
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 26 July 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.00987
This article is part of the Research Topic Insect Olfactory Proteins (From Gene Identification to Functional Characterization) View all 39 articles
Insect attractants are important prevention tools for managing populations of the Oriental fruit fly, Bactrocera dorsalis (Hendel), which is a highly destructive agricultural pest with health implications in tropical and subtropical countries. Methyl eugenol (ME) is still considered the gold standard of B. dorsalis attractants. Mature male flies use their olfactory system to detect ME, but the molecular mechanism underlying their olfactory detection of ME largely remains unclear. Here, we showed that ME activates the odorant receptors OR63a-1 and OR88a in mature B. dorsalis males antennae by RNA-Seq and qRT-PCR analysis. Interestingly, ME only elicited robust responses in the BdorOR88a/BdorOrco-expressing Xenopus oocytes, thus suggesting that BdorOR88a is necessary for ME reception and tropism in B. dorsalis. Next, our indoor behavioral assays demonstrated that BdorOR63a-1 knockdown had no significant effects on ME detection and tropism. By contrast, reducing the BdorOR88a transcript levels led to a significant decrease in the males’ responsiveness to ME. Taken together, our results gave novel insight in the understanding of the olfactory background to the Oriental fruit fly’s attraction toward ME.
Insects rely primarily on sophisticated olfactory reception systems to detect and discriminate many exogenous chemical signals, and odorant receptors (ORs) are at the core of odorant detection (Hallem and Carlson, 2006; Missbach et al., 2014; Zhang et al., 2016; Fleischer et al., 2017; Wang et al., 2017). Apparently, ORs show a high degree of sensitive and play critical roles in detecting long-range volatile odorants and triggering the transduction of chemical signals into electric signals (Mitsuno et al., 2008; Leal, 2013). For example, an odorant receptor isolated from Plutella xylostella, PxylOR1, was narrowly tuned to the main component of the sex pheromone, (11Z)-hexadecenal (Z11-16Ald) (Sakurai et al., 2011). Interestingly, the direct activation of CquiOR136, is necessary for DEET (N,N-diethyl-3-methylbenzamide) reception and repellency effects in Culex quinquefasciatus (Xu et al., 2014). Additionally, knockdown of AjapOR35 in Anastatus japonicus reduced its antennal response to two oviposition attractants, β-caryophyllene and (E)-α-farnesene (Wang et al., 2017). In this respect, using a “computational reverse chemical ecology" strategy will likely generate new insights for the rapid screening of potentially effective semiochemicals that modify the behavioral patterns of insects (Siderhurst and Jang, 2006; Wu et al., 2015).
The oriental fruit fly, Bactrocera dorsalis (Hendel), is among the most destructive fruit/vegetable-eating agricultural pests in the world (Zheng et al., 2013; Liu et al., 2016). Due to polyphagia in the larval stages and high fecundity of the adults, B. dorsalis can cause serious damage to more than 250 species of commercially grown vegetables and fruits (Clarke et al., 2005; Stephens et al., 2007; Shen et al., 2012). Plant damage caused by B. dorsalis consists of oviposition stings to host fruit tissues by adult females and the subsequent larval feeding and decaying in the fruit pulp. Controlling the male adult fly’s behavior is the main method to reduce the damage caused by this pest (Clarke et al., 2005). Methyl eugenol (ME), a highly potent phytochemical lure, has been exploited in the male annihilation technique (MAT) systems for detecting, monitoring, and luring B. dorsalis male individuals (Vargas and Prokopy, 2006; Pagadala et al., 2012; Shelly, 2017). Particularly noteworthy is the fact that ME has its risks, however: it is carcinogenic to humans and its attractiveness is limited to mature males (Smith et al., 2002; Khrimian et al., 2009; Zheng et al., 2012). Although novel attractants of B. dorsalis are diligently being developed, their progress toward better and more affordable attractants has nonetheless been slow (Khrimian et al., 1994, 2006, 2009; Jang et al., 2011). Design of attractants to target specific ORs may promote the development of new baits for pest management (Di et al., 2017; Mitchell et al., 2017). However, ME receptors ORs in B. dorsalis are hitherto unknown. Therefore, exploring the molecular mechanism underlying olfactory detection of ME in B. dorsalis is of great importance for developing sustainable pest control strategies based on manipulating insect chemosensory communication.
In trying to identify the chemosensory genes responsible for detecting ME, recent studies have identified and confirmed that BdorOrco, BdorOBP83a-2, and BdorOBP2 actively participate in the process of ME detection by B. dorsalis male adults (Zheng et al., 2012; Wu et al., 2016; Liu H. et al., 2017). Nevertheless, the specific ORs and their molecular functional involvement in the mature male fly’s reception of ME remain a mystery. Here, we used an RNA-Seq approach coupled to RNA interference silencing, supplemented by in vivo expression in Xenopus laevis oocytes, to investigate the functional roles of BdorOR63a-1 and BdorOR88a; both genes are abundantly expressed in ME-treated male antennae and were hypothesized to encode possible receptors for ME. Based on the results presented here, we infer that BdorOR88a likely plays an essential role in the molecular mechanism underlying B. dorsalis olfactory detection of ME.
No specific permits were required in our studies of this widespread agriculture pest. We confirm that the study locations were not privately owned or protected. This work did not involve endangered or protected species. To avoid chemical hazards, always observe safety laboratory practice when operating the ME.
The B. dorsalis genetic sexing strain (GSS) used in this study was reared in a laboratory for more than 30 generations at the South China Agricultural University, Guangzhou, China. The male pupae are brown and the female pupae are white. Insects were reared under a photoperiod cycle of 14 h light/10 h dark at 27 ± 1°C, 75 ± 1% relative humidity (RH). Larvae were reared on an artificial diet that included yeast (15.06%), sugar (8.99%), nipagen (0.15%), sodium benzoate (0.15%), citric acid (1.70%), wheat germ oil (0.15%), and water (73.81%) (Chang et al., 2006; Liu H. et al., 2017). Adult flies were maintained in 35 cm × 35 cm × 35 cm wooden cages and fed a diet consisting of sugar: yeast extract at 1:1 (w/w) (Liu H. et al., 2017).
The ME solution used consisted of 1:1 dilution with mineral oil (MO) (Energy Chemical Company, Shanghai, China), of which 0.5-ml amounts were used to coat the inner wall of 500-ml conical flasks. Next, sample of 200 mature males (15-day-old) were randomly selected and placed in each flask. Flies in the control group were likewise handled but put into flasks containing an equal volume of MO only. After being treated for 1 h, all the antennae were dissected and flash frozen in liquid nitrogen, and were then immediately transferred to a -80°C freezer pending RNA extraction. Three independent biological replicates were performed for use in the transcriptome analyses. Experiments were conducted between 9 and 11 a.m. During these experiments, RH and temperature in the laboratory were maintained at 75 ± 1% RH and 27 ± 1°C, respectively.
Under an RNA-free environment, antennal total RNAs were extracted by using a RNA extraction kit (Takara Biotechnology Co., Ltd., Japan), following the manufacturers protocol. Each sample consisted of 200 males’ antennae. The purity of all RNA samples was assessed at absorbance ratios of OD260/230 and OD260/280, while the integrity of RNA was verified through 1%-RNase-free agarose gel electrophoresis. The concentration of RNA was quantified by measuring their absorbance at 260 nm in a spectrophotometer (Thermo Nano DropTM 2000c; Santa Clara, CA, United States) and qualified using an Agilent 2,100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, United States). High-quality RNA from each replication of the ME treatment and control groups was used for the next suite of steps: mRNA was first isolated from total RNA using magnetic beads with oligo (dT) and sheared into short fragments in a fragmentation buffer. Then complementary DNA (cDNA) was synthesized from the mRNA fragments, by using SuperScript III Reverse Transcriptase (Invitrogen). A cDNA library was constructed for each sample. The library preparations were sequenced on an Illumina HiSeq 4000TM platform (Illumina Inc., San Diego, CA, United States) with 125-nucleotide (nt) paired-end reads; this final step carried out by the Gene Denovo Technology Company (in Guangzhou, China).
For quality control, the raw sequencing data in the FASTQ format were processed by in-house Perl scripts to obtain clean data reads. Before assembly, any adapter sequences were removed from the raw reads. Short or low-quality reads – those reads containing an adaptor, reads containing >5% unknown nt “N,” and reads with >20% quality value ≤10 – were removed from raw data to obtain more reliable results. The Q20, Q30, and GC contents of the cleaned data were calculated. Next, the clean reads were de novo-assembled by using Trinity software (version v2013-02-25) (Trinity Software, Inc., Plymouth, MA, United States) and clustered with TGICL Clustering tools (Version 2.1) (The Institute for Genomic Research, Rockville, MD, United States) (Pertea et al., 2003; Grabherr et al., 2011). Functional annotation of these assembled unigenes was performed with BLASTx1, an integrated Gene Ontology (GO) annotation and data mining tool that assigns GO based on four publically available databases: the National Center for Biotechnology Information (NCBI) non-redundant protein database (Nr), the Kyoto Encyclopedia of Genes and Genomes (KEGG2), the Eukaryotic Ortholog Groups (KOG3), and the Swiss-Prot protein database4. All searches were performed with an E-value < 10-5. We used the Blast2GO5 program to do the GO functional classification for all unigenes, according to their molecular function, biological process, and cellular component (Ashburner et al., 2000; Conesa et al., 2005).
The sequenced reads for each sample were remapped onto the reference sequences with RSEM software v1.2.12 (Li et al., 2011; Zhang et al., 2016). Gene expression levels were estimated using the Fragments Per Kilobase of transcript per Million fragments (FPKM) method that is based on the number of uniquely mapped reads (Trapnell et al., 2010). Genes differentially expressed between the male antennae from ME treatment and MO groups were identified based on their FPKM values (Mortazavi et al., 2008; Anders and Huber, 2010). The false discovery rate (FDR) adjustment was made to correct the P-value threshold in these multiple tests and analyses (Benjamini and Hochberg, 1995). An FDR-adjusted P-value < 0.05 and an absolute value of the log2 ratio > 1 were set a priori as the significance threshold for gene differential expression in this study. For convenience, the differential expression genes showing higher expression levels in the ME than in the MO group were designated as “up-regulated,” whereas those displaying lower expression levels were designated as “down-regulated.”
Differentially expressed genes were also annotated using the GO database, and the numbers of DEGs in each GO term were calculated. To determine, which GO terms were significantly enriched in the DEGs, we performed a GO enrichment analysis that used a hypergeometric test to map all differentially expressed proteins to the GO terms in the database. This test used the following equation (Blüthgen et al., 2005; Tu et al., 2015):
where, N is the number of all genes with a GO annotation; n is the number of DEGs in N; M is the number of all genes annotated to specific GO terms; and m is the number of DEGs in M (M–m ≥ 0). The calculated P-value was first subjected to a Bonferroni correction, taking a corrected P-value of 0.05 as a threshold for statistical significance. GO terms fulfilling this condition were defined as significantly enriched GO terms in the DEGs.
All identified genes were mapped to pathways in the KEGG database by using the KEGG Automatic Annotation Server software. To identify significantly enriched metabolic pathways or signal transduction pathways in DEGs, we used the same formula calculation as in the GO analysis. Here, N represented the number of all genes with a KEGG annotation, n is the number of DEGs in N, M is the number of all genes annotated to specific pathways, and m is the number of DEGs in M.
To verify our RNA-Seq results, 16 genes related to insect olfactory transport process that showed different expression levels, as revealed via RNA sequencing, were randomly selected for validation in a qRT-PCR analysis. In addition, other independent sampling experiments were conducted in order to obtain new biological replicates by ME and MO treatment. Those experiments were performed as described above. A RNA extraction kit (Takara Biotechnology Co., Ltd., Japan) was used to extract antennal total RNA from the ME treatment and control groups of male flies according to the manufacturer’s protocol, and a gDNA eliminator spin column removed genomic DNA. Approximately 1 μg of total RNA from each sample was used to synthesize cDNA, by using a PrimeScriptTM RT Reagent Kit (Takara Biotechnology Co., Ltd., Japan), which then served as a template for qRT-PCR. The gene-specific primers were designed according to the gene sequences in Primer v5.0 software (Premier, Canada) were listed in Supplementary Table S1. RT-PCR was performed to test whether all primers could amplify the correct products. Amplification efficiencies of all primers were validated before the gene expression analysis.
To perform the qRT-PCR reactions, a SYBR Premix ExTaq Kit (Tiangen, Guangzhou, China) was used following the manufacturer’s instructions, and run on a Stratagene Mx3000P thermal cycler (Agilent Technologies, Wilmington, Germany). qRT-PCR was carried out according to the protocol reported in our previous study (Liu H. et al., 2017). The α-tubulin gene of B. dorsalis was amplified to serve as the internal control (GenBank accession number: XM_011212814) (Shen et al., 2010; Yang et al., 2013; Liu et al., 2015; Gui et al., 2016). Dissociation curve analyses were performed to ensure amplification specificity. Three independent biological and three technical replicates were used and performed for each gene, respectively. The relative gene expression levels were calculated by using the 2-ΔΔCT method as described previously (refer to Livak and Schmittgen, 2001).
To identify the characteristics of B. dorsalis OR genes, particularly the significant differently expression ORs (BdorOR63a-1, GenBank accession number: KP743726 and BdorOR88a, GenBank accession number: KP743732), and their relationship to other Dipteran insects. To do this, a maximum likelihood tree for the ORs was constructed using the amino acid sequences derived from B. dorsalis and the published sequences of two Dipteran species: Ceratitis capitata (Wiedemann) and Drosophila melanogaster (Meigen). All the information on the amino acid sequences of ORs was obtained from the NCBI database. Alignments of OR amino acid sequences were performed using the program ClustalW2. The maximum likelihood tree was constructed in MEGA v7.0 software (Molecular Evolutionary Genetics Analysis, v4.0, Sudhir Kumar, United States) and with the Interactive Tree Of Life (iTOL) web tool6.
Bioassays were performed in the laboratory following a method similar to that of Karunaratne and Karunaratne (2012) and Liu H. et al. (2017). For the assessment of effects of age on the male responses to ME, each sample of 2-day- and 10-day-old males (200 of each) were randomly selected as the test subjects and released into a screened cage (1.0 m × 1.0 m × 1.0 m) without a trap. Approximately 30 min later, a fly trap containing 1.0 mL of pure ME was placed inside the screened cage. For the control, a trap was likewise placed inside the cage but without any ME. After trapping for 2 h, we removed the traps and counted the number of attracted flies. This bioassay was conducted between 9 and 12 a.m. under daylight conditions. Three independent biological replicates were performed.
To determine whether the mature male response to ME varied throughout the day, the responses of 10-day-old male B. dorsalis were assayed at 9 a.m., 1 p.m., and 5 p.m., as described above. For this, three biological replicates of flies were used per ME treatment and per control group for each time point. Additionally, from the untested individuals, the antennae from the 2-day- and 10-day-old males, and the mature males at 9 a.m. (morning), 1 p.m. (early afternoon), and 5 p.m. (near dusk), were dissected and immediately frozen in liquid nitrogen. Then, their total RNAs were extracted and reverse transcribed into single-chain cDNAs. Next, the expression of BdorOR63a-1 and BdorOR88a was evaluated by qRT-PCR quantitative technique. Each treatment was replicated three times.
BdorOR88a, BdorOR63a-1, and BdorOrco (GenBank accession number: EU621792) were amplified using specific primers (Supplementary Table S2). The purified PCR products were ligated into the pCS2+ vector; then, linearized modified pCS2+ vectors were used to synthesize cRNAs by using the mMESSAGE mMACHINE SP6 Kit (Ambion, Austin, TX, United States) following the manufacturer’s instructions. The purified cRNAs were re-suspended in nuclease-free water at 2000 ng/μL.
The oocyte microinjection and two-electrode voltage clamp recording were performed following published protocols (see Xu et al., 2014; Li et al., 2017; Liu F. et al., 2017). Briefly, mature healthy X. laevis oocytes (stage V–VII) were treated with 2 mg/ml of collagenase I (GIBCO, Carlsbad, CA, United States) in a washing buffer (96 mM NaCl, 5 mM MgCl2, 2 mM KCl, and 5 mM HEPES [pH = 7.6]) for ca. 1 h at room temperature. Next, the oocytes were microinjected with 27.6 ng of BdorOR cRNAs and 27.6 ng of BdorOrco cRNA, and then incubated at 18°C for 3–8 days in 1 × Ringer’s solution (96 mM NaCl, 5 mM MgCl2, 5 mM HEPES, 2 mM KCl, and 0.8 mM CaCl2 [pH = 7.6]). Stock solutions (1 M) of ME and MO were prepared in DMSO and they were subsequently diluted to the indicated concentrations with 1 × Ringer’s buffer. The two-electrode voltage-clamp (TEVC) technique measured the odorant-induced currents in Xenopus oocytes. Whole-cell currents were recorded and amplified by an OC-725C amplifier (Warner Instruments, Hamden, CT, United States) at a holding potential of -80 mV, low-pass-filtered at 50 Hz, and digitized at 1 kHz. Oocytes with nuclease-free water injection served as the negative control. Data acquisition and analysis were conducted with Digidata 1440A and pCLAMP10 software (Axon Instruments Inc., Union City, CA, United States).
An RNA interference experiment was performed to demonstrate the roles of BdorOR88a and BdorOR63a-1 in ME detection by male flies. To prepare the double-stranded RNA (dsRNA), we used a template cDNA generated by PCR-targeting fragments. Those primers with T7 promoter sequences used to synthesize dsRNA are listed in Supplementary Table S3. The GFP gene served as the control dsRNA (dsGFP) (GenBank accession number: AHE38523). Based on the manufacturer’s protocol, the dsRNAs were synthesized and purified by the MEGAscript® RNAi Kit (Thermo Fisher Scientific, United States); then, their concentrations were quantified on a Nanodrop 1,000 (Thermo Fisher Scientific, United States) and their integrity was determined by 2%-agarose gel electrophoresis.
Subsequently, expression of the dsRNAs and the injection procedure for the male flies were carried out following established techniques (Liu et al., 2015; Dong et al., 2016; Hou et al., 2017; Liu H. et al., 2017). Each sample of 50 mature males (15-day-old) was randomly selected and placed into a 35 cm × 35 cm × 35 cm cage. Needles were prepared with a puller at 60°C (PC-10, Narishige, Tokyo Japan). Microinjection was performed using an Eppendorf Microinjector (Eppendorf Ltd., Germany). The injection condition was set to an injection pressure (Pi) of 600 hPa and a timing setting (Ti) of 0.6 s. For each treated male fly, 400 nL of dsBdorOR88a or dsBdorOR63a-1 (2,000 ng/μL) was injected into its body cavity between the second and third abdominal segments. Males were injected with an equal volume of dsGFP served as negative control groups. The blank control group consisted of male flies that were fed normally. Injected and non-injected flies were reared on the artificial diet in the cages as described above. The respective numbers of dead files were counted after treatment for 24 h and 48 h. For the bioassay, males from the dsBdorOR88a or dsBdorOR63a-1 treatment groups, the dsGFP treatment group, and the blank control group, were placed separately inside a 1.0 m × 1.0 m × 1.0 m screen cage equipped with ME as a trap. The lured males were counted after 2 h, and the silencing efficiency of BdorOR88a or BdorOR63a-1 was detected by qRT-PCR. Each bioassay experiment was replicated five times.
All analyses were carried out by SAS v9.20 software (SAS Institute Inc. Cary, NC, United States). Results from the experimental replicates were expressed as the mean ± SE. The responses of immature and mature male flies to ME, diurnal pattern of mature male responsiveness to ME, and the expression pattern of ORs were analyzed by independent Student’s t-test (P = 0.05). Cases of P-values < 0.05 were considered to be statistically significant. Datasets of the attractiveness of ME to mature male, adult mortality, and gene silencing efficiencies were checked for normality of distribution and homogeneity of variances with Shapiro-Wilk’s and Levene’s tests, respectively. If data were normally distributed and had similar variances, then the means were compared by one-way analysis of variance (ANOVA). Following a significant ANOVA result, multiple comparisons among five groups were assessed by Duncan’s multiple range test (DMRT, P = 0.05). Non-normally distributed data were analyzed with the non-parametric Kruskal–Wallis test to compare medians (P = 0.05), followed by a Mann–Whitney test for follow-up pairwise comparisons. All results were plotted with Origin v9.0 software.
The RNA taken from the ME-treated and control (MO) males was used for RNA-Seq – with three experimental replicates per treatment – generated 23,552,399,343 raw reads. In general, all the libraries were of good quality, with average Q20 and Q30 percentages of over 95.60 and 89.41%, respectively. After removing the low-quality reads and trimming the adapter sequences, 157,514,454 clean reads were obtained for sequencing from the six samples (Tables 1, 2). These clean reads were ultimately assembled into 36,215 unigenes that had a mean length of 1,147 bp, an N50 of 2,362 bp, and a GC content of 41.61% (Table 2). The transcriptome data were deposited into the NCBI Short Reads Archive (SRA) database under the accession number SRP124917.
To determine the effects of ME exposure on antennal gene expression in male B. dorsalis adults, their DEGs were identified using the FPKM method. A total of 4,433 DEGs were detected from the ME treatment and MO (control) groups (|log2FC| > 1, P-value < 0.05; FDR < 0.05). Of these, 3,813 (86.01%) DEGs were up-regulated and 620 (13.99%) were down-regulated in the ME-treated male antennae (Figure 1A). The global expression changes of all genes with RNA-Seq ratios are shown in Figure 1B; the green and red circles are genes having a differential expression pattern in ME-treated males compared with the control (MO group). Figure 1C shows the hierarchical clustering analysis of their 4,433 DEGs.
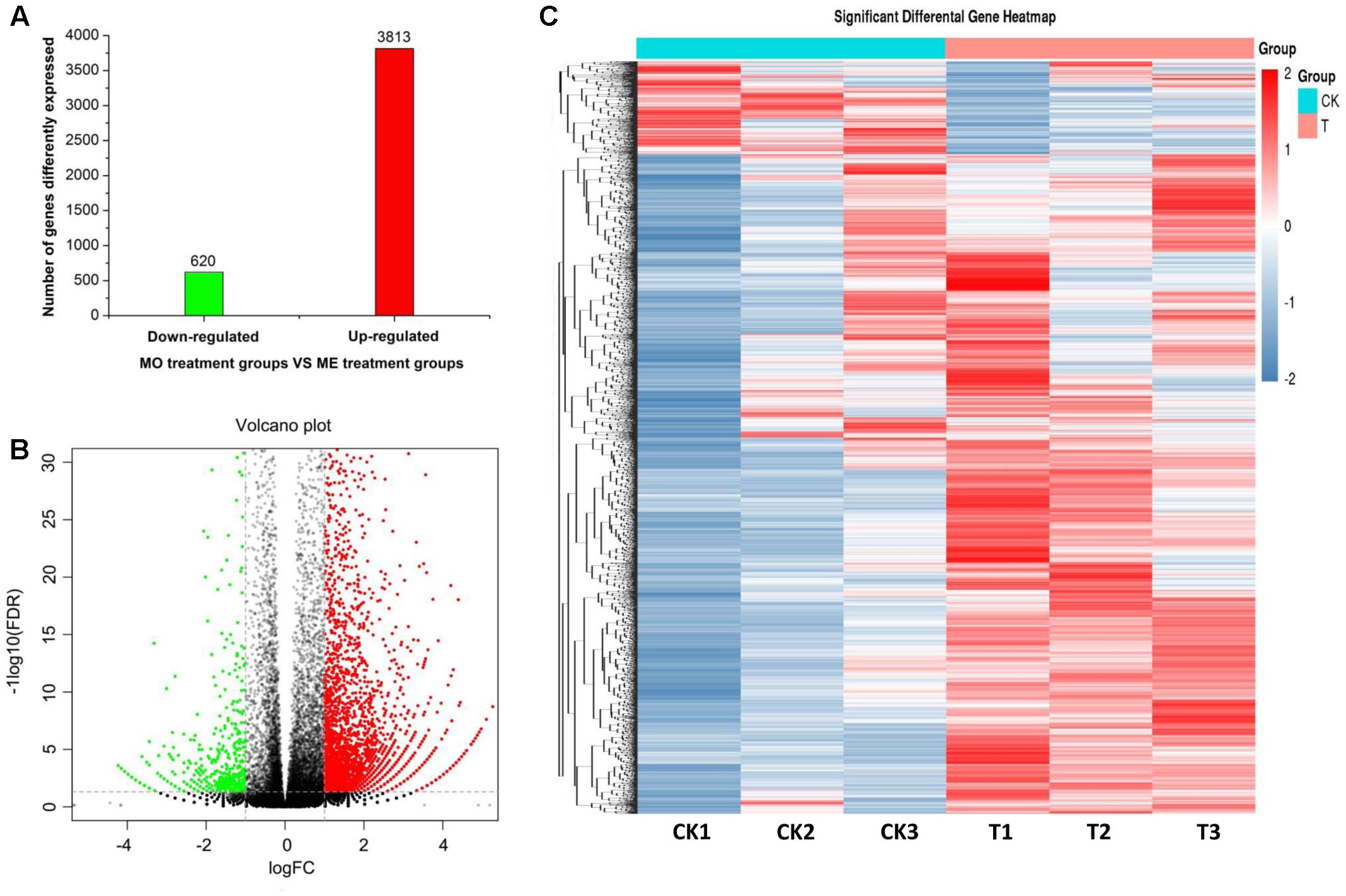
FIGURE 1. Statistical analysis of the differential expression of genes in male antennae of Bactrocera dorsalis flies from the MO (mineral oil) and ME (methyl eugenol) treatment groups. (A) Classification for the differential abundance of genes. (B) Volcano plots of differentially expressed genes (DEGs) between the ME and MO treatments. Y axis represents -log10 significance. X axis represents logFC (fold change). Red points indicate up-regulated expression of DEGs; green points indicate down-regulated expression of DEGs; and black points show non-differentially expressed genes. Genes with a P-value < 0.05 and fold change ≥1.0 were considered as DEGs in this study. (C) Hierarchical clustering graph of differential gene expression profiles in the ME and MO treatments. Clustering was done using RNA-Seq data derived from the six samples based on log10FPKM values. The column and row indicate the sample and gene, respectively. CK and T represent MO- and ME-treatment, respectively. Red and blue bands indicate, respectively, those genes that were significantly up-regulated and down-regulated in the ME-treated males.
With the GO annotation results in hand, the DEGs were classified and categorized into 83 functional groups (Supplementary Figure S1). Specifically, the GO enrichment analysis (P-value < 0.05) revealed that the most enriched biological process terms were cellular, followed by metabolic, single-organism, biological regulation, and response to stimulus; while binding and catalytic activity were the most enriched terms for the molecular function category; finally, under the cellular component category, cell and cell part were the most enriched terms (Supplementary Figure S2A). To investigate the biological pathways actively involved in ME detection by male flies, the DEGs were assigned to reference canonical pathways in KEGG: this revealed 20 significantly enriched pathways (Supplementary Figure S2B). The annotations presented here provide a valuable information for investigating the specific processes, pathways, and functions involved in the ME detection process in B. dorsalis, and perhaps other Diptera, too.
Many DEGs associated with response to stimulus, catalytic activity, binding, biological adhesion, molecular transducer activity, and transporter activity may contribute to ME detection in adult B. dorsalis males. Based on the literature and our GO/KEGG enrichment analyses, we identified putative DEGs encoding proteins involved in insect olfactory transport, viz: three DEGs for odorant binding proteins (BdorOBP57c, BdorOBP5, and BdorOBP2), two for ORs (BdorOR88a and BdorOR63a-1), one encoding an ionotropic receptor (BdorIR92a), and one encoding a sensory neuron membrane protein (BdorSNMP1-1) (Figure 2). Notably, relative to the control (MO) group, both BdorOR88a and BdorOR63a-1 were up-regulated by 6.33- and 2.06-fold, respectively, in the antennae of males exposed to ME. Additionally, the expression levels of carboxylesterase, esterase B1, cytochrome P450-6a14,-313a, and UDP-glucuronosyltransferase 2A3, 3A1 – genes that encode candidate odorant-degrading enzymes (ODEs) – were significantly down-regulated at the transcriptional level.
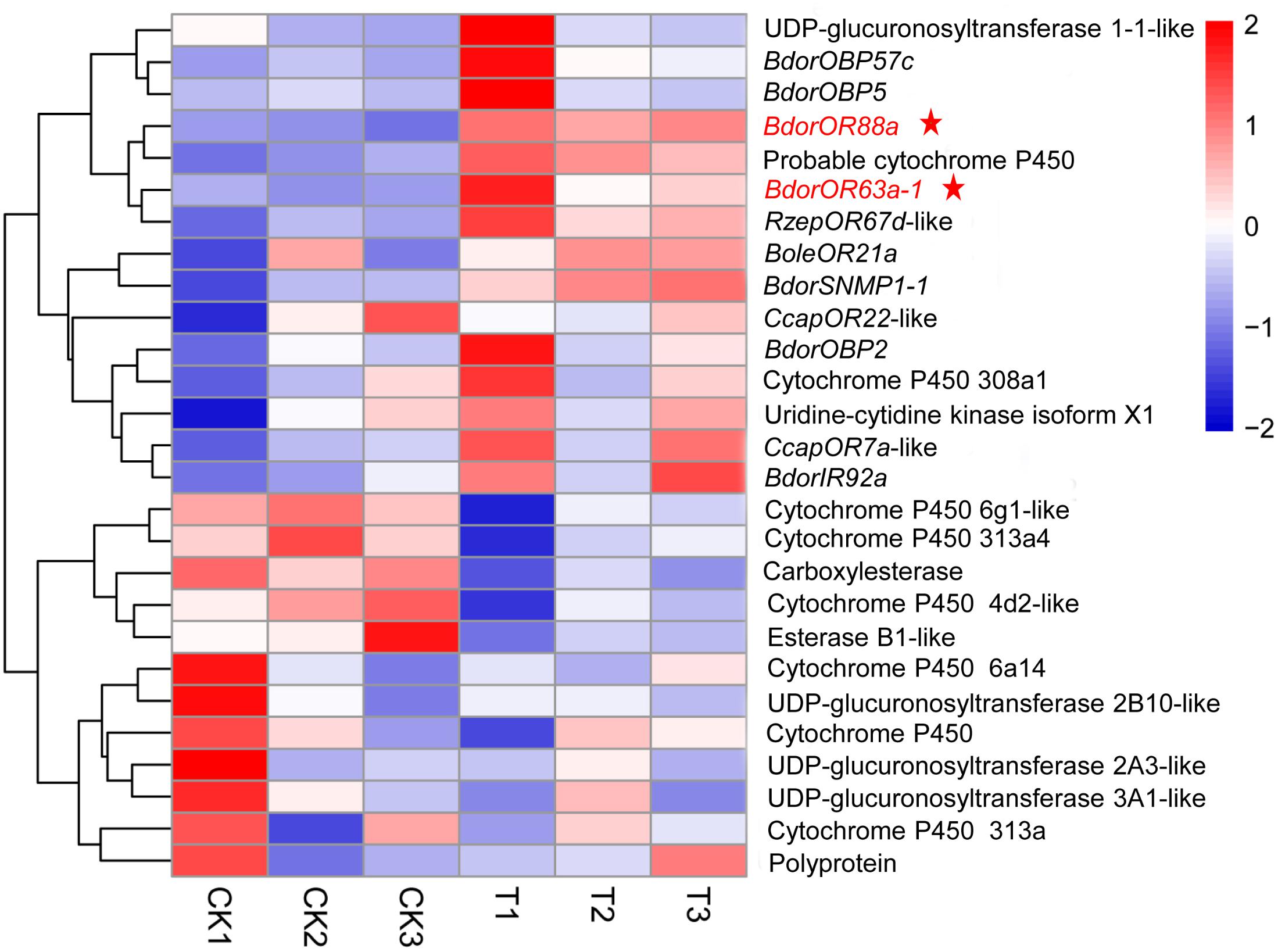
FIGURE 2. Heatmap representing the expression patterns of DEGs related to olfactory transport in male antennae from the ME (methyl eugenol) and MO (mineral oil) treatment groups. Clustering was done using RNA-Seq data derived from the six samples based on log10FPKM values. The column and row indicate the sample and gene, respectively. CK and T represent MO- and ME-treatment, respectively. Blue, white, and red indicate low, medium, and high levels of gene expression, respectively. Three biological replicates were conducted for each treatment.
We used qRT-PCR analysis to validate the results of differential gene expression obtained from the RNA-sequencing data (Figure 3). Two genes, BdorOBP57c and BdorOBP5 expressed no significant difference in qRT-PCR analysis, which were inconsistent with RNA-Seq results. However, of the 16 selected genes, 14 agreed with our RNA-Seq results. For example, BdorOR88a, BdorOR63a-1, BdorIR92a, and BdorSNMP1-1 were all significantly up-regulated in the ME-treated male antennae, as found in the RNA-Seq analysis, and multiple cytochrome P450 and carboxylesterase encoding genes were down-regulated in the ME-treated males in both the RNA-Seq and RT-qPCR analyses, with a similar fold change detected. For the other genes tested – BdorOR43a-1, BdorOR43b, BdorOR7a-2, BdorOR7a-3, BdorOR7a-5, BdorOR67c, BdorOR59a, and BdorOR69a – they expressed no significant differences according to the qRT-PCR test, not unlike the RNA-Seq results. Hence, the qRT-PCR analysis revealed up- or down-regulated gene expression profiles that were consistent with the RNA-Seq data, confirming that our comparative transcriptome analysis was robust and reliable.
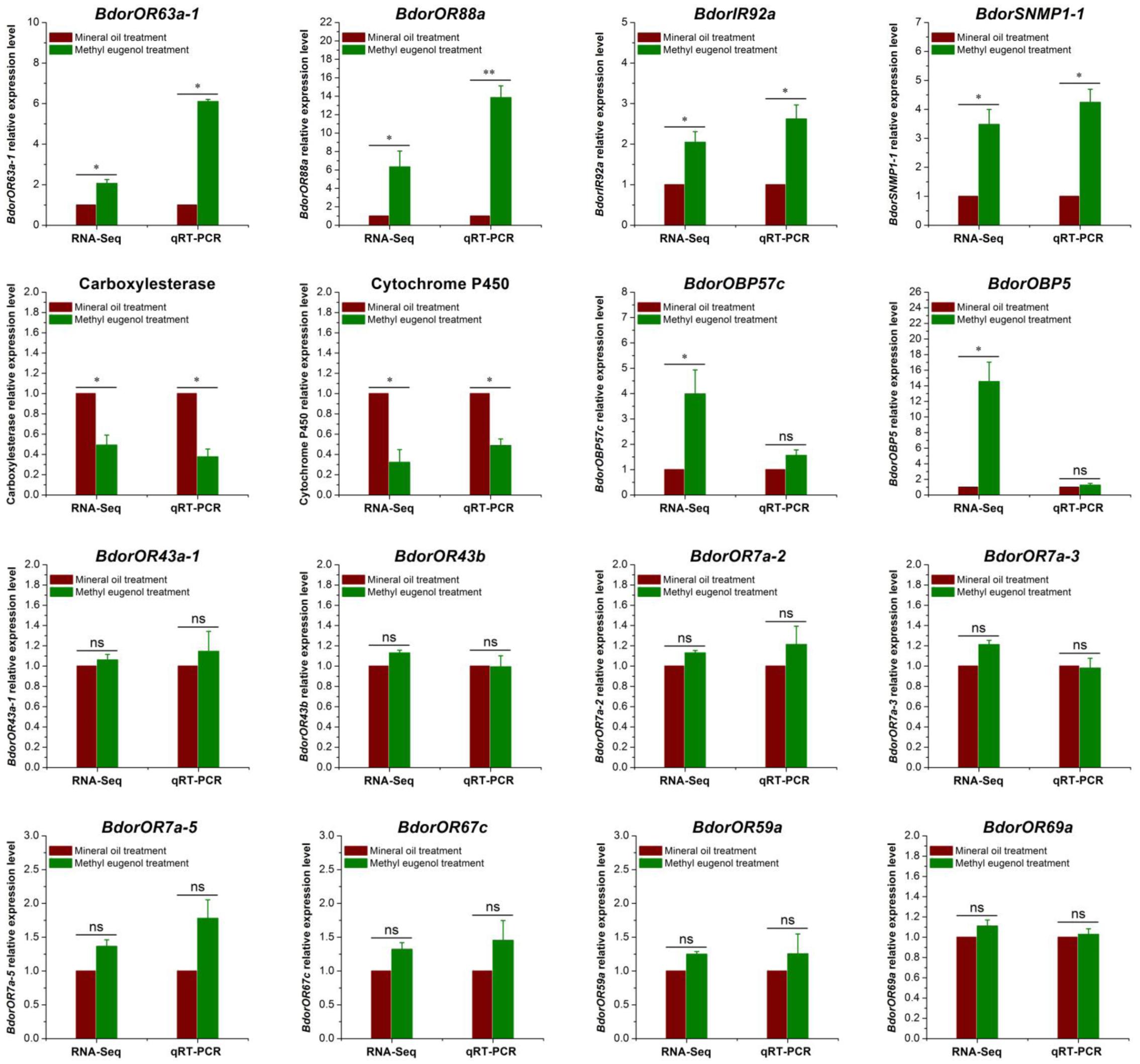
FIGURE 3. Validation of antennal DEGs (differentially expressed genes) in Bactrocera dorsalis male flies using quantitative real-time PCR (qRT-PCR). The α-tubulin gene was used as an internal control and three biological replicates were performed. The results were evaluated using a 2-ΔΔCT method, and the 2-ΔΔCT value of calibrant equals to 1.0. Bars represent mean ± SE values. ns, not significant, ∗P < 0.05, ∗∗P < 0.01 (t-tests).
The full-length BdorOR88a and BdorOR63a-1 cDNA segments consisted of 1,245 and 1,248 nt, encoding a polypeptide of 414 and 415 amino acids, respectively. To determine the phylogenetic relationship between BdorORs and the other ORs reported in C. capitata and D. melanogaster, a maximum likelihood tree was constructed. The BdorORs clustered together with the orthologous ORs from two Dipteran species with the best BLASTP hit. The ligand binding ORs from B. dorsalis shared phylogenetic relationships with the OR homologs of both Dipteran species. Notably, BdorOR88a clustered in a branch with DmelOR88a, but clustered in a different group than BdorOR63a-1 (Figure 4).
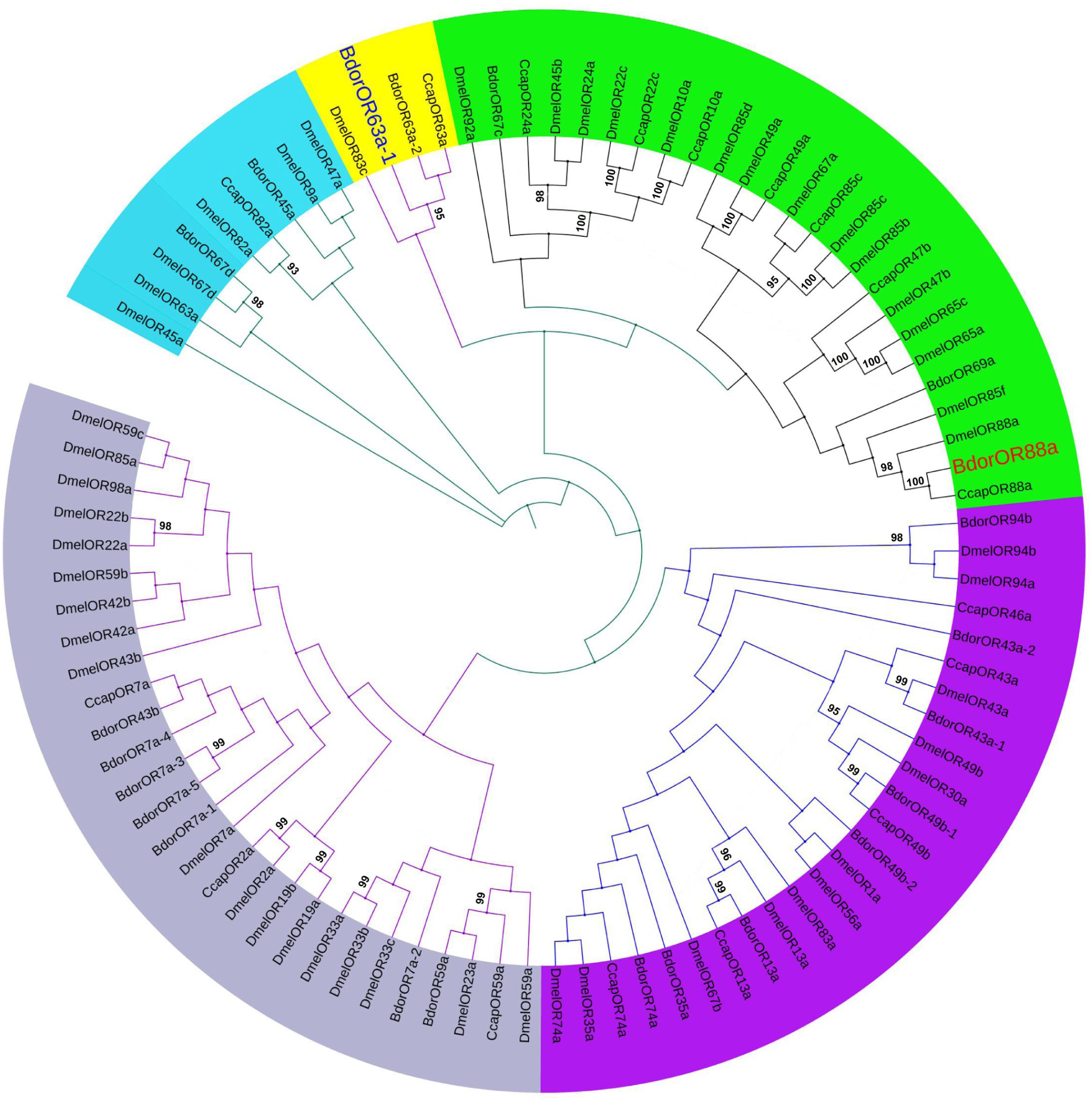
FIGURE 4. Maximum likelihood dendrogram based on protein sequences of candidate odorant receptors (ORs) in Bactrocera dorsalis and other insects. Evolutionary history was inferred using the maximum likelihood method. Bootstrap values greater than 90% (n = 1,000 replications) are displayed. ORs from Bactrocera dorsalis (Bdor), Ceratitis capitata (Ccap), and Drosophila melanogaster (Dmel) were included.
Under laboratory conditions, the male flies’ taxis to ME had a profile similar to their sexual development. As Figure 5A shows, fly responsiveness increased with age, with newly emerged males (2-day-old) presenting the lowest taxis, but by the time they were 10-day-old the males had become highly attracted to ME. The 10-day-old male responders numbered 173.67 ± 3.84, which was about 17 times higher than the abundance of 2-day-old males (11.67 ± 1.00) (t = 32.40; df = 2; P = 0.001). Furthermore, in contrast to the low expression observed in the 2-day-old male flies’ antennae, BdorOR63a-1 and BdorOR88a were highly expressed in 10-day-old males, corresponding to a 2.40-fold (t = 5.20; df = 2; P = 0.035) and 4.57-fold (t = 13.01; df = 2; P = 0.0059) increase, respectively (Figures 5B,C).
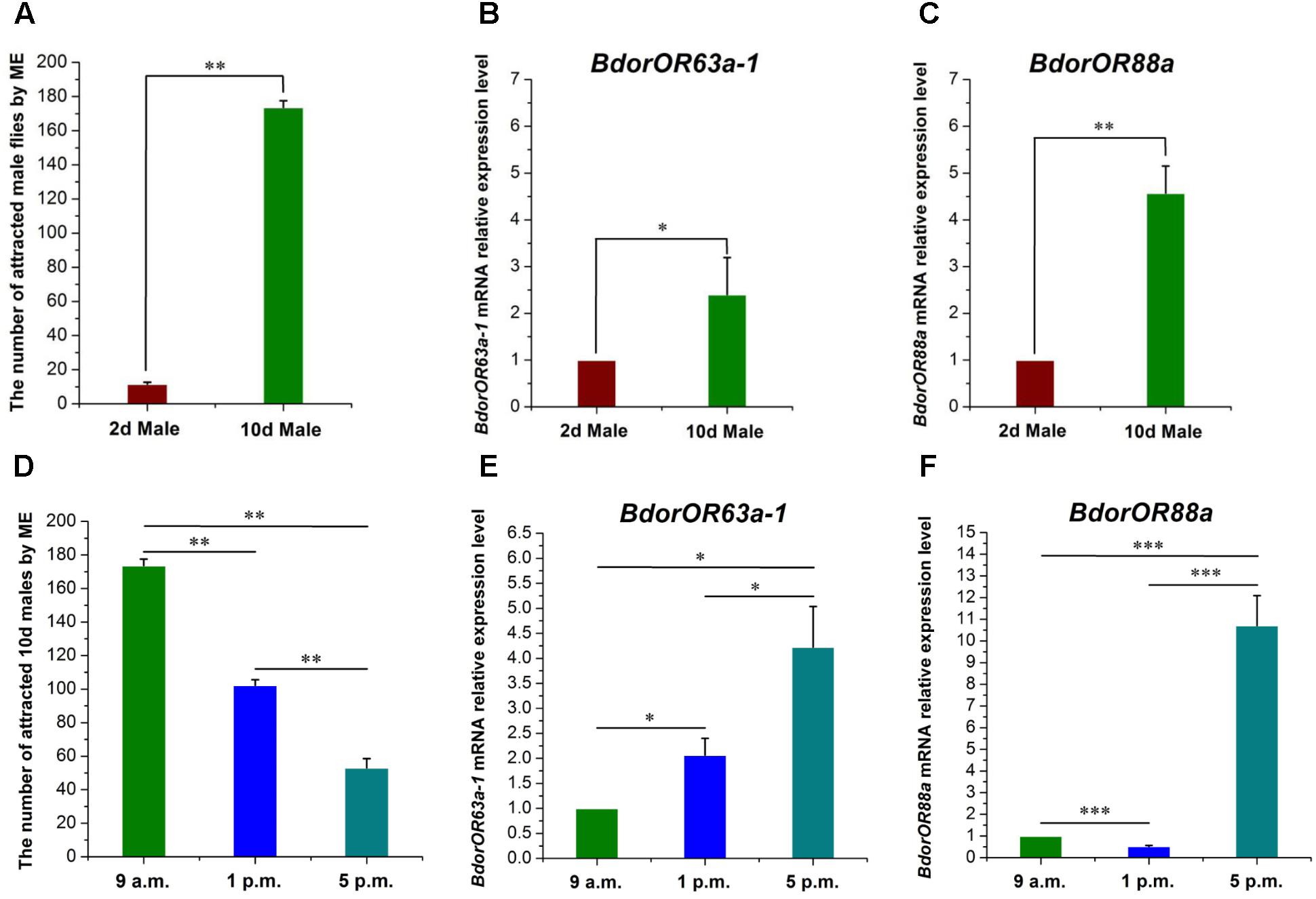
FIGURE 5. The attractiveness effect of ME (methyl eugenol) to Bactrocera dorsalis male flies and the expression pattern of BdorOR63a-1 and BdorOR88a in the males’ antennae. (A) Responses of immature and mature male flies to ME. (B,C) BdorOR63a-1 and BdorOR88a expressed at higher levels in mature males than immature males antennae. (D) Diurnal pattern of mature male responsiveness to ME. (E,F) Diurnal pattern of BdorOR63a-1 and BdorOR88a expression levels in the mature males antennae. All the experiments were performed in triplicate. Transcription levels of the BdorOR63a-1 and BdorOR88a gene were normalized by α-tubulin gene. The results were evaluated using a 2-ΔΔCT method, and the 2-ΔΔCT value of calibrant equals to 1.0. Bars represent mean ± SE values. ns, not significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (t-tests).
The diurnal pattern of male responsiveness to ME is illustrated in Figure 5D. The male fly response to ME was highest during the morning (173.67 ± 3.84 males), declining to a lower level in the early afternoon (102.33 ± 3.28), but dropped markedly near dusk (53.00 ± 5.51) (F = 312.49; df = 2; P < 0.0001). Furthermore, the expression levels of BdorOR63a-1 and BdorOR88a were not uniform throughout the day. Specifically, the BdorOR88a expression level was significantly reduced in the afternoon (p.m.) male adult flies compared with their morning (a.m.) counterparts (t = 18.57; df = 2; P < 0.0001) (Figure 5F). Interestingly, the transcript levels of BdorOR63a-1 and BdorOR88a at dusk were dramatically higher than those in the morning and early afternoon males (Figures 5E,F).
To verify whether ME olfactory detection in the B. dorsalis flies is mediated by BdorOR63a-1 or BdorOR88a, we expressed these putative receptors along with co-receptor BdorOrco, by using the Xenopus oocytes and two-electrode voltage clamping recording system. We found that H2O injected oocytes did not generate detectable currents when challenged with either MO or ME (Figure 6A). BdorOR63a-1/BdorOrco-expressed oocytes did not respond to MO and only weakly generated a ∼30 nA current in response to ME, even at concentrations as high as 1 × 10-3 M (Figure 6B). By contrast, the BdorOR88a/BdorOrco-expressing oocytes were clearly activated by ME, with no response to MO. Additionally, ME elicited dose-dependent currents from the BdorOR88a/BdorOrco-expressing oocytes. This analysis also indicated that the lowest measurable response was observed at a concentration of 1 × 10-8 M (Figure 6C) and the EC50 value – half the maximal effective concentration refers to the concentration of odorant, which induces a response halfway between the baseline and maximum – was 2.83 × 10-5 M (Figure 6D). Therefore, it is conceivable that BdorOR88a is likely to play a role in the reception of ME by B. dorsalis males.
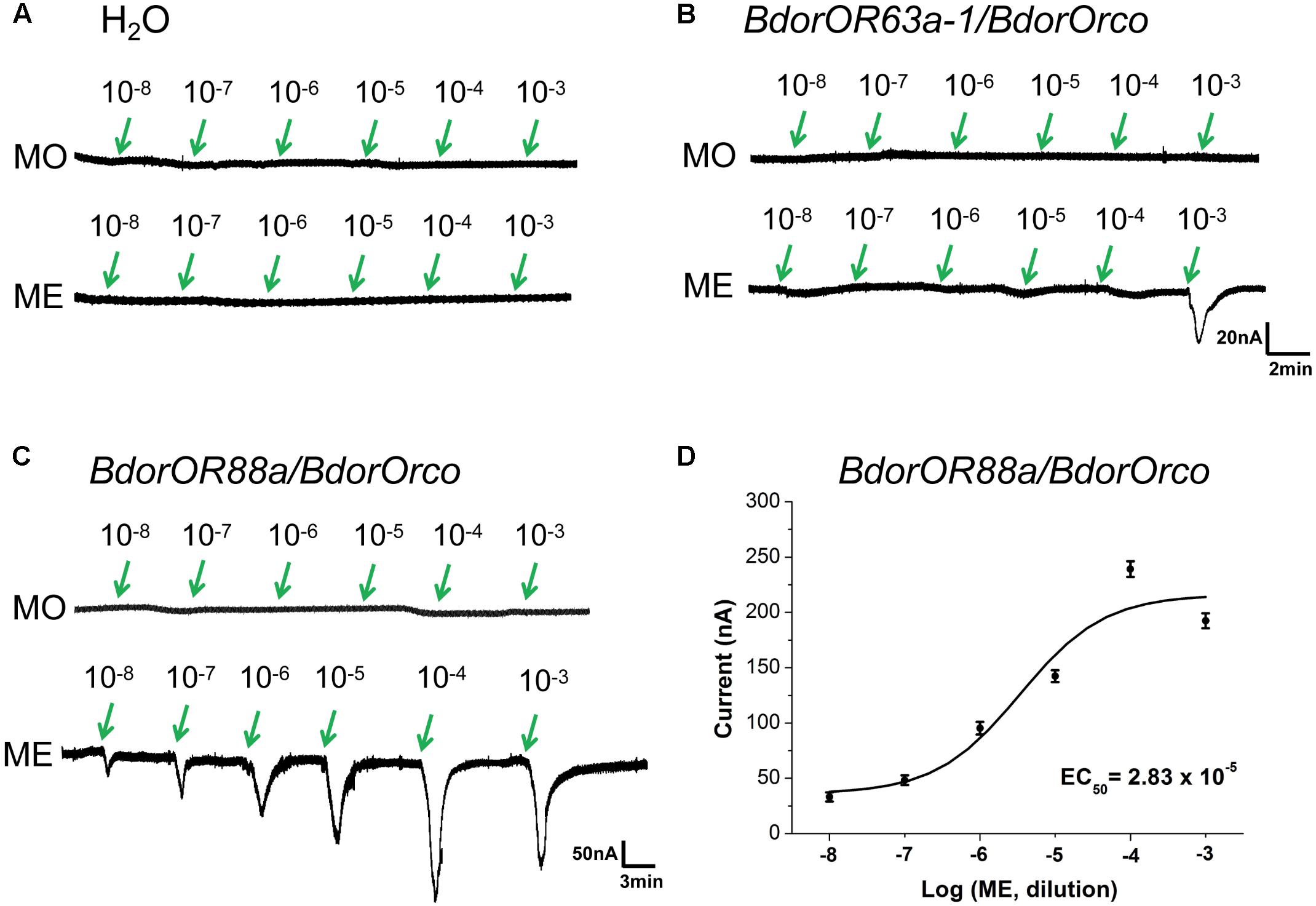
FIGURE 6. Responses of Xenopus oocytes with co-expressed BdorOR88a/BdorOrco or BdorOR63a-1/BdorOrco to stimulation with ME and MO compounds. (A) H2O-injected Xenopus oocytes failed to respond to the ME (methyl eugenol) and MO (mineral oil) odorants. (B) BdorOR63a-1/BdorOrco only slightly responded to the 10-3 M solution of ME. (C) BdorOR88a/BdorOrco Xenopus oocytes were stimulated with a range of ME concentrations. (D) Dose-dependent curve of BdorOR88a to ME using doses of 1.0 × 10-8 to 1.0 × 10-3 M. EC50 = 2.83 × 10-5. Symbols show the electrical current responses from the BdorOR88a/BdorOrco complex presented as the mean ± SE (n = 8). The dose-response curve shown was fitted with a sigmoidal model that had a variable slope (in Origin v9.0 software).
Microinjection had a clear and negative influence on B. dorsalis mature male flies’ survival. As Figures 7A,B show, the average mortalities of flies in the dsBdorOR63a-1, dsBdorOR88a, and dsGFP treatment groups at 24 h were 9.20 ± 0.49, 7.60 ± 0.75, and 8.40 ± 0.75%, respectively, and this mortality increased to 15.20 ± 1.02, 12.80 ± 1.36, and 13.60 ± 0.75% at 48 h (Figures 7A,B). However, the mortalities of the blank controls were comparatively much lower: 2.40 ± 0.75% at 24 h (F = 16.83; df = 3; P = 0.0001) and 3.60 ± 0.75% at 48 h (F = 24.56; df = 3; P < 0.0001). Notably, mortality of the dsBdorOR63a-1- and dsBdorOR88a-treated male flies was not significantly different from that of the dsGFP-treated male flies.
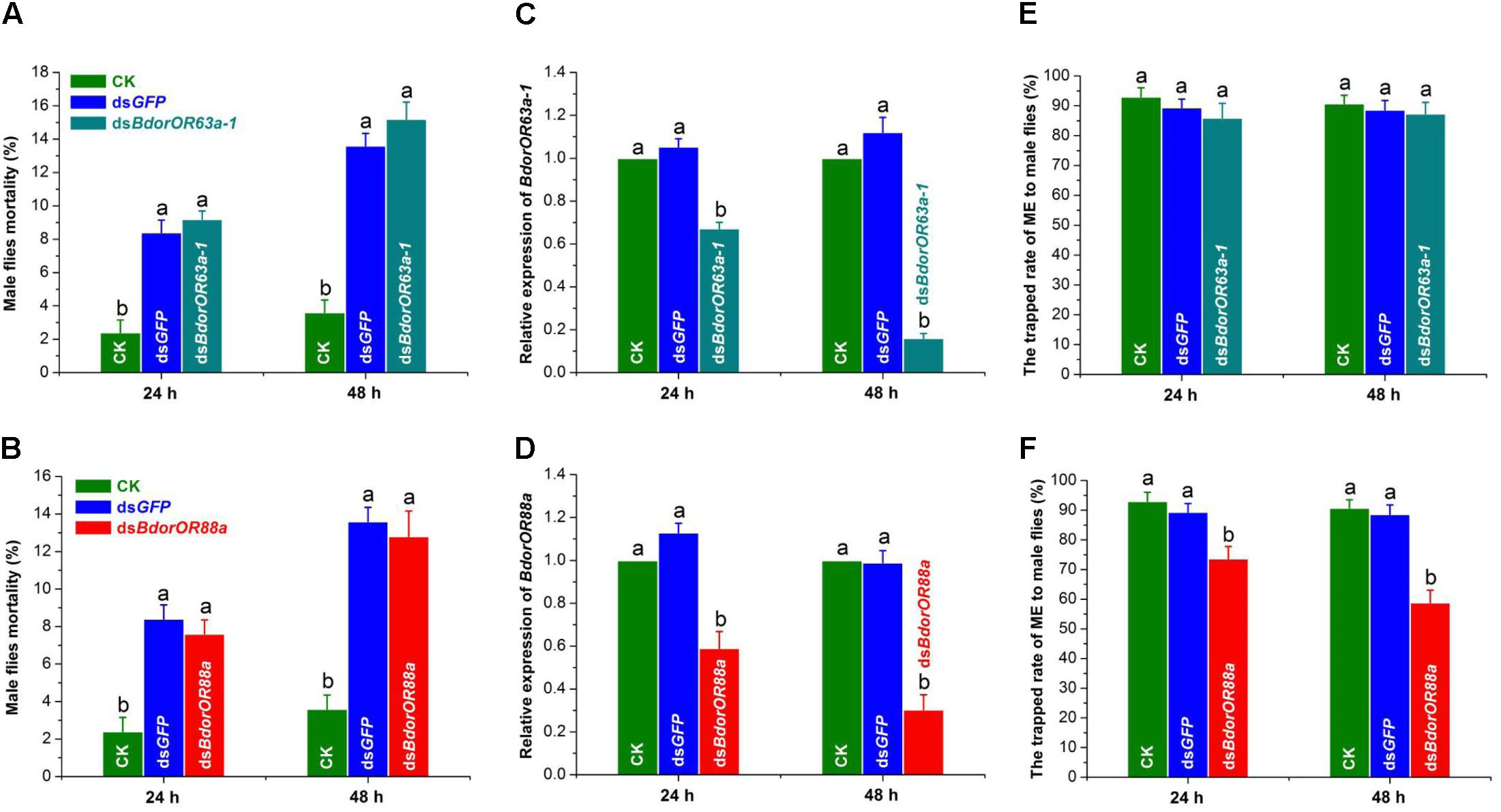
FIGURE 7. Effects of RNA interference on the mortality of Bactrocera dorsalis male flies, BdorOR63a-1 and BdorOR88a expression in their antennae, and fly responsiveness to ME. (A,B) Mortality of male flies in the dsBdorOR63a-1 and dsBdorOR88a treatments in the 24 h and 48 h bioassays were calculated. Negative control males were injected with the same amount of dsGFP. Blank control groups (CK) were normally reared. (C,D) The RNAi efficiency was measured at 24 and 48 h by qRT-PCR after the dsRNA injection. α-tubulin gene was used as an internal reference gene. The results were evaluated using a 2-ΔΔCT method, and the 2-ΔΔCT value of calibrant equals to 1.0. (E,F) The attractiveness effect of ME (methyl eugenol) to males after BdorOR63a-1 and BdorOR88a genes were silenced by RNAi. Different letters among the columns within a time point indicate significant differences (ANOVA, P < 0.05). Five biological replicates were used. Bars represent mean ± SE values.
Consequently, transcript levels of BdorOR63a-1 and BdorOR88a in the adult males were significantly reduced in the dsRNAs-injected flies compared with the two control groups (Figures 7C,D). As Figure 7C shows, BdorOR63a-1 expression was down-regulated in the dsBdorOR63a-1-treated males, to a level approximately 1.49-fold lower than that of the blank control and dsGFP treatment groups at 24 h (F = 55.62; df = 2; P < 0.0001), a significant difference that reached c. 6.25-fold at 48 h (F = 154.08; df = 2; P < 0.0001). Similarly, after treatment for 24 h and 48 h, there was a pronounced decrease in BdorOR88a expression of approximately 1.69-fold (F = 31.72; df = 2; P = 0.0002) and 3.29-fold (F = 46.69; df = 2; P < 0.0001) when compared with that of the dsGFP and blank control groups, respectively (Figure 7D).
We then analyzed the effects of RNAi of the BdorOR63a-1 and BdorOR88a gene transcript on male responsiveness to ME. Only the dsBdorOR88a-treated males were much less trapped by ME than were the dsGFP-treated and blank control flies. After 24 h of the treatment, the proportion of males trapped in the dsGFP-treated (89.38 ± 2.83%) and blank control (92.97 ± 3.06%) groups were both significantly higher than recorded in the dsBdorOR88a treatment (73.60 ± 4.16%; F = 53.37; df = 2; P < 0.0001). Furthermore, this difference was greatest at 48 h, when the proportion of ME-trapped males was 58.80 ± 4.25% in the dsBdorOR88a treatment group, significantly less than in the dsGFP-treated (88.63 ± 3.13%) and control (90.71 ± 2.82%) groups (F = 114.81; df = 2; P < 0.0001) (Figure 7F). By contrast, males were similarly trapped in the dsBdorOR63a-1 and dsGFP treatment groups and the blank control at 24 h (F = 2.83; df = 2; P = 0.1180) and 48 h (F = 2.87; df = 2; P = 0.1151) (Figure 7E).
Here, we propose a model describing how chemosensory proteins might be used by a mature male to distinguish the ME odorant from a noisy environment (Figure 8). Once the ME odorant penetrates the pore tubules of the antennae, the protein products of BdorOBP83a-2 and BdorOBP2 are posited to assist in transporting the ME odorant across the aqueous sensillum lymph. After its release from these proteins, the ME odorant is then transferred to the protein encoded by BdorOR88a. With the ME odorant now bound, the BdorOR88a/BdorOrco complexes are activated to trigger the signals that lead to neural spikes generated in the fly’s brain, evoking its characteristic response behavior to ME. After the activation of the odorant receptor, ME odorant is deactivated and degraded by the ODEs.
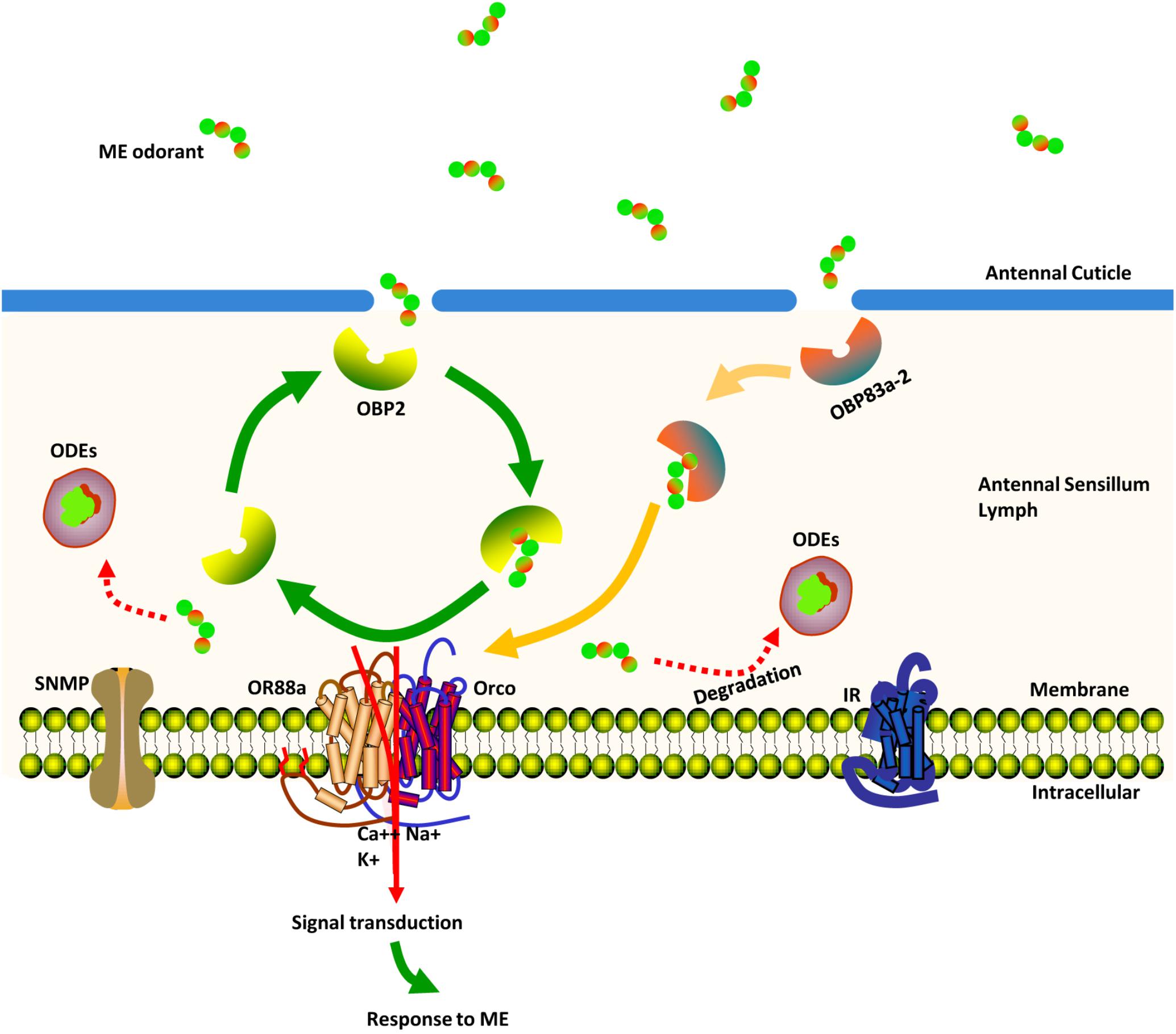
FIGURE 8. Schematic view of the ME (methyl eugenol) odorant detection process in the antennae of mature Bactrocera dorsalis male flies. Once ME odorants penetrate pore tubules of the sensillum, they are bound and solubilized by BdorOBP83a-2 and BdorOBP2, transported through the sensillum lymph and finally reach sensory dendrites, where they activate membrane-bound BdorOR88a. Signal transduction evokes a response behavior to the detected ME. ME odorants are rapidly degraded and inactivated by odorant-degrading enzymes.
Because ME is a potent attractant of B. dorsalis mature males, this parapheromone has been widely used for decades as a sexual lure in the detection and control of male B. dorsalis field populations worldwide (Smith et al., 2002; Shelly et al., 2010; Pagadala et al., 2012). However, until now, we do not know how or why it works. Based on this study’s RNA-Seq and qRT-PCR results, two odorant receptor genes (BdorOR63a-1 and BdorOR88a) were found abundantly expressed in mature males’ antennae after ME stimulation. Next, we functionally characterized these ORs in a heterologous expression system. In contrast to BdorOR63a-1, BdorOR88a co-expressed with BdorOrco in the Xenopus oocytes elicited dose-dependent inward currents upon application of ME. Additionally, silencing BdorOR63a-1 via the injection of dsRNA had no significant effect on the males’ attraction to ME, whereas silencing BdorOR88a significantly reduced the number of males attracted to ME. As such, we conclude that BdorOR88a is necessary for explaining the observed attraction of mature males toward ME. Hence, our present results further improve the current knowledge of the molecular mechanism underpinning ME detection by B. dorsalis mature male flies.
Male fly responsiveness to ME was clearly age-dependent. Males were strongly attracted to ME when 10-day-old, while immature male flies (2-day-old) were not attracted to ME. This result is not unlike that found in prior studies (Karunaratne and Karunaratne, 2012; Liu H. et al., 2017). Accordingly, in contrast to BdorOR63a-1, BdorOR88a was extreme abundantly expressed in the mature male antennae. ME functions as a precursor for B. dorsalis male synthesis of sex pheromone components (Shelly et al., 2008; McInnis et al., 2011; Liu H. et al., 2017). ME-acquired males produced a more attractive sexual pheromonal signal and enjoyed a higher mating success than ME-deprived males (Nishida et al., 1997; Shelly et al., 2000). Decades of publications have suggested that ingestion of ME enhances male mating competitiveness and thus demonstrated that this effect underlies the strong attraction of males to ME (Shelly, 2010). Prior studies had already revealed that male responsiveness to ME was not uniform throughout the day: it peaks in the morning, declines in the afternoon, and drops markedly at dusk (Ibrahim and Hashim, 1980; Tan et al., 1986; Karunaratne and Karunaratne, 2012). Our present study agrees rather well with these observations. The daily fluctuation in male flies responsiveness to ME displays a negative correlation with the daily rhythms of their courting and mating behavior (Karunaratne and Karunaratne, 2012; Liu H. et al., 2017). The attractiveness of a volatile depends on both the chemical properties of volatile and the physiological status of insect (Anton et al., 2007; Gadenne et al., 2016). Generally, responses to sex pheromones are switched on during courting and mating, whereas responses to oviposition-site cues or food odors are switched off. B. dorsalis mature males were very active at dusk, but extremely low numbers were attracted to the ME source (Karunaratne and Karunaratne, 2012). During the courting and mating at dusk, B. dorsalis males may transiently stop responding to the ME until the next morning and begin responding to sex pheromone in search of a mating partner.
Olfactory plasticity is a powerful evolutionary strategy that optimizes critical resources for insect survival (Gadenne et al., 2016). Recently, study on olfactory plasticity in insects, such as Agrotis ipsilon, Spodoptera littoralis, D. melanogaster, Anopheles gambiae, C. capitata, and B. dorsalis, is becoming one of the hot topics (Jang, 1995; Zhou et al., 2009; Barrozo et al., 2011; Deisig et al., 2012; Rund et al., 2013a,b; Kromann et al., 2015; Jin et al., 2017). Furthermore, olfactory plasticity enables insects to modify their response to semiochemical according to their physiological conditions, such as, feeding state, circadian rhythm, age, and mating status (Jin et al., 2017). Interestingly, olfactory-guided behaviors vary according to the time of day, which help insects respond to environmental chemical stimulus at the right moment (Gadenne et al., 2016). Considerable literature indicated that antennal and olfactory sensory neurons (OSNs) sensitivity of many insects tends to be regulated by the body endogenous rhythm or biological clock (Krishnan et al., 1999; Page and Koelling, 2003; Tanoue et al., 2004; Jin et al., 2017). Clock genes in the Drosophila antennae allow autonomous rhythmicity of environmental cues detection (Krishnan et al., 1999; Tanoue et al., 2004). In Anopheles gambiae mosquitoes, OBPs gene expression rhythms are driven in part by the endogenous circadian clock (Rund et al., 2013a,b). The chemosensory receptors gene expression levels of B. dorsalis gravid female flies fluctuate rhythmically at different times of the day (Jin et al., 2017). In our study, the expression level of BdorOR63a-1 increased gradually from morning to dusk in the mature male antennae. Nonetheless, BdorOR88a was abundantly expressed during the morning, but much less so in the afternoon. Yet more remarkably, BdorOR88a expression level increased considerably at the dusk period. B. dorsalis adults mating activity is restricted to dusk. The accurate detection and recognition of a potential mating partner is the key step in insect courtship and subsequently mating (Sayin et al., 2018). The peak in expression of BdorOR63a-1 and BdorOR88a at dusk may correspond to the time of increasing male chemosensory activity to female-emitted sex pheromone blend (containing several components).
Comparative phylogenetic analyses of the OR repertoire of insects can provide useful information on the evolutionary origin of OR families and their expansion in insect lineages (Missbach et al., 2014; Koenig et al., 2015). More strikingly, in our phylogenetic analysis, BdorOR88a was distributed within a distinct cluster of ORs with DmelOR88a, suggesting that they probably perform the same molecular function in the neuronal circuitry. Intriguingly, in Drosophila, the olfactory receptor OR88a attuned to semiochemicals has been identified. OSNs expressing the olfactory receptor OR88a housed in trichoid sensilla of Drosophila antennae can mediate responses to unidentified odors in the fly’s male and female body wash extracts (van Naters and Carlson, 2007). More recently, D. melanogaster OR88a has been characterized as a receptor of the fly-produced odorants methyl myristate, methyl palmitate, and methyl laurate that mediated copulation and attraction (Dweck et al., 2015). Analogous to other fly OR types, the role of DmelOR88a has been considered that of a pheromone receptor (PR) (Dweck et al., 2015; Fleischer et al., 2017). Therefore, we strongly suspect that B. dorsalis mature males’ possible use BdorOR88a to detect female sex-pheromones and locate their mates at dusk, although further functional experiments are needed to confirm this hypothesis.
From chemical stimulus to behavioral response, the olfactory process involves the capture, binding, transport, and inactivation of odors. The initial steps in odor detection involve the binding of an odor to the ORs positioned on the dendritic membrane of the OSNs within antennae. OBPs are able to bind various hydrophobic odorant molecules in the environment (Sato et al., 2008; Taylor et al., 2008; Silbering et al., 2011; Siciliano et al., 2014). Once semiochemicals are bound, the OR/Orco heteromeric complexes are activated to trigger signals leading to characteristic spikes generated in the brain, thereby producing a behavioral response in the insect (Neuhaus et al., 2005; Benton et al., 2006; Hallem and Carlson, 2006; Sato et al., 2008; Croset et al., 2010; Silbering et al., 2011; Di et al., 2017; Fleischer et al., 2017). Gene silencing, via the ingestion or microinjection of dsRNA, recently confirmed that BdorOrco, BdorOBP83a-2, and BdorOBP2 actively had critical roles in mediating the taxis of B. dorsalis males to ME (Zheng et al., 2012; Wu et al., 2016; Liu H. et al., 2017). Considering our results alongside those of prior research to date, we preliminarily posit that the ME odor molecules bind to either BdorOBP2 or BdorOBP83a-2, after which they are transferred to OSNs, where BdorOR88a coordinates with BdorOrco to bind to the ME odor molecules – only then is the olfactory signal transduction pathway finally activated. Our findings from this study thus represent an important breakthrough in our mechanistic understanding of how B. dorsalis mature male flies are attracted to ME. Hopefully, this will advance how attractants are designed to target this specific olfactory pathway, which should promote the development of better and more affordable attractants for B. dorsalis management in the field.
Combinatorial coding, that is individual OR can be activated by multiple odorants and a specific odor ligand can be detected by multiple ORs, is the primary coding mode of the insect olfaction system (Malnic et al., 1999; Hallem et al., 2004; Suh et al., 2014; Andersson et al., 2015; Fleischer et al., 2017). In D. melanogaster, olfactory system sensitivity seems to be enhanced by a combinatorial coding, with specific groups of ORs detecting the same chemical cues (Leal, 2013). Yet more remarkably, knock-down BdorOR88a gene did not lead to the complete disappearance of the mature male flies’ responsiveness to ME. Therefore, presumably, in addition to BdorOR88a, there are further ORs and other receptors, which may also contribute to detect ME. However, due to the lack of B. dorsalis genomic information, many genes cannot yet be annotated and their functions remain unknown, especially for those olfactory-related genes. In their detection of odorant signals, insects use several families of chemosensory receptors, including the ORs, ionotropic receptors (IRs), gustatory receptors (GRs), and sensory neuron membrane proteins (SNMPs) (Benton et al., 2007, 2009; Jin et al., 2008; Suh et al., 2014). The identification of a new family of IRs, complementary to the ORs family yet expressed in different olfactory neurons, has provided new insight into the molecular mechanisms of odor detection in insect (Benton et al., 2009; Silbering et al., 2011). IR92a mediates attraction behavior to ammonia and volatile amines in D. melanogaster (Min et al., 2013). SNMPs are highly conserved in multiple insect species and involved in pheromone-based chemical communication (Nichols and Vogt, 2008). One subfamily in particular, SNMP1, when co-expressed with PRs, is believed to contribute to the sensitivity of pheromone detection in insects (Rogers et al., 1997; Benton et al., 2007; Vogt et al., 2009; Liu et al., 2014; Pregitzer et al., 2014). Study has demonstrated that SNMP1 played a vital role in detecting the sex pheromone Z11-18OAc in D. melanogaster (Benton et al., 2007; Jin et al., 2008). Accordingly, in moths, SNMP1 is considered indicative of sex pheromone-responsive neurons in antennae (Forstner et al., 2008; Thode et al., 2008). In our present study, the consistency between the RNA-Seq results and the mRNA expression from the qRT-PCR analysis implies that BdorIR92a and BdorSNMP1-1 possibly participate in the processing of ME detection by mature B. dorsalis male flies. Nonetheless, knowledge of the precise functional relevance of BdorIR92a and BdorSNMP1-1 for ME signaling remains elusive. To conclude, we deduce that these genes are involved in how B. dorsalis males detect ME, but this awaits further investigation and testing.
HL and Y-YL conceived and designed the experiments and wrote the manuscript. HL, Z-SC, and D-JZ performed the experiments. HL analyzed the data. Z-SC and D-JZ provided material support. All authors discussed the results and reviewed the final manuscript.
This research was supported by Guangdong Province College High-Quality Professional Foundation (No. 246), National Key Research and Development Program (No. 2016YFC1201200), and Guangzhou Science and Technology Project, China (No. 201601010179). The funders have no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We sincerely thank the Charlesworth Group for correction of the manuscript language and the Guangzhou Gene Denovo Co., Ltd., for technology support and helpful advice. We are grateful to the members of our laboratory for their cooperation in oriental fruit fly rear and treatment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00987/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andersson, M. N., Löfstedt, C., and Newcomb, R. D. (2015). Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 3:53. doi: 10.3389/fevo.2015.00053
Anton, S., Dufour, M. C., and Gadenne, C. (2007). Plasticity of olfactory-guided behaviour and its neurobiological basis: lessons from moths and locusts. Entomol. Exp. Appl. 123, 1–11. doi: 10.1111/j.1570-7458.2007.00516.x
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 2000–2009. doi: 10.1038/75556
Barrozo, R. B., Jarriault, D., Deisig, N., Gemeno, C., Monsempes, C., Lucas, P., et al. (2011). Mating-induced differential coding of plant odour and sex pheromone in a male moth. Eur. J. Neurosci. 33, 1841–1850. doi: 10.1111/j.1460-9568.2011.07678.x
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery ratea practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300.
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). A typical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio-0040020
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell-2008-12-001
Benton, R., Vannice, K. S., and Vosshall, L. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293. doi: 10.1038/nature06328
Blüthgen, N., Brand, K., Cajavec, B., Swat, M., Herzel, H., and Beule, D. (2005). Biological profiling of gene groups utilizing Gene Ontology. Genome Inform. 16, 106–115.
Chang, C. L., Vargas, R. I., Caceres, C., Jang, E., and Cho, I. K. (2006). Development and assessment of a liquid larval diet for Bactrocera dorsalis (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 99, 1191–1198. doi: 10.1603/0013-8746200699
Clarke, A. R., Armstrong, K. F., Carmichael, A. E., Milne, J. R., Raghu, S., Yeates, D. K., et al. (2005). Invasive phytophagous pests arising through a recent tropical evolutionary radiation: the Bactrocera dorsalis complex of fruit flies. Annu. Rev. Entomol. 50, 293–319. doi: 10.1146/annurev-ento-50-071803-130428
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Deisig, N., Kropf, J., Vitecek, S., Pevergne, D., Rouyar, A., Sandoz, J. C., et al. (2012). Differential interactions of sex pheromone and plant odour in the olfactory pathway of a male moth. PLoS One 7:e33159. doi: 10.1371/journal.pone.0033159
Di, C., Ning, C., Huang, L. Q., and Wang, C. Z. (2017). Design of larval chemical attractants based on odorant response spectra of odorant receptors in the cotton bollworm. Insect Biochem. Mol. Biol. 84, 48–62. doi: 10.1016/j.ibmb.2017-03-007
Dong, Y. C., Wang, Z. J., Chen, Z. Z., Clarke, A. R., and Niu, C. Y. (2016). Bactrocera dorsalis male sterilization by targeted RNA interference of spermatogenesis: empowering sterile insect technique programs. Sci. Rep. 6:35750. doi: 10.1038/Srep35750
Dweck, H. K. M., Ebrahim, S. A. M., Thoma, M., Mohamed, A. A. M., Keesey, I. W., Trona, F., et al. (2015). Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, 2829–2835. doi: 10.1073/pnas.1504527112
Fleischer, J., Pregitzer, P., Breer, H., and Krieger, J. (2017). Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 75, 485–508. doi: 10.1007/s00018-017-2627-5
Forstner, M., Gohl, T., Gondesen, I., Raming, K., Breer, H., and Krieger, J. (2008). Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem. Senses. 33, 291–299. doi: 10.1093/chemse/bjm087
Gadenne, C., Barrozo, R. B., and Anton, S. (2016). Plasticity in insect olfaction: to smell or not to smell? Annu. Rev. Entomol. 61, 317–333. doi: 10.1146/annurev-ento-010715-023523
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gui, S. H., Jiang, H. B., Liu, X. Q., Xu, L., and Wang, J. J. (2016). Molecular characterizations of natalisin and its roles in modulating mating in the oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Mol. Biol. 26, 103–112. doi: 10.1111/imb.12274
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Hou, Q. L., Chen, E. H., Jiang, H. B., Wei, D. D., Gui, S. H., Wang, J. J., et al. (2017). Adipokinetic hormone receptor gene identification and its role in triacylglycerol mobilization and sexual behavior in the oriental fruit fly (Bactrocera dorsalis). Insect Biochem. Mol. Biol. 90, 1–13. doi: 10.1016/j.ibmb.2017-09-006
Ibrahim, A. G., and Hashim, A. G. (1980). Efficacy of methyl eugenol as male attractant for Dacus dorsalis (Hendel). Pertanika 3, 108–112.
Jang, E. B. (1995). Effects of mating and accessory gland injections on olfactory-mediated behavior in the female mediterranean fruit fly, Ceratitis capitata. J. Insect Physiol. 41, 705–710. doi: 10.1016/0022-1910(95)00015-M
Jang, E. B., Khrimian, A., and Siderhurst, M. S. (2011). Di- and Tri-fluorinated analogs of methyl eugenol: attraction to and metabolism in the oriental fruit fly, Bactrocera dorsalis (Hendel). J. Chem. Ecol. 37, 553–564. doi: 10.1007/s10886-011-9963-y
Jin, S., Zhou, X., Gu, F., Zhong, G., and Yi, X. (2017). Olfactory plasticity: variation in the expression of chemosensory receptors in Bactrocera dorsalis in different physiological states. Front. Physiol. 8:672. doi: 10.3389/fphys.2017.00672
Jin, X., Ha, T. S., and Smith, D. P. (2008). SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 10996–11001. doi: 10.1073/pnas.0803309105
Karunaratne, S., and Karunaratne, U. K. P. R. (2012). Factors influencing the responsiveness of male oriental fruit fly, Bactrocera dorsalis, to methyl eugenol (3,4-dimethoxyalyl benzene). Trop. Agric. Res. Exten. 15, 92–97. doi: 10.4038/tare.v15i4.5258
Khrimian, A., Jang, E. B., Nagata, J., and Carvalho, L. (2006). Consumption and metabolism of 1, 2-Dimethoxy-4- (3-Fluoro-2-Propenyl) benzene, a fluorine analog of methyl eugenol, in the oriental fruit fly, Bactrocera dorsalis (Hendel). J. Chem. Ecol. 32, 1513–1526. doi: 10.1007/s10886-006-9066-3
Khrimian, A., Siderhurst, M. S., Mcquate, G. T., Liquido, N. J., Nagata, J., Carvalho, L., et al. (2009). Ring-fluorinated analog of methyl eugenol: attractiveness to and metabolism in the oriental fruit fly, Bactrocera dorsalis (Hendel). J. Chem. Ecol. 35, 209–218. doi: 10.1016/S0278-6915(02)00012-1
Khrimian, A. P., Demilo, A. B., Waters, R. M., Liqyido, N. J., and NicholsonI, J. M. (1994). Monofluoro analogs of eugenol methyl ether as novel attractants for the oriental fruit fly. J. Org. Chem. 59, 8034–8039. doi: 10.1021/jo00105a019
Koenig, C., Hirsh, A., Bucks, S., Klinner, C., Vogel, H., Shukla, A., et al. (2015). A reference gene set for chemosensory receptor genes of Man-duca sexta. Insect Biochem. Mol. Biol. 66, 51–63. doi: 10.1016/j.ibmb.2015.09.007
Krishnan, B., Dryer, S. E., and Hardin, P. E. (1999). Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature 400, 375–378. doi: 10.1038/22566
Kromann, S. H., Saveer, A. M., Binyameen, M., Bengtsson, M., Birgersson, G., Hansson, B. S., et al. (2015). Concurrent modulation of neuronal and behavioural olfactory responses to sex and host plant cues in a male moth. Proc. Biol. Sci. 282:20141884. doi: 10.1098/rspb.2014.1884
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, B., Dewey, C. N., Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, Z. Q., Luo, Z. X., Cai, X. M., Bian, L., Xin, Z. J., Liu, Y., et al. (2017). Chemosensory gene families in Ectropis grisescens and candidates for detection of type-II sex pheromones. Front. Physiol. 8:953. doi: 10.3389/fphys-2017-00953
Liu, C., Zhang, J., Liu, Y., Wang, G., and Dong, S. (2014). Expression of SNMP1 and SNMP2 genes in antennal sensilla of Spodoptera exigua (Hubner). Arch. Insect Biochem. Physiol. 85, 114–126. doi: 10.1002/arch.21150
Liu, F., Xia, X., and Liu, N. (2017). Molecular basis of N, N-Diethyl-3-Methylbenzamide (DEET) in repelling the common Bed Bug, Cimex lectularius. Front. Physiol. 8:418. doi: 10.3389/fphys-2017-00418
Liu, G., Wu, Q., Li, J., Zhang, G., and Wan, F. (2015). RNAi-mediated knock-down of transformer and transformer 2 to generate male-only progeny in the oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS One 10:e0128892. doi: 10.1371/journal.pone.0128892
Liu, H., Zhao, X. F., Fu, L., Han, Y. Y., Chen, J., and Lu, Y. Y. (2017). BdorOBP2 plays an indispensable role in the perception of methyl eugenol by mature males of Bactrocera dorsalis (Hendel). Sci. Rep. 7:15894. doi: 10.1038/s41598-017-15893-6
Liu, Z., Smagghe, G., Lei, Z., and Wang, J. J. (2016). Identification of male- and female-specific olfaction genes in antennae of the oriental fruit fly (Bactrocera dorsalis). PLoS One 11:e0147783. doi: 10.1371/jorunal.pone.0147783
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Malnic, B., Hirono, J., Sato, T., and Buck, L. B. (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. doi: 10.1016/S0092-8674(00)80581-4
McInnis, D., Kurashima, R., Shelly, T., Komatsu, J., Edu, J., and Pahio, E. (2011). Prerelease exposure to methyl eugenol increases the mating competitiveness of sterile males of the oriental fruit fly (Diptera: Tephritidae) in a Hawaiian orchard. J. Econ. Entomol. 104, 1969–1978. doi: 10.1603/EC11134
Min, S., Ai, M., Shin, S. A., and Suh, G. S. (2013). Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, 1321–1329. doi: 10.1073/pnas
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.7554/eLife.02115
Mitchell, R. F., Hall, L. P., Reagel, P. F., McKenna, D. D., Baker, T. C., and Hildebrand, J. G. (2017). Odorant receptors and antennal lobe morphology offer a new approach to understanding olfaction in the Asian longhorned beetle. J. Comp. Physiol. A 203, 99–109. doi: 10.1007/s00359-016-1138-4
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Nishioka, T., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Neuhaus, E. M., Gisselmann, G., Zhang, W., Dooley, R., Stortkuhl, K., and Hatt, H. (2005). Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 8, 15–17. doi: 10.1038/nn1371
Nichols, Z., and Vogt, R. G. (2008). The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 398–415. doi: 10.1016/j.ibmb.2007.11.003
Nishida, R., Shelly, T. E., and Kaneshiro, K. Y. (1997). Acquisition of female-attracting fragrance from a Hawaiian lei flower, Fagraea berteriana, by males of the Oriental fruit fly. J. Chem. Ecol. 23, 2275–2285. doi: 10.1023/B:JOEC.0000006673.27221.2e
Pagadala, D. K. J., Christine, M. W., John, C., Michael, A. B., and Toby, J. A. B. (2012). Isolation and identification of host cues from Mango, Mangifera indica, that attract gravid female oriental fruit fly, Bactrocera dorsalis. J. Chem. Ecol. 38, 361–369. doi: 10.1007/s10886-012-0093-y
Page, T. L., and Koelling, E. (2003). Circadian rhythm in olfactory response in the antennae controlled by the optic lobe in the cockroach. J. Insect Physiol. 49, 697–707. doi: 10.1016/S0022-1910(03)00071-4
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Pregitzer, P., Greschista, M., Breer, H., and Krieger, J. (2014). The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol. Biol. 23, 733–742. doi: 10.1111/imb.12119
Rogers, M. E., Sun, M., Lerner, M. R., and Vogt, R. G. (1997). Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J. Biol. Chem. 272, 14792–14799. doi: 10.1074/jbc.272-23-14792
Rund, S. S., Bonar, N. A., Champion, M. M., Ghazi, J. P., Houk, C. M., Leming, M. T., et al. (2013a). Daily rhythms in antennal protein and olfactory sensitivity in the malaria mosquito Anopheles gambiae. Sci. Rep. 3:2494. doi: 10.1038/srep02494
Rund, S. S., Gentile, J. E., and Duffield, G. E. (2013b). Extensive circadian and light regulation of the transcriptome in the malaria mosquito Anopheles gambiae. BMC Genomics 14:218. doi: 10.1186/1471-2164-14-218
Sakurai, T., Mitsuno, H., Haupt, S. S., Uchino, K., Yokohari, F., Kanzaki, R., et al. (2011). A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet. 7:e1002115. doi: 10.1371/jorunal.pgen.1002115
Sato, K., Pellegrino, M., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Sayin, S., Boehm, A. C., Kobler, J., and Grunwald Kadow, I. C. (2018). Internal state dependent odor processing and perception-the role of neuromodulation in the fly olfactory system. Front. Cell. Neurosci. 12:11. doi: 10.3389/fincel-2018-00011
Shelly, T. E. (2010). Effects of methyl eugenol and raspberry ketone/cue lure on the sexual behavior of Bactrocera species (Diptera: Tephritidae). Appl. Entomol. Zool. 45, 349–361. doi: 10.1117/1.601355
Shelly, T. E. (2017). Zingerone and the mating success and field attraction of male melon flies (Diptera: Tephritidae). J. Asia Pac. Entomol. 20, 175–178. doi: 10.1016/j.aspen.2016.12.013
Shelly, T. E., Edu, J., and McInnis, D. (2010). Prerelease consumption of methyl eugenol increases the mating competitiveness of sterile males of the oriental fruit fly, Bactrocera dorsalis, in large field enclosures. J. Insect Sci. 10, 1–16. doi: 10.1673/031-010-0801
Shelly, T. E., Edu, J., Pahio, E., Wee, S. L., and Nishida, R. (2008). Re-examining the relationship between sexual maturation and age of response to methyl eugenol in males of the oriental fruit fly. Entomol. Exp. Appl. 128, 380–388. doi: 10.1111/j.1570-7458.2008.00710.x
Shelly, T. E., McCombs, S. D., and McInnis, D. O. (2000). Mating competitiveness of male oriental fruit flies from a translocation strain (Diptera: Tephritidae). Environ. Entomol. 29, 1152–1156. doi: 10.1603/0046-225X-29.6.1152
Shen, G. M., Jiang, H. B., Wang, X. N., and Wang, J. J. (2010). Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Mol. Biol. 11:76. doi: 10.1186/1471-2199-11-76
Shen, G. M., Wang, X. N., Dou, W., and Wang, J. J. (2012). Biochemical and molecular characterization of acetylcholinesterase in four field populations of Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). Pest Manag. Sci. 68, 1553–1563. doi: 10.1002/ps.3340
Siciliano, P., He, X. L., Woodcock, C., Field, L. M., Kalinova, B., Scolaru, F., et al. (2014). Identification of pheromone components and their binding affinity to the odorant binding protein CcapOBP83a-2 of the Mediterranean fruit fly, Ceratitis capitata. Insect Biochem. Mol. Biol. 48, 51–62. doi: 10.1016/j.ibmb-2014-02-005
Siderhurst, M. S., and Jang, E. B. (2006). Female-biased attraction of Oriental fruit fly, Bactrocera dorsalis (Hendel), to a blend of host fruit volatiles from Terminalia catappa L. J. Chem. Ecol. 32, 2513–2524. doi: 10.1007/s10886-006-9160-6
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Benton, R., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila Olfactory subsystems. J. Neurosci. 31, 13357–13375. doi: 10.1523/JNEUROSCI-2360-11-2011
Smith, R. L., Adams, T. B., Doull, J., Feron, V. J., Goodman, J. I., Marnett, L. J., et al. (2002). Safety assessment of allylalkoxybenzene derivatives used as flavouring substances—methyl eugenol and estragole. Food Chem. Toxicol. 40, 851–870. doi: 10.1016/S0278-6915(02)00012-1
Stephens, A., Kriticos, D., and Leriche, A. (2007). The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull. Entomol. Res. 97, 369–378. doi: 10.1017/S0007485307005044
Suh, E., Bohbot, J., and Zwiebel, L. J. (2014). Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 6, 86–92. doi: 10.1016/j.cois.2014.10.006
Tan, K. H., Kirton, L. G., and Serit, M. (1986). “Age response of Dacus dorsalis (Hendel) to methyl eugenol in a wind tunnel and traps set in a village and its implication in population estimation,” in Proceedings of the 2nd International Symposium, Colymbari (Greece), ed. A. P. Economopoulos (Athens: G. Tsiveriotis Ltd), 425–432.
Tanoue, S., Krishnan, P., Krishnan, B., Dryer, S. E., and Hardin, P. E. (2004). Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr. Biol. 14, 638–649. doi: 10.1016/j.cub.2004.04.009
Taylor, A. J., Cook, D. J., and Scott, D. J. (2008). Role of odorant binding proteins: comparing hypothetical mechanisms with experimental data. Chemosens. Percept. 1, 153–162. doi: 10.1007/s12078-008-9016-2
Thode, A. B., Kruse, S. W., Nix, J. C., and Jones, D. N. (2008). The role of multiple hydrogen bonding groups in specific alcohol binding sites in proteins: insights from structural studies of LUSH. J. Mol. Biol. 376, 1360–1376. doi: 10.1016/j.jmb.2007.12.063
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tu, X., Wang, J., Hao, K., Whitman, D. W., Fan, Y., Cao, G., et al. (2015). Transcriptomic and proteomic analysis of pre-diapause and non-diapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Sci. Rep. 5:11402. doi: 10.1038/srep11402
van Naters, V. D. G., and Carlson, J. R. (2007). Receptors and neurons for fly odors in Drosophila. Curr. Biol. 17, 606–612. doi: 10.1016/j.cub-2007-02-043
Vargas, R. I., and Prokopy, R. (2006). Attraction and feeding responses of melon flies and oriental fruit flies (Diptera: Tephritidae) to various protein baits with and without toxicants. Hawaii Entomol. Soc. 38, 49–60. doi: 10.10125/178
Vogt, R. G., Miller, N. E., Litvack, R., Fandino, R. A., Sparks, J., Staples, J., et al. (2009). The insect SNMP gene family. Insect Biochem. Mol. Biol. 39, 448–456. doi: 10.1016/j.ibmb.2009-03-007
Wang, Y. L., Chen, Q., Guo, J. Q., Li, J., Wang, J. T., Wen, M., et al. (2017). Molecular basis of peripheral olfactory sensing during oviposition in the behavior of the parasitic wasp Anastatus japonicus. Insect Biochem. Mol. Biol. 89, 58–70. doi: 10.1016/j.ibmb.2017.09.001
Wu, Z. Z., Lin, J. T., Zhang, H., and Zeng, X. N. (2016). BdorOBP83a-2 mediates responses of the oriental fruit fly to semiochemicals. Front. Physiol. 7:452. doi: 10.3389/fphys-2016-00452
Wu, Z. Z., Zhang, H., Wang, Z. B., Bin, S. Y., He, H. L., and Lin, J. T. (2015). Discovery of chemosensory genes in the oriental fruit fly, Bactrocera dorsalis. PLoS One 10:e0129794. doi: 10.1371/jorunal.pone.0129794
Xu, P., Choo, Y. M., De La Rosa, A., and Leal, W. S. (2014). Mosquito odorant receptor for DEET and methyl jasmonate. Proc. Natl. Acad. Sci. U.S.A. 111, 16592–16597. doi: 10.1073/pnas.1417244111
Yang, W. J., Xu, K. K., Cong, L., and Wang, J. J. (2013). Identification, mRNA Expression, and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in Oriental fruit fly, Bactrocera dorsalis. Int. J. Biol. Sci. 9, 331–342. doi: 10.7150/ijbs.6022
Zhang, X. L., Liu, Y. M., Fang, Z. Y., Li, Z. S., Yang, L. M., Zhuang, M., et al. (2016). Comparative transcriptome analysis between Broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa Guss.) in response to Plasmodiophora brassicae during different infection stages. Front. Plant Sci. 7:1929. doi: 10.3389/fpls.2016.01929
Zheng, W., Zhu, C., Peng, T., and Zhang, H. (2012). Odorant receptor co-receptor Orco is upregulated by methyl eugenol in male Bactrocera dorsalis (Diptera: Tephritidae). J. Insect Physiol. 58, 1122–1127. doi: 10.1016/j.jinsphys-2012-05-011
Zheng, W. W., Peng, W., Zhu, C. P., Zhang, Q., Saccone, G., and Zhang, H. Y. (2013). Identification and expression profile analysis of odorant binding proteins in the Oriental fruit fly Bactrocera dorsalis. Int. J. Mol. Sci. 14, 14936–14949. doi: 10.3390/ijms140714936
Keywords: Bactrocera dorsalis, methyl eugenol, transcriptomic analysis, olfactory, odorant receptor, Xenopus oocytes
Citation: Liu H, Chen Z-S, Zhang D-J and Lu Y-Y (2018) BdorOR88a Modulates the Responsiveness to Methyl Eugenol in Mature Males of Bactrocera dorsalis (Hendel). Front. Physiol. 9:987. doi: 10.3389/fphys.2018.00987
Received: 28 February 2018; Accepted: 05 July 2018;
Published: 26 July 2018.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
William Benjamin Walker III, Swedish University of Agricultural Sciences, SwedenCopyright © 2018 Liu, Chen, Zhang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Yue Lu, bHV5b25neXVlQHNjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.