQ domain within the first 155 aa downstream of the proximal C-terminus in Cav1, Cav2, Nav1 and Nav2 channels, but is absent in Cav3 T-Type channels or NALCN channels (Figures 7, 10). The calmodulin binding in the C-terminus puts a calcium ion sensor within the pathway of calcium entry through the channel pore of calcium channels (Lee et al., 1985; Zuhlke et al., 1999) and resides in a similar position in sodium channels (Mori et al., 2000; Potet et al., 2009; Sarhan et al., 2009) to potentially regulate channel gating. The conserved III-IV linker and the C-terminal domain join to form a globular domain in the cryo-electron microscopy of single calcium channel (Wu et al., 2015, 2016) and sodium channel (Shen et al., 2017; Yan et al., 2017) nanoparticles.
The globular domain formed by the III-IV linker and proximal C-terminus generates a qualitatively different regulation of gating in different calcium and sodium channels, based on sequence differences. A highly conserved IQ motif among L-type channels contributes to a canonical calcium-sensitive inactivation shared amongst invertebrate Cav1 and vertebrate Cav1.2 and Cav1.3 channels (Taiakina et al., 2013). This conservation extends to single cell protozoans, including Paramecium L-type currents which possesses a calcium-calmodulin dependent inactivation (Brehm et al., 1980). A less conserved proximal C-terminal IQ motif engenders a calcium-dependent facilitation of vertebrate Cav2.1 and Cav2.2 channels involving calmodulin kinase activation (Christel and Lee, 2012). A calcium dependent facilitation and relief of G-protein βγ subunit inhibition by repetitive nerve activity are uniquely featured in vertebrate synaptic Cav2 calcium channels (Zamponi and Currie, 2013), and such modulation is lacking in homologous invertebrate Cav2 channels (Dunn et al., 2018). Invertebrates Cav2 channels are regulated by G-protein coupled receptor signaling that involves a non-voltage dependent form of regulation of activity involving phosphorylation by SRC kinase (Dunn et al., 2018) or cAMP dependent protein kinase A (Huang et al., 2010).
All sodium channels have a highly conserved proximal C-terminal IQ motif, including Nav2 channels in single cell, choanoflagellate Nav2 (Figure 10; Mehta, 2016). The proximal C-terminal IQ motif contributes to a calcium-sensitive regulation that is variable amongst different vertebrate Nav1 channels (Mori et al., 2000; Potet et al., 2009; Sarhan et al., 2009), with a potential secondary binding site for calmodulin in the III-IV linker (Sarhan et al., 2009, 2012). Cav3 T-Type channels differ from other calcium and sodium channels in possessing an equivalent high affinity binding site for calmodulin at their unique “gating brake” located in the proximal I-II linker at homologous location where the accessory Cavβ subunits normally associate with Cav1 and Cav2 channels (Chemin et al., 2017).
Nav1 sodium channels are differentiated from calcium channels in possessing a very rapid gating that is required for generating high frequency trains of action potential spikes milliseconds in length, observed in nervous systems (Hodgkin and Huxley, 1952). Faster sodium channel gates contribute to a faster overshooting action potential, while slower and higher voltage-activated calcium channels contribute to a plateau potential following the overshoot, such as in the vertebrate ventricular action potential (Hille, 2001).
A key player which differentiates fast Nav1 channel gating is in the III-IV linker (Ulbricht, 2005) (Figure 10). Glycine residues flanking the III-IV linker create a flexible hinge, permitting movement of the III-IV linker, and rapid inactivation, within milliseconds of sodium channel opening (Kellenberger et al., 1997). A key “IFM” motif in the center of the III-IV linker, serves as a critical “latch” for the “hinged lid” (West et al., 1992), The IFM motif forms a plug inserting in the corner surrounded by the outer S4-S5 and inner S6 segments in repeats III and IV in three dimensional structure, indicating a likely allosteric blocking mechanism for fast inactivation gating in Nav1 channels (Yan et al., 2017). The “IFM” motif, and its surrounding conserved sequence of the III-IV linker is absent in Nav2 channels, and appears to be a critical structural feature responsible for the slower inactivation gating of Nav2 channels in choanoflagellates, according to our mutagenesis studies (Mehta, 2016). Other fine tuning for fast channel gating in Nav1 channels include likely changes to the speed of voltage-sensor gating charge movements and the coupling efficiency of the voltage-sensors to pore gates (Capes et al., 2013), as well as roles for the proximal C-terminus which includes a calcium sensor in a calmodulin-binding IQ motif (Christel and Lee, 2012), found in sodium and calcium channels.
The region of greatest sequence divergence between Nav2 and Nav1 channels is in the cytoplasmic I-II and II-III linkers, and these differences appear to relate to the increasing specialization of Nav1 channels for more sophisticated signaling, that reaches its apex in vertebrate tissues (Figures 10, 11). Cytoplasmic regions linking the four domains created in the first 4 × 6 TM channels, diverged and specialized for the more complex and diverse tissue environments for different genes isoforms as numbers grew from singleton gene homologs of the simplest single cell organisms to the ten sodium and ten calcium channel genes available in the vertebrate body plan.
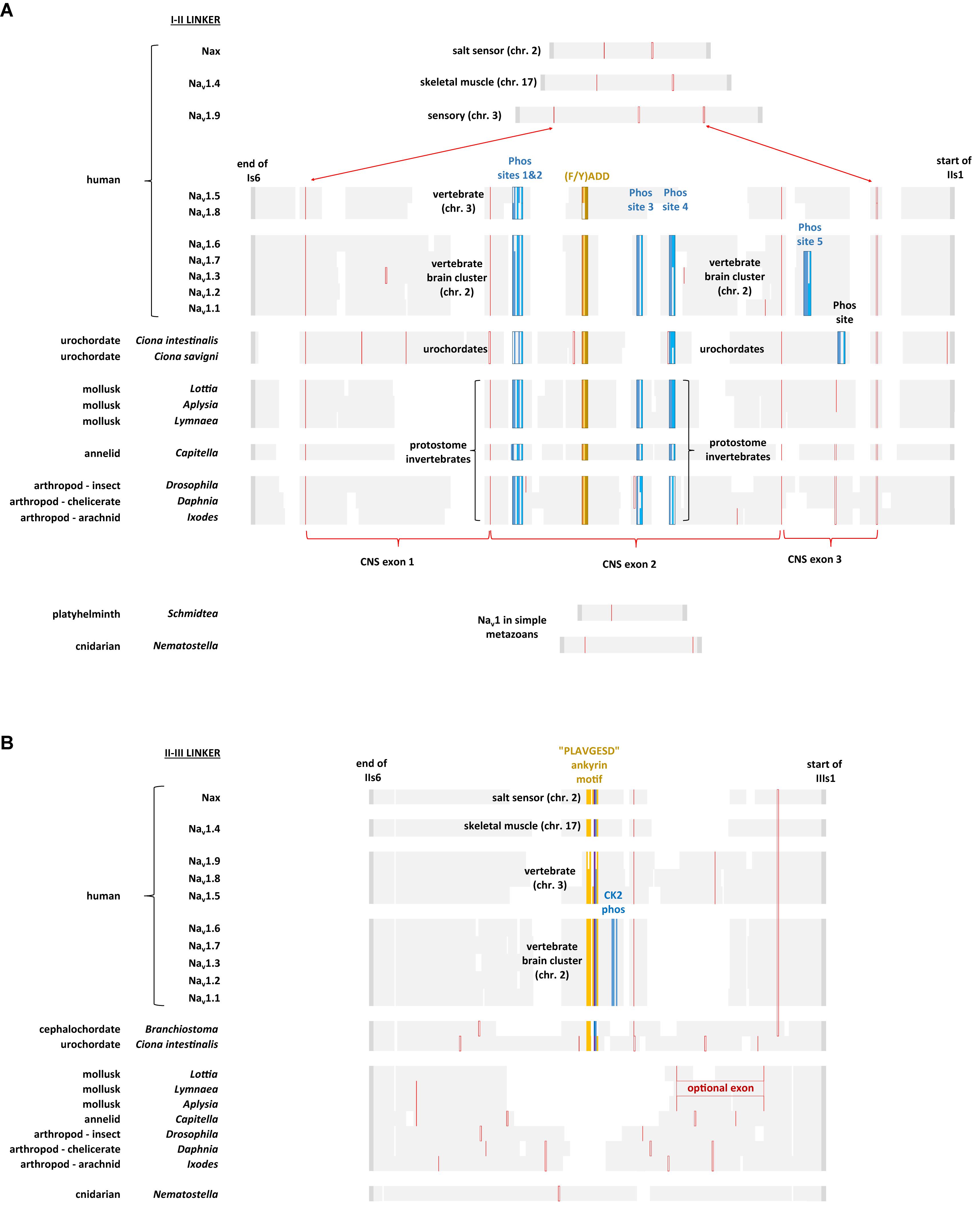
FIGURE 11. Alignment of amino acid sequences coding for cytoplasmic I-II linker (A) and II-III linker (B) of invertebrate and vertebrate Nav1 channels. Note the expanded size of the cytoplasmic I-II linker in protostome invertebrate Nav1 and vertebrate brain cluster channels (Nav1.x, where x = 1,2,3,6, and 7) containing four conserved phosphorylation sites at serine residues. Two of these four phosphorylation sites are subject to phosphorylation in mammalian Nav1.2 channels by cAMP dependent protein kinase A (serine: 573 and 623) (Smith and Goldin, 1996) and one of the four is a protein kinase C regulatory site (serine 576) (Cantrell et al., 2002). Other vertebrate sodium channels, such as Nav1.5 and Nav1.8 retain two out of four of these phosphorylation sites (serine: 573 and 576) while Nav1.4, Nav1.9 and Nax lack any of these phosphorylation sites. While all chordate Nav1 channels possess the ankyrin G binding motif, only the nervous system specific cluster of vertebrate sodium channels (Nav1.1, Nav1.2, Nav1.3, Nav1.6, and Nav1.7) possess a CK2 serine/threonine kinase phosphorylation site downstream of the ankyrin G binding site in the II-III linker, that greatly increases affinity and required for the formation of stable ankyrin G – sodium channel complexes upon phosphorylation (Xu and Cooper, 2015). Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
With increasing animal complexity, there is adjustments and fine-tuning of cellular function within signaling networks involving neurotransmitters, hormones and other modulators to up-and-down regulate cellular activity and excitability. One of the cellular targets for regulation is Nav1 channels, both in their control of gene expression and changes to ion channel gating. The Nav1 channel in protostome invertebrates, but not those in simple metazoans (e.g., cnidarians, flatworms) possess a greatly expanded cytoplasmic I-II linker (coded in exon 11), containing four conserved phosphorylation sites at serine residues (Figure 11A) (Fux et al., 2016). Two of these four phosphorylation sites are subject to phosphorylation in mammalian Nav1.2 channels by cAMP dependent protein kinase A (serine: 573 and 623) (Smith and Goldin, 1996, 1997) and one of the four is a protein kinase C regulatory site (serine 576) (Cantrell et al., 2002) (Figure 11A). Phosphorylation by protein kinase A (Li et al., 1992) or protein kinase C (Numann et al., 1991), downregulates sodium channel activity. These phosphorylation sites are shared amongst the single Nav1 channel homolog expressed in nervous systems of protostome invertebrates and in the cluster of related sodium channels specifically expressed in the vertebrate central and peripheral nervous systems, Nav1.1, Nav1.2, Nav1.3, Nav1.6, and Nav1.7 (Figure 11A; Fux et al., 2016). Other vertebrate sodium channels, such as Nav1.5 and Nav1.8 retain two out of four of these phosphorylation sites (serine: 573 and 576) while Nav1.4, Nav1.9, and Nax lack any of these phosphorylation sites (Figure 11A). The conservation of sequences for neurotransmitter and G protein coupled regulation of sodium channel activity supports the hypothesis that Nav1 channels evolved in conjunction with appearance of nervous tissue, with a closer structural kinship of the nervous system specific (Nav1.1, Nav1.2, Nav1.3, Nav1.6, and Nav1.7) sodium channels in vertebrates, with the singleton nervous system-specific Nav1 sodium channel within the protostome invertebrates.
Further specialization of sodium channels for nervous systems is evident in the emergence of unique sequences in chordate Nav1 channels, related to the specific targeting and clustering of sodium channels to Axon Initial Segments (AIS) and Nodes of Ranvier (Hill et al., 2008). The singleton Nav1 channels in basal chordates including the urochordates (e.g., C. intestinalis) and cephalochordates (Branchiostoma floridae) and the ten vertebrate Nav1 channels, share a conserved anchoring motif that binds ankyrin G, an adapter protein that tethers the Nav1 channels and KCNQ2/3 potassium channels to the spectrin-actin cytoskeleton (Hill et al., 2008). The anchoring motif is a 9 amino acid sequence V/A-P-I/L-A-x-x-E-S/D-D, where x can be any amino acid (Hill et al., 2008). The ankyrin G binding motif is located in the II-III linker, a cytoplasmic linker that is ∼50% shorter than the I-II linker of vertebrate neuronal Nav1 channels (Figures 10, 11B). While all chordate Nav1 channels possess the ankyrin G binding motif, only the nervous system specific cluster of vertebrate sodium channels (Nav1.1, Nav1.2, Nav1.3, Nav1.6, and Nav1.7) possess a CK2 serine/threonine kinase phosphorylation site downstream of the ankyrin G binding site in the II-III linker (Figures 10, 11B), that greatly increases affinity and required for the formation of stable ankyrin G – sodium channel complexes upon phosphorylation (Xu and Cooper, 2015).
The presence of an AnkyrinG-specific Na channel clustering motif correlates with the greater specialization and differentiation of axons from dendrites in chordates, often lacking in invertebrates. Nav1 channels in chordates cluster at the proximate ends of axons in AIS, creating fidelity in nerve impulse generation, by ensuring that depolarizing synaptic inputs safely reach threshold necessary for firing action potentials. Another major invention is in structures that increases the speed of nerve propagation. This takes the form of unique invertebrate giant axons (mm in diameter) of the Atlantic squid, for example, which lowers the internal resistance to ionic current flow, and creates a large surface area for sodium channels to populate and contribute to rapid nerve impulses (Hartline and Colman, 2007). An alternative process, and favorable to the limited space for neurons within a cranium and energy efficiency, is myelination, where insulation layers of lipid wrappings of myelin surround axonal membranes. Myelination is omnipresent in craniates, vertebrates which includes all the bony fish and contemporary sharks (gnathostomes) and most basally appearing in a placoderm-like ancestor (Hill et al., 2008). Wraps of insulating myelin, increases conduction speeds by shielding axons from current loss, by increasing transmembrane resistance to current flow, and decreasing the need for input currents to charge the membrane capacitance. Ankyrin-G specific Nav1 channel are clustered in periodic bare regions lacking myelination (Nodes of Ranvier), provides a voltage-boost to perpetuate the nerve signal between myelinated internodes lacking Nav1 channels (Freeman et al., 2016). Nav1 channel localization is inhibited under myelinated regions, and quickly populate along axonal membranes of freshly denervated nerve fibers (England et al., 1990). The close and inverse relationship between the localization of Nav1 channels and myelination in axons, relates to an axonal specialization in nervous systems of craniates. The jawless fishes, represented by Petromyzon marinus, appear to be the transition group, which rely on squid-like giant axons instead of myelinated axons of the craniates for fast axonal conduction velocities (Teravainen and Rovainen, 1971; Bullock et al., 1984), yet possess the Ankyrin-G specific targeting motif for Nav1 channel clustering (Hill et al., 2008) which is lacking in invertebrates.
It should be noted though that myelination isn’t strictly a vertebrate innovation, with some invertebrate lineages possessing myelinated axons (internodes) and bare nodes resembling that of vertebrate axons. Examples of myelinated invertebrate axons are found in crustaceans (malacostraca, including cecapod shrimp and copepods) and annelids (polychaetes and oligochaetes) (Hartline and Colman, 2007). There are also nodal-like clusters of Nav1 sodium channels along unmyelinated fibers of the marine snail, Aplysia californica (Johnston et al., 1996). Although lacking a vertebrate-like Ankyrin G dependent clustering mechanisms, different invertebrate groups must possess a parallel mechanisms to cluster Nav1 channels, given that invertebrates possess clustered sodium channels in unmyelinated axons and vertebrate-like Nodes of Ranvier within myelinated axons. Populations of sodium channels strategically localized along axons is necessary to generate the high fidelity, unidirectional action potentials that are found in both invertebrate and vertebrate nervous systems (Freeman et al., 2016).
The very ends of the C-termini of particular classes of calcium and sodium channels, like other ion channels or receptors (e.g., Kv channels or NMDA receptors), can possess conserved sequence motifs that are known to associate with different classes of PDZ domains containing proteins, which are known to play supporting roles in the anchoring of ion channels or receptors to proteins associated with the cytoskeleton (Figure 12; Lee and Zheng, 2010; Hui et al., 2013). Nav2 channels, including homologs in the earliest branching representatives, apusozoan (Thecamonas) and choanoflagellate (Salpingoeca) possess a common PDZ binding motif: xxxxL (Figure 12). The xxxxL motif is shared amongst most Cav1 channel homologs too including invertebrate (Cav1) and vertebrate (Cav1.1 to Cav1.4), suggesting that the C-terminal xxxxL motif may have appeared in the first common ancestors of Nav2/Cav1 channels in a single cell choanoflagellate-like ancestor (Figure 12). Cav2 channels, evolved as a separate calcium channel class from Cav1 channels to service the presynaptic release of transmitters at nerve synapses, and possess a different C-terminal motif of xxxWC that is shared amongst invertebrate Cav2 and vertebrate Cav2.1 and Cav2.2 channels (Figure 12).
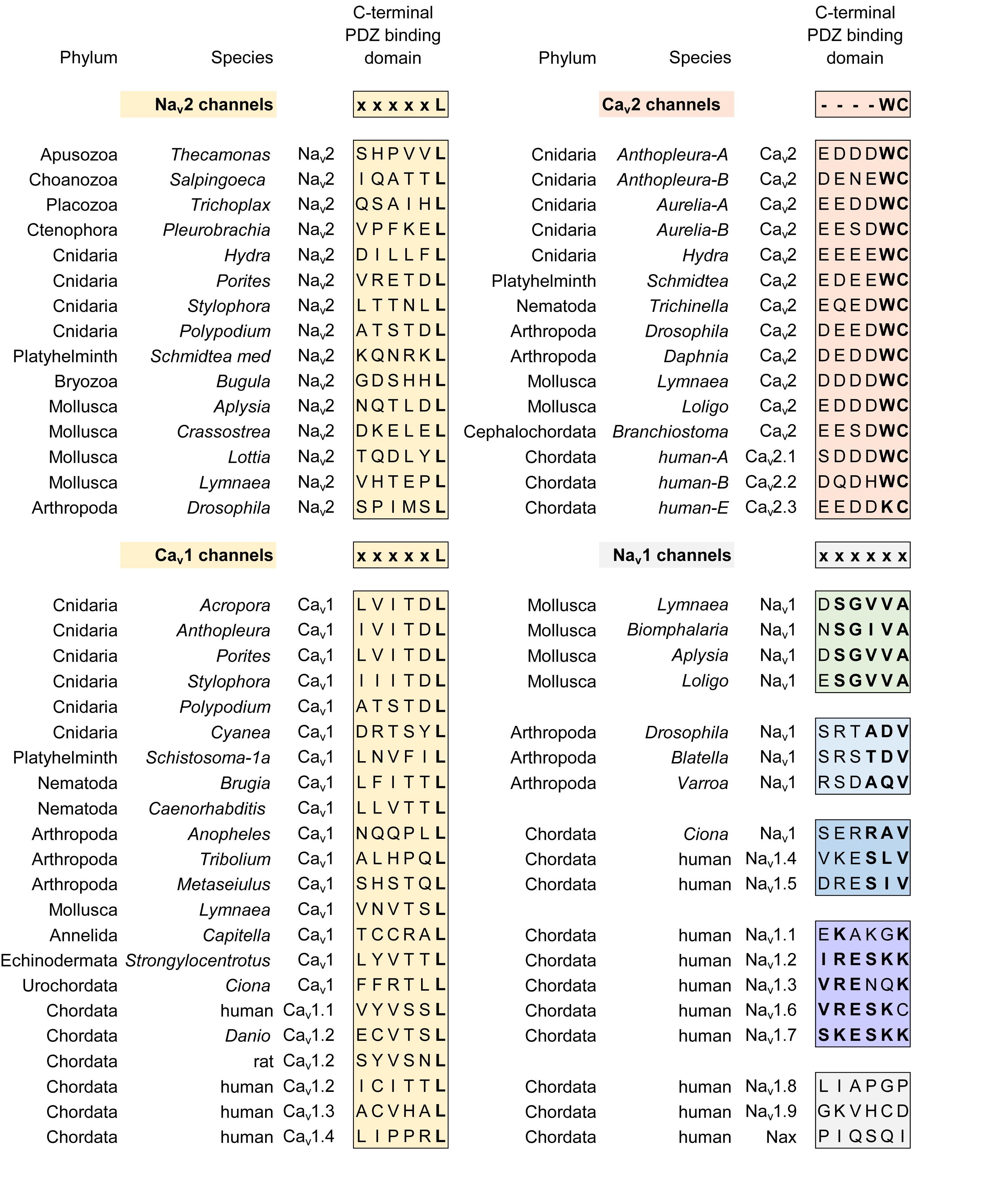
FIGURE 12. Alignment of amino acid sequences coding for the C-terminal ends of calcium channels (Cav1 and Cav2) and sodium channels (Nav2 and Nav1) where there are conserved sequence motifs associated with binding to different classes of PDZ domain containing proteins. There is a surprising consistency in a xxxxL motif conserved between Nav2 and Cav1 channels (yellow color), both of which constitute the basal complement of high voltage-activated calcium channels and sodium channels present in genomes of single cell choanoflagellates such as S. rosetta. The Nav1 and Cav2 channels, which are likely derived from Nav2 and Cav1 channels, respectively, possess different sets of PDZ binding domain motifs (denoted by orange, green, blue, purple colors). PDZ binding motifs uniquely diverge within the Nav1 channel class where molluscan, arthropod, and different human sodium channels associate with separate classes of PDZ domain containing proteins, contributing to their specific localization within their unique cellular environments. The “SLV/SIV” C-terminal PDZ binding motif in mammalian Nav1.4 and Nav1.5, for example, aids in the complexing of these Nav1 channels to syntrophins and SAP97, for the cellular targeting within skeletal and cardiac muscle, respectively. Cav3 channels lack conservation in a C-terminal PDZ domain. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).
Different Nav1 homologs of invertebrate groups and different vertebrate Nav1 isoforms, have variable C-terminal PDZ binding motifs than the xxxxL motif common to the Nav2 and Cav1 channels. The different PDZ binding motifs likely reflect a difference in PDZ domain containing proteins that supporting the scaffolding of Nav1 channels to the cytoskeleton in different invertebrate groups and amongst different vertebrate Nav1 isoforms. Arthropod Nav1 channels have a common PDZ binding motif of xxA/TDV, while molluscan Nav1 channels have a common PDZ binding motif of SGVVA (Figure 12). Vertebrate Nav1 channel PDZ binding motifs have a common motif that loosely resembles R/K ESKK in nervous system related sodium channels (Nav1.1, Nav1.2, Nav1.4, Nav1.6, and Nav1.7) while skeletal muscle specific sodium channel (Nav1.4) and cardiac-specific sodium channel (Nav1.5) have a PDZ binding motif of R/K ES L/I V (Figure 12). A role for PDZ binding motifs in Nav channels has been illustrated for the last three residues (SIV) of the PDZ domain binding motif in the association of syntrophins and SAP97 to Nav1.5 and Kv channels. PDZ domain containing scaffolding proteins are required for the protein targeting of Nav1.5 and Kv channels to the lateral cardiomyocyte membrane specifically, without influencing their location at intercalated disks (Shy et al., 2014). This C-terminal PDZ binding motif also appears to protect Nav1.5 channels from proteosomal degradation within cardiomyocytes (Shy et al., 2014). C-terminal PDZ binding motifs are not always conserved in different homologs of Nav1, Nav2, Cav1, and Cav2 channels and a consensus sequence is notably absent at the C-terminal ends of Cav3 channels. The conservation of C-terminal PDZ binding motifs amongst different calcium and sodium channel classes is suggestive of their importance, but their lack of ubiquity suggest that they are not obligatory factors involved in the regulation of sodium and calcium channels.
An apparently obligatory association for different Nav1 sodium channel representatives is the accessory, Navβ subunits of Nav1 channels. The Navβ subunits of Nav1 channels are non-homologous in structure between different animal groups, suggestive of differing protein interactions and cellular environments for sodium channel localization in tissues of different animal groups (see Table 1). The differing Navβ subunits for Nav1 channels in different animal groups is consistent with a similar divergence of C-terminal PDZ binding motifs amongst differing invertebrate Nav1 channels (e.g., arthropod versus mollusk) and vertebrate Nav1 channels (nervous vs. muscle tissue specific isoforms).
Most ion channels are complexed with auxiliary subunits that can serve a chaperoning role to help traffic and target ion channels to specific membrane locales and prevent their proteosomal degradation (Dolphin, 2012; Calhoun and Isom, 2014). Auxiliary subunits also modulate channel gating, such as the kinetic rate of gating or shifts in the voltage-sensitivity of activation or inactivation properties of voltage-gated channels (Dolphin, 2012; Calhoun and Isom, 2014). An important context for assessing the subset of sodium channel specific accessory subunits, is to compare them with the accessory subunits of calcium channels. High voltage-activated Cav1 and Cav2 calcium channels form a multi-protein complex with Cavα2δ and Cavβ subunit homologs (Dolphin, 2012). The high homology of these accessory subunits is evident in the ability to mix and match the four mammalian Cavβ1 to Cavβ4 subunits with singleton Cavβ homologs from invertebrate species, like jellyfish (Jeziorski et al., 1999) or snail (Dawson et al., 2014) or squid (Kimura and Kubo, 2003), and these different accessory subunits will alter the biophysical properties or expression features of invertebrate or mammalian Cav1 or Cav2 channels. Both an accessory Cavα2δ and Cavβ subunit are required for expressing calcium channels in vitro, even for the Cav1 homolog from single cell choanoflagellate, S. rosetta (Mehta, 2016). The importance of the Cavα2δ and Cavβ subunit for calcium channels is evident in the ubiquity of these accessory subunit in genomes of known species with calcium channel homologs, including the basal representative in S. rosetta, while the Cavα2δ and Cavβ subunits are not found in apusozoan Thecamonas trahens or similar choanoflagellate, Monosiga brevicollis, which possess an Nav2 but are lacking a calcium channel homolog in their genome (Liebeskind et al., 2011). Cavβ subunits possess conserved guanylate kinase (GK), and Src-homology 3 (SH3) domains, which are featured in the related family membrane-associated guanylate kinase (MAGUK) family (Buraei and Yang, 2010). MAGUKs serve as protein scaffolds in the organization of multiprotein complexes at specialized membrane locales such as synapses or other cellular junctions (Zhu et al., 2016).
Known sodium channel β subunits (Navβ) resemble cell adhesion molecules (CAMs) but are not structural homologs amongst different animal groups, unlike the calcium channel α2δ and β, which are homologous, cross-compatible subunits from single cell choanoflagellates to humans. A comparison of features of accessory sodium and calcium subunits is provided in Table 1.
The four vertebrate Navβ1 to Navβ4 subunits resemble CAMs of the immunoglobulin (Ig) superfamily with an extracellular Ig V-fold domain (Isom et al., 1995). These Navβ subunits play critical roles in nerve outgrowth, the migration of neurites, and trafficking of Nav1 channels to specialized positions in nerve membranes (O′Malley and Isom, 2015). CAM domains of Navβ1 and Navβ2 subunit isoforms are reported to contact and adhere CAM domains of Navβ subunits and with other CAM domain containing proteins within particular cells or between CAM domains linking adjacent cells (Malhotra et al., 2000). Navβ facilitates the recruitment of Ankyrin-G to points of cell–cell contact, and tether vertebrate nervous system-specific Nav1 channels to AIS and nodes of Ranvier (Chen et al., 2004).
The equivalent to the vertebrate Navβ subunits were discovered as TipE (Temperature-induced paralytic E) gene (Hodges et al., 2002) and closely related TipE-homologous genes (TEH1-4) (Derst et al., 2006) in fruit fly (Drosophila). Fly mutants of the accessory Navβ subunit TipE generates a similar temperature-sensitive paralysis resembling the mutant para (short for paralytic) phenotype resulting from impaired nerve conduction in fruit flies lacking their functional Nav1 channel (para) homolog. Double mutants of TipE and para genes generates a lethal phenotype in fruit flies (Ganetzky, 1986). Like the vertebrate Navβ subunits, co-expression of TipE is necessary to reconstitute the fast channel kinetics of the fruit fly Nav1 channel, and greatly enhances the membrane expression of fruit fly Nav1 channels in vitro (Warmke et al., 1997). TipE and TEH homologs don’t resemble vertebrate Navβ subunits and more resemble the structure of accessory subunits to BK (Slo, maxi-K, KCa1.1) channels, with two transmembrane domains separated by an extracellular loop containing disulphide bridges (Derst et al., 2006). Extracellular loops in some TEH homologs can resemble CAMs within calcium-binding epidermal growth factor (EGF)-like domains (Derst et al., 2006). Recently, we discovered a family of calcium-binding, CUB domain-contain proteins in Lymnaea pond snails, identified as Navβ subunits in the protein complex bound to Nav1 sodium channels (Fux et al., 2016). Like the vertebrate Navβ subunits which can function as CAMs, CUB domain containing proteins have various roles such as in developmental patterning, tissue repair, angiogenesis and cell signaling (Gaboriaud et al., 2011). The most closely related vertebrate CUB domain protein to snail Navβ subunits is neuropilin-1 and neuropilin-2, which possess CAMs and serve as co-receptors for class-3 semaphorins, which contribute to axon guidance in the vertebrate nervous system (Nakamura and Goshima, 2002). Other CUB domain containing proteins can be auxiliary subunits, such Sol-1 and Sol-2 that are auxiliary subunits of AMPA glutamate receptors in nematode, Caenorhabditis elegans (Wang et al., 2012). Similar Neto1 and Neto2, CUB domain containing proteins, co-assemble with vertebrate glutamate receptors (NMDA and kainite) to modulate their channel gating and enhance their membrane trafficking (Wyeth et al., 2014). The common thread between the Navβ subunits is cellular adhesion involving extracellular motifs, but in different structural forms within CUB domains (gastropod snails) (Fux et al., 2016), EGF-like domains (insects) (Derst et al., 2006), or Ig V-fold (vertebrates) (Isom et al., 1995; see Table 1).
Navβ subunit homologs possess a highly restricted distribution within animal sub-phyla. We could identify close homologs to the CUB-domain containing Navβ subunits from giant pond snail (Lymnaea stagnalis) in a closely related pulmonate freshwater pond snail, Biomphalaria glabrata. We could not find Navβ subunit homologs outside of pulmonate snails, including the gastropod snail genomes of Aplysia californica (California sea hare, marine snail) or Lottia gigantea (giant owl limpet), or any non-snail species within the Phylum Mollusca (Fux et al., 2016). Similarly, Tip-E and TEH subunit homologs first identified in fruit flies, are limited to insect and crustacean genomes within the Phylum Arthropoda, and not identifiable, within other arthropods such as chelicerates or myriapods (Li et al., 2011). The vertebrate Navβ subunits are similarly restricted in distribution to the subphylum Vertebrata, and absent for example, in genomes of non-vertebrate chordates.
Analogous Navβ subunits likely appeared alongside Nav1 channels within the first nervous systems, in a common ancestor to extant cnidarians. The more ancestral sodium channel isoform, Nav2, likely requires no accessory Navβ subunit. Nav2 from single cell choanoflagellate, S. rosetta expresses well in a vertebrate surrogate cell (such as HEK-293T cell lines or Xenopus oocytes) without a co-expressed accessory Navβ subunit (Mehta, 2016). Almost all invertebrate Nav1 channels do not reach detectable levels for electrophysiological recording using standard in vitro expression system in vertebrate cells, outside of insect [fruit fly (Warmke et al., 1997), mosquito (Du et al., 2013), honeybee (Gosselin-Badaroudine et al., 2016), bumblebee (Wu et al., 2017), or cockroach (Du et al., 2009a)] or chelicerate (varroa mite) (Du et al., 2009b). The generally poor expressibility of invertebrate Nav1 channels outside their native environment and their requirements for very different structures representing Navβ subunits, is consistent with the highly variable composition of invertebrate nervous systems, and the appearance of Nav1 channels - protein complexes adapted for the unique neuronal environments within different animal sub-phyla. This is different from calcium channels where all the invertebrate Cav1 (Dawson et al., 2014), Cav2 (Huang et al., 2010) and Cav3 (Senatore and Spafford, 2010) homologs are readily expressible in vitro, such as those derived from pond snail, L. stagnalis.
Sodium channels rapidly expand in gene numbers from the solitary Nav1 channel gene in most invertebrates and non-vertebrate chordates to nine vertebrate genes, Nav1.1 to Nav1.9 (Alexander et al., 2017). Vertebrate isoforms contribute to the primary action potential generation and conduction in neurons (Nav1.1, Nav1.2, Nav1.3, Nav1.6, Nav1.7, and Nav1.8) or in skeletal or heart muscle, respectively (Nav1.4 or Nav1.5) (Alexander et al., 2017). Most of the vertebrate channel isoforms (Alexander et al., 2017), (with notable exceptions) generate native currents with characteristics resembling the typical invertebrate (Cole and Moore, 1960; Hille, 1968; Grigoriev et al., 1996; Spafford et al., 1996; Warmke et al., 1997; Mehta, 2016) sodium currents (e.g., midpoint of activation = -30 mV to -20 mV, a rapid inactivation decay (τ = ∼1 ms) and possess a nanomolar drug sensitivity to tetrodotoxin) (Figure 9A and Table 2). Each vertebrate sodium channel gene varies in their pattern of expression during development, and in their tissue and sub-cellular localization (Alexander et al., 2017). Nav1.3, for example, specializes as an embryonic and neonatal sodium channel that re-expresses in adults after nerve axotomy or injury (Hains and Waxman, 2007). An L858H sodium channel mutation reported in Nav1.7 generates a depolarized resting membrane potential in human patients with erythromelalgia (Rush et al., 2006). Expression of the higher threshold sodium channel (Nav1.8) creates a hyper-excitability phenotype in sympathetic neurons and its absence generates a hypo-excitability phenotype in dorsal root ganglion neurons (Rush et al., 2006). The higher threshold Nav1.8 channels uniquely drives a greater excitability in the more depolarized membranes of sympathetic neurons in the diseased condition (Rush et al., 2006). The lower threshold sodium channels normally responsible for the action potential upstroke (Nav1.1 and Nav1.6) are refractory or inactivated due to the more depolarized resting membrane potential of the diseased condition in dorsal root ganglion neurons (Rush et al., 2006). The differing consequences in this example of a single mutation in human Nav1.7 illustrate how the cellular phenotype critically depends on the unique properties of the ensemble of sodium channel genes expressed in different neuron types (e.g., sympathetic neurons vs. dorsal root ganglion subtypes).
There are examples of apparent redundancies suggesting that the expansion of vertebrate gene isoforms does not always fulfill a requirement for functional diversity. Nav1.6 is the dominantly expressed, adult isoform in AIS and nodes of Ranvier (Van Wart and Matthews, 2006). Despite the critical role that it plays, Nav1.6 is remarkably well-compensated by Nav1.2 channels expressed in neocortical pyramidal neurons in response to the tissue-selective knockdown of Nav1.6 (Katz et al., 2018). The only difference in Nav1.6-null compared to wild type neocortical neurons is a reduction in the size of the persistent sodium current (Katz et al., 2018).
“Persistent” or “late” sodium currents are residual (<5%), steady-state, sub-threshold sodium currents that are often generated from the incomplete inactivation of Nav1 channels after the fast transient sodium spike, or as a trickle of “window current” of open and non-inactivated Nav1 channels at rest (Crill, 1996). Persistent sodium currents can drive pace-making (Bevan and Wilson, 1999), burst firing (Azouz et al., 1996) or amplify excitatory post-synaptic potentials (Deisz et al., 1991). Persistent sodium currents have been identified across neurons of every major phyla (invertebrate and vertebrate) (Kiss, 2008) and vertebrate cardiac muscle (Moreno and Clancy, 2012) (see Figure 9B). Persistent sodium currents are readily generated with as little as a single amino acid change, presence of a gating modifying toxin or after co-expression of a Navβ subunit isoform in vertebrates which can influence the voltage-dependence or kinetics of inactivation of Nav1 channels (Qu et al., 2001; Aman et al., 2009; Savio-Galimberti et al., 2012).
A striking example of the independent evolution of persistent sodium currents is in the specialized Nav1.4 channel isoforms for the muscle-derived electric organs of weakly-electric fish, which can generate electrical pulses in excesses of 1 kHz for sensing their nocturnal environment and the electro-communication with other fish (Thompson et al., 2018). The electromotor neurons of weakly-electric fish can fire at a faster rate of any known animal neuron, using a persistent sodium current generated from the modification of its structural elements for inactivation (S4-S5 linker of Domain IV) of their Nav1.4 channels (Thompson et al., 2018).
A different type of inter-spike sodium current contributing to pace-making is the “resurgent” sodium current (Figure 9C) (Raman and Bean, 1997). The resurgent sodium current is likely an adaptation limited to vertebrates, reported for particular neuronal cell types including the cerebellum and brainstem, globus pallidus, the hippocampus and dorsal root ganglia (Lewis and Raman, 2014). The “resurgent” current flows through the same channels which generate the “transient” and “persistent” components of the Nav1 channel currents (Lewis and Raman, 2014; Figure 9C). The “resurgent” sodium current is characterized by a rise, peak and subsequent fall of sodium current during the repolarization phase of the action potential, after the action potential overshoot and repolarization of the membrane below zero mV (Figure 9C) (Raman and Bean, 1997). A fraction of the Nav1 channel pores are considered to be under an “open channel” blockade during membrane depolarization, with a visible “resurgent” current appearing at the rate of unblocking of channel pores by the “blocking particle” upon action potential repolarization, with a surge of inward current enhanced by the greater driving force for ion flow through Nav1 channels at the more hyperpolarized membrane potentials during repolarization (Lewis and Raman, 2014). One of the likely contributors as the “blocking particle” is the unique C-terminal insert in the vertebrate Navβ4 subunit which contains positively charged and hydrophobic residues that can reconstitute resurgent currents in neurons that do not exhibit an endogenous resurgent current (Grieco et al., 2002). Not all neurons containing resurgent sodium currents express Navβ4 subunit and co-expression of Navβ4 subunit alone does not reconstitute the resurgent sodium current in vitro (Lewis and Raman, 2014). There are likely other mechanisms alongside Navβ4 subunits which contribute to generating resurgent sodium currents in vertebrate neurons. The regulation by Navβ4 is reminiscent of the blockade of the inward-rectifying potassium channels by endogenous polyamines, namely spermine, as well as magnesium ions that plug the channel pore at positive potentials (Baronas and Kurata, 2014). It also has similarities in the adaptations of different accessory β subunits which mimic the positively charged and hydrophobic N-terminal “ball” of the “ball-and-chain”, contributing to the fast N-type inactivation in different voltage-gated potassium channels (Bentrop et al., 2001).
Teasing apart the different ion channel contributions in the electrical signaling of vertebrate tissues has relied on parallel approaches in analyzing expressed genes in surrogate expression systems (e.g., mammalian cell lines or Xenopus oocytes) (Tapper et al., 2003). While convenient, in vitro expression studies are usually insufficient in recapitulating the condition of the native cell such as the many gene splicing variants expressed natively in cells, complex of associated, accessory subunits, post-translational regulation, and other myriad of critical interactions required for ion channel subunits within specific subcellular locales of different tissues. Surrogate expression systems also can possess endogenous, contaminating sodium currents (He and Soderlund, 2010; Terhag et al., 2010). A parallel approach is to examine a simple eukaryotic model to better understand the functions of voltage-gated sodium and calcium channels, where there are limitations in gene complexity, such as in the choanoflagellates where there are only five gene homologs (Cav1, Cavα2δ, Cavβ, Cav3, and Nav2). From an evaluation of their structural and functional characteristics, it is evident that these five choanoflagellates genes are homologs of the 28 equivalent human sodium and calcium channel genes, packaged in a living organism consisting of a single eukaryotic cell.
The groundwork for the appearance of nervous systems and Nav1 channels is evident in the first multicellular organisms. Sponge and Trichoplax have no traditional nerve structures such as neurons or synapses, but do possess the building blocks for nervous systems including protein homologs for synaptic and scaffolding elements, and cell-cell communication elements that involves many of the classical ionic channels and neurotransmitter receptors of higher organisms (Moroz and Kohn, 2016). The coelenterates were considered a grouping of basal, radial symmetrical animals that superficially resembled each other with rudimentary diffuse nervous nets serving as nervous systems, including the cnidarians (hydra, corals, sea anemones, jellyfish, sea pens) and the ctenophores (comb jellies) (Zapata et al., 2015). Delving into genome comparisons between cnidarians and ctenophores has reveal that the genomic blueprints are starkly different, where ctenophores appear to have an early and parallel evolution to cnidarians and other bilateral animals (Moroz et al., 2014), and lack many of the classical neural elements, including major voltage-gated cation channel classes, including the classical Nav1 channels required for fast sodium channel spikes along nerve axons, and Cav2 and Cav3 calcium channels involved in mediating neurotransmitter secretion across nerve synapses or required for pace-making of action potentials, respectively. Single cell choanoflagellates, while more basal to the multicellular ctenophores, possess a complement of single homologs of the major sodium (Nav2) and calcium channel subunits (Cav1 and Cav3), plus contain accessory calcium channel subunits (α2δ, Cavβ) (Mehta, 2016). It is in the single cell choanoflagellates that we are likely able to resolve the structural and functional basis of which lead to the specialization of electrical signaling in voltage-gated cation channels (see summary flow chart, Figure 13). Species like S. rosetta contain both solitary, highly mobile life stages as well as colonial, and more sessile life stages (Dayel et al., 2011), where we can evaluate the fundamental underpinnings for the first appearances of 4 × 6 TM, voltage-gated sodium and calcium channels, which we mostly attribute to specialized roles in the rapid electrical and chemical signaling within multicellular organisms.
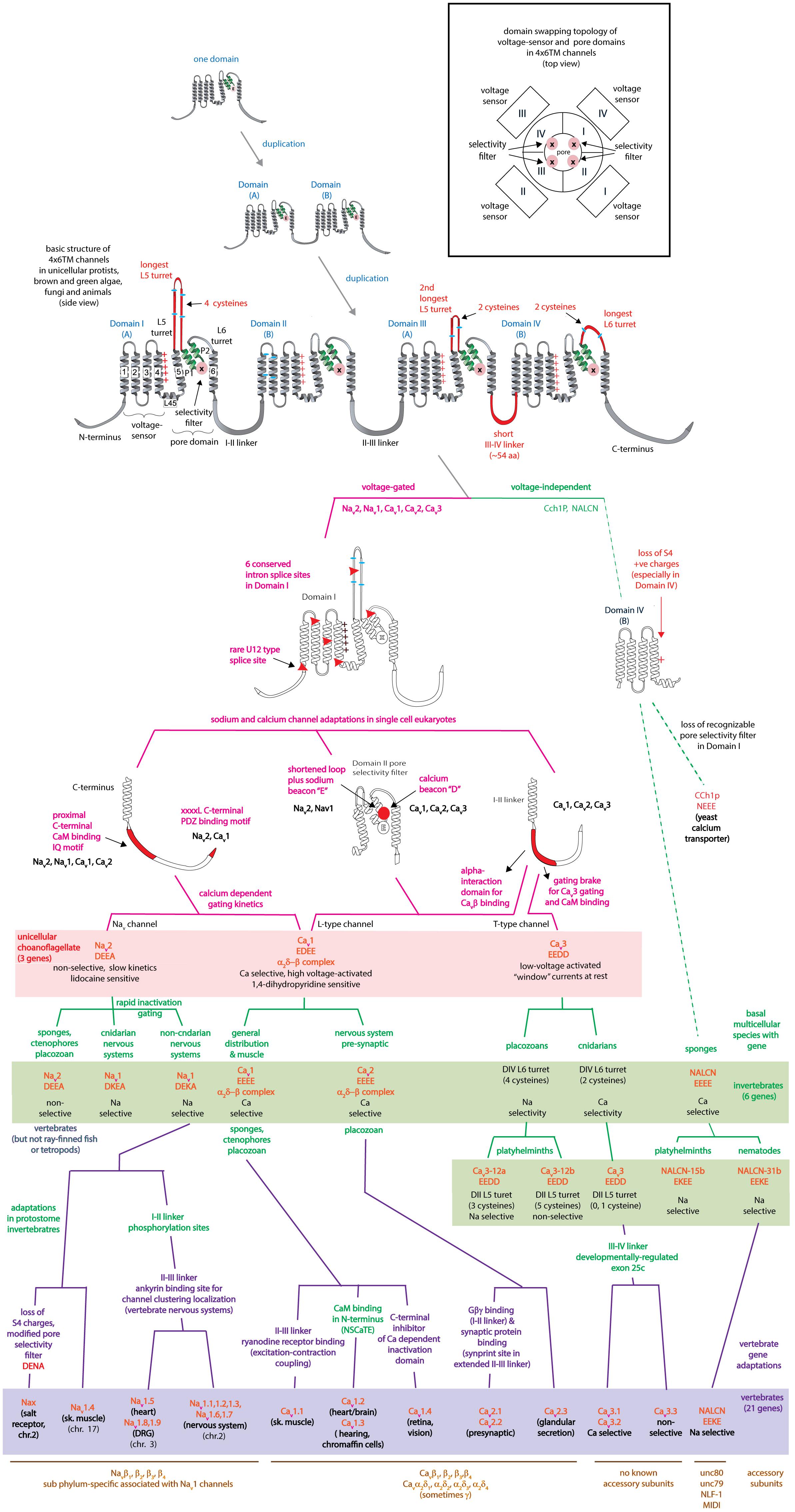
FIGURE 13. Flow chart illustrating progression of features evolving within 4 × 6 TM cation channels, including the three primary voltage-gated channels contained in single cell choanoflagellates (Nav2, Cav1, and Cav3) (pink color, 3 genes) and their diversification and specialization within protostome invertebrates (green color, 6 genes) and vertebrates (purple color, 21 genes).
JF, AM, and JM carried out the experiments for the work that this review is based on. JS wrote the text and made the figures for the manuscript.
JS was grateful to the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Heart and Stroke Foundation of Canada for support of operating grants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate the helpful feedback from Dr. Harold H. Zakon on an earlier draft of this manuscript.
Alexander, S. P., Striessnig, J., Kelly, E., Marrion, N. V., Peters, J. A., Faccenda, E., et al. (2017). The concise guide to pharmacology 2017/18: voltage-gated ion channels. Br. J. Pharmacol. 174(Suppl. 1), S160–S194. doi: 10.1111/bph.13884
Alzheimer, C., Schwindt, P. C., and Crill, W. E. (1993). Postnatal development of a persistent Na + current in pyramidal neurons from rat sensorimotor cortex. J. Neurophysiol. 69, 290–292. doi: 10.1152/jn.1993.69.1.290
Aman, T. K., Grieco-Calub, T. M., Chen, C., Rusconi, R., Slat, E. A., Isom, L. L., et al. (2009). Regulation of persistent Na current by interactions between beta subunits of voltage-gated Na channels. J. Neurosci. 29, 2027–2042. doi: 10.1523/JNEUROSCI.4531-08.2009
Apweiler, R., Hermjakob, H., and Sharon, N. (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8. doi: 10.1016/S0304-4165(99)00165-8
Azouz, R., Jensen, M. S., and Yaari, Y. (1996). Ionic basis of spike after-depolarization and burst generation in adult rat hippocampal CA1 pyramidal cells. J. Physiol. 492( Pt 1), 211–223.
Bagneris, C., Naylor, C. E., McCusker, E. C., and Wallace, B. A. (2015). Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J. Gen. Physiol. 145, 5–16. doi: 10.1085/jgp.201411242
Baronas, V. A., and Kurata, H. T. (2014). Inward rectifiers and their regulation by endogenous polyamines. Front. Physiol. 5:325. doi: 10.3389/fphys.2014.00325
Baycin-Hizal, D., Gottschalk, A., Jacobson, E., Mai, S., Wolozny, D., Zhang, H., et al. (2014). Physiologic and pathophysiologic consequences of altered sialylation and glycosylation on ion channel function. Biochem. Biophys. Res. Commun. 453, 243–253. doi: 10.1016/j.bbrc.2014.06.067
Ben-Johny, M., Dick, I. E., Sang, L., Limpitikul, W. B., Kang, P. W., Niu, J., et al. (2015). Towards a unified theory of calmodulin regulation (calmodulation) of voltage-gated calcium and sodium channels. Curr. Mol. Pharmacol. 8, 188–205. doi: 10.2174/1874467208666150507110359
Ben-Johny, M., Yang, P. S., Niu, J., Yang, W., Joshi-Mukherjee, R., and Yue, D. T. (2014). Conservation of Ca2 + /calmodulin regulation across Na and Ca2 + channels. Cell 157, 1657–1670. doi: 10.1016/j.cell.2014.04.035
Bennett, E. S. (2002). Isoform-specific effects of sialic acid on voltage-dependent Na + channel gating: functional sialic acids are localized to the S5-S6 loop of domain I. J. Physiol. 538, 675–690. doi: 10.1113/jphysiol.2001.013285
Bentrop, D., Beyermann, M., Wissmann, R., and Fakler, B. (2001). NMR structure of the ”ball-and-chain” domain of KCNMB2, the beta 2-subunit of large conductance Ca2 + - and voltage-activated potassium channels. J. Biol. Chem. 276, 42116–42121. doi: 10.1074/jbc.M107118200
Bevan, M. D., and Wilson, C. J. (1999). Mechanisms underlying spontaneous oscillation and rhythmic firing in rat subthalamic neurons. J. Neurosci. 19, 7617–7628. doi: 10.1523/JNEUROSCI.19-17-07617.1999
Bonigk, W., Loogen, A., Seifert, R., Kashikar, N., Klemm, C., Krause, E., et al. (2009). An atypical CNG channel activated by a single cGMP molecule controls sperm chemotaxis. Sci. Signal. 2:ra68. doi: 10.1126/scisignal.2000516
Boone, A. N., Senatore, A., Chemin, J., Monteil, A., and Spafford, J. D. (2014). Gd3 + and calcium sensitive, sodium leak currents are features of weak membrane-glass seals in patch clamp recordings. PLoS One 9:e98808. doi: 10.1371/journal.pone.0098808
Brehm, P., Eckert, R., and Tillotson, D. (1980). Calcium-mediated inactivation of calcium current in Paramecium. J. Physiol. 306, 193–203. doi: 10.1113/jphysiol.1980.sp013391
Brezina, V., Evans, C. G., and Weiss, K. R. (1994). Characterization of the membrane ion currents of a model molluscan muscle, the accessory radula closer muscle of Aplysia californica. III. Depolarization-activated Ca current. J. Neurophysiol. 71, 2126–2138. doi: 10.1152/jn.1994.71.6.2126
Bullock, T. H., Moore, J. K., and Fields, R. D. (1984). Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci. Lett. 48, 145–148. doi: 10.1016/0304-3940(84)90010-7
Buraei, Z., and Yang, J. (2010). The beta subunit of voltage-gated Ca2 + channels. Physiol. Rev. 90, 1461–1506. doi: 10.1152/physrev.00057.2009
Cai, X. (2012). Ancient origin of four-domain voltage-gated Na + channels predates the divergence of animals and fungi. J. Membr. Biol. 245, 117–123. doi: 10.1007/s00232-012-9415-9
Calhoun, J. D., and Isom, L. L. (2014). The role of non-pore-forming beta subunits in physiology and pathophysiology of voltage-gated sodium channels. Handb. Exp. Pharmacol. 221, 51–89. doi: 10.1007/978-3-642-41588-3_4
Cantrell, A. R., Tibbs, V. C., Yu, F. H., Murphy, B. J., Sharp, E. M., Qu, Y., et al. (2002). Molecular mechanism of convergent regulation of brain Na( + ) channels by protein kinase C and protein kinase A anchored to AKAP-15. Mol. Cell. Neurosci. 21, 63–80. doi: 10.1006/mcne.2002.1162
Capes, D. L., Goldschen-Ohm, M. P., Arcisio-Miranda, M., Bezanilla, F., and Chanda, B. (2013). Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J. Gen. Physiol. 142, 101–112. doi: 10.1085/jgp.201310998
Catterall, W. A., Goldin, A. L., and Waxman, S. G. (2005a). International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57, 397–409.
Catterall, W. A., Perez-Reyes, E., Snutch, T. P., and Striessnig, J. (2005b). International union of pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol. Rev. 57, 411–425.
Catterall, W. A., and Swanson, T. M. (2015). Structural basis for pharmacology of voltage-gated sodium and calcium channels. Mol. Pharmacol. 88, 141–150. doi: 10.1124/mol.114.097659
Cavalier-Smith, T., and Chao, E. E. (2010). Phylogeny and evolution of apusomonadida (protozoa: apusozoa): new genera and species. Protist 161, 549–576. doi: 10.1016/j.protis.2010.04.002
Chau, R., Kalaitzis, J. A., and Neilan, B. A. (2011). On the origins and biosynthesis of tetrodotoxin. Aquat. Toxicol. 104, 61–72. doi: 10.1016/j.aquatox.2011.04.001
Chemin, J., Taiakina, V., Monteil, A., Piazza, M., Guan, W., Stephens, R. F., et al. (2017). Calmodulin regulates CaV3 T-type channels at their gating brake. J. Biol. Chem. 292, 20010–20031. doi: 10.1074/jbc.M117.807925
Chen, C., Westenbroek, R. E., Xu, X., Edwards, C. A., Sorenson, D. R., Chen, Y., et al. (2004). Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J. Neurosci. 24, 4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004
Chojnacki, S., Cowley, A., Lee, J., Foix, A., and Lopez, R. (2017). Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553. doi: 10.1093/nar/gkx273
Christel, C., and Lee, A. (2012). Ca2 + -dependent modulation of voltage-gated Ca2 + channels. Biochim. Biophys. Acta 1820, 1243–1252. doi: 10.1016/j.bbagen.2011.12.012
Cochet-Bissuel, M., Lory, P., and Monteil, A. (2014). The sodium leak channel, NALCN, in health and disease. Front. Cell Neurosci. 8:132. doi: 10.3389/fncel.2014.00132
Cole, K. S., and Moore, J. W. (1960). Ionic current measurements in the squid giant axon membrane. J. Gen. Physiol. 44, 123–167. doi: 10.1085/jgp.44.1.123
Collet, C., and Belzunces, L. (2007). Excitable properties of adult skeletal muscle fibres from the honeybee Apis mellifera. J. Exp. Biol. 210, 454–464. doi: 10.1242/jeb.02667
Colmers, W. F., Lewis, D. V. Jr., and Wilson, W. A. (1982). Cs + loading reveals Na + -dependent persistent inward current and negative slope resistance region in Aplysia giant neurons. J. Neurophysiol. 48, 1191–1200. doi: 10.1152/jn.1982.48.5.1191
Crill, W. E. (1996). Persistent sodium current in mammalian central neurons. Annu. Rev. Physiol. 58, 349–362. doi: 10.1146/annurev.ph.58.030196.002025
Dawson, T. F., Boone, A. N., Senatore, A., Piticaru, J., Thiyagalingam, S., Jackson, D., et al. (2014). Gene splicing of an invertebrate beta subunit (LCaVbeta) in the N-terminal and HOOK domains and its regulation of LCaV1 and LCaV2 calcium channels. PLoS One 9:e92941. doi: 10.1371/journal.pone.0092941
Dayel, M. J., Alegado, R. A., Fairclough, S. R., Levin, T. C., Nichols, S. A., McDonald, K., et al. (2011). Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82. doi: 10.1016/j.ydbio.2011.06.003
Deisz, R. A., Fortin, G., and Zieglgansberger, W. (1991). Voltage dependence of excitatory postsynaptic potentials of rat neocortical neurons. J. Neurophysiol. 65, 371–382. doi: 10.1152/jn.1991.65.2.371
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic.Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Derst, C., Walther, C., Veh, R. W., Wicher, D., and Heinemann, S. H. (2006). Four novel sequences in Drosophila melanogaster homologous to the auxiliary Para sodium channel subunit TipE. Biochem. Biophys. Res. Commun. 339, 939–948. doi: 10.1016/j.bbrc.2005.11.096
Dolphin, A. C. (2012). Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat. Rev. Neurosci. 13, 542–555. doi: 10.1038/nrn3311
Doyle, D. A., Morais, C. J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., et al. (1998). The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science 280, 69–77. doi: 10.1126/science.280.5360.69
Du, Y., Nomura, Y., Liu, Z., Huang, Z. Y., and Dong, K. (2009a). Functional expression of an arachnid sodium channel reveals residues responsible for tetrodotoxin resistance in invertebrate sodium channels. J. Biol. Chem. 284, 33869–33875. doi: 10.1074/jbc.M109.045690
Du, Y., Nomura, Y., Luo, N., Liu, Z., Lee, J. E., Khambay, B., et al. (2009b). Molecular determinants on the insect sodium channel for the specific action of type II pyrethroid insecticides. Toxicol. Appl. Pharmacol. 234, 266–272. doi: 10.1016/j.taap.2008.10.006
Du, Y., Nomura, Y., Satar, G., Hu, Z., Nauen, R., He, S. Y., et al. (2013). Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc. Natl. Acad. Sci. U.S.A. 110, 11785–11790. doi: 10.1073/pnas.1305118110
Dunn, T. W., Fan, X., Ase, A. R., Seguela, P., and Sossin, W. S. (2018). The CaV2alpha1 EF-hand F helix tyrosine, a highly conserved locus for GPCR inhibition of CaV2 channels. Sci. Rep. 8:3263. doi: 10.1038/s41598-018-21586-5
Edel, K. H., Marchadier, E., Brownlee, C., Kudla, J., and Hetherington, A. M. (2017). The evolution of calcium-based signalling in plants. Curr. Biol. 27, R667–R679. doi: 10.1016/j.cub.2017.05.020
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ednie, A. R., and Bennett, E. S. (2012). Modulation of voltage-gated ion channels by sialylation. Compr. Physiol. 2, 1269–1301. doi: 10.1002/cphy.c110044
Ednie, A. R., Harper, J. M., and Bennett, E. S. (2015). Sialic acids attached to N- and O-glycans within the Nav1.4 D1S5-S6 linker contribute to channel gating. Biochim. Biophys. Acta 1850, 307–317. doi: 10.1016/j.bbagen.2014.10.027
England, J. D., Gamboni, F., Levinson, S. R., and Finger, T. E. (1990). Changed distribution of sodium channels along demyelinated axons. Proc. Natl. Acad. Sci. U.S.A. 87, 6777–6780. doi: 10.1073/pnas.87.17.6777
Fechner, S., Alvarez, L., Bonigk, W., Muller, A., Berger, T. K., Pascal, R., et al. (2015). A K( + )-selective CNG channel orchestrates Ca(2 + ) signalling in zebrafish sperm. Elife 4:e07624. doi: 10.7554/eLife.07624
Feldman, C. R., Brodie, E. D. Jr., Brodie, E. D. III, and Pfrender, M. E. (2012). Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proc. Natl. Acad. Sci. U.S.A. 109, 4556–4561. doi: 10.1073/pnas.1113468109
Feng, G., Deak, P., Chopra, M., and Hall, L. M. (1995). Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell 82, 1001–1011. doi: 10.1016/0092-8674(95)90279-1
Finol-Urdaneta, R. K., Wang, Y., Al-Sabi, A., Zhao, C., Noskov, S. Y., and French, R. J. (2014). Sodium channel selectivity and conduction: prokaryotes have devised their own molecular strategy. J. Gen. Physiol. 143, 157–171. doi: 10.1085/jgp.201311037
Freeman, S. A., Desmazieres, A., Fricker, D., Lubetzki, C., and Sol-Foulon, N. (2016). Mechanisms of sodium channel clustering and its influence on axonal impulse conduction. Cell. Mol. Life Sci. 73, 723–735. doi: 10.1007/s00018-015-2081-1
Fux, J. E. (2014). Primary and Auxiliary Subunits of Sodium Channel Nav1 in Lymnaea Stagnalis. UWSPACE. Master’s thesis, Department of Biology, University of Waterloo, Waterloo.
Gaboriaud, C., Gregory-Pauron, L., Teillet, F., Thielens, N. M., Bally, I., and Arlaud, G. J. (2011). Structure and properties of the Ca(2 + )-binding CUB domain, a widespread ligand-recognition unit involved in major biological functions. Biochem. J. 439, 185–193. doi: 10.1042/BJ20111027
Ganetzky, B. (1986). Neurogenetic analysis of Drosophila mutations affecting sodium channels: synergistic effects on viability and nerve conduction in double mutants involving tip-E. J. Neurogenet. 3, 19–31. doi: 10.3109/01677068609106892
Geffeney, S. L., Fujimoto, E., Brodie, E. D. III, Brodie, E. D. Jr., and Ruben, P. C. (2005). Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature 434, 759–763. doi: 10.1038/nature03444
Gosselin-Badaroudine, P., Moreau, A., Delemotte, L., Cens, T., Collet, C., Rousset, M., et al. (2015). Characterization of the honeybee AmNav1 channel and tools to assess the toxicity of insecticides. Sci. Rep. 5:12475. doi: 10.1038/srep12475
Gosselin-Badaroudine, P., Moreau, A., Simard, L., Cens, T., Rousset, M., Collet, C., et al. (2016). Biophysical characterization of the honeybee DSC1 orthologue reveals a novel voltage-dependent Ca2 + channel subfamily: CaV4. J. Gen. Physiol. 148, 133–145. doi: 10.1085/jgp.201611614
Grieco, T. M., Afshari, F. S., and Raman, I. M. (2002). A role for phosphorylation in the maintenance of resurgent sodium current in cerebellar purkinje neurons. J. Neurosci. 22, 3100–3107. doi: 10.1523/JNEUROSCI.22-08-03100.2002
Grigoriev, N. G., Spafford, J. D., Przysiezniak, J., and Spencer, A. N. (1996). A cardiac-like sodium current in motor neurons of a jellyfish. J. Neurophysiol. 76, 2240–2249. doi: 10.1152/jn.1996.76.4.2240
Groome, J. R. (2014). The voltage sensor module in sodium channels. Handb. Exp. Pharmacol. 221, 7–31. doi: 10.1007/978-3-642-41588-3_2
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gur Barzilai, M., Reitzel, A. M., Kraus, J. E., Gordon, D., Technau, U., Gurevitz, M., et al. (2012). Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep. 2, 242–248. doi: 10.1016/j.celrep.2012.06.016
Hagiwara, S., Naka, K. I., and Chichibu, S. (1964). Membrane properties of barnacle muscle fiber. Science 143, 1446–1448. doi: 10.1126/science.143.3613.1446
Hains, B. C., and Waxman, S. G. (2007). Sodium channel expression and the molecular pathophysiology of pain after SCI. Prog. Brain Res. 161, 195–203. doi: 10.1016/S0079-6123(06)61013-3
Hartline, D. K., and Colman, D. R. (2007). Rapid conduction and the evolution of giant axons and myelinated fibers. Curr. Biol. 17, R29–R35. doi: 10.1016/j.cub.2006.11.042
He, B., and Soderlund, D. M. (2010). Human embryonic kidney (HEK293) cells express endogenous voltage-gated sodium currents and Na v 1.7 sodium channels. Neurosci. Lett. 469, 268–272. doi: 10.1016/j.neulet.2009.12.012
Hedrich, R., and Marten, I. (2011). TPC1-SV channels gain shape. Mol. Plant 4, 428–441. doi: 10.1093/mp/ssr017
Heinemann, S. H., Terlau, H., Stuhmer, W., Imoto, K., and Numa, S. (1992). Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 356, 441–443. doi: 10.1038/356441a0
Hill, A. S., Nishino, A., Nakajo, K., Zhang, G., Fineman, J. R., Selzer, M. E., et al. (2008). Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 4:e1000317. doi: 10.1371/journal.pgen.1000317
Hille, B. (1968). Pharmacological modifications of the sodium channels of frog nerve. J. Gen. Physiol. 51, 199–219. doi: 10.1085/jgp.51.2.199
Hille, B. (1972). The permeability of the sodium channel to metal cations in myelinated nerve. J. Gen. Physiol. 59, 637–658. doi: 10.1085/jgp.59.6.637
Hille, B. (2001). Ion Channels of Excitable Membranes, 3rd Edition. Sunderland, MA: Sinauer Associates.
Hite, R. K., and MacKinnon, R. (2017). Structural titration of Slo2.2, a Na( + )-dependent K( + ) channel. Cell 168, 390–399.e11. doi: 10.1016/j.cell.2016.12.030
Hodges, D. D., Lee, D., Preston, C. F., Boswell, K., Hall, L. M., and O’Dowd, D. K. (2002). Tipe regulates Na + -dependent repetitive firing in Drosophila neurons. Mol. Cell. Neurosci. 19, 402–416. doi: 10.1006/mcne.2001.1088
Hodgkin, A. L., and Huxley, A. F. (1952). The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J. Physiol. 116, 497–506. doi: 10.1113/jphysiol.1952.sp004719
Hong, C. S., and Ganetzky, B. (1994). Spatial and temporal expression patterns of two sodium channel genes in Drosophila. J. Neurosci. 14, 5160–5169. doi: 10.1523/JNEUROSCI.14-09-05160.1994
Huang, X., Senatore, A., Dawson, T. F., Quan, Q., and Spafford, J. D. (2010). G-proteins modulate invertebrate synaptic calcium channel (LCaV2) differently from the classical voltage-dependent regulation of mammalian Cav2.1 and Cav2.2 channels. J. Exp. Biol. 213, 2094–2103. doi: 10.1242/jeb.042242
Hui, S., Xing, X., and Bader, G. D. (2013). Predicting PDZ domain mediated protein interactions from structure. BMC Bioinformatics 14:27. doi: 10.1186/1471-2105-14-27
Isom, L. L., Ragsdale, D. S., De Jongh, K. S., Westenbroek, R. E., Reber, B. F., Scheuer, T., et al. (1995). Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell 83, 433–442. doi: 10.1016/0092-8674(95)90121-3
Jeziorski, M. C., Greenberg, R. M., and Anderson, P. A. (1997). Cloning of a putative voltage-gated sodium channel from the turbellarian flatworm Bdelloura candida. Parasitology 115( Pt 3), 289–296. doi: 10.1017/S0031182097001388
Jeziorski, M. C., Greenberg, R. M., and Anderson, P. A. (1999). Cloning and expression of a jellyfish calcium channel beta subunit reveal functional conservation of the alpha1-beta interaction. Receptors Channels 6, 375–386.
Johnson, D., Montpetit, M. L., Stocker, P. J., and Bennett, E. S. (2004). The sialic acid component of the beta1 subunit modulates voltage-gated sodium channel function. J. Biol. Chem. 279, 44303–44310. doi: 10.1074/jbc.M408900200
Johnston, W. L., Dyer, J. R., Castellucci, V. F., and Dunn, R. J. (1996). Clustered voltage-gated Na + channels in Aplysia axons. J. Neurosci. 16, 1730–1739. doi: 10.1523/JNEUROSCI.16-05-01730.1996
Jost, M. C., Hillis, D. M., Lu, Y., Kyle, J. W., Fozzard, H. A., and Zakon, H. H. (2008). Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 25, 1016–1024. doi: 10.1093/molbev/msn025
Kalia, J., Milescu, M., Salvatierra, J., Wagner, J., Klint, J. K., King, G. F., et al. (2015). From foe to friend: using animal toxins to investigate ion channel function. J. Mol. Biol. 427, 158–175. doi: 10.1016/j.jmb.2014.07.027
Karmazinova, M., Beyl, S., Stary-Weinzinger, A., Suwattanasophon, C., Klugbauer, N., Hering, S., et al. (2010). Cysteines in the loop between IS5 and the pore helix of Ca(V)3.1 are essential for channel gating. Pflugers Arch. 460, 1015–1028. doi: 10.1007/s00424-010-0874-5
Katz, E., Stoler, O., Scheller, A., Khrapunsky, Y., Goebbels, S., Kirchhoff, F., et al. (2018). Role of sodium channel subtype in action potential generation by neocortical pyramidal neurons. Proc. Natl. Acad. Sci. U.S.A. 115, E7184–E7192. doi: 10.1073/pnas.1720493115
Kellenberger, S., West, J. W., Catterall, W. A., and Scheuer, T. (1997). Molecular analysis of potential hinge residues in the inactivation gate of brain type IIA Na + channels. J. Gen. Physiol. 109, 607–617. doi: 10.1085/jgp.109.5.607
Kim, D. M., and Nimigean, C. M. (2016). Voltage-gated potassium channels: a structural examination of selectivity and gating. Cold Spring Harb. Perspect. Biol. 8:a029231. doi: 10.1101/cshperspect.a029231
Kimura, T., and Kubo, T. (2003). Cloning and functional characterization of squid voltage-dependent Ca2 + channel beta subunits: involvement of N-terminal sequences in differential modulation of the current. Neurosci. Res. 46, 105–117. doi: 10.1016/S0168-0102(03)00038-5
Kiss, T. (2008). Persistent Na-channels: origin and function. A review. Acta Biol. Hung. 59(Suppl. 2), 1–12. doi: 10.1556/ABiol.59.2008.Suppl.1
Kulkarni, N. H., Yamamoto, A. H., Robinson, K. O., Mackay, T. F., and Anholt, R. R. (2002). The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics 161, 1507–1516.
Lazniewska, J., and Weiss, N. (2017). Glycosylation of voltage-gated calcium channels in health and disease. Biochim. Biophys. Acta 1859, 662–668. doi: 10.1016/j.bbamem.2017.01.018
Lee, C. H., and MacKinnon, R. (2017). Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120.e11. doi: 10.1016/j.cell.2016.12.023
Lee, H. J., and Zheng, J. J. (2010). PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 8:8. doi: 10.1186/1478-811X-8-8
Lee, K. S., Marban, E., and Tsien, R. W. (1985). Inactivation of calcium channels in mammalian heart cells: joint dependence on membrane potential and intracellular calcium. J. Physiol. 364, 395–411. doi: 10.1113/jphysiol.1985.sp015752
Leung, A. T., Imagawa, T., and Campbell, K. P. (1987). Structural characterization of the 1,4-dihydropyridine receptor of the voltage-dependent Ca2 + channel from rabbit skeletal muscle. Evidence for two distinct high molecular weight subunits. J. Biol. Chem. 262, 7943–7946.
Lewis, A. H., and Raman, I. M. (2014). Resurgent current of voltage-gated Na( + ) channels. J. Physiol. 592, 4825–4838. doi: 10.1113/jphysiol.2014.277582
Leys, S. P. (2015). Elements of a ’nervous system’ in sponges. J. Exp. Biol. 218, 581–591. doi: 10.1242/jeb.110817
Li, J., Waterhouse, R. M., and Zdobnov, E. M. (2011). A remarkably stable TipE gene cluster: evolution of insect Para sodium channel auxiliary subunits. BMC Evol. Biol. 11:337. doi: 10.1186/1471-2148-11-337
Li, M., West, J. W., Lai, Y., Scheuer, T., and Catterall, W. A. (1992). Functional modulation of brain sodium channels by cAMP-dependent phosphorylation. Neuron 8, 1151–1159. doi: 10.1016/0896-6273(92)90135-Z
Li, M., Zhou, X., Wang, S., Michailidis, I., Gong, Y., Su, D., et al. (2017). Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65. doi: 10.1038/nature20819
Liebeskind, B. J., Hillis, D. M., and Zakon, H. H. (2011). Evolution of sodium channels predates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. U.S.A. 108, 9154–9159. doi: 10.1073/pnas.1106363108
Liu, P., Ge, Q., Chen, B., Salkoff, L., Kotlikoff, M. I., and Wang, Z. W. (2011). Genetic dissection of ion currents underlying all-or-none action potentials in C. elegans body-wall muscle cells. J. Physiol. 589, 101–117. doi: 10.1113/jphysiol.2010.200683
Locke, E. G., Bonilla, M., Liang, L., Takita, Y., and Cunningham, K. W. (2000). A homolog of voltage-gated Ca(2 + ) channels stimulated by depletion of secretory Ca(2 + ) in yeast. Mol. Cell. Biol. 20, 6686–6694. doi: 10.1128/MCB.20.18.6686-6694.2000
Long, S. B., Campbell, E. B., and Mackinnon, R. (2005). Crystal structure of a mammalian voltage-dependent Shaker family K + channel. Science 309, 897–903. doi: 10.1126/science.1116269
Lopreato, G. F., Lu, Y., Southwell, A., Atkinson, N. S., Hillis, D. M., Wilcox, T. P., et al. (2001). Evolution and divergence of sodium channel genes in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 98, 7588–7592. doi: 10.1073/pnas.131171798
Loughney, K., Kreber, R., and Ganetzky, B. (1989). Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58, 1143–1154. doi: 10.1016/0092-8674(89)90512-6
Malhotra, J. D., Kazen-Gillespie, K., Hortsch, M., and Isom, L. L. (2000). Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J. Biol. Chem. 275, 11383–11388. doi: 10.1074/jbc.275.15.11383
Mehta, A. (2016). Cloning and Characterization of Voltage-Gated Sodium and Calcium Channel Homologs from the Single-Cell Choanoflagellate, Salpingoeca rosetta. UWSPACE. Master’s thesis, Department of Biology, University of Waterloo, Waterloo.
Mellquist, J. L., Kasturi, L., Spitalnik, S. L., and Shakin-Eshleman, S. H. (1998). The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37, 6833–6837. doi: 10.1021/bi972217k
Moran, Y., Barzilai, M. G., Liebeskind, B. J., and Zakon, H. H. (2015). Evolution of voltage-gated ion channels at the emergence of Metazoa. J. Exp. Biol. 218, 515–525. doi: 10.1242/jeb.110270
Moreno, J. D., and Clancy, C. E. (2012). Pathophysiology of the cardiac late Na current and its potential as a drug target. J. Mol. Cell Cardiol. 52, 608–619. doi: 10.1016/j.yjmcc.2011.12.003
Mori, M., Konno, T., Ozawa, T., Murata, M., Imoto, K., and Nagayama, K. (2000). Novel interaction of the voltage-dependent sodium channel (VDSC) with calmodulin: does VDSC acquire calmodulin-mediated Ca2 + -sensitivity? Biochemistry 39, 1316–1323.
Moroz, L. L., Kocot, K. M., Citarella, M. R., Dosung, S., Norekian, T. P., Povolotskaya, I. S., et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. doi: 10.1038/nature13400
Moroz, L. L., and Kohn, A. B. (2016). Independent origins of neurons and synapses: insights from ctenophores. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150041. doi: 10.1098/rstb.2015.0041
Mounier, Y., and Vassort, G. (1975). Initial and delayed membrane currents in crab muscle fibre under voltage-clamp conditions. J. Physiol. 251, 589–608. doi: 10.1113/jphysiol.1975.sp011110
Nakamura, F., and Goshima, Y. (2002). Structural and functional relation of neuropilins. Adv. Exp. Med. Biol. 515, 55–69. doi: 10.1007/978-1-4615-0119-0_5
Noda, M., and Hiyama, T. Y. (2015). Sodium sensing in the brain. Pflugers Arch. 467, 465–474. doi: 10.1007/s00424-014-1662-4
Noda, M., Shimizu, S., Tanabe, T., Takai, T., Kayano, T., Ikeda, T., et al. (1984). Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature 312, 121–127. doi: 10.1038/312121a0
Numann, R., Catterall, W. A., and Scheuer, T. (1991). Functional modulation of brain sodium channels by protein kinase C phosphorylation. Science 254, 115–118. doi: 10.1126/science.1656525
O′Malley, H. A., and Isom, L. L. (2015). Sodium channel beta subunits: emerging targets in channelopathies. Annu. Rev. Physiol. 77, 481–504. doi: 10.1146/annurev-physiol-021014-071846
Parada, G. E., Munita, R., Cerda, C. A., and Gysling, K. (2014). A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 42, 10564–10578. doi: 10.1093/nar/gku744
Payandeh, J., Scheuer, T., Zheng, N., and Catterall, W. A. (2011). The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358. doi: 10.1038/nature10238
Peng, G., Shi, X., and Kadowaki, T. (2015). Evolution of TRP channels inferred by their classification in diverse animal species. Mol. Phylogenet. Evol. 84, 145–157. doi: 10.1016/j.ympev.2014.06.016
Perez-Reyes, E., Cribbs, L. L., Daud, A., Lacerda, A. E., Barclay, J., Williamson, M. P., et al. (1998). Molecular characterization of a neuronal low-voltage-activated T-type calcium channel. Nature 391, 896–900. doi: 10.1038/36110
Potet, F., Chagot, B., Anghelescu, M., Viswanathan, P. C., Stepanovic, S. Z., Kupershmidt, S., et al. (2009). Functional interactions between distinct sodium channel cytoplasmic domains through the action of calmodulin. J. Biol. Chem. 284, 8846–8854. doi: 10.1074/jbc.M806871200
Qu, Y., Curtis, R., Lawson, D., Gilbride, K., Ge, P., DiStefano, P. S., et al. (2001). Differential modulation of sodium channel gating and persistent sodium currents by the beta1, beta2, and beta3 subunits. Mol. Cell Neurosci. 18, 570–580. doi: 10.1006/mcne.2001.1039
Rahman, T., Cai, X., Brailoiu, G. C., Abood, M. E., Brailoiu, E., and Patel, S. (2014). Two-pore channels provide insight into the evolution of voltage-gated Ca2 + and Na + channels. Sci. Signal. 7:ra109. doi: 10.1126/scisignal.2005450
Raman, I. M., and Bean, B. P. (1997). Resurgent sodium current and action potential formation in dissociated cerebellar Purkinje neurons. J. Neurosc. 17, 4517–4526. doi: 10.1523/JNEUROSCI.17-12-04517.1997
Rush, A. M., Dib-Hajj, S. D., Liu, S., Cummins, T. R., Black, J. A., and Waxman, S. G. (2006). A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc. Natl. Acad. Sci. U.S.A. 103, 8245–8250. doi: 10.1073/pnas.0602813103
Salkoff, L. B., and Wyman, R. J. (1983). Ion currents in Drosophila flight muscles. J. Physiol. 337, 687–709. doi: 10.1113/jphysiol.1983.sp014649
Sarhan, M. F., Tung, C. C., Van Petegem, F., and Ahern, C. A. (2012). Crystallographic basis for calcium regulation of sodium channels. Proc.Natl.Acad.Sci.U.S.A. 109, 3558–3563. doi: 10.1073/pnas.1114748109
Sarhan, M. F., Van Petegem, F., and Ahern, C. A. (2009). A double tyrosine motif in the cardiac sodium channel domain III-IV linker couples calcium-dependent calmodulin binding to inactivation gating. J. Biol. Chem. 284, 33265–33274. doi: 10.1074/jbc.M109.052910
Savio-Galimberti, E., Gollob, M. H., and Darbar, D. (2012). Voltage-gated sodium channels: biophysics, pharmacology, and related channelopathies. Front. Pharmacol. 3:124. doi: 10.3389/fphar.2012.00124
Schlief, T., Schonherr, R., Imoto, K., and Heinemann, S. H. (1996). Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur. Biophys. J. 25, 75–91. doi: 10.1007/s002490050020
Senatore, A., Guan, W., Boone, A. N., and Spafford, J. D. (2014). T-type channels become highly permeable to sodium ions using an alternative extracellular turret region (S5-P) outside the selectivity filter. J. Biol. Chem. 289,11952–11969. doi: 10.1074/jbc.M114.551473
Senatore, A., Monteil, A., van Minnen, J., Smit, A. B., and Spafford, J. D. (2013). NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PLoS One 8:e55088. doi: 10.1371/journal.pone.0055088
Senatore, A., Raiss, H., and Le, P. (2016). Physiology and evolution of voltage-gated calcium channels in early diverging animal phyla: cnidaria, placozoa, porifera and ctenophora. Front. Physiol. 7:481. doi: 10.3389/fphys.2016.00481
Senatore, A., Reese, T. S., and Smith, C. L. (2017). Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J. Exp. Biol. 220, 3381–3390. doi: 10.1242/jeb.162396
Senatore, A., and Spafford, J. D. (2010). Transient and big are key features of an invertebrate T-type channel (LCaV3) from the central nervous system of Lymnaea stagnalis. J. Biol. Chem. 285, 7447–7458. doi: 10.1074/jbc.M109.090753
Senatore, A., and Spafford, J. D. (2012). Gene transcription and splicing of T-type channels are evolutionarily-conserved strategies for regulating channel expression and gating. PLoS One 7:e37409. doi: 10.1371/journal.pone.0037409
Senatore, A., and Spafford, J. D. (2013). A uniquely adaptable pore is consistent with NALCN being an ion sensor. Channels 7, 60–68. doi: 10.4161/chan.23981
Shcheglovitov, A., Kostyuk, P., and Shuba, Y. (2007). Selectivity signatures of three isoforms of recombinant T-type Ca2 + channels. Biochim. Biophys. Acta 1768, 1406–1419. doi: 10.1016/j.bbamem.2007.02.017
Shen, H., Li, Z., Jiang, Y., Pan, X., Wu, J., Cristofori-Armstrong, B., et al. (2018). Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science doi: 10.1126/science.aau2596 [Epub ahead of print] doi: 10.1126/science.aau2596
Shen, H., Zhou, Q., Pan, X., Li, Z., Wu, J., and Yan, N. (2017). Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355:eaal4326. doi: 10.1126/science.aal4326
Shy, D., Gillet, L., Ogrodnik, J., Albesa, M., Verkerk, A. O., Wolswinkel, R., et al. (2014). PDZ domain-binding motif regulates cardiomyocyte compartment-specific Nav1.5 channel expression and function. Circulation 130, 147–160. doi: 10.1161/CIRCULATIONAHA.113.007852
Singh, A. K., Saotome, K., and Sobolevsky, A. I. (2017). Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 7:10669. doi: 10.1038/s41598-017-10993-9
Smith, R. D., and Goldin, A. L. (1996). Phosphorylation of brain sodium channels in the I–II linker modulates channel function in Xenopus oocytes. J. Neurosci. 16, 1965–1974. doi: 10.1523/JNEUROSCI.16-06-01965.1996
Smith, R. D., and Goldin, A. L. (1997). Phosphorylation at a single site in the rat brain sodium channel is necessary and sufficient for current reduction by protein kinase A. J. Neurosci. 17, 6086–6093. doi: 10.1523/JNEUROSCI.17-16-06086.1997
Spafford, J., Grigoriev, N., and Spencer, A. (1996). Pharmacological properties of voltage-gated Na + currents in motor neurones from a hydrozoan jellyfish Polyorchis penicillatus. J. Exp. Biol. 199, 941–948.
Spafford, J. D., Spencer, A. N., and Gallin, W. J. (1998). A putative voltage-gated sodium channel alpha subunit (PpSCN1) from the hydrozoan jellyfish, Polyorchis penicillatus: structural comparisons and evolutionary considerations. Biochem. Biophys. Res. Commun. 244, 772–780. doi: 10.1006/bbrc.1998.8332
Spafford, J. D., Spencer, A. N., and Gallin, W. J. (1999). Genomic organization of a voltage-gated Na + channel in a hydrozoan jellyfish: insights into the evolution of voltage-gated Na + channel genes. Receptors Channels 6, 493–506.
Stephens, R. F., Guan, W., Zhorov, B. S., and Spafford, J. D. (2015). Selectivity filters and cysteine-rich extracellular loops in voltage-gated sodium, calcium, and NALCN channels. Front. Physiol. 6:153. doi: 10.3389/fphys.2015.00153
Strong, M., Chandy, K. G., and Gutman, G. A. (1993). Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol. Biol. Evol. 10, 221–242.
Stühmer, W., Methfessel, C., Sakmann, B., Noda, M., and Numa, S. (1987). Patch clamp characterization of sodium channels expressed from rat brain cDNA. Eur. Biophys. J. 14, 131–138. doi: 10.1007/BF00253837
Taiakina, V., Boone, A. N., Fux, J., Senatore, A., Weber-Adrian, D., Guillemette, J. G., et al. (2013). The calmodulin-binding, short linear motif, NSCaTE is conserved in L-type channel ancestors of vertebrate Cav1.2 and Cav1.3 channels. PLoS One 8:e61765. doi: 10.1371/journal.pone.0061765
Takeshita, K., Sakata, S., Yamashita, E., Fujiwara, Y., Kawanabe, A., Kurokawa, T., et al. (2014). X-ray crystal structure of voltage-gated proton channel. Nat. Struct. Mol. Biol. 21, 352–357. doi: 10.1038/nsmb.2783
Tang, L., Gamal El-Din, T. M., Payandeh, J., Martinez, G. Q., Heard, T. M., Scheuer, T., et al. (2014). Structural basis for Ca2 + selectivity of a voltage-gated calcium channel. Nature 505, 56–61. doi: 10.1038/nature12775
Tao, X., Hite, R. K., and MacKinnon, R. (2017). Cryo-EM structure of the open high-conductance Ca(2 + )-activated K( + ) channel. Nature 541, 46–51. doi: 10.1038/nature20608
Tapper, A. R., and George, A. L. Jr. (2003). Heterologous expression of ion channels. Methods Mol. Biol. 217, 285–294.
Tasaki, I., and Spyropoulos, C. S. (1958). Membrane conductance and current-voltage relation in the squid axon under voltage-clamp. Am. J. Physiol. 193, 318–327. doi: 10.1152/ajplegacy.1958.193.2.318
Taylor, A. R. (2009). A fast Na + /Ca2 + -based action potential in a marine diatom. PLoS One 4:e4966. doi: 10.1371/journal.pone.0004966
Teravainen, H., and Rovainen, C. M. (1971). Fast and slow motoneurons to body muscle of the sea lamprey. J. Neurophysiol. 34, 990–998. doi: 10.1152/jn.1971.34.6.990
Terhag, J., Cavara, N. A., and Hollmann, M. (2010). Cave Canalem: how endogenous ion channels may interfere with heterologous expression in Xenopus oocytes. Methods 51, 66–74. doi: 10.1016/j.ymeth.2010.01.034
Terlau, H., Heinemann, S. H., Stuhmer, W., Pusch, M., Conti, F., Imoto, K., et al. (1991). Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 293, 93–96. doi: 10.1016/0014-5793(91)81159-6
Thompson, A., Infield, D. T., Smith, A. R., Smith, G. T., Ahern, C. A., and Zakon, H. H. (2018). Rapid evolution of a voltage-gated sodium channel gene in a lineage of electric fish leads to a persistent sodium current. PLoS Biol. 16:e2004892. doi: 10.1371/journal.pbio.2004892
Tikhonov, D. B., and Zhorov, B. S. (2011). Possible roles of exceptionally conserved residues around the selectivity filters of sodium and calcium channels. J. Biol. Chem. 286, 2998–3006. doi: 10.1074/jbc.M110.175406
Tikhonov, D. B., and Zhorov, B. S. (2012). Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol. Pharmacol. 82, 97–104. doi: 10.1124/mol.112.078212
Tikhonov, D. B., and Zhorov, B. S. (2018). Predicting structural details of the sodium channel pore basing on animal toxin studies. Front. Pharmacol. 9:880. doi: 10.3389/fphar.2018.00880
Toledo, G., Hanifin, C., Geffeney, S., and Brodie, E. D. III (2016). Convergent evolution of tetrodotoxin-resistant sodium channels in predators and prey. Curr. Top. Membr. 78, 87–113. doi: 10.1016/bs.ctm.2016.07.006
Ulbricht, W. (2005). Sodium channel inactivation: molecular determinants and modulation. Physiol. Rev. 85, 1271–1301. doi: 10.1152/physrev.00024.2004
Van den Steen, P., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (1998). Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208. doi: 10.1080/10409239891204198
Van Wart, A., and Matthews, G. (2006). Impaired firing and cell-specific compensation in neurons lacking Nav1.6 sodium channels. J. Neurosci. 26, 7172–7180. doi: 10.1523/JNEUROSCI.1101-06.2006
Verret, F., Wheeler, G., Taylor, A. R., Farnham, G., and Brownlee, C. (2010). Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 187, 23–43. doi: 10.1111/j.1469-8137.2010.03271.x
Wang, R., Mellem, J. E., Jensen, M., Brockie, P. J., Walker, C. S., Hoerndli, F. J., et al. (2012). The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron 75, 838–850. doi: 10.1016/j.neuron.2012.06.038
Warmke, J. W., Reenan, R. A., Wang, P., Qian, S., Arena, J. P., Wang, J., et al. (1997). Functional expression of Drosophila para sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J. Gen. Physiol. 110, 119–133. doi: 10.1085/jgp.110.2.119
West, J. W., Patton, D. E., Scheuer, T., Wang, Y., Goldin, A. L., and Catterall, W. A. (1992). A cluster of hydrophobic amino acid residues required for fast Na( + )-channel inactivation. Proc. Natl. Acad. Sci. U.S.A. 89, 10910–10914. doi: 10.1073/pnas.89.22.10910
Whicher, J. R., and MacKinnon, R. (2016). Structure of the voltage-gated K( + ) channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–669. doi: 10.1126/science.aaf8070
Wu, J., Yan, Z., Li, Z., Qian, X., Lu, S., Dong, M., et al. (2016). Structure of the voltage-gated calcium channel Cav1.1 at 3.6 a resolution. Nature 537, 191–196. doi: 10.1038/nature19321
Wu, J., Yan, Z., Li, Z., Yan, C., Lu, S., Dong, M., et al. (2015). Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350:aad2395. doi: 10.1126/science.aad2395
Wu, L. J., Sweet, T. B., and Clapham, D. E. (2010). International union of basic and clinical pharmacology. LXXVI. Current progress in the mammalian TRP ion channel family. Pharmacol. Rev. 62, 381–404. doi: 10.1124/pr.110.002725
Wu, M., Ye, N., Sengupta, B., and Zakon, H. H. (2013). A naturally occurring amino acid substitution in the voltage-dependent sodium channel selectivity filter affects channel gating. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 199, 829–842. doi: 10.1007/s00359-013-0845-3
Wu, S., Nomura, Y., Du, Y., Zhorov, B. S., and Dong, K. (2017). Molecular basis of selective resistance of the bumblebee BiNav1 sodium channel to tau-fluvalinate. Proc. Natl. Acad. Sci. U.S.A. 114, 12922–12927. doi: 10.1073/pnas.1711699114
Wyeth, M. S., Pelkey, K. A., Petralia, R. S., Salter, M. W., McInnes, R. R., and McBain, C. J. (2014). Neto auxiliary protein interactions regulate kainate and NMDA receptor subunit localization at mossy fiber-CA3 pyramidal cell synapses. J. Neurosci. 34, 622–628. doi: 10.1523/JNEUROSCI.3098-13.2014
Xu, M., and Cooper, E. C. (2015). An ankyrin-G N-terminal gate and protein kinase CK2 dually regulate binding of voltage-gated sodium and KCNQ2/3 potassium channels. J. Biol. Chem. 290, 16619–16632. doi: 10.1074/jbc.M115.638932
Yan, Z., Zhou, Q., Wang, L., Wu, J., Zhao, Y., Huang, G., et al. (2017). Structure of the Nav1.4-beta1 complex from electric Eel. Cell 170, 470–482.e11. doi: 10.1016/j.cell.2017.06.039
Yue, L., Navarro, B., Ren, D., Ramos, A., and Clapham, D. E. (2002). The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 120, 845–853. doi: 10.1085/jgp.20028699
Zakon, H. H. (2012). Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc. Natl. Acad. Sci. U.S.A. 109(Suppl. 1), 10619–10625. doi: 10.1073/pnas.1201884109
Zakon, H. H., Li, W., Pillai, N. E., Tohari, S., Shingate, P., Ren, J., et al. (2017). Voltage-gated sodium channel gene repertoire of lampreys: gene duplications, tissue-specific expression and discovery of a long-lost gene. Proc. Biol. Sci. 284:20170824. doi: 10.1098/rspb.2017.0824
Zamponi, G. W., and Currie, K. P. (2013). Regulation of Ca(V)2 calcium channels by G protein coupled receptors. Biochim. Biophys. Acta 1828, 1629–1643. doi: 10.1016/j.bbamem.2012.10.004
Zapata, F., Goetz, F. E., Smith, S. A., Howison, M., Siebert, S., Church, S. H., et al. (2015). Phylogenomic analyses support traditional relationships within Cnidaria. PLoS One 10:e0139068. doi: 10.1371/journal.pone.0139068
Zhang, J., Mao, W., Ren, Y., Sun, R. N., Yan, N., and Gong, H. (2018). Simulating the ion permeation and ion selection for a eukaryotic voltage-gated sodium channel NaVPaS. Protein Cell 9, 580–585.
Zhang, T., Liu, Z., Song, W., Du, Y., and Dong, K. (2011). Molecular characterization and functional expression of the DSC1 channel. Insect Biochem. Mol. Biol. 41, 451–458. doi: 10.1016/j.ibmb.2011.04.010
Zhang, T., Wang, Z., Wang, L., Luo, N., Jiang, L., Liu, Z., et al. (2013). Role of the DSC1 channel in regulating neuronal excitability in Drosophila melanogaster: extending nervous system stability under stress. PLoS Genet. 9:e1003327. doi: 10.1371/journal.pgen.1003327
Zhang, Z. Q. (2013). Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (Addenda 2013). Zootaxa 3703, 1–82. doi: 10.11646/zootaxa.3703.1.1
Zhou, W., Chung, I., Liu, Z., Goldin, A. L., and Dong, K. (2004). A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron 42, 101–112. doi: 10.1016/S0896-6273(04)00148-5
Zhu, J., Shang, Y., and Zhang, M. (2016). Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat. Rev. Neurosci. 17, 209–223. doi: 10.1038/nrn.2016.18
Keywords: sodium channels, calcium channels, auxiliary beta subunits, evolution, ion selectivity, NALCN, patch clamp electrophysiology, U12-type splice site
Citation: Fux JE, Mehta A, Moffat J and Spafford JD (2018) Eukaryotic Voltage-Gated Sodium Channels: On Their Origins, Asymmetries, Losses, Diversification and Adaptations. Front. Physiol. 9:1406. doi: 10.3389/fphys.2018.01406
Received: 20 June 2018; Accepted: 14 September 2018;
Published: 21 November 2018.
Edited by:
Mauricio Antonio Retamal, Universidad del Desarrollo, ChileReviewed by:
Gabriel Stölting, Charité – Universitätsmedizin Berlin, GermanyCopyright © 2018 Fux, Mehta, Moffat and Spafford. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. David Spafford, c3BhZmZvcmRAdXdhdGVybG9vLmNh
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.