
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 31 August 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01189
 Cecilia S. Vieira1
Cecilia S. Vieira1 Otacílio C. Moreira2
Otacílio C. Moreira2 Kate K. S. Batista1
Kate K. S. Batista1 Norman A. Ratcliffe3,4
Norman A. Ratcliffe3,4 Daniele P. Castro1,5
Daniele P. Castro1,5 Patrícia Azambuja1,5*
Patrícia Azambuja1,5*Rhodnius prolixus is an insect vector of Trypanosoma cruzi, the causative agent of Chagas disease in Latin America. Nuclear factor-κB (NF-κB) transcription factors (TF) are conserved components of the innate immune system in several multicellular organisms including insects. The drug IMD-0354 [N-(3,5-bis-trifluoromethyl-phenyl)-5-chloro-2-hydroxy-benzamide] is a selective inhibitor of IκB kinases. It blocks IκBα phosphorylation thus preventing nuclear translocation of the NF-κb TF. In humans, NF-κB is involved in several biological processes such as inflammation, cell proliferation and immunity. In insects, the activation of the immune system upon microbial challenge can be controlled by signaling pathways such as the immune deficiency (IMD) and Toll, to combat infection. These activated pathways signal to downstream NF-κB TF to stimulate specific immune genes, triggering the synthesis of several molecules such as the antimicrobial peptides. In Drosophila melanogaster, the activation and regulation of NF-κB TF have been elucidated, while in triatomines these mechanisms are not fully understood Therefore, the present study investigated the effects of oral administration of the drug IMD-0354 on the R. prolixus immune response to challenge with bacteria and T. cruzi, as well as the impact on the gut bacterial microbiota. R. prolixus were fed with rabbit blood containing IMD-0354 and Escherichia coli, Staphylococcus aureus, or T. cruzi. The effects of IMD-0354 on insect mortality and antimicrobial activity in insect midgut samples, as well as the relative expression of R. prolixus immune genes were recorded. The bacterial microbiota was analyzed, and viable parasites were counted in insect midgut samples. The IMD-0354 treatment modulated antibacterial activity and the gene expression patterns of defensin A, defensin B, defensin C, and prolixicin, and the genes involved in the IMD and Toll pathways. Additionally, there was an increase of bacterial microbiota in treated insects. Insects treated with IMD-0354 and concomitantly infected with bacteria or T. cruzi through the blood meal had increased mortality, while the T. cruzi population in R. prolixus midgut was reduced. The inhibitory effect of IMD-0354 indicates the importance of NF-κB TF in the innate immune responses involved in the control of bacteria and parasite infections in the R. prolixus midgut.
The invertebrate immune system relies on innate responses and lacks the classical adaptive immunity observed in vertebrates (Hoffmann and Reichhart, 2002). However, innate immunity is very effective as the first-line of host defense against infection in all eukaryotic organisms (Fearon and Locksley, 1996; Hoffmann and Reichhart, 2002). Nuclear Factor-Kappa B (NF-κB) transcription factors (TF) are important and conserved multi-components from pathways that regulate the expression of many genes including those responsible for the innate immune system (Silverman and Maniatis, 2001; Li and Verma, 2002; Kumar et al., 2004). NF-κB proteins have been identified in a range of multicellular organisms including cnidarians (Sullivan et al., 2007; Sinkovics, 2017), insects (Hetru and Hoffmann, 2009; Ganesan et al., 2011; Dong et al., 2012; Mesquita et al., 2015) and humans (Hayden and Ghosh, 2004; Senegas et al., 2015). Thus, these pathways emerged early in evolution, more than 500 million years ago, so that and the basic mechanisms of recognition and activation of response against pathogens are conserved in the animal kingdom (Hoffmann et al., 1999; Palmer and Jiggins, 2015).
In naïve animal cells, NF-κB TF are usually associated with inhibitory IκB proteins (such as cactus protein in insects) which retain the TF in the cytoplasm (Belvin et al., 1995; Silverman and Maniatis, 2001; Häcker and Karin, 2006). In insects, the recognition of an invading microorganism activates immune signaling pathways, such as the IMD and Toll pathways, inducing the phosphorylation and cleavage of the IκB inhibitory proteins (Li and Verma, 2002; Häcker and Karin, 2006) allowing NF-κB TF (relish, dorsal) to translocate to the nucleus, triggering the transcription of several effector molecules such as the antimicrobial peptides (AMPs) (Ferrandon et al., 2007; Hetru and Hoffmann, 2009) to combat infection (Häcker and Karin, 2006; Lemaitre and Hoffmann, 2007). Additionally, κB motifs have been detected in promoter regions of several AMPs genes from various insects such as Drosophila melanogaster (Kappler et al., 1993; Ganesan et al., 2011), Hyalophora cecropia (Engström et al., 1993), Glossina morsitans (Wang et al., 2008), and Anopheles gambiae (Christophides et al., 2002; Dong et al., 2012).
The drug IMD-0354 [N-(3,5-bis-trifluoromethyl-phenyl)-5-chloro-2-hydroxy-benzamide] is a selective molecular inhibitor of IκK-2 (Tanaka et al., 2005; Sugita et al., 2009). It blocks IκBα phosphorylation thus preventing nuclear translocation of the transcription factor NF-κB (Uota et al., 2012; Kong et al., 2015). Rhodnius prolixus is a triatomine species of medical importance vectoring Trypanosoma cruzi, the causative agent of Chagas disease in Latin America (Chagas, 1909; Vallejo et al., 2009; Coura, 2015; World Health Organization WHO, 2017). Therefore, studying aspects of R. prolixus immune system is relevant, for providing potential targets for insect control. In D. melanogaster, although the processes of activation and regulation of NF-κB TF are well-known (Ferrandon et al., 2007; Ganesan et al., 2011; Buchon et al., 2014; Imler, 2014), in triatomines these pathways are not fully understood. For example, the gene encoding the protein Relish, a conserved TF related to IMD signaling pathway (Ferrandon et al., 2007), was identified in R. prolixus as RpRelish, but its inhibitory protein Caspar was not detected (Mesquita et al., 2015). In addition, Dorsal, a NF-κB TF involved in the Toll pathway and its inhibitory protein, Cactus, a member of IκB-family protein (Geisler et al., 1992; Ferrandon et al., 2007) were detected in R. prolixus (Ursic-Bedoya et al., 2009; Ribeiro et al., 2014). Regarding effector molecules regulated by immune signaling pathways, AMPs have been isolated from R. prolixus midgut and fat body, including defensins (Lopez et al., 2003), lysozymes (Ursic-Bedoya et al., 2008) and prolixicin (Ursic-Bedoya et al., 2011). The induction of these effector molecules has been studied in R. prolixus challenged with different microorganisms. AMPs are differentially regulated, depending upon whether infection occurs with Gram-positive or Gram-negative bacteria or protozoans (Vieira et al., 2014, 2015, 2016).
Therefore, the main objective of the present work was to determine the effect of IMD-0354 in the regulation of genes involved in NF-κB pathways (relish, dorsal and cactus, respectively) and effector AMPs (defensins and prolixicin) in R. prolixus challenged with T. cruzi or bacteria. The results indicate that R. prolixus NF-κB TF have a pivotal role in the modulation of the innate immune system and in the ability of triatomines to deal with different infections.
Staphylococcus aureus 9518 and Escherichia coli K12 4401, both obtained from the National Collections of Industrial and Marine Bacteria (NCIMB), Aberdeen, United Kingdom, and Serratia marcescens RPH, previously isolated from R. prolixus midgut (Azambuja et al., 2004) were used in all experiments. Bacteria were maintained frozen at −70°C in brain heart infusion (BHI) plus 10% glycerol until use. For all experimental procedures, bacteria were cultivated with shaking (90 revolutions per minute) in 20 mL of tryptone soy broth (TSB) for 17 h at 30°C, and then 10 mL of fresh TSB were inoculated with 100 μL of the respective bacterial culture and incubated for a further 4 h under the same conditions. The bacteria were then washed in phosphate buffered saline (PBS, 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride, pH 7.4) and diluted in TSB for use.
Trypanosoma cruzi Dm28c clone (COLPROT 0010) were provided from the Coleção de Protozoários da Fundação Oswaldo Cruz, Rio de Janeiro, Brazil (Fiocruz, COLPROT)1. Epimastigotes were cultivated at 28°C in modified brain heart infusion (BHI) media (Sigma–Aldrich), containing hemin and supplemented with 10% heat-inactivated bovine fetal serum (Azambuja and Garcia, 1997).
The R. prolixus colony was maintained at Laboratório de Bioquímica e Fisiologia de Insetos, IOC/FIOCRUZ, at a relative humidity of 50–60% and at 27°C. For the experiments, R. prolixus 5th instar nymphs were randomly chosen and then fed with defibrinated rabbit blood through a membrane feeding device (Azambuja and Garcia, 1997). IMD-0354 (Sigma–Aldrich) was diluted in dimethyl sulfoxide (DMSO) (20 mg/mL) as a stock solution, according to the manufacturer’s instructions. For assaying the effects of IMD-0354 on bacterial and parasite infections insects were fed with defibrinated rabbit blood containing 5 or 10 μg of IMD-0354/mL. Additionally, the insects treated with IMD-0354 were concomitantly orally infected with bacteria, E. coli or S. aureus at a final concentration of 104 bacteria/mL of blood or with T. cruzi epimastigotes at 1 × 106 parasites/mL of blood (Vieira et al., 2014). IMD-0354 untreated control groups, infected or not, were fed on blood containing the same final concentration of the solvent DMSO (0,004%) as the treated groups. Parasite quantification (detailed below) was assessed in a Neubauer chamber under an optical microscope. Insects not fully engorged were discarded.
Anterior midgut samples were collected from 5th instar nymphs of R. prolixus 7 days after feeding (DAF) on blood (control untreated group) or blood plus 5 μg of IMD-0354/mL (Sigma–Aldrich). From each group, 10 insect midguts were individually collected in 1.5 mL reaction tubes and diluted in 200 μL ultrapure water. The samples were homogenized, centrifuged at 10,000 ×g for 10 min at 4°C and the supernatants filtered using Millipore PVDF (0.22 μm) and maintained at −20 °C until use (Vieira et al., 2014, 2016). Midgut samples from control and treated insects were incubated for 19 h at 37°C with E. coli, S. aureus or S. marcescens (Vieira et al., 2014). The midgut antibacterial activity was measured by the turbidimetric assays (TB) using a Spectra Max 190 Plate Reader (Molecular Devices, Sunnyvale, CA, United States), as described previously (Castro et al., 2012; Vieira et al., 2016).
Temporal relative expression of R. prolixus antimicrobial peptides genes, defensins and prolixicin (defA, defB, defC, prol) and Toll and IMD pathway related genes (RpDorsal, RpCactus, and RpRelish) were analyzed through reverse transcription quantitative PCR. The specific primers for the genes of interest, as well as R. prolixus housekeeping genes (α-tubulin and GAPDH), were designed and used as previously published: α-tubulin and GAPDH (Paim et al., 2012), defA, defB and defC (Lopez et al., 2003; Vieira et al., 2016); prol (Ursic-Bedoya et al., 2011; Vieira et al., 2016), RpCactus (Ribeiro et al., 2014), RpRelish, RpDorsal (Mesquita et al., 2015) (Supplementary Material). The relative quantification of gene expression was estimated using untreated insects (controls) as calibrators. Untreated control and IMD-0354-treated R. prolixus 5th instar nymphs were dissected at different days after feeding to collect and separate midgut samples in three pools containing five anterior midguts each (Vieira et al., 2016). Total RNA was extracted and quantified using a NucleoSpin® RNA II Kit (Macherey-Nagel, Düren, Germany) and a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, United States), respectively. Synthesis of cDNA was with a First-Strand cDNA Synthesis Kit (GE Healthcare, Buckinghamshire, United Kingdom) using 2.5 μg of total RNA and the pd(N)6 primer. Quantification of cDNA was assessed by fluorescence in a Qubit Fluorimeter (Life Technologies) with the ssDNA assay kit. Real-time quantitative polymerase chain reactions (RT-qPCR) were conducted in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems) at the FIOCRUZ facilities (Real-Time PCR Platform RPT-09A) using GoTaq® qPCR Master Mix (PROMEGA). Gene expression assays and analyses were performed as previously described by Vieira et al. (2016).
Total midguts from R. prolixus 5th instar nymphs were collected 7 days after feeding with blood with or without IMD-0354. It was performed three independent experiments with 10 insects from each group (n = 30). Cultivable bacteria microbiota population were quantified using the colony forming units (CFU) procedure. The samples were serially tenfold diluted with sterile PBS in 1.5 mL reaction tubes. Then 20 μL of each dilution was spread on BHI agar (Sigma–Aldrich) plates and immediately incubated at 30°C for 24 h and the CFU counted. Autoclaved PBS was also plated and incubated to assess the sterility of all experiments. Additionally, RT-qPCR was performed to analyze relative expression of 16S-rRNA from S. marcescens, Rhodococcus rhodnii and Enterococcaceae using cDNA from 3 pools of 10 anterior midguts from insects: untreated; treated with IMD-0354; infected with T. cruzi epimastigotes; treated with IMD-0354 simultaneously infected with T. cruzie pimastigotes.
To determine the toxicity of IMD-0354 in R. prolixus 5th instar nymphs, mortalities were evaluated 10 days after feeding. The groups analyzed were: control (fed only on blood), insects fed on blood containing 5 μg IMD-0354/mL, insects fed on blood containing 5 μg IMD-0354/mL plus T. cruzi Dm28c or E. coli or S. aureus. Each blood meal type Was replicated twice, and 30 insects Were used for each replicate.
Fifth instar R. prolixus nymphs fed With T. cruzi epimastigotes Were dissected and the total digestive tract Was collected 20 DAF in 1.5 mL microtubes and homogenized in 1.0 mL of sterile PBS. Parasite number Was determined by counting in a Neubauer haemocytometer and expressed as parasites/mL. Parasites quantification Was performed in three independent experiments With five individually insects each (n = 15).
The statistical analysis of data was performed using one-way ANOVA, two-way ANOVA, Student’s T, Tukey’s or Mann–Whitney tests on GraphPad Prism 5 software, depending on data distribution. Significant differences between groups are displayed in each figure and legends in the present study and were considered statistically different when p < 0.05.
The colony of R. prolixus was maintained in controlled environmental conditions at Laboratório de Bioquímica e Fisiologia de Insetos of IOC-FIOCRUZ and fed with defibrinated rabbit blood obtained at the Instituto de Ciência e Tecnologia em Biomodelos (ICTB), according to the Ethical Principles in Animal Experimentation approved by the Comitê de Ética de Experimentação Animal (CEUA/FIOCRUZ, under the protocol number LW019/17). The protocol was developed by CONCEA/MCT2, which is associated with the American Association for Animal Science (AAAS), the Federation of European Laboratory Animal Science Associations (FELASA), the International Council for Animal Science (ICLAS) and the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
To evaluate IMD-0454 toxicity, initial experiments, tested feeding 5 or 10 μg IMD-0354/mL blood to insects. At the 10 μg level, R. prolixus mortality rate was greater than 60%, a fact that impaired the infection experiments (data not shown). So, at 5 μg IMD-0354/mL a similar mortality rate (15%) occurred as in the control insects (12%) (data not shown) then, that dosage was used in all subsequent experiments. Additionally, at 5 μg IMD-0354/mL had no direct effect on parasite and bacteria cultures after 24 h of incubation.
To test if oral administration of the drug IMD-0354 (5ug/mL) alters the regulation of the NF-κB TF genes in the R. prolixus midgut, quantification of the temporal gene expression of TF RpRelish, RpDorsal and the inhibitory TF gene RpCactus, as well as some AMPs, was performed by RT-qPCR. The IMD-0354 treatment induced a significant downregulation of TF RpRelish gene expression and an upregulation of RpCactus and RpDorsal 1 DAF in comparison to control insects (Figure 1A, p < 0.05; Figure 1B, p < 0.05; and Figure 1C, p < 0.01). At 5 DAF, however, both RpRelish and RpCactus expressions are similar to untreated control insects while Dorsal expression is higher in treated insects (p < 0.05). In contrast, at 7 DAF, RpRelish levels were significantly higher, RpCactus mRNA levels were significantly lower than control insects (Figures 1A,B, p < 0.05). RpDorsal levels in treated or untreated insects are the same at 7 DAF (Figure 1C). The IMD-0354 treatment induced a significantly downregulation of the defA, defB and defC genes in the anterior midgut compared with control insects at 1 and 7 DAF (Figure 2A, p < 0.001, p < 0.01; Figure 2B, p < 0.001, p < 0.001; and Figure 2C, p < 0.01, p < 0.01). Prol expression was also lower at 1DAF in treated insects in comparison to control insects 1 DAF (Figure 2D, p < 0.01) but, in contrast, at 7 DAF, prol was higher than the control group (Figure 2D, p < 0.001).
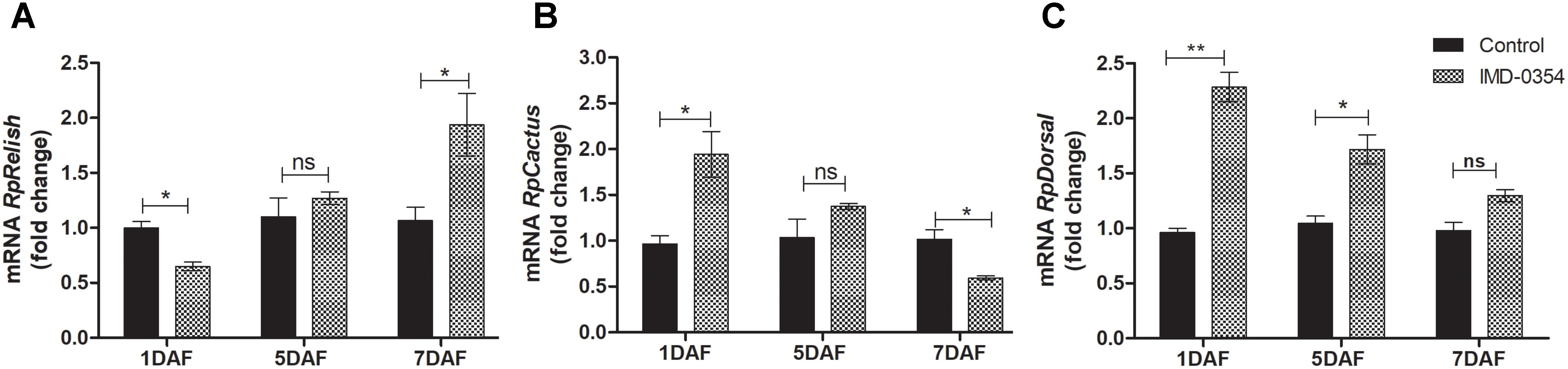
FIGURE 1. IMD-0354 effects on the relative gene expression of Relish, Cactus and Dorsal in the anterior midgut of Rhodnius prolixus. Relish, Cactus and Dorsal gene expressions were analyzed using the anterior midgut of fifth instar R. prolixus nymphs at different days after feeding on blood containing IMD-0354 (5 μg/ml). Data were quantified using the gene expression of untreated control insects as the calibrator (black columns) and shown as the relative expression of (A) RpRelish, (B) RpCactus, (C) RpDorsal on the 1st, 5th, and 7th days after feeding (DAF). Bars represent the mean ± SEM of 3 independent experiments with 3 pools of insects (n = 3). Means were compared using Student’s T-test; ∗p < 0.05, ∗∗p < 0.01, ns, non-significant.
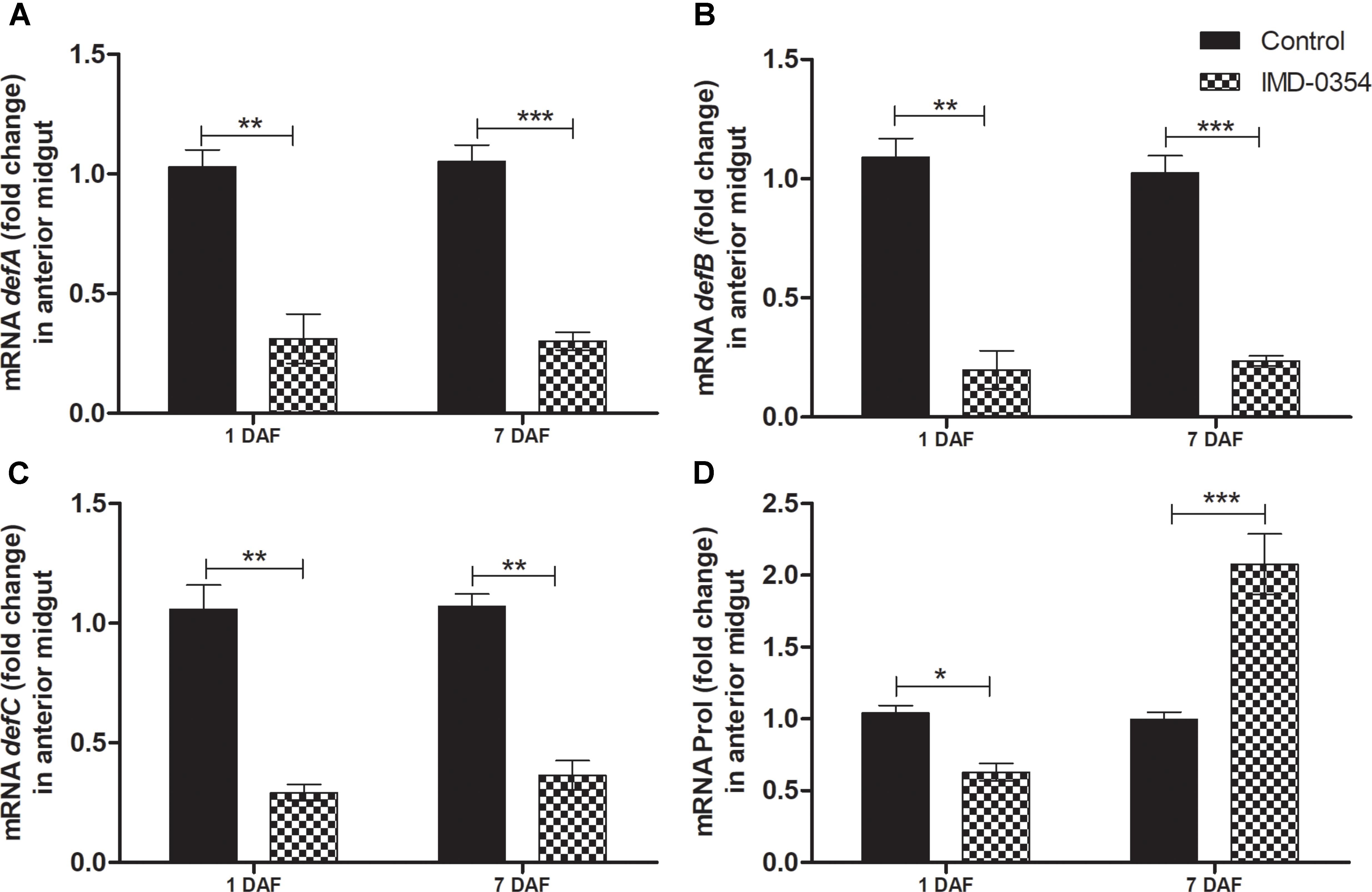
FIGURE 2. IMD-0354 effects on the relative gene expression of antimicrobial peptides in the anterior midgut of Rhodnius prolixus. Fifth instar nymphs of R. prolixus were fed on blood containing IMD-0354 (5 μg/ml). Data were quantified using the gene expression of untreated control insects as the calibrator (black columns). The grid columns show the relative expression of the antimicrobial peptide genes at the 1st and 7th days after feeding with IMD-0354. Relative expression of: (A) defA; (B) defB (C) defC (D) Prol at the 1st and 7th days after feeding. Treatments: black columns – insects fed with blood (control); grid columns – insects fed with blood containing the drug IMD-0354. Each bar represents 3 independent experiments performed in duplicate (n = 6). Means were compared using one-way ANOVA and Student T-test; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.1.
The antibacterial assays performed with midgut samples from insects fed on blood with or without IMD-0354 was an attempt to corroborate AMP activity and gene expression. In fact, the IMD-0354 treatment modulated both AMP gene expression and antibacterial activity in the R. prolixus midgut (Figure 3). Anterior midgut samples from treated insects showed significantly lower activity against S. aureus and S. marcescens in comparison to the untreated control (Figure 3B, p < 0.001, Figure 3C, p < 0.001). In contrast, a higher antibacterial activity against E. coli in vitro was detected in treated insects compared to the controls (Figure 3A, p < 0.01).
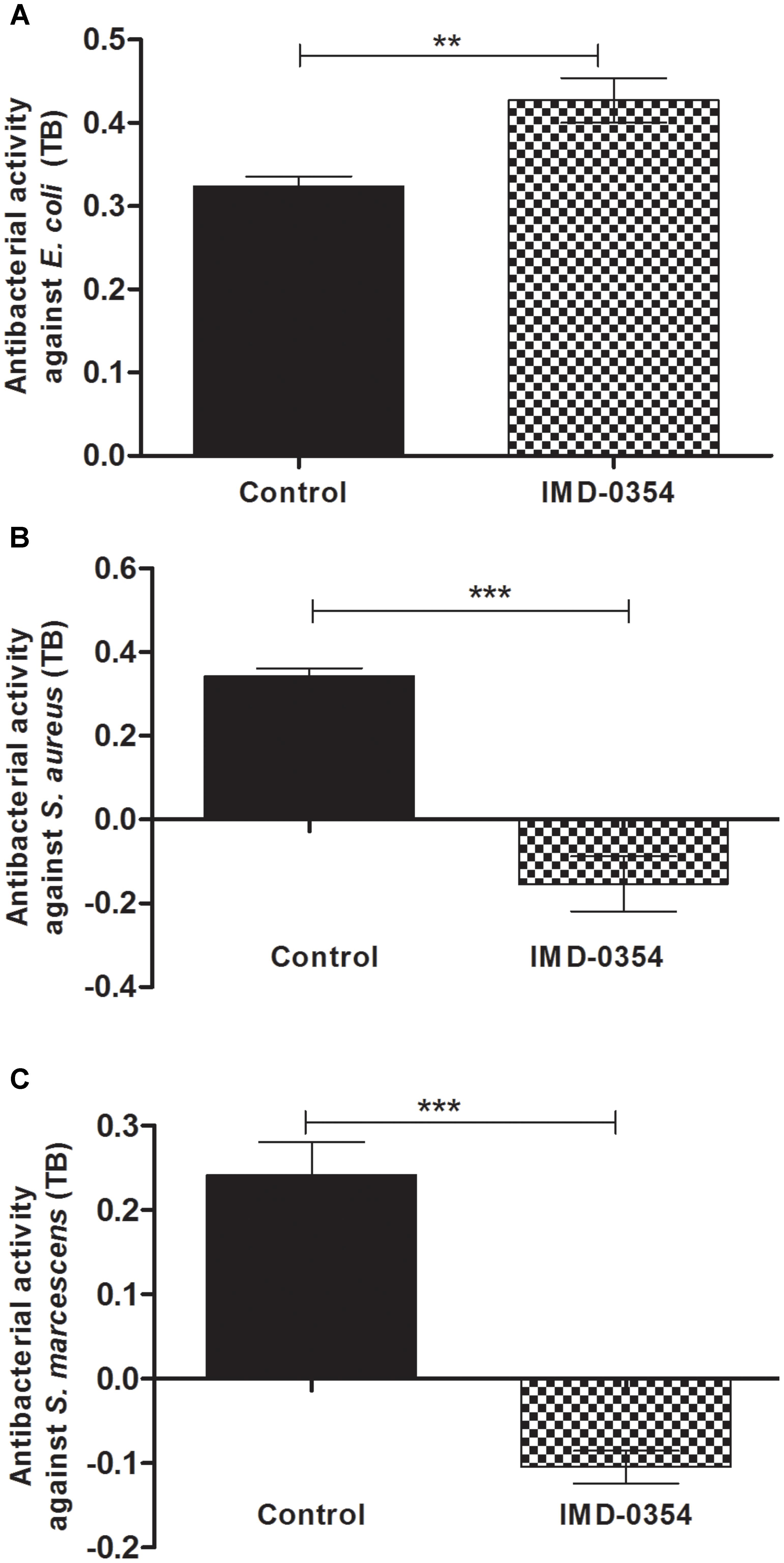
FIGURE 3. IMD-0354 effects on antibacterial activity in the anterior midgut of Rhodnius prolixus. Fifth instar nymphs of R. prolixus were fed on blood containing IMD-0354 (5 μg/ml). The antibacterial activities were measured in vitro using the anterior midgut samples 7 days after feeding and tested against (A) E. coli, (B) S. aureus, and (C) S. marcescens through the turbidimetric assay (OD550 nm) after 19 h incubation. Black column – untreated control insects fed only on blood; grid column – insects fed on blood containing IMD-0354. Bars represent the mean ± SEM of three independent experiments with nine pools of insects (n = 9). Means were compared using one-way ANOVA and Student’s T-test; ∗∗∗p < 0.001, ∗∗p < 0.01, NS, not significant.
To investigate whether the treatment with a NF-κB TF inhibitor could impact natural midgut homeostasis, the cultivable bacterial population was analyzed in the R. prolixus midgut at different times after drug ingestion. Insects fed with blood containing IMD-0354 had higher CFU counts than control insects at 1 and 7 DAF (p < 0.01) (Figure 4A). Furthermore, analysis of 16S RNA confirmed a significant increase of S. marcescens and R. rhodnii population in insects treated with IMD-0354 comparing to control insects (p < 0.001, p < 0.05) (Figure 4B). Additionally, in insects treated with IMD-0354 and concomitantly infected with T. cruzi, the population of S. marcescens and Enterococcaceae were significantly higher than untreated T. cruzi infected insects (p < 0.001, p < 0.001) (Figure 4B).
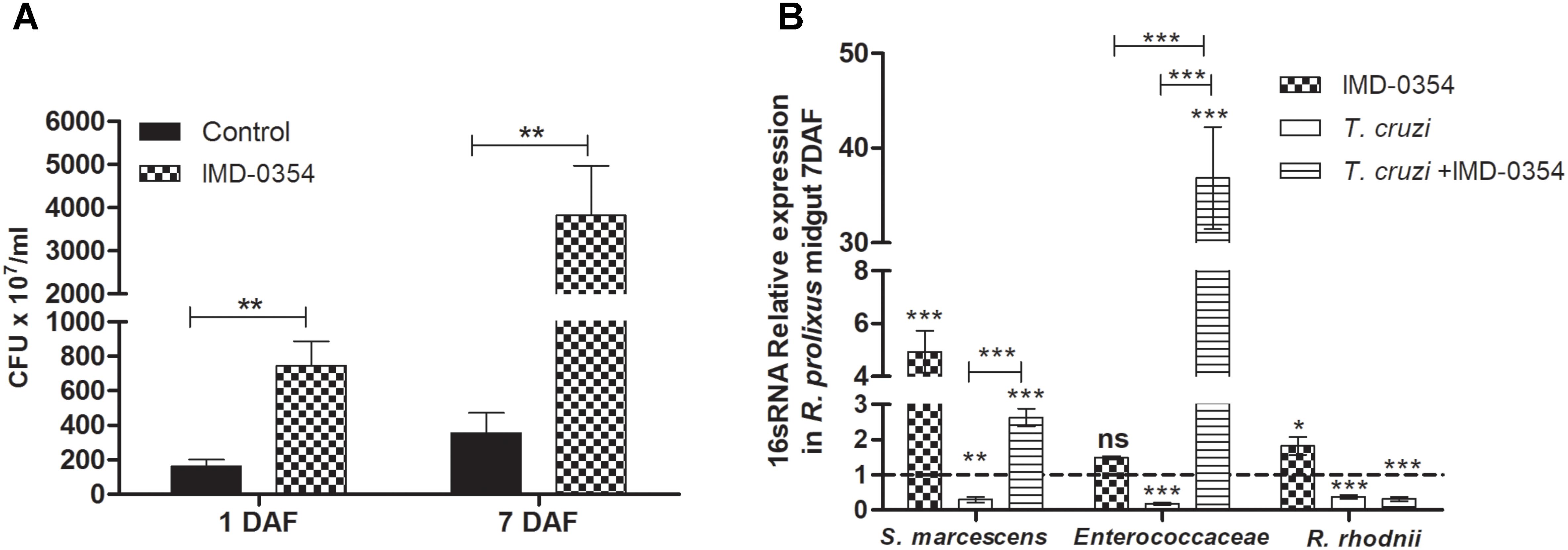
FIGURE 4. IMD-0354 effects on microbiota population in Rhodnius prolixus anterior midgut. Determination of bacterial load in the midgut of 5th instar nymphs of R. prolixus fed with blood containing IMD-0354 (5 μg/ml). (A) Colony forming units (CFU) were counted in the anterior midgut samples 1 and 7 days after feeding. (B) Relative expression of 16S-rRNA of Serratia marcescens, Enterococcaceae and Rhodococcus rhodnii analyzed by RT-qPCR. Data was normalized to the R. prolixus 18S RNA gene and quantified using the gene expression of untreated control insects as the calibrator represented by dotted lines on each graph. Black column – untreated control insects fed on blood; grid column – insects fed on blood containing IMD-0354; white column – insects fed on blood containing T. cruzi epimastigotes (106 parasites/mL of blood); striped columns - insects fed on blood containing IMD-0354 plus T. cruzi epimastigotes (106 parasites/mL of blood). Bars represent the mean ± SEM of three independent experiments with 10 insects (n = 30). Means were compared using Student’s T-test or Mann–Whitney test; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
The importance of NF-κB TF in R. prolixus survival after a microbial challenge was analyzed by recording mortalities 72 h after a blood meal. Adding T. cruzi or bacteria to the blood of IMD-0354 treated insects increased the mortality to 31, 42, and 59% after T. cruzi, E. coli, and S. aureus oral infection, respectively (Figure 5). Insects infected with E. coli, S. aureus or T. cruzi without IMD-0354 treatment, had a similar mortality rate to control insects, lower than 18% (data not shown).
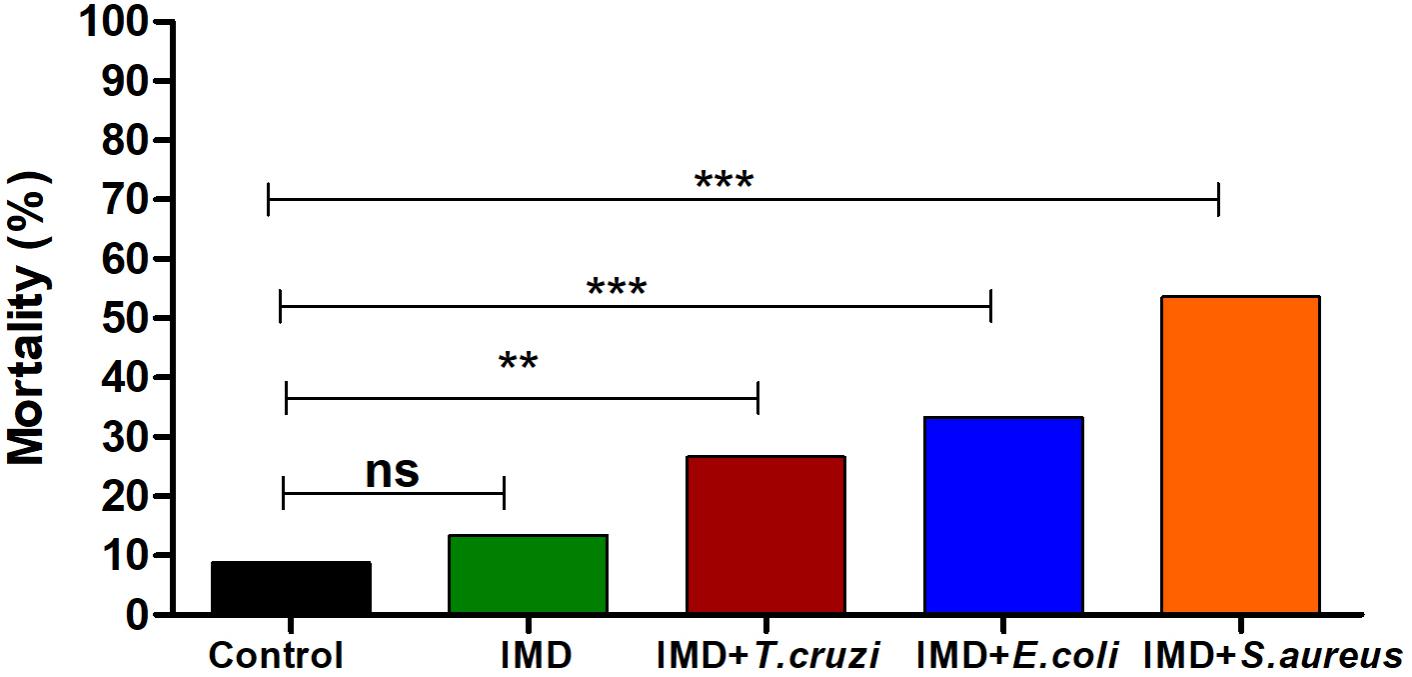
FIGURE 5. IMD-0354 effects on the mortality of Rhodnius prolixus infected with bacteria or Trypanosoma cruzi. R. prolixus 5th instar nymphs were fed on blood containing IMD-0354 (5 μg/ml) and E. coli or S. aureus at a final concentration of 1x 104 bacteria/ml or T. cruzi at 1 × 106 epimastigotes/ml. Numbers of dead insects were counted 7 days after the blood meal. Treatments: black column – untreated control insects fed on blood; green column - insect feed on blood with –IMD-0354; red column –IMD-0354 plus T. cruzi; blue column –IMD-0354 plus Escherichia coli.; orange column - IMD-0354 plus Staphylococcus aureus. Bars represent the percentage of dead insects (among 30 insects in each treatment) in 3 independent experiments. Means were compared using one-way ANOVA and Tukey’s post-test; ns ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
To investigate the role of NF-κB TF in the establishment of T. cruzi in R. prolixus gut, insects were concomitantly treated with IMD-0354 and infected with T. cruzi epimastigotes through the blood meal. The parasite population was quantified in the digestive tract collected from 15 insects 20 DAF. The T. cruzi population in the gut of insects treated with IMD-0354 was significantly lower in comparison to untreated infected insects (Figure 6, p < 0.001). Moreover, there was a decrease of 50% in the percentage of insects infected after IMD-0354 treatment.
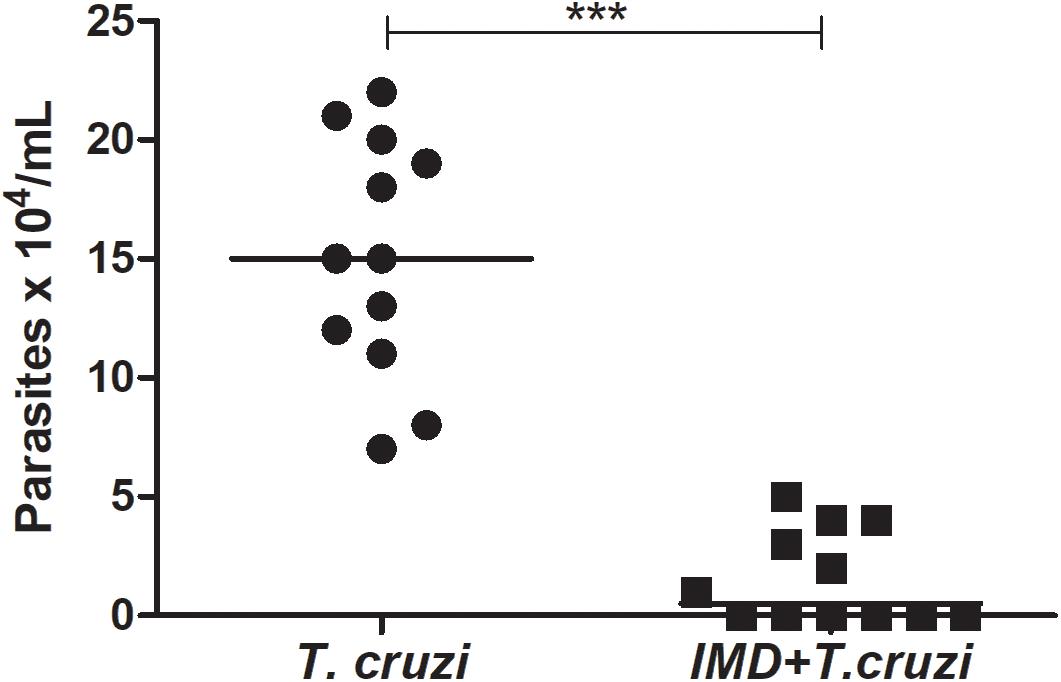
FIGURE 6. IMD-0354 effect on Trypanosoma cruzi population in the digestive tract of Rhodnius prolixus. R. prolixus 5th instar nymphs were fed on blood containing IMD-0354 (5 μg/ml of blood) and T. cruzi epimastigotes at a final concentration of 106 parasites/mL of blood. Number of T. cruzi were estimated in R. prolixus 5th instar nymphs in whole digestive tracts at 20 days after feeding (DAF). Each point represents the number of parasites in an individual insect, and bars indicate the median. Means were compared using Student T-test; ∗∗∗p < 0.001.
After observing that IMD-0354 impedes T. cruzi infection in R. prolixus midgut whether in these infected insects the drug altered the expression of defC and prol was tested. Defensin C and prolixicin genes were chosen for these experiments since previous results demonstrated the upregulation of these genes after T. cruzi infection (Vieira et al., 2016). Quantification of gene expression was examined at 1 and 7 DAF. At 1 DAF, T. cruzi infected insects concomitantly treated with IMD-0354 had a higher prol expression in comparison to controls and to T. cruzi infected untreated insects (Figure 7A, p < 0.01, p < 0.001, respectively). In contrast, on 7DAF, prol levels in the midgut of infected insects concomitantly treated with IMD-0354 were 8-fold lower in comparison to T. cruzi infected untreated insects (Figure 7A, p < 0.05). Unlike the prol gene expression, that of the defC gene at 1DAF showed no differences between infected insects and insects infected with T. cruzi and concomitantly treated with IMD-0354. However, on the 7 DAF, defC levels were lower in T. cruzi infected/IMD-0354 treated insects in comparison to infected but untreated insects (Figure 7B, p < 0.05). Both prol and defC genes were also significantly upregulated after T. cruzi infection in comparison to control insects 7DAF (Figure 7A, p < 0.01; Figure 7B, p < 0.001, respectively).
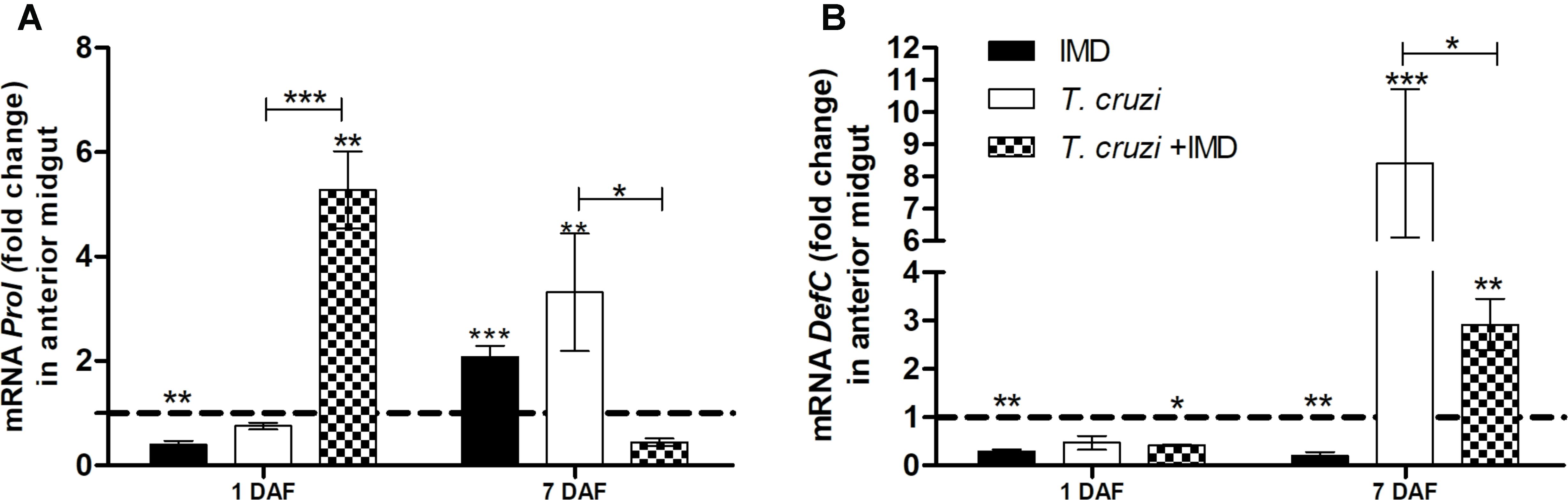
FIGURE 7. IMD-0354 effects on relative gene expression of antimicrobial peptides in the midgut of Rhodnius prolixus infected with Trypanosoma cruzi. R. prolixus 5th instar nymphs were fed on blood containing IMD-0354 and T. cruzi epimastigotes at a final concentration of 106 parasites/mL. Data were quantified using the gene expression of untreated control insects as the calibrator represented by horizontal dotted lines on each graph to show the relative expression of antimicrobial peptide. (A) Prolixicin relative expression; (B) Defensin C relative expression on the 1st and 7th days after feeding. Treatments: Black column- insect feed on blood containing IMD-0354 alone; white columns – fed on T. cruzi alone; grid columns – insects fed on IMD-0354 and T. cruzi. Each bar represents 3 independent experiments performed in duplicate (n = 6). Means were compared using two-way ANOVA and Student T-test; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
The innate immune system represents the first line of defense against infection by pathogens. For decades, insect innate immunity has commonly been studied using the genetic malleable model, D. melanogaster. This model elucidated details of the genes involved in the Toll and IMD pathways, as well as their specific roles in combating infection, by challenging with Gram-positive and negative bacteria and fungi (Lemaitre and Hoffmann, 2007; Ferrandon et al., 2007; Hetru and Hoffmann, 2009).
Nowadays, attention has also turned to the immune systems of insect vectors of disease in order to identify potential targets to develop new control strategies. Regarding the triatomine vector, R. prolixus, only recently its whole genome sequence was published (Mesquita et al., 2015). With these available data now, it is possible to study the pattern recognition mechanisms and genes involved in immune signaling pathways as well as the effector genes regulated by these pathways. In this context, in the present work, the compound IMD-0354 was used as an immunosuppressive drug, since it is selective NF-κB TF inhibitor, preventing the transcription of several antimicrobial peptides in mammals (Kamon et al., 2004; Tanaka et al., 2005; Sugita et al., 2009). The use of this drug in arthropods, as far as we are aware, has not previously been documented to study components of insect vector immunity. Through drug incorporation in the insect blood meal, the importance of genes regulated by NF-κB pathways in the activation and modulation of the R. prolixus immune genes against infection by different microorganisms can be evaluated, as well as the influence of Toll and IMD TF in the establishment of T. cruzi infections.
The lack of some canonical elements from the IMD pathway was observed in the R. prolixus genome, with the absence of some genes such as IMD, FADD (Fas-Associated protein with dead domain) and Caspar, the inhibitor of the IMD pathway (Mesquita et al., 2015). The gene encoding the TF Relish, however, was detected and its knockdown induced a decrease of some AMPs and allowed the proliferation of the intestinal microbiota in R. prolixus (Mesquita et al., 2015), indicating the presence of a functional IMD pathway. Some disparity in the IMD signaling pathway elements is also observed in other insects such as in Pediculus humanus (Kim et al., 2011), Acyrthosiphon pisum (Gerardo et al., 2010) and in the ticks, Ixodes scapularis (Shaw et al., 2017), Rhipicephalus microplus (Rosa et al., 2016) suggesting some plasticity in the IMD pathway among arthropods (Palmer and Jiggins, 2015; Shaw et al., 2017).
When R. prolixus 5th instar nymphs were fed with blood containing IMD-0354, defA, defB, and defC expression and activity were downregulated, suggesting that NF-κB TF activation and its translocation to the nucleus is essential to trigger the transcription of, at least, the defensins genes in R. prolixus. In contrast, the NF-κB inhibitor induced an increase in prol levels at 7 DAF. This discrepancy may be due to the fact that the R. prolixus defensins, and those of other triatomines such as Triatoma brasiliensis and Panstrongylus megistus, are regulated by NF-κB TF (Lopez et al., 2003; Waniek et al., 2009; de Araújo et al., 2015), whereas NF-κB binding sites are absent in the prolixicin gene sequence, which appears to be regulated by GATA transcription factors (Ursic-Bedoya et al., 2011). Further experiments are necessary to fully understand the signaling pathways that regulate prolixicin in R. prolixus and in other hemipterans. The increase of prolixicin mRNA levels in the R. prolixus midgut after NF-κB inhibition by IMD-0354 could represent an indirect compensatory effect that implicates other pathways, such as those controlled by GATA TF, for regulating the genes.
The low antibacterial activity detected in the midgut samples of insects treated with IMD-0354, against the Gram-positive S. aureus and the Gram-negative S. marcescens might be related to the negative regulation of defA, defB, and defC. In contrast, the increased antibacterial activity against E. coli which could be related to the enhancement of the prol expression. Previously, in in vitro studies, of purified prolixicin from R. prolixus fat body have been shown to have strong activity against E. coli (Ursic-Bedoya et al., 2011). In addition, the increase of cultivable bacterial numbers, as well S. marcescens, R. rhodnii and Enterococcaceae in the midgut in IMD-0354-treated insects suggests microbiota regulation by the defensins, highlighting the importance of NF-κB TF in R. prolixus intestinal homeostasis.
Moreover, knowledge of the role of immune signaling pathways and effector proteins (AMPs) in the regulation of insect intestinal bacterial microbiota is still fragmentary. Since AMP synthesis can inhibit the development of pathogens in their insect vectors (Vizioli et al., 2001a,b; Boulanger et al., 2002, 2004; Telleria et al., 2013; Das De et al., 2018), innumerable microorganisms present in the intestinal tract are able to induce the insect immune system (Cirimotich et al., 2011; Eappen et al., 2013; Vieira et al., 2014), consequently interfering in the life cycle of the parasites. S. marcescens, one of the most abundant bacteria found in R. prolixus gut (da Mota et al., 2012), possesses lytic mechanisms that drastically reduce the success of T. cruzi infection in the R. prolixus digestive tract (Azambuja et al., 2004; Castro et al., 2007, 2012; Vieira et al., 2016). It was suggested that the normal S. marcescens population growth is inhibited by R. prolixus AMPs, especially by defensins (Vieira et al., 2014, 2016). T. cruzi Dm28c infection in R. prolixus seems to overcome the trypanolytic effect of S. marcescens, inducing a strong upregulation of defC and prol (Vieira et al., 2015, 2016) which in turn decrease the S. marcescens population.
In the present study, IMD-0354 affected the normal signaling requirements needed for AMP transcription and protein synthesis since the drug prevents the translocation of the TF to the nucleus (Tanaka et al., 2005). A transitory modulation in transcriptional levels of NF-kB TFs (RpRelish, RpDorsal)and of IkB RpCactus was observed in IMD-0354 treated insects. Since insects treated withIMD-0354recordedan increased population of some bacterial species in the midgut, this effect might be associated with the activation of the Toll pathway, marked by an enhancementinmRNA levels of Cactus (IkB) and Dorsal(NF-kBTF). Theopposite occurs in IMD pathways, in which the TF, Relish, is downregulated. Several studies using different insect species have demonstrated the role of the gut microbiota in stimulating immune signaling pathways (Eappen et al., 2013; Eleftherianos et al., 2013; Jiménez-Cortés et al., 2018). After the recognition of bacterial components by specific receptors, the Toll pathway is activated, requiring the degradation of the Cactus protein to release Dorsal protein, allowing its nuclear translocation to initiate the transcription of effector genes (Ferrandon et al., 2007; Ganesan et al., 2011). These processes are followed by the transcription of both Cactus and Dorsal mRNAs, in order to rescue and retain these regulatory proteins in the cytoplasm (Geisler et al., 1992; Nicolas et al., 1998; Rutschmann et al., 2002). It is known that the drug IMD-0354 acts downstream in microbial recognition and upstream of NF-kB TF (Relish and Dorsal) nuclear translocation, only impairing the phosphorylation of IkB proteins (Cactus) but not the transcription of the genes (NF-kB TF and IkB) (Tanaka et al., 2005). Thus, the alterations in Relish, Cactus and Dorsal gene expressions detected here could represent secondary effects of IMD-0354 treatment in R. prolixus.
The main effect of IMD-0354 treatment is to inhibit AMPs transcription. In the present work, the three defensins studied were suppressed in IMD-0354 treated insects. Downregulation of the AMP genes confirmed by the lower antibacterial activity detected in R. prolixus midgut samples was one of the possible reasons that allowed an intense proliferation of S. marcescens, R. rhodnii and Enterococcaceae. Thereby, defensins and its related NF-κB signaling pathways seem to play an essential role in regulating microbiota homeostasis in R. prolixus midgut. In this sense, the negative impact on the T. cruzi population was observed in insects fed with the NF-κB inhibitor could be related to the increased bacterial population of S. marcescens and Enterococcaceae. As discussed above, S. marcescens presents trypanolytic activity (Azambuja et al., 2004; Castro et al., 2007) and large amounts of this bacteria in the midgut probably interfered in T. cruzi development in IMD-0453 treated insects. Additionally, a decrease in the defC and prol levels in insects fed with IMD-0354 concurrently infected with T. cruzi, reinforcing the importance of the activation of these genes for the establishment of T. cruzi infection in R. prolixus. Together these results highlight the relevance of genes regulated by NF-κB TF in the R. prolixus immune system and gut homeostasis, as well as in the development of the parasite in the insect vector. Studying the genes involved in the signaling pathways regulated by NF-κB TF is essential for a broad knowledge of the mechanisms of activation and modulation of the immune response. More experiments are necessary to understand how different microorganisms selectively activate immune signaling pathways and different NF-κB TF molecules. Investigation of NF-κB TF genes and genomic analysis of AMPs promoter regions could provide relevant insights into the function of each pathway in controlling effector molecules against specific invading microorganisms.
CSV conceived and designed the experiments, performed all the experiments, participated in the analysis and interpretation of the data, wrote the manuscript, and corrected and approved the final manuscript. OCM conceived and designed the PCR experiments, participated in the analysis and interpretation of the data, and read, corrected, and approved the final manuscript. KSB performed the experiments of feeding and infection in insects, parasite quantification in insect midgut, participated in the analysis and interpretation of the data, and read, corrected, and approved the final manuscript. NAR conceived and designed the experiments, wrote the manuscript, and corrected and approved the final manuscript. DPC participated in the physiological and molecular experiments, analysis and interpretation of the data, and read, corrected, and approved the final manuscript. PA conceived and designed the experiments, contributed with reagents and materials, participated in the analysis and interpretation of the data, and read, corrected, and approved the final manuscript.
This work was sponsored by Brazilian Research Agencies FIOCRUZ, FAPERJ, and CNPq.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Zildo Alves da Cruz and José Carlos Nascimento de Oliveira for Rhodnius prolixus colony maintenance and Maria Angelica Cardoso for technical support at Real-Time PCR Platform Rpt-09A at the Fiocruz facilities. We thank the Program for Technological Development in Inputs for Health (Pdtis-Fiocruz).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01189/full#supplementary-material
Azambuja, P., Feder, D., and Garcia, E. S. (2004). Isolation of Serratia marcescens in the midgut of Rhodnius prolixus: impact on the establishment of the parasite Trypanosoma cruzi in the vector. Exp. Parasitol. 107, 89–96. doi: 10.1016/j.exppara.2004.04.007
Azambuja, P., and Garcia, E. S. (1997). “Care and maintenance of triatomine colonies,” in Molecular Biology of Insect Disease Vectors: a Methods Manual, eds J. M. Crampton, C. B. Beard, and C. Louis (London: Chapman and Hall), 56–64. doi: 10.1007/978-94-009-1535-0_6
Belvin, M. P., Jin, Y., and Anderson, K. V. (1995). Cactusprotein degradationmediates Drosophila dorsal-ventral signaling. Genes Dev. 9, 783–793. doi: 10.1101/gad.9.7.783
Boulanger, N., Brun, R., Ehret-Sabatier, L., Kunz, C., and Bulet, P. (2002). Immunopeptides in the defense reactions of Glossina morsitans to bacterial and Trypanosoma brucei brucei infections. Insect Biochem. Mol. Biol. 32, 369–375. doi: 10.1016/S0965-1748(02)00029-2
Boulanger, N., Lowenberger, C., Volf, P., Ursic, R., Sigutova, L., Sabatier, L., et al. (2004). Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect. Immun. 72, 7140–7146. doi: 10.1128/IAI.72.12.7140-7146.2004
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunityin Drosophila melanogaster-from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi: 10.1038/nri3763
Castro, D., Moraes, C., Gonzalez, M., Ratcliffe, N., Azambuja, P., and Garcia, E. (2012). Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS One 7:e36591. doi: 10.1371/journal.pone.0036591
Castro, D. P., Seabra, S. H., Garcia, E. S., de Souza, W., and Azambuja, P. (2007). Trypanosoma cruzi: ultrastructural studies of adhesion, lysis and biofilm formation by Serratia marcescens. Exp. Parasitol. 117, 201–207. doi: 10.1016/j.exppara.2007.04.014
Chagas, C. (1909). Nova tripanosomíase humana. Estudos sobre a morphologia e o ciclo evolutivo do schizotrypanum cruzi, agente da nova entidade mórbida do homem. Mem. Inst. Oswaldo Cruz 1, 159–218. doi: 10.1590/S0074-02761909000200008
Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., et al. (2002). Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165. doi: 10.1126/science.1077136
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Coura, J. R. (2015). The main sceneries of chagas disease transmission. The vectors, blood and oral transmissions–a comprehensive review. Mem. Inst. Oswaldo Cruz 110, 277–282. doi: 10.1590/0074-0276140362
da Mota, F. F., Marinho, L. P., Moreira, C. J., Lima, M. M., Mello, C. B., Garcia, E. S., et al. (2012). Cultivation-independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Negl. Trop. Dis. 6:e1631. doi: 10.1371/journal.pntd.0001631
Das De T, Sharma, P., Thomas, T., Singla, D., Tevatiya, S., Kumari, S., et al. (2018). Interorgan molecular communication strategies of ”Local” and ”Systemic” innate immune responses in mosquito Anopheles stephensi. Front. Immunol. 9:148. doi: 10.3389/fimmu.2018.00148
de Araújo, C. A., Lima, A. C., Jansen, A. M., Galvão, C., Jurberg, J., Costa, J., et al. (2015). Genes encoding defensins of important Chagas disease vectors used for phylogenetic studies. Parasitol. Res. 114, 4503–4511. doi: 10.1007/s00436-015-4694-6
Dong, Y., Cirimotich, C. M., Pike, A., Chandra, R., and Dimopoulos, G. (2012). Anopheles NF-κB-regulated splicing factors direct pathogen-specific repertoires of the hypervariable pattern recognition receptor AgDscam. Cell Host Microbe 12, 521–530. doi: 10.1016/j.chom.2012.09.004
Eappen, A. G., Smith, R. C., and Jacobs-Lorena, M. (2013). Enterobacter-activated mosquito immune responses to Plasmodium involve activationofSRPN6 in Anopheles stephensi. PLoS One 8:e62937. doi: 10.1371/journal.pone.0062937
Eleftherianos, I., Atri, J., Accetta, J., and Castillo, J. C. (2013). Endosymbiotic bacteria in insects: guardians of the immune system? Front. Physiol. 4:46. doi: 10.3389/fphys.2013.00046
Engström, Y., Kadalayil, L., Sun, S. C., Samakovlis, C., Hultmark, D., and Faye, I. (1993). kappa B-like motifs regulate the induction of immune genes in Drosophila. J. Mol. Biol. 232, 327–333. doi: 10.1006/jmbi.1993.1392
Fearon, D. T., and Locksley, R. R. (1996). The instructive role of innate immunity in the acquired immune response. Science 272, 50–53. doi: 10.1126/Science.272.5258.50
Ferrandon, D., Imler, J. L., Hetru, C., and Hoffmann, J. A. (2007). The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874. doi: 10.1038/nri2194
Ganesan, S., Aggarwal, K., Paquette, N., and Silverman, N. (2011). NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 349, 25–60. doi: 10.1007/82_2010_107
Geisler, R., Bergmann, A., Hiromi, Y., and Niisslein-volhard, C. (1992). Cactus, a gene involved in dorsoventral pattern formation of Drosophila is related to the IkB gene family of vertebrates. Cell 71, 613–621. doi: 10.1016/0092-8674(92)90595-4
Gerardo, N. M., Altincicek, B., Anselme, C., Atamian, H., Barribeau, S. M., de Vos, M., et al. (2010). Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 11:R21. doi: 10.1186/gb-2010-11-2-r21
Häcker, H., and Karin, M. (2006). Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006:re13. doi: 10.1126/stke.3572006re13
Hayden, M. S., and Ghosh, S. (2004). Signaling to NF-kappaB. Genes Dev. 18, 2195–2224. doi: 10.1101/gad.1228704
Hetru, C., and Hoffmann, J. (2009). NF-kB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol. 1:a000232. doi: 10.1101/cshperspect.a000232
Hoffmann, J. A., Kafatos, F. C., Janeway, C. A., and Ezekowitz, R. A. (1999). Phylogenetic perspectives in innate immunity. Science 284, 1313–1318. doi: 10.1126/science.284.5418.1313
Hoffmann, J. A., and Reichhart, J. M. (2002). Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3, 121–126. doi: 10.1038/ni0202-121
Imler, J. L. (2014). Overview of Drosophila immunity: a historical perspective. Dev. Comp. Immunol. 42, 3–15. doi: 10.1016/j.dci.2013.08.018
Jiménez-Cortés, J. G., García-Contreras, R., Bucio-Torres, M. I., Cabrera-Bravo, M., Córdoba-Aguilar, A., Benelli, G., et al. (2018). Bacterial symbionts in human blood-feeding arthropods: patterns, general mechanisms and effects of global ecological changes. Acta Trop. 186, 69–101. doi: 10.1016/j.actatropica.2018.07.005
Kamon, J., Yamauchi, T., Muto, S., Takekawa, S., Ito, Y., Hada, Y., et al. (2004). A novel IKK beta inhibitor stimulates adiponectin levels and ameliorates obesity-linked insulin resistance. Biochem. Biophys. Res. Commun. 323, 242–248. doi: 10.1016/j.bbrc.2004.08.083
Kappler, C., Meister, M., Lagueux, M., Gateff, E., Hoffmann, J. A., and Reichhart, J. M. (1993). Insect immunity. Two 17 bp repeats nesting a kappa B-related sequence confer inducibility to the diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. EMBO J. 12, 1561–1568.
Kim, J. H., Min, J. S., Kang, J. S., Kwon, D. H., Yoon, K. S., Strycharz, J., et al. (2011). Comparison of the humoral and cellular immune responses between body and head lice following bacterial challenge. Insect Biochem. Mol. Biol. 41, 332–339. doi: 10.1016/j.ibmb.2011.01.011
Kong, X. J., Duan, L. J., Qian, X. Q., Xu, D., Liu, H. L., Zhu, Y. J., et al. (2015). Tumor-suppressive microRNA-497 targets IKKβ to regulate NF-κB signaling pathway in human prostate cancer cells. Am. J. Cancer Res. 5, 1795–1804.
Kumar, A., Takada, Y., Boriek, A. M., and Aggarwal, B. B. (2004). Nuclear factor-kappaB: its role in health and disease. J. Mol. Med. 82, 434–448. doi: 10.1007/s00109-004-0555-y
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Li, Q., and Verma, I. M. (2002). NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734. doi: 10.1038/nri910
Lopez, L., Morales, G., Ursic, R., Wolff, M., and Lowenberger, C. (2003). Isolation and characterization of a novel insect defensin from Rhodnius prolixus, a vector of chagas disease. Insect Biochem. Mol. Biol. 33, 439–447. doi: 10.1016/S0965-1748(03)00008-0
Mesquita, R. D., Vionette-Amaral, R. J., Lowenberger, C., Rivera-Pomar, R., Monteiro, F. A., Minx, P., et al. (2015). Genome of Rhodnius prolixus, an insect vector of chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. U.S.A. 112, 14936–14941. doi: 10.1073/pnas.1506226112
Nicolas, E., Reichhart, J. M., Hoffmann, J., and Lemaitre, B. (1998). In vivo regulation of the IkB homologue cactus during the immune response of Drosophila. J. Biol. Chem. 273, 10463–10469. doi: 10.1074/jbc.273.17.10463
Paim, R. M., Pereira, M. H., Di Ponzio, R., Rodrigues, J. O., Guarneri, A. A., Gontijo, N. F., et al. (2012). Validation of reference genes for expression analysis in the salivary gland and the intestine of Rhodnius prolixus (Hemiptera, Reduviidae) under different experimental conditions by quantitative real-time PCR. BMC Res. Notes 5:128. doi: 10.1186/1756-0500-5-128
Palmer, W. J., and Jiggins, F. M. (2015). Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol. Biol. Evol. 32, 2111–2129. doi: 10.1093/molbev/msv093
Ribeiro, J. M., Genta, F. A., Sorgine, M. H., Logullo, R., Mesquita, R. D., Paiva-Silva, G. O., et al. (2014). An insight into the transcriptome of the digestive tract of the bloodsucking bug. Rhodnius prolixus. PLoS Negl. Trop. Dis. 8:e2594. doi: 10.1371/journal.pntd.0002594
Rosa, R. D., Capelli-Peixoto, J., Mesquita, R. D., Kalil, S. P., Pohl, P. C., Braz, G. R., et al. (2016). Exploring the immune signalling pathway-related genes of the cattle tick Rhipicephalus microplus: from molecular characterization to transcriptional profile upon microbial challenge. Dev. Comp. Immunol. 59, 1–14. doi: 10.1016/j.dci.2015.12.018
Rutschmann, S., Kilinc, A., and Ferrandon, D. (2002). Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J. Immunol. 168, 1542–1546. doi: 10.4049/jimmunol.168.4.1542
Senegas, A., Gautheron, J., Dit Maurin, A. G., and Courtois, G. (2015). IKK-related genetic diseases: probing NF-κB functions in humans and other matters. Cell. Mol. Life Sci. 72, 1275–1287. doi: 10.1007/s00018-014-1793-y
Shaw, D. K., Wang, X., Brown, L. J., Chávez, A. S., Reif, K. E., Smith, A. A., et al. (2017). Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat. Commun. 8:14401. doi: 10.1038/ncomms14401
Silverman, N., and Maniatis, T. (2001). NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342. doi: 10.1101/gad.909001
Sinkovics, J. G. (2017). The cnidarian origin of the proto-oncogenes NF-κB/STAT and WNT-like oncogenic pathway drives the ctenophores. Int. J. Oncol. 50:338. doi: 10.3892/ijo.2016.3762
Sugita, A., Ogawa, H., Azuma, M., Muto, S., Honjo, A., Yanagawa, H., et al. (2009). Antiallergic and anti-inflammatory effects of a novel IκB kinase β Inhibitor, IMD-0354, in a mouse model of allergic inflammation. Int. Arch. Allergy Immunol. 148, 186–198. doi: 10.1159/000161579
Sullivan, J. C., Kalaitzidis, D., Gilmore, T. D., and Finnerty, J. R. (2007). Rel homology domain-containing transcription factors in the cnidarian Nematostella vectensis. Dev. Genes Evol. 217, 63–72. doi: 10.1007/s00427-006-0111-6
Tanaka, A., Konno, M., Muto, S., Kambe, N., Morii, E., Nakahata, T., et al. (2005). A novel NF-kappaB inhibitor,IMD-0354, suppresses neoplastic proliferation of human mast cells with constitutively activated c-kit receptors. Blood 105, 2324–2331. doi: 10.1182/blood-2004-08-3247
Telleria, E. L., Sant’Anna, M. R., Alkurbi, M. O., Pitaluga, A. N., Dillon, R. J., and Traub-Csekö, Y. M. (2013). Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit. Vectors 6:12. doi: 10.1186/1756-3305-6-12
Uota, S., Zahidunnabi-Dewan, M., Saitoh, Y., Muto, S., Itai, A., Utsunomiya, A., et al. (2012). An IkB kinase 2 inhibitor IMD-0354 suppresses the survival of adult T-cell leukemia cells. Cancer Sci. 103, 100–106. doi: 10.1111/j.1349-7006.2011.02110.x
Ursic-Bedoya, R., Buchhop, J., Joy, J. B., Durvasula, R., and Lowenberger, C. (2011). Prolixicin: a novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect Mol. Biol. 20, 775–786. doi: 10.1111/j.1365-2583.2011.01107.x
Ursic-Bedoya, R., Buchhop, J., and Lowenberger, C. (2009). Cloning and characterization of Dorsal homologues in the hemipteran Rhodnius prolixus. Insect. Mol. Biol. 18, 681–689. doi: 10.1111/j.1365-2583.2009.00909.x
Ursic-Bedoya, R. J., Nazzari, H., Cooper, D., Triana, O., Wolff, M., and Lowenberger, C. (2008). Identification and characterization of two novel lysozymes from Rhodnius prolixus, a vector of Chagas disease. J. Insect Physiol. 54, 593–603. doi: 10.1016/j.jinsphys.2007.12.009
Vallejo, G. A., Guhl, F., and Schaub, G. A. (2009). Triatominae-Trypanosoma cruzi/T. rangeli: vector-parasite interactions. Acta Trop. 110, 137–147. doi: 10.1016/j.actatropica.2008.10.001
Vieira, C. S., Mattos, D. P., Waniek, P. J., Santangelo, J. M., Figueiredo, M. B., Gumiel, M., et al. (2015). Rhodnius prolixus interaction with Trypanosoma rangeli: modulation of the immune system and microbiota population. Parasit. Vectors 8:135. doi: 10.1186/s13071-015-0736-2
Vieira, C. S., Waniek, P. J., Castro, D. P., Mattos, D. P., Moreira, O. C., and Azambuja, P. (2016). Impact of Trypanosoma cruzi on antimicrobial peptide gene expression and activity in the fat body and midgut of Rhodnius prolixus. Parasit. Vectors 9:119. doi: 10.1186/s13071-016-1398-4
Vieira, C. S., Waniek, P. J., Mattos, D. P., Castro, D. P., Mello, C. B., Ratcliffe, N. A., et al. (2014). Humoral responses in Rhodnius prolixus: bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit. Vectors 7:232. doi: 10.1186/1756-3305-7-232
Vizioli, J., Bulet, P., Hoffmann, J. A., Kafatos, F. C., Müller, H. M., and Dimopoulos, G. (2001a). Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 98, 12630–12635. doi: 10.1073/pnas.221466798
Vizioli, J., Richman, A. M., Uttenweiler-Joseph, S., Blass, C., and Bulet, P. (2001b). The defensin peptide of the malaria vector mosquito Anopheles gambiae: antimicrobial activities and expression in adult mosquitoes. Insect Biochem. Mol. Biol. 31, 241–248.
Wang, J., Hu, C., Wu, Y., Stuart, A., Amemiya, C., Berriman, M., et al. (2008). Characterization of the antimicrobial peptide attacinloci from Glossina morsitans. Insect Mol. Biol. 17, 293–302. doi: 10.1111/j.1365-2583.2008.00805.x
Waniek, P. J., Castro, H. C., Sathler, P. C., Miceli, L., Jansen, A. M., and Araújo, C. A. (2009). Two novel defensin-encoding genes of the chagas disease vector Triatoma brasiliensis (Reduviidae, Triatominae): gene expression and peptide-structure modeling. J. Insect Physiol. 55, 840–848. doi: 10.1016/j.jinsphys.2009.05.015
Keywords: insect immune system, Rhodnius prolixus, antimicrobial peptides, Nf-κB transcription factor, Trypanosoma cruzi, microbiota
Citation: Vieira CS, Moreira OC, Batista KKS, Ratcliffe NA, Castro DP and Azambuja P (2018) The NF-κB Inhibitor, IMD-0354, Affects Immune Gene Expression, Bacterial Microbiota and Trypanosoma cruzi Infection in Rhodnius prolixus Midgut. Front. Physiol. 9:1189. doi: 10.3389/fphys.2018.01189
Received: 08 May 2018; Accepted: 07 August 2018;
Published: 31 August 2018.
Edited by:
Xanthe Vafopoulou, York University, CanadaReviewed by:
Andréa Cristina Fogaça, Universidade de São Paulo, BrazilCopyright © 2018 Vieira, Moreira, Batista, Ratcliffe, Castro and Azambuja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrícia Azambuja, YXphbWJ1amEucEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.