
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 07 August 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01067
Chilo supprressalis is one of the most important rice pests that causes serious damage to production in the rice growth area of Asia. Vetiver grass (Vetiveria zizanioides) was previously found to effectively attract female adults of C. suppressalis laying eggs on vetiver leaves, while the larvae cannot complete their life cycles by feeding on vetiver, indicating a potential means of controlling this pest. In the present study, the transcriptomes of midguts of rice-fed and vetiver-fed C. suppressalis larvae were profiled, which aimed to clarify the molecular mechanism of vetiver as a dead-end trap plant preliminarily. We found that ingestion of vetiver provoked a robust transcriptional response in the larval midguts, and a total of 1,849 differentially expressed UniGenes were identified. We focused on 12 digestion-related genes, four immune-related genes and three detoxification-related genes. Most of these genes were significantly down regulated in the larval midguts at 6, 8, and 10 days after feeding on vetiver compared to on rice. Transcriptional dynamics suggested that these genes might be involved in toxicity responses following exposure to vetiver. Taken together, this study provides an initial molecular framework for developing biological control strategies for C. suppressalis in an effort to protect economically important rice crops.
Rice (Oryza sativa L.) is the staple food for half the world’s population. It is frequently attacked by rice stem borers, which are persistent and chronic pests occurring in nearly all paddy fields during each growing season throughout the rice producing area of Asia. Among these stem borers, the striped stem borer Chilo suppressalis (Walker) (Lepidoptera: Crambidae) is widely distributed. It is responsible for huge annual yield losses for rice (Qu et al., 2003). C. suppressalis has developed resistance to numerous insecticides, which has led to frequent control failures in paddy fields (Lu et al., 2017b). Hence, considerable attention has been paid to trap plants as a means of biological control of C. suppressalis.
Vetiver grass (Vetiveria zizanioides L.), which is typically planted to stop soil erosion on hills and roads, repels many insect species. Up to now, most studies on vetiver have been carried out with vetiver oil (Zhu et al., 2001; Weyerstahl et al., 2006; Wedler et al., 2016). However, insect infestations of vetiver grass have been reported, and this grass has been identified as a potential trap plant for some crop pests. Our previous results have shown that adult female C. suppressalis preferred to lay eggs in vetiver grass, but their larvae cannot survive while feeding on this plant (Zheng et al., 2009; Lu et al., 2017a). Hence, vetiver is a bone fide dead-end trap plant [The term “dead-end trap plant” refers to plants that insects lay eggs on, but their larval offspring cannot survive on the plant as a food source (Shelton and Badenes-Perez, 2006)] for this insect species, and it offers a very promising approach for controlling C. suppressalis populations in rice paddies in China (Zheng et al., 2009; Liang et al., 2015; Lu et al., 2015). In fact, vetiver has already been successfully deployed on rice farms to date (Lu et al., 2015), and its nutriological and toxicological mechanisms against the larvae of C. suppressalis have been reported recently (Lu et al., 2017a). However, the molecular mechanisms underpinning vetiver’s lethal effects on C. suppressalis larvae are currently unclear.
The C. suppressalis larval midgut is likely to be important for mediating toxic responses triggered by vetiver ingestion and functions in nutrient uptake and utilization. It plays a vital role in xenobiotic metabolism. Several detoxification enzymes have been identified in the C. suppressalis midgut, including carboxylesterase, cytochrome P450 and glutathione S-transferase (Huang et al., 2009). Except digestive and detoxification functions, the midgut epithelium serves as a barrier between the internal and external environment. This barrier protects the host against exogenous stresses and maintains intestinal immune homeostasis (Buchon et al., 2009; Bao et al., 2012). In spite of the importance of midgut, little is known about the physiological and immunological responses of this tissue to vetiver in C. suppressalis.
Recently, transcriptional profiling has successfully elucidated mechanisms underpinning the insect defense response following exposure to toxic plants (Crava et al., 2016). In the current study, we likewise utilized genome-wide high-throughput RNA sequencing to generate a complete transcriptional profile of the midgut of C. suppressalis larvae after ingestion of rice and vetiver. This comparison profiling facilitated the investigation of the complex molecular responses of these larvae to vetiver. Candidate digestion-, detoxification-, and immune-related genes with different expression patterns were selected for additional study and their transcriptional dynamics following vetiver exposure were validated by quantitative real-time PCR (qRT-PCR). Cumulatively, this research provides a transcriptomic resource for further study of the complex responses of the C. suppressalis larval midgut after feeding on vetiver, and it offers a rich panel of genes to functionally test for their roles in the toxicity response. This work will be important for the development of effective approaches for sustainable biological control of C. suppressalis using vetiver as a trap plant in the economically vital rice production.
Chilo suppressalis individuals were collected in 2014 from a rice paddy field in Xiaoshan (120°12′E, 30°04′N), Hangzhou, Zhejiang province. Samples were immediately transferred to the laboratory to maintain genetic diversity in the population. The C. suppressalis population was reared en masse in an Artificial Atmospheric Phenomena Simulator (Ningbo Jiangnan Instrument Factory, China). Rearing conditions: temperature, 27 ± 1°C; relative humidity (RH), 70-80%; photoperiod of 16 h: 8 h light: dark (L: D). Larvae were reared on an artificial diet (Hu et al., 2012), and adults were fed a 10% honey solution. No specific permissions were required because C. suppressalis is not a protected species.
Vetiver grass was propagated from stem cuttings. The vetiver stems were cut from green house and these vetiver grasses were planted in the same time. TN1 rice (Oryza sativa L.) was transplanted in pots from 14-day-old seedlings. The TN1 seeds were donated by the International Rice Research Institute, Philippines. All seedlings were grown in a standard potting mix in a climate-controlled room (25 ± 1°C; 16 h: 8 h L: D; 70–80% RH). The stems from 40-day-old plants (vetiver grass and rice) were used for feeding experiments. The health of the vetiver grass and rice seedlings was grown well and not damaged by pests.
Third-instar larvae of C. suppressalis were selected for the feeding experiments. Experimental larvae were reared on vetiver stems, and control larvae were fed rice stems. All treated larvae were incubated under controlled climate conditions (27 ± 1°C; 70–80% RH; 16 h: 8 h L: D). At 2, 4, 6, 8, and 10 days after feeding, at least 40 larvae were dissected to obtain the midgut for further processing. Both treatments were repeated three times. The samples obtained after 2 days of feeding were used for transcriptional profiling and qRT-PCR validation. All samples were used for qRT-PCR analysis.
Total RNA was extracted from 30 C. suppressalis midguts using the miRNeasy Mini Kit (Qiagen, United States) according to manufacturer’s protocol. Aimed to check the RNA quality, the absorbance at 260 nm/280 nm (A260/A280) of RNA was measured by a ND-2000 spectrophotometer (ThermoFisher, United States). The purified RNA concentration was determined using a Qubit® 2.0 Fluorometer (Invitrogen, United States). RNA integrity was assessed using the Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). The 260/280 ratios of all the RNA samples were 1.9–2.0 and the RIN numbers were >8.0. We collected 3.0 μg of total RNA from each sample, which was midgut sample from C. suppressalis larvae after 2 days feeding on rice or vetiver, for RNA-Seq library construction and sequencing. After mRNA was purified and fragmented, the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, United States) was utiltized to establish the cDNA libraires. Six samples were then clustered and sequenced with 2 × 125 bp read lengths on an Illumina Hiseq 2500 platform (Illumina, United States) with TruSeq SBS Kit v4-HS (250-cycles) sequencing kit (Song et al., 2016). All the samples were sequenced on the same lane.
RNA-Seq reads were assessed for quality control with FastQC (version 0.10.1; Babraham Bioinformatics, Cambridge, United Kingdom). All reads were assembled by trinity (trinityrnaseq_r20131110, Grabherr et al., 2011) with default parameters. The transcript abundances were measured as fragments per kilobase of exon per million fragments mapped (FPKM) by RSEM follow trintiy script. Cuffdiff (Trapnell et al., 2013) was then used to determine differential expression (FDR ≤ 0.05) with blind dispersion methods. The raw RNAseq data were submitted to SRA (PRJNA477756).
The raw data were filtered using Fqfilter -q 20 -p 30 -Q 33 and fqtrimmer_paired -t 30 -Q 33 -l 50 program. The RSeQC (RSeQC-2.3.6) were used to determine the GC content and sequence duplication level. The list of differentially expressed transcripts generated from Cufflinks-Cuffdiff (v2.0.2) was divided into up-regulated and down-regulated lists of transcripts. Gene Ontology and KEGG pathways were determined to be over-represented using the Fisher exact test with a false discovery rate (FDR) correction (FDR ≤ 0.05). 1E-5 setting was used for the BLASTx analyses.
To validate the transcriptomic data, a subset of differentially expressed genes was quantified by qRT-PCR. The genes were selected based on our transcriptomic data and previous study (Lu et al., 2017a), however, only some related genes were focused on owing to a lot of differential expressed genes identified. Total RNA from each sample was extracted and all RNA samples were treated with DNaseI (ThermoFisher Scientific) following the manufacturer’s protocols. 3.0 μg of total RNA was used to synthesize first-strand cDNA template using a First Strand cDNA Synthesis Kit (ThermoFisher Scientific) with oligo (dT)18 as the primer. qRT-PCR on samples prepared with iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, United States) were performed using the CFX96 Real-Time System (Bio-Rad). The following PCR cycling condition was used: one cycle of 95°C for 30 s; 40 cycles of 95°C for 30 s, and 60°C for 35 s. Melting curve analysis was applied determine the specificity and quantity of the PCR product. Gene-specific primer sequences are listed in Table 1. β-tubulin (EU429675) served as a reference gene. Relative gene expression values were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Larval midguts dissected at 2, 4, 6, 8, and 10 days after feeding were also analyzed by qRT-PCR.
QRT-PCR data are expressed as the mean ± standard error of triplicate experiments, and statistical significance was determined by t-test in the larvae fed on rice and vetiver. Values of P < 0.0.5 were considered significant in all treatments.
A complete transcriptome was generated by high-throughput RNA sequencing. After removing low-quality and adaptor sequences, 51,810,574 and 53,910,888 clean reads were generated from the midguts of C. suppressalis larvae after they were fed for 2 days with rice and vetiver, respectively. The GC-content for each set of reads was about 39.6%, and 76.2–77.0% of the reads mapped to unique sequences. After assembly using the Trinity method, 48,943 UniGenes with an N50 value of 1,874 base pairs (bp) and a mean length of 994.3 bp were obtained (Table 2). The largest proportion of UniGenes identified were 200–300 bp in length (26.1%). Size distribution analysis showed that 13,795 UniGenes (28.2%) were more than 1,000 bp, 10,730 UniGenes (21.9%) were 500–1,000 bp and 24,418 UniGenes (49.9%) were 200–500 bp (Figure 1).
To annotate these UniGenes, we searched reference sequences using BLASTx within the non-redundant (NR) NCBI nucleotide database using a cut-off E-value of 10−5. A total of 28,942 UniGenes (59.1%) did not match any annotated sequences due to both a short nucleotide length and the lack of a sequenced genome for C. suppressalis. However, 20,001 UniGenes (40%) displayed annotated BLASTx hits. The E-value distributions for the 20,001 annotated UniGenes showed that 36.3% of the sequences had significant homology matches (<1E10−100) in the NCBI database, while 63.7% of the sequences ranged from 1E10−5 to 1E10−100 (Figure 2A). The similarity distribution showed that 4.69% of the sequences had greater than 95% homology, 50.23% of the sequences had 60–95% homology, 31.89% of the sequences had 40–60% homology and 13.18% of the sequences had less than 40% homology (Figure 2B). A species distribution analysis revealed that 46.21% and 7.11% of the sequences most closely resembled Danaus plexippus and Bombyx mori, respectively, followed by Tribolium castaneum (4.40%) and Nasonia vitripennis (4.04%). Approximately 57.0% of the UniGenes were top homology matches for Lepidopterans, including D. plexippus, B. mori, Papilio xuthus and Papilio polytes. Only 0.81% of the UniGenes matched C. suppressalis sequences previously deposited in the NCBI database (Figure 2C).
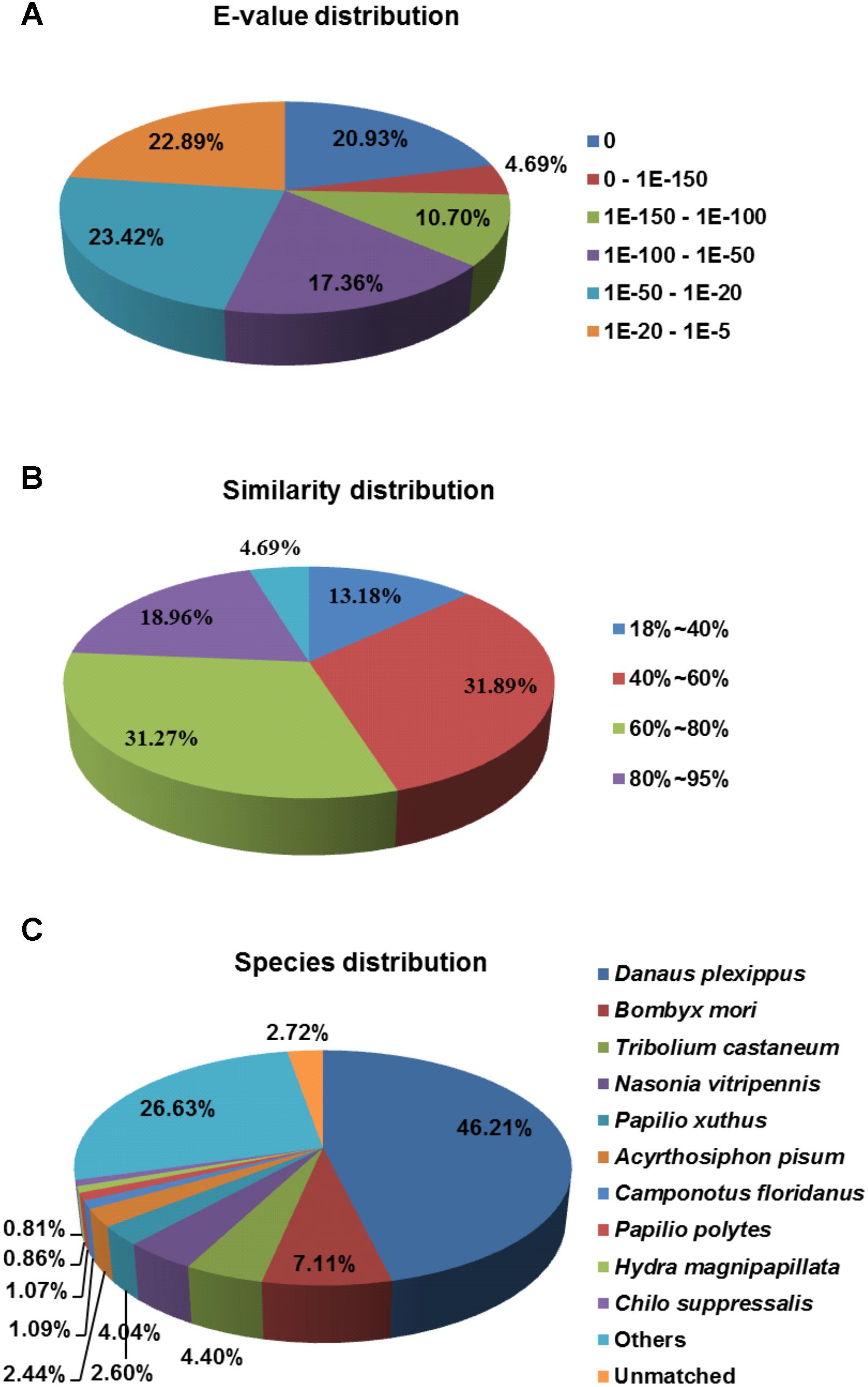
FIGURE 2. Homology analysis of the C. suppressalis midgut UniGenes. All unique gene sequences (20,001) that had BLAST annotations within the NR database with a cut-off E-value ≤ 10–5 were analyzed for (A) E-value distribution, (B) similarity distribution, and (C) species distribution.
UniGenes were annotated using the following databases: NCBI non-redundant protein database (NR), Universal Protein (UniProt), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). A total of 20,818 UniGenes (42.53%) were successfully annotated, including 20,001 (40.87%) from the NR database, 13,980 (28.56%) from the UniProt database, 7,943 (16.23%) from the KEGG database and 8,239 (16.83%) from the GO database. The GO distributions for the UniGenes were classified into three categories (45 subcategories): cellular components (11 subcategories), biological processes (22 subcategories) and molecular function (12 subcategories) (Figure 3). For the cellular components category, the clusters relating to “cell part” had an enrichment of 2,222 genes and “cell” had an enrichment of 2,222 genes. For the biological processes category, clusters relating to “metabolic processes” were enriched for 3,450 genes, and those relating to “cellular processes” were enriched for 3,237 genes. For the molecular functions category, clusters relating to “catalytic activity” and “binding” were highly represented (2,849 and 4,867 genes, respectively) (Figure 3).
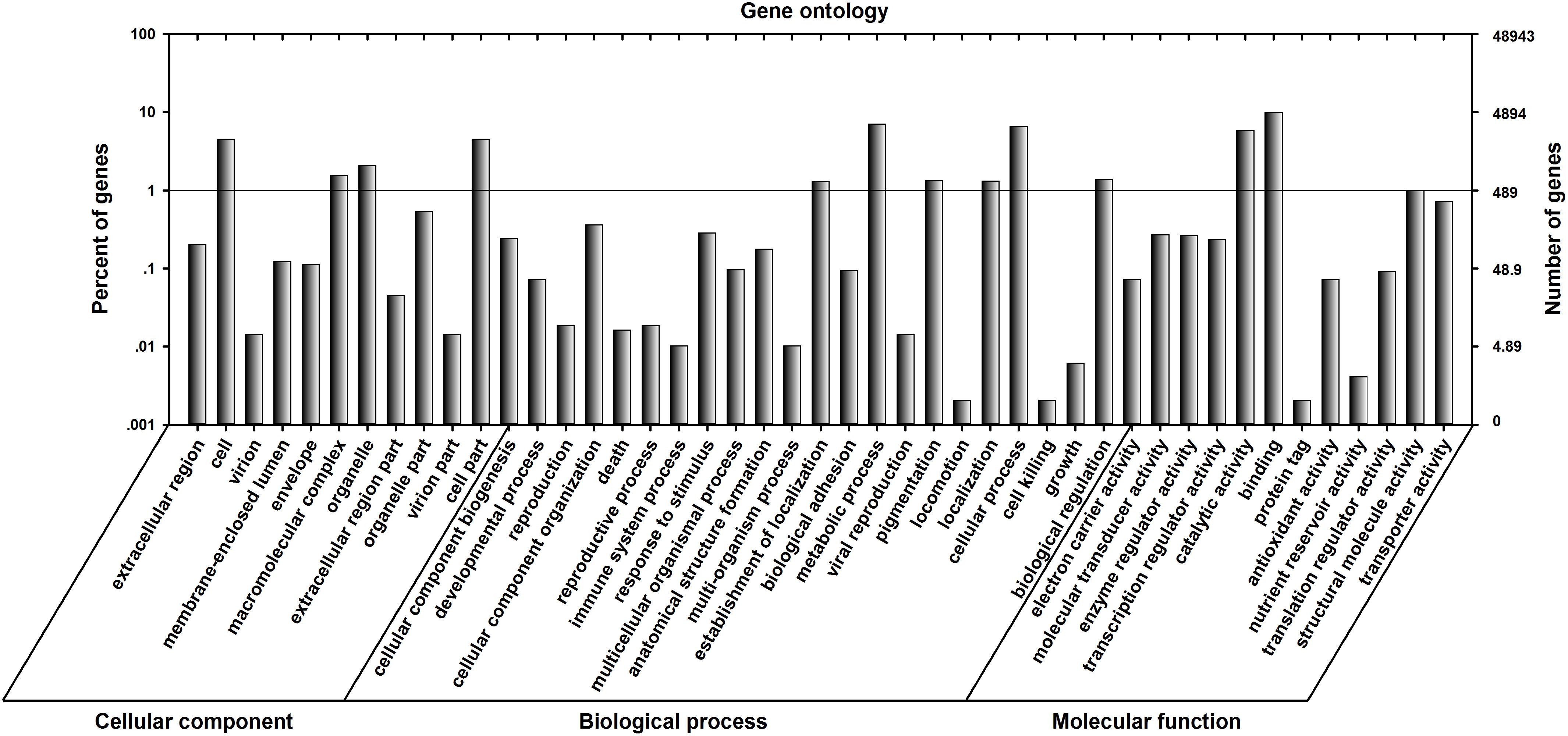
FIGURE 3. Gene Ontology (GO) classification for the C. suppressalis midgut UniGenes. Of the 48,943 UniGenes, 8,239 sequences could be annotated within the GO database into three main categories: cellular component, biological process, and molecular function. The left and right y-axes show the percentage and number of genes in each category, respectively.
To further evaluate the effectiveness of annotation, we examined the annotated UniGenes that possessed COG classifications. Of the 20,001 UniGenes, 10,772 sequences had a COG annotation (Figure 4). Among the COG categories, the cluster for “General function prediction” represented the largest group (1,989 UniGenes, 18.5%), and followed by “Post-translational modification, protein turnover and chaperones” (1,100 UniGenes, 10.2%) and “Translation, ribosomal structure and biogenesis” (923 UniGenes, 8.6%). The clusters for “RNA processing and modification,” “Defence mechanisms” and “Cell motility” displayed similar abundance (36–79 UniGenes, 0.3–0.7%). In contrast, “Nuclear structure” was the smallest cluster, containing only nine UniGenes (Figure 4).
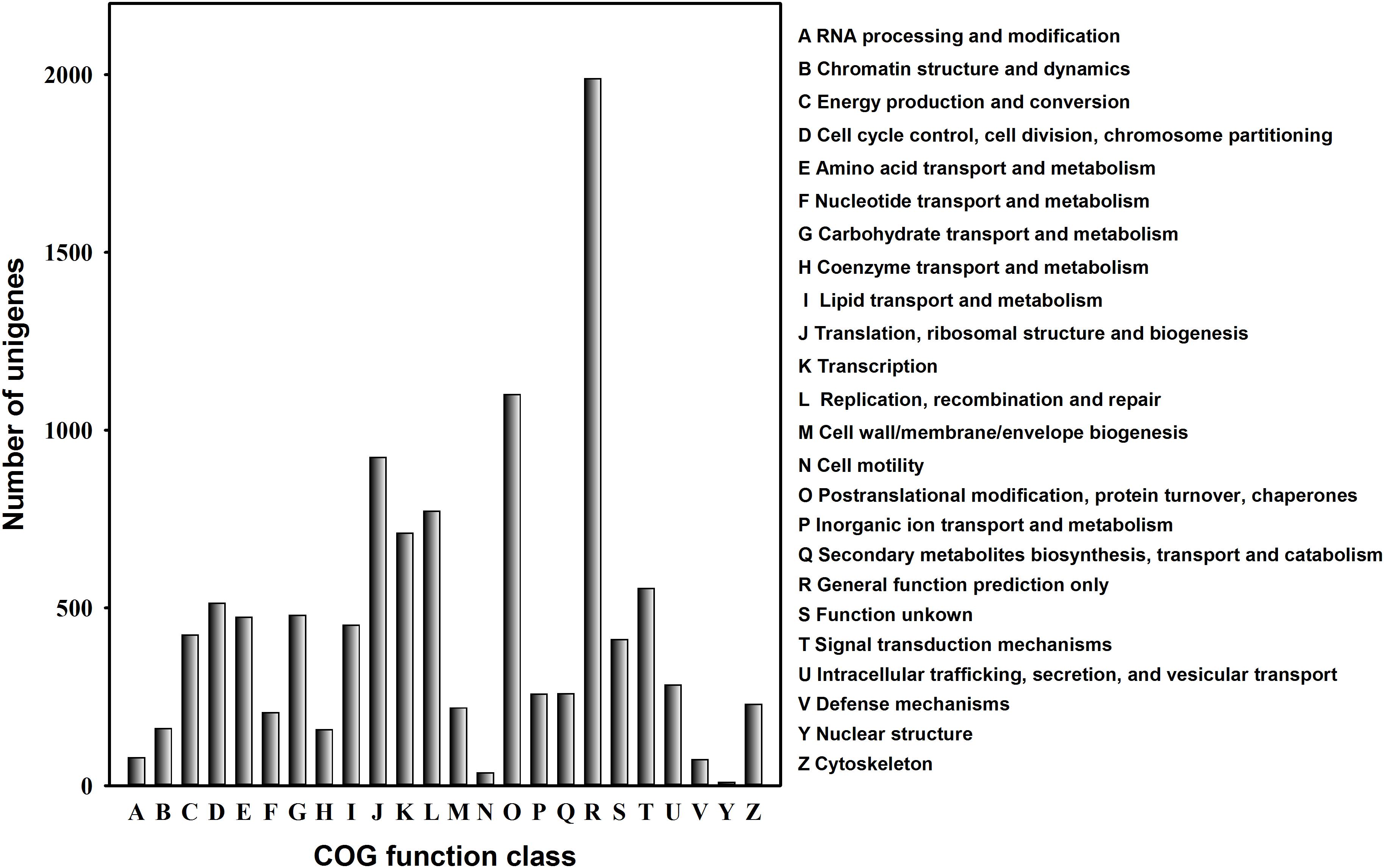
FIGURE 4. Clusters of Orthologous Groups (COG) classification for the C. suppressalis midgut UniGenes. About one-quarter of all UniGenes (10,772) have a COG annotation among the 24 categories.
A total of 1,849 differentially expressed UniGenes were detected in the midguts of C. suppressalis larvae that were fed vetiver compared to those that were fed rice. The number of up-regulated genes (1,110) was greater than the number of down-regulated genes (739). In order to study the complex response of the C. suppressalis midgut to vetiver exposure, the KEGG database, GO annotations, Genefamily database, and Pfam database were utilized to characterize these differentially expressed UniGenes. Using the KEGG annotations, the differentially expressed UniGenes were enriched for ribosomal, metabolic, and proteasomal pathways (Supplementary Table S1). Using GO annotations, the differentially expressed UniGenes were enriched for such categories as translation, ribosomes, intracellular components, structural constituents of the cuticle and chitin binding (Supplementary Table S2). Using Genefamily annotations, the differentially expressed UniGenes were enriched for histone folding proteins, DNA/RNA polymerases, serine protease inhibitors and invertebrate chitin-binding proteins (Supplementary Table S3). Finally, using Pfam annotations, the differentially expressed UniGenes were enriched for insect cuticle proteins, trypsin inhibitor-like cysteine-rich domain-containing proteins, chitin binding peritrophin-A domain-containing proteins and lipases (Supplementary Table S4).
To validate the transcriptomic data, qRT-PCR was performed for 19 genes that were significantly differentially expressed in the C. suppressalis midguts following ingestion of vetiver and rice. These included 12 digestion-related genes, four immune response-related genes and three detoxification-related genes. The results of the qRT-PCR validation are presented as fold-changes normalized to the β-tublin gene. A comparison of the mean values for the qRT-PCR results and the original transcriptome data is summarized (Table 3). This qRT-PCR analysis validated the results of the transcriptomic analysis. In addition, we generated heatmaps for digestion related genes, immune response-related genes and three detoxification-related genes (Figure 5).

TABLE 3. Potential digestion-related, immune response-related, and detoxification-related genes that respond to the ingestion of vetiver grass in the C. suppressalis midgut.

FIGURE 5. Heatmaps for digestion-related genes (A), immune response-related genes (B), and detoxification-related genes (C).
To begin exploring the mechanism of lethality induced by vetiver in larvae of C. suppressalis, we randomly selected 12 digestion-related genes from the list of differentially expressed UniGenes and analyzed their expression dynamics in larval midguts at 2, 4, 6, 8, and 10 days after feeding on vetiver or rice (Figure 6). The expression levels of three serine proteases, chymotrypsin-like protease (000273) and trypsin-like proteinase (000399) were initially up-regulated and then down-regulated in the larvae feeding on vetiver compared to rice-fed controls. Meanwhile, serine protease 20 (001033) was up-regulated in the larvae feeding on vetiver compared to controls. The expression levels of carboxypeptidase genes (005074, 00365), dipeptidyl peptidase (000540), and glucosidase (000582) were all significantly reduced in larvae at 8 and 10 days after feeding on vetiver compared to the rice-fed controls. Pancreatic lipase (006146) displayed significantly higher expression levels in all larvae that ingested vetiver compared to controls. In contrast, neutral lipase (000272) displayed significantly higher expression levels at 2 and 4 days after feeding on vetiver, but this expression returned to levels comparable to rice-fed control larvae. Two alpha amylases (000092, 000242) showed almost similar expression kinetics in the larvae fed vetiver on different days. However, the expression of aminopeptidase N2 (001125) was not significantly changed in the larvae fed vetiver compared to those fed rice (Figure 6).
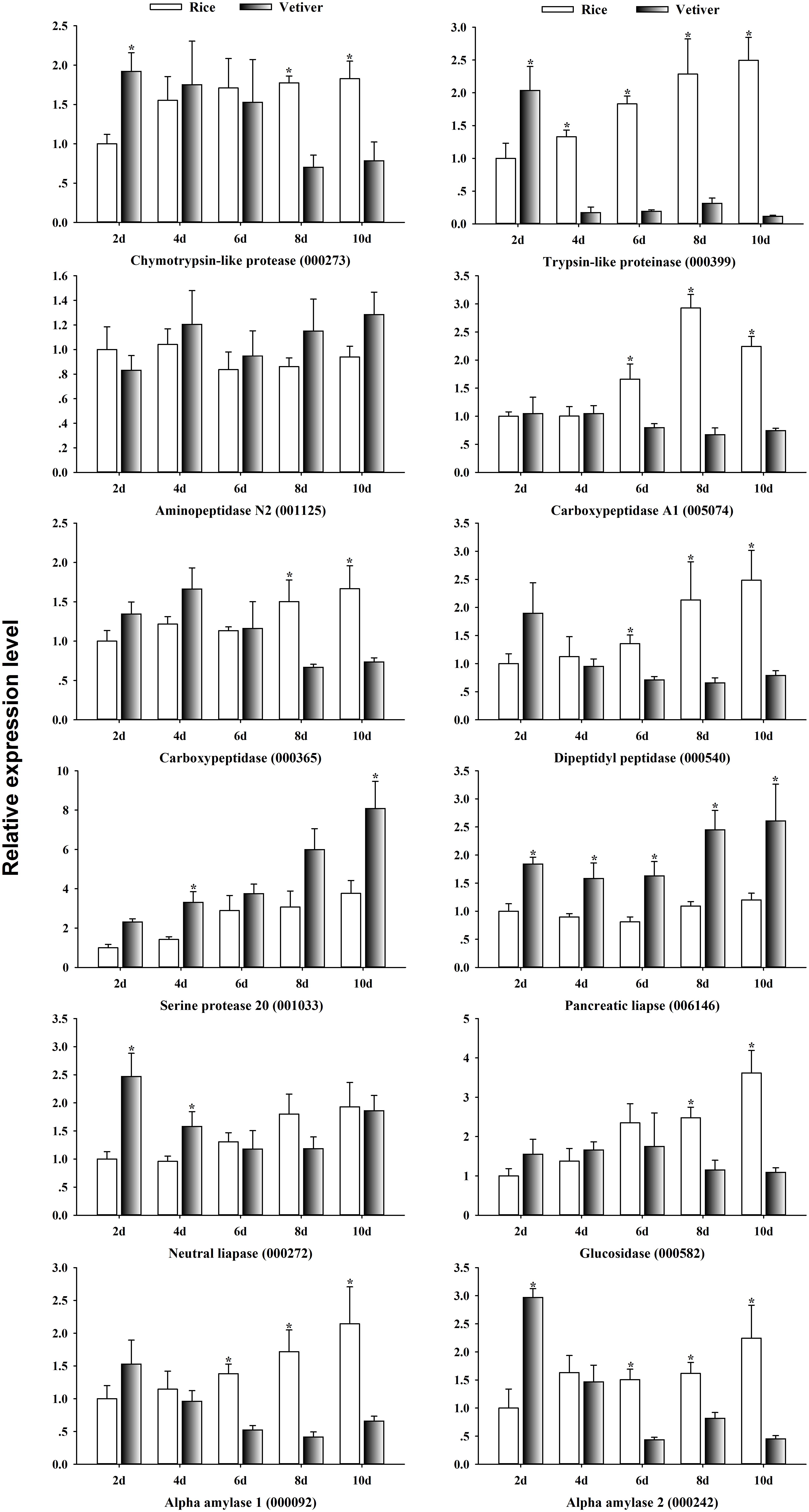
FIGURE 6. The digestion-related gene expression dynamics in C. suppressalis larvae after feeding on either rice or vetiver. Larvae were fed for 2, 4, 6, 8, and 10 days. First-strand cDNA was used as template for each qRT-PCR reaction. The reactions were performed with gene-specific primers to amplify digestion-related genes. The relative expression levels of each gene in each tissue were normalized using C. suppressalis β-tubulin (EU429675). Threshold cycle (Ct) values were obtained from reactions run on the same plate. Two technical replicates and three biological replicates were analyzed, and the mean ± SEM was calculated to determine the relative transcript levels using the 2−ΔΔCt method. The number in parentheses represents the UniGene IDs. Asterisks indicated significant differences between the gene expressions in the larvae fed on rice and vetiver (t-test, P < 0.05).
Insects rely on innate immunity for their defense against environmental stresses, including host plants. To clarify the mechanism of lethality of vetiver in the larvae of C. suppressalis, we randomly selected four immune-related genes from the list of differentially expressed UniGenes. We assayed their expression kinetics in the larval midgut at 2, 4, 6, 8, and 10 days after feeding on either vetiver or rice using qRT-PCR. Peptidoglycan recognition protein (001314) displayed significantly higher expression in larvae 2 days after feeding on vetiver compared to rice-fed controls; however, this expression subsequently returned to the levels observed in the controls. Glucan recognition protein (004227) exhibited up-regulated and then down-regulated expression kinetics in the larvae feeding on vetiver compared to the controls. Finally, at 4 days after feeding on vetiver, larvae displayed elevated expression of Hdd 1 protein (005250) and reduced expression of Hdd 13 protein (000203) compared to rice-fed controls (Figure 7).
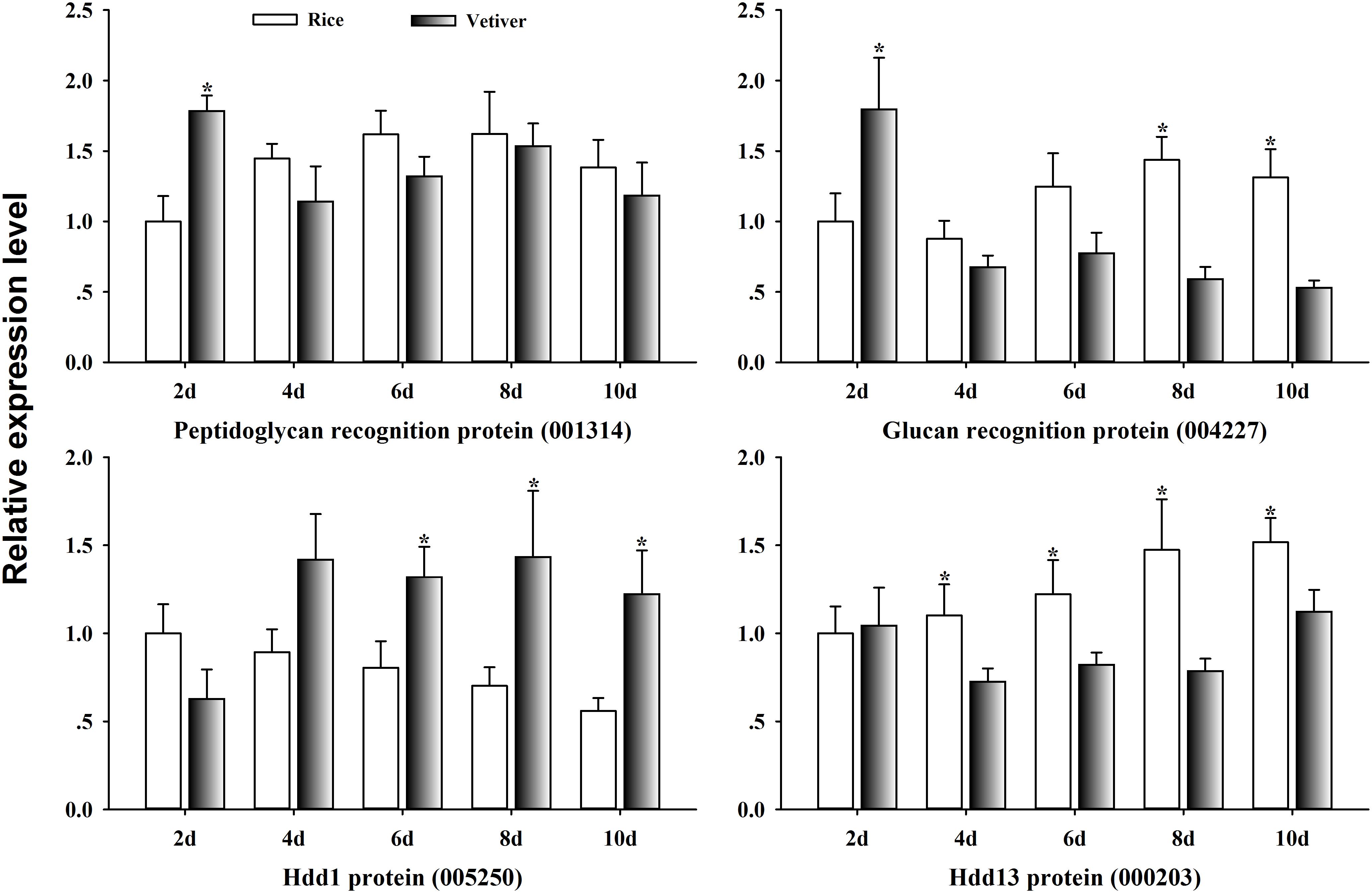
FIGURE 7. Immune response-related gene expression dynamics in C. suppressalis larvae after feeding on either rice or vetiver. Larvae were fed for 2, 4, 6, 8, and 10 days. The qRT-PCR procedure and statistical analysis were performed as described in Figure 6. Asterisks indicated significant differences between the gene expressions in the larvae fed on rice and vetiver (t-test, P < 0.05).
Cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs), and carboxylesterases are members of the three major multigene enzyme families primarily responsible for xenobiotic metabolism (i.e., insecticides and plant-toxic allelochemicals) (Ranson et al., 2002; Bao et al., 2012). To investigate the mechanism of lethality of vetiver in larvae, we randomly selected three detoxification-related genes from the list of differentially expressed UniGenes. We assessed their expression kinetics in the midgut of C. suppressalis larvae by qRT-PCR at 2, 4, 6, 8, and 10 days after feeding on vetiver or rice. CYP306A1 (002963) exhibited significantly lower expression levels at 4, 6, 8, and 10 days after feeding on vetiver compared to the rice-fed controls. GST-like protein (000438) displayed significantly lower expression levels at 6, 8, and 10 days after feeding on vetiver compared to controls. Finally, carboxylesterase (000471) displayed significantly higher expression levels in vetiver-fed larvae at all timepoints compared to controls (Figure 8).
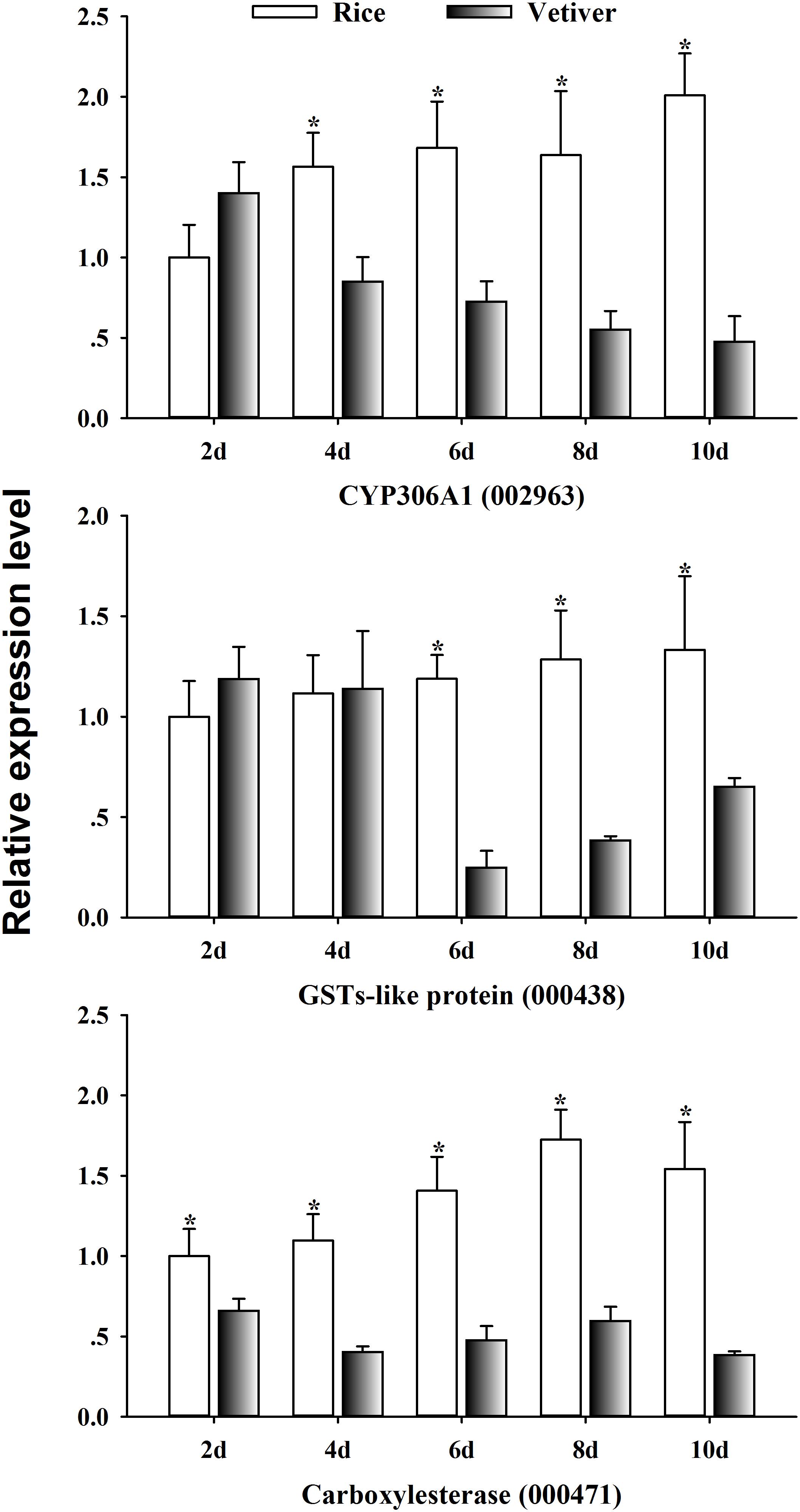
FIGURE 8. The detoxification-related gene expression dynamics in C. suppressalis larvae after feeding on either rice or vetiver. Larvae were fed for 2, 4, 6, 8, and 10 days. The qRT-PCR procedure and statistical analysis were performed as described in Figure 6. Asterisks indicated significant differences between the gene expressions in the larvae fed on rice and vetiver (t-test, P < 0.05).
Recently, transcriptome sequencing has become a crucial research method due to its low cost and high throughput nature (Kang et al., 2013; Song et al., 2016). Here, we report a midgut transcriptomic study of C. suppressalis. We identified a large number of high-quality sequences, and investigated the complex responses of the larval midgut after ingestion of vetiver. Due to the lack of a reference genome for C. suppressalis, UniGenes were identified and annotated using publicly available protein databases, including the NR, GO, UniProt, KEGG, Swiss-Prot, and Pfam databases. Recently, GO terms have been utilized for the functional annotation and visualization of UniGenes. A framework for categorizing genes has been established, which includes categories consisting of biological process, molecular function, and cellular compartment. In the present study, 8,239 UniGenes were assigned to one or more ontologies. KEGG analysis was utilized for assigning enriched biological pathways to 7,943 UniGenes. Overall, the data provide a useful resource as a C. suppressalis transcriptome, yielding insights into the potential genetic cascades that respond to exposure to vetiver.
Previous studies have demonstrated that vetiver effectively attracts female C. suppressalis adults, which lay eggs on the plant. However, the larvae cannot complete their life cycle by feeding on vetiver (Zheng et al., 2009). There are two possibly that explain this lethality. First, vetiver may lack certain nutrients present in rice, which leads to digestive dysfunction and, ultimately, larval death. Alternatively, vetiver might contain toxic substances that inhibit the activities of larval detoxification enzymes, leading to fatal metabolic dysfunction (Lu et al., 2017a). Therefore, we explored the potential mechanisms underpinning the observed lethality in C. suppressalis larvae. Using qRT-PCR, we investigated the expression dynamics of 12 digestion-related, four immune responsive-related and three detoxification-related UniGenes that were differentially expressed, such as some up-regulated genens, in vetiver-fed larvae compared to rice-fed controls. Digestion is a critical facet of insect physiology (Zibaee et al., 2008), and our results of transcriptome indicated that the expression of numerous digestion-related genes was disrupted in C. suppressalis larvae after feeding on vetiver. The expression of most of these selected genes was reduced in larvae at 6 and 8 days after feeding on vetiver, except for aminopeptidase N2 (001125), serine protease 20 (001033), pancreatic lipase (006146) and neutral lipase (000272). This suggested that these digestive enzymes may function in the breakdown of vetiver in the larval midgut, and they may play important roles in absorption and utilization of nutrients from this plant. To date, most studies have focused on the effects that host plants have on the digestive enzymes on insects that feed on them. In particular, host plants have been shown to have significant effects on the activities of insect amylase, pectinase and pepsin (Yan et al., 2012). The amylase and lipase activities of Spodoptera exigua, for example, depend on the host plant, which includes maize, sugar beets and cucumber (Zhang et al., 2009). Furthermore, when the action of these digestive enzymes is inhibited, the ability of the insect to derive nutrition from the plant becomes impaired, and larval growth and development are retarded. This eventually leads to starvation and death (Zibaee et al., 2008). Therefore, we suggest that the lethality induced by vetiver in C. suppressalis larvae may be related to the inhibition of digestive enzymes. However, a detailed molecular mechanism must be elucidated in order to determine whether impaired digestive enzymes cause lethality in vetiver-fed larvae.
Insects rely on innate immunity for their defense against environmental stresses. Peptidoglycans, beta-1,3-glucan and lipopolysaccharides frequently elicit immune responses in insects (Bao et al., 2012). Accordingly, a variety of pattern recognition proteins mediate immune responses and induce the expression of immune-responsive genes (Cerenius et al., 2010; Bao et al., 2012). However, even though C. suppressalis is one of the most serious agricultural pests, little is known about its immune responses to vetiver. This is critical for identifying potential target molecules in this insect’s immune system. Beta-glucan recognition protein is a particularly important pattern recognition molecule in the insect immune system (Bao et al., 2012). In this study, we found that the expression level of beta-glucan recognition protein (004227) significantly decreased 8 days after the larvae were fed vetiver compared to the controls. This suggests that beta-glucan recognition protein might be inhibited in the midgut by the presence of vetiver. Furthermore, among the immune response-related UniGenes, Hdd1, and Hdd13 showed opposing expression dynamics. Hdd1 (005250) displayed very high transcript levels in larvae 6 days after feeding on vetiver, while Hdd13 (000203) displayed low transcript levels in larvae 4 days after feeding on vetiver. This suggested that these two genes might have different functions in C. suppressalis larvae during the digestion of vetiver.
Most insect species metabolize xenobiotics, such as secondary plant chemicals and insecticides, using a suite of detoxification enzymes. These include P450s, GSTs and carboxylesterases (Karatolos et al., 2011; Bao et al., 2012). In this study, CYP306A1 (002963), GST-like protein (000438), and carboxylesterase (000471) exhibited significantly reduced expression in larvae 6, 8, and 10 days after feeding on vetiver. This finding supported our previous results, suggesting that vetiver contains toxic substances that have lethal effects on C. suppressalis larvae. These substances likely inhibited the activities of detoxification enzymes, leading to disruptions in larval metabolism and toxicity responses (Lu et al., 2017a).
In this study, a transcriptome for C. suppressalis has been generated, and the overall midgut responses of the larvae ingestion of vetiver have been described for the first time. We validated this transcriptome by examining the expression of 12 digestion-related, four immune response-related, and three detoxification-related UniGenes by qRT-PCR in larvae that were fed on vetiver. Our results suggest that these genes are possibly involved in the mechanism of lethality induced by trap plant-vetiver in C. suppressalis larvae. These transcriptomic data establish a foundation for the identification and functional validation of differentially expressed genes of C. suppressalis after vetiver exposure. Overall, our findings provide a molecular framework for the development of a sustainable control approach for C. suppressalis, which has potentially beneficial implications for the rice production worldwide.
YL, YZ, HL, and ZL conceived and designed the experiments and analyzed the data. YL, YZ, HL, QB, and YY performed the experiments. YL, XZ, and ZL contributed reagents and materials. YL and ZL wrote the manuscript.
This work was supported by grants from the General Program of National Natural Science Foundation of China (No. 31672050), the National Key Research and Development Program of China (No. 2016YFD0200800-04), the Zhejiang Key Research and Development Program (No. 2015C02014), and the Zhejiang Academy of Agricultural Sciences Young Personnel Training Project to YL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01067/full#supplementary-material
Bao, Y. Y., Wang, Y., Wu, W. J., Zhao, D., Xue, J., Zhang, B. Q., et al. (2012). De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics 99, 256–264. doi: 10.1016/j.ygeno.2012.02.002
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S., and Lemaitre, B. (2009). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211. doi: 10.1016/j.chom.2009.01.003
Cerenius, L., Kawabata, S., Lee, B. L., Nonaka, M., and Söderhäll, K. (2010). Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 35, 575–583. doi: 10.1016/j.tibs.2010.04.006
Crava, C. M., Brütting, C., and Baldwin, I. T. (2016). Transcriptome profiling reveals differential gene expression of detoxification enzymes in a hemimetabolous tobacco pest after feeding on jasmonate-silenced Nicotiana attenuata plants. BMC Genomics 17:1005. doi: 10.1186/s12864-016-3348-0
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hu, Y., Fu, Q., Luo, J., and Lai, F. (2012). Artificial diet of Chilo suppressalis and preparation and mass rearing method. Patent CN 102356811 A.
Huang, C. H., Fang, Q., Ye, G. Y., Yao, H. W., Guo, J. Y., and Cheng, J. A. (2009). Tissue and subcellular distribution of insecticide-resistance related enzymes in larvae of striped stem borer, Chilo suppressalis (Walker). Guangxi Agric. Sci. 40, 153–158.
Kang, C., Darwish, O., Geretz, A., Shahan, R., Alkharouf, N., and Liu, Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25, 1960–1978. doi: 10.1105/tpc.113.111732
Karatolos, N., Pauchet, Y., Wilkinson, P., Chauhan, R., Denholm, I., Gorman, K., et al. (2011). Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes. BMC Genomics 12:56. doi: 10.1186/1471-2164-12-56
Liang, Q., Lu, Y. H., He, X. C., Zheng, X. S., Xu, H. X., Yang, Y. J., et al. (2015). Mini review of the significance of trap crop in insect pest management. J. Biosaf. 24, 184–193.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Y. H., Gao, G. C., Zheng, X. S., and Lu, Z. X. (2017a). The lethal mechanism of trap plant Vetiveria zizanioides against the larvae of Chilo suppressalis. Sci. Agric. Sin. 50, 486–495.
Lu, Y. H., Wang, G. R., Zhong, L. Q., Zhang, F. C., Bai, Q., Zheng, X. S., et al. (2017b). Resistance monitoring of Chilo suppressalis (Walker) (Lepidoptera: Crambidae) to chlorantraniliprole in eight field populations from east and central China. Crop Prot. 100, 196–202. doi: 10.1016/j.cropro.2017.07.006
Lu, Z. X., Zhu, P. Y., Gurr, G. M., Zheng, X. S., Chen, G. H., and Kong, L. H. (2015). Rice Pest Management by Ecological Engineering: A Pioneering Attempt in China. Berlin: Springer.
Qu, M. J., Han, Z. J., Xu, X. J., and Yue, L. N. (2003). Triazophos resistance mechanisms in the rice stem borer (Chilo suppressalis Walker). Pestic. Biochem. Physiol. 77, 99–105. doi: 10.1016/j.pestbp.2003.09.003
Ranson, R., Claudianos, C., Ortelli, F., Abgrall, C., Hemingway, J., Sharakhova, M. V., et al. (2002). Evolution of supergene families associated with insecticide resistance. Science 298, 179–181. doi: 10.1126/science.1076781
Shelton, A., and Badenes-Perez, F. (2006). Concepts and applications of trap cropping in pest management. Annu. Rev. Entomol. 51, 285–308. doi: 10.1146/annurev.ento.51.110104.150959
Song, F., Chen, C., Wu, S., Shao, E., Li, M., Xiong, G., et al. (2016). Transcriptional profiling analysis of Spodoptera litura larvae challenged with Vip3Aa toxin and possible involvement of trypsin in the toxin activation. Sci. Rep. 6:23861. doi: 10.1038/srep23861
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pacher, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Wedler, H. B., Newman, T., and Tantillo, D. J. (2016). Decarboxylation facilitated by carbocation formation and rearrangement during steam distillation of vetiver oil. J. Nat. Prod. 79, 2744–2748. doi: 10.1021/acs.jnatprod.6b00348
Weyerstahl, P., Marschall, H., Splittgerber, U., and Wolf, D. (2006). New sesquiterpene ethers from vetiver oil. Liebigs Ann. Chem. 1996, 1195–1199. doi: 10.1002/jlac.199619960720
Yan, X. F., Liu, J., and Liu, Y. H. (2012). Effect of different host plants on the activitiy of digestive enzyme in Anoplophora glabripennis motschulsky adults. Hubei Agric. Sci. 51, 4010–4013.
Zhang, N., Guo, J. Y., Wan, F. H., and Wu, G. (2009). Effects of three kinds of host plants on development and some digestive enzyme activities of beet armyworm Spodoptera exigua. Acta Phytophylacica Sin. 36, 146–150.
Zheng, X. S., Xu, H. X., Chen, G. H., Wu, J. X., and Lu, Z. X. (2009). Potential function of Sudan grass and vetiver grass as trap crops for suppressing population of stripped stem borer, Chilo suppressalis in rice. Chin. J. Biol. Control 25, 299–303.
Zhu, B. C., Henderson, G., Chen, F., Fei, H., and Laine, R. A. (2001). Evaluation of vetiver oil and seven insect-active essential oils against the formosan subterranean termite. J. Chem. Ecol. 27, 1617–1625. doi: 10.1023/A:1010410325174
Keywords: Chilo suppressalis, vetiver, midgut transcriptome, digestion, immunity, detoxification
Citation: Lu Y, Zhao Y, Lu H, Bai Q, Yang Y, Zheng X and Lu Z (2018) Midgut Transcriptional Variation of Chilo suppressalis Larvae Induced by Feeding on the Dead-End Trap Plant, Vetiveria zizanioides. Front. Physiol. 9:1067. doi: 10.3389/fphys.2018.01067
Received: 03 May 2018; Accepted: 17 July 2018;
Published: 07 August 2018.
Edited by:
Andrzej Stasiak, Université de Lausanne, SwitzerlandReviewed by:
Kumar Saurabh Singh, University of Exeter, United KingdomCopyright © 2018 Lu, Zhao, Lu, Bai, Yang, Zheng and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongxian Lu, bHV6eG1oQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.