- 1MOE Key Laboratory of Marine Genetics and Breeding, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Sex determination and differentiation have long been a research hotspot in metazoans. However, little is known about when and how sex differentiation occurs in most mollusks. In this study, we conducted a combined morphological and molecular study on sex differentiation in the Yesso scallop Patinopecten yessoensis. Histological examination on gonads from 5- to 13-month-old juveniles revealed that the morphological sex differentiation occurred at 10 months of age. To determine the onset of molecular sex differentiation, molecular markers were screened for early identification of sex. The gonadal expression profiles of eight candidate genes for sex determination or differentiation showed that only two genes displayed sexually dimorphic expression, with FOXL2 being abundant in ovaries and DMRT1L in testes. In situ hybridization revealed that both of them were detected in germ cells and follicle cells. We therefore developed LOG10(DMRT1L/FOXL2) for scallop sex identification and confirmed its feasibility in differentiated individuals. By tracing its changes in 5- to 13-month-old juveniles, molecular sex differentiation time was determined: some scallops differentiate early in September when they are 7 months old, and some do late in December when they are 10 months old. Two kinds of coexpression patterns were found between FOXL2 and DMRT1L: expected antagonism after differentiation and unexpected coordination before differentiation. Our results revealed that scallop sex differentiation co-occurs with the formation of follicles, and molecular sex differentiation is established prior to morphological sex differentiation. Our study will assist in a better understanding of the molecular mechanism underlying bivalve sex differentiation.
Introduction
Sexual reproduction is one of the most universal phenomena that widely exist in animals. As a focus of this research area, sexual development encompasses sex determination and differentiation. The former is defined as the process by which sex is established under the influence of genetic or environmental factors and the latter as the process that an undifferentiated gonad uses to transform into an ovary or a testis (Bull, 1983; Penman and Piferrer, 2008; Piferrer and Guiguen, 2008). Therefore, sexual development is a complex network that is initiated by a sex-determining trigger mediating the expression of sex differentiation genes, which ultimately gives rise to the phenotypic differences between sexes (Heule et al., 2014).
Sex determination and differentiation have received much research attention in various species. For example, in mammals, genotypic sex is defined by the presence or absence of sex-specific chromosome Y that carries the dominant male determinant SRY (Swain and Lovell-Badge, 1999; Kocer et al., 2009). In chickens that possess a ZZ/ZW sex chromosome system, DMRT1 is required for testis determination (Zarkower, 2001; Smith et al., 2009). In Caenorhabditis elegans and Drosophila melanogaster, a cascade of sex switch genes is controlled by the ratio of X chromosomes to sets of autosomes (the X:A ratio; Zarkower, 2001; Goodwin and Ellis, 2002). It seems sex is determined by sex chromosomes and controlled by some master switch genes in many organisms.
Mollusca represents the second largest phylum of invertebrates after Arthropoda. Bivalves are a large group of mollusks that exhibit different reproductive strategies: some are gonochoric, some are hermaphroditic, and some are capable of sex changes. Although much research has been done on many different bivalves, there is no clear evidence for the existence of sex chromosomes in these animals. However, some progress has been made regarding the identification of sex determination or differentiation genes. Several key sex-related genes in model species have been characterized in bivalves, including FOXL2 (Naimi et al., 2009a; Liu et al., 2012), WNT4 (Li et al., 2013; Yang et al., 2015), FST (follistatin; Ni et al., 2012), β-catenin (Li et al., 2014b; Santerre et al., 2014), DMRT (Naimi et al., 2009b; Feng et al., 2010; Yu et al., 2011; Shi et al., 2014), DAX1 (Li et al., 2014a), and SOXE (Santerre et al., 2014). Many of them display sexually dimorphic expression patterns in the gonads and are regarded as participants in the sex differentiation cascade. Meanwhile, gonadal transcriptome analyses identified some key candidate genes for sex determination or differentiation, such as FOXL2, WNT4, β-catenin, DMRT, DAX1, SOXE, and SOXH (Teaniniuraitemoana et al., 2014; Zhang et al., 2014; Tong et al., 2015; Li et al., 2016; Patnaik et al., 2016). These studies suggest that sex determination and differentiation genes may be deeply conserved in animals.
Yesso scallop Patinopecten yessoensis is a commercially important species widely cultured in China and Japan. It is predominantly gonochoric, with scarce hermaphroditism. Due to its commercial importance, much work has been performed to obtain an understanding of its reproductive process (Osada et al., 2004; Tanabe et al., 2010; Nagasawa et al., 2015). Recently, our group completed whole genome sequencing and gonadal transcriptome analysis of P. yessoensis (Li et al., 2016; Wang et al., 2017), which provide valuable resources for unraveling the molecular mechanisms underlying scallop sex differentiation. In the present study, morphological and molecular sex differentiation was examined in the Yesso scallop. Eight candidate genes for sex determination or differentiation were chosen to establish markers for sex identification and the onset of molecular sex differentiation was determined; we also compared the molecular changes in the undifferentiated and differentiated gonads. This study will pave the way for a better understanding of the regulatory network in bivalve sex differentiation.
Materials and Methods
Sample Collection
To obtain adult scallops with known sexes, 15-month-old mature female and male individuals were selected in May 2015, cultured in separate cages, and transported to the laboratory every month from August 2015 to March 2016. Meanwhile, juvenile scallops 5–13 months of age were obtained from July 2015 to March 2016. All of these scallops were collected from the Dalian Zhangzidao Fishery Group Corporation (Liaoning Province, China) and acclimated in filtered and aerated seawater for 1 week at the temperature at which they were collected. For each month, gonads of 50 adults and/or 50 juveniles were dissected: the majority were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction, and the remainder were fixed in 4% paraformaldehyde overnight, dehydrated with serial methanol (25, 50, 75, and 100%) diluted in 0.01 M phosphate-buffered saline and then stored at −20°C for paraffin sectioning and in situ hybridization. Our experiments were conducted according to the guidelines and regulations established by the Ocean University of China and the local government.
Histology
The samples were transferred to ethanol, cleared in xylene, embedded in paraffin wax, and cut into 5-μm-thick sections on a rotary microtome (Leica, Wetzlar, Germany). Serial sections were tiled on glass slides, deparaffinized with xylene, hydrated with graded ethanol to water, and stained with hematoxylin. After that, the glass slides were counterstained with eosin, dehydrated with ethanol, cleared with xylene, mounted with neutral balsam, and covered with coverslips. Finally, the sections were observed under a Nikon’s Eclipse E600 research microscope.
Candidate Genes Identification
Four ovary-related genes (FOXL2, WNT4, FST, β-catenin) and four testis-related genes (DMRT1L, DAX1, SOXE, and SOXH) were selected for analysis. To identify these genes, protein sequences from other organisms were collected from NCBI and used as queries against the genome (Wang et al., 2017) and gonadal transcriptomes (Li et al., 2016) by TBLASTN with an e-value threshold of 1e−5. The resultant scallop sequences were further confirmed by BLASTX against the NCBI protein sequence database.
Phylogenetic Analyses
Full-length protein sequences encoding FOXL2/3 and DMRT homologs were downloaded from NCBI. Multiple sequence alignments (Supplementary Material) were conducted using ClustalW2 program with default parameters (Larkin et al., 2007). The neighbor-joining phylogenetic trees were constructed using MEGA 6 (Tamura et al., 2013), with a Poisson model and assuming uniform rates among sites. Bootstrapping with 1000 replications was conducted to evaluate the phylogenetic tree.
RNA Extraction
Total RNA was isolated using the conventional guanidinium isothiocyanate method and digested with DNase I (TaKaRa, Shiga, Japan) to remove potential DNA contamination. RNA concentration and purity were determined by Nanovue Plus spectrophotometer (GE Healthcare, Piscataway, NJ, United States), and RNA integrity was verified by agarose gel electrophoresis. Only RNA samples with clear bands corresponding to 18S and 28S rRNA on the gel, an OD260/OD280 ratio between 1.8–2.0, and an OD260/OD230 ratio higher than 2.0 were used for subsequent experiments.
Reverse Transcription Quantitative-PCR (RT-qPCR)
First-strand cDNA was synthesized from 2 μg total RNA using oligo(dT)18 and MMLV reverse transcriptase (TaKaRa, Shiga, Japan) in a volume of 20 μl. The reaction was performed at 42°C for 90 min and terminated by heating at 70°C for 10 min. Finally, the cDNA products were diluted to 10 ng/μl and stored at −20°C. Gene-specific primers were designed using Primer Premier 5.0 and listed in Table 1. Amplification efficiency of each primer pair was calculated based on the standard curve generated from a twofold dilution series spanning six orders of magnitude. Quantitative-PCR was conducted using Light Cycler 480 SYBR Green I Master on a Light Cycler 480 Real-time PCR System (Roche Diagnostics, Mannheim, Germany) with the following program: 94°C for 10 min, followed by 40 cycles of 94°C for 15 s and 60°C for 1 min. For each month, six to eight samples were assayed and all reactions were conducted in triplicate. To ensure that the RT-qPCR ran properly, negative controls, including NTCs (no-template controls) and no-reverse transcription controls, and positive controls were included in each run. Melting curve analysis was performed to verify that each primer set amplified a single product (Supplementary Figure S1). EF1A (elongation factor 1-alpha), which was stably expressed throughout the entire experiment, was used as an endogenous control for the normalization of gene expression (Santerre et al., 2013; Teaniniuraitemoana et al., 2015; Li et al., 2016). The relative expression level of each gene was calculated using the 2−ΔΔCt method. Statistical analysis was performed by Welch’s t-tests. Pearson’s correlation coefficient was calculated to explore the relationships between the two genes. P-values lower than 0.05 were considered statistically significant.
In situ Hybridization
To prevent cross-detection of other FOX and DMRT genes, cDNA fragments avoiding conserved DNA binding domains were amplified with specific primers (Table 2) containing a 5′ T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′). Purified PCR products were used as templates for in vitro transcription. Digoxigenin-labeled sense and anti-sense probes were generated using the DIG RNA Labeling Mix (Roche, Mannheim, Germany) and T7 RNA polymerase (Thermo, Waltham, MA, United States). Sections of the gonadal tissues were serially rehydrated in PBST (phosphate-buffered saline plus 0.1% Tween-20) and digested with 2 μg/ml proteinase K at 37°C for 15 min. After pre-hybridization at 60°C for 4 h, hybridization was performed with 1 μg/ml denatured RNA probe in hybridization buffer (50% formamide, 5× SSC, 100 μg/ml yeast tRNA, 1.5% blocking reagent, 0.1% Tween-20) at 60°C for 16 h. Then, the probes were washed away, and antibody incubation was performed in a fresh solution of anti-digoxigenin-AP Fab fragments (Roche, Mannheim, Germany) coupled with blocking buffer (diluted 1:2000) at 4°C for 16 h. After extensive washing with maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl, 0.1% Tween-20, pH = 7.5), sections were incubated with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrate solution and counterstained with 1% neutral red solution.
Results
Histological Analysis of the Juvenile Gonads
Gonads were examined histologically in juveniles aged 5 (shell height ∼10 mm) to 13 months old (shell height ∼60 mm). Figures 1A,B show the morphology of 5- and 6-month-old gonads, respectively. As seen, the majority of the gonads are intestine, surrounded by gonadal tissues. In the 7-month-old gonad, some small-sized follicles have formed (Figure 1C). In the 8-month-old gonad, the follicles grow bigger but are basically empty, containing some follicle cells and sexually indistinguishable gonia (Figure 1D). The 10-month-old gonads are already morphologically differentiated, with oocytes (Figure 1E) or spermatocytes (Figure 1F) scattering in the follicles. Oogonia, oocytes, and a few mature oocytes coexist in the 11-month-old ovary (Figure 1G), and several layers of spermatogonia and spermatocytes are found in the 11-month-old testis (Figure 1H), indicating that the gonads are at the growing stage. As shown in Figures 1I,J, the 13-month-old gonads have reached full maturity. In the ovary, the follicles are filled with mature oocytes with a polygonal shape due to packing (Figure 1I). In the follicles of the testis, diverse germ cells can be detected, including spermatogonia, spermatocytes, spermatids, and spermatozoa (Figure 1J).
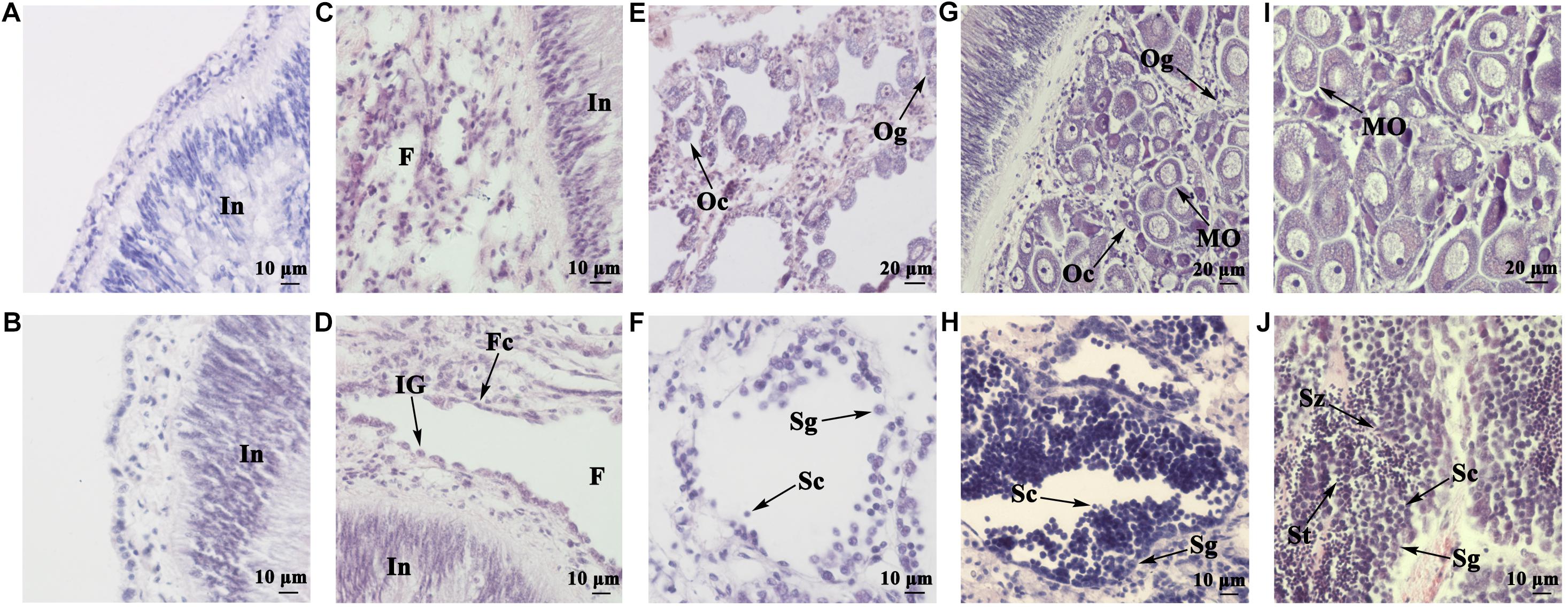
FIGURE 1. Histological observation of juvenile gonads of different ages. (A) Gonad of a 5-month-old juvenile. (B) Gonad of a 6-month-old juvenile. (C) Gonad of a 7-month-old juvenile. (D) Gonad of an 8-month-old juvenile. (E) Ovary of a 10-month-old juvenile. (F) Testis of a 10-month-old juvenile. (G) Ovary of an 11-month-old juvenile. (H) Testis of an 11-month-old juvenile. (I) Ovary of a 13-month-old juvenile. (J) Testis of a 13-month-old juvenile. In, intestine; F, follicle; Fc, follicle cell; IG, indistinguishable gonium; Og, oogonium; Oc, oocyte; MO, mature oocyte; Sg, spermatogonium; Sc, spermatocyte; St, spermatid; Sz, spermatozoon.
Selection of Candidate Genes for Sex Analysis
To determine the onset of molecular sex differentiation, molecular markers were screened for sex identification. All eight candidate genes (FOXL2, WNT4, FST, β-catenin, DMRT1L, DAX1, SOXE, and SOXH) were identified in the scallop genome. Their expression patterns in ovaries and testes were examined by RT-qPCR. In order to obtain genes that show sexually dimorphic expression in somatic and/or gonia cells and eliminate the effects of gametogenesis, we only used gonads at the resting stage for gene expression analysis (Yu et al., 2017). As shown in Figure 2A, only FOXL2 and DMRT1L displayed significant (P < 0.01) sexually dimorphic expression, with FOXL2 being abundant in ovaries, and DMRT1L in testes. Therefore, LOG10(DMRT1L/FOXL2) is a potential marker for scallop sex identification, which led us to further investigate its values in the other three reproductive stages (proliferative, growing, and maturation stages). The results also confirmed that LOG10(DMRT1L/FOXL2) could be used to determine the sex of differentiated scallops throughout the reproductive cycle: the value was always lower than 0 for ovaries and higher than 2 for testes (Figure 2B).
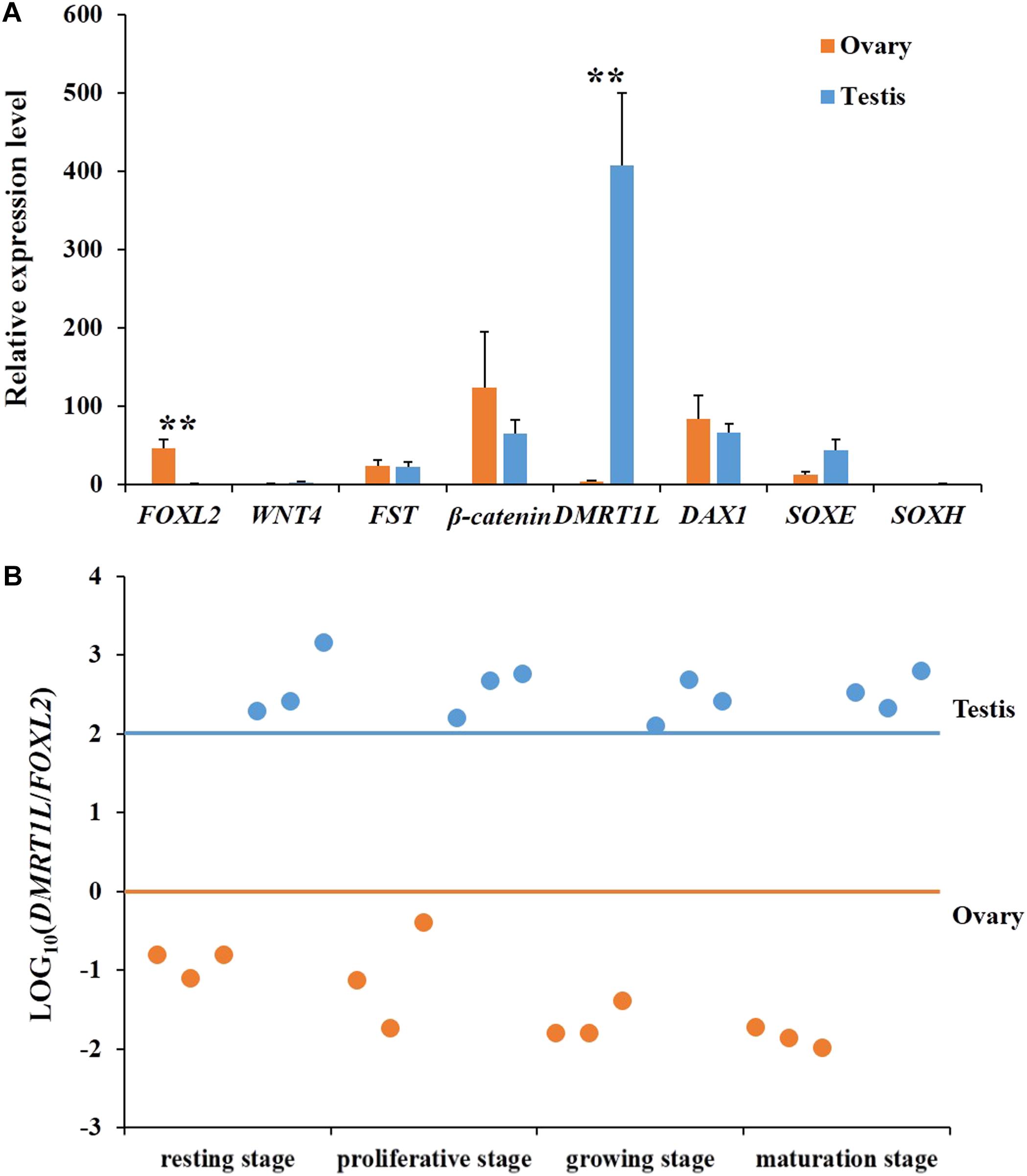
FIGURE 2. Screening of key genes responsible for scallop sex differentiation. (A) Expression profiles of eight candidate genes (FOXL2, WNT4, FST, β-catenin, DMRT1L, DAX1, SOXE, and SOXH) in scallop ovaries (orange) and testes (blue) at the resting stage. The vertical bars represent the means ± SE (N = 7). “∗∗” indicates differences that are statistically significant (P < 0.01). (B) The LOG10(DMRT1L/FOXL2) in ovaries (orange) and testes (blue) at the resting stage, proliferative stage, growing stage, and maturation stage. For each stage, six gonads, including three ovaries and three testes, were assayed. The horizontal lines indicate the threshold for sex differentiation, i.e., orange for ovary and blue for testis, respectively.
Phylogenetic Analyses of FOXL2 and DMRT1L
To determine the identity of the two sexually dimorphic expressed genes, phylogenetic analysis was conducted. As shown in Figure 3A, Yesso scallop FOXL2 clustered with FOXL2 from other bivalves and gastropods, and then clustered with vertebrate FOXL2 and FOXL3. In Figure 3B, the selected DMRT proteins were clustered into several major groups. The Yesso scallop DMRT, together with DMRT from Mimachlamys nobilis and Haliotis asinina forms a new group, namely, DMRT1L.
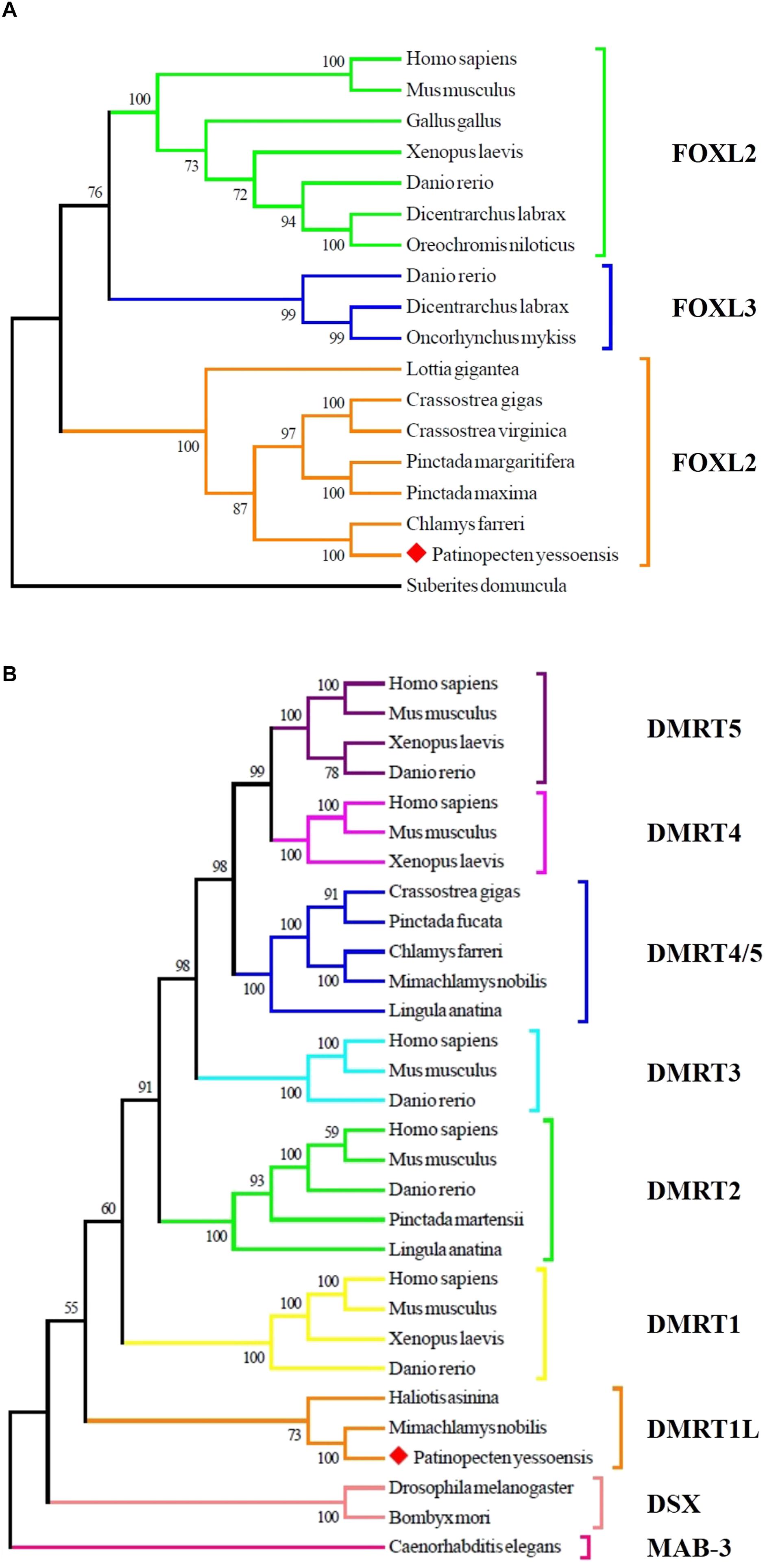
FIGURE 3. Phylogenetic analyses of FOXL2 (A) and DMRT (B) proteins. The numbers show the bootstrap percentages (1000 replicates) obtained using neighbor-joining (NJ) method. Different branch colors denote different groups, and red diamonds indicate FOXL2 and DMRT1L of P. yessoensis. The protein sequences used for phylogenetic analyses include: FOXL2 of Homo sapiens (AAY21823.1), Mus musculus (NP_036150.1), Gallus gallus (NP_001012630.1), Xenopus laevis (BAG69484.1), Danio rerio (AAI16586.1), Dicentrarchus labrax (AGS36082.1), Oreochromis niloticus (AAT36328.1), Lottia gigantea (BAQ19215.1), Crassostrea gigas (ACN80999.1), Crassostrea virginica (XP_022345405.1), Pinctada margaritifera (AIE16098.1), Pinctada maxima (ATJ00808.1), Chlamys farreri (AFB35647.1), Suberites domuncula (CAE51212.1); FOXL3 of Danio rerio (AAI62838.1), Dicentrarchus labrax (AFV13295.1), Oncorhynchus mykiss (NP_001117956.1); MAB-3 of Caenorhabditis elegans (NP_871909.1); DSX of Drosophila melanogaster (NP_731197.1), Bombyx mori (AGS48306.1); DMRT1 of Homo sapiens (NP_068770.2), Mus musculus (NP_056641.2), Xenopus laevis (BAE45870.1), Danio rerio (AAU04562.1); DMRT1L of Haliotis asinina (ACC94178.1), Mimachlamys nobilis (AHW85419.1); DMRT2 of Homo sapiens (AAD40475.1), Mus musculus (NP_665830.1), Danio rerio (NP_571027.1), Pinctada martensii (ADD97887.1), Lingula anatina (XP_013397304.1); DMRT3 of Homo sapiens (NP_067063.1), Mus musculus (NP_796334.2), Danio rerio (AAU89440.1); DMRT4 of Homo sapiens (AAI30436.1), Mus musculus (AAN77234.1), Xenopus laevis (AAV66322.1); DMRT5 of Homo sapiens (Q96SC8.2), Mus musculus (AAN10254.1), Xenopus laevis (AAI70170.1), Danio rerio (AAU85258.1); DMRT4/5 of Crassostrea gigas (ABS88697.1), Pinctada fucata (AIW04133.1), Chlamys farreri (ADK55063.1), Mimachlamys nobilis (AHW85420.1), Lingula anatina (XP_013419494.1).
Localization of FOXL2 and DMRT1L mRNA
Spatial expression of FOXL2 and DMRT1L was detected in the ovary and testis at the proliferative stage after sex differentiation. For both genes, the signals were present in germ cells and follicle cells. Specifically, the anti-sense probe of FOXL2 was detected in the cytoplasm of oogonia, oocytes, and follicle cells (Figure 4A) and faint in testis (Figure 4B). DMRT1L transcripts were located clearly in all germ cells of the ovary (Figure 4C) and testis (Figure 4D), but the signal intensity was higher in the testis than in the ovary. No signal was detected in the ovary or testis with the sense probes (Figures 4E–H).
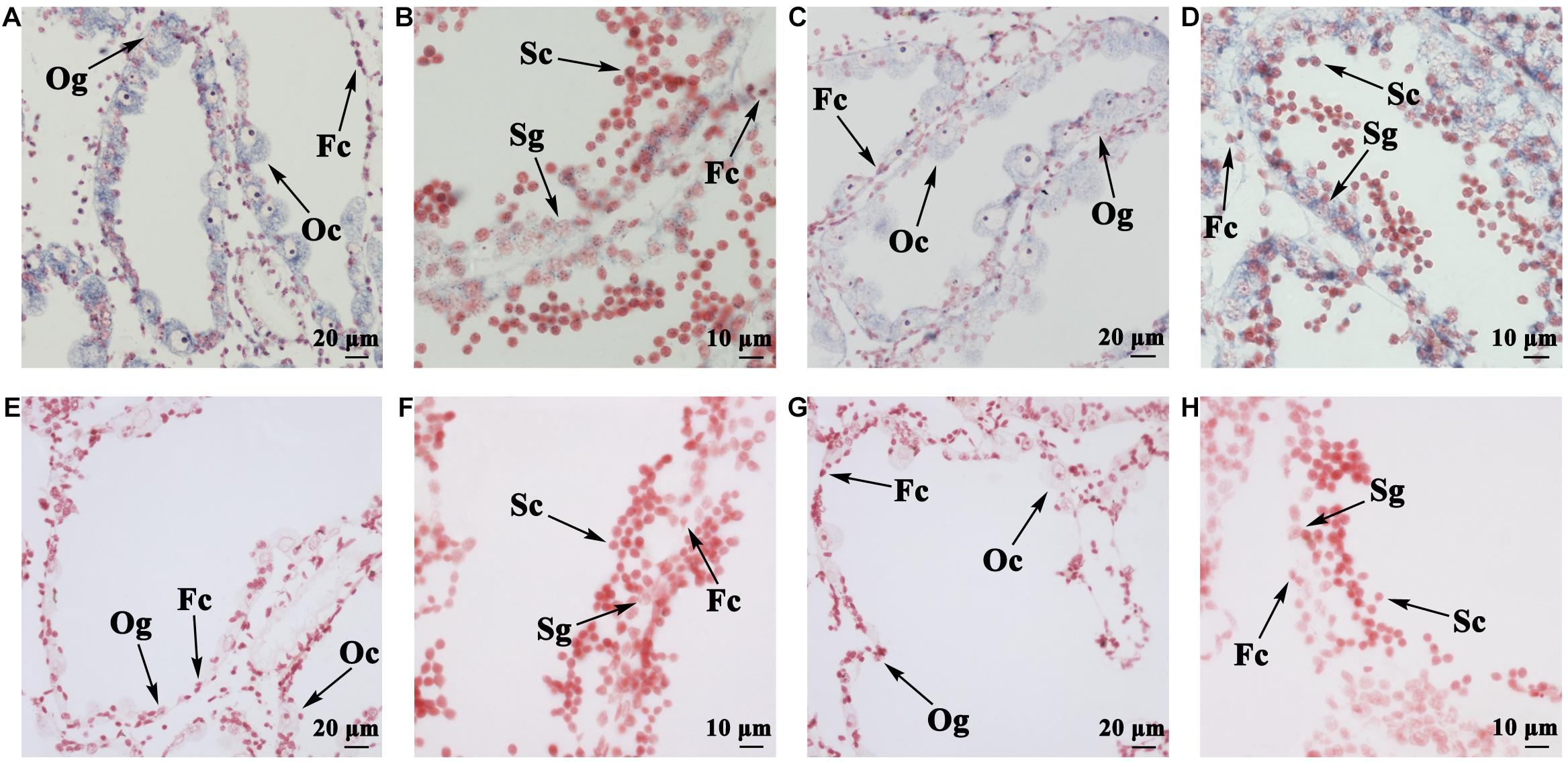
FIGURE 4. Localization of FOXL2 and DMRT1L in gonads by in situ hybridization. (A) Localization of FOXL2 with an anti-sense probe in ovaries. (B) Localization of FOXL2 with an anti-sense probe in testes. (C) Localization of DMRT1L with an anti-sense probe in ovaries. (D) Localization of DMRT1L with an anti-sense probe in testes. (E) Localization of FOXL2 with a sense probe in ovaries. (F) Localization of FOXL2 with a sense probe in testes. (G) Localization of DMRT1L with a sense probe in ovaries. (H) Localization of DMRT1L with a sense probe in testes. Fc, follicle cell; Og, oogonium; Oc, oocyte; Sg, spermatogonium; Sc, spermatocyte.
Determining the Timing of Molecular Sex Differentiation
The feasibility of LOG10(DMRT1L/FOXL2) for accurate sex identification enables us to determine the timing of gonadal sex differentiation in the Yesso scallop. Here, we traced the dynamic changes of LOG10(DMRT1L/FOXL2) in juveniles aged 5–13 months old. According to Figure 5A, LOG10(DMRT1L/FOXL2) values of young individuals (5 and 6 months old) are between 0 and 1, suggesting that these scallops are still undifferentiated. In 7- to 10-month-old scallops, we found a decreasing number of LOG10(DMRT1L/FOXL2) falling within 0–1. This indicates that sex differentiation does not occur simultaneously in all individuals, with some scallops differentiating early when they are 7 months old and some not differentiating until they are 10 months old. However, after reaching 11 months of age, all investigated gonads had differentiated into ovaries or testes. The histogram in Figure 5B shows the corresponding differentiation rate for each month based on the LOG10(DMRT1L/FOXL2) values. It reveals an increase in the differentiation rate from 0 in 5- and 6-month-old individuals to 100% in 11- to 13-month-old scallops.
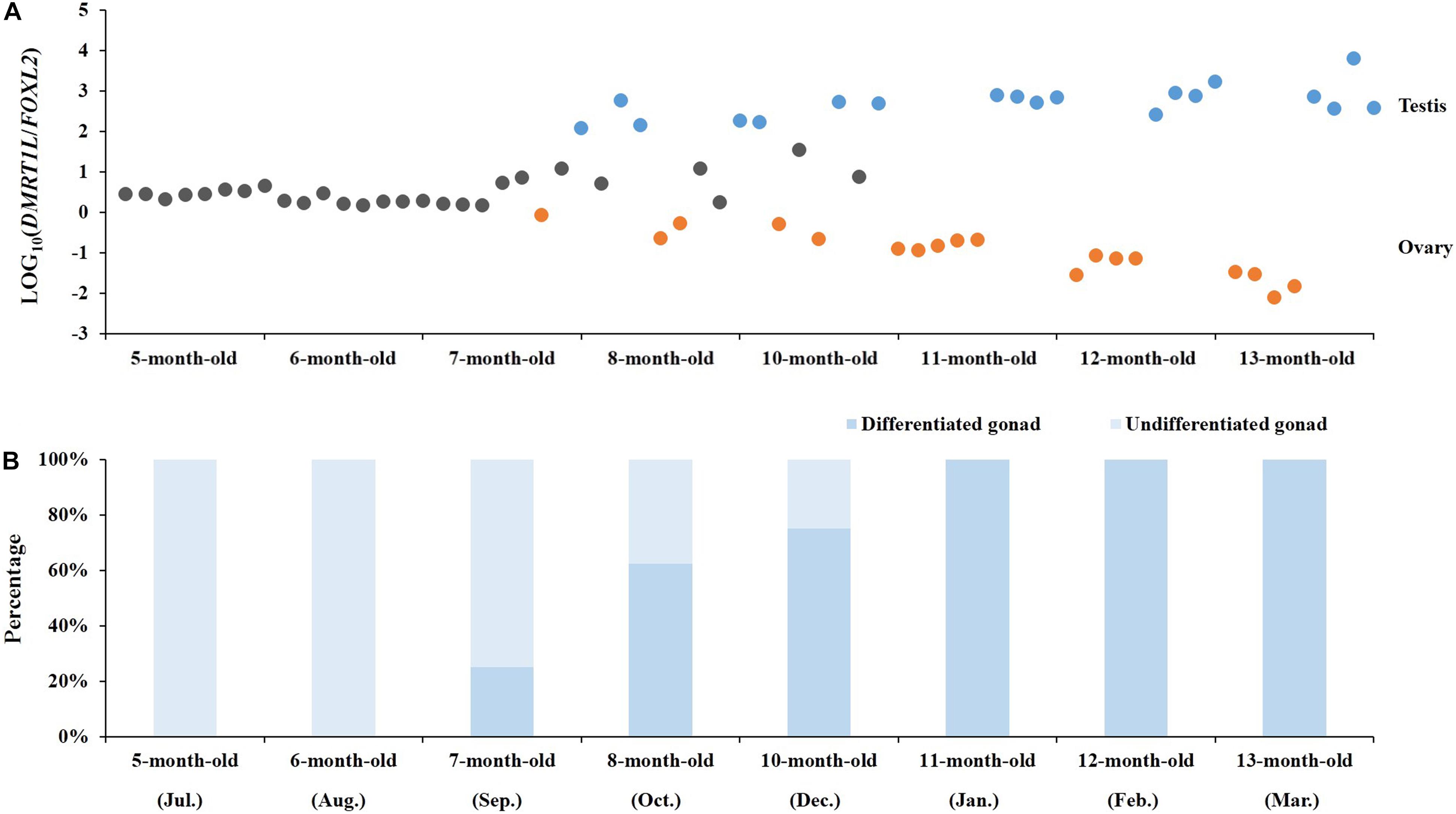
FIGURE 5. Determination of the timing of molecular sex differentiation in juvenile scallops. (A) The dynamic changes in LOG10(DMRT1L/FOXL2) in 5- to 13-month-old juvenile gonads (N = 8 for each month). Gray, orange, and blue dots stand for undifferentiated gonads, ovaries and testes, respectively. (B) The histogram indicates the corresponding percentage of undifferentiated (light blue) and differentiated gonads (blue) in each month.
Coexpression of FOXL2 and DMRT1L in the Juvenile Gonads
Figure 6A shows the temporal expression profiles of both FOXL2 and DMRT1L in juveniles of 5- to 13-month-olds. According to the results, FOXL2 and DMRT1L are relatively low in 5- to 10-month-old scallops, but expression of FOXL2 in ovaries and DMRT1L in testes was dramatically increased in 11- to 13-month-old scallops. During the eight investigated months, there existed two major coexpression patterns between FOXL2 and DMRT1L: (i) a significantly positive correlation for the two undifferentiated stages (r = 0.955–0.980, P < 0.01), with the regression equation of y = 1.018x + 0.370 (Figure 6B); (ii) significantly negative correlation after the completion of sex differentiation in the last three time points (r = −0.791 to −0.957, P < 0.05), with the regression equation of y = −0.684x + 1.584 (Figure 6C). Between the undifferentiated and fully differentiated months are three partially differentiated ones, in which correlation coefficients displayed a transition from significantly positive (0.860) to negative (−0.587).
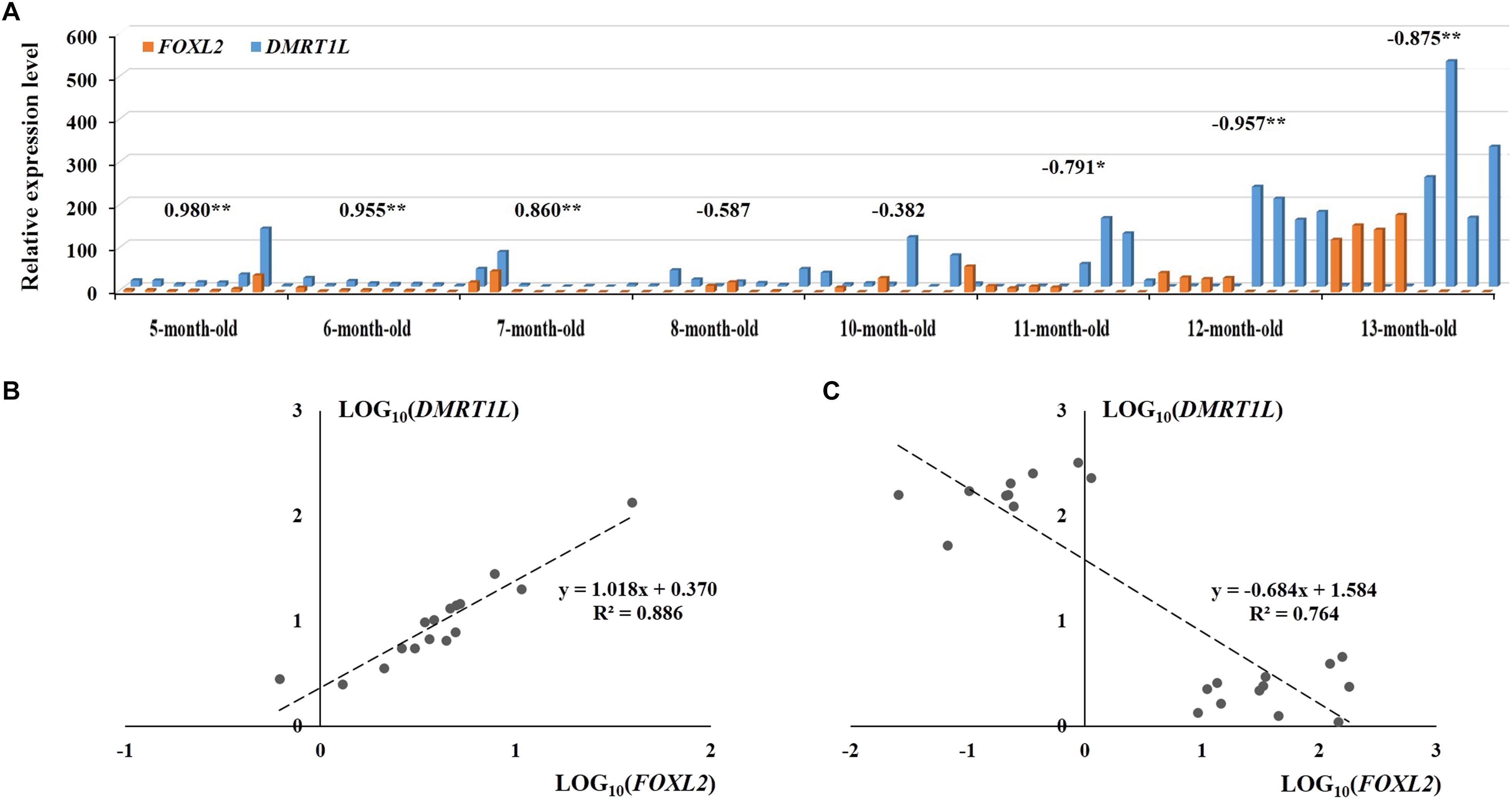
FIGURE 6. The expression patterns of FOXL2 and DMRT1L in juvenile scallops 5–13 months old. (A) The samples correspond to those in Figure 5A. The numbers above the bars are the correlation coefficients between LOG10(FOXL2) and LOG10(DMRT1L) in the particular months. “∗” indicates P < 0.05; “∗∗” indicates P < 0.01. (B) The positive correlation between LOG10(FOXL2) and LOG10(DMRT1L) for 5- to 6-month-old undifferentiated gonads. (C) The negative correlation between LOG10(FOXL2) and LOG10(DMRT1L) for 11- to 13-month-old differentiated gonads.
Discussion
By examining the gonadal expression of eight candidate sex determination or differentiation genes in the resting stage, we found that only two genes (FOXL2 and DMRT1L) displayed sexually dimorphic expression. Both genes are transcription factors, with the former being a forkhead transcription factor and the latter encoding a DM domain-containing protein. Specific sequence information, including conserved domain alignments, has been described previously (Li et al., 2016). According to the transcriptomes of nine adult tissues/organs (Wang et al., 2017), FOXL2 and DMRT1L seem to be specifically expressed in scallop gonads, and no evidence of splice isoforms was found for DMRT1L despite that splicing of DMRT is common in many animals (Nothiger et al., 1987; Kopp, 2012). Interestingly, sexually dimorphic expression of FOXL2 (Liu et al., 2012; Teaniniuraitemoana et al., 2014; Zhang et al., 2014) and DMRT1L (Shi et al., 2014) occurs not only in the Yesso scallop but also in other bivalves. Therefore, FOXL2 and DMRT1L, like Yin and Yang, may be crucial genes for female and male gonadal differentiation in bivalves. Similarly, in mammals, FOXL2 is an ovarian marker that suppresses genes involved in testicular differentiation from the early embryonic gonad throughout adult life (Cocquet et al., 2002; Ottolenghi et al., 2007; Uhlenhaut et al., 2009; Liu et al., 2014). Although DMRT genes have diverse roles in development and physiology (Huang et al., 2005; Hong et al., 2007; Reitzel et al., 2016), DMRT1 is a deeply conserved gene that is involved in testis differentiation and development in various animal taxa (Naimi et al., 2009b; Kopp, 2012; Matson and Zarkower, 2012; Picard et al., 2015). In mammals, a battle between FOXL2 and DMRT1 for sex control has been proposed (Herpin and Schartl, 2011; Matson et al., 2011). We assume that this battle may be extended to invertebrates, but with different DMRT members.
Spatial expression of FOXL2 and DMRT1L in the ovary and testis reveals that both genes are distributed in germ cells and follicle cells, consistent with previous research in other marine bivalves (Naimi et al., 2009a; Liu et al., 2012). The signal intensity also agrees with the RT-qPCR data. The expression patterns of FOXL2 and DMRT1L make LOG10(DMRT1L/FOXL2) an alternative method to histological examination for scallop sex identification. By tracking LOG10(DMRT1L/FOXL2) at four gonadal developmental stages, we found the value was always lower than 0 for ovaries and higher than 2 for testes, which indicates that in the Yesso scallop, (i) there is no sex reversal once the gonads differentiate, (ii) FOXL2 and DMRT1L may play important roles in both sex maintenance and gametogenesis, and (iii) LOG10(DMRT1L/FOXL2) is an effective solution for sex identification during the resting stage, which is usually indistinguishable by morphology- or histology-based approaches. Since FOXL2 and DMRT1L also show sexually dimorphic expression patterns in other bivalves (Liu et al., 2012; Shi et al., 2014; Teaniniuraitemoana et al., 2014; Zhang et al., 2014), using LOG10(DMRT1L/FOXL2) for sex identification may be applicable in these organisms. For oysters that are capable of sex reversal (Teaniniuraitemoana et al., 2014; Zhang et al., 2014), sex-reversing individuals may be easily screened out by examining the LOG10(DMRT1L/FOXL2) values that fall between the ovary and testis thresholds, which could contribute to a better understanding of the molecular mechanisms underlying sex reversal.
Based on the histological analysis, we found that the developmental process of juvenile gonads can be classified into four stages, namely, undifferentiated, differentiated, growing, and maturation stages, which are consistent with the documentation by Shumway and Parsons (2016). Combined with the dynamic changes of LOG10(DMRT1L/FOXL2) in juvenile scallops, we found that sex differentiation occurs earlier at the molecular level than at the histological level, which is similar to the findings in other organisms (Siegfried, 2010; Haugen et al., 2012; Ayers et al., 2013; Robledo et al., 2015; Tao et al., 2018). In addition, our study shows that although the onset of molecular sex differentiation varies between individuals, it always occurs from 7 to 10 months of age, and almost all of the scallops investigated completed differentiation before 11 months of age. This falls within the time frame of gonadal differentiation documented by Shumway and Parsons (2016) (between 4 months old and 1 year old). It suggests that LOG10(DMRT1L/FOXL2) allows early identification of sex, and is a convenient and accurate way to assess the molecular sex differentiation period. This opens a gate for unraveling the regulatory network of sex differentiation in the Yesso scallop.
The coexpression patterns of FOXL2 and DMRT1L are different before and after gonadal differentiation. Before differentiation, FOXL2 and DMRT1L are relatively low but display unexpected coordinated expression patterns. Combined with the histological results showing that the follicles have not yet formed at this stage, we assume these two genes may coexpress in GSCs that are dispersed in the connective tissues. Sex differentiation co-occurs with the formation of follicles, in which relative expression of FOXL2 vs DMRT1L changes, possibly leading to the differentiation of undifferentiated germ cells into spermatogonia or oogonia. Therefore, we speculate that in Yesso scallops, the undifferentiated gonads directly differentiate into either ovaries or testes. This challenges a previous opinion regarding the existence of sex reversal from male to female through an undifferentiated stage (Maru, 1978). In particular, some researchers claimed that Yesso scallops grow as males at less than one year of age; sex is thereafter reversed to female through an undifferentiated or ambiguous status in half of the scallops, resulting in a sex ratio close to 1:1; after sex reversal, the sex may become stable throughout successive periods of life (Mori et al., 1977; Maru, 1978; Otani et al., 2017). Considering that these studies are generally based on macroscopic observation of gonadal color (testis with white color, ovary with orange or pink color), which could be misleading in some cases, we assume that findings of sex reversal in Yesso scallops may be an artifact. However, since environmental factors have been demonstrated to be involved in the sex determination of many bivalves, we cannot rule out the possibility that the Yesso scallop does undergo sex reversal in Japan.
Author Contributions
LinZ and SW conceived and designed the experiments. RL, WL, and YZ performed the experiments. YL, MZ, LiaZ, and XH collected the samples. RL and LinZ analyzed the data. RL, LinZ, SW, and ZB wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31572600, U1706203, and 2018A07) and Fundamental Research Funds for the Central Universities (Grant Nos. 201762001 and 201841001).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01166/full#supplementary-material
References
Ayers, K. L., Davidson, N. M., Demiyah, D., Roeszler, K. N., Grützner, F., Sinclair, A. H., et al. (2013). RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol. 14:R26. doi: 10.1186/gb-2013-14-3-r26
Bull, J. J. (1983). Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin/Cummings Publishing Company, Inc.
Cocquet, J., Pailhoux, E., Jaubert, F., Servel, N., Xia, X., Pannetier, M., et al. (2002). Evolution and expression of FOXL2. J. Med. Genet. 39, 916–921. doi: 10.1136/jmg.39.12.916
Feng, Z., Shao, M., Sun, D., and Zhang, Z. (2010). Cloning, characterization and expression analysis of Cf-dmrt4-like gene in Chlamys farreri. J. Fish. Sci. China 17, 930–940.
Goodwin, E. B., and Ellis, R. E. (2002). Turning clustering loops: sex determination in Caenorhabditis elegans. Curr. Biol. 12, R111–R120. doi: 10.1016/S0960-9822(02)00675-9
Haugen, T., Almeida, F. F., Andersson, E., Bogerd, J., Male, R., Skaar, K. S., et al. (2012). Sex differentiation in Atlantic cod (Gadus morhua L.): morphological and gene expression studies. Reprod. Biol. Endocrinol. 10:47. doi: 10.1186/1477-7827-10-47
Herpin, A., and Schartl, M. (2011). Sex determination: switch and suppress. Curr. Biol. 21, R656–R659. doi: 10.1016/j.cub.2011.07.026
Heule, C., Göppert, C., Salzburger, W., and Böhne, A. (2014). Genetics and timing of sex determination in the East African cichlid fish Astatotilapia burtoni. BMC Genet. 15:140. doi: 10.1186/s12863-014-0140-5
Hong, C. S., Park, B. Y., and Saintjeannet, J. P. (2007). The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 310, 1–9. doi: 10.1016/j.ydbio.2007.07.035
Huang, X., Hong, C. S., O’Donnell, M., and Saintjeannet, J. P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. U.S.A. 102, 11349–11354. doi: 10.1073/pnas.0505106102
Kocer, A., Reichmann, J., Best, D., and Adams, I. R. (2009). Germ cell sex determination in mammals. Mol. Hum. Reprod. 15, 205–213. doi: 10.1093/molehr/gap008
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175–184. doi: 10.1016/j.tig.2012.02.002
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R. M., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W. clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, H., Liu, J., Huang, X., Wang, D., and Zhang, Z. (2014a). Characterization, expression and function analysis of DAX1 gene of scallop (Chlamys farreri Jones and Preston 1904) during its gametogenesis. J. Ocean Univ. China 13, 696–704. doi: 10.1007/s11802-014-2299-9
Li, H., Zhang, Z., Bi, Y., Yang, D., Zhang, L., and Liu, J. (2014b). Expression characteristics of β-catenin in scallop Chlamys farreri gonads and its role as a potential upstream gene of Dax1 through canonical Wnt signalling pathway regulating the spermatogenesis. PLoS One 9:e115917. doi: 10.1371/journal.pone.0115917
Li, H., Liu, J., Liu, X., and Zhang, Z. (2013). Molecular cloning and expression analysis of wnt4 cDNA from the Zhikong scallop Chlamys farreri. J. Fish. Sci. China 20, 260–268.
Li, Y., Zhang, L., Sun, Y., Ma, X., Wang, J., Li, R., et al. (2016). Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop patinopecten yessoensis. Mar. Biotechnol. 18, 453–465. doi: 10.1007/s10126-016-9706-8
Liu, X. L., Li, Y., Liu, J. G., Cui, L. B., and Zhang, Z. F. (2014). Gonadogenesis in scallop Chlamys farreri and Cf-foxl2 expression pattern during gonadal sex differentiation. Aquac. Res. 47, 1605–1611. doi: 10.1111/are.12621
Liu, X.-L., Zhang, Z.-F., Shao, M.-Y., Liu, J.-G., and Muhammad, F. (2012). Sexually dimorphic expression of foxl2 during gametogenesis in scallop Chlamys farreri, conserved with vertebrates. Dev. Genes Evol. 222, 279–286. doi: 10.1007/s00427-012-0410-z
Maru, K. (1978). Studies on the reproduction of a scallop, Patinopecten yessoensis (Jay), 2: gonad development in 1-year-old scallops. Sci. Rep. Hokkaido Fish. Exp. Stn. 20, 13–26.
Matson, C. K., Murphy, M. W., Sarver, A. L., Griswold, M. D., Bardwell, V. J., and Zarkower, D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104. doi: 10.1038/nature10239
Matson, C. K., and Zarkower, D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163–174. doi: 10.1038/nrg3161
Mori, K., Sato, R., and Osanai, K. (1977). Seasonal gonad changes in scallops under culture in Toni Bay, Iwate Prefecture [Japan]. Bull. Jpn. Soc. Sci. Fish. 43, 1–8. doi: 10.2331/suisan.43.1
Nagasawa, K., Oouchi, H., Itoh, N., Takahashi, K. G., and Osada, M. (2015). In Vivo administration of scallop gnrh-like peptide influences on gonad development in the yesso Scallop, Patinopecten yessoensis. PLoS One 10:e0129571. doi: 10.1371/journal.pone.0129571
Naimi, A., Martinez, A.-S., Specq, M.-L., Diss, B., Mathieu, M., and Sourdaine, P. (2009a). Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 134–142. doi: 10.1016/j.cbpb.2009.05.011
Naimi, A., Martinez, A.-S., Specq, M.-L., Mrac, A., Diss, B., Mathieu, M., et al. (2009b). Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 152, 189–196. doi: 10.1016/j.cbpa.2008.09.019
Ni, J., Zeng, Z., Han, G., Huang, H., and Ke, C. (2012). Cloning and characterization of the follistatin gene from Crassostrea angulata and its expression during the reproductive cycle. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 163, 246–253. doi: 10.1016/j.cbpb.2012.06.006
Nothiger, R., Leuthold, M., Andersen, N., Gerschwiler, P., Gruter, A., Keller, W., et al. (1987). Genetic and developmental analysis of the sex-determining gene ‘double sex’ (dsx) of Drosophila melanogaster. Genet. Res. 50, 113–123. doi: 10.1017/S001667230002351X
Osada, M., Harata, M., Kishida, M., and Kijima, A. (2004). Molecular cloning and expression analysis of vitellogenin in scallop, Patinopecten yessoensis (Bivalvia, mollusca). Mol. Reprod. Dev. 67, 273–281. doi: 10.1016/j.dci.2014.12.004
Otani, A., Nakajima, T., Okumura, T., Fujii, S., and Tomooka, Y. (2017). Sex reversal and analyses of possible involvement of sex steroids in scallop gonadal development in newly established organ-culture systems. Zoolog. Sci. 34, 86–92. doi: 10.2108/zs160070
Ottolenghi, C., Pelosi, E., Tran, J., Colombino, M., Douglass, E., Nedorezov, T., et al. (2007). Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet. 16, 2795–2804. doi: 10.1093/hmg/ddm235
Patnaik, B. B., Wang, T. H., Kang, S. W., Hwang, H.-J., Park, S. Y., Park, E. B., et al. (2016). Sequencing, de novo assembly, and annotation of the transcriptome of the endangered freshwater pearl bivalve, Cristaria plicata, provides novel insights into functional genes and marker discovery. PLoS One 11:e0148622. doi: 10.1371/journal.pone.0148622
Penman, D. J., and Piferrer, F. (2008). Fish gonadogenesis. Part I: genetic and environmental mechanisms of sex determination. Rev. Fish. Sci. 16, 16–34. doi: 10.1080/10641260802324610
Picard, M. A. L., Cosseau, C., Mouahid, G., Duval, D., Grunau, C., Toulza, È, et al. (2015). The roles of Dmrt (double sex/male-abnormal-3 related transcription factor) genes in sex determination and differentiation mechanisms: ubiquity and diversity across the animal kingdom. C. R. Biol. 338, 451–462. doi: 10.1016/j.crvi.2015.04.010
Piferrer, F., and Guiguen, Y. (2008). Fish gonadogenesis. Part II: molecular biology and genomics of sex differentiation. Rev. Fish. Sci. 16, 35–55. doi: 10.1080/10641260802324644
Reitzel, A. M., Pang, K., and Martindale, M. Q. (2016). Developmental expression of “germline”- and “sex determination”-related genes in the ctenophore Mnemiopsis leidyi. Evodevo 7:17. doi: 10.1186/s13227-016-0051-9
Robledo, D., Ribas, L., Cal, R., Sánchez, L., Piferrer, F., Martínez, P., et al. (2015). Gene expression analysis at the onset of sex differentiation in turbot (Scophthalmus maximus). BMC Genomics 16:973. doi: 10.1186/s12864-015-2142-8
Santerre, C., Sourdaine, P., Adeline, B., and Martinez, A.-S. (2014). Cg-SoxE and Cg-β-catenin, two new potential actors of the sex-determining pathway in a hermaphrodite lophotrochozoan, the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 167, 68–76. doi: 10.1016/j.cbpa.2013.09.018
Santerre, C., Sourdaine, P., Marc, N., Mingant, C., Robert, R., and Martinez, A.-S. (2013). Oyster sex determination is influenced by temperature—first clues in spat during first gonadic differentiation and gametogenesis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 165, 61–69. doi: 10.1016/j.cbpa.2013.02.007
Shi, Y., Wang, Q., and He, M. (2014). Molecular identification of dmrt2 and dmrt5 and effect of sex steroids on their expressions in Chlamys nobilis. Aquaculture 426, 21–30. doi: 10.1016/j.aquaculture.2014.01.021
Shumway, S. E., and Parsons, G. J. (2016). Scallops: Biology, Ecology, Aquaculture, and Fisheries. New York, NY: Elsevier.
Siegfried, K. R. (2010). In search of determinants: gene expression during gonadal sex differentiation. J. Fish Biol. 76, 1879–1902. doi: 10.1111/j.1095-8649.2010.02594.x
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271. doi: 10.1038/nature08298
Swain, A., and Lovell-Badge, R. (1999). Mammalian sex determination: a molecular drama. Genes Dev. 13, 755–767. doi: 10.1101/gad.13.7.755
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanabe, T., Yuan, Y., Nakamura, S., Itoh, N., Takahashi, K. G., and Osada, M. (2010). The role in spawning of a putative serotonin receptor isolated from the germ and ciliary cells of the gonoduct in the gonad of the Japanese scallop, Patinopecten yessoensis. Gen. Comp. Endocrinol. 166, 620–627. doi: 10.1016/j.ygcen.2010.01.014
Tao, W., Chen, J., Tan, D., Yang, J., Sun, L., Wei, J., et al. (2018). Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genomics 19:363. doi: 10.1186/s12864-018-4756-0
Teaniniuraitemoana, V., Huvet, A., Levy, P., Gaertner-Mazouni, N., Gueguen, Y., and Le Moullac, G. (2015). Molecular signatures discriminating the male and the female sexual pathways in the pearl oyster Pinctada margaritifera.PLoS One 10:e0122819. doi: 10.1371/journal.pone.0122819
Teaniniuraitemoana, V., Huvet, A., Levy, P., Klopp, C., Lhuillier, E., Gaertner-Mazouni, N., et al. (2014). Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics 15:491. doi: 10.1186/1471-2164-15-491
Tong, Y., Zhang, Y., Huang, J., Xiao, S., Zhang, Y., Li, J., et al. (2015). Transcriptomics analysis of Crassostrea hongkongensis for the discovery of reproduction-related genes. PLoS One 10:e0134280. doi: 10.1371/journal.pone.0134280
Uhlenhaut, N. H., Jakob, S., Anlag, K., Eisenberger, T., Sekido, R., Kress, J., et al. (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142. doi: 10.1016/j.cell.2009.11.021
Wang, S., Zhang, J., Jiao, W., Li, J., Xun, X., Sun, Y., et al. (2017). Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1:0120. doi: 10.1038/s41559-017-0120
Yang, M., Xu, F., and Liu, J. (2015). Molecular cloning and expression of Wnt4 gene in pacific oyster Crassostrea gigas. Oceanol. Limnol. Sin. 46, 35–42.
Yu, F.-F., Wang, M.-F., Zhou, L., Gui, J.-F., and Yu, X.-Y. (2011). Molecular cloning and expression characterization of Dmrt2 in Akoya pearl oysters, Pinctada martensii. J. Shellfish Res. 30, 247–254. doi: 10.2983/035.030.0208
Yu, J., Zhang, L., Li, Y., Li, R., Zhang, M., Li, W., et al. (2017). Genome-wide identification and expression profiling of the SOX gene family in a bivalve mollusc Patinopecten yessoensis. Gene 627, 530–537. doi: 10.1016/j.gene.2017.07.013
Zarkower, D. (2001). Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2, 175–185. doi: 10.1038/35056032
Keywords: Yesso scallop, sex differentiation, FOXL2, DMRT1L, LOG10(DMRT1L/FOXL2)
Citation: Li R, Zhang L, Li W, Zhang Y, Li Y, Zhang M, Zhao L, Hu X, Wang S and Bao Z (2018) FOXL2 and DMRT1L Are Yin and Yang Genes for Determining Timing of Sex Differentiation in the Bivalve Mollusk Patinopecten yessoensis. Front. Physiol. 9:1166. doi: 10.3389/fphys.2018.01166
Received: 08 May 2018; Accepted: 03 August 2018;
Published: 22 August 2018.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, United StatesZhigang Shen, Huazhong Agricultural University, China
Copyright © 2018 Li, Zhang, Li, Zhang, Li, Zhang, Zhao, Hu, Wang and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingling Zhang, bGluZ2xpbmc4MEBvdWMuZWR1LmNu
†These authors have contributed equally to this work
 Ruojiao Li1,2†
Ruojiao Li1,2† Lingling Zhang
Lingling Zhang Meiwei Zhang
Meiwei Zhang Shi Wang
Shi Wang Zhenmin Bao
Zhenmin Bao
